
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci., 01 December 2022
Sec. Molecular Diagnostics and Therapeutics
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.1073797
This article is part of the Research TopicImpact of Genome Instability on Human HealthView all 5 articles
DNA damage response (DDR) deficiencies result in genome instability, which is one of the hallmarks of cancer. Poly (ADP-ribose) polymerase (PARP) enzymes take part in various DDR pathways, determining cell fate in the wake of DNA damage. PARPs are readily druggable and PARP inhibitors (PARPi) against the main DDR-associated PARPs, PARP1 and PARP2, are currently approved for the treatment of a range of tumor types. Inhibition of efficient PARP1/2-dependent DDR is fatal for tumor cells with homologous recombination deficiencies (HRD), especially defects in breast cancer type 1 susceptibility protein 1 or 2 (BRCA1/2)-dependent pathway, while allowing healthy cells to survive. Moreover, PARPi indirectly influence the tumor microenvironment by increasing genomic instability, immune pathway activation and PD-L1 expression on cancer cells. For this reason, PARPi might enhance sensitivity to immune checkpoint inhibitors (ICIs), such as anti-PD-(L)1 or anti-CTLA4, providing a rationale for PARPi-ICI combination therapies. In this review, we discuss the complex background of the different roles of PARP1/2 in the cell and summarize the basics of how PARPi work from bench to bedside. Furthermore, we detail the early data of ongoing clinical trials indicating the synergistic effect of PARPi and ICIs. We also introduce the diagnostic tools for therapy development and discuss the future perspectives and limitations of this approach.
Cancer is a large group of diseases characterized by the uncontrollable growth of abnormal cells. There are 19.3 milion new cancer diagnoses each year worldwide, with an estimated 10 milion cancer-related deaths occurring in 2020, placing an enormous burden on healthcare systems (Sung et al., 2021).
Although the targeted cancer therapies have advanced noticeably in recent years, chemotherapy remains the most commonly used treatment in many kinds of cancer. Unfortunately, the side effects are inevitable, as chemotherapy is unable to differentiate malignant and non-malignant cells. Severe side effects of cancer treatment like vomiting (>90% of patients require antiemetics during the treatment), fatigue, generalized pain or gastrointestinal disturbances are common in patients (Lorusso et al., 2017) (Pearce et al., 2017). For this reason, the development of new therapies focuses on acting directly on cancer-specific targets, which, in theory, allows increased efficacy against cancer cells while minimizing side effects. Currently available treatments that meet the above requirements include small-molecule drugs (used for the targets inside the cells, such as proteasome inhibitors and signal transduction inhibitors) and monoclonal antibodies (designed to attach to specific targets on cancer cells). Generally, targeted therapies counter cancer through different mechanisms, such as inhibition of angiogenesis, blocking of the cell cycle, delivering cytotoxic substances directly into cancer cells etc.
One of the most dynamically developing targeted therapies are poly (ADP-ribose) polymerase (PARP) inhibitors (PARPi). Although this term in principle refers to inhibitors of any member of the PARP family of enzymes, most relevant for cancer therapy are those targeting PARP1 and PARP2, which are primarily involved in DNA repair (Lüscher et al., 2021). Therefore, inhibition of PARP1/2 results in genome instability that destroys cancer cells while allowing non-malignant cell survival. Since 2014, there are four PARPi approved for clinical use (olaparib, rucaparib, niraparib, talazoparib), indicated for the treatment of ovarian, fallopian tube and primary peritoneal carcinoma, HER2-negative breast cancer, metastatic pancreatic cancer and prostate cancer–especially (but not exclusively) when these cancers harbor breast cancer type 1 susceptibility protein 1 or 2 (BRCA1/2) mutations. In addition, there are many promising clinical trials at various stages that investigate other PARPi and their diverse indications. Moreover, PARPi are showing encouraging results in combined therapies, especially with immune checkpoint inhibitors (ICIs), holding great promise for many cancer patients. Although PARPi are thought to be relatively specific blocking agents and do not show frequent and severe side effects, PARP1/2 enzymes play many roles at different cell cycle stages. Therefore, drawing the interaction network of PARP1/2 in the cell is crucial for navigating the new possible therapies and determining possible side effects that may appear on the way. In this review, we try to place PARPi in that network, discuss recent combined therapies of PARPi and ICIs, and point out the possible future perspectives that come into view en route.
PARP proteins were discovered in the early 1960s by Chambon et al. (1963). Subsequent research showed that PARP family takes part in several cellular processes (Krishnakumar and Kraus, 2010), the most intriguing being the interaction with DNA (Nobori et al., 1989), which contributes to cellular recovery from DNA damage (Durkacz et al., 1980).
First discoveries of possible functions of PARPs in the cell on the molecular level went hand in hand with dissecting its structure. To date the family of PARP proteins has 17 members that share structural and functional similarities while constituting a diverse and remarkable group of proteins. PARP family members are defined by their shared ADP-ribosyl transferase (ART) domain but otherwise differ in length and domain composition. PARPs generally function by modifying other proteins with single ADP-ribose units or poly (ADP-ribose) (PAR) chains (derived from nicotamide adenine dinucleotide–NAD+), which affect protein activity, interactions, localization and half-life. Moreover, PARP family members can also catalyse the reversible ADP-ribosylation of phosphorylated DNA and RNA ends, which was discussed by Groslambert et al. (2021). The 90% of total cellular PARylation is produced by PARP1—the most abundant and best studied member of the PARP family (Kameshita et al., 1984). The basic structure of the PARP1 molecule and its role in DNA repair was summarized in Figure 1. PARP1 consists of three main domains: the DNA Binding Domain (DBD) at the N-terminus, the central Automodification Domain (AMD), and the Catalytic Domain (CD) at the C-terminus (Hakmé et al., 2008). The most remarkable features of the DBD are the nuclear localization sequence (NLS) and three zinc fingers (Zn1/2/3). The latter are–together with the WGR motif in the CD–the primary sensors of DNA breaks, both Double Strand Breaks (DSBs) and Single Strand Breaks (SSBs) (Langelier et al., 2011), driving the collective assembly of PARP1 domains around the break in a way that stimulates the catalytic activity of this enzyme (Langelier et al., 2012) (Eustermann et al., 2015).
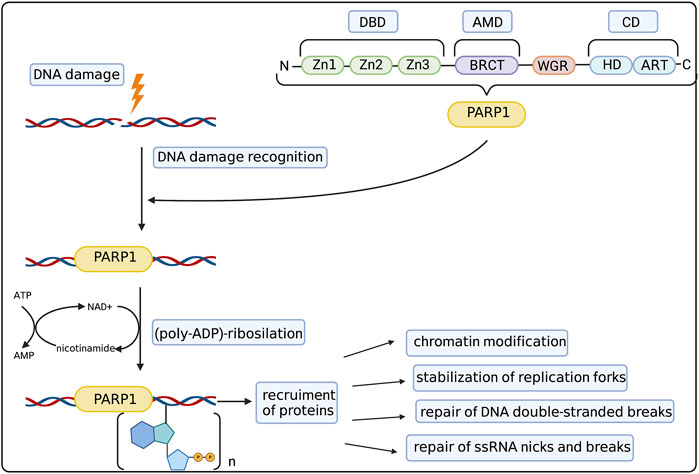
FIGURE 1. PARP1 structure and mechanism of action. PARP1 molecule consists of 3 main domains: DNA binding domain (DBD) with zinc fingers (Zn1-Zn3), Automodification Domain (AMD) with BRCA1 C-terminus (BRCT) subdomain and catalytic domain (CD) with helical domain (HD) and C-terminal ADP-ribosyl transferase (ART) subdomain. The WGR domain consists of the Trp-Gly-Arg motif. Such structure allows PARP1 to recognize and bind to the DNA damage site. Then it catalyzes the (poly-ADP)-ribosylation process, allowing the recruitment of auxiliary proteins responsible for chromatin modification, stabilization of replication forks, repair of DSBs and repair of ssDNA nicks and breaks. Created with BioRender.com.
AMD is composed of a globular BRCA1 C-terminus (BRCT) motif and a linker region that comprises the main automodification (ADP-ribosylation) sites (Prokhorova et al., 2021b). The BRCT motif has been implicated in interactions with proteins and DNA alike (Yelamos et al., 2011) (Hottiger et al., 2010) (Rudolph et al., 2021a).
The main motif of the CD domain is called the signature motif of the ART subdomain. It is composed of six beta-sheets and one alpha-helix (Otto et al., 2005) and includes both an NAD+ binding pocket and the key catalytic residue, Glu988 (Schreiber et al., 2006). The regulatory helical domain (HD) consists of helices in the absence of DNA and becomes unfolded when PARP1 binds to damaged DNA (Dawicki-McKenna et al., 2015). The HD domain is thought to inhibit the access of NAD+ to the active site in the absence of DNA but allow NAD+ access upon DNA break-induced unfolding.
Upstream of the HD domain, the WGR (Trp-Gly-Arg) motif is the key regulator of catalytic activity in response to DNA damage by interacting with DNA (Langelier et al., 2012).
In addition to PARP1, two other PARPs have been implicated in DNA damage response (DDR): PARP2 and PARP3. These paralogues are shorter than PARP1, lacking the N-terminal portion comprising zinc finger and BRCT motifs. The WGR motif of PARP2 and PARP3 is responsible for DNA binding as well as sensing the nature of DNA breaks, especially phosphorylation (Langelier et al., 2014). In contrast, PARP1 is relatively insensitive to the phosphorylation state of DNA breaks that it binds to and is activated by (Obaji et al., 2018) (Bilokapic et al., 2020). Specifically in the case of PARP2, the WGR domain is apparently capable of simultaneously binding to two blunt DNA ends, possibly in order to bridge them for subsequent ligation (Gaullier et al., 2020)
The product of PARP activity, the PAR chain, is a large, negatively charged polymer composed of ADP-ribose monomers connected through glycosidic ribose-ribose bonds. The chains can be linear or branched and are typically attached to protein substrates. PARPs can also modify proteins with only a single ADP-ribose unit (mono (ADP-ribose) or MAR. PAR and MAR modification occurs mostly on Ser, Glu, and Asp residues in proteins, and can be enzymatically removed by specific ADP-ribosyl hydrolases (Palazzo et al., 2018) (Larsen et al., 2018) (Hendriks et al., 2019).
Enzymatic DDR mechanisms correct the damage that occurs spontaneously in the cell during metabolic changes under the influence of both exogenous and endogenous (physical and chemical) factors. These mechanisms act during different phases of the cell cycle, both during replication and during the intervals between divisions. If the defects are not recognized and removed, they may lead to permanent changes in DNA and mutations in subsequent cycles. Due to the variety of factors influencing DNA integrity, multiple repair mechanisms have developed in cells. The most significant DDR pathways include:
1) Nucleotide excision repair (NER), which is a highly conserved DDR pathway that corrects a wide range of genomic lesions, especially double helix-distorting bulky lesions (Spivak, 2015),
2) Base excision repair (BER), occurring mainly during the G1 phase of the cell cycle, repairing forms of damage that do not significantly distort the DNA helix (Dianov and Hübscher, 2013). To note, the BER pathway generates SSBs, which are the most common lesions occurring in the cell (Caldecott, 2008). They are also generated by various endogenous or exogenous DNA-damaging agents, such as ionizing radiation, free radicals, topoisomerase I (TOP1) (Caldecott, 2008). SSBs may cause the blockade or collapse of DNA replication forks during the S phase, leading to the formation of DSBs (Kouzminova and Kuzminov, 2006) (Kuzminov, 2001). SSBs in non-proliferating cells may stall RNA polymerase progression during transcription, leading to cell death (Bendixen et al., 1990). SSBs are repaired in the SSB Repair (SSBR) process (Dianov and Hübscher, 2013), which is sometimes considered as a subpathway of BER (Abbotts and Wilson, 2017). SSBR is orchestrated by X-ray repair cross complementing protein 1 (XRCC1) and PARP1 as the key proteins and generally consists of SSB detection (by PARP1), DNA end processing, DNA gap-filling and DNA ligation (Abbotts and Wilson, 2017).
3) Mismatch repair (MMR) removes errors that were not corrected by DNA polymerases during replication (Kunkel and Erie, 2015),
4) Homologous recombination (HR) dominates the late S and G2 phases (Branzei and Foiani, 2008). Upon damage detection, BRCA1, BRCA2 and a partner and localizer of BRCA2, PALB2, recruit the RAD51 recombinase, which is the end effector in HR that performs DSB repair using a homologous chromatid as a template. DSBs are especially dangerous for the cell and can easily lead to apoptosis. In addition to HR, they can also be repaired via different pathways depending on cell cycle phase in which they occur (Chapman et al., 2012).
5) Non-homologous end joining recombination (NHEJ) is an alternative pathway for repairing DSBs, preferred during the G1 phase of the cell cycle. (Branzei and Foiani, 2008) (Lee and Dutta, 2021). DSBs are recognized by the Ku70-Ku80 heterodimer, which allows recruitment of DNA-dependent protein kinase (DNA-PK), DNA ligase 4 (LIG4) and the X-ray repair cross-complementing protein 4 (XRCC4) factor. These proteins activate the process, stabilize DNA and orientate it during LIG4-mediated ligation. If the ends of the DNA are incompatible, they can be adjusted for ligation by the nuclease Artemis or by DNA polymerase mu (Polμ), DNA polymerase lambda (Polλ), and terminal deoxynucleotidyl transferase (TdT) polymerases. NHEJ can also be divided into two main pathways of common characteristics: classical (cNHEJ) and alternative (aNHEJ). While cNHEJ comprises most of the features described above and attributed to NHEJ in general, aNHEJ is a more recently discovered mechanism of DSB repair, serving as a less efficient backup reaction. Its activity has been noticed during cNHEJ deficiency or impairment (Zhao et al., 2020).
In this complex scenery of proteins engaged in the diverse repertoire of DNA repair pathways, one of the most intriguing characters belong to the PARP family, which is known to act in various pathways of the cell (Figure 2); however, their most remarkable feature is undoubtedly initiation of the DDR pathway (Ray Chaudhuri and Nussenzweig, 2017) (Langelier et al., 2014).
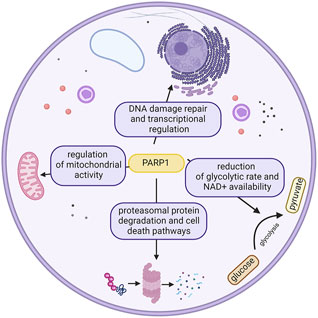
FIGURE 2. Different roles of PARP1 in the cell. This ubiquitousness defines the potential effects of its inhibition. Created with BioRender.com.
In the DDR process, PARP1 is involved in three general steps: 1) detection of DNA damage, 2) recruitment of co-factors and 3) regulation of biochemical activities. Initially, PARP was mostly known to be involved in the BER and SSBR pathways, involving such factors as XRCC1 and DNA ligase 3 (LIG3) (Fisher et al., 2007) (El-Khamisy, 2003) (Dantzer et al., 2000). Later studies showed the role of PARP1 in NER (Pines et al., 2012) (Robu et al., 2013), cNHEJ (Luijsterburg et al., 2016) and aNHEJ (Mansour et al., 2010), microhomology-mediated end joining (MMEJ) (Dutta et al., 2017), HR (Hochegger et al., 2006) (Hu et al., 2014), MMR (Liu et al., 2011), and maintenance of replication fork stability (Ronson et al., 2018) (Yang et al., 2004). Notably, recent publications point out the connection between PARP1 and Okazaki fragments (Hanzlikova et al., 2018) (Vaitsiankova et al., 2022).
Regarding the function in replication fork stability, PARP1 and PARP2 act in concert to detect disrupted replication forks, recruit the repair protein Mre11, stimulate recombination repair and restart replication (Bryant et al., 2009). Mre11 is activated by PARP at the stalled fork and restarts the Mre11-dependent replication and recombination (Kim et al., 2005).
The recruitment of different binding proteins to PARP1 is possible because of the zinc fingers 1–3. However the presence of zinc finger domains seems not to be necessary, as PARP2 and PARP3 are also able to recognize specific DNA breaks featuring 5’ phosphate groups, despite lacking the zinc finger domains. This process also leads to DDR initiation (Langelier et al., 2014). In this case the WGR domain, present also in PARP1, allows PARP2 and PARP3 to interact with DNA. Also the N-terminus of PARP2 manifests DNA-binding activity. It plays a role in the activation of PARP2 on SSBs (Riccio et al., 2016). PARP1 and PARP2 molecules remain mostly monomeric while in free form, but they form temporary dimer formations while binding to nicked sites or gaps (Eustermann et al., 2011).
DNA damage sensing is a clocklike mechanism and requires precise mechanism of break site detection as well as fast transport to the damaged site on DNA and between such sites. One recent speculative model for PARP1’s movement along DNA is called the ‘monkey bar’ mechanism (inter-segment transfer) (Rudolph et al., 2018). In this mechanism, PARP1 moves from one DNA break to the next in such a way that it always holds on to at least one DNA break, like a monkey that is holding a branch with one hand until it grasps the next branch with the other hand. This is due to the fact that binding to a new DNA break is required in order to stimulate release from the previous break. This mechanism is thought to enhance the ability of PARP1 to effectively scan genome and detect the level of DNA damage (Rudolph et al., 2018). The conformational change of the WGR domain was shown to be crucial in this process (Rudolph et al., 2020).
As described above, engagement with a DNA break through zinc finger domains and WGR is also key to the allosteric activation, which is mediated by the destabilization of the HD subdomain (Dawicki-McKenna et al., 2015) (Langelier et al., 2008).
As the PAR balance is crucial for the cell, the processes of PARylation and dePARylation are tightly controlled to maintain the equilibrium (D’Amours et al., 1999) (Fouquerel et al., 2014). PAR removal is carried out by poly (ADP-ribose) glycohydrolase (PARG) and ADP-ribosyl-acceptor hydrolase 3 (ARH3) (Rack et al., 2020) (Fontana et al., 2017) PARG and ARH3 suppression were noted to be synthetically lethal due to the increase in PARylation. (Prokhorova et al., 2021a).
PARP enzymes are able to covalently modify various targets, like transcription factors (involving transcriptional repressor CTCF, activator protein 1 (AP1), yin yang 1 (YY1), nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB), nuclear enzymes (aurora B kinase) (Kraus, 2008) (Monaco et al., 2005), and histone H1, H2A and H2B, regulating the chromatin structure (Kim et al., 2005). PARPs also tend to modify themselves (automodification), which is particularly well established for PARP1 that ADP-ribosylates a set of conserved residues in the AD. PARylation is known to play an important role in the epigenetic regulation of chromatin structure, as well as gene expression, under physiological conditions, with the maintenance of DNA integrity (Caiafa et al., 2009). On the epigenetic level, histone PARylation factor 1 (HPF1) is one of the most essential proteins interacting with PARP1 and PARP2 enzymes (Gibbs-Seymour et al., 2016). Recent findings of Suskiewicz et al. describe the role of HPF1 as a molecule complementing the PARP1/2 active site essential for the addition of ADP-ribose moieties in the DDR process (Suskiewicz et al., 2020). It turns out that the most common cellular acceptor of ADP-ribosylation–protein serine residues–cannot be efficiently targeted by PARP1 or PARP2 alone, and HPF1 must be present.
Besides ribosomal and nuclear DNA, PARP1 was also reported to be involved in the epigenetic regulation of mtDNA repair and transcription (Lapucci et al., 2011).
PARP enzymes are NAD + consumers with the capacity to globally affect the cellular energy pool and metabolic state, potentially having dramatic cellular consequences. Thus, their enzymatic activity is tightly controlled (Andrabi et al., 2014). PARP1 is also known to influence mitochondrial function and oxidative metabolism. Studies on a 129/SvImJ mouse knockout showed that PARP1 deletion led to increased food intake (Devalaraja-Narashimha and Padanilam, 2010). Further investigation showed the reduction in the glycolytic rate, which has been linked to a reduction in NAD+ availability over the years (Bai et al., 2011). PARP1 over-activation was shown to be able to reduce hexokinase activity, thus resulting in global carbohydrate metabolism perturbations (Houtkooper et al., 2010) (Andrabi et al., 2014)
PARPs have been reported to take part in several kinds of cell death pathways. At moderate levels of DNA damage apoptosis is the key feature, with Caspase3 and Caspase7 cutting PARP1 molecule into 2 fragments after D214, between first two zinc finger domains and Zn3 (Soldani and Scovassi, 2002) (Germain et al., 1999) (Kaufmann et al., 1993) (Tewari et al., 1995). At highly elevated levels of DNA damage, PARP1 overactivation is observed, resulting in necrotic cell death (caused by depletion of NAD+ and ATP) (Sosna et al., 2014). Meanwhile PARP1 detecting low levels of DNA damage steers the fate of cells into survival and DNA repair (Bouchard et al., 2003).
PARP1, along with apoptosis inducing factor (AIF), has been suggested to play a crucial role in another kind of caspase-independent cell death, parthanatos (Yu et al., 2006). Parthanatos was thought to be the ultimate effect of PARP overactivation leading to increased NAD+ consumption and subsequent loss of ATP (Ha and Snyder, 1999), until the decrease of NAD+ and ATP was associated with glycolysis inhibition through PAR chain binding on the PAR-binding motif (PBM) of hexokinase (Andrabi et al., 2014) (Fouquerel et al., 2014). It can be specifically observed in stroke, diabetes, and Parkinson disease (Andrabi et al., 2008).
Besides this, PARP1 is engaged in DNA damage-dependent autophagy (Muñoz-Gámez et al., 2009) and shows a cytoprotective role in oxidative stress-induced necrotic cell death. PARP1 also stimulates the unfolded protein response (Shoab Mansuri and Singh, 2014).
PARP1 may facilitate HR by recruitment of such proteins as ataxia telangiectasia mutated (ATM), nibrin (NBS1), and Mre11 to sites of DSB of DNA (Haince et al., 2008), however its major role in the HR repair involves localization of BRCA1/2. BRCA1 is involved in the surveillance of DNA damage and transduction of DNA repair responses, while BRCA2 is directly engaged in DNA DSBR via modulation of Rad51 by HR (Tutt and Ashworth, 2002). Their mutations are able to prevent DNA repair mechanisms and increase the risk of malignances (Hennessy et al., 2010).
PARP1 overexpression has been demonstrated in many cancers, which indicates its importance in cancerogenesis and may be an independent prognostic factor. In colorectal cancer level of PARP1 expression was positively correlated with tumor size and histopathological features according to TNM classification system (Nosho et al., 2006). Increased levels of PARP1 have also been observed in breast cancer (Siraj et al., 2018), gastric cancer (Afzal et al., 2019), ovarian cancer (Kummar et al., 2012), pancreatic cancer (Xu et al., 2019), and liver cancer (Li et al., 2017) The dependence of prostate cancer on PARP1 activity was shown in the reduction of AR to PARP1 inhibition (Espinoza, 2013).
PARP1 is also relevant for diseases other than cancer. There is evidence that it is associated with the pathogenesis of diseases such as rheumatoid arthritis (García et al., 2006) (Li et al., 2016), chronic gastritis (Lee et al., 2016), acute and chronic inflammatory bowel disease (Sánchez-Fidalgo et al., 2007), allergic responses and asthma (Ghonim et al., 2015) (Oumouna et al., 2006), oxidative/nitrosative stress following infarction-reperfusion and septic shock (Soriano et al., 2006) (Kang et al., 2010) (Pazzaglia and Pioli, 2019).
Considering PARP1’s role in the DDR and having in mind that genome instability is one of the hallmarks of cancer, the idea of developing PARPi arose in oncology studies. Almost 3 decades ago, Satoh et al. (1994) showed that as much as 90% of PARP activity must be stopped to impair the DNA repair process. Hence, potential PARPi have to manifest both high specificity and effectiveness, to bear a potential for clinical application (Bryant et al., 2005) (Farmer et al., 2005) (Calvert and Azzariti, 2011) (Plummer et al., 2008) (Calabrese et al., 2003). The PARPi currently developed and used in clinics are mainly active against PARP1 and its closest homologue, PARP2, but only weakly against other PARPs (Rudolph et al., 2022). The attempts to develop PARPi led to two different approaches: 1) targeting cells which are predisposed to cell death when PARP activity is lost; 2) searching for a combination therapy with a different type of DNA-damaging agents (Papeo et al., 2013). First of these ideas is presented by PARPi’s role in the potential therapy of cells with deficiency in the BRCA1/2-dependent HR pathway–a process known as a synthetic lethality (Lord and Ashworth, 2017). This approach has medical implications, establishing PARPi as potential drugs in BRCA1/2-mutated (BRCA1/2m) cancers, as has been validated both in vitro and in vivo (Bryant et al., 2005) (Farmer et al., 2005). Later on, PARPi were successfully translated into clinical treatment of BRCA1/2m cancers such as breast (Robson et al., 2017) and ovarian cancer (Coleman et al., 2019). To date, a large number of research studies, trying to apply PARPi in other cancers, is in progress (Table 2).
The antitumor potential of PARPi—which has been widely observed and confirmed in multiple studies—is an intensively explored area today, so numerous hypotheses are emerging, but the exact mechanism of action of PARPi is still not fully understood.
The first described mechanism of PARPi action was associated with the well-known role of PARP1 in SSBR. It linked PARP1 inhibition with blocking this pathway and therefore triggering a process known as synthetic lethality (Bryant et al., 2005). It occurs when the combination of at least two genetic or molecular events results in a death of a cell or an organism (Nijman, 2011). PARP1 hyperactivation was noted in HR-defective cells, making them more sensitive to PARPi (Gottipati et al., 2010). However, later studies investigating the genetic knockout (Horton et al., 2014), or molecular silencing with siRNA (Patel et al., 2011) of a marker molecule of PARP1 activity—XRCC1, showed confounding results depending on whether it was genetic or molecular context of depletion. Disfunctional SSBR could lead to accumulation of DSBs, which, ultimately, can only be repaired by HR or NHEJ pathways, with NHEJ being much more error-prone, while HR needs BRCA1/2 genes to proceed (Isono et al., 2017) (Holloman, 2011) (Lieber, 2008) (Ashworth, 2008). In BRCA1/2-deficient tumors, the only way to repair the DSB upon PARP1 inhibition is NHEJ, which, due to its potency to generate mutations, leads to a genomic catastrophe. In addition to the SSBR connection, the synthetic lethality between PARPi and HR deficiencies could also be explained by the roles of both PARP1 and BRCA1/2 in replication fork stability and restart (Bryant et al., 2009) (Koppensteiner et al., 2014) or DDR.
Besides the fact that PARP1 inhibition favours the NHEJ pathway by blocking the alternative HR, the interactions between PARP1 and NHEJ are much more complex. It was suggested that PARPi treatment increases the phosphorylation of DNA-dependent protein kinase (DNA-PK) substrates and therefore increases NHEJ activity (Patel et al., 2011). Indeed, inhibition of PARP1 ultimately blocks its interactions with Ku70 and Ku80, which are negative regulators of this pathway, therefore leading to increase in NHEJ activity (Wang et al., 2006) (Hochegger et al., 2006) (Paddock et al., 2011) (Yang et al., 2018).
Recent studies suggest that PARP inhibition-associated effects may be the result of DSBs occurring as a result of high-speed replication (Maya-Mendoza et al., 2018) (Quinet and Vindigni, 2018). This might in turn cause the accumulation of cytotoxic replication-associated single stranded DNA (ssDNA) gaps (Cong et al., 2021), subsequently leading to reversal of stalled replication forks. This theory is relatively unexplored and new; however, there is existing evidence supporting this mechanism of action of PARPi (Cong et al., 2021). If PARPi function solely by blocking the action of PARP1, then their effect should not be greater than that of the genetic deletion of the PARP1 gene. However, several authors have reported that inhibiting PARP1 is more cytotoxic than deleting it, even in chicken cells, which lack the PARP2 orthologue that PARPi inhibit in addition to PARP1 in human cells (Murai et al., 2012) (Patel et al., 2012) (Pettitt et al., 2013). This implies that PARP1 that is blocked by PARPi not only fails to perform its physiological role(s) but also acquires a new, toxic function. This line of reasoning—together with the observed accumulation of PARP1 on chromatin upon PARPi treatment—led to the hypothesis of PARP trapping proposed by Murai et al. (2012), whereby inhibited PARP1 persists on DNA damage and actively interferes with vital cellular processes. In general, PARP1 is thought to undergo a cycle whereby it associates with DNA breaks, becomes catalytically activated, and then modifies both other substrates and itself. Automodification of PARP1 prevents PARP1’s association with a DNA break, so PARP1 dissociates from DNA and eventually becomes dePARylated by PARG, terminating the cycle. This cycle was elaborated early on in publications by Ferro and Olivera (Ferro and Olivera, 1982), Zahradka and Ebisuzaki. (1982) and, especially, Satoh and Lindahl. (1992). Murai et al. proposed that interference with PARP1’s ability to automodify could explain the trapping effect of PARPi (Murai et al., 2012). Additionally, these authors hypothesized that some PARPi might have an allosteric effect, making PARP1 more tightly associated with DNA independent of automodification inhibition, but this has not been confirmed for the current clinical PARPi in recent publications (Zandarashvili et al., 2020) (Rudolph et al., 2022) (Hopkins et al., 2015) (Xue et al., 2022). Another revision to Murai et al.‘s model was recently offered by Shao et al. (Shao et al., 2020), who showed that PARP1, even in the “trapped state,” is not physically stalled on DNA but rather undergoes constant exchange. Nonetheless, PARP1 molecules that are exchanging on DNA might, considered as a population, could effectively outcompete other factors from binding to DNA, e.g., those required for an efficient replication fork restart or those engaged in DNA repair progression etc., explaining in part the particularly toxic effect of PARP inhibition.
Although in cancer therapies PARP trapping is a desired mechanism, it is an obstacle in conditions characterized by PARP hyperactivation, i.e. neurodegenerative diseases or ischemia-reperfusion injury. An interesting solution might be group of PROTAC PARP degraders, which are able to inhibit PARP1 without trapping, mimicking PARP1 depletion and protecting the cell from genotoxic stress-induced cell death (Wang et al., 2019a).
Finally, the role of PARP1 in histone modification and chromatin structure regulation is widely discussed and briefly reviewed here. Discovering these actions of PARP1 naturally leads to formation of a new hypothesis encompassing the epigenetic aspect of PARP1. Inhibition of PARP1 could also lead to inhibition of important oncogenes that are controlled by PARP1-dependent histone modification. This theory was already explored in Ewing sarcoma (Brenner et al., 2012) and BRCA1/2-deficient breast cancer patients (Kim et al., 2019).
Based on their presumed mechanism of action, PARP inhibitors are currently approved for the treatment of breast, ovarian, pancreatic, and prostate cancers carrying BRCA1/2 mutations. However, their application is limited by the relatively low percentage of BRCA1/2m occurring in 10%–15% of breast and ovarian, 4%–7% of pancreatic and 1.5% of prostate tumors (Bryant et al., 2005) (Iqbal et al., 2012). On the other hand, recent studies showed PARPi might be effective in tumors which do not carry any BRCA1/2m, but have different alternative HR deficiencies or other DDR gene alterations (Bryant et al., 2005) (Turner et al., 2008) (Jonsson et al., 2019). Moreover, tumor cells face both oxidative and replicative stress, which, as studies suggest, makes them more sensitive to blocking DNA repair pathways i.e. using PARPi monotherapy (Majuelos-Melguizo et al., 2015) (Schoonen et al., 2017) (Michelena et al., 2018). These studies broaden the possible application of PARPi in tumor therapies.
All four PARPi approved for treatment (olaparib, talazoparib, niraparib, and rucaparib) share similarities on the molecular level. The common element of their chemical structure is an aromatic system that mimics nicotinamide (a part of NAD+), allowing PARPi to compete with NAD+ for PARP binding. The precise way in which a given PARPi engages with the NAD+-binding pocket determines how efficient it is at outcompeting NAD+ and thus blocking the catalytic activity (including automodification). Additionally, PARPi extend to a varying degree towards the HD domain and can affect the position and folding of HD helices. Since the HD is allosterically coupled—via the WGR—with DNA breaks, PARPi can in principle allosterically modulate DNA binding, in addition to affecting it indirectly via PARP automodification inhibition. In terms of the allosteric effects, PARPi can be divided into threegroups: 1) promoting allosteric retention on DNA (including several PARPi not yet used in the clinic), 2) allostery-neutral drugs (olaparib, talazoparib), and 3) drugs having an allosteric pro-release effect (rucaparib, niraparib, veliparib), as proposed (Zandarashvili et al., 2020). However, later studies of Lunger’s group (Rudolph et al., 2022) (Rudolph et al., 2021b) showed the affinity of PARPi depends rather on known trapping properties than the allosteric effect, which is additional and either neutral or negative in all existing clinical PARPi. By determining both the affinity for the NAD+ pocket and any potential allosteric effects, the molecular structure of PARPi is key to determining their molecular properties including trapping potential. Here, we briefly introduce the quartette of PARPi, more detailed description covering the FDA/EMA recommendations can be found in Table 1.
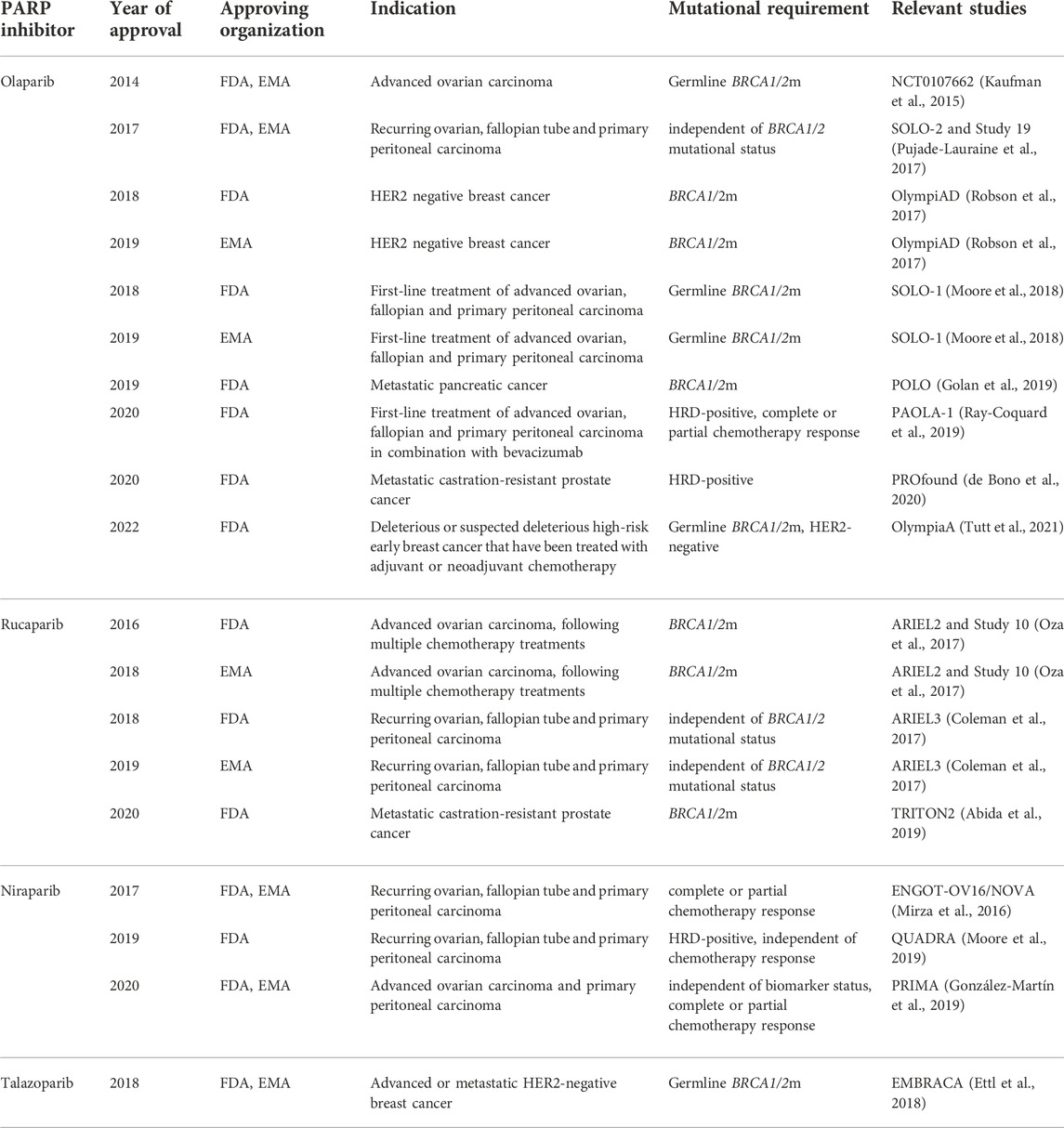
TABLE 1. History of PARPi in the clinic–European Medicines Agency (EMA) and Food and Drug Administration (FDA) approvals. Full access to the research studies description is available on ClinicalTrials.gov.
Olaparib (Lynparza) was the first drug to be approved in 2014 by EMA and FDA in clinical use as monotherapy for the treatment of advanced germline BRCA1/2m ovarian cancer (Wiggans et al., 2015) (Deeks, 2015).
Since the first approval of olaparib in clinic, several next-generation PARPi (i.e. talazoparib, niraparib, and rucaparib) have been tested in clinical trials (Murai et al., 2012). Talazoparib (Talzenna), a drug targeting both PARP1 and PARP2 (Shen et al., 2013), was approved in 2018 for the treatment of the germline BRCA1/2m-advanced or metastatic HER2-negative breast cance (Ettl et al., 2018) (Hoy, 2018). Talazoparib has an exceptionally high affinity for PARP1 (up to 100 fold higher trapping efficiency than olaparib) and therefore requires very low concentration to produce an effect that would require a considerably higher concentration of olaparib. Nevertheless, since 2018, talazoparib has not been approved for any further treatment. The most selective PARP1 and PARP2 inhibitor in clinical use is considered to be niraparib (Thorsell et al., 2017). Niraparib (Zejula) was approved in 2017 in the US and the EU for maintenance treatment of reoccurring ovarian, fallopian, and primary peritoneal carcinomas, regardless of their BRCA1/2m status, in patients that show complete or partial response to chemotherapy (Mirza et al., 2016) (Del Campo et al., 2019) (Scott, 2017).
Unlike olaparib and niraparib, rucaparib (Rubraca) inhibits PARP3 in addition to PARP1 and PARP2. As PARP3 has been suggested to activate the enzymatic activity of PARP1 in the absence of DNA, rucaparib’s ability to inhibit PARP3 may potentiate its effects compared to olaparib or niraparib (Loseva et al., 2010). It was first approved by FDA in 2016 for somatic and germline BRCA1/2m advanced ovarian carcinomas in patients following multiple chemotherapy trials (Oza et al., 2017) (Syed, 2017).
To date, all four clinically-approved PARPi have been tested in various clinical trials to broaden their application into different cancers (Table 2). The highly specific mechanism of action of PARPi does not exclude their toxicity, especially considering PARPs comprehensive role and its omnipresence in the cell. PARPi show side effects that are characteristic for the class and for each drug separately that should be considered while making clinical decisions (LaFargue et al., 2019). (Supplementary Table S1).
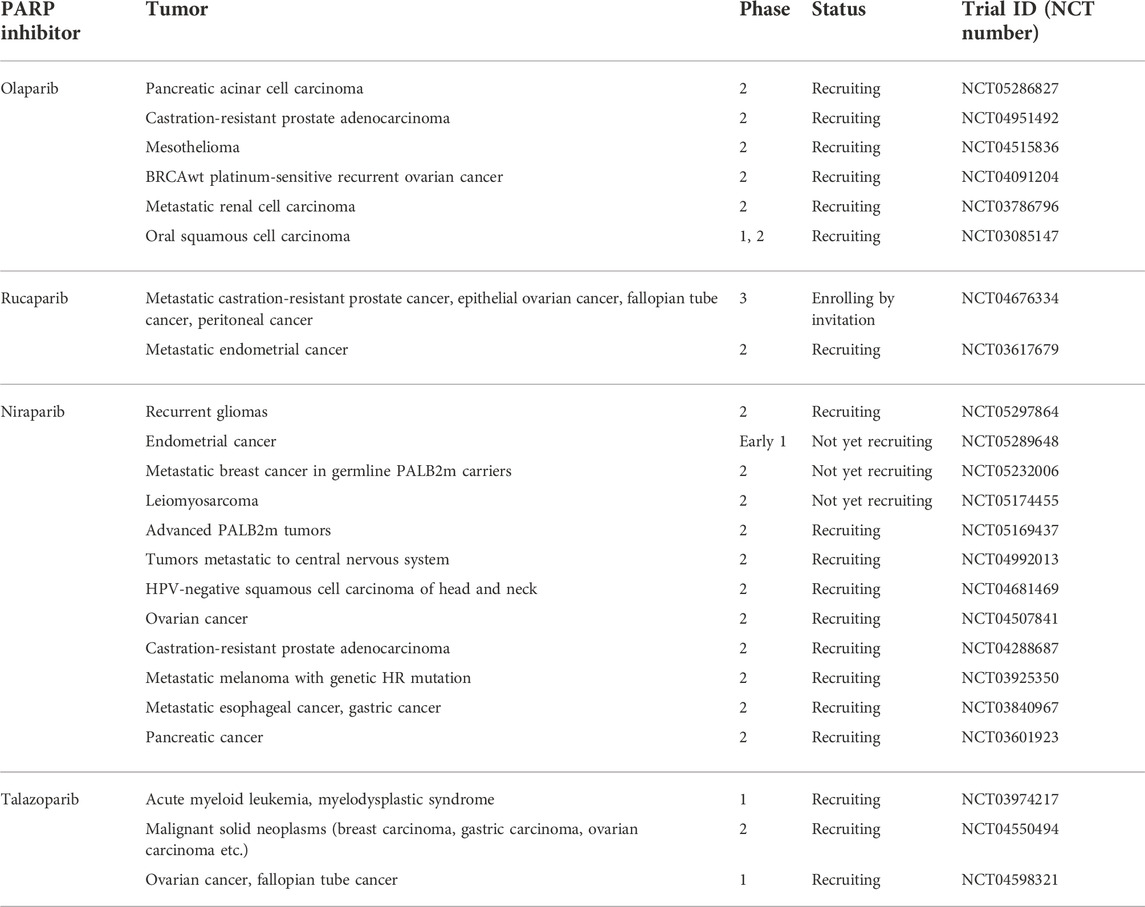
TABLE 2. List of selected PARP inhibitor monotherapy trials. Full access to the research studies description is available on ClinicalTrials.gov.
One of the biggest obstacles, which must be faced in the successful translation of PARPi into the clinic as an anti-cancer therapy, is frequently observed tumor resistance. The most well-known mechanism of DNA repair developed by tumor cells is the restoration of HR activity (Noordermeer and van Attikum, 2019). The mechanisms responsible for this are: recreation of BRCA1/2 activity, observed in clinical trials in patients with BRCA1/2m cancers (Kondrashova et al., 2017) (reverse mutations (Domchek, 2017) or gene fusion under the transcriptional control of heterologous promoter (Ter Brugge et al., 2016)) and suppression of NHEJ (caused by specific 53BP1 mutation in BRCA1 protein sequence) (Bouwman et al., 2010) (Hurley et al., 2019).
Downregulation of poly (ADP-ribose) glycohydrolase (PARG) protein levels is another hypothesized mechanism of PARPi resistance. Some studies suggest that depletion of PARG leads to PARPi resistance in BRCA2-deficient mouse mammary tumor models, which results in increased PAR levels even when PARP1 is largely or partially inhibited, thus counteracting PARP1 trapping and promoting PARPi resistance (Miwa et al., 1974) (Gogola et al., 2018). Moreover, miRNA expression patterns (especially miRNA-622 level) and drug efflux are also known to act as resistance mechanisms in PARPi therapy (Choi et al., 2016) (Rottenberg et al., 2008) (Vaidyanathan et al., 2016).
The observed resistance toward PARPi triggered development of strategies combining PARPi with other therapies. The beforementioned mechanism of PARP trapping is known to sensitize cells to an alkylating agent temozolomide, while the catalytic inhibition enhances the effect of topoisomerase inhibitors (Murai et al., 2012) (Murai et al., 2014). The results of combining chemotherapy with PARPi are confounding, as the dose-limiting tissue toxicity is often reported. Also, due to the overlapping mechanisms of action, the mechanisms of resistance can also be shared and become a prominent limiting factor. This top of the iceberg of mechanistic complexity of action of PARPi and chemotherapy agents resulted in relatively slow development of combination therapies (Dréan et al., 2016) (Lu et al., 2018). Despite this, a recent meta-analysis shows a promising, yet cautious, view on this topic (Ren et al., 2021).
An interesting direction of development of new combined therapies seems to be combination of PARPi with immunotherapies, one of the most promising being immune checkpoint inhibitors (ICIs). ICIs block surface proteins: T lymphocyte-associated antigen-4 (CTLA4) and programmed cell death receptor-1 (PD1) expressed by activated T-cells, and its ligand PD-L1 (Carlino et al., 2021). The mechanism of action of ICIs is based on enhancement of the immune response against cancer, namely the activation of T cells, which are stimulated by their surface receptors TCR and a costimulatory signal provided by CD28 (Lucas et al., 1995). PD-1 and CTLA4 are vital transmembrane receptor proteins engaged in the downregulation of T cells. PD1 binds to its PD-L1 ligand, which is present on tumor cells and antigen-presenting cells, causing a cascade of intracellular reactions leading to inactivation of the CD28 protein and inhibition of T cell activation (Walunas et al., 1996) (Brown et al., 2003) (Ishida et al., 2002) (Freeman et al., 2000). The anti-stimulatory effect of CTLA4 is slightly weaker. It is based on the competitive binding of CD28 ligands—B7-1 (CD80) and B7-2 (CD86) - located on antigen presenting cells (APC) with higher affinity (Peeraphatdit et al., 2020) (Peyraud and Italiano, 2020) (Wu et al., 2021) (Walunas et al., 1994) (Schweitzer and Sharpe, 1998). Malignant cells can create immunosuppressive tumor microenvironment (TME). TME arising relates to recruitment of regulatory T-cells (Treg) (Li et al., 2020). In TME PD1 and CTLA4 on T-cells and PD-L1 on cancer cells expression is upregulated. That prevents the effective anti-tumor immune response. Blocking these molecules by ICIs allows eliminating local suppression and inducing cancer-cell killing by CD8 positive T cells producing interferon gamma (IFN-γ) and tumor necrosis factor α (TNF-α) (Carlino et al., 2021) (Rameshbabu et al., 2021). The simplified rationale for combination of PARPi and ICIs was shown in Figure 3.
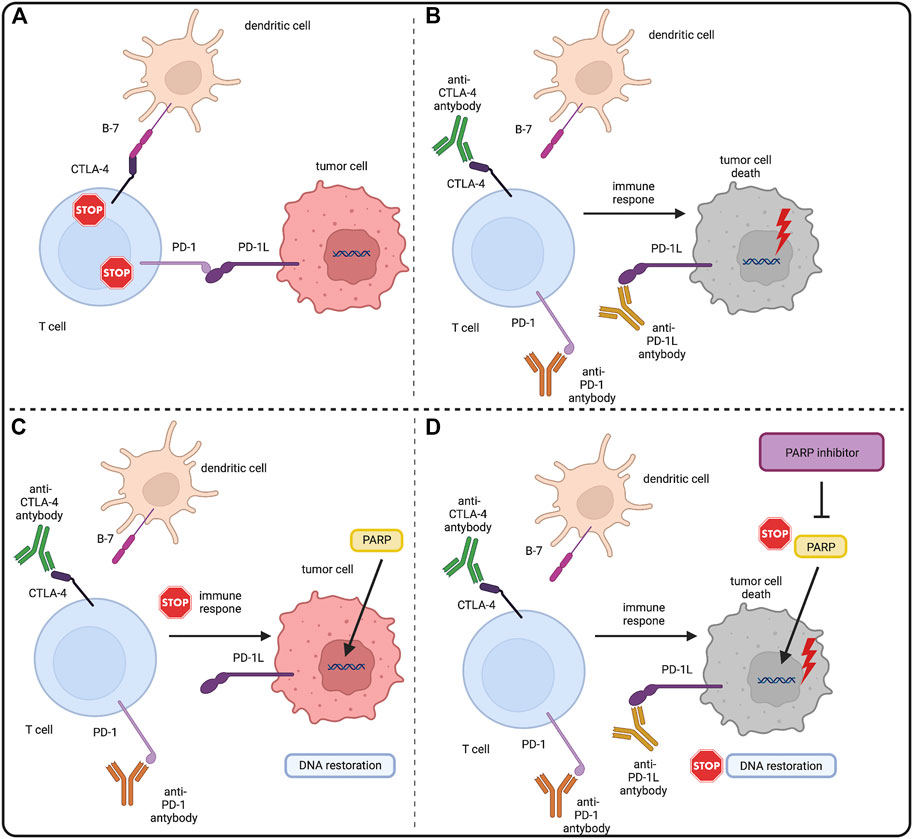
FIGURE 3. PARP inhibitors and immune checkpoint inhibitors–molecular rationale. (A) CTLA4 and PD1 are surface molecules present on T-cells. CTLA4 binds with the B7 molecule and PD1 binds with PD-L1 present on antigen presenting cells (APCs) and tumor cells. This downregulates the immunogenic response of both T-cells, allowing tumor cells to grow. (B) Immune checkpoint inhibitors (ICIs) by binding with CTLA4 and PD(L)1 block the downregulation of T-cells allowing them to eliminate tumor cells with the immune response. Also tumor cells genome instability makes them also more prone to the tumor mutation burden (TMB) and mutation overload leads to their death. (C) PARP role in DNA restoration allows tumor cells to repair DNA lesions and mitigate the TMB, eventually enabling them to evade the immune response. (D) PARP inhibitors by blocking the PARP DNA repair pathways in tumor cells stop the DNA restoration processes, exposing tumor cells to the immune response of T-cells under the treatment of ICIs. Created with BioRender.com.
The application of ICIs into clinical cancer immunotherapy has a huge impact on patients' life and cancer response to applied therapy. Since 2011, many ICIs have been approved by FDA. Presently available ICIs are targeting CTLA4 (ipilimumab), PD1 (nivolumab, pembrolizumab, camipilimab) and PD-L1 (atezolizumab, avelumab, durvalumab). Their indications include melanoma (Carlino et al., 2021) (Larkin et al., 2015), renal cell carcinoma, non-small cell lung carcinoma (NSCLC), head and neck squamous cell carcinoma (HNSCC), metastatic urothelial carcinoma, gastric cancer, metastatic triple negative breast cancer, hepatocellular carcinoma (HCC), solid tumors, Merkel cell carcinoma, colorectal cancer, classical Hodgkin’s lymphoma. Many clinical trials are currently underway, so the list of approved drugs and their indications can be expected to expand (Twomey and Zhang, 2021) (Lee et al., 2022). (Supplementary Table S2)
Combining PARPi therapy with agents interacting with PD1/PD-L1 pathways was based on observations that DNA-damaging agents lead to the activation of interferon pathways due to DNA damage (Bakhoum et al., 2018). It has also been observed that the level of interferon expression has an impact on levels of PD-L1 (Garcia-Diaz et al., 2017) and that PARPi themselves cause upregulation of PD-L1 (Jiao et al., 2017). The combination of these three pieces of information suggests the potential of combining PARP inhibitors as a DNA-damaging agent and drugs interacting with PD1/PD-L1 pathways. Moreover, numerous mutations occurring in tumor cells, particularly the non-synonymous single nucleotide variants (nsSNVs), inevitably lead to the tumor mutation burden (TMB) and may lead to the increase of immunogenic peptides, which are known to be significantly correlated with the ICI response (Snyder et al., 2014) (Rizvi et al., 2015) (Hellmann et al., 2018). TMB is also thought to be correlated with the neoantigen load on the tumor cells, being an important predictive factor of the therapeutic response of ICIs (Schumacher and Schreiber, 2015) (Lee et al., 2018) (McGranahan et al., 2016). Highly mutated tumors often exhibit deficiencies in DDR pathways, which are also suggested to be closely related to the TMB (Mouw et al., 2017). Thus, a possible novel therapeutic approaches that combine ICIs with DDR blocking agents have been considered. Here we focus on the combination of ICIs and PARPi, the latter being one of the most effective agents in their field.
Based on the promising results of various preclinical studies providing a rationale for combining PARPi with immunotherapy in cancer patients, (Jiao et al., 2017) (Wang et al., 2019b) (Ding et al., 2018) (Pantelidou et al., 2019) (Shen et al., 2019), several clinical trials have been conducted (Table 3). A significant group of the available clinical trials focused on non-small cell lung carcinoma (NSCLC). A study, evaluating dose and safety of veliparib combined with nivolumab and platinum doublet chemotherapy (pemetrexed and paclitaxel) focused on patients with metastatic and advanced NSCLC [NCT02944396]1. The overall response rate (ORR) reached 27% for cohort with pemetrexed and 17% for paclitaxel. This trial also confirmed the anticipated safety signals with no additional toxicity upon adding veliparib to these regimens (Clarke et al., 2021).
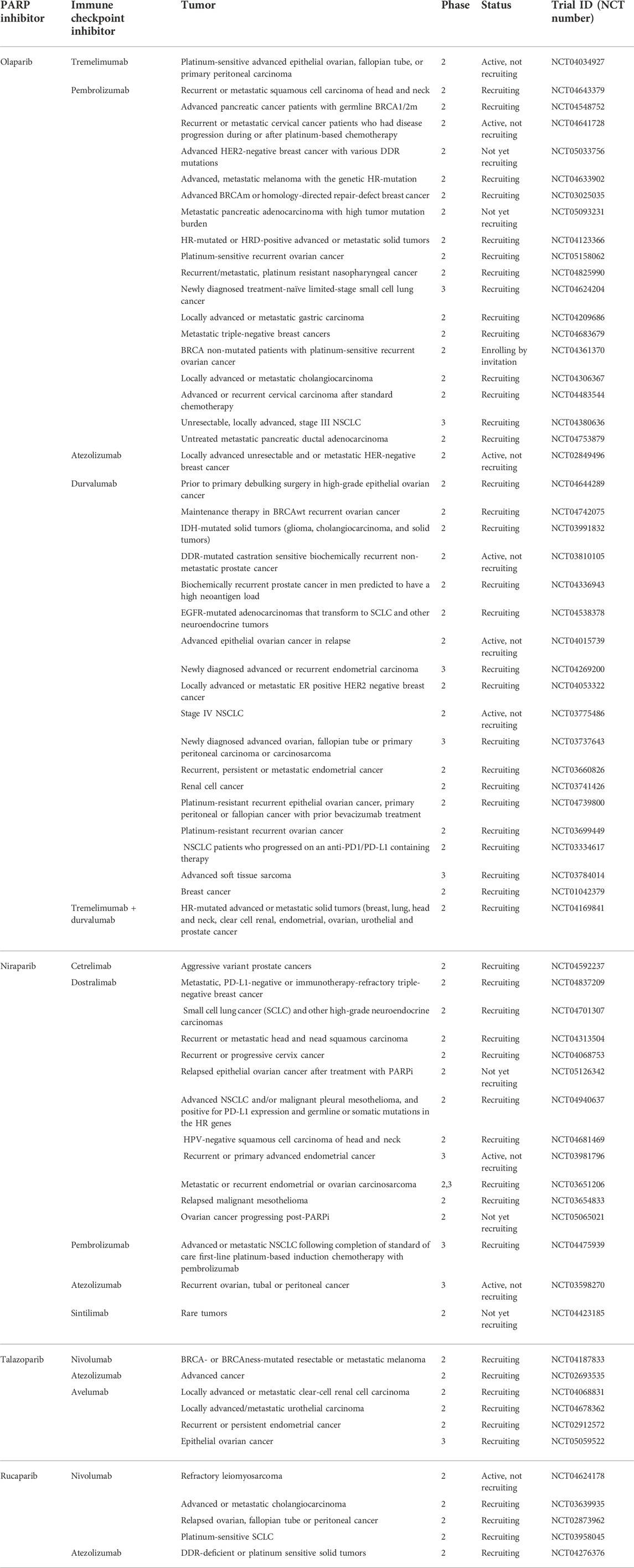
TABLE 3. List of selected PARP inhibitors and immune checkpoint inhibitors combination therapies. Full access to the research studies description is available on ClinicalTrials.gov.
PD-L1 is known target for most of the ICIs in use as monotherapies, therefore the impact of PD-L1 expression differences between individuals on the trials involving ICIs and PARPi was probed in an interventional JASPER phase II study [NCT03308942]2 tested the combination of niraparib and pembrolizumab or dostarlimab on a group of chemotherapy-naïve patients with locally advanced or metastatic NSCLC with no prior PD-(L1) chemotherapy. The group was divided into two cohorts regarding the PD-L1 expression status of patients: PD-L1-rich (tumor proportion score–TPS ≥ 50%) and PD-L1-poor (TPS <50%). The ORR, duration of response (DOR), progression-free survival (PFS), and safety were assessed as endpoints. The study demonstrated that the combination of niraparib and pembrolizumab induces durable responses in patients with NSCLC, with larger effects in the PD-L1-rich cohort. Moreover, the combination showed no new safety signals (Ramalingam et al., 2022). As this combination was shown to be active and well tolerated, the ZEAL-1L phase III study [NCT04475939]3 was launched to compare the efficacy and safety of maintenance of niraparib + pembrolizumab versus pembrolizumab + placebo in patients with NSCLC (Ramalingam et al., 2021).
Nonetheless, the combination of another PARPi, olaparib, and durvalumab in a phase II study [NCT02484404]4 applied to patients with relapsed SCLC did not meet the present bar for efficiency. The tumor responses were predicted by the preexisting TILs level, which suggests an immune-mediated response as a predictive marker. Therefore, identification of patients with inflamed phenotype at the baseline may help to identify those most likely to respond to ICIs, although the predictive value of the preexisting CD8+ T-cell infiltrates must be confirmed in larger cohorts (Thomas et al., 2019).
Although in case of prostate cancer, ICIs have been shown to be ineffective as single agents, the results from a cohort analysis of the phase II CheckMate 9KD trial [NCT03338790]5 suggest that the treatment with nivolumab plus rucaparib may produce positive results in patients with HRD-positive chemotherapy naive metastatic castration-resistant prostate cancer (mCRPC). However, there was limited clinical activity of the combination therapy in patients with HRD-negative tumors. The confirmed ORR among the patients with HRD-positive tumors was 25% in comparison with 5.3% for the HRD-negative patients (Fizazi et al., 2022).
Moreover, the combination of olaparib (agent that demonstrated an improvement in median PFS in patients with prostate cancer) and durvalumab evaluated in the castration-resistant prostate cancer in a phase I/II MEDI4736 study [NCT02484404] showed eight out of 17 patients exhibited radiographic and/or PSA responses. The efficacy was noted particularly in men with DDR abnormalities (12-month PFS probability of DDR-deficient reached 83,3%, vs. 36,4% in DDR-proficient patients). Those with fewer peripheral myeloid-derived suppressor cells (pMDSC) were also more likely to respond. This suggests DDR deficiency and pMDSC level as predictive markers of the response (Karzai et al., 2018).
In order to identify biomarkers of a promising response to a combined PARPi and ICI, immunogenomic profiling and single-cell imaging have been performed on tumor samples subjected to such regimens. In a phase I/II trial [NCT02657889]6 of niraparib and pembrolizumab in ovarian cancer two determinants of response were identified: mutational signature 3 (correlating with the HR in DDR), and positive immune score as a function of interferon-primed exhausted CD8+ T cells in the tumor microenvironment. The interactions of exhausted CD8+ T-cells, PD-L1+ macrophages, and PD-L1 tumor cells with each other were noted in the single-cell spatial analysis, and PD-L1 tumor cells were shown to be the mechanistic response determinants, confirming the observations from previous studies (Färkkilä et al., 2020).
Patients with various solid tumors, including ovarian, breast and gastric cancer, were investigated in a phase I/II study MEDIOLA [NCT02734004]7 evaluating the effect of the combination of olaparib and durvalumab. In a germline BRCA1/2m metastatic breast cancer group, 80% of patients had disease control at 12 weeks and 50% at 28 weeks. Higher ORR and longer overall survival (OS) were observed in patients who had no prior line of chemotherapy in comparison to those with two prior lines (78% ORR and 21,3 months OS vs. 50% ORR and 16,9 months OS respectively). The investigated combination of agents exhibited promising activity and safety consistent with the profiles of individual agents (Domchek et al., 2020). In another cohort of this study enrolling patients with germline BRCA1/2m platinum-sensitive relapsed ovarian cancer, showed ORR of 63% and a 12-week DCR of 81%. The combination was well tolerated and the tumor responses in this initial analysis were higher in comparison with those reported for single-agent therapy with PARPi (Drew et al., 2018). However the results of another cohort of this study compassing patients with relapsed gastric cancer were less promising. The ORR was 10% and the disease control rate (DCR) at 26%. The combination was tolerable, with no unexpected adverse events. The durable responses after the combination of olaparib and durvalumab suggest synergistic treatment effect of the combination in some patients. Nonetheless, DCR value did not meet the target because of the high rate of early progressive diseases (PDs) occuring after the olaparib run in. Therefore, due to the initial treatment failures, an addition of new more effective therapies to the combination should be taken into account (Bang et al., 2019).
Besides this, several different phase I/II clinical trials were performed to verify the safety, toxicity and tolerability of the combination of PARPi and ICIs on patients with advanced solid tumors. In phase Ib IOLite study [NCT03307785]8 dostarlimab in combination with niraparib and niraparib + bevacizumab was shown to be well tolerated. Besides this, the study also evaluated combination of chemotherapy with PARPi and ICI, also with good tolerability. None of the combination agents used in this study altered the pharmacokinetics of dostarlimab nor niraparib. Moreover, no new safety signals were noted (Gabrail et al., 2019). In another phase I/II TOPACIO trial [NCT02657889]9 therapy combining niraparib and pembrolizumab in patients with ovarian and triple negative breast carcinoma showed general clinical improvement. However, the ovarian carcinoma cohort did not meet the primary endpoint with ORR of 18%, and the median duration of the response was not reached as well (Konstantinopoulos et al., 2019). The significantly higher response rates in patients with BRCA1/2m tumors was observed only in the breast cancer arm, with ORR of BRCA1/2m of 74% vs. wtBRCA1/2 11%. The PFS and DCR were 8.3 months and 80% for BRCAm, and 2.1 month with 33% for wtBRCA1/2, respectively (Vinayak et al., 2019). Finally, an ongoing JAVELIN PARP Mendley phase Ib/II study [NCT03330405]10 enrolled patients with advanced breast cancer cohorts treated with avelumab plus talazoparib. It showed preliminary antitumor activity and safety profile comparable to that of these agents used as monotherapies (Yap et al., 2020).
The clinical trials regarding combined therapies of PARPi and ICIs on patients with advanced solid tumors comprise also a novel agents. Pamiparib is an experimental selective PARP1/2i recently approved in China for the treatment of germline BRCA1/2m-associated recurrent advanced ovarian, fallopian tube or primary peritoneal cancer previously treated with two or more lines of chemotherapy [NCT03333915]11 (Markham, 2021). The safety of combination of pamiparib and tiselizumab was explored in a phase Ia/b trial [NCT02660034]12 on patients with advanced solid tumor. The combination treatment achieved an ORR of 20% and was well-tolerated, although the higher rate of immune-related hepatitis was noted in 8% of patients (Friedlander et al., 2019).
Even though the standard of care for muscle-invasive bladder cancer (MIBC) remains the relatively curative radical cystectomy, new non-surgery treatment approaches, including PARPi and ICIs, are being developed. The preliminary data from the phase II NEODURVARIB trial [NCT03534492]13 suggest that durvalumab in combination with olaparib administered prior to the surgery could be active and tolerated neoadjuvant treatment for MIBC, with the pathological complete response rate of 44,5% (Rodriguez-Moreno et al., 2020).
Numerous clinical trials are still ongoing in a vast range of cancers that will help to probe the features of the PARPi plus anti-PD-(L)1 combination therapy.
In contrast to the anti-PD(-L)1 combination, which is investigated with much attention, the combination therapies of anti-CTLA4 and PARPi are largely unexplored. Despite the fact that previous studies demonstrated that tumors harboring BRCA1/2 dysfunction and treated with PARPi could increase tumor immunogenicity, therefore sensitizing tumor cells to anti-CTLA4 agents, the whole process remains understudied (Snyder et al., 2014) (Clarke et al., 2009) (McAlpine et al., 2012) (Wen and Leong, 2019) (Brown et al., 2016) (Higuchi et al., 2015).
Nonetheless, few important trials are currently conducted. The combination of olaparib and tremelimumab was verified in a phase I/II study [NCT02571725]14 involving women with BRCA1/2-deficient recurrent ovarian cancer. The preliminary results demonstrated a significant therapeutic effect along with acceptable tolerability (Adams et al., 2017).
This combination of olaparib plus tremelimumab is also under investigation in an ongoing phase II trial for patients with recurrent ovarian, fallopian tube or peritoneal cancer [NCT04034927]15.
In an ongoing phase II study [NCT04169841]16 the efficacy of olaparib combined with double immunotherapy of durvalumab and tremelimumab is being evaluated on patients with solid cancers, who were selected for this study based on their HR repair mutation profile and response after a previous olaparib treatment. This is the first clinical trial evaluating the combination of PARPi with ICI double therapy. Nonetheless, the combination of durvalumab plus tremelimumab itself has been studied before. To date, it has not shown significant advantages in comparison with the durvalumab monotherapy but displayed certain advantages over the traditional chemotherapies in some tumors, although more data of higher quality is required in this area (Arru et al., 2021).
Despite a relative shortage of published results regarding the combination of PARPi and anti-CTLA4 antibodies, the ongoing clinical trials may help to revive the promising antitumor activity of this approach.
The successful introduction of PARPi into current oncology treatment methods causes an urgent need to develop modern techniques, which can help to predict tumor sensitivity to this type of therapy. A useful role in the selection process could be played by some specific biomarkers.
The most predictive marker at present are BRCA1/2m (Ganguly et al., 2016). Latest research studies demonstrated that a general testing of ovarian cancer patients for the presence of BRCA1/2m could be useful in planning anticancer therapy (Vos et al., 2020). Prediction of PARPi sensitivity is facilitated by several interesting approaches. One of them is BRCAnalysis CDx, which offers the analysis of germlineBRCA1/2m–unfortunately, this method does not measure HR deficiency and might miss some of the patients who could have benefited from PARPi treatment (Gunderson and Moore, 2015). Another one is FoundationOne Liquid CDx, which offers tests of 324 genes including BRCA1 and BRCA2 (Woodhouse et al., 2020). Unfortunately, the status of BRCA1/does not always correlate with tumor sensitivity to PARPi therapy (Jonsson et al., 2019). Luckily, some other mutations have been described as potential biomarkers to predict therapeutic efficiency (Criscuolo et al., 2019). Also, a HR deficiency score was presented as a possible biomarker of chemotherapy efficacy in some tumors (Telli et al., 2016), and it could conceivably be extended to predicting the possible cancer response to PARPi therapy. Furthermore, specific methylation patterns could also be helpful in prediction of therapy effectiveness. Recent research studies presented hypermethylation of RAD51 (Kondrashova et al., 2018) or hypermetylation of HOXA9 (Montavon et al., 2012) as such promising markers.
Other potentially valuable approaches are focused on the assessment of the expression levels and modulation of various PARP1 regulators. One of these proteins is HPF1. Depletion of this protein makes PARPi more effective and induces tumor cell sensitivity to treatment with PARPi and other DNA-interacting drugs (Gibbs-Seymour et al., 2016). Another one is Y-box-binding protein (YB1), which inhibits PAR degradation by PARG (Alemasova et al., 2016). YB1 was presented as a protein playing a crucial role in tumor cell chemoresistance and a beneficial role of combining therapy with PARP inhibitors and DNA damaging agents was suggested as a potential answer to YB1 activity (Alemasova et al., 2018). The assessment of the YB1 level in patients with tumors could also be used as a promising biomarker in PARPi treatment. Moreover, depletion of PARP1-interacting Src-associated substrate during mitosis 68 kDA (Sam68), leads to impaired PARP1 activation, establishing Sam68 as a potential PARP1 activator (Fu et al., 2016). Using Sam68 as a biomarker could boost potential cancer therapy (Zhang et al., 2015) (Zhang et al., 2009). The other tested proteins were Barrier to Autointegration Factor 1 (Banf1)—a negative regulator of PARP1 activity (Bolderson et al., 2019) and TRIP12—ubiquitin E3 ligase, which regulates PARP1 stability (Gatti et al., 2020). These two proteins could also play an important role in the prediction of success in PARPi therapy and need to be studied further.
Recent years have brought plethora of studies exploring the combination of PARPi with other agents. The efficacy of the combination therapy of PARPi and ICIs, which we discussed in this review, is being tested in numerous clinical trials.
Nonetheless clinical trials themselves are not fully conclusive. Most of them rely on ORR or DCR, the early endpoints, which provide information only about the initial phase of therapy, while the prolonged monitoring data are still mostly undetermined. Moreover, all current clinical trials are non-randomized, allowing only cross-trial comparison. Thus, PARPi and ICI treatment strategies should be optimized by recruiting randomized controlled multi arms phase III trials, which are designed to enable the interpretation of the effect of each drug alone or in combination.
Another blind spot of clinical studies involving PARPi and ICIs is that most of the tumor types where the combination strategy was tested already had demonstrated significant improvement from PARPi monotherapy and limited benefit from ICI addition. Therefore, it would be reasonable to investigate the new combination of PARPi and ICIs in a group of patients that have an unmet anticancer clinical need, rather than in those with well-established therapies. Thus, studies involving patients who do not respond well to PARPi or ICI monotherapies should be taken into account. Finally, the schedule and timing of these studies should be optimized to preserve tolerability and the associated impact on health care costs should also be considered in terms of the therapy duration.
Although the mechanism of the combinatorial effect of PARPi and ICIs is now intensively investigated and several mechanisms have been proposed to be involved in this process in HR-deficient patients, like those with BRCA1/2 dysfunction, the rationale for using this therapy on patients with functional HR mechanisms is yet to be determined. Uncovering the rationale of this combining therapy in non-HRD patients and more preclinical in vivo trials will be crucial to pick up the target group of patients that would benefit most of the therapy.
The crucial factor in identifying the optimal target population of patients are biomarkers. Although in tumors with BRCA1/2m or HRD the effect is beneficial, HR-proficient patient markers are yet to be determined. Elucidating the molecular profile of this population will allow answering the question whether the tumors could be sensitized to ICIs by PARPi. The deeper understanding of molecular mechanisms of PARPi and ICI pathways should result in a collection of biomarkers that could help to understand the potential sensitivity and resistance.
PARPi monotherapy has proved to be a milestone in the treatment of many BRCA1/2m cancers, bringing patients hope of an effective therapy. Throughout the years many therapies combining PARPi with different agents were proposed, ICIs being the promising among them, Currently, numerous clinical trials are ongoing, validating the combined therapies with PARPi and ICIs. Although the studies do not encompass the whole complexity of these agents, the available data is promising. The key to comprehend the true power of PARPi- ICI combination is an in-depth understanding of their molecular mechanism. Identifying such biomarkers might also facilitate the search for adequate biomarkers that could help to guide the doctor’s hand to treat the patients in the most effective way and find the most suitable place for the combination of PARPi and ICIs in the clinic.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This work has been supported by a grant 013/RID/2018/19 (Regional Initiative for Excellence) from the Polish Ministry of Education and Science, project budget 12,000,000 PLN.
All of the illustrations in the manuscript were created with Biorender.com. We gratefully thank Paweł Matryba and Łukasz Komorowski (Department of Immunology, Medical University of Warsaw), and Karolina Wójcik (Department of Biomaterials and Composites, AGH University of Science and Technology, Kraków) for helpful editorial insights and guidance preceding the submission of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.1073797/full#supplementary-material
1Phase 2, Multi-Arm Study of Niraparib Administered Alone and in Combination With a PD-1 Inhibitor in Patients With Non-Small Cell Lung Cancer. Available online at: https://clinicaltrials.gov/ct2/show/NCT03308942 (accessed 17 October 2022).
2Phase 2, Multi-Arm Study of Niraparib Administered Alone and in Combination With a PD-1 Inhibitor in Patients With Non-Small Cell Lung Cancer, Available online at: https://clinicaltrials.gov/ct2/show/NCT03308942 (accessed 17 October 2022).
3Phase 3, Randomized, Double-Blind, Placebo-Controlled, Multicenter Study Comparing Niraparib Plus Pembrolizumab Versus Placebo Plus Pembrolizumab as Maintenance Therapy in Participants Whose Disease Has Remained Stable or Responded to First-Line Platinum Based Chemotherapy With Pembrolizumab for Stage IIIB/IIIC or IV Non-Small Cell Lung Cancer (ZEAL-1L). Available online at: https://clinicaltrials.gov/ct2/show/NCT04475939 (accessed 17 October 2022).
4Phase I/II Study of the Anti-Programmed Death Ligand-1 Antibody Durvalumab (MEDI4736) in Combination With Olaparib and/or Cediranib for Advanced Solid Tumors and Advanced or Recurrent Ovarian, Triple Negative Breast, Lung, Prostate and Colorectal Cancers, Available online at: https://www.clinicaltrials.gov/ct2/show/NCT02484404 (accessed 17 October 2022).
5A Phase 2 Study of Nivolumab in Combination With Either Rucaparib, Docetaxel, or Enzalutamide in Men With Castration-resistant Metastatic Prostate Cancer. Available online at: https://clinicaltrials.gov/ct2/show/NCT03338790 (accessed 17 October 2022).
6Phase 1/2 Clinical Study of Niraparib in Combination With Pembrolizumab (MK-3475) in Patients With Advanced or Metastatic Triple-Negative Breast Cancer and in Patients With Recurrent Ovarian Cancer. Available online at: https://clinicaltrials.gov/ct2/show/NCT02657889 (accessed 17 October 2022).
7A Phase I/II Study of MEDI4736 (Anti-PD-L1 Antibody) in Combination With Olaparib (PARP Inhibitor) in Patients With Advanced Solid Tumors. Available online at: https://clinicaltrials.gov/ct2/show/NCT02734004 (accessed 17 October 2022).
8Phase 1b Dose-Finding Study of Niraparib, TSR-022, Bevacizumab, and Platinum-Based Doublet Chemotherapy in Combination With TSR-042 in Patients With Advanced or Metastatic Cancer. Available online at: https://www.clinicaltrials.gov/ct2/show/NCT03307785 (accessed 17 October 2022).
9Phase 1/2 Clinical Study of Niraparib in Combination With Pembrolizumab (MK-3475) in Patients With Advanced or Metastatic Triple-Negative Breast Cancer and in Patients With Recurrent Ovarian Cancer. Available online at: https://clinicaltrials.gov/ct2/show/NCT02657889 (accessed 17 October 2022).
10Phase 1/2 Clinical Study of Niraparib in Combination With Pembrolizumab (MK-3475) in Patients With Advanced or Metastatic Triple-Negative Breast Cancer and in Patients With Recurrent Ovarian Cancer. Available online at: https://clinicaltrials.gov/ct2/show/NCT02657889 (accessed 17 October 2022).
11An Open Label, Multi-Center Phase I/II Study to Evaluate Efficacy and Safety of BGB-290 in Chinese Subjects With Advanced Ovarian Cancer, Fallopian Cancer, and Primary Peritoneal Cancer or Advanced Triple Negative Breast Cancer. Available online at: https://clinicaltrials.gov/ct2/show/NCT03333915 (accessed 17 October 2022).
12A Phase 1/1b, Open Label, Multiple Dose, Dose Escalation and Expansion Study to Investigate the Safety, Pharmacokinetics and Antitumor Activity of the Anti-PD-1 Monoclonal Antibody BGB-A317 in Combination With the PARP Inhibitor BGB-290 in Subjects With Advanced Solid Tumors. Available online at: https://clinicaltrials.gov/ct2/show/NCT02660034 (accessed 17 October 2022).
13Impact of the Combination of Durvalumab (MEDI4736) Plus Olaparib (AZD2281) Administered Prior to Surgery in the Molecular Profile of Resectable Urothelial Bladder Cancer. Available online at: https://clinicaltrials.gov/ct2/show/NCT03534492 (accessed 17 October 2022).
14Phase 1-2 Study of the Combination of Olaparib and Tremelimumab, in BRCA1 and BRCA2 Mutation Carriers With Recurrent Ovarian Cancer. Available online at: https://clinicaltrials.gov/ct2/show/NCT02571725 (accessed 17 October 2022).
15A Phase II Randomized Trial of Olaparib Versus Olaparib Plus Tremelimumab in Platinum-Sensitive Recurrent Ovarian Cancer. Available online at: https://clinicaltrials.gov/ct2/show/NCT04034927 (accessed 17 October 2022).
16Precision Medicine Phase II Study Evaluating the Efficacy of a Double Immunotherapy by Durvalumab and Tremelimumab Combined With Olaparib in Patients With Solid Cancers and Carriers of Homologous Recombination Repair Genes Mutation in Response or Stable After Olaparib Treatment. Available online at: https://clinicaltrials.gov/ct2/show/NCT04169841 (accessed 17 October 2022).
Abbotts, R., and Wilson, D. M. (2017). Coordination of DNA single strand break repair. Free Radic. Biol. Med. 107, 228–244. doi:10.1016/j.freeradbiomed.2016.11.039
Abida, W., Campbell, D., Patnaik, A., Sautois, B., Shapiro, J., Vogelzang, N. J., et al. (2019). Preliminary results from the TRITON2 study of rucaparib in patients (pts) with DNA damage repair (DDR)-deficient metastatic castration-resistant prostate cancer (mCRPC): Updated analyses. Ann. Oncol. 30, v327–v328. doi:10.1093/annonc/mdz248.003
Adams, S. F., Rixe, O., Lee, J.-H., McCance, D. J., Westgate, S., Eberhardt, S. C., et al. (2017). Phase I study combining olaparib and tremelimumab for the treatment of women with BRCA-deficient recurrent ovarian cancer. J. Clin. Oncol. 35, e17052. doi:10.1200/JCO.2017.35.15_suppl.e17052
Afzal, H., Yousaf, S., Rahman, F., Ahmed, M. W., Akram, Z., Akhtar Kayani, M., et al. (2019). PARP1: A potential biomarker for gastric cancer. Pathol. Res. Pract. 215, 152472. doi:10.1016/j.prp.2019.152472
Alemasova, E. E., Moor, N. A., Naumenko, K. N., Kutuzov, M. M., Sukhanova, M. V., Pestryakov, P. E., et al. (2016). Y-box-binding protein 1 as a non-canonical factor of base excision repair. Biochim. Biophys. Acta 1864, 1631–1640. doi:10.1016/j.bbapap.2016.08.012
Alemasova, E. E., Naumenko, K. N., Kurgina, T. A., Anarbaev, R. O., and Lavrik, O. I. (2018). The multifunctional protein YB-1 potentiates PARP1 activity and decreases the efficiency of PARP1 inhibitors. Oncotarget 9, 23349–23365. doi:10.18632/oncotarget.25158
Andrabi, S. A., Dawson, T. M., and Dawson, V. L. (2008). Mitochondrial and nuclear cross talk in cell death: Parthanatos. Ann. N. Y. Acad. Sci. 1147, 233–241. doi:10.1196/annals.1427.014
Andrabi, S. A., Umanah, G. K. E., Chang, C., Stevens, D. A., Karuppagounder, S. S., Gagné, J.-P., et al. (2014). Poly(ADP-ribose) polymerase-dependent energy depletion occurs through inhibition of glycolysis. Proc. Natl. Acad. Sci. U. S. A. 111, 10209–10214. doi:10.1073/pnas.1405158111
Arru, C., De Miglio, M. R., Cossu, A., Muroni, M. R., Carru, C., Zinellu, A., et al. (2021). Durvalumab plus tremelimumab in solid tumors: A systematic review. Adv. Ther. 38, 3674–3693. doi:10.1007/s12325-021-01796-6
Ashworth, A. (2008). A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J. Clin. Oncol. 26, 3785–3790. doi:10.1200/JCO.2008.16.0812
Bai, P., Cantó, C., Oudart, H., Brunyánszki, A., Cen, Y., Thomas, C., et al. (2011). PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell. Metab. 13, 461–468. doi:10.1016/j.cmet.2011.03.004
Bakhoum, S. F., Ngo, B., Laughney, A. M., Cavallo, J.-A., Murphy, C. J., Ly, P., et al. (2018). Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472. doi:10.1038/nature25432
Bang, Y.-J., Kaufman, B., Geva, R., Stemmer, S. M., Hong, S.-H., Lee, J.-S., et al. (2019). An open-label, phase II basket study of olaparib and durvalumab (MEDIOLA): Results in patients with relapsed gastric cancer. J. Clin. Oncol. 37, 140. doi:10.1200/JCO.2019.37.4_suppl.140
Bendixen, C., Thomsen, B., Alsner, J., and Westergaard, O. (1990). Camptothecin-stabilized topoisomerase I-DNA adducts cause premature termination of transcription. Biochemistry 29, 5613–5619. doi:10.1021/bi00475a028
Bilokapic, S., Suskiewicz, M. J., Ahel, I., and Halic, M. (2020). Bridging of DNA breaks activates PARP2–HPF1 to modify chromatin. Nature 585, 609–613. doi:10.1038/s41586-020-2725-7
Bolderson, E., Burgess, J. T., Li, J., Gandhi, N. S., Boucher, D., Croft, L. V., et al. (2019). Barrier-to-autointegration factor 1 (Banf1) regulates poly [ADP-ribose] polymerase 1 (PARP1) activity following oxidative DNA damage. Nat. Commun. 10, 5501. doi:10.1038/s41467-019-13167-5
Bouchard, V. J., Rouleau, M., and Poirier, G. G. (2003). PARP-1, a determinant of cell survival in response to DNA damage. Exp. Hematol. 31, 446–454. doi:10.1016/s0301-472x(03)00083-3
Bouwman, P., Aly, A., Escandell, J. M., Pieterse, M., Bartkova, J., van der Gulden, H., et al. (2010). 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 17, 688–695. doi:10.1038/nsmb.1831
Branzei, D., and Foiani, M. (2008). Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell. Biol. 9, 297–308. doi:10.1038/nrm2351
Brenner, J. C., Feng, F. Y., Han, S., Patel, S., Goyal, S. V., Bou-Maroun, L. M., et al. (2012). PARP-1 inhibition as a targeted strategy to treat Ewing’s sarcoma. Cancer Res. 72, 1608–1613. doi:10.1158/0008-5472.CAN-11-3648
Brown, J. A., Dorfman, D. M., Ma, F.-R., Sullivan, E. L., Munoz, O., Wood, C. R., et al. (2003). Blockade of programmed death-1 ligands on dendritic cells enhances T cell activation and cytokine production. J. Immunol. 170, 1257–1266. doi:10.4049/jimmunol.170.3.1257
Brown, J. S., Kaye, S. B., and Yap, T. A. (2016). PARP inhibitors: The race is on. Br. J. Cancer 114, 713–715. doi:10.1038/bjc.2016.67
Bryant, H. E., Petermann, E., Schultz, N., Jemth, A.-S., Loseva, O., Issaeva, N., et al. (2009). PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 28, 2601–2615. doi:10.1038/emboj.2009.206
Bryant, H. E., Schultz, N., Thomas, H. D., Parker, K. M., Flower, D., Lopez, E., et al. (2005). Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917. doi:10.1038/nature03443
Caiafa, P., Guastafierro, T., and Zampieri, M. (2009). Epigenetics: poly(ADP-ribosyl)ation of PARP-1 regulates genomic methylation patterns. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 23, 672–678. doi:10.1096/fj.08-123265
Calabrese, C. R., Batey, M. A., Thomas, H. D., Durkacz, B. W., Wang, L.-Z., Kyle, S., et al. (2003). Identification of potent nontoxic poly(ADP-ribose) polymerase-1 inhibitors: Chemopotentiation and pharmacological studies. Clin. Cancer Res. 9, 2711–2718.
Caldecott, K. W. (2008). Single-strand break repair and genetic disease. Nat. Rev. Genet. 9, 619–631. doi:10.1038/nrg2380
Calvert, H., and Azzariti, A. (2011). The clinical development of inhibitors of poly(ADP-ribose) polymerase. Ann. Oncol. 22 (1), i53–i59. doi:10.1093/annonc/mdq667
Carlino, M. S., Larkin, J., and Long, G. V. (2021). Immune checkpoint inhibitors in melanoma. Lancet 398, 1002–1014. doi:10.1016/S0140-6736(21)01206-X
Chambon, P., Weill, J. D., and Mandel, P. (1963). Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem. Biophys. Res. Commun. 11, 39–43. doi:10.1016/0006-291X(63)90024-X
Chapman, J. R., Taylor, M. R. G., and Boulton, S. J. (2012). Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 47, 497–510. doi:10.1016/j.molcel.2012.07.029
Choi, Y. E., Meghani, K., Brault, M.-E., Leclerc, L., He, Y. J., Day, T. A., et al. (2016). Platinum and PARP inhibitor resistance due to overexpression of MicroRNA-622 in BRCA1-mutant ovarian cancer. Cell. Rep. 14, 429–439. doi:10.1016/j.celrep.2015.12.046
Clarke, B., Tinker, A. V., Lee, C.-H., Subramanian, S., van de Rijn, M., Turbin, D., et al. (2009). Intraepithelial T cells and prognosis in ovarian carcinoma: Novel associations with stage, tumor type, and BRCA1 loss. Mod. Pathol. 22, 393–402. doi:10.1038/modpathol.2008.191
Clarke, J. M., Patel, J. D., Robert, F., Kio, E. A., Thara, E., Ross Camidge, D., et al. (2021). Veliparib and nivolumab in combination with platinum doublet chemotherapy in patients with metastatic or advanced non-small cell lung cancer: A phase 1 dose escalation study. Lung Cancer 161, 180–188. doi:10.1016/j.lungcan.2021.09.004
Coleman, R. L., Fleming, G. F., Brady, M. F., Swisher, E. M., Steffensen, K. D., Friedlander, M., et al. (2019). Veliparib with irst-line chemotherapy and as maintenance therapy in ovarian cancer. N. Engl. J. Med. 381, 2403–2415. doi:10.1056/NEJMoa1909707
Coleman, R. L., Oza, A. M., Lorusso, D., Aghajanian, C., Oaknin, A., Dean, A., et al. (2017). ARIEL3 investigatorsRucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 390, 1949–1961. doi:10.1016/S0140-6736(17)32440-6
Cong, K., Peng, M., Kousholt, A. N., Lee, W. T. C., Lee, S., Nayak, S., et al. (2021). Replication gaps are a key determinant of PARP inhibitor synthetic lethality with BRCA deficiency. Mol. Cell. 81, 3128–3144.e7. e7. doi:10.1016/j.molcel.2021.06.011
Criscuolo, D., Morra, F., Giannella, R., Cerrato, A., and Celetti, A. (2019). Identification of novel biomarkers of homologous recombination defect in DNA repair to predict sensitivity of prostate cancer cells to PARP-inhibitors. Int. J. Mol. Sci. 20, E3100. doi:10.3390/ijms20123100
D’Amours, D., Desnoyers, S., D’Silva, I., and Poirier, G. G. (1999). Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 342 (2), 249–268. doi:10.1042/0264-6021:3420249
Dantzer, F., de la Rubia, G., Ménissier-de Murcia, J., Hostomsky, Z., de Murcia, G., and Schreiber, V. (2000). Base excision repair is impaired in mammalian cells lacking poly(ADP-ribose) polymerase-1. Biochemistry 39, 7559–7569. doi:10.1021/bi0003442
Dawicki-McKenna, J. M., Langelier, M.-F., DeNizio, J. E., Riccio, A. A., Cao, C. D., Karch, K. R., et al. (2015). PARP-1 activation requires local unfolding of an autoinhibitory domain. Mol. Cell. 60, 755–768. doi:10.1016/j.molcel.2015.10.013
de Bono, J., Mateo, J., Fizazi, K., Saad, F., Shore, N., Sandhu, S., et al. (2020). Olaparib for metastatic castration-resistant prostate cancer. N. Engl. J. Med. 382, 2091–2102. doi:10.1056/NEJMoa1911440
Deeks, E. D. (2015). Olaparib: First global approval. Drugs 75, 231–240. doi:10.1007/s40265-015-0345-6
Del Campo, J. M., Matulonis, U. A., Malander, S., Provencher, D., Mahner, S., Follana, P., et al. (2019). Niraparib maintenance therapy in patients with recurrent ovarian cancer after a partial response to the last platinum-based chemotherapy in the ENGOT-OV16/NOVA trial. J. Clin. Oncol. 37, 2968–2973. doi:10.1200/JCO.18.02238
Devalaraja-Narashimha, K., and Padanilam, B. J. (2010). PARP1 deficiency exacerbates diet-induced obesity in mice. J. Endocrinol. 205, 243–252. doi:10.1677/JOE-09-0402
Dianov, G. L., and Hübscher, U. (2013). Mammalian base excision repair: The forgotten archangel. Nucleic Acids Res. 41, 3483–3490. doi:10.1093/nar/gkt076
Ding, L., Kim, H.-J., Wang, Q., Kearns, M., Jiang, T., Ohlson, C. E., et al. (2018). PARP inhibition elicits STING-dependent antitumor immunity in brca1-deficient ovarian cancer. Cell. Rep. 25, 2972–2980. e5. doi:10.1016/j.celrep.2018.11.054
Domchek, S. M., Postel-Vinay, S., Im, S.-A., Park, Y. H., Delord, J.-P., Italiano, A., et al. (2020). Olaparib and durvalumab in patients with germline BRCA-mutated metastatic breast cancer (MEDIOLA): An open-label, multicentre, phase 1/2, basket study. Lancet. Oncol. 21, 1155–1164. doi:10.1016/S1470-2045(20)30324-7
Domchek, S. M. (2017). Reversion mutations with clinical use of PARP inhibitors: Many genes, many versions. Cancer Discov. 7, 937–939. doi:10.1158/2159-8290.CD-17-0734
Dréan, A., Lord, C. J., and Ashworth, A. (2016). PARP inhibitor combination therapy. Crit. Rev. Oncol. Hematol. 108, 73–85. doi:10.1016/j.critrevonc.2016.10.010
Drew, Y., de Jonge, M., Hong, S. H., Park, Y. H., Wolfer, A., Brown, J., et al. (2018). An open-label, phase II basket study of olaparib and durvalumab (MEDIOLA): Results in germline BRCA -mutated ( gBRCA m) platinum-sensitive relapsed (PSR) ovarian cancer (OC). Gynecol. Oncol. 149, 246–247. doi:10.1016/j.ygyno.2018.04.555
Durkacz, B. W., Omidiji, O., Gray, D. A., and Shall, S. (1980). (ADP-ribose)n participates in DNA excision repair. Nature 283, 593–596. doi:10.1038/283593a0
Dutta, A., Eckelmann, B., Adhikari, S., Ahmed, K. M., Sengupta, S., Pandey, A., et al. (2017). Microhomology-mediated end joining is activated in irradiated human cells due to phosphorylation-dependent formation of the XRCC1 repair complex. Nucleic Acids Res. 45, 2585–2599. doi:10.1093/nar/gkw1262
El-Khamisy, S. F., Masutani, M., Suzuki, H., and Caldecott, K. W. (2003). A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 31, 5526–5533. doi:10.1093/nar/gkg761
Espinoza, L. A. (2013). “The role of PARP activation in prostate cancer,” in Advances in prostate cancer. Editor G. Hamilton (Rijeka: IntechOpen). doi:10.5772/53297
Ettl, J., Quek, R. G. W., Lee, K.-H., Rugo, H. S., Hurvitz, S., Gonçalves, A., et al. (2018). Quality of life with talazoparib versus physician’s choice of chemotherapy in patients with advanced breast cancer and germline BRCA1/2 mutation: Patient-reported outcomes from the EMBRACA phase III trial. Ann. Oncol. 29, 1939–1947. doi:10.1093/annonc/mdy257
Eustermann, S., Videler, H., Yang, J.-C., Cole, P. T., Gruszka, D., Veprintsev, D., et al. (2011). The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger. J. Mol. Biol. 407, 149–170. doi:10.1016/j.jmb.2011.01.034
Eustermann, S., Wu, W.-F., Langelier, M.-F., Yang, J.-C., Easton, L. E., Riccio, A. A., et al. (2015). Structural basis of detection and signaling of DNA single-strand breaks by human PARP-1. Mol. Cell. 60, 742–754. doi:10.1016/j.molcel.2015.10.032
Färkkilä, A., Gulhan, D. C., Casado, J., Jacobson, C. A., Nguyen, H., Kochupurakkal, B., et al. (2020). Immunogenomic profiling determines responses to combined PARP and PD-1 inhibition in ovarian cancer. Nat. Commun. 11, 1459. doi:10.1038/s41467-020-15315-8
Farmer, H., McCabe, N., Lord, C. J., Tutt, A. N. J., Johnson, D. A., Richardson, T. B., et al. (2005). Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921. doi:10.1038/nature03445
Ferro, A. M., and Olivera, B. M. (1982). Poly(ADP-ribosylation) in vitro. Reaction parameters and enzyme mechanism. J. Biol. Chem. 257, 7808–7813. doi:10.1016/s0021-9258(18)34453-3
Fisher, A. E. O., Hochegger, H., Takeda, S., and Caldecott, K. W. (2007). Poly(ADP-Ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol. Cell. Biol. 27, 5597–5605. doi:10.1128/MCB.02248-06
Fizazi, K., González Mella, P., Castellano, D., Minatta, J. N., Rezazadeh Kalebasty, A., Shaffer, D., et al. (2022). Nivolumab plus docetaxel in patients with chemotherapy-naïve metastatic castration-resistant prostate cancer: Results from the phase II CheckMate 9KD trial. Eur. J. Cancer 160, 61–71. doi:10.1016/j.ejca.2021.09.043
Fontana, P., Bonfiglio, J. J., Palazzo, L., Bartlett, E., Matic, I., and Ahel, I. (2017). Serine ADP-ribosylation reversal by the hydrolase ARH3. eLife 6, e28533. doi:10.7554/eLife.28533
Fouquerel, E., Goellner, E. M., Yu, Z., Gagné, J.-P., Barbi de Moura, M., Feinstein, T., et al. (2014). ARTD1/PARP1 negatively regulates glycolysis by inhibiting hexokinase 1 independent of NAD + depletion. Cell. Rep. 8, 1819–1831. doi:10.1016/j.celrep.2014.08.036
Freeman, G. J., Long, A. J., Iwai, Y., Bourque, K., Chernova, T., Nishimura, H., et al. (2000). Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J. Exp. Med. 192, 1027–1034. doi:10.1084/jem.192.7.1027
Friedlander, M., Meniawy, T., Markman, B., Mileshkin, L., Harnett, P., Millward, M., et al. (2019). Pamiparib in combination with tislelizumab in patients with advanced solid tumours: Results from the dose-escalation stage of a multicentre, open-label, phase 1a/b trial. Lancet. Oncol. 20, 1306–1315. doi:10.1016/S1470-2045(19)30396-1
Fu, K., Sun, X., Wier, E. M., Hodgson, A., Liu, Y., Sears, C. L., et al. (2016). Sam68/KHDRBS1 is critical for colon tumorigenesis by regulating genotoxic stress-induced NF-κB activation. eLife 5, e15018. doi:10.7554/eLife.15018
Gabrail, N. Y., Bessudo, A., Hamilton, E. P., Sachdev, J. C., Patel, M. R., Rodon Ahnert, J., et al. (2019). IOLite: Multipart, phase 1b, dose-finding study of the PD-1 inhibitor dostarlimab in combination with the PARP inhibitor niraparib ± bevacizumab (bev), or with platinum-based chemotherapy ± bev for advanced cancer. J. Clin. Oncol. 37, 2560. doi:10.1200/JCO.2019.37.15_suppl.2560
Ganguly, B., Dolfi, S. C., Rodriguez-Rodriguez, L., Ganesan, S., and Hirshfield, K. M. (2016). Role of biomarkers in the development of PARP inhibitors. Biomark. Cancer 8, 15–25. doi:10.4137/BIC.S36679
García, S., Bodaño, A., González, A., Forteza, J., Gómez-Reino, J. J., and Conde, C. (2006). Partial protection against collagen antibody-induced arthritis in PARP-1 deficient mice. Arthritis Res. Ther. 8, R14. doi:10.1186/ar1865
Garcia-Diaz, A., Shin, D. S., Moreno, B. H., Saco, J., Escuin-Ordinas, H., Rodriguez, G. A., et al. (2017). Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell. Rep. 19, 1189–1201. doi:10.1016/j.celrep.2017.04.031
Gatti, M., Imhof, R., Huang, Q., Baudis, M., and Altmeyer, M. (2020). The ubiquitin ligase TRIP12 limits PARP1 trapping and constrains PARP inhibitor efficiency. Cell. Rep. 32, 107985. doi:10.1016/j.celrep.2020.107985
Gaullier, G., Roberts, G., Muthurajan, U. M., Bowerman, S., Rudolph, J., Mahadevan, J., et al. (2020). Bridging of nucleosome-proximal DNA double-strand breaks by PARP2 enhances its interaction with HPF1. PloS One 15, e0240932. doi:10.1371/journal.pone.0240932
Germain, M., Affar, E. B., D’Amours, D., Dixit, V. M., Salvesen, G. S., and Poirier, G. G. (1999). Cleavage of automodified poly(ADP-ribose) polymerase during apoptosis. Evidence for involvement of caspase-7. J. Biol. Chem. 274, 28379–28384. doi:10.1074/jbc.274.40.28379
Ghonim, M. A., Pyakurel, K., Ibba, S. V., Wang, J., Rodriguez, P., Al-Khami, A. A., et al. (2015). PARP is activated in human asthma and its inhibition by olaparib blocks house dust mite-induced disease in mice. Clin. Sci. 129, 951–962. doi:10.1042/CS20150122
Gibbs-Seymour, I., Fontana, P., Rack, J. G. M., and Ahel, I. (2016). HPF1/C4orf27 is a PARP-1-interacting protein that regulates PARP-1 ADP-ribosylation activity. Mol. Cell. 62, 432–442. doi:10.1016/j.molcel.2016.03.008
Gogola, E., Duarte, A. A., de Ruiter, J. R., Wiegant, W. W., Schmid, J. A., de Bruijn, R., et al. (2018). Selective loss of PARG restores PARylation and counteracts PARP inhibitor-mediated synthetic lethality. Cancer Cell. 33, 1078–1093. e12. doi:10.1016/j.ccell.2018.05.008
Golan, T., Hammel, P., Reni, M., Van Cutsem, E., Macarulla, T., Hall, M. J., et al. (2019). Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N. Engl. J. Med. 381, 317–327. doi:10.1056/NEJMoa1903387
González-Martín, A., Pothuri, B., Vergote, I., DePont Christensen, R., Graybill, W., Mirza, M. R., et al. (2019). PRIMA/ENGOT-OV26/GOG-3012 InvestigatorsNiraparib in patients with newly diagnosed advanced ovarian cancer. N. Engl. J. Med. 381, 2391–2402. doi:10.1056/NEJMoa1910962
Gottipati, P., Vischioni, B., Schultz, N., Solomons, J., Bryant, H. E., Djureinovic, T., et al. (2010). Poly(ADP-ribose) polymerase is hyperactivated in homologous recombination-defective cells. Cancer Res. 70, 5389–5398. doi:10.1158/0008-5472.CAN-09-4716
Groslambert, J., Prokhorova, E., and Ahel, I. (2021). ADP-ribosylation of DNA and RNA. DNA Repair 105, 103144. doi:10.1016/j.dnarep.2021.103144
Gunderson, C. C., and Moore, K. N. (2015). BRACAnalysis CDx as a companion diagnostic tool for Lynparza. Expert Rev. Mol. diagn. 15, 1111–1116. doi:10.1586/14737159.2015.1078238
Ha, H. C., and Snyder, S. H. (1999). Poly(ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proc. Natl. Acad. Sci. U. S. A. 96, 13978–13982. doi:10.1073/pnas.96.24.13978
Haince, J.-F., McDonald, D., Rodrigue, A., Déry, U., Masson, J.-Y., Hendzel, M. J., et al. (2008). PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 283, 1197–1208. doi:10.1074/jbc.M706734200
Hakmé, A., Wong, H.-K., Dantzer, F., and Schreiber, V. (2008). The expanding field of poly(ADP-ribosyl)ation reactions. “Protein modifications: Beyond the usual suspects” review series. EMBO Rep. 9, 1094–1100. doi:10.1038/embor.2008.191
Hanzlikova, H., Kalasova, I., Demin, A. A., Pennicott, L. E., Cihlarova, Z., and Caldecott, K. W. (2018). The importance of poly(ADP-ribose) polymerase as a sensor of unligated Okazaki fragments during DNA replication. Mol. Cell. 71, 319–331. e3. doi:10.1016/j.molcel.2018.06.004
Hellmann, M. D., Callahan, M. K., Awad, M. M., Calvo, E., Ascierto, P. A., Atmaca, A., et al. (2018). Tumor mutational burden and efficacy of nivolumab monotherapy and in combination with ipilimumab in small-cell lung cancer. Cancer Cell. 33, 853–861. e4. doi:10.1016/j.ccell.2018.04.001
Hendriks, I. A., Larsen, S. C., and Nielsen, M. L. (2019). An advanced strategy for comprehensive profiling of ADP-ribosylation sites using mass spectrometry-based proteomics. Mol. Cell. Proteomics 18, 1010–1026. doi:10.1074/mcp.TIR119.001315
Hennessy, B. T. J., Timms, K. M., Carey, M. S., Gutin, A., Meyer, L. A., Flake, D. D., et al. (2010). Somatic mutations in BRCA1 and BRCA2 could expand the number of patients that benefit from poly (ADP ribose) polymerase inhibitors in ovarian cancer. J. Clin. Oncol. 28, 3570–3576. doi:10.1200/JCO.2009.27.2997
Higuchi, T., Flies, D. B., Marjon, N. A., Mantia-Smaldone, G., Ronner, L., Gimotty, P. A., et al. (2015). CTLA-4 blockade synergizes therapeutically with PARP inhibition in BRCA1-deficient ovarian cancer. Cancer Immunol. Res. 3, 1257–1268. doi:10.1158/2326-6066.CIR-15-0044
Hochegger, H., Dejsuphong, D., Fukushima, T., Morrison, C., Sonoda, E., Schreiber, V., et al. (2006). Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. EMBO J. 25, 1305–1314. doi:10.1038/sj.emboj.7601015
Holloman, W. K. (2011). Unraveling the mechanism of BRCA2 in homologous recombination. Nat. Struct. Mol. Biol. 18, 748–754. doi:10.1038/nsmb.2096
Hopkins, T. A., Shi, Y., Rodriguez, L. E., Solomon, L. R., Donawho, C. K., DiGiammarino, E. L., et al. (2015). Mechanistic dissection of PARP1 trapping and the impact on in vivo tolerability and efficacy of PARP inhibitors. Mol. Cancer Res. 13, 1465–1477. doi:10.1158/1541-7786.MCR-15-0191-T
Horton, J. K., Stefanick, D. F., Prasad, R., Gassman, N. R., Kedar, P. S., and Wilson, S. H. (2014). Base excision repair defects invoke hypersensitivity to PARP inhibition. Mol. Cancer Res. 12, 1128–1139. doi:10.1158/1541-7786.MCR-13-0502
Hottiger, M. O., Hassa, P. O., Lüscher, B., Schüler, H., and Koch-Nolte, F. (2010). Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends biochem. Sci. 35, 208–219. doi:10.1016/j.tibs.2009.12.003
Houtkooper, R. H., Williams, R. W., and Auwerx, J. (2010). Metabolic networks of longevity. Cell. 142, 9–14. doi:10.1016/j.cell.2010.06.029
Hoy, S. M. (2018). Talazoparib: First global approval. Drugs 78, 1939–1946. doi:10.1007/s40265-018-1026-z
Hu, Y., Petit, S. A., Ficarro, S. B., Toomire, K. J., Xie, A., Lim, E., et al. (2014). PARP1-Driven poly-ADP-ribosylation regulates BRCA1 function in homologous recombination–mediated DNA repair. Cancer Discov. 4, 1430–1447. doi:10.1158/2159-8290.CD-13-0891
Hurley, R. M., Wahner Hendrickson, A. E., Visscher, D. W., Ansell, P., Harrell, M. I., Wagner, J. M., et al. (2019). 53BP1 as a potential predictor of response in PARP inhibitor-treated homologous recombination-deficient ovarian cancer. Gynecol. Oncol. 153, 127–134. doi:10.1016/j.ygyno.2019.01.015
Iqbal, J., Ragone, A., Lubinski, J., Lynch, H. T., Moller, P., Ghadirian, P., et al. (2012). Hereditary Breast Cancer Study GroupThe incidence of pancreatic cancer in BRCA1 and BRCA2 mutation carriers. Br. J. Cancer 107, 2005–2009. doi:10.1038/bjc.2012.483
Ishida, M., Iwai, Y., Tanaka, Y., Okazaki, T., Freeman, G. J., Minato, N., et al. (2002). Differential expression of PD-L1 and PD-L2, ligands for an inhibitory receptor PD-1, in the cells of lymphohematopoietic tissues. Immunol. Lett. 84, 57–62. doi:10.1016/s0165-2478(02)00142-6
Isono, M., Niimi, A., Oike, T., Hagiwara, Y., Sato, H., Sekine, R., et al. (2017). BRCA1 directs the repair pathway to homologous recombination by promoting 53BP1 dephosphorylation. Cell. Rep. 18, 520–532. doi:10.1016/j.celrep.2016.12.042
Jiao, S., Xia, W., Yamaguchi, H., Wei, Y., Chen, M.-K., Hsu, J.-M., et al. (2017). PARP inhibitor upregulates PD-L1 expression and enhances cancer-associated immunosuppression. Clin. Cancer Res. 23, 3711–3720. doi:10.1158/1078-0432.CCR-16-3215
Jonsson, P., Bandlamudi, C., Cheng, M. L., Srinivasan, P., Chavan, S. S., Friedman, N. D., et al. (2019). Tumour lineage shapes BRCA-mediated phenotypes. Nature 571, 576–579. doi:10.1038/s41586-019-1382-1
Kameshita, I., Matsuda, Z., Taniguchi, T., and Shizuta, Y. (1984). Poly (ADP-Ribose) synthetase. Separation and identification of three proteolytic fragments as the substrate-binding domain, the DNA-binding domain, and the automodification domain. J. Biol. Chem. 259, 4770–4776. doi:10.1016/S0021-9258(17)42913-9
Kang, X., Kim, H.-J., Ramirez, M., Salameh, S., and Ma, X. (2010). The septic shock-associated IL-10 -1082 A > G polymorphism mediates allele-specific transcription via poly(ADP-Ribose) polymerase 1 in macrophages engulfing apoptotic cells. J. Immunol. 184, 3718–3724. doi:10.4049/jimmunol.0903613
Karzai, F., VanderWeele, D., Madan, R. A., Owens, H., Cordes, L. M., Hankin, A., et al. (2018). Activity of durvalumab plus olaparib in metastatic castration-resistant prostate cancer in men with and without DNA damage repair mutations. J. Immunother. Cancer 6, 141. doi:10.1186/s40425-018-0463-2
Kaufman, B., Shapira-Frommer, R., Schmutzler, R. K., Audeh, M. W., Friedlander, M., Balmaña, J., et al. (2015). Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J. Clin. Oncol. 33, 244–250. doi:10.1200/JCO.2014.56.2728
Kaufmann, S. H., Desnoyers, S., Ottaviano, Y., Davidson, N. E., and Poirier, G. G. (1993). Specific proteolytic cleavage of poly(ADP-ribose) polymerase: An early marker of chemotherapy-induced apoptosis. Cancer Res. 53, 3976–3985.
Kim, D.-S., Camacho, C. V., Nagari, A., Malladi, V. S., Challa, S., and Kraus, W. L. (2019). Activation of PARP-1 by snoRNAs controls ribosome biogenesis and cell growth via the RNA helicase DDX21. Mol. Cell. 75, 1270–1285. e14. doi:10.1016/j.molcel.2019.06.020
Kim, M. Y., Zhang, T., and Kraus, W. L. (2005). Poly(ADP-ribosyl)ation by PARP-1: `PAR-laying’ NAD + into a nuclear signal. Genes. Dev. 19, 1951–1967. doi:10.1101/gad.1331805
Kondrashova, O., Nguyen, M., Shield-Artin, K., Tinker, A. V., Teng, N. N. H., Harrell, M. I., et al. AOCS Study Group (2017). Secondary somatic mutations restoring RAD51C and RAD51D associated with acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 7, 984–998. doi:10.1158/2159-8290.CD-17-0419
Kondrashova, O., Topp, M., Nesic, K., Lieschke, E., Ho, G.-Y., Harrell, M. I., et al. (2018). Australian Ovarian Cancer Study (AOCS), deFazio, AMethylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat. Commun. 9, 3970. doi:10.1038/s41467-018-05564-z
Konstantinopoulos, P. A., Waggoner, S., Vidal, G. A., Mita, M., Moroney, J. W., Holloway, R., et al. (2019). Single-arm phases 1 and 2 trial of niraparib in combination with pembrolizumab in patients with recurrent platinum-resistant ovarian carcinoma. JAMA Oncol. 5, 1141–1149. doi:10.1001/jamaoncol.2019.1048
Koppensteiner, R., Samartzis, E. P., Noske, A., von Teichman, A., Dedes, I., Gwerder, M., et al. (2014). Effect of MRE11 loss on PARP-inhibitor sensitivity in endometrial cancer in vitro. PLoS ONE 9, e100041. doi:10.1371/journal.pone.0100041
Kouzminova, E. A., and Kuzminov, A. (2006). Fragmentation of replicating chromosomes triggered by uracil in DNA. J. Mol. Biol. 355, 20–33. doi:10.1016/j.jmb.2005.10.044
Kraus, W. L. (2008). Transcriptional control by PARP-1: Chromatin modulation, enhancer-binding, coregulation, and insulation. Curr. Opin. Cell. Biol. 20, 294–302. doi:10.1016/j.ceb.2008.03.006
Krishnakumar, R., and Kraus, W. L. (2010). The PARP side of the nucleus: Molecular actions, physiological outcomes, and clinical targets. Mol. Cell. 39, 8–24. doi:10.1016/j.molcel.2010.06.017
Kummar, S., Chen, A., Parchment, R. E., Kinders, R. J., Ji, J., Tomaszewski, J. E., et al. (2012). Advances in using PARP inhibitors to treat cancer. BMC Med. 10, 25. doi:10.1186/1741-7015-10-25
Kunkel, T. A., and Erie, D. A. (2015). Eukaryotic mismatch repair in relation to DNA replication. Annu. Rev. Genet. 49, 291–313. doi:10.1146/annurev-genet-112414-054722
Kuzminov, A. (2001). Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc. Natl. Acad. Sci. U. S. A. 98, 8241–8246. doi:10.1073/pnas.131009198
LaFargue, C. J., Dal Molin, G. Z., Sood, A. K., and Coleman, R. L. (2019). Exploring and comparing adverse events between PARP inhibitors. Lancet. Oncol. 20, e15–e28. doi:10.1016/S1470-2045(18)30786-1
Langelier, M.-F., Planck, J. L., Roy, S., and Pascal, J. M. (2011). Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: Structural and functional insights into DNA-dependent PARP-1 activity. J. Biol. Chem. 286, 10690–10701. doi:10.1074/jbc.M110.202507
Langelier, M.-F., Planck, J. L., Roy, S., and Pascal, J. M. (2012). Structural basis for DNA damage–dependent poly(ADP-ribosyl)ation by human PARP-1. Science 336, 728–732. doi:10.1126/science.1216338
Langelier, M.-F., Riccio, A. A., and Pascal, J. M. (2014). PARP-2 and PARP-3 are selectively activated by 5′ phosphorylated DNA breaks through an allosteric regulatory mechanism shared with PARP-1. Nucleic Acids Res. 42, 7762–7775. doi:10.1093/nar/gku474
Langelier, M.-F., Servent, K. M., Rogers, E. E., and Pascal, J. M. (2008). A third zinc-binding domain of human poly(ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation. J. Biol. Chem. 283, 4105–4114. doi:10.1074/jbc.M708558200
Lapucci, A., Pittelli, M., Rapizzi, E., Felici, R., Moroni, F., and Chiarugi, A. (2011). Poly(ADP-ribose) polymerase-1 is a nuclear epigenetic regulator of mitochondrial DNA repair and transcription. Mol. Pharmacol. 79, 932–940. doi:10.1124/mol.110.070110
Larkin, J., Chiarion-Sileni, V., Gonzalez, R., Grob, J. J., Cowey, C. L., Lao, C. D., et al. (2015). Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 373, 23–34. doi:10.1056/NEJMoa1504030
Larsen, S. C., Hendriks, I. A., Lyon, D., Jensen, L. J., and Nielsen, M. L. (2018). Systems-wide analysis of serine ADP-ribosylation reveals widespread occurrence and site-specific overlap with phosphorylation. Cell. Rep. 24, 2493–2505. e4. doi:10.1016/j.celrep.2018.07.083
Lee, C.-H., Yelensky, R., Jooss, K., and Chan, T. A. (2018). Update on tumor neoantigens and their utility: Why it is good to Be different. Trends Immunol. 39, 536–548. doi:10.1016/j.it.2018.04.005
Lee, J. B., Kim, H. R., and Ha, S.-J. (2022). Immune checkpoint inhibitors in 10 Years: Contribution of basic research and clinical application in cancer immunotherapy. Immune Netw. 22, e2. doi:10.4110/in.2022.22.e2
Lee, K. Y., and Dutta, A. (2021). Chk1 promotes non-homologous end joining in G1 through direct phosphorylation of ASF1A. Cell. Rep. 34, 108680. doi:10.1016/j.celrep.2020.108680
Lee, W.-P., Hou, M.-C., Lan, K.-H., Li, C.-P., Chao, Y., Lin, H.-C., et al. (2016). Helicobacter pylori-induced chronic inflammation causes telomere shortening of gastric mucosa by promoting PARP-1-mediated non-homologous end joining of DNA. Arch. Biochem. Biophys. 606, 90–98. doi:10.1016/j.abb.2016.07.014
Li, C., Jiang, P., Wei, S., Xu, X., and Wang, J. (2020). Regulatory T cells in tumor microenvironment: New mechanisms, potential therapeutic strategies and future prospects. Mol. Cancer 19, 116. doi:10.1186/s12943-020-01234-1
Li, G., Cunin, P., Wu, D., Diogo, D., Yang, Y., Okada, Y., et al. (2016). The rheumatoid arthritis risk variant CCR6DNP regulates CCR6 via PARP-1. PLoS Genet. 12, e1006292. doi:10.1371/journal.pgen.1006292
Li, J., Dou, D., Li, P., Luo, W., Lv, W., Zhang, C., et al. (2017). PARP-1 serves as a novel molecular marker for hepatocellular carcinoma in a Southern Chinese Zhuang population. Tumour Biol. 39, 1010428317706914. doi:10.1177/1010428317706914
Lieber, M. R. (2008). The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 283, 1–5. doi:10.1074/jbc.R700039200
Liu, Y., Kadyrov, F. A., and Modrich, P. (2011). PARP-1 enhances the mismatch-dependence of 5′-directed excision in human mismatch repair in vitro. DNA Repair 10, 1145–1153. doi:10.1016/j.dnarep.2011.08.012
Lord, C. J., and Ashworth, A. (2017). PARP inhibitors: Synthetic lethality in the clinic. Science 355, 1152–1158. doi:10.1126/science.aam7344
Lorusso, D., Bria, E., Costantini, A., Di Maio, M., Rosti, G., and Mancuso, A. (2017). Patients’ perception of chemotherapy side effects: Expectations, doctor-patient communication and impact on quality of life - an Italian survey. Eur. J. Cancer Care (Engl.) 26, e12618. doi:10.1111/ecc.12618
Loseva, O., Jemth, A.-S., Bryant, H. E., Schüler, H., Lehtiö, L., Karlberg, T., et al. (2010). PARP-3 is a mono-ADP-ribosylase that activates PARP-1 in the absence of DNA. J. Biol. Chem. 285, 8054–8060. doi:10.1074/jbc.M109.077834
Lu, Y., Liu, Y., Pang, Y., Pacak, K., and Yang, C. (2018). Double-barreled gun: Combination of PARP inhibitor with conventional chemotherapy. Pharmacol. Ther. 188, 168–175. doi:10.1016/j.pharmthera.2018.03.006
Lucas, P. J., Negishi, I., Nakayama, K., Fields, L. E., and Loh, D. Y. (1995). Naive CD28-deficient T cells can initiate but not sustain an in vitro antigen-specific immune response. J. Immunol. 154, 5757–5768.
Luijsterburg, M. S., de Krijger, I., Wiegant, W. W., Shah, R. G., Smeenk, G., de Groot, A. J. L., et al. (2016). PARP1 links CHD2-mediated chromatin expansion and H3.3 deposition to DNA repair by non-homologous end-joining. Mol. Cell. 61, 547–562. doi:10.1016/j.molcel.2016.01.019
Lüscher, B., Ahel, I., Altmeyer, M., Ashworth, A., Bai, P., Chang, P., et al. (2021). ADP-ribosyltransferases, an update on function and nomenclature. FEBS J. doi:10.1111/febs.16142
Majuelos-Melguizo, J., Rodríguez, M. I., López-Jiménez, L., Rodríguez-Vargas, J. M., Martí Martín-Consuegra, J. M., Serrano-Sáenz, S., et al. (2015). PARP targeting counteracts gliomagenesis through induction of mitotic catastrophe and aggravation of deficiency in homologous recombination in PTEN-mutant glioma. Oncotarget 6, 4790–4803. doi:10.18632/oncotarget.2993
Mansour, W. Y., Rhein, T., and Dahm-Daphi, J. (2010). The alternative end-joining pathway for repair of DNA double-strand breaks requires PARP1 but is not dependent upon microhomologies. Nucleic Acids Res. 38, 6065–6077. doi:10.1093/nar/gkq387
Markham, A. (2021). Correction to: Avatrombopag: A review in thrombocytopenia. Drugs 81, 2169. doi:10.1007/s40265-021-01651-6
Maya-Mendoza, A., Moudry, P., Merchut-Maya, J. M., Lee, M., Strauss, R., and Bartek, J. (2018). High speed of fork progression induces DNA replication stress and genomic instability. Nature 559, 279–284. doi:10.1038/s41586-018-0261-5
McAlpine, J. N., Porter, H., Köbel, M., Nelson, B. H., Prentice, L. M., Kalloger, S. E., et al. (2012). BRCA1 and BRCA2 mutations correlate with TP53 abnormalities and presence of immune cell infiltrates in ovarian high-grade serous carcinoma. Mod. Pathol. 25, 740–750. doi:10.1038/modpathol.2011.211
McGranahan, N., Furness, A. J. S., Rosenthal, R., Ramskov, S., Lyngaa, R., Saini, S. K., et al. (2016). Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351, 1463–1469. doi:10.1126/science.aaf1490
Michelena, J., Lezaja, A., Teloni, F., Schmid, T., Imhof, R., and Altmeyer, M. (2018). Analysis of PARP inhibitor toxicity by multidimensional fluorescence microscopy reveals mechanisms of sensitivity and resistance. Nat. Commun. 9, 2678. doi:10.1038/s41467-018-05031-9
Mirza, M. R., Monk, B. J., Herrstedt, J., Oza, A. M., Mahner, S., Redondo, A., et al. (2016). ENGOT-OV16/NOVA InvestigatorsNiraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N. Engl. J. Med. 375, 2154–2164. doi:10.1056/NEJMoa1611310
Miwa, M., Tanaka, M., Matsushima, T., and Sugimura, T. (1974). Purification and properties of a glycohydrolase from calf thymus splitting ribose-ribose linkages of poly(adenosine diphosphate ribose). J. Biol. Chem. 249, 3475–3482. doi:10.1016/s0021-9258(19)42597-0
Monaco, L., Kolthur-Seetharam, U., Loury, R., Murcia, J. M., de Murcia, G., and Sassone-Corsi, P. (2005). Inhibition of Aurora-B kinase activity by poly(ADP-ribosyl)ation in response to DNA damage. Proc. Natl. Acad. Sci. U. S. A. 102, 14244–14248. doi:10.1073/pnas.0506252102
Montavon, C., Gloss, B. S., Warton, K., Barton, C. A., Statham, A. L., Scurry, J. P., et al. (2012). Prognostic and diagnostic significance of DNA methylation patterns in high grade serous ovarian cancer. Gynecol. Oncol. 124, 582–588. doi:10.1016/j.ygyno.2011.11.026
Moore, K., Colombo, N., Scambia, G., Kim, B.-G., Oaknin, A., Friedlander, M., et al. (2018). Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N. Engl. J. Med. 379, 2495–2505. doi:10.1056/NEJMoa1810858
Moore, K. N., Secord, A. A., Geller, M. A., Miller, D. S., Cloven, N., Fleming, G. F., et al. (2019). Niraparib monotherapy for late-line treatment of ovarian cancer (QUADRA): A multicentre, open-label, single-arm, phase 2 trial. Lancet. Oncol. 20, 636–648. doi:10.1016/S1470-2045(19)30029-4
Mouw, K. W., Goldberg, M. S., Konstantinopoulos, P. A., and D’Andrea, A. D. (2017). DNA damage and repair biomarkers of immunotherapy response. Cancer Discov. 7, 675–693. doi:10.1158/2159-8290.CD-17-0226
Muñoz-Gámez, J. A., Rodríguez-Vargas, J. M., Quiles-Pérez, R., Aguilar-Quesada, R., Martín-Oliva, D., de Murcia, G., et al. (2009). PARP-1 is involved in autophagy induced by DNA damage. Autophagy 5, 61–74. doi:10.4161/auto.5.1.7272
Murai, J., Huang, S.-Y. N., Renaud, A., Zhang, Y., Ji, J., Takeda, S., et al. (2014). Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol. Cancer Ther. 13, 433–443. doi:10.1158/1535-7163.MCT-13-0803
Murai, J., Huang, S. N., Das, B. B., Renaud, A., Zhang, Y., Doroshow, J. H., et al. (2012). Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 72, 5588–5599. doi:10.1158/0008-5472.CAN-12-2753
Nijman, S. M. B. (2011). Synthetic lethality: General principles, utility and detection using genetic screens in human cells. FEBS Lett. 585, 1–6. doi:10.1016/j.febslet.2010.11.024
Nobori, T., Yamanaka, H., and Carson, D. A. (1989). Poly(ADP-ribose) polymerase inhibits DNA synthesis initiation in the absence of NAD. Biochem. Biophys. Res. Commun. 163, 1113–1118. doi:10.1016/0006-291X(89)92336-X
Noordermeer, S. M., and van Attikum, H. (2019). PARP inhibitor resistance: A tug-of-war in BRCA-mutated cells. Trends Cell. Biol. 29, 820–834. doi:10.1016/j.tcb.2019.07.008
Nosho, K., Yamamoto, H., Mikami, M., Taniguchi, H., Takahashi, T., Adachi, Y., et al. (2006). Overexpression of poly(ADP-ribose) polymerase-1 (PARP-1) in the early stage of colorectal carcinogenesis. Eur. J. Cancer 42, 2374–2381. doi:10.1016/j.ejca.2006.01.061
Obaji, E., Haikarainen, T., and Lehtiö, L. (2018). Structural basis for DNA break recognition by ARTD2/PARP2. Nucleic Acids Res. 46, 12154–12165. doi:10.1093/nar/gky927
Otto, H., Reche, P. A., Bazan, F., Dittmar, K., Haag, F., and Koch-Nolte, F. (2005). In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs). BMC Genomics 6, 139. doi:10.1186/1471-2164-6-139
Oumouna, M., Mustapha, O., Datta, R., Oumouna-Benachour, K., Suzuki, Y., Hans, C., et al. (2006). Poly(ADP-ribose) polymerase-1 inhibition prevents eosinophil recruitment by modulating Th2 cytokines in a murine model of allergic airway inflammation: A potential specific effect on IL-5. J. Immunol. 177, 6489–6496. doi:10.4049/jimmunol.177.9.6489
Oza, A. M., Tinker, A. V., Oaknin, A., Shapira-Frommer, R., McNeish, I. A., Swisher, E. M., et al. (2017). Antitumor activity and safety of the PARP inhibitor rucaparib in patients with high-grade ovarian carcinoma and a germline or somatic BRCA1 or BRCA2 mutation: Integrated analysis of data from Study 10 and ARIEL2. Gynecol. Oncol. 147, 267–275. doi:10.1016/j.ygyno.2017.08.022
Paddock, M. N., Bauman, A. T., Higdon, R., Kolker, E., Takeda, S., and Scharenberg, A. M. (2011). Competition between PARP-1 and Ku70 control the decision between high-fidelity and mutagenic DNA repair. DNA Repair 10, 338–343. doi:10.1016/j.dnarep.2010.12.005
Palazzo, L., Leidecker, O., Prokhorova, E., Dauben, H., Matic, I., and Ahel, I. (2018). Serine is the major residue for ADP-ribosylation upon DNA damage. eLife 7, e34334. doi:10.7554/eLife.34334
Pantelidou, C., Sonzogni, O., De Oliveria Taveira, M., Mehta, A. K., Kothari, A., Wang, D., et al. (2019). PARP inhibitor efficacy depends on CD8(+) T-cell recruitment via intratumoral STING pathway activation in BRCA-deficient models of triple-negative breast cancer. Cancer Discov. 9, 722–737. doi:10.1158/2159-8290.CD-18-1218
Papeo, G., Casale, E., Montagnoli, A., and Cirla, A. (2013). PARP inhibitors in cancer therapy: An update. Expert Opin. Ther. Pat. 23, 503–514. doi:10.1517/13543776.2013.768615
Patel, A. G., Flatten, K. S., Schneider, P. A., Dai, N. T., McDonald, J. S., Poirier, G. G., et al. (2012). Enhanced killing of cancer cells by poly(ADP-ribose) polymerase inhibitors and topoisomerase I inhibitors reflects poisoning of both enzymes. J. Biol. Chem. 287, 4198–4210. doi:10.1074/jbc.M111.296475
Patel, A. G., Sarkaria, J. N., and Kaufmann, S. H. (2011). Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc. Natl. Acad. Sci. U. S. A. 108, 3406–3411. doi:10.1073/pnas.1013715108
Pazzaglia, S., and Pioli, C. (2019). Multifaceted role of PARP-1 in DNA repair and inflammation: Pathological and therapeutic implications in cancer and non-cancer diseases. Cells 9, 41. doi:10.3390/cells9010041
Pearce, A., Haas, M., Viney, R., Pearson, S.-A., Haywood, P., Brown, C., et al. (2017). Incidence and severity of self-reported chemotherapy side effects in routine care: A prospective cohort study. PLOS ONE 12, e0184360. doi:10.1371/journal.pone.0184360
Peeraphatdit, T. B., Wang, J., Odenwald, M. A., Hu, S., Hart, J., and Charlton, M. R. (2020). Hepatotoxicity from immune checkpoint inhibitors: A systematic review and management recommendation. Hepatology 72, 315–329. doi:10.1002/hep.31227
Pettitt, S. J., Rehman, F. L., Bajrami, I., Brough, R., Wallberg, F., Kozarewa, I., et al. (2013). A genetic screen using the PiggyBac transposon in haploid cells identifies Parp1 as a mediator of olaparib toxicity. PloS One 8, e61520. doi:10.1371/journal.pone.0061520
Peyraud, F., and Italiano, A. (2020). Combined PARP inhibition and immune checkpoint therapy in solid tumors. Cancers 12, E1502. doi:10.3390/cancers12061502
Pines, A., Vrouwe, M. G., Marteijn, J. A., Typas, D., Luijsterburg, M. S., Cansoy, M., et al. (2012). PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. J. Cell. Biol. 199, 235–249. doi:10.1083/jcb.201112132
Plummer, R., Jones, C., Middleton, M., Wilson, R., Evans, J., Olsen, A., et al. (2008). Phase I study of the poly(ADP-ribose) polymerase inhibitor, AG014699, in combination with temozolomide in patients with advanced solid tumors. Clin. Cancer Res. 14, 7917–7923. doi:10.1158/1078-0432.CCR-08-1223
Prokhorova, E., Agnew, T., Wondisford, A. R., Tellier, M., Kaminski, N., Beijer, D., et al. (2021a). Unrestrained poly-ADP-ribosylation provides insights into chromatin regulation and human disease. Mol. Cell. 81, 2640–2655.e8. e8. doi:10.1016/j.molcel.2021.04.028
Prokhorova, E., Zobel, F., Smith, R., Zentout, S., Gibbs-Seymour, I., Schützenhofer, K., et al. (2021b). Serine-linked PARP1 auto-modification controls PARP inhibitor response. Nat. Commun. 12, 4055. doi:10.1038/s41467-021-24361-9
Pujade-Lauraine, E., Ledermann, J. A., Selle, F., Gebski, V., Penson, R. T., Oza, A. M., et al. (2017). SOLO2/ENGOT-Ov21 investigatorsOlaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): A double-blind, randomised, placebo-controlled, phase 3 trial. Lancet. Oncol. 18, 1274–1284. doi:10.1016/S1470-2045(17)30469-2
Quinet, A., and Vindigni, A. (2018). Superfast DNA replication causes damage in cancer cells. Nature 559, 186–187. doi:10.1038/d41586-018-05501-6
Rack, J. G. M., Palazzo, L., and Ahel, I. (2020). (ADP-ribosyl)hydrolases: Structure, function, and biology. Genes. Dev. 34, 263–284. doi:10.1101/gad.334631.119
Ramalingam, S., Arora, S., Whipple Neibauer, M., Zhou, J., Hazard, S., Frenkl, T., et al. (2021). P83.02 niraparib + pembrolizumab (pembro) versus placebo + pembro 1L maintenance therapy in advanced NSCLC: ZEAL-1L phase III study. J. Thorac. Oncol. 16, S653–S654. doi:10.1016/j.jtho.2021.01.1197
Ramalingam, S. S., Thara, E., Awad, M. M., Dowlati, A., Haque, B., Stinchcombe, T. E., et al. (2022). Jasper: Phase 2 trial of first-line niraparib plus pembrolizumab in patients with advanced non-small cell lung cancer. Cancer 128, 65–74. doi:10.1002/cncr.33885
Rameshbabu, S., Labadie, B. W., Argulian, A., and Patnaik, A. (2021). Targeting innate immunity in cancer therapy. Vaccines 9, 138. doi:10.3390/vaccines9020138
Ray Chaudhuri, A., and Nussenzweig, A. (2017). The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell. Biol. 18, 610–621. doi:10.1038/nrm.2017.53
Ray-Coquard, I., Pautier, P., Pignata, S., Pérol, D., González-Martín, A., Berger, R., et al. (2019). Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N. Engl. J. Med. 381, 2416–2428. doi:10.1056/NEJMoa1911361
Ren, N., Zhang, L., Yu, J., Guan, S., Dai, X., Sun, L., et al. (2021). Efficacy and safety of PARP inhibitor combination therapy in recurrent ovarian cancer: A systematic review and meta-analysis. Front. Oncol. 11, 638295. doi:10.3389/fonc.2021.638295
Riccio, A. A., Cingolani, G., and Pascal, J. M. (2016). PARP-2 domain requirements for DNA damage-dependent activation and localization to sites of DNA damage. Nucleic Acids Res. 44, 1691–1702. doi:10.1093/nar/gkv1376
Rizvi, N. A., Hellmann, M. D., Snyder, A., Kvistborg, P., Makarov, V., Havel, J. J., et al. (2015). Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128. doi:10.1126/science.aaa1348
Robson, M., Im, S.-A., Senkus, E., Xu, B., Domchek, S. M., Masuda, N., et al. (2017). Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N. Engl. J. Med. 377, 523–533. doi:10.1056/NEJMoa1706450
Robu, M., Shah, R. G., Petitclerc, N., Brind’Amour, J., Kandan-Kulangara, F., and Shah, G. M. (2013). Role of poly(ADP-ribose) polymerase-1 in the removal of UV-induced DNA lesions by nucleotide excision repair. Proc. Natl. Acad. Sci. U. S. A. 110, 1658–1663. doi:10.1073/pnas.1209507110
Rodriguez-Moreno, J. F., de Velasco, G., Bravo Fernandez, I., Alvarez-Fernandez, C., Fernandez, R., Vazquez-Estevez, S., et al. (2020). Impact of the combination of durvalumab (MEDI4736) plus olaparib (AZD2281) administered prior to surgery in the molecular profile of resectable urothelial bladder cancer: NEODURVARIB trial. J. Clin. Oncol. 38, 542. doi:10.1200/JCO.2020.38.6_suppl.542
Ronson, G. E., Piberger, A. L., Higgs, M. R., Olsen, A. L., Stewart, G. S., McHugh, P. J., et al. (2018). PARP1 and PARP2 stabilise replication forks at base excision repair intermediates through Fbh1-dependent Rad51 regulation. Nat. Commun. 9, 746. doi:10.1038/s41467-018-03159-2
Rottenberg, S., Jaspers, J. E., Kersbergen, A., van der Burg, E., Nygren, A. O. H., Zander, S. A. L., et al. (2008). High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc. Natl. Acad. Sci. U. S. A. 105, 17079–17084. doi:10.1073/pnas.0806092105
Rudolph, J., Jung, K., and Luger, K. (2022). Inhibitors of PARP: Number crunching and structure gazing. Proc. Natl. Acad. Sci. U. S. A. 119, e2121979119. doi:10.1073/pnas.2121979119
Rudolph, J., Mahadevan, J., Dyer, P., and Luger, K. (2018). Poly(ADP-ribose) polymerase 1 searches DNA via a ‘monkey bar’ mechanism. eLife 7, e37818. doi:10.7554/eLife.37818
Rudolph, J., Mahadevan, J., and Luger, K. (2020). Probing the conformational changes associated with DNA binding to PARP1. Biochemistry 59, 2003–2011. doi:10.1021/acs.biochem.0c00256
Rudolph, J., Muthurajan, U. M., Palacio, M., Mahadevan, J., Roberts, G., Erbse, A. H., et al. (2021a). The BRCT domain of PARP1 binds intact DNA and mediates intrastrand transfer. Mol. Cell. 81, 4994–5006.e5. e5. doi:10.1016/j.molcel.2021.11.014
Rudolph, J., Roberts, G., and Luger, K. (2021b). Histone Parylation factor 1 contributes to the inhibition of PARP1 by cancer drugs. Nat. Commun. 12, 736. doi:10.1038/s41467-021-20998-8
Sánchez-Fidalgo, S., Villegas, I., Martín, A., Sánchez-Hidalgo, M., and Alarcón de la Lastra, C. (2007). PARP inhibition reduces acute colonic inflammation in rats. Eur. J. Pharmacol. 563, 216–223. doi:10.1016/j.ejphar.2007.01.070
Satoh, M. S., and Lindahl, T. (1992). Role of poly(ADP-ribose) formation in DNA repair. Nature 356, 356–358. doi:10.1038/356356a0
Satoh, M. S., Poirier, G. G., and Lindahl, T. (1994). Dual function for poly(ADP-ribose) synthesis in response to DNA strand breakage. Biochemistry 33, 7099–7106. doi:10.1021/bi00189a012
Schoonen, P. M., Talens, F., Stok, C., Gogola, E., Heijink, A. M., Bouwman, P., et al. (2017). Progression through mitosis promotes PARP inhibitor-induced cytotoxicity in homologous recombination-deficient cancer cells. Nat. Commun. 8, 15981. doi:10.1038/ncomms15981
Schreiber, V., Dantzer, F., Ame, J.-C., and de Murcia, G. (2006). Poly(ADP-ribose): Novel functions for an old molecule. Nat. Rev. Mol. Cell. Biol. 7, 517–528. doi:10.1038/nrm1963
Schumacher, T. N., and Schreiber, R. D. (2015). Neoantigens in cancer immunotherapy. Science 348, 69–74. doi:10.1126/science.aaa4971
Schweitzer, A. N., and Sharpe, A. H. (1998). Studies using antigen-presenting cells lacking expression of both B7-1 (CD80) and B7-2 (CD86) show distinct requirements for B7 molecules during priming versus restimulation of Th2 but not Th1 cytokine production. J. Immunol. 161, 2762–2771.
Scott, L. J. (2017). Niraparib: First global approval. Drugs 77, 1029–1034. doi:10.1007/s40265-017-0752-y
Shao, Z., Lee, B. J., Rouleau-Turcotte, É., Langelier, M.-F., Lin, X., Estes, V. M., et al. (2020). Clinical PARP inhibitors do not abrogate PARP1 exchange at DNA damage sites in vivo. Nucleic Acids Res. 48, 9694–9709. doi:10.1093/nar/gkaa718
Shen, J., Zhao, W., Ju, Z., Wang, L., Peng, Y., Labrie, M., et al. (2019). PARPi triggers the STING-dependent immune response and enhances the therapeutic efficacy of immune checkpoint blockade independent of BRCAness. Cancer Res. 79, 311–319. doi:10.1158/0008-5472.CAN-18-1003
Shen, Y., Rehman, F. L., Feng, Y., Boshuizen, J., Bajrami, I., Elliott, R., et al. (2013). BMN 673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin. Cancer Res. 19, 5003–5015. doi:10.1158/1078-0432.CCR-13-1391
Shoab Mansuri, M., and Singh, M. (2014). Could er stress Be A major link between oxidative stress and autoimmunity in vitiligo? J. Pigment. Disord. 01. doi:10.4172/2376-0427.1000123
Siraj, A. K., Pratheeshkumar, P., Parvathareddy, S. K., Divya, S. P., Al-Dayel, F., Tulbah, A., et al. (2018). Overexpression of PARP is an independent prognostic marker for poor survival in Middle Eastern breast cancer and its inhibition can be enhanced with embelin co-treatment. Oncotarget 9, 37319–37332. doi:10.18632/oncotarget.26470
Snyder, A., Makarov, V., Merghoub, T., Yuan, J., Zaretsky, J. M., Desrichard, A., et al. (2014). Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 371, 2189–2199. doi:10.1056/NEJMoa1406498
Soldani, C., and Scovassi, A. I. (2002). Poly(ADP-ribose) polymerase-1 cleavage during apoptosis: An update. Apoptosis 7, 321–328. doi:10.1023/a:1016119328968
Soriano, F. G., Nogueira, A. C., Caldini, E. G., Lins, M. H., Teixeira, A. C., Cappi, S. B., et al. (2006). Potential role of poly(adenosine 5’-diphosphate-ribose) polymerase activation in the pathogenesis of myocardial contractile dysfunction associated with human septic shock. Crit. Care Med. 34, 1073–1079. doi:10.1097/01.CCM.0000206470.47721.8D
Sosna, J., Voigt, S., Mathieu, S., Lange, A., Thon, L., Davarnia, P., et al. (2014). TNF-induced necroptosis and PARP-1-mediated necrosis represent distinct routes to programmed necrotic cell death. Cell. Mol. Life Sci. 71, 331–348. doi:10.1007/s00018-013-1381-6
Spivak, G. (2015). Nucleotide excision repair in humans. DNA Repair 36, 13–18. doi:10.1016/j.dnarep.2015.09.003
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Suskiewicz, M. J., Zobel, F., Ogden, T. E. H., Fontana, P., Ariza, A., Yang, J.-C., et al. (2020). HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation. Nature 579, 598–602. doi:10.1038/s41586-020-2013-6
Syed, Y. Y. (2017). Rucaparib: First global approval. Drugs 77, 585–592. doi:10.1007/s40265-017-0716-2
Telli, M. L., Timms, K. M., Reid, J., Hennessy, B., Mills, G. B., Jensen, K. C., et al. (2016). Homologous recombination deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin. Cancer Res. 22, 3764–3773. doi:10.1158/1078-0432.CCR-15-2477
Ter Brugge, P., Kristel, P., van der Burg, E., Boon, U., de Maaker, M., Lips, E., et al. (2016). Mechanisms of therapy resistance in patient-derived xenograft models of BRCA1-deficient breast cancer. JNCI. J. Natl. Cancer Inst. 108, djw148. doi:10.1093/jnci/djw148
Tewari, M., Quan, L. T., O’Rourke, K., Desnoyers, S., Zeng, Z., Beidler, D. R., et al. (1995). Yama/CPP32 beta, a mammalian homolog of CED-3, is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase. Cell. 81, 801–809. doi:10.1016/0092-8674(95)90541-3
Thomas, A., Vilimas, R., Trindade, C., Erwin-Cohen, R., Roper, N., Xi, L., et al. (2019). Durvalumab in combination with olaparib in patients with relapsed SCLC: Results from a phase II study. J. Thorac. Oncol. 14, 1447–1457. doi:10.1016/j.jtho.2019.04.026
Thorsell, A.-G., Ekblad, T., Karlberg, T., Löw, M., Pinto, A. F., Trésaugues, L., et al. (2017). Structural basis for potency and promiscuity in poly(ADP-ribose) polymerase (PARP) and tankyrase inhibitors. J. Med. Chem. 60, 1262–1271. doi:10.1021/acs.jmedchem.6b00990
Turner, N. C., Lord, C. J., Iorns, E., Brough, R., Swift, S., Elliott, R., et al. (2008). A synthetic lethal siRNA screen identifying genes mediating sensitivity to a PARP inhibitor. EMBO J. 27, 1368–1377. doi:10.1038/emboj.2008.61
Tutt, A., and Ashworth, A. (2002). The relationship between the roles of BRCA genes in DNA repair and cancer predisposition. Trends Mol. Med. 8, 571–576. doi:10.1016/s1471-4914(02)02434-6
Tutt, A. N. J., Garber, J. E., Kaufman, B., Viale, G., Fumagalli, D., Rastogi, P., et al. (2021). Adjuvant olaparib for patients with BRCA1 - or BRCA2 -mutated breast cancer. N. Engl. J. Med. 384, 2394–2405. doi:10.1056/NEJMoa2105215
Twomey, J. D., and Zhang, B. (2021). Cancer immunotherapy update: FDA-approved checkpoint inhibitors and companion diagnostics. AAPS J. 23, 39. doi:10.1208/s12248-021-00574-0
Vaidyanathan, A., Sawers, L., Gannon, A.-L., Chakravarty, P., Scott, A. L., Bray, S. E., et al. (2016). ABCB1 (MDR1) induction defines a common resistance mechanism in paclitaxel- and olaparib-resistant ovarian cancer cells. Br. J. Cancer 115, 431–441. doi:10.1038/bjc.2016.203
Vaitsiankova, A., Burdova, K., Sobol, M., Gautam, A., Benada, O., Hanzlikova, H., et al. (2022). PARP inhibition impedes the maturation of nascent DNA strands during DNA replication. Nat. Struct. Mol. Biol. 29, 329–338. doi:10.1038/s41594-022-00747-1
Vinayak, S., Tolaney, S. M., Schwartzberg, L., Mita, M., McCann, G., Tan, A. R., et al. (2019). Open-label clinical trial of niraparib combined with pembrolizumab for treatment of advanced or metastatic triple-negative breast cancer. JAMA Oncol. 5, 1132–1140. doi:10.1001/jamaoncol.2019.1029
Vos, J. R., Fakkert, I. E., de Hullu, J. A., van Altena, A. M., Sie, A. S., Ouchene, H., et al. OPA Working Group (2020). Universal tumor DNA BRCA1/2 testing of ovarian cancer: Prescreening PARPi treatment and genetic predisposition. J. Natl. Cancer Inst. 112, 161–169. doi:10.1093/jnci/djz080
Walunas, T. L., Bakker, C. Y., and Bluestone, J. A. (1996). CTLA-4 ligation blocks CD28-dependent T cell activation. J. Exp. Med. 183, 2541–2550. doi:10.1084/jem.183.6.2541
Walunas, T. L., Lenschow, D. J., Bakker, C. Y., Linsley, P. S., Freeman, G. J., Green, J. M., et al. (1994). CTLA-4 can function as a negative regulator of T cell activation. Immunity 1, 405–413. doi:10.1016/1074-7613(94)90071-x
Wang, M., Wu, Weizhong, Wu, Wenqi, Rosidi, B., Zhang, L., Wang, H., et al. (2006). PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 34, 6170–6182. doi:10.1093/nar/gkl840
Wang, S., Han, L., Han, J., Li, P., Ding, Q., Zhang, Q.-J., et al. (2019a). Uncoupling of PARP1 trapping and inhibition using selective PARP1 degradation. Nat. Chem. Biol. 15, 1223–1231. doi:10.1038/s41589-019-0379-2
Wang, Z., Sun, K., Xiao, Y., Feng, B., Mikule, K., Ma, X., et al. (2019b). Niraparib activates interferon signaling and potentiates anti-PD-1 antibody efficacy in tumor models. Sci. Rep. 9, 1853. doi:10.1038/s41598-019-38534-6
Wen, W. X., and Leong, C.-O. (2019). Association of BRCA1- and BRCA2-deficiency with mutation burden, expression of PD-L1/PD-1, immune infiltrates, and T cell-inflamed signature in breast cancer. PLOS ONE 14, e0215381. doi:10.1371/journal.pone.0215381
Wiggans, A. J., Cass, G. K. S., Bryant, A., Lawrie, T. A., and Morrison, J. (2015). Poly(ADP-ribose) polymerase (PARP) inhibitors for the treatment of ovarian cancer. Cochrane Database Syst. Rev., CD007929. doi:10.1002/14651858.CD007929.pub3
Woodhouse, R., Li, M., Hughes, J., Delfosse, D., Skoletsky, J., Ma, P., et al. (2020). Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PloS One 15, e0237802. doi:10.1371/journal.pone.0237802
Wu, Q., Jiang, L., Li, S.-C., He, Q.-J., Yang, B., and Cao, J. (2021). Small molecule inhibitors targeting the PD-1/PD-L1 signaling pathway. Acta Pharmacol. Sin. 42, 1–9. doi:10.1038/s41401-020-0366-x
Xu, F., Sun, Y., Yang, S.-Z., Zhou, T., Jhala, N., McDonald, J., et al. (2019). Cytoplasmic PARP-1 promotes pancreatic cancer tumorigenesis and resistance. Int. J. Cancer 145, 474–483. doi:10.1002/ijc.32108
Xue, H., Bhardwaj, A., Yin, Y., Fijen, C., Ephstein, A., Zhang, L., et al. (2022). A two-step mechanism governing PARP1-DNA retention by PARP inhibitors. Sci. Adv. 8, eabq0414. doi:10.1126/sciadv.abq0414
Yang, G., Liu, C., Chen, S.-H., Kassab, M. A., Hoff, J. D., Walter, N. G., et al. (2018). Super-resolution imaging identifies PARP1 and the Ku complex acting as DNA double-strand break sensors. Nucleic Acids Res. 46, 3446–3457. doi:10.1093/nar/gky088
Yang, Y.-G., Cortes, U., Patnaik, S., Jasin, M., and Wang, Z.-Q. (2004). Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene 23, 3872–3882. doi:10.1038/sj.onc.1207491
Yap, T. A., Konstantinopoulos, P., Telli, M. L., Saraykar, S., Beck, J. T., Galsky, M. D., et al. (2020). Abstract P1-19-03: JAVELIN PARP medley, a phase 1b/2 study of avelumab plus talazoparib: Results from advanced breast cancer cohorts. Cancer Res. 80, P1–P19-03-P1-19-P03. doi:10.1158/1538-7445.SABCS19-P1-19-0319-03-P1-19–03
Yelamos, J., Farres, J., Llacuna, L., Ampurdanes, C., and Martin-Caballero, J. (2011). PARP-1 and PARP-2: New players in tumour development. Am. J. Cancer Res. 1, 328–346.
Yu, S.-W., Andrabi, S. A., Wang, H., Kim, N. S., Poirier, G. G., Dawson, T. M., et al. (2006). Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc. Natl. Acad. Sci. U. S. A. 103, 18314–18319. doi:10.1073/pnas.0606528103
Zahradka, P., and Ebisuzaki, K. (1982). A shuttle mechanism for DNA-protein interactions. The regulation of poly(ADP-ribose) polymerase. Eur. J. Biochem. 127, 579–585. doi:10.1111/j.1432-1033.1982.tb06912.x
Zandarashvili, L., Langelier, M.-F., Velagapudi, U. K., Hancock, M. A., Steffen, J. D., Billur, R., et al. (2020). Structural basis for allosteric PARP-1 retention on DNA breaks. Science 368, eaax6367. doi:10.1126/science.aax6367
Zhang, L. L., Cao, F. F., Wang, Y., Meng, F. L., Zhang, Y., Zhong, D. S., et al. (2015). The protein kinase C (PKC) inhibitors combined with chemotherapy in the treatment of advanced non-small cell lung cancer: meta-analysis of randomized controlled trials. Clin. Transl. Oncol. 17, 371–377. doi:10.1007/s12094-014-1241-3
Zhang, Z., Li, J., Zheng, H., Yu, C., Chen, J., Liu, Z., et al. (2009). Expression and cytoplasmic localization of SAM68 is a significant and independent prognostic marker for renal cell carcinoma. Cancer Epidemiol. Biomarkers Prev. 18, 2685–2693. doi:10.1158/1055-9965.EPI-09-0097
Keywords: PARP inhibitors, immune checkpoint inhibitors, immunotherapy, synthetic lethality, DNA repair, BRCA, cancer
Citation: Hunia J, Gawalski K, Szredzka A, Suskiewicz MJ and Nowis D (2022) The potential of PARP inhibitors in targeted cancer therapy and immunotherapy. Front. Mol. Biosci. 9:1073797. doi: 10.3389/fmolb.2022.1073797
Received: 18 October 2022; Accepted: 15 November 2022;
Published: 01 December 2022.
Edited by:
Huiming Lu, University of Texas Southwestern Medical Center, United StatesReviewed by:
Shuai Wang, University of Texas Southwestern Medical Center, United StatesCopyright © 2022 Hunia, Gawalski, Szredzka, Suskiewicz and Nowis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jaromir Hunia, amFyb21pcmhAaW50ZXJpYS5wbA==; Karol Gawalski, a2Fyb2wuZ2F3YWxza2lAd3VtLmVkdS5wbA==; Aleksandra Szredzka, YWxla3NhbmRyYS5zenJlZHprYUBnbWFpbC5jb20=; Marcin J. Suskiewicz, bWFyY2luLnN1c2tpZXdpY3pAY25ycy1vcmxlYW5zLmZy; Dominika Nowis, ZG9taW5pa2Eubm93aXNAd3VtLmVkdS5wbA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.