
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Mol. Biosci. , 06 October 2022
Sec. RNA Networks and Biology
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.1008921
This article is part of the Research Topic Insights in RNA Networks and Biology View all 11 articles
The regulation of mRNA translation plays an essential role in neurons, contributing to important brain functions, such as brain plasticity and memory formation. Translation is conducted by ribosomes, which at their core consist of ribosomal proteins (RPs) and ribosomal RNAs. While translation can be regulated at diverse levels through global or mRNA-specific means, recent evidence suggests that ribosomes with distinct configurations are involved in the translation of different subsets of mRNAs. However, whether and how such proclaimed ribosome heterogeneity could be connected to neuronal functions remains largely unresolved. Here, we postulate that the existence of heterologous ribosomes within neurons, especially at discrete synapses, subserve brain plasticity. This hypothesis is supported by recent studies in rodents showing that heterogeneous RP expression occurs in dendrites, the compartment of neurons where synapses are made. We further propose that sleep, which is fundamental for brain plasticity and memory formation, has a particular role in the formation of heterologous ribosomes, specialised in the translation of mRNAs specific for synaptic plasticity. This aspect of our hypothesis is supported by recent studies showing increased translation and changes in RP expression during sleep after learning. Thus, certain RPs are regulated by sleep, and could support different sleep functions, in particular brain plasticity. Future experiments investigating cell-specific heterogeneity in RPs across the sleep-wake cycle and in response to different behaviour would help address this question.
Neurons are the primary cell type in the nervous system (the other being glia, which provide structural and metabolic support), responsible for transmission of electrochemical signals in the form of action potentials (APs). On average, the volume of a neuron is more than 10,000 times greater than most mammalian cells. Neurons have a complex and polarised morphology with processes (i.e., neurites) which are divided into different compartments called dendrites and axons, that can extend millimetres to meters away from the cell body containing the nucleus. Axons transmit APs to other neurons and dendrites receive APs and release neurotransmitters onto little protrusions called spines (Donato et al., 2019) (Figure 1A). The site of connection between axon termini and dendritic spines is called the synapse (Figure 1A). One neuron can host up to hundred thousand spines on its dendrites and thus receive thousands of contacts from different axons, maximising the number of possible synapses between neurons (Chidambaram et al., 2019; Nishiyama, 2019; Megías et al., 2001; Spruston, 2008).
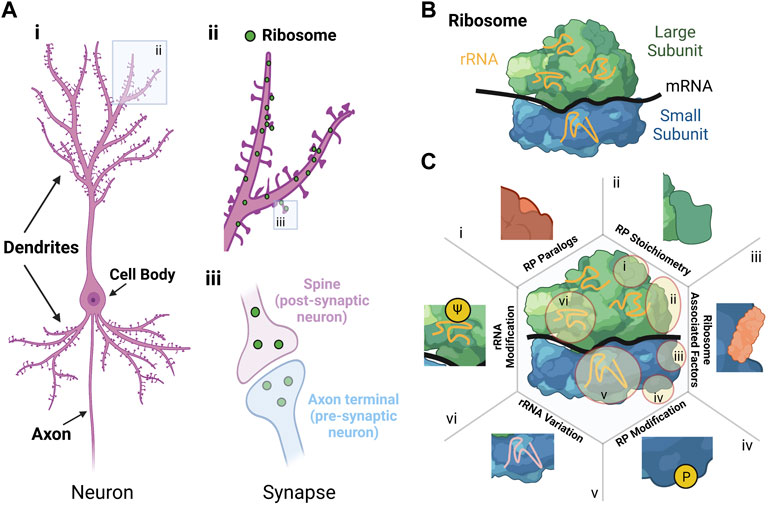
FIGURE 1. Sources and effect of ribosomal heterogeneity in neurons. (A) Morphology of a neuron. i) Schematic of a neuron showing the cell body, dendrites and axon. ii) Close-up schematic of dendrites with ribosomes shown as green dots located both in and adjacent to synapses. iii) Close-up schematic of the synapse with ribosomes shown as green dots in the cytoplasm in both the post-synaptic neuron (i.e., spine in purple) and pre-synaptic neuron (i.e., axon terminal in blue). (B) Structure of the ribosome with the large subunit (green), small subunit (blue), ribosomal RNA (rRNA in orange), and messenger RNA (mRNA in black). For the large and small subunit, the different colours indicate different ribosomal proteins. (C) Schematic showing six sources of ribosomal variation, with illustrative examples of how these changes manifest: i) RP paralogs, ii) RP stoichiometry, iii) Ribosomal associated factors, iv) RP modification, v) rRNA variation, and vi) rRNA modification (adapted from (Norris et al., 2021)). Created with BioRender.com.
In neurons, timely production of proteins in individual neurites and synapses is critical for spatial control of cellular function (Perez et al., 2021b). Indeed, using high resolution imaging of neurons in vivo and in vitro, it has been shown that most pre-synaptic axon termini and post-synaptic spines contain ribosomes, translation factors, and mRNAs (Cajigas et al., 2012; Kitamura et al., 2015; Holt et al., 2019; Sun et al., 2021) (Figure 1A). Local protein synthesis in neurons is required for growth and remodelling of synapses (i.e., synaptic plasticity) (Sutton and Schuman, 2006), which enables critical processes such as brain development, brain plasticity, learning and memory (Banko and Klann, 2008) and recovery from brain injury by promoting synapse-specific production of functionally distinct groups of proteins.
For example, strengthening and weakening of individual synaptic contacts relies on the synthesis of different pools of proteins which ultimately increase or decrease efficiency of communication (Zukin et al., 2009). Local translation of proteins critical for long-term increases in synaptic transmission includes α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors which increases the sensitivity of dendrites to incoming stimuli and strengthens individual synapses (Perez et al., 2021b). Similarly, local translation of proteins with key roles in synaptic structure, such as Arc, actin, PSD95 and α/βCaMKII, participates in the structural modification of dendritic spines during plasticity processes (Nakahata and Yasuda, 2018; Newpher et al., 2018).
As modulation of synaptic communication is naturally linked to memory formation, translation regulation is also important for memory (Sutton and Schuman, 2006; Banko and Klann, 2008), as shown in Drosophila (Ashraf et al., 2006) and rodents. Inhibition of translation initiation (i.e., mTOR signalling) in the rodent hippocampus, a brain structure important for memory formation, impairs performance in a widely used single trial inhibitory avoidance memory task in rodents (Bekinschtein et al., 2007). While in vivo manipulation of local translation at specific synapses remains challenging, experimental and computational studies support the specific involvement of molecular changes in dendrites (Kastellakis and Poirazi, 2019) and spines (Hayashi-Takagi et al., 2015) for memory formation. Altogether, localised mRNA translation within neurons plays a critical role in many brain functions involving the growth and remodelling of synapses. However, a complete picture of the forms and functions of various translation regulatory mechanisms contributing to brain function in neurons remains largely unexplored.
The eukaryotic ribosome is a large ribonucleoprotein complex that consists of a large (60S) and a small (40S) subunit that assemble during the initiation step of translation to form complete 80S ribosomes (Figure 1B). The two subunits share four ribosomal RNAs (rRNAs) (Ford, 2020) and 80 ribosomal proteins RPs, 33 of them allocated to the small (RPS) and 47 to the large (RPL) subunit (De La Cruz et al., 2015). Translation can be regulated at diverse levels (e.g., initiation, elongation, or termination) but major impact is given at the initiation step. Thereby, translation can be controlled at a global level or for specific mRNAs, for example through modification of translation initiation factors and specific RNA binding proteins (RBPs) or microRNAs, respectively (Sonenberg and Hinnebusch, 2009; King and Gerber, 2016).
Beside the regulation of translation mediated through those accessory factors, the view that ribosomes are all identical prevailed for decades. However, increasing evidence over the last decade suggests that cell-specific heterogeneous populations of ribosomes could exist and may result in different preferences of individual ribosomes (or “specialised ribosomes”) for the translation of diverse subsets of mRNAs or in the modulation of translation elongation rates (Genuth and Barna, 2018; Gay et al., 2022). Such heterogenous populations of ribosomes can be formed by (Figure 1C, (Norris et al., 2021)) 1) the exchange or substitution of RP paralogues (e.g., RPL39 and RPL39L) (Komili et al., 2007), 2) altered RP stoichiometry based on differences in RP expression (Emmott et al., 2019); 3) different ribosome-associated proteins (Simsek et al., 2017), 4) post-translational modifications of RPs (Carroll et al., 2008); 5) rRNA composition through variation of rRNA gene sequences (Locati et al., 2017); and 6) rRNA modifications, such as ribose-methylation or pseudouridylation (ψ) (Natchiar et al., 2017). Consequently, these factors could lead to a cell-specific array of ribosome variants, some of them having specialised functions in translation (Li and Wang, 2020; Gay et al., 2022).
A well-described example of ribosome specialisation concerns RPL38 (Xue et al., 2015). Ribosomes containing RPL38 are required for translation of certain Homeobox (Hox) mRNAs that code for proteins defining the body axis and structures (Kondrashov et al., 2011). In developing mice embryos, Rpl38 transcripts were enriched in certain regions including the face, eye, neural tube (brain and spinal cord precursor) and importantly somites, which are precursors of the axial skeleton. These locations tended to overlap with regions where tissue patterning defects were observed in mice with the “Tail short” (Ts) mutation (which display a short and curled tail, an anteroposterior skeletal patterning defect and several other skeletal abnormalities), a phenotype thought to be caused by Rpl38 gene mutation (Kondrashov et al., 2011). RPL38 has been reported to control cap-independent translation of Hox mRNAs via specific internal ribosome entry sites (IRES) (Xue et al., 2015), although results from a recent study suggest that transcriptional promoters or splice sites may instead be responsible for the putative IRES activity in Hox genes (Akirtava et al., 2022). However, selective translation through IRES specificity is likely replicated with other RPs (Hertz et al., 2013; Shi et al., 2017; Kampen et al., 2019). Selective capture of ribosomes containing two specific RPs, RPS25, and RPL10A, combined with ribosome profiling, showed that those RPs preferentially translated unique sets of transcripts (Shi et al., 2017). Specifically, RPS25-containing ribosomes were preferentially associated with transcripts coding for proteins involved in the cell cycle and vitamin B12 pathway, while RPL10A-enriched transcripts were associated with extracellular matrix organisation, system development and steroid metabolism (Shi et al., 2017). In the same way that RPL38 is thought to be required for IRES-dependent translation of Hox mRNAs (Xue et al., 2015), Shi et al. (2017) found that RPL10A is required for the translation of three mRNAs known to contain IRES elements (Igf2, App, and Chmp2a). IRES-dependent translation of these transcripts was significantly reduced upon knockdown of Rpl10a, but not the control Rpl29, indicating that translation of Igf2, App and Chmp2a is attributed to RPL10A.
Further support for the model of ribosome specialisation is provided by examples of human diseases such as ribosomopathies and fragile-X syndrome. Ribosomopathies are pathologies which result from mutations in certain RP genes (Orgebin et al., 2020), with as many as 20 RP genes, including RPS19, involved in Diamond-Blackfan anaemia (DBA) (Leger-Silvestre et al., 2005; Da Costa et al., 2020). Besides a deficiency of erythroblasts, about 50% of DBA patients experience other congenital anomalies such as growth retardation, cardiac and urogenital abnormalities, increased risk of cancer, cephalic malformations and learning difficulties (Da Costa et al., 2020; Panda et al., 2020). Fragile-X syndrome is another well-characterised neurodevelopmental disorder associated with intellectual disability and learning difficulties. Excessive translation due to the loss of the RBP fragile-X mental retardation protein (FMRP) caused by CGG triplet repeat expansions in the FMR1 gene is a key mechanism in the disease (Richter et al., 2015). In this regard, a recent study suggests that excessive translation of RPs in neurons reduces the translation of longer-length transcripts coding for proteins contributing to synaptic stability (Seo et al., 2022), supporting the established link between the loss of FMRP and aberrant synaptic plasticity (Sidorov et al., 2013).
While research is needed to determine the biological functions of ribosome heterogeneity, some studies have highlighted its importance in specific cellular processes, in particular during development (Li and Wang, 2020; Norris et al., 2021). Whether ribosome heterogeneity exists and has functions in the brain remains under-investigated.
Although specific RPs (e.g., RPL38, RPS25, and RPL10A) have functional roles in different cell types (see above) (Li and Wang, 2020), still very little is known about whether RPs are differentially regulated or expressed in different areas of the brain or in specific brain cells. For example, RP heterogeneity between brain regions has been reported in normal and brain cancer samples (Panda et al., 2020). Conversely, ageing does apparently not drastically influence RP stoichiometry in the cortex, cerebellum and hippocampus when assessed in a mixed cell preparation (Amirbeigarab et al., 2019); this does not, however, exclude the possibility for heterogeneity associated with specific cell types forming those tissues, including neurons and glial cells. In the following sections, we further focus our considerations on ribosome heterogeneity and functions in neurons, which are the brain cells responsible for activity in the brain and are central for information processing supporting cognitive functions.
The recent implementation of various transcriptomics and proteomics approaches applied in vitro and in vivo (Figure 2C) revealed that neurons show differential expression of RPs within the dendritic and axonal sub-compartment compared to a mixed cell population (Poulopoulos et al., 2019; Biever et al., 2020; Perez et al., 2021a) (Figure 2A) and suggest a compartment-specific heterogeneity in RP expression within neurons (Figure 2B). Additionally, an influential study into ribosome specialisation was recently carried out in Xenopus laevis showing that remodelling of ribosomes occurred in axons by exchange of locally synthesised RPs (Shigeoka et al., 2019). While ribosome biogenesis typically occurs in the nucleus, this study provides further evidence that ribosomes are dynamic structures.
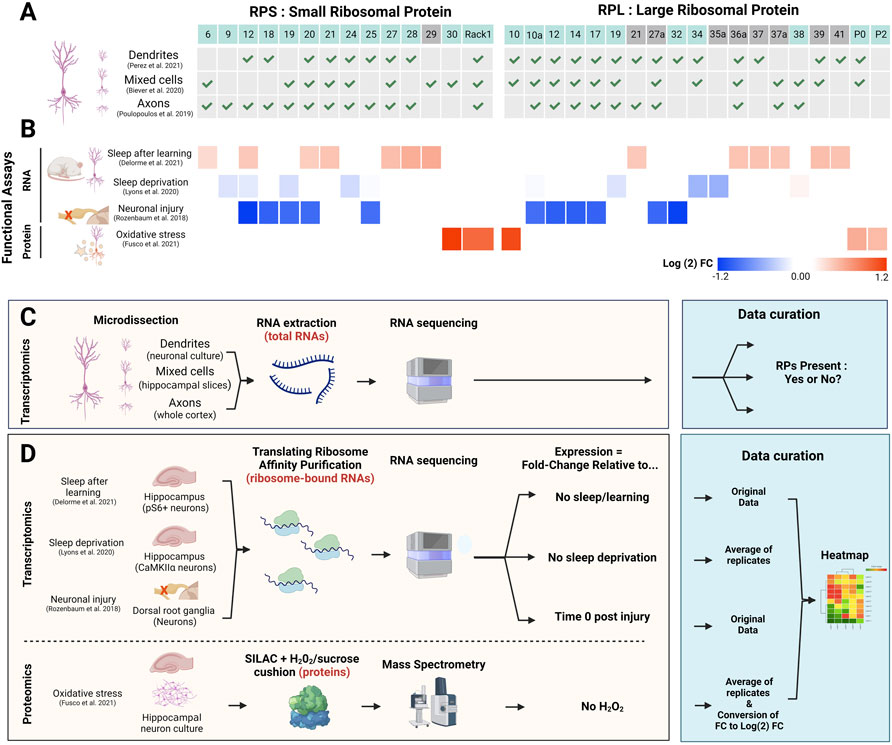
FIGURE 2. Contribution of RPs to ribosome heterogeneity in neurons and functional significance. (A) Table indicating the presence (green check mark) or absence of select RP (top line) mRNAs in whole neurons vs. dendrites or axons, according to previous studies (Poulopoulos et al., 2019; Biever et al., 2020; Perez et al., 2021a). RP location (non-surface: grey boxes; surface: blue boxes) was determined using human ribosome structure (PDB: 4V6X). (B) Heatmap (Morpheus, https://software.broadinstitute.org/morpheus) representing heterogeneous expression of selected RPs in neurons from in vivo studies in rodents or in vitro studies. The colour bar depicts log (2) fold changes (FC) of the particular conditions (indicated on the left) against control condition values (see D and (Rozenbaum et al., 2018; Lyons et al., 2020; Delorme et al., 2021; Fusco et al., 2021) for experimental details). From top to bottom: 1) 13 RP transcripts on membrane-bound ribosomes in activated (pS6+) neurons in the hippocampus following contextual fear conditioning and subsequent sleep in mice (Delorme et al., 2021). 2) 7 RP mRNAs affected by acute (5 h) sleep deprivation in excitatory neurons (CamKIIα+) in the mouse hippocampus (Lyons et al., 2020). 3) The five largest changes for small and large ribosomal subunits (10 RPs) in dorsal root ganglion (DRG) neurons following sciatic nerve injury, 4 h after injury (Rozenbaum et al., 2018). 4) 5 RPs that are significantly upregulated after 0.1 mM H2O2 treatment of primary neuronal culture; values are average of three biological replicates (Fusco et al., 2021). Additionally, log (2)FC values for RPs previously mentioned in the text and associated with ribosome specialisation (RPS19, RPS25, RPL10A and RPL38) were included for each study if they were present in the original data. (C,D) Schematic experimental approaches of the studies performed in neuronal compartments (C) relates to (A) and functional assays (D) relates to (B). The blue boxes specify the type of analysis performed for each study for data visualisation. Created with BioRender.com.
Several studies have been using the translating ribosome affinity purification (TRAP) method to study neuron-specific changes in the translatome (i.e., pool of translated mRNA). Those studies have provided evidence for differential expression of ribosomal components and RPs in neurons, in particular upon changing conditions/neuronal activation (Figures 2B,D). For instance, more than 1,600 differentially expressed transcripts were identified in dorsal root ganglion (DRG) neurons following sciatic nerve injury (Rozenbaum et al., 2018). Importantly, in lumbar DRG neurons, mRNAs for several RPs were significantly decreased 4 h after injury, with the greatest decrease in expression seen for RPS12 and RPL32 (Figure 2B), and recovery of RP expression after 12 h. Another study in rats utilised single-molecule fluorescence in situ hybridisation to show variable expression of 29 different RP mRNAs in dendrites in both hippocampal slices and primary neuronal culture (Fusco et al., 2021). Using an elegant approach combining heavy amino acid labelling (dynamic SILAC) followed by mass spectrometry to label and identify newly synthesised RPs in translating ribosomes, the authors revealed that a subset of 12 RPs showed a dynamic profile of association/dissociation with ribosomes in dendrites, including RPL38 described above. The majority of those 12 RPs are located at the surface of the ribosome, making those proteins susceptible to exchange or post-translational modifications (PTMs) that could—in principle—modulate the function/activity of ribosomes. In fact, many of the RPs showing compartment-specific differences and functional changes are located on the surface of the ribosome (Figure 2). An example of such PTMs of RPs is the phosphorylation of RPS6 in response to a variety of neuronal stimuli, adding the possibility for introduction of compartment specific and activity specific PTM of RPs, propagating functional impact for translation of mRNA subsets (Knight et al., 2012). Finally, changes in the environment (i.e., oxidative stress induced by hydrogen peroxide exposure) elicit five of those 12 RPs to increase their association with the ribosome (Fusco et al., 2021) (Figure 2B), suggesting that physiological stress modifies ribosome composition and function, which could directly affect translation of specific mRNAs. Therefore, other changes in physiological states could also have a significant impact on RP ribosomal protein heterogeneity in neurons.
Altogether, we propose that the existence of heterologous ribosomes within neurons, especially at discrete synapses, may contribute to specific functions, including synaptic plasticity. Dynamic incorporation of RPs to alter ribosome stoichiometry could facilitate rapid formation of specialised ribosomes and enable the translation of subsets of mRNAs involved in the remodelling of synapses. We further postulate that sleep, which is accompanied by major physiological changes in the brain and is an important regulator of the synaptic proteome (Noya et al., 2019), translation and brain plasticity (Seibt and Frank, 2019), could use changes in ribosome heterogeneity and specialised ribosomes to regulate the translation of mRNAs important for synaptic remodelling.
The role of sleep in brain plasticity and memory is well established; mounting evidence shows that sleep enhances the physiological and behavioural changes associated with new experiences (Abel et al., 2013; Rasch and Born, 2013; Raven et al., 2018; Puentes-Mestril et al., 2019). For example, performance in various types of memory tasks in humans and animals is significantly increased when sleep occurs right after learning (Alger et al., 2015; Schmid et al., 2020). Similarly, new sensory experience changes perception in a sleep-dependent manner during development (Frank et al., 2001) and adulthood (Durkin and Aton, 2019). Furthermore, work in the last decade, using high-resolution imaging techniques, has provided strong evidence that sleep influences changes in dendritic spine structure, linked to brain plasticity and memory (Yang et al., 2014; Li et al., 2017; Seibt et al., 2017; Zhou et al., 2020; Aime et al., 2022). Finally, the establishment of our basic sensorimotor system in the central nervous system is thought to largely depend on sleep during early development (Blumberg, 2015). Since long-term changes associated with synaptic plasticity and memory require protein synthesis to persist over time (Davis and Squire, 1984; Costa-Mattioli et al., 2009), sleep may support brain plasticity via translation regulation, including specialised ribosomes, for translation of particular subsets of mRNAs.
The current view suggests that experience-dependent transcription (e.g., immediate early genes) (Yap and Greenberg, 2018) occurs preferentially during wakefulness in the nucleus, while mRNA translation occurs mostly during sleep in a distributed manner across neurites (Seibt and Frank, 2019). This is supported by evidence showing that translation rates are increased during sleep in the brain in various species (Ramm and Smith, 1990; Nakanishi et al., 1997) and the expression of regulators of translation initiation and elongation occurs preferentially during sleep (Cirelli et al., 2004; Mackiewicz et al., 2007; Seibt et al., 2012). Moreover, sleep deprivation leads to a decrease in translation initiation with associated memory deficits (Tudor et al., 2016) and pharmacological disruption of translation initiation during sleep impairs experience-dependent synaptic plasticity in vivo (Seibt et al., 2012), further suggesting the importance of translation during sleep for brain plasticity and memory consolidation. The underlying pathways for translational control during sleep are still not well-characterised, but global control of translation initiation, via the mTORC-1 signalling pathway, seems to be specifically activated during sleep (Seibt et al., 2012; Tudor et al., 2016). (Gingras et al., 1999; Seibt et al., 2012; Tudor et al., 2016) Other factors such as RBPs, microRNAs or non-coding RNAs (ncRNAs) are all potential mechanisms involved in translational control of specific mRNAs during sleep. There is some evidence that sleep deprivation differentially impacts the expression of groups of microRNAs in different parts of the brain, with a trend toward decreased expression in the cortex and increased expression in the hippocampus (Davis et al., 2007). Although this supports a region-specific regulation of translation by the sleep-wake cycle via microRNAs, the data on this remain isolated.
Besides global and specific control through signalling pathways and RBPs/ncRNAs, respectively, whether the formation of specialised ribosomes could also contribute to selected translation during sleep remains unclear. However, sleep-dependent translational changes were examined within neurons in the hippocampus after learning in mice (Delorme et al., 2021) (Figure 2C). Using TRAP, ribosome-associated transcripts were identified from different subcellular fractions of neurons (i.e., cytosolic vs. membrane-associated ribosomes). Sleep deprivation primarily affected mRNA translated in the cytosol, while learning mainly altered transcripts on membrane-bound ribosomes, suggesting a first level of translational specificity within neurons. Importantly, sleep after learning showed increased translation of membrane-bound transcripts, including mRNAs of 13 RPs, such as Rps27 and Rps28, with ∼50% of the RPs located at the ribosome surface (Figure 2B) (Delorme et al., 2021). These changes were specifically allocated to sleep as RP mRNA expression did not increase if sleep was prevented after learning. Differential RP translation during sleep may thus support compartmentalised heterogenous populations of ribosomes occurring through exchange and incorporation of RPs at the surface of ribosomes. Another, indirect, evidence supporting a role for sleep in increased translation of RPs was provided by a study investigating the impact of sleep deprivation (SD) on hippocampal neurons using TRAP in mice (Lyons et al., 2020). Following 5 h of SD, 198 mRNAs showed differential association with ribosomes compared to sleep control mice (Lyons et al., 2020). Certain RPs showed a trend toward decreased expression; transcripts for RPs previously associated with ribosome specialisation such as Rps25, Rpl10a and Rpl38 displayed little to no change, but other RP mRNAs like Rpl34 and Rpl35a were more affected (Figure 2B). Although none of these changes were found to be significant, the data nevertheless suggests that sleep may promote the expression of particular RPs as short sleep deprivation tends to reduce their translation (Lyons et al., 2020).
Although the data are still sparse, they do support a specific effect of sleep on differential RP expression in brain regions important for memory. The variability of changes observed in the various physiological and behavioural paradigms align with the idea that different neuronal functions are accompanied by expression of different ribosomes, which may favour formation of specialised ribosomes.
Several studies revealed differences in RP expression in neurons, some of them specifically during sleep. Furthermore, many RPs are located on the ribosomal surface, adding the possibility for alternative integration or exchange with other RPs. Changes in RP expression and stoichiometry could contribute to the remodelling of neuronal networks and other processes that benefit from sleep, such as general metabolism and membrane repair (Mackiewicz et al., 2007; Anafi et al., 2019), energy conservation (Roth et al., 2010), mood and stress restoration (Goldstein and Walker, 2014), or the clearance of toxins (Xie et al., 2013; van Alphen et al., 2021). While the incorporation and presence of specialised ribosomes in neurons needs to be shown, it may allow for translation of different subsets of mRNAs across individual neuronal compartments and sleep stages.
Experience-dependent plasticity, including memory, leads to the formation or strengthening of certain synapses, whereas others are weakened or even removed. (Yang et al., 2009; Sanders et al., 2012; El-Boustani et al., 2018). Consequently, within the same dendrites, some spines grow while others retract. These dynamic processes involve different mechanisms and proteins, which are - at least in part - instructed by the synthesis of process-relevant proteins. Furthermore, sleep is composed of two different stages, rapid-eye movement (REM) and non-REM sleep, which alternate within minutes in rodents (i.e., one NREM-REM cycle ∼5–10 min) (Trachsel et al., 1991). Interestingly, those phases are coupled with the formation and removal of synapses (Bellesi and De Vivo, 2020; Sun et al., 2020), occurring preferentially during NREM and REM sleep phases, respectively (Yang et al., 2014; Zhou et al., 2020). Hence, assembly of specialised ribosomes during NREM and/or REM sleep could be well-suited to quickly adjust mRNA translation, thereby promoting and/or consolidating the bi-directional plasticity at synapses. How specialised ribosomes are established, controlled, and could become selectively activated during particular sleep phases remains to be uncovered and may be linked to specific brain waves during NREM and REM sleep, known to reactivate selected circuits and enhance memory (Poe, 2017; Ngo et al., 2020; Zhou et al., 2020; Skelin et al., 2021).
The currently available data relate mostly to differential mRNA expression of RPs (Figure 2). However, the data does not necessarily show that the RPs are also synthesised and incorporated into active ribosomes, possibly contributing to ribosome heterogeneity. Thus, besides establishing and monitoring specific RP synthesis in neurons, several questions need to be addressed in the future: 1) Does RP heterogeneity take place in neuronal sub-compartments? 2) How does RP heterogeneity in specific neuronal compartments impact the translation of subsets of mRNAs? 3) How do brain states modulate ribosome heterogeneity to generate specialised ribosomes? Finally, besides neurons there are other important brain cells, such as glia (e.g., astrocytes, microglia) and the vasculature, where ribosome heterogeneity may apply and entail specialised functions (Knight et al., 2012; Bellesi et al., 2015).
In the future, we expect that fundamental questions will be addressed in vitro using either brain slices or primary neuronal cultures. For instance, the application of proximity-based labelling techniques could allow the isolation of proteins/RNAs in the vicinity of a target molecule (e.g., RPs and mRNAs) (Padron et al., 2019; Ramanathan et al., 2019), which should identify the spatial partners present under different physiological conditions to help understand function. Furthermore, we have shown evidence of translating RPs in different physiological states (Figure 2). Combining puromycylation (Tom Dieck et al., 2015), to tag newly synthesised proteins, with specific antibodies (i.e. RPs) could help understand the spatial location of these newly translated RPs, to further understand RNA/protein interactions (Weissinger et al., 2021) in different brain cells and neuronal compartments in vitro. Obtaining functional in vivo data remains the gold standard for understanding molecular mechanisms linked to behaviour, including sleep. However, due to technical challenges of in vivo pharmacology (e.g., diffusion, dilution, biochemical reactions), the application of the above-mentioned methods remains difficult (Uezu et al., 2016) and further advances for the relevant techniques are required. For example, in vivo imaging of small RPs and RNA (Tatavarty et al., 2012; Tutucci et al., 2018; Cawte et al., 2020) is currently difficult, but not impossible (Grimm et al., 2017; Wegner et al., 2017; Hrabetova et al., 2018). Technical improvements such as brighter and more photostable fluorophores, higher resolution imaging, and better access to the tissue of interest (Tutucci et al., 2018; Das et al., 2021) may be key developments to advance the field, allowing better monitoring of RP localisation in cells. Furthermore, improved methods for genetic manipulations (Raguram et al., 2022), to target RPs with fewer off-target side effects, may allow tagging of different ribosomal components at the same time, facilitating isolation of different ribosome complexes from the same cell with TRAP. At the end, those approaches could be combined with behavioural and sleep manipulations, opening the paths towards fundamental understanding of the functional impact of ribosome heterogeneity in complex physiological processes.
The original contributions presented in the study are included in the article; further enquiries can be directed to the corresponding authors.
IB performed data analysis. TS and JS made the figures. All authors made intellectual contributions and wrote the manuscript.
This work was supported by a grant from the Leverhulme Trust to JS and APG (RPG-2020-340), and a Wolfson Research Merit Award from the Royal Society to APG (WM170026). We thank the University of Surrey for defraying publication costs.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abel, T., Havekes, R., Saletin, J. M., and Walker, M. P. (2013). Sleep, plasticity and memory from molecules to whole-brain networks. Curr. Biol. 23, R774–R788. doi:10.1016/j.cub.2013.07.025
Aime, M., Calcini, N., Borsa, M., Campelo, T., Rusterholz, T., Sattin, A., et al. (2022). Paradoxical somatodendritic decoupling supports cortical plasticity during REM sleep. Science 376, 724–730. doi:10.1126/science.abk2734
Akirtava, C., May, G. E., and Mcmanus, C. J. (2022). False-positive IRESes from Hoxa9 and other genes resulting from errors in mammalian 5' UTR annotations. Proc. Natl. Acad. Sci. U. S. A. 119, e2122170119. doi:10.1073/pnas.2122170119
Alger, S. E., Chambers, A. M., Cunningham, T., and Payne, J. D. (2015). The role of sleep in human declarative memory consolidation. Curr. Top. Behav. Neurosci. 25, 269–306. doi:10.1007/7854_2014_341
Amirbeigarab, S., Kiani, P., Sanchez, A. V., Krisp, C., Kazantsev, A., Fester, L., et al. (2019). Invariable stoichiometry of ribosomal proteins in mouse brain tissues with aging. Proc. Natl. Acad. Sci. U. S. A. 116, 22567–22572. doi:10.1073/pnas.1912060116
Anafi, R. C., Kayser, M. S., and Raizen, D. M. (2019). Exploring phylogeny to find the function of sleep. Nat. Rev. Neurosci. 20, 109–116. doi:10.1038/s41583-018-0098-9
Ashraf, S. I., Mcloon, A. L., Sclarsic, S. M., and Kunes, S. (2006). Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell 124, 191–205. doi:10.1016/j.cell.2005.12.017
Banko, J. L., and Klann, E. (2008). Cap-dependent translation initiation and memory. Prog. Brain Res. 169, 59–80. doi:10.1016/S0079-6123(07)00004-0
Bekinschtein, P., Katche, C., Slipczuk, L. N., Igaz, L. M., Cammarota, M., Izquierdo, I., et al. (2007). mTOR signaling in the hippocampus is necessary for memory formation. Neurobiol. Learn. Mem. 87, 303–307. doi:10.1016/j.nlm.2006.08.007
Bellesi, M., and De Vivo, L. (2020). Structural synaptic plasticity across sleep and wake. Curr. Opin. Physiol. 15, 74–81. doi:10.1016/j.cophys.2019.12.007
Bellesi, M., De Vivo, L., Tononi, G., and Cirelli, C. (2015). Transcriptome profiling of sleeping, waking, and sleep deprived adult heterozygous Aldh1L1 - eGFP-L10a mice. Genom. Data 6, 114–117. doi:10.1016/j.gdata.2015.08.031
Biever, A., Glock, C., Tushev, G., Ciirdaeva, E., Dalmay, T., Langer, J. D., et al. (2020). Monosomes actively translate synaptic mRNAs in neuronal processes. Science 367, eaay4991. doi:10.1126/science.aay4991
Blumberg, M. S. (2015). Developing sensorimotor systems in our sleep. Curr. Dir. Psychol. Sci. 24, 32–37. doi:10.1177/0963721414551362
Cajigas, I. J., Tushev, G., Will, T. J., Tom Dieck, S., Fuerst, N., and Schuman, E. M. (2012). The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron 74, 453–466. doi:10.1016/j.neuron.2012.02.036
Carroll, A. J., Heazlewood, J. L., Ito, J., and Millar, A. H. (2008). Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification. Mol. Cell. Proteomics 7, 347–369. doi:10.1074/mcp.M700052-MCP200
Cawte, A. D., Unrau, P. J., and Rueda, D. S. (2020). Live cell imaging of single RNA molecules with fluorogenic Mango II arrays. Nat. Commun. 11, 1283. doi:10.1038/s41467-020-14932-7
Chidambaram, S. B., Rathipriya, A. G., Bolla, S. R., Bhat, A., Ray, B., Mahalakshmi, A. M., et al. (2019). Dendritic spines: Revisiting the physiological role. Prog. Neuropsychopharmacol. Biol. Psychiatry 92, 161–193. doi:10.1016/j.pnpbp.2019.01.005
Cirelli, C., Gutierrez, C. M., and Tononi, G. (2004). Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron 41, 35–43. doi:10.1016/s0896-6273(03)00814-6
Costa-Mattioli, M., Sossin, W. S., Klann, E., and Sonenberg, N. (2009). Translational control of long-lasting synaptic plasticity and memory. Neuron 61, 10–26. doi:10.1016/j.neuron.2008.10.055
Da Costa, L., Leblanc, T., and Mohandas, N. (2020). Diamond-Blackfan anemia. Blood 136, 1262–1273. doi:10.1182/blood.2019000947
Das, S., Vera, M., Gandin, V., Singer, R. H., and Tutucci, E. (2021). Intracellular mRNA transport and localized translation. Nat. Rev. Mol. Cell Biol. 22, 483–504. doi:10.1038/s41580-021-00356-8
Davis, C. J., Bohnet, S. G., Meyerson, J. M., and Krueger, J. M. (2007). Sleep loss changes microRNA levels in the brain: A possible mechanism for state-dependent translational regulation. Neurosci. Lett. 422, 68–73. doi:10.1016/j.neulet.2007.06.005
Davis, H. P., and Squire, L. R. (1984). Protein synthesis and memory: A review. Psychol. Bull. 96, 518–559. doi:10.1037/0033-2909.96.3.518
De La Cruz, J., Karbstein, K., and Woolford, J. L. (2015). Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu. Rev. Biochem. 84, 93–129. doi:10.1146/annurev-biochem-060614-033917
Delorme, J., Wang, L., Kodoth, V., Wang, Y., Ma, J., Jiang, S., et al. (2021). Hippocampal neurons' cytosolic and membrane-bound ribosomal transcript profiles are differentially regulated by learning and subsequent sleep. Proc. Natl. Acad. Sci. U. S. A. 118, e2108534118. doi:10.1073/pnas.2108534118
Donato, A., Kagias, K., Zhang, Y., and Hilliard, M. A. (2019). Neuronal sub-compartmentalization: A strategy to optimize neuronal function. Biol. Rev. Camb. Philos. Soc. 94, 1023–1037. doi:10.1111/brv.12487
Durkin, J. M., and Aton, S. J. (2019). How sleep shapes thalamocortical circuit function in the visual system. Annu. Rev. Vis. Sci. 5, 295–315. doi:10.1146/annurev-vision-091718-014715
El-Boustani, S., Ip, J. P. K., Breton-Provencher, V., Knott, G. W., Okuno, H., Bito, H., et al. (2018). Locally coordinated synaptic plasticity of visual cortex neurons in vivo. Science 360, 1349–1354. doi:10.1126/science.aao0862
Emmott, E., Jovanovic, M., and Slavov, N. (2019). Ribosome stoichiometry: From form to function. Trends biochem. Sci. 44, 95–109. doi:10.1016/j.tibs.2018.10.009
Ford, D. (2020). Ribosomal heterogeneity - a new inroad for pharmacological innovation. Biochem. Pharmacol. 175, 113874. doi:10.1016/j.bcp.2020.113874
Frank, M. G., Issa, N. P., and Stryker, M. P. (2001). Sleep enhances plasticity in the developing visual cortex. Neuron 30, 275–287. doi:10.1016/s0896-6273(01)00279-3
Fusco, C. M., Desch, K., Dorrbaum, A. R., Wang, M., Staab, A., Chan, I. C. W., et al. (2021). Neuronal ribosomes exhibit dynamic and context-dependent exchange of ribosomal proteins. Nat. Commun. 12, 6127. doi:10.1038/s41467-021-26365-x
Gay, D. M., Lund, A. H., and Jansson, M. D. (2022). Translational control through ribosome heterogeneity and functional specialization. Trends biochem. Sci. 47, 66–81. doi:10.1016/j.tibs.2021.07.001
Genuth, N. R., and Barna, M. (2018). The discovery of ribosome heterogeneity and its implications for gene regulation and organismal life. Mol. Cell 71, 364–374. doi:10.1016/j.molcel.2018.07.018
Gingras, A. C., Gygi, S. P., Raught, B., Polakiewicz, R. D., Abraham, R. T., Hoekstra, M. F., et al. (1999). Regulation of 4E-BP1 phosphorylation: A novel two-step mechanism. Genes Dev. 13, 1422–1437. doi:10.1101/gad.13.11.1422
Goldstein, A. N., and Walker, M. P. (2014). The role of sleep in emotional brain function. Annu. Rev. Clin. Psychol. 10, 679–708. doi:10.1146/annurev-clinpsy-032813-153716
Grimm, J. B., Muthusamy, A. K., Liang, Y., Brown, T. A., Lemon, W. C., Patel, R., et al. (2017). A general method to fine-tune fluorophores for live-cell and in vivo imaging. Nat. Methods 14, 987–994. doi:10.1038/nmeth.4403
Hayashi-Takagi, A., Yagishita, S., Nakamura, M., Shirai, F., Wu, Y. I., Loshbaugh, A. L., et al. (2015). Labelling and optical erasure of synaptic memory traces in the motor cortex. Nature 525, 333–338. doi:10.1038/nature15257
Hertz, M. I., Landry, D. M., Willis, A. E., Luo, G., and Thompson, S. R. (2013). Ribosomal protein S25 dependency reveals a common mechanism for diverse internal ribosome entry sites and ribosome shunting. Mol. Cell. Biol. 33, 1016–1026. doi:10.1128/MCB.00879-12
Holt, C. E., Martin, K. C., and Schuman, E. M. (2019). Local translation in neurons: Visualization and function. Nat. Struct. Mol. Biol. 26, 557–566. doi:10.1038/s41594-019-0263-5
Hrabetova, S., Cognet, L., Rusakov, D. A., and Nagerl, U. V. (2018). Unveiling the extracellular space of the brain: From super-resolved microstructure to in vivo function. J. Neurosci. 38, 9355–9363. doi:10.1523/JNEUROSCI.1664-18.2018
Kampen, K. R., Sulima, S. O., Verbelen, B., Girardi, T., Vereecke, S., Rinaldi, G., et al. (2019). The ribosomal RPL10 R98S mutation drives IRES-dependent BCL-2 translation in T-ALL. Leukemia 33, 319–332. doi:10.1038/s41375-018-0176-z
Kastellakis, G., and Poirazi, P. (2019). Synaptic clustering and memory formation. Front. Mol. Neurosci. 12, 300. doi:10.3389/fnmol.2019.00300
King, H. A., and Gerber, A. P. (2016). Translatome profiling: Methods for genome-scale analysis of mRNA translation. Brief. Funct. Genomics 15, 22–31. doi:10.1093/bfgp/elu045
Knight, Z. A., Tan, K., Birsoy, K., Schmidt, S., Garrison, J. L., Wysocki, R. W., et al. (2012). Molecular profiling of activated neurons by phosphorylated ribosome capture. Cell 151, 1126–1137. doi:10.1016/j.cell.2012.10.039
Komili, S., Farny, N. G., Roth, F. P., and Silver, P. A. (2007). Functional specificity among ribosomal proteins regulates gene expression. Cell 131, 557–571. doi:10.1016/j.cell.2007.08.037
Kondrashov, N., Pusic, A., Stumpf, C. R., Shimizu, K., Hsieh, A. C., Ishijima, J., et al. (2011). Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145, 383–397. doi:10.1016/j.cell.2011.03.028
Kozak, M. (2005). Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361, 13–37. doi:10.1016/j.gene.2005.06.037
Leger-Silvestre, I., Caffrey, J. M., Dawaliby, R., Alvarez-Arias, D. A., Gas, N., Bertolone, S. J., et al. (2005). Specific role for yeast homologs of the Diamond blackfan anemia-associated Rps19 protein in ribosome synthesis. J. Biol. Chem. 280, 38177–38185. doi:10.1074/jbc.M506916200
Li, D., and Wang, J. (2020). Ribosome heterogeneity in stem cells and development. J. Cell Biol. 219, e202001108. doi:10.1083/jcb.202001108
Li, W., Ma, L., Yang, G., and Gan, W. B. (2017). REM sleep selectively prunes and maintains new synapses in development and learning. Nat. Neurosci. 20, 427–437. doi:10.1038/nn.4479
Locati, M. D., Pagano, J. F. B., Girard, G., Ensink, W. A., Van Olst, M., Van Leeuwen, S., et al. (2017). Expression of distinct maternal and somatic 5.8S, 18S, and 28S rRNA types during zebrafish development. RNA 23, 1188–1199. doi:10.1261/rna.061515.117
Lyons, L. C., Chatterjee, S., Vanrobaeys, Y., Gaine, M. E., and Abel, T. (2020). Translational changes induced by acute sleep deprivation uncovered by TRAP-Seq. Mol. Brain 13, 165. doi:10.1186/s13041-020-00702-5
Mackiewicz, M., Shockley, K. R., Romer, M. A., Galante, R. J., Zimmerman, J. E., Naidoo, N., et al. (2007). Macromolecule biosynthesis: A key function of sleep. Physiol. Genomics 31, 441–457. doi:10.1152/physiolgenomics.00275.2006
Megías, M., Emri, Z., Freund, T. F., and Gulyás, A. I. (2001). Total number and distribution of inhibitory and excitatory synapses on hippocampal CA1 pyramidal cells. Neuroscience 102, 527–540.
Nakahata, Y., and Yasuda, R. (2018). Plasticity of spine structure: Local signaling, translation and cytoskeletal reorganization. Front. Synaptic Neurosci. 10, 29. doi:10.3389/fnsyn.2018.00029
Nakanishi, H., Sun, Y., Nakamura, R. K., Mori, K., Ito, M., Suda, S., et al. (1997). Positive correlations between cerebral protein synthesis rates and deep sleep in Macaca mulatta. Eur. J. Neurosci. 9, 271–279. doi:10.1111/j.1460-9568.1997.tb01397.x
Natchiar, S. K., Myasnikov, A. G., Kratzat, H., Hazemann, I., and Klaholz, B. P. (2017). Visualization of chemical modifications in the human 80S ribosome structure. Nature 551, 472–477. doi:10.1038/nature24482
Newpher, T. M., Harris, S., Pringle, J., Hamilton, C., and Soderling, S. (2018). Regulation of spine structural plasticity by Arc/Arg3.1. Semin. Cell Dev. Biol. 77, 25–32. doi:10.1016/j.semcdb.2017.09.022
Ngo, H. V., Fell, J., and Staresina, B. (2020). Sleep spindles mediate hippocampal-neocortical coupling during long-duration ripples. Elife 9, e57011. doi:10.7554/eLife.57011
Nishiyama, J. (2019). Plasticity of dendritic spines: Molecular function and dysfunction in neurodevelopmental disorders. Psychiatry Clin. Neurosci. 73, 541–550. doi:10.1111/pcn.12899
Norris, K., Hopes, T., and Aspden, J. L. (2021). Ribosome heterogeneity and specialization in development, Wiley Interdiscip. Rev. RNA, 12. e1644. doi:10.1002/wrna.1644
Noya, S. B., Colameo, D., Bruning, F., Spinnler, A., Mircsof, D., Opitz, L., et al. (2019). The forebrain synaptic transcriptome is organized by clocks but its proteome is driven by sleep. Science 366, eaav2642. doi:10.1126/science.aav2642
Orgebin, E., Lamoureux, F., Isidor, B., Charrier, C., Ory, B., Lezot, F., et al. (2020)., 9. Cells, E2080. doi:10.3390/cells9092080Ribosomopathies: New therapeutic perspectivesCells
Padron, A., Iwasaki, S., and Ingolia, N. T. (2019). Proximity RNA labeling by APEX-seq reveals the organization of translation initiation complexes and repressive RNA granules. Mol. Cell 75, 875–887. doi:10.1016/j.molcel.2019.07.030
Panda, A., Yadav, A., Yeerna, H., Singh, A., Biehl, M., Lux, M., et al. (2020). Tissue- and development-stage-specific mRNA and heterogeneous CNV signatures of human ribosomal proteins in normal and cancer samples. Nucleic Acids Res. 48, 7079–7098. doi:10.1093/nar/gkaa485
Perez, J. D., Dieck, S. T., Alvarez-Castelao, B., Tushev, G., Chan, I. C., and Schuman, E. M. (2021a). Subcellular sequencing of single neurons reveals the dendritic transcriptome of GABAergic interneurons. Elife 10, e63092. doi:10.7554/eLife.63092
Perez, J. D., Fusco, C. M., and Schuman, E. M. (2021b). A functional dissection of the mRNA and locally synthesized protein population in neuronal dendrites and axons. Annu. Rev. Genet. 55, 183–207. doi:10.1146/annurev-genet-030321-054851
Poe, G. R. (2017). Sleep is for forgetting. J. Neurosci. 37, 464–473. doi:10.1523/JNEUROSCI.0820-16.2017
Poulopoulos, A., Murphy, A. J., Ozkan, A., Davis, P., Hatch, J., Kirchner, R., et al. (2019). Subcellular transcriptomes and proteomes of developing axon projections in the cerebral cortex. Nature 565, 356–360. doi:10.1038/s41586-018-0847-y
Puentes-Mestril, C., Roach, J., Niethard, N., Zochowski, M., and Aton, S. J. (2019). How rhythms of the sleeping brain tune memory and synaptic plasticity. Sleep 42, zsz095. doi:10.1093/sleep/zsz095
Raguram, A., Banskota, S., and Liu, D. R. (2022). Therapeutic in vivo delivery of gene editing agents. Cell 185, 2806–2827. doi:10.1016/j.cell.2022.03.045
Ramanathan, M., Porter, D. F., and Khavari, P. A. (2019). Methods to study RNA-protein interactions. Nat. Methods 16, 225–234. doi:10.1038/s41592-019-0330-1
Ramm, P., and Smith, C. T. (1990). Rates of cerebral protein synthesis are linked to slow wave sleep in the rat. Physiol. Behav. 48, 749–753. doi:10.1016/0031-9384(90)90220-x
Rasch, B., and Born, J. (2013). About sleep's role in memory. Physiol. Rev. 93, 681–766. doi:10.1152/physrev.00032.2012
Raven, F., Van Der Zee, E. A., Meerlo, P., and Havekes, R. (2018). The role of sleep in regulating structural plasticity and synaptic strength: Implications for memory and cognitive function. Sleep. Med. Rev. 39, 3–11. doi:10.1016/j.smrv.2017.05.002
Richter, J. D., Bassell, G. J., and Klann, E. (2015). Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat. Rev. Neurosci. 16, 595–605. doi:10.1038/nrn4001
Roth, T. C., Rattenborg, N. C., and Pravosudov, V. V. (2010). The ecological relevance of sleep: The trade-off between sleep, memory and energy conservation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 945–959. doi:10.1098/rstb.2009.0209
Rozenbaum, M., Rajman, M., Rishal, I., Koppel, I., Koley, S., Medzihradszky, K. F., et al. (2018). Translatome regulation in neuronal injury and axon regrowth. eNeuro 5, ENEURO.0276–17.2018. doi:10.1523/ENEURO.0276-17.2018
Sanders, J., Cowansage, K., Baumgartel, K., and Mayford, M. (2012). Elimination of dendritic spines with long-term memory is specific to active circuits. J. Neurosci. 32, 12570–12578. doi:10.1523/JNEUROSCI.1131-12.2012
Schmid, D., Erlacher, D., Klostermann, A., Kredel, R., and Hossner, E. J. (2020). Sleep-dependent motor memory consolidation in healthy adults: A meta-analysis. Neurosci. Biobehav. Rev. 118, 270–281. doi:10.1016/j.neubiorev.2020.07.028
Seibt, J., Dumoulin, M. C., Aton, S. J., Coleman, T., Watson, A., Naidoo, N., et al. (2012). Protein synthesis during sleep consolidates cortical plasticity in vivo. Curr. Biol. 22, 676–682. doi:10.1016/j.cub.2012.02.016
Seibt, J., and Frank, M. G. (2019). Primed to sleep: The dynamics of synaptic plasticity across brain states. Front. Syst. Neurosci. 13, 2. doi:10.3389/fnsys.2019.00002
Seibt, J., Richard, C. J., Sigl-Glockner, J., Takahashi, N., Kaplan, D. I., Doron, G., et al. (2017). Publisher Correction: Cortical dendritic activity correlates with spindle-rich oscillations during sleep in rodents. Nat. Commun. 8, 1838. doi:10.1038/s41467-017-01652-8
Seo, S. S., Louros, S. R., Anstey, N., Gonzalez-Lozano, M. A., Harper, C. B., Verity, N. C., et al. (2022). Excess ribosomal protein production unbalances translation in a model of Fragile X Syndrome. Nat. Commun. 13, 3236. doi:10.1038/s41467-022-30979-0
Shi, Z., Fujii, K., Kovary, K. M., Genuth, N. R., Rost, H. L., Teruel, M. N., et al. (2017). Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol. Cell 67, 71–83. doi:10.1016/j.molcel.2017.05.021
Shigeoka, T., Koppers, M., Wong, H. H., Lin, J. Q., Cagnetta, R., Dwivedy, A., et al. (2019). On-site ribosome remodeling by locally synthesized ribosomal proteins in axons. Cell Rep. 29, 3605–3619. doi:10.1016/j.celrep.2019.11.025
Sidorov, M. S., Auerbach, B. D., and Bear, M. F. (2013). Fragile X mental retardation protein and synaptic plasticity. Mol. Brain 6, 15. doi:10.1186/1756-6606-6-15
Simsek, D., Tiu, G. C., Flynn, R. A., Byeon, G. W., Leppek, K., Xu, A. F., et al. (2017). The mammalian ribo-interactome reveals ribosome functional diversity and heterogeneity. Cell 169, 1051–1065. doi:10.1016/j.cell.2017.05.022
Skelin, I., Zhang, H., Zheng, J., Ma, S., Mander, B. A., Kim Mcmanus, O., et al. (2021). Coupling between slow waves and sharp-wave ripples engages distributed neural activity during sleep in humans. Proc. Natl. Acad. Sci. U. S. A. 118, e2012075118. doi:10.1073/pnas.2012075118
Sonenberg, N., and Hinnebusch, A. G. (2009). Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136, 731–745. doi:10.1016/j.cell.2009.01.042
Spruston, N. (2008). Pyramidal neurons: dendritic structure and synaptic integration. Nat. Rev. Neurosci. 9, 206–21.
Sun, C., Nold, A., Fusco, C. M., Rangaraju, V., Tchumatchenko, T., Heilemann, M., et al. (2021). The prevalence and specificity of local protein synthesis during neuronal synaptic plasticity. Sci. Adv. 7, eabj0790. doi:10.1126/sciadv.abj0790
Sun, L., Zhou, H., Cichon, J., and Yang, G. (2020). Experience and sleep-dependent synaptic plasticity: From structure to activity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 375, 20190234. doi:10.1098/rstb.2019.0234
Sutton, M. A., and Schuman, E. M. (2006). Dendritic protein synthesis, synaptic plasticity, and memory. Cell 127, 49–58. doi:10.1016/j.cell.2006.09.014
Tatavarty, V., Ifrim, M. F., Levin, M., Korza, G., Barbarese, E., Yu, J., et al. (2012). Single-molecule imaging of translational output from individual RNA granules in neurons. Mol. Biol. Cell 23, 918–929. doi:10.1091/mbc.E11-07-0622
Tom Dieck, S., Kochen, L., Hanus, C., Heumuller, M., Bartnik, I., Nassim-Assir, B., et al. (2015). Direct visualization of newly synthesized target proteins in situ. Nat. Methods 12, 411–414. doi:10.1038/nmeth.3319
Trachsel, L., Tobler, I., Achermann, P., and Borbély, A. A. (1991). Sleep continuity and the REM-nonREM cycle in the rat under baseline conditions and after sleep deprivation. Physiol. Behav. 49, 575–580. doi:10.1016/0031-9384(91)90283-t
Tudor, J. C., Davis, E. J., Peixoto, L., Wimmer, M. E., Van Tilborg, E., Park, A. J., et al. (2016). Sleep deprivation impairs memory by attenuating mTORC1-dependent protein synthesis. Sci. Signal. 9, ra41. doi:10.1126/scisignal.aad4949
Tutucci, E., Livingston, N. M., Singer, R. H., and Wu, B. (2018). Imaging mRNA in vivo, from birth to death. Annu. Rev. Biophys. 47, 85–106. doi:10.1146/annurev-biophys-070317-033037
Uezu, A., Kanak, D. J., Bradshaw, T. W., Soderblom, E. J., Catavero, C. M., Burette, A. C., et al. (2016). Identification of an elaborate complex mediating postsynaptic inhibition. Science 353, 1123–1129. doi:10.1126/science.aag0821
Van Alphen, B., Semenza, E. R., Yap, M., Van, S. B., and Allada, R. (2021). A deep sleep stage in Drosophila with a functional role in waste clearance. Sci. Adv. 7, eabc2999. doi:10.1126/sciadv.abc2999
Wegner, W., Ilgen, P., Gregor, C., Van Dort, J., Mott, A. C., Steffens, H., et al. (2017). In vivo mouse and live cell STED microscopy of neuronal actin plasticity using far-red emitting fluorescent proteins. Sci. Rep. 7, 11781. doi:10.1038/s41598-017-11827-4
Weissinger, R., Heinold, L., Akram, S., Jansen, R. P., and Hermesh, O. (2021). RNA proximity labeling: A new detection tool for RNA-protein interactions. Molecules 26, 2270. doi:10.3390/molecules26082270
Xie, L., Kang, H., Xu, Q., Chen, M. J., Liao, Y., Thiyagarajan, M., et al. (2013). Sleep drives metabolite clearance from the adult brain. Science 342, 373–377. doi:10.1126/science.1241224
Xue, S., Tian, S., Fujii, K., Kladwang, W., Das, R., and Barna, M. (2015). RNA regulons in Hox 5' UTRs confer ribosome specificity to gene regulation. Nature 517, 33–38. doi:10.1038/nature14010
Yang, G., Lai, C. S., Cichon, J., Ma, L., Li, W., and Gan, W. B. (2014). Sleep promotes branch-specific formation of dendritic spines after learning. Science 344, 1173–1178. doi:10.1126/science.1249098
Yang, G., Pan, F., and Gan, W. B. (2009). Stably maintained dendritic spines are associated with lifelong memories. Nature 462, 920–924. doi:10.1038/nature08577
Yap, E. L., and Greenberg, M. E. (2018). Activity-regulated transcription: Bridging the gap between neural activity and behavior. Neuron 100, 330–348. doi:10.1016/j.neuron.2018.10.013
Zhou, Y., Lai, C. S. W., Bai, Y., Li, W., Zhao, R., Yang, G., et al. (2020). REM sleep promotes experience-dependent dendritic spine elimination in the mouse cortex. Nat. Commun. 11, 4819. doi:10.1038/s41467-020-18592-5
Keywords: ribosomal protein, ribosome heterogeneity, neuron, neurites, sleep, brain plasticity, synapse
Citation: Buchanan IM, Smith TM, Gerber AP and Seibt J (2022) Are there roles for heterogeneous ribosomes during sleep in the rodent brain?. Front. Mol. Biosci. 9:1008921. doi: 10.3389/fmolb.2022.1008921
Received: 01 August 2022; Accepted: 20 September 2022;
Published: 06 October 2022.
Edited by:
Barbara Bardoni, UMR7275 Institut de Pharmacologie Moléculaire et Cellulaire (IPMC), FranceReviewed by:
Shilpi Minocha, Indian Institute of Technology Delhi, IndiaCopyright © 2022 Buchanan, Smith, Gerber and Seibt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: André P. Gerber, YS5nZXJiZXJAc3VycmV5LmFjLnVr; Julie Seibt, ai5zZWlidEBzdXJyZXkuYWMudWs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.