- 1Department of Biomedical Science, Iowa State University, Ames, IA, United States
- 2Roy J. Carver Department of Biochemistry, Biophysics and Molecular Biology, Iowa State University, Ames, IA, United States
Humans contain two nearly identical copies of Survival Motor Neuron genes, SMN1 and SMN2. Deletion or mutation of SMN1 causes spinal muscular atrophy (SMA), one of the leading genetic diseases associated with infant mortality. SMN2 is unable to compensate for the loss of SMN1 due to predominant exon 7 skipping, leading to the production of a truncated protein. Antisense oligonucleotide and small molecule-based strategies aimed at the restoration of SMN2 exon 7 inclusion are approved therapies of SMA. Many cis-elements and transacting factors have been implicated in regulation of SMN exon 7 splicing. Also, several structural elements, including those formed by a long-distance interaction, have been implicated in the modulation of SMN exon 7 splicing. Several of these structures have been confirmed by enzymatic and chemical structure-probing methods. Additional structures formed by inter-intronic interactions have been predicted by computational algorithms. SMN genes generate a vast repertoire of circular RNAs through inter-intronic secondary structures formed by inverted Alu repeats present in large number in SMN genes. Here, we review the structural context of the exonic and intronic cis-elements that promote or prevent exon 7 recognition. We discuss how structural rearrangements triggered by single nucleotide substitutions could bring drastic changes in SMN2 exon 7 splicing. We also propose potential mechanisms by which inter-intronic structures might impact the splicing outcomes.
Introduction
Survival Motor Neuron (SMN) protein is an essential housekeeping protein involved in multiple processes, including DNA replication and repair, transcription, pre-mRNA splicing, translation, macromolecular trafficking, stress granule formation, cell cycle regulation, signal transduction and maintenance of cytoskeletal dynamics (Singh R. N. et al., 2017). Low levels of SMN due to deletion or mutation of SMN1 gene causes spinal muscular atrophy (SMA), one of the leading genetic diseases associated with infant mortality (Wirth et al., 2020; Singh et al., 2021). SMN2, a nearly identical copy of SMN1, cannot compensate for the loss of SMN1 due to predominant skipping of exon 7 (Lefebvre et al., 1995; Cho and Dreyfuss 2010). Functions of SMN2 remain unknown, although high demand of SMN in testis is partially met through an adult-specific switch in SMN2 exon 7 splicing (Ottesen et al., 2016). While not intensively investigated, low levels of SMN also affects male fertility in adults suffering from mild SMA (Ottesen, Howell, Singh, Seo, Whitley and Singh 2016; Lipnick et al., 2019). Two currently approved therapies of SMA are based on the restoration of SMN2 exon 7 inclusion (Singh et al., 2020b). Due to the fact that SMA is predominantly linked to defects in SMN1 and that SMN2, a nearly identical copy of SMN1, is “available” in SMA patients, regulation of exon 7 splicing has been intensively investigated. Several cis-elements and transacting factors have been implicated in the regulation of SMN exon 7 splicing (Singh and Singh 2018) (Figure 1).
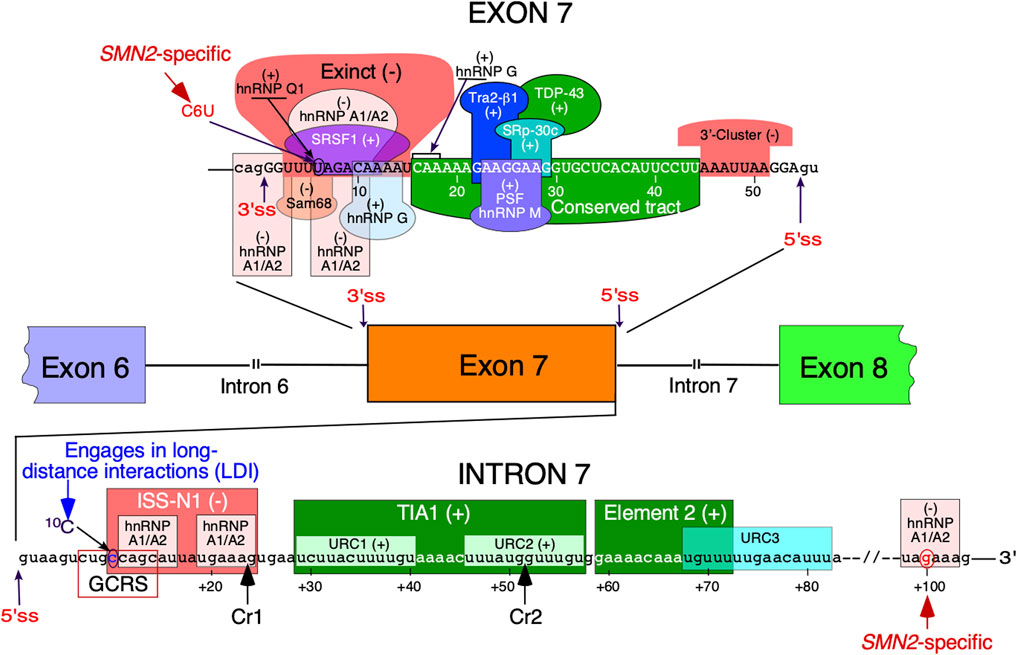
FIGURE 1. Regulation of SMN exon 7 splicing. Diagrammatic representation of intronic and exonic cis-elements as well as trans-acting factors that modulate SMN exon 7 splicing. Upper-case letters signify exonic sequences, small-case letters, intronic sequences. Exons and introns are also shown as colored boxes and lines, respectively. Numbering of nucleotides, neutral and positive, starts from the first exonic and intronic position, respectively. The 5 and 3′ splice sites (5′ss and 3′ss) are indicated by the arrows. Exinct, conserved tract and the 3′-Cluster are cis-elements revealed by in vivo selection as described in (Singh et al., 2004b). Cr1 and Cr2 represent cryptic 5′ splice sites as described in (Singh et al., 2017a). Negative and positive regulators of exon 7 splicing are indicated by (−) and (+), respectively. Abbreviations: Exinct, extended inhibitory context, GCRS, GC-rich region; LDI, long-distance interaction; URC, Uridine-rich clusters.
Splicing regulation is a complex process requiring precise definition of the splice sites (Shenasa and Hertel 2019). Recognition of the 5′ss by U1 snRNP is one of the earliest steps of assembly of the spliceosome that catalyzes the splicing reaction (Charenton et al., 2019). Once recruited, U1 snRNP can also define the upstream 3′ss through cross-exon interactions (De Conti et al., 2013). Similarly, U2 snRNP recruited at the 3′ss promotes recruitment of U1 snRNP at the downstream 5′ss through cross-exon interaction (De Conti et al., 2013). A critical C-to-T mutation at the 6th exonic position of exon 7 (C6U substitution in RNA) was found to be the primary cause of SMN2 exon 7 skipping (Lorson et al., 1999; Monani et al., 1999). Being close to the 3′ss, the C6U was assumed to weaken the 3′ss of the exon (Lim and Hertel 2001). Initially, it was proposed that C6U abrogates an enhancer associated with ASF/SF2 (Cartegni and Krainer 2002) (Figure 1). However, this claim was promptly challenged and a competing hypothesis suggesting that C6U creates a silencer associated with hnRNP A1 was put forward (Kashima and Manley 2003) (Figure 1). Our earlier work showed that C6U strengthens an extended inhibitory context (Exinct) at the 3′ss of exon 7 (Singh et al., 2004a; Singh et al., 2004c) (Figure 1). Additional factors that interact with C6U or in its vicinity were subsequently identified (Pedrotti et al., 2010; Singh and Singh 2018) (Figure 1).
The inhibitory nature of C6U was independently validated by in vivo selection in which relative significance of all 54 positions of SMN exon 7 was probed simultaneously (Singh et al., 2004b). Results of in vivo selection of exon 7 confirmed the presence of “Exinct” in the beginning of the exon 7 and revealed two additional regulatory regions termed as the “Conserved tract” and “3′-Cluster”. Located in the middle of exon 7, the “Conserved tract” exerts a positive effect on exon 7 splicing. The “3′-Cluster” is located towards the end of exon 7 and exerts a negative effect on exon 7 splicing (Figure 1). The most surprising finding of in vivo selection was the suboptimal nature of the 5′ss of exon 7 (Singh et al., 2004b; Singh 2007). In humans, the last exonic position in most cases is represented by a G residue. This G residue base pairs with a C residue of U1 snRNA (Lund and Kjems 2002). In addition, during catalytic core formation this G residue forms a base pair with a C residue of U5 snRNP (Lund and Kjems 2002). In vivo selection of the entire exon 7 revealed that an A residue at the last position of exon 7 constitutes the most inhibitory nucleotide that contributes to exon 7 skipping. Consistently, an A-to-G mutation at the last position of exon 7 (A54G substitution) fully restored SMN2 exon 7 inclusion. A GA-rich enhancer in the middle of exon 7 has been found to be critical for SMN2 exon 7 inclusion (Hofmann et al., 2000; Hofmann and Wirth 2002; Young et al., 2002). The enhancer constitutes a binding site for Tra2β1 and its associated factors (Figure 1). Of note, the effect of the A54G substitution was so strong that it promoted SMN2 exon 7 inclusion even in the absence of this GA-rich enhancer (Singh et al., 2004b). A subsequent study confirmed that A54G destabilizes a terminal stem-loop structure (TSL2) and helps recruit U1 snRNP through extended based pairing with U1 snRNA at the 5′ss of exon 7 (Singh et al., 2007). It should be noted that risdiplam also interacts with A54 and helps recruit U1 snRNP (Campagne et al., 2019). It is worth mentioning that recruitment of engineered U1 snRNAs to sequences located downstream of the 5′ss of exon seven can also promote exon 7 inclusion (Singh et al., 2017a; Singh RN. and Singh NN. 2019).
Several studies have focused on the role of regulatory elements within SMN intron 7, the last intron of SMN genes (Singh and Singh 2018) (Figure 1). The discovery of the 15-nucleotide long intronic splicing silencer N1 (ISS-N1) spanning the region from the 10th to 24th positions of intron 7 revealed (for the first time) the strong inhibitory impact of the intronic element on SMN2 exon 7 splicing (Figure 1). Deletion or an ASO-directed blocking of ISS-N1 fully restored SMN2 exon 7 inclusion (Singh et al., 2006). Subsequent studies confirmed that among many potential targets for an ASO-based therapy for SMA, ISS-N1 was the leading contender (Hua et al., 2008). Upon successful completion of clinical trials, the ISS-N1-targeting ASO nusinersen (commercial name: Spinraza) was approved as the first drug for the treatment of SMA (Singh et al., 2017b; Bennett et al., 2019). ISS-N1 is a complex regulatory element that harbors two putative sites contacted by a single hnRNP A1 molecule (Beusch et al., 2017). The first five nucleotides of ISS-N1 also overlaps with an upstream 8-nucleotide-long GC-rich motif, sequestration of which by a short ASO promoted SMN2 exon 7 inclusion and provided therapeutic benefits in mouse models of SMA (Singh et al., 2009; Keil et al., 2014). Another splicing silencer element located in intron 7 is created by a SMN2-specific A-to-G substitution at the 100th position of intron 7 (Kashima et al., 2007). Intron 7 also contains two positive regulatory elements downstream of ISS-N1 (Miyaso et al., 2003; Singh et al., 2011). One of these elements is a binding site for TIA1 that is known to stimulate recruitment of U1 snRNP at the 5′ss (Singh et al., 2011). We have demonstrated that TIA1 is indeed a modifier of SMA in a gender-specific manner (Howell et al., 2017a). Here, we review the structural context of SMN exon 7 and its flanking introns 6 and 7. We focus on both probed and predicted secondary structures that have been implicated in regulation of SMN exon 7 splicing. We also discuss the structural context of mutations that profoundly impact SMN exon 7 splicing.
Structure of SMN Exon 7
Secondary structure of SMN exon 7 and the downstream intron 7 probed by enzymatic and chemical methods reveals the presence of two terminal stem-loop structures, TSL1 and TSL2 (Singh et al., 2004a; Singh et al. 2007; Singh et al. 2013) (Figure 2). Located at the 5′-end of exon 7, TSL1 sequesters several presumed cis-elements that define the 3′ss of exon 7. SMN2 specific C6U is predicted to stabilize TSL1 by increasing the size of the stem in TSL1. However, results of enzymatic structure probing showed that the U residue at the 6th exonic position is highly accessible. Hence, the six-nucleotide loop of TSL1 encompasses the UAGAC motif that may serve as a binding site of hnRNP A1, a negative regulator of SMN2 exon 7 splicing. A recent study in the context of telomerase RNA has shown that hnRNP A1 preferentially interacts with its motif presented in the loop (Liu et al., 2017). The findings of this study are instructive as they highlight the viewpoint that a structural context plays a pivotal role in deciding the “fate” of RNA-protein interactions in a living cell, where limited protein supply requires binding of a given protein to its best RNA target. Supporting the inhibitory nature of TSL1, mutations predicted to abrogate this secondary structure stimulated SMN2 exon 7 inclusion (Singh et al., 2004a). However, results of mutations should be interpreted with caution as the stimulatory effect of a given mutation on splicing could be due to accidental creation of an enhancer element and/or abrogation of a silencer element.
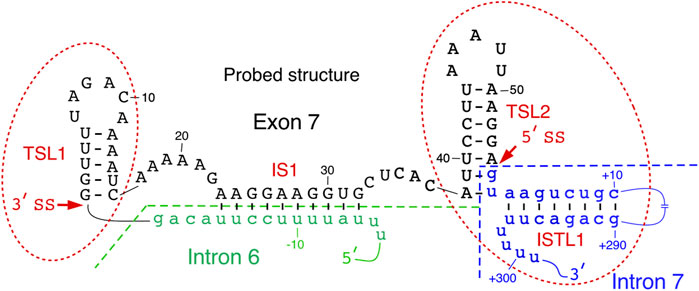
FIGURE 2. Local structure of SMN exon 7 and adjacent upstream/downstream intronic sequences. Existence of TSL2 and its effect on exon 7 splicing was also confirmed by mutational analysis. Intron 6 and intron 7 sequences are shown in small-case green and blue letters, respectively. Exon 7 sequence is shown in upper-case black letters. Numbering of nucleotides, neutral, positive and negative, starts from the first position in exon 7, first position of intron 7 and the last position in intron 6, respectively. The splice sites of exon 7 are indicated by the arrows. IS1 structure is boxed. Abbreviation: IL, internal loop; IS, internal stem; TSL, terminal stem loop.
TSL2 is one of the most scrutinized structures of exon 7. TSL2 is formed by sequences at the 3′-end of exon 7. Compared to TSL1, the structure of TSL2 is more rigid due to its longer stem. Importantly, TSL2 sequesters the first two intronic positions that define the 5′ss of exon 7 (Figure 2). In principle, U1 snRNA has a potential to form an 11-bp long RNA:RNA duplex with the 5′ss of an exon formed between U1 snRNA and the last three exonic and the first eight intronic positions (Lund and Kjems 2002). Engineered U1 snRNAs capable of forming a “perfect” 11-nucleotide-long duplex with the 5′ss of exon 7 or downstream sequences have been shown to promote SMN2 exon 7 inclusion (Singh et al., 2007; Singh et al., 2017a). In the case of SMN exon 7, U1 snRNP is predicted to form only 6 base pairs with the 5′ss of exon 7, deeming it suboptimal. Partial sequestration of the 5′ss of exon 7 by TSL2 imparts a strong inhibitory effect on splicing of SMN2 exon 7. Consistently, mutations that disrupted TSL2 promoted SMN2 exon 7 inclusion (Singh et al., 2007). Hence, a strong stimulatory effect of A54G substitution revealed by in vivo selection of the entire exon 7 could be attributed, at least in part, to the disruption of TSL2 (Singh 2007). Confirming the inhibitory role of TSL2, compensatory mutations that reinstated TSL2 restored skipping of SMN2 exon 7 splicing. Interestingly, dinucleotide substitutions that strengthened TSL2 promoted exon 7 skipping, even in the context of SMN1 (Singh et al., 2004a). These findings support the idea that transfactors interacting with splicing enhancers present within SMN1 exon 7 are insufficient to promote exon 7 inclusion in the context of a rigid secondary structure sequestering the 5′ss of exon 7.
TSL1 and TSL2 are separated from each other by an internal stem, IS1, formed between sequences in the middle of exon 7 and the 3′-end of intron 6 (Figure 2). IS1 partially sequesters the polypyrimidine tract with a potential negative impact on the recognition of the 3′ss of exon 7 by U2 snRNP. IS1 also sequesters the “Conserved tract”, a positive regulator of exon 7 splicing (Singh et al., 2004b). Supporting the negative impact of IS1 on exon 7 splicing, substitutions at several positions within IS1 were selected for in our “in vivo selection of the entire exon 7” experiment (Singh et al., 2004b). In addition, stimulatory mutations in the 5′ strand of TSL1 stem are also predicted to disrupt IS1 by forming alternative structures. We hypothesize that an abrogated IS1 might make interactions between a positive transfactor(s) and the “Conserved tract” possible, which in turn will lead to destabilization of TSL2 and improved recruitment of U1 snRNP at the 5′ss of exon 7. IS1 may also serve as an “anchor” for an interaction(s) with splicing-modulating small molecules. For instance, risdiplam, the recently approved small molecule for SMA therapy has been shown to interact with an AG-rich motif located within IS1 (Sivaramakrishnan et al., 2017; Ratni et al., 2018; Singh NN. and Singh RN. 2019; Singh et al., 2020a).
Structure of SMN Intron 7
The secondary structure of intron 7 of SMN2 probed using SHAPE revealed five terminal stem loops (TSL3, TSL4, TSL5, TSL6 and TSL7) and three internal stems formed by long-distance interactions (ISTL1, ISTL2 and ISTL3) (Singh et al., 2013; Singh et al., 2015b) (Figure 3). Among structures formed by intron 7, ISTL1 has been intensively investigated. ISTL1 is an 8-bp long duplex that sequesters the remaining portion of the 5′ss of exon 7. The cytosine residue at the 10th intronic position (10C) is the last nucleotide of the 5′-strand of ISTL1. 10C also happens to be located at the first position of the 15-nucleotide-long ISS-N1 (Figure 3). The functional significance of ISTL1 was uncovered due to an unexpected finding that two 14-nucleotide ASOs, F14 and L14, produced an opposite effect on SMN2 exon 7 splicing (Singh et al., 2010). While F14 promoted SMN2 exon 7 inclusion by sequestering the first 14 nucleotides of ISS-N1, L14 triggered SMN2 exon 7 skipping by sequestering the last 14 nucleotides of ISS-N1. The annealing positions of F14 and L14 differed by a single nucleotide as F14 sequestered 10C and L14 did not. Subsequent experiments revealed that F14 and L14 destabilize and stabilize ISTL1, respectively (Singh et al., 2013). The finding that L14 triggers SMN2 exon 7 skipping by stabilizing ISTL1 shows that an ASO has a potential to drastically alter the structural context outside its annealing positions.
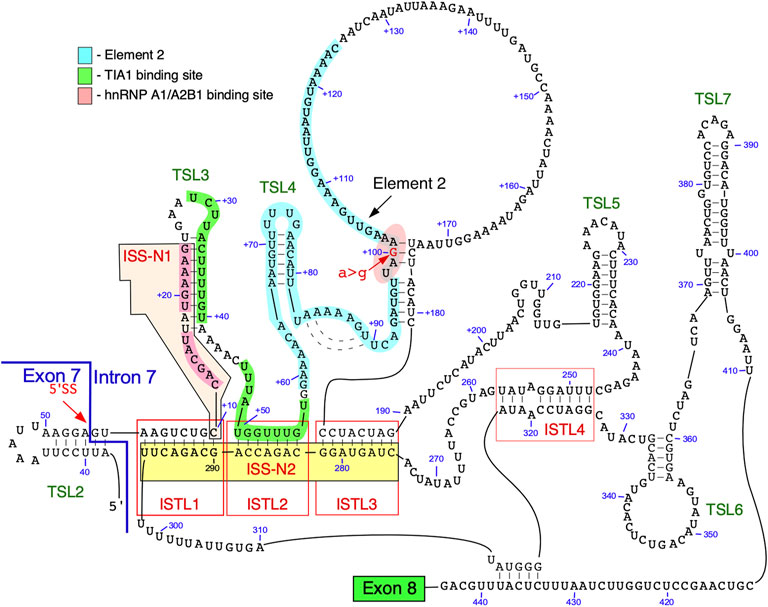
FIGURE 3. Secondary structure of SMN intron 7. The structure is based on combined probing by enzymatic and chemical methods. An exon 7/intron 7 junction as well as the 5′ss are indicated. Exon 8 is represented by a green box. Numbering of nucleotides, neutral and. positive, starts from the first position of exon 7 and the first position of intron 7, respectively. Binding sites of hnRNP A1/A2 and TIA1 are highlighted in pink and green, respectively. Element 2 sequence is highlighted in light blue. ISTL, internal stem formed by a long-distance interaction; TSL, terminal stem-loop; ISS-N2, intronic splicing silencer 2.
Two strands of ISTL1 are separated from each other by 279 nucleotides. Supporting the inhibitory nature of ISTL1, mutations that disrupted ISTL1 promoted SMN2 exon 7 inclusion (Singh et al., 2013). Compensatory mutations that reinstate the disrupted ISTL1 restored the inhibitory effect of ISTL1. ISTL2 and ISTL3 are additional structures formed by long-distance interactions, they share a continuous 3′-strand with ISTL1. The continuous sequence encompassing the 3′-strands of ISTL1, ISTL2 and ISTL3 was termed ISS-N2 (Figure 3) (Singh et al., 2013). ASOs blocking different regions of ISS-N2 stimulate SMN2 exon 7 inclusion, supporting the inhibitory nature of ISTL1, ISTL2, and ISTL3 on SMN2 exon 7 splicing. The stimulatory effect of an ISS-N2-targeting ASO was maximal when ASO sequestered the 3′-strand of ISTL1. An ISS-N2-targeting ASO also showed therapeutic benefit in a mouse model of SMA (Howell et al., 2017b). These results underscored that deep intronic sequences associated with RNA structure could be exploited for therapeutic purposes.
The structural context of intron 7 has significance for a better understanding of RNA-protein interactions and their role in SMN2 exon 7 splicing (Singh et al., 2015b). For instance, TSL3 and ISTL2 sequester the binding sites of TIA1 that is known to stimulate SMN2 exon 7 inclusion (Singh et al., 2011). One of the two putative binding motifs of hnRNP A1 present within ISS-N1 is located within a loop (Figure 3). Based on the structural context, we hypothesize that the strong binding site for hnRNP A1 presented in the loop of the stem-loop structure enables more efficient recruitment of this protein, which in turn renders the 5′ss of exon 7 inaccessible for the recruitment of U1 snRNP (Singh et al., 2015a). Furthermore, an ISS-N1 targeting ASO not only blocks the binding site of hnRNP A1 but also makes the TIA1 binding site “available” for interaction with TIA1 due to disruption of TSL3. Similarly, an ISS-N2-targeting ASO makes U1 snRNP and TIA1 binding sites accessible by disrupting ISTL1 and ISTL2, respectively.
Element 2, a positive regulator of SMN exon 7 splicing, is located downstream of the TIA1 binding site within intron 7 (Figures 1, 3) (Miyajima et al., 2002; Miyaso et al., 2003). The probed structure of intron 7 places Element 2 in both the structured region and the loop (Figure 3). Interestingly, the SMN2-specific A-to-G mutation at the 100th position of intron 7 falls within the stem region of Element 2 (Figure 3). Of note, A-to-G mutation at the 100th intronic position has been suggested to create a binding site for hnRNP A1, a negative regulator of SMN exon 7 splicing (Kashima et al., 2007). At the same time, deletion of Element 2 (together with A100G) has been shown to have a negative effect on SMN exon 7 splicing (Miyaso et al., 2003). These contradictory findings could be explained by the presence of multiple overlapping cis-elements within Element 2, a 66 nucleotide-long sequence (Miyaso et al., 2003). The strongest stimulatory effect associated with the region within Element 2 corresponds to TSL4 that harbors a U-rich loop (Figure 3). An alternative structure in this region is predicted to form a different stem-loop structure with an A-rich sequence within the loop (Figure 4) (Miyaso et al., 2003). However, the significance of any of these structures have not yet been investigated. Of note, transfactor(s) that interact with Element 2 remain unknown.
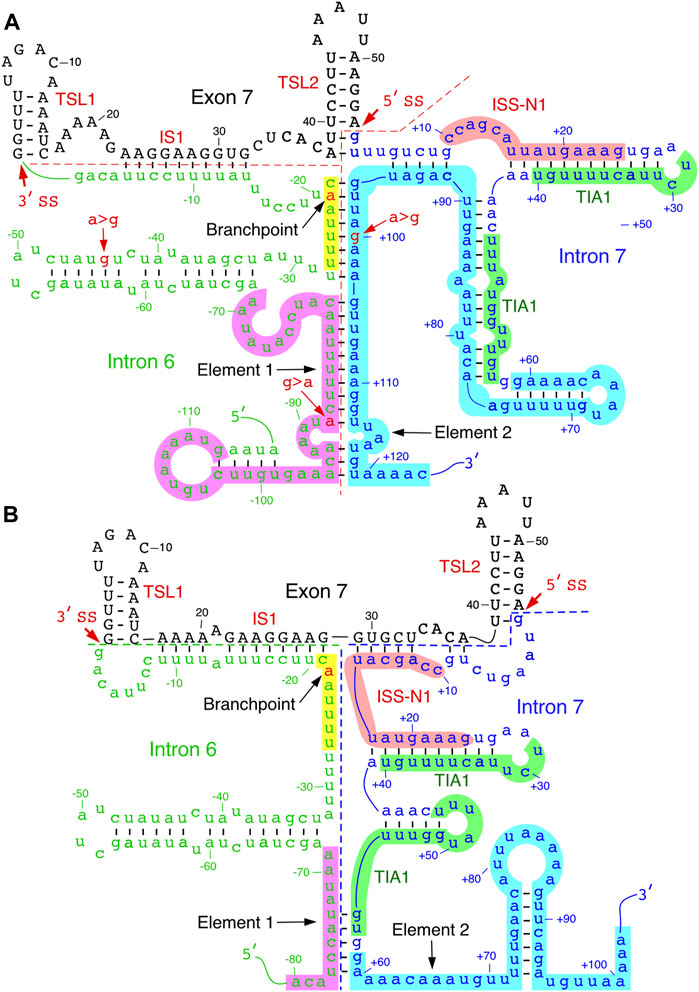
FIGURE 4. Predicted inter-intronic structures. (A). Secondary structure formed between elements 1 and 2 located within introns 6 and 7, respectively. The structure sequesters both TIA1 binding sites. Numbering of nucleotides, negative, neutral and negative, starts from the last position of intron 6, first position of exon 7 and the first position of intron 7, respectively. Element 1 is highlighted in purple; branch point sequence, in yellow, with “A” indicated in red. Other markings and abbreviations are same as shown in Figure 3. (B). An alternative secondary structure of element 2. The structural context changes the positioning of the TIA1 binding sites. Markings and abbreviation are the same as in panel A.
Inter-Intronic Structures
Folding algorithms, including mfold, RNAfold and ScanFold, can predict secondary structures with high confidence (Wang et al., 2008; Rouse et al., 2022); notably, the latter program provides metrics and models describing base pairs with likely functionality (Andrews et al., 2018). Secondary structures of SMN exon 7 and downstream intron 7 predicted by the above algorithms have generally agreed with the probed structures. Considering the structure of SMN intron 6 has not yet been probed, one can gain valuable insights from the predicted secondary structure. SMN intron 6 harbors fourteen copies of Alu-like elements, some of them are present as inverted repeats (Ottesen et al., 2017). One of these Alu elements is used as an exon, although it is predominantly skipped or/and degraded via nonsense-mediated decay (Seo et al., 2016). Secondary structures formed by inverted Alu repeats within intron 6 have been implicated in the generation of SMN circRNAs (Ottesen et al., 2019; Ottesen and Singh 2020). At the same time, a large deletion encompassing all Alu-elements of intron 6 was found to have no impact on splicing of SMN exon 7 (Singh et al., 2004a). However, sequence motifs close to the 3′-end of intron 6 have been shown to modulate SMN exon 7 splicing (Singh and Singh 2018). One such motif is Element 1 that imparts a negative impact on SMN exon 7 (Miyajima et al., 2002). Deletion or an ASO-mediated sequestration of Element 1 has been shown to promote SMN2 exon 7 inclusion (Miyajima et al., 2002; Osman et al., 2016). One of the predicted structures of exon 7 and its flanking intronic sequences places the middle of Element 1 in an internal stem formed with sequences of intron 7 (Figure 4A). Interestingly, the 3′-strand of this internal stem comes from a portion of Element 2. Hence, it is likely that this inter-intronic RNA:RNA duplex suppresses the positive effect of Element 2 on exon 7 inclusion.
Formation of an RNA:RNA duplex between two neighboring introns leads to looping out of an exon which might trigger exon skipping. In addition to the inter-intronic structure that involves sequences of Elements 1 and 2, other predicted structures reveal additional RNA:RNA duplexes formed between introns 6 and 7. One such structure is created by base pairing between a sequence located downstream of Element 1 and the sequence located in the middle of Element 2. Factors such as hnRNP A1/A2 have been implicated in exon skipping through a looping out mechanism (Martinez-Contreras et al., 2006). Considering inter-intronic interactions/structures have potential to bring different hnRNP A1/A2 binding sites of in close proximity, it is likely that skipping of SMN2 exon 7 is facilitated, at least in part, by a looping-out mechanism aided by inter-intronic RNA structures. An additional hypothesis would be that the inter-intronic RNA:RNA duplexes enforce the neighboring intra-intronic structures that sequester the positive regulatory elements in the vicinity of the splice sites of exon 7. The “looping out” hypothesis and “sequestration of splice sites due to RNA structure” hypothesis do not have to be mutually exclusive. The last fifty nucleotides of intron 6 harbor critical splicing regulatory elements, including the polypyrimidine tract and branchpoint. The predicted secondary structure in this region places most of the polypyrimidine tract in the internal stems. The precise location of SMN intron 6 branchpoint has not been identified yet. Nonetheless, the UUUUAAC motif corresponding to the consensus mammalian branchpoint motif YNYURAY is locked in a predicted RNA:RNA duplex (Figure 4A) (Gao et al., 2008). In an alternative predicted structure, the UUUUAAC motif is placed in the loop region, while positioning of ISS-N1 and the TIA1-binding sites is drastically changed (Gao et al., 2022) (Figure 4B). In this alternative structure, the first half of ISS-N1 interacts with exon 7 and blocks a portion of the “Conserved tract”, the positive regulator of exon 7 inclusion (Figure 4B). Mutations within the 5′ half of ISS-N1 are known to promote exon 7 inclusion (Supplementary Figure S1). Future experiments will reveal if the stimulatory effect of these mutations is linked, at least in part, to the RNA structure.
Assessing Propensity for Secondary Structure Formation
To assess potential for additional functional RNA structural elements, we performed preliminary scans of the pre-mRNA sequence of SMN2 using the ScanFold tool (Andrews et al., 2018) (Figure 5A). Within SMN2, ScanFold identified 84 significantly stable structures (containing base pairs with <-2 average z-score) suggesting these structures have a functional role to play (e.g., the SMN2 intronic structure in Figure 5B). Only two of these 84 highlighted structures overlap exonic sequence, while the rest resides in introns. Notably, exon 7 is spanned by one of the significantly stable ScanFold predicted exonic structures (Figure 5C) and the results recapitulate portions of the two previously determined hairpin structures TSL1 and TSL2 (Singh et al., 2007; Singh et al., 2013).
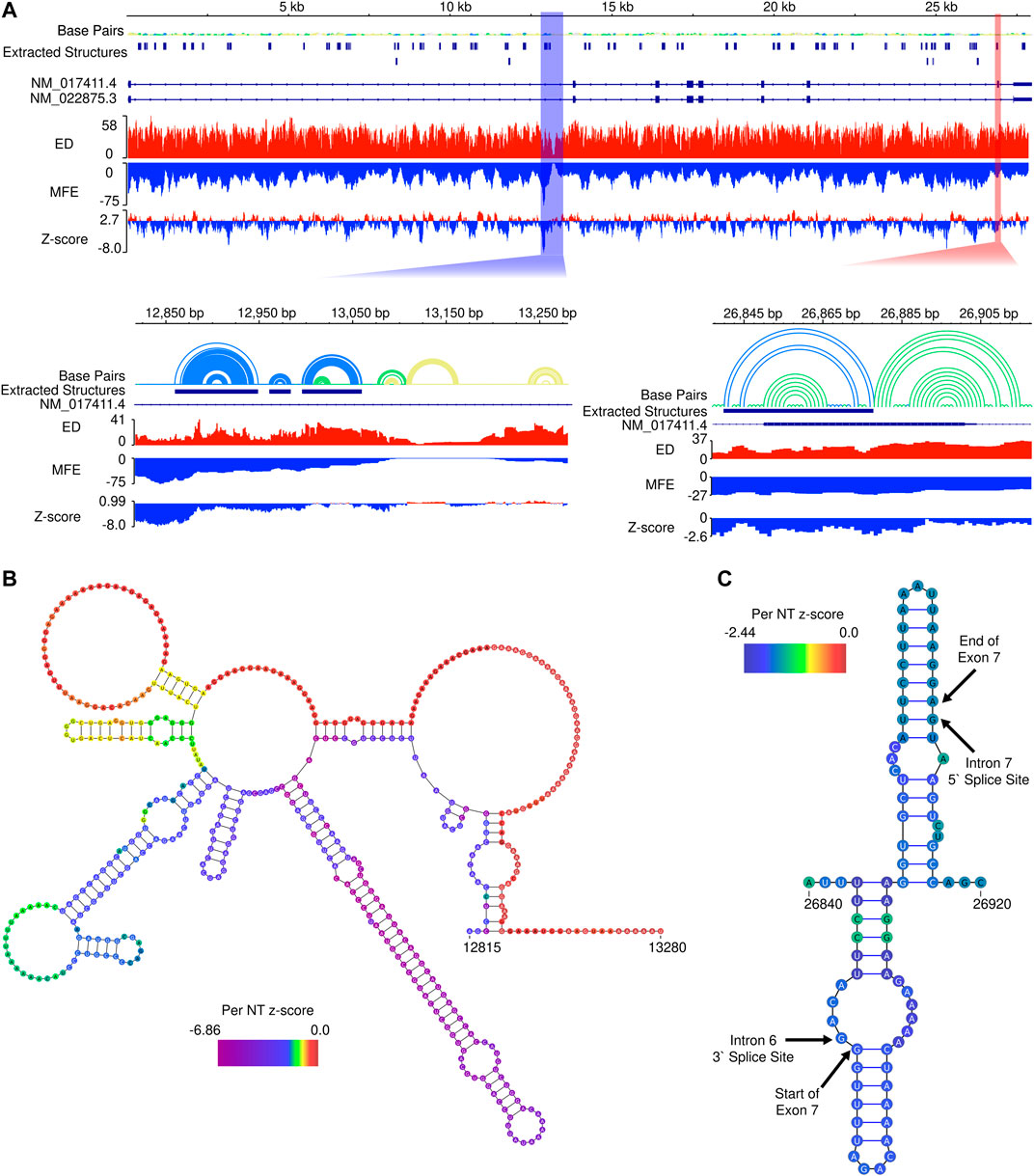
FIGURE 5. In silico ScanFold results for the pre-mRNA of SMN2. (A). At the top, an IGV representation of the whole SMN2 pre-mRNA transcript with 6 data tracks: a base pair (arc diagram) track, a track showing ScanFold extracted structures where base pairs with z-score < -2, -1 and 0 are indicated in blue, green and yellow, respectively; a track of transcripts (with introns as lines and exons as boxes), an ensemble diversity (ED) track, a minimum free energy (MFE) track, and a ΔG z-score track. Below the whole transcript are two zoomed in regions. Highlighted by the blue box is a region of SMN2 with the lowest z-score and both the lowest and highest MFE regions in the transcript. Highlighted by the red box is Exon 7, which contains one of the two extracted structures present in an exonic region. (B). The ScanFold informed 2D model of the blue highlighted region from panel A is shown with the per nucleotide (NT) z-score overlaid on the model. (C). The ScanFold informed 2D model of the red highlighted region from panel A is shown with the per nucleotide (NT) z-score overlaid on the model. Here, the boundaries of intron splice sites and the start and stop site of exon 7 are labelled.
Interestingly, the region of SMN2 which had the lowest ΔG z-score also had the lowest (most negative) minimum free energy (MFE), which is not always the case, as some relatively unstable (ΔG) regions may have ordered stability bias. Additionally, this region is adjacent to the region of highest (least stable) MFE predictions in SMN2 (Figure 5B). Notably, the region downstream of this identified structural motif also had the lowest ensemble diversity (ED). In this case, the lack of conformational diversity indicated by low ED suggests that the nucleotides here are likely to be single-stranded in most or all conformations, possibly to facilitate intermolecular interactions with regulatory transfactors.
We also employed the ScanFold program to examine the local structural context of the 5-nucleotide motifs occupying the 11th to 15th positions of SMN2 intron 7. This region falls within the ISS-N1 sequence and has recently been intensively investigated (Gao et al., 2022). One of the putative hnRNP A1 motifs, CAGCA, happens to occupy the 11th to 15th positions of SMN2 intron 7 (Hua et al., 2008). Local structure predicted by ScanFold places the last four residues of the inhibitory CAGCA motif within a stem (Supplementary Figure S1). Six mutants that promoted SMN2 exon 7 inclusion by abrogating the CAGCA motif bring noticeable changes in the local structural context (Gao et al., 2022) (Supplementary Figure S1). The stimulatory effect of these mutations could be due to both, abrogation of the negative hnRNP A1 motif and creation of a novel positive motif with the enhanced accessibility in the newly formed loop. We also observed changes in the local structural context in case of other mutants that maintained the inhibitory context while abrogating the putative motif associated with hnRNP A1 (Supplementary Figure S1). Interestingly, a single U-to-C substitution (from UUUUU to UUUCU) transformed a stimulatory motif into an inhibitory motif (Gao et al., 2022). This change retains the positioning of these 5-nucleotide motifs within the loop (Supplementary Figure S1). These results suggest that both positive and negative regulatory transfactors preferentially interact with their cognate binding sites in the single-stranded region. Future studies will reveal if additional features of high-order structure provide secondary contacts for RNA-protein interactions. Of note, the role of secondary interactions for an enhanced affinity has been reported for a factor known to promote self-splicing of a group II intron (Wank et al., 1999; Singh et al., 2002).
The ScanFold program identified 84 regions with significant thermodynamic stability throughout the pre-mRNA of SMN2. These regions have an ordered sequence arrangement, which displays higher than expected stability compared to randomized sequences of identical nucleotide composition. SMN2 contains 42 Alu elements located within its intronic sequences that represent ∼39% of the SMN2 gene (Ottesen et al., 2017). The huge repertoire of circRNAs produced by SMN genes is attributed to the RNA duplexes formed between inverted Alu repeats (Ottesen et al., 2019). Of the 84 significantly stable regions identified by ScanFold, 54 overlapped Alu elements, including 47 regions that are fully contained within an Alu element. Of the 42 Alu elements located within SMN2 pre-mRNA, 30 overlapped regions of significant stability. These regions of ordered sequence and enhanced stability are prime candidates for future investigations in structure-function mechanisms associated with Alu elements present in SMN2. Overall, these preliminary results strongly indicate that RNA secondary structure may be playing functional roles, most significantly in processing pre-mRNA into a mature transcript(s).
Concluding Remarks
The role of RNA structure in pre-mRNA splicing is a topic of growing significance. Local RNA structures are formed instantaneously as soon as a transcript emerges from the RNA polymerase. Initial RNA structures transition to more favorable structures formed by long-range interactions. However, the assessment of structural transitions in the cell remains a challenging task: for example, RNA helicases break certain secondary structures with high specificity. Protein factors tightly interacting with RNA may also impede the transition from one RNA structure into another. Potentially hundreds of proteins could be recruited during the removal of a single intron. Some of these RNA-protein interactions are expected to be non-specific due to the propensity for negatively charged RNA molecules to form electrostatic interactions with positively charged (basic) residues presented by proteins. In contrast, other RNA-protein interactions rely on certain RNA motifs. RNA structures may provide specificity by placing a given motif into a unique context. A previous report uncovered the structural context of several splicing factors that were initially thought to have preference for small linear motifs (Dominguez et al., 2018). Findings summarized in this study represent the tip of the iceberg as methods to uncover genome-wide RNA-protein interactions in the cell are still at their infancy.
Two steps of transesterification involved in the removal of every intron during pre-mRNA splicing are RNA catalyzed reactions resembling those of the self-splicing group II introns (Smathers and Robart 2019). However, unlike in self-splicing group II introns where an intronic structure alone brings and holds the splice sites together, the role of intronic sequences in pre-mRNA splicing has been assigned to recruiting snRNPs that bring and hold the splice sites together. Owing to the diverse nature of intronic sequences, the mechanism of snRNPs recruitment differs from one intron to another. Currently, there is no explanation why certain introns are removed more efficiently than others despite the comparable strength of their splice sites. The answer to this question may partly lie in the RNA structures that may sequester the splice sites and/or fold in a manner that brings the splice sites of an intron in close proximity. The generation of circRNAs provides the most convincing example of how RNA structures bring a downstream 5′ss to an upstream 3′ss for backsplicing. The process of circRNA generation competes with the forward splicing that produces the linear transcripts.
Aberrant splicing is associated with many genetic disorders. SMA happens to be one of the model diseases in which modulation of splicing employing ASOs and small molecules have conferred therapeutic benefits. The therapeutic compounds utilized in treating SMA were selected based on their ability to restore SMN2 exon 7 inclusion. Several cis-elements and transacting factors have roles implicated in regulation of SMN exon 7 splicing. Probed RNA structures place the 5′ss of SMN exon 7 in a highly sequestered structural context encompassing TSL2 and ISTL1. TSL2 and ISTL1 represent examples of an inhibitory local stem-loop structure and an inhibitory structure formed by long-distance interactions, respectively. The formation of TSL2 and ISTL1 is proposed to provide a platform for the recruitment of negative regulatory factors, including hnRNP A1/A2. Another inhibitory structure, TSL1, formed at the 3′ss of SMN2 exon 7 shifts the hnRNP A1/A2 binding site into the loop. An additional hnRNP A1/A2 binding site is located in the partially structured region of SMN2 intron 7. Overall structural context of SMN2 exon 7 and its flanking intronic sequences renders binding sites inaccessible for stimulatory transfactors that recruit U1 and U2 snRNPs at the 5 and 3′ ss of exon 7, respectively. Abrogation of a long-distance interaction similar to that of ISTL1 has been associated with the X-linked leukodystrophy Pelizaeus–Merzbacher disease (Taube et al., 2014). There is a growing interest to predict such structures as they may offer novel therapeutic targets for a number of diseases (Bernat and Disney 2015; Pervouchine 2018). Although beyond the scope of this review, recent studies on the short- and long-range RNA-RNA interactome provide interesting insights into replication of viruses, including SARS-CoV-2 (Smyth et al., 2018; Ziv et al., 2020). Future studies will reveal if these structures could be exploited for therapeutic purposes.
The roles of inter-intronic structures are the least appreciated aspects of SMN exon 7 splicing. Folding algorithms support formation of structures between introns 6 and 7 with potential to prime exon 7 to skipping if these structures are not immediately disrupted. One mechanism to avoid inter-intronic interactions is through fast removal of either intron 6 or intron 7 before structures are formed. The order in which SMN introns are removed remain largely unknown. In general, large introns are removed later than the smaller introns (Kim et al., 2017). Considering SMN intron 6 is > 13-fold larger than intron 7, its removal is expected to happen after the removal of intron 7. It is possible that a delayed removal of intron 7 further delays the removal of intron 6 leading to skipping of exon 7. We hypothesize that ASOs and small molecules that promote SMN2 exon 7 inclusion preferentially stimulate a fast removal of intron 7 through strengthening of the 5′ss of exon 7. Future studies will determine if the disruption of inter-intronic structures present therapeutic avenues for the treatment of SMA.
Available tools of RNA structure prediction and methods of RNA structure probing have provided important insights into our understanding of SMN exon 7 splicing. Yet, much remains to be learned about how RNA structures decide the fate of RNA-protein interactions in the context of SMN exon 7 splicing. Several mutations in the intron 7 have been recently shown to restore SMN2 exon 7. It will be interesting to see if RNA structure provides a mechanistic basis to explain the consequences of these mutations. Forward splicing affects backsplicing and introns used for backsplicing are generally spliced at the end (Kim et al., 2017). Considering that the 3′ss of exon 6 is used for backsplicing (Ottesen et al., 2019), it will be important to know how backsplicing-associated inter-intronic structures between intron 5 and downstream introns impact the forward splicing events, including removal of introns 6 and 7. Splicing is coupled to transcription elongation as many factors are recruited to the nascent pre-mRNA by RNA polymerase II before the termination of transcription (Saldi et al., 2016). The structure of a nascent RNA affects transcription elongation and vice-versa (Saldi et al., 2016). Hence, compounds that promote SMN exon 7 splicing through RNA structures formed during transcription elongation may provide yet another avenue for SMA therapy. Despite tremendous progress, currently approved therapies do not fully meet the needs of SMA patients (Singh 2019). A proper understanding of an RNA structure of SMN pre-mRNA will make a profound contribution to our understanding of splicing regulation of SMN genes. Novel findings emerging from the studies pertaining to the structure of SMN pre-mRNA will also shape the future therapeutic development of SMA and other diseases amenable by splicing modulation.
Author Contributions
CO, NS, RS, TE and WM analyzed data and generated figures. NS, RS and WM conceived idea, reviewed literature and wrote the manuscript.
Funding
This work was supported by grants from the National Institutes of Health R01 NS055925 and R21 NS101312 to RS; and R01GM133810 to WM.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
While authors have attempted to include most contributions on role of RNA structure in splicing regulation of SMA gene, they regret for not being able to include several related references due to the lack of space.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.928581/full#supplementary-material
References
Andrews, R. J., Roche, J., and Moss, W. N. (2018). ScanFold: an Approach for Genome-wide Discovery of Local RNA Structural Elements-Applications to Zika Virus and HIV. PeerJ 6, e6136. doi:10.7717/peerj.6136
Bennett, C. F., Krainer, A. R., and Cleveland, D. W. (2019). Antisense Oligonucleotide Therapies for Neurodegenerative Diseases. Annu. Rev. Neurosci. 42, 385–406. doi:10.1146/annurev-neuro-070918-050501
Bernat, V., and Disney, M. D. (2015). RNA Structures as Mediators of Neurological Diseases and as Drug Targets. Neuron 87, 28–46. doi:10.1016/j.neuron.2015.06.012
Beusch, I., Barraud, P., Moursy, A., Cléry, A., and Allain, F. H. (2017). Tandem hnRNP A1 RNA Recognition Motifs Act in Concert to Repress the Splicing of Survival Motor Neuron Exon 7. Elife 6, 6. doi:10.7554/eLife.25736
Campagne, S., Boigner, S., Rüdisser, S., Moursy, A., Gillioz, L., Knörlein, A., et al. (2019). Structural Basis of a Small Molecule Targeting RNA for a Specific Splicing Correction. Nat. Chem. Biol. 15 (15), 1191–1198. doi:10.1038/s41589-019-0384-5
Cartegni, L., and Krainer, A. R. (2002). Disruption of an SF2/ASF-dependent Exonic Splicing Enhancer in SMN2 Causes Spinal Muscular Atrophy in the Absence of SMN1. Nat. Genet. 30, 377–384. doi:10.1038/ng854
Charenton, C., Wilkinson, M. E., and Nagai, K. (2019). Mechanism of 5' Splice Site Transfer for Human Spliceosome Activation. Science 364 (364), 362–367. doi:10.1126/science.aax3289
Cho, S., and Dreyfuss, G. (2010). A Degron Created by SMN2 Exon 7 Skipping Is a Principal Contributor to Spinal Muscular Atrophy Severity. Genes Dev. 24, 438–442. doi:10.1101/gad.1884910
De Conti, L., Baralle, M., and Buratti, E. (2013). Exon and Intron Definition in Pre-mRNA Splicing. WIREs RNA 4, 49–60. doi:10.1002/wrna.1140
Dominguez, D., Freese, P., Alexis, M. S., Su, A., Hochman, M., Palden, T., et al. (2018). Sequence, Structure, and Context Preferences of Human RNA Binding Proteins. Mol. Cell 70, 854–e9. doi:10.1016/j.molcel.2018.05.001
Gao, K., Masuda, A., Matsuura, T., and Ohno, K. (2008). Human Branch Point Consensus Sequence Is yUnAy. Nucleic Acids Res. Apr 36, 2257–2267. doi:10.1093/nar/gkn073
Gao, Y., Lin, K. T., Jiang, T., Yang, Y., Rahman, M. A., Gong, S., et al. (2022). Systematic Characterization of Short Intronic Splicing-Regulatory Elements in SMN2 Pre-mRNA. Nucleic Acids Res. 01 25 (50), 731–749. doi:10.1093/nar/gkab1280
Hofmann, Y., Lorson, C. L., Stamm, S., Androphy, E. J., and Wirth, B. (2000). Htra2-β 1 Stimulates an Exonic Splicing Enhancer and Can Restore Full-Length SMN Expression to Survival Motor Neuron 2 ( SMN2 ). Proc. Natl. Acad. Sci. U.S.A. 97, 9618–9623. doi:10.1073/pnas.160181697
Hofmann, Y., and Wirth, B. (2002). hnRNP-G Promotes Exon 7 Inclusion of Survival Motor Neuron (SMN) via Direct Interaction with Htra2-Beta1. Hum. Mol. Genet. Aug 11, 2037–2049. doi:10.1093/hmg/11.17.2037
Howell, M. D., Ottesen, E. W., Singh, N. N., Anderson, R. L., Seo, J., Sivanesan, S., et al. (2017a). TIA1 Is a Gender-specific Disease Modifier of a Mild Mouse Model of Spinal Muscular Atrophy. Sci. Rep. 7 (7), 7183. doi:10.1038/s41598-017-07468-2
Howell, M. D., Ottesen, E. W., Singh, N. N., Anderson, R. L., and Singh, R. N. (2017b). Gender-Specific Amelioration of SMA Phenotype upon Disruption of a Deep Intronic Structure by an Oligonucleotide. Mol. Ther. 25 (25), 1328–1341. doi:10.1016/j.ymthe.2017.03.036
Hua, Y., Vickers, T. A., Okunola, H. L., Bennett, C. F., and Krainer, A. R. (2008). Antisense Masking of an hnRNP A1/A2 Intronic Splicing Silencer Corrects SMN2 Splicing in Transgenic Mice. Am. J. Hum. Genet. 82, 834–848. doi:10.1016/j.ajhg.2008.01.014
Kashima, T., Rao, N., and Manley, J. L. (2007). An Intronic Element Contributes to Splicing Repression in Spinal Muscular Atrophy. Proc. Natl. Acad. Sci. U. S. A. 104 (104), 3426–3431. doi:10.1073/pnas.0700343104
Kashima, T., and Manley, J. L. (2003). A Negative Element in SMN2 Exon 7 Inhibits Splicing in Spinal Muscular Atrophy. Nat. Genet. 34, 460–463. doi:10.1038/ng1207
Keil, J. M., Seo, J., Howell, M. D., Hsu, W. H., Singh, R. N., and DiDonato, C. J. (2014). A Short Antisense Oligonucleotide Ameliorates Symptoms of Severe Mouse Models of Spinal Muscular Atrophy. Mol. Ther. - Nucleic Acids 3, e174. doi:10.1038/mtna.2014.23
Kim, S. W., Taggart, A. J., Heintzelman, C., Cygan, K. J., Hull, C. G., Wang, J., et al. (2017). Widespread Intra-dependencies in the Removal of Introns from Human Transcripts. Nucleic Acids Res. 45, 9503–9513. doi:10.1093/nar/gkx661
Lefebvre, S., Bürglen, L., Reboullet, S., Clermont, O., Burlet, P., Viollet, L., et al. (1995). Identification and Characterization of a Spinal Muscular Atrophy-Determining Gene. Cell 80, 155–165. doi:10.1016/0092-8674(95)90460-3
Lim, S. R., and Hertel, K. J. (2001). Modulation of Survival Motor Neuron Pre-mRNA Splicing by Inhibition of Alternative 3' Splice Site Pairing. J. Biol. Chem. 276 (276), 45476–45483. doi:10.1074/jbc.M107632200
Lipnick, S. L., Agniel, D. M., Aggarwal, R., Makhortova, N. R., Finlayson, S. G., Brocato, A., et al. (2019). Systemic Nature of Spinal Muscular Atrophy Revealed by Studying Insurance Claims. PLoS One 14, e0213680. doi:10.1371/journal.pone.0213680
Liu, X., Ishizuka, T., Bao, H. L., Wada, K., Takeda, Y., Iida, K., et al. (2017). Structure-Dependent Binding of hnRNPA1 to Telomere RNA. J. Am. Chem. Soc. 139 (139), 7533–7539. doi:10.1021/jacs.7b01599
Lorson, C. L., Hahnen, E., Androphy, E. J., and Wirth, B. (1999). A Single Nucleotide in the SMN Gene Regulates Splicing and Is Responsible for Spinal Muscular Atrophy. Proc. Natl. Acad. Sci. U.S.A. 96, 6307–6311. doi:10.1073/pnas.96.11.6307
Lund, M., and Kjems, J. R. (2002). Defining a 5??? Splice Site by Functional Selection in the Presence and Absence of U1 snRNA 5??? End. Rna 8, 166–179. doi:10.1017/s1355838202010786
Martinez-Contreras, R., Fisette, J.-F., Nasim, F.-u. H., Madden, R., Cordeau, M., and Chabot, B. (2006). Intronic Binding Sites for hnRNP A/B and hnRNP F/H Proteins Stimulate Pre-mRNA Splicing. PLoS Biol. 4, e21. doi:10.1371/journal.pbio.0040021
Miyajima, H., Miyaso, H., Okumura, M., Kurisu, J., and Imaizumi, K. (2002). Identification of a Cis-Acting Element for the Regulation of SMN Exon 7 Splicing. J. Biol. Chem. 277 (277), 23271–23277. doi:10.1074/jbc.M200851200
Miyaso, H., Okumura, M., Kondo, S., Higashide, S., Miyajima, H., and Imaizumi, K. (2003). An Intronic Splicing Enhancer Element in Survival Motor Neuron (SMN) Pre-mRNA. J. Biol. Chem. 278, 15825–15831. doi:10.1074/jbc.m209271200
Monani, U. R., Lorson, C. L., Parsons, D. W., Prior, T. W., Androphy, E. J., Burghes, A. H., et al. (1999). A Single Nucleotide Difference that Alters Splicing Patterns Distinguishes the SMA Gene SMN1 from the Copy Gene SMN2. Hum. Mol. Genet. Jul 8, 1177–1183. doi:10.1093/hmg/8.7.1177
Moore, M. J., and Proudfoot, N. J. (2009). Pre-mRNA Processing Reaches Back toTranscription and Ahead to Translation. Cell 136, 688–700. doi:10.1016/j.cell.2009.02.001
Osman, E. Y., Washington, C. W., Kaifer, K. A., Mazzasette, C., Patitucci, T. N., Florea, K. M., et al. (2016). Optimization of Morpholino Antisense Oligonucleotides Targeting the Intronic Repressor Element1 in Spinal Muscular Atrophy. Mol. Ther. 0924, 1592–1601. doi:10.1038/mt.2016.145
Ottesen, E. W., Howell, M. D., Singh, N. N., Seo, J., Whitley, E. M., and Singh, R. N. (2016). Severe Impairment of Male Reproductive Organ Development in a Low SMN Expressing Mouse Model of Spinal Muscular Atrophy. Sci. Rep. 6, 20193. doi:10.1038/srep20193
Ottesen, E. W., Luo, D., Seo, J., Singh, N. N., and Singh, R. N. (2019). HumanSurvival Motor Neurongenes Generate a Vast Repertoire of Circular RNAs. Nucleic Acids Res. 47, 2884–2905. doi:10.1093/nar/gkz034
Ottesen, E. W., Seo, J., Singh, N. N., and Singh, R. N. (2017). A Multilayered Control of the Human Survival Motor Neuron Gene Expression by Alu Elements. Front. Microbiol. 8, 2252. doi:10.3389/fmicb.2017.02252
Ottesen, E. W., and Singh, R. N. (2020). Characteristics of Circular RNAs Generated by Human Survival Motor Neuron Genes. Cell. Signal. 73, 109696. doi:10.1016/j.cellsig.2020.109696
Pedrotti, S., Bielli, P., Paronetto, M. P., Ciccosanti, F., Fimia, G. M., Stamm, S., et al. (2010). The Splicing Regulator Sam68 Binds to a Novel Exonic Splicing Silencer and Functions in SMN2 Alternative Splicing in Spinal Muscular Atrophy. Embo J. 29, 1235–1247. doi:10.1038/emboj.2010.19
Pervouchine, D. D. (2018). Towards Long-Range RNA Structure Prediction in Eukaryotic Genes. Genes (Basel) 9, 9. doi:10.3390/genes9060302
Ratni, H., Ebeling, M., Baird, J., Bendels, S., Bylund, J., Chen, K. S., et al. (2018). Discovery of Risdiplam, a Selective Survival of Motor Neuron-2 (SMN2) Gene Splicing Modifier for the Treatment of Spinal Muscular Atrophy (SMA). J. Med. Chem. 61, 6501–6517. doi:10.1021/acs.jmedchem.8b00741
Rouse, W. B., Andrews, R. J., Booher, N. J., Wang, J., Woodman, M. E., Dow, E. R., et al. (2022). Prediction and Analysis of Functional RNA Structures within the Integrative Genomics Viewer. Nar. Genom Bioinform. Mar. 4. doi:10.1093/nargab/lqab127
Saldi, T., Cortazar, M. A., Sheridan, R. M., and Bentley, D. L. (2016). Coupling of RNA Polymerase II Transcription Elongation with Pre-mRNA Splicing. J. Mol. Biol. 06428, 2623–2635. doi:10.1016/j.jmb.2016.04.017
Seo, J., Singh, N. N., Ottesen, E. W., Lee, B. M., and Singh, R. N. (2016). A Novel Human-specific Splice Isoform Alters the Critical C-Terminus of Survival Motor Neuron Protein. Sci. Rep. 6 (6), 30778. doi:10.1038/srep30778
Shenasa, H., and Hertel, K. J. (2019). Combinatorial Regulation of Alternative Splicing. Biochimica Biophysica Acta (BBA) - Gene Regul. Mech. 1862, 194392. doi:10.1016/j.bbagrm.2019.06.003
Singh, N. K., Singh, N. N., Androphy, E. J., and Singh, R. N. (2006). Splicing of a Critical Exon of Human Survival Motor Neuron Is Regulated by a Unique Silencer Element Located in the Last Intron. Mol. Cell Biol. 26, 1333–1346. doi:10.1128/mcb.26.4.1333-1346.2006
Singh, N. N., Hoffman, S., Reddi, P. P., and Singh, R. N. (2021). Spinal Muscular Atrophy: Broad Disease Spectrum and Sex-specific Phenotypes. Biochim. Biophys. Acta Mol. Basis Dis. 1867, 166063. doi:10.1016/j.bbadis.2020.166063
Singh, N. N., Singh, R. N., and Androphy, E. J. (2007). Modulating Role of RNA Structure in Alternative Splicing of a Critical Exon in the Spinal Muscular Atrophy Genes. Nucleic Acids Res. 35, 371–389. doi:10.1093/nar/gkl1050
Singh, N. N., and Singh, R. N. (2019a). How RNA Structure Dictates the Usage of a Critical Exon of Spinal Muscular Atrophy Gene. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 194403. doi:10.1016/j.bbagrm.2019.07.004
Singh, N. N., Androphy, E. J., and Singh, R. N. (2004a). An Extended Inhibitory Context Causes Skipping of Exon 7 of SMN2 in Spinal Muscular Atrophy. Biochem. Biophysical Res. Commun. 315, 381–388. doi:10.1016/j.bbrc.2004.01.067
Singh, N. N., Androphy, E. J., and Singh, R. N. (2004b). In Vivo selection Reveals Combinatorial Controls that Define a Critical Exon in the Spinal Muscular Atrophy Genes. Rna 10, 1291–1305. doi:10.1261/rna.7580704
Singh, N. N., Androphy, E. J., and Singh, R. N. (2004c). The Regulation and Regulatory Activities of Alternative Splicing of the SMN Gene. Crit. Rev. Eukaryot. Gene Expr. 14, 271–286. doi:10.1615/critreveukaryotgeneexpr.v14.i4.30
Singh, N. N., Del Rio-Malewski, J. B., Luo, D., Ottesen, E. W., Howell, M. D., and Singh, R. N. (2017a). Activation of a Cryptic 5′ Splice Site Reverses the Impact of Pathogenic Splice Site Mutations in the Spinal Muscular Atrophy Gene. Nucleic Acids Res. Dec 45, 12214–12240. doi:10.1093/nar/gkx824
Singh, N. N., Hollinger, K., Bhattacharya, D., and Singh, R. N. (2010). An Antisense Microwalk Reveals Critical Role of an Intronic Position Linked to a Unique Long-Distance Interaction in Pre-mRNA Splicing. Rna 16, 1167–1181. doi:10.1261/rna.2154310
Singh, N. N., Howell, M. D., Androphy, E. J., and Singh, R. N. (2017b). How the Discovery of ISS-N1 Led to the First Medical Therapy for Spinal Muscular Atrophy. Gene Ther. 24, 520–526. doi:10.1038/gt.2017.34
Singh, N. N., Lawler, M. N., Ottesen, E. W., Upreti, D., Kaczynski, J. R., and Singh, R. N. (2013). An Intronic Structure Enabled by a Long-Distance Interaction Serves as a Novel Target for Splicing Correction in Spinal Muscular Atrophy. Nucleic Acids Res. Sep. 41, 8144–8165. doi:10.1093/nar/gkt609
Singh, N. N., Lee, B. M., DiDonato, C. J., and Singh, R. N. (2015a). Mechanistic Principles of Antisense Targets for the Treatment of Spinal Muscular Atrophy. Future Med. Chem. 7, 1793–1808. doi:10.4155/fmc.15.101
Singh, N. N., Lee, B. M., and Singh, R. N. (2015b). Splicing Regulation in Spinal Muscular Atrophy by an RNA Structure Formed by Long-Distance Interactions. Ann. N.Y. Acad. Sci. 1341, 176–187. doi:10.1111/nyas.12727
Singh, N. N., Seo, J., Ottesen, E. W., Shishimorova, M., Bhattacharya, D., and Singh, R. N. (2011). TIA1 Prevents Skipping of a Critical Exon Associated with Spinal Muscular Atrophy. Mol. Cell Biol. 31, 935–954. doi:10.1128/mcb.00945-10
Singh, N. N., Shishimorova, M., Cao, L. C., Gangwani, L., and Singh, R. N. (2009). A Short Antisense Oligonucleotide Masking a Unique Intronic Motif Prevents Skipping of a Critical Exon in Spinal Muscular Atrophy. RNA Biol. 6, 341–350. doi:10.4161/rna.6.3.8723
Singh, R. N. (2019). More Is Needed to Complement the Available Therapies of Spinal Muscular Atrophy. Future Med. Chem. 11 (11), 2873–2876. doi:10.4155/fmc-2019-0239
Singh, R. N., Ottesen, E. W., and Singh, N. N. (2020a). The First Orally Deliverable Small Molecule for the Treatment of Spinal Muscular Atrophy. Neurosci. Insights 15, 2633105520973985. doi:10.1177/2633105520973985
Singh, R. N., Seo, J., and Singh, N. N. (2020b). RNA in Spinal Muscular Atrophy: Therapeutic Implications of Targeting. Expert Opin. Ther. Targets 24, 731–743. doi:10.1080/14728222.2020.1783241
Singh, R. N., and Singh, N. N. (2019b). A Novel Role of U1 snRNP: Splice Site Selection from a Distance. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 1862634–1862642. doi:10.1016/j.bbagrm.2019.04.004
Singh, R. N., Howell, M. D., Ottesen, E. W., and Singh, N. N. (2017c). Diverse Role of Survival Motor Neuron Protein. Biochimica Biophysica Acta (BBA) - Gene Regul. Mech. 1860, 299–315. doi:10.1016/j.bbagrm.2016.12.008
Singh, R. N., Saldanha, R. J., D'Souza, L. M., and Lambowitz, A. M. (2002). Binding of a Group II Intron-Encoded Reverse Transcriptase/maturase to its High Affinity Intron RNA Binding Site Involves Sequence-specific Recognition and Autoregulates Translation. J. Mol. Biol. 318, 287–303. doi:10.1016/s0022-2836(02)00054-2
Singh, R. N., and Singh, N. N. (2018). Mechanism of Splicing Regulation of Spinal Muscular Atrophy Genes. Adv. Neurobiol. 20, 31–61. doi:10.1007/978-3-319-89689-2_2
Singh, R. N. (2007). Unfolding the Mystery of Alternative Splicing through a Unique Method of In Vivo Selection. Front. Biosci. 12, 3263–3272. doi:10.2741/2310
Sivaramakrishnan, M., McCarthy, K. D., Campagne, S., Huber, S., Meier, S., Augustin, A., et al. (2017). Binding to SMN2 Pre-mRNA-protein Complex Elicits Specificity for Small Molecule Splicing Modifiers. Nat. Commun. 8 (8), 1476. doi:10.1038/s41467-017-01559-4
Smathers, C. M., and Robart, A. R. (2019). The Mechanism of Splicing as Told by Group II Introns: Ancestors of the Spliceosome. Biochimica Biophysica Acta (BBA) - Gene Regul. Mech. 1862, 194390. doi:10.1016/j.bbagrm.2019.06.001
Smyth, R. P., Negroni, M., Lever, A. M., Mak, J., and Kenyon, J. C. (2018). RNA Structure-A Neglected Puppet Master for the Evolution of Virus and Host Immunity. Front. Immunol. 9, 2097. doi:10.3389/fimmu.2018.02097
Taube, J. R., Sperle, K., Banser, L., Seeman, P., Cavan, B. C., Garbern, J. Y., et al. (2014). PMD Patient Mutations Reveal a Long-Distance Intronic Interaction that Regulates PLP1/DM20 Alternative Splicing. Hum. Mol. Genet. 23 (23), 5464–5478. doi:10.1093/hmg/ddu271
Wang, Q., Barr, I., Guo, F., and Lee, C. (2008). Evidence of a Novel RNA Secondary Structurein the Coding Region of HIV-1 Pol Gene. Rna 14, 2478–2488. doi:10.1261/rna.1252608
Wank, H., SanFilippo, J., Singh, R. N., Matsuura, M., and Lambowitz, A. M. (1999). A Reverse Transcriptase/maturase Promotes Splicing by Binding at its Own Coding Segment in a Group II Intron RNA. Mol. Cell 4, 239–250. doi:10.1016/s1097-2765(00)80371-8
Wirth, B., Karakaya, M., Kye, M. J., and Mendoza-Ferreira, N. (2020). Twenty-Five Years of Spinal Muscular Atrophy Research: From Phenotype to Genotype to Therapy, and what Comes Next. Annu. Rev. Genomics Hum. Genet. 21, 231–261. doi:10.1146/annurev-genom-102319-103602
Young, P. J., DiDonato, C. J., Hu, D., Kothary, R., Androphy, E. J., and Lorson, C. L. (2002). SRp30c-dependent Stimulation of Survival Motor Neuron (SMN) Exon 7 Inclusion Is Facilitated by a Direct Interaction with hTra2beta1. Hum. Mol. Genet. Mar. 01 11, 577–587. doi:10.1093/hmg/11.5.577
Keywords: RNA structure, splicing, small molecule, spinal muscular atrophy, SMA, survival motor neuron, SMN, ISS-N1
Citation: Singh NN, O'Leary CA, Eich T, Moss WN and Singh RN (2022) Structural Context of a Critical Exon of Spinal Muscular Atrophy Gene. Front. Mol. Biosci. 9:928581. doi: 10.3389/fmolb.2022.928581
Received: 10 May 2022; Accepted: 13 June 2022;
Published: 01 July 2022.
Edited by:
Agnieszka Kiliszek, Institute of Bioorganic Chemistry (PAS), PolandReviewed by:
Yiliang Ding, John Innes Centre, United KingdomSebastien Campagne, INSERM U1212 Régulations Naturelles et Artificielles (ARNA), France
Copyright © 2022 Singh, O'Leary, Eich, Moss and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ravindra N. Singh, c2luZ2hyQGlhc3RhdGUuZWR1
 Natalia N. Singh1
Natalia N. Singh1 Collin A. O'Leary
Collin A. O'Leary Walter N. Moss
Walter N. Moss Ravindra N. Singh
Ravindra N. Singh