
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci., 30 September 2022
Sec. Molecular Diagnostics and Therapeutics
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.1024764
This article is part of the Research TopicCancer Stem Cells: Hidden Modulators of Tumor Immune ResponseView all 5 articles
 Sibhghatulla Shaikh1,2†
Sibhghatulla Shaikh1,2† Shahid Ali1,2†
Shahid Ali1,2† Jeong Ho Lim1,2
Jeong Ho Lim1,2 Hee Jin Chun1
Hee Jin Chun1 Khurshid Ahmad1,2
Khurshid Ahmad1,2 Syed Sayeed Ahmad1,2
Syed Sayeed Ahmad1,2 Ye Chan Hwang1
Ye Chan Hwang1 Ki Soo Han3
Ki Soo Han3 Na Ri Kim3
Na Ri Kim3 Eun Ju Lee1,2*
Eun Ju Lee1,2* Inho Choi1,2*
Inho Choi1,2*Type 2 diabetes mellitus (T2DM) is a growing global public health issue, and dipeptidyl peptidase-4 (DPP-4) is a potential therapeutic target in T2DM. Several synthetic anti-DPP-4 medications can be used to treat T2DM. However, because of adverse effects, there is an unmet demand for the development of safe and effective medications. Natural medicines are receiving greater interest due to the inherent safety of natural compounds. Glycyrrhiza uralensis (licorice) is widely consumed and used as medicine. In this study, we investigated the abilities of a crude water extract (CWE) of G. uralensis and two of its constituents (licochalcone A (LicA) and licochalcone B (LicB)) to inhibit the enzymatic activity of DPP-4 in silico and in vitro. In silico studies showed that LicA and LicB bind tightly to the catalytic site of DPP-4 and have 11 amino acid residue interactions in common with the control inhibitor sitagliptin. Protein-protein interactions studies of LicA-DPP4 and LicB-DPP4 complexes with GLP1 and GIP reduced the DPP-4 to GLP1 and GIP interactions, indicated that these constituents might reduce the degradations of GLP1 and GIP. In addition, molecular dynamics simulations revealed that LicA and LicB stably bound to DPP-4 enzyme. Furthermore, DPP-4 enzyme assay showed the CWE of G. uralensis, LicA, and LicB concentration-dependently inhibited DPP-4; LicA and LicB had an estimated IC50 values of 347.93 and 797.84 μM, respectively. LicA and LicB inhibited DPP-4 at high concentrations, suggesting that these compounds could be used as functional food ingredients to manage T2DM.
The prevalence of diabetes continues to increase rapidly worldwide. Some 463 million people were affected by the disease in 2019, which is expected to increase to 578 million by 2030 and 700 million by 2045 (Saeedi et al., 2019). Moreover, diabetes has been reported to be responsible for around 10% of all fatalities (Reed et al., 2021). Type 2 diabetes mellitus (T2DM) is the most common form of the disease and accounts for ∼90% of cases. T2DM is characterized by inadequate pancreatic insulin production and insulin resistance in peripheral tissues (Chatterjee et al., 2017), and diabetes-related chronic hyperglycemia can result in long-term damage to and the failure of vital organs, such as eyes, kidneys, nerves, and heart (Dodds, 2017; Park et al., 2019).
The term “incretin” refers to a group of hormones that include glucagon-like peptide-1 (GLP-1) and glucose-dependent insulinotropic polypeptide (GIP). When a meal is ingested, these hormones are produced in gut and stimulate insulin production by acting on pancreatic β cells (Nauck and Meier, 2018). Dipeptidyl peptidase-4 (DPP-4) degrades circulating GLP-1 and GIP with a half-live of around a minute, and thus, DPP-4 inhibitors enhance active GLP-1 and GIP levels (Pathak and Bridgeman, 2010). Therefore, DPP-4 inhibitors are considered a novel means of treating T2DM. However, currently available DPP-4 inhibitors have several adverse effects such as arthritis, pancreatitis, diarrhea, and congestive heart failure that limit their practical applications (Mascolo et al., 2016; Packer, 2018; Padron et al., 2020), and thus, new DPP-4 inhibitors are needed. Computational techniques had shown to be effective in finding novel drugs and their development in the therapeutic development process (Katsila et al., 2016; Ali et al., 2022).
Glycyrrhiza uralensis is a well-known medicinal plant from the Leguminosae family and has been reported to contain various biologically active compounds, which include various triterpene saponins, flavonoids, and licochalcones (Asl and Hosseinzadeh, 2008; Zhang and Ye, 2009; Lee et al., 2021). However, although G. uralensis and its biologically active components have been shown to have a variety of pharmacological actions, their effects on DPP-4 inhibition have not been explored. Here, we investigated the ability of the crude water extract (CWE) of G. uralensis and two of its bioactive components, namely, licochalcone A (LicA) and licochalcone B (LicB), to inhibit DPP-4 using computational docking simulation analyses and an in vitro DPP-4 enzyme assay.
The crystal structure of DPP-4 (PDB ID: 4FFW) was obtained from the Protein Databank and the 3-D structures of LicA (CID: 5318998) and LicB (CID: 5318999) were retrieved from the PubChem database.
AutoDock 4.2 was used to dock ligands to DPP-4, and the MMFF94 force field was used to energy minimization of the compounds. Affinity (grid) maps sized 40 × 40 × 40 were generated using an auto grid tool to target grid coordinates in the DPP-4 catalytic site. The x, y, and z values were set at 34.36, 48.98, and 40.19, respectively. The initial positions, orientations, and torsions of ligands were determined randomly. Each docking experiment consisted of 100 separate runs, and each run was programmed to end after a maximum of 2,500,000 energy evaluations. Final figures were produced using Discovery Studio (DS) Visualizer 2021.
PatchDock (https://bioinfo3d.cs.tau.ac.il/PatchDock/) was used to run protein–protein docking simulations, which were then refined using FireDock (http://bioinfo3d.cs.tau.ac.il/FireDock/). PatchDock produced 100 predictions for each interaction, which were then submitted to FireDock to select the top ten solutions based on global energy (GE).
Hidden biological functions and complex mechanisms can be revealed by studying the internal movements of proteins. Molecular dynamics (MD) simulations of DPP4-LicA, DPP4-LicB, and DPP4-sitagliptin were performed using GROMACS (2019.6) at 300 K (Van Der Spoel et al., 2005) using the GROMOS96 43a1 force-field (Pol-Fachin et al., 2009). The PRODRG server was used to generate the compound topology and force-field parameters (Schuttelkopf and van Aalten, 2004). To neutralize ions in the solution, appropriate charges were introduced. To solvate the protein, the Simple Point Charge (spc216) water model was used. The particle-mesh Ewald method was used to investigate interactions between DPP4 and LicA, LicB, and sitagliptin using energy-grps in the MDs parameters (mdp) file. MD systems were then minimized using the steepest descent method (1500 steps) and the NVT and NPT ensembles were used for equilibration. The final production phase (100 ns) was then performed at 300 K. GROMACS analysis modules were employed to examine MD trajectories, and DS 2021 and PyMOL to create graphical representations of 3D models.
A DPP-4 Inhibitor Screening Kit (Sigma Aldrich, St. Louis, MO, United States) was used to confirm the inhibitory effects of the CWE of G. uralensis, LicA, and LicB in vitro. Fluorescence was measured in a microwell plate reader (λexcitation = 360 nm, λemission = 460 nm). Fluorescence emissions were recorded in kinetic mode for 30 min, and relative percentage inhibitions were calculated using the formula below. ΔF/ΔT is the rate of change of fluorescence. All experiments were performed using 6 replicates.
LicA and LicB were found to bind strongly to DPP-4. LicA interacted with the Arg123, His124, Glu203, Glu204, Ile205, Phe206, Gly207, Phe355, Arg356, Tyr548, Tyr663, Tyr667, Arg670, and Asn711 residues of DPP-4, and the Glu203 residue H-bonded with LicA (Figure 1A). LicB interacted with the Arg123, Glu203, Glu204, Ile205, Phe206, Gly207, Phe355, Arg356, Tyr548, Ser631, Tyr663, Tyr667, Arg670, Asn711, and His741 residues of DPP-4, and the Arg123, Ser631, Arg670, and His741 residues H-bonded with LicB (Figure 1B). The binding energies of LicA and LicB with DPP-4 were −6.16 and −6.29 kcal/mol, respectively (Table 1). Further, sitagliptin (the positive control) interacted with the Arg123, Glu203, Glu204, Ile205, Phe206, Gly207, Phe355, Arg356, Ser631, Tyr632, Tyr663, Tyr667, and Asn711 residues of DPP-4 (Figure 1C) with a binding energy of −6.70 kcal/mol (Table 1). ‘Molecular Overlay’ visualization revealed that the binding patterns and conformational alignments of LicA and LicB in the catalytic pocket of DPP-4 were similar to those of sitagliptin (Supplementary Figure S1).
LicA-DPP4, LicB-DPP4, and sitagliptin-DPP4 complexes were subjected to PPI with GLP1 and GIP. LicA, LicB, and sitagliptin DPP-4 complexes reduced the DPP-4 to GLP1 and GIP interactions, which suggested LicA, LicB, and sitagliptin might reduce the in vivo degradations of GLP1 and GIP (Figure 2). For example, DPP-4 bound to GLP1 with a GE of −84.60, and this was reduced to −74.53, −78.52, and −55.14 in the presence of LicA, LicB, or sitagliptin, respectively. Similarly, DPP-4 bound to GIP with a GE of −62.42, which was reduced to −23.17, −22.00, and −45.30 in the presence of LicA, LicB, or sitagliptin, respectively (Figure 2).
The stability profiles of LicA, LicB, and sitagliptin DPP-4 complexes were monitored to assess their relative root-mean-square deviation (RMSD) values throughout simulation runs. RMSD is commonly used to infer the extents of spatial deviations of groups of atoms (proteins, ligands, or ligand–protein complexes) from initial reference structure. Here, MD provided RMSD values with respect to the backbone of DPP-4. LicA-DPP4 and LicB-DPP4 complexes had average RMSD values of 0.251 and 0.250 nm, respectively, and sitagliptin-DPP4 complex had a slightly higher average value of 0.335 nm (Figure 3A). We also explored ligand dynamics in the catalytic pocket of DPP4. Visual inspections of complex trajectories revealed that all LicA, LicB, and sitagliptin displayed similar interaction patterns. Interestingly, LicA-DPP4 and sitagliptin-DPP4 complexes had more stable binding in catalytic pocket of DPP-4 (Figure 3B and Movie clip). Further, we assessed the compactness of complexes using the radius of gyration (Rg). The Rg average values for LicA-DPP4, LicB-DPP4 and sitagliptin-DPP4 complexes was found to be 2.59, 2.58, and 2.59 nm respectively. It was determined that there was little difference between the DPP4 enzyme and compound complexes, implying that the enzyme was stable with these compounds (Figure 3C).
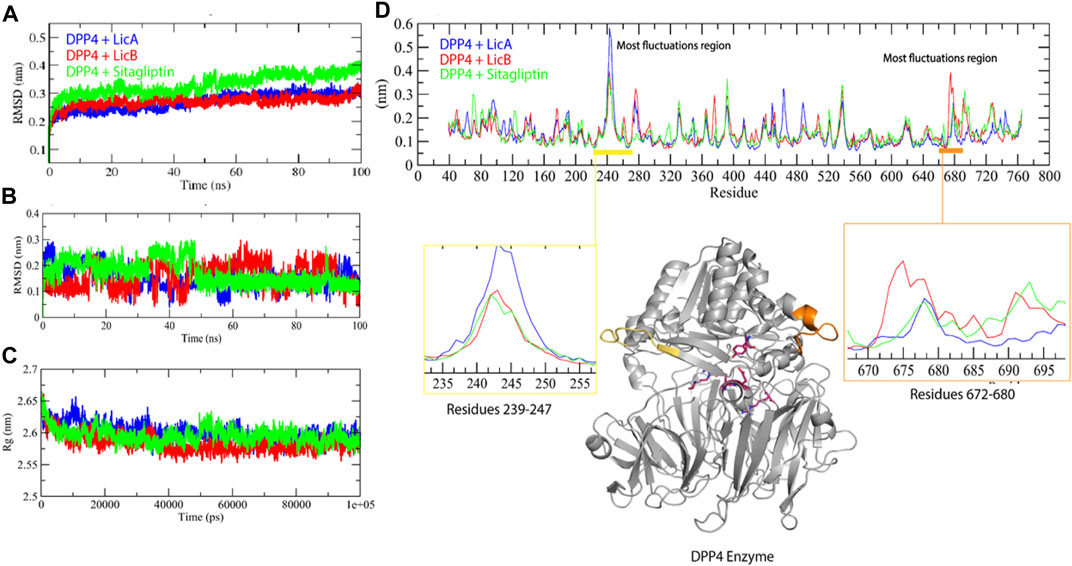
FIGURE 3. Molecular dynamics simulation studies of DPP-4 with LicA, LicB, and sitagliptin. RMSD backbone of DPP-4 enzyme in complexes (A), RMSD of ligands (B), Rg plot (C), and RMSF plot (D), DDP-4 is shown in gray color.
Root-mean-square fluctuation (RMSF) plot was used to determine specific residues interacting motion with compounds, and showed that two regions of DPP4, that is, residues 239-247 and 672-680, have a higher fluctuation region. DPP4-LicA complex exhibited fluctuation in the region 239-247, whereas DPP4-LicB complex has high fluctuation in the region 672-680 (Figure 3D). Collectively, these results indicated that DPP4 was stable with binding of LicA, LicB, and sitagliptin.
Furthermore, Solvent accessible surface area (SASA) has been assumed to be an important factor in molecular stability and folding studies. The LicA-DPP4, LicB-DPP4, and sitagliptin-DPP4 complexes were reported to have average SASA values of 293.74, 291.87, and 285.53 nm2, respectively. The LicA-DPP4, LicB-DPP4, and sitagliptin-DPP4 complexes had solvation energies of 373.21, 387.55, and 370.60 kJ/mol/nm2, respectively (Figures 4A,B). The LicA-DPP4 complex showed more SASA than LicB-DPP4 and sitagliptin-DPP4. It was inferred that DPP4 enzyme residues were more expose to water molecules.
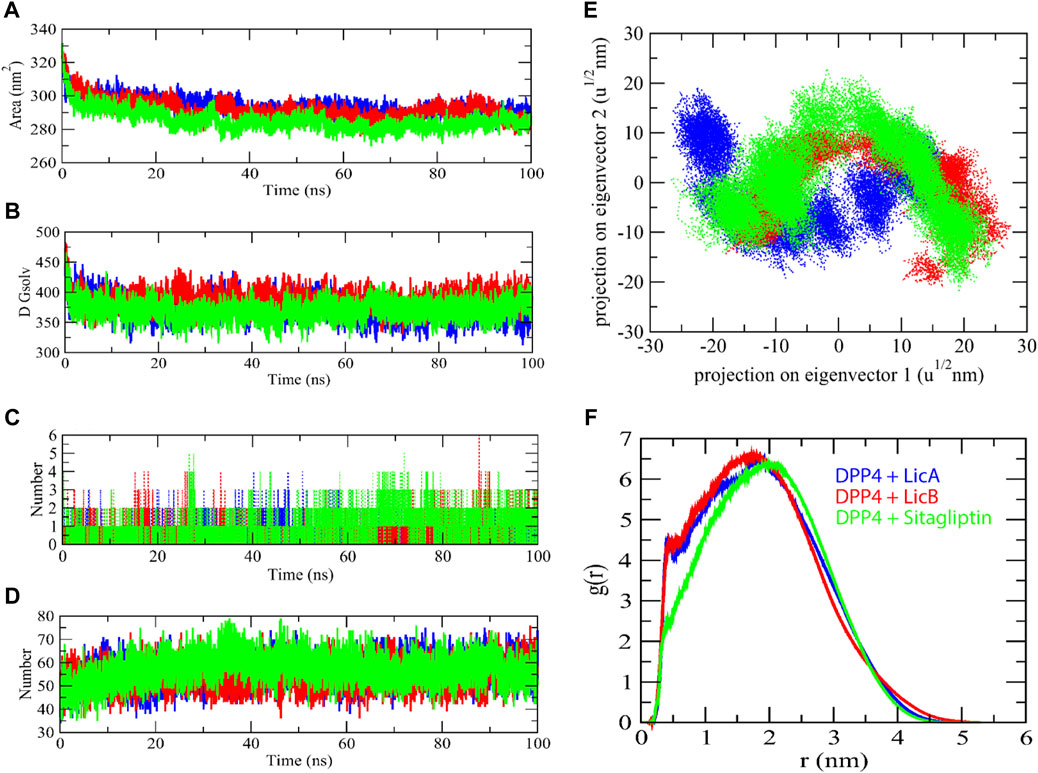
FIGURE 4. The solvent accessible surface area of complexes. SASA (A), Free energy of solvation (B), Number of H-bond with complexes (C), Number of H-bond between enzyme and water molecules (D), 2D projection of eigenvectors (E), and radial distribution function of complexes (F).
H-bond analysis was used to determine the binding interaction pattern of compounds with the DPP4 enzyme. The H-bond is essential for the ligand-protein complex’s stability. The complexes LicA-DPP4 and LicB-DPP4 bind to the DPP4 active pocket with 3–5 H-bonds, whereas sitagliptin-DPP4 binds with 1–3 H-bonds. H-bond analysis with protein and water found that sitagliptin-DPP4 complex had more H-bonds than LicA-DPP4 and LicB-DPP4 complexes (Figures 4C,D). Furthermore, the 2D projections of trajectories on eigenvectors revealed a variety of projections of LicA, LicB, and sitagliptin compounds. There was a significant variation in trajectory projections in the case of the LicA-DPP4 complex. The differences in atom position in LicA are quite distinct from those in LicB and sitagliptin (Figure 4E). This could be due to the fact that different conformations of during MD simulations and radial distribution function revealed that LicA-DPP4 and LicB-DPP4 were more stable than sitagliptin-DPP4 (Figure 4F).
In addition, the Gibbs’ free energy (GFE) landscape was estimated using GROMACS analysis modules and projections of their first (PC1) and second (PC2) eigenvectors. The energy represented by the Comparable GFE contour map with darker blue hues is lower. During the simulations, the complexes interacting with the DPP4 enzyme cause a fluctuation in the global minimum of DPP4. The projections of the LicB-DPP4 and sitagliptin-DPP4 complexes were similar, but the global minima of the LicA-DPP4 complex was different (Figures 5A–C).
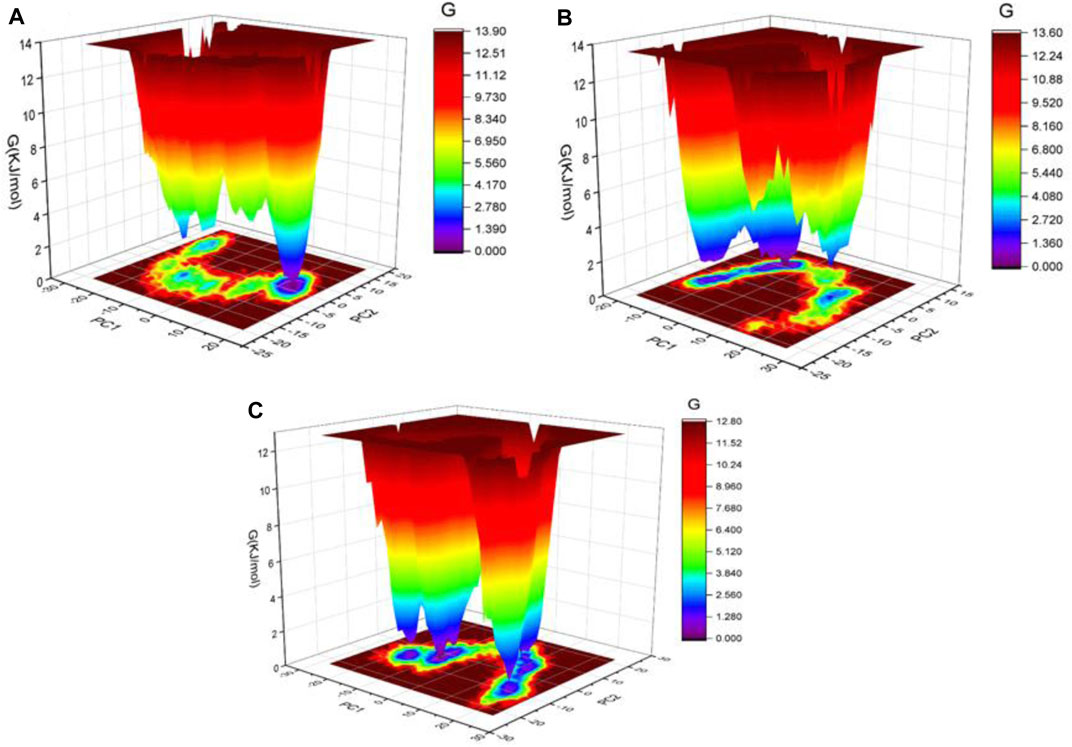
FIGURE 5. Gibbs’ free energy landscape plot of LicA-DPP4 (A), LicB-DPP4 (B), and sitagliptin-DPP4 (C) complexes.
The abilities of the CWE of G. uralensis, LicA, and LicB to inhibit DPP-4 were investigated at different concentrations. At high concentration (800 μg/ml), CWE inhibited DPP-4 by 21.6%. LicA and LicB inhibited DPP-4 activity concentration-dependently with an estimated IC50 values of 347.93 and 797.84 μM, respectively (Figure 6).
Licorice root is obtained from perennial herbs native to the Mediterranean, central to southern Russia, and some Asian regions, and the roots of G. glabra and G. uralensis are commonly used in cosmetics, meals, tobacco, and for several applications in the food and pharmaceutical sectors (Pastorino et al., 2018). Licorice and its bioactive constituents have a variety of beneficial health effects due to their antibacterial, antioxidant, anti-inflammatory, and immunomodulatory activities (Asl and Hosseinzadeh, 2008; Kim et al., 2014; Maria Pia et al., 2019; Wang et al., 2019). This study was performed to assess the possible anti-diabetic effects of the CWE of G. uralensis, LicA, and LicB by examining their inhibitory effects on DPP-4.
Molecular docking has become a valuable tool for drug development as it can accurately identify binding modes between drugs and their target proteins (Pinzi and Rastelli, 2019; Ahmad et al., 2021; Lee et al., 2021). In this study, LicA and LicB were found to bind to DPP-4 in silico, and an enzyme assay showed that they inhibited DPP-4 in vitro. Docking simulation revealed that LicA and LicB inhibit DPP-4 by binding to its catalytic sites. To deep insight into DPP-4 interacting residue with LicA and LicB, we analyzed the DPP-4 residues that interact with its co-crystallized inhibitor sitagliptin (PDB ID: 4FFW) by re-docking sitagliptin with DPP-4 within a defined grid, which revealed that the Arg123, Glu203, Glu204, Ile205, Phe206, Gly207, Phe355, Arg356, Ser631, Tyr632, Tyr663, Tyr667, and Asn711 residues of DPP-4 were involved in sitagliptin binding. Consistent with this, Arg123, Glu203, Glu204, Ile205, Phe206, Gly207, Phe355, Arg356, Tyr663, Tyr667, and Asn711 were the common interacting residues with LicA, LicB, and the sitagliptin. In addition, Glu203 was the common H-bonded residue of DPP-4 with LicA and sitagliptin; while Arg123, and Arg356 were the common H-bonded residues of DPP-4 with LicB and sitagliptin. Overall, our results showed LicA and LicB bind to DPP-4 in the same binding pocket as sitagliptin.
DPP-4 inhibitors are used to treat T2DM because of their ability to increase plasma GLP-1 and GIP levels, and thus, increase insulin production and improve blood glucose control (Deacon, 2020). The degree of interaction between two proteins is assessed in terms of GE, and a high negative GE indicates the strength of the interaction between a protein and its receptor (Lee et al., 2022). We found, DPP-4 interacted with GLP1 with a GE of −84.60, and this was reduced to −74.53 and −78.52 in the presence of LicA and LicB, respectively. On the other hand, DPP-4 interacted with GIP with a GE of −62.42, which was reduced to −23.17 and -22.00 in the presence of LicA, and LicB, respectively. These differences in GEs indicate that LicA and LicB weaken interactions between DPP-4 and GLP1 and GIP and possibly reduce the degradations of these hormones.
The stabilities of LicA, LicB, and sitagliptin DPP-4 complexes were demonstrated by MD simulation, which is a computationally intensive means of simulating the physiological environment of proteins and investigating changes in tertiary structure in biological settings (Adcock and McCammon, 2006). RMSD average values and Rg plots revealed that LicA, LicB, and sitagliptin stably bound to DPP-4 enzyme. Although high fluctuations were observed in some domains of DPP-4 after binding to LicA or LicB, these fluctuations did not occur in DPP-4 binding pocket residues. Furthermore, the MD trajectory movie clip demonstrates that LicA bound to the catalytic pocket of DPP-4 more strongly than LicB, which agrees with our DPP-4 enzyme assay result that LicA inhibits DPP-4 more potentially and has a lower IC50 value than LicB.
DPP-4 inhibition is a well-established glucose-lowering treatment in T2DM. Given the various side effects of currently available DPP-4 inhibitors (or ‘gliptins’), which include hypersensitivity reactions, gastrointestinal discomfort, pancreatitis, diarrhea, urinary tract infections, arthritis, and congestive heart failure (Chen et al., 2015; Mascolo et al., 2016; Packer, 2018), the development of natural product-like DPP-4 inhibitors with fewer adverse effects is of considerable importance. Natural products are well-known for providing new molecular entities for treating many diseases (Patridge et al., 2016; Shaikh et al., 2021a; Shaikh et al., 2021b). In addition, phenolics are well known for their ability to alleviate diabetes by lowering blood glucose levels (Rauter et al., 2010). Interestingly, we found that the natural phenols LicA and LicB inhibit DPP-4 activity, and thus, would be expected to have anti-diabetic effects. LicA more potently inhibited DPP-4 with an estimated IC50 of 347.93 μM compared to that of the LicB (IC50 = 797.84 μM).
The search for natural bioactive compounds in plants has recently been viewed as a priority by the food and pharmaceutical industries due to their commercial potentials (Atanasov et al., 2015). Diet or dietary supplements plays an important role in the prevention and management of diseases such as obesity, diabetes, and cancer (Lee et al., 2013) and increase healthspan (Chen et al., 2022). In this study, LicA, and LicB were found to inhibit DPP-4 at high concentrations, which suggests these agents could be used as functional food ingredients for the management of T2DM.
LicA and LicB from G. uralensis potently inhibit DPP-4 by binding to its catalytic pocket and have several amino acid residues interactions in common with the sitagliptin. MD simulation studies revealed that LicA and LicB bound stably with DPP-4. In addition, DPP-4 in vitro enzyme assay showed that LicA, and LicB concentration-dependently inhibited DPP-4. These findings suggest that LicA and LicB may be responsible for the anti-diabetic effects of G. uralensis and can be used as functional food ingredients to manage T2DM. Further in vivo studies are required to determine whether LicA and LicB can improve glycemic control in animal models of diabetes.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Conceptualization, SS and IC; Formal analysis, SS, SA, KA, EJL, and IC; Funding acquisition, EJL and IC; Methodology, SS, SA, and JHL; Writing—original draft, SS and SA; Writing—review & editing, HJC, KA, SSA, YCH, KSH, and NRK.
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (2020R1A6A1A03044512) and by the National Research Foundation of Korea (NRF), and funded by the Korean government (NRF-2021R1A2C2004177 and NRF-2019R1C1C1006542).
KSH and NRK were employed by the company Neo Cremar Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.1024764/full#supplementary-material
CWE, crude water extract; DPP-4, Dipeptidyl peptidase-4; DS, Discovery studio; GE, Global energy; GFE, Gibbs’ free energy; GLP-1, Glucagon-like peptide-1; GIP, Glucose-dependent insulinotropic polypeptide; LicA, licochalcone A; LicB, licochalcone B; MD, Molecular dynamics; PPIs, Protein-protein interactions; Rg, Radius of gyration; RMSD, Root-mean-square deviation; RMSF, Root-mean-square fluctuation, SASA, Solvent accessible surface area; T2DM, Type 2 diabetes mellitus.
Adcock, S. A., and McCammon, J. A. (2006). Molecular dynamics: Survey of methods for simulating the activity of proteins. Chem. Rev. 106, 1589–1615. doi:10.1021/cr040426m
Ahmad, S. S., Khan, M. B., Ahmad, K., Lim, J. H., Shaikh, S., Lee, E. J., et al. (2021). Biocomputational screening of natural compounds against acetylcholinesterase. Molecules 26, 92641. doi:10.3390/molecules26092641
Ali, S., Ahmad, K., Shaikh, S., Lim, J. H., Chun, H. J., Ahmad, S. S., et al. (2022). Identification and evaluation of traditional Chinese medicine natural compounds as potential myostatin inhibitors: An in silico approach. Molecules 27, 34303. doi:10.3390/molecules27134303
Asl, M. N., and Hosseinzadeh, H. (2008). Review of pharmacological effects of glycyrrhiza sp. and its bioactive compounds. Phytother. Res. 22, 709–724. doi:10.1002/ptr.2362
Atanasov, A. G., Waltenberger, B., Pferschy-Wenzig, E. M., Linder, T., Wawrosch, C., Uhrin, P., et al. (2015). Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnol. Adv. 33, 1582–1614. doi:10.1016/j.biotechadv.2015.08.001
Chatterjee, S., Khunti, K., and Davies, M. J. (2017). Type 2 diabetes. Lancet 389, 2239–2251. doi:10.1016/s0140-6736(17)30058-2
Chen, X. W., He, Z. X., Zhou, Z. W., Yang, T., Zhang, X., Yang, Y. X., et al. (2015). Clinical pharmacology of dipeptidyl peptidase 4 inhibitors indicated for the treatment of type 2 diabetes mellitus. Clin. Exp. Pharmacol. Physiol. 42, 999–1024. doi:10.1111/1440-1681.12455
Chen, Y., Hamidu, S., Yang, X., Yan, Y., Wang, Q., Li, L., et al. (2022). Dietary supplements and natural products: An update on their clinical effectiveness and molecular mechanisms of action during accelerated biological aging. Front. Genet. 13, 880421. doi:10.3389/fgene.2022.880421
Deacon, C. F. (2020). Dipeptidyl peptidase 4 inhibitors in the treatment of type 2 diabetes mellitus. Nat. Rev. Endocrinol. 16, 642–653. doi:10.1038/s41574-020-0399-8
Dodds, S. (2017). The how-to for type 2. Nurs. Clin. N. Am. 52, 513–522. doi:10.1016/j.cnur.2017.07.002
Katsila, T., Spyroulias, G. A., Patrinos, G. P., and Matsoukas, M. T. (2016). Computational approaches in target identification and drug discovery. Comput. Struct. Biotechnol. J. 14, 177–184. doi:10.1016/j.csbj.2016.04.004
Kim, J. S., Park, M. R., Lee, S. Y., Kim, D. K., Moon, S. M., Kim, C. S., et al. (2014). Licochalcone A induces apoptosis in KB human oral cancer cells via a caspase-dependent FasL signaling pathway. Oncol. Rep. 31, 755–762. doi:10.3892/or.2013.2929
Lee, E. J., Shaikh, S., Ahmad, K., Ahmad, S. S., Lim, J. H., Park, S., et al. (2021). Isolation and characterization of compounds from glycyrrhiza uralensis as therapeutic agents for the muscle disorders. Int. J. Mol. Sci. 22, 20876. doi:10.3390/ijms22020876
Lee, E. J., Shaikh, S., Baig, M. H., Park, S. Y., Lim, J. H., Ahmad, S. S., et al. (2022). MIF1 and MIF2 myostatin peptide inhibitors as potent muscle mass regulators. Int. J. Mol. Sci. 23. doi:10.3390/ijms23084222
Lee, H., Lee, I. S., and Choue, R. (2013). Obesity, inflammation and diet. Pediatr. Gastroenterol. Hepatol. Nutr. 16, 143–152. doi:10.5223/pghn.2013.16.3.143
Lee, S. J., Reed, L. A., Davies, M. V., Girgenrath, S., Goad, M. E., Tomkinson, K. N., et al. (2005). Regulation of muscle growth by multiple ligands signaling through activin type II receptors. Proc. Natl. Acad. Sci. U.S.A. 102, 18117–18122. doi:10.1073/pnas.0505996102
Maria Pia, G. D., Sara, F., Mario, F., and Lorenza, S. (2019). Biological effects of licochalcones. Mrmc 19, 647–656. doi:10.2174/1389557518666180601095420
Mascolo, A., Rafaniello, C., Sportiello, L., Sessa, M., Cimmaruta, D., Rossi, F., et al. (2016). Dipeptidyl peptidase (DPP)-4 inhibitor-induced arthritis/arthralgia: A review of clinical cases. Drug Saf. 39, 401–407. doi:10.1007/s40264-016-0399-8
Nauck, M. A., and Meier, J. J. (2018). Incretin hormones: Their role in health and disease. Diabetes Obes. Metab. 20 1, 5–21.doi:10.1111/dom.13129
Packer, M. (2018). Worsening heart failure during the use of DPP-4 inhibitors. JACC Heart Fail. 6, 445–451. doi:10.1016/j.jchf.2017.12.016
Padron, S., Rogers, E., Demory Beckler, M., and Kesselman, M. (2020). Republished: DPP-4 inhibitor (sitagliptin)-induced seronegative rheumatoid arthritis. Dtb 58, 12–15. doi:10.1136/dtb.2019.228981rep
Park, S., Kang, H. J., Jeon, J. H., Kim, M. J., and Lee, I. K. (2019). Recent advances in the pathogenesis of microvascular complications in diabetes. Arch. Pharm. Res. 42, 252–262. doi:10.1007/s12272-019-01130-3
Pastorino, G., Cornara, L., Soares, S., Rodrigues, F., and Oliveira, M. (2018). Liquorice (Glycyrrhiza glabra): A phytochemical and pharmacological review. Phytotherapy Res. 32, 2323–2339. doi:10.1002/ptr.6178
Pathak, R., and Bridgeman, M. B. (2010). Dipeptidyl peptidase-4 (DPP-4) inhibitors in the management of diabetes. P T 35, 509–513.
Patridge, E., Gareiss, P., Kinch, M. S., and Hoyer, D. (2016). An analysis of FDA-approved drugs: Natural products and their derivatives. Drug Discov. Today 21, 204–207. doi:10.1016/j.drudis.2015.01.009
Pinzi, L., and Rastelli, G. (2019). Molecular docking: Shifting paradigms in drug discovery. Int. J. Mol. Sci. 20, 84331. doi:10.3390/ijms20184331
Pol-Fachin, L., Fernandes, C. L., and Verli, H. (2009). GROMOS96 43a1 performance on the characterization of glycoprotein conformational ensembles through molecular dynamics simulations. Carbohydr. Res. 344, 491–500. doi:10.1016/j.carres.2008.12.025
Rauter, A. P., Martins, A., Borges, C., Mota-Filipe, H., Pinto, R., Sepodes, B., et al. (2010). Antihyperglycaemic and protective effects of flavonoids on streptozotocin-induced diabetic rats. Phytother. Res. 24 Suppl 2 (2), S133–S138. doi:10.1002/ptr.3017
Reed, J., Bain, S., and Kanamarlapudi, V. (2021). A review of current trends with type 2 diabetes epidemiology, aetiology, pathogenesis, treatments and future perspectives. Dmso 14, 3567–3602. doi:10.2147/dmso.s319895
Saeedi, P., Petersohn, I., Salpea, P., Malanda, B., Karuranga, S., Unwin, N., et al. (2019). Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the international diabetes federation diabetes atlas, 9th edition. Diabetes Res. Clin. Pract. 9157, 107843. doi:10.1016/j.diabres.2019.107843
Schüttelkopf, A. W., and van Aalten, D. M. (2004). Prodrg: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D. Biol. Cryst. 60, 1355–1363. doi:10.1107/s0907444904011679
Shaikh, S., Ahmad, K., Ahmad, S. S., Lee, E. J., Lim, J. H., Beg, M. M. A., et al. (2021a). Natural products in therapeutic management of multineurodegenerative disorders by targeting autophagy. Oxid. Med. Cell. Longev. 2021, 6347792. doi:10.1155/2021/6347792
Shaikh, S., Lee, E. J., Ahmad, K., Ahmad, S. S., Lim, J. H., and Choi, I. (2021b). A Comprehensive review and perspective on natural sources as dipeptidyl peptidase-4 Inhibitors for management of diabetes, Pharm. (Basel) 14, 591. doi:10.3390/ph14060591
Van Der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A. E., and Berendsen, H. J. (2005). Gromacs: Fast, flexible, and free. J. Comput. Chem. 26, 1701–1718. doi:10.1002/jcc.20291
Wang, J., Liao, A. M., Thakur, K., Zhang, J. G., Huang, J. H., and Wei, Z. J. (2019). Licochalcone B extracted from glycyrrhiza uralensis fisch induces apoptotic effects in human hepatoma cell HepG2. J. Agric. Food Chem. 67, 3341–3353. doi:10.1021/acs.jafc.9b00324
Keywords: type 2 diabetes mellitus, dipeptidyl peptidase-4, Glycyrrhiza uralensis, natural compounds, licochalcone A
Citation: Shaikh S, Ali S, Lim JH, Chun HJ, Ahmad K, Ahmad SS, Hwang YC, Han KS, Kim NR, Lee EJ and Choi I (2022) Dipeptidyl peptidase-4 inhibitory potentials of Glycyrrhiza uralensis and its bioactive compounds licochalcone A and licochalcone B: An in silico and in vitro study. Front. Mol. Biosci. 9:1024764. doi: 10.3389/fmolb.2022.1024764
Received: 22 August 2022; Accepted: 13 September 2022;
Published: 30 September 2022.
Edited by:
Mustafa Aziz Hatiboglu, Bezmiâlem Vakıf Üniversitesi, TurkeyReviewed by:
Qazi Mohammad Sajid Jamal, Qassim University, Saudi ArabiaCopyright © 2022 Shaikh, Ali, Lim, Chun, Ahmad, Ahmad, Hwang, Han, Kim, Lee and Choi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eun Ju Lee, Z29yYXBhZG9jMDMxNUBoYW5tYWlsLm5ldA==; Inho Choi, aW5ob2Nob2lAeW51LmFjLmty
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.