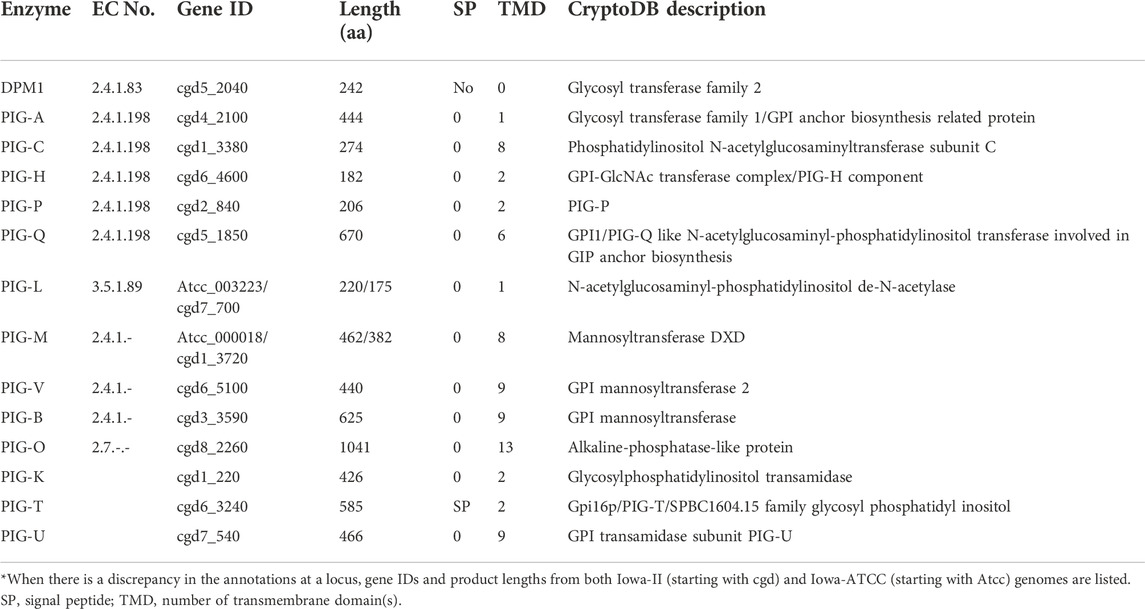
- Key Laboratory of Zoonosis Research of the Ministry of Education, Institute of Zoonosis, College of Veterinary Medicine, Jilin University, Changchun, China
Cryptosporidium is a genus of apicomplexan parasites infecting humans or other vertebrates. The majority of the Cryptosporidium species live in host intestines (e.g., C. parvum, C. hominis and C. ubiquitum), but there are a few gastric species (e.g., C. muris and C. andersoni). Among them, C. parvum is the most important zoonotic species, for which a number of glycoproteins have been reported for being involved in the interacting with host cells. However, little is known on the cryptosporidium glycobiology. Information on the glycosylation pathways in Cryptosporidium parasites remains sketchy and only a few studies have truly determined the glycoforms in the parasites. Here we reanalyzed the Cryptosporidium genomes and reconstructed the glycosylation pathways, including the synthesis of N- and O-linked glycans and GPI-anchors. In N-glycosylation, intestinal Cryptosporidium possesses enzymes to make a simple precursor with two terminal glucoses on the long arm (i.e., Glc2Man5GlcNAc2 vs. Glc3Man9GlcNAc2 in humans), but gastric species only makes a simpler precursor containing only the “core” structure (i.e., Man3GlcNAc2). There is an ortholog of glucosidase II (GANAB) in all Cryptosporidium species, for which the authenticity is questioned because it contains no signal peptide and exist in gastric species lacking terminal glucoses for the enzyme to act on. In O-linked glycosylation, all Cryptosporidium species may attach one-unit HexNAc (GalNAc and GlcNAc) and two-unit Fuc-type (Man-Fuc) glycans to the target proteins. Cryptosporidium lacks enzymes to further process N- and O-glycans in the Golgi. The glycosylphosphatidylinositol (GPI)-anchor in Cryptosporidium is predicted to be unbranched and unprocessed further in the Golgi. Cryptosporidium can synthesize limited nucleotide sugars, but possesses at least 12 transporters to scavenge nucleotide sugars or transport them across the ER/Golgi membranes. Overall, Cryptosporidium makes much simpler glycans than the hosts, and the N-glycoforms further differ between intestinal and gastric species. The Cryptosporidium N- and O-glycans are neutrally charged and have limited capacity to absorb water molecules in comparison to the host intestinal mucins that are negatively charged and highly expandable in waters.
Introduction
Cryptosporidium is a genus of protozoan parasites under the Phylum Apicomplexa that contains many pathogens of medical and veterinary importance (e.g., Toxoplasma, Cyclospora, Eimeria, Plasmodium, Babesia and Theileria). Among more than 40 known Cryptosporidium species, C. parvum is the most important zoonotic pathogen infecting humans and many other mammals including domesticated bovids (Checkley et al., 2015; Ryan et al., 2016; Pumipuntu and Piratae, 2018; Innes et al., 2020). Humans and animals with weakened or compromised immunity are more vulnerable to severe to deadly cryptosporidial infection. As a member of Apicomplexa, Cryptosporidium possesses an apical complex comprised of a set of unique apical cytoskeletal structures (e.g., apical rings, conoid and microtubes) and secretory organelles (i.e., rhoptries, micronemes and dense granules). Although the morphology and lifecycle of Cryptosporidium assemble intestinal coccidia (e.g., Eimeria, Cyclospora and Isospora), Cryptosporidium species are in fact highly divergent from the coccidia and other apicomplexans. Evolutionarily, they form a single phylogenetic clade at the base of the Apicomplexa, rather than a sister to the coccidia (Zhu et al., 2000a; Madern et al., 2004). Morphologically, they lack an apicoplast and a classic mitochondrion as well as their organellar genomes, but possess two unique central microtubular filaments (Zhu et al., 2000b; Templeton et al., 2010; Wang et al., 2021). Metabolically, Cryptosporidium lacks de novo synthetic pathways for any nutrients (e.g., nucleosides, amino acids, fatty acids and isoprenoids) (Abrahamsen et al., 2004; Xu et al., 2004; Rider and Zhu, 2010; Baptista et al., 2022).
Protein glycosylation, including the attachment of glycans to the asparagine (i.e., N-linked glycosylation) or the serine/threonine (i.e., O-linked glycosylation) residues and that of glycosylphosphatidylinositol (GPI)-anchor to the C-terminus of a protein, is one of the major post-translational modifications (Cherepanova et al., 2016; Kinoshita and Fujita, 2016; Corfield, 2017; Reily et al., 2019; Kinoshita, 2020; Toustou et al., 2022). Most glycosylated proteins (glycoproteins) are secretory, being distributed to the cell surface as transmembrane proteins or discharged as extracellular proteins. In the hosts (humans and animals), glycoproteins are known to be involved in every biological process in or around the cells, such as functioning in the structure, reproduction, immune system, hormones, intercellular communications and protection of cells and organisms (Moremen et al., 2012; Varki, 2017; Schjoldager et al., 2020). In the parasites in general, glycosylated and GPI-anchored proteins are more noticeable for their roles in the host-parasite interaction, including parasite adhesion/attachment to host cells and stimulation/evasion of the host immune responses (Anantharaman et al., 2007; Paschinger and Wilson, 2019; Lin et al., 2020; Mule et al., 2020).
In Cryptosporidium, a number of glycoproteins have been identified as, or predicted to be, glycosylated (e.g., gp900, gp15, gp40 and a number of mucin-like glycoproteins) (Wanyiri and Ward, 2006; Lendner and Daugschies, 2014). The presence of glycans in the immunodominant antigen gp900 was confirmed earlier by deglycosylation with N-glycosidase F (Petersen et al., 1992). Mass spectrometry-based studies of proteins from C. parvum oocysts have detected the presence of O-linked glycans in four proteins (i.e., gp900, gp40, gp15 and gp20) and N-linked glycans in 16 proteins (e.g., gp900, POWP1, GOP50 and seven other unique Cryptosporidium glycoproteins) (Haserick et al., 2017a; Haserick et al., 2017b). Among them, gp40 and gp15 are derived from precursor gp60 by the cleavage of furin-like protease. The C-segment gp15 (aka Cp17 in some earlier studies) and a low molecular weight antigen were found to be attached to the membrane by GPI-anchor (Priest et al., 2001; Priest et al., 2003). These studies implied the importance of protein glycosylation in the parasite based on the roles of these proteins in interacting with host cells during the parasite invasion and/or in serving as immunodominant antigens (Wanyiri and Ward, 2006; Lendner and Daugschies, 2014). Some vaccine candidates for cryptosporidiosis are also glycoproteins (Ifeonu et al., 2016; Haserick et al., 2017a). Glycosylation at the binding sites affects the protein-ligand interactions, including those between two proteins, proteins and small molecules, as well as antigen/epitopes and antibodies. Therefore, understanding the glycoforms in Cryptosporidium is important in the study of molecular interactions during the parasite invasion and development, host immunological responses and vaccine development.
The C. parvum genomes encode around 4,000 proteins, or 3,994 proteins to be exact for the Iowa-II reference strain, based on the annotated at the CryptoDB (https://www.CryptoDB.org) (Abrahamsen et al., 2004; Xu et al., 2004; Amos et al., 2022). These include some known glycoproteins and a large number of candidate proteins for glycosylation. Our reanalysis of the annotated proteins with Phobius have identified over one-third of them contain N-terminal signal peptide (SP) and/or transmembrane domains (TMDs) for targeting to the endoplasmic reticulum (ER). Around 800 proteins contain SP but lack TMD, or as type I (one TMD and SP) or type II/III (one TMD but lacking SP) transmembrane proteins that are candidates of extracellular proteins with high probability for glycosylation. Enzymes for glycosylation can also be identified in the annotated genomes for potentially synthesizing O- and N-glycans and the GPI-anchors. Mass spectrometry-based assays also showed that the glycans in C. parvum were much simpler than those in humans and animals (Haserick et al., 2017a; Haserick et al., 2017b). Additionally, significant differences in the glycosylation pathways were known between apicomplexans and the hosts and among various apicomplexan lineages (Liu et al., 2016; Florin-Christensen et al., 2021; Toustou et al., 2022). However, information on the glycosylation pathways in Cryptosporidium is fragmented in the literature. There are also variations in the annotations of certain genes in the pathways in the databases (e.g., KEGG and CryptoDB) that needs attention for the glyco-parasitology community.
In this manuscript, we analyzed the Cryptosporidium genomes with focus on C. parvum and C. muris that represent intestinal and gastric species, reconstructed the pathways related to glycosylation pathways including the synthesis of N- and O-linked glycans and GPI-anchors. We assembled glycosylation-related pathways to serve as a comprehensive but concise resource for the glyco-parasitology and cryptosporidium research communities. Features of glycosylation in cryptosporidium parasites and biological implications were also discussed.
Reconstruction of the glycosylation from the Cryptosporidium genomes
Several approaches were employed to identify genes encoding protein glycosylation enzymes, including those for the biosynthesis nucleotide-sugars, O- and N-linked glycans and GPI-anchors. We focused on C. parvum and C. muris that represented intestinal and gastric species that show major differences in the N-glycoforms. All protein sequences predicted from the genomes of C. parvum (Iowa-II and ATCC strains) and C. muris (RN66 strain) were fetched from the CryptoDB (https://www.cryptodb.org/; release 5.5) and used as queries for K-number assignment using BlastKOALA tool (https://www.kegg.jp/blastkoala/) at the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa et al., 2022). Pathways reconstructed with the assigned K numbers were compared with these already annotated by KEGG for C. parvum. Enzymes mapped to the glycosylation-related pathways were subjected to analysis for protein families and domains by InterProScan (https://www.ebi.ac.uk/interpro/search/sequence/) (Mitchell et al., 2019). Transmembrane domain (TMD) topology and signal peptides (SPs) were predicted using Phobius (https://phobius.sbc.su.se) (Kall et al., 2007). Some protein sequences were further analyzed by BLAST searches for information on their homologs and conserved domains against the NCBI protein databases and conserved domain databases (CDD). Potential nucleotide sugar transporters were identified by combining results from InterProScan and TransportDB (http://www.membranetransport.org/transportDB2/) (Elbourne et al., 2017), together with a recent annotation of transporters of the re-sequenced C. parvum genome (Baptista et al., 2022).
Cryptosporidium genomes encode enzymes for synthesizing N-linked glycans that are much simpler than those in the hosts and differ between intestinal and gastric species
In mammals and yeasts, the N-glycan synthetic pathway starts with the synthesis of glycan precursor (Glc3Man9GlcNAc2-PP-Dol) on the endoplasmic reticulum membrane (ER), followed by the transfer of the precursor to a nascent protein at selected Asn residues (Glc3Man9GlcNAc2-Asn). Terminal sugars are trimmed in the ER and the Golgi into a core structure (i.e., Man3GlcNAc2) that is then extended in the Golgi to form complex glycans (Chung et al., 2017; Corfield, 2017; Toustou et al., 2022). In the synthesis of precursor, polyprenol (unsaturated long-chain isoprenoid alcohol) is first reduced by polyprenol reductase (PPR) to dolichol (Dol). Dol is phosphorylated by dolichol kinase (DolK) to form Dol-P on the cytosolic side of the ER membrane. Dol-P is extended with oligosaccharides by a set of asparagine-linked glycosylation (ALG) enzymes. These include ALG7 and ALG13/ALG14 to add two N-acetyl-D-glucosamines (GlcNAc2), and ALG1 and ALG2 to add three mannoses (Man3) to form the “core” structure (i.e., Man3GlcNAc2-PP-Dol). The core is extended by ALG11 by adding two more mannoses to one of the two arms. The saccharide chain on the PP-Dol is then translocated into the ER luminal side for adding four more mannoses to form three branches by ALG3, ALG9 and ALG12. Three glucoses are then added to one of the branch by ALG6, ALG8 and ALG10 to form the precursor Glc3Man9GlcNAc2-PP-Dol (see Figure 1A for illustration of N-glycan precursors in humans and apicomplexans).
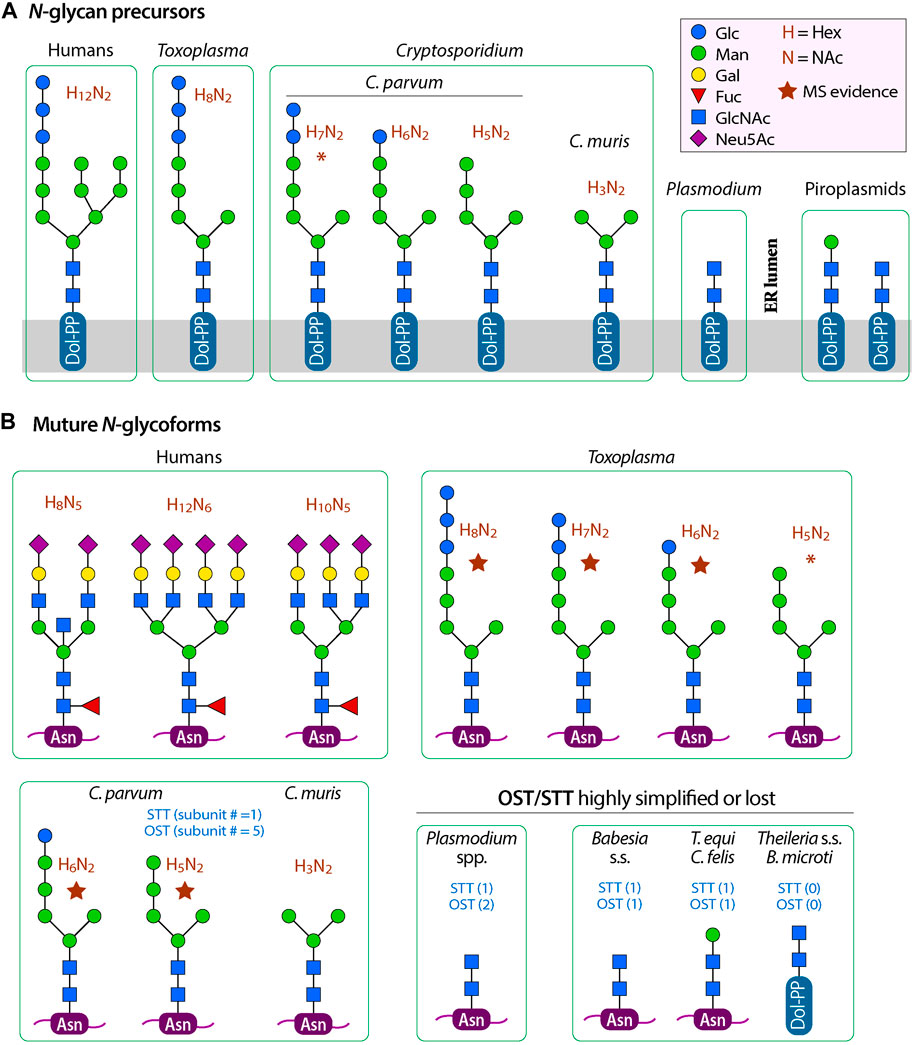
FIGURE 1. Comparison of N-glycan precursors (A) and typical mature glycoforms (B) between humans, Cryptosporidium species and other apicomplexans. Five-point purple stars indicate that the glycoforms have been detected by mass spectrometric analysis in the literature. Asterisks (*) indicate: 1) the glycan precursor H7N2 (Hex7NAc2) in C. parvum is undetected by mass spectrometric analysis in the literature, but the form may exist if ALG8 performs its function; or 2) the glycoform H5N2 (Hex5NAc2) in T. gondii is undetected by mass spectrometric analysis in the literature, but the form may exist if GANAB performs two consecutive trimming of the terminal glucoses. OST, oligosaccharyltransferase complex; STT3, oligosaccharyltransferase STT3 subunit (catalytic).
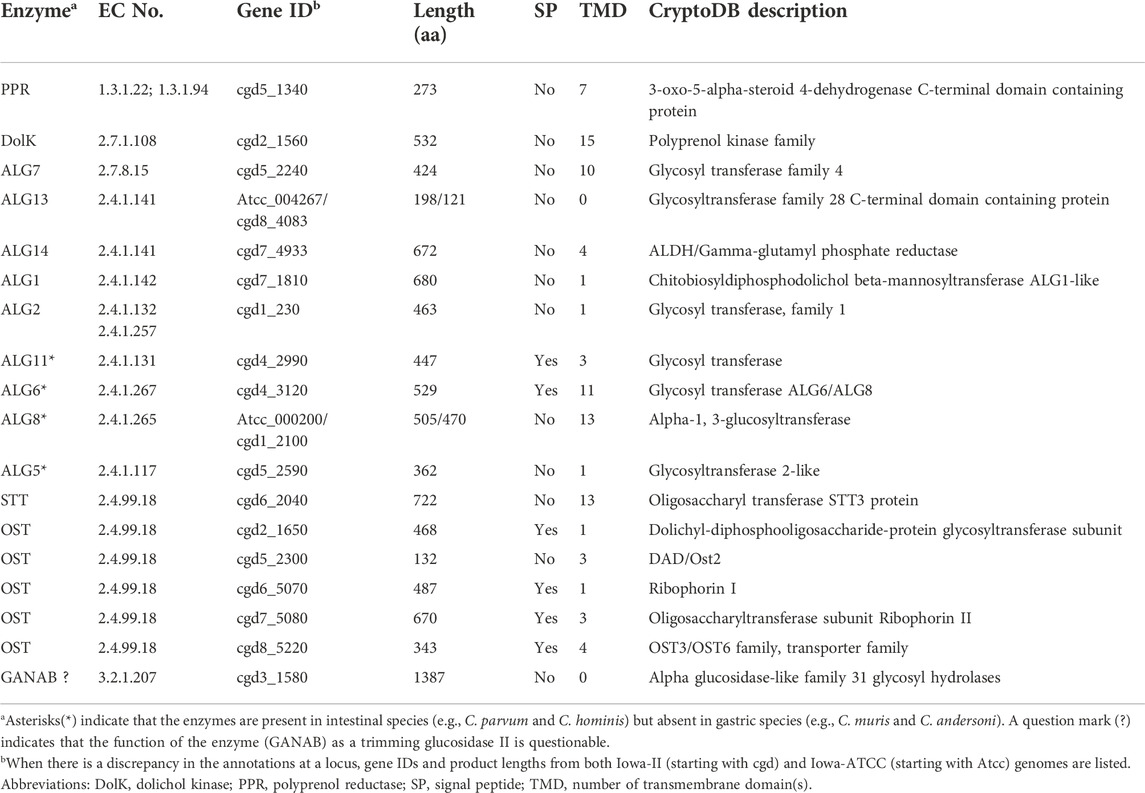
In Cryptosporidium, all enzyme orthologs responsible for the synthesis of the “core” could be identified from their genomes. In the reference genome based on Iowa-II strain, these enzymes and gene IDs are PPR (cgd5_1340), DolK (cgd2_1560), ALG7 (cgd5_2240), AGL13/ALG14 (cgd8_4083 and cgd7_4933), ALG1 (cgd7_1810) and ALG2 (cgd1_230) (Table 1 and Figure 2). Enzymes for the extension of the long arm, up to the addition of two mannoses and two terminal glucoses, are present in the intestinal Cryptosporidium species (e.g., C. parvum and C. hominis). These include ALG11 (cgd4_2990), ALG6 (cgd4_3120) and ALG8 (cgd1_2100). Therefore, the predicted N-glycan precursor in C. parvum and other intestinal species is Glc2Man5GlcNAc2, which is simpler than that in the hosts (Figure 1A).
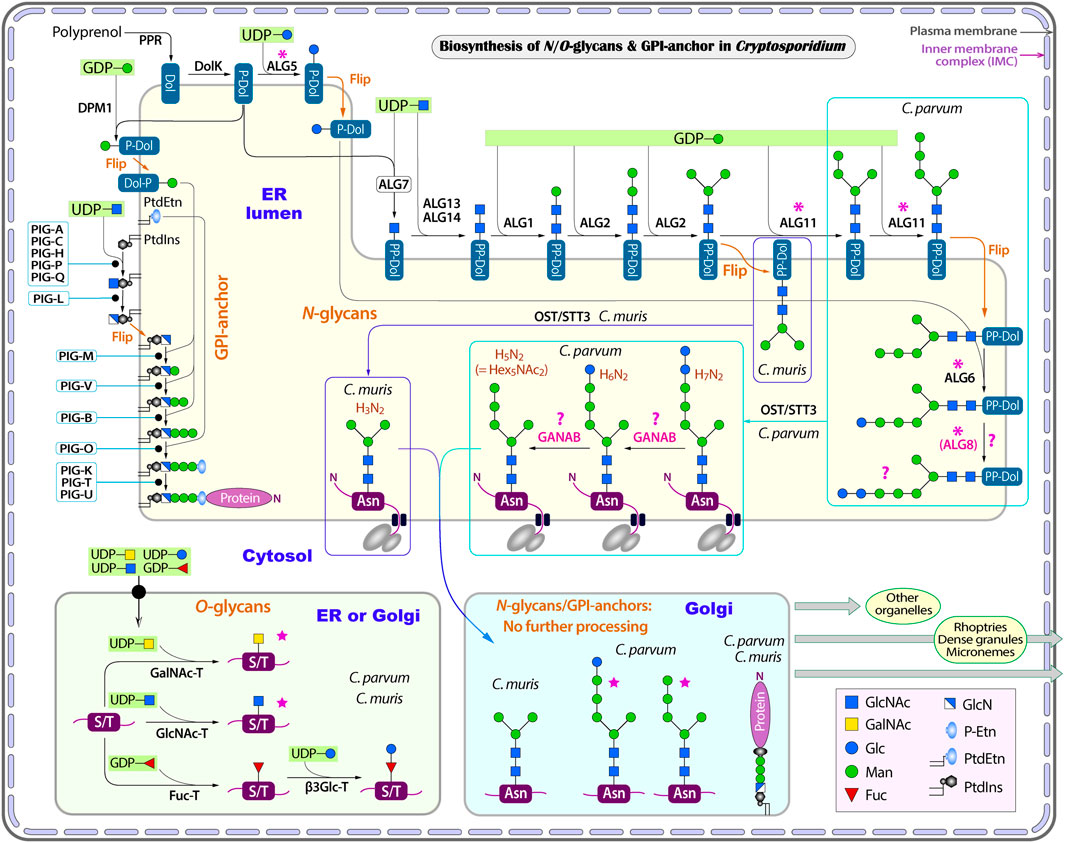
FIGURE 2. Illustration of glycosylation pathways in Cryptosporidium as exemplified with C. parvum (intestinal species) and C. muris (gastric species). These include the biosynthesis of N- and O-linked glycans and glycosylphosphatidylinositol (GPI) anchor in the endoplasmic reticulum (ER) and the Golgi apparatus. Five-point purple stars indicate that the glycoforms have been detected by mass spectrometric analysis in the literature. Asterisks indicate enzymes that are present in the intestinal Cryptosporidium species (e.g., C. parvum) but absent in gastric species (e.g., C. muris). Question marks indicate questionable enzymes or glycoforms. Enzyme abbreviations: ALG, asparagine-linked glycosylation enzyme series; β3Glc-T, UDP-glucose:O-linked fucose beta-1,3-glucosyltransferase; DolK, dolichol kinase; DPM1, dolichol-phosphate mannosyltransferase 1; Fuc-T, peptide-O-fucosyltransferase; GANAB, trimming glucosidase II (mannosyl-oligosaccharide alpha-1,3-glucosidase GANAB); GlcNAc-T, polypeptide N-acetylglucosaminyltransferase; GalNAc-T, polypeptide N-acetylgalactosaminyltransferase; OST, oligosaccharyltransferase complex; PIG, phosphatidylinositol glycan enzyme series; PPR, polyprenol reductase; STT3, oligosaccharyltransferase STT3 subunit (catalytic). Compound abbreviations: Fuc, fucose; Gal, Galactose; GalNAc, N-acetylgalactosamine; Glc, glucose; GlcN, glucosamine; GlcNAc, N-acetylglucosamine; Man, mannose; P-Etn, phosphoethanolamine; PtdEtn, phosphatidylethanolamine; PtdIns, phosphatidylinositol.
However, the three enzymes for further extension of the core (i.e., ALG11, ALG6 and ALG8), and the enzyme ALG5 for synthesizing Glc-P-Dol, are absent in C. muris and C. andersoni (Figure 2 and Table 1, marked with asterisks), suggesting that the N-glycan precursor is further simplified in the gastric Cryptosporidium species (i.e., Man3GlcNAc2). It is intriguing to see a significant structural diversity in the N-glycan precursors within a single genus. Structural diversity in the N-glycan precursors has also been found in the piroplasmids (Florin-Christensen et al., 2021). In fact, the N-glycosylation seems to be highly amendable for the protozoan parasites to adapt to varied parasitic lifestyles in diverse host environments (Mule et al., 2020; Toustou et al., 2022). For comparison with other apicomplexans, the N-glycan precursors in the cyst-forming coccidian T. gondii is predicted to be similar to that in C. parvum, but containing three terminal glucoses in the long arm (Figure 1A) (Fauquenoy et al., 2011; Haserick et al., 2017b). Overall, the predicted N-glycan precursors in Cryptosporidium parasites are much simpler than those in humans and animals, and there is a significant diversity in the precursors among different apicomplexan lineages.
It is noticed that ALG13 is missing in the current KEGG pathway maps for C. parvum, while our analysis showed the presence of a candidate ortholog for encoding ALG13 (cgd8_4083). This is due to the fact that KEGG pathway maps for C. parvum were based on an earlier version of the annotation of the parasite genome, in which the cgd8_4083 locus was not identified as protein-coding site due to the presence of introns. There are also variations in the annotations at the ALG13, ALG14 and ALG18 loci between the Iowa-II and Iowa-ATCC genomes in the CryptoDB. For ALG13, cgd8_4083 (Iowa-II) was predicted to contain two introns and encodes 121 amino acids, while CPATCC_004267 (Iowa-ATCC) has three introns and encode 198 amino acids. For ALG14, cgd7_4933 (Iowa-II) contains three introns and encodes 672 amino acids. However at the same region, the Iowa-ATCC genome is separated into two genes, i.e., CPATCC_003712 (two introns) and CPATCC_003713 (no intron) that encode 226 and 443 amino acids, respectively. All three predicted products are annotated as “ALDH/Gamma-glutamyl phosphate reductase” at the CryptoDB. At the ALG8 locus, cgd1_2100 (Iowa-II) contains two introns near the 5′-end, predicting a 470 aa product, while CPATCC_000200 contains three introns near the 5′-end and encodes a 505 aa product. Overall, variances in intron prediction is the cause of discrepancies in annotations, which could be clarified by careful sequencing the RT-PCR products derived from their transcripts.
Cryptosporidium possesses a full set of oligosaccharyltransferase subunits to transfer N-glycan precursors to proteins, but lacks enzymes for further processing
Both gastric and intestinal Cryptosporidium species possess a set of genes encoding subunits of oligosaccharyltransferase (OST) complex responsible for the transfer of glycan precursors to the Asn residue of targeted proteins (Figure 2 and Table 1). These include orthologs for subunits alpha (ribophorin I; cgd6_5070), beta (Wbp1p; cgd2_1650), gamma (cgd8_5220), delta (ribophorin II; cgd7_5080), epsilon (DAD1/Ost2-like; cgd5_2300) and STT3 (cgd6_2040). The OST complex is slightly simpler than that in humans that contains two catalytic STT3 subunits (i.e., STT3A and STT3B), but more complex than those in the piroplasmids (Shrimal and Gilmore, 2019; Florin-Christensen et al., 2021).
However, Cryptosporidium parasites lacks enzymes to further process the N-glycans in the ER and Golgi, including those to trim the terminal sugars that takes place in the ER and Golgi and those to extend the core to build more complex glycans in the Golgi (Figure 2). It is noticed that an ortholog of the alpha glucosidase-like family 31 glycosyl hydrolase is present in both intestinal and gastric Cryptosporidium parasites (e.g., cgd5_2300 in C. parvum and CMU_011750 in C. muris). The ortholog from C. parvum is annotated in the KEGG as mannosyl-oligosaccharide alpha-1,3-glucosidase (GANAB; aka glucosidase 2). In humans, GANAB is responsible for trimming the second and third glucoses after the removal of the first glucose on the long arm of the glycan precursor. However, both cgd5_2300 and CMU_011750 gene products lack an N-terminal SP and transmembrane domains (TMDs), indicating that these orthologs are not targeted to the ER, thus unable to function as GANAB. Additionally, the present of orthologs in C. muris and C. andersoni also implies that these family 31 glycosyl hydrolase orthologs unlikely function as GANAB, as gastric species lack any terminal glucoses for GANAB to act on. For comparison, GANAB in humans (UniProt: Q14697) and T. gondii (Gene ID: TGME49_253030) contains a signal peptide for targeting to the ER. For discussion, the “GANAB” ortholog is listed in Table 1 and Figure 2, but marked with question marks.
As mentioned earlier, Cryptosporidium parasites lacks enzymes to further extend N-glycans in the Golgi. If they truly lack GANAB to trim the two terminal glucoses, the predicted final N-glycoform in C. parvum glycoproteins would contain two terminal glucoses (i.e., Glc2Man5GlcNAc2) (Figure 1B). However, a mass spectrometry-based analysis of N-glycans in glycoproteins isolated from C. parvum oocysts has detected only two N-glycoforms (Haserick et al., 2017b). The most abundant form is Hex6HexNAc2, suggesting the presence of a single terminal glucose on the long arm (i.e., Glc1Man5GlcNAc2). The less abundant form is Hex5HexNAc2, suggesting the absence of any glucose on the long arm (i.e., Man5GlcNAc2). The authors speculated “that ALG8, which adds the second glucose to the N-glycan precursor, is not active, or a glucose residue is rapidly removed by glucosidase 2 from Glc2Man5GlcNAc2 after it is transferred to the nascent peptide”. The ALG8 ortholog in C. parvum (based on CPATCC_000200 product) is predicted to be functional based on the sequence similarity with ALG6/ALG8 enzymes from other species and the presence of catalytic residue Asp in the first non-cytosolic domain between transmembrane domains 1 and 2.
Considering that the OST complexes are highly homologous within the Cryptosporidium genus and that in the gastric species could transfer a smaller core Man3GlcNAc2, it is possible that the OST complex in the intestinal C. parvum is a relatively “sloppy” enzyme, capable of transferring varied N-glycan precursors to proteins. Precursors might be transferred to proteins quickly after Man5GlcNAc2 is translocated from the cytosolic side of the ER membrane to the luminal face and/or after the addition of one glucose to the long arm (GlcMan5GlcNAc2) before the action of ALG8. The presence of multiple N-glycoforms was also observed in T. gondii, in which mass spectrometry analysis detected both Hex8NexNAc2 and Hex7NexNAc2 in GAP50 protein (Fauquenoy et al., 2011). The predicted precursor in T. gondii is Hex9NexNAc2, so that the presence of both Hex8NexNAc2 and Hex7NexNAc2 would be a result of incomplete trimming of terminal glucoses and/or the quick transfer of non-glucosylated or partially glucosylated precursors. Nonetheless, these speculations need experimental validation, such as the confirmation of the presence and subcellular localization of GANAB, ALG8 and other enzymes and their substrate specificities. Considering that the two mature glycoforms detected by Haserick and colleagues (i.e., Hex6HexNAc2 and Hex5HexNAc2) were derived from a limited number of glycoproteins from the oocysts, one could not exclude the possible presence of Hex7HexNAc2 in other glycoproteins in the oocysts or in the glycoproteins from other developmental stages of the parasite. Because of the uncertainty of the final forms of precursors, we list three hypothetic precursors in C. parvum (Figures 1, 2).
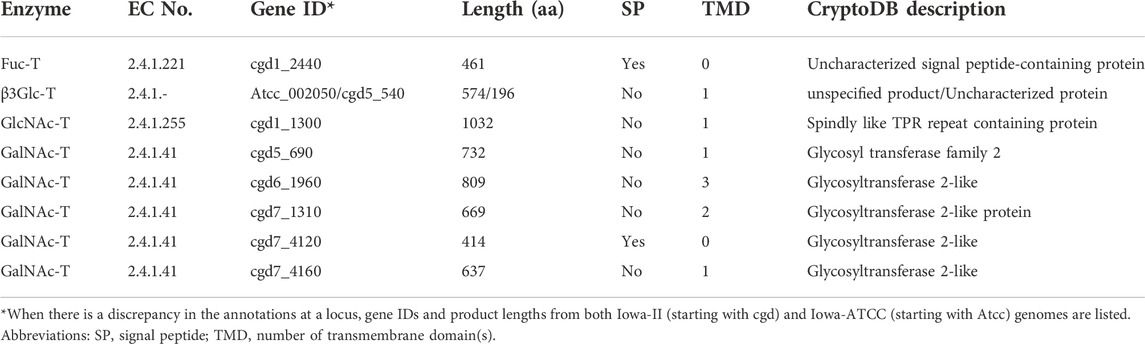
Cryptosporidium parasites produce mainly one-unit O-linked glycans
In eukaryotes, O-glycosylation occurs in the ER and Golgi (Van den Steen et al., 1998; Joshi et al., 2018). Humans and animals make a diverse of O-glycans attached to the hydroxyl groups of Ser or Thr (S/T) residues in a protein, including mucin-type, mannose-type and other types of O-glycans. The oligosaccharides of O-glycans can be single-chained or branched. Both gastric and intestinal Cryptosporidium species contain three types of O-glycan transferases (Figure 2 and Table 2): protein O-fucosyltransferase (Fuc-T; cgd1_2440), protein O-GlcNAc transferase (GlcNAc-T; cgd1_1300) and five candidates for polypeptide GalNAc transferase (GalNAc-T; cgd5_690, cgd6_1960, cgd7_1310, cgd7_4120 and cgd7_4160). These transferases possess either a signal peptide or one to two TMDs for targeting to the ER/Golgi where O-glycosylation occurs (Table 2). Cryptosporidium lacks enzymes to add more sugars to the one-unit O-GlcNAc and O-GalNAc glycans, but possesses a putative UDP-glucose:O-linked fucose beta-1,3-glucosyltransferase (β3Glc-T; cgd5_540) to potentially extend the O-Fuc with a glucose. Therefore, Cryptosporidium parasites are predicted to make only three types of simple O-glycoforms: GlcNAc(β1)-S/T, GalNAc(β1)-S/T and Glc(β1-3)Fuc(α3)-S/T (Figure 2).
The annotation of cgd5_540 and orthologs are less certain, though. At this locus, the earliest C. parvum (Iowa-II) genome project predicted a 507-aa long partial “hypothetical protein” that contains a domain homologous to galactosyltransferase (GenBank: XP_001388252; two introns). The current annotation of the Iowa-II genome in the CryptoDB predicted a 196 aa protein (no introns). The annotation of the Iowa-ATCC genome predicted (CPATCC_002050; four introns) a 574 aa “unspecified product”. The CPATCC_002050 product is the longest of the three predictions, containing a Fringe domain at the C-terminal half of the protein (InterProScan ID: IPR017374). Fringe proteins are a family of glycosyltransferases, including beta-1,3-glucosyltransferase (i.e., β3Glc-T) that glucosylates O-linked fucosylglycan on thrombospondin type 1 repeat domains in humans (Sato et al., 2006). The C. parvum ortholog contains no signal peptide, but has a TMD very close to the N-terminus (amino acid positions from 6 to 22). The architecture resembles human β3Glc-T (UniProt: Q6Y288; GenBank: NP_919299), but the long domain is predicted to be cytosolic, rather than non-cytoplasmic by Phobius and InterProScan. Collectively, it is certain that the cdg5_540 locus encodes a glycosyltransferase, likely β3Glc-T, but its true identity and function need experimental validation.
O-glycans in proteins from the C. parvum oocysts were detected by mass spectrometry-based analysis that identified HexNAc from four glycoprotein (i.e., gp40, gp15, gp20 and gp900) (Haserick et al., 2017a). The acetylhexosamine was thought to be likely GalNAc by the authors based on earlier report on the presence of four GalNAc-Ts. The presence of a putative GlcNAc-T in the C. parvum genome suggests that both GalNAc and GlcNAc might exit. The same study failed to detect Glc-Fuc or Fuc, which might be due to the fact that only four highly abundant glycoproteins from a single developmental stage (oocysts) were analyzed. Further experiments are needed to validate the presence/absence and the scale of O-fucosylation in the parasite.
Cryptosporidium parasites synthesize only three nucleotide-sugars de novo as glycan substrates, but possess at least 12 transporters with substrate preference towards nucleotide sugars or phosphorylated sugars
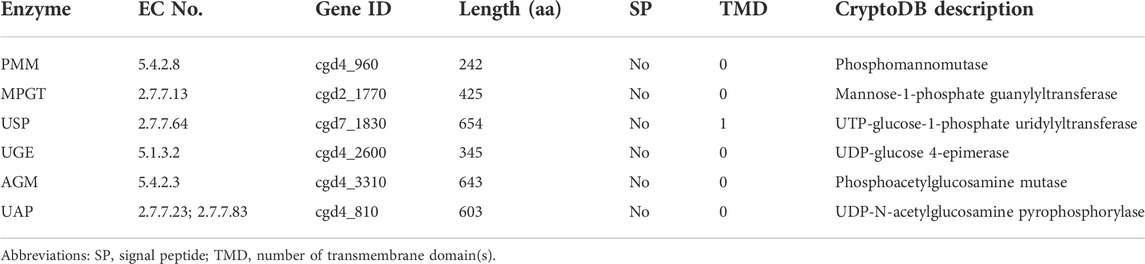
While humans and animals synthesize all nucleotide sugar substrates for use in glycosylation, Cryptosporidium only makes GDP-Man, UDP-Glc and UDP-Gal from the glycolytic pathway (Figure 3 and Table 3). The parasite lacks enzymes to make other nucleotide sugars de novo, including GDP-Fuc, UDP-GlcNAc and UDP-GalNAc. It contains an ortholog of dual functional UDP-N-acetylglucosamine/UDP-N-acetylgalactosamine diphosphorylase (UAP; cgd4_810) that may potentially convert phosphorylated N-acetylated amino sugars (e.g., GlcNAc-1P and GalNAc-1P) into UDP-sugars (e.g., UDP-GlcNAc and UDP-GalNAc). In Cryptosporidium, GDP-Man is used in synthesizing both N-glycans and GPI-anchor (on the cytosolic and luminal side of the ER, respectively) (Figure 3), while other nucleotide sugars are used in synthesizing N- and O-glycans. Cryptosporidium lacks enzymes to make GDP-Fuc from either fucose or GDP-Man (vs. humans possessing both pathways), so that the parasite has to scavenge GDP-Fuc from the host for use in O-fucosylation. It is worth to mention that Cryptosporidium is unable to synthesize nucleosides and pentoses de novo, but contains enzymes to produce various nucleotides from nucleosides (e.g., GDP and UDP) (Abrahamsen et al., 2004; Xu et al., 2004; Rider and Zhu, 2010).
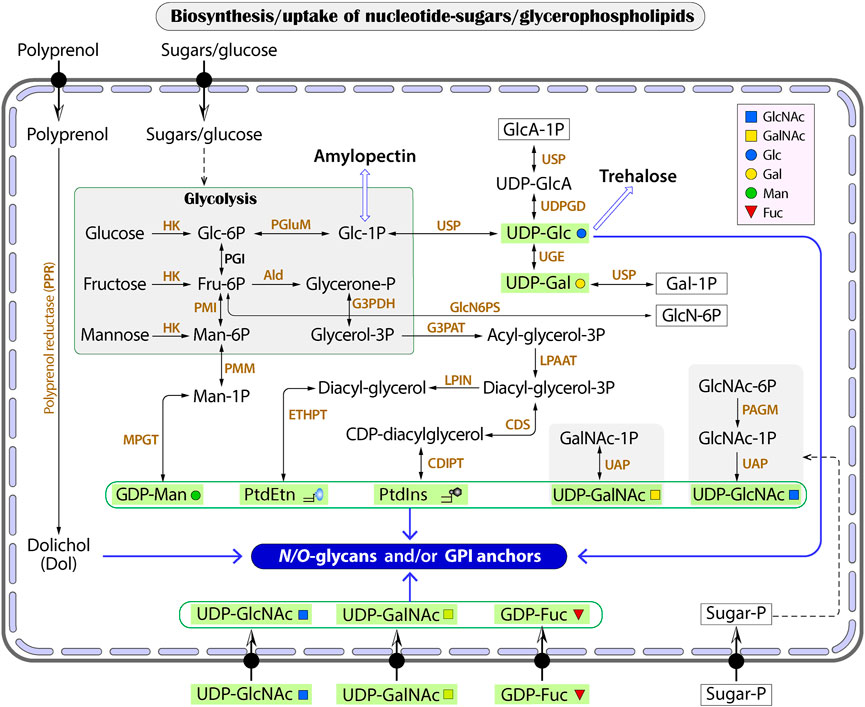
FIGURE 3. The biosynthesis of glycan substrates derived from glycolysis or conversions between phosphorylated HexNAc (GalNAc-1P and GlcNAc-1P) and UDP-HexNAc. Transporters for nucleotide sugars and phosphorylated sugars are illustrated without naming of specific proteins. Wide arrows indicate multi-step reactions. Enzyme abbreviations: Ald, aldolase; CDIPT, CDP-diacylglycerol-inositol 3-phosphatidyltransferase (phosphatidylinositol synthase); CDS, CDP-diacylglycerol synthase; ETHPT, ethanolaminephosphotransferase; G3PAT, glycerol-3-phosphate acyltransferase; G3PDH, glycerol-3-phosphate dehydrogenase; HK, hexokinase; LPIN, phosphatidate phosphatase LPIN; LPAAT, lysophosphatidic acid-acyltransferase (LPA acyltransferase); PAGM, phosphoacetylglucosamine mutase; PGI, phosphoglucose isomerase; PGluM, phosphoglucose mutase; PMGD, mannose-1P guanylyltransferase; PMI, phosphomannose isomerase; PMM, phosphomannose mutase; UAP, UDP-N-acetylglucosamine/UDP-N-acetylgalactosamine diphosphorylase; UDPDG, UDP-glucose 6-dehydrogenase; UGE, UDP-glucose 4-epimerase; USP, UDP-sugar pyrophosphorylase (UDP-N-acetylglucosamine pyrophosphorylase). Compound abbreviations: Fru, fructose; Gal, galactose; GalNAc, N-acetylgalactosamine; Glc, glucose; GlcA, glucuronic acid; GlcN, glucosamine; GlcNAc, N-acetylglucosamine; Man, mannose; PtdEtn, phosphatidylethanolamine; PtdIns, phosphatidylinositol.
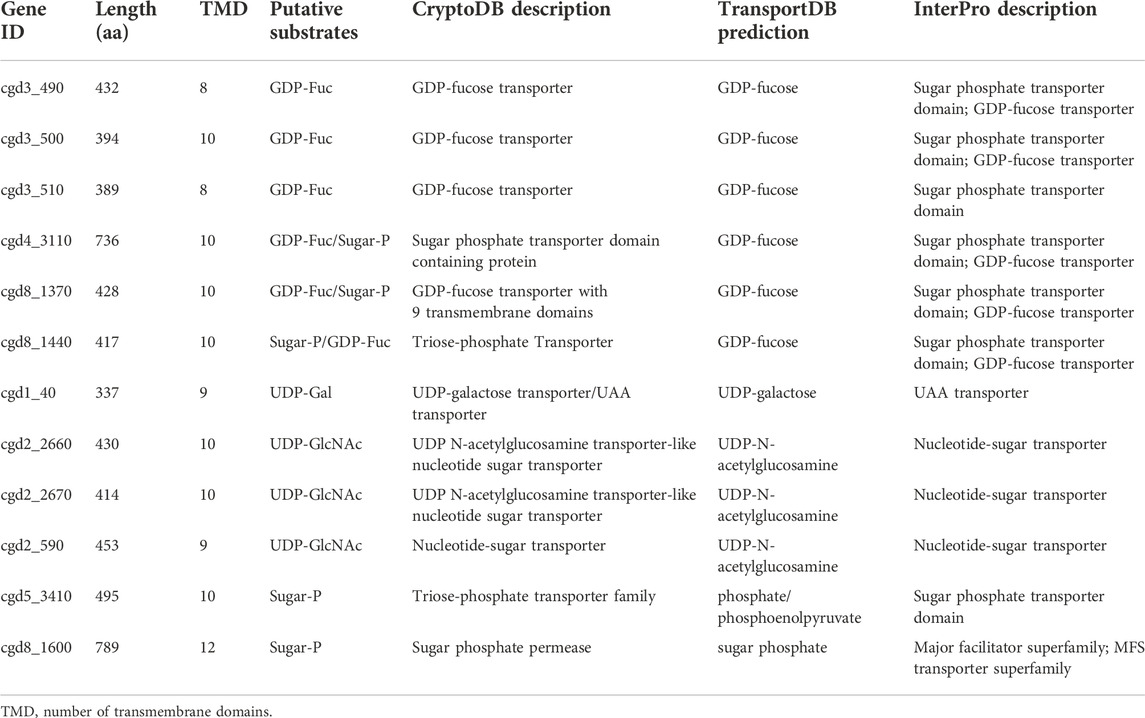
On the other hand, Cryptosporidium possesses at least 12 transporters with predicted substrate specificity on nucleotide-sugars based on genome analysis with InterProScan and TransportDB, together with the annotation of transporters on the Iowa-ATCC genome of C. parvum by other investigators (Table 4). These include six putative GDP-Fuc transporters, in which three are arranged in tandem in chromosome 3 (i.e., cgd3_490, cgd3_500 and cgd3_510). There are four UDP-sugar transporters for UDP-GlcNAc or UDP-Gal, in which two are arranged in tandem in chromosome 2 (i.e., cgd2_2660 and cgd2_2670). There are two putative transporters for phosphorylated sugars. There is a need to determine the subcellular localizations of these transporters to confirm whether they are responsible for scavenging substrates from the host and/or for transporting substrates into the ER/Golgi lumens. The true substrates of these putative transporters also remain to be elucidated.
The Cryptosporidium genomes also encode a “mannose-P-dolichol utilization defect 1” (MPDU1; cgd2_550) that was found to be required for Dol-P-monosaccharide-dependent glycosyltransferase activities in mammals (Anand et al., 2001; van Tol et al., 2019). In Cryptosporidium, MPDU1 would play roles in assisting the synthesis of N-glycans and GPI-anchors that utilize Dol-P-Glc and Dol-P-Man as substrates, respectively.
Cryptosporidium parasites synthesize unbranched GPI-anchors
The GPI-anchor in Cryptosporidium is predicted to be unbranched. Both gastric and intestinal Cryptosporidium species contain a set of genes encoding various classes of phosphatidylinositol glycan (PIG) protein (Table 5 and Figure 2). The GPI-anchor biosynthesis starts with the action of phosphatidylinositol N-acetylglucosaminyltransferase complex that catalyzes the attachment of GlcNAc to the phosphatidylinositol (PtdIns) on the cytosolic side of the ER membrane (Kinoshita and Fujita, 2016; Morotti et al., 2019). This complex in Cryptosporidium consists of five subunits, including PIG-A (cgd4_2100), PIG-C (cgd1_3380), PIG-H (cgd6_4600), PIG-P (cgd2_840) and PIG-Q (cgd5_1850). The parasite contains PIG-L (cgd7_700) to deacetylate the attached GlcNAc to GlcN (glucosamine), also on the cytosolic side of the ER membrane. PtdIns-GlcN is translocated into the ER luminal side, from which three mannoses (Man1, Man2 and Man3) from Dol-P-Man are sequentially attached to GlcN by PIG-M (cgd1_3720), PIG-V (cgd6_5100) and PIG-B (cgd3_3590), respectively. Dol-P-Man is synthesized by Dol-P-Man synthase (DPM1; cgd5_2040). In humans and animals, Dol-P-Man is also a donor of mannoses in synthesizing complex N-glycans (e.g., high-mannose N-glycans) that are absent in Cryptosporidium. Therefore, Dol-P-Man seems to be solely used as a substrate in GPI-anchor synthesis in the parasite.
In the final two steps, a “bridging” ethanolamine-phosphate (Etn-P) is transferred from phosphatidylethanolamine (PtdEtn) to the 6-position of Man3 by PIG-O (cgd8_2260) (Figure 2). PtdEtn can be derived from the glycolytic pathway in Cryptosporidium that contains a full set of five enzymes to convert glycerone-P to PtdEtn. These include glycerol-3-phosphate dehydrogenase (G3PDH; cgd2_210), glycerol-3-phosphate acyltransferase (G3PAT; cgd6_1270), two lysophosphatidic acid-acyltransferases (LPAAT1; cgd2_4050 and LPAAT2; cgd8_1400), phosphatidate phosphatase (LPIN; cgd3_3210), and ethanolamine-phosphotransferase (ETHPT; cgd4_2790). Finally, GPI transamidase consisting of three subunits, i.e., PIG-K (cgd1_220), PIG-T (cgd6_3240) and PIG-U (cgd7_540), catalyzes the attachment of GPI to a targeted protein. In this reaction, PIG-K acts as a caspase-like cysteine peptidase that cleaves off the C-terminal GPI attachment signal peptide.
There are some discrepancies in the annotation of three genes between the two C. parvum genomes. 1) DPM1 gene in the Iowa-II genome (cgd5_2040) contains four introns and encodes a 242 aa product, while that in Iowa-ATCC (CPATCC_002211) contains four introns and encodes a 219 aa product. The intron predictions are identical in both annotations, but the open reading frame of cgd5_2040 contains 69 extra coding sequences in the upstream of the gene. 2) PIG-L in Iowa-II (cgd7_700) contains two introns and encodes a 175 aa, while in Iowa-ATCC (CPATCC_003223) it contains four introns and encodes a 220 aa product. 3) PIG-M in Iowa-II (cgd1_3720) is predicted to have one intron and encode a 382 aa product, while that in Iowa-ATCC (CPATCC_000018) contains two introns and encodes a 462 aa product.
In comparison to humans and animals whose GPI-anchors are branched with two individual Etn-P molecules attached to Man1 (2-position) and Man3 (6-position), Cryptosporidium lacks PIG-N and PIG-G to add any Etn-P to the side of the GPI-anchor. Cryptosporidium also lacks PIG-W to attach a long fatty acyl chain to PtdIns that occurs in the humans immediately after the translocation of PtdIns-GlcN to the luminal face of ER. The GPI-anchor in Cryptosporidium is not processed further in the Golgi by adding additional sugars (e.g., GalNAc, Gal or sialic acid). Therefore, the final form of the GPI in the parasite is simple and unbranched, differing significantly from the hosts.
Implication on the parasite biology
The importance of glycoproteins in Cryptosporidium has been indicated by their presence in all stages of the parasite, including the oocyst walls (e.g., the cryptosporidial oocyst wall proteins (COWPs) and molecules tethering sporozoites on the inner side of the oocyst wall), the surface of sporozoites (e.g., gp40/gp15, CpMuc4) and the PVM (recognizable by certain lectins such as VVL) (Petersen et al., 1992; Winter et al., 2000; Priest et al., 2001; Wanyiri and Ward, 2006; Paluszynski et al., 2014; Haserick et al., 2017a; Haserick et al., 2017b; Tandel et al., 2019; Li et al., 2022). Structural glycoproteins play protective and lubricative roles, such as those in the oocyst walls and those on the plasma membranes of zoites and PVM. These structures protect the parasite from hazardous factors in the environments or the gastrointestinal tracts. Glycoproteins distributed on the surface of zoites or discharged from the zoites also interact with host cells during the parasite invasion of host cells.
While the N-glycoform in the intestinal Cryptosporidium is of simplicity, that in the gastric species is further simplified to contain only the core structure. The further reduction of complexity of the N-glycoform in species of the same genus is an intriguing subject of study in the evolution of glycosylation pathways and the adaptation of parasitic lifestyles from the intestine to the stomach. The lack of enzymes to further process N-glycans in the Golgi is in fact a general feature in the apicomplexans based on the KEGG annotations and mass-spectrometric evidence for Cryptosporidium, Toxoplasma and Plasmodium (Priest et al., 2003; Fauquenoy et al., 2011; Haserick et al., 2017a; Haserick et al., 2017b). Hematozoa (e.g., Plasmodium, Babesia and Theileria) even make highly truncated N-glycans (i.e., NAc2 or Man1NAc2), in which most Theileria species and B. microti also lost all OST/STT subunits (Figure 3) (Morotti et al., 2019; Florin-Christensen et al., 2021). On the other hand, other alveolates (e.g., ciliates and dinoflagellates) may possess some enzymes to process N-glycans in the Golgi (e.g., various types of mannosidases and GlcNAc transferases), although not as extensive as those in vertebrates and other higher eukaryotes, based on KEGG pathway annotations and literature (Tivey et al., 2020; Toustou et al., 2022). These imply that the apicomplexan ancestor might process glycans in the Golgi, at least to a certain degree, but the ability was lost in adaptation of the intracellular parasitic lifestyle. Because apicomplexans possess an apicoplast derived from an algae via a secondary endosymbiosis (although it has been lost in the Cryptosporidium lineage), there was a hypothesis for “Darwinian selection against N-glycans in nucleus-encoded proteins that must pass through the ER prior to threading into the apicoplast” (Bushkin et al., 2010). The selection against glycosylation would affect non-apicoplast proteins as well (e.g., secretory proteins) because the process would apply to both apicoplast and non-apicoplast proteins in the ER or Golgi.
Another interesting feature is the presence of malectin in Cryptosporidium (cgd6_110 and orthologs), but absent in other apicomplexan lineages (Wu et al., 2022). Malectin in humans and animals binds to Glc2Man9GlcNAc2 N-glycan after the trimming of the terminal glucose from Glc3Man9GlcNAc2 to facilitate the recruitment of glucosidase II (Schallus et al., 2008; Schallus et al., 2010; Galli et al., 2011; Takeda et al., 2014). However, gastric Cryptosporidium lacks any terminal glucoses or mannoses but possesses malectin, indicating that Cryptosporidium malectin plays a function that differs from humans and animals. This notion is supported by a recent study that showed difference in binding properties between human and Cryptosporidium malectins (Wu et al., 2022), although the exact binding partner remains to be elucidated.
The physiochemical properties of glycans differ between Cryptosporidium and hosts due to the differences in their compositions and structures. These need to be considered in studying the roles of the parasite glycoproteins. The host gastrointestinal tracts are covered by a layer of mucosa consisting of a network of mucins (mainly mucin-2) that is negatively charged due to the presence of sialic acid (Boegh and Nielsen, 2015; Wagner et al., 2018). The negative charge allows the mucins to bind to positively charged molecules on microbes, thus restricting the movement of microbes. This is one of the mechanisms of protection from free access of pathogens to the epithelial cells. On the other hand, the glycans in Cryptosporidium are not only simplified, but also neutrally charged by lacking sialic acid. Therefore, the glycans themselves would make no or little contribution to the adhesion of the parasite glycoproteins to the host cell extracellular structures, which has been proposed for the gp900 (Li et al., 2022). Instead, glycans in the parasite mainly play a lubrication role and/or are responsible for binding to certain lectins (if any) on the host cells.
The genetically trackable T. gondii has been used as a surrogate system to study C. parvum glycoproteins (e.g., CpGP40/15) (O'Connor et al., 2003). Knowing the difference between the native glycoforms of C. parvum and T. gondii would allow investigators to consider manipulate the pathway in T. gondii to make true cryptosporidium-type glycans. Many vaccine candidates are also glycoproteins (Mead, 2014), in which glycans are a source of antigenicity in triggering immunological responses in the hosts. However, recombinant proteins expressed in prokaryotes are not glycosylated. Eukaryotic expression systems (e.g., yeasts, insect cell-based baculovirus or mammalian cells) might glycosylate heterogeneous proteins, but in different glycoforms. Knowing the glycoforms in specified vaccine candidates for cryptosporidiosis would guide investigators in protein glycoengineering or manipulating the glycosylation pathway in prokaryotic or eukaryotic expression systems to produce immunogens with correct glycoforms (Anyaogu and Mortensen, 2015; Hamilton and Zha, 2015; Naegeli and Aebi, 2015; Ma et al., 2020; Schon et al., 2021).
Author contributions
DW, CW, and GZ performed the datamining, genome analysis and pathway reconstruction. GZ and DW wrote the manuscript draft. CW rechecked annotations of individual enzymes. GZ wrote the final version of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The authors are partly funded by a grant from the National Natural Science Foundation of China (project number 32250710141).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abrahamsen, M. S., Templeton, T. J., Enomoto, S., Abrahante, J. E., Zhu, G., Lancto, C. A., et al. (2004). Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304, 441–445. doi:10.1126/science.1094786
Amos, B., Aurrecoechea, C., Barba, M., Barreto, A., Basenko, E. Y., Bazant, W., et al. (2022). VEuPathDB: The eukaryotic pathogen, vector and host bioinformatics resource center. Nucleic Acids Res. 50, D898–D911. doi:10.1093/nar/gkab929
Anand, M., Rush, J. S., Ray, S., Doucey, M. A., Weik, J., Ware, F. E., et al. (2001). Requirement of the Lec35 gene for all known classes of monosaccharide-P-dolichol-dependent glycosyltransferase reactions in mammals. Mol. Biol. Cell 12, 487–501. doi:10.1091/mbc.12.2.487
Anantharaman, V., Iyer, L. M., Balaji, S., and Aravind, L. (2007). Adhesion molecules and other secreted host-interaction determinants in Apicomplexa: Insights from comparative genomics. Int. Rev. Cytol. 262, 1–74. doi:10.1016/S0074-7696(07)62001-4
Anyaogu, D. C., and Mortensen, U. H. (2015). Manipulating the glycosylation pathway in bacterial and lower eukaryotes for production of therapeutic proteins. Curr. Opin. Biotechnol. 36, 122–128. doi:10.1016/j.copbio.2015.08.012
Baptista, R. P., Li, Y., Sateriale, A., Sanders, M. J., Brooks, K. L., Tracey, A., et al. (2022). Long-read assembly and comparative evidence-based reanalysis of Cryptosporidium genome sequences reveal expanded transporter repertoire and duplication of entire chromosome ends including subtelomeric regions. Genome Res. 32, 203–213. doi:10.1101/gr.275325.121
Boegh, M., and Nielsen, H. M. (2015). Mucus as a barrier to drug delivery - understanding and mimicking the barrier properties. Basic Clin. Pharmacol. Toxicol. 116, 179–186. doi:10.1111/bcpt.12342
Bushkin, G. G., Ratner, D. M., Cui, J., Banerjee, S., Duraisingh, M. T., Jennings, C. V., et al. (2010). Suggestive evidence for Darwinian Selection against asparagine-linked glycans of Plasmodium falciparum and Toxoplasma gondii. Eukaryot. Cell 9, 228–241. doi:10.1128/EC.00197-09
Checkley, W., White, A. C., Jaganath, D., Arrowood, M. J., Chalmers, R. M., Chen, X. M., et al. (2015). A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet. Infect. Dis. 15, 85–94. doi:10.1016/S1473-3099(14)70772-8
Cherepanova, N., Shrimal, S., and Gilmore, R. (2016). N-linked glycosylation and homeostasis of the endoplasmic reticulum. Curr. Opin. Cell Biol. 41, 57–65. doi:10.1016/j.ceb.2016.03.021
Chung, C. Y., Majewska, N. I., Wang, Q., Paul, J. T., and Betenbaugh, M. J. (2017). SnapShot: N-glycosylation processing pathways across kingdoms. Cell 171, 258–258 e251. doi:10.1016/j.cell.2017.09.014
Corfield, A. (2017). Eukaryotic protein glycosylation: A primer for histochemists and cell biologists. Histochem. Cell Biol. 147, 119–147. doi:10.1007/s00418-016-1526-4
Elbourne, L. D., Tetu, S. G., Hassan, K. A., and Paulsen, I. T. (2017). TransportDB 2.0: A database for exploring membrane transporters in sequenced genomes from all domains of life. Nucleic Acids Res. 45, D320–D324. doi:10.1093/nar/gkw1068
Fauquenoy, S., Hovasse, A., Sloves, P. J., Morelle, W., Dilezitoko Alayi, T., Slomianny, C., et al. (2011). Unusual N-glycan structures required for trafficking Toxoplasma gondii GAP50 to the inner membrane complex regulate host cell entry through parasite motility. Mol. Cell. Proteomics 10, M111.008953008953. doi:10.1074/mcp.M111.008953
Florin-Christensen, M., Rodriguez, A. E., Suarez, C. E., Ueti, M. W., Delgado, F. O., Echaide, I., et al. (2021). N-glycosylation in piroplasmids: Diversity within simplicity. Pathogens 10, 50. doi:10.3390/pathogens10010050
Galli, C., Bernasconi, R., Solda, T., Calanca, V., and Molinari, M. (2011). Malectin participates in a backup glycoprotein quality control pathway in the mammalian ER. PLoS One 6, e16304. doi:10.1371/journal.pone.0016304
Hamilton, S. R., and Zha, D. (2015). Progress in yeast glycosylation engineering. Methods Mol. Biol. 1321, 73–90. doi:10.1007/978-1-4939-2760-9_6
Haserick, J. R., Klein, J. A., Costello, C. E., and Samuelson, J. (2017a). Cryptosporidium parvum vaccine candidates are incompletely modified with O-linked-N-acetylgalactosamine or contain N-terminal N-myristate and S-palmitate. PLoS One 12, e0182395. doi:10.1371/journal.pone.0182395
Haserick, J. R., Leon, D. R., Samuelson, J., and Costello, C. E. (2017b). Asparagine-linked glycans of Cryptosporidium parvum contain a single long arm, are barely processed in the endoplasmic reticulum (ER) or Golgi, and show a strong bias for sites with threonine. Mol. Cell. Proteomics 16, S42–S53. doi:10.1074/mcp.M116.066035
Ifeonu, O. O., Simon, R., Tennant, S. M., Sheoran, A. S., Daly, M. C., Felix, V., et al. (2016). Cryptosporidium hominis gene catalog: A resource for the selection of novel cryptosporidium vaccine candidates. Oxford: Database. doi:10.1093/database/baw137
Innes, E. A., Chalmers, R. M., Wells, B., and Pawlowic, M. C. (2020). A one health approach to tackle cryptosporidiosis. Trends Parasitol. 36, 290–303. doi:10.1016/j.pt.2019.12.016
Joshi, H. J., Narimatsu, Y., Schjoldager, K. T., Tytgat, H. L. P., Aebi, M., Clausen, H., et al. (2018). SnapShot: O-glycosylation pathways across kingdoms. Cell 172, 632–632 e632. doi:10.1016/j.cell.2018.01.016
Kall, L., Krogh, A., and Sonnhammer, E. L. (2007). Advantages of combined transmembrane topology and signal peptide prediction-the Phobius web server. Nucleic Acids Res. 35, W429–W432. doi:10.1093/nar/gkm256
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M., and Ishiguro-Watanabe, M. (2022). KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res., gkac963. doi:10.1093/nar/gkac963
Kinoshita, T. (2020). Biosynthesis and biology of mammalian GPI-anchored proteins. Open Biol. 10, 190290. doi:10.1098/rsob.190290
Kinoshita, T., and Fujita, M. (2016). Biosynthesis of GPI-anchored proteins: Special emphasis on GPI lipid remodeling. J. Lipid Res. 57, 6–24. doi:10.1194/jlr.R063313
Lendner, M., and Daugschies, A. (2014). Cryptosporidium infections: Molecular advances. Parasitology 141, 1511–1532. doi:10.1017/S0031182014000237
Li, X., Yin, J., Wang, D., Gao, X., Zhang, Y., Wu, M., et al. (2022). The mucin-like, secretory type-I transmembrane glycoprotein GP900 in the apicomplexan Cryptosporidium parvum is cleaved in the secretory pathway and likely plays a lubrication role. Parasit. Vectors 15, 170. doi:10.1186/s13071-022-05286-8
Lin, B., Qing, X., Liao, J., and Zhuo, K. (2020). Role of protein glycosylation in host-pathogen interaction. Cells 9. doi: doi:10.3390/cells9041022
Liu, S., Roellig, D. M., Guo, Y., Li, N., Frace, M. A., Tang, K., et al. (2016). Evolution of mitosome metabolism and invasion-related proteins in Cryptosporidium. BMC Genomics 17, 1006. doi:10.1186/s12864-016-3343-5
Ma, B., Guan, X., Li, Y., Shang, S., Li, J., and Tan, Z. (2020). Protein glycoengineering: An approach for improving protein properties. Front. Chem. 8, 622. doi:10.3389/fchem.2020.00622
Madern, D., Cai, X., Abrahamsen, M. S., and Zhu, G. (2004). Evolution of Cryptosporidium parvum lactate dehydrogenase from malate dehydrogenase by a very recent event of gene duplication. Mol. Biol. Evol. 21, 489–497. doi:10.1093/molbev/msh042
Mead, J. R. (2014). Prospects for immunotherapy and vaccines against Cryptosporidium. Hum. Vaccin. Immunother. 10, 1505–1513. doi:10.4161/hv.28485
Mitchell, A. L., Attwood, T. K., Babbitt, P. C., Blum, M., Bork, P., Bridge, A., et al. (2019). InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47, D351–D360. doi:10.1093/nar/gky1100
Moremen, K. W., Tiemeyer, M., and Nairn, A. V. (2012). Vertebrate protein glycosylation: Diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 13, 448–462. doi:10.1038/nrm3383
Morotti, A. L. M., Martins-Teixeira, M. B., and Carvalho, I. (2019). Protozoan parasites glycosylphosphatidylinositol anchors: Structures, functions and trends for drug discovery. Curr. Med. Chem. 26, 4301–4322. doi:10.2174/0929867324666170727110801
Mule, S. N., Saad, J. S., Fernandes, L. R., Stolf, B. S., Cortez, M., and Palmisano, G. (2020). Protein glycosylation in Leishmania spp. Mol. Omics 16, 407–424. doi:10.1039/d0mo00043d
Naegeli, A., and Aebi, M. (2015). Current approaches to engineering N-linked protein glycosylation in bacteria. Methods Mol. Biol. 1321, 3–16. doi:10.1007/978-1-4939-2760-9_1
O'connor, R. M., Kim, K., Khan, F., and Ward, H. D. (2003). Expression of cpgp40/15 in Toxoplasma gondii: A surrogate system for the study of cryptosporidium glycoprotein antigens. Infect. Immun. 71, 6027–6034. doi:10.1128/IAI.71.10.6027-6034.2003
Paluszynski, J., Monahan, Z., Williams, M., Lai, O., Morris, C., Burns, P., et al. (2014). Biochemical and functional characterization of CpMuc4, a Cryptosporidium surface antigen that binds to host epithelial cells. Mol. Biochem. Parasitol. 193, 114–121. doi:10.1016/j.molbiopara.2014.03.005
Paschinger, K., and Wilson, I. B. H. (2019). Comparisons of N-glycans across invertebrate phyla. Parasitology 146, 1733–1742. doi:10.1017/S0031182019000398
Petersen, C., Gut, J., Doyle, P. S., Crabb, J. H., Nelson, R. G., and Leech, J. H. (1992). Characterization of a > 900, 000-M(r) Cryptosporidium parvum sporozoite glycoprotein recognized by protective hyperimmune bovine colostral immunoglobulin. Infect. Immun. 60, 5132–5138. doi:10.1128/iai.60.12.5132-5138.1992
Priest, J. W., Mehlert, A., Arrowood, M. J., Riggs, M. W., and Ferguson, M. A. (2003). Characterization of a low molecular weight glycolipid antigen from Cryptosporidium parvum. J. Biol. Chem. 278, 52212–52222. doi:10.1074/jbc.M306835200
Priest, J. W., Xie, L. T., Arrowood, M. J., and Lammie, P. J. (2001). The immunodominant 17-kDa antigen from Cryptosporidium parvum is glycosylphosphatidylinositol-anchored. Mol. Biochem. Parasitol. 113, 117–126. doi:10.1016/s0166-6851(00)00386-8
Pumipuntu, N., and Piratae, S. (2018). Cryptosporidiosis: A zoonotic disease concern. Vet. World 11, 681–686. doi:10.14202/vetworld.2018.681-686
Reily, C., Stewart, T. J., Renfrow, M. B., and Novak, J. (2019). Glycosylation in health and disease. Nat. Rev. Nephrol. 15, 346–366. doi:10.1038/s41581-019-0129-4
Rider, S. D., and Zhu, G. (2010). Cryptosporidium: Genomic and biochemical features. Exp. Parasitol. 124, 2–9. doi:10.1016/j.exppara.2008.12.014
Ryan, U., Zahedi, A., and Paparini, A. (2016). Cryptosporidium in humans and animals-a one health approach to prophylaxis. Parasite Immunol. 38, 535–547. doi:10.1111/pim.12350
Sato, T., Sato, M., Kiyohara, K., Sogabe, M., Shikanai, T., Kikuchi, N., et al. (2006). Molecular cloning and characterization of a novel human beta1, 3-glucosyltransferase, which is localized at the endoplasmic reticulum and glucosylates O-linked fucosylglycan on thrombospondin type 1 repeat domain. Glycobiology 16, 1194–1206. doi:10.1093/glycob/cwl035
Schallus, T., Fehér, K., Sternberg, U., Rybin, V., and Muhle-Goll, C. (2010). Analysis of the specific interactions between the lectin domain of malectin and diglucosides. Glycobiology 20, 1010–1020. doi:10.1093/glycob/cwq059
Schallus, T., Jaeckh, C., Feher, K., Palma, A. S., Liu, Y., Simpson, J. C., et al. (2008). Malectin: A novel carbohydrate-binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein N-glycosylation. Mol. Biol. Cell 19, 3404–3414. doi:10.1091/mbc.E08-04-0354
Schjoldager, K. T., Narimatsu, Y., Joshi, H. J., and Clausen, H. (2020). Global view of human protein glycosylation pathways and functions. Nat. Rev. Mol. Cell Biol. 21, 729–749. doi:10.1038/s41580-020-00294-x
Schon, K., Lepenies, B., and Goyette-Desjardins, G. (2021). Impact of protein glycosylation on the design of viral vaccines. Adv. Biochem. Eng. Biotechnol. 175, 319–354. doi:10.1007/10_2020_132
Shrimal, S., and Gilmore, R. (2019). Oligosaccharyltransferase structures provide novel insight into the mechanism of asparagine-linked glycosylation in prokaryotic and eukaryotic cells. Glycobiology 29, 288–297. doi:10.1093/glycob/cwy093
Takeda, K., Qin, S. Y., Matsumoto, N., and Yamamoto, K. (2014). Association of malectin with ribophorin I is crucial for attenuation of misfolded glycoprotein secretion. Biochem. Biophys. Res. Commun. 454, 436–440. doi:10.1016/j.bbrc.2014.10.102
Tandel, J., English, E. D., Sateriale, A., Gullicksrud, J. A., Beiting, D. P., Sullivan, M. C., et al. (2019). Life cycle progression and sexual development of the apicomplexan parasite Cryptosporidium parvum. Nat. Microbiol. 4, 2226–2236. doi:10.1038/s41564-019-0539-x
Templeton, T. J., Enomoto, S., Chen, W. J., Huang, C. G., Lancto, C. A., Abrahamsen, M. S., et al. (2010). A genome-sequence survey for Ascogregarina taiwanensis supports evolutionary affiliation but metabolic diversity between a Gregarine and Cryptosporidium. Mol. Biol. Evol. 27, 235–248. doi:10.1093/molbev/msp226
Tivey, T. R., Parkinson, J. E., Mandelare, P. E., Adpressa, D. A., Peng, W., Dong, X., et al. (2020). N-linked surface glycan biosynthesis, composition, inhibition, and function in Cnidarian-dinoflagellate symbiosis. Microb. Ecol. 80, 223–236. doi:10.1007/s00248-020-01487-9
Toustou, C., Walet-Balieu, M. L., Kiefer-Meyer, M. C., Houdou, M., Lerouge, P., Foulquier, F., et al. (2022). Towards understanding the extensive diversity of protein N-glycan structures in eukaryotes. Biol. Rev. Camb. Philos. Soc. 97, 732–748. doi:10.1111/brv.12820
Van Den Steen, P., Rudd, P. M., Dwek, R. A., and Opdenakker, G. (1998). Concepts and principles of O-linked glycosylation. Crit. Rev. Biochem. Mol. Biol. 33, 151–208. doi:10.1080/10409239891204198
Van Tol, W., Ashikov, A., Korsch, E., Abu Bakar, N., Willemsen, M. A., Thiel, C., et al. (2019). A mutation in mannose-phosphate-dolichol utilization defect 1 reveals clinical symptoms of congenital disorders of glycosylation type I and dystroglycanopathy. JIMD Rep. 50, 31–39. doi:10.1002/jmd2.12060
Wagner, C. E., Wheeler, K. M., and Ribbeck, K. (2018). Mucins and their role in shaping the functions of mucus barriers. Annu. Rev. Cell Dev. Biol. 34, 189–215. doi:10.1146/annurev-cellbio-100617-062818
Wang, C., Wang, D., Nie, J., Gao, X., Yin, J., and Zhu, G. (2021). Unique tubulin-based structures in the zoonotic apicomplexan parasite Cryptosporidium parvum. Microorganisms 9, 1921. doi:10.3390/microorganisms9091921
Wanyiri, J., and Ward, H. (2006). Molecular basis of Cryptosporidium-host cell interactions: Recent advances and future prospects. Future Microbiol. 1, 201–208. doi:10.2217/17460913.1.2.201
Winter, G., Gooley, A. A., Williams, K. L., and Slade, M. B. (2000). Characterization of a major sporozoite surface glycoprotein of Cryptosporidum parvum. Funct. Integr. Genomics 1, 207–217. doi:10.1007/s101420000028
Wu, M., Yang, B., Wang, D., Zhang, Y., Li, X., Zhi, Y., et al. (2022). Zoonotic Cryptosporidium parasites possess a unique carbohydrate-binding protein (malectin) that is absent in other apicomplexan lineages. Zoonoses 2, 12. doi:10.15212/zoonoses-2022-0011
Xu, P., Widmer, G., Wang, Y., Ozaki, L. S., Alves, J. M., Serrano, M. G., et al. (2004). The genome of Cryptosporidium hominis. Nature 431, 1107–1112. doi:10.1038/nature02977
Zhu, G., Keithly, J. S., and Philippe, H. (2000a). What is the phylogenetic position of Cryptosporidium? Int. J. Syst. Evol. Microbiol. 50, 1673–1681. doi:10.1099/00207713-50-4-1673
Keywords: cryptosporidium, glycosylation pathway, N-glycans, O-glycans, GPI anchor biosynthesis
Citation: Wang D, Wang C and Zhu G (2022) Genomic reconstruction and features of glycosylation pathways in the apicomplexan Cryptosporidium parasites. Front. Mol. Biosci. 9:1051072. doi: 10.3389/fmolb.2022.1051072
Received: 22 September 2022; Accepted: 03 November 2022;
Published: 17 November 2022.
Edited by:
Kentaro Kato, Nagasaki University, JapanCopyright © 2022 Wang, Wang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guan Zhu, Y3J5cHRvc3BvcmlkYUBnbWFpbC5jb20=
 Dongqiang Wang
Dongqiang Wang Chenchen Wang
Chenchen Wang Guan Zhu
Guan Zhu