
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci., 05 December 2022
Sec. RNA Networks and Biology
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.1077968
This article is part of the Research TopicTumorigenesis Regulated by miRNAs, Volume IIView all 5 articles
 Soudeh Ghafouri-Fard1
Soudeh Ghafouri-Fard1 Arian Askari2
Arian Askari2 Bashdar Mahmud Hussen3,4
Bashdar Mahmud Hussen3,4 Mohammed Fatih Rasul5
Mohammed Fatih Rasul5 Sevak Hatamian6
Sevak Hatamian6 Mohammad Taheri7,8*
Mohammad Taheri7,8* Arda Kiani9*
Arda Kiani9*miR-671 is encoded by a gene on 7q36.1 and contributes to the pathogenesis of a variety of disorders, including diverse types of cancers, atherosclerosis, ischemic stroke, liver fibrosis, osteoarthritis, Parkinson’s disease, rheumatoid arthritis, acute myocardial infarction and Crohn’s disease. In the context of cancer, different studies have revealed opposite roles for this miRNA. In brief, it has been shown to be down-regulated in pancreatic ductal carcinoma, ovarian cancer, gastric cancer, osteosarcoma, esophageal squamous cell carcinoma and myelodysplastic syndromes. Yet, miR-671 has been up-regulated in glioma, colorectal cancer, prostate cancer and hepatocellular carcinoma. Studies in breast, lung and renal cell carcinoma have reported inconsistent results. The current review aims at summarization of the role of miR-671 in these disorders focusing on its target mRNA in each context and dysregulated signaling pathways. We also provide a summary of the role of this miRNA as a prognostic factor in malignancies.
microRNAs (miRNAs) are small-sized non-coding RNAs that partake in the post-transcriptional regulation of gene expression through influencing the stability and translation of transcripts. They are transcribed by RNA polymerase II. The pri-miRNAs produced by this enzyme is capped and polyadenylated. This transcript undergoes a series of cleavage by the Drosha and cytoplasmic Dicer ribonuclease enzymes to produce the stem-loop precursor miRNA and mature miRNA, respectively. The latter is embraced into a RNA-induced silencing complex which can recognize target mRNAs and suppress its translation or destabilize it (Macfarlane and Murphy, 2010). miRNAs participate in the pathoetiology of several disorders through modulation of expression of genes (Hussen et al., 2021), altering signaling pathways (Hussen et al., 2022) or interactions with other types of non-coding RNAs (Ghafouri-Fard et al., 2021a; Taheri et al., 2022).
miR-671 is encoded by a gene on 7q36.1 and involved in the pathogenesis of a range of disorders, including diverse types of cancers, atherosclerosis, ischemic stroke, liver fibrosis, osteoarthritis, Parkinson’s disease, rheumatoid arthritis, acute myocardial infarction and Crohn’s disease. There is not sufficient data about the role of this miRNA in normal physiological processes. However, differential expression of this miRNA in the visceral adipose tissues of patients with non-alcoholic fatty liver disease (Estep et al., 2010) indicates its possible role in metabolic pathways. Moreover, miR-671 has been shown to down-regulate the CDR1 (Cerebellar Degeneration-Related protein 1) gene through an Ago2-slicer-dependent mechanism (Hansen et al., 2011). Moreover, this miRNA has been found to be mainly localized in the nucleus (Hansen et al., 2011). There is no clear evidence about differential expression or functional roles of miR-671-3p versus miR-671-5p. The current review aims at summarization of the role of miR-671 in these disorders focusing on its target mRNA in each context and dysregulated signaling pathways. We also provide a summary of the role of this miRNA as a prognostic factor in malignancies.
The influence of miR-671 in the carcinogenesis has been valued by a number of studies in cancer cell lines, animal models of cancers and samples obtained from affected individuals. In the succeeding sections, we define the role of miR-671 in the carcinogenesis based on these three lines of evidence.
Studies in colorectal cancer cell lines have shown down-regulation of circ_PTPRA. Exosomal circ_PTPRA has been shown to induce cell cycle arrest and inhibit proliferation of colorectal cancer cells. In addition, exosomal circ_PTPRA could promote sensitivity of these cells to radiation, resulting in inhibition of colony formation and induction of apoptosis. Mechanistically, circ_PTPRA functions as a sponge for miR-671-5p to increase SMAD4 levels. Taken together, circ_PTPRA inhibits growth and radioresistance of colorectal cancer cells through down-regulation of miR-671-5p levels. Moreover, suppression of miR-671-5p has also blocked growth and radioresistance of these cells through enrichment of expression of SMAD4 (Yang et al., 2022b). Another study in this type of cancer has shown overexpression of a circular RNA, namely circGLIS2. This circRNA is sponged by miR-671. Over-expression of circGLIS2 has led to activation of NF-ƙB pathway and induction of production of pro-inflammatory chemokines leading to stimulation of tumor-associated inflammatory responses via recruitment of leukocytes. Taken together, circGLIS2 activates NF-ƙB signaling and promotes migratory ability of colorectal cancer cells through adsorbing miR-671 (Figure 1) (Chen et al., 2020a). Another functional study in colorectal cancer cells has shown the effect of miR-671-5p up-regulation in enhancement of cell proliferation, migratory capacity, and invasiveness of these cells, whereas its downregulation has led to reverse effects. Therefore, miR-671-5p has been suggested as an oncogenic miRNA in colon cancer which exerts its effects through targeting Tripartite Motif Containing 67 (TRIM67) (Jin et al., 2019), a gene, that is, possibly involved in zinc ion binding activity, regulation of protein localization and negative regulation of Ras protein signal transduction (https://www.genecards.org/cgi-bin/carddisp.pl?gene=TRIM67).
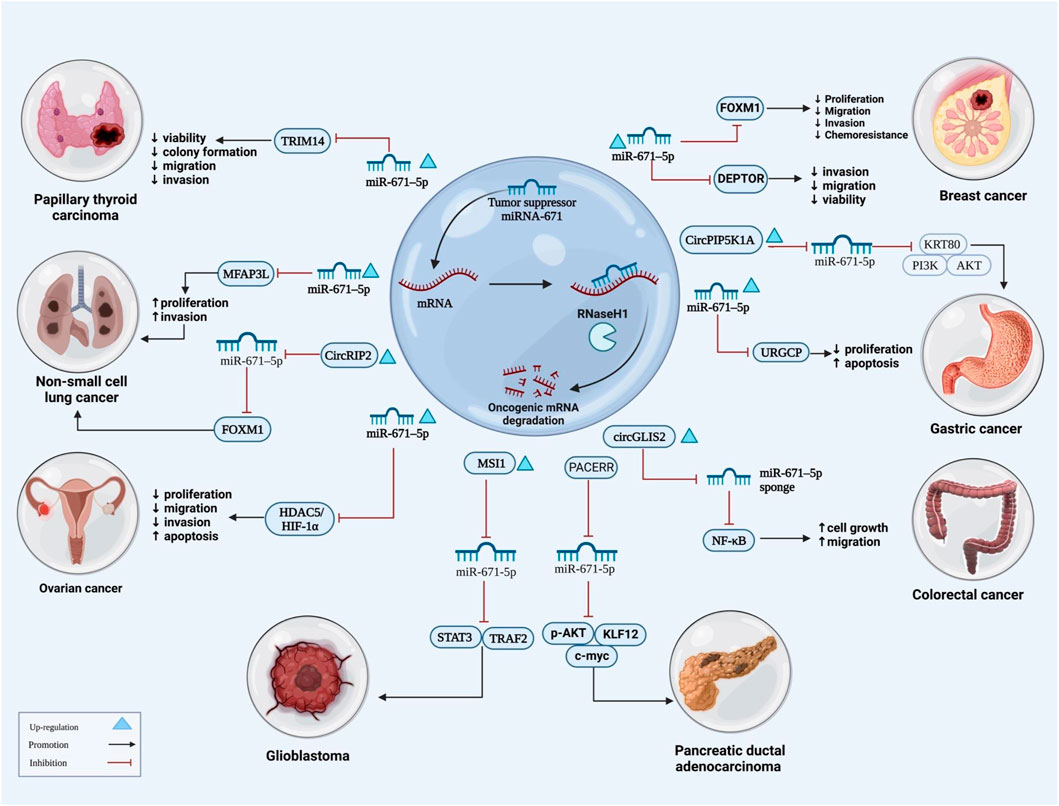
FIGURE 1. The illustration shows signaling pathways underlying the role of miRNA-671 is as a tumor suppressor miRNA in cancers. miRNA-671 inhibits many signaling pathways and carcinogenic mRNAs, resulting in increased apoptosis while lowering proliferation, migration, and invasion of cancer cells.
miR-671-3p has also been shown to exert oncogenic roles in glioma cells through targeting CKAP4 (Lu et al., 2018). Moreover, it has been demonstrated to be sponged by the tumor suppressor circRNA circDLC1 in these cells (Wu et al., 2022a). A single study in lung cancer cells has shown that miR-671-3p enhances progression of lung cancer through blocking expression of FOXP2 expression in lung cancer (Li et al., 2019b), thus referring to an oncogenic role for this miRNA in lung cancer.
Two independent studies in glioblastoma cell lines have revealed that miR-671-5p has transforming roles. Firstly, more than two-fold upregulated levels of miR-671-5p reduced levels of CDR1-AS/VSNL1 in glioblastoma cell lines A172, CAS-1 and DBTRG. This phenomenon is associated with increased migration and proliferation (Barbagallo et al., 2016). In another study it was demonstrated that if upregulated, miR-671-5p has oncogenic roles, but with competing endogenous features of Circular RNA circ_0001946, this miRNA is suppressed and its suppression is in favor of benign properties (Li and Diao, 2019).
Prostate cancer related bioinformatics analysis has shown that miR-671-5p is amongst top differentially expressed miRNAs (Zhu et al., 2020). miR-671-5p has a binding site on the 3′-UTR region of NFIA (Zhu et al., 2020). According to Yang et al., NFIA acts as a tumor suppressor gene in glioma and squamous carcinoma (Yang et al., 2018). Upregulation of miR-671-5p in prostate cancer cell lines reduces NFIA/CRYAB levels and contributes to malignant features like increased proliferation, migration and invasion (Figure 2) (Zhu et al., 2020).
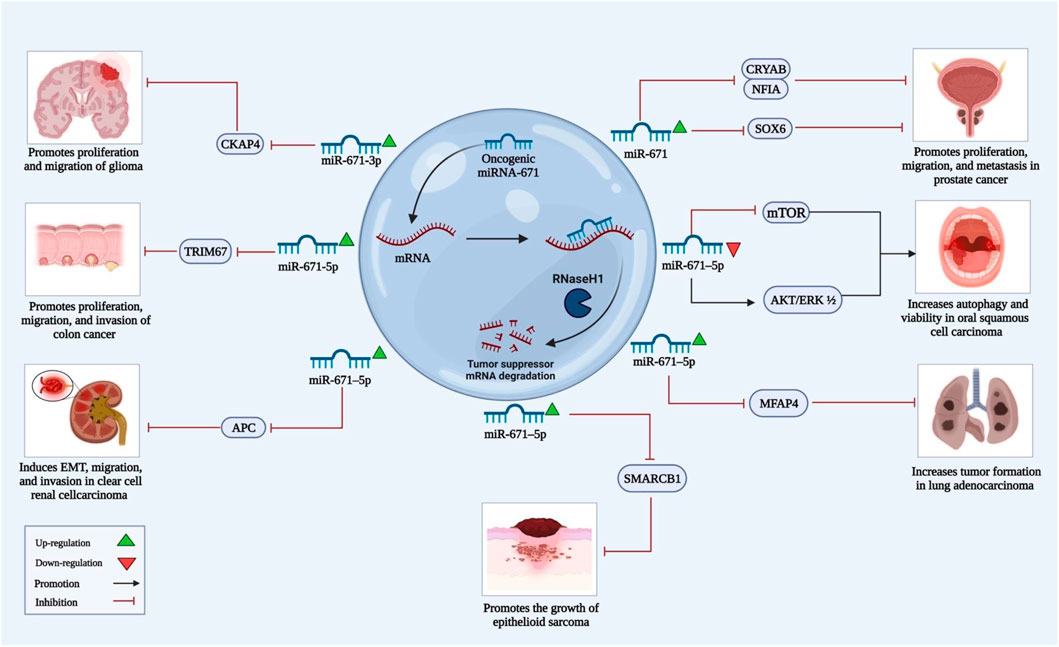
FIGURE 2. The above illustration shows the roles of miRNA-671, which acts as an oncogene and stimulates cancer growth in several types of cancer. MiRNA-671 can target different tumor suppressor mRNAs and, through inhibition of translation, increase proliferation, migration, and invasion of cancer cells.
In kidney cancers category, miR-671-5p has been shown to be overexpressed patterns in clear cell renal cell carcinoma (ccRCC) cell lines (786-O and CAKI-1) (Chi et al., 2020). Its overexpression is regulated by HMGA1, which involves in chromatin remodeling (Chiefari et al., 2018). Upregulated levels of miR-671-5p targets APC (a tumor suppressor gene) and gives rise to invasiveness of ccRCC cells (Chi et al., 2020).
The lncRNA PACERR that sponges miR-671 has been shown to increase the number of M2-polarized cells and enhance proliferation, invasiveness and migration of pancreatic cancer cells. From a mechanistical point of view, PACERR has a role in activation of KLF12/p-AKT/c-myc pathway through sponging miR-671-3p. In fact, this lncRNA is regarded as a regulator of tumor-associated macrophages in pancreatic ductal carcinoma microenvironment (Liu et al., 2022b). Moreover, circ_0092314 has been identified as another non-coding RNA that sponges miR-671 in pancreatic cancer cells, thus increasing expression of S100P and inducing epithelial-mesenchymal transition (EMT) (Shen et al., 2021). These two studies have designated a tumor suppressor effect for miR-671 in pancreatic cancer.
The miR-671-sponging circRNA Circ_0001946 has been shown to be over-expressed in tamoxifen resistant breast cancer cells. This circRNA has been shown to be activated by YY1 in these cells. miR-671-5p mimics could partially reverse the effects of circ_0001946 up-regulation in enhancement of proliferation and invasive properties of drug-resistant breast cancer cells. EGFR has been shown to be the downstream target of miR-671-5p in these cells (Gao et al., 2022). Another study has shown the sponging effect of circSLC8A1 on miR-671 and the impact of this miRNA in the regulation of PTEN/PI3k/AKT pathway (Zhu et al., 2021). Moreover, miR-671-3p has been shown to suppress proliferation and invasiveness of breast cancer cells through modulation of expression of the MTOR-interacting protein DEPTOR (Xia et al., 2020).
Lung cancer cells have also been the subject of functional studies on the role of miR-671. As an example of these studies, Liu et al. (2022a) have shown that the oncogenic role of circRIP2 in this type of cancer is exerted through sequestering miR-671-5p and increasing expression of FOXM1. Moreover, miR-671-5p has been found to inhibit proliferation, migration and invasive aptitude of lung cancer cells through targeting MFAP3L (Ye et al., 2022).
In esophageal squamous cell carcinoma cell lines (including different subtypes of KYSE), elevated levels of FGFR2 activates ERK and AKT signaling pathway and contributes to the malignancy (Li et al., 2019a). Interestingly, miR-671-5p level has shown to be downregulated, hence acting as a tumor suppressor (Li et al., 2019a). Forced expression of this miRNA contributes to diminished levels of FGFR phosphorylation, thus reversing malignant features like proliferation and migration (Li et al., 2019a).
Downregulated levels of miR-671 have also been shown in gastric cancer. In a study conducted by Qiu et al. (2018), reduced level of miR-671-5p has been demonstrated in MKN28 cells compared with normal gastric cells, suggesting an anti-tumor role. Elevating its expression yields decreased ratio of Bcl-2/Bax (increase in BAX), thus promoting apoptosis (Qiu et al., 2018). miR-671-5p targets URGCP and inhibits its expression in MKN28 cells (Qiu et al., 2018). Considering the roles of Up regulator Of Cell Proliferation (URGCP) in the carcinogenesis (Xie et al., 2012; Cai et al., 2015), there is no surprise that targeting it by miR-671-5p has shifted MKN28 cells to normal cell features (Qiu et al., 2018).
Detailed information about the roles of miR-671 in different cancer cell lines is shown in Table 1.
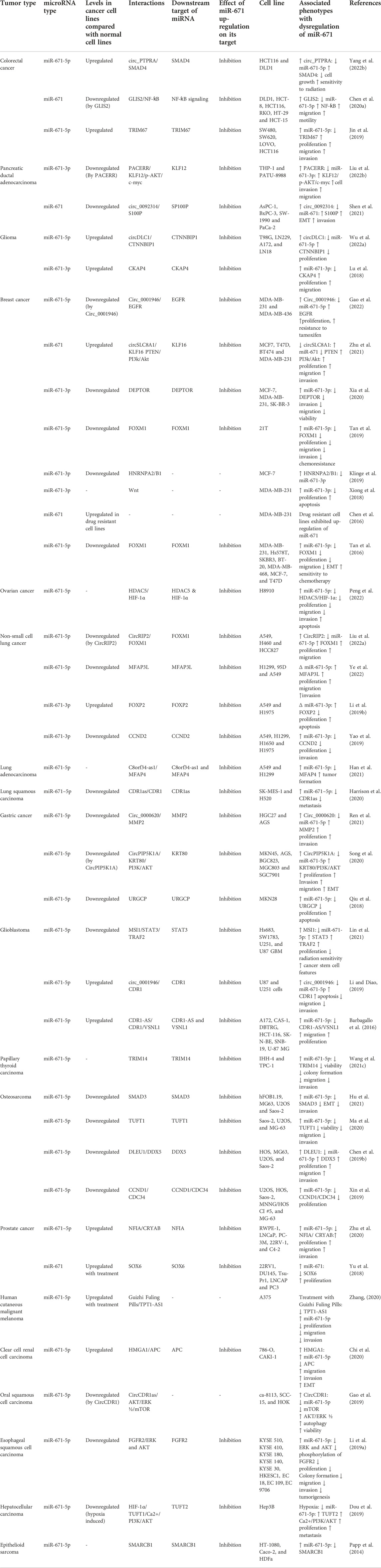
TABLE 1. Function of miR-671 in cancer cell lines (Arrows indicate the effects of changes in the expression of mentioned genes (either endogenous or exogenous). ∆: knockdown or downregulation, MPP+: 1-methyl-4-phenylpyridinium).
Different animal studies have been performed to evaluate the impact of miR-671 dysregulation on the course of tumor formation. Moreover, a number of other studies have focused on circRNAs that act as molecular sponges for miR-671. For instance, up-regulation of circ_00923 in pancreatic cancer cells has led to down-regulation of miR-671 in tissues of affected animals and enhancement of tumor growth (Shen et al., 2021). On the other hand, over-expression of circ_0001946 has resulted in reduction of glioma growth in animal models (Li and Diao, 2019) Similar to cell line studies, studies in xenograft models of cancers have indicated different results regarding the oncogenic versus tumor suppressor effect of miR-671 (Table 2). For instance, in pancreatic cancer models, down-regulation of miR-671 has been associated with enhancement of tumor growth (Shen et al., 2021). Similar results have been obtained in xenograft models of ovarian cancer (Peng et al., 2022). On the other hand, studies in animal models of colorectal cancer have reported opposite results (Yang et al., 2022b). Detailed information about the role of miR-671 in animal models of cancer is presented in Table 2.
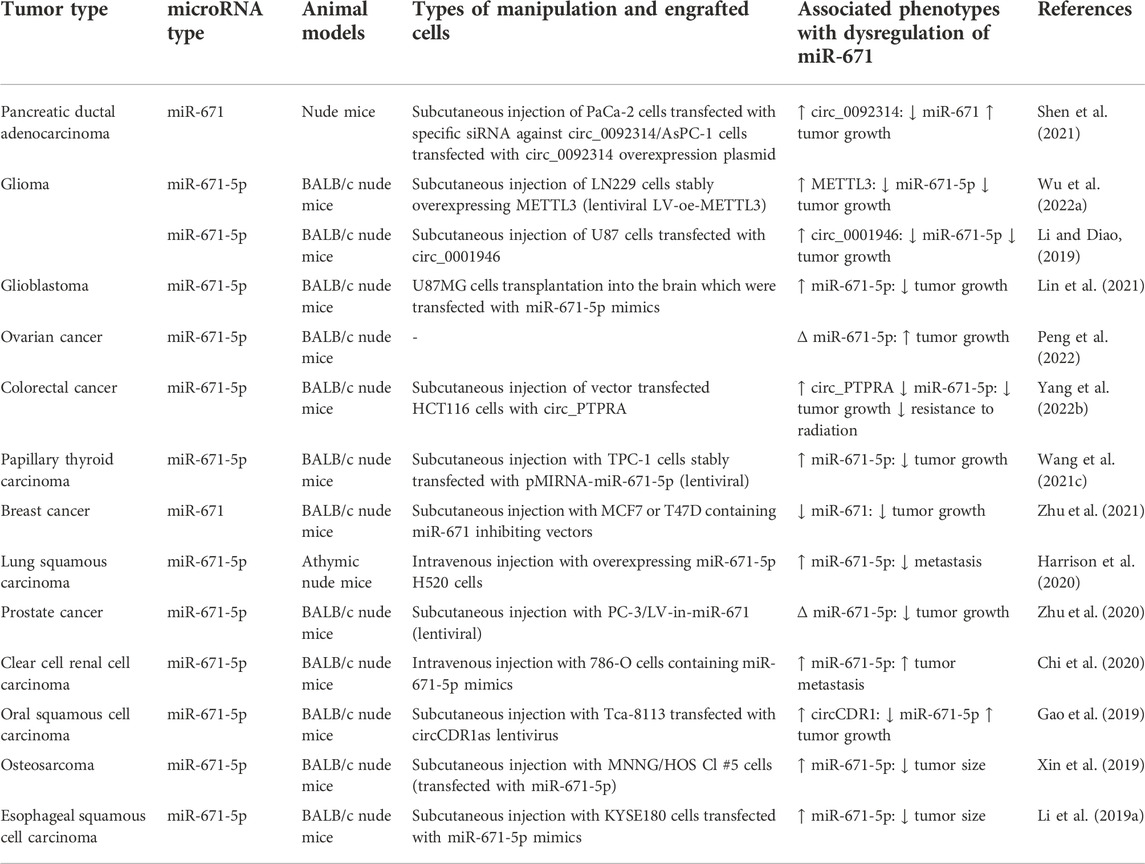
TABLE 2. Effect of miR-671 in cancer development based on research in animal models. (∆: knockdown or downregulation).
Expression of miR-671-5p has been increased in colon cancer tissues. Notably, up-regulation of miR-671-5p in this type of cancer has been associated with involvement of lymph nodes, TNM stage, and low overall survival time of affected individuals (Jin et al., 2019). In tumor associated macrophages of pancreatic cancer patients, the lncRNA PACERR that sponges miR-7671 has been shown to be over-expressed in association with poor prognosis of patients (Liu et al., 2022b).
Studies in clinical samples of breast cancer have reported different results regarding the expression of miR-671. First, the miR-671-sponging circRNA circ_0001946 has been shown to be over-expressed in breast cancer tissues, leading to down-regulation of miR-671 (Gao et al., 2022). Although two other studies have reported down-regulation of miR-671-3p (Xiong et al., 2018) and miR-671-5p (Tan et al., 2016) in breast cancer samples, another study has demonstrated up-regulation of miR-671 in another cohort of breast cancer patients (Zhu et al., 2021).
Several studies have shown the impact of miR-671 dysregulation on survival of patients with different kinds of cancer, including ovarian, colorectal and lung cancers as well as osteosarcoma (Table 3). However, a single study in breast cancer patients has reported lack of association between expression levels of miR-671 and median survival of patients (Xiong et al., 2018). Moreover, abnormal expression of miR-671 has been associated with tumor size, TNM stage or metastasis in some kind of cancers, such as colorectal cancer (Jin et al., 2019), lung cancer (Ye et al., 2022) and renal cell carcinoma (Chi et al., 2020). In prostate cancer, up-regulation of miR-671-5p has been associated with higher Gleason score, and BCR status and poor prognosis, but not with tumor stage and lymph node metastasis (Zhu et al., 2020).
Association between miR-671 variants and risk of soft tissue sarcomas has been assessed in a population of Chinese patients and healthy controls. The results of this study has shown association between miR-671 rs1870238 GC + CC and miR-671 rs2446065 CG + GG genotypes and risk of this type of tumor after adjustment for age and smoking (Zhang et al., 2022a).
Experiments in ox-LDL-treated HUVECs have shown down-regulation of miR-671-5p and up-regulation of circPTPRA expression. These two transcripts have been shown to interact with each other. While circPTPRA silencing has reversed ox-LDL-induced decrease in viability of HUVECs, miR-671-5p downregulation could abolish this effect. Cumulatively, circPTPRA silencing can protect against ox-LDL-associated HUVECs damage through enhancing expression of miR-671-5p (Luo and Zhou, 2022).
Another study has shown that the effects of ANRIL silencing in alleviation of neuroinflammatory responses in ischemia is mediated through influencing the miR-671-5p/NF-κB axis (Figure 3) (Deng et al., 2022). Moreover, miR-671-5p could attenuates neuroinflammation through suppression of NF-κB levels (Deng et al., 2021).
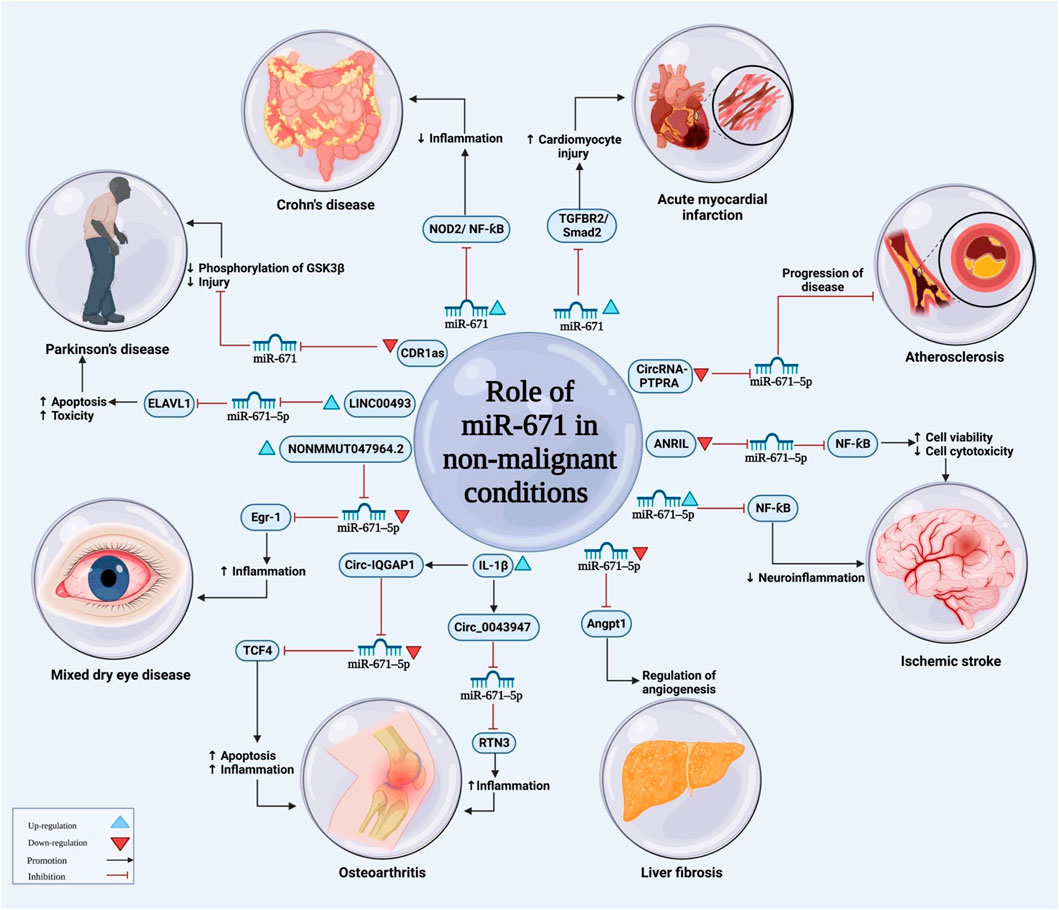
FIGURE 3. The illustration represents the main functions of miRNA-671 in non-malignant disorders. The level of miRNA-671 expression influences the development of a wide range of disorders by modulation of several signaling pathways.
miR-671-5p expression has been revealed to be reduced in S1P-induced hepatic stellate cells and TGFβ1-activated hepatic sinusoidal endothelial cells. Moreover, its expression has been negatively correlated with levels of Angpt1 and VWF. Mechanistically, miR-671-5p could target Angpt1 and VWF (Yang et al., 2022b).
miR-671-5p has also been shown to facilitate the effect of lncRNA DLEU1 in the regulation of chondrocytes proliferation, inflammatory responses, and degradation of extracellular matrix (Wu et al., 2022b). Moreover, the sponging effect of circ_0043947 on miR-671-5p is involved in the pathoetiology of IL1β-induced chondrocyte damage and pathogenesis of osteoarthritis (He et al., 2022). Table 4 summarizes the role of miR-671 in the pathogenesis of non-malignant conditions based on the results of cell line studies.
Expression of miR-671-5p has been down-regulated in the mouse fibrotic liver. Notably, its levels have been negatively correlated with expressions of Angpt1, VWF, sphingosine kinase-1, TGFβ1, HIF1α, HIF2α, and markers of fibrosis. Moreover, expression of miR-671-5p has been lower in hepatic sinusoidal endothelial cells and hepatic stellate cells of CCl4 mice compared with control mice. Administration of miR-671-5p agomir could decrease expressions of Anpgt1 and VWF mRNA and protein levels, and attenuate angiogenesis and fibrosis in the liver of animal models (Yang et al., 2022b). Other investigations in animal models of ischemic stroke, mixed dry eye disease, podocyte injury, acute myocardial infarction and osteoarthritis have verified the role of miR-671 in the pathogenesis of these disorders (Table 5).
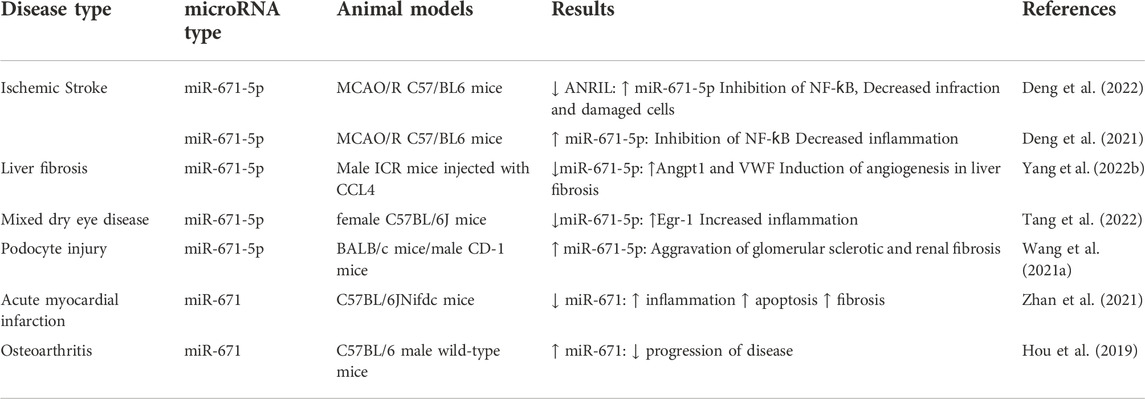
TABLE 5. Animal studies on the role of miR-671 in non-malignant conditions (MCAO: middle cerebral artery occlusion-reperfusion).
A high throughput sequencing study in pseudoexfoliation syndrome has led to identification of four aberrantly expressed miRNAs among them being miR-671-3p (Tomczyk-Socha et al., 2022). miR-671-5p has also been among miRNAs participating in the pathogenesis of periodontitis through establishment of ceRNA regulatory network regulating autophagy (Bian et al., 2022). miR-671 has also been found to be down-regulated in patients with rheumatoid arthritis (Tang et al., 2019), hand, foot, and mouth disease (Lin et al., 2020), placenta accreta spectrum (Chen et al., 2020b), coronary artery disease (Zhong et al., 2020), Parkinson’s disease (Uwatoko et al., 2019) and Kawasaki disease (Zhang et al., 2018). Table 6 shows the detailed information about the role of this miRNA in human diosrders.
Expression levels of miR-671 can be used as diagnostic marker in placenta accreta spectrum, osteoarthritis and hand, foot, and mouth disease (Table 7). The best AUC values have been obtained in extremely severe cases of hand, foot, and mouth disease where mir-671 levels could differentiate this condition from healthy status with AUC value of 1.00 (Jia et al., 2014).
miR-671 is a miRNA with various roles in human disorders. In the context of cancer, different studies have revealed opposite roles for this miRNA. In brief, it has been shown to be down-regulated in pancreatic ductal carcinoma, ovarian cancer, gastric cancer, osteosarcoma, esophageal squamous cell carcinoma and myelodysplastic syndromes. Yet, miR-671 has been up-regulated in glioma, colorectal cancer, prostate cancer and hepatocellular carcinoma. Studies in breast, lung and renal cell carcinoma have reported inconsistent results which cannot be explained by the differences in the roles of miR-671-3p or miR-671-5p. It is possible that this miRNA exert stage- or grade-specific roles in the carcinogenesis.
miR-671 has functional interactions with circ_PTPRA, circ_0092314, circDLC1, circ_0001946, circSLC8A1, circRIP2, circ_0000620, circPIP5K1A and circCDR1as. In fact, these circRNAs act as molecular sponges for miR-671 to influence expression of miR-671 targets. NF-ƙB, EGFR, PTEN/PI3K/AKT, Wnt, HIF-1α, STAT3 and AKT/ERK/mTOR signaling pathways are among those being influenced by dysregulation of miR-671 in different cancers. Moreover, miR-671 has a role in the regulation of EMT in different tissues. This finding is based on functional studies on the role of this miRNA or circRNAs that sponge this miRNA. Thus, miR-671-targetin therapies might affect progression of cancer, invasiveness and metastatic ability of malignant cells.
miR-671 has also been suggested to predict course of cancers originated from different tissues. This speculation is based on the observed associations between dysregulation of this miRNA and survival of patients as well as correlation between its expression levels and clinicopathological data. However, the role of miR-671 as a diagnostic marker for cancers should be investigated in future. Based on the inconsistencies regarding the exact effects of miR-671 in the development and progression of different cancers, it is not expected that miR-671-targetted therapies enter the clinics in near future. More researches are needed to assign a definite role for this miRNA in each type of cancer.
The impact of miR-671 polymorphisms on risk of cancers has only assessed in sarcoma. Similar studies should be conducted to evaluate the association between these polymorphisms and risk of other cancers.
miR-671 has also a fundamental role in the pathophysiology of non-malignant conditions such as atherosclerosis, ischemic stroke, liver fibrosis, osteoarthritis, Parkinson’s disease, rheumatoid arthritis, acute myocardial infarction and Crohn’s disease. Moreover, it has a potential to be used as a diagnostic marker for placenta accreta spectrum, osteoarthritis and hand, foot, and mouth disease. However, dysregulation of miR-671 in malignant and non-malignant disorders originated from a certain tissue complicates the diagnostic application of this miRNA. Meanwhile, contribution of miR-671 to the pathogenesis of both malignant and non-malignant diseases is best explained by the prominent role of this miRNA in the regulation of activity of signaling pathways the control cell proliferation and apoptosis.
Taken together, miR-671 is a miRNA that can affect several target mRNAs and influence activity of signaling pathways that are involved in a variety of human disorders. However, several questions should be answered in order to propose miR-671-targeted therapies as efficient therapies for human disorders.
SG-F wrote the draft and revised it. MT designed and supervised the study. AA, BH, and AK collected the data and designed the figures and tables. All the authors read the submitted version and approved it.
This study was financially supported by Shahid Beheshti University of Medical Sciences.
The authors would like to thank the clinical research Development Unit (CRDU) of Loghman Hakim Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran for their support, cooperation and assistance throughout the period of study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ali, S. A., Gandhi, R., Potla, P., Keshavarzi, S., Espin-Garcia, O., Shestopaloff, K., et al. (2020). Sequencing identifies a distinct signature of circulating microRNAs in early radiographic knee osteoarthritis. Osteoarthr. Cartil. 28, 1471–1481. doi:10.1016/j.joca.2020.07.003
Barbagallo, D., Condorelli, A., Ragusa, M., Salito, L., Sammito, M., Banelli, B., et al. (2016). Dysregulated miR-671-5p/CDR1-AS/CDR1/VSNL1 axis is involved in glioblastoma multiforme. Oncotarget 7, 4746–4759. doi:10.18632/oncotarget.6621
Bian, M., Wang, W., Song, C., Pan, L., Wu, Y., and Chen, L. (2022). Autophagy-related genes predict the progression of periodontitis through the ceRNA network. J. Inflamm. Res. 15, 1811–1824. doi:10.2147/JIR.S353092
Borze, I., Scheinin, I., Siitonen, S., Elonen, E., Juvonen, E., and Knuutila, S. (2011). miRNA expression profiles in myelodysplastic syndromes reveal Epstein-Barr virus miR-BART13 dysregulation. Leuk. Lymphoma 52, 1567–1573. doi:10.3109/10428194.2011.568652
Cai, J., Li, R., Xu, X., Zhang, L., Wu, S., Yang, T., et al. (2015). URGCP promotes non-small cell lung cancer invasiveness by activating the NF-κB-MMP-9 pathway. Oncotarget 6, 36489–36504. doi:10.18632/oncotarget.5351
Chen, B., Gao, T., Yuan, W., Zhao, W., Wang, T. H., and Wu, J. (2019a). Prognostic value of survival of MicroRNAs signatures in non-small cell lung cancer. J. Cancer 10, 5793–5804. doi:10.7150/jca.30336
Chen, J., Yang, X., Liu, R., Wen, C., Wang, H., Huang, L., et al. (2020a). Circular RNA GLIS2 promotes colorectal cancer cell motility via activation of the NF-κB pathway. Cell. Death Dis. 11, 788. doi:10.1038/s41419-020-02989-7
Chen, L., Huang, H., Chen, L., Xu, L., Chen, J., and Lu, Q. (2021). circ-PTTG1IP/miR-671-5p/TLR4 axis regulates proliferation, migration, invasion and inflammatory response of fibroblast-like synoviocytes in rheumatoid arthritis. Gen. Physiol. Biophys. 40, 207–219. doi:10.4149/gpb_2021014
Chen, R., Chen, M., Xiao, Y., Liang, Q., Cai, Y., Chen, L., et al. (2018). Bioinformatics analysis of microRNAs related to blood stasis syndrome in diabetes mellitus patients. Biosci. Rep. 38, BSR20171208. doi:10.1042/BSR20171208
Chen, S., Pang, D., Li, Y., Zhou, J., Liu, Y., Yang, S., et al. (2020b). Serum miRNA biomarker discovery for placenta accreta spectrum. Placenta 101, 215–220. doi:10.1016/j.placenta.2020.09.068
Chen, X., Lu, P., Wang, D. D., Yang, S. J., Wu, Y., Shen, H. Y., et al. (2016). The role of miRNAs in drug resistance and prognosis of breast cancer formalin-fixed paraffin-embedded tissues. Gene 595, 221–226. doi:10.1016/j.gene.2016.10.015
Chen, X., Zhang, C., and Wang, X. (2019b). Long noncoding RNA DLEU1 aggravates osteosarcoma carcinogenesis via regulating the miR-671-5p/DDX5 axis. Artif. Cells Nanomed. Biotechnol. 47, 3322–3328. doi:10.1080/21691401.2019.1648285
Chi, X. G., Meng, X. X., Ding, D. L., Xuan, X. H., Chen, Y. Z., Cai, Q., et al. (2020). HMGA1-mediated miR-671-5p targets APC to promote metastasis of clear cell renal cell carcinoma through Wnt signaling. Neoplasma 67, 46–53. doi:10.4149/neo_2019_190217N135
Chiefari, E., Foti, D. P., Sgarra, R., Pegoraro, S., Arcidiacono, B., Brunetti, F. S., et al. (2018). Transcriptional regulation of glucose metabolism: The emerging role of the HMGA1 chromatin factor. Front. Endocrinol. 9, 357. doi:10.3389/fendo.2018.00357
Chuang, A. Y., Chuang, J. C., Zhai, Z., Wu, F., and Kwon, J. H. (2014). NOD2 expression is regulated by microRNAs in colonic epithelial HCT116 cells. Inflamm. Bowel Dis. 20, 126–135. doi:10.1097/01.MIB.0000436954.70596.9b
della Vittoria Scarpati, G., Falcetta, F., Carlomagno, C., Ubezio, P., Marchini, S., de Stefano, A., et al. (2012). A specific miRNA signature correlates with complete pathological response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Int. J. Radiat. Oncol. Biol. Phys. 83, 1113–1119. doi:10.1016/j.ijrobp.2011.09.030
Deng, L., Guo, Y., Liu, J., Wang, X., Chen, S., Wang, Q., et al. (2021). miR-671-5p attenuates neuroinflammation via suppressing NF-κB expression in an acute ischemic stroke model. Neurochem. Res. 46, 1801–1813. doi:10.1007/s11064-021-03321-1
Deng, L., Jiang, J., Chen, S., Lin, X., Zuo, T., Hu, Q., et al. (2022). Long non-coding RNA ANRIL downregulation alleviates neuroinflammation in an ischemia stroke model via modulation of the miR-671-5p/NF-κB pathway. Neurochem. Res. 47, 2002–2015. doi:10.1007/s11064-022-03585-1
Dou, C., Zhou, Z., Xu, Q., Liu, Z., Zeng, Y., Wang, Y., et al. (2019). Hypoxia-induced TUFT1 promotes the growth and metastasis of hepatocellular carcinoma by activating the Ca(2+)/PI3K/AKT pathway. Oncogene 38, 1239–1255. doi:10.1038/s41388-018-0505-8
Estep, M., Armistead, D., Hossain, N., Elarainy, H., Goodman, Z., Baranova, A., et al. (2010). Differential expression of miRNAs in the visceral adipose tissue of patients with non-alcoholic fatty liver disease. Aliment. Pharmacol. Ther. 32, 487–497. doi:10.1111/j.1365-2036.2010.04366.x
Gao, G., Li, X., Zhang, J., and Yu, H. (2022). YY1 as a promoter regulating the circ_0001946/miR-671-5p/EGFR axis to promote chemotherapy resistance in breast cancer cells. Am. J. Transl. Res. 14, 2550–2566.
Gao, L., Dou, Z. C., Ren, W. H., Li, S. M., Liang, X., and Zhi, K. Q. (2019). CircCDR1as upregulates autophagy under hypoxia to promote tumor cell survival via AKT/ERK(½)/mTOR signaling pathways in oral squamous cell carcinomas. Cell. Death Dis. 10, 745. doi:10.1038/s41419-019-1971-9
Ge, Y. Z., Xu, L. W., Zhou, C. C., Lu, T. Z., Yao, W. T., Wu, R., et al. (2017). A BAP1 mutation-specific MicroRNA signature predicts clinical outcomes in clear cell renal cell carcinoma patients with wild-type BAP1. J. Cancer 8, 2643–2652. doi:10.7150/jca.20234
Ghafouri-Fard, S., Glassy, M. C., Abak, A., Hussen, B. M., Niazi, V., and Taheri, M. (2021a). The interaction between miRNAs/lncRNAs and Notch pathway in human disorders. Biomed. Pharmacother. 138, 111496. doi:10.1016/j.biopha.2021.111496
Ghafouri-Fard, S., Hussen, B. M., Nicknafs, F., Nazer, N., Sayad, A., and Taheri, M. (2021b). Expression analysis of Protein inhibitor of activated STAT in inflammatory demyelinating polyradiculoneuropathy. Front. Immunol. 12, 659038. doi:10.3389/fimmu.2021.659038
Han, P., Yang, H., Li, X., Wu, J., Wang, P., Liu, D., et al. (2021). Identification of a novel cancer stemness-associated ceRNA Axis in lung adenocarcinoma via stemness indices analysis. Oncol. Res. 28, 715–729. doi:10.3727/096504020X16037124605559
Hansen, T. B., Wiklund, E. D., Bramsen, J. B., Villadsen, S. B., Statham, A. L., Clark, S. J., et al. (2011). miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 30, 4414–4422. doi:10.1038/emboj.2011.359
Harrison, E. B., Porrello, A., Bowman, B. M., Belanger, A. R., Yacovone, G., Azam, S. H., et al. (2020). A circle RNA regulatory Axis promotes lung squamous metastasis via CDR1-mediated regulation of golgi trafficking. Cancer Res. 80, 4972–4985. doi:10.1158/0008-5472.CAN-20-1162
He, M., Jia, Z., Wen, Y., and Chen, X. (2022). Circ_0043947 contributes to interleukin 1β-induced injury in chondrocytes by sponging miR-671-5p to up-regulate RTN3 expression in osteoarthritis pathology. J. Orthop. Surg. Res. 17, 177. doi:10.1186/s13018-022-02970-4
Hou, J. C., Xu, Z., Zhong, S. L., Zhang, H. D., Jiang, L. H., Chen, X., et al. (2019). Circular RNA circASS1 is downregulated in breast cancer cells MDA-MB-231 and suppressed invasion and migration. Epigenomics 11, 199–213. doi:10.2217/epi-2017-0167
Hu, Y., Liang, D., Chen, X., Chen, L., Bai, J., Li, H., et al. (2021). MiR-671-5p negatively regulates SMAD3 to inhibit migration and invasion of osteosarcoma cells. Nan Fang. Yi Ke Da Xue Xue Bao 41, 1562–1568. doi:10.12122/j.issn.1673-4254.2021.10.16
Hussen, B. M., Hidayat, H. J., Salihi, A., Sabir, D. K., Taheri, M., and Ghafouri-Fard, S. (2021). MicroRNA: A signature for cancer progression. Biomed. Pharmacother. 138, 111528. doi:10.1016/j.biopha.2021.111528
Hussen, B. M., Salihi, A., Abdullah, S. T., Rasul, M. F., Hidayat, H. J., Hajiesmaeili, M., et al. (2022). Signaling pathways modulated by miRNAs in breast cancer angiogenesis and new therapeutics. Pathol. Res. Pract. 230, 153764. doi:10.1016/j.prp.2022.153764
Jia, H. L., He, C. H., Wang, Z. Y., Xu, Y. F., Yin, G. Q., Mao, L. J., et al. (2014). MicroRNA expression profile in exosome discriminates extremely severe infections from mild infections for hand, foot and mouth disease. BMC Infect. Dis. 14, 506. doi:10.1186/1471-2334-14-506
Jin, W., Shi, J., and Liu, M. (2019). Overexpression of miR-671-5p indicates a poor prognosis in colon cancer and accelerates proliferation, migration, and invasion of colon cancer cells. Onco. Targets. Ther. 12, 6865–6873. doi:10.2147/OTT.S219421
Kim, H., Yang, J. M., Jin, Y., Jheon, S., Kim, K., Lee, C. T., et al. (2017). MicroRNA expression profiles and clinicopathological implications in lung adenocarcinoma according to EGFR, KRAS, and ALK status. Oncotarget 8, 8484–8498. doi:10.18632/oncotarget.14298
Klinge, C. M., Piell, K. M., Tooley, C. S., and Rouchka, E. C. (2019). HNRNPA2/B1 is upregulated in endocrine-resistant LCC9 breast cancer cells and alters the miRNA transcriptome when overexpressed in MCF-7 cells. Sci. Rep. 9, 9430. doi:10.1038/s41598-019-45636-8
Li, X., and Diao, H. (2019). Circular RNA circ_0001946 acts as a competing endogenous RNA to inhibit glioblastoma progression by modulating miR-671-5p and CDR1. J. Cell. Physiol. 234, 13807–13819. doi:10.1002/jcp.28061
Li, X., Nie, C., Tian, B., Tan, X., Han, W., Wang, J., et al. (2019a). miR-671-5p blocks the progression of human esophageal squamous cell carcinoma by suppressing FGFR2. Int. J. Biol. Sci. 15, 1892–1904. doi:10.7150/ijbs.32429
Li, Z. Y., Zhang, Z. Z., Bi, H., Zhang, Q. D., Zhang, S. J., Zhou, L., et al. (2019b). Upregulated microRNA-671-3p promotes tumor progression by suppressing forkhead box P2 expression in non-small-cell lung cancer. Mol. Med. Rep. 20, 3149–3159. doi:10.3892/mmr.2019.10563
Lin, J. C., Kuo, C. Y., Tsai, J. T., and Liu, W. H. (2021). miR-671-5p inhibition by MSI1 promotes glioblastoma tumorigenesis via radioresistance, tumor motility and cancer stem-like cell properties. Biomedicines 10, 21. doi:10.3390/biomedicines10010021
Lin, S., Yang, L., Wang, S., Weng, B., and Lin, M. (2020). Bioinformatics analysis of key micro-RNAs and mRNAs under the hand, foot, and mouth disease virus infection. Pol. J. Microbiol. 69, 479–490. doi:10.33073/pjm-2020-052
Liu, Y., Feng, X., Kang, S., Lv, F., Ni, Y., and Wu, H. (2022a). CircRIP2 promotes NSCLC progression by sponging for miR-671-5p to regulate FOXM1 expression. Histol. Histopathol. 37, 117–124. doi:10.14670/HH-18-360
Liu, Y., Shi, M., He, X., Cao, Y., Liu, P., Li, F., et al. (2022b). LncRNA-PACERR induces pro-tumour macrophages via interacting with miR-671-3p and m6A-reader IGF2BP2 in pancreatic ductal adenocarcinoma. J. Hematol. Oncol. 15, 52. doi:10.1186/s13045-022-01272-w
Liu, Z., Chen, S., Yang, Y., Lu, S., Zhao, X., Hu, B., et al. (2019). MicroRNA-671-3p regulates the development of knee osteoarthritis by targeting TRAF3 in chondrocytes. Mol. Med. Rep. 20, 2843–2850. doi:10.3892/mmr.2019.10488
Lu, G. F., You, C. Y., Chen, Y. S., Jiang, H., Zheng, X., Tang, W. W., et al. (2018). MicroRNA-671-3p promotes proliferation and migration of glioma cells via targeting CKAP4. Onco. Targets. Ther. 11, 6217–6226. doi:10.2147/OTT.S177325
Luo, X., and Zhou, X. (2022). CircRNA-PTPRA knockdown inhibits atherosclerosis progression by repressing ox-LDL-induced endothelial cell injury via sponging of miR-671-5p. Biochem. Genet. doi:10.1007/s10528-022-10256-x
Ma, C., Nie, Z. K., Guo, H. M., and Kong, Y. (2020). MiR-671-5p plays a promising role in restraining osteosarcoma cell characteristics through targeting TUFT1. J. Biochem. Mol. Toxicol. 34, e22490. doi:10.1002/jbt.22490
Ma, L., Zhang, X. Q., Zhou, D. X., Cui, Y., Deng, L. L., Yang, T., et al. (2016). Feasibility of urinary microRNA profiling detection in intrahepatic cholestasis of pregnancy and its potential as a non-invasive biomarker. Sci. Rep. 6, 31535. doi:10.1038/srep31535
Macfarlane, L. A., and Murphy, P. R. (2010). MicroRNA: Biogenesis, function and role in cancer. Curr. Genomics 11, 537–561. doi:10.2174/138920210793175895
Malgulwar, P. B., Pathak, P., Singh, M., Kale, S. S., Suri, V., Sarkar, C., et al. (2017). Downregulation of SMARCB1/INI1 expression in pediatric chordomas correlates with upregulation of miR-671-5p and miR-193a-5p expressions. Brain Tumor Pathol. 34, 155–159. doi:10.1007/s10014-017-0295-7
Mcdonald, A. C., Vira, M., Shen, J., Sanda, M., Raman, J. D., Liao, J., et al. (2018). Circulating microRNAs in plasma as potential biomarkers for the early detection of prostate cancer. Prostate 78, 411–418. doi:10.1002/pros.23485
Nardelli, C., Iaffaldano, L., Pilone, V., Labruna, G., Ferrigno, M., Carlomagno, N., et al. (2017). Changes in the MicroRNA profile observed in the subcutaneous adipose tissue of obese patients after laparoscopic adjustable gastric banding. J. Obes. 2017, 6754734. doi:10.1155/2017/6754734
Ntoumou, E., Tzetis, M., Braoudaki, M., Lambrou, G., Poulou, M., Malizos, K., et al. (2017). Serum microRNA array analysis identifies miR-140-3p, miR-33b-3p and miR-671-3p as potential osteoarthritis biomarkers involved in metabolic processes. Clin. Epigenetics 9, 127. doi:10.1186/s13148-017-0428-1
Nunes, S., Silva, I. B., Ampuero, M. R., de Noronha, A. L. L., de Souza, L. C. L., Correia, T. C., et al. (2018). Integrated analysis reveals that miR-193b, miR-671, and TREM-1 correlate with a good response to treatment of human localized cutaneous leishmaniasis caused by leishmania braziliensis. Front. Immunol. 9, 640. doi:10.3389/fimmu.2018.00640
Papp, G., Krausz, T., Stricker, T. P., Szendrői, M., and Sápi, Z. (2014). SMARCB1 expression in epithelioid sarcoma is regulated by miR-206, miR-381, and miR-671-5p on Both mRNA and protein levels. Genes. Chromosom. Cancer 53, 168–176. doi:10.1002/gcc.22128
Peng, D., Wu, T., Wang, J., Huang, J., Zheng, L., Wang, P., et al. (2022). microRNA-671-5p reduces tumorigenicity of ovarian cancer via suppressing HDAC5 and HIF-1α expression. Chem. Biol. Interact. 355, 109780. doi:10.1016/j.cbi.2021.109780
Qiu, T., Wang, K., Li, X., and Jin, J. (2018). miR-671-5p inhibits gastric cancer cell proliferation and promotes cell apoptosis by targeting URGCP. Exp. Ther. Med. 16, 4753–4758. doi:10.3892/etm.2018.6813
Quan, H., Chen, Q., Wang, K., Wang, Q., Lu, M., Zhang, Y., et al. (2021). Exendin-4 reversed the PC12 cell damage induced by circRNA CDR1as/miR-671/gsk3β signaling pathway. J. Mol. Neurosci. 71, 778–789. doi:10.1007/s12031-020-01698-2
Ren, J., Pan, G., Yang, J., Xu, N., Zhang, Q., and Li, W. (2021). Circ_0000620 acts as an oncogenic factor in gastric cancer through regulating MMP2 expression via sponging miR-671-5p. J. Biol. Res. 28, 23. doi:10.1186/s40709-021-00154-5
Sadeghi, M., Ranjbar, B., Ganjalikhany, M. R., Khan, F, M., Schmitz, U., Wolkenhauer, O., et al. (2016). MicroRNA and transcription factor gene regulatory network analysis reveals key regulatory elements associated with prostate cancer progression. PLoS One 11, e0168760. doi:10.1371/journal.pone.0168760
Shen, Q., Zheng, G., Zhou, Y., Tong, J., Xu, S., Gao, H., et al. (2021). CircRNA circ_0092314 induces epithelial-mesenchymal transition of pancreatic cancer cells via elevating the expression of S100P by sponging miR-671. Front. Oncol. 11, 675442. doi:10.3389/fonc.2021.675442
Singh, A. K., Rooge, S. B., Varshney, A., Vasudevan, M., Bhardwaj, A., Venugopal, S. K., et al. (2018). Global microRNA expression profiling in the liver biopsies of Hepatitis B virus-infected patients suggests specific microRNA signatures for viral persistence and hepatocellular injury. Hepatology 67, 1695–1709. doi:10.1002/hep.29690
Song, H., Xu, Y., Xu, T., Fan, R., Jiang, T., Cao, M., et al. (2020). CircPIP5K1A activates KRT80 and PI3K/AKT pathway to promote gastric cancer development through sponging miR-671-5p. Biomed. Pharmacother. 126, 109941. doi:10.1016/j.biopha.2020.109941
Sun, L., Hu, J., Xiong, W., Chen, X., Li, H., and Jie, S. (2013). MicroRNA expression profiles of circulating microvesicles in hepatocellular carcinoma. Acta Gastroenterol. belg. 76, 386–392.
Taheri, M., Mahmud Hussen, B., Tondro Anamag, F., Shoorei, H., Dinger, M. E., and Ghafouri-Fard, S. (2022). The role of miRNAs and lncRNAs in conferring resistance to doxorubicin. J. Drug Target. 30, 1–21. doi:10.1080/1061186X.2021.1909052
Tan, X., Fu, Y., Chen, L., Lee, W., Lai, Y., Rezaei, K., et al. (2016). miR-671-5p inhibits epithelial-to-mesenchymal transition by downregulating FOXM1 expression in breast cancer. Oncotarget 7, 293–307. doi:10.18632/oncotarget.6344
Tan, X., Li, Z., Ren, S., Rezaei, K., Pan, Q., Goldstein, A. T., et al. (2019). Dynamically decreased miR-671-5p expression is associated with oncogenic transformation and radiochemoresistance in breast cancer. Breast Cancer Res. 21, 89. doi:10.1186/s13058-019-1173-5
Tang, X., Wang, J., Xia, X., Tian, J., Rui, K., Xu, H., et al. (2019). Elevated expression of ciRS-7 in peripheral blood mononuclear cells from rheumatoid arthritis patients. Diagn. Pathol. 14, 11. doi:10.1186/s13000-019-0783-7
Tang, Z., Zhang, Y., Wu, H., Liu, C., Lian, Y., Ling, H., et al. (2022). Integrated analysis of lncRNA-miRNA-mRNA ceRNA network in mixed dry eye disease. Contrast Media Mol. Imaging 2022, 1534142. doi:10.1155/2022/1534142
Tomczyk-Socha, M., Kręcicka, J., Misiuk-Hojło, M., and Turno-Kręcicka, A. (2022). MicroRNA expression in pseudoexfoliation syndrome with the use of next-generation sequencing. Genes. (Basel) 13, 582. doi:10.3390/genes13040582
Uwatoko, H., Hama, Y., Iwata, I. T., Shirai, S., Matsushima, M., Yabe, I., et al. (2019). Identification of plasma microRNA expression changes in multiple system atrophy and Parkinson's disease. Mol. Brain 12, 49. doi:10.1186/s13041-019-0471-2
Wang, B. H., Zhao, Y. F., Shen, L. R., and Zhuang, Q. (2019). Differential screening and functional prediction analysis of miRNA expression profiles in periodontitis. Shanghai Kou Qiang Yi Xue 28, 408–411.
Wang, C., Liu, J., Zhang, X., Chen, Q., Bai, X., Hong, X., et al. (2021a). Role of miRNA-671-5p in mediating wnt/β-catenin-triggered podocyte injury. Front. Pharmacol. 12, 784489. doi:10.3389/fphar.2021.784489
Wang, C., Su, Z., Sanai, N., Xue, X., Lu, L., Chen, Y., et al. (2012). microRNA expression profile and differentially-expressed genes in prolactinomas following bromocriptine treatment. Oncol. Rep. 27, 1312–1320. doi:10.3892/or.2012.1690
Wang, Q., Luo, S., Yang, J., Li, J., Huan, S., She, G., et al. (2021b). Circ_0114876 promoted IL-1β-induced chondrocyte injury by targeting miR-671/TRAF2 axis. Biotechnol. Lett. 43, 791–802. doi:10.1007/s10529-020-03070-1
Wang, W. J., Yuan, Y., Zhang, D., Liu, P., and Liu, F. (2021c). miR-671-5p repressed progression of papillary thyroid carcinoma via TRIM14. Kaohsiung J. Med. Sci. 37, 983–990. doi:10.1002/kjm2.12424
Warnecke-Eberz, U., Chon, S. H., Hölscher, A. H., Drebber, U., and Bollschweiler, E. (2015). Exosomal onco-miRs from serum of patients with adenocarcinoma of the esophagus: Comparison of miRNA profiles of exosomes and matching tumor. Tumour Biol. 36, 4643–4653. doi:10.1007/s13277-015-3112-0
Wu, Q., Yin, X., Zhao, W., Xu, W., and Chen, L. (2022a). Molecular mechanism of m(6)A methylation of circDLC1 mediated by RNA methyltransferase METTL3 in the malignant proliferation of glioma cells. Cell. Death Discov. 8, 229. doi:10.1038/s41420-022-00979-6
Wu, X., Yin, S., Yan, L., Liu, Y., Shang, L., and Liu, J. (2022b). lncRNA DLEU1 modulates proliferation, inflammation, and extracellular matrix degradation of chondrocytes through regulating miR-671-5p. J. Immunol. Res. 2022, 1816217. doi:10.1155/2022/1816217
XI, P., Zhang, C. L., Wu, S. Y., Liu, L., Li, W. J., and Li, Y. M. (2021). CircRNA circ-IQGAP1 knockdown alleviates interleukin-1β-induced osteoarthritis progression via targeting miR-671-5p/TCF4. Orthop. Surg. 13, 1036–1046. doi:10.1111/os.12923
Xia, W., Gong, D., Qin, X., and Cai, Z. (2020). MicroRNA-671-3p suppresses proliferation and invasion of breast cancer cells by targeting DEPTOR. Nan Fang. Yi Ke Da Xue Xue Bao 40, 42–48. doi:10.12122/j.issn.1673-4254.2020.01.07
Xiao, S., Zhou, Y., Liu, Q., Zhang, T., and Pan, D. (2021). Identification of pivotal MicroRNAs and target genes associated with persistent atrial fibrillation based on bioinformatics analysis. Comput. Math. Methods Med. 2021, 6680211. doi:10.1155/2021/6680211
Xie, C., Song, L. B., Wu, J. H., Li, J., Yun, J. P., Lai, J. M., et al. (2012). Upregulator of cell proliferation predicts poor prognosis in hepatocellular carcinoma and contributes to hepatocarcinogenesis by downregulating FOXO3a. PLoS One 7, e40607. doi:10.1371/journal.pone.0040607
Xin, C., Lu, S., Li, Y., Zhang, Y., Tian, J., Zhang, S., et al. (2019). miR-671-5p inhibits tumor proliferation by blocking cell cycle in osteosarcoma. DNA Cell. Biol. 38, 996–1004. doi:10.1089/dna.2019.4870
Xiong, D. D., Chen, H., He, R. Q., Lan, A. H., Zhong, J. C., Chen, G., et al. (2018). MicroRNA-671-3p inhibits the development of breast cancer: A study based on in vitro experiments, in-house quantitative polymerase chain reaction and bioinformatics analysis. Int. J. Oncol. 52, 1801–1814. doi:10.3892/ijo.2018.4339
Yang, B., Zhou, Z. H., Chen, L., Cui, X., Hou, J. Y., Fan, K. J., et al. (2018). Prognostic significance of NFIA and NFIB in esophageal squamous carcinoma and esophagogastric junction adenocarcinoma. Cancer Med. 7, 1756–1765. doi:10.1002/cam4.1434
Yang, Y., Lin, S., Yang, Z., Huang, Y., and Zhan, F. (2022a). Circ_0001947 promotes cell proliferation, invasion, migration and inflammation and inhibits apoptosis in human rheumatoid arthritis fibroblast-like synoviocytes through miR-671-5p/STAT3 axis. J. Orthop. Surg. Res. 17, 54. doi:10.1186/s13018-022-02939-3
Yang, Y., Yang, N., and Jiang, J. (2022b). Exosomal circ_PTPRA inhibits tumorigenesis and promotes radiosensitivity in colorectal cancer by enriching the level of SMAD4 via competitively binding to miR-671-5p. Cytotechnology 74, 51–64. doi:10.1007/s10616-021-00506-y
Yao, Y., Zhou, Y., and Fu, X. (2019). miR-671-3p is downregulated in non-small cell lung cancer and inhibits cancer progression by directly targeting CCND2. Mol. Med. Rep. 19, 2407–2412. doi:10.3892/mmr.2019.9858
Ye, J., Luo, W., Luo, L., Zhai, L., and Huang, P. (2022). MicroRNA-671-5p inhibits cell proliferation, migration and invasion in non-small cell lung cancer by targeting MFAP3L. Mol. Med. Rep. 25, 30. doi:10.3892/mmr.2021.12546
Yu, Y., Wang, Z., Sun, D., Zhou, X., Wei, X., Hou, W., et al. (2018). miR-671 promotes prostate cancer cell proliferation by targeting tumor suppressor SOX6. Eur. J. Pharmacol. 823, 65–71. doi:10.1016/j.ejphar.2018.01.016
Zhan, Y., Jiang, L., Jin, X., Ying, S., Wu, Z., Wang, L., et al. (2021). Inhibiting RRM2 to enhance the anticancer activity of chemotherapy. Biomed. Pharmacother. 133, 110996. doi:10.1016/j.biopha.2020.110996
Zhang, B. (2020). Guizhi Fuling pills inhibit the proliferation, migration and invasion of human cutaneous malignant melanoma cells by regulating the molecular axis of LncRNA TPT1-AS1/miR-671-5p. Cell. Mol. Biol. 66, 148–154. doi:10.14715/cmb/2020.66.5.26
Zhang, C., Bai, N., Huang, W., Zhang, P., Luo, Y., Men, S., et al. (2016). The predictive value of selected serum microRNAs for acute GVHD by TaqMan MicroRNA arrays. Ann. Hematol. 95, 1833–1843. doi:10.1007/s00277-016-2781-0
Zhang, P., Li, X., Huang, L., Hu, F., Niu, X., Sun, Y., et al. (2022a). Association between microRNA 671 polymorphisms and the susceptibility to soft tissue sarcomas in a Chinese population. Front. Oncol. 12, 960269. doi:10.3389/fonc.2022.960269
Zhang, X., Luan, N., and Shi, J. (2022b). A novel LINC00943/miR-671-5p/ELAVL1 ceRNA crosstalk regulates MPP(+) toxicity in SK-N-SH cells. Metab. Brain Dis. 37, 2349–2362. doi:10.1007/s11011-022-01034-0
Zhang, X., Xin, G., and Sun, D. (2018). Serum exosomal miR-328, miR-575, miR-134 and miR-671-5p as potential biomarkers for the diagnosis of Kawasaki disease and the prediction of therapeutic outcomes of intravenous immunoglobulin therapy. Exp. Ther. Med. 16, 2420–2432. doi:10.3892/etm.2018.6458
Zhong, Z., Zhong, W., Zhang, Q., Zhang, Q., Yu, Z., and Wu, H. (2020). Circulating microRNA expression profiling and bioinformatics analysis of patients with coronary artery disease by RNA sequencing. J. Clin. Lab. Anal. 34, e23020. doi:10.1002/jcla.23020
Zhu, J., Ma, X., Zhang, Y., Ni, D., Ai, Q., Li, H., et al. (2016). Establishment of a miRNA-mRNA regulatory network in metastatic renal cell carcinoma and screening of potential therapeutic targets. Tumor Biol. 37, 15649–15663. doi:10.1007/s13277-016-5135-6
Zhu, Q., Zhang, X., Zai, H. Y., Jiang, W., Zhang, K. J., He, Y. Q., et al. (2021). circSLC8A1 sponges miR-671 to regulate breast cancer tumorigenesis via PTEN/PI3k/Akt pathway. Genomics 113, 398–410. doi:10.1016/j.ygeno.2020.12.006
Keywords: mir-671, cancer, biomarker, expression, prognostic
Citation: Ghafouri-Fard S, Askari A, Hussen BM, Rasul MF, Hatamian S, Taheri M and Kiani A (2022) A review on the role of miR-671 in human disorders. Front. Mol. Biosci. 9:1077968. doi: 10.3389/fmolb.2022.1077968
Received: 23 October 2022; Accepted: 25 November 2022;
Published: 05 December 2022.
Edited by:
Wei Ye, Guangdong Academy of Science, ChinaReviewed by:
Meredith Tennis, University of Colorado Denver, United StatesCopyright © 2022 Ghafouri-Fard, Askari, Hussen, Rasul, Hatamian, Taheri and Kiani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Taheri, Mohammad.taheri@uni-jena.de; Arda Kiani, ardakiani@sbmu.ac.ir
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.