
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Mol. Biosci. , 01 December 2022
Sec. RNA Networks and Biology
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.1066650
This article is part of the Research Topic The Why of RNA Granules: Form, Function, and Regulation View all 7 articles
Stress granules (SGs) are non-membrane bound cytoplasmic condensates that form in response to a variety of different stressors. Canonical SGs are thought to have a cytoprotective role, reallocating cellular resources during stress by activation of the integrated stress response (ISR) to inhibit translation and avoid apoptosis. However, different stresses result in compositionally distinct, non-canonical SG formation that is likely pro-apoptotic, though the exact function(s) of both SGs subtypes remain unclear. A unique non-canonical SG subtype is triggered upon exposure to ultraviolet (UV) radiation. While it is generally agreed that UV SGs are bona fide SGs due to their dependence upon the core SG nucleating protein Ras GTPase-activating protein-binding protein 1 (G3BP1), the localization of other key components of UV SGs are unknown or under debate. Further, the dynamics of UV SGs are not known, though unique properties such as cell cycle dependence have been observed. This Perspective compiles the available information on SG subtypes and on UV SGs in particular in an attempt to understand the formation, dynamics, and function of these mysterious stress-specific complexes. We identify key gaps in knowledge related to UV SGs, and examine the unique aspects of their formation. We propose that more thorough knowledge of the distinct properties of UV SGs will lead to new avenues of understanding of the function of SGs, as well as their roles in disease.
Stress granules (SGs) are evolutionarily conserved cytoplasmic biomolecular condensates that form in response to a variety of environmental stressors (Nover et al., 1989; Kedersha et al., 1999; Kedersha et al., 2002). Since the discovery of SGs in mammalian cells in 1999 (Kedersha et al., 1999) an ever-broadening array of stressors have been identified as inducers of SGs. While it is known that different stressors can give rise to compositionally and functionally distinct SGs (Aulas et al., 2017; Advani and Ivanov, 2020), our understanding of the components of SG subtypes, and their functional consequences, is understudied. The study of SGs is of intense interest due to the role SGs likely play in various diseases, especially protein aggregation diseases such as Alzheimer’s Disease (AD) and Amyotrophic Lateral Sclerosis (ALS) (Liu-Yesucevitz et al., 2010; Li et al., 2013; Ash et al., 2014; Boyd et al., 2014; Wolozin, 2014; Wolozin and Ivanov, 2019). Because SG function may be determined both by composition and by the environmental conditions under which they form, gaining insights into compositionally distinct SG subtypes will be important for understanding and manipulating SG formation in the context of disease.
One of the most distinct and least understood SG subtypes are those induced by ultraviolet radiation (UV). In countless reports and reviews of SG biology, UV is listed among the litany of stresses that induce SG formation. Humans experience UV daily in the environment, and its genotoxic and carcinogenic effects have been well studied (Roy, 2017). However, published evidence documenting the composition, dynamics, and function of UV-induced SGs is surprisingly limited. We do not yet understand the mechanism(s) driving UV SG assembly, nor the functional consequences of their formation. In this Perspective, we assemble the available evidence on the biology of UV SGs. We believe that this unique SG subtype, which unlike most SGs seems to lack mRNA, is also the only known example of G1-specific cell cycle-controlled SG formation. We propose that the unique properties of UV SGs represent an untapped source for new hypotheses to contribute broadly to the fields of SG biology, aggregation-mediated disease, and biomolecular condensation.
As one of the earliest discovered and most broadly studied SG types, SGs induced by acute high-dose exposure to sodium arsenite have become the canon by which SGs are defined (Kedersha et al., 1999; Kedersha et al., 2002; Aulas and vande Velde, 2015; Aulas et al., 2017; Advani and Ivanov, 2020; Riggs et al., 2020). So-called “canonical” or arsenite-like SGs—caused by many stresses including heat shock (Kedersha et al., 1999), thapsigargin (Aulas et al., 2017), and bisphenol A (Fay et al., 2021) among others—are associated with the inhibition of global protein synthesis and the preferential translation of stress-induced transcripts (Harding et al., 2000a). Bulk translation is inhibited through two main pathways (Advani and Ivanov, 2019): 1) the phosphorylation of the alpha subunit of eukaryotic initiation factor 2 alpha (P-eIF2α), which occurs in mammalian cells via one of four serine/threonine kinases: (heme-regulated eIF2α kinase (HRI) (McEwen et al., 2005), protein kinase R (PKR) (Srivastava et al., 1998), PKR-like ER kinase (PERK) (Harding et al., 2000b) and general control nonderepressible 2 (GCN2) (Wek et al., 1995); and 2) the inhibition of the mammalian target of rapamycin (mTOR) (Proud, 2019). The former mechanism inhibits recycling of the eIF2/tRNAiMet/GTP ternary complex required for translation initiation (Wek, 2018), while the latter mechanism results in hypophosphorylated eIF4E binding protein (4E-BP) which binds eIF4E and inhibits translation (Proud, 2019). When translation is inhibited, 48S translation pre-initiation complexes and untranslated mRNAs accumulate and aggregate in the cytoplasm, favoring a liquid-liquid phase separation (LLPS) event (Guillén-Boixet et al., 2020; Sanders et al., 2020; Yang et al., 2020) and assembling into SGs. Canonical SGs are in dynamic equilibrium with polysomes, as drugs that inhibit polysome disassembly (e.g., emetine, cycloheximide) inhibit SG formation, while drugs that induce premature polysome disassembly (e.g., puromycin) promote SG assembly (Kedersha et al., 2000; Kedersha et al., 2002).
The first protein markers associated with SGs were the RNA-binding proteins TIA-1 and TIAR, as well as the cytoplasmic poly(A)-binding protein (PABPC1) (Kedersha et al., 1999). Soon thereafter, the Ras GTPase-activating protein-binding protein 1 (G3BP1) was identified as a key modulator of SG assembly (Tourrière et al., 2003). It is now firmly established that G3BP1 is a master regulator of SG assembly, and most known SG subtypes require G3BP1 or its close homolog G3BP2 for their assembly (Kedersha et al., 2016; Guillén-Boixet et al., 2020; Sanders et al., 2020; Yang et al., 2020). Related to their relationship with eIF2α-mediated translational control, canonical SGs contain poly(A)+ mRNAs as well as stalled translation 48S-preinitiation complexes including the eIF3 complex, eIF4G, and the small ribosomal subunit (Kedersha et al., 2002; Aulas et al., 2017) (Table 1).
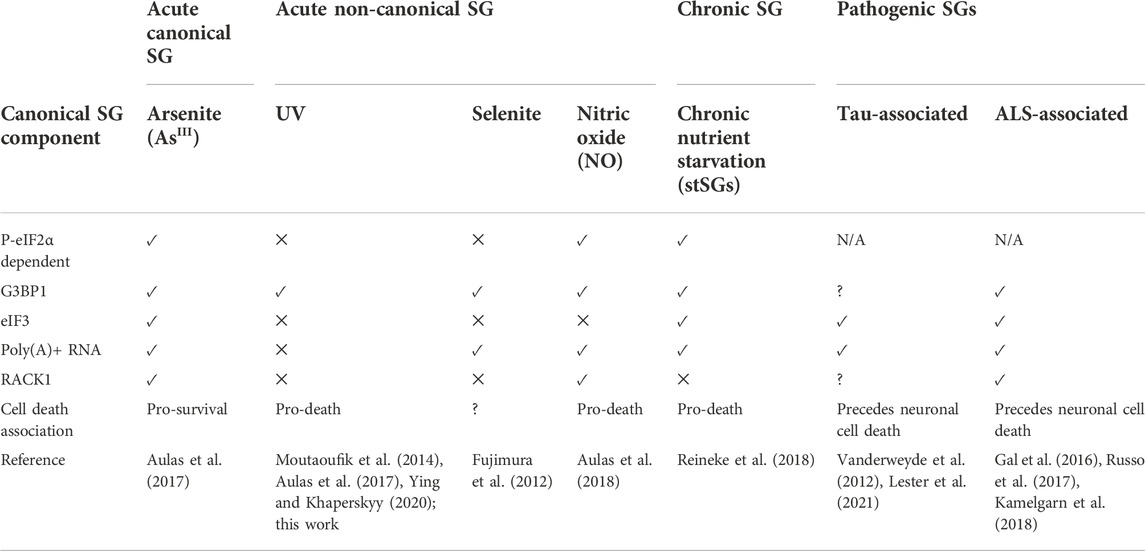
TABLE 1. A comparison of key features of canonical, non-canonical, chronic, and pathogenic SG subtypes.
While the composition of canonical SGs have been intensively catalogued through affinity labeling and biochemical purification studies (Jain et al., 2016; Markmiller et al., 2018; Youn et al., 2019; Marmor-Kollet et al., 2020; An et al., 2022), the functional consequences of SG formation remain unclear. One proposed function for SGs is the modulation of apoptosis12,13 (Arimoto et al., 2008; Arimoto-Matsuzaki et al., 2016; Park et al., 2020). In a now classic report, Arimoto et al. showed that SGs caused by overexpression of G3BP1 conferred resistance to GADD45-mediated apoptosis (Arimoto et al., 2008). They further demonstrated that hypoxia, which triggers SGs, protected cells from etoposide-mediated apoptosis. In both cases, they confirmed that sequestration of the signaling scaffold protein RACK1 at the SG was essential for protection from apoptosis (Arimoto et al., 2008). Other putative functions for SGs include mRNA triage (Kedersha et al., 2005) and stress-induced translational control (Dey et al., 2010); while SGs likely play a role, the idea that SG formation is absolutely essential or even largely responsible for such processes has largely been discredited (Mateju et al., 2020; Glauninger et al., 2022; Mateju and Chao, 2022).
SGs are dynamic. Canonical SGs rapidly exchange factors with the surrounding cytoplasm, though some factors exchange more rapidly than others (Kedersha et al., 2000). The extent to which factors move in and out of SGs is related to the structure of the granule, which appears to have a less dynamic core surrounded by a more dynamic shell (Wheeler et al., 2016). SGs that are more liquid-like also tend to be more dynamic, whereas those associated with gel or solid-like states (including some non-canonical and pathogenic subtypes, described below) are less dynamic (Kroschwald et al., 2015; Zhang et al., 2019).
The so-called “non-canonical” SG is a category used to collect SGs that do not mirror the formation, composition or function of canonical SGs as primarily understood from studies of arsenite and heat shock SGs (Advani and Ivanov, 2020). This SG subtype was originally dubbed non-canonical based on the lack of the canonical SG protein eukaryotic initiation factor 3 (eIF3) (Advani and Ivanov, 2020; Hofmann et al., 2021) in SGs triggered by sodium selenite (Fujimura et al., 2012), which suggested that the 48S preinitiation complex was not recruited to these SGs. Like canonical SGs, non-canonical SGs form acutely (in 4 h or less of stress), require G3BP1 or G3BP2 for their formation (Yang et al., 2020), and are associated with translational arrest, though they may or may not be associated with eIF2α phosphorylation depending upon the stress (Aulas et al., 2017; Advani and Ivanov, 2020; Hofmann et al., 2021). Non-canonical SGs are reported to form in response to many stresses including sodium selenite (Fujimura et al., 2012), nitric oxide (NO) (Aulas et al., 2018), the eIF4A inhibitor Rocaglamide A (RocA) (Aulas et al., 2017), hydrogen peroxide (Emara et al., 2012), and UV radiation (Kedersha et al., 1999; Pothof et al., 2009). There is broad variation within this non-canonical category as to which components do and do not localize at the SG, examples of which are described in Table 1. Others have attempted to further subdivide canonical and non-canonical SGs into designations including Types I, II, and III (Hofmann et al., 2021), however there is no consensus and some conflicting characterizations among these classifications (Advani and Ivanov, 2020; Hofmann et al., 2021), and thus we will address all acute non-canonical SG subtypes holistically in this Perspective.
Non-canonical SGs are reported to be functionally distinct from canonical SGs in that they are believed to be cytotoxic and less dynamic (Advani and Ivanov, 2020). However, there are few published studies that experimentally validate these characterizations. Relative to cell death, the most thoroughly characterized non-canonical SGs are those caused by nitric oxide (NO). NO SGs were determined to be triggering a non-apoptotic and potentially necrotic cell death by assaying a combination of propidium iodide and trypan blue staining, ATP levels and Caspase-3 cleavage (Aulas et al., 2018). For other non-canonical stresses like selenite, RocA, and UV, a decrease in cell viability was associated with SGs, but a mechanism of death was not elucidated (Fujimura et al., 2012; Aulas et al., 2018). Interestingly, while canonical arsenite-induced SGs are believed to be anti-apoptotic (Arimoto et al., 2008), the inverse relationship does not appear to be a universal feature of non-canonical SG subtypes, in that the lack of sequestration of pro-apoptotic factors like RACK1 at non-canonical SGs cannot simply be interpreted to mean that those pro-apoptotic factors are therefore active and inducing apoptotic cell death associated with non-canonical SG formation. Relative to their dynamics, the only report of which we are aware that directly measured dynamic behavior of a protein component of a non-canonical SG is in the context of NO-induced SGs (Aulas et al., 2018). G3BP1 recovery to photobleached NO SGs was approximately 10% lower than in arsenite SGs (Aulas et al., 2018). To our knowledge, this summarizes the extent to which the cytotoxic and dynamic properties of acute non-canonical SGs have been directly assessed in the literature.
The canonical and non-canonical labels are most frequently associated with acute SG formation. Chronic stresses and disease states are associated long-term SG formation. Chronic starvation induced SGs (stSGs) appear after prolonged (8–16 h) starvation of glucose, serum, glutamine and pyruvate (Reineke et al., 2018). These stSGs require both G3BP1 and eIF2α phosphorylation, and are in dynamic equilibrium with polysomes. StGSs appear to be pro-apoptotic, as assessed by Annexin V staining (Reineke et al., 2018), but the dynamic properties of these stSGs remain unknown. Acute metabolic stress, such as complete glucose starvation, glycolysis inhibitors (2-deoxy-D-glucose) and inhibitors of mitochondrial respiration and ATP synthesis (CCCP, oligomycin) are also known to cause stress granules (Kedersha et al., 2002; Wang et al., 2019; Amen and Kaganovich 2020). It is unclear whether or how prolonged metabolic stress relates to acute metabolic stress with respect to SG composition and function, though future investigation in this area could provide a better understanding of the evolution of SGs over prolonged stress conditions.
Pathological SGs (pSGs) are chronic SGs that form in diseased cells and are hypothesized to seed the accumulation of irreversible and toxic disease aggregates. pSGs have been most closely associated to date with ALS and Alzheimer’s Disease (Vanderweyde et al., 2012; Vanderweyde et al., 2016; Ash et al., 2014; Aulas and vande Velde, 2015; Apicco et al., 2018; Zhang et al., 2019; Marmor-Kollet et al., 2020; An et al., 2022). Several excellent reviews have been written on the subject (Wolozin, 2014; Aulas and vande Velde, 2015; Wolozin and Ivanov, 2019), including one within this Research Topic (Rhine et al., 2022). Interestingly, stSGs and pSGs can contain many of the components of canonical granules including poly(A)+ RNA, eIF3, and eIF4G (Table 1), though unlike canonical SGs they are clearly associated with cell death (Liu-Yesucevitz et al., 2010; Vanderweyde et al., 2012; Reineke et al., 2018). This observation raises the question of whether the categorization of SGs into canonical and non-canonical groups based primary on composition data will remain a useful proxy for the breadth of SG function as we continue to discover SGs in new contexts.
Within the past several years, several research groups have noted specific relationships between SGs and membranous organelles (Nicchitta 2022). SGs caused by activation of the unfolded protein response (UPR) can form directly on the endoplasmic reticulum (ER) and incorporate ER-associated mRNAs (Child et al., 2021), which are underrepresented in the transcriptome of SGs caused by arsenite (Khong et al., 2017). These ER-associated SGs contained G3BP1 and were inhibited by cycloheximide, suggesting they are bona fide SGs. SGs were also recently reported to associate with the ER in an autophagy-independent disassembly process that was specific for SGs caused by heat shock (Gwon et al., 2021). It is unknown whether SGs caused by other stresses can associate with the ER, however it has been observed that the ER acts as a facilitator of fission of both SGs and processing bodies (PBs) caused by arsenite stress (Lee et al., 2020). Relationships to other membranous organelles have also been noted. SG nucleation localized to the plasma membrane has been observed in yeast in response to starvation, and was linked to protein kinase C signaling under stress (Amen and Kaganovich 2020). Lysosomal damage was found to trigger SG formation, and a subset of those granules were physically associated with the damaged lysosome (Jia et al., 2022). Similarly, significant SG association with the mitochondrial membrane and with peroxisomes has been reported (Amen and Kaganovich 2021). The composition and function of these membrane-associated SGs remains unclear, but their observation adds an interesting dimension to our consideration of the role of SGs in cellular physiology.
UV SGs were first reported in 1999 by Kedersha et al. (1999) (as data not shown) as containing the SG marker proteins TIA-1, TIAR, and PABPC1. The first published evidence of UV-induced cytoplasmic aggregation was in 2005, when Teixeira et al. were investigating processing bodies [PBs, a cytoplasmic mRNP complex associated with mRNA decay (Luo et al., 2018)], in the budding yeast S. cerevisiae (Teixeira et al., 2005). Then in 2008, UV-induced poly(A)+ RNA-containing bodies were reported in S. cerevisiae by Gaillard and Aguilera (2008), but were not thought to be bona fide SGs due to the lack of co-localization of putative SG homologs. At that time however, SGs had not yet been discovered in S. cerevisiae, and thus core SG markers had not been established in that species.
Finally in 2009, Pothof et al. (2009) investigated the DNA damage response (DDR) to UV in HeLa cells, and discovered that the cytoplasmic condensates they observed in response to UV are SGs, based on the co-localization of TIA-1 and Ago2, and that UV SGs may be involved in microRNA-mediated silencing in response to DNA damage. Several more recent publications characterize UV SGs as non-canonical SGs lacking eIF3 and eIF4G (Table 1) (Moutaoufik et al., 2014; Aulas et al., 2017; Ying and Khaperskyy, 2020). We confirm here that UV SGs also lack the pro-apoptotic scaffolding protein RACK1, 4 h after UV treatment of U2OS human osteosarcoma cells (Figure 1A). Like other non-canonical subtypes, UV SGs are G3BP-dependent, eIF2α-independent, and cytotoxic (Pothof et al., 2009; Moutaoufik et al., 2014; Aulas et al., 2017; Ying and Khaperskyy, 2020). Surprisingly however, there are conflicting reports about whether UV SGs contain poly(A)+ RNA (Moutaoufik et al., 2014; Aulas et al., 2017).
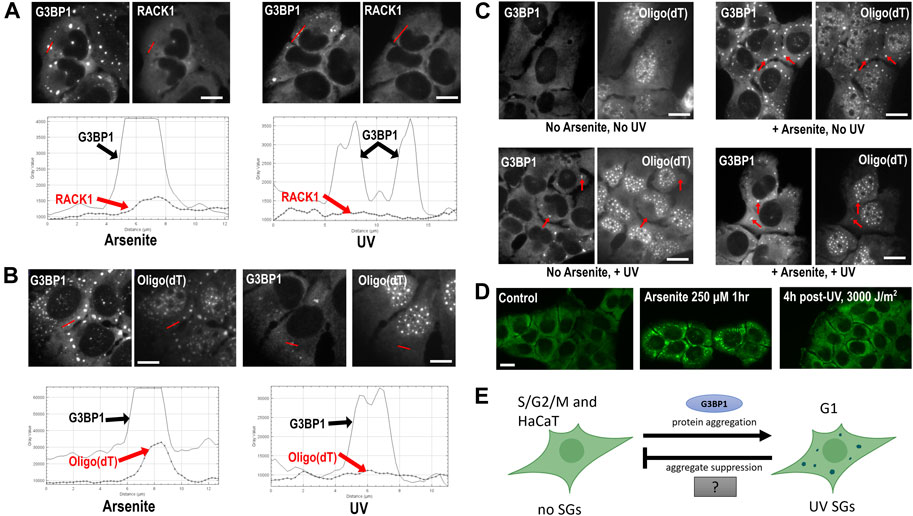
FIGURE 1. Composition and formation of UV SGs. (A) RACK1 does not localize to UV-induced SGs. U2OS treated with arsenite (500 μM, left panels) or UV (15 J/m2, then assayed at 4 h post-UV, right panels) and co-stained with antibodies to G3BP1 and RACK1. Profile intensity plots over the red line from each image were compiled using ImageJ. (B) U2OS treated with arsenite (500 μM, left panels) or UV (15 J/m2, then assayed at 4 h post-UV, right panels) and co-stained with antibodies to G3BP1 and Oligo (dT). Profile intensity plots over the red line from each image were compiled using ImageJ. (C) Poly(A)+ RNA SGs can form after UV treatment. U2OS were untreated or treated with UV (15 J/m2) for 3 h, then arsenite (500 µM) was added where indicated for a 1 h, then cells were fixed and co-stained with antibodies to G3BP1 and Oligo (dT) FISH. Red arrows point to prominent SGs in each treatment condition. (D) HaCaT cells do not form UV SGs. HaCaT cells were exposed to arsenite 250 µM for 1 h, or exposed to 3,000 J/m2 UV (4 h post-exposure), then stained with antibody for G3BP1 to assess SG formation. Detailed methods for all panels are available as Supplementary Material online. (E) A model for UV SG suppression. See Discussion section for details.
It was widely assumed in the early years of their discovery that all SGs contained mRNA. SGs are defined as mRNA-containing bodies or mRNP aggregates in many articles and reviews (Anderson and Kedersha, 2006; Riggs et al., 2020; Hofmann et al., 2021). In 2014, Moutaoufik et al. (2014) observed the colocalization of poly(A)+ RNA at UV SGs by co-localizing an oligo (dT) probe with the Fragile X Mental Retardation Protein (FMRP) as a SG marker in NIH-3T3 cells 18 h post-UV exposure. This report is, to our knowledge, the first published account of RNA localization to UV SGs in a mammalian cell type. However, a study published in 2017 by Aulas et al. (2017) found no poly(A)+ RNA localization at UV SGs, using G3BP1 as a SG marker in Hap1 human haploid cells, 2 h post-UV.
To independently validate the observation of poly(A)+ RNA at UV SGs, we treated U2OS cells with 15 J/m2 UVC, and used fluorescence in situ hybridization (FISH) to examine poly(A)+ RNA localization to UV SGs using G3BP1 as a SG marker. We find, as did Aulas et al. (2017), that poly(A)+ RNA is not enriched at UV SGs at 4 h post-UV exposure, as detectable by FISH (Figure 1B). It is tempting to speculate that perhaps UV light degrades cellular pools of mRNA, which could explain why poly(A)+ RNA FISH signal does not accumulate at UV SGs. However, the literature actually suggests that UV stabilizes and inhibits the degradation of mRNAs (Bollig et al., 2002; Gowrishankar et at., 2005; Gaillard and Aguilera, 2008) Click or tap here to enter text. mRNAs are sequestered in the nucleus upon UV treatment (Burgess et al., 2011) which reduces the cytoplasmic mRNA pool and may contribute to the loss of signal at UV SGs. PABP1 and PABP4 also relocalize to the nucleus, however the poly(A)+ RNA nuclear retention and PABP relocalization under UV stress appear to be independent (Burgess et al., 2011), which may support a model by which PABPs dissociate from their RNA targets under UV stress. Although it is reported UV can stabilize mRNAs, it is also known that UV induces reactive oxygen species and that oxidized RNAs can be targeted for degradation (Wurtmann and Wolin, 2009), meaning that some mRNA degradation may also be possible. Arsenite SGs have been shown to contain reactive oxygen species (ROS), which may further enhance RNA damage and subsequent degradation (Hu et al., 2021), however whether UV SGs contain ROS remains unknown.
To determine whether it was likely that mass degradation of mRNA was responsible for the lack of poly(A)+ RNA we observed at UV SGs, we treated U2OS with UV, then used arsenite to induce SGs at 3 h post-UV exposure. We observe that arsenite SGs induced in UV irradiated cells contain poly(A)+ RNA, though the intensity of localization is decreased compared to arsenite SGs in non-UV treated cells (Figure 1C). Therefore, we submit that mass degradation of mRNA cannot fully explain the lack of poly(A)+ RNA signal at UV SGs, as we show it is possible to assemble poly(A)+-containing SGs after UV treatment. We conclude that poly(A)+ RNAs are either not enriched or not detectable by FISH in UV SGs, though the mechanism of this phenomenonis unknown. As UV SGs are generally smaller than other canonical SGs, it may be the case that the RNA they contain cannot be detected above the cytoplasmic background using FISH. The results do not preclude the possibility that deadenylated mRNAs, non-coding mRNAs and/or small RNAs may still be present at UV SGs, and this possibility warrants further investigation. UV-induced damage may also affect the detection of poly(A)+ RNA using this method. If, however, it is the case that UV SGs are not RNA granules, it would mean that UV SGs do not play any role in the RNA-associated putative functions of SGs such as mRNA triage and translational control.
The finding that UV SGs are depleted of mRNA may also have implications for the assembly of SGs in general. Numerous reports have documented the role of RNA in driving LLPS during SG formation (van Treeck et al., 2018; Guillén-Boixet et al., 2020; Yang et al., 2020; Matheny et al., 2021). If indeed UV SGs do not contain mRNA, then one concludes that the interactions that drive UV SG assembly, unlike most other SG subtypes, must be primarily protein-driven. The lack of facilitation by RNA in LLPS of UV SG formation may also explain why UV SGs have been reported to be smaller and less numerous than other SG subtypes (Moutaoufik et al., 2014). Antiviral ribonucleases generally suppress SGs and have been used to demonstrate the importance of RNAs in SG formation (Burke et al., 2019; Burke et al., 2020); a similar approach could be applied in the case of UV SGs to further confirm or reject a role for RNA in UV SG assembly. Another reasonable prediction is that UV SGs may be more solid or gel-like than canonical or mRNA-containing SGs, as RNA is known to increase the liquid-like properties of biomolecular condensates (Guillén-Boixet et al., 2020; Yang et al., 2020; Bevilacqua et al., 2022). If indeed UV SG assembly is primarily protein-driven, then UV SGs may be a useful model for the specific study of essential protein-protein interactions within SGs. The dynamic properties of UV SGs have not yet been measured, but will no doubt be the topic of future investigation.
Because SGs have been implicated in seeding pathological protein aggregation (Wolozin and Ivanov, 2019), mechanisms of SG suppression are of intense interest. For example, SG induced by arsenite, a strong oxidizer, can be suppressed by strong antioxidants such as N-acetylcysteine (Szaflarski et al., 2016; Aulas et al., 2018) as well as by preconditioning that likely enhances cellular resilience to oxidative stress (Glass and Wente, 2019; Fay et al., 2021). SGs are also known to be suppressed in mitotic cells, because stalled translation elongation prohibits polysome disassembly in M phase (Gilad et al., 2007). While assessing the DNA damage response, Pothof et al. noted that UV-induced SGs did not form in cells that were positive for cyclin A, a marker of S phase (Pothof et al., 2009). By synchronizing their cells prior to irradiation, it was determined that UV SGs only appeared in cells that were irradiated in G2, passed through mitosis, and were in G1 phase at the assay timepoint. This observation of cell cycle dependence likely explains why only ∼10%–30% of cells in an asynchronous culture form UV SGs (Pothof et al., 2009; Aulas et al., 2017). The mechanism for the G1 specificity of UV SGs is unknown, however the ATM and ATR DNA damage checkpoint kinases were not required for UV SG formation (Pothof et al., 2009).
The observation of cell cycle dependence of UV SGs was confirmed in 2014 by Moutaoufik et al. (2014), who discovered that a dose of 10 J/m2 UV resulted in complete G1 arrest. These researchers used FACS profiling to assess the cell cycle, while concurrently assessing SG formation by immunofluorescence microscopy, and reported that G1 arrest correlated with the highest SG formation, and resumption of the cell cycle corresponded with SG clearance. Although this implies UV SGs may be dissolvable, it is also possible that the cells with UV SGs underwent cell death, leaving behind SG-free cells to resume the cell cycle. Thus, suppression of UV SGs in S and G2 phases is a unique feature of UV SGs.
Because skin is the organ most directly affected by UV, we decided to examine UV SG formation in an untransformed human keratinocyte cell line, HaCaT. To our great surprise, we were completely unable to induce UV SGs in HaCaT cells. We applied increasing amounts of UV, from our starting dose of 15 J/M2 up to 3,000 J/M2, and observed zero UV SG formation in HaCaTs by G3BP1 staining (Figure 1D). We do not yet know if this intriguing observation of UV SG suppression is a common property of all keratinocytes or specific to HaCaT, nor do we know whether it is related to the mechanisms of G2, S, and M phase suppression described above. We should note here that all studies of UV SGs reported to date, including the data reported herein, use UVC (254 nm) light sources. UVC is well known to cause damage to human tissues (Roy, 2017), but does not penetrate the Earth’s atmosphere. It remains unknown whether UVA (400–315 nm) and UVB (315–280 nm) rays can trigger UV SG formation in any cell type, including keratinocytes. However, UVA causes significantly more reactive oxygen species in human tissues than other UV wavelengths (Karran and Brem, 2016), which would cause an increase in cellular stress over other wavelengths.
The observation that UV SGs are RNA-depleted, protein-driven, and cell cycle dependent bodies makes them unique among known SG subtypes, and therefore they are fascinating research targets. Because UV SGs do not appear to contain much if any RNA (Figure 1B), we hypothesize that they primarily represent the effects of protein-protein interaction-driven condensation. In this way, UV SGs are a platform for studying the protein-specific drivers and suppressors of SG phase separation. The observations of SG suppression in S/G2/M phases, as well as in keratinocytes (Figure 1D), coupled with the apparent lack of poly(A)+ RNA (Figure 1B), informs our model for UV SG suppression (Figure 1E). In response to UV, which may cause nuclear retention and/or RNA-binding protein dissociation from RNA (Burgess et al., 2011), the protein drivers of SG formation including G3BP1/2 are just barely capable of surpassing a critical threshold and achieving protein-driven phase separation. In S/G2/M phases, and in certain UV-prone cell types such as keratinocytes, yet unknown cytoplasmic factors suppress phase separation by shifting the equilibrium to disfavor protein-driven condensation. This model is supported by the observation that G3BP1 condensation is significantly facilitated by the presence of RNA both in vitro and in vivo (Guillén-Boixet et al., 2020; Yang et al., 2020). Thus, in the absence of RNA, cytoplasmic factors may play a greater role in favoring or disfavoring biomolecular condensation in a way that would not be detectable if RNA were there to push the balance toward condensate formation. The “mystery SG suppressor” may not be a single gene, but a set of cytoplasmic environmental conditions associated with cell cycle progression. Cross-referencing proteomic, transcriptomic, and metabolomic differences between G1 and the rest of the cell cycle with similar data from keratinocytes is a starting point to identify the hidden factor(s) regulating UV SG suppression.
The unique nature of UV SGs may have revealed to us a path forward to identify suppressors of other protein aggregates. Many reviews have built powerful arguments for the relationship of SGs to protein aggregation diseases including ALS and AD (Li et al., 2013; Ash et al., 2014; Wolozin, 2014; Aulas and vande Velde, 2015; Wolozin and Ivanov, 2019). We suggest that the identification of UV SG suppressors, as modulators of a protein-driven condensation process, represent a new Frontier for research in neurodegeneration. If SGs can become pathological and can seed toxic aggregation of proteins like tau and TDP-34, then suppressors of SG formation may prevent or even reverse these seeds and thereby prevent disease aggregates from forming. It is also worth noting that the affected cells in neurodegenerative disease—neurons—are perpetually frozen in G0; UV SGs are repressed in S/G2/M phases. If we can understand the cellular conditions underlying cell cycle-dependent UV SG suppression, we could potentially apply this knowledge to create a G2-like state in neurons and thereby suppress pathological aggregates. In light of their potential to illuminate new paths for protein aggregation disease research, addressing these questions about an oft-neglected SG subtype takes on a fresh urgency.
Many key gaps in knowledge about non-canonical SGs in general, and UV SGs in particular, remain to be addressed. It is still entirely unclear how UV irradiation triggers SG formation in the first place. The lack of poly(A)+ RNA suggests that perhaps G3BP1, and other RNA-binding proteins like PABPs (Burgess et al., 2011) may dissociate from their mRNA targets in response to UV stress. But the pathways that would lead to such dissociation are unknown. Identifying the components of UV SGs is a key step to understanding these unusual SGs. Biochemical purification strategies and proximity labeling studies have enabled us to understand the proteomic and transcriptomic landscapes of canonical SGs (Jain et al., 2016; Khong et al., 2017; Markmiller et al., 2018; Namkoong et al., 2018; Padrón et al., 2019; Youn et al., 2019; Marmor-Kollet et al., 2020; Matheny et al., 2021; Vu et al., 2021; An et al., 2022). However, parallel studies on UV SGs, or any non-canonical subtype, remain to be performed. The dynamics of UV SGs have not been directly measured, and while the evidence presented here leads us to predict that UV SGs have a more solid or gel-like than liquid like behavior, for now their dynamic nature is unclear. It further has not been investigated whether UV SGs, like canonical subtypes, are also organized into core and shell subdomains, or whether their substructure is substantially distinct form known SGs. Finally, while we have not addressed the topic here, most canonical and non-canonical SGs are believed to be reversible and to dissolve upon the resolution of stress, facilitated by the autophagy pathway (Buchan et al., 2013). The mechanisms of UV SG dissolution, if they are in fact reversible, remain unknown.
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
NF conceived of the project. All authors participated in experimentation that led to the results reported in Figure 1. AC assembled Table 1. NF assembled Figure 1. AC and DC wrote the initial draft. NF edited the draft. All authors participated in the final preparation of the manuscript for publication. NF provided supervisory and financial support.
This research is supported by NIH R03AG077140 to NF, and new faculty start up funds from WPI to NF.
The authors thank Nancy Kedersha, Paul Anderson, and Pavel Ivanov (Brigham and Women’s Hospital, Boston, MA) for U2OS cells; Louis Roberts (WPI) for HaCaT cells; members of the Farny lab for helpful discussions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.1066650/full#supplementary-material
Advani, V. M., and Ivanov, P. (2020). Stress granule subtypes: An emerging link to neurodegeneration. Cell. Mol. Life Sci. 77, 4827–4845. doi:10.1007/s00018-020-03565-0
Advani, V. M., and Ivanov, P. (2019). Translational control under stress: Reshaping the translatome. BioEssays 41, 1900009. doi:10.1002/bies.201900009
Amen, T., and Kaganovich, D. (2021). Stress granules inhibit fatty acid oxidation by modulating mitochondrial permeability. Cell Rep. 35 (11), 109237. doi:10.1016/j.celrep.2021.109237
Amen, T., and Kaganovich, D. (2020). Stress granules sense metabolic stress at the plasma membrane and potentiate recovery by storing active Pkc1. Sci. Signal. 13 (623), eaaz6339. doi:10.1126/scisignal.aaz6339
An, H., Litscher, G., Watanabe, N., Wei, W., Hashimoto, T., Iwatsubo, T., et al. (2022). ALS-linked cytoplasmic FUS assemblies are compositionally different from physiological stress granules and sequester hnRNPA3, a novel modifier of FUS toxicity. Neurobiol. Dis. 162, 105585. doi:10.1016/j.nbd.2021.105585
Anderson, P., and Kedersha, N. (2006). RNA granules. J. Cell Biol. 172, 803–808. doi:10.1083/jcb.200512082
Apicco, D. J., Ash, P. E. A., Maziuk, B., LeBlang, C., Medalla, M., al Abdullatif, A., et al. (2018). Reducing the RNA binding protein TIA1 protects against tau-mediated neurodegeneration in vivo. Nat. Neurosci. 21, 72–80. doi:10.1038/s41593-017-0022-z
Arimoto, K., Fukuda, H., Imajoh-Ohmi, S., Saito, H., and Takekawa, M. (2008). Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell Biol. 10, 1324–1332. doi:10.1038/ncb1791
Arimoto-Matsuzaki, K., Saito, H., and Takekawa, M. (2016). TIA1 oxidation inhibits stress granule assembly and sensitizes cells to stress-induced apoptosis. Nat. Commun. 7, 10252. doi:10.1038/ncomms10252
Ash, P. E., Vanderweyde, T. E., Youmans, K. L., Apicco, D. J., and Wolozin, B. (2014). Pathological stress granules in Alzheimer’s disease. Brain Res. 1584, 52–58. doi:10.1016/j.brainres.2014.05.052
Aulas, A., Fay, M. M., Lyons, S. M., Achorn, C. A., Kedersha, N., Anderson, P., et al. (2017). Stress-specific differences in assembly and composition of stress granules and related foci. J. Cell Sci. 130, 927–937. doi:10.1242/jcs.199240
Aulas, A., Lyons, S. M., Fay, M. M., Anderson, P., and Ivanov, P. (2018). Nitric oxide triggers the assembly of “type II” stress granules linked to decreased cell viability. Cell Death Dis. 9, 1129. doi:10.1038/s41419-018-1173-x
Aulas, A., and vande Velde, C. (2015) Alterations in stress granule dynamics driven by TDP-43 and FUS: a link to pathological inclusions in ALS? Front. Cell. Neurosci. 9:423, doi:10.3389/fncel.2015.00423
Bevilacqua, P. C., Williams, A. M., Chou, H-L., and Assmann, S. M. (2022). RNA multimerization as an organizing force for liquid–liquid phase separation. RNA 28, 16–26. doi:10.1261/rna.078999.121
Bollig, F., Winzen, R., Kracht, M., Ghebremedhin, B., Ritter, B., Wilhelm, A., et al. (2002). Evidence for general stabilization of mRNAs in response to UV light. Eur. J. Biochem. 269 (23), 5830–5839. doi:10.1046/j.1432-1033.2002.03300.x
Boyd, J. D., Lee-Armandt, J. P., Feiler, M. S., Zaarur, N., Liu, M., Kraemer, B., et al. (2014). A high-content screen identifies novel compounds that inhibit stress-induced TDP-43 cellular aggregation and associated cytotoxicity. J. Biomol. Screen. 19, 44–56. doi:10.1177/1087057113501553
Buchan, J. R., Kolaitis, R-M., Taylor, J. P., and Parker, R. (2013). Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153, 1461–1474. doi:10.1016/j.cell.2013.05.037
Burgess, H. M., Richardson, W. A., Anderson, R. C., Salaun, C., Graham, S. V., and Gray, N. K. (2011). Nuclear relocalisation of cytoplasmic poly (A)-binding proteins PABP1 and PABP4 in response to UV irradiation reveals mRNA-dependent export of metazoan PABPs. J. Cell Sci. 124, 3344–3355. doi:10.1242/jcs.087692
Burke, J. M., Lester, E. T., Tauber, D., and Parker, R. (2020). RNase L promotes the formation of unique ribonucleoprotein granules distinct from stress granules. J. Biol. Chem. 295 (6), 1426–1438. doi:10.1074/jbc.RA119.011638
Burke, J. M., Moon, S. L., Matheny, T., and Parker, R. (2019). RNase L reprograms translation by widespread mRNA turnover escaped by antiviral mRNAs. Mol. Cell 1975 (6), 1203–1217. doi:10.1016/j.molcel.2019.07.029
Child, J. R., Chen, Q., Reid, D. W., Jagannathan, S., and Nicchitta, C. V. (2021). Recruitment of endoplasmic reticulum-targeted and cytosolic mRNAs into membrane-associated stress granules. RNA 27 (10), 1241–1256. doi:10.1261/rna.078858.121
Dey, S., Baird, T. D., Zhou, D., Palam, L. R., Spandau, D. F., and Wek, R. C. (2010). Both transcriptional regulation and translational control of ATF4 are central to the integrated stress response. J. Biol. Chem. 285, 33165–33174. doi:10.1074/jbc.M110.167213
Emara, M. M., Fujimura, K., Sciaranghella, D., Ivanova, V., Ivanov, P., and Anderson, P. (2012). Hydrogen peroxide induces stress granule formation independent of eIF2α phosphorylation. Biochem. Biophys. Res. Commun. 423, 763–769. doi:10.1016/j.bbrc.2012.06.033
Fay, M. M., Columbo, D., Cotter, C., Friend, C., Henry, S., Hoppe, M., et al. (2021). Bisphenol A promotes stress granule assembly and modulates the integrated stress response. Biol. Open 10, bio057539. doi:10.1242/bio.057539
Fujimura, K., Sasaki, A. T., and Anderson, P. (2012). Selenite targets eIF4E-binding protein-1 to inhibit translation initiation and induce the assembly of non-canonical stress granules. Nucleic Acids Res. 40, 8099–8110. doi:10.1093/nar/gks566
Gaillard, H., and Aguilera, A. (2008). A novel class of mRNA-containing cytoplasmic granules are produced in response to UV-irradiation. Mol. Biol. Cell 19, 4980–4992. doi:10.1091/mbc.e08-02-0193
Gal, J., Kuang, L., Barnett, K. R., Zhu, B. Z., Shissler, S. C., Korotkov, K. V., et al. (2016). ALS mutant SOD1 interacts with G3BP1 and affects stress granule dynamics. Acta Neuropathol. 132, 563–576. doi:10.1007/s00401-016-1601-x
Gilad, S., Nancy, K., and Orna, E-S. (2007). Ribosomal slowdown mediates translational arrest during cellular division. Mol. Cell Biol. 27, 6639–6646. doi:10.1128/MCB.00798-07
Glass, L., and Wente, S. R. (2019). Gle1 mediates stress granule-dependent survival during chemotoxic stress. Adv. Biol. Regul. 71, 156–171. doi:10.1016/j.jbior.2018.09.007
Glauninger, H., Wong Hickernell, C. J., Bard, J. A. M., and Drummond, D. A. (2022). Stressful steps: Progress and challenges in understanding stress-induced mRNA condensation and accumulation in stress granules. Mol. Cell 82, 2544–2556. doi:10.1016/j.molcel.2022.05.014
Gowrishankar, G., Winzen, R., Bollig, F., Ghebremedhin, B., Redich, N., Ritter, B., et al. (2005). Inhibition of mRNA deadenylation and degradation by ultraviolet light. Biol. Chem. 386 (12), 1287–1293. doi:10.1515/BC.2005.146
Guillén-Boixet, J., Kopach, A., Holehouse, A. S., Wittmann, S., Jahnel, M., Schlubler, R., et al. (2020). RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181, 346–361. doi:10.1016/j.cell.2020.03.049
Gwon, Y., Maxwell, B. A., Kolaitis, R. M., Zhang, P., Kim, H. J., and Taylor, J. P. (2021). Ubiquitination of G3BP1 mediates stress granule disassembly in a context-specific manner. Science 372 (6549), 346eabf6548–361. doi:10.1126/science.abf6548
Harding, H. P., Novoa, I., Zhang, Y., Zeng, H., Wek, R., Schapira, M., et al. (2000a). Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108. doi:10.1016/s1097-2765(00)00108-8
Harding, H. P., Zhang, Y., Bertolotti, A., Zeng, H., and Ron, D. (2000b). Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5, 897–904. doi:10.1016/s1097-2765(00)80330-5
Hofmann, S., Kedersha, N., Anderson, P., and Ivanov, P. (2021). Molecular mechanisms of stress granule assembly and disassembly. Biochimica Biophysica Acta Mol. Cell Res. 1868, 118876. doi:10.1016/j.bbamcr.2020.118876
Hu, K., Relton, E., Locker, N., Phan, N. T., and Ewing, A. G. (2021). Electrochemical measurements reveal reactive oxygen species in stress granules. Angew. Chem. Int. Ed. 60 (28), 15302–15306. doi:10.1002/anie.202104308
Jain, S., Wheeler, J. R., Walters, R. W., Agrawal, A., Barsic, A., and Parker, R. (2016). ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498. doi:10.1016/j.cell.2015.12.038
Jia, J., Wang, F., Bhujabal, Z., Peters, R., Mudd, M., Duque, T., et al. (2022). Stress granules and mTOR are regulated by membrane atg8ylation during lysosomal damage. J. Cell Biol. 221 (11), e202207091. doi:10.1083/jcb.202207091
Kamelgarn, M., Chen, J., Kuang, L., Jin, H., Kasarskis, E. J., and Zhu, H. (2018). ALS mutations of FUS suppress protein translation and disrupt the regulation of nonsense-mediated decay. Proc. Natl. Acad. Sci. U. S. A. 115, E11904–E11913. doi:10.1073/pnas.1810413115
Karran, P., and Brem, R. (2016). Protein oxidation, UVA and human DNA repair. DNA Repair (Amst) 44, 178–185. doi:10.1016/j.dnarep.2016.05.024
Kedersha, N., Chen, S., Gilks, N., Li, W., Miller, I. J., Stahl, J., et al. (2002). Evidence that ternary complex (eIF2-GTP-tRNA i met )-Deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell 13, 195–210. doi:10.1091/mbc.01-05-0221
Kedersha, N., Cho, M. R., Li, W., Yacono, P. W., Chen, S., Gilks, N., et al. (2000). Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151, 1257–1268. doi:10.1083/jcb.151.6.1257
Kedersha, N., Panas, M. D., Achorn, C. A., Lyons, S., Tisdale, S., Hickman, T., et al. (2016). G3BP–Caprin1–USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 212, 845–860. doi:10.1083/jcb.201508028
Kedersha, N., Stoecklin, G., Ayodele, M., Yacono, P., Lykke-Andersen, J., Fritzler, M. J., et al. (2005). Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871–884. doi:10.1083/jcb.200502088
Kedersha, N. L., Gupta, M., Li, W., Miller, I., and Anderson, P. (1999). RNA-binding proteins tia-1 and tiar link the phosphorylation of eif-2α to the assembly of mammalian stress granules. J. Cell Biol. 147, 1431–1442. doi:10.1083/jcb.147.7.1431
Khong, A., Matheny, T., Jain, S., Mitchell, S. F., Wheeler, J. R., and Parker, R. (2017). The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol. Cell 68, 808–820. e5. doi:10.1016/j.molcel.2017.10.015
Kroschwald, S., Maharana, S., Mateju, D., Malinovska, L., Nüske, E., Poser, I., et al. (2015). Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. Elife 4, e06807. doi:10.7554/eLife.06807
Lee, J. E., Cathey, P. I., Wu, H., Parker, R., and Voeltz, G. K. (2020). Endoplasmic reticulum contact sites regulate the dynamics of membraneless organelles. Science 31 (6477), 367eaay7108. doi:10.1126/science.aay7108
Lester, E., Ooi, F. K., Bakkar, N., Ayers, J., Woerman, A. L., Wheeler, J., et al. (2021). Tau aggregates are RNA-protein assemblies that mislocalize multiple nuclear speckle components. Neuron 109, 1675–1691. e9. doi:10.1016/j.neuron.2021.03.026
Li, Y. R., King, O. D., Shorter, J., and Gitler, A. D. (2013). Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 201, 361–372. doi:10.1083/jcb.201302044
Liu-Yesucevitz, L., Bilgutay, A., Zhang, Y-J., Vanderweyde, T., Citro, A., Mehta, T., et al. (2010). Tar DNA binding protein-43 (TDP-43) associates with stress granules: Analysis of cultured cells and pathological brain tissue. PLoS One 5, e13250. doi:10.1371/journal.pone.0013250
Luo, Y., Na, Z., and Slavoff, S. A. (2018). P-Bodies: Composition, properties, and functions. Biochemistry 57, 2424–2431. doi:10.1021/acs.biochem.7b01162
Markmiller, S., Soltanieh, S., Server, K. L., Mak, R., Jin, W., Fang, M. Y., et al. (2018). Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172, 590–604. doi:10.1016/j.cell.2017.12.032
Marmor-Kollet, H., Siany, A., Kedersha, N., Knafo, N., Rivkin, N., Danino, Y. M., et al. (2020). Spatiotemporal proteomic analysis of stress granule disassembly using APEX reveals regulation by SUMOylation and links to ALS pathogenesis. Mol. Cell 80, 876–891. doi:10.1016/j.molcel.2020.10.032
Mateju, D., and Chao, J. A. (2022). Stress granules: Regulators or by-products? FEBS J. 289, 363–373. doi:10.1111/febs.15821
Mateju, D., Eichenberger, B., Voigt, F., Eglinger, J., Roth, G., and Chao, J. A. (2020). Single-molecule imaging reveals translation of mRNAs localized to stress granules. Cell 183, 1801–1812. doi:10.1016/j.cell.2020.11.010
Matheny, T., van Treeck, B., Huynh, T. N., and Parker, R. (2021). RNA partitioning into stress granules is based on the summation of multiple interactions. RNA 27, 174–189. doi:10.1261/rna.078204.120
McEwen, E., Kedersha, N., Song, B., Scheuner, D., Gilks, N., Han, A., et al. (2005). Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 280, 16925–16933. doi:10.1074/jbc.M412882200
Moutaoufik, M. T., el Fatimy, R., Nassour, H., Gareau, C., Lang, J., Tanguay, R. M., et al. (2014). UVC-induced stress granules in mammalian cells. PLoS One 9, e112742. doi:10.1371/journal.pone.0112742
Namkoong, S., Ho, A., Woo, Y. M., Kwak, H., and Lee, J. H. (2018). Systematic characterization of stress-induced RNA granulation. Mol. Cell 70, 175–187. doi:10.1016/j.molcel.2018.02.025
Nicchitta, C. V. (2022). An emerging role for the endoplasmic reticulum in stress granule biogenesis, Semin. Cell Dev. Biol. S1084-9521(22)00275-0. doi:10.1016/j.semcdb.2022.09.013
Nover, L., Scharf, K-D., and Neumann, D. (1989). Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol. Cell. Biol. 9, 1298–1308. doi:10.1128/mcb.9.3.1298
Padrón, A., Iwasaki, S., and Ingolia, N. T. (2019). Proximity RNA labeling by APEX-seq reveals the organization of translation initiation complexes and repressive RNA granules. Mol. Cell 75, 875–887. e5. doi:10.1016/j.molcel.2019.07.030
Park, Y-J., Choi, D. W., Cho, S. W., Han, J., Yang, S., and Choi, C. Y. (2020). Stress granule formation attenuates RACK1-mediated apoptotic cell death induced by morusin. Int. J. Mol. Sci. 21, 5360. doi:10.3390/ijms21155360
Pothof, J., Verkaik, N. S., Hoeijmakers, J. H. J., and van Gent, D. C. (2009). MicroRNA responses and stress granule formation modulate the DNA damage response. Cell Cycle 8, 3462–3468. doi:10.4161/cc.8.21.9835
Proud, C. G. (2019). Phosphorylation and signal transduction pathways in translational control. Cold Spring Harb. Perspect. Biol. 11, a033050. doi:10.1101/cshperspect.a033050
Reineke, L. C., Cheema, S. A., Dubrulle, J., and Neilson, J. R. (2018). Chronic starvation induces noncanonical pro-death stress granules. J. Cell Sci. 131, jcs220244. doi:10.1242/jcs.220244
Rhine, K., Al-Azzam, N., Yu, T., and Yeo, G. W. (2022). Aging RNA granule dynamics in neurodegeneration. Front. Mol. Biosci. 9, 991641. doi:10.3389/fmolb.2022.991641
Riggs, C. L., Kedersha, N., Ivanov, P., and Anderson, P. (2020). Mammalian stress granules and P bodies at a glance. J. Cell Sci. 133, jcs242487. doi:10.1242/jcs.242487
Roy, S. (2017). “Impact of UV radiation on genome stability and human health,” in Ultraviolet light in human health, diseases and environment. Editors S. I. Ahmad (Cham: Springer International Publishing), 207–219.
Russo, A., Scardigli, R., la Regina, F., Murray, M. E., Romano, N., Dickson, D. W., et al. (2017). Increased cytoplasmic TDP-43 reduces global protein synthesis by interacting with RACK1 on polyribosomes. Hum. Mol. Genet. 26, 1407–1418. doi:10.1093/hmg/ddx035
Sanders, D. W., Kedersha, N., Lee, D. S. W., Strom, A. R., Drake, V., Riback, J. A., et al. (2020). Competing protein-RNA interaction Networks control multiphase intracellular organization. Cell 181, 306–324. e28. doi:10.1016/j.cell.2020.03.050
Srivastava, S. P., Kumar, K. U., and Kaufman, R. J. (1998). Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J. Biol. Chem. 273, 2416–2423. doi:10.1074/jbc.273.4.2416
Szaflarski, W., Fay, M. M., Kedersha, N., Zabel, M., Anderson, P., and Ivanov, P. (2016). Vinca alkaloid drugs promote stress-induced translational repression and stress granule formation. Oncotarget 7, 30307–30322. doi:10.18632/oncotarget.8728
Teixeira, D., Sheth, U., Valencia-Sanchez, M. A., Brengues, M., and Parker, R. (2005). Processing bodies require RNA for assembly and contain nontranslating mRNAs. Rna 11, 371–382. doi:10.1261/rna.7258505
Tourrière, H., Chebli, K., Zekri, L., Courselaud, B., Blanchard, J. M., Bertrand, E., et al. (2003). The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160, 823–831. doi:10.1083/jcb.200212128
van Treeck, B., Protter, D. S. W., Matheny, T., Khong, A., Link, C. D., and Parker, R. (2018). RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc. Natl. Acad. Sci. U. S. A. 115, 2734–2739. doi:10.1073/pnas.1800038115
Vanderweyde, T., Apicco, D. J., Youmans-Kidder, K., Ash, P. E. A., Cook, C., da Rocha, E. L., et al. (2016). Interaction of tau with the RNA-binding protein TIA1 regulates tau pathophysiology and toxicity. Cell Rep. 15, 1455–1466. doi:10.1016/j.celrep.2016.04.045
Vanderweyde, T., Yu, H., Varnum, M., Liu-Yesucevitz, L., Citro, A., Ikezu, T., et al. (2012). Contrasting pathology of the stress granule proteins TIA-1 and G3BP in tauopathies. J. Neurosci. 32, 8270–8283. doi:10.1523/JNEUROSCI.1592-12.2012
Vu, L., Ghosh, A., Tran, C., Tebung, W. A., Sidibé, H., Garcia-Mansfield, K., et al. (2021). Defining the caprin-1 interactome in unstressed and stressed conditions. J. Proteome Res. 20, 3165–3178. doi:10.1021/acs.jproteome.1c00016
Wang, S., Kwon, S. H., Su, Y., and Dong, Z. (2019). Stress granules are formed in renal proximal tubular cells during metabolic stress and ischemic injury for cell survival. Am. J. Physiol. Ren. Physiol. 317 (1), F116–F123. doi:10.1152/ajprenal.00139.2019
Wek, R. C. (2018). Role of eIF2α kinases in translational control and adaptation to cellular stress. Cold Spring Harb. Perspect. Biol. 10, a032870. doi:10.1101/cshperspect.a032870
Wek, S. A., Zhu, S., and Wek, R. C. (1995). The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell. Biol. 15, 4497–4506. doi:10.1128/mcb.15.8.4497
Wheeler, J. R., Matheny, T., Jain, S., Abrisch, R., and Parker, R. (2016). Distinct stages in stress granule assembly and disassembly. Elife 5, e18413. doi:10.7554/eLife.18413
Wolozin, B., and Ivanov, P. (2019). Stress granules and neurodegeneration. Nat. Rev. Neurosci. 20, 649–666. doi:10.1038/s41583-019-0222-5
Wolozin, B. (2014). Physiological protein aggregation run amuck: Stress granules and the Genesis of neurodegenerative disease. Discov. Med. 17, 47–52.
Wurtmann, E., and Wolin, S. (2009). RNA under attack: Cellular handling of RNA damage. Crit. Rev. Biochem. Mol. Biol. 44 (1), 34–49. doi:10.1080/10409230802594043
Yang, P., Mathieu, C., Kolaitis, R. M., Zhang, P., Messing, J., Yurtsever, U., et al. (2020). G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181, 325–345. e28. doi:10.1016/j.cell.2020.03.046
Ying, S., and Khaperskyy, D. A. (2020). UV damage induces G3BP1-dependent stress granule formation that is not driven by mTOR inhibition-mediated translation arrest. J. Cell Sci. 133, jcs248310. doi:10.1242/jcs.248310
Youn, J. Y., Dyakov, B. J. A., Zhang, J., Knight, J. D. R., Vernon, R. M., Forman-Kay, J. D., et al. (2019). Properties of stress granule and P-body proteomes. Mol. Cell 76, 286–294. doi:10.1016/j.molcel.2019.09.014
Keywords: stress granules, ultraviolet radiation (UV), biomolecular condensation, poly(A)+ RNA, cell cycle, neurodegeneration
Citation: Cabral AJ, Costello DC and Farny NG (2022) The enigma of ultraviolet radiation stress granules: Research challenges and new perspectives. Front. Mol. Biosci. 9:1066650. doi: 10.3389/fmolb.2022.1066650
Received: 11 October 2022; Accepted: 17 November 2022;
Published: 01 December 2022.
Edited by:
Matthew Brook, University of Edinburgh, United KingdomReviewed by:
Hannah Burgess, University of Surrey, United KingdomCopyright © 2022 Cabral, Costello and Farny. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Natalie G. Farny, bmZhcm55QHdwaS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.