- 1Department of Functional Materials, Institute of Physics, Czech Academy of Sciences, Prague, Czechia
- 2Department of Neurology and Centre of Clinical Neuroscience, First Faculty of Medicine and General University Hospital in Prague, Charles University, Prague, Czechia
During past decades, enormous progress in understanding the mechanisms of the intermolecular interactions between the protein and surface at the single-molecule level has been achieved. These advances could only be possible by the ongoing development of highly sophisticated experimental methods such as atomic force microscopy, optical microscopy, surface plasmon resonance, ellipsometry, quartz crystal microbalance, conventional mass spectrometry, and, more recently, the nanomechanical systems. Here, we highlight the main findings of recent studies on the label-free single-molecule (protein) detection by nanomechanical systems including those focusing on the protein adsorption on various substrate surfaces. Since the nanomechanical techniques are capable of detecting and manipulating proteins even at the single-molecule level, therefore, they are expected to open a new way of studying the dynamics of protein functions. It is noteworthy that, in contrast to other experimental methods, where only given protein properties like molecular weight or protein stiffness can be determined, the nanomechanical systems enable a real-time measurement of the multiple protein properties (e.g., mass, stiffness, and/or generated surface stress), making them suitable for the study of protein adsorption mechanisms. Moreover, we also discuss the possible future trends in label-free detection and analysis of dynamics of protein complexes with these nanomechanical systems.
1 Introduction
Proteins play a critical role in all living organisms. For example, they are responsible for intercellular transport and cell communication, or they also provide cells a given shape and mechanical resistance to deformation and contribute to DNA synthesis/replication (Satzer et al., 2015; Wingert et al., 2021). Moreover, many biological processes such as protein aggregation, transmembrane signaling, inflammation, or even the blood coagulation cascade are initiated by the adsorption/interaction of proteins with various (bio-) material surfaces (Deng et al., 2021). Hence, the understanding of protein adsorption mechanisms is of great importance to not only the fundamental science but also the majority of biomedical applications including orthopedic implants, stents, or ultrasensitive diagnostic tools (Talha et al., 2019). Up to date, many experimental methods to analyze protein adsorption have been developed (Deng et al., 2021). However, most of these methods either provide only the statistically averaged information on the single-protein adsorption (e.g., quartz crystal microbalance method), need a sophisticated experimental setup (e.g., X-ray photoelectron spectrometry), or require the knowledge of protein properties a priori own measurement (e.g., for surface plasmon resonance) (Kubiak-Ossowska et al., 2019). In contrast, mass spectrometers (MS) offer a high degree of sensitivity and selectivity; therefore, they have become a fundamental experimental tool used for the simultaneous identification and analysis of a larger number of proteins, often referred to as proteomics (Aebersold and Mann, 2003). MS utilize the ionization of the sample of interest which is then brought to the gas phase and subsequently analyzed by comparing the mass of individual ions obtained from the observed mass-to-charge ratio with the available protein database as also shown in Figure 1A (Johnson et al., 2020). Unfortunately, the necessity of performing destructive ionization of the investigated sample and especially for the large protein molecules to link the observed complex pattern with the relevant protein(s) is a formidable challenge that restricts the application of the conventional MS to only a relatively light protein molecules; that is, MS is limited to samples with mass ranging from Da to kDa (see Figure 1A) (Angel et al., 2012).
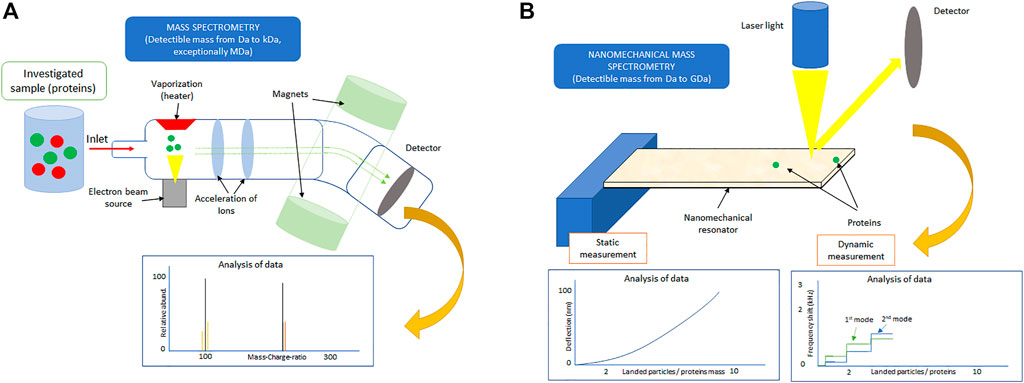
FIGURE 1. Sketch of (A) the conventional mass spectrometry and (B) nanomechanical mass spectrometry with commonly detectable analyte (protein) mass.
More than a decade ago, alternative methods to conventional MS that do not require ionization of the investigated sample were proposed (Eom et al., 2011; Sage et al., 2015). These methods identify studied protein(s) indirectly based on the observed changes in the dynamic and/or static response of the nanomechanical resonators (NRs) (Boisen et al., 2011; Hanay et al., 2012; Stachiv et al., 2012). We remind the reader that the static mode measurement is usually used to identify proteins that form a thin-layer film on the surface of a nanomechanical sensor (Stachiv et al., 2014), while the dynamic mode enables the identification of a single protein adsorbed in an arbitrary position on the NR (Naik et al., 2009). Because of its superior mass resolution (Jensen et al., 2008), the dynamic mode, that is, detection of the resonant frequency shift(s), is preferable in nanomechanical mass spectrometry. The commonly achievable mass sensitivities of static and dynamic mode measurement methods are illustrated in Figure 1B. The simplest and also the most often used dynamic mode measurement method models the protein molecule adsorbed on the surface of NR as a point particle (Dohn et al., 2007). Then, by measuring frequency shift in at least three (two) consecutive resonant frequencies of the cantilever (suspended) NR, the “protein” mass and its position of attachment can be determined (Stachiv et al., 2012). A very good agreement between theory and experiments enabled to confirm the validity of this multimode frequency shift measurement method (Schmid et al., 2010; Hanay et al., 2012; Sage et al., 2018; Ma et al., 2022). Nevertheless, this “simplest” method does not provide a sufficient amount of information needed to gain insights into the mechanisms of the protein adsorption on the surface of NR; that is, this method is primarily developed for nanoparticle mass sensing (Stachiv et al., 2012; Sage et al., 2018; Ma et al., 2022). Hence, there is an ongoing dispute on the application limits of the NRs, especially the multimode method, in the identification and analysis of a single or multiple protein(s) adsorbed on the resonator surface (Tamayo et al., 2013). Interestingly, recent studies have shown that NRs hold promise in the identification of even the larger masses (>GDa) (Dominguez-Medina et al., 2018; Stachiv and Gan., 2019; Stachiv et al., 2020; Erdogan et al., 2022). Moreover, there is a consensus among researchers that in order to gain more necessary information on the protein adsorption beyond its mass and position of attachment, a symbiosis of experiments with advanced theoretical modeling would be needed (Malvar et al., 2016; Stassi et al., 2019). In this study, we discuss the current findings, future trends, and perspectives in research and development of NRs for the identification of protein molecules beyond the simple molecular weighting. We also note that a systematic review of articles relevant to the considered topic of the present study was performed using the main electronic databases via Scopus, Google Scholar, and PubMed, which account for the majority of the journals focusing on the label-free detection and analysis of protein adsorption.
2 Nanomechanical mass spectrometry: From Da to GDa masses
The earlier studies on NR-based mass spectrometry have aimed to reach sensitivity comparable to that of commercial MS. We remind the reader that the achievable sensitivity of the NR depends on the resonant frequencies; that is, the higher operating frequencies of the NR yield the higher mass sensitivity (Ekinci et al., 2004; Gruber et al., 2019). To date, the most promising strategies to achieve the ultra-high operating resonant frequencies and, correspondingly, to enhance the mass sensitivity include the miniaturization of NRs (Yang et al., 2006; Xu et al., 2022), detection of the higher vibrational modes (Dohn et al., 2005; Ghatkesar et al., 2007) or, more recently, the non-linear oscillations (Yuksel et al., 2019; Lyu et al., 2020). It is noteworthy that during the past decade, the superb, that is, a Dalton, mass sensitivity has been experimentally demonstrated by using the one-dimensional carbon nanotube-based NR (Chaste et al., 2012), while in the same period of time the mass spectrometry community were trying to improve the capability of the conventional MS for measurement of larger (>MDa) masses including the protein complexes (Snijder et al., 2013). Hence, nowadays, the research attention of the nanotechnology community has also shifted toward the identification of larger masses, that is, in the GDa range, a goal that cannot still be reached by the conventional MS (Keifer et al., 2017).
Unfortunately, the unprecedented mass sensitivity of NRs has been confirmed for a large number of various nanoparticles (Yang et al., 2006; Jensen et al., 2008; Chaste et al., 2012), but for the protein molecules adsorbed on the resonator surface, the obtained results were inconsistent. Some studies showed that the point-mass approximation yields the accurately estimated masses (Naik et al., 2009; Hanay et al., 2012), whereas others revealed that this approximation fails to correctly determine the molecular mass (Gupta et al., 2006; Malvar et al., 2016). As a result, many theoretical models have already been developed to explain the effect of the bio-molecule adsorption on the NR frequency response (Dorignac et al., 2006; Tamayo et al., 2006; Lachut and Sader, 2007; Naik et al., 2009; Yi and Duan, 2009; Karabalin et al., 2012; Stachiv 2014). The majority of earlier studies suggested that during the molecule adsorption, the surface stress is generated causing changes in the resonant frequencies of NR, and, consequently, it can also result in the incorrectly estimated molecular mass (Dorignac et al., 2006; Yi and Duan, 2009). However, more recently, the surface stress effect has been proven to be insufficient to explain the inconsistency in the experimentally determined molecular masses (Karabalin et al., 2012; Lachut and Sader, 2012; Stachiv, 2014).
A different approach to gaining an understanding of the impact of biomolecules on the resonant frequencies was proposed by Tamayo et al. (2006). In their model, the adsorbate molecule was modeled as a cuboid with a given mass and stiffness. Then, based on the obtained computational results, they suggested that for larger masses or ultrathin resonators, where the adsorbed molecule and resonator thicknesses are of the same order, the molecule stiffness must be accounted when analyzing experimental data; that is, the point-mass approximation can result in the incorrectly identified molecular mass (Ruz et al., 2014). The validity of their model for mass sensing of larger molecules has been reinforced by comparing predictions with experiments carried out on gold nanoparticles and E. coli bacteria cells adsorbed on the cantilever-based NR (Malvar et al., 2016). It is noteworthy that the correctness of the point-mass approximation in MS of larger masses with arrays of NRs, each of distinct resonant frequencies obtained by altering the resonators’ dimensions, has also been successfully demonstrated (Sage et al., 2018). Whether the point-mass approximation or Tamayo’s model or even both models enable the accurate identification of larger masses was under extensive debate. Surprisingly, the application limits of both models have only been recently revealed (Stachiv et al., 2022a). In this work, they found that there exist three different sensing regimes, namely, ultra-light, light, and heavy, depending on the molecule-to-resonator mass ratio, for which the effect of the molecule properties (mass and stiffness) on the resonant frequency can be easily separated. Interestingly, they showed that the molecule stiffness plays an important role for NR in the ultra-light regime, that is, with the mass ratio <∼10−3, while for the light regime the point-mass approximation is always accurate, and, finally, for the heavy regime, that is, with the mass ratio >2 10−2, the molecule mass has a strong impact on the vibrational characteristic of NR, making the point-mass approximation inaccurate. The validity of these theoretical findings has been confirmed by comparing model predictions with the recent experimental results, and, as such, they can help to explain the observed inconsistencies in the molecule mass sensing. For instance, in the study of Malvar et al. (2016), the NR operates in the ultra-light regime, and correspondingly, the stiffness effect must be included when analyzing experimental data. Similarly, for Sage et al. (2018), the considered NRs operate in the light regime, enabling an accurate identification of the molecule mass even without accounting for its stiffness. In addition, the easily accessible procedure of mass sensing by NR in the heavy regime has recently been proposed (Stachiv et al., 2022b). Main findings of the aforementioned discussed key studies on mass spectrometry by the NR are, for the reader convenience, summarized in Table 1.
TABLE 1. Main findings of important experimental/theoretical studies focusing on the NR-based mass spectrometry in chronological order.
We emphasize here that NR-based spectrometry proteomics is still not fully developed. Nevertheless, it can be expected that with the recent findings and continuous fast progress in NR-based spectrometry the full potential of these devices in proteomics will be achieved in a near future. We foresee that to fulfill a full potential of the nanomechanical systems in MS would require a significant simplification of the experimental apparatus, that is, these devices are still difficult to be fabricated and operated (Sansa et al., 2020), and development of the advanced computational models (Rocha et al., 2021; Xia et al., 2021) and approaches to nanomechanical mass sensing in also gaseous or aqueous solution (Urbanska et al., 2020). Furthermore, it is important to note that, as it was demonstrated by, for instance, Karabalin et al. (2012), Malvar et al. (2016), and Sader et al. (2018), the NR can already provide an additional information on the molecule adsorption than just the simple mass; that is, NR can be used to detect the generated surface stress and/or the molecule stiffness.
3 What other properties can be determined beyond the molecular weight?
It is a well-known fact that protein stiffness plays a crucial role in a large number of processes within living organisms (Senese et al., 2018; Li et al., 2021). For example, protein stiffness is responsible for the catalytic efficiency of many enzymes (Richard, 2019), and, additionally, even some mutation processes can be associated with relatively small changes in protein stiffness (Pfeifer et al., 2017). The mechanical stress contributes to the folding and unfolding of proteins, which is important in the modulation of a variety of cellular functions such as cell replication and metabolisms or ion transport (Eisner et al., 2018). Measurements of both the mechanical stress and protein stiffness are usually performed in vitro using either a combination of several experimental apparatus such as optical and/or magnetic tweezers combined with the atomic force microscope (Yang et al., 2020) or a highly sophisticated devices including the centrifuge force microscope (Yang et al., 2016), unconfined force testing machines (Su et al., 2019) or in situ TEM/SEM nanoindentation (Shen et al., 2020).
Nanomechanical sensors can detect numerous physical quantities such as mass, temperature, pressure, or force; therefore, they have been considered a suitable candidate for measurement of the mechanical properties and stress of the adsorbate protein molecule (a detailed discussion on nanomechanical sensors can be found in the review of Ruz et al. (2021)). The biomolecule adsorption-induced surface stress is usually measured using the static mode of the NRs, that is, by using the optical (Kidane et al., 2020) or resistive (Minami et al., 2021) measurement methods. Note that prior measurements of the functionalization of the surface of NR are necessary to immobilize the molecule of interest on the resonator surface (Zhang et al., 2018; Zhao et al., 2019). Once the molecule is adsorbed on the functionalized resonator surface, it generates surface stress, which, in turn, causes deformation of the NR. Hence, by detecting changes in the deflection of NR, the surface stress can easily be estimated (Boisen et al., 2011; Minami and Yoshikawa, 2021). Importantly, a theoretical model, which accounts for the molecule and effects of functional film viscoelastic properties on the adsorption-induced stress and/or strain, has only recently been developed (Minami et al., 2021). It is expected that this model will help researchers to gain insights into the mechanisms of the biomolecule adsorption-induced stress, and, correspondingly, the cellular functions could be studied.
Gil-Santos et al. (2010), in their pioneering work, demonstrated the extraordinary capability of carbon nanotube-based NR in mass sensing and stiffness spectrometry. They showed that by monitoring the nanotube vibrations in two directions, the adsorbate mass and its stiffness can simultaneously be determined with high accuracy, even for the protein stiffness, that is, the error in stiffness is within .1%. As mentioned previously, the first mass and stiffness spectrometry on larger molecules by means of NRs was realized in the study of Malvar et al. (2016). More recently, the same group showed that the ultra-high operating disk NRs may be used to retrieve other information on the protein, including the molecule hydration (Gil-Santos et al., 2020). The main drawback of these methods is the requirement of solving the complex inverse problem while taking into account the non-trivial molecule–resonator surface binding effects, molecule size, and stiffness (Malvar et al., 2016; Gil-Santos et al., 2020). Another limitation of this NR is the limited capturing and sensing area. This problem has already been solved by using an array of the high-frequency operating NRs (Sage et al., 2018; Lamberti et al., 2022). Gil-Santos et al. (2020) also showed that the resonant frequencies of the NR must be close to those of the investigated analyte (molecule). Only in this case, the analyte vibrational properties can be estimated. It is evident from the present discussion that this measurement would require the NRs with a tunable spectrum of the resonant frequencies (Liu et al., 2015). Frequency modulation by means of the piezoelectric effect (Kozinsky et al., 2006) or electrostatic actuation (Zhang et al., 2015) has been developed for frequency tuning of resonators.
Alternatively, the nanomechanical resonators with the NiTi shape memory alloy-based thin films have been shown having an extraordinary frequency tunability (Stachiv et al., 2017). This superb tunability is achieved by the diffusionless martensitic transformation in NiTi, while the high operating frequencies are due to the elastic substrate. It has also been demonstrated that NiTi-based NR can be used to measure mass, stiffness, and adsorption-induced surface stress (Stachiv and Sittner, 2018). This measurement utilizes a phase transformation in the shape memory alloys, which causes changes in the resonator stiffness; that is, at a low temperature NiTi is in the martensite phase with E ≈ 25 GPa, while at the high temperature NiTi transforms to the austenite phase with E up to 80 GPa and altering the interlayer stress, that is, the stress between NiTi and substrate can change more than four times (Stachiv et al., 2021). It can be easily expected that the shape memory alloy-based resonators would play a significant role in the next-generation nanomechanical mass spectrometers. These smart materials will enable resonator mechanical properties and interlayer stress to be adjusted based on the investigated analyte (protein molecule). In addition, shape memory materials can sustain a large recoverable deformation of more than 5% in the case of NiTi films (Juan et al., 2008). Hence, the shape memory alloy-based NRs can be also designed to change its shape before or during own measurements. We foresee that these resonators would enable manipulation of the molecule of interest simply by changing the resonator shape.
4 Nanomechanical mass spectrometry in fluids
Extraordinary high sensitivity of NRs can only be obtained in the vacuum, whereas in aqueous solution, which is relevant to the majority of biological systems and, therefore is also of emergent interests from the life science community, the mass sensitivity strongly degrades. Hence, by immersing the NR into fluid, the quality factor and mass sensitivity notably decrease, that is, sensitivity is usually > MDa (Eom et al., 2011). Burg et al. (2007), in their pioneer study, suggested using the hollow resonator, where the measured molecule flows in the fluid inside the resonator. This resonator is then entirely operating in the vacuum. As a result, they achieved a mass sensitivity of tens of kDa. More recently, this technique has notably been improved, and the sensitivity of kDa has been demonstrated (Kim et al., 2016; De Pastina et al., 2018). Other approaches to mass sensing in fluids utilize measurement of quality factor changes (Stachiv et al., 2020). In this alternative approach, the changes in quality factor caused by the bound analyte are used to extract the analyte mass. Unfortunately, up-to-date measurement in gaseous or aqueous solutions has not reached sensitivity yet comparable to that of NRs operating in the vacuum. Nevertheless, it can also be expected that with further improvements and optimization of the hollow resonators, the Dalton sensitivity could possibly be reached.
Overall, we highlight here recent progress in nanomechanical mass spectrometry, current challenges, and possible future trends in research.
Author contributions
IS designed the study. IS, C-YK, and WL collected materials. The manuscript was prepared by IS and C-YK with the help of WL. All authors contributed to the final version of the article and approved its submission.
Funding
The current study was supported by the Czech Science Foundation under Project No. 21-12994J and by the Operational Programme Research, Development, and Education financed by the European Structural and Investment Funds and the Czech Ministry of Education, Youth and Sports (Project No. SOLID21—CZ.02.1.01/0.0/0.0/16_019/0000760).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aebersold, R., and Mann, M. (2003). Mass spectrometry-based proteomics. Nature 422, 198–207. doi:10.1038/nature01511
Angel, T. E., Aryal, U. K., Hengel, S. M., Baker, E. S., Kelly, R. T., Robinson, E. W., et al. (2012). Mass spectrometry-based proteomics: Existing capabilities and future directions. Chem. Soc. Rev. 41, 3912–3928. doi:10.1039/c2cs15331a
Boisen, A., Dohn, S., keller, S. S., Schmid, S., and Tenje, M. (2011). Cantilever-like micromechanical sensors. Rep. Prog. Phys. 74, 036101. doi:10.1088/0034-4885/74/3/036101
Burg, T. P., Godin, M., Knudsen, S. M., Shen, W., Carlson, G., Foster, J. S., et al. (2007). Weighing of biomolecules, single cells and single nanoparticles in fluid. Nature 446, 1066–1069. doi:10.1038/nature05741
Chaste, J., Eichler, A., Moser, J., Ceballos, G., Rurali, R., Bachtold, A., et al. (2012). A nanomechanical mass sensor with yoctogram resolution. Nat. Nanotechnol. 7, 301–304. doi:10.1038/nnano.2012.42
De Pastina, A., Maillard, D., and Villanueva, L. G. (2018). Fabrication of suspended microchannel resonators with integrated piezoelectric transduction. Microelectron. Eng. 192, 83–87. doi:10.1016/j.mee.2018.02.011
Deng, F., Zhai, W., Yin, Y., Peng, C., and Ning, C. (2021). Advanced protein adsorption properties of a novel silicate-based bioceramic: A proteomic analysis. Bioact. Mat. 6, 208–218. doi:10.1016/j.bioactmat.2020.08.011
Dohn, S., Sandberg, R., Svendsen, W., and Boisen, A. (2005). Enhanced functionality of cantilever based mass sensors using higher modes. Appl. Phys. Lett. 86, 233501. doi:10.1063/1.1948521
Dohn, S., Schmid, S., Amiot, F., and Boisen, A. (2007). Position and mass determination of multiple particles using cantilever based mass sensors. Appl. Phys. Lett. 97, 044103. doi:10.1063/1.3473761
Dominguez-Medina, S., Fostner, S., Defoort, M., Sansa, M., Stark, A.-K., Halim, M. A., et al. (2018). Neutral mass spectrometry of virus capsids above 100 megadaltons with nanomechanical resonators. Science 362, 918–922. doi:10.1126/science.aat6457
Dorignac, J., Kalinowski, A., Erramilli, S., and Mohanty, P. (2006). Dynamical response of nanomechanical oscillators in immiscible viscous fluid for in vitro biomolecular recognition. Phys. Rev. Lett. 96, 186105. doi:10.1103/PhysRevLett.96.186105
Eisner, V., Picard, M., and Hajnóczky, G. (2018). Mitochondrial dynamics in adaptive and maladaptive cellular stress responses. Nat. Cell. Biol. 20, 755–765. doi:10.1038/s41556-018-0133-0
Ekinci, K., Yang, Y. T., and Roukes, M. L. (2004). Ultimate limits to inertial mass sensing based upon nanoelectromechanical systems. J. Appl. Phys. 95, 2682–2689. doi:10.1063/1.1642738
Eom, K., Park, H-S., Yoon, D. S., and Kwon, T. (2011). Nanomechanical resonators and their applications in biological/chemical detection: Nanomechanics principles. Phys. Rep. 503, 115–163. doi:10.1016/j.physrep.2011.03.002
Erdogan, R. T., Alkhaled, M., Kaynak, B. E., Alhmoud, H., Pisheh, H. S., Kellici, M., et al. (2022). Atmospheric pressure mass spectrometry of single viruses and nanoparticles by nanoelectromechanical systems. ACS Nano 16, 3821–3833. doi:10.1021/acsnano.1c08423
Ghatkesar, M. K., Barwich, V., Braun, T., Ramseyer, J.-P., Gerber, C., Hegner, M., et al. (2007). Higher modes of vibration increase mass sensitivity in nanomechanical microcantilevers. Nanotechnology 18, 445502. doi:10.1088/0957-4484/18/44/445502
Gil-Santos, E., Ramos, D., Martínez, F., RegúlezGarcía, J. M. R., San Paulo, A., Garcia, R., et al. (2010). Nanomechanical mass sensing and stiffness spectrometry based on two-dimensional vibrations of resonant nanowires. Nat. Nanotech. 5, 641–645. doi:10.1038/nnano.2010.151
Gil-Santos, E., Ruz, J. J., Malvar, O., Favero, I., Lemaître, A., Kosaka, P. M., et al. (2020). Optomechanical detection of vibration modes of a single bacterium. Nat. Nanotech. 15, 469–474. doi:10.1038/s41565-020-0672-y
Gruber, G., Urgell, C., Tavernarakis, A., Stavrinadis, A., Tepsic, S., magen, C., et al. (2019). Mass sensing for the advanced fabrication of nanomechanical resonators. Nano Lett. 19, 6987–6992. doi:10.1021/acs.nanolett.9b02351
Gupta, A. K., Nair, P. R., Akin, D., Ladisch, M. R., Broyles, S., Alam, M. A., et al. (2006). Anomalous resonance in a nanomechanical biosensor. Proc. Natl. Acad. Sci. U. S. A. 5, 13362–13367. doi:10.1073/pnas.0602022103
Hanay, M. S., Kelber, S., Naik, A. K., Chi, D., Hentz, S., Bullard, E., et al. (2012). Single-protein nanomechanical mass spectrometry in real time. Nat. Nanotechnol. 7, 602–608. doi:10.1038/nnano.2012.119
Jensen, K., Kim, K., and Zettl, A. (2008). An atomic-resolution nanomechanical mass sensor. Nat. Nanotechnol. 3, 533–537. doi:10.1038/nnano.2008.200
Johnson, E. C. B., Dammer, E. B., Duong, D. M., Ping, L., Zhou, M., Yin, L., et al. (2020). Large-scale proteomic analysis of Alzheimer’s disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat. Med. 26, 769–780. doi:10.1038/s41591-020-0815-6
Juan, J. M. S., No, M. L., and Schuh, C. A. (2008). Superelasticity and shape memory in micro- and nanometer-scale pillars. Adv. Mat. 20, 272–278. doi:10.1002/adma.200701527
Karabalin, R. B., Villanueva, L. G., Matheny, M. H., Sader, J. E., and Roukes, M. L. (2012). Stress-induced variations in the stiffness of micro- and nanocantilever beams. Phys. Rev. Lett. 108, 236101. doi:10.1103/PhysRevLett.108.236101
Keifer, D. Z., Pierson, E. E., and Jarrold, M. F. (2017). Charge detection mass spectrometry: Weighing heavier things. Analyst 142, 1654–1671. doi:10.1039/C7AN00277G
Kidane, S., Ishida, H., Sawada, K., and Takahashi, K. (2020). A suspended-graphene based optical interferometric surface stress sensor for selective biomolecular detection. Nanoscale Adv. 2, 1431–1436. doi:10.1039/C9NA00788A
Kim, J., Song, J., Kim, K., Kim, S., Song, J., Kim, N., et al. (2016). Hollow microtube resonators via silicon self-assembly toward subattogram mass sensing applications. Nano Lett. 16, 1537–1545. doi:10.1021/acs.nanolett.5b03703
Kozinsky, I., Postma, H. W. Ch., Bargatin, I., and Roukes, M. L. (2006). Tuning nonlinearity, dynamic range, and frequency of nanomechanical resonators. Appl. Phys. Lett. 88, 253101. doi:10.1063/1.2209211
Kubiak-Ossowska, K., Jachimska, B., Al Qaraghuli, M., and Mulheran, P. A. (2019). Protein interactions with negatively charged inorganic surfaces. Curr. Opin. Colloid Interface Sci. 41, 104–117. doi:10.1016/j.cocis.2019.02.001
Lachut, M. J., and Sader, J. E. (2007). Effect of surface stress on the stiffness of cantilever plates. Phys. Rev. Lett. 99, 206102. doi:10.1103/PhysRevLett.99.206102
Lachut, M. J., and Sader, J. E. (2012). Effect of surface stress on the stiffness of thin elastic plates and beams. Phys. Rev. B 85, 085440. doi:10.1103/PhysRevB.85.085440
Lamberti, F-R., Palanchoke, U., Geurts, T. P. J., Gely, M., Regord, S., Banniard, L., et al. (2022). Real-time sensing with multiplexed optomechanical resonators. Nano Lett. 22, 1866–1873. doi:10.1021/acs.nanolett.1c04017
Li, M.-L., Wang, W., and Jin, Z.-B. (2021). Circular RNAs in the central nervous system. Front. Mol. Biosci. 8, 629593. doi:10.3389/fmolb.2021.629593
Liu, W., Wang, J., Yu, Y., Chang, Y., Tang, N., Qu, H., et al. (2015). Tuning the resonant frequency of resonators using molecular surface self-assembly approach. ACS Appl. Mat. Interfaces 7, 950–958. doi:10.1021/am507640g
Lyu, M., Zhao, J., Kacem, N., Liu, P., Tang, B., Xiong, Z., et al. (2020). Exploiting nonlinearity to enhance the sensitivity of mode-localized mass sensor based on electrostatically coupled MEMS resonators. Int. J. Non-Linear Mech. 121, 103455. doi:10.1016/j.ijnonlinmec.2020.103455
Ma, S., Li, M., Wang, S., Liu, H., Wang, H., Ren, L., et al. (2022). Multiple particle identification by sequential frequency-shift measurement of a micro-plate. Int. J. Mech. Sci. 231, 107587. doi:10.1016/j.ijmecsci.2022.107587
Malvar, O., Ruz, J. J., Kosaka, P. M., Dominguez, C. M., Gil-Santos, E., Calleja, M., et al. (2016). Mass and stiffness spectrometry of nanoparticles and whole intact bacteria by multimode nanomechanical resonators. Nat. Commun. 11, 13452. doi:10.1038/ncomms13452
Minami, K., Shiba, K., and Yoshikawa, G. (2021). Sorption-induced static mode nanomechanical sensing with viscoelastic receptor layers for multistep injection-purge cycles. J. Appl. Phys. 129, 124503. doi:10.1063/5.0039045
Minami, K., and Yoshikawa, G. (2021). Effects of partial attachment at the interface between receptor and substrate on nanomechanical cantilever sensing. Sens. Act. A Phys. 319, 112533. doi:10.1016/j.sna.2020.112533
Naik, A. K., Hanay, M. S., Hiebert, W. K., Feng, X. L., and Roukes, M. L. (2009). Towards single-molecule nanomechanical mass spectrometry. Nat. Nanotechnol. 4, 445–450. doi:10.1038/nnano.2009.152
Pfeifer, C. R., Alveya, C. M., Iriantoa, J., and Dischera, D. E. (2017). Genome variation across cancers scales with tissue stiffness – An invasion-mutation mechanism and implications for immune cell infiltration. Curr. Opin. Syst. Biol. 2, 103–114. doi:10.1016/j.coisb.2017.04.005
Richard, P. J. (2019). Protein flexibility and stiffness enable efficient enzymatic catalysis. J. Am. Chem. Soc. 141, 3320–3331. doi:10.1021/jacs.8b10836
Rocha, R. T., Alfosail, F., Zhao, W., Younis, M. I., and Masri, S. F. (2021). Nonparametric identification of a micro-electromechanical resonator. Mech. Syst. Sig. Proc. 161, 107932. doi:10.1016/j.ymssp.2021.107932
Ruz, J. J., Malvar, O., Gil-Santos, E., Ramos, D., Calleja, M., and Tamayo, J. (2021). A review on theory and modelling of nanomechanical sensors for biological applications. Process 9, 164. doi:10.3390/pr9010164
Ruz, J. J., Tamayo, J., Pini, V., Kosaka, P. M., and Calleja, M. (2014) Physics of nanomechanical spectrometry of viruses. Sci. Rep. 4, 6051; doi:10.1038/srep06051
Sader, J. E., Hanay, M. S., Neumann, A. P., and Roukes, M. L. (2018). Mass spectrometry using nanomechanical systems: Beyond the point-mass approximation. Nano Lett. 18, 1608–1614. doi:10.1021/acs.nanolett.7b04301
Sage, E., Brenac, A., Alava, T., Morel, R., Dupré, C., Hanay, M. S., et al. (2015). Neutral particle mass spectrometry with nanomechanical systems. Nat. Commun. 6, 6482. doi:10.1038/ncomms7482
Sage, E., Sansa, M., Fostner, S., Defoort, M., Gely, M., Naik, A. K., et al. (2018). Single-particle mass spectrometry with arrays of frequency-addressed nanomechanical resonators. Nat. Commun. 9, 3283. doi:10.1038/s41467-018-05783-4
Sansa, M., Defoort, M., Brenac, A., Hermouet, M., Banniard, L., Fafin, A., et al. (2020). Optomechanical mass spectrometry. Nat. Commun. 11, 3781. doi:10.1038/s41467-020-17592-9
Satzer, P., Svec, F., Sekot, G., and Jungbauer, A. (2015). Protein adsorption onto nanoparticles induces conformational changes: Particle size dependency, kinetics, and mechanisms. Eng. Life. Sci. 16, 238–246. doi:10.1002/elsc.201500059
Schmid, S., Dohn, S., and Boisen, A. (2010). Real-time particle mass spectrometry based on resonant micro strings. Sensors 10, 8092–8100. doi:10.3390/s100908092
Senese, N. B., Rasenick, M. M., and Traynor, J. R. (2018). The role of G-proteins and G-protein regulating proteins in depressive disorders. Front. Pharmacol. 9, 1289. doi:10.3389/fphar.2018.01289
Shen, L., de Sousa, F. B., Tay, N. B., Lang, T. S., Kaixin, V. L., Han, J., et al. (2020). Deformation behavior of normal human enamel: A study by nanoindentation. J. Mech. Behav. Biomed. Mat. 108, 103799. doi:10.1016/j.jmbbm.2020.103799
Snijder, J., Rose, R. J., Veesler, D., Johnson, J. E., and Heck, A. J. R. (2013). Studying 18MDa virus assemblies with native mass spectrometry. Angew. Chem. Int. Ed. 52, 4020–4023. doi:10.1002/anie.201210197
Stachiv, I., Alarcon, E., and Lamac, M. (2021). Shape memory alloys and polymers for MEMS/NEMS applications: Review on recent findings and challenges in design, preparation, and characterization. Metals 11, 415. doi:10.3390/met11030415
Stachiv, I., Fedorchenko, A. I., and Chen, Y-L. (2012). Mass detection by means of the vibrating nanomechanical resonators. Appl. Phys. Lett. 100, 093110. doi:10.1063/1.3691195
Stachiv, I., and Gan, L. (2019). Hybrid shape memory alloy-based nanomechanical resonators for ultrathin film elastic properties determination and heavy mass spectrometry. Materials 12, 3593. doi:10.3390/ma12213593
Stachiv, I., Gan, L., Kuo, C.-Y., Sittner, P., and Sevecek, O. (2020). Mass spectrometry of heavy analytes and large biological aggregates by monitoring changes in the quality factor of nanomechanical resonators in air. ACS Sens. 5, 2128–2135. doi:10.1021/acssensors.0c00756
Stachiv, I. (2014). Impact of surface and residual stresses and electro-/magnetostatic axial loading on the suspended nanomechanical based mass sensors: A theoretical study. J. Appl. Phys. 115, 214310. doi:10.1063/1.4880396
Stachiv, I., Machu, Z., Sevecek, O., Jeng, Y.-R., Li, W.-L., Kotoul, M., et al. (2022b). Achievable accuracy of resonating nanomechanical systems for mass sensing of larger analytes in GDa range. Int. J. Mech. Sci. 224, 107353. doi:10.1016/j.ijmecsci.2022.107353
Stachiv, I., Machu, Z., Sevecek, O., Tuhovcak, O., Kotoul, M., and Jeng, Y.-R. (2022a). Resolving measurement of large (∼ GDa) chemical/biomolecule complexes with multimode nanomechanical resonators. Sens. Act. B Chem. 353, 131062. doi:10.1016/j.snb.2021.131062
Stachiv, I., and Sittner, P. (2018). Nanocantilevers with adjustable static deflection and significantly tunable spectrum resonant frequencies for applications in nanomechanical mass sensors. Nanomaterials 8, 116. doi:10.3390/nano8020116
Stachiv, I., Sittner, P., Olejnicek, J., Landa, M., and Heller, L. (2017). Exploiting NiTi shape memory alloy films in design of tunable high frequency microcantilever resonators. Appl. Phys. Lett. 111, 213105. doi:10.1063/1.4998006
Stachiv, I., Zapomel, J., and Chen, Y-L. (2014). Simultaneous determination of the elastic modulus and density/thickness of ultrathin films utilizing micro-/nanoresonators under applied axial force. J. Appl. Phys. 115, 124304. doi:10.1063/1.4869415
Stassi, S., De Laurentis, G., Chakraborty, D., Bejtka, K., Chiodoni, A., Sader, J. E., et al. (2019). Large-scale parallelization of nanomechanical mass spectrometry with weakly-coupled resonators. Nat. Commun. 10, 3647. doi:10.1038/s41467-019-11647-2
Su Renay, S-C., Gill, E. E., Kim, Y., and Liu, J. C. (2019). Characterization of resilin-like proteins with tunable mechanical properties. J. Mech. Behav. Biomed. Mat. 91, 68–75. doi:10.1016/j.jmbbm.2018.11.015
Talha, M., Ma, Y., Kumar, P., Lin, Y., and Singh, A. (2019). Role of protein adsorption in the bio corrosion of metallic implants – a review. Colloids Surf. B Biointerfaces 176, 494–506. doi:10.1016/j.colsurfb.2019.01.038
Tamayo, J., Kosaka, P. M., Ruz, J. J., San Paulo, A., and Calleja, M. (2013). Biosensors based on nanomechanical systems. Chem. Soc. Rev. 42, 1287–1311. doi:10.1039/C2CS35293A
Tamayo, J., Ramos, D., Mertens, J., and Calleja, M. (2006). Effect of the adsorbate stiffness on the resonance response of microcantilever sensors. Appl. Phys. Lett. 89, 224104. doi:10.1063/1.2388925
Urbanska, M., Muñoz, H. E., Shaw, B., Otto, O., Manalis, S. R., Di Carlo, D., et al. (2020). A comparison of microfluidic methods for high-throughput cell deformability measurements. Nat. Methods 17, 587–593. doi:10.1038/s41592-020-0818-8
Wingert, B., Krieger, J., Li, H., and Bahar, I. (2021). Adaptability and specificity: How do proteins balance opposing needs to achieve function? Curr. Opin. Struct. Biol. 67, 25–32. doi:10.1016/j.sbi.2020.08.009
Xia, C., Wang, D. F., Ono, T., Itoh, T., and Esashi, M. (2021). Internal resonance in coupled oscillators – Part I: A double amplification mass sensing scheme without duffing nonlinearity. Mech. Syst. Sig. Proc. 159, 107886. doi:10.1016/j.ymssp.2021.107886
Xu, B., Zhang, P., Zhu, J., Liu, Z., Eichler, A., Zheng, X.-Q., et al. (2022). Nanomechanical resonators: Toward atomic scale. ACS Nano 16, 15545–15585. (accepted). doi:10.1021/acsnano.2c01673
Yang, B., Liu, Z., Liu, H., and Nash, M. A. (2020). Next generation methods for single-molecule force spectroscopy on polyproteins and receptor-ligand complexes. Front. Mol. Biosci. 7, 85. doi:10.3389/fmolb.2020.00085
Yang, D., Ward, A., Halvorsen, K., and Wong, W. P. (2016). Multiplexed single-molecule force spectroscopy using a centrifuge. Nat. Commun. 7, 11026. doi:10.1038/ncomms11026
Yang, Y. T., Callegari, C., Feng, X. L., Ekinci, K. L., and Rooukes, M. L. (2006). Zeptogram-scale nanomechanical mass sensing. Nano Lett. 6, 583–586. doi:10.1021/nl052134m
Yi, X., and Duan, H. L. (2009). Surface stress induced by interactions of adsorbates and its effect on deformation and frequency of microcantilever sensors. J. Mech. Phys. Solids 57, 1254–1266. doi:10.1016/j.jmps.2009.04.010
Yuksel, M., Orhan, E., Yanik, C., Ari, A. B., Demir, A., and Hanay, M. S. (2019). Nonlinear nanomechanical mass spectrometry at the single-nanoparticle level. Nano Lett. 19, 3583–3589. doi:10.1021/acs.nanolett.9b00546
Zhang, G., Li, C., Wu, S., and Zhang, Q. (2018). Label-free aptamer-based detection of microcystin-LR using a microcantilever array biosensor. Sens. Act. B Chem. 260, 42–47. doi:10.1016/j.snb.2017.12.112
Zhang, W.-M., Hu, K.-M., Peng, Z.-K., and Meng, G. (2015). Tunable micro- and nanomechanical resonators. Sensors 15, 26478–26566. doi:10.3390/s151026478
Keywords: protein adsorption, molecule mechanical properties, molecular weight, protein detection, mass spectrometry, mass sensing
Citation: Stachiv I, Kuo C-Y and Li W (2023) Protein adsorption by nanomechanical mass spectrometry: Beyond the real-time molecular weighting. Front. Mol. Biosci. 9:1058441. doi: 10.3389/fmolb.2022.1058441
Received: 30 September 2022; Accepted: 14 December 2022;
Published: 05 January 2023.
Edited by:
Prashanth Asuri, Santa Clara University, United StatesReviewed by:
Mahmoud Hussein Hadwan, University of Babylon, IraqCopyright © 2023 Stachiv, Kuo and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ivo Stachiv, c3RhY2hpdkBmenUuY3o=
 Ivo Stachiv
Ivo Stachiv Chih-Yun Kuo
Chih-Yun Kuo Wei Li1
Wei Li1