
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci. , 28 January 2022
Sec. Molecular Diagnostics and Therapeutics
Volume 8 - 2021 | https://doi.org/10.3389/fmolb.2021.813175
This article is part of the Research Topic The Upcoming Complications of COVID-19 on Recovered Patients: Molecular Mechanisms and Therapeutic Opportunities View all 13 articles
 Ilnaz Rahimmanesh1†
Ilnaz Rahimmanesh1† Laleh Shariati2,3†
Laleh Shariati2,3† Nasim Dana1
Nasim Dana1 Yasaman Esmaeili4
Yasaman Esmaeili4 Golnaz Vaseghi5
Golnaz Vaseghi5 Shaghayegh Haghjooy Javanmard1*
Shaghayegh Haghjooy Javanmard1*Previous studies suggested that patients with comorbidities including cancer had a higher risk of mortality or developing more severe forms of COVID-19. The interaction of cancer and COVID-19 is unrecognized and potential long-term effects of COVID-19 on cancer outcome remain to be explored. Furthermore, whether COVID‐19 increases the risk of cancer in those without previous history of malignancies, has not yet been studied. Cancer progression, recurrence and metastasis depend on the complex interaction between the tumor and the host inflammatory response. Extreme proinflammatory cytokine release (cytokine storm) and multi‐organ failure are hallmarks of severe COVID‐19. Besides impaired T-Cell response, elevated levels of cytokines, growth factors and also chemokines in the plasma of patients in the acute phase of COVID-19 as well as tissue damage and chronic low‐grade inflammation in “long COVID‐19” syndrome may facilitate cancer progression and recurrence. Following a systemic inflammatory response syndrome, some counterbalancing compensatory anti-inflammatory mechanisms will be activated to restore immune homeostasis. On the other hand, there remains the possibility of the integration of SARS- CoV-2 into the host genome, which potentially may cause cancer. These mechanisms have also been shown to be implicated in both tumorigenesis and metastasis. In this review, we are going to focus on potential mechanisms and the molecular interplay, which connect COVID-19, inflammation, and immune-mediated tumor progression that may propose a framework to understand the possible role of COVID-19 infection in tumorgenesis and cancer progression.
Over the previous decades, viral infections have posed significant challenges for cancer management. Several oncogenic viruses are known to cause cancer. However, there is no evidence linking between cancer subtypes and Severe Acute Respiratory Syndrome-Coronavirus-2 (SARS-CoV-2) infections in clinical and pre-clinical studies. Cancer patients are more susceptible to SARS-CoV-2 infection with possible poor prognosis than normal population due to their systemic immunosuppressive state caused by the cancer and anticancer treatments, such as active chemotherapy. The perpetually growing numbers of COVID-19 infections and the increase in numbers of diagnosed and undiagnosed cancer patients have warranted apprehending the interrelationship between COVID-19 and cancer. COVID-19 is concomitant with activation of oncogenic pathways, including The Janus kinase signal transducer and activator of transcription)JAK-STAT (, mitogen-activated protein kinase (MAPK), and Nuclear factor kappa B (NF-κB), which potentially can increase the risk of cancer (Li et al., 2020a).
On the other hand, COVID-19 and cancer look alike in several ways such as inappropriate T-cell responses. Another similarity is antigenic stimulation caused by damage-associated molecular pattern (DAMP) and pathogen-associated molecular pattern (PAMP) molecules occurs in both cancer and infectious disease (Hotchkiss and Moldawer, 2014). DAMP and PAMP figure out inflammation, leading to the release of various cytokines, increased levels of reactive oxygen and nitrogen species, tissues damage, and apoptosis. Moreover, hypoxia as well as hypoxic microenvironment secondary to inflammation or virus‐induced angiotensin‐converting enzyme 2 depletion provoke oxidative stress and probable malignant transformation (Saini and Aneja, 2021). In addition, the hypoxic microenvironment results in lysyl oxidase (LOX) production, which increases tumor cells invasion and facilitates migration and metastasis (Ye et al., 2020).
Immune responses in acute phase of COVID-19 patients, called cytokine storm are arranged by proinflammatory cytokines, which are also recognized to promote tumorigenesis (Del Valle et al., 2020).
Furthermore, the consecutive low-grade inflammation seen in COVID-19 patients after acute phase may result in a constant cycle of inflammation-induced organ injury and injury-induced inflammation. Although, cancer development is never the consequence of an insulated event; it could be hypothesized that COVID-19 may predispose the human to cancer development and accelerate cancer progression and metastasis. In this review, we discuss the pathways and hypotheses that may expose patients with COVID-19 to cancer in the future.
The recent coronaviruses epidemics have rapidly spread with irreparable consequences (Guan et al., 2020; Sohrabi et al., 2020). As of 28 October 2021, the World Health Organization (WHO) reported a total of 245M confirmed cases with 4.97M deaths, making it as one of the deadliest crisis in history (Dikid et al., 2020).
Coronaviruses can cause multiple organs infection especially respiratory tract infection. The clinical signs and symptoms are typically including fever, dry cough, muscle pain, diarrhea, and breathing difficulties (Abdel-Moneim and Hosni, 2021). Sever cases of COVID-19 represent acute respiratory distress syndrome (ARDS), sepsis and septic shock, multiorgan failure leading to death (Zafer et al., 2021). Indeed, in these cases, viruses can evade the immune system and spread to other organs, such as cardiovascular system, gastrointestinal system, central nervous system, liver and kidney, where they cause a variety of serious diseases (Bajaj et al., 2021; Soltani et al., 2021). Apart from the consequences created by the coronaviruses, underlying medical conditions such as cancer, cardiovascular diseases, renal diseases, and type I/II diabetes, increase the risk for severe COVID-19 infection (Haybar et al., 2020).
SARS-CoV-2 is a positively-sensed, and single-stranded RNA-enveloped virus with spherical capsids (120–160 nm) (Hoffmann et al., 2020; Nakagawa and Makino, 2021). The coronaviruse genome (GenBank no. MN908947) is about 26.4–31.7 kb long, making the largest among RNA viruses encoding 9,860 amino acids (Woo et al., 2010). Both structural and nonstructural proteins are found in gene fragments. Non-structural proteins such as 3-chymotrypsin-like protease, papain-like protease, and RNA-dependent RNA polymerase that are encoded by open reading frame (ORF) region, while structural proteins such as spike protein (S), envelope protein (E), membrane protein (M) and nucleocapsid protein (N).
SARS-CoV-2 surface is covered by the S protein, a large glycosylated transmembrane protein (1,160–1,400 aa) that binds to the host cell receptor angiotensin-converting enzyme 2 (ACE2) and mediates viral cell entrance (de Wit et al., 2016; Wu et al., 2020). Since the pathogenesis of coronaviruses has not been completely understood, the precise molecular mechanism of entry of viruses into cells remains unclear (Glebov, 2020). Coronaviruses are thought to enter the host cells by two main routes: direct genome into the cytosol via fusion with the host cell membrane, and endocytosis (Pelkmans and Helenius, 2003) (Figure 1). Typically, when the S protein attaches to the receptor, proteases on the host cell membrane, such as transmembrane protease serine 2 (TMPRSS2) and airway trypsin-like protease TMPRSS11D, promote virus entry into the cell by activating the S protein. The viral RNA genome is translated into two polyproteins and structural proteins in the cytoplasm once the virus enters the cell, facilitating the construction of virus progeny (Perlman and Netland, 2009). Replication and transcription of the viral RNA genome occur through protein cleavage with continuation/discontinuation of RNA synthesis that is mediated by a replicase-transcriptase complex. Eventually, structural proteins are synthesized, assembled, and packaged in the endoplasmic reticulum-Golgi intermediate compartment of the host cell (Li et al., 2020b).
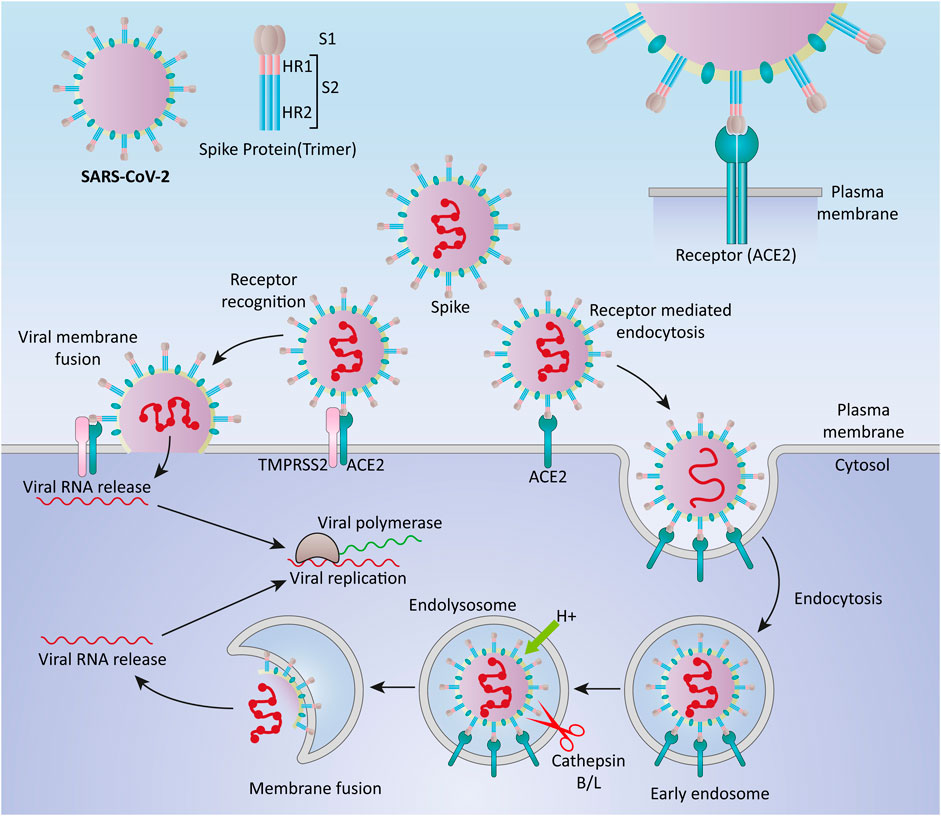
FIGURE 1. Cell entry mechanisms of SARS-CoV-2. Coronaviruses are believed to enter the host cells via two main routes, through direct delivery of the genome into the cytosol via fusion with the host cell membrane, and through endocytosis.
Furthermore, it has been proposed that SARS-CoV-2 entered cells through clathrin-mediated endocytosis (Bayati et al., 2020). SARS-CoV-2, like SARS-CoV and MERS-CoV, may use numerous pathways to successfully enter the cytosol of the host cell (Hartenian et al., 2020; Yang and Shen, 2020).
The immune system responses to SARS-CoV-2 are considered to be critical in controlling the pathogenicity and clinical symptoms of the patient (Yang et al., 2020). After virus infection, the immune system uses multiple mechanisms for recognition and defends against viruses (Koch et al., 2013). The body uses both the immune responses (innate and adaptive immunity), to eliminate viral infection (Ye et al., 2021) (Figure 2). During the first hours and days, the innate immune response to viruses begins with innate immune cells, like phagocytic cells (neutrophils, macrophages) and dendritic cells (Zhou and Ye, 2021). These cells express different types of Pattern-recognition receptors (PRRs) to recognize DAMPs and PAMPs (Li and Wu, 2021). In COVID-19 patients, NK cells can kill virus-infected cells by releasing cytotoxic granules or through participating in antibody-dependent cellular cytotoxicity and producing different cytokines and chemokines (Ma et al., 2021).
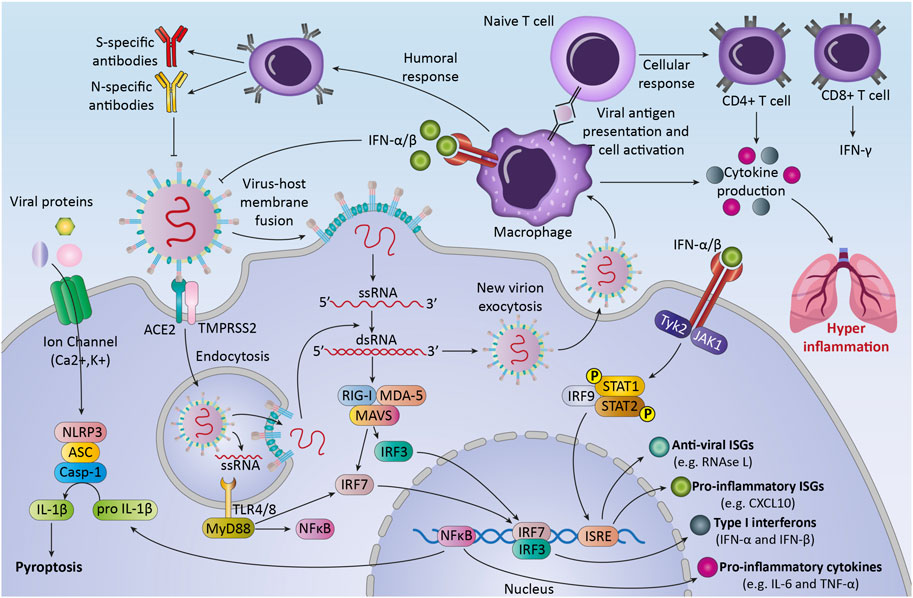
FIGURE 2. The immune response against SARS-CoV-2.The first line of host defense against SARS-CoV2, innate immunity, is the response against the virus by recognizing Pattern associated molecular patterns (PAMPs) through transmembrane or intracellular pattern recognition receptors (PRRs). Recognition of viral components leads to the activation of immune cells and transcription factors that lead to the production of different cytokines, chemokines, and anti-viral proteins. These processes can lead to activation of adaptive immune response that consists of three major cell types (B cells, CD4+ T cells, and CD8+ T cells). After pathogen elimination, adaptive immunity regulates innate immunity to avoid unnecessary host cell damage. Unbalanced response and immune system overactivation can cause collateral damage to host tissues and exacerbate disease severity.
After the fusion of the virus into the cells, viral genome starts replication and transcription (Astuti and Ysrafil, 2020). Then immune system recognized RNA viruses by cytosolic and endosomal RNA receptors, such as RIG-I-like receptors and toll-like receptors (TLR3, TLR7, and TLR8) (Hosseini et al., 2020). Within hours, RNA virus recognition by these receptors can activate some of the transcription factors like nuclear factor kappa-light-chain- enhancer of activated B cells (NF-κB), to produce pro-inflammatory cytokines and chemokines and activation of immune cells (Kircheis et al., 2020). The result of these processes is the production of interferons -λ (type III IFN) and type I IFN, pro-inflammatory cytokines (for example, interleukin-6, interleukin-18, and interleukin-1) and some chemokines such as chemokine (C-C motif) ligand 2 (CCL2) and chemokine (C-C motif) ligand 7 (CCL7) by immune cells (Stetson and Medzhitov, 2006).
Moreover, the infected cells may undergo the process of inflammatory cell death (pyroptosis) and release DAMPs, such as viral nucleic acids and oligomers (Yap et al., 2020).
The second line of defense, the adaptive immune response, happens several days later after the innate immune response occurs. T cells and B cells, two classes of lymphocytes, carry out the adaptive immune response against COVID-19. In the majority of infected patients, B cells at first produce IgM and IgA antibodies and then IgG, that is measurable between weeks two and five after infection (Emmerich et al., 2021). Various structural proteins of the virus can be targeted with these antibodies (Pan et al., 2021).
The other adaptive immune cells, CD8+ T cells, directly kill cells that are infected with the virus and stop virus spreading further (de Candia et al., 2021). The other type of T cells, CD4+ T cells, produce different types of cytokines and chemokines that influence the function of B and T cells, and other immune cells such as macrophages (Chen et al., 2010).
After pathogen elimination, adaptive immunity regulates innate immunity to avoid unnecessary host cell damage (Zhou et al., 2020). Notably, while a highly organized immune response to viral infection is important, an unbalanced response and immune system overactivation inadvertently targeting healthy cells and lead to host tissues damage (Catanzaro et al., 2020). The imbalance of the immune system in fighting against SARS-CoV-2 can induce inflammation, lymphopenia, and cytokine storm in patients and follow that septic shock, acute lung injury, ARDS, and multi-organs failure may occur, that increases patient’s mortality rate (Jamal et al., 2021). Early studies indicated that COVID-19 patients have higher neutrophil and lower lymphocyte counts in blood compared with healthy controls (Eslamijouybari et al., 2020). Also, CD8+ lymphocytes and NK cells show an exhausted phenotype and significant reduction in severe COVID-19 patients compared with patients with mild SARS-CoV-2 -infection and healthy controls (Zheng et al., 2020). These changes can disrupt the immune system and the antiviral effects of immune cells. On the other hand, cytokine storm can occur as a result of hyperactivity of infected macrophages and other immune cells that cause the production of various inflammatory cytokines (Zheng et al., 2020).
Increasing NF-κB has an important effect on cytokine storms created by an acute respiratory RNA virus. Activation of TLRs by recognition of virus PAMPs leads to downstream signaling pathways like NF-κB and interferon regulatory factor 3 (IRF3) (Hariharan et al., 2021). NF-κB plays key roles in regulating different cellular functions, such as cell growth and cell death, immune responses, and inflammation (Barnabei et al., 2021). It has been shown that COVID-19 is able to activate TLR4-mediated NF-κB signaling (Kircheis et al., 2020). Overactivation of NF-κB targets genes in the context of inflammation and affect the activation of adhesion molecules and inflammatory cells that all can cause or worsen lung disease and is a sign of chronic inflammation (Liu et al., 2017). A growing body of evidence suggests that infection and chronic inflammation as a risk factor can lead to cancer and develop some other diseases (Rossi et al., 2021). Therefore, further investigation about the interaction among the immune system and coronavirus can be effective to reduce its adverse consequences.
Infection with SARS-CoV-2 causes innate immune responses, which result in a diverse set of immune mediators and the activation of some signaling pathways, including Janus kinase/signal transducer and activator of transcription (JAK/STAT), nuclear factor kappa B (NF-κB), interferon response factor 3 (IRF3), and IRF7. The signaling cascade that results from these pathways in infected cells increases the production of pro-inflammatory cytokines (Li et al., 2020c; Prompetchara et al., 2020).
In COVID-19 related cytokine storm syndrome (COVID-CSS), different inflammatory cytokines such as Interleukin-1 (IL-1), Interleukin-10 (IL-10), and tumor necrosis factor (TNF)-α are elevated about 2-100 fold above normative values. In some patients with COVID-19, IL-6 levels rise more than 1,000 fold above normal range (Blanco-Melo et al., 2020; Herold et al., 2020; Laing et al., 2020; Al-Baadani et al., 2021). IL-6 signals through both classic and trans-signaling pathways, activates JAK-STAT3 signaling. The IL-6/JAK/STAT3 pathway is also abnormally activated in many types of cancer, which is generally associated with a poor clinical prognosis (Johnson et al., 2018). In the tumor microenvironment, IL-6/JAK/STAT3 signaling pathway plays a pivotal role in regulating the growth, survival, invasiveness, metastasis, and development of many cancers. In addition, the IL-6/JAK/STAT3 acts as a key component in suppressing the antitumor immune response (Bromberg and Wang, 2009; Kumari et al., 2016). Previous research identified the role of IL-6 as a driver of tumorigenesis and anti-apoptosis signaling and a vital biomarker of cancer diagnosis, risk, and prognosis (Vargas and Harris, 2016). Also, IL-6 enhanced the metastasis rate in breast cancer through the positive effect on tumor stem cell self-renewal and epithelial-to-mesenchymal transition (EMT) process (Bharti et al., 2016; Xiao et al., 2017).
STAT3 hyperactivation in tumor cells can occur as a result of increased IL-6 levels in the serum and/or the tumor microenvironment, or as a result of loss-of-function mutations affecting STAT3 negative regulators. IL-6 is produced by different cell types including stromal cells, tumor-infiltrating immune cells, and the tumor cells (Nozawa et al., 2006; Walter et al., 2009; Nagasaki et al., 2014). IL-6 directly stimulates the expression of STAT3 downstream targets in tumor cells. STAT3 signaling controls the expression of genes involved in tumor growth, survival, invasion, and angiogenesis. The ability of STAT3 to promote IL6 gene expression is via binding to the IL6 promoter, which results in a positive autocrine feedback loop (Chang et al., 2013). STAT3 also promotes angiogenesis, invasiveness, and/or metastasis, as well as immunosuppression (Teng et al., 2014; Huynh et al., 2017).
In addition to direct effects on tumor cells, IL-6 and JAK/STAT3 signaling can have a profound effect on tumor-infiltrating immune cells. STAT3 is often hyperactivated in tumor-infiltrating immune cells and negatively regulates natural killer (NK) cells, neutrophils, effector T cells, and dendritic cells (DCs). Taken together, these data suggest that, STAT3 activation in immune cells probably leads to down-modulation of antitumor immunity response (Harris et al., 2007; Yu et al., 2007; Herrmann et al., 2010; Iwata-Kajihara et al., 2011). Collectively, IL-6/JAK/STAT signaling pathway plays a fundamental role in tumorgenesis and immunosuppressive tumor microenvironment (Kortylewski et al., 2005; Yu et al., 2009).
The hyper-activation of NF-κB pathway has been implicated in the pathogenesis of the severe/critical COVID-19 phenotype (Hirano and Murakami, 2020). The excessive NF-κB activation subsequent to viral proteins detection via the innate immune system, probably has a causative role in covid-19 cytokine storm, extrapulmonary manifestations of COVID-19, and fatality rate (Liao et al., 2005; Oeckinghaus and Ghosh, 2009; DeDiego et al., 2014). This ‘rapid acting’ primary transcription factor seems to exert its effects by stimulating the gene expression of a wide variety of cytokines, adhesion molecules, chemokines, acute phase proteins, and inducible effector enzymes (Liang et al., 2004; Zhang et al., 2017).
In a similar way, NFκB is an important signaling pathway involved in the pathogenesis of tumors, and the potential role of excessive activation of this signaling pathway in oncogenesis has been confirmed. In various tumor types, intervention in this signaling pathway for targeted cancer therapy has been reported. NFκB signaling is involved in cell biogenic activities such as inflammation, cellular immunity, and stress (Salazar et al., 2014; Sau et al., 2016; Song et al., 2017). In lymphatic cancer, breast cancer, and colon cancer, the excessive activation of the NFκB-signaling pathway leads to uncontrolled and abnormal cell proliferation, differentiation, and apoptosis, as well as metastasis, and treatment resistance. The NFκB pathway is often altered in both solid and hematopoietic malignancies, promoting tumor-cell proliferation and survival (Hayden and Ghosh, 2008; Walther et al., 2015; Forlani et al., 2016; Sau et al., 2016). Therefore, the activation of NFκB pathway, as a shared pathway between COVID-19 and some cancers, plays an important role in disease progression.
IFNs are members of a large family of cytokines that are currently divided into three groups (type I, II, and III IFNs) (Negishi et al., 2018). Type I IFNs are involved in the development of innate and adaptive immune responses against both cancer and infectious diseases (Bonjardim, 2005). Similar to IL-6, IFN-Is signaling activates the JAK/STAT pathway (Schindler et al., 2007). Recent studies have demonstrated that IFN signaling could be a key mechanism in tumor proliferation. Inhibition of IFN pathway induction and dysregulated IFN signaling promote tumor cell senescence and death (Sangfelt et al., 1999; Fuertes et al., 2011; Schreiber et al., 2011). Type I interferons are crucial for restricting responses during the early stage of SARS-CoV-2 infection, however, IFN-I signaling was dampened in patients with severe COVID-19 (Blanco-Melo et al., 2020), and low level of IFNs along with an increase in IL-6, have been detected in the peripheral blood samples or lungs of patients (Hadjadj et al., 2020). These data illustrate the possible role of impaired INF-I signaling induced by SARS-CoV-2 infection, in inefficient antitumor response, which leads to tumor progression. On the other hand, lymphopenia, functional exhaustion, and impaired cellular immunity responses are associated with IFN production suppression. This notion is supported by evidence, which highlights the role of IFNs in effective T cell proliferation and survival (Marrack et al., 1999). The continued production of pro-inflammatory mediators due to viral persistence has a negative effect on natural killer cells and CD8+ T-cell activation in the case of COVID-19 infection. The CD8+ T-cell population undergoes quantitative and qualitative changes. Decreased in NK cells, CD4+ T cells, and CD8+ T cell population and impaired activation phenotypes are frequently observed, particularly in the severe form of COVID-19 disease (Ai et al., 2020; Pedersen and Ho, 2020). In COVID-19 patients specific cell population, the cytotoxic CD3−CD56dimCD16+, was significantly reduced, and more importantly, elevated expression of regulatory molecules such as CD244 and programmed death-1 (PD-1) on NK cells and T cells, as well as decreased serum cytotoxic effector molecules such as perforin and granzyme A, were noticed. Previous research has also found exhaustion phenotypes of CD8+ T cells in patients with severe COVID-19, which is caused by the upregulation of inhibitory receptors (IRs), which may impair host defences and lead to poor disease outcomes (Caruso et al., 2020; De Biasi et al., 2020; Diao et al., 2020; Laing et al., 2020; Schultheiß et al., 2020; Song et al., 2020; Zheng et al., 2020).
However, immune dysregulation may result in the COVID-19 severity, this functional exhaustion and subset alteration of NK and T cells may have a negative effect on efficient cell function during anti-tumor immune responses (McKinney and Smith, 2016; Wang et al., 2017; He et al., 2019).
Although the role of immune dysregulation and robust production of cytokines in COVID-19 pathogenesis has been well documented, future studies will be vital to investigate the shared molecular pathways and possible interaction between COVID-19 and the development and progression of cancer.
The World Health Organization, considers the infections as inducers for 15.4% of all cancers, with the viruses constitute up to 9.9% of all cancer causig infectious agents. The most prominent viral carcinogenic agent known to be hepatitis B virus, hepatitis C virus, human papillomavirus, and Epstein-Barr virus (Plummer et al., 2016). Integration of the viral genome into the host genome has been illustrated to be an unintended mechanism that may lead to the clonal growth and cancer develpemnt. In head-and-neck, cervical cancer, and liver cancer, the integration of viral genome into the host genom has been proven. Most of the geneome integrations were observed to be distributed across chromosomes, and a significant number of viral genome integrations occurred in the intronic (40%) regions, whereas only 3.4% of integrations were recognized in gene coding regions (Zapatka et al., 2020). In RNA viruses, the RNA genome is reverse-transcribed by the viral reverse transcriptase which subsequently integrates into the host chromosome shortly after infection, and expressed under the control of viral transcriptional regulatory elements. Following integration process, oncogenesis occurs due to activation or inactivation of the host genes, oncoproteins, and tumor suppressors (McLaughlin-Drubin and Munger, 2008). Several hypotheses have been put forward regarding integration of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) into the human genome (Dai et al., 2020; Zhang et al., 2021) (Figure 3). Prolonged SARS-CoV-2 RNA shedding, and recurrence of PCR-positive tests, 60 days after the initial positive PCR test, have been widely demonstrated in non-infectious patients. These patients, howevere, do not appear to shed any infectious viruses (Bullard et al., 2020; Li et al., 2020d; Hosseini et al., 2020; Mina et al., 2020). Consequently, researchers have investigated the capability of SARS-CoV-2 as positive-stranded RNA viruses, for integration into human genome subsequent to reverse-transcription (RT). Hence, the transcription of the integrated sequences, provides logical evidence fo recurrence of PCR-positive tests (Dai et al., 2020). However, several investigations refuse the integration of SARS-CoV-2 into the human genome (Briggs et al., 2021; Kazachenka and Kassiotis, 2021; Parry et al., 2021; Smits et al., 2021).
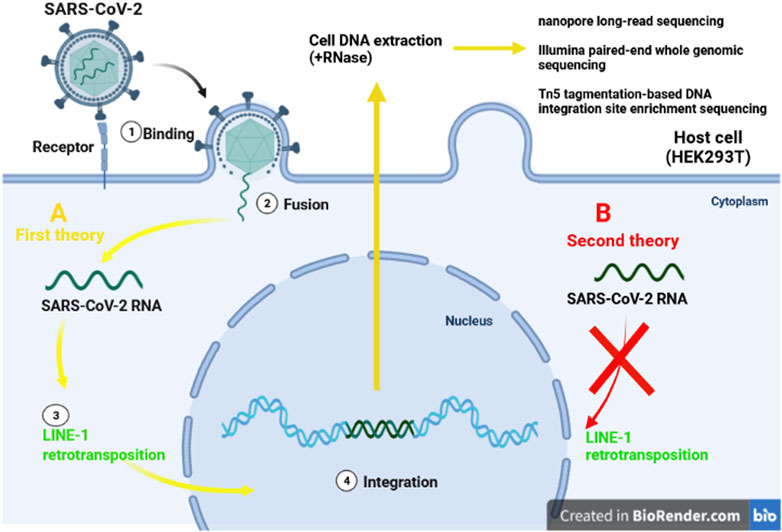
FIGURE 3. Two different theories of integration SARS-CoV-2 RNA. The first idea claims that LINE-1 retrotransposon proteins (A/yellow lines) can incorporate SARS-CoV-2 into human DNA. Despite the presence of the LINE-1 protein machinery (B/red line), the second explanation finds no evidence of virus genomic integration.
To investigate the integration of SARS-CoV-2 RNA into the genome of infected human cells in vitro, Zhang et al. applied three different procedures: nanopore long-read sequencing, Illumina paired-end whole genomic sequencing, and Tn5 tagmentation-based DNA integration site enrichment sequencing (Zhang et al., 2020).
Recent studies provided enough evidences for the possibility of SARS-CoV2 integration into the human genome. The viral RNA found to be reverse transcribed in human cells by RT from LINE-1 elements or by HIV-1 RT. It has found the endogenous LINE-1 in human cells was induced to be overexpressed upon SARS-CoV-2 infection or following SARS CoV2-associated cytokine storm in vitro. The upregulation of host cell LINE-1 RT seems supportive for long-term positivity of PCR tests, even after recovery (Kazazian et al., 2017; Zhang et al., 2021). Although, Smits et al. applied long-read DNA sequencing to cultured HEK293T cells infected with SARS-CoV2 which provided no evidence for genomic integration of the SARS-CoV-2 into the host genome (Smits et al., 2021).
In another experiment, Briggs et al. examined 768 COVID patient’s nasal swabs in order to evaluate the positive role of viral genome integration on diagnostic PCR test for COVID-19. In this study, LINE-1-mediated retrotranscription of SARS-CoV2 RNA genome into the host DNA found to be a rare event with no practical impact on RT-PCR-based diagnostic capability. These data indicated the supposed SARS-CoV-2 integrations are likely artefactual, stemming from amplicon DNA contaminations or other unintended processes (Briggs et al., 2021).
As a consequence, the correlation between the integration of SARS- CoV-2 into the host genome, and cancer has remained unknown. Moreover, further studies on different species of SARS-CoV-2 are needed for more comprehensive conclusion.
In clinically COVID-19 suspected patients, computed tomography (CT) is the preferred imaging modality, and it is useful for monitoring patients during treatment. A dose reduction procedure is an essential requirement given the increased number of chest CT scans requested by referring physicians and the need to reduce the possible dangers posed to patients by ionizing radiation (Radpour et al., 2020; Azadbakht et al., 2021). Several works reported that the chest CT had higher sensitivity (around 97%) for COVID-19 pneumonia diagnosis compared to standard methods such as real-time PCR and NGS (Ai et al., 2020; Caruso et al., 2020; Fang et al., 2020). One of the major concerns is that CT ionising radiation may increase the risk of leukaemia and solid tumors incidence (Power et al., 2016). Some organ systems are highly radiosensitive, whereas others have stronger defences against the effects of ionizing radiation. Organs like the oesophagus, breast, and bladder, for example, are particularly vulnerable, although the rectum, pancreas, and prostate are far less so (Power et al., 2016). A study from February 24th to April 28th 2020 including 3,224 high-resolution thorax CT showed a very low risk estimation of cancer despite the impressive increment in thoracic CT examinations due to COVID-19 cases (Ghetti et al., 2020).
It has been suggested that one CT scan is equivalent to 300-400 chest X-rays, and that having multiple CT scans at a young age may raise the risk of cancer later in life. CT scans have two major dangers, according to the FDA. First, the test findings reveal a benign or coincidental finding, prompting unnecessary, perhaps invasive follow-up examinations that could pose further dangers. Second, x-ray radiation exposure increases the risk of cancer induction. The infection risk from contamination of surface, aerosolization during the CT acquisition of a Covid-19 patient, putting healthcare workers working in the CT suite at an increased risk of contracting this infection, and the CT imaging facility becoming a Covid-19 infection source to other patients undergoing CT examination are some of its limitations.
Future studies should focus on the disposition of patients recovering from the novel coronavirus for cancer. This increased risk to cancer could be related to the cumulative effect of different distinct aspects of coronavirus infection yet to be elucidated. Some of these aspects include the immune dysregulation, intensive production of cytokines, organ damage secondry to infection or other factors such as CT scans, and induced mutations as the result of the viral genome integration into the host genome.
While both innate and adaptive immune systems are indispensible to combat SARS-CoV-2 infection, uncontrolled host immune responses are among main COVID- related pathogenicity. Therefore, these two opposite aspects of the immune system should be addressed before putting forth fututre solutions to reduce post-infection side effects.
Future studies are vital to investigate shared molecular pathways and possible interactions between COVID-19 and the development and progression of cancer and to investigate whether the virus can be regarded as an etiological factor in the development of cancer. Our knowledge about coronavirus is relentlessly being updated and this review should be interpreted in light of future reliable findings.
SH and LS contributed in the conception of the work, revising the draft, approval of the final version of the manuscript, and agreed for all aspects of the work. IR, ND, GV, and YE contributed in the providing and revising the draft and agreed for all aspects of the work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, orclaim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdel-Moneim, A., and Hosni, A. (2021). “Insights into the Possible Impact of COVID-19 on the Endocrine System,” in COVID-19 Pandemic and Male Fertility: Clinical Manifestations and Pathogenic Mechanisms (Archives of Physiology and Biochemistry), 1–9. doi:10.1080/13813455.2021.1890131
Ai, T., Yang, Z., Hou, H., Zhan, C., Chen, C., Lv, W., et al. (2020). Correlation of Chest CT and RT-PCR Testing for Coronavirus Disease 2019 (COVID-19) in China: a Report of 1014 Cases. Radiology 296 (2), E32–E40. doi:10.1148/radiol.2020200642
Al-Baadani, A., Eltayeb, N., Alsufyani, E., Albahrani, S., Basheri, S., Albayat, H., et al. (2021). Efficacy of Tocilizumab in Patients with Severe COVID-19: Survival and Clinical Outcomes. J. Infect. Public Health 14 (8), 1021–1027. doi:10.1016/j.jiph.2021.05.015
Astuti, I., and Ysrafil, (2020). Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An Overview of Viral Structure and Host Response. Diabetes Metab. Syndr. Clin. Res. Rev. 14 (4), 407–412. doi:10.1016/j.dsx.2020.04.020
Azadbakht, J., Khoramian, D., Lajevardi, Z. S., Elikaii, F., Aflatoonian, A. H., Farhood, B., et al. (2021). A Review on Chest CT Scanning Parameters Implemented in COVID-19 Patients: Bringing Low-Dose CT Protocols into Play. Egypt. J. Radiol. Nucl. Med. 52 (1), 13. doi:10.1186/s43055-020-00400-1
Bajaj, V., Gadi, N., Spihlman, A. P., Wu, S. C., Choi, C. H., and Moulton, V. R. (2021). Aging, Immunity, and COVID-19: How Age Influences the Host Immune Response to Coronavirus Infections. Front. Physiol. 11, 1793. doi:10.3389/fphys.2020.571416
Barnabei, L., Laplantine, E., Mbongo, W., Rieux-Laucat, F., and Weil, R. (2021). NF-κB: At the Borders of Autoimmunity and Inflammation. Front. Immunol. 12, 716469. doi:10.3389/fimmu.2021.716469
Bayati, A., Kumar, R., Francis, V., and McPherson, P. S. (2020). SARS-CoV-2 Uses Clathrin-Mediated Endocytosis to Gain Access into Cells, BioRxiv. J. Biol. Chem. 296, 100306. doi:10.1016/j.jbc.2021.100306
Bharti, R., Dey, G., and Mandal, M. (2016). Cancer Development, Chemoresistance, Epithelial to Mesenchymal Transition and Stem Cells: A Snapshot of IL-6 Mediated Involvement. Cancer Lett. 375 (1), 51–61. doi:10.1016/j.canlet.2016.02.048
Blanco-Melo, D., Nilsson-Payant, B. E., Liu, W.-C., Uhl, S., Hoagland, D., Møller, R., et al. (2020). Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 181 (5), 1036–1045. doi:10.1016/j.cell.2020.04.026
Bonjardim, C. A. (2005). Interferons (IFNs) Are Key Cytokines in Both Innate and Adaptive Antiviral Immune Responses-And Viruses Counteract IFN Action. Microbes Infect. 7 (3), 569–578. doi:10.1016/j.micinf.2005.02.001
Briggs, E., Ward, W., Rey, S., Law, D., Nelson, K., Bois, M., et al. (2021). Assessment of Potential SARS-CoV-2 Virus Integration into Human Genome Reveals No Significant Impact on RT-qPCR COVID-19 Testing. Proc. Natl. Acad. Sci. 118 (44), e2113065118. doi:10.1073/pnas.2113065118
Bromberg, J., and Wang, T. C. (2009). Inflammation and Cancer: IL-6 and STAT3 Complete the Link. Cancer cell 15 (2), 79–80. doi:10.1016/j.ccr.2009.01.009
Bullard, J., Dust, K., Funk, D., Strong, J. E., Alexander, D., Garnett, L., et al. (2020). Predicting Infectious Severe Acute Respiratory Syndrome Coronavirus 2 from Diagnostic Samples. Clin. Infect. Dis. 71 (10), 2663–2666. doi:10.1093/cid/ciaa638
Caruso, D., Zerunian, M., Polici, M., Pucciarelli, F., Polidori, T., Rucci, C., et al. (2020). Chest CT Features of COVID-19 in Rome, Italy. Radiology 296, E79–E85. –E85. doi:10.1148/radiol.2020201237
Catanzaro, M., Fagiani, F., Racchi, M., Corsini, E., Govoni, S., and Lanni, C. (2020). Immune Response in COVID-19: Addressing a Pharmacological challenge by Targeting Pathways Triggered by SARS-CoV-2. Sig Transduct Target. Ther. 5 (1), 84. doi:10.1038/s41392-020-0191-1
Chang, Q., Bournazou, E., Sansone, P., Berishaj, M., Gao, S. P., Daly, L., et al. (2013). The IL-6/JAK/Stat3 Feed-Forward Loop Drives Tumorigenesis and Metastasis. Neoplasia 15 (7), 848–IN45. doi:10.1593/neo.13706
Chen, J., Lau, Y. F., Lamirande, E. W., Paddock, C. D., Bartlett, J. H., Zaki, S. R., et al. (2010). Cellular Immune Responses to Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) Infection in Senescent BALB/c Mice: CD4 + T Cells Are Important in Control of SARS-CoV Infection. J. Virol. 84 (3), 1289–1301. doi:10.1128/JVI.01281-09
Dai, M., Liu, D., Liu, M., Zhou, F., Li, G., Chen, Z., et al. (2020). Patients with Cancer Appear More Vulnerable to SARS-COV-2: a Multi-center Study during the COVID-19 Outbreak. Cancer Discov. 10 (6), 20. doi:10.1158/2159-8290.CD-20-0422
De Biasi, S., Meschiari, M., Gibellini, L., Bellinazzi, C., Borella, R., Fidanza, L., et al. (2020). Marked T Cell Activation, Senescence, Exhaustion and Skewing towards TH17 in Patients with COVID-19 Pneumonia. Nat. Commun. 11 (1), 3434. doi:10.1038/s41467-020-17292-4
de Candia, P., Prattichizzo, F., Garavelli, S., and Matarese, G. (2021). T Cells: Warriors of SARS-CoV-2 Infection. Trends Immunol. 42 (1), 18–30. doi:10.1016/j.it.2020.11.002
de Wit, E., van Doremalen, N., Falzarano, D., and Munster, V. J. (2016). SARS and MERS: Recent Insights into Emerging Coronaviruses. Nat. Rev. Microbiol. 14, 523–534. doi:10.1038/nrmicro.2016.81
DeDiego, M. L., Nieto-Torres, J. L., Regla-Nava, J. A., Jimenez-Guardeño, J. M., Fernandez-Delgado, R., Fett, C., et al. (2014). Inhibition of NF- B-Mediated Inflammation in Severe Acute Respiratory Syndrome Coronavirus-Infected Mice Increases Survival. J. Virol. 88 (2), 913–924. doi:10.1128/jvi.02576-13
Del Valle, D. M., Kim-Schulze, S., Huang, H.-H., Beckmann, N. D., Nirenberg, S., Wang, B., et al. (2020). An Inflammatory Cytokine Signature Predicts COVID-19 Severity and Survival. Nat. Med. 26 (10), 1636–1643. doi:10.1038/s41591-020-1051-9
Diao, B., Wang, C., Tan, Y., Chen, X., Liu, Y., Ning, L., et al. (2020). Reduction and Functional Exhaustion of T Cells in Patients with Coronavirus Disease 2019 (COVID-19). Front. Immunol. 11, 827. doi:10.3389/fimmu.2020.00827
Dikid, T., Chaudhary, S., Goel, K., Padda, P., Sahu, R., Kumar, T., et al. (2020). Responding to COVID-19 Pandemic: Why a strong Health System Is Required. Indian J. Med. Res. 151 (2), 140–145. doi:10.4103/ijmr.IJMR_761_20
Emmerich, P., Possel, R., Hemmer, C. J., Fritzsche, C., Geerdes‐Fenge, H., Menge, B., et al. (2021). Longitudinal Detection of SARS‐CoV‐2‐specific Antibody Responses with Different Serological Methods. J. Med. Virol. 93 (10), 5816–5824. doi:10.1002/jmv.27113
Eslamijouybari, M., Heydari, K., Maleki, I., Moosazadeh, M., Hedayatizadeh-Omran, A., Vahedi, L., et al. (2020). Neutrophil-to-Lymphocyte and Platelet-To-Lymphocyte Ratios in COVID-19 Patients and Control Group and Relationship with Disease Prognosis. Caspian J. Intern. Med. 11 (Suppl. 1), 531–535. doi:10.22088/cjim.11.0.531
Fang, Y., Fang, Y., Zhang, H., Xie, J., Lin, M., Ying, L., et al. (2020). Sensitivity of Chest CT for COVID-19: Comparison to RT-PCR. Radiology 296 (2), E115–E117. doi:10.1148/radiol.2020200432
Forlani, G., Abdallah, R., Accolla, R. S., and Tosi, G. (2016). The Major Histocompatibility Complex Class II Transactivator CIITA Inhibits the Persistent Activation of NF-Κb by the Human T Cell Lymphotropic Virus Type 1 Tax-1 Oncoprotein. J. Virol. 90 (7), 3708–3721. doi:10.1128/jvi.03000-15
Fuertes, M. B., Kacha, A. K., Kline, J., Woo, S.-R., Kranz, D. M., Murphy, K. M., et al. (2011). Host Type I IFN Signals Are Required for Antitumor CD8+ T Cell Responses through CD8α+ Dendritic Cells. J. Exp. Med. 208 (10), 2005–2016. doi:10.1084/jem.20101159
Ghetti, C., Ortenzia, O., Maddalo, M., Altabella, L., and Sverzellati, N. (2020). Dosimetric and Radiation Cancer Risk Evaluation of High Resolution Thorax CT during COVID-19 Outbreak. Physica Med. 80, 119–124. doi:10.1016/j.ejmp.2020.10.018
Glebov, O. O. (2020). Understanding SARS‐CoV‐2 Endocytosis for COVID‐19 Drug Repurposing. FEBS J. 287, 3664–3671. doi:10.1111/febs.15369
Guan, W.-j., Ni, Z.-y., Hu, Y., Liang, W.-h., Ou, C.-q., He, J.-x., et al. (2020). Clinical Characteristics of 2019 Novel Coronavirus Infection in China. MedRxiv.
Hadjadj, J., Yatim, N., Barnabei, L., Corneau, A., Boussier, J., Smith, N., et al. (2020). Impaired Type I Interferon Activity and Inflammatory Responses in Severe COVID-19 Patients. Science 369 (6504), 718–724. doi:10.1126/science.abc6027
Hariharan, A., Hakeem, A. R., Radhakrishnan, S., Reddy, M. S., and Rela, M. (2021). The Role and Therapeutic Potential of NF-Kappa-B Pathway in Severe COVID-19 Patients. Inflammopharmacol 29 (1), 91–100. doi:10.1007/s10787-020-00773-9
Harris, T. J., Grosso, J. F., Yen, H.-R., Xin, H., Kortylewski, M., Albesiano, E., et al. (2007). Cutting Edge: An In Vivo Requirement for STAT3 Signaling in TH17 Development and TH17-dependent Autoimmunity. J. Immunol. 179 (7), 4313–4317. doi:10.4049/jimmunol.179.7.4313
Hartenian, E., Nandakumar, D., Lari, A., Ly, M., Tucker, J. M., and Glaunsinger, B. A. (2020). The Molecular Virology of Coronaviruses. J. Biol. Chem. 295 (37), 12910–12934. doi:10.1074/jbc.rev120.013930
Haybar, H., Kazemnia, K., and Rahim, F. (2020). Underlying Chronic Disease and COVID-19 Infection: a State-Of-The-Art Review. Jundishapur J. Chronic Dis. Care 9 (2). doi:10.5812/jjcdc.103452
Hayden, M. S., and Ghosh, S. (2008). Shared Principles in NF-Κb Signaling. Cell 132 (3), 344–362. doi:10.1016/j.cell.2008.01.020
He, Q.-F., Xu, Y., Li, J., Huang, Z.-M., Li, X.-H., and Wang, X. (2019). CD8+ T-Cell Exhaustion in Cancer: Mechanisms and New Area for Cancer Immunotherapy. Brief. Funct. Genomics 18 (2), 99–106. doi:10.1093/bfgp/ely006
Herold, T., Jurinovic, V., Arnreich, C., Lipworth, B. J., Hellmuth, J. C., von Bergwelt-Baildon, M., et al. (2020). Elevated Levels of IL-6 and CRP Predict the Need for Mechanical Ventilation in COVID-19. J. Allergy Clin. Immunol. 146 (1), 128–136. doi:10.1016/j.jaci.2020.05.008
Herrmann, A., Kortylewski, M., Kujawski, M., Zhang, C., Reckamp, K., Armstrong, B., et al. (2010). Targeting Stat3 in the Myeloid Compartment Drastically Improves the In Vivo Antitumor Functions of Adoptively Transferred T Cells. Cancer Res. 70 (19), 7455–7464. doi:10.1158/0008-5472.can-10-0736
Hirano, T., and Murakami, M. (2020). COVID-19: a New Virus, but a Familiar Receptor and Cytokine Release Syndrome. Immunity 52 (5), 731–733. doi:10.1016/j.immuni.2020.04.003
Hoffmann, M., Kleine-Weber, H., Krüger, N., Mueller, M. A., Drosten, C., and Pöhlmann, S. (2020). The Novel Coronavirus 2019 (2019-nCoV) Uses the SARS-Coronavirus Receptor ACE2 and the Cellular Protease TMPRSS2 for Entry into Target Cells. BioRxiv.
Hosseini, A., Hashemi, V., Shomali, N., Asghari, F., Gharibi, T., Akbari, M., et al. (2020). Innate and Adaptive Immune Responses against Coronavirus. Biomed. Pharmacother. 132, 110859. doi:10.1016/j.biopha.2020.110859
Hotchkiss, R. S., and Moldawer, L. L. (2014). Parallels between Cancer and Infectious Disease. N. Engl. J. Med. 371 (4), 380–383. doi:10.1056/NEJMcibr1404664
Huynh, J., Etemadi, N., Hollande, F., Ernst, M., and Buchert, M. (2017). The JAK/STAT3 axis: A Comprehensive Drug Target for Solid Malignancies. Semin. Cancer Biol. 45, 13–22. Elsevier. doi:10.1016/j.semcancer.2017.06.001
Iwata-Kajihara, T., Sumimoto, H., Kawamura, N., Ueda, R., Takahashi, T., Mizuguchi, H., et al. (2011). Enhanced Cancer Immunotherapy Using STAT3-Depleted Dendritic Cells with High Th1-Inducing Ability and Resistance to Cancer Cell-Derived Inhibitory Factors. J.I. 187 (1), 27–36. doi:10.4049/jimmunol.1002067
Jamal, M., Bangash, H. I., Habiba, M., Lei, Y., Xie, T., Sun, J., et al. (2021). Immune Dysregulation and System Pathology in COVID-19. Virulence 12 (1), 918–936. doi:10.1080/21505594.2021.1898790
Johnson, D. E., O'Keefe, R. A., and Grandis, J. R. (2018). Targeting the IL-6/JAK/STAT3 Signalling axis in Cancer. Nat. Rev. Clin. Oncol. 15 (4), 234–248. doi:10.1038/nrclinonc.2018.8
Kazachenka, A., and Kassiotis, G. (2021). SARS-CoV-2-Host Chimeric RNA-Sequencing Reads Do Not Necessarily Arise from Virus Integration into the Host DNA. Front. Microbiol. 12 (1367). doi:10.3389/fmicb.2021.676693
Kazazian, H. H., Moran, J. V., and Moran, J. V. (2017). Mobile DNA in Health and Disease. N. Engl. J. Med. 377 (4), 361–370. doi:10.1056/NEJMra1510092
Kircheis, R., Haasbach, E., Lueftenegger, D., Heyken, W. T., Ocker, M., and Planz, O. (2020). NF-κB Pathway as a Potential Target for Treatment of Critical Stage COVID-19 Patients. Front. Immunol. 11, 598444. doi:10.3389/fimmu.2020.598444
Koch, J., Steinle, A., Watzl, C., and Mandelboim, O. (2013). Activating Natural Cytotoxicity Receptors of Natural Killer Cells in Cancer and Infection. Trends Immunol. 34 (4), 182–191. doi:10.1016/j.it.2013.01.003
Kortylewski, M., Kujawski, M., Wang, T., Wei, S., Zhang, S., Pilon-Thomas, S., et al. (2005). Inhibiting Stat3 Signaling in the Hematopoietic System Elicits Multicomponent Antitumor Immunity. Nat. Med. 11 (12), 1314–1321. doi:10.1038/nm1325
Kumari, N., Dwarakanath, B. S., Das, A., and Bhatt, A. N. (2016). Role of Interleukin-6 in Cancer Progression and Therapeutic Resistance. Tumor Biol. 37 (9), 11553–11572. doi:10.1007/s13277-016-5098-7
Laing, A. G., Lorenc, A., del Molino del Barrio, I., Das, A., Fish, M., Monin, L., et al. (2020). A Dynamic COVID-19 Immune Signature Includes Associations with Poor Prognosis. Nat. Med. 26 (10), 1623–1635. doi:10.1038/s41591-020-1038-6
Li, D., and Wu, M. (2021). Pattern Recognition Receptors in Health and Diseases. Sig Transduct Target. Ther. 6 (1), 291. doi:10.1038/s41392-021-00687-0
Li, G., Fan, Y., Lai, Y., Han, T., Li, Z., Zhou, P., et al. (2020). Coronavirus Infections and Immune Responses. J. Med. Virol. 92 (4), 424–432. doi:10.1002/jmv.25685
Li, G., Fan, Y., Lai, Y., Han, T., Li, Z., Zhou, P., et al. (2020). Coronavirus Infections and Immune Responses. J. Med. Virol. 92 (4), 424–432. doi:10.1002/jmv.25685
Li, N., Wang, X., and Lv, T. (2020). Prolonged SARS‐CoV‐2 RNA Shedding: Not a Rare Phenomenon. J. Med. Virol. 92 (11), 2286–2287. doi:10.1002/jmv.25952
Li, X., Geng, M., Peng, Y., Meng, L., and Lu, S. (2020). Molecular Immune Pathogenesis and Diagnosis of COVID-19. J. Pharm. Anal. 10, 102–108. doi:10.1016/j.jpha.2020.03.001
Liang, Y., Zhou, Y., and Shen, P. (2004). NF-kappaB and its Regulation on the Immune System. Cell Mol Immunol 1 (5), 343–350.
Liao, Q.-J., Ye, L.-B., Timani, K. A., Zeng, Y.-C., She, Y.-L., Ye, L., et al. (2005). Activation of NF-kappaB by the Full-Length Nucleocapsid Protein of the SARS Coronavirus. Acta Biochim. Biophys. Sinica 37 (9), 607–612. doi:10.1111/j.1745-7270.2005.00082.x
Liu, T., Zhang, L., Joo, D., and Sun, S.-C. (2017). NF-κB Signaling in Inflammation. Sig Transduct Target. Ther. 2, 2. doi:10.1038/sigtrans.2017.23
Ma, L., Li, Q., Cai, S., Peng, H., Huyan, T., and Yang, H. (2021). The Role of NK Cells in Fighting the Virus Infection and Sepsis. Int. J. Med. Sci. 18 (14), 3236–3248. doi:10.7150/ijms.59898
Marrack, P., Kappler, J., and Mitchell, T. (1999). Type I Interferons Keep Activated T Cells Alive. J. Exp. Med. 189 (3), 521–530. doi:10.1084/jem.189.3.521
McKinney, E. F., and Smith, K. G. (2016). T Cell Exhaustion and Immune-Mediated Disease-The Potential for Therapeutic Exhaustion. Curr. Opin. Immunol. 43, 74–80. doi:10.1016/j.coi.2016.09.005
McLaughlin-Drubin, M. E., and Munger, K. (2008). Viruses Associated with Human Cancer. Biochim. Biophys. Acta (Bba) - Mol. Basis Dis. 1782 (3), 127–150. doi:10.1016/j.bbadis.2007.12.005
Mina, M. J., Parker, R., and Larremore, D. B. (2020). Rethinking Covid-19 Test Sensitivity - A Strategy for Containment. N. Engl. J. Med. 383 (22), e120. doi:10.1056/nejmp2025631
Nagasaki, T., Hara, M., Nakanishi, H., Takahashi, H., Sato, M., and Takeyama, H. (2014). Interleukin-6 Released by colon Cancer-Associated Fibroblasts Is Critical for Tumour Angiogenesis: Anti-interleukin-6 Receptor Antibody Suppressed Angiogenesis and Inhibited Tumour-Stroma Interaction. Br. J. Cancer 110 (2), 469–478. doi:10.1038/bjc.2013.748
Nakagawa, K., and Makino, S. (2021). Mechanisms of Coronavirus Nsp1-Mediated Control of Host and Viral Gene Expression. Cells 10 (2), 300. doi:10.3390/cells10020300
Negishi, H., Taniguchi, T., and Yanai, H. (2018). The Interferon (IFN) Class of Cytokines and the IFN Regulatory Factor (IRF) Transcription Factor Family. Cold Spring Harb Perspect. Biol. 10 (11), a028423. doi:10.1101/cshperspect.a028423
Nozawa, H., Chiu, C., and Hanahan, D. (2006). Infiltrating Neutrophils Mediate the Initial Angiogenic Switch in a Mouse Model of Multistage Carcinogenesis. Proc. Natl. Acad. Sci. 103 (33), 12493–12498. doi:10.1073/pnas.0601807103
Oeckinghaus, A., and Ghosh, S. (2009). The NF- B Family of Transcription Factors and its Regulation. Cold Spring Harbor Perspect. Biol. 1 (4), a000034. doi:10.1101/cshperspect.a000034
Pan, Y., Jiang, X., Yang, L., Chen, L., Zeng, X., Liu, G., et al. (2021). SARS-CoV-2-specific Immune Response in COVID-19 Convalescent Individuals. Signal. Transduction Targeted Ther. 6, 256. doi:10.1038/s41392-021-00686-1
Parry, R., Gifford, R. J., Lytras, S., Ray, S. C., and Coin, L. J. M. (2021). No Evidence of SARS-CoV-2 Reverse Transcription and Integration as the Origin of Chimeric Transcripts in Patient Tissues. Proc. Natl. Acad. Sci. USA 118 (33), e2109066118. doi:10.1073/pnas.2109066118
Pedersen, S. F., and Ho, Y.-C. (2020). SARS-CoV-2: a Storm Is Raging. J. Clin. Invest. 130 (5), 2202–2205. doi:10.1172/jci137647
Pelkmans, L., and Helenius, A. (2003). Insider Information: what Viruses Tell Us about Endocytosis. Curr. Opin. Cel. Biol. 15 (4), 414–422. doi:10.1016/s0955-0674(03)00081-4
Perlman, S., and Netland, J. (2009). Coronaviruses post-SARS: Update on Replication and Pathogenesis. Nat. Rev. Microbiol. 7 (6), 439–450. doi:10.1038/nrmicro2147
Plummer, M., de Martel, C., Vignat, J., Ferlay, J., Bray, F., and Franceschi, S. (2016). Global burden of Cancers Attributable to Infections in 2012: a Synthetic Analysis. Lancet Glob. Health 4 (9), e609–e616. doi:10.1016/s2214-109x(16)30143-7
Power, S. P., Moloney, F., Twomey, M., James, K., O’Connor, O. J., and Maher, M. M. (2016). Computed Tomography and Patient Risk: Facts, Perceptions and Uncertainties. Wjr 8 (12), 902–915. doi:10.4329/wjr.v8.i12.902
Prompetchara, E., Ketloy, C., and Palaga, T. (2020). Immune Responses in COVID-19 and Potential Vaccines: Lessons Learned from SARS and MERS Epidemic. Asian Pac. J. Allergy Immunol. 38 (1), 1–9. doi:10.12932/AP-200220-0772
Radpour, A., Bahrami-Motlagh, H., Taaghi, M. T., Sedaghat, A., Karimi, M. A., Hekmatnia, A., et al. (2020). COVID-19 Evaluation by Low-Dose High Resolution CT Scans Protocol. Acad. Radiol. 27 (6), 901. doi:10.1016/j.acra.2020.04.016
Rossi, J.-F., Lu, Z. Y., Massart, C., and Levon, K. (2021). Dynamic Immune/Inflammation Precision Medicine: The Good and the Bad Inflammation in Infection and Cancer. Front. Immunol. 12, 595722. doi:10.3389/fimmu.2021.595722
Saini, G., and Aneja, R. (2021). Cancer as a Prospective Sequela of Long COVID‐19. Bioessays 43 (6), 2000331. doi:10.1002/bies.202000331
Salazar, L., Kashiwada, T., Krejci, P., Meyer, A. N., Casale, M., Hallowell, M., et al. (2014). Fibroblast Growth Factor Receptor 3 Interacts with and Activates TGFβ-Activated Kinase 1 Tyrosine Phosphorylation and NFκB Signaling in Multiple Myeloma and Bladder Cancer. PLoS One 9 (1), e86470. doi:10.1371/journal.pone.0086470
Sangfelt, O., Erickson, S., Castro, J., Heiden, T., Gustafsson, A., Einhorn, S., et al. (1999). Molecular Mechanisms Underlying Interferon-Alpha-Induced G0/G1 Arrest: CKI-Mediated Regulation of G1 Cdk-Complexes and Activation of Pocket Proteins. Oncogene 18 (18), 2798–2810. doi:10.1038/sj.onc.1202609
Sau, A., Lau, R., Cabrita, M. A., Nolan, E., Crooks, P. A., Visvader, J. E., et al. (2016). Persistent Activation of NF-Κb in BRCA1-Deficient Mammary Progenitors Drives Aberrant Proliferation and Accumulation of DNA Damage. Cell stem cell 19 (1), 52–65. doi:10.1016/j.stem.2016.05.003
Schindler, C., Levy, D. E., and Decker, T. (2007). JAK-STAT Signaling: from Interferons to Cytokines. J. Biol. Chem. 282 (28), 20059–20063. doi:10.1074/jbc.r700016200
Schreiber, R. D., Old, L. J., and Smyth, M. J. (2011). Cancer Immunoediting: Integrating Immunity's Roles in Cancer Suppression and Promotion. Science 331 (6024), 1565–1570. doi:10.1126/science.1203486
Schultheiß, C., Paschold, L., Simnica, D., Mohme, M., Willscher, E., von Wenserski, L., et al. (2020). Next-Generation Sequencing of T and B Cell Receptor Repertoires from COVID-19 Patients Showed Signatures Associated with Severity of Disease. Immunity 53 (2), 442e4–455. doi:10.1016/j.immuni.2020.06.024
Smits, N., Rasmussen, J., Bodea, G. O., Amarilla, A. A., Gerdes, P., Sanchez-Luque, F. J., et al. (2021). No Evidence of Human Genome Integration of SARS-CoV-2 Found by Long-Read DNA Sequencing. Cel Rep. 36 (7), 109530. doi:10.1016/j.celrep.2021.109530
Sohrabi, C., Alsafi, Z., O’Neill, N., Khan, M., Kerwan, A., Al-Jabir, A., et al. (2020). World Health Organization Declares Global Emergency: A Review of the 2019 Novel Coronavirus (COVID-19). Int. J. Surg. 76, 71–76. doi:10.1016/j.ijsu.2020.02.034
Soltani, S., Tabibzadeh, A., Zakeri, A., Zakeri, A. M., Latifi, T., Shabani, M., et al. (2021). COVID-19 Associated central Nervous System Manifestations, Mental and Neurological Symptoms: a Systematic Review and Meta-Analysis. Rev. Neurosciences 32 (3), 351–361. doi:10.1515/revneuro-2020-0108
Song, J.-W., Zhang, C., Fan, X., Meng, F.-P., Xu, Z., Xia, P., et al. (2020). Immunological and Inflammatory Profiles in Mild and Severe Cases of COVID-19. Nat. Commun. 11 (1), 3410. doi:10.1038/s41467-020-17240-2
Song, W., Mazzieri, R., Yang, T., and Gobe, G. C. (2017). Translational Significance for Tumor Metastasis of Tumor-Associated Macrophages and Epithelial-Mesenchymal Transition. Front. Immunol. 8, 1106. doi:10.3389/fimmu.2017.01106
Stetson, D. B., and Medzhitov, R. (2006). Type I Interferons in Host Defense. Immunity 25 (3), 373–381. doi:10.1016/j.immuni.2006.08.007
Teng, Y., Ross, J. L., and Cowell, J. K. (2014). The Involvement of JAK-STAT3 in Cell Motility, Invasion, and Metastasis. Jak-stat 3 (1), e28086. doi:10.4161/jkst.28086
Vargas, A. J., and Harris, C. C. (2016). Biomarker Development in the Precision Medicine Era: Lung Cancer as a Case Study. Nat. Rev. Cancer 16 (8), 525–537. doi:10.1038/nrc.2016.56
Walter, M., Liang, S., Ghosh, S., Hornsby, P. J., and Li, R. (2009). Interleukin 6 Secreted from Adipose Stromal Cells Promotes Migration and Invasion of Breast Cancer Cells. Oncogene 28 (30), 2745–2755. doi:10.1038/onc.2009.130
Walther, W., Kobelt, D., Bauer, L., Aumann, J., and Stein, U. (2015). Chemosensitization by Diverging Modulation by Short-Term and Long-Term TNF-α Action on ABCB1 Expression and NF-Κb Signaling in colon Cancer. Int. J. Oncol. 47 (6), 2276–2285. doi:10.3892/ijo.2015.3189
Wang, X., Lu, X.-J., and Sun, B. (2017). The Pros and Cons of Dying Tumour Cells in Adaptive Immune Responses. Nat. Rev. Immunol. 17 (9), 591. doi:10.1038/nri.2017.87
Woo, P. C. Y., Huang, Y., Lau, S. K. P., and Yuen, K.-Y. (2010). Coronavirus Genomics and Bioinformatics Analysis. viruses 2 (8), 1804–1820. doi:10.3390/v2081803
Wu, F., Zhao, S., Yu, B., Chen, Y.-M., Wang, W., Song, Z.-G., et al. (2020). A New Coronavirus Associated with Human Respiratory Disease in China. Nature 579 (7798), 265–269. doi:10.1038/s41586-020-2008-3
Xiao, J., Gong, Y., Chen, Y., Yu, D., Wang, X., Zhang, X., et al. (2017). IL-6 Promotes Epithelial-To-Mesenchymal Transition of Human Peritoneal Mesothelial Cells Possibly through the JAK2/STAT3 Signaling Pathway. Am. J. Physiology-Renal Physiol. 313 (2), F310–F318. doi:10.1152/ajprenal.00428.2016
Yang, L., Liu, S., Liu, J., Zhang, Z., Wan, X., Huang, B., et al. (2020). COVID-19: Immunopathogenesis and Immunotherapeutics. Sig Transduct Target. Ther. 5 (1), 128. doi:10.1038/s41392-020-00243-2
Yang, N., and Shen, H.-M. (2020). Targeting the Endocytic Pathway and Autophagy Process as a Novel Therapeutic Strategy in COVID-19. Int. J. Biol. Sci. 16 (10), 1724–1731. doi:10.7150/ijbs.45498
Yap, J. K. Y., Moriyama, M., and Iwasaki, A. (2020). Inflammasomes and Pyroptosis as Therapeutic Targets for COVID-19. J.I. 205 (2), 307–312. doi:10.4049/jimmunol.2000513
Ye, C.-H., Hsu, W.-L., Peng, G.-R., Yu, W.-C., Lin, W.-C., Hu, S., et al. (2021). Role of the Immune Microenvironment in SARS-CoV-2 Infection. Cel Transpl. 30, 096368972110106. doi:10.1177/09636897211010632
Ye, M., Song, Y., Pan, S., Chu, M., Wang, Z.-W., and Zhu, X. (2020). Evolving Roles of Lysyl Oxidase Family in Tumorigenesis and Cancer Therapy. Pharmacol. Ther. 215, 107633. doi:10.1016/j.pharmthera.2020.107633
Yu, H., Kortylewski, M., and Pardoll, D. (2007). Crosstalk between Cancer and Immune Cells: Role of STAT3 in the Tumour Microenvironment. Nat. Rev. Immunol. 7 (1), 41–51. doi:10.1038/nri1995
Yu, H., Pardoll, D., and Jove, R. (2009). STATs in Cancer Inflammation and Immunity: a Leading Role for STAT3. Nat. Rev. Cancer 9 (11), 798–809. doi:10.1038/nrc2734
Zafer, M. M., El-Mahallawy, H. A., and Ashour, H. M. (2021). Severe COVID-19 and Sepsis: Immune Pathogenesis and Laboratory Markers. Microorganisms 9 (1), 159. doi:10.3390/microorganisms9010159
Zapatka, M., Borozan, I., Borozan, I., Brewer, D. S., Iskar, M., Grundhoff, A., et al. (2020). The Landscape of Viral Associations in Human Cancers. Nat. Genet. 52 (3), 320–330. doi:10.1038/s41588-019-0558-9
Zhang, L., Richards, A., Barrasa, M. I., Hughes, S. H., Young, R. A., and Jaenisch, R. (2021). Reverse-transcribed SARS-CoV-2 RNA Can Integrate into the Genome of Cultured Human Cells and Can Be Expressed in Patient-Derived Tissues. Proc. Natl. Acad. Sci. 118 (21), e2105968118. doi:10.1073/pnas.2105968118
Zhang, L., Richards, A., Khalil, A., Wogram, E., Ma, H., Young, R. A., et al. (2020). SARS-CoV-2 RNA Reverse-Transcribed and Integrated into the Human Genome. bioRxiv.
Zhang, Q., Lenardo, M. J., and Baltimore, D. (2017). 30 Years of NF-Κb: A Blossoming of Relevance to Human Pathobiology. Cell 168 (1-2), 37–57. doi:10.1016/j.cell.2016.12.012
Zheng, M., Gao, Y., Wang, G., Song, G., Liu, S., Sun, D., et al. (2020). Functional Exhaustion of Antiviral Lymphocytes in COVID-19 Patients. Cel Mol Immunol 17 (5), 533–535. doi:10.1038/s41423-020-0402-2
Zhou, T., Su, T. T., Mudianto, T., and Wang, J. (2020). Immune Asynchrony in COVID-19 Pathogenesis and Potential Immunotherapies. J. Exp. Med. 217 (10). doi:10.1084/jem.20200674
Keywords: COVID-19 sequelae, SARS-CoV-2, cancer, long COVID-19, immune homeostasis
Citation: Rahimmanesh I, Shariati L, Dana N, Esmaeili Y, Vaseghi G and Haghjooy Javanmard S (2022) Cancer Occurrence as the Upcoming Complications of COVID-19. Front. Mol. Biosci. 8:813175. doi: 10.3389/fmolb.2021.813175
Received: 11 November 2021; Accepted: 21 December 2021;
Published: 28 January 2022.
Edited by:
Rana Jahanban-Esfahlan, Tabriz University of Medical Sciences, IranReviewed by:
Roopa Biswas, Uniformed Services University of the Health Sciences, United StatesCopyright © 2022 Rahimmanesh, Shariati, Dana, Esmaeili, Vaseghi and Haghjooy Javanmard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaghayegh Haghjooy Javanmard, c2hhZ2hheWVnaC5oYWdoam9vQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.