
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci. , 09 March 2022
Sec. Structural Biology
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.808036
This article is part of the Research Topic Nucleic Acid Polymerases: The Two-Metal-Ion Mechanism and Beyond View all 11 articles
 Joon Park1,2‡
Joon Park1,2‡ Noe Baruch-Torres3†‡
Noe Baruch-Torres3†‡ Shigenori Iwai4
Shigenori Iwai4 Geoffrey K. Herrmann1,2
Geoffrey K. Herrmann1,2 Luis G. Brieba3*
Luis G. Brieba3* Y. Whitney Yin1,2*
Y. Whitney Yin1,2*Human mitochondrial DNA contains more UV-induced lesions than the nuclear DNA due to lack of mechanism to remove bulky photoproducts. Human DNA polymerase gamma (Pol γ) is the sole DNA replicase in mitochondria, which contains a polymerase (pol) and an exonuclease (exo) active site. Previous studies showed that Pol γ only displays UV lesion bypassing when its exonuclease activity is obliterated. To investigate the reaction environment on Pol γ translesion activity, we tested Pol γ DNA activity in the presence of different metal ions. While Pol γ is unable to replicate through UV lesions on DNA templates in the presence of Mg2+, it exhibits robust translesion DNA synthesis (TLS) on cyclobutane pyrimidine dimer (CPD)-containing template when Mg2+ was mixed with or completely replaced by Mn2+. Under these conditions, the efficiency of Pol γ′s TLS opposite CPD is near to that on a non-damaged template and is 800-fold higher than that of exonuclease-deficient Pol γ. Interestingly, Pol γ exhibits higher exonuclease activity in the presence of Mn2+ than with Mg2+, suggesting Mn2+-stimulated Pol γ TLS is not via suppressing its exonuclease activity. We suggest that Mn2+ ion expands Pol γ′s pol active site relative to Mg2+ so that a UV lesion can be accommodated and blocks the communication between pol and exo active sites to execute translesion DNA synthesis.
Ultraviolet (UV) exposure of DNA causes dimerization of two adjacent pyrimidines, forming cyclobutane pyrimidine dimers (CPDs) or (6-4) pyrimidine-pyrimidones ((6-4)PP), which stalls replicating polymerases (Varghese and Wang, 1967a; Varghese and Wang, 1967b; Brash and Haseltine, 1982). Human mitochondria lack nucleotide excision repair or photolyase that removes the photoproducts (Maher et al., 1976; Maher et al., 1979; Maher et al., 1982); therefore, mitochondrial DNA replication machinery would inevitably encounter the UV photoproducts. Increasing sun exposure is correlated to a large spectrum of mitochondrial DNA (mtDNA) deletions in dermis, which, albeit unknown in mechanism, has been used as a biomarker for UV exposure and chronological aging (Birch-Machin et al., 1998; Kawasaki et al., 2000; Ray et al., 2000; Harbottle et al., 2010; Bharti et al., 2014). Deletions and mutations of mtDNA are also associated with skin cancers (Durham et al., 2003; Yang et al., 2004).
Human mitochondria DNA is replicated by DNA polymerase gamma (Pol γ), which consists of a catalytic subunit Pol γA and a dimeric accessory subunit Pol γB. Pol γ possesses activities of 5′-3′ polymerization (pol) for DNA synthesis, 3′-5′ exonuclease (exo) for proofreading, and 5′-deoxyribose phosphate lyase for DNA repair. Pol γB has no intrinsic enzymatic activity, but it accelerates polymerization rate, increases affinity to DNA, and enhances processivity of the holoenzyme (Johnson et al., 2000; Johnson and Johnson, 2001; Lee et al., 2010). Pol γA belongs to the A-family polymerases that are single polypeptide enzymes structurally resembling a right-hand shape and containing fingers, palm, and thumb domains (Steitz, 1999; Garcia-Diaz and Bebenek, 2007; Bębenek and Ziuzia-Graczyk, 2018). Other replicative A-family DNA polymerase members include bacterial DNA polymerase I and phage T7 DNA polymerase.
All known DNA polymerases require divalent metal ions to catalyze DNA synthesis. A general mechanism of two-metal-ion catalysis established a foundation for DNA and RNA polymerases’ enzymatic reactions (Steitz and Steitz, 1993). An exonuclease-deficient Pol γ variant was shown to have limited translesion DNA synthesis (TLS) activity on UV lesion-containing DNA in the presence of Mg2+, suggesting that Pol γ possesses intrinsic TLS ability (Kasiviswanathan et al., 2012). Mitochondria are the main storage site of intracellular Mn2+ ion (Maynard and Cotzias, 1955; Liccione and Maines, 1988; Gavin et al., 1990; Gavin et al., 1992), which raises an important question on impact of Mn2+ or Mg2+/Mn2+ mixture on Pol γ mtDNA replication as well as TLS.
We report here studies of the A-family member Pol γ lesion bypassing across UV lesions in the presence of different metal ions. We show that Pol γ is unable to replicate past the CPD- or (6-4)PP-containing DNA template in the presence of Mg2+, but displays efficient TLS ability comparable to that on the non-damaged template in the presence of Mn2+ or Mg2+/Mn2+ mixture. Mn2+ promotes TLS activity without involvement of the polymerase’s exonucleolytic activity. The Mn2+-mediated trans-UV lesion DNA synthesis appears uniquely to mitochondrial DNA polymerase and is absent in other A-family DNA polymerases tested. These results also give a novel insight into the metal-regulated error recognition and communication between the pol and exo active sites in Pol γ.
DNA templates (49 nt) containing either a CPD (T-CPD) or (6-4)PP (T-(6-4)PP) (Table 1) at the 28th position from the 3′-end was synthesized as described in Methods. Pol γ translesion replication was first tested on a 27 nt primer (PN) annealed to the T-CPD where the 3′- thymine (3′- T) in the CPD serves as the coding base (Figure 1A, lanes 9–16, Table 1). Time-dependent (0–20 min) primer extension assay was conducted in the presence of Mg2+. As a control, a parallel assay was carried out on a non-damaged template (T-ND) with identical sequence except that the CPD is replaced by 2 dT (Figure 1A, lanes 1–8, Table 1). In anticipation of higher polymerase activity on T-ND, the Pol γ concentration was reduced (see Methods). For convenience, only the full-length product formation rate at normalized enzyme concentration was used for rate calculation. Full-length formation rate of Pol γ on T-ND was 4.5 nM/s (Figure 1C). However, on T-CPD, the primer was not extended, but instead was degraded to PN-1 to PN-3 over time (Figure 1A lanes 9–16).
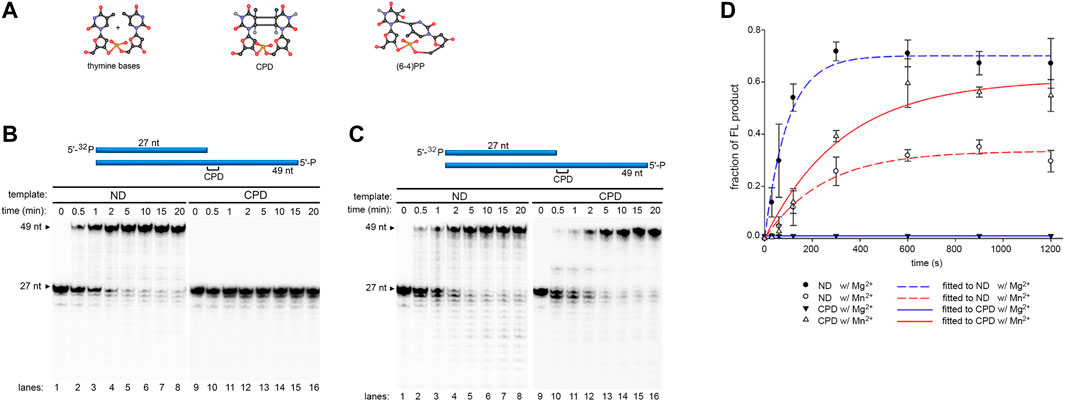
FIGURE 1. DNA synthesis of wild-type Pol γ on non-damaged and CPD-containing templates in the presence of Mg2+ or Mn2+. (A) Structure of thymine bases, CPD, and (6-4)PP where nitrogen is represented in blue spheres, oxygen in red spheres, carbon in black spheres, and phosphorus in golden spheres. (B) Time-dependent DNA synthesis of wild-type Pol γ on 5′-32P-Primer N (PN) annealed to a non-damaged template (T-ND) (lanes 1–8) or CPD-containing template (T-CPD) (lanes 9–16) in the presence of Mg2+. (C) Time-dependent DNA synthesis of wild-type Pol γ on 5′-32P-PN annealed to T-ND (lanes 1–8) or T-CPD (lanes 9–16) in the presence of Mn2+. (D) Quantification of normalized full-length (FL) products from (B) and (C) over time in seconds. The results are presented as mean and standard deviation from three independent experiments.
To evaluate the effect of high Mn2+ content in mitochondria on Pol γ activity, we substitute Mg2+ with Mn2+ and repeated the above experiments. In stark contrast to results with Mg2+, Pol γ exhibited strong TLS across CPD in the presence of Mn2+ (Figure 1B, lanes 9–16). The rates of full-length product formation by Pol γ on T-CPD were calculated to be 0.58 nM/s, which is only 1.7-fold lower than that on T-ND at 1 nM/s (Figure 1C). Interestingly, Pol γ is 4.5-fold less efficient on the non-damaged template in the presence of Mn2+ than in the presence of Mg2+.
Previous studies showed that exonuclease-deficient (exo-) Pol γ displayed limited intrinsic CPD-bypassing ability (Kasiviswanathan et al., 2012). To assess whether Mn2+ plays a redundant role with exonuclease activity in Pol γ TLS activity, Pol γ exo- (D198A/E200A) (Spelbrink et al., 2000) was analyzed on T-ND and T-CPD. In the presence of Mg2+, Pol γ exo- extended a small amount of the primer PN over the CPD site to full-length product, consistent with the previously reported data (Kasiviswanathan et al., 2012). However, a predominant amount of primer was extended by only a single nucleotide and formed product PN+1 (Figure 2A, lanes 1–8). Accumulation of the PN+1 indicates that silencing exonuclease enables the polymerase to synthesize across the 3′-T but not 5′-T of the CPD, suggesting that synthesis using the 5′-T as a coding base is the rate-limiting step of Pol γ TLS.

FIGURE 2. DNA synthesis of exonuclease-deficient Pol γ on non-damaged and CPD-containing templates in the presence of Mg2+ or Mn2+. (A) Time-dependent DNA synthesis of exonuclease-deficient (exo-) Pol γ on 5′-32P-PN annealed to T-ND in the presence of Mg2+ (lanes 1–8) or Mn2+ (lanes 9–16) (B) Time-dependent DNA synthesis of Pol γ exo-on 5′-32P-PN annealed to T-CPD in the presence of Mg2+ (lanes 1–8) or Mn2+ (lanes 9–16). (C) Quantification of normalized full-length (FL) products from (A) and (B) over time in seconds. The results are presented as mean and standard deviation from three independent experiments.
In the presence of Mn2+, Pol γ exo- exhibited significantly elevated TLS activity (Figure 2B, lanes 9–16). The rate of Pol γ exo- full-length product formation on T-CPD was calculated to be 0.81 nM/s with Mn2+, which is more than 800-fold increase than that with Mg2+ (0.001 nM/s) and exceeds that of wild-type (0.58 nM/s) (Figure 2C), confirming that suppressing exo activity indeed increases TLS in the presence of either metal ion. PN+1 product was drastically diminished, indicating Mn2+ facilitates Pol γ to overcome the energy barrier at the rate-limiting step. Similar to wild-type enzyme, Mn2+ exhibits inferior activity (eightfold lower) on Pol γ exo- synthesis on T-ND than Mg2+.
When translesion synthesis was tested on (6-4)PP lesion containing template, wild-type Pol γ showed no activity in the presence of either Mg2+ or Mn2+ (Figure 3, lanes 4 and 5). However, Pol γ exo- extended the primer to PN+1 with Mg2+, similarly to that on the T-CPD (Figure 3, lanes 7 and 8). Similarly, no full-length product was formed in the presence of Mn2+, and only PN+1 and a small amount of PN+2 was formed (Figure 3, lanes 9–10). Taken together, these results show that exonuclease activity contributes to the Pol γ′s lack of TLS activity, and Mn2+ further promotes Pol γ′s TLS ability independent of exonuclease activity.
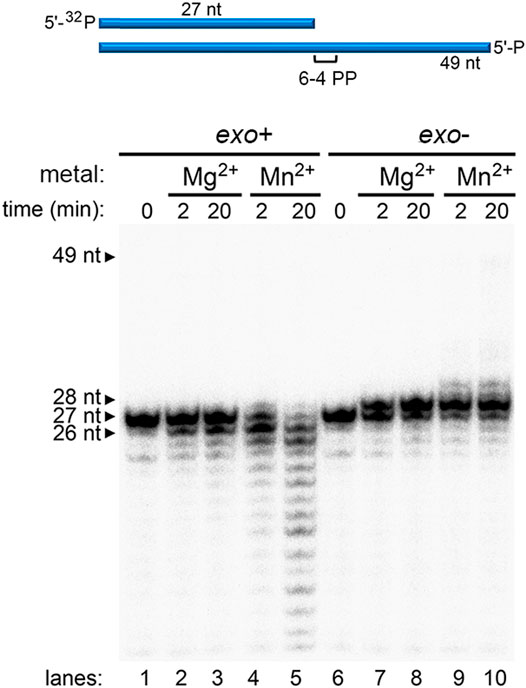
FIGURE 3. Metal-dependent DNA synthesis of wild-type and exonuclease-deficient Pol γ on a (6-4)PP-containing template. Time-dependent polymerization assay of wild-type Pol γ (exo+) on 5′-32P-PN annealed to T-(6–4)PP in the presence of Mg2+ (lanes 2 and 3) or in the presence of Mn2+ (lanes 4 and 5) and of Pol γ (exo-) on identical DNA substrates in the presence of Mg2+ (lanes 7 and 8) or Mn2+ (lanes 9 and 10).
The results thus far showed Mn2+ promotes TLS ability to both Pol γ wild-type and exo- variant, suggesting Mn2+ functions independent of exonuclease activity. To test the conclusion, we tested the exo activity of Pol γ with Mn2+ and compared it to that with Mg2+. We defined the rate of exonucleolysis as disappearance of the substrate and exonuclease events following the initial reaction were discounted, thus it is an underestimate of the actual rate. The rate of exonucleolysis of Pol γ on single-stranded DNA PN is 12.6 min−1 with Mn2+, sevenfold faster than that of Mg2+ at 1.8 min−1 (Figures 4A,C). Pol γ exo activity was also assayed on duplex PN/T-ND (Figure 4B). The excision rate of the duplex PN/T-ND in the presence of Mn2+ is at 3.9 min−1, which is about eightfold higher than Mg2+ at 0.5 min−1 (Figure 4C). Our results are consistent with a previous report where Mn2+ stimulates porcine liver Pol γ excision of terminal mismatch (Longley and Mosbaugh, 1991). As Mn2+ increased exo activity of Pol γ on PN/N-ND, which is free of error, it suggests that primer is sent more frequently from the pol to the exo site. As suppressing exo activity as well as addition of Mn2+ that increases exo activity both promote Pol γ′s TLS, we predict that Mn2+ must reduce error recognition ability of reading correct Watson-Crick base pairing geometry, which in turn promotes erroneous shuttling of the primer strand to the exo site.
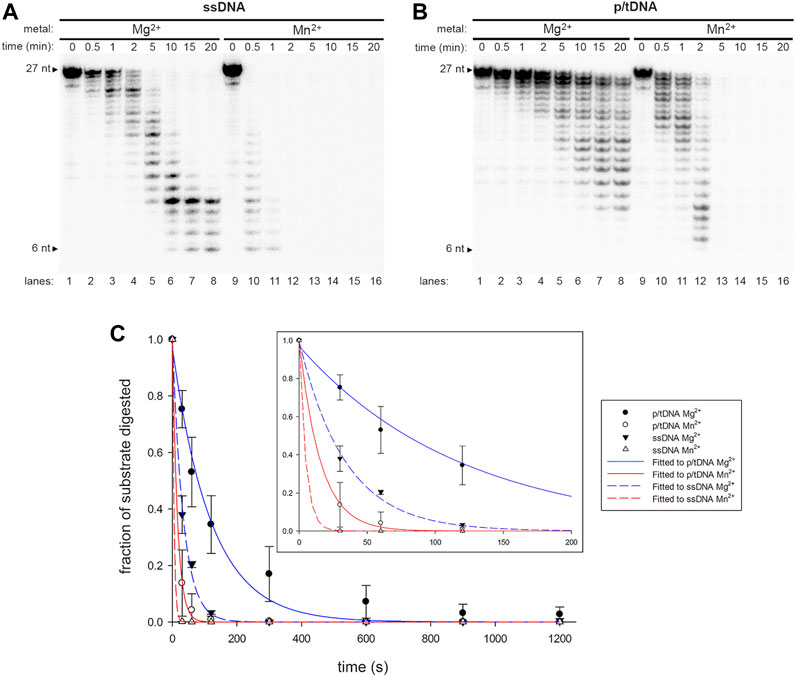
FIGURE 4. Metal-regulated exonuclease activity of wild-type Pol γ. (A) Exonuclease assay of Pol γ on a single-stranded DNA, 5′-32P-PN, in the presence of Mg2+ (lanes 1–8) or in the presence of Mn2+ (lanes 9–16). (B) Exonuclease assay Pol γ on primer/template, 5′-32P-PN/T-ND, in the presence of Mg2+ (lanes 1–8) or in the presence of Mn2+ (lanes 9–16) (C). Quantification of normalized fraction of substrate remaining in (A) and (B) over time in seconds shown. The results are presented as mean and standard deviation from three independent experiments. Shorter time points (0–200 s) are shown as an inset.
We show that Mg2+ and Mn2+ play opposite roles on Pol γ TLS. Since both ions are present in mitochondria, we tested the effects of metal ion mixture. Pol γ′s translesion synthesis assays were carried out at a fixed, near physiological concentration of one metal ion and varying the concentrations of the other. Specifically, Mg2+ was kept constant at 1.0 mM, and Mn2+ varied from 0.1 to 10 mM (Figure 6A); Mn2+ was kept constant at 0.4 mM, and Mg2+ varied from 0.1 to 10 mM (Figure 6C). Titrations of a single metal were also performed (Figure 5). As each dNTP is capable of binding to one of either metal ion and with higher affinity to Mn2+, the precise concentrations of free Mg2+ and Mn2+ ions for catalysis cannot be accurately calculated, but, in the presence of a total 0.8 mM dNTPs, it should be significantly lower than the initial concentrations.
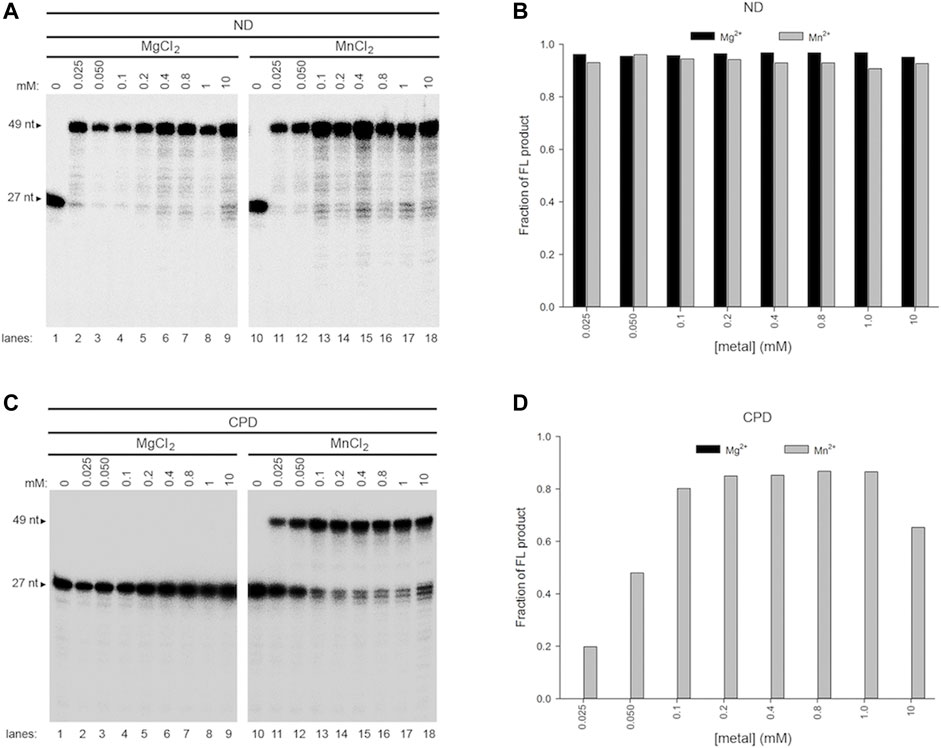
FIGURE 5. Effect of a single metal ion on wild-type Pol γ DNA activity on non-damaged and CPD-containing templates. (A) Wild-type Pol γ primer extension on T-ND annealed to 5′-32P-PN in the presence of Mg2+(lanes 1–9) or Mn2+ (lanes 10-18). (B) Quantification of the full-length product on T-ND vs. a gradient of Mg2+ or Mn2+ shown in (A). (C) Wild-type Pol γ primer extension on T-CPD annealed to 5′-32P-PN in the presence of Mg2+ (lanes 1–9) or Mn2+ (lanes 10-18). (D) Quantification of the full-length product on T-CPD vs. a gradient of Mg2+ or Mn2+ shown in (C).
In the presence of Mg2+ alone, Pol γ showed no TLS activity on CPD at any concentration tested (Figure 5C, lanes 1-9, 5D), whereas, in the presence of Mn2+ alone, it produced full-length (FL) TLS product from 0.025 to 10 mM (Figure 5C, lanes 10–18, 5D).
The TLS activity with mixed metal ions differs from that with a single metal ion alone. For example, assays with constant 1.0 mM Mg2+ and increasing Mn2+, Pol γ began to show TLS activity at 0.1 mM Mn2+ (Figure 6A, lane 10–18, 6B). Nonetheless, different from Pol γ producing 97% FL product with 0.1 mM Mn2+ on T-ND (Figure 5B), it only produced 5% FL TLS product with 0.1 mM Mn2+/1.0 mM Mg2+ on T-CPD (Figure 6B); the polymerase produced 95% FL product with 0.2 mM Mn2+ alone on T-ND (Figure 5B) but no activity with 1.0 mM Mg2+ on T-CPD (Figure 5C, lanes1-9), yet producing 12% FL TLS product with metal mixture (Figure 6B). These results suggest that Pol γ′s metal binding sites are unlikely occupied by the same metal ion. We thus hypothesize that both metals are bound to the polymerase simultaneously.
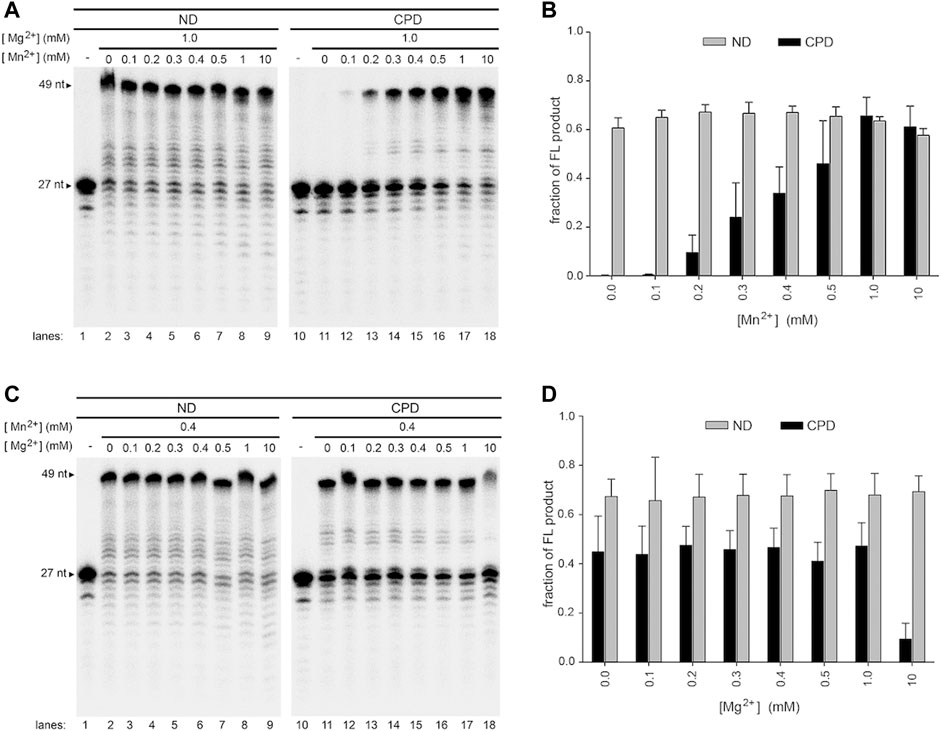
FIGURE 6. Effect of mixed metal ions on wild-type Pol γ DNA synthesis on non-damaged and CPD-containing templates in a mixture of Mg2+ and Mn2+. (A) Wild-type Pol γ primer extension on T-ND (lanes 1–9) or T-CPD (lanes 10–18) annealed to the 5′-32P-PN at a constant concentration of Mg2+ and varied concentrations of Mn2+. (B) Quantification of the full-length product vs. concentration of Mn2+ shown in (A). (C) Wild-type Pol γ primer extension on T-ND (lanes 1–9) or T-CPD (lanes 10–18) annealed to the 5′-32P-PN at constant concentration of Mn2+ and varied concentrations of Mg2+. (D) Quantification of the full-length product vs. concentration of Mg2+ shown in (C).
A similar conclusion can be drawn from assays with fixed Mn2+ and varying Mg2+. In the presence of 0.4 mM Mn2+ and substoichiometric (0.1–0.3 mM), equimolar (0.4–0.5 mM) to slightly excess stoichiometric among Mg2+ (1.0 mM), Pol γ generated 35–40% FL TLS product (Figure 6C, lanes 10–17, 6D). Even with the addition of 25-fold excess, 10 mM Mg2+, Pol γ still displays 10% of TLS activity (Figure 6C, lane 18, 6D), suggesting high probability of Pol γ binding to both metal ions.
We should note that under the reaction condition Pol γ TLS activity was observed (0.2 mM Mn2+ and 1.0 mM Mg2+, Figure 6A, lane 13), and assume dNTPs exhibit equal affinity to Mg2+ and Mn2+ ions and ignore metal binding to DNA, the free Mn2+ ions available for Pol γ catalysis is ∼67 µM
Previous studies show that Mn2+ does not stimulate TLS activity in other A-family DNA polymerase members, such as E. coli Pol I and Klenow Fragment (Moore et al., 1981; Rabkin et al., 1983). To further evaluate the uniqueness of Mn2+ activity on Pol γ, we analyzed Mn2+ function on another A-family member, T7 DNA polymerase (T7 DNAP), a structural homolog of Pol γ. Neither Mg2+ nor Mn2+ enables wild-type T7 DNAP to bypass CPD (Figures 7A,B, lanes 8–14), while both metals support activity on T-ND (Figures 7A,B lanes 1–7). Similar to Pol γ, T7 DNAP exonuclease activity appeared to be higher in the presence of Mn2+ than Mg2+. The results suggest Mn2+ exclusively stimulates Pol γ′s TLS activity among replicative A-family polymerases.
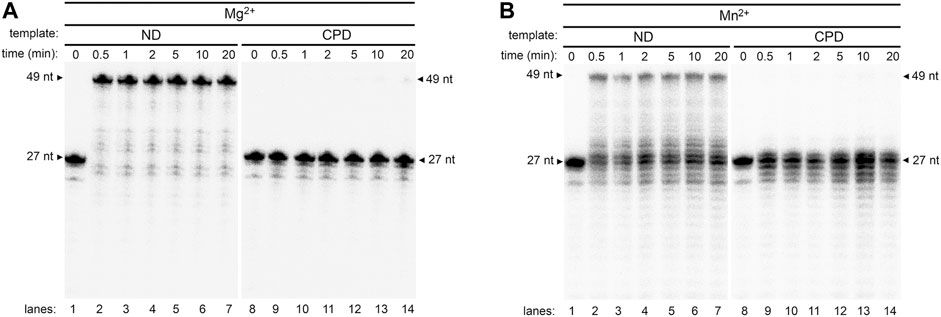
FIGURE 7. Evaluation of wild-type T7 polymerase activity on a non-damaged and CPD substrates in the presence of Mg2+ or Mn2+. (A) Time course activity assay of wild-type T7 DNA polymerase on T-ND (lanes 1–8) and on T-CPD (lanes 9–16) annealed to 5′- 32P-PN in the presence of Mg2+. (B) Time course activity assay of wild-type T7 DNA polymerase on T-ND (lanes 1–8) and on T-CPD (lanes 9–16) annealed to 5′-32P-PN in the presence of Mn2+.
Perhaps due to lack of UV lesion repair mechanism, mtDNA bares more pyrimidine dimers than chromosomal DNA (Birch-Machin et al., 2013). Nuclear DNA polymerases assume distinct functions in replication or lesion bypassing, but mitochondrial DNA polymerase, Pol γ, is the sole replicase responsible for synthesis on normal as well as lesion-containing mtDNA. Thus, the mitochondrial replicase will unavoidably encounter these bulky photoproducts during mtDNA replication. We present here studies of Pol γ replicating on CPD- and (6-4)PP-containing DNA templates. We showed that Pol γ exerts robust metal dependent activity on lesioned templates under physiological concentrations of Mg2+ and Mn2+ mixture or Mn2+ alone. The metal dependent UV dimer bypassing under a wide range of Mg2+ and Mn2+ concentrations is unique to Pol γ and has not been found in other A-family DNA polymerases.
Manganese is an essential element for biological functions and many ubiquitous enzymatic reactions in mitochondria. Mn2+ is rapidly transported into mitochondria via the uniporter with activated Ca2+ but slowly exported (Chance, 1965; Gavin et al., 1992), thus the organelle concentration is higher than that in cytosol. Mitochondrial concentration of Mn2+ is reported at 0.1–0.35 mM with an upper limit of ∼1 mM in brain, heart, and liver (Konji et al., 1985; Gunter et al., 2004), and a concentration of Mg2+ was 0.37–0.95 mM with upper limit of ∼1.5 mM in the heart (Corkey et al., 1986; Romani, 2011). We showed that, under the sub-stoichiometric concentrations of Mg2+/Mn2+, Pol γ displayed significant trans-CPD synthesis ability and presented features of binding to both metal ions. Based on these results, we hypothesize that Pol γ could bind to both metals in vivo and is therefore capable of replicating on non-damaged and CPD-containing templates. At very low Mg2+ or Mn2+ concentration, Pol γ is still able to replicate on both non-damaged and CPD-containing templates. Although the exact affinities of Mg2+ and Mn2+ to Pol γ are not known, it must be higher than the affinities toward dNTPs. Mg2+ or Mn2+ will first bind to metal A site in Pol γ, then Pol γ will selectively recruit incoming nucleotide pre-bound to metal ion, or metal B site. This feature of Pol γ partially compensates for lack of UV-lesion repair mechanism and TLS polymerase in mitochondria.
It is interesting to speculate Pol γ replication fidelity in the presence of Mg2+/Mn2+ mixture. At a high concentration of Mn2+ (above mM), many DNA polymerases become mutagenic and exhibit elevated replication errors with Mg2+ (Sirover and Loeb, 1976). A simplistic hypothesis would be that Mn2+ suppresses the proofreading exonuclease activity in DNA polymerases. However, others (Longley and Mosbaugh, 1991) and we show here that Pol γ displayed increased exo activity in the presence of Mn2+ relative to that of Mg2+. Thus, high concentrations in Mn2+-induced replication errors are not by direct inhibition of exo activity, rather by hindrance of error recognition. Nevertheless, at low concentrations of Mn2+, DNA polymerases can replicate accurately. For example, at 2 µM free Mn2+ concentration, E. coli Pol I was shown to synthesize DNA with similar error frequency as with Mg2+ (Beckman et al., 1985). We predict Pol γ would also replicate with high fidelity at a physiological concentration of Mn2+ and Mg2+. Needless to say, the hypothesis should be rigorously tested experimentally in future investigations. Lack of TLS activity on (6-4)PP-containing template by Pol γ suggests two possible outcomes: there may be another TLS polymerase that can overcome this UV lesion, or the damaged mtDNA is eliminated by a mitochondrial degradation mechanism as observed in C. elegans and primary human fibroblasts upon UV radiation damage (Bess et al., 2012; Bess et al., 2013).
Pol γ is the only A-family polymerase found to have an intrinsic TLS activity across UV lesions as a wild-type enzyme. Studies from the Copeland and Meyer groups showed that silencing Pol γ′s exonuclease activity led to limited CPD bypassing activity, despite the diminished activity for wild-type enzymes (Miller and Grollman, 1997; Kasiviswanathan et al., 2012). We show that the TLS activities of both wild-type and exonuclease-deficient Pol γ are greatly stimulated by the presence of Mn2+. Additionally, plant and yeast mitochondrial DNA polymerases also exhibited TLS activity similar to human Pol γ (unpublished results). However, such activities are not found in other replicative A-family DNA polymerases. E. coli Pol I and Klenow Fragment lack intrinsic TLS activity and only insert one nucleotide against 3′-T in the CPD in presence of Mn2+ (Moore et al., 1981; Rabkin et al., 1983; Smith et al., 1998). We showed here that wild-type T7 DNAP does not display Mn2+-stimulated TLS activity. The Mn2+-stimulated TLS activity appears to be reserved only to mitochondrial DNA polymerases, perhaps to take advantage of higher Mn2+ concentration in the mitochondria.
Nevertheless, Mn2+-dependent activity is observed in certain TLS polymerases (Putrament et al., 1975; Kunkel and Loeb, 1979; Goodman et al., 1983). For example, Dpo4 bypasses abasic sites and CPD much more efficiently with Mn2+ than with Mg2+ (Vaisman et al., 2005), and DNA polymerase ι bypasses abasic sites, benzopyrenes, CPD, and (6-4)PP only in the presence of Mn2+ (Frank and Woodgate, 2007). In addition, PrimPol bypasses more lesions when bound to Mn2+ relative to Mg2+ (Tokarsky et al., 2017; Makarova et al., 2018).
A distinct structural difference between high fidelity DNA polymerase such as Pol γ and bona fide TLS polymerases such as Pol η is the pol active site conformation and polymerase-enforced template DNA conformation. Crystal structures of Pol γ showed that Pol γ belongs to the A-family polymerases with characteristic fingers, thumb, and palm domains (Lee et al., 2009). Upon binding to a correct incoming nucleotide, the fingers domain undergoes large open-closed conformational changes to align the γ-phosphate of dNTP, 3′-OH of the primer, and catalytic metal ions for optimal catalysis for phosphodiester bond formation (Lee et al., 2009; Szymanski et al., 2015; Sohl et al., 2015). Significantly, the template bends 90° at the coding base (n) and downstream neighboring residue (n+1) (Figure 8, right). DNA template bending in Pol γ is accomplished by the helices O and O1 in the finger domain. Such template strand bending is thought to prevent template ‘slippage’ and ensuring replication fidelity. A non-bendable dimer is deterred from entering the pol site and stalls replication (Li et al., 2004). Indeed, the structure of T7 DNA polymerase, a Pol γ homolog, complexed with CPD lesion (Li et al., 2004) shows that after incorporating a nucleotide opposing the 3′-T of CPD, base pairing with the 5′-T of CPD is severely crooked, preventing replication at the n+1 position.
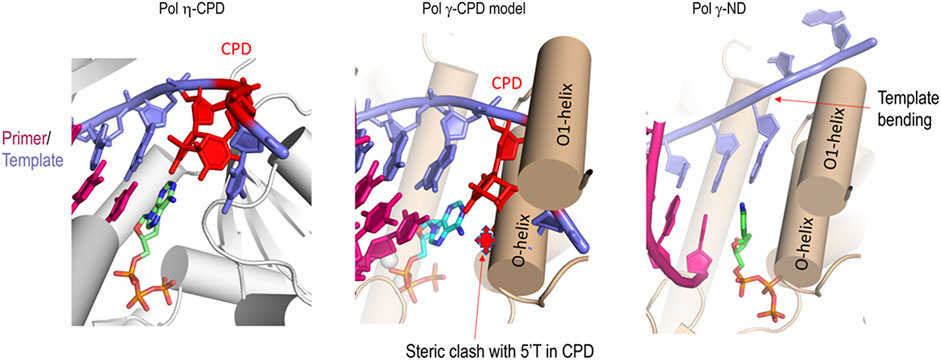
FIGURE 8. Comparison of pol site conformation of Pol η and Pol γ. Left, Pol η ternary complex with CPD (red)-containing template (blue) and primer (magenta) and incoming nucleotide (green) (PDB: 3MR3). Middle, modeled Pol γ-CPD containing primer/template DNA showing the CPD (red) steric clashes with the O helices. Right, Pol γ ternary complex with primer/template and incoming nucleotide showing the bending of the template by O- and O1-helix.
Contrary to the closed pol site in high-fidelity polymerases ternary conformation, the pol active site is more open in Y-family TLS polymerases Pol η. Structure of DNA polymerase η (Pol η) complexed with a CPD-containing template and primer duplex reveals that the bulky CPD can be easily accommodated in the pol site due to the preformed, spacious pol active site and highly mobile little-fingers and thumb domains, shared features in Y-family DNA polymerases (Figure 8, left) (Yang and Woodgate, 2007; Biertümpfel et al., 2010). Unlike replicative DNA polymerases, the fingers subdomain in Y-family DNA polymerases does not go through large conformational changes. Rather, the enzyme is already poised for DNA synthesis even in the absence of incoming nucleotides. Little-finger and thumb domains go through the largest conformational changes and seem to confer specific TLS activities (Boudsocq et al., 2004; Yang and Woodgate, 2007).
To provide a structural interpretation for Pol γ metal-dependent TLS activity, we composited a ternary complex of Pol γ, primer/CPD-template, and an incoming nucleotide by docking CPD-containing substrate from T7 DNA polymerase complex (PDB: 1SL2) onto Pol γ ternary complex (PDB: 4ZTZ) after superposition on the two pol active sites (Figure 8, middle). The resulting complex showed that the CPD would sterically clash with the O1 helix fingers domain severely if no conformational change occurs. For Pol γ to accommodate the bulky CPD, the fingers domain would have to adopt a more open configuration resembling the Pol γ-DNA binary complex. In addition, Mn2+ may also enlarge the pol site in a way that no longer bends the template strand, allowing the bulky thymine dimers to enter effortlessly. We thus hypothesize that Mn2+ may alter Pol γ structure from high-fidelity configuration to that of TLS polymerases. Under this idea, when Pol γ replicates on a non-damaged DNA, the catalytic site is not challenged for which a catalytic site reshaping is not needed (Figure 9A). But on a CPD-containing substrate, due to its volume, Pol γ′s catalytic site is reshaped in the presence of Mn2+ or a mixture with Mg2+ (Figure 9B). More studies should be addressed to better understand this mechanism.
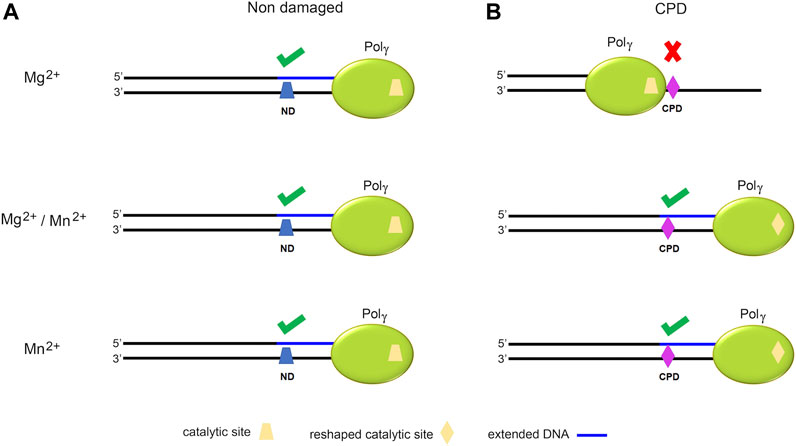
FIGURE 9. Schematic model of the hypothetical function of Mg2+ and Mn2+ in reshaping Pol γ′s active site. Illustration of Pol γ (green) on an ND- (blue trapezoid) or CPD-containing template (violet rhomboid). (A) Representation of DNA replication by Pol γ on an ND-containing substrate in the presence of Mg2+, Mg2+/Mn2+, or Mn2+ showing full extension in all cases without significant changes in the catalytic site (yellow trapezoid). (B) Representation of DNA replication by Pol γ on a CPD-containing template illustrating a change of the Pol γ′s catalytic site (yellow trapezoid) in the presence of Mn2+ upon encountering CPD to trigger TLS activity.
Pol ι, another Y-family DNA polymerase, has better TLS activity across UV lesions in the presence of Mn2+ compared to Mg2+ (Frank and Woodgate, 2007). The mechanism behind Mn2+-mediated TLS in Pol ι is not by large conformational changes of protein. Crystal structures of Pol ι do not display noticeable overall structural changes in the presence of Mn2+ compared to Mg2+ except for small changes near divalent cations in the active site (Choi et al., 2016). Mn2+ supports more optimal octahedral coordination compared to Mg2+, explaining the higher activity and fidelity in presence of Mn2+. In addition, Pol ι does not go through large conformational change upon dNTP binding like replicative DNA polymerases do, which gives molecular explanation behind its ability to tolerate bulkier lesions in its wide and rigid active site (Nair et al., 2006). For replicative polymerases like Pol γ, finger domain movement upon dNTP binding is essential. It is possible that Mn2+ binding induces sufficient structural changes to accommodate CPD.
Mn2+-induced conformational changes are seen in other polymerases. The pol active site of Pol η widens when bound to Mn2+ relative to being bound to Mg2+ and Ca2+ (Yang and Woodgate, 2007; Ling et al., 2003; Rechkoblit et al., 2006; Weng et al., 2018). The proteolysis pattern for a catalytic subunit of herpes simplex virus DNA polymerase (UL30) bound to platinated DNA differs in the presence of Mn2+ than in the presence of Mg2+, implying that UL30 undergoes conformational changes between Mn2+ to Mg2+ bound states (Villani et al., 2002). If Mn2+ actually alters Pol γ pol site and the fingers domain resemble that in Pol η, Pol γ would be able to accommodate the rigid thymine dimer in the enlarged pol site and to incorporate incoming nucleotide opposite to the dimer without stringent geometry check for W-C base pairing complementarity. Crystal structures of RB69 DNA polymerase, replicative B-family DNA polymerase, in the presence of Mg2+ or Mn2+ do not display significant conformational changes (Xia et al., 2011). It is likely the same for Pol γ when it is replicating on T-ND. However, when Pol γ is challenged with bulky lesion like CPD, the Mn2+-mediated flexibility in the pol site may allow enlargement to accommodate CPD (Figure 8). Currently, structural studies revealed only two catalytic metal ions in the pol site; however, EPR study of E. coli Pol I showed at least 25 Mn2+ binding sites were present (Slater et al., 1972). Whether the catalytic metal ions or additional metal ions are involved in reshaping the pol site warrants further investigation.
His-tagged Pol γA and Pol γB subunits were expressed in Sf9 insect cells and E. coli Rosetta (DE3) cells, respectively, and purified as previously described (Lee et al., 2009). Wild-type Pol γA subunit and exonuclease-deficient (exo-) variant containing double mutation D198A/E200A lack N-terminal 25 residues (the putative mitochondrial localization sequence) as well as 10 of the 13 sequential glutamines (residues 43–52). Briefly, both Pol γA wild-type and exonuclease deficient variant were purified sequentially using TALON (Cytiva, Marlborough, MA) and gel filtration column Superdex 200. Pol γB subunit lacking N-terminal 25 residues was purified using Ni-NTA (Qiagen, Germantown, MD) and Mono S affinity chromatography. Purified Pol γA and Pol γB were complexed at 1:2 M ratio, respectively, then applied to a gel filtration column Superdex 200. After gel filtration, pure fractions were pooled and concentrated in a buffer containing 20 mM HEPES (pH 7.5), 140 mM KCl, 5% glycerol, 1 mM EDTA (pH 8,0), and 10 mM BME. Fractions were aliquoted and stored at −80°C.
Wild-type T7 DNA polymerase bound to thioredoxin was purchased from New England Biolab (Ipswich, MA). Concentrations of the proteins were determined on the NanoDrop™ spectrophotometer (Thermo Scientific, Waltham, MA) based on the absorbance at 280 nm. All experiments reported in this study were carried out with the same batch purified proteins. The protein concentrations are determined by A280 absorption and specific extinction coefficients, not active site concentration. The active site concentrations of purified Pol γ are routinely measured to be 80–84%.
All oligonucleotides are synthesized by Integrated DNA Technologies (Morrisville, NC) except for templates containing CPD and (6-4)PP (Table 1). The oligonucleotides containing the CPD and the (6-4)PP were synthesized as described previously (Murata et al., 1990; Iwai et al., 1996). Complementary oligonucleotides were mixed in solution containing 50 mM Tris (pH 7.8), 50 mM NaCl, and 1 mM EDTA (pH 8), then were annealed by heating at 95°C for 5 min followed by slowly cooling to room temperature overnight. For all activity assays, 5′-end of the primer strands were labeled using T4 polynucleotide kinase (New England Biolab, Ipswich, MA) using γ-32P-ATP (PerkinElmer, Waltham, MA).
Three primer/template duplexes were formed by primer PN (27 nt) annealed to a 49 nt non-damaged template (T-ND), a CPD-containing template (T-CPD), or a (6-4)PP-containing template (T-(6–4)PP) (Table 1). For UV product-containing DNA, 200 nM wild-type or exonuclease-deficient Pol γ was pre-incubated with 1000 nM PN/T-CPD or PN/T-(6–4)PP in Buffer N (25 mM HEPES pH 7.5, 140 mM KCl, 1 mM EDTA, 5% glycerol, and 100 μg/ml BSA) at 37°C for 5 min. For T-ND, 100 nM wild-type or exonuclease-deficient Polγ was preincubated with 1000 nM PN/T-ND in Buffer N. The reaction was started by the addition of equal volume of Buffer C (Buffer N supplemented with 400 µM dNTP mix and 40 mM MgCl2 or MnCl2) to a final concentration of 200 µM dNTP mix and 20 mM MgCl2 or MnCl2. Reaction was quenched at the indicated times by adding ninefold excess of Buffer Q (80% Formamide, 50 mM EDTA pH 8, 0.1% SDS, 5% glycerol, and 0.02% bromophenol blue). Quenched samples were heated at 95°C for 5 min and resolved on a 23% polyacrylamide gel containing 7 M urea. Gels were soaked in solution containing 50% methanol and 20% glycerol prior to drying overnight under vacuum at 50°C. Gels were exposed on a phosphor screen, which was imaged using Amersham Typhoon RGB scanner (Cytiva, Marlborough, MA). The intensities of bands were quantified using ImageQuant TL 8.2 (Cytiva, Marlborough, MA). Graphs were plotted using the mean and standard deviation in SigmaPlot 14 (Systat Software, San Jose, CA). To obtain the rate of full-length product formation, we used the following equation:
For polymerase activity assays using wild-type T7 DNA polymerase, a total of 400 nM T7 DNA polymerase was pre-incubated with 200 nM of PN/T-ND or PN/T-CPD in Buffer N at 37°C for 5 min. The reaction was started by addition of equal volume of Buffer C to a final concentration of 200 µM dNTP mix and 20 mM MgCl2 or MnCl2. Quench step, electrophoresis, and autoradiography were carried out identically to polymerase activity assays using PN.
A total of 400 nM Polγ was pre-incubated with 1000 nM single stranded primer PN or duplex PN/T-ND in Buffer N, and the reaction was started by addition of an equal volume of Buffer C without dNTPs to a final concentration of 20 mM MgCl2 or MnCl2. Quench step, electrophoresis, autoradiography, and quantification were carried out identically to polymerase activity assays. The experimental data were fitted to an equation
For polymerase assay using mixed metal ions, 1000 nM wild-type Polγ was pre-incubated with 1000 nM PN/T-ND or PN/T-CPD in Buffer N at 37°C for 5 min. The reaction was started by addition of an equal volume of Buffer C to final concentrations of either constant 1 mM MgCl2 and varied MnCl2 (0.1, 0.2, 0.3 0.4, 0.5, 1, and 10 mM) concentrations, or constant 0.4 mM MnCl2 and varied MgCl2 (0.1, 0.2, 0.4, 0.4, 0.5, 1, and 10 mM) concentrations. Reaction was quenched 5 min after adding ninefold excess of Buffer Q. Electrophoresis, autoradiography, and quantification were carried out identically to polymerase activity assays.
For polymerase assay using single metal ions, 200 nM wild-type Polγ was pre-incubated with 200 nM PN/T-ND or PN/T-CPD in Buffer N at 37°C for 5 min. Reaction was started by addition of an equal volume of Buffer C containing MgCl2 or MnCl2 (0.025, 0.05, 0.1 0.2, 0.4, 0.8, 1.0, and 10 mM). Reaction was quenched 5 min after adding ninefold excess of Buffer Q. Electrophoresis, autoradiography, and quantification were carried out identically to polymerase activity assays.
Experiments with corresponding graphs containing error bars were repeated three times. The rest of the experiments with corresponding graphs not containing error bars were repeated twice.
The data supporting the conclusions of this article will be made available by the corresponding authors, without undue reservation.
JP, NB-T, LB, and YWY designed experiments, JP, NB-T, SI, and GH executed the experiments, JP and NB-T wrote the first draft, and all authors contributed to the final manuscript.
The work is supported in parts by grants from NIH to YWY and SJT (R01AI134611 and R01NS095747), the James W. McLaughlin Fellowship Fund to JP, and an endowment from the Sealy and Smith Foundation to the Sealy Center for Structural Biology and Molecular Biophysics at UTMB.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Satya Prakash and Louise Prakash for helpful discussion.
Bębenek, A., and Ziuzia-Graczyk, I. (2018). Fidelity of DNA Replication-A Matter of Proofreading. Curr. Genet. 64, 985–996. doi:10.1007/s00294-018-0820-1
Beckman, R. A., Mildvan, A. S., and Loeb, L. A. (1985). On the Fidelity of DNA Replication: Manganese Mutagenesis In Vitro. Biochemistry 24, 5810–5817. doi:10.1021/bi00342a019
Bess, A. S., Crocker, T. L., Ryde, I. T., and Meyer, J. N. (2012). Mitochondrial Dynamics and Autophagy Aid in Removal of Persistent Mitochondrial DNA Damage in Caenorhabditis elegans. Nucleic Acids Res. 40, 7916–7931. doi:10.1093/nar/gks532
Bess, A. S., Ryde, I. T., Hinton, D. E., and Meyer, J. N. (2013). UVC-induced Mitochondrial Degradation via Autophagy Correlates with mtDNA Damage Removal in Primary Human Fibroblasts. J. Biochem. Mol. Toxicol. 27, 28–41. doi:10.1002/jbt.21440
Bharti, S. K., Sommers, J. A., Zhou, J., Kaplan, D. L., Spelbrink, J. N., Mergny, J.-L., et al. (2014). DNA Sequences Proximal to Human Mitochondrial DNA Deletion Breakpoints Prevalent in Human Disease Form G-Quadruplexes, a Class of DNA Structures Inefficiently Unwound by the Mitochondrial Replicative Twinkle Helicase. J. Biol. Chem. 289, 29975–29993. doi:10.1074/jbc.m114.567073
Biertümpfel, C., Zhao, Y., Kondo, Y., Ramón-Maiques, S., Gregory, M., Lee, J. Y., et al. (2010). Structure and Mechanism of Human DNA Polymerase η. Nature 465, 1044–1048. doi:10.1038/nature09196
Birch-Machin, M. A., Russell, E. V., and Latimer, J. A. (2013). Mitochondrial DNA Damage as a Biomarker for Ultraviolet Radiation Exposure and Oxidative Stress. Br. J. Dermatol. 169, 9–14. doi:10.1111/bjd.12207
Birch-Machin, M. A., Tindall, M., Turner, R., Haldane, F., and Rees, J. L. (1998). Mitochondrial DNA Deletions in Human Skin Reflect Photo- rather Than Chronologic Aging. J. Invest. Dermatol. 110, 149–152. doi:10.1046/j.1523-1747.1998.00099.x
Boudsocq, F., Kokoska, R. J., Plosky, B. S., Vaisman, A., Ling, H., Kunkel, T. A., et al. (2004). Investigating the Role of the Little finger Domain of Y-Family DNA Polymerases in Low Fidelity Synthesis and Translesion Replication. J. Biol. Chem. 279, 32932–32940. doi:10.1074/jbc.m405249200
Brash, D. E., and Haseltine, W. A. (1982). UV-induced Mutation Hotspots Occur at DNA Damage Hotspots. Nature 298, 189–192. doi:10.1038/298189a0
Chance, B. (1965). The Energy-Linked Reaction of Calcium with Mitochondria. J. Biol. Chem. 240, 2729–2748. doi:10.1016/s0021-9258(18)97387-4
Choi, J.-Y., Patra, A., Yeom, M., Lee, Y.-S., Zhang, Q., Egli, M., et al. (2016). Kinetic and Structural Impact of Metal Ions and Genetic Variations on Human DNA Polymerase ι. J. Biol. Chem. 291, 21063–21073. doi:10.1074/jbc.m116.748285
Corkey, B. E., Duszynski, J., Rich, T. L., Matschinsky, B., and Williamson, J. R. (1986). Regulation of Free and Bound Magnesium in Rat Hepatocytes and Isolated Mitochondria. J. Biol. Chem. 261, 2567–2574. doi:10.1016/s0021-9258(17)35825-8
Durham, S. E., Krishnan, K. J., Betts, J., and Birch-Machin, M. A. (2003). Mitochondrial DNA Damage in Non-melanoma Skin Cancer. Br. J. Cancer 88, 90–95. doi:10.1038/sj.bjc.6600773
Frank, E. G., and Woodgate, R. (2007). Increased Catalytic Activity and Altered Fidelity of Human DNA Polymerase ι in the Presence of Manganese. J. Biol. Chem. 282, 24689–24696. doi:10.1074/jbc.m702159200
Garcia-Diaz, M., and Bebenek, K. (2007). Multiple Functions of DNA Polymerases. Crit. Rev. Plant Sci. 26, 105–122. doi:10.1080/07352680701252817
Gavin, C. E., Gunter, K. K., and Gunter, T. E. (1990). Manganese and Calcium Efflux Kinetics in Brain Mitochondria. Relevance to Manganese Toxicity. Biochem. J. 266, 329–334. doi:10.1042/bj2660329
Gavin, C. E., Gunter, K. K., and Gunter, T. E. (1992). Mn2+ Sequestration by Mitochondria and Inhibition of Oxidative Phosphorylation. Toxicol. Appl. Pharmacol. 115, 1–5. doi:10.1016/0041-008x(92)90360-5
Goodman, M. F., Keener, S., Guidotti, S., and Branscomb, E. W. (1983). On the Enzymatic Basis for Mutagenesis by Manganese. J. Biol. Chem. 258, 3469–3475. doi:10.1016/s0021-9258(18)32685-1
Gunter, T. E., Miller, L. M., Gavin, C. E., Eliseev, R., Salter, J., Buntinas, L., et al. (2004). Determination of the Oxidation States of Manganese in Brain, Liver, and Heart Mitochondria. J. Neurochem. 88, 266–280. doi:10.1046/j.1471-4159.2003.02122.x
Harbottle, A., Maki, J., Reguly, B., Wittock, R., Robinson, K., Parr, R., et al. (2010). Real-time Polymerase Chain Reaction Analysis of a 3895-bp Mitochondrial DNA Deletion in Epithelial Swabs and its Use as a Quantitative Marker for Sunlight Exposure in Human Skin. Br. J. Dermatol. 163, 1291–1295. doi:10.1111/j.1365-2133.2010.10001.x
Iwai, S., Shimizu, M., Kamiya, H., and Ohtsuka, E. (1996). Synthesis of a Phosphoramidite Coupling Unit of the Pyrimidine (6-4) Pyrimidone Photoproduct and its Incorporation into Oligodeoxynucleotides. J. Am. Chem. Soc. 118, 7642–7643. doi:10.1021/ja9603158
Johnson, A. A., and Johnson, K. A. (2001). Exonuclease Proofreading by Human Mitochondrial DNA Polymerase. J. Biol. Chem. 276, 38097–38107. doi:10.1074/jbc.m106046200
Johnson, A. A., Tsai, Y.-c., Graves, S. W., and Johnson, K. A. (2000). Human Mitochondrial DNA Polymerase Holoenzyme: Reconstitution and Characterization. Biochemistry 39, 1702–1708. doi:10.1021/bi992104w
Kasiviswanathan, R., Gustafson, M. A., Copeland, W. C., and Meyer, J. N. (2012). Human Mitochondrial DNA Polymerase γ Exhibits Potential for Bypass and Mutagenesis at UV-Induced Cyclobutane Thymine Dimers. J. Biol. Chem. 287, 9222–9229. doi:10.1074/jbc.m111.306852
Kawasaki, K., Suzuki, T., Ueda, M., Ichihashi, M., Reguer, G., and Yamasaki, H. (2000). CC to TT Mutation in the Mitochondrial DNA of normal Skin: Relationship to Ultraviolet Light Exposure. Mutat. Research/Genetic Toxicol. Environ. Mutagenesis 468, 35–43. doi:10.1016/s1383-5718(00)00038-3
Konji, V., Montag, A., Sandri, G., Nordenbrand, K., and Ernster, L. (1985). Transport of Ca2+ and Mn2+ by Mitochondria from Rat Liver, Heart and Brain. Biochimie 67, 1241–1250. doi:10.1016/s0300-9084(85)80133-4
Kunkel, T. A., and Loeb, L. A. (1979). On the Fidelity of DNA Replication. Effect of Divalent Metal Ion Activators and Deoxyrionucleoside Triphosphate Pools on In Vitro Mutagenesis. J. Biol. Chem. 254, 5718–5725. doi:10.1016/s0021-9258(18)50474-9
Lee, Y.-S., Kennedy, W. D., and Yin, Y. W. (2009). Structural Insight into Processive Human Mitochondrial DNA Synthesis and Disease-Related Polymerase Mutations. Cell 139, 312–324. doi:10.1016/j.cell.2009.07.050
Lee, Y.-S., Lee, S., Demeler, B., Molineux, I. J., Johnson, K. A., and Yin, Y. W. (2010). Each Monomer of the Dimeric Accessory Protein for Human Mitochondrial DNA Polymerase Has a Distinct Role in Conferring Processivity. J. Biol. Chem. 285, 1490–1499. doi:10.1074/jbc.m109.062752
Li, Y., Dutta, S., Doublié, S., Bdour, H. M. d., Taylor, J.-S., and Ellenberger, T. (2004). Nucleotide Insertion Opposite a Cis-Syn Thymine Dimer by a Replicative DNA Polymerase from Bacteriophage T7. Nat. Struct. Mol. Biol. 11, 784–790. doi:10.1038/nsmb792
Liccione, J. J., and Maines, M. D. (1988). Selective Vulnerability of Glutathione Metabolism and Cellular Defense Mechanisms in Rat Striatum to Manganese. J. Pharmacol. Exp. Ther. 247, 156–161.
Ling, H., Boudsocq, F., Plosky, B. S., Woodgate, R., and Yang, W. (2003). Replication of a Cis-Syn Thymine Dimer at Atomic Resolution. Nature 424, 1083–1087. doi:10.1038/nature01919
Longley, M. J., and Mosbaugh, D. W. (1991). Properties of the 3' to 5' Exonuclease Associated with Porcine Liver DNA Polymerase Gamma. Substrate Specificity, Product Analysis, Inhibition, and Kinetics of Terminal Excision. J. Biol. Chem. 266, 24702–24711. doi:10.1016/s0021-9258(18)54287-3
Maher, V. M., Dorney, D. J., Mendrala, A. L., Konze-Thomas, B., and McCormick, J. J. (1979). DNA Excision-Repair Processes in Human Cells Can Eliminate the Cytotoxic and Mutagenic Consequences of Ultraviolet Irradiation. Mutat. Research/Fundamental Mol. Mech. Mutagenesis 62, 311–323. doi:10.1016/0027-5107(79)90087-3
Maher, V. M., Ouellette, L. M., Curren, R. D., and McCormick, J. J. (1976). Frequency of Ultraviolet Light-Induced Mutations Is Higher in Xeroderma Pigmentosum Variant Cells Than in normal Human Cells. Nature 261, 593–595. doi:10.1038/261593a0
Maher, V. M., Rowan, L. A., Silinskas, K. C., Kateley, S. A., and McCormick, J. J. (1982). Frequency of UV-Induced Neoplastic Transformation of Diploid Human Fibroblasts Is Higher in Xeroderma Pigmentosum Cells Than in normal Cells. Proc. Natl. Acad. Sci. 79, 2613–2617. doi:10.1073/pnas.79.8.2613
Makarova, A. V., Boldinova, E. O., Belousova, E. A., and Lavrik, O. I. (2018). In Vitro lesion Bypass by Human PrimPol. DNA Repair 70, 18–24. doi:10.1016/j.dnarep.2018.07.009
Maynard, L. S., and Cotzias, G. C. (1955). The Partition of Manganese Among Organs and Intracellular Organelles of the Rat. J. Biol. Chem. 214, 489–495. doi:10.1016/s0021-9258(18)70986-1
Miller, H., and Grollman, A. P. (1997). Kinetics of DNA Polymerase I (Klenow Fragment Exo-) Activity on Damaged DNA Templates: Effect of Proximal and Distal Template Damage on DNA Synthesis. Biochemistry 36, 15336–15342. doi:10.1021/bi971927n
Moore, P. D., Bose, K. K., Rabkin, S. D., and Strauss, B. S. (1981). Sites of Termination of In Vitro DNA Synthesis on Ultraviolet- and N-Acetylaminofluorene-Treated Phi X174 Templates by Prokaryotic and Eukaryotic DNA Polymerases. Proc. Natl. Acad. Sci. 78, 110–114. doi:10.1073/pnas.78.1.110
Murata, T., Iwai, S., and Ohtsuka, E. (1990). Synthesis and Characterization of a Substrate for T4 Endonuclease V Containing a Phosphorodithioate Linkage at the Thymine Dimer Site. Nucl. Acids Res. 18, 7279–7286. doi:10.1093/nar/18.24.7279
Nair, D. T., Johnson, R. E., Prakash, L., Prakash, S., and Aggarwal, A. K. (2006). An Incoming Nucleotide Imposes an Anti to Syn Conformational Change on the Templating Purine in the Human DNA Polymerase-ι Active Site. Structure 14, 749–755. doi:10.1016/j.str.2006.01.010
Putrament, A., Baranowska, H., Ejchart, A., and Prazmo, W. (1975). Manganese Mutagenesis in Yeast. A Practical Application of Manganese for the Induction of Mitochondrial Antibiotic-Resistant Mutations. J. Gen. Microbiol. 90, 265–270. doi:10.1099/00221287-90-2-265
Rabkin, S. D., Moore, P. D., and Strauss, B. S. (1983). In Vitro bypass of UV-Induced Lesions by Escherichia coli DNA Polymerase I: Specificity of Nucleotide Incorporation. Proc. Natl. Acad. Sci. 80, 1541–1545. doi:10.1073/pnas.80.6.1541
Ray, A. J., Turner, R., Nikaido, O., Rees, J. L., and Birch-Machin, M. A. (2000). The Spectrum of Mitochondrial DNA Deletions Is a Ubiquitous Marker of Ultraviolet Radiation Exposure in Human Skin. J. Invest. Dermatol. 115, 674–679. doi:10.1046/j.1523-1747.2000.00092.x
Rechkoblit, O., Malinina, L., Cheng, Y., Kuryavyi, V., Broyde, S., Geacintov, N. E., et al. (2006). Stepwise Translocation of Dpo4 Polymerase during Error-free Bypass of an oxoG Lesion. Plos Biol. 4, e11. doi:10.1371/journal.pbio.0040011
Romani, A. M. P. (2011). Cellular Magnesium Homeostasis. Arch. Biochem. Biophys. 512, 1–23. doi:10.1016/j.abb.2011.05.010
Sirover, M. A., and Loeb, L. A. (1976). Infidelity of DNA Synthesis In Vitro: Screening for Potential Metal Mutagens or Carcinogens. Science 194, 1434–1436. doi:10.1126/science.1006310
Slater, J. P., Tamir, I., Loeb, L. A., and Mildvan, A. S. (1972). The Mechanism of Escherichia coli Deoxyribonucleic Acid Polymerase I. J. Biol. Chem. 247, 6784–6794. doi:10.1016/s0021-9258(19)44655-3
Smith, C. A., Baeten, J., and Taylor, J.-S. (1998). The Ability of a Variety of Polymerases to Synthesize Past Site-specific Cis-Syn, Trans-syn-II, (6-4), and Dewar Photoproducts of Thymidylyl-(3′→5′)-Thymidine. J. Biol. Chem. 273, 21933–21940. doi:10.1074/jbc.273.34.21933
Sohl, C. D., Szymanski, M. R., Mislak, A. C., Shumate, C. K., Amiralaei, S., Schinazi, R. F., et al. (2015). Probing the Structural and Molecular Basis of Nucleotide Selectivity by Human Mitochondrial DNA Polymerase γ. Proc. Natl. Acad. Sci. USA 112, 8596–8601. doi:10.1073/pnas.1421733112
Spelbrink, J. N., Toivonen, J. M., Hakkaart, G. A. J., Kurkela, J. M., Cooper, H. M., Lehtinen, S. K., et al. (2000). In Vivo functional Analysis of the Human Mitochondrial DNA Polymerase POLG Expressed in Cultured Human Cells. J. Biol. Chem. 275, 24818–24828. doi:10.1074/jbc.m000559200
Steitz, T. A. (1999). DNA Polymerases: DNA Polymerases: Structural Diversity and Common Mechanisms. J. Biol. Chem. 274, 17395–17398. doi:10.1074/jbc.274.25.17395
Steitz, T. A., and Steitz, J. A. (1993). A General Two-Metal-Ion Mechanism for Catalytic RNA. Proc. Natl. Acad. Sci. 90, 6498–6502. doi:10.1073/pnas.90.14.6498
Szymanski, M. R., Kuznetsov, V. B., Shumate, C., Meng, Q., Lee, Y. S., Patel, G., et al. (2015). Structural Basis for Processivity and Antiviral Drug Toxicity in Human Mitochondrial DNA Replicase. EMBO J. 34, 1959–1970. doi:10.15252/embj.201591520
Tokarsky, E. J., Wallenmeyer, P. C., Phi, K. K., and Suo, Z. (2017). Significant Impact of Divalent Metal Ions on the Fidelity, Sugar Selectivity, and Drug Incorporation Efficiency of Human PrimPol. DNA Repair 49, 51–59. doi:10.1016/j.dnarep.2016.11.003
Vaisman, A., Ling, H., Woodgate, R., and Yang, W. (2005). Fidelity of Dpo4: Effect of Metal Ions, Nucleotide Selection and Pyrophosphorolysis. EMBO J. 24, 2957–2967. doi:10.1038/sj.emboj.7600786
Varghese, A. J., and Wang, S. Y. (1967a). cis-syn Thymine Homodimer from Ultraviolet Irradiated Calf Thymus DNA. Nature 213, 909–910. doi:10.1038/213909a0
Varghese, A. J., and Wang, S. Y. (1967b). Ultraviolet Irradiation of DNA In Vitro and In Vivo Produces a Third Thymine-Derived Product. Science 156, 955–957. doi:10.1126/science.156.3777.955
Villani, G., Tanguy Le Gac, N., Wasungu, L., Burnouf, D., Fuchs, R. P., and Boehmer, P. E. (2002). Effect of Manganese on In Vitro Replication of Damaged DNA Catalyzed by the Herpes Simplex Virus Type-1 DNA Polymerase. Nucleic Acids Res. 30, 3323–3332. doi:10.1093/nar/gkf463
Weng, P. J., Gao, Y., Gregory, M. T., Wang, P., Wang, Y., and Yang, W. (2018). Bypassing a 8,5′-Cyclo-2′-Deoxyadenosine Lesion by Human DNA Polymerase η at Atomic Resolution. Proc. Natl. Acad. Sci. USA 115, 10660–10665. doi:10.1073/pnas.1812856115
Xia, S., Wang, M., Blaha, G., Konigsberg, W. H., and Wang, J. (2011). Structural Insights into Complete Metal Ion Coordination from Ternary Complexes of B Family RB69 DNA Polymerase. Biochemistry 50, 9114–9124. doi:10.1021/bi201260h
Yang, J. H., Lee, H. C., Chung, J. G., and Wei, Y. H. (2004). Mitochondrial DNA Mutations in Light-Associated Skin Tumors. Anticancer Res. 24, 1753–1758.
Keywords: mitochondrial DNA, DNA polymerase gamma, UV lesion, metal-dependence, TLS
Citation: Park J, Baruch-Torres N, Iwai S, Herrmann GK, Brieba LG and Yin YW (2022) Human Mitochondrial DNA Polymerase Metal Dependent UV Lesion Bypassing Ability. Front. Mol. Biosci. 9:808036. doi: 10.3389/fmolb.2022.808036
Received: 02 November 2021; Accepted: 11 January 2022;
Published: 09 March 2022.
Edited by:
Gianluca Molla, University of Insubria, ItalyReviewed by:
Sue Cotterill, St George’s University of London, United KingdomCopyright © 2022 Park, Baruch-Torres, Iwai, Herrmann, Brieba and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luis G. Brieba, bHVpcy5icmllYmFAY2ludmVzdGF2Lm14; Y. Whitney Yin, eXd5aW5AdXRtYi5lZHU=
†Present address: Department of Biochemistry and Molecular Biology, University of Texas Medical Branch, Galveston, TX, United States
‡These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.