- 1Department of Molecular Medicine, Morsani College of Medicine, University of South Florida, Tampa, FL, United States
- 2Department of Chemistry, College of Arts and Sciences, University of South Florida, Tampa, FL, United States
- 3Global and Planetary Health, College of Public Health, University of South Florida, Tampa, FL, United States
5-Aminolevulinate synthase (ALAS; E.C. 2.3.1.37) is a pyridoxal 5′-phosphate (PLP)-dependent enzyme that catalyzes the key regulatory step of porphyrin biosynthesis in metazoa, fungi, and α-proteobacteria. ALAS is evolutionarily related to transaminases and is therefore classified as a fold type I PLP-dependent enzyme. As an enzyme controlling the key committed and rate-determining step of a crucial biochemical pathway ALAS is ideally positioned to be subject to allosteric feedback inhibition. Extensive kinetic and mutational studies demonstrated that the overall enzyme reaction is limited by subtle conformational changes of a hairpin loop gating the active site. These findings, coupled with structural information, facilitated early prediction of allosteric regulation of activity via an extended C-terminal tail unique to eukaryotic forms of the enzyme. This prediction was subsequently supported by the discoveries that mutations in the extended C-terminus of the erythroid ALAS isoform (ALAS2) cause a metabolic disorder known as X-linked protoporphyria not by diminishing activity, but by enhancing it. Furthermore, kinetic, structural, and molecular modeling studies demonstrated that the extended C-terminal tail controls the catalytic rate by modulating conformational flexibility of the active site loop. However, the precise identity of any such molecule remains to be defined. Here we discuss the most plausible allosteric regulators of ALAS activity based on divergences in AlphaFold-predicted ALAS structures and suggest how the mystery of the mechanism whereby the extended C-terminus of mammalian ALASs allosterically controls the rate of porphyrin biosynthesis might be unraveled.
Introduction
5-Aminolevulinate synthase (ALAS; EC 2.3.1.37) catalyzes the initial and key regulatory step of heme biosynthesis in metazoa, fungi, and the α-subclass of proteobacteria (Stojanovski et al., 2019; Taylor and Brown, 2022). Pyridoxal 5′-phosphate (PLP) is an essential cofactor for the reaction, which involves the condensation of the α-carbon of glycine with the succinyl group of succinyl-Coenzyme A (SCoA) to produce 5-aminolevulinate (ALA), carbon dioxide, and Coenzyme A (Hunter and Ferreira, 2011) (Supplementary Figure S1). In metazoa and fungi, ALAS is translated as a precursor with an N-terminal signal sequence that codes for import into the mitochondrial matrix. Following import, the signal sequence is cleaved, and the mature enzyme has access to the substrate SCoA, which is produced in mitochondria as part of the citric acid cycle. The requirement of SCoA as a substrate integrates heme biosynthesis with oxidative respiration, and as a result the two pathways are synchronized under normal healthy conditions. ALAS activity is additionally synchronized with cellular iron transport as porphyrin biosynthesis and iron transport unite in the final step of heme production wherein the enzyme ferrochelatase inserts ferrous iron into protoporphyrin IX to yield heme (Kafina and Paw, 2017; Poli et al., 2021). As a result of the central position of ALAS in these fundamental biochemical pathways ALAS activity is highly regulated and new modes of ALAS regulation continue to be discovered (Tanimura et al., 2016; Zhang et al., 2017; Liu et al., 2018; Peoc'h et al., 2019; Bailey et al., 2020; Nomura et al., 2021; Rondelli et al., 2021).
Vertebrate genomes encode two chromosomally distinct copies of the ALAS gene: ALAS1, which acts as a “housekeeping” gene and initiates heme biosynthesis in all cells for production of cytochromes and other heme-binding proteins, and ALAS2, which is expressed only in developing erythrocytes and produces, almost exclusively, the much larger quantities of heme required for hemoglobin formation (Riddle et al., 1989; Peoc'h et al., 2019). The catalytic cores of human ALAS1 and ALAS2 are 75% identical and 94% similar in terms of amino acid sequences, suggesting gene duplication and similar enzymology despite the different metabolic functionalities of the gene products. The high degree of similarity in the catalytic cores of ALAS1 and ALAS2 is lessened in the extended N- and C-termini of the enzymes (Supplementary Figure S2) but the precise extent to which the mature mitochondrial enzymes might be differentially regulated is still an open question. The monomeric primary structures of prokaryotic and vertebrate ALASs are illustrated schematically in Figure 1A.
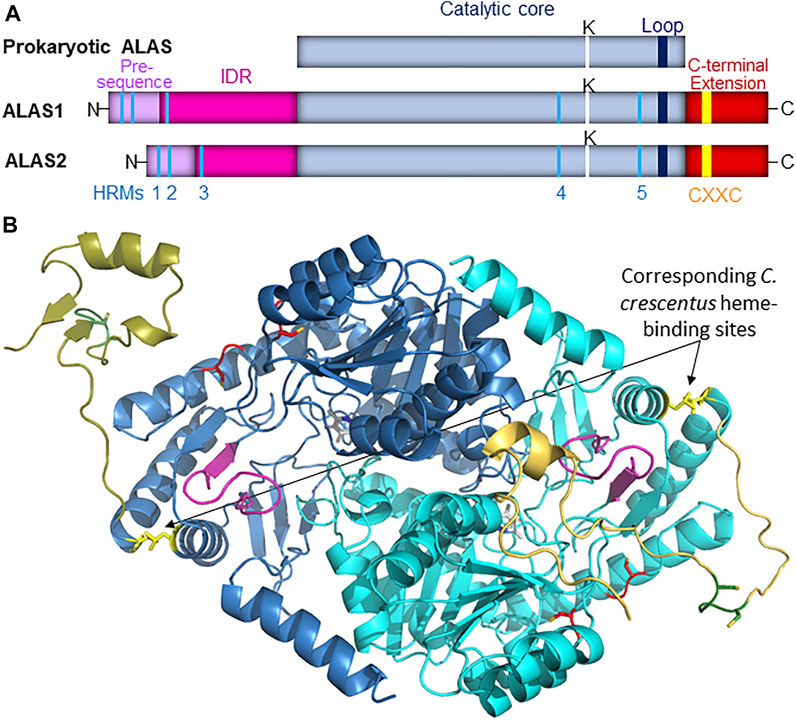
FIGURE 1. (A). Schematic representation of ALAS monomeric structure. In vertebrate species the ALAS gene is duplicated, and the protein catalytic core (light blue) observed in prokaryotes is bracketed by extended N- and C-termini. The mitochondrial targeting sequence is illustrated in light purple, while the intrinsically disordered N-terminal extension (IDR) is in magenta, and the C-terminal extension is in dark red. Five conserved Heme Regulatory Motifs (HRMs, colored in cyan) are conserved in vertebrate ALAS isozymes, as is a CXXC motif (yellow) in the extended C-terminus. The position of the active site lysine residue that binds PLP in the active site is denoted by a white line, and the loop that gates the active site is represented by a dark blue line. (B). The position corresponding to the heme-binding site in C. crescentus ALAS modeled into mammalian ALAS2. AlphaFold-predicted structures for human (light blue and gold; AlphaFold entry P22557) and murine (dark blue and gold; AlphaFold entry P08680) ALAS2 were aligned with R. capsulatus ALAS crystal structure (PDB code 2bwn; not shown) using Pymol. The modeled site depicted in yellow here is not expected to bind heme in mammals due to evolutionary divergences, and this site is illustrated solely for perspective on its spatial relationship to the mammalian ALAS2 active site loop (purple), the C-terminal extension (shades of gold), and HRMs 4 and 5 (red). Additionally, the CXXC motifs are in green with the cysteines shown as sticks.
The ALAS-catalyzed reaction not only represents the first committed step of heme production, but also the rate-determining step of porphyrin biosynthesis, as most poignantly evinced by the consistent observation that exogenous ALA administration to mammalian cells leads to rapid protoporphyrin IX accumulation (Hunter and Ferreira, 2011; Nokes et al., 2013). This is clinically important because it means aberrations in ALAS activity can change the overall rate of porphyrin production and cause porphyrin biosynthesis to decouple from oxidative respiration and iron transport, resulting in metabolic imbalances (Taylor and Brown, 2022). For instance, certain liver toxins, such as allylisopropylacetamide, have long been known to elevate ALAS1 activity beyond the rate of iron transport, resulting in porphyrin accumulation and chemically induced porphyria (Goldberg and Rimington, 1955; Granick, 1966). Conversely, genetic defects in ALAS2 that lead to lower enzymatic activity have been identified as the basis for X-linked sideroblastic anemia, a condition characterized by accumulation of iron in erythroblast mitochondria (Abu-Zeinah and DeSancho, 2020). Remarkably, however, loss-of-function mutations are not the only cause of ALAS2-associated metabolic disorder. A limited number of mutations causing premature truncation or frameshifts in the extreme C-terminal extension of ALAS2 lead to variants with increased catalytic efficiencies and a disorder known as X-linked protoporphyria (Whatley et al., 2008; Ducamp et al., 2013; Wang et al., 2020). Interestingly, mutations in ALAS1 have not been associated with any disorder (Stenson et al., 2003).
5-Aminolevulinate Synthase is a Fold Type I Pyridoxal 5′-Phosphate-dependent Enzyme With a Distinct Active Site Loop
PLP-dependent enzymes are structurally classified into seven different fold types, of which fold type I, sometimes referred to as the transaminase family, is by far the largest, with over 170 different Enzyme Classification numbers currently assigned (Percudani and Peracchi, 2009). Like other members of the PLP-dependent fold type I family ALAS is a homodimer with the active site buried near the center of the enzyme at the interface between the two monomers, with residues from each monomer being critical for substrate recognition (Brown et al., 2018; Stojanovski et al., 2019). Even though fold type I PLP-dependent enzymes have very little overall primary sequence similarity the active sites are highly conserved and facilitate phylogenetic analyses demonstrating function-based evolutionary relationships (Catazaro et al., 2014). It is thus informative to compare the structure of aspartate aminotransferase (AATase), which has been extensively characterized and is generally considered to be a model for the fold type I family (Toney, 2014), with the ALAS catalytic core, as seen in Supplementary Figure S3. The aligned structures of AATase in the open and closed conformations reveal the structure collapses inwards towards the PLP cofactor upon substrate binding (McPhalen et al., 1992a; McPhalen et al., 1992b) A short active site loop (green and gold in Supplementary Figure S3A) closes inward over the active site cleft upon substrate binding, culminating in an arginine residue that is highly conserved in fold type I enzymes, and functions to form an ionic bond with the carboxylate group of the amino acid substrate (Tan et al., 1998; Liang et al., 2019). In AATase, this arginine is one of only two amino acids that has been designated as a “closure-inducing residue”, meaning it is essential for substrate-induced conformational change from the open to the closed state in which catalysis is optimized (Hayward, 2004). Comparison of these structures to analogous structures of Rhodobacter capsulatus ALAS (Supplementary Figure S3B) reveals that in ALAS substrate-induced conformational changes are largely limited to the active site loop, which has become longer and is turned more inward over the active site cleft relative to AATase.
Detailed mutational, kinetic, and molecular modeling studies have found that the rate of ALAS catalysis, and hence the rate of porphyrin production, are controlled by the slow opening of this active site loop, which allows the products to rapidly dissociate from the enzyme (Hunter and Ferreira, 1999; Hunter and Ferreira, 2011; Hunter et al., 2007; Stojanovski et al., 2019). This rate-dependence on conformational dynamics would seem to be an ideal situation for allosteric feedback inhibition of the heme biosynthesis pathway via a mechanism wherein effector binding to ALAS would modulate the active site loop conformational dynamics, as we previously suggested (Hunter et al., 2007).
5-Aminolevulinate Synthase Structural Features Reveal important Clues to the Possibility of Allosteric Regulation
Feedback inhibition of ALAS activity by heme has been known for over 50 years (Granick, 1966), and since then this regulation has been found to occur at a variety of levels, including gene transcription (Yamamoto et al., 1982), transport into mitochondria (Lathrop and Timko, 1993; Munakata et al., 2004), and targeting for degradation (Cable et al., 1996; Yoshino et al., 2007; Tian et al., 2011; Nomura et al., 2021). However, as of this writing direct binding of heme leading to allosteric feedback inhibition of ALAS has only been reported for the enzyme from the prokaryote Caulobacter crescentus, in which axial heme binding by H340 and C398 near the C-terminus of the enzyme causes PLP dissociation (Ikushiro et al., 2018) (Figure 1B). While the authors reported that these residues are conserved in some other α-proteobacteria and did confirm that recombinant R. capsulatus ALAS could also be isolated as a mixture of PLP- and heme-bound forms, these residues are not conserved in eukaryotes, so if allosteric feedback inhibition of ALAS in higher species occurs it must be via a different site. The recently resolved crystal structure for human ALAS2 revealed that the extended C-terminus might act as an autoinhibitory element by folding directly over the active site cleft, clearly implying the existence of some allosteric modulator that alters the conformational dynamics about the C-terminus to allow substrates to access the active site (Bailey et al., 2020), and yet the identity of this effector remains a mystery.
Each of the vertebrate ALAS isozymes contains five heme-regulatory motifs (HRMs), consensus sequences containing a cysteine-proline dipeptide with the cysteine functioning as a ligand to Fe3+-heme (Figure 1A) (Carter et al., 2017; Fleischhacker et al., 2020). HRMs are important in regulating the activity of a wide variety of enzymes controlling gene transcription (Hou et al., 2006; Fleischhacker et al., 2018; Arunachalam et al., 2021), protein synthesis (Igarashi et al., 2008), circadian rhythms (Yang et al., 2008), iron homeostasis (Nishitani et al., 2019), signal transduction (Shen et al., 2014; Schmalohr et al., 2021), and heme degradation (Fleischhacker et al., 2015; Fleischhacker et al., 2018). The first two ALAS HRMs reside in the mitochondrial import signal sequence, where they are positioned to bind excess labile heme and form a complex that is not imported into mitochondria, thus providing a form of feedback inhibition (Lathrop and Timko, 1993; Munakata et al., 2004). Following import the signal sequences are proteolytically removed to produce mature enzymes with intrinsically disordered N-termini (Stojanovski et al., 2016; Nomura et al., 2021). This N-terminal extension contains a third conserved HRM that feedback inhibits ALAS1 by binding heme to form a complex targeting ALAS1 for proteolysis by the matrix peptidase chaperone subunit ClpX (Nomura et al., 2021). ClpX also controls ALAS2 turnover (Rondelli et al., 2021), and since HRM 3 is conserved in ALAS2, it seems likely that HRM 3 also mediates ClpX degradation of ALAS2 in a heme-dependent fashion, although this remains to be conclusively demonstrated.
The catalytic core of mammalian ALAS, which is approximately 44 kD in size, contains two additional conserved HRMs, which we designate HRMs 4 and 5. To the best of our knowledge, no studies have yet examined their potential biochemical significance. Along with the human ALAS2 crystal structure, the mammalian ALAS1 and ALAS2 AlphaFold-predicted structures reveal that even though HRMs 4 and 5 are ∼132 amino acids apart in the primary sequence, in the three-dimensional structures the cysteine α-carbons are only 11 Å apart, and most importantly, they are near or at the enzyme surface in proximity to both the active site loop and the extended C-terminus, in conspicuous positions for heme-mediated feedback regulation of the mature enzyme (Figure 2). The positions of HRMs 4 and 5 in the AlphaFold-predicted ALAS structures are virtually indistinguishable from those in the human ALAS2 crystal structure (Figure 2B).
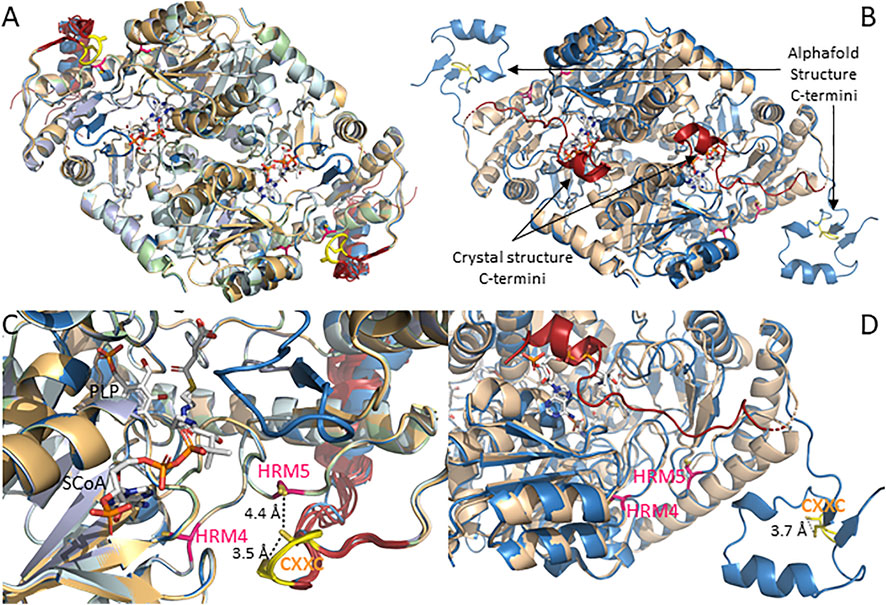
FIGURE 2. AlphaFold structures for mammalian ALAS1 and ALAS2 reveal C-terminal divergences from the human ALAS2 crystal structure. (A). Alignment of AlphaFold-predicted structures of ALAS1 from human (UniProt accession # P13196), orangutan (UniProt accession Q5R9R9), bovine (UniProt accession A6QLI6), beluga whale (UniProt accession Q9XS79), mouse (UniProt accession Q8VC19), and rat (UniProt accession # P13195). (B). Alignment of human ALAS2 crystal and AlphaFold-predicted structures. AlphaFold-predicted structure (UniProt accession # P22557; blue) and crystal structure (PDB code 6HRH; beige with red C-termini). (C). Zoom of panel (A). (D). Zoom of panel (B).
There are, however, important differences in the relative positions of the extended C-termini of the ALAS1 and ALAS2 isozymes as it relates to HRMs 4 and 5. As seen in Figures 2A,C, all six of the currently available AlphaFold-predicted structures for mammalian ALAS1 position the extended C-terminus such that the CXXC motif forms a hairpin loop that brings the cysteine sulfur atoms within ∼ 3.5 Å of each other, suggesting disulfide bond formation and a possible redox sensing role. Furthermore, the CXXC loop is positioned almost directly over HRM5.
In contrast to the consensus positioning of the ALAS1 extended C-terminus over HRMs 4 and 5, the AlphaFold-predicted mammalian ALAS2 structures have more conformational heterogeneity about the C-terminal extension (Figures 2B,D). Moreover, none of the ALAS2 C-terminal extensions align with the ALAS1 C-terminus. Instead, the ALAS2 C-terminal extensions fall into one of three different conformations. In the AlphaFold-predicted structures for orangutan, bovine, beluga whale, and rat ALAS2s, the extended C-terminus folds over the active site to form an “autoinhibited” structure, in excellent alignment with the recently solved human ALAS2 crystal structure (Bailey et al., 2020), but the AlphaFold-predicted human ALAS2 structure places the extended C-terminus away from the catalytic core in what would presumably correspond to an active enzyme conformation. Meanwhile, in the mouse ALAS2, the extended C-terminus adopts a conformation between these two extremes. In all cases the cysteines of the ALAS2 CXXC motifs, like those of the ALAS1 CXXC motifs, are in sufficient proximity to reversibly form disulfides, and thus potentially act as redox sensors. But unlike ALAS1, HRMs 4 and 5 of ALAS2 are not occluded by the C-terminal extension and are thus more available to bind heme in what would presumably be a feedback-inhibited complex.
A Case for Differential Regulation by the C-Terminal Extensions
Remarkably, in ten out of twelve different mammalian ALAS mitochondrial import presequences AlphaFold predicts the side chains of the cysteines in HRMs 1 and 2 to be almost ideally positioned to act as axial ligands for heme (Supplementary Figure S4). This agrees with experimental evidence demonstrating HRMs 1 and 2 bind heme to feedback inhibit mitochondrial import (Lathrop and Timko, 1993; Goodfellow et al., 2001; Munakata et al., 2004). Further, it leads us to suggest that the predicted conformational differences in the extended C-termini might in turn be experimentally revealed to be accurate predictors of important structural/functional divergences between the two ALAS isozymes.
The AlphaFold structural database currently has nearly a million protein structures available, including complete proteomes for Homo sapiens and 47 other species (Jumper et al., 2021; Tunyasuvunakool et al., 2021). These structures are rapidly facilitating an unprecedented understanding of structural biology (Hegedus et al., 2022; Porta-Pardo et al., 2022; Varadi et al., 2022; Wehrspan et al., 2022). Yet, the accuracy of AlphaFold in terms of predicting otherwise unsolved structures is relatively untested since it only became publicly available less than a year ago. AlphaFold is reported to accurately predict not just the highly organized structures observed in crystallized proteins, but also the extent of conformational dynamics or even intrinsic disorder in individual residues or peptides by calculating a per residue confidence score referred to as a predicted local distance difference test (pLDDT) (Tunyasuvunakool et al., 2021). The current interpretation of this score is that it predicts the extent to which a residue is unstructured, meaning a low score should be seen not so much as an indication the structure is inaccurate, but more as an accurate indication of greater conformational dynamics. Because of this AlphaFold should provide important insight into dynamic regulatory structures that have been difficult to crystallize.
The ALAS1/2 conserved CXXC motif is of particular interest since similar motifs act as allosteric redox switches via reversible formation of a disulfide bond in many enzymes, including the PLP-dependent enzymes cystathionine β-synthase and human mitochondrial branched chain aminotransferase (Conway et al., 2004; Wouters et al., 2010; Niu et al., 2018; Herbert et al., 2020). The CXXC motif-containing region was only partially resolved in the human ALAS2 crystal structure, implying a high degree of conformational mobility. The AlphaFold pLDDT scores for the six mammalian ALAS2 (and six ALAS1) structures in the public database agree, as they drop from very high confidence to low or even very low for the corresponding amino acids in all species except human ALAS2 (Supplementary Figure S5), in which the extended C-terminus adopts what is presumably an activated enzyme conformation. In this “activated” ALAS2 structure the scores for the CXXC motif are mostly confident, indicating greater structural organization, and with the cysteine side chain sulfur atoms within 3.7 Å of each other, disulfide bond formation is possible. Given all these considerations, if the CXXC motif in the extended C-terminus of ALAS2 acts as a redox switch we would predict that the “activated” structure would be oxidized to the disulfide, while the more disordered autoinhibited structure would be reduced.
The positioning of the human ALAS2 extended C-terminus over the active site leads us to raise the questions as to what the active conformation might look like and how the interconversion between the inhibited and activated conformations might be triggered. The corresponding AlphaFold structure appears to provide a plausible answer to the first of these two questions, but only hints at the answer to the second. Binding of the β-subunit of succinyl-CoA synthetase (Furuyama and Sassa, 2000; Bishop et al., 2012; Bishop et al., 2013) and/or other heme biosynthetic enzymes might promote activation (Medlock et al., 2015). A novel, but certainly not mutually exclusive, possibility supported by the structures analyzed here is that the CXXC motif acts as a redox sensor to modulate conformational dynamics about the extended C-terminus.
In contrast to ALAS2, a crystal structure for ALAS1 has not yet been reported, and the AlphaFold-predicted structures indicate only one conformation for the ALAS1 extended C-terminus. Yet, the CXXC motif is conserved in ALAS1, and if it has a redox switching function then some degree of conformational perturbation presumably occurs to form an autoinhibited conformation or to alter the dynamics about the active site loop, which controls the catalytic rate. This latter possibility is attractive as it would be consistent with the anti-correlation between the active site loop and C-terminal extension of ALAS2 during molecular dynamics simulations (Na et al., 2018). Additionally, the shielding of the otherwise solvent exposed HRMs 4 and 5 by the ALAS1 C-terminal extension suggests an alternative conformation that would allow heme access to feedback inhibit the enzyme. Given these considerations we posit that the ALAS1 structures represent an activated form wherein the CXXC motif is oxidized to the disulfide and positioned to prevent allosteric feedback inhibition by heme. Reduction of the CXXC motif would then facilitate a conformation change allowing heme to allosterically feedback inhibit ALAS1 via HRMs 4 and/or 5. A more prominent role of redox sensing in ALAS1 is in part attractive due to the role of ALAS1 in producing heme specifically for hemoproteins catalyzing redox chemistry, such as cytochrome P450 enzymes, catalase, and superoxide dismutase.
Conclusion and Outlook
In summary, based upon the alignment of the ALAS1 structures we put forth the following postulates: 1) HRMs 4 and/or 5 facilitate feedback inhibition of ALAS1; 2) under oxidizing conditions, the CXXC motif forms a disulfide bond that causes the C-terminal extension to fold over HRMs 4 and 5 such that it sterically prevents hemin binding and feedback inhibition; 3) under non-oxidizing conditions, the CXXC motif is reduced and adopts an alternative conformation wherein HRMs 4 and 5 are exposed to provide feedback inhibition by excess heme. Stated more concisely, feedback inhibition of ALAS1 by heme is dependent upon cellular redox status.
Based on the alignment of the ALAS2 structures we put forth the following postulates: 1) the C-terminal extension of ALAS2 adopts two different conformations, neither of which prevents feedback regulation via heme binding to HRMs 4 and 5. 2) In ALAS2 oxidizing conditions cause disulfide bond formation in the CXXC motif and movement of the extended C-terminus not over HRMs 4&5 but instead to a more equatorial and activated position relative to the enzyme, thereby relieving the autoinhibition observed when the extended C-terminus folds over the active site. Stated more succinctly, heme and redox status independently regulate ALAS2 activity.
These postulates are not incompatible with the possibility of protein-protein interactions regulating activity. Of course, experimental data will be required to further support or refine the views presented here, but whatever the outcome the remarkably divergent structures discussed here will likely represent a key test of the capacity of AlphaFold to discern fine structural differences and facilitate prediction of allostery in all enzymes, including those dependent upon PLP for functionality.
Author Contributions
GH performed the structural analyses and analyzed the data. GH and GF conceptualized, wrote, and edited the manuscript. GH and GF approved the submitted version of the manuscript.
Funding
During the writing of this review, the authors were partially supported by a grant from the Florida Department of Health, Bankhead-Coley Cancer Research Program (#9BC14).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.920668/full#supplementary-material
References
Abu-Zeinah, G., and DeSancho, M. T. (2020). Understanding Sideroblastic Anemia: An Overview of Genetics, Epidemiology, Pathophysiology and Current Therapeutic Options. Jbm 11, 305–318. doi:10.2147/jbm.s232644
Arunachalam, A., Lakshmanan, D. K., Ravichandran, G., Paul, S., Manickam, S., Kumar, P. V., et al. (2021). Regulatory Mechanisms of Heme Regulatory Protein BACH1: A Potential Therapeutic Target for Cancer. Med. Oncol. 38, 122. doi:10.1007/s12032-021-01573-z
Bailey, H. J., Bezerra, G. A., Marcero, J. R., Padhi, S., Foster, W. R., Rembeza, E., et al. (2020). Human Aminolevulinate Synthase Structure Reveals a Eukaryotic-Specific Autoinhibitory Loop Regulating Substrate Binding and Product Release. Nat. Commun. 11, 2813. doi:10.1038/s41467-020-16586-x
Bishop, D. F., Tchaikovskii, V., Hoffbrand, A. V., Fraser, M. E., and Margolis, S. (2012). X-Linked Sideroblastic Anemia Due to Carboxyl-Terminal ALAS2 Mutations that Cause Loss of Binding to the β-Subunit of Succinyl-CoA Synthetase (SUCLA2). J. Biol. Chem. 287, 28943–28955. doi:10.1074/jbc.m111.306423
Bishop, D. F., Tchaikovskii, V., Nazarenko, I., and Desnick, R. J. (2013). Molecular Expression and Characterization of Erythroid-Specific 5-Aminolevulinate Synthase Gain-Of-Function Mutations Causing X-Linked Protoporphyria. Mol. Med. 19, 18–25. doi:10.2119/molmed.2013.00003
Brown, B. L., Kardon, J. R., Sauer, R. T., and Baker, T. A. (2018). Structure of the Mitochondrial Aminolevulinic Acid Synthase, a Key Heme Biosynthetic Enzyme. Structure 26, 580–589. doi:10.1016/j.str.2018.02.012
Cable, E. E., Gildemeister, O. S., Pepe, J. A., Donohue, S. E., Lambrecht, R. W., and Bonkovsky, H. L. (1996). Hepatic 5-Aminolevulinic Acid Synthase mRNA Stability Is Modulated by Inhibitors of Heme Biosynthesis and by Metalloporphyrins. Eur. J. Biochem. 240, 112–117. doi:10.1111/j.1432-1033.1996.0112h.x
Carter, E. L., Ramirez, Y., and Ragsdale, S. W. (2017). The Heme-Regulatory Motif of Nuclear Receptor Rev-Erbβ Is a Key Mediator of Heme and Redox Signaling in Circadian Rhythm Maintenance and Metabolism. J. Biol. Chem. 292, 11280–11299. doi:10.1074/jbc.m117.783118
Catazaro, J., Caprez, A., Guru, A., Swanson, D., and Powers, R. (2014). Functional Evolution of PLP-Dependent Enzymes Based on Active-Site Structural Similarities. Proteins 82, 2597–2608. doi:10.1002/prot.24624
Conway, M. E., Poole, L. B., and Hutson, S. M. (2004). Roles for Cysteine Residues in the Regulatory CXXC Motif of Human Mitochondrial Branched Chain Aminotransferase Enzyme. Biochemistry 43, 7356–7364. doi:10.1021/bi0498050
Ducamp, S., Schneider-Yin, X., de Rooij, F., Clayton, J., Fratz, E. J., Rudd, A., et al. (2013). Molecular and Functional Analysis of the C-Terminal Region of Human Erythroid-Specific 5-Aminolevulinic Synthase Associated with X-Linked Dominant Protoporphyria (XLDPP). Hum. Mol. Genet. 22, 1280–1288. doi:10.1093/hmg/dds531
Fleischhacker, A. S., Carter, E. L., and Ragsdale, S. W. (2018). Redox Regulation of Heme Oxygenase-2 and the Transcription Factor, Rev-Erb, Through Heme Regulatory Motifs. Antioxidants Redox Signal. 29, 1841–1857. doi:10.1089/ars.2017.7368
Fleischhacker, A. S., Gunawan, A. L., Kochert, B. A., Liu, L., Wales, T. E., Borowy, M. C., et al. (2020). The Heme-Regulatory Motifs of Heme Oxygenase-2 Contribute to the Transfer of Heme to the Catalytic Site for Degradation. J. Biol. Chem. 295, 5177–5191. doi:10.1074/jbc.ra120.012803
Fleischhacker, A. S., Sharma, A., Choi, M., Spencer, A. M., Bagai, I., Hoffman, B. M., et al. (2015). The C-Terminal Heme Regulatory Motifs of Heme Oxygenase-2 Are Redox-Regulated Heme Binding Sites. Biochemistry 54, 2709–2718. doi:10.1021/acs.biochem.5b00266
Furuyama, K., and Sassa, S. (2000). Interaction Between Succinyl CoA Synthetase and the Heme-Biosynthetic Enzyme ALAS-E Is Disrupted in Sideroblastic Anemia. J. Clin. Invest. 105, 757–764. doi:10.1172/jci6816
Goldberg, A., and Rimington, C. (1955). Experimentally Produced Porphyria in Animals. Proc. R. Soc. Lond B Biol. Sci. 143, 257–279. doi:10.1098/rspb.1955.0009
Goodfellow, B. J., Dias, J. S., Ferreira, G. C., Henklein, P., Wray, V., and Macedo, A. L. (2001). The Solution Structure and Heme Binding of the Presequence of Murine 5-Aminolevulinate Synthase. FEBS Lett. 505, 325–331. doi:10.1016/s0014-5793(01)02818-6
Granick, S. (1966). The Induction In Vitro of the Synthesis of δ-Aminolevulinic Acid Synthetase in Chemical Porphyria: A Response to Certain Drugs, Sex Hormones, and Foreign Chemicals. J. Biol. Chem. 241, 1359–1375. doi:10.1016/s0021-9258(18)96783-9
Hayward, S. (2004). Identification of Specific Interactions That Drive Ligand-Induced Closure in Five Enzymes with Classic Domain Movements. J. Mol. Biol. 339, 1001–1021. doi:10.1016/j.jmb.2004.04.004
Hegedűs, T., Geisler, M., Lukács, G. L., and Farkas, B. (2022). Ins and Outs of AlphaFold2 Transmembrane Protein Structure Predictions. Cell Mol. Life Sci. 79, 73. doi:10.1007/s00018-021-04112-1
Herbert, D., Gibbs, S., Riddick, A., Conway, M., and Dong, M. (2020). Crystal Structure of an Oxidized Mutant of Human Mitochondrial Branched-Chain Aminotransferase. Acta Cryst. Sect. F. 76, 14–19. doi:10.1107/s2053230x19016480
Hou, S., Reynolds, M. F., Horrigan, F. T., Heinemann, S. H., and Hoshi, T. (2006). Reversible Binding of Heme to Proteins in Cellular Signal Transduction. Acc. Chem. Res. 39, 918–924. doi:10.1021/ar040020w
Hunter, G. A., and Ferreira, G. C. (2011). Molecular Enzymology of 5-Aminolevulinate Synthase, the Gatekeeper of Heme Biosynthesis. Biochimica Biophysica Acta (BBA) - Proteins Proteomics 1814, 1467–1473. doi:10.1016/j.bbapap.2010.12.015
Hunter, G. A., and Ferreira, G. C. (1999). Pre-Steady-State Reaction of 5-Aminolevulinate Synthase. J. Biol. Chem. 274, 12222–12228. doi:10.1074/jbc.274.18.12222
Hunter, G. A., Zhang, J., and Ferreira, G. C. (2007). Transient Kinetic Studies Support Refinements to the Chemical and Kinetic Mechanisms of Aminolevulinate Synthase. J. Biol. Chem. 282, 23025–23035. doi:10.1074/jbc.m609330200
Igarashi, J., Murase, M., Iizuka, A., Pichierri, F., Martinkova, M., and Shimizu, T. (2008). Elucidation of the Heme Binding Site of Heme-Regulated Eukaryotic Initiation Factor 2α Kinase and the Role of the Regulatory Motif in Heme Sensing by Spectroscopic and Catalytic Studies of Mutant Proteins. J. Biol. Chem. 283, 18782–18791. doi:10.1074/jbc.m801400200
Ikushiro, H., Nagami, A., Takai, T., Sawai, T., Shimeno, Y., Hori, H., et al. (2018). Heme-Dependent Inactivation of 5-Aminolevulinate Synthase from Caulobacter crescentus. Sci. Rep. 8, 14228. doi:10.1038/s41598-018-32591-z
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly Accurate Protein Structure Prediction with AlphaFold. Nature 596, 583–589. doi:10.1038/s41586-021-03819-2
Kafina, M. D., and Paw, B. H. (2017). Intracellular Iron and Heme Trafficking and Metabolism in Developing Erythroblasts. Metallomics 9, 1193–1203. doi:10.1039/c7mt00103g
Lathrop, J. T., and Timko, M. P. (1993). Regulation by Heme of Mitochondrial Protein Transport Through a Conserved Amino Acid Motif. Science 259, 522–525. doi:10.1126/science.8424176
Liang, J., Han, Q., Tan, Y., Ding, H., and Li, J. (2019). Current Advances on Structure-Function Relationships of Pyridoxal 5′-Phosphate-Dependent Enzymes. Front. Mol. Biosci. 6, 4. doi:10.3389/fmolb.2019.00004
Liu, J., Li, Y., Tong, J., Gao, J., Guo, Q., Zhang, L., et al. (2018). Long Non-Coding RNA-Dependent Mechanism to Regulate Heme Biosynthesis and Erythrocyte Development. Nat. Commun. 9, 4386. doi:10.1038/s41467-018-06883-x
McPhalen, C. A., Vincent, M. G., and Jansonius, J. N. (1992a). X-Ray Structure Refinement and Comparison of Three Forms of Mitochondrial Aspartate Aminotransferase. J. Mol. Biol. 225, 495–517. doi:10.1016/0022-2836(92)90935-d
McPhalen, C. A., Vincent, M. G., Picot, D., Jansonius, J. N., Lesk, A. M., and Chothia, C. (1992b). Domain Closure in Mitochondrial Aspartate Aminotransferase. J. Mol. Biol. 227, 197–213. doi:10.1016/0022-2836(92)90691-c
Medlock, A. E., Shiferaw, M. T., Marcero, J. R., Vashisht, A. A., Wohlschlegel, J. A., Phillips, J. D., et al. (2015). Identification of the Mitochondrial Heme Metabolism Complex. PLoS One 10, e0135896. doi:10.1371/journal.pone.0135896
Munakata, H., Sun, J. Y., Yoshida, K., Nakatani, T., Honda, E., Hayakawa, S., et al. (2004). Role of the Heme Regulatory Motif in the Heme-Mediated Inhibition of Mitochondrial Import of 5-Aminolevulinate Synthase. J. Biochem. 136, 233–238. doi:10.1093/jb/mvh112
Na, I., Catena, D., Kong, M. J., Ferreira, G. C., and Uversky, V. N. (2018). Anti-Correlation Between the Dynamics of the Active Site Loop and C-Terminal Tail in Relation to the Homodimer Asymmetry of the Mouse Erythroid 5-Aminolevulinate Synthase. Int. J. Mol. Sci. 19. doi:10.3390/ijms19071899
Nishitani, Y., Okutani, H., Takeda, Y., Uchida, T., Iwai, K., and Ishimori, K. (2019). Specific Heme Binding to Heme Regulatory Motifs in Iron Regulatory Proteins and its Functional Significance. J. Inorg. Biochem. 198, 110726. doi:10.1016/j.jinorgbio.2019.110726
Niu, W., Wang, J., Qian, J., Wang, M., Wu, P., Chen, F., et al. (2018). Allosteric Control of Human Cystathionine β-Synthase Activity by a Redox Active Disulfide Bond. J. Biol. Chem. 293, 2523–2533. doi:10.1074/jbc.ra117.000103
Nokes, B., Apel, M., Jones, C., Brown, G., and Lang, J. E. (2013). Aminolevulinic Acid (ALA): Photodynamic Detection and Potential Therapeutic Applications. J. Surg. Res. 181, 262–271. doi:10.1016/j.jss.2013.02.002
Nomura, K., Kitagawa, Y., Aihara, M., Ohki, Y., Furuyama, K., and Hirokawa, T. (2021). Heme‐Dependent Recognition of 5‐Aminolevulinate Synthase by the Human Mitochondrial Molecular Chaperone ClpX. FEBS Lett. 595, 3019–3029. doi:10.1002/1873-3468.14214
Peoc'h, K., Nicolas, G., Schmitt, C., Mirmiran, A., Daher, R., Lefebvre, T., et al. (2019). Regulation and Tissue-Specific Expression of δ-Aminolevulinic Acid Synthases in Non-Syndromic Sideroblastic Anemias and Porphyrias. Mol. Genet. Metabolism 128, 190–197. doi:10.1016/j.ymgme.2019.01.015
Percudani, R., and Peracchi, A. (2009). The B6 Database: A Tool for the Description and Classification of Vitamin B6-Dependent Enzymatic Activities and of the Corresponding Protein Families. BMC Bioinforma. 10, 273. doi:10.1186/1471-2105-10-273
Poli, A., Schmitt, C., Moulouel, B., Mirmiran, A., Puy, H., Lefebvre, T., et al. (2021). Iron, Heme Synthesis and Erythropoietic Porphyrias: A Complex Interplay. Metabolites 11 (12), 798. doi:10.3390/metabo11120798
Porta-Pardo, E., Ruiz-Serra, V., Valentini, S., and Valencia, A. (2022). The Structural Coverage of the Human Proteome Before and After AlphaFold. PLoS Comput. Biol. 18, e1009818. doi:10.1371/journal.pcbi.1009818
Riddle, R. D., Yamamoto, M., and Engel, J. D. (1989). Expression of Delta-Aminolevulinate Synthase in Avian Cells: Separate Genes Encode Erythroid-Specific and Nonspecific Isozymes. Proc. Natl. Acad. Sci. U.S.A. 86, 792–796. doi:10.1073/pnas.86.3.792
Rondelli, C. M., Perfetto, M., Danoff, A., Bergonia, H., Gillis, S., O'Neill, L., et al. (2021). The Ubiquitous Mitochondrial Protein Unfoldase CLPX Regulates Erythroid Heme Synthesis by Control of Iron Utilization and Heme Synthesis Enzyme Activation and Turnover. J. Biol. Chem. 297, 100972. doi:10.1016/j.jbc.2021.100972
Schmalohr, B. F., Mustafa, A. H. M., Krämer, O. H., and Imhof, D. (2021). Structural Insights into the Interaction of Heme with Protein Tyrosine Kinase JAK2**. Chembiochem 22, 861–864. doi:10.1002/cbic.202000730
Shen, J., Sheng, X., Chang, Z., Wu, Q., Wang, S., Xuan, Z., et al. (2014). Iron Metabolism Regulates P53 Signaling Through Direct Heme-P53 Interaction and Modulation of P53 Localization, Stability, and Function. Cell Rep. 7, 180–193. doi:10.1016/j.celrep.2014.02.042
Stenson, P. D., Ball, E. V., Mort, M., Phillips, A. D., Shiel, J. A., Thomas, N. S. T., et al. (2003). Human Gene Mutation Database (HGMD): 2003 Update. Hum. Mutat. 21, 577–581. doi:10.1002/humu.10212
Stojanovski, B. M., Breydo, L., Uversky, V. N., and Ferreira, G. C. (2016). Murine Erythroid 5-Aminolevulinate Synthase: Truncation of a Disordered N-Terminal Extension Is Not Detrimental for Catalysis. Biochimica Biophysica Acta (BBA) - Proteins Proteomics 1864, 441–452. doi:10.1016/j.bbapap.2016.02.002
Stojanovski, B. M., Hunter, G. A., Na, I., Uversky, V. N., Jiang, R. H. Y., and Ferreira, G. C. (2019). 5-Aminolevulinate Synthase Catalysis: The Catcher in Heme Biosynthesis. Mol. Genet. Metabolism 128, 178–189. doi:10.1016/j.ymgme.2019.06.003
Tan, D., Harrison, T., Hunter, G. A., and Ferreira, G. C. (1998). Role of Arginine 439 in Substrate Binding of 5-aminolevulinate Synthase. Biochemistry 37, 1478–1484. doi:10.1021/bi971928f
Tanimura, N., Miller, E., Igarashi, K., Yang, D., Burstyn, J. N., Dewey, C. N., et al. (2016). Mechanism Governing Heme Synthesis Reveals a GATA Factor/heme Circuit that Controls Differentiation. EMBO Rep. 17, 249–265. doi:10.15252/embr.201541465
Taylor, J. L., and Brown, B. L. (2022). Structural Basis for Dysregulation of Aminolevulinic Acid Synthase in Human Disease. J. Biol. Chem. 298 (3), 101643. doi:10.1016/j.jbc.2022.101643
Tian, Q., Li, T., Hou, W., Zheng, J., Schrum, L. W., and Bonkovsky, H. L. (2011). Lon Peptidase 1 (LONP1)-Dependent Breakdown of Mitochondrial 5-Aminolevulinic Acid Synthase Protein by Heme in Human Liver Cells. J. Biol. Chem. 286, 26424–26430. doi:10.1074/jbc.m110.215772
Toney, M. D. (2014). Aspartate Aminotransferase: an Old Dog Teaches New Tricks. Archives Biochem. Biophysics 544, 119–127. doi:10.1016/j.abb.2013.10.002
Tunyasuvunakool, K., Adler, J., Wu, Z., Green, T., Zielinski, M., Žídek, A., et al. (2021). Highly Accurate Protein Structure Prediction for the Human Proteome. Nature 596, 590–596. doi:10.1038/s41586-021-03828-1
Varadi, M., Anyango, S., Deshpande, M., Nair, S., Natassia, C., Yordanova, G., et al. (2022). AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 50, D439–D444. doi:10.1093/nar/gkab1061
Wang, T., Wang, Y., Dong, Q., Xu, C., Zhou, X., Ouyang, Y., et al. (2020). X-Linked Dominant Protoporphyria in a Chinese Pedigree Reveals a Four-Based Deletion of ALAS2. Ann. Transl. Med. 8, 344. doi:10.21037/atm.2020.02.80
Wehrspan, Z. J., McDonnell, R. T., and Elcock, A. H. (2022). Identification of Iron-Sulfur (Fe-S) Cluster and Zinc (Zn) Binding Sites within Proteomes Predicted by DeepMind's AlphaFold2 Program Dramatically Expands the Metalloproteome. J. Mol. Biol. 434, 167377. doi:10.1016/j.jmb.2021.167377
Whatley, S. D., Ducamp, S., Gouya, L., Grandchamp, B., Beaumont, C., Badminton, M. N., et al. (2008). C-Terminal Deletions in the ALAS2 Gene Lead to Gain of Function and Cause X-Linked Dominant Protoporphyria Without Anemia or Iron Overload. Am. J. Hum. Genet. 83, 408–414. doi:10.1016/j.ajhg.2008.08.003
Wouters, M. A., Fan, S. W., and Haworth, N. L. (2010). Disulfides as Redox Switches: From Molecular Mechanisms to Functional Significance. Antioxidants Redox Signal. 12, 53–91. doi:10.1089/ars.2009.2510
Yamamoto, M., Hayashi, N., and Kikuchi, G. (1982). Evidence for the Transcriptional Inhibition by Heme of the Synthesis of δ-Aminolevulinate Synthase in Rat Liver. Biochem. Biophysical Res. Commun. 105, 985–990. doi:10.1016/0006-291x(82)91067-1
Yang, J., Kim, K. D., Lucas, A., Drahos, K. E., Santos, C. S., Mury, S. P., et al. (2008). A Novel Heme-Regulatory Motif Mediates Heme-Dependent Degradation of the Circadian Factor Period 2. Mol. Cell Biol. 28, 4697–4711. doi:10.1128/mcb.00236-08
Yoshino, K., Munakata, H., Kuge, O., Ito, A., and Ogishima, T. (2007). Haeme-Regulated Degradation of -Aminolevulinate Synthase 1 in Rat Liver Mitochondria. J. Biochem. 142, 453–458. doi:10.1093/jb/mvm159
Keywords: 5-aminolevulinate synthase, pyridoxal 5′-phosphate, heme regulatory motif, allostery, redox sensor, porphyrin, regulation, AlphaFold
Citation: Hunter GA and Ferreira GC (2022) An Extended C-Terminus, the Possible Culprit for Differential Regulation of 5-Aminolevulinate Synthase Isoforms. Front. Mol. Biosci. 9:920668. doi: 10.3389/fmolb.2022.920668
Received: 14 April 2022; Accepted: 30 May 2022;
Published: 14 July 2022.
Edited by:
Robert Stephen Phillips, University of Georgia, United StatesReviewed by:
Andrea Mozzarelli, University of Parma, ItalyTim Mueser, University of Toledo, United States
Giada Rossignoli, University of Padua, Italy
Copyright © 2022 Hunter and Ferreira. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gregory A. Hunter, Z2h1bnRlckB1c2YuZWR1; Gloria C. Ferreira, Z2ZlcnJlaXJAdXNmLmVkdQ==
 Gregory A. Hunter
Gregory A. Hunter Gloria C. Ferreira
Gloria C. Ferreira