- Department of Biosciences, Durham University, Durham, United Kingdom
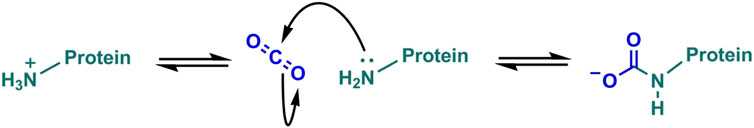
Carbon dioxide is essential for life. It is at the beginning of every life process as a substrate of photosynthesis. It is at the end of every life process as the product of post-mortem decay. Therefore, it is not surprising that this gas regulates such diverse processes as cellular chemical reactions, transport, maintenance of the cellular environment, and behaviour. Carbon dioxide is a strategically important research target relevant to crop responses to environmental change, insect vector-borne disease and public health. However, we know little of carbon dioxide’s direct interactions with the cell. The carbamate post-translational modification, mediated by the nucleophilic attack by carbon dioxide on N-terminal α-amino groups or the lysine ɛ-amino groups, is one mechanism by which carbon dioxide might alter protein function to form part of a sensing and signalling mechanism. We detail known protein carbamates, including the history of their discovery. Further, we describe recent studies on new techniques to isolate this problematic post-translational modification.
Introduction
Since its discovery in gas exhaled from the lung in 1757 (West, 2004, 2014) carbon dioxide (CO2) has been recognised as a critical component of biological processes throughout the biosphere. Its contribution to the essential physiological processes of metabolism, photosynthesis, chemosensing, and cellular homeostasis (Cummins et al., 2020) has meant that organisms across the three domains of life had evolved mechanisms to sense, transport, and respond to CO2 (Cummins et al., 2014). Although a great deal of knowledge exists about the physiological processes where CO2 is produced or consumed, less is known about the direct mechanisms of CO2 interactions with biomolecules.
One way in which CO2 has been shown to interact with protein directly is through carbamylation of neutral N-terminal -amino or lysine ɛ-amino groups (Figure 1). This carbamate post-translational modification (PTM) is critical to regulating oxygen-binding in haemoglobin and the activation of the CO2-fixing enzyme RuBisCO. It has been suggested that protein carbamylation could form the basis of a widespread mechanism for biological regulation (Morrow et al., 1974; Lorimer, 1983). Computational studies predict that carbamates may be found in more than 1.3% of large proteins (Jimenez-Morales et al., 2014).
In this review, we bring together contextual examples of carbamylation and explore recent computational and experimental approaches with the potential to uncover the distribution of protein carbamylation within proteomes.
CO2 and Haemoglobin
The linked processes of ventilation and metabolism are essential to survival in higher animals. Following ventilation, gas transfer enables oxygen to be provided promptly to the cells and their mitochondria, which is used in the final process of aerobic cellular respiration (oxidative phosphorylation). Oxidative phosphorylation coordinates with the tricarboxylic acid (TCA) cycle to form adenosine triphosphate (ATP) and the metabolites required by the organism for survival, producing CO2 as a waste product (Martínez-Reyes and Chandel, 2020). CO2 exits the cells to the bloodstream and is transported to the lungs for excretion, thus contributing to pH homeostasis (Cummins et al., 2020).
The link between ventilation and metabolism was first made in 1777 by Antoine Lavoisier with the observation that “Eminently respirable air [oxygen] that enters the lung, leaves it in the form of chalky aeroform acids [carbon dioxide]… in almost equal volume….” (West, 2014). At this time, it was presumed that the process of metabolism (“slow combustion”) was performed within the lung (West, 2014). By the mid to late 1800s it was clear that O2 was transported via the blood to tissues (where metabolism occurred) by the formation of a loose, dissociable interaction with haemoglobin (oxyhaemoglobin) (Barcroft and Hill, 1910; Saha et al., 2014), and CO2 was returned to the lung by similar means (Severinghaus and Bradley, 1958; Giegé, 2013).
In 1904 Bohr, Hasselbalch and Krogh measured haemoglobin oxygenation in canine blood and described the sigmoidal (rather than hyperbolic) nature of the oxyhaemoglobin dissociation curve (Bohr et al., 1904). This experiment demonstrated that increasing pCO2 resulted in a lowered affinity of haemoglobin for O2 (known as the Bohr effect). Conversely, Christiensen et al. (1914) described that increasing pO2 resulted in a decreased affinity of haemoglobin for CO2 (known as the Haldane effect). The Bohr and Haldane effects were reversible, and observed in various mammalian systems (Christiansen et al., 1914; West, 2019). Combined, the Bohr-Haldane effect results in haemoglobin being an efficient O2 transporter from the lungs to tissues and CO2 from the tissues to the lungs (Eaton et al., 1999). The nature of the oxyhaemoglobin dissociation curve led to the hypothesis that multiple O2 binding sites on haemoglobin acted cooperatively (Barcroft and Hill, 1910; Hill, 1913).
Haemoglobins belongs to a large family of proteins with members distributed across all three domains of life. The first structures (myoglobin and equine haemoglobin) were determined by X-ray crystallography in the 1950s (Perutz et al., 1960, 1964). Human adult haemoglobin is a tetramer consisting of two α and two β subunits similar in structure and size. The α and β subunits are formed of seven and eight helixes, respectively (A–H), joined by non-helical segments. Each subunit binds a heme group consisting of a porphyrin ring that coordinate a Fe2+ ion (capable of binding to O2) by four nitrogen atoms at its centre. The oxygenated and deoxygenated haemoglobin quaternary structures differ. The gap between two polypeptide chains in the haemoglobin molecule narrows when O2 binds to the Fe2+ (Paoli et al., 1996; Park et al., 2006; Perutz et al., 1960, 1964). The binding of the first O2 to the haemoglobin subunit enhances the ability of subsequent O2 molecules to bind to the remaining subunits (Adair, 1925; Pauling, 1935). This knowledge was used to develop the Monod Wyman and Changeux “two-state concerted” model for allostery where deoxygenated haemoglobin exists in a tense (T) state (with relatively low O2 affinity). When Fe2+ binds O2, there is a movement of Fe2+ into the heme plane, which triggers a transition to the relaxed (R) state. In this R state, the remaining binding sites are more exposed and have an increased O2 affinity (Monod and Jacob, 1961; Bringas et al., 2017). Additional allosteric sites on haemoglobin were available for binding allosteric modulators, including H+, CO2, 2,3-diphosphoglycerate (2,3-DPG) and Cl− (Perutz, 1970; Perrella and Russo, 2003; Yuan et al., 2015).
In the 1930s, CO2 was shown to combine rapidly and reversibly with haemoglobin to form carbaminohaemoglobin (Ferguson and Roughton, 1934; Stadie, 1935; Stadie and O’Brien, 1936). It was suggested that carbaminohaemoglobin formation was possible at multiple amino sites on haemoglobin (Stadie and O’Brien, 1936). Confirmation of carbamate formation at the N-terminal of the valines of each of the four human deoxyhaemoglobin chains was performed using cyanate-based blocking, which inhibited the uptake of CO2 by haemoglobin (Kilmartin and Rossi-Bernardi, 1969) and confirmed that CO2-binding was O2-linked (at constant pCO2 and pH, deoxyhemoglobin forms more haemoglobin-CO2 than haemoglobin-O2). The CO2 binding site was demonstrated to occur at the Val-1β site and linked to the O2-binding state of the β-chain (Matthew et al., 1977). Perrella et al., 1975 found that the adduct formed on the β chain is more prominent than the α chain. Physiologically, the reaction between the α-amino group and CO2 stabilises the protein’s deoxygenated form. Morrow et al., 1974 suggested that carbamino formation may be a general and functionally important phenomenon throughout biology and not limited to haemoglobin (Morrow et al., 1974).
CO2 and Ribulose 1,5-Bisphosphate Carboxylase-Oxygenase (RuBisCO)
Thought to have emerged about three billion years ago during the Archaean aeon and before the Great Oxygenation Events (Kacar et al., 2017; Banda et al., 2020), the RuBisCO family of proteins is one of the most abundant on the planet (Ellis, 1979). Initially discovered in the 1940s by Wildman and Bonner (Wildman, 2002), these enzymes are represented across the three domains of life and grouped as structurally distinct operational forms based on protein sequence and secondary and tertiary structure (Duff et al., 2000; Schneider et al., 1990, 1992). Forms I, II and III catalyse the carboxylation (and oxygenation) of ribulose 1,5-bisphosphate, while form IV contains RuBisCO-like proteins (RLP), which although sequentially and structurally similar, perform distinct biological functions (Watson et al., 1999; Tabita et al., 2008; Kono et al., 2017).
RuBisCO I is found in plants, cyanobacteria, algae, and some proteobacteria and is responsible for the vast majority of atmospheric CO2 fixation through the Calvin-Benson-Bassham (CBB) reductive pentose phosphate pathway (Andersson and Backlund, 2008; the dark reactions of photosynthesis). Most commonly, Form I is a hexadecamer of eight large and eight small subunits (L8S8). There are four Form I subtypes: A and B in cyanobacteria, eukaryotic algae, and higher plants; C and D are found in nongreen algae and phototropic bacteria (Stec, 2012). It has been demonstrated that although operational RuBisCO forms differ in overall structure, all share a similar catalytic subunit dimer (Duff et al., 2000; Schneider et al., 1990; Tabita et al., 2007, 2008), formed from the interaction of the large subunits (the C-terminus of the first interacting with the N-terminus of the second to form two active sites with residues from both). It is thought that RuBisCO remains in the open state when receiving substates and eliminating products. Still, when catalytic events occur, it is in a closed state, essentially sequestering the active site from the bulk solvent (Duff et al., 2000).
RuBisCO requires the catalytic site to be activated (Tabita et al., 2007, 2008) by sequential carbamylation of a specific lysine side chain followed by Mg2+ binding (Stec, 2012) (Figure 2). Once activated, RuBisCO’s role within the CBB is in catalysing the initial stage of carboxylation of ribulose 1,5-bisphosphate (RuBP) and cleavage to form two molecules of 3-phosphoglycerate (3-PGA) (one of which is used in RuBP regeneration, the other diverted to sustain biosynthesis of sugars and other high energy compounds). However, once activated, RuBisCO also catalyses a competitive oxygenation reaction, leading to the formation of one molecule of 3-PGA and one molecule of 2-phosphoglycolate (2-PG), which inhibits central carbon metabolism (Tcherkez, 2016). RuBisCO is a relatively inefficient enzyme with a low turnover rate (Bathellier et al., 2018). The turnover rate is further reduced by competition from the oxygenation reaction with the potential to reduce carbon fixation by up to 50% (Andersson and Taylor, 2003; Andersson, 2007). Therefore, optimisation of the carboxylation action of RuBisCO has been explored to improve crop efficiency and climate-resilient photosynthesis (Andersson and Backlund, 2008; Erb and Zarzycki, 2018; Valegård et al., 2018).
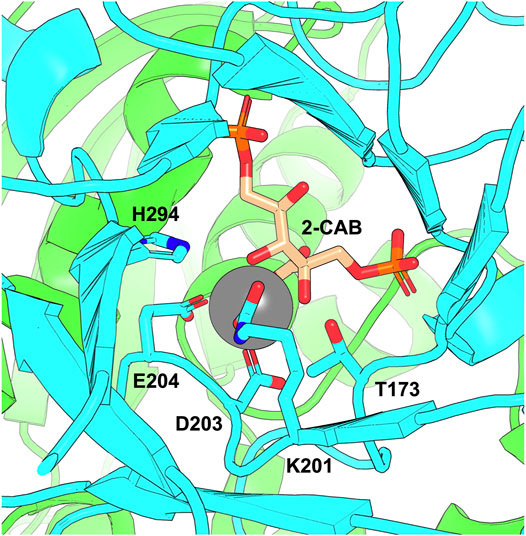
FIGURE 2. The RuBisCO active site showing the K201 carbamate. The figure shows the active site of Arabidopsis thaliana RuBisCO (PDB: 5IU0) with two large subunit chains shown in green and cyan and bound transition-state analogue 2-carboxy-D-arabinitol-1,5-bisphosphate (2-CAB). D203 and E204 coordinate Mg2+ co-factor (grey sphere) in the active site. The carbamate forms on K201 and is stabilised through interactions that include H294 and T173.
Identification of the RuBisCO catalytic site and its activation were, in most part, uncovered through kinetic and physical investigations in the 1960s and 70s. It became apparent that the sequential addition of CO2 (Lorimer et al., 1976) and Mg2+ (Miziorko and Mildvan, 1974; Lorimer et al., 1976) was required for activation. It was further demonstrated that this occurred through CO2.Mg2+.Enzyme complex formation (Miziorko and Mildvan, 1974). Further experiments demonstrated that the CO2 involved in the activation process was distinct from the substrate CO2 used in the subsequent carboxylase reaction (Miziorko, 1979; Lorimer and Miziorko, 1980) and that the catalytic site was located on the RuBisCO large subunit (Nishimura and Akazawa, 1974; Lorimer and Miziorko, 1980; Tabita et al., 2007). The hypothesis that activation was through carbamate formation on the ε-amino group of a lysine residue (Lorimer et al., 1976) was confirmed by Lorimer and Miziorko (1980), building on the work of (Ephraim Katchalski et al., 1951) and (Akoyunoglou et al., 1967). This finding was followed by identifying the carbamylation site on Lys-201 (Lorimer, 1981) close to the Mg2+ co-factor binding site (Pierce and Reddy, 1986). Based on the observed carbamylation of Haemoglobin and RuBisCO, George Lorimer proposed carbamate modification of N-terminal α-amino and lysine ε-amino groups as the basis of a widespread mechanism for CO2 detection. However, as observed by Professor Lorimer, there was no available method for trapping these carbamates on protein to enable their identification. The carbamate modification was, therefore, largely forgotten.
Discovery of Other Carbamates
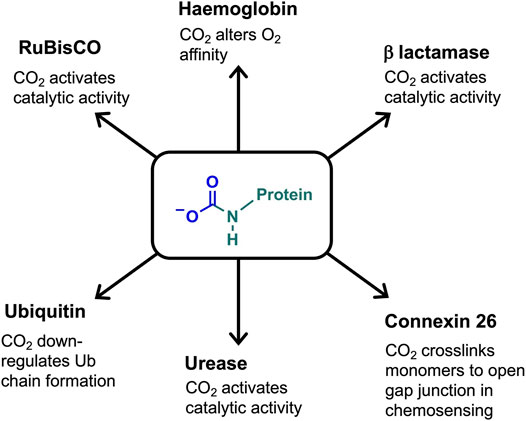
Carbamates have been observed on specific lysine side chains in the crystal structures of several proteins, e.g. class D β-lactamase (Golemi et al., 2001), urease (Yamaguchi and Hausinger, 1997), alanine racemase (Morollo et al., 1999), and transcarboxylase 5 S (Hall et al., 2004). Lysine carbamylation can be critical to protein/enzyme activity. Examples include having a direct role in catalytic activity (class D β-lactamase, (Golemi et al., 2001), being a co-catalytic determinant, which bridges metal ions, (e.g. urease, (Yamaguchi and Hausinger, 1997), or in promoting (through hydrogen bonding) active site rearrangements for protein function (e.g. alanine racemase; (Morollo et al., 1999). Figure 3 summarises the carbamate functional role in haemoglobin, RuBisCO and other proteins discussed below.
Class D β-lactamase
β-lactam antibiotics are essential in treating bacterial infection, inhibiting bacterial growth by acylating an active-site serine in essential penicillin-binding proteins and preventing the cross-linking of peptide chains to form peptidoglycan. β-Lactamases enable resistance to β-lactam antibiotics. These enzymes hydrolyse compounds containing a β-lactam ring and are classified based on sequence motifs and mechanism of hydrolysis (Class A-D). Class D OXA enzymes are the most diverse and least well-understood β-lactamases (Maveyraud et al., 2000; Tooke et al., 2019). The class is widely dispersed, found in gram-negative and gram-positive bacterial species and mobilised horizontally predominantly by plasmids and integrons (Bush, 2018; Tooke et al., 2019). In 2000, the first crystal structures of OXA-10 of Pseudomonas aeruginosa were published (Maveyraud et al., 2000; Strynadka et al., 2000). Maveyraud et al., 2000 discovered that OXA-10 is a dimeric beta-lactamase. However, the overall topology of OXA-10 class D β-lactamase is like class A, although amino acid sequence identity is low. Significant structural differences were found in the active site of OXA-10 compared to corresponding regions in class A (and C β-lactamases). In its native state, OXA-10 Lys-70 is carbamylated. It was suggested that this carbamylation offered a possible relationship between enzyme activation by CO2 and anion inhibition (Maveyraud et al., 2000). Golemi et al., 2001 used activity assays and fluorescence-based approaches to confirm that Lys-70 carbamylation is reversible and that OXA-10 β-lactamase depends on Lys-70 for enzyme acylation and deacylation steps in catalysis. This finding was supported by the inability of Lys-70 mutants to support deacylation. It was suggested that the hydrophobicity of the OXA-10 active site lowered the Lys-70 pKa favouring carbamylation (Golemi et al., 2001; Sun, 2003).
Urease
Urease plays an essential role in nitrogen metabolism in archaea, bacteria, fungi, plants and invertebrates (Dixon et al., 1975; Krajewska, 2009; Rutherford, 2014). It catalyses urea hydrolysis to form carbonic acid (H2CO3) and ammonia (NH3) and is important in human disease, plant metabolism, and agricultural ammonia emissions (Krajewska, 2009; Svane et al., 2020). In bacteria, ureases’ active site and primary structure are well studied in organisms such as Proteus mirabilis, Helicobacter pylori, and Klebsiella aerogenes (Jabri et al., 1995; Jabri and Karplus, 1996). The mechanism of urease activation is CO2 dependent in vitro (Pearson et al., 1998). This dependence has been explained by the active site containing two Ni2+ ions, which are bridged (stabilised) by the carboxyl group of a carbamylated lysine residue (Lys-217) that is essential for urease activation (Jabri and Karplus, 1996; I.-S. Park and Hausinger, 1995).
Computational Approaches for the Identification of Carbamate Sites
To enhance protein carbamate discovery, Jimenez-Morales et al. (2014) developed a model to predict the uncarboxylated and carboxylated status of lysine residues in proteins. They initially used a training set of 251 proteins (identified by X-ray crystallography) which contained at least one protein subunit with a carboxylated lysine residue (KCX) to investigate the characteristics of the carbamate microenvironment when compared to uncarboxylated (LYS) sites. They observed that the crucial feature of the KCX microenvironment “was the large numbers of packed atoms, water molecules and ions found in proximity to the KCX site chain.” Additionally, all KCX sites were buried (inaccessible from the surface), with residues converging structurally at the KCX site dispersed along the protein’s primary sequence (so no sequence motif was associated with lysine carboxylation). The KCX residue was often in contact with positively charged ions with mono-nuclear or bi-nuclear interactions with divalent ions (Zn2+, Mg2+, Co2+, Fe2+, Ni2+, Mn2+). His and Asp residues were present in all analyses of KCX containing metal-binding sites. No more than one KCX site was observed on any single protein chain. The RuBisCO carbamylated lysine (used in the training set) is typical of a structure where the carbamate binds Mg2+ coordinated by Asp and Glue residues (Figure 2).
This information was used to develop a naïve Bayesian model (to predict potential KCX and LYS sites (Predictor of Lysine Carboxylation: PreLysCarb)). The 251-protein data set was used for training/testing (along with sub-sets with redundancy reduction implemented at 40 and 90% sequence identity). They carried out “leave-one-out cross-validation tests” on the three data sets. At 90% sequence identity PreLysCar correctly classified 54/62 KCX sites (87% sensitivity) and 4255/4259 LYS sites (99.7% specificity). Investigating false-positive rates in high-resolution protein structures, the model indicated that 11 to 19/575 proteins were incorrectly predicted to have a KCX residue (false positive rate of between 1.9 and 3.3%). When PreLysCar was applied to a subset of solved protein structures from the PDB (structures greater than 200 residues, solved by X-ray crystallography, containing 14,261 protein chains after 90% redundancy removal), it predicted that at least 1.3% of proteins with more than 200 amino acids in the PDB could potentially be subject to spontaneous lysine carboxylation. As the model has been trained using previously identified stable carbamates, which are predominantly buried, it may only represent a subset of possible KCX sites (Linthwaite et al., 2020).
What is clear from this analysis is that no consensus sequence enables easy CO2-binding site prediction as has proven so successful for phosphorylation sites, for example. This observation is borne out by analysing the growing numbers of experimentally observed carbamates (see following sections) with no clear primary consensus sequence. Future machine learning approaches as experimental data sets increase in size will, therefore, be essential as predictive tools.
Mass Spectrometry Approaches for the Identification of Carbamate Sites
Mass spectrometry-based proteomics is a powerful tool for discovering and exploring PTMs. Although carbamylation of neutral N-terminal α-amino or lysine ε-amino groups may be a “general and functionally important phenomenon throughout biology” (Morrow et al., 1974) and “form the basis for a widespread mechanism of biological regulation” (Lorimer, 1983), the transient and readily reversible nature of this PTM all but renders impossible the use of mass spectrometry-based proteomics workflows (even when soft ionisation techniques are utilised; Terrier and Douglas, 2010). This ready reversibility limits our discovery of the distribution and contribution of carbamylation to biological functions.
Linthwaite et al., 2018 developed a chemical mechanism of covalently “trapping” carbamates under physiologically relevant conditions to overcome this limitation. In this technique, the triethyloxonium ion (TEO; a crystalline salt soluble under aqueous conditions) stabilises pre-formed carbamate modifications (formed through incubation of proteins or cellular lysates with CO2) by transferring an ethyl group from the oxonium ion to the negatively charged carbamate, forming a covalent bond which can withstand downstream protease digestion and enables CO2-binding protein identification by HPLC-ESI-MS/MS analysis. This approach has been validated for non-buried exchangeable carbamate binding sites (where presumably the carbamate is labile and the bound CO2 exchangeable into the bulk solvent) using individual amino acids and single proteins (previously known to be carbamylated; Linthwaite et al., 2018). This approach has recently contributed to studies of CO2 interaction with connexin 26 and ubiquitin (Sections 6.1 and 6.2) and shows potential in carbamate discovery through screening cell lysates (Section 6.3).
CO2 and Connexin 26
Connexins (Cx) have an essential role in intercellular and extracellular communication and, therefore, homeostasis in multicellular organisms. These transmembrane proteins form hexameric connexons or hemichannels (HCs) in the plasma membrane, allowing low molecular weight molecules to transfer across the plasma membrane (Meigh et al., 2014). In closely apposed membranes, two HCs can dock together to form homomeric or heteromeric gap junctions (GJs), which mediate intercellular communication through the transfer of low molecular weight molecules (<1.1.5 kD), e.g. ions, metabolites and second messengers, between cells (Giepmans, 2004; Meigh et al., 2014; Willebrords et al., 2016). GJ intercellular communication can be regulated by pH (closed by acidification), transmembrane voltage (opened by voltage potential greater than −20 mV) and calcium concentration (opened by removal of extracellular Ca2+). PTMs such as S-nitrosylation, sumoylation and phosphorylation can directly regulate GJ opening (Willebrords et al., 2016).
Connexin 26 (Cx26) HCs and GJs are found in cells throughout the body, including in the cochlear epithelial network of the ear, keratinocytes of the skin, alveolar epithelium of the lungs, epithelial cells of the GI tract, and chemosensory areas of the brain (Cohen-Salmon et al., 2002; Huckstepp et al., 2010a; Huckstepp et al., 2010b; Meigh et al., 2014; Willebrords et al., 2016; Srinivas et al., 2018; van de Wiel et al., 2020). Mutations in the gene encoding Cx26 (GJB2) are relatively common (frequency of carriers ∼2–4% of the human population) and are linked to nine non-syndromic and syndromic deafness disorders (SDD; Cook et al., 2019; Srinivas et al., 2018). SDDs are associated with visual impairment and dermatological abnormalities, and in some instances, Keratitis-Ichthyosis-Deafness syndrome is underpinned by Cx26-A88V and Cx26-G45E missense mutations (Meigh et al., 2014).
Huckstepp et al., 2010b and Wenker et al., 2012 suggested that Cx26 has a role in mediating the central CO2-dependant drive to breathe with HCs enabling ATP release from the medulla oblongata in the absence of extracellular acidification. Exploring the potential molecular mechanism, Huckstepp et al., 2010a found that increasing pCO2 at fixed pH opens Cx26 HCs (and HCs of two related beta-connexins, Cx30 and Cx32). Meigh et al., 2013 compared the amino acid sequences of Cx26, Cx30 and Cx32 with Cx31 (a connexin that has no sensitivity to pCO2) and hypothesised a carbamylation motif present in Cx26, 30, 32 that was absent from Cx31. Using the existing Cx26 crystal structure (Maeda et al., 2009), the authors noted that the carbamylation “motif” contained K125 at the end of a subunit’s alpha helix where K125 is oriented towards R104 on a neighbouring subunit of the hexamer. Therefore, they hypothesised that if K125 was carbamylated, it could feasibly form a salt bridge (“carbamate bridge”) with R104, linking the subunits, and preventing hemichannel closure. By inserting the identified carbamylation “motif” into Cx31, they demonstrated that it was sufficient to form a CO2-sensitive hemichannel. If the K125 residue was substituted for an amino acid that could not be carbamylated, CO2 sensitivity was lost. Using Cx26, they confirmed that K125 and R104 were essential for forming the carbamate bridge. Meigh et al., 2015 used a similar mutation-based approach to explore the potential for alternative mechanisms of bridge formation between residues 125 and 104 in adjacent hexamer subunits. They found they could convert the CO2-sensitive hemichannels to a NO/NO2− sensitive hemichannel using Cx26-K125C with Cx26-R104 or a redox-sensitive hemichannel using the combination of Cx26-K125C and Cx26-R104C, thus suggesting that distinct mechanisms of bridging involving residues 125 and 104 on adjacent hexamer subunits of Cx26 was possible (Meigh et al., 2015).
Using a combination of carbamate trapping, high-resolution cryo-EM and classification of particles, Brotherton et al., 2020 proposed that under physiologically relevant high pCO2 conditions (90 mmHg), a carbamate was formed on Lys-125, and additionally at two other positions Lys-108 and Lys-122, but not under physiologically relevant low pCO2 conditions (20 mmHg). They suggested that the shared environment within the cytoplasmic TM2 and TM3 regions of the Cx26 mobile loop favoured CO2 modification. Classification of particles indicated that the positions of TM2 and TM3 could influence the conformation of the N-terminal helix and found that under high pCO2 conditions, the N-terminus was more defined than under low pCO2 conditions. The authors hypothesised that gating is mediated by the movement of the N-terminal helix and its ability to plug the channel under physiologically relevant conditions (Brotherton et al., 2020).
Recent work by Nijjar et al., 2020 has shown in contrast to the opening of HCs at high physiological CO2 that intact Cx26 GJs connecting HeLa cells are closed when pCO2 is increased from 35 to 55 mmHg. This closing effect is dependent on the same residues (K125 and R104) involved in the CO2-dependant opening of Cx26 hemichannels. Further, the action is also directly attributed to a change in pCO2 rather than to changes in pH. They explained the contrasting action based on the free energy difference between CO2-bound and unbound states for HC and GJ (Nijjar et al., 2020). Specifically, it was energetically more favourable for HCs to bind CO2 in the open state and less energetically unfavourable to close. In contrast, it was energetically more favourable for the gap junction to bind CO2 in the closed state and then energetically unfavourable to open. The docking of two connexons as an HC provided a close interaction that constrained the conformation of hemichannels docked in a gap junction. Niijar et al., 2020 proposed the need for more information about the structures of Cx26 as free HCs and as components of GJs to investigate conformational differences and provide a greater understanding of the differential modulation of hemichannels and gap junctions by CO2. What is clear from these studies is that CO2 can have a signalling role via carbamate formation.
CO2 and Ubiquitin
Ubiquitin (Ub) is a highly conserved protein found in all eukaryotic cells. It plays a crucial role in regulating protein activity and degradation through Ub protein covalent conjugation to a lysine side chain on target proteins (Komander, 2009). Ubiquitylation conjugation occurs through the sequential activity of Ub-activating enzymes (E1), Ub-conjugating enzymes (E2), and Ub ligases (E3) (Yau and Rape, 2016; Heap et al., 2017) and can be reversed by deubiquitylating enzymes (DUBs). Ub can be conjugated into poly-Ub chains through the N-terminus (M1) or at any of the seven conserved lysine residues (K6, K11, K27, K29, K33, K48, and K63) forming single, mixed or branched Ub-chains, resulting in a diverse range of Ub-chain structures (Swatek and Komander, 2016; Yau and Rape, 2016; Heap et al., 2017). Distinct Ub-chains are involved in different biological functions (Heap et al., 2017). The most abundant and well-studied of these are K48 single linked Ub-chains which target proteins for proteasomal degradation (Heap et al., 2017), and K63 single linked Ub-chains involved in cell signalling, trafficking and lysosomal degradation (Ohtake et al., 2015; Swatek and Komander, 2016; Michel et al., 2017). Roles of other less abundant single and mixed linked Ub-chains include regulation of enzymatic activity (K33), cell cycle regulation (K11), inflammation and immune response (M1, K63/M1, K48) and post-replication repair (K6 and K33) (Komander, 2009; Michel et al., 2017).
Ub activity can be further regulated by additional PTMs, e.g. acetylation, phosphorylation and SUMOylation (Koyano et al., 2014; Morimoto and Shirakawa, 2016; Lamoliatte et al., 2017; Lacoursiere et al., 2020). For example, all Ub lysine residues (except K29) are acetylated under differing cellular conditions (Choudhary et al., 2009; Elia et al., 2015; Lacoursiere et al., 2020). Ohtake et al., 2015 demonstrated that when endogenous Ub is acetylated at K6 and K48, this does not affect the ability of Ub to conjugate with substrate protein but inhibits the elongation of K11, K48 and K63-linked Ub-chains by several E2 enzymes (by neutralising the lysine residue positive charge involved in non-covalent interaction of Ub with specific E2s). This acetylation results in the accumulation of monoubiquitylated substrates in the cell. Using a SILAC based approach, Ohtake et al., 2015 confirmed that acetylation of K6 and K48 was linked to the enrichment of chromosome or chromatin related factors, including histone H2B, and that monoubiquitylated H2B was stabilised by the expression of acetylated Ub (Ohtake et al., 2015; Morimoto and Shirakawa, 2016; Michel et al., 2017). It has recently been suggested that Ub PTMs add an extra layer of complexity to the ‘ubiquitin code’ and that this extends beyond currently identified PTMs (Ohtake and Tsuchiya, 2016).
Linthwaite et al., 2021 recently explored the potential modification of human Ub by CO2. They demonstrated that Ub K33 and K48 could be carbamylated under physiological conditions by CO2 (25 mM CO2/HCO3 at pH 7.4), using the “trapping” method (Figure 4) outlined in Linthwaite et al., 2018. Ub carbamylation was confirmed independently by 13C-NMR, identifying K6 and K63 as additional carbamylation sites. Using in vitro conjugation assays, Linthwaite et al. (2021) found that di-Ub formation at K48 was significantly decreased when the CO2 concentration was increased from the normal physiological reference range (1.8–2.3 mM dissolved CO2) to the hypercapnic range (>2.3 mM CO2).
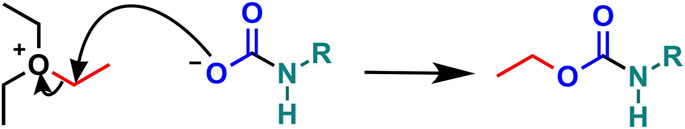
FIGURE 4. Trapping a protein carbamate with TEO. TEO transfers an ethyl group (red) to the anionic carbamate derived from CO2 (blue) and protein primary amine (green).
The effect of elevated CO2 on Ub-dependant processes was explored and focused on the regulation of nuclear factor κB (NF-κB) (Iwai, 2014). NF-κB is a transcription factor central to inflammation, innate and acquired immune responses, nervous system function, and cell survival (Iwai, 2014; Albensi, 2019; Song and Li, 2021). Under elevated pCO2, NF-kB mediated transcription is suppressed (Cummins et al., 2010). NF-κB is inactive in resting cells, bound in the cytoplasm to the inhibitor of NF-KBs (IκBs) proteins, blocking its transport to the nucleus. There are two known pathways of NF-κB activation, the canonical and non-canonical pathways. In the canonical pathway, the IκB kinase complex (IKK1, IKK2 and NF-κB essential modulator (NEM)) is activated upon interaction with, e.g., inflammatory cytokines or Toll-like receptor (TLR) ligands. This activation results in specific phosphorylation of IκB Ser residues within IκBs. Phosphorylated IκBs are recognised by the Ub ligase complex SCFβTrCPs to generate K48-linked poly-Ub chains and are subsequently degraded by the proteasome. NF-κB is released from IκBs and translocates to the nucleus, where it binds to the DNA consensus sequence of a target gene. TNF-receptor family proteins activate the non-canonical pathway. This activation results in the stabilisation of NIK and its phosphorylation of IKKα. This phosphorylated IKKα induces further phosphorylation of NF-κB2/p100, which forms a complex with RelB. Once phosphorylated NF-κB/p100 is ubiquitinated by SCFβTrCPs E3s generating K48-linked poly-Ub chains. This ubiquitinylation is followed by the partial degradation to p52 by the proteasome. The resulting RelB/p52 heterodimer is translocated to the nucleus (Iwai, 2014).
When cells carrying an NF-κB dependent GFP reporter were exposed to increasing TNFα concentrations under normal (5% (v/v) CO2 in air) and elevated CO2 conditions (10% (v/v) CO2 in air), NF-kB dependent GFP reporter activity was significantly decreased under elevated CO2 (Linthwaite et al., 2021). Transfection of the same cells with plasmids encoding wild-type Ub, mutant K48R Ub, mutant K63R Ub, or an empty vector was used to address the hypothesis that overexpression of K48R Ub would alter the relative response of the NF-kB pathway to elevated CO2. Cells transfected with an empty vector, Wt Ub or K63R Ub, showed an unaltered CO2 response. In contrast, when cells were transfected with K48R Ub, the effect of increased CO2 on the inhibition of the NF-κB reporter was ablated, suggesting that Ub K48 may be the target for CO2 in the NF-KB-dependent transcriptional response to hypercapnia in human cells.
Mass Spectrometry-Based Approaches for Discovery of Carbamate Sites in the Proteome
The TEO-based carbamate trapping method combined with HPLC-MS/MS has also been used to discover carbamylated proteins in whole-cell lysates. A screen of an Arabidopsis thaliana cell lysate, which corresponded to 6% of the total proteome (3614 proteins/25,000 proteins), identified eight CO2-binding sites (i.e. lipid-transfer protein (Lys-K65), Rubisco Large Subunit (Lys-185), Peroxidase (Lys-262 and Lys-268), FBA1 (Lys-293), eukaryotic aspartyl protease family protein (Lys-251), PSBQA (Lys-109), and Fe Superoxide dismutase 1 (Lys-208) (Linthwaite et al., 2018)). Similarly, a screen of an Escherichia coli lysates corresponding to 14% of the total proteome (294/4300 proteins) identified six CO2 -binding sites (Linthwaite and Cann, 2021). In this instance, these included proteins involved in cellular processes identified as responsive to CO2, i.e. assisting in the refolding of stress-denatured proteins, i.e. 60 kDa chaperone, carbamylated at Lys-34; (Kerner et al., 2005), preventing denaturation of DNA under extreme conditions (histone-like DNA HU-α, Lys-67; (Oberto et al., 2009), and proteins not previously identified in cellular processes responsive to CO2, i.e. glutamine-binding periplasmic protein (Lys-127), ribose import binding protein RbsB (Lys-45 and Lys-285), and tryptophanase (Lys-121). In the future, this approach could be optimised for rapid screening of proteomes for non-buried exchangeable CO2-binding sites by including protein and/or peptide fractionation steps to increase proteome coverage and through the development of mechanisms of carbamate enrichment.
Conclusion
We can draw some clear conclusions from the studies presented. First, carbamylation is a non-enzymatic PTM that can also be readily reversible. Second, the biological consequences of the carbamate PTM can vary discretely with changing CO2. Third, the carbamate PTM is more widespread among proteomes than suspected. Several future challenges arise from these conclusions. First, the extent of carbamylation in a proteome is unknown. More extensive proteomics analyses can address this challenge. However, such studies might benefit from developing enrichment methods and parallel computational approaches for carbamate prediction. Second, once the complement of carbamates in a proteome is known, how do we identify those that might have a functional role in CO2 sensing and signalling instead of forming through standard physicochemical mechanisms and being functionally neutral? Third, is carbamylation the sole mechanism for CO2 detection? If not, what other means exist, and how do we identify them? It is clear; there is plenty more to do in CO2 detection.
Author Contributions
LB and MC conceived, designed and wrote the review manuscript.
Funding
This review was supported by the Biotechnology and Biological Sciences Research Council grant no. BB/S015132/1.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Tim Blower for assistance with analysis of the RuBisCO structure.
References
Adair, G. (1925). A Critical Study of the Direct Method of Measuring the Osmotic Pressure of Hæmoglobin. Proc. R. Soc. Lond. Ser. B 98 (692), 523. doi:10.1098/rspb.1925.0054
Akoyunoglou, G., Argyroudi-Akoyunoglou, J.-H., and Methenitou, H. (1967). Studies on the Active Site of the Enzyme Ribulosediphosphate Carboxylase. Biochim. Biophys. Acta (Bba) - Enzymol. 132 (2), 481–491. doi:10.1016/0005-2744(67)90166-0
Albensi, B. C. (2019). What Is Nuclear Factor Kappa B (NF-Κb) Doing in and to the Mitochondrion? Front. Cel Develop. Biol. 7, 154. doi:10.3389/fcell.2019.00154
Andersson, I., and Backlund, A. (2008). Structure and Function of Rubisco. Plant Physiol. Biochem., 46(3). doi:10.1016/j.plaphy.2008.01.001
Andersson, I. (2007). Catalysis and Regulation in Rubisco. J. Exp. Bot. 59 (7), 1555–1568. doi:10.1093/jxb/ern091
Andersson, I., and Taylor, T. C. (2003). Structural Framework for Catalysis and Regulation in Ribulose-1,5-Bisphosphate Carboxylase/oxygenase. Arch. Biochem. Biophys. 414 (2), 130–140. doi:10.1016/S0003-9861(03)00164-4
Banda, D. M., Pereira, J. H., Liu, A. K., Orr, D. J., Hammel, M., He, C., et al. (2020). Novel Bacterial Clade Reveals Origin of Form I Rubisco. Nat. Plants 6 (9), 1158–1166. doi:10.1038/s41477-020-00762-4
Barcroft, J., and Hill, A. V. (1910). The Nature of Oxyhaemoglobin, with a Note on its Molecular Weight. J. Physiol. 39 (39), 411–428. doi:10.1113/jphysiol.1910.sp001350
Bathellier, C., Tcherkez, G., Lorimer, G. H., and Farquhar, G. D. (2018). Rubisco Is Not Really So Bad. Plant Cel Environ 41 (4), 705–716. doi:10.1111/pce.13149
Bohr, C., Hasselbalch, K., and Krogh, A. (1904). Ueber einen in biologischer Beziehung wichtigen Einfluss, den die Kohlensäurespannung des Blutes auf dessen Sauerstoffbindung übt. Acta Physiol. 16, 401–412. doi:10.1111/j.1748-1716.1904.tb01382.x
Bringas, M., Petruk, A. A., Estrin, D. A., Capece, L., and Martí, M. A. (2017). Tertiary and Quaternary Structural Basis of Oxygen Affinity in Human Hemoglobin as Revealed by Multiscale Simulations. Sci. Rep. 7 (1), 10926. doi:10.1038/s41598-017-11259-0
Brotherton, D. H., Savva, C. G., Ragan, T. J., Linthwaite, V. L., Cann, M. J., Dale, N., et al. (2020). Conformational Changes and Channel Gating Induced by CO2 Binding to Connexin26. BioRxiv. doi:10.1101/2020.08.11.243964
Bush, K. (2018). Past and Present Perspectives on β-Lactamases. Antimicrob. Agents Chemother. 62, 18. doi:10.1128/AAC.01076-18
Choudhary, C., Kumar, C., Gnad, F., Nielsen, M. L., Rehman, M., Walther, T. C., et al. (2009). Lysine Acetylation Targets Protein Complexes and Co-regulates Major Cellular Functions. Science 325 (5942), 834–840. doi:10.1126/science.1175371
Christiansen, J., Douglas, C. G., and Haldane, J. S. (1914). The Absorption and Dissociation of Carbon Dioxide by Human Blood. J. Physiol. 48 (48), 244–271. doi:10.1113/jphysiol.1914.sp001659
Cohen-Salmon, M., Ott, T., Michel, V., Hardelin, J.-P., Perfettini, I., Eybalin, M., et al. (2002). Targeted Ablation of Connexin26 in the Inner Ear Epithelial Gap Junction Network Causes Hearing Impairment and Cell Death. Curr. Biol. 12 (13), 1106–1111. doi:10.1016/s0960-9822(02)00904-1
Cook, J., de Wolf, E., and Dale, N. (2019). Cx26 Keratitis Ichthyosis Deafness Syndrome Mutations Trigger Alternative Splicing of Cx26 to Prevent Expression and Cause Toxicity In Vitro. R. Soc. Open Sci. 6, 191128(8). doi:10.1098/rsos.191128
Cummins, E. P., Oliver, K. M., Lenihan, C. R., Fitzpatrick, S. F., Bruning, U., Scholz, C. C., et al. (2010). NF-κB Links CO2 Sensing to Innate Immunity and Inflammation in Mammalian Cells. J. Immunol. 185, 4439–4445. doi:10.4049/jimmunol.1000701
Cummins, E. P., Selfridge, A. C., Sporn, P. H., Sznajder, J. I., and Taylor, C. T. (2014). Carbon Dioxide-Sensing in Organisms and its Implications for Human Disease. Cell. Mol. Life Sci. 71 (5), 831–845. doi:10.1007/s00018-013-1470-6
Cummins, E. P., Strowitzki, M. J., and Taylor, C. T. (2020). Mechanisms and Consequences of Oxygen and Carbon Dioxide Sensing in Mammals. Physiol. Rev. 100 (1), 463–488. doi:10.1152/physrev.00003.2019
Dixon, N. E., Gazzola, C., Blakeley, R. L., and Zerner, B. (1975). Jack Bean Urease (EC 3.5.1.5). Metalloenzyme. Simple Biological Role for Nickel. J. Am. Chem. Soc. 97 (14), 4131–4133. doi:10.1021/ja00847a045
Duff, A. P., Andrews, T. J., and Curmi, P. M. (2000). The Transition between the Open and Closed States of Rubisco Is Triggered by the Inter-phosphate Distance of the Bound Bisphosphate. J. Mol. Biol. 298, 903–916. doi:10.1006/jmbi.2000.3724
Eaton, W. A., Henry, E. R., Eric, J., and Mozzarelli, A. (1999). Is Cooperative Oxygen Binding by Hemoglobin Really Understood? Nat. Struct. Biol. 6 (4), 351–358. doi:10.1038/7586
Elia, A. E. H., Boardman, A. P., Wang, D. C., Huttlin, E. L., Everley, R. A., Dephoure, N., et al. (2015). Quantitative Proteomic Atlas of Ubiquitination and Acetylation in the DNA Damage Response. Mol. Cel 59 (5), 867–881. doi:10.1016/j.molcel.2015.05.006
Ellis, R. J. (1979). The Most Abundant Protein in the World. Trends Biochem. Sci. 4 (11), 241–244. doi:10.1016/0968-0004(79)90212-3
Erb, T. J., and Zarzycki, J. (2018). A Short History of RubisCO: the Rise and Fall (?) of Nature's Predominant CO2 Fixing Enzyme. Curr. Opin. Biotechnol. 49, 100–107. doi:10.1016/j.copbio.2017.07.017
Ferguson, J. K. W., and Roughton, F. J. W. (1934). The Direct Chemical Estimation of Carbamino Compounds of CO2with Haemoglobin. J. Physiol. 83 (1), 68–86. doi:10.1113/jphysiol.1934.sp003212
Giegé, R. (2013). A Historical Perspective on Protein Crystallization from 1840 to the Present Day. Febs J. 280 (24), 6456–6497. doi:10.1111/febs.12580
Giepmans, B. (2004). Gap Junctions and Connexin-Interacting Proteins. Cardiovasc. Res. 62 (2), 233–245. doi:10.1016/j.cardiores.2003.12.009
Golemi, D., Maveyraud, L., Vakulenko, S., Samama, J.-P., and Mobashery, S. (2001). Critical Involvement of a Carbamylated Lysine in Catalytic Function of Class D -lactamases. Proc. Natl. Acad. Sci. 98 (25), 14280–14285. doi:10.1073/pnas.241442898
Hall, P. R., Zheng, R., Antony, L., Pusztai-Carey, M., Carey, P. R., and Yee, V. C. (2004). Transcarboxylase 5S Structures: Assembly and Catalytic Mechanism of a Multienzyme Complex Subunit. Embo J. 23 (18), 3621–3631. doi:10.1038/sj.emboj.7600373
Heap, R. E., Gant, M. S., Lamoliatte, F., Peltier, J., and Trost, M. (2017). Mass Spectrometry Techniques for Studying the Ubiquitin System. Biochem. Soc. Trans. 45 (5), 1137–1148. doi:10.1042/BST20170091
Hill, A. V. (1913). The Combinations of Haemoglobin with Oxygen and with Carbon Monoxide. I. Biochem. J. 7 (5), 471–480. doi:10.1042/bj0070471
Huckstepp, R. T. R., Eason, R., Sachdev, A., and Dale, N. (2010a). CO2-dependent Opening of Connexin 26 and Related β Connexins. J. Physiol. 588 (20), 3921–3931. doi:10.1113/jphysiol.2010.192096
Huckstepp, R. T. R., Id Bihi, R., Eason, R., Spyer, K. M., Dicke, N., Willecke, K., et al. (2010b). Connexin Hemichannel-Mediated CO2-dependent Release of ATP in the Medulla Oblongata Contributes to central Respiratory Chemosensitivity. J. Physiol. 588 (20), 3901–3920. doi:10.1113/jphysiol.2010.192088
Iwai, K. (2014). Diverse Roles of the Ubiquitin System in NF-Κb Activation. Biochim. Biophys. Acta (Bba) - Mol. Cel Res. 1843 (1), 129–136. doi:10.1016/j.bbamcr.2013.03.011
Jabri, E., Carr, M., Hausinger, R., and Karplus, P. (1995). The crystal Structure of Urease from Klebsiella Aerogenes. Science 268 (5213), 998–1004. doi:10.1126/science.7754395
Jabri, E., and Karplus, P. A. (1996). Structures of the Klebsiella Aerogenes Urease Apoenzyme and Two Active-Site Mutants,. Biochemistry 35 (33), 10616–10626. doi:10.1021/bi960424z
Jimenez-Morales, D., Adamian, L., Shi, D., and Liang, J. (2014). Lysine Carboxylation: Unveiling a Spontaneous post-translational Modification. Acta Cryst. D Biol. Crystallogr. 70 (1), 48–57. doi:10.1107/S139900471302364X
Kacar, B., Hanson-Smith, V., Adam, Z. R., and Boekelheide, N. (2017). Constraining the Timing of the Great Oxidation Event within the Rubisco Phylogenetic Tree. Geobiology 15 (5), 628–640. doi:10.1111/gbi.12243
Katchalski, E., Berliner-Klibanski, C., and Berger, A. (1951). The Chemical Structure of Some Diamine Carbamates1. J. Am. Chem. Soc. 73 (4), 1829–1831. doi:10.1021/ja01148a124
Kerner, M. J., Naylor, D. J., Ishihama, Y., Maier, T., Chang, H.-C., Stines, A. P., et al. (2005). Proteome-wide Analysis of Chaperonin-dependent Protein Folding in Escherichia coli. Cell 122 (2), 209–220. doi:10.1016/j.cell.2005.05.028
Kilmartin, J. V., and Rossi-Bernardi, L. (1969). Inhibition of CO2 Combination and Reduction of the Bohr Effect in Haemoglobin Chemically Modified at its α-Amino Groups. Nature 222 (5200), 1243–1246. doi:10.1038/2221243a0
Komander, D. (2009). The Emerging Complexity of Protein Ubiquitination. Biochem. Soc. Trans. 37 (5), 937. doi:10.1042/BST0370937
Kono, T., Mehrotra, S., Endo, C., Kizu, N., Matusda, M., Kimura, H., et al. (2017). A RuBisCO-Mediated Carbon Metabolic Pathway in Methanogenic Archaea. Nat. Commun. 8, 14007. doi:10.1038/ncomms14007
Koyano, F., Okatsu, K., Kosako, H., Tamura, Y., Go, E., Kimura, M., et al. (2014). Ubiquitin Is Phosphorylated by PINK1 to Activate Parkin. Nature 510, 162. doi:10.1038/nature13392
Krajewska, B. (2009). Ureases I. Functional, Catalytic and Kinetic Properties: A Review. J. Mol. Catal. B: Enzymatic 59 (1–3), 9–21. doi:10.1016/j.molcatb.2009.01.003
Lacoursiere, R. E., O’Donoghue, P., and Shaw, G. S. (2020). Programmed Ubiquitin Acetylation Using Genetic Code Expansion Reveals Altered Ubiquitination Patterns. FEBS Lett. 594 (7), 1226–1234. doi:10.1002/1873-3468.13702
Lamoliatte, F., McManus, F. P., Maarifi, G., Chelbi-Alix, M. K., and Thibault, P. (2017). Uncovering the SUMOylation and Ubiquitylation Crosstalk in Human Cells Using Sequential Peptide Immunopurification. Nat. Commun. 8, 14109(1). doi:10.1038/ncomms14109
Linthwaite, V. L., Janus, J. M., Brown, A. P., Wong-Pascua, D., O'Donoghue, A. C., Porter, A., et al. (2018). The Identification of Carbon Dioxide Mediated Protein post-translational Modifications. Nat. Commun. 9, 3092. doi:10.1038/s41467-018-05475-z
Linthwaite, V. L., Pawloski, W., Pegg, H. B., Townsend, P. D., Thomas, M. J., So, V. K. H., et al. (2021). Ubiquitin Is a Carbon Dioxide-Binding Protein. Sci. Adv. 7, eabi5507. doi:10.1126/sciadv.abi5507
Linthwaite, V. L., and Cann, M. J. (2021). A Methodology for Carbamate post-translational Modification Discovery and its Application in Escherichia coli. Interf. Focus 11 (2), 28. doi:10.1098/rsfs.2020.0028
Lorimer, G. H., Badger, M. R., and Andrews, T. J. (1976). The Activation of Ribulose-1,5-Bisphosphate Carboxylase by Carbon Dioxide and Magnesium Ions. Equilibria, Kinetics, a Suggested Mechanism, and Physiological Implications. Biochemistry 15 (3), 529–536. doi:10.1021/bi00648a012
Lorimer, G. H. (1983). Carbon Dioxide and Carbamate Formation: the Makings of a Biochemical Control System. Trends Biochem. Sci. 8 (2), 65–68. doi:10.1016/0968-0004(83)90393-6
Lorimer, G. H., and Miziorko, H. M. (1980). Carbamate Formation on the .epsilon.-amino Group of a Lysyl Residue as the Basis for the Activation of Ribulosebisphosphate Carboxylase by Carbon Dioxide and Magnesium(2+). Biochemistry 19 (23), 5321–5328. doi:10.1021/bi00564a027
Lorimer, G. H. (1981). The Carboxylation and Oxygenation of Ribulose 1,5-Bisphosphate: The Primary Events in Photosynthesis and Photorespiration. Annu. Rev. Plant Physiol. 32 (1), 349–382. doi:10.1146/annurev.pp.32.060181.002025
Maeda, S., Nakagawa, S., Suga, M., Yamashita, E., Oshima, A., Fujiyoshi, Y., et al. (2009). Structure of the Connexin 26 gap junction Channel at 3.5 Å Resolution. Nature 458 (7238), 597–602. doi:10.1038/nature07869
Martínez-Reyes, I., and Chandel, N. S. (2020). Mitochondrial TCA Cycle Metabolites Control Physiology and Disease. Nat. Commun. 102, 3. doi:10.1038/s41467-019-13668-3
Matthew, J. B., Morrow, J. S., Wittebort, R. J., and Gurd, F. R. (1977). Quantitative Determination of Carbamino Adducts of Alpha and Beta Chains in Human Adult Hemoglobin in Presence and Absence of Carbon Monoxide and 2,3-diphosphoglycerate. J. Biol. Chem. 252 (7), 2234–2244. doi:10.1016/S0021-9258(17)40546-1
Maveyraud, L., Golemi, D., Kotra, L. P., Tranier, S., Vakulenko, S., Mobashery, S., et al. (2000). Insights into Class D β-Lactamases Are Revealed by the Crystal Structure of the OXA10 Enzyme from Pseudomonas aeruginosa. Structure 8 (12), 7. doi:10.1016/S0969-2126(00)00534-7
Meigh, L., Cook, D., Zhang, J., and Dale, N. (2015). Rational Design of New NO and Redox Sensitivity into Connexin26 Hemichannels. Open Biol. 5, 140208. doi:10.1098/rsob.140208
Meigh, L., Greenhalgh, S. A., Rodgers, T. L., Cann, M. J., Roper, D. I., and Dale, N. (2013). CO₂directly Modulates Connexin 26 by Formation of Carbamate Bridges between Subunits. ELife 2, e01213. doi:10.7554/elife.01213
Meigh, L., Hussain, N., Mulkey, D. K., and Dale, N. (2014). Connexin26 Hemichannels with a Mutation that Causes KID Syndrome in Humans Lack Sensitivity to CO2. ELife 25 (3), e04249. doi:10.7554/eLife.04249
Michel, M. A., Swatek, K. N., Hospenthal, M. K., and Komander, D. (2017). Ubiquitin Linkage-specific Affimers Reveal Insights into K6-Linked Ubiquitin Signaling. Mol. Cel 68 (1), 20. doi:10.1016/j.molcel.2017.08.020
Miziorko, H. M., and Mildvan, A. S. (1974). Electron Paramagnetic Resonance, 1H, and 13C Nuclear Magnetic Resonance Studies of the Interaction of Manganese and Bicarbonate with Ribulose 1,5-Diphosphate Carboxylase. J. Biol. Chem. 249 (9), 2743–2750. doi:10.1016/S0021-9258(19)42692-6
Miziorko, H. M. (1979). Ribulose-1,5-biphosphate Carboxylase. Evidence in Support of the Existence of Distinct CO2 Activator and CO2 Substrate Sites. J. Biol. Chem. 254 (2), 270–272. doi:10.1016/S0021-9258(17)37912-7
Monod, J., and Jacob, F. (1961). General Conclusions: Teleonomic Mechanisms in Cellular Metabolism, Growth, and Differentiation. Cold Spring Harbor Symposia Quantitative Biol. 26, 389–401. doi:10.1101/sqb.1961.026.01.048
Morimoto, D., and Shirakawa, M. (2016). The Evolving World of Ubiquitin: Transformed Polyubiquitin Chains. Biomol. Concepts 7 (3), 157–167. doi:10.1515/bmc-2016-0009
Morollo, A. A., Petsko, G. A., and Ringe, D. (1999). Structure of a Michaelis Complex Analogue: Propionate Binds in the Substrate Carboxylate Site of Alanine Racemase,. Biochemistry 38 (11), 3293–3301. doi:10.1021/bi9822729
Morrow, J. S., Keim, P., and Gurd, F. R. N. (1974). CO2 Adducts of Certain Amino Acids, Peptides, and Sperm Whale Myoglobin Studied by Carbon 13 and Proton Nuclear Magnetic Resonance. J. Biol. Chem. 249 (23), 7484–7494. doi:10.1016/s0021-9258(19)81264-4
Nijjar, S., Maddison, D., Meigh, L., Wolf, E., Rodgers, T., Cann, M. J., et al. (2020). Opposing Modulation of Cx26 gap Junctions and Hemichannels by CO 2. J. Physiol. 599 (1), 103–118. doi:10.1113/jp280747
Nishimura, M., and Akazawa, T. (1974). Structure and Function of Chloroplast Proteins. J. Biochem. 76 (1), 169–176. doi:10.1093/oxfordjournals.jbchem.a130542
Oberto, J., Nabti, S., Jooste, V., Mignot, H., and Rouviere-Yaniv, J. (2009). The HU Regulon Is Composed of Genes Responding to Anaerobiosis, Acid Stress, High Osmolarity and SOS Induction. PLoS ONE 4 (2), e4367. doi:10.1371/journal.pone.0004367
Ohtake, F., Saeki, Y., Sakamoto, K., Ohtake, K., Nishikawa, H., Tsuchiya, H., et al. (2015). Ubiquitin Acetylation Inhibits Polyubiquitin Chain Elongation. EMBO Rep. 16, 192. doi:10.15252/embr.201439152
Ohtake, F., and Tsuchiya, H. (2016). The Emerging Complexity of Ubiquitin Architecture. J. Biochem. doi:10.1093/jb/mvw088
Paoli, M., Liddington, R., Tame, J., Wilkinson, A., and Dodson, G. (1996). Crystal Structure of T State Haemoglobin with Oxygen Bound at All Four Haems. J. Mol. Biol. 256 (4), 775–792. doi:10.1006/jmbi.1996.0124
Park, I.-S., and Hausinger, R. P. (1995). Requirement of Carbon Dioxide for In Vitro Assembly of the Urease Nickel Metallocenter. Science 267 (5201), 1156–1158. doi:10.1126/science.7855593
Park, S.-Y., Yokoyama, T., Shibayama, N., Shiro, Y., and Tame, J. R. H. (2006). 1.25 Å Resolution Crystal Structures of Human Haemoglobin in the Oxy, Deoxy and Carbonmonoxy Forms. J. Mol. Biol. 360 (3), 690–701. doi:10.1016/j.jmb.2006.05.036
Pauling, L. (1935). The Oxygen Equilibrium of Hemoglobin and its Structural Interpretation. Proc. Natl. Acad. Sci. 21 (4), 186–191. doi:10.1073/pnas.21.4.186
Pearson, M. A., Schaller, R. A., Michel, L. O., Karplus, P. A., and Hausinger, R. P. (1998). Chemical Rescue of Klebsiella Aerogenes Urease Variants Lacking the Carbamylated-Lysine Nickel Ligand,. Biochemistry 37 (17), 6214–6220. doi:10.1021/bi980021u
Perrella, M., Kilmartin, J. V., Fogg, J., and Rossi-bernardi, L. (1975). Identification of the High and Low Affinity CO2-binding Sites of Human haemoglobinIdentification of the High and Low Affinity CO2-binding Sites of Human Haemoglobin. Nature 256 (5520), 759–761. doi:10.1038/256759a0
Perrella, M., and Russo, R. (2003). Allosteric Proteins: Lessons to Be Learned from the Hemoglobin Intermediates. Physiology 18 (6), 232–236. doi:10.1152/nips.01451.2003
Perutz, M. F., Bolton, W., Diamond, R., Muirhead, H., and Watson, H. C. (1964). Structure of Hæmoglobin : An X-ray Examination of Reduced Horse Hæmoglobin. Nature 203 (4946), 687–690. doi:10.1038/203687a0
Perutz, M. F., Rossmann, M. G., Cullis, A. F., Muirhead, H., Will, G., and North, A. C. T. (1960). Structure of Hæmoglobin: A Three-Dimensional Fourier Synthesis at 5.5-Å. Resolution, Obtained by X-Ray Analysis. Nature 185 (4711), 416–422. doi:10.1038/185416a0
Perutz, M. F. (1970). Stereochemistry of Cooperative Effects in Haemoglobin: Haem-Haem Interaction and the Problem of Allostery. Nature 228 (5273), 726–734. doi:10.1038/228726a0
Pierce, J., and Reddy, G. S. (1986). The Sites for Catalysis and Activation of Ribulosebisphosphate Carboxylase Share a Common Domain. Arch. Biochem. Biophys. 245 (2), 483–493. doi:10.1016/0003-9861(86)90241-9
Rutherford, J. C. (2014). The Emerging Role of Urease as a General Microbial Virulence Factor. Plos Pathog. 10 (5), e1004062. doi:10.1371/journal.ppat.1004062
Saha, D., Patgaonkar, M., Shroff, A., Ayyar, K., Bashir, T., and Reddy, K. V. R. (2014). Hemoglobin Expression in Nonerythroid Cells: Novel or Ubiquitous? Int. J. Inflamm. 2014, 1–8. doi:10.1155/2014/803237
Schneider, G., Knight, S., Andersson, I., Brändén, C. I., Lindqvist, Y., and Lundqvist, T. (1990). Comparison of the crystal Structures of L2 and L8S8 Rubisco Suggests a Functional Role for the Small Subunit. EMBO J. 9 (7), 2045–2050. doi:10.1002/j.1460-2075.1990.tb07371.x
Schneider, G., Lindqvist, Y., and Branden, C. I. (1992). RUBISCO: Structure and Mechanism. Annu. Rev. Biophys. Biomol. Struct. 21 (1), 119–143. doi:10.1146/annurev.bb.21.060192.001003
Severinghaus, J. W., and Bradley, A. F. (1958). Electrodes for Blood pO2 and pCO2 Determination. J. Appl. Physiol. 13, 515–520. doi:10.1152/jappl.1958.13.3.515
Song, K., and Li, S. (2021). The Role of Ubiquitination in NF-kB Signaling during Virus Infection. Viruses 13 (2), 145. doi:10.3390/v13020145
Srinivas, M., Verselis, V. K., and White, T. W. (2018). Human Diseases Associated with Connexin Mutations. Biochim. Biophys. Acta (Bba) - Biomembranes 1860 (1), 192–201. doi:10.1016/j.bbamem.2017.04.024
Stadie, W. C., and O'Brien, H. (1936). The Carbamate Equilibrium. J. Biol. Chem. 112 (2), 723–758. doi:10.1016/S0021-9258(18)74954-5
Stadie, W. C. (1935). The Rôle of the Carbamino Compounds in the Transport of CO 2 by the Blood. Science 81 (2095), 207–208. doi:10.1126/science.81.2095.207
Stec, B. (2012). Structural Mechanism of RuBisCO Activation by Carbamylation of the Active Site Lysine. Proc. Natl. Acad. Sci. 109 (46), 18785–18790. doi:10.1073/pnas.1210754109
Strynadka, N. C. J., Paetzel, M., Danel, F., de Castro, L., Mosimann, S. C., and Page, M. G. P. (2000). Crystal Structure of the Class D β-lactamase OXA-10. Nat. Struct. Biol. 7 (10), 918–925. doi:10.1038/79688
Sun, T. (2003). Comparison of -lactamases of Classes A and D: 1.5-A Crystallographic Structure of the Class D OXA-1 Oxacillinase. Protein Sci. 12 (1), 82–91. doi:10.1110/ps.0224303
Svane, S., Sigurdarson, J. J., Finkenwirth, F., Eitinger, T., and Karring, H. (2020). Inhibition of Urease Activity by Different Compounds Provides Insight into the Modulation and Association of Bacterial Nickel Import and Ureolysis. Sci. Rep. 10 (1), 8503. doi:10.1038/s41598-020-65107-9
Swatek, K. N., and Komander, D. (2016). Ubiquitin Modifications. Cel Res. 26 (4), 39. doi:10.1038/cr.2016.39
Tabita, F. R., Satagopan, S., Hanson, T. E., Kreel, N. E., and Scott, S. S. (2008). Distinct Form I, II, III, and IV Rubisco Proteins from the Three Kingdoms of Life Provide Clues about Rubisco Evolution and Structure/function Relationships. J. Exp. Bot. 59 (7), 1515–1524. doi:10.1093/jxb/erm361
Tabita, F. R., Hanson, T. E., Li, H., Satagopan, S., Singh, J., and Chan, S. (2007). Function, Structure, and Evolution of the RubisCO-like Proteins and Their RubisCO Homologs. Microbiol. Mol. Biol. Rev. 71 (4), 576–599. doi:10.1128/MMBR.00015-07
Tcherkez, G. (2016). The Mechanism of Rubisco-Catalysed Oxygenation. Plant Cel Environ. 39 (5), 983–997. doi:10.1111/pce.12629
Terrier, P., and Douglas, D. J. (2010). Carbamino Group Formation with Peptides and Proteins Studied by Mass Spectrometry. J. Am. Soc. Mass. Spectrom. 21 (9), 1500–1505. doi:10.1016/j.jasms.2010.05.008
Tooke, C. L., Hinchliffe, P., Bragginton, E. C., Colenso, C. K., Hirvonen, V. H. A., Takebayashi, Y., et al. (2019). β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 431 (18), 3472–3500. doi:10.1016/j.jmb.2019.04.002
Valegård, K., Andralojc, P. J., Haslam, R. P., Pearce, F. G., Eriksen, G. K., Madgwick, P. J., et al. (2018). Structural and Functional Analyses of Rubisco from Arctic Diatom Species Reveal Unusual Posttranslational Modifications. J. Biol. Chem. 293, 13033. doi:10.1074/jbc.RA118.003518
van de Wiel, J., Meigh, L., Bhandare, A., Cook, J., Nijjar, S., Huckstepp, R., et al. (2020). Connexin26 Mediates CO2-dependent Regulation of Breathing via Glial Cells of the Medulla Oblongata. Commun. Biol. 3 (1), 521. doi:10.1038/s42003-020-01248-x
Watson, G. M. F., Yu, J.-P., and Tabita, F. R. (1999). Unusual Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase of Anoxic Archaea. J. Bacteriol. 181 (5), 1569–1575. doi:10.1128/jb.181.5.1569-1575.1999
Wenker, I. C., Sobrinho, C. R., Takakura, A. C., Moreira, T. S., and Mulkey, D. K. (2012). Regulation of Ventral Surface CO2/H+-sensitive Neurons by Purinergic Signalling. J. Physiol. 590 (9), 2137–2150. doi:10.1113/jphysiol.2012.229666
West, J. B. (2014). Joseph Black, Carbon Dioxide, Latent Heat, and the Beginnings of the Discovery of the Respiratory Gases. Am. J. Physiol. Lung Cel Mol Physiol 306, L1057–L1063. doi:10.1152/ajplung.00020.2014
West, J. B. (2004). A Century of Pulmonary Gas Exchange. Am. J. Respir. Crit. Care Med. 169 (8), 897–902. doi:10.1164/rccm.200312-1781oe
West, J. B. (2019). Three Classical Papers in Respiratory Physiology by Christian Bohr (1855-1911) Whose Work Is Frequently Cited but Seldom Read. Am. J. Physiology-Lung Cell Mol. Physiol. 316, L585–L588. doi:10.1152/ajplung.00527.2018
Wildman, S. G. (2002). Along the Trail from Fraction I Protein to Rubisco (Ribulose Bisphosphate Carboxylase-Oxygenase). Photosynthesis Res. 73 (1/3), 243–250. doi:10.1023/a:1020467601966
Willebrords, J., Crespo Yanguas, S., Maes, M., Decrock, E., Wang, N., Leybaert, L., et al. (2016). Connexins and Their Channels in Inflammation. Crit. Rev. Biochem. Mol. Biol. 51 (6), 413–439. doi:10.1080/10409238.2016.1204980
Yamaguchi, K., and Hausinger, R. P. (1997). Substitution of the Urease Active Site Carbamate by Dithiocarbamate and Vanadate. Biochemistry 36 (49), 15118–15122. doi:10.1021/bi971767f
Yau, R., and Rape, M. (2016). The Increasing Complexity of the Ubiquitin Code. Nat. Cel Biol 18, 579. doi:10.1038/ncb3358
Keywords: carbon dioxide, post-translational modification, carbamate, haemoglobin, rubisco, connexin, ubiquitin
Citation: Blake LI and Cann MJ (2022) Carbon Dioxide and the Carbamate Post-Translational Modification. Front. Mol. Biosci. 9:825706. doi: 10.3389/fmolb.2022.825706
Received: 01 December 2021; Accepted: 03 February 2022;
Published: 01 March 2022.
Edited by:
Xiangming Zha, University of Missouri–Kansas City, United StatesReviewed by:
Nilmar Silvio Moretti, Federal University of São Paulo, BrazilRoland G Huber, Bioinformatics Institute (A∗STAR), Singapore
Copyright © 2022 Blake and Cann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lynsay I. Blake, TC5JLkJsYWtlQGR1cmhhbS5hYy51aw==; Martin J. Cann, TS5KLkNhbm5AZHVyaGFtLmFjLnVr
 Lynsay I. Blake
Lynsay I. Blake Martin J. Cann
Martin J. Cann