- Department of Cell Biology and Physiology, Washington University School of Medicine, St. Louis, MO, United States
Gene expression is regulated at multiple levels in eukaryotic cells. Regulation at the post-transcriptional level is modulated by various trans-acting factors that bind to specific sequences in the messenger RNA (mRNA). The binding of different trans factors influences various aspects of the mRNA such as degradation rate, translation efficiency, splicing, localization, etc. MicroRNAs (miRNAs) are short endogenous ncRNAs that combine with the Argonaute to form the microRNA-induced silencing complex (miRISC), which uses base-pair complementation to silence the target transcript. RNA-binding proteins (RBPs) contribute to post-transcriptional control by influencing the mRNA stability and translation upon binding to cis-elements within the mRNA transcript. RBPs have been shown to impact gene expression through influencing the miRISC biogenesis, composition, or miRISC-mRNA target interaction. While there is clear evidence that those interactions between RBPs, miRNAs, miRISC and target mRNAs influence the efficiency of miRISC-mediated gene silencing, the exact mechanism for most of them remains unclear. This review summarizes our current knowledge on gene expression regulation through interactions of miRNAs and RBPs.
Introduction
The central dogma of biology follows the flow of genetic information; DNA is transcribed into RNA and RNA is translated into protein. Correct gene expression in a timely and quantitative way is essential for maintaining cellular homeostasis as dysregulated protein production can lead to various diseased states. RNA is not just a simple intermediate for conveying the genetic code, but it also regulates when, where, and how much protein will be produced. RNA has a complex, multistage lifecycle starting in the nucleus, where it is transcribed from DNA. After maturation, which includes co- and post-transcriptional processing, including 5′ capping, polyadenylation, and splicing, the mRNA is exported into the cytoplasm from the nucleus. In the cytoplasm ribosomes translate coding mRNAs into protein. From its initial transcription until its degradation, mRNAs are highly regulated at both a global and individual level. Individual mRNAs are regulated by various trans-acting factors that bind to specific cis-regulatory elements within the mRNA and influence the stability, localization, modifications, and translation of the information encoded in the mRNA (Gebauer and Hentze, 2004; Fukao et al., 2021).
Several classes of small noncoding RNAs (ncRNAs) have been shown to bind to specific sequences within mRNAs, mainly in the 3′ untranslated region (UTR), and influence the extent of expression of encoded genes. The three main classes of small ncRNAs, within 19–31 nucelotide length, are microRNAs (miRNA), small interfering RNAs (siRNAs), and piwi-interacting RNAs (piRNA). Each class of these small ncRNAs associates with distinct sets of effector proteins to carry out their function, both miRNAs and siRNAs associated with the Ago-clade, while piwi-RNAs associate with the PIWI-clade (Parker and Barford, 2006; Peters and Meister, 2007; Farazi et al., 2008; Thomson and Lin, 2009; Juliano et al., 2011). miRNAs and siRNAs are the most well understood and similar of the three; they are mostly derived from endogenous encoded double-stranded hairpin-shaped RNA. The main difference between miRNAs and siRNAs is their complementarity; siRNAs are derived from stem-loop with perfect complementarity while miRNAs contain imperfect complementarity. This review focuses on miRNAs, but siRNAs and piRNAs have been well-documented in recent reviews (Farazi et al., 2008; Carthew and Sontheimer, 2009; Thomson and Lin, 2009; Dana et al., 2017; Czech et al., 2018).
miRNAs are conserved, endogenous, short (19–22 nt) ncRNAs that combine with Argonaute (Ago) proteins to form the microRNA-induced silencing complex (miRISC). The miRISC uses imperfect base pair complementarity to bind to specific sequences found mostly in the 3′UTR of target mRNAs and repress the gene expression of that transcript (Siegel et al., 2011; Lin and Gregory, 2015; Duchaine and Fabian, 2019). While there were hints of miRNAs in the 1980’s in developmental and cell lineage screens (Horvitz and Sulston, 1980), the first miRNA, lin-4, was discovered in 1993 by the Ambros lab (Lee et al., 1993). Lin-4 was found to bind to the lin-14 mRNA and post-transcriptionally repress the expression of the Lin-14 protein. Lin-14 is necessary for proper timing of larval development in Caenorhabditis elegans (C. elegans) as loss-of-function lin-4 mutants revert to an early developmental stage later in their development. Seven years later the, second miRNA, let-7, was discovered (Reinhart et al., 2000). As let-7 was also found to be a heterochromatic switching factor, it was first thought that these short, hence the name micro, ncRNAs must target mRNAs that code for developmental genes. However, as more miRNAs continued to be discovered, their roles became more diverse, suggesting a much broader role in biological processes (Bartel, 2018). Over 60% of human protein-coding genes have been shown to be targeted by miRNAs (Friedman et al., 2008). As miRNAs modulate the expression of genes involved in cellular differentiation, division, growth, and apoptosis it comes to no surprise that the miRNAs themselves must be highly regulated to avoid diseased states (Siomi and Siomi, 2010). In addition to small ncRNAs, RNA-binding proteins (RBPs) are another class of trans-factors that bind to cis elements within mRNA transcripts. RBPs regulate many aspects of processes associated with RNAs; splicing, transcription, modification, localization, translation, and decay (Díaz-Muñoz and Turner, 2018; Hentze et al., 2018). RBPs have been found to greatly influence both miRNA biogenesis and miRNA-mediated gene silencing. This review focuses on miRNA-mediated gene silencing and investigates the influence of RBPs on the modulation of miRISC function (Gebauer et al., 2021).
miRNA Biogenesis in Eukaryotes
As miRNAs play a major role in controlling protein abundance, the expression and number of miRNAs is highly important. As depicted in Figure 1A, prior to silencing their target transcripts, miRNAs are transcribed in the nucleus mostly by RNA polymerase II, forming the primary (pri) miRNA. The pri-miRNA is composed of a local stem loop containing 3 helical stems that are flanked by basal and apical junctions at both ends (Ha and Kim, 2014; Nguyen et al., 2015; Bartel, 2018; Duchaine and Fabian, 2019). Figure 1A shows how the pri-miRNA is processed in the nucleus to form the shorter pre-miRNA. The pre-miRNA is created through two sequential processing reactions; first, the pri-miRNA hairpin is recognized and cleaved from the transcript by the microprocessor, which comprises Drosha and the double-strand RNA (dsRNA) binding protein DGCR8 (DiGeorge Critical Regulator 8). DGCR8 helps provide the affinity for the microprocessor as it acts as a molecular anchor to position Drosha’s catalytic site at the desired distance from the stem flanking region (Bartel, 2004, 2018). DGCR8 interacts with the stem and apical portion of the stem-loop structure in the pri-miRNA and positions Drosha for cleavage (Nguyen et al., 2015; Figure 1A). Drosha, a member of the Rnase III family, localizes at the pri-miRNA basal junction and cleaves the stem-loop from the rest of the transcript, creating a pre-miRNA product ∼60–75 nucleotides long (Ha and Kim, 2014). There are multiple transcripts with the ability to fold backward and form hairpins, but not all are selected to become pri-miRNAs and enter the miRNA biogenesis pathway. The microprocessor is the gate-keeper of this process and seems to have a preference for pri-miRNAs containing hairpins with a stem that is ∼35 base pairs long, an unstructured apical loop that is over 10 nucleotides long, single-stranded sequences flanking the hairpin, and there are 4 sequence motifs at sites correlating to the position of the microprocessor. These sequence motifs include a basal UG motif, an apical UGU motif, a CNNC flanking motif, and a mismatched GHG motif with 6 nucleotides of the basal stem (Fang and Bartel, 2020; Shang et al., 2020). However, not all pri-miRNAs are optimal substrates of the microprocessor so they rely on neighboring canonical pri-miRNAs (Fang and Bartel, 2020; Shang et al., 2020).
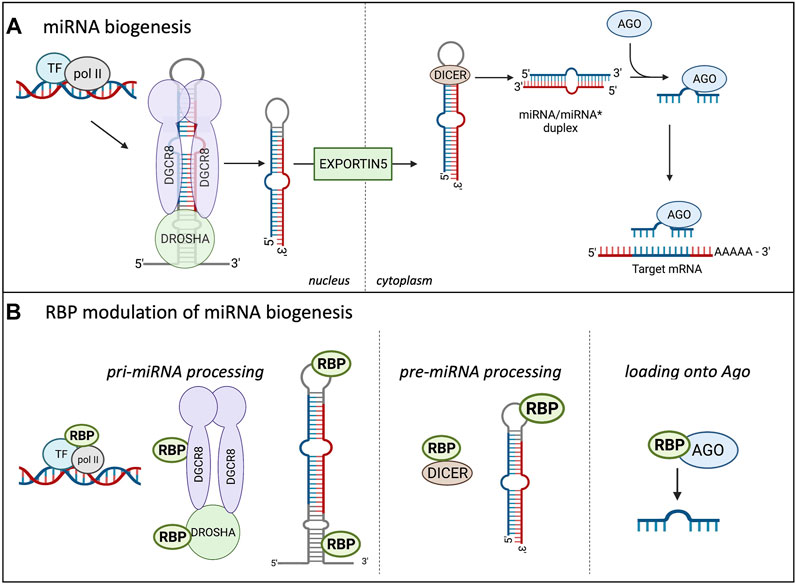
FIGURE 1. Canonical miRNA biogenesis in Eukaryotes and the influence of RBPs. (A) shows the miRNA biogenesis in eukaryotes. miRNAs are transcribed in the nucleus by RNA polymerase II (pol II), creating the pri-miRNA, two sequential cleavage reactions follow. The microprocessor consists of Drosha and DGCR8 and performs the first cleavage reaction in the nucleus, creating the pre-miRNA. The pre-miRNA is transported into the cytoplasm through Exportin5 where the second cleavage reaction occurs. Dicer cleaves the terminal loop of the pre-miRNA, creating the miRNA/miRNA* duplex. The miRNAs are incorporated into the Ago protein, forming the minimal effector RNA induced silencing complex (miRISC) and target mRNA sequences. (B) Highlights the modulation of miRNA biogenesis by RBPs. RBPs can bind to the promoter region of certain miRNAs and influence their transcription. RBPs modulate miRNA expression at the pri-miRNA processing level through binding to Drosha and enhance or repress the cleavage. RBPs can also bind to the terminal loop or other sequences in the pri- and pre-miRNAs to influence the cleavage reactions. Additionally, RBPs can bind to DICER and influence this cleavage reaction through modulating DICER expression and availability. Lastly, RBPs can bind to AGO and increase the miRNA loading onto the AGO, increasing the miRISC silencing.
The pre-miRNA is transported to the cytoplasm via exportin V where it undergoes the second cleavage reaction to form the mature miRNA/miRNA* duplex, which consists of the guide and passenger (*) strand (reviewed in Duchaine and Fabian, 2019; Loffreda et al., 2015). Dicer functions in miRNA maturation and helps to facilitate loading of the mature miRNA onto Argonaute (Foulkes et al., 2014). Dicer cleaves the terminal loop from the pre-miRNA stem creating the mature duplex that is 18–21 base pairs long and contains 3′ overhangs (Murphy et al., 2008). Dicer contains 2 catalytic RNAse III domains, C-terminal dsRBD, ATPase/RNA helicase, Piwi-Argonaute-Zwille (PAZ) domain, and the DUF283 domain (Foulkes et al., 2014; Gebauer et al., 2021). The PAZ domain recognizes the 5′ terminal phosphate of authentic pre-miRNAs and acts as a ruler to measure the cleavage site at the 3′ overhang where the RNAse III domains will cut a strand to create the mature miRNA duplex (Connerty et al., 2015). The mature miRNA duplex associates with the Ago protein to form the minimal effector miRNA-induced silencing complex (miRISC). The guide strand from the duplex becomes associated with the Ago protein and is unwound while the passenger strand is lost. This process is referred to as differential strand retention, which is based on the thermodynamic stability of the ends of the duplex (Bartel, 2018). The miRNA biogenesis involving the microprocessor is the canonical pathway and is depicted in Figure 1A. Research has shown that there are microprocessor-independent, or non-canonical, pathways for miRNA biogenesis (Miyoshi et al., 2010; Connerty et al., 2015; Bartel, 2018). Probably the most well-defined alternative miRNA biogenesis pathway is the one which combines intron splicing with dicing of the miRNA. These miRNAs are known as “mirtrons” and they are processed from RNAs that are both pre-miRNAs and introns (Westholm and Lai, 2011). Upon splicing of introns from transcribed RNAs, the processing of pri-miRNAs and pre-miRNAs is preceded by linearization of intron lariat by debranching enzymes. While it was originally thought that mirtrons are Drosophila and C. elegans specific pathways further studies found mirtrons throughout the animal kingdom (Siomi and Siomi, 2010; Salim et al., 2021). There are also miRNAs that are dicer-independent, but rely on the nuclear canonical machinery for miRNA biogenesis usually through cluster assistance, which will be discussed in detail in the next section (Fang and Bartel, 2020; Shang et al., 2020).
While this review focuses on miRNAs in the context of mammalian cells, it is important to note that miRNA biogenesis in plants is very similar to mammals, however, there are some key differences that should be highlighted. Similar to animal miRNA biogenesis, pri-miRNAs are transcribed in the nucleus by RNA polymerase II, followed by stabilization in a region known as the D-body (Voinnet, 2009; Wang et al., 2019). While in the D-body region the pri-miRNA interacts with a complex composed of zinc finger protein serrate (SE), a double-stranded binding protein hyponastic leaves 1 (Hyl1), dicer like 1 (Dcl1), and other accessory proteins depending on the specific type of miRNA (Wang et al., 2019). Dc11 functions much like Dicer in animal miRNA biogenesis, dc11 performs two cleavage reactions on the pri-miRNA; the first creating the shorter pre-miRNA, which then proceeds to another round of processing by dc11 to form the mature miRNA/miRNA* duplex. The mature duplex is exported out of the nucleus through HASTY (homolog to the animal exportin-V) (Bartel., 2004; Voinnet, 2009). Once in the cytoplasm, the guide strand interacts with Ago to guide the RISC to its target mRNA through near-perfect complementarity to silence the gene through direct cleavage and/or translational inhibition.
RBPs in Control of miRNA Biogenesis
As the proper expression of miRNAs is essential for maintaining cellular homeostasis, their biogenesis is highly regulated at all steps, as highlighted in Figure 1B (Treiber et al., 2017; Nussbacher and Yeo, 2018). Analyses of crosslinking immunoprecipitation (CLIP) data with proteins involved in the miRNA biogenesis has helped identify specific RBPs involved in miRNA processing (Ramanathan et al., 2019). Post-transcriptional modifications of the RNA, as well as post-translational control of the biogenesis machinery and effector proteins involved in these processes, can impact the production of miRNAs and formation of the miRISC. Multiple modifications take place on miRNAs and the majority of them are important for their biogenesis, function, and stability; in addition, ubiquitination and phosphorylation of Ago proteins can play a role in miRISC formation (Peters and Meister, 2007; Meister, 2013; Müller et al., 2020; Wu et al., 2020). Some modifications have been extensively studied, including uridylation, editing of adenosine to inosine, or methylation of miRNAs, but many of them still need to be functionally characterized (Wyman et al., 2011; Michlewski and Cáceres, 2019). As an example, uridylation of pre-let7a miRNA by TUT4 or TUT7 blocks its processing and marks the miRNA for degradation (Heo et al., 2012; Michlewski and Cáceres, 2019). Deamination of specific adenosine to inosine by ADAR in pri-miR-142 (Yang et al., 2006) and pri-miR-151 (Kawahara et al., 2007) targets these miRNAs for degradation or blocks their processing, respectively. It was also shown that N6-methyladenosine (m6A) methylation by both mammalian or plant METTL3 homolog affects proper levels of mature microRNA (miRNA) biogenesis in human tissue cultures (Alarcón et al., 2015) and Arabidopsis (Bhat et al., 2020). The proposed mechanism involves specific methylation of a set of pri-miRNAs affecting proper folding of the RNA and recruitment of the microprocessor for proper and efficient pri-miRNA processing. In this case it’s the loss of the modification (m6A) in pri-miRNA that leads to the reduction of pri-miRNA-microprocessor interactions and reduction in the levels of mature miRNAs in both mammalian and plant studies. Additionally, experiments with an introduction of m6A marks by in vitro transcription in pri-miRNAs indicated more efficient processing of the modified pri-miRNA by the microprocessor, thus confirming the role of m6a modifications in miRNA biogenesis (Alarcón et al., 2015). Certain RBPs have been shown to increase or decrease efficiency of miRNA biogenesis through direct binding to the miRNA precursor and/or altering the machinery involved in the biogenesis. RBPs can influence pre-miRNAs through directly binding to the miRNA sequence or indirectly through binding to DICER and impacting its expression and/or function (Bicker et al., 2013; Connerty et al., 2015; Loffreda et al., 2015). PACT and TRBP are two RBPs that have been shown to bind to and stabilize the expression of DICER, thus indirectly increasing the fidelity of miRNA biogenesis (Peters and Meister, 2007; Ha and Kim, 2014). At the pri-miRNA level, RBPs have been shown to bind to the miRNA’s terminal loop. hnRNPA1 can bind to the stem loop of the pri-miR-18a (Díaz-Muñoz and Turner, 2018; Kooshapur et al., 2018). This binding alters the secondary structure, creating a more relaxed loop conformation which provides more accessibility for the microprocessor. On the other hand, RBPs can bind to the pri-miRNA and inhibit the biogenesis reaction through blocking the binding site for the microprocessor or other RBPs that are necessary for miRNA biogenesis (Loffreda et al., 2015; Michlewski and Cáceles, 2019). As depicted in Figure 1B the production of miRNAs is highly regulated by RBPs at multiple levels. As shown in Figure 1B, the two cleavage reactions that take place during miRNA biogenesis require the assistance of multiple RNA-binding proteins (RBPs). Fused in Sarcoma (FUS) protein is a ubiquitously expressed RBP that has been shown to directly interact with the microprocessor and recruit it to the transcription site of the miRNA (Morlando et al., 2012). Besides the FUS protein, a TAR DNA-binding protein-43 (TDP-43) has been shown to interact with both the microprocessor and Dicer complexes promoting the biogenesis of specific subsets of miRNAs and playing a role in neuronal differentiation (Kawahara and Mieda-Sato, 2012; Di Carlo et al., 2013). The RBP Ewing Sarcoma Protein (EWS) can inhibit the expression of DROSHA, likely through direct binding to the promoter (Ouyang et al., 2017), and thus regulate the overall efficiency of miRNA biogenesis. Another RBP that has been shown to interact with Drosha to influence miRNA biogenesis is SRSF3, a serine arginine splicing factor. SRSF3 regulates a large portion of canonical miRNAs (Kim et al., 2018). The CNNC motif lies about 17 nucleotides from the microprocessor and has been shown to interact with SRSF3 to aid in stimulating the processing of pri-miRNAs. It is thought that SRSF3 binding to the CNNC motif helps Drosha bind to the basal junction, thus aiding in pri-miRNA processing (Kim et al., 2018). Treiber and others performed a systematic analysis of pre-, pri- and mature miRNAs and identified a set of 72 human pre-miRNAs and found that 180 RBPs had preferential binding to a single or multiple miRNA precursor (Treiber et al., 2017). While it’s clear RBPs play a critical role in the biogenesis of miRNAs, most of these studies have been performed in vitro which could result in unphysiological binding. It will be important to investigate these interactions of RBPs with miRNA biogenesis pathways in vivo to identify their physiological significance as well as connect this type of regulation to cell type specific biogenesis of miRNAs.
There are multiple miRNAs that are made from pri-miRNAs that contain multiple clustered stem loop structures that were originally thought to be treated as independent units and thus individually cleaved by the microprocessor. However, it is becoming clear that certain pri-miRNAs that are poor substrates of the microprocessor are dependent on a close pri-miRNA neighbor for proper miRNA biogenesis, this process is known as cluster assistance. miR-451 is a miRNA that is dicer-independent but requires the canonical nuclear miRNA processing for its biogenesis as indicated with decreased miR-451 expression upon knockout of Drosha (Shang et al., 2020). miR-451 is a poor substrate for the microprocessor due to its short step loop, however, despite this, miR-451 is somehow a substrate of the microprocessor. Interestingly, miR-451 has been found to be tightly clustered by miR-144 throughout evolution (Fang and Bartel., 2020; Shang et al., 2020). miR-144 is an optimal substrate of the microprocessor and several groups have demonstrated that miR-451 biogenesis is dependent on its close proximity to miR-144. Shang and colleauges replaced miR-144 with miR-7a and miR-454, which are optimal substrates of the microprocessor, and miR-451 biogenesis was still promoted. This suggests a general mechanism whereby miRNAs that are suboptimal substrates of the microprocessor require canonical miRNAs in close proximity for their biogenesis (Shang et al., 2020). Additionally, several groups are identifying RBPs that are shown to assist in this clustering process (Fang and Bartel, 2020; Hutter et al., 2020; Kwon et al., 2020). Enhancer of rudimentary homolog (ERH) is a recently discovered component of the microprocessor that has been shown to aid in the cluster assistance process of miR-451 and miR-144 (Fang and Bartel., 2020; Hutter et al., 2020; Kwon et al., 2020). During cluster assistance the microprocessor is loaded onto the poor substrate with the aid of its neighboring high affinity miRNA substrate and ERH increases the processing of the suboptimal miRNA substrate through binding to the N-terminus of DGCR8 (Kwon et al., 2020).
Using a CRISPR/Cas9 LOF screen Hutter and colleuges found ERH and SAFB2 to be critical factors for cluster-mediated assistance (Hutter et al., 2020). Scaffold attachment factor B2 (SAFB2) is an RBP that has been found to be an accessory protein of the microprocessor in mammals (Hutter et al., 2020). The study by Hutter et al. looked at the miR-15a-16-1 cluster. Due to a large unpaired region in its basal stem, miR-15a is a suboptimal substrate of the microprocessor and therefore cannot be efficiently processed without the assistance from an optimal miRNA neighbor. Through use of a CRISPR/Cas9 screen they were able to identify SAFB2 as an essential cofactor for the efficient cleavage of pri-miR-15a through miR-16-1 assistance. Cluster assistance has also been observed in plants. In Arabidopsis, MAC5 is a component of the MOS4-associated complex which is needed for immunity and development (Palma et al., 2007; Li et al., 2020). MAC5 is an RBP that binds to stem loops and appears to help with cluster assistance. MAC5 is essential for plant development, as loss of function mutants of MAC5a and MAC5b have been shown to be embryonic lethal. Recently, MAC5 was shown to interact with the SE to promote pri-miRNA processing in plants via protecting the pri-miRNA from SE-dependent exoribonuclease activity (Li et al., 2020). Interestingly there is a human counterpart to MAC5, but it remains to be tested whether or not it interacts with the microprocessor in humans.
miRISC Function and Nature of the miRISC
As mentioned above, once the miRNA combines with the Ago protein, the functional miRISC is formed. The Ago protein is the minimal effector needed for the miRNA to carry out its silencing mechanism (Connerty et al., 2015). Ago is made up of 4 domains; the N-terminal domain, Piwi-Argonaute-Zwille (PAZ) domain, the middle domain (MID), and the p-element induced wimpy testis (PIWI) domain (Song et al., 2004; Carthew and Sontheimer, 2009; Djuranovic et al., 2011; Sheu-Gruttadauria and MacRae, 2017). The N-terminal domain facilitates small RNA loading and unwinding of the duplex. The PAZ domain recognizes and anchors to the 3′ ends of miRNA. The MID domain binds the 5′ terminal monophosphate moiety and the 5′ terminal nucleotide of the miRNA-guide strand. Finally, the PIWI domain shows extensive homology to RNase H. Of note, it is important to mention that mammalian genomes encode 4 Ago proteins, Ago one to four, with Ago2 being most highly expressed and the only one to have endonucleolytic activity allowing it to cleave target mRNAs with full complementarity to miRNAs (Bhattacharyya et al., 2006; Gebert and MacRae, 2019; Kakumani et al., 2021). Plants and C. elegans have multiple Ago proteins that are further specialized in their cellular function and association with particular miRNA/small RNAs based on their length or 5′ nucleotide (Lim et al., 2003; Wang et al., 2019). The structure and function of Ago proteins have been well characterized (Hutvagner and Simard, 2008; Meister, 2013; Müller et al., 2020; Wu et al., 2020). The miRISC functions to identify the target transcripts first and then silence the gene expression of the target mRNA. The miRISC uses imperfect base complementarity to identify the target sequence in the mRNA. The target sites for the miRNA are usually located in the 3′UTR of the mRNA. Target site prediction is strengthened based on complementarity to the seed region. The seed region of the miRNA is from nucleotide 2 to 7 from the 5′ end, and its complementarity is one of the main criteria for target-site prediction (Bartel, 2009).
Unlike siRNA, miRNAs have imperfect base pair complementarity making target mRNA cleavage a rare event for mammalian miRNAs (Gebauer et al., 2021). miRNAs employ their silencing function mostly through translational repression and mRNA decay (Braun et al., 2012; Djuranovic et al., 2011; Fabian and Sonenberg, 2012; Bartel, 2018). While the exact mechanism of silencing remains a topic of debate, the “default” mechanism agreed upon includes inhibition of translation, followed by deadenylation, decapping, and decay of the mRNA transcript, as shown in Figure 2A (Djuranovic et al., 2011; Braun et al., 2013; Gardiner et al., 2015; Nawalpuri et al., 2020). Even though research has shown translational repression occurs first and might be one mode of controlling gene expression, it is the mRNA decay that ultimately consolidates and silences the target mRNAs (Bazzini et al., 2012; Bêthune et al., 2012; Djuranovic et al., 2011; Hu and Coller, 2012). However, it is important to note that this model of miRISC function will vary depending on additional factors as depicted in Figure 2B. There are several proposed models for miRISC-mediated translational repression, and they are not mutually exclusive (Gu and Kay, 2010; Fabian and Sonenberg, 2012). While the mature miRNA and Ago protein are part of the minimal miRISC, this association alone is insufficient to carry out miRISC-mediated translational repression. The more complete miRISC, including GW182, or its mammalian homologs TNRC6, is needed for translational repression (Eulalio et al., 2009; Huntzinger et al., 2013). GW182 is also a bridging factor between Ago proteins and the poly(A) binding protein (PABP) (Duchaine and Fabian, 2019), binding both of these proteins through the N-terminal domain. However, it is the carboxy terminal domain of GW182, which is referred to as the silencing domain, that recruits effector proteins such as deadenylases (PAN2-PAN3 and CCR4-NOT) and mRNA decapping factors (DCP1/2) (Eulalio et al., 2009; Braun et al., 2012; Wilczynska and Bushell, 2015). In a sequential series of actions, the miRISC is thought to induce translational repression of targeted mRNAs, which are later deadenylated by action of deadenylases, decapped by DCP1/2, and then degraded by the exosome and XRN1 (Rehwinkel et al., 2005; Braun et al., 2012). The presence of PABP aids in regulating both mRNA translation and mRNA turnover since it enhances miRNA-mediated deadenylation (Roy and Jacobson, 2013; Wigington et al., 2014). However, the presence of PABP on targeted mRNAs or its interaction with the miRISC is not necessary for translational repression (Djuranovic et al., 2011). Recently, 4EHP and GIGYF2 have been shown to bind to a conserved proline rich region of GW182 and impact miRISC-induced translational repression potentially at the step of 5′ mRNA cap binding (Chapat et al., 2017; Schopp et al., 2017). Regardless of whether there are multiple modes or one unifying model of translational repression by miRISC, it is becoming clearer that miRISC composition may influence outcome of the miRISC-target mRNA interaction. Both Ago and GW182 proteins may serve as molecular hubs for multiple ribonucleoprotein complexes (RNPs) or other enzymes involved in RNA metabolism further define their role in gene expression control.
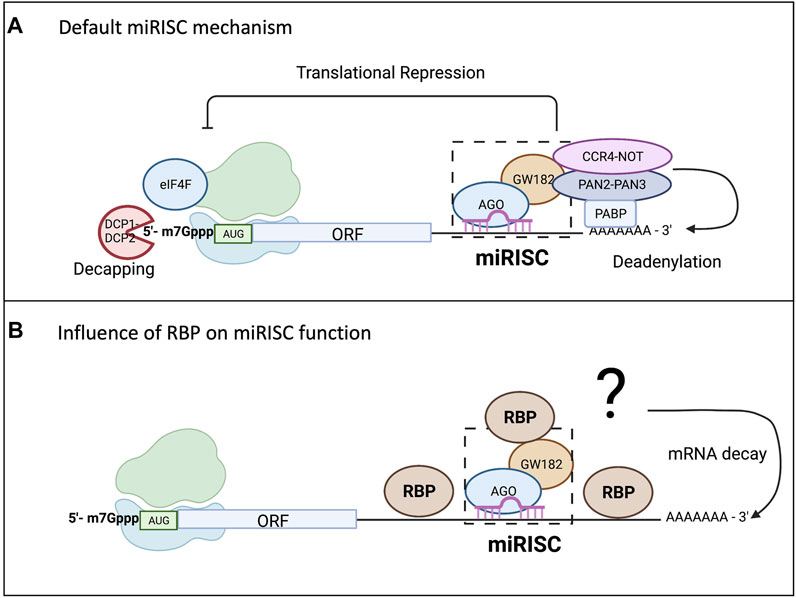
FIGURE 2. miRISC-mediated gene repression and the influence from RBPs. While there are conflicting models of miRISC-mediated gene silencing that are not mutually exclusive, scientists have agreed upon a “default” mechanism as all the proposed mechanisms for miRNA-mediated repression involve repression of translation and mRNA decay. As shown in (A), Ago interacts with the PABP complex to promote mRNA deadenylation through recruitment of poly(A) nuclease deadenylation complex subunit 2 (PAN2)-PAN3 and carbon catabolite repressor protein 4 (CCR4)-NOT. Deadenylation promotes decapping by the mRNA-decapping enzyme subunit DCP1-DCP2, making the mRNA vulnerable to degradation by exoribonuclease 1 (XRN1). (B) Highlights the influence RBP binding in the 3′UTR can have on miRISC-mediated gene repression. The RBP can bind up or downstream of the miRISC and either enhance the repression, usually through increasing mRNA degradation, or it could reduce the silencing efficiency of the miRISC.
RBPs and a Functional miRISC
Multiple RBPs fulfill their gene regulation function independently of the miRISC but may impact the actions of the miRISC on target mRNAs. These RBPs can directly influence the functional outcome of the miRISC by modulating the actual composition of the miRISC or indirectly through interactions with the translational machinery involved in miRISC regulation. As previously mentioned, the FUS protein is an RBP that has been shown to directly associate with the core miRISC and influence the downstream function (Zhang et al., 2021). It is proposed that FUS facilitates the association between miRISC components such as the Ago protein, a set of mature miRNAs, and interacting with their mRNA targets, thus increasing efficiency of miRNA-mediated silencing. Since the interaction of FUS with the Ago protein is miRNA-independent, it may impact global miRNA regulation. FUS-enabled and selective miRISC targeting through direct interactions with “preferred” miRNAs and mRNA targets could create an even bigger challenge for “non-preferred” miRISC complexes and their ability to locate targets among the other RNAs in the cell.
In a very similar fashion, Smaug, an important RBP in the early development of Drosophila, has been shown to recruit the minimal miRISC complex or Ago proteins, regardless of the miRNA targeting (Pinder and Smibert, 2013). Smaug interaction with the miRISC or Ago proteins is driven by a direct protein-protein interaction (Smaug-Ago1/2) and recruitment of such a complex to the 3′UTR of targeted mRNAs is purely driven by Smaug-recognition elements (SREs). As such, the Drosophila Smaug-Ago1 complex does not require miRNAs for translational repression of Nanos mRNA. This direct recruitment of miRISC without the involvement of miRNA-mRNA target recognition has not been found in other cases, but several members of the Hu family of RBPs have been found to reduce miRISC function by preventing the formation of the repressive miRISC on target mRNAs or directly competing for components of translation machinery targeted by the miRISC (Fukao et al., 2015). The HuD member of the Hu family is known to stimulate eIF4A activity on bound mRNAs and thus prevent potential miRISC translational repression through the translation initiation scanning mechanism (Fukao et al., 2015). The ability of other members of Hu family, as well as other AU-rich element-binding proteins (ARE-BPs) to oligomerize on target mRNAs can also abrogate miRISC-mRNA interaction (as discussed in the next section).
There are multiple reports that the miRISC is indeed not a homogenous complex. The factors that make miRISC variations are associated with cell types as well as cellular processes such as cellular stress, growth, and differentiation. The heterogeneity of the miRISC complex allows for diversity in its function (Sheu-Gruttadauria and MacRae, 2017; Dallaire et al., 2018; Nawalpuri et al., 2020). The different cell-specific cofactors will contribute to the miRISC function, potentially changing the function of the same miRNA-Ago complex depending on the cell type. A recent study looked at miRNAs in somatic and germline cells and found that they formed distinct miRISC depending on the cell type (Dallaire et al., 2018). They used an in vivo fluorescent reporter with binding sites for miR-228 and germline- and somatic-specific promoters. They observed that in intestinal cells the miR-228 reporter was repressed at both the protein and mRNA level. However, they discovered stabilization of the miR-228 reporter in germline cells, suggesting a different mechanism where translational repression is uncoupled from mRNA destabilization. Using RNA affinity assays to purify specific miRISCs they identified a GW182-independent silencing mechanism used in germline cells of C. elegans compared to somatic cells. As such a single mRNA can have flexible regulation that changes based on cellular and developmental context, RBP presence, and miRISC composition, adding another layer of complexity to miRISC-mediated mechanisms and differential target regulation.
Crosstalk Between miRNAs and RBPs on Target mRNAs
Historically miRNA research has focused on one individual miRNA, however, in an endogenous system there can be multiple miRNA binding sites in the 3′UTR of a single target mRNA. Currently miRbase has identified 1917 precursor and 2654 mature miRNAs. Additionally, over 1,000 RBPs have been identified in the human genome, so it is no surprise that their binding sites can be right next to each other, or even overlapping as depicted in Figures 3A,B (van Kouwenhove et al., 2011; Jiang et al., 2013; Cottrell et al., 2018). It has become apparent that in order to understand miRNA-mediated gene silencing fully, the miRNA must be studied in combination with other miRNAs and RBPs. Computational analysis has been helpful to predict crosstalk between RBPs and miRNAs (Jiang et al., 2013; Loffreda et al., 2015). PAR-CLIP and RIP-Seq experiments have been critical for identifying enrichment of RBP binding sites next to or overlapping with miRNA recognition target sites (Jiang and Coller, 2012; Iadevaia and Gerber, 2015; Ramanathan et al., 2019).
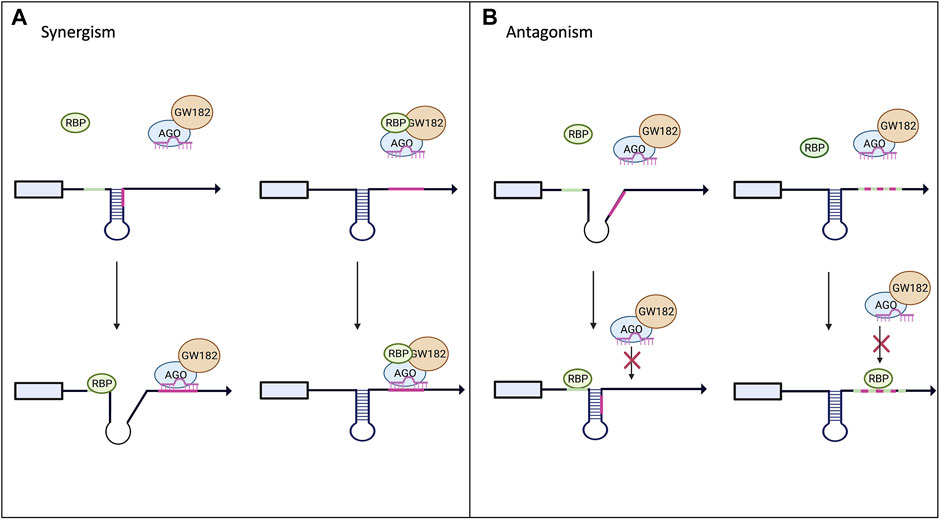
FIGURE 3. Interplay between RBPs and miRNAs on miRISC-mediated gene expression. Panel (A) shows potential models of synergism between RBPs and miRNAs. Upon binding to the mRNA target the RBP can alter the secondary structure and increase the exposure of the miRNA binding site allowing for increased miRISC binding. The RBP could also bind to the miRISC and increase its binding affinity to its target mRNA. (B) Highlights the possible antagonistic mechanism between RBPs and miRNAs. RBP binding to its target could alter the secondary structure, decreasing the miRISC’s access to its binding site. RBPs and miRNA can also compete for the same binding site and the RBP can outcompete and block the miRISC from binding to its site (Loffreda et al., 2015).
RBPs can have an antagonistic effect on miRISC gene silencing when competing for the same or nearby binding site within the 3′UTR of the target mRNA as demonstrated in Figure 3B. The best-known examples for this are Hu and ARE-BP family members (Jiang and Coller, 2012). Multiple groups have found that the CAT1 mRNA can be targeted by miR-122 and HuR (Filipowicz et al., 2008; Fabian and Sonenberg, 2012; Kundu et al., 2012). Under normal conditions, miR-122 targets CAT1 and represses its expression. However, it has been shown that when stimulated with stress, HuR can rescue CAT1 from miRNA-mediated repression by miR-122 (Filipowicz et al., 2008; Kundu et al., 2012). The mechanism by which HuR rescues CAT1 expression remains unclear; it could be through dissociation of miRNPs from the mRNA or prevention of the miRISC from binding to its target site in the 3′UTR. Another example of competition between miRNAs and RBPs is with HuR and miR-16. They both have binding sites in the 3′UTR of prostaglandin synthase cyclooxtgenesis-2 (COX-2). miR-16 normally binds to the 3′UTR of COX-2 and promotes rapid repression and degradation of the transcript. However, when HuR levels are increased, HuR can outcompete miR-16 for the binding site and stabilize COX2, thus increasing its expression (Young et al., 2012). Similarly, AU-rich element-binding protein 1 (AUF1) binds to AU-rich elements (AREs) in the 3′UTR of target mRNAs and potentially oligomerizes, preventing or enabling miRISC binding (Wilson et al., 1999; Zucconi et al., 2010). However, scientists have shown that AUF1 also has a high affinity for the let-7b miRNA. When AUF1 binds to the miRNA, let-7b, it facilitates its transfer onto Ago2, thus enhancing the miRNA-mediated repression (Yoon et al., 2014) in a way similar to FUS.
On the other hand, RBPs can cooperate with miRNAs to enhance the silencing of the target mRNA as shown in Figure 3A. This can be done through recruitment of the miRISC to the binding site on the target mRNA (example of Smaug and FUS above) or the binding of the RBP can change the secondary structure of the mRNA to increase the binding accessibility for the miRISC. Pumilio, a member of the Puf family, is an RBP that has been shown to enhance miRISC activity through unwinding the mRNA 3′UTR, thus promoting miRISC binding (Kedde et al., 2010; Miles et al., 2012; Nawalpuri et al., 2020). Pumilio is known to promote cell-cycle re-entry of quiescent cells upon binding to the 3′UTR of the mRNA that encodes for the tumor suppressor p27. Binding of Pumilo to the Pumilio-recognition elements (PREs) in the 3′UTR of p27 mRNA induces a change in the secondary structure of p27 mRNA that increases the accessibility of the target sites for miR-221 and miR-222, thus enabling the repression of this mRNA (Kedde et al., 2010). The similar mode of miRISC binding regulation and activity by Pumilio was found in a 3′UTR of transcription factors important for regulating cell proliferation, such as E2F3 (Miles et al., 2012).
While scientists have found examples showing antagonism and synergism between RBPs and miRNAs on miRISC gene silencing, it is important to note that all of these actions are co-occurring inside of the cell. Table 1 highlights certain known RBPs and miRISC interactions on common target mRNAs and you can see that a single RBP can either enhance or repress miRNA-mediated repression depending on the miRISC and RBPs binding sites. This suggests that there cannot be a universal model for the crosstalk between RBPs and miRNAs. A single mRNA can have binding sites for multiple RBPs and miRNAs, therefore the additive or countering effects of the different interactions will determine the final outcome of miRISC silencing efficiency.
Positional-Dependent Interactions of RBPs and miRNAs on miRISC Silencing
Many studies have indicated the influence of RBPs on miRISC-mediated gene silencing, but as different cells have varying RBP expression profiles, the same miRNA may have a different silencing mechanism or the transcript may be under control of both miRNAs and RBPs, depending on the specific cell type (Jiang et al., 2013; Cottrell et al., 2018). Many groups have investigated combined effects of miRNAs and RBPs, but studies are usually limited to a single mRNA or reporter 3′UTRs. As such, it is hard to conclude whether the exact mechanism that causes the antagonistic or synergistic effect of RBPs and miRISC on gene repression in a single 3′UTR is applicable in global analyses of other regulated genes. The answer to this question may lie in the positions of the miRNA and RBP binding sites in relation to one another in the 3′UTR of the target mRNA as well as in cell types. In an endogenous system the RBP binding site can be close to or far away from the miRNA binding site.
Recently two groups have identified that proximity of the binding sites influences the cross talk between RBPs and miRNAs (Cottrell et al., 2018). These studies identified that the closer the RBP and miRNA binding sites were to one another on target mRNA, the larger the influence of RBPs on miRISC gene targeting. One study indicated that nearby RBP binding was associated with enhanced miRISC targeting, which could potentially mean an increase in gene silencing. The other study used a massively parallel reporter (MPRA) assay with an eGFP plasmid library containing synthetic or endogenously encoded 3′UTRs, covering all possible combinations of several repressive translation elements (let-7 miRNA binding sites, AREs, PREs and SREs). Authors discovered positional effects between miRNA binding sites and AREs. AREs positioned upstream of the miRNA binding site of let-7 caused an increase in miRISC-silencing efficiency, while a decrease in let-7 silencing was observed when the AREs were downstream of the let-7 binding site (Cottrell et al., 2018). This effect was not only specific for miRNA binding sites but it was also seen in the combination of Pumilio binding sites (PREs) and AREs.
Interestingly, a study that looked at the ARE-BP, HuR, found that this RBP was able to mediate de-repression of Cat1 mRNA from miRISC silencing by binding to AREs located next to miRNA binding sites (Kundu et al., 2012). Given that HuR and other ARE-BPs are known to oligomerize upon binding to their target sites the group sought to investigate if the multimerization of HuR contributed to its ability to rescue Cat1 from miRNA repression. They created several mutants of HuR and found that the mutants with compromised multimerization of HuR did not interfere with the miRISC activity arguing for a steric occlusion model as a potential mechanism (Kundu et al., 2012). Such a model suggests the positional effect and distance-dependence through directional oligomerization of the RBPs may play a role in ARE-dependent modulation of miRISC activity. However, not all RBPs are known to oligomerize so this would not explain the mechanism for all RBP-miRNA crosstalk. Another possibility is that the RNA structure within the 3′UTR alters which would change the availability of the binding sites for both the miRISC and RBP, as described in models for Pumilio and miRISC on p27 and E2F3 mRNAs (Kedde et al., 2010; Miles et al., 2012).
A study in the zebrafish model demonstrated the impact of the RNA structure and RBP-binding sequence motifs during the maternal-to-zygotic transition (MZT) (Beaudoin et al., 2018; Vejnar et al., 2019). They found that certain AREs, U-rich and C-rich motifs, and miR-430 activity are responsible for variation in gene expression seen during the development. They identified multiple sequence and RNA structural elements can have antagonistic effects on the same mRNAs. Combination of these elements on the same mRNAs such as stabilizing U-rich motifs and destabilizing miR-430 target sites leads to differential temporal or spatial regulation and creates specific patterns of gene expression. The transcripts would be stabilized by maternally provided poly(U)-binding proteins and then deadenylated and degraded later in development. These actions are carried by combinatorial effects of the miR-430 miRISC and ARE-BPs by a dose-dependent mechanism established through either maternally deposited or newly synthesized RBPs and RNPs. It is clear the outcome of the interaction between RBPs and miRNAs can vary depending on their position from one another, primarily the distance between the binding sites and the local mRNA structure, thus further enforcing the idea that there is no single mechanism of miRISC function due to the these variations in mRNA targets. Scientists will need to identify methods to predict how certain miRNAs and RBPs interact with one another and identify their impact on gene expression regulation and then validate these results experimentally in endogenous targets. Additionally, other factors will be also at play, such as mRNA modifications or alternative splicing, that influence the structure within the 3′UTR, which in turn would alter miRISC and/or RBPs activity.
Disease-Associated States Resulting From Interactions Between RBPs and miRNAs
Upon their initial discovery, miRNAs were believed to play a role in just development, but as more miRNAs were uncovered, scientists recognized their roles were much more diverse (Bartel, 2018). miRNAs likely play a role in every single biological process, and their dysregulation is seen in many diseased states, such as cancer and neurodegenerative disorders. Timed and correct expression of miRNAs in specific cell types is critical as mRNAs that are not silenced through miRNA-mediated repression will be translated into proteins and may impact cell growth, proliferation, or differentiation. The above-mentioned interplay between RBPs and miRNAs on certain targeted mRNA transcripts can easily contribute to pathogenesis if the levels of RBPs and miRNAs are altered. Most cancer cells see a decrease in global miRNA levels, but individual miRNAs have been shown to increase and accumulate in cancers (Peng and Croce, 2016; Vos et al., 2019; Zhang et al., 2021). Such variation in miRNA levels can indeed lead to tumorigenesis. For example, if there were an increase in miRNA regulators that target tumor suppressors, then there would be a decrease in the tumor suppressor genes (Cheng et al., 2017). In the opposite scenario, an increase in oncogenes can occur if there is a decrease in levels of their regulatory miRNAs. While the importance of miRNAs in cancer is evident, their role in cancer progression should be studied in parallel with RBPs. RBPs may be one of the key factors in determining miRNA function and mutations or changes in the expression of RBPs can impair miRNA biogenesis and miRISC activity. For instance, DEAD-Box 5 (DDX5) and DDX17 RNA helicases are RBPs that help regulate the Drosha-mediated cleavage to produce pri-miRNA. Studies have shown that there is an increase in DDX5 and DDX17 expression levels in breast, cervix, colon, and prostate cancer (Connerty et al., 2015; Shen and Hung, 2015; Khan et al., 2019). Knockdown of both DDX5 and DDX17, in human cervical carcinoma cells suppressed cellular proliferation indicating their association with abnormal cell growth. On the other hand, overexpression of DDX5 caused a proliferation of keratinocytes (van Kouwenhove et al., 2011) thus confirming its role in tumor phenotypes of these cells. It is worth mentioning that DDX5 exerts helicase activity when in the cytoplasm, and it facilitates miRISC loading by unwinding the let-7 precursor duplex.
Another example is an increased expression of COX-2 gene as a hallmark of colorectal cancer (Asting et al., 2011; Young et al., 2012). Under normal physiological conditions, miR-16 degrades COX-2, but miR-16 is decreased by about two-fold in colorectal cancer cells, thus its likelihood of binding and regulating COX-2 mRNA is drastically decreased. HuR expression is increased in colorectal cancer cells and since HuR also binds in the 3′UTR of COX-2 mRNA, additional stabilization of this mRNA and overexpression of COX-2 protein is warranted through the multimerization of HuR, a mechanism described in Kundu et al. (2012).
As Dicer is a major modulator of miRNA biogenesis it comes to no surprise that its level is important. Low levels of Dicer have been identified in several cancers associated with a poor outcome, such as breast, endometrial, lung, and ovarian cancer (Foulkes et al., 2014). However, there is also an observed increase in Dicer1 levels in the metastatic lesions in prostate cancer (Foulkes et al., 2014). This inconsistency in Dicer1 levels led scientist to focus on the specific mutations in the DICER gene rather than the levels. Both germline and somatic mutations in the DICER gene have been found in various cancers (Foulkes et al., 2014). Dicer1 syndrome is a rare genetic condition where specific mutations in the dicer gene predispose the patient for hereditary cancers (Caroleo et al., 2020). Only a third of Dicer1 pathogenic variant carriers present neoplasms during their life, suggesting there may be multiple additive events needed to create the neoplasm (Foulkes et al.,. 2014; Stewart et al., 2019; Caroleo et al., 2020). As the type of mutation in DICER gene can vary, future work will be needed to uncover how the specific type of mutation will alter the role of Dicer in recognition, binding, and processing of pre-miRNAs in addition to identifying the other events that may increase the chances of neoplasms occurring.
Neuronal development is another process that requires the function of genes whose expression is highly dependent on miRNAs. The proper expression of miRNAs is necessary for normal neuronal development, as altered levels of miRNA are observed for numerous neurodegenerative diseases (Juźwik et al., 2019). As we mentioned above, the components of the miRISC determine the function and mechanism of the miRISC and disruptions to these components, which are often due to RBPs, are observed in multiple neurodegenerative disorders. For example, brain atrophy, neurodegeneration, and gliosis are observed when Dicer is depleted in certain regions of the brain (Júzwik et al., 2019). FUS is involved in several biological processes and mutations in this RBP are observed in various neurodegenerative diseases, such as Amyotrophic Lateral Sclerosis (ALS) and Fronto-Temporal Lobar Degeneration (FTLD) (Loffreda et al., 2015; Nakaya et al., 2013). FUS has been shown to enhance the processing of certain miRNA by binding to the terminal loop of the pre-miRNA, these include miR-9, miR-125, and miR-132 (Morlando et al., 2012) which are all known to play important roles in neuronal functions (Loffreda et al., 2015). Cyclin-dependent kinases regulate the cell cycle and can initiate cell death. Their misregulation is often observed in neurodegenerative diseases, such as ALS, Parkinson’s and Alzheimer’s (Nguyen et al., 2002). CDK4 and CDK6 are known targets of the miRNA, miR-663a. miR-663a promotes cellular senescence by decreasing CDK4 and CDK6 expression (Kinoshita et al., 2021). Recent reports have found increased levels of CDK4 and CDK6 in the blood of ALS patients and connected this observation with combinatorial miRNA and RBP regulation (Katerina et al., 2018; Li et al., 2020; Kinoshita et al., 2021). Authors indicate that hnRNPH is an RBP that has been shown to indirectly decrease the expression of miR-663a. hnRNPH binds to RP11-670E13.6, which is a long noncoding RNA that, upon activation from hnRNPH binds to miR-663a and prevents miR-663a from binding to and repressing CDK4 and CDK6 mRNAs (Li et al., 2020:; Kinoshita et al., 2021). Exactly how increased levels of CDK4 and CDK6 contribute to ALS remains unclear, but it is evident that the interplay of RBPs and miRNAs contributes to this observation.
While there is clear evidence that disruption of miRISC machinery is seen in many neurodegenerative diseases, there is still a lack in understanding whether these disruptions of machinery cause the diseased state, or perhaps the disruption is a side-effect from another mishap that caused the disease (Kinoshita et al., 2021). Similarly, as an increase in pre-miRNAs is observed in tumor cells compared to normal cells, and might be a cause for tumorigenesis or metastasis, it would seem useful to study how changes in RBP expression influences miRNA biogenesis in certain cell types and cancers. Such data would clearly be beneficial in disentangling the complicated interactions between miRISC and RBPs.
Conclusion
miRNAs play an essential role in gene regulation as they control the expression of genes involved in nearly all biological processes. Dysregulation of miRISC-mediated gene silencing is prevalent in human diseases, especially in neurological disorders and cancers. The importance of miRNAs is evident, and it’s becoming clear that in order to understand the biogenesis and function of miRNAs fully, RBPs, developmental stage, and cell types must be considered. Translational repression, modulated by RBPs and miRISCs, has been shown to be reversible (Bhattacharyya et al., 2006) so it is important to consider such regulation in spatial-temporal relations. The reversibility of translational repression modulated by RBPs and miRISCs allows for flexible control of the expression of targeted genes in a wide pool of mRNAs in a timely manner. This is, as mentioned earlier, important in cases such as cellular stress, cellular growth, or proliferation as well as during cellular specification (Nawalpuri et al., 2020). In some cases, “interruptions” in translation induced by miRISC and RBPs translational control will be enough to buffer cellular stress or developmental transition states. In other cases, additional gene expression control of transcribed but “unwanted” mRNAs would be further enforced by the miRISCs associated with mRNA decay factors or by interaction with RBPs. Establishing these connections between RBPs and miRISC components will be important as they will determine mechanisms by which miRISC complexes and RBPs regulate gene expression in biological systems. These studies require the novel design of experimental setups and new biochemical, genetic, and bioinformatics methodologies. Previous methods used to study the mechanism of miRISC mediated gene silencing focused on a single miRNA or a single RBP, looking at either single reporters or multiple endogenous genes but without a good overview of miRISC and RBP interactions. Current and future methods focus on uncovering RBPs and miRNAs’ combinatorial mechanism during miRISC- or RPB-mediated gene regulation. RNA element selection assays (RESA) have been useful for selecting RNA elements based on their activity in vivo, followed by high throughput sequencing to measure their regulatory function (Yartseva et al., 2017). The use of massively parallel reporter assay libraries (MPRAs) will be critical to study the individual and combined effects of miRNAs and RBPs both in vitro and in vivo. As there are infinite combinations of miRNAs and RBPs, the use of computational analysis for predicting the interactions of RBPs and miRNAs, followed by experimental validation, will help uncover the mechanism of certain RBPs and novel RNPs. Deciphering this crosstalk between RBPs and the miRISC in development stages and disease will be critical to identify new therapeutics.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
Authors are funded by NIH T32GM007067-46 (CJ), NIH R01 GM112824 (CJ and SD), NIH R01 GM136823 (SD), NIMH R01 MH116999 (SD), The Simons Foundation (571009 to SD) and Siteman Investment Program funds (823924 to SD).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are thankful to the members of the Djuranovic lab for their help in revising this review.
References
Alarcón, C. R., Lee, H., Goodarzi, H., Halberg, N., and Tavazoie, S. F. (2015). N6-methyladenosine marks Primary microRNAs for Processing. Nature 519 (7544), 482–485. doi:10.1038/nature14281
Asting, A. G., Carén, H., Andersson, M., Lönnroth, C., Lagerstedt, K., and Lundholm, K. (2011). COX-2 Gene Expression in colon Cancer Tissue Related to Regulating Factors and Promoter Methylation Status. BMC Cancer 11 (1), 238. doi:10.1186/1471-2407-11-238
Bartel, D. P. (2004). MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 116 (2), 281–297. doi:10.1016/s0092-8674(04)00045-5
Bartel, D. P. (2009). MicroRNAs: Target Recognition and Regulatory Functions. Cell 136 (2), 215–233. doi:10.1016/j.cell.2009.01.002
Bazzini, A. A., Lee, M. T., and Giraldez, A. J. (2012). Ribosome Profiling Shows that miR-430 Reduces Translation before Causing mRNA Decay in Zebrafish. Science 336 (6078), 233–237. doi:10.1126/science.1215704
Beaudoin, J.-D., Novoa, E. M., Vejnar, C. E., Yartseva, V., Takacs, C. M., Kellis, M., et al. (2018). Analyses of mRNA Structure Dynamics Identify Embryonic Gene Regulatory Programs. Nat. Struct. Mol. Biol. 25 (8), 677–686. doi:10.1038/s41594-018-0091-z
Béthune, J., Artus‐Revel, C. G., and Filipowicz, W. (2012). Kinetic Analysis Reveals Successive Steps Leading to miRNA‐mediated Silencing in Mammalian Cells. EMBO Rep. 13 (8), 716–723. doi:10.1038/embor.2012.82
Bhat, S. S., Bielewicz, D., Gulanicz, T., Bodi, Z., Yu, X., Anderson, S. J., et al. (2020). mRNA Adenosine Methylase (MTA) Deposits m6A on pri-miRNAs to Modulate miRNA Biogenesis inArabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 117 (35), 21785–21795. doi:10.1073/pnas.2003733117
Bhattacharyya, S. N., Habermacher, R., Martine, U., Closs, E. I., and Filipowicz, W. (2006). Relief of microRNA-Mediated Translational Repression in Human Cells Subjected to Stress. Cell 125 (6), 1111–1124. doi:10.1016/j.cell.2006.04.031
Bicker, S., Khudayberdiev, S., Weiss, K., Zocher, K., Baumeister, S., and Schratt, G. (2013). The DEAH-Box Helicase DHX36 Mediates Dendritic Localization of the Neuronal Precursor-microRNA-134. Genes Dev. 27 (9), 991–996. doi:10.1101/gad.211243.112
Braun, J. E., Huntzinger, E., and Izaurralde, E. (2012). A Molecular Link between miRISCs and Deadenylases Provides New Insight into the Mechanism of Gene Silencing by MicroRNAs. Cold Spring Harbor Perspect. Biol. 4 (12), a012328. doi:10.1101/cshperspect.a012328
Braun, J. E., Huntzinger, E., and Izaurralde, E. (2013). The Role of GW182 Proteins in miRNA-Mediated Gene Silencing. Adv. Exp. Med. Biol. 768, 147–163. doi:10.1007/978-1-4614-5107-5_9
Caroleo, A. M., De Ioris, M. A., Boccuto, L., Alessi, I., Del Baldo, G., Cacchione, A., et al. (2020). DICER1 Syndrome and Cancer Predisposition: From a Rare Pediatric Tumor to Lifetime Risk. Front. Oncol. 10, 614541. doi:10.3389/fonc.2020.614541
Carthew, R. W., and Sontheimer, E. J. (2009). Origins and Mechanisms of miRNAs and siRNAs. Cell 136 (4), 642–655. doi:10.1016/j.cell.2009.01.035
Chapat, C., Jafarnejad, S. M., Matta-Camacho, E., Hesketh, G. G., Gelbart, I. A., Attig, J., et al. (2017). Cap-binding Protein 4EHP Effects Translation Silencing by microRNAs. Proc. Natl. Acad. Sci. USA 114 (21), 5425–5430. doi:10.1073/pnas.1701488114
Cheng, J., Maier, K. C., Avsec, Ž., Rus, P., and Gagneur, J. (2017). Cis-regulatory Elements Explain Most of the mRNA Stability Variation across Genes in Yeast. Rna 23 (11), 1648–1659. doi:10.1261/rna.062224.117
Connerty, P., Ahadi, A., and Hutvagner, G. (2015). RNA Binding Proteins in the miRNA Pathway. Ijms 17 (1), 31. doi:10.3390/ijms17010031
Cottrell, K. A., Chaudhari, H. G., Cohen, B. A., and Djuranovic, S. (2018). PTRE-seq Reveals Mechanism and Interactions of RNA Binding Proteins and miRNAs. Nat. Commun. 9 (1), 301. doi:10.1038/s41467-017-02745-0
Czech, B., Munafò, M., Ciabrelli, F., Eastwood, E. L., Fabry, M. H., Kneuss, E., et al. (2018). piRNA-Guided Genome Defense: From Biogenesis to Silencing. Annu. Rev. Genet. 52 (1), 131–157. doi:10.1146/annurev-genet-120417-031441
Dallaire, A., Frédérick, P.-M., and Simard, M. J. (2018). Somatic and Germline MicroRNAs Form Distinct Silencing Complexes to Regulate Their Target mRNAs Differently. Develop. Cel 47 (2), 239–247. e4. doi:10.1016/j.devcel.2018.08.022
Dana, H., Chalbatani, G. M., Mahmoodzadeh, H., Karimloo, R., Rezaiean, O., Moradzadeh, A., et al. (2017). Molecular Mechanisms and Biological Functions of siRNA. Int. J. Biomed. Sci. 13 (2), 48–57.
Di Carlo, V., Grossi, E., Laneve, P., Morlando, M., Dini Modigliani, S., Ballarino, M., et al. (2013). TDP-43 Regulates the Microprocessor Complex Activity during In Vitro Neuronal Differentiation. Mol. Neurobiol. 48 (3), 952–963. doi:10.1007/s12035-013-8564-x
Díaz-Muñoz, M. D., and Turner, M. (2018). Uncovering the Role of RNA-Binding Proteins in Gene Expression in the Immune System. Front. Immunol. 9, 1094. doi:10.3389/fimmu.2018.01094
Djuranovic, S., Nahvi, A., and Green, R. (2011). A Parsimonious Model for Gene Regulation by miRNAs. Science 331 (6017), 550–553. doi:10.1126/science.1191138
Duchaine, T. F., and Fabian, M. R. (2019). Mechanistic Insights into MicroRNA-Mediated Gene Silencing. Cold Spring Harb Perspect. Biol. 11 (3), a032771. doi:10.1101/cshperspect.a032771
Elcheva, I., Goswami, S., Noubissi, F. K., and Spiegelman, V. S. (2009). CRD-BP Protects the Coding Region of βTrCP1 mRNA from miR-183-Mediated Degradation. Mol. Cel 35 (2), 240–246. doi:10.1016/j.molcel.2009.06.007
Eulalio, A., Huntzinger, E., Nishihara, T., Rehwinkel, J., Fauser, M., and Izaurralde, E. (2009). Deadenylation is a Widespread Effect of miRNA Regulation. RNA. 15 (1), 21–32. doi:10.1261/rna.1399509
Fabian, M. R., and Sonenberg, N. (2012). The Mechanics of miRNA-Mediated Gene Silencing: A Look under the Hood of miRISC. Nat. Struct. Mol. Biol. 19 (6), 586–593. doi:10.1038/nsmb.2296
Fang, W., and Bartel, D. P. (2020). MicroRNA Clustering Assists Processing of Suboptimal MicroRNA Hairpins through the Action of the ERH Protein. Mol. Cel 78 (2), 289–302. e6. doi:10.1016/j.molcel.2020.01.026
Farazi, T. A., Juranek, S. A., and Tuschl, T. (2008). The Growing Catalog of Small RNAs and Their Association with Distinct Argonaute/Piwi Family Members. Development (Cambridge, England) 135 (7), 1201–1214. doi:10.1242/dev.005629
Filipowicz, W., Bhattacharyya, S. N., and Sonenberg, N. (2008). Mechanisms of post-transcriptional Regulation by microRNAs: Are the Answers in Sight?. Nat. Rev. Genet. 9 (2), 102–114. doi:10.1038/nrg2290
Foulkes, W. D., Priest, J. R., and Duchaine, T. F. (2014). DICER1: Mutations, microRNAs and Mechanisms. Nat. Rev. Cancer 14 (10), 662–672. doi:10.1038/nrc3802
Friedman, R. C., Farh, K. K.-H., Burge, C. B., and Bartel, D. P. (2008). Most Mammalian mRNAs Are Conserved Targets of microRNAs. Genome Res. 19 (1), 92–105. doi:10.1101/gr.082701.108
Fukao, A., Aoyama, T., and Fujiwara, T. (2015). The Molecular Mechanism of Translational Control via the Communication Between the MicroRNA Pathway and RNA-binding Proteins. RNA Biology. 12 (9), 922–926. doi:10.1080/15476286.2015.1073436
Fukao, A., Tomohiro, T., and Fujiwara, T. (2021). Translation Initiation Regulated by RNA-Binding Protein in Mammals: The Modulation of Translation Initiation Complex by Trans-acting Factors. Cells 10 (7), 1711. doi:10.3390/cells10071711
Gardiner, A., Twiss, J., and Perrone-Bizzozero, N. (2015). Competing Interactions of RNA-Binding Proteins, MicroRNAs, and Their Targets Control Neuronal Development and Function. Biomolecules 5 (4), 2903–2918. doi:10.3390/biom5042903
Gebauer, F., and Hentze, M. W. (2004). Molecular Mechanisms of Translational Control. Nat. Rev. Mol. Cel Biol 5 (10), 827–835. doi:10.1038/nrm1488
Gebauer, F., Schwarzl, T., Valcárcel, J., and Hentze, M. W. (2021). RNA-binding Proteins in Human Genetic Disease. Nat. Rev. Genet. 22 (3), 185–198. doi:10.1038/s41576-020-00302-y
Gebert, L. F. R., and MacRae, I. J. (2019). Regulation of microRNA Function in Animals. Nat. Rev. Mol. Cel Biol 20 (1), 21–37. doi:10.1038/s41580-018-0045-7
Gu, S., and Kay, M. A. (2010). How Do miRNAs Mediate Translational Repression?. Silence 1 (1), 11. doi:10.1186/1758-907X-1-11
Ha, M., and Kim, V. N. (2014). Regulation of microRNA Biogenesis. Nat. Rev. Mol. Cel Biol 15 (8), 509–524. doi:10.1038/nrm3838
Hentze, M. W., Castello, A., Schwarzl, T., and Preiss, T. (2018). A Brave New World of RNA-Binding Proteins. Nat. Rev. Mol. Cel Biol 19 (5), 327–341. doi:10.1038/nrm.2017.130
Heo, I., Ha, M., Lim, J., Yoon, M-J., Park, J-E., Kwon, S. C., et al. (2012). Mono-Uridylation of Pre-MicroRNA as a Key Step in the Biogenesis of Group II let-7 MicroRNAs. Cell 151 (3), 521–532. doi:10.1016/j.cell.2012.09.022
Horvitz, H. R., and Sulston, J. E. (1980). Isolation and Genetic Characterization of Cell-Lineage Mutants of the Nematode Caenorhabditis Elegans. Genetics 96 (2), 435–454. doi:10.1093/genetics/96.2.435
Hu, W., and Coller, J. (2012). What Comes First: Translational Repression or mRNA Degradation? the Deepening Mystery of microRNA Function. Cell Res 22 (9), 1322–1324. doi:10.1038/cr.2012.80
Huntzinger, E., Kuzuoğlu-Öztürk, D., Braun, J. E., Eulalio, A., Wohlbold, L., and Izaurralde, E. (2013). The Interactions of GW182 Proteins with PABP and Deadenylases Are Required for Both Translational Repression and Degradation of miRNA Targets. Nucleic Acids Res. 41 (2), 978–994. doi:10.1093/nar/gks1078
Hutter, K., Lohmüller, M., Jukic, A., Eichin, F., Avci, S., Labi, V., et al. (2020). SAFB2 Enables the Processing of Suboptimal Stem-Loop Structures in Clustered Primary miRNA Transcripts. Mol. Cel 78 (5), 876–889. e6. doi:10.1016/j.molcel.2020.05.011
Hutvagner, G., and Simard, M. J. (2008). Argonaute Proteins: Key Players in RNA Silencing. Nat. Rev. Mol. Cel Biol 9 (1), 22–32. doi:10.1038/nrm2321
Iadevaia, V., and Gerber, A. (2015). Combinatorial Control of mRNA Fates by RNA-Binding Proteins and Non-coding RNAs. Biomolecules 5 (4), 2207–2222. doi:10.3390/biom5042207
Jiang, P., and Coller, H. (2012). Functional Interactions between microRNAs and RNA Binding Proteins. Mirna 1 (1), 70–79. doi:10.2174/2211536611201010070
Jiang, P., Singh, M., and Coller, H. A. (2013). Computational Assessment of the Cooperativity between RNA Binding Proteins and MicroRNAs in Transcript Decay. Plos Comput. Biol. 9 (5), e1003075. doi:10.1371/journal.pcbi.1003075
Jing, Q., Huang, S., Guth, S., Zarubin, T., Motoyama, A., Chen, J., et al. (2005). Involvement of microRNA in AU-Rich Element-Mediated mRNA Instability. Cell 120 (5), 623–634. doi:10.1016/j.cell.2004.12.038
Juliano, C., Wang, J., and Lin, H. (2011). Uniting Germline and Stem Cells: The Function of Piwi Proteins and the piRNA Pathway in Diverse Organisms. Annu. Rev. Genet. 45 (1), 447–469. doi:10.1146/annurev-genet-110410-132541
Juźwik, C. A., Drake, S. S., Zhang, Y., Paradis-Isler, N., Sylvester, A., Amar-Zifkin, A., et al. (2019). microRNA Dysregulation in Neurodegenerative Diseases: A Systematic Review. Prog. Neurobiol. 182, 101664. doi:10.1016/j.pneurobio.2019.101664
Kakumani, P. K., Guitart, T., Houle, F., Harvey, L.-M., Goyer, B., Germain, L., et al. (2021). CSDE1 Attenuates microRNA-Mediated Silencing of PMEPA1 in Melanoma. Oncogene 40 (18), 3231–3244. doi:10.1038/s41388-021-01767-9
Kanellopoulou, C., Muljo, S. A., Kung, A. L., Ganesan, S., Drapkin, R., Jenuwein, T., et al. (2005). Dicer-deficient Mouse Embryonic Stem Cells Are Defective in Differentiation and Centromeric Silencing. Genes Dev. 19 (4), 489–501. doi:10.1101/gad.1248505
Kawahara, Y., and Mieda-Sato, A. (2012). TDP-43 Promotes microRNA Biogenesis as a Component of the Drosha and Dicer Complexes. Proc. Natl. Acad. Sci. 109 (9), 3347–3352. doi:10.1073/pnas.1112427109
Kawahara, Y., Zinshteyn, B., Chendrimada, T. P., Shiekhattar, R., and Nishikura, K. (2007). RNA Editing of the microRNA‐151 Precursor Blocks Cleavage by the Dicer-TRBP Complex. EMBO Rep. 8 (8), 763–769. doi:10.1038/sj.embor.7401011
Kedde, M., van Kouwenhove, M., Zwart, W., Oude Vrielink, J. A. F., Elkon, R., and Agami, R. (2010). A Pumilio-Induced RNA Structure Switch in P27-3′ UTR Controls miR-221 and miR-222 Accessibility. Nat. Cel Biol 12 (10), 1014–1020. doi:10.1038/ncb2105
Khan, S., Ayub, H., Khan, T., and Wahid, F. (2019). MicroRNA Biogenesis, Gene Silencing Mechanisms and Role in Breast, Ovarian and Prostate Cancer. Biochimie 167, 12–24. doi:10.1016/j.biochi.2019.09.001
Kim, H. H., Kuwano, Y., Srikantan, S., Lee, E. K., Martindale, J. L., and Gorospe, M. (2009). HuR Recruits Let-7/RISC to Repress C-Myc Expression. Genes Dev. 23 (15), 1743–1748. doi:10.1101/gad.1812509
Kim, K., Nguyen, T. D., Li, S., and Nguyen, T. A. (2018). SRSF3 Recruits DROSHA to the Basal junction of Primary microRNAs. Rna 24 (7), 892–898. doi:10.1261/rna.065862.118
Kinoshita, C., Kubota, N., and Aoyama, K. (2021). Interplay of RNA-Binding Proteins and microRNAs in Neurodegenerative Diseases. Ijms 22 (10), 5292. doi:10.3390/ijms22105292
Kooshapur, H., Choudhury, N. R., Simon, B., Mühlbauer, M., Jussupow, A., Fernandez, N., et al. (2018). Structural Basis for Terminal Loop Recognition and Stimulation of Pri-miRNA-18a Processing by hnRNP A1. Nat. Commun. 9 (1), 2479. doi:10.1038/s41467-018-04871-9
Kundu, P., Fabian, M. R., Sonenberg, N., Bhattacharyya, S. N., and Filipowicz, W. (2012). HuR Protein Attenuates miRNA-Mediated Repression by Promoting miRISC Dissociation from the Target RNA. Nucleic Acids Res. 40 (11), 5088–5100. doi:10.1093/nar/gks148
Kwon, S. C., Jang, H., Shen, S., Baek, S. C., Kim, K., Yang, J., et al. (2020). ERH Facilitates microRNA Maturation through the Interaction with the N-Terminus of DGCR8. Nucleic Acids Res. 48 (19), 11097–11112. doi:10.1093/nar/gkaa827
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell 75 (5), 843–854. doi:10.1016/0092-8674(93)90529-y
Li, S., Li, M., Liu, K., Zhang, H., Zhang, S., Zhang, C., et al. (2020). MAC5, an RNA-Binding Protein, Protects Pri-miRNAs from SERRATE-dependent Exoribonuclease Activities. Proc. Natl. Acad. Sci. USA 117 (38), 23982–23990. doi:10.1073/pnas.2008283117
Lim, L. P., Lau, N. C., Weinstein, E. G., Abdelhakim, A., Yekta, S., Rhoades, M. W., et al. (2003). The microRNAs ofCaenorhabditis Elegans. Genes Dev. 17 (8), 991–1008. doi:10.1101/gad.1074403
Lin, S., and Gregory, R. I. (2015). MicroRNA Biogenesis Pathways in Cancer. Nat. Rev. Cancer 15 (6), 321–333. doi:10.1038/nrc3932
Loffreda, A., Rigamonti, A., Barabino, S., and Lenzken, S. (2015). RNA-binding Proteins in the Regulation of miRNA Activity: A Focus on Neuronal Functions. Biomolecules 5 (4), 2363–2387. doi:10.3390/biom5042363
Martin, J. C., Tippie, M. A., McGee, D. P. C., and Verheyden, J. P. H. (1987). Synthesis and Antiviral Activity of Various Esters of 9-[(1,3-Dihydroxy-2-Propoxy)methyl]guanine. J. Pharm. Sci. 76 (2), 180–184. doi:10.1002/jps.2600760221
Meister, G. (2013). Argonaute Proteins: Functional Insights and Emerging Roles. Nat. Rev. Genet. 14 (7), 447–459. doi:10.1038/nrg3462
Michlewski, G., and Cáceres, J. F. (2019). Post-transcriptional Control of miRNA Biogenesis. RNA 25 (1), 1–16. doi:10.1261/rna.068692.118
Michlewski, G., Guil, S., Semple, C. A., and Cáceres, J. F. (2008). Posttranscriptional Regulation of miRNAs Harboring Conserved Terminal Loops. Mol. Cel 32 (3), 383–393. doi:10.1016/j.molcel.2008.10.013
Miles, W. O., Tschöp, K., Herr, A., Ji, J.-Y., and Dyson, N. J. (2012). Pumilio Facilitates miRNA Regulation of the E2F3 Oncogene. Genes Dev. 26 (4), 356–368. doi:10.1101/gad.182568.111
Miyoshi, K., Miyoshi, T., and Siomi, H. (2010). Many Ways to Generate microRNA-like Small RNAs: Non-canonical Pathways for microRNA Production. Mol. Genet. Genomics 284 (2), 95–103. doi:10.1007/s00438-010-0556-1
Morlando, M., Dini Modigliani, S., Torrelli, G., Rosa, A., Di Carlo, V., Caffarelli, E., et al. (2012). FUS Stimulates microRNA Biogenesis by Facilitating Co-transcriptional Drosha Recruitment. EMBO J. 31 (24), 4502–4510. doi:10.1038/emboj.2012.319
Müller, M., Fazi, F., and Ciaudo, C. (2020). Argonaute Proteins: From Structure to Function in Development and Pathological Cell Fate Determination. Front. Cel Dev. Biol. 7, 360. doi:10.3389/fcell.2019.00360
Murphy, D., Dancis, B., and Brown, J. R. (2008). The Evolution of Core Proteins Involved in microRNA Biogenesis. BMC Evol. Biol. 8 (1), 92. doi:10.1186/1471-2148-8-92
Nakaya, T., Alexiou, P., Maragkakis, M., Chang, A., and Mourelatos, Z. (2013). FUS Regulates Genes Coding for RNA-Binding Proteins in Neurons by Binding to Their Highly Conserved Introns. RNA 19 (4), 498–509. doi:10.1261/rna.037804.112
Nawalpuri, B., Ravindran, S., and Muddashetty, R. S. (2020). The Role of Dynamic miRISC during Neuronal Development. Front. Mol. Biosci. 7, 8. doi:10.3389/fmolb.2020.00008
Nguyen, M. D., Mushynski, W. E., and Julien, J.-P. (2002). Cycling at the Interface between Neurodevelopment and Neurodegeneration. Cell Death Differ 9 (12), 1294–1306. doi:10.1038/sj.cdd.4401108
Nguyen, T. A., Jo, M. H., Choi, Y-G., Park, J., Kwon, S. C., Hohng, S., et al. (2015). Functional Anatomy of the Human Microprocessor. Cell 161 (6), 1374–1387. doi:10.1016/j.cell.2015.05.010
Nolde, M. J., Saka, N., Reinert, K. L., and Slack, F. J. (2007). The Caenorhabditis elegans Pumilio Homolog, Puf-9, Is Required for the 3′UTR-Mediated Repression of the Let-7 microRNA Target Gene, Hbl-1. Develop. Biol. 305 (2), 551–563. doi:10.1016/j.ydbio.2007.02.040hbl-1. 25
Nussbacher, J. K., and Yeo, G. W. (2018). Systematic Discovery of RNA Binding Proteins that Regulate MicroRNA Levels. Mol. Cel 69 (6), 1005–1016. e7. doi:10.1016/j.molcel.2018.02.012
Ouyang, H., Zhang, K., Fox-Walsh, K., Yang, Y., Zhang, C., Huang, J., et al. (2017). The RNA Binding Protein EWS Is Broadly Involved in the Regulation of Pri-miRNA Processing in Mammalian Cells. Nucleic Acids Res. 45 (21), 12481–12495. doi:10.1093/nar/gkx912
Palma, K., Zhao, Q., Cheng, Y. T., Bi, D., Monaghan, J., Cheng, W., et al. (2007). Regulation of Plant Innate Immunity by Three Proteins in a Complex Conserved across the Plant and Animal Kingdoms. Genes Dev. 21 (12), 1484–1493. doi:10.1101/gad.1559607
Parker, J. S., and Barford, D. (2006). Argonaute: A Scaffold for the Function of Short Regulatory RNAs. Trends Biochem. Sci. 31 (11), 622–630. doi:10.1016/j.tibs.2006.09.010
Peng, Y., and Croce, C. M. (2016). The Role of MicroRNAs in Human Cancer. Sig Transduct Target. Ther. 1 (1), 15004. doi:10.1038/sigtrans.2015.4
Peters, L., and Meister, G. (2007). Argonaute Proteins: Mediators of RNA Silencing. Mol. Cel 26 (5), 611–623. doi:10.1016/j.molcel.2007.05.001
Pinder, B. D., and Smibert, C. A. (2013). microRNA‐Independent Recruitment of Argonaute 1 to Nanos mRNA through the Smaug RNA‐binding Protein. EMBO Rep. 14 (1), 80–86. doi:10.1038/embor.2012.192
Ramanathan, M., Porter, D. F., and Khavari, P. A. (2019). Methods to Study RNA-Protein Interactions. Nat. Methods 16 (3), 225–234. doi:10.1038/s41592-019-0330-1
Rehwinkel, J., Behm-Ansmant, I., Gatfield, D., and Izaurralde, E. (2005). A Crucial Role for GW182 and the DCP1:DCP2 Decapping Complex in miRNA-Mediated Gene Silencing. RNA 11 (11), 1640–1647. doi:10.1261/rna.2191905
Reinhart, B. J., Slack, F. J., Basson, M., Pasquinelli, A. E., Bettinger, J. C., Rougvie, A. E., et al. (2000). The 21-nucleotide Let-7 RNA Regulates Developmental Timing in Caenorhabditis elegans. Nature 403 (6772), 901–906. doi:10.1038/35002607
Roy, B., and Jacobson, A. (2013). The Intimate Relationships of mRNA Decay and Translation. Trends Genet. 29 (12), 691–699. doi:10.1016/j.tig.2013.09.002
Salim, U., Kumar, A., Kulshreshtha, R., and Vivekanandan, P. (2021). Biogenesis, Characterization, and Functions of Mirtrons. WIREs RNA 13 (1), e1680. doi:10.1002/wrna.1680
Schopp, I. M., Amaya Ramirez, C. C., Debeljak, J., Kreibich, E., Skribbe, M., Wild, K., et al. (2017). Split-BioID a Conditional Proteomics Approach to Monitor the Composition of Spatiotemporally Defined Protein Complexes. Nat. Commun. 8 (1), 15690. doi:10.1038/ncomms15690
Shang, R., Baek, S. C., Kim, K., Kim, B., Kim, V. N., and Lai, E. C. (2020). Genomic Clustering Facilitates Nuclear Processing of Suboptimal Pri-miRNA Loci. Mol. Cel 78 (2), 303–316. e4. doi:10.1016/j.molcel.2020.02.009
Shenoy, A., and Blelloch, R. H. (2014). Regulation of microRNA Function in Somatic Stem Cell Proliferation and Differentiation. Nat. Rev. Mol. Cel Biol 15 (9), 565–576. doi:10.1038/nrm3854
Sheu-Gruttadauria, J., and MacRae, I. J. (2017). Structural Foundations of RNA Silencing by Argonaute. J. Mol. Biol. 429 (17), 2619–2639. doi:10.1016/j.jmb.2017.07.018
Shih, S.-C., and Claffey, K. P. (1999). Regulation of Human Vascular Endothelial Growth Factor mRNA Stability in Hypoxia by Heterogeneous Nuclear Ribonucleoprotein L. J. Biol. Chem. 274 (3), 1359–1365. doi:10.1074/jbc.274.3.1359
Siegel, G., Saba, R., and Schratt, G. (2011). microRNAs in Neurons: Manifold Regulatory Roles at the Synapse. Curr. Opin. Genet. Develop. 21 (4), 491–497. doi:10.1016/j.gde.2011.04.008
Siomi, H., and Siomi, M. C. (2010). Posttranscriptional Regulation of MicroRNA Biogenesis in Animals. Mol. Cel 38 (3), 323–332. doi:10.1016/j.molcel.2010.03.013
Song, J.-J., Smith, S. K., Hannon, G. J., and Joshua-Tor, L. (2004). Crystal Structure of Argonaute and its Implications for RISC Slicer Activity. Science 305 (5689), 1434–1437. doi:10.1126/science.1102514
Stewart, D. R., Best, A. F., Williams, G. M., Harney, L. A., Carr, A. G., Harris, A. K., et al. (2019). Neoplasm Risk Among Individuals with a Pathogenic Germline Variant in DICER1. Jco 37 (8), 668–676. doi:10.1200/JCO.2018.78.4678
Sun, G., Li, H., and Rossi, J. J. (2010). Sequence Context outside the Target Region Influences the Effectiveness of miR-223 Target Sites in the RhoB 3′UTR. Nucleic Acids Res. 38 (1), 239–252. doi:10.1093/nar/gkp870
Thomson, T., and Lin, H. (2009). The Biogenesis and Function of PIWI Proteins and piRNAs: Progress and prospect. Annu. Rev. Cel Dev. Biol. 25, 355–376. doi:10.1146/annurev.cellbio.24.110707.175327
Treiber, T., Treiber, N., Plessmann, U., Harlander, S., Daiß, J.-L., Eichner, N., et al. (2017). A Compendium of RNA-Binding Proteins that Regulate MicroRNA Biogenesis. Mol. Cel 66 (2), 270–284. e13. doi:10.1016/j.molcel.2017.03.014
van Kouwenhove, M., Kedde, M., and Agami, R. (2011). MicroRNA Regulation by RNA-Binding Proteins and its Implications for Cancer. Nat. Rev. Cancer 11 (9), 644–656. doi:10.1038/nrc3107
Vejnar, C. E., Abdel Messih, M., Takacs, C. M., Yartseva, V., Oikonomou, P., Christiano, R., et al. (2019). Genome Wide Analysis of 3′ UTR Sequence Elements and Proteins Regulating mRNA Stability during Maternal-To-Zygotic Transition in Zebrafish. Genome Res. 29 (7), 1100–1114. doi:10.1101/gr.245159.118
Voinnet, O. (2009). Origin, Biogenesis, and Activity of Plant MicroRNAs. Cell 136 (4), 669–687. doi:10.1016/j.cell.2009.01.046
Vos, P. D., Leedman, P. J., Filipovska, A., and Rackham, O. (2019). Modulation of miRNA Function by Natural and Synthetic RNA-Binding Proteins in Cancer. Cell. Mol. Life Sci. 76 (19), 3745–3752. doi:10.1007/s00018-019-03163-9
Vrabec, K., Boštjančič, E., Koritnik, B., Leonardis, L., Dolenc Grošelj, L., Zidar, J., et al. (2018). Differential Expression of Several miRNAs and the Host Genes AATK and DNM2 in Leukocytes of Sporadic ALS Patients. Front. Mol. Neurosci. 11, 106. doi:10.3389/fnmol.2018.00106
Wang, J., Mei, J., and Ren, G. (2019). Plant microRNAs: Biogenesis, Homeostasis, and Degradation. Front. Plant Sci. 10, 360. doi:10.3389/fpls.2019.00360
Westholm, J. O., and Lai, E. C. (2011). Mirtrons: MicroRNA Biogenesis via Splicing. Biochimie 93 (11), 1897–1904. doi:10.1016/j.biochi.2011.06.017
Wigington, C. P., Williams, K. R., Meers, M. P., Bassell, G. J., and Corbett, A. H. (2014). Poly(A) RNA-Binding Proteins and Polyadenosine RNA: New Members and Novel Functions. WIREs RNA 5 (5), 601–622. doi:10.1002/wrna.1233
Wilczynska, A., and Bushell, M. (2015). The Complexity of miRNA-Mediated Repression. Cel Death Differ 22 (1), 22–33. doi:10.1038/cdd.2014.112
Wilson, G. M., Sun, Y., Lu, H., and Brewer, G. (1999). Assembly of AUF1 Oligomers on U-Rich RNA Targets by Sequential Dimer Association. J. Biol. Chem. 274 (47), 33374–33381. doi:10.1074/jbc.274.47.33374
Wu, J. e., Yang, J., Cho, W. C., and Zheng, Y. (2020). Argonaute Proteins: Structural Features, Functions and Emerging Roles. J. Adv. Res. 24, 317–324. doi:10.1016/j.jare.2020.04.017
Wyman, S. K., Knouf, E. C., Parkin, R. K., Fritz, B. R., Lin, D. W., Dennis, L. M., et al. (2011). Post-transcriptional Generation of miRNA Variants by Multiple Nucleotidyl Transferases Contributes to miRNA Transcriptome Complexity. Genome Res. 21 (9), 1450–1461. doi:10.1101/gr.118059.110
Yang, W., Chendrimada, T. P., Wang, Q., Higuchi, M., Seeburg, P. H., Shiekhattar, R., et al. (2006). Modulation of microRNA Processing and Expression through RNA Editing by ADAR Deaminases. Nat. Struct. Mol. Biol. 13 (1), 13–21. doi:10.1038/nsmb1041
Yartseva, V., Takacs, C. M., Vejnar, C. E., Lee, M. T., and Giraldez, A. J. (2017). RESA Identifies mRNA-Regulatory Sequences at High Resolution. Nat. Methods 14 (2), 201–207. doi:10.1038/nmeth.4121
Yoon, J.-H., De, S., Srikantan, S., Abdelmohsen, K., Grammatikakis, I., Kim, J., et al. (2014). PAR-CLIP Analysis Uncovers AUF1 Impact on Target RNA Fate and Genome Integrity. Nat. Commun. 5, 5248. doi:10.1038/ncomms6248
Young, L. E., Moore, A. E., Sokol, L., Meisner-Kober, N., and Dixon, D. A. (2012). The mRNA Stability Factor HuR Inhibits MicroRNA-16 Targeting of COX-2. Mol. Cancer Res. 10 (1), 167–180. doi:10.1158/1541-7786.MCR-11-0337
Zhang, Y., Wei, Z., Dong, H., Zhou, J., Yuan, J., Ni, B., et al. (2021). Regulation of mRNA Stability by RBPs and Noncoding RNAs Contributing to the Pathogenicity of Th17 Cells. RNA Biol. 18 (5), 647–656. doi:10.1080/15476286.2020.1862567
Zucconi, B. E., Ballin, J. D., Brewer, B. Y., Ross, C. R., Huang, J., Toth, E. A., et al. (2010). Alternatively Expressed Domains of AU-Rich Element RNA-Binding Protein 1 (AUF1) Regulate RNA-Binding Affinity, RNA-Induced Protein Oligomerization, and the Local Conformation of Bound RNA Ligands. J. Biol. Chem. 285 (50), 39127–39139. doi:10.1074/jbc.M110.180182
Keywords: miRNA, miRISC, RNA binding protein, mRNA, miRISC activity, RBP binding models
Citation: Jungers CF and Djuranovic S (2022) Modulation of miRISC-Mediated Gene Silencing in Eukaryotes. Front. Mol. Biosci. 9:832916. doi: 10.3389/fmolb.2022.832916
Received: 10 December 2021; Accepted: 18 January 2022;
Published: 14 February 2022.
Edited by:
Deepika Vasudevan, University of Pittsburgh, United StatesReviewed by:
Jeffrey Vedanayagam, Memorial Sloan Kettering Cancer Center, United StatesUbaldo Gioia, IFOM - The FIRC Institute of Molecular Oncology, Italy
Copyright © 2022 Jungers and Djuranovic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sergej Djuranovic, c2VyZ2VqLmRqdXJhbm92aWNAd3VzdGwuZWR1
 Courtney F. Jungers
Courtney F. Jungers Sergej Djuranovic
Sergej Djuranovic