
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci., 25 March 2022
Sec. Protein Biochemistry for Basic and Applied Sciences
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.868568
This article is part of the Research TopicCarbohydrate-Active Enzymes and Their Inhibitors - Volume IIView all 4 articles
Numerous plants, including cereals, contain seed proteins able to inhibit amylolytic enzymes. Some of these inhibitors, the CM-proteins (soluble in chloroform:methanol mixtures)—also referred to as cereal-type inhibitors (CTIs)—are the topic of this review. CM-proteins were first reported 75 years ago. They are small sulfur-rich proteins of the prolamine superfamily embracing bifunctional α-amylase/trypsin inhibitors (ATIs), α-amylase inhibitors (AIs), limit dextrinase inhibitors (LDIs), and serine protease inhibitors. Phylogenetically CM-proteins are predicted across poaceae genomes and many isoforms are identified in seed proteomes. Their allergenicity and hence adverse effect on humans were recognized early on, as were their roles in plant defense. Generally, CTIs target exogenous digestive enzymes from insects and mammals. Notably, by contrast LDI regulates activity of the endogenous starch debranching enzyme, limit dextrinase, during cereal seed germination. CM-proteins are four-helix bundle proteins and form enzyme complexes adopting extraordinarily versatile binding modes involving the N-terminal and different loop regions. A number of these inhibitors have been characterized in detail and here focus will be on target enzyme specificity, molecular recognition, forces and mechanisms of binding as well as on three-dimensional structures of CM-protein–enzyme complexes. Lastly, prospects for CM-protein exploitation, rational engineering and biotechnological applications will be discussed.
Proteinaceous α-amylase inhibitors belonging to different protein families; knottins, defensins, Kunitz-type inhibitors, CM-proteins, legume lectins, γ-thionins, lipid-transfer proteins, xylanase-α-amylase inhibitory proteins, and thaumatin-like inhibitors are mostly found in plants, although some occur in mollusks and microorganisms. This kind of α-amylase inhibitors were recognized long ago. They have been collectively covered in several reviews as well as in recent publications on specific inhibitors presenting rather different levels of structural and mechanistic insights (Kneen and Sandstedt, 1946; Garcia-Olmedo et al., 1987; Blanco et al., 1991; Carbonero and García-Olmedo, 1999; Svensson et al., 2004; Juge and Svensson, 2006; de Oliveira Carvalho and Gomes, 2009; Rehm et al., 2009; dos Santos et al., 2010; Kumar et al., 2010; Wang et al., 2014; Gadge et al., 2015; Sun et al., 2015; da Silva et al., 2018; Panwar et al., 2018; Tysoe and Withers, 2018; Tsvetkov and Yarullina, 2019; Juhász et al., 2020; Rane et al., 2020; Aguieiras et al., 2021; Geisslitz et al., 2021). The biological role of the plant hydrolase inhibitors is primarily in defense against insect pests and pathogenic fungi, whereas they are rarely involved in regulation of the activity of endogenous plant enzymes. Hydrolase inhibitors of certain protein families can be bifunctional and act both on amylolytic enzymes of glycoside hydrolase family 13 (GH13) (Drula et al., 2022) and serine proteases, while other members of the same families only inhibit either amylolytic enzymes of GH13 or serine proteases (Barber et al., 1986; Carbonero and Garcia-Olmedo, 1999; di Maro et al., 2011). In several cases, the dual enzyme inhibition has been experimentally confirmed along with corresponding three-dimensional structures and models of enzyme–double-headed plant inhibitor complexes (Mundy et al., 1983; Maskos et al., 1996; Strobl et al., 1998; Vallée et al., 1998; Micheelsen et al., 2008; Grosse-Holz and van der Hoorn, 2016). The topic of the present review is the family of cereal-type inhibitors (CTIs), in particular inhibitors of amylolytic enzymes, which have been first described 75 years ago (Kneen and Sandstedt, 1946). CTIs are all found in cereals and other grass species and can amount to 2–4% of the seed protein content. These inhibitors belong to the prolamine superfamily of plant proteins and are called CM-proteins after their solubility in chloroform:methanol mixtures (Carbonero and García-Olmedo, 1999; Mills et al., 2004; Geisslitz et al., 2021). Some CM-proteins, referred to a-amylase/trypsin inhibitors—or ATIs for short—display bifunctionality and have two target enzymes. This protein family also contains monofunctional inhibitors against α-amylases from insects and mammals as well as the starch debranching enzyme limit dextrinase, which all belong to GH13, and similarly other members only inhibit serine proteases. Notably, CM-proteins also receive major attention due to their behavior as antinutrients and allergens harmful to human health including non-celiac wheat sensitivity (NCWS) and Bakers’ asthma (Wang et al., 2014; Reig-Otero et al., 2018) (for a review see Geisslitz et al., 2021). However, our focus will be on biochemical and structural properties of CM-protein inhibitors, i.e. their target enzyme specificity and inhibition kinetics, affinity and mechanism of enzyme binding as well as on three-dimensional structures of complexes with enzymes of family GH13. Some of these cases can provide a basis for using rational protein engineering to develop improved inhibitors for various potential applications.
The CM-proteins are small proteins of 12–16 kDa containing four to five well-conserved disulfide bonds and can be either monomeric or composed of two or four subunits (Carbonero and García-Olmedo, 1999). They are found in seeds of a wide range of cereal crops; wheat, barley, oats, rye, finger millet, barnyard millet, corn, rice, and sorghum (Garcia-Olmedo et al., 1987; Feng et al., 1991; Chen et al., 1992; Maskos et al., 1996; Carbonero and García-Olmedo, 1999; Altenbach et al., 2011; Wang et al., 2014; Gadge et al., 2015; Gazza et al., 2016; Panwar et al., 2018; Sagu et al., 2020). Many isoforms have been identified for example in barley and wheat (Østergaard et al., 2004; Guo et al., 2016; Bose et al., 2020; di Francesco et al., 2020; Geisslitz et al., 2020). In wheat the more prominent ones are 0.28 (monomeric), 0.19 and 0.53 (both homodimeric) (named based on electrophoretic mobility), CM1, CM2, CM3, CM16 and CM17 (all heterotetrameric) (for names and numbering see Rodriguez-Loperena et al., 1975; Carbonero and Garcia-Olmedo, 1999; Geisslitz et al., 2021). A time lag between ATI accumulation during wheat grain filling and detection of the biological activity suggested that assembly into dimers and tetramers determined the inhibitory potential (Call et al., 2021). Nineteen ATI isoforms from the wheat cultivar Butte 86 (Altenbach et al., 2011) and 33 proteoforms of ATIs across different bread wheat cultivars are reported (Bose et al., 2020; Geisslitz et al., 2021). These comprehensive analyses of wheat reflect the interest in CM-proteins due to the impact they may have on human health, albeit some address their role in plant defense as α-amylase or protease inhibitors, while the studies on barley also concerned protein mapping of cultivars for malting and beer brewing (Østergaard et al., 2004; Iimure et al., 2015; Guo et al., 2016; Perlikowski et al., 2016; Bose et al., 2019). Different proteoforms of CM-proteins in wheat and food products in light of possible implications in NCWS were determined by using advanced mass spectrometry (Bose et al., 2020; Geisslitz et al., 2020). Related to consumers’ interest in ancient cultivars, it has been noted that the CM-protein contents in old and modern Italian durum wheat genotypes, showed most isoforms to be shared, although a couple were only identified in an ancient cultivar (di Francesco et al., 2020). As mentioned above, the isoforms occur in different states of oligomerisation, the monomeric show high inhibitory activity on insect α-amylases, the homodimeric inhibitors react well with both insect and mammalian α-amylases, while the heterotetrameric inhibitors are highly active towards insect a-amylases (Table 1) (Carbonero and García-Olmedo, 1999; Franco et al., 2002). In addition to the CTIs inhibiting α-amylases, a small group of CTIs only act on serine proteases. WCI (wheat chymotrypsin inhibitor) is a strong inhibitor of bovine pancreatic chymotrypsin as well as of chymotryptic-like activities isolated from cotton bollworm and yellow mealworm (Tenebrio molitor), while no inhibition was detected against bovine pancreatic trypsin, or α-amylases from yellow mealworm (TMA) and human saliva (HSA) (di Maro et al., 2011). Barley CMc (equivalent to WCI) and CMe inhibited trypsin, but not TMA. Only CMa inhibited TMA among the CMa–e proteins from barley (Barber et al., 1986). Because a large number of different CM-proteins and posttranslationally modified forms thereof are present in seeds, it is difficult to purify any of the proteins to a highly homogenous state from natural sources for characterization of structure and function. Therefore, selected CM-proteins have been produced recombinantly in microbial hosts, Escherichia coli (CM2, CM3, CM16, 0.28, corn Hageman factor inhibitor, bifunctional α-amylase/trypsin inhibitor, and rye BIII) and Pichia pastoris (LDI, CM3, CM16, and 0.28) (García-Maroto et al., 1991; Strobl et al., 1995; Behnke et al., 1998; Kusaba-Nakayama et al., 2001; Dias et al., 2005; Jensen et al., 2011; Tundo et al., 2018) or in lentivirus transfected human embryonic kidney cells (CM3, the most prominent isoform in wheat) (Thiel et al., 2020). While recombinant CTIs were not applied in clinical testing (Geisslitz et al., 2021), evaluation of allergenicity has been performed in cellular assays (Tundo et al., 2018) and the effect on gut microbiota in Drosophila melanogaster (Thiel et al., 2020). Studies in rats and using caco-2 cells showed enhanced absorption rate for the abundant isoform CM3 as compared to CM16 and 0.28 from wheat (Kusaba-Nakayama et al., 2001). Phylogenetic analyses reveal that the monomeric and dimeric wheat CTIs are closely related (Figure 1), while the bifunctional CTIs are more related to the LDI-like CTIs than the other CTI groups (Behnke et al., 1998; Møller et al., 2015; Geisslitz et al., 2021). The sequence conservation of the CTIs is very low (Figure 1), but comparison of the four CTIs for which 3D structures have been determined shows that the structural conservation is very high (Figure 2A).
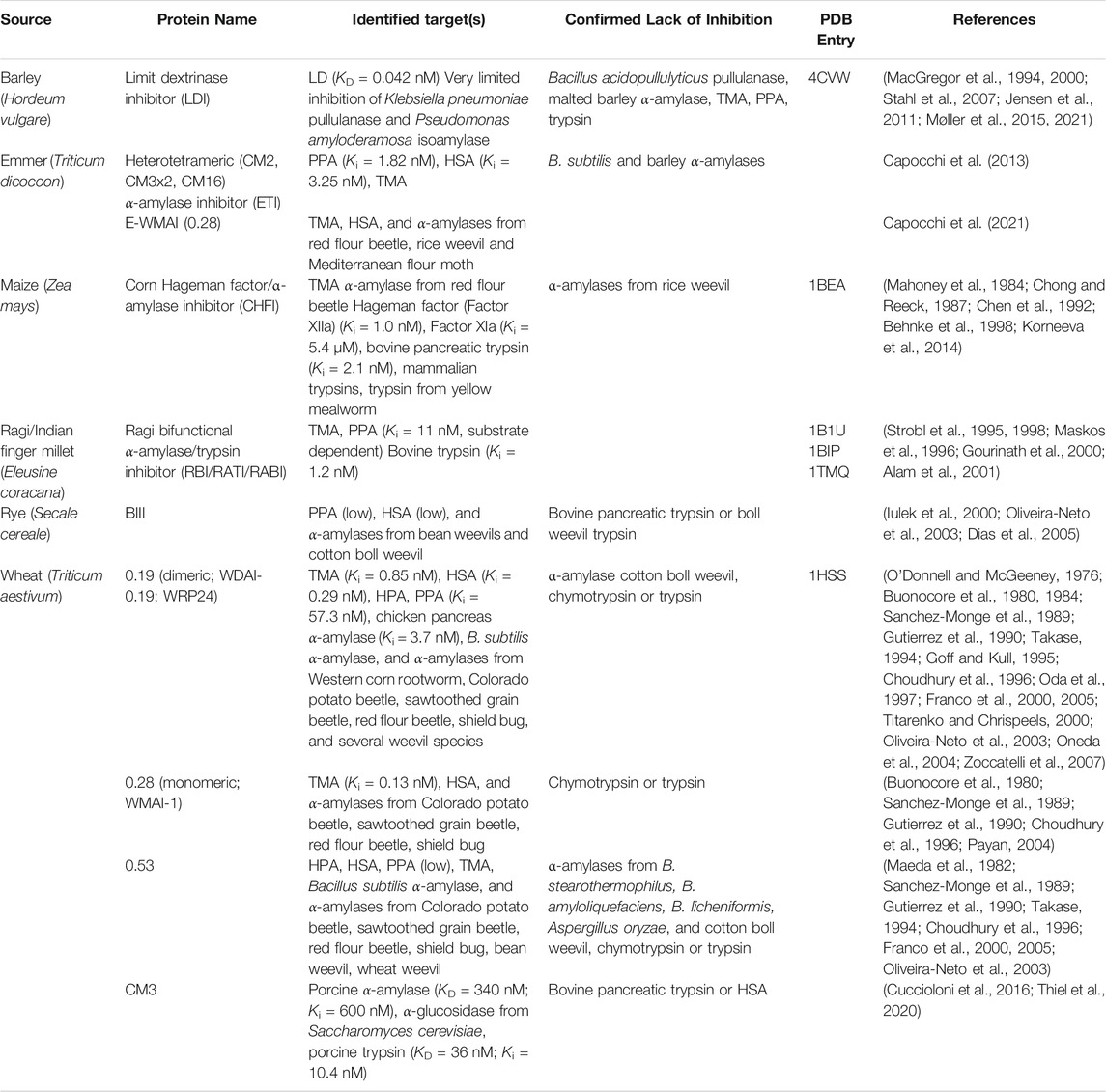
TABLE 1. Biochemically well-characterized cereal type inhibitors. The Protein Data Bank (PDB) entries are given for structure-determined proteins. Abbreviations of enzymes mentioned in the table: HPA, human pancreatic α-amylase; HSA, human salivary α-amylase; LD, barley limit dextrinase; PPA, porcine pancreatic α-amylase; TMA, yellow mealworm α-amylase.
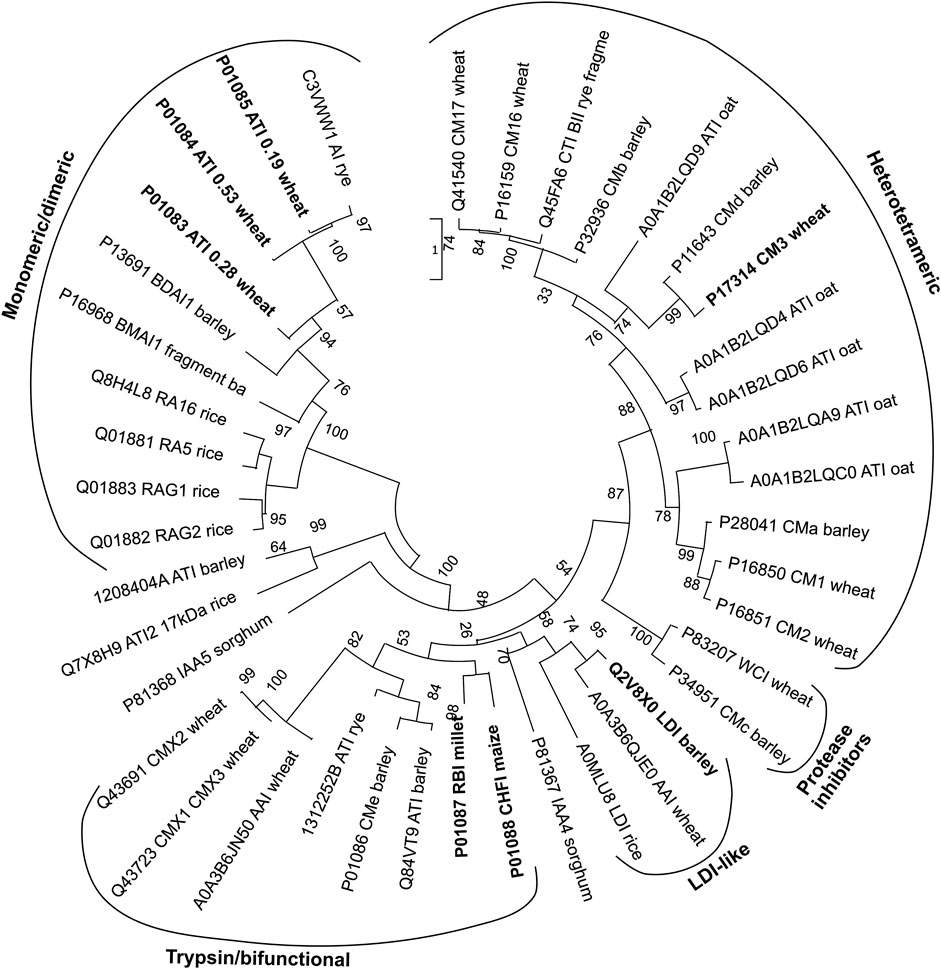
FIGURE 1. Phylogenetic analysis of characterized CTIs and homologues from other plants. The well-characterized proteins included in Table 1 are shown in bold. Names and origin of the proteins are indicated. The sequences were retrieved from UniProt database (1 February 2022). Software used: Promals3D (Pei et al., 2008) for structure-guided multiple alignment and MEGA 11 (Kumar et al., 2018) for Maximum likelihood for phylogeny analysis. Bootstrap values for 1,000 replicates are shown.
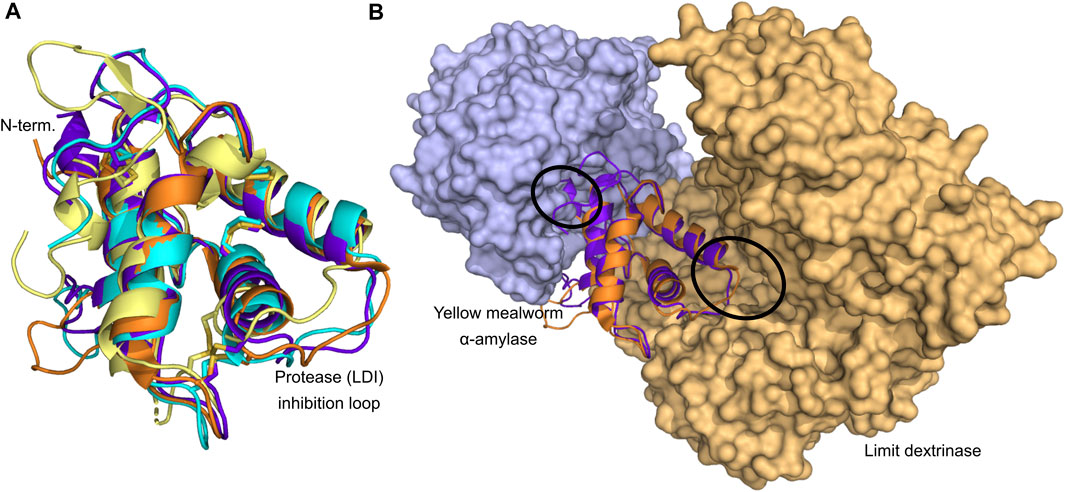
FIGURE 2. Structure determined CTIs. (A) Structural superposition of the four structure determined CTIs; LDI (orange; PDB entry 4CWV), RBI (purple; PDB entry 1TMQ), CHFI (cyan; PDB entry 1BEA), and 0.19 (yellow; PDB entry 1HSS). Disulfide bonds are shown as sticks. (B) Comparison of the inhibitor orientation of the two available CTI–enzyme complexes. LDI (orange; PDB entry 4CWV) and RBI (purple; PDB entry 1TMQ) are superposed revealing that opposite sides of the inhibitors are involved in the inhibition. The active sites of the enzymes are encircled. Structures were retrieved from the Protein Data Bank (PDB; www.rcsb.org).
Several CTIs originating from different cereals have been biochemically characterized to some degree during the past decades. While their target enzyme specificity and selectivity have been tested, actual inhibition constant (Ki) or binding constant (KD) values have only been reported for very few (Table 1). This is in most cases because the proteins were purified from their original source; hence, either the purity or the yield or both have been low. Recombinant protein production has enabled mutational analysis of structure/function relationships of a few CM-protein inhibitors (García-Maroto et al., 1991; Alam et al., 2001; Møller et al., 2015, 2021) and three-dimensional structures have been determined for four members including some in complex with target enzymes, namely wheat 0.19 dimeric inhibitor (Oda et al., 1997), ragi bifunctional a-amylase/trypsin inhibitor (RBI) (Strobl et al., 1995, 1998; Gourinath et al., 2000), corn Hageman factor/α-amylase inhibitor (CHFI) (Behnke et al., 1998), and barley limit dextrinase inhibitor (LDI) (Møller et al., 2015) (Table 1). Most characterized CTIs that inhibit hydrolases, except LDI, target α-1,4-glucan endo-acting a-amylases from GH13. By contrast, LDI from barley exclusively inhibits a debranching enzyme, limit dextrinase also belonging to GH13, that hydrolyses α-1,6-branch points in starch and glycogen and, in particular, α-limit dextrins obtained from these two branched α-glucans (Table 1). In addition, LDI is a special case for another reason, namely that it inhibits and hence regulates an endogenous enzyme, rather than exogenous enzymes typically from insect pests. Most CM-protein α-amylase inhibitors show specificity with regard to the target α-amylase, mainly explained by small differences in the architecture around the active site in the α-amylases (Rane et al., 2020). Usually, the CTIs act to a different degree against mammalian and insect digestive a-amylases (Table 1). Their inhibition of α-amylases has been shown to be influenced by the presence and type of substrate (Alam et al., 2001), and in general CTIs are not capable of completely inhibiting their target a-amylases (O’Donnell and McGeeney, 1976; Takase, 1994; Maskos et al., 1996; Titarenko and Chrispeels, 2000; Alam et al., 2001). Notably, RBI has been shown to bind to starch, which makes it unable to inhibit its target enzyme TMA (Alam et al., 2001). Among the characterized CTIs, LDI is the most potent. It binds its target enzyme, barley limit dextrinase, with a KD of 42 pM mainly owing to an extremely slow off rate (Møller et al., 2015). The complex formation between LDI and barley limit dextrinase is driven by a free energy change (ΔG° = –57 kJ/mol) originating from equally favorable entropy and enthalpy changes (Møller et al., 2015). Wheat 0.19 AI inhibited porcine pancreatic a-amylase (PPA) activity by an inhibition constant (Ki) of 57.3 nM, and the interaction was found to be endothermic and driven by a large increase in entropy (Oneda et al., 2004). Generally, CTIs are very stable proteins as inherent to their disulfide bonds connecting the four a-helix bundle (Figure 2A). The activation energy for the thermal inactivation of 0.19 AI was determined to be 87.0 kJ/mol, and T50, here the temperature causing 50% inactivation by 30 min incubation at pH 6.9, was 88.1°C (Oneda et al., 2004). RBI is stable in 8 M urea and 6 M guanidine-HCl. Notably, in 150 mM NaCl, thermal denaturation does not occur up to 90°C. However, RBI is irreversibly denatured in 5 mM NaCl if heated above 73°C. The acidic denaturation of RBI is reversible in both high and low salt conditions (Alagiri and Singh, 1993). LDI from barley is stable in the pH 2–12 range, and, at pH 6.5, its half-life is 53 and 33 min at 90 and 93 °C, respectively (Jensen et al., 2011). Furthermore, the melting temperature (Tm) is 97.4 °C at pH 6.5 and the unfolding is irreversible. Notably, the inhibitor had a stabilizing effect on its target enzyme, barley limit dextrinase. The free enzyme has a Tm of 65.9°C, while the Tm of the complex is 77.4 °C (Møller et al., 2015). It is not known how LDI is released from limit dextrinase in vivo, but in vitro studies have shown that LDI can be inactivated by barley thioredoxin-catalysed disulfide reduction resulting in conformational destabilization and loss of function. Furthermore, the destabilized structure is more susceptible to protease degradation (Jensen et al., 2012).
The four structure-determined members; LDI, RBI, wheat 0.19, and CHFI are among the best characterized CTIs (Table 1). Especially, the complex structures between LDI (Møller et al., 2015) and RBI (Strobl et al., 1998) and their respective target enzymes give unique insights into the structural basis for the function of CTIs from the different crop cereals. Besides these two published complex structures, the structure of the complex between TMA and the wheat 0.28 α-amylase inhibitor has been determined (Payan, 2004), but the coordinates of this structure have not been published. Prior to the determination of the structure of the complex between RBI and TMA, it was known that RBI contained two separate binding sites; one for α-amylase and one for protease (Maskos et al., 1996), and, moreover, that the N-terminal segment of the wheat 0.28 α-amylase inhibitor was crucial for its inhibitory activity (García-Maroto et al., 1991). In the RBI–TMA complex indeed, the N-terminal segment of RBI was a key element in the α-amylase binding site (Figure 2B), while a loop between two of the α-helices served as the protease-binding site (Strobl et al., 1998). Comparisons between the structures of free RBI (Protein Data Bank www.rcsb.org, PDB, entries 1B1U and 1BIP), solved both by NMR and X-ray crystallography, and RBI in complex with TMA (PDB entry 1TMQ) revealed that the N-terminal segment undergoes a conformational change upon complexation, adopting a 310-helix structure, whereas it is highly flexible in the free inhibitor (Strobl et al., 1998). Notably, the unpublished complex structure between the wheat 0.28 CM-protein and TMA was reported to show the same binding features (Payan, 2004). Lastly, the protease inhibition site of RBI is a canonical substrate-like conformational region (Gly32–Thr37) situated at the opposite side of the protein. Hence, RBI can bind an a-amylase and a protease simultaneously. The complex structure analysis between LDI and limit dextrinase (PDB entry 4CVW) displayed an unexpected binding mode in which, unlike the other characterized CTI-α-amylase complexes, the N-terminal region of LDI is not interacting with the target enzyme (Figure 2B) (Møller et al., 2015). This was also confirmed by LDI N-terminal truncations showing no influence on the inhibition of limit dextrinase (Møller et al., 2015). Moreover, site-directed mutagenesis established that a hydrophobic cluster situated on the second LDI α-helix flanked by ionic interactions at the protein-protein interface was important for the picomolar affinity of the enzyme complex (Møller et al., 2015). Furthermore, computer-guided thorough mutational analysis of the complex revealed that LDI–limit dextrinase intermolecular contacts as well as intramolecular interactions in LDI play a role for the ultra-high affinity (Møller et al., 2021). Remarkably, the inhibitor–enzyme complexation does not rely on an interface-centered hotspot constituted by a few residues, as in the case of the other CTI α-amylase inhibitors. Rather LDI residues across the protein interface contributed importantly to binding, hence making the complex more robust to mutational drift in evolution (Møller et al., 2021).
Among the CTIs targeting hydrolases, only LDI and RBI have been subjected to protein engineering attempts. The complex structure between LDI and limit dextrinase provided as mentioned above a starting point for rational and computer-guided engineering of LDI showing the potential of LDI as a backbone for engineering (Møller et al., 2015, 2021). Limit dextrinase plays a key role in malting and mashing together with other endogenous amylolytic enzymes in germinated barley seeds. Thus, LDI could be considered as unwanted because it inhibits limit dextrinase and hence decreases the degradation of starch to fermentable sugars, but as mentioned above LDI also protects limit dextrinase from thermal inactivation during mashing. Besides the engineering of LDI, it has been shown that a structure-guided point mutation in barley limit dextrinase, importantly reduced the affinity for LDI without affecting the activity of the enzyme (Møller et al., 2015). Alam et al. investigated N-terminal fragments and various peptides (7–11 residues) homologous to the N-terminal sequence of RBI for their potential to inhibit PPA. The peptides all inhibited PPA catalyzed hydrolysis of the substrate p-nitrophenyl-α-D-maltoside more weakly as compared to RBI. Notably, however, unlike RBI, these peptides did not interact with larger substrates like starch and actually exerted a clear competitive inhibition of the hydrolysis of starch by PPA, which confirmed the potential for design of simple a-amylase inhibitors (Alam et al., 2001).
The individual target enzyme specificities of CTIs towards digestive a-amylases from insects and mammals have motivated profiling of inhibitor contents for potential cultivar selection or enrichment by using gene editing of plants to reinforce their defense against primarily insect pests (Tsvetkov and Yarullina, 2019). A recent review addresses the inhibition of different insect a-amylases by plant proteinaceous inhibitors including CM-proteins from wheat, rye, corn and barley (da Lage, 2018). A more practical approach consists in application of artificial diets containing either recombinantly produced inhibitors or efficient plant fractions to reduce viability of herbivorous insects (Dias et al., 2005; da Silva et al., 2013; Sagu et al., 2021). For crops, the backside of this strategy can be negative consequences of higher levels of CTIs on human health and in feed for livestock. The awareness on NCWS is important (Geisslitz et al., 2021) and durum wheat recently has been gene edited to reduce CM3 and CM16 (Camerlengo et al., 2020). Notably, CM3 treatment reduced the lifespan of Drosophila melanogaster fruit flies and led to bacterial overgrow in their gut, which can also be seen in humans and leading to symptoms reminiscent to NCWS (Thiel et al., 2020). On a related note, it has been reported that sourdough fermentation can degrade wheat ATIs and reduce pro-inflammatory activity (Huang et al., 2020). Quantitation using targeted multi-reaction-monitoring LC-MS/MS of ATIs in the sourdough, during proofing, and after baking, respectively, demonstrated that ATI contents were much reduced by baking (Won et al., 2021). Interestingly, recently CM-proteins are suggested to slow starch digestion rate of cooked pasta (Zou et al., 2019) in line with previous reports of wheat amylase inhibitors reducing postprandial plasma glucose concentrations (Kodama et al., 2005; Ninomiya et al., 2018) and CM-protein inhibitors 0.19, 0.28 and 0.53 being shown to effectively inhibit human pancreatic α-amylase (HPA) secreted into the duodenum (Choudhury et al., 1996). In brewing, the barley CM-proteins have been associated with beer-haze formation (Ye et al., 2011) and foam stability to be significantly improved by BDAI-1 (Iimure et al., 2015). Thioredoxin reduction of disulphide bonds in CTIs may influence their activity (Wong et al., 2004; Jensen et al., 2012). Barley thioredoxin h preferably reduced different disulfides in the two CM-protein inhibitors monomeric and dimeric amylase inhibitors (BMAI and BDAI), which probably has no implications for the malting and mashing, as these are directed towards exogenous α-amylases (Maeda et al., 2004, 2005; Jensen et al., 2012). By contrast, the inactivating reduction of disulfides in LDI (Jensen et al., 2012) may play a role during mashing, as α-1,6-glucosidic bond hydrolysis by limit dextrinase will be able to occur to a greater extent.
In a few cases, the structure as well as biochemical and biophysical parameters or enzyme complexation have been determined providing a basis for rational engineering of CTIs to be directed towards specific enzymes. One area of interest would be to be able to engineer CTIs to control and arrest catalysis by new enzyme targets with selected activities in cocktails of liquefying, saccharifying and debranching enzymes of family GH13, used for productions of syrups and maltooligosaccharides from starch.
Another area of emerging application is breeding and gene editing for crops to lower CTI contents to avoid allergies and still maintain resistance against pathogens. In this context, it deserves mentioning that many open questions remain to the causative role of CTIs in NCWS pathophysiology triggered by our diets (Geisslitz et al., 2021). Towards improved understanding one may use antibodies raised against reombinantly produced individual CTIs (Tundo et al., 2018). The example of sourdough baking reducing ATIs (Won et al., 2021) draws attention to examination of various cooking and other food-processing practices as a way to further develop the management of ATI contents.
MSM and BS conceived the content of this review and wrote the manuscript.
The work described has been funded by Independent Research Fund Denmark | Technology and Production Sciences and Natural Sciences (DFF-1337-00158 and DFF-1335-00769 to MSM; and DFF-0602-01153B and DFF-6108-00476B to BS).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
AI, α-amylase inhibitor; ATI, α-amylase/trypsin inhibitor; BDAI, barley dimeric α-amylase inhibitor; BMAI, barley monomeric α-amylase inhibitor; CHFI, corn Hageman factor/α-amylase inhibitor; CM1 (2, 3, 16, 17), CM-protein 1 (2, 3, 16, 17) from wheat; CMa-e, CM-protein a-e from barley; CM-proteins, Chloroform:methanol soluble proteins; CTI, cereal type inhibitor; GH13, glycoside hydrolase family 13; HPA, human pancreatic α-amylase; HSA, human salivary α-amylase; LD, limit dextrinase; LDI, limit dextrinase inhibitor; NCWS, non-celiac wheat sensitivity; PDB, Protein Data Bank; PPA, porcine pancreatic α-amylase; RBI, ragi bifunctional α-amylase/trypsin inhibitor; TMA, Tenebrio molitor α-amylase; WCI, wheat chymotrypsin inhibitor
Aguieiras, M. C. L., Resende, L. M., Souza, T. A. M., Nagano, C. S., Chaves, R. P., Taveira, G. B., et al. (2021). Potent Anti-candida Fraction Isolated from Capsicum chinense Fruits Contains an Antimicrobial Peptide that Is Similar to Plant Defensin and Is Able to Inhibit the Activity of Different α-Amylase Enzymes. Probiotics Antimicro. Prot. 13, 862–872. doi:10.1007/s12602-020-09739-3
Alagiri, S., and Singh, T. P. (1993). Stability and Kinetics of a Bifunctional Amylase/trypsin Inhibitor. Biochim. Biophys. Acta (Bba) - Protein Struct. Mol. Enzymol. 1203, 77–84. doi:10.1016/0167-4838(93)90038-S
Alam, N., Gourinath, S., Dey, S., Srinivasan, A., and Singh, T. P. (2001). Substrate−Inhibitor Interactions in the Kinetics of α-Amylase Inhibition by Ragi α-amylase/Trypsin Inhibitor (RATI) and its Various N-Terminal Fragments. Biochemistry 40, 4229–4233. doi:10.1021/bi002537v
Altenbach, S. B., Vensel, W. H., and Dupont, F. M. (2011). The Spectrum of Low Molecular Weight Alpha-Amylase/protease Inhibitor Genes Expressed in the US Bread Wheat Cultivar Butte 86. BMC Res. Notes 4, 242. doi:10.1186/1756-0500-4-242
Barber, D., Sanchez-Monge, R., Mendez, E., Lazaro, A., Garcia-Olmedo, F., and Salcedo, G. (1986). New α-amylase and Trypsin Inhibitors Among the CM-Proteins of Barley (Hordeum vulgare). Biochim. Biophys. Acta (Bba) - Protein Struct. Mol. Enzymol. 869, 115–118. doi:10.1016/0167-4838(86)90318-3
Behnke, C. A., Yee, V. C., Trong, I. L., Pedersen, L. C., Stenkamp, R. E., Kim, S.-S., et al. (1998). Structural Determinants of the Bifunctional Corn Hageman Factor Inhibitor: X-ray Crystal Structure at 1.95 Å Resolution,. Biochemistry 37, 15277–15288. doi:10.1021/bi9812266
Blanco, F. J., Jimenez, M. A., Rico, M., Santoro, J., Herranz, J., and Nieto, J. L. (1991). Tendamistat (12-26) Fragment. NMR Characterization of Isolated β-Turn Folding Intermediates. Eur. J. Biochem. 200, 345–351. doi:10.1111/j.1432-1033.1991.tb16191.x
Bose, U., Byrne, K., Howitt, C. A., and Colgrave, M. L. (2019). Targeted Proteomics to Monitor the Extraction Efficiency and Levels of Barley α-amylase Trypsin Inhibitors that Are Implicated in Non-coeliac Gluten Sensitivity. J. Chromatogr. A 1600, 55–64. doi:10.1016/j.chroma.2019.04.043
Bose, U., Juhász, A., Broadbent, J. A., Byrne, K., Howitt, C. A., and Colgrave, M. L. (2020). Identification and Quantitation of Amylase Trypsin Inhibitors across Cultivars Representing the Diversity of Bread Wheat. J. Proteome Res. 19, 2136–2148. doi:10.1021/acs.jproteome.0c00059
Buonocore, V., Giardina, P., Parlamenti, R., Poerio, E., and Silano, V. (1984). Characterisation of Chicken Pancreas α-amylase Isozymes and Interaction with Protein Inhibitors from Wheat Kernel. J. Sci. Food Agric. 35, 225–232. doi:10.1002/jsfa.2740350216
Buonocore, V., Gramenzi, F., Pace, W., Petrucci, T., Poerio, E., and Silano, V. (1980). Interaction of Wheat Monomeric and Dimeric Protein Inhibitors with α-amylase from Yellow Mealworm (Tenebrio molitor L. Larva). Biochem. J. 187, 637–645. doi:10.1042/bj1870637
Call, L., Haider, E., D’Amico, S., Reiter, E., and Grausgruber, H. (2021). Synthesis and Accumulation of Amylase-Trypsin Inhibitors and Changes in Carbohydrate Profile during Grain Development of Bread Wheat (Triticum aestivum L.). BMC Plant Biol. 21, 113. doi:10.1186/s12870-021-02886-x
Camerlengo, F., Frittelli, A., Sparks, C., Doherty, A., Martignago, D., Larré, C., et al. (2020). CRISPR-Cas9 Multiplex Editing of the α-Amylase/Trypsin Inhibitor Genes to Reduce Allergen Proteins in Durum Wheat. Front. Sustain. Food Syst. 4, 104. doi:10.3389/fsufs.2020.00104
Capocchi, A., Athanassiou, C. G., Benelli, G., Muccilli, V., Kavallieratos, N. G., Cunsolo, V., et al. (2021). A New Monomeric α-amylase Inhibitor from the Tetraploid Emmer Wheat Is Mostly Active against Stored Product Pests. J. Pest Sci. doi:10.1007/s10340-021-01447-3
Capocchi, A., Muccilli, V., Cunsolo, V., Saletti, R., Foti, S., and Fontanini, D. (2013). A Heterotetrameric Alpha-Amylase Inhibitor from Emmer (Triticum dicoccon Schrank) Seeds. Phytochemistry 88, 6–14. doi:10.1016/j.phytochem.2012.12.010
Carbonero, P., and García-Olmedo, F. (1999). “A Multigene Family of Trypsin/α-Amylase Inhibitors from Cereals,” in Seed Proteins (Dordrecht: Springer Netherlands), 617–633. doi:10.1007/978-94-011-4431-5_26
Carvalho, A. d. O., and Gomes, V. M. (2009). Plant Defensins-Prospects for the Biological Functions and Biotechnological Properties. Peptides 30, 1007–1020. doi:10.1016/j.peptides.2009.01.018
Chen, M.-S., Feng, G., Zen, K. C., Richardson, M., Valdes-Rodriguez, S., Reeck, G. R., et al. (1992). α-Amylases from Three Species of Stored Grain Coleoptera and Their Inhibition by Wheat and Corn Proteinaceous Inhibitors. Insect Biochem. Mol. Biol. 22, 261–268. doi:10.1016/0965-1748(92)90063-K
Chong, G. L., and Reeck, G. R. (1987). Interaction of Trypsin, β-factor XIIa, and Plasma Kallikrein with a Trypsin Inhibitor Isolated from Barley Seeds: A Comparison with the Corn Inhibitor of Activated Hageman Factor. Thromb. Res. 48, 211–221. doi:10.1016/0049-3848(87)90418-X
Choudhury, A., Maeda, K., Murayama, R., and DiMagno, E. (1996). Character of a Wheat Amylase Inhibitor Preparation and Effects on Fasting Human Pancreaticobiliary Secretions and Hormones. Gastroenterology 111, 1313–1320. doi:10.1053/gast.1996.v111.pm8898646
Cuccioloni, M., Mozzicafreddo, M., Ali, I., Bonfili, L., Cecarini, V., Eleuteri, A. M., et al. (2016). Interaction between Wheat Alpha-Amylase/trypsin Bi-functional Inhibitor and Mammalian Digestive Enzymes: Kinetic, Equilibrium and Structural Characterization of Binding. Food Chem. 213, 571–578. doi:10.1016/j.foodchem.2016.07.020
da Lage, J.-L. (2018). The Amylases of Insects. Int. J. Insect Sci. 10, 117954331880478–14. doi:10.1177/1179543318804783
da Silva, F. C. V., do Nascimento, V. V., Machado, O. L. T., Pereira, L. d. S., Gomes, V. M., and de Oliveira Carvalho, A. (2018). Insight into the α-Amylase Inhibitory Activity of Plant Lipid Transfer Proteins. J. Chem. Inf. Model. 58, 2294–2304. doi:10.1021/acs.jcim.8b00540
di Francesco, A., Saletti, R., Cunsolo, V., Svensson, B., Muccilli, V., Vita, P. D., et al. (2020). Qualitative Proteomic Comparison of Metabolic and CM-like Protein Fractions in Old and Modern Wheat Italian Genotypes by a Shotgun Approach. J. Proteomics 211, 103530. doi:10.1016/j.jprot.2019.103530
di Maro, A., Farisei, F., Panichi, D., Severino, V., Bruni, N., Ficca, A. G., et al. (2011). WCI, a Novel Wheat Chymotrypsin Inhibitor: Purification, Primary Structure, Inhibitory Properties and Heterologous Expression. Planta 234, 723–735. doi:10.1007/s00425-011-1437-5
Dias, S. C., Franco, O. L., Magalhães, C. P., Oliveira-Neto, O. B. d., Laumann, R. A., Figueira, E. L. Z., et al. (2005). Molecular Cloning and Expression of an α-Amylase Inhibitor from Rye with Potential for Controlling Insect Pests. Protein J. 24, 113–123. doi:10.1007/s10930-004-1518-4
dos Santos, I. S., Carvalho, A. d. O., de Souza-Filho, G. A., do Nascimento, V. V., Machado, O. L. T., and Gomes, V. M. (2010). Purification of a Defensin Isolated from Vigna unguiculata Seeds, its Functional Expression in Escherichia coli, and Assessment of its Insect α-amylase Inhibitory Activity. Protein Expr. Purif. 71, 8–15. doi:10.1016/j.pep.2009.11.008
Drula, E., Garron, M.-L., Dogan, S., Lombard, V., Henrissat, B., and Terrapon, N. (2022). The Carbohydrate-Active Enzyme Database: Functions and Literature. Nucl. Acids Res. 50, D571–D577. doi:10.1093/nar/gkab1045
Feng, G.-H., Chen, M., Kramer, K. J., and Reeck, G. R. (1991). α-Amylase Inhibitors from rice: Fractionation and Selectivity towards, Insects, Mammalian and Bacterial α-amylases. Cereal Chem. 68, 516–521.
Franco, O. L., Melo, F. R., Mendes, P. A., Paes, N. S., Yokoyama, M., Coutinho, M. V., et al. (2005). Characterization of Two Acanthoscelides obtectus α-Amylases and Their Inactivation by Wheat Inhibitors. J. Agric. Food Chem. 53, 1585–1590. doi:10.1021/jf049343x
Franco, O. L., Rigden, D. J., Melo, F. R., and Grossi-de-Sá, M. F. (2002). Plant α-amylase Inhibitors and Their Interaction with Insect α-amylases. Eur. J. Biochem. 269, 397–412. doi:10.1046/j.0014-2956.2001.02656.x
Franco, O. L., Rigden, D. J., R. Melo, F., Bloch, C., Silva, C. P., and Grossi de Sá, M. F. (2000). Activity of Wheat α-amylase Inhibitors towards Bruchid α-amylases and Structural Explanation of Observed Specificities. Eur. J. Biochem. 267, 2166–2173. doi:10.1046/j.1432-1327.2000.01199.x
Gadge, P. P., Wagh, S. K., Shaikh, F. K., Tak, R. D., Padul, M. V., and Kachole, M. S. (2015). A Bifunctional α-amylase/trypsin Inhibitor from Pigeonpea Seeds: Purification, Biochemical Characterization and its Bio-Efficacy against Helicoverpa armigera. Pestic. Biochem. Physiol. 125, 17–25. doi:10.1016/j.pestbp.2015.06.007
García-Maroto, F., Carbonero, P., and García-Olmedo, F. (1991). Site-directed Mutagenesis and Expression in Escherichia coli of WMAI-1, a Wheat Monomeric Inhibitor of Insect α-amylase. Plant Mol. Biol. 17, 1005–1011. doi:10.1007/BF00037140
Garcia-Olmedo, F., Salcedo, G., Sanchez-Monge, R., Gómez, L., Royo, J., and Carbonero, P. (1987). “Plant Proteinaceous Inhibitors of Proteinases and α-Amylases,” in Oxford Surveys of Plant Molecular and Cell Biology (Oxford, United Kingdom: Oxford University Press), 275–334.
Gazza, L., Gazzelloni, G., Taddei, F., Latini, A., Muccilli, V., Alfieri, M., et al. (2016). The Starch-Bound Alpha-Amylase/trypsin-Inhibitors in Avena. Mol. Genet. Genomics 291, 2043–2054. doi:10.1007/s00438-016-1238-4
Geisslitz, S., Longin, C. F. H., Koehler, P., and Scherf, K. A. (2020). Comparative Quantitative LC-MS/MS Analysis of 13 Amylase/trypsin Inhibitors in Ancient and Modern Triticum Species. Sci. Rep. 10, 14570. doi:10.1038/s41598-020-71413-z
Geisslitz, S., Shewry, P., Brouns, F., America, A. H. P., Caio, G. P. I., Daly, M., et al. (2021). Wheat ATIs: Characteristics and Role in Human Disease. Front. Nutr. 8, 667370. doi:10.3389/fnut.2021.667370
Goff, D. J., and Kull, F. J. (1995). The Inhibition of Human Salivary α-Amylase by Type II α-Amylase Inhibitor from Triticum aestivum is Competitive, Slow and Tight-Binding. J. Enzyme Inhib. 9, 163–170. doi:10.3109/14756369509042815
Gourinath, S., Alam, N., Srinivasan, A., Betzel, C., and Singh, T. P. (2000). Structure of the Bifunctional Inhibitor of Trypsin and α-amylase from Ragi Seeds at 2.2 Å Resolution. Acta Crystallogr. D Biol. Cryst. 56, 287–293. doi:10.1107/S0907444999016601
Grosse‐Holz, F. M., and Hoorn, R. A. L. (2016). Juggling Jobs: Roles and Mechanisms of Multifunctional Protease Inhibitors in Plants. New Phytol. 210, 794–807. doi:10.1111/nph.13839
Guo, B., Luan, H., Lin, S., Lv, C., Zhang, X., and Xu, R. (2016). Comparative Proteomic Analysis of Two Barley Cultivars (Hordeum vulgare L.) with Contrasting Grain Protein Content. Front. Plant Sci. 7, 542. doi:10.3389/fpls.2016.00542
Gutierrez, C., Sanchez-Monge, R., Gomez, L., Ruiz-Tapiador, M., Castañera, P., and Salcedo, G. (1990). α-Amylase Activities of Agricultural Insect Pests Are Specifically Affected by Different Inhibitor Preparations from Wheat and Barley Endosperms. Plant Sci. 72, 37–44. doi:10.1016/0168-9452(90)90184-P
Huang, X., Schuppan, D., Rojas Tovar, L. E., Zevallos, V. F., Loponen, J., and Gänzle, M. (2020). Sourdough Fermentation Degrades Wheat Alpha-Amylase/trypsin Inhibitor (ATI) and Reduces Pro-inflammatory Activity. Foods 9, 943. doi:10.3390/foods9070943
Iimure, T., Kihara, M., Sato, K., and Ogushi, K. (2015). Purification of Barley Dimeric α-amylase Inhibitor-1 (BDAI-1) and Avenin-like Protein-A (ALP) from Beer and Their Impact on Beer Foam Stability. Food Chem. 172, 257–264. doi:10.1016/j.foodchem.2014.09.012
Iulek, J., Franco, O. L., Silva, M., Slivinski, C. T., Bloch, C., Rigden, D. J., et al. (2000). Purification, Biochemical Characterisation and Partial Primary Structure of a New α-amylase Inhibitor from Secale cereale (rye). Int. J. Biochem. Cell Biol. 32, 1195–1204. doi:10.1016/S1357-2725(00)00053-4
Jensen, J. M., Hägglund, P., Christensen, H. E. M., and Svensson, B. (2012). Inactivation of Barley Limit Dextrinase Inhibitor by Thioredoxin-Catalysed Disulfide Reduction. FEBS Lett. 586, 2479–2482. doi:10.1016/j.febslet.2012.06.009
Jensen, J. M., Vester-Christensen, M. B., Møller, M. S., Bønsager, B. C., Christensen, H. E. M., Hachem, M. A., et al. (2011). Efficient Secretory Expression of Functional Barley Limit Dextrinase Inhibitor by High Cell-Density Fermentation of Pichia pastoris. Protein Expr. Purif. 79, 217–222. doi:10.1016/j.pep.2011.04.009
Juge, N., and Svensson, B. (2006). Proteinaceous Inhibitors of Carbohydrate-Active Enzymes in Cereals: Implication in Agriculture, Cereal Processing and Nutrition. J. Sci. Food Agric. 86, 1573–1586. doi:10.1002/jsfa.2454
Juhász, J., Gáspári, Z., and Pongor, S. (2020). Structure and Oxidative Folding of AAI, the Major Alfa-Amylase Inhibitor from Amaranth Seeds. Front. Chem. 8, 180. doi:10.3389/fchem.2020.00180
Kneen, E., and Sandstedt, R. M. (1946). Distribution and General Properties of an Amylase Inhibitor in Cereals. Arch. Biochem. 9, 235–49. Available at: http://www.ncbi.nlm.nih.gov/pubmed/21015716.
Kodama, T., Miyazaki, T., Kitamura, I., Suzuki, Y., Namba, Y., Sakurai, J., et al. (2005). Effects of Single and Long-Term Administration of Wheat Albumin on Blood Glucose Control: Randomized Controlled Clinical Trials. Eur. J. Clin. Nutr. 59, 384–392. doi:10.1038/sj.ejcn.1602085
Korneeva, V. A., Trubetskov, M. M., Korshunova, A. V., Lushchekina, S. V., Kolyadko, V. N., Sergienko, O. V., et al. (2014). Interactions outside the Proteinase-Binding Loop Contribute Significantly to the Inhibition of Activated Coagulation Factor XII by its Canonical Inhibitor from Corn. J. Biol. Chem. 289, 14109–14120. doi:10.1074/jbc.M114.553735
Kumar, S., Singh, N., Mishra, B., Dube, D., Sinha, M., Singh, S. B., et al. (2010). Modulation of Inhibitory Activity of Xylanase - α-amylase Inhibitor Protein (XAIP): Binding Studies and crystal Structure Determination of XAIP- II from Scadoxus multiflorus at 1.2 Å Resolution. BMC Struct. Biol. 10, 41. doi:10.1186/1472-6807-10-41
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi:10.1093/molbev/msy096
Kusaba-Nakayama, M., Ki, M., Kawada, E., Sato, M., Ikeda, I., Mochizuki, T., et al. (2001). Intestinal Absorbability of Wheat Allergens, Subunits of a Wheat α-Amylase Inhibitor, Expressed by Bacteria. Biosci. Biotechnol. Biochem. 65, 2448–2455. doi:10.1271/bbb.65.2448
MacGregor, A. W., Macri, L. J., Schroeder, S. W., and Bazin, S. L. (1994). Purification and Characterisation of Limit Dextrinase Inhibitors from Barley. J. Cereal Sci. 20, 33–41. doi:10.1006/jcrs.1994.1042
MacGregor, E. A., Bazin, S. L., Ens, E. W., Lahnstein, J., Macri, L. J., Shirley, N. J., et al. (2000). Structural Models of Limit Dextrinase Inhibitors from Barley. J. Cereal Sci. 31, 79–90. doi:10.1006/jcrs.1999.0284
Maeda, K., Finnie, C., and Svensson, B. (2004). Cy5 Maleimide Labelling for Sensitive Detection of Free Thiols in Native Protein Extracts: Identification of Seed Proteins Targeted by Barley Thioredoxin H Isoforms. Biochem. J. 378, 497–507. doi:10.1042/bj20031634
Maeda, K., Finnie, C., and Svensson, B. (2005). Identification of Thioredoxin h Reducible Disulphides in Proteomes by Differential Labelling of Cysteines: Insight into Recognition and Regulation of Proteins in Barley Seeds by Thioredoxin h Proteomics 5, 1634–1644. doi:10.1002/pmic.200401050
Maeda, K., Takamori, Y., and Oka, O. (1982). Isolation and Properties of an α-Amylase Inhibitor (0.53) from Wheat (Triticum aestivum). Agric. Biol. Chem. 46, 2873–2875. doi:10.1080/00021369.1982.10865528
Mahoney, W. C., Hermodson, M. A., Jones, B., Powers, D. D., Corfman, R. S., and Reeck, G. R. (1984). Amino Acid Sequence and Secondary Structural Analysis of the Corn Inhibitor of Trypsin and Activated Hageman Factor. J. Biol. Chem. 259, 8412–8416. doi:10.1016/s0021-9258(17)39746-6
Maskos, K., Huber-Wunderlich, M., and Glockshuber, R. (1996). RBI, a One-Domain α-amylase/trypsin Inhibitor with Completely Independent Binding Sites. FEBS Lett. 397, 11–16. doi:10.1016/S0014-5793(96)01131-3
Micheelsen, P. O., Vévodová, J., de Maria, L., Østergaard, P. R., Friis, E. P., Wilson, K., et al. (2008). Structural and Mutational Analyses of the Interaction between the Barley α-Amylase/Subtilisin Inhibitor and the Subtilisin Savinase Reveal a Novel Mode of Inhibition. J. Mol. Biol. 380, 681–690. doi:10.1016/j.jmb.2008.05.034
Mills, E. N. C., Jenkins, J. A., Alcocer, M. J. C., and Shewry, P. R. (2004). Structural, Biological, and Evolutionary Relationships of Plant Food Allergens Sensitizing via the Gastrointestinal Tract. Crit. Rev. Food Sci. Nutr. 44, 379–407. doi:10.1080/10408690490489224
Møller, M. S., Olesen, S. V., and André, I. (2021). An Ultra‐high Affinity Protein-Protein Interface Displaying Sequence‐robustness. Protein Sci. 30, 1144–1156. doi:10.1002/pro.4080
Møller, M. S., Vester-Christensen, M. B., Jensen, J. M., Hachem, M. A., Henriksen, A., and Svensson, B. (2015). Crystal Structure of Barley Limit Dextrinase-Limit Dextrinase Inhibitor (LD-LDI) Complex Reveals Insights into Mechanism and Diversity of Cereal Type Inhibitors. J. Biol. Chem. 290, 12614–12629. doi:10.1074/jbc.M115.642777
Mundy, J., Svendsen, I., and Hejgaard, J. (1983). Barley α-amylase/subtilisin Inhibitor. I. Isolation and Characterization. Carlsberg Res. Commun. 48, 81–90. doi:10.1007/BF02906171
Ninomiya, K., Ina, S., Hamada, A., Yamaguchi, Y., Akao, M., Shinmachi, F., et al. (2018). Suppressive Effect of the α-Amylase Inhibitor Albumin from Buckwheat (Fagopyrum esculentum Moench) on Postprandial Hyperglycaemia. Nutrients 10, 1503. doi:10.3390/nu10101503
Oda, Y., Matsunaga, T., Fukuyama, K., Miyazaki, T., and Morimoto, T. (1997). Tertiary and Quaternary Structures of 0.19 α-Amylase Inhibitor from Wheat Kernel Determined by X-ray Analysis at 2.06 Å Resolution,. Biochemistry 36, 13503–13511. doi:10.1021/bi971307m
O’Donnell, M. D., and McGeeney, K. F. (1976). Purification and Properties of an α-amylase Inhibitor from Wheat. Biochim. Biophys. Acta Enzymol. 422, 159–169. doi:10.1016/0005-2744(76)90016-4
Oliveira-Neto, O. B., Batista, J. A. N., Rigden, D. J., Franco, O. L., Falcão, R., Fragoso, R. R., et al. (2003). Molecular Cloning of α-Amylases from Cotton Boll Weevil, Anthonomus grandis and Structural Relations to Plant Inhibitors: An Approach to Insect Resistance. J. Protein Chem. 22, 77–87. doi:10.1023/a:1023024012657
Oneda, H., Lee, S., and Inouye, K. (2004). Inhibitory Effect of 0.19 α-Amylase Inhibitor from Wheat Kernel on the Activity of Porcine Pancreas α-Amylase and its Thermal Stability. J. Biochem. 135, 421–427. doi:10.1093/jb/mvh050
Østergaard, O., Finnie, C., Laugesen, S., Roepstorff, P., and Svennson, B. (2004). Proteome Analysis of Barley Seeds: Identification of Major Proteins from Two-Dimensional Gels (pI 4-7). Proteomics 4, 2437–2447. doi:10.1002/pmic.200300753
Panwar, P., Verma, A. K., and Dubey, A. (2018). Purification, Developmental Expression, and In Silico Characterization of α-amylase Inhibitor from Echinochloa frumentacea. 3 Biotech. 8, 227. doi:10.1007/s13205-018-1260-9
Payan, F. (2004). Structural Basis for the Inhibition of Mammalian and Insect α-amylases by Plant Protein Inhibitors. Biochim. Biophys. Acta (Bba) - Proteins Proteomics 1696, 171–180. doi:10.1016/j.bbapap.2003.10.012
Pei, J., Tang, M., and Grishin, N. V. (2008). PROMALS3D Web Server for Accurate Multiple Protein Sequence and Structure Alignments. Nucleic Acids Res. 36, W30–W34. doi:10.1093/nar/gkn322
Perlikowski, D., Wiśniewska, H., Kaczmarek, J., Góral, T., Ochodzki, P., Kwiatek, M., et al. (2016). Alterations in Kernel Proteome after Infection with Fusarium culmorum in Two Triticale Cultivars with Contrasting Resistance to Fusarium Head Blight. Front. Plant Sci. 7, 1–10. doi:10.3389/fpls.2016.01217
Rane, A. S., Joshi, R. S., and Giri, A. P. (2020). Molecular Determinant for Specificity: Differential Interaction of α-amylases with Their Proteinaceous Inhibitors. Biochim. Biophys. Acta (Bba) - Gen. Subjects 1864, 129703. doi:10.1016/j.bbagen.2020.129703
Rehm, S., Han, S., Hassani, I., Sokocevic, A., Jonker, H. R. A., Engels, J. W., et al. (2009). The High Resolution NMR Structure of Parvulustat (Z-2685) from Streptomyces parvulus FH-1641: Comparison with Tendamistat from Streptomyces tendae 4158. ChemBioChem 10, 119–127. doi:10.1002/cbic.200800547
Reig-Otero, Y., Mañes, J., and Manyes, L. (2018). Amylase-Trypsin Inhibitors in Wheat and Other Cereals as Potential Activators of the Effects of Nonceliac Gluten Sensitivity. J. Med. Food 21, 207–214. doi:10.1089/jmf.2017.0018
Rodriguez-Loperena, M. A., Aragoncillo, C., Carbonero, P., and Garcia-Olmedo, F. (1975). Heterogeneity of Wheat Endosperm Proteolipids (CM Proteins). Phytochemistry 14, 1219–1223. doi:10.1016/S0031-9422(00)98598-4
Sagu, S. T., Landgräber, E., Henkel, I. M., Huschek, G., Homann, T., Bußler, S., et al. (2021). Effect of Cereal α-Amylase/Trypsin Inhibitors on Developmental Characteristics and Abundance of Digestive Enzymes of Mealworm Larvae (Tenebrio molitor L.). Insects 12, 454. doi:10.3390/insects12050454
Sagu, S. T., Landgräber, E., Rackiewicz, M., Huschek, G., and Rawel, H. (2020). Relative Abundance of Alpha-Amylase/trypsin Inhibitors in Selected Sorghum Cultivars. Molecules 25, 5982. doi:10.3390/molecules25245982
Sanchez-Monge, R., Gomez, L., Garcia-olmedo, F., and Salcedo, G. (1989). New Dimeric Inhibitor of Heterologous α-Amylases Encoded by a Duplicated Gene in the Short Arm of Chromosome 3B of Wheat (Triticum aestivum L.). Eur. J. Biochem. 183, 37–40. doi:10.1111/j.1432-1033.1989.tb14893.x
Silva, M. C. M. d., del Sarto, R. P., Lucena, W. A., Rigden, D. J., Teixeira, F. R., Bezerra, C. d. A., et al. (2013). Employing In Vitro Directed Molecular Evolution for the Selection of α-amylase Variant Inhibitors with Activity toward Cotton Boll Weevil Enzyme. J. Biotechnol. 167, 377–385. doi:10.1016/j.jbiotec.2013.07.016
Stahl, Y., Alexander, R. D., Coates, S., Bryce, J. H., Jenkinson, H. R., and Morris, P. C. (2007). The Barley Limit Dextrinase Inhibitor: Gene Expression, Protein Location and Interaction with 14-3-3 Protein. Plant Sci. 172, 452–461. doi:10.1016/j.plantsci.2006.10.008
Strobl, S., Maskos, K., Wiegand, G., Huber, R., Gomis-Rüth, F. X., and Glockshuber, R. (1998). A Novel Strategy for Inhibition of α-amylases: Yellow Meal Worm α-amylase in Complex with the Ragi Bifunctional Inhibitor at 2.5 Å Resolution. Structure 6, 911–921. doi:10.1016/S0969-2126(98)00092-6
Strobl, S., Muehlhahn, P., Bernstein, R., Wiltscheck, R., Maskos, K., Wunderlich, M., et al. (1995). Determination of the Three-Dimensional Structure of the Bifunctional α-Amylase/Trypsin Inhibitor from Ragi Seeds by NMR Spectroscopy. Biochemistry 34, 8281–8293. doi:10.1021/bi00026a009
Sun, Z., Lu, W., Liu, P., Wang, H., Huang, Y., Zhao, Y., et al. (2015). Isolation and Characterization of a Proteinaceous α-amylase Inhibitor AAI-CC5 from Streptomyces sp. CC5, and its Gene Cloning and Expression. Antonie van Leeuwenhoek 107, 345–356. doi:10.1007/s10482-014-0333-y
Svensson, B., Fukuda, K., Nielsen, P. K., and Bønsager, B. C. (2004). Proteinaceous α-amylase Inhibitors. Biochim. Biophys. Acta (Bba) - Proteins Proteomics 1696, 145–156. doi:10.1016/j.bbapap.2003.07.004
Takase, K. (1994). Site-Directed Mutagenesis Reveals Critical Importance of the Catalytic Site in the Binding of α-Amylase by Wheat Proteinaceous Inhibitor. Biochemistry 33, 7925–7930. doi:10.1021/bi00191a020
Thiel, A.-L., Ragab, M., Wagner, A. E., Divanovic, S., Derer, S., and Sina, C. (2020). Purification and Functional Characterization of the Chloroform/methanol-Soluble Protein 3 (CM3) from Triticum aestivum in Drosophila melanogaster. Front. Nutr. 7, 607937. doi:10.3389/fnut.2020.607937
Titarenko, E., and J. Chrispeels, M. (2000). cDNA Cloning, Biochemical Characterization and Inhibition by Plant Inhibitors of the α-amylases of the Western Corn Rootworm, Diabrotica virgifera virgifera. Insect Biochem. Mol. Biol. 30, 979–990. doi:10.1016/S0965-1748(00)00071-0
Tsvetkov, V. O., and Yarullina, L. G. (2019). Structural and Functional Characteristics of Hydrolytic Enzymes of Phytophagon Insects and Plant Protein Inhibitors (Review). Appl. Biochem. Microbiol. 55, 460–469. doi:10.1134/S0003683819050156
Tundo, S., Lupi, R., Lafond, M., Giardina, T., Larré, C., Denery-Papini, S., et al. (2018). Wheat ATI CM3, CM16 and 0.28 Allergens Produced in Pichia pastoris Display a Different Eliciting Potential in Food Allergy to Wheat ‡. Plants 7, 101. doi:10.3390/plants7040101
Tysoe, C., and Withers, S. G. (2018). Structural Dissection of Helianthamide Reveals the Basis of its Potent Inhibition of Human Pancreatic α-Amylase. Biochemistry 57, 5384–5387. doi:10.1021/acs.biochem.8b00825
Vallée, F., Kadziola, A., Bourne, Y., Juy, M., Rodenburg, K. W., Svensson, B., et al. (1998). Barley α-Amylase Bound to its Endogenous Protein Inhibitor BASI: crystal Structure of the Complex at 1.9 Å Resolution. Structure 6, 649–59. doi:10.1016/S0969-2126(98)00066-5
Wang, J., Yang, L., Zhao, X., Li, J., and Zhang, D. (2014). Characterization and Phylogenetic Analysis of Allergenic Tryp_alpha_amyl Protein Family in Plants. J. Agric. Food Chem. 62, 270–278. doi:10.1021/jf402463w
Won, S., Curtis, J., and Gänzle, M. (2021). LC‐MS/MS Quantitation of α‐amylase/trypsin Inhibitor CM3 and Glutathione during Wheat Sourdough Breadmaking. J. Appl. Microbiol. 1–10. doi:10.1111/jam.15346
Wong, J. H., Cai, N., Tanaka, C. K., Vensel, W. H., Hurkman, W. J., and Buchanan, B. B. (2004). Thioredoxin Reduction Alters the Solubility of Proteins of Wheat Starchy Endosperm: An Early Event in Cereal Germination. Plant Cell Physiol 45, 407–415. doi:10.1093/pcp/pch044
Ye, L., Dai, F., Qiu, L., Sun, D., and Zhang, G. (2011). Allelic Diversity of a Beer Haze Active Protein Gene in Cultivated and Tibetan Wild Barley and Development of Allelic Specific Markers. J. Agric. Food Chem. 59, 7218–7223. doi:10.1021/jf200419k
Zoccatelli, G., Dalla Pellegrina, C., Mosconi, S., Consolini, M., Veneri, G., Chignola, R., et al. (2007). Full-fledged Proteomic Analysis of Bioactive Wheat Amylase Inhibitors by a 3-D Analytical Technique: Identification of New Heterodimeric Aggregation States. Electrophoresis 28, 460–466. doi:10.1002/elps.200600348
Keywords: CM-proteins, proteinaceous inhibitor, enzyme complexes, binding constant, x-ray crystallography, insect pests, limit dextrinase, food and nutrition
Citation: Møller MS and Svensson B (2022) Structure, Function and Protein Engineering of Cereal-Type Inhibitors Acting on Amylolytic Enzymes. Front. Mol. Biosci. 9:868568. doi: 10.3389/fmolb.2022.868568
Received: 02 February 2022; Accepted: 08 March 2022;
Published: 25 March 2022.
Edited by:
Tian Liu, Dalian University of Technology, ChinaReviewed by:
Balakumaran Chandrasekar, Birla Institute of Technology and Science, IndiaCopyright © 2022 Møller and Svensson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Birte Svensson, YmlzQGJpby5kdHUuZGs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.