- 1Centre for Informational Biology, School of Life Science and Technology, University of Electronic Science and Technology of China, Chengdu, China
- 2Centre for Advanced Materials Research, Advanced Institute of Natural Sciences, Beijing Normal University at Zhuhai, Zhuhai, China
The Hedgehog (Hh) signaling pathway is essential in cell development and regeneration, which is activated by the ligand Sonic hedgehog (Shh). The binding of Shh to its receptor Patched1 (PTCH1) releases the inhibitory effect on the downstream protein Smoothened (SMO), a G-protein-coupled-receptor (GPCR) protein. Cholesterol was supposed to function as a secondary messenger between PTCH1 and SMO. However, the molecular mechanism of this regulation process is still unclear. Therefore, microsecond coarse-grained molecular dynamics simulations were performed to investigate the protein-lipid interactions of the PTCH1 monomer and dimer-Shh complex. It was observed that the binding of cholesterols to the monomer is more stable than that to the dimer-Shh complex. It is regulated by the enrichment of Ganglioside lipids around proteins and the conformation of Y446, a residue in the sterol-sensing domain (SSD). The regulation of Shh on the dynamics of PTCH1 was further analyzed to explore the allosteric communication pathways between the Shh and the SSD. Our study provides structural and dynamic details of an additional perspective on the regulation of Hh signaling pathway through the lipid micro-environments of PTCH1.
Introduction
The Hedgehog (Hh) signaling pathway transmits information between cells, and is critical for embryogenesis and tissue regeneration (Ingham et al., 2011; Briscoe and Therond, 2013). Defects in Hh pathway may lead to birth defects such as holoprosencephaly (Roessler et al., 1996), whereas abnormally activated Hh pathway leads to cancers, most commonly basal cell carcinoma and medulloblastoma (Teglund and Toftgard, 2010). Numerous biophysical and biochemical studies have been reported to explore the molecular mechanism of the Hh pathway (Kong et al., 2019).
The transmitting of Hh signals across the cell membrane is accomplished by two membrane proteins, Patched 1 (PTCH1) and Smoothened (SMO) (Ingham and McMahon, 2001; Haycraft et al., 2005; Wen et al., 2010). PTCH1 works as a receptor, receiving extracellular Hh signals, and then passes to SMO. SMO, a G-protein-coupled-receptor (GPCR), transduces signals across the cell membrane. In the absence of Hh signals, PTCH1 inhibits SMO. The reception of Hh signals, such as the Sonic hedgehog (Shh), activates the Hh signaling pathway by inhibiting PTCH1 and releasing the inhibition of SMO. SMO was not found to have directly interactions with PTCH1 (Taipale et al., 2002), and is activated by cholesterols (CHOLs) (Radhakrishnan et al., 2020), as most GPCRs do (Kiriakidi et al., 2019). Therefore, different mechanisms of the regulation have been proposed on how PTCH1 regulates the activity of SMO, but converged at accessible CHOL working as a second messenger between PTCH1 and SMO (Radhakrishnan et al., 2020).
The Hh ligand, Shh, is modified by N-terminal palmitoylation and C-terminal cholesterylation (Qi et al., 2018a). Shh binds two PTCH1 receptors (PTCH1-A and PTCH1-B) asymmetrically, forming the PTCH1 dimer-Shh complex. PTCH1 contains two extended ectodomains (ECDs, ECD1 and ECD2) and a transmembrane domain (TMD) with 12 TM helices (TM 1-12) (Gong et al., 2018; Qi et al., 2019) (Figure 1A). TMs 1-6 and TMs 7-12 form two pseudo-symmetrical segments. A sterol-sensing domain (SSD), conserved in several cholesterol-related proteins (Davies and Ioannou, 2000), was identified at TMs 2-6, and an SSD-like (SSDL) domain at TMs 8-12 (Figure 1B; Supplementary Figure S1). CHOLs were found to interact at multiple sites of both SSD and SSDL (Qi et al., 2019) with different binding affinities (Ansell et al., 2021). Two ECDs are located between TMs 1 and 2, and TMs 7 and 8 respectively, and involved in ligand binding. Densities of sterol-like molecules in the ECD suggest a hydrophobic conduit (Zhang et al., 2018). The structures of the 2:1 PTCH1 and Shh complex (termed dimer-Shh here) (Qi et al., 2018a; Qian et al., 2019; Rudolf et al., 2019) show that Shh binds ECDs of two PTCH1s with its C-terminal CHOL bound in the ECD1, and the N-terminal palmitate and 15 residues inserted into the ECD cavity of PTCH1-A (Figure 1A).
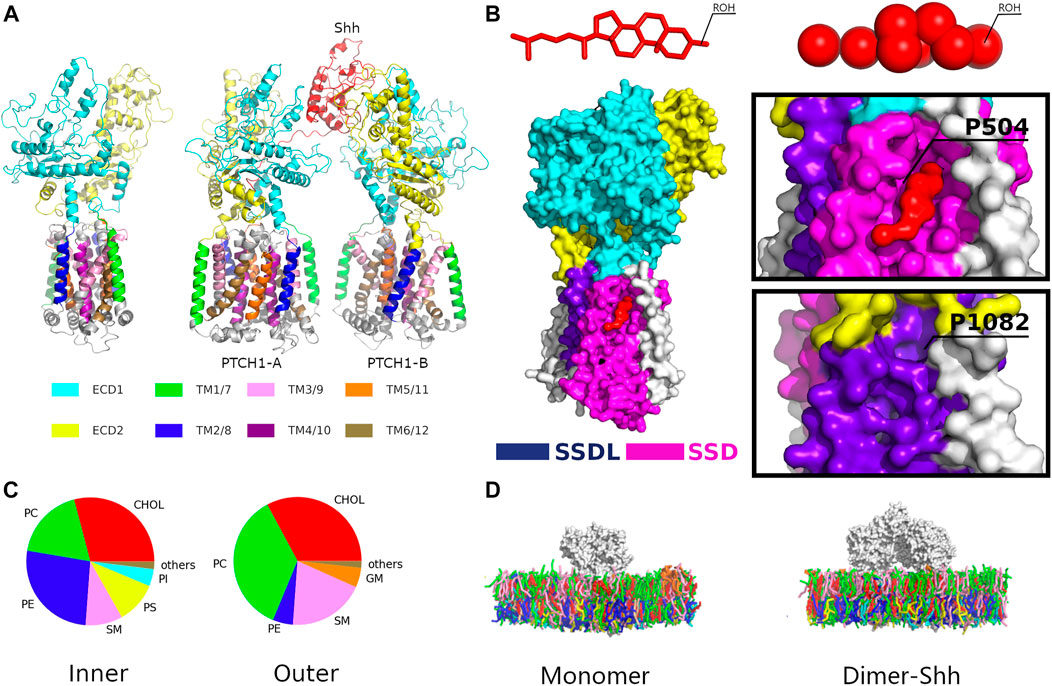
FIGURE 1. The simulation system. (A) Structures of the PTCH1 monomer and dimer-Shh. (B) The locations of the SSD and SSDL sites with a close view of Cholesterol binding to the SSD pocket. (C) The lipid compositions of the outer and inner leaflets of the membrane model. Lipid types included are phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), sphingomyelin (SM), phosphatidylinositol (PI), ganglioside (GM), Phosphatidic acid (PA), and cholesterol (CHOL). (D) The Martini CG models of PTCH1 monomer and dimer-Shh systems.
PTCH1 has homology with Niemann–Pick C1 (NPC1), a lysosomal membrane protein that transports CHOL from the lumen of the lysosome to the cytoplasm, and resistance-nodulation-division (RND)-family transporters, which efflux toxic molecules out of gram-negative bacteria (Gong et al., 2016; Pfeffer, 2019). Supported by the structural evidence (Qi et al., 2018b; Zhang et al., 2018), PTCH1 was supposed to be a potential CHOL transporter that transfers CHOLs from the cell membrane to the outside of the cell, to reduce accessible CHOLs (Zhang et al., 2018; Kinnebrew et al., 2019). The insertion of the N-terminal palmitate of Shh would block the hydrophobic tunnel, thus inactivating PTCH1. However, Shh, lacking of the N-terminal palmitate and C-terminal CHOL, can also inhibit PTCH1, although the potency is significantly reduced (Pepinsky et al., 1998; Chen et al., 2004). It indicates that the PTCH1-Shh interactions play a role in the regulation of Hh signaling pathway.
In this study, therefore, we resorted to molecular dynamics (MD) simulations to elucidate how the protein-protein interactions of PTCH1 and Shh regulate the accessible CHOL. MD simulations with the Martini Coarse-Grained (CG) force field (Marrink et al., 2007) have been widely used to get insight of the structural details of protein-lipid interactions (Corradi et al., 2018; Corradi et al., 2019; Sejdiu and Tieleman, 2020). Microsecond (μs) CG MD simulations were performed for the PTCH1 monomer and dimer-Shh systems inserted in asymmetric plasma membranes. Differences in lipid micro-environments and CHOL binding between the two systems may illustrate how the binding of Shh regulates accessible CHOLs.
Methods
System setup. The cryo-EM structure of human PTCH1 (PDB ID: 6DMB) (Gong et al., 2018) and human PTCH1-Shh-N complex (PDB ID: 6E1H) (Qi et al., 2018a) were used for the initial structures of PTCH1 monomer and dimer-Shh respectively. In order to investigate the binding of CHOLs in the cell membrane to the protein, the bound cholesterols were removed. The N-terminal palmitate moiety of Shh was not included in the simulation system too, to study the function of the PTCH1-Shh interactions.
The autopsf tool of VMD (Humphrey et al., 1996) was used to repair the missing side chains. Then the atomistic models were converted to Martini CG models using martinize.py, and then CG models were inserted into the CG membrane by insane.py (Wassenaar et al., 2015) with the lipid composition according to the plasma membrane (Ingolfsson et al., 2014) (Figures 1C,D; Supplementary Figure S2; Supplementary Table S1). Finally, the systems were solvated with Martini water models, and neutralized and ionized with 0.15 m/L NaCl. The final simulation systems consist of approximate 55,000 Martini beads.
Simulation details. All simulations were carried out with GROMACS version 2019.2 (Van Der Spoel et al., 2005) and Martini 2.2 force field (Marrink et al., 2007). The systems were firstly minimized for 2000 steps with steepest descent method, and then equilibrated by 20 ns NVT simulations with incremental time steps of 2 fs, 5 fs, 10 fs, 15 fs, and 20 fs. Production runs were performed with the NPT ensemble of the constant temperature of 300 K by velocity-rescaling thermostat (Bussi et al., 2007) and constant pressure (1atm) via Parrinello-Rahman barostat (Berendsen et al., 1984). A cutoff of 12 Å with a shift of 10 Å was used for calculations of van der Waals and electrostatic interactions. Both PTCH1 monomer and dimer-Shh simulations were performed for a total simulation time of 200 μs, 5 replicates of 40 μs, respectively. All the analyses were performed using VMD (Humphrey et al., 1996).
Protein-lipid interactions. The protein-lipid interaction was considered when the headgroup of a lipid is within 0.65 nm of any residue of the protein. For CHOL binding to the SSD/SSDL, the cutoff was set to 1 nm instead of 0.65 nm, because CHOL could be mobile even when interacting at specific interaction sites (Rouviere et al., 2017; Hedger et al., 2019; Duncan et al., 2020). All analyses were performed on the last 10 μs of each simulation.
Allosteric communication pathways. The Network View plugin (Sethi et al., 2009) of VMD were used to identify allosteric communication networks. The dynamic networks were constructed using our MD trajectories, each sampled every 1ns, with residues represented by the backbone beads. Two nodes (excluding neighboring nodes) within a distance of 6 Å for more than 75% of MD trajectories were defined as contact residue pairs and used to calculate the local-contact matrix. The correlation between two nodes was calculated as
The length of a path is the sum of the weights along this path, and calculated as
Results
In total of 400 μs simulations were produced. Firstly, we analyzed the CHOL binding and interactions in the SSD pocket. Then, the lipid micro-environments were characterized for both systems. Finally, the conformational dynamic couplings were calculated to reveal the regulation of Shh binding on PTCH1.
CHOL binding to the SSD and SSDL. As above-mentioned, the SSD of PTCH1 appears a V-shaped cavity, usually occupied by sterol molecules in reported structures (Gong et al., 2018). Comparatively, the SSDL pocket is shallow, and showed low CHOL binding affinity (Supplementary Figure S1). Here, the CHOL binding to the SSD was defined by the distance between the polar hydroxyl group of CHOL (termed ROH in the CG model) and P504, a conserved residue mapping the bottom of the SSD pocket (Boutet et al., 2003; Li et al., 2016; Gong et al., 2018) (Figure 1B). P504 has direct interactions with the ligand and it is involved in all interaction sites of the SSD (Supplementary Figure S1), and the mutant PTCH1-P504L may result in Gorlin syndrome (Boutet et al., 2003). As for the SSDL, the corresponding residue P1082 was used. When the distance is less than 1 nm (the black dotted line in Figure 2; Supplementary Figure S3), it is considered that CHOL binds to the SSD/SSDL. Distinct CHOL affinities of the SSD and SSDL pockets were also observed in our simulations (Figure 2; Supplementary Figure S3). CHOL binding to the SSDL is remarkably weaker and shorter than that to the SSD. There is a CHOL binding to the SSD in about 60%/40% of the simulation time for the monomer and dimer-Shh systems respectively. While, in case of SSDL, the percentage is 11%/18%.
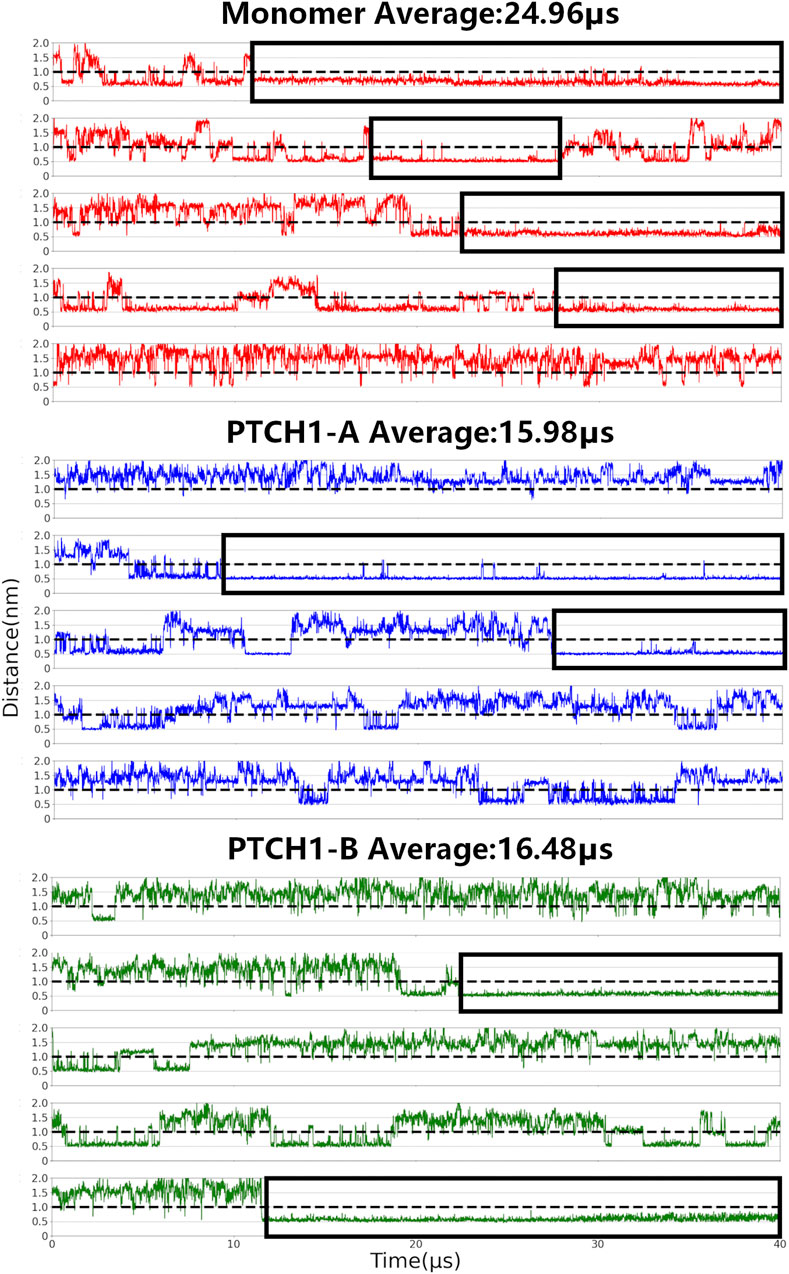
FIGURE 2. CHOL binding to the SSD. Center-of-mass distances between P504 and its nearest CHOL ROH bead were calculated for all simulations of the PTCH1 monomer and dimer-Shh (PTCH1-A and PTCH1-B) systems.
Our simulations also show that CHOL binding to the SSD is also different between the monomer and dimer-Shh simulation systems. For the monomer system, CHOLs bind to the SSD for an average of 25 μs in the 40 μs simulations, and in 4 out of 5 simulations, the binding time of one CHOL lasts more than 10 μs. In simulations of the dimer-Shh system, comparatively, the binding time of CHOLs to the SSD is about 16 μs on average, with stable interactions lasting for more than 10 μs appearing in 2 out of 5 simulations. It is worth to note that CHOL binding affinity to SSD in the PTCH1-A and PTCH1-B is very similar. The populations of the number of residues, that have interactions with CHOLs, also show that in the monomer system, CHOLs have more interactions with the protein than that in the dimer-Shh system (Supplementary Figure S4). In summary, our simulations illustrate that the PTCH1 monomer presents a stronger CHOL binding to SSD than the dimer-Shh.
Interactions between the CHOL and SSD pocket. The frequencies of interactions between CHOLs and residues involved in the SSD were further calculated, to illustrate the participations of different residues (Figure 3A). For both the monomer and dimer-Shh systems, the residues of TM3 (residues 465-490) and TM4 (residues 495-520), making up the bottom of the SSD cavity, contribute the most in interacting with the polar head of CHOL. The difference presents the contributions of residues on the edge of the SSD, and the hydrophobic tail of CHOL shows distinct binding preferences, termed subsites A and B (Figures 3A; Supplementary Figure S5) in simulations of the monomer and dimer-Shh systems. The subsite A locates at the left edge of the SSD in Figure 3A, involving the residues of TM2 (residues 430–455), while the subsite B locates at the right edge of the SSD in Figure 3A, involving the residues of TM1 (residues 100–125). In simulations of the monomer system, CHOLs prefer the subsite A. In contrast, in the dimer-Shh simulations, CHOL binding to the subsite B is favored.
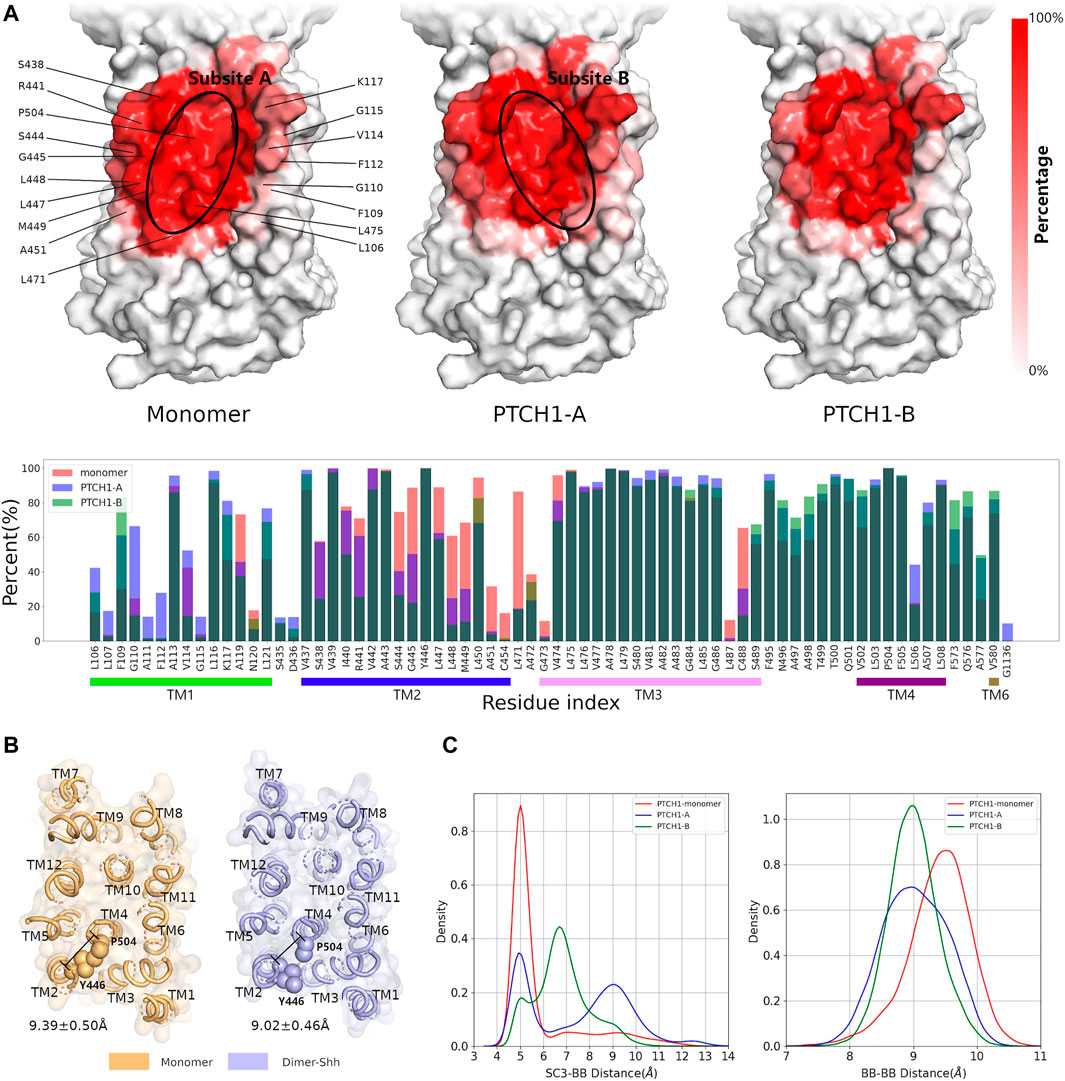
FIGURE 3. Interactions between CHOL and the SSD. (A) Frequencies of residues in the SSD involved in interacting CHOLs for the monomer and dimer-Shh (PTCH1-A and PTCH1-B) systems. The darker the red, the higher the frequency. (B) The conformation of Y446 in monomer and dimer-Shh systems. (C) Distributions of distances between sidechains (SC3 bead) of Y446 and the backbone (BB) of P504, and backbones of Y446 and P504.
The conformational changes of the SSD pocket in the monomer and dimer-Shh simulations were further compared. The critical conformational change of Y446 was identified to play a key role in CHOL binding. Mutations near Y446, such as inserting an alanine between Y446 and L447, cause serious diseases (Gu et al., 2006). In monomer simulations, the distances between the backbones of Y446 and P504 are slightly larger than those in dimer-Shh simulations, with the peak of the distance population moving from about 9.5 Å to 9.0 Å (Figures 3B,C). On the contrary, the distribution of distances between the sidechains of Y446 and P504 presents a sharp peak at about 5.0 Å in monomer simulations, while in dimer-Shh simulations, the distribution is wide and ranges from 4.0 Å to 13.0 Å (Figures 3B,C). In monomer simulations, the conformation of Y446 sidechain may benefit the π-π interactions between Y446 and CHOL. However, in dimer-Shh simulations, the Y446 sidechain rotates and points to the center of the SSD pocket, which makes CHOL binding to subsite A unfavorable.
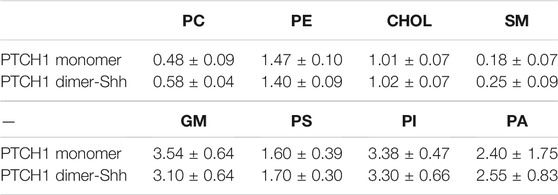
Lipid micro-environments of the PTCH1 monomer and dimer-Shh systems. The lipid micro-environments are fingerprints of membrane proteins (Corradi et al., 2018). The depletion enrichment (D-E) index was defined to describe the tendency of lipids to be enriched (>1) or depleted (<1) around membrane proteins (Corradi et al., 2018; Sejdiu and Tieleman, 2020). The number of lipids in contact with proteins converges after about 30 µs (Supplementary Figure S6), so the last 10 μs trajectories were used to calculate the D-E indices (Table 1; Supplementary Tables S2,S3).
Within the first lipid shell, we observed the depletion of PC and SM, and the enrichment of PE, GM, PI, PS, and PA in both systems. Highly unsaturated lipids are enriched around the protein, correspondingly, lipids with low unsaturation are depleted. In spite of the overall consistency, there are notable differences of the D-E indices of SM and GM. The D-E index of SM in the dimer-Shh system is about 30% larger than that in monomer system, while the enrichment of GM in the dimer-Shh system is lower than that in the monomer system.
Two-dimensional (2D) density maps of GM show clearly the enrichments of GMs around proteins, and different patterns of GM distributions of the PTCH1 monomer and dimer-Shh systems (Figure 4A). In particular, in the dimer-Shh system, around the SSD appears no enrichment of GMs, which looks more intuitive via frequencies of residues involved in interacting the head groups of GMs and the snapshots of the GM-PTCH1 interactions (Figures 4B,C; Supplementary Figure S7). Frequencies are mapped to the surface of the PTCH1 structure, presenting that in the PTCH1 monomer system, GMs have extensive interactions with the residues of SSD, but GMs seldomly distribute around the TM1 in the dimer-Shh system. The enrichment of GMs around the SSD allows the forming of GM clusters, the tails of which interact with the bound CHOL extensively, favoring the stable CHOL binding (Figure 4C).
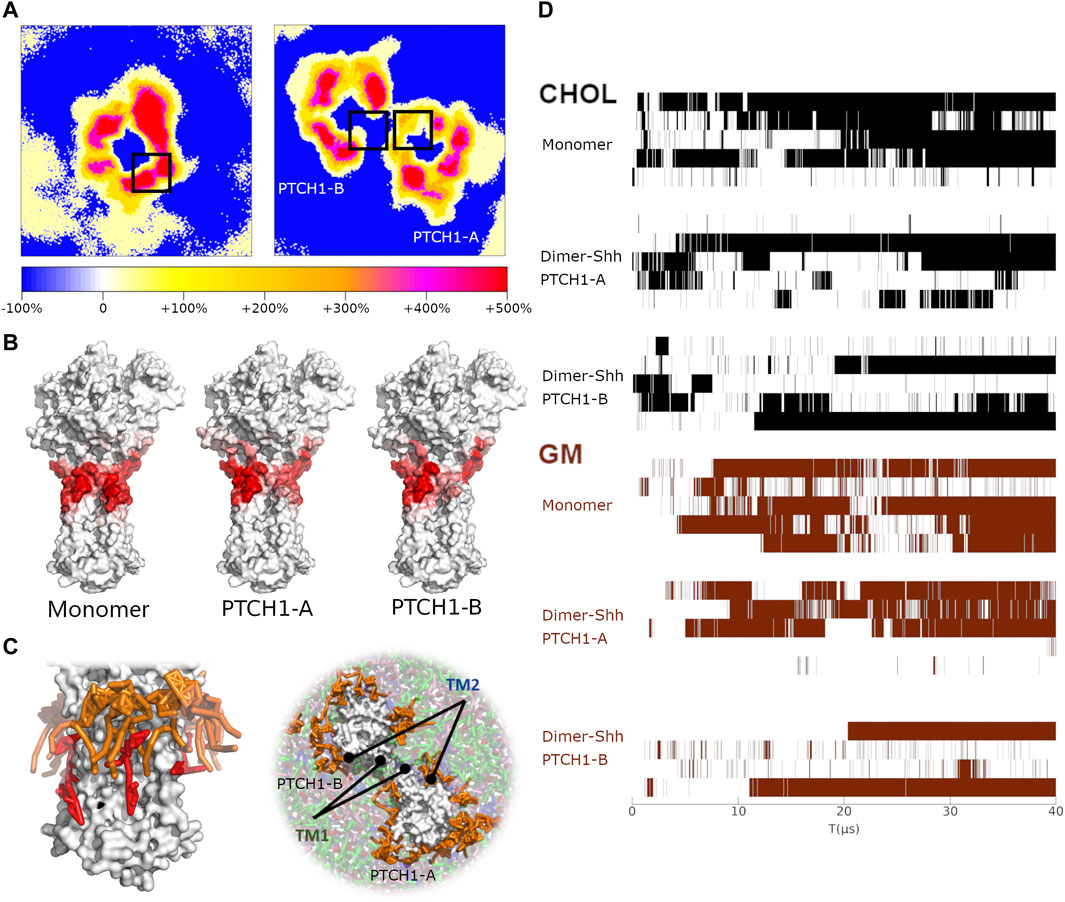
FIGURE 4. The correlation of GM clusters and CHOL binding. (A) 2D density maps of GM for the PTCH1 monomer and dimer-Shh systems, calculated by averaging the last 10μs simulations. The black boxes identify the location of SSDs. (B) The hotspots maps of the GM-PTCH1 interactions in the monomer and dimer-Shh (PTCH1-A and PTCH1-B) systems. (C) Snapshots of the monomer and dimer-Shh systems. GMs are colored in orange, and CHOL in red. (D) Variations of the CHOL binding to the SSD and the GM interacting with the SSD over the simulation time. For the CHOL binding, when the distance of P504-ROH is less than 1 nm, it is marked in black. For the GM interactions, when the residues of TM1 and TM2 have interactions simultaneously with GMs (minimum distance less than 0.65 nm), it is marked in brown.
We also noticed a close relationship between the existence of GMs around the protein and the CHOL binding in our simulations (Figure 4D). The long-term CHOL binding is always accompanied by the strong interactions of GM-PTCH1. It indicates that the lack of GMs around the SSDs in the dimer-Shh system may be one of the factors of the weak CHOL binding to the SSD.
Regulation of the Shh on the conformational dynamics of the PTCH1. Finally, conformational changes induced by the Shh were investigated. Elastic networks were used for the Martini protein models to maintain secondary structures, so the overall structures of the monomer and dimer-Shh are stable in all simulations, even the relative motion between PTCH1-A and PTCH1-B is limited (Supplementary Figure S8). However, average dynamical cross-correlation matrices (DCCMs) of the PTCH1 monomer and dimer-Shh systems show different patterns (Figure 5A; Supplementary Figure S9). The strong negative coupling of ECD2 and TMD in the PTCH1 monomer is weakened in the PTCH1 dimer-Shh. PTCH1-A is negatively coupled with PTCH1-B in general, but TMs 1-6 of PTCH1-A show weak positive correlation with TMs 1-6 and ECD2 of PTCH1-B. As for the Shh, it is negatively coupled with PTCH1-A, but positively coupled with PTCH1-B in general.
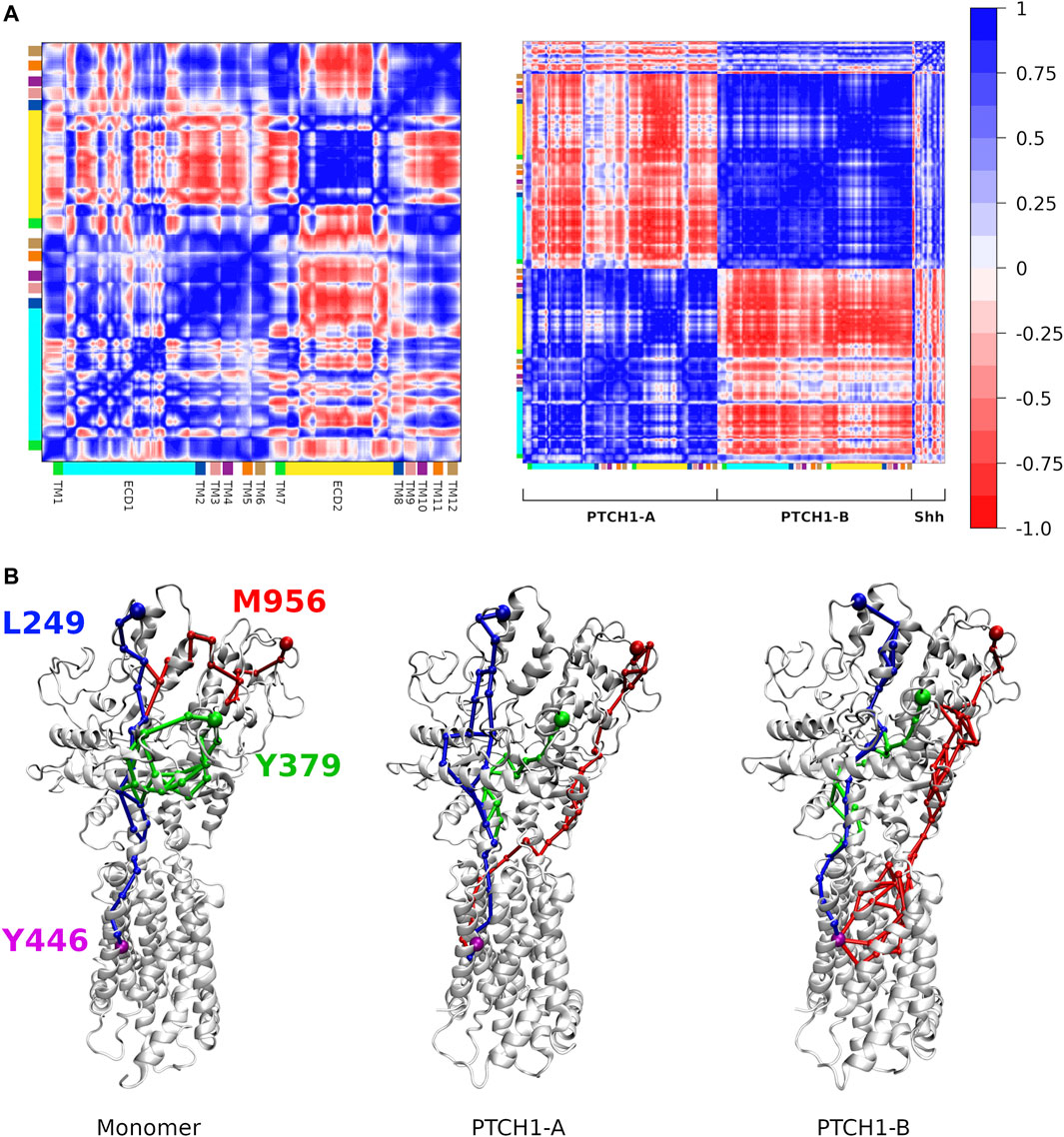
FIGURE 5. Dynamic communications in the PTCH1. (A) Dynamical cross-correlation matrices of backbone beads of the PTCH1 monomer (left) and dimer-Shh (right) systems. (B) Allosteric communication pathways between L249/Y379/M956 and Y446, colored in blue, green and red respectively.
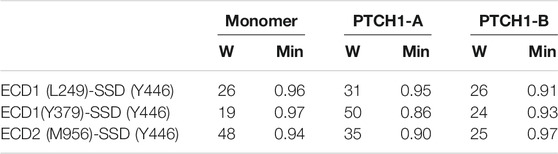
Based on the DCCMs, the allosteric communication pathways between Shh-ECD interface and the SSD were calculated. Three residues at the Shh-ECD interface, L249, Y379, and M956, were selected to represent for the ECD end, while Y446 for the SSD end. Two parameters, the overall weight of the path (
Discussions
How Shh transmits the signal to SMO through PTCH1 is one of the key questions in the Hh signaling pathway, and is still controversial. In this study, microsecond Martini CG MD simulations were performed for the PTCH1 monomer and dimer-Shh systems, to investigate their characteristic lipid micro-environments. The distinct CHOL binding affinities to SSD and binding sites were observed between two systems. The conformation of a critical residue, Y446, may play an important role in CHOL binding. The lipid micro-environments of the two systems are very similar, but with noticeable difference on the distributions of SM and GM. In the monomer system, GMs are enriched around the SSD, and have extensive interactions with the bound CHOL. However, the enrichment of GMs around the SSD is lack in the dimer-Shh system. Finally, the different patterns of the ECD-TMD coupling were also identified.
As most of GPCRs do, SMO is activated by CHOLs (Kinnebrew et al., 2019; Radhakrishnan et al., 2020). In addition, direct contacts of PTCH1 and SMO are not required for the signaling. Therefore, one of the main proposals is that accessible CHOLs function as a second messenger to pass signals from PTCH1 to SMO (Kinnebrew et al., 2019; Radhakrishnan et al., 2020). It was proposed that PTCH1 regulates accessible CHOLs in two ways: one is to change the distribution/number of accessible CHOLs, the other is to transfer CHOLs out of the cell membrane. Our simulations provide structural details about the regulation of PTCH1 on accessible CHOLs.
Firstly, in the monomer system, CHOL binding to the SSD is more stable and lasts for a longer time than that in the dimer-Shh system. It indicates that the Shh-induced dimerization may release the sequestered CHOLs by PTCH1 monomers, and increase the number of accessible CHOLs in the cell membrane. The stability of CHOL binding to the SSD is related to the population of GMs around the SSD and the conformation of Y446. The tails of GMs interact extensively with CHOLs, which could stabilize the bound CHOL. However, our simulations show clearly that the formation of dimer restricts the approach of GMs to the SSD. On the other hand, the conformation of Y446 in the monomer may form π-π interactions with the bound CHOL, while its conformational change in the dimer-Shh system blocks the subsite A, and destabilizes the CHOL binding. The conformational change of Y446 may be allosterically regulated by the Shh binding. Secondly, we also noticed that more SMs distribute around the dimer-Shh than the monomer. It is known that SMs sequester CHOLs by forming complexes (McConnell and Radhakrishnan, 2003; Das et al., 2014; Endapally et al., 2019). Depletion of SMs in the cell membrane could also activate SMO (Kinnebrew et al., 2019). Interacting with proteins could also reduce the SM level in the cell membrane, liberating accessible CHOLs from the SM-CHOL complexes. CHOLs in the cell membrane could be divided into three pools: essential, SM-sequestered, and accessible. It is clear that in both the two ways mentioned above, Shh regulates PTCH1 by adjusting the number of accessible CHOLs.
The structural homology of PTCH1 with NPC1 suggests that PTCH1 may work as a transporter, and pass CHOLs out of the cell membrane to an unknown CHOL receptor. However, no CHOL entering the PTCH1 putative hydrophobic tunnel was observed in our simulations. This may be because elastic networks are used for the Martini protein models, and restrict the large-scale conformational changes of proteins (Periole et al., 2009). Further simulations, with smaller force constant of elastic networks or with atomistic force fields, are required to explore PTCH1’s function as a CHOL transporter.
In conclusion, the analysis of protein-lipid interactions, in this study, provides structural details for the improved understanding of how Shh inhibits PTCH1 by regulation of accessible CHOLs, highlights the regulation of the lipid micro-environments on the function of PTCH1, and may open a new avenue for experimental design to reveal the molecular mechanism of the Hh signaling pathway.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.jianguoyun.com/p/DSbNbZwQ27-LChjV2Z8E.
Author Contributions
BW supervised and designed the research. CZ performed the simulations. CZ and BW analyzed the data and wrote the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Nos 31971176 and 31800616) and the Fundamental Research Funds for the Central Universities (No. A03018023601045).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.831891/full#supplementary-material
References
Ansell, T. B., Curran, L., Horrell, M. R., Pipatpolkai, T., Letham, S. C., Song, W., et al. (2021). Relative Affinities of Protein-Cholesterol Interactions from Equilibrium Molecular Dynamics Simulations. J. Chem. Theor. Comput. 17, 6548–6558. doi:10.1021/acs.jctc.1c00547
Berendsen, H. J. C., Postma, J. P. M., van Gunsteren, W. F., Dinola, A., and Haak, J. R. (1984). Molecular Dynamics with Coupling to an External bath. J. Chem. Phys. 81 (8), 3684–3690. doi:10.1063/1.448118
Boutet, N., Bignon, Y.-J., Drouin-Garraud, V., Sarda, P., Longy, M., Lacombe, D., et al. (2003). Spectrum of PTCH1 Mutations in French Patients with Gorlin Syndrome. J. Invest. Dermatol. 121 (3), 478–481. doi:10.1046/j.1523-1747.2003.12423.x
Briscoe, J., and Thérond, P. P. (2013). The Mechanisms of Hedgehog Signalling and its Roles in Development and Disease. Nat. Rev. Mol. Cel Biol 14 (7), 416–429. doi:10.1038/nrm3598
Brown, D. K., Penkler, D. L., Sheik Amamuddy, O., Ross, C., Atilgan, A. R., Atilgan, C., et al. (2017). MD-TASK: a Software Suite for Analyzing Molecular Dynamics Trajectories. Bioinformatics 33 (17), 2768–2771. doi:10.1093/bioinformatics/btx349
Bussi, G., Donadio, D., and Parrinello, M. (2007). Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 126, 014101. doi:10.1063/1.2408420
Chen, M.-H., Li, Y.-J., Kawakami, T., Xu, S.-M., and Chuang, P.-T. (2004). Palmitoylation Is Required for the Production of a Soluble Multimeric Hedgehog Protein Complex and Long-Range Signaling in Vertebrates. Genes Dev. 18 (6), 641–659. doi:10.1101/gad.1185804
Corradi, V., Mendez-Villuendas, E., Ingólfsson, H. I., Gu, R.-X., Siuda, I., Melo, M. N., et al. (2018). Lipid-Protein Interactions Are Unique Fingerprints for Membrane Proteins. ACS Cent. Sci. 4 (6), 709–717. doi:10.1021/acscentsci.8b00143
Corradi, V., Sejdiu, B. I., Mesa-Galloso, H., Abdizadeh, H., Noskov, S. Y., Marrink, S. J., et al. (2019). Emerging Diversity in Lipid-Protein Interactions. Chem. Rev. 119 (9), 5775–5848. doi:10.1021/acs.chemrev.8b00451
Das, A., Brown, M. S., Anderson, D. D., Goldstein, J. L., and Radhakrishnan, A. (2014). Three Pools of Plasma Membrane Cholesterol and Their Relation to Cholesterol Homeostasis. Elife 3, e02882. doi:10.7554/eLife.02882
Davies, J. P., and Ioannou, Y. A. (2000). Topological Analysis of Niemann-Pick C1 Protein Reveals that the Membrane Orientation of the Putative Sterol-Sensing Domain Is Identical to Those of 3-Hydroxy-3-Methylglutaryl-CoA Reductase and Sterol Regulatory Element Binding Protein Cleavage-Activating Protein. J. Biol. Chem. 275 (32), 24367–24374. doi:10.1074/jbc.M002184200
Duncan, A. L., Corey, R. A., and Sansom, M. S. P. (2020). Defining How Multiple Lipid Species Interact with Inward Rectifier Potassium (Kir2) Channels. Proc. Natl. Acad. Sci. USA 117 (14), 7803–7813. doi:10.1073/pnas.1918387117
Endapally, S., Frias, D., Grzemska, M., Gay, A., Tomchick, D. R., and Radhakrishnan, A. (2019). Molecular Discrimination between Two Conformations of Sphingomyelin in Plasma Membranes. Cell 176 (5), 1040–1053. doi:10.1016/j.cell.2018.12.042
Floyd, R. W. (1962). Algorithm 97: Shortest Path. J. Commun. Acm. 345 (6), 5. doi:10.1145/367766.368168
Gong, X., Qian, H., Cao, P., Zhao, X., Zhou, Q., Lei, J., et al. (2018). Structural Basis for the Recognition of Sonic Hedgehog by Human Patched1. Science 361 (6402), eaas8935. doi:10.1126/science.aas8935
Gong, X., Qian, H., Zhou, X., Wu, J., Wan, T., Cao, P., et al. (2016). Structural Insights into the Niemann-Pick C1 (NPC1)-Mediated Cholesterol Transfer and Ebola Infection. Cell 165 (6), 1467–1478. doi:10.1016/j.cell.2016.05.022
Gu, X.-M., Zhao, H.-S., Sun, L.-S., and Li, T.-J. (2006). PTCH Mutations in Sporadic and Gorlin-Syndrome-Related Odontogenic Keratocysts. J. Dent Res. 85 (9), 859–863. doi:10.1177/154405910608500916
Haycraft, C. J., Banizs, B., Aydin-Son, Y., Zhang, Q., Michaud, E. J., and Yoder, B. K. (2005). Gli2 and Gli3 Localize to Cilia and Require the Intraflagellar Transport Protein Polaris for Processing and Function. Plos Genet. 1 (4), e53. doi:10.1371/journal.pgen.0010053
Hedger, G., Koldsø, H., Chavent, M., Siebold, C., Rohatgi, R., and Sansom, M. S. P. (2019). Cholesterol Interaction Sites on the Transmembrane Domain of the Hedgehog Signal Transducer and Class F G Protein-Coupled Receptor Smoothened. Structure 27 (3), 549–559. doi:10.1016/j.str.2018.11.003
Humphrey, W., Dalke, A., and Schulten, K. (1996). VMD: Visual Molecular Dynamics. J. Mol. Graphics 14 (1), 33–38. doi:10.1016/0263-7855(96)00018-5
Ingham, P. W., and McMahon, A. P. (2001). Hedgehog Signaling in Animal Development: Paradigms and Principles. Genes Dev. 15 (23), 3059–3087. doi:10.1101/gad.938601
Ingham, P. W., Nakano, Y., and Seger, C. (2011). Mechanisms and Functions of Hedgehog Signalling across the Metazoa. Nat. Rev. Genet. 12 (6), 393–406. doi:10.1038/nrg2984
Ingólfsson, H. I., Melo, M. N., van Eerden, F. J., Arnarez, C., Lopez, C. A., Wassenaar, T. A., et al. (2014). Lipid Organization of the Plasma Membrane. J. Am. Chem. Soc. 136 (41), 14554–14559. doi:10.1021/ja507832e
Kinnebrew, M., Iverson, E. J., Patel, B. B., Pusapati, G. V., Kong, J. H., Johnson, K. A., et al. (2019). Cholesterol Accessibility at the Ciliary Membrane Controls Hedgehog Signaling. Elife 8, e50051. doi:10.7554/eLife.50051
Kiriakidi, S., Kolocouris, A., Liapakis, G., Ikram, S., Durdagi, S., and Mavromoustakos, T. (2019). Effects of Cholesterol on GPCR Function: Insights from Computational and Experimental Studies. Adv. Exp. Med. Biol. 1135, 89–103. doi:10.1007/978-3-030-14265-0_5
Kong, J. H., Siebold, C., and Rohatgi, R. (2019). Biochemical Mechanisms of Vertebrate Hedgehog Signaling. Development 146 (10), dev166892. doi:10.1242/dev.166892
Li, X., Wang, J., Coutavas, E., Shi, H., Hao, Q., and Blobel, G. (2016). Structure of Human Niemann-Pick C1 Protein. Proc. Natl. Acad. Sci. USA 113 (29), 8212–8217. doi:10.1073/pnas.1607795113
Marrink, S. J., Risselada, H. J., Yefimov, S., Tieleman, D. P., and de Vries, A. H. (2007). The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. J. Phys. Chem. B 111 (27), 7812–7824. doi:10.1021/jp071097f
McConnell, H. M., and Radhakrishnan, A. (2003). Condensed Complexes of Cholesterol and Phospholipids. Biochim. Biophys. Acta (Bba) - Biomembranes 1610 (2), 159–173. doi:10.1016/s0005-2736(03)00015-4
Pepinsky, R. B., Zeng, C., Wen, D., Rayhorn, P., Baker, D. P., Williams, K. P., et al. (1998). Identification of a Palmitic Acid-Modified Form of Human Sonic Hedgehog. J. Biol. Chem. 273 (22), 14037–14045. doi:10.1074/jbc.273.22.14037
Periole, X., Cavalli, M., Marrink, S.-J., and Ceruso, M. A. (2009). Combining an Elastic Network with a Coarse-Grained Molecular Force Field: Structure, Dynamics, and Intermolecular Recognition. J. Chem. Theor. Comput. 5 (9), 2531–2543. doi:10.1021/ct9002114
Pfeffer, S. R. (2019). NPC Intracellular Cholesterol Transporter 1 (NPC1)-Mediated Cholesterol export from Lysosomes. J. Biol. Chem. 294 (5), 1706–1709. doi:10.1074/jbc.TM118.004165
Qi, C., Di Minin, G., Vercellino, I., Wutz, A., and Korkhov, V. M. (2019). Structural Basis of Sterol Recognition by Human Hedgehog Receptor PTCH1. Sci. Adv. 5 (9), eaaw6490. doi:10.1126/sciadv.aaw6490
Qi, X., Schmiege, P., Coutavas, E., and Li, X. (2018a). Two Patched Molecules Engage Distinct Sites on Hedgehog Yielding a Signaling-Competent Complex. Science 362 (6410), eaas8843. doi:10.1126/science.aas8843
Qi, X., Schmiege, P., Coutavas, E., Wang, J., and Li, X. (2018b). Structures of Human Patched and its Complex with Native Palmitoylated Sonic Hedgehog. Nature 560 (7716), 128–132. doi:10.1038/s41586-018-0308-7
Qian, H., Cao, P., Hu, M., Gao, S., Yan, N., and Gong, X. (2019). Inhibition of Tetrameric Patched1 by Sonic Hedgehog through an Asymmetric Paradigm. Nat. Commun. 10 (1), 2320. doi:10.1038/s41467-019-10234-9
Radhakrishnan, A., Rohatgi, R., and Siebold, C. (2020). Cholesterol Access in Cellular Membranes Controls Hedgehog Signaling. Nat. Chem. Biol. 16 (12), 1303–1313. doi:10.1038/s41589-020-00678-2
Roessler, E., Belloni, E., Gaudenz, K., Jay, P., Berta, P., Scherer, S. W., et al. (1996). Mutations in the Human Sonic Hedgehog Gene Cause Holoprosencephaly. Nat. Genet. 14 (3), 357–360. doi:10.1038/ng1196-357
Rouviere, E., Arnarez, C., Yang, L., and Lyman, E. (2017). Identification of Two New Cholesterol Interaction Sites on the A2A Adenosine Receptor. Biophysical J. 113 (11), 2415–2424. doi:10.1016/j.bpj.2017.09.027
Rudolf, A. F., Kinnebrew, M., Kowatsch, C., Ansell, T. B., El Omari, K., Bishop, B., et al. (2019). The Morphogen Sonic Hedgehog Inhibits its Receptor Patched by a Pincer Grasp Mechanism. Nat. Chem. Biol. 15 (10), 975–982. doi:10.1038/s41589-019-0370-y
Sejdiu, B. I., and Tieleman, D. P. (2020). Lipid-Protein Interactions Are a Unique Property and Defining Feature of G Protein-Coupled Receptors. Biophysical J. 118 (8), 1887–1900. doi:10.1016/j.bpj.2020.03.008
Sethi, A., Eargle, J., Black, A. A., and Luthey-Schulten, Z. (2009). Dynamical Networks in tRNA:protein Complexes. Proc. Natl. Acad. Sci. 106 (16), 6620–6625. doi:10.1073/pnas.0810961106
Taipale, J., Cooper, M. K., Maiti, T., and Beachy, P. A. (2002). Patched Acts Catalytically to Suppress the Activity of Smoothened. Nature 418 (6900), 892–896. doi:10.1038/nature00989
Teglund, S., and Toftgård, R. (2010). Hedgehog beyond Medulloblastoma and Basal Cell Carcinoma. Biochim. Biophys. Acta (Bba) - Rev. Cancer 1805 (2), 181–208. doi:10.1016/j.bbcan.2010.01.003
Van Der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A. E., and Berendsen, H. J. C. (2005). GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 26 (16), 1701–1718. doi:10.1002/jcc.20291
Wassenaar, T. A., Ingólfsson, H. I., Böckmann, R. A., Tieleman, D. P., and Marrink, S. J. (2015). Computational Lipidomics with Insane: A Versatile Tool for Generating Custom Membranes for Molecular Simulations. J. Chem. Theor. Comput. 11 (5), 2144–2155. doi:10.1021/acs.jctc.5b00209
Wen, X., Lai, C. K., Evangelista, M., Hongo, J.-A., de Sauvage, F. J., and Scales, S. J. (2010). Kinetics of Hedgehog-dependent Full-Length Gli3 Accumulation in Primary Cilia and Subsequent Degradation. Mol. Cel Biol 30 (8), 1910–1922. doi:10.1128/MCB.01089-09
Keywords: Hedgehog signaling pathway, sterol-sensing domain, Martini coarse-grained model, protein-lipid interactions, allosteric communication pathways
Citation: Zhong C and Wang B (2022) Regulation of Cholesterol Binding to the Receptor Patched1 by its interactions With the Ligand Sonic Hedgehog (Shh). Front. Mol. Biosci. 9:831891. doi: 10.3389/fmolb.2022.831891
Received: 09 December 2021; Accepted: 11 January 2022;
Published: 02 February 2022.
Edited by:
Ruo-Xu Gu, Shanghai Jiao Tong University, ChinaCopyright © 2022 Zhong and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beibei Wang, YmJ3YW5nQGJudS5lZHUuY24=
 Changqing Zhong
Changqing Zhong Beibei Wang
Beibei Wang