
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci., 06 July 2022
Sec. Protein Biochemistry for Basic and Applied Sciences
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.926959
This article is part of the Research TopicMolecular Determinants of Protein Assemblies in Health and DiseaseView all 9 articles
Candida Als family adhesins mediate adhesion to biological and abiotic substrates, as well as fungal cell aggregation, fungal-bacterial co-aggregation and biofilm formation. The activity of at least two family members, Als5 and Als1, is dependent on amyloid-like protein aggregation that is initiated by shear force. Each Als adhesin has a ∼300-residue N-terminal Ig-like/invasin region. The following 108-residue, low complexity, threonine-rich (T) domain unfolds under shear force to expose a critical amyloid-forming segment 322SNGIVIVATTRTV334 at the interface between the Ig-like/invasin domain 2 and the T domain of Candida albicans Als5. Amyloid prediction programs identified six potential amyloidogenic sequences in the Ig-like/invasin region and three others in the T domain of C. albicans Als5. Peptides derived from four of these sequences formed fibrils that bound thioflavin T, the amyloid indicator dye, and three of these revealed atomic-resolution structures of cross-β spines. These are the first atomic-level structures for fungal adhesins. One of these segments, from the T domain, revealed kinked β-sheets, similarly to LARKS (Low-complexity, Amyloid-like, Reversible, Kinked segments) found in human functional amyloids. Based on the cross-β structures in Als proteins, we use evolutionary arguments to identify functional amyloidogenic sequences in other fungal adhesins, including adhesins from Candida auris. Thus, cross-β structures are often involved in fungal pathogenesis and potentially in antifungal therapy.
Potential amyloid-like segments in fungal adhesins are functional in the sense that they mediate microbial adhesion, aggregation, and biofilm formation (Lipke et al., 2017; Tayeb-Fligelman et al., 2017; Evans et al., 2018; Jain and Chapman, 2019). They are amyloid in the sense that they mediate protein assembly into structured β-rich aggregates and fibrils, the same molecular interactions as those in amyloids in neurodegenerative diseases. Amyloid definition is based on the formation of cross-β fibrils composed of tightly mated β-sheets and interdigitating side chains (Shewmaker et al., 2011; Eisenberg and Jucker, 2012; Eisenberg and Sawaya, 2017). Many microbial aggregates and biofilms display these dye-binding properties, and several adhesins form amyloid fibers in vitro (Ramsook et al., 2010; Garcia et al., 2011; Lipke et al., 2017). Among the best characterized, gram negative curli are pili composed of proteins assembled through cross-β amyloid-like interactions, and they mediate bacterial adhesion and biofilm formation (Tayeb-Fligelman et al., 2017; Evans et al., 2018; Jain and Chapman, 2019). In vitro, anti-amyloid compounds inhibit biofilm formation in bacterial and fungal model systems at concentrations similar to those that inhibit formation of amyloids associated with neurodegenerative diseases and serum amyloidosis (Cegelski et al., 2009; Garcia et al., 2011; Landau et al., 2011; Perov et al., 2019). Thus, the characteristics of microbial functional amyloid mirror those of the better-known pathological amyloids.
Classical amyloids including pathological amyloids in Alzheimer’s disease and serum amyloidoses form fibers that have a characteristic cross-β pattern under X-ray diffraction (Eisenberg and Sawaya, 2017). This pattern shows strong orthogonal reflections of β-strands and β-sheets, with the β-strands orthogonal to the fiber axis and the sheets parallel to the axis. The β-sheets are stacked, and at least one interface between β-sheets is composed of tightly packed interdigitated sidechains, interacting through van der Waals forces and anhydrous H-bonds. This close interdigitation and dry interface give rise to the name “steric zipper,” and the crystalline cross-β structures of short segments are called “amyloid spines” (Eisenberg and Sawaya, 2017).
A second type of homologous crystalline assembly has been recently described: Low-complexity, Amyloid-like, Reversible, Kinked segments (LARKS) (Hughes et al., 2018). These structures also have a cross-β like arrangement, but the β-strands are kinked, the sheet interactions are dependent on smaller interfaces, and the structures are less stable and more evanescent. These LARKs spines are common in low-complexity unstructured regions of proteins (Hughes et al., 2018). Low complexity domains are regions in protein sequences that differ from the composition and complexity of most globular proteins. Such LARKS sequences are involved in RNA-dependent phase separations that influence cellular stress response (Hughes et al., 2018; Hughes et al., 2021); Moreover, LARKS have not been previously reported in adhesins.
Among the best studied fungal adhesins are the paralogs Als1, Als3, and Als5 from Candida albicans (Hoyer, 2001; Lipke et al., 2017). Als1 and Als3 are important for fungal virulence in mice, and Als5 can attenuate immune response in the host (Garcia-Sherman et al., 2015; Behrens et al., 2019). Als adhesins are covalently attached to the cell wall glycans and are encoded at 8 loci in the genome (Hoyer et al., 1998; Hoyer, 2001). They are typically 1,200–2,200 residues long (Hoyer, 2001). The N-terminal region is distal to the wall and consists of a 300-residue β-sheet-rich Ig-like/invasin region with two subdomains with Ig-like Greek key folds stabilized by disulfide bonds (Salgado et al., 2011; Lin et al., 2014). In each Als protein, the Ig-like/invasin region is followed by a 108-residue Threonine-rich low-complexity domain (T domain) that is highly conserved in all paralogs and includes a strong amyloidogenic sequence GIVIVA (Otoo et al., 2008). This strong amyloid signature is part of a larger segment, 322SNGIVIVATTRTV334, which was shown crucial for Als5p amyloid formation (Garcia et al., 2011; Lipke et al., 2017). The entire T region is essential for secretion and processing in yeast (Rauceo et al., 2006). C-terminal to T regions are 5–30 copies of a 36-residue tandem repeat. This region mediates hydrophobic effect interactions with abiotic substrates and with homologous structures. The Als adhesin stalk regions includes the C-terminal 500–1,000 residues and are also of low complexity. These regions are rich in O-glycosylated Ser and Thr residues, as well as in N-glycosylated Asn-Xaa-Ser/Thr motifs. Therefore, the stalks extend up to 200 nm from the surface of the wall. The proteins are synthesized with C-terminal glycosyl phosphatidyl inositol (GPI) anchors, which are subsequently modified to form covalent crosslinks to cell wall glucans (Lipke and Kurjan, 1992; Kapteyn et al., 2000; Roh et al., 2002).
Als5 and other fungal adhesins have characteristics of functional amyloids activated by physical stress. Under extension force applied by liquid flow or atomic force microscopy (AFM) extension, Als adhesin T domains unfold and expose the amyloidogenic sequence GIVIVA from the interface between the Ig/invasin region and the T domain (Lipke et al., 2017). Peptides with this sequence form amyloid fibers, as do soluble forms of Als5, but peptides of fragments with a non-amyloid substitution GINIVA do not form amyloids (Otoo et al., 2008; Ramsook et al., 2010). When anchored to the cell surface, exposure of the GIVIVA sequence leads to formation of high avidity patches of adhesins on the cell surface (Alsteens et al., 2010). These patches consist of adhesins aggregated through amyloid-like bonds, and as a result, cells expressing these adhesins bind amyloidophilic dyes strongly (nM concentrations of thioflavins) (Garcia et al., 2011). Furthermore, aggregates of cells expressing these adhesins are birefringent in polarized light, a phenomenon not seen in the controls (Otoo et al., 2008; Ramsook et al., 2010). Recent evidence also shows that cell-to-cell binding is mediated by amyloid-like interactions between cells for Als5 and Als1 (Dehullu et al., 2019a; Dehullu et al., 2019b; Ho et al., 2019). Thus, formation of amyloid-like interactions mediates strong bonding between fungal cells, especially in biofilms formed under flow. Furthermore, amyloid-like structures coat the surface of fungi in fatal invasive fungal infections caused by many species. In superficial and invasive candidiasis, Als proteins are major components of these surface amyloid-like structures (Garcia-Sherman et al., 2014; Garcia-Sherman et al., 2015; Klotz et al., 2016). The fungal surface amyloid attenuates macrophage response to the invading fungi (Behrens et al., 2019). Thus, extensive amyloid-like interactions are common on adhering fungi and in fungal biofilms and can influence host response.
Amyloidogenic sequences in fungal adhesins have been widely predicted, and force-dependent formation of amyloid-like surface nanodomains has been demonstrated in several cases (Chan and Lipke, 2014; Lipke et al., 2017; Lipke, 2018; Bouyx et al., 2021). However, no atomic-level structures of amyloid spines or steric zippers from this type of protein have been reported. The adhesin Als5 conforms to the general model in its sequence, with an Ig/invasin region (residues 20–325), a 108-residue T region, six tandem repeats, and a 750-residue unstructured stalk. The tandem repeats and stalk are low complexity sequences, rich in serine and threonine residues, and are glycosylated on up to a third of all residues (Rauceo et al., 2004; Lipke, 2018). Such high glycosylation is not compatible with amyloid formation through cross-β structures, and therefore we restricted our analysis to the Ig-like/invasin and T regions. We have screened the Als5 Ig-like/invasin and T domains for predicted amyloidogenic sequences and determined structural characteristics of several of these. We have also analyzed the conservation and potential in vivo roles of these sequence segments in Als5 and homologous proteins.
Peptide segments 156NTVTFN161, 168SIAVNF173, 196IATLYV201, 324GIVIVA329 and 369TSYVGV374 from Als5 (UniProt accession number Q5A8T7) were synthesized at >98% purity and purchased from GL Biochem (Shanghai) Ltd. The peptides were synthesized with unmodified termini for crystallography or with fully capped termini (acetylated in the N-terminus and amidated in the C structural terminus) for fibrillation assays. Thioflavin T (ThT) was purchased from Sigma-Aldrich. Dimethyl sulfoxide (DMSO) was purchased from Merck. Ultra-pure water was purchased from Biological Industries.
Amyloidogenic propensities of Als5 segments were predicted using combined information from several computational methods, including TANGO (Fernandez-Escamilla et al., 2004), AmylPred 2 (Tsolis et al., 2013) and FiSH Amyloid (Gasior and Kotulska, 2014). TANGO and FiSH Amyloid were used in their default settings, and AmylPred 2 was used with the default consensus option of only 4 of 9 predictors (TANGO and Amyloid Mutants were omitted from its prediction). We used TANGO, AmylPred 2, and FiSH Amyloid because they were the most sensitive and consistent predictors with quantifiable scores. The spine segments were defined as short segments of 6 residues, detected by at least two methods out of three at the epicenter of the predicted amyloidogenic area in the sequence (like in the case of 369TSYVGV374). Only in the case of the segment 322SNGIVIVATTRTV334, which is known to be critical to fibrillation of the Als5 protein (Garcia et al., 2011), we worked with both its full length and a shorter version of 6 residues.
The three-dimensional (3D) model of the entire Als5 sequence from Candida albicans, taken directly from the UniProt accession number Q5A8T7 was generated by AlphaFold v2.1.0 pipeline (Jumper et al., 2021; Varadi et al., 2022). Using Chimera (Goddard et al., 2007), we focused only on residues 20–433 which include the Ig-like/invasin and T domains. The per-residue confidence score (pLDDT) of the Ig-like domain ranged above 90 indicating a reliable prediction (Mariani et al., 2013). For the T domain, the per-residue confidence score ranged between 70 and 90 indicating lower confidence in the prediction.
We used ConSurf (Landau et al., 2005; Ashkenazy et al., 2016) with default settings to examine the evolutionary conservation patterns of Candida albicans Als5 residues 20–433 which include the Ig-like and T domains. The calculation was run using ConSeq (Berezin et al., 2004) (no structure was provided). One hundred and twenty-one unique sequences of homologs with 35%–95% sequence identity were retrieved from the UniRef90 database. Homologs with large gaps (more than 5 residues) in their alignment were filtered using Jalview2 multiple sequence alignment editing program (Waterhouse et al., 2009). The alignment of the remaining 71 homologs was used for the calculations presented here. The alignment is accessible at https://drive.google.com/file/d/1jWLOldYEx5aZz5VnrlnYv7lvYcN6DgkO/view?usp=sharing. The alignment of the seven orthologs used to generate the information presented in Figure 5 is accessible at https://drive.google.com/file/d/1SFTarMIUj762evoOPkWXi-S56jTezG45/view?usp=sharing.
Comparison between the segments 156NTVTFN161, 196IATLYV201, 369TSYVGV374, and 322SNGIVIVATTRTV334 evolutionary conservation was done by averaging the amino acid conservation scores attained from the ConSurf results for each segment. The segments 177TVDQSG182 and 238VNDWNH243 were taken as controls due to their lack of amyloidogenic propensity and their different conservation values, namely low and high respectively. The 71 retrieved homologs were ranked by their sequence similarity to C. albicans Als5, with the lowest e-value corresponding to the closer homologs of C. albicans Als5. Out of the 71 homologs we selected one along every five ranked homologs (in descending order of similarity to C. albicans Als5) for a total of fourteen sequences representing diverse sequences in the multiple sequence alignment. We then calculated the amyloidogenic propensity for each of the fourteen homologs using TANGO (Fernandez-Escamilla et al., 2004), AmylPred 2 (Tsolis et al., 2013), and FiSH Amyloid (Gasior and Kotulska, 2014) (Figures 4B, 5). The multiple sequence alignment was used, via Jalview2 (Waterhouse et al., 2009), to identify equivalent regions the segments of C. albicans discussed here. We then calculated the average predicted amyloidogenic propensity values in these corresponding sequences in each of the 14 homologs. TANGO averages were calculated by dividing the sum of each residue’s amyloidogenic propensity by its length. AmylPred 2 averages were calculated by assigning to each residue of the segments either a Boolean value of zero or one (based on consensus recognition of the method) and calculating the average similarly to TANGO averages. Using the default sliding window settings of the method, FiSH Amyloid averages were calculated by dividing the sum of the values assigned to all residues covering the segment by its length.
For the calculations of the amyloidogenic propensity of the entire Ig-like/invasin and T domains, we selected seven orthologs (homologs with a common ancestral parent in different species). Six of those were chosen along the ranked 71 homologs in gaps of 10 (starting from C. dubliniensis orthologs located five places away from the original C. albicans Als5). To these six, we added the emerging pathogen Candida auris, which was not a part of the homologs collected by the ConSurf webserver due to low sequence identity to the C. albicans Als5p (Willaert, 2018; Willaert et al., 2021). The multiple sequence alignment of the 71 homologs along with the C. auris sequence was obtained by reproducing the ConSurf search with similar parameters but lower sequence identity of 25%–95%, and then filtering out the irrelevant homologs using Jalview2.
Thioflavin T (ThT) is a commonly used for identifying and investigating the formation of amyloid fibrils in vivo and in vitro. In the presence of ThT, fibrillation curves commonly show delayed nucleation followed by rapid aggregation. Fibrillation kinetics of the peptides 156NTVTFN161, 196IATLYV201, 369TSYVGV374, and 322SNGIVIVATTRTV334 were monitored using ThT. All of the above peptides were synthesized with fully capped termini (acetylated in the N-terminus and amidated in the C structural terminus) to mimic its chemical nature in the full-length protein, and were dissolved to 10 mM in DMSO. Each of the freshly dissolved peptides was mixed with 50 mM Tris-HCl buffer with pH 7.3 and with 2 mM filtered ThT stock (made in ultra-pure water) to reach final concentrations of 300 μM of peptide and 20 μM ThT in the final volume of 100 μl reaction. The reaction mixture was carried out in a black 96-well flat-bottom plate (Greiner bio-one) covered with a thermal seal film (EXCEL scientific) and incubated in a plate reader (CLARIOstar, BMG Labtech), at 37°C, with orbital shaking at 300 rpm, for 30 s before each measurement. ThT fluorescence was recorded every 2 minutes for a total time of ∼48 h (we show only 5 h of the measurements) using an excitation of 438 ± 20 nm and an emission of 490 ± 20 nm. The measurements were conducted in triplicates, and the entire experiment was repeated at least three times.
TEM was used to visualize fibrils. Samples for TEM were taken directly from the ThT kinetic assay plate, which was left to incubate at 37°C for 2 days with 300 rpm shaking in the plate reader. The TEM grids were prepared by applying 5 µl samples of each 300 μM sample of peptide on 400 mesh copper grids with support films of Formvar/Carbon (Ted Pella), that were charged by high-voltage, alternating current glow-discharge (PELCO easiGlow, Ted Pella) immediately before use. Samples were allowed to adhere for 1 min followed by negative staining with 1% uranyl acetate for 1 min. Micrographs were recorded using a FEI Tecnai G2 T20 S-Twin transmission electron microscope at an accelerating voltage of 200 KeV at the MIKA electron microscopy center of the Department of Material Science & Engineering at the Technion.
The peptide SNGIVIVATTRTV was dissolved to 10 mg/ml in ultra-pure water. A few microliters were applied between two sealed glass capillaries until completely dried. X-ray diffraction of the samples was collected at the micro-focused beam P14 at the high brilliance 3rd Generation Synchrotron Radiation Source at DESY: PETRA III, Hamburg, Germany.
Peptides synthesized with free (unmodified) termini were used for crystallization experiments to facilitate crystal contacts. All peptides were dissolved to 10 mM in ultrapure water if possible, and in DMSO in case they were water-insoluble. 196IATLYV201 was dissolved in 95% DMSO, 156NTVTFN161 168SIAVNF173 and 324GIVIVA329 were dissolved in 100% DMSO. 369TSYVGV374 was dissolved in 100% ultrapure water. Peptide solution drops (100 nl) were dispensed onto crystallization screening plates, using the Mosquito automated liquid dispensing robot (TTP Labtech, United Kingdom) located at the Technion Center for Structural Biology (TCSB). Crystallization using the hanging drop method, was performed in 96-well plates, with 100 µl solution in each well. The drop volumes were 150–300 nl. All plates were incubated in a Rock imager 1,000 robot (Formulatrix), at 293K. Micro-crystals grew after few days and were mounted on glass needles glued to brass pins. No cryogenic protection was used. Crystals were kept at room temperature prior to data collection. Structures were obtained from drops that were a mixture of the following peptide and reservoir solutions: 196IATLYV201: 10 mM IATLYV, 0.1 M tri-Sodium citrate pH 5.6 and 1.0 M Ammonium phosphate; 156NTVTFN161: 10mM NTVTFN, 0.1 M HEPES pH 7.5, 0.8 M NaH2PO4, 0.8 M KH2PO4; 369TSYVGV374: 10 mM TSYVGV, 0.2 M Sodium chloride, 0.1 M Sodium acetate pH 4.6, 30% (v/v) 2-Methyl-2,4-pentanediol (MPD).
X-ray diffraction data were collected at 100K, using 5° oscillation. The X-ray diffraction data were collected at the micro-focus beamline ID23-EH2 of the European Synchrotron Radiation Facility (ESRF) in Grenoble, France; wavelength of data collection was 0.8729 Å. Data indexation, integration and scaling were performed using XDS/XSCALE (Kabsch, 2010). Molecular replacement solutions for all segments were obtained using the program Phaser within the CCP4 suite (Murshudov et al., 1997; McCoy et al., 2007; Winn et al., 2011). The search models consisted of geometrically idealized β-strands. Crystallographic refinements were performed with the program Refmac5 (Murshudov et al., 1997). Model building was performed with Coot (Emsley et al., 2010) and illustrated with Chimera (Goddard et al., 2007). There were no residues that fell in the disallowed region of the Ramachandran plot. Crystallographic statistics are listed in Table 1.
The Lawrence and Colman’s shape complementarity index (Lawrence and Colman, 1993) was used to calculate the shape complementarity between pairs of sheets forming the dry interface (Supplementary Table S1). The buried surface area was calculated with Chimera (UCSF) (Goddard et al., 2007), with a default probe radius and vertex density of 1.4 Å and 2.0/Å2, respectively. The number of solvent accessible buried surface areas was calculated as the average area buried of one strand within two β-sheets (total area buried from both sides is therefore double the reported number in Supplementary Table S1.
A prediction of the multimeric assembly of Als5 Ig-like and T-domains was done with AlphaFoldv2 Advanced pipeline (modified AlphaFold v2.1.0 Multimer pipeline) (Jumper et al., 2021) (Richard Evans et al. bioRxiv 2021). The pipeline was executed via the Google Colab service available online to each AlphaFold program (AlphaFold v2.1.0—https://colab.research.google.com/github/deepmind/alphafold/blob/main/notebooks/AlphaFold.ipynb, AlphaFold v2.1.0 advanced—https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/beta/AlphaFold2_advanced.ipynb#scrollTo=pc5-mbsX9PZC). The prediction was based on the sequence of Als5 20–433 residues (UniProt accession number Q5A8T7). A total of five homodimers of Als5 20–433 were retrieved and were ranked based on their per-residue confidence score. Models of homodimers were illustrated, colored, and analyzed using UCSF Chimera (Goddard et al., 2007).
Sequence- and structure-based amyloid predictors use a variety of criteria including solubility and physicochemical properties, geometry, homology, and secondary structure propensity to predict occurrence of amyloidogenic core regions that can form cross-β aggregates and steric zippers. Accordingly, we scanned the primary structure of Als5 Ig/invasin and T domains (residues 20–433) with three amyloid prediction programs. TANGO has been useful for predictions in Als adhesins, is a thermodynamics state-based predictor (Fernandez-Escamilla et al., 2004; Lipke et al., 2017). AmylPred 2 is a consensus method based on eleven predictors (Tsolis et al., 2013). Lastly, FiSH Amyloid detects co-occurrence patterns in sequence data based on machine-learning techniques (Gasior and Kotulska, 2014). Nine segments scored above threshold in at least two of the three predictors, marked on the Als5 sequence in Figure 1A. Six segments are in the Ig-like/invasin region: 65DTFILN70, 132IAFNVG137, 156NTVTFN161, 168SIAVNF173, 196IATLYV201, and 259FGISIT264, and three sequences are in the T domain: 324GIVIVA329, a section from the longer segment 322SNGIVIVATTRTV334 at the Ig/T domain interface, 369TSYVGV374, and 388TATVIV393. These segments were mapped onto an AlphaFold Monomer v2.0 model of Als5p (Jumper et al., 2021; Varadi et al., 2022) (Figure 1). AlphaFoldv2 predicted a folded structure for the Ig/T domains and the first four tandem repeats regions. As expected, there was no structural prediction for the highly glycosylated low complexity C-terminal stalk region, which is unstructured and in extended conformation in vivo (Lipke et al., 2017).
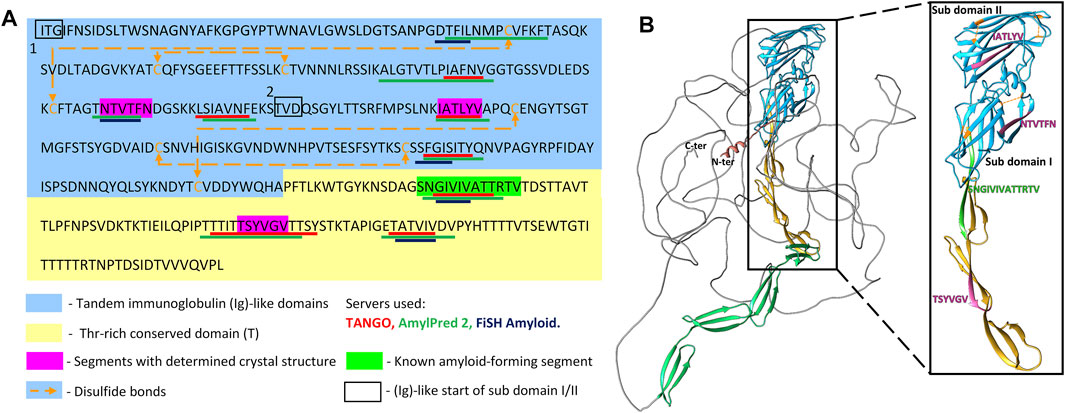
FIGURE 1. Predicted amyloidogenic segments within Als5 sequence and modeled structure (A) Annotated N-terminal residues 20–433 of the Candida albicans Als5 protein containing the Ig-like/invasin and T regions, indicated by blue and yellow backgrounds, respectively. Segments with strong amyloidogenic predictions are denoted with colored bars under the sequence: TANGO (Fernandez-Escamilla et al., 2004) in red, AmylPred 2 (Tsolis et al., 2013) in green, and FiSH Amyloid in blue (Gasior and Kotulska, 2014). Segments for which the crystal structure was determined are marked magenta, and the known amyloid segment 303SNGIVIVATTRTV315 is marked green. Disulfide bonds are illustrated by orange dashed arrow lines. (B) A three-dimensional (3D) model of the Als5 predicted by AlphaFold Monomer v2.0 (Jumper et al., 2021; Varadi et al., 2022). Alphafoldv2 predicted a folded structure for the Ig/T domains (blue and yellow) and four subdomains of the TR region (green), but not for the rest two tandem repeats and the C-terminal 850-residue glycosylated stalk region (grey line). Right inset is an enlarged image of residues 20–433 with the two Ig-fold subdomains indicated.
We determined the atomic structures of 156NTVTFN161, 196IATLYV201 and 369TSYVGV374 segments (PDB IDs 6RHA, 6RHB, and 6RHD, respectively; Figure 2, Table 1, and Supplementary Table S1). The locations of the amyloid segments on the AlphaFoldv2 predicted structure of Als5 are shown in Figure 1B. The crystal structure of the 156NTVTFN161 spine segment from the Ig-like domain formed a canonical steric-zipper structure of tightly mated parallel β-sheets with a dry interface. The 196IATLYV201 spine segment, also from the Ig-like subdomain 2, revealed a partial mating between antiparallel β-sheets. The 369TSYVGV374 spine segment from the T domain of Als5 exhibited an atypical amyloid structure, with antiparallel β-sheets and kinked β-strands (Figure 2 and Supplementary Figure S1).
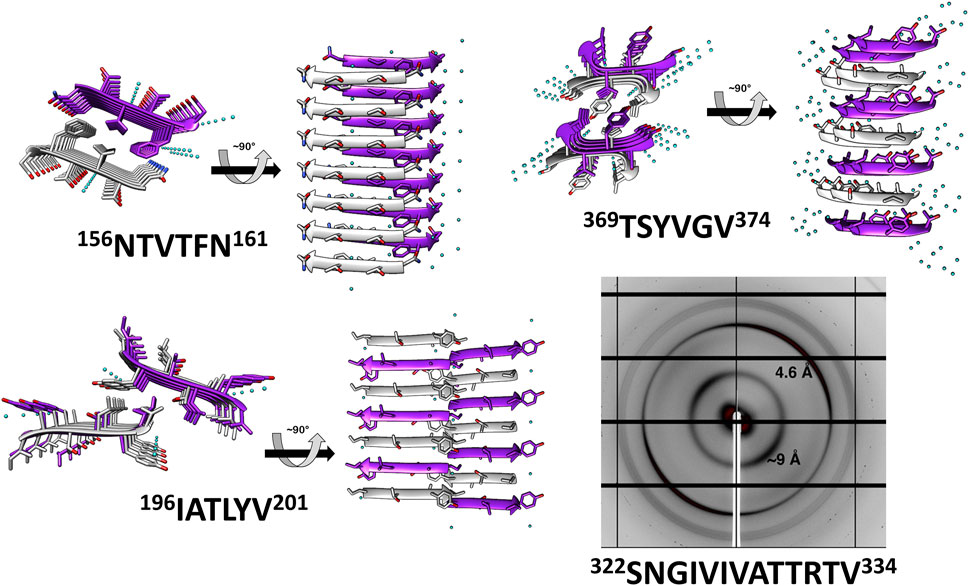
FIGURE 2. Crystal structures and X-ray fiber diffraction of Als5 amyloidogenic segments. High-resolution crystal structure of three predicted amyloidogenic segments of Als5 and the known amyloid segment 322SNGIVIVATTRTV334. The left panel of each structure is viewed down the fibril axis, with β-strands shown as ribbons and residues as sticks, while the right panel is viewed perpendicular to the fibril axis displaying β-strands running horizontally. Fibrils contain thousands of layers of β-strands, but only seven layers are shown here. Segment backbones are colored either gray or purple; heteroatoms are colored according to their atom type (nitrogen in blue, oxygen in red) and water molecules in cyan. The X-ray fiber diffraction pattern with major reflections labeled showing a cross-β pattern with orthogonal reflections at 4.6 Å and ∼9 Å spacings. Some reflections originating from ice also appear.
These peptides induced development of thioflavin-T (ThT) fluorescence typical of amyloid aggregation (Figure 3A) and formed amyloid-like fibrils visualized by transmission electron microscopy (Figure 3B). The peptides 156NTVTFN161 and 196IATLYV201 each formed abundant and long fibrils, although without the typical lag associated with amyloid formation, possibly due to presence of seeds in the original sample. 369TSYVGV374 formed abundant short plump bundled fibrils. The longer peptide 322SNGIVIVATTRTV334 segment, known to be critical for biological activity of the adhesin and crucial for fibrillation of soluble forms of Als5 protein, formed long and relatively straight fibrils that also induced ThT fluorescence (Lipke et al., 2017). This sequence contains a strongly predicted amyloid core sequence 324GIVIVA329. Although we did not successfully crystallize either the 6-residue or the 13-residue segments, the X-ray fiber diffraction of 322SNGIVIVATTRTV334 showed a canonical amyloid cross-β pattern with orthogonal reflection arches at 4.6 and ∼9 Å (Figure 2).
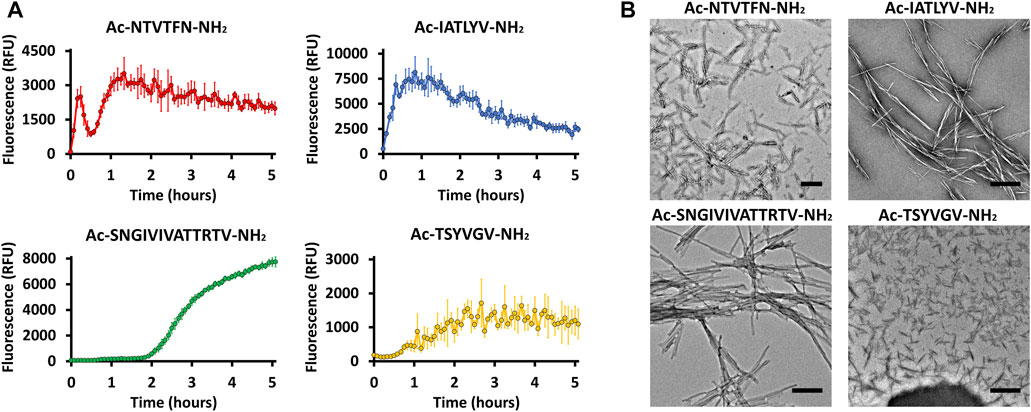
FIGURE 3. Predicted segments exhibiting amyloidogenic properties (A) Segments for which the crystal structure was determined and 322SNGIVIVATTRTV334 show fibrillation when incubated with ThT. Measurements were conducted in 50 mM Tris-HCl buffer, 20 mM ThT, and 300 µM of peptide. The graph shows the average of three technical repeats, with error bars representing standard deviations. (B) Electron micrographs of the four segments show fibril formation after 48 h of incubation. Scale bar represents 300 nm.
Because steric-zipper fibrils are unusual in that pairs of β-sheets mate more closely than the adjoining surfaces in other protein complexes, quantitative measures of amyloid stability are based on solvent-accessible surface area buried at the interface between the mating sheets, and shape complementarity indicating on the closeness of fit of two protein surfaces (Tayeb-Fligelman and Landau, 2017). The shape complementarity, inter-strand distance, and solvent-exposed surface area buried were calculated for Als5 spine structures and compared to the NNQQNY segment from yeast prion Sup35 steric zipper (PDB ID 1YJO) (Lawrence and Colman, 1993). The NNQQNY structure was chosen for this comparison as it shows one of the highest values of shape complementarity and surface area buried among steric zipper structures (Eisenberg and Sawaya, 2017). The shape complementarity was similar for all Als5 spines segment, all lower than prion NNQQYY (Supplementary Table S1). However, the T domain 369TSYVGV374 spine segment, displaying kinked β-strands, showed a significantly lower solvent-exposed surface area buried compared to the other structures (Supplementary Table S1), indicating a less stable structure that might also suggest reversible fibril formation.
The 156NTVTFN161 and 196IATLYV201 segments in the Ig-like/invasin region are each close to a disulfide bonded Cys residue (Figure 1). Therefore, it is likely that these sequences would not have the conformational freedom to lead to solvent exposure or interaction necessary for amyloid formation. In contrast, the amyloid-forming segments in the T region, which is conformationally variable in vivo, are not constrained by disulfides or other structured regions. Thus, the amyloid propensity in this region is likely to be expressed in vivo, as demonstrated before for 322SNGIVIVATTRTV334 (Lipke et al., 2017).
Evolution can shift and alter traits in all biological systems. Selection pressure will work to conserve traits that possess an important beneficial function. It is also true for functional amyloids, where purifying selection acts to conserve the amyloidogenic potential, amino acid sequence, and accessible conformation; however, the amyloidogenic potential and amino acid sequence are not always conserved equally. Occasionally, amino acid sequences can change considerably without greatly affecting their amyloidogenic potential (Maji et al., 2009). In light of this difference, we have examined the evolutionary conservation patterns of the amyloidogenic segments correlated with the trajectory of the change in amyloidogenic potential from close to more distant homologs of C. albicans Als5.
To evaluate the conservation of the amyloidogenic segments in protein homologous to Als5 we used the ConSurf webserver (Berezin et al., 2004; Ashkenazy et al., 2016). First, we searched for homologs for Als5 residues 20–433 which include the Ig-like/invasin and T domains. Seventy-one unique sequences of homologs with 35%–95% sequence identity were retrieved from the UniProt (UniRef90) database. We excluded homologs with large gaps in the multiple sequence alignment. The segments 156NTVTFN161, 196IATLYV201, 322SNGIVIVATTRTV334, 324GIVIVA329 and 369TSYVGV374 showed diverse pattern of sequence conservation across the Als5 homologs. In the Ig-like/invasin region, the sequence 156NTVTFN161 was remarkably conserved across the homologs (Figure 4A), with five strongly conserved positions (Supplementary Figure S2), implying a conserved functional or structural role. In contrast, 196IATLYV201 was not well conserved and showed low average conservation value with four highly variable positions in its sequence. In the T domain, the 322SNGIVIVATTRTV334 region showed intermediate to above-average conservation. This observation is in agreement with previous reports showing that the functional amyloid-forming sequence 322SNGIVIVA329 is strongly conserved within the C. albicans ALS gene family (Otoo et al., 2008; Ho et al., 2019). The LARKS-like sequence 369TSYVGV374 also showed intermediate average conservation value with three highly conserved positions, with highest value at Gly373.
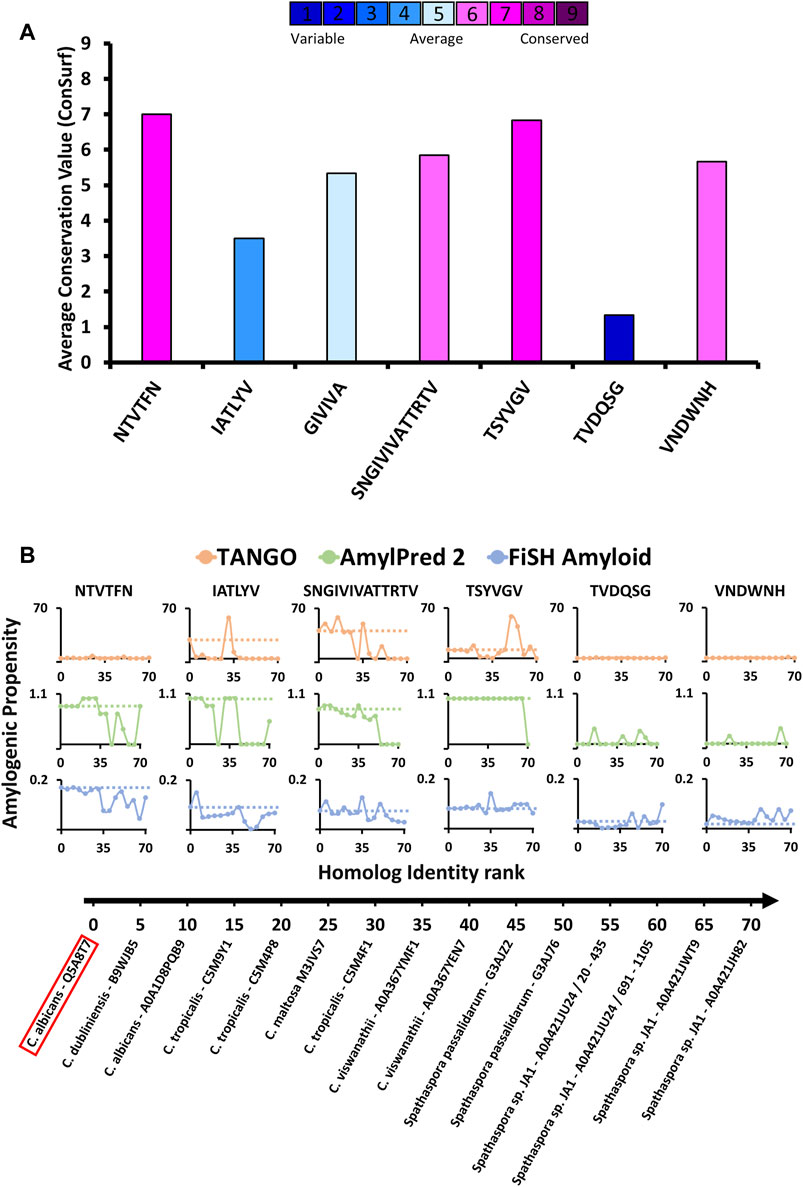
FIGURE 4. Amyloidogenic propensity and the evolutionary conservation of C. albicans Als5 segments. (A) Evolutionary conservation of Als5 segments. The graph displays evolutionary conservation values for seven segments (three segments with resolved crystal structures, known amyloid spine 322SNGIVIVATTRTV334, and two control segments) from C. albicans Als5. These values were calculated based on a multiple sequence alignment of 71 homologs of Als5 generated using the ConSurf webserver (Berezin et al., 2004; Landau et al., 2005; Ashkenazy et al., 2016). The columns’ colors correspond to the average evolutionary conservation score (1 being the least conserved and 9 being the most conserved). (B) The amyloidogenic propensities of the above seven segments in Als5 orthologs (homologs from different species). Fourteen representative orthologs of Als5p were selected to represent a diverse group (as described in the Method section). The orthologs are plotted on the X-axis by their sequence similarity to C. albicans (evaluated based on ConSurf e-value parameter) from most to least similar homolog. The Y-axis shows the average amyloidogenic propensity of each segment in the fourteen orthologs. The amyloidogenic values were calculated using three servers and displayed using different colors: TANGO (orange) (Fernandez-Escamilla et al., 2004), AmylPred2 (green) (Tsolis et al., 2013), FiSH Amyloid (blue) (Gasior and Kotulska, 2014). The different methods have a different score range, thus in each graph, a horizontal dashed line indicates the amyloidogenic value of the relevant segment from C. albicans Als5.
We also evaluated the conservation of amyloidogenic potential within the Als5p sequence homologs (Figure 4B). The spine-forming segments 156NTVTFN161, 196IATLYV201, 322SNGIVIVATTRTV334, and 369TSYVGV374 presented a wide range of differences in amyloidogenic potential calculated by three predictors: TANGO, AmylPred 2, and FiSH Amyloid. For further analyses, we selected as controls two segments with a predicted low amyloidogenic propensity, 177TVDQSG182 and 238VNDWNH243, showing low and intermediate evolutionary conservation, respectively. The 322SNGIVIVATTRTV334 segment already implicated in Als5 function (Lipke et al., 2017) showed a high amyloidogenic propensity in close homologs, which declined in more distant homologs. The LARKS-like sequence 369TSYVGV374, which is relatively conserved (Figure 4A), showed the highest stability in maintaining amyloidogenic potential across homologs according to Amylpred2 and FiSH amyloid, with more fluctuations in propensity towards more distant homologs (Figure 4B). In contrast, the Ig-like/invasin region segments 156NTVTFN161 and 196IATLYV201 show a less clear pattern of amyloidogenic propensity across homologs, similar to the control sequences 177TVDQSG182 and 238VNDWNH243. Control sequence 177TVDQSG182 showed high fluctuations in amyloidogenic propensity, despite its low value in the C. albicans proteins. This finding corresponds to its low sequence conservation (Figure 4). Overall, the 69TSYVGV374 and 322SNGIVIVATTRTV334 segments in the Als5 T domain appear to have the most conserved amyloidogenic propensity among examined segments, at least across close homologs of C. albicans.
We compared the amyloidogenic propensity of the entire Als5 Ig-like/invasin and T domains in Als5 and homologs from seven species: Candida dubliniensis, C. tropicalis, C. maltosa, C. viswanathii, Spathaspora passalidarum, Spathaspora sp. JA1, and C. auris (Muñoz et al., 2021). The amyloidogenic propensities predicted by AmylPred2 are summarized in Figure 5, with amyloidogenic sequences marked in purple. Each of the adhesins was predicted to have amyloid-forming sequence at the equivalent region to the 322SNGIVIVATTRTV334 segment of C. albicans Als5 (marked green), except for one homolog from Spathaspora sp. JA1. The equivalent region to the LARKS-like segment 369TSYVGV374 also showed a strong amyloidogenic propensity (marked dark yellow). In the C. auris homolog, there was no equivalent sequence to 369TSYVGV374, but other segments in this region (residues 343–370) were predicted to be amyloidogenic (marked purple). In the Ig-like/invasin region, the equivalent sequence to 196IATLYV201 (marked blue in Figure 5) showed no amyloidogenic propensity in C. maltosa and S. passalidarum or Spathaspora sp. JA1, but this segment did show some amyloidogenic propensity in C. auris. The predicted amyloidogenic propensity of this region was the least conserved among the distant homologs examined in Figure 4B. The segment equivalent to 156NTVTFN161 showed amyloidogenic propensity predictions (marked red in Figure 5) across the homologs except for S. passalidarum and C. auris. Overall, each ortholog shown in Figure 5 contains various regions with strong prediction for amyloidogenic propensity. Among the identified segments of Als5, the T-domain segments displayed more conserved amyloidogenic traits compared to segment in the Ig-like/invasin region.
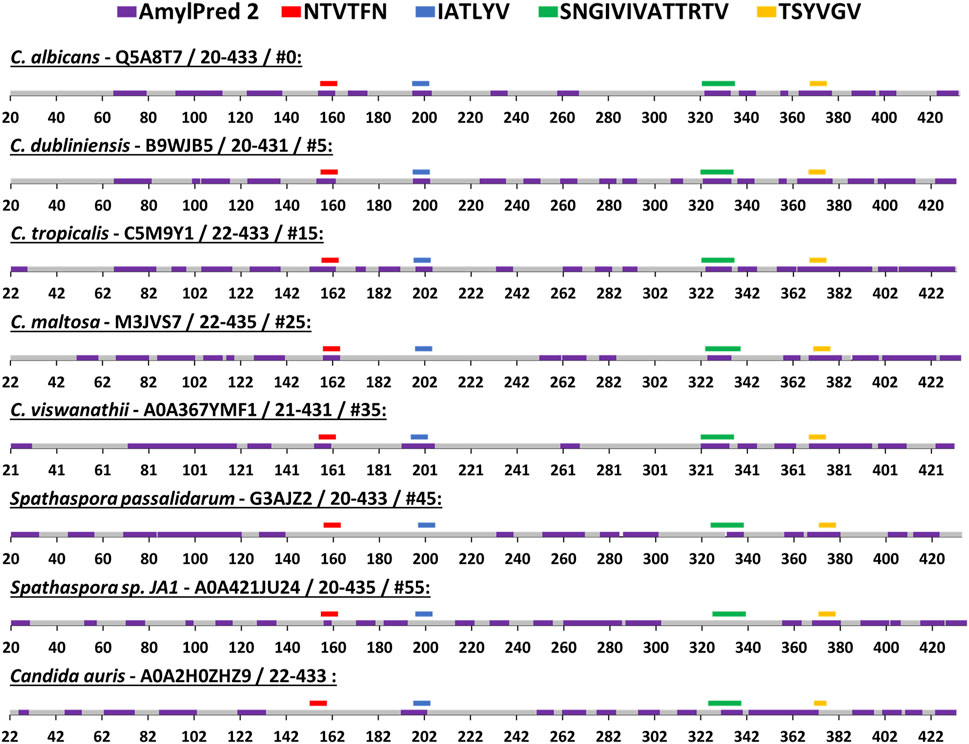
FIGURE 5. Predicted segments with amyloidogenic propensities in the Als5 Ig-like/invasin and T regions of C. albicans and seven orthologs. The amyloidogenic propensity, calculated using the AmylPred2 server (Tsolis et al., 2013), of Als5 Ig-like/invasin and T domains of C. albicans and seven orthologs. These specific orthologs were chosen as a diverse group among the 71 homologs collected by the ConSurf webserver along with C. auris, as described in the Method section. Each ortholog is identified by the name of the species, their Swissprot ID, residue range of the Ig-like/invasin and T regions and their similarity to C. albicans ranked among 71 homologs (determined using the ConSurf-generated multiple sequence alignment and marked with #). The regions with predicted amyloidogenic propensity are marked purple on the sequences, represented as a linear stretch with residue position marks. Regions equivalent to the C. albicans sequences are marked above the sequence with different colors as indicated: 156NTVTFN161 (red), 196IATLYV201 (blue), and 369TSYVGV374 (yellow) and 322SNGIVIVATTRTV334 (green).
Als5 polymerizes in solution to form initial oligomers which convert to SDS-resistant oligomers, and then to amyloid fibers (Otoo et al., 2008). To gain insight into these interactions, we used AlphaFoldv2 Advanced (modification of AlphaFold v2.1.0 Multimer pipeline) to model the formation of Als5 dimers in the Ig-like/invasin and TR regions (Jumper et al., 2021) to model interactions between two copies of the Ig-like/invasin and T domains Als5. The model suggested dimerization of the folded monomers with an interface roughly parallel to long axis of the protein (Figure 6). In the three highest-ranked models, the major interface was mediated by the T domains, with minor contacts between the Ig-like/invasin domains. The highest-ranked model had the most extensive interface. The predicted amyloidogenic segments are not a part of the interface, but those in the T domain are close to their equivalent segment in the facing monomer. The models are based on the folded structure of the Als5, and do not show partial misfolding into an amyloid state. It is possible that future development of AlphaFold and other methods will allow modeling the cross-β fibrils.
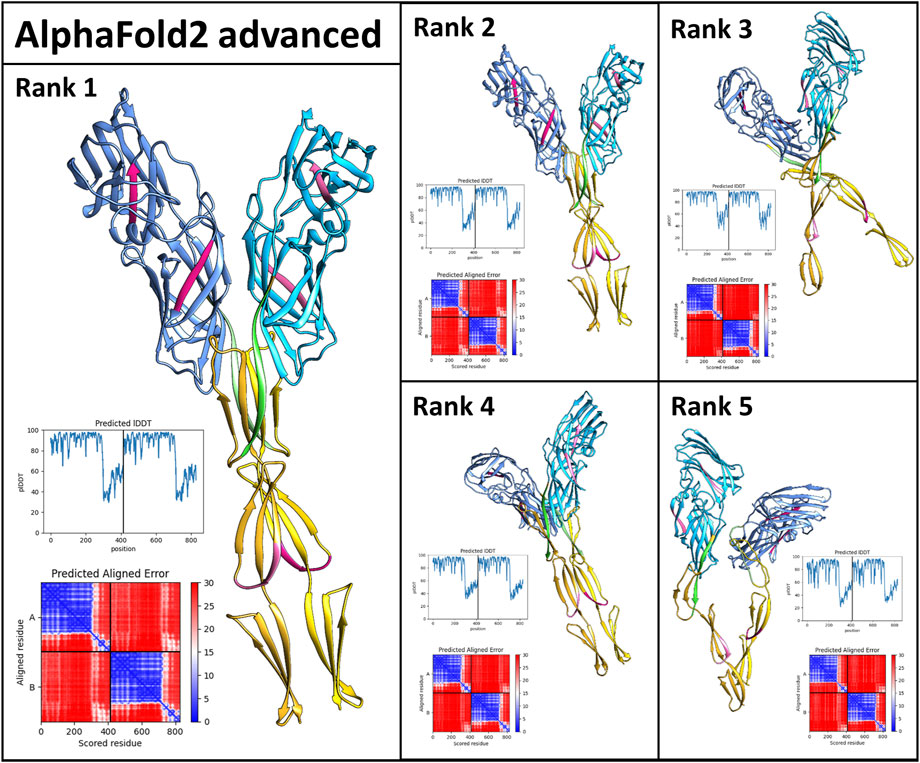
FIGURE 6. AlphaFold2_Advanced models of Als5 dimers. The model is based on the sequences of the Ig/T domains, colored as in Figure 1. The ranking is based on AlphaFold confidence score. The scores are quantified by the graphs showing the confidence score lDDT plotted against the sequence and the alignment plots (Mariani et al., 2013).
The structures and analyses presented here support the predictions of functional amyloid formation for fungal adhesins. Of the segments identified as potentially forming amyloids, we argue on the bases of structural and evolutionary constraints that two of them in the T domain are likely to function in vivo. We note that Als5 might have main and minor aggregation sites, with the latter taking over in case of interference or mutation in the main site, as previously suggested for Tau involved in Alzheimer’s disease and for Fap amyloids involved in the biofilm of Pseudomonas (Christensen et al., 2019; Annadurai et al., 2022). We also showed atomic-level cross-β spine structures with known and novel geometries, including one LARKS-like structure reminiscent of functional and reversible human amyloids (Guenther et al., 2018; Hughes et al., 2018).
Sequence- and structure-based amyloid predictors generally identify amyloidogenic or β-aggregation potential in many protein sequences. Indeed, systematic studies show that the majority of small peptides with high potential amyloidogenic sequences do form amyloids in vitro (Porat et al., 2003; Eisenberg and Sawaya, 2017; Hughes et al., 2018; Bera et al., 2019). However, only few of the proteins containing these sequences form amyloids in vivo, probably because peptides are constrained by being part of a stable protein fold in situ. Formation of cross-β structures depends on conformation, solvent accessibility, and compatible geometry in flanking regions (Shewmaker et al., 2011; Eisenberg and Sawaya, 2017; Bera et al., 2019). Therefore, amyloids can often form only after proteins denature to expose the amyloidogenic sequences and allow flexibility in the peptide chains. In Als5, we identified 9 segments with high potential to form amyloid-like cross-β structures. We argue here two of these sequences are involved in functional amyloid formation, while 6 others probably do not form cross-β structures in situ in the protein. The arguments are based on structural and evolutionary arguments, and on positive evidence for functionality of one segment.
Of the six potential amyloidogenic β-aggregating sequences in the Ig-like/invasin domain of Als5, three are within disulfide-stapled regions of the protein and so these segments are unlikely to be solvent-exposed and flexible enough to form cross-β structures (Figure 1). Of the three potential amyloidogenic sequences between that are not disulfide-constrained, 156NTVFN160 and 196IATLYV201 formed cross-β spines in vitro. However, these segments are unlikely to do so in vivo. Although the sequence 156NTVTFN161 is relatively highly evolutionary conserved (Figure 4A and Supplementary Figure S2), its TANGO amyloidogenic propensity is low and not conserved across homologs. It has greater potential in the other two predictors (Figures 4B, 5). In Ig/invasin subdomain II, 156NTVTFN161 seals the edge of a β-sheet (Figure 1), and so it contributes to the hydrophobic core of the fold. Therefore, this sequence is expected to be key for the high stability of this region, and this region of the protein unfolds only at high extension forces > 350 pN (Alsteens et al., 2009; Alsteens et al., 2013). In addition, 156NTVTFN161 is near disulfide-bonded Cys150 as well as the domain I/II interface, which both constrain its flexibility and geometry in the native protein. Therefore, this sequence is unlikely to form a cross-β structure in vivo in the intact protein.
The sequence 196IATLYV201 is in a β-strand (Figure 1B) and is poorly conserved in sequence (Figure 4A). Its equivalent region is predicted to have amyloidogenic propensity in some of the homologs, but without the specific trend of amyloidogenic propensity conservation along relatively close and distant homologs (Figures 4B, 5). Therefore, it seems unlikely that this segment participates in the formation of functional β-aggregates in Als adhesins. The segment is extremely close to disulfide bonded Cys204, and so it is likely to be too geometrically constrained in vivo to form cross-β structures.
The four other potential amyloid sequences in the Ig-like/invasin region failed to form cross-β spines. Therefore, there is little support for formation of cross-β structures in situ within the Ig/invasin region of the protein.
Two of the potential amyloid sequences in the T domain show conservation of their sequence and amyloidogenic propensity. Figures 4B, 5 demonstrate the strong cross-β potential of 322SNGIVIVATTRTV334 and the conservation of this propensity in this region across homologs. Similarly, the LARKS-like segment 369TSYVGV374 also shows high conservation of amyloidogenic propensity (Figures 4B, 5) and conservation in sequence as well (Figure 4A and Supplementary Figure S2). Both of these segments showed strong cross-β properties (Figures 2, 3). Also contrasting with the Ig/Invasin region, this region is unconstrained by disulfide bonds and becomes unstructured under low shear stress. Thus, these sequences are likely to be exposed and relatively unconstrained in vivo (Alsteens et al., 2009; Alsteens et al., 2010; Lipke et al., 2017). These features are in accord with the known T domain functional amyloid formation.
The T domain and the sequence 322SNGIVIVATTRTV334 are essential for many activities of Als5 and its close paralog Als1 (Lipke et al., 2017; Ho et al., 2019). This sequence is necessary for strong cell-to-cell binding, as well as for clustering the adhesins on the cell surface. A single site mutation V326N reduces the TANGO β-aggregation potential by about 20-fold and also reduces aggregation activity in Als5 without affecting ligand binding to the Ig/invasin domain (Garcia et al., 2011). Furthermore, adhesion activity is enhanced in the presence of a homologous peptide, and activity is reduced in the presence of a homologous non-amyloid peptide with the Val to Asn substitution. These are properties expected from a sequence interacting through β-aggregation. The T region unfolds easily under shear, including under physiological shear stresses (Chan and Lipke, 2014; Lipke et al., 2017). Thus, extension forces unfold the T domain, rendering it unstructured giving the conformational flexibility needed for formation of cross-β structures. Accordingly, we have published extensively on the importance of the 322SNGIVIVATTRTV334 sequence (Garcia et al., 2011; Lipke et al., 2017; Dehullu et al., 2019a). We have also found that the T region is essential for secretion and processing in yeast (Rauceo et al., 2006). This result implies that the deletion affects the entire fold of the domain.
The discovery of the highly-conserved T-region 369TSYVGV374 segment that can form a cross-β structure explains several observations. Specifically, Als5 and Als1 show residual aggregation activity and thioflavin binding after mutations that abolish the amyloid potential of the major T domain sequence 322SNGIVIVATTRTV334 (Alsteens et al., 2010; Garcia et al., 2011; Chan and Lipke, 2014; Dehullu et al., 2019a; Dehullu et al., 2019b; Ho et al., 2019). Similarly, there is 25%–40% residual activity after treatment with a specific peptide that inhibits cross-β formation of the 322SNGIVIVATTRTV334 segment (Garcia et al., 2011; Alsteens et al., 2013; Chan and Lipke, 2014; Dehullu et al., 2019a; Dehullu et al., 2019b). The residual activity has characteristics of cross-β structures because the cell surface remains somewhat thioflavin T fluorescent. Also consistent with cross-β functional bonds, the activity can be inhibited by concentrations of Congo red or thioflavins 1000-fold higher than those used in fluorescence. At these concentrations, these dyes are sequence-independent inhibitors of amyloid formation (Garcia et al., 2011; Landau et al., 2011; Harris, 2012; Chan and Lipke, 2014). These observations support the idea that another cross-β forming sequence mediates the residual activity. In general, LARKS-like sequences generate transient, low energy cross-β aggregates (Guenther et al., 2018; Hughes et al., 2021). These sequences often mediate reversible liquid-liquid phase separations (Hughes et al., 2018). On the surface of stimulated cells, Als proteins form mobile surface nanodomains visible in atomic force microscopy (AFM) and by in vivo staining with thioflavins (Rauceo et al., 2004; Alsteens et al., 2010; Garcia et al., 2011). These nanodomains would be a two-dimensional equivalent of such reversible liquid-like associations.
Therefore, evolutionary conservation, the presence of liquid-like surface nanodomains, the residual amyloid-like behavior in fluorescence and AFM are consistent with the LARKS-like sequence 369TSYVGV374 providing a supportive role in adherence. Thus, the use of our screens for cross-β sequences has resulted in discovery of a new cross-β segment that appears to contribute to the activity of amyloid-like sequences in the T domain of Candida Als adhesins.
In vivo, the T domain is unfolded by flow-induced shear stress over mucosal surfaces as low as 0.1–10 (dyne/cm2) (Chan and Lipke, 2014). These values are well-within the range of shear stresses in mucosal flow and in the blood stream. Shear-triggered unfolding could thus lead to peptide extension and exposure of segments with cross-β potential, and then to the formation of cross-β aggregates in vivo. Models for the transition from the well-folded state to cross-β of Als have been recently published (Lipke et al., 2017; Lipke et al., 2021).
Atomic resolution structures show that sequences in Als5 can form amyloid-like cross-β aggregates. Of the 4 sequences for which we have structural information, two in the Ig-like/invasin region have characteristics of fortuitous amyloid-formers, i.e., these segments have key roles in a hydrophobic domain core, but they are close to disulfide bonds and one of them has poor conservation of amyloid potential across homologs. In contrast, two sequences in the T domain show characteristics of force-dependent functional amyloids: conservation of sequence and amyloid potential, and the 322SNGIVIVATTRTV334 segment has demonstrated roles in activity of the protein Als5 and Als1 (Lipke et al., 2017; Ho et al., 2019). The newly discovered LARKS sequence 369TSYVGV374 is also highly conserved and can explain the residual cross-β activity of Als5 that lacks 322SNGIVIVATTRTV334 activity. These results demonstrate a role for conservation of amyloid potential as well as sequence in determining functionality of naturally occurring sequences that form β-aggregates. The results illustrate that evolutionary considerations for both sequence and amyloid potential can identify sequences that have high amyloid potential and can potentially distinguish between those that form amyloids in vivo and those that do not.
The datasets presented in this study can be found in online repositories. The coordinates of the atomic structures are available at the PDB via the accession codes 6RHA (156-NTVTFN-161), 6RHB (196-IATLYV-201), and 6RHD (369-TSYVGV-374).
ML and PL conceived the study. ML provided all support. ML and SS-P collected X-ray data and determined the crystal structures. SS-P initiated amyloid segment analysis. NG conducted all bioinformatics and biophysical analyses and prepared figures. All authors generated data, analyzed results, and contributed to writing and editing.
ML acknowledges the Israel Science Foundation (Grant No. 2111/20), Israel Ministry of Science, Technology and Space (Grant No. 78567), U.S.-Israel Binational Science Foundation (BSF) (Grant No. 2017280), BioStruct-X, funded by FP7, and the iNEXT consortium of Instruct-ERIC. We are also grateful for the support from NIH P41-GM103311 (UCSF).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We acknowledge technical support provided by Yael Pazy-Benhar and Dikla Hiya at the Technion Center for Structural Biology (TCSB). We acknowledge support from Yaron Kauffmann from the MIKA electron microscopy center of the Department of Material Science and Engineering at the Technion, and Na’ama Koifman from the Russell Berrie Electron Microscopy Center of Soft Matter at the Technion, Israel. Molecular graphics and analyses performed with UCSF Chimera, UCSF. The synchrotron MX data collection experiments were performed at beamlines ID23-EH2 at the European Synchrotron Radiation Facility (ESRF), Grenoble, France, and at beamline P14, operated by EMBL Hamburg at the PETRA III storage ring (DESY, Hamburg, Germany). We are grateful to the teams at ESRF and EMBL Hamburg for their assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.926959/full#supplementary-material
Alsteens, D., Dupres, V., Klotz, S. A., Gaur, N. K., Lipke, P. N., and Dufrêne, Y. F. (2009). Unfolding Individual Als5p Adhesion Proteins on Live Cells. ACS Nano 3, 1677–1682. doi:10.1021/nn900078p
Alsteens, D., Garcia, M. C., Lipke, P. N., and Dufrêne, Y. F. (2010). Force-induced Formation and Propagation of Adhesion Nanodomains in Living Fungal Cells. Proc. Natl. Acad. Sci. U.S.A. 107, 20744–20749. doi:10.1073/pnas.1013893107
Alsteens, D., Van Dijck, P., Lipke, P. N., and Dufrêne, Y. F. (2013). Quantifying the Forces Driving Cell-Cell Adhesion in a Fungal Pathogen. Langmuir 29, 13473–13480. doi:10.1021/la403237f
Annadurai, N., Malina, L., Malohlava, J., Hajdúch, E. J., and Das, V. (2022). Tau R2 and R3 are Essential Regions for Tau Aggregation, Seeding and Propagation. Biochimie 200, 79–86. doi:10.1016/J.BIOCHI.2022.05.013
Ashkenazy, H., Abadi, S., Martz, E., Chay, O., Mayrose, I., Pupko, T., et al. (2016). ConSurf 2016: an Improved Methodology to Estimate and Visualize Evolutionary Conservation in Macromolecules. Nucleic Acids Res. 44, W344–W350. doi:10.1093/nar/gkw408
Behrens, N. E., Lipke, P. N., Pilling, D., Gomer, R. H., and Klotz, S. A. (2019). Serum Amyloid P Component Binds Fungal Surface Amyloid and Decreases Human Macrophage Phagocytosis and Secretion of Inflammatory Cytokines. MBio 10, 1–14. doi:10.1128/mBio.00218-19
Bera, S., Arad, E., Schnaider, L., Shaham-Niv, S., Castelletto, V., Peretz, Y., et al. (2019). Unravelling the Role of Amino Acid Sequence Order in the Assembly and Function of the Amyloid-β Core. Chem. Commun. 55, 8595–8598. doi:10.1039/c9cc03654g
Berezin, C., Glaser, F., Rosenberg, J., Paz, I., Pupko, T., Fariselli, P., et al. (2004). ConSeq: the Identification of Functionally and Structurally Important Residues in Protein Sequences. Bioinformatics 20, 1322–1324. doi:10.1093/bioinformatics/bth070
Bouyx, C., Schiavone, M., Teste, M. A., Dague, E., Sieczkowski, N., Julien, A., et al. (2021). The Dual Role of Amyloid-β-Sheet Sequences in the Cell Surface Properties of FLO11-Encoded Flocculins in Saccharomyces cerevisiae. Elife 10. doi:10.7554/eLife.68592
Cegelski, L., Pinkner, J. S., Hammer, N. D., Cusumano, C. K., Hung, C. S., Chorell, E., et al. (2009). Small-molecule Inhibitors Target Escherichia coli Amyloid Biogenesis and Biofilm Formation. Nat. Chem. Biol. 5 (5), 913–919. doi:10.1038/nchembio.242
Chan, C. X. J., and Lipke, P. N. (2014). Role of Force-Sensitive Amyloid-like Interactions in Fungal Catch Bonding and Biofilms. Eukaryot. Cell 13, 1136–1142. doi:10.1128/ec.00068-14
Christensen, L. F. B., Jensen, K. F., Nielsen, J., Vad, B. S., Christiansen, G., and Otzen, D. E. (2019). Reducing the Amyloidogenicity of Functional Amyloid Protein FapC Increases Its Ability To Inhibit α-Synuclein Fibrillation. ACS Omega 4, 4029–4039. doi:10.1021/acsomega.8b03590
Dehullu, J., Valotteau, C., Herman-Bausier, P., Garcia-Sherman, M., Mittelviefhaus, M., Vorholt, J. A., et al. (2019). Fluidic Force Microscopy Demonstrates that Homophilic Adhesion by Candida Albicans Als Proteins Is Mediated by Amyloid Bonds between Cells. Nano Lett. 19, 3846–3853. doi:10.1021/acs.nanolett.9b01010
Dehullu, J., Vorholt, J. A., Lipke, P. N., and Dufrêne, Y. F. (2019). Fluidic Force Microscopy Captures Amyloid Bonds between Microbial Cells. Trends Microbiol. 27, 728–730. doi:10.1016/j.tim.2019.06.001
Diederichs, K., and Karplus, P. A. (1997). Improved R-Factors for Diffraction Data Analysis in Macromolecular Crystallography. Nat. Struct. Mol. Biol. 4, 269–275. doi:10.1038/nsb0497-269
Eisenberg, D., and Jucker, M. (2012). The Amyloid State of Proteins in Human Diseases. Cell 148, 1188–1203. doi:10.1016/j.cell.2012.02.022
Eisenberg, D. S., and Sawaya, M. R. (2017). Structural Studies of Amyloid Proteins at the Molecular Level. Annu. Rev. Biochem. 86, 69–95. doi:10.1146/annurev-biochem-061516-045104
Emsley, P., Lohkamp, B., Scott, W. G., and Cowtan, K. (2010). Features and Development ofCoot. Acta Crystallogr. D. Biol. Cryst. 66, 486–501. doi:10.1107/s0907444910007493
Evans, M. L., Gichana, E., Zhou, Y., and Chapman, M. R. (2018). Bacterial Amyloids. Methods Mol. Biol. 1779, 267–288. doi:10.1007/978-1-4939-7816-8_17
Fernandez-Escamilla, A.-M., Rousseau, F., Schymkowitz, J., and Serrano, L. (2004). Prediction of Sequence-dependent and Mutational Effects on the Aggregation of Peptides and Proteins. Nat. Biotechnol. 22, 1302–1306. doi:10.1038/nbt1012
Garcia, M. C., Lee, J. T., Ramsook, C. B., Alsteens, D., Dufrêne, Y. F., and Lipke, P. N. (2011). A Role for Amyloid in Cell Aggregation and Biofilm Formation. PLoS One 6, e17632. doi:10.1371/journal.pone.0017632
Garcia-Sherman, M. C., Lundberg, T., Sobonya, R. E., Lipke, P. N., and Klotz, S. A. (2015). A Unique Biofilm in Human Deep Mycoses: Fungal Amyloid Is Bound by Host Serum Amyloid P Component. NPJ biofilms microbiomes 1. 15009 doi:10.1038/npjbiofilms.2015.9
Garcia-Sherman, M. C., Lysak, N., Filonenko, A., Richards, H., Sobonya, R. E., Klotz, S. A., et al. (2014). Peptide Detection of Fungal Functional Amyloids in Infected Tissue. PLoS One 9, e86067. doi:10.1371/journal.pone.0086067
Gasior, P., and Kotulska, M. (2014). FISH Amyloid - a New Method for Finding Amyloidogenic Segments in Proteins Based on Site Specific Co-occurrence of Aminoacids. BMC Bioinforma. 15, 54–58. doi:10.1186/1471-2105-15-54
Goddard, T. D., Huang, C. C., and Ferrin, T. E. (2007). Visualizing Density Maps with UCSF Chimera. J. Struct. Biol. 157, 281–287. doi:10.1016/j.jsb.2006.06.010
Guenther, E. L., Cao, Q., Trinh, H., Lu, J., Sawaya, M. R., Cascio, D., et al. (2018). Atomic Structures of TDP-43 LCD Segments and Insights into Reversible or Pathogenic Aggregation. Nat. Struct. Mol. Biol. 25, 463–471. doi:10.1038/s41594-018-0064-2
Harris, J. R. (2012). Protein Aggregation and Fibrillogenesis in Cerebral and Systemic Amyloid Disease. Sub-cellular Biochemistry 65, 4. doi:10.1007/978-94-007-5416-4
Ho, V., Herman-Bausier, P., Shaw, C., Conrad, K. A., Garcia-Sherman, M. C., Draghi, J., et al. (2019). An Amyloid Core Sequence in the Major Candida Albicans Adhesin Als1p Mediates Cell-Cell Adhesion. MBio 10. e01766–19. doi:10.1128/mBio.01766-19
Hoyer, L. L., Payne, T. L., Bell, M., Myers, A. M., and Scherer, S. (1998). Candida Albicans ALS3 and Insights into the Nature of the ALS Gene Family. Curr. Genet. 33, 451–459. doi:10.1007/s002940050359
Hoyer, L. L. (2001). The ALS Gene Family of Candida Albicans. Trends Microbiol. 9, 176–180. doi:10.1016/s0966-842x(01)01984-9
Hughes, M. P., Goldschmidt, L., and Eisenberg, D. S. (2021). Prevalence and Species Distribution of the Low-Complexity, Amyloid-like, Reversible, Kinked Segment Structural Motif in Amyloid-like Fibrils. J. Biol. Chem. 297, 101194. doi:10.1016/j.jbc.2021.101194
Hughes, M. P., Sawaya, M. R., Boyer, D. R., Goldschmidt, L., Rodriguez, J. A., Cascio, D., et al. (2018). Atomic Structures of Low-Complexity Protein Segments Reveal Kinked β Sheets that Assemble Networks. Science 359, 698–701. doi:10.1126/science.aan6398
Jain, N., and Chapman, M. R. (2019). Bacterial Functional Amyloids: Order from Disorder. Biochimica Biophysica Acta (BBA) - Proteins Proteomics 1867, 954–960. doi:10.1016/j.bbapap.2019.05.010
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly Accurate Protein Structure Prediction with AlphaFold. Nature 596, 583–589. doi:10.1038/s41586-021-03819-2
Kabsch, W. (2010). XDS. Acta Crystallogr. D. Biol. Cryst. 66, 125–132. doi:10.1107/s0907444909047337
Kapteyn, J. C., Hoyer, L. L., Hecht, J. E., Müller, W. H., Andel, A., Verkleij, A. J., et al. (2000). The Cell Wall Architecture of Candida Albicans Wild-type Cells and Cell Wall-Defective Mutants. Mol. Microbiol. 35, 601–611. doi:10.1046/j.1365-2958.2000.01729.x
Karplus, P. A., and Diederichs, K. (2012). Linking Crystallographic Model and Data Quality. Science 336 (6084), 1030–1033. doi:10.1126/science.1218231
Klotz, S. A., Sobonya, R. E., Lipke, P. N., and Garcia-Sherman, M. C. (2016). Serum Amyloid P Component and Systemic Fungal Infection: Does it Protect the Host or Is it a Trojan Horse? Open forum Infect. Dis. 3, ofw166. doi:10.1093/ofid/ofw166
Landau, M., Mayrose, I., Rosenberg, Y., Glaser, F., Martz, E., Pupko, T., et al. (2005). ConSurf 2005: the Projection of Evolutionary Conservation Scores of Residues on Protein Structures. Nucleic Acids Res. 33, W299–W302. doi:10.1093/nar/gki370
Landau, M., Sawaya, M. R., Faull, K. F., Laganowsky, A., Jiang, L., Sievers, S. A., et al. (2011). Towards a Pharmacophore for Amyloid. PLOS Biol. 9, e1001080. doi:10.1371/journal.pbio.1001080
Lawrence, M. C., and Colman, P. M. (1993). Shape Complementarity at Protein/protein Interfaces. J. Mol. Biol. 234, 946–950. doi:10.1006/jmbi.1993.1648
Lin, J., Oh, S.-H., Jones, R., Garnett, J. A., Salgado, P. S., Rusnakova, S., et al. (2014). The Peptide-Binding Cavity Is Essential for Als3-Mediated Adhesion of Candida Albicans to Human Cells. J. Biol. Chem. 289, 18401–18412. doi:10.1074/jbc.m114.547877
Lipke, P. N., Klotz, S. A., Dufrene, Y. F., Jackson, D. N., and Garcia-Sherman, M. C. (2017). Amyloid-Like β-Aggregates as Force-Sensitive Switches in Fungal Biofilms and Infections. Microbiol. Mol. Biol. Rev. 82 (1), e00035–17. doi:10.1128/MMBR.00035-17
Lipke, P. N., Mathelié-Guinlet, M., Viljoen, A., and Dufrêne, Y. F. (2021). A New Function for Amyloid-like Interactions: Cross-Beta Aggregates of Adhesins Form Cell-To-Cell Bonds. Pathog 10, 1013. 1013 10. doi:10.3390/pathogens10081013
Lipke, P. N. (2018). What We Do Not Know about Fungal Cell Adhesion Molecules. J. fungi (Basel, Switz. 4 (2), 59. doi:10.3390/jof4020059
Lipke, P. N., and Kurjan, J. (1992). Sexual Agglutination in Budding Yeasts: Structure, Function, and Regulation of Adhesion Glycoproteins. Microbiol. Rev. 56, 180–194. doi:10.1128/mr.56.1.180-194.1992
Maji, S. K., Wang, L., Greenwald, J., and Riek, R. (2009). Structure-activity Relationship of Amyloid Fibrils. FEBS Lett. 583, 2610–2617. doi:10.1016/j.febslet.2009.07.003
Mariani, V., Biasini, M., Barbato, A., and Schwede, T. (2013). lDDT: a Local Superposition-free Score for Comparing Protein Structures and Models Using Distance Difference Tests. Bioinformatics 29, 2722–2728. doi:10.1093/bioinformatics/btt473
McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C., and Read, R. J. (2007). Phasercrystallographic Software. J. Appl. Cryst. 40, 658–674. doi:10.1107/s0021889807021206
Muñoz, J. F., Welsh, R. M., Shea, T., Batra, D., Gade, L., Howard, D., et al. (2021). Clade-specific Chromosomal Rearrangements and Loss of Subtelomeric Adhesins in Candida Auris. Genetics 218 (1), iyab029. doi:10.1093/genetics/iyab029
Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997). Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Cryst. D. 53, 240–255. doi:10.1107/s0907444996012255
Otoo, H. N., Lee, K. G., Qiu, W., and Lipke, P. N. (2008). Candida Albicans Als Adhesins Have Conserved Amyloid-Forming Sequences. Eukaryot. Cell 7, 776–782. doi:10.1128/ec.00309-07
Perov, S., Lidor, O., Salinas, N., Golan, N., Tayeb-Fligelman, E., Deshmukh, M., et al. (2019). Structural Insights into Curli CsgA Cross-β Fibril Architecture Inspire Repurposing of Anti-amyloid Compounds as Anti-biofilm Agents. PLoS Pathog. 15, e1007978. doi:10.1371/journal.ppat.1007978
Porat, Y., Stepensky, A., Ding, F.-X., Naider, F., and Gazit, E. (2003). Completely Different Amyloidogenic Potential of Nearly Identical Peptide Fragments. Biopolymers 69, 161–164. doi:10.1002/bip.10386
Ramsook, C. B., Tan, C., Garcia, M. C., Fung, R., Soybelman, G., Henry, R., et al. (2010). Yeast Cell Adhesion Molecules Have Functional Amyloid-Forming Sequences. Eukaryot. Cell 9, 393–404. doi:10.1128/ec.00068-09
Rauceo, J. M., De Armond, R., Otoo, H., Kahn, P. C., Klotz, S. A., Gaur, N. K., et al. (2006). Threonine-Rich Repeats Increase Fibronectin Binding in the Candida albicans Adhesin Als5p. Eukaryot. Cell 5, 1664–1673. doi:10.1128/EC.00120-06
Rauceo, J. M., Gaur, N. K., Lee, K.-G., Edwards, J. E., Klotz, S. A., and Lipke, P. N. (2004). Global Cell Surface Conformational Shift Mediated by a Candida Albicans Adhesin. Infect. Immun. 72, 4948–4955. doi:10.1128/iai.72.9.4948-4955.2004
Roh, D.-H., Bowers, B., Riezman, H., and Cabib, E. (2002). Rho1p Mutations Specific for Regulation of β(1→3)glucan Synthesis and the Order of Assembly of the Yeast Cell Wall. Mol. Microbiol. 44, 1167–1183. doi:10.1046/j.1365-2958.2002.02955.x
Salgado, P. S., Yan, R., Taylor, J. D., Burchell, L., Jones, R., Hoyer, L. L., et al. (2011). Structural Basis for the Broad Specificity to Host-Cell Ligands by the Pathogenic Fungus Candida Albicans. Proc. Natl. Acad. Sci. U.S.A. 108, 15775–15779. doi:10.1073/pnas.1103496108
Shewmaker, F., McGlinchey, R. P., and Wickner, R. B. (2011). Structural Insights into Functional and Pathological Amyloid. J. Biol. Chem. 286, 16533–16540. doi:10.1074/jbc.r111.227108
Tayeb-Fligelman, E., and Landau, M. (2017). X-Ray Structural Study of Amyloid-like Fibrils of Tau Peptides Bound to Small-Molecule Ligands. Methods Mol. Biol. 1523, 89–100. doi:10.1007/978-1-4939-6598-4_5
Tayeb-Fligelman, E., Tabachnikov, O., Moshe, A., Goldshmidt-Tran, O., Sawaya, M. R., Coquelle, N., et al. (2017). The Cytotoxic Staphylococcus aureus PSMα3 Reveals a Cross-α Amyloid-like Fibril. Science 355, 831–833. doi:10.1126/science.aaf4901
Tsolis, A. C., Papandreou, N. C., Iconomidou, V. A., and Hamodrakas, S. J. (2013). A Consensus Method for the Prediction of 'Aggregation-Prone' Peptides in Globular Proteins. PLoS One 8, e54175. doi:10.1371/journal.pone.0054175
Varadi, M., Anyango, S., Deshpande, M., Nair, S., Natassia, C., Yordanova, G., et al. (2022). AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 50, D439–D444. doi:10.1093/nar/gkab1061
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M., and Barton, G. J. (2009). Jalview Version 2--a Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 25, 1189–1191. doi:10.1093/bioinformatics/btp033
Willaert, R. G. (2018). Adhesins of Yeasts: Protein Structure and Interactions. J. fungi (Basel, Switz. 4 (4), 119. doi:10.3390/jof4040119
Willaert, R. G., Kayacan, Y., and Devreese, B. (2021). The Flo Adhesin Family. Pathogens. Pathogens 10 (11), 1397. doi:10.3390/pathogens10111397
Keywords: cross-beta, biofilm adhesin, Candida pathogenesis, cell wall mannoproteins, functional amyloids
Citation: Golan N, Schwartz-Perov S, Landau M and Lipke PN (2022) Structure and Conservation of Amyloid Spines From the Candida albicans Als5 Adhesin. Front. Mol. Biosci. 9:926959. doi: 10.3389/fmolb.2022.926959
Received: 23 April 2022; Accepted: 09 June 2022;
Published: 06 July 2022.
Edited by:
Sonja Kroschwald, ETH Zürich, SwitzerlandReviewed by:
Vladimir N. Uversky, University of South Florida, United StatesCopyright © 2022 Golan, Schwartz-Perov, Landau and Lipke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meytal Landau, bWxhbmRhdUB0ZWNobmlvbi5hYy5pbA==; Peter N. Lipke, UExpcGtlQGJyb29rbHluLmN1bnkuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.