
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci. , 01 December 2022
Sec. Protein Folding, Misfolding and Degradation
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.1054408
This article is part of the Research Topic A Focus on Chaperone Clients View all 6 articles
Proteostasis mechanisms significantly contribute to the sculpting of the proteomes of all living organisms. ClpXP is a central AAA+ chaperone-protease complex present in both prokaryotes and eukaryotes that facilitates the unfolding and subsequent degradation of target substrates. ClpX is a hexameric unfoldase ATPase, while ClpP is a tetradecameric serine protease. Substrates of ClpXP belong to many cellular pathways such as DNA damage response, metabolism, and transcriptional regulation. Crucially, disruption of this proteolytic complex in microbes has been shown to impact the virulence and infectivity of various human pathogenic bacteria. Loss of ClpXP impacts stress responses, biofilm formation, and virulence effector protein production, leading to decreased pathogenicity in cell and animal infection models. Here, we provide an overview of the multiple critical functions of ClpXP and its substrates that modulate bacterial virulence with examples from several important human pathogens.
Antimicrobial resistance to common treatments continues to be one of the largest threats to global public health. Therefore, new antibiotics with new modes of action are in great demand. Molecular chaperones and proteases play a critical role in maintaining cellular protein homeostasis (proteostasis). Dysregulation of these proteostasis mechanisms has been shown to disrupt major cellular pathways and is lethal for many organisms (Ingmer and Brondsted, 2009; Frees et al., 2013; Bhandari et al., 2018). Due to their importance in proteostasis and the lack of preexisting resistance associated with them, chaperones and proteases have recently emerged as promising targets for the development of novel antimicrobial compounds (Raju et al., 2012a; Brӧtz-Oesterhelt and Sass, 2014).
Here, we discuss one such candidate for antibiotic targeting, the caseinolytic protease (ClpP) that is conserved across many kingdoms of life. ClpP is a serine protease that functions with cognate unfoldase ATPases. In bacteria, it acts as one of the major ATP-dependent cellular proteases along with Lon, HslUV, and FtsH (Culp and Wright, 2017). ClpP functions in the removal of damaged and unwanted proteins and also in the degradation of regulatory proteins. Additionally, ClpP is integral to many important cellular pathways across different bacterial species. Its substrates include proteins involved in cell cycle regulation, stress tolerance, virulence factor production, biofilm formation, antibiotic tolerance, and metabolism (Gottesman, 2019; Kirsch et al., 2021). Though it plays important roles across a variety of bacterial pathogens, its influence varies between different species. The role of ClpP in the pathogenesis of select species is explored below along with a discussion on factors that determine its substrates.
ClpP is a self-compartmentalizing serine protease. It is composed of fourteen subunits that typically assemble into a stack of two heptameric rings to create a hollow barrel-like structure. Each protomer typically contains a Ser-His-Asp catalytic triad facing the interior of the complex, forming the catalytic chamber. A ClpP subunit is composed of three subdomains or regions: N-terminal loops, the head domain, and the handle region (Figure 1A). The N-terminal loops are also called the axial loops, which line the entrance of the axial pore of the tetradecameric complex and protrude from the apical surface of ClpP (Figure 1A). The pore lining is comprised of hydrophobic residues that are stabilized by interactions with the head domain, whereas the axial protrusion is comprised of hydrophilic or charged residues. The head domain forms the main body of the degradation chamber, while the handle region facilitates the interaction between the two heptameric rings by intercalation. When ClpP is not associated with its cognate ATPase, the N-terminal regions are typically disordered and partially block the entrance to the degradation chamber (Moreno-Cinos et al., 2019; Kahne and Darwin, 2021; Mabanglo and Houry, 2022). This ensures that cellular proteins are protected from uncontrolled proteolysis by the catalytic residues within the ClpP degradation chamber. On its own, the ClpP tetradecamer can degrade short peptides that are small enough to pass through the axial pores. Proteolysis of larger proteins requires the formation of a complex with a Clp ATPase chaperone (Figures 1B,C) (Liu et al., 2014).
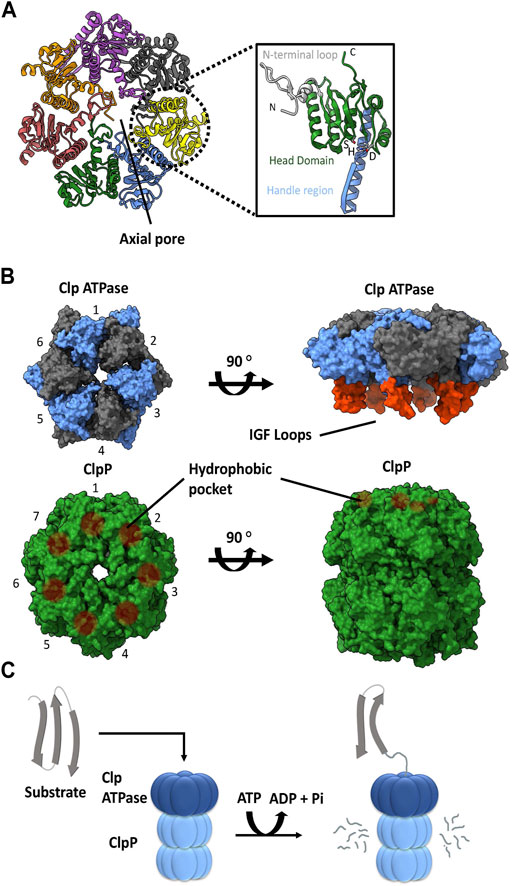
FIGURE 1. ClpP and Clp ATPases general architecture. (A) Top view of the E. coli ClpP cylinder (PDB ID: 1YG6). In the illustration on the right, the N-terminal loop (silver), head domain (green), handle region (blue) and catalytic triad (S, H, D) of the ClpP monomer are indicated (PDB ID 1YG6). (B) The association of a Clp ATPase hexamer (PDB ID 6SFW) with a ClpP tetradecamer (PDB ID 6SFX). Clp ATPase subunits (blue and grey) with their corresponding IGF loops (orange) are shown. ClpP subunits (green) and their hydrophobic pockets are denoted as brown circles. (C) General schematic of the proteolytic degradation cycle mediated by ClpP with its cognate Clp ATPase. Substrates are unfolded by the Clp ATPase and then translocated in an ATP-dependent manner into the ClpP catalytic chamber for proteolysis.
A variety of Clp ATPases belonging to the Hsp100 class of the AAA+ superfamily (ATPases associated with various cellular activities) have been identified. These include ClpA, ClpB, ClpC, ClpE, ClpL, and ClpX chaperones. They typically form a hexameric structure with a central pore (Figures 1B). They usually contain an N-terminal domain followed by one or more ATPase domains. As such, the Clp ATPases serve two functions: selection and binding of a substrate protein, followed by its unfolding and subsequent translocation into the ClpP catalytic chamber (Olivares et al., 2016) (Figure 1C).
With the exception of ClpB, all of the other Clp ATPases can associate with a ClpP. The ability to do so is dependent on the presence of ClpP recognition loops. These loops contain the IGF tripeptide motif in gamma-proteobacteria, but they can be MGF or LGF in other bacterial phyla (Amor et al., 2019). The 6 M/L/IGF loops of the Clp ATPases dock onto specific hydrophobic pockets on the surface of ClpP (Figures 1B). These hydrophobic pockets are located near the outer edge of the apical surface between the ClpP protomer subunits. The 6:7 Clp ATPase to ClpP symmetry mismatch has been a topic of debate for decades. However, recently three Cryo-EM structures of bacterial ClpXP were solved which show that ClpX docks in an offset manner from ClpP in a tilted position. This allows 6 of the 7 hydrophobic pockets in ClpP to interact with the IGF loops of ClpX, thereby leaving one hydrophobic pocket empty (Gatsogiannis et al., 2019; Fei et al., 2020b; Ripstein et al., 2020). Consequently, this 6:7 symmetry mismatch seems to be required for target substrate unfolding and translocation into the ClpP catalytic chamber (Figures 1C); however, the molecular basis and consequences of this symmetry mismatch on ClpXP activity is not clear.
The number and type of Clp ATPases found in different species varies even among closely related organisms (Figure 2A). However, some trends have been observed. ClpX is the most widespread ATPase and can be found in almost all bacteria. ClpA is generally found among the Gram-negative proteobacteria. ClpC is present in the firmicutes and actinobacteria groups, as well as in species of cyanobacteria, while the presence of ClpE is limited to some species of the firmicutes group (Kress et al., 2009).
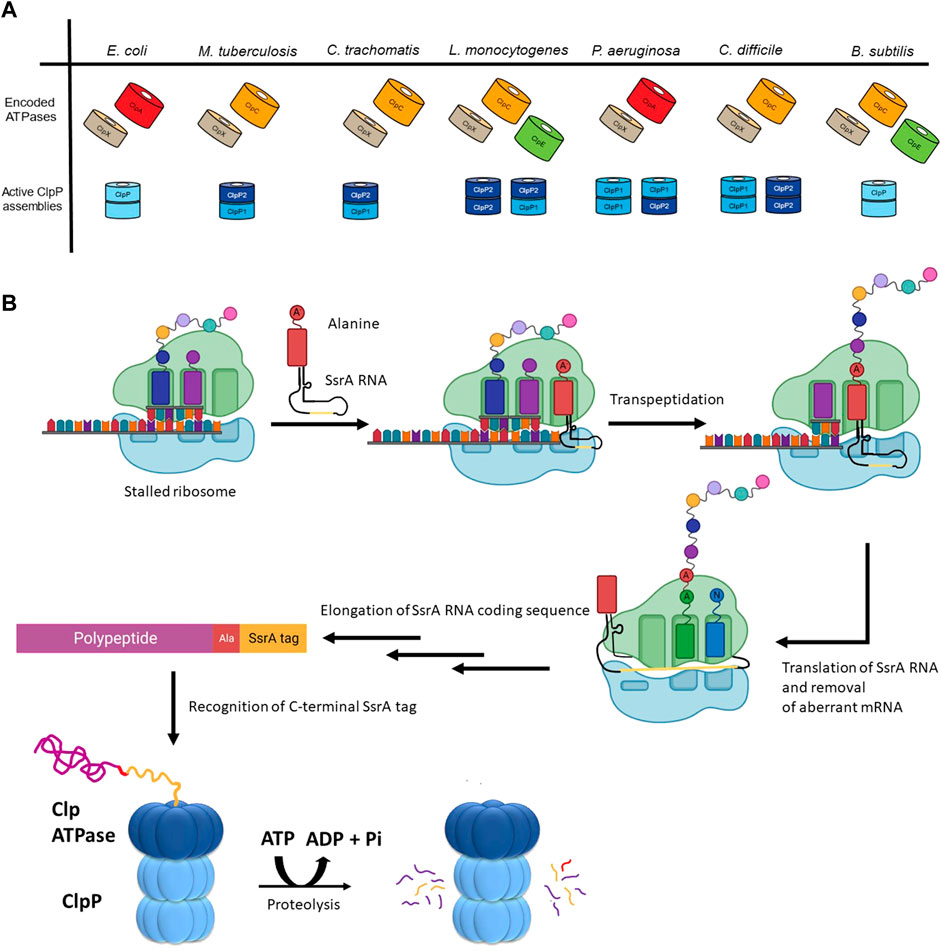
FIGURE 2. The diversity of ClpP substrate regulation. (A) The Clp ATPases and ClpPs of a select group of bacterial pathogens are shown. The tetradecameric organization for the active form(s) of the ClpP protease for each of these species is also shown. (B) Shown is the mechanism of SsrA tagging of nascent chains as means of targeting them for ClpP-mediated degradation. During translation of aberrant mRNAs lacking a stop codon, the ribosome stalls, resulting in the recruitment of a charged alanine-SsrA RNA (tmRNA) to the A site of the ribosome. Following transpeptidation of the charged alanine-residue to the nascent chain, the aberrant mRNA open reading frame is replaced with the RNA carried by the tmRNA, resulting in the translation of the SsrA RNA coding sequence (yellow). This C-terminal tag placed on the stalled nascent chains acts as a degron that is recognized by the Clp ATPase (e.g., ClpX or ClpA) enabling ClpP-mediated degradation.
Substrate selection is not performed by the ClpP protease but rather by its partner ATPase. The diversity of these ATPases correlates with the diversity of substrates selected for proteolysis by ClpP. For example, Bacillus subtilis (though not a pathogenic bacterium) encodes the protease ClpP as well as three ATPases, which have been well-characterized: ClpX, ClpE, and ClpC (Figure 2A) (Gerth et al., 2004). These ATPases have all been shown to act on heat-damaged protein aggregates for subsequent ClpP-mediated turnover (Kruger et al., 1994; Derre et al., 1999; Kruger et al., 2000). Unlike ClpX, both ClpE and ClpC are under the transcriptional control of CtsR (class III stress gene repressor), which negatively regulates their expression (Derre et al., 1999; Kirstein and Turgay, 2005). ClpE is the most tightly repressed of the three ATPases and is only expressed under severe heat shock conditions (Miethke et al., 2006). Additionally, ClpE is further regulated through its turnover by ClpCP (Gerth et al., 2004).
As suggested above, a separation of functions is seen between the Clp ATPase chaperones. ClpX appears to be constitutively expressed and is available for maintenance of protein quality control at all times during the cell cycle, specifically for the removal of proteins whose translation has stalled (Flynn et al., 2001). Other ATPases are dedicated for degradation of substrates during stress, such as ClpC, which is the primary ATPase responsible for the degradation of non-native proteins (Kruger et al., 1994; Rouquette et al., 1996; Chatterjee et al., 2005). The presence of multiple substrate-selecting ATPases appears to be advantageous for the cell. Selective expression presumably allows the cell to diversify its use of the ClpP protease to meet the demands of the changes in the conditions imparted by a variety of environmental stresses.
In addition to the control resulting from encoding for multiple ATPases, cells possess the ability to modulate ClpP substrate selection co-translationally. One prominent example is the SsrA tagging system in prokaryotes, which is employed, for example, when truncated mRNAs are generated during aberrant or premature termination of transcription (Figure 2B) (Keiler et al., 1996). At the centre of this system is a transfer-messenger RNA (tmRNA), which is a tRNA that encodes for its own message; the message codes for a hydrophobic 8–35 residues long SsrA tag that is added to the C-terminus of incomplete nascent chains. During translation of truncated mRNAs, tmRNA binds to the A-site of a stalled ribosome and, subsequently, this leads to the translation of the complete SsrA tag that is added to the C-terminus of the nascent polypeptide (Keiler, 2015). In Escherichia coli, this tag is AANDENYALAA-(COO-) (Keiler et al., 1996), which is recognized by ClpX, leading to the degradation of the protein by ClpXP (Flynn et al., 2001; Farrell et al., 2007; Martin et al., 2008; Fei et al., 2020a). Proteins labelled with an SsrA tag can also be degraded by ClpAP, FtsH, and Lon (Gottesman et al., 1998; Herman et al., 1998; Choy et al., 2007).
The selectivity of ATPase chaperones is modulated by dedicated adaptors. Some adaptors are essential to ATPase function, such as in the case of ClpC in B. subtilis, where the binding of MecA, McsB, or YpbH adaptors (Schlothauer et al., 2003; Frees et al., 2007) is required for the assembly of an active hexameric chaperone (Kirstein et al., 2006). Recent data suggests that ClpC in Staphylococcus aureus and B. subtilis first assemble into a resting state comprised of a helical decamer; however, upon binding to an adaptor, the decamer reorganizes into an active hexameric state which can assemble with the ClpP tetradecamer (Carroni et al., 2017; Morreale et al., 2022). Other adaptors can modulate substrate selection but are not essential for ATPase function. This is true of ClpX that functions with several adaptor proteins. SspB (stringent starvation protein B) is one such adaptor that modulates the ClpX substrate pool by enhancing the degradation of SsrA tagged substrates (Levchenko et al., 2000; Dougan et al., 2003; Wah et al., 2003).
There is also diversity in the ClpP proteases themselves as shown in Figure 2A. The E. coli ClpP protease provides a baseline standard: a tetradecamer protein composed of two identical heptameric rings, where each heptameric ring of the active complex can associate with a Clp ATPase (Porankiewicz et al., 1999). However, several pathogenic organisms have two paralogs of ClpP encoded as: ClpP1 and ClpP2. For example, in Pseudomonas aeruginosa, ClpP1 can form a functional homotetradecamer protease, while ClpP2 cannot (Hall et al., 2017). Rather, ClpP2 assembles into a heptameric ring to associate with a ClpP1 heptamer. Thus, two active versions of the protease complex exist, ClpP114 and ClpP17P27 (Figure 2A) (Hall et al., 2017). In Mycobacterium tuberculosis, ClpP114 and ClpP214 homotetradecamers can form but are inactive. It is only in the presence of activators that these subunits reassociate to form a ClpP17P27 heterotetradecamer capable of proteolysis (Akopian et al., 2012). Furthermore, the food-borne pathogen Listeria monocytogenes can form a functional ClpP214 homotetradecamer and ClpP17P27 heterotetradecamer while the ClpP114 is inactive (Zeiler et al., 2013) (Figure 2A). Additionally, in the anaerobic spore-forming gut bacterium Clostridioides difficile, both ClpP114 and ClpP214 homotetradecamers assemble into a functional protease (Lavey et al., 2019) (Figure 2A). Finally, in Chlamydia trachomatis, neither ClpP1 nor ClpP2 can form a homotetradecamer but rather associate with one another to create a ClpP17P27 heterotetradecamer capable of proteolytic activity (Pan et al., 2019) (Figure 2A). Currently, little is known about the advantages of producing multiple ClpP paralogs, however, ClpP complex asymmetry, and ATPase binding and peptidase specificity might provide additional levels of regulation for the protease (Personne et al., 2013; Leodolter et al., 2015; Nagpal et al., 2019).
The pathogenicity of a bacterium is critically dependent on its ability to survive in a host. ClpP is known to influence many pathways involved in maintaining normal cellular functions. Classic ClpP trapping experiments have been used to identify substrates of ClpP (Flynn et al., 2003). In these experiments, ClpPTrap, an active site serine mutant of the protease that is unable to degrade proteins, is used. Substrates entering the ClpPTrap will not be degraded but will be trapped inside the ClpP cylinder and are subsequently identified by pulling down ClpPTrap followed by mass spectrometry analysis to identify trapped proteins. Using this approach in S. aureus, for example, central transcriptional and stress regulatory proteins were identified to be the main targets of the protease including Spx (global transcription regulator), CodY (transcriptional repressor) and FtsZ (essential Z-ring forming protein at site of cell division) (Michel et al., 2006; Frees et al., 2012; Feng et al., 2013; Panasenko et al., 2020) (Table 1). RecA (DNA damage repair protein), PerR (regulator of peroxide inducible genes), and DnaK (heat-shock protein 70) (Table 1) were also among the substrates of S. aureus ClpP, highlighting the key role ClpP plays in maintaining central biological functions such as DNA repair, cell division, and protein homeostasis (Krysiak et al., 2017; Kirsch et al., 2021). Additionally, the role of ClpP impacting pathogen survivability was seen in M. tuberculosis, since ClpP depletion led to a reduction in colony forming units both in vitro and in a mouse model (Raju et al., 2012b). One essential substrate of M. tuberculosis ClpP that was identified was Whib1 (Table 1), a transcriptional repressor whose lack of turnover and subsequent accumulation led to cell toxicity and thereby increased cell death (Raju et al., 2014).
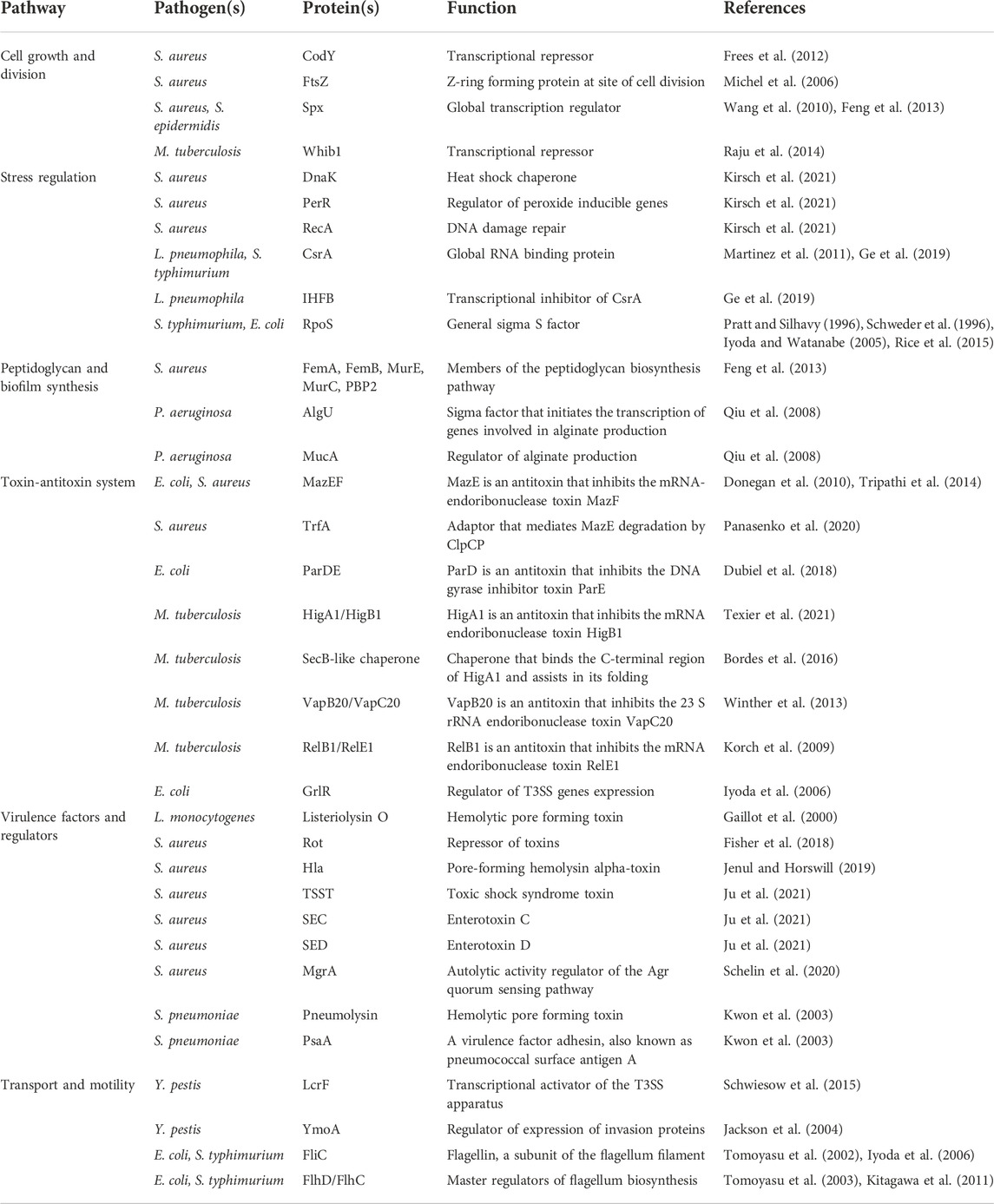
TABLE 1. Bacterial proteins and virulence factors regulated by the ClpP system discussed in this review.
Below we discuss the role of ClpP and ClpX in pathways that are pertinent to a pathogen’s ability to survive in a host organism. Table 1 lists bacterial proteins that are discussed here, which are regulated by the ClpP system in different pathogens.
Maintaining the integrity of the cell wall is vital for bacterial survival, which is needed to prevent cytolysis when bacteria encounter the turgor pressure of the host cell’s cytoplasm. Loss of ClpXP activity has been linked to increased susceptibility to cell wall stress in S. aureus. It is thought that antibiotics targeting the cell wall result in protein damage and misfolding, and lead to increased production of chaperones and proteases to aid in protein turnover and repair (McGillivray et al., 2012; Michiels et al., 2016). Thus, inhibiting ClpX or ClpP likely hinders the clearance of such damaged proteins. This hypothesis is supported by observations in S. aureus, since ΔclpP, ΔclpX, or ΔclpC mutants showed an increase in peptidoglycan cross-linking and thicker cell walls (Baek et al., 2014). Indeed, a separate study purified ClpPTrap from S. aureus and identified several enzymes involved in the peptidoglycan-synthesis pathway such as FemA, FemB, MurE, MurC, and PBP2 (Feng et al., 2013) (Table 1). While further research is required to better define the specific pathways modulated by ClpP, these results suggest that ClpP and its associated ATPases play an important role in maintaining the integrity of the cell wall.
In addition to peptidoglycan regulation, clpP deletion has been shown to dysregulate biofilm formation in P. aeruginosa (Qiu et al., 2008), Staphylococcus epidermidis (Wang et al., 2007) and S. aureus (Frees et al., 2004). For P. aeruginosa, overproduction of alginate, a central component of biofilms, leads to a mucoid phenotype, which indicates the onset of chronic lung infection in cystic fibrosis. A global genome-wide transposon mutagenesis screen for non-mucoid isolates of a mucoid strain led to the identification of ClpX, ClpP1, and ClpP2 as regulators of alginate production. Null mutants of clpX, clpP1 or clpP2 resulted in reduced alginate production in the non-mucoid strain, thereby implicating the role of Clp proteins in biofilm formation. It was found that ClpXP is required for the degradation of MucA, a negative regulator of the sigma factor AlgU (Table 1). By preventing the repression of AlgU by MucA, AlgU is available to bind the algD operon and initiate the transcription of genes involved in alginate production (Qiu et al., 2008).
Similarly, in an S. epidermidis ΔclpP strain, a decrease in virulence and biofilm formation was observed in a rat model of intravascular catheter-associated infection (Wang et al., 2007). Further investigation attributed this effect to increased levels of Spx, which also regulates exopolysaccharide production (Table1). It is thought that increased Spx levels leads to defects in the pathogen’s ability to initiate attachment, necessary for the early stages of biofilm establishment (Wang et al., 2010). Interestingly, in S. aureus, clpP deletion appears to enhance biofilm formation (Frees et al., 2004). However, S. aureus ΔclpP mutant CFU counts in a mouse model 7 days post infection were significantly reduced relative to wildtype S. aureus, suggesting that increased biofilm formation cannot compensate for the reduction in ClpP-mediated bacterial virulence and dissemination in the host (Liu et al., 2017).
ClpP deletion appears to decrease the effectiveness of a pathogen’s innate stress responses against temperature changes. For example, in Legionella pneumophila, the causative agent of Legionnaires’ disease, ΔclpP mutants exhibited growth deficiency at 42°C, as well as reduced tolerance to heat shock treatment compared to the wild type bacterium (Li et al., 2010). These cells presented incomplete division and exhibited compromised colony formation. Similar findings were seen in a Streptococcus pneumoniae clpP knockout strain, where bacterial growth was severely impaired at 30°C, 37°C, and 43°C. Considering that this pathogen undergoes a drastic change in temperature as it enters the host bloodstream from the nasopharynx, it was thought that the survival of mice challenged with ΔclpP S. pneumoniae was ameliorated due to the role of ClpP in the heat shock response (Kwon et al., 2003; LaBreck et al., 2017). Furthermore, ClpP was also required for growth at 42°C in Campylocabter jejuni, a food-borne pathogen, whereby a ΔclpP mutant displayed an increase in the levels of misfolded protein aggregates (Cohn et al., 2007). Like S. pneumoniae, C. jejuni must maintain proper proteostasis to be able to overcome a drastic shift in temperatures during its life cycle to survive in the environment, as well as in avian carriers (42°C) and human hosts (37°C). Finally, in Salmonella typhimurium, the etiological agent of gastroenteritis, ΔclpP mutant growth was impaired in experiments subjecting the bacterium to high and low temperatures, high salt, and low pH conditions (Thomsen et al., 2002; Knudsen et al., 2014). Overall, the above observations appear to implicate the role of ClpP in removing misfolded proteins during abrupt temperature variations, thereby promoting bacterial virulence and resilience in the host.
A common phenotype seen amongst pathogens that are deficient in protein homeostasis mechanisms is a lowered tolerance towards pH changes. Such pathogens are particularly susceptible to the phagocytic macrophages, which are part of the innate immune response. Upon the engulfment of a pathogen, macrophages acidify the phagosome and produce reactive oxygen species for bacterial killing (Westman and Grinstein, 2020). There are many studies that implicate the role of ClpP in the viability of intracellular pathogens following internalization by host cells. For instance, it was shown that S. typhimurium (Yamamoto et al., 2001), L. monocytogenes (Gaillot et al., 2000), L. pneumophila (Zhao et al., 2016), S. aureus (Kim et al., 2020) and S. pneumoniae (Kwon et al., 2004) deleted of clpP exhibited impaired growth in macrophages, suggesting that ClpP is required for intracellular survival. Further studies on S. pneumoniae indicated that ΔclpP mutants exhibited increased sensitivity to oxidative stress and that treatment of macrophages by the nitric oxide synthase inhibitor S-methylisothiourea sulfate, led to a significant increase in ΔclpP S. pneumoniae viability (Lee et al., 2010). In addition, ΔclpP S. pneumoniae were found to stimulate apoptosis in dendritic cells less than their wildtype counterpart (Cao et al., 2013). Dendritic cells are antigen presenting cells that bridge the innate and adaptive immune responses together and are crucial for conferring long-term protection against pathogens (Banchereau et al., 2000). Thus, disruption of this network along with resistance against oxidative stress sheds light on the role of ClpP in promoting S. pneumoniae survival in the host. Interestingly, a similar observation was made for S. aureus, whereby a genome wide transposon screen revealed that ΔclpP strains showed a decrease in neutrophil lysis and that ClpP was necessary for growth and survival in a zebrafish embryo infection model (Yang et al., 2019). Therefore, ClpP seems to be implicated in modulating host cell immunity, thus disrupting the function of this protease in various intracellular pathogens confers protection against disease in the host.
The bacterial toxin-antitoxin (TA) system consists of a constitutively active toxin and its cognate inhibitory, albeit labile antitoxin. TA modules are highly complex with eight systems characterized thus far (Types I-VIII) based on the composition of the antitoxin (RNA or protein) (Singh et al., 2021). During regular physiological conditions, the antitoxin is stable and inhibits the function of the toxin. However, during stress conditions, the antitoxin is degraded allowing the toxin to interact with effector proteins or DNA rendering a bacterial cell in a persistent dormant-like state. This is thought to assist the pathogen in resisting assault from antibiotic treatments, phage infections, and host immune defense mechanisms. (Harms et al., 2018; Singh et al., 2021). In type II TA modules, proteases such as ClpP stimulate this system by degrading the antitoxin, which causes the unrepressed toxin to inhibit translation and DNA replication, thereby promoting growth arrest (Page and Peti, 2016). One of the most well-documented type II TA modules is the MazEF system, which is comprised of the MazE antitoxin that inhibits the mRNA-cleaving endoribonuclease toxin MazF (Figure 3A; Table 1) (Zhang et al., 2003). In E. coli, ClpAP was shown to degrade MazE during starvation (Aizenman et al., 1996) to promote persister cell formation and survival following antibiotic treatment (Tripathi et al., 2014). In S. aureus, ClpCP degrades MazE (Donegan et al., 2010) but requires the adaptor TrfA to mediate this degradation (Figure 3A; Table 1) (Panasenko et al., 2020). Nevertheless, the downstream effects are the same as for E. coli, whereby the mazEF module in S. aureus is stimulated during oxidative and antibiotic stress to promote bacteriostasis (Fu et al., 2009; Panasenko et al., 2020). Deletion of either gene in the mazEF module or in the clpCP operon resulted in a significant reduction in persister cell formation and viability following antibiotic treatment (Schuster et al., 2015; Springer et al., 2016), demonstrating the role of ClpP-mediated proteostasis during stress.
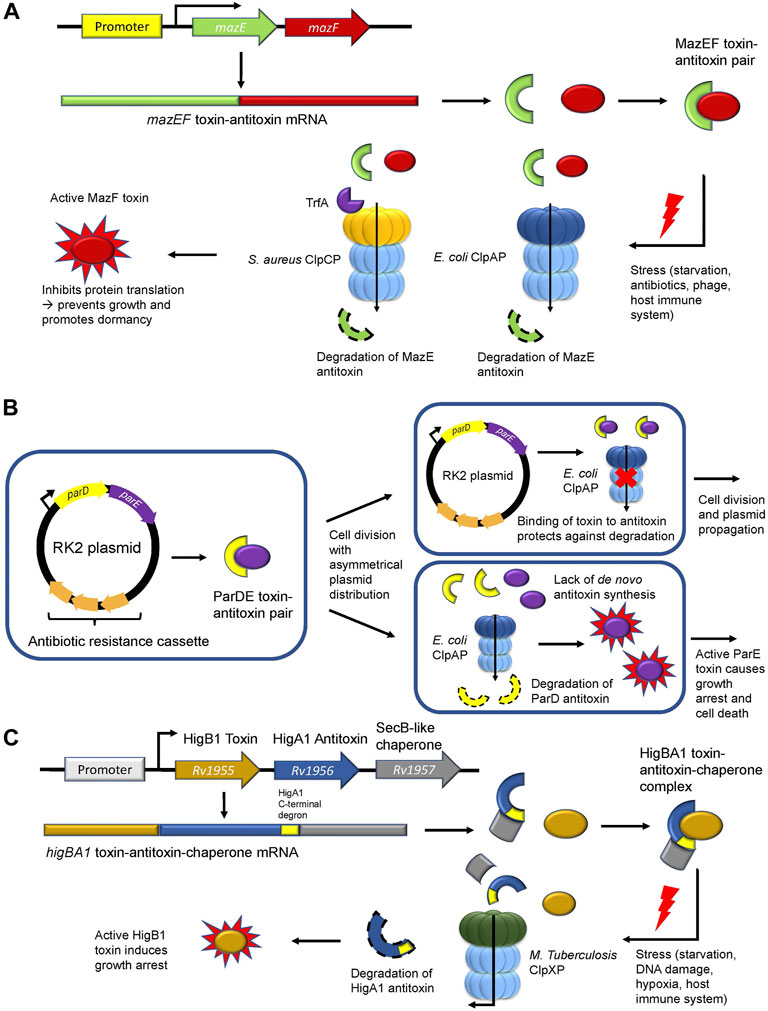
FIGURE 3. ClpP regulates the type II toxin-antitoxin system. (A) The MazEF toxin-antitoxin module. The MazE antitoxin (green) binds and inhibits the MazF toxin (red). During stress, ClpAP (E. coli) or ClpCP in association with TrfA (S. aureus) degrades the MazE antitoxin. Free MazF, which is an mRNA endoribonuclease, cleaves various intracellular targets leading to bacterial growth arrest. (B) The parDE toxin-antitoxin module. The RK2 plasmid encodes for the ParD antitoxin (yellow) and the ParE toxin (purple) and harbors an antibiotic resistance cassette (orange). Following asymmetrical cell division with unequal plasmid distribution to daughter cells, one of the progenies may not inherit any RK2 plasmid copies. Without the ability to synthesize the ParD antitoxin de novo, the antitoxin is degraded by ClpAP leading to free ParE toxin, which causes growth arrest. (C) The toxin-antitoxin-chaperone (TAC) module in M. tuberculosis. The SecB-like chaperone (grey) binds the C-terminal region (yellow) of the HigA1 antitoxin (blue) and assists in its folding and stabilization. HigA1 in turn binds and inhibits the HigB1 toxin (light brown). During stress, the SecB-like chaperone dissociates from HigA1 via an unknown signalling mechanism causing M. tuberculosis ClpXP to degrade the HigA1 antitoxin via the recognition of the now unmasked C-terminal degron. Free HigB1 toxin induces growth arrest causing the bacteria to enter a state of dormancy.
The parDE system found in RK2 plasmids is another Type II TA module in E. coli that promotes bacterial persistence indirectly through plasmid maintenance in the bacterial population (Figure 3B). This system is comprised of the antitoxin ParD and the DNA gyrase inhibitor toxin ParE (Jiang et al., 2002) (Table 1). Following unequal cell division, a daughter cell may receive excess RK2 plasmids while the other daughter cell would be devoid of any RK2 plasmids. Given that these cells cannot synthesize the ParD antitoxin de novo, ClpAP degrades the remaining inherited ParD antitoxin, thereby causing ParE to inhibit DNA gyrase and promote growth arrest or cell death (Karlowicz et al., 2016; Dubiel et al., 2018). Maintenance of the RK2 plasmid ensures that the antibiotic resistance cassette encoding for kanamycin, ampicillin, and tetracycline found in this plasmid is retained in the population, thus enabling bacterial resistance against multi-drug treatments (Thomas et al., 1980; Villarroel et al., 1983).
Recently, the role of ClpXP in the regulation of the rv1955-rv1957 TA system in M. tuberculosis was elucidated (Texier et al., 2021). The rv1955-rv1957 operon encodes for an atypical toxin-antitoxin-chaperone (TAC) tripartite module consisting of HigB1 (toxin), HigA1 (antitoxin) and a SecB-like chaperone (Figure 3C; Table 1). The latter recognizes and binds a C-terminal region of the HigA1 antitoxin and assists in its folding (Bordes et al., 2016). In doing so, folded HigA1 antitoxin binds and inhibits the HigB1 toxin, preventing it from exerting its toxic effect. The C-terminal region also encompasses the degron sequence by which ClpXP recognizes HigA1 for degradation. Therefore, the binding of the chaperone to the C-terminal region of HigA1 masks the degron from recognition by ClpXP, thereby, inhibiting HigA1 turnover (Texier et al., 2021). However, following stress and through an unknown signalling mechanism, the SecB-like chaperone disengages from HigA1. Subsequently, ClpXP recognizes the C-terminal degron sequence on HigA1 and degrades it, thereby freeing HigB1 to exert its downstream toxic effect (Figure 3C) (Texier et al., 2021). The consequence of the HigB1 toxin activity includes growth inhibition (Schuessler et al., 2013). This TA module was shown to be upregulated in cell persisters (Keren et al., 2011), during starvation (Betts et al., 2002), DNA damage (Rand et al., 2003), hypoxia, and engulfment by phagocytes (Ramage et al., 2009), allowing the pathogen to withstand extreme environments. Such pathogens can become reactivated when conditions are favorable (Wayne and Sohaskey, 2001) and cause disease relapse, thereby highlighting the crucial role of this TA module during M. tuberculosis infection and its regulation by ClpXP.
In a recent proteomic screen and follow-up in vitro degradation assays, M. tuberculosis ClpCP was shown to degrade the VapB20 and RelB1 antitoxins, which bind and inhibit the VapC20 and RelE1 toxins, respectively (Ziemski et al., 2021) (Table 1). VapC20 enables growth arrest by cleavage of the Sarcin-ricin loop found on the 23 S ribosomal RNA, thereby, inhibiting protein translation (Winther et al., 2013). On the other hand, the RelE1 toxin is upregulated post-macrophage engulfment and nitrogen starvation (Korch et al., 2009; Korch et al., 2015). The activity of RelE1 compromises the structural integrity of the mycobacterial envelope and degrades mRNA, thereby, inhibiting protein translation and altering the proteome. Overall, ClpCP and ClpXP are essential in enabling cell persistence of M. tuberculosis by regulating the steady-state levels of antitoxins under a variety of environmental and biological stresses.
ClpP plays an important role in facilitating the switch between the different life stages of certain bacteria. Pathogens such as L. pneumophila greatly rely on ClpP to transition between a replicative phase (RP) and an effector secreting virulent transmissive phase (TP) (Ge et al., 2022). One such regulator of this transition is CsrA (Table 1), whereby its accumulation in ΔclpP L. pneumophila mutants both inhibited the initiation of the non-virulent phase and reduced invasiveness to Acanthamoeba castellanii amoebae in vitro. Further investigation showed that the temporally expressed IHFB (Table 1), the transcriptional inhibitor of csrA, is degraded in a ClpP-dependent manner during the non-virulent phase. Thus, ClpP appears to be involved in regulating the transition both into and out of the virulent stage of L. pneumophila (Ge et al., 2019).
A similar switch between two life stages also occurs in the obligate intracellular pathogen C. trachomatis, which transitions between elementary bodies (EBs) and reticulate bodies (RBs). EBs are the non-dividing infectious form, while RBs are the replicative non-infectious form (Elwell et al., 2016). It was found that transcripts of clp genes (clpP1, clpP2, clpX, and clpC) were greatly upregulated during the RB stage of the pathogen, suggesting that they play a role in cell cycle progression (Wood et al., 2019). A follow-up study showed that a ΔclpP2 mutant disrupted the transition from the RB stage to the EB stage, while a ΔclpX mutant caused a morphological defect in EBs and decreased their viability (Wood et al., 2020).
Moreover, while no regulatory substrates were identified, ClpP was found to be required for sporulation in C. difficile (Bishop et al., 2022). The virulence and resilience of this pathogen to environmental stresses is critically dependent on spore formation (Shen, 2020). Spores formed from C. difficile clpP1 and clpP2 null mutants were found to be generally heat labile, morphologically abnormal, and they germinated significantly less than their wildtype counterpart (Bishop et al., 2022). Therefore, ClpP appears to be involved in regulating the transition between different life stages of several pathogens to modulate their virulence.
ClpP can affect virulence by regulating the transcription of specific virulence factors. In L. monocytogenes, it was found that ClpP2 is required specifically for the expression of Listeriolysin O (Table 1), a hemolytic toxin that forms pores in phagocytic vacuoles, allowing the pathogen to escape and propagate (Figure 4A) (Gaillot et al., 2000). Untangling these pathways into a linear “cause and effect” is a difficult task. However, it is likely that the reduced stress resistance in ΔclpP2 L. monocytogenes was the cause of the pathogen’s poor growth in mice, while reduced expression of Listeriolysin O resulted in its inability to propagate and cause infection. Similar transcriptional regulatory pathways have been shown to be modulated by ClpP in S. pneumoniae, whereby deletion of clpP reduced survival in infected mice (Kwon et al., 2003). Transcriptional analysis revealed that the ΔclpP mutants demonstrated decreased expression of the hemolytic pore-forming pneumolysin toxin and the adhesin pneumococcal surface antigen A (PsaA) (Table 1), suggesting that ClpP is a positive regulator of these virulence factors.
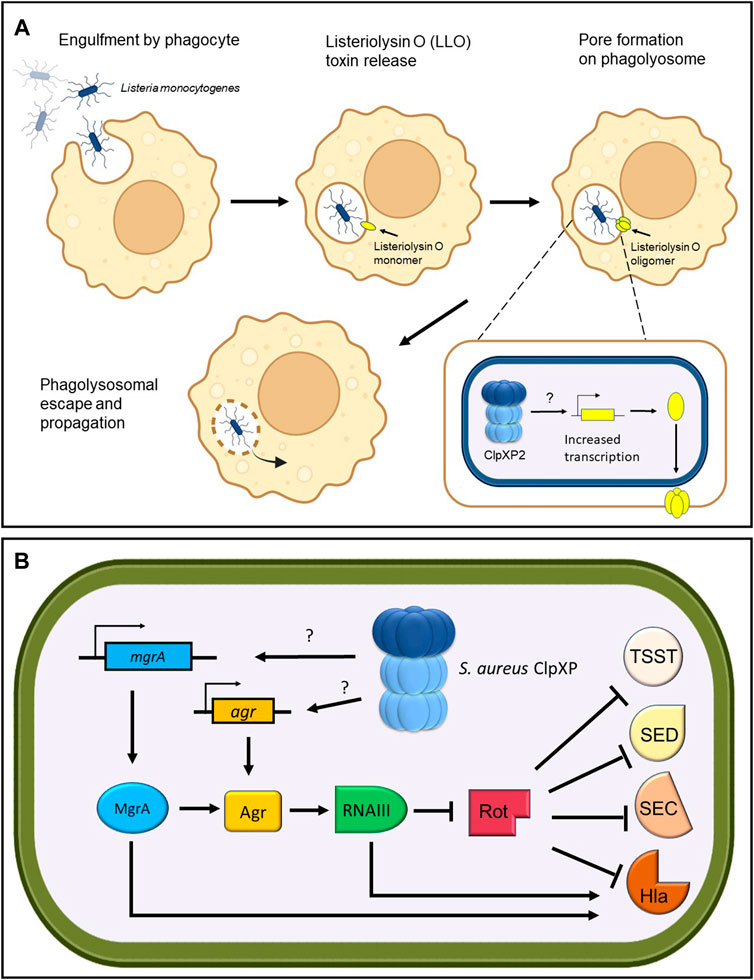
FIGURE 4. ClpP regulates toxin production. (A) Listeria monoctyogenes ClpXP2 transcriptionally increases the production of Listeriolysin O toxin for phagolysosomal escape and propagation. (B) S. aureus ClpXP regulates the production of toxic shock syndrome toxin (TSST), enterotoxin D (SED), enterotoxin C (SEC) and hemolysin alpha-toxin (Hla) through transcriptional control of the global master regulator mgrA and the agr quorum sensor. Through an unknown target, S. aureus ClpXP stimulates the transcription of mgrA and agr leading to RNAIII upregulation, which in turn inhibits the repressor of toxins (Rot). The inhibition of Rot enables the production of Tsst, Sed, Sec, and Hla toxins. In addition, MgrA and RNAIII can directly activate the production of Hla.
One of the most well-studied examples that implicates ClpP within a complex regulatory network that controls virulence factor expression stems from extensive studies on S. aureus. The Agr quorum sensor in S. aureus controls the expression of virulence factors ultimately through the transcription of RNAIII, a regulatory RNA effector of the agr regulon (Figure 4B). RNAIII in turn blocks the expression of the repressor of toxins (Rot) and stimulates production of toxins such as the pore-forming hemolysin alpha-toxin (Hla) (Table 1). Repression of rot also allows the transcription of virulence factor genes such as toxic shock syndrome toxin (tsst), enterotoxin C (sec) and enterotoxin D (sed) (Fisher et al., 2018; Jenul and Horswill, 2019; Ju et al., 2021) (Table 1). S. aureus ΔclpXP mutants showed attenuated virulence in a murine skin abscess model, whereby hemolysin alpha-toxin protein and expression levels were reduced concomitant with a reduction in agr and RNAIII transcripts (Frees et al., 2003; Stahlhut et al., 2017). Therefore, this suggests that ClpXP positively regulates hla expression through the agr locus (Frees et al., 2003). Interestingly, in S. aureus clpXP null mutants, expression of tsst, sed and sec were not affected despite the reduction in Rot protein levels. However, the global master regulator mgrA was downregulated (Schelin et al., 2020) (Table 1). MgrA is known to modulate the activity of more than 350 genes (Luong et al., 2006) and can stimulate Hla production in a dual fashion-agr dependent and agr independent (Ingavale et al., 2005). Therefore, this implies that tsst, sed, and sec expression might be postively regulated by ClpXP through the upregulation of mgrA, but further investigation is required.
Virulence factors can be secreted into the extracellular environment or directly into host cells. Typically, this is accomplished through specialized transport and secretion systems or both. These secretion systems (Types I-VII) use a single energy-coupled step to transport proteins across membranes and are important for injecting bacterial virulence factors out into the extracellular environment or directly into host cells, causing a signal transduction cascade. The consequence of this may include disruption of essential signalling pathways and cytoskeletal remodelling (Asrat et al., 2015; Green and Mecsas, 2016). Expression and turnover of these bacterial secretion systems are regulated by ClpP in several pathogens. Recently, clpP deletion in L. pneumophila was shown to downregulate and upregulate an extensive array of type IVB secretion system and effector proteins. These proteins were differentially expressed depending on the life cycle stage of the bacterium (replicative or transmissive). Notably, of the 428 differentially expressed proteins, 316 were found to be modulated in a ClpP-dependent manner suggesting that ClpP plays a major role in manipulating host cell machinery through these secretion systems and effector proteins (Ge et al., 2022).
Additionally, ClpP appears to directly regulate Type III secretion system (T3SS) genes in Yersinia pestis (Jackson et al., 2004) (Figure 5). In this pathogen, expression of the T3SS apparatus is mediated by the transcriptional activator LcrF and is repressed by a histone-like protein termed YmoA (Schwiesow et al., 2015) (Table 1). Deletion of clpP in Y. pestis caused the pathogen to express a non-functional T3SS, which was revealed to be, in part, due to the upregulation of YmoA. Therefore, this suggests that YmoA is a target of ClpXP, and that its proteolytic degradation enables LcrF to express the genes found in the T3SS apparatus (Figure 5) (Jackson et al., 2004).
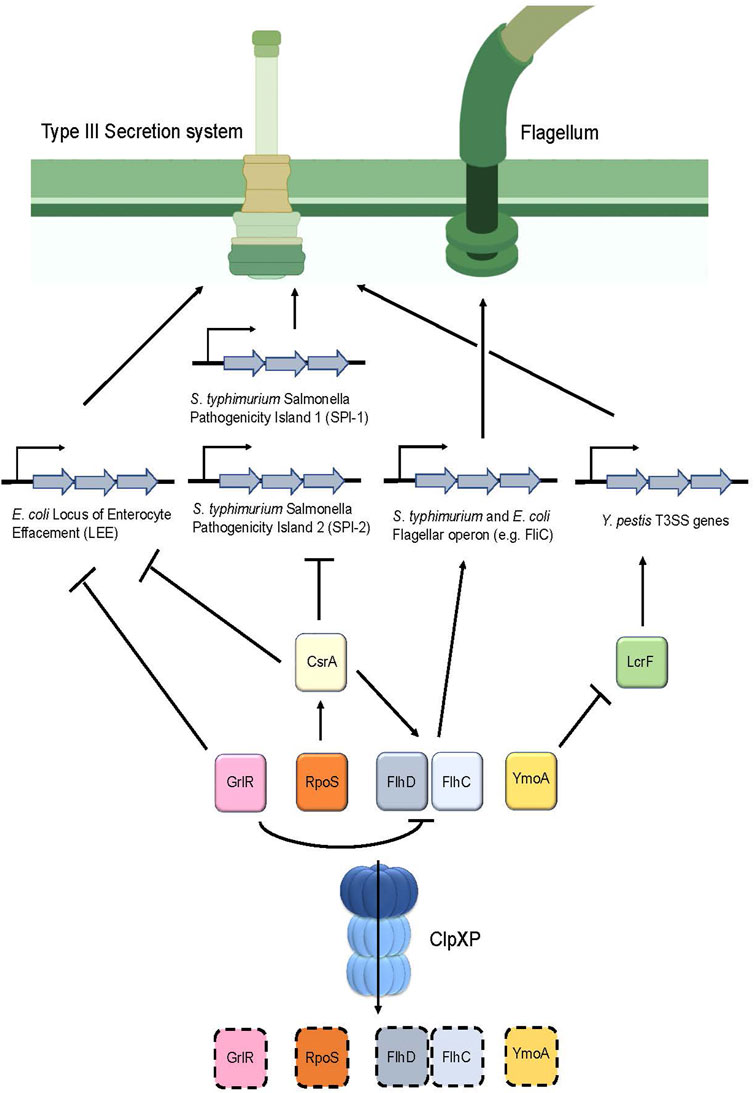
FIGURE 5. ClpP regulates the assembly of the Type III secretion system as well as flagellum synthesis in various pathogens. Degradation of the repressor GrlR by ClpXP removes the inhibition of transcription of the Locus of the Enterocyte Effacement (LEE) genes, which encode for the T3SS apparatus in E. coli. In addition, degradation of GrlR prevents repression of FlhD and FlhC, the master regulators of flagellar genes, thereby causing flagellum synthesis to occur. Furthermore, RpoS turnover by ClpXP prevents upregulation of CsrA, thereby causing expression of LEE in E. coli and Salmonella Pathogenicity Islands 1 and 2 in S. typhimurium. Additionally, CsrA positively regulates FlhD and FlhC expression, therefore, turnover of RpoS by ClpXP ultimately results in FlhD and FlhC downregulation through CsrA, leading to decreased flagellar synthesis. In Y. pestis, degradation of the repressor YmoA enables LcrF to stimulate production of the T3SS apparatus.
A similar mechanism involving the regulation of the T3SS by ClpP was found in S. typhimurium (Figure 5). This bacterium possesses two distinct T3SS apparatuses that are encoded by two separate Salmonella pathogenicity islands (SPI-1 and SPI-2) for invasion and pathogenesis, respectively (Pizarro-Cerda and Cossart, 2006). There are several regulators that control these islands; noteworthy are the two negative regulators: the sigma factor RpoS (Rice et al., 2015) and the global RNA binding protein CsrA (Martinez et al., 2011) (Table 1). ClpP deletion in S. typhimurium showed that rpoS and csrA mRNA expression were upregulated. Further, csrA mRNA expression was reduced in the rpoS and clpP/rpoS double deletion mutants, suggesting that csrA is modulated by RpoS (Knudsen et al., 2013). Based on these results, and the observation that RpoS is a substrate of ClpXP in E. coli, (Pratt and Silhavy, 1996; Schweder et al., 1996), it is thought that ClpP in S. typhimurium positively controls the expression of the T3SS by regulating csrA levels indirectly through RpoS turnover (Figure 5).
In E. coli, attaching and effacing lesions in the intestinal mucosa produced by this pathogen are encoded on a pathogenicity island termed the locus of enterocyte effacement (LEE). These genes encode for chaperones, structural and effector proteins that comprise the E. coli T3SS (Furniss and Clements, 2018). LEE appears to be positively regulated by ClpXP as evidenced by a significant reduction of the LEE-encoded Esp proteins following clpXP deletion. Further analysis indicated that ClpXP modulates the expression of LEE via two distinct mechanisms (Figure 5). First, deletion of the LEE negative regulator grlR showed a comparable increase of esp expression in both the ΔgrlR mutants and in the ΔgrlR/ΔclpXP mutants, indicating that grlR expression is under the control of ClpXP (Iyoda and Watanabe, 2005). Second, deletion of the sigma factor rpoS, a known substrate of ClpXP (Schweder et al., 1996) in the clpXP mutants, resulted in a partial increase in esp expression (Iyoda and Watanabe, 2005). rpoS overexpression through insertion of a multicopy plasmid in these ΔrpoS/ΔclpXP mutants significantly repressed esp expression. Taken together, these results suggest that ClpXP positively modulates LEE expression through RpoS and GrlR turnover (Figure 5; Table 1).
Bacterial motility is essential for pathogens to navigate through the environment to obtain nutrients and to seek a host for infection and colonization. Motility strongly contributes to the disease-causing capabilities of a pathogen, with those lacking such mechanisms exhibiting attenuated virulence (Josenhans and Suerbaum, 2002). Given that the T3SS injectisome largely resembles the supramolecular structure of the flagellar complex, the T3SS is likely derived from a flagellar common ancestor (Diepold and Armitage, 2015). It is postulated that ClpXP regulates motility by modulating flagellum synthesis. Indeed, clpXP deletions in S. typhimurium, resulted in cells exhibiting a “hyperflagellated” phenotype characterized by hypermobility and an increased rate of fliC transcription that encodes for flagellin, a subunit of the flagellum filament (Tomoyasu et al., 2002) (Figure 5; Table 1). Further analysis showed that clpXP deletions increased and stabilized the half-lives of the FlhD/FlhC proteins, which are the master regulators that encode for all the genes in the flagellum complex. Therefore, ClpXP acts as a negative regulator of flagellum synthesis by post-translationally regulating FlhD/FlhC turnover (Tomoyasu et al., 2003) (Figure 5; Table 1). The role of ClpXP in regulating flagellar synthesis was also reported in E. coli. ClpXP deletion led to an upregulation of fliC expression (Iyoda et al., 2006), as well as FlhD/FlhC accumulation and stabilization (Kitagawa et al., 2011). Moreover, deletion of grlR led to the downregulation of flhD transcripts and, correspondingly, fliC expression (Iyoda et al., 2006). Given that ClpXP was also shown to downregulate GrlR protein levels (Iyoda and Watanabe, 2005), this suggests that ClpXP negatively regulates flagellar synthesis directly through FlhD/FlhC and GrlR turnover in E. coli (Figure 5).
Currently there is a lack of mechanistic studies implicating ClpXP in P. aeruginosa motility. Nevertheless, phenotypic effects were observed upon clpXP deletion whereby twitching, swimming and swarming motility were impaired (Shanks et al., 2006; Fernandez et al., 2012). Importantly, ClpP1 but not ClpP2 is required for both swimming and twitching motility (Hall et al., 2017). Swimming motility refers to the individual movement of bacteria, whereas swarming motility refers to bacteria that migrate en masse; both types mediated by flagellar rotation. In twitching motility, bacteria migrate on surfaces through attachment and retraction of the pili appendages (Kearns, 2010). Overall, ClpXP appears to be involved in the regulation of flagellar and pilus synthesis in P. aeruginosa, but further studies are needed to elucidate signalling pathways and regulatory mechanisms that are at play.
In the previous sections, the role of ClpP and its cognate ATPases in pathogenesis was highlighted, demonstrating that the protease not only influences a pathogen’s survivability, but also its infectivity potential and virulence. As such, much research has gone into the discovery and optimization of compounds that can interfere with the regulatory roles of ClpP and its associating ATPases.
Chemical alteration of ClpP activity has been shown to affect the virulence and infectivity of several pathogens (Conlon et al., 2013; Brӧtz-Oesterhelt and Sass, 2014; Silber et al., 2020). One strategy employed is the use of compounds such as β-lactones/lactams, phenyl esters, peptide boronic acids, and clipibicyclene as ClpP inhibitors (Gersch et al., 2013; Akopian et al., 2015; Hackl et al., 2015; Culp et al., 2022). These compounds typically bind to the active site serine of ClpP to prevent proteolysis. The mechanism of action of β-lactones/lactams, phenyl esters and peptide boronic acids on ClpP were comprehensively discussed in these reviews from our group and others (Culp and Wright, 2017; Bhandari et al., 2018; Moreno-Cinos et al., 2019; Mabanglo and Houry, 2022).
The unique structure and regulation of the ATPase-ClpP complexes allows for an additional mode of chemical interference of ClpP activity. Acyldepsipeptides (ADEPs) and activators of self-compartmentalizing proteases (ACPs) have been shown to bind the hydrophobic pockets of ClpP that are normally occupied by the ATPase’s IGF loops (Brӧtz-Oesterhelt et al., 2005; Martin et al., 2007; Lee et al., 2010; Leung et al., 2011; Goodreid et al., 2014; Goodreid et al., 2016; Mabanglo et al., 2019; Binepal et al., 2020) (Figure 1B). This binding results in the widening of the tetradecameric ClpP axial pores, which facilitates substrate entry and increases proteolytic activity (Li et al., 2010a). Association of the drugs to ClpP disrupts the chaperone-ClpP interaction while keeping ClpP in its active conformation, resulting in unregulated proteolysis (Mroue et al., 2019; Jacques et al., 2020). These compounds have been shown to induce promiscuous degradation of non-substrate proteins both in vitro and in vivo, thereby resulting in cell death (Kirstein et al., 2009; Li et al., 2010a) and validating ClpP as a promising antimicrobial target. It is worth noting that in firmicutes (e.g., Bacillus, Staphylococcus, and Streptococcus), ClpP is not essential for cell viability. The bactericidal activity of ADEPs is mainly due to unregulated proteolysis of substrates by ClpP such as FtsZ, which is required for cell division (Sass et al., 2011). However in M. tuberculosis, clpP1 deletion leads to cell death making it an essential component of the proteostasis network (Ollinger et al., 2012); accordingly, the mechanism by which ADEPs induce cell death in mycobacteria is different. While ADEPs can stimulate degradation of peptides and full-length proteins in M. tuberculosis, its effect is weaker than in firmicutes and other bacteria. Rather, the binding of ADEPs inhibits the association of ClpP1P2 with the ATPases ClpX or ClpC1 to prevent essential protein turnover in mycobacteria, thus, leading to cell death (Famulla et al., 2016).
It may seem like a negative trait for an antibacterial target to possess multiple paralogs and regulatory pathways that can compensate for each other. However, this can be advantageous because disruption of the ClpP-chaperone complex’s activity can increase the bacterium’s susceptibility to the host’s immune system and to other antibiotics without imposing strong evolutionary pressure, which gives rise to resistant mutants. Indeed, co-administering traditional antibiotics with ClpP dysregulators has been shown to be effective at killing otherwise resistant mutants. Incubating high density cultures of methicillin resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus faecalis (VRE) with linezolid, ampicillin, oxacillin, or ADEPs individually, showed that none of the compounds had bactericidal activity against either of the two species. However, co-incubation of linezolid, ampicillin, or oxacillin with ADEPs was found to be effective at killing and preventing regrowth of the pathogens, with no evidence of developing resistant mutants (Mroue et al., 2019).
However, targeting ClpP function as an effective anti-virulence strategy with small molecule compounds has some caveats. In addition to the above-mentioned S. aureus infection model, a study treating L. monocytogenes with β-lactones in mouse macrophages showed a significant reduction in intracellular replication. Follow up analyses confirmed that this was due to the compound binding to ClpP (Bottcher and Sieber, 2009). Here, disruption of ClpP function was seen to decrease virulence. In contrast, deletion of clpX and clpP from MRSA led to increased resistance to β-lactam antibiotics (Baek et al., 2014). These results suggested that ClpXP controls one or more of the pathways modulating β-lactam resistance in S. aureus. MRSA expresses a peptidoglycan transpeptidase, penicillin binding protein 2a (PBP2a), which has decreased affinity for β-lactams compared to other PBPs enabling resistance against β-lactam antibiotics (Baek et al., 2014). In the clpXP knockout mutants, cellular levels of PBP2a were found to be unchanged suggesting that PBP2a mediated resistance against β-lactam antibiotics is still intact. Nevertheless, it was noted that these mutants possessed generally thicker cell walls consisting of an altered muropeptide composition and increased levels of crosslinking (Baek et al., 2014). Therefore, it is likely that the role of ClpXP in the peptidoglycan synthesis pathway is multifaceted, which led to this undesirable phenotype (Baek et al., 2014).
Discrepancies in drug effectiveness like the ones described above may also in part be due to the various functional forms of ClpPs found within different bacterial species. Specific ClpP paralogs are known to serve different functions, and the expression levels of each paralog have been shown to fluctuate through various growth stages for some pathogens. For example, in P. aeruginosa, clpP1 and clpX were observed to be constitutively expressed throughout the cell growth cycle, while clpP2 expression was shown to increase during the stationary phase (Hall et al., 2017). Similar differences between paralogs have been recorded for other species such as L. monocytogenes and C. difficile (Balogh et al., 2017; Lavey et al., 2019; Mawla et al., 2021). Furthermore, variations in encoded ATPases could contribute to these discrepancies by minimizing the effects of dysregulating any specific chaperone. The redundancy of ClpC and ClpE suggests that many of the pathways dysregulated by inhibiting ClpC could be compensated for by the presence of ClpE except in M. tuberculosis, which lacks the latter Clp ATPase. ClpC1 dysregulators in M. tuberculosis such as ecumicin and lassomycin possess bactericidal properties by enhancing the ATPase activity of ClpC1 and uncoupling its proteolytic activity from ClpP1P2 (Gavrish et al., 2014; Gao et al., 2015). By dysregulating proteostasis and causing cell death, ClpC1 appears to be a viable drug target against M. tuberculosis. Nevertheless, how the overlapping functions of multiple paralogs or chaperones factor into the efficacy of antibiotics in other species is not well understood. Further research is needed to design and administer effective antibiotics that are specific towards particular pathogens, accounting for variations in ClpP and chaperones.
An additional consideration with respect to drug design is the presence of a ClpP in the mitochondrion of eukaryotes. Novel antibiotic inhibitors and activators therefore must be highly specific for prokaryotic ClpP, so as not to target human ClpP. Indeed, it has been shown that mitochondrial ClpP is also dysregulated by ADEPs (Lowth et al., 2012; Wong et al., 2018).
In summary, ClpP and its cognate ATPases affect bacterial virulence through its involvement in peptidoglycan and biofilm formation, cell stress tolerance, motility, defense against host immune responses, the shift to a virulent life stage, and the production and transport of virulence factors. Although the presence of multiple ClpPs and the redundancy of Clp chaperones suggest that the protease is not essential for pathogen survival, chemical compounds that dysregulate ClpP have been shown to induce anti-virulence effects and increase bacterial susceptibility towards other antibiotics. It is worth noting that most contemporary antimicrobial compounds targeting ClpP were tested using S. aureus or E. coli, necessitating further investigation into other pathogens as well. Nevertheless, ClpP is a desirable druggable target and future research should be aimed at optimizing ClpP antimicrobial compounds in order to validate their use in animal models and eventually humans.
MEA-writing, editing, making figures. MMB-conceptualization, writing, editing. EM-conceptualization, writing, editing. VB-writing, making Figure 2A. WAH-conceptualization, writing, editing, securing funding.
MEA is the recipient of the Canadian Institutes of Health Research (CIHR) Canada Graduate Scholarships – Master’s program (CGS M) graduate fellowship. MMB is supported by an Ontario Graduate Scholarship (OGS) and a Dorthy Sterling Dow Walsh Award from the University of Toronto. VB is the recipient of an OGS and previously held the Natural Sciences and Engineering Research Council of Canada’s (NSERC) Postgraduate Scholarship-Doctoral (PGS-D) award and a Jaro Sodek Award—Ontario Student Opportunity Trust Fund (OSOTF) fellowship from the Department of Biochemistry at the University of Toronto. This work was supported by a CIHR Project grant (PJT-148564).
The literature review process was in part facilitated by a text mining technique done by Dr. Sadhna Phanse, which correlated word frequency profiles in PubMed. “ClpP” and “infection” or “pathogen” or “virulence” were found to have direct connections in the correlation profiles ranging from 0.2 to 0.3. Approximately 400 articles were found in this manner, aiding in compiling, to our knowledge, a complete representation of the field. The following figures were drawn in PowerPoint: the ClpXP cartoon in the Graphical abstract, Figure 1C, Figure 2A, the ClpXP cartoon in Figure 2B, Figure 3, the ClpXP cartoon in Figures 4A,B, and Figure 5. The following figures were generated using UCSF ChimeraX: Figure 1A,B. The following figures were made using BioRender (https://biorender.com): Figure 2B, the plasmid in Figure 3B, Figure 4A, and the flagellum and T3SS apparatus in Figure 5.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aizenman, E., Engelberg-Kulka, H., and Glaser, G. (1996). An Escherichia coli chromosomal "addiction module" regulated by guanosine [corrected] 3', 5'-bispyrophosphate: A model for programmed bacterial cell death. Proc. Natl. Acad. Sci. U. S. A. 93, 6059–6063. doi:10.1073/pnas.93.12.6059
Akopian, T., Kandror, O., Raju, R. M., Unnikrishnan, M., Rubin, E. J., and Goldberg, A. L. (2012). The active ClpP protease from M. tuberculosis is a complex composed of a heptameric ClpP1 and a ClpP2 ring. EMBO J. 31, 1529–1541. doi:10.1038/emboj.2012.5
Akopian, T., Kandror, O., Tsu, C., Lai, J. H., Wu, W., Liu, Y., et al. (2015). Cleavage specificity of Mycobacterium tuberculosis ClpP1P2 protease and identification of novel peptide substrates and boronate inhibitors with anti-bacterial activity. J. Biol. Chem. 290, 11008–11020. doi:10.1074/jbc.M114.625640
Amor, A. J., Schmitz, K. R., Baker, T. A., and Sauer, R. T. (2019). Roles of the ClpX IGF loops in ClpP association, dissociation, and protein degradation. Protein Sci. 28, 756–765. doi:10.1002/pro.3590
Asrat, S., Davis, K. M., and Isberg, R. R. (2015). Modulation of the host innate immune and inflammatory response by translocated bacterial proteins. Cell. Microbiol. 17, 785–795. doi:10.1111/cmi.12445
Baek, K. T., Grundling, A., Mogensen, R. G., Thogersen, L., Petersen, A., Paulander, W., et al. (2014). β-Lactam resistance in methicillin-resistant Staphylococcus aureus USA300 is increased by inactivation of the ClpXP protease. Antimicrob. Agents Chemother. 58, 4593–4603. doi:10.1128/AAC.02802-14
Balogh, D., Dahmen, M., Stahl, M., Poreba, M., Gersch, M., Drag, M., et al. (2017). Insights into ClpXP proteolysis: Heterooligomerization and partial deactivation enhance chaperone affinity and substrate turnover in Listeria monocytogenes. Chem. Sci. 8, 1592–1600. doi:10.1039/c6sc03438a
Banchereau, J., Briere, F., Caux, C., Davoust, J., Lebecque, S., Liu, Y. J., et al. (2000). Immunobiology of dendritic cells. Annu. Rev. Immunol. 18, 767–811. doi:10.1146/annurev.immunol.18.1.767
Betts, J. C., Lukey, P. T., Robb, L. C., McAdam, R. A., and Duncan, K. (2002). Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43, 717–731. doi:10.1046/j.1365-2958.2002.02779.x
Bhandari, V., Wong, K. S., Zhou, J. L., Mabanglo, M. F., Batey, R. A., and Houry, W. A. (2018). The role of ClpP protease in bacterial pathogenesis and human diseases. ACS Chem. Biol. 13, 1413–1425. doi:10.1021/acschembio.8b00124
Binepal, G., Mabanglo, M. F., Goodreid, J. D., Leung, E., Barghash, M. M., Wong, K. S., et al. (2020). Development of antibiotics that dysregulate the neisserial ClpP protease. ACS Infect. Dis. 6, 3224–3236. doi:10.1021/acsinfecdis.0c00599
Bishop, C. E., Shadid, T. M., Lavey, N. P., Kempher, M. L., Ballard, J. D., and Duerfeldt, A. S. (2022). Identification of ClpP dual isoform disruption as an antisporulation strategy for Clostridioides difficile. J. Bacteriol. 204, e0041121. doi:10.1128/JB.00411-21
Bordes, P., Sala, A. J., Ayala, S., Texier, P., Slama, N., Cirinesi, A. M., et al. (2016). Chaperone addiction of toxin-antitoxin systems. Nat. Commun. 7, 13339. doi:10.1038/ncomms13339
Bottcher, T., and Sieber, S. A. (2009). Beta-lactones decrease the intracellular virulence of Listeria monocytogenes in macrophages. ChemMedChem 4, 1260–1263. doi:10.1002/cmdc.200900157
Brötz-Oesterhelt, H., Beyer, D., Kroll, H. P., Endermann, R., Ladel, C., Schroeder, W., et al. (2005). Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat. Med. 11, 1082–1087. doi:10.1038/nm1306
Brӧtz-Oesterhelt, H., and Sass, P. (2014). Bacterial caseinolytic proteases as novel targets for antibacterial treatment. Int. J. Med. Microbiol. 304, 23–30. doi:10.1016/j.ijmm.2013.09.001
Cao, J., Gong, Y., Dong, S., Zhang, L., Lai, X., Zhang, X., et al. (2013). Pneumococcal ClpP modulates the maturation and activation of human dendritic cells: Implications for pneumococcal infections. J. Leukoc. Biol. 93, 737–749. doi:10.1189/jlb.0812428
Carroni, M., Franke, K. B., Maurer, M., Jager, J., Hantke, I., Gloge, F., et al. (2017). Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control. Elife 6, e30120. doi:10.7554/eLife.30120
Chatterjee, I., Becker, P., Grundmeier, M., Bischoff, M., Somerville, G. A., Peters, G., et al. (2005). Staphylococcus aureus ClpC is required for stress resistance, aconitase activity, growth recovery, and death. J. Bacteriol. 187, 4488–4496. doi:10.1128/JB.187.13.4488-4496.2005
Choy, J. S., Aung, L. L., and Karzai, A. W. (2007). Lon protease degrades transfer-messenger RNA-tagged proteins. J. Bacteriol. 189, 6564–6571. doi:10.1128/JB.00860-07
Cohn, M. T., Ingmer, H., Mulholland, F., Jorgensen, K., Wells, J. M., and Brondsted, L. (2007). Contribution of conserved ATP-dependent proteases of Campylobacter jejuni to stress tolerance and virulence. Appl. Environ. Microbiol. 73, 7803–7813. doi:10.1128/AEM.00698-07
Conlon, B. P., Nakayasu, E. S., Fleck, L. E., LaFleur, M. D., Isabella, V. M., Coleman, K., et al. (2013). Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503, 365–370. doi:10.1038/nature12790
Culp, E. J., Sychantha, D., Hobson, C., Pawlowski, A. C., Prehna, G., and Wright, G. D. (2022). ClpP inhibitors are produced by a widespread family of bacterial gene clusters. Nat. Microbiol. 7, 451–462. doi:10.1038/s41564-022-01073-4
Culp, E., and Wright, G. D. (2017). Bacterial proteases, untapped antimicrobial drug targets. J. Antibiot. 70, 366–377. doi:10.1038/ja.2016.138
Derre, I., Rapoport, G., Devine, K., Rose, M., and Msadek, T. (1999). ClpE, a novel type of HSP100 ATPase, is part of the CtsR heat shock regulon of Bacillus subtilis. Mol. Microbiol. 32, 581–593. doi:10.1046/j.1365-2958.1999.01374.x
Diepold, A., and Armitage, J. P. (2015). Type III secretion systems: The bacterial flagellum and the injectisome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20150020. doi:10.1098/rstb.2015.0020
Donegan, N. P., Thompson, E. T., Fu, Z., and Cheung, A. L. (2010). Proteolytic regulation of toxin-antitoxin systems by ClpPC in Staphylococcus aureus. J. Bacteriol. 192, 1416–1422. doi:10.1128/JB.00233-09
Dougan, D. A., Weber-Ban, E., and Bukau, B. (2003). Targeted delivery of an ssrA-tagged substrate by the adaptor protein SspB to its cognate AAA+ protein ClpX. Mol. Cell 12, 373–380. doi:10.1016/j.molcel.2003.08.012
Dubiel, A., Wegrzyn, K., Kupinski, A. P., and Konieczny, I. (2018). ClpAP protease is a universal factor that activates the parDE toxin-antitoxin system from a broad host range RK2 plasmid. Sci. Rep. 8, 15287. doi:10.1038/s41598-018-33726-y
Elwell, C., Mirrashidi, K., and Engel, J. (2016). Chlamydia cell biology and pathogenesis. Nat. Rev. Microbiol. 14, 385–400. doi:10.1038/nrmicro.2016.30
Famulla, K., Sass, P., Malik, I., Akopian, T., Kandror, O., Alber, M., et al. (2016). Acyldepsipeptide antibiotics kill mycobacteria by preventing the physiological functions of the ClpP1P2 protease. Mol. Microbiol. 101, 194–209. doi:10.1111/mmi.13362
Farrell, C. M., Baker, T. A., and Sauer, R. T. (2007). Altered specificity of a AAA+ protease. Mol. Cell 25, 161–166. doi:10.1016/j.molcel.2006.11.018
Fei, X., Bell, T. A., Barkow, S. R., Baker, T. A., and Sauer, R. T. (2020a). Structural basis of ClpXP recognition and unfolding of ssrA-tagged substrates. Elife 9, e61496. doi:10.7554/eLife.61496
Fei, X., Bell, T. A., Jenni, S., Stinson, B. M., Baker, T. A., Harrison, S. C., et al. (2020b). Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate. Elife 9, e52774. doi:10.7554/eLife.52774
Feng, J., Michalik, S., Varming, A. N., Andersen, J. H., Albrecht, D., Jelsbak, L., et al. (2013). Trapping and proteomic identification of cellular substrates of the ClpP protease in Staphylococcus aureus. J. Proteome Res. 12, 547–558. doi:10.1021/pr300394r
Fernandez, L., Breidenstein, E. B., Song, D., and Hancock, R. E. (2012). Role of intracellular proteases in the antibiotic resistance, motility, and biofilm formation of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 56, 1128–1132. doi:10.1128/AAC.05336-11
Fisher, E. L., Otto, M., and Cheung, G. Y. C. (2018). Basis of virulence in enterotoxin-mediated staphylococcal food poisoning. Front. Microbiol. 9, 436. doi:10.3389/fmicb.2018.00436
Flynn, J. M., Levchenko, I., Seidel, M., Wickner, S. H., Sauer, R. T., and Baker, T. A. (2001). Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc. Natl. Acad. Sci. U. S. A. 98, 10584–10589. doi:10.1073/pnas.191375298
Flynn, J. M., Neher, S. B., Kim, Y. I., Sauer, R. T., and Baker, T. A. (2003). Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell 11, 671–683. doi:10.1016/s1097-2765(03)00060-1
Frees, D., Andersen, J. H., Hemmingsen, L., Koskenniemi, K., Baek, K. T., Muhammed, M. K., et al. (2012). New insights into Staphylococcus aureus stress tolerance and virulence regulation from an analysis of the role of the ClpP protease in the strains Newman, COL, and SA564. J. Proteome Res. 11, 95–108. doi:10.1021/pr200956s
Frees, D., Brondsted, L., and Ingmer, H. (2013). Bacterial proteases and virulence. Subcell. Biochem. 66, 161–192. doi:10.1007/978-94-007-5940-4_7
Frees, D., Chastanet, A., Qazi, S., Sorensen, K., Hill, P., Msadek, T., et al. (2004). Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 54, 1445–1462. doi:10.1111/j.1365-2958.2004.04368.x
Frees, D., Qazi, S. N., Hill, P. J., and Ingmer, H. (2003). Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol. Microbiol. 48, 1565–1578. doi:10.1046/j.1365-2958.2003.03524.x
Frees, D., Savijoki, K., Varmanen, P., and Ingmer, H. (2007). Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol. Microbiol. 63, 1285–1295. doi:10.1111/j.1365-2958.2007.05598.x
Fu, Z., Tamber, S., Memmi, G., Donegan, N. P., and Cheung, A. L. (2009). Overexpression of MazFsa in Staphylococcus aureus induces bacteriostasis by selectively targeting mRNAs for cleavage. J. Bacteriol. 191, 2051–2059. doi:10.1128/JB.00907-08
Furniss, R. C. D., and Clements, A. (2018). Regulation of the locus of enterocyte effacement in attaching and effacing pathogens. J. Bacteriol. 200, 003366-e417. doi:10.1128/JB.00336-17
Gaillot, O., Pellegrini, E., Bregenholt, S., Nair, S., and Berche, P. (2000). The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 35, 1286–1294. doi:10.1046/j.1365-2958.2000.01773.x
Gao, W., Kim, J. Y., Anderson, J. R., Akopian, T., Hong, S., Jin, Y. Y., et al. (2015). The cyclic peptide ecumicin targeting ClpC1 is active against Mycobacterium tuberculosis in vivo. Antimicrob. Agents Chemother. 59, 880–889. doi:10.1128/AAC.04054-14
Gatsogiannis, C., Balogh, D., Merino, F., Sieber, S. A., and Raunser, S. (2019). Cryo-EM structure of the ClpXP protein degradation machinery. Nat. Struct. Mol. Biol. 26, 946–954. doi:10.1038/s41594-019-0304-0
Gavrish, E., Sit, C. S., Cao, S., Kandror, O., Spoering, A., Peoples, A., et al. (2014). Lassomycin, a ribosomally synthesized cyclic peptide, kills mycobacterium tuberculosis by targeting the ATP-dependent protease ClpC1P1P2. Chem. Biol. 21, 509–518. doi:10.1016/j.chembiol.2014.01.014
Ge, Z. H., Long, Q. S., Yuan, P. B., Pan, X., Shen, D., and Lu, Y. J. (2019). The temporal expression of global regulator protein CsrA is dually regulated by ClpP during the biphasic life cycle of Legionella pneumophila. Front. Microbiol. 10, 2495. doi:10.3389/fmicb.2019.02495
Ge, Z., Yuan, P., Chen, L., Chen, J., Shen, D., She, Z., et al. (2022). New global insights on the regulation of the biphasic life cycle and virulence via ClpP-dependent proteolysis in Legionella pneumophila. Mol. Cell. Proteomics. 21, 100233. doi:10.1016/j.mcpro.2022.100233
Gersch, M., Gut, F., Korotkov, V. S., Lehmann, J., Bottcher, T., Rusch, M., et al. (2013). The mechanism of caseinolytic protease (ClpP) inhibition. Angew. Chem. Int. Ed. Engl. 52, 3009–3014. doi:10.1002/anie.201204690
Gerth, U., Kirstein, J., Mostertz, J., Waldminghaus, T., Miethke, M., Kock, H., et al. (2004). Fine-tuning in regulation of Clp protein content in Bacillus subtilis. J. Bacteriol. 186, 179–191. doi:10.1128/jb.186.1.179-191.2004
Goodreid, J. D., Janetzko, J., Santa Maria, J. P., Wong, K. S., Leung, E., Eger, B. T., et al. (2016). Development and characterization of potent cyclic acyldepsipeptide analogues with increased antimicrobial activity. J. Med. Chem. 59, 624–646. doi:10.1021/acs.jmedchem.5b01451
Goodreid, J. D., Wong, K., Leung, E., McCaw, S. E., Gray-Owen, S. D., Lough, A., et al. (2014). Total synthesis and antibacterial testing of the A54556 cyclic acyldepsipeptides isolated from Streptomyces hawaiiensis. J. Nat. Prod. 77, 2170–2181. doi:10.1021/np500158q
Gottesman, S., Roche, E., Zhou, Y., and Sauer, R. T. (1998). The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12, 1338–1347. doi:10.1101/gad.12.9.1338
Gottesman, S. (2019). Trouble is coming: Signaling pathways that regulate general stress responses in bacteria. J. Biol. Chem. 294, 11685–11700. doi:10.1074/jbc.REV119.005593
Green, E. R., and Mecsas, J. (2016). Bacterial secretion systems: An overview. Microbiol. Spectr. 4. doi:10.1128/microbiolspec.VMBF-0012-2015
Hackl, M. W., Lakemeyer, M., Dahmen, M., Glaser, M., Pahl, A., Lorenz-Baath, K., et al. (2015). Phenyl esters are potent inhibitors of caseinolytic protease P and reveal a stereogenic switch for deoligomerization. J. Am. Chem. Soc. 137, 8475–8483. doi:10.1021/jacs.5b03084
Hall, B. M., Breidenstein, E. B. M., de la Fuente-Nunez, C., Reffuveille, F., Mawla, G. D., Hancock, R. E. W., et al. (2017). Two isoforms of clp peptidase in Pseudomonas aeruginosa control distinct aspects of cellular physiology. J. Bacteriol. 199, 005688-e616. doi:10.1128/JB.00568-16
Harms, A., Brodersen, D. E., Mitarai, N., and Gerdes, K. (2018). Toxins, targets, and triggers: An overview of toxin-antitoxin biology. Mol. Cell 70, 768–784. doi:10.1016/j.molcel.2018.01.003
Herman, C., Thévenet, D., Bouloc, P., Walker, G. C., and D'Ari, R. (1998). Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH). Genes Dev. 12, 1348–1355. doi:10.1101/gad.12.9.1348
Ingavale, S., van Wamel, W., Luong, T. T., Lee, C. Y., and Cheung, A. L. (2005). Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in Staphylococcus aureus. Infect. Immun. 73, 1423–1431. doi:10.1128/IAI.73.3.1423-1431.2005
Ingmer, H., and Brondsted, L. (2009). Proteases in bacterial pathogenesis. Res. Microbiol. 160, 704–710. doi:10.1016/j.resmic.2009.08.017
Iyoda, S., Koizumi, N., Satou, H., Lu, Y., Saitoh, T., Ohnishi, M., et al. (2006). The GrlR-GrlA regulatory system coordinately controls the expression of flagellar and LEE-encoded type III protein secretion systems in enterohemorrhagic Escherichia coli. J. Bacteriol. 188, 5682–5692. doi:10.1128/JB.00352-06
Iyoda, S., and Watanabe, H. (2005). ClpXP protease controls expression of the type III protein secretion system through regulation of RpoS and GrlR levels in enterohemorrhagic Escherichia coli. J. Bacteriol. 187, 4086–4094. doi:10.1128/JB.187.12.4086-4094.2005
Jackson, M. W., Silva-Herzog, E., and Plano, G. V. (2004). The ATP-dependent ClpXP and Lon proteases regulate expression of the Yersinia pestis type III secretion system via regulated proteolysis of YmoA, a small histone-like protein. Mol. Microbiol. 54, 1364–1378. doi:10.1111/j.1365-2958.2004.04353.x
Jacques, S., van der Sloot, A. M., C, C. H., Coulombe-Huntington, J., Tsao, S., Tollis, S., et al. (2020). Imipridone anticancer compounds ectopically activate the ClpP protease and represent a new scaffold for antibiotic development. Genetics 214, 1103–1120. doi:10.1534/genetics.119.302851
Jenul, C., and Horswill, A. R. (2019). Regulation of Staphylococcus aureus virulence. Microbiol. Spectr. 7. doi:10.1128/microbiolspec.GPP3-0031-2018
Jiang, Y., Pogliano, J., Helinski, D. R., and Konieczny, I. (2002). ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 44, 971–979. doi:10.1046/j.1365-2958.2002.02921.x
Josenhans, C., and Suerbaum, S. (2002). The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291, 605–614. doi:10.1078/1438-4221-00173
Ju, Y., An, Q., Zhang, Y., Sun, K., Bai, L., and Luo, Y. (2021). Recent advances in Clp protease modulation to address virulence, resistance and persistence of MRSA infection. Drug Discov. Today 26, 2190–2197. doi:10.1016/j.drudis.2021.05.014
Kahne, S. C., and Darwin, K. H. (2021). Structural determinants of regulated proteolysis in pathogenic bacteria by ClpP and the proteasome. Curr. Opin. Struct. Biol. 67, 120–126. doi:10.1016/j.sbi.2020.09.012
Karlowicz, A., Wegrzyn, K., Dubiel, A., Ropelewska, M., and Konieczny, I. (2016). Proteolysis in plasmid DNA stable maintenance in bacterial cells. Plasmid 86, 7–13. doi:10.1016/j.plasmid.2016.05.002
Kearns, D. B. (2010). A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8, 634–644. doi:10.1038/nrmicro2405
Keiler, K. C. (2015). Mechanisms of ribosome rescue in bacteria. Nat. Rev. Microbiol. 13, 285–297. doi:10.1038/nrmicro3438
Keiler, K. C., Waller, P. R., and Sauer, R. T. (1996). Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271, 990–993. doi:10.1126/science.271.5251.990
Keren, I., Minami, S., Rubin, E., and Lewis, K. (2011). Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. mBio 2, e00100–e00111. doi:10.1128/mBio.00100-11
Kim, G. L., Akoolo, L., and Parker, D. (2020). The ClpXP protease contributes to Staphylococcus aureus pneumonia. J. Infect. Dis. 222, 1400–1404. doi:10.1093/infdis/jiaa251
Kirsch, V. C., Fetzer, C., and Sieber, S. A. (2021). Global inventory of ClpP- and ClpX-regulated proteins in Staphylococcus aureus. J. Proteome Res. 20, 867–879. doi:10.1021/acs.jproteome.0c00668
Kirstein, J., Hoffmann, A., Lilie, H., Schmidt, R., Rubsamen-Waigmann, H., Brӧtz-Oesterhelt, H., et al. (2009). The antibiotic ADEP reprogrammes ClpP, switching it from a regulated to an uncontrolled protease. EMBO Mol. Med. 1, 37–49. doi:10.1002/emmm.200900002
Kirstein, J., Schlothauer, T., Dougan, D. A., Lilie, H., Tischendorf, G., Mogk, A., et al. (2006). Adaptor protein controlled oligomerization activates the AAA+ protein ClpC. EMBO J. 25, 1481–1491. doi:10.1038/sj.emboj.7601042
Kirstein, J., and Turgay, K. (2005). A new tyrosine phosphorylation mechanism involved in signal transduction in Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 9, 182–188. doi:10.1159/000089646
Kitagawa, R., Takaya, A., and Yamamoto, T. (2011). Dual regulatory pathways of flagellar gene expression by ClpXP protease in enterohaemorrhagic Escherichia coli. Microbiol. Read. 157, 3094–3103. doi:10.1099/mic.0.051151-0
Knudsen, G. M., Nielsen, M. B., Thomsen, L. E., Aabo, S., Rychlik, I., and Olsen, J. E. (2014). The role of ClpP, RpoS and CsrA in growth and filament formation of Salmonella enterica serovar Typhimurium at low temperature. BMC Microbiol. 14, 208. doi:10.1186/s12866-014-0208-4
Knudsen, G. M., Olsen, J. E., Aabo, S., Barrow, P., Rychlik, I., and Thomsen, L. E. (2013). ClpP deletion causes attenuation of Salmonella Typhimurium virulence through mis-regulation of RpoS and indirect control of CsrA and the SPI genes. Microbiol. Read. 159, 1497–1509. doi:10.1099/mic.0.065797-0
Korch, S. B., Contreras, H., and Clark-Curtiss, J. E. (2009). Three Mycobacterium tuberculosis Rel toxin-antitoxin modules inhibit mycobacterial growth and are expressed in infected human macrophages. J. Bacteriol. 191, 1618–1630. doi:10.1128/JB.01318-08
Korch, S. B., Malhotra, V., Contreras, H., and Clark-Curtiss, J. E. (2015). The Mycobacterium tuberculosis relBE toxin:antitoxin genes are stress-responsive modules that regulate growth through translation inhibition. J. Microbiol. 53, 783–795. doi:10.1007/s12275-015-5333-8
Kruger, E., Volker, U., and Hecker, M. (1994). Stress induction of clpC in Bacillus subtilis and its involvement in stress tolerance. J. Bacteriol. 176, 3360–3367. doi:10.1128/jb.176.11.3360-3367.1994
Kruger, E., Witt, E., Ohlmeier, S., Hanschke, R., and Hecker, M. (2000). The clp proteases of Bacillus subtilis are directly involved in degradation of misfolded proteins. J. Bacteriol. 182, 3259–3265. doi:10.1128/jb.182.11.3259-3265.2000
Krysiak, J., Stahl, M., Vomacka, J., Fetzer, C., Lakemeyer, M., Fux, A., et al. (2017). Quantitative map of beta-lactone-induced virulence regulation. J. Proteome Res. 16, 1180–1192. doi:10.1021/acs.jproteome.6b00705
Kwon, H. Y., Kim, S. W., Choi, M. H., Ogunniyi, A. D., Paton, J. C., Park, S. H., et al. (2003). Effect of heat shock and mutations in ClpL and ClpP on virulence gene expression in Streptococcus pneumoniae. Infect. Immun. 71, 3757–3765. doi:10.1128/iai.71.7.3757-3765.2003
Kwon, H. Y., Ogunniyi, A. D., Choi, M. H., Pyo, S. N., Rhee, D. K., and Paton, J. C. (2004). The ClpP protease of Streptococcus pneumoniae modulates virulence gene expression and protects against fatal pneumococcal challenge. Infect. Immun. 72, 5646–5653. doi:10.1128/IAI.72.10.5646-5653.2004
LaBreck, C. J., May, S., Viola, M. G., Conti, J., and Camberg, J. L. (2017). The protein chaperone ClpX targets native and non-native aggregated substrates for remodeling, disassembly, and degradation with ClpP. Front. Mol. Biosci. 4, 26. doi:10.3389/fmolb.2017.00026
Lavey, N. P., Shadid, T., Ballard, J. D., and Duerfeldt, A. S. (2019). Clostridium difficile ClpP homologues are capable of uncoupled activity and exhibit different levels of susceptibility to acyldepsipeptide modulation. ACS Infect. Dis. 5, 79–89. doi:10.1021/acsinfecdis.8b00199
Lee, B. G., Park, E. Y., Lee, K. E., Jeon, H., Sung, K. H., Paulsen, H., et al. (2010). Structures of ClpP in complex with acyldepsipeptide antibiotics reveal its activation mechanism. Nat. Struct. Mol. Biol. 17, 471–478. doi:10.1038/nsmb.1787
Leodolter, J., Warweg, J., and Weber-Ban, E. (2015). The Mycobacterium tuberculosis ClpP1P2 protease interacts asymmetrically with its ATPase partners ClpX and ClpC1. PLoS One 10, e0125345. doi:10.1371/journal.pone.0125345
Leung, E., Datti, A., Cossette, M., Goodreid, J., McCaw, S. E., Mah, M., et al. (2011). Activators of cylindrical proteases as antimicrobials: Identification and development of small molecule activators of ClpP protease. Chem. Biol. 18, 1167–1178. doi:10.1016/j.chembiol.2011.07.023
Levchenko, I., Seidel, M., Sauer, R. T., and Baker, T. A. (2000). A specificity-enhancing factor for the ClpXP degradation machine. Science 289, 2354–2356. doi:10.1126/science.289.5488.2354
Li, D. H., Chung, Y. S., Gloyd, M., Joseph, E., Ghirlando, R., Wright, G. D., et al. (2010a). Acyldepsipeptide antibiotics induce the formation of a structured axial channel in ClpP: A model for the ClpX/ClpA-bound state of ClpP. Chem. Biol. 17, 959–969. doi:10.1016/j.chembiol.2010.07.008
Li, X. H., Zeng, Y. L., Gao, Y., Zheng, X. C., Zhang, Q. F., Zhou, S. N., et al. (2010b). The ClpP protease homologue is required for the transmission traits and cell division of the pathogen Legionella pneumophila. BMC Microbiol. 10, 54. doi:10.1186/1471-2180-10-54
Liu, K., Ologbenla, A., and Houry, W. A. (2014). Dynamics of the ClpP serine protease: A model for self-compartmentalized proteases. Crit. Rev. Biochem. Mol. Biol. 49, 400–412. doi:10.3109/10409238.2014.925421
Liu, Q., Wang, X., Qin, J., Cheng, S., Yeo, W. S., He, L., et al. (2017). The ATP-dependent protease ClpP inhibits biofilm formation by regulating agr and cell wall hydrolase Sle1 in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 7, 181. doi:10.3389/fcimb.2017.00181
Lowth, B. R., Kirstein-Miles, J., Saiyed, T., Brӧtz-Oesterhelt, H., Morimoto, R. I., Truscott, K. N., et al. (2012). Substrate recognition and processing by a Walker B mutant of the human mitochondrial AAA+ protein CLPX. J. Struct. Biol. 179, 193–201. doi:10.1016/j.jsb.2012.06.001
Luong, T. T., Dunman, P. M., Murphy, E., Projan, S. J., and Lee, C. Y. (2006). Transcription profiling of the mgrA regulon in Staphylococcus aureus. J. Bacteriol. 188, 1899–1910. doi:10.1128/JB.188.5.1899-1910.2006
Mabanglo, M. F., and Houry, W. A. (2022). Recent structural insights into the mechanism of ClpP protease regulation by AAA+ chaperones and small molecules. J. Biol. Chem. 298, 101781. doi:10.1016/j.jbc.2022.101781
Mabanglo, M. F., Leung, E., Vahidi, S., Seraphim, T. V., Eger, B. T., Bryson, S., et al. (2019). ClpP protease activation results from the reorganization of the electrostatic interaction networks at the entrance pores. Commun. Biol. 2, 410. doi:10.1038/s42003-019-0656-3
Martin, A., Baker, T. A., and Sauer, R. T. (2007). Distinct static and dynamic interactions control ATPase-peptidase communication in a AAA+ protease. Mol. Cell 27, 41–52. doi:10.1016/j.molcel.2007.05.024
Martin, A., Baker, T. A., and Sauer, R. T. (2008). Diverse pore loops of the AAA+ ClpX machine mediate unassisted and adaptor-dependent recognition of ssrA-tagged substrates. Mol. Cell 29, 441–450. doi:10.1016/j.molcel.2008.02.002
Martinez, L. C., Yakhnin, H., Camacho, M. I., Georgellis, D., Babitzke, P., Puente, J. L., et al. (2011). Integration of a complex regulatory cascade involving the SirA/BarA and Csr global regulatory systems that controls expression of the Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol. Microbiol. 80, 1637–1656. doi:10.1111/j.1365-2958.2011.07674.x
Mawla, G. D., Hall, B. M., Carcamo-Oyarce, G., Grant, R. A., Zhang, J. J., Kardon, J. R., et al. (2021). ClpP1P2 peptidase activity promotes biofilm formation in Pseudomonas aeruginosa. Mol. Microbiol. 115, 1094–1109. doi:10.1111/mmi.14649
McGillivray, S. M., Tran, D. N., Ramadoss, N. S., Alumasa, J. N., Okumura, C. Y., Sakoulas, G., et al. (2012). Pharmacological inhibition of the ClpXP protease increases bacterial susceptibility to host cathelicidin antimicrobial peptides and cell envelope-active antibiotics. Antimicrob. Agents Chemother. 56, 1854–1861. doi:10.1128/AAC.05131-11
Michel, A., Agerer, F., Hauck, C. R., Herrmann, M., Ullrich, J., Hacker, J., et al. (2006). Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 188, 5783–5796. doi:10.1128/JB.00074-06
Michiels, J. E., Van den Bergh, B., Verstraeten, N., Fauvart, M., and Michiels, J. (2016). In vitro emergence of high persistence upon periodic aminoglycoside challenge in the ESKAPE pathogens. Antimicrob. Agents Chemother. 60, 4630–4637. doi:10.1128/AAC.00757-16
Miethke, M., Hecker, M., and Gerth, U. (2006). Involvement of Bacillus subtilis ClpE in CtsR degradation and protein quality control. J. Bacteriol. 188, 4610–4619. doi:10.1128/JB.00287-06
Moreno-Cinos, C., Goossens, K., Salado, I. G., Van Der Veken, P., De Winter, H., and Augustyns, K. (2019). ClpP protease, a promising antimicrobial target. Int. J. Mol. Sci. 20, E2232. doi:10.3390/ijms20092232
Morreale, F. E., Kleine, S., Leodolter, J., Junker, S., Hoi, D. M., Ovchinnikov, S., et al. (2022). BacPROTACs mediate targeted protein degradation in bacteria. Cell 185, 2338–2353.e18. e2318. doi:10.1016/j.cell.2022.05.009
Mroue, N., Arya, A., Brown Gandt, A., Russell, C., Han, A., Gavrish, E., et al. (2019). Pharmacodynamics of ClpP-activating antibiotic combinations against gram-positive pathogens. Antimicrob. Agents Chemother. 64, 015544-e1619. doi:10.1128/AAC.01554-19
Nagpal, J., Paxman, J. J., Zammit, J. E., Alhuwaider, A., Truscott, K. N., Heras, B., et al. (2019). Molecular and structural insights into an asymmetric proteolytic complex (ClpP1P2) from Mycobacterium smegmatis. Sci. Rep. 9, 18019. doi:10.1038/s41598-019-53736-8
Olivares, A. O., Baker, T. A., and Sauer, R. T. (2016). Mechanistic insights into bacterial AAA+ proteases and protein-remodelling machines. Nat. Rev. Microbiol. 14, 33–44. doi:10.1038/nrmicro.2015.4
Ollinger, J., O'Malley, T., Kesicki, E. A., Odingo, J., and Parish, T. (2012). Validation of the essential ClpP protease in Mycobacterium tuberculosis as a novel drug target. J. Bacteriol. 194, 663–668. doi:10.1128/JB.06142-11
Page, R., and Peti, W. (2016). Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 12, 208–214. doi:10.1038/nchembio.2044
Pan, S., Malik, I. T., Thomy, D., Henrichfreise, B., and Sass, P. (2019). The functional ClpXP protease of Chlamydia trachomatis requires distinct clpP genes from separate genetic loci. Sci. Rep. 9, 14129. doi:10.1038/s41598-019-50505-5
Panasenko, O. O., Bezrukov, F., Komarynets, O., and Renzoni, A. (2020). YjbH solubility controls spx in Staphylococcus aureus: Implication for MazEF toxin-antitoxin system regulation. Front. Microbiol. 11, 113. doi:10.3389/fmicb.2020.00113
Personne, Y., Brown, A. C., Schuessler, D. L., and Parish, T. (2013). Mycobacterium tuberculosis ClpP proteases are co-transcribed but exhibit different substrate specificities. PLoS One 8, e60228. doi:10.1371/journal.pone.0060228
Pizarro-Cerda, J., and Cossart, P. (2006). Bacterial adhesion and entry into host cells. Cell 124, 715–727. doi:10.1016/j.cell.2006.02.012
Porankiewicz, J., Wang, J., and Clarke, A. K. (1999). New insights into the ATP-dependent clp protease: Escherichia coli and beyond. Mol. Microbiol. 32, 449–458. doi:10.1046/j.1365-2958.1999.01357.x
Pratt, L. A., and Silhavy, T. J. (1996). The response regulator SprE controls the stability of RpoS. Proc. Natl. Acad. Sci. U. S. A. 93, 2488–2492. doi:10.1073/pnas.93.6.2488
Qiu, D., Eisinger, V. M., Head, N. E., Pier, G. B., and Yu, H. D. (2008). ClpXP proteases positively regulate alginate overexpression and mucoid conversion in Pseudomonas aeruginosa. Microbiol. Read. 154, 2119–2130. doi:10.1099/mic.0.2008/017368-0
Raju, R. M., Goldberg, A. L., and Rubin, E. J. (2012a). Bacterial proteolytic complexes as therapeutic targets. Nat. Rev. Drug Discov. 11, 777–789. doi:10.1038/nrd3846
Raju, R. M., Jedrychowski, M. P., Wei, J. R., Pinkham, J. T., Park, A. S., O'Brien, K., et al. (2014). Post-translational regulation via Clp protease is critical for survival of Mycobacterium tuberculosis. PLoS Pathog. 10, e1003994. doi:10.1371/journal.ppat.1003994
Raju, R. M., Unnikrishnan, M., Rubin, D. H., Krishnamoorthy, V., Kandror, O., Akopian, T. N., et al. (2012b). Mycobacterium tuberculosis ClpP1 and ClpP2 function together in protein degradation and are required for viability in vitro and during infection. PLoS Pathog. 8, e1002511. doi:10.1371/journal.ppat.1002511
Ramage, H. R., Connolly, L. E., and Cox, J. S. (2009). Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: Implications for pathogenesis, stress responses, and evolution. PLoS Genet. 5, e1000767. doi:10.1371/journal.pgen.1000767
Rand, L., Hinds, J., Springer, B., Sander, P., Buxton, R. S., and Davis, E. O. (2003). The majority of inducible DNA repair genes in Mycobacterium tuberculosis are induced independently of RecA. Mol. Microbiol. 50, 1031–1042. doi:10.1046/j.1365-2958.2003.03765.x
Rice, C. J., Ramachandran, V. K., Shearer, N., and Thompson, A. (2015). Transcriptional and post-transcriptional modulation of SPI1 and SPI2 expression by ppGpp, RpoS and DksA in Salmonella enterica sv typhimurium. PLoS One 10, e0127523. doi:10.1371/journal.pone.0127523
Ripstein, Z. A., Vahidi, S., Houry, W. A., Rubinstein, J. L., and Kay, L. E. (2020). A processive rotary mechanism couples substrate unfolding and proteolysis in the ClpXP degradation machinery. Elife 9, e52158. doi:10.7554/eLife.52158
Rouquette, C., Ripio, M. T., Pellegrini, E., Bolla, J. M., Tascon, R. I., Vazquez-Boland, J. A., et al. (1996). Identification of a ClpC ATPase required for stress tolerance and in vivo survival of Listeria monocytogenes. Mol. Microbiol. 21, 977–987. doi:10.1046/j.1365-2958.1996.641432.x
Sass, P., Josten, M., Famulla, K., Schiffer, G., Sahl, H. G., Hamoen, L., et al. (2011). Antibiotic acyldepsipeptides activate ClpP peptidase to degrade the cell division protein FtsZ. Proc. Natl. Acad. Sci. U. S. A. 108, 17474–17479. doi:10.1073/pnas.1110385108
Schelin, J., Cohn, M. T., Frisk, B., and Frees, D. (2020). A functional ClpXP protease is required for induction of the accessory toxin genes, tst, sed, and sec. Toxins (Basel) 12, E553. doi:10.3390/toxins12090553
Schlothauer, T., Mogk, A., Dougan, D. A., Bukau, B., and Turgay, K. (2003). MecA, an adaptor protein necessary for ClpC chaperone activity. Proc. Natl. Acad. Sci. U. S. A. 100, 2306–2311. doi:10.1073/pnas.0535717100
Schuessler, D. L., Cortes, T., Fivian-Hughes, A. S., Lougheed, K. E., Harvey, E., Buxton, R. S., et al. (2013). Induced ectopic expression of HigB toxin in Mycobacterium tuberculosis results in growth inhibition, reduced abundance of a subset of mRNAs and cleavage of tmRNA. Mol. Microbiol. 90, 195–207. doi:10.1111/mmi.12358
Schuster, C. F., Mechler, L., Nolle, N., Krismer, B., Zelder, M. E., Gotz, F., et al. (2015). The MazEF toxin-antitoxin system Alters the beta-lactam susceptibility of Staphylococcus aureus. PLoS One 10, e0126118. doi:10.1371/journal.pone.0126118
Schweder, T., Lee, K. H., Lomovskaya, O., and Matin, A. (1996). Regulation of Escherichia coli starvation sigma factor (sigma s) by ClpXP protease. J. Bacteriol. 178, 470–476. doi:10.1128/jb.178.2.470-476.1996
Schwiesow, L., Lam, H., Dersch, P., and Auerbuch, V. (2015). Yersinia type III secretion system master regulator LcrF. J. Bacteriol. 198, 604–614. doi:10.1128/JB.00686-15
Shanks, R. M., Caiazza, N. C., Hinsa, S. M., Toutain, C. M., and O'Toole, G. A. (2006). Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl. Environ. Microbiol. 72, 5027–5036. doi:10.1128/AEM.00682-06
Shen, A. (2020). Clostridioides difficile spore formation and germination: New insights and opportunities for intervention. Annu. Rev. Microbiol. 74, 545–566. doi:10.1146/annurev-micro-011320-011321
Silber, N., Pan, S., Schakermann, S., Mayer, C., Brӧtz-Oesterhelt, H., and Sass, P. (2020). Cell division protein FtsZ is unfolded for N-terminal degradation by antibiotic-activated ClpP. mBio 11, e01006-e01020. doi:10.1128/mBio.01006-20
Singh, G., Yadav, M., Ghosh, C., and Rathore, J. S. (2021). Bacterial toxin-antitoxin modules: Classification, functions, and association with persistence. Curr. Res. Microb. Sci. 2, 100047. doi:10.1016/j.crmicr.2021.100047
Springer, M. T., Singh, V. K., Cheung, A. L., Donegan, N. P., and Chamberlain, N. R. (2016). Effect of clpP and clpC deletion on persister cell number in Staphylococcus aureus. J. Med. Microbiol. 65, 848–857. doi:10.1099/jmm.0.000304
Stahlhut, S. G., Alqarzaee, A. A., Jensen, C., Fisker, N. S., Pereira, A. R., Pinho, M. G., et al. (2017). The ClpXP protease is dispensable for degradation of unfolded proteins in Staphylococcus aureus. Sci. Rep. 7, 11739. doi:10.1038/s41598-017-12122-y
Texier, P., Bordes, P., Nagpal, J., Sala, A. J., Mansour, M., Cirinesi, A. M., et al. (2021). ClpXP-mediated degradation of the TAC antitoxin is neutralized by the SecB-like chaperone in Mycobacterium tuberculosis. J. Mol. Biol. 433, 166815. doi:10.1016/j.jmb.2021.166815
Thomas, C. M., Meyer, R., and Helinski, D. R. (1980). Regions of broad-host-range plasmid RK2 which are essential for replication and maintenance. J. Bacteriol. 141, 213–222. doi:10.1128/JB.141.1.213-222.1980
Thomsen, L. E., Olsen, J. E., Foster, J. W., and Ingmer, H. (2002). ClpP is involved in the stress response and degradation of misfolded proteins in Salmonella enterica serovar Typhimurium. Microbiol. Read. 148, 2727–2733. doi:10.1099/00221287-148-9-2727
Tomoyasu, T., Ohkishi, T., Ukyo, Y., Tokumitsu, A., Takaya, A., Suzuki, M., et al. (2002). The ClpXP ATP-dependent protease regulates flagellum synthesis in Salmonella enterica serovar typhimurium. J. Bacteriol. 184, 645–653. doi:10.1128/jb.184.3.645-653.2002
Tomoyasu, T., Takaya, A., Isogai, E., and Yamamoto, T. (2003). Turnover of FlhD and FlhC, master regulator proteins for Salmonella flagellum biogenesis, by the ATP-dependent ClpXP protease. Mol. Microbiol. 48, 443–452. doi:10.1046/j.1365-2958.2003.03437.x
Tripathi, A., Dewan, P. C., Siddique, S. A., and Varadarajan, R. (2014). MazF-induced growth inhibition and persister generation in Escherichia coli. J. Biol. Chem. 289, 4191–4205. doi:10.1074/jbc.M113.510511
Villarroel, R., Hedges, R. W., Maenhaut, R., Leemans, J., Engler, G., Van Montagu, M., et al. (1983). Heteroduplex analysis of P-plasmid evolution: The role of insertion and deletion of transposable elements. Mol. Gen. Genet. 189, 390–399. doi:10.1007/BF00325900
Wah, D. A., Levchenko, I., Rieckhof, G. E., Bolon, D. N., Baker, T. A., and Sauer, R. T. (2003). Flexible linkers leash the substrate binding domain of SspB to a peptide module that stabilizes delivery complexes with the AAA+ ClpXP protease. Mol. Cell 12, 355–363. doi:10.1016/s1097-2765(03)00272-7
Wang, C., Fan, J., Niu, C., Wang, C., Villaruz, A. E., Otto, M., et al. (2010). Role of spx in biofilm formation of Staphylococcus epidermidis. FEMS Immunol. Med. Microbiol. 59, 152–160. doi:10.1111/j.1574-695X.2010.00673.x
Wang, C., Li, M., Dong, D., Wang, J., Ren, J., Otto, M., et al. (2007). Role of ClpP in biofilm formation and virulence of Staphylococcus epidermidis. Microbes Infect. 9, 1376–1383. doi:10.1016/j.micinf.2007.06.012
Wayne, L. G., and Sohaskey, C. D. (2001). Nonreplicating persistence of mycobacterium tuberculosis. Annu. Rev. Microbiol. 55, 139–163. doi:10.1146/annurev.micro.55.1.139
Westman, J., and Grinstein, S. (2020). Determinants of phagosomal pH during host-pathogen interactions. Front. Cell Dev. Biol. 8, 624958. doi:10.3389/fcell.2020.624958
Winther, K. S., Brodersen, D. E., Brown, A. K., and Gerdes, K. (2013). VapC20 of Mycobacterium tuberculosis cleaves the sarcin-ricin loop of 23S rRNA. Nat. Commun. 4, 2796. doi:10.1038/ncomms3796
Wong, K. S., Mabanglo, M. F., Seraphim, T. V., Mollica, A., Mao, Y. Q., Rizzolo, K., et al. (2018). Acyldepsipeptide analogs dysregulate human mitochondrial ClpP protease activity and cause apoptotic cell death. Cell Chem. Biol. 25, 1017–1030. doi:10.1016/j.chembiol.2018.05.014
Wood, N. A., Blocker, A. M., Seleem, M. A., Conda-Sheridan, M., Fisher, D. J., and Ouellette, S. P. (2020). The ClpX and ClpP2 orthologs of Chlamydia trachomatis perform discrete and essential functions in organism growth and development. mBio 11, e02016–e02020. doi:10.1128/mBio.02016-20
Wood, N. A., Chung, K. Y., Blocker, A. M., Rodrigues de Almeida, N., Conda-Sheridan, M., Fisher, D. J., et al. (2019). Erratum for Wood et al., "Initial Characterization of the Two ClpP Paralogs of Chlamydia trachomatis Suggests Unique Functionality for Each". J. Bacteriol. 201, e00613-e00619. doi:10.1128/JB.00613-19
Yamamoto, T., Sashinami, H., Takaya, A., Tomoyasu, T., Matsui, H., Kikuchi, Y., et al. (2001). Disruption of the genes for ClpXP protease in Salmonella enterica serovar Typhimurium results in persistent infection in mice, and development of persistence requires endogenous gamma interferon and tumor necrosis factor alpha. Infect. Immun. 69, 3164–3174. doi:10.1128/IAI.69.5.3164-3174.2001
Yang, D., Ho, Y. X., Cowell, L. M., Jilani, I., Foster, S. J., and Prince, L. R. (2019). A genome-wide screen identifies factors involved in S. Aureus-Induced human neutrophil cell death and pathogenesis. Front. Immunol. 10, 45. doi:10.3389/fimmu.2019.00045
Zeiler, E., List, A., Alte, F., Gersch, M., Wachtel, R., Poreba, M., et al. (2013). Structural and functional insights into caseinolytic proteases reveal an unprecedented regulation principle of their catalytic triad. Proc. Natl. Acad. Sci. U. S. A. 110, 11302–11307. doi:10.1073/pnas.1219125110
Zhang, Y., Zhang, J., Hoeflich, K. P., Ikura, M., Qing, G., and Inouye, M. (2003). MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12, 913–923. doi:10.1016/s1097-2765(03)00402-7
Zhao, B. B., Li, X. H., Zeng, Y. L., and Lu, Y. J. (2016). ClpP-deletion impairs the virulence of Legionella pneumophila and the optimal translocation of effector proteins. BMC Microbiol. 16, 174. doi:10.1186/s12866-016-0790-8
Keywords: ClpP protease, virulence, pathogenesis, ATP-dependent proteases, substrates
Citation: Aljghami ME, Barghash MM, Majaesic E, Bhandari V and Houry WA (2022) Cellular functions of the ClpP protease impacting bacterial virulence. Front. Mol. Biosci. 9:1054408. doi: 10.3389/fmolb.2022.1054408
Received: 26 September 2022; Accepted: 15 November 2022;
Published: 01 December 2022.
Edited by:
Abdussalam Azem, Tel Aviv University, IsraelReviewed by:
David A. Dougan, La Trobe University, AustraliaCopyright © 2022 Aljghami, Barghash, Majaesic, Bhandari and Houry. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Walid A. Houry, d2FsaWQuaG91cnlAdXRvcm9udG8uY2E=
†These authors have contributedequally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.