- 1Department of Medical Biochemistry and Molecular Biology, Faculty of Medicine, Fayoum University, Fayoum, Egypt
- 2Department of Medical Microbiology and Immunology, Faculty of Medicine, Fayoum University, Fayoum, Egypt
- 3Departments of Rheumatology and Rehabilitation, Faculty of Medicine, Al-Azhar University, Cairo, Egypt
- 4Department of Internal Medicine, Faculty of Medicine, Fayoum University, Fayoum, Egypt
- 5Department of Clinical Pharmacy, Faculty of Pharmacy, Fayoum University, Fayoum, Egypt
- 6Department of Clinical Pathology, Faculty of Medicine, Fayoum University, Fayoum, Egypt
Background: Behçet’s disease (BD) is a chronic autoimmune disease. The early diagnosis of BD is very important to avoid serious and/or fatal complications such as eye damage, severe neurological involvement, and large vessel occlusion. New, sensitive biomarkers would aid in rapid diagnosis, the monitoring of disease activity, and the response to treatment.
Methods: This study’s aim is to identify two immune system-related BD biomarkers. We measured long non-coding RNAs (lncRNAs) NEAT1 (nuclear-enriched abundant transcript 1), and lnc-DC (lncRNA in dendritic cells) in serum by real-time polymerase chain reaction (RT-PCR) in 52 BD patients and 52 controls. We analyzed the association between NEAT1 and lnc-DC and the clinical parameters of BD. Receiver operating characteristic (ROC) curve analysis was performed to explore the diagnostic performance of the studied genes.
Results: Compared to controls, the significant upregulation of NEAT1 {median [interquartile range (IQR)] = 1.68 (0.38–7.7), p < 0.0001} and downregulation of lnc-DC [median (IQR) = 0.2 (0.12–1.39), p = 0.03] were detected in the sera collected from BD patients. Higher serum expression levels of NEAT1 and lnc-DC were significantly associated with the following clinical presentations: cutaneous lesions, vascular manifestations, articular manifestations, neurological manifestations, and higher disease activity score. Also, high NEAT1 levels were significantly associated with a negative pathergy test, while higher lnc-DC was significantly associated with a positive family history. ROC curves showed that NEAT1 and lnc-DC levels in serum could be used as predictors of BD with high specificity and fair sensitivity. NEAT1 had an area under the curve (AUC) of 0.692 (95% CI: 0.591–0.794, p = 0.001), and lnc-DC had an AUC of 0.615 (95% CI: 0.508–0.723, p = 0.043).
Conclusion: Serum lncRNAs NEAT1 and lnc-DC are biomarkers for BD.
Introduction
Behçet’s disease (BD) is a chronic multi-systemic inflammatory disease with a wide spectrum of clinical presentations varying from mucocutaneous ulcerations and ocular inflammation to more severe and potentially fatal manifestations such as the vascular system, nervous system, and gastrointestinal tract lesions. Different theories posit that BD is a result of immune system dysfunction due to environmental or infectious triggering factors in genetically susceptible individuals. To date, there is no specific laboratory test for BD diagnosis, and so the identification of essential genes underlying the susceptibility, activity status, or manifestations of the disease is of great importance and may provide new regimens for controlling or treating the disease (Leccese and Alpsoy 2019).
Long non-coding RNAs (lncRNAs) are a group of RNAs that are longer than 200 nucleotides in length and do not encode proteins (Wei et al., 2017). Although the function of most lncRNAs is unclear, numerous studies have documented pivotal functions for lncRNAs in human immune diseases through the activation, differentiation, and imbalanced expression of immune cells (T cells, B cells, macrophages, and natural killer cells) that are implicated in the pathogenesis of various autoimmune diseases such as psoriasis, rheumatoid arthritis (RA), and systemic lupus erythematosus (SLE) (Sigdel et al., 2015). Among these biomarkers are lncRNA NEAT1 (nuclear-enriched abundant transcript 1) (Imamura et al., 2014; Zhang et al., 2016; Huang et al., 2017; Pandey et al., 2017; Huang et al., 2018; Prinz et al., 2019; Zhang et al., 2019), and lnc-DC (lncRNA in dendritic cells) (Wu et al., 2017)].
NEAT1 is an lncRNA that localizes to and plays a critical role in maintaining the formation of paraspeckles, an interchromatin nuclear body with unknown function (Zhang et al., 2019). Various stimuli that activate inflammasomes can also trigger the expression of NEAT1, such as viral infections and tumor suppressor p53 (Adriaens et al., 2016; Zhang et al., 2019). Inflammasome-inducing signals stimulate the release of NEAT1 from paraspeckles and their translocation to the cytoplasm, where they participate in inflammasome activity. NEAT1 stimulates one or more innate pattern recognition receptors (PRRs) (Zhang et al., 2019). In addition, NEAT1 is crucial for inflammasomes and caspase-1 activation, cytokine production, and pyroptotic cell death. NEAT1 also regulates the expression of a group of chemokines and cytokines, including IL-6 and CXCL10, through the MAPK pathway (Sigdel et al., 2015; Zhang et al., 2019), suggesting a role in the pathogenesis of BD.
Lnc-DC is expressed in dendritic cells (DCs) and mediates DC maturation by phosphorylating signal transducer and activator of transcription 3 (STAT3) (Zhang et al., 2020b). As such, lnc-DC can induce the differentiation and maturation of DCs, the transcription of immune response genes, and the production of related cytokines, such as TNF-a, IFN-c, IL-13, IL-5, IL-4, and IL-12 (Wang et al., 2014).
Based on this background, we investigated the relationship between the expression levels of two immunity-related lncRNAs, NEAT1 and lnc-DC, with BD susceptibility, activity, and clinical manifestations. We also determined the diagnostic performance of NEAT1 and lnc-DC in differentiating BD patients from healthy controls.
Subject and Methods
Patients and Controls
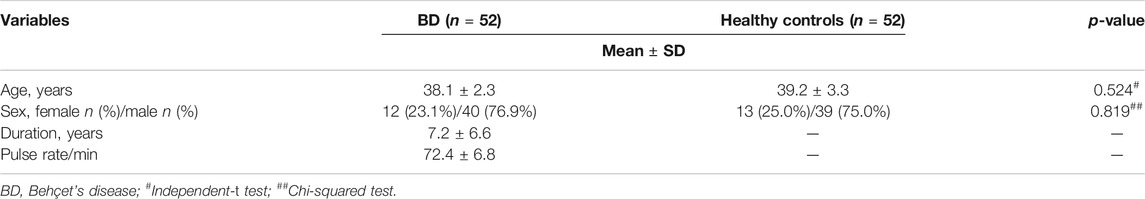
This study was conducted per the Declaration of Helsinki and was approved by the Ethical Committee of the Faculty of Medicine, Fayoum University (protocol code M400). Written consent was obtained from all participants after the declaration of all involved procedures and steps. Participants were categorized into two groups. For Group I, we selected 40 male and 12 female patients fulfilling the criteria of the International Study Group (ISG) for the diagnosis of BD (INTERNATIONAL STUDY GROUP FOR BEHC 1990). Participants were recruited from outpatients’ clinics, patients admitted to the Rheumatology and Rehabilitation Department and/or the Internal Medicine Department of Fayoum University Hospital (Fayoum, Egypt) from June 2019 to January 2021. Patients with other concurrent diseases such as hepatic, renal dysfunction, autoimmune diseases, atherosclerosis, coronary artery disease, essential hypertension, and diabetes were excluded. For Group II, we included 52 age- and sex-matched healthy controls without acute or chronic disease in the study.
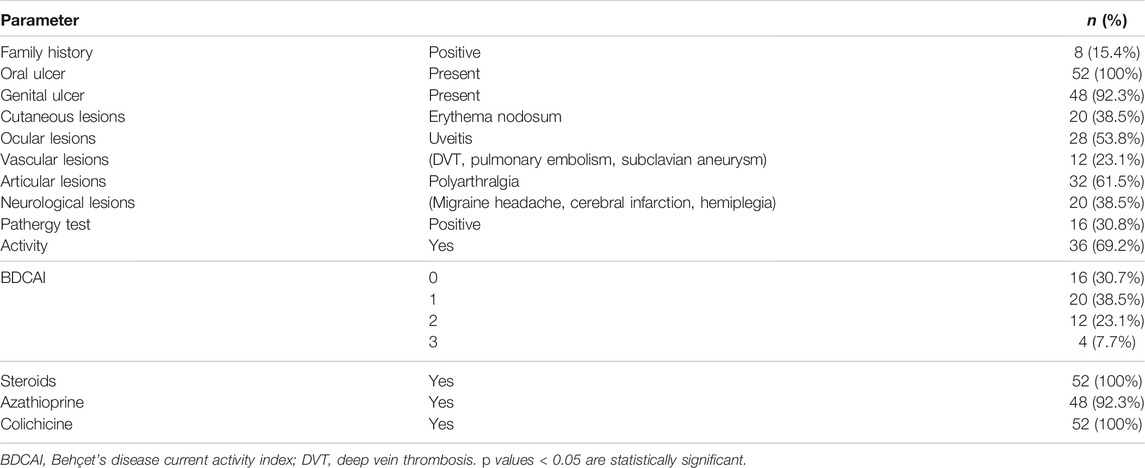
Medical history and clinical examination were performed for all patients to detect disease features and complications. Patients underwent a complete medical history (age, sex, duration of the disease, family history), general examination, specific examination to detect the symptoms and signs of BD disease, involving oral or genital ulcers, the presence of cutaneous lesions, Duplex and color-coded Doppler examination on arterial and venous systems, and fundus examination by slit lamp. Central nervous system (CNS) and joint examination by computed tomography and magnetic resonance imaging were performed when indicated to detect any vascular, ocular, CNS, or articular lesions. The pathergy test was done for all patients, and a positive pathergy test was indicated by the formation of a small papule or pustule (3–10 mm) within 1 or 2 days at the site of sterile needle prick (El Boghdady and Shaker 2019). All patients received steroids and colchicine, while 92.3% received azathioprine. Patients were assessed for disease activity using the Behçet’s disease Current Activity Index (BDCAI) score, which is calculated based on the answers to 12 questions regarding disease manifestations over the previous 4 weeks. Active disease scores range from 0 to 12. A higher score indicates a higher level of disease activity (Bhakta et al., 1999). Accordingly, patients were divided into active (patients who met at least one criterion of the ISG for BD) or inactive (no lesions for the previous 4 weeks or more) patients at the time of the study. In the current study, 36 patients (69.3%) were in active disease, of which 38.5% had an activity score of 1, 23.1% had an activity score of 2, and 7.7% had an activity score of 3.
Sample Collection, RNA Extraction, cDNA Synthesis, RT-PCR Performance, and Calculation of Target Genes’ Fold Changes
Venous blood samples were obtained from all patients and healthy individuals under sterile conditions. The collected venous blood was kept undisturbed at room temperature to allow clotting. The samples were subsequently subjected to centrifugation for serum separation. The clear supernatant present in the upper part of the tube (serum) was transferred to a clean tube and stored in the freezer at −80°C until RNA extraction.
Total RNA, including NEAT1 and lnc-DC, was extracted from the serum using the miRNeasy extraction kit (Qiagen, Valencia, CA, United States) by following the manufacturer’s protocol.
The steps of RNA extraction include the following: first, we cleaned the working place with 70% ethanol. Then, we added 1 ml QIAzol lysis reagent to a 200 μl sample and incubated it for 5 min at room temperature. Next, we added 200 μl chloroform, vortexed the tube for 15 s, and incubated the mixture at room temperature for 2 min. Afterwards, we used cool centrifugation to avoid the destruction of RNA at 120,00× g for 15 min at 4°C. Next, we transported the upper aqueous phase to a new collection tube, followed by the addition of 1.5 times its volume of 100% ethanol. After that, we pipetted 750 μl of the mix to the 2 ml RNeasy Mini spin column and centrifuged it at 8,000 ×g at room temperature for 15 s. When the mixture had totally passed the column, we discarded the flow through water and reused the tube to repeat this step with the remaining part of the mix. Then, we used the washing buffers in the kit (RWT and RPF) as follows: firstly, we added 700 μl of the RWT buffer to each spin column. Then, we centrifuged it at 8,000 ×g at room temperature for 15 s, discarded the flow through, and reused the column for the next step. Secondly, we pipetted a 500 μl buffer RPE to the spin column and again centrifuged it at 8,000 ×g at room temperature for 15 s, discarded the flow through water, and repeated the previous step. To finish, we transported the spin column to the new collecting tube and centrifuged the tube for 2 min at full speed for dehydration, and for elusion, the column was transferred to a clean Eppendorf tube and 50 ul Rnase-free water was pipetted directly onto the column and centrifuged for 1 min at 8,000 ×g. A NanoDrop (ND)-1000 spectrophotometer (NanoDrop Technologies, Inc. Wilmington, DE, USA) was used to quantify extracted RNA purity and concentration; about 1–2 μl of eluted RNA samples were sufficient, and the ratio of absorbance at 260 and 280 nm is used to assess the purity of RNA; a ratio of ∼2.0 is generally accepted as “pure” for RNA.
Reverse transcription to obtain cDNA was performed using the RT2 First Strand Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol in a final volume of 20 μl (10 μl reverse-transcription mix was added to each tube containing a genomic DNA elimination mix that has the extracted RNA and DNA elimination buffer).
The expression of lncRNAs, including target genes, in serum, has been previously documented (Shaker et al., 2019a; Shaker et al., 2019b). The expression of lncRNAs NEAT1 and lnc-DC in serum was measured using the RT2 SYBR Green PCR kit (Qiagen, Germantown, MD, USA) using predesigned primers obtained for NEAT1 (Qiagen, Valencia, CA, USA, Catalog no: 330701 LPH15809A, Accession no: NR_028272.1), lnc-DC (Catalog no: 330701 LPH23184A, Accession no: NR_030732.1), and GAPDH as an internal housekeeping gene (Adriaens et al., 2016) (Catalog no: 330701 LPH31725A, Accession no: ENST00000496049.0).
Quantitative real-time PCR was carried out by the Rotor-gene Q real-time PCR system (Qiagen, USA) under the following conditions: 95°C for 10 min, followed by 45 cycles at 95°C for 15 s and 60°C for 60 s. The relative fold changes of the target genes in patients affected by BD compared to the controls were calculated using the 2−ΔΔCt equation (Livak and Schmittgen 2001). For the control sample, ΔΔCt equals zero, and 20 equals one (Shaker et al., 2019b).
Sample Size
The sample size was calculated using G power version 3.0.10. The minimum sample size of patients needed to get a power level of 0.80, an alpha level of 0.05, and a medium effect size of 0.50 for NEAT1 and lnc-DC was 52 in each group.
Statistical Analysis
The collected data were organized, tabulated, and statistically analyzed using SPSS software package version 22 (SPSS Inc., Chicago, IL, USA). Age and other basic data were presented as mean and standard deviation (SD). Independent t-test was used to test significance. NEAT1 and lnc-DC data were presented as median and interquartile range (IQR). The Mann–Whitney U and Kruskal–Wallis tests were used to compare between any two or three groups, respectively. Spearman correlation was performed to identify any relationships between NEAT1 and lnc-DC with study parameters among patient cases. ROC curve analysis was used to determine the cut-off point differentiating between case and control with the highest sensitivity and specificity for NEAT1 and lnc-DC. Categorical data were presented as frequencies and percentages, and chi-square (χ2) was used as a test of significance. For the interpretation of results, the significance was determined to be p ≤ 0.05.
Results
Basic Characteristics of BD Patients and Controls
The demographic data of the studied groups are presented in Table 1. This case-control study was conducted on 52 (76.9% males, 23.1% females) BD patients aged 38.1 ± 2.3 and with a disease duration of 7.2 ± 6.6 years compared with 52 healthy controls with matched age (39.2 ± 3.3, p = 0.524) and sex (75% males, 25% females, p = 0.819).
Clinical Characteristics of Patients
Family history, clinical data, activity index, and treatment options are presented in Table 2. Out of 52 BD patients, eight (15.4%) had a positive family history, 16 (30.8%) had a positive pathergy test, and 36 (69.3%) had an active disease, with a score of 1 (38.5%), 2 (23.1%), or 3 (7.7%) on the activity index. All patients suffered from oral ulcers, 92.3% from genital ulcers, 38.5% from cutaneous lesions (erythema nodosum), and 61.5% from joint manifestations (polyarthralgia). Ocular examination revealed that 53.8% of patients had uveitis. Vascular and CNS assessments showed that 23.1% of patients had vascular manifestations (deep venous thrombosis, pulmonary embolism, or subclavian aneurysm), while 38.5% had neurological manifestations (migraine headache, hemiplegia, or cerebral infarctions).
Expression of NEAT1 and lnc-DC in the BD Patient Group Relative to Healthy Controls
Relative target gene expression is shown in Figure 1. Significantly more NEAT1 [median (IQR) = 1.68 (0.38–7.7), p < 0.0001] and significantly less lnc-DC [median (IQR) = 0.2 (0.12–1.39), p = 0.03] were detected in the sera collected from BD patients relative to the controls.
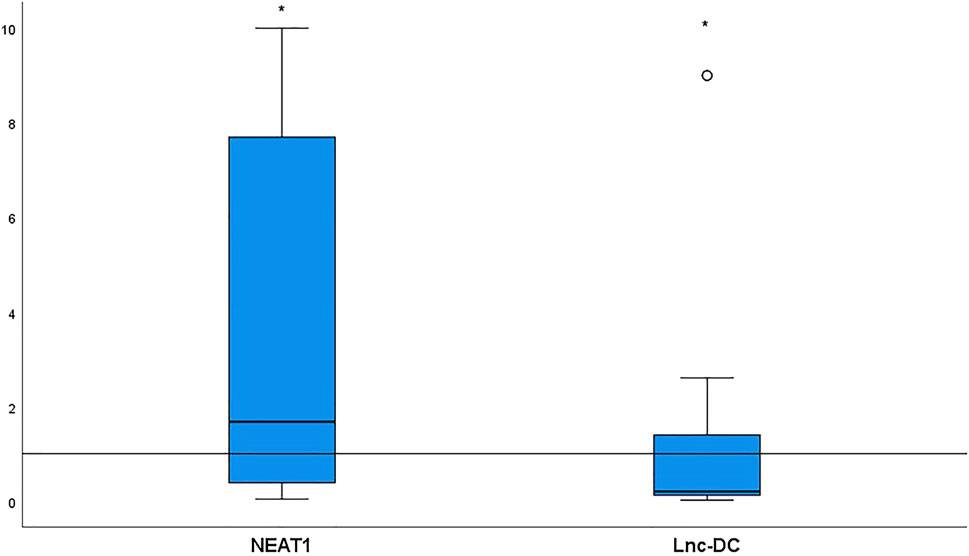
FIGURE 1. NEAT1 and lnc-DC expressions in serum from 52 BD patients relative to healthy controls. Data are represented by box plot (median, upper, and lower quartiles). The horizontal dotted line represents transcript expression in the control group, which is 1 according to the 2−ΔΔCt equation. The increase of NEAT1 (p < 0.0001) and the decrease of lnc-DC (p = 0.03) were detected in total BD patients vs. healthy controls. *Significant.
Relationship Between NEAT1, lnc-DC, and Patient Demographics and Clinical Data
The relationships between NEAT1, lnc-DC, and patient demographics and clinical data were analyzed. We determined that higher serum expression levels of NEAT1 and lnc-DC were significantly associated with the following clinical presentations: cutaneous lesions, vascular manifestations, articular manifestations, neurological manifestations, and higher activity score. NEAT1 was also significantly associated with a negative pathergy test, and higher lnc-DC was significantly associated with a positive family history (Table 3).
Correlation Analysis Between NEAT1 and lnc-DC and Patient Numerical Data
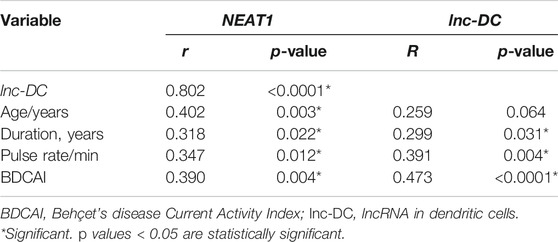
There was a strong positive correlation between the two studied genes (r = 0.802, p < 0.0001). NEAT1 and lnc-DC expressions also showed significant positive correlation with disease duration (r = 0.318, p = 0.022 for NEAT1, r = 0.299, p = 0.031 for lnc-DC), pulse rate (r = 0.347, p = 0.012 for NEAT1, r = 0.391, p = 0.004 for lnc-DC), and with BDCAI activity score (r = 0.390, p = 0.004 for NEAT1, r = 0.473, p < 0.0001 for lnc-DC). However, the patient’s age was only significantly positively correlated with NEAT1 (r = 0.402, p = 0.003) (Table 4).
ROC Analysis to Determine the Diagnostic Performance of NEAT1 and lnc-DC
The expression of NEAT1 and lnc-DC in serum had diagnostic value. RNA levels of these two genes could be used to distinguish BD patients from healthy controls. For NEAT1, AUC = 0.692, 95% confidence interval (CI) = 0.591–0.794, cut-off point = 1.09, p = 0.001, with sensitivity = 69.2%, and specificity = 100%. For lnc-DC, AUC = 0.615, 95% CI = 0.508–0.723, cut-off point = 0.66, p = 0.043, with sensitivity = 61.5%, and specificity = 100% (Table 5; Figure 2).
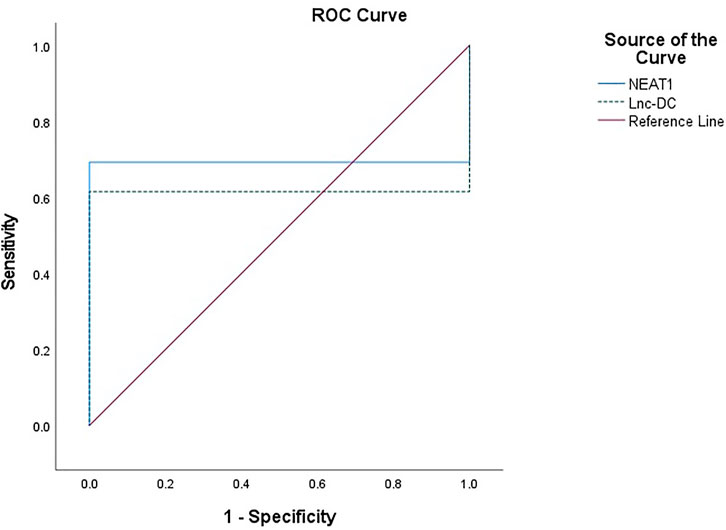
FIGURE 2. ROC curve to predict the diagnostic performance of NEAT1 and lnc-DC found in serum for differentiating BD patients from controls. ROC curves showed that the expression of NEAT1 and lnc-DC in serum could be used as predictors of BD. NEAT1 had an area under the curve (AUC) of 0.692 (95% CI: 0.591–0.794, p = 0.001), and lnc-DC had an AUC of 0.615 (95% CI: 0.508–0.723, p = 0.043).
Discussion
BD is a rare systemic autoimmune vasculitis disease with self-limiting recurrent inflammatory attacks. The recurrent attacks involve vital organs, leading to secondary serious consequences such as uveitis, optic atrophy and blindness, cerebral venous thrombosis, aseptic meningitis, hemiplegia, deep venous thrombosis, pulmonary embolism, and major artery aneurysms. An early diagnosis of BD is very important to avoid fatal complications, and as there is no specific diagnostic test for BD, there is a pressing need to identify sensitive biomarkers to aid in rapid diagnosis and serve in monitoring disease activity and response to treatment (Zeidan et al., 2016; Leccese and Alpsoy 2019).
Recently, increasing evidence indicates that the lncRNAs found in blood serum are dysregulated in numerous autoimmune diseases and could be used as the identifying the biomarkers and novel therapeutic targets of those diseases (Santoro et al., 2016; Shaker et al., 2019b). Up to now, little was known about the importance of lncRNAs in BD, and no studies had been performed to uncover any role(s) that may be played by NEAT1 and/or lnc-DC in BD.
In the present study, we showed that the NEAT1 expression level was increased significantly in the serum from patients with BD relative to controls.
Previous studies have demonstrated a robust relationship between NEAT1 and the inflammatory process. The upregulation of NEAT1 has been shown to promote the release of several inflammasomes (NLRP3, NLRC4, and AIM2) and pro-inflammatory cytokines (CXCL10, IL-6, and IL-1β), resulting in an immune response in inflammatory and immune diseases (Imamura et al., 2014; Zhang et al., 2016; Huang et al., 2017; Pandey et al., 2017; Huang et al., 2018; Prinz et al., 2019; Zhang et al., 2019). For example, the upregulation of NEAT1 was recently reported in bronchial asthma (Li, Ye, and Lu 2020) and SLE (Zhang et al., 2016). In addition, several studies have shown that NEAT1 overexpression results in elevated levels of reactive oxygen species, which worsen immune reactions and exacerbate inflammatory reactions (Wang et al., 2019). Because of its role(s) in immune response, it has been suggested that NEAT1 could be used as a therapeutic target for inflammatory and autoimmune diseases (Imamura et al., 2014; Zhang et al., 2016; Huang et al., 2017; Pandey et al., 2017; Prinz et al., 2019; Zhang et al., 2019).
Considering the previous evidence, we hypothesized that NEAT1 may be implicated in the immune and inflammatory responses occurring in BD.
Our results showed that the expression of NEAT1 is markedly elevated in patients who have skin lesions, neurological symptoms, vascular involvement, and articular involvement (arthritis). Moreover, it is interesting that NEAT1 positively correlated with disease activity score (BDCAI), demonstrating that increased inflammation may lead to higher expression of NEAT1.
Our findings agree with Zhang et al. (2016) who found a positive correlation between the NEAT1 expression and disease activity in SLE. In addition, Wang et al. (2017) reported that the inhibition of NEAT1 results in the improvement of skin lesions associated with herpes simplex infection. NEAT1 is also hypothesized to contribute to neurodegenerative diseases (Michelhaugh et al., 2011; Nishimoto et al., 2013; Zhong et al., 2017). Interestingly, NEAT1 is also abundant in the rat model of osteoarthritis (OA) and contributes to the disease via different mechanisms that result in the increased production of inflammatory cytokines and the increased apoptosis of chondrocytes (Wang et al., 2017; Zhang et al., 2020a).
In the present study, our results showed that the lnc-DC expression in serum is markedly reduced in BD patients relative to controls. However, higher levels of lnc-DC are significantly associated with the presence of cutaneous, vascular, articular, and neurological lesions; the presence of family history; and a higher activity score (BDCAI). The serum level of lnc-DC was also positively correlated with the duration of disease and pulse rate.
lnc-DC is exclusively expressed in human conventional DCs, promoting their differentiation and increasing the capacity of DCs to stimulate T cell activation. lnc-DC mediates these effects by activating the transcription factor STAT3 (Wang et al., 2014). STAT3 regulates IL-21 expression, and STAT3-deficient T helper (Th) cells fail to produce IL-21 mRNA and protein. Previous literature suggests that IL-21 may contribute to the pathogenesis of BD by disrupting the equilibrium between Th17 cells and regulatory T cells (Tregs). triggering an autoimmune response in BD patients (Zeidan et al., 2016). This is because Th17 cells cause autoimmunity and inflammation, whereas Treg cells inhibit these phenomena and maintain immune homeostasis (Lee 2018). In turn, we propose that lnc-DC is implicated in the pathogenesis of BD because increased lnc-DC was related to more active diseases and the presence of their manifestations.
The reduced lnc-DC in serum from patients in our study relative to the controls could be explained by the reduced number of DC cells present in the blood of patients with autoimmune diseases compared with that of healthy controls, while concurrently, the number of DCs in target tissues of individuals with autoimmune disease increases (Coutant and Miossec 2016). The aforementioned findings suggest that circulating DCs and, in turn, lnc-DC are reduced in the blood obtained from patients with autoimmune diseases. Another explanation for this finding is that the lncRNAs in serum could be fragmented because of their very long sequence lengths and so are unlikely to exist in a full-length form in the serum (Ren et al., 2013; Wang et al., 2015).
In recent publications’ results, lnc-DC levels were shown to be markedly downregulated in patients with RA and SLE (Li et al., 2017; Zhang et al., 2019) which is consistent with our results concerning BD patients. Meanwhile, Shaker et al. (2019) suggest that lnc-DC can be used as a biomarker for the diagnosis of multiple sclerosis (MS) since they observed a high level of lnc-DC in the serum of patients with MS. The discrepancy between upregulation vs. downregulation of lnc-DC in different autoimmune diseases is hard to definitively clarify as few studies examine the role(s) of lnc-DC in the pathogenesis of autoimmune diseases. The differences in lncRNA expression may be due to tissue specificity in the expression of some lncRNAs (Cabili et al., 2011).
Higher lnc-DC expression was associated with positive family history. Possible explanations for this relationship include that the familial accumulation of BD has been documented in 1–18% of the population with a higher incidence of familial association in juvenile patients (Zeidan et al., 2016) or that the loci transcribing novel lncRNAs co-localize with leukocyte transcriptional enhancers near genetic variants associated with autoimmune disease risk, Moreover, the modifications to enhance function, including lncRNA expression, produced by genetics and environmental stimulants can change cellular phenotypes contributing to disease susceptibility and pathogenesis (Aune et al., 2017).
A positive correlation between NEAT1 and lnc-DC enforces their synergistic effects. ROC curve analysis was performed to explore the diagnostic value of lncRNAs NEAT1 and lnc-DC as predictors for discrimination between BD patients and controls. At cut-off points of 1.07 and 0.66 for NEAT1 and lnc-DC, respectively, both biomarkers had an excellent specificity (100% for both) and fair sensitivity (69.2 and 61.5% for NEAT1 and lnc-DC, respectively). The AUC for NEAT1 and lnc-DC was 0.692 and 0.615, respectively.
The results presented here offer a potential new significance for lncRNA NEAT1 and lnc-DC in the prediction of BD. Future studies are required to verify our results and elucidate the involvement of NEAT1 and lnc-DC in the pathogenesis of BD. Such future studies are needed to overcome the limitations of the current study, which include 1) small sample size due to the rare pattern of the disease and 2) the lack of scientific literature illustrating the exact role or underlying molecular contributions of lncRNA generally and of NEAT1 and lnc-DC specifically in the pathogenesis, propagation, and prognosis of BD. Larger-scaled studies that explore the exact functions of these genes in BD pathogenesis are required.
Conclusion
We showed that NEAT1 and lnc-DC can be used as novel biomarkers for BD. In addition, their high transcript levels were associated with disease activity score and could be targets of therapy in patients with BD.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethical Committee of Faculty of Medicine, Fayoum University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.797689/full#supplementary-material
References
Adriaens, C., Standaert, L., Barra, J., Latil, M., Verfaillie, A., Kalev, P., et al. (2016). P53 Induces Formation of NEAT1 LncRNA-Containing Paraspeckles that Modulate Replication Stress Response and Chemosensitivity. Nat. Med. 22 (8), 861–868. doi:10.1038/nm.4135
Aune, T. M., Crooke, P. S., Patrick, A. E., Tossberg, J. T., Olsen, N. J., and Spurlock, C. F. (2017). Expression of Long Non-Coding RNAs in Autoimmunity and Linkage to Enhancer Function and Autoimmune Disease Risk Genetic Variants. J. Autoimmun. 81, 99–109. doi:10.1016/j.jaut.2017.03.014
Bhakta, B. B., Brennan, P., James, T. E., Chamberlain, M. A., Noble, B. A., and Silman, A. J. (1999). Behçet's disease: Evaluation of a New Instrument to Measure Clinical Activity. Rheumatology 38 (8), 728–733. doi:10.1093/rheumatology/38.8.728
Cabili, M. N., Trapnell, C., Goff, L., Koziol, M., Tazon-Vega, B., Regev, A., et al. (2011). Integrative Annotation of Human Large Intergenic Noncoding RNAs Reveals Global Properties and Specific Subclasses. Genes Dev. 25 (18), 1915–1927. doi:10.1101/gad.17446611
Coutant, F., and Miossec, P. (2016). Altered Dendritic Cell Functions in Autoimmune Diseases: Distinct and Overlapping Profiles. Nat. Rev. Rheumatol. 12 (12), 703–715. doi:10.1038/nrrheum.2016.147
Davatchi, F. (2012). Diagnosis/Classification Criteria for Behcet's Disease. Pathol. Res. Int. 2012, 607921. doi:10.1155/2012/607921
El Boghdady, N. A., and Shaker, O. G. (2019). Role of Serum miR-181b, Proinflammatory Cytokine, and Adhesion Molecules in Behçet's Disease. J. Interferon Cytokine Res. 39 (6), 347–354. doi:10.1089/jir.2018.0116
Huang, Q., Huang, C., Luo, Y., He, F., and Zhang, R. (2018). Circulating LncRNA NEAT1 Correlates with Increased Risk, Elevated Severity and Unfavorable Prognosis in Sepsis Patients. Am. J. Emerg. Med. 36 (9), 1659–1663. doi:10.1016/j.ajem.2018.06.008
Huang, S., Qian, K., Zhu, Y., Huang, Z., Luo, Q., and Qing, C. (2017). Diagnostic Value of the LncRNA NEAT1 in Peripheral Blood Mononuclear Cells of Patients with Sepsis. Dis. Markers 2017, 7962836. doi:10.1155/2017/7962836
Imamura, K., Imamachi, N., Akizuki, G., Kumakura, M., Kawaguchi, A., Nagata, K., et al. (2014). Long Noncoding RNA NEAT1-Dependent SFPQ Relocation from Promoter Region to Paraspeckle Mediates IL8 Expression upon Immune Stimuli. Mol. Cell 53 (3), 393–406. doi:10.1016/j.molcel.2014.01.009
INTERNATIONALSTUDYGROUPFORBEHC (1990). Criteria for Diagnosis of Behcet’s Disease. The Lancet 335 (8697). doi:10.1016/0140-6736(90)92643-V
Leccese, P., and Alpsoy, E. (2019). Behçet's Disease: An Overview of Etiopathogenesis. Front. Immunol. 10, 1067. doi:10.3389/fimmu.2019.01067
Lee, G. (2018). The Balance of Th17 versus Treg Cells in Autoimmunity. Int. J. Mol. Sci. 19, 730. doi:10.3390/ijms19030730
Li, J., Wu, G.-C., Zhang, T.-P., Yang, X.-K., Chen, S.-S., Li, L.-J., et al. (2017). Association of Long Noncoding RNAs Expression Levels and Their Gene Polymorphisms with Systemic Lupus Erythematosus. Sci. Rep. 7 (1). doi:10.1038/s41598-017-15156-4
Li, X., Ye, S., and Lu, Y. (2020). Long Non‐coding RNA NEAT1 Overexpression Associates with Increased Exacerbation Risk, Severity, and Inflammation, as Well as Decreased Lung Function through the Interaction with microRNA‐124 in Asthma. J. Clin. Lab. Anal. 34 (1). doi:10.1002/jcla.23023
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25 (4), 402–408. doi:10.1006/meth.2001.1262
Michelhaugh, S. K., Lipovich, L., Blythe, J., Jia, H., Kapatos, G., and Bannon, M. J. (2011). Mining Affymetrix Microarray Data for Long Non-Coding RNAs: Altered Expression in the Nucleus Accumbens of Heroin Abusers. J. Neurochem. 116 (3), 459–466. doi:10.1111/j.1471-4159.2010.07126.x
Nishimoto, Y., Nakagawa, S., Hirose, T., Okano, H., Takao, M., Shibata, S., et al. (2013). The Long Non-Coding RNA Nuclear-Enriched Abundant Transcript 1_2 Induces Paraspeckle Formation in the Motor Neuron during the Early Phase of Amyotrophic Lateral Sclerosis. Mol. Brain 6 (1), 31. doi:10.1186/1756-6606-6-31
Pandey, A. D., Goswami, S., Shukla, S., Das, S., Ghosal, S., Pal, M., et al. (2017). Correlation of Altered Expression of a Long Non-Coding RNA, NEAT1, in Peripheral Blood Mononuclear Cells with Dengue Disease Progression. J. Infect. 75 (6), 541–554. doi:10.1016/j.jinf.2017.09.016
Prinz, F., Kapeller, A., Pichler, M., and Klec, C. (2019). The Implications of the Long Non-Coding RNA NEAT1 in Non-Cancerous Diseases. Int. J. Mol. Sci. 20 (3), 627. doi:10.3390/ijms20030627
Ren, S., Wang, F., Shen, J., Sun, Y., Xu, W., Lu, J., et al. (2013). Long Non-Coding RNA Metastasis Associated in Lung Adenocarcinoma Transcript 1 Derived MiniRNA as a Novel Plasma-Based Biomarker for Diagnosing Prostate Cancer. Eur. J. Cancer 49 (13), 2949–2959. doi:10.1016/j.ejca.2013.04.026
Santoro, M., Nociti, V., Lucchini, M., de Fino, C., Losavio, F. A., and Mirabella, M. (2016). Expression Profile of Long Non-Coding RNAs in Serum of Patients with Multiple Sclerosis. J. Mol. Neurosci. 59 (1), 18–23. doi:10.1007/s12031-016-0741-8
Shaker, O. G., Ali, M. A., Ahmed, T. I., Zaki, O. M., Ali, D. Y., Hassan, E. A., et al. (2019). Association between LINC00657 andmiR‐106aserum Expression Levels and Susceptibility to Colorectal Cancer, Adenomatous Polyposis, and Ulcerative Colitis in Egyptian Population. IUBMB Life 71 (9), 1322–1335. doi:10.1002/iub.2039
Shaker, O. G., Mahmoud, R. H., Abdelaleem, O. O., Ahmed, T. I., Fouad, N. A., Hussein, H. A., et al. (2019a). Expression Profile of Long Noncoding RNAs, Lnc-Cox2, and HOTAIR in Rheumatoid Arthritis Patients. J. Interferon Cytokine Res. 39 (3), 174–180. doi:10.1089/jir.2018.0117
Shaker, O. G., Mahmoud, R. H., Abdelaleem, O. O., Ibrahem, E. G., Mohamed, A. A., Zaki, O. M., et al. (2019b). LncRNAs, MALAT1 and Lnc-DC as Potential Biomarkers for Multiple Sclerosis Diagnosis. Biosci. Rep. 39 (1). doi:10.1042/BSR20181335
Sigdel, K. R., Cheng, A., Wang, Y., Duan, L., and Zhang, Y. (20152015). The Emerging Functions of Long Noncoding RNA in Immune Cells: Autoimmune Diseases. J. Immunol. Res. 2015, 1–9. doi:10.1155/2015/848790
Wang, P., Xue, Y., Han, Y., Lin, L., Wu, C., Xu, S., et al. (2014). The STAT3-Binding Long Noncoding RNA Lnc-DC Controls Human Dendritic Cell Differentiation. Science 344 (6181), 310–313. doi:10.1126/science.1251456
Wang, Q., Wang, W., Zhang, F., Deng, Y., and Long, Z. (2017). NEAT1/miR‐181c Regulates Osteopontin (OPN)‐Mediated Synoviocyte Proliferation in Osteoarthritis. J. Cell. Biochem. 118 (11), 3775–3784. doi:10.1002/jcb.26025
Wang, X., Shen, C., Zhu, J., Shen, G., Li, Z., and Dong, J. (2019). Long Noncoding RNAs in the Regulation of Oxidative Stress. Oxidative Med. Cell Longevity 2019, 1318795. doi:10.1155/2019/1318795
Wang, Y., Li, Z., Zheng, S., Zhou, Y., Zhao, L., Ye, H., et al. (2015). Expression Profile of Long Non-Coding RNAs in Pancreatic Cancer and Their Clinical Significance as Biomarkers. Oncotarget 6 (34), 35684–35698. doi:10.18632/oncotarget.5533
Wei, J.-W., Huang, K., Yang, C., and Kang, C.-S. (2017). Non-Coding RNAs as Regulators in Epigenetics. Oncol. Rep. 37 (1), 3–9. doi:10.3892/or.2016.5236
Wu, G.-C., Li, J., Leng, R.-X., Li, X.-P., Li, X.-M., Wang, D.-G., et al. (2017). Identification of Long Non-Coding RNAs GAS5, Linc0597 and Lnc-DC in Plasma as Novel Biomarkers for Systemic Lupus Erythematosus. Oncotarget 8 (14), 23650–23663. doi:10.18632/oncotarget.15569
Zeidan, M. J., Saadoun, D., Garrido, M., Klatzmann, D., Six, A., and Cacoub, P. (2016). Behçet's Disease Physiopathology: a Contemporary Review. Autoimmun. Highlights 7 (1), 1–12. doi:10.1007/s13317-016-0074-1
Zhang, D., Song, L., and Wang, X. (2020a). NEAT1 Attenuates Osteoarthritis Development by Sponging miR-424-5p and Up-Regulating SMAD7 Expression. Res. Square. doi:10.21203/Rs.2.20874/V1
Zhang, F., Wu, L., Qian, J., Qu, B., Xia, S., La, T., et al. (2016). Identification of the Long Noncoding RNA NEAT1 as a Novel Inflammatory Regulator Acting through MAPK Pathway in Human Lupus. J. Autoimmun. 75, 96–104. doi:10.1016/j.jaut.2016.07.012
Zhang, P., Cao, L., Zhou, R., Yang, X., and Wu, M. (2019). The LncRNA Neat1 Promotes Activation of Inflammasomes in Macrophages. Nat. Commun. 10 (1), 1495. doi:10.1038/s41467-019-09482-6
Zhang, T.-P., Zhu, B.-Q., Tao, S.-S., Fan, Y.-G., Li, X.-M., Pan, H.-F., et al. (2019). Long Non-Coding RNAs Genes Polymorphisms and Their Expression Levels in Patients with Rheumatoid Arthritis. Front. Immunol. 10, 2529. doi:10.3389/fimmu.2019.02529
Zhang, W., Yang, M., Yu, L., Hu, Y., Deng, Y., Liu, Y., et al. (2020b). Long Non‐coding RNA Lnc‐DC in Dendritic Cells Regulates Trophoblast Invasion via p‐STAT3‐mediated TIMP/MMP Expression. Am. J. Reprod. Immunol. 83 (6). doi:10.1111/aji.13239
Keywords: Behcet’s disease, lnc-DC, NEAT1, RT-PCR, lncRNAs
Citation: Mohammed SR, Abdelaleem OO, Ahmed FA, Abdelaziz AA, Hussein HA, Eid HM, Kamal M, Ezzat MA and Ali MA (2022) Expression of lncRNAs NEAT1 and lnc-DC in Serum From Patients With Behçet’s Disease Can Be Used as Predictors of Disease. Front. Mol. Biosci. 8:797689. doi: 10.3389/fmolb.2021.797689
Received: 19 October 2021; Accepted: 06 December 2021;
Published: 19 January 2022.
Edited by:
Xin Zhang, Jiangmen Central Hospital, ChinaReviewed by:
Kamel Hamzaoui, Tunis El Manar University, TunisiaAlexandre Wagner Silva De Souza, Federal University of São Paulo, Brazil
Copyright © 2022 Mohammed, Abdelaleem, Ahmed, Abdelaziz, Hussein, Eid, Kamal, Ezzat and Ali. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marwa A. Ali, ZHJfbWFyd2FnQHlhaG9vLmNvbQ==, bWFnMDRARmF5b3VtLmVkdS5lZw==
†These authors have contributed equally to this work and share first authorship
 Shereen Rashad Mohammed1†
Shereen Rashad Mohammed1† Omayma O. Abdelaleem
Omayma O. Abdelaleem Marwa Kamal
Marwa Kamal Marwa A. Ali
Marwa A. Ali