
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Mol. Biosci. , 14 November 2022
Sec. Lipids, Membranes and Membranous Organelles
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.1035574
This article is part of the Research Topic In Celebration of Women in Science: Lipids, Membranes, and Membranous Organelles View all 11 articles
Lipids play a major role in bacterial cells. Foremost, lipids are the primary constituents of the cell membrane bilayer, providing structure and separating the cell from the surrounding environment. This makes the lipid bilayer a prime target for antimicrobial peptides and membrane-acting antibiotics such as daptomycin. In response, bacteria have evolved mechanisms by which the membrane can be adapted to resist attack by these antimicrobial compounds. In this review, we focus on the membrane phospholipid changes associated with daptomycin resistance in enterococci, Staphylococcus aureus, and the Viridans group streptococci.
The cell membrane is a vital component of the bacterial cell, serving as a part of the protective barrier against the surrounding environment and as a scaffold for metabolic and regulatory proteins. Bacterial membranes are primarily composed of a bilayer of phospholipids with varying headgroups, acyl chain lengths, and acyl saturation which can influence membrane properties such as fluidity or charge. The major lipids in Gram-positive bacteria, particularly the firmicutes, are anionic phospholipids (APLs) [e.g., phosphatidylglycerol (PG), cardiolipin (CL)] and their derivatives (lysyl-PG, alanyl-PG), zwitterionic phospholipids, as well as other lipid classes like glycolipids and diacylglycerols (Sohlenkamp and Geiger, 2016). While the traditional “fluid mosaic model” describes a uniform bilayer whereby lipids and proteins are free to diffuse throughout the space, there is increasing evidence of the existence of distinct domains within the membrane (Matsumoto et al., 2006). These phospholipid domains have been described across several clinically important species of Gram-positive organisms, including “functional membrane microdomains” in S. aureus (García-Fernández et al., 2017), APL microdomains in Enterococcus faecalis (Tran et al., 2013a), or the ExPortal of Streptococcus spp. (Vega and Caparon, 2013). Importantly, alterations in these domains have been associated with specific roles in the bacterial response to antibiotics and antimicrobial peptides active at the cell envelope.
The relative accessibility and essential functions of the bacterial membrane components make them effective targets for antimicrobials. The rise of multidrug resistant organisms such as methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE) (Munita et al., 2015; Khan et al., 2018) spurred interest in antibiotics with alternative mechanisms of action that could bypass resistance to available agents. Daptomycin (DAP) has emerged as a treatment option that retained in vitro activity against resistant Gram-positive organisms and has seen increasing use against invasive infections due to MRSA and particularly VRE (Jorgensen et al., 2003; Mortin et al., 2007). Resistance to DAP is being reported with increasing frequency in clinical isolates, approaching 15%–28% in centers with heavy use of DAP (Kamboj et al., 2011; Munita et al., 2015; DiPippo et al., 2017; Wang et al., 2018). This review will address the role of the cell membrane in DAP resistance (DAP-R), with a focus on the alteration and adaptation of membrane phospholipids in Gram-positive bacteria of clinical importance.
DAP is a lipopeptide antibiotic originally isolated from Streptomyces roseosporus in the 1980s (Eisenstein et al., 2010). DAP possesses a cyclic peptide core linked to a fatty acyl chain and requires calcium to adopt an amphipathic conformation that facilitates oligomerization and insertion into the bacterial membrane. In addition, complex formation with calcium masks the lipopeptides’s negative charge and increases its affinity for the membrane lipid PG, the major constituent of Gram-positive membranes (typically comprising 50%–65% of the total APL content in S. aureus and E. faecalis (Müller et al., 2016; DeMars et al., 2020; Kotsogianni et al., 2021; Woodall et al., 2021). Recent structural analysis of Ca2+-DAP in lipid bilayers containing PG shows that DAP forms tetramers within the outer leaflet (Beriashvili et al., 2020). These tetramers can reversibly flip between the outer and inner leaflets and associate with one another to form a complex that spans the entire membrane (Zhang et al., 2014), resulting in membrane leakage and depolarization in cells after prolonged incubation with Ca2+-DAP (Silverman et al., 2003).
Multiple DAP mechanisms of action have been proposed based on the different reported cellular responses to DAP exposure (Figure 1), including membrane permeabilization, inhibition of cell wall synthesis, and physical alteration of membrane fluidity or curvature (Mengin-Lecreulx et al., 1990; Silverman et al., 2003; Pogliano et al., 2012; Müller et al., 2016). A recent study published by Grein et al. demonstrated that in S. aureus Ca2+-DAP oligomers form a tripartite complex with PG and lipid II (or other undecaprenyl cell envelope precursors, UDP), which are primarily located at the cell septum (Figure 1). The DAP-PG-lipid II complex not only sequesters lipid II substrate, but also disrupts the localization and assembly of the peptidoglycan synthesis machinery (Grein et al., 2020). In Bacillus subtilis, the insertion of DAP complexes in the membrane and subsequent alteration in membrane fluidity leads to mislocalization of crucial membrane proteins (Müller et al., 2016). After prolonged incubation, the DAP-UDP-PG complexes spread throughout the membrane, compromising envelope integrity and leading to cell death (Grein et al., 2020). The ability of DAP to disrupt cell wall biosynthesis is also supported by multiple in vitro and ex vivo studies where synergy or re-sensitization of DAP was observed upon combination with cell-wall acting antibiotics such as β-lactams (Dhand et al., 2011; Sakoulas et al., 2013; Sakoulas et al., 2014; Smith et al., 2015a; Smith et al., 2015b; Werth et al., 2015; Yim et al., 2017; Kebriaei et al., 2019; Johnson et al., 2021). Additionally, DAP has been shown to induce production of reactive oxygen species in S. aureus via binding to the universal stress response protein Usp2, a membrane protein which has been postulated to mediate the response to oxidative stress (Po et al., 2021). Thus, the binding and action of DAP is tied to the specific properties of the target bacterial membrane.
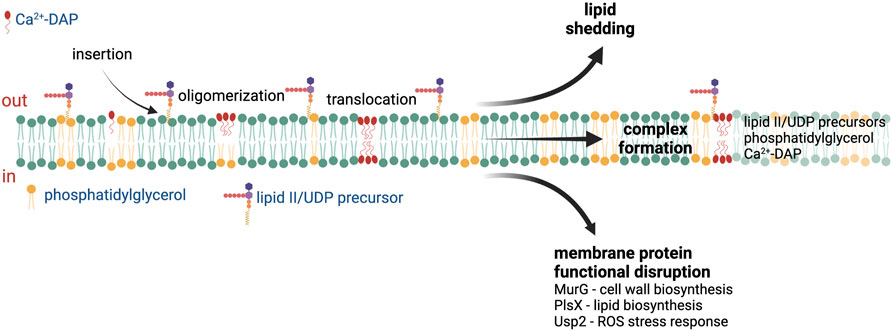
FIGURE 1. Proposed mechanisms of action of daptomycin. After insertion into the cell membrane, the Ca2+-daptomycin complex oligomerize in the outer leaflet of the cell membrane. Ca2+-daptomycin oligomers are translocated to the inner leaflet of the cell membrane which result in lipid shedding, tripartite formation with phosphatidylglycerol and lipid II/UDP precursors, or membrane protein functional disruption. See text for details. ROS—reactive oxygen species, UDP—undecaprenyl. Figure produced with BioRender.
A recurring theme in the emergence of DAP resistance across species is the presence of mutations leading to changes in both proteins of the lipid metabolic pathways and two-component sensors (TCS) involved in regulating cell envelope homeostasis (Tran et al., 2015). These changes can lead to alterations in the synthesis and modification of membrane lipid species and acyl-groups, thus impacting the composition and properties of the membrane (Table 1). In addition to de novo synthesis, bacteria can utilize exogenous fatty acids from their environment, including from human hosts, and importantly these exogenous acyl-chain profiles may be substantially different from those achievable by de novo synthesis in bacteria (Parsons et al., 2014a; Parsons et al., 2014b; Saito et al., 2014). These findings are also seen in model membranes where phospholipid head group and fatty acid residues in phospholipids can influence the membrane properties and alter the sensitivity to DAP (Beriashvili et al., 2018). The specific mechanisms by which these alterations impact the membrane are the subject of ongoing investigation, but shifts in membrane lipid content, fatty acid saturation, and alteration of membrane surface charge are common features of resistant isolates (Table 1).
E. faecalis and E. faecium are important opportunistic human pathogens that cause a variety of infections ranging from skin and soft tissue infections to bacteremia and endocarditis (Krawczyk et al., 2021). DAP has emerged as a front-line agent against complicated VRE infections; however, emergence of DAP-R has threatened its utility in clinical practice. Characterization of both lab-evolved and clinical DAP-R strains has shed light on the genetic and phenotypic changes associated with DAP-R in enterococci (Arias et al., 2011; Palmer et al., 2011; Tran et al., 2013b; Miller et al., 2013; Diaz et al., 2014; Wang et al., 2018; Miller et al., 2019). In E. faecalis, there is an observable re-distribution of APL microdomains, while in E. faecium a DAP repulsion phenotype similar to S. aureus is seen (Ernst et al., 2009; Tran et al., 2013a; Diaz et al., 2014; Khan et al., 2019) (Figure 2).
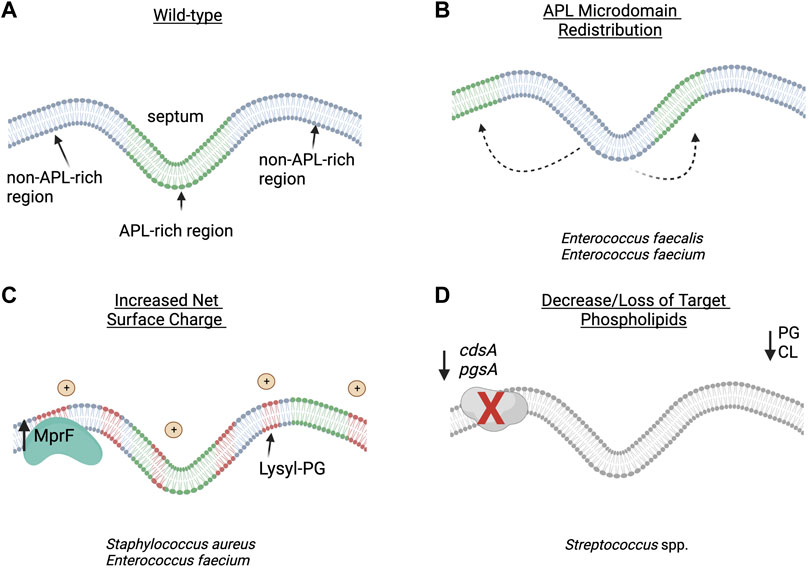
FIGURE 2. Proposed strategies of daptomycin resistance in Gram-positive pathogens: (A) wild-type bacterial cell membrane with anionic phospholipid (APL) microdomains clustered at the division septum. (B) In Enterococcus faecalis, redistribution of APL diverts daptomycin (DAP) away from the critical division septum. (C) In Staphylococcus aureus and E. faecium, changes in membrane phospholipid content and net surface charge lead to decreased daptomycin binding and oligomerization to the cell membrane. (D) In Viridans group streptococci, the decrease or loss of target phospholipid, PG and CL, are associated with overall decreased binding to the cell membrane or hyperaccumulation of daptomycin in a small subset of the bacterial population. APL–anionic phospholipid; CL–cardiolipin; PG–phosphatidylglycerol. Figure produced with BioRender.
Enterococci predominantly contain the APL PG in their cell membranes, in addition to CL, cationic PG derivatives such as lysyl-PG, and various glycolipids and diacylglycerols. In DAP-R enterococcal isolates, several general patterns in membrane lipid changes begin to emerge. Two studies using the clinical strain pair E. faecalis S613 (DAP-S) and R712 (DAP-R) grown to stationary phase found that the DAP-R isolate had a significant reduction in the amount of membrane PG as compared to its DAP-S parent, and an increase in glycerolphospho-diglycodiacylglycerol (GP-DGDAG) (Mishra et al., 2012; Hines et al., 2017). Further, an analysis of the laboratory strain E. faecalis OG1RF and two derivative strains rendered DAP-R via in vitro passage showed decreases in PG and lysyl-PG in the resistant isolates during mid-logarithmic phase growth (Rashid et al., 2017). In paired isolates of E. faecium, the DAP-R isolate showed a significant decrease in PG (14% vs. 33%) and increases in GP-DGDAG (23% vs. 12%) as compared to the susceptible parent strain (Mishra et al., 2012). Taken together, these data appear to indicate that a relative loss of PG, the primary phospholipid involved in DAP binding, may contribute to the resistant phenotype. Interestingly, a different analysis of membrane lipids of the DAP-R E. faecalis strain performed by 2D-TLC during exponential growth showed a significant increase in PG, a reduction in CL, and no differences in lysyl-PG as compared to S613, while levels of GP-DGDAG were not reported (Khan et al., 2019). Thus, lipid alterations other than a decrease in membrane PG are likely to also contribute to DAP-R. A recent study showed that the absence of CL in E. faecalis OG1RF, achieved by deletion of both CL synthases, increased sensitivity to DAP and other cell damaging agents such as SDS, while the absence of lysyl-PG, resulting from deletion of mprF2, did not change sensitivity to DAP but rendered the mutant more resistant to SDS (Woodall et al., 2021). The triple mutant, which lacked CL and lysyl-PG, restored membrane tolerance to DAP and SDS to the level of parental strain OG1RF. These results further demonstrate the complexity of membrane lipid adaptation to DAP and other cell membrane damaging compounds.
The importance of cell surface charge in the DAP-R phenotype can be inferred from direct measurement and the pattern of mutations in resistant isolates. Both the clinical DAP-R strains E. faecalis and E. faecium displayed increased positive surface charge relative to their DAP-S counterparts (Arias et al., 2011; Mishra et al., 2012). Mutations impacting the dlt operon and mprF have been associated with DAP-R in E. faecium (Diaz et al., 2014). Further, in vitro adaptation of E. faecium to DAP led to the identification of additional genes associated with increases in cell surface charge, including yvcRS, oatA, and divIVA (Prater et al., 2019). Despite these observations, levels of lysyl-PG do not show a predictive trend in net surface charge in DAP-R strains (Mishra et al., 2012).
Alteration of membrane fluidity has also been observed in association with changes in DAP susceptibility. Differences in the length, saturation, and cyclization of fatty acyl chains of individual lipid species influence fluidity by altering lipid packing, and DAP has been reported to preferentially locate to regions of increased fluidity (Müller et al., 2016). Thus, changes in fluidity may alter DAP insertion and oligomerization in the membrane. Decreases in membrane fluidity (i.e., more rigid membranes) are associated with DAP-R resistance in enterococci. In E. faecium, this phenotype was associated with a decrease in the total proportion of unsaturated fatty acids and an increase in cyclic fatty acids which may be associated with mutations in cfa (cyclopropane fatty acid synthase) (Tran et al., 2013b; Diaz et al., 2014). Increased membrane rigidity has also been described in E. faecalis in association with changes in genes involved in the lipid metabolic pathway, including cls, gdpD (glycerophosphodiester phosphodiesterase), and dak (encoding a homologue of Fak, the staphylococcal fatty acid kinase) (Miller et al., 2019).
Further evidence supporting the potential importance of acyl-chain composition comes from a series of experiments examining the influence of exogenous fatty acids on the enterococcal membrane. Using laboratory isolates of E. faecalis, Harp and colleagues demonstrated that supplementation of growth media with oleic and linoleic acid induced tolerance to membrane stress, including protecting against DAP mediated killing (Saito et al., 2014; Harp et al., 2016). Subsequent studies profiling the changes in E. faecalis membranes associated with supplementation of a range of both saturated and unsaturated fatty acids found that exogenous fatty acids were rapidly incorporated into the acyl-chains of phospholipids. Unlike oleic acid, supplementation with saturated fatty acids and the native enterococcal monounsaturated fatty acid cis-vaccenic acid did not confer a survival advantage in the presence of DAP (Saito et al., 2017) while other combinations of “protective” fatty acids could induce DAP tolerance (Brewer et al., 2020). Thus, it is likely that variations in acyl-chains, as well as overall lipid species, play a role in protecting the membrane against daptomycin induced stress.
S. aureus is an important pathogen that causes a wide range of infections including cellulitis, bacteremia, and infective endocarditis (Tong et al., 2015). Vancomycin has been the mainstay of therapy for infections due to methicillin-resistant MRSA, however DAP has seen increasing use as salvage therapy for recalcitrant MRSA infections (Lee et al., 2018). While DAP retains activity against the vast majority of MRSA isolates, the overlap of genetic pathways leading to vancomycin-intermediate S. aureus (VISA) and heterogenous VISA isolates and decreased susceptibility to DAP has contributed to the emergence of DAP-R on therapy (Patel et al., 2006; Julian et al., 2007; Kelley et al., 2011).
Similar to enterococci, resistance to DAP has been linked to mutations in genes encoding TCS (vraSR, yycFG) and lipid biosynthetic enzymes (pgsA, cls, and mprF), with subsequent alteration of membrane composition and surface charge associated with reduced binding of DAP (Table 1) (Friedman et al., 2006; Muthaiyan et al., 2008; Mehta et al., 2012; Peleg et al., 2012; Bayer et al., 2013). It has been noted that DAP-R associated mutations in yycG (also known as WalK, a histidine kinase sensor involved in cell wall biosynthesis) could play a role in modulating fatty acid biosynthesis and potentially membrane fluidity as shown by the role of the YycFG homologue in S. pneumoniae (Mohedano et al., 2005). Interestingly, S. aureus has also been shown to exhibit “lipid shedding,” where DAP exposure triggered active release of membrane phospholipids that were able to inactivate DAP and protect from bacterial killing (Pader et al., 2016). This phenomenon has since been shown in other organisms, including in E. faecalis and Streptococcus spp. (Ledger et al., 2022).
Although a variety of lipids exist in staphylococcal membranes, the major phospholipids include PG, CL, and lysyl-PG, with PG and lysyl-PG being the most abundant (DeMars et al., 2020). Like enterococci, membranes from DAP-R S. aureus tend to have decreased amounts of PG, with increases in lysyl-PG which are closely correlated with mutations in mprF. Mutations in mprF are thought to lead to a gain-of-function, with increased production and/or flipping of lysyl-PG into the outer leaflet of the membrane (Ernst et al., 2009; Slavetinsky et al., 2022). In a DAP-S/DAP-R clinical strain pair, Jones et al. showed increased levels of lysyl-PG with decreased levels of PG in the resistant isolate (Jones et al., 2008). Other independent studies which analyzed MRSA strain pairs containing mutations in mprF, yycG and/or cls2 showed similar increased levels of lysyl-PG in the DAP-R strains (Peleg et al., 2012; Mishra and Bayer, 2013). Conversely, a DAP-R MRSA strain containing mutations in pgsA (encoding PG synthase), mprF, and yycG amongst others showed reduced levels of PG, but also reduced levels of lysyl-PG and CL (Hines et al., 2017). This may be explained by the hypothesis that, in addition to electrostatic repulsion, MprF-mediated resistance may also reduce the available pool of PG for DAP to target, which has been supported through biochemical work using large unilamellar vesicles (Kilelee et al., 2010). Mutations in dltABCD (involved in D-alanylation of teichoic acids) have also been associated with DAP-R in S. aureus (Bayer et al., 2013; Bayer et al., 2016). Gain-of-function mutations in this operon are proposed to increase overall cell surface charge through alanylation of lipoteichoic acids and wall teichoic acids (Table 1; Figure 2), and deletion of dlt results in increased susceptibility to cationic antimicrobial peptides (Peschel et al., 1999).
Mutations in cls2, one of the two staphylococcal CL synthases, have been implicated in DAP-R. Alterations of the enzyme resulted in increased biosynthetic activity, increased levels of CL, decreased levels of PG, and no changes in lysyl-PG levels. CL-rich membranes have shown to have increased thickness by neutron reflectometry, which was associated with decrease in penetration and aggregation of DAP. This impaired ability of DAP insertion and translocation, which was postulated as the as mechanism underlying DAP-R in these strains (Jiang et al., 2019). Increases in CL and decreased levels of PG has also been linked to DAP-tolerance in serum-adapted strains of S. aureus (Ledger et al., 2022).
In addition, changes in membrane fluidity also correlate with decreased DAP bactericidal activity. In contrast to enterococci, clinical isolates of S. aureus have in general shown increases in membrane fluidity associated with DAP-R (Jones et al., 2008; Mishra et al., 2011a; Mishra and Bayer, 2013). While no significant differences in fatty acid content in saturation levels, length, or branching, the fluidity changes may be explained by membrane carotenoid content of S. aureus. In many clinical isolates, there was a statistically significant decrease in the membrane carotenoid staphyloxanthin accompanied by a decrease in membrane fluidity. Staphyloxanthin is expressed by the majority of S. aureus isolated from infections and has previously been implicated in protecting the bacteria from DAP and antimicrobial peptide mediated killing, although prior studies in laboratory strains associated resistance with increased staphyloxanthin content and more rigid membranes (Mishra et al., 2011b). Interestingly, a more rigid membrane phenotype has also been observed in isolates arising from in vitro adaptation experiments or exposure to other cell envelope active compounds (such as the lipoglycopeptide dalbavancin) for which there was also cross resistance to DAP (Mishra et al., 2009; Zhang et al., 2022). These changes were related to differences in the ratio of long and short-chain fatty acids; however, differences in strain background mutations and growth media prevent a direct comparison. It is not clear if the observed shifts in fluidity are mechanistically important in disrupting the binding or translocation of DAP in the membrane, or merely a consequence of the alterations of membrane composition.
Viridans group streptococci (VGS) include a variety of species (i.e., S. mitis, S. oralis, S. anginosus, among others) that can cause severe infections including infective endocarditis, and resistance to commonly used antibiotics such as β-lactams is increasing (Doern et al., 1996; Marron et al., 2001; Prabhu et al., 2004). Unfortunately, high-level DAP-R [minimum inhibitory concentration (MIC) ≥ 256 μg/ml] can rapidly emerge in VGS upon DAP exposure (García-De-La-Mària et al., 2013; Akins et al., 2015).
While wild-type DAP-S cells contain PG and CL as the predominant phospholipids in the membrane, DAP-R VGS strains show no detectable PG or CL. Instead, cell membranes of resistant derivatives contain increased levels of the phospholipid precursor phosphatidic acid (PA) and decreased levels of phosphatidylcholine (not found in E. faecalis or S. aureus) (Adams et al., 2017; Mishra et al., 2017; Tran et al., 2019). This correlates with the identification of loss-of-function mutations in cdsA, a gene that encodes the phosphatidate cytidyltransferase enzyme (which generates the substrate CDP-diacylglycerol from PA for downstream phospholipid synthesis). The disappearance of CL and PG in various VGS strains harboring mutations in cdsA and/or pgsA was confirmed by lack of fluorescence in binding studies with 10-N-nonyl acridine orange NAO (Mishra et al., 2017), a fluorescent dye which binds APLs (Mileykovskaya and Dowhan, 2000).
Despite the lack of PG and CL in the membrane, several patterns of DAP binding have been observed in DAP-R streptococci (Figure 2). In S. oralis strains that developed DAP-R in association with mutations in pgsA, binding of DAP to the cell membrane appears to be uniform, but with significantly less binding overall (Tran et al., 2019). Conversely, strains that developed DAP-R via mutations in cdsA demonstrated hyperaccumulation of DAP in a small population of the cells (Mishra et al., 2017). The authors of this study hypothesized that a minority of bacterial cells may sequester DAP and allow the larger population to survive the antibiotic exposure, although the viability of cells as determined by propidium iodide uptake did not correlate with DAP hyperaccumulation. The discordance between a lack of PG and overall binding of DAP in VGS strains remains unexplained. While lack of PG and CL are consistent with many DAP-R VGS strains which have been adapted by serial passage or in an ex vivo simulated endocarditis vegetation model (SEV), there are other strains that showed no significant changes in lipid content (Kebriaei et al., 2019).
Overall, alterations in cell membrane fluidity have not been a consistent or predictable phenotype of DAP-R in VGS. Initial studies showed an increase in membrane fluidity was associated with resistance in S. mitis/oralis strains that were obtained from serial passage (Mishra et al., 2017; Mishra et al., 2020). These findings were later confirmed by Kebriaei et al. in an SEV model using the same strain. However, these changes may be isolate specific, as adaptation of a different strain background identified no differences in fluidity in the DAP-R derivative, and displayed a similar membrane lipid content relative to its DAP-S parent (Kebriaei et al., 2019). In a separate study, Tran et al. found no changes in fluidity between parental strains and evolved DAP-R derivatives of S. mitis or S. oralis strains despite the complete disappearance of PG and CL (Tran et al., 2019).
Similarly, alterations in cell surface charge have varied across DAP-R VGS. In S. mitis and S. oralis strains which developed DAP-R via mutations in pgsA or cdsA, Tran et al. found no changes in surface charge between parental strains and evolved DAP-R derivatives (Tran et al., 2019). In a serial passage experiment, DAP-R in S. mitis/oralis SF100 was associated with a decrease in net surface positive charge while maintaining the similar overall DAP binding profiles between the DAP-R and DAP-S strain (Mishra et al., 2020). However, in the SEV model Kebriaei et al. found no difference in surface charge in the DAP-R SF100 strain (Kebriaei et al., 2019). Furthermore, DAP-R in S. anginosus has been linked to changes in capsular polysaccharide and other cell surface modification genes that may affect surface charge. Substitutions in genes encoding cls, yycG and the dlt operon were identified in the DAP-R strain (Rahman et al., 2016) however, the exact contribution of surface charge to DAP-R in VGS remains unclear.
In summary, changes in lipid content associated with DAP-R in S. mitis/oralis seem to be dependent on strain and the type of mutation present despite a similar phenotype of high-level DAP-R (MIC ≥256 μg/ml). The precise mechanism of DAP-R in VGS or how this group of bacteria maintains cell envelope function and integrity despite the absence of PG and CL remains unclear. Further evaluation is warranted to determine the impact of membrane changes on DAP-R in VGS.
The bacterial cell membrane is a dynamic structure that has evolved to adapt to changing environmental conditions. Understanding how bacteria alter their membrane in the face of external stress is critical to preserve the usefulness of membrane active antibiotics such as DAP. While the precise role of membrane lipid changes in the mechanism of resistance to DAP has yet to be completely explored, a deeper understanding of this process can be leveraged to overcome the limitations of current therapeutics.
AN, KH, EM, WM, and TT conceptualized the manuscript. AN and KH prepared the manuscript. EM, WM, and TT revised and edited the manuscript. All authors contributed to the article and approved the final version.
KH is supported by a training fellowship from the Gulf Coast Consortia, on the Texas Medical Center Training Program in Antimicrobial Resistant (TPAMR) (NIH Grant No. T32AI141349). WM is supported by National Institute of Allergy and Infectious Disease of the National Institutes of Health (Award Number K08AI135093).
WM has received grants and/or honoraria from Entasis Therapeutics and UpToDate.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adams, H. M., Joyce, L. R., Guan, Z., Akins, R. L., and Palmer, K. L. (2017). Streptococcus mitis and S. oralis lack a requirement for CdsA, the enzyme required for synthesis of major membrane phospholipids in bacteria. Antimicrob. Agents Chemother. 61 (5), e02552-16. doi:10.1128/AAC.02552-16
Akins, R. L., Katz, B. D., Monahan, C., and Alexander, D. (2015). Characterization of high-level daptomycin resistance in viridans group streptococci developed upon in vitro exposure to daptomycin. Antimicrob. Agents Chemother. 59 (4), 2102–2112. doi:10.1128/AAC.04219-14
Arias, C. A., Panesso, D., Mcgrath, D. M., Qin, X., Mojica, M. F., Miller, C., et al. (2011). Genetic basis for in vivo daptomycin resistance in enterococci. N. Engl. J. Med. 365, 892–900. doi:10.1056/NEJMoa1011138
Bayer, A. S., Mishra, N. N., Cheung, A. L., Rubio, A., and Yang, S. J. (2016). Dysregulation of mprF and dltABCD expression among daptomycin-non-susceptible MRSA clinical isolates. J. Antimicrob. Chemother. 71 (8), 2100–2104. doi:10.1093/jac/dkw142
Bayer, A. S., Schneider, T., and Sahl, H. G. (2013). Mechanisms of daptomycin resistance in Staphylococcus aureus: Role of the cell membrane and cell wall. Ann. N. Y. Acad. Sci. 1277 (1), 139–158. doi:10.1111/J.1749-6632.2012.06819.X
Beriashvili, D., Spencer, N. R., Dieckmann, T., Overduin, M., and Palmer, M. (2020). Characterization of multimeric daptomycin bound to lipid nanodiscs formed by calcium-tolerant styrene-maleic acid copolymer. Biochim. Biophys. Acta. Biomembr. 1862 (6), 183234. doi:10.1016/J.BBAMEM.2020.183234
Beriashvili, D., Taylor, R., Kralt, B., Abu Mazen, N., Taylor, S. D., and Palmer, M. (2018). Mechanistic studies on the effect of membrane lipid acyl chain composition on daptomycin pore formation. Chem. Phys. Lipids 216, 73–79. doi:10.1016/j.chemphyslip.2018.09.015
Brewer, W., Harrison, J., Saito, H. E., and Fozoa, E. M. (2020). Induction of daptomycin tolerance in Enterococcus faecalis by fatty acid combinations. Appl. Environ. Microbiol. 86 (20), e01178–20. doi:10.1128/AEM.01178-20
DeMars, Z., Singh, V. K., and Bose, J. L. (2020). Exogenous fatty acids remodel staphylococcus aureus lipid composition through fatty acid kinase. J. Bacteriol. 202 (14), e00128-20. doi:10.1128/JB.00128-20
Dhand, A., Bayer, A. S., Pogliano, J., Yang, S. J., Bolaris, M., Nizet, V., et al. (2011). Use of antistaphylococcal beta-lactams to increase daptomycin activity in eradicating persistent bacteremia due to methicillin-resistant Staphylococcus aureus: Role of enhanced daptomycin binding. Clin. Infect. Dis. 53 (2), 158–163. doi:10.1093/cid/cir340
Diaz, L., Tran, T. T., Munita, J. M., Miller, W. R., Rincon, S., Carvajal, L. P., et al. (2014). Whole-genome analyses of Enterococcus faecium isolates with diverse daptomycin MICs. Antimicrob. Agents Chemother. 58 (8), 4527–4534. doi:10.1128/AAC.02686-14
DiPippo, A. J., Tverdek, F. P., Tarrand, J. J., Munita, J. M., Tran, T. T., Arias, C. A., et al. (2017). Daptomycin non-susceptible Enterococcus faecium in leukemia patients: Role of prior daptomycin exposure. J. Infect. 74 (3), 243–247. doi:10.1016/j.jinf.2016.11.004
Doern, G. V., Ferraro, M. J., Brueggemann, A. B., and Ruoff, K. L. (1996). Emergence of high rates of antimicrobial resistance among Viridans group streptococci in the United States. Antimicrob. Agents Chemother. 40 (4), 891–894. doi:10.1128/AAC.40.4.891
Eisenstein, B. I., Oleson, F. B., and Baltz, R. H. (2010). Daptomycin: From the mountain to the clinic, with essential help from francis tally, MD. Clin. Infect. Dis. 50 (1), S10–S15. doi:10.1086/647938
Ernst, C. M., Staubitz, P., Mishra, N. N., Yang, S. J., Hornig, G., Kalbacher, H., et al. (2009). The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 5 (11), e1000660. doi:10.1371/journal.ppat.1000660
Friedman, L., Alder, J. D., and Silverman, J. A. (2006). Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 50 (6), 2137–2145. doi:10.1128/AAC.00039-06
García-De-La-Mària, C., Pericas, J. M., Del Río, A., Castaneda, X., Vila-Farres, X., Armero, Y., et al. (2013). Early in vitro and in vivo development of high-level daptomycin resistance is common in mitis group streptococci after exposure to daptomycin. Antimicrob. Agents Chemother. 57 (5), 2319–2325. doi:10.1128/AAC.01921-12
García-Fernández, E., Koch, G., Wagner, R. M., Fekete, A., Stengel, S. T., Schneider, J., et al. (2017). Membrane microdomain disassembly inhibits MRSA antibiotic resistance. Cell 171 (6), 1354–1367. doi:10.1016/j.cell.2017.10.012
Grein, F., Müller, A., Scherer, K. M., Liu, X., Ludwig, K. C., Klöckner, A., et al. (2020). Ca2+-Daptomycin targets cell wall biosynthesis by forming a tripartite complex with undecaprenyl-coupled intermediates and membrane lipids. Nat. Commun. 11 (1), 1455. doi:10.1038/s41467-020-15257-1
Harp, J. R., Saito, H. E., Bourdon, A. K., Reyes, J., Arias, C. A., Campagna, S. R., et al. (2016). Exogenous fatty acids protect Enterococcus faecalis from daptomycin induced membrane stress independent of the response regulator LiaR. Appl. Environ. Microbiol. 82, 4410–4420. doi:10.1128/AEM.00933-16
Hines, K. M., Waalkes, A., Penewit, K., Holmes, E. A., Salipante, S. J., Werth, B. J., et al. (2017). Characterization of the mechanisms of daptomycin resistance among Gram- positive bacterial pathogens by multidimensional lipidomics. mSphere 2 (6), e00492-17. doi:10.1128/mSphere.00492-17
Jiang, J. H., Bhuiyan, M. S., Shen, H. H., Cameron, D. R., Rupasinghe, T. W. T., Wu, C. M., et al. (2019). Antibiotic resistance and host immune evasion in Staphylococcus aureus mediated by a metabolic adaptation. Proc. Natl. Acad. Sci. U. S. A. 116 (9), 3722–3727. doi:10.1073/pnas.1812066116
Johnson, T. M., Molina, K. C., Miller, M. A., Kiser, T. H., Huang, M., and Mueller, S. W. (2021). Combination ceftaroline and daptomycin salvage therapy for complicated methicillin-resistant Staphylococcus aureus bacteraemia compared with standard of care. Int. J. Antimicrob. Agents 57 (4), 106310. doi:10.1016/j.ijantimicag.2021.106310
Jones, T., Yeaman, M. R., Sakoulas, G., Yang, S. J., Proctor, R. A., Sahl, H. G., et al. (2008). Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob. Agents Chemother. 52 (1), 269–278. doi:10.1128/AAC.00719-07
Jorgensen, J. H., Crawford, S. A., Kelly, C. C., and Patterson, J. E. (2003). In vitro activity of daptomycin against vancomycin-resistant enterococci of various Van types and comparison of susceptibility testing methods. Antimicrob. Agents Chemother. 47 (12), 3760–3763. doi:10.1128/AAC.47.12.3760-3763.2003
Julian, K., Kosowska-Shick, K., Whitener, C., Roos, M., Labischinski, H., Rubio, A., et al. (2007). Characterization of a daptomycin-nonsusceptible vancomycin-intermediate Staphylococcus aureus strain in a patient with endocarditis. Antimicrob. Agents Chemother. 51 (9), 3445–3448. doi:10.1128/AAC.00559-07
Kamboj, M., Cohen, N., Gilhuley, K., Babady, N. E., Seo, S. K., and Sepkowitz, K. A. (2011). Emergence of daptomycin-resistant VRE: Experience of a single institution. Infect. Control Hosp. Epidemiol. 32 (4), 391–394. doi:10.1086/659152
Kebriaei, R., Rice, S. A., Stamper, K. C., Seepersaud, R., Garcia-de-la-Maria, C., Mishra, N. N., et al. (2019). Daptomycin dose-ranging evaluation with single-dose versus multidose ceftriaxone combinations against streptococcus mitis/oralis in an ex vivo simulated endocarditis vegetation model. Antimicrob. Agents Chemother. 63 (6), e00386‐19. doi:10.1128/AAC.00386-19
Kelley, P. G., Gao, W., Ward, P. B., and Howden, B. P. (2011). Daptomycin non-susceptibility in vancomycin-intermediate Staphylococcus aureus (VISA) and heterogeneous-VISA (hVISA): Implications for therapy after vancomycin treatment failure. J. Antimicrob. Chemother. 66 (5), 1057–1060. doi:10.1093/JAC/DKR066
Khan, A., Davlieva, M., Panesso, D., Rincon, S., Miller, W. R., Diaz, L., et al. (2019). Antimicrobial sensing coupled with cell membrane remodeling mediates antibiotic resistance and virulence in Enterococcus faecalis. Proc. Natl. Acad. Sci. U. S. A. 116 (52), 26925–26932. doi:10.1073/pnas.1916037116
Khan, A., Miller, W. R., and Arias, C. A. (2018). Mechanisms of antimicrobial resistance among hospital-associated pathogens. Expert Rev. anti. Infect. Ther. 16 (4), 269–287. doi:10.1080/14787210.2018.1456919
Kilelee, E., Pokorny, A., Yeaman, M. R., and Bayer, A. S. (2010). Lysyl-phosphatidylglycerol attenuates membrane perturbation rather than surface association of the cationic antimicrobial peptide 6W-RP-1 in a model membrane system: Implications for daptomycin resistance. Antimicrob. Agents Chemother. 54 (10), 4476–4479. doi:10.1128/AAC.00191-10
Kotsogianni, I., Wood, T. M., Alexander, F. M., Cochrane, S. A., and Martin, N. I. (2021). Binding studies reveal phospholipid specificity and its role in the calcium-dependent mechanism of action of daptomycin. ACS Infect. Dis. 7 (9), 2612–2619. doi:10.1021/acsinfecdis.1c00316
Krawczyk, B., Wityk, P., Gałęcka, M., and Michalik, M. (2021). The many faces of Enterococcus spp.—commensal, probiotic and opportunistic pathogen. Microorganisms 9 (9), 1900. doi:10.3390/MICROORGANISMS9091900
Ledger, E. V. K., Mesnage, S., and Edwards, A. M. (2022). Human serum triggers antibiotic tolerance in Staphylococcus aureus. Nat. Commun. 13 (1), 2041–2119. doi:10.1038/s41467-022-29717-3
Ledger, E., Mesnage, S., and Edwards, A. M. (2022). Human serum triggers antibiotic tolerance in Staphylococcus aureus. Nat. Commun. 13 (1), 2401. doi:10.1038/s41467-022-29717-3
Lee, A. S., de Lencastre, H., Garau, J., Kluytmans, J., Malhotra-Kumar, S., Peschel, A., et al. (2018). Methicillin-resistant Staphylococcus aureus. Nat. Rev. 4, 18033. doi:10.1038/NRDP.2018.33
Marron, A., Carratalà, J., Alcaide, F., Fernández-Sevilla, A., Gudiol, F., and Fernandez-SevillA, A. (2001). High rates of resistance to cephalosporins among viridans-group streptococci causing bacteraemia in neutropenic cancer patients. J. Antimicrob. Chemother. 47 (1), 87–91. doi:10.1093/jac/47.1.87
Matsumoto, K., Kusaka, J., Nishibori, A., and Hara, H. (2006). Lipid domains in bacterial membranes. Mol. Microbiol. 61 (5), 1110–1117. doi:10.1111/j.1365-2958.2006.05317.x
Mehta, S., Cuirolo, A. X., Plata, K. B., Riosa, S., Silverman, J. A., Rubio, A., et al. (2012). VraSR two-component regulatory system contributes to mprF-mediated decreased susceptibility to daptomycin in in vivo-selected clinical strains of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 56 (1), 92–102. doi:10.1128/AAC.00432-10
Mengin-Lecreulx, D., Allen, N. E., Hobbs, J. N., and van Heijenoort, J. (1990). Inhibition of peptidoglycan biosynthesis in Bacillus megaterium by daptomycin. FEMS Microbiol. Lett. 57 (3), 245–248. doi:10.1016/0378-1097(90)90074-Z
Mileykovskaya, E., and Dowhan, W. (2000). Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J. Bacteriol. 182 (4), 1172–1175. doi:10.1128/JB.182.4.1172-1175.2000
Miller, C., Kong, J., Tran, T. T., Arias, C. A., Saxer, G., and Shamoo, Y. (2013). Adaptation of Enterococcus faecalis to daptomycin reveals an ordered progression to resistance. Antimicrob. Agents Chemother. 57 (11), 5373–5383. doi:10.1128/AAC.01473-13
Miller, W. R., Tran, T. T., Diaz, L., Rios, R., Khan, A., Reyes, J., et al. (2019). LiaR-independent pathways to daptomycin resistance in Enterococcus faecalis reveal a multilayer defense against cell envelope antibiotics. Mol. Microbiol. 111 (3), 811–824. doi:10.1111/MMI.14193
Mishra, N. N., and Bayer, A. S. (2013). Correlation of cell membrane lipid profiles with daptomycin resistance in methicillin-resistant staphylococcus aureus. Antimicrob. Agents Chemother. 57 (2), 1082–1085. doi:10.1128/AAC.02182-12
Mishra, N. N., Bayer, A. S., Tran, T. T., Shamoo, Y., Mileykovskaya, E., Dowhan, W., et al. (2012). Daptomycin resistance in enterococci is associated with distinct alterations of cell membrane phospholipid content. PLoS One 7 (8), e43958. doi:10.1371/JOURNAL.PONE.0043958
Mishra, N. N., Liu, G. Y., Yeaman, M. R., Nast, C. C., Proctor, R. A., McKinnell, J., et al. (2011). Carotenoid-related alteration of cell membrane fluidity impacts Staphylococcus aureus susceptibility to host defense peptides. Antimicrob. Agents Chemother. 55 (2), 526–531. doi:10.1128/AAC.00680-10
Mishra, N. N., McKinnell, J., Yeaman, M. R., Rubio, A., Nast, C. C., Chen, L., et al. (2011). In vitro cross-resistance to daptomycin and host defense cationic antimicrobial peptides in clinical methicillin-resistant Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 55 (9), 4012–4018. doi:10.1128/AAC.00223-11
Mishra, N. N., Tran, T. T., Arias, C. A., Seepersaud, R., Sullam, P. M., and Bayer, A. S. (2020). Strain-specific adaptations of Streptococcus mitis-oralis to serial in vitro passage in daptomycin (DAP): Genotypic and phenotypic characteristics. Antibiotics 9 (8), 520. doi:10.3390/ANTIBIOTICS9080520
Mishra, N. N., Tran, T. T., Seepersaud, R., Garcia-de-la-Maria, C., Faull, K., Yoon, A., et al. (2017). Perturbations of phosphatidate cytidylyltransferase (CdsA) mediate daptomycin resistance in Streptococcus mitis/oralis by a novel mechanism. Antimicrob. Agents Chemother. 61 (4), e02435-16. doi:10.1128/AAC.02435-16
Mishra, N. N., Yang, S. J., Sawa, A., Rubio, A., Nast, C. C., Yeaman, M. R., et al. (2009). Analysis of cell membrane characteristics of in vitro-selected daptomycin-resistant strains of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53 (6), 2312–2318. doi:10.1128/AAC.01682-08
Mohedano, M. L., Overweg, K., De La Fuente, A., Reuter, M., Altabe, S., Mulholland, F., et al. (2005). Evidence that the essential response regulator YycF in Streptococcus pneumoniae modulates expression of fatty acid biosynthesis genes and alters membrane composition. J. Bacteriol. 187 (7), 2357–2367. doi:10.1128/JB.187.7.2357-2367.2005
Mortin, L. I., Li, T., Van Praagh, A. D. G., Zhang, S., Zhang, X. X., and Alder, J. D. (2007). Rapid bactericidal activity of daptomycin against methicillin-resistant and methicillin-susceptible Staphylococcus aureus peritonitis in mice as measured with bioluminescent bacteria. Antimicrob. Agents Chemother. 51 (5), 1787–1794. doi:10.1128/AAC.00738-06
Müller, A., Wenzel, M., Strahl, H., Grein, F., Saaki, T. N. V., Kohl, B., et al. (2016). Daptomycin inhibits cell envelope synthesis by interfering with fluid membrane microdomains. Proc. Natl. Acad. Sci. U. S. A. 113 (45), E7077–E7086. doi:10.1073/PNAS.1611173113
Munita, J. M., Bayer, A. S., and Arias, C. A. (2015). Evolving resistance among gram-positive pathogens. Clin. Infect. Dis. doi:10.1093/cid/civ523
Muthaiyan, A., Silverman, J. A., Jayaswal, R. K., and Wilkinson, B. J. (2008). Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. Antimicrob. Agents Chemother. 52 (3), 980–990. doi:10.1128/AAC.01121-07
Pader, V., Hakim, S., Painter, K. L., Wigneshweraraj, S., Clarke, T. B., and Edwards, A. M. (2016). Staphylococcus aureus inactivates daptomycin by releasing membrane phospholipids. Nat. Microbiol. 2, 16194. doi:10.1038/NMICROBIOL.2016.194
Palmer, K. L., Daniel, A., Hardy, C., Silverman, J., and Gilmore, M. S. (2011). Genetic basis for daptomycin resistance in enterococci. Antimicrob. Agents Chemother. 55 (7), 3345–3356. doi:10.1128/AAC.00207-11
Parsons, J. B., Broussard, T. C., Bose, J. L., Rosch, J. W., Jackson, P., Subramanian, C., et al. (2014). Identification of a two-component fatty acid kinase responsible for host fatty acid incorporation by Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 111 (29), 10532–10537. doi:10.1073/PNAS.1408797111
Parsons, J. B., Frank, M. W., Jackson, P., Subramanian, C., and Rock, C. O. (2014). Incorporation of extracellular fatty acids by a fatty acid kinase-dependent pathway in Staphylococcus aureus. Mol. Microbiol. 92 (2), 234–245. doi:10.1111/MMI.12556
Patel, J. B., Jevitt, L. A., Hageman, J., McDonald, L. C., and Tenover, F. C. (2006). An association between reduced susceptibility to daptomycin and reduced susceptibility to vancomycin in Staphylococcus aureus. Clin. Infect. Dis. 42 (11), 1652–1653. doi:10.1086/504084
Peleg, A. Y., Miyakis, S., Ward, D. V., Earl, A. M., Rubio, A., Cameron, D. R., et al. (2012). Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. PLoS One 7 (1), e28316. doi:10.1371/journal.pone.0028316
Peschel, A., Jack, R. W., Otto, M., Collins, L. V., Staubitz, P., Nicholson, G., et al. (2001). Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J. Exp. Med. 193 (9), 1067–1076. doi:10.1084/jem.193.9.1067
Peschel, A., Otto, M., Jack, R. W., Kalbacher, H., Jung, G., and Götz, F. (1999). Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274 (13), 8405–8410. doi:10.1074/jbc.274.13.8405
Po, K. H. L., Chow, H. Y., Cheng, Q., Chan, B. K., Deng, X., Wang, S., et al. (2021). Daptomycin exerts bactericidal effect through induction of excessive ROS production and blocking the function of stress response protein Usp2. Nat. Sci. 1 (2), e10023. doi:10.1002/ntls.10023
Pogliano, J., Pogliano, N., and Silverman, J. A. (2012). Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J. Bacteriol. 194 (17), 4494–4504. doi:10.1128/JB.00011-12
Prabhu, R. M., Piper, K. E., Baddour, L. M., Steckelberg, J. M., Wilson, W. R., and Patel, R. (2004). Antimicrobial susceptibility patterns among viridans group streptococcal isolates from infective endocarditis patients from 1971 to 1986 and 1994 to 2002. Antimicrob. Agents Chemother. 48 (11), 4463–4465. doi:10.1128/AAC.48.11.4463-4465.2004
Prater, A. G., Mehta, H. H., Kosgei, A. J., Miller, W. R., Tran, T. T., Arias, C. A., et al. (2019). Environment shapes the accessible daptomycin resistance mechanisms in Enterococcus faecium. Antimicrob. Agents Chemother. 63 (10), e00790-19. doi:10.1128/AAC.00790-19
Rahman, M., Nguyen, S. V., McCullor, K. A., King, C. J., Jorgensen, J. H., and Michael McShan, W. (2016). Comparative genome analysis of the daptomycin-resistant Streptococcus anginosus strain J4206 associated with breakthrough bacteremia. Genome Biol. Evol. 8 (11), 3446–3459. doi:10.1093/GBE/EVW241
Rashid, R., Cazenave-Gassiot, A., Gao, I. H., Nair, Z. J., Kumar, J. K., Gao, L., et al. (2017). Comprehensive analysis of phospholipids and glycolipids in the opportunistic pathogen Enterococcus faecalis. PLoS One 12 (4), e0175886. doi:10.1371/journal.pone.0175886
Saito, H. E., Harp, J. R., and Fozo, E. M. (2017). Enterococcus faecalis responds to individual exogenous fatty acids independently of their degree of saturation or chain length. Appl. Environ. Microbiol. 84 (1), e01633-17. doi:10.1128/AEM.01633-17
Saito, H. E., Harp, J. R., and Fozo, E. M. (2014). Incorporation of exogenous fatty acids protects Enterococcus faecalis from membrane-damaging agents. Appl. Environ. Microbiol. 80 (20), 6527–6538. doi:10.1128/AEM.02044-14
Sakoulas, G., Nonejuie, P., Nizet, V., Pogliano, J., Crum-Cianflone, N., and Haddad, F. (2013). Treatment of high-level gentamicin-resistant Enterococcus faecalis endocarditis with daptomycin plus ceftaroline. Antimicrob. Agents Chemother. 57 (8), 4042–4045. doi:10.1128/AAC.02481-12
Sakoulas, G., Rose, W., Nonejuie, P., Olson, J., Pogliano, J., Humphries, R., et al. (2014). Ceftaroline restores daptomycin activity against daptomycin-nonsusceptible vancomycin-resistant Enterococcus faecium. Antimicrob. Agents Chemother. 58 (3), 1494–1500. doi:10.1128/AAC.02274-13
Silverman, J. A., Perlmutter, N. G., and Shapiro, H. M. (2003). Correlation of daptomycin bactericidal activity and membrane depolarization in Staphylococcus aureus. Antimicrob. Agents Chemother. 47 (8), 2538–2544. doi:10.1128/AAC.47.8.2538-2544.2003
Slavetinsky, C. J., Hauser, J. N., Gekeler, C., Slavetinsky, J., Geyer, A., Kraus, A., et al. (2022). Sensitizing Staphylococcus aureus to antibacterial agents by decoding and blocking the lipid flippase MprF. Elife 11, e66376. doi:10.7554/ELIFE.66376
Smith, J. R., Barber, K. E., Raut, A., Aboutaleb, M., Sakoulas, G., and Rybak, M. J. (2015). β-Lactam combinations with daptomycin provide synergy against vancomycin-resistant Enterococcus faecalis and Enterococcus faecium. J. Antimicrob. Chemother. 70 (6), 1738–1743. doi:10.1093/jac/dkv007
Smith, J. R., Barber, K. E., Raut, A., and Rybak, M. J. (2015). β-Lactams enhance daptomycin activity against vancomycin-resistant Enterococcus faecalis and Enterococcus faecium in in vitro pharmacokinetic/pharmacodynamic models. Antimicrob. Agents Chemother. 59 (5), 2842–2848. doi:10.1128/AAC.00053-15
Sohlenkamp, C., and Geiger, O. (2016). Bacterial membrane lipids: Diversity in structures and pathways. FEMS Microbiol. Rev. 40 (1), 133–159. doi:10.1093/femsre/fuv008
Tong, S. Y. C., Davis, J. S., Eichenberger, E., Holland, T. L., and Fowler, V. G. (2015). Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28 (3), 603–661. doi:10.1128/CMR.00134-14
Tran, T. T., Mishra, N. N., Seepersaud, R., Diaz, L., Rios, R., Dinh, A. Q., et al. (2019). Mutations in cdsA and pgsA Correlate with Daptomycin Resistance in Streptococcus mitis and S. oralis. Antimicrob. Agents Chemother. 63 (2018), e01531-18–e01536. doi:10.1128/AAC.01531-18
Tran, T. T., Munita, J. M., and Arias, C. A. (2015). Mechanisms of drug resistance: Daptomycin resistance. Ann. N. Y. Acad. Sci. 1354 (1), 32–53. doi:10.1111/NYAS.12948
Tran, T. T., Panesso, D., Gao, H., Roh, J. H., Munita, J. M., Reyes, J., et al. (2013b). Whole-genome analysis of a daptomycin-susceptible enterococcus faecium strain and its daptomycin-resistant variant arising during therapy. Antimicrob. Agents Chemother. 57 (1), 261–268. doi:10.1128/AAC.01454-12
Tran, T. T., Panesso, D., Mishra, N. N., Mileykovskaya, E., Guan, Z., Munita, J. M., et al. (2013a). Daptomycin-resistant Enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids. mBio 4 (4e00281-13), e00281–13. doi:10.1128/mBio.00281-13
Vega, L. A., and Caparon, M. G. (2013). Cationic antimicrobial peptides disrupt the Streptococcus pyogenes ExPortal. Mol. Microbiol. 85 (6), 1119–1132. doi:10.1111/j.1365-2958.2012.08163.x
Wang, G., Yu, F., Lin, H., Murugesan, K., Huang, W., Hoss, A. G., et al. (2018). Evolution and mutations predisposing to daptomycin resistance in vancomycin-resistant Enterococcus faecium ST736 strains. PLoS One 13 (12), e0209785. doi:10.1371/JOURNAL.PONE.0209785
Werth, B. J., Barber, K. E., Tran, K. N. T., Nonejuie, P., Sakoulas, G., Pogliano, J., et al. (2015). Ceftobiprole and ampicillin increase daptomycin susceptibility of daptomycin-susceptible and -resistant VRE. J. Antimicrob. Chemother. 70 (2), 489–493. doi:10.1093/jac/dku386
Woodall, B. M., Harp, J. R., Brewer, W. T., Tague, E. D., Campagna, S. R., and Fozo, E. M. (2021). Enterococcus faecalis readily adapts membrane phospholipid composition to environmental and genetic perturbation. Front. Microbiol. 12. doi:10.3389/fmicb.2021.616045
Yim, J., Smith, J. R., Singh, N. B., Rice, S., Stamper, K., Garcia de la Maria, C., et al. (2017). Evaluation of daptomycin combinations with cephalosporins or gentamicin against Streptococcus mitis group strains in an in vitro model of simulated endocardial vegetations (SEVs). J. Antimicrob. Chemother. 72 (8), 2290–2296. doi:10.1093/jac/dkx130
Zhang, R., Polenakovik, H., Barreras Beltran, I. A., Waalkes, A., Salipante, S. J., Xu, L., et al. (2022). Emergence of dalbavancin, vancomycin, and daptomycin nonsusceptible Staphylococcus aureus in a patient treated with dalbavancin: Case report and isolate characterization. Clin. Infect. Dis. 1, ciac341. doi:10.1093/CID/CIAC341
Keywords: daptomycin, resistance, phospholipids, enterococci, Staphylococcus aureus, streptococci
Citation: Nguyen AH, Hood KS, Mileykovskaya E, Miller WR and Tran TT (2022) Bacterial cell membranes and their role in daptomycin resistance: A review. Front. Mol. Biosci. 9:1035574. doi: 10.3389/fmolb.2022.1035574
Received: 02 September 2022; Accepted: 26 October 2022;
Published: 14 November 2022.
Edited by:
Isabel María López-Lara, UNAM Campus Morelos, National Autonomous University of Mexico, MexicoReviewed by:
Elizabeth Fozo, The University of Tennessee, Knoxville, United StatesCopyright © 2022 Nguyen, Hood, Mileykovskaya, Miller and Tran. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Truc T. Tran, dHR0cmFuNEBob3VzdG9ubWV0aG9kaXN0Lm9yZw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.