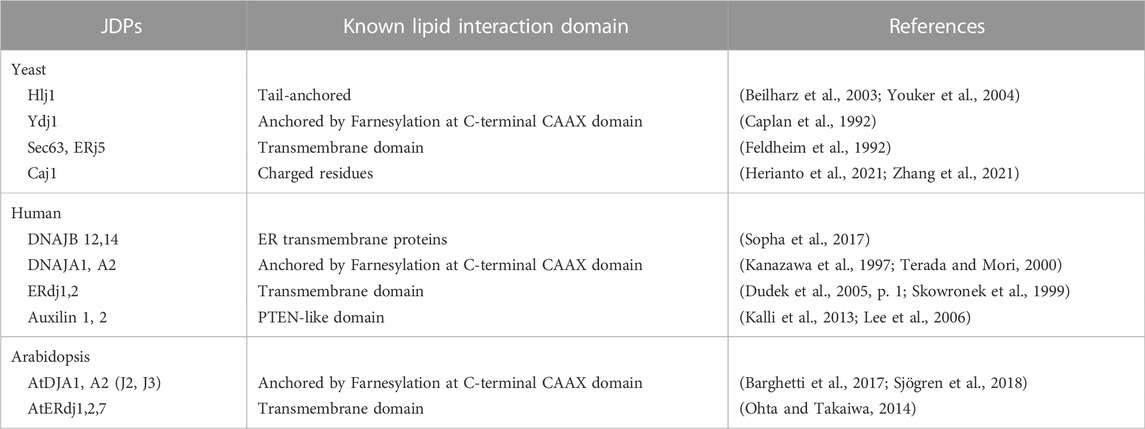
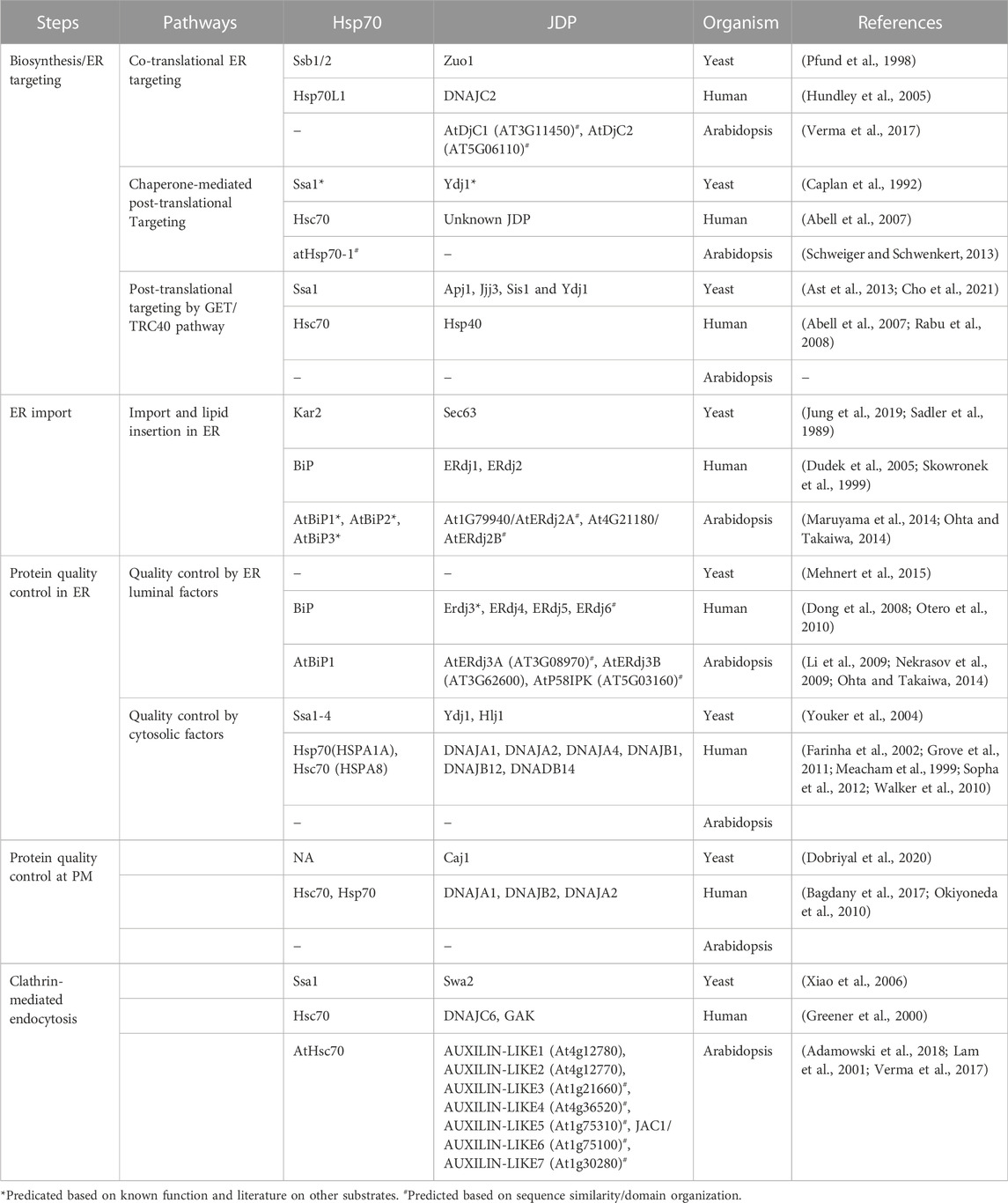
- Department of Biological Sciences, IISER Bhopal, Bhopal, India
The function, stability, and turnover of plasma membrane (PM) proteins are crucial for cellular homeostasis. Compared to soluble proteins, quality control of plasma membrane proteins is extremely challenging. Failure to meet the high quality control standards is detrimental to cellular and organismal health. J-domain proteins (JDPs) are among the most diverse group of chaperones that collaborate with other chaperones and protein degradation machinery to oversee cellular protein quality control (PQC). Although fragmented, the available literature from different models, including yeast, mammals, and plants, suggests that JDPs assist PM proteins with their synthesis, folding, and trafficking to their destination as well as their degradation, either through endocytic or proteasomal degradation pathways. Moreover, some JDPs interact directly with the membrane to regulate the stability and/or functionality of proteins at the PM. The deconvoluted picture emerging is that PM proteins are relayed from one JDP to another throughout their life cycle, further underscoring the versatility of the Hsp70:JDP machinery in the cell.
Introduction
The plasma membrane (PM) is a multi-protein collage where proteins are frequently clustered and embedded in the lipid bilayer. PM proteins broadly consist of integral and peripheral proteins. Together, they perform numerous cellular functions, such as cell anchoring and cell-cell interactions, regulating cellular nutrient uptake and ion homeostasis, as well as integrating cellular physiology with the environment. Moreover, they are critical for the structural integrity of the PM (Alberts B et al., 2002; Bernardino de la Serna et al., 2016). Therefore, preserving the functionality and the heterogeneity of these cell surface proteins is crucial. To accomplish this, cells have an intricate internal membrane system that mediates their insertion and removal from the plasma membrane (Okiyoneda et al., 2011; MacGurn, 2014; Sardana and Emr, 2021).
PM proteins are extremely hydrophobic and are embedded, partially or completely in the lipid bilayer. Hence, the biogenesis, trafficking, stability, and degradation of PM proteins are far more challenging than soluble cytosolic proteins. Cells are extremely sensitive to conformationally compromised PM proteins, and often this leads to loss of cell integrity or death (Zhao et al., 2013; Juarez-Navarro et al., 2020). To circumvent this, cells employ efficient, multi-layered cellular protein quality control (PQC) machineries which act as stringent checkpoints to tightly regulate the synthesis, folding, and degradation of PM proteins (Reviewed in (Apaja et al., 2010; Okiyoneda et al., 2011; Babst, 2014; MacGurn, 2014; Sardana and Emr, 2021; Vasconcelos-Cardoso et al., 2022). At each of these steps, PM proteins must pass the “fit to move forward” test. In case they do not meet the quality control (QC) standards, they are appropriately handled by one or the other component(s) of the cellular PQC machinery (Anelli and Sitia, 2008; Okiyoneda et al., 2011; Sun and Brodsky, 2019; Phillips et al., 2020; Schwabl and Teis, 2022). At the PM, too, these proteins are constantly at risk of misfolding or aggregation. Furthermore, their stability and/or abundance at the PM is also very tightly regulated; for example, for many nutrient transporters, mild temperature shifts or availability of substrate promotes their endocytosis and degradation (Apaja et al., 2010; Ghaddar et al., 2014; Rodriguez-Furlan et al., 2019).
Molecular chaperones, mainly the Hsp70:J-domain protein (JDP) machinery (see Box 1), play a major role in regulating cellular proteostasis (Rosenzweig et al., 2019; Liu et al., 2020; Zhang et al., 2022). In the last two decades, the essential requirement of chaperones for various PQC pathways has been extensively highlighted. Paradoxically, even though PM proteins pose greater challenges to PQC machinery compared to cytosolic proteins, the involvement of chaperones in the QC of PM proteins is less understood. Nevertheless, different Hsp70s and JDPs have been implicated in assisting PM proteins in their biogenesis, maturation, trafficking, and degradation in different organisms. Moreover, the ability of selected Hsp70s and JDPs to directly associate with lipids and membranes sheds light on their possible role in the PM as well (Caplan et al., 1992; Kanazawa et al., 1997; Beilharz et al., 2003; Kalli et al., 2013; Radons, 2016; Sopha et al., 2017; Sjögren et al., 2018; Dobriyal et al., 2020; De Maio and Hightower, 2021; Zhang et al., 2021) (Table 1). The importance of different chaperones and other PQC factors in diseases associated with PM proteins is discussed elsewhere (Juarez-Navarro et al., 2020). In this review, we collate data available from multiple organisms, including yeast, plants, and mammalian models, and come up with a unifying model to show that, like cytosolic proteins, the versatile Hsp70:JDP systems are guardians of PM proteins as well, throughout their odyssey in the cell (Table 2).
PM proteins: The PQC machinery’s nightmare
Maintaining the proper QC of PM proteins is one of the major challenges faced by cellular PQC machinery. Like other membrane proteins, PM proteins are folded into different conformations and topologies governed by their primary structures. Many membrane proteins are known to have dual or multiple topologies, which adds to the complexity of the plasma membrane PQC (PMPQC) (von Heijne, 2006; Woodall et al., 2015). There are different types of membrane proteins, including transmembrane proteins, which are amphipathic. They harbor one or more hydrophobic segments, spanning the lipid bilayer, and hydrophilic domains, that stay exposed to the cytosol and extracellular space (Deol et al., 2004). Several other PM proteins are anchored to the lipid bilayer either through a hydrophobic segment a covalent lipid (fatty acid chain or a prenyl group), or a sugar modification. PM proteins have a complex life cycle (shown in Figure 1). They follow the secretory pathway through the cellular endomembrane system to reach their destination. En route, they pass through different sub-cellular environments with varying pH, ionic concentrations, and redox states (Anelli and Sitia, 2008; Delic et al., 2013). PM proteins are synthesized on the ribosomes and are co- or post-translationally targeted to the ER. Then they are incorporated into the ER membrane to undergo folding and maturation, which involves the participation of both ER and cytosolic factors (Sun and Brodsky, 2019; Phillips et al., 2020). ER provides an oxidative environment for the disulfide bond formation and proper folding. ER-residing enzymes are essential for adding GPI anchors to the anchored PM proteins. The assembly and maturation of proteins are followed by their packaging into vesicles and targeting to the Golgi apparatus for further post-translational modifications (PTMs) and sorting to PM. These organelles have different pH and provide a protective environment to the proteins until they reach the cell surface. The Golgi apparatus and vesicles are slightly acidic compared to the cytosol and ER (pH 7-7.5). Any change in the pH (pH 6.2-6.8 at cis-Golgi to pH 6.0-6.3 at the trans-Golgi network) or ion (H+, Ca2+, Mg2+, Mn2+) concentrations can lead to perturbed Golgi functions (Shen et al., 2013; Kellokumpu, 2019).
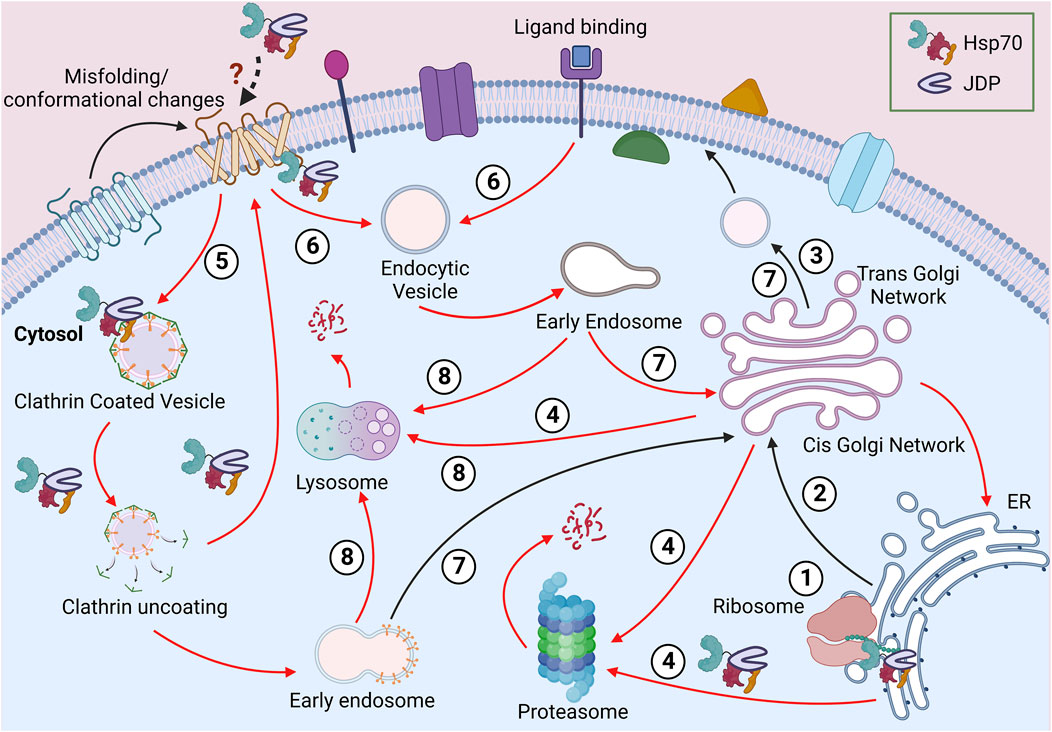
FIGURE 1. Journey of a PM protein. Soon after the ribosomal biosynthesis, the newly synthesized PM protein translocates to the ER (1). The well-folded proteins are packed in vesicles and transported to the PM via the Golgi apparatus (2-3). The misfolded proteins from the endomembrane system can be directed for proteasomal or lysosomal degradation (4). Due to misfolding or stimuli-based conformational change at the PM, proteins can be packaged in clathrin-independent (5) or clathrin-dependent (6) vesicles. These proteins can be recycled to PM (7) or targeted for lysosomal degradation (8). Different steps requiring Hsp70:JDPs are shown in the diagram.
Being the cellular periphery, at the PM, proteins are often exposed to various physical, mechanical, or chemical stress, risking their native structure and function. Additionally, the lipid composition of the PM too plays a vital role in maintaining the local structure, dynamics, stability, and functionality of PM proteins (Lenaz, 1987; Lee, 2004; Bukiya and Dopico, 2019; Fratti, 2021). For example, the localized lipids interact with the G-protein coupled receptors (GPCR) and affect conformation, stability and signaling at the membrane (Mahmood et al., 2013; Desai and Miller, 2018). Similarly, polyunsaturated fatty acids (PUFA) interact with several voltage-gated ion channels to regulate their activity (Elinder and Liin, 2017). Apart from the direct interactions, localized lipid rafts also form membrane compartments and, in turn, regulate the function, stability, sorting, maturation, and turnover of proteins (Casares et al., 2019; Lujan and Campelo, 2021). Even the final stages of PM proteins’ life cycle are not simple (Figure 1). The removal of proteins from PM follows the endocytic route, where the proteins are packaged into vesicles and targeted to early endosomes (Zhao et al., 2013; Ghaddar et al., 2014; Claus et al., 2018). These proteins are further targeted to lysosome/vacuole for degradation. However, endocytosis does not ensure degradation as, in some cases, PM proteins are modified to their native structure and recycled back to the PM (Claus et al., 2018; Rodriguez-Furlan et al., 2019; Sardana and Emr, 2021).
Hence the complex life cycle of PM proteins counts on the proper functioning of multiple PQC machineries that work in parallel in different subcellular compartments. It is very likely that these heterogeneous groups of proteins require dedicated cellular factors during the complex journey inside the cell.
Targeting to ER
Almost all PM proteins follow the secretory pathway to reach their destination. This begins with the translocation of PM proteins to the endoplasmic reticulum (ER); either co-translationally, where protein synthesis is coupled with protein import into the ER or post-translationally, where protein translocation takes place after the complete polypeptide is synthesized (reviewed by (Rapoport, 2007; Cross et al., 2009; Hegde and Keenan, 2011; Zimmermann et al., 2011; Aviram and Schuldiner, 2017; Linxweiler et al., 2017). The mode of ER import depends on the amino acid composition of the polypeptide. Different Hsp70:JDP machineries assist these pathways to stabilize the translation elongation, prevent misfolding, aggregation, or facilitate interaction with the ER translocon complex (Ast et al., 2013; Aviram and Schuldiner, 2017; Chen et al., 2022) (Figure 2). During protein synthesis, the Ssb1/2 (yeast)/Hsp70L1 (mammals), together with Ribosome Associated Complex (RAC), functions to regulate the folding and targeting of nascent polypeptides to the ER. RAC is a heterodimer composed of Hsp70, Ssz1 (HSPA14 in mammals), and JDP, Zuo1 (DNAJC2 in mammals) (Hundley et al., 2005; Otto et al., 2005). Upon nascent chain elongation, Ssz1 and Zuo1 undergo a conformational change, thus making the J-domain of Zuo1 accessible to binding with another Hsp70, Ssb1/2 (Chen et al., 2022). Ssb interacts with a large number of ribosome-associated nascent chains as well as the cytosolic factor, signal recognition particle (SRP), to facilitate efficient translation and ER targeting of proteins (Pfund et al., 1998; Willmund et al., 2013; Döring et al., 2017; Shiber et al., 2018). Together, Ssb and Zuo1 help in co-translational protein folding and their targeting to the ER (Kramer et al., 2019; Chen et al., 2022). Although deletion of Ssb leads to widespread misfolding and aggregation of newly synthesized polypeptides (Koplin et al., 2010), its requirement in the biogenesis or maturation of membrane proteins is unknown. While Ssb preferentially interacts with cytosolic and nuclear proteins and less with membrane proteins (Willmund et al., 2013), the sensitivity of yeast cells lacking Ssb1/2 or the partner JDP, Zuo1, to aminoglycosides is linked to an increase in cation influx as a consequence of altered maturation of membrane proteins (Kim and Craig, 2005). It suggests that Ssb1/2, along with Zuo1 may regulate the biogenesis of PM proteins as well (Peisker et al., 2010).
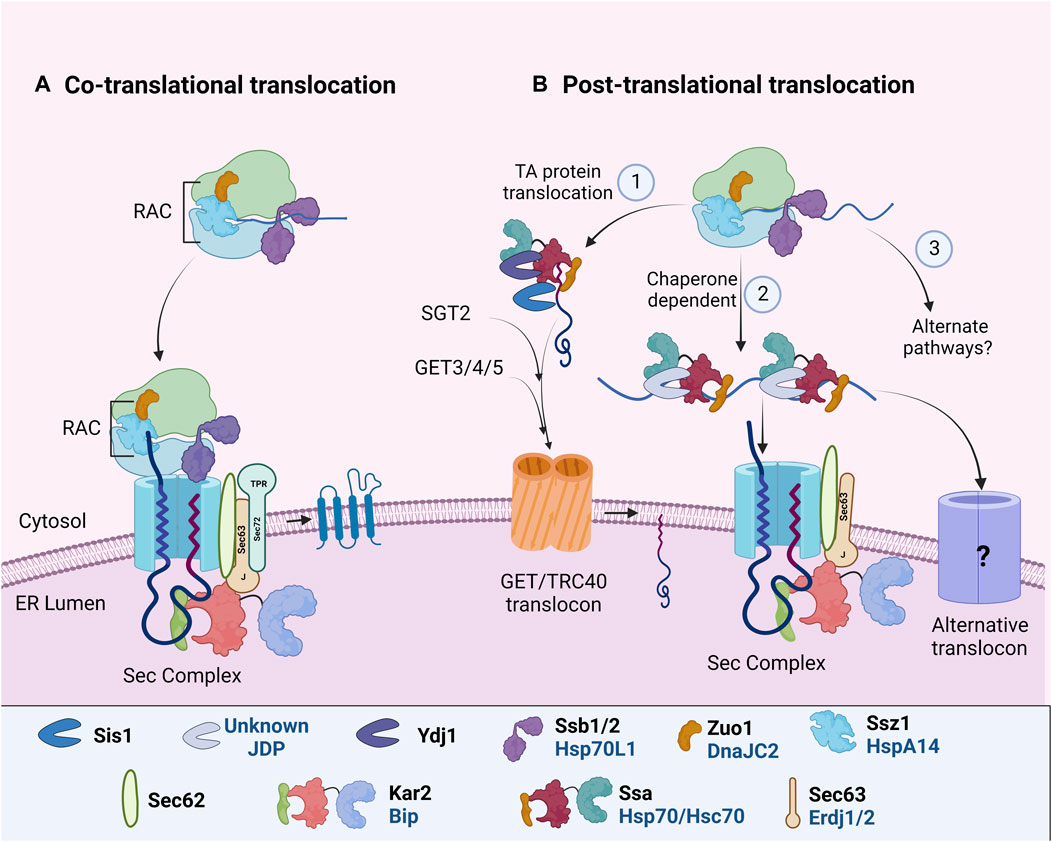
FIGURE 2. ER targeting and import of PM proteins. In different organisms, specific Hsp70: JDPs mediate the ER targeting of PM proteins by (A) CTT or (B) PTT. During both CTT and PTT, ribosome-associated Hsp70:JDPsY,H interact and protect the nascent polypeptides (Otto et al., 2005; Döring et al., 2017; Chen et al., 2022). PTT occurs through multiple pathways (1) GET/TRC40 mdiatedY, (2) Chaperone mediatedH or (3) alternate pathwaysN (Rabu et al., 2009; Casson et al., 2017; Cho et al., 2021). Further, the ER-localized Hsp70:JDPsY, H, A perform the protein import and lateral insertion of PM proteins (Ohta and Takaiwa, 2014; Pobre et al., 2019; Itskanov et al., 2021, p. 63). Y Literature available in Yeast; H Humans; A Arabidopsis. N No literature available. Names of Hsp70 and JDPs are denoted in different colors; Black for Yeast; Blue for Human.
Co-translational translocation
Many of the transmembrane PM proteins undergo co-translational translocation (Figure 2A). Co-translationally translocated proteins usually harbor a cleavable hydrophobic N-terminal signal sequence (SS) or a non-cleavable transmembrane (TM) domain. As soon as the polypeptide emerges from the ribosome, SRP recognizes and binds the ribosome nascent chain complex (RNC) and targets it to the Sec61 complex at the ER membrane (Pool et al., 2002; Halic et al., 2004; Linxweiler et al., 2017). As the RNC interacts with the translocon, the peptide is inserted co-translationally into the ER until the synthesis of a hydrophobic TM segment (i.e., stop transfer sequence) terminates nascent chain translocation. TM segments move laterally out of the translocon as they integrate into the lipid bilayer (Figure 2). In recent years, an alternate ER targeting mechanism involving Snd2p and Snd3p (SRP-iNDependent targeting) has been reported to target substrates to the Sec61 complex. It can partially compensate for the loss of both SRP-mediated and post-translationally targeted substrates. However, whether it operates co-translationally or post-translationally is yet to be understood (Aviram et al., 2016; Haßdenteufel et al., 2017; Lei et al., 2020). The co-translational translocation pathways prevent the exposure of nascent polypeptides to the cytosol. Although ribosome-associated Hsp70:JDPs regulate the process, there is no report of the involvement of additional cytosolic chaperones.
Post-translational translocation
In post-translational translocation (PTT), fully translated polypeptides are targeted to the ER translocon. Proteins remain unfolded for their translocation through ER. Hence cytosolic factors, specifically Hsp70s and JDPs, associate with the polypeptide to prevent intermolecular interaction and aggregation, as well as facilitate interaction with the translocon complex (Figure 2B). A substantial number of proteins are translocated post-translationally into the ER in yeast as well as in complex eukaryotes (Ast et al., 2013; Ast and Schuldiner, 2013; Aviram and Schuldiner, 2014). For PTT of membrane proteins, specifically tail-anchored (TA) proteins, at least two distinct yet overlapping pathways have been reported. First is the GET/TRC40 pathway, targeting TA proteins to the GET/TRC40 receptor complex at ER, and second, the chaperone-mediated pathway, targeting some secretory and TA proteins to ER Sec translocon complex.
GET/TRC40 pathway
Tail-anchored (TA) proteins harbor a single transmembrane domain at their C-terminus (Ast et al., 2013; Casson et al., 2017). A major pathway for TA protein targeting is by the guided entry of tail-anchored proteins (GET) or transmembrane recognition complex of 40 kDa (TRC40/ASNA1) pathway in yeast and mammals respectively (Favaloro et al., 2010, 2008; Schuldiner et al., 2008). In Arabidopsis, multiple GET pathway components have been identified and predicted to be involved in the translocation of TA proteins (Srivastava et al., 2017; Xing et al., 2017; Anderson et al., 2021; Asseck et al., 2021), but our understanding of the Hsp70:JDPs in this process is far from complete. A pre-targeting complex composed of cytosolic chaperone Sgt2 (yeast)/SGTA (mammals) and the Get4-Get5 heterodimer (yeast)/TRC35-UBL4A-BAG6 complex (mammals) recognizes the nascent TA polypeptide that further interacts with the ATP-bound Get3 (yeast)/TRC40 (mammals). This transfers the TA protein to Get3/TRC40 translocon complex for ER insertion (Jonikas et al., 2009; Mariappan et al., 2011; Wang et al., 2011).
In yeast and mammals, JDP (Ydj1) and Hsp/c70 physically interact with Sgt2/SGTA (Wu et al., 2001; Liou et al., 2007; Chartron et al., 2011). Recent studies in yeast have identified Hsp70:JDP machinery as the upstream component of this pathway (Cho et al., 2021) (Figure 2B). Hsp70, Ssa1, captures the TA polypeptide, which is further transferred to the chaperone component, Sgt2. While Ydj1 helps Ssa1 to capture nascent TA polypeptides and prevent their aggregation, both Ydj1 and Sis1 (another cytosolic JDP) enhance the transfer of TA polypeptides to Sgt2. The initial client capture by Ydj1 requires both the functional J-domain as well as the client binding domain, while only J-domain is sufficient for TA polypeptide’s transfer to Sgt2. The study clearly suggests that JDPs not only capture substrates but also induce conformational changes to facilitate their transfer to downstream factors and subsequently to the ER. Ydj1 needs special mention here as it is tethered to the ER membrane through c-terminal farnesyl modification, which is important for targeting proteins to the ER (Caplan et al., 1992; Ast et al., 2013). Additionally, yeast cytosolic JDPs, Apj1 and Jjj3, are also implicated in targeting GPI-anchored protein Gas1 to ER (Ast et al., 2013) through unknown mechanisms.
Chaperone-mediated PTT
In humans, the insertion of TA proteins seems to be more complex and substrate-specific. The TRC40/GET mediated pathway, although specific, is not essential for targeting all TA proteins (Coy-Vergara et al., 2019). Alternative pathways involving Hsc70:JDPs, signal recognition particle (SRP), or Sndp (SRP-iNDependent targeting), execute client targeting to the ER (Abell et al., 2007; Rabu et al., 2009, 2008; Aviram et al., 2016; Casson et al., 2017; Farkas and Bohnsack, 2021) (Figure 2B). Hsc70 functions with JDPs to promote the ATP-dependent membrane targeting of the TA protein, Sec61β, and other TA precursors in vitro (Abell et al., 2007). However, inhibition of Hsc70 affected only a subset of TA proteins’ integration in Hela cells ER, suggesting functional redundancy with other Hsp70s or other parallelly operating pathways (Rabu et al., 2008).
The Hsp70:JDP does not only interact with clients but also with the ER translocon. Studies reveal that yeast Ssa1, through its c-term EEVD motif, and Ssb1, through its nucleotide-binding domain, interact with the tetratricopeptide repeat (TPR) domain of Sec72 translocon (Tripathi et al., 2017). Similarly, in Arabidopsis, constitutively expressed AtHsp70-1 interacts with the ER translocation machinery component AtTPR7 (Schweiger et al., 2012; Schweiger and Schwenkert, 2013). Although metazoans do not have a Sec72 ortholog, one of the ER membrane JDPs, Erdj1, regulates translation and protein import into the ER (Blau et al., 2005). However, the precise mechanism of how exactly the JDPs regulate PTT in humans, is still unknown. Several questions remain unanswered; for example, 1) at which step Hsc70:JDPs act on the proteins, 2) what drives its substrate specificity, 3) whether it is in parallel or in the same pathways as TRC40, and 4) how it interacts with the ER translocon in the absence of Sec72.
Import into the ER
ER is the hub of protein folding and maturation, and it also acts as the first PQC checkpoint for PM proteins. Unsurprisingly, the number and complexity of the Hsp70:JDP network in the ER is only next to the cytosol. Moreover, the ER-localized Hsp70:JDP complex is conserved across yeast, mammals, and plants. Budding yeast has a single Hsp70 (Kar2) and four JDPs, Scj1, Jem1, Erj5 and Sec63 (Nishikawa et al., 2005), and mammals have 7 JDPs Erdj1-7 working with Hsp70, BiP. Out of all these, yeast Sec63 and mammalian ERdj1/2 aid in the protein import across ER membrane, while the other JDPs are proposed to maintain the ER PQC (Kampinga and Craig, 2010; Otero et al., 2010; Melnyk et al., 2015). The JDP, Sec63/Erdj2, is a transmembrane protein and is an integral part of the conserved Sec61 translocon complex. It partners with Hsp70, Kar2/BiP, through its luminal J-domain to facilitate ATP-dependent unidirectional import of proteins (Corsi and Schekman, 1997; Misselwitz et al., 1999, 1999; Melnyk et al., 2015; Pobre et al., 2019; Daverkausen-Fischer and Pröls, 2021) (Figure 2). Sec63 lacking the N-term residues (Sec63_ΔN39) is severely impaired in the sorting of membrane proteins (Jung et al., 2019). Along with import, Sec63 also helps in the lateral gate opening of the Sec61 channel, thus helping in the lateral insertion of proteins into the ER membrane (Itskanov et al., 2021). Hence, it is an essential component of the ER translocon pathway. Sec63 deletion in yeast is lethal, and in humans, mutations in Sec63/Erdj2 are linked to autosomal dominant polycystic liver disease (PCLD) (Sadler et al., 1989; Davila et al., 2004; Janssen et al., 2012).
Apart from Erdj2, Erdj1 is also membrane-localized with an N-terminal luminal J-domain and C-terminal cytosolic domain, wherein the cytosolic domain binds ribosomes and inhibits translation (Dudek et al., 2005). Interestingly, this regulatory inhibition is released when Erdj1 binds to BiP in the ER lumen. It has been hypothesized that Erdj1 regulates translation in response to ER stress through its interaction with BiP (Blau et al., 2005; Benedix et al., 2010). However, under unstressed conditions, it may have protein import functions, possibly redundant with Erdj2.
In Arabidopsis there are six ER-localized JDPs (AtP58IPK, AtERdj2A, AtERdj2B, AtERdj3A, AtERdj3B, and AtERdj7). The ER luminal Hsp70, AtBiP, is present as three isoforms in Arabidopsis, AtBiP1, AtBiP2, and AtBiP3 (Maruyama et al., 2010). They probably interact with the J-domain of AtERdj2A/B (Sec63 orthologs) to perform the energy-dependent protein import in the ER (Yamamoto et al., 2008; Ohta and Takaiwa, 2014). However, it is important to further understand the redundancy and/or specificities of the Hsp70:JDP network in Arabidopsis. Although both AtERdj2A and AtERdj2B mediate protein import, they may interact with different substrates or different AtBiP paralogs, leading to diverse consequences. More detailed studies will help us elucidate their substrate specificity and novel mechanisms of protein import and their insertion in the ER membrane in plants. Non-etheless, the increased number of JDPs in ER from yeast to humans, and plants posits that besides functional redundancy, and making the system more robust, JDPs may also have unique functions as well (Blau et al., 2005; Dudek et al., 2005; Yamamoto et al., 2008; Benedix et al., 2010).
Protein quality control in the ER
Once proteins reach the ER, they undergo folding and maturation. They require a variety of ER chaperones and co-chaperones for the same. Consistent with their structural heterogeneity, PM proteins undergo several maturation steps that require the formation of cis-trans prolyl isomers, disulfide bonds in the oxidizing ER environment, and the adoption of complex transmembrane topologies. Also, the addition, formation, and/or modification of sugar or lipid moieties is crucial for the maturation of several PM proteins (Braakman and Bulleid, 2011). The ER quality control (ERQC) machinery provides a suitable environment for their maturation and regulates the stoichiometry/concentration of subunits to favor correct assembly. Apart from the quality of proteins, ERQC also regulates the quantity of proteins by regulating their secretion according to the physiological requirements of the cell. Once the protein is folded and assembled, it is packaged into vesicles and transported to the Golgi apparatus.
Role of luminal JDPs
In the ER lumen, yeast Kar2, Jem1, and Scj1; Human BiP and ERdj3-6, and Arabidopsis AtBiP1-3, AtERdjs maintain the QC of the luminal domains of trans-membrane proteins and soluble luminal proteins. They not only interact with protein substrates but also regulate the ER stress signaling pathways. For e.g., ERdj4 interacts with the key UPR (Unfolded Protein Response) (Box 2) signal activator, Ire1, and recruits BiP to suppress the UPR signaling in Chinese hamster ovary (CHO) cells (Amin-Wetzel et al., 2017). Similarly, in yeast and Arabidopsis, Kar2/AtBiP1-3 regulates UPR induction (Noh et al., 2003; Moreno et al., 2012). Although none of the Arabidopsis JDPs are yet identified to directly regulate Ire1, AtERdj3B interacts with SDF2, another crucial UPR regulator (Nekrasov et al., 2009). AtBiP1-3 and AtERdjs regulate the folding and maturation of a variety of PM proteins, specifically pattern-recognition receptors (PRRs), thus regulating their surface expression (Li et al., 2009; Saijo, 2010; Gupta and Tuteja, 2011; Park and Seo, 2015). Moreover, AtERdj3 interacts with AtBiP1-3 to regulate the biogenesis of surface receptor EFR (PRRs for EF-Tu), thereby playing an important role in regulating plant immune response (Nekrasov et al., 2009).
The role of luminal Hsp70:JDPs are not restricted to the ER. Human ERdj3, 4, and 6 show dual topology, which may also tether them to the ER membrane to perform different functions (Araki and Nagata, 2011; Daverkausen-Fischer and Pröls, 2021). ERdj3 is predicted to have two transmembrane domains and it interacts with clients as well as the Sec translocon. Interestingly, under UPR stress, Erdj3 is reported to be overexpressed and exported to the extracellular space to maintain proteostasis (Genereux et al., 2015). Also, under stress or apoptosis, BiP and the lectin calreticulin are trafficked to the PM (Zhang et al., 2010; Genereux et al., 2015), further underscoring the versatility of ER luminal Hsp70s and JDPs.
Role of cytosolic JDPs
Transmembrane proteins with an extended cytosolic domain are acted upon by cytosolic chaperones. Hsp70, JDP, and Hsp90 are involved in the early biogenesis of multiple PM transporters by interacting with their cytosolic domains (Kim and Skach, 2012; Donnelly et al., 2013) (Figure 3). They sequentially pass through three complexes with different co-chaperone compositions to achieve their active conformation. First, Hsp70:JDPs interact with the substrates to form the “early complex”. It prevents the aggregation and misfolding of substrates and also targets the misfolded proteins for degradation via ERAD (Box 3). The substrate is then transferred from Hsp70 to Hsp90 by HOP, forming an “intermediate complex”. Interaction with Hsp90 leads to the formation of a “folding complex” and favors substrate maturation and stabilization (Figure 3).
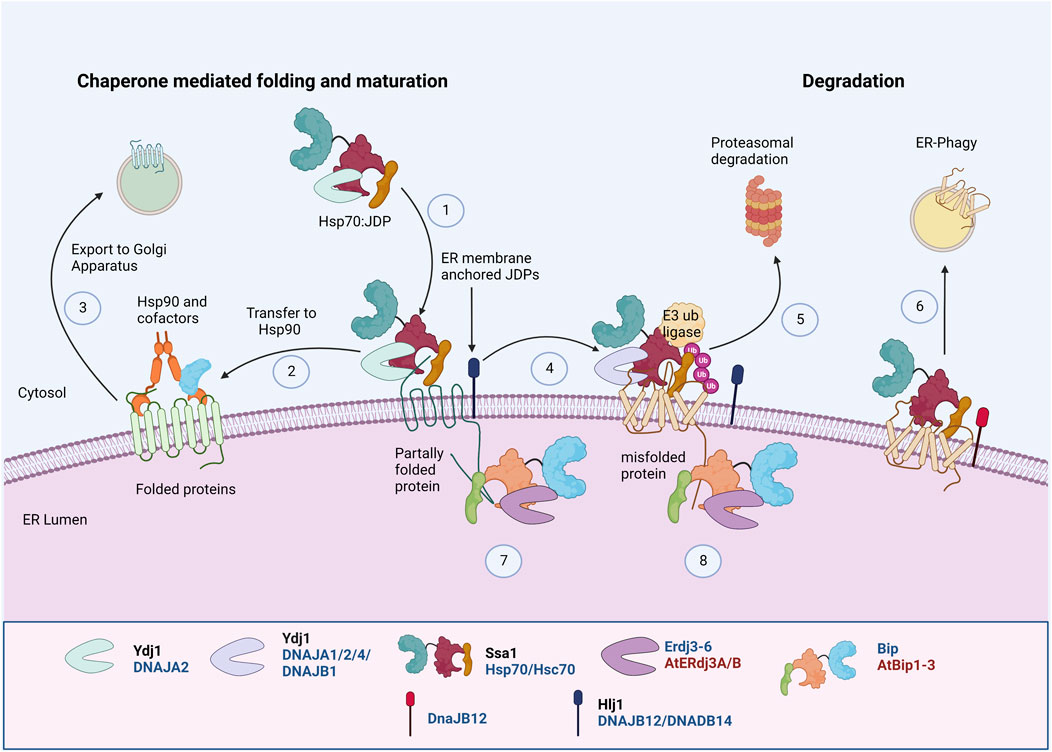
FIGURE 3. Protein Quality Control of PM proteins in ER membrane. PM proteins’ folding and maturation at ER involves both cytosolic and ER luminal Hsp70:JDP machineries. The cytosolic Hsp70:JDPs regulate the PQC by interacting with unfolded/partially folded proteins at the cytosolic domainY, H (1), mediating the interaction with Hsp90 for folding Y, H (2) and further trafficking to Golgi (3) (Youker et al., 2004; Donnelly et al., 2013; Li et al., 2017). Defect in proper folding and maturation, can re-direct them for ubiquitinationY, H (4) and further proteasomal Y, H (5) or lysosomal degradationH (6) (Nakatsukasa et al., 2008; Kim and Skach, 2012; He et al., 2021). Similarly, the luminal Hsp70:JDPs regulate the folding, maturation A, H (7) and degradationA, H (8) of PM proteins by interacting at the luminal domains (Park and Seo, 2015; Ji et al., 2016). Y Literature available in Yeast; H Humans; A Arabidopsis. Names of Hsp70 and JDPs are denoted in different colors; Black for Yeast; Blue for Human.
The early folding of mammalian chloride channel CFTR at the ER requires Hsp90, Hsp70:DNAJB1/Hdj1, and Hsc70:DNAJA1/Hdj2 (Meacham et al., 1999; Farinha et al., 2002; Farinha and Amaral, 2005). Similarly, in HEK293T cells, Hsp70 (HSPA1A) and Hsc70 (HSPA8), along with JDP, DNAJA1, regulate the early QC of thiazide-sensitive NaCl cotransporter (NCC), which further undergo Hsp90 mediated folding and maturation (Donnelly et al., 2013). Hsp70:JDPs also aid in the oligomerization and maturation of several membrane proteins (Li et al., 2017; Needham et al., 2019). Mammalian JDPs, DNAJB12 and DNAJB14, stabilize and promote tetrameric assembly of UNC-103, hERG, and Kv4.2 K+ channel subunits in vitro through an HSP70-independent mechanism. The oligomerization of DNAJB12 is important for its functions (Li et al., 2017). These observations reinforce the idea that cytosolic chaperones are equally important and work as surrogates to augment ER chaperones in regulating the PQC of PM proteins.
Protein quality control in Golgi
After the PM proteins pass through the ER, they are packaged into vesicles and transported to the Golgi apparatus, which acts as the second QC checkpoint. Only a few misfolded proteins escape the ER PQC, and Golgi helps in 1) the retrotranslocation of defective proteins to the ER, 2) targeting of protein for proteasomal degradation, or 3) lysosomal degradation. Golgi is a central hub for protein sorting and modification of secretory and PM proteins. Some unassembled PM proteins, which escape the ER PQC, and reach the cis-Golgi, are retrieved back to the ER by “retention in ER sorting receptor 1” (Rer1), which mediates ER-retrieval (these proteins might undergo ERAD). Some mutant PM proteins further escape ER PQC and Rer1 retrieval and reach the Golgi. There, they undergo Golgi-PQC and ESCRT (endosomal sorting complex required for transport)-dependent vacuolar degradation. Apart from vacuolar degradation, some proteins also undergo proteasomal degradation by “endosome and Golgi associated degradation (EGAD)”. However, the mechanism of substrate recognition and the decision of ER retrieval vs degradation remains unclear (Rosquete et al., 2018; Hellerschmied et al., 2019; Benyair et al., 2022; Schwabl and Teis, 2022). Although likely, the involvement of an Hsp70:JDP in this process is not reported as yet.
Folding and stability at the PM
PM proteins, passing through multiple subcellular organelles, finally reach their destination, the PM, where various extrinsic, as well as intrinsic factors regulate their function/stability. Moreover, PM is extremely dynamic, with continuous fusion and fission of membrane components. Further, the lipid and protein components of the PM are immensely variable depending on cellular physiology. PM proteins undergo conformational changes due to various perturbations and stimuli. These conformational changes are detected by the plasma membrane PQC (PMPQC) machinery and if required, the proteins are targeted for degradation (Nikko and Pelham, 2009; Zhao et al., 2013; Ghaddar et al., 2014). Little information is available on refolding of these proteins at the PM (Bagdany et al., 2017). Hsp70, along with Hsp90, are well known to regulate the conformational rearrangement and modulate the ligand binding activity of several cytosolic steroid receptors (Johnson and Craig, 2000; Pratt and Toft, 2003). It is likely that they regulate the stability and activity of PM proteins as well. A recent study underlined the role of Hsp70s in refolding and stabilizing the misfolded CFTR (Bagdany et al., 2017). Additionally, Hsp90 and Hsc70 stabilize the mature ΔF508CFTR in post-Golgi compartments. They reduce its protease susceptibility and shift the conformation towards native folding. Interestingly, active Hsc70 and DNAJA2, but not DNAJA1, are required for the conformational stabilization of the PM ΔF508CFTR at restrictive temperatures (Bagdany et al., 2017).
At the PM, the extracellular domains of the PM proteins are exposed and communicate with the outside environment. Defects in the extracellular domains are linked to various diseases (Accili et al., 1991; Amagai et al., 2015; Binder et al., 2018; Guo et al., 2019). Although the presence of proteases and chaperones, including Hsp70, BiP, and ERdj3, are reported in the extracellular space, the mechanism by which they regulate the proteins is yet to be established (De Maio, 2014; Wilson et al., 2022). Additionally, several Hsp70s are known to associate with lipids, interact with cell membranes, and even get inserted into lipid bilayers (Guidon and Hightower, 1986a; 1986b) reviewed in (De Maio and Hightower, 2021). Hsp70 (HSP1A) is known to interact with phosphatidylserine (PS) and insert into the PM during stress recovery to regulate a variety of cellular processes (Bilog et al., 2019). It has also been shown to induce pores in lipid membranes and insert into lipid bilayers to form stable ion conductors (Arispe and De Maio, 2000; Macazo and White, 2014). Only time will tell if these perform some canonical chaperone functions at the PM.
Endocytosis
PM proteins, once misfolded or no longer required, are ubiquitinated and removed from the PM. They can be further targeted for lysosomal degradation or can be recycled back to the PM after de-ubiquitination. PM proteins undergo endocytosis by several pathways, broadly categorized into clathrin-dependent or clathrin-independent mechanisms (Mayor and Pagano, 2007) (Figure 4). Clathrin-mediated endocytosis is the major and best-understood pathway for PM protein endocytosis in yeast, mammals, and plants (Lu et al., 2016; Kaksonen and Roux, 2018; Narasimhan et al., 2020). The process starts with the association of cytoskeletal factors and adaptor proteins near the substrate/cargo. It is followed by clathrin coat assembly and the release of the mature vesicle from the PM. Soon after, the vesicles are uncoated and fuse with endosomes. Some proteins in the early endosomes are deubiquitinated and are recycled back to the PM directly or via the trans-Golgi network. If not recycled, the ESCRT machinery targets them and forms MVBs, committing the proteins to lysosomal degradation.
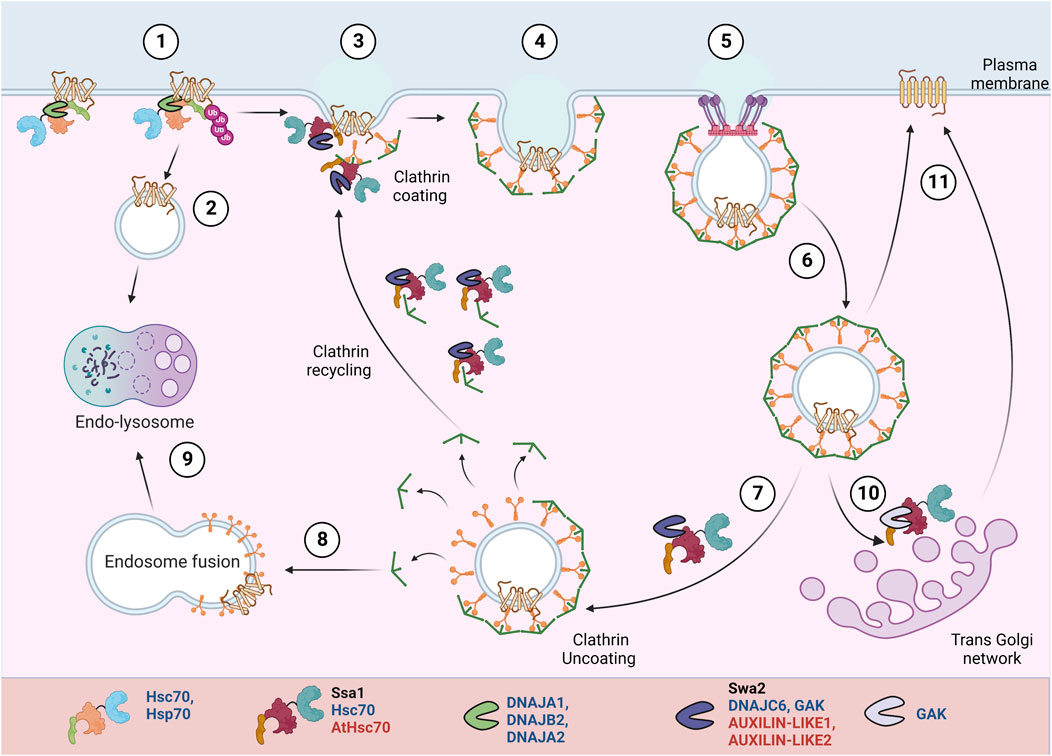
FIGURE 4. PM protein recycling/degradation by clathrin mediated endocytosis. In response to misfolding or stimuli mediated conformational changes, Hsp70:JDPs mediate PM proteins’ ubiquitination H (1) (Bagdany et al., 2017). The proteins undergo clathrin-independent (2) or clathrin-dependent (3-6) endocytosis. They can be targeted for lysosomal degradation (8-9) or recycled back to PM (10-11). A specific set of Hsp70:JDPs regulate clathrin-dependent endocytosis Y,H,A, by mediating the membrane invagination, clathrin exchange, uncoating and recycling of clathrin to new vesicles (Greener et al., 2000; Xiao et al., 2006; Adamowski et al., 2018). Y Literature available in Yeast; H Humans; A Arabidopsis.Names of Hsp70 and JDPs are denoted in different colors; Black for Yeast; Blue for Human; Red for Arabidopsis.
Hsc70:Auxilin are essential for clathrin mediated endocytosis. During vesicle invagination, Hsc70:Auxilin are necessary for ATP-dependent clathrin exchange, thus mediating rearrangement of clathrin coat. At the end of each cycle, they moderate clathrin dissociation and possibly prevent their aggregation in the cytosol. Also, auxilin and Hsc70 may separately chaperone adaptor proteins, further helping to generate new clathrin coated pits (CCPs) (Jiang et al., 2000; Wu et al., 2001; Eisenberg and Greene, 2007; Sousa and Lafer, 2015).Yeast contains a single JDP, auxilin (Swa2), while in humans, Auxilin (Aux1/DNAJC6) and cyclin-G-associated kinase (GAK) Auxilin-2, regulate the clathrin dynamics (Xiao et al., 2006; Krantz et al., 2013; He et al., 2020). Auxilin (Aux1/DNAJC6) is nerve specific, while its homolog cyclin-G-associated kinase (GAK) or Auxilin-2 is ubiquitously expressed. Unlike auxilin, GAK is also enriched in the Golgi (Greener et al., 2000).
In yeast, normal cell development and endocytosis depend on the interaction of Swa2 (by J-domain and TPR-domain) with Hsp70, while Swa2-mediated clathrin binding is expendable (Gall et al., 2000; Xiao et al., 2006; Krantz et al., 2013; Tuo et al., 2013). In HeLa cells, transient GAK knockdown affected the transferrin uptake in a J-domain-dependent manner. The lack of J-domain did not affect its interaction with clathrin or other endocytic factors but disabled GAK’s ability to uncoat clathrin (Zhang et al., 2005). Stable depletion of GAK inhibits the uptake of epidermal growth factor (EGF), resulting in changes in downstream EGFR signaling and a 50-fold increase in the expression levels of EGF receptor (EGFR) (Zhang et al., 2004). Depletion of GAK reduces the trans-Golgi-associated perinuclear clathrin and PM CCPs. Besides clathrin uncoating, the depletion of GAK also partially inhibited trafficking between the trans-Golgi network (TGN) and lysosomes. It alters the Golgi morphology, probably by affecting the clathrin adaptors (Zhang et al., 2005, 2004; Lee et al., 2005). In plants as well, the role of Hsp70:JDP in uncoating of CCV (Clathrin coated vesicles) is reported. AUXILIN-LIKE1 stimulates vesicle uncoating in the presence of HSC70 and interacts with SH3P1 and clathrin in vitro (Lam et al., 2001). In planta studies showed that overexpression of both AUXILIN-LIKE1 and 2 leads to inhibition of endocytosis, most likely by preventing clathrin recruitment to endocytic pits. They result in an arrest in seedling growth and development. However, auxilin-like1/2 loss-of-function mutant does not result in endocytic or developmental defects, suggesting a possible redundant role of AUXILIN-LIKE 3-7 proteins (Adamowski et al., 2018).
Degradation of PM proteins
Genetic mutations, biosynthetic errors or cellular stress can cause misfolding and aggregation of proteins. Clearing of non-functional or cytotoxic PM proteins by different cellular degradation machineries is essential for maintaining cellular homeostasis. Luckily, for PM proteins, such PQC checkpoints operate right from their synthesis in the cytosol, their movement through the endomembrane system to their final destination in the PM. Hsp70s and JDPs have a major role in the solubilization of protein aggregates and the degradation of unfolded proteins (Rosenzweig et al., 2019). Interaction of Hsp70:JDPs with the degradation-competent clients may act as a signal for the recruitment of E3 ubiquitin ligases, thereby targeting proteins for protein degradation (Buchberger et al., 2010). Additionally, Hsp70:JDPs interact with the substrate to avoid protein aggregation or keep it in a competent state for ubiquitination and further targeting to proteasomal degradation (Vembar and Brodsky, 2008; Shiber et al., 2013). Below we discuss examples where Hsp70:JDPs systems target specific PM proteins for degradation, if deemed unfit.
Failure to enter the ER
Failure of PM proteins to enter the ER results in ubiquitination, either co- or post-translationally, followed by their proteasomal degradation (Wang et al., 2013; Tian et al., 2021). Yeast cytosolic Hsp70:JDP (Ydj1) mediate proteasomal degradation of import-incompetent proteins (Park et al., 2007). Similarly, in human MCF-7 cell lines, Hsp70 and DNAJB1 mediate rapid nascent polypeptide degradation during heat stress (Tian et al., 2021). The mammalian Hsp70 and DNAJA1 interact and facilitate the degradation of the PM protein, Apolipoprotein B (ApoB) (Kumari et al., 2022). In lipid-deficient conditions, ApoB does not enter the ER and is deposited in the cytoplasm, where it is targeted for degradation by Hsp70, Hsp90, Hsp110, and the ER membrane-associated JDP, p58IPK (Zhou et al., 1998; Kumari and Brodsky, 2021).
Defects in folding and maturation in the ER
The ER harbors a very stringent PQC network where the quality and quantity of proteins are tightly regulated. However, defects in protein folding or maturation often lead to its degradation by ER-associated degradation (ERAD) (Box 3) or selective ER autophagy (Anelli and Sitia, 2008; Chino and Mizushima, 2020). Hsp70:JDPs regulate the folding, maturation as well as degradation of PM proteins both in the cytosol and the ER lumen (Figure 3). A vast portion of PM proteins are transmembrane proteins and are acted upon by both cytosolic and luminal PQC components. The Hsp70:JDPs interact with the misfolded proteins to mediate their ubiquitination and maintain them in a retro-translocation competent state (Silberstein et al., 1998; Gillece et al., 1999; Nishikawa et al., 2001; Vembar and Brodsky, 2008; Xie and Ng, 2010; Shiber et al., 2013; Pobre et al., 2019). Upon ubiquitination, the ERAD substrates are retrotranslocated to the cytosol by the AAA-ATPase, Cdc48, and targeted for proteasomal degradation (Wolf and Stolz, 2012). In yeast, Hsp70 (Ssa1), Hsp42, and Hsp100 interact with the substrates to prevent aggregation (Shiber et al., 2013; Doonan et al., 2019).
Role of luminal JDPs
In budding yeast, luminal JDP, Scj1, interacts with E3 ubiquitin ligase, Hrd1, for the degradation of soluble proteins. However, the luminal Hsp70:JDPs have little effect on the degradation of membrane proteins (Plemper and Wolf, 1999; Nishikawa et al., 2001; Mehnert et al., 2015). In higher eukaryotes, the role of ER luminal JDPs in regulating PM proteins is more evident. In human m17 neuroblastoma cells, Hsp70, BiP, targets the GPI-anchored mutant prion protein, PrP Q217R, for proteasomal degradation (Jin et al., 2000). A study in mice showed that BiP regulates the degradation of pre-B cell receptors (pre-BCR), thus regulating pre-BCR signaling (Ji et al., 2016). Similarly, in HEK293 cells, BiP, along with ERdj4 and ERdj5, regulate the ERAD of misfolded surfactant protein C (SP-C) proprotein associated with human interstitial lung disease (Dong et al., 2008). Erdj4 (DNAJB9) also physically interacts and regulates the expression of cystic fibrosis transmembrane conductance regulator (CFTR) at the cell surface (Huang et al., 2019).
Interestingly, luminal JDPs can also regulate the degradation of substrates, independent of Hsp70. One of the ERAD substrates, the mammalian Epithelial sodium channel (ENaC), is composed of α-, β-, and γ-subunits. Nearly 75% of each subunit resides in the ER lumen and membrane, and ∼25% resides in the cytoplasm. In the kidney, the β- and γ-subunits are constitutively expressed at much higher levels than the α-subunit. In the absence of the α-subunit, the β- and γ-subunits undergo degradation. Jem1, Scj1 (in yeast), and Erdj3, Erdj4 (in Xenopus oocyte) can function independently of luminal Hsp70 to facilitate ENaC degradation (Buck et al., 2010).
Role of cytosolic JDPs
Often the large, multimeric PM proteins harbor an extended cytosolic domain. Cytosolic Hsp70:JDPs often interact with these domains to regulate their maturation or degradation. In budding yeast, two JDPs, namely Hlj1 and Ydj1, regulate the degradation of various misfolded PM proteins, including mammalian protein CFTR, ABC transporter Ste6* and Pma1-D378S mutant (Zhang et al., 2001; Huyer et al., 2004; Youker et al., 2004; Han et al., 2007; Nakatsukasa et al., 2008). Both Hlj1 and Ydj1 associate with the ER and act in an Ssa1-dependent manner. Interestingly, the majorly cytosolic JDP, Sis1, also regulates the ubiquitination of a subset of ERAD substrates; however, its Hsp70 co-chaperone activity seems to be dispensable for the function (Shiber et al., 2013). It is suggested that depending on the extent of misfolding, the role of Ssa1 might be dispensable for substrate ubiquitination (Shiber et al., 2013). This is not particularly surprising, as many JDPs carry out chaperone functions that do not require their J-domain (Ajit Tamadaddi and Sahi, 2016). JDPs, Cwc23, and Jid1, are also reported to affect the ERAD of certain substrates in unknown ways (Taxis et al., 2003). However, later studies found Jid1 to be localized to mitochondria and Cwc23 to have nuclear functions (Bursać and Lithgow, 2009; Sahi et al., 2010). Hence, the observed effects might not be direct.
In mammalian systems, CFTR is one of the most extensively studied PM proteins. The most prevalent CFTR mutation, ∆F508, is found in ∼90% of cystic fibrosis (CF) patients, where it impairs CFTR folding, inhibits channel gating, and decreases PM stability. Cytosolic (DNAJB1/Hdj1, DNAJA1/Hdj2) and ER membrane JDPs (DNAJB12, DNAJB14) work with cytosolic and luminal Hsp70s and Hsc70, respectively, to oversee the ER-PQC of CFTR (Meacham et al., 1999; Choo-Kang and Zeitlin, 2001; Farinha et al., 2002; Grove et al., 2011; Sopha et al., 2012; Li et al., 2017). In vitro studies suggest that the co-translational interaction of Hsc70 makes CFTR less susceptible to degradation, while post-translational interaction favors degradation (Matsumura et al., 2011). Co-overexpression of Hsp70 and DNAJB1 in Baby hamster kidney (BHK) cells stabilized the immature form of wildtype CFTR but not mutant ∆F508 CFTR (Farinha et al., 2002). In HeLa cells, DNAJA1 and Hsc70 interact with the cytosolic domain of misfolded (∆F508 CFTR) as well as wild-type CFTR. While the misfolded protein is targeted for degradation, chaperones interact with the wild-type CFTR only until it attains its tertiary structure. The levels of complex formation between ∆F508 CFTR and DNAJA1/Hdj2-Hsp70 are approximately 2-fold higher than those with wild-type CFTR (Meacham et al., 1999; Grove et al., 2011), suggesting additional chaperone assistance required by the mutant CFTR as compared to the wild-type protein. Another mammalian channel protein, ENaC, is differentially regulated by Hsc70 and Hsp70. While overexpression of Hsc70 favors the degradation, Hsp70 stabilizes the protein, thus modulating its trafficking and surface expression (Chanoux et al., 2013; Kang and Jeon, 2021). The Hsp70:DNAJA1 regulate the degradation of CFTR and other proteins via the CHIP E3 Ub ligase mediated ubiquitination. Similarly, NaCl cotransporter (NCC), whose loss of function results in Gitelman syndrome, is regulated by cytosolic Hsp70-JDPs. HEK293T cells expressing Gitelman mutants of NCC showed enhanced association with Hsp70 and Hsp40 as compared to the wild-type protein leading to higher CHIP-mediated degradation (Donnelly et al., 2013).
The ER membrane anchored JDPs, DNAJB12 and DNAJB14, also regulate the QC of PM proteins at their cytosolic domain. Modest elevation of DNAJB12 (but not its J-domain mutant, DNAJB12-QPD) decreased the accumulation of both wild-type and mutant (ΔF508) CFTR by increasing its association with Hsc70 and the ubiquitin E3 ligase, RMA1. Depletion of DNAJB12 increased CFTR folding efficiency up to three-fold and permitted a pool of ΔF508 CFTR to fold and escape the ER (Grove et al., 2011). It is suggested that DNAJB12/14 enhance the protein degradation in an Hsp70-dependent manner to regulate the maturation of proteins in cells. DNAJB12/14, although involved in ERAD of CFTR and gonadotropin-releasing hormone receptor mutant, S168R-GnRHR (Houck et al., 2014), they do not participate in ERAD of misfolded hERG (K+ channel) proteins (Li et al., 2017). Rather, DNAJA1, DNAJA2, and DNAJA4 overexpression in HEK-293 and HeLa cells inhibited the hERG maturation and trafficking in an Hsc70-dependent manner (Walker et al., 2010). It suggests the specificity of JDPs in handling different PM proteins.
Besides the ERAD pathways, multiple PM proteins are also degraded by selective ER-autophagy (Figure 3). Often, large misfolded membrane proteins can accumulate in complex tertiary structures, which are difficult to unfold and retrotranslocate. These ERAD-resistant clients are degraded by ER autophagy, for e.g., misfolded gonadotropin-releasing hormone receptor (GnRHR) E90K-GnRHR and N1303K-CFTR (Houck et al., 2014; He et al., 2021). Intuitively, inefficient ERAD also leads to the autophagy of some substrates. Although the mechanism is not completely understood, Hsp70 and DNAJB12 have been shown to interact with the autophagy machinery in the presence of specific complex substrates. Depletion of DNAJB12 inhibits the lysosomal targeting of N1303K-CFTR in HEK293 cells. Also, the overexpression of the DNAJB12-QPD mutant (J-domain mutant) prevented ER phagy. Hence, it acts with the partner Hsp70 to regulate ER phagy of complex substrates (He et al., 2021). However, at this point, the involvement of other Hsp70:JDP machinery in ER autophagy cannot be ruled out.
Removal of proteins from PM
PM Proteins destined to be degraded are recognized at their exposed cytosolic domain. Cytosolic adaptors (in yeast) or chaperones (in mammals) interact with the substrates, recognize the conformational changes, and recruit the E3 ubiquitin ligases (Babst, 2014; MacGurn, 2014). Ubiquitination of proteins ensures their endocytosis and fusion with early endosomes. Further, they can be deubiquitinated and recycled back to the PM directly or through Golgi; else, they are targeted for MVB sorting and undergo lysosomal degradation (Figure 4).
In yeast, the major components of the PMPQC pathway include the ART-Rsp5 system, which recognizes, ubiquitinates, and targets proteins for degradation. Although the turnover of multiple PM proteins is regulated by E3-ubiquitin ligase, Rsp5, the degradation of misfolded PM proteins in yeast is poorly understood. To this end, it is even proposed that in budding yeast, PM proteins with an exposed misfolded cytosolic domain might be escaping the PMPQC network and may not be as efficiently degraded (Lewis and Pelham, 2009). The multi-functional JDP Ydj1, although it operates with Rsp5 for cytosolic heat-denatured substrates, it is not involved in the QC of PM proteins (Fang et al., 2014). It is still an open question if another JDP operates with Rsp5 for the degradation of PM proteins. Nevertheless, Caj1, a cytosolic JDP in budding yeast, negatively regulates the Rsp5-mediated degradation of tryptophan transporters Tat1 and Tat2 (Welsch et al., 2003; Hernández-López et al., 2011; Dobriyal et al., 2020). Interestingly, Caj1 also associates with different lipids, reportedly phosphatidic acid and phosphatidylinositol-5-phosphate. Besides being majorly cytosolic, Caj1 is partially localized to the PM as well (Dobriyal et al., 2020; Herianto et al., 2021; Zhang et al., 2021). Overexpression of Caj1 (but not Caj1 H34Q mutant) stabilized tryptophan permeases Tat1 and Tat2, suggesting a possible role of Hsp70:JDP machinery in regulating the stability/turnover of PM proteins in budding yeast.
In mammals, a more elaborate PMPQC network has been deciphered that regulates the turnover of misfolded proteins. Several molecular chaperones and co-chaperones recognize and decide the fate of the misfolded PM proteins (Okiyoneda et al., 2011). Similar to the chaperone-mediated substrate handling at the ER, defective PM protein degradation is regulated by JDP-Hsp70-Hsp90s. However, Hsp70/HSPA1A, Hsc70/HSPA8, DNAJA1, and DNAJB2 are among the major components of PMPQC, deciding the fate of misfolded proteins (Okiyoneda et al., 2011). They help in the ubiquitination of proteins by the recruitment of the E3 ubiquitin ligase, CHIP (C-terminal Hsp70 interacting protein) (Connell et al., 2001). Hsp70, through its C-terminal EEVD motif, directly interacts with the N-terminal TPR domain of CHIP. The interaction stalls protein folding and concomitantly facilitates the ubiquitination of Hsp70-bound substrates. In this process, the JDP stimulates the chaperone activity of Hsp70 and CHIP and increases client ubiquitination (Rosser et al., 2007; Zhang et al., 2015).
Several misfolded PM proteins undergo Hsp/c70-JDP-CHIP mediated degradation. For example, the mature, glycosylated ΔF508CFTR interacts with Hsp70 and DNAJB2/Hdj1 under restrictive conditions in post-Golgi compartments in BHK-21 cells (Bagdany et al., 2017). Si-RNA mediated downregulation of Hsc70 (HSPA8), DNAJB2 (Hdj1), DNAJA1 (Hdj2), and DNAJC7 (TPR2) caused the accumulation of mutant ΔF508CFTR at the PM of HeLa cell lines (Okiyoneda et al., 2010). Similarly, JDP, Hsp70, Hsc70, and Hsp90, along with CHIP, are known to interact with the unfolded protein CD4T-λC at the PM but not with the native protein to facilitate ubiquitination and degradation (Apaja et al., 2010). CHIP-mediated PQC was also found for dopamine D4.4 receptor (DRD4) and vasopressin V2 receptor (V2R) mutants, which escape the secretory pathway PQC, and undergo rapid degradation at the PM (Apaja et al., 2010). In yet another case, defects in the turnover of the hERG K+ channel, which leads to long QT syndrome 2 (LQT2), is also regulated by Hsp70-CHIP. Disruption of Hsp70’s interaction with CHIP prevented substrate-based rapid endocytosis of both wild-type as well as unstable mutant hERG from the PM (Apaja et al., 2013). In the human collecting duct (mpkDCT and mpkCCD cells), CHIP-Hsc70 directly interact and regulate the degradation of water channel aquaporin (AQP2), thus maintaining the urine osmolarity and renal water handling (Lu et al., 2007; Wu et al., 2018). A JDP co-chaperoning Hsp70 in these processes is yet to be identified.
Conclusion and perspectives
PM proteins are essential for life. During their lifetime, they pass through a series of rigorous PQC checkpoints that ensures their proper folding, stability, functionality, and finally, their abundance in the cell. Hsp70:JDP networks in the cytosol, the ER, as well as at the PM are emerging as the key players that assist in the trafficking, biogenesis, maturation as well as degradation of PM proteins. As they navigate inside the cell, PM proteins are relayed through many different JDPs. And depending on the type of client, JDPs may have pro-folding or pro-degradation functions. Understanding how Hsp70:JDP machinery manages to perform this task is the Achilles heel of this field. Interestingly, multiple JDPs sequentially operate with a single pool of Hsp70 to perform rather unique PQC functions on a single PM protein, suggesting highly intricate and regulated interactions between JDPs with client proteins and with Hsp70. Finally, the discovery of lipid interacting and PM-associated Hsp70s and JDPs is bound to bring about a paradigm shift in our understanding of how these versatile chaperones cater to the needs of PM proteins at the plasma membrane.
BOX 1 Hsp70: J-domain protein (JDP) machinery
Hsp70s, the 70 KDa molecular chaperones, are the key players in cellular proteostasis. They work in collaboration with two other components of the chaperone network, i.e., Hsp40s (also called J-domain proteins or JDPs) and Nucleotide exchange factors (NEFs) (Mayer and Bukau, 2005; Young, 2010; Rosenzweig et al., 2019). Hsp70s have a C-terminal Substrate Binding Domain (SBD), through which they bind to the hydrophobic regions of their client proteins, and an N-terminal Nucleotide Binding Domain (NBD), which binds to ATP and hydrolyzes it to ADP. The ATPase activity of Hsp70 is central to all its chaperone functions as this allosterically regulates its client-binding properties. However, the weak intrinsic ATPase activity of Hsp70s is not sufficient for its chaperone functions. JDPs, the obligate co-chaperones of Hsp70s, have a conserved J-domain with an invariant HPD motif responsible for stimulating the ATPase activity of Hsp70s, thereby potentiating its chaperone functions. The immense versatility of Hsp70 stems from their ability to operate with multiple JDPs in any given compartment, where, besides stimulating the ATPase activity of Hsp70s, they also determine the multi-valency of Hsp70 chaperones (Qiu et al., 2006; Craig and Marszalek, 2017; Rosenzweig et al., 2019; Liu et al., 2020). Furthermore, JDPs can also specify Hsp70s function by tethering Hsp70s to a specific subcellular localization (Kampinga and Craig, 2010). JDPs are highly diverse in their structure, function as well as their cellular abundance. Different cellular compartments harbor multiple JDPs performing general or highly specialized functions, ranging from folding, transport, and degradation of proteins, to mediating protein-protein interactions and assembly or disassembly of protein complexes. While JDPs form obligate partnerships with the Hsp70 chaperone, there is increasing evidence that some JDPs may have functions outside this association as well.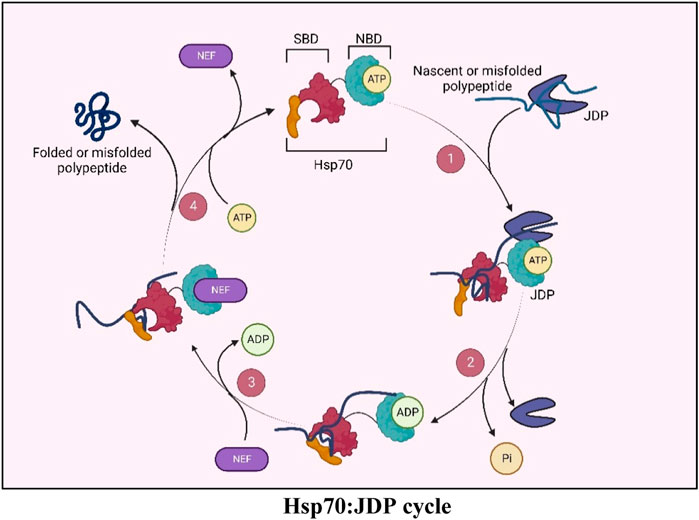
BOX 2 UPR
Unfolded protein response (UPR) is an evolutionarily conserved ER (Endoplasmic reticulum) stress response pathway to regulate the cell’s optimal capacity for protein synthesis and maturation. It is primarily induced by the accumulation of aberrant proteins in the ER due to various environmental stimuli, activation of other cellular stress response pathways, or lack of efficient protein folding machinery in the ER. UPR signaling pathways communicate the ER-stress to the gene expression machinery in the nucleus and cytosol, and regulate cellular transcription and translation. UPR restores ER homeostasis by 1) expanding the ER membrane and increasing ER abundance to meet protein folding demands, 2) positively regulating protein folding machinery, 3) activating the ERAD (ER-associated degradation) pathway etc. Further, under aggravated and long-term ER stress, UPR signaling may lead to apoptotic cell death (Walter and Ron, 2011; Hetz et al., 2020; Read and Schröder, 2021).
BOX 3 ERAD
The Endoplasmic reticulum-associated degradation (ERAD) is an extensive PQC network that oversees the regulated degradation of ER proteins. ER is the primary site for the folding and maturation of endomembrane, secretory and PM proteins, and failure to reach mature confirmation targets these proteins for degradation. ERAD pathway mediates the removal of unfolded, misfolded or unwanted proteins from the ER (Vembar and Brodsky, 2008). ERAD has three major branches based on the site of protein misfolding. Proteins with a lesion on the luminal, transmembrane, and cytosolic domain are targeted by ERAD-L, ERAD-M, and ERAD-C pathways, respectively. Different classes of molecular chaperones, ubiquitination machinery, and other PQC factors regulate these pathways. It involves recognition of misfolded substrates by chaperones, its ubiquitination by E2 ub conjugating enzymes and E3 ub ligases, extraction out of ER by retrotranslocons, and finally, targeting to the proteasome for degradation (Kumari and Brodsky, 2021).
Author contributions
PS and CS conceived the work. PS and KY wrote the manuscript. CS, PS, and KY edited the manuscript.
Acknowledgments
The authors would like to thank SERB (project no. CRG/2018/002024) and IISER Bhopal for funding. PS and KY thank CSIR for fellowship.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abell, B. M., Rabu, C., Leznicki, P., Young, J. C., and High, S. (2007). Post-translational integration of tail-anchored proteins is facilitated by defined molecular chaperones. J. Cell. Sci. 120, 1743–1751. doi:10.1242/jcs.002410
Accili, D., Mosthaf, L., Ullrich, A., and Taylor, S. I. (1991). A mutation in the extracellular domain of the insulin receptor impairs the ability of insulin to stimulate receptor autophosphorylation. J. Biol. Chem. 266, 434–439. doi:10.1016/S0021-9258(18)52453-4
Adamowski, M., Narasimhan, M., Kania, U., Glanc, M., De Jaeger, G., and Friml, J. (2018). A functional study of AUXILIN-LIKE1 and 2, two putative clathrin uncoating factors in Arabidopsis. Plant Cell. 30, 700–716. doi:10.1105/tpc.17.00785
Ajit Tamadaddi, C., and Sahi, C. (2016). J domain independent functions of J proteins. Cell. Stress Chaperones 21, 563–570. doi:10.1007/s12192-016-0697-1
Alberts, B., Johnson, A., and Lewis, J. (2002). “Membrane proteins,” in Molecular biology of the cell. New York, NY: Garland Science. Available at: https://www.ncbi.nlm.nih.gov/books/NBK26878/.
Amagai, Y., Matsuda, A., Jung, K., Oida, K., Jang, H., Ishizaka, S., et al. (2015). A point mutation in the extracellular domain of KIT promotes tumorigenesis of mastcells via ligand-independent auto-dimerization. Sci. Rep. 5, 9775. doi:10.1038/srep09775
Amin-Wetzel, N., Saunders, R. A., Kamphuis, M. J., Rato, C., Preissler, S., Harding, H. P., et al. (2017). A J-protein Co-chaperone recruits BiP to monomerize IRE1 and repress the unfolded protein response. Cell. 171, 1625–1637. e13. doi:10.1016/j.cell.2017.10.040
Anderson, S. A., Satyanarayan, M. B., Wessendorf, R. L., Lu, Y., and Fernandez, D. E. (2021). A homolog of GuidedEntry of Tail-anchored proteins3 functions in membrane-specific protein targeting in chloroplasts of Arabidopsis. Plant Cell. 33, 2812–2833. doi:10.1093/plcell/koab145
Anelli, T., and Sitia, R. (2008). Protein quality control in the early secretory pathway. EMBO J. 27, 315–327. doi:10.1038/sj.emboj.7601974
Apaja, P. M., Foo, B., Okiyoneda, T., Valinsky, W. C., Barriere, H., Atanasiu, R., et al. (2013). Ubiquitination-dependent quality control of hERG K + channel with acquired and inherited conformational defect at the plasma membrane. Mol. Biol. Cell. 24, 3787–3804. doi:10.1091/mbc.e13-07-0417
Apaja, P. M., Xu, H., and Lukacs, G. L. (2010). Quality control for unfolded proteins at the plasma membrane. J. Cell. Biol. 191, 553–570. doi:10.1083/jcb.201006012
Araki, K., and Nagata, K. (2011). Protein folding and quality control in the ER. Cold Spring Harb. Perspect. Biol. 3, a007526. doi:10.1101/cshperspect.a007526
Arispe, N., and De Maio, A. (2000). ATP and ADP modulate a cation channel formed by Hsc70 in acidic phospholipid membranes. J. Biol. Chem. 275, 30839–30843. doi:10.1074/jbc.M005226200
Asseck, L. Y., Mehlhorn, D. G., Monroy, J. R., Ricardi, M. M., Breuninger, H., Wallmeroth, N., et al. (2021). Loss of GET pathway orthologs in Arabidopsis thaliana causes root hair growth defects and affects SNARE abundance. Proc. Natl. Acad. Sci. U. S. A. 118, E1544-E1553. doi:10.1073/pnas.1619525114
Ast, T., Cohen, G., and Schuldiner, M. (2013). A network of cytosolic factors targets SRP-independent proteins to the endoplasmic reticulum. Cell. 152, 1134–1145. doi:10.1016/j.cell.2013.02.003
Ast, T., and Schuldiner, M. (2013). All roads lead to rome (but some may be harder to travel): SRP-independent translocation into the endoplasmic reticulum. Crit. Rev. Biochem. Mol. Biol. 48, 273–288. doi:10.3109/10409238.2013.782999
Aviram, N., Ast, T., Costa, E. A., Arakel, E. C., Chuartzman, S. G., Jan, C. H., et al. (2016). The SND proteins constitute an alternative targeting route to the endoplasmic reticulum. Nature 540, 134–138. doi:10.1038/nature20169
Aviram, N., and Schuldiner, M. (2014). Embracing the void—How much do we really know about targeting and translocation to the endoplasmic reticulum? Curr. Opin. Cell. Biol. 29, 8–17. doi:10.1016/j.ceb.2014.02.004
Aviram, N., and Schuldiner, M. (2017). Targeting and translocation of proteins to the endoplasmic reticulum at a glance. J. Cell. Sci. 130, 4079–4085. doi:10.1242/jcs.204396
Babst, M. (2014). Quality control: Quality control at the plasma membrane: One mechanism does not fit all. J. Cell. Biol. 205, 11–20. doi:10.1083/jcb.201310113
Bagdany, M., Veit, G., Fukuda, R., Avramescu, R. G., Okiyoneda, T., Baaklini, I., et al. (2017). Chaperones rescue the energetic landscape of mutant CFTR at single molecule and in cell. Nat. Commun. 8, 398. doi:10.1038/s41467-017-00444-4
Barghetti, A., Sjögren, L., Floris, M., Paredes, E. B., Wenkel, S., and Brodersen, P. (2017). Heat-shock protein 40 is the key farnesylation target in meristem size control, abscisic acid signaling, and drought resistance. Genes. Dev. 31, 2282–2295. doi:10.1101/gad.301242.117
Beilharz, T., Egan, B., Silver, P. A., Hofmann, K., and Lithgow, T. (2003). Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J. Biol. Chem. 278, 8219–8223. doi:10.1074/jbc.M212725200
Benedix, J., Lajoie, P., Jaiswal, H., Burgard, C., Greiner, M., Zimmermann, R., et al. (2010). BiP modulates the affinity of its Co-chaperone ERj1 for ribosomes. J. Biol. Chem. 285, 36427–36433. doi:10.1074/jbc.M110.143263
Benyair, R., Eisenberg-Lerner, A., and Merbl, Y. (2022). Maintaining Golgi homeostasis: A balancing act of two proteolytic pathways. Cells 11, 780. doi:10.3390/cells11050780
Bernardino de la Serna, J., Schütz, G. J., Eggeling, C., and Cebecauer, M. (2016). There is No simple model of the plasma membrane organization. Front. Cell. Dev. Biol. 4, 106. doi:10.3389/fcell.2016.00106
Bilog, Smulders, Oliverio, Labanieh, Stahelin, Nikolaidis, Labanieh, C., Zapanta, J., Stahelin, R. V., et al. (2019). Membrane localization of HspA1A, a stress inducible 70-kDa heat-shock protein, depends on its interaction with intracellular phosphatidylserine. Biomolecules 9, 152. doi:10.3390/biom9040152
Binder, Z. A., Thorne, A. H., Bakas, S., Wileyto, E. P., Bilello, M., Akbari, H., et al. (2018). Epidermal growth factor receptor extracellular domain mutations in glioblastoma present opportunities for clinical imaging and therapeutic development. Cancer Cell. 34, 163–177. e7. doi:10.1016/j.ccell.2018.06.006
Blau, M., Mullapudi, S., Becker, T., Dudek, J., Zimmermann, R., Penczek, P. A., et al. (2005). ERj1p uses a universal ribosomal adaptor site to coordinate the 80S ribosome at the membrane. Nat. Struct. Mol. Biol. 12, 1015–1016. doi:10.1038/nsmb998
Braakman, I., and Bulleid, N. J. (2011). Protein folding and modification in the mammalian endoplasmic reticulum. Annu. Rev. Biochem. 80, 71–99. doi:10.1146/annurev-biochem-062209-093836
Buchberger, A., Bukau, B., and Sommer, T. (2010). Protein quality control in the cytosol and the endoplasmic reticulum: Brothers in arms. Mol. Cell. 40, 238–252. doi:10.1016/j.molcel.2010.10.001
Buck, T. M., Kolb, A. R., Boyd, C. R., Kleyman, T. R., and Brodsky, J. L. (2010). The endoplasmic reticulum–associated degradation of the epithelial sodium channel requires a unique complement of molecular chaperones. Mol. Biol. Cell. 21, 1047–1058. doi:10.1091/mbc.e09-11-0944
Bukiya, A. N., and Dopico, A. M. (2019). “Regulation of BK channel activity by cholesterol and its derivatives,” in Cholesterol modulation of protein function, advances in experimental medicine and biology. Editors A. Rosenhouse-Dantsker, and A. N. Bukiya (Cham: Springer International Publishing), 53–75. doi:10.1007/978-3-030-04278-3_3
Bursać, D., and Lithgow, T. (2009). Jid1 is a J-protein functioning in the mitochondrial matrix, unable to directly participate in endoplasmic reticulum associated protein degradation. FEBS Lett. 583, 2954–2958. doi:10.1016/j.febslet.2009.08.005
Caplan, A. J., Cyr, D. M., and Douglas, M. G. (1992). YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell. 71, 1143–1155. doi:10.1016/S0092-8674(05)80063-7
Casares, D., Escribá, P. V., and Rosselló, C. A. (2019). Membrane lipid composition: Effect on membrane and organelle structure, function and compartmentalization and therapeutic avenues. Int. J. Mol. Sci. 20, 2167. doi:10.3390/ijms20092167
Casson, J., McKenna, M., Haßdenteufel, S., Aviram, N., Zimmerman, R., and High, S. (2017). Multiple pathways facilitate the biogenesis of mammalian tail-anchored proteins. J. Cell. Sci. 130, 3851–3861. doi:10.1242/jcs.207829
Chanoux, R. A., Shubin, C. B., Robay, A., Suaud, L., and Rubenstein, R. C. (2013). Hsc70 negatively regulates epithelial sodium channel trafficking at multiple sites in epithelial cells. Am. J. Physiol. Cell. Physiol. 305, C776–C787. doi:10.1152/ajpcell.00059.2013
Chartron, J. W., Gonzalez, G. M., and Clemons, W. M. (2011). A structural model of the Sgt2 protein and its interactions with chaperones and the get4/get5 complex. J. Biol. Chem. 286, 34325–34334. doi:10.1074/jbc.M111.277798
Chen, Y., Tsai, B., Li, N., and Gao, N. (2022). Structural remodeling of ribosome associated Hsp40-Hsp70 chaperones during co-translational folding. Nat. Commun. 13, 3410. doi:10.1038/s41467-022-31127-4
Chino, H., and Mizushima, N. (2020). ER-Phagy: Quality control and turnover of endoplasmic reticulum. Trends Cell. Biol. 30, 384–398. doi:10.1016/j.tcb.2020.02.001
Cho, H., Shim, W. J., Liu, Y., and Shan, S.-O. (2021). J-domain proteins promote client relay from Hsp70 during tail-anchored membrane protein targeting. J. Biol. Chem. 296, 100546. doi:10.1016/j.jbc.2021.100546
Choo-Kang, L. R., and Zeitlin, P. L. (2001). Induction of HSP70 promotes DeltaF508 CFTR trafficking. Am. J. Physiol. Lung Cell. Mol. Physiol. 281, L58–L68. doi:10.1152/ajplung.2001.281.1.L58
Claus, L. A. N., Savatin, D. V., and Russinova, E. (2018). The crossroads of receptor-mediated signaling and endocytosis in plants: Endocytosis and signaling in plants. J. Integr. Plant Biol. 60, 827–840. doi:10.1111/jipb.12672
Connell, P., Ballinger, C. A., Jiang, J., Wu, Y., Thompson, L. J., Höhfeld, J., et al. (2001). The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell. Biol. 3, 93–96. doi:10.1038/35050618
Corsi, A. K., and Schekman, R. (1997). The lumenal domain of Sec63p stimulates the ATPase activity of BiP and mediates BiP recruitment to the translocon in Saccharomyces cerevisiae. J. Cell. Biol. 137, 1483–1493. doi:10.1083/jcb.137.7.1483
Coy-Vergara, J., Rivera-Monroy, J., Urlaub, H., Lenz, C., and Schwappach, B. (2019). A trap mutant reveals the physiological client spectrum of TRC40. J. Cell. Sci. 132, 230094. doi:10.1242/jcs.230094
Craig, E. A., and Marszalek, J. (2017). How do J-proteins get Hsp70 to do so many different things? Trends biochem. Sci. 42, 355–368. doi:10.1016/j.tibs.2017.02.007
Cross, B. C. S., Sinning, I., Luirink, J., and High, S. (2009). Delivering proteins for export from the cytosol. Nat. Rev. Mol. Cell. Biol. 10, 255–264. doi:10.1038/nrm2657
Daverkausen-Fischer, L., and Pröls, F. (2021). Dual topology of co-chaperones at the membrane of the endoplasmic reticulum. Cell. Death Discov. 7, 203. doi:10.1038/s41420-021-00594-x
Davila, S., Furu, L., Gharavi, A. G., Tian, X., Onoe, T., Qian, Q., et al. (2004). Mutations in SEC63 cause autosomal dominant polycystic liver disease. Nat. Genet. 36, 575–577. doi:10.1038/ng1357
De Maio, A. (2014). Extracellular Hsp70: Export and function. Curr. Protein Pept. Sci. 15, 225–231. doi:10.2174/1389203715666140331113057
De Maio, A., and Hightower, L. (2021). The interaction of heat shock proteins with cellular membranes: A historical perspective. Cell. Stress Chaperones 26, 769–783. doi:10.1007/s12192-021-01228-y
Delic, M., Valli, M., Graf, A. B., Pfeffer, M., Mattanovich, D., and Gasser, B. (2013). The secretory pathway: Exploring yeast diversity. FEMS Microbiol. Rev. 37, 872–914. doi:10.1111/1574-6976.12020
Deol, S. S., Bond, P. J., Domene, C., and Sansom, M. S. P. (2004). Lipid-protein interactions of integral membrane proteins: A comparative simulation study. Biophys. J. 87, 3737–3749. doi:10.1529/biophysj.104.048397
Desai, A. J., and Miller, L. J. (2018). Changes in the plasma membrane in metabolic disease: Impact of the membrane environment on G protein-coupled receptor structure and function. Br. J. Pharmacol. 175, 4009–4025. doi:10.1111/bph.13943
Dobriyal, N., Sagarika, P., Shrivastava, A., Verma, A. K., Islam, Z., Gupta, P., et al. (2020). Over-expression of Caj1, a plasma membrane associated J-domain protein in Saccharomyces cerevisiae, stabilizes amino acid permeases. Biochim. Biophys. Acta. Biomembr. 1862, 183435. doi:10.1016/j.bbamem.2020.183435
Dong, M., Bridges, J. P., Apsley, K., Xu, Y., and Weaver, T. E. (2008). ERdj4 and ERdj5 are required for endoplasmic reticulum-associated protein degradation of misfolded surfactant protein C. Mol. Biol. Cell. 19, 2620–2630. doi:10.1091/mbc.e07-07-0674
Donnelly, B. F., Needham, P. G., Snyder, A. C., Roy, A., Khadem, S., Brodsky, J. L., et al. (2013). Hsp70 and Hsp90 multichaperone complexes sequentially regulate thiazide-sensitive cotransporter endoplasmic reticulum-associated degradation and biogenesis. J. Biol. Chem. 288, 13124–13135. doi:10.1074/jbc.M113.455394
Doonan, L. M., Guerriero, C. J., Preston, G. M., Buck, T. M., Khazanov, N., Fisher, E. A., et al. (2019). Hsp104 facilitates the endoplasmic-reticulum-associated degradation of disease-associated and aggregation-prone substrates. Protein Sci. 28, 1290–1306. doi:10.1002/pro.3636
Döring, K., Ahmed, N., Riemer, T., Suresh, H. G., Vainshtein, Y., Habich, M., et al. (2017). Profiling ssb-nascent chain interactions reveals principles of hsp70-assisted folding. Cell. 170, 298–311. e20. doi:10.1016/j.cell.2017.06.038
Dudek, J., Greiner, M., Müller, A., Hendershot, L. M., Kopsch, K., Nastainczyk, W., et al. (2005). ERj1p has a basic role in protein biogenesis at the endoplasmic reticulum. Nat. Struct. Mol. Biol. 12, 1008–1014. doi:10.1038/nsmb1007
Eisenberg, E., and Greene, L. E. (2007). Multiple roles of auxilin and Hsc70 in clathrin-mediated endocytosis. Traffic 8, 640–646. doi:10.1111/j.1600-0854.2007.00568.x
Elinder, F., and Liin, S. I. (2017). Actions and mechanisms of polyunsaturated fatty acids on voltage-gated ion channels. Front. Physiol. 8, 43. doi:10.3389/fphys.2017.00043
Fang, N. N., Chan, G. T., Zhu, M., Comyn, S. A., Persaud, A., Deshaies, R. J., et al. (2014). Rsp5/Nedd4 is the main ubiquitin ligase that targets cytosolic misfolded proteins following heat stress. Nat. Cell. Biol. 16, 1227–1237. doi:10.1038/ncb3054
Farinha, C. M., and Amaral, M. D. (2005). Most F508del-CFTR is targeted to degradation at an early folding checkpoint and independently of calnexin. Mol. Cell. Biol. 25, 5242–5252. doi:10.1128/MCB.25.12.5242-5252.2005
Farinha, C. M., Nogueira, P., Mendes, F., Penque, D., and Amaral, M. D. (2002). The human DnaJ homologue (Hdj)-1/heat-shock protein (Hsp) 40 co-chaperone is required for the in vivo stabilization of the cystic fibrosis transmembrane conductance regulator by Hsp70. Biochem. J. 366, 797–806. doi:10.1042/bj20011717
Farkas, Á., and Bohnsack, K. E. (2021). Capture and delivery of tail-anchored proteins to the endoplasmic reticulum. J. Cell. Biol. 220, e202105004. doi:10.1083/jcb.202105004
Favaloro, V., Spasic, M., Schwappach, B., and Dobberstein, B. (2008). Distinct targeting pathways for the membrane insertion of tail-anchored (TA) proteins. J. Cell. Sci. 121, 1832–1840. doi:10.1242/jcs.020321
Favaloro, V., Vilardi, F., Schlecht, R., Mayer, M. P., and Dobberstein, B. (2010). Asna1/TRC40-mediated membrane insertion of tail-anchored proteins. J. Cell. Sci. 123, 1522–1530. doi:10.1242/jcs.055970
Feldheim, D., Rothblatt, J., and Schekman, R. (1992). Topology and functional domains of Sec63p, an endoplasmic reticulum membrane protein required for secretory protein translocation. Mol. Cell. Biol. 12, 3288–3296. doi:10.1128/mcb.12.7.3288
Fratti, R. A. (2021). Editorial: Effects of membrane lipids on protein function. Front. Cell. Dev. Biol. 9, 675264. doi:10.3389/fcell.2021.675264
Gall, W. E., Higginbotham, M. A., Chen, C.-Y., Ingram, M. F., Cyr, D. M., and Graham, T. R. (2000). The auxilin-like phosphoprotein Swa2p is required for clathrin function in yeast. Curr. Biol. 10, 1349–1358. doi:10.1016/S0960-9822(00)00771-5
Genereux, J. C., Qu, S., Zhou, M., Ryno, L. M., Wang, S., Shoulders, M. D., et al. (2015). Unfolded protein response-induced ERdj3 secretion links ER stress to extracellular proteostasis. EMBO J. 34, 4–19. doi:10.15252/embj.201488896
Ghaddar, K., Merhi, A., Saliba, E., Krammer, E.-M., Prévost, M., and André, B. (2014). Substrate-induced ubiquitylation and endocytosis of yeast amino acid permeases. Mol. Cell. Biol. 34, 4447–4463. doi:10.1128/MCB.00699-14
Gillece, P., Luz, J. M., Lennarz, W. J., de la Cruz, F. J., and Römisch, K. (1999). Export of a cysteine-free misfolded secretory protein from the endoplasmic reticulum for degradation requires interaction with protein disulfide isomerase. J. Cell. Biol. 147, 1443–1456. doi:10.1083/jcb.147.7.1443
Greener, T., Zhao, X., Nojima, H., Eisenberg, E., and Greene, L. E. (2000). Role of cyclin G-associated kinase in uncoating clathrin-coated vesicles from non-neuronal cells. J. Biol. Chem. 275, 1365–1370. doi:10.1074/jbc.275.2.1365
Grove, D. E., Fan, C.-Y., Ren, H. Y., and Cyr, D. M. (2011). The endoplasmic reticulum-associated Hsp40 DNAJB12 and Hsc70 cooperate to facilitate RMA1 E3-dependent degradation of nascent CFTRDeltaF508. Mol. Biol. Cell. 22, 301–314. doi:10.1091/mbc.E10-09-0760
Guidon, P. T., and Hightower, L. E. (1986b). Purification and initial characterization of the 71-kilodalton rat heat-shock protein and its cognate as fatty acid binding proteins. Biochemistry 25, 3231–3239. doi:10.1021/bi00359a023
Guidon, P. T., and Hightower, L. E. (1986a). The 73 kilodalton heat shock cognate protein purified from rat brain contains nonesterified palmitic and stearic acids. J. Cell. Physiol. 128, 239–245. doi:10.1002/jcp.1041280215
Guo, R., Gu, D., Zhao, J., Zhu, L., Chen, M., Liu, S., et al. (2019). EGFR extracellular domain mutation in patients with lung cancer. JCO 37, e20532. doi:10.1200/JCO.2019.37.15_suppl.e20532
Gupta, D., and Tuteja, N. (2011). Chaperones and foldases in endoplasmic reticulum stress signaling in plants. Plant Signal. Behav. 6, 232–236. doi:10.4161/psb.6.2.15490
Halic, M., Becker, T., Pool, M. R., Spahn, C. M. T., Grassucci, R. A., Frank, J., et al. (2004). Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427, 808–814. doi:10.1038/nature02342
Han, S., Liu, Y., and Chang, A. (2007). Cytoplasmic Hsp70 promotes ubiquitination for endoplasmic reticulum-associated degradation of a misfolded mutant of the yeast plasma membrane ATPase, PMA1. J. Biol. Chem. 282, 26140–26149. doi:10.1074/jbc.M701969200
Haßdenteufel, S., Sicking, M., Schorr, S., Aviram, N., Fecher-Trost, C., Schuldiner, M., et al. (2017). hSnd2 protein represents an alternative targeting factor to the endoplasmic reticulum in human cells. FEBS Lett. 591, 3211–3224. doi:10.1002/1873-3468.12831
He, K., Song, E., Upadhyayula, S., Dang, S., Gaudin, R., Skillern, W., et al. (2020). Dynamics of Auxilin 1 and GAK in clathrin-mediated traffic. J. Cell. Biol. 219, e201908142. doi:10.1083/jcb.201908142
He, L., Kennedy, A. S., Houck, S., Aleksandrov, A., Quinney, N. L., Cyr-Scully, A., et al. (2021). DNAJB12 and Hsp70 triage arrested intermediates of N1303K-CFTR for endoplasmic reticulum-associated autophagy. Mol. Biol. Cell. 32, 538–553. doi:10.1091/mbc.E20-11-0688
Hegde, R. S., and Keenan, R. J. (2011). Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat. Rev. Mol. Cell. Biol. 12, 787–798. doi:10.1038/nrm3226
Hellerschmied, D., Serebrenik, Y. V., Shao, L., Burslem, G. M., and Crews, C. M. (2019). Protein folding state-dependent sorting at the Golgi apparatus. Mol. Biol. Cell. 30, 2296–2308. doi:10.1091/mbc.E19-01-0069
Herianto, S., Rathod, J., Shah, P., Chen, Y.-Z., Wu, W.-S., Liang, B., et al. (2021). Systematic analysis of phosphatidylinositol-5-phosphate-interacting proteins using yeast proteome microarrays. Anal. Chem. 93, 868–877. doi:10.1021/acs.analchem.0c03463
Hernández-López, M. J., García-Marqués, S., Randez-Gil, F., and Prieto, J. A. (2011). Multicopy suppression screening of Saccharomyces cerevisiae identifies the ubiquitination machinery as a main target for improving growth at low temperatures. Appl. Environ. Microbiol. 77, 7517–7525. doi:10.1128/AEM.00404-11
Hetz, C., Zhang, K., and Kaufman, R. J. (2020). Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell. Biol. 21, 421–438. doi:10.1038/s41580-020-0250-z
Houck, S. A., Ren, H. Y., Madden, V. J., Bonner, J. N., Conlin, M. P., Janovick, J. A., et al. (2014). Quality control autophagy degrades soluble ERAD-resistant conformers of the misfolded membrane protein GnRHR. Mol. Cell. 54, 166–179. doi:10.1016/j.molcel.2014.02.025
Huang, Y., Arora, K., Mun, K. S., Yang, F., Moon, C., Yarlagadda, S., et al. (2019). Targeting DNAJB9, a novel ER luminal co-chaperone, to rescue ΔF508-CFTR. Sci. Rep. 9, 9808. doi:10.1038/s41598-019-46161-4
Hundley, H. A., Walter, W., Bairstow, S., and Craig, E. A. (2005). Human mpp11 J protein: Ribosome-tethered molecular chaperones are ubiquitous. Science 308, 1032–1034. doi:10.1126/science.1109247
Huyer, G., Piluek, W. F., Fansler, Z., Kreft, S. G., Hochstrasser, M., Brodsky, J. L., et al. (2004). Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J. Biol. Chem. 279, 38369–38378. doi:10.1074/jbc.M402468200
Itskanov, S., Kuo, K. M., Gumbart, J. C., and Park, E. (2021). Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62. Nat. Struct. Mol. Biol. 28, 162–172. doi:10.1038/s41594-020-00541-x
Janssen, M. J., Salomon, J., te Morsche, R. H. M., and Drenth, J. P. H. (2012). Loss of heterozygosity is present in SEC63 germline carriers with polycystic liver disease. PLoS ONE 7, e50324. doi:10.1371/journal.pone.0050324
Ji, Y., Kim, H., Yang, L., Sha, H., Roman, C. A., Long, Q., et al. (2016). The Sel1L-hrd1 endoplasmic reticulum-associated degradation complex manages a key checkpoint in B cell development. Cell. Rep. 16, 2630–2640. doi:10.1016/j.celrep.2016.08.003
Jiang, R., Gao, B., Prasad, K., Greene, L. E., and Eisenberg, E. (2000). Hsc70 chaperones clathrin and primes it to interact with vesicle membranes. J. Biol. Chem. 275, 8439–8447. doi:10.1074/jbc.275.12.8439
Jin, T., Gu, Y., Zanusso, G., Sy, M., Kumar, A., Cohen, M., et al. (2000). The chaperone protein BiP binds to a mutant prion protein and mediates its degradation by the proteasome. J. Biol. Chem. 275, 38699–38704. doi:10.1074/jbc.M005543200
Johnson, J. L., and Craig, E. A. (2000). A role for the Hsp40 Ydj1 in repression of basal steroid receptor activity in yeast. Mol. Cell. Biol. 20, 3027–3036. doi:10.1128/MCB.20.9.3027-3036.2000
Jonikas, M. C., Collins, S. R., Denic, V., Oh, E., Quan, E. M., Schmid, V., et al. (2009). Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323, 1693–1697. doi:10.1126/science.1167983
Juarez-Navarro, K., Ayala-Garcia, V. M., Ruiz-Baca, E., Meneses-Morales, I., Rios-Banuelos, J. L., and Lopez-Rodriguez, A. (2020). Assistance for folding of disease-causing plasma membrane proteins. Biomolecules 10, 728. doi:10.3390/biom10050728
Jung, S., Jung, Y., and Kim, H. (2019). Proper insertion and topogenesis of membrane proteins in the ER depend on Sec63. Biochim. Biophys. Acta. Gen. Subj. 1863, 1371–1380. doi:10.1016/j.bbagen.2019.06.005
Kaksonen, M., and Roux, A. (2018). Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell. Biol. 19, 313–326. doi:10.1038/nrm.2017.132
Kalli, A. C., Morgan, G., and Sansom, M. S. P. (2013). Interactions of the auxilin-1 PTEN-like domain with model membranes result in nanoclustering of phosphatidyl inositol phosphates. Biophys. J. 105, 137–145. doi:10.1016/j.bpj.2013.05.012
Kampinga, H. H., and Craig, E. A. (2010). The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell. Biol. 11, 579–592. doi:10.1038/nrm2941
Kanazawa, M., Terada, K., Kato, S., and Mori, M. (1997). HSDJ, a human homolog of DnaJ, is farnesylated and is involved in protein import into mitochondria. J. Biochem. 121, 890–895. doi:10.1093/oxfordjournals.jbchem.a021670
Kang, J. A., and Jeon, Y. J. (2021). How is the fidelity of proteins ensured in terms of both quality and quantity at the endoplasmic reticulum? Mechanistic insights into E3 ubiquitin ligases. Int. J. Mol. Sci. 22, 2078. doi:10.3390/ijms22042078
Kellokumpu, S. (2019). Golgi pH, ion and redox homeostasis: How much do they really matter? Front. Cell. Dev. Biol. 7, 93. doi:10.3389/fcell.2019.00093
Kim, S.-Y., and Craig, E. A. (2005). Broad sensitivity of Saccharomyces cerevisiae lacking ribosome-associated chaperone Ssb or Zuo1 to cations, including aminoglycosides. Eukaryot. Cell. 4, 82–89. doi:10.1128/EC.4.1.82-89.2005
Kim, S. J., and Skach, W. R. (2012). Mechanisms of CFTR folding at the endoplasmic reticulum. Front. Pharmacol. 3, 201. doi:10.3389/fphar.2012.00201
Koplin, A., Preissler, S., Ilina, Y., Koch, M., Scior, A., Erhardt, M., et al. (2010). A dual function for chaperones SSB–RAC and the NAC nascent polypeptide–associated complex on ribosomes. J. Cell. Biol. 189, 57–68. doi:10.1083/jcb.200910074
Kramer, G., Shiber, A., and Bukau, B. (2019). Mechanisms of cotranslational maturation of newly synthesized proteins. Annu. Rev. Biochem. 88, 337–364. doi:10.1146/annurev-biochem-013118-111717
Krantz, K. C., Puchalla, J., Thapa, R., Kobayashi, C., Bisher, M., Viehweg, J., et al. (2013). Clathrin coat disassembly by the yeast hsc70/ssa1p and auxilin/swa2p proteins observed by single-particle burst analysis spectroscopy. J. Biol. Chem. 288, 26721–26730. doi:10.1074/jbc.M113.491753
Kumari, D., and Brodsky, J. L. (2021). The targeting of native proteins to the endoplasmic reticulum-associated degradation (ERAD) pathway: An expanding repertoire of regulated substrates. Biomolecules 11, 1185. doi:10.3390/biom11081185
Kumari, D., Fisher, E. A., and Brodsky, J. L. (2022). Hsp40s play distinct roles during the initial stages of apolipoprotein B biogenesis. Mol. Biol. Cell. 33, ar15. ar15. doi:10.1091/mbc.E21-09-0436
Lam, B. C.-H., Sage, T. L., Bianchi, F., and Blumwald, E. (2001). Role of SH3 domain–containing proteins in clathrin-mediated vesicle trafficking in Arabidopsis. Plant Cell. 13, 2499–2512. doi:10.1105/tpc.010279
Lee, A. G. (2004). How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta 1666, 62–87. doi:10.1016/j.bbamem.2004.05.012
Lee, D., Wu, X., Eisenberg, E., and Greene, L. E. (2006). Recruitment dynamics of GAK and auxilin to clathrin-coated pits during endocytosis. J. Cell. Sci. 119, 3502–3512. doi:10.1242/jcs.03092
Lee, D., Zhao, X., Zhang, F., Eisenberg, E., and Greene, L. E. (2005). Depletion of GAK/auxilin 2 inhibits receptor-mediated endocytosis and recruitment of both clathrin and clathrin adaptors. J. Cell. Sci. 118, 4311–4321. doi:10.1242/jcs.02548
Lei, P., Xiao, Y., Li, P., Xie, P., Wang, H., Huang, S., et al. (2020). hSnd2/TMEM208 is an HIF-1α-targeted gene and contains a WH2 motif. Acta Biochim. Biophys. Sin. 52, 328–331. doi:10.1093/abbs/gmz157
Lenaz, G. (1987). Lipid fluidity and membrane protein dynamics. Biosci. Rep. 7, 823–837. doi:10.1007/BF01119473
Lewis, M. J., and Pelham, H. R. B. (2009). Inefficient quality control of thermosensitive proteins on the plasma membrane. PLoS ONE 4, e5038. doi:10.1371/journal.pone.0005038
Li, J., Zhao-Hui, C., Batoux, M., Nekrasov, V., Roux, M., Chinchilla, D., et al. (2009). Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc. Natl. Acad. Sci. U. S. A. 106, 15973–15978. doi:10.1073/pnas.0905532106
Li, K., Jiang, Q., Bai, X., Yang, Y.-F., Ruan, M.-Y., and Cai, S.-Q. (2017). Tetrameric assembly of K+ channels requires ER-located chaperone proteins. Mol. Cell. 65, 52–65. doi:10.1016/j.molcel.2016.10.027
Linxweiler, M., Schick, B., and Zimmermann, R. (2017). Let’s talk about Secs: Sec61, Sec62 and Sec63 in signal transduction, oncology and personalized medicine. Signal Transduct. Target. Ther. 2, 17002. doi:10.1038/sigtrans.2017.2
Liou, S.-T., Cheng, M.-Y., and Wang, C. (2007). SGT2 and MDY2 interact with molecular chaperone YDJ1 in Saccharomyces cerevisiae. Cell. Stress Chaperones 12, 59–70. doi:10.1379/CSC-220R.1
Liu, Q., Liang, C., and Zhou, L. (2020). Structural and functional analysis of the Hsp70/Hsp40 chaperone system. Protein Sci. 29, 378–390. doi:10.1002/pro.3725
Lu, H. A. J., Sun, T.-X., Matsuzaki, T., Yi, X.-H., Eswara, J., Bouley, R., et al. (2007). Heat shock protein 70 interacts with aquaporin-2 and regulates its trafficking. J. Biol. Chem. 282, 28721–28732. doi:10.1074/jbc.M611101200
Lu, R., Drubin, D. G., and Sun, Y. (2016). Clathrin-mediated endocytosis in budding yeast at a glance. J. Cell. Sci. 129, 1531–1536. doi:10.1242/jcs.182303
Lujan, P., and Campelo, F. (2021). Should I stay or should I go? Golgi membrane spatial organization for protein sorting and retention. Arch. Biochem. Biophys. 707, 108921. doi:10.1016/j.abb.2021.108921
Macazo, F. C., and White, R. J. (2014). Monitoring charge flux to quantify unusual ligand-induced ion channel activity for use in biological nanopore-based sensors. Anal. Chem. 86, 5519–5525. doi:10.1021/ac500832a
MacGurn, J. A. (2014). Garbage on, garbage off: New insights into plasma membrane protein quality control. Curr. Opin. Cell. Biol. 29, 92–98. doi:10.1016/j.ceb.2014.05.001
Mahmood, Md.I., Liu, X., Neya, S., and Hoshino, T. (2013). Influence of lipid composition on the structural stability of G-protein coupled receptor. Chem. Pharm. Bull. 61, 426–437. doi:10.1248/cpb.c12-01059
Mariappan, M., Mateja, A., Dobosz, M., Bove, E., Hegde, R. S., and Keenan, R. J. (2011). The mechanism of membrane-associated steps in tail-anchored protein insertion. Nature 477, 61–66. doi:10.1038/nature10362
Maruyama, D., Endo, T., and Nishikawa, S. (2010). BiP-mediated polar nuclei fusion is essential for the regulation of endosperm nuclei proliferation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 107, 1684–1689. doi:10.1073/pnas.0905795107
Maruyama, D., Yamamoto, M., Endo, T., and Nishikawa, S. (2014). Different sets of ER-resident J-proteins regulate distinct polar nuclear-membrane fusion events in Arabidopsis thaliana. Plant Cell. Physiol. 55, 1937–1944. doi:10.1093/pcp/pcu120
Matsumura, Y., David, L. L., and Skach, W. R. (2011). Role of Hsc70 binding cycle in CFTR folding and endoplasmic reticulum-associated degradation. Mol. Biol. Cell. 22, 2797–2809. doi:10.1091/mbc.E11-02-0137
Mayer, M. P., and Bukau, B. (2005). Hsp70 chaperones: Cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62, 670–684. doi:10.1007/s00018-004-4464-6
Mayor, S., and Pagano, R. E. (2007). Pathways of clathrin-independent endocytosis. Nat. Rev. Mol. Cell. Biol. 8, 603–612. doi:10.1038/nrm2216
Meacham, G. C., Lu, Z., King, S., Sorscher, E., Tousson, A., and Cyr, D. M. (1999). The Hdj-2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J. 18, 1492–1505. doi:10.1093/emboj/18.6.1492
Mehnert, M., Sommermeyer, F., Berger, M., Kumar Lakshmipathy, S., Gauss, R., Aebi, M., et al. (2015). The interplay of Hrd3 and the molecular chaperone system ensures efficient degradation of malfolded secretory proteins. Mol. Biol. Cell. 26, 185–194. doi:10.1091/mbc.E14-07-1202
Melnyk, A., Rieger, H., and Zimmermann, R. (2015). “Co-Chaperones of the mammalian endoplasmic reticulum,” in The networking of chaperones by Co-chaperones, subcellular biochemistry. Editors G. L. Blatch, and A. L. Edkins (Cham: Springer International Publishing), 179–200. doi:10.1007/978-3-319-11731-7_9
Misselwitz, B., Staeck, O., Matlack, K. E. S., and Rapoport, T. A. (1999). Interaction of BiP with the J-domain of the Sec63p component of the endoplasmic reticulum protein translocation complex. J. Biol. Chem. 274, 20110–20115. doi:10.1074/jbc.274.29.20110
Moreno, A. A., Mukhtar, M. S., Blanco, F., Boatwright, J. L., Moreno, I., Jordan, M. R., et al. (2012). IRE1/bZIP60-Mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS ONE 7, e31944. doi:10.1371/journal.pone.0031944
Nakatsukasa, K., Huyer, G., Michaelis, S., and Brodsky, J. L. (2008). Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 132, 101–112. doi:10.1016/j.cell.2007.11.023
Narasimhan, M., Johnson, A., Prizak, R., Kaufmann, W. A., Tan, S., Casillas-Pérez, B., et al. (2020). Evolutionarily unique mechanistic framework of clathrin-mediated endocytosis in plants. eLife 9, e52067. doi:10.7554/eLife.52067
Needham, P. G., Guerriero, C. J., and Brodsky, J. L. (2019). Chaperoning endoplasmic reticulum-associated degradation (ERAD) and protein conformational diseases. Cold Spring Harb. Perspect. Biol. 11, a033928. doi:10.1101/cshperspect.a033928
Nekrasov, V., Li, J., Batoux, M., Roux, M., Chu, Z.-H., Lacombe, S., et al. (2009). Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 28, 3428–3438. doi:10.1038/emboj.2009.262
Nikko, E., and Pelham, H. R. B. (2009). Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic 10, 1856–1867. doi:10.1111/j.1600-0854.2009.00990.x
Nishikawa, S., Brodsky, J. L., and Nakatsukasa, K. (2005). Roles of molecular chaperones in endoplasmic reticulum (ER) quality control and ER-associated degradation (ERAD). J. Biochem. 137, 551–555. doi:10.1093/jb/mvi068
Nishikawa, S., Fewell, S. W., Kato, Y., Brodsky, J. L., and Endo, T. (2001). Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell. Biol. 153, 1061–1070. doi:10.1083/jcb.153.5.1061
Noh, S.-J., Kwon, C. S., Oh, D.-H., Moon, J. S., and Chung, W.-I. (2003). Expression of an evolutionarily distinct novel BiP gene during the unfolded protein response in Arabidopsis thaliana. Gene 311, 81–91. doi:10.1016/s0378-1119(03)00559-6
Ohta, M., and Takaiwa, F. (2014). Emerging features of ER resident J-proteins in plants. Plant Signal. Behav. 9, e28194. doi:10.4161/psb.28194
Okiyoneda, T., Apaja, P. M., and Lukacs, G. L. (2011). Protein quality control at the plasma membrane. Curr. Opin. Cell. Biol. 23, 483–491. doi:10.1016/j.ceb.2011.04.012
Okiyoneda, T., Barrière, H., Bagdány, M., Rabeh, W. M., Du, K., Höhfeld, J., et al. (2010). Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science 329, 805–810. doi:10.1126/science.1191542
Otero, J. H., Lizák, B., and Hendershot, L. M. (2010). Life and death of a BiP substrate. Semin. Cell. Dev. Biol. 21, 472–478. doi:10.1016/j.semcdb.2009.12.008
Otto, H., Conz, C., Maier, P., Wölfle, T., Suzuki, C. K., Jenö, P., et al. (2005). The chaperones MPP11 and Hsp70L1 form the mammalian ribosome-associated complex. Proc. Natl. Acad. Sci. U. S. A. 102, 10064–10069. doi:10.1073/pnas.0504400102
Park, C.-J., and Seo, Y.-S. (2015). Heat shock proteins: A review of the molecular chaperones for plant immunity. Plant Pathol. J. 31, 323–333. doi:10.5423/PPJ.RW.08.2015.0150
Park, S.-H., Bolender, N., Eisele, F., Kostova, Z., Takeuchi, J., Coffino, P., et al. (2007). The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin–proteasome system. Mol. Biol. Cell. 18, 153–165. doi:10.1091/mbc.e06-04-0338
Peisker, K., Chiabudini, M., and Rospert, S. (2010). The ribosome-bound Hsp70 homolog Ssb of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1803, 662–672. doi:10.1016/j.bbamcr.2010.03.005
Pfund, C., Lopez-Hoyo, N., Ziegelhoffer, T., Schilke, B. A., Lopez-Buesa, P., Walter, W. A., et al. (1998). The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J. 17, 3981–3989. doi:10.1093/emboj/17.14.3981
Phillips, B. P., Gomez-Navarro, N., and Miller, E. A. (2020). Protein quality control in the endoplasmic reticulum. Curr. Opin. Cell. Biol. 65, 96–102. doi:10.1016/j.ceb.2020.04.002
Plemper, R. K., and Wolf, D. H. (1999). Endoplasmic reticulum degradation. Reverse protein transport and its end in the proteasome. Mol. Biol. Rep. 26, 125–130. doi:10.1023/A:1006913215484
Pobre, K. F. R., Poet, G. J., and Hendershot, L. M. (2019). The endoplasmic reticulum (ER) chaperone BiP is a master regulator of ER functions: Getting by with a little help from ERdj friends. J. Biol. Chem. 294, 2098–2108. doi:10.1074/jbc.REV118.002804
Pool, M. R., Stumm, J., Fulga, T. A., Sinning, I., and Dobberstein, B. (2002). Distinct modes of signal recognition particle interaction with the ribosome. Science 297, 1345–1348. doi:10.1126/science.1072366
Pratt, W. B., and Toft, D. O. (2003). Regulation of signaling protein function and trafficking by the hsp90/hsp70-Based chaperone machinery. Exp. Biol. Med. 228, 111–133. doi:10.1177/153537020322800201
Qiu, X.-B., Shao, Y.-M., Miao, S., and Wang, L. (2006). The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 63, 2560–2570. doi:10.1007/s00018-006-6192-6
Rabu, C., Schmid, V., Schwappach, B., and High, S. (2009). Biogenesis of tail-anchored proteins: The beginning for the end? J. Cell. Sci. 122, 3605–3612. doi:10.1242/jcs.041210
Rabu, C., Wipf, P., Brodsky, J. L., and High, S. (2008). A precursor-specific role for hsp40/hsc70 during tail-anchored protein integration at the endoplasmic reticulum. J. Biol. Chem. 283, 27504–27513. doi:10.1074/jbc.M804591200
Radons, J. (2016). The human HSP70 family of chaperones: Where do we stand? Cell. Stress Chaperones 21, 379–404. doi:10.1007/s12192-016-0676-6
Rapoport, T. A. (2007). Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450, 663–669. doi:10.1038/nature06384
Read, A., and Schröder, M. (2021). The unfolded protein response: An overview. Biology 10, 384. doi:10.3390/biology10050384
Rodriguez-Furlan, C., Minina, E. A., and Hicks, G. R. (2019). Remove, recycle, degrade: Regulating plasma membrane protein accumulation. Plant Cell. 31, 2833–2854. doi:10.1105/tpc.19.00433
Rosenzweig, R., Nillegoda, N. B., Mayer, M. P., and Bukau, B. (2019). The Hsp70 chaperone network. Nat. Rev. Mol. Cell. Biol. 20, 665–680. doi:10.1038/s41580-019-0133-3
Rosquete, M. R., Davis, D. J., and Drakakaki, G. (2018). The plant trans-golgi network: Not just a matter of distinction. Plant Physiol. 176, 187–198. doi:10.1104/pp.17.01239
Rosser, M. F. N., Washburn, E., Muchowski, P. J., Patterson, C., and Cyr, D. M. (2007). Chaperone functions of the E3 ubiquitin ligase CHIP. J. Biol. Chem. 282, 22267–22277. doi:10.1074/jbc.M700513200
Sadler, I., Chiang, A., Kurihara, T., Rothblatt, J., Way, J., and Silver, P. (1989). A yeast gene important for protein assembly into the endoplasmic reticulum and the nucleus has homology to DnaJ, an Escherichia coli heat shock protein. J. Cell. Biol. 109, 2665–2675. doi:10.1083/jcb.109.6.2665
Sahi, C., Lee, T., Inada, M., Pleiss, J. A., and Craig, E. A. (2010). Cwc23, an essential J protein critical for pre-mRNA splicing with a dispensable J domain. Mol. Cell. Biol. 30, 33–42. doi:10.1128/MCB.00842-09
Saijo, Y. (2010). ER quality control of immune receptors and regulators in plants: ER quality control of plant immune receptors. Cell. Microbiol. 12, 716–724. doi:10.1111/j.1462-5822.2010.01472.x
Sardana, R., and Emr, S. D. (2021). Membrane protein quality control mechanisms in the endo-lysosome system. Trends Cell. Biol. 31, 269–283. doi:10.1016/j.tcb.2020.11.011
Schuldiner, M., Metz, J., Schmid, V., Denic, V., Rakwalska, M., Schmitt, H. D., et al. (2008). The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 134, 634–645. doi:10.1016/j.cell.2008.06.025
Schwabl, S., and Teis, D. (2022). Protein quality control at the Golgi. Curr. Opin. Cell. Biol. 75, 102074. doi:10.1016/j.ceb.2022.02.008
Schweiger, R., Müller, N. C., Schmitt, M. J., Soll, J., and Schwenkert, S. (2012). AtTPR7 is a chaperone docking protein of the Sec translocon in Arabidopsis. J. Cell. Sci. 125, 5196–5207. doi:10.1242/jcs.111054
Schweiger, R., and Schwenkert, S. (2013). AtTPR7 as part of the Arabidopsis Sec post-translocon. Plant Signal. Behav. 8, e25286. doi:10.4161/psb.25286
Shen, J., Zeng, Y., Zhuang, X., Sun, L., Yao, X., Pimpl, P., et al. (2013). Organelle pH in the Arabidopsis endomembrane system. Mol. Plant 6, 1419–1437. doi:10.1093/mp/sst079
Shiber, A., Breuer, W., Brandeis, M., and Ravid, T. (2013). Ubiquitin conjugation triggers misfolded protein sequestration into quality control foci when Hsp70 chaperone levels are limiting. Mol. Biol. Cell. 24, 2076–2087. doi:10.1091/mbc.E13-01-0010
Shiber, A., Döring, K., Friedrich, U., Klann, K., Merker, D., Zedan, M., et al. (2018). Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature 561, 268–272. doi:10.1038/s41586-018-0462-y
Silberstein, S., Schlenstedt, G., Silver, P. A., and Gilmore, R. (1998). A role for the DnaJ homologue Scj1p in protein folding in the yeast endoplasmic reticulum. J. Cell. Biol. 143, 921–933. doi:10.1083/jcb.143.4.921
Sjögren, L., Floris, M., Barghetti, A., Völlmy, F., Linding, R., and Brodersen, P. (2018). Farnesylated heat shock protein 40 is a component of membrane-bound RISC in Arabidopsis. J. Biol. Chem. 293, 16608–16622. doi:10.1074/jbc.RA118.003887
Skowronek, M. H., Rotter, M., and Haas, I. G. (1999). Molecular characterization of a novel mammalian DnaJ-like Sec63p homolog. Biol. Chem. 380, 1133–1138. doi:10.1515/BC.1999.142
Sopha, P., Kadokura, H., Yamamoto, Y., Takeuchi, M., Saito, M., Tsuru, A., et al. (2012). A novel mammalian ER-located J-protein, DNAJB14, can accelerate ERAD of misfolded membrane proteins. Cell. Struct. Funct. 37, 177–187. doi:10.1247/csf.12017
Sopha, P., Ren, H. Y., Grove, D. E., and Cyr, D. M. (2017). Endoplasmic reticulum stress–induced degradation of DNAJB12 stimulates BOK accumulation and primes cancer cells for apoptosis. J. Biol. Chem. 292, 11792–11803. doi:10.1074/jbc.M117.785113
Sousa, R., and Lafer, E. M. (2015). The role of molecular chaperones in clathrin mediated vesicular trafficking. Front. Mol. Biosci. 2, 26. doi:10.3389/fmolb.2015.00026
Srivastava, R., Zalisko, B. E., Keenan, R. J., and Howell, S. H. (2017). The GET system inserts the tail-anchored protein, SYP72, into endoplasmic reticulum membranes. Plant Physiol. 173, 1137–1145. doi:10.1104/pp.16.00928
Sun, Z., and Brodsky, J. L. (2019). Protein quality control in the secretory pathway. J. Cell. Biol. 218, 3171–3187. doi:10.1083/jcb.201906047
Taxis, C., Hitt, R., Park, S.-H., Deak, P. M., Kostova, Z., and Wolf, D. H. (2003). Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of ERAD. J. Biol. Chem. 278, 35903–35913. doi:10.1074/jbc.M301080200
Terada, K., and Mori, M. (2000). Human DnaJ homologs dj2 and dj3, and bag-1 are positive cochaperones of hsc70. J. Biol. Chem. 275, 24728–24734. doi:10.1074/jbc.M002021200
Tian, G., Hu, C., Yun, Y., Yang, W., Dubiel, W., Cheng, Y., et al. (2021). Dual roles of HSP70 chaperone HSPA1 in quality control of nascent and newly synthesized proteins. EMBO J. 40, e106183. doi:10.15252/embj.2020106183
Tripathi, A., Mandon, E. C., Gilmore, R., and Rapoport, T. A. (2017). Two alternative binding mechanisms connect the protein translocation Sec71-Sec72 complex with heat shock proteins. J. Biol. Chem. 292, 8007–8018. doi:10.1074/jbc.M116.761122
Tuo, S., Nakashima, K., and Pringle, J. R. (2013). Role of endocytosis in localization and maintenance of the spatial markers for bud-site selection in yeast. PLoS One 8, e72123. doi:10.1371/journal.pone.0072123
Vasconcelos-Cardoso, M., Batista-Almeida, D., Rios-Barros, L. V., Castro-Gomes, T., and Girao, H. (2022). Cellular and molecular mechanisms underlying plasma membrane functionality and integrity. J. Cell. Sci. 135, jcs259806. doi:10.1242/jcs.259806
Vembar, S. S., and Brodsky, J. L. (2008). One step at a time: Endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell. Biol. 9, 944–957. doi:10.1038/nrm2546
Verma, A. K., Diwan, D., Raut, S., Dobriyal, N., Brown, R. E., Gowda, V., et al. (2017). Evolutionary conservation and emerging functional diversity of the cytosolic Hsp70:J protein chaperone network of Arabidopsis thaliana. G3 7, 1941–1954. doi:10.1534/g3.117.042291
von Heijne, G. (2006). Membrane-protein topology. Nat. Rev. Mol. Cell. Biol. 7, 909–918. doi:10.1038/nrm2063
Walker, V. E., Wong, M. J. H., Atanasiu, R., Hantouche, C., Young, J. C., and Shrier, A. (2010). Hsp40 chaperones promote degradation of the hERG potassium channel. J. Biol. Chem. 285, 3319–3329. doi:10.1074/jbc.M109.024000
Walter, P., and Ron, D. (2011). The unfolded protein response: From stress pathway to homeostatic regulation. Science 334, 1081–1086. doi:10.1126/science.1209038
Wang, F., Durfee, L. A., and Huibregtse, J. M. (2013). A cotranslational ubiquitination pathway for quality control of misfolded proteins. Mol. Cell. 50, 368–378. doi:10.1016/j.molcel.2013.03.009
Wang, F., Whynot, A., Tung, M., and Denic, V. (2011). The mechanism of tail-anchored protein insertion into the ER membrane. Mol. Cell. 43, 738–750. doi:10.1016/j.molcel.2011.07.020
Welsch, C. A., Hagiwara, S., Goetschy, J. F., and Movva, N. R. (2003). Ubiquitin pathway proteins influence the mechanism of action of the novel immunosuppressive drug FTY720 in Saccharomyces cerevisiae. J. Biol. Chem. 278, 26976–26982. doi:10.1074/jbc.M213144200
Willmund, F., del Alamo, M., Pechmann, S., Chen, T., Albanèse, V., Dammer, E. B., et al. (2013). The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell. 152, 196–209. doi:10.1016/j.cell.2012.12.001
Wilson, M. R., Satapathy, S., Jeong, S., and Fini, M. E. (2022). Clusterin, other extracellular chaperones, and eye disease. Prog. Retin. Eye Res. 89, 101032. doi:10.1016/j.preteyeres.2021.101032
Wolf, D. H., and Stolz, A. (2012). The Cdc48 machine in endoplasmic reticulum associated protein degradation. Biochim. Biophys. Acta 1823, 117–124. doi:10.1016/j.bbamcr.2011.09.002
Woodall, N. B., Yin, Y., and Bowie, J. U. (2015). Dual-topology insertion of a dual-topology membrane protein. Nat. Commun. 6, 8099. doi:10.1038/ncomms9099
Wu, Q., Moeller, H. B., Stevens, D. A., Sanchez-Hodge, R., Childers, G., Kortenoeven, M. L. A., et al. (2018). CHIP regulates aquaporin-2 quality control and body water homeostasis. J. Am. Soc. Nephrol. 29, 936–948. doi:10.1681/ASN.2017050526
Wu, X., Zhao, X., Baylor, L., Kaushal, S., Eisenberg, E., and Greene, L. E. (2001). Clathrin exchange during clathrin-mediated endocytosis. J. Cell. Biol. 155, 291–300. doi:10.1083/jcb.200104085
Xiao, J., Kim, L. S., and Graham, T. R. (2006). Dissection of swa2p/auxilin domain requirements for cochaperoning Hsp70 clathrin-uncoating activity in vivo. Mol. Biol. Cell. 17, 3281–3290. doi:10.1091/mbc.e06-02-0106
Xie, W., and Ng, D. T. W. (2010). ERAD substrate recognition in budding yeast. Semin. Cell. Dev. Biol. 21, 533–539. doi:10.1016/j.semcdb.2010.02.007
Xing, S., Mehlhorn, D. G., Wallmeroth, N., Asseck, L. Y., Kar, R., Voss, A., et al. (2017). Loss of GET pathway orthologs in Arabidopsis thaliana causes root hair growth defects and affects SNARE abundance. Proc. Natl. Acad. Sci. U. S. A. 114, E1544-E1553. doi:10.1073/pnas.1619525114
Yamamoto, M., Maruyama, D., Endo, T., and Nishikawa, S. (2008). Arabidopsis thaliana has a set of J proteins in the endoplasmic reticulum that are conserved from yeast to animals and plants. Plant Cell. Physiol. 49, 1547–1562. doi:10.1093/pcp/pcn119
Youker, R. T., Walsh, P., Beilharz, T., Lithgow, T., and Brodsky, J. L. (2004). Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol. Biol. Cell. 15, 4787–4797. doi:10.1091/mbc.e04-07-0584
Young, J. C. (2010). Mechanisms of the Hsp70 chaperone system. Biochem. Cell. Biol. 88, 291–300. doi:10.1139/O09-175
Zhang, C. X., Engqvist-Goldstein, Å. E. Y., Carreno, S., Owen, D. J., Smythe, E., and Drubin, D. G. (2005). Multiple roles for cyclin G-associated kinase in clathrin-mediated sorting events: GAK in clathrin-mediated trafficking. Traffic 6, 1103–1113. doi:10.1111/j.1600-0854.2005.00346.x
Zhang, H., Amick, J., Chakravarti, R., Santarriaga, S., Schlanger, S., McGlone, C., et al. (2015). A bipartite interaction between Hsp70 and CHIP regulates ubiquitination of chaperoned client proteins. Structure 23, 472–482. doi:10.1016/j.str.2015.01.003
Zhang, L., Gjoerup, O., and Roberts, T. M. (2004). The serine/threonine kinase cyclin G-associated kinase regulates epidermal growth factor receptor signaling. Proc. Natl. Acad. Sci. U. S. A. 101, 10296–10301. doi:10.1073/pnas.0403175101
Zhang, R., Malinverni, D., Cyr, D. M., Rios, P. D. L., and Nillegoda, N. B. (2022). J-domain protein chaperone circuits in proteostasis and disease. Trends Cell. Biol. doi:10.1016/j.tcb.2022.05.004
Zhang, X. X., Young, J. W., Foster, L. J., and Duong, F. (2021). Nanodisc-based proteomics identify Caj1 as an Hsp40 with affinity for phosphatidic acid lipids. J. Proteome Res. 20, 4831–4839. doi:10.1021/acs.jproteome.1c00503
Zhang, Y., Liu, R., Ni, M., Gill, P., and Lee, A. S. (2010). Cell surface relocalization of the endoplasmic reticulum chaperone and unfolded protein response regulator GRP78/BiP. J. Biol. Chem. 285, 15065–15075. doi:10.1074/jbc.M109.087445
Zhang, Y., Nijbroek, G., Sullivan, M. L., McCracken, A. A., Watkins, S. C., Michaelis, S., et al. (2001). Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol. Biol. Cell. 12, 1303–1314. doi:10.1091/mbc.12.5.1303
Zhao, Y., MacGurn, J. A., Liu, M., and Emr, S. (2013). The ART-Rsp5 ubiquitin ligase network comprises a plasma membrane quality control system that protects yeast cells from proteotoxic stress. eLife 2, e00459. doi:10.7554/eLife.00459
Zhou, M., Fisher, E. A., and Ginsberg, H. N. (1998). Regulated Co-translational ubiquitination of apolipoprotein B100. A new paradigm for proteasomal degradation of a secretory protein. J. Biol. Chem. 273, 24649–24653. doi:10.1074/jbc.273.38.24649
Keywords: J-domain proteins (JDPs), Hsp70, plasma membrane proteins, protein quality control (PQC), secretory pathway
Citation: Sagarika P, Yadav K and Sahi C (2022) Volleying plasma membrane proteins from birth to death: Role of J-domain proteins. Front. Mol. Biosci. 9:1072242. doi: 10.3389/fmolb.2022.1072242
Received: 17 October 2022; Accepted: 02 December 2022;
Published: 15 December 2022.
Edited by:
Matthias Christof Truttmann, Michigan Medicine, University of Michigan, United StatesReviewed by:
Tessa Sinnige, Utrecht University, NetherlandsYvonne Nyathi, University of Bradford, United Kingdom
Copyright © 2022 Sagarika, Yadav and Sahi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chandan Sahi, c2FoaUBpaXNlcmIuYWMuaW4=
 Preeti Sagarika
Preeti Sagarika Kirpa Yadav
Kirpa Yadav Chandan Sahi
Chandan Sahi