
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci., 14 March 2022
Sec. Glycoscience
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.799703
This article is part of the Research TopicAdvances in GlycobiotechnologyView all 4 articles
 Ying Sheng1,2†
Ying Sheng1,2† Anita Vinjamuri1†
Anita Vinjamuri1† Michael Russelle S. Alvarez3
Michael Russelle S. Alvarez3 Yixuan Xie1
Yixuan Xie1 Marisa McGrath4
Marisa McGrath4 Siyu Chen1
Siyu Chen1 Mariana Barboza1,5
Mariana Barboza1,5 Matthew Frieman4
Matthew Frieman4 Carlito B. Lebrilla1,2*
Carlito B. Lebrilla1,2*Glycans on the host cell membrane and viral proteins play critical roles in pathogenesis. Highly glycosylated epithelial cells represent the primary boundary separating embedded host tissues from pathogens within the respiratory and intestinal tracts. SARS-CoV-2, the causative agent for the COVID-19 pandemic, reaches into the respiratory tract. We found purified human milk oligosaccharides (HMOs) inhibited the viral binding on cells. Spike (S) protein receptor binding domain (RBD) binding to host cells were partly blocked by co-incubation with exogenous HMOs, most by 2-6-sialyl-lactose (6′SL), supporting the notion that HMOs can function as decoys in defense against SARS-Cov2. To investigate the effect of host cell glycocalyx on viral adherence, we metabolically modified and confirmed with glycomic methods the cell surface glycome to enrich specific N-glycan types including those containing sialic acids, fucose, mannose, and terminal galactose. Additionally, Immunofluorescence studies demonstrated that the S protein preferentially binds to terminal sialic acids with α-(2,6)-linkages. Furthermore, site-specific glycosylation of S protein RBD and its human receptor ACE2 were characterized using LC-MS/MS. We then performed molecular dynamics calculations on the interaction complex to further explore the interactive complex between ACE2 and the S protein. The results showed that hydrogen bonds mediated the interactions between ACE2 glycans and S protein with desialylated glycans forming significantly fewer hydrogen bonds. These results supported a mechanism where the virus binds initially to glycans on host cells preferring α-(2,6)-sialic acids and finds ACE2 and with the proper orientation infects the cell.
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the causative agent of COVID-19 ((Zhu et al., 2020)), encodes an extensively glycosylated spike (S) protein that protrudes from the viral surface and binds angiotensin-converting enzyme 2 (ACE2) on host cells ((Hoffmann et al., 2020; Walls et al., 2020; Wan et al., 2020; Wrapp et al., 2020; Gstöttner et al., 2021)). This novel SARS-CoV-2 was found to share similarities with the SARS-CoV, which was responsible for the SARS pandemic that occurred in 2002 ((Peiris et al., 2003; Viruses, 2020)). ACE2 serves as the entry point for several coronaviruses into cells, including SARS-CoV and SARS-CoV-2 ((Li et al., 2003; Letko et al., 2020)). The receptor binding domain (RBD) of SARS-CoV-2 S protein has been limited to amino acid residues Arg319 to Phe541 ((Lan et al., 2020; Tai et al., 2020; Tian et al., 2020)). In vitro binding measurements also showed that the SARS-CoV-2 RBD binds to ACE2 with an affinity in the low nanomolar range, indicating that the RBD is a key functional component within the S1 subunit responsible for the binding of SARS-CoV-2 to ACE2 ((Tian et al., 2020; Walls et al., 2020)). The plasma membrane protein ACE2 is abundantly expressed in humans tissues, including respiratory and intestinal epithelia, liver arteries, heart and kidney ((Hamming et al., 2004)).
Mammalian epithelial cells are highly glycosylated ((Park et al., 2015; Park et al., 2017)) due to glycoproteins and glycolipids found on the cell membrane. Both the ACE2 receptor and the S protein are similarly extensively glycosylated. Several glycosylation sites are found near the binding interface ((Park et al., 2015; Park et al., 2017), (Shajahan et al., 2020; Watanabe et al., 2020; Shajahan et al., 2021)). The role of glycosylation in the interaction between human ACE2 and SARS-CoV-2 S protein has been extensively studied, primarily using molecular dynamics (MD) simulations ((Shajahan et al., 2020; Watanabe et al., 2020; Shajahan et al., 2021), (Zhao et al., 2020; Mehdipour and Hummer, 2021)). Human ACE2 variants have also been modeled, characterized, and examined for susceptibility to coronavirus interactions ((Chan et al., 2020; Eric et al., 2021)). Among ACE2 glycosylation sites, one of the most characterized position for its role in S protein binding and viral infectivity is the asparagine on position 90 (N90). Recent genetic and biochemical studies showed that mutations that removed glycosylation on N90 site directly increased the susceptibility to SARS-CoV-2 infection ((Chan et al., 2020; Zhao et al., 2020)). In contrast, glycans present on N322 and N90 have the opposite effects on S protein binding. The N322 glycan interacts tightly with the RBD of the ACE2-bound S protein and strengthens the complex ((Mehdipour and Hummer, 2021)). The S protein also contains glycosaminoglycan (GAG) binding motifs so that host surface GAGs contribute to cell entry by SARS-CoV-2 (Kim et al., 2020). Additionally, heparan sulfate has also been shown to promote spike-ACE2 interaction (Clausen et al., 2020).
Pathogen adhesion is often mediated by highly specific lectin-glycan interactions. For example, Escherichia coli with type 1 fimbriae binds to cell surfaces exhibiting preference for high mannose glycans, while Escherichia coli with type S fimbriae has binding specificity for α-(2,3)-linked sialic acids. Cell surface glycans have also been shown to act as a shield to mask its identity as a viable host to the pathogen. It was recently proposed that HMOs can prevent viral adhesion to intestinal epithelial cells via binding to the epithelial surface, causing structural changes in the receptor thereby impeding the virus from hijacking the host cell ((Moore et al., 2021)). Breast-fed infants have significant amounts of HMOs lining the mucosal surface of their gastrointestinal tract. While the viral binding to glycans and HMO in particular have been studied, the direct interaction between the virus and host glycans remain relatively unexplored.
In this study, the role of host glycosylation and its effect on S protein binding was examined by identifying the host glycans that are involved in the binding. The study began with HMOs in a rapid assay to determine the broad details of the oligosaccharide that bind the virus. We then examined the impact of host cell glycosylation on S protein binding, by modifying the host glycosylation while leaving protein expression unchanged using transferase inhibitors. Using newly developed methods glycomic tools, we found that specific glycans on the host cell facilitate S protein binding and that binding depends more on the nature of glycans than it does on the membrane proteins.
HMOs were obtained from breast milk samples using previously reported methods ((Wu et al., 2010; Wu et al., 2011)). Briefly, breast milk samples from seven mothers were pooled. Pooled sample was defatted through centrifugation, proteins were precipitated with ethanol, and the resulting glycans were reduced with sodium borohydride (Sigma-Aldrich, St. Louis, MO, United States). Solid phase extraction was performed on 25 mg graphitized carbon cartridges (ThermoFisher). Solvents were dried in vacuo using miVac (SP Scientific, PA, United States) and purified HMOs were reconstituted and diluted prior to analysis. The individual HMO compounds 2′-fucosyllactose (GKAD-02001; Agilent), 6′- sialyllactose (GKAD-02013; Agilent), and lacto-N-neotetraose (GKAD-02005; Agilent), were tested in their native state.
All HMO screens were performed with Vero E6 cells. Cells were plated in 96 well plates at 5e3 cells/well one day prior to infection. HMOs were diluted from stock to 50 μM and an 8-point 1:2 dilution series was prepared in duplicate in Vero Media. Every compound dilution and control were normalized to contain the same concentration of HMO vehicle (e.g., DMSO). Cell plates were pre-treated with the HMO for 2 h at 37°C (5% CO2) prior to infection with diluted SARS-CoV-2 GFP for a final MOI of 0.1. In addition to plates that were infected, parallel plates were left uninfected to monitor cytotoxicity of HMO alone, measured by CellTiter-Glo (CTG) assays as per the manufacturer’s instructions (Promega, Madison, WI, United States). Plates were then incubated at 37°C (5% CO2) for 48 h, followed by fixation with 4.0% paraformaldehyde, nuclear staining with Hoechst (Invitrogen, Carlsbad, CA, United States), and data acquisition on a Celigo 5-channel Imaging Cytometer (Nexcelom Bioscience, Lawrence, MA, United States). The percent of infected cells was determined for each well based on GFP expression by manual gating using the Celigo software. For the CTG assays, luminescence was read on a BioTek Synergy HTX plate reader (BioTek Instruments Inc., Winooski, VT, United States) using the Gen5 software (v7.07; Biotek Instruments Inc., Winooski, VT, United States).
Human liver hepatocellular carcinoma HepG2, lung carcinoma epithelial Calu-3, urinary bladder epithelial RT4 cells were obtained from American Type Culture Collection (ATCC, VA, United States). HepG2 and Calu-3 cells were grown in Eagle’s Minimum Essential Medium (EMEM). RT4 cells were cultured in McCoy’s 5a Medium. All media were supplemented with 10% (v/v) fetal bovine serum and 100 U ml−1 penicillin and streptomycin. Cells were subcultured at 90% confluency and maintained at 37°C in a humidified incubator with 5% CO2. At 50% cell confluency, the cells were either treated with 150 μM kifunensine, 2-fluoro-L-fucose, or 3-fluorinated sialic acid for 48 h.
Recombinant human angiotensin-converting enzyme 2 (ACE2) (RayBiotech, Georgia, Product Number 230-30165), SARS-CoV-2 spike protein S1 Subunit RBD (Arg319-Phe541) (RayBiotech, Georgia, Product Number 230-30162) and spike protein S1 subunit (Val16-Arg685) (Sino Biological, China, Product Number 40591-V08H) were all derived from transfected human HEK293 cells. The recombinant proteins had C-terminal His-tags but were not conjugated to the fluorophore.
The cells were seeded into FluoroDish™ cell culture dishes (WPI, FL) coated with poly-d-lysine with appropriate density using EMEM cell culture media. At 40% confluency, cells were treated with media either supplemented with 150 μM kifunensine, 2-fluoro-L-fucose, or 3-fluorinated sialic acid for 48 h. Control cell culture without treatment and treated cells were rinsed with phosphate-buffered saline (PBS), and fixed with 4% paraformaldehyde (Affymetrix, OH). Recombinant SARS-CoV-2 spike protein RBD and S1 subunits were conjugated to a fluorescent label with Alexa Fluor™ 555 according to manufacturing instructions (Microscale Protein Labeling Kit, Invitrogen, MA, United States). Fixed control and glyco-modified cells were then incubated with fluorescent labelled S proteins or Anti-ACE2 antibody (Santa Cruz Biotechnology, TX, United States) in PBS at 4°C for 18 h. Cells were stained for the nucleus with 1.6 μM Hoechst 33342 (Thermo Fisher Scientific, MA, United States) followed by the staining for the plasma membrane with 1000-fold diluted CellMask™ Deep Red Plasma Membrane Stain (Thermo Fisher Scientific, MA, United States), respectively at 37°C for 10 min. Fluorescence images were captured using a Leica TCS SP8 STED 3X Super-Resolution Confocal Microscope (Wetzlar, Germany). Fluorescence intensity was quantified for selected cell area. Quantification was performed with software ImageJ.
Cell membrane fractions were prepared as previously described (Park et al., 2015; Li et al., 2019; Li et al., 2020). Briefly, control and glycoengineered cells were collected and resuspended in homogenization buffer containing 0.25 M sucrose, 20 mM HEPES-KOH (pH 7.4), and protease inhibitor mixture (1:100; Calbiochem/EMD Chemicals). Cells were lysed on coolrack (Corning, MA, United States) with five alternating on and off pulses in 5 and 10 s intervals using a probe sonicator (Qsonica, CT, United States). Nuclear and mitochondrial fractions and cellular debris were pelleted by centrifugation at 2000 × g for 10 min. The supernatants were then submitted for ultra-centrifugation at 200,000 × g for 45 min at 4°C to extract the plasma membrane.
Details of the glycomic sample preparation have been described previously ((Wu et al., 2010; Wu et al., 2011)). Extracted cell membrane fractions or protein (RNase B) were suspended with 100 μl of 100 mM NH4HCO3 in 5 mM dithiothreitol and heated in boiling water for 2 min to denature the proteins. Solutions of with 2 μl of peptide N-glycosidase F (New England Biolabs, MA, United States) were added to the samples, and the resulting solutions were then incubated in a microwave reactor (CEM Corporation, NC, United States) at 20 W, 37°C for 10 min. The samples were further placed in a 37°C water bath for 18 h. Ultracentrifugation at 200,000 × g for 45 min was performed to precipitate proteins, and the supernatant containing N-glycans was collected and desalted using porous graphitic carbon (PGC) on a 96-well SPE plate (Grace, IL, United States). The plate was equilibrated with 80% (v/v) acetonitrile containing 0.1% (v/v) trifluoroacetic acid. Then the samples were loaded onto the plate and washed with nanopure water. N-Glycans were eluted with a solution of 40% (v/v) acetonitrile containing 0.05% (v/v) trifluoroacetic acid, and dried in vacuo using miVac (SP Scientific, PA, United States) prior to further analysis.
Details of the protein digestion have been described previously ((Wu et al., 2010; Wu et al., 2011)). Extracted cell membrane proteins were reconstituted in 60 μl of 8 M urea. Recombinant proteins and dissolved cell membrane proteins were reduced with 2 μl of 550 mM dithiothreitol, and then alkylated with 4 μl of 450 mM iodoacetamide. A 420 μl of 50 mM ammonium bicarbonate solution was added to dilute the urea concentration and to adjust the pH value. The samples were incubated with trypsin at 37°C for 18 h. The resulting peptides were concentrated in vacuo using miVac (SP Scientific, PA, United States). Glycopeptides were enriched by solid-phase extraction using iSPE®-HILIC cartridges (HILICON, Sweden). The cartridges were conditioned with 0.1% (v/v) trifluoroacetic acid in acetonitrile, followed by 1% (v/v) trifluoroacetic acid and 80% (v/v) acetonitrile in water. The samples were loaded and washed with 1% (v/v) trifluoroacetic acid and 80% (v/v) acetonitrile in water. The enriched glycopeptides were eluted with water containing 0.1% (v/v) trifluoroacetic acid and dried prior to mass spectrometric analysis.
Details of the glycomic MS analysis have been described previously ((Wu et al., 2010; Wu et al., 2011)). Glycan samples were reconstituted with 30 μl nanopure water and analyzed using an Agilent 6520 Accurate Mass Q-TOF LC/MS equipped with a PGC nano-chip (Agilent Technologies, CA, United States). The glycan separation was performed at a constant flow rate of 300 nl min −1, and a binary gradient was applied using (A) 0.1% (v/v) formic acid in 3% acetonitrile and (B) 1% (v/v) formic acid in 90% acetonitrile: 0–2 min, 0–0% (B); 2–20 min, 0–16% (B); 20–40 min, 16%–72% (B); 40–42 min, 72–100% (B); 42–52 min, 100–100% (B); 52–54 min, 100–0% (B); 54–65 min, 0–0% (B). MS spectra within the mass range of m/z 600–2000 were collected at a rate of 1.5 s per spectrum in positive ionization mode. The most abundant precursor ions in each MS1 spectrum were subjected to fragmentation through collision-induced dissociation (CID) based on the equation V collision = 1.8 × (m/z)/100–2.4 V.
Details of the glycomic data analysis have been described previously ((Wu et al., 2010; Wu et al., 2011)). Extraction of the compound chromatographs of glycans from cells was obtained via the MassHunter Qualitative Analysis B08 software (Agilent, CA, United States). N-Glycan compositions were identified according to accurate masses using an in-house library constructed based on the knowledge of N-glycan biosynthetic pathways and previously obtained in-house structures of N-glycans. Relative abundances were determined by integrating peak areas for observed glycan masses and normalizing to the summed peak areas of all glycans detected.
Details of this analysis have been described previously ((Wu et al., 2010; Wu et al., 2011)). The enriched glycopeptide samples were reconstituted with nanopure water and directly characterized using UltiMate™ WPS-3000RS nanoLC 980 system coupled to the Nanospray Flex ion source of an Orbitrap Fusion Lumos Tribrid Mass Spectrometer system (Thermo Fisher Scientific, MA, United States). The analytes were separated on an Acclaim™ PepMap™ 100 C18 LC Column (3 μm, 0.075 mm × 150 mm, ThermoFisher Scientific). A binary gradient was applied using 0.1% (v/v) formic acid in (A) water and (B) 80% acetonitrile: 0–5 min, 4–4% (B); 5–133 min, 4–32% (B); 133–152 min, 32%–48% (B); 152–155 min, 48–100% (B); 155–170 min, 100–100% (B); 170–171 min, 100–4% (B); 171–180 min, 4–4% (B). The instrument was run in data-dependent mode with 1.8 kV spray voltage, 275°C ion transfer capillary temperature, and the acquisition was performed with the full MS scanned from 700 to 2000 in positive ionization mode. Stepped higher-energy C-trap dissociation (HCD) at 30 ± 10% was applied to obtain tandem MS/MS spectra with m/z values starting from 120.
Glycopeptide fragmentation spectra were annotated using Byonic software (Protein Metrics, CA, United States) against the reviewed UniProt protein database. Common modifications, including including cysteine carbamidomethyl, methionine oxidation, asparagine deamidation and glutamine deamidation were assigned. The glycan database used for the search were previously published ((Wu et al., 2010; Wu et al., 2011)).
The 3D structure of S protein and ACE2 complex was obtained from PDB (PDB code 7DF4) (Xu et al., 2021). The most abundant glycans for each ACE2 glycosite were modeled and attached to the protein using CHARMM-GUI((Park et al., 2019)). Additionally, the fully-desialylated glycans were modeled and attached to generate a fully-desialylated homolog of the ACE2 glycoprotein. The models were solvated using the TIP3P water model, and counterions were added to neutralize the system. The CHARMM carbohydrate force field ((Guvench et al., 2011)) and CHARMM36m force field ((Huang et al., 2017)) were used for the carbohydrate and protein structures. Equilibration was performed at 303.15 K over 10 ps. Molecular dynamics simulation was performed using NAMD software package version 2.13 ((Acun et al., 2018)) at 303.15 K under NPT conditions over 5 ns with an output every 10 ps. Long-range electrostatics were evaluated using the particle-mesh Ewald (PME) method ((Mallajosyula et al., 2015)). Covalent bonds involving hydrogen were constrained with the SHAKE algorithm ((Jo et al., 2017)). After dynamics simulations, trajectories were loaded onto VMD for visualization and analysis ((Humphrey et al., 1996)). Specifically, the intermolecular hydrogen-bonding interactions (donor-acceptor distance 3.0 Å, angle cutoff 20°) of each glycan in the fully-sialylated and desialylated forms were compared over the simulation period.
Human milk oligosaccharides (HMOs) contain a number of unique structures that can be used to rapidly screen the glycan specificity of the virus. We tested whether SARS-CoV-2 virus could be inhibited by HMOs. We first examined whether pooled samples of purified HMOs from seven different mothers could affect the binding of SARS-CoV-2 virus on Vero E6 cells. Figure 1A showed that the binding capability was affected by the HMO mixture to about 25%. HMOs contain compounds with terminal fucose, sialic acid and galactose. To identify the functional components that could specifically affect binding, we further tested native individual compounds that contained these terminal saccharides. The HMOs 2′-fucosyllactose (2′-FL), 6′- sialyllactose (6′-SL), and lacto-N-neotetraose (LNnT) were selected for this study because they represent many of the structures and are abundant in mothers’ milk. 2′-FL and 6′-SL were produced by adding fucose or a N‐acetylneuraminic acid (Neu5Ac) (Castanys-Muñoz et al., 2013) to the lactose core, respectively. Lacto-N-neotetraose (LNnT) is a neutral HMO with a galactose terminus and contained neither fucose nor sialic acid. The infection studies showed that 2′-FL did not diminish infection, while both 6′-SL and LNnT showed some diminished infection with the latter being slightly more effective than the former and to a similar extent as the pooled sample (Figure 1). However, the variations at the different concentrations are large, particularly for LNnT.
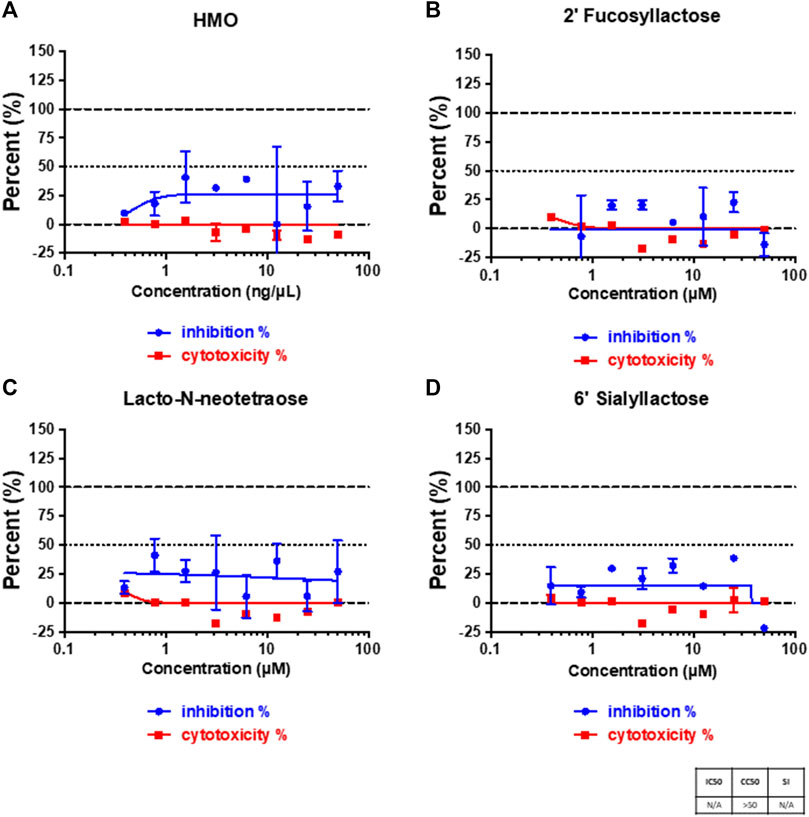
FIGURE 1. Viral infection on cells and cytotoxicity assays. Cell plates were pre-treated with pooled HMOs (A), 2′-FL (B), LNnT (C), and 6′-SL (D), respectively. The treatment was performed for 2 h at 37°C (5% CO2) prior to infection. The percent of infected cells was determined for each well based on GFP expression. All samples were run in triplicate on both an assay plate and a toxicity plate.
Due to limitations with working on the whole intact virus, we moved the research towards using the S protein as a surrogate for the virus. To validate this model, we performed the experiments on the S protein using the fluorescent labeling and immunofluorescence imaging. SARS-CoV-2 enters host cells via the angiotensin-converting enzyme 2 (ACE2) receptor, which binds the receptor binding domain (RBD) of the S protein (12). The Human Protein Atlas (HPA), a website resource for protein expression profiles in cells, tissues and organs (https://www.proteinatlas.org/) (Thul and Lindskog, 2018; Digre and Lindskog, 2021) was used to select the host cell with ACE2 expression. HepG2 was selected after confirming ACE2 expression with labeled antibody and immunofluorescence on the cell membrane (Supplementary Figure S1).
In order to verify further whether HMOs block viral adhesion, we tested the ability of the selected HMO compounds to inhibit RBD binding to HepG2 cells with immunofluorescence. Preincubating HepG2 cells with HMOs did not decrease the binding between the RBD and the cells suggesting that the HMOs did not block binding sites on the host cell surface (Supplementary Figure S2). We then tested whether the HMOs could block or alter the RBD of the virus by preincubating the RBD and the HMOs before introduction to HepG2 cells. Fluorescently labelled RBD was preincubated with 2′-FL, LNnT and 6′-SL separately then allowed to interact with host cells (Figure 2A). Quantitation of fluorescent signal intensity showed that HMOs blocked binding of RBD to cells presumably reflecting the behavior of the intact virus. The RBD was blocked only slightly by 2′FL (not statistically significant), more by LNnT (significant), and the most by 6′SL (Figure 2B). Comparison of LNnT and 6′SL showed that latter one is more effective (significant). The data further showed that HMOs can potentially function as decoys to affect SARS-CoV-2 adherence.
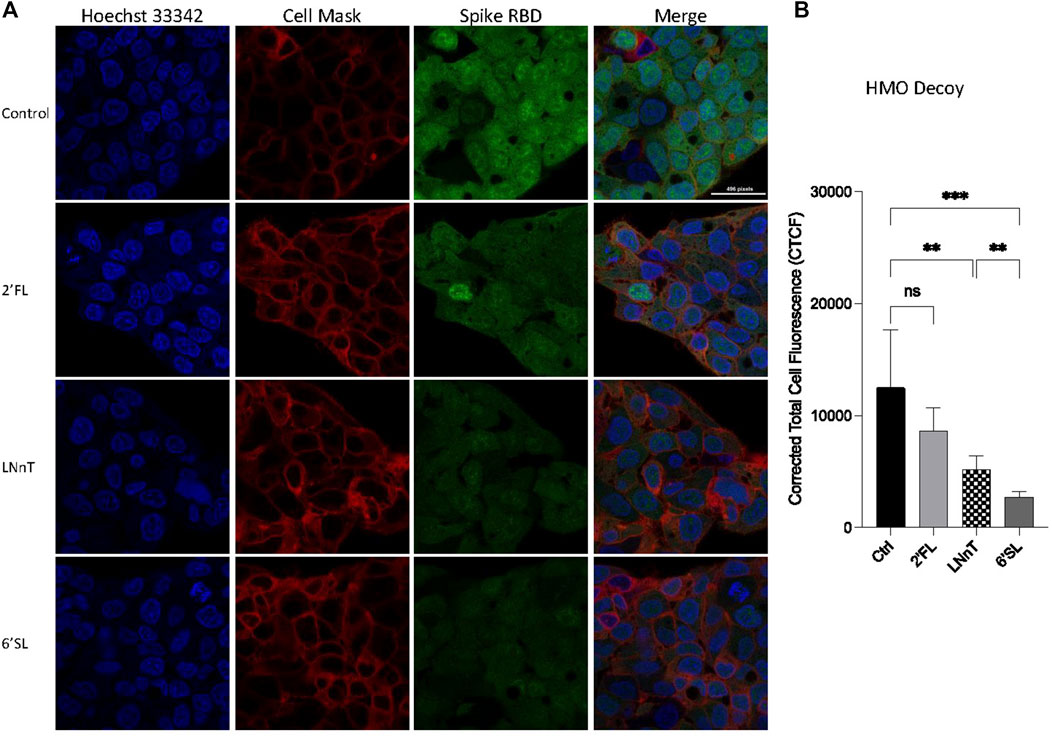
FIGURE 2. Inhibition of HMOs on the binding between HepG2 cells and Spike protein RBD. Fluorescent labelled proteins were preincubated with 1 mg per ml 2′-FL, 6′-SL, and LNnT, respectively. The preincubation was performed at room temperature for 30 min. (A) Immunofluorescence for S protein RBD binding. The columns (from left to right) show staining of nuclear acid (Hoechst 33342), plasma membrane (CellMask™ Deep Red), S protein RBD, and merged image. Scale bar, 496 pixels. (B) Quantification of fluorescent intensity of spike protein RBD binding. Fluorescence intensity was quantified for selected cell area. Quantification was performed with software ImageJ. Asterisks indicate the statistical significance between groups compared (**p< 0.01%; ***p< 0.001%; ns p < 0.05).
The notable decrease in binding caused by 6′-SL drew our attention to sialic acids as potential receptors on the cell surface. To further investigate the effects of cell surface glycans on RBD binding, we altered the cell membrane glycans through transferase inhibitors. We first characterized the glycan of the cell membrane and ACE2 on the native cell line. For this analysis, complex and hybrid type glycans were combined to distinguish them from oligomannose type. The N-glycan profile shows a notable abundance of sialylated and sialyfucosylated structures (Figure 3A). The most abundant N-glycan compositions had multiple fucose and sialic acid (N-acetylneuraminic acid or Neu5Ac) residues such as Hex6HexNAc5Fuc2NeuAc3, Hex6HexNAc5Fuc1NeuAc3 and Hex5HexNAc4Fuc1NeuAc2. Glycoproteomic analysis of the cell membrane revealed seven glycosites on the ACE2 protein of HepG2 cells. The N-glycoforms of the ACE2 protein extracted from HepG2 cells were diverse and the most common structures were both fucosylated and sialylated (Figure 3B and Supplementary Table S1). For comparison, we analyzed the glycosylation of commercial recombinant ACE2 protein expressed from HEK293 (Supplementary Figure S2 and Supplementary Table S2) and found them to be similar to those expressed by HepG2 (Table 1). Both proteins were highly sialylated and fucosylated with limited amounts of high-mannose glycans.
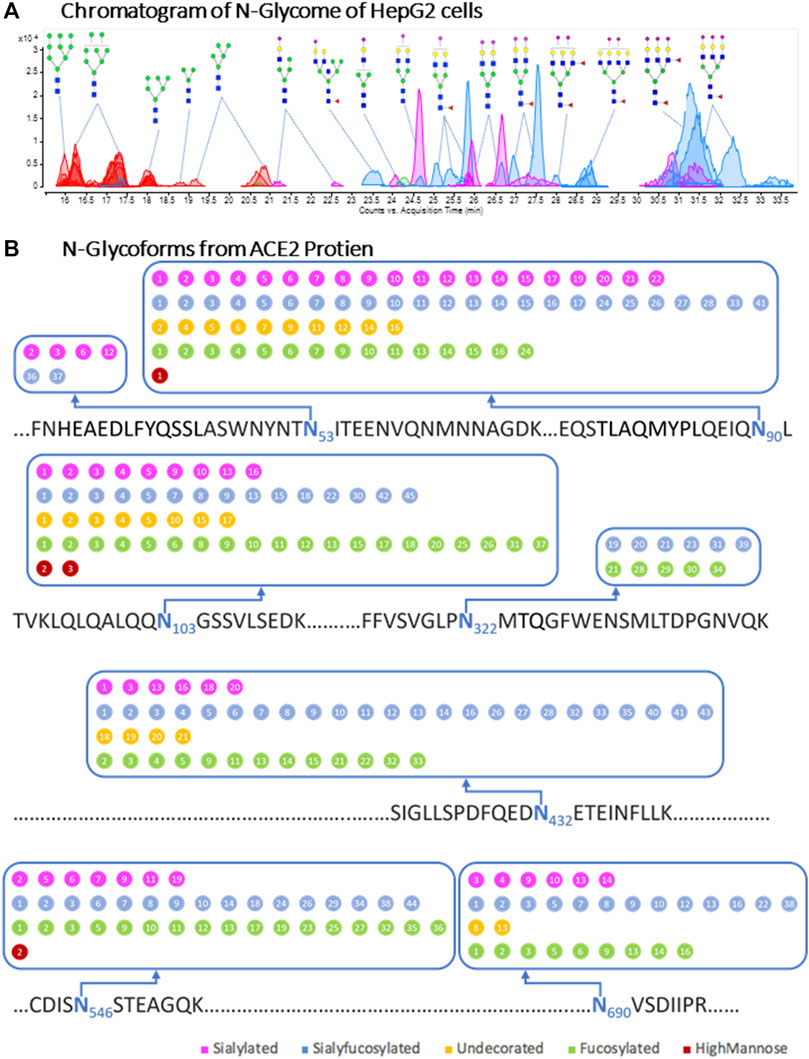
FIGURE 3. Cell membrane N-glycome and site-specific occupancy of ACE2 receptor in HepG2 cells. (A) Individual N-glycan species of HepG2 host cells. LC-MS peaks were color coded to assign glycan subtype. Abundant peaks are annotated with putative structures. Symbol nomenclature is used for representing glycan structures (https://www.ncbi.nlm.nih.gov/glycans/snfg.html). (B) Site-specific occupancy of ACE2 receptor in HepG2 cells. The N-glycoforms from ACE2 protien extracted from HepG2 cells are distributed on seven glycosites. The labeled numbers inside dots denote identified individual glycan and the details were shown in Supplementary Table S2.
In order to explore the effects of glycocalyx on RBD binding, we metabolically altered the cell surface glycome by treating the cells with inhibitors. To diminish fucosylation on the HepG2 cell surface, we employed a fucosyltransferase inhibitor, 2-fluoro-L-fucose (2F-Fucose). To inhibit sialylation, a sialyltransferase inhibitor 3-fluorinated sialic acid (3-F-Sia) was used. To enrich high-mannose glycans, kifunensine (Kif) was applied to prevent mannose trimming. To determine whether these changes in glycosylation affected ACE2 expression on the cell membrane, we probed the cells with fluorescently labeled antibodies (Supplementary Figure S4). These experiments showed no significant changes in protein expression for ACE2 in any of the glycan modification procedures.
The predicted behavior of each substrate are shown in Figure 4A. Compositional profiles were generated for the modified cells, using the sum of the intensities for similar glycan types from the LC-MS analysis. These inhibitors have recently been applied for altering cell surface glycosylation to yield similar results (Zhou et al., 2021) (Figure 4B). 2F-Fucose inhibits fucosylation by being converted to the sugar nucleotide GDP-2F-Fuc ((Villalobos et al., 2015)). It then accumulates in the cell and binds to the transferase and prohibits the enzyme from adding fucose to the nascent chain, thereby decreasing fucose expression on the cell surface (Zhou et al., 2017). The sialyfucosylated N-glycans decreased from 75 to 10% after inhibition with 2F-Fucose treatment. The sialyfucosylated N-glycans were converted to sialylated (only) ones. For example, the abundant sialyfucosylated compound Hex5HexNAc4Fuc1NeuAc decreased (9.6–1.9%, relative abundance) relative to the unfucosylated species Hex5HexNAc4Fuc0NeuAc2 which increased in abundance from 3.7 to 19% (Supplementary Table S3). The sialylation pathway was inhibited using 3-F-Sia, a fluorinated sialic acid substrate [cytidine monophosphate (CMP)–SiaFAc]((Suzuki et al., 2015)), which binds more strongly to the enzyme thereby prohibiting the transfer of sialic acids. Treatment with 3-F-Sia decreased the relative abundances of all sialyfucosylated N-glycans from 75 to 34%. Simultaneously, the relative abundance of fucosylated (only) species increased from 1 to 27%. Thus, it appears that the inhibitors are highly effective diminishing fucosylated and sialylated structures, respectively.
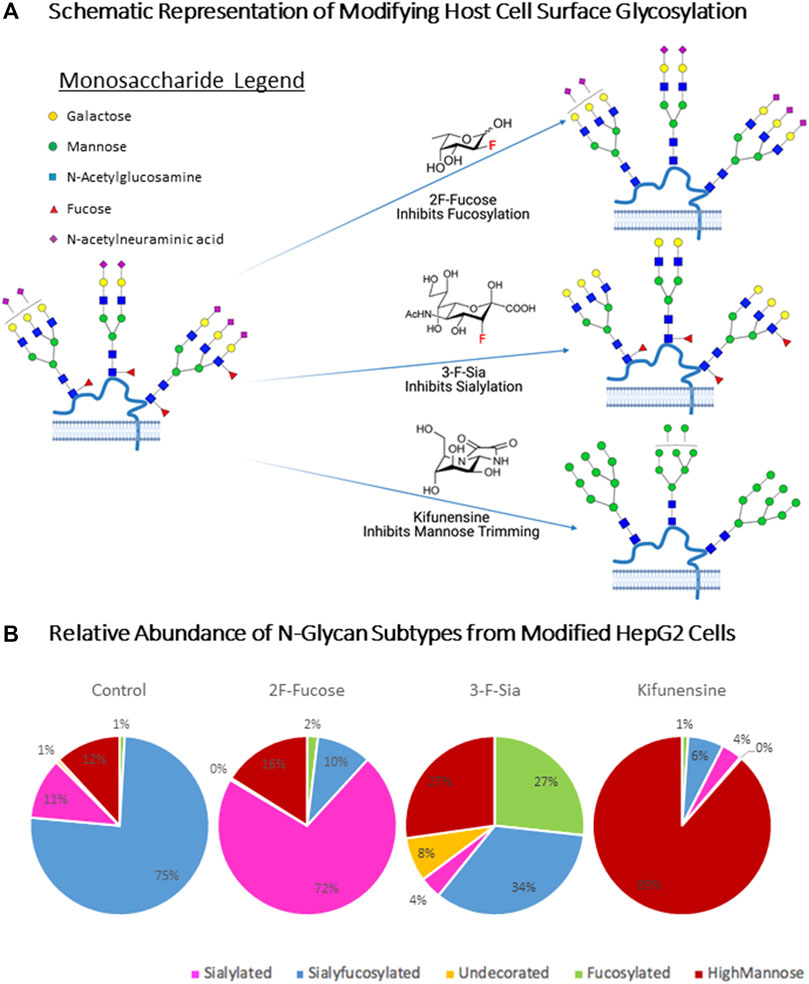
FIGURE 4. Host Cell Surface Glycome Modification. 2F-Fucose (Fucosyltransferase Inhibitor); 3-F-Sia (Sialyltransferase Inhibitor). (A) Metabolic engineering stargey for altering host cell glycosulation. Symbol nomenclature is used for representing glycan structures (https://www.ncbi.nlm.nih.gov/glycans/snfg.html). (B) N-Glycome Profiles of unmodified and modified HepG2 cells from LC-MS analysis. Compound list and details are shown in Supplementary Table S3. Pie charts were color coded to assign glycan subtype. Numbers inside pie charts denote the relative abundance of each identified glycan subtype.
After confirming that glycan alteration had taken place in host cells, immunofluorescence analysis was used to observe the effect of host glycome alterations on viral binding. Treatment of 2F-Fucose did not affect RBD binding to the HepG2 cell significantly as observed by immunofluorescence imaging (Figure 5A). However, inhibition of sialylation by 3-F-Sia decreased the S protein RBD binding with HepG2 cells by 64% (Figure 5B), indicating that the binding was likely mediated by sialic acid residues on the host cell surface. Similar trends were observed in other cell lines with ACE2 expression, namely Calu3 and RT4 (Figure 5B). Desialylation inhibited the binding from S protein RBD significantly, and decreased fucosylation did not change the extent of the binding.
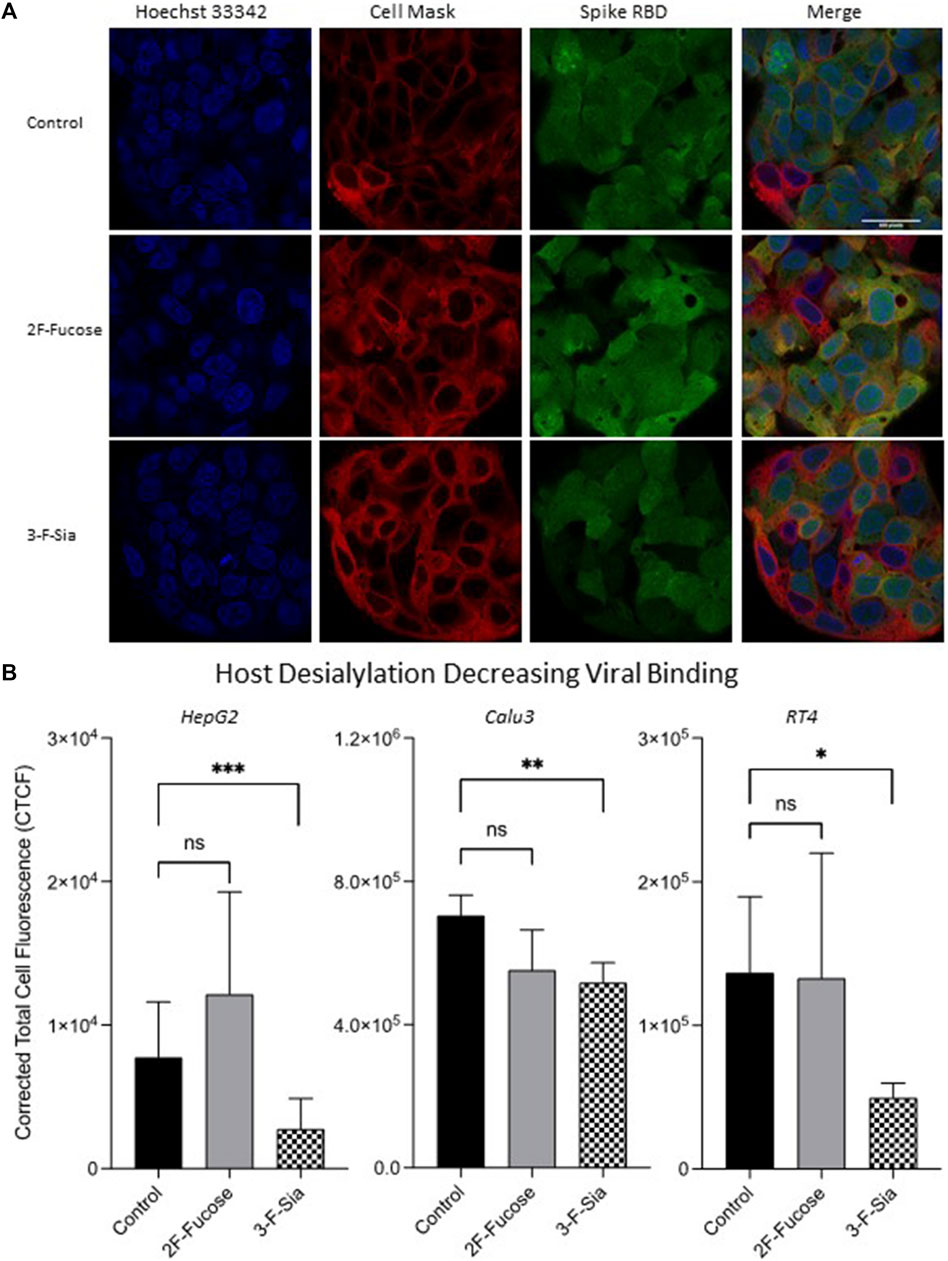
FIGURE 5. Remodeling host glycome alters binding between host cells and spike protein RBD. 2F-Fucose (Fucosyltransferase Inhibitor); 3-F-Sia (Sialyltransferase Inhibitor). (A) Immunofluorescence for S protein RBD binding with modified HepG2 cells. The columns (from left to right) show staining of nuclear acid (Hoechst 33342), plasma membrane (CellMask™ Deep Red), S protein RBD, and merged image. Scale bar, 600 pixels. (B) Quantification of fluorescent intensity of spike protein RBD binding. Fluorescence intensity was quantified for selected cell area. Quantification was performed with software ImageJ. Asterisks indicate the statistical significance between groups compared (*p< 0.05%; **p< 0.01%; ***p< 0.001%; ns p < 0.05).
In mammalian cells, terminal sialic acids are commonly found in α-glycosidic linkage to the C-3 or C-6 hydroxyl of galactose via α-(2,3)- or α-(2,6)-linkage for N-glycans (Varki A et al., 2015). In nasal mucosa, α-(2-6)-sialic acids are dominant ((Shinya et al., 2006)) with significantly less detected in the lung ((Nicholls et al., 2007)). We further investigated linkage specificities for RBD binding by preincubating the RBD with sialylated HMOs. A significant decrease of RBD intensity was observed after preincubation with 6′‐SL (Figure 6), confirming S protein RBD binds with sialic acids. Comparsion of α-(2-3) with α-(2-6) showed a slight preference against α-(2-6), however the difference was not statistically significant.
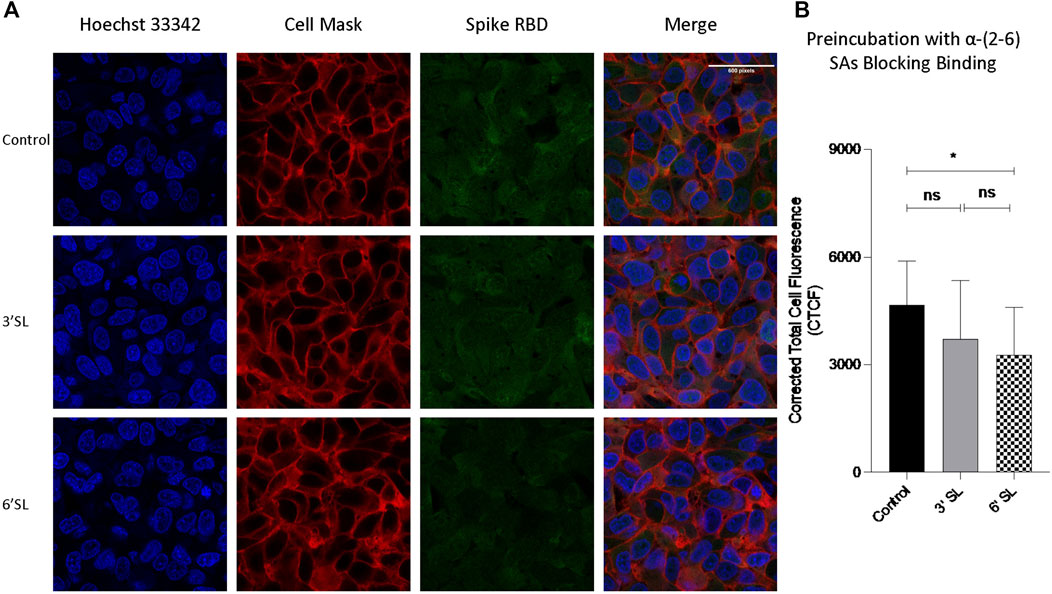
FIGURE 6. Sialylated HMOs diminished binding of spike protein RBD to HepG2 Cells. Fluorescent labelled proteins were preincubated with 1 mg per ml 3′-FL (3′-sialyllactose) and 6′-SL (6′-sialyllactose) respectively at room temperature. (A) Immunofluorescence for S protein RBD binding with cells. The columns (from left to right) show staining of nuclear acid (Hoechst 33342), plasma membrane (CellMask™ Deep Red), S protein RBD, and merged image. Scale bar, 600 pixels. (B) Quantification of fluorescent intensity of spike protein RBD binding. Fluorescence intensity was quantified for selected cell area. Quantification was performed with software ImageJ. Asterisks indicate the statistical significance between groups compared (*p< 0.05%; ns p < 0.05).
Fucosylated glycans were also observed on ACE2 proteins in HepG2 cells (Supplementary Table S1). Terminal α-(1,2) and α-(1,3)-fucose residues are commonly found in mammalian cells ((Ma et al., 2006; Schneider et al., 2017)). To confirm that fucosylation is less important, 2′-FL and 3′-fucosyllactose (3′-FL), components of HMOs, were used ((Ma et al., 2006; Schneider et al., 2017)). Preincubating the S protein RBD with 2′-FL or 3′-FL did not significantly alter binding (Supplementary Figure S5). The S protein RBD again showed little affinity to terminal fucose residues on host cells.
Glycosylation of HepG2 included primarily complex and hybrid type structures with fewer high mannose structures. The latter have been reported as important mediators in host-virus binding for human coronaviruses HKU1((Ma et al., 2006; Schneider et al., 2017)) and severe acute respiratory syndrome (SARS) (Han et al., 2007). We remodeled the cell surface to produce primarily oligomannose and determined its effects on SARS-CoV-2 binding. Kifunensine (Kif) is commonly used to inhibit the α-mannosidase-I ((Kommineni et al., 2019)), thereby preventing mannose trimming to increase oligomannose-type glycans ((Han et al., 2007; Choi et al., 2018)). Our LC/MS data also proved its increasing the relative abundance of oligomannose to 89% in whole cell N-glycome as shown in Figure 4. Introduction of Kif to the cell resulted in a fourfold increase in the binding as measured by immunofluorescence imaging (Figures 7A,B). N-Glycans, released from RNase B, was also employed to examine high mannose type binding. Preincubation with the oligomannose decreased the binding of S protein RBD with host cells (Figure 7C). This effect was dose dependent with higher concentrations preventing binding more strongly. High mannose glycans on host cell surface can therefore increase the adherence of S protein RBD.
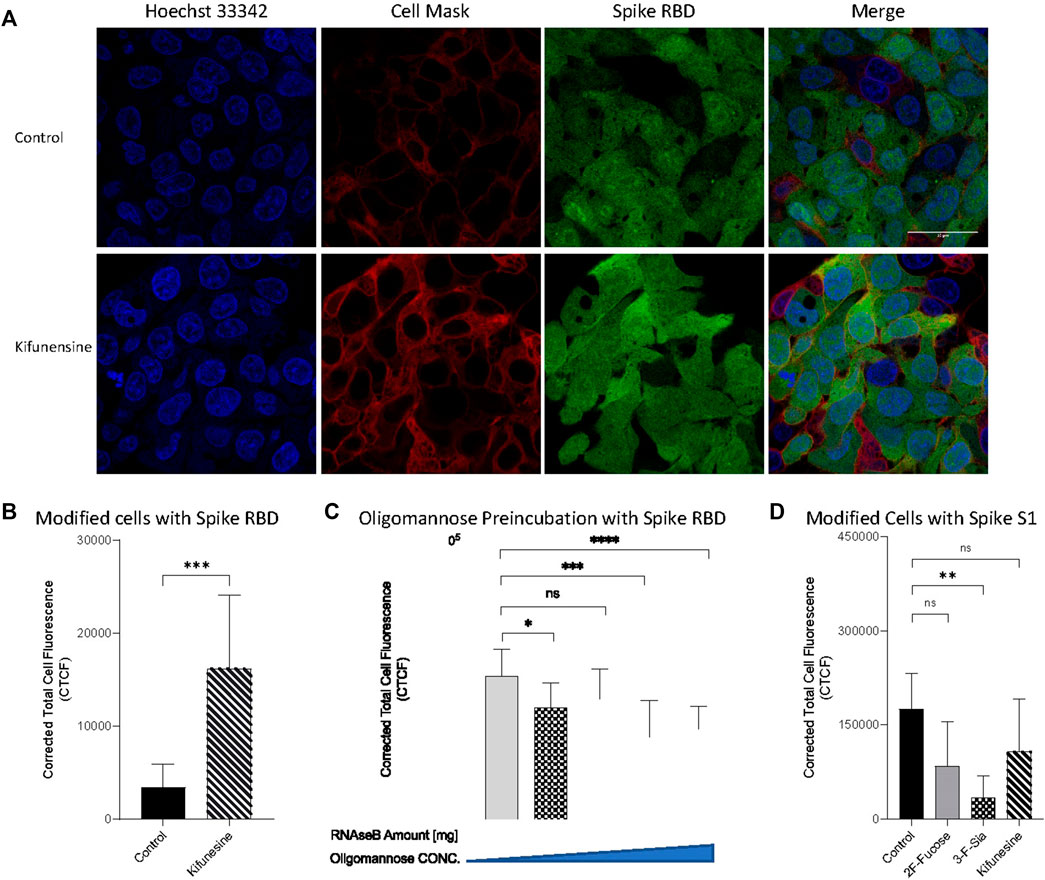
FIGURE 7. Introducing high mannose glycans into viral binding. (A) Immunofluorescence for S protein RBD binding with modified cells. The columns (from left to right) show staining of nuclear acid (Hoechst 33342), plasma membrane (CellMask™ Deep Red), S protein RBD, and merged image. Scale bar, 600 pixels. Quantification of fluorescent intensity of spike protein RBD (B,C) or S1 subunit (D) binding. (C) Fluorescent labelled proteins were preincubated with purified high mannose at room temperature for 30 min before binding. Fluorescence intensity was quantified for selected cell area. Quantification was performed with software ImageJ. Asterisks indicate the statistical significance between groups compared (*p< 0.05%; **p< 0.01%; ***p< 0.001%; ****p< 0.0001%; ns p < 0.05).
To further validate the binding of the spike protein with the host glycocalyx, we used spike protein S1 subunit, a longer polypeptide segment of the S protein and includes the sequence of RBD. Treatment of the cell line with 2F-Fucose did not change the binding between S1 subunit and host cells (Figure 7D). Similarly, the use of 3-F-Sia significant decreased the fluorescent intensity of the assay demonstrating again that the spike protein binds to sialic acids. Surprisingly, the use of Kif in the cell culture no longer increased binding with the S1 subunit. The binding studies showed that there was no significant change in binding relative to the control.
To gain further insight into the interactions between the primary receptor ACE2 (Lan et al., 2020; Nguyen et al., 2021; Wong et al., 2004) and the SARS-CoV-2 S protein, we performed molecular dynamics calculations on the interacting complex. Based on the glycoproteomic results for ACE2 from the HepG2 cell line (Supplementary Table S1), we constructed a model with selected glycoforms on ACE2. ACE2 contained seven occupied N-glycan sites corresponding to Asn 53, 90, 103, 322, 432, 546, and 690 (Figure 8A). From the quantitative glycoproteomic results and the Protein Data Bank-derived complex (PDB ID: 7DF4) (Xu et al., 2021), the most abundant glycan at each site were modelled with CHARMM-GUI(32). The resulting structure, shown in the “up” conformation, was selected because it represented the activated complex prior to invasion. Molecular dynamics simulations were performed on the complex with solvent and associated ions for 5 ns (See Methods and Materials Section). After the simulations, the number of hydrogen bonds formed between the ACE2 glycans and S protein determined. For comparison, the same calculations were performed on the fully desialylated ACE2 homologs (Figure 8B). The results showed that many of the glycans on ACE2 interacted with the S protein through hydrogen bonding interactions. Comparison of the fully sialylated and desialylated glycans showed significantly lower number of hydrogen bonds (based on 3 Å, donor-acceptor distances) particularly on Asn 90 (22 hydrogen bonds by glycan) and Asn 322 (51 hydrogen bonds by glycan) of the desialylated homolog (Figure 9A). These results are consistent with earlier simulations performed by Zhao et al. on ACE2 - S who noted that both glycan sites were also the most interactive in the complex ((Han et al., 2007; Choi et al., 2018)). Furthermore, when the sialic acids were considered relative to other monosaccharide residues (3 by sialic acid at Asn 90, 15 by sialic acid at Asn 322), their contributions to the overall interactions were proportionally larger (Figure 9B).
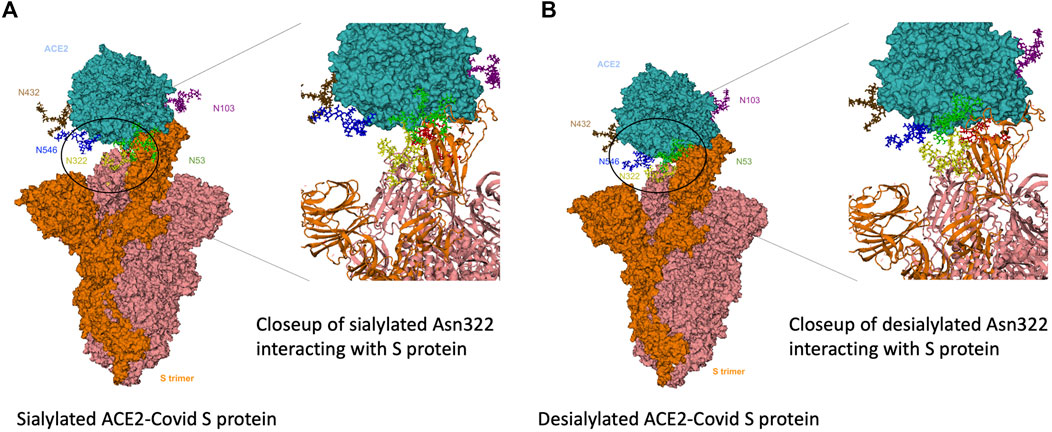
FIGURE 8. Modelled sialylated and desialylated ACE2-Covid S protein complexes. 3D structural modeling of glycosylated ACE2 interacting with S-protein. Results from glycomics and glycoproteomics of HEPG2 cell lines were used to generate (A) fully-desialylated and (B) fully-sialylated homologs of ACE2, interacting with S-protein.
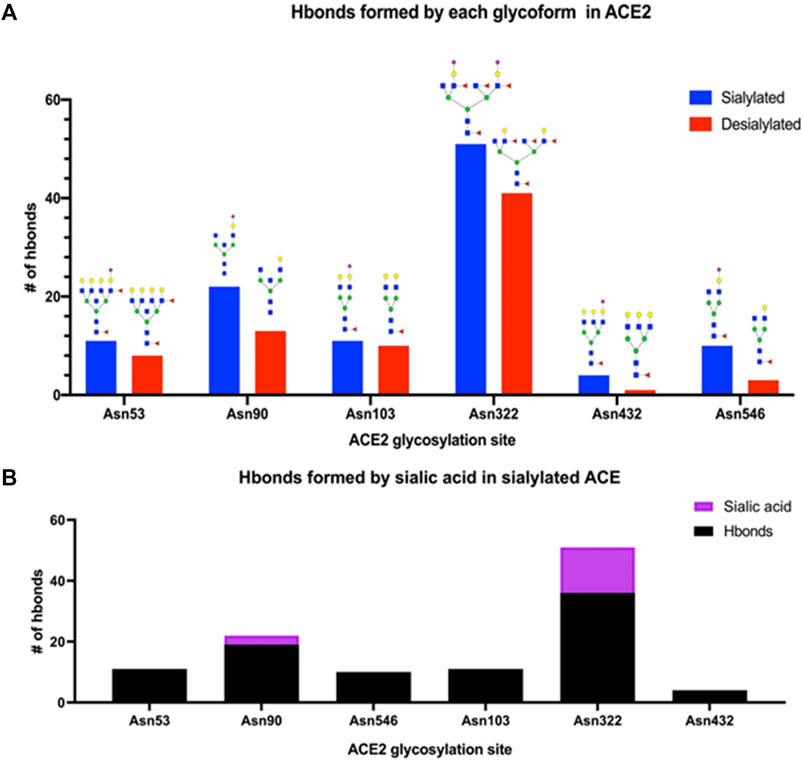
FIGURE 9. Interactions of glycosylated ACE2 and S-protein were revealed using molecular dynamics simulations. (A) The number of intramolecular hydrogen bonds was drastically higher for each fully-sialylated N-glycan compared to the desialylated glycoform. (B) For Asn 90 and Asn 322, the sialic acid residue in the glycoform accounted for ∼10% of hydrogen bonds.
Glycans on the host cell membrane and on viral proteins play key roles in the infection of SARS-CoV-2. Viral glycosylation has been the primary focus of glycomic studies related to the virus. Indeed, the spike protein is highly glycosylated with at least 17 N-glycosylation and two O-glycosylation sites identified ((Shajahan et al., 2020; Sanda et al., 2021)). We found two occupied N-glycosites on spike RBD (Supplementary Table S4) consistent with earlier findings. However, the host cells were also highly glycosylated. The LC-MS glycomic profile of HepG2 shows cell membrane with an abundance of high mannose-type glycans as well as complex-type structures with a high degree of sialylation. These structures are also branched with a combination of bi, tri, and higher antennary structures. The HepG2 cell lines was selected for its expression of ACE2, and these highly sialylated branched structures were similarly present in the protein further alluding to the importance of sialylation in at least the host-virus adhesion process.
The results showed that sialic acid in human milk oligosaccharides (HMOs) can block the binding of virus on the cell membrane. These results are further supported by recent findings that show similar deflecting properties of sialylated HMOs toward the S protein of SARS-CoV-2 ((Han et al., 2007; Choi et al., 2018)) and illustrating further the protective nature of human milk against these pathogens. HMOs are more similar to O-glycans in structure, however N-glycans on membrane proteins similarly provide sialic acid on their termini. Altering the glycans on the cell membrane, while maintaining the expression levels of proteins such as ACE2, further shows that sialic acid on the cell surface induces stronger binding to the virus. ACE2 is itself highly sialylated, in the cell line used in this study and from commercial sources (mainly from HEK293). ACE2 expressed recombinantly in other cell lines have similar glycosylation profiles that are similarly rich in sialylation. Deeper structural analysis showed that the binding prefers a specific linkage, namely α-(2,6)-sialic acids. Interestingly, the human influenza virus has a similar preference for binding ((Trebbien et al., 2011; Wu et al., 2015; Sieben et al., 2020)). Perhaps not coincidently, human epithelial in the nasal mucosa is rich in α-(2,6)-sialic acid, which is also more abundant than the isomer α-(2,3)-sialic acid, the binding site of avian bird flu ((Shajahan et al., 2020; Sanda et al., 2021)).
The binding of sialic acid point to specific protective measures by the host. In breast fed infants, HMOs can provide some protection. Human milk is also full of proteins that are highly sialylated such as the immunoglobulins and lactoferrins ((Almond et al., 2013; Smilowitz et al., 2013; Jorgensen et al., 2017; Morniroli et al., 2021)). In adults, pathogen deflection is performed by the mucus layer. SARS-CoV-2 is a respiratory disease reaching deep into the respiratory tract and the lungs. It also infects the intestine (Wang et al., 2020), with both types of tissues protected by a mucus layer constructed around high molecular weight glycoproteins called mucins ((Hansson, 2019)). Mucin are expressed in epithelial surfaces of gastrointestinal, genitourinary, and respiratory tracts, where they also shield the surface against chemical and physical damages ((Trebbien et al., 2011; Wu et al., 2015; Sieben et al., 2020)). While mucins are covered primarily by O-glycans that are similar to human milk oligosaccharides, they contain the same sialic acid termini as N-glycans. The mucus layer therefore presents a myriad of potential binding sites for commensal and pathogenic microbes ((Tran and Ten Hagen, 2013; Tailford et al., 2015)), and shedding mucins is a defense strategy against pathogen infection.
The high mannose glycans were also strongly bound in the shorter version (RBD) of the S protein. However, in the longer homolog (S1 subunit) this binding was diminished. These results suggest that that there is a high mannose binding site on the S protein that is potentially shielded in the longer homolog. On the other hand, high mannose glycans are typically not found on epithelial cells ((Tran and Ten Hagen, 2013; Tailford et al., 2015)) and are not abundant in the blood. However, they are much more abundant in the tissue samples compared to serum. These glycans are found in cancer cells (Radcliffe et al., 2007; Möginger et al., 2018) and stem cells ((An et al., 2012)). The levels of several oligomannose type glycans are upregulated in tumor tissue ((Balog et al., 2012; Ruhaak et al., 2015)). The role of mannose residue as a host receptor has been studied and proved in the microbe-host interactions, such as Salmonella enterica subsp. enterica serovar typhimurium (S. typhimurium) (Park et al., 2016), influenza virus ((Reading et al., 2000; Upham et al., 2010)), dengue virus ((Miller et al., 2008)) and human immunodeficiency virus (HIV) (Nguyen and Hildreth, 2003). Mannan is usually employed for studying mannose binding with virus ((Han et al., 2007; Upham et al., 2010; Hulswit et al., 2019; Routhu et al., 2019)). The mannans are highly heterogeneous in length and branching. The repeating α-(1,6)-linked mannose backbone is usually branched by short chains of α-(1-2) and α-(1-3)-linked mannose structures ((Varki et al., 2015)). In this study, we used oligomannose released form RNase B ((Roberts et al., 2000)) instead of mannan. The released high mannose glycans were determined with mass spectrometry (Supplementary Figure S6), and all those structures have been found in human cell glycomes.
The integrated method developed here, which includes alteration of cell surface glycan products through specific inhibitors, coupled with the enrichment of the membrane proteins and extensive glycomic and glycoproteomic analysis provides a new platform for obtaining structural specificity in host-microbe interactions. Glycans are common targets for many commensals and pathogens alike. This method will have great utility in identifying glycan targets of individual microbes and even toxins that bind glycans. The method is made possible by recent advancements in novel glycosyl transferase inhibitors that produce specifically glycosylated membrane proteins. We noted that the conversion to a glycan type is never fully complete. There are residual endogenous glycans due to the differences in turnover of different glycoproteoform ((Wong et al., 2020)). However, the ability to perform glycomic profiling with LC-MS provides a guiding assay to examine the extent of the glycomic transformation.
The study supports a mechanism for binding of SARS-CoV-2 to the cell membrane that is primarily mediated by glycans. The preferred target of the S protein is sialylated glycans with α-(2,6)-sialic acids on the termini positions. The virus likely binds to cells and tissues rich in sialylated glycans, whether N-, O-, and potentially even glycolipids that are found in the surface of the epithelial surface. The airway epithelial is rich in sialic acids and in particularly α-(2,6)-sialic acids. In this regard, the human influenza virus and SARS-CoV-2 have the same binding preference in the host membranes. Invasion of SARS-CoV-2 likely occurs when the virus fortuitously binds to the ACE2 protein, which itself is highly sialylated. The alignment between the S and the ACE2 protein is further facilitated by hydrogen binding interactions between the sialylated glycans of the host cell and the polypeptide of the S protein.
The data presented in the study are deposited in the MassIVE data repository, accession number MSV000088722.
YS, AV, YX, MB, and CL designed the experiments. AV and MA performed the HMO enrichment and the LC-MS analysis. MM and MF performed HMO inhibition assays against SARS-CoV-2. SC purified the oligomannose compounds. YS wrote the led the team in writing the manuscript. All were involved in writing, reviewing, and editing.
This work was supported by grants from the NIH (R01GM049077 to CL).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The molecular dynamics calculations were carried out using the High-Performance Computing Cluster, COARE Facility of the Department of Science and Technology – Advance Science and Technology Institute, Philippines.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.799703/full#supplementary-material
Acun, B., Hardy, D. J., Kale, L. V., Li, K., Phillips, J. C., and Stone, J. E. (2018). Scalable Molecular Dynamics with NAMD on the Summit System. IBM J. Res. Dev. 62 (6), 1–4. doi:10.1147/jrd.2018.2888986
Almond, R. J., Flanagan, B. F., Antonopoulos, A., Haslam, S. M., Dell, A., Kimber, I., et al. (2013). Differential Immunogenicity and Allergenicity of Native and Recombinant Human Lactoferrins: Role of Glycosylation. Eur. J. Immunol. 43 (1), 170–181. doi:10.1002/eji.201142345
An, H. J., Gip, P., Kim, J., Wu, S., Park, K. W., McVaugh, C. T., et al. (2012). Extensive Determination of Glycan Heterogeneity Reveals an Unusual Abundance of High Mannose Glycans in Enriched Plasma Membranes of Human Embryonic Stem Cells. Mol. Cell Proteomics 11 (4), M111–1010660. doi:10.1074/mcp.M111.010660
Balog, C. I. A., Stavenhagen, K., Fung, W. L. J., Koeleman, C. A., McDonnell, L. A., Verhoeven, A., et al. (2012). N-glycosylation of Colorectal Cancer Tissues. Mol. Cell Proteomics 11 (9), 571–585. doi:10.1074/mcp.M111.011601
Castanys-Muñoz, E., Martin, M. J., and Prieto, P. A. (2013). 2′-fucosyllactose: an Abundant, Genetically Determined Soluble Glycan Present in Human Milk. Nutr. Rev. 71 (12), 773–789. doi:10.1111/nure.12079
Chan, K. K., Dorosky, D., Sharma, P., Abbasi, S. A., Dye, J. M., Kranz, D. M., et al. (2020). Engineering Human ACE2 to Optimize Binding to the Spike Protein of SARS Coronavirus 2. Science 369 (6508), 1261–1265. doi:10.1126/science.abc0870
Choi, H.-Y., Park, H., Hong, J. K., Kim, S.-D., Kwon, J.-Y., You, S., et al. (2018). N-glycan Remodeling Using Mannosidase Inhibitors to Increase High-Mannose Glycans on Acid α-Glucosidase in Transgenic Rice Cell Cultures. Sci. Rep. 8 (1), 16130. doi:10.1038/s41598-018-34438-z
Clausen, T. M., Sandoval, D. R., Spliid, C. B., Pihl, J., Perrett, H. R., Painter, C. D., et al. (2020). SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell 183 (4), 1043e15–1057. doi:10.1016/j.cell.2020.09.033
C. R. Varki A, J. D. Esko, R. D. Cummings, G. Darvill, W. Hart, M. Aebiet al. (2015). Essentials of Glycobiology. 3rd Edn.
Digre, A., and Lindskog, C. (2021). The Human Protein Atlas-Spatial Localization of the Human Proteome in Health and Disease. Protein Sci. 30 (1), 218–233. doi:10.1002/pro.3987
Gstöttner, C., Zhang, T., Resemann, A., Ruben, S., Pengelley, S., Suckau, D., et al. (2021). Structural and Functional Characterization of SARS-CoV-2 RBD Domains Produced in Mammalian Cells. Anal. Chem. 93 (17), 6839–6847. doi:10.1021/acs.analchem.1c00893
Guvench, O., Mallajosyula, S. S., Raman, E. P., Hatcher, E., Vanommeslaeghe, K., Foster, T. J., et al. (2011). CHARMM Additive All-Atom Force Field for Carbohydrate Derivatives and its Utility in Polysaccharide and Carbohydrate-Protein Modeling. J. Chem. Theor. Comput. 7 (10), 3162–3180. doi:10.1021/ct200328p
Hamming, I., Timens, W., Bulthuis, M., Lely, A., Navis, G., and van Goor, H. (2004). Tissue Distribution of ACE2 Protein, the Functional Receptor for SARS Coronavirus. A First Step in Understanding SARS Pathogenesis. J. Pathol. 203 (2), 631–637. doi:10.1002/path.1570
Han, D. P., Lohani, M., and Cho, M. W. (2007). Specific Asparagine-Linked Glycosylation Sites Are Critical for DC-SIGN- and L-SIGN-Mediated Severe Acute Respiratory Syndrome Coronavirus Entry. J. Virol. 81 (21), 12029–12039. doi:10.1128/JVI.00315-07
Hansson, G. C. (2019). Mucus and Mucins in Diseases of the Intestinal and Respiratory Tracts. J. Intern. Med. 285 (5), 479–490. doi:10.1111/joim.12910
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., et al. (2020). SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181 (2), 271e8–280. doi:10.1016/j.cell.2020.02.052
Huang, J., Rauscher, S., Nawrocki, G., Ran, T., Feig, M., de Groot, B. L., et al. (2017). CHARMM36m: an Improved Force Field for Folded and Intrinsically Disordered Proteins. Nat. Methods 14 (1), 71–73. doi:10.1038/nmeth.4067
Hulswit, R. J. G., Lang, Y., Bakkers, M. J. G., Li, W., Li, Z., Schouten, A., et al. (2019). Human Coronaviruses OC43 and HKU1 Bind to 9-O-Acetylated Sialic Acids via a Conserved Receptor-Binding Site in Spike Protein Domain A. Proc. Natl. Acad. Sci. USA 116 (7), 2681–2690. doi:10.1073/pnas.1809667116
Humphrey, W., Dalke, A., and Schulten, K. (1996). VMD: Visual Molecular Dynamics. J. Mol. Graphics 14 (1), 3327–3388. doi:10.1016/0263-7855(96)00018-5
Jo, S., Cheng, X., Lee, J., Kim, S., Park, S.-J., Patel, D. S., et al. (2017). CHARMM-GUI 10 Years for Biomolecular Modeling and Simulation. J. Comput. Chem. 38 (15), 1114–1124. doi:10.1002/jcc.24660
Jorgensen, J. M., Arnold, C., Ashorn, P., Ashorn, U., Chaima, D., Cheung, Y. B., et al. (2017). Lipid-Based Nutrient Supplements during Pregnancy and Lactation Did Not Affect Human Milk Oligosaccharides and Bioactive Proteins in a Randomized Trial. J. Nutr. 147 (10), 1867–1874. doi:10.3945/jn.117.252981
Kim, S. Y., Jin, W., Sood, A., Montgomery, D. W., Grant, O. C., Fuster, M. M., et al. (2020). Characterization of Heparin and Severe Acute Respiratory Syndrome-Related Coronavirus 2 (SARS-CoV-2) Spike Glycoprotein Binding Interactions. Antiviral Res. 181, 104873. doi:10.1016/j.antiviral.2020.104873
Kommineni, V., Markert, M., Ren, Z., Palle, S., Carrillo, B., Deng, J., et al. (2019). In Vivo Glycan Engineering via the Mannosidase I Inhibitor (Kifunensine) Improves Efficacy of Rituximab Manufactured in Nicotiana Benthamiana Plants. Ijms 20 (1), 194. doi:10.3390/ijms20010194
Lan, J., Ge, J., Yu, J., Shan, S., Zhou, H., Fan, S., et al. (2020). Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor. Nature 581 (7807), 215–220. doi:10.1038/s41586-020-2180-5
Letko, M., Marzi, A., and Munster, V. (2020). Functional Assessment of Cell Entry and Receptor Usage for SARS-CoV-2 and Other Lineage B Betacoronaviruses. Nat. Microbiol. 5 (4), 562–569. doi:10.1038/s41564-020-0688-y
Li, Q., Xie, Y., Wong, M., Barboza, M., and Lebrilla, C. B. (2020). Comprehensive Structural Glycomic Characterization of the Glycocalyxes of Cells and Tissues. Nat. Protoc. 15 (8), 2668–2704. doi:10.1038/s41596-020-0350-4
Li, Q., Xie, Y., Xu, G., and Lebrilla, C. B. (2019). Identification of Potential Sialic Acid Binding Proteins on Cell Membranes by Proximity Chemical Labeling. Chem. Sci. 10 (24), 6199–6209. doi:10.1039/c9sc01360a
Li, W., Moore, M. J., Vasilieva, N., Sui, J., Wong, S. K., Berne, M. A., et al. (2003). Angiotensin-converting Enzyme 2 Is a Functional Receptor for the SARS Coronavirus. Nature 426 (6965), 450–454. doi:10.1038/nature02145
Ma, B., Simala-Grant, J. L., and Taylor, D. E. (2006). Fucosylation in Prokaryotes and Eukaryotes. Glycobiology 16 (12), 158R–184R. doi:10.1093/glycob/cwl040
Mallajosyula, S. S., Jo, S., Im, W., and MacKerell, A. D. (2015). Molecular Dynamics Simulations of Glycoproteins Using CHARMM. Methods Mol. Biol. 1273, 407–429. doi:10.1007/978-1-4939-2343-4_25
Mehdipour, A. R., and Hummer, G. (2021). Dual Nature of Human ACE2 Glycosylation in Binding to SARS-CoV-2 Spike. Proc. Natl. Acad. Sci. USA 118 (19), e2100425118. doi:10.1073/pnas.2100425118
Miller, J. L., deWet, B. J. M., Martinez-Pomares, L., Radcliffe, C. M., Dwek, R. A., Rudd, P. M., et al. (2008). The Mannose Receptor Mediates Dengue Virus Infection of Macrophages. Plos Pathog. 4 (2), e17. doi:10.1371/journal.ppat.0040017
Möginger, U., Grunewald, S., Hennig, R., Kuo, C.-W., Schirmeister, F., Voth, H., et al. (2018). Alterations of the Human Skin N- and O-Glycome in Basal Cell Carcinoma and Squamous Cell Carcinoma. Front. Oncol. 8, 70. doi:10.3389/fonc.2018.00070
Moore, R. E., Xu, L. L., and Townsend, S. D. (2021). Prospecting Human Milk Oligosaccharides as a Defense against Viral Infections. ACS Infect. Dis. 7, 254–263. doi:10.1021/acsinfecdis.0c00807
Morniroli, D., Consales, A., Crippa, B. L., Vizzari, G., Ceroni, F., Cerasani, J., et al. (2021). The Antiviral Properties of Human Milk: A Multitude of Defence Tools from Mother Nature. Nutrients 13 (2), 694. doi:10.3390/nu13020694
Nguyen, D. G., and Hildreth, J. E. K. (2003). Involvement of Macrophage Mannose Receptor in the Binding and Transmission of HIV by Macrophages. Eur. J. Immunol. 33 (2), 483–493. doi:10.1002/immu.200310024
Nguyen, L., McCord, K. A., Bui, D. T., Bouwman, K. M., Kitova, E. N., Kumawat, D., et al. (2021). Sialic Acid-dependent Binding and Viral Entry of SARS-CoV-2. bioRxiv 2021, 0308–434228. doi:10.1101/2021.03.08.434228
Nicholls, J. M., Bourne, A. J., Chen, H., Guan, Y., and Peiris, J. M. (2007). Sialic Acid Receptor Detection in the Human Respiratory Tract: Evidence for Widespread Distribution of Potential Binding Sites for Human and Avian Influenza Viruses. Respir. Res. 8, 73. doi:10.1186/1465-9921-8-73
Park, D., Arabyan, N., Williams, C. C., Song, T., Mitra, A., Weimer, B. C., et al. (2016). Salmonella Typhimurium Enzymatically Landscapes the Host Intestinal Epithelial Cell (IEC) Surface Glycome to Increase Invasion. Mol. Cell Proteomics 15 (12), 3653–3664. doi:10.1074/mcp.M116.063206
Park, D., Brune, K. A., Mitra, A., Marusina, A. I., Maverakis, E., and Lebrilla, C. B. (2015). Characteristic Changes in Cell Surface Glycosylation Accompany Intestinal Epithelial Cell (IEC) Differentiation: High Mannose Structures Dominate the Cell Surface Glycome of Undifferentiated Enterocytes. Mol. Cell Proteomics 14 (11), 2910–2921. doi:10.1074/mcp.M115.053983
Park, D., Xu, G., Barboza, M., Shah, I. M., Wong, M., Raybould, H., et al. (2017). Enterocyte Glycosylation Is Responsive to Changes in Extracellular Conditions: Implications for Membrane Functions. Glycobiology 27 (9), 847–860. doi:10.1093/glycob/cwx041
Park, S.-J., Lee, J., Qi, Y., Kern, N. R., Lee, H. S., Jo, S., et al. (2019). CHARMM-GUIGlycan Modelerfor Modeling and Simulation of Carbohydrates and Glycoconjugates. Glycobiology 29 (4), 320–331. doi:10.1093/glycob/cwz003
Peiris, J., Lai, S., Poon, L., Guan, Y., Yam, L., Lim, W., et al. (2003). Coronavirus as a Possible Cause of Severe Acute Respiratory Syndrome. The Lancet 361 (9366), 1319–1325. doi:10.1016/s0140-6736(03)13077-2
Radcliffe, C. M., Arnold, J. N., Suter, D. M., Wormald, M. R., Harvey, D. J., Royle, L., et al. (2007). Human Follicular Lymphoma Cells Contain Oligomannose Glycans in the Antigen-Binding Site of the B-Cell Receptor. J. Biol. Chem. 282 (10), 7405–7415. doi:10.1074/jbc.M602690200
Reading, P. C., Miller, J. L., and Anders, E. M. (2000). Involvement of the Mannose Receptor in Infection of Macrophages by Influenza Virus. J. Virol. 74 (11), 5190–5197. doi:10.1128/jvi.74.11.5190-5197.2000
Roberts, G., Tarelli, E., Homer, K. A., Philpott-Howard, J., and Beighton, D. (2000). Production of an Endo-β- N -Acetylglucosaminidase Activity Mediates Growth of Enterococcus faecalis on a High-mannose-type Glycoprotein. J. Bacteriol. 182 (4), 882–890. doi:10.1128/jb.182.4.882-890.2000
Routhu, N. K., Lehoux, S. D., Rouse, E. A., Bidokhti, M. R. M., Giron, L. B., Anzurez, A., et al. (2019). Glycosylation of Zika Virus Is Important in Host-Virus Interaction and Pathogenic Potential. Ijms 20 (20), 5206. doi:10.3390/ijms20205206
Ruhaak, L. R., Taylor, S. L., Stroble, C., Nguyen, U. T., Parker, E. A., Song, T., et al. (2015). Differential N-Glycosylation Patterns in Lung Adenocarcinoma Tissue. J. Proteome Res. 14 (11), 4538–4549. doi:10.1021/acs.jproteome.5b00255
Sanda, M., Morrison, L., and Goldman, R. (2021). N- and O-Glycosylation of the SARS-CoV-2 Spike Protein. Anal. Chem. 93 (4), 2003–2009. doi:10.1021/acs.analchem.0c03173
Schneider, M., Al-Shareffi, E., and Haltiwanger, R. S. (2017). Biological Functions of Fucose in Mammals. Glycobiology 27 (7), 601–618. doi:10.1093/glycob/cwx034
Shajahan, A., Archer-Hartmann, S., Supekar, N. T., Gleinich, A. S., Heiss, C., and Azadi, P. (2021). Comprehensive Characterization of N- and O- Glycosylation of SARS-CoV-2 Human Receptor Angiotensin Converting Enzyme 2. Glycobiology 31 (4), 410–424. doi:10.1093/glycob/cwaa101
Shajahan, A., Supekar, N. T., Gleinich, A. S., and Azadi, P. (2020). Deducing the N- and O-Glycosylation Profile of the Spike Protein of Novel Coronavirus SARS-CoV-2. Glycobiology 30 (12), 981–988. doi:10.1093/glycob/cwaa042
Shinya, K., Ebina, M., Yamada, S., Ono, M., Kasai, N., and Kawaoka, Y. (2006). Influenza Virus Receptors in the Human Airway. Nature 440 (7083), 435–436. doi:10.1038/440435a
Sieben, C., Sezgin, E., Eggeling, C., and Manley, S. (2020). Influenza A Viruses Use Multivalent Sialic Acid Clusters for Cell Binding and Receptor Activation. Plos Pathog. 16 (7), e1008656. doi:10.1371/journal.ppat.1008656
Smilowitz, J. T., Totten, S. M., Huang, J., Grapov, D., Durham, H. A., Lammi-Keefe, C. J., et al. (2013). Human Milk Secretory Immunoglobulin a and Lactoferrin N-Glycans Are Altered in Women with Gestational Diabetes Mellitus. J. Nutr. 143 (12), 1906–1912. doi:10.3945/jn.113.180695
Suryamohan, K., Diwanji, D., Stawiski, E. W., Gupta, R., Miersch, S., Liu, J., et al. (2021). Human ACE2 Receptor Polymorphisms Predict SARS-CoV-2 Susceptibility. Commun. Biol. 4 (1), 475. doi:10.1038/s42003-021-02030-3
Suzuki, K., Daikoku, S., Son, S.-H., Ito, Y., and Kanie, O. (2015). Synthetic Study of 3-fluorinated Sialic Acid Derivatives. Carbohydr. Res. 406, 1–9. doi:10.1016/j.carres.2014.12.010
Tai, W., He, L., Zhang, X., Pu, J., Voronin, D., Jiang, S., et al. (2020). Characterization of the Receptor-Binding Domain (RBD) of 2019 Novel Coronavirus: Implication for Development of RBD Protein as a Viral Attachment Inhibitor and Vaccine. Cell Mol Immunol 17 (6), 613–620. doi:10.1038/s41423-020-0400-4
Tailford, L. E., Crost, E. H., Kavanaugh, D., and Juge, N. (2015). Mucin Glycan Foraging in the Human Gut Microbiome. Front. Genet. 6, 81. doi:10.3389/fgene.2015.00081
Thul, P. J., and Lindskog, C. (2018). The Human Protein Atlas: A Spatial Map of the Human Proteome. Protein Sci. 27 (1), 233–244. doi:10.1002/pro.3307
Tian, X., Li, C., Huang, A., Xia, S., Lu, S., Shi, Z., et al. (2020). Potent Binding of 2019 Novel Coronavirus Spike Protein by a SARS Coronavirus-specific Human Monoclonal Antibody. Emerging Microbes & Infections 9 (1), 382–385. doi:10.1080/22221751.2020.1729069
Tran, D. T., and Ten Hagen, K. G. (2013). Mucin-type O-Glycosylation during Development. J. Biol. Chem. 288 (10), 6921–6929. doi:10.1074/jbc.R112.418558
Trebbien, R., Larsen, L. E., and Viuff, B. M. (2011). Distribution of Sialic Acid Receptors and Influenza A Virus of Avian and Swine Origin in Experimentally Infected Pigs. Virol. J. 8, 434. doi:10.1186/1743-422X-8-434
Upham, J. P., Pickett, D., Irimura, T., Anders, E. M., and Reading, P. C. (2010). Macrophage Receptors for Influenza A Virus: Role of the Macrophage Galactose-type Lectin and Mannose Receptor in Viral Entry. J. Virol. 84 (8), 3730–3737. doi:10.1128/JVI.02148-09
Villalobos, J. A., Yi, B. R., and Wallace, I. S. (2015). 2-Fluoro-L-Fucose Is a Metabolically Incorporated Inhibitor of Plant Cell Wall Polysaccharide Fucosylation. PLoS One 10 (9), e0139091. doi:10.1371/journal.pone.0139091
Viruses, C. S. G. o. t. I. C. o. T. o. (2020). The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-nCoV and Naming it SARS-CoV-2. Nat. Microbiol. 5 (4), 536–544. doi:10.1038/s41564-020-0695-z
Walls, A. C., Park, Y.-J., Tortorici, M. A., Wall, A., McGuire, A. T., and Veesler, D. (2020). Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 183 (6), 1735. doi:10.1016/j.cell.2020.11.032
Wan, Y., Shang, J., Graham, R., Baric, R. S., and Li, F. (2020). Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 94 (7). doi:10.1128/JVI.00127-20
Wang, X., Zhou, Y., Jiang, N., Zhou, Q., and Ma, W.-L. (2020). Persistence of Intestinal SARS-CoV-2 Infection in Patients with COVID-19 Leads to Re-admission after Pneumonia Resolved. Int. J. Infect. Dis. 95, 433–435. doi:10.1016/j.ijid.2020.04.063
Watanabe, Y., Allen, J. D., Wrapp, D., McLellan, J. S., and Crispin, M. (2020). Site-specific Glycan Analysis of the SARS-CoV-2 Spike. Science 369 (6501), 330–333. doi:10.1126/science.abb9983
Wong, M., Xu, G., Barboza, M., Maezawa, I., Jin, L.-W., Zivkovic, A., et al. (2020). Metabolic Flux Analysis of the Neural Cell Glycocalyx Reveals Differential Utilization of Monosaccharides. Glycobiology 30 (11), 859–871. doi:10.1093/glycob/cwaa038
Wong, S. K., Li, W., Moore, M. J., Choe, H., and Farzan, M. (2004). A 193-amino Acid Fragment of the SARS Coronavirus S Protein Efficiently Binds Angiotensin-Converting Enzyme 2. J. Biol. Chem. 279 (5), 3197–3201. doi:10.1074/jbc.C300520200
Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C.-L., Abiona, O., et al. (2020). Cryo-EM Structure of the 2019-nCoV Spike in the Prefusion Conformation. Science 367 (6483), 1260–1263. doi:10.1126/science.abb2507
Wu, N.-H., Meng, F., Seitz, M., Valentin-Weigand, P., and Herrler, G. (2015). Sialic Acid-dependent Interactions between Influenza Viruses and Streptococcus Suis Affect the Infection of Porcine Tracheal Cells. J. Gen. Virol. 96 (9), 2557–2568. doi:10.1099/jgv.0.000223
Wu, S., Grimm, R., German, J. B., and Lebrilla, C. B. (2011). Annotation and Structural Analysis of Sialylated Human Milk Oligosaccharides. J. Proteome Res. 10 (2), 856–868. doi:10.1021/pr101006u
Wu, S., Tao, N., German, J. B., Grimm, R., and Lebrilla, C. B. (2010). Development of an Annotated Library of Neutral Human Milk Oligosaccharides. J. Proteome Res. 9 (8), 4138–4151. doi:10.1021/pr100362f
Xu, C., Wang, Y., Liu, C., Zhang, C., Han, W., Hong, X., et al. (2021). Conformational Dynamics of SARS-CoV-2 Trimeric Spike Glycoprotein in Complex with Receptor ACE2 Revealed by Cryo-EM. Sci. Adv. 7 (1). doi:10.1126/sciadv.abe5575
Zhao, P., Praissman, J. L., Grant, O. C., Cai, Y., Xiao, T., Rosenbalm, K. E., et al. (2020). Virus-Receptor Interactions of Glycosylated SARS-CoV-2 Spike and Human ACE2 Receptor. Cell Host & Microbe 28 (4), 586e6–601. doi:10.1016/j.chom.2020.08.004
Zhou, Q., Xie, Y., Lam, M., and Lebrilla, C. B. (2021). N-glycomic Analysis of the Cell Shows Specific Effects of Glycosyl Transferase Inhibitors. Cells 10 (9), 2318. doi:10.3390/cells10092318
Zhou, Y., Fukuda, T., Hang, Q., Hou, S., Isaji, T., Kameyama, A., et al. (2017). Inhibition of Fucosylation by 2-fluorofucose Suppresses Human Liver Cancer HepG2 Cell Proliferation and Migration as Well as Tumor Formation. Sci. Rep. 7 (1), 11563. doi:10.1038/s41598-017-11911-9
Keywords: SARS-CoV2, glycocalyx, LC/MS, remodeling glycome, host-virus interaction, spike (S) protein, ACE2 glycosylation
Citation: Sheng Y, Vinjamuri A, Alvarez MRS, Xie Y, McGrath M, Chen S, Barboza M, Frieman M and Lebrilla CB (2022) Host Cell Glycocalyx Remodeling Reveals SARS-CoV-2 Spike Protein Glycomic Binding Sites. Front. Mol. Biosci. 9:799703. doi: 10.3389/fmolb.2022.799703
Received: 21 October 2021; Accepted: 31 January 2022;
Published: 14 March 2022.
Edited by:
Michael C. Jewett, Northwestern University, United StatesReviewed by:
Wengang Chai, Imperial College London, United KingdomCopyright © 2022 Sheng, Vinjamuri, Alvarez, Xie, McGrath, Chen, Barboza, Frieman and Lebrilla. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carlito B. Lebrilla, Y2JsZWJyaWxsYUB1Y2RhdmlzLmVkdQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.