
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci. , 03 November 2022
Sec. Cellular Biochemistry
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.1009402
 Leon Lufkin1,2
Leon Lufkin1,2 Ankita Samanta3
Ankita Samanta3 DeVaun Baker3,2
DeVaun Baker3,2 Sina Lufkin3,2
Sina Lufkin3,2 JesslynHope Schulze3
JesslynHope Schulze3 Benjamin Ellis3
Benjamin Ellis3 Jillian Rose3
Jillian Rose3 Thomas Lufkin3
Thomas Lufkin3 Petra Kraus3*
Petra Kraus3*Regenerative medicine aims to repair degenerate tissue through cell refurbishment with minimally invasive procedures. Adipose tissue (FAT)-derived stem or stromal cells are a convenient autologous choice for many regenerative cell therapy approaches. The intervertebral disc (IVD) is a suitable target. Comprised of an inner nucleus pulposus (NP) and an outer annulus fibrosus (AF), the degeneration of the IVD through trauma or aging presents a substantial socio-economic burden worldwide. The avascular nature of the mature NP forces cells to reside in a unique environment with increased lactate levels, conditions that pose a challenge to cell-based therapies. We assessed adipose and IVD tissue-derived stromal cells through in vitro transcriptome analysis in 2D and 3D culture and suggested that the transcription factor Glis1 and metabolite oxaloacetic acid (OAA) could provide NP cells with survival tools for the harsh niche conditions in the IVD.
Intervertebral disc degeneration (IVDD) through accidental or lifestyle-induced trauma or aging presents a substantial socio-economic burden worldwide, affecting all genders (DePalma et al., 2011; Gatchel, 2015). As a crucial shock-absorbing organ in the axial skeleton of vertebrates, the IVD protects the spine while facilitating its mobility. The IVD consists of a central nucleus pulposus (NP) surrounded by an inner and outer annulus fibrosus (AF) and is flanked by rigid vertebral bodies and their cartilaginous endplates (CEP) (Eyre, 1979; Bayliss et al., 1988; Humzah and Soames, 1988; Oegema, 1993; Bibby and Urban, 2004; Sivakamasundari and Lufkin, 2012; Sivakamasundari et al., 2013; Bedore et al., 2014; Erwin and Hood, 2014) (Figure 1A). The NP is unique in being the largest avascular tissue among vertebrate organs. Cells in the NP of a mature IVD are sparse in numbers and embedded in vast amounts of extracellular matrix (ECM). Cells in the AF are longitudinally stretched in shape and located within the compacted, lamellar organization of densely packed ECM (Trout et al., 1982a; Kraus et al., 2017; Li et al., 2019a). In a healthy mature IVD, vasculature and innervation only reach the CEPs and outer AF, whereas NP cells depend on diffusion for nutrient access and waste product clearance (Figure 1A). Therefore, NP cells reside in a unique and naturally harsh environment, low in oxygen, reduced in nutrients (1–5 mM glucose), and slightly acidic pH of 7.1–7.4, with lactate concentrations of 16 mM in the center from lactic acid fermentation alongside proton retention by negatively charged proteoglycans (PG) in the NP-ECM. The pH drops below 7 in a degenerated disc. This harsh environment impacting the NP cell function alongside lifestyle choices, obesity, and other reasons, is considered a contributor to IVDD, eventually leading to pain and mobility impairment (Diamant et al., 1968; Nachemson, 1969; Urban et al., 1977; Bartels et al., 1998; Razaq et al., 2000; Razaq et al., 2003; Urban and Roberts, 2003; Bibby and Urban, 2004; Urban et al., 2004; Bibby et al., 2005; Wuertz et al., 2008; Wuertz et al., 2009; Gilbert et al., 2016; Wang et al., 2021a).
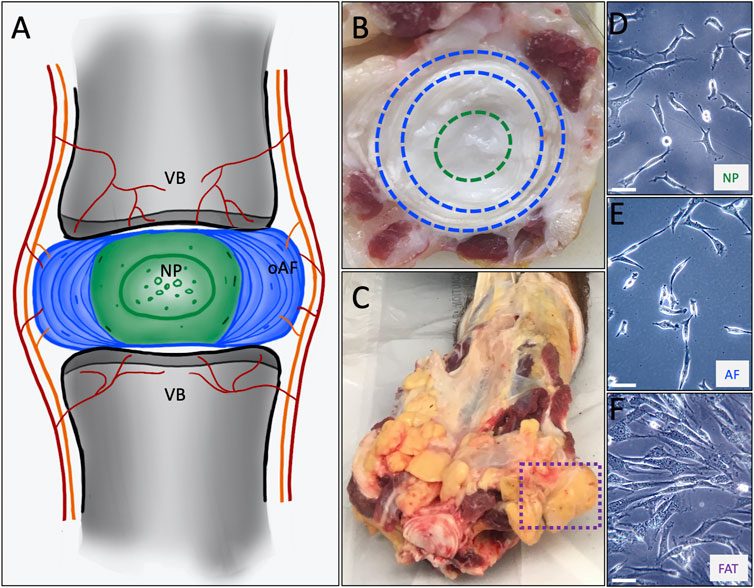
FIGURE 1. (A) Schematic representing the intervertebral disc consisting of an outer annulus fibrosus (oAF) and inner nucleus pulposus (NP) sandwiched between the cartilaginous endplates of the adjacent vertebral bodies (VB). (B) A cut through a bovine tail showing a coccygeal IVD. The outer AF as seen between the blue dashed lines and the NP within the green dashed circle served as tissue source to derive AF and NP cell lines. (C) The box with the dashed purple line shows coccygeal, subcutaneous adipose tissue used as source to derive FAT cells. The primary cell lines are shown at p0 as they attach to a plastic surface for NP (D), AF (E) and FAT (F) cells. The scale bar represents 50 μM.
Regenerative medicine is a highly intricate field that proposes ways to repair degenerated tissue with minimally invasive procedures using cell-based therapies, among other strategies. The cell is the fundamental unit of all living organisms, which develops based on its genomic blueprint. Starting from a zygote, the spatiotemporally coordinated transcription of coding and non-coding DNA results in the different cell types, tissues, and organs of a multicellular organism. This regulation is modulated by transcription and signaling factors alongside a variety of non-coding RNAs discovered in recent years (Kraus et al., 2013; Kraus et al., 2019a) and epigenetic encoding of histone molecules controlling DNA access (Portela and Esteller, 2010). Differential transcription guides descendants of the initially totipotent zygote toward ectoderm, mesoderm, or endoderm. Mesoderm, the middle layer of the gastrula stage embryo, gives rise to many body cell lineages after transcriptome-dependent specialization. Terminal differentiation can eventually culminate in cell death (Kraus and Lufkin, 2017). However, some cells, referred to as stem, progenitor, or stromal cells depending on their stage of pluripotency, remain in mature tissues and retain a degree of “stemness” throughout the organism’s lifespan with a persisting degree of multipotency (Pittenger et al., 1999; Lander, 2009; Kraus and Lufkin, 2017). Based on this “stemness,” regenerative cell therapies are built.
IVDD cell therapies long engaged in the idea of using stem cells to replace ailing NP cells, increase ECM production, and restore IVD function (Wuertz et al., 2008; Sivakamasundari and Lufkin, 2013; Pennicooke et al., 2016; Kraus and Lufkin, 2017; Sakai and Schol, 2017; Xia et al., 2019; Wang et al., 2021b). Mesenchymal stem or stromal cells (MSCs) have attracted attention given their multi-over pluripotency, with the benefit of reduced tumorigenicity (Kraus et al., 2021). Among several stromal cell sources in the context of IVDD, the AF tissue would be more easily accessible than NP tissue, whereas subcutaneous adipose MSCs are an established source (Rodriguez et al., 2004; Rodriguez et al., 2005) (Table 1). The progenitor cell potential of IVD cells was suggested previously (Risbud et al., 2007; Henriksson et al., 2009; Risbud et al., 2015; Tekari et al., 2016; Thorpe et al., 2016; Kraus et al., 2017). NP cells originate from the notochord (NC) (Christ and Wilting, 1992; Choi et al., 2008; Choi and Harfe, 2011; Choi et al., 2012; McCann et al., 2012; Lawson and Harfe, 2015; McCann and Seguin, 2016), a transient embryonic signaling center in vertebrates derived from axial mesoderm. The NC was identified as the origin of NP cells through transgenic experiments in mice, identifying Sonic hedgehog (Shh), Noto, and Brachyury (Tbx1 or T-box) as important for NC cell development (Choi et al., 2008; Choi and Harfe, 2011; Choi et al., 2012). Progressive loss or trans-differentiation of NC cells in the human IVD coincides with the onset of IVDD (Choi et al., 2008; McCann and Seguin, 2016). This loss of NC cells is also seen in the mature bovine IVD but not the mature rodent or porcine IVD (Trout et al., 1982b; Urban and Roberts, 2003; Vujovic et al., 2006; Lawson and Harfe, 2015; McCann and Seguin, 2016; Kraus et al., 2017). Coccygeal bovine IVDs are, therefore, an accepted model organism to represent cells of a healthy human IVD (Li et al., 2019b). The AF is formed by sclerotome cells originating from the ventral somite and is, therefore, of mesodermal origin (Christ and Wilting, 1992; Sivakamasundari and Lufkin, 2012). Cells for tissue regeneration are typically of shared developmental origin. Among the types of mesenchymal stromal cells, adipose tissue (FAT)-derived autologous stem cells have gained popularity. Adipose cells, too, are of mesoderm origin, with various discrete deposits present in mammals (Duan et al., 2007; Berry et al., 2013). Of those, white FAT can be located as visceral fat deposits in the abdominal cavity associated with internal organs or subcutaneously. FAT-derived stromal or stem cells are a by-product of cosmetic liposuction procedures and a convenient autologous choice for regenerative cell therapy approaches (Table 1). The naturally harsh NP environment poses a challenge to cell refurbishment with any cell type, including adipose stromal cells. Metabolic studies characterized the impact of nutrients, oxygen, and pH on NP cell survival in the niche (Diamant et al., 1968; Nachemson, 1969; Urban et al., 1977; Bartels et al., 1998; Razaq et al., 2000; Razaq et al., 2003; Urban and Roberts, 2003; Bibby and Urban, 2004; Urban et al., 2004; Bibby et al., 2005; Wuertz et al., 2008; Wuertz et al., 2009; Gilbert et al., 2016; Wang et al., 2021a) and NP cells appear unusually resilient in vitro, even for extended culture times (Kraus et al., 2017). We show that acquired niche survival strategies distinguish NP stromal cells from AF and adipose stromal cells on a molecular level and propose that lactate metabolism is key to survival in the NP niche. These molecular adaptions could be exploited for regenerative applications.
Cells were isolated from coccygeal bovine tissue sources of four different healthy mature animals as previously described (Kraus et al., 2017) (Figures 1B–F, 2A). Briefly, tails were retrieved from local abattoirs within 2 h of euthanasia and kept on ice until tissue dissection. After skin removal, the tissue was disinfected with a betadine solution. Subcutaneous fat, tendon, and muscle were removed before dissecting the NP and outer AF of IVDs, omitting the transition zone. Each tissue was briefly dipped into 70% ethanol (EtOH) and minced into small pieces. The pieces were moved to an untreated 35 mm polystyrene tissue culture dish (Corning/Falcon #353001) and covered with 100% heat-inactivated fetal bovine serum (HI-FBS) (Gibco # 10082147). No enzymatic treatment was applied for cell isolation. Tissue pieces were incubated for 48 h at 5% CO2 before the FBS was replaced with Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% HI-FBS, 1X non-essential amino acids (MEM-NEAA) (Gibco), 1X GlutaMAX™ (Gibco), 1 mM sodium pyruvate (Gibco), 48 μG/ml gentamicin (Gibco), and 0.12 mM β-mercaptoethanol buffered with sodium bicarbonate (Gibco), thereafter referred to as DMEM with 10% FBS or standard medium (D10S). The medium composition was based on our embryonic stem cell work (Chatterjee et al., 2013; Sivakamasundari et al., 2017). Tissue pieces were removed after 5 days or when cells were attached to the plate (p0) (Figure 2B).
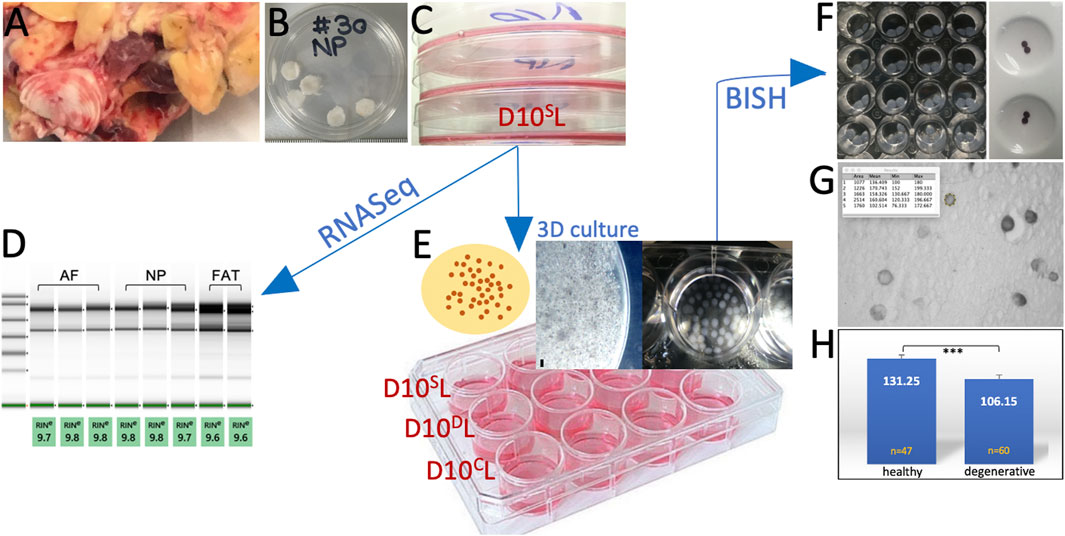
FIGURE 2. Experimental workflow showing the bovine tail as cell source (A). Nucleus pulposus (NP) explants in 1X PBS prior to cell isolation (B). Initial primary cell expansion in monolayer culture (C). Transcriptome analysis through RNASeq after RNA quality assessment (D). Cells were embedded in alginate for 3D culture as shown in the schematic. A high-magnification image of such a bead is shown next to beads collected in a 24-well plate (E) under standard (D10SL), degenerate (D10DL), and chondrogenic (D10CL) conditions. Phenotyping was carried out by RNA in situ hybridization on beads (BISH) (F). Beads were sectioned (G) and submitted to ImageJ analysis (H).
For passaging, monolayer cultured cells were rinsed with 1X phosphate-buffered saline (PBS) and disassociated with 0.05% Trypsin/EDTA (Gibco) for no more than 5 min once ∼90% confluency was reached. To assess glucose and FBS needs, primary NP, AF, and FAT cell lines from two donors were maintained in monolayer culture (Figures 1B–F, 2) supplementing glucose-free DMEM (Corning Cellgro #90-113-PB), as described earlier by adding different concentrations of glucose at 10% FBS [4.5 g/L (high (H)), 1 g/L (5.5 mM, low (L)), 0.5 g/L or 0 g/L], and different volumes of FBS [10%, 5%, 1%, and 0.2% at low (1 g/L) glucose. Three technical replicates per cell line and condition were seeded in 96-well plates at ∼300 cells/well. Once attached (0 h), cells were washed twice with 1X PBS and fixed immediately (0 h) with 4% (w/v) paraformaldehyde (PFA) or cultured for 5 days in DMEM with described glucose or FBS concentrations. Fixed cells were subjected to staining with propidium iodide following the manufacturer’s instructions (Thermo Fisher Scientific) for a total cell count using a SpectraMax i39 (Molecular Devices) multi-mode microplate reader. The ratio of average cell count was expressed as a percentage of the initial cell count (0 h). Bootstrapping was applied to test for the significance of the mean ratios of average cell counts, assuming mean counts before and after the experiment were normally distributed. The Benjamini–Yekutieli correction to control the false discovery rate (FDR) was set at 5%. Adjusted p-values less than 0.05 are denoted with *, those less than 0.01 with **, and those less than 0.001 with ***. Based on these observations, subsequent monolayer (2D) cell culture was carried out with DMEM/10% FBS and 1 g/L glucose (D10L) medium as the base. For 3D culture, ∼ 105 cells were embedded per 1 ml alginate solution (Figure 2E) (Maldonado and Oegema, 1992; May et al., 2020). Alginate solution was prepared at 1.2% under sterile conditions by resuspending alginate powder in sterile 0.9% (w/v) NaCl until a homogeneous solution was achieved (Gilbert et al., 2016) and filtered through a 0.45-μm syringe filter. The alginate/cell solution was dripped into sterile 102 mM CaCl2 under agitation using a 211/2-gauge needle. After curation, the beads were rinsed twice with sterile 0.9% NaCl and transferred to a 6-well dish with D10L-based culture medium for 10 days, changing the medium after 5 days (Figure 2E). D10L medium was modified to achieve three different media conditions: pH 7.1 (normal, D10SL), supplemented with 50 μG/ml ascorbic acid and 40 μG/ml proline (chondrogenic, D10CL) (Andriamanalijaona et al., 2008; Kraus et al., 2017), or adjusted to lactic acid concentration based degenerate conditions (∼pH 6.5, D10DL) (Nachemson, 1969; Razaq et al., 2000; Wuertz et al., 2009; Gilbert et al., 2016; Wang et al., 2021a) using 3 μl/ml lactic acid (13.4 M, Sigma) to a final concentration of ∼40 mM based on initial pH measurement. Induced pH changes remained steady during incubation based on color changes of the pH indicator phenol red. All cell culture was carried out under atmospheric O2 and 5% CO2.
NP, AF, and FAT primary cell lines were derived from the same donor to minimize genetic bias. To minimize technical bias triplicates from NP and AF and duplicates from FAT, tissue-derived cell lines were cultured and handled simultaneously in monolayer culture using D10SL media as described. RNA extraction was carried out at passage 2 (p2) from ∼106 cells per sample with the RNEasy Micro Kit (Qiagen) following the manufacturer’s instructions (Figure 2E). All RNA was stored at −80°C. RNA concentration was determined with a NanoDrop® spectrophotometer (Thermo Fisher Scientific), and RNA quality was assessed with an Agilent 2,100 Bioanalyzer (Agilent Technologies Inc.). RNASeq was carried out by Genewiz to identify significantly differentially expressed genes from total RNA (500 ng) of three biological replicates per NP and AF sample and two biological replicates per FAT sample as per the Genewiz recommendation. RNA sequencing libraries were prepared using the NEBNext Ultra RNA Library Prep Kit (NEB) following kit protocol, fragmented, reverse-transcribed, and ligated to universal adapters, followed by index addition and library enrichment via limited-cycle PCR. The sequencing libraries were validated on an Agilent TapeStation and quantified with a Qubit 2.0 Fluorometer and quantitative PCR (KAPA Biosystems), clustered on a single lane of a flowcell and loaded on an Illumina HiSeq4000 instrument. Following 2 × 150-bp paired-end sequencing, image analysis and base calling were conducted by the HiSeq Control Software (HCS). Raw sequence data (.bcl files) generated from Illumina HiSeq were converted into fastq files and de-multiplexed using Illumina’s bcl2fastq 2.17 software. One mismatch was allowed for index sequence identification. The bioinformatic workflow followed the Genewiz protocol using GeneSCF v.1.1-p2, along with PANTHER.db and ToppFun (part of the ToppGene Suite) for Gene Ontology (GO) annotation of biological pathways, molecular mechanisms, and pathways. Graphs were generated using ggplot2 in R. ToppCluster was used for network generation of transcripts with Bonferroni correction and a p-value cutoff of 0.05. Generated networks were visualized and analyzed in Cytoscape.
RNA in situ hybridisation (RISH) was used for targeted transcript-based cell phenotyping on sectioned tissue (SISH) (Kraus et al., 2001; Kraus et al., 2014; Kraus et al., 2019a; Kraus et al., 2019b), cells cultured on glass coverslips (CISH) (Li et al., 2021), or in alginate beads (BISH). Briefly, the SISH tissues from more than three donors were fixed in 4% (w/v) PFA overnight at 4°C and then washed 3 × in 1X PBS for 5 min. Samples were dehydrated through rising ethanol gradients followed by 3 × 100% ethanol and 3 × HistoClear for 10 mins each, embedded in paraffin, and sectioned at 7 μM. For CISH, coverslips containing cells of three donors were fixed in 4% PFA post-culture and subjected to the RISH procedure below. BISH cells in alginate beads from one donor were fixed in 4% PFA for at least 2 h at 4°C and briefly rinsed in bead buffer (BB) prepared as 50 mM CaCl2 and 150 mM NaCl in nuclease-free (nf) water, followed by 150 mM NaClnf (Figure 2F). Prior to RISH, the sections on polylysine-coated slides were de-waxed in HistoClear, rehydrated, and post-fixed with 4% PFA. Prehybridization for 2–3 h was followed by overnight hybridization, both at 62°C. Antisense digoxygenin (DIG)-labeled RNA probes based on PCR amplified templates of the bovine genome (Table 2) were in vitro transcribed using the DIG RNA labeling mix (Roche/Sigma), purified with RNA Spin columns, and used as 500–800 ng/ml in pre-hybridization solution (Kraus et al., 2017). Washes included Solution I (50% formamide, 5X SSC, 1% SDS; 62°C), TNT (10 mM Tris-HCl, pH 7.5, 0.5 M NaCl, 0.1% Tween 20; RT), TNT:Solution II (1:1), and Solution II (50% formamide, 2X SSC, 0.2% SDS; 58°C). Before blocking with Superblock (Thermo Fisher Scientific), samples were rinsed with 1X PBS or BB and probed with alkaline phosphatase (AP)-conjugated anti-DIG antibody diluted 2,000-fold in blocking solution. Samples were exposed overnight to an anti-DIG-AP antibody (Roche). After washes with 1X PBS or BB and 150 mM NaCl, samples were conditioned in NTMT (0.1 M Tris-HCl, pH 9.5, 0.1 M NaCl, 0.05 M MgCl2, 0.1% Tween 20) before the NBT/BCIP (Roche) substrate was added for color development at 4°C in the dark. All BISH experiments were performed under the same conditions for triplicates of alginate beads per probe. Beads were dehydrated as described and sectioned at 7 μM. Outcomes were documented on a Zeiss Primovert microscope (Kraus et al., 2017; Kraus et al., 2018; Amirdelfan et al., 2021). BISH data were quantified via ImageJ at single-cell resolution and assessed for statistically significant differences in expression levels on a population level using Student’s t-test. The Benjamini–Yekutieli correction to control the FDR was set at 5%. Adjusted p-values less than 0.05 are denoted with *, those less than 0.01 with **, and those less than 0.001 with ***. All BISH data acquisition was performed by the same researcher to avoid inter-person variability. Graphs were generated using ggplot2 in R.
Acidic β-galactosidase staining was conducted using a staining solution of 1 mG/ml X-gal, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 150 mM NaCl, and 2 mM MgCl2 in 40 mM citric acid/sodium phosphate buffer at pH 6 (Price et al., 2002; Debacq-Chainiaux et al., 2009). The proportion of senescent, SA-β-gal-positive cells was quantitated by scoring an average of 126 cells for each condition and cell line as determined from six non-overlapping images for each condition. Data acquisition was performed by the same researcher to avoid inter-person variability. Significance was established using the proportion Z-test. The Benjamini–Yekutieli correction to control the FDR was set at 5%. Adjusted p-values less than 0.05 are denoted with *, those less than 0.01 with **, and those less than 0.001 with ***. Graphs were generated using ggplot2 in R. For supporting histological analysis, Mallory’s tetrachrome was used, as previously described (Kraus et al., 2017).
Primary NP, AF, and FAT stromal cells derived from two donors were cultured for 5 days and analyzed as three technical replicates for different glucose or serum concentrations after normalization for the initial cell count at 0 h (Figure 3). All cell lines proliferated under reduced glucose concentrations at 10% FBS, and low glucose (1 g/L; D10SL) was generally tolerated well. Growth factors and other stimulants for mammalian cells in vitro are typically provided via fetal bovine serum (FBS) (Kraus et al., 2015). HI-FBS by Gibco used here contains ∼134 mg/dl glucose, which adds additional 13.4% glucose to the low glucose medium if used as a 10% medium supplement. When challenged under low glucose conditions, all cell lines tolerated an FBS reduction to 5%, with AF cells appearing most sensitive based on a total cell count. However, no live/dead assessment was conducted (Figure 3B). The results prompted us to use a low glucose medium (1 g/L) supplemented with 10% FBS (D10SL) as our standard to accommodate all three cell lines for the remaining experiments.
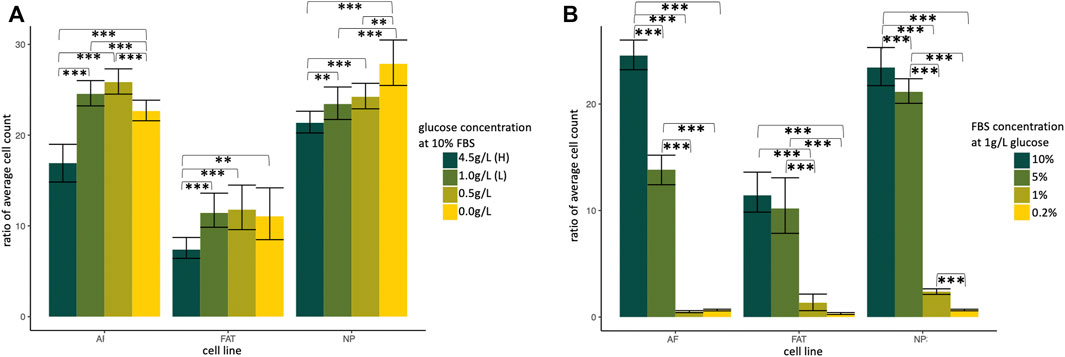
FIGURE 3. Normalized ratio of average cell count indicating cell proliferation of primary cells isolated from annulus fibrosus (AF), nucleus pulposus (NP), and subcutaneous adipose tissue (FAT). Cells were cultured in supplemented DMEM and subjected to a glucose (A) and growth factor (B) challenge. H, high; L, low; FBS, fetal bovine serum. Cells were isolated from two different donors and plated as three technical replicates for each cell line and condition.
RNASeq transcriptome analysis was based on three replicates for AF and NP cells and two replicates for FAT cells. All cells were derived from the same donor to avoid genotype-related expression differences. Focusing on the NP cell phenotype, we found a nearly equal number of transcripts higher or lower in NP compared to the AF and FAT cells. More genes were differentially expressed between NP and FAT compared to NP and AF cells in vitro (Figure 4A). Functional enrichment analysis (Figure 4B, Supplementary Figure S2) showed that transcripts identified as higher in the NP were predominantly involved with structural components of the ECM (GO:0005201), extracellular structure organization (GO:0043062), and cell adhesion (GO:0007155). Higher transcripts in NP over FAT cells showed enrichment for DNA binding transcription activator activity (GO:0001228). Amidst the lowest transcripts in NP over FAT cells were those involved in neurogenesis (GO: 0022008) and neuron differentiation (GO:0030182). Comparing NP to AF cells, transcripts higher in NP cells were associated with the molecular function of ECM structural constituent (GO:0005201) and Ca2+ ion binding (GO:0005509). A numerical summary of selected transcripts is provided in Figure 4C. Col2a1, Pax1, and Acan were among the top 30 transcripts higher in NP over FAT cells, Pax1 and Acan were in the top 30 transcripts lower in FAT over AF cells, and Col2a1was the second highest differentially expressed gene in NP over AF cells (Figure 4D). The top transcript higher in NP over FAT cells was Glis1. The lowest transcript levels in this grouping had Slc32A1. The highest transcript levels in FAT over AF cells were noted for Glp1r, and the lowest were noted for Brinp3. Selected transcripts were validated by RISH on cells in vitro and in vivo (Figure 4E), including Klhdc7a, a transcript higher in NP cells, encoding a not well-characterized Kelch-domain containing protein and two collagen transcripts, Col9a1 and Col11a2, also higher in NP cells (Figure 4E, Figure 5 and Supplementary Table S1).
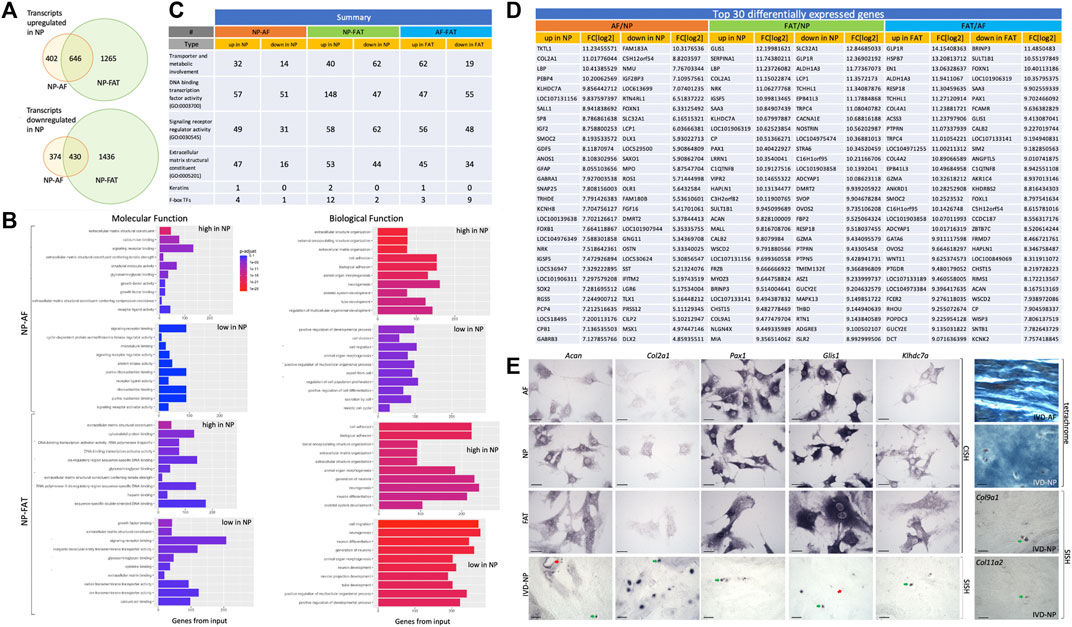
FIGURE 4. Overview of differentially expressed transcripts from annulus fibrosus (AF), nucleus pulposus (NP), and subcutaneous adipose (FAT)-derived primary cells (A). Bar graphs indicate the 10 top categories after functional enrichment for molecular or biological functions. Colors indicate the Benjamini–Yekutieli adjusted p-values. (B). Summary of RNASeq results (C). Top 30 differentially expressed transcripts (D) and validation of selected targets through RNA in situ hybridization and alkaline phosphatase reporter gene expression on cultured cells (CISH) and sections through the nucleus pulposus of the IVD (SISH) where green arrows point to positive and red arrows to negative cells. Tetrachrome staining of IVD sections provides ECM histology for reference (E). Scale bar represents 50 μm. Cells were isolated from a single donor to avoid the genotype effect. Cells were analyzed as three replicates for AF and NP and two replicates for FAT cells.
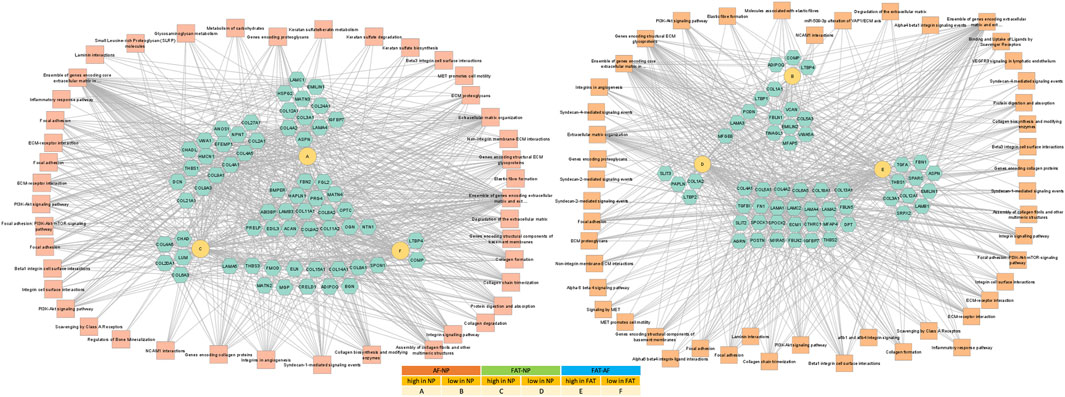
FIGURE 5. Network of differentially expressed transcripts between annulus fibrosus (AF) and nucleus pulposus (NP) cells of the IVD and adipose (FAT) stromal cells associated with the functional enrichment term extracellular matrix structural constituent (GO:0005201). Networks were generated with ToppCluster and analyzed in Cytoscape. Cells were isolated from a single donor to avoid the genotype effect. Cells were analyzed as three replicates for AF and NP and two replicates for FAT cells. Nodes are identified in the legend.
One of the top-ranking terms after functional enrichment analysis of the differentially regulated transcripts was ECM structural component (GO:0005201) (Figure 4B, Figure 5 Supplementary Table S1), with collagen as a major contributor. We found many collagen transcripts higher in NP cells upon moving into 2D culture [n = 15 (AF), n = 16 (FAT)] and fewer were lower [n = 2 (AF) and n = 9 (FAT)]. Col2a1, was found to be >2,000-fold higher in NP cells. Col1a1 was reduced in NP over AF cells, and Col1a2 expression was lower in NP versus FAT cells. A similar number of collagen transcripts was higher (n = 9) and lower (n = 7) in FAT over AF cells, with 7 of the higher transcripts identical for both IVD cell types, including two isoforms each for collagen IV and collagen V and Col4a1 and Col4a2 transcripts being the highest ECM related transcripts in FAT cells. Acan expression was 1,000x higher in NP over FAT cells and slightly elevated over AF cells, placing it in a group of transcripts higher in NP/IVD over FAT cells (Figure 5, Supplementary Table S1). Furthermore, in that group are transcripts for the large PG lubricin (Prg4) and other PG transcripts such as Npnt, Matn4, and Fbn2, along with transcripts for members of small leucine-rich proteoglycans (SLRPs), such as osteoglycin (Ogn) and prolargin (PRELP) (Figure 5, Supplementary Table S1). Transcripts for Abi3bp, a protein with glycosaminoglycan (GAG) binding affinity, and transcripts for the proteoglycan-link protein Hapln1 are considerable, and those for the SLRPs biglycan (Bgn) and fibromodulin (Fmod) were slightly lower in FAT cells. Other transcripts with a fold-change of ∼10 or higher in NP over FAT cells included Anos1 and Bmper. Vwa5a transcripts were higher in FAT and down in NP cells.
Pathway association-based gene regulatory trends were analyzed between IVD (NP, AF) and subcutaneous FAT-derived primary cells (Figure 6, Supplementary Table S2). Pparg was over 50x elevated in FAT over IVD cells. Transcripts higher in NP/IVD over FAT cells included Sox5, Sox6, Sox9, Bapx1 (Nkx3.2), Lmx1b, and Ntn1. Glis1 showed the highest transcript level in NP over FAT cells. Gli1 was higher in IVD over FAT cells and Gli2 in NP cells (Figure 6, Supplementary Table S2). Tbx1 (Tbox) transcripts were higher in NP than AF (10x) and FAT (239x) cells. We also found transcripts of six members of the Hox A, C, and D cluster higher in NP over FAT cells along with Pax9, involved in IVD patterning (Peters et al., 1999; Sivakamasundari and Lufkin, 2012; Sivakamasundari et al., 2017). Dlx1/Dlx2 transcripts were lower in NP/IVD over FAT cells and Msx1/Msx2 transcripts in NP over AF and FAT cells. Runx2 transcript levels were higher in FAT over IVD-derived cells, with a more than fivefold increase compared to NP cells (Supplementary Table S2). Sox10 transcripts were higher in NP cells. Pax3 and Pax6 transcripts were generally elevated in AF cells along with Foxn1 transcripts. Pax9 expression was lower in FAT cells (Figure 6, Supplementary Table S2). Furthermore, higher transcript levels in IVD over FAT cells were found for the semaphorins Sema3b and Sema4d, along with Mia and Gas6 transcripts. Tnfsr11b transcript levels were up to 478x higher in IVD over FAT cells. Another group of transcripts was lower in NP/IVD over FAT cells, with some of the highest fold changes observed for Sfrp2, Vgf, Fgf18, and Cxcl3 (Figure 7, Supplementary Table S3).
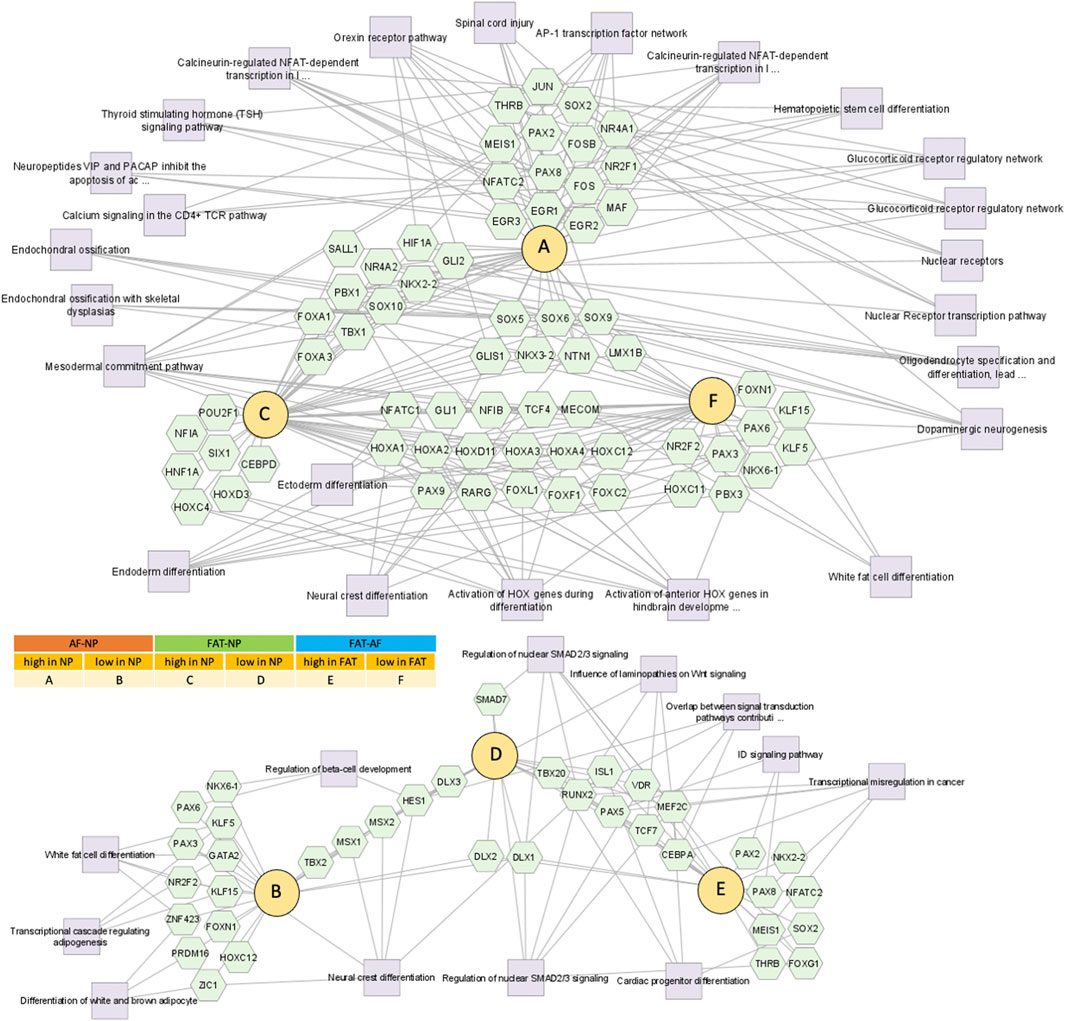
FIGURE 6. Network of differentially expressed transcripts between annulus fibrosus (AF) and nucleus pulposus (NP) cells of the IVD and adipose (FAT) stromal cells associated with the functional enrichment term DNA binding transcription factor activity (GO:0003700). Networks were generated with ToppCluster and analyzed in Cytoscape. Cells were isolated from a single donor to avoid the genotype effect. Cells were analyzed as three replicates for AF and NP and two replicates for FAT cells. Nodes are identified in the legend.
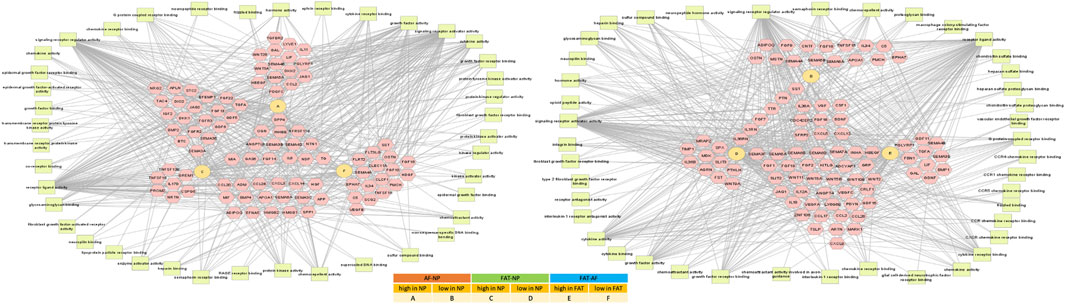
FIGURE 7. Network of differentially expressed transcripts between annulus fibrosus (AF) and nucleus pulposus (NP) cells of the IVD and adipose (FAT) stromal cells associated with the functional enrichment term signaling receptor regulator activity (GO:0030545). Networks were generated with ToppCluster and analyzed in Cytoscape. Cells were isolated from a single donor to avoid the genotype effect. Cells were analyzed as three replicates for AF and NP and two replicates for FAT cells. Nodes are identified in the legend.
The highest number of differentially regulated transcripts was found between NP and FAT cells (Supplementary Table S4. Transcript levels for Il12A, Il18 and its receptor, and Il21R, as well as Il1RL1, were increased in FAT cells. Expression of stromal cell-derived factor 1 (Cxcl12) was reduced in NP over FAT cells. Transcripts encoding the cytokines Il6, Cxcl14, and Cxcr4 were increased in IVD over FAT cells (Supplementary Table S4). Differentially expressed transcripts for metalloproteinases, such as matrix metalloproteinase (Mmp), disintegrin, and metalloproteinases (Adam), and those with thrombospondin motifs (Adamts) higher in IVD/NP over FAT cells included Adamdec1, Adam23, Mmp3, and Mmp13. More transcripts were higher in FAT cells such as Adamst14, Mmp9, Mmp12, and Mmp23b (Supplementary Table S5).
An up to a 5x increase in Slc2a1 transcripts encoding Glut1 was noted in NP over AF cells along with Slc2a3, Slc2a10, and Slc2a11 encoding further glucose transporters. FAT cells showed an up to 15x increase in Slc2a3 transcript levels over IVD cells (Figure 8; Table 3). An over 2x increase in G6pd expression in NP over AF cells was noted. Tktl1 transcripts were highest comparing NP to AF cells (2,400x) and increased over FAT cells (200x) (Figure 8A; Table 3). Furthermore, higher transcripts levels were noted in NP over FAT encoding two fructose-1,6-bisphosphate (F1,6PP) aldolase isoforms AldoA (2x) and AldoB (>4x). Transcripts of Eno2 encoding for an enolase isoform involved in generating phosphoenol-pyruvate (PEP) were lower in NP cells over AF and FAT cells (Table 3). Pyruvate kinase (Pkm) transcript levels were not significantly different. Transcripts for different isoforms of lactate dehydrogenase (Ldh) were differentially expressed in the three cell types (Figure 8A; Table 3). LdhA transcripts doubled in AF over FAT cells, whereas LdhC transcription was doubled in FAT over AF cells. NP cells showed an up to 18x increase in LdhD transcripts. Monocarboxylic transporter proteins (Mct) are encoded by the Slc16 gene family. Expression of Slc16a2 (Mct8) and Slc16a3 (Mct4) was increased >4x in NP over AF cells, and Slc16a2, Slc16a3, Slc16a6 (Mct6), and Slc16a7 (Mct2) transcript levels were higher in NP over FAT cells, with Slc16a2 transcripts being increased by 247x (Figure 8; Table 3). Transcripts for the gluconeogenesis associated fructo-bi-phosphatase 2 (Fbp2) were over 700x lower in NP versus FAT cells (Figure 8A; Table 3). A threefold increase in transcripts encoding pyruvate carboxylase (Pc) was seen in NP over FAT cells. However, transcripts for many enzymes converting the various metabolites of the Krebs cycle were lower in NP cells (Figure 8A; Table 3). Transcripts for the mitochondrial phosphoenolpyruvate carboxykinase 2 (Pck2) were reduced in NP over AF and/or FAT cells (Table 3). Me3 transcripts encoding a pyruvic-malic carboxylase (malic enzyme) were also lower in NP cells, and an up to 40x reduction of transcripts for the glutamate aspartate transporter 1 (Glast1) encoded by Slc1a3 was noted in NP cells. In contrast, transcript levels for ATP-citrate lyase (Acly) and malate dehydrogenase (Mdh) were not significantly different (Figure 8; Table 3). Transcripts for glutamate transporters (Glt) were higher in NP over FAT cells [Slc1a1 (5x), Slc1a2 (10x)]. Several other Glt transcripts were up to 17x higher in FAT over NP cells (Table 3). Transcripts for glutamate dehydrogenase (Glutd) were not increased in NP cells, nor were those for oxoglutarate dehydrogenase (Ogdh).
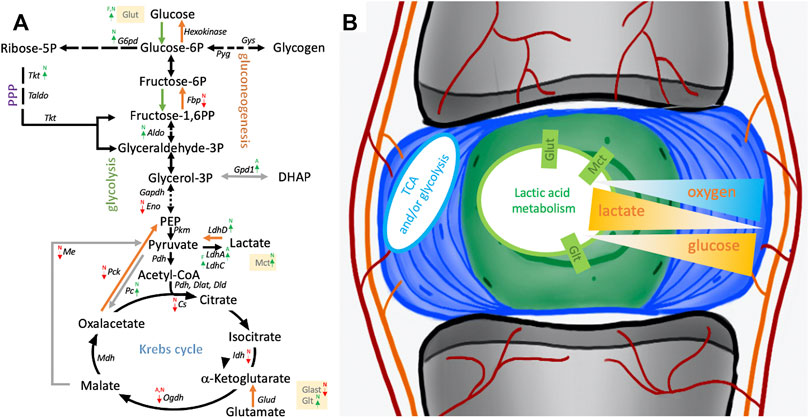
FIGURE 8. Schematic of pathways involving glucose and lactate metabolism. (A) Arrows indicate differentially expressed transcripts encoding for an enzyme or transporter in cells derived from the nucleus pulposus (N), annulus fibrosus (A), or adipose (F) tissue. (B) Illustrates NP niche conditions, metabolic pathways and gradients of crucial contributors to the NP niche condition alongside their transporters. Cells were isolated from a single donor to avoid the genotype effect. Cells were analyzed as three replicates for AF and NP and two replicates for FAT cells. P, phosphate; PPP, pentose phosphate pathway; TCA, tricarboxylic acid cycle.
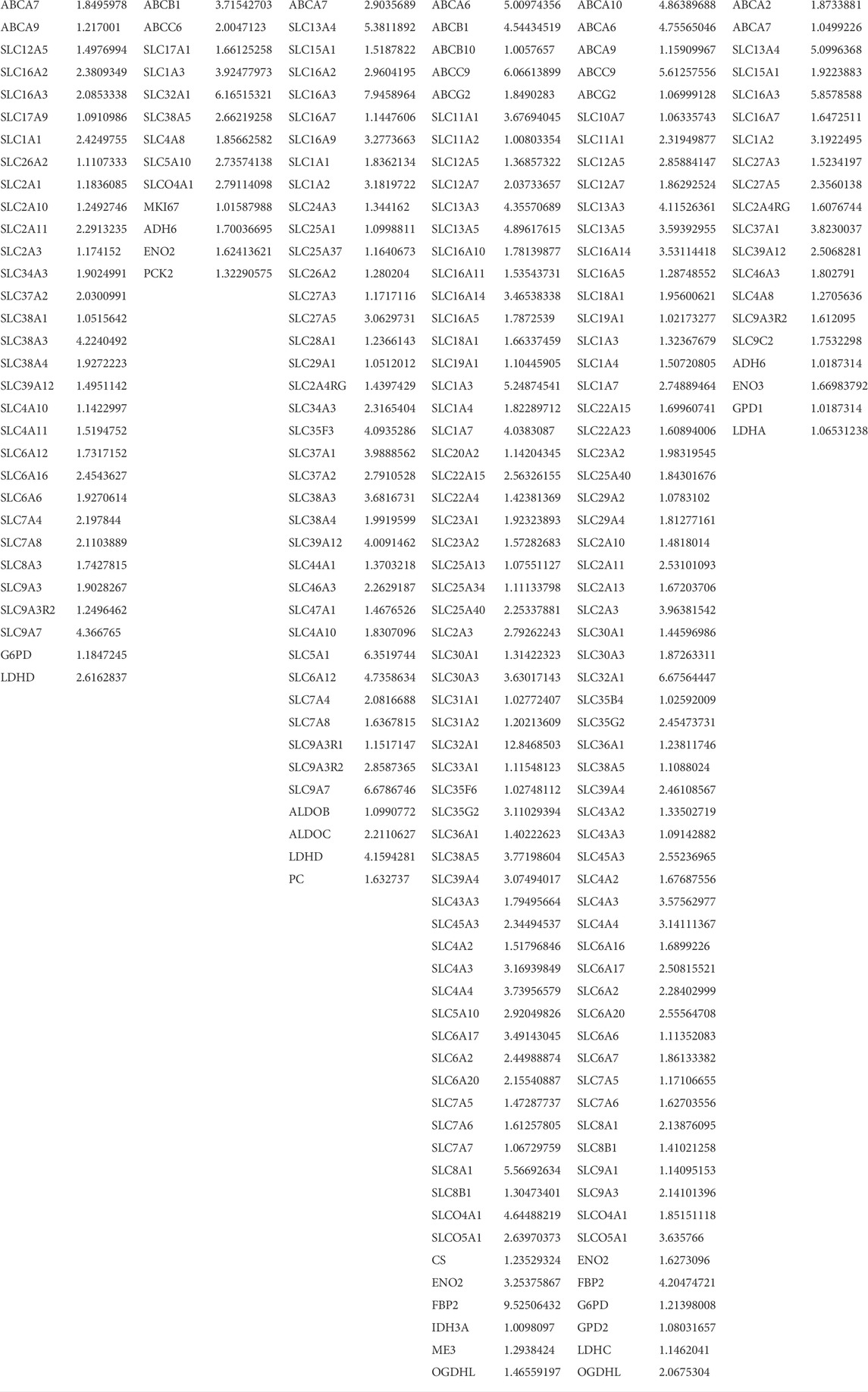
TABLE 3. List of differentially expressed genes between annulus fibrosus (AF) and nucleus pulposus (NP) cells of the IVD and adipose (FAT) stromal cells of the same donor associated with transporter or enzymatic function as it is related to glucose and lactate metabolism. Differences in transcript levels are displayed as log2 fold changes (FC[log2]).
We did not see a significant difference in Oct4 transcripts in 2D culture. However, transcript levels for its heterodimer partner Sox2 were over 128x higher in NP and FAT over AF cells. We also noted an increase in Lif expression in NP and FAT over AF cells and increased transcripts for the Lif receptor in NP over FAT cells (Table 5, Supplementary Table S2). Transcripts for Sox2/Oct4 downstream targets, such as Klf4 and Top2a, were slightly higher in NP over FAT cells, whereas Mras was slightly higher in FAT over IVD cells (data not shown). Transcript levels of Glis1 were high in IVD, especially NP over FAT cells (Figure 4D, Supplementary Table S2) when maintained in 2D culture with standard media. This was supported by CISH results (Figure 4E). Glis1 was expressed in a subpopulation of NP cells in sections of the IVD, as shown by SISH (Figure 4E, green arrow).
Twelve Fox-TF had higher transcript levels in NP over FAT cells, with FoxO4 levels doubled, alongside the transcript Prkaa2. No significant difference was seen for FoxO4 transcript levels between NP and AF cells (Figure 3D; Table 4). Fam183A was lowest in NP compared to AF cells (Figure 3D; Table 4). FoxF2 and FoxA3 transcription levels were higher in NP cells (Figure 9; Table 5). FoxP2 and FoxC2 were higher in NP over FAT cells, and FoxL1 was the highest Fox transcript in IVD cells, with a 410-fold increase over FAT cells. In this context, we found that P53 and P73 transcripts at least doubled in IVD over FAT cells (Table 5). P21 was not differentially expressed between the cell lines in 2D. A positive correlation between P53, P73, and Ki67 transcript levels was noted in all cell types, with higher levels in IVD over FAT cells, whereas P53-induced protein 11(TP53I11 or Pig11) showed a negative correlation and was lower in IVD over FAT cells. Perp and Ros1 expression were also lower in NP over FAT cells in 2D culture (Table 5).
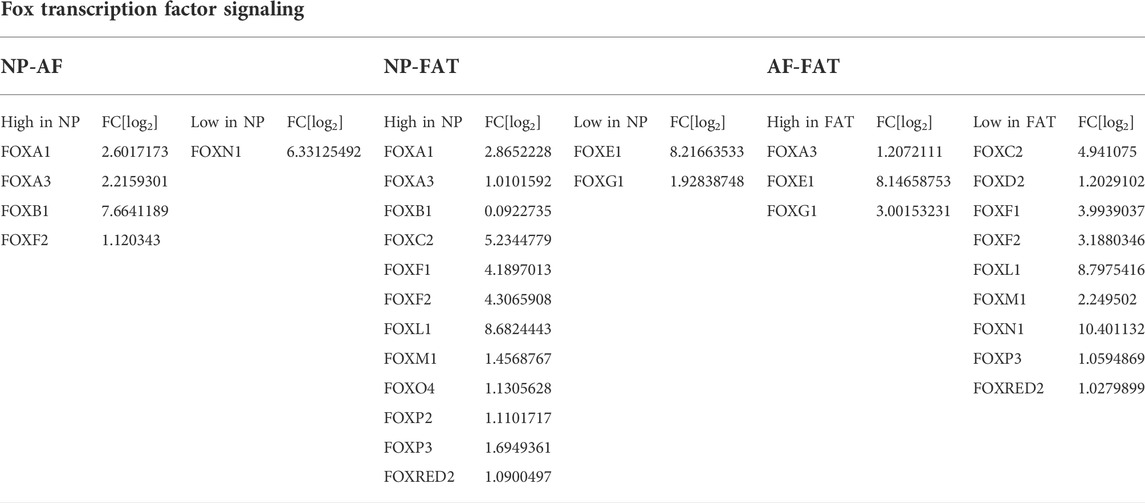
TABLE 4. Differentially expressed Fox transcription factors between annulus fibrosus (AF) and nucleus pulposus (NP) cells of the IVD and adipose (FAT) stromal cells of the same donor displayed by the log2 fold changes (FC[log2].
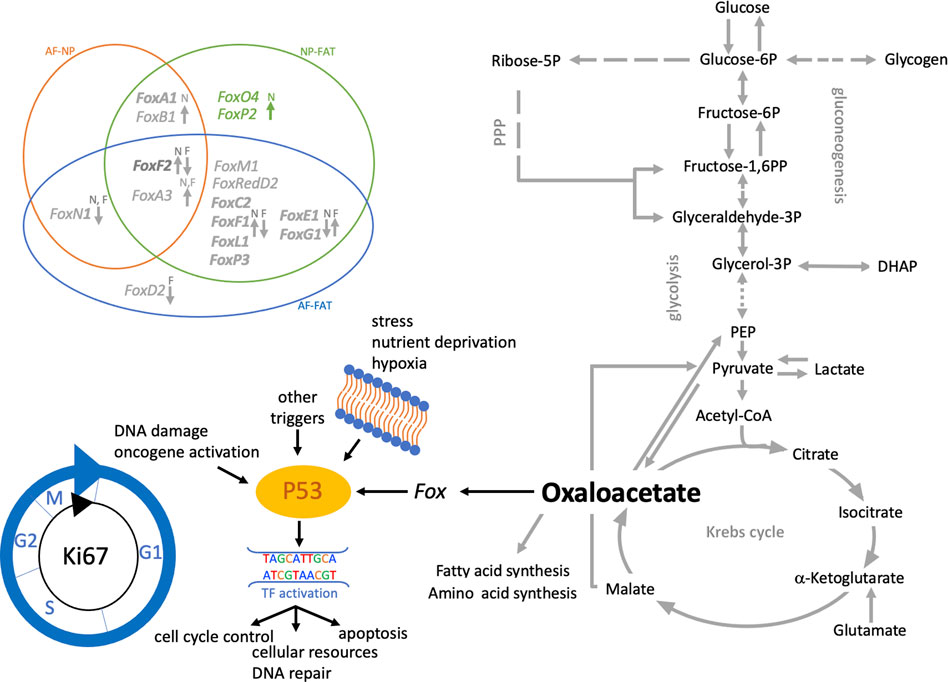
FIGURE 9. Schematic of pathways involving glucose and lactate metabolism leading to oxalacetate production, which modulates cell function through Fox transcription factors. Venn diagram shows the differentially expressed Fox genes between IVD (AF, NP) and adipose (FAT) derived cells. Those with a fold change of 3 or higher in NP over FAT. Arrows indicate up- or downregulation. The color scheme follows Table 3. Cells were isolated from a single donor to avoid the genotype effect. Cells were analyzed as three replicates for AF and NP and two replicates for FAT cells.
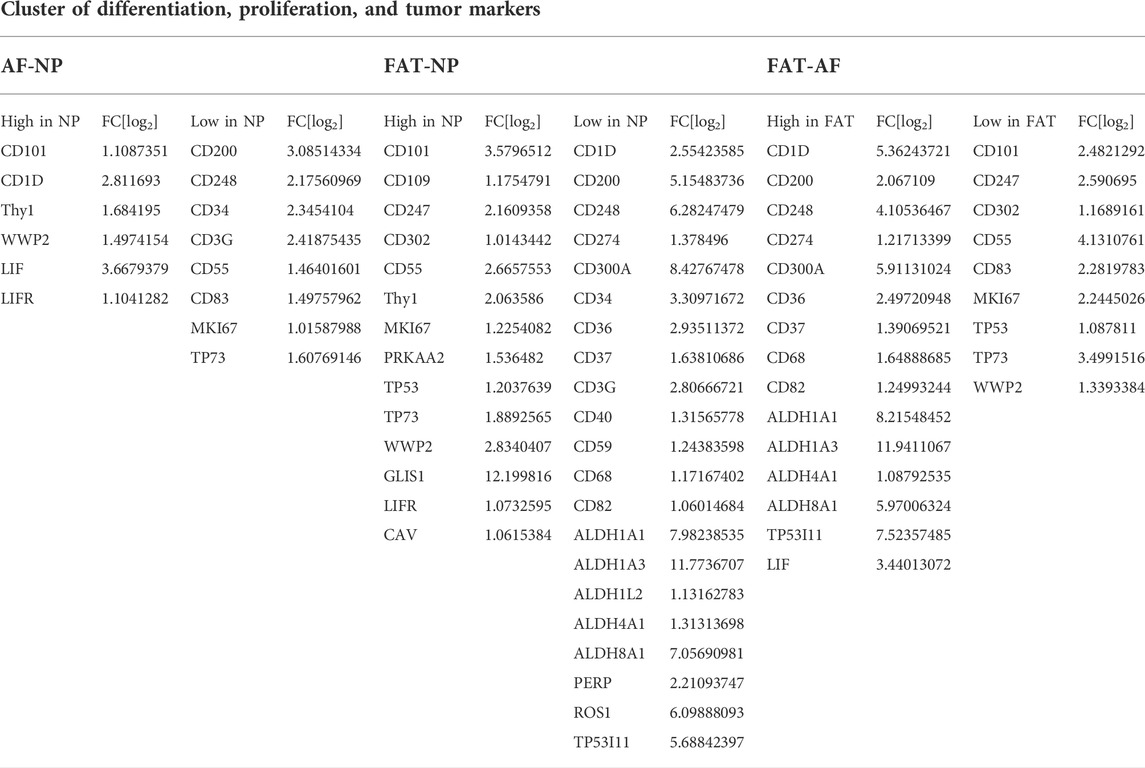
TABLE 5. Differentially expressed genes related to cell proliferation and differentiation between annulus fibrosus (AF) and nucleus pulposus (NP) cells of the IVD and adipose (FAT) stromal cells of the same donor displayed by the log2 fold changes (FC[log2]).
Several clusters of differentiation (CD) marker transcripts were higher in IVD over FAT-derived cells (Table 5). CD90 transcripts were fourfold higher in NP over FAT cells and twofold increased over AF cells. Transcripts encoding CD101 were higher in NP cells compared to AF and FAT cells (Table 5). CD109 transcript levels were higher in NP over FAT cells. Transcript levels for CD36, CD200, CD248, and CD300 were reduced in NP and/or IVD over FAT cells (Table 5). Lastly, aldehyde dehydrogenase (Aldh) transcripts encoding isoforms of Aldh1a1, Aldh1a3, and Aldh8a1 were ∼60–4,000x increased in FAT over IVD cells (Table 5).
NP and FAT cells in 3D culture were exposed to standard media (D10SL), degenerate conditions mimicking the IVD niche pH (D10DL), and chondrogenic conditions (D10CL) through supplements added to the D10SL media as described and subjected to BISH gene expression analysis. Cells were isolated from the same donor animal to avoid genotype-related expression differences. Three beads were used as technical replicates, and a minimum of 25 cells were analyzed for each gene, cell type, and condition (Figure 10A).
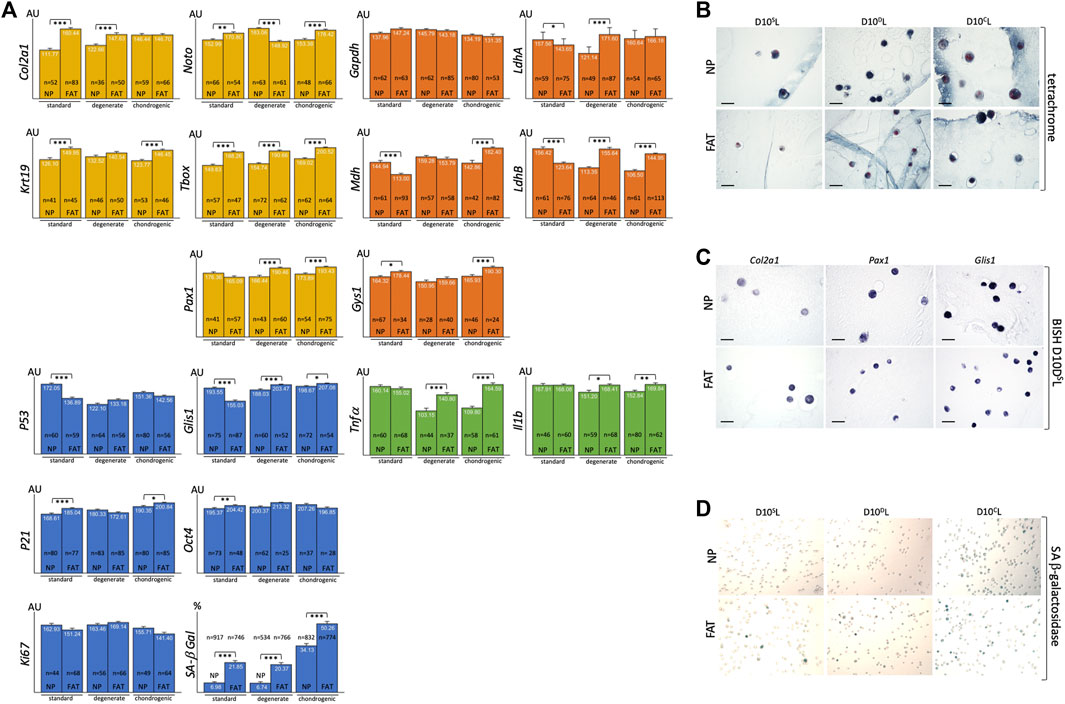
FIGURE 10. Bar graphs indicate RNA levels after BISH and ImageJ analysis of NP and FAT cells after 10 days of 3D alginate bead culture under different culture conditions for genes related to the nucleus pulposus (NP) (yellow), glucose and lactate metabolism (orange), inflammatory markers (green), cell proliferation and senescence markers, and SA-beta galactosidase staining (blue). Cells were isolated from a single donor to avoid genotype effects. A minimum of 25 cells were analyzed after subjecting three beads per probe, cell line, and condition to BISH (A). Tetrachrome-based histology of 7 μm paraffin sections of NP and FAT cells in alginate beads after 10 days of 3D alginate bead culture under different culture conditions shows the round cell morphology (B). Examples of 7 μm paraffin sections of NP and FAT cells after 10 days of 3D alginate bead culture under different culture conditions and BISH show AP-reporter gene activity for Col2a1, Pax1, and Glis1 expression (C). Example of images used for a cell count of NP and FAT cells after 10 days of 3D alginate bead culture under different culture conditions and subjection to SA-β-galactosidase stain (D). AU, arbitrary units; S, standard media D10SL. D, Media D10DL mimicking degenerate conditions. C, chondrogenic D10CL media. Significance is indicated as *** (p < 0.001); **(p < 0.01); *(p < 0.05); n indicates the number of individual cells analyzed for BISH. Scale bar represents 50 μm.
Using 1.2% alginate does not promote AF cell morphology in vitro (Figure 3E). The latter were therefore excluded from this comparison. A morphological change from flat, spread-out, to round was noted upon moving NP and FAT cells into the 3D alginate-based environment (Figure 4E, Figure 10B). In 3D culture using standard D10SL media, Col2a1 and Krt19 expression along with TFs such as T-box and Noto was higher in FAT over NP cells (Figure 10A). Transcripts for the metabolic enzyme Gys1 were also elevated in FAT over NP cells (Figure 10A). Transcript levels for LdhA, particularly LdhB and Mdh, were higher in NP over FAT cells (Figure 8, Figure 9, Figure 10A). Expression of Glis1 remained higher in NP over FAT cells, but Oct4 levels were now increased in FAT cells (Figure 4E, Figures 10A,C). P53 levels remained higher, and P21 levels were lower in NP cells. A higher proportion of senescent cells was noted in the FAT cell lineage in 3D culture as identified by SA β-galactosidase staining (Figures 10A,D). Transcript levels for the inflammatory markers Tnfα and Il1b were not significantly different between the two cell lines in 3D under standard conditions (Figure 10A).
To mimic some attributes of the degenerate NP, lactic acid was added to the culture medium for pH adjustment to pH6.5 (D10DL). Expression of Col2a1, T-box, and Pax1 was significantly higher in FAT cells, whereas Noto was now higher in NP cells. Resulting from a significant increase in FAT Pax1, yet a decrease in Noto expression between standard and degenerate conditions and an increase in NP Noto (Figure 10A, Supplementary Figure S1). Upon changing from standard to degenerate media, no significant changes in Glis1, Oct4, or Ki67 levels were noted in NP cells, whereas an increase was noted for all three transcripts in FAT cells, raising Glis1 and Oct4 expression in FAT cells above those in NP cells. The proportion of senescent cells remained significantly higher in FAT cells with no significant difference in P53, P21, or Ki67 transcripts between the cell lines. However, the addition of lactic acid reduced P53 and increased P21 expression in NP cells, whereas P21 levels decreased and Ki67 levels increased in FAT cells (Figures 10A,D, Supplementary Figure S1). For glucose/lactate metabolism-related enzymes, a significant increase for LdhA/B was noted in FAT over NP cells due to reduced expression in NP and an increase thereof in FAT cells when changing from standard to degenerate medium (Figure 10A, Supplementary Figure S1). Expression of Tnfa decreased significantly in NP cells between normal and degenerate conditions, with both inflammatory markers (Tnfα, Il1b) being significantly higher in FAT over NP cells (Figure 10A, Supplementary Figure S1).
Although Col2a1 transcript levels were similar between NP and FAT cells in chondrogenic (D10CL) conditions, they were at their highest for NP cells (Supplementary Figure S1). Krt19, Noto, T-box, and Pax1 expression was increased in FAT over NP cells, and the three TFs were at their highest level in FAT cells (Figure 10A, Supplementary Figure S1). From a metabolic perspective, transcript levels for Gys1, Mdh, and LdhB were significantly lower in NP over FAT cells (Figure 10A), and Mdh and LdhA/B transcripts were significantly increased in the FAT lineage between standard and chondrogenic conditions, whereas LdhB decreased in the NP lineage (Supplementary Figure S1). The proportion of senescent cells remained higher in the FAT lineage. However, for both cell lines, the highest proportion of senescent cells and P21 transcript levels was noted under chondrogenic conditions (Figure 10A, Supplementary Figure S1). Glis1 expression was slightly higher in FAT over NP cells (Figure 10A). Transcript levels for the inflammatory markers Tnfα and IL1b were significantly higher in FAT over NP cells under chondrogenic conditions.
Regenerative medicine employs cell-based therapies amidst other strategies (Kraus et al., 2021). Stem or progenitor cells are a valuable but sparse resource and typically require expansion in culture for quality control and enrichment purposes prior to therapeutic applications. For practical reasons, this expansion is most feasible in 2D monolayer culture, an often-scrutinized approach, as it represents an unnatural environment for cells removed from complex ECM-rich tissues. The use of adult mesenchymal stem or stromal cells is appealing, especially if environmental cues can be used efficiently for somatic cell reprogramming and differentiation. However, these processes occur in a lineage-specific manner, and more insight into the complicated, interrelated lineage relationship of adult stem cells is needed because their ability to differentiate into a particular cell lineage and long-term survival in an aging tissue are important (Zhou et al., 2019). Settling into an existing tissue niche challenges transplanted cells, especially if the niche is not a safe harbor.
Serum, typically FBS, delivers growth factors, some glucose, and other undefined components to promote life in vitro. Compositions vary between suppliers and production lots and can impact experimental outcomes (Kraus et al., 2018). A connection between growth factors, their effect on glucose metabolism, and cell proliferation exists (Vander Heiden et al., 2001). Glucose is an energy source for cells in vivo and in vitro under aerobic and anaerobic (lactic acid fermentation) conditions and serves as a building block for macromolecules; hence, the culture medium is typically enriched with glucose. However, the NP of a healthy mature IVD is avascular, with few cells residing in an abundance of ECM under low-nutrient conditions where lactic acid production lowers the pH (Diamant et al., 1968; Nachemson, 1969; Urban et al., 1977; Razaq et al., 2000; Razaq et al., 2003; Urban and Roberts, 2003; Bibby and Urban, 2004; Urban et al., 2004; Bibby et al., 2005; Wuertz et al., 2008; Wuertz et al., 2009; Gilbert et al., 2016). For survival, NP cells likely adapted their metabolism to these conditions. Previous findings in a different context showed that constant glucose availability and lactate removal alongside a likely oversupply of growth factors through routine medium changes does not reflect the in vivo situation of cells, especially not those of NP cells (Cantor et al., 2017). Nutrient concentrations of commonly used media often differ from those in human plasma (Cantor et al., 2017) and are frequently high in glucose (4.5 g/L), yet many stem or stromal cells, including those of the NP, perform better under low glucose conditions (Saki et al., 2013; Liu et al., 2020; Pang et al., 2020). Low glucose medium (1 g/L, 5.5 mM), including the glucose contribution from 10% FBS, is not yet considered pre-diabetic (10 mM) (Johnson et al., 2008; Wuertz et al., 2008; Turner et al., 2016; Lyu, 2022). In our limited and simplistic 5-day culture experiment with daily medium changes, NP cells could thrive on only the small amount of glucose provided by the serum, confirming studies by others (Wuertz et al., 2008). All cell lines tolerated a reduction to 5% FBS at low glucose concentrations (1 g/L); however, only a total cell count and no live/dead assay was performed. Reduced proliferation below 5% FBS at 1 g/L glucose is likely attributable to the reduced growth factors.
Observations in rat cells suggested higher proliferation of IVD cells than adipose stromal cells in 2D (Han et al., 2014). Here, using RNASeq transcriptome profiling on bovine cells, higher transcript levels in NP over FAT cells for P53, P73, Ki67, Glis1, Cav1, and Lif1R reflect this on a molecular level. Caveolin-1 (Cav1) can stimulate Akt signaling and increase glycolysis in cancer cells. It was suggested that low Cav1 expression correlates with high reliance on glucose metabolism (Nwosu et al., 2016). Hence, our 2D transcription data suggest that NP cells depend less on glucose than FAT cells and, therefore, would be at an advantage in the NP niche. Similarly, the expression of the cell proliferation marker Ki67 in standard and degenerate conditions alongside a lower percentage of senescent cells, as demonstrated by SA-β galactosidase staining in the NP lineage in 3D, also suggests a higher proliferation rate for NP cells.
Considered the two most abundant circulating carbon metabolites, glucose (5 mM) and lactate (1 mM) can be interconverted via glycolysis and gluconeogenesis (Rabinowitz and Enerback, 2020). The lower senescence rate of NP cells (7.15%) compared to FAT and AF cells (both ∼21%, AF data not shown) at 40 mM lactate further supports that NP cells are well adapted to the NP niche environment. AF cells challenged with 20 mM lactate showed modest decrease in viability in a similar experiment (Wang et al., 2021a). It was shown that lactate acts as a carbon source for various cell types (Hui et al., 2017; Wang et al., 2021a). Based on our existing data, we propose that NP cells in their unique niche developed an efficient metabolic adaption catering to the use of lactic acid over glucose and the Krebs cycle. High glucose might interfere with shared enzymes involved in these metabolic reactions. To further elevate lactate from waste to fuel status, it was recently shown that rabbit AF cells metabolize NP-produced lactate (Wang et al., 2021a). Recent findings in rat NP cells demonstrated that lactate-derived pyruvate is also introduced into the Krebs cycle (Silagi et al., 2020). In this context and given the increasing recognition of lactate as a metabolic driver (Warburg et al., 1927; Rabinowitz and Enerback, 2020; Wang et al., 2021a; Chen et al., 2021), maybe a reciprocal in vivo scenario might be envisioned where inner AF cells could fuel NP cells through glycolysis and lactate production.
Cells in the IVD and FAT are of mesoderm origin, yet terminal differentiation follows different paths. Advanced omics technologies in IVD research allow for transcriptome profiling at a single and cell population average level (Hickman et al., 2022). Despite the lack of natural signaling cues in monolayer culture, transcripts of the three cell lines (NP, AF, and FAT) in early 2D culture reflect their mesoderm origin alongside their lineage profile through overlapping and distinct molecular signatures. Our in vitro transcriptome data largely support biomarkers previously identified for NP and AF cells in vivo (Li et al., 2019a; Li et al., 2019b), except that Snap25 was higher in NP over AF cells in vitro, which could be a 2D culture effect (Minogue et al., 2010; Li et al., 2019b).
Transcription factors (TF) are primary regulators of gene expression. Their profile indicates cell lineage commitment and responses to environmental cues. As protagonists of chondrogenesis, the Sox-trio (Sox5, Sox6, and Sox9), hedgehog, and Tgfβ signaling antagonize adipogenesis (Lefebvre et al., 2001; Ikeda et al., 2004; Lee et al., 2017; Zhou et al., 2019). Transcripts of the Sox-trio along those of Shh-mediators Gli1 and Gli2 were higher in NP cells along with Tbx1 transcripts encoding the T-box containing protein brachyury, a clinical marker for chordoma (Dai et al., 1999; Vujovic et al., 2006; Chen et al., 2020), indicating NP cell linage commitment alongside higher transcript levels for Bapx1 (Nkx3.2) an important player in skeletal development (Tribioli and Lufkin, 1999; Lefebvre et al., 2001; Chatterjee et al., 2014; Lee et al., 2017); Lmx1b known for its role in dorsal limb mesenchyme patterning (Loomis et al., 1998; Kraus et al., 2001) and Ntn1 encoding a protein involved in tumor progression and apoptosis control (Mazelin et al., 2004). Dlx1/Dlx2 and Msx1/Msx2 transcripts, regulators of cell lineage commitment during nervous system and bone formation (Kraus and Lufkin, 1999), were lower in NP cells, as was Pparg, which is crucial for adipogenesis (Rosen and MacDougald, 2006) and higher in FAT over IVD cells. Surprisingly, in 2D culture, Runx2 transcript levels were higher in FAT over IVD-derived cells. Runx2 is important for the proliferation of osteoblast progenitors (Kawane et al., 2018), and its loss-of-function in primary chondrocytes facilitates differentiation along the adipogenic lineage in vitro (Enomoto et al., 2004). Elevated Runx2 expression could indicate a phenotypical adaption of FAT cells to the monolayer culture condition as a stretched morphology appears to favor differentiation toward an osteoblast cell type (Lu et al., 2007). Sox10 transcripts encoding a TF implicated with neural crest and peripheral NS system development acting synergistic with Pax3 were higher in NP cells. However, Pax3 and Pax6 transcripts were generally elevated in AF cells along with Foxn1 encoding a TF that, together with Pax1, plays a role in thymus development. Pax1 and Pax9 are well known for their role in IVD patterning (Wilm et al., 1998; Peters et al., 1999; Sivakamasundari and Lufkin, 2012; Sivakamasundari et al., 2013; Sivakamasundari et al., 2017), and Pax9 was generally downregulated in FAT cells.
Signaling factors are both at the giving and receiving end of differential gene expression. Responding to extracellular cues and channeling the information to the nucleus, they trigger transcriptome changes to direct cell function. Transcripts encoding the semaphorins Sema3b and Sema4d, cell signaling peptides involved in tissue homeostasis (Janssen et al., 2010); Mia, a growth factor involved with ECM organization and neural crest differentiation (Blesch et al., 1994); Gas6 a stimulator of cell proliferation (Gomes et al., 2019); and Tnfsr11b known to inhibit the formation of osteoclasts (Yamaguchi et al., 1998) were all higher in NP over FAT cells. In contrast, transcripts encoding the nerve growth factor VGF; transcripts for Fgf18, a pleiotropic growth factor involved in neurite outgrowth in vitro; and Cxcl3, a cytokine promoting angiogenesis and connective tissue reorganization, were lower in NP cells, reflecting their non-vascularized and non-innervated source tissue.
Importantly, the mature cells have not lost the ability to produce ECM in vitro. Many of the transcripts higher in NP/IVD over FAT cells were associated with the GO terms ECM components, ECM homeostasis, and ECM remodeling, alongside suppression of neurogenesis and angiogenesis, as one would expect given the natural environment of NP cells. Col1a1, predominantly in the AF in vivo, was reduced in NP over AF cells and Col2a1, predominantly the NP in vivo, was higher in NP cells as expected (Li et al., 2019b). Transcripts for proteoglycans (PG) such as aggrecan (Acan) and other large and small proteoglycans remained higher in the NP/IVD over FAT cells in monolayer culture. PGs are heavily glycosylated proteins and crucial ECM components alongside collagens, especially in the IVD (Eyre and Muir, 1976; Bushell et al., 1977). Many ECM-PGs have signaling and biomechanical properties (Wilda et al., 2000; Chen et al., 2017). Being of negative charge and involved in maintaining osmotic pressure, especially the bottlebrush PG Acan, plays an important role in the IVD niche (Bibby et al., 2001; Singh et al., 2009). SLRPs are also bioactive components of the IVD. Ogn, with one of the highest transcript levels in NP over FAT cells, impacts proliferation and apoptosis in fibroblasts (Deckx et al., 2016). Other transcripts with higher levels in NP over FAT but less studied in the context of the IVD ECM were Anos1, encoding anosmin-1 associated with tumor progression (Choy et al., 2014), and Bmper, encoding for a protein that inhibits Bmp signaling, therefore impacting many important signaling cascades critical in chondrogenesis (Yoon and Lyons, 2004; Manzari-Tavakoli et al., 2022). Vwa5a transcripts encoding a tumor suppressor (Zhou et al., 2009) were upregulated in FAT over NP cells. Aside from impacting ECM organization and collagen fibrillogenesis, some of these proteins engage in inflammatory response pathways and the phosphatidylinositol-3-kinase (PI3K)/Akt and mammalian target of rapamycin (mTOR) signaling pathway critical for cell cycle regulation (Ryu and Lee, 2017). Surprisingly, transcripts lower in NP over FAT/AF cells were associated with cell migration, despite NP cells being very mobile in monolayer culture (Kraus et al., 2019b).
Fernandes et al. (2020) were among the pioneers applying single-cell RNASeq to analyze the human IVD, identifying SFRP1, BIRC5*, CYTL1, ESM1, and CCNB2* as AF specific and COL2A1*, DSC3, COL9A3*, COL11A1*, and ANGPTL7 as NP-specific after enzymatic cell isolation and 7–8 day culture in DMEM/F12/10% FBS supplemented with 20 μG/ml ascorbic acid. Our data are derived after non-enzymatic cell isolation and in vitro expansion without ascorbic acid. Markers identified by Fernades and marked with an asterisk were also expressed with significant differences between AF/NP cells in our bovine data set, alongside 9/20 AF and 6/20 NP markers of their top 20 lists. Very recent single-cell RNASeq data from human NP cells isolated from healthy and degenerate human NP tissue after a multi-step isolation protocol to clarify chondrocyte fate and differentiation advocates that NC lineage cells evolve and take on the cell fate of different chondrocyte subgroups once the IVD undergoes degeneration (Han et al., 2022). Due to a fundamentally different approach, we cannot directly compare expression patterns. However, our transcriptome data suggest that NP cells, often referred to as chondrocyte-like, indeed share chondrocyte attributes but continue to express the NC cell marker Tbx1 in 2D and 3D culture, reflecting their NC origin (Herrmann et al., 1990; Tang et al., 2012; Nibu et al., 2013). It remains to be investigated if our procedure of isolating NP stromal cells selects for NCNP cells. A comparison with markers of another recent single-cell RNASeq project highlighting the heterogeneous nature of NP cells is hampered due to their differential expression analysis between NP and inner AF tissue, also referred to as the transition zone (Cherif et al., 2022). In summary, the expression of key transcription and signaling factors, along with ECM components, allowed distinguishing the three lineages based on distinct transcript signatures.
Metalloproteinases, such as Mmp, Adam, and Adamst, are important in ECM remodeling and associated with cancer and other diseases (Edwards et al., 2008; Jablonska-Trypuc et al., 2016). Differentially regulated transcripts upregulated in IVD/NP over FAT cells included Adamdec1, encoding a secreted catalytic protein involved in cell competition (Yako et al., 2018); Adam23, encoding a non-catalytic metalloprotease-like protein involved in cell–matrix and cell–cell interaction; Mmp3, encoding an enzyme degrading several collagens, PGs, fibronectin, and laminin (Wan et al., 2021); and Mmp13, encoding Mmp cleaving predominant type II collagens. More transcripts were differentially upregulated in FAT cells, such as Adamst14, encoding Adamst involved in type I procollagen cleavage (Bekhouche and Colige, 2015); Mmp9, encoding an enzyme that degrades type IV and type V collagens and can activate Il1b (Forsyth et al., 1999; Van den Steen et al., 2002); Mmp12, encoding an inactivator of Ifnγ (Collison, 2018); and Mmp23b, encoding a protein associated with GPCR pathways (Supplementary Table S5). This suggests that in vitro, during early 2D culture, the association between generated ECM components and the associated remodeling enzyme is preserved on a transcription level.
ECM remodeling and homeostasis are important aspects of developing and mature organisms. Metalloproteases were expressed by all three cell lines and enabled this process, some specific to the type of ECM molecule they produce. Interestingly, Adamdec1 upregulated in NP cells is considered to provide a competitive edge when adjacent cells fight for survival and space and could be a further indicator that NP cells did develop unique survival mechanisms (Yako et al., 2018).
Degenerative diseases, such as IVDD, rheumatoid, or osteoarthritis, are of complex origin, including multiple comorbidities and other factors, eventually resulting in a chronic inflammatory pain-inducing stage (Klodzinski and Wislowska, 2018; Swain et al., 2020; Lufkin et al., 2021; Parenteau et al., 2021). Regenerative cell therapies targeting these diseases should not further aggravate such a situation. The generation of pro-inflammatory cytokines, such as Il1β, Il6, and Tnfα, that are linked to pathological pain should be avoided (Zhang and An, 2007). However, cytokines with immune suppressive effects could be beneficial. MSCs are attributed to anti-inflammatory effects (Ghannam et al., 2010), whereas no such evidence was previously available for IVD/NP cells (Lyu, 2022). We provide data based on differential RNA expression analysis in 2D and 3D culture for cells from the same donor that IVD cells, with a focus on NP cells, also have beneficial features.
Cytokines mediate inflammatory responses in various ways. Transcripts for the pro-inflammatory cytokine Il12A, a mediator of graft versus host disease (Bastian et al., 2019), Il18 and its receptor, Il21r, and Il1rl1 were lower in NP over FAT cells. Il21 signaling involves the Janus kinase and Signal Transducer and Activator of Transcription (JAK-STAT), phosphoinositide 3-kinase (PI3K), and mitogen-activated protein kinase (MAPK) pathway and is considered to have immune suppressive effects (Leonard and Wan, 2016). The Toll-like receptor Il1rl1 appears to convey anti-inflammatory effects in collagen-induced arthritis (Xu et al., 2008). Moreover, transcripts for the angiogenesis promoting and MSC recruiting stromal cell-derived factor 1 (Cxcl12) were lower in NP cells (Takano et al., 2014), whereas transcripts encoding the cytokine Il6 with impact on inflammation, bone homeostasis, and angiogenesis; Cxcl14, acting as an angiogenesis inhibitor (Hromas et al., 1999; Shellenberger et al., 2004); and Cxcr4, encoding the receptor for Cxcl12, were higher in NP cells. This likely renders monolayer cultured adipose stromal cells preferable over NP-derived cells for cell replacement therapies. However, increased expression of Tnfα and IL1b was noted in 3D culture in FAT versus NP stromal cells under degenerate or chondrogenic conditions, which puts adipose stromal cells in a less favorable position for use in NP niche refurbishment.
While the physiological pH of most body fluids is in the neutral range (pH 7.4) (Razaq et al., 2003), the IVD niche is slightly acidic (∼pH 7.1) owing to anaerobic lactic acid fermentation and PG-mediated proton retention. This value drops further in the degenerate stage (∼pH 6.5) (Diamant et al., 1968; Nachemson, 1969; Urban et al., 1977; Bartels et al., 1998; Razaq et al., 2000; Razaq et al., 2003; Urban and Roberts, 2003; Bibby and Urban, 2004; Urban et al., 2004; Bibby et al., 2005; Wuertz et al., 2008; Wuertz et al., 2009; Gilbert et al., 2016; Wang et al., 2021a). The presence of stem or progenitor cells in the IVD was demonstrated previously (Risbud et al., 2007; Kraus et al., 2017), and the acidic microenvironment was generally found to negatively impact cell survival, proliferation, and ECM production. Most studies were conducted for the time frame of about 1 week with HCl/NaOH-mediated pH adjustment analyzing various MSC sources, including subcutaneous FAT, bone marrow, and the IVD from diverse host organisms with effects likely mediated through acid-sensing ion channels (ASICs) (Razaq et al., 2003; Wuertz et al., 2008; Wuertz et al., 2009; Li et al., 2012; Han et al., 2014; Gilbert et al., 2016; Liu et al., 2017). As reviewed recently in detail by Lyu (2022), differences in coping with pH-related and other challenges exist between MSC and IVD cells. However, acidity impairs the function of both cell types. None of these findings are disputed here. However, we did not identify differentially expressed transcripts for ASICs between the three stromal cell lines in standard 2D culture. We noted higher levels of NP transcripts associated with the molecular function of Ca2+ binding, supporting a rising interest in ion channel signaling in the IVD (Sadowska et al., 2019).
Lyu further mentioned that insufficient clearance in the IVD results in lactic acid waste accumulation. However, due to sparse information about the response of IVD progenitors, conclusions had to be drawn from MSC (Lyu, 2022). Wang et al. (2021a) suggested that lactic acid clearance, in part, might happen through lactate uptake and metabolism by neighboring AF cells. Our analysis in 3D alginate beads used lactic acid for pH adjustment. With lactic acid as a proton and carbon source, we noted an increase in Col2a1 transcripts in NP and a decrease in FAT cells. We further noted changes in the expression of key metabolic enzymes as described below and proposed that such an adaption might support the production of OAA in NP cells, a metabolite and signaling molecule linked to longevity (Lehmann et al., 2003; Williams et al., 2009; Martins et al., 2016).
Intrigued by the low glucose preference, a natural solute gradient in the IVD (Holm et al., 1981), and the hardiness of NP-derived cells, molecular cues might identify metabolic adaptions that facilitate NP cell survival in the NP niche (Figure 8). The nutrient import starts with members of the solute carrier (Slc) superfamily of transporters. The Slc2 gene family encodes for Glut integral membrane proteins that facilitate hexose and sugar alcohol transport across membranes (Mueckler and Thorens, 2013). Glut1, encoded by Slc2a1, transports glucose, hexoses, glucosamine, and reduced ascorbic acid and is present in NP cells (Rajpurohit et al., 2002; Carruthers et al., 2009). One might assume that levels are tightly regulated as Glut1 haploinsufficiency is pathological in humans, whereas overexpression was linked to tumorigenesis (Yamamoto et al., 1990). Despite the glucose transporter Glut1 being highly expressed in NP tissue (Richardson et al., 2008), recent work has suggested that Glut1 is dispensable and that neither glutamine nor fatty acid oxidation is used as an alternative under low glucose conditions. This further supports the idea of lactate as a key metabolite in NP cells (Hui et al., 2017). While Slc2a1 transcripts were higher in NP over AF cells, Slc2a3 transcripts encoding for Glut3, with a high affinity for D-glucose (Manolescu et al., 2007), were higher in FAT cells, indicating and supporting higher dependence of FAT cells on glucose. Important cell conditions affecting PPP activity are a high proliferation rate and the need for NADPH. Numerous studies have revealed a significant upregulation of glucose-6-phosphate dehydrogenase (G6pd) in tumor cells (Kowalik et al., 2017). G6pd transcripts were higher in NP over AF cells. The enzyme drives the first rate-limiting step of the oPPP, providing the electron donor NADPH and initiating the conversion of glucose-6-phosphate (G6P) to ribose-5-phosphate (R5P), an important building block for nucleotides and as such nucleic acids, especially in proliferating cells. The noPPP leads to metabolites, including glyceraldehyde-3-phosphate (G3P), that can feed into glycolysis. Transketolase (Tkt) and transaldolase (Taldo) are key enzymes of the noPPP (Jin and Zhou, 2019). Tktl1 showed the highest transcript levels in NP over AF cells and was higher over FAT cells along with transcripts encoding two fructose-1,6-bisphosphate (F1,6 PP) aldolase isoforms AldoA and AldoB, enzymes that mediate the conversion of F1,6PP to G3P and dihydroxyacetone-phosphate (DHAP) during glycolysis and fructose metabolism (Gamblin et al., 1991). Eno2 transcripts, encoding for an enolase isoform involved in generating PEP, were lower in NP cells, whereas Pyruvate kinase (Pkm) transcripts were not affected (Figure 8).
Lactate, a less oxidized form of pyruvate, is classically considered a waste product of glucose metabolism but recently gained attention as a circulating carbon source and metabolic driver in a niche environment (Warburg et al., 1927; Valvona et al., 2016; Hui et al., 2017; Rabinowitz and Enerback, 2020; Wang et al., 2021a; Chen et al., 2021). Transcripts for different isoforms of lactate dehydrogenase (Ldh) were differentially expressed in the three cell types, with NP cells showing higher LdhD transcripts. LdhA has a higher affinity to pyruvate over lactate, whereas LdhD converts lactate to pyruvate (LeVan and Goldberg, 1991; Valvona et al., 2016). The Ldh activity is associated with the monocarboxylic transporter protein (Mct) activity, which transports lactic acid and pyruvate across plasma membranes. The transporters are encoded by the Slc16 gene family (Jitrapakdee et al., 2006). Several Mct encoding transcripts were higher in NP cells, especially Slc16a2. Gluconeogenesis-associated fructo-bi-phosphatase 2 (Fbp2) transcripts were over 700-fold lower in NP versus FAT cells, suggesting gluconeogenesis from lactate is not a prominent feature of NP cells. Interestingly, higher transcripts encoding pyruvate carboxylase (Pc) in NP over FAT cells could aid NP cells in refueling the Krebs cycle; however, transcripts for many enzymes converting the various metabolites of the Krebs cycle were lower in NP cells. Similarly, transcripts encoding enzymes that would allow for alternative pathways or shuttle mechanisms were lower in NP cells (Figure 8).
Generated by condensation of pyruvate or PEP with CO2, the oxidation of malate or deamination of aspartate, the metabolite OAA wears multiple hats. However, malate conversion to OAA was described as a thermodynamically unfavorable step in the Krebs cycle (Wang et al., 2021a). Being an intermediate in the Krebs cycle and participating in gluconeogenesis, OAA could serve as an energy source. Nevertheless, based on carbon tracing, this does not appear to be its major function in NP cells (Silagi et al., 2020). Our transcriptome-based data, however, suggest that metabolic adaptions in NP over FAT and AF cells might channel some lactate into OAA production. Transcripts for the mitochondrial phosphoenolpyruvate carboxykinase 2 (Pck2) facilitating the conversion of OAA to PEP when producing glucose from lactate were lower in NP over AF and/or FAT cells. Me3 transcripts encoding a pyruvic-malic carboxylase (malic enzyme) were also lower in NP cells along with Slc1a3 transcripts encoding the glutamate aspartate transporter 1 (Glast1), whereas transcript levels for ATP-citrate lyase (Acly) and malate dehydrogenase (Mdh) showed no significant difference between the cell lines, altogether supporting reduced importance of mitochondrial activity in NP cells (Gan et al., 2003; Madhu et al., 2020) and an adaption that favors a lactate based metabolism. Since most amino acids were provided through the medium, the need for shuttle mechanisms might not have arisen. Transcripts for glutamate dehydrogenase (Glutd) were not increased in NP cells, nor were those for oxoglutarate dehydrogenase (Ogdh). However, existing levels might be sufficient, or many of those enzymes could be regulated at the protein level. From a transcriptome point, metabolic adaptions in the NP cells appear to channel lactate into OAA production based on higher Pc transcript levels in NP cells, encoding pyruvate carboxylase, an enzyme that converts pyruvate to OAA (Figure 8).
An OAA-induced lifespan extension through the FoxO/Ampk signaling pathway was suggested in C. elegans. FoxO proteins have an important role in development, are connected to many signaling pathways, and have a complex role in cell longevity and tumor suppression (Lehmann et al., 2003; Williams et al., 2009; Martins et al., 2016). OAA can trigger Fox expression, which impacts P53/P21 signaling and possibly enables NP cells to survive under NP niche conditions (Figure 9). Fox-TFs showed higher transcript levels in NP cells, including FoxO4 alongside Prkaa2, encoding a subunit of the AMP-activated protein kinase (Ampk) in vertebrates (Hardie and Carling, 1997). Fam183A, the top downregulated transcript in NP over AF cells, is speculated to be a target for FoxO4 binding. FoxP2 is associated with Wnt, Hedgehog, and Notch signaling. FoxF2 has inhibitory effects in many tumors and might bind to promoters of ECM-related genes such as Acan and Col2a1 during vertebrate development (He et al., 2020), both NP biomarkers in vitro and in vivo (Li et al., 2019b). FoxC2 is a TF involved in mesenchyme development and tumor progression (Mani et al., 2007). FoxA3 is associated with glucose metabolism homeostasis and longevity (Ma et al., 2014). FoxL1 was the most upregulated Fox transcript in IVD over FAT cells. Although FoxL1 overexpression is associated with gliomas (Chen et al., 2019), the FoxL group of TFs is less well studied, but a connection between FoxL1 and cell survival was established through the PI3K/Akt/mTor signaling pathway, which was impacted upon through the differential expression of ECM molecules such as Col9a1 (high in NP cells) and Col1a1 and LamC2 (both lower in NP cells) (Ryu and Lee, 2017). Crosstalk between Fox-TFs and P53 was suggested (Dijkers et al., 2000; You and Mak, 2005). In this context, we found that P53 and P73 transcripts at least doubled in IVD over FAT cells. As a tumor suppressor, P53 protects cells by controlling progression through the cell cycle and allowing DNA repair (Figure 9) (Levine, 1997; May and May, 1999). Cell cycle arrest is arranged through transcriptional activation of P21 by arresting cells in G1, promoting differentiation over proliferation (Harada and Ogden, 2000). P21 was not differentially expressed in 2D. In adult stem cells, reduced P53 aids in reprogramming (Marion et al., 2009; Maeso-Alonso et al., 2021), whereas P73 aids in controlling cell cycle progression and plays additional rolls in cell–adhesion and cell–ECM interactions (Maeso-Alonso et al., 2021). A positive correlation between P53, P73, and Ki67 transcript levels was noted in all cell types, with higher levels in IVD over FAT cells, whereas P53-induced protein 11 transcripts (TP53I11, Pig11) (Gu et al., 2021) showed a negative correlation and were downregulated in IVD over FAT cells. Perp encoding an effector in the TP53-dependent apoptotic pathway was also downregulated in NP over FAT cells in 2D culture. In this context, the upregulation of Ki67, a cell proliferation marker with gradually increasing transcript and protein levels throughout the cell cycle (Li et al., 2021), in NP over FAT cells might indicate that P53/P73 expression in IVD cells is not inducing the apoptotic pathway; instead, it is slowing down cell cycle progression, potentially facilitating somatic cell reprograming (Figure 9). Ros1 transcripts for a proto-oncogene upregulated in many tumor cell lines were lower in NP over FAT cells, suggesting that increased proliferation in NP cells is not due to an aberrant phenotype.
Ideally, OAA concentration measurements would support our findings. However, currently available enzymatic tests are not suitable for using DMEM-based medium according to the manufacturer’s instructions (BioAssay Systems). In this context, a detailed metabolic assessment studying lactate efflux through carbon isotope labeling in rat NP cells indicated, based on isotope recovery in glutamate, that the majority of lactate-derived pyruvate is introduced into the Krebs cycle via Pdh and acetyl-CoA without detectable Pc and OAA contribution to glutamate production (Silagi et al., 2020). This challenges our transcriptome-based findings, which identified higher Pc transcripts in NP over FAT cells in vitro; yet reduced transcription of enzymes of the PDH complex and Krebs cycle or Pck. In support of our hypothesis that OAA could promote longevity in NP cells, the differential expression of transcripts for the aforementioned metabolic enzymes, along with differential Fox gene expression and a generally lower proportion of senescent cells among NP-derived stromal cells in all tested conditions, suggests such a mechanism as feasible. Therefore, we suggest that molecular mechanisms for the generation of OAA as a signaling molecule through extended lactate metabolism could be in place in NP cells. This could be a minor contribution irrelevant or not favored for glutamate synthesis. OAA was recognized as a bioenergetic medicine agent to treat neurodegenerative diseases (Wilkins et al., 2014). OAA, as a signaling molecule, could provide survival benefits to NP progenitor cells that adipose stromal cells lack. There was no indication of the OAA-Fox connection in FAT cells from the 2D differential expression analysis and the higher proportion of senescence seen in FAT cells might pose a challenge when using adipose stromal cells to repopulate a degenerate NP niche. This finding is supported by recent studies in rats (Han et al., 2014) and the abovementioned single-cell RNASeq data from healthy and degenerated human NP cells recommending the use of NP progenitor cells for IVDD treatment (Han et al., 2022).
Stem cell presence has been suggested for most vascularized tissue of the body (Crisan et al., 2008), yet the NP is avascular. We have previously demonstrated that NP stromal cells have chondrogenic and osteogenic potential (Kraus et al., 2017). In the larger context of cell reprogramming, CD markers can provide information about the differentiation status of a cell line. CD markers are based on cell surface proteins initially used for immunophenotyping (Bernard and Boumsell, 1984). Over time, many were characterized in their function and helped characterize mesenchymal stem or stromal cells (Dominici et al., 2006; Lv et al., 2014). Minimum criteria of MSCs include plastic-adherence under standard culture conditions alongside the expression of CD105, CD73, and CD90 and the absence of CD45, CD34, CD14 or CD11b, CD79a or CD19, and HLA-DR and the ability to differentiate into the three-mesoderm lineage in vitro. Other surface antigens generally expressed by MSCs include CD10, CD13, CD29, and CD44 (Dominici et al., 2006; Lv et al., 2014). The NP, AF, and FAT stromal cells investigated here were isolated based on their ability to attach to an untreated plastic surface. NP cells showed higher transcript levels for the CD90 (Thy1) cell surface glycoprotein and lower levels for CD34 transcripts, whereas other markers were not differentially expressed between the cell lines. Increased Thy1 expression for NP over AF cells in vitro differs from our previous observations in vivo (Li et al., 2019b). Higher transcript levels in the NP were also found for CD101 encoding the transmembrane glycoprotein V7, a prognostic marker for gliomas (Rivas et al., 1995; Liu et al., 2022a), and CD109 encoding a protein that negatively regulates TGFβ signaling and is involved in tumorigenesis (Li et al., 2016; Lee et al., 2020). Transcript levels higher in FAT cells included those encoding CD36, a glycoprotein receptor involved in cytokine and inflammasome production (Stewart et al., 2010); CD200 with a role in immunosuppression and regulation of anti-tumor activity (Khan et al., 2021); CD248 (endosialin) with predicted involvement in migration and ECM binding (Rettig et al., 1992); and CD300, a hypoxia-inducible regulator of VEGF production involved in immune response (Raggi et al., 2014; Cao et al., 2021). Aldh enzymes oxidize aldehydes to carboxylic acids, yet some members are discussed as cancer stem cell markers (Nakahata et al., 2015). Several transcripts we found higher in FAT over IVD cells. A benefit of adult stromal cells is their multipotent over pluripotent differentiation potential, a restriction rendering them less tumorigenic than induced pluripotent or embryonic stem cells, however, potentially hampering their plasticity in replacing ailing cells (Pittenger et al., 1999; Kraus et al., 2021). Oct4 is known as a stemness and reprogramming marker but also has a role in lineage specification (Zeineddine et al., 2014). We did not see transcripts for the pluripotency factor Oct4 differentially expressed between the cell types in 2D culture. However, transcripts for its partner Sox2 were higher in NP and FAT over AF cells. Sox2 encoded one of the reprogramming factors important for stem cell maintenance and cell fate determination (Takahashi et al., 2007; Feng et al., 2009) and was suggested as a biomarker to distinguish between NP and AF cells in vivo (Li et al., 2019b). We also noted an increase in Lif expression in NP and FAT over AF cells and increased transcripts for the Lif receptor in NP over FAT cells. Lif signaling is considered crucial for maintaining pluripotency in naïve stem cells. Interestingly, Sox2/Oct4 downstream, such as Klf4 and Top2a, were slightly higher in NP over FAT cells, whereas Mras was slightly higher in FAT over IVD cells (data not shown) (Rizzino, 2013; Zeineddine et al., 2014).
Glis1, a zinc finger TF, recognized as a somatic cell reprogramming factor involved in cell differentiation by restricting lineage commitment of multipotent cells (Maekawa et al., 2011; Scoville et al., 2017; Jetten, 2018; Li et al., 2020) could be a key player in NP cell phenotype adaptions. Glis1 showed the highest transcript levels in NP over FAT cells, and transcripts higher in NP over FAT cells were enriched for DNA binding transcription activator activity. This suggests a more progressive role of NP cells in adapting to the 2D environment in a standard medium through active reprogramming. High levels of Glis1 in NP over FAT cells and the presence of Glis1 in a subpopulation of NP cells in vivo in combination with increased transcripts for other reprogramming affiliated markers further suggest a higher progenitor cell potential in NP over FAT cells. Expression of Glis1 remained higher in NP over FAT cells in 3D culture in standard medium, but Oct4 levels were higher in FAT cells, perhaps due to a structurally influenced component triggering somatic cell reprogramming of adipose cells (Figure 4E, Figures 10A,C). P53 levels remained higher, and P21 levels were lower in NP cells in 3D culture. P53 is primarily considered a transcriptional activator, with transcriptional repression mediated through its target P21, among other mechanisms (Sullivan et al., 2018). Increased P21 levels could be one reason for a higher proportion of senescent cells in the FAT cell lineage in 3D culture as identified by SA β-galactosidase staining (Figures 10A,D). Transcript levels for the inflammatory markers Tnfα and Il1b were not significantly different between the two cell lines in 3D culture under standard conditions (Figure 10A).
Monolayer culture and adherence to plastic do not reflect the typical environment of mammalian cells, which are usually embedded in 3D ECM of various stiffness, composition, and density. Bio-scaffold development, including the use of alginate, has proven helpful in the field of IVD research (May et al., 2020). Cell shape and surface stiffness are important determinants for adipose stem cell differentiation into a chondrogenic phenotype. Stem cells maintained with a round morphology differentiated into adipocytes even in osteogenic media (Lu et al., 2007). Morphological changes from flat to round in the 3D alginate-based environment could be seen by histological staining, and a BISH protocol for the analysis of selected transcripts was developed. Upon 3D culture using standard D10SL media, Col2a1 and Krt19 expression along with TFs such as Tbox and Noto were now higher in FAT over NP cells indicating adjustment of adipose stromal cells to matrix cues of a scaffold that was developed for IVD cell culture (Maldonado and Oegema, 1992; May et al., 2020). Noto, a homeobox TF acting downstream of FoxA2 and brachyury (Tbox) during the NC development can direct induced pluripotent stem cell-derived progenitor cells toward an NC fate (Colombier et al., 2020). Noto showed no significant difference in 2D, suggesting some form of somatic cell reprogramming upon transitioning from 2D to 3D.
FAT is a convenient source for autologous stem/stromal cells in regenerative medicine. However, the degenerated NP offers a harsh environment for settlement (Lyu, 2022). Some attributes of the degenerate NP were partially mimicked through lactic acid at a concentration of ∼40 mM. This is higher than plasma levels (1 mM) and higher than previously described for the center of an IVD (16 mM) (Bartels et al., 1998; Sugimoto et al., 2012; Hui et al., 2017; Wang et al., 2021a). The presence of lactic acid could impact cells in several ways: a breakdown of ECM molecules, as described in vivo, could modify ECM signaling cues, lactic acid as a carbon source could impact cell metabolism, and lactylation could promote epigenetic cell reprogramming. As a novel concept, somatic cell-reprogramming through lactylation or lactate-induced histone modification recently moved into the spotlight (Zhang et al., 2019). Metabolic reprogramming is best studied in cancer cells as an adaption to hypoxia and hypo-nutrient conditions (Yoshida, 2015; Zhang et al., 2019; Chen et al., 2021), a condition all too familiar for NP cells. NP cells are in possession of molecular tools for lactate transport and metabolism. Adding lactic acid to 3D culture significantly increased Glis1 expression in FAT cells beyond levels in NP cells, suggesting Glis1 as a mediator of lactylation-based somatic cell reprogramming. We do not claim that the stromal cells undergo reprogramming toward a pluripotent state. Somatic cell reprogramming and cancer development share a metabolic switch toward glycolysis, and concern exists based on data from induced pluripotent cells that partial reprogramming could be tumorigenic (Folmes et al., 2011; Ohnishi et al., 2014). It is unclear if partial somatic reprogramming might negatively impact stromal cell performance. We noticed an increase in tumor-promoting or a decrease in anti-tumorigenic transcripts, alongside an increased inflammatory potential associated with high Glis1 transcript levels in 2D. Glis1, a P53-independent somatic reprogramming factor, promotes but is not required for Oct4/Sox2/Klf4- (OSK-) dependent reprogramming (Maekawa et al., 2011; Scoville et al., 2017), which could explain why the relative Oct4 and Glis1 transcript levels vary depending on the culture condition. Based on Glis1/Sox2 expression in 2D, we propose that NP stromal cells have a higher degree of stemness than FAT stromal cells, but a 3D environment and exposure to lactic acid appear to facilitate stemness in FAT cells and might elevate the risk of tumorigenesis for FAT stromal cells in the degenerate NP niche as indicated by higher Tbox (Tbx1) levels, a prognostic chordoma marker. Initiation of somatic reprogramming is further indicated by a now higher expression of Col2a1 and Pax1. NP cells find themselves under “homeroom” conditions, prompting no significant changes in Glis1 but increased Oct4 levels. P53, P21, and Ki67 transcripts levels agree with the concept of transcriptional activation of P21, leading to G1 arrest that would promote differentiation over proliferation, as in FAT cells reduced P21 expression upon lactic acid exposure correlated with increased Ki67 expression, indicating cell proliferation (Harada and Ogden, 2000; Marion et al., 2009; Maeso-Alonso et al., 2021) (Supplementary Figure S1). For glucose/lactate metabolism-related enzymes, a significant increase in LdhA/B was noted in FAT over NP cells (Figure 10A, Supplementary Figure S1). Possibly other Ldh isoforms, as seen in the transcriptome profiling of cells in 2D culture, could play a role here. Inflammatory markers, Tnfα and Il1b, were significantly higher in FAT over NP cells in these limited NP niche conditions (Figure 10A, Supplementary Figure S1).
Although the NP cells are of NC origin, they are often considered chondrocyte-like (Trout et al., 1982b; Hunter et al., 2004). Hence, media for IVD cells is often supplemented with ascorbic acid and proline (Kraus et al., 2017). Ascorbic acid (vitamin C), among several functions, is involved in collagen biosynthesis on a transcriptional and posttranslational level (Pinnell, 1985; Combs and McClung, 2016), and proline is an important constitute of collagen (Karna et al., 2020). Glut1 transporters enable the glucose and ascorbic acid to pass the plasma membrane. Although Col2a1 transcript levels were similar between NP and FAT cells in chondrogenic (D10CL) conditions, they were at their highest for NP cells (Supplementary Figure S1). Krt19, Noto, Tbox, and Pax1 expression was increased in FAT over NP cells. From a metabolic perspective, transcript levels for Gys1, Mdh, and LdhB were significantly lower in NP over FAT cells. The proportion of senescent cells remained higher in FAT cells. However, for both cell lines, the highest proportion of senescent cells and P21 transcript levels was noted under chondrogenic conditions (Figure 10A, Supplementary Figure S1). Glis1 expression was slightly higher in FAT over NP cells. Transcript levels for the inflammatory markers, Tnfα and IL1b, were significantly higher in FAT over NP cells (Figure 10A). Chondrogenic conditions triggered somatic reprogramming in both cell lines, potentially mediated through the presence of ascorbic acid as recently a role of ascorbic acid in epigenetic reprogramming was suggested (Young et al., 2015; Liu et al., 2022b). Aside from an increase in P21 transcripts, it is unclear what caused the increase in senescence in both cell lines. A high senescence rate might counteract some potentially deleterious tumorigenic reprogramming effects but would likely impact the efficiency of transplanted cells to contribute to a chondrogenic niche. In summary, we propose that changes in extracellular (lactic) acid promote cell type-dependent Glis1-mediated somatic cell reprogramming in NP and FAT-derived stromal cells.
Ideally, cells used for cell therapy in IVDD can maintain themselves without disturbing the unique cell/ECM relation, can replenish the correct ECM macromolecules, and will not further aggravate the condition, simply hanging in there and doing the job for as long as possible. It is not an easy task in an unwelcoming environment and not an easy task to assess, given the nature of the IVD. Herein, we aimed to understand the ability of IVD and adipose stromal cells to serve the IVDD niche from a transcriptome point of view. While adipose stromal cells have the benefit of accessibility and can adapt to a NP-cell like phenotype, key differences remain. Lactic or ascorbic acid appears to reprogram stromal cells through Glis1. High levels of Glis1 appear to be associated with an increased risk for inflammation and tumorigenesis. 3D adipose stromal cells show a higher risk of aggravating inflammatory conditions and a higher senescence rate compared to NP cells. We further propose that NP cells developed mechanisms to use lactic acid to their advantage, potentially even for “in-house” production of a longevity supplement.
The original contributions presented in the study are publicly available. This data can be found in NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/), accession number GSE216377.
PK conceived the project and conducted all cell culture, transcriptome experiments, and development of the BISH protocol. TL helped conceive, oversee, and fund the project. LL conducted all statistical analyses. AS assisted with data analysis, generated Cytoscape networks, and conducted clinic trial research. SL assisted with data analysis and imaging. DB, BE, and JR contributed to the data analysis. All authors provided intellectual input and contributed to writing the manuscript.
This work was supported by the Bayard and Virginia Clarkson Endowment Fund and NIH HD099588 granted to TL.
We are grateful to Darren Sipes and Shantanu Sur for their support and to Tritown Meat Packing (Brasher Falls, NY) and Woodcrest Dairy (Lisbon, NY) for providing us with bovine specimens.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.1009402/full#supplementary-material
Supplementary Figure S1 | Bar graphs indicate RNA levels after BISH and ImageJ analysis of NP and FAT cells after 10 days of 3D alginate bead culture under different culture conditions for genes related to the nucleus pulposus (NP) (yellow), glucose and lactate metabolism (orange), inflammatory markers (green), cell proliferation and senescence markers, and SA-beta galactosidase staining (blue). Cells were isolated from a single donor to avoid the genotype effect. A minimum of 25 cells were analyzed after subjecting three beads per probe, cell line, and condition to BISH. S, standard media D10SL; D, media D10DL mimicking degenerate conditions; C, chondrogenic D10CL media. Significance is indicated as *** (p < 0.001); **(p < 0.01); *(p < 0.05).
Supplementary Figure S2 | Top three Gene Ontology (GO) categories after functional enrichment for molecular or biological functions.
Amirdelfan, K., Bae, H., McJunkin, T., DePalma, M., Kim, K., Beckworth, W. J., et al. (2021). Allogeneic mesenchymal precursor cells treatment for chronic low back pain associated with degenerative disc disease: A prospective randomized, placebo-controlled 36-month study of safety and efficacy. Spine J. 21 (2), 212–230. doi:10.1016/j.spinee.2020.10.004
Andriamanalijaona, R., Duval, E., Raoudi, M., Lecourt, S., Vilquin, J. T., Marolleau, J. P., et al. (2008). Differentiation potential of human muscle-derived cells towards chondrogenic phenotype in alginate beads culture. Osteoarthr. Cartil. 16 (12), 1509–1518. doi:10.1016/j.joca.2008.04.018
Bartels, E. M., Fairbank, J. C., Winlove, C. P., and Urban, J. P. (1998). Oxygen and lactate concentrations measured in vivo in the intervertebral discs of patients with scoliosis and back pain. Spine 23 (1), 1–7. doi:10.1097/00007632-199801010-00001
Bastian, D., Wu, Y., Betts, B. C., and Yu, X. Z. (2019). The IL-12 cytokine and receptor family in graft-vs.-host disease. Front. Immunol. 10, 988. doi:10.3389/fimmu.2019.00988
Bayliss, M. T., Johnstone, B., and O'Brien, J. P. (1988). 1988 Volvo award in basic science. Proteoglycan synthesis in the human intervertebral disc. Variation with age, region and pathology. Spine 13 (9), 972–981. doi:10.1097/00007632-198809000-00003
Bedore, J., Leask, A., and Seguin, C. A. (2014). Targeting the extracellular matrix: Matricellular proteins regulate cell-extracellular matrix communication within distinct niches of the intervertebral disc. Matrix Biol. 37, 124–130. doi:10.1016/j.matbio.2014.05.005
Bekhouche, M., and Colige, A. (2015). The procollagen N-proteinases ADAMTS2, 3 and 14 in pathophysiology. Matrix Biol. 44-46, 46–53. doi:10.1016/j.matbio.2015.04.001
Bernard, A., and Boumsell, L. (1984). Human leukocyte differentiation antigens. Presse Med. 13 (38), 2311–2316.
Berry, D. C., Stenesen, D., Zeve, D., and Graff, J. M. (2013). The developmental origins of adipose tissue. Development 140 (19), 3939–3949. doi:10.1242/dev.080549
Bibby, S. R., Jones, D. A., Lee, R. B., Yu, J., and Urban, J. P. G. (2001). The pathophysiology of the intervertebral disc. Jt. Bone Spine 68 (6), 537–542. doi:10.1016/s1297-319x(01)00332-3
Bibby, S. R., Jones, D. A., Ripley, R. M., and Urban, J. P. (2005). Metabolism of the intervertebral disc: Effects of low levels of oxygen, glucose, and pH on rates of energy metabolism of bovine nucleus pulposus cells. Spine 30 (5), 487–496. doi:10.1097/01.brs.0000154619.38122.47
Bibby, S. R., and Urban, J. P. (2004). Effect of nutrient deprivation on the viability of intervertebral disc cells. Eur. Spine J. 13 (8), 695–701.
Blesch, A., Bosserhoff, A. K., Apfel, R., Behl, C., Hessdoerfer, B., Schmitt, A., et al. (1994). Cloning of a novel malignant melanoma-derived growth-regulatory protein, MIA. Cancer Res. 54 (21), 5695–5701.
Bushell, G. R., Ghosh, P., Taylor, T. F., and Akeson, W. H. (1977). Proteoglycan chemistry of the intervertebral disks. Clin. Orthop. Relat. Res. 129 (129), 115–123. doi:10.1097/00003086-197711000-00013
Cantor, J. R., Abu-Remaileh, M., Kanarek, N., Freinkman, E., Gao, X., Louissaint, A., et al. (2017). Physiologic medium rewires cellular metabolism and reveals uric acid as an endogenous inhibitor of UMP synthase. Cell 169 (2), 258–272. doi:10.1016/j.cell.2017.03.023
Cao, Y., Ao, T., Wang, X., Wei, W., Fan, J., and Tian, X. (2021). CD300a and CD300f molecules regulate the function of leukocytes. Int. Immunopharmacol. 93, 107373. doi:10.1016/j.intimp.2021.107373
Carruthers, A., DeZutter, J., Ganguly, A., and Devaskar, S. U. (2009). Will the original glucose transporter isoform please stand up. Am. J. Physiol. Endocrinol. Metab. 297 (4), E836–E848. doi:10.1152/ajpendo.00496.2009
Chatterjee, S., Kraus, P., Sivakamasundari, V., Xing, X., Yap, S. P., Jie, S., et al. (2013). A conditional mouse line for lineage tracing of Sox9 loss-of-function cells using enhanced green fluorescent protein. Biotechnol. Lett. 35 (12), 1991–1996. doi:10.1007/s10529-013-1303-6
Chatterjee, S., Sivakamasundari, V., Yap, S. P., Kraus, P., Kumar, V., Xing, X., et al. (2014). In vivo genome-wide analysis of multiple tissues identifies gene regulatory networks, novel functions and downstream regulatory genes for Bapx1 and its co-regulation with Sox9 in the mammalian vertebral column. BMC genomics 15, 1072. doi:10.1186/1471-2164-15-1072
Chen, A., Zhong, L., and Lv, J. (2019). FOXL1 overexpression is associated with poor outcome in patients with glioma. Oncol. Lett. 18 (1), 751–757. doi:10.3892/ol.2019.10351
Chen, A. N., Luo, Y., Yang, Y. H., Fu, J. T., Geng, X. M., Shi, J. P., et al. (2021). Lactylation, a novel metabolic reprogramming code: Current status and prospects. Front. Immunol. 12, 688910. doi:10.3389/fimmu.2021.688910
Chen, L., Liao, J., Klineberg, E., Leung, V. Y., and Huang, S. (2017). Small leucine-rich proteoglycans (SLRPs): Characteristics and function in the intervertebral disc. J. Tissue Eng. Regen. Med. 11 (3), 602–608. doi:10.1002/term.2067
Chen, M., Wu, Y., Zhang, H., Li, S., Zhou, J., and Shen, J. (2020). The roles of embryonic transcription factor BRACHYURY in tumorigenesis and progression. Front. Oncol. 10, 961. doi:10.3389/fonc.2020.00961
Cherif, H., Mannarino, M., Pacis, A. S., Ragoussis, J., Rabau, O., Ouellet, J. A., et al. (2022). Single-cell RNA-seq analysis of cells from degenerating and non-degenerating intervertebral discs from the same individual reveals new biomarkers for intervertebral disc degeneration. Int. J. Mol. Sci. 23 (7), 3993. doi:10.3390/ijms23073993
Choi, K. S., Cohn, M. J., and Harfe, B. D. (2008). Identification of nucleus pulposus precursor cells and notochordal remnants in the mouse: Implications for disk degeneration and chordoma formation. Dev. Dyn. 237 (12), 3953–3958. doi:10.1002/dvdy.21805
Choi, K. S., and Harfe, B. D. (2011). Hedgehog signaling is required for formation of the notochord sheath and patterning of nuclei pulposi within the intervertebral discs. Proc. Natl. Acad. Sci. U. S. A. 108 (23), 9484–9489. doi:10.1073/pnas.1007566108
Choi, K. S., Lee, C., and Harfe, B. D. (2012). Sonic hedgehog in the notochord is sufficient for patterning of the intervertebral discs. Mech. Dev. 129 (9-12), 255–262. doi:10.1016/j.mod.2012.07.003
Choy, C. T., Kim, H., Lee, J. Y., Williams, D. M., Palethorpe, D., Fellows, G., et al. (2014). Anosmin-1 contributes to brain tumor malignancy through integrin signal pathways. Endocr. Relat. Cancer 21 (1), 85–99. doi:10.1530/ERC-13-0181
Christ, B., and Wilting, J. (1992). From somites to vertebral column. Ann. Anat. 174 (1), 23–32. doi:10.1016/s0940-9602(11)80337-7
Collison, J. (2018). MMP12 makes the cut. Nat. Rev. Rheumatol. 14 (9), 501. doi:10.1038/s41584-018-0056-y
Colombier, P., Halgand, B., Chedeville, C., Chariau, C., Francois-Campion, V., Kilens, S., et al. (2020). NOTO transcription factor directs human induced pluripotent stem cell-derived mesendoderm progenitors to a notochordal fate. Cells 9 (2), E509. doi:10.3390/cells9020509
Combs, G. F., and McClung, J. P. (2016). The vitamins: Fundamental aspects in nutrition and health. Academic Press.
Comella, K., Silbert, R., and Parlo, M. (2017). Erratum to: Effects of the intradiscal implantation of stromal vascular fraction plus platelet rich plasma in patients with degenerative disc disease. J. Transl. Med. 15 (1), 108. doi:10.1186/s12967-017-1217-5
Crisan, M., Yap, S., Casteilla, L., Chen, C. W., Corselli, M., Park, T. S., et al. (2008). A perivascular origin for mesenchymal stem cells in multiple human organs. Cell stem cell 3 (3), 301–313. doi:10.1016/j.stem.2008.07.003
Dai, P., Akimaru, H., Tanaka, Y., Maekawa, T., Nakafuku, M., and Ishii, S. (1999). Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J. Biol. Chem. 274 (12), 8143–8152. doi:10.1074/jbc.274.12.8143
Debacq-Chainiaux, F., Erusalimsky, J. D., Campisi, J., and Toussaint, O. (2009). Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc. 4 (12), 1798–1806. doi:10.1038/nprot.2009.191
Deckx, S., Heymans, S., and Papageorgiou, A. P. (2016). The diverse functions of osteoglycin: A deceitful dwarf, or a master regulator of disease? FASEB J. 30 (8), 2651–2661. doi:10.1096/fj.201500096R
DePalma, M. J., Ketchum, J. M., and Saullo, T. (2011). What is the source of chronic low back pain and does age play a role? Pain Med. 12 (2), 224–233. doi:10.1111/j.1526-4637.2010.01045.x
Diamant, B., Karlsson, J., and Nachemson, A. (1968). Correlation between lactate levels and pH in discs of patients with lumbar rhizopathies. Experientia 24 (12), 1195–1196. doi:10.1007/BF02146615
Dijkers, P. F., Medema, R. H., Pals, C., Banerji, L., Thomas, N. S., Lam, E. W., et al. (2000). Forkhead transcription factor FKHR-L1 modulates cytokine-dependent transcriptional regulation of p27(KIP1). Mol. Cell. Biol. 20 (24), 9138–9148. doi:10.1128/mcb.20.24.9138-9148.2000
Dominici, M., Le Blanc, K., Mueller, I., Slaper-Cortenbach, I., Marini, F., Krause, D., et al. (2006). Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 8 (4), 315–317. doi:10.1080/14653240600855905
Duan, S. Z., Ivashchenko, C. Y., Whitesall, S. E., D'Alecy, L. G., Duquaine, D. C., Brosius, F. C., et al. (2007). Hypotension, lipodystrophy, and insulin resistance in generalized PPARgamma-deficient mice rescued from embryonic lethality. J. Clin. Invest. 117 (3), 812–822. doi:10.1172/JCI28859
Edwards, D. R., Handsley, M. M., and Pennington, C. J. (2008). The ADAM metalloproteinases. Mol. Asp. Med. 29 (5), 258–289. doi:10.1016/j.mam.2008.08.001
Enomoto, H., Furuichi, T., Zanma, A., Yamana, K., Yoshida, C., Sumitani, S., et al. (2004). Runx2 deficiency in chondrocytes causes adipogenic changes in vitro. J. Cell Sci. 117 (3), 417–425. doi:10.1242/jcs.00866
Erwin, W. M., and Hood, K. E. (2014). The cellular and molecular biology of the intervertebral disc: A clinician's primer. J. Can. Chiropr. Assoc. 58 (3), 246–257.
Eyre, D. R. (1979). Biochemistry of the intervertebral disc. Int. Rev. Connect. Tissue Res. 8, 227–291. doi:10.1016/b978-0-12-363708-6.50012-6
Eyre, D. R., and Muir, H. (1976). Types I and II collagens in intervertebral disc. Interchanging radial distributions in annulus fibrosus. Biochem. J. 157 (1), 267–270. doi:10.1042/bj1570267
Feng, B., Jiang, J., Kraus, P., Ng, J. H., Heng, J. C., Chan, Y. S., et al. (2009). Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat. Cell Biol. 11 (2), 197–203. doi:10.1038/ncb1827
Fernandes, L. M., Khan, N. M., Trochez, C. M., Duan, M., Diaz-Hernandez, M. E., Presciutti, S. M., et al. (2020). Single-cell RNA-seq identifies unique transcriptional landscapes of human nucleus pulposus and annulus fibrosus cells. Sci. Rep. 10 (1), 15263. doi:10.1038/s41598-020-72261-7
Folmes, C. D., Nelson, T. J., Martinez-Fernandez, A., Arrell, D. K., Lindor, J. Z., Dzeja, P. P., et al. (2011). Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 14 (2), 264–271. doi:10.1016/j.cmet.2011.06.011
Forsyth, P. A., Wong, H., Laing, T. D., Rewcastle, N. B., Morris, D. G., Muzik, H., et al. (1999). Gelatinase-A (MMP-2), gelatinase-B (MMP-9) and membrane type matrix metalloproteinase-1 (MT1-MMP) are involved in different aspects of the pathophysiology of malignant gliomas. Br. J. Cancer 79 (11-12), 1828–1835. doi:10.1038/sj.bjc.6690291
Gamblin, S. J., Davies, G. J., Grimes, J. M., Jackson, R. M., Littlechild, J. A., and Watson, H. C. (1991). Activity and specificity of human aldolases. J. Mol. Biol. 219 (4), 573–576. doi:10.1016/0022-2836(91)90650-u
Gan, J. C., Ducheyne, P., Vresilovic, E. J., and Shapiro, I. M. (2003). Intervertebral disc tissue engineering II: Cultures of nucleus pulposus cells. Clin. Orthop. Relat. Res. 411 (411), 315–324. doi:10.1097/01.blo.0000063797.98363.d3
Gatchel, R. J. (2015). The continuing and growing epidemic of chronic low back pain. Healthc. (Basel) 3 (3), 838–845. doi:10.3390/healthcare3030838
Ghannam, S., Bouffi, C., Djouad, F., Jorgensen, C., and Noel, D. (2010). Immunosuppression by mesenchymal stem cells: Mechanisms and clinical applications. Stem Cell Res. Ther. 1 (1), 2. doi:10.1186/scrt2
Gilbert, H. T. J., Hodson, N., Baird, P., Richardson, S. M., and Hoyland, J. A. (2016). Acidic pH promotes intervertebral disc degeneration: Acid-sensing ion channel -3 as a potential therapeutic target. Sci. Rep. 6, 37360. doi:10.1038/srep37360
Gomes, A. M., Carron, E. C., Mills, K. L., Dow, A. M., Gray, Z., Fecca, C. R., et al. (2019). Stromal Gas6 promotes the progression of premalignant mammary cells. Oncogene 38 (14), 2437–2450. doi:10.1038/s41388-018-0593-5
Gu, J., Zhang, S., He, X., Chen, S., and Wang, Y. (2021). High expression of PIG11 correlates with poor prognosis in gastric cancer. Exp. Ther. Med. 21 (3), 249. doi:10.3892/etm.2021.9680
Han, B., Wang, H. C., Li, H., Tao, Y. Q., Liang, C. Z., Li, F. C., et al. (2014). Nucleus pulposus mesenchymal stem cells in acidic conditions mimicking degenerative intervertebral discs give better performance than adipose tissue-derived mesenchymal stem cells. Cells Tissues Organs 199 (5-6), 342–352. doi:10.1159/000369452
Han, S., Zhang, Y., Zhang, X., Zhang, H., Meng, S., Kong, M., et al. (2022). Single-cell RNA sequencing of the nucleus pulposus reveals chondrocyte differentiation and regulation in intervertebral disc degeneration. Front. Cell Dev. Biol. 10, 824771. doi:10.3389/fcell.2022.824771
Harada, K., and Ogden, G. R. (2000). An overview of the cell cycle arrest protein, p21(WAF1). Oral Oncol. 36 (1), 3–7. doi:10.1016/s1368-8375(99)00049-4
Hardie, D. G., and Carling, D. (1997). The AMP-activated protein kinase-fuel gauge of the mammalian cell? Eur. J. Biochem. 246 (2), 259–273. doi:10.1111/j.1432-1033.1997.00259.x
He, W., Kang, Y., Zhu, W., Zhou, B., Jiang, X., Ren, C., et al. (2020). FOXF2 acts as a crucial molecule in tumours and embryonic development. Cell Death Dis. 11 (6), 424. doi:10.1038/s41419-020-2604-z
Henriksson, H., Thornemo, M., Karlsson, C., Hagg, O., Junevik, K., Lindahl, A., et al. (2009). Identification of cell proliferation zones, progenitor cells and a potential stem cell niche in the intervertebral disc region: A study in four species. Spine 34 (21), 2278–2287. doi:10.1097/BRS.0b013e3181a95ad2
Herrmann, B. G., Labeit, S., Poustka, A., King, T. R., and Lehrach, H. (1990). Cloning of the T gene required in mesoderm formation in the mouse. Nature 343 (6259), 617–622. doi:10.1038/343617a0
Hickman, T. T., Rathan-Kumar, S., and Peck, S. H. (2022). Development, pathogenesis, and regeneration of the intervertebral disc: Current and future insights spanning traditional to omics methods. Front. Cell Dev. Biol. 10, 841831. doi:10.3389/fcell.2022.841831
Holm, S., Maroudas, A., Urban, J. P., Selstam, G., and Nachemson, A. (1981). Nutrition of the intervertebral disc: Solute transport and metabolism. Connect. Tissue Res. 8 (2), 101–119. doi:10.3109/03008208109152130
Hromas, R., Broxmeyer, H. E., Kim, C., Nakshatri, H., Christopherson, K., Azam, M., et al. (1999). Cloning of BRAK, a novel divergent CXC chemokine preferentially expressed in normal versus malignant cells. Biochem. Biophys. Res. Commun. 255 (3), 703–706. doi:10.1006/bbrc.1999.0257
Hui, S., Ghergurovich, J. M., Morscher, R. J., Jang, C., Teng, X., Lu, W., et al. (2017). Glucose feeds the TCA cycle via circulating lactate. Nature 551 (7678), 115–118. doi:10.1038/nature24057
Humzah, M. D., and Soames, R. W. (1988). Human intervertebral disc: Structure and function. Anat. Rec. 220 (4), 337–356. doi:10.1002/ar.1092200402
Hunter, C. J., Matyas, J. R., and Duncan, N. A. (2004). Cytomorphology of notochordal and chondrocytic cells from the nucleus pulposus: A species comparison. J. Anat. 205 (5), 357–362. doi:10.1111/j.0021-8782.2004.00352.x
Ikeda, T., Kamekura, S., Mabuchi, A., Kou, I., Seki, S., Takato, T., et al. (2004). The combination of SOX5, SOX6, and SOX9 (the SOX trio) provides signals sufficient for induction of permanent cartilage. Arthritis Rheum. 50 (11), 3561–3573. doi:10.1002/art.20611
Jablonska-Trypuc, A., Matejczyk, M., and Rosochacki, S. (2016). Matrix metalloproteinases (MMPs), the main extracellular matrix (ECM) enzymes in collagen degradation, as a target for anticancer drugs. J. Enzyme Inhib. Med. Chem. 31 (1), 177–183. doi:10.3109/14756366.2016.1161620
Janssen, B. J., Robinson, R. A., Perez-Branguli, F., Bell, C. H., Mitchell, K. J., Siebold, C., et al. (2010). Structural basis of semaphorin-plexin signalling. Nature 467 (7319), 1118–1122. doi:10.1038/nature09468
Jetten, A. M. (2018). GLIS1-3 transcription factors: Critical roles in the regulation of multiple physiological processes and diseases. Cell. Mol. Life Sci. 75 (19), 3473–3494. doi:10.1007/s00018-018-2841-9
Jin, L., and Zhou, Y. (2019). Crucial role of the pentose phosphate pathway in malignant tumors. Oncol. Lett. 17 (5), 4213–4221. doi:10.3892/ol.2019.10112
Jitrapakdee, S., Vidal-Puig, A., and Wallace, J. C. (2006). Anaplerotic roles of pyruvate carboxylase in mammalian tissues. Cell. Mol. Life Sci. 63 (7-8), 843–854. doi:10.1007/s00018-005-5410-y
Johnson, W. E., Stephan, S., and Roberts, S. (2008). The influence of serum, glucose and oxygen on intervertebral disc cell growth in vitro: Implications for degenerative disc disease. Arthritis Res. Ther. 10 (2), R46. doi:10.1186/ar2405
Karna, E., Szoka, L., Huynh, T. Y. L., and Palka, J. A. (2020). Proline-dependent regulation of collagen metabolism. Cell. Mol. Life Sci. 77 (10), 1911–1918. doi:10.1007/s00018-019-03363-3
Kawane, T., Qin, X., Jiang, Q., Miyazaki, T., Komori, H., Yoshida, C. A., et al. (2018). Runx2 is required for the proliferation of osteoblast progenitors and induces proliferation by regulating Fgfr2 and Fgfr3. Sci. Rep. 8 (1), 13551. doi:10.1038/s41598-018-31853-0
Khan, I. Z., Del Guzzo, C. A., Shao, A., Cho, J., Du, R., Cohen, A. O., et al. (2021). The cd200-cd200r Axis promotes squamous cell carcinoma metastasis via regulation of cathepsin K. Cancer Res. 81 (19), 5021–5032. doi:10.1158/0008-5472.CAN-20-3251
Klodzinski, L., and Wislowska, M. (2018). Comorbidities in rheumatic arthritis. Reumatologia 56 (4), 228–233. doi:10.5114/reum.2018.77974
Kowalik, M. A., Columbano, A., and Perra, A. (2017). Emerging role of the pentose phosphate pathway in hepatocellular carcinoma. Front. Oncol. 7, 87. doi:10.3389/fonc.2017.00087
Kraus, P., Fraidenraich, D., and Loomis, C. A. (2001). Some distal limb structures develop in mice lacking Sonic hedgehog signaling. Mech. Dev. 100 (1), 45–58. doi:10.1016/s0925-4773(00)00492-5
Kraus, P., Kocsis, V., Williams, C., Youngs, B., and Lufkin, T. (2015). Plate in situ hybridization (PISH) as a time and cost effective RNA expression assay to study phenotypic heterogeneity in a population of cultured murine cells at single cell resolution. Biotechnol. Lett. 37 (8), 1573–1577. doi:10.1007/s10529-015-1833-1
Kraus, P., Li, K., Sipes, D., Varden, L., Yerden, R., Henderson, A., et al. (2019). “Single-cell phenotyping of complex heterogeneous tissue,” in Handbook of single cell technologies. Editors T. S. Santra, and F-G. Tseng (Singapore: Springer Singapore), 1–17.
Kraus, P., and Lufkin, T. (2017). Implications for a stem cell regenerative medicine based approach to human intervertebral disk degeneration. Front. Cell Dev. Biol. 5, 17. doi:10.3389/fcell.2017.00017
Kraus, P., and Lufkin, T. (1999). Mammalian Dlx homeobox gene control of craniofacial and inner ear morphogenesis. J. Cell. Biochem. 32-33, 133–140. doi:10.1002/(sici)1097-4644(1999)75:32+<133::aid-jcb16>3.0.co;2-e
Kraus, P., Samanta, A., Lufkin, S., and Lufkin, T. (2021). Stem cells in intervertebral disc regeneration-more talk than action? Biocell. 46 (6), 893–898. doi:10.32604/biocell.2022.018432
Kraus, P., Sivakamasundari, V., Lim, S. L., Xing, X., Lipovich, L., and Lufkin, T. (2013). Making sense of Dlx1 antisense RNA. Dev. Biol. 376 (2), 224–235. doi:10.1016/j.ydbio.2013.01.035
Kraus, P., Sivakamasundari, V., Olsen, V., Villeneuve, V., Hinds, A., and Lufkin, T. (2019). Klhl14 antisense RNA is a target of key skeletogenic transcription factors in the developing intervertebral disc. Spine 44 (5), E260–E8. doi:10.1097/BRS.0000000000002827
Kraus, P., Sivakamasundari, V., Yu, H. B., Xing, X., Lim, S. L., Adler, T., et al. (2014). Pleiotropic functions for transcription factor zscan10. PloS one 9 (8), e104568. doi:10.1371/journal.pone.0104568
Kraus, P., Yerden, R., Kocsis, V., and Lufkin, T. (2017). RNA in situ hybridization characterization of non-enzymatic derived bovine intervertebral disc cell lineages suggests progenitor cell potential. Acta Histochem. 119 (2), 150–160. doi:10.1016/j.acthis.2016.12.004
Kraus, P., Yerden, R., Sipes, D., Sur, S., and Lufkin, T. (2018). A quantitative and qualitative RNA expression profiling assay for cell culture with single cell resolution. Cytotechnology 70 (1), 185–192. doi:10.1007/s10616-017-0132-1
Kumar, H., Ha, D. H., Lee, E. J., Park, J. H., Shim, J. H., Ahn, T. K., et al. (2017). Safety and tolerability of intradiscal implantation of combined autologous adipose-derived mesenchymal stem cells and hyaluronic acid in patients with chronic discogenic low back pain: 1-year follow-up of a phase I study. Stem Cell Res. Ther. 8 (1), 262. doi:10.1186/s13287-017-0710-3
Lander, A. D. (2009). The 'stem cell' concept: Is it holding us back? J. Biol. 8 (8), 70. doi:10.1186/jbiol177
Lawson, L., and Harfe, B. D. (2015). Notochord to nucleus pulposus transition. Curr. Osteoporos. Rep. 13 (5), 336–341. doi:10.1007/s11914-015-0284-x
Lee, K. Y., Shueng, P. W., Chou, C. M., Lin, B. X., Lin, M. H., Kuo, D. Y., et al. (2020). Elevation of CD109 promotes metastasis and drug resistance in lung cancer via activation of EGFR-AKT-mTOR signaling. Cancer Sci. 111 (5), 1652–1662. doi:10.1111/cas.14373
Lee, W. J., Chatterjee, S., Yap, S. P., Lim, S. L., Xing, X., Kraus, P., et al. (2017). An integrative developmental genomics and systems biology approach to identify an in vivo Sox trio-mediated gene regulatory network in murine embryos. Biomed. Res. Int. 2017, 8932583. doi:10.1155/2017/8932583
Lefebvre, V., Behringer, R. R., and de Crombrugghe, B. L. (2001). L-Sox5, Sox6 and Sox9 control essential steps of the chondrocyte differentiation pathway. Osteoarthr. Cartil. 9 (1), S69–S75. doi:10.1053/joca.2001.0447
Lehmann, O. J., Sowden, J. C., Carlsson, P., Jordan, T., and Bhattacharya, S. S. (2003). Fox's in development and disease. Trends Genet. 19 (6), 339–344. doi:10.1016/S0168-9525(03)00111-2
LeVan, K. M., and Goldberg, E. (1991). Properties of human testis-specific lactate dehydrogenase expressed from Escherichia coli. Biochem. J. 273 (3), 587–592. doi:10.1042/bj2730587
Levine, A. J. (1997). p53, the cellular gatekeeper for growth and division. Cell 88 (3), 323–331. doi:10.1016/s0092-8674(00)81871-1
Li, C., Hancock, M. A., Sehgal, P., Zhou, S., Reinhardt, D. P., and Philip, A. (2016). Soluble CD109 binds TGF-beta and antagonizes TGF-beta signalling and responses. Biochem. J. 473 (5), 537–547. doi:10.1042/BJ20141488
Li, H., Liang, C., Tao, Y., Zhou, X., Li, F., Chen, G., et al. (2012). Acidic pH conditions mimicking degenerative intervertebral discs impair the survival and biological behavior of human adipose-derived mesenchymal stem cells. Exp. Biol. Med. 237 (7), 845–852. doi:10.1258/ebm.2012.012009
Li, K., Kapper, D., Mondal, S., Lufkin, T., and Kraus, P. (2019). Quantitative single-cell transcript assessment of biomarkers supports cellular heterogeneity in the bovine IVD. Vet. Sci. 6 (2), E42. doi:10.3390/vetsci6020042
Li, K., Kapper, D., Youngs, B., Kocsis, V., Mondal, S., Kraus, P., et al. (2019). Potential biomarkers of the mature intervertebral disc identified at the single cell level. J. Anat. 234 (1), 16–32. doi:10.1111/joa.12904
Li, K., Varden, L., Henderson, A., Lufkin, T., and Kraus, P. (2021). Simultaneous detection of multiple mRNAs and proteins in bovine IVD cells and tissue with single cell resolution. Biotechnol. Lett. 43 (1), 13–24. doi:10.1007/s10529-020-02997-9
Li, L., Chen, K., Wang, T., Wu, Y., Xing, G., Chen, M., et al. (2020). Glis1 facilitates induction of pluripotency via an epigenome-metabolome-epigenome signalling cascade. Nat. Metab. 2 (9), 882–892. doi:10.1038/s42255-020-0267-9
Liu, J., Tao, H., Wang, H., Dong, F., Zhang, R., Li, J., et al. (2017). Biological behavior of human nucleus pulposus mesenchymal stem cells in response to changes in the acidic environment during intervertebral disc degeneration. Stem Cells Dev. 26 (12), 901–911. doi:10.1089/scd.2016.0314
Liu, X., Khan, A., Li, H., Wang, S., Chen, X., and Huang, H. (2022). Ascorbic acid in epigenetic reprogramming. Curr. Stem Cell Res. Ther. 17 (1), 13–25. doi:10.2174/1574888X16666210714152730
Liu, Y., Li, Y., Nan, L. P., Wang, F., Zhou, S. F., Wang, J. C., et al. (2020). The effect of high glucose on the biological characteristics of nucleus pulposus-derived mesenchymal stem cells. Cell biochem. Funct. 38 (2), 130–140. doi:10.1002/cbf.3441
Liu, Y., Yao, R., Shi, Y., Liu, Y., Liu, H., Liu, J., et al. (2022). Identification of CD101 in glioma: A novel prognostic indicator expressed on M2 macrophages. Front. Immunol. 13, 845223. doi:10.3389/fimmu.2022.845223
Loomis, C. A., Kimmel, R. A., Tong, C. X., Michaud, J., and Joyner, A. L. (1998). Analysis of the genetic pathway leading to formation of ectopic apical ectodermal ridges in mouse Engrailed-1 mutant limbs. Development 125 (6), 1137–1148. doi:10.1242/dev.125.6.1137
Lu, Z. F., Zandieh Doulabi, B., Wuisman, P. I., Bank, R. A., and Helder, M. N. (2007). Differentiation of adipose stem cells by nucleus pulposus cells: Configuration effect. Biochem. Biophys. Res. Commun. 359 (4), 991–996. doi:10.1016/j.bbrc.2007.06.002
Lufkin, L., Budisic, M., Mondal, S., and Sur, S. (2021). A bayesian model to analyze the association of rheumatoid arthritis with risk factors and their interactions. Front. Public Health 9, 693830. doi:10.3389/fpubh.2021.693830
Lv, F. J., Tuan, R. S., Cheung, K. M., and Leung, V. Y. (2014). Concise review: The surface markers and identity of human mesenchymal stem cells. Stem cells 32 (6), 1408–1419. doi:10.1002/stem.1681
Lyu, F. J. (2022). Impact of microenvironmental changes during degeneration on intervertebral disc progenitor cells: A comparison with mesenchymal stem cells. Bioeng. (Basel) 9 (4), 148. doi:10.3390/bioengineering9040148
Ma, X., Xu, L., Gavrilova, O., and Mueller, E. (2014). Role of forkhead box protein A3 in age-associated metabolic decline. Proc. Natl. Acad. Sci. U. S. A. 111 (39), 14289–14294. doi:10.1073/pnas.1407640111
Madhu, V., Boneski, P. K., Silagi, E., Qiu, Y., Kurland, I., Guntur, A. R., et al. (2020). Hypoxic regulation of mitochondrial metabolism and mitophagy in nucleus pulposus cells is dependent on HIF-1α-BNIP3 Axis. J. Bone Min. Res. 35 (8), 1504–1524. doi:10.1002/jbmr.4019
Maekawa, M., Yamaguchi, K., Nakamura, T., Shibukawa, R., Kodanaka, I., Ichisaka, T., et al. (2011). Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature 474 (7350), 225–229. doi:10.1038/nature10106
Maeso-Alonso, L., Lopez-Ferreras, L., Marques, M. M., and Marin, M. C. (2021). p73 as a tissue architect. Front. Cell Dev. Biol. 9, 716957. doi:10.3389/fcell.2021.716957
Maldonado, B. A., and Oegema, T. R. (1992). Initial characterization of the metabolism of intervertebral disc cells encapsulated in microspheres. J. Orthop. Res. 10 (5), 677–690.
Mani, S. A., Yang, J., Brooks, M., Schwaninger, G., Zhou, A., Miura, N., et al. (2007). Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc. Natl. Acad. Sci. U. S. A. 104 (24), 10069–10074. doi:10.1073/pnas.0703900104
Manolescu, A. R., Augustin, R., Moley, K., and Cheeseman, C. (2007). A highly conserved hydrophobic motif in the exofacial vestibule of fructose transporting SLC2A proteins acts as a critical determinant of their substrate selectivity. Mol. Membr. Biol. 24 (5-6), 455–463. doi:10.1080/09687680701298143
Manzari-Tavakoli, A., Babajani, A., Farjoo, M. H., Hajinasrollah, M., Bahrami, S., and Niknejad, H. (2022). The cross-talks among bone morphogenetic protein (BMP) signaling and other prominent pathways involved in neural differentiation. Front. Mol. Neurosci. 15, 827275. doi:10.3389/fnmol.2022.827275
Marion, R. M., Strati, K., Li, H., Murga, M., Blanco, R., Ortega, S., et al. (2009). A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460 (7259), 1149–1153. doi:10.1038/nature08287
Martins, R., Lithgow, G. J., and Link, W. (2016). Long live FOXO: Unraveling the role of FOXO proteins in aging and longevity. Aging Cell 15 (2), 196–207. doi:10.1111/acel.12427
May, P., and May, E. (1999). Twenty years of p53 research: Structural and functional aspects of the p53 protein. Oncogene 18 (53), 7621–7636. doi:10.1038/sj.onc.1203285
May, R. D., Frauchiger, D. A., Albers, C. E., Hofstetter, W., and Gantenbein, B. (2020). Exogenous stimulation of human intervertebral disc cells in 3-dimensional alginate bead culture with BMP2 and L51P: Cytocompatibility and effects on cell phenotype. Neurospine 17 (1), 77–87. doi:10.14245/ns.2040002.001
Mazelin, L., Bernet, A., Bonod-Bidaud, C., Pays, L., Arnaud, S., Gespach, C., et al. (2004). Netrin-1 controls colorectal tumorigenesis by regulating apoptosis. Nature 431 (7004), 80–84. doi:10.1038/nature02788
McCann, M. R., and Seguin, C. A. (2016). Notochord cells in intervertebral disc development and degeneration. J. Dev. Biol. 4 (1), 3. doi:10.3390/jdb4010003
McCann, M. R., Tamplin, O. J., Rossant, J., and Seguin, C. A. (2012). Tracing notochord-derived cells using a noto-cre mouse: Implications for intervertebral disc development. Dis. Model. Mech. 5 (1), 73–82. doi:10.1242/dmm.008128
Minogue, B. M., Richardson, S. M., Zeef, L. A., Freemont, A. J., and Hoyland, J. A. (2010). Transcriptional profiling of bovine intervertebral disc cells: Implications for identification of normal and degenerate human intervertebral disc cell phenotypes. Arthritis Res. Ther. 12 (1), R22. doi:10.1186/ar2929
Mueckler, M., and Thorens, B. (2013). The SLC2 (GLUT) family of membrane transporters. Mol. Asp. Med. 34 (2-3), 121–138. doi:10.1016/j.mam.2012.07.001
Nachemson, A. (1969). Intradiscal measurements of pH in patients with lumbar rhizopathies. Acta Orthop. Scand. 40 (1), 23–42. doi:10.3109/17453676908989482
Nakahata, K., Uehara, S., Nishikawa, S., Kawatsu, M., Zenitani, M., Oue, T., et al. (2015). Aldehyde dehydrogenase 1 (ALDH1) is a potential marker for cancer stem cells in embryonal rhabdomyosarcoma. PloS one 10 (4), e0125454. doi:10.1371/journal.pone.0125454
Nibu, Y., Jose-Edwards, D. S., and Di Gregorio, A. (2013). From notochord formation to hereditary chordoma: The many roles of brachyury. Biomed. Res. Int. 2013, 826435. doi:10.1155/2013/826435
Nwosu, Z. C., Ebert, M. P., Dooley, S., and Meyer, C. (2016). Caveolin-1 in the regulation of cell metabolism: A cancer perspective. Mol. Cancer 15 (1), 71. doi:10.1186/s12943-016-0558-7
Oegema, T. R. (1993). Biochemistry of the intervertebral disc. Clin. Sports Med. 12 (3), 419–438. doi:10.1016/s0278-5919(20)30404-x
Ohnishi, K., Semi, K., Yamamoto, T., Shimizu, M., Tanaka, A., Mitsunaga, K., et al. (2014). Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell 156 (4), 663–677. doi:10.1016/j.cell.2014.01.005
Pang, L., Yang, K., and Zhang, Z. (2020). High-glucose environment accelerates annulus fibrosus cell apoptosis by regulating endoplasmic reticulum stress. Biosci. Rep. 40 (7), BSR20200262. doi:10.1042/BSR20200262
Parenteau, C. S., Lau, E. C., Campbell, I. C., and Courtney, A. (2021). Prevalence of spine degeneration diagnosis by type, age, gender, and obesity using Medicare data. Sci. Rep. 11 (1), 5389. doi:10.1038/s41598-021-84724-6
Pennicooke, B., Moriguchi, Y., Hussain, I., Bonssar, L., and Hartl, R. (2016). Biological treatment approaches for degenerative disc disease: A review of clinical trials and future directions. Cureus 8 (11), e892. doi:10.7759/cureus.892
Peters, H., Wilm, B., Sakai, N., Imai, K., Maas, R., and Balling, R. (1999). Pax1 and Pax9 synergistically regulate vertebral column development. Development 126 (23), 5399–5408. doi:10.1242/dev.126.23.5399
Pinnell, S. R. (1985). Regulation of collagen biosynthesis by ascorbic acid: A review. Yale J. Biol. Med. 58 (6), 553–559.
Pittenger, M. F., Mackay, A. M., Beck, S. C., Jaiswal, R. K., Douglas, R., Mosca, J. D., et al. (1999). Multilineage potential of adult human mesenchymal stem cells. Science 284 (5411), 143–147. doi:10.1126/science.284.5411.143
Portela, A., and Esteller, M. (2010). Epigenetic modifications and human disease. Nat. Biotechnol. 28 (10), 1057–1068. doi:10.1038/nbt.1685
Price, J. S., Waters, J. G., Darrah, C., Pennington, C., Edwards, D. R., Donell, S. T., et al. (2002). The role of chondrocyte senescence in osteoarthritis. Aging Cell 1 (1), 57–65. doi:10.1046/j.1474-9728.2002.00008.x
Rabinowitz, J. D., and Enerback, S. (2020). Lactate: The ugly duckling of energy metabolism. Nat. Metab. 2 (7), 566–571. doi:10.1038/s42255-020-0243-4
Raggi, F., Blengio, F., Eva, A., Pende, D., Varesio, L., and Bosco, M. C. (2014). Identification of CD300a as a new hypoxia-inducible gene and a regulator of CCL20 and VEGF production by human monocytes and macrophages. Innate Immun. 20 (7), 721–734. doi:10.1177/1753425913507095
Rajpurohit, R., Risbud, M. V., Ducheyne, P., Vresilovic, E. J., and Shapiro, I. M. (2002). Phenotypic characteristics of the nucleus pulposus: Expression of hypoxia inducing factor-1, glucose transporter-1 and MMP-2. Cell Tissue Res. 308 (3), 401–407. doi:10.1007/s00441-002-0563-6
Razaq, S., Urban, J. P., and Wilkins, R. J. (2000). Regulation of intracellular pH by bovine intervertebral disc cells. Cell. Physiol. biochem. 10 (1-2), 109–115. doi:10.1159/000016340
Razaq, S., Wilkins, R. J., and Urban, J. P. (2003). The effect of extracellular pH on matrix turnover by cells of the bovine nucleus pulposus. Eur. spine J. 12 (4), 341–349.
Rettig, W. J., Garin-Chesa, P., Healey, J. H., Su, S. L., Jaffe, E. A., and Old, L. J. (1992). Identification of endosialin, a cell surface glycoprotein of vascular endothelial cells in human cancer. Proc. Natl. Acad. Sci. U. S. A. 89 (22), 10832–10836. doi:10.1073/pnas.89.22.10832
Richardson, S. M., Knowles, R., Tyler, J., Mobasheri, A., and Hoyland, J. A. (2008). Expression of glucose transporters GLUT-1, GLUT-3, GLUT-9 and HIF-1alpha in normal and degenerate human intervertebral disc. Histochem. Cell Biol. 129 (4), 503–511. doi:10.1007/s00418-007-0372-9
Risbud, M. V., Guttapalli, A., Tsai, T. T., Lee, J. Y., Danielson, K. G., Vaccaro, A. R., et al. (2007). Evidence for skeletal progenitor cells in the degenerate human intervertebral disc. Spine 32 (23), 2537–2544. doi:10.1097/BRS.0b013e318158dea6
Risbud, M. V., Schoepflin, Z. R., Mwale, F., Kandel, R. A., Grad, S., Iatridis, J. C., et al. (2015). Defining the phenotype of young healthy nucleus pulposus cells: Recommendations of the spine research interest group at the 2014 annual ORS meeting. J. Orthop. Res. 33 (3), 283–293.
Rivas, A., Ruegg, C. L., Zeitung, J., Laus, R., Warnke, R., Benike, C., et al. (1995). V7, a novel leukocyte surface protein that participates in T cell activation. I. Tissue distribution and functional studies. J. Immunol. 154 (9), 4423–4433.
Rizzino, A. (2013). Concise review: The sox2-oct4 connection: Critical players in a much larger interdependent network integrated at multiple levels. Stem cells 31 (6), 1033–1039. doi:10.1002/stem.1352
Rodriguez, A. M., Elabd, C., Amri, E. Z., Ailhaud, G., and Dani, C. (2005). The human adipose tissue is a source of multipotent stem cells. Biochimie 87 (1), 125–128. doi:10.1016/j.biochi.2004.11.007
Rodriguez, A. M., Elabd, C., Delteil, F., Astier, J., Vernochet, C., Saint-Marc, P., et al. (2004). Adipocyte differentiation of multipotent cells established from human adipose tissue. Biochem. Biophys. Res. Commun. 315 (2), 255–263. doi:10.1016/j.bbrc.2004.01.053
Rosen, E. D., and MacDougald, O. A. (2006). Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 7 (12), 885–896. doi:10.1038/nrm2066
Ryu, D., and Lee, C. (2017). Expression quantitative trait loci for PI3K/AKT pathway. Med. Baltim. 96 (1), e5817. doi:10.1097/MD.0000000000005817
Sadowska, A., Hitzl, W., Karol, A., Jaszczuk, P., Cherif, H., Haglund, L., et al. (2019). Differential regulation of TRP channel gene and protein expression by intervertebral disc degeneration and back pain. Sci. Rep. 9 (1), 18889. doi:10.1038/s41598-019-55212-9
Sakai, D., and Schol, J. (2017). Cell therapy for intervertebral disc repair: Clinical perspective. J. Orthop. Transl. 9, 8–18. doi:10.1016/j.jot.2017.02.002
Saki, N., Jalalifar, M. A., Soleimani, M., Hajizamani, S., and Rahim, F. (2013). Adverse effect of high glucose concentration on stem cell therapy. Int. J. Hematol. Oncol. Stem Cell Res. 7 (3), 34–40.
Scoville, D. W., Kang, H. S., and Jetten, A. M. (2017). GLIS1-3: Emerging roles in reprogramming, stem and progenitor cell differentiation and maintenance. Stem Cell Investig. 4, 80. doi:10.21037/sci.2017.09.01
Shellenberger, T. D., Wang, M., Gujrati, M., Jayakumar, A., Strieter, R. M., Burdick, M. D., et al. (2004). BRAK/CXCL14 is a potent inhibitor of angiogenesis and a chemotactic factor for immature dendritic cells. Cancer Res. 64 (22), 8262–8270. doi:10.1158/0008-5472.CAN-04-2056
Silagi, E. S., Novais, E. J., Bisetto, S., Telonis, A. G., Snuggs, J., Le Maitre, C. L., et al. (2020). Lactate efflux from intervertebral disc cells is required for maintenance of spine Health. J. Bone Min. Res. 35 (3), 550–570. doi:10.1002/jbmr.3908
Singh, K., Masuda, K., Thonar, E. J., An, H. S., and Cs-Szabo, G. (2009). Age-related changes in the extracellular matrix of nucleus pulposus and anulus fibrosus of human intervertebral disc. Spine 34 (1), 10–16. doi:10.1097/BRS.0b013e31818e5ddd
Sivakamasundari, V., Kraus, P., Jie, S., and Lufkin, T. (2013). Pax1(EGFP): New wildtype and mutant EGFP mouse lines for molecular and fate mapping studies. Genesis 51 (6), 420–429. doi:10.1002/dvg.22379
Sivakamasundari, V., Kraus, P., Sun, W., Hu, X., Lim, S. L., Prabhakar, S., et al. (2017). A developmental transcriptomic analysis of Pax1 and Pax9 in embryonic intervertebral disc development. Biol. Open 6 (2), 187–199. doi:10.1242/bio.023218
Sivakamasundari, V., and Lufkin, T. (2012). Bridging the gap: Understanding embryonic intervertebral disc development. Cell Dev. Biol. 1 (2), 103.
Sivakamasundari, V., and Lufkin, T. (2013). Stemming the degeneration: IVD stem cells and stem cell regenerative therapy for degenerative disc disease. Adv. stem cells, 2013, 724547. doi:10.5171/2013.724547
Stewart, C. R., Stuart, L. M., Wilkinson, K., van Gils, J. M., Deng, J., Halle, A., et al. (2010). CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nat. Immunol. 11 (2), 155–161. doi:10.1038/ni.1836
Sugimoto, M., Ikeda, S., Niigata, K., Tomita, M., Sato, H., and Soga, T. (2012). Mmmdb: Mouse multiple tissue metabolome database. Nucleic Acids Res. 40 (1), D809–D814. doi:10.1093/nar/gkr1170
Sullivan, K. D., Galbraith, M. D., Andrysik, Z., and Espinosa, J. M. (2018). Mechanisms of transcriptional regulation by p53. Cell Death Differ. 25 (1), 133–143. doi:10.1038/cdd.2017.174
Swain, S., Sarmanova, A., Coupland, C., Doherty, M., and Zhang, W. (2020). Comorbidities in osteoarthritis: A systematic review and meta-analysis of observational studies. Arthritis Care Res. Hob. 72 (7), 991–1000. doi:10.1002/acr.24008
Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., et al. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131 (5), 861–872. doi:10.1016/j.cell.2007.11.019
Takano, T., Li, Y. J., Kukita, A., Yamaza, T., Ayukawa, Y., Moriyama, K., et al. (2014). Mesenchymal stem cells markedly suppress inflammatory bone destruction in rats with adjuvant-induced arthritis. Lab. Invest. 94 (3), 286–296. doi:10.1038/labinvest.2013.152
Tang, X., Jing, L., and Chen, J. (2012). Changes in the molecular phenotype of nucleus pulposus cells with intervertebral disc aging. PloS one 7 (12), e52020. doi:10.1371/journal.pone.0052020
Tekari, A., Chan, S. C., Sakai, D., Grad, S., and Gantenbein, B. (2016). Angiopoietin-1 receptor Tie2 distinguishes multipotent differentiation capability in bovine coccygeal nucleus pulposus cells. Stem Cell Res. Ther. 7 (1), 75. doi:10.1186/s13287-016-0337-9
Thorpe, A. A., Binch, A. L., Creemers, L. B., Sammon, C., and Le Maitre, C. L. (2016). Nucleus pulposus phenotypic markers to determine stem cell differentiation: Fact or fiction? Oncotarget 7 (3), 2189–2200. doi:10.18632/oncotarget.6782
Tribioli, C., and Lufkin, T. (1999). The murine Bapx1 homeobox gene plays a critical role in embryonic development of the axial skeleton and spleen. Development 126 (24), 5699–5711. doi:10.1242/dev.126.24.5699
Trout, J. J., Buckwalter, J. A., Moore, K. C., and Landas, S. K. (1982). Ultrastructure of the human intervertebral disc. I. Changes in notochordal cells with age. Tissue Cell 14 (2), 359–369. doi:10.1016/0040-8166(82)90033-7
Trout, J. J., Buckwalter, J. A., and Moore, K. C. (1982). Ultrastructure of the human intervertebral disc: II. Cells of the nucleus pulposus. Anat. Rec. 204 (4), 307–314. doi:10.1002/ar.1092040403
Turner, S. A., Wright, K. T., Jones, P. N., Balain, B., and Roberts, S. (2016). Temporal analyses of the response of intervertebral disc cells and mesenchymal stem cells to nutrient deprivation. Stem Cells Int. 2016, 5415901. doi:10.1155/2016/5415901
Urban, J. P., Holm, S., Maroudas, A., and Nachemson, A. (1977). Nutrition of the intervertebral disk. An in vivo study of solute transport. Clin. Orthop. Relat. Res. 129 (129), 101–114. doi:10.1097/00003086-197711000-00012
Urban, J. P., and Roberts, S. (2003). Degeneration of the intervertebral disc. Arthritis Res. Ther. 5 (3), 120–130. doi:10.1186/ar629
Urban, J. P., Smith, S., and Fairbank, J. C. (2004). Nutrition of the intervertebral disc. Spine 29 (23), 2700–2709. doi:10.1097/01.brs.0000146499.97948.52
Valvona, C. J., Fillmore, H. L., Nunn, P. B., and Pilkington, G. J. (2016). The regulation and function of lactate dehydrogenase A: Therapeutic potential in brain tumor. Brain Pathol. 26 (1), 3–17. doi:10.1111/bpa.12299
Van den Steen, P. E., Dubois, B., Nelissen, I., Rudd, P. M., Dwek, R. A., and Opdenakker, G. (2002). Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9). Crit. Rev. Biochem. Mol. Biol. 37 (6), 375–536. doi:10.1080/10409230290771546
Vander Heiden, M. G., Plas, D. R., Rathmell, J. C., Fox, C. J., Harris, M. H., and Thompson, C. B. (2001). Growth factors can influence cell growth and survival through effects on glucose metabolism. Mol. Cell. Biol. 21 (17), 5899–5912. doi:10.1128/mcb.21.17.5899-5912.2001
Vujovic, S., Henderson, S., Presneau, N., Odell, E., Jacques, T. S., Tirabosco, R., et al. (2006). Brachyury, a crucial regulator of notochordal development, is a novel biomarker for chordomas. J. Pathol. 209 (2), 157–165. doi:10.1002/path.1969
Wan, J., Zhang, G., Li, X., Qiu, X., Ouyang, J., Dai, J., et al. (2021). Matrix metalloproteinase 3: A promoting and destabilizing factor in the pathogenesis of disease and cell differentiation. Front. Physiol. 12, 663978. doi:10.3389/fphys.2021.663978
Wang, D., Hartman, R., Han, C., Zhou, C. M., Couch, B., Malkamaki, M., et al. (2021). Lactate oxidative phosphorylation by annulus fibrosus cells: Evidence for lactate-dependent metabolic symbiosis in intervertebral discs. Arthritis Res. Ther. 23 (1), 145. doi:10.1186/s13075-021-02501-2
Wang, H., Yang, Y., Liu, J., and Qian, L. (2021). Direct cell reprogramming: Approaches, mechanisms and progress. Nat. Rev. Mol. Cell Biol. 22 (6), 410–424. doi:10.1038/s41580-021-00335-z
Warburg, O., Wind, F., and Negelein, E. (1927). The metabolism of tumors in the body. J. Gen. Physiol. 8 (6), 519–530. doi:10.1085/jgp.8.6.519
Wilda, M., Bachner, D., Just, W., Geerkens, C., Kraus, P., Vogel, W., et al. (2000). A comparison of the expression pattern of five genes of the family of small leucine-rich proteoglycans during mouse development. J. Bone Min. Res. 15 (11), 2187–2196. doi:10.1359/jbmr.2000.15.11.2187
Wilkins, H. M., Harris, J. L., Carl, S. M., Roy, N., Lu, J., Eva Selfridge, J., et al. (2014). Oxaloacetate activates brain mitochondrial biogenesis, enhances the insulin pathway, reduces inflammation and stimulates neurogenesis. Hum. Mol. Genet. 23 (24), 6528–6541. doi:10.1093/hmg/ddu371
Williams, D. S., Cash, A., Hamadani, L., and Diemer, T. (2009). Oxaloacetate supplementation increases lifespan in Caenorhabditis elegans through an AMPK/FOXO-dependent pathway. Aging Cell 8 (6), 765–768. doi:10.1111/j.1474-9726.2009.00527.x
Wilm, B., Dahl, E., Peters, H., Balling, R., and Imai, K. (1998). Targeted disruption of Pax1 defines its null phenotype and proves haploinsufficiency. Proc. Natl. Acad. Sci. U. S. A. 95 (15), 8692–8697. doi:10.1073/pnas.95.15.8692
Wuertz, K., Godburn, K., and Iatridis, J. C. (2009). MSC response to pH levels found in degenerating intervertebral discs. Biochem. Biophys. Res. Commun. 379 (4), 824–829. doi:10.1016/j.bbrc.2008.12.145
Wuertz, K., Godburn, K., Neidlinger-Wilke, C., Urban, J., and Iatridis, J. C. (2008). Behavior of mesenchymal stem cells in the chemical microenvironment of the intervertebral disc. Spine 33 (17), 1843–1849. doi:10.1097/BRS.0b013e31817b8f53
Xia, K., Gong, Z., Zhu, J., Yu, W., Wang, Y., Wang, J., et al. (2019). Differentiation of pluripotent stem cells into nucleus pulposus progenitor cells for intervertebral disc regeneration. Curr. Stem Cell Res. Ther. 14 (1), 57–64. doi:10.2174/1574888X13666180918095121
Xu, D., Jiang, H. R., Kewin, P., Li, Y., Mu, R., Fraser, A. R., et al. (2008). IL-33 exacerbates antigen-induced arthritis by activating mast cells. Proc. Natl. Acad. Sci. U. S. A. 105 (31), 10913–10918. doi:10.1073/pnas.0801898105
Yako, Y., Hayashi, T., Takeuchi, Y., Ishibashi, K., Kasai, N., Sato, N., et al. (2018). ADAM-like Decysin-1 (ADAMDEC1) is a positive regulator of Epithelial Defense against Cancer (EDAC) that promotes apical extrusion of RasV12-transformed cells. Sci. Rep. 8 (1), 9639. doi:10.1038/s41598-018-27469-z
Yamaguchi, K., Kinosaki, M., Goto, M., Kobayashi, F., Tsuda, E., Morinaga, T., et al. (1998). Characterization of structural domains of human osteoclastogenesis inhibitory factor. J. Biol. Chem. 273 (9), 5117–5123. doi:10.1074/jbc.273.9.5117
Yamamoto, T., Seino, Y., Fukumoto, H., Koh, G., Yano, H., Inagaki, N., et al. (1990). Over-expression of facilitative glucose transporter genes in human cancer. Biochem. Biophys. Res. Commun. 170 (1), 223–230. doi:10.1016/0006-291x(90)91263-r
Yoon, B. S., and Lyons, K. M. (2004). Multiple functions of BMPs in chondrogenesis. J. Cell. Biochem. 93 (1), 93–103. doi:10.1002/jcb.20211
Yoshida, G. J. (2015). Metabolic reprogramming: The emerging concept and associated therapeutic strategies. J. Exp. Clin. Cancer Res. 34, 111. doi:10.1186/s13046-015-0221-y
You, H., and Mak, T. W. (2005). Crosstalk between p53 and FOXO transcription factors. Cell Cycle 4 (1), 37–38. doi:10.4161/cc.4.1.1401
Young, J. I., Zuchner, S., and Wang, G. (2015). Regulation of the epigenome by vitamin C. Annu. Rev. Nutr. 35, 545–564. doi:10.1146/annurev-nutr-071714-034228
Zeineddine, D., Hammoud, A. A., Mortada, M., and Boeuf, H. (2014). The Oct4 protein: More than a magic stemness marker. Am. J. Stem Cells 3 (2), 74–82.
Zhang, D., Tang, Z., Huang, H., Zhou, G., Cui, C., Weng, Y., et al. (2019). Metabolic regulation of gene expression by histone lactylation. Nature 574 (7779), 575–580. doi:10.1038/s41586-019-1678-1
Zhang, J. M., and An, J. (2007). Cytokines, inflammation, and pain. Int. Anesthesiol. Clin. 45 (2), 27–37. doi:10.1097/AIA.0b013e318034194e
Zhou, S., Chen, S., Jiang, Q., and Pei, M. (2019). Determinants of stem cell lineage differentiation toward chondrogenesis versus adipogenesis. Cell. Mol. Life Sci. 76 (9), 1653–1680. doi:10.1007/s00018-019-03017-4
Keywords: intervertebral disc, nucleus pulposus, annulus fibrosus, adipose, stromal cell, 3D culture, transcriptome analysis, regenerative medicine
Citation: Lufkin L, Samanta A, Baker D, Lufkin S, Schulze J, Ellis B, Rose J, Lufkin T and Kraus P (2022) Glis1 and oxaloacetate in nucleus pulposus stromal cell somatic reprogramming and survival. Front. Mol. Biosci. 9:1009402. doi: 10.3389/fmolb.2022.1009402
Received: 13 August 2022; Accepted: 10 October 2022;
Published: 03 November 2022.
Edited by:
Benjamin Gantenbein, University of Bern, SwitzerlandCopyright © 2022 Lufkin, Samanta, Baker, Lufkin, Schulze, Ellis, Rose, Lufkin and Kraus. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Petra Kraus, cGtyYXVzQGNsYXJrc29uLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.