- 1Department of Urology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 2Institute of Urology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
Erectile dysfunction (ED) is a common sexual dysfunction in males, with multifactorial alterations which consist of psychological and organic. Diabetes mellitus (DM) induced erectile dysfunction (DMED) is a disconcerting and critical complication of DM, and remarkably different from non-diabetic ED. The response rate of phosphodiesterase type 5 inhibitor (PDE5i), a milestone for ED therapy, is far from satisfactory in DMED. Unfortunately, the contributing mechanisms of DMED remains vague. Hence, It is urgent to seek for novel prospective biomarkers or targets of DMED. Numerous studies have proved that non-coding RNAs (ncRNAs) play essential roles in the pathogenesis process of DM, which comprise of long non-coding RNAs (lncRNAs) and small non-coding RNAs (sncRNAs) like microRNAs (miRNAs), PIWI-interacting RNAs (piRNAs) and circular RNAs (circRNAs). However, the implications of ncRNAs in DMED are still understudied. This review highlights the pathophysiology of DMED, summarizes identified mechanisms of ncRNAs associated with DMED and covers the topic of perspectives for ncRNAs in DMED.
Introduction
Diabetes mellitus (DM) has evolved as one of the most severe and widespread chronic conditions, leading in life threatening, debilitating and high-cost complications, as well as a reduction in life expectancy (Heald et al., 2020). Over the last three decades, the worldwide prevalence of DM has risen fast and reached pandemic proportions, with the 10th edition of the International Diabetes Federation indicating a prevalence of 536.6 million individuals (Sun et al., 2022). However, lowering diabetes mortality as a consequence of improved medical treatment, as well as rises in diabetes incidence in certain countries as a result of rising prevalence of diabetes risk factors, particularly obesity, are also significant drivers of increased prevalence (Magliano et al., 2019; Chan et al., 2020). It was concurrent with an increase in the prevalence of microvascular and macrovascular complications of DM (Graves and Donaghue, 2020).
Erectile dysfunction (ED) is a common and frequently occurring disease, characterized as the inability to attain or sustain an erection adequate for enjoyable sexual performance (Shamloul and Ghanem, 2013; Yafi et al., 2016). The incidence of ED is reported to be as high as 75% in diabetics, more than three times that in non-diabetics (Thorve et al., 2011). Numerous pathological alterations affecting the corpus cavernosum, including as endothelial dysfunction and nitric oxide (NO) bioactivity, have been implicated in the development of ED (Lue, 2000). Sildenafil citrate, the first successful oral medication, is a selective phosphodiesterase type 5 inhibitor (PDE5i). It is generally acknowledged as the first-line treatment for erectile dysfunction (ED). The PDE5i were developed based on the crucial function of NO in penile cavernous smooth muscle relaxation, which results in effective erections in 63% of men with general ED. However, its response rate in DMED is far from optimal, only about 44% in patients with inadequate glycemic control (Liu et al., 2010). As a result, DMED is a current research hotspot. Regrettably, the processes behind DMED remains vague.
Profiling of various cell lines using high-throughput sequencing found that 74% of the human genome is transcribed, although only 2% of it contains protein-coding genes (Esteller, 2011; Djebali et al., 2012). As a result, the vast majority of the human transcriptome is composed of non-coding RNAs (ncRNAs). ncRNAs are characterized as regulatory RNAs that do not comprise a protein-coding region (Carninci et al., 2005). Based on their length, ncRNAs are classified as small non-coding RNAs (200 nt) and long non-coding RNAs (lncRNAs, >200 nt) (Matsui and Corey, 2017). However, according to localization and functions, ncRNAs can be divided into lncRNAs, miRNAs, circular RNAs (circRNAs), small nucleolar RNAs (snoRNAs), small nuclear RNAs (snRNAs) and PIWI-interacting RNAs (piRNAs). So, it is vital not to be dogmatic with regard to terminology, as this may impede data interpretation and mechanistic understanding (Matsui and Corey, 2017). In recent years, it has become clearer that ncRNAs play a critical role in normal development and physiology, as well as in diseases (Chi et al., 2021; de Goede et al., 2021). The functional significance of ncRNAs is most clear in the case of miRNAs. It has been demonstrated that human illnesses frequently exhibit epigenetic and genetic alterations in miRNAs and their processing machinery (Ji and Guo, 2019; Mori et al., 2019; Agbu and Carthew, 2021). However, miRNAs are just the tip of the iceberg; other ncRNAs also have a role in the development of a variety of other illnesses (Bridges et al., 2021). There is mounting evidence that ncRNAs grease the wheels of the development of DM and associated complications (Feng et al., 2019; Thomas et al., 2019). There is considerable interest in medicinal methods that target these ncRNAs disruptions (Matsui and Corey, 2017).
Here, we begin by discussing the pathophysiology of DMED. On top of that, we consider in greater detail the growing evidence for the roles of miRNAs and lncRNAs, which have been implicated in several cellular processes. Finally, this Review covers the potential for adopting innovative treatment techniques to target these ncRNAs disruptions.
Diabetes Mellitus Induced Erectile Dysfunction
DMED is considered to have a multifactorial etiology (Malavige and Levy, 2009). Numerous physical causes are assumed to be essential, but psychological and relationship concerns frequently coincide (Muneer et al., 2014; Najari and Kashanian, 2016). During an erection, increased blood flow to penile corpora cavernosa is caused by nerve signals that relax the vascular and corpus cavernosum smooth muscle cells (CCSMCs) (McMahon, 2019). This is primarily mediated by NO, which is generated by parasympathetic nonadrenergic noncholinergic neurons and cholinergic neurons when they stimulate vascular endothelial cells (VECs), initiating a biochemical cascade that leads in smooth muscle relaxation and vasodilation (Udelson, 2007; Mitidieri et al., 2020). Increasing blood flow compresses subtunical venules to impede venous return, hence preserving the erection (Yafi et al., 2016) (Figure 1). The pathophysiology of DMED is primarily characterized by functional and structural alterations in two dimensions. Early alterations that were mostly functional in nature: long-term hyperglycemia stimulation results in dysfunction of the penile artery and endothelium (Castela and Costa, 2016), increased reactive oxygen species (Yuan et al., 2020) and decreased NO production and bioavailability (Zhou et al., 2021). Simultaneously, the generation of contractile substances like as angiotensin and endothelin raise the concentration of Ca2+ in CCSMCs and activates RohA/ROCK signaling pathways (Yuan et al., 2020), resulting in the impairment of smooth muscle’s diastolic function. As the disease progresses, a high concentration of reactive oxygen species and fibrogenic factors are produced in the cavernosum of the penis, resulting in excessive endothelial cell apoptosis, smooth muscle atrophy, structural changes such as fibrosis, and eventually cavernous venous closure dysfunction (Li et al., 2021).
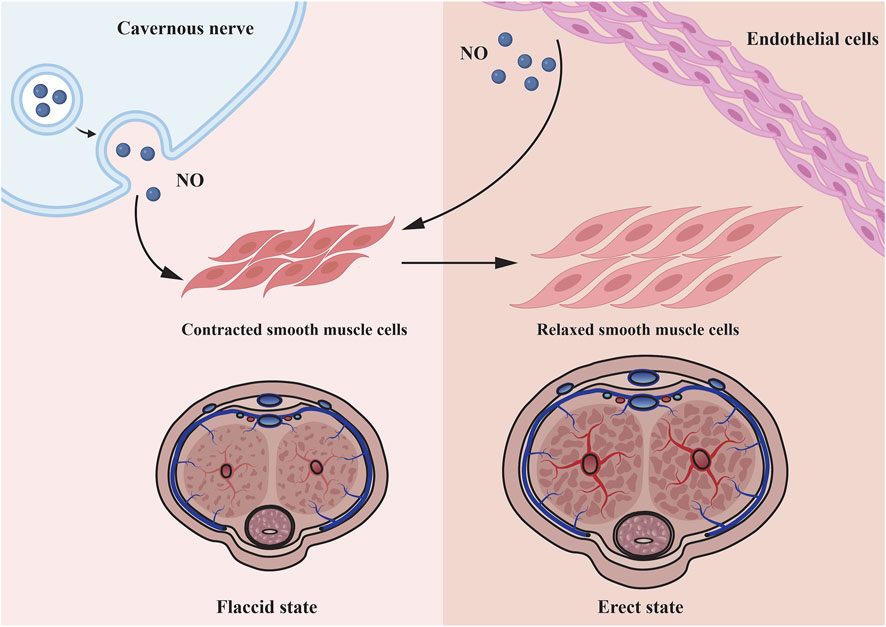
FIGURE 1. Microscopic mechanisms underlying penile smooth muscle relaxation. Cavernous nerve and endothelial cell all can secrete NO to relax corpus cavernosum smooth muscle cells. As the smooth muscle relaxes, blood fills the lacunar spaces, resulting to compression of the subtunical venules, thereby restricting the venous outflow.
miRNAs in Diabetes Mellitus Induced Erectile Dysfunction
For a long period of time, the function of ncRNAs were unknown, and they were not regarded as critical as transcripts of protein-coding genes. Our current understanding of the functions which some research hotspots in ncRNAs, like miRNAs and lncRNAs, play within cells is expanding rapidly, and we now know more than ever before about their impact on a wide range of physiological processes and illnesses (Pandolfini et al., 2019). One of the most well-studied classes of ncRNAs is miRNAs, which function to silence genes post-transcriptionally by interfering with mRNA translation into proteins. Although the discovery is only a few years old, it is of great significance for our understanding of post-transcriptional regulation of genes (Bartel, 2018). In contrast to certain miRNAs, which target particular genes, others might act as master regulators of processes, regulating hundreds of genes concurrently and cooperating with other miRNAs (Agbu and Carthew, 2021).
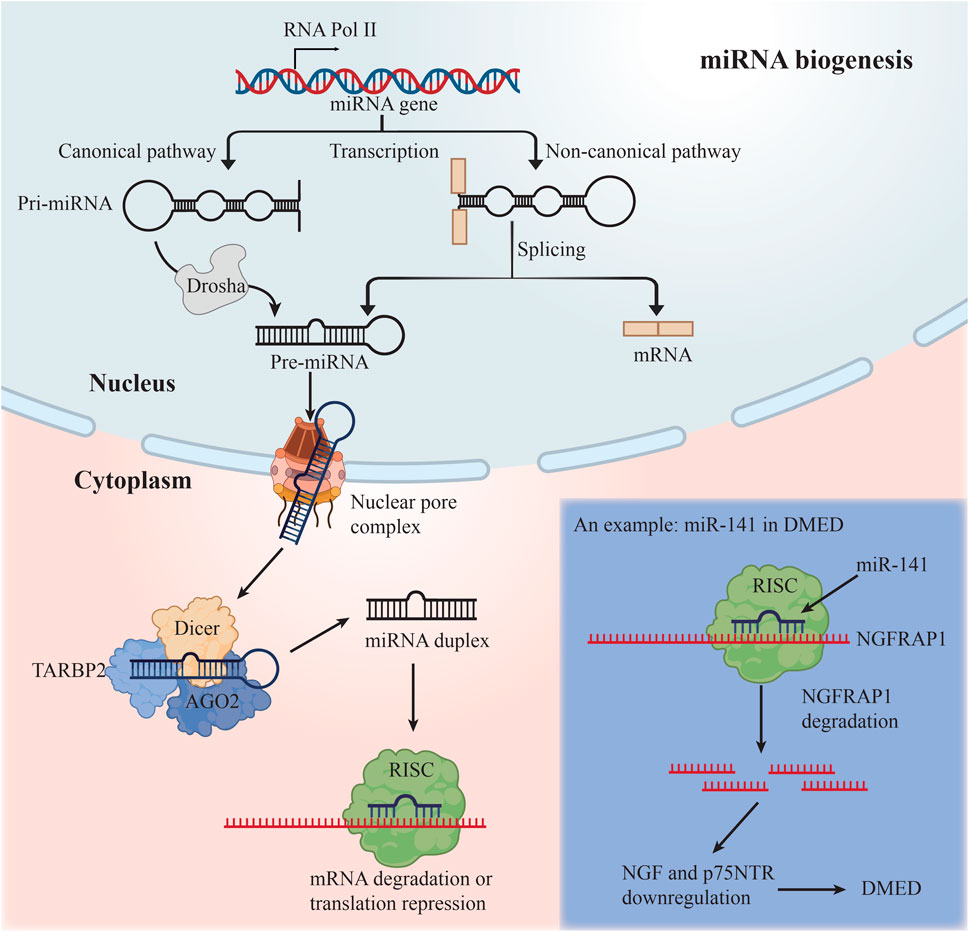
In order to produce mature miRNAs, the RNase III enzymes Drosha and Dicer participate in a multi-step biogenesis process (Krol et al., 2010) (Figure 2). The Dicer–TARBP2 (TAR RNA-binding protein 2) complex loads these molecules into a member of the Argonaute protein subfamily, which serves as the catalytic endonuclease component, to create the RNA-induced silencing complex (RISC). RISC recognizes a complementary region in the 3′UTR of the targeted mRNA and guides the regulation of mRNA. There is a great deal of control over the loading of miRNAs into RISC (Gebert and MacRae, 2019) and the operation of the miRNA machinery itself (Krol et al., 2010). miRNAs impede the translation of mRNA by mRNA degradation and translation start inhibition.
The first miRNA profile for DMED patients was conducted by (Jiang et al., 2015). They assessed the serum miRNA content in normal individuals, non-diabetic ED, and DMED patients, and discovered that the serum miRNA content of DMED patients was considerably elevated for miR-93, miR-320, and miR-16 (Jiang et al., 2015). Recently, Xu et al. (2021) applied miRNA sequencing form the serum of diabetic and DMED patients and discovered that the serum levels of let-7e-5p, miR-30d-5p, miR-199b-5p, and miR-342-3p were considerably higher in DMED patients. It is indicated the presence of miRNAs in serum may aid in the early detection of DMED.
Wei et al. (2011) discovered decreased expression of miR-145 in the corpus cavernosum of DMED rats, establishing the first relationship between miRNAs and DMED. It wasn’t until 2016 that the first miRNA profile for erectile dysfunction in mice with type 2 diabetes was published. eNOS/cGMP/PKG pathway and the contraction of vascular smooth muscle may all be affected by specific miRNAs, such as miR-18a, miR206, and miR-122 and miR-133. The researchers hypothesized that these miRNAs could have a significant impact on the endothelium and smooth muscle in the corpus cavernosum (Pan et al., 2016). Additionally, miR-328 antagomir has been shown to enhance erectile performance in diabetic rats by downregulating the expression of advanced glycation end products and increasing the levels of DKK3, cGMP and eNOS (Li et al., 2017). According to a recent research, elevation of NGFRAP1, NGF, and p75NTR in DMED is related with decreased expression of miR-141, and the overexpression of miR-141 can improve erectile function in DMED rats (Wen et al., 2018). Furthermore, Wen et al. (2019) discovered that DMED rats had significantly higher levels of miR-205 expression than normal rats, and further research revealed that miR-205 could directly act on androgen receptors, causing fibrosis and apoptosis of corpus cavernosum smooth muscle cells (CCSMCs), ultimately leading to DMED. This work presented a novel method by which miRNAs influence AR expression in target cells, which is predicted to give a new technique for treating DMED in the clinic by modulating AR expression. As demonstrated by Huo et al. (2020a), the pathophysiology of DMED rats may be connected to the downregulation of the miR-874-3p after an increase in methylation at the promoter region, followed by the upregulation of the Nupr1/Chop pathway, thereby speeding the death of CCSMCs in rats and impairing erectile function. Bioinformatics is a critical tool for predicting the presence of ED-related miRNAs. Kazemi et al. (2021) attempted to identify the conserved site for miRNAs, and revealed that the conserved miR-29-3p binding site is found in the 3’UTR of genes related with ED.
Research by Gonçalves et al. (2018) and Tiraboschi et al. (2021) created three distinct types of ED models in rats: diabetic, alcoholic, or both. Their outcomes, however, are distinct. Gonçalves et al. (2018) discovered that in alcoholic and alcoholic–diabetic conditions, reduced expression of miR-199 and miR-155 boosted endothelin receptor (ETA and ERB) expression. Then, in another study, miR-15b, miR-16, miR-138, miR-221, and miR-222 could be considered prospective biomarkers for diabetic alcoholic ED (Tiraboschi et al., 2021).
Adipose tissue-derived stem cells (ADSCs) can enhance erectile function in diabetic rats by altering the microarchitecture of the corpus cavernosum. A second option is ADSC-exosomes, which is derived from ADSCs. Zhu et al. (2018) showed that ADSCs-Exosomes had a greater expression of various pro-angiogenic or antifibrotic miRNAs, including miR-126, miR-130a, miR-132, let7b, and let7c, than ADSCs. Ouyang et al. (2019) revealed that some pro-angiogenic miRNAs, including miR-21-5p, the let-7 family, the miR-10 family, the miR-30 family, and miR-148a-3p, are expressed at a greater level in ADSCs-exosomes than in ADSCs. Taken together, these distinctions between ADSCs and ADSCs-exosomes may contribute to ADSC-exosomes superior therapeutic efficacy over ADSCs. Furthermore, Huo et al. (2020b) verified that exosomal miR-21-5p generated from bone marrow-derived stem cells (BMSCs) inhibited PDCD4 expression and ED in rats with DM.
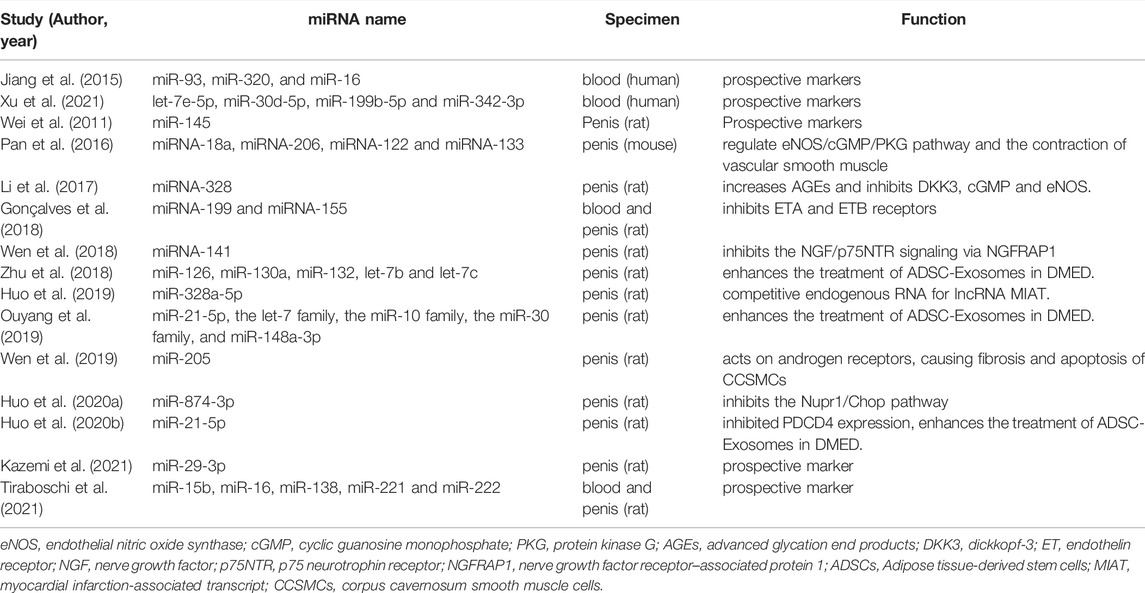
Table 1 summarizes the miRNAs in DMED and their possible mechanism. Clearly, the majority of miRNAs appear to be intimately linked to smooth muscle contraction or apoptosis. Thus, it is beneficial to regulate miRNAs in order to optimize smooth muscle performance.
lncRNAs in Diabetes Mellitus Induced Erectile Dysfunction
IncRNAs has some structural characteristics of mRNA, including 3′poly(A) tails and terminal 5′caps, but lacks open reading frame, so it does not encode protein (Statello et al., 2020). Rapid advancements in high-throughput sequencing technology have resulted in the identification of an increasing number of differentially expressed lncRNAs (Fang et al., 2018; Uszczynska-Ratajczak et al., 2018). They are abundant in tissues, urine, and serum, with expression patterns that vary according to cell type, tissue, and developmental stage (Encode Project Consortium, 2012). lncRNAs are functionally involved in a variety of complicated biological processes via a variety of methods. These include transcription factor titration (Luo et al., 2016), splicing modification, miRNA sponging (Fatica and Bozzoni, 2014), and chromatin modification enzyme recruitment (Isoda et al., 2017; Mumbach et al., 2019). Additionally, there is mounting evidence that lncRNAs have a role in the onset and progression of a variety of disorders, including cardiovascular diseases (Wang and Sun, 2020), metabolic syndrome (Sun L.-Y. et al., 2018), renal fibrosis (Yang et al., 2019), and malignancies (Tan et al., 2021).
Huo et al. (2019) has demonstrated that the lncRNA myocardial infarction-associated transcript (MIAT) is a competitive endogenous RNA for miR-328a-5p. Additionally, by suppressing miR-328a-5p, excessive levels of MIAT/lipoprotein lipase (LPL) pathway can induce damage and death in vascular endothelial cells (VECs). Thus, the MIAT/miR-328a-5p/LPL signaling pathway may provide therapeutic targets for DMED attenuation if the expression of the three critical sites is reversed.
Although stem cell therapy is widely accepted as an effective treatment for erectile dysfunction, the underlying processes are still a mystery. By promoting the degradation of FOXM1 protein and decreasing VEGF expression, lncRNA MEG3 plays a vital role in the differentiation of bone marrow-derived mesenchymal stem cells (BM-MSCs) into VECs (Sun X. et al., 2018). VEGF expression was upregulated by another lncRNA, MALAT1, which worked as a sponge for miR-206 and aided in the differentiation process (Sun et al., 2020).
In general, researchers specializing in the pathophysiology of DMED do not appear to devote sufficient attention to lncRNAs (Chen et al., 2020). Other studies, on the other hand, may discover the mechanism through which BM-MSCs increase erectile function, hence improving therapy options for DMED.
Therapies Targeting ncRNAs
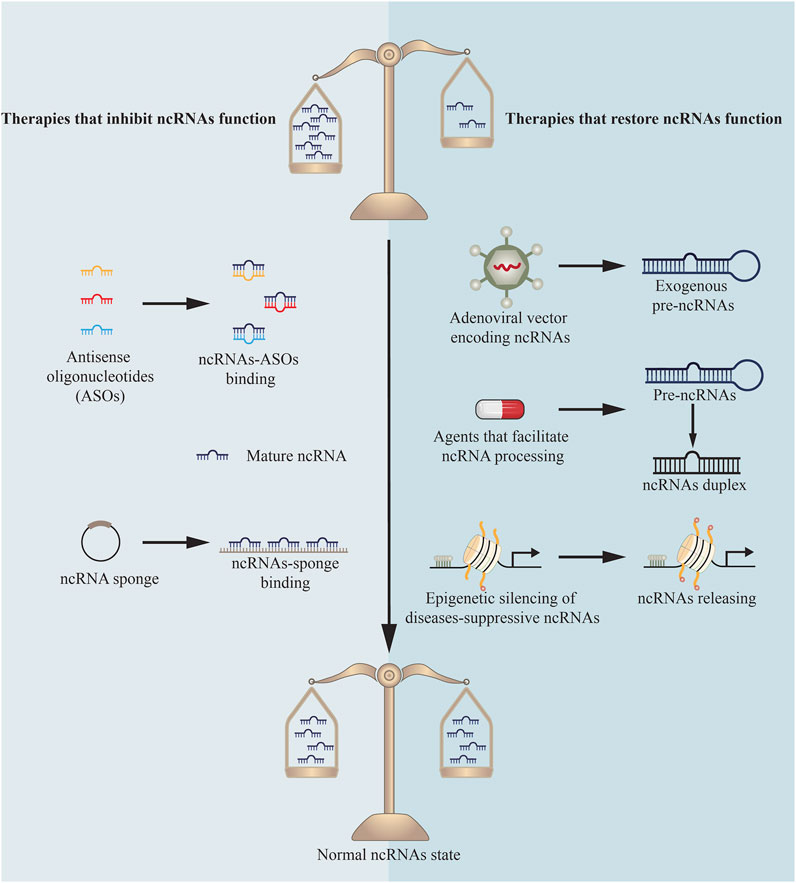
NcRNAs and the protein machinery involved in their synthesis or function have been identified as targets for innovative treatment strategies and tested in clinical practices (Figure 3). Until now, the majority of research in this field has focused on miRNAs’ function in cancer to repress tumor growth (Matsui and Corey, 2017; Mollaei et al., 2019). Additionally, antisense oligonucleotides (ASOs) complementary to miR-122 are being developed to treat Hepatitis C virus (Thakral and Ghoshal, 2015) and miR-21 mimics are being applied to treat cutaneous and pulmonary fibrosis (Matsui and Corey, 2017). While this research is still in its infancy, there is tremendous interest in extending comparable methodologies to different types of diseases, both for miRNAs and other ncRNAs.
Therapies That Inhibit miRNAs Function
Because miRNAs control their targets via base pairing, ASOs have been developed to therapeutically decrease miRNA activity. Through base pair complementarity, ASOs block miRNA targets. Three distinct classes of ASOs have been developed: locked nucleic acids (LNAs) (Elmén et al., 2008), anti-miRNA oligonucleotides (AMOs) (Yu et al., 2020), and antagomirs (Krützfeldt et al., 2005). Each of these classes incorporates a variety of chemical changes to boost stability and efficacy. For example, antagomirs were first synthesized as miRNA silencing agents in 2005, and they are chemically modified, cholesterol-conjugated oligonucleotides complementary to or the same as miRNAs (Krützfeldt et al., 2005; Mohr and Mott, 2015). Antagomirs bind on the 3’ untranslated region of targeted mRNA strands, which is expected to avoid other miRNAs positioning as to inhibit miRNAs function (Krützfeldt et al., 2005).
ASOs have been shown to be effective in some instances. Antagomirs have been used to target miR-328 in the setting of DMED (Li et al., 2017). Systemic dosing of these antagomirs has been demonstrated to inhibit miR-328 activity in the penis and reduce advanced glycation end products to improve erectile function. However, because diseases are pleiotropic and diverse in their biology, silencing a single miRNA may not always be adequate. Recent studies in this field reveal that single ASOs targeting multiple miRNAs can suppress several miRNAs at once (Lu et al., 2009). A multiple-target anti-miRNA antisense oligodeoxyribonucleotide (MTg-AMO) is the result of this strategy. In a wide variety of malignancies, miR-21, miR-155 and miR-17-5p are overexpressed, and one MTg-AMO was created to target these three oncogenic microRNAs. Individual AMOs that target a single miRNA and combinations of AMOs that target several miRNAs were shown to be less effective than this MTg-AMO (Lu et al., 2009). In the future, MTg-AMOs might be engineered to concurrently block the activity of miRNAs that have a role in DMED.
Another novel approach is to develop competitive inhibitors of miRNA activity. Vectors having several artificial miRNA binding sites are called “miRNA sponges”. They are used to create enormous amounts of transcript under the direction of powerful promoters and keep homologous miRNAs from binding to their native targets as a sponge (Cohen, 2009; Hansen et al., 2013). This method was utilized to decrease miR-141 and miR-205 expression, revealing their function in DMED (Wen et al., 2018; Wen et al., 2019).
Therapies That Restore miRNAs Function
Numerous techniques for reactivating miRNAs with disease-suppressive effects that are downregulated in DMED have been proposed. A new study shows that “miRNA replacement therapy”, a method for restoring miR-874-3p expression in DMED, has been successfully used (Huo et al., 2020a). This miRNA was delivered by an f in a rat model of DMED, which resulted in improved erectile function and decreased apoptosis. Conventional gene therapy procedures, on the other hand, have the same issues in delivering protein-coding genes via viral delivery (Kay, 2011).
A substantial body of research indicates that the majority of human diseases are characterized by miRNA production abnormalities that result in a global decrease in miRNA levels. As a result, a therapeutic benefit might be gained by returning the global miRNAome to a normal state. A new “miRNAome-based” approach has been proposed as a result of these discoveries. By binding to TARBP2, the small chemical enoxacin facilitates RNAi and miRNA processing (Shan et al., 2008). Following therapy with enoxacin, a global restoration of downregulated miRNAs to more normal miRNA expression patterns has been demonstrated to prevent tumor progression (Melo et al., 2011). The medication had no effect on healthy cells and was not hazardous to mice. Other methods for reviving the global miRNAome include the use of histone acetylase inhibitors and DNA demethylating agents. Epigenetic silencing of diseases-suppressive miRNAs is released by these chemicals, which have demonstrated therapeutic effectiveness and have been approved for the treatment of some hematological malignancies despite their lack of target specificity (Rodríguez-Paredes and Esteller, 2011; Zhang and Zhang, 2020). However, no comparable research exists in DMED up to now.
Targeting Other Types of ncRNAs
It’s possible that similar techniques outlined before for improving deregulated miRNAs may be applicable to additional ncRNAs, hence expanding the therapeutic target pool. Even while lncRNAs can be targeted using siRNAs (Zhang et al., 2021), they are more challenging to block than miRNAs because of their complex secondary structures (Tsai et al., 2010). Our growing understanding of other ncRNAs is also being leveraged to generate innovative therapeutic strategies for a variety of diseases (Matsui and Corey, 2017). While further research is necessary, cell lines, mouse models and non-human primate investigations have yielded encouraging findings so far. Clinical applications of ncRNAs-based therapeutics are still a long way off, but researchers are optimistic.
Perspectives for ncRNAs in Diabetes Mellitus Induced Erectile Dysfunction
There has been considerable interest in ncRNAs recently, but more research is needed to properly understand their function and the mechanisms through which they exert their effects. One significant hurdle will be identifying all of the human genome’s functional ncRNAs, for which developing genomic, epigenomic, and bioinformatic techniques will be critical. ENCODE, for example, is making significant progress in its mission to catalog all of the human genome’s functional components (Birney et al., 2007; Encode Project Consortium, 2012). Second-generation sequencing methods, like RNA sequencing, will yield a more complete view of the human ncRNA transcriptome (Pareek et al., 2011). The use of bioinformatics methods to find ncRNAs that may be useful will also be critical (Chen et al., 2019). Due of the complex secondary structures that ncRNAs form to function, sequence-based alignments alone may be insufficient to detect ncRNAs. Albeit plenty of algorithms have been established to predict ncRNAs with potential function (Wang et al., 2020; Singh et al., 2021; Fu et al., 2022), only a few kinds of ncRNAs can be identified so far.
The identification of ncRNA defects in human diseases has boosted expectations in the therapeutic field. Inhibition of crucial DMED-promoting genes that were previously not regarded feasible conventional therapeutic targets is now possible with siRNAs or miRNAs, which expand the universe of “druggable” targets. Clinical studies utilizing compounds derived from ncRNAs are currently being conducted. However, novel techniques, such as small molecule inhibitors of the miRNA machinery, are on the horizon. Many hurdles must be overcome before this strategy may be beneficial in treating DMED in a way that standard treatments haven't yet been able to. Furthermore, except for miRNAs and lncRNAs, there are currently no research examining the involvement of additional ncRNAs in DMED. New discoveries are predicted to be made as the field evolves. Exciting times lie ahead for us (Esteller, 2011).
Conclusion
Since their identification as functional RNAs, ncRNAs have been regarded as the milestone in the treatment of numerous diseases, even though therapeutic applications are still in their infancy. Similarly, the discovery of ncRNAs has ushered in a new age in DMED, one that may provide a slew of novel biomarkers or therapeutic targets and revolutionize how DMED is diagnosed and treated.
Author Contributions
WX: writing, editing, visualization and revision of the manuscript. HJ: writing, funding acquisition. JL and HL: conceptualization, funding acquisition, critical revision. All authors approved the manuscript.
Funding
This study was supported by grants from the National Natural Science Foundation of China (No. 82001536 and No. 81873831) and the Tongji Hospital Funding (No. 2021A14).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We acknowledge the BioRender for use.
References
Agbu, P., and Carthew, R. W. (2021). MicroRNA-mediated Regulation of Glucose and Lipid Metabolism. Nat. Rev. Mol. Cell. Biol. 22 (6), 425–438. doi:10.1038/s41580-021-00354-w
Birney, E., Birney, E., Stamatoyannopoulos, J. A., Dutta, A., Guigó, R., Gingeras, T. R., et al. (2007). Identification and Analysis of Functional Elements in 1% of the Human Genome by the ENCODE Pilot Project. Nature 447 (7146), 799–816. doi:10.1038/nature05874
Bridges, M. C., Daulagala, A. C., and Kourtidis, A. (2021). LNCcation: lncRNA Localization and Function. J. cell Biol. 220 (2). doi:10.1083/jcb.202009045
Carninci, P., Kasukawa, T., Katayama, S., Gough, J., Frith, M. C., Maeda, N., et al. (2005). The Transcriptional Landscape of the Mammalian Genome. Science 309 (5740), 1559–1563. doi:10.1126/science.1112014
Castela, Â., and Costa, C. (2016). Molecular Mechanisms Associated with Diabetic Endothelial-Erectile Dysfunction. Nat. Rev. Urol. 13 (5), 266–274. doi:10.1038/nrurol.2016.23
Chan, J. C. N., Lim, L.-L., Wareham, N. J., Shaw, J. E., Orchard, T. J., Zhang, P., et al. (2020). The Lancet Commission on Diabetes: Using Data to Transform Diabetes Care and Patient Lives. Lancet 396 (10267), 2019–2082. doi:10.1016/s0140-6736(20)32374-6
Chen, L., Heikkinen, L., Wang, C., Yang, Y., Sun, H., and Wong, G. (2019). Trends in the Development of miRNA Bioinformatics Tools. Briefings Bioinforma. 20 (5), 1836–1852. doi:10.1093/bib/bby054
Chen, S., Sun, X., Wu, S., Jiang, J., Zhu, C., Xu, K., et al. (2020). Role of Identified Noncoding RNA in Erectile Dysfunction. Andrologia 52 (7), e13596. doi:10.1111/and.13596
Chi, T., Lin, J., Wang, M., Zhao, Y., Liao, Z., and Wei, P. (2021). Non-Coding RNA as Biomarkers for Type 2 Diabetes Development and Clinical Management. Front. Endocrinol. 12, 630032. doi:10.3389/fendo.2021.630032
Cohen, S. M. (2009). Use of microRNA Sponges to Explore Tissue-specific microRNA Functions In Vivo. Nat. Methods 6 (12), 873–874. doi:10.1038/nmeth1209-873
de Goede, O. M., Nachun, D. C., Ferraro, N. M., Gloudemans, M. J., Rao, A. S., Smail, C., et al. (2021). Population-scale Tissue Transcriptomics Maps Long Non-coding RNAs to Complex Disease. Cell. 184 (10), 2633–e19. doi:10.1016/j.cell.2021.03.050
Djebali, S., Davis, C. A., Merkel, A., Dobin, A., Lassmann, T., Mortazavi, A., et al. (2012). Landscape of Transcription in Human Cells. Nature 489 (7414), 101–108. doi:10.1038/nature11233
Elmén, J., Lindow, M., Schütz, S., Lawrence, M., Petri, A., Obad, S., et al. (2008). LNA-mediated microRNA Silencing in Non-human Primates. Nature 452 (7189), 896–899. doi:10.1038/nature06783
Encode Project Consortium (2012). An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature 489 (7414), 57–74. doi:10.1038/nature11247
Esteller, M. (2011). Non-coding RNAs in Human Disease. Nat. Rev. Genet. 12 (12), 861–874. doi:10.1038/nrg3074
Fang, S., Zhang, L., Guo, J., Niu, Y., Wu, Y., Li, H., et al. (2018). NONCODEV5: a Comprehensive Annotation Database for Long Non-coding RNAs. Nucleic acids Res. 46 (D1), D308–D314. doi:10.1093/nar/gkx1107
Fatica, A., and Bozzoni, I. (2014). Long Non-coding RNAs: New Players in Cell Differentiation and Development. Nat. Rev. Genet. 15 (1), 7–21. doi:10.1038/nrg3606
Feng, Y., Xu, W., Zhang, W., Wang, W., Liu, T., and Zhou, X. (2019). LncRNA DCRF Regulates Cardiomyocyte Autophagy by Targeting miR-551b-5p in Diabetic Cardiomyopathy. Theranostics 9 (15), 4558–4566. doi:10.7150/thno.31052
Fu, L., Cao, Y., Wu, J., Peng, Q., Nie, Q., and Xie, X. (2022). UFold: Fast and Accurate RNA Secondary Structure Prediction with Deep Learning. Nucleic acids Res. 50 (3), e14. doi:10.1093/nar/gkab1074
Gebert, L. F. R., and MacRae, I. J. (2019). Regulation of microRNA Function in Animals. Nat. Rev. Mol. Cell. Biol. 20 (1), 21–37. doi:10.1038/s41580-018-0045-7
Gonçalves, F. Z., Lizarte Neto, F. S., Novais, P. C., Gattas, D., Lourenço, L. G., de Carvalho, C. A. M., et al. (2018). Expression Profile of Endothelin Receptors (ETA and ETB) and microRNAs-155 and -199 in the Corpus Cavernosum of Rats Submitted to Chronic Alcoholism and Diabetes Mellitus. Braz J. Med. Biol. Res. 51 (3), e6329. doi:10.1590/1414-431x20176329
Graves, L. E., and Donaghue, K. C. (2020). Vascular Complication in Adolescents with Diabetes Mellitus. Front. Endocrinol. 11, 370. doi:10.3389/fendo.2020.00370
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013). Natural RNA Circles Function as Efficient microRNA Sponges. Nature 495 (7441), 384–388. doi:10.1038/nature11993
Heald, A. H., Stedman, M., Davies, M., Livingston, M., Alshames, R., Lunt, M., et al. (2020). Estimating Life Years Lost to Diabetes: Outcomes from Analysis of National Diabetes Audit and Office of National Statistics Data. Cardiovasc. Endocrinol. metabolism 9 (4), 183–185. doi:10.1097/XCE.0000000000000210
Huo, W., Hou, Y., Li, Y., and Li, H. (2019). Downregulated lncRNA-MIAT Confers Protection against Erectile Dysfunction by Downregulating Lipoprotein Lipase via Activation of miR-328a-5p in Diabetic Rats. Biochimica Biophysica Acta (BBA) - Mol. Basis Dis. 1865 (6), 1226–1240. doi:10.1016/j.bbadis.2019.01.018
Huo, W., Li, H., Zhang, Y., and Li, H. (2020a). Epigenetic Silencing of microRNA‐874‐3p Implicates in Erectile Dysfunction in Diabetic Rats by Activating the Nupr1/Chop‐mediated Pathway. FASEB J. 34 (1), 1695–1709. doi:10.1096/fj.201902086R
Huo, W., Li, Y., Zhang, Y., and Li, H. (2020b). Mesenchymal Stem Cells‐derived Exosomal microRNA‐21‐5p Downregulates PDCD4 and Ameliorates Erectile Dysfunction in a Rat Model of Diabetes Mellitus. FASEB J. 34 (10), 13345–13360. doi:10.1096/fj.202000102RR
Isoda, T., Moore, A. J., He, Z., Chandra, V., Aida, M., Denholtz, M., et al. (2017). Non-coding Transcription Instructs Chromatin Folding and Compartmentalization to Dictate Enhancer-Promoter Communication and T Cell Fate. Cell. 171 (1), 103–119. doi:10.1016/j.cell.2017.09.001
Ji, C., and Guo, X. (2019). The Clinical Potential of Circulating microRNAs in Obesity. Nat. Rev. Endocrinol. 15 (12), 731–743. doi:10.1038/s41574-019-0260-0
Jiang, X., Luo, Y., Zhao, S., Chen, Q., Jiang, C., Dai, Y., et al. (2015). Clinical Significance and Expression of microRNA in Diabetic Patients with Erectile Dysfunction. Exp. Ther. Med. 10 (1), 213–218. doi:10.3892/etm.2015.2443
Kay, M. A. (2011). State-of-the-art Gene-Based Therapies: the Road Ahead. Nat. Rev. Genet. 12 (5), 316–328. doi:10.1038/nrg2971
Kazemi, E., Zargooshi, J., Kaboudi, M., Heidari, P., Kahrizi, D., Mahaki, B., et al. (2021). A Genome-wide Association Study to Identify Candidate Genes for Erectile Dysfunction. Brief. Bioinform 22 (4). doi:10.1093/bib/bbaa338
Krol, J., Loedige, I., and Filipowicz, W. (2010). The Widespread Regulation of microRNA Biogenesis, Function and Decay. Nat. Rev. Genet. 11 (9), 597–610. doi:10.1038/nrg2843
Krützfeldt, J., Rajewsky, N., Braich, R., Rajeev, K. G., Tuschl, T., Manoharan, M., et al. (2005). Silencing of microRNAs In Vivo with 'antagomirs'. Nature 438 (7068), 685–689. doi:10.1038/nature04303
Liu, D. F., Jiang, H., Hong, K., Zhao, L. M., Tang, W. H., and Ma, L. L. (2010). Influence of Erectile Dysfunction Course on its Progress and Efficacy of Treatment with Phosphodiesterase Type 5 Inhibitors. Chin. Med. J. Engl. 123(22), 3258–3261. doi:10.3760/cma.j.issn.0366-6999.2010.22.016
Li, D.-S., Feng, L., Luo, L.-H., Duan, Z.-F., Li, X.-L., Yin, C.-H., et al. (2017). The Effect of microRNA-328 Antagomir on Erectile Dysfunction in Streptozotocin-Induced Diabetic Rats. Biomed. Pharmacother. 92, 888–895. doi:10.1016/j.biopha.2017.05.071
Li, H., Xu, W., Liu, X., Wang, T., Wang, S., Liu, J., et al. (2021). JAK2 Deficiency Improves Erectile Function in Diabetic Mice through Attenuation of Oxidative Stress, Apoptosis, and Fibrosis. Andrology 9 (5), 1662–1671. doi:10.1111/andr.13061
Lu, Y., Xiao, J., Lin, H., Bai, Y., Luo, X., Wang, Z., et al. (2009). A Single Anti-microRNA Antisense Oligodeoxyribonucleotide (AMO) Targeting Multiple microRNAs Offers an Improved Approach for microRNA Interference. Nucleic acids Res. 37 (3), e24. doi:10.1093/nar/gkn1053
Lue, T. F. (2000). Erectile Dysfunction. N. Engl. J. Med. 342 (24), 1802–1813. doi:10.1056/nejm200006153422407
Luo, S., Lu, J. Y., Liu, L., Yin, Y., Chen, C., Han, X., et al. (2016). Divergent lncRNAs Regulate Gene Expression and Lineage Differentiation in Pluripotent Cells. Cell. stem cell 18 (5), 637–652. doi:10.1016/j.stem.2016.01.024
Magliano, D. J., Islam, R. M., Barr, E. L. M., Gregg, E. W., Pavkov, M. E., Harding, J. L., et al. (2019). Trends in Incidence of Total or Type 2 Diabetes: Systematic Review. BMJ 366, l5003. doi:10.1136/bmj.l5003
Malavige, L. S., and Levy, J. C. (2009). Erectile Dysfunction in Diabetes Mellitus. J. Sex. Med. 6 (5), 1232–1247. doi:10.1111/j.1743-6109.2008.01168.x
Matsui, M., and Corey, D. R. (2017). Non-coding RNAs as Drug Targets. Nat. Rev. Drug Discov. 16 (3), 167–179. doi:10.1038/nrd.2016.117
McMahon, C. G. (2019). Current Diagnosis and Management of Erectile Dysfunction. Med. J. Aust. 210 (10), 469–476. doi:10.5694/mja2.50167
Melo, S., Villanueva, A., Moutinho, C., Davalos, V., Spizzo, R., Ivan, C., et al. (2011). Small Molecule Enoxacin Is a Cancer-specific Growth Inhibitor that Acts by Enhancing TAR RNA-Binding Protein 2-mediated microRNA Processing. Proc. Natl. Acad. Sci. U.S.A. 108 (11), 4394–4399. doi:10.1073/pnas.1014720108
Mitidieri, E., Cirino, G., d’Emmanuele di Villa Bianca, R., and Sorrentino, R. (2020). Pharmacology and Perspectives in Erectile Dysfunction in Man. Pharmacol. Ther. 208, 107493. doi:10.1016/j.pharmthera.2020.107493
Mohr, A., and Mott, J. (2015). Overview of microRNA Biology. Semin. Liver Dis. 35 (1), 003–011. doi:10.1055/s-0034-1397344
Mollaei, H., Safaralizadeh, R., and Rostami, Z. (2019). MicroRNA Replacement Therapy in Cancer. J. Cell. Physiology 234 (8), 12369–12384. doi:10.1002/jcp.28058
Mori, M. A., Ludwig, R. G., Garcia-Martin, R., Brandão, B. B., and Kahn, C. R. (2019). Extracellular miRNAs: From Biomarkers to Mediators of Physiology and Disease. Cell. metab. 30 (4), 656–673. doi:10.1016/j.cmet.2019.07.011
Mumbach, M. R., Granja, J. M., Flynn, R. A., Roake, C. M., Satpathy, A. T., Rubin, A. J., et al. (2019). HiChIRP Reveals RNA-Associated Chromosome Conformation. Nat. Methods 16 (6), 489–492. doi:10.1038/s41592-019-0407-x
Muneer, A., Kalsi, J., Nazareth, I., and Arya, M. (2014). Erectile Dysfunction. BMJ 348, g129. doi:10.1136/bmj.g129
Najari, B. B., and Kashanian, J. A. (2016). Erectile Dysfunction. JAMA 316 (17), 1838. doi:10.1001/jama.2016.12284
Ouyang, B., Xie, Y., Zhang, C., Deng, C., Lv, L., Yao, J., et al. (2019). Extracellular Vesicles from Human Urine-Derived Stem Cells Ameliorate Erectile Dysfunction in a Diabetic Rat Model by Delivering Proangiogenic MicroRNA. Sex. Med. 7 (2), 241–250. doi:10.1016/j.esxm.2019.02.001
Pan, F., You, J., Liu, Y., Qiu, X., Yu, W., Ma, J., et al. (2016). Differentially Expressed microRNAs in the Corpus Cavernosum from a Murine Model with Type 2 Diabetes Mellitus-Associated Erectile Dysfunction. Mol. Genet. Genomics 291 (6), 2215–2224. doi:10.1007/s00438-016-1250-8
Pandolfini, L., Barbieri, I., Bannister, A. J., Hendrick, A., Andrews, B., Webster, N., et al. (2019). METTL1 Promotes Let-7 MicroRNA Processing via m7G Methylation. Mol. cell 74 (6), 1278–1290. doi:10.1016/j.molcel.2019.03.040
Pareek, C. S., Smoczynski, R., and Tretyn, A. (2011). Sequencing Technologies and Genome Sequencing. J. Appl. Genet. 52 (4), 413–435. doi:10.1007/s13353-011-0057-x
Rodríguez-Paredes, M., and Esteller, M. (2011). Cancer Epigenetics Reaches Mainstream Oncology. Nat. Med. 17 (3), 330–339. doi:10.1038/nm.2305
Shamloul, R., and Ghanem, H. (2013). Erectile Dysfunction. Lancet 381 (9861), 153–165. doi:10.1016/s0140-6736(12)60520-0
Shan, G., Li, Y., Zhang, J., Li, W., Szulwach, K. E., Duan, R., et al. (2008). A Small Molecule Enhances RNA Interference and Promotes microRNA Processing. Nat. Biotechnol. 26 (8), 933–940. doi:10.1038/nbt.1481
Singh, J., Paliwal, K., Zhang, T., Singh, J., Litfin, T., and Zhou, Y. (2021). Improved RNA Secondary Structure and Tertiary Base-Pairing Prediction Using Evolutionary Profile, Mutational Coupling and Two-Dimensional Transfer Learning. Bioinformatics, 37. Oxford, England, 2589–2600. doi:10.1093/bioinformatics/btab165
Statello, L., Guo, C.-J., Chen, L.-L., and Huarte, M. (2020). Gene Regulation by Long Non-coding RNAs and its Biological Functions. Nat. Rev. Mol. Cell. Biol. 22, 96–118. doi:10.1038/s41580-020-00315-9
Sun, H., Saeedi, P., Karuranga, S., Pinkepank, M., Ogurtsova, K., Duncan, B. B., et al. (2022). IDF Diabetes Atlas: Global, Regional and Country-Level Diabetes Prevalence Estimates for 2021 and Projections for 2045. Diabetes Res. Clin. Pract. 183, 109119. doi:10.1016/j.diabres.2021.109119
Sun, L.-Y., Li, X.-J., Sun, Y.-M., Huang, W., Fang, K., Han, C., et al. (2018a). LncRNA ANRIL Regulates AML Development through Modulating the Glucose Metabolism Pathway of AdipoR1/AMPK/SIRT1. Mol. Cancer 17 (1), 127. doi:10.1186/s12943-018-0879-9
Sun, X., Luo, L.-h., Feng, L., and Li, D.-s. (2018b). Down-regulation of lncRNA MEG3 Promotes Endothelial Differentiation of Bone Marrow Derived Mesenchymal Stem Cells in Repairing Erectile Dysfunction. Life Sci. 208, 246–252. doi:10.1016/j.lfs.2018.07.024
Sun, X., Luo, L., and Li, J. (2020). LncRNA MALAT1 Facilitates BM-MSCs Differentiation into Endothelial Cells via Targeting miR-206/VEGFA axis. Cell. Cycle 19, 3018–3028. doi:10.1080/15384101.2020.1829799
Tan, Y. T., Lin, J. F., Li, T., Li, J. J., Xu, R. H., and Ju, H. Q. (2021). LncRNA‐mediated Posttranslational Modifications and Reprogramming of Energy Metabolism in Cancer. Cancer Commun. 41 (2), 109–120. doi:10.1002/cac2.12108
Thakral, S., and Ghoshal, K. (2015). miR-122 Is a Unique Molecule with Great Potential in Diagnosis, Prognosis of Liver Disease, and Therapy Both as miRNA Mimic and Antimir. Cgt 15 (2), 142–150. doi:10.2174/1566523214666141224095610
Thomas, A. A., Biswas, S., Feng, B., Chen, S., Gonder, J., and Chakrabarti, S. (2019). lncRNA H19 Prevents Endothelial-Mesenchymal Transition in Diabetic Retinopathy. Diabetologia 62 (3), 517–530. doi:10.1007/s00125-018-4797-6
Thorve, V. S., Kshirsagar, A. D., Vyawahare, N. S., Joshi, V. S., Ingale, K. G., and Mohite, R. J. (2011). Diabetes-induced Erectile Dysfunction: Epidemiology, Pathophysiology and Management. J. diabetes its Complicat. 25 (2), 129–136. doi:10.1016/j.jdiacomp.2010.03.003
Tiraboschi, R. B., Neto, F. S. L., da Cunha Tirapelli, D. P., de Bessa, J., Miranda, E. P., de Assis Cirino, M. L., et al. (2021). Expression of MicroRNAs (miR-15b, miR-16, miR-138, miR-221, and miR-222) as Biomarkers of Endothelial Corpus Cavernosum Dysfunction in a Diabetic Alcoholic Murine Model. Sex. Med. 9 (2), 100326. doi:10.1016/j.esxm.2021.100326
Tsai, M.-C., Manor, O., Wan, Y., Mosammaparast, N., Wang, J. K., Lan, F., et al. (2010). Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 329 (5992), 689–693. doi:10.1126/science.1192002
Udelson, D. (2007). Biomechanics of Male Erectile Function. J. R. Soc. Interface. 4 (17), 1031–1048. doi:10.1098/rsif.2007.0221
Uszczynska-Ratajczak, B., Lagarde, J., Frankish, A., Guigó, R., and Johnson, R. (2018). Towards a Complete Map of the Human Long Non-coding RNA Transcriptome. Nat. Rev. Genet. 19 (9), 535–548. doi:10.1038/s41576-018-0017-y
Wang, Y., Liu, Y., Wang, S., Liu, Z., Gao, Y., Zhang, H., et al. (2020). ATTfold: RNA Secondary Structure Prediction with Pseudoknots Based on Attention Mechanism. Front. Genet. 11, 612086. doi:10.3389/fgene.2020.612086
Wang, Y., and Sun, X. (2020). The Functions of LncRNA in the Heart. Diabetes Res. Clin. Pract. 168, 108249. doi:10.1016/j.diabres.2020.108249
Wei, A. Y., Yang, Y., He, S. H., Luo, X. G., Zhang, T., and Liu, Y. (2011). Role of miR-145 in Erectile Dysfunction in Diabetic Rats. Nan Fang. Yi Ke Da Xue Xue Bao 31 (6), 1051–1054.
Wen, Y., Liu, G., Jia, L., Ji, W., and Li, H. (2018). MicroRNA‐141 Binds to the Nerve Growth Factor Receptor Associated Protein 1 Gene and Restores the Erectile Function of Diabetic Rats through Down‐regulating the Nerve Growth Factor/neurotrophin Receptor P75 (NGF/p75NTR) Signaling. J Cell. Biochem. 120, 7940–7951. doi:10.1002/jcb.28071
Wen, Y., Liu, G., Zhang, Y., and Li, H. (2019). MicroRNA‐205 Is Associated with Diabetes Mellitus‐induced Erectile Dysfunction via Down‐regulating the Androgen Receptor. J. Cell. Mol. Med. 23 (5), 3257–3270. doi:10.1111/jcmm.14212
Xu, H., Zhao, B., Zhong, W., Teng, P., and Qiao, H. (2021). Identification of miRNA Signature Associated with Erectile Dysfunction in Type 2 Diabetes Mellitus by Support Vector Machine-Recursive Feature Elimination. Front. Genet. 12, 762136. doi:10.3389/fgene.2021.762136
Yafi, F. A., Jenkins, L., Albersen, M., Corona, G., Isidori, A. M., Goldfarb, S., et al. (2016). Erectile Dysfunction. Nat. Rev. Dis. Prim. 2, 16003. doi:10.1038/nrdp.2016.3
Yang, Z., Jiang, S., Shang, J., Jiang, Y., Dai, Y., Xu, B., et al. (2019). LncRNA: Shedding Light on Mechanisms and Opportunities in Fibrosis and Aging. Ageing Res. Rev. 52, 17–31. doi:10.1016/j.arr.2019.04.001
Yu, A.-M., Choi, Y. H., and Tu, M.-J. (2020). RNA Drugs and RNA Targets for Small Molecules: Principles, Progress, and Challenges. Pharmacol. Rev. 72 (4), 862–898. doi:10.1124/pr.120.019554
Yuan, P., Ma, D., Gao, X., Wang, J., Li, R., Liu, Z., et al. (2020). Liraglutide Ameliorates Erectile Dysfunction via Regulating Oxidative Stress, the RhoA/ROCK Pathway and Autophagy in Diabetes Mellitus. Front. Pharmacol. 11. doi:10.3389/fphar.2020.01257
Zhang, P., and Zhang, M. (2020). Epigenetic Alterations and Advancement of Treatment in Peripheral T-Cell Lymphoma. Clin. Epigenet 12 (1), 169. doi:10.1186/s13148-020-00962-x
Zhang, Q., Li, D., Dong, X., Zhang, X., Liu, J., Peng, L., et al. (2021). LncDACH1 Promotes Mitochondrial Oxidative Stress of Cardiomyocytes by Interacting with Sirtuin3 and Aggravates Diabetic Cardiomyopathy. Sci. China Life Sci. doi:10.1007/s11427-021-1982-8
Zhou, B., Chen, Y., Yuan, H., Wang, T., Feng, J., Li, M., et al. (2021). NOX1/4 Inhibitor GKT-137831 Improves Erectile Function in Diabetic Rats by ROS Reduction and Endothelial Nitric Oxide Synthase Reconstitution. J. Sex. Med. 18 (12), 1970–1983. doi:10.1016/j.jsxm.2021.09.007
Keywords: erectile dysfunction, diabetes mellitus, non-coding RNA, miRNA, lncRNA
Citation: Xu W, Jiang H, Liu J and Li H (2022) Non-Coding RNAs: New Dawn for Diabetes Mellitus Induced Erectile Dysfunction. Front. Mol. Biosci. 9:888624. doi: 10.3389/fmolb.2022.888624
Received: 03 March 2022; Accepted: 01 June 2022;
Published: 22 June 2022.
Edited by:
Shilpy Sharma, Savitribai Phule Pune University, IndiaReviewed by:
Giuseppe Fallara, San Raffaele Hospital (IRCCS), ItalyGilberto De Nucci, State University of Campinas, Brazil
Copyright © 2022 Xu, Jiang, Liu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jihong Liu, amhsaXVAdGpoLnRqbXUuZWR1LmNu; Hao Li, dGptd2xoQGh1c3QuZWR1LmNu
 Wenchao Xu
Wenchao Xu Hongyang Jiang
Hongyang Jiang Jihong Liu
Jihong Liu Hao Li
Hao Li