
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci. , 08 February 2022
Sec. Cellular Biochemistry
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.821146
This article is part of the Research Topic Genetically Mobile Elements Repurposed by Nature and Biotechnologists View all 10 articles
Multidrug-resistant pathogens are of significant concern in recent years. Hence new antifungal and anti-bacterial drug targets are urgently needed before the situation goes beyond control. Inteins are polypeptides that self-splice from exteins without the need for cofactors or external energy, resulting in joining of extein fragments. Inteins are present in many organisms, including human pathogens such as Mycobacterium tuberculosis, Cryptococcus neoformans, C. gattii, and Aspergillus fumigatus. Because intein elements are not present in human genes, they are attractive drug targets to develop antifungals and antibiotics. Thus far, a few inhibitors of intein splicing have been reported. Metal-ions such as Zn2+ and Cu2+, and platinum-containing compound cisplatin inhibit intein splicing in M. tuberculosis and C. neoformans by binding to the active site cysteines. A small-molecule inhibitor 6G-318S and its derivative 6G-319S are found to inhibit intein splicing in C. neoformans and C. gattii with a MIC in nanomolar concentrations. Inteins have also been used in many other applications. Intein can be used in activating a protein inside a cell using small molecules. Moreover, split intein can be used to deliver large genes in experimental gene therapy and to kill selected species in a mixed population of microbes by taking advantage of the toxin-antitoxin system. Furthermore, split inteins are used in synthesizing cyclic peptides and in developing cell culture model to study infectious viruses including SARS-CoV-2 in the biosafety level (BSL) 2 facility. This mini-review discusses the recent research developments of inteins in drug discovery and therapeutic research.
Prolonged use of drugs can lead to drug-resistant strains of pathogens and is a major challenge in treating the diseases. Drug resistance have been reported in Mycobacterium tuberculosis, Cryptococcosis neoformans and C. gattii that are causative agents of tuberculosis (TB) and cryptococcosis, respectively. In fact, antimicrobial resistance has been determined as one of the top 10 global public health threats by the World Health Organization (WHO) (WHO, 2021a). Although TB has been declining over the years, around 10 million people caught the disease with 1.5 million deaths reported in 2020 (Furin et al., 2019; WHO, 2021b). Since the start of antibiotic treatment in 1943, some strains of bacteria developed resistance to first-line anti-TB drugs, isoniazid and rifampin. The majority of the multi-drug resistant strains of M. tuberculosis are of Beijing lineage (Parwati et al., 2010; Stoffels et al., 2013). Similarly, there are reports of drug resistance to frontline antifungal drugs such as amphotericin B, fluconazole and 5-fluorocytosine (Bandara et al., 2020; Billmyre et al., 2020; Carolus et al., 2020). Although many antifungals are in the pipeline (Gintjee et al., 2020), the discovery of new targets will help to develop alternative strategies to combat the diseases (Su et al., 2018).
Inteins are small mobile elements within a host protein. Inteins can self-splice without external energy or cofactors and ligate the host protein fragments to generate active proteins (Pinto et al., 2020). Intein was first reported from the protein vacuolar membrane H+-translocating adenosine triphosphatase (VMA1 or TFP1) gene of Saccharomyces cerevisiae (Hirata et al., 1990). Inteins can be classified by its structural components or by splicing mechanisms. There are mainly three types of inteins based on the structural components prior to splicing (Lin et al., 2013). The max-intein has a homing endonuclease (HE) domain (Singh et al., 2009) which can hydrolyze genomic DNA within the cells (Chevalier and Stoddard, 2001), whereas mini-inteins do not have HE (Volkmann and Mootz, 2013). The third type is split inteins which have two fragments; the N terminal fragment joins to the N-terminal extein, whereas the C-terminal fragment links to the C terminal extein (Wu et al., 1998). Once the two fragments of split intein assemble, the split intein performs regular splicing activity (Beyer et al., 2020).
The splicing of intein is a rapid reaction. Hence rarely the precursor protein is observed in its native form. The active site is formed by folding of intein within the precursor, resulting in splice junctions (Tori et al., 2010). The intein and the first C-extein amino acid (aa) together act as a single turnover enzyme for splicing. Based on the splicing mechanisms, there are three classes of inteins (Tori et al., 2010; Nanda et al., 2020). Most inteins fall under class I inteins that most commonly have Ser, Thr, Cys, or Asn as essential residues that act as nucleophiles during splicing. The class I intein splicing consists of four coordinated nucleophilic displacement reactions. The reactions are: 1) amide (thio)ester and rearrangement, 2) transesterification and branch formation, 3) Asn cyclization and branch resolution, 4) acyl rearrangement or succinimide hydrolysis (Mills et al., 2014). Class II and III inteins have an Ala at their N-termini (Basturea, 2021). These two class inteins also form branched intermediate present in class I intein, but do so by different pathways as the N-terminal Ala cannot form linear thioester intermediates. The class II inteins skip the first step of class I intein splicing in which the intein N-terminus Cys/Ser residue directly attacks the N-terminal splice site amide bond to form a Block G branched intermediate (Mills et al., 2014). The class III inteins form a specific Block F branched intermediate with Cys at block F as the branch point before arrive at the Block G branched intermediate (Tori and Perler, 2011a). The active site residues and positions within the intein vary in each classes (Tori and Perler, 2011b).
The presence or absence of intein is species dependent. A total of 2,709 intein-containing genomes were found from the NCBI database, out of which 56% were found in eukaryotes, 19.8% in archaea, 6.64% in eubacteria and 17.4% in viruses (Nanda et al., 2020). Most of the inteins are located at conserved sites of housekeeping proteins with important functions, such as aminoacyl tRNA synthetases, DNA and RNA polymerases, recombinases, helicases topoisomerases, and spliceosomal components (Novikova et al., 2014; Fernandes et al., 2016).
Many human pathogens have intein elements in their genes (Table 1A). Among infectious diseases, TB is one of the significant causes of human death worldwide. The causative agent M. tuberculosis has one intein in each of the three M. tuberculosis proteins: replicative helicase (DnaB), recombinase (RecA), and an iron-sulfur cluster assembly (SufB) (Topilina et al., 2015). In contrast, closely related nonpathogenic M. smagmatis has only two inteins Dnabi1 and Dnabi2, in DnaB (Kelley et al., 2018). The DnaB helicase unwinds DNA from 5′ to 3′ direction at the replication fork, which is critical in replication initiation (LeBowitz and McMacken, 1986; Bailey et al., 2007). M. laprae, the causative agent of leprosy, has one intein in each of its DnaB, RecA, SufB and GyrA proteins (Table 1A). Although the RecA protein of M. leprae is structurally analogous to that of M. tuberculosis, it functions differently from its counterpart in M. tuberculosis (Patil et al., 2011). RecA deletion studies in E. coli (Kurnit, 1989) and M. smegmatis (Papavinasasundaram et al., 1998) indicate that RecA is not necessary for survival. In contrast, both DnaB and SufB (Huet et al., 2006) are essential for M. tuberculosis.
Coxiella burnetii is considered a re-emerging zoonosis in many countries. C. burnetii naturally infects livestock animals, such as goats, sheep, and cattle (Abdel-Moein and Zaher, 2021). It causes Q fever in human. In 2019, 178 acute and 34 chronic Q fever cases were reported in USA (CDC, 2021). C. burnetii has a DnaB intein with an approximate size of 16 kDa (Raghavan et al., 2008).
In the fungal kingdom, the pre mRNA processing factor 8 (Prp8) intein is the most widespread (Theodoro et al., 2013). Active Prp8 is critical for eukaryotic spliceosome responsible for pre-mRNA splicing (Grainger, 2005). The human version of Prp8 is also known by other names such as PRPF8, PRPC8, p220, and 220K in literature (Grainger, 2005). Some fungal pathogens such as C. neoformans and C. gattii have mini inteins without the HE domains in their Prp8 proteins, whereas A. fumigatus and Histoplasma capsulatum have the HE domains in their inteins (Table 1A).
Emergomycosis is an emerging disease caused by a novel dimorphic fungus Emergomyces species in immunocompromised individuals (Samaddar and Sharma, 2021). Due to taxonomic similarity, Emergomyces was under genus Emmonsia. As per recent classification, Emergomyces genera include E. pasteurianus, E. africanus, E. canadensis, E. orientalis, and E. europaeus (Jiang et al., 2018; Schwartz et al., 2019), many of which have inteins in their Prp8 proteins (Garcia Garces et al., 2020) (Table 1A).
Many human pathogens contain inteins in some of their proteins that are crucial for survival. Therefore, inhibition of intein splicing will be an attractive strategy, especially when drug resistance is reported for frontline therapeutic agents. The absence of inteins in human proteins is an added advantage in targeting pathogens. Below is a summary of currently known intein inhibitors (Table 1B).
Biologically relevant metal ions Cu2+ and Zn2+ at 0.5 mM and 2 mM respectively could inhibit splicing of the C. neoformans Prp8 intein in an in vitro Prp8 intein splicing assay, whereas Mg2+ at 0.5 mM and 2 mM did not (Green et al., 2019). ZnSO4 binds with the Prp8 intein of C. neoformans with a binding affinity KD of 1 ± 0.8 nM in an isothermal titration calorimetry assay. The crystal structure of the Prp8 intein in complex with Zn2+ shows that C1, H65, H170, N171 are involved in Zn2+ binding (Figure 1A). The mechanism of action of Zn2+ and Cu2+ seems different. Cu2+ likely stimulates reversible modifications on catalytically active cysteine, whereas Zn2+ binds at the terminal asparagine and the critical cysteine, resulting in inhibition of splicing (Green et al., 2019).
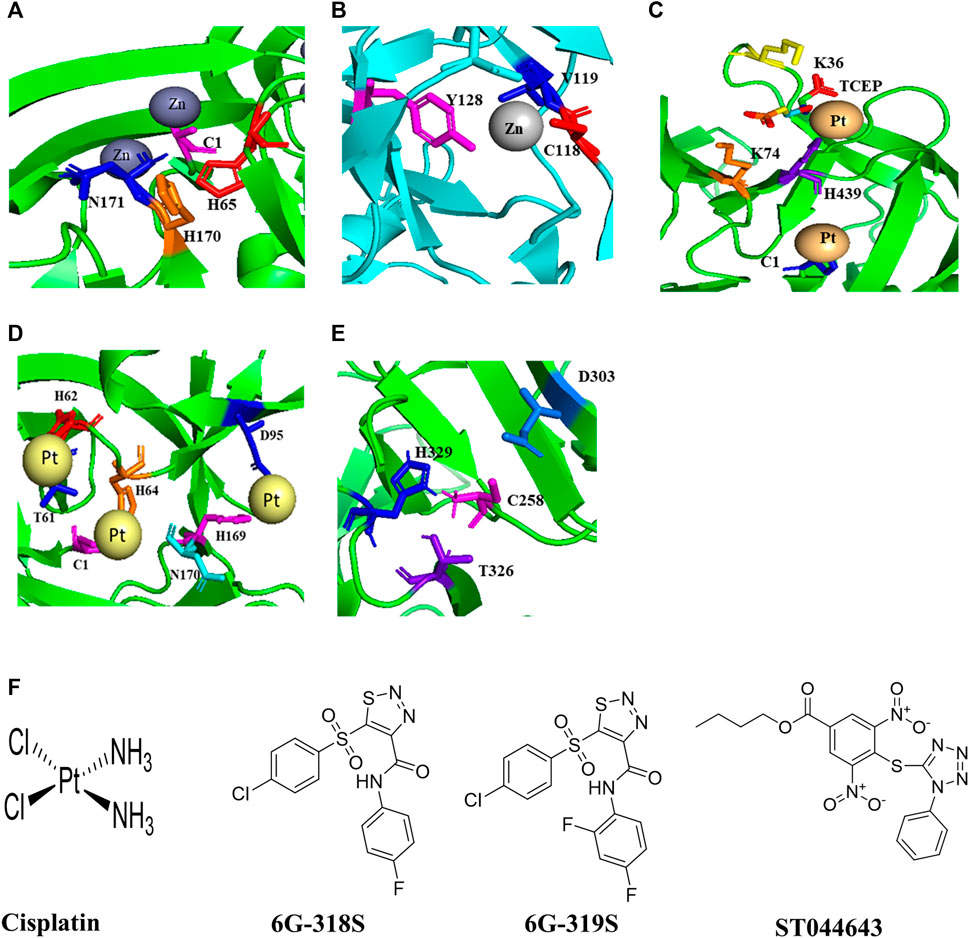
FIGURE 1. Structures of inteins, intein-ligand complexes, and small molecule inhibitors of intein splicing and hedgehog cholesterolysis. (A) The Prp8 intein of C. neoformans and Zn2+ (Green et al., 2019). (B) The DnaBi1 intein of M. smegmatis and Zn2+ (Woods et al., 2020). (C), The RecA intein of M. tuberculosis in complex with platinum and TCEP (Chan et al., 2016). (D) The Prp8 intein of C. gattii and cisplatin (Li et al., 2019). (E) The C-terminal 17 kDa fragment of drosophila hedgehog showing catalytic residues in cholesterolysis (Hall et al., 1997). (F) Structures of small molecules that inhibit intein splicing and hedgehog cholesterolysis.
Zn2+ also reversibly inhibits the splicing of DnaBi1 of M. smegmatis. In a splicing assay where the intein is placed between MBP and GFP (MIG), complete inhibition of DnaBi1splicing was observed at 10 µM of Zn2+. The metal-chelator EDTA could reverse the splicing (Woods et al., 2020). Zn2+ also inhibits splicing of the DnaB intein of M. laprae (Woods et al., 2020) which is homologous to the M. smegmatis DnaBi1 (Kelley et al., 2018). Additionally, Zn2+ could inhibit splicing in vivo in M. smegmatis. DnaBi1 and the M. leprae DnaB intein share 68.0% sequence identity. DnaBi2 and the M. tuberculosis DnaB intein have 61.0% amino acid identity (Kelley et al., 2018). A 1.95 Å crystal structure of M. smegmatis DnaBi1 indicates that the sulfur on C118, peptide backbone atoms N and O of V119, and the hydroxyl group of Y128 contributed to the coordination of the Zn2+ ions (Figure 1B) (Woods et al., 2020). The DnaBi1 of M. smegmatis responds to oxidative stress, whereas DnaBi2 is not (Kelley et al., 2018). Cd2+, Ni2+ and Co2+ have also shown inhibitory activity, although weaker than Zn2+, in splicing of the RecA intein from M. tuberculosis and of naturally split DnaE intein from Synechocystis sp., whereas there is no inhibition by Mg2+ and Ca2+ (Mills and Paulus, 2001; Panda et al., 2021).
Cisplatin, a platinum-containing anticancer drug, inhibits the RecA intein splicing with an IC50 of 2 µM in an in vitro splicing assay. The minimum inhibitory concentration (MIC) of cisplatin against M. tuberculosis was 40 µM (Zhang et al., 2011). Platinum-based compounds Pttfbz and Zeise’s salt have similar IC50 values (1.97, and 1.18 μm, respectively) as that of cisplatin (1.67 µM) in split GFP based assay, which employs a minimized M. tuberculosis RecA intein (Chan et al., 2016). Cisplatin and Pttfbz bind to intein in the presence of TCEP, whereas Zeise’s salt binding is independent of TCEP (Chan et al., 2016). The interaction of TCEP with cisplatin is also reported previously (Boal and Rosenzweig, 2009; Chen et al., 2013). The crystal structure of an HE-less RecA in complex with cisplatin and TCEP was solved at 1.50 Å (Figure 1C) (Chan et al., 2016). The cisplatin may modify one of the cysteine residues in the active site of the RecA intein (Zhang et al., 2011).
Cisplatin also inhibits the C. neoformans Prp8 intein splicing with IC50 of 2.5 µM in an in vitro splicing assay based on split Renilla luciferase; and the MIC90 was 4.5–20 μg/ml in various strains of C. neoformans and C. gattii (Li et al., 2019). It was reported that cisplatin treatment at 8 mg/kg once daily for 4 days led to two log-order reductions of C. neoformans in the lungs of BALB/c mice that were challenged with H99 at 1 × 107 CFU/mouse (Li et al., 2019). The crystal structure of the C. gatti Prp8 intein in complex with cisplatin indicates that the residues C1, T61, H62, H64, H169, N170, D95 are involved in the complex formation (Figure 1D) (Li et al., 2019). Mutagenesis of active site residues C1 and H169 abolished the binding of cisplatin to the Prp8 intein.
In addition to metal-containing inhibitors, reactive nitrogen species compounds (DEA NONOate at 1.2 mM and 12 mM) and Angeli’s salt at 2 mM and 20 mM) were found to inhibit splicing of the C. neoformans Prp8 intein in an in vitro Prp8 intein splicing assay, whereas H2O2 (.8 and 8 mM) did not (Green et al., 2019). In contrast, H2O2 at 8 mM concentration inhibited the splicing of M. smegmatis DnaBi1 in the MIG splicing assay in vitro (Kelley et al., 2018). Moreover, H2O2 at 5 mM abolished splicing of the DnaB precursor in vivo in S. smegmatis in a western blot assay using an anti-DnaB extein antibody (Kelley et al., 2018).
From a small-scale screening of small molecules using split luciferase and split GFP-based C. neoformans Prp8 intein splicing assays, a compound 6G-318 was found as an inhibitor of intein splicing with an IC50 of 5.8 µM and 11.2 µM, respectively (Li et al., 2021) (Figure 1F). When tested in C. neoformans, the compound displayed MIC values of 0.62–1 μg/ml. Out of the four commercially available derivatives, 6G-319S, 12G-305S, 6G-313S, and 12G-315S, only the fluoride derivative 6G-319 was active against C. neoformans with MIC of 1.3 μg/ml (Figure 1F). 6G-318S has shown a synergistic effect with amphotericin B. The cytotoxicity CC50 was determined for the adenocarcinomic human alveolar basal epithelial cells (A549) cells. The ratio of CC50 to MIC for 6G-318S and 6G-319S were 22.4 and 34.5, respectively. The inhibition of intein splicing was further demonstrated by an in vitro MIG splicing assay and in vivo in C. neoformans. The binding of 6G-318S to the Prp8 intein was demonstrated by thermal shift assay, mass-spectrometry, and surface plasmon resonance (SPR) assays. The dissociation constant KD of 6G-318S was .36 µM for wild type Prp8 intein in the SPR assay (Li et al., 2021).
Split-inteins express as two separate polypeptides at the ends of two host proteins and catalyze their trans-splicing, resulting in the formation of a single larger polypeptide (Li, 2015). Translation of two genes happens separately. After translation, two intein parts ligate their flanking protein parts to each other, resulting in mature protein (Caspi et al., 2003). Naturally, split-inteins are found only in the DNA polymerase III alpha subunit (polC or dnaE gene) of some cyanobacteria (Caspi et al., 2003). The DnaE intein family of cyanobacteria are the largest known class of split inteins (Caspi et al., 2003).
Inteins are being used for various biotechnological applications (Shah and Muir, 2014). One such application is gene therapy which is gaining importance. Gene therapy is promising for various diseases. Adeno-associated viral (AAV) vectors mediated retinal gene therapy promises to treat inherited and non-inherited eye diseases. The delivery of genes that are large (>5 kb) is a challenge. AAV vectors with fragments of target proteins flanked by short split inteins result in protein trans-splicing, leading to reconstitution of full-length protein in the retina of mice and pigs, and in human retinal organoids (Tornabene et al., 2019) (Figure 2A).
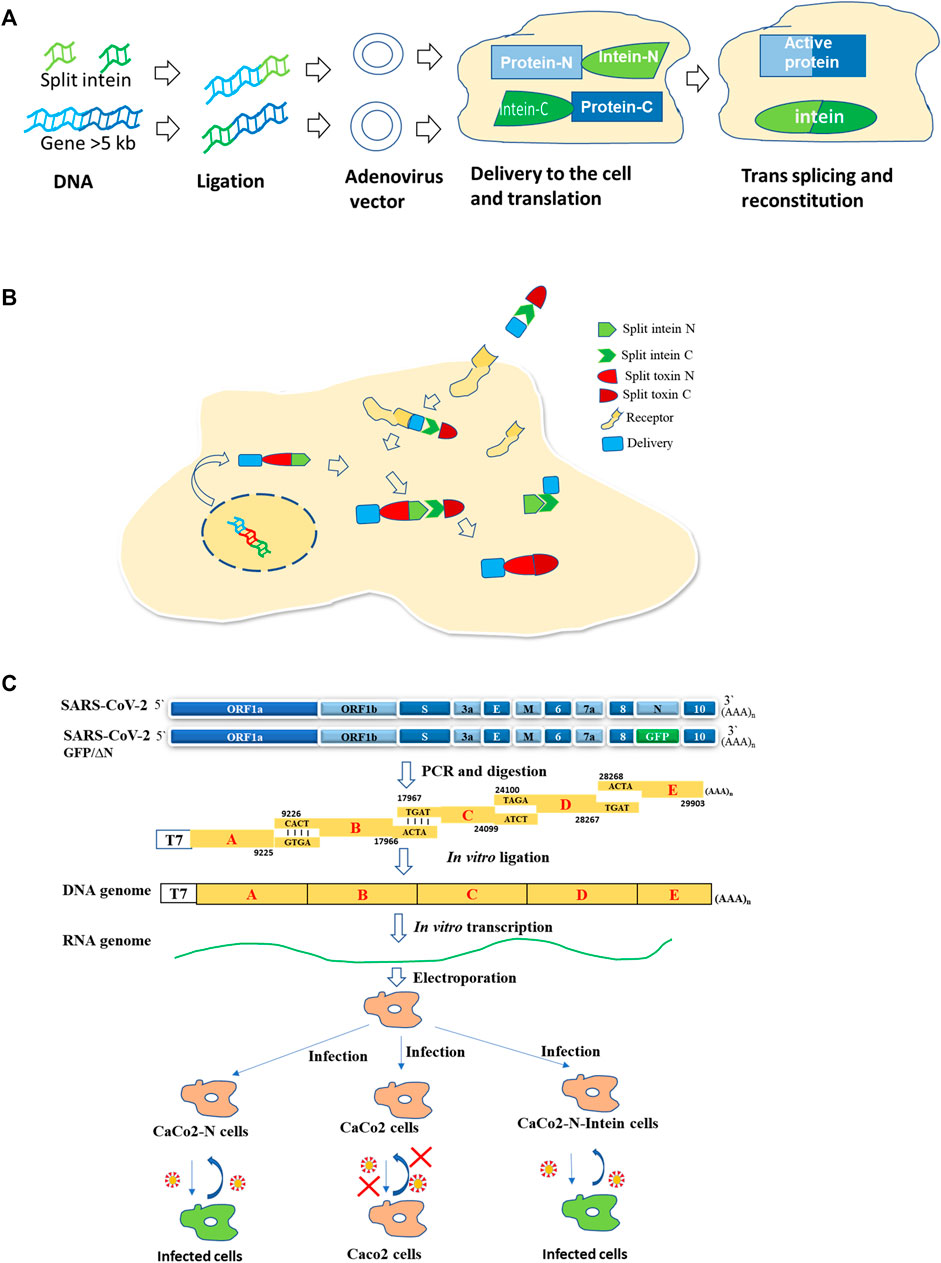
FIGURE 2. Use of split intein in therapy and pathogenesis studies. (A) Large gene delivery in gene therapy (Tornabene et al., 2019; Lim et al., 2020). (B) Intein mediated toxin reconstitution inside the cells for selective cell killing in tumor therapy (Purde et al., 2020). (C) Development of a BSL2 cell culture model for SARS-COV-2 in CaCo2 cells (Ju et al., 2021).
A similar methodology was used for expressing large genes in several studies. Using CRISPR/Cas9, one can target any genes, but the size of Cas9 is a limitation. The coding sequence for Cas9 is divided into two parts on a dual vector having split intein fragments, which will get reconstituted post-translationally without affecting its endonuclease activity (Truong et al., 2015). Furthermore, the methodology was used for CRISPR-based editors to treat amyotrophic lateral sclerosis (ALS), which is due to mutations in the superoxide dismutase 1 (SOD1) gene in a G93A-SOD1 mouse model of ALS (Lim et al., 2020). Dual AAV particles encoding split intein engineered to trans-splice and substitute a mutant SOD1 gene resulted in prolonged survival, slow disease progression, reduced muscle atrophy, improved neuromuscular function, and 40% fewer SOD1 immunoreactive inclusions as compared to control mice (Lim et al., 2020). Similar treatments were made using split inteins in dual AVV1 vectors to replace the mutated dystrophin gene in Duchenne muscular dystrophy (DMD) (Li et al., 2008). DMD is a common and lethal childhood muscle disorder, affecting 1 in every 3,500 male births (Koenig et al., 1987). After confirming the Becker-form dystrophin protein that has X chromosome-linked mutation in cell culture in vitro, AAV1 vectors were transferred into the muscle of Duchenne muscular dystrophy mouse model, resulting in therapeutic gene expression and benefits (Li et al., 2008).
The split inteins were also used for gene editing. Point mutations are seen in pathogenic human genetic variants. Adenine base editors (ABEs) catalyzes target A·T base pairs to G·C; and cytosine base editors (CBEs) converts target C·G base pairs to T·A. Although many studies have been done with base editors (Porto et al., 2020), the size of base editors is a limitation. To solve the issue, both CBE and ABE can be divided and fused to split intein fragments in an AAV vector system to form full base editors after trans splicing reconstitutes. This type of editing was done in somatic tissues liver, heart, muscle, retina, and brain that have relevance in therapeutic (Levy et al., 2020). Moreover, dual-AAV split-intein base editors have been used to treat Niemann-Pick disease type C that affects the central nervous system in a mouse model (Levy et al., 2020). In another study, the clotting factor VIII fused with split inteins was delivered into HEK293 and Cos-7 cells as a proof of concept. Lack of clotting factor 8 is seen in a hereditary bleeding disorder called Hemophilia A (Zhu et al., 2010). Split inteins were also used for treating experimental retinal Stargardt macular dystrophy (SMD), an inherited macular dystrophy in humans. In SMD, the ATP binding cassette subfamily A member 4 (ABCA4) is mutated. ABCA4 is involved in the clearance of photoisomerized all-trans-retinal from the photoreceptor disk lumen. Mutated ABCA4 results in a buildup of lipofuscin pigments in the retinal pigment epithelial cells, causing vision loss in patients with Stargardt disease (Tornabene et al., 2021). In order to repair the ABCA4 mutation, the ABCA4 gene is split into two; and each half is linked to split inteins which are separated, loaded onto AVV vectors, and delivered to one-month-old Abca4−/− mice subretinally. As expected, lipofuscin was observed to accumulate in the retinal pigmented epithelium of the vehicle-treated Abca4−/− mice. In contrast, there was a significant reduction in lipofuscin, in addition to improvement in other phenotypes for the mice treated with ABCA4-split intein after 3 months (Tornabene et al., 2021). Similarly, studies with the retina of pig and human retinal organoids also showed improvement for the ABCA4-split-intein group compared with the vehicle controls (Tornabene et al., 2021).
One of the issues is that non-mammalian origin components of AAV vectors could elicit immune and toxic responses in target cells or raise regulatory concerns for clinical use. To overcome this issue, a degron can be included in the trans-splicing system. The specific signals which turn the protein susceptible to ubiquitin-mediated proteasomal degradation are called degrons (Varshavsky, 1991). Inclusion of degron E. coli dihydrofolate reductase (ecDHFR) in the N-intein results in selective degradation of excised inteins from the AVV vector that is used for delivery of the ABCA4 gene for retinal therapy (Tornabene et al., 2021). The degradation ability of ecDHFR is inhibited by a small stabilizing ligand, trimethoprim.
Split intein was used to reconstitute a toxin inside selected cells enabling selective cell killing in mixed populations and tumor xenografts (Purde et al., 2020). The Diphtheria toxin catalytic domain (DTA) was split into two, with each fused to split intein from N. punctiforme. Split-toxinN encoding DNA is delivered by transfection/viral transduction, while recombinant split toxin C is delivered via a specific receptor by the anthrax toxin translocation system. The active toxin will be formed intracellularly by intein-mediated trans-splicing of two split-toxin parts (Purde et al., 2020) (Figure 2B). The delivery of split toxin via cell surface-specific receptors will help in the killing of specific cell populations. An attempt of HER2 receptor-mediated delivery was also made to deliver a split Pseudomonas aeruginosa exotoxin, although it was reconstituted first in vitro outside the cell (Wang et al., 2019).
One of the drawbacks of broad-spectrum antibiotics is dysbiosis and concomitant health sequelae. Human gut has around 100 trillion microbes from over 1,000 species (Zhang and Chen, 2019). Targeted killing of harmful bacteria without harming beneficial ones can reduce dysbiosis and drug resistance. With the help of split intein, López-Igual et al. (2019) developed a toxin against V. cholerae using a toxin-antitoxin (TA) system. Various TA systems are present in most of prokaryotes. Toxins are proteins that reduce metabolism, whereas antitoxins are either RNA or proteins that counteract toxins (Song and Wood, 2020). TA is involved in the stabilization and fitness of mobile DNA, genome stabilization, and phage protection. The M. tuberculosis has 85 TA modules, whereas nonpathogenic M. smegmatis has only 5 TA modules (Yu et al., 2020). The TA toxins are different from endotoxin and exotoxins. The TA toxins function only inside the cells that produce TA and are not secreted outside the cells (Singh et al., 2021). The TA system is grouped into Type I to Type VI based on the mode of action of antitoxin (Harms et al., 2018). Over expression of toxins is bactericidal. The CcdA/CcdB Type II TA system is one example of the bacterial TAs. In order to design a toxin–intein antimicrobial which is V. cholerae specific, the type II toxin gyrase poison CcdB is split and fused to the split intein DnaE of N. punctiforme. Antibiotic-resistant V. cholerae bacteria receiving the toxin-intein containing plasmid are completely killed at its hosts such as zebrafish and crustacean Artemia salina (López-Igual et al., 2019).
The protein trans-splicing is also utilized to generate a novel cell culture model for SARS-COV2, which consists of viral RNA without N capsid and a producer cell line expressing viral N protein. In this system, two fragments of N are linked to split intein fragments. Ligation takes place to produce full-length N protein (Ju et al., 2021) (Figure 2C). This model will help in the study of viral pathogenesis as well as in screening antivirals in a BSL2 facility.
Engineered full-length inteins can also be employed to activate protein of interest using small molecules such as rapamycin (Mootz and Muir, 2002) and 4-hydroxytamoxifen (4-HT). 4-HT is an agonist of estrogen receptor (ER) (Sasson and Notides, 1988) and is cell-permeable. By inserting a natural ligand-binding domain (LBD) into a mini-intein can destroy splicing activity (Buskirk et al., 2004). A modified RecA can get spliced in the presence of 4-HT. The HE of the RecA is replaced with LBD of ER, which can be placed in any protein to facilitate ligand-dependent splicing using 4-HT. The splicing was 50–90% in the presence of 4-HT and <5% in the absence of 4-HT (Peck et al., 2011). These inteins may help in the modulation of protein activities post-translationally in living systems such as mammalian cells and help in vivo activation of intein-fused therapeutic proteins.
Split-intein circular ligation of peptides and proteins (SICLOPPS) is used to develop macrocyclic peptides inside cells and to phenotypically screen cells for them. The split inteins are fused to C and N termini of the target peptide. Upon trans splicing, circular peptide is formed (Scott et al., 1999). Circular peptides are more resistant to protease activity and may be suitable for oral administration as a drug (Tavassoli, 2017; Muttenthaler et al., 2021).
Drug resistant strains of pathogens are reported in many disease outbreaks. In the year of 2018, there were about half a million new cases of rifampicin-resistant TB globally, the majority of which have multi-drug resistant TB (MDR-TB), a TB form resistant to two or more anti-TB drugs. TB was one of the major infectious killer worldwide prior to the COVID-19 pandemic (WHO, 2021b).
The distribution of intein is species-specific. It is still not clear why intein persists for millions of years of evolution (Novikova et al., 2016; Nanda et al., 2020). Although the exact functions of inteins remain elusive, splicing of intein from exteins is essential for activity of the intein-harboring protein. Many bacterial and fungal pathogens harbor inteins in their essential proteins. Since DnaB is an essential protein, inhibition of intein splicing can inhibit the growth of M. tuberculosis. Nonpathogenic M. smegmatis is frequently used as a model to study M. tuberculosis. Presence of the DnaB intein in M. smegmatis will facilitate the research on intein as it can be done in a BSL1 facility.
Another group of disease-causing organisms that have inteins are fungal pathogens. Although immunocompromised individuals are major concern for fungal infection, immunocompetent individuals are also susceptible to fungal species such as C. gattii (Andreou et al., 2019). Thus, it is essential to develop more antifungals with novel mechanisms of action such as via inhibition of intein splicing. Significant progress has been made recently in identification of inhibitors of intein splicing. Non-metal small molecule inhibitors started to emerge, showing inhibition of intein splicing both in vitro and in vivo (Li et al., 2021). Further studies will be required to identify more potent candidate intein splicing inhibitors and/or to optimize existing inhibitors to achieve clinical significance to combat infections of intein-containing microbes. It is noted that intein splicing inhibitors may be limited to intein-containing microbes, and may not be suitable for non-intein containing fungal pathogens such as Candida albicans (Yapar, 2014; Fernandes et al., 2016).
No intein has been reported in humans. However, an autoprocessing mechanism is found for cholesterolysis of the human Hedgehog (Hh) protein, which mimics intein splicing. The Hh protein is synthesized as a 45 kDa precursor, undergoes auto-processing to yield a 25 kDa C-terminal fragment and a 20 kDa N-terminal fragment with cholesterol moiety covalently attached to it. Therefore, it is necessary to check the off-target activity of the intein inhibitors (Perler, 1998) with human Hh protein. The crystal structure of a 17 kDa Hh fragment active in the initiation of auto-processing is solved (Figure 1E). The Hh residues Cys-258, His-329, Thr-326, and Asp-303 are involved in auto-processing (Hall et al., 1997). Mutants of these residues were tested for autoprocessing activity by inducing with dithiothreitol (DTT) and/or cholesterol. The DTT was used for the initial steps in the auto-processing reaction of Hh, the thioester formation, whereas cholesterol was used for the next step, the cholesterol transfer. Alanine mutants of Cys-258, His-329, Thr-326 lost the activity in both DTT and cholesterol assays, whereas D303A mutant was only active with DTT but not with cholesterol (Hall et al., 1997).
The compound ST044643 can be used as a positive control as it is active with an IC50 of 5 µM in a cholesterolysis assay based on protein fluorescence resonance energy transfer (FRET) (Owen et al., 2015). Nucleophiles such as DTT, glutathione, and hydroxylamine, can stimulate Hh autoprocessing in vitro, similar to cholesterol (Hall et al., 1997). The hedgehog autoprocessing is also inhibited by Zn2+ with a Ki of 2 μM in vitro and at the cellular environment in primary rat astrocyte culture (Xie et al., 2015).
Another area of research that employs the intein, especially the split intein, is gene therapy and gene delivery system. Although AAV-mediated gene delivery is approved, the size of the gene is a limiting factor. The split intein-based techniques could deliver genes above 5 kb in size by a dual vector. The use of split intein to deliver larger genes are gaining importance recently. The immune response due to presence of external intein fragments can be overcome by adding degrons which facilitate the ubiquitin-mediated degradation of the spliced intein.
The Nanoarchaeum equitans DNA polymerase (Pol) and Pyrococcus sp (GBD strain) Pol inteins are the only 2 reported cases of cysteine-less split inteins. They have inferior splicing properties under native and ambient conditions (Bhagawati et al., 2019). Because of the cysteine residues, the cysteine-containing split intein splicing requires a reducing environment. The reducing agents such as DTT and TCEP may affect the fused protein such as antibodies. The activity of cysteine-dependent inteins may be limited by oxidizing environments like the endoplasmic reticulum, bacterial periplasmic space, or the in vivo extracellular milieu. In order to overcome this issue, a cysteine-less split intein was designed and active at ambient temperatures and in the absence of reducing agents, without requiring a denaturation step (Bhagawati et al., 2019; Basturea, 2021). The point of splitting of the target protein to fuse with split intein fragments is also important to get active protein after trans-splicing (Purde et al., 2020). Care must be taken to expose the least hydrophobic residues.
To conclude, although intein was discovered many years ago, the use of it in drug discovery, therapeutics and as a biotechnological tool is of recent origin and is very promising. More efforts are needed to screen and optimize lead intein splicing inhibitors and to develop split inteins in clinical applications.
AT wrote the initial draft. ZL gave suggestions. HL edited the manuscript, suggested changes, and included modifications.
HL was supported by NIH grants: AI161845, AI131669, AI140406, AI141178, and AI140726, by University of Arizona College of Pharmacy faculty startup fund, and by R. Ken and Donna Coit Endowed Chair fund in Drug Discovery.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
4-HT, 4-hydroxytamoxifen; A549, adenocarcinomic human alveolar basal epithelial cells; AAV, Adeno-associated viral; AAV, Adeno-associated virus vector; ABCA4, ATP Binding Cassette Subfamily A Member 4; ABEs, Adenine base editors; ALS, Amyotrophic lateral sclerosis; CBEs, cytosine base editors; DTA, Diphtheria toxin catalytic domain; DTT, Dithiothreitol; ecDHFR, E. coli dihydrofolate reductase; ER, Estrogen receptor; FRET, Fluorescence resonance energy transfer; GFP, Green fluorescent protein; GLT1, glutamate synthase; HE, homing endonuclease; HER2/ERBB2, Human epidermal growth factor receptor 2; Hh, Hedgehog; KD, Dissociation constant; MDR, multi-drug resistant; MIC, minimum inhibitory concentration; SDS-PAGE, Sodium dodecyl sulphate–polyacrylamide gel electrophoresis; SICLOPPS, Split-intein circular ligation of peptides and proteins; SOD1, Superoxide dismutase 1; SPR, Surface plasmon resonance; TA, Toxin-antitoxin; TB, tuberculosis; TCEP, Tris (2-carboxyethyl) phosphine; ThrRS, Threonyl-tRNA synthetase; VMA, Vacuolar ATPase subunit.
Abdel-Moein, K. A., and Zaher, H. M. (2021). Parturient Cat as a Potential Reservoir for Coxiella Burnetii: A Hidden Threat to Pet Owners. Vector-Borne Zoonotic Dis. 21, 264–268. doi:10.1089/vbz.2020.2714
Andreou, M., Cogliati, M., Kolonitsiou, F., Stroumpos, C., Stamouli, V., Ravazoula, P., et al. (2020). Cryptococcus Gattii Infection in an Immunocompetent Host in Greece. Med. Mycol. Case Rep. 27, 1–3. doi:10.1016/j.mmcr.2019.12.002
Bailey, S., Eliason, W. K., and Steitz, T. A. (2007). Structure of Hexameric DnaB Helicase and its Complex with a Domain of DnaG Primase. Science 318, 459–463. doi:10.1126/science.1147353
Bandara, H. M. H. N., Wood, D. L. A., Vanwonterghem, I., Hugenholtz, P., Cheung, B. P. K., and Samaranayake, L. P. (2020). Fluconazole Resistance in Candida Albicans Is Induced by Pseudomonas aeruginosa Quorum Sensing. Sci. Rep. 10, 7769. doi:10.1038/s41598-020-64761-3
Bastos, R. W., Valero, C., Silva, L. P., Schoen, T., Drott, M., Brauer, V., et al. (2020). Functional Characterization of Clinical Isolates of the Opportunistic Fungal Pathogen Aspergillus nidulans. mSphere 5, e00153–20. doi:10.1128/mSphere.00153-20
Basturea, G. N. (2021). Inteins. Materials and Methods. Available at: https://www.labome.com/method/Inteins.html (Accessed November 14, 2021).
Beyer, H. M., Mikula, K. M., Li, M., Wlodawer, A., and Iwaï, H. (2020). The crystal Structure of the Naturally Split Gp41‐1 Intein Guides the Engineering of Orthogonal Split Inteins Fromcis‐splicing Inteins. FEBS J. 287, 1886–1898. doi:10.1111/febs.15113
Bhagawati, M., Terhorst, T. M. E., Füsser, F., Hoffmann, S., Pasch, T., Pietrokovski, S., et al. (2019). A Mesophilic Cysteine-Less Split Intein for Proteintrans-Splicing Applications under Oxidizing Conditions. Proc. Natl. Acad. Sci. USA 116, 22164–22172. doi:10.1073/pnas.1909825116
Billmyre, R. B., Applen Clancey, S., Li, L. X., Doering, T. L., and Heitman, J. (2020). 5-fluorocytosine Resistance Is Associated with Hypermutation and Alterations in Capsule Biosynthesis in Cryptococcus. Nat. Commun. 11, 127. doi:10.1038/s41467-019-13890-z
Boal, A. K., and Rosenzweig, A. C. (2009). Crystal Structures of Cisplatin Bound to a Human Copper Chaperone. J. Am. Chem. Soc. 131, 14196–14197. doi:10.1021/ja906363t
Buskirk, A. R., Ong, Y.-C., Gartner, Z. J., and Liu, D. R. (2004). Directed Evolution of Ligand Dependence: Small-Molecule-Activated Protein Splicing. Proc. Natl. Acad. Sci. 101, 10505–10510. doi:10.1073/pnas.0402762101
Butler, M. I., Gray, J., Goodwin, T. J., and Poulter, R. T. (2006). The Distribution and Evolutionary History of the PRP8 Intein. BMC Evol. Biol. 6, 42. doi:10.1186/1471-2148-6-42
Butler, M. I., and Poulter, R. T. M. (2005). The PRP8 Inteins in Cryptococcus Are a Source of Phylogenetic and Epidemiological Information. Fungal Genet. Biol. 42, 452–463. doi:10.1016/j.fgb.2005.01.011
Carolus, H., Pierson, S., Lagrou, K., and Van Dijck, P. (2020). Amphotericin B and Other Polyenes-Discovery, Clinical Use, Mode of Action and Drug Resistance. JoF 6, 321. doi:10.3390/jof6040321
Caspi, J., Amitai, G., Belenkiy, O., and Pietrokovski, S. (2003). Distribution of Split DnaE Inteins in Cyanobacteria. Mol. Microbiol. 50, 1569–1577. doi:10.1046/j.1365-2958.2003.03825.x
CDC (2021). Q Fever Epidemiology and Statistics | CDC. Available at: https://www.cdc.gov/qfever/stats/index.html (Accessed November 17, 2021).
Chan, H., Pearson, C. S., Green, C. M., Li, Z., Zhang, J., Belfort, G., et al. (2016). Exploring Intein Inhibition by Platinum Compounds as an Antimicrobial Strategy. J. Biol. Chem. 291, 22661–22670. doi:10.1074/jbc.M116.747824
Chaturvedi, V., and de Hoog, G. S. (2020). Onygenalean Fungi as Major Human and Animal Pathogens. Mycopathologia 185, 1–8. doi:10.1007/s11046-020-00428-x
Chen, S., Jiang, H., Wei, K., and Liu, Y. (2013). Tris-(2-carboxyethyl) Phosphine Significantly Promotes the Reaction of Cisplatin with Sp1 Zinc finger Protein. Chem. Commun. 49, 1226–1228. doi:10.1039/C2CC38517A
Chevalier, B. S., and Stoddard, B. L. (2001). Homing Endonucleases: Structural and Functional Insight into the Catalysts of Intron/intein Mobility. Nucleic Acids Res. 29, 3757–3774. doi:10.1093/nar/29.18.3757
Daugelat, S., and Jacobs, W. R. (1999). The Mycobacterium tuberculosis recA Intein Can Be Used in an ORFTRAP to Select for Open reading Frames. Protein Sci. 8, 644–653. doi:10.1110/ps.8.3.644
Davis, E. O., Thangaraj, H. S., Brooks, P. C., and Colston, M. J. (1994). Evidence of Selection for Protein Introns in the recAs of Pathogenic Mycobacteria. EMBO J. 13, 699–703. doi:10.1002/j.1460-2075.1994.tb06309.x
Dukik, K., Muñoz, J. F., Jiang, Y., Feng, P., Sigler, L., Stielow, J. B., et al. (2017). Novel Taxa of Thermally Dimorphic Systemic Pathogens in theAjellomycetaceae(Onygenales). Mycoses 60, 296–309. doi:10.1111/myc.12601
Eiglmeier, K., Honoré, N., Woods, S. A., Caudron, B., and Cole, S. T. (1993). Use of an Ordered Cosmid Library to Deduce the Genomic Organization of Mycobacterium leprae. Mol. Microbiol. 7, 197–206. doi:10.1111/j.1365-2958.1993.tb01111.x
Fernandes, J. A. L., Prandini, T. H. R., Castro, M. d. C. A., Arantes, T. D., Giacobino, J., Bagagli, E., et al. (2016). Evolution and Application of Inteins in Candida Species: A Review. Front. Microbiol. 7, 1585. doi:10.3389/fmicb.2016.01585
Fsihi, H., Vincent, V., and Cole, S. T. (1996). Homing Events in the gyrA Gene of Some Mycobacteria. Proc. Natl. Acad. Sci. 93, 3410–3415. doi:10.1073/pnas.93.8.3410
Furin, J., Cox, H., and Pai, M. (2019). Tuberculosis. The Lancet 393, 1642–1656. doi:10.1016/S0140-6736(19)30308-3
Garcia Garces, H., Hamae Yamauchi, D., Theodoro, R. C., and Bagagli, E. (2020). PRP8 Intein in Onygenales: Distribution and Phylogenetic Aspects. Mycopathologia 185, 37–49. doi:10.1007/s11046-019-00355-6
Gintjee, T. J., Donnelley, M. A., and Thompson, G. R. (2020). Aspiring Antifungals: Review of Current Antifungal Pipeline Developments. JoF 6, 28. doi:10.3390/jof6010028
Grainger, R. J., and Beggs, J. D. (2005). Prp8 Protein: At the Heart of the Spliceosome. RNA 11, 533–557. doi:10.1261/rna.2220705
Green, C. M., Li, Z., Smith, A. D., Novikova, O., Bacot-Davis, V. R., Gao, F., et al. (2019). Spliceosomal Prp8 Intein at the Crossroads of Protein and RNA Splicing. Plos Biol. 17, e3000104. doi:10.1371/journal.pbio.3000104
Hall, T. M. T., Porter, J. A., Young, K. E., Koonin, E. V., Beachy, P. A., and Leahy, D. J. (1997). Crystal Structure of a Hedgehog Autoprocessing Domain: Homology between Hedgehog and Self-Splicing Proteins. Cell 91, 85–97. doi:10.1016/S0092-8674(01)80011-8
Harms, A., Brodersen, D. E., Mitarai, N., and Gerdes, K. (2018). Toxins, Targets, and Triggers: An Overview of Toxin-Antitoxin Biology. Mol. Cell 70, 768–784. doi:10.1016/j.molcel.2018.01.003
Hirata, R., Ohsumk, Y., Nakano, A., Kawasaki, H., Suzuki, K., and Anraku, Y. (1990). Molecular Structure of a Gene, VMA1, Encoding the Catalytic Subunit of H(+)-translocating Adenosine Triphosphatase from Vacuolar Membranes of Saccharomyces cerevisiae. J. Biol. Chem. 265, 6726–6733. doi:10.1016/s0021-9258(19)39210-5
Huet, G., Castaing, J.-P., Fournier, D., Daffé, M., and Saves, I. (2006). Protein Splicing of SufB Is Crucial for the Functionality of the Mycobacterium tuberculosis SUF Machinery. J. Bacteriol. 188, 3412–3414. doi:10.1128/JB.188.9.3412-3414.2006
Jiang, Y., Dukik, K., Muñoz, J. F., Sigler, L., Schwartz, I. S., Govender, N. P., et al. (2018). Phylogeny, Ecology and Taxonomy of Systemic Pathogens and Their Relatives in Ajellomycetaceae (Onygenales): Blastomyces, Emergomyces, Emmonsia, Emmonsiellopsis. Fungal Divers. 90, 245–291. doi:10.1007/s13225-018-0403-y
Ju, X., Zhu, Y., Wang, Y., Li, J., Zhang, J., Gong, M., et al. (2021). A Novel Cell Culture System Modeling the SARS-CoV-2 Life Cycle. Plos Pathog. 17, e1009439. doi:10.1371/journal.ppat.1009439
Kelley, D. S., Lennon, C. W., Li, Z., Miller, M. R., Banavali, N. K., Li, H., et al. (2018). Mycobacterial DnaB Helicase Intein as Oxidative Stress Sensor. Nat. Commun. 9, 4363. doi:10.1038/s41467-018-06554-x
Klabunde, T., Sharma, S., Telenti, A., Jacobs, W. R., and Sacchettini, J. C. (1998). Crystal Structure of GyrA Intein from Mycobacterium Xenopi Reveals Structural Basis of Protein Splicing. Nat. Struct. Mol. Biol. 5, 31–36. doi:10.1038/nsb0198-31
Koenig, M., Hoffman, E. P., Bertelson, C. J., Monaco, A. P., Feener, C., and Kunkel, L. M. (1987). Complete Cloning of the Duchenne Muscular Dystrophy (DMD) Cdna and Preliminary Genomic Organization of the DMD Gene in Normal and Affected Individuals. Cell 50 (3), 509–517. doi:10.1016/0092-8674(87)90504-6
Kurnit, D. M. (1989). Escherichia coli recA Deletion Strains that Are Highly Competent for Transformation and for In Vivo Phage Packaging. Gene 82, 313–315. doi:10.1016/0378-1119(89)90056-5
LeBowitz, J. H., and McMacken, R. (1986). The Escherichia coli dnaB Replication Protein Is a DNA Helicase. J. Biol. Chem. 261, 4738–4748. doi:10.1016/s0021-9258(17)38564-2
Lennon, C. W., Wahl, D., Goetz, J. R., and Weinberger, J. (2021). Reactive Chlorine Species Reversibly Inhibit DnaB Protein Splicing in Mycobacteria. Microbiol. Spectr. 9, e00301–21. doi:10.1128/Spectrum.00301-21
Levy, J. M., Yeh, W.-H., Pendse, N., Davis, J. R., Hennessey, E., Butcher, R., et al. (2020). Cytosine and Adenine Base Editing of the Brain, Liver, Retina, Heart and Skeletal Muscle of Mice via Adeno-Associated Viruses. Nat. Biomed. Eng. 4, 97–110. doi:10.1038/s41551-019-0501-5
Li, J., Sun, W., Wang, B., Xiao, X., and Liu, X.-Q. (2008). ProteinTrans-Splicing as a Means for Viral Vector-MediatedIn VivoGene Therapy. Hum. Gene Ther. 19, 958–964. doi:10.1089/hum.2008.009
Li, Y. (2015). Split-inteins and Their Bioapplications. Biotechnol. Lett. 37, 2121–2137. doi:10.1007/s10529-015-1905-2
Li, Z., Fu, B., Green, C. M., Liu, B., Zhang, J., Lang, Y., et al. (2019). Cisplatin Protects Mice from challenge of Cryptococcus Neoformans by Targeting the Prp8 Intein. Emerging Microbes & Infections 8, 895–908. doi:10.1080/22221751.2019.1625727
Li, Z., Tharappel, A. M., Xu, J., Lang, Y., Green, C. M., Zhang, J., et al. (2021). Small-molecule Inhibitors for the Prp8 Intein as Antifungal Agents. Proc. Natl. Acad. Sci. USA 118, e2008815118. doi:10.1073/pnas.2008815118
Lim, C. K. W., Gapinske, M., Brooks, A. K., Woods, W. S., Powell, J. E., and Zeballos C., M. A. (2020). Treatment of a Mouse Model of ALS by In Vivo Base Editing. Mol. Ther. 28, 1177–1189. doi:10.1016/j.ymthe.2020.01.005
Lin, Y., Li, M., Song, H., Xu, L., Meng, Q., and Liu, X.-Q. (2013). Protein Trans-splicing of Multiple Atypical Split Inteins Engineered from Natural Inteins. PLoS One 8, e59516. doi:10.1371/journal.pone.0059516
Liu, X.-Q., and Yang, J. (2004). Prp8 Intein in Fungal Pathogens: Target for Potential Antifungal Drugs. FEBS Lett. 572, 46–50. doi:10.1016/j.febslet.2004.07.016
López-Igual, R., Bernal-Bayard, J., Rodríguez-Patón, A., Ghigo, J.-M., and Mazel, D. (2019). Engineered Toxin–Intein Antimicrobials Can Selectively Target and Kill Antibiotic-Resistant Bacteria in Mixed Populations. Nat. Biotechnol. 37, 755–760. doi:10.1038/s41587-019-0105-3
Mills, K. V., Johnson, M. A., and Perler, F. B. (2014). Protein Splicing: How Inteins Escape from Precursor Proteins. J. Biol. Chem. 289, 14498–14505. doi:10.1074/jbc.R113.540310
Mills, K. V., and Paulus, H. (2001). Reversible Inhibition of Protein Splicing by Zinc Ion. J. Biol. Chem. 276, 10832–10838. doi:10.1074/jbc.M011149200
Mootz, H. D., and Muir, T. W. (2002). Protein Splicing Triggered by a Small Molecule. J. Am. Chem. Soc. 124, 9044–9045. doi:10.1021/ja026769o
Muttenthaler, M., King, G. F., Adams, D. J., and Alewood, P. F. (2021). Trends in Peptide Drug Discovery. Nat. Rev. Drug Discov. 20, 309–325. doi:10.1038/s41573-020-00135-8
Nanda, A., Nasker, S. S., Mehra, A., Panda, S., and Nayak, S. (2020). Inteins in Science: Evolution to Application. Microorganisms 8, 2004. doi:10.3390/microorganisms8122004
Neupane, K., and Kaswan, D. (2021). “Coxiella Burnetii,” in StatPearls (Treasure Island (FL): StatPearls Publishing). Available at: http://www.ncbi.nlm.nih.gov/books/NBK557893/(Accessed November 17, 2021).
Novikova, O., Jayachandran, P., Kelley, D. S., Morton, Z., Merwin, S., Topilina, N. I., et al. (2016). Intein Clustering Suggests Functional Importance in Different Domains of Life. Mol. Biol. Evol. 33, 783–799. doi:10.1093/molbev/msv271
Novikova, O., Topilina, N., and Belfort, M. (2014). Enigmatic Distribution, Evolution, and Function of Inteins. J. Biol. Chem. 289, 14490–14497. doi:10.1074/jbc.R114.548255
Owen, T. S., Ngoje, G., Lageman, T. J., Bordeau, B. M., Belfort, M., and Callahan, B. P. (2015). FRET-based Cholesterolysis Assay Identifies a Novel Hedgehog Inhibitor. Anal. Biochem. 488, 1–5. doi:10.1016/j.ab.2015.06.021
Panda, S., Nanda, A., Nasker, S. S., Sen, D., Mehra, A., and Nayak, S. (2021). Metal Effect on Intein Splicing: A Review. Biochimie 185, 53–67. doi:10.1016/j.biochi.2021.03.006
Papavinasasundaram, K. G., Colston, M. J., and Davis, E. O. (1998). Construction and Complementation of a recA Deletion Mutant of Mycobacterium Smegmatis Reveals that the Intein in Mycobacterium tuberculosis recA Does Not Affect RecA Function. Mol. Microbiol. 30, 525–534. doi:10.1046/j.1365-2958.1998.01083.x
Parwati, I., van Crevel, R., and van Soolingen, D. (2010). Possible Underlying Mechanisms for Successful Emergence of the Mycobacterium tuberculosis Beijing Genotype Strains. Lancet Infect. Dis. 10, 103–111. doi:10.1016/S1473-3099(09)70330-5
Patil, K. N., Singh, P., Harsha, S., and Muniyappa, K. (2011). Mycobacterium leprae RecA Is Structurally Analogous but Functionally Distinct from Mycobacterium tuberculosis RecA Protein. Biochim. Biophys. Acta (Bba) - Proteins Proteomics 1814, 1802–1811. doi:10.1016/j.bbapap.2011.09.011
Paulus, H. (2003). Inteins as Targets for Potential Antimycobacterial Drugs. Front. Biosci. 8, s1157–1165. doi:10.2741/1195
Peck, S. H., Chen, I., and Liu, D. R. (2011). Directed Evolution of a Small Molecule-Triggered Intein with Improved Splicing Properties in Mammalian Cells. Chem. Biol. 18, 619–630. doi:10.1016/j.chembiol.2011.02.014
Perler, F. B. (1998). Protein Splicing of Inteins and Hedgehog Autoproteolysis: Structure, Function, and Evolution. Cell 92, 1–4. doi:10.1016/S0092-8674(00)80892-2
Pinto, F., Thornton, E. L., and Wang, B. (2020). An Expanded Library of Orthogonal Split Inteins Enables Modular Multi-Peptide Assemblies. Nat. Commun. 11, 1529. doi:10.1038/s41467-020-15272-2
Porto, E. M., Komor, A. C., Slaymaker, I. M., and Yeo, G. W. (2020). Base Editing: Advances and Therapeutic Opportunities. Nat. Rev. Drug Discov. 19, 839–859. doi:10.1038/s41573-020-0084-6
Purde, V., Kudryashova, E., Heisler, D. B., Shakya, R., and Kudryashov, D. S. (2020). Intein-mediated Cytoplasmic Reconstitution of a Split Toxin Enables Selective Cell Ablation in Mixed Populations and Tumor Xenografts. PNAS 117, 22090–22100. doi:10.1073/pnas.2006603117
Raghavan, R., Hicks, L. D., and Minnick, M. F. (2008). Toxic Introns and Parasitic Intein in Coxiella Burnetii: Legacies of a Promiscuous Past. J. Bacteriol. 190, 5934–5943. doi:10.1128/JB.00602-08
Roque, W. (2020). Pulmonary Disease Caused by mycobacterium Xenopi in a Patient with Dm and Copd. CHEST 158, A442. doi:10.1016/j.chest.2020.08.427
Salazar-Leal, J. I., Ramírez-Montelongo, S. M., and Luis, B. A. L. (2019). Clinical Significance of Nosocomial Cryptococcus Laurentii in Urine: A Case Series. Infect. Control. Hosp. Epidemiol. 40, 1442–1444. doi:10.1017/ice.2019.263
Samaddar, A., and Sharma, A. (2021). Emergomycosis, an Emerging Systemic Mycosis in Immunocompromised Patients: Current Trends and Future Prospects. Front. Med. (Lausanne) 8, 670731. doi:10.3389/fmed.2021.670731
Sasson, S., and Notides, A. C. (1988). Mechanism of the Estrogen Receptor Interaction with 4-hydroxytamoxifen. Mol. Endocrinol. 2, 307–312. doi:10.1210/mend-2-4-307
Saves, I., Lewis, L.-A., Westrelin, F., Warren, R., Daffé, M., and Masson, J.-M. (2002). Specificities and Functions of the recA and Pps1 Intein Genes of Mycobacterium tuberculosis and Application for Diagnosis of Tuberculosis. J. Clin. Microbiol. 40, 943–950. doi:10.1128/JCM.40.3.943-950.2002
Schwartz, I. S., Govender, N. P., Sigler, L., Jiang, Y., Maphanga, T. G., Toplis, B., et al. (2019). Emergomyces: The Global Rise of New Dimorphic Fungal Pathogens. Plos Pathog. 15, e1007977. doi:10.1371/journal.ppat.1007977
Scott, C. P., Abel-Santos, E., Wall, M., Wahnon, D. C., and Benkovic, S. J. (1999). Production of Cyclic Peptides and Proteins In Vivo. Proc. Natl. Acad. Sci. U S A. 96, 13638–13643. doi:10.1073/pnas.96.24.13638
Seshadri, R., Paulsen, I. T., Eisen, J. A., Read, T. D., Nelson, K. E., Nelson, W. C., et al. (2003). Complete Genome Sequence of the Q-Fever Pathogen Coxiella Burnetii. PNAS 100, 5455–5460. doi:10.1073/pnas.0931379100
Shah, N. H., and Muir, T. W. (2014). Inteins: Nature’s Gift to Protein Chemists. Chem. Sci. 5, 446–461. doi:10.1039/C3SC52951G
Singh, G., Yadav, M., Ghosh, C., and Rathore, J. S. (2021). Bacterial Toxin-Antitoxin Modules: Classification, Functions, and Association with Persistence. Curr. Res. Microb. Sci. 2, 100047. doi:10.1016/j.crmicr.2021.100047
Singh, P., Tripathi, P., Silva, G. H., Pingoud, A., and Muniyappa, K. (2009). Characterization of Mycobacterium leprae RecA Intein, a LAGLIDADG Homing Endonuclease, Reveals a Unique Mode of DNA Binding, Helical Distortion, and Cleavage Compared with a Canonical LAGLIDADG Homing Endonuclease. J. Biol. Chem. 284, 25912–25928. doi:10.1074/jbc.M109.042861
Smith, D. R., Richterich, P., Rubenfield, M., Rice, P. W., Butler, C., Lee, H.-M., et al. (1997). Multiplex Sequencing of 1.5 Mb of the Mycobacterium leprae Genome. Genome Res. 7, 802–819. doi:10.1101/gr.7.8.802
Song, S., and Wood, T. K. (2020). A Primary Physiological Role of Toxin/Antitoxin Systems Is Phage Inhibition. Front. Microbiol. 11, 1895. doi:10.3389/fmicb.2020.01895
Stoffels, K., Allix-Béguec, C., Groenen, G., Wanlin, M., Berkvens, D., Mathys, V., et al. (2013). From Multidrug- to Extensively Drug-Resistant Tuberculosis: Upward Trends as Seen from a 15-Year Nationwide Study. PLoS One 8, e63128. doi:10.1371/journal.pone.0063128
Su, H., Han, L., and Huang, X. (2018). Potential Targets for the Development of New Antifungal Drugs. J. Antibiot. 71, 978–991. doi:10.1038/s41429-018-0100-9
Tavassoli, A. (2017). SICLOPPS Cyclic Peptide Libraries in Drug Discovery. Curr. Opin. Chem. Biol. 38, 30–35. doi:10.1016/j.cbpa.2017.02.016
Telenti, A., Southworth, M., Alcaide, F., Daugelat, S., Jacobs, W. R., and Perler, F. B. (1997). The Mycobacterium Xenopi GyrA Protein Splicing Element: Characterization of a Minimal Intein. J. Bacteriol. 179, 6378–6382. doi:10.1128/jb.179.20.6378-6382.1997
Theodoro, R. C., Scheel, C. M., Brandt, M. E., Kasuga, T., and Bagagli, E. (2013). PRP8 Intein in Cryptic Species of Histoplasma Capsulatum: Evolution and Phylogeny. Infect. Genet. Evol. 18, 174–182. doi:10.1016/j.meegid.2013.05.001
Theodoro, R. C., Volkmann, G., Liu, X.-Q., and Bagagli, E. (2011). PRP8 Intein in Ajellomycetaceae Family Pathogens: Sequence Analysis, Splicing Evaluation and Homing Endonuclease Activity. Fungal Genet. Biol. 48, 80–91. doi:10.1016/j.fgb.2010.07.010
Topilina, N. I., Green, C. M., Jayachandran, P., Kelley, D. S., Stanger, M. J., Piazza, C. L., et al. (2015). SufB Intein of Mycobacterium tuberculosis as a Sensor for Oxidative and Nitrosative Stresses. PNAS 112, 10348–10353. doi:10.1073/pnas.1512777112
Tori, K., Dassa, B., Johnson, M. A., Southworth, M. W., Brace, L. E., Ishino, Y., et al. (2010). Splicing of the Mycobacteriophage Bethlehem DnaB Intein. J. Biol. Chem. 285, 2515–2526. doi:10.1074/jbc.M109.069567
Tori, K., and Perler, F. B. (2011a). Expanding the Definition of Class 3 Inteins and Their Proposed Phage Origin. J. Bacteriol. 193, 2035–2041. doi:10.1128/JB.01407-10
Tori, K., and Perler, F. B. (2011b). The Arthrobacter Species FB24 Arth_1007 (DnaB) Intein Is a Pseudogene. PLoS One 6, e26361. doi:10.1371/journal.pone.0026361
Tornabene, P., Trapani, I., Centrulo, M., Marrocco, E., Minopoli, R., Lupo, M., et al. (2021). Inclusion of a Degron Reduces Levelsof Undesired Inteins after AAV-Mediated Proteintrans-Splicing in the Retina. Mol. Ther. - Methods Clin. Dev. 23, 448–459. doi:10.1016/j.omtm.2021.10.004
Tornabene, P., Trapani, I., Minopoli, R., Centrulo, M., Lupo, M., de Simone, S., et al. (2019). Intein-mediated Protein Trans-splicing Expands Adeno-Associated Virus Transfer Capacity in the Retina. Sci. Transl Med. 11, eaav4523. doi:10.1126/scitranslmed.aav4523
Truong, D.-J. J., Kühner, K., Kühn, R., Werfel, S., Engelhardt, S., Wurst, W., et al. (2015). Development of an Intein-Mediated Split-Cas9 System for Gene Therapy. Nucleic Acids Res. 43, 6450–6458. doi:10.1093/nar/gkv601
Volkmann, G., and Mootz, H. D. (2013). Recent Progress in Intein Research: from Mechanism to Directed Evolution and Applications. Cell. Mol. Life Sci. 70, 1185–1206. doi:10.1007/s00018-012-1120-4
Wang, J., Han, L., Chen, J., Xie, Y., Jiang, H., and Zhu, J. (2019). Reduction of Non-specific Toxicity of Immunotoxin by Intein Mediated Reconstitution on Target Cells. Int. Immunopharmacol 66, 288–295. doi:10.1016/j.intimp.2018.11.039
WHO (2021a). Antimicrobial Resistance. Available at: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (Accessed November 13, 2021).
WHO (2021b). Tuberculosis (TB). Available at: https://www.who.int/news-room/fact-sheets/detail/tuberculosis (Accessed December 18, 2021).
Woods, D., Vangaveti, S., Egbanum, I., Sweeney, A. M., Li, Z., Bacot-Davis, V., et al. (2020). Conditional DnaB Protein Splicing Is Reversibly Inhibited by Zinc in Mycobacteria. mBio 11, e01403–20. doi:10.1128/mBio.01403-20
Wu, H., Hu, Z., and Liu, X.-Q. (1998). Protein Trans-splicing by a Split Intein Encoded in a Split DnaE Gene of Synechocystis Sp. PCC6803. PNAS 95, 9226–9231. doi:10.1073/pnas.95.16.9226
Xie, J., Owen, T., Xia, K., Singh, A. V., Tou, E., Li, L., et al. (2015). Zinc Inhibits Hedgehog Autoprocessing. J. Biol. Chem. 290, 11591–11600. doi:10.1074/jbc.M114.623264
Yapar, N. (2014). Epidemiology and Risk Factors for Invasive Candidiasis. Ther. Clin. Risk Manag. 10, 95–105. doi:10.2147/TCRM.S40160
Yu, X., Gao, X., Zhu, K., Yin, H., Mao, X., Wojdyla, J. A., et al. (2020). Characterization of a Toxin-Antitoxin System in Mycobacterium tuberculosis Suggests Neutralization by Phosphorylation as the Antitoxicity Mechanism. Commun. Biol. 3, 1–15. doi:10.1038/s42003-020-0941-1
Zhang, L., Xiao, N., Pan, Y., Zheng, Y., Pan, Z., Luo, Z., et al. (2010). Binding and Inhibition of Copper Ions to RecA Inteins from Mycobacterium tuberculosis. Chemistry 16, 4297–4306. doi:10.1002/chem.200903584
Zhang, L., Zheng, Y., Callahan, B., Belfort, M., and Liu, Y. (2011). Cisplatin Inhibits Protein Splicing, Suggesting Inteins as Therapeutic Targets in Mycobacteria. J. Biol. Chem. 286, 1277–1282. doi:10.1074/jbc.M110.171124
Zhang, S., and Chen, D.-C. (2019). Facing a New challenge: the Adverse Effects of Antibiotics on Gut Microbiota and Host Immunity. Chin. Med. J. (Engl) 132, 1135–1138. doi:10.1097/CM9.0000000000000245
Keywords: intein, inhibitor, drug target, therapeutic tool, anti-microbial
Citation: Tharappel AM, Li Z and Li H (2022) Inteins as Drug Targets and Therapeutic Tools. Front. Mol. Biosci. 9:821146. doi: 10.3389/fmolb.2022.821146
Received: 23 November 2021; Accepted: 10 January 2022;
Published: 08 February 2022.
Edited by:
Brian Callahan, Binghamton University, United StatesReviewed by:
Sasmita Nayak, KIIT University, IndiaCopyright © 2022 Tharappel, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongmin Li, aGxpMUBwaGFybWFjeS5hcml6b25hLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.