
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci. , 23 May 2022
Sec. Molecular Recognition
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.888721
 Yue Pan1
Yue Pan1 Jinxiao Bao1
Jinxiao Bao1 Xingyi Zhang2
Xingyi Zhang2 Hui Ni2
Hui Ni2 Yue Zhao1
Yue Zhao1 Fengdong Zhi1
Fengdong Zhi1 Bohuan Fang1
Bohuan Fang1 Xiao He1,3*
Xiao He1,3* John Z. H. Zhang1,3,4*
John Z. H. Zhang1,3,4* Lujia Zhang1,3*
Lujia Zhang1,3*Aromatic aldehydes are important industrial raw materials mainly synthesized by anti-Markovnikov (AM) oxidation of corresponding aromatic olefins. The AM product selectivity remains a big challenge. P450 aMOx is the first reported enzyme that could catalyze AM oxidation of aromatic olefins. Here, we reported a rational design strategy based on the “butterfly” model of the active site of P450 aMOx. Constrained molecular dynamic simulations and a binding energy analysis of key residuals combined with an experimental alanine scan were applied. As a result, the mutant A275G showed high AM selectivity of >99%. The results also proved that the “butterfly” model is an effective design strategy for enzymes.
Aromatic aldehydes are important intermediate compounds in the industry and have a wide range of applications in chemical, pharmaceutical, and daily chemical fields (Dubrovskiy et al., 2018). The direct synthesis of aromatic aldehydes from aromatic olefins via the anti-Markovnikov pathway greatly simplifies the synthesis procedure in industrial production (Dong et al., 2015; Wu et al., 2019). However, due to the complex mechanism, the AM selectivity is still a challenge. Currently, metal organics are mainly used as catalysts for the oxidation of olefins to synthesize such substances (Beller et al., 2004). Although some achievements have been made in related research studies (Chen et al., 2011; Nguyen et al., 2019), the complexity of the structural modification of metal-derivative catalysts still remains to be addressed.
Compared with metal-organic catalysts, biocatalysts have the advantages of abundant sources, high reactivity, and environmental friendliness (Musa and Phillips, 2011; Sheldon and Woodley, 2018). Among biocatalysts, the widely distributed NAD(P)H-dependent P450 mono-oxygenases can be used to catalyze hydroxylation, epoxidation, and nitration (Barry et al., 2012; Guengerich and Munro, 2013; Dodani et al., 2016; Girvan and Munro, 2016). P450s have become a hotspot for biocatalyst research, owing to the diversity of their catalytic reactions (Sono et al., 1996). In 2017, Arnold et al. reported that P450 aMOx, a Class IV self-sufficient cytochrome P450 (Munro et al., 2007), from directed evolution could catalyze the oxidation of aromatic olefin styrene to phenylethylaldehyde with a high selectivity at 81% (Hammer et al., 2017). Many achievements have been realized via direct evolution (Reetz et al., 2004; Kan et al., 2016; Buller et al., 2018), and the mutations from directed evolution consist of a mutagenesis library for further study (Acevedo-Rocha et al., 2018). However, directed evolution is laborious, time-consuming, costly (Yang et al., 2019), and lack a regular enzyme design modification method. Hence, this study used a combination of rational designs and experimental methods to redesign aMOx in order to further improve the AM selectivity for the catalysis of aromatic compounds.
The complete aMOx gene sequence was obtained from a previous report (Hammer et al., 2017). Synthesis of gene sequence, mutant primers, and sequencing were assigned to Shanghai Sunny Biotechnology Co. (Shanghai, China). The pET 22b (Invitrogen, Carlsbad, CA, United States) was used as the vector, and the target gene was located between Nde I and Hind III enzymatic sites with a C-terminal 6*His Tag. The mutants were constructed by using a seamless cloning kit from Beyotime Biotechnology (Shanghai, China). Escherichia coli DH5α (Weidi Biotechnology Co., Ltd., Shanghai, China) was used as the vector cloning host, and E. coli BL21 (DE3) (Weidi Biotechnology Co., Ltd., Shanghai, China) was used as the expression host. All chemicals applied were of AR grade.
Bacteria harboring both aMOx and its mutants were cultivated and expressed in an LB medium containing 100 µg/ml ampicillin at 37°C and allowed for shaking at 250 rpm for 3 h, for protein expression. When the OD600 value of the bacterial solution was between 0.6 and 0.8, the final concentrations of 0.1 mM IPTG and 0.5 mM 5-aminolevulinic acid (5-ALA) were added. The system was induced at 20°C, 250 rpm for 12 h. The system was centrifuged at 8,000 rpm for 5 min to collect the bacteria. The pellet was treated with a crushing solution at the ratio of 10 ml of crushing solution (0.1 M PBS, 150 mM NaCl, and 10% glycerol; pH 8.0) to 1 g of wet bacterium and re-suspended on ice with ultrasonic crushing for 30 min. The crushing solution was centrifuged at 20,000 g for 15 min to take the supernatant. The target protein was purified using 5 ml Ni-NTA and AKTA PURE system (Cytiva, Boston, MA, United States). The target proteins were eluted with 0.1 M PBS; 150 mM NaCl; 10% glycerol; and 20, 50, 200, and 500 mM imidazole; each gradient elution volume had five columns as the volume. Target proteins of aMOx and mutants were eluted by 200 mM imidazole. The buffer was replaced by using 30kD ultrafiltration tubes with 0.1 M PBS, 150 mM NaCl, and 10% glycerol, at pH 8.0, frozen in liquid nitrogen and stored in the refrigerator at −80°C.
Because the substrate selectivity of P450 is exclusively related to its oxidant domain, this study specifically targeted the oxidative structural domain (450 aa) of aMOx for structural analysis. The Protein BLAST module of the NCBI database was used for homologous sequence alignment of aMOx amino acid sequences, and CYP116B5 (Ciaramella et al., 2019) (PDB ID: 6RO8) with 82% sequence identity was used as the template. The homology modeling module of Discovery Studio was used for homology modeling of the oxidant domain of aMOx.
The Glide module of Maestro was used for molecular docking of the substrate styrene to aMOx to obtain possible structures of the substrate binding complex. The docking was centered on the heme oxygen, the grid size was set to 20 Å, and the distance between the olefin reaction site of the constrained styrene and the heme oxygen was 4 Å for constrained docking. The structure with the highest score was selected for subsequent studies.
The specific activity of aMOx and mutants was determined by evaluating the consumption of NADPH (Liu et al., 2021). The Bradford (1976) method was used to determine the protein concentration using a Bradford protein quantification kit (YEASEN Biotechnology Co., Ltd., Shanghai, China). The reaction conditions were as follows: 0.1 μM purified enzyme, 0.1 mM styrene, and 0.2 mM NADPH in 25 mM Tris-HCl, at pH 8.0. The reaction volume was 200 μl. The reaction system was shaken for 30 min at 25°C. Then, a Multiskan Sky microplate spectrophotometer (Thermo Scientific, Waltham, MA, United States) was used to detect the change of absorbance value at 340 nm within 30 min.
Purpald (Hopps, 2000) (4-amino-5-hydrazino-1, 2, 4-triazole-3-thiol) is a class of chromogenic agents that reacts specifically with aldehydes to form purple complexes, which are complexation products with aldehydes that have a characteristic absorption at 528 nm (Scheme 1). In this study, Purpald was used as a derivatization reagent for the quantification of aMOx and the AM selectivity of the mutant based on this property. Purpald showed good linearity for 0–5 mM of phenylethylaldehyde. The reaction conditions were as follows: 100 μl lysis enzyme (approximately 1 μM aMOx), 5 mM substrate, 10 mM NADPH, and 25 mM Tris-HCl at pH 8.0 to make up the reaction volume of 200 μl, 20°C, 900 rpm for 2 h. The reaction was terminated by adding 50 μl 31 mM Purpald and incubated for 30 min at 25°C, and the absorbance value at 538 nm was measured.
Molecular dynamics simulations (MDs) were performed with Amber18 and AmberTools19 software applications. The ff14SB force field was used for protein, the gaff force field and the AM1-BCC charge model were used for organic ligands, the parameters provided by Shaik et al. (2010) and Shahrokh et al.( 2012) were used for the Cpd I group, and the TIP3P waters and the Li/Merz monovalent ion parameters were also used. The water box was set to be truncated ortho-octahedrally, and the minimum distance between complex atoms and box boundaries was set to be 12.0 Å; Na+ and Cl− ions were added according Shahrokh et al. (2012) to neutralize the system, and the ionic strength was made near 0.15 mol/L.
First, the initial system was minimized with the 5,000-step steepest descent method (SD), followed by the 5,000-step conjugate gradient method (CG) with harmonic Cartesian space restraints on all complex atoms; the restraint weight was set to be 10.0 kcal mol−1 Å−2. Next, the Cartesian space restraints were removed, and the whole system was further minimized with 10,000-step SD and 10,000-step CG. Subsequently, the system was progressively heated from 0 K to the target temperature (300.0 K) within 300 ps under NVT ensemble and then equilibrated to the target pressure (1.0 bar) within 1.0 ns under NPT ensemble. The Langevin thermostat with the collision frequency set as 2.0 ps−1 and the Monte Carlo barostat with the relaxation time set to 2.0 ps were used. Harmonic Cartesian space restraints were added on all complex atoms during heating and equilibration with the weight set to 5.0 and 2.0 kcal mol−1 Å−2, respectively. During the MDs, the SHAKE method was used to constraint bonds involving hydrogen atoms, and the time step was set to 2.0 fs; the PME method was used to calculate the long-range electrostatic energy, and the cutoff on the long-range van der Waals (VDW) energy was set to 10.0 Å.
To retain the substrate’s vinyl group near the oxygen of Cpd I, during the minimization, heating, and equilibration periods, the NMR type of distance restraints were added between the vinyl carbons and the Cpd I oxygen. When the actual distance was less than the target distance, the restraint weight was zero, whereas when the actual distance was larger than the target distance, the restraint weight was set to 3.0 kcal mol−1 Å−2.
To analyze the receptor residue binding energy contribution to the substrate when the substrate is close to the reaction center, the NMR distance restraint was added between position 1 of the substrate and the Cpd I oxygen with the target distance set to 3.0 Å. After the equilibration, the Cartesian space restraints were removed, and the system was produced with different random seeds five times with the NMR distance restraint kept. The simulation duration of each production was set to 100 ns, and snapshots were recorded every 100 ps to form the trajectory for further analysis. The molecular mechanics generalized born surface area (MMGBSA) calculations were performed on the last 800 frames (80 ns) of each trajectory, and the binding energy was decomposed to each receptor residue (except for the Cpd I group). During the calculation, the GBOBC model I (igb = 2) was used as the implicit solvent model, the interior dielectric constant was set to 3.0, the ionic strength was set to 0.1 mol/L, the LCPO method was used to calculate the solvent accessible surface area, and the surface tension was set to 0.0005. The corresponding values of all the five trajectories were averaged to get the final results.
To analyze the contact between the substrate vinyl carbons and the Cpd I oxygen of different protein mutants, the NMR distance restraints were added both on the position 1 carbon and position 2 carbon of the substrate to the Cpd I oxygen during minimization, heating, and equilibration with the target distance set to 3.5 Å. Then, the system was further equilibrated during 5 ns with only the NMR distance restraint kept and then equilibrated for 5 ns with no restraints. After these equilibrations, the accelerated molecular dynamics (aMD) were performed on these systems. The system’s averaged total potential energy and dihedral energy were calculated among the last 4 ns of the equilibrations. The aMD parameter: alphaD was set to be 0.7 times the number of the complex residues, and alphaP was set to be 0.2 times the number of system total atoms. Both the system’s total potential energy and the dihedral energy were boosted during the aMD calculation, and the boost energy on the dihedral energy term was set to be twice of alphaD. The time step was set to 1.5 fs during aMD, and the relative geometrical tolerance of SHAKE and the dsum_tol parameter of PME were both set to 0.000001. Snapshots were recorded every 7.5 ps to form the trajectory. Each system was calculated with different random seeds ten times, and the aMD duration of each production was set to 30 ns.
P450 aMOx is a self-sufficient P450 mono-oxygenase produced by 10 rounds of directed evolution of P450LaMO (Yin et al., 2014). The predicted structure of P450 aMOx was determined by homology modeling (Figure 2A). Since the Cpd I is a highly active intermediate of the P450 aMOx catalyzing cycle (Rittle and Green, 2010; Krest et al., 2013), its homology-modeled structure was both Cpd I and the substrate styrene, through molecular docking (Figure 2B). A previous report (Hammer et al., 2017) has described the possible mechanism of the aMOx-catalyzed AM oxidation reaction as a 1, 2-σ migration rearrangement reaction (Bonneau et al., 1989; Kim et al., 2003). A possible reaction mechanism is as follows: the substrate approaches the oxygen atom on heme, the active center of the aMOx reaction; next, the oxygen atom attacks position 1 of the aromatic olefin substrate, and the olefin’s double bond opens to form a carbon positive ion at position 2; this process induces the hydrogen ion at position 1 and migrates to position 2; simultaneously, the double bond formed by heme iron and oxygen breaks; the reaction is completed after the oxygen atom forms a double bond with carbon at position1 (Scheme 2).
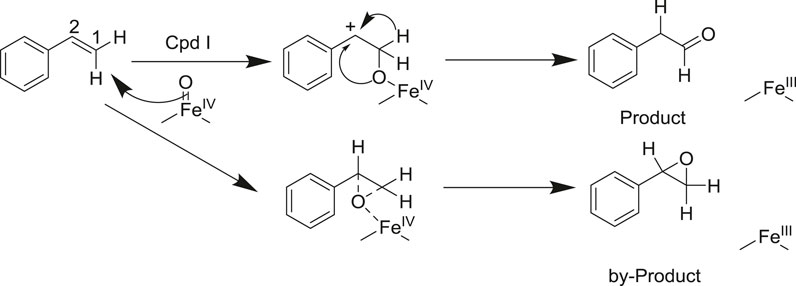
SCHEME 2. Reaction mechanism of the aMOx-catalyzed olefin Markovnikov oxidation reaction (supposed).
The reaction distance between the reaction site and electrophilic reagent in AM oxidation was 3.0–4.0 Å in our pre-MD simulations. The near attack configuration (NAC) angle of the nucleophilic reagent, which is C7/C8-O-Fe, affects the selectivity of the AM oxidation reaction (Feng et al., 2019) (Figure 2B). We first analyzed the results of directed evolution reported by Arnold et al. via MD simulations in order to investigate the effects of these factors on AM selectivity and to establish a catalytic model and an efficient strategy for aMOx-catalyzed AM reaction. Among the 10 rounds of the directed evolution results, round 2 yielded a mutant with the highest increase (from 45% to 55%) in AM selectivity. Round 8 had 76% AM selectivity, which differed from the reported final selectivity of 81% by only 5%. Therefore, we selected round 1 (P450LaMO), round 2, round 8, and aMOx for a molecular dynamics simulation analysis. Mutations of residues could alter the steric hindrance of the active pocket and affect the way the reaction substrate contacts the active site, which is considered as one of the common causes of altered reaction selectivity due to residue mutations (Chen et al., 2018; Wang et al., 2020). The connection between the carbon atom on the vinyl of the substrate and the oxygen atom in Cpd I was investigated by aMD, an enhanced sampling tool (Hamelberg et al., 2004).
Distances between the carbons at positions 1 (C8-O) and 2 (C7-O) and the oxygen on Cpd I, in the respective 10–30 ns aMD simulation trajectories of the aforementioned systems, as well as time-related variation in the angle made by these two carbons with oxygen and iron atoms in Cpd I, respectively, are shown in Supplementary Figures S1, S2.
It is supposed that spatial positional selectivity also lead to a significant influence in enzyme-catalyzed reactions. The frequency of reactive site 1 and non-reactive site 2 appearing within a 3–5 Å radius of the oxygen atom of Cpd I was further analyzed. We calculated the number of frames in which at least one of C7 and C8 of round 1, round 2, round 8, and aMOx has a distance to the oxygen on Cpd I smaller than a specific threshold as a percentage of the total number of frames of the corresponding trajectory [denoted as p (C7 or C8), Figure 1, orange bar], while the horizontal coordinates of the graph indicate the corresponding distance threshold. We also analyzed the percentage of the total number of frames in the substrate with the distance from the carbon at site 1 to the oxygen on Cpd I only less than this threshold [notated as p (C8 only)] at the corresponding threshold (Figure 1, blue bar). The corresponding ratio of p (C8 only) top (C7 or C8) is recorded in parentheses in Figure 1. We focused on the ratio of p (C8 only) to p (C7 or C8) at the reaction distance of 3.6–4.0 Å. The p (C8 only)/p (C7 or C8) values for round 1, round 2, round 8, and wild type are 0.34, 0.34, 0.35, and 0.42, respectively, when the distance threshold is 3.6 Å; when the distance threshold is 3.8 Å, the p (C8 only)/p (C7 or C8) values of round 1, round 2, round 8, and wild type are 0.29, 0.30, 0.33, and 0.36, respectively; when the distance threshold is 3.8 Å, the p (C8 only)/p (C7 or C8) values were 0.25, 0.26, 0.30, and 0.30, respectively. p (C8 only)/p (C7 or C8) trends for the three different thresholds were consistent with the trends of the inverse martensite rule product selectivity.
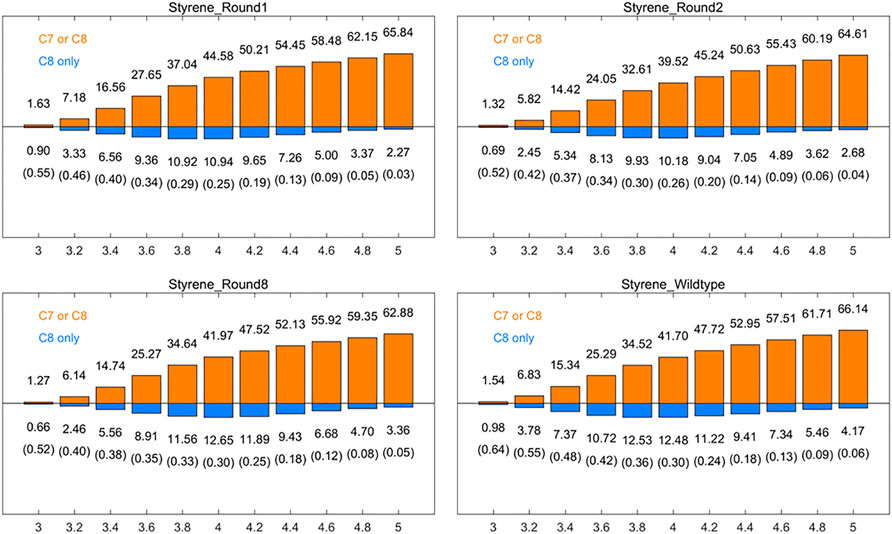
FIGURE 1. Percentages [p (C7 or C8), orange] of the frames that have at least one carbon (of styrene’s position 1 and position 2 carbons) within the given distance (x-axis, in Å) to the Cpd I oxygen among all the aMD frames, and the percentages [p (C8only), blue] of the frames where only position 1 carbon of styrene vinyl is within the given distance. The ratios of p (C8 only)/p (C7 or C8) are also listed in the brackets. All results are the averages of the corresponding ten 30 ns aMD trajectories.
Inspired by the results of the distance analysis, we analyzed the NAC angle (C7/C8-O-Fe) via the same method. We first considered the angle as the standard and then calculated the number of frames in which at least one of the NAC angles is more than the given angle [denoted as p (C7 or C8)] and the number of frames in which only C8-O-Fe is more than the given angle [denoted as p (C8only)] of all track frames that meet the angle standard under different distance indexes. The consistency between p (C8 only)/p (C7 or C8) of different systems and their product selectivity is the same when the distance is 3.6, 3.8, 4.0, 4.2, and 4.4 Å, and 90°, 105°, 120°, and 135° are used as angle standards (Supplementary Figures S3–S8). The Spearman rank correlation was better with the angle standard of 120°, while the rank correlation decreased with the angle standard of 135° (Supplementary Table S1).Then, we considered the distance as the standard and then calculated the number of frames in which at least one of C7 or C8 is within the given distance [denoted as p (C7 or C8)], and only C8 is within the given distance [denoted as p (C8 only)] of all the track frames that meet the distance standard under different angle indicators (Supplementary Figures S9–S12). When the angle standard is 90°, 105°, 120°, and 135°, there is a good agreement between p (C8 only)/p (C7 or C8) and its product selectivity in the range of 3.6–4.0 (Supplementary Table S1), which is consistent with the optimal reaction distance obtained by the distance analysis. The results of the NAC angle analysis indicate that the tolerance of this kind of reaction to the angle is relatively broad, so the influence of distance on the occurrence of reaction is more important. Therefore, considering both distance and angle is not a significant improvement compared with only considering distance in this research, so the NAC angle is not a key factor that affects reaction selectivity.
These data mentioned previously indicate that the following two key factors determine the AM selectivity of aMOx: the p (C8 only)/p (C7 or C8) and the distance from the reaction site C8 to the oxygen atom of the reaction active center Cpd I (C8-O). The ratio of p (C8 only)/p (C7 or C8) also showed a consistent upward trend when the AM selectivity increased. The distance from the reaction site C8 to the oxygen atom of the reaction active center Cpd I was maintained within 3–5 Å when the reaction occurred. Consequently, we proposed a butterfly catalysis model built on the aMOx reaction active site because the structure of the substrate and Cpd I was like a butterfly (Figure 2B). We hope that our model will provide a visual understanding of our rational design mechanism for experimental scientists.
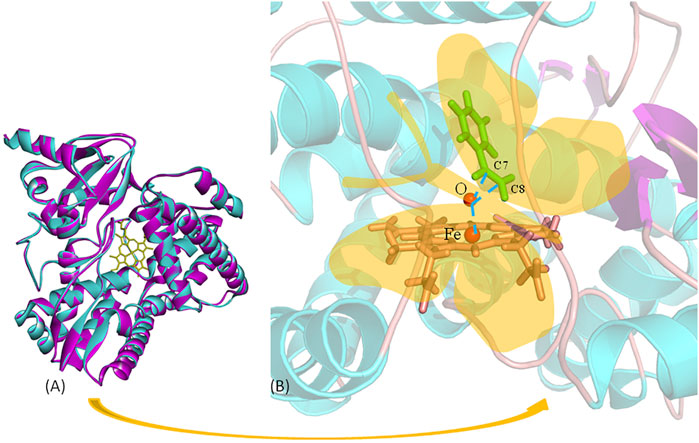
FIGURE 2. Butterfly model-based structure and computational analysis. (A) Overlay structure for aMOx (cyan) and the model (purple) CYP116B5 (PDB No.: 6RO8). (B) “Butterfly” catalytic model for P450 aMOx.
The whole procedure of the aMOx rational design is shown in Figure 3. According to the results of the preliminary computational analysis, C8-O and p (C8 only)/p (C7 or C8) are important factors. Then, we investigated the binding energy contribution of each residue in aMOx to the substrate molecule when the substrate was close to the reaction center by introducing a distance constraint so that C8 in the substrate is at a distance of 3 Å from the oxygen atom in Cpd I during the simulation, and the results are shown in Table 1 by the MMGBSA binding energy decomposition method.
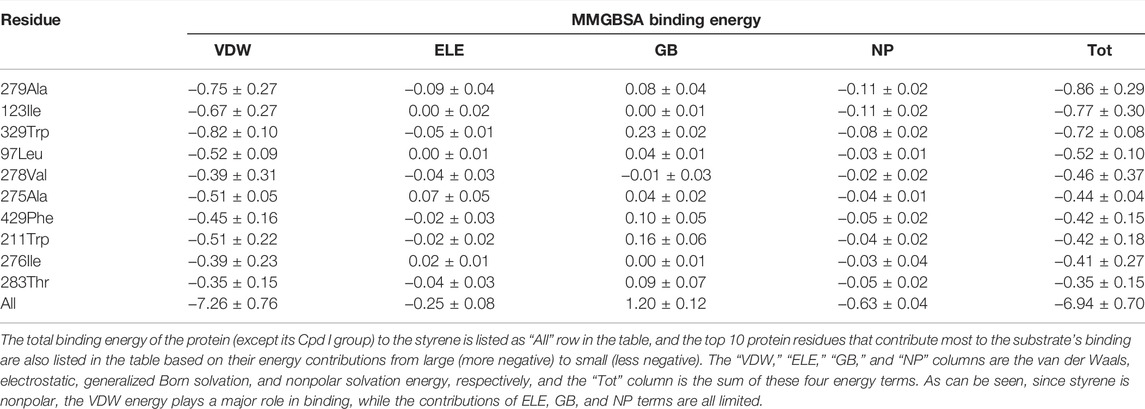
TABLE 1. MMGBSA binding energy decomposition of the CYP450 wild type binding with styrene (all units are in kcal/mol).
The key residuals that affect the AM selectivity of the aMOx are shown in Figure 4B. The resolution of the substrate binding channel of the template CYP116B5 used in the homology modeling part has been reported by previous studies. We performed a structural comparison between the binding pocket of aMOx and CYP116B5. The results indicated that the substrate-binding channel of aMOx is largely identical to that of CYP116B5, both being a four-layered substrate binding channel composed of non-polar hydrophobic amino acids (Figure 4A). In addition, Ile123, His206, Trp211, Ala275, Val278, Trp329, and Phe429 were all on the substrate-binding channel among the key sites affecting the substrate binding conformation of styrene analyzed by MMGBSA mentioned previously. Overall, it was confirmed that AM selectivity of P450 aMOx could be altered by altering its substrate binding channel.
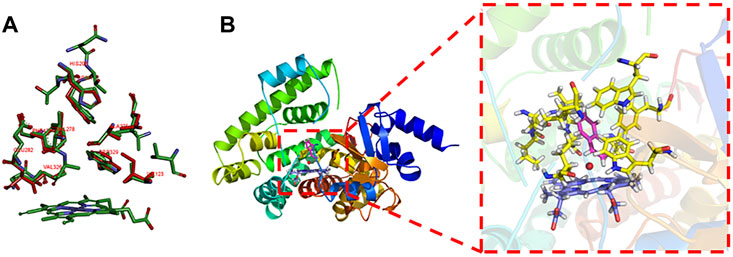
FIGURE 4. (A) Analysis of the aMOx substrate’s binding pocket. Residuals consisting of the aMOx substrate-binding channel are shown in red and those of CYP116B5 are shown in green. (B) Key residuals that affect the AM selectivity of aMOx. Key residuals are marked in yellow.
To verify the AM selectivity of aMOx and its variants, we performed an experimental alanine scan for the aforementioned substrate-binding key residues. Seamless cloning was achieved for aMOx A279G, I123A, W329A, L97A, V278A, A275G, F429A, W211A, I276A, and T283A. The primers used are listed in Table 2.
As all of the aforementioned mutations were soluble expressions, we examined the specific activity of these mutants before the assay for mutant anti-Markovnikov selectivity (Figure 5). All 10 mutations had a specific activity. Compared to the wild type, V278A, L97A, A275G, and W329A all had elevated specific activities of 6%, 33%, 19%, and 29%, respectively. We performed quantitative assays of the AM selectivity products for all mutants. The AM selectivity of the aforementioned mutants was experimentally verified with styrene as the substrate and Purpald derivatization reaction, and the experimental results showed that the AM selectivity of A275G appeared to be significantly enhanced to >99% (Table 3).
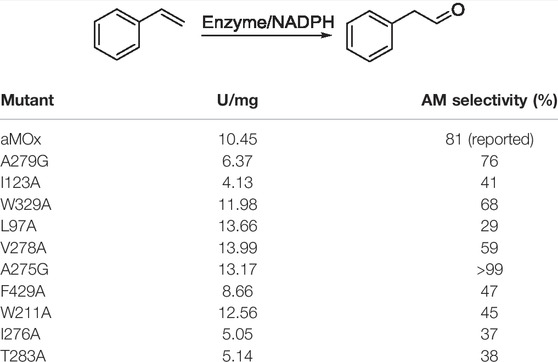
TABLE 3. Specific activity and AM selectivity of aMOx and its variants. All samples were performed at least thrice.
A275G is located on the substrate-binding channel of aMOx (Figure 4). To analyze the increased AM selectivity of A275G, we performed molecular dynamic simulations for A275G (Figure 6). p (C8 only)/p (C7 or C8) values for A275G were 0.53, 0.47, and 0.42, whereas the distance thresholds were 3.6, 3.8, and 4.0 Å. When analyzed in conjunction with round 1, round 2, round 8, and aMOx in the previous design, these ratios have a strong correlation with the AM selectivity of the corresponding systems. As Table 4 shows, the Spearman ranking correlation coefficient between the two systems was 1 with a 3.8 Å distance threshold. The Spearman ranking correlation coefficient was 0.97. This revealed that the AM selectivity of round 1, round 2, round 8, aMOx, and A275G changed in the same trend, which also proved that p (C8 only)/p (C7 or C8) and the distance between C8 and heme oxygen are two key factors in the AM selective design of an aMOx product. Hence, this strategy provides guidance for the rational design of this class of enzymes.
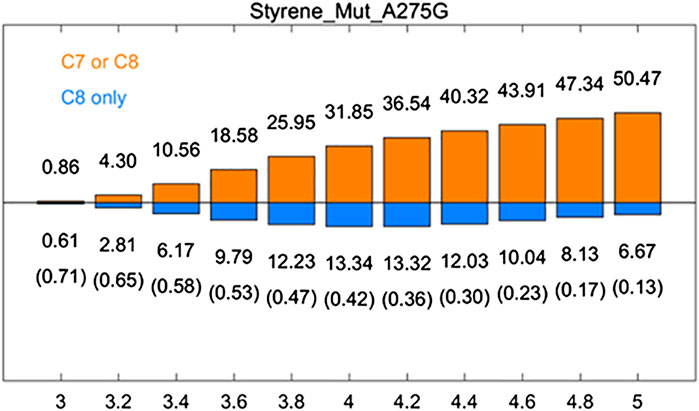
FIGURE 6. Rate of p (C8 only)/p (C7 or C8) of aMOx A275G. p (C8 only) are shown in the blue bar, and p (C7 or C8) are shown in the orange bar. The ratios of p (C8 only)/p (C7 or C8) are also listed in the brackets. All results are the averages of the corresponding ten 30 ns aMD trajectories.
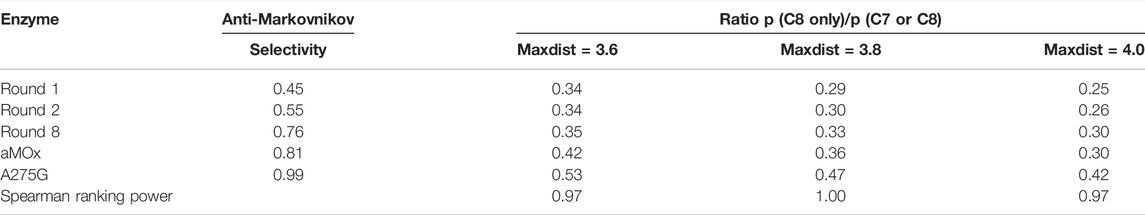
TABLE 4. Inverse martensite selectivity of the system and the ratio of p (C8 only)/p (C7 or C8) in the simulated trajectories of aMD at different distance thresholds.
The AM oxidation of aromatic olefins into corresponding aromatic aldehydes is an important synthetic route in the industry. Using computational methods combined with experiments, we revealed two key factors that influence the substrate’s AM selectivity of P450 aMOx as follows: 1) distance and 2) frequency of substrate reaction sites within a reasonable distance from the reaction. Based on these key factors, we have proposed a “butterfly” catalytic model. Finally, we successfully designed the mutant aMOx A275G with an AM selectivity of more than 99%. The results of this study are expected to contribute to the rational design of AM selectivity for enzyme-catalyzed AM oxidation reactions.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
YP: performed the experiments, data processing and analysis, and wrote the manuscript. JB: performed molecular dynamics simulations, data processing and analysis, and wrote the computation part of the manuscript. XZ, YZ, and FZ: performed part of the experiments. BF and XH: reviewed the manuscript. LZ and JZ: gave the support of funding and supervised the project, conceived and designed the study, and edited the manuscript critically. All authors have given approval to the final version of the manuscript.
This work was supported by the National Key R&D Program of China (Grant No. 2021YFC2102400), the National Natural Science Foundation of China (Grant Nos. U1805235, 21933010), and the NYU-ECNU Center for Computational Chemistry at NYU Shanghai.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.888721/full#supplementary-material
Acevedo-Rocha, C. G., Gamble, C., Lonsdale, R., Li, A., Nett, N., Hoebenreich, S., et al. (2018). P450-Catalyzed Regio- and Diastereoselective Steroid Hydroxylation: Efficient Directed Evolution Enabled by Mutability Landscaping. Acs Catal. 8, 3395–3410. doi:10.1021/acscatal.8b00389
Barry, S. M., Kers, J. A., Johnson, E. G., Song, L., Aston, P. R., Patel, B., et al. (2012). Cytochrome P450-Catalyzed L-Tryptophan Nitration in Thaxtomin Phytotoxin Biosynthesis. Nat. Chem. Biol. 8 (10), 814–816. doi:10.1038/nchembio.1048
Beller, M., Seayad, J., Tillack, A., and Jiao, H. (2004). Catalytic Markovnikov and Anti-Markovnikov Functionalization of Alkenes and Alkynes: Recent Developments and Trends. Angew. Chem. Int. Ed. 43 (26), 3368–3398. doi:10.1002/anie.200300616
Bonneau, R., Liu, M., and Rayez, M. T. (1989). Rates for 1,2-migration in Alkylchlorocarbenes. J. Am. Chem. Soc. 111, 5973–5974. doi:10.1021/ja00197a086
Bradford, M. M. (1976). A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 72 (1–2), 248–254. doi:10.1006/abio.1976.9999
Buller, A. R., Roye, P. V., Cahn, J., Scheele, R. A., Herger, M., and Arnold, F. H. (2018). Directed Evolution Mimics Allosteric Activation by Stepwise Tuning of the Conformational Ensemble. J. Am. Chem. Soc. 140, 7256–7266. doi:10.1021/jacs.8b03490
Chen, F.-F., Zheng, G.-W., Liu, L., Li, H., Chen, Q., Li, F.-L., et al. (2018). Reshaping the Active Pocket of Amine Dehydrogenases for Asymmetric Synthesis of Bulky Aliphatic Amines. ACS Catal. 8 (3), 2622–2628. doi:10.1021/acscatal.7b04135
Chen, G.-Q., Xu, Z.-J., Zhou, C.-Y., and Che, C.-M. (2011). Selective Oxidation of Terminal Aryl and Aliphatic Alkenes to Aldehydes Catalyzed by Iron(III) Porphyrins with Triflate as a Counter Anion. Chem. Commun. 47 (39), 10963–10965. doi:10.1039/c1cc13574k
Ciaramella, A., Catucci, G., Gilardi, G., and Di Nardo, G. (2019). Crystal Structure of Bacterial CYP116B5 Heme Domain: New Insights on Class VII P450s Structural Flexibility and Peroxygenase Activity. Int. J. Biol. Macromol. 140, 577–587. doi:10.1016/j.ijbiomac.2019.08.141
Dodani, S. C., Kiss, G., Cahn, J. K. B., Su, Y., Pande, V. S., and Arnold, F. H. (2016). Discovery of a Regioselectivity Switch in Nitrating P450s Guided by Molecular Dynamics Simulations and Markov Models. Nat. Chem. 8 (5), 419–425. doi:10.1038/nchem.2474
Dong, J. J., Browne, W. R., and Feringa, B. L. (2015). Palladium-catalyzed Anti-markovnikov Oxidation of Terminal Alkenes. Angew. Chem. Int. Ed. 54 (3), 734–744. doi:10.1002/anie.201404856
Dubrovskiy, A. V., Kesharwani, T., Markina, N. A., Pletnev, A. A., Raminelli, C., Yao, T., et al. (2018). Comprehensive Organic Transformations, 4 Volume Set: A Guide to Functional Group Preparations. Hoboken, New Jersey, U.S.: John Wiley & Sons.
Feng, L., Zhou, B., and Lu, G.-p. (2019). A DFT Study on the Mechanism of Rhodium-Catalyzed Regioselective Hydrothiolation of the Allyl Amine. Mol. Catal. 468, 62–74. doi:10.1016/j.mcat.2019.02.012
Girvan, H. M., and Munro, A. W. (2016). Applications of Microbial Cytochrome P450 Enzymes in Biotechnology and Synthetic Biology. Curr. Opin. Chem. Biol. 31, 136–145. doi:10.1016/j.cbpa.2016.02.018
Guengerich, F. P., and Munro, A. W. (2013). Unusual Cytochrome P450 Enzymes and Reactions. J. Biol. Chem. 288 (24), 17065–17073. doi:10.1074/jbc.R113.462275
Hamelberg, D., Mongan, J., and McCammon, J. A. (2004). Accelerated Molecular Dynamics: a Promising and Efficient Simulation Method for Biomolecules. J. Chem. Phys. 120 (24), 11919–11929. doi:10.1063/1.1755656
Hammer, S. C., Kubik, G., Watkins, E., Huang, S., Minges, H., and Arnold, F. H. (2017). Anti-Markovnikov Alkene Oxidation by Metal-Oxo-Mediated Enzyme Catalysis. Science 358 (6360), 215–218. doi:10.1126/science.aao1482
Hopps, H. B. (2000). Purpald®: A Reagent that Turns Aldehydes Purple. Aldrichim. Acta 33 (1), 28–30.
Kan, S. B. J., Lewis, R. D., Chen, K., and Arnold, F. H. (2016). Directed Evolution of Cytochrome C for Carbon-Silicon Bond Formation: Bringing Silicon to Life. Science 354 (6315), 1048–1051. doi:10.1126/science.aah6219
Kim, T. J., Kel'in, V. A., and Gevorgyan, V. (2003). 1,2-Migration of the Thio Group in Allenyl Sulfides: Efficient Synthesis of 3-Thio-Substituted Furans and Pyrroles. ChemInform 34 (19), 98–101. doi:10.1002/anie.200390064
Krest, C. M., Onderko, E. L., Yosca, T. H., Calixto, J. C., Karp, R. F., Livada, J., et al. (2013). Reactive Intermediates in Cytochrome P450 Catalysis. J. Biol. Chem. 288 (24), 17074–17081. doi:10.1074/jbc.R113.473108
Liu, Y., Cong, Y., Zhang, C., Fang, B., Pan, Y., Li, Q., et al. (2021). Engineering the Biomimetic Cofactors of NMNH for Cytochrome P450 BM3 Based on Binding Conformation Refinement. RSC Adv. 11, 12036–12042. doi:10.1039/d1ra00352f
Munro, W. A., Girvan, H. M., and McLean, K. J. (2007). Variations on a (T)heme — Novel Mechanisms, Redox Partners and Catalytic Functions in the Cytochrome P450 Superfamily. ChemInform 38 (36), 585–609. doi:10.1039/b604190f
Musa, M. M., and Phillips, R. S. (2011). Recent Advances in Alcohol Dehydrogenase-Catalyzed Asymmetric Production of Hydrophobic Alcohols. Catal. Sci. Technol. 1 (8), 1311. doi:10.1039/c1cy00160d
Nguyen, J., Chong, A., and Lalic, G. (2019). Nickel-catalyzed Anti-markovnikov Hydroarylation of Alkenes. Chem. Sci. 10 (11), 3231–3236. doi:10.1039/c8sc05445b
Reetz, T. M., Brunner, B., Schneider, T., Schulz, F., Clouthier, C. M., and Kayser, M. M. (2004). Directed Evolution as a Method to Create Enantioselective Cyclohexanone Monooxygenases for Catalysis in Baeyer–Villiger Reactions. Angew. Chem. Int. Ed. 43, 4075–4078. doi:10.1002/anie.200460272
Rittle, J., and Green, M. T. (2010). Cytochrome P450 Compound I: Capture, Characterization, and C-H Bond Activation Kinetics. Science 330 (6006), 933–937. doi:10.1126/science.1193478
Shahrokh, K., Orendt, A., Yost, G. S., and Cheatham, T. E. (2012). Quantum Mechanically Derived AMBER-Compatible Heme Parameters for Various States of the Cytochrome P450 Catalytic Cycle. J. Comput. Chem. 33 (2), 119–133. doi:10.1002/jcc.21922
Shaik, S., Cohen, S., Wang, Y., Chen, H., Kumar, D., and Thiel, W. (2010). P450 Enzymes: Their Structure, Reactivity, and Selectivity-Modeled by QM/MM Calculations. Chem. Rev. 110 (2), 949–1017. doi:10.1021/cr900121s
Sheldon, R. A., and Woodley, J. M. (2018). Role of Biocatalysis in Sustainable Chemistry. Chem. Rev. 118 (2), 801–838. doi:10.1021/acs.chemrev.7b00203
Sono, M., Roach, M. P., Coulter, E. D., and Dawson, J. H. (1996). Heme-Containing Oxygenases. Chem. Rev. 96 (7), 2841–2888.
Wang, F., Zhu, M., Song, Z., Li, C., Wang, Y., Zhu, Z., et al. (2020). Reshaping the Binding Pocket of Lysine Hydroxylase for Enhanced Activity. ACS Catal. 10 (23), 13946–13956. doi:10.1021/acscatal.0c03841
Wu, S., Zhou, Y., and Li, Z. (2019). Biocatalytic Selective Functionalisation of Alkenesviasingle-step and One-Pot Multi-step Reactions. Chem. Commun. 55 (7), 883–896. doi:10.1039/c8cc07828a
Yang, K. K., Wu, Z., and Arnold, F. H. (2019). Machine-Learning-Guided Directed Evolution for Protein Engineering. Nat. Methods 16 (8), 687–694. doi:10.1038/s41592-019-0496-6
Keywords: anti-Markovnikov (AM) oxidation, product selectivity, rational design, molecular dynamic simulations, experimental alanine scan, “butterfly” model
Citation: Pan Y, Bao J, Zhang X, Ni H, Zhao Y, Zhi F, Fang B, He X, Zhang JZH and Zhang L (2022) Rational Design of P450 aMOx for Improving Anti-Markovnikov Selectivity Based on the “Butterfly” Model. Front. Mol. Biosci. 9:888721. doi: 10.3389/fmolb.2022.888721
Received: 03 March 2022; Accepted: 15 April 2022;
Published: 23 May 2022.
Edited by:
Thomas Simonson, École Polytechnique, FranceReviewed by:
Xiang Sheng, Tianjin Institute of Industrial Biotechnology (CAS), ChinaCopyright © 2022 Pan, Bao, Zhang, Ni, Zhao, Zhi, Fang, He, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao He, eGlhb2hlQHBoeS5lY251LmVkdS5jbg==; John Z. H. Zhang, am9obi56aGFuZ0BueXUuZWR1; Lujia Zhang, bGp6aGFuZ0BjaGVtLmVjbnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.