- 1Department of Spine Surgery, The Affiliated Hospital of Qingdao University, Qingdao, China
- 2Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore, Singapore
- 3Department of Anaesthesia and Intensive Care and Peter Hung Pain Research Institute, The Chinese University of Hong Kong, Hong Kong, China
- 4State Key Laboratory of Digestive Diseases, LKS Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong, China
Low back pain, a common medical condition, could result in severe disability and inflict huge economical and public health burden. Its pathogenesis is attributed to multiple etiological factors, including intervertebral disc degeneration (IDD). Emerging evidence suggests that circular RNAs (circRNAs), a major type of regulatory non-coding RNA, play critical roles in cellular processes that are pertinent to IDD development, including nucleus pulposus cell proliferation and apoptosis as well as extracellular matrix deposition. Increasing number of translational studies also indicated that circRNAs could serve as novel biomarkers for the diagnosis of IDD and/or predicting its clinical outcomes. Our review aims to discuss the recent progress in the functions and mechanisms of newly discovered IDD-related circRNAs.
1 Introduction
Low back pain (LBP) is one of the most common medical conditions. It can cause severe incapacity at the individual level and impact the workforce and the health care system at the societal level (Abbasi et al., 2021; Aransay et al., 2020; Martín-Corrales et al., 2020). Much effort has been taken to study the mechanism underlying the pathogenesis of LBP and to improve its treatment (Shi et al., 2020; Mera et al., 2021; Umimura et al., 2021). The etiology of LBP involves multiple factors, among which intervertebral disc degeneration (IDD) is a widely known cause (Abdollah et al., 2020; Ishitani et al., 2020; Shi et al., 2020; Huang et al., 2021a; Zhang et al., 2021a; Liu et al., 2021). IDD is a chronic process resulting in structural failure of the intervertebral disc, with accelerated or advanced signs of aging. Etiologically, IDD may be hastened by co-morbidities (e.g., diabetes and obesity), certain lifestyles (e.g., smoking, occupation, alcohol consumption), aging, and genetic predisposition (Tang et al., 2020; Tsingas et al., 2020; Yang et al., 2021a; Baldia et al., 2021; Shao et al., 2021; Zhao et al., 2021). The mainstay of treatment for IDD-related diseases is surgical intervention. Nucleus pulposus (NP), which is a core structural component of the intervertebral disc, is consisted of extracellular matrix (ECM) and NP cells (Jiang et al., 2019a; Bach et al., 2019; Kang et al., 2019; Ohnishi et al., 2019; Hanaei et al., 2020). Deregulated functions of NP cells, such as aberrant cell apoptosis, proliferation, and ECM degradation/synthesis, have been demonstrated to contribute to IDD development (Li et al., 2020a; Li et al., 2020b; Gao et al., 2020; He et al., 2020; Zhao et al., 2021).
Circular RNAs (circRNAs) are a major type of non-coding regulatory RNAs that are produced by non-canonical back-splicing events (Li et al., 2019a; Liu et al., 2020; Ma et al., 2020; Li et al., 2021a; Li et al., 2021b). CircRNAs are known to modulate gene expression principally via sponging microRNAs (miRNAs) (Li et al., 2020c; Pan et al., 2020; Ni et al., 2021). In addition, some circRNAs could mediate their biological functions through direct interactions with proteins or inhibiting mature mRNA formation (Tian et al., 2019; Tu et al., 2020; Li et al., 2021c). A minor subset of circRNAs also harbor coding potential for translation into protein through rolling circle amplification (Jiang et al., 2019b; Jin et al., 2019; Long et al., 2020). Growing evidence suggests that circRNAs play critical roles in different cellular processes including cell proliferation, apoptosis, differentiation, migration, and metabolism (Cai et al., 2018; Zhang et al., 2018; Liang et al., 2019; Yu et al., 2019; Guo et al., 2020a; Li et al., 2020d). A myriad of circRNAs have also been identified to be deregulated in human diseases, such as cancers, cardiovascular diseases, congenital diseases, and IDD (Chen et al., 2019; Pan et al., 2019; Li et al., 2020c). Translational studies also indicate that circRNAs could act as novel diagnostic and prognostic biomarkers for early disease detection and predicting the clinical outcomes of some diseases, including IDD (Liu et al., 2019; Li et al., 2020e; Yang et al., 2021b; Guo et al., 2021).
We previously reviewed the involvement of circRNAs in NP cell biology and IDD pathogenesis (Li et al., 2019b). Since then, more than a dozen of circRNAs had been identified to be associated with IDD. In the present work, we would like to summarize the functions and mechanisms of these newly discovered IDD-related circRNAs that have not been covered by our previous review. The potential prognostic and therapeutic utilities of these newly identified circRNAs in IDD will also been discussed.
2 Common Approaches for the Identification and Functional Characterization of IDD-Related circRNAs
Transcriptome sequencing is the most widely used method for the identification of disease-related circRNAs. Nevertheless, due to the overlap with sequence of the linear RNA transcribed from the same gene, additional processing workflows (e.g., linear RNA removal through exonuclease digestion) and specific computational algorithms (e.g., CIRI2, DCC, Sailfish-cir, CIRIquant) are required to such back-spliced reads (Cheng et al., 2016; Li et al., 2017; Gao et al., 2018; Zhang et al., 2020). Microarrays with probes specifically targeting the back-splice sites have also been developed for circRNA profiling (Li et al., 2019c) (29415187). Following the initial discovery, the differential expression of circRNAs can be confirmed by reverse transcription-quantitative PCR (RT-qPCR) using divergent primers spanning the back-splice junction sequence (Panda and Gorospe, 2018). In IDD studies, most commonly used samples for differentially expressed circRNA discovery are degenerative discs from IDD patients with normal discs from cadaveric donors or those from patients suffering from vertebral fracture as controls. Some investigators also profiled circRNAs in the cartilage endplates. For functional characterization of the identified circRNAs, gain-of-function (i.e., overexpression) and loss-of-function (i.e., silencing with small interfering RNA) approaches have been done in cultured NP cells or chondrocytes with or without further validation in animal models of IDD.
3 Functions and Mechanisms of Action of Newly Discovered circRNAs in IDD
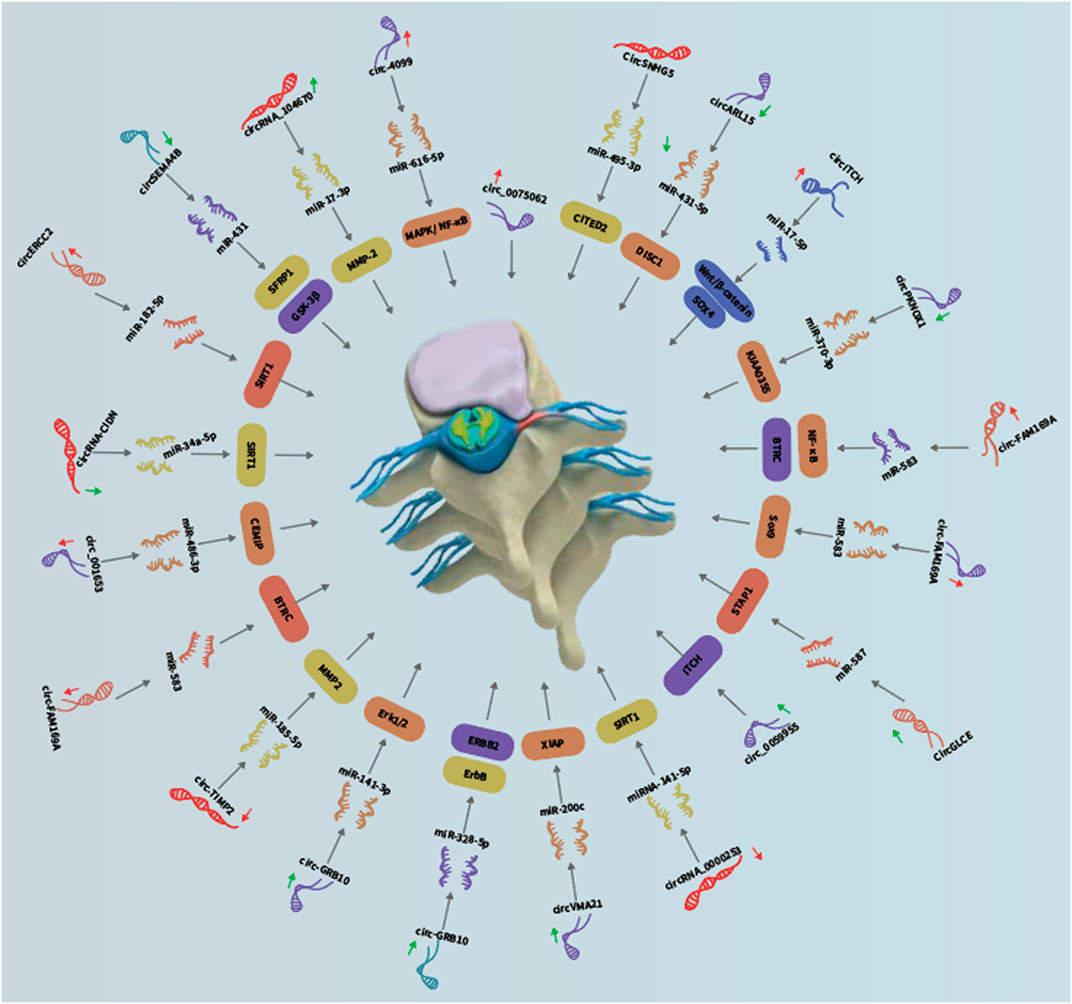
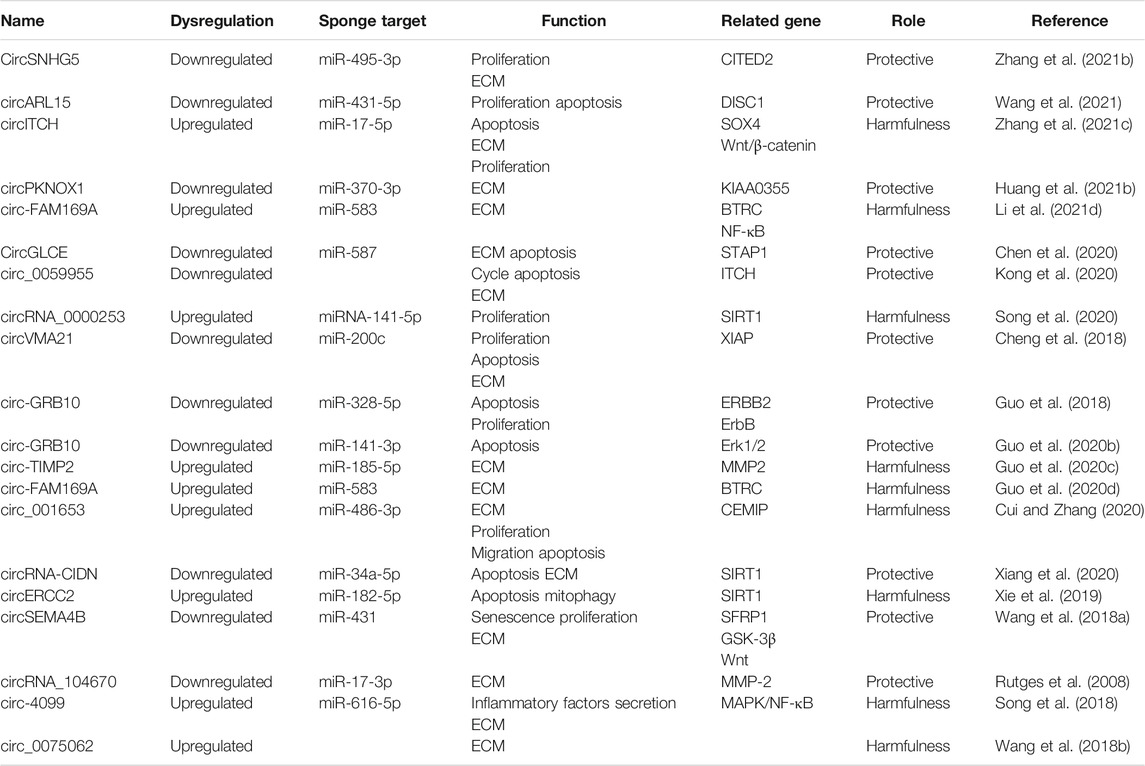
3.1 CircSNHG5
Like the NP cells, dysfunction of chondrocytes in the cartilage endplates has been implicated in IDD. Zhang et al. (2021b) studied the functional role of circSNHG5 in IDD-associated cartilage endplates. The authors showed that circSNHG5 was downregulated in the degenerative cartilage endplates as compared with healthy cartilage endplates. Functionally, knockdown of circSNHG5 inhibited chondrocyte proliferation and drove the degradation of collagen II and aggrecan. Moreover, they showed that circSNHG5 sponged miR-495-3p expression to derepress the downstream gene CITED2. Their data suggested that deregulation of the circSNHG5/miR-495-3p/CITED2 axis contributes to IDD development.
3.2 CircARL15
The loss of balance between proliferation and apoptosis of NP cells could contribute to NP cell loss during IDD. Wang et al. (2021) showed that circARL15 was one of the most downregulated circRNAs within the competing endogenous RNA (ceRNA) network in IDD. CircARL15 level was decreased whereas miR-431-5p level was increased in IDD samples, in which their expression showed significant negative correlation with each other. Ectopic expression of circARL15 enhanced NP cell proliferation and suppressed NP cell apoptosis. Mechanistically, circARL15 was shown to sponge miR-431-5p to disinhibit DISC1 to mediate its protective effects on NP cells. These data suggested that the aberrant downregulation of circARL15 contributes to IDD through promoting NP cell death via the miR-431-5p/DISC1 pathway.
3.3 CircITCH
Aberrant degradation of disc ECM components, such as aggrecan and type II collagen, is a hallmark of IDD. Zhang et al. (2021c) studied the role of circITCH in the degradation of ECM during IDD. They found that circITCH was overexpressed in the NP tissues from IDD patients compared to the control NP samples. Overexpression of circITCH suppressed NP cell proliferation and induced NP cell apoptosis. Enforced expression of circITCH also shifted the balance from ECM production to ECM degradation, characterized by decreased aggrecan and collagen II expression and increased MMP13 and ADAMTS4 expression. Mechanistically, circITCH sponged miR-17-5p to derepress SOX4 that promoted the Wnt/β-catenin signaling to accelerate ECM degradation in NP cells. Accordingly, miR-17-5p inhibitor, SOX4 overexpression and LiCl (a β-catenin/Wnt signaling activator) reversed ECM degradation and apoptosis induced by circITCH knockdown. These data suggested that circITCH induced ECM degradation through inducing the Wnt/β-catenin pathway via the miR-17-5p/SOX4 axis.
3.4 circPKNOX1
Similar to SOX4, SOX9 plays a key role in regulating NP cell function. Huang et al. (2021b) demonstrated that circPKNOX1 was downregulated in IDD cells as compared with control NP cells. Importantly, enforced expression of circPKNOX1 increased the levels of SOX9, aggrecan and collagen II and suppressed the expression of ECM-degrading enzymes, namely MMP13, MMP3, ADAMTS-5, and ADAMTS4. Furthermore, circPKNOX1 sponged miR-370-3p to restore the expression of KIAA0355. Their data suggested that reduced expression of circPKNOX1 promotes IDD development via promoting ECM degradation through the miR-370-3p/KIAA0355 axis. Thus, these data demonstrated that miR-370-3p may serve as a therapeutic target for IDD treatment.
3.5 Circ-FAM169A
The nuclear factor (NF)-κB-mediated signaling is an important pro-inflammatory pathway. Li et al. (2021d) showed that circ-FAM169A was overexpressed in degenerative NP samples compared to control NP tissues. Overexpression of circ-FAM169A significantly increased ECM degradation and inhibited ECM synthesis in NP cells. Moreover, they found that circ-FAM169A sequestered miR-583 to restore BTRC expression, which is an inducer of the NF-κB pathway. These data suggested that circ-FAM169A may induce IDD progression through miR-583/BTRC signaling. Concordantly, Li et al. (2021d) found that circ-FAM169A was overexpressed whereas miR-583 was downregulated in the degenerative NP tissues, in which their expression showed significant negative correlation with each other. They then reconstructed the circ-FAM169A/miR-583/mRNA ceRNA network and suggested that the circ-FAM169A/miR-583 pathway may play important roles in regulating apoptosis and ECM metabolism of NP cells. Furthermore, they showed that circ-FAM169A directly sponged miR-583 to derepress SOX9. These two studies suggested that the circ-FAM169A-miR-583 axis is involved in IDD pathogenesis.
3.6 CircGLCE
Chen et al. (2020) showed that circGLCE was localized in the cytoplasm of NP cells and it was decreased in the IDD tissues. Functionally, silencing of circGLCE was found to promote the expression of ECM-degrading enzymes and induce apoptosis of NP cells. Mechanistically, circGLCE sponged miR-587 expression in NP cells to derepress STAP1. The protective effect of circGLCE was also confirmed in vivo. Collectively, these data showed that circGLCE could suppress IDD development through inhibiting NP cell apoptosis and ECM degradation via targeting the miR-587/STAP1 axis.
3.7 Circ_0059955
Like circITCH, circ_0059955 is another circRNA transcribed from the host gene ITCH (Itchy E3 ubiquitin protein ligase). Kong et al. (2020) demonstrated that circ_0059955 was significantly downregulated in IDD samples. Knockdown of circ_0059955 suppressed ITCH expression and inhibited proliferation, induced cell cycle arrest and promoted apoptosis of NP cells. Furthermore, they demonstrated that enforced expression of circ_0059955 ameliorated IDD development in a rat model. These data suggested that circ_0059955 could protect against IDD.
3.8 CircRNA_0000253
Exosomes are a major means for intercellular transfer of non-coding RNAs. Song et al. (2020) identified circRNA_0000253 as one of the most upregulated circRNAs in the exosomes released from the degenerative NP cells. Functionally, circRNA_0000253 could promote IDD development via downregulating SIRT1 and sponging miRNA-141-5p in vitro and in vivo. Their data suggested that the aberrant upregulation of exosomal circRNA_0000253 may contribute to IDD.
3.9 CircVMA21
XIAP (X linked inhibitor-of-apoptosis protein) is a potent inhibitor of apoptosis by binding to caspases 3, 7, and 9. Cheng et al. (2018) found that XIAP was downregulated in the degenerative NP samples and inflammatory cytokine-induced NP cells, where XIAP downregulation was correlated with the deregulated balance between ECM synthesis and degradation as well as excessive apoptosis. miR-200c could undermine NP cell viability via suppressing XIAP expression. circVMA21 was identified to sponge miR-200c to promote NP cell function via derepressing XIAP. Moreover, they demonstrated that circVMA21 alleviated IDD in the rat model. Their data suggested that circVMA21 may counteract inflammatory cytokine-induced imbalance between the catabolism and anabolism of ECM and NP cell apoptosis via regulating the miR-200c-XIAP axis.
3.10 Circ-GRB10
ERBB2 is a member of the human epidermal growth factor receptor family that transduces mitogenic and pro-survival signal. Guo et al. (2018) showed that circ-GRB10 was decreased in NP samples from IDD patients as compared to normal NP samples. Enforced expression of circ-GRB10 suppressed NP cell apoptosis. Mechanistically, circ-GRB10 targeted miR-328-5p to derepress ERBB2 to positively regulate cell proliferation. Circ-GRB10 suppressed IDD development in the rat model. Moreover, the upstream mechanism underlying circ-GRB10 dysregulation was elucidated. Guo et al. (2020b) found that FUS could promote circ-GBR10 biosynthesis in the NP cells in which FUS expression was regulated by miR-141-3p. Downregulation of ERBB2 was also found to suppress Erk1/2 phosphorylation that regulated miR-141-3p expression in NP cells, highlighting FUS and miR-141-3p as important modulators of circ-GRB10 synthesis. Their data suggested that circ-GRB10 could protect against IDD development and the downstream miR-328-5p might serve as a potential therapeutic target for IDD.
3.11 Circ-TIMP2
The gene TIMP2 (Tissue Inhibitor Of Metalloproteinases 2) encodes a protein that serves as a natural inhibitor of the matrix metalloproteinases. Guo et al. (2020c) showed that circ-TIMP2, also transcribed from TIMP2, was overexpressed in IDD samples as compared to normal NP samples. Upregulation of circ-TIMP2 inhibited ECM synthesis and induced ECM degradation. In contrast, miR-185-5p suppressed ECM degradation induced by interleukin (IL)-1β and tumor necrosis factor (TNF)-α. Bioinformatic analysis showed that MMP2 was a potential target of miR-185-5p. Consistently, MMP2 was upregulated after exposure to IL-1β and TNF-α in NP cells, which could be rescued by transfecting with miR-185-5p mimic. Importantly, they demonstrated that circ-TIMP2 sponged miR-185-5p to induce ECM degradation. These data suggested that circ-TIMP2 could promote ECM degradation through the miR-185-5p/MMP2 signaling in NP cells.
3.12 circ_001653
Cui and Zhang (2020) showed that circ_001653 was upregulated in the degenerative NP cells and samples compared to control groups and was closely related to IDD severity. Overexpression of circ_001653 promoted NP cell apoptosis and resulted in an imbalance between catabolic and anabolic factors of ECM whereas miR-486-3p enhanced NP cell viability by suppressing CEMIP. Circ_001653 was found to sponge miR-486-3p to induce CEMIP to contribute to NP cell death. In mice, knockdown of circ_001653 alleviated IDD development. Their data suggested that targeting circ_001653 may be a novel therapeutic strategy the delay IDD development.
3.13 Circ-CIDN
Excessive mechanical loading can be transduced by NP cells. Xiang et al. (2020) showed that circ-CIDN was decreased in the compressed NP cells compared to control NP cells. Functionally, enforced expression of circRNA-CIDN rescued compression-induced NP cell apoptosis and ECM degradation. CircRNA-CIDN was found to sponge miR-34a-5p that could promote compression-induced damage via inhibiting SIRT1 expression. The protective effect of circ-CIDN against IDD was also confirmed in the IDD model. Their data suggested that circ-CIDN played an important role in mitigating mechanical loading-induced NP cell damage through regulating the miR-34a-5p/SIRT1 axis.
3.14 CircERCC2
Removal of damaged mitochondria through the autophagy (i.e., mitophagy) is key to cell survival. Xie et al. (2019) demonstrated that circERCC2 was one of the most downregulated circRNAs in IDD. Knockdown of circERCC2 promoted miR-182-5p expression and suppressed SIRT1 expression in the degenerative NP samples and tert-Butyl hydroperoxide (TBHP; oxidative stress inducer)-exposed NP cells, in which knockdown of SIRT1 inhibited mitophagy and promoted apoptosis. Moreover, miR-182-5p was found to regulate the mitophagy and NP cell apoptosis by targeting SIRT1. The protective effects of circERCC2 on NP cells and IDD rat model were mediated by the miR-182-5p/SIRT1 axis. This study indicated circERCC2 could ameliorate IDD via sponging miR-182 to derepress SIRT1 for activating mitophagy and inhibiting NP cell apoptosis. Restoring circERCC2 expression might be a potential therapeutic approach for IDD.
3.15 CircSEMA4B
Aberrant Wnt signaling is linked to IDD. Wang et al. (2018a) showed that circSEMA4B was decreased in the IDD samples compared to control tissues. Overexpression of circSEMA4B attenuated the effect of IL-1β on NP cell senescence, proliferation, and aggrecan degradation through the Wnt signaling in IDD. CircSEMA4B was found to sponge miR-431 to disinhibit GSK-3β and SFRP1. The effect of circSEMA4B inhibition on NP cells was partially rescued by the inhibition of miR-431. These findings suggested that circSEMA4B could suppressing IL-1β-induced degeneration via Wnt signaling in NP cells.
3.16 CircRNA_104670
Increased MMP2 activity has been reported in IDD (Rutges et al., 2008). Song et al. (2018) demonstrated that circRNA_104670 was upregulated in IDD NP tissues compared to normal tissues. circRNA_104670 and its downstream target could distinguish IDD from healthy NP with an area under the receiver-operating characteristic curve of 0.96 and 0.91, respectively. In additional, there are significant positive and negative correlation of Pfirrmann disc grade with the expression of circRNA_104670 and miR-17-3p, respectively. Functionally, knockdown of circRNA_104670 inhibited NP cell apoptosis and reduced MMP2 expression accompanied by enhanced ECM formation. CircRNA_104670 inhibition in mice also resulted in lower IDD grade, whereas miRNA-17-3p inhibition or circRNA_104670 overexpression in mice led to higher IDD grade. These data suggested that circRNA_104670 could serve as a prognostic marker and a therapeutic target in IDD.
3.17 Circ-4099
Aside from functioning as pathogenic mediators, differential expression of circRNAs could serve as an autoprotective mechanism. Wang et al. (2018b) demonstrated that circ-4099 was upregulated in the degenerated NP samples. Interestingly, enforced expression of circ-4099 induced aggrecan and collagen II expression and suppressed the release of pro-inflammatory factors, including prostaglandin E2, TNF-α and IL-1β. Upstream, TNF-α increased the level of circ-4099 in NP cells via upregulating GRP78, in which MAPK/NF-κB shRNAs or inhibitors abolished the induction of circ-4099 by TNF-α. Downstream, circ-4099 sponged miR-616-5p to derepress SOX9. These results suggested that circ-4099 acted as an autoprotective circRNA in IDD development.
3.18 circ_0075062
Insufficient supply of nutrition to NP cells has been implicated in IDD pathogenesis (De Geer, 2018). Chang et al. (2021) showed that circ_0075062 was overexpressed in the IDD NP samples and glucose-deprived NP cells. Using RNase tolerance assay coupled with sequencing, they found that circ_0075062 was confirmed to be a circular transcript. Knockdown of circ_0075062 alleviated ECM degradation in glucose-deprived NP cells. These results suggested that circ_0075062 may be a target for IDD.
4 Conclusion
IDD is contributed by deregulated NP cell functions, including aberrant apoptosis, proliferation, and ECM degradation/synthesis. Emerging studies have pointed to the involvement of non-coding RNAs, including miRNAs, long non-coding RNAs and circRNAs (Figures 1, 2, 3 and 4; Table 1), in IDD through their extensive crosstalk with NP cell function-related signaling pathways, such as Wnt and NF-κB signaling. Transcription regulators/factors, such as CITED2, SIRT1, and SOX4/9, also appeared to be heavily deregulated by circRNA-miRNA networks in IDD. To date, the upstream mechanism mediating the deregulation of most circRNAs in NP cells and their relationship with IDD risk factors, including mechanical loading, aging, smoking and genetic predisposition, remain largely unknown. Moreover, reports on deregulated circRNAs in other cell types pertinent to IDD, such as cartilage endplate chondrocytes, are scarce. For clinical translation, although some tissue circRNAs were shown to be able to discriminate IDD samples from healthy tissues, the use of circulating circRNAs as diagnostic markers have not been reported. Future research efforts should therefore be put forward to large-scale screening and validation of diagnosis/prognosis-related circulating IDD-related circRNAs. As therapeutic targets, upregulated circRNAs of pathogenic significance might be inhibited by RNA interference or antisense oligos. Disinhibited miRNAs downstream to downregulated circRNAs might also be targeted to achieve therapeutic effect. Nevertheless, tissue- or cell type-specific delivery of RNA-targeting agents remains technically challenging. In this connection, NP cell-targeting approaches, for instance with nanoparticles, are urgently needed. With these, it is hopeful that novel circRNA-based therapeutics and biomarkers will emerge to improve the clinical management of IDD.
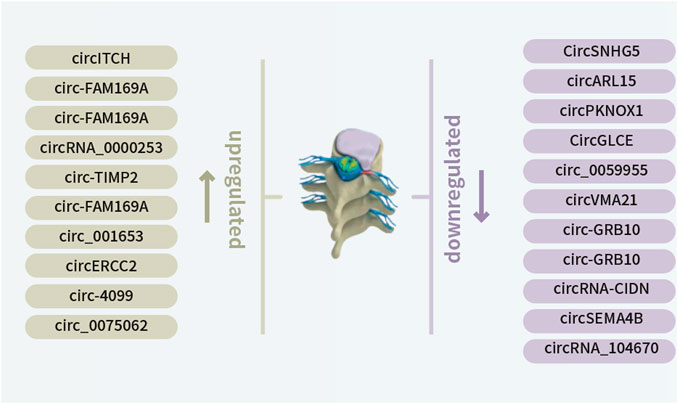
FIGURE 1. Newly identified differentially expressed circRNAs in IDD, including 10 downregulated and 11 upregulated circRNAs.
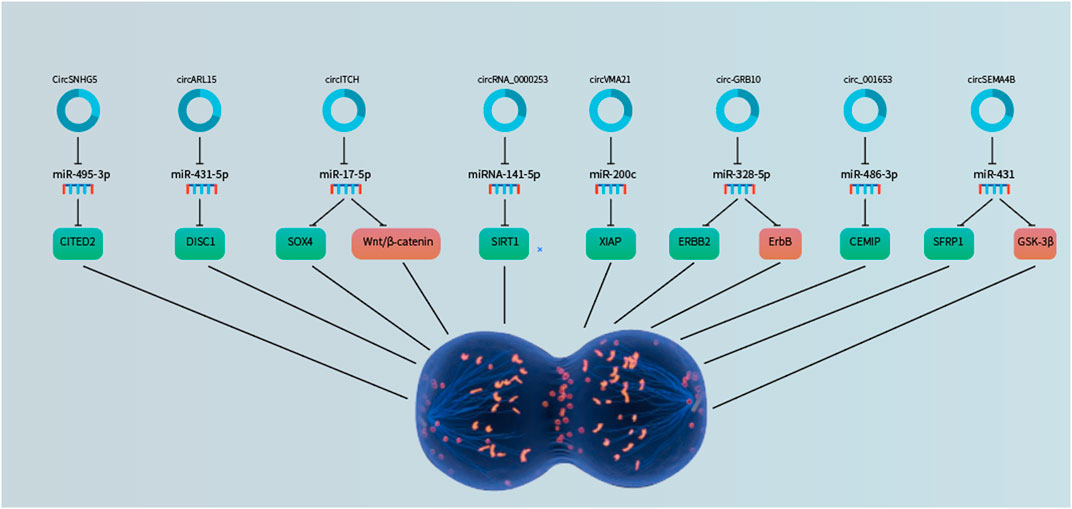
FIGURE 2. CircRNAs as regulators of NP cell proliferation through sponging microRNAs to derepress the downstream target genes.
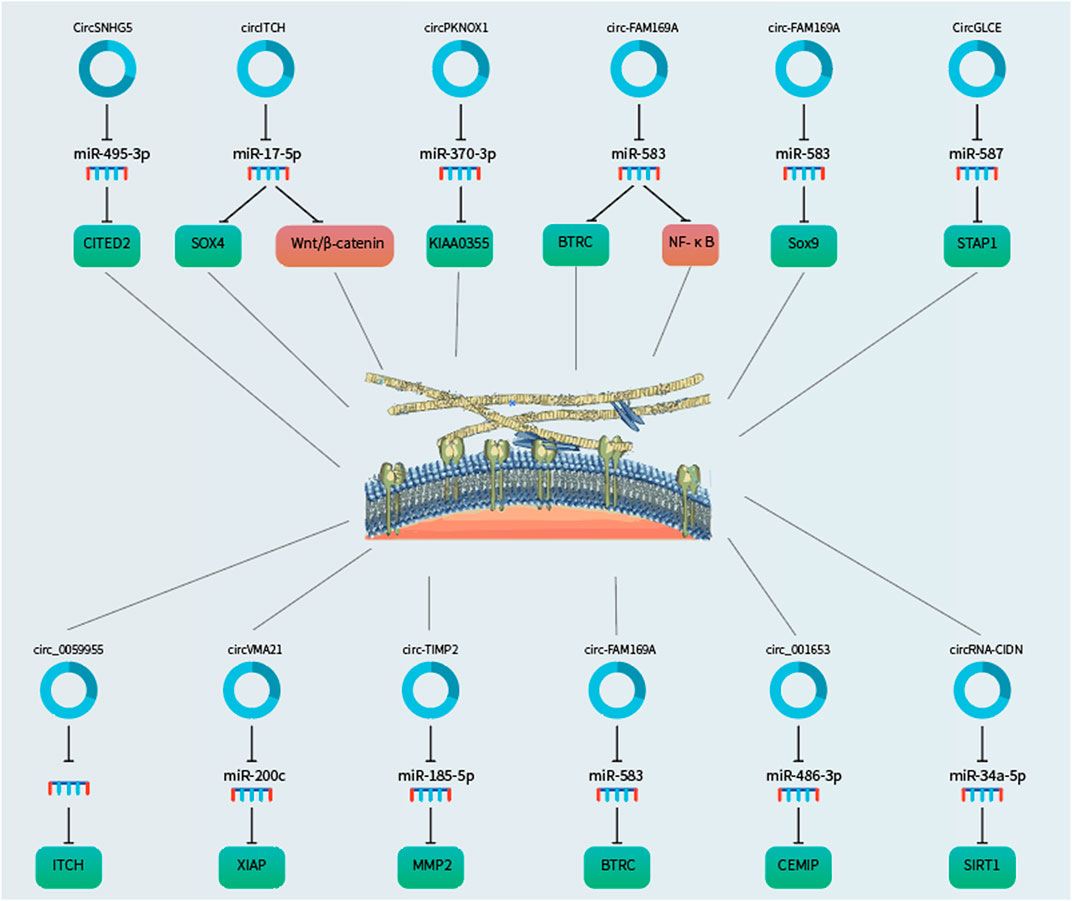
FIGURE 3. CircRNAs as regulators of ECM remodelling in the NP cells via sponging microRNAs to derepress the downstream target genes.
Author Contributions
DX, XM, CS, JH, and CZ drafted and wrote the manuscript. MC and WW revised the manuscript. DX and CZ participated in the design of the review. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbasi, S., Hadian Rasanani, M. R., Olyaei, G. R., Ghotbi, N., and Rasouli, O. (2021). Reliability of Ultrasound Measurement of the Lateral Abdominal and Lumbar Multifidus Muscles in Individuals with Chronic Low Back Pain: A Cross-Sectional Test-Retest Study. J. Bodywork Mov. Therapies 26, 394–400. doi:10.1016/j.jbmt.2020.12.029
Abdollah, V., Parent, E. C., Su, A., Wachowicz, K., and Battié, M. C. (2020). Could Compression and Traction Loading Improve the Ability of Magnetic Resonance Imaging to Identify Findings Related to Low Back Pain? Musculoskelet. Sci. Pract. 50, 102250. doi:10.1016/j.msksp.2020.102250
Aransay, Á. L. S., Valladares, Á. C., Muñoz, R. C., Parrilla, Á. R. P., Muñiz, I. P., Cuello, L. G., et al. (2020). Prospective Analysis of Radiofrequency Denervation in Patients with Chronic Low Back Pain. J. Spine Surg. 6 (4), 703–712. doi:10.21037/jss-20-599
Bach, F. C., Rooij, K. M., Riemers, F. M., Snuggs, J. W., Jong, W. A. M., Zhang, Y., et al. (2019). Hedgehog Proteins and Parathyroid Hormone‐related Protein Are Involved in Intervertebral Disc Maturation, Degeneration, and Calcification. JOR Spine 2 (4), e1071. doi:10.1002/jsp2.1071
Baldia, M., Mani, S., Walter, N., Kumar, S., Srivastava, A., and Prabhu, K. (2021). Development of a Unique Mouse Intervertebral Disc Degeneration Model Using a Simple Novel Tool. Asian Spine J. 15 (4), 415–423. doi:10.31616/asj.2020.0366
Cai, H., Hu, B., Ji, L., Ruan, X., and Zheng, Z. (2018). Hsa_circ_0103809 Promotes Cell Proliferation and Inhibits Apoptosis in Hepatocellular Carcinoma by Targeting miR-490-5p/SOX2 Signaling Pathway. Am. J. Transl Res. 10 (6), 1690–1702.
Chang, H., Wang, H., Yang, X., You, K., Jiang, M., Cai, F., et al. (2021). Comprehensive Profile Analysis of Differentially Expressed circRNAs in Glucose Deprivation-Induced Human Nucleus Pulposus Cell Degeneration. Biomed. Res. Int. 2021, 1–14. doi:10.1155/2021/4770792
Chen, T., Yang, Z., Liu, C., Wang, L., Yang, J., Chen, L., et al. (2019). Circ_0078767 Suppresses Non‐small‐cell Lung Cancer by Protecting RASSF1A Expression via Sponging miR‐330‐3p. Cell Prolif 52 (2), e12548. doi:10.1111/cpr.12548
Chen, Z., Zhang, W., Deng, M., Li, Y., and Zhou, Y. (2020). CircGLCE Alleviates Intervertebral Disc Degeneration by Regulating Apoptosis and Matrix Degradation through the Targeting of miR-587/STAP1. Aging 12 (21), 21971–21991. doi:10.18632/aging.104035
Cheng, J., Metge, F., and Dieterich, C. (2016). Specific Identification and Quantification of Circular RNAs from Sequencing Data. Bioinformatics 32 (7), 1094–1096. doi:10.1093/bioinformatics/btv656
Cheng, X., Zhang, L., Zhang, K., Zhang, G., Hu, Y., Sun, X., et al. (2018). Circular RNA VMA21 Protects against Intervertebral Disc Degeneration through Targeting miR-200c and X Linked Inhibitor-Of-Apoptosis Protein. Ann. Rheum. Dis. 77 (5), 770–779. doi:10.1136/annrheumdis-2017-212056
Cui, S., and Zhang, L. (2020). circ_001653 Silencing Promotes the Proliferation and ECM Synthesis of NPCs in IDD by Downregulating miR-486-3p-Mediated CEMIP. Mol. Ther. - Nucleic Acids 20, 385–399. doi:10.1016/j.omtn.2020.01.026
De Geer, C. M. (2018). Intervertebral Disk Nutrients and Transport Mechanisms in Relation to Disk Degeneration: A Narrative Literature Review. J. Chiropractic Med. 17 (2), 97–105. doi:10.1016/j.jcm.2017.11.006
Gao, D., Hao, L., and Zhao, Z. (2020). Long Non-coding RNA PART1 Promotes Intervertebral Disc Degeneration through Regulating the miR-93/MMP2 P-athway in N-ucleus P-ulposus C-ells. Int. J. Mol. Med. 46 (1), 289–299. doi:10.3892/ijmm.2020.4580
Gao, Y., Zhang, J., and Zhao, F. (2018). Circular RNA Identification Based on Multiple Seed Matching. Brief Bioinform 19 (5), 803–810. doi:10.1093/bib/bbx014
Guo, F., Li, S., Guo, C., Xu, X., Zhou, X., Ma, D., et al. (2020a). Circular RNA circMAGI3 Accelerates the Glycolysis of Non-small Cell Lung Cancer through miR-515-5p/HDGF. Am. J. Transl Res. 12 (7), 3953–3963.
Guo, S., Hu, C., Zhai, X., and Sun, D. (2021). Circular RNA 0006602 in Plasma Exosomes: a New Potential Diagnostic Biomarker for Hepatocellular Carcinoma. Am. J. Transl Res. 13 (6), 6001–6015.
Guo, W., Mu, K., Zhang, B., Sun, C., Zhao, L., Dong, Z.-Y., et al. (2020d). The Circular RNA FAM169A Functions as a Competitive Endogenous RNA and Regulates Intervertebral Disc Degeneration by Targeting miR-583 and BTRC. Cell Death Dis 11 (5), 315. doi:10.1038/s41419-020-2543-8
Guo, W., Mu, K., Zhang, B., Sun, C., Zhao, L., Li, H.-R., et al. (2020b). The Circular RNA Circ-GRB10 Participates in the Molecular Circuitry Inhibiting Human Intervertebral Disc Degeneration. Cel Death Dis 11 (8), 612. doi:10.1038/s41419-020-02882-3
Guo, W., Zhang, B., Mu, K., Feng, S.-Q., Dong, Z.-Y., Ning, G.-Z., et al. (2018). Circular RNA GRB10 as a Competitive Endogenous RNA Regulating Nucleus Pulposus Cells Death in Degenerative Intervertebral Disk. Cel Death Dis 9 (3), 319. doi:10.1038/s41419-017-0232-z
Guo, W., Zhang, B., Sun, C., Duan, H. Q., Liu, W. X., Mu, K., et al. (2020c). Circular RNA Derived from TIMP2 Functions as a Competitive Endogenous RNA and Regulates Intervertebral Disc Degeneration by Targeting miR-185-5p and M-atrix M-etalloproteinase 2. Int. J. Mol. Med. 46 (2), 621–632. doi:10.3892/ijmm.2020.4621
Hanaei, S., Abdollahzade, S., Sadr, M., Mirbolouk, M. H., Fattahi, E., Khoshnevisan, A., et al. (2020). The Role of Interleukin 4 and IL-4RA in Intervertebral Disc Degeneration: Investigation of Single Nucleotide Polymorphisms in Genes and a Systematic Review & Meta-Analysis of IL-4 Expression Level. Br. J. Neurosurg. 34 (1), 66–71. doi:10.1080/02688697.2019.1698010
He, D., Zhou, M., Bai, Z., Wen, Y., Shen, J., and Hu, Z. (2020). Propionibacterium Acnes Induces Intervertebral Disc Degeneration by Promoting Nucleus Pulposus Cell Pyroptosis via NLRP3-dependent Pathway. Biochem. Biophysical Res. Commun. 526 (3), 772–779. doi:10.1016/j.bbrc.2020.03.161
Huang, B., Liu, J., Wei, X., Li, S., Xiang, Y., Wu, H., et al. (2021a). Damage to the Human Lumbar Cartilage Endplate and its Clinical Implications. J. Anat. 238 (2), 338–348. doi:10.1111/joa.13321
Huang, Y., Gao, J., Wang, J., Ye, H., Yao, T., Xu, Y., et al. (2021b). Inhibition of Intervertebral Disc Disease Progression via the circPKNOX1-miR-370-3p-Kiaa0355 axis. Cell Death Discov. 7 (1), 39. doi:10.1038/s41420-021-00420-4
Ishitani, H., Tamura, T., Kanaya, S., and Fujimoto, H. (2020). Examination of the Regression Model to Quantify the Degree of Low Back Pain and Lower Limb Symptoms in Patients with Lumbar Disc Herniation by the Japanese Orthopaedic Association Back Pain Evaluation Questionnaire (JOABPEQ). PLoS One 15 (12), e0243861. doi:10.1371/journal.pone.0243861
Jiang, C., Guo, Q., Jin, Y., Xu, J.-J., Sun, Z.-M., Zhu, D.-C., et al. (2019a). Inhibition of EZH2 Ameliorates Cartilage Endplate Degeneration and Attenuates the Progression of Intervertebral Disc Degeneration via Demethylation of Sox-9. EBioMedicine 48, 619–629. doi:10.1016/j.ebiom.2019.10.006
Jiang, Y., Wei, T., Li, W., Zhang, R., and Chen, M. (2019b). Circular RNA Hsa_circ_0002024 Suppresses Cell Proliferation, Migration, and Invasion in Bladder Cancer by Sponging miR-197-3p. Am. J. Transl Res. 11 (3), 1644–1652.
Jin, C., Shi, L., Li, Z., Liu, W., Zhao, B., Qiu, Y., et al. (2019). Circ_0039569 Promotes Renal Cell Carcinoma Growth and Metastasis by Regulating miR-34a-5p/CCL22. Am. J. Transl Res. 11 (8), 4935–4945.
Kang, L., Xiang, Q., Zhan, S., Song, Y., Wang, K., Zhao, K., et al. (2019). Restoration of Autophagic Flux Rescues Oxidative Damage and Mitochondrial Dysfunction to Protect against Intervertebral Disc Degeneration. Oxidative Med. Cell Longevity 2019, 1–27. doi:10.1155/2019/7810320
Kong, D., Gu, R., Zhang, C., and Yin, R. (2020). Knockdown of Hsa_circ_0059955 Induces Apoptosis and Cell Cycle Arrest in Nucleus Pulposus Cells via Inhibiting Itchy E3 Ubiquitin Protein Ligase. Dddt Vol. 14, 3951–3963. doi:10.2147/DDDT.S253293
Li, J., Song, Y., Wang, J., and Huang, J. (2020e). Plasma Circular RNA Panel Acts as a Novel Diagnostic Biomarker for Colorectal Cancer Detection. Am. J. Transl Res. 12 (11), 7395–7403.
Li, M., Xie, X., Zhou, J., Sheng, M., Yin, X., Ko, E.-A., et al. (2017). Quantifying Circular RNA Expression from RNA-Seq Data Using Model-Based Framework. Bioinformatics 33 (14), 2131–2139. doi:10.1093/bioinformatics/btx129
Li, S., Jiang, C., Tan, J., Zhou, Q., Yin, J., and He, Y. (2021b). Sodium Arsenite-Mediated Upregulation of circDHX34 Promotes Apoptosis in Hormone-independent Breast Cancer Cells by Regulating Apoptotic Genes. Environ. Sci. Pollut. Res.. doi:10.1007/s11356-021-15891-2
Li, S., Teng, S., Xu, J., Su, G., Zhang, Y., Zhao, J., et al. (2019c). Microarray Is an Efficient Tool for circRNA Profiling. Brief Bioinform 20 (4), 1420–1433. doi:10.1093/bib/bby006
Li, X., Gao, F., Fan, Y., Xie, S., Li, C., Meng, L., et al. (2021a). A Novel Identified Circ-ANKHD1 Targets the miR-27a-3p/SFRP1 Signaling Pathway and Modulates the Apoptosis of Granulosa Cells. Environ. Sci. Pollut. Res. 28, 57459–57469. doi:10.1007/s11356-021-14699-4
Li, Y., Pan, D., Liu, S., Xing, X., Zhou, H., Zhang, B., et al. (2021d). Identification of Circ-Fam169a Sponges miR-583 Involved in the Regulation of Intervertebral Disc Degeneration. J. Orthopaedic Translation 26, 121–131. doi:10.1016/j.jot.2020.07.007
Li, Y., Zhang, T., Tian, W., Hu, H., Xin, Z., Ma, X., et al. (2020b). Loss of TIMP3 Expression Induces Inflammation, Matrix Degradation, and Vascular Ingrowth in Nucleus Pulposus: A New Mechanism of Intervertebral Disc Degeneration. FASEB j. 34 (4), 5483–5498. doi:10.1096/fj.201902364RR
Li, Z., Chen, Z., Feng, Y., Hu, G., and Jiang, Y. (2020d). CircMMP11 Acts as a Ce-circRNA in Breast Cancer Progression by Regulating miR-1204. Am. J. Transl Res. 12 (6), 2585–2599.
Li, Z., Chen, X., Xu, D., Li, S., Chan, M. T. V., and Wu, W. K. K. (2019b). Circular RNAs in Nucleus Pulposus Cell Function and Intervertebral Disc Degeneration. Cel Prolif 52 (6), e12704. doi:10.1111/cpr.12704
Li, Z., Li, X., Shen, J., Zhang, L., Chan, M. T. V., and Wu, W. K. K. (2020c). Emerging Roles of Non‐coding RNAs in Scoliosis. Cel Prolif 53 (2), e12736. doi:10.1111/cpr.12736
Li, Z., Li, X., Xu, D., Chen, X., Li, S., Zhang, L., et al. (2021c). An Update on the Roles of Circular RNAs in Osteosarcoma. Cel Prolif 54 (1), e12936. doi:10.1111/cpr.12936
Li, Z., Ma, J., Shen, J., Chan, M. T. V., Wu, W. K. K., and Wu, Z. (2019a). Differentially Expressed Circular RNAs in Air Pollution-Exposed Rat Embryos. Environ. Sci. Pollut. Res. 26 (33), 34421–34429. doi:10.1007/s11356-019-06489-w
Li, Z., Shao, Z., Chen, S., Huang, D., Peng, Y., Chen, S., et al. (2020a). TIGAR Impedes Compression‐induced Intervertebral Disc Degeneration by Suppressing Nucleus Pulposus Cell Apoptosis and Autophagy. J. Cel Physiol 235 (2), 1780–1794. doi:10.1002/jcp.29097
Liang, M., Huang, G., Liu, Z., Wang, Q., Yu, Z., Liu, Z., et al. (2019). Elevated Levels of Hsa_circ_006100 in Gastric Cancer Promote Cell Growth and Metastasis via miR‐195/GPRC5A Signalling. Cel Prolif 52 (5), e12661. doi:10.1111/cpr.12661
Liu, L., He, J., Liu, C., Yang, M., Fu, J., Yi, J., et al. (2021). Cartilage Intermediate Layer Protein Affects the Progression of Intervertebral Disc Degeneration by Regulating the Extracellular Microenvironment (Review). Int. J. Mol. Med. 47 (2), 475–484. doi:10.3892/ijmm.2020.4832
Liu, P., Li, X., Guo, X., Chen, J., Li, C., Chen, M., et al. (2019). Circular RNA DOCK1 Promotes Bladder Carcinoma Progression via Modulating circDOCK1/hsa‐miR‐132‐3p/Sox5 Signalling Pathway. Cel Prolif 52 (4), e12614. doi:10.1111/cpr.12614
Liu, R., Liu, X., Bai, X., Xiao, C., and Dong, Y. (2020). Identification and Characterization of circRNA in Longissimus Dorsi of Different Breeds of Cattle. Front. Genet. 11, 565085. doi:10.3389/fgene.2020.565085
Long, M. Y., Chen, J. W., Zhu, Y., Luo, D. Y., Lin, S. J., Peng, X. Z., et al. (2020). Comprehensive Circular RNA Profiling Reveals the Regulatory Role of circRNA_0007694 in Papillary Thyroid Carcinoma. Am. J. Transl Res. 12 (4), 1362–1378.
Ma, C., Shi, T., Qu, Z., Zhang, A., Wu, Z., Zhao, H., et al. (2020). CircRNA_ACAP2 Suppresses EMT in Head and Neck Squamous Cell Carcinoma by Targeting the miR-21-5p/STAT3 Signaling Axis. Front. Oncol. 10, 583682. doi:10.3389/fonc.2020.583682
Martín-Corrales, C., Bautista, I. V., Méndez-Mera, J. E., Fernández-Matías, R., Achalandabaso-Ochoa, A., Gallego-Izquierdo, T., et al. (2020). Benefits of Adding Gluteal Dry Needling to a Four-Week Physical Exercise Program in a Chronic Low Back Pain Population. A Randomized Clinical Trial. Pain Med. 21 (11), 2948–2957. doi:10.1093/pm/pnaa279
Mera, Y., Teraguchi, M., Hashizume, H., Oka, H., Muraki, S., Akune, T., et al. (2021). Association between Types of Modic Changes in the Lumbar Region and Low Back Pain in a Large Cohort: the Wakayama Spine Study. Eur. Spine J. 30 (4), 1011–1017. doi:10.1007/s00586-020-06618-x
Ni, W., Jiang, C., Wu, Y., Zhang, H., Wang, L., Yik, J. H. N., et al. (2021). CircSLC7A2 Protects against Osteoarthritis through Inhibition of the miR‐4498/TIMP3 axis. Cel Prolif 54 (6), e13047. doi:10.1111/cpr.13047
Ohnishi, T., Yamada, K., Iwasaki, K., Tsujimoto, T., Higashi, H., Kimura, T., et al. (2019). Caspase-3 Knockout Inhibits Intervertebral Disc Degeneration Related to Injury but Accelerates Degeneration Related to Aging. Sci. Rep. 9 (1), 19324. doi:10.1038/s41598-019-55709-3
Pan, G., Mao, A., Liu, J., Lu, J., Ding, J., and Liu, W. (2020). Circular RNA Hsa_circ_0061825 (circ‐TFF1) Contributes to Breast Cancer Progression through Targeting miR‐326/TFF1 Signalling. Cel Prolif 53 (2), e12720. doi:10.1111/cpr.12720
Pan, R. Y., Zhao, C. H., Yuan, J. X., Zhang, Y. J., Jin, J. L., Gu, M. F., et al. (2019). Circular RNA Profile in Coronary Artery Disease. Am. J. Transl Res. 11 (11), 7115–7125.
Panda, A., and Gorospe, M. (2018). Detection and Analysis of Circular RNAs by RT-PCR. Bio-protocol 8 (6), 2775. doi:10.21769/BioProtoc.2775
Rutges, J., Kummer, J., Oner, F., Verbout, A., Castelein, R., Roestenburg, H., et al. (2008). Increased MMP-2 Activity during Intervertebral Disc Degeneration Is Correlated to MMP-14 Levels. J. Pathol. 214 (4), 523–530. doi:10.1002/path.2317
Shao, Z., Ni, L., Hu, S., Xu, T., Meftah, Z., Yu, Z., et al. (2021). RNA‐binding Protein HuR Suppresses Senescence through Atg7 Mediated Autophagy Activation in Diabetic Intervertebral Disc Degeneration. Cel Prolif 54 (2), e12975. doi:10.1111/cpr.12975
Shi, W., Agbese, E., Solaiman, A. Z., Leslie, D. L., and Gater, D. R. (2020). Performance of Pain Interventionalists from Different Specialties in Treating Degenerative Disk Disease-Related Low Back Pain. Arch. Rehabil. Res. Clin. Translation 2 (3), 100060. doi:10.1016/j.arrct.2020.100060
Song, J., Chen, Z.-H., Zheng, C.-J., Song, K.-H., Xu, G.-Y., Xu, S., et al. (2020). Exosome-Transported circRNA_0000253 Competitively Adsorbs MicroRNA-141-5p and Increases IDD. Mol. Ther. - Nucleic Acids 21, 1087–1099. doi:10.1016/j.omtn.2020.07.039
Song, J., Wang, H.-L., Song, K.-H., Ding, Z.-W., Wang, H.-L., Ma, X.-S., et al. (2018). CircularRNA_104670 Plays a Critical Role in Intervertebral Disc Degeneration by Functioning as a ceRNA. Exp. Mol. Med. 50 (8), 1–12. doi:10.1038/s12276-018-0125-y
Tang, N., Dong, Y., Xiao, T., and Zhao, H. (2020). LncRNA TUG1 Promotes the Intervertebral Disc Degeneration and Nucleus Pulposus Cell Apoptosis Though Modulating miR-26a/HMGB1 axis and Regulating NF-Κb Activation. Am. J. Transl Res. 12 (9), 5449–5464.
Tian, F., Yu, C., Wu, M., Wu, X., Wan, L., and Zhu, X. (2019). MicroRNA‐191 Promotes Hepatocellular Carcinoma Cell Proliferation by has_circ_0000204/miR‐191/KLF6 axis. Cel Prolif 52 (5), e12635. doi:10.1111/cpr.12635
Tsingas, M., Ottone, O. K., Haseeb, A., Barve, R. A., Shapiro, I. M., Lefebvre, V., et al. (2020). Sox9 Deletion Causes Severe Intervertebral Disc Degeneration Characterized by Apoptosis, Matrix Remodeling, and Compartment-specific Transcriptomic Changes. Matrix Biol. 94, 110–133. doi:10.1016/j.matbio.2020.09.003
Tu, F. L., Guo, X. Q., Wu, H. X., He, Z. Y., Wang, F., Sun, A. J., et al. (2020). Circ-0001313/miRNA-510-5p/AKT2 axis Promotes the Development and Progression of colon Cancer. Am. J. Transl Res. 12 (1), 281–291.
Umimura, T., Orita, S., Inage, K., Shiga, Y., Maki, S., Inoue, M., et al. (2021). Percutaneously-quantified Advanced Glycation End-Products (AGEs) Accumulation Associates with Low Back Pain and Lower Extremity Symptoms in Middle-Aged Low Back Pain Patients. J. Clin. Neurosci. 84, 15–22. doi:10.1016/j.jocn.2020.12.005
Wang, H., He, P., Pan, H., Long, J., Wang, J., Li, Z., et al. (2018b). Circular RNA Circ-4099 Is Induced by TNF-α and Regulates ECM Synthesis by Blocking miR-616-5p Inhibition of Sox9 in Intervertebral Disc Degeneration. Exp. Mol. Med. 50 (4), 1–14. doi:10.1038/s12276-018-0056-7
Wang, H., Zhu, Y., Cao, L., Guo, Z., Sun, K., Qiu, W., et al. (2021). circARL15 Plays a Critical Role in Intervertebral Disc Degeneration by Modulating miR-431-5p/DISC1. Front. Genet. 12, 669598. doi:10.3389/fgene.2021.669598
Wang, X., Wang, B., Zou, M., Li, J., Lü, G., Zhang, Q., et al. (2018a). CircSEMA4B Targets miR-431 Modulating IL-1β-induced Degradative Changes in Nucleus Pulposus Cells in Intervertebral Disc Degeneration via Wnt Pathway. Biochim. Biophys. Acta (Bba) - Mol. Basis Dis. 1864 (11), 3754–3768. doi:10.1016/j.bbadis.2018.08.033
Xiang, Q., Kang, L., Wang, J., Liao, Z., Song, Y., Zhao, K., et al. (2020). CircRNA-CIDN Mitigated Compression Loading-Induced Damage in Human Nucleus Pulposus Cells via miR-34a-5p/SIRT1 axis. EBioMedicine 53, 102679. doi:10.1016/j.ebiom.2020.102679
Xie, L., Huang, W., Fang, Z., Ding, F., Zou, F., Ma, X., et al. (2019). CircERCC2 Ameliorated Intervertebral Disc Degeneration by Regulating Mitophagy and Apoptosis through miR-182-5p/SIRT1 axis. Cel Death Dis 10 (10), 751. doi:10.1038/s41419-019-1978-2
Yang, L., Liu, Z., Ma, J., Wang, H., Gao, D., Zhang, C., et al. (2021b). CircRPPH1 Serves as a Sponge for miR-296-5p to Enhance Progression of Breast Cancer by Regulating FOXP4 Expression. Am. J. Transl Res. 13 (7), 7556–7573.
Yang, R. Z., Xu, W. N., Zheng, H. L., Zheng, X. F., Li, B., Jiang, L. S., et al. (2021a). Involvement of Oxidative Stress‐induced Annulus Fibrosus Cell and Nucleus Pulposus Cell Ferroptosis in Intervertebral Disc Degeneration Pathogenesis. J. Cel Physiol 236 (4), 2725–2739. doi:10.1002/jcp.30039
Yu, C., Tian, F., Liu, J., Su, M., Wu, M., Zhu, X., et al. (2019). Circular RNA cMras Inhibits Lung Adenocarcinoma Progression via Modulating miR‐567/PTPRG Regulatory Pathway. Cel Prolif 52 (3), e12610. doi:10.1111/cpr.12610
Zhang, F., Lin, F., Xu, Z., and Huang, Z. (2021c). Circular RNA ITCH Promotes Extracellular Matrix Degradation via Activating Wnt/β-Catenin Signaling in Intervertebral Disc Degeneration. Aging 13 (10), 14185–14197. doi:10.18632/aging.203036
Zhang, J., Chen, S., Yang, J., and Zhao, F. (2020). Accurate Quantification of Circular RNAs Identifies Extensive Circular Isoform Switching Events. Nat. Commun. 11 (1), 90. doi:10.1038/s41467-019-13840-9
Zhang, J., Hu, H., Zhao, Y., and Zhao, Y. (2018). CDR1as Is Overexpressed in Laryngeal Squamous Cell Carcinoma to Promote the Tumour's Progression via miR-7 Signals. Cel Prolif 51 (6), e12521. doi:10.1111/cpr.12521
Zhang, J., Hu, S., Ding, R., Yuan, J., Jia, J., Wu, T., et al. (2021b). CircSNHG5 Sponges Mir-495-3p and Modulates CITED2 to Protect Cartilage Endplate from Degradation. Front. Cel Dev. Biol. 9, 668715. doi:10.3389/fcell.2021.668715
Zhang, S., Hu, B., Liu, W., Wang, P., Lv, X., Chen, S., et al. (2021a). The Role of Structure and Function Changes of Sensory Nervous System in Intervertebral Disc-Related Low Back Pain. Osteoarthritis and Cartilage 29 (1), 17–27. doi:10.1016/j.joca.2020.09.002
Keywords: CircRNAs, lower back pain, ncRNAs, intervertebral disc degeneration, CircITCH
Citation: Xu D, Ma X, Sun C, Han J, Zhou C, Wong SH, Chan MTV and Wu WKK (2022) Circular RNAs in Intervertebral Disc Degeneration: An Updated Review. Front. Mol. Biosci. 8:781424. doi: 10.3389/fmolb.2021.781424
Received: 22 September 2021; Accepted: 13 December 2021;
Published: 06 January 2022.
Edited by:
Zhen Fan, Tongji University, ChinaReviewed by:
Rodrigo Paolo Flores Abuna, University of Sao Paulo, BrazilGuangjian Fan, Shanghai General Hospital, China
Copyright © 2022 Xu, Ma, Sun, Han, Zhou, Wong, Chan and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuanli Zhou, anVzdGluXzUyNTdAaG90bWFpbC5jb20=
 Derong Xu1
Derong Xu1 Chuanli Zhou
Chuanli Zhou