- 1Intercollegiate Faculty of Biotechnology, University of Gdansk and Medical University of Gdansk, Gdansk, Poland
- 2Department of Biochemistry, University of Wisconsin—Madison, Madison, WI, United States
In cells molecular chaperone systems consisting of Hsp70 and its obligatory J-domain protein (JDP) co-chaperones transiently interact with a myriad of client proteins—with JDPs typically recruiting their partner Hsp70 to interact with particular clients. The fundamentals of this cyclical interactions between JDP/Hsp70 systems and clients are well established. Much less is known about other aspects of JDP/Hsp70 system function, including how such systems evolved over time. Here we discuss the JDP/Hsp70 system involved in the biogenesis of iron-sulfur (FeS) clusters. Interaction between the client protein, the scaffold on which clusters are built, and its specialized JDP Hsc20 has stayed constant. However, the system’s Hsp70 has changed at least twice. In some species Hsc20’s Hsp70 partner interacts only with the scaffold, in others it has many JDP partners in addition to Hsc20 and interacts with many client proteins. Analysis of this switching of Hsp70 partners has provided insight into the insulation of JDP/Hsp70 systems from one another that can occur when more than one Hsp70 is present in a cellular compartment, as well as how competition among JDPs is balanced when an Hsp70 partner is shared amongst a number of JDPs. Of particularly broad relevance, even though the scaffold’s interactions with Hsc20 and Hsp70 are functionally critical for the biogenesis of FeS cluster-containing proteins, it is the modulation of the Hsc20-Hsp70 interaction per se that allows Hsc20 to function with such different Hsp70 partners.
Introduction
Via their cyclic interaction with client proteins, Hsp70-based molecular chaperone systems are key players, not only in general protein homeostasis, but also in central cellular pathways (Clerico et al., 2019; Mayer and Gierasch, 2019; Rosenzweig et al., 2019; Balchin et al., 2020; Liu et al., 2020). This review is focused on such an Hsp70-dependent pathway—the biogenesis of iron-sulfur (FeS) clusters, cofactors necessary for activities of many proteins (Py and Barras, 2015; Maio and Rouault, 2020; Braymer et al., 2021; Esquilin-Lebron et al., 2021). In this pathway, which was inherited by eukaryotic cells from the bacterial progenitor of mitochondria, clusters are assembled on the client scaffold protein, called IscU/Isu in bacteria/mitochondria, and then transferred to recipient proteins. However, regardless of the exact client or cellular function, the Hsp70-client interaction cycle depends on the same conformational changes of Hsp70, which are triggered by binding and hydrolysis of ATP (Figure 1). ATP hydrolysis profoundly affects Hsp70’s affinity for client—resulting in a non-equilibrium enhancement of client affinity (often called ultra-affinity) by minimizing escape of the interacting client (Barducci and De Los Rios, 2015). Client release is driven by subsequent ADP dissociation. This cycle is driven by cochaperones. Via action of its J-domain, a J-domain protein (JDP) cochaperone activates the ATPase of Hsp70; ADP release is typically induced by a nucleotide exchange factor (NEF) cochaperone (Mayer and Gierasch, 2019; Liu et al., 2020).
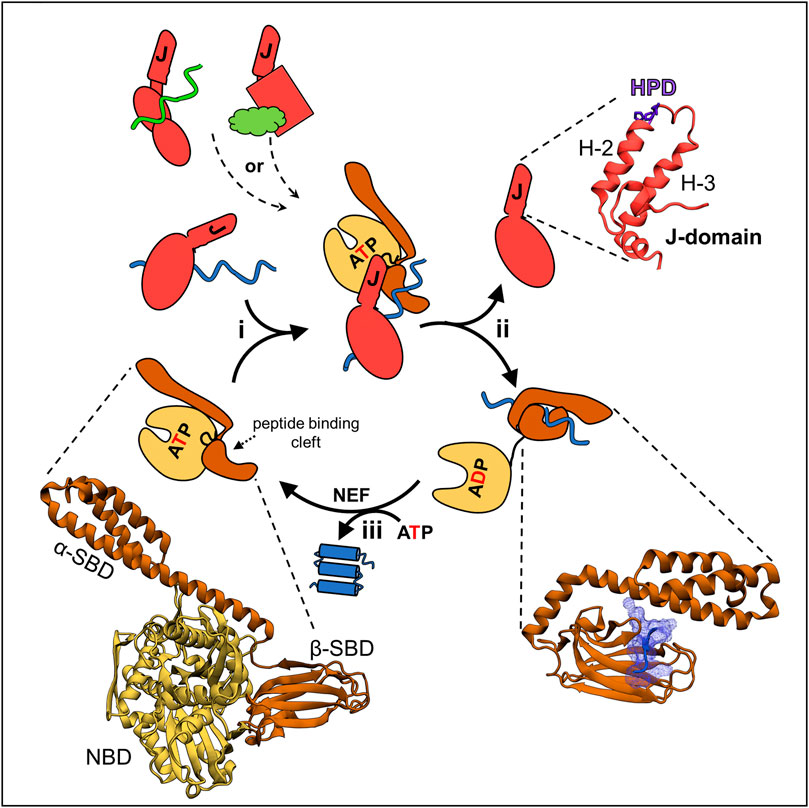
FIGURE 1. JDP/Hsp70 client binding cycle and structures of key components. (upper left) Different JDPs (red) can deliver different clients, indicated by different colors and shapes, to the same Hsp70 by interacting with them directly via distinct client binding domains, indicated by different shapes. (center) This scheme depicts the cycle involved in folding of a client protein. The same cycle occurs when JDP/Hsp70 systems are involved in other cellular processes, including FeS cluster biogenesis (see Figure 3): (i) JDP-client complex interacts with ATP bound Hsp70. In this “open” conformation the nucleotide binding domain (NBD, beige) is docked on to the substrate binding domain (SBD, brown) and the interdomain flexible linker (black) is bound in a crevice of the NBD. This conformation allows easy access of client to the peptide binding cleft of β-SBD, as α-SBD is retracted from it, and enables binding of the J-domain, which interacts at the NBD-β-SBD interface. It also allows simultaneous, transient interactions of the J-domain and JDP-bound client, and thus synergistic stimulation of ATP hydrolysis by Hsp70. (ii) ATP hydrolysis triggers conformational changes resulting in separation of the NBD and SBD, only connected by a flexible linker. The α-SBD subdomain covers the peptide binding cleft of β-SBD, stabilizing its interaction with the client. Separation of the SBD and NBD also results in loss of the J-domain binding face, and thus JDP release. (iii) ADP exchange for ATP, often involving a nucleotide exchange factor (NEF), reverting Hsp70 to the ATP bound state, causing client release. Structures of critical components of the cycle from E. coli DnaK and DnaJ—the best studied JDP/Hsp70 system—are depicted (bottom left) the ATP bound conformation of DnaK (PDB ID: 4B9Q). (Upper right) J-domain architecture is conserved with helices 2 and 3 (H-2, H-3) forming a finger like structure. Loop between these helices contains His Pro Asp (HPD) sequence present in all J-domains and required for the stimulation of Hsp70s’ ATPase. (bottom right) Hsp70’s SBD interacting with a peptide derived from a client protein (PDB ID: 1DKZ). Note helical α-SBD subdomain covering the binding cleft of β-SBD, but without contacting the client.
JDPs are key to the functional diversity of Hsp70 systems, in good part by influencing what clients Hsp70 binds (Kampinga and Craig, 2010; Craig and Marszalek, 2017; Zhang et al., 2022). As the name implies, membership in this diverse family of proteins requires only a J-domain. All J-domains, regardless of their role in the cell or their partner Hsp70, have a His, Pro, Asp tripeptide (HPD) in a loop connecting two helices (Figure 1). The HPD plays a critical role in stimulating ATPase activity, while the two flanking helices form the extended Hsp70 binding face. Other JDP domains often bind client polypeptides. These are directly targeted to Hsp70 via interaction of the JDP’s J-domain, which then stabilizes the client-Hsp70 interaction by stimulating ATPase activity. J-domains act synergistically with clients, as robust ATPase stimulation occurs only when client interacts in Hsp70’s peptide binding cleft. The JDP involved in the conserved FeS cluster biogenesis system (Hsc20) is highly specialized. Its only client is the Isu/IscU scaffold on which the FeS clusters are built (Vickery and Cupp-Vickery, 2007; Dutkiewicz and Nowak, 2018).
Unlike JDPs, all Hsp70s are structurally similar—having an N-terminal nucleotide binding domain (NBD) and a C-terminal substrate binding domain (SBD) composed of α and β subdomains. β-SBD contains the hydrophobic peptide binding cleft formed by two four-stranded β-sheets and two pairs of upward loops. It binds an extended polypeptide segment of the client—five residues enriched in hydrophobic amino acids (Clerico et al., 2015; Mayer and Gierasch, 2019; Liu et al., 2020; Clerico et al., 2021). α-SBD acts as a lid covering the peptide binding cleft, thus stabilizing client interaction (Figure 1). Initiation of client binding occurs when Hsp70 is in the ATP-bound conformation—in which both SBD subdomains and the linker that separates the SBD from the NBD are docked on the NBD, leaving the peptide binding cleft very accessible. The HPD and Helices 2 and 3 of the J-domain form a binding interface with Hsp70 sequences in the NBD, β-SBD and the linker.
Typically, JDPs function within networks consisting of a limited number of Hsp70s, sometimes just one, cooperating with a larger number of JDPs, including both those involved in general proteostasis and those involved in a singular cellular pathway (Craig and Marszalek, 2017; Barriot et al., 2020; Piette et al., 2021). Such networks are highly dynamic, as the number of JDPs and Hsp70s constituting them, as well their partnerships, vary across evolving lineages (Kominek et al., 2013; Powers and Balch, 2013; Rebeaud et al., 2021). Although such changes are observed in both prokaryotic and eukaryotic lineages, an overall trend is apparent: networks are composed of a larger number of JDP/Hsp70 systems in complex multicellular eukaryotes, i.e., more in animals and plants than in bacteria, archaea or unicellular eukaryotes (Powers and Balch, 2013; Rebeaud et al., 2021). Little is known, however, about the molecular mechanisms underlying expansion, contraction and reorganization of these networks.
Most Hsp70s bind many different substrates. However, there are examples of specialized Hsp70s. Hsp70s that partner with JDP Hsc20 in FeS cluster biogenesis in bacteria and a subset of fungi are of this type (Schilke et al., 2006; Vickery and Cupp-Vickery, 2007). Here we focus on the client-JDP/Hsp70 system involved in FeS cluster biogenesis. This system is biologically important—the client is the central component of a crucial biological pathway present in both prokaryotes and eukaryotes. Moreover, across the tree of life, the dedicated JDP of the system sometimes functions with a specialized Hsp70 and sometimes a multifunctional Hsp70. Consequently, it has proven key to understanding how networks composed of one Hsp70 cooperating with a number of JDPs are able to effectively carry out so many functions in the cell.
The client Isu/IscU, the scaffold for biogenesis of FeS clusters
The conserved Isu/IscU scaffold protein on which FeS clusters are built is present in the bacteria and in mitochondria of eukaryotes (Maio and Rouault, 2020; Braymer et al., 2021; Esquilin-Lebron et al., 2021). It is a small polypeptide with a compact fold—five α-helices and three β-strands (Kim et al., 2012b; Adrover et al., 2015; Tanaka et al., 2019; Kunichika et al., 2021). It coordinates a 2Fe-2S cluster utilizing three invariant Cys residues and one or more additional residue(s) whose identity is not yet clear (Figure 2). Nuclear Magnetic Resonance (NMR) analyses of bacterial IscU revealed that the scaffold is structurally highly dynamic, populating two conformational states—a structured conformation that resembles the conformation when in complex with an FeS cluster and a disordered conformation (Kim et al., 2012b; Kim et al., 2012c).
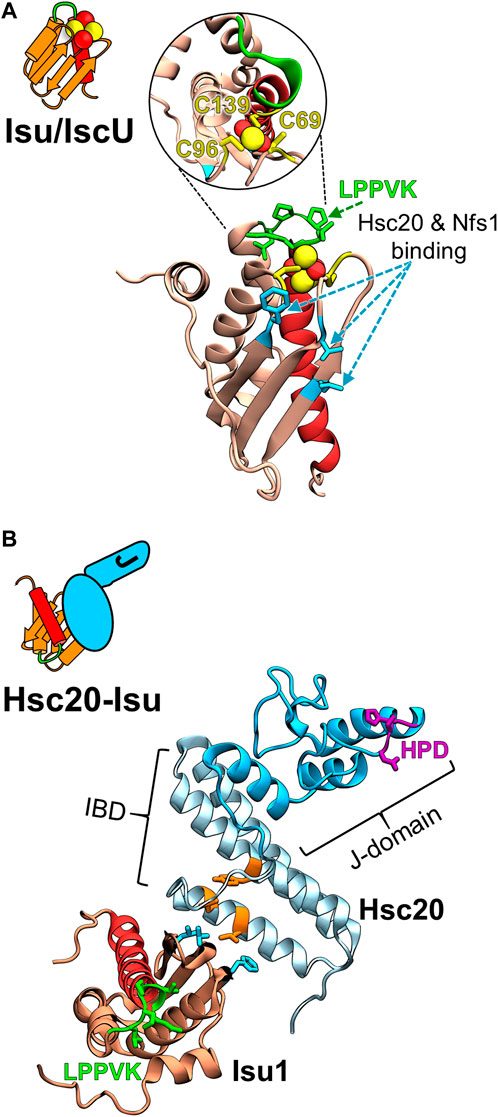
FIGURE 2. Structural models of the FeS cluster scaffold and its complex with Hsc20. (A) Cartoon depicting key structural elements of FeS cluster loaded (yellow/red balls) Isu/IscU scaffold and homology model of S. cerevisiae Isu1 with bound FeS cluster based on the crystal structure of FeS cluster bound IscU from Aquifex aeolicus (PDB ID: 2Z7E) at left and right, respectively. Insert within the circle is the structure tilted 90° to visualize the three Cys residues (yellow) that coordinate a 2Fe-2S cluster (yellow/red balls). LPPVK sequence (green) localized on a loop prior to C-terminal helix 5 (red) is the binding site for Hsp70. Hot spot residues involved in the Isu1 interactions with both Hsc20 and cysteine desulfurase Nfs1 are indicated (cyan). Note: the Hsc20 and Hsp70 binding sites on Isu1 do not overlap. (B) Cartoon representing J-domain protein Hsc20 in complex with Isu/IscU and structural model of the Hsc20 of S. cerevisiae (PDB ID: 3UO2 and 3UO3) in complex with Isu1 at left and right, respectively. Hsc20 is a simple JDP consisting of the N-terminal J-domain (dark cyan) with HPD motif (magenta) and C-terminal Isu binding domain, IBD (light cyan), which interacts with Isu via a contact surface involving hot spot residues (orange).
During the process of assembly of the cluster and its subsequent transfer onto target proteins, Isu/IscU interacts with several proteins (Figure 3) (Bonomi et al., 2011; Boniecki et al., 2017; Gervason et al., 2019; Lin et al., 2020). Hsc20 and the cysteine desulfurase which donates sulfur for cluster assembly have overlapping binding sites on a face of the structured Isu/IscU conformation (Figure 2) (Shi et al., 2010; Majewska et al., 2013). These interfaces are conserved across bacterial and eukaryotic systems (Füzéry et al., 2011; Ciesielski et al., 2012; Boniecki et al., 2017; Fox et al., 2019). Hsp70 binds an invariant LPPVK pentapeptide in the flexible loop that is just prior to the C-terminal α-helix (Hoff et al., 2002; Hoff et al., 2003; Dutkiewicz et al., 2004; Bonomi et al., 2011). This short loop has been reported to also be involved in binding of frataxin, a key component of the FeS synthesis complex assembled around cysteine desulfurase (Prischi et al., 2010; Manicki et al., 2014; Fox et al., 2019). Moreover, Isu/IscU is a substrate for the ATP dependent protease LON (known as Pim1 in Saccharomyces cerevisiae) (Figure 3) (Ciesielski et al., 2016; Ciesielski and Craig, 2017). Consistent with its structural instability, Isu of S. cerevisiae has a LON/Pim1-dependent fast turnover rate in vivo (Andrew et al., 2008; Song et al., 2012). Whereas the physiological importance of the high turnover rate is not yet clear, evidence indicates that the high levels of Isu observed under stress conditions are beneficial for the cell. How such regulation may occur is not known.
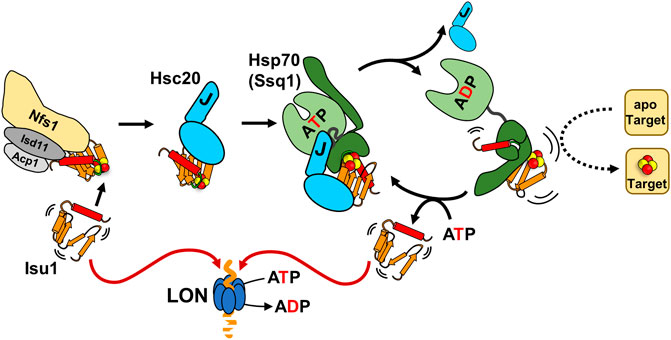
FIGURE 3. The role of Hsc20/Hsp70 scaffold binding cycle in FeS cluster biogenesis. Interaction of the Isu/IscU scaffold, which is structurally highly dynamic (indicated by agitrons) with a FeS cluster assembly complex (Nfs1/Isd11/Acp1 and other accessory proteins: frataxin, ferredoxin-not shown) is required for de novo cluster synthesis. The cluster loaded scaffold interacts with the dedicated JDP Hsc20. The scaffold interactions with both the cluster assembly complex and Hsc20 protect it from degradation by LON protease. Hsc20-scaffold complex interacts with its Hsp70 partner in the ATP bound conformation. Synergistic stimulation of Hsp70’s ATPase by the J-domain of Hsc20 and the scaffold triggers conformational changes that leads to Hsc20 release and stabilization of an Hsp70-scaffold complex. Formation of this complex is required for effective cluster transfer onto apo-target proteins. Exchange of ADP for ATP results in scaffold release. It is not clear what regulates whether a scaffold is reused in another round of cluster assembly/transfer or proteolyzed.
Importance of Hsc20 and Hsp70 interaction with the Isu/IscU scaffold for FeS cluster biogenesis
Strong evidence exists for the importance of the binding of Hsc20 and its partner Hsp70 to Isu for functioning of the FeS cluster biogenesis pathway (Figure 3). Studies of S. cerevisiae—which unlike bacteria, has no alternative, Hsc20/Hsp70-independent FeS cluster biogenesis pathway (Py and Barras, 2015; Esquilin-Lebron et al., 2021)—demonstrated that the Hsc20-Isu interaction is critical in vivo (Ciesielski et al., 2012). Cells are inviable when the binding face of either Hsc20 or Isu is severely compromised (Majewska et al., 2013). When the interaction is partially defective, cells grow slowly and have reduced activities of enzymes that required FeS clusters for catalysis (i.e., succinate dehydrogenase and aconitase). Like the Hsc20-Isu interaction, Isu binding by Hsp70 is critical (Dutkiewicz et al., 2004). S. cerevisiae cells expressing variants having either amino acid substitutions in the LPPVK of Isu or the peptide binding cleft of Hsp70 have a “null phenotype” (Dutkiewicz et al., 2004; Knieszner et al., 2005). Partial inhibition of the interaction results in slow growth and reduced activity of enzymes dependent on an FeS cluster for function (Dutkiewicz et al., 2004; Knieszner et al., 2005). That the Isu-Hsc20 interaction is important for delivery of Isu to Hsp70 by the typical client-JDP-Hsp70 interaction cycle is supported by in vitro data (Ciesielski et al., 2012; Dutkiewicz et al., 2017). As simultaneous interaction of a J-domain and client are required for robust ATPase stimulation, client-JDP complex formation would be expected to increase the efficiency of stimulation as it increases the chance of both being near Hsp70 at the same time. Consistent with this idea, when the Isu-Hsc20 interaction is compromised, Hsp70’s ATPase activity is stimulated only weakly (Ciesielski et al., 2012). Furthermore, consistent with the typical requirement for ATP hydrolysis for stable Hsp70-client binding, stable Hsp70-Isu complex formation is very inefficient when the Isu-Hsc20 interaction is compromised (Dutkiewicz et al., 2017).
That interactions of Hsc20 and partner Hsp70 with Isu/IscU are important for the transfer of cluster from Isu/IscU onto recipient protein(s) is supported by results of both in vivo and in vitro experiments. Depletion of either Hsc20 or Hsp70 Ssq1 in S. cerevisiae results in increased amounts of FeS cluster bound Isu and reduced activity of FeS cluster requiring enzymes (Mühlenhoff et al., 2003). Similar results were obtained when Hsc20 and Hsp70 Ssq1 variants defective in interactions were tested (Dutkiewicz et al., 2006). The most extensive in vitro experiments have been carried out using the bacterial system—transferring cluster from Escherichia coli IscU onto recipient apo-proteins including apo-ferredoxin (Fdx) and apo-glutaredoxin 5 (Grx5) (Chandramouli and Johnson, 2006; Bonomi et al., 2008, 2011; Iametti et al., 2015). Transfer did occur in absence of Hsc20 (HscB) and partner Hsp70 (HscA), but at a rate considered lower than expected. The presence of both HscB and HscA accelerated the rate. This acceleration was dependent on ATP hydrolysis, as expected for chaperones using the canonical client binding cycle. It has been hypothesized that Hsp70 binding facilitates cluster release by inducing major conformational change of IscU (Bonomi et al., 2011)—consistent with NMR results demonstrating that HscA binding shifts the IscU population into the disordered conformation (Kim et al., 2012a). But how Hsp70 binding to the LPPVK sequence mechanistically translates into accelerated FeS cluster transfer from Isu onto recipient protein(s) is not yet clear, as structural information is lacking. However, it is not difficult to imagine, given how Hsp70s bind substrates, that Isu would undergo a conformational change that would destabilize the cluster from Isu/IscU—especially considering that the Hsp70 binding site lies in a loop just prior to the C-terminal α-helix that contains a cysteine required for cluster coordination (Bonomi et al., 2011).
Hsc20 functions with three different Hsp70 partners across the tree of life
Although the overall client interaction cycle utilized by Hsc20 and its Hsp70 partner in the FeS cluster biogenesis pathway is typical, it is unusual in that Hsc20 partners with different Hsp70s across evolving lineages (Figure 4). Two of the Hsp70 partners are specialized, interacting with only one client—Isu/IscU; one is multifunctional, interacting with many clients, including Isu/IscU (Pukszta et al., 2010; Delewski et al., 2016; Kleczewska et al., 2020). In bacteria, Hsc20 partners with the specialized Hsp70, HscA (Hesterkamp and Bukau, 1998; Silberg et al., 1998). In S. cerevisiae and closely related species Hsc20 functions with a specialized mitochondrial Hsp70, Ssq1 (Schilke et al., 1999; Schilke et al., 2006). However, in mitochondria of most eukaryotes there is only one Hsp70 (mtHsp70). Hsc20 and all other JDPs partner with mtHsp70 (Srivastava et al., 2019; Maio and Rouault, 2020), which is a descendent of DnaK, the multifunctional Hsp70 of bacteria, not HscA (Huynen et al., 2001; Kleczewska et al., 2020). Like DnaK, it binds a wide array of clients, including Isu (Schilke et al., 2006; Srivastava et al., 2019).
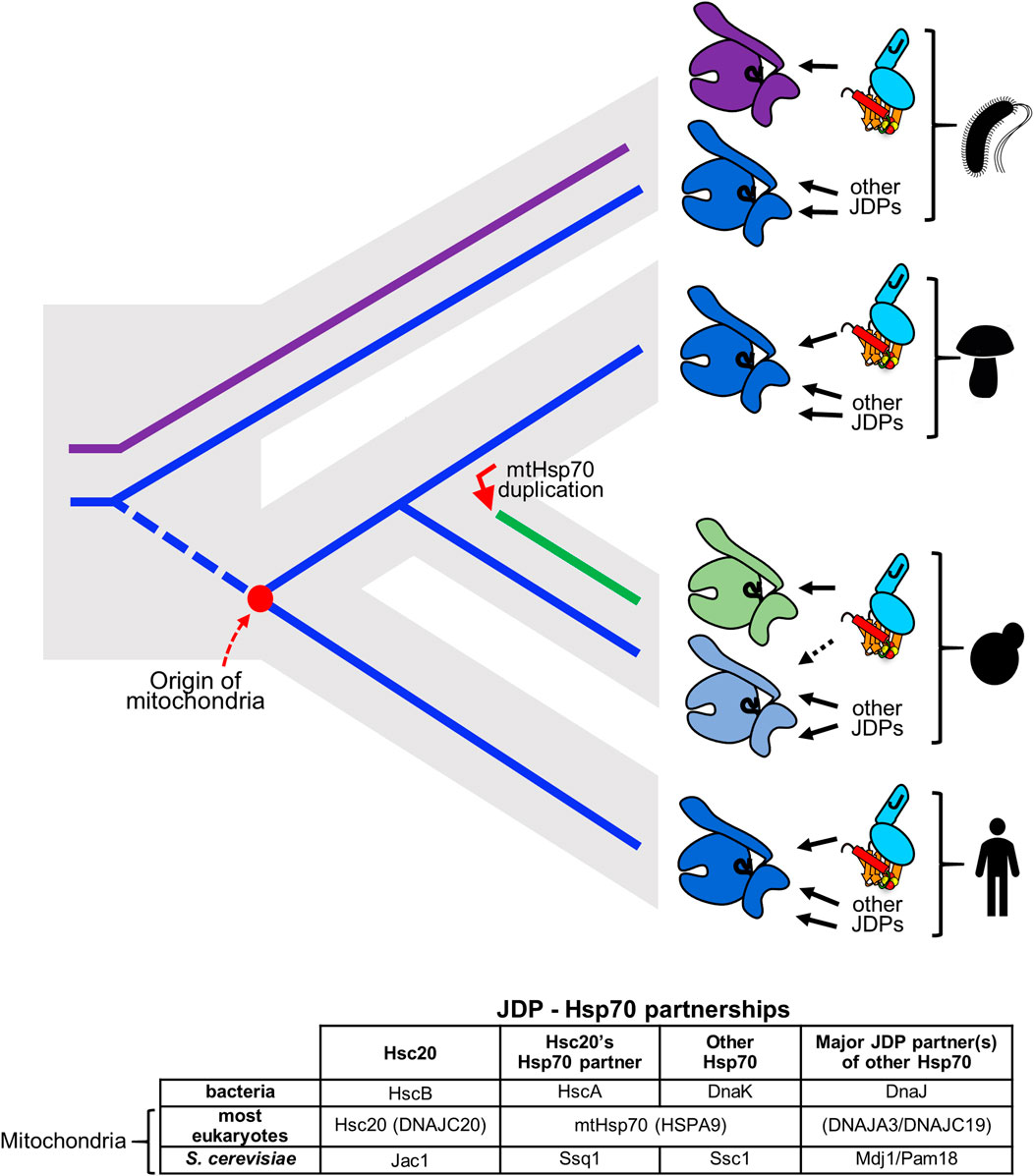
FIGURE 4. Hsc20 has changed Hsp70 partners twice across the tree of life. (top) Gray contour indicates phylogenetic relationships among evolving lineages: bacteria, fungi, metazoa. Solid lines indicate evolution of Hsp70s HscA (magenta) and DnaK (blue); broken blue line indicates the eukaryotic lineage prior to the origin of mitochondria (red dot). JDP Hsc20 is found in bacteria and mitochondria of eukaryotes (cyan, shown bound to client Isu/IscU in brown with FeS cluster). Solid arrows indicate its Hsp70 partner. In bacteria Hsc20 (HscB) partners with FeS cluster specialized Hsp70 HscA (magenta). This system coexists with multifunctional Hsp70 DnaK (dark blue). In mitochondria of eukaryotic cells Hsc20 partners with mtHsp70 e.g., most fungi and metazoans (indicated by mushroom and human pictograms) or with FeS cluster specialized Hsp70 Ssq1 (green), which evolved after an Hsp70 gene duplication (red arrow) in a progenitor of the S. cerevisiae lineage (budding yeast pictogram). The other duplicate, Ssc1 (light blue) remained multifunctional partnering with multiple JDPs, but only minimally with Hsc20, as indicated by dotted line. Following the origin of mitochondria Hsc20 was maintained, but HscA was not. Its function in FeS cluster biogenesis was substituted by mtHsp70, a descendant of DnaK (dark blue). (bottom) Names of JDPs and Hsp70s functioning in FeS cluster biogenesis. Those in parentheses are the name of human proteins.
Why/how did such shifts in Hsc20/Hsp70 partnership occur? As HscA orthologues have not been found in eukaryotic genomes (Kleczewska et al., 2020), most likely loss of an Hsp70 drove the change to Hsc20 functioning with multifunctional mtHsp70. This shift could have occurred during or before the emergence of mitochondria. HscA, but not Hsc20, may have been lost during the massive transfer of genes from the bacterial endosymbiont to the host cell (Roger et al., 2017; Sloan et al., 2018). Alternatively, mitochondria may have evolved from a bacterium that already lacked HscA, as two bacterial genomes have been found to harbor HscB, but not HscA (Barriot et al., 2020). While the timing of the switch to multifunctional Hsp70 in mitochondria remains elusive, it is clear that the specialist Hsp70 Ssq1 arose during fungal evolution from a duplication of the gene encoding mtHsp70 (Schilke et al., 2006; Kominek et al., 2013; Kleczewska et al., 2020). Thus, though Ssq1 resembles bacterial HscA in its specialization, evolutionarily Ssq1 is not directly related to HscA. Following this duplication, which took place in a common ancestor of S. cerevisiae and Candida albicans, Ssq1 evolved toward being an Isu-binding, FeS cluster biogenesis specialist. Its paralog, termed Ssc1, maintained the multifunctionality typical of mtHsp70s, binding many clients and its ATPase being activated by several different JDPs (Figure 4).
That either a specialized or a multifunctional Hsp70 can partner effectively with Hsc20 in the FeS cluster biogenesis pathway is not only interesting in its own right, it also provides tools to address some fundamental questions regarding the functioning of JDP/Hsp70 networks in general. Two are discussed in the next sections. First, is the question of insulation of JDP/Hsp70 systems. When more than one Hsp70 is present in a cellular compartment, sometimes JDP/Hsp70 systems function independently from one another. What is the basis of such insulation? And, more fundamentally, how is any network fine-tuned to allow many different JDPs to effectively function with a single Hsp70? Second, what determines the substrate specificity of a specialized Hsp70? How does such specificity arise?
Hsc20 J-domain interaction with Hsp70 is responsible for functional insulation of specialized systems
Even though they share the same cellular space, the specialized and multifunctional JDP/Hsp70 systems of E. coli and the mitochondria of S. cerevisiae are functionally insulated from one another. The insulation is more extreme in E. coli. Specialized Hsc20 HscB does not function at all with multifunctional DnaK, consistent with the fact that HscB does not stimulate the ATPase activity of DnaK. Nor can DnaJ, the main JDP that functions with DnaK, partner with HscA, even though it can weakly stimulate its ATPase activity (Silberg et al., 1998). Insulation of the systems in mitochondria is not as complete. Hsc20 can work, albeit poorly, with multifunctional Hsp70 Ssc1 (Kim et al., 2001; Pukszta et al., 2010). Although Hsc20 is essential (Voisine et al., 2001), Ssq1 is not (Schilke et al., 1996; Knight et al., 1998). Cells lacking Ssq1 grow very slowly, and even this poor growth requires the upregulated expression of Isu that occurs upon deletion of Ssq1 (Andrew et al., 2008; Song et al., 2012; Dong et al., 2017). However, the reverse is not the case. Ssq1 does not work with other mitochondrial JDPs—Ssq1’s ATPase is stimulated only by Hsc20, not by other mitochondrial JDPs (D'Silva et al., 2003; Dutkiewicz et al., 2003).
Recent data suggests that the key to this functional insulation of JDP/Hsp70 systems specialized in FeS cluster biogenesis is the J-domian-Hsp70(ATP) interaction (Pukszta et al., 2010; Delewski et al., 2016). Overall, the mode of J-domain interaction across systems—via the HPD motif and helices 2 and 3—is conserved (Figure 5A) (Kityk et al., 2018; Wang et al., 2021). However, while the HPD is invariant, the residues of helix 2 and 3 forming the Hsp70 binding face are not conserved. Critical interfacial positions are occupied by very different amino acids, even in relatively closely related species (Figures 5B,C) (Delewski et al., 2016; Tomiczek et al., 2020). Perhaps most importantly, these interfaces are networks of interconnecting residues—key residues on one side of an interface form contacts with more than one residue on the other side (Figure 5B). Such contact networks are known to be highly flexible (Keskin et al., 2008; Aakre et al., 2015). During the course of evolution contacts can be added, deleted or replaced, as long as enough contacts are maintained across the interface that interaction remains (Levin et al., 2009). They enable coevolution between the interacting partners—change(s) on one side of the interface can be compensated by a complementary change(s) on the other (Szurmant and Weigt, 2018; Cong et al., 2019).
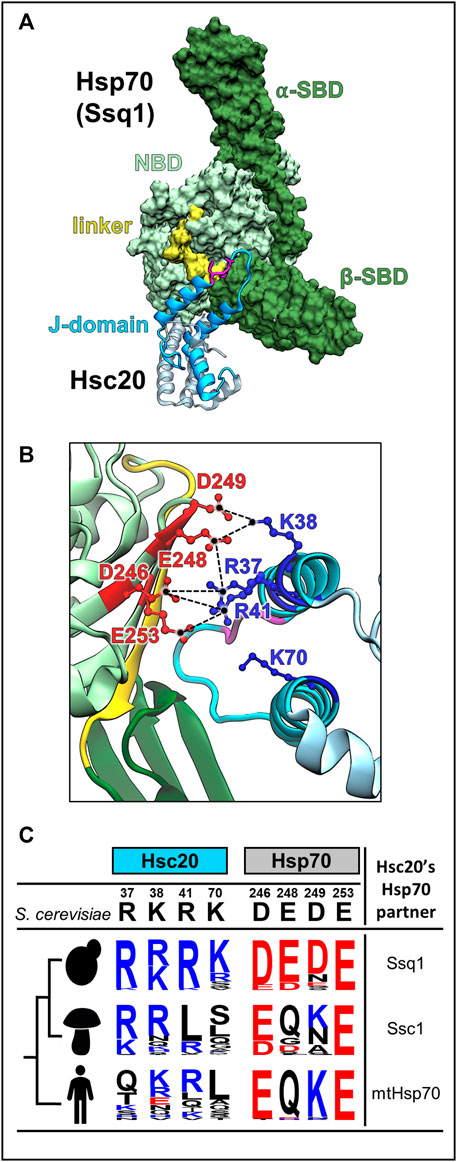
FIGURE 5. J-domain-Hsp70 interaction involves evolutionary variable interface. (A) Structural model of the S. cerevisiae Hsc20-Ssq1 (ATP) complex (Tomiczek et al., 2020). As in all JDP-Hsp70 interactions, the J-domain binds at the interface of NBD, β-SBD and interdomain linker formed when Ssq1 is in the ATP-bound conformation. Hsc20 in cyan with J-domain dark, Isu binding domain, light; HPD motif (magenta). Ssq1: NBD (light green), α- and β-SBDs (dark green); interdomain linker (yellow) is bound to the NBD. (B) The interconnected network of contacts between positively charged residues of helix 2 of the J-domain (blue) and negatively charged residues of Ssq1 (red) is critical for J-domain interaction with Ssq1; electrostatic interactions are depicted as broken lines (black). Note that one residue on the J-domain side interacts with more than one residue on the Hsp70 side e.g., J-domain’s R41 contacts D246 and E253 of Ssq1. K70 of helix 3 is also critical for Hsc20-Ssq1 complex formation, though its binding partner(s) was not identified. (C) The key residues involved in the J-domain-Ssq1 interaction are evolutionary variable. Sequence logos represent the amino acid frequency at positions of Hsc20 and Hsp70, which are homologous to the hot spot positions of the S. cerevisiae Hsc20 J-domain-Ssq1 interface; positively charged (blue), negatively charged (red) and uncharged (black). The analysis was based on Hsc20 and Hsp70 sequences from 23 yeast species, which diverged after Hsp70 gene duplication leading to Ssq1, 47 pre-duplication fungi species (harboring only mtHsp70) and 20 metazoan species (harboring mtHsp70), designated by human figure. Note: in contrast to the invariant HPD motif hot spot residues involved in J-domain/Ssq1 interaction in S. cerevisiae are evolutionary variable, particularly outside the post-duplication species.
Such changes could explain how new partnerships between JDPs and Hsp70s have formed and how functionally insulated JDP/Hsp70 systems have emerged. For example, the emergence of the insulated Hsc20/Ssq1 system could have occurred in the following way: Immediately following the gene duplication, sister copies of mtHsp70 were identical, and Hsc20 was able to interact and function with both. However, via J-domain/Hsp70 coevolution the affinity of Hsc20 for one paralogue (Ssq1) increased such that a new, more specific, partnership was formed. This new partnership was advantageous as it allowed Hsc20 to avoid competing with other JDPs for Hsp70 access. Thus, strengthening of this partnership was likely driven by positive selection, accelerating the coevolutionary changes. Furthermore, a gain of high specificity toward the LPPVK by Ssq1, as discussed in the next section, made reversion to multifunctionality unlikely (Pukszta et al., 2010; Delewski et al., 2016; Tomiczek et al., 2020).
The existence of the insulated JDP/Hsp70 systems in bacteria and mitochondria raises the question of whether other functionally insulated JDP/Hsp70 systems exist. The best documented example is the specialized Hsp70 (Ssb) in the cytosol of S. cerevisiae (James et al., 1997) (Huang et al., 2005; Zhang et al., 2017). Ssb arose via gene duplication from the multifunctional Hsp70 (Ssa) during the emergence of fungi. It functions with a specialized JDP termed Zuo1. In metazoans, including humans, the orthologue of Zuo1 functions with the multifunctional Hsp70, which is closely related to Ssa—thus, resembling the situation of mitochondrial Hsc20/mtHsp70 system (Hundley et al., 2005). Considering the high copy number dynamics observed for both Hsp70s and JDPs across the tree of life, it is likely that other functionally insulated JDP/Hsp70 systems await discovery (Barriot et al., 2020; Rebeaud et al., 2021; Zhang et al., 2022).
Specialization of Hsp70 client binding arose from restricting its ability to bind multiple clients
HscA and DnaK bind the LPPVK peptide derived from IscU with similar affinities. The mode of binding by the two Hsp70s is similar and fundamentally resembles that of β-SBDs with other client peptides—in an extended conformation in the peptide binding cleft with the α-helical lid (α-SBD) covering it without directly interacting (Cupp-Vickery et al., 2004; Clerico et al., 2015; Clerico et al., 2021) (Figure 1). However, while most peptides bind Hsp70 with the central position of the cleft occupied by a Leu, Val, or Phe, in the case of LPPVK it is occupied by the second Pro residue (Cupp-Vickery et al., 2004). Furthermore, LPPVK binds in the reverse orientation from that of most peptides—C to N backbone direction (Tapley et al., 2005; Tapley et al., 2006), rather than the more typical N to C (Clerico et al., 2021).
HscA, however, does not bind the peptide NRLLLTG that is most commonly used in studies of DnaK and other multifunctional Hsp70s (Zhu et al., 1996). There are only a small number of amino acid differences between HscA and DnaK β-SBDs in or near the peptide binding cleft (Tapley et al., 2006; Vickery and Cupp-Vickery, 2007). DnaK-like substitutions at these positions in HscA allow interaction with the NRLLLTG peptide, suggesting that the client specificity of HscA evolved by remodeling the peptide binding cleft, not by changing the mechanism of peptide binding per se. Although no structural data is available for the Ssq1 and Ssc1 Hsp70 pair, their SBDs, like those of HscA and DnaK, bind the LPPVK peptide with very similar affinities. Furthermore, the SBDs of Ssq1 do not bind other peptides recognized by Ssc1 (Schilke et al., 2006). Thus, like HscA, Ssq1 likely evolved high specificity toward LPPVK by restricting its ability to accommodate sequences other than LPPVK and provides an explanation for why this increased specificity is not accompanied by an increased affinity.
Considering that multifunctional mtHsp70 binds LPPVK and other pentapeptide substrates with similar affinity it is possible that the system could tolerate changes in LPPVK without disrupting function. Why then has this sequence remains invariant across all species, even those engaging multifunctional mtHsp70 in FeS cluster biogenesis. A possible explanation is provided by biochemical experiments demonstrating the direct involvement of LPPVK in Isu’s interaction with frataxin (Manicki et al., 2014). Since frataxin is a key player in FeS cluster biogenesis in all eukaryotes, evolutionary conservation of LPPVK may have been driven by its dual function (Srour et al., 2020; Braymer et al., 2021; Esquilin-Lebron et al., 2021).
Additional roles of Hsc20-Isu binding beyond Hsp70 cochaperone activity in FeS cluster transfer
Though Hsc20 is a simple, small JDP, consisting only of a J-domain and an Isu binding domain (Cupp-Vickery and Vickery, 2000; Bitto et al., 2008; Ciesielski et al., 2012), the results of both in vivo and in vitro experiments have raised the possibility that Hsc20 binding to Isu/IscU has effects in addition to targeting it to Hsp70 for binding. These results are discussed below as they raise the intriguing question of whether JDPs can evolve to be intimately intertwined in cellular pathways, and more generally how the diverse JDP family of proteins evolves.
Hsc20 binding affects both Isu/IscU’s structural stability and its susceptibility to LON protease. HscB binding promotes the shift of IscU towards the structured conformation (Kim et al., 2012a) and binding of HscB to cluster loaded IscU in the absence of HscA reduces the rate of cluster transfer below that measured for IscU alone (Iametti et al., 2015). Overproduction of Hsc20 in S. cerevisiae leads to elevated levels of Isu (Ciesielski et al., 2016). This in vivo effect on Isu levels may go beyond structural stabilization; it may directly affect recognition of Isu by LON (Ciesielski and Craig, 2017). Hydrophobic residues serve as LON recognition sequences (Pinti et al., 2016). Replacement of hydrophobic residues in the Hsc20 binding site of Isu with serines slows down degradation of Isu by LON in vitro (Ciesielski and Craig, 2017).
The overlap of Hsc20 and desulfurase binding sites on Isu/IscU, and thus mutual exclusivity of binding, places Hsc20 at the center of the FeS cluster biogenesis pathway—at the transition from cluster assembly to transfer (Figure 3) (Majewska et al., 2013). It is tempting to speculate that the Hsc20/Hsp70 system plays roles in controlling both physiological levels of cluster loaded Isu and flow of FeS clusters through the biogenesis pathway. Such regulatory roles would likely involve Hsc20-dependent displacement of cluster-loaded Isu from the complex with cysteine desulfurase and facilitation of FeS cluster transfer onto recipient proteins via delivery of cluster loaded Isu to Hsp70. But they could also involve regulating the levels of Isu. Not surprisingly, since the Isu binding site of cysteine desulfurase that provides the sulfur for the cluster assembly overlaps that of Hsc20 (Majewska et al., 2013), desulfurase also protects Isu from LON degradation (Andrew et al., 2008; Song et al., 2012; Ciesielski et al., 2016).
In thinking about the possible roles of Hsc20 in the complex FeS cluster biogenesis pathway, it is potentially informative to consider how they may have evolved. The original Hsc20 progenitor may have had no J-domain or connection to Hsp70s at all. Its role may have been to control FeS cluster biogenesis potential, that is IscU levels by reversibly interacting and protecting it against proteolytic degradation. However, this protective function may have been in adaptive conflict with cluster transfer, as stronger interaction with IscU would have been good for protection but bad for transfer and vice versa. This adaptive conflict could have been resolved by recruitment of a J-domain because interaction with Hsp70 facilitated release of IscU from the complex, thus lifting its inhibitory effect on the cluster transfer. Moreover, Hsp70 interaction with IscU further accelerated the cluster transfer rate. Similar scenarios could explain evolution of other specialized JDPs involved in other cellular pathways, e.g., splicing (Sahi et al., 2010), histone biogenesis (Hammond et al., 2021; Piette et al., 2021) and gene silencing (Ichino et al., 2021).
Conclusion
Studies on the specialized Isu/IscU-Hsc20/Hsp70 FeS cluster biogenesis systems have provided insights into previously underappreciated aspects of client-chaperone interactions that are broadly applicable to JDP/Hsp70 systems. First, the plasticity of the J-domain-Hsp70 interaction allows modulation of the JDP-Hsp70 interactions and thereby drives the ability of one Hsp70 to productively function with several JDPs, as well as the insulation of networks when more than one Hsp70 is present. Second, some JDPs may be more involved in the underlying biological pathway in which they are involved than generally recognized—having evolved by either gaining a J-domain or by gaining other interactions after already being an active JDP cochaperone for Hsp70.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
JM: National Science Center, Poland (OPUS 21 2021/41/B/NZ1/00449) EC: National Institutes of Health (R35 GM127009).
Acknowledgments
We thank Igor Grochowina for help with figures and for preparing the structural representations of proteins and their complexes.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aakre, C. D., Herrou, J., Phung, T. N., Perchuk, B. S., Crosson, S., and Laub, M. T. (2015). Evolving new protein-protein interaction specificity through promiscuous intermediates. Cell 163 (3), 594–606. doi:10.1016/j.cell.2015.09.055
Adrover, M., Howes, B. D., Iannuzzi, C., Smulevich, G., and Pastore, A. (2015). Anatomy of an iron-sulfur cluster scaffold protein: Understanding the determinants of [2Fe-2S] cluster stability on IscU. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1853 (6), 1448–1456. doi:10.1016/j.bbamcr.2014.10.023
Andrew, A. J., Song, J.-Y., Schilke, B., and Craig, E. A. (2008). Posttranslational regulation of the scaffold for Fe-S cluster biogenesis, Isu. MBoC 19 (12), 5259–5266. doi:10.1091/mbc.e08-06-0622
Balchin, D., Hayer-Hartl, M., and Hartl, F. U. (2020). Recent advances in understanding catalysis of protein folding by molecular chaperones. FEBS Lett. 594 (17), 2770–2781. doi:10.1002/1873-3468.13844
Barducci, A., and De Los Rios, P. (2015). Non-equilibrium conformational dynamics in the function of molecular chaperones. Curr. Opin. Struct. Biol. 30, 161–169. doi:10.1016/j.sbi.2015.02.008
Barriot, R., Latour, J., Castanié-Cornet, M.-P., Fichant, G., and Genevaux, P. (2020). J-domain proteins in bacteria and their viruses. J. Mol. Biol. 432 (13), 3771–3789. doi:10.1016/j.jmb.2020.04.014
Bitto, E., Bingman, C. A., Bittova, L., Kondrashov, D. A., Bannen, R. M., Fox, B. G., et al. fnm (2008). Structure of human J-type co-chaperone HscB reveals a tetracysteine metal-binding domain. J. Biol. Chem. 283(44), 30184–30192. doi:10.1074/jbc.m804746200
Boniecki, M. T., Freibert, S. A., Mühlenhoff, U., Lill, R., and Cygler, M. (2017). Structure and functional dynamics of the mitochondrial Fe/S cluster synthesis complex. Nat. Comm. 8 (1), 1287. doi:10.1038/s41467-017-01497-1
Bonomi, F., Iametti, S., Morleo, A., Ta, D., and Vickery, L. E. (2011). Facilitated transfer of IscU-[2Fe2S] clusters by chaperone-mediated ligand exchange. Biochemistry 50 (44), 9641–9650. doi:10.1021/bi201123z
Bonomi, F., Iametti, S., Morleo, A., Ta, D., and Vickery, L. E. (2008). Studies on the mechanism of catalysis of Iron−Sulfur cluster transfer from IscU[2Fe2S] by HscA/HscB chaperones. Biochemistry 47 (48), 12795–12801. doi:10.1021/bi801565j
Braymer, J. J., Freibert, S. A., Rakwalska-Bange, M., and Lill, R. (2021). Mechanistic concepts of iron-sulfur protein biogenesis in Biology. Biochim. Biophys. Acta. Mol. Cell Res. 1868 (1), 118863. doi:10.1016/j.bbamcr.2020.118863
Chandramouli, K., and Johnson, M. K. (2006). HscA and HscB stimulate [2Fe-2S] cluster transfer from IscU to apoferredoxin in an ATP-dependent reaction. Biochemistry 45 (37), 11087–11095. doi:10.1021/bi061237w
Ciesielski, S. J., and Craig, E. A. (2017). Posttranslational control of the scaffold for Fe-S cluster biogenesis as a compensatory regulatory mechanism. Curr. Genet. 63 (1), 51–56. doi:10.1007/s00294-016-0618-y
Ciesielski, S. J., Schilke, B. A., Osipiuk, J., Bigelow, L., Mulligan, R., Majewska, J., et al. (2012). Interaction of J-protein co-chaperone Jac1 with Fe-S scaffold Isu is indispensable in vivo and conserved in evolution. J. Mol. Biol. 417 (1-2), 1–12. doi:10.1016/j.jmb.2012.01.022
Ciesielski, S. J., Schilke, B., Marszalek, J., and Craig, E. A. (2016). Protection of scaffold protein Isu from degradation by the Lon protease Pim1 as a component of Fe-S cluster biogenesis regulation. Mol. Biol. Cell 27 (7), 1060–1068. doi:10.1091/mbc.e15-12-0815
Clerico, E. M., Meng, W., Pozhidaeva, A., Bhasne, K., Petridis, C., and Gierasch, L. M. (2019). Hsp70 molecular chaperones: Multifunctional allosteric holding and unfolding machines. Biochem. J. 476 (11), 1653–1677. doi:10.1042/bcj20170380
Clerico, E. M., Pozhidaeva, A. K., Jansen, R. M., Özden, C., Tilitsky, J. M., and Gierasch, L. M. (2021). Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein. Proc. Natl. Acad. Sci. USA. 118 (41), e2016962118. doi:10.1073/pnas.2016962118
Clerico, E. M., Tilitsky, J. M., Meng, W., and Gierasch, L. M. (2015). How hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J. Mol. Biol. 427 (7), 1575–1588. doi:10.1016/j.jmb.2015.02.004
Cong, Q., Anishchenko, I., and Baker, D. (2019). Protein interaction networks revealed by proteome coevolution. Science 365, 185–189. doi:10.1126/science.aaw617810.1126/science.aaw6718
Craig, E. A., and Marszalek, J. (2017). How do J-proteins get Hsp70 to do so many different things? Trends biochem. Sci. 42 (5), 355–368. doi:10.1016/j.tibs.2017.02.007
Cupp-Vickery, J. R., Peterson, J. C., Ta, D. T., and Vickery, L. E. (2004). Crystal structure of the molecular chaperone HscA substrate binding domain complexed with the IscU recognition peptide ELPPVKIHC. J. Mol. Biol. 342 (4), 1265–1278. doi:10.1016/j.jmb.2004.07.025
Cupp-Vickery, J. R., and Vickery, L. E. (2000). Crystal structure of Hsc20, a J-type Co-chaperone from Escherichia coli. J. Mol. Biol. 304 (5), 835–845. doi:10.1006/jmbi.2000.4252
D'Silva, P. D., Schilke, B., Walter, W., Andrew, A., and Craig, E. A. (2003). J protein cochaperone of the mitochondrial inner membrane required for protein import into the mitochondrial matrix. Proc. Natl. Acad. Sci. USA. 100 (24), 13839–13844. doi:10.1073/pnas.1936150100
Delewski, W., Paterkiewicz, B., Manicki, M., Schilke, B., Tomiczek, B., Ciesielski, S. J., et al. (2016). Iron-sulfur cluster biogenesis chaperones: Evidence for emergence of mutational robustness of a highly specific protein-protein interaction. Mol. Biol. Evol. 33 (3), 643–656. doi:10.1093/molbev/msv254
Dong, Y., Zhang, D., Yu, Q., Zhao, Q., Xiao, C., Zhang, K., et al. (2017). Loss of Ssq1 leads to mitochondrial dysfunction, activation of autophagy and cell cycle arrest due to iron overload triggered by mitochondrial iron-sulfur cluster assembly defects in Candida albicans. Int. J. Biochem. Cell Biol. 85, 44–55. doi:10.1016/j.biocel.2017.01.021
Dutkiewicz, R., Marszalek, J., Schilke, B., Craig, E. A., Lill, R., and Mühlenhoff, U. (2006). The Hsp70 chaperone Ssq1p is dispensable for iron-sulfur cluster formation on the scaffold protein Isu1p. J. Biol. Chem. 281 (12), 7801–7808. doi:10.1074/jbc.m513301200
Dutkiewicz, R., Nowak, M., Craig, E. A., and Marszalek, J. (2017). Fe-S cluster Hsp70 chaperones: The ATPase cycle and protein interactions. Methods Enz 595, 161–184. doi:10.1016/bs.mie.2017.07.004
Dutkiewicz, R., and Nowak, M. (2018). Molecular chaperones involved in mitochondrial iron-sulfur protein biogenesis. J. Biol. Inorg. Chem. 23 (4), 569–579. doi:10.1007/s00775-017-1504-x
Dutkiewicz, R., Schilke, B., Cheng, S., Knieszner, H., Craig, E. A., and Marszalek, J. (2004). Sequence-specific interaction between mitochondrial Fe-S scaffold protein Isu and Hsp70 Ssq1 is essential for their in vivo function. J. Biol. Chem. 279 (28), 29167–29174. doi:10.1074/jbc.m402947200
Dutkiewicz, R., Schilke, B., Knieszner, H., Walter, W., Craig, E. A., and Marszalek, J. (2003). Ssq1, a mitochondrial Hsp70 involved in iron-sulfur (Fe/S) center biogenesis. Similarities to and differences from its bacterial counterpart. J. Biol. Chem. 278 (32), 29719–29727. doi:10.1074/jbc.m303527200
Esquilin-Lebron, K., Dubrac, S., Barras, F., and Boyd, J. M. (2021). Bacterial approaches for assembling iron-sulfur proteins. mBio 12 (6), e02425–e02421. doi:10.1128/mbio.02425-21
Fox, N. G., Yu, X., Feng, X., Bailey, H. J., Martelli, A., Nabhan, J. F., et al. (2019). Structure of the human frataxin-bound iron-sulfur cluster assembly complex provides insight into its activation mechanism. Nat. Comm. 10 (1), 2210. doi:10.1038/s41467-019-09989-y
Füzéry, A. K., Oh, J. J., Ta, D. T., Vickery, L. E., and Markley, J. L. (2011). Three hydrophobic amino acids in Escherichia coli HscB make the greatest contribution to the stability of the HscB-IscU complex. BMC Biochem. 12 (1), 3. doi:10.1186/1471-2091-12-3
Gervason, S., Larkem, D., Mansour, A. B., Botzanowski, T., Müller, C. S., Pecqueur, L., et al. (2019). Physiologically relevant reconstitution of iron-sulfur cluster biosynthesis uncovers persulfide-processing functions of ferredoxin-2 and frataxin. Nat. Comm. 10 (1), 3566. doi:10.1038/s41467-019-11470-9
Hammond, C. M., Bao, H., Hendriks, I. A., Carraro, M., García-Nieto, A., Liu, Y., et al. (2021). DNAJC9 integrates heat shock molecular chaperones into the histone chaperone network. Mol. Cell 81 (12), 2533–2548. e2539. doi:10.1016/j.molcel.2021.03.041
Hesterkamp, T., and Bukau, B. (1998). Role of the DnaK and HscA homologs of Hsp70 chaperones in protein folding in E.coli. EMBO J. 17 (16), 4818–4828. doi:10.1093/emboj/17.16.4818
Hoff, K. G., Cupp-Vickery, J. R., and Vickery, L. E. (2003). Contributions of the LPPVK motif of the iron-sulfur template protein IscU to interactions with the Hsc66-Hsc20 chaperone system. J. Biol. Chem. 278 (39), 37582–37589. doi:10.1074/jbc.m305292200
Hoff, K. G., Ta, D. T., Tapley, T. L., Silberg, J. J., and Vickery, L. E. (2002). Hsc66 substrate specificity is directed toward a discrete region of the iron-sulfur cluster template protein IscU. J. Biol. Chem. 277 (30), 27353–27359. doi:10.1074/jbc.m202814200
Huang, P., Gautschi, M., Walter, W., Rospert, S., and Craig, E. A. (2005). The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nat. Struct. Mol. Biol. 12 (6), 497–504. doi:10.1038/nsmb942
Hundley, H. A., Walter, W., Bairstow, S., and Craig, E. A. (2005). Human mpp11 J protein: Ribosome-tethered molecular chaperones are ubiquitous. Science 308 (5724), 1032–1034. doi:10.1126/science.1109247
Huynen, M. A., Snel, B., Bork, P., and Gibson, T. J. (2001). The phylogenetic distribution of frataxin indicates a role in iron-sulfur cluster protein assembly. Hum. Mol. Genet. 10 (21), 2463–2468. doi:10.1093/hmg/10.21.2463
Iametti, S., Barbiroli, A., and Bonomi, F. (2015). Functional implications of the interaction between HscB and IscU in the biosynthesis of FeS clusters. J. Biol. Inorg. Chem. 20 (6), 1039–1048. doi:10.1007/s00775-015-1285-z
Ichino, L., Boone, B. A., Strauskulage, L., Harris, C. J., Kaur, G., Gladstone, M. A., et al. (2021). MBD5 and MBD6 couple DNA methylation to gene silencing through the J-domain protein SILENZIO. Science 372 (6549), 1434–1439. doi:10.1126/science.abg6130
James, P., Pfund, C., and Craig, E. A. (1997). Functional specificity among Hsp70 molecular chaperones. Science 275 (5298), 387–389. doi:10.1126/science.275.5298.387
Kampinga, H. H., and Craig, E. A. (2010). The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11 (8), 579–592. doi:10.1038/nrm2941
Keskin, O., Gursoy, A., Ma, B., and Nussinov, R. (2008). Principles of Protein−Protein interactions: What are the preferred ways for proteins to interact? Chem. Rev. 108 (4), 1225–1244. doi:10.1021/cr040409x
Kim, J. H., Tonelli, M., Frederick, R. O., Chow, D. C. F., and Markley, J. L. (2012a). Specialized Hsp70 chaperone (HscA) binds preferentially to the disordered form, whereas J-protein (HscB) binds preferentially to the structured form of the iron-sulfur cluster scaffold protein (IscU). J. Biol. Chem. 287 (37), 31406–31413. doi:10.1074/jbc.m112.352617
Kim, J. H., Tonelli, M., Kim, T., and Markley, J. L. (2012b). Three-dimensional structure and determinants of stability of the iron–sulfur cluster scaffold protein IscU from Escherichia coli. Biochemistry 51 (28), 5557–5563. doi:10.1021/bi300579p
Kim, J. H., Tonelli, M., and Markley, J. L. (2012c). Disordered form of the scaffold protein IscU is the substrate for iron-sulfur cluster assembly on cysteine desulfurase. Proc. Natl. Acad. Sci. U. S. A. 109 (2), 454–459. doi:10.1073/pnas.1114372109
Kim, R., Saxena, S., Gordon, D. M., Pain, D., and Dancis, A. (2001). J-domain protein, Jac1p, of yeast mitochondria required for iron homeostasis and activity of Fe-S cluster proteins. J. Biol. Chem. 276 (20), 17524–17532. doi:10.1074/jbc.m010695200
Kityk, R., Kopp, J., and Mayer, M. P. (2018). Molecular mechanism of J-domain-triggered ATP hydrolysis by Hsp70 chaperones. Mol. Cell 69 (2), 227–237. e224. doi:10.1016/j.molcel.2017.12.003
Kleczewska, M., Grabinska, A., Jelen, M., Stolarska, M., Schilke, B., Marszalek, J., et al. (2020). Biochemical convergence of mitochondrial Hsp70 system specialized in iron–sulfur cluster biogenesis. Int. J. Mol. Sci. 21 (9), 3326. doi:10.3390/ijms21093326
Knieszner, H., Schilke, B., Dutkiewicz, R., D'Silva, P., Cheng, S., Ohlson, M., et al. (2005). Compensation for a defective interaction of the hsp70 ssq1 with the mitochondrial Fe-S cluster scaffold isu. J. Biol. Chem. 280 (32), 28966–28972. doi:10.1074/jbc.m503031200
Knight, S. A., Sepuri, N. B., Pain, D., and Dancis, A. (1998). Mt-Hsp70 homolog, Ssc2p, required for maturation of yeast frataxin and mitochondrial iron homeostasis. J. Biol. Chem. 273 (29), 18389–18393. doi:10.1074/jbc.273.29.18389
Kominek, J., Marszalek, J., Neuvéglise, C., Craig, E. A., and Williams, B. L. (2013). The complex evolutionary dynamics of Hsp70s: A genomic and functional perspective. Genome Biol. Evol. 5 (12), 2460–2477. doi:10.1093/gbe/evt192
Kunichika, K., Nakamura, R., Fujishiro, T., and Takahashi, Y. (2021). The structure of the dimeric state of IscU harboring two adjacent [2Fe-2S] clusters provides mechanistic insights into cluster conversion to [4Fe-4S]. Biochemistry 60 (20), 1569–1572. doi:10.1021/acs.biochem.1c00112
Levin, K. B., Dym, O., Albeck, S., Magdassi, S., Keeble, A. H., Kleanthous, C., et al. (2009). Following evolutionary paths to protein-protein interactions with high affinity and selectivity. Nat. Struct. Mol. Biol. 16 (10), 1049–1055. doi:10.1038/nsmb.1670
Lin, C.-W., McCabe, J. W., Russell, D. H., and Barondeau, D. P. (2020). Molecular mechanism of ISC iron-sulfur cluster biogenesis revealed by high-resolution native mass spectrometry. J. Am. Chem. Soc. 142 (13), 6018–6029. doi:10.1021/jacs.9b11454
Liu, Q., Liang, C., and Zhou, L. (2020). Structural and functional analysis of the Hsp70/Hsp40 chaperone system. Protein Sci. 29 (2), 378–390. doi:10.1002/pro.3725
Maio, N., and Rouault, T. A. (2020). Outlining the complex pathway of mammalian Fe-S cluster biogenesis. Trends biochem. Sci. 45 (5), 411–426. doi:10.1016/j.tibs.2020.02.001
Majewska, J., Ciesielski, S. J., Schilke, B., Kominek, J., Blenska, A., Delewski, W., et al. (2013). Binding of the chaperone Jac1 protein and cysteine desulfurase Nfs1 to the iron-sulfur cluster scaffold Isu protein is mutually exclusive. J. Biol. Chem. 288 (40), 29134–29142. doi:10.1074/jbc.m113.503524
Manicki, M., Majewska, J., Ciesielski, S., Schilke, B., Blenska, A., Kominek, J., et al. (2014). Overlapping binding sites of the frataxin homologue assembly factor and the heat shock protein 70 transfer factor on the Isu iron-sulfur cluster scaffold protein. J. Biol. Chem. 289 (44), 30268–30278. doi:10.1074/jbc.m114.596726
Mayer, M. P., and Gierasch, L. M. (2019). Recent advances in the structural and mechanistic aspects of Hsp70 molecular chaperones. J. Biol. Chem. 294 (6), 2085–2097. doi:10.1074/jbc.rev118.002810
Mühlenhoff, U., Gerber, J., Richhardt, N., and Lill, R. (2003). Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p. EMBO J. 22 (18), 4815–4825. doi:10.1093/emboj/cdg446
Piette, B. L., Alerasool, N., Lin, Z.-Y., Lacoste, J., Lam, M. H. Y., Qian, W. W., et al. (2021). Comprehensive interactome profiling of the human Hsp70 network highlights functional differentiation of J domains. Mol. Cell. 81, 2549–2565. e8. doi:10.1016/j.molcel.2021.04.012
Pinti, M., Gibellini, L., Nasi, M., Biasi, S. D., Bortolotti, C. A., Iannone, A., et al. (2016). Emerging role of Lon protease as a master regulator of mitochondrial functions. Biochim. Biophys. Acta (BBA) - Bioenerg. 1857 (8), 1300–1306. doi:10.1016/j.bbabio.2016.03.025
Powers, E. T., and Balch, W. E. (2013). Diversity in the origins of proteostasis networks--a driver for protein function in evolution. Nat. Rev. Mol. Cell Biol. 14 (4), 237–248. doi:10.1038/nrm3542
Prischi, F., Konarev, P. V., Iannuzzi, C., Pastore, C., Adinolfi, S., Martin, S. R., et al. (2010). Structural bases for the interaction of frataxin with the central components of iron-sulphur cluster assembly. Nat. Comm. 1 (1), 95. doi:10.1038/ncomms1097
Pukszta, S., Schilke, B., Dutkiewicz, R., Kominek, J., Moczulska, K., Stepien, B., et al. (2010). Co-evolution-driven switch of J-protein specificity towards an Hsp70 partner. EMBO Rep. 11 (5), 360–365. doi:10.1038/embor.2010.29
Py, B., and Barras, F. (2015). Genetic approaches of the Fe-S cluster biogenesis process in bacteria: Historical account, methodological aspects and future challenges. Biochim. Biophys. Acta 1853 (6), 1429–1435. doi:10.1016/j.bbamcr.2014.12.024
Rebeaud, M. E., Mallik, S., Goloubinoff, P., and Tawfik, D. S. (2021). On the evolution of chaperones and cochaperones and the expansion of proteomes across the Tree of Life. Proc. Natl. Acad. Sci. U. S. A. 118 (21). doi:10.1073/pnas.2020885118
Roger, A. J., Muñoz-Gómez, S. A., and Kamikawa, R. (2017). The origin and diversification of mitochondria. Curr. Biol. CB 27 (21), R1177–R1192. doi:10.1016/j.cub.2017.09.015
Rosenzweig, R., Nillegoda, N. B., Mayer, M. P., and Bukau, B. (2019). The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 20 (11), 665–680. doi:10.1038/s41580-019-0133-3
Sahi, C., Lee, T., Inada, M., Pleiss, J. A., and Craig, E. A. (2010). Cwc23, an essential J protein critical for pre-mRNA splicing with a dispensable J domain. Mol. Cell. Biol. 30 (1), 33–42. doi:10.1128/mcb.00842-09
Schilke, B., Forster, J., Davis, J., James, P., Walter, W., Laloraya, S., et al. (1996). The cold sensitivity of a mutant of Saccharomyces cerevisiae lacking a mitochondrial heat shock protein 70 is suppressed by loss of mitochondrial DNA. J. Cell Biol. 134 (3), 603–613. doi:10.1083/jcb.134.3.603
Schilke, B., Voisine, C., Beinert, H., and Craig, E. (1999). Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 96 (18), 10206–10211. doi:10.1073/pnas.96.18.10206
Schilke, B., Williams, B., Knieszner, H., Pukszta, S., D'Silva, P., Craig, E. A., et al. (2006). Evolution of mitochondrial chaperones utilized in Fe-S cluster biogenesis. Curr. Biol. 16 (16), 1660–1665. doi:10.1016/j.cub.2006.06.069
Shi, R., Proteau, A., Villarroya, M., Moukadiri, I., Zhang, L., Trempe, J.-F., et al. (2010). Structural basis for Fe–S cluster Assembly and tRNA thiolation mediated by IscS protein–protein interactions. PLoS Biol. 8 (4), e1000354. doi:10.1371/journal.pbio.1000354.t004
Silberg, J. J., Hoff, K. G., and Vickery, L. E. (1998). The hsc66-hsc20 chaperone system in Escherichia coli: Chaperone activity and interactions with the DnaK-DnaJ-grpE system. J. Bacteriol. 180 (24), 6617–6624. doi:10.1128/jb.180.24.6617-6624.1998
Sloan, D. B., Warren, J. M., Williams, A. M., Wu, Z., Abdel-Ghany, S. E., Chicco, A. J., et al. (2018). Cytonuclear integration and co-evolution. Nat. Rev. Genet. 19 (10), 635–648. doi:10.1038/s41576-018-0035-9
Song, J.-Y., Marszalek, J., and Craig, E. A. (2012). Cysteine desulfurase Nfs1 and Pim1 protease control levels of Isu, the Fe-S cluster biogenesis scaffold. Proc. Natl. Acad. Sci. U. S. A. 109 (26), 10370–10375. doi:10.1073/pnas.1206945109
Srivastava, S., Vishwanathan, V., Birje, A., Sinha, D., and D'Silva, P. (2019). Evolving paradigms on the interplay of mitochondrial Hsp70 chaperone system in cell survival and senescence. Crit. Rev. Biochem. Mol. Biol. 54 (6), 517–536. doi:10.1080/10409238.2020.1718062
Srour, B., Gervason, S., Monfort, B., and D'Autréaux, B. (2020). Mechanism of iron–sulfur cluster Assembly: In the intimacy of iron and sulfur encounter. Inorganics 8 (10), 55–38. doi:10.3390/inorganics8100055
Szurmant, H., and Weigt, M. (2018). ScienceDirect inter-residue, inter-protein and inter-family coevolution: Bridging the scales. Curr. Opin. Struct. Biol. 50, 26–32. doi:10.1016/j.sbi.2017.10.014
Tanaka, N., Yuda, E., Fujishiro, T., Hirabayashi, K., Wada, K., and Takahashi, Y. (2019). Identification of IscU residues critical for de novo iron-sulfur cluster assembly. Mol. Microbiol. 112 (6), 1769–1783. doi:10.1111/mmi.14392
Tapley, T. L., Cupp-Vickery, J. R., and Vickery, L. E. (2005). Sequence-dependent peptide binding orientation by the molecular chaperone DnaK. Biochemistry 44 (37), 12307–12315. doi:10.1021/bi051145r
Tapley, T. L., Cupp-Vickery, J. R., and Vickery, L. E. (2006). Structural determinants of HscA peptide-binding specificity. Biochemistry 45 (26), 8058–8066. doi:10.1021/bi0606187
Tomiczek, B., Delewski, W., Nierzwicki, L., Stolarska, M., Grochowina, I., Schilke, B., et al. (2020). Two-step mechanism of J-domain action in driving Hsp70 function. PLOS Comp. Biol. 16 (6), e1007913. doi:10.1371/journal.pcbi.1007913
Vickery, L. E., and Cupp-Vickery, J. R. (2007). Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in iron-sulfur protein maturation. Crit. Rev. Biochem. Mol. Biol. 42 (2), 95–111. doi:10.1080/10409230701322298
Voisine, C., Cheng, Y. C., Ohlson, M., Schilke, B., Hoff, K., Beinert, H., et al. (2001). Jac1, a mitochondrial J-type chaperone, is involved in the biogenesis of Fe/S clusters in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 98 (4), 1483–1488. doi:10.1073/pnas.98.4.1483
Wang, W., Liu, Q., Liu, Q., and Hendrickson, W. A. (2021). Conformational equilibria in allosteric control of Hsp70 chaperones. Mol. Cell 81 (19), 3919–3933. e7. doi:10.1016/j.molcel.2021.07.039
Zhang, R., Malinverni, D., Cyr, D. M., Rios, P. D. L., and Nillegoda, N. B. (2022). J-domain protein chaperone circuits in proteostasis and disease. Trends Cell Biol. 1, 1. doi:10.1016/j.tcb.2022.05.004
Zhang, Y., Sinning, I., and Rospert, S. (2017). Two chaperones locked in an embrace: Structure and function of the ribosome-associated complex RAC. Nat. Struct. Mol. Biol. 24 (8), 611–619. doi:10.1038/nsmb.3435
Keywords: molecular chaperones, FeS cluster biogenesis, Isu/IscU scaffold, Hsp70-client interaction cycle, protein interactions, protein evolution, post-duplication functional divergence
Citation: Marszalek J and Craig EA (2022) Interaction of client—the scaffold on which FeS clusters are build—with J-domain protein Hsc20 and its evolving Hsp70 partners. Front. Mol. Biosci. 9:1034453. doi: 10.3389/fmolb.2022.1034453
Received: 01 September 2022; Accepted: 26 September 2022;
Published: 12 October 2022.
Edited by:
Abdussalam Azem, Tel Aviv University, IsraelReviewed by:
Johannes M. Herrmann, University of Kaiserslautern, GermanyRina Rosenzweig, Weizmann Institute of Science, Israel
Copyright © 2022 Marszalek and Craig. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jaroslaw Marszalek, amFyb3NsYXcubWFyc3phbGVrQHVnLmVkdS5wbA==; Elizabeth A. Craig, ZWNyYWlnQHdpc2MuZWR1
†These authors have contributed equally to this work and share senior authorship
 Jaroslaw Marszalek
Jaroslaw Marszalek Elizabeth A. Craig
Elizabeth A. Craig