
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci., 16 March 2022
Sec. Cellular Biochemistry
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.855511
This article is part of the Research TopicGenetically Mobile Elements Repurposed by Nature and BiotechnologistsView all 10 articles
Inteins catalyze their removal from a host protein through protein splicing. Inteins that contain an additional site-specific endonuclease domain display genetic mobility via a process termed “homing” and thereby act as selfish DNA elements. We elucidated the crystal structures of two archaeal inteins associated with an active or inactive homing endonuclease domain. This analysis illustrated structural diversity in the accessory domains (ACDs) associated with the homing endonuclease domain. To augment homing endonucleases with highly specific DNA cleaving activity using the intein scaffold, we engineered the ACDs and characterized their homing site recognition. Protein engineering of the ACDs in the inteins illuminated a possible strategy for how inteins could avoid their extinction but spread via the acquisition of a diverse accessory domain.
Protein-splicing intervening sequences often include a homing endonuclease (HEN) domain, which is embedded within inteins containing the Hedgehog/INTein (HINT) domain (Perler, 1998). The HINT domain catalyzes the protein splicing reaction, whereas HEN domains often function independently of the HINT domain (Figure 1) (Derbyshire et al., 1997). Inteins are generally considered selfish genetic elements, frequently invading conserved coding sequences across many unicellular host organisms. In this scenario, inteins make use of homing endonuclease domains for efficient invasion by directing sequence insertion via horizontal gene transfer (HGT) initiated by DNA-strand breaks in intein-less host alleles (Figure 1B) (Barzel et al., 2011). HENs themselves are selfish genetic elements that exist free-standing (without intein or intron) or associated with inteins or introns, e.g., in group I introns (Dujon et al., 1989; Derbyshire and Belfort, 1998; Burt and Koufopanou, 2004). However, being an integral component of inteins enables HENs to invade coding sequences, which are usually more preserved than noncoding regions such as introns (Barzel et al., 2011). This association with the HINT domain becomes possible due to the unique autocatalytic protein splicing activity of inteins leading to self-removal from the host protein and ligation of the flanking protein sequences (Figure 1A). Through the association between the HINT and HEN, the latter benefits from a conserved homing environment while inteins take advantage of rapid dissemination across alleles in a given genome or population (Liu, 2000; Burt and Koufopanou, 2004; Barzel et al., 2011). Many HENs within inteins belong to the most diverse LAGLIDADG family with an extensive phylogenetic distribution (Dalgaard et al., 1997). LAGLIDADG homing endonucleases (LHEs) recognize about 14–40 bp pseudo palindromic or asymmetric target DNA sites (homing sites) and contain conserved LAGLIDADG motifs (Chevalier and Stoddard, 2001). The relatively long recognition sequence supposedly warrants high cleavage specificity, thereby reducing possible toxic effects to the host. Importantly, in contrast to most endonucleases, LHEs tolerate a certain degree of sequence variation within their homing site, an essential property for maintaining their propagation along evolutionary drifts (Argast et al., 1998).
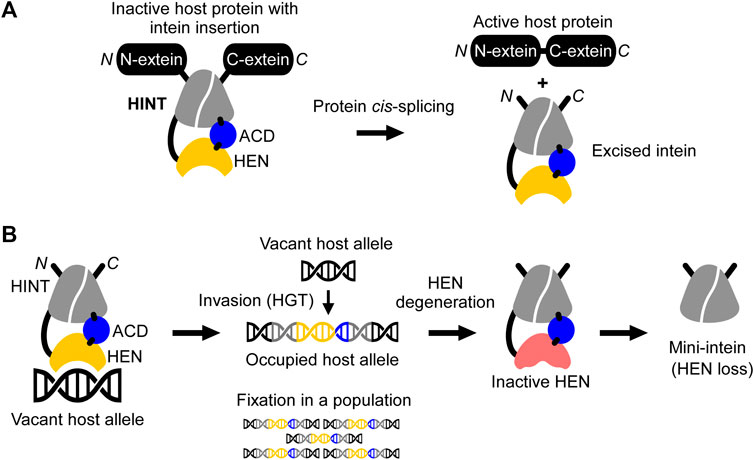
FIGURE 1. Schematic mechanisms of homing endonuclease (HEN) and Hedgehog/INTein (HINT) domains in inteins. (A) The HINT domain catalyzes self-excision of the intein (here consisting of HINT, HEN, and an accessory domain (ACD)) while covalently ligating the flanking extein sequences of the host protein during the protein cis-splicing reaction. (B) The nested HEN domain of the intein promotes gene conversion by introducing DNA double-strand breaks at the homing site into a vacant host allele followed by invasion via horizontal gene transfer (HGT) and fixation into the organism or population. Saturation of occupied alleles may cause HEN degeneration and loss.
During recent years, inteins have increasingly become important as a robust protein engineering platform thanks to their peptide-bond forming catalytic activity (Figure 1A) (Volkmann and Iwaï, 2010; Wood and Camarero, 2014). In particular, natural mini- and split inteins lacking HEN domains as well as the feasibility of splitting inteins into halves, have fostered this development (Southworth et al., 1998; Aranko et al., 2014). HEN-free mini-inteins are prevalent and have presumably emerged from HEN-associated inteins that have lost their HEN domain through size reduction (Barzel et al., 2011; Aranko et al., 2013; Novikova et al., 2016). According to the homing cycle model, HEN-less inteins may emerge after an intein has invaded and occupied all vacant alleles of a host population (“Fixation,” Figure 1B) (Burt and Koufopanou, 2004). After the fixation, HEN suffers target-site depletion and degeneration because HEN-associated inteins do not provide any benefits to host organisms, and the HEN activity is required only for invasion while protein-splicing activity is constantly selected by the production of active host proteins (Burt and Koufopanou, 2004; Barzel et al., 2011). Thus, degenerative mutations accumulate, eventually resulting in the loss of the HEN, thereby creating a mini-intein (Iwaï et al., 2017). To avoid the loss of HENs in inteins, some HEN domains might have developed a mutualism with HINT (Barzel et al., 2011; Iwaï et al., 2017). This mutualism emerged, albeit HEN and HINT were long thought as functionally independent, as seen with mini-inteins that lack HENs (Derbyshire et al., 1997). Artificially deleting HEN domains in several inteins impaired their protein splicing activity, suggesting that HEN domains, regardless of their nuclease activities, could assist in the protein splicing reaction of HINT. This domain interplay thereby might provide the selection to ensure the persistence of the HEN domain in inteins (Iwaï et al., 2017). Thus, structural and functional studies of HEN-associated inteins could shed light on the evolutionary history of individual inteins and contribute to the development of novel reagents as genomic and protein engineering tools.
In this study, we elucidated crystal structures of HEN-associated archaeal inteins inserted at the same insertion site (VMA-b), which is located within the A subunit P-loop of the vascular-type ATP synthase (VMA) from Thermococcus litoralis (Tli) and Pyrococcus horikoshii (Pho). The two three-dimensional structures highlighted a modular architecture consisting of HINT, HEN, and an accessory domain (ACD). The structures of the ACDs are diverse, even among the known three-dimensional structures of HEN-containing inteins. We further investigated the structural role of the ACD in DNA recognition of inteins by engineering the ACDs. These results suggest that the ACDs modulate DNA cleavages by the HEN-associated inteins. We speculate that acquiring a diverse ACD in HEN-associated inteins could be a general strategy to avoid their eventual extinction by promoting further spread into more distant insertion sites.
To understand the molecular evolution of inteins, we are interested in elucidating three-dimensional structures of various inteins with a presumable HEN domain. The first intein was identified as an intervening sequence within the yeast vacuolar membrane ATPase (VMA), subunit A (Hirata et al., 1990). The majority of inteins among eukaryotes reside at the highly conserved insertion site within the Vacuolar ATPase (VMA-a insertion site) (Swithers et al., 2009). The extensively investigated VMA intein from Saccharomyces cerevisiae (SceVMA) defines a proto-typical intein possessing homing endonuclease activity, also called PI-SceI, as a rare cutting DNA endonuclease (Grindl et al., 1998). Whereas yeast inteins are inserted at the highly conserved insertion site (VMA-a site), archaeal inteins commonly target a region approximately 20-residue downstream of the VMA-a insertion site (VMA-b insertion site), located at the P-loop motif of ATPases (Swithers et al., 2009). The VMA intein from P. horikoshii (PhoVMA) consists of 376 amino acids, which is considerably smaller than canonical HEN-associated inteins, e.g., SceVMA consisting of 454 residues but more similar to the size of the TFIIB intein from Methanococcus jannaschii (MjaTFIIB, 335 residues). The structure of the MjaTFIIB intein could previously not be determined together with the HEN domain by protein crystallography (Iwaï et al., 2017). Inteins share conserved amino acid sequence stretches designated as Blocks A-G (Pietrokovski, 1994; Perler et al., 1997) (Figure 2A). Blocks C, D, E, and H denote the HEN domain, out of which Blocks C and E represent the eponymous conserved LAGLIDADG helices bearing the acidic catalytic residues (Perler et al., 1997). The sequence alignment of the archaeal inteins also suggests that PhoVMA intein lacks homing endonuclease activity due to the absence of the active site residues in Blocks C and E (Figure 2B). We were successful in producing the PhoVMA intein and obtaining diffracting crystals. We solved the crystal structure of the PhoVMA intein at the 2.5 Å-resolution (Figure 2C; Supplementary Table S1). The crystal structure of PhoVMA intein revealed the typical HINT domain of thermophilic inteins, which contains a β-strand insertion and the HEN domain structure (Figure 2C) (Aranko et al., 2014). As expected from the primary structure, the PhoVMA intein lacks the presumed HEN active site residues in both usually conserved LAGLIDADG helices (Blocks C and E). It shows a partial truncation in Block E, hypothetically indicating progressive HEN degeneration (Figures 2A–C).
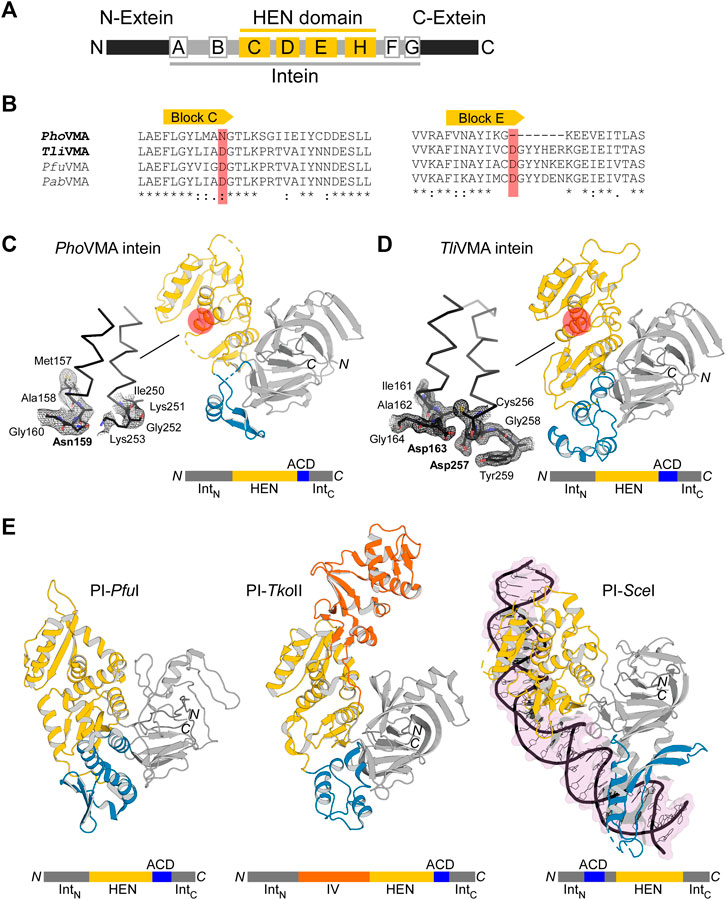
FIGURE 2. Structures of degenerated and active VMA inteins. (A) General domain organization and conservation in inteins. The HEN domain resides within the intein while the intein resides within a host protein (N- and C-exteins). Conserved sequence Blocks A–H are indicated. Host protein, black; intein, gray; HEN, yellow. (B) Sequence comparison around Blocks C and E corresponding to the active site-carrying LAGLIDADG helices of the HEN domains. Comparison of the intein orthologs of Pyrococcus horikoshii (PhoVMA), Thermococcus litoralis (TliVMA), Pyrococcus furiosus (PfuVMA), and Pyrococcus abyssi (PabVMA). The position of the catalytic aspartates in Blocks C and E are highlighted in red. (C) Crystal structure of PhoVMA intein. (D) Crystal structure of TliVMA intein. For (C,D), the locations of the active sites are highlighted by red circles. The close-ups of the active sites are shown to the left with electron density maps at 1.0 σ contour level. (E) The previously reported three crystal structures of PI-PfuI (PDB: 1dq3) (Ichiyanagi et al., 2000)(Ichiyanagi et al., 2000), PI-TkoII (PDB: 2cw7) (Matsumura et al., 2006), and PI-SceI (PDB: 1lws) (Moure et al., 2002). In (C–E), HINT, HEN, and ACD domains are colored in gray, yellow, and blue, respectively. PI-TkoII contains an additional domain IV indicated in orange. IntN and IntC indicate the N- and C-terminal parts of the HINT domain, which are separated by the HEN. The domain arrangement is schematically illustrated below each structure. N and C denote the termini.
The length of inteins considerably varies from 123 to >1,000 residues due to various insertions such as HENs and sequence deletions (Green et al., 2018). Large intein sequences generally indicate the presence of an active or inactive nested HEN. Therefore, we were interested in elucidating the structures of other VMA inteins inserted at the same VMA-b site to reveal a possible structural basis directing inteins of diverse sizes to the same target insertion site within host genomes. Among VMA inteins inserted at the VMA-b insertion site, we could obtain crystals of the VMA intein from Thermococcus litoralis (Tli). The TliVMA intein comprises 429 residues and is larger than the PhoVMA intein (376 residues) but similar to the size of PI-SceI (454 residues). To prevent self-cleavages during protein production, we expressed both inteins in E. coli with alanine substitutions of the catalytic cysteines 1 (Cys1). We used the N-terminal small ubiquitin-like modifier (SUMO) fusion to facilitate protein purification of PhoVMA intein (Guerrero et al., 2015). However, TliVMA intein required an N-terminal MBP fusion in addition to SUMO (H6-MBP-SUMO-TliVMA intein) for successful soluble expression (Guerrero et al., 2015). Unlike PhoVMA intein with the presumably degenerated HEN domain, TliVMA intein also required a high salt buffer composition, compensating for the lack of nucleic acids to maintain solubility after proteolytic removal of the fusion tag.
We solved the structure of TliVMA intein at the 1.6 Å-resolution (Figure 2D, Supplementary Table S1). The crystal structures of the TliVMA intein revealed a very similar overall structure as found in the PhoVMA intein, including the three-domain architecture known from the three other reported HEN-containing inteins (Figures 2C–E) (Duan et al., 1997; Ichiyanagi et al., 2000; Moure et al., 2002; Matsumura et al., 2006). The Hedgehog/Intein domains (HINT, gray) are composed of the N- and C-terminal fragments (IntN and IntC) with the HEN domains (yellow) inserted into the common intein split-site located between the two pseudo-two-fold symmetrical units forming a horseshoe-like fold common to all HINT domains (Eryilmaz et al., 2014; Iwaï et al., 2017). The HINT domain of the TliVMA intein also contains the β-strand extension found among thermophilic inteins (Figure 2C) (Aranko et al., 2014; Beyer et al., 2019; Hiltunen et al., 2021).
Not surprisingly, the HINT domains of PhoVMA andTliVMA inteins show a virtually identical three-dimensional structure with a 86% sequence identity (Figure 2; Supplementary Table S4). The HINT domains connect to the first of the two LAGLIDADG helices of the HEN domains via unstructured loops of 28–32 residues, located distant from the DNA-binding interfaces. We have identified “accessory domains” (ACD, shown in blue) residing between the HEN domains and the C-terminal part of the HINT domains, where we observed the most notable differences. The striking contrast between both inteins is the divergence of their ACDs, showing the least structural homology between the two molecules (33% pairwise sequence identity, Figures 2C,D; Supplementary Figure S3). The 53-residue difference in the lengths between the PhoVMA andTliVMA inteins can be mainly attributed to the difference in the ACDs. Even though ACDs at the intersections between HINT and HEN domain were identified in other reported HEN-associated inteins, their biological functions remain unclear (Figure 2E) (Duan et al., 1997; Ichiyanagi et al., 2000; Moure et al., 2002; Matsumura et al., 2006).
As for the nested HEN domains, the deletion in Block E in the PhoVMA intein (Figure 2B) causes a truncation of the second LAGLIDADG helix by one turn, thereby removing one of the catalytic aspartate residues (Figures 2B–D). Another obvious consequence of the degeneration in the PhoVMA intein appeared in the structure of the DNA-binding interfaces of the HEN mediated by two stretches of β-sheets, each originating from one copy of the two-fold pseudo symmetric LAGLIDADG elements (Supplementary Figures S1A,B) (Moure et al., 2002). The electrostatic surface potential of the HEN domains is very different between the two VMA inteins, which is in line with their binding to DNA fragments (see below).
Based on the three-dimensional structures, we deleted the HEN domain (residues 123-335 for PhoVMA intein and 123-388 for TliVMA intein) and connected with an “NG” sequence linker, resulting in 165-residue cis-splicing PhoVMAΔHEN and TliVMAΔHEN inteins. We modeled the structure of the two deletion variants with the RoseTTAFold software using the deep-learning algorithm (Supplementary Figures S1C,D) (Baek et al., 2021). Both structures appear identical with high confidence scores and an r.m.s.d. of 0.7 Å for 165 Cα atoms.
The PhoVMAΔHEN intein still retained the protein splicing activity, indicating that the HINT domain of PhoVMA intein is functionally independent of the nested HEN domain without having developed a mutualism (Supplementary Figure S1C) (Iwaï et al., 2017). However, the protein splicing activity of the TliVMAΔHEN intein largely diminished, presumably because of the mutualism developed between the HINT and HEN domains (Supplementary Figure S1D) (Iwaï et al., 2017). Even though the two three-dimensional structures are predicted to be almost identical to the original HINT domain (Supplementary Figures S1C,D), the HEN domain of TliVMA intein likely contributes to the protein splicing activity, as it has also been suggested for MvuTFIIB intein (Iwaï et al., 2017).
The primary structures and the three-dimensional crystal structures of the VMA inteins suggest that the HEN activity of PhoVMA intein has most likely degenerated during evolution and is inactive due to the lack of active site aspartate residues. However, TliVMA intein probably carries a catalytically active HEN domain capable of binding to DNA and introducing DNA double-strand breaks (Figures 2A,B).
To experimentally validate these assumptions, we performed in vitro DNA-binding and cleavage studies. First, we generated DNA substrates containing the theoretical homing sites of the inteins, that is, the coding DNA sequence of the Tli and Pho vma genes without the intein coding region (Figure 3A). These reconstructed intein-less DNA fragments should resemble the allelic situation before invasion by the inteins (Figure 3A). We generated 750-bp linear double-strand DNA fragments asymmetrically harboring the reconstituted homing site by amplifying their respective sequences from the genomic DNA by PCR and tested the cleavage of the DNA fragments by the inteins. Indeed, we observed that TliVMA intein cleaved its reconstituted homing site accompanied by a strong DNA binding affinity as indicated by an electrophoretic mobility shift (EMSA) of the substrate- and product-DNA fragments (Figure 3B). In contrast, PhoVMA intein was neither able to process its theoretical homing site, nor did it show any detectable DNA binding affinity (Figure 3C). Thus, the DNA substrates with the reconstituted homing sites validated our assumptions derived from the structures of Tli and Pho VMA inteins.
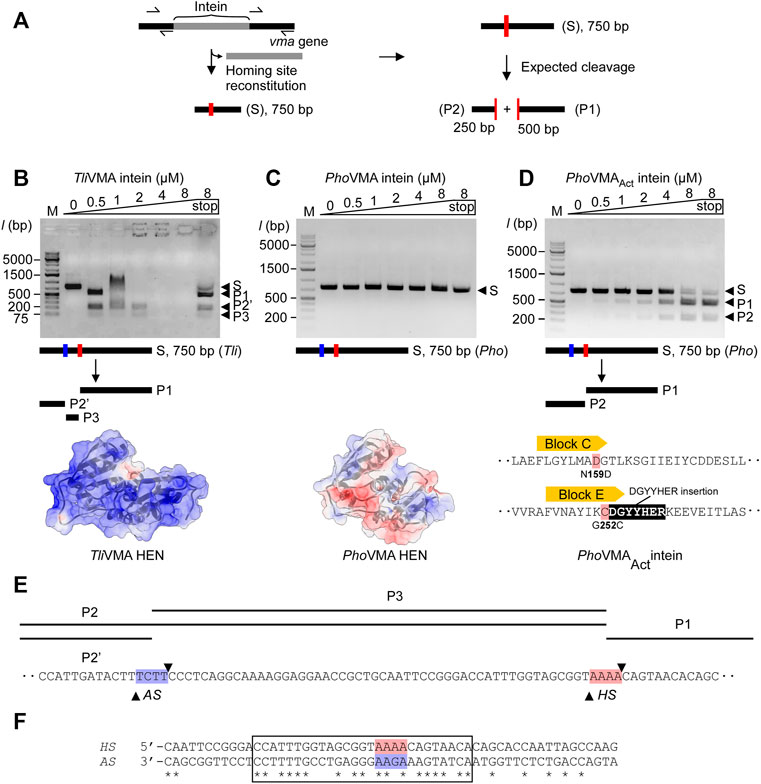
FIGURE 3. DNA-binding and cleavages of the theoretical homing sites by TliVMA and PhoVMA intein variants. (A) PCR-based construction of linear DNA substrates from Tli and Pho genomic DNA and expected cleavage pattern. The homing site was reconstituted by deleting the intein coding sequence from the vma gene while adjusting the size to 750 bp asymmetrically harboring the homing sites to generate the expected 250- and 500-bp cleavage products. (B) DNA-binding and cleavage of the 750-bp Tli DNA substrate by incubation with increasing concentrations of TliVMA intein at 80°C for 2 h. The electrostatic surface potential with an isoelectric point of 10.23 is shown below with a view of the DNA-binding interface. Positive, blue; neutral, white; negative, red. (C) Experiment as in (B) but using the 750-bp Pho DNA substrate and PhoVMA intein. The isoelectric point of the electrostatic surface potential is 5.45. The electrostatic surface potential model was generated using an alternative coordinate file without gaps in the HEN domain. (D) Activity test of the reactivated PhoVMAAct intein. Experiment as in (C) but using PhoVMAAct intein. The reconstitution of active site regions in the PhoVMA intein by grafting the sequence from Block C and E regions of the TliVMA intein is illustrated below. In panels (B–D), S, substrate; P1, 500-bp product, P2, 250-bp product, P2′, ∼200-bp product, P3, ∼50-bp product. M stands for the DNA ladder, “stop” indicates the addition of SDS-containing stop solution after incubation. (E) Distance of homing site (HS) and alternative site (AS) in the T. litoralis DNA substrate. Arrowheads indicate the positions where strand cleavage occurs. (F) The alignment of TliVMA intein homing (HS) and alternative (AS, reversed) sites. A region of 27 bp with high sequence similarity between the HS and the reversed AS is indicated by a box. The central four base pairs where cleavage occurs are highlighted in red (HS) and blue (AS).
We attributed the catalytic inactivity of the HEN in PhoVMA intein to the loss of presumed active-site residues. The differences in the electrostatic surface potential distributions of the HEN domains between Tli and Pho VMA inteins further might support the weaker DNA-binding affinity of PhoVMA compared with TliVMA intein (Figures 3B,C). Because the architecture of the HEN domain in PhoVMA intein was retained intact despite degenerative mutations and deletion, we wondered whether the nuclease activity of PhoVMA intein could be restored by protein engineering to reverse the evolutional process. To validate our hypothesis, we engineered the inactive PhoVMA intein by grafting the active sites in the LAGLIDADG helices from the sequences of the TliVMA intein (Figures 2B, 3D). Indeed, the engineered PhoVMA intein (PhoVMAAct intein) with the restored catalytic residues cleaved the DNA substrate containing the reconstituted homing site, albeit less efficiently not attaining the complete substrate digestion as observed with TliVMA intein (TliVMA intein, Figure 3B; PhoVMAAct intein, Figure 3D).
We designed and generated the DNA substrates for the DNA cleavage assay from Tli and Pho genomic DNA using PCR. Removing the intein coding sequences from the vma genes restored the theoretical homing site within a linear DNA of 750 bp containing the 250- and 500-bp fragments of the genomic sequences upstream and downstream of the reconstituted homing site, respectively (Figure 3A). The DNA cleavage at the homing site by the VMA inteins should produce 250- and 500-bp products. While the engineered PhoVMAAct intein produced the expected two fragments (Figure 3D), TliVMA intein exhibited an unexpected pattern of the products (Figure 3B). The disappearance of the DNA fragments at higher concentrations of TliVMA intein without SDS-treated denaturation is presumably due to the strong affinity to the DNA molecule (“end-holding”). Interestingly, besides the expected 500-bp fragment, a product of ∼200 bp and a third one shorter than 75 bp appeared with TliVMA intein. The analysis of the cleavage products by DNA sequencing revealed that PhoVMAAct intein cleaved precisely at the expected homing site (Supplementary Figure S2A).
In contrast, TliVMA intein cleaved at two different sites. One site was indeed at the theoretical homing site (HS) with the central four base pairs of the sequence 5′-AAAA-3′, while the other alternative site (AS) is located 52 bp upstream of the reconstituted homing site and contains the central sequence 5′-TCTT-3′ (Supplementary Figure S2B). We assume that recognition and cleavage of the AS by TliVMA intein occur on the opposite strand of the HS. The sequence on the opposite strand corresponds to the sequence of 5′-AAGA-3′, reminiscent of the reconstituted homing site of PhoVMAAct intein (Figures 3E,F; Supplementary Figure S2C) and bearing a single substitution to the central four base pairs of the homing site of TliVMA intein. Indeed, the alignment of the DNA sequence against the reverse strand of the alternative site revealed a striking 63% identity encompassing a 30 bp region surrounding the two cleavage sites (Figure 3F). Overall, the DNA substrates reconstituted from Tli and Pho genomic DNA have sufficient similarity to assume that both contain the AS next to the HS (Supplementary Figure S2C). However, PhoVMAAct intein could exclusively process the homing site (HS), leaving the AS unaffected (Figure 3D). We could conclude that the activated PhoVMAAct intein is more specific toward recognizing the reconstituted homing site (HS) despite its lower affinity.
The lengthy DNA sequences recognized by homing endonucleases (HENs) attracted protein engineering of HENs for genomic application because the high specificity of HENs could facilitate various in vitro and in vivo applications (Stoddard, 2011). However, the number of HENs that recognize different DNA sequences which could be used for broad applications is small. Although dozens of intein structures have been deposited to the protein data bank (PDB), only three of those contain a nested HEN domain. Moreover, exclusively the intein structure of PI-SceI from Saccharomyces cerevisiae was elucidated as the DNA/intein complex (Duan et al., 1997; Moure et al., 2002). The limited structural information of HEN-associated inteins hinders our understanding of inteins as site-specific DNA endonucleases, impeding further development of HEN-associated inteins by protein engineering as genetic engineering tools. Other reported HEN-associated intein structures are archaeal inteins from Thermococcus kodakaraensis (PI-TkoII) (Matsumura et al., 2006) and Pyrococcus furiosus (PI-PfuI) (Ichiyanagi et al., 2000). Just like the VMA inteins from T. litoralis and P. horikoshii, these inteins have an accessory domain (ACD) in addition to HINT and HEN domains (Figure 2D). Furthermore, in the case of PI-TkoII, an additional domain, termed domain IV, was reported (Matsumura et al., 2006).
It is believed that ACDs in HEN-associated inteins might generally contribute to interactions with DNA. For PI-SceI, where the ACD is referred to as DNA recognition region (DRR), this role has been demonstrated, although the location of the ACD in PI-SceI is different from other reported HEN-associated intein structures (Figures 2C–E; Supplementary Figure S3). The ACD (DDR) can be seen as an insertion into the HINT domain rather than a connection of HEN and HINT domains (Moure et al., 2002) (Figure 2E; Supplementary Figure S3). However, HENs also exist free-standing without being embedded in inteins or introns. They are known to be among the most sequence-specific endonucleases due to their relatively long sequence recognition motif (Chevalier and Stoddard, 2001). Some of such HENs do not contain ACDs. Thus, it remains elusive why some HEN-associate inteins require ACDs and cannot define sufficient DNA sequence specificity with their intrinsic DNA recognition capability.
To investigate the structural and functional roles of ACDs in HEN-associated inteins, we decided to delete the ACD region from the TliVMA intein based on the three-dimensional structure. We could also validate our deletion design by determining the crystal structure of the deletion variant, termed TliVMAΔACD intein (Figure 4A; Supplementary Table S1). The crystal structure of TliVMAΔACD intein confirmed that the deletion of the ACD did not influence the HINT and HEN domain folds, nor their relative orientation toward each other (Figure 4A). In the DNA cleavage and binding assays, TliVMAΔACD intein similarly cleaved the same substrate as the wild-type TliVMA intein did, albeit with reduced DNA-binding (Figures 3B, 4B). To our surprise, the deletion of the ACD from the TliVMA intein changed the cleavage profile. The substrate cleavage profile by TliVMAΔACD intein resembled that of the PhoVMAAct intein, producing two main products as opposed to three products generated by TliVMA intein (Figure 4C). The DNA sequencing chromatogram of the smaller cleavage product generated by TliVMAΔACD supported that TliVMAΔACD intein did not cleave the alternative site as observed for the wild-type TliVMA intein, similar to the digestion pattern of the PhoVMAAct intein (Figure 4F). Moreover, we found that TliVMA intein cleaved the reconstituted DNA substrate from the Pyrococcus horikoshii (Pho) genome at the alternative cleavage site (Figure 4E).
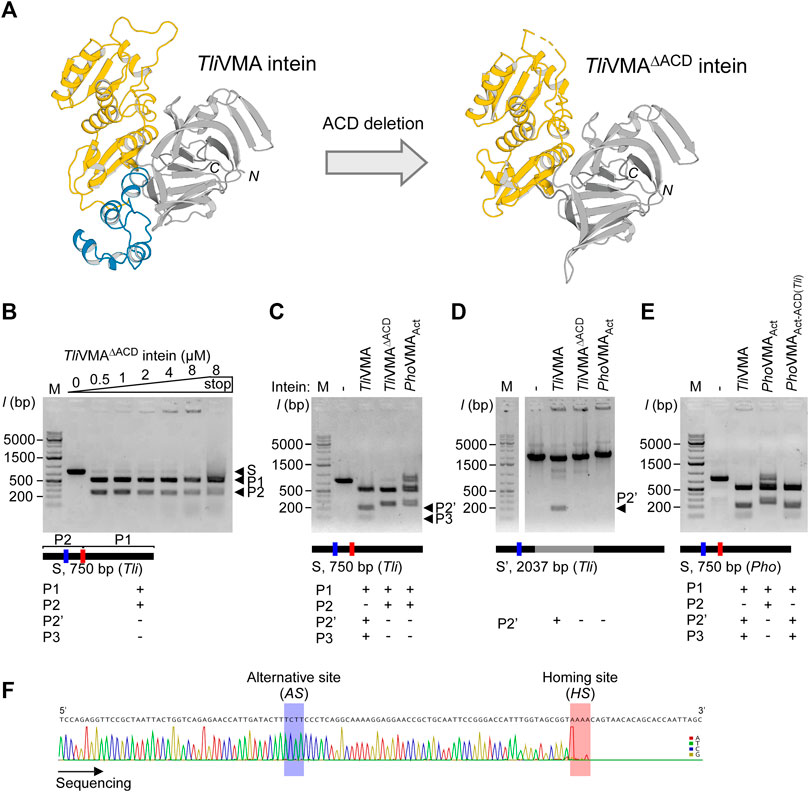
FIGURE 4. Deletion and grafting of the ACD in the TliVMA intein. (A) The crystal structures of TliVMA intein (left) and TliVMAΔACD intein (right) without ACD, connecting the HEN domain and the C-terminal part of the HINT domain. HEN, ACD, and HINT are colored in yellow, blue, and gray, respectively. (B) DNA-binding and cleavages of the reconstituted 750-bp DNA fragment with the homing site from Thermococcus litoralis genome (Tli) by TliVMAΔACD intein. The cleavages were analyzed on agarose gels after the incubation with increasing concentrations at 80°C for 2 h. (C) Comparison of DNA cleavages of the reconstituted 750-bp Tli DNA fragment by TliVMA, TliVMAΔACD, and PhoVMAAct inteins. (D) Comparison of the DNA cleavages of the 2037 bp DNA fragment, including the TliVMA intein coding sequence at the homing site by the three inteins as in C. This fragment lacked the reconstituted homing site (HS) due to the TliVMA intein coding sequence, hence only possessed the alternative site (AS). (E) DNA cleavages of the 750-bp Pho DNA fragment by the reactivated PhoVMAAct intein with the ACD from TliVMA intein (PhoVMAAct-ACD(Tli)) and the comparison with TliVMA and PhoVMAAct inteins. In (B–E), M stands for the DNA ladder, “stop” indicates the addition of SDS-containing stop solution after incubation. The migration height for the 750-bp substrate (S) and the 500 bp (P1) and 250 bp (P2) products are indicated in (B), P2’ (200 bp), and P3 (50 bp) are shown in (C,D). (F) The Sanger sequencing chromatogram of the 250-bp cleavage product generated by the TliVMAΔACD intein lacking the ACD.
Similarly, PhoVMAAct intein was able to digest the homing site within the Tli genome, albeit less efficiently as its cognate homing site (Figure 4C; Supplementary Figure S4). This cross-activity between TliVMA and PhoVMAAct inteins is presumably due to the close homology between the two substrate sequences created from Pho and Tli genome (Supplementary Figures S2C, S6A). The deletion of ACDs suggests that ACDs could play critical roles in increasing the cleavage specificity of HEN-associated inteins as well as making them more promiscuous by adding the capability to recognize an alternative cleavage site.
Next, we were interested in how the ACD in TliVMA intein influences the DNA recognition specificity. We speculated two possible scenarios: direct recognition of the alternative site sequence mediated by the ACD or indirect recognition via a cooperative binding effect. The binding of the intein to the homing site could guide the recognition of the alternative site separated by only 52 bp from the homing site by cooperative domain interaction with a second intein molecule involving the ACD. The DNA substrate containing only the alternative cleavage (AS) site (intein coding sequence remained inserted into the homing site) indicated that only TliVMA intein bearing the ACD was capable of digesting the DNA substrate, whereas TliVMAΔACD intein was not (Figure 4D). These results revealed that the TliVMA intein cleaved the alternative site (AS) independent of the homing site (HS) but depended on the presence of the ACD domain. We also performed DNA-binding tests using the isolated ACD domain of the TliVMA intein and revealed that the ACD seemingly does not contribute to the overall DNA affinity (Supplementary Figure S5).
To further validate the role of the ACD in TliVMA intein as a modulator of the DNA recognition responsible for the alternative site, we tested whether grafting of the ACD in PhoVMAAct intein from TliVMA intein would confer alternative site recognition. We thus created PhoVMAAct-ACD(Tli) intein having the grafted ACD from TliVMA intein (ACD (Tli)). Indeed, PhoVMAAct-ACD(Tli) could process both homing and alternative sites of the DNA substrate generated from the Pho genome, reminiscent of the cleavage profile produced by the TliVMA intein (Figure 4E). Furthermore, the swapping of the ACD rendered PhoVMAAct-ACD(Tli) intein more efficient in processing the DNA substrate without altering the apparent overall DNA affinity (Figure 3D; Supplementary Figure S6B). The weaker activity of PhoVMAAct-ACD(Tli) intein also allowed resolving a preferentiality of the homing site over the alternative site as the latter required a higher enzyme concentration (Supplementary Figure S6B).
The crystal structures of Tli and PhoVMA inteins, inserted at the same VMA-b insertion site of their host proteins, revealed a notable structural difference in their ACDs, largely deviating from each other (Figures 2C, 5A,B). The structural difference prompted us to investigate the functional role of ACDs. Our results demonstrated that the ACD in TliVMA intein induced a second cleavage site in addition to the theoretical homing site (Figure 4C). Interestingly, engineering the reactivated PhoVMA intein by grafting the ACD from TliVMA intein triggered cleavage at the alternative site (AS) adjacent to the homing site (HS), suggesting that the ACD is responsible for the cleavage at the AS (Figure 4E). The ACD of TliVMA intein strongly resembles the helix-turn-helix motif common for many DNA binding proteins, such as transcriptional regulator proteins (Figures 5A,B; Supplementary Table S5) (Anderson et al., 1981; Brennan and Matthews, 1989). The homology to DNA binding proteins suggests that ACDs mediate contacts with the DNA substrate.
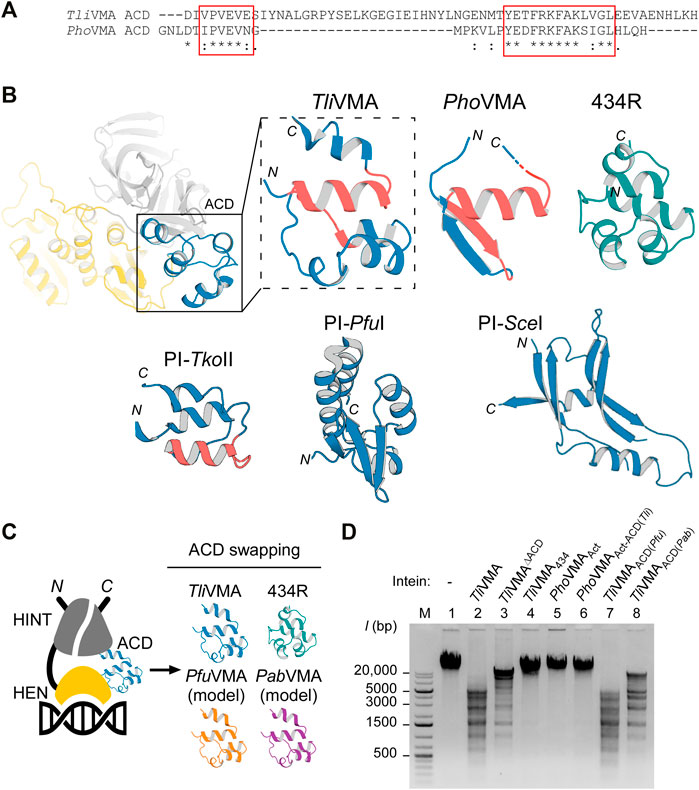
FIGURE 5. Effect of swapping ACDs in the VMA inteins on DNA recognition. (A) Primary structure comparison of ACDs in Tli and Pho VMA inteins. Regions with high similarity are highlighted with red rectangles. (B) The ACD structures of Tli and Pho VMA inteins and a comparison with the bacteriophage 434 repressor (434R, PDB: 2or1) (Aggarwal et al., 1988) and the ACDs of PI-TkoII (PDB: 2cw7), PI-PfuI (PDB: 1dq3), and PI-SceI (PDB: 1lws). The regions highlighted with red rectangles in (B) are colored in red. (C) Schematic illustration of the swapping experiments using different ACDs. The ACD in the TliVMA intein was replaced with the respective ACDs from the VMA intein of P. furiosus (Pfu), P. abyssi (Pab), or the 434-bacteriophage repressor domain. (D) DNA cleavages of λ-phage DNA using Tli and Pho VMA intein variants carrying different ACDs. Cleavage of λ-phage DNA was tested by overnight incubation with the indicated intein variants at 80°C. Agarose gel analysis of the digestion reactions. Lane 1, λ-phage DNA (48k bp) without intein; lane 2, the wild-type TliVMA intein; lane 3, TliVMA intein with deletion of ACD (TliVMAΔACD); lane 4, TliVMA intein with phage 434 repressor as ACD (TliVMA434); lane 5, reactivated PhoVMAAct intein; lane 6, the reactivated PhoVMA intein with ACD from TliVMA intein (PhoVMAAct-ACD(Tli)); lane 7, TliVMA with ACD from PfuVMA (TliVMAACD(Pfu)); lane 8, TliVMA intein with ACD from PabVMA (TliVMAACD(Pab)). M stands for the DNA size ladder.
Next, we wanted to test whether TliVMA intein is a promiscuous endonuclease and could cut unrelated substrates. We, therefore, tested digestion of λ-phage DNA by incubating overnight with TliVMA intein or TliVMAΔACD intein lacking the ACD (Figures 5C,D). To our surprise, we identified multiple cleavages in line with our observations using the model DNA substrates generated from T. litoralis genomic DNA. Furthermore, similar to our model DNA substrate, deletion of the ACD in TliVMA intein indeed reduced cleavage of λ-phage DNA, supporting our hypothesis that the ACD renders the intein endonuclease more promiscuous. In contrast, the activated PhoVMAAct intein with the endogenous ACD and the activated PhoVMAAct-ACD(Tli) with the grafted ACD from TliVMA intein (ACD (Tli)) did not produce any detectable λ-phage DNA cleavage, presumably due to the much lower affinity to the DNA substrate (Figures 3D, 4E, 5D).
We wondered how ACDs from other homologous inteins and an unrelated DNA-binding domain of the bacteriophage 434 repressor (434R) would affect cleavages of λ-phage DNA by TliVMA intein (Aggarwal et al., 1988). We engineered TliVMA intein by ACD-swapping the 434R domain and found that the engineered TliVMA intein decreased λ-phage DNA processing. However, we could still detect some extent of cleavages (Figure 5D). Replacing the ACD inTliVMA intein with an ACD from the more related inteins like VMA inteins from P. furiosus (PfuVMA) and P. abyssi (PabVMA) had a milder effect on the cleavage of λ-phage DNA (Figure 5D). Whereas the TliVMA intein variant carrying the ACD from PfuVMA intein (TliVMAACD(Pfu)) produced a restriction pattern very similar to the wild-type TliVMA intein, the variant with the ACD from Pab (TliVMAACD(Pab)) exhibited a less similar pattern (Figure 5D). The difference in the digestion profiles might arise from the fact that the ACD from PfuVMA intein has eight mutations, while the ACD from PabVMA intein contains 12-residue changes relative to the 55-residue region of the ACD in the TliVMA intein. Interestingly, replacing the ACD in the TliVMA intein with an unrelated DNA binding domain of phage 434R nearly abolished the cleavage of λ-phage DNA by the TliVMA intein (TliVMA434), indicating that grafting of 434R might disrupt the functional structure completely or create steric hindrances due to the poor protein engineering.
Homing endonucleases as rare cutting DNA endonucleases have sparked great interest in gene targeting and genome engineering (Stoddard, 2011). Currently, four classes of targetable DNA cleavage enzymes exist: zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), CRISPR/Cas RNA-guided nucleases (RGNs), and LAGLIDADG homing endonucleases (LHEs), the latter also termed “Meganucleases.” These enzymes can assist in targeted gene modification (Carroll, 2014). Engineering of rare cutting DNA endonucleases with novel desired recognition sites could open a myriad of in vitro and in vivo applications targeting specific DNA sequences. Whereas the modular architectures of TALENs and ZFNs facilitate their protein engineering attempts to recognize novel sequences (Maeder et al., 2008; Cermak et al., 2011), LAGLIDADG-type homing endonucleases (LHEs) have been the most difficult enzymes to engineer for altered DNA recognition (Taylor et al., 2012).
In this study, we determined the crystal structures of two archaeal inteins inserted at the same VMA-b site, revealing their molecular architecture consisting of HINT, HEN, and ACD. We found that the three-dimensional structures of ACDs were highly diverse among the five solved three-dimensional structures of inteins with nested HEN domains. Moreover, two ACDs from TliVMA intein and PI-TkoII resemble typical DNA-binding proteins containing the helix-turn-helix motif (Figures 2D,E). The modular structures of the HEN-containing inteins motivated us to engineer the nested HEN-associated inteins with altered DNA specificities for cleaving novel target sites by engineering the ACDs. We originally assumed that the presence of the ACD provided a higher specificity by additional DNA binding mediated by the ACD.
Contrary to our expectation, the deletion of the ACD from TliVMA intein and grafting of the ACD from TliVMA intein to PhoVMA intein indicated that the ACD enables recognizing an additional cleavage site (AS), thereby rendering the homing endonuclease domain more promiscuous (Figures 4C,E). However, grafting of ACDs from other archaeal VMA inteins and an unrelated phage DNA binding domain resulted in different digestion profiles of λ-phage DNA. Protein engineering of ACDs suggests the potential of HEN-associated inteins as a scaffold for creating novel meganucleases capable of recognizing novel target sites. Further detailed characterization of DNA recognition mechanisms by HEN-associated inteins could open the possibility to develop novel reagents with modulated DNA recognition specificities (Pâques and Duchateau, 2007; Carroll, 2014).
Inteins do not impact the host protein function because protein-splicing produces intact functional host proteins by self-excision of the inteins. Inteins, therefore, are found inserted into essential enzymes such as Vacuolar-type ATPase to ensure their selection. Abrogated inteins that accumulated mutations could result in inactive host proteins detrimental to the host organism. Therefore, protein splicing is required for the integrity of host proteins and establishes the selection. The homing endonuclease activity of inteins, however, is only required for invasion. Once the intein element occupies all target sites and is fixed in the population, the homing endonuclease activity degenerates and eventually becomes extinct, establishing the homing cycle (Figure 6A) (Burt and Koufopanou, 2004). In some inteins, HENs have developed a mutualism with HINT by making HINT dependent on the presence of the HEN scaffold for protein splicing (Iwaï et al., 2017). However, the mutualism between HINT and HEN could only slow down the eventual loss of HENs.
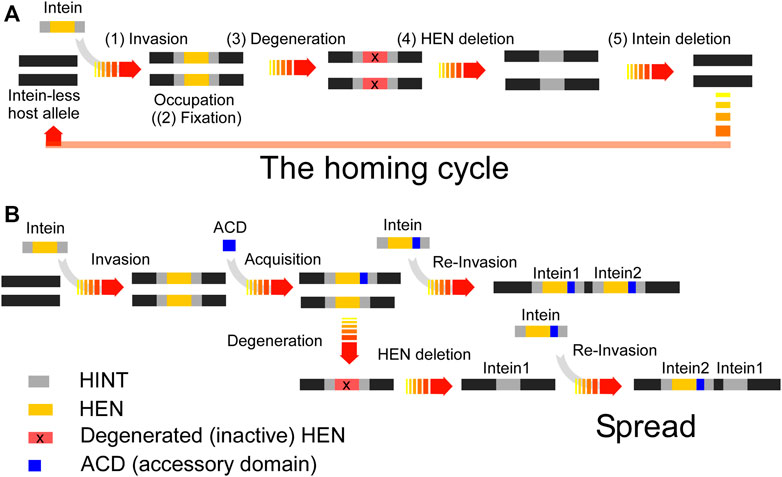
FIGURE 6. (A) The intein homing cycle model. The intein homing cycle model starts with the (1) Invasion of a HEN-containing intein via horizontal gene transfer followed by (2) Fixation into vacant alleles within the population. Depletion of HEN homing sites causes (3) Degeneration of the HEN due to accumulation of mutations tolerated by the lack of selection. Degenerated HENs are prone to (4) Deletion, rendering the intein incapable of competing with intein-free alleles, which might cause (5) Intein-loss upon interbreeding with strains providing an intein-free allele. (B) Intein spread model. Inteins might obtain an ACD modulating the HEN specificity, e.g., via changes in the ACD which can lower the HEN specificity to find novel insertion sites. Thus, the acquisition of a diverse ACD provides a spreading mechanism to prevent degeneration and extinction.
Our studies on ACDs in archaeal VMA inteins suggest that ACDs play an essential role in directing inteins to new alternative homing sites by acquiring diverse ACDs, presumably to avoid the extinction of HEN and HEN-associated inteins (Figure 6B). We hypothesize that HEN-associated inteins obtain an ACD from other genes such as transcription factors containing DNA-binding domains by yet unknown mechanisms to avoid the fixation (Figure 6A). The observed diversity in the structures of ACDs implies the divergent evolution and might support our hypothesis. Moreover, in nature, many genes host multiple inteins. For example, DNA polymerase from Thermococcus kodakaraensis hosts the two inteins PI-TkoI and PI-TkoII, separated by 85 amino acid residues in the host protein (Ichiyanagi et al., 2000). Cell division control protein 21 (CDC-21) in Pyrococcus abyssi also contains two mini-inteins separated by 48 amino-acid residues (Beyer et al., 2019). Thus, the prevalence of genes harboring multiple inteins in nature could support our hypothesis that inteins exploit ACDs for expanding the homing site to spread. However, there might still be other unknown advantages of having alternative cleavage sites by HEN-associated inteins (Figure 6B).
The structural basis of DNA recognition by HEN-associated inteins still awaits experimental elucidation of the high-resolution structure of DNA/inteins complexes. Such structural information of various HEN-associated inteins could shed light on the evolutionary histories of individual inteins and open a new avenue to develop a novel genetic engineering tool, which is smaller than RNA-guided nucleases for biotechnological applications.
All plasmids, oligonucleotides, and synthetic DNA substrate molecules used in this study are described in Supplementary Table S2. All recombinant proteins were produced in the E. coli strain T7 Express (New England Biolabs, USA). Expression details are given in Supplementary Table S3. All inteins carry a substitution of the catalytic cysteine 1 to alanine (C1A) to enable purification as fusion proteins except for those used in protein splicing tests. Residue numbering starts with 1 for this catalytic intein amino acid position and proceeds toward the C-terminus. Intein preceding residues are given negative indices.
Expression cultures were harvested by centrifugation at 4,700g for 10 min, 4°C. Pelleted cells from 1 or 2 L cultures were lysed in buffer A (50 mM sodium phosphate, pH 8.0, 300 mM NaCl) using continuous passaging through an EmulsiFlex-C3 homogenizer (Avestin, Canada) at 15,000 psi, 4°C for 10 min. Lysates were cleared by centrifugation at 38,000g for 60 min, 4°C. Proteins were purified in two steps using 5 ml HisTrap HP columns (GE Healthcare Life Sciences, USA) as previously described, including the removal of the hexahistidine tag and MBP and SUMO fusion domains (Guerrero et al., 2015).
After proteolytic removal of the fusion domains using Ubiquitin-like-specific protease 1 (UlpI), thermostable PhoVMAAct intein (pHBRSF067) and TliVMAΔACD intein (pHBRSF075) were heat fractionated at 80°C for 20 min before application to the Ni2+-NTA HisTrap column. After purification, all proteins were dialyzed overnight at 8°C against the following buffers: TliVMA intein (pHBRSF063) was first dialyzed against 10 mM Tris-HCl pH 8.0, resulting in precipitation of the target protein. Precipitated protein was resolubilized by the addition of 500 mM KCl followed by dialysis against 10 mM Tris-HCl pH 8.0, 400 mM KCl. TliVMAΔACD intein was treated like TliVMA intein, but 800 mM KCl was used for resolubilization, and the last step of dialysis was omitted. PhoVMA intein (pCARSF54) was dialyzed against deionized water. PhoVMAAct and PhoVMAAct-ACD(Tli) intein (pHBRSF079) were dialyzed against 10 mM Tris-HCl pH 8.0, 300 mM KCl. The isolated ACD of Tli (pHBRSF082) was dialyzed against 20 mM Tris-HCl pH 8.0, 300 mM KCl. The TliVMA intein variants containing exchanged ACDs (pHBRSF083, PfuVMA ACD; pHBRSF084, PabVMA ACD; pHBRSF161, 434-repressor domain) were purified like the TliVMA intein (pHBRSF063). After proteolytic removal of the H6-MBP-SUMO purification tag, sample and purification buffers were supplemented with 350 and 200 mM KCl, respectively. After dialysis against salt-free buffers, the three proteins were resolubilized by addition of 475, 600, and 700 mM KCl, respectively.
Proteins were subsequently concentrated using Macrosep® Advance Centrifugal Devices 10K MWCO (PALL). For enzymatic assays, proteins were diluted to 50 µM with 50 mM Tris-HCl pH 8.0, 300 mM KCl, 10% (v/v) glycerol, 1 mM dithiothreitol (DTT) and stored at −80°C for further use.
All diffracting crystals were obtained at room temperature using the sitting drop vapour diffusion method by mixing 100 nL concentrated PhoVMA, TliVMA, and TliVMAΔACD inteins with 100 nL of the following mother liquors: PhoVMA, (100 mM HEPES pH 8.0, 5 mM cadmium chloride, 5 mM magnesium chloride, 5 mM nickel (II) chloride, 10% (w/v) polyethylene glycol (PEG) 3,350); TliVMA, (100 mM magnesium formate, 15% (w/v) PEG 3350); TliVMAΔACD, (100 mM HEPES pH 7.5, 70% (v/v) 2-methyl-2,4-pentanediol (MPD)). Data were collected at beamline i04 (PhoVMA and TliVMAΔACD inteins) at Diamond Light Source (Didcot, United Kingdom) and beamline ID30A-1 (MASSIF-1, TliVMA intein) at ESRF (Grenoble, France) (Bowler et al., 2015). Data were processed using XDS (Kabsch, 2010). Structures were solved by molecular replacement (MR) starting from a PhoVMA intein search model generated using SWISS-MODEL (Arnold et al., 2006) with the intein homing endonuclease II of Thermococcus kodakarensis DNA polymerase (PDB: 2cw7) as a template. Initial models were obtained using the MR pipeline of Auto-Rickshaw (Panjikar et al., 2005) and ARP/wARP (Langer et al., 2008). Model-building and refinement were performed with COOT (Emsley et al., 2010) and PHENIX (Adams et al., 2002). The obtained structures were validated with Molprobity (Chen et al., 2010). Please refer to the Supporting Materials and Methods for a detailed description. Figures presenting three-dimensional coordinates were generated using PyMOL (The PyMOL Molecular Graphics System, Version 2.2.0, Schrödinger, LLC.). Electrostatic surface distributions were calculated and visualized using UCSF Chimera 1.13.1 (Pettersen et al., 2004) with ABS 1.3 (Jurrus et al., 2018) after model preparation with PDB2PQR 2.2.1 (Dolinsky et al., 2004) using a PARSE force field. Sequence alignments were performed with Clustal Omega 1.2.4. (Sievers et al., 2011).
If not indicated otherwise, electrophoretic mobility shift and enzymatic cleavage assays were performed in a total volume of 10 µL in 10 mM Tris-HCl pH 8.0, 100 mM KCl, 10 mM MgCl2, 1 mM DTT using 0.05 µM (750 bp), or 0.01 µM (2037 bp) linear dsDNA substrates containing the desired intein homing site (Supplementary Table S2). Typically, reactions contained 0.5 µM of TliVMA- or 8 µM of PhoVMA-derived inteins and were incubated at 80°C for 2 h. Where indicated, reactions were stopped by the addition of 1 µL endonuclease stop solution (5% (w/v) SDS, 250 mM EDTA, 100 mM Tris-HCl pH 7.5). Mobility shifts and cleavage products were visualized on 1.2% agarose gels.
For the determination of HEN cleavage sites, 1.5 µg of substrate DNA were digested overnight using the respective endonuclease with the above-described buffers, temperature, and concentrations. For the TliVMA intein, a stop solution was used to dissociate the HEN from the restriction products. Products were gel-purified and sequenced via Eurofins Genomics GmbH using the exterior oligonucleotides as used for the generation of the DNA substrates (Supplementary Table S2). For the digestion of λ-phage DNA, 1 µg substrate was incubated overnight with the indicated intein variants as described above.
Protein cis-splicing of intein variants was tested by expressing the indicated intein variants flanked by two B1 domains of the IgG-binding protein G in 5 ml cultures of E. coli strain T7 Express (New England Biolabs, USA) and purified using immobilized metal affinity chromatography as described elsewhere (Beyer et al., 2020). The used plasmids are listed in Supplementary Table S2. The experiments were performed at 30–37°C and the expression period lasted 3–4 h. Protein splicing was analyzed by SDS-PAGE using 16.5% gels and Coomassie Blue staining.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material. Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data bank under accession numbers 7QSS, 7QST, and 7QSU for TliVMA intein (C1A), PhoVMA intein (C1A), and TliVMAΔACD intein, respectively.
HI and HB conceptualized the project and contributed to the design of the studies. HB established and produced the recombinant proteins and performed the assays. HB and HI solved the structures. HB wrote the first draft of the manuscript. All authors contributed to the manuscript revision and approved the submitted version.
This work is supported by grants from the Novo Nordisk foundation (NNF17OC0025402 to HB, NNF17OC0027550 to HI), the Academy of Finland (137995, 277335 to HI), and the “Freigeist” fellowship of the Volkswagen Foundation to HB. Helsinki University Library supported funding for the open access charge.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank S. M. Backlund and C. Albert for technical assistance and K. M. Mikula and the crystallization facility for crystallization and data collections. We acknowledge the Finnish Block Allocation Group for access to the synchrotron light sources.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.855511/full#supplementary-material
Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., et al. (2002). PHENIX: Building New Software for Automated Crystallographic Structure Determination. Acta Crystallogr. D Biol. Cryst. 58 (11), 1948–1954. doi:10.1107/s0907444902016657
Aggarwal, A. K., Rodgers, D. W., Drottar, M., Ptashne, M., and Harrison, S. C. (1988). Recognition of a DNA Operator by the Repressor of Phage 434: a View at High Resolution. Science 242 (4880), 899–907. doi:10.1126/science.3187531
Anderson, W. F., Ohlendorf, D. H., Takeda, Y., and Matthews, B. W. (1981). Structure of the Cro Repressor from Bacteriophage λ and its Interaction with DNA. Nature 290 (5809), 754–758. doi:10.1038/290754a0
Aranko, A. S., Oeemig, J. S., Kajander, T., and Iwaï, H. (2013). Intermolecular Domain Swapping Induces Intein-Mediated Protein Alternative Splicing. Nat. Chem. Biol. 9 (10), 616–622. doi:10.1038/nchembio.1320
Aranko, A. S., Wlodawer, A., and Iwaï, H. (2014). Nature's Recipe for Splitting Inteins. Protein Eng. Des. Selection 27 (8), 263–271. doi:10.1093/protein/gzu028
Argast, G. M., Stephens, K. M., Emond, M. J., and Monnat, R. J. (1998). I- Ppo I and I- Cre I Homing Site Sequence Degeneracy Determined by Random Mutagenesis and Sequential In Vitro Enrichment 1 1Edited by G. Smith. J. Mol. Biol. 280 (3), 345–353. doi:10.1006/jmbi.1998.1886
Arnold, K., Bordoli, L., Kopp, J., and Schwede, T. (2006). The SWISS-MODEL Workspace: a Web-Based Environment for Protein Structure Homology Modelling. Bioinformatics 22 (2), 195–201. doi:10.1093/bioinformatics/bti770
Baek, M., DiMaio, F., Anishchenko, I., Dauparas, J., Ovchinnikov, S., Lee, G. R., et al. (2021). Accurate Prediction of Protein Structures and Interactions Using a Three-Track Neural Network. Science 373 (6557), 871–876. doi:10.1126/science.abj8754
Barzel, A., Naor, A., Privman, E., Kupiec, M., and Gophna, U. (2011). Homing Endonucleases Residing within Inteins: Evolutionary Puzzles Awaiting Genetic Solutions. Biochem. Soc. Trans. 39 (1), 169–173. doi:10.1042/bst0390169
Beyer, H. M., Mikula, K. M., Kudling, T. V., and Iwaï, H. (2019). Crystal Structures of CDC21-1 Inteins from Hyperthermophilic Archaea Reveal the Selection Mechanism for the Highly Conserved Homing Endonuclease Insertion Site. Extremophiles 23 (6), 669–679. doi:10.1007/s00792-019-01117-4
Beyer, H. M., Mikula, K. M., Li, M., Wlodawer, A., and Iwaï, H. (2020). The crystal Structure of the Naturally Split Gp41‐1 Intein Guides the Engineering of Orthogonal Split Inteins Fromcis‐splicing Inteins. FEBS J. 287 (9), 1886–1898. doi:10.1111/febs.15113
Bowler, M. W., Nurizzo, D., Barrett, R., Beteva, A., Bodin, M., Caserotto, H., et al. (2015). MASSIF-1: a Beamline Dedicated to the Fully Automatic Characterization and Data Collection from Crystals of Biological Macromolecules. J. Synchrotron Radiat. 22 (6), 1540–1547. doi:10.1107/s1600577515016604
Brennan, R. G., and Matthews, B. W. (1989). The helix-turn-helix DNA Binding Motif. J. Biol. Chem. 264 (4), 1903–1906. doi:10.1016/s0021-9258(18)94115-3
Burt, A., and Koufopanou, V. (2004). Homing Endonuclease Genes: the Rise and Fall and Rise Again of a Selfish Element. Curr. Opin. Genet. Dev. 14 (6), 609–615. doi:10.1016/j.gde.2004.09.010
Carroll, D. (2014). Genome Engineering with Targetable Nucleases. Annu. Rev. Biochem. 83, 409–439. doi:10.1146/annurev-biochem-060713-035418
Cermak, T., Doyle, E. L., Christian, M., Wang, L., Zhang, Y., Schmidt, C., et al. (2011). Efficient Design and Assembly of Custom TALEN and Other TAL Effector-Based Constructs for DNA Targeting. Nucleic Acids Res. 39 (12), e82. doi:10.1093/nar/gkr218
Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., et al. (2010). MolProbity: All-Atom Structure Validation for Macromolecular Crystallography. Acta Crystallogr. D Biol. Cryst. 66 (1), 12–21. doi:10.1107/s0907444909042073
Chevalier, B. S., and Stoddard, B. L. (2001). Homing Endonucleases: Structural and Functional Insight into the Catalysts of Intron/intein Mobility. Nucleic Acids Res. 29 (18), 3757–3774. doi:10.1093/nar/29.18.3757
Dalgaard, J., Klar, A. J., Moser, M. J., Holley, W. R., Chatterjee, A., and Mian, I. S. (1997). Statistical Modeling and Analysis of the LAGLIDADG Family of Site- Specific Endonucleases and Identification of an Intein that Encodes a Site-specific Endonuclease of the HNH Family. Nucleic Acids Res. 25 (22), 4626–4638. doi:10.1093/nar/25.22.4626
Derbyshire, V., and Belfort, M. (1998). Lightning Strikes Twice: Intron-Intein Coincidence. Proc. Natl. Acad. Sci. 95 (4), 1356–1357. doi:10.1073/pnas.95.4.1356
Derbyshire, V., Wood, D. W., Wu, W., Dansereau, J. T., Dalgaard, J. Z., and Belfort, M. (1997). Genetic Definition of a Protein-Splicing Domain: Functional Mini-Inteins Support Structure Predictions and a Model for Intein Evolution. Proc. Natl. Acad. Sci. 94 (21), 11466–11471. doi:10.1073/pnas.94.21.11466
Dolinsky, T. J., Nielsen, J. E., McCammon, J. A., and Baker, N. A. (2004). PDB2PQR: an Automated Pipeline for the Setup of Poisson-Boltzmann Electrostatics Calculations. Nucleic Acids Res. 32 (Web Server issue), W665–W667. doi:10.1093/nar/gkh381
Duan, X., Gimble, F. S., and Quiocho, F. A. (1997). Crystal Structure of PI-SceI, a Homing Endonuclease with Protein Splicing Activity. Cell 89 (4), 555–564. doi:10.1016/s0092-8674(00)80237-8
Dujon, B., Beifort, M., Butow, R. A., Jacq, C., Lemieux, C., Perlman, P. S., et al. (1989). Mobile Introns: Definition of Terms and Recommended Nomenclature. Gene 82 (1), 115–118. doi:10.1016/0378-1119(89)90035-8
Emsley, P., Lohkamp, B., Scott, W. G., and Cowtan, K. (2010). Features and Development of Coot. Acta Crystallogr. D Biol. Cryst. 66 (4), 486–501. doi:10.1107/s0907444910007493
Eryilmaz, E., Shah, N. H., Muir, T. W., and Cowburn, D. (2014). Structural and Dynamical Features of Inteins and Implications on Protein Splicing. J. Biol. Chem. 289 (21), 14506–14511. doi:10.1074/jbc.r113.540302
Green, C. M., Novikova, O., and Belfort, M. (2018). The Dynamic Intein Landscape of Eukaryotes. Mobile DNA 9, 4. doi:10.1186/s13100-018-0111-x
Grindl, W., Wende, W., Pingoud, V., and Pingoud, A. (1998). The Protein Splicing Domain of the Homing Endonuclease PI-sceI Is Responsible for Specific DNA Binding. Nucleic Acids Res. 26 (8), 1857–1862. doi:10.1093/nar/26.8.1857
Guerrero, F., Ciragan, A., and Iwaï, H. (2015). Tandem SUMO Fusion Vectors for Improving Soluble Protein Expression and Purification. Protein Expr. Purif. 116, 42–49. doi:10.1016/j.pep.2015.08.019
Hiltunen, M. K., Beyer, H. M., and Iwaï, H. (2021). Mini-Intein Structures from Extremophiles Suggest a Strategy for Finding Novel Robust Inteins. Microorganisms 9 (6), 1226. doi:10.3390/microorganisms9061226
Hirata, R., Ohsumk, Y., Nakano, A., Kawasaki, H., Suzuki, K., and Anraku, Y. (1990). Molecular Structure of a Gene, VMA1, Encoding the Catalytic Subunit of H(+)-translocating Adenosine Triphosphatase from Vacuolar Membranes of Saccharomyces cerevisiae. J. Biol. Chem. 265 (12), 6726–6733. doi:10.1016/s0021-9258(19)39210-5
Ichiyanagi, K., Ishino, Y., Ariyoshi, M., Komori, K., and Morikawa, K. (2000). Crystal Structure of an Archaeal Intein-Encoded Homing Endonuclease PI-PfuI. J. Mol. Biol. 300 (4), 889–901. doi:10.1006/jmbi.2000.3873
Iwaï, H., Mikula, K. M., Oeemig, J. S., Zhou, D., Li, M., and Wlodawer, A. (2017). Structural Basis for the Persistence of Homing Endonucleases in Transcription Factor IIB Inteins. J. Mol. Biol. 429 (24), 3942–3956. doi:10.1016/j.jmb.2017.10.016
Jurrus, E., Engel, D., Star, K., Monson, K., Brandi, J., Felberg, L. E., et al. (2018). Improvements to the APBS Biomolecular Solvation Software Suite. Protein Sci. 27 (1), 112–128. doi:10.1002/pro.3280
Kabsch, W. (2010). XDS. Acta Crystallogr. D Biol. Cryst. 66 (2), 125–132. doi:10.1107/s0907444909047337
Langer, G., Cohen, S. X., Lamzin, V. S., and Perrakis, A. (2008). Automated Macromolecular Model Building for X-ray Crystallography Using ARP/wARP Version 7. Nat. Protoc. 3 (7), 1171–1179. doi:10.1038/nprot.2008.91
Liu, X.-Q. (2000). Protein-splicing Intein: Genetic Mobility, Origin, and Evolution. Annu. Rev. Genet. 34, 61–76. doi:10.1146/annurev.genet.34.1.61
Maeder, M. L., Thibodeau-Beganny, S., Osiak, A., Wright, D. A., Anthony, R. M., Eichtinger, M., et al. (2008). Rapid "Open-Source" Engineering of Customized Zinc-finger Nucleases for Highly Efficient Gene Modification. Mol. Cel 31 (2), 294–301. doi:10.1016/j.molcel.2008.06.016
Matsumura, H., Takahashi, H., Inoue, T., Yamamoto, T., Hashimoto, H., Nishioka, M., et al. (2006). Crystal Structure of Intein Homing Endonuclease II Encoded in DNA Polymerase Gene from Hyperthermophilic Archaeon Thermococcus Kodakaraensis Strain KOD1. Proteins 63 (3), 711–715. doi:10.1002/prot.20858
Moure, C. M., Gimble, F. S., and Quiocho, F. A. (2002). Crystal Structure of the Intein Homing Endonuclease PI-SceI Bound to its Recognition Sequence. Nat. Struct. Biol. 9 (10), 764–770. doi:10.1038/nsb840
Novikova, O., Jayachandran, P., Kelley, D. S., Morton, Z., Merwin, S., Topilina, N. I., et al. (2016). Intein Clustering Suggests Functional Importance in Different Domains of Life. Mol. Biol. Evol. 33 (3), 783–799. doi:10.1093/molbev/msv271
Panjikar, S., Parthasarathy, V., Lamzin, V. S., Weiss, M. S., and Tucker, P. A. (2005). Auto-Rickshaw: an Automated crystal Structure Determination Platform as an Efficient Tool for the Validation of an X-ray Diffraction experiment. Acta Crystallogr. D Biol. Cryst. 61 (4), 449–457. doi:10.1107/s0907444905001307
Pâques, F., and Duchateau, P. (2007). Meganucleases and DNA Double-Strand Break-Induced Recombination: Perspectives for Gene Therapy. Curr. Gene Ther. 7 (1), 49–66. doi:10.2174/156652307779940216
Perler, F. B. (1998). Protein Splicing of Inteins and Hedgehog Autoproteolysis: Structure, Function, and Evolution. Cell 92 (1), 1–4. doi:10.1016/s0092-8674(00)80892-2
Perler, F., Olsen, G. J., and Adam, E. (1997). Compilation and Analysis of Intein Sequences. Nucleic Acids Res. 25 (6), 1087–1093. doi:10.1093/nar/25.6.1087
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera?A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 25 (13), 1605–1612. doi:10.1002/jcc.20084
Pietrokovski, S. (1994). Conserved Sequence Features of Inteins (Protein Introns) and Their Use in Identifying New Inteins and Related Proteins. Protein Sci. 3 (12), 2340–2350. doi:10.1002/pro.5560031218
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, Scalable Generation of High‐quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 7, 539. doi:10.1038/msb.2011.75
Southworth, M. W., Adam, E., Panne, D., Byer, R., Kautz, R., and Perler, F. B. (1998). Control of Protein Splicing by Intein Fragment Reassembly. EMBO J. 17 (4), 918–926. doi:10.1093/emboj/17.4.918
Stoddard, B. L. (2011). Homing Endonucleases: from Microbial Genetic Invaders to Reagents for Targeted DNA Modification. Structure 19 (1), 7–15. doi:10.1016/j.str.2010.12.003
Swithers, K. S., Senejani, A. G., Fournier, G. P., and Gogarten, J. P. (2009). Conservation of Intron and Intein Insertion Sites: Implications for Life Histories of Parasitic Genetic Elements. BMC Evol. Biol. 9, 303. doi:10.1186/1471-2148-9-303
Taylor, G. K., Petrucci, L. H., Lambert, A. R., Baxter, S. K., Jarjour, J., and Stoddard, B. L. (2012). LAHEDES: the LAGLIDADG Homing Endonuclease Database and Engineering Server. Nucleic Acids Res. 40 (Web Server issue), W110–W116. doi:10.1093/nar/gks365
Volkmann, G., and Iwaï, H. (2010). Protein Trans-splicing and its Use in Structural Biology: Opportunities and Limitations. Mol. Biosyst. 6 (11), 2110–2121. doi:10.1039/c0mb00034e
Keywords: intein structures, horizontal gene transfer, protein splicing, DNA recognition, intein, meganuclease, homing endonuclease
Citation: Beyer HM and Iwaï H (2022) Structural Basis for the Propagation of Homing Endonuclease-Associated Inteins. Front. Mol. Biosci. 9:855511. doi: 10.3389/fmolb.2022.855511
Received: 26 January 2022; Accepted: 08 February 2022;
Published: 16 March 2022.
Edited by:
Christopher Lennon, Murray State University, United StatesReviewed by:
Barry Stoddard, Fred Hutchinson Cancer Research Center, United StatesCopyright © 2022 Beyer and Iwaï. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hideo Iwaï, aGlkZW8uaXdhaUBoZWxzaW5raS5maSBvcg==, aXdhaUBhbHVtbmkuZXRoei5jaA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.