
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci. , 16 February 2022
Sec. Cellular Biochemistry
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.744901
This article is part of the Research Topic Tumorigenesis Regulated by miRNAs, Volume II View all 5 articles
Chlamydia trachomatis (C. trachomatis) is a major etiological agent of sexually transmitted infection. Some stressing conditions can result in persistent chlamydial infection, which is thought to be associated with severe complications including ectopic pregnancy and tubal factor infertility. Long noncoding RNAs (lncRNAs) have been identified as key modulators in many biological processes. Nevertheless, the role of lncRNAs in persistent chlamydial infection is still unclear. In this study, we used lncRNA and mRNA microarray to identify the global lncRNAs and mRNAs expression in penicillin-induced persistent chlamydial infection in HeLa cells as well as the control group (HeLa cells without C. trachomatis infection). Among 1005 differentially expressed lncRNAs, 585 lncRNAs were upregulated and 420 downregulated in persistent chlamydial infection, while 410 mRNAs were identified to express differentially, of which 113 mRNAs were upregulated and 297 downregulated. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis with differentially expressed genes were performed. We then constructed the lncRNA-miRNA-mRNA competing endogenous RNAs (ceRNAs) network. Four mRNAs were validated to be changed by quantitative real-time PCR which were correlated with the microarray result. Integration of protein-protein interaction network was constructed and hub genes were identified. These findings provide a new perspective on the molecular mechanisms of penicillin-induced persistent chlamydial infection.
Chlamydia trachomatis (C. trachomatis), as a Gram-negative obligate intracellular bacterium, causes various diseases and sequelae in human beings. Different chlamydial serovars cause different diseases. Serovars D to K are the major etiological pathogens of the most common sexually transmitted infection, while serovars L1 to L3 lead to lymphatic system infection (Witkin et al., 2017). Females infected with C. trachomatis in the genital tract usually show asymptomatic. Diseases such as cervicitis, pelvic inflammatory disease (PID), and severe complications including ectopic pregnancy and tubal factor infertility (TFI) probably happen after the asymptomatic infection without a timely treatment (Tsevat et al., 2017).
The developmental cycle of C. trachomatis alternates between two forms, infectious non-replicative elementary bodies (EBs) and replicative, non-infectious reticulate bodies (RBs) (Abdelrahman and Belland, 2005; Stephens et al., 2011). The bacterium develops and replicates in vesicles called inclusions. When the normal lifecycle is disturbed by stress conditions, including amino acid deficiency (Beatty et al., 1994), nutrient depletion (Capmany and Damiani, 2010), antibiotics (Zhu et al., 2014; Xue et al., 2017), immunological factors, and interferon-gamma (IFN-γ) (Beatty et al., 1993), the inclusions become smaller containing aberrant reticulate bodies (ABs), with slow metabolism and weakened infectivity. It results in a persistent chlamydial infection, which is believed to be associated with female infertility (Witkin et al., 2017). After removing the stressful conditions, persistent infection can be reactivated to acute infection. However, the persistent infection of C. trachomatis is still a public health problem because of the difficulty of diagnosis and resistance to antibiotics (Patton et al., 1994). Although there were some studies have found the aberrant reticulate bodies in female endocervix via electron microscopy to diagnose the persistent chlamydial infection (Bragina et al., 2001; Lewis et al., 2014), the diagnosis by electron microscopy still cannot be used widespread in clinical cases. In the previous study, we have used penicillin to induce persistent chlamydial infection. We discovered the different structural changes in the Golgi apparatus between persistent and acute chlamydial infections, which indicated the low requirements of lipid in persistent infection (Zhu et al., 2014).
Noncoding RNA (ncRNA) is thought to be a new regulatory layer in transcriptional and posttranscriptional gene regulation (Akhade et al., 2017). Studies based on high-throughput transcriptomics show that more than two-thirds of the mammalian genome is transcribed encoding millions of different classes of small and long noncoding RNAs (lncRNAs). LncRNAs (ncRNAs that are >200 nt long) now have been identified as key modulators in many biological processes, including cell proliferation, cell cycle, differentiation, apoptosis, metabolism, and maintenance of pluripotency, etc (Wang et al., 2011; Geisler and Coller, 2013). LncRNAs exert their harbor sequences complementary to microRNA (miRNA) sequences to sequester them and prevent them from binding to their targets. Such lncRNAs can be derived from pseudogenes or have a similar form with circular RNAs or be common intergenic lncRNAs possessing miRNA binding sites (Akhade et al., 2017). Nowadays lncRNAs are emerging as a hotspot in cancer, diagnosis, and therapy (Bhan et al., 2017). However, in persistent chlamydial infection, the function of lncRNAs is still unknown.
In the study, we used microarray to identify the differentially expressed lncRNAs and messenger RNAs (mRNAs) between persistent chlamydial infection and uninfected cells. We constructed a network for these differentially expressed genes (DEGs) to clarify the relationship between lncRNA and mRNA. To investigate the potential regulatory roles, Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were processed. We then predicted the competing endogenous RNAs (ceRNAs) network among the DEGs. The differential expression levels of 4 mRNAs were validated by quantitative real-time PCR. Besides, we identified hub genes via the integration of the protein-protein interaction (PPI) network. These findings provide a new perspective on the molecular mechanism of penicillin-induced persistent chlamydial infection.
HeLa cells were cultured in RPMI 1640 medium (Gibco, United States) supplemented with 10% fetal bovine serum (Gibco, United States) at 37°C in 5% CO2. The cells were transferred into six-well plates and cultured for 24 h under the same condition. Chlamydia trachomatis serovar D at an MOI of 2 was incubated with HeLa cells by centrifugation at 3,000 rpm/min at 37°C for 1 h and then incubated for another hour at 37°C in 5% CO2. Extracellular bacteria in the supernatant were aspirated followed by the addition of fresh RPMI 1640 medium supplemented with 10% fetal bovine serum, 0.5% glucose, and 100 U/ml penicillin G (Sigma, United States) (Skilton et al., 2009). HeLa cells without chlamydial infection treated with the same medium containing 100U/ml penicillin G were used as control mock cells. The cells were maintained at 37°C in 5% CO2 (Zhu et al., 2014).
The immunofluorescence staining was performed as followed: samples were fixed with 4% paraformaldehyde, permeabilized in 0.5% (v/v) Triton X-100 (Sigma, United States), and then blocked with 1% (w/v) BSA (Thermo Scientific, United States). Cells were then incubated with goat polyclonal to Chlamydia trachomatis major outer-membrane protein (MOMP) coupled to FITC (Abcam, ab30951, United States). The samples were counterstained with DAPI (Beyotime, China). Immunofluorescence images were acquired by fluorescence microscopy (Olympus, Japan).
Based on the previous study (Zhu et al., 2014), the total RNA was extracted from the HeLa cells with or without chlamydial infection at 40 h post-infection using TRIzol reagent (Invitrogen, United States) according to the manufacturer’s specifications. The yield of RNA was determined using a NanoDrop 2000 spectrophotometer (Thermo Scientific, United States), and the integrity was evaluated using agarose gel electrophoresis stained with ethidium bromide.
The Agilent Human ceRNA Microarray 2019 (4*180k, Design ID:086188) was used in this experiment and data analysis of the six samples was conducted by OE Biotechnology Co., Ltd., (Shanghai, China). The steps of sample labeling, microarray hybridization and washing were performed following the manufacturer’s standard protocols. In brief, total RNA was transcribed to double-strand cDNA, then synthesized into cRNA and labeled with Cyanine-3-CTP. The labeled cRNAs were hybridized onto the microarray. After washing steps, the arrays were scanned by the Agilent Scanner G2505C (Agilent Technologies, United States). Feature Extraction software (version10.7.1.1, Agilent Technologies, United States) was used to analyze array images to get raw data. Secondary, the raw data were normalized with the quantile algorithm. The probes that at least one condition out of 2 conditions have flags in “Detected” were chosen for further data analysis. Differentially expressed genes (DEGs) were identified through fold change as well as p values calculated with t-test. The threshold set for up-and down-regulated genes was a fold change ≥1.5 and a p value < 0.01. Afterward, Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were applied to determine the roles of these differentially expressed genes (DEGs). Finally, Hierarchical Clustering was performed to display the distinguishable genes’ expression pattern among samples.
The competing endogenous RNAs (ceRNAs) network was constructed based on the theory that lncRNA can affect miRNA acting as miRNA sponges to further regulate mRNA (Guo et al., 2015). This analysis selects the differential lncRNAs that are significantly positively correlated with the differential mRNAs as the target lncRNA of the ceRNA analysis. Potential miRNAs were searched for in the miRBase22. The predicted interactions of miRNA-mRNA and miRNA-lncRNA were analyzed. The miRNA–lncRNA and miRNA–mRNA pairs sharing the same miRNA were merged into a lncRNA–miRNA–mRNA interaction as a ceRNA relationship.
To validate the results of microarray analysis, four mRNAs were selected. Quantification was performed with reverse transcription (RT) and PCR. Each RT reaction consisted of 0.5 μg RNA, 2 μL of 5×TransScript All-in-one SuperMix for qPCR and 0.5 μL of gDNA Remover, in a total volume of 10 μL. Reactions were performed in a GeneAmp® PCR System 9700 (Applied Biosystems, United States) for 15 min at 42°C, 5 s at 85°C. The 10 μL RT reaction mix was then diluted × 10 in nuclease-free water and held at −20°C.
Real-time PCR was performed using LightCycler® 480 Ⅱ Real-time PCR Instrument (Roche, Swiss) with 10 μL PCR reaction mixture that included 1 μL of cDNA, 5 μL of 2×PerfectStartTM Green qPCR SuperMix, 0.2 μL of forward primer, 0.2 μL of reverse primer and 3.6 μL of nuclease-free water. Reactions were incubated in a 384-well optical plate (Roche, Swiss) at 94°C for 30 s, followed by 45 cycles of 94°C for 5 s, 60°C for 30 s. Each sample was run in triplicate for analysis. At the end of the PCR cycles, a melting curve analysis was performed to validate the specific generation of the expected PCR product. The primer sequences were designed in the laboratory and synthesized by TsingKe Biotech based on the mRNA sequences obtained from the NCBI database. The expression levels of mRNAs were normalized to GAPDH and were calculated using the 2-ΔΔCt method. Characteristics and primers of these RNAs are listed in Table 1.
The Search Tool for the Retrieval of Interacting Genes (STRING, http://string.embl.de/) (von Mering et al., 2003) is a biological database for predicting protein-protein interaction (PPI) information. The DEGs were mapped to STRING to evaluate the interactive relationships, with a confidence score >0.9 defined as significant. Then, Cytoscape (Shannon et al., 2003), a biological graph visualization tool for integrated models of biologic molecular interaction networks software, was used to construct PPI networks. The CytoHubba (Chin et al., 2014), a plugin for Cytoscape, was used to rank nodes in a network by their network features. The top 10 essential nodes ranked by Maximal Clique Centrality (MCC) scores were selected.
All statistical analyses were performed using SPSS 25.0 software (SPSS, Chicago, IL, United States). Graphs were created using Graphpad Prism 8.0 (GraphPad Software, La Jolla, CA). Data were presented as the mean ± SD. Statistical analysis for comparison of two groups was subject to a two-tailed Student’s t-test. For comparison of multiple groups, one-way ANOVA followed by Student–Newman–Keuls post-hoc test was performed. p < 0.05 was considered statistically significant.
According to the result of previous experiments, a persistent infection model was successfully established by inoculating with C. trachomatis at MOI of 2 and then induced in medium containing 100 U/ml penicillin for 40 h. Compared with chlamydial infection in penicillin-free medium (acute infection), the inclusions in persistent chlamydial infection were relatively small (Figure 1).
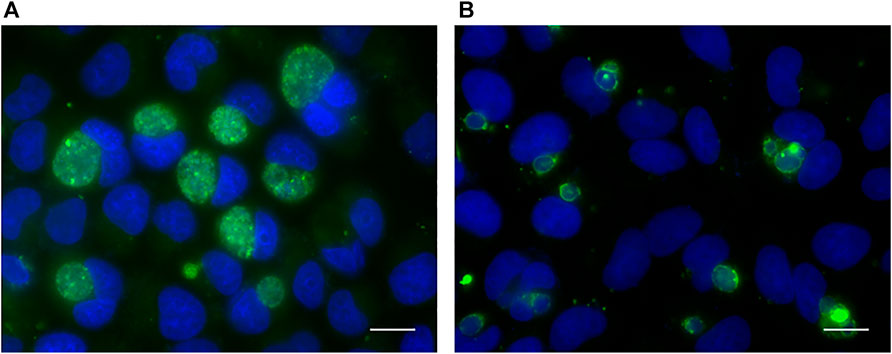
FIGURE 1. Immunofluorescence analysis of acute (A) and persistent (B) chlamydial infection in HeLa cell. The chlamydial inclusions were stained with goat polyclonal to Chlamydia trachomatis MOMP coupled to FITC (green). DNA was stained with DAPI (blue). Scale bar = 50 μm.
According to the microarray results, a total of 1005 lncRNAs were identified to express differentially in penicillin-induced persistent chlamydial infection (over 1.5-fold changes, p < 0.01), of which 585 lncRNAs were upregulated and 420 downregulated (Figure 2A). Up- and down-regulated lncRNAs were mapped in a volcano plot (Figure 2B).
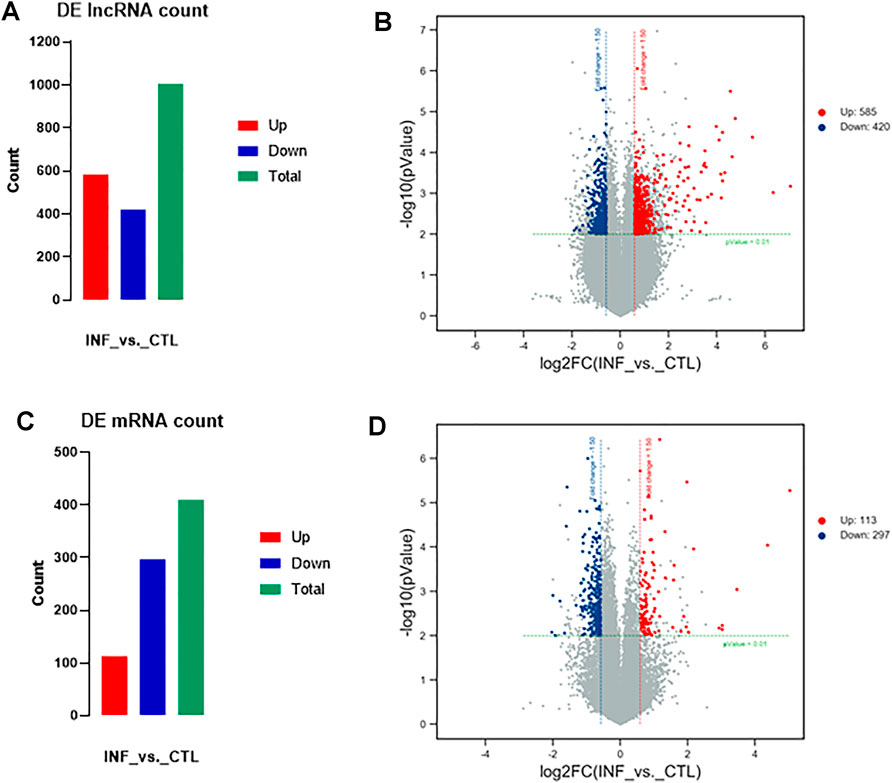
FIGURE 2. Variation in lncRNAs and mRNAs expression in persistent chlamydial infection. (A) Summarizes the lncRNAs that were differentially expressed. In total of 1005 differentially expressed lncRNAs, 585 were upregulated and 420 were downregulated. (B) Volcano plot of up- and downregulated lncRNAs mapped via log_2 (fold change). (C) Summarizes the mRNAs that were differentially expressed. In total of 410 differentially expressed mRNAs, 113 were upregulated and 297 were downregulated. (D) Volcano plot of up- and downregulated mRNAs mapped via
In terms of mRNA analysis, a total of 410 mRNAs were identified to express differentially in penicillin-induced persistent chlamydial infection (over 1.5-fold changes, p < 0.01), of which 113 mRNAs were upregulated and 297 downregulated (Figure 2C). Up- and downregulated mRNAs were mapped in a volcano plot (Figure 2D).
Among the 585 upregulated lncRNAs, most came from chromosome 1 (8.71%) and chromosome 2 (8.21%), while the percentages of chromosome 18 (1.88%), chromosome 21 (1.88%), and chromosome Y (0.00%) were <2% (Figure 3A). The distribution of 420 downregulated lncRNAs was also mainly from chromosome 1 (10.00%) and chromosome 2 (9.76%), while those from chromosome 18 (1.90%), chromosome 20 (1.19%), chromosome 21 (1.67%) and chromosome Y (0.24%) were <2% (Figure 3B).
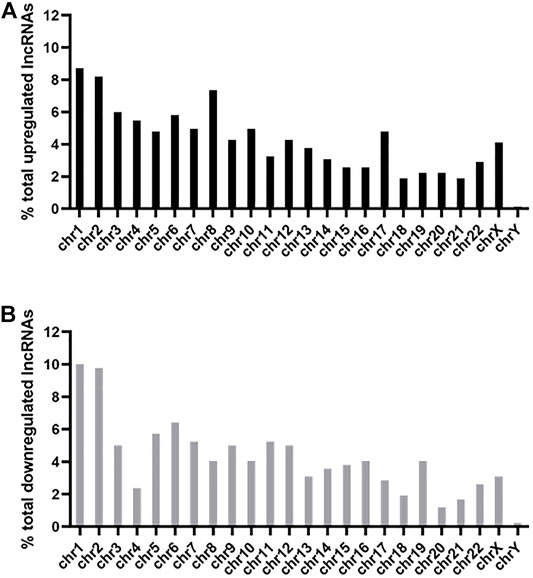
FIGURE 3. Chromosomal distribution of differentially expressed lncRNAs. The upregulated (A) and downregulated (B) lncRNAs were widely distributed among all chromosomes, including the sex chromosome X and chromosome Y.
GO gene functional classification was performed to classify the possible function of the differential expressed genes (DEGs). Based on GO analysis, the DEGs were classified into three main categories: molecular function (MF), cellular component (CC), and biological process (BP). The top ten in each category were presented in Figure 4. In molecular function, changes were significantly enriched in “transcription regulatory region DNA binding,” “carboxylic acid binding,” and “MAP kinase tyrosine/serine/threonine phosphatase activity.” In cellular component, changes were involved in “nucleosome,” “nucleus,” and “endosome.” In biological process, the top three changes were “protein refolding,” “nucleosome assembly,” and “hydrogen ion transmembrane transport” (Figure 4).
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis identified 13 significant pathways with DEGs (p < 0.05). Figure 5 showed the top 30 pathways. The top three pathways included Steroid hormone biosynthesis (TermID: path: hsa00140), Legionellosis (TermID: path: hsa05134), and Viral carcinogenesis (TermID: path: hsa05203) (Figure 5).
To validate the reliability of the microarray results, four mRNAs were selected to exam their expression by quantitative real-time PCR. As shown in Figure 6, the expression of STX5, DUSP4, BICDL1, and RAB30 were downregulated which were correlated with the microarray results.
To better understand the interaction of lncRNA, miRNA and mRNA, we constructed a lncRNA-associated ceRNA network by integrating the expression profiles and regulatory relationships of the lncRNAs, miRNAs and mRNAs from the sequencing data of the six samples. A total of 101334 ceRNA relationships was identified from the interaction of 783 differentially expressed lncRNAs, 395 mRNAs, and 814 miRNAs. The network containing the top 100 relationships ranked by correlation coefficient was shown in Figure 7A. In the network, LINC00926:3 and NONHSAT173474.1 were the lncRNAs that link to most nodes, regulating BICDL1 and DUSP4 respectively. Besides, we selected two pairs of ceRNA to exam the expression change by quantitative real-time PCR: LINC00926:3- has-miR-1207-5P- BICDL1, and NONHSAT173474.1- has-miR-5088-5P- DUSP4. It is well-established that lncRNAs may function as ceRNA by competitively binding miRNAs to regulate the downstream mRNA. Therefore, the lncRNA and its downstream mRNA have the same changing trend, while the microRNA has an opposite one. According to the results, the expression change of NONHSAT173474.1- has-miR-5088-5P- DUSP4 was in line with the changing trend of ceRNA network (Figures 7B,C). These data showed us the potential role of ceRNA regulatory networks in the pathogenesis of penicillin-induced persistent chlamydial infection.
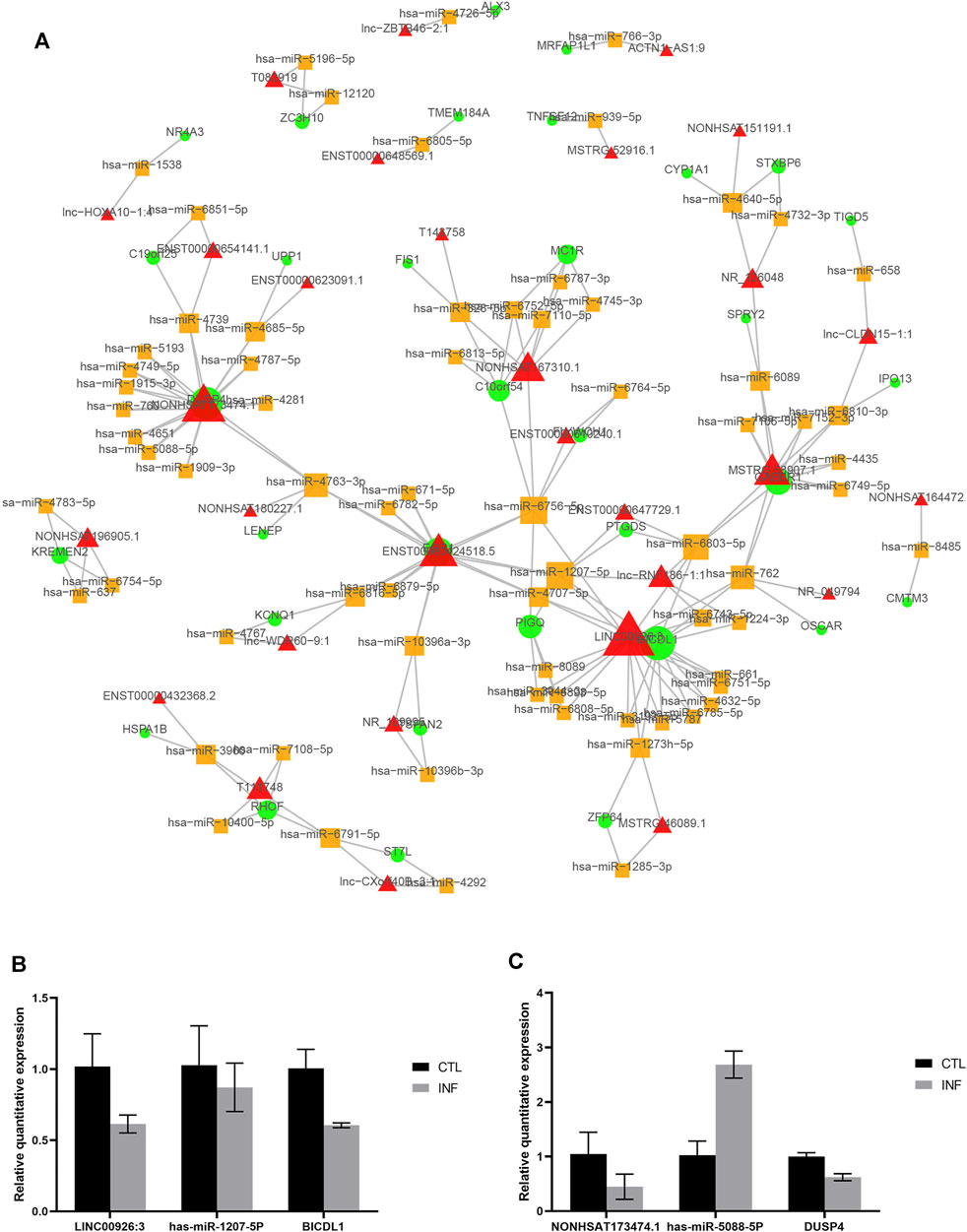
FIGURE 7. CeRNA network in the persistent chlamydial infection. (A) The top 100 lncRNA-miRNA-mRNA competing endogenous RNA (ceRNA) network. The triangle node represents lncRNA, the circular node represents mRNA, and the square node represents miRNA. The node size proportional to the number of connected nodes. (B,C) Quantitative real-time PCR results of ceRNA.
To identify the hub genes that play an essential role in persistent chlamydial infection, a PPI network was constructed. The PPI network of DEGs consisted of 373 nodes and 505 edges constructed in the STRING database (version 11.0) and visualized using Cytoscape software. The top 10 essential nodes ranked by Maximal Clique Centrality (MCC) scores were selected and the network was shown in Figure 8. The detailed information of these 10 hub genes was presented in Table 2. Among these genes, centromere protein A (CENPA) showed the highest score of 11064.
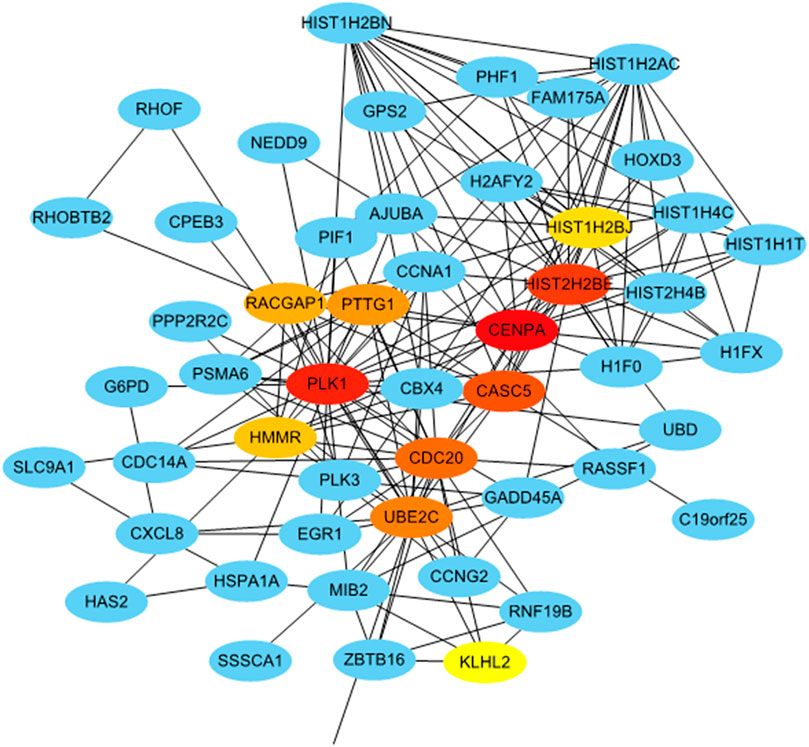
FIGURE 8. Identification of hub genes from the PPI network with the CytoHubba. Top ten hub genes were ranked by MCC score.
C. trachomatis is an obligate intracellular bacterium. It acquires nutrients, including amino acids, nucleotides, and lipids, from the host cells (van Ooij et al., 2000; Moore et al., 2008; Saka and Valdivia, 2010; Mehlitz et al., 2017). Therefore, C. trachomatis may change the biological process of the host cells by a molecular mechanism to support its development, replication, and inclusion growth.
It has been estimated that miRNAs target 20–30% of human mRNAs that affect various aspects, including transcription, transduction, growth, and fatty acid metabolism. MiRNAs have been thought to be immune modulators serving as a connection between innate and adaptive immune responses, and dysregulation of miRNA expression plays a role in various diseases, such as cardiovascular disease, cancer, and infectious and metabolic diseases (Dai and Ahmed, 2011; Ha, 2011; O'Connell et al., 2012). Several miRNAs have been elucidated the role in chlamydial infection in the previous studies, involving dampening fibrosis, transcriptional regulation of cytokine responses, and relationship with the degree of clinical inflammation (Igietseme et al., 2015; Derrick et al., 2016; Gupta et al., 2016; Chowdhury et al., 2017; Yeruva et al., 2017). The latest research found that miR-193b may serve as a potential serum biomarker for C. trachomatis infection (Dzakah et al., 2021). Besides, circRNAs have been identified to play a role in chlamydial infection (Liu et al., 2019). However, there was little research about the lncRNA-mRNA interaction in persistent chlamydial infection.
In this study, we used microarray to investigate the differentially expressed lncRNAs and mRNAs in persistent chlamydial infection. We identified a total of 1005 differentially expressed lncRNAs, of which 585 lncRNAs were upregulated and 420 downregulated. In terms of mRNA analysis, a total of 410 differentially expressed mRNAs were identified, of which 113 mRNAs were upregulated and 297 downregulated.
Based on the data, genes producing the differentially expressed lncRNAs were widely distributed in all chromosomes including sex chromosome X and chromosome Y. Surprisingly, no matter up-or downregulated, the differentially expressed lncRNAs were mainly from chromosome one and chromosome 2, which are the largest and second-largest human chromosomes and consist of numerous genes. It may explain the reason why most of the differentially expressed genes are on these two chromosomes. However, there has not been proven yet.
GO analysis and KEGG pathway annotation were conducted to determine the functions of differentially expressed mRNAs between persistent chlamydial infection and control mock cells. GO enrichment data revealed the differentially expressed genes that are mainly involved in the regulation of biological processes, cellular components, and molecular functions. The most significant GO items were nucleosome, protein refolding, transcription regulatory region DNA binding, indicating that such genes relating to the regulation of gene expression via chromatin structure and production of proteins may contribute to the mechanism of persistent chlamydial infection. It is interesting to note that most genes from these items are downregulating in the persistent-infected cells. This may be associated with the reduction of cell division with persistent chlamydial infection, which is basically consistent with previous studies (Gerard et al., 2001; Klos et al., 2009). In the KEGG pathway analysis, thirteen significant pathways were identified. Among these pathways, the MAPK signaling pathway plays an important role in chlamydial infection, including mediating the inflammation or anti-apoptosis (Krüll et al., 2004; Vignola et al., 2010; Kun et al., 2013; Zhou et al., 2013; Jia et al., 2019; Liu et al., 2019; Wen et al., 2020). However, whether this pathway has a different regulatory function between acute and persistent chlamydial infection is still unclear. Furthermore, we reanalyzed the data from the GEO dataset (GSE158814) that contained the persistent chlamydial infection in HeLa cells with IFN-γ treatment for 44 h (Dzakah et al., 2021). The threshold set for up-and down-regulated genes was a fold change ≥2 and a p value ≤ 0.05. Based on the criteria, we found a total of 1024 differentially expressed mRNAs, of which 213 mRNAs were upregulated and 811 downregulated. We found that 40 differentially expressed genes were overlapped in both penicillin and IFN-γ treatment, indicating that these DEGs may be crucial in the persistent chlamydial infection. We then performed GO gene functional classification and KEGG pathway analysis on these 40 DEGs (Supplementary Figures S1, S2). The most significant GO item was protein refolding. KEGG pathway analysis identified 13 significant pathways with DEGs, while Legionellosis and MAPK signaling pathway were also included. Therefore, we believe that our results provide the data to screen out the crucial genes in the persistent chlamydial infection for future study.
Previous studies suggest that lncRNAs could play a sponge/decoy role, competing with other genes for miRNA binding and therefore reducing the regulatory effect of miRNAs on targeted mRNAs (Klein et al., 2010; Wang et al., 2010; Paraskevopoulou and Hatzigeorgiou, 2016). To fully investigate the potential regulatory mechanism of differentially expressed lncRNAs, the ceRNAs network was constructed. A total of 101334 ceRNA relationships was identified from the interaction of 783 differentially expressed lncRNAs, 395 mRNAs, and 814 miRNAs. In the top 100 relationships, LINC00926:3 and NONHSAT173474.1 were the lncRNAs that link to most nodes. The downregulation of LINC00926:3 could act as a sponge for several miRNAs, which influence the function of BICDL1. BICDL1 may be the component of secretory vesicle machinery regulating the transport of Rab6-containing vesicles (Schlager et al., 2010), while Rab6 has been demonstrated to be a regulator of Chlamydia development (Rejman Lipinski et al., 2009). Thus, the downregulation of BICDL1 in persistent chlamydial infection suggested the slow development of C. trachomatis in the persistent stage. In addition, NONHSAT173474.1 may regulate the function of DUSP4 through the downstream miRNAs. The protein encoded by DUSP4 is a member of the dual-specificity protein phosphatase subfamily. These phosphatases dephosphorylate both the phosphoserine/threonine and phosphotyrosine residues to inactivate their target kinases. They negatively regulate members of the mitogen-activated protein (MAP) kinase superfamily (MAPK/ERK, SAPK/JNK, p38) that are associated with cellular proliferation and differentiation. The decreased expression of DUSP4 in cells persistently infected with C. trachomatis may contribute to chlamydial suppression of host cell proliferation. We then used quantitative real-time PCR to confirm the expression change of a ceRNA interaction, NONHSAT173474.1- has-miR-5088-5P- DUSP4, in persistent-infected cells. Taken together, ceRNAs have an important influence on regulating gene expression at the post-transcriptional level and our results presented a potential regulatory network in persistent chlamydial infection.
A recent study showed that penicillin-binding proteins (PBP) regulate multiple steps in the polarized cell division process of Chlamydia (Cox et al., 2020). Peptidoglycan regulates at least two distinct steps in the polarized division of C. trachomatis and Chlamydia muridarum. Peptidoglycan crosslinking in cells treated with penicillin was prevented by PBP3. Thereby, cells can initiate polarized division, but the process arrests at an early stage of daughter cell growth, indicating that penicillin has an adverse effect on cell division in persistent chlamydial infection. We selected 10 hub genes from the PPI network, and CENPA is the gene with the highest score. This gene encodes a centromere protein which contains a histone H3 related histone fold domain that is required for targeting the centromere, while centromeres are the differentiated chromosomal domains that specify the mitotic behavior of chromosomes. Its downregulation suggested decreased mitosis. Most of the hub genes related to cell division and cell cycle and most of them were detected to be downregulated in persistently infected cells. In persistent infection, Chlamydia slows down DNA replication and continues to transcribe genes, but stops dividing, becoming viable but non-cultivable (Muramatsu et al., 2016). Taken together, our results suggested that the inhibition of cell division might be an important biological phenomenon in the persistent chlamydial infection induced by penicillin.
In summary, our microarray data revealed the dysregulation of lncRNAs and mRNAs in penicillin-induced persistent chlamydial infection in HeLa cells. GO and KEGG pathway analyses were performed to analyze the potential functions of dysregulated mRNAs. LncRNA-miRNA-mRNA networks indicated that the alterations in lncRNA may affect the mRNA transcription and protein translation of vital pathways during the pathogenesis of persistent chlamydial infection. Ten hub genes were selected from the PPI network. Our results provide newly found information regarding the crucial role of lncRNAs in persistent chlamydial infection, which could be beneficial to understand more about the function of lncRNAs and may provide novel insight into the molecular mechanisms during the pathogenesis of persistent chlamydial infection. However, our study has only shown the profile of differentially expressed lncRNAs and mRNAs and screened some potential genes and pathways via bioinformatics analysis. Further studies need to be carried out to identify the function of the differentially expressed lncRNAs that may become novel diagnostic biomarkers for persistent chlamydial infection. We hope that these genes can be validated in human tissues and used to assist the diagnosis of persistent chlamydial infection in humans.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, GSE180478.
XH and QL were the main investigators of the study and drafted the manuscript. RX, XC, and ML contributed in performing the experiment and analyzing the data. JH contributed to the conception and design of the work. HG and CM were the corresponding authors who contributed to the conception and design of the work, funding, and revising the manuscript. All authors approved the version to be published and agreed to be accountable for all aspects of the work.
This study was supported by the Science and Technology Projects in Guangzhou, China (grant number 201807010081).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.744901/full#supplementary-material
Abdelrahman, Y. M., and Belland, R. J. (2005). The Chlamydial Developmental Cycle: Figure 1. FEMS Microbiol. Rev. 29, 949–959. doi:10.1016/j.femsre.2005.03.002
Akhade, V. S., Pal, D., and Kanduri, C. (2017). Long Noncoding RNA: Genome Organization and Mechanism of Action. Adv. Exp. Med. Biol. 1008, 47–74. doi:10.1007/978-981-10-5203-3_2
Beatty, W. L., Belanger, T. A., Desai, A. A., Morrison, R. P., and Byrne, G. I. (1994). Tryptophan Depletion as a Mechanism of Gamma Interferon-Mediated Chlamydial Persistence. Infect. Immun. 62, 3705–3711. doi:10.1128/iai.62.9.3705-3711.1994
Beatty, W. L., Byrne, G. I., and Morrison, R. P. (1993). Morphologic and Antigenic Characterization of Interferon Gamma-Mediated Persistent Chlamydia trachomatis Infection In Vitro. Proc. Natl. Acad. Sci. 90, 3998–4002. doi:10.1073/pnas.90.9.3998
Bhan, A., Soleimani, M., and Mandal, S. S. (2017). Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 77, 3965–3981. doi:10.1158/0008-5472.CAN-16-2634
Bragina, E., Gomberg, M., and Dmitriev, G. (2001). Electron Microscopic Evidence of Persistent Chlamydial Infection Following Treatment. J. Eur. Acad. Dermatol. Venerol 15, 405–409. doi:10.1046/j.1468-3083.2001.00342.x
Capmany, A., and Damiani, M. T. (2010). Chlamydia trachomatis Intercepts Golgi-Derived Sphingolipids through a Rab14-Mediated Transport Required for Bacterial Development and Replication. PLoS One 5, e14084. doi:10.1371/journal.pone.0014084
Chin, C. H., Chen, S. H., Wu, H. H., Ho, C. W., Ko, M. T., and Lin, C. Y. (2014). cytoHubba: Identifying Hub Objects and Sub-networks from Complex Interactome. Bmc Syst. Biol. 8 Suppl 4, S11. doi:10.1186/1752-0509-8-S4-S11
Chowdhury, S. R., Reimer, A., Sharan, M., Kozjak-Pavlovic, V., Eulalio, A., Prusty, B. K., et al. (2017). Chlamydia Preserves the Mitochondrial Network Necessary for Replication via microRNA-dependent Inhibition of Fission. J. Cel Biol. 216, 1071–1089. doi:10.1083/jcb.201608063
Cox, J. V., Abdelrahman, Y. M., and Ouellette, S. P. (2020). Penicillin-binding Proteins Regulate Multiple Steps in the Polarized Cell Division Process of Chlamydia. Sci. Rep. 10, 12588. doi:10.1038/s41598-020-69397-x
Dai, R., and Ahmed, S. A. (2011). MicroRNA, a New Paradigm for Understanding Immunoregulation, Inflammation, and Autoimmune Diseases. Translational Res. 157, 163–179. doi:10.1016/j.trsl.2011.01.007
Derrick, T., Last, A. R., Burr, S. E., Roberts, C. H., Nabicassa, M., Cassama, E., et al. (2016). Inverse Relationship between microRNA-155 and -184 Expression with Increasing Conjunctival Inflammation during Ocular Chlamydia trachomatis Infection. BMC Infect. Dis. 16, 60. doi:10.1186/s12879-016-1367-8
Dzakah, E. E., Huang, L., Xue, Y., Wei, S., Wang, X., Chen, H., et al. (2021). Host Cell Response and Distinct Gene Expression Profiles at Different Stages of Chlamydia trachomatis Infection Reveals Stage-specific Biomarkers of Infection. BMC Microbiol. 21, 3. doi:10.1186/s12866-020-02061-6
Geisler, S., and Coller, J. (2013). RNA in Unexpected Places: Long Non-coding RNA Functions in Diverse Cellular Contexts. Nat. Rev. Mol. Cel Biol 14, 699–712. doi:10.1038/nrm3679
Gérard, H. C., Krausse-Opatz, B., Wang, Z., Rudy, D., Rao, J. P., Zeidler, H., et al. (2001). Expression of Chlamydia trachomatis Genes Encoding Products Required for DNA Synthesis and Cell Division during Active versus Persistent Infection. Mol. Microbiol. 41, 731–741. doi:10.1046/j.1365-2958.2001.02550.x
Guo, L.-L., Song, C. H., Wang, P., Dai, L. P., Zhang, J. Y., and Wang, K. J. (2015). Competing Endogenous RNA Networks and Gastric Cancer. Wjg 21, 11680–11687. doi:10.3748/wjg.v21.i41.11680
Gupta, R., Arkatkar, T., Keck, J., Koundinya, G. K. L., Castillo, K., Hobel, S., et al. (2016). Antigen Specific Immune Response in Chlamydia Muridarum Genital Infection Is Dependent on Murine microRNAs-155 and -182. Oncotarget 7, 64726–64742. doi:10.18632/oncotarget.11461
Ha, T.-Y. (2011). The Role of MicroRNAs in Regulatory T Cells and in the Immune Response. Immune Netw. 11, 11–41. doi:10.4110/in.2011.11.1.11
Igietseme, J. U., Omosun, Y., Stuchlik, O., Reed, M. S., Partin, J., He, Q., et al. (2015). Role of Epithelial-Mesenchyme Transition in Chlamydia Pathogenesis. PLoS One 10, e0145198. doi:10.1371/journal.pone.0145198
Jia, L., Sun, F., Wang, J., Gong, D., and Yang, L. (2019). Chlamydia trachomatis Ct143 Stimulates Secretion of Proinflammatory Cytokines via Activating the P38/MAPK Signal Pathway in THP-1 Cells. Mol. Immunol. 105, 233–239. doi:10.1016/j.molimm.2018.12.007
Klein, U., Lia, M., Crespo, M., Siegel, R., Shen, Q., Mo, T., et al. (2010). The DLEU2/miR-15a/16-1 Cluster Controls B Cell Proliferation and its Deletion Leads to Chronic Lymphocytic Leukemia. Cancer Cell 17, 28–40. doi:10.1016/j.ccr.2009.11.019
Klos, A., Thalmann, J., Peters, J., Gã©rard, H. C., and Hudson, A. P. (2009). The Transcript Profile of persistentChlamydophila(Chlamydia)pneumoniae in Vitrodepends on the Means by Which Persistence Is Induced. Fems Microbiol. Lett. 291, 120–126. doi:10.1111/j.1574-6968.2008.01446.x
Krüll, M., Kramp, J., Petrov, T., Klucken, A. C., Hocke, A. C., Walter, C., et al. (2004). Differences in Cell Activation by Chlamydophila Pneumoniae and Chlamydia trachomatis Infection in Human Endothelial Cells. Infect. Immun. 72, 6615–6621. doi:10.1128/iai.72.11.6615-6621.2004
Kun, D., Xiang-Lin, C., Ming, Z., and Qi, L. (2013). Chlamydia Inhibit Host Cell Apoptosis by Inducing Bag-1 via the MAPK/ERK Survival Pathway. Apoptosis 18, 1083–1092. doi:10.1007/s10495-013-0865-z
Lewis, M. E., Belland, R. J., AbdelRahman, Y. M., Beatty, W. L., Aiyar, A. A., Zea, A. H., et al. (2014). Morphologic and Molecular Evaluation of Chlamydia trachomatis Growth in Human Endocervix Reveals Distinct Growth Patterns. Front. Cel. Infect. Microbiol. 4, 71. doi:10.3389/fcimb.2014.00071
Liu, Y., Hu, C., Sun, Y., Wu, H., Chen, X., and Liu, Q. (2019). Identification of Differentially Expressed Circular RNAs in HeLa Cells Infected with Chlamydia trachomatis. Pathog. Dis. 77, ftz062. doi:10.1093/femspd/ftz062
Mehlitz, A., Eylert, E., Huber, C., Lindner, B., Vollmuth, N., Karunakaran, K., et al. (2017). Metabolic Adaptation ofChlamydia Trachomatisto Mammalian Host Cells. Mol. Microbiol. 103, 1004–1019. doi:10.1111/mmi.13603
Mering, C. v., Huynen, M., Jaeggi, D., Schmidt, S., Bork, P., and Snel, B. (2003). STRING: a Database of Predicted Functional Associations between Proteins. Nucleic Acids Res. 31, 258–261. doi:10.1093/nar/gkg034
Moore, E. R., Fischer, E. R., Mead, D. J., and Hackstadt, T. (2008). The Chlamydial Inclusion Preferentially Intercepts Basolaterally Directed Sphingomyelin-Containing Exocytic Vacuoles. Traffic 9, 2130–2140. doi:10.1111/j.1600-0854.2008.00828.x
Muramatsu, M. K., Brothwell, J. A., Stein, B. D., Putman, T. E., Rockey, D. D., and Nelson, D. E. (2016). Beyond Tryptophan Synthase: Identification of Genes that Contribute to Chlamydia trachomatis Survival during Gamma Interferon-Induced Persistence and Reactivation. Infect. Immun. 84, 2791–2801. doi:10.1128/Iai.00356-16
O'Connell, R. M., Rao, D. S., and Baltimore, D. (2012). microRNA Regulation of Inflammatory Responses. Annu. Rev. Immunol. 30, 295–312. doi:10.1146/annurev-immunol-020711-075013
Paraskevopoulou, M. D., and Hatzigeorgiou, A. G. (2016). Analyzing MiRNA-LncRNA Interactions. Methods Mol. Biol. 1402, 271–286. doi:10.1007/978-1-4939-3378-5_21
Patton, D. L., Askienazy-Elbhar, M., Henry-Suchet, J., Campbell, L. A., Cappuccio, A., Tannous, W., et al. (1994). Detection of Chlamydia trachomatis in Fallopian Tube Tissue in Women with Postinfectious Tubal Infertility. Am. J. Obstet. Gynecol. 171, 95–101. doi:10.1016/s0002-9378(94)70084-2
Rejman Lipinski, A., Heymann, J., Meissner, C., Karlas, A., Brinkmann, V., Meyer, T. F., et al. (2009). Rab6 and Rab11 Regulate Chlamydia trachomatis Development and Golgin-84-dependent Golgi Fragmentation. Plos Pathog. 5, e1000615. doi:10.1371/journal.ppat.1000615
Saka, H. A., and Valdivia, R. H. (2010). Acquisition of Nutrients by Chlamydiae: Unique Challenges of Living in an Intracellular Compartment. Curr. Opin. Microbiol. 13, 4–10. doi:10.1016/j.mib.2009.11.002
Schlager, M. A., Kapitein, L. C., Grigoriev, I., Burzynski, G. M., Wulf, P. S., Keijzer, N., et al. (2010). Pericentrosomal Targeting of Rab6 Secretory Vesicles by Bicaudal-D-Related Protein 1 (BICDR-1) Regulates Neuritogenesis. EMBO J. 29, 1637–1651. doi:10.1038/emboj.2010.51
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 13, 2498–2504. doi:10.1101/gr.1239303
Skilton, R. J., Cutcliffe, L. T., Barlow, D., Wang, Y., Salim, O., Lambden, P. R., et al. (2009). Penicillin Induced Persistence in Chlamydia trachomatis: High Quality Time Lapse Video Analysis of the Developmental Cycle. PLoS One 4, e7723. doi:10.1371/journal.pone.0007723
Stephens, A. J., Aubuchon, M., and Schust, D. J. (2011). Antichlamydial Antibodies, Human Fertility, and Pregnancy Wastage. Infect. Dis. Obstet. Gynecol. 2011, 1–9. doi:10.1155/2011/525182
Tsevat, D. G., Wiesenfeld, H. C., Parks, C., and Peipert, J. F. (2017). Sexually Transmitted Diseases and Infertility. Am. J. Obstet. Gynecol. 216, 1–9. doi:10.1016/j.ajog.2016.08.008
van Ooij, C., Kalman, L., van Ijzendoorn, S., Nishijima, M., Hanada, K., Mostov, K., et al. (2000). Host Cell-Derived Sphingolipids Are Required for the Intracellular Growth of Chlamydia trachomatis. Cell Microbiol 2, 627–637. doi:10.1046/j.1462-5822.2000.00077.x
Vignola, M. J., Kashatus, D. F., Taylor, G. A., Counter, C. M., and Valdivia, R. H. (2010). cPLA2 Regulates the Expression of Type I Interferons and Intracellular Immunity to Chlamydia trachomatis. J. Biol. Chem. 285, 21625–21635. doi:10.1074/jbc.M110.103010
Wang, J., Liu, X., Wu, H., Ni, P., Gu, Z., Qiao, Y., et al. (2010). CREB Up-Regulates Long Non-coding RNA, HULC Expression through Interaction with microRNA-372 in Liver Cancer. Nucleic Acids Res. 38, 5366–5383. doi:10.1093/nar/gkq285
Wang, X., Song, X., Glass, C. K., and Rosenfeld, M. G. (2011). The Long Arm of Long Noncoding RNAs: Roles as Sensors Regulating Gene Transcriptional Programs. Cold Spring Harbor Perspect. Biol. 3, a003756. doi:10.1101/cshperspect.a003756
Wen, Y., Luo, F., Zhao, Y., Su, S., Shu, M., and Li, Z. (2020). Chlamydia trachomatis Plasmid-Encoded Protein pORF5 Activates Unfolded Protein Response to Induce Autophagy via MAPK/ERK Signaling Pathway. Biochem. Biophysical Res. Commun. 527, 805–810. doi:10.1016/j.bbrc.2020.04.117
Witkin, S. S., Minis, E., Athanasiou, A., Leizer, J., and Linhares, I. M. (2017). Chlamydia trachomatis: the Persistent Pathogen. Clin. Vaccin. Immunol 24, e00203–17. doi:10.1128/CVI.00203-17
Xue, Y., Zheng, H., Mai, Z., Qin, X., Chen, W., Huang, T., et al. (2017). An In Vitro Model of Azithromycin-Induced Persistent Chlamydia trachomatis Infection. FEMS Microbiol. Lett. 364, 1–8. doi:10.1093/femsle/fnx145
Yeruva, L., Pouncey, D. L., Eledge, M. R., Bhattacharya, S., Luo, C., Weatherford, E. W., et al. (2017). MicroRNAs Modulate Pathogenesis Resulting from Chlamydial Infection in Mice. Infect. Immun. 85, e00768–16. doi:10.1128/IAI.00768-16
Zhou, H., Huang, Q., Li, Z., Wu, Y., Xie, X., Ma, K., et al. (2013). PORF5 Plasmid Protein of Chlamydia trachomatis Induces MAPK-Mediated Pro-inflammatory Cytokines via TLR2 Activation in THP-1 Cells. Sci. China Life Sci. 56, 460–466. doi:10.1007/s11427-013-4470-8
Keywords: lncRNAs, mRNAs, Chlamydia trachomatis, persistent infection, microarray, bioinformatics analysis
Citation: Huang X, Liufu Q, Xu R, Chen X, Liu M, Han J, Guan H and Ma C (2022) Integrating lncRNAs and mRNAs Expression Profiles in Penicillin-Induced Persistent Chlamydial Infection in HeLa Cells. Front. Mol. Biosci. 9:744901. doi: 10.3389/fmolb.2022.744901
Received: 21 July 2021; Accepted: 31 January 2022;
Published: 16 February 2022.
Edited by:
Prasun K. Datta, Tulane University, United StatesReviewed by:
Guangming Zhong, The University of Texas Health Science Center at San Antonio, United StatesCopyright © 2022 Huang, Liufu, Xu, Chen, Liu, Han, Guan and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunguang Ma, bWFjaHVuZ0BtYWlsLnN5c3UuZWR1LmNu; Hongyu Guan, Z2hvbmd5QG1haWwuc3lzdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.