- 1Institute of Pharmaceutical Science, King’s College London, London, United Kingdom
- 2Institute for Pharmaceutical and Medicinal Chemistry, Heinrich-Heine-University Düsseldorf, Duesseldorf, Germany
- 3Department of Biotechnology, Delft University of Technology, Delft, Netherlands
- 4Magnetic Resonance Center (CERM), University of Florence and Consorzio Interuniversitario Risonanze Magnetiche di Metalloproteine (CIRMMP), Florence, and Department of Chemistry, University of Florence, Florence, Italy
- 5Sir William Dunn School of Pathology, University of Oxford, Oxford, United Kingdom
- 6Instituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa, Av. da República–EAN, Oeiras, Portugal
- 7Institute of Chemistry, Technische Universität Berlin, Berlin, Germany
- 8Université Grenoble Alpes, Grenoble, France
- 9Department of Chemistry, Molecular Sciences Research Hub, Imperial College London, London, United Kingdom
- 10Centre for Molecular and Structural Biochemistry, School of Chemistry, University of East Anglia, Norwich, United Kingdom
Main text
In 1789, the influential French chemist Antoine-Laurent Lavoisier described his view of science and its langague in his book Traité élémentaire de chimie. According to the Robert Kerr’s translation it states (Lavoisier, 1790): “As ideas are preserved and communicated by means of words, it necessarily follows that we cannot improve the language of any science without at the same time improving the science itself; neither can we, on the other hand, improve a science without improving the language or nomenclature which belongs to it.” This view reminds us of Confucius’s earlier doctrine, the rectification of names (Steinkraus, 1980; Lau, 2000). Confucius believed that rectification of names is imperative. He explained (Steinkraus, 1980; Lau, 2000): “If language is incorrect, then what is said does not concord with what was meant, what is to be done cannot be affected. If what is to be done cannot be affected, then rites and music will not flourish. If rites and music do not flourish, then mutilations and lesser punishments will go astray. And if mutilations and lesser punishments go astray, then the people have nowhere to put hand or foot. Therefore the gentleman uses only such language as is proper for speech, and only speaks of what it would be proper to carry into effect. The gentleman in what he says leaves nothing to mere chance.” Inspired by these views, we make the analogy that the progress of science and the language used to describe it are two entangled electrons. This entanglement highlights the importance of introducing systemic names for enzymes using EC classification and the ever-growing problem of protein names (McDonald and Tipton, 2021). Here, we tackle one specific case of iron-sulfur ([FeS]) enzymes. We show that the language used to describe a conserved [FeS] enzyme of the innate immune system, i.e., viperin or RSAD2, is now inadequate and disentangled from its science. We discuss that the enzyme has cellular functions beyond its antiviral activity and that eukaryotic and prokaryotic enzymes catalyse the same chemical reactions. To prevent bias towards antiviral activity while studying various biochemical activities of the enzyme and using scientifically incorrect terms like “prokaryotic viperins,” we rectify the language describing the enzyme. Based on NC-IUBMB recommendations, we introduce the nomenclature S-adenosylmethionine (SAM) dependent Nucleotide Dehydratase (SAND).
Firstly, considering the progress in understanding the biology of the enzyme in humans (Figure 1), the name “viperin” is no longer adequate and should be avoided. In 1997, Hua Z., et al. found that in response to human cytomegalovirus infection, the mRNA level of a novel protein was elevated in human cells (Zhu et al., 1997). The gene related to this mRNA was named cytomegalovirus-induced human gene-5 (cig-5). In 2001, Chin and Cresswell showed that interferons (IFNs) induce the expression of the protein product of cig-5 (Chin and Cresswell, 2001). This induction restricted the replication of human cytomegalovirus, and the protein was localised to the cytoplasmic face of the endoplasmic reticulum (ER) (Chin and Cresswell, 2001). Because, at the time, nothing was known about the chemistry of the enzyme, an abbreviation based on the cellular localisation and antiviral activity was introduced, “viperin” (virus inhibitory protein, endoplasmic reticulum-associated, interferon-inducible) (Chin and Cresswell, 2001). Subsequent studies showed that the expression of the protein affects the life-cycle of many RNA and DNA viruses, including Influenza (Wang et al., 2007), HIV-1 (Nasr et al., 2012), Hepatitis C (Wang et al., 2012; Ghosh et al., 2020), Zika (Van der Hoek et al., 2017; Panayiotou et al., 2018), and tick-borne encephalitis (Panayiotou et al., 2018), among others. However, for each virus, different mechanisms were proposed (Figure 1A). For example, the enzyme affects lipid rafts (lipid microdomains on the cellular membrane and enriched in cholesterol and sphingolipids (Ripa et al., 2021)) and inhibits influenza virus (Wang et al., 2007) or HIV-1 (Nasr et al., 2012) release. In the case of the Hepatitis C virus, viperin expression appears to interfere with the binding of the viral nonstructural protein NS5A with host hVAP22 (Wang et al., 2012) or promotes proteasomal-dependent degradation of viral NS5A (Ghosh et al., 2020). In the case of Zika and tick-borne encephalitis viruses, the enzyme appears to induce proteasomal degradation of the viral nonstructural protein NS3 (Panayiotou et al., 2018). Finally, in the case of tick-borne encephalitis virus and Dengue virus type-2, the enzyme restricts viral RNA reproduction (Helbig et al., 2013; Upadhyay et al., 2014).
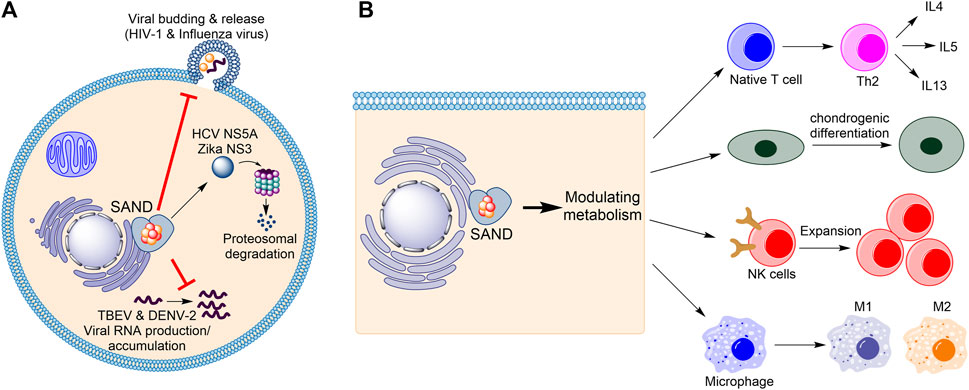
FIGURE 1. Human SAND has functions beyond its antiviral activity. (A) Various mechanisms of antiviral activity are proposed for human SAND. (B) Expression of SAND affects the function and differentiation of various types of cells. HCV, hepatitis C virus; TBEV, tick-born encephalitis virus; DENV-2, Dengue type-2 virus; NK, natural killer; IL, interleukin.
Despite the established antiviral activity, many studies have reported biological functions inconsistent with or unrelated to the biology defined by the nomenclature “viperin”. Cresswell and colleagues showed that the enzyme localises to lipid droplets (Hinson and Cresswell, 2009) and enhances human cytomegalovirus infection (Seo et al., 2011). In addition to interferons, lipopolysaccharides were found to induce protein expression (Olofsson et al., 2005). The proposal of multiple mechanisms of antiviral activity prompted us to postulate that the protein’s enzymatic activity regulates metabolism to affect various cellular processes causing broad-spectrum antiviral activity (Ebrahimi, 2018) (Figure 1B). This effect of the enzyme on metabolism suggests a cellular function beyond its antiviral activity. Indeed, many studies corroborate this proposal and demonstrate that human SAND has a role in modulating metabolism, regulating the activity/maturation of the immune cells, and inducing the expression of immune genes (Figure 1B). For example, the activity of SAND modulates central carbon metabolism (Ebrahimi et al., 2020c), regulates thermogenesis in adipose tissues (Eom et al., 2019), inhibits thiolase activity of the trifunctional enzyme complex (a mitochondrial enzyme complex with three activities: enoyl-CoA hydratase, 3-hydroxyacyl-CoA dehydrogenase, and 3-ketoacyl-CoA thiolase) (Dumbrepatil et al., 2020), and modulates cholesterol metabolism (Tang et al., 2016; Grunkemeyer et al., 2021). It is required for optimal T helper two cell response (Qiu et al., 2009) and chondrogenic differentiation via CXCL10 protein secretion (Steinbusch et al., 2019). It has a role in the innate system (Ebrahimi et al., 2022) and modules immune cell function and maturation e.g., expansion of natural killer cells (Wiedemann et al., 2020), dendritic cell maturation (Jang et al., 2018), B cell hyperactivity (Zhu et al., 2021), and polarisation of macrophages (Eom et al., 2018). Additionally, the enzyme’s expression induces the expression of many immune genes (Zhang et al., 2014).
Secondly, the nomenclature RSAD2 should be revised to fully describe the chemistry of the enzyme relevant to its biological function. By 2010, it became clear that human SAND has a CxxxCxxC motif coordinating a [4Fe-4S] cluster, similar to many members of the radical S-adenosylmethionine (SAM) enzymes (Duschene and Broderick, 2010; Shaveta et al., 2010). As a result, the HUGO Gene Nomenclature Committee suggested the name RSAD2 (radical-SAM domain containing 2) around this time. This name can be easily confused with another radical-SAM enzyme of unknown function (RSAD1), and it only partially describes the SAM-dependent chemistry of the enzyme. In 2017, the structure of mouse SAND was solved (Fenwick et al., 2017), confirming that it is a radical-SAM enzyme. It was shown that the cytosolic iron-sulfur biogenesis machinery is required to deliver and insert the [4Fe-4S] cluster into the enzyme (Upadhyay et al., 2017). The expression of human SAND in E. coli changed the cells’ morphology, suggesting the enzyme’s substrate is a metabolite common between eukaryotic and prokaryotic cells (Nelp et al., 2017), and initial structural studies proposed that the substrate is a nucleotide (Fenwick et al., 2017). Subsequently, it was revealed that eukaryotic SAND could catalyse the dehydration of CTP or UTP to 3ʹ-deoxy-3ʹ, 4ʹ-didehydro (ddh) analogues (Figure 2A) (Fenwick et al., 2020). In human macrophages, the enzyme was found to produce ddhCTP (Gizzi et al., 2018; Ebrahimi et al., 2020b). This novel nucleotide analogue metabolite may act as a chain-terminator to inhibit viral replication (IC50 values
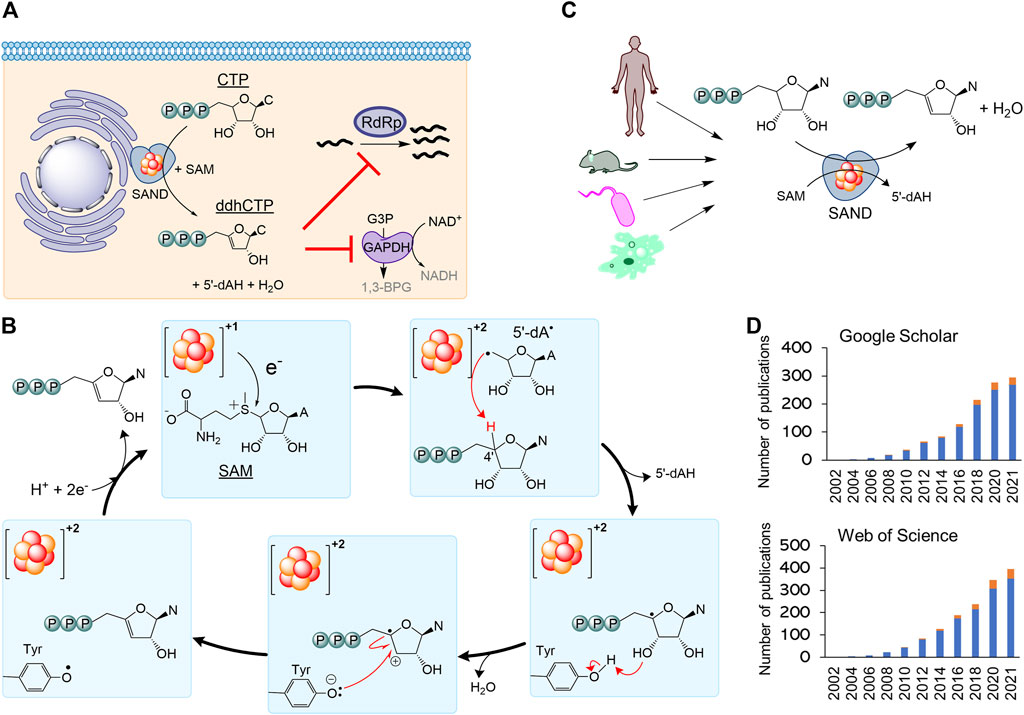
FIGURE 2. The nomenclature SAND (SAM-dependent Nucleotide Dehydratase) defines chemistry relevant to biology across all domains of life. (A) SAND produces the nucleoside triphosphate analogue ddhCTP in humans. ddhCTP modulates metabolism affecting cell function and restricting viral replication. (B) The proposed mechanism of dehydration of nucleoside triphosphates by SAND. The mechanism shows the transfer of a proton and an electron from a conserved tyrosine. Alternatively, it is possible that proton transfer occurs via another amino acid residue. It is not clear if the transfer of proton and electron occurs simultaneously (proton-coupled electron transfer). (C) SANDs from various organisms produce diverse ddh analogues. (D) An increasing number of investigators study SANDs. The data were obtained from a search of nomenclature viperin and RSAD2 in the title of articles. Google Scholar (scholar.google.com) and Web of Science search engines were used. N, nucleobase; C, cytosine; A, adenine; 5′-dAH, 5′-deoxyadenosine, 5′-dA·, 5′-deoxyadenosyl radical.
Finally, the use of the outdated nomenclature “viperin” can introduce scientifically incorrect terms such as “prokaryotic viperin.” Before 2017 little was done to isolate fungal and microbial SANDs and characterise the chemical reaction catalysed by them. In 2017, a thermostable fungal SAND from Thielavia terrestris was isolated and characterised (Ebrahimi et al., 2017). It was hypothesised that the fungal enzyme produces antiviral natural products and is a suitable candidate for the biotechnological production of antiviral lead molecules. The fungal SAND has promiscuous activity and catalyses the dehydration of diverse nucleoside triphosphates (NTPs), e.g., CTP, UTP, and 5-bromo-UTP, to their ddh analogues via a mechanism requiring the transfer of an electron and a proton (Figure 2B) (Ebrahimi et al., 2020b). Next, a number of other groups characterised some microbial enzymes and showed that they catalyse dehydration of various NTPs to their ddh analogues (Bernheim et al., 2021; Lachowicz et al., 2021) (Figure 2C). While the cellular function of these microbial proteins is not fully understood, the chemical reaction catalysed by SANDs can inhibit the activity of phage T7 RNA polymerase in E. coli (Bernheim et al., 2021). These findings suggest that the enzyme might have a cellular function and act as an antimicrobial/antiviral defence system. The fungal enzyme was named TtRSAD2 (Ebrahimi et al., 2020b) due to the lack of a proper name, and studies with bacterial enzymes (Bernheim et al., 2021) introduced a new nomenclature, i.e., “prokaryotic viperin,” to describe prokaryotic enzymes producing ddh analogues with antiviral activity (Bernheim et al., 2021; Wein and Sorek, 2022). The term “prokaryotic viperin” is not fit for purpose because it implies that bacteria and archaea have endoplasmic reticulum, and interferons activate their immune system. This assertion questions our fundamental understanding of biology, i.e., prokaryotes do not have an endoplasmic reticulum and interferon-mediated antiviral response.
A growing number of investigators are studying this new class of enzymes across all domains of life (Figure 2D). Consequently, different nomenclatures like RSAD2, viperin, prokaryotic viperin, or viperin-like enzymes are being used by various investigators, including us, to describe eukaryotic or microbial enzymes. As discussed above, none of the existing nomenclatures accurately describe the cellular function or chemistry in prokaryotes or eukaryotes. Additionally, using various terminologies for enzymes performing the same chemical reaction is confusing. Hence, we strongly suggest the classification of the enzyme as a nucleoside triphosphate dehydratase (NTPD, EC 4.2.1) and the nomenclature SAND describing the SAM-dependent chemistry across all domains of life. This classification and abbreviation to rectify the naming of an ancient iron-sulfur enzyme should help the increasing number of investigators studying the cellular function or biotechnological application of these enzymes and the discovery of new enzymes performing novel chemistries.
Author contributions
KE conceived the idea and wrote the manuscript together with all the other authors.
Acknowledgments
All authors acknowledge the support from the European Cooperation in Science and Technology (COST) Action CA21115.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.1032220/full#supplementary-material
References
Bernheim, A., Millman, A., Ofir, G., Meitav, G., Avraham, C., Shomar, H., et al. (2021). Prokaryotic viperins produce diverse antiviral molecules. Nature 589, 120–124. doi:10.1038/s41586-020-2762-2
Chin, K.-C., and Cresswell, P. (2001). Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 98, 15125–15130. doi:10.1073/pnas.011593298
Dumbrepatil, A. B., Zegalia, K. A., Sajja, K., Kennedy, R. T., and Marsh, E. N. G. (2020). Targeting viperin to the mitochondrion inhibits the thiolase activity of the trifunctional enzyme complex. J. Biol. Chem. 295, 2839–2849. doi:10.1074/jbc.RA119.011526
Duschene, K. S., and Broderick, J. B. (2010). The antiviral protein viperin is a radical SAM enzyme. FEBS Lett. 584, 1263–1267. doi:10.1016/j.febslet.2010.02.041
Ebrahimi, K. H. (2018). A unifying view of the broad-spectrum antiviral activity of RSAD2 (viperin) based on its radical-SAM chemistry. Metallomics 10, 539–552. doi:10.1039/C7MT00341B
Ebrahimi, K. H., Carr, S. B., McCullagh, J., Wickens, J., Rees, N. H., Cantley, J., et al. (2017). The radical‐SAM enzyme Viperin scatalyses reductive addition of a 5′‐deoxyadenosyl radical to UDP‐glucose in vitro. FEBS Lett. 591, 2394–2405.
Ebrahimi, K. H., Diofi, S., Hagedoorn, P. L., Nicolet, Y., Le Brun, N. E., Hagen, W. R., et al. (2022). Iron-sulphur clusters as inhibitors and catalysts of viral replication. Nat. Chem. 14, 253–266.
Ebrahimi, K. H., Gilbert-Jaramillo, J., James, W. S., and McCullagh, J. S. O. (2021). Interferon-stimulated gene products as regulators of central carbon metabolism. FEBS J. 288, 3715–3726. doi:10.1111/febs.15625
Ebrahimi, K. H., Howie, D., Rowbotham, J., McCullagh, J., Armstrong, F., and James, W. S. (2020a). Viperin, through its radical-SAM activity, depletes cellular nucleotide pools and interferes with mitochondrial metabolism to inhibit viral replication. FEBS Lett. 594, 1624–1630. doi:10.1002/1873-3468.13761
Ebrahimi, K. H., Rowbotham, J., McCullagh, J., and James, W. S. (2020b). Mechanism of diol dehydration by a promiscuous radical‐SAM enzyme homologue of the antiviral enzyme viperin (RSAD2). ChemBioChem 21, 1605–1612. doi:10.1002/cbic.201900776
Ebrahimi, K. H., Vowles, J., Browne, C., McCullagh, J., and James, W. S. (2020c). ddhCTP produced by the radical‐SAM activity of RSAD2 (viperin) inhibits the NAD+‐dependent activity of enzymes to modulate metabolism. FEBS Lett. 594, 1631–1644. doi:10.1002/1873-3468.13778
Eom, J., Kim, J. J., Yoon, S. G., Jeong, H., Son, S., Lee, J. B., et al. (2019). Intrinsic expression of viperin regulates thermogenesis in adipose tissues. Proc. Natl. Acad. Sci. U. S. A. 116, 17419–17428. doi:10.1073/pnas.1904480116
Eom, J., Yoo, J., Kim, J. J., Lee, J. B., Choi, W., Park, C. G., et al. (2018). Viperin deficiency promotes polarization of macrophages and secretion of M1 and M2 cytokines. Immune Netw. 18, e32. doi:10.4110/in.2018.18.e32
Fenwick, M. K., Li, Y., Cresswell, P., Modis, Y., and Ealick, S. E. (2017). Structural studies of viperin, an antiviral radical SAM enzyme. Proc. Natl. Acad. Sci. U. S. A. 114, 6806–6811. doi:10.1073/pnas.1705402114
Fenwick, M. K., Su, D., Dong, M., Lin, H., and Ealick, S. E. (2020). Structural basis of the substrate selectivity of viperin. Biochemistry 59, 652–662. doi:10.1021/acs.biochem.9b00741
Ghosh, S., Patel, A. M., Grunkemeyer, T. J., Dumbrepatil, A. B., Zegalia, K. A., Kennedy, R. T., et al. (2020). Interactions between viperin, vesicle-associated membrane protein A and Hepatitis C virus protein NS5A modulate viperin activity and NS5A degradation. Biochemistry 59, 780–789. doi:10.1021/acs.biochem.9b01090
Gizzi, A. S., Grove, T. L., Arnold, J. J., Jose, J., Jangra, R. K., Garforth, S. J., et al. (2018). A naturally occurring antiviral ribonucleotide encoded by the human genome. Nature 558, 610–614. doi:10.1038/s41586-018-0238-4
Grunkemeyer, T. J., Ghosh, S., Patel, A. M., Sajja, K., Windak, J., Basrur, V., et al. (2021). The antiviral enzyme viperin inhibits cholesterol biosynthesis. J. Biol. Chem. 297, 100824. doi:10.1016/j.jbc.2021.100824
Helbig, K. J., Carr, J. M., Calvert, J. K., Wati, S., Clarke, J. N., Nicholase, S. E., et al. (2013). Viperin is induced following Dengue virus type-2 (DENV-2) infection and has anti-viral actions requiring the C-terminal end of viperin. PLoS Negl. Trop. Dis. 7, e2178. doi:10.1371/journal.pntd.0002178
Hinson, E. R., and Cresswell, P. (2009). The antiviral protein, viperin, localizes to lipid droplets via its N-terminal amphipathic alpha-helix. Proc. Natl. Acad. Sci. U. S. A. 106, 20452–20457. doi:10.1073/pnas.0911679106
Jang, J.-S., Lee, J.-H., Jung, N.-C., Choi, S.-Y., Park, S.-Y., Yoo, J.-Y., et al. (2018). Rsad2 is necessary for mouse dendritic cell maturation via the IRF7-mediated signaling pathway. Cell. Death Dis. 9, 823. doi:10.1038/s41419-018-0889-y
Lachowicz, J. C., Gizzi, A. S., Almo, S. C., and Grove, T. L. (2021). Structural insight into the substrate scope of viperin and viperin-like enzymes from three domains of life. Biochemistry 60, 2116–2129. doi:10.1021/acs.biochem.0c00958
McDonald, A. G., and Tipton, K. F. (2021). Enzyme nomenclature and classification: The state of the art. FEBS J. doi:10.1111/febs.16274
Nasr, N., Maddocks, S., Turville, S. G., Harman, A. N., Woolger, N., Helbig, K. J., et al. (2012). HIV-1 infection of human macrophages directly induces viperin which inhibits viral production. Blood `20, 778–788. doi:10.1182/blood-2012-01-407395
Nelp, M. T., Young, A. P., Stepanski, B. M., and Bandarian, V. (2017). Human viperin causes radical SAM-dependent elongation of Escherichia coli, hinting at its physiological role. Biochemistry 56, 3874–3876. doi:10.1021/acs.biochem.7b00608
Olofsson, P. S., Jatta, K., Wagsater, D., Gredmark, S., Hedin, U., Paulsson-Berne, G., et al. (2005). The antiviral cytomegalovirus inducible gene 5/viperin is expressed in atherosclerosis and regulated by proinflammatory agents. Arterioscler. Thromb. Vasc. Biol. 25, e113–e116. doi:10.1161/01.ATV.0000170130.85334.38
Panayiotou, C., Lindqvist, R., Kurhade, C., Vonderstein, K., Pasto, J., Edlund, K., et al. (2018). Viperin restricts Zika virus and tick-borne encephalitis virus replication by targeting NS3 for proteasomal degradation. J. Virol. 92, 020544–e2117. doi:10.1128/JVI.02054-17
Qiu, L.-Q., Cresswell, P., and Chin, K.-C. (2009). Viperin is required for optimal Th2 responses and T-cell receptor-mediated activation of NF-kappaB and AP-1. Blood 113, 3520–3529. doi:10.1182/blood-2008-07-171942
Ripa, I., Andreu, S., López-Guerrero, J. A., and Bello-Morales, R. (2021). Membrane rafts: Portals for viral entry. Front. Microbiol. 12, 631274. doi:10.3389/fmicb.2021.631274
Seo, J.-Y., Yaneva, R., Hinson, E. R., and Cresswell, P. (2011). Human cytomegalovirus directly induces the antiviral protein viperin to enhance infectivity. Sci. (80-. ) 332, 1093–1097. doi:10.1126/science.1202007
Shaveta, G., Shi, J., Chow, V. T. K., and Song, J. (2010). Structural characterization reveals that viperin is a radical S-adenosyl-L-methionine (SAM) enzyme. Biochem. Biophys. Res. Commun. 391, 1390–1395. doi:10.1016/j.bbrc.2009.12.070
Steinbusch, M. M. F., Caron, M. M. J., Surtel, D. A. M., van den Akker, G. G. H., van Dijk, P. J., Friedrich, F., et al. (2019). The antiviral protein viperin regulates chondrogenic differentiation via CXCL10 protein secretion. J. Biol. Chem. 294, 5121–5136. doi:10.1074/jbc.RA119.007356
Steinkraus, W. E. (1980). Socrates, Confucius, and the rectification of names. Philos. East West 30, 261–264. doi:10.2307/1398850
Tang, H.-B., Lu, Z.-L., Wei, X.-K., Zhong, T.-Z., Ouyang, L.-X., Luo, Y., et al. (2016). Viperin inhibits rabies virus replication via reduced cholesterol and sphingomyelin and is regulated upstream by TLR4. Sci. Rep. 6, 30529. doi:10.1038/srep30529
Upadhyay, A. S., Stehling, O., Panayiotou, C., Rosser, R., Lill, R., and Overby, A. K. (2017). Cellular requirements for iron-sulfur cluster insertion into the antiviral radical SAM protein viperin. J. Biol. Chem. 292, 13879–13889. doi:10.1074/jbc.M117.780122
Upadhyay, A. S., Vonderstein, K., Pichlmair, A., Stehling, O., Bennett, K. I., Dobler, G., et al. (2014). Viperin is an iron–sulfur protein that inhibits genomesynthesis of tick-borne encephalitis virus via radical SAM domain activity. Cell. Microbiol. 16, 834–848. doi:10.1111/cmi.12241
Van der Hoek, K. H., Eyre, N. S., Shue, B., Khantisitthiporn, O., Glab-Ampi, K., Carr, J. M., et al. (2017). Viperin is an important host restriction factor in control of Zika virus infection. Sci. Rep. 7, 4475. doi:10.1038/s41598-017-04138-1
Wang, S., Wu, X., Pan, T., Song, W., Wang, Y., Zhang, F., et al. (2012). Viperin inhibits hepatitis C virus replication by interfering with binding of NS5A to host protein hVAP-33. J. Gen. Virol. 93, 83–92. doi:10.1099/vir.0.033860-0
Wang, X., Hinson, E. R., and Cresswell, P. (2007). The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell. Host Microbe 2, 96–105. doi:10.1016/j.chom.2007.06.009
Wein, T., and Sorek, R. (2022). Bacterial origins of human cell-autonomous innate immune mechanisms. Nat. Rev. Immunol. 22, 629–638. doi:10.1038/s41577-022-00705-4
Wiedemann, G. M., Geary, C. D., Lau, C. M., and Sun, J. C. (2020). Cutting edge: STAT1-Mediated epigenetic control of Rsad2 promotes clonal expansion of antiviral NK cells. J. Immunol. 205, 21–25. doi:10.4049/jimmunol.2000086
Zhang, B., Zhang, J., Xiao, Z. Z., and Sun, L. (2014). Rock bream (Oplegnathus fasciatus) viperin is a virus-responsive protein that modulates innate immunity and promotes resistance against megalocytivirus infection. Dev. Comp. Immunol. 45, 35–42. doi:10.1016/j.dci.2014.02.001
Zhu, H., Cong, J.-P., and Shenk, T. (1997). Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: Induction of interferon-responsive RNAs. Proc. Natl. Acad. Sci. U. S. A. 94, 13985–13990. doi:10.1073/pnas.94.25.13985
Keywords: antiviral, innate immun system, dehydratase, nucleotide analogue, iron-sulfur [FeS] cluster
Citation: Ji Y, Wei L, Da A, Stark H, Hagedoorn P-L, Ciofi-Baffoni S, Cowley SA, Louro RO, Todorovic S, Mroginski MA, Nicolet Y, Roessler MM, Le Brun NE, Piccioli M, James WS, Hagen WR and Ebrahimi KH (2022) Radical-SAM dependent nucleotide dehydratase (SAND), rectification of the names of an ancient iron-sulfur enzyme using NC-IUBMB recommendations. Front. Mol. Biosci. 9:1032220. doi: 10.3389/fmolb.2022.1032220
Received: 30 August 2022; Accepted: 07 October 2022;
Published: 21 October 2022.
Edited by:
Marie-Pierre Golinelli, UPR2301 Institut de Chimie des Substances Naturelles (ICSN CNRS), FranceReviewed by:
Teresita Padilla-Benavides, Wesleyan University, United StatesSoumi Ghosh, Massachusetts Institute of Technology, United States
Copyright © 2022 Ji, Wei, Da, Stark, Hagedoorn, Ciofi-Baffoni, Cowley, Louro, Todorovic, Mroginski, Nicolet, Roessler, Le Brun, Piccioli, James, Hagen and Ebrahimi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kourosh H. Ebrahimi, S291cm9zaC5lYnJhaGltaUBrY2wuYWMudWs=
 Yuxuan Ji1
Yuxuan Ji1 Holger Stark
Holger Stark Peter-Leon Hagedoorn
Peter-Leon Hagedoorn Simone Ciofi-Baffoni
Simone Ciofi-Baffoni Sally A. Cowley
Sally A. Cowley Ricardo O. Louro
Ricardo O. Louro Maria Andrea Mroginski
Maria Andrea Mroginski Yvain Nicolet
Yvain Nicolet Maxie M. Roessler
Maxie M. Roessler Nick E. Le Brun
Nick E. Le Brun Mario Piccioli
Mario Piccioli William S. James
William S. James Kourosh H. Ebrahimi
Kourosh H. Ebrahimi