- 1Department of Clinical Epidemiology, Jiangsu Province Geriatric Institute, Geriatric Hospital of Nanjing Medical University, Nanjing, China
- 2Department of Epidemiology, School of Public Health, Nanjing Medical University, Nanjing, China
- 3Department of Neurobiology, Nanjing Medical University, Nanjing, China
- 4Department of Medical Psychology, Huai’an Third Hospital, Huai’an, China
- 5Department of Medical Laboratory, Huai’an Third Hospital, Huai’an, China
Objective: Abnormal lipid metabolism has a close link to the pathophysiology of schizophrenia (SZ). This study mainly aimed to evaluate the association of variants at apolipoprotein A1 (APOA1) and APOA4 with SZ in a Chinese Han population.
Methods: The rs5072 of APOA1 and rs1268354 of APOA4 were examined in a case–control study involving 2,680 patients with SZ from the hospital and 2,223 healthy controls screened by physical examination from the community population. The association was estimated with the odds ratio (OR) and 95% confidence intervals (95% CIs) by logistic regression. The APOA1 and APOA4 messenger RNA (mRNA) in peripheral blood leukocytes were measured by real-time PCR and compared between SZ cases and controls. Serum apoA1 levels were detected by turbidimetric inhibition immunoassay and high-density lipoprotein cholesterol (HDL-C) levels were detected by the homogeneous method.
Results: Both of the rs5072 of APOA1 and rs1268354 of APOA4 had statistically significant associations with SZ. After adjustment for age and sex, ORs (95% CIs) of the additive model of rs5072 and rs1268354 were 0.82 (0.75–0.90) and 1.120 (1.03–1.23), and p-values were 3.22 × 10−5 and 0.011, respectively. The association of rs5072 with SZ still presented statistical significance even after Bonferroni correction (p-value×6). SZ patients during the episode presented lower levels of apoA1, HDL-C, mRNA of APOA1 common variants and transcript variant 4, and APOA4 mRNA than controls (p < 0.01) while SZ patients in remission showed a significantly decreased APOA1 transcript variant 3 expression level and increased APOA4 mRNA expression level (p < 0.01). mRNA expression levels of APOA1 transcript variant 4 significantly increased with the variations of rs5072 in SZ during the episode (ptrend = 0.017). After the SZ patients received an average of 27.50 ± 9.90 days of antipsychotic treatment, the median (interquartile) of serum apoA1 in the SZ episode significantly increased from 1.03 (1.00.1.20) g/L to 1.08 (1.00.1.22) g/L with the p-value of 0.044.
Conclusion: Our findings suggest that the genetic variations of APOA1 rs5072 and APOA4 rs1268354 contribute to the susceptibility of SZ, and the expression levels of APOA1 and APOA4 mRNA of peripheral blood leukocytes decreased in SZ patients during the episode while APOA4 increased after antipsychotic treatment.
Introduction
Schizophrenia (SZ) is a severe chronic psychiatric disorder characterized by distorted thinking processes, hallucinations, delusions, and functional deterioration (Barnett, 2018; Charlson et al., 2019). Lifetime risk for developing SZ, in general, is estimated to be approximately 1% (McGrath et al., 2008; Huang et al., 2019). The global age-standardized point prevalence of schizophrenia in 2016 was estimated to be 0.28% (95% UI: 0.24–0.31) (Charlson et al., 2018). This disorder accounted for 7.4% (5.0%–9.8%) of the fifth leading disorder in the disability-adjusted life year (DALY) (Collins et al., 2011). People suffering from schizophrenia often experience a decline in the quality of life, are prone to suicide, have an increased risk of comorbidities, and have a high mortality rate (Rossler et al., 2005; Tan et al., 2021). Due to its early age of onset and permanent disability, SZ invariably brings emotional and financial devastation to its victims and their families. Thus, SZ is considered to be one of the most catastrophic mental illnesses (Ayalew et al., 2012). However, to date, there is still no effective method available for the prevention and treatment of SZ.
SZ is a complex psychosis caused by genetic and environmental factors (Perzel Mandell et al., 2021; Robinson and Bergen, 2021). Previous studies suggest that abnormal lipid metabolism is closely related to the dysfunction of monoamine neurotransmitters (Lewis et al., 2010) and may contribute to the pathophysiology of SZ (La et al., 2007; Keeney et al., 2013; Song et al., 2014). Low levels and activity of ApoA1 might be a cause and consequence of immune-inflammatory and oxidative stress pathways in SZ patients (Morris et al., 2021). ApoA1 is the major apolipoprotein of HDL-C, which plays a key role in the process of cholesterol reverse transport, phospholipid efflux, and lipoprotein metabolism(Boiko et al., 2019). Several studies have shown that the decreased level of plasma, serum, or cerebrospinal fluid (CSF) apoA1is associated with a variety of neurodegenerative diseases, such as Alzheimer’s disease (AD), Parkinson’s disease (PD), and SZ (Liu et al., 2006; Huertas et al., 2017).
ApoA1 is encoded by the APOA1 gene (Saito et al., 2004). In the human body, the APOA1 gene is localized at chromosome 11q23 (11q23.3), which has been a popular locus for SZ research (Mack et al., 2017). Some studies have suggested that the abnormalities of chromosome 11q23 increase the risk for SZ (Gurling et al., 2001; Demirhan and Tastemir, 2003; Lewis et al., 2003).
Apolipoprotein A4 (apoA4), a component of lipoprotein particles, has been suggested to play an important role in brain metabolism. In 1977, apoA4 was first described to be a component of chylomicrons and HDL-C in the rat (Swaney et al., 1977) and detected in astrocytes of the rat. In fasting plasma, apoA4 is mainly found in HDL-C (Green et al., 1980). APOA4 is also localized at chromosome 11q23.3. In the meantime, it has been suggested that APOA1 and APOA4 genes, which are transcribed in the same direction, have an evolutionary relationship and come from the same ancestral sequence (Karathanasis, 1985).
Although genome-wide association studies (GWAS) of SZ have not identified susceptible loci linked to APOA1 and APOA4 yet, the aforementioned evidence suggests that APOA1 and APOA4 may be involved in SZ pathophysiology whereas the molecular mechanism is still unclear. Therefore, this study aimed to investigate the association of variants at APOA1 and APOA4 with SZ, compare APOA1 and APOA4 mRNA expression levels in peripheral blood leukocytes between SZ cases and controls, and evaluate the influence of APOA1 and APOA4 variations on the mRNA expression level.
Methods
Study Population
A total of 2,680 patients with SZ (1,229 men and 1,451 women) were consecutively recruited from the Third People’s Hospital of Huai’an, Jiangsu Province, China, from the year 2009–2018. All patients were diagnosed with SZ according to the Diagnostic and Statistical Manual fourth edition (DSM-IV), and were diagnosed by at least two experienced psychiatrists. A total of 2,223 healthy controls (including 1,365 men and 858 women) screened by the physical examination were recruited from the local community, and those who had a personal or family history of psychiatric disorders were excluded. All participants were free of cardiovascular diseases, cerebral vascular diseases, serious infections, and surgery. The controls had a higher average age [(43.71 ± 8.79) years] than SZ cases [(34.69 ± 13.00) years]. Thus, the controls would have less risk to develop into SZ, and that would help to enrich the chance to find susceptible genetic loci.
This study was done in full compliance with the Helsinki Declaration. The protocol and consent form were approved by the Institutional Review Board of Nanjing Medical University, China. All participants or their guardians provided signed informed consents.
Data Collection
Questionnaire interviews were conducted to collect information on participants’ age, sex, nationality, history of chronic diseases, and family status.
Total cholesterol (TC), low-density lipoprotein cholesterol (LDL-C), apoA1, high-density lipoprotein cholesterol (HDL-C), triglycerides (TG), and glucose (GLU) were detected by Total Cholesterol Kit (CHOD-PAP Method), Low-Density Lipoprotein Cholesterol Kit (Clearance Method), Turbidimetric inhibition immunoassay, High-Density Lipoprotein Cholesterol Kit (Clearance Method), enzymatic method, and Glucose oxidase, respectively. Total bilirubin (TBIL) and direct bilirubin (DBIL) were examined by the vanadate oxidation method. Indirect bilirubin (IBIL) is the difference between total bilirubin (TBIL) and direct bilirubin (DBIL). All biochemistry indices were detected in the Third People’s Hospital of Huai’an.
SNP Selection
APOA1 [GenBank ID:335 also known as apo(a)] and APOA4 (GeneBank ID:337), encoding apoA1 and apoA4 protein, respectively, were both mapped on chromosome 11p23.3. We searched for seven SNPs (rs5072, rs2070665, rs10750098, rs632153, rs12718462, and rs12718464 of APOA1, and rs1268354 of APOA4) covering the APOA1 and APOA4 from the upstream 2 kb to the downstream 1 kb, and further selected tag SNPs (tagSNPs) from the database of the Genome Variation Server 147 (GVS) (http://gvs.gs.washington.edu/GVS147/).
SNPs with minor allele frequency (MAF) > 0.05 will be included as candidate SNPs, and tagSNPs were selected for the criterion of linkage disequilibrium (LD) r2 ≥ 0.8 in the GVS. Furthermore, the role of transcription, regulating, or splicing was evaluated on the website of selection tool for SNPs (SNPINFO, https://snpinfo.niehs.nih.gov/) and Regulome (http://www.regulomedb.org.). Thus, the SNPs with predicted biological functions of rs5072of APOA1 (TF binding and DNase peak) and rs1268354 of APOA4 (eQTL, TF binding and DNase peak) were preferentially included in this study. The biological information analysis for regions, alleles, MAFs, predicted functions, and Regulome DB score of the seven SNPs are shown in Supplementary Table S1. Primers and probes of rs5072 and rs1268354 are summarized in Supplementary Table S2.
DNA Isolating and Genotyping
Genomic DNA was isolated from leukocytes of venous blood by proteinase K digestion and phenol-chloroform extraction. The primers and probes of TaqMan® assays were designed using Primer Express Oligo Design software v2.0 (ABI PRISM), and available upon request as TaqMan® Pre-Designed SNP Genotyping Assays. From randomly selected samples, 5% were genotyped again for quality control with complete concordance. PCR reactions and conditions are described at the Supplementary Information Genotyping PCR reactions and conditions.
Determination of APOA1 and APOA4 mRNA in Peripheral Blood Leukocytes
A total of 61 patients with SZ during the acute episode lack systemic and effective anti-psychotic treatment, 64 patients with SZ in remission after a period of antipsychotic treatment and identified by clinicians, and 84 controls were selected to detect the expression level of APOA1 and APOA4 mRNAs. Fasting blood samples over 12 h of SZ patients on admission and on discharge were collected. Healthy controls were recruited from the local community population, and fasting blood samples over 12 h were collected. EDTA-containing blood sample was mixed with blood preservation solution (Eaglink Cat#EGEN2024, NANJING YININGFUSHENG Biotech. Co., Ltd. Nanjing, China) by 1:3. The total RNA was isolated from an 800-µl mixture using RNA Blood Kit (Cat#Yu-B02-1, Yuan Corp. Wuxi, China) according to the manual instructions. RNA quality and integrity were evaluated using electrophoresis on a 1.0% agarose gel and agarose gel and NanoDrop® ND ND-2000 spectrophotometer (NanoDrop, Wilmington, DE), respectively. cDNA was synthesized using TAKARA reverse transcription (RT) kits (RR047A Takara PrimeScript™ RT reagent Kit with gDNA Eraser, Japan). The quantitative real-time PCR (qPCR) reactions were performed in a 10-μl reaction mixture including 2 μl of cDNA and 5 μl of HieffTM qPCR SYBR® GEEN Master Mix. Primer sequences of APOA1, APOA4, and GAPDH (control) cDNAs are listed in Supplementary Table S3. The qPCR instrument used in this study was ABI RT-PCR 7900 (Applied Biosystems; Thermo Fisher Scientific, Inc.). Three parallel samples were set for each sample. The CT value results were read in the SDS2.4 software. The standard deviation of CT values between three parallel samples was less than 0.5, and finally we took the average number. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control, and that could address blood sample-associated RNA quality issues in qPCR. The CT values among different plates had good and stable expressions. All the samples of SZ patients during the episode and in remission, and all control samples were arranged in one 384 plate to control variations. The relative expression of mRNA was calculated by 2-∆∆CT (∆∆CT case = ∆CT case - ∆CT control average value, ∆∆CT control = ∆CT control - ∆CT control average value, ∆CT = CT target gene - CT housekeeper gene). All the reverse transcription and qPCR reactions, as well as conditions, are described at the Supplementary Information Reverse transcription reactions and conditions, and qPCR reactions and conditions.
Statistical Analysis
Quantitative variables with normal distribution were expressed as mean ± standard deviation, and differences between cases and controls were assessed with unpaired Student’s t-test. ApoA1 concentration did not follow the normal distribution and thus was expressed as median (interquartile, IRQ). The differences of mRNA expression, apoA1, and HDL-C between SZ cases and controls, as well as among genotypes, were assessed with Mann–Whitney U test and Jonckheere–Terpstra test, respectively. Comparison of apoA1 and HDL-C levels before and after antipsychotic treatment was assessed by Wilcoxon test. Partial correlation between apoA1, HDL-C, and mRNA expression levels was estimated with adjustment for age and sex. Qualitative variables, the genotype, and allele frequency distributions between cases and controls were compared via the two-sided chi-square (χ2) test. Among controls, genotype frequencies for each SNP were tested by Fisher’s exact χ2 test using the program Hardy–Weinberg equilibrium (HWE). The association between loci and SZ was estimated by logistic regression. Odds ratios (OR) and 95% confidence intervals (CIs) were calculated. Then, we conducted a propensity matching analysis. We matched the SZ cases and controls by propensity score matching model of age and sex. Bonferroni correction was used for correcting the p-values for multiple comparisons. Statistical significance was set at two-tailed p < 0.05. All statistical analyses were performed with SPSS 18.0 (SPSS, Inc. Chicago, United States).
Results
Clinical Characteristics of Study Population
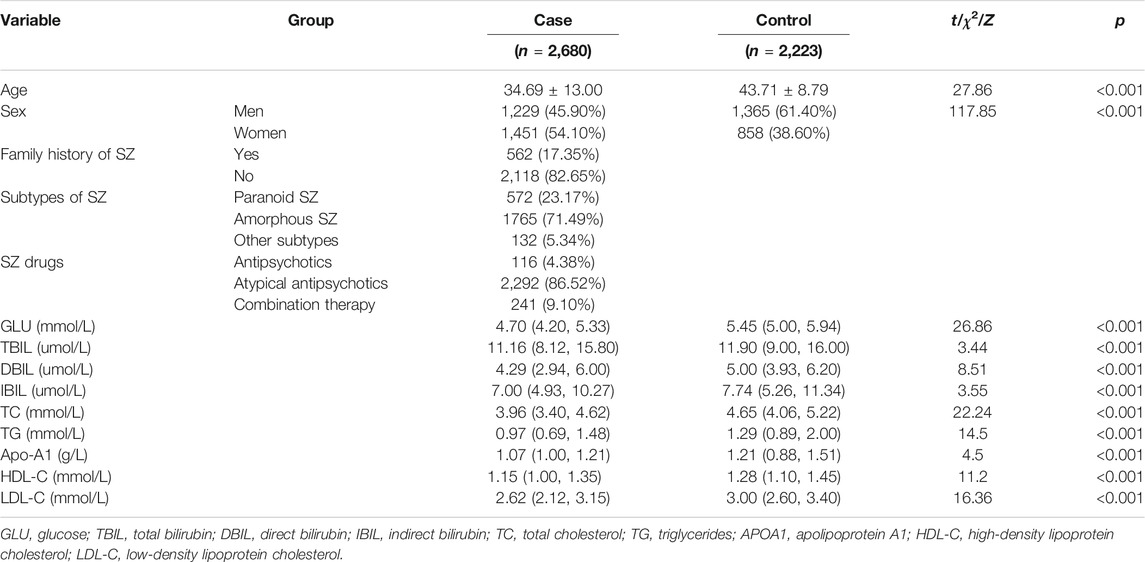
The general demographic characteristics of participants are summarized in Table 1. There were significant differences in age between SZ patients and controls (t = 27.86, p < 0.001). Sex differed significantly between the two groups (χ2 = 117.85, p < 0.001). A total of 562 SZ patients (17.35%) had a family history of SZ and 2,118 patients (82.65%) did not. Among SZ patients, 116 (4.38%) patients were mainly treated with antipsychotics, 2,292 (86.52%) patients were treated with atypical antipsychotics, and 241 (9.10%) patients were treated with the combination of antipsychotics and atypical antipsychotics. All the biochemical indices were statistically lower in the cases than in the controls (p < 0.005).
The clinical characteristics of the participants detected mRNA are summarized in Supplementary Table S4. There were significant differences in age among SZ patients during the episode [(37.25 ± 13.39) years], patients in remission [(31.11 ± 7.82) years], and controls [(34.61 ± 10.87) years] (F = 5.01, p = 0.01). In addition, there were significant differences in GLU, TBIL, DBIL, IBIL, TG, and HDL-C among the three groups (all p < 0.01). There were no significant differences in sex, TC, Apo-A1, and LDL-C among these three groups (all p > 0.05).
Association Analysis of Case–Control Study for SZ
The genotype and allele frequency distributions of rs5072 of APOA1 and rs1268354 of APOA4 between SZ cases and controls are summarized in Supplementary Table S5 in the Supplement. The allele frequency distributions of the two SNPs were consistent with the HWE (p > 0.1) in the controls.
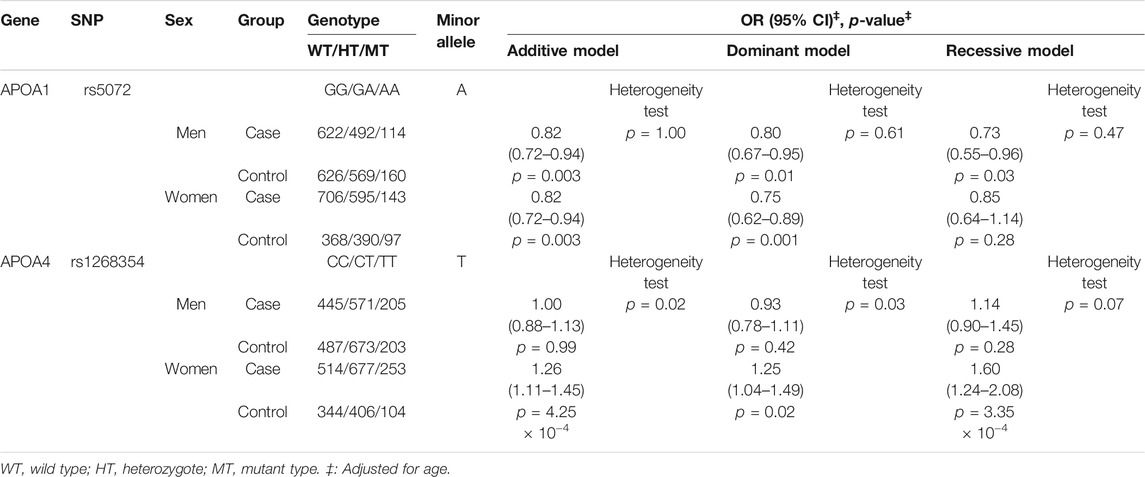
Both rs5072 of APOA1 and rs1268354 of APOA4 were significantly associated with SZ even after adjustment for age and sex. The adjusted ORs (95% CIs) of the additive model for rs5072 (GG vs. GA vs. AA) and rs1268354 (CC vs. CT vs. TT) were 0.82 (0.75–0.90) and 1.12 (1.03–1.23), respectively, and p-values were 3.22 × 10−5 and 0.011. The associations of rs5072 with SZ still presented statistical significance even after Bonferroni correction (p-value×6).
Sensitivity Analysis by Propensity Score Matching Model
We conducted a sensitivity analysis using a propensity score matching model to match SZ cases and controls by age and sex. There were no difference in average age and sex between SZ case and controls (p > 0.30). The serum levels of Glu, DBIL, TC, TG, ApoA1, HDL-C, and LDL-C were significantly lower in SZ cases than those of controls with all p values less than 0.01 (Supplementary Table S6).
Both rs5072 of APOA1 and rs1268354 of APOA4 were significantly associated with SZ even after adjustment for age and sex. The adjusted OR (95% CI) of the additive model for rs5072 was 0.84 (0.75–0.94) with p-values of 0.003. The association of rs5072 of APOA1 and SZ still presented statistical significance even after Bonferroni correction (p-value×6). OR (95% CI) of the recessive model (TT vs. CT + CC) for rs1268354 was 1.31 (1.07–1.61) with a p value of 0.010. The OR of the subjects selected by the propensity score matching model was not different from that of the whole study population (less than 3%) (Supplementary Table S7).
Stratification Analysis by Sex
Further stratified analysis by sex indicated that rs5072 was significantly associated with SZ both in men and women, and the association was homogeneous. rs1268354 of APOA4 was significantly associated with SZ in women but not in men with p-value of heterogeneity test less than 0.05. In women, OR (95% CI) of the additive model for rs1268354 was 1.26 (1.11–1.45) with a p-value of 4.25 × 10−4, after adjustment for age (Table 2).
Comparing mRNA Levels Between SZ Cases and Controls
The medians (IRQs) of common variants and transcript variant 4 levels (2-∆∆CT) of APOA1 mRNA in SZ patients during the episode (0.57 [0.31.0.99]; 0.73 [0.38, 1.15]) were significantly lower than those of controls (1.07 [0.62.1.80]; 0.93 [0.66.1.34]); both the p-values were less than 0.01 (Figure 1). The median (IRQ) of common variants level of APOA4 mRNA was significantly lower in SZ patients during the episode [0.16 (0.08.1.29)] than controls [1.34 (0.33.3.63)]; p-value was less than 0.001.
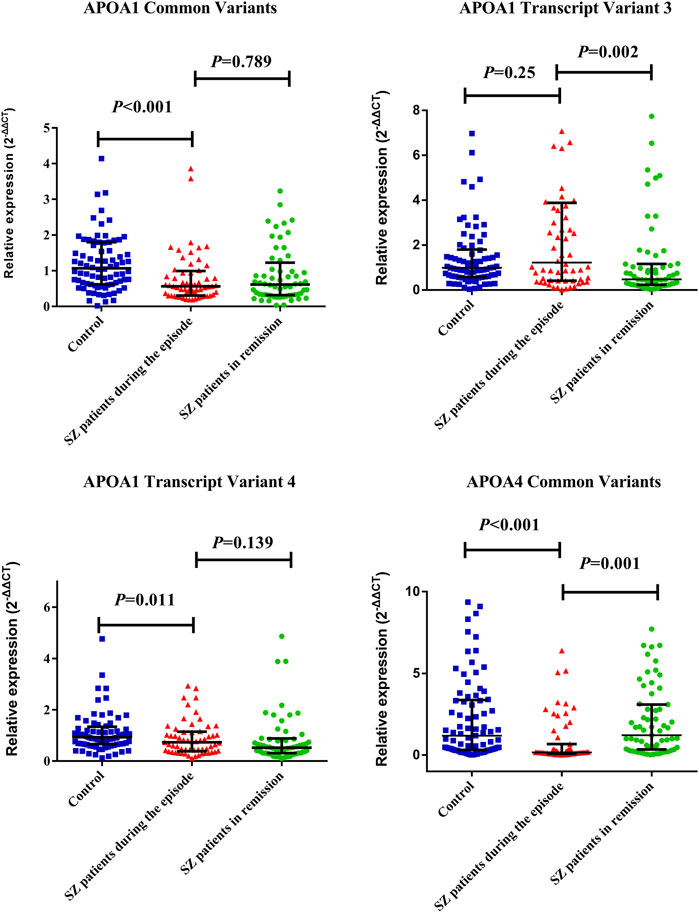
FIGURE 1. Distribution of APOA1 common transcript variants and transcript variant 4 of APOA1 among SZ patients during the episode, patients in remission, and controls. The dots represent individual data of mRNA levels (2−ΔΔCT). The upper line and lower line represent 75th percentage and 25th percentage, respectively, and the medium line represents the median. The medians (IRQs) of common variants and transcript variant 4 levels (2-∆∆CT) of APOA1 mRNA in SZ during the episode (0.57 [0.31.0.99] (0.73 [0.38, 1.15]) were significantly lower than that of controls (1.07 [0.62.1.80] (0.93 [0.66.1.34]). Both the p-values were less than 0.05. SZ patients in remission showed a significantly decreased APOA1 transcript variant 3 expression level (0.47 [0.24.1.16]) and increased APOA4 mRNA expression level (1.35 [0.35.3.12]) than patients during the episode as above (p < 0.01).
SZ patients in remission showed a significantly decreased APOA1 transcript variant 3 expression level (0.47 [0.24.1.16]) and increased APOA4 mRNA expression level (1.35 [0.35.3.12]) than patients during the episode as above (p < 0.01). The data are listed in Supplementary Table S8.
Comparing mRNA Expression Levels Among Genotypes
The distribution of APOA1 gene relative expression levels (2-∆∆CT) among the genotypes of rs5072 and rs1268354 is summarized in Supplementary Table S9 and Supplementary Table S10. The transcript variant 4 expression levels of APOA1 mRNA significantly increased with the variations of rs5072 in SZ patients during the episode (ptrend = 0.017) but not in controls; the results are shown in Supplementary Table S9 in the Supplement.
Partial Correlation Analysis of Serum apoA1, HDL-C, and mRNA Levels
Partial correlation analysis showed that apoA1 level was significantly correlated with HDL-C level in the controls (r = 0.89), SZ patients during the episode (r = 0.82), and patients in remission (r = 0.86) (all the p-values less than 0.001) after adjusting age and sex. However, mRNA levels of APOA4 showed no correlation with APOA1 in SZ cases and controls (Supplementary Table S11). In controls, APOA1 transcript 3 was significantly correlated with apoA1 and HDL-C levels (r = −0.37, p = 0.001; r = −0.30, p = 0.006) and the correlations were found in women (r = −0.46, p < 0.001; r = −0.40, p = 0.006) but not in men (Supplementary Table S11), whereas the correlations were not observed in SZ cases.
However, no significant correlation of APOA1 mRNA and APOA4 mRNA was observed in SZ cases and controls (Supplementary Table S12).
Comparing ApoA1 and HDL-C Levels Between SZ Cases and Controls
The serum apoA1 [median (IQR)] was significantly lower in SZ patients during the episode [1.07 (1.00.1.21) g/L] than controls [1.21 (0.88.1.51) g/L], Z = 4.04, p < 0.001 (Figure 2A). Similarly, HDL-C level in SZ patients during the episode [1.14 (0.99.1.34) mmol/L] was lower than controls [1.28 (1.10.1.45) mmol/L], Z = 9.02, p < 0.001 (Figure 2B). All data are listed in Supplementary Table S12 in detail.

FIGURE 2. Distribution of apoA1 (A) and HDL-C (B) levels between SZ patients during the episode and controls. The dots represent individual data of apoA1 and HDL-C levels. The upper line and lower line represent 75th percentage and 25th percentage respectively, and the medium line represents the median. The serum apoA1 [median (IQR)] was significantly lower in SZ patients during the episode [1.07 (1.00.1.21) g/L] than controls [1.21 (0.88.1.51) g/L], Z = 4.04, p < 0.001 (Figure 2A). Similarly, HDL-C level in SZ during the episode [1.14 (0.99.1.34) mmol/L] was lower than controls [1.28 (1.10.1.45) mmol/L], Z = 9.02, p < 0.001 (Figure 2B).
Comparison of ApoA1 and HDL-C Levels Before and After Antipsychotic Treatment
The apoA1 levels of 209 SZ patients during the episode [1.03 (1.00.1.20) g/L] significantly increased after receiving an average 27.50 ± 9.90 days antipsychotic treatment [1.08 (1.00.1.22) g/L]; p-value was 0.04 (Figure 3). However, the serum HDL-C level was not varied after treatment (p > 0.05). All the data are shown in Supplementary Table S13.
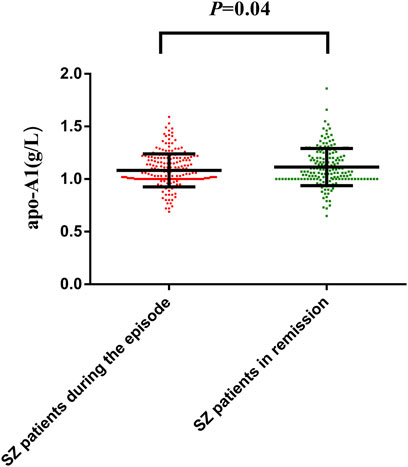
FIGURE 3. Distribution of apoA1 levels in SZ patients during the episode and patients in remission with antipsychotic treatment. The dots represent individual data of apoA1. The upper line and lower line represent 75th percentage and 25th percentage, respectively, and the medium line represents the median. Comparison of apoA1 and HDL-C levels before and after antipsychotic treatment was assessed by Wilcoxon test. The apoA1 levels of 209 SZ patients during the episode [1.03 (1.00.1.20) g/L] significantly increased after receiving an average 27.50 ± 9.90 days antipsychotic treatment [1.08 (1.00.1.22) g/L], p-value was 0.04 (Figure 3).
Comparison of Serum ApoA1 and HDL-C Levels Among the Genotypes
In SZ patients during the episode, serum HDL-C levels [median (IRQ)] showed significant difference among the genotypes of rs5072 GG [1.10 (0.97, 1.33) mmol/L], GA [1.20 (1.00.1.35) mmol/L], and AA [1.10 (0.99.1.27) mmol/L], p < 0.05 (Supplementary Table S14). There was no significant difference in serum apoA1 level among the genotypes of rs5072 and rs1268354 (Supplementary Table S15) in SZ cases or controls.
Discussion
In this study, we reported for the first time that variants at rs5072 of APOA1 and rs1268354 of APOA4 were significantly associated with SZ in a Chinese Han population. It is remarkable that the expression of APOA1 mRNA was significantly down-regulated in peripheral blood leukocytes in the SZ patients during the episode.
APOA1 gene encodes the apoA1 protein that belongs to the apolipoprotein A1/A4/E family (Wang et al., 2009). Rs5072 (G/A) is located in the intron region and is predicted to affect the binding of the transcription factor c-Myb (proximal promoter DNA-binding transcription activator activity), which regulates the expression of APOA1 (http://alggen.lsi.upc.es/, last accessed September 1, 2018). The results of this study indicated that mRNA expression levels of APOA1 transcript variant 4 significantly increased with the variations of rs5072 in SZ cases during the episode. Therefore, further functional research on the role of rs5072 and c-Myb in the regulation of mRNA expression would be warranted.
In this study, we observed lower serum apoA1 levels in SZ patients during the episode, compared with controls. This result is consistent with previous reports that apoA1 level decreased in the cerebrospinal fluid (CSF), liver, peripheral red blood cells, serum, and plasma in SZ patients (La et al., 2007; Huang et al., 2008). These findings suggest that the decreased apoA1 is involved in the pathology of SZ. It is well known that apoA1 is the primary core component required for normal HDL-C synthesis. The result in the present study indicated that apoA1 level is significantly correlated with HDL-C level. Moreover, we found that HDL-C levels were also lower in SZ patients during the episode. Antipsychotic agents can increase apoA1 levels as part of their therapeutic actions (Martins-De-Souza et al., 2010). In this study, the serum apoA1 levels of SZ patients during the episode significantly increased with the atypical antipsychotic treatment for a period of time, suggesting the potential adverse effect of antipsychotic treatment on the apoA1 levels. Since serum apoA1 varied with APOA1 mRNA expression and clinic antipsychotic treatment, serum apoA1 would be suggested to detect and evaluate SZ prognosis besides HDL-C detection.
ApoA1 and HDL-C were mainly synthesized in the liver. The evidence of this study indicated that the APOA1 mRNA expression in peripheral blood leukocytes has a population-based association with serum the apoA1 and HDL-C, suggesting a possible regulatory mechanism involved in apolipoprotein synthesis in the liver. On the other hand, serum lipid may be independent of the brain, but an unfavorable circulating lipid profile may cause hyperlipidemia and subtle changes in brain lipid metabolism (Salter, 1992). The exploration of the molecular mechanism of SZ supported the role of glutamate, γ-aminobutyric acid (GABA) and 5-hydroxytryptamine (5-HT) (Meyer and MacCabe, 2012; Owen et al., 2016). Thus, the relationship between dyslipidemia and dysfunction of neurotransmitters also received widespread attention (Salter, 1992; Terao et al., 1997). The impact of ApoA1 in the process of cholesterol efflux(Remaley et al., 2001; Zeng et al., 2004; Nofer et al., 2006; Ji et al., 2011) and phospholipid efflux (Remaley et al., 2001) may affect the dysfunction of 5-HT involved in the pathology of SZ.
The genetic ablation of APOA4 may accelerate AD pathogenesis in an APP/PS1 transgenic mouse model (Cui et al., 2011). Csazar et al. reported that the APOA4 codon 360 mutation (C > T) is associated with the increased risk of AD patients (Csaszar et al., 1997; Deng et al., 2015). This study found a significant association of rs1268354 C to T variation with SZ, particularly in women. However, no significant association of the levels of APOA4 mRNA and the variations of rs1268354 was observed. rs1268354 does not have a linkage locus nearby APOA1, and the largest r2 (with rs10750098) is 0.474. Thus, further study on epigenetics would be warranted to investigate the role of APOA4 genetic variation in the pathogenesis of SZ.
This study has several potential limitations. First, after adjustment for age and sex, potential confounding effect remains to influence the association because of the differences in sex and age between SZ cases and controls. Nevertheless, the sensitivity analysis by propensity matching for age and sex validated the main significance (OR alterations less than 3%). Thus, we would accept the main findings of this study as well as the rationality of the primary study design for the SZ genetic association study. Second, the SZ patients during the episode with mRNA expression data were different from those in remission, so the selection bias is inevitable. Third, we failed to design an allele-specific primer for transcript 1 and transcript 2 of APOA1 mRNA. Therefore, the possible effect of missing transcripts on SZ needs to be verified. Last, the effects of antipsychotic treatment and disease duration on mRNA expression warrant further investigation.
Conclusion
In conclusion, the genetic variations of APOA1 rs5072 and APOA4 rs1268354 were significantly associated with SZ, and the expression levels of APOA1 and APOA4 mRNAs in peripheral blood leukocytes decreased in SZ patients during the episode while APOA1 transcript variant 3 decreased and APOA4 increased after antipsychotic treatment. These findings support the contribution of the genetic variations of APOA1 and APOA4 to the susceptibility to SZ with inhibiting mRNA expression in peripheral blood leukocytes. Further research on the role of apoA1 and apoA4 metabolism in molecular mechanism SZ is still warranted.
Data Availability Statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by the Institutional Review Board of Nanjing Medical University, China. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
CS and YX designed the study; YF, WY, and TZ performed the experiments; YL and XC collected the serum samples; YF and TZ analyzed and interpreted the data; YF wrote the manuscript; JG and CS edited the manuscript. All authors commented and approved the final manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (Grant Nos. 81872686 and 81573232). Jiangsu Provincial Fourth “333 Project”, and the Priority Academic Program for the Development of Jiangsu Higher Education Institutions (Public Health and Preventive Medicine), Clinical Research Project of Huai’an Technology Project Bureau (Grant No. HAS2011023). The funders had no role in study design, data collection, data analysis, decision to publish, and preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Hankun Xie and Solim Essomandan Clemence Bafei for helping to edit the English language.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.785445/full#supplementary-material
References
Ayalew, M., Le-Niculescu, H., Levey, D. F., Jain, N., Changala, B., Patel, S. D., et al. (2012). Convergent Functional Genomics of Schizophrenia: from Comprehensive Understanding to Genetic Risk Prediction. Mol. Psychiatry 17, 887–905. doi:10.1038/mp.2012.37
Boiko, A. S., Mednova, I. A., Kornetova, E. G., Semke, A. V., Bokhan, N. A., Loonen, A. J. M., et al. (2019). Apolipoprotein Serum Levels Related to Metabolic Syndrome in Patients with Schizophrenia. Heliyon 5, e02033. doi:10.1016/j.heliyon.2019.e02033
Charlson, F. J., Ferrari, A. J., Santomauro, D. F., Diminic, S., Stockings, E., Scott, J. G., et al. (2018). Global Epidemiology and Burden of Schizophrenia: Findings from the Global Burden of Disease Study 2016. Schizophr. Bull. 44, 1195–1203. doi:10.1093/schbul/sby058
Charlson, F., Van Ommeren, M., Flaxman, A., Cornett, J., Whiteford, H., and Saxena, S. (2019). New WHO Prevalence Estimates of Mental Disorders in Conflict Settings: a Systematic Review and Meta-Analysis. Lancet 394, 240–248. doi:10.1016/s0140-6736(19)30934-1
Collins, P. Y., Patel, V., Joestl, S. S., March, D., Insel, T. R., Daar, A. S., et al. (2011). Grand Challenges in Global Mental Health. Nature 475, 27–30. doi:10.1038/475027a
Császár, A., Kálmán, J., Szalai, C., Janka, Z., and Romics, L. (1997). Association of the Apolipoprotein A-IV Codon 360 Mutation in Patients with Alzheimer's Disease. Neurosci. Lett. 230, 151–154. doi:10.1016/s0304-3940(97)00500-4
Cui, Y., Huang, M., He, Y., Zhang, S., and Luo, Y. (2011). Genetic Ablation of Apolipoprotein A-IV Accelerates Alzheimer's Disease Pathogenesis in a Mouse Model. Am. J. Pathol. 178, 1298–1308. doi:10.1016/j.ajpath.2010.11.057
Demirhan, O., and Taştemir, D. (2003). Chromosome Aberrations in a Schizophrenia Population. Schizophr. Res. 65, 1–7. doi:10.1016/s0920-9964(02)00504-2
Deng, X., Walker, R. G., Morris, J., Davidson, W. S., and Thompson, T. B. (2015). Role of Conserved Proline Residues in Human Apolipoprotein A-IV Structure and Function. J. Biol. Chem. 290, 10689–10702. doi:10.1074/jbc.m115.637058
Green, P. H. R., Glickman, R. M., Riley, J. W., and Quinet, E. (1980). Human Apolipoprotein A-IV. J. Clin. Invest. 65, 911–919. doi:10.1172/jci109745
Gurling, H. M. D., Kalsi, G., Brynjolfson, J., Sigmundsson, T., Sherrington, R., Mankoo, B. S., et al. (2001). Genomewide Genetic Linkage Analysis Confirms the Presence of Susceptibility Loci for Schizophrenia, on Chromosomes 1q32.2, 5q33.2, and 8p21-22 and Provides Support for Linkage to Schizophrenia, on Chromosomes 11q23.3-24 and 20q12.1-11.23. Am. J. Hum. Genet. 68, 661–673. doi:10.1086/318788
Huang, J. T.-J., Wang, L., Prabakaran, S., Wengenroth, M., Lockstone, H. E., Koethe, D., et al. (2008). Independent Protein-Profiling Studies Show a Decrease in Apolipoprotein A1 Levels in Schizophrenia CSF, Brain and Peripheral Tissues. Mol. Psychiatry 13, 1118–1128. doi:10.1038/sj.mp.4002108
Huang, Y., Wang, Y., Wang, H., Liu, Z., Yu, X., Yan, J., et al. (2019). Prevalence of Mental Disorders in China: a Cross-Sectional Epidemiological Study. Lancet Psychiatry 6, 211–224. doi:10.1016/s2215-0366(18)30511-x
Huertas, I., Jesús, S., García-Gómez, F. J., Lojo, J. A., Bernal-Bernal, I., Bonilla-Toribio, M., et al. (2017). Genetic Factors Influencing Frontostriatal Dysfunction and the Development of Dementia in Parkinson's Disease. PLoS One 12, e0175560. doi:10.1371/journal.pone.0175560
Ji, A., Meyer, J. M., Cai, L., Akinmusire, A., De Beer, M. C., Webb, N. R., et al. (2011). Scavenger Receptor SR-BI in Macrophage Lipid Metabolism. Atherosclerosis 217, 106–112. doi:10.1016/j.atherosclerosis.2011.03.017
Karathanasis, S. K. (1985). Apolipoprotein Multigene Family: Tandem Organization of Human Apolipoprotein AI, CIII, and AIV Genes. Proc. Natl. Acad. Sci. 82, 6374–6378. doi:10.1073/pnas.82.19.6374
Keeney, J. T. R., Swomley, A. M., Förster, S., Harris, J. L., Sultana, R., and Butterfield, D. A. (2013). Apolipoprotein A-I: Insights from Redox Proteomics for its Role in Neurodegeneration. Proteomices. Clin. Appl. 7, 109–122. doi:10.1002/prca.201200087
La, Y. J., Wan, C. L., Zhu, H., Yang, Y. F., Chen, Y. S., Pan, Y. X., et al. (2007). Decreased Levels of Apolipoprotein A-I in Plasma of Schizophrenic Patients. J. Neural Transm. 114, 657–663. doi:10.1007/s00702-006-0607-2
Lewis, C. M., Levinson, D. F., Wise, L. H., Delisi, L. E., Straub, R. E., Hovatta, I., et al. (2003). Genome Scan Meta-Analysis of Schizophrenia and Bipolar Disorder, Part II: Schizophrenia. Am. J. Hum. Genet. 73, 34–48. doi:10.1086/376549
Lewis, T. L., Cao, D., Lu, H., Mans, R. A., Su, Y. R., Jungbauer, L., et al. (2010). Overexpression of Human Apolipoprotein A-I Preserves Cognitive Function and Attenuates Neuroinflammation and Cerebral Amyloid Angiopathy in a Mouse Model of Alzheimer Disease. J. Biol. Chem. 285, 36958–36968. doi:10.1074/jbc.m110.127829
Liu, H.-C., Hu, C.-J., Chang, J.-G., Sung, S.-M., Lee, L.-S., Yuan, R.-Y., et al. (2006). Proteomic Identification of Lower Apolipoprotein A-I in Alzheimer's Disease. Dement. Geriatr. Cogn. Disord. 21, 155–161. doi:10.1159/000090676
Mack, S., Coassin, S., Rueedi, R., Yousri, N. A., Seppälä, I., Gieger, C., et al. (2017). A Genome-wide Association Meta-Analysis on Lipoprotein (A) Concentrations Adjusted for Apolipoprotein (A) Isoforms. J. Lipid Res. 58, 1834–1844. doi:10.1194/jlr.m076232
Martins-De-Souza, D., Wobrock, T., Zerr, I., Schmitt, A., Gawinecka, J., Schneider-Axmann, T., et al. (2010). Different Apolipoprotein E, Apolipoprotein A1 and Prostaglandin-H2 D-Isomerase Levels in Cerebrospinal Fluid of Schizophrenia Patients and Healthy Controls. World J. Biol. Psychiatry 11, 719–728. doi:10.3109/15622971003758748
McGrath, J., Saha, S., Chant, D., and Welham, J. (2008). Schizophrenia: a Concise Overview of Incidence, Prevalence, and Mortality. Epidemiol. Rev. 30, 67–76. doi:10.1093/epirev/mxn001
Meyer, N., and MacCabe, J. H. (2012). Schizophrenia. Medicine 40, 586–590. doi:10.1016/j.mpmed.2012.08.002
Morris, G., Puri, B. K., Bortolasci, C. C., Carvalho, A., Berk, M., Walder, K., et al. (2021). The Role of High-Density Lipoprotein Cholesterol, Apolipoprotein A and Paraoxonase-1 in the Pathophysiology of Neuroprogressive Disorders. Neurosci. Biobehavior. Rev. 125, 244–263. doi:10.1016/j.neubiorev.2021.02.037
Nofer, J.-R., Remaley, A. T., Feuerborn, R., Wolinnéska, I., Engel, T., Von Eckardstein, A., et al. (2006). Apolipoprotein A-I Activates Cdc42 Signaling through the ABCA1 Transporter. J. Lipid Res. 47, 794–803. doi:10.1194/jlr.m500502-jlr200
Owen, M. J., Sawa, A., and Mortensen, P. B. (2016). Schizophrenia. Lancet 388, 86–97. doi:10.1016/s0140-6736(15)01121-6
Perzel Mandell, K. A., Eagles, N. J., Wilton, R., Price, A. J., Semick, S. A., Collado-Torres, L., et al. (2021). Genome-wide Sequencing-Based Identification of Methylation Quantitative Trait Loci and Their Role in Schizophrenia Risk. Nat. Commun. 12, 5251. doi:10.1038/s41467-021-25517-3
Remaley, A. T., Stonik, J. A., Demosky, S. J., Neufeld, E. B., Bocharov, A. V., Vishnyakova, T. G., et al. (2001). Apolipoprotein Specificity for Lipid Efflux by the Human ABCAI Transporter. Biochem. Biophys. Res. Commun. 280, 818–823. doi:10.1006/bbrc.2000.4219
Robinson, N., and Bergen, S. E. (2021). Environmental Risk Factors for Schizophrenia and Bipolar Disorder and Their Relationship to Genetic Risk: Current Knowledge and Future Directions. Front. Genet. 12, 686666. doi:10.3389/fgene.2021.686666
Rössler, W., Joachim Salize, H., Van Os, J., and Riecher-Rössler, A. (2005). Size of burden of Schizophrenia and Psychotic Disorders. Eur. Neuropsychopharmacol. 15, 399–409. doi:10.1016/j.euroneuro.2005.04.009
Saito, H., Lund-Katz, S., and Phillips, M. C. (2004). Contributions of Domain Structure and Lipid Interaction to the Functionality of Exchangeable Human Apolipoproteins. Prog. Lipid Res. 43, 350–380. doi:10.1016/j.plipres.2004.05.002
Song, X., Li, X., Gao, J., Zhao, J., Li, Y., Fan, X., et al. (2014). APOA-I: a Possible Novel Biomarker for Metabolic Side Effects in First Episode Schizophrenia. PLoS One 9, e93902. doi:10.1371/journal.pone.0093902
Swaney, J. B., Braithwaite, F., and Eder, H. A. (1977). Characterization of the Apolipoproteins of Rat Plasma Lipoproteins. Biochemistry 16, 271–278. doi:10.1021/bi00621a018
Tan, X. W., Lee, E. S., Toh, M. P. H. S., Lum, A. W. M., Seah, D. E. J., Leong, K. P., et al. (2021). Comparison of Mental-Physical Comorbidity, Risk of Death and Mortality Among Patients with Mental Disorders - A Retrospective Cohort Study. J. Psychiatr. Res. 142, 48–53. doi:10.1016/j.jpsychires.2021.07.039
Terao, T., Yoshimura, R., Ohmori, O., Takano, T., Takahashi, N., Iwata, N., et al. (1997). Effect of Serum Cholesterol Levels on Meta-Chlorophenylpiperazine-Evoked Neuroendocrine Responses in Healthy Subjects. Biol. Psychiatry 41, 974–978. doi:10.1016/s0006-3223(96)00213-2
Wang, X., Dai, S., Zhang, Z., Liu, L., Wang, J., Xiao, X., et al. (2009). Characterization of Apolipoprotein A-I as a Potential Biomarker for Cholangiocarcinoma. Eur. J. Cancer Care (Engl) 18, 625–635. doi:10.1111/j.1365-2354.2008.00965.x
Keywords: schizophrenia, APOA1 gene, APOA4 gene, mRNA, case–control study, apoA1, HDLC
Citation: Fan Y, Gao J, Li Y, Chen X, Zhang T, You W, Xue Y and Shen C (2021) The Variants at APOA1 and APOA4 Contribute to the Susceptibility of Schizophrenia With Inhibiting mRNA Expression in Peripheral Blood Leukocytes. Front. Mol. Biosci. 8:785445. doi: 10.3389/fmolb.2021.785445
Received: 29 September 2021; Accepted: 10 November 2021;
Published: 06 December 2021.
Edited by:
Yanan Wang, Xi’an Jiaotong University Health Science Center, ChinaReviewed by:
Yi Wu, Xi’an Jiaotong University, ChinaAlessandro Trentini, University of Ferrara, Italy
Copyright © 2021 Fan, Gao, Li, Chen, Zhang, You, Xue and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chong Shen, c2NAbmptdS5lZHUuY24=; Yong Xue, eHVleW9uZzNAMTI2LmNvbQ==
†These authors have contributed equally to this work and share the first authorship
 Yao Fan
Yao Fan Jun Gao
Jun Gao Yinghui Li4†
Yinghui Li4† Weiyan You
Weiyan You Chong Shen
Chong Shen