- 1Department of Obstetrics and Gynecology, International Peace Maternity and Child Health Hospital, Shanghai Key Laboratory of Embryo Original Diseases, and Institute of Birth Defects and Rare Diseases, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 2Shanghai Epiprobe Biotechnology Co., Ltd, Shanghai, China
- 3Department of Obstetrics and Gynecology, Jinshan Hospital of Fudan University, Shanghai, China
- 4Shanghai Public Health Clinical Center and Department of General Surgery, Huashan Hospital, Cancer Metastasis Institute and Laboratory of RNA Epigenetics, Institutes of Biomedical Sciences, Shanghai Medical College, Fudan University, Shanghai, China
Endometrial cancer (EC) is one of the most common gynecologic cancers in developed countries. Presently, it is imperative to develop a reliable, noninvasive, or minimally invasive detection method for EC. We explored the possibility of using DNA methylation marker from endometrial brush samples (with a “Tao brush”) and cervical scrapes (with a “Pap brush”) for early detection of EC. We analyzed the methylation data of EC and normal endometrial tissues from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) data sets. An optimized methylation-sensitive restriction enzyme combined with real-time fluorescent quantitative PCR (MSRE-qPCR) was used for methylation detection. Included in the training set were 143 endometrial tissues, 103 Tao, and 109 Pap brush samples. The validation set included 110 Tao and 112 Pap brush samples. PCDHGB7 was significantly hypermethylated in EC compared with normal endometrial tissues in the TCGA and GEO data sets (AUC >0.95), which was verified in clinical samples. In the Pap brush samples, the AUC was 0.86 with 80.65% sensitivity and 82.81% specificity, whereas the Tao brush samples exhibited higher specificity (95.31%). The combination of Tao and Pap brush samples significantly increased the sensitivity to 90.32%. In the validation set, the final model yielded a sensitivity of 98.61%, specificity of 60.53%, positive predictive value of 82.56%, and negative predictive value of 95.83%. These results demonstrate the potential application of the novel methylation marker, hypermethylated PCDHGB7, in cervical scrapings and endometrial brush, which provides a viable, noninvasive, or minimally invasive method for early endometrial cancer detection across different clinical features and histologies to supplement current hysteroscopy diagnosis.
Highlights
• This work reveals that PCDHGB7 is hypermethylated in EC by bioinformatic analysis and clinical validation.
• We systematically evaluated the performance of hypermethylated PCDHGB7 as a novel biomarker for EC detection in endometrial tissues and Tao and Pap brush samples.
• PCDHGB7 hypermethylation functions as an early stage biomarker for its emergence at the early stage of EC progression.
• The robustness of hypermethylated PCDHGB7 enables Tao and Pap brush samples to be minimally invasive or even noninvasive samples for early EC detection.
Introduction
Endometrial cancer (EC) is one of the most common malignant tumors of the female reproductive tract worldwide (Sung et al., 2021). In view of the increasing prevalence of EC risk factors, such as diabetes and obesity in the general population, EC’s incidence is expected to increase to 82,000 and 122,000 new cases per year in 2020 and 2030, respectively (Rahib et al., 2014). EC is highly curable in the early stages with a 5-year overall survival of 95% for stage I disease, which is only 19% in stage IV. Although most EC occurs in postmenopausal women, there has been a recent significant increase of EC occurrence ranging from 2% to14% in women aged 40 years or younger (Duska et al., 2001; Ota et al., 2005; Nagase et al., 2019). For young women of childbearing age, if diagnosed in the early stage of EC without myometrial invasion and extrauterine spread, they still hold the opportunity to retain the uterus and/or ovaries. Therefore, early diagnosis of EC is crucial, which reduces female mortality and strives for treatment opportunities in younger patients to retain fertility or reproductive endocrine function.
Presently, effective early detection or screening methods for EC are urgently needed. The current clinical evaluation is not carried out until the patient has symptoms, such as abnormal uterine bleeding (AUB) or postmenopausal bleeding (PMB) (Clarke et al., 2018). Due to the lack of noninvasive or minimally invasive triage diagnosis, women with AUB or PMB have to undergo invasive procedures to obtain endometrial tissues by curettage and hysteroscopic biopsy to exclude EC. Most ECs appear to develop from endometrial hyperplasia (EH) to atypical hyperplasia (AH) with progress stretching for years. During the long course of disease progression, AUB and PMB patients endure invasive endometrial biopsies repeatedly. Only 5%–10% of them have an underlying EC or an EC precursor (Clarke et al., 2018), resulting in physical, psychological, and economic pressures.
The most common detection method for EC in postmenopausal women without clinical symptoms is transvaginal ultrasound (TVS), which measures endometrium thickness. However, it is not reliable to distinguish between benign and malignant endometrium due to its high false-positive rate (Jacobs et al., 2011). Considering serous ECs (SECs) with a poor prognosis can be present in the atrophic endometrium, which is often diagnosed at an advanced stage (Urick and Bell, 2019), EC can also occur in women without endometrial thickening. Therefore, it is necessary to develop a noninvasive or minimally invasive and effective molecular method for early detection of EC among symptomatic and asymptomatic high-risk populations, such as women with Lynch syndrome or increased BMI and tamoxifen users (Costas et al., 2019) to avoid a large number of unnecessary invasive diagnostic workups for most women with benign endometrium.
It is well-known that genomic and epigenomic alterations participate in carcinogenesis in various human organs (Baylin and Jones, 2016; Jones et al., 2016). DNA methylation changes are among essential epigenomic alterations leading to chromosomal instability and aberrant expression of tumor-related genes (Kanai, 2010; Ahuja et al., 2016). Accumulating evidence reveals DNA methylation as a promising cancer biomarker (Widschwendter et al., 2018). Previously, our group found that PCDHGB7 was hypermethylated in various cancer types compared with their corresponding normal tissues, and hypermethylated PCDHGB7 was identified as a universal cancer only marker (UCOM). (Dong et al., 2019; Dong et al., 2021). However, hypermethylated PCDHGB7 has not been systematically investigated in EC until now. The shedding of EC cells and/or cell-free EC DNA into the lower genital tract provides an opportunity to leverage more sensitive molecular testing and less invasive biospecimen sampling methods comprising endometrial brushes (Wentzensen et al., 2014; Bakkum-Gamez et al., 2015), cervical scrapes (De Strooper et al., 2014; Huang et al., 2017; Liew et al., 2019), vaginal swabs, and vaginal tampons (Fiegl et al., 2004; Bakkum-Gamez et al., 2015; Sangtani et al., 2020).
Optimizing sample collection methods and identifying the molecular markers with the greatest sensitivity to detect EC and its precursors are all critical in the development of early detection among symptomatic and asymptomatic patients. In terms of sampling location, endometrial brush sampling is the closest to the neoplastic foci and can collect the most amount of DNA content in theory, and this has higher absolute methylation percentages compared with the cervical scrape samples also taken by physicians. However, the endometrial brush’s main flaw is that it is to be inserted into the uterine cavity with an unsuccessful insertion rate of 20% in nulliparous and 8% in parous women (Williams et al., 2008). Cervical scrape sampling is a relatively noninvasive and more convenient method, and it may be more accepted even for postmenopausal women.
The present study aimed to identify PCDHGB7 methylation in EC and to further compare the performance of PCDHGB7 hypermethylation in the detection of EC by cervical scrapes (with a “Pap brush”) with that of endometrial brush sampling (with a “Tao brush”) so as to provide evidence for adopting a more suitable, noninvasive or minimally invasive sampling method of DNA methylation markers in clinical settings for women with AUB or PMB and within the asymptomatic detection population.
Materials and Methods
The Cancer Genome Atlas and Gene Expression Omnibus DNA Methylation Data Analysis
The Illumina 450 K methylation array data from The Cancer Genome Atlas (TCGA) database was downloaded from the UCSC Xena browser (https://xenabrowser.net/). The absolute methylation values were calculated from the β values of a 450 K methylation array [methylation value = (β value +0.5) ×100%]. The only six probes (cg13933262, cg14011639, cg10435816, cg23563234, cg08938584, cg17011276) within the CpG island in the PCDHGB7 promoter region were used, the final methylation value of each sample was calculated by the average of these six probes. The basic information and methylation levels of all samples from TCGA project are listed in Supplementary Table S1. The methylation array data sets (from the GSE136791, GSE93589, GSE67116, GSE33422, and GSE40032 series) from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) were downloaded. The absolute methylation values were calculated by the same method as above. A total of 11 probes (cg14011639, cg27487435, cg08938584, cg05558169, cg14378860, cg23563234, cg13933262, cg10435816, cg01643675, cg03510378, cg17011276) of EPIC methylation array, six probes (cg13933262, cg14011639, cg10435816, cg23563234, cg08938584, cg17011276) of 450 K methylation array, and two probes (cg14011639, cg23563234) of 27 K methylation array within the CpG island in the PCDHGB7 promoter region were used, respectively, and the final methylation value of each sample was calculated by the average of all used probes. The basic information and methylation level of all samples from GEO database are listed in Supplementary Table S2.
Clinical Samples Collection
A total of 577 samples, including 355 samples in the training set and 222 samples in the validation set (Supplementary Table S3), were collected from the International Peace Maternity and Child Health Hospital, School of Medicine, Shanghai Jiaotong University, Shanghai, China. The study was approved by the Ethics Committee of International Peace Maternity and Child Health Hospital. Informed consent was acquired from all patients and control subjects.
The benign endometrium (BE) group (normal endometrium (NE) and EH patients) was enrolled before hysteroscopy or hysterectomy, including women with an endometrium pathology diagnosis by hysteroscopy because of PMB, AUB, increased endometrial thickness, or by hysterectomy for benign indications. The EC and AH patients newly diagnosed by hysteroscopy or curettage and endometrial biopsy were prospectively enrolled prior to hysterectomy. Women were excluded if they had recurrent EC, other known cancers, or received neoadjuvant chemotherapy or radiotherapy. In the cases of EC and AH, patients underwent cervical scrape sampling with a Cervex-Brush® (Rovers Medical Devices B.V., Oss, Netherlands) and endometrial brush sampling with a Tao brush (Cook Medical, Bloomington IN) by the physician before the day of hysterectomy or hysteroscopy, which were rinsed in SurePathTM Preservative Fluid (TriPath Imaging®, Inc., Burlington, United States), respectively, and used for subsequent testing. Moreover, control group specimens were collected before their hysterectomy or hysteroscopy by the same method. Fresh-frozen tissue specimens were collected immediately after hysterectomy or hysteroscopy.
DNA Extraction
Genomic DNA (gDNA) from endometrial tissue samples was extracted with the TIANamp Genomic DNA Kit (Tiangen Biotech, DP304), and gDNA from Pap brush and Tao brush samples was extracted with the EP Genomic DNA Kit (Epiprobe Biotech, K-21), both following the manufacturer’s protocol. The quantity and quality of gDNA were determined with Nanodrop 2000 (Thermo Scientific, Waltham, Massachusetts, United States).
MSRE-qPCR and DNA Methylation Evaluation
Methylation-sensitive restriction enzyme-combined real-time fluorescent quantitative PCR (MSRE-qPCR) was used to detect PCDHGB7 methylation as described previously (Dong et al., 2021). GAPDH gene-related region absence of any cutting sites was selected for normalization. For each digestion reaction, 100 ng gDNA was taken as input, and every two units of the HhaI (NEB, R0139) and HpaII (NEB, R0171) were added, making the final volume 25 μL, followed by digestion at 37°C for 30 min and heat inactivation at 95°C for 5 min. The subsequent dual real-time PCR was performed on the 7500 Real-Time PCR System (Life Technologies) with a program as follows: initiation at 95°C for 10 min, then 45 cycles of 94°C for 20 s and 60°C for 60 s. The DNA methylation level for each sample was evaluated by ΔCt = Ct_PCDHGB7 – Ct_GAPDH.
Statistical Analysis
p values were calculated using the two-tailed nonparametric Mann–Whitney test by GraphPad Prism 7.0. ROC analysis was conducted using GraphPad Prism 7.0. The sensitivity, specificity, positive predictive values (PPVs), and negative predictive values (NPVs) were calculated by clinical calculator 1 (http://vassarstats.net/clin1.html).
Results
PCDHGB7 Hypermethylation as a Biomarker in EC Detection
In our previous study, PCDHGB7 was found to be hypermethylated in various types of cancer and identified as a universal cancer only marker (UCOM). Dong et al. (2019); Dong et al., 2021). Therefore, it is intriguing to investigate its potential application in alternative carcinoma detection sequentially. Methylation microarray data from the TCGA and GEO databases were analyzed. As expected, the methylation level of PCDHGB7 in EC endometrial tissue samples was higher compared with normal tissue samples in 478 samples from the TCGA database and in 269 samples from the GEO database (Figures 1A,B). In addition, we evaluated the performance of PCDHGB7 methylation as a biomarker for EC by receiver operating characteristic (ROC) analysis, and the results showed that the area under the curve (AUC) was 0.98 and 0.95, respectively (Figure 1C), suggesting that PCDHGB7 hypermethylation holds potential to be a diagnostic marker for EC.

FIGURE 1. Hypermethylated PCDHGB7 acted as a potential marker in EC in TCGA and GEO data sets. (A) PCDHGB7 was hypermethylated in 432 EC endometrial tissues compared with 46 normal endometrial tissues in the TCGA database. (B) PCDHGB7 was hypermethylated in 246 EC endometrial tissues compared with 23 normal endometrial tissues in GEO database. (C) The ROC curve of hypermethylated PCDHGB7 in endometrial tissues from TCGA database and GEO database. (D) PCDHGB7 methylation level in 468 endometrial tissues with determined stage from TCGA database. (E) The ROC curve of hypermethylated PCDHGB7 in endometrial tissues with different stages from the TCGA database. In both (A, B) and (D), p values were calculated using the two-tailed unpaired parametric test by GraphPad Prism 7.0. ****, p < .0001.
Moreover, to examine its probable application in early stage detection, we sorted 468 primary endometrial tissue samples with stages determined from the TCGA database and conducted the ROC curve analysis. The analysis shows significant differences of PCDHGB7 methylation levels between normal and EC tissue samples of Stages I and II∼IV (Figure 1D). Meanwhile, the AUC of Stage I, II, III, and IV was 0.93, 0.95, 0.98, and 1.00, respectively (Figure 1E), suggesting that hypermethylation of PCDHGB7 is a powerful marker for early stage ECs.
Validation of PCDHGB7 Hypermethylation in Endometrial Tissue Samples From EC Patients
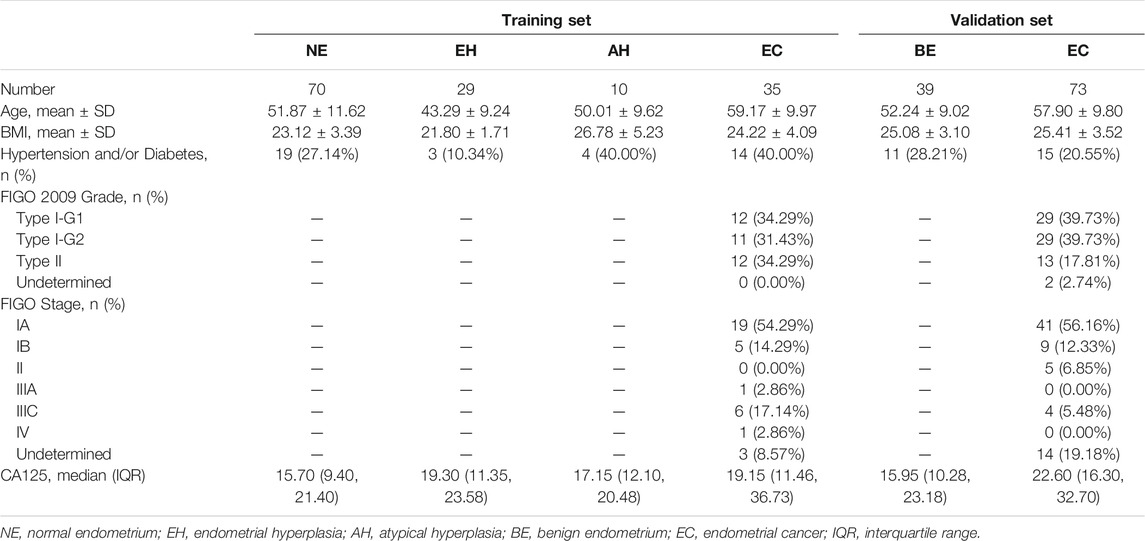
Overall, 355 samples of the training set, including endometrial tissue samples, Pap brush samples, and Tao brush samples from 144 individuals, were included in this study, including 70 patients with NE, 29 patients with EH, 10 patients with AH, and 35 patients with EC, whereas 24 of 35 EC patients were in the early stage (Stage I). The statistics of samples refers to Supplementary Table S3. The age, BMI, histopathologic diagnosis, stage, and other clinics’ pathologic information for the participants are shown in Table 1. To assess whether the differences in age and BMI affect the methylation level of PCDHGB7, we analyzed the correlation between PCDHGB7 methylation and age/BMI in both the TCGA database and clinical samples enrolled in the study. It turned out that the methylation level of PCDHGB7 showed no relevance to age or BMI regardless of pathological groups (Supplementary Figure S1).
The flowchart for the study is shown in Figure 2A. To evaluate the bioinformatics analysis results, we validated the methylation level of PCDHGB7 in 33 endometrial cancer samples from EC patients, and 105 BE or tumor-adjacent endometrial tissue samples using MSRE-qPCR (Supplementary Table S4). As expected, the methylation level of PCDHGB7 was found to be significantly higher in endometrial tissues from EC patients than in endometrial tissue samples of the BE group (Figure 2B). Additionally, we also found that PCDHGB7 methylation was significantly higher in tissue samples from patients with EC/AH than patients with NE or EH (Figure 2C), demonstrating that detection of hypermethylated PCDHGB7 using MSRE-qPCR contributes to early stage detection of EC by distinguishing AH from EH in endometrial tissue samples.
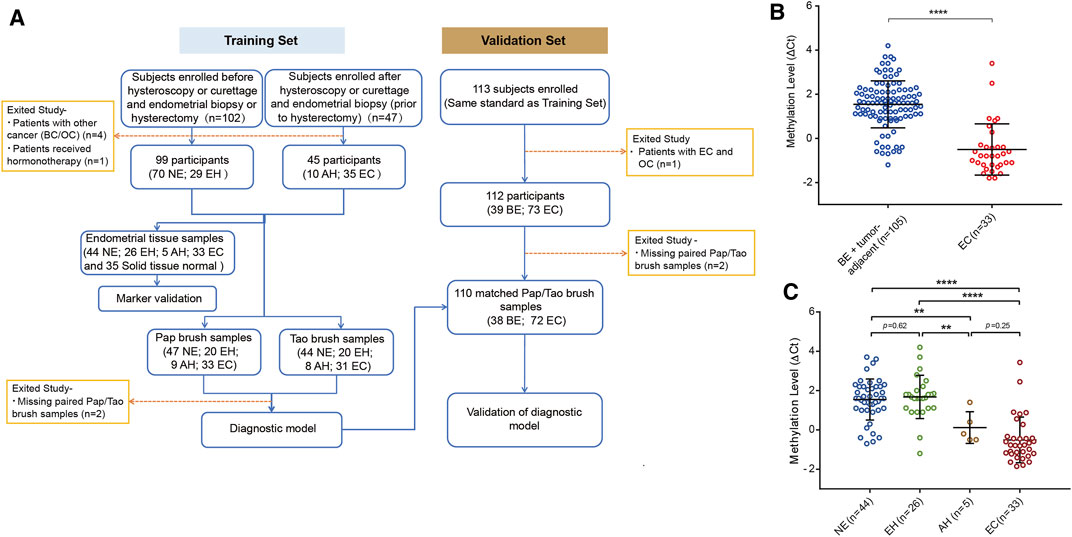
FIGURE 2. Methylation status of PCDHGB7 had the capacity to make distinctions between BEs and ECs in clinical endometrial tissue samples. (A) The workflow of this study. NE, Normal endometrium; EH, Endometrial hyperplasia without atypia; BE, benign endometrium (NE + EH); AH, Atypical hyperplasia; EC, Endometrial cancer; BC, Breast cancer; OC, Ovarian cancer. (B) PCDHGB7 methylation level in 138 endometrial tissue samples by MSRE-qPCR. (C) PCDHGB7 methylation level in NE, EH, AH, and EC groups. In both (B, C), p values were calculated using the two-tailed unpaired parametric test by GraphPad Prism 7.0. **, p < .01; ****, p < .0001.
Evaluation of PCDHGB7 Hypermethylation in Pap Brush and Tao Brush Samples From Patients With EC and AH
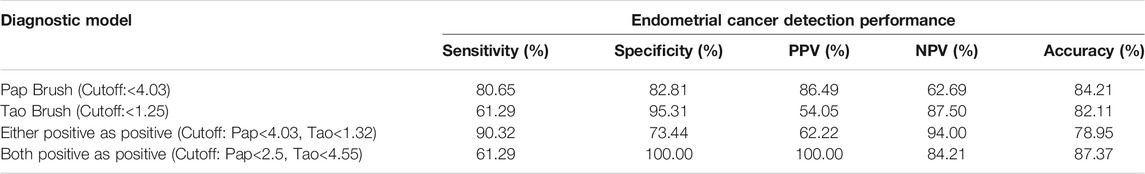
To make our EC detection method more applicable and cost-effective, we applied our assay in Tao brush samples of 44 patients with NE, 20 patients with EH, 8 patients with AH, and 31 patients with EC as well as Pap brush samples of 47 patients with NE, 20 patients with EH, 9 patients with AH, and 33 patients with EC (Supplementary Tables S3, S4). We found that PCDHGB7 methylation was significantly higher in Tao brush samples from patients with EC than in NE and EH, whereas the difference between AH and EH showed no statistical significance (Figure 3A). However, significant differences between PCDHGB7 methylation levels were found between Pap brush samples of AH and EH (Figure 3B). Furthermore, the clinical performance of PCDHGB7 hypermethylation in the detection of EC from Pap brush and Tao brush samples were calculated. In Tao brush samples, PCDHGB7 achieved ROC curves of 0.83 (95% CI = 0.73–0.93), and in Pap brush samples, it resulted in ROC curves of 0.86 (95% CI = 0.77–0.95) (Figure 3C). Meanwhile, the sensitivity and specificity of PCDHGB7 methylation were 80.65% and 82.81% in Pap brush samples, whereas in Tao brush samples, the specificity (95.31%) was higher though with 61.29% sensitivity in the detection of EC (Figure 3D). Considering early stage EC is operable and largely curable, it is crucial to evaluate the robustness of hypermethylated PCDHGB7 in early stage EC detection. As shown in Figure 3C, the performance of PCDHGB7 hypermethylation in detection of Stage I EC (AUC = 0.88 in Pap brush and AUC = 0.83 in Tao brush) was comparable of it in detection of all EC samples, which was also supported by the sensitivity and specificity data in Figure 3D.
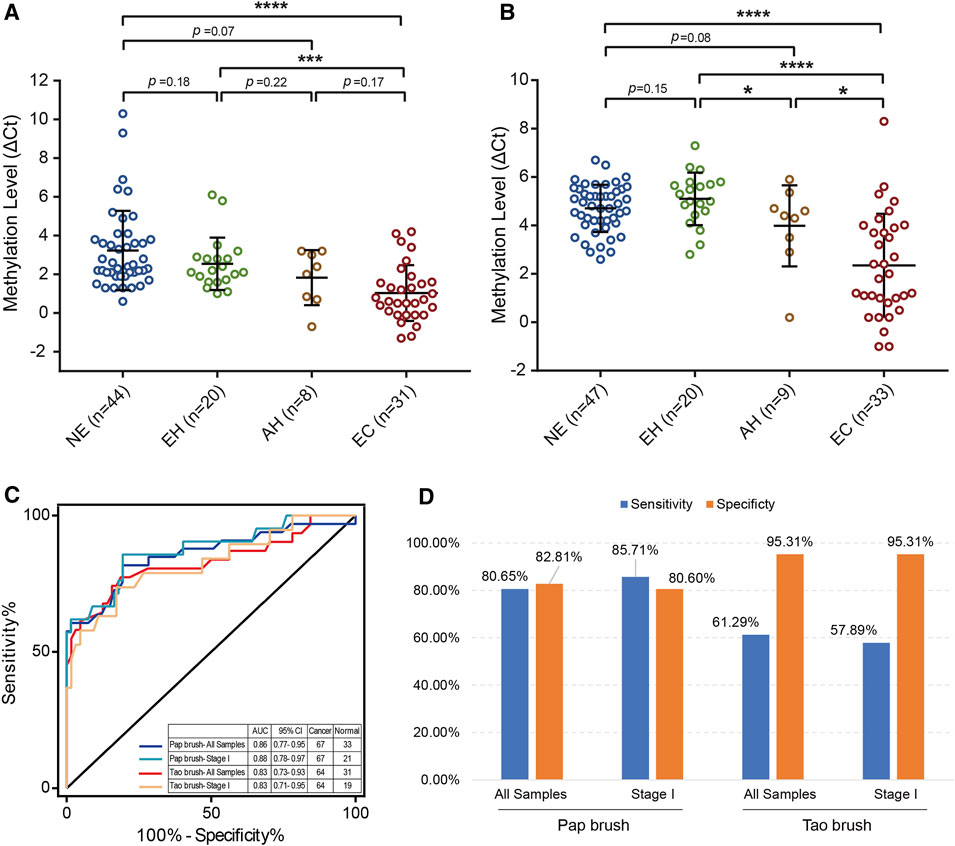
FIGURE 3. Detection of EC by using hypermethylated PCDHGB7 in Pap brush and Tao brush. (A) PCDHGB7 methylation level in 103 Tao brush samples by MSRE-qPCR. (B) PCDHGB7 methylation level in 109 Pap brush samples by MSRE-qPCR. (C) The ROC curves of hypermethylated PCDHGB7 in Pap brush and Tao brush samples for detecting ECs and early stage ECs. (D) The sensitivity and specificity of PCDHGB7 hypermethylation in Pap brush and Tao brush samples. In both (A, B), p values were calculated using the two-tailed unpaired parametric test by GraphPad Prism 7.0. *, p < .1; ***, p < .001; ****, p < .0001.
Moreover, similar to the TCGA database results, significant differences in PCDHGB7 methylation levels can be found between EC and BE groups of early stage and early grade in Tao brush and Pap brush samples (Supplementary Figures S2A–S2D). Moreover, the methylation level of PCDHGB7 was also significantly different in patients with tumor size less than 2 cm compared with BE groups (NEs and EHs) (Supplementary Figures S2E, S2F). Additionally, hypermethylation of PCDHGB7 in Tao brush and Pap brush has also been observed in patients without myometrial invasion, vascular invasion, or AUB/PMB (Figure Supplementary Figures S2G–2L). These results support that detecting PCDHGB7 hypermethylation in Tao brush and Pap brush samples are applicable in the early diagnosis of EC.
To further assess its application value in detecting EC, we compared the performance of hypermethylated PCDHGB7 and the widely implemented EC biomarker, serum CA125 (Farias-Eisner et al., 2010). It demonstrated that serum CA125 of normal and cancer samples were largely overlapped (Supplementary Figure S3A). The inability of discriminating between normal and cancer samples indicates serum CA125 is a terrible marker for EC detection, which is further supported by the low AUC (0.61) in detecting EC (Supplementary Figure S3B). Under the cutoff (CA125 = 35 U/ml), the sensitivity of serum CA125 was 24.78%, which was far from satisfaction, though the specificity was 92.31%. The performance of hypermethylated PCDHGB7 in both Pap and Tao brushes significantly outperformed serum CA125, suggesting that hypermethylated PCDHGB7 was a robust marker for noninvasive or minimally invasive detection of EC.
Construction and Validation of the Diagnostic Model for the Detection of Early Stage EC From Tao and Pap Brush Samples
We assessed whether the combination of hypermethylated PCDHGB7 in Tao brush and Pap brush might provide better performance for EC early stage detection in terms of sensitivity and specificity. As shown in Table 2, the sensitivity and specificity (either positive as positive) of combined assay were 90.32% and 73.44%, respectively (PPV = 62.22%, NPV = 94.00%, accuracy = 78.95%). Furthermore, the sensitivity and specificity of both positive as positive were 61.29% and 100%, respectively (PPV = 100%, NPV = 84.21%, accuracy = 87.37%). Hypermethylated PCDHGB7 was designated to be used for the early detection (screening or auxiliary diagnosis) of EC by using Pap or Tao brushes in the current study. In these application scenarios, sensitivity was the overarching priority; thus, the first model was chosen as the final diagnostic model.
In addition, the results of the training set were further validated in an independent testing set of 112 patients that included 73 EC and 39 BE groups (Supplementary Tables S3, S5). The EC group included 50 patients (68.49%) in the early stage (41 Stage IA, 9 Stage IB). The methylation level of PCDHGB7 in Tao brush (Figure 4A) and Pap brush (Figure 4B) were both higher than those in controls. The clinical performance of PCDHGB7 was calculated in the detection of EC from Tao brush and Pap brush, separately and collectively. The diagnostic model resulted in sensitivity of 98.61% and specificity of 60.53% (PPV = 82.56%, NPV = 95.83%, accuracy = 85.45%) (Figure 4C).
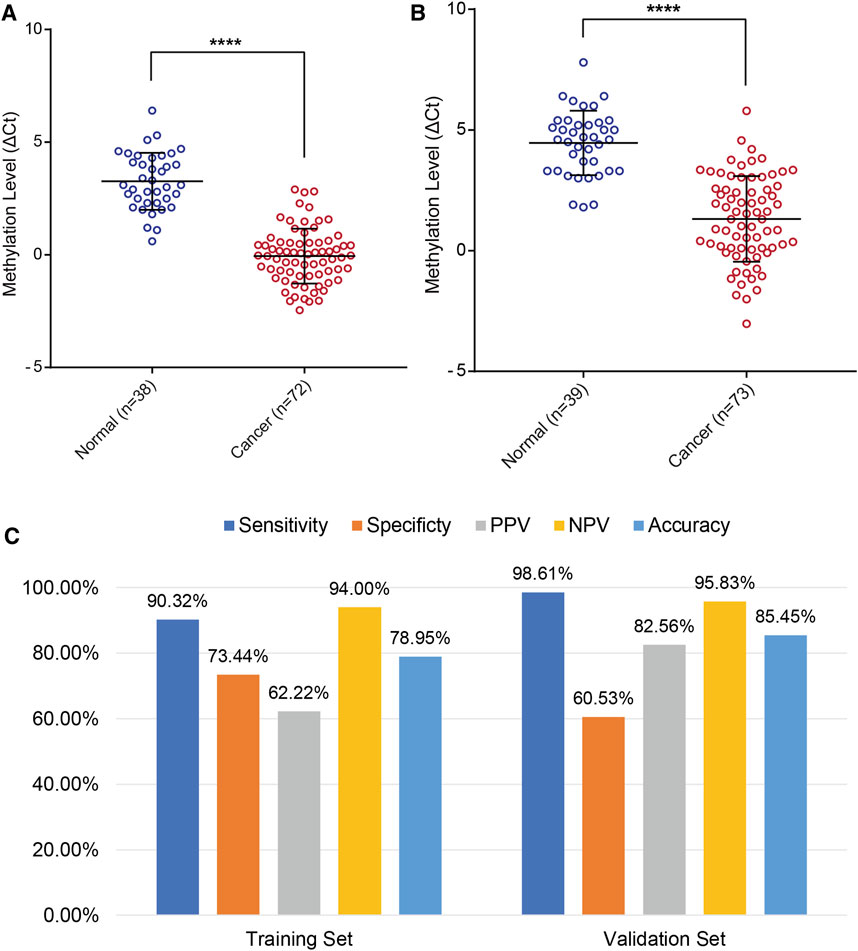
FIGURE 4. Validation of PCDHGB7 performance for the detection of EC in validation set. (A) PCDHGB7 methylation level in 68 Tao brush samples by MSRE-qPCR. (B), PCDHGB7 methylation level in 69 Pap brush samples by MSRE-qPCR. (C) The performance of PCDHGB7 hypermethylation diagnostic model in training set and validation set. In both (A, B), p values were calculated using the two-tailed unpaired parametric test by GraphPad Prism 7.0. ****, p < .0001.
Discussion
PCDHGB7 is a member of protocadherins, which plays an inhibitory role in tumorigenesis and cancer progression by inducing cell cycle arrest and apoptosis (Hou et al., 2019). Previous studies demonstrate that PCDHGB7 is crucial in the process of self-recognition and mutual recognition between synapses, the movement of synapses, and the establishment of the nervous system network (Sano et al., 1993; Frank and Kemler, 2002; Junghans et al., 2005; Morishita and Yagi, 2007). PCDHGB7 is reported to be significantly hypermethylated in non-Hodgkin’s lymphoma (Shi et al., 2007). Besides, PCDHGB7 hypermethylation gene was detected in approximately 80% of breast cancer, and PCDHGB7 expression was reduced in breast cancer tissue (Shan et al., 2016a; Shan et al., 2016b). Importantly, we raised the concept of universal cancer only marker (UCOM) and identified hypermethylated PCDHGB7 as a UCOM (Dong et al., 2019; Dong et al., 2021). This is the first study that enables PCDHGB7 methylation in endometrial tissue and Pap/Tao brush samples for early EC detection. The great performance of UCOM, hypermethylated PCDHGB7, in noninvasive early detection of EC not only extends it clinical applications, but also proves the robustness of UCOM once again. The potential applications of UCOM in other cancer types deserve further investigation.
In this study, we verify that hypermethylated PCDHGB7 can be used to early detect EC by Pap and Tao brush sampling even in EC patients of stage IA without myometrial invasion, G1, tumor < 2 cm in greatest dimension, lymphovascular space invasion (LVSI) (-) and without AUB/PMB (Additional file 2: Supplementary Figure S2). However, we did not observe different methylation levels of PCDHGB7 in EC among various tumor sizes, histologic subtypes, and stages with or without myometrial invasion and LVSI, which reports similar results in previous studies (Bakkum-Gamez et al., 2015; Liew et al., 2019). It is indicated that ECs may share common epigenetic events, and that DNA methylation changes regardless of genetic heterogeneity and clinicopathology (Liew et al., 2019). However, it was encouraging that significant hypermethylated PCDHGB7 could be detected via Pap or Tao brush in early low-risk EC patients of stage IA without myometrial invasion, G1, tumor < 2 cm in greatest dimension and LVSI (-), which are suitable for a fertility preservation procedure. Thus, DNA methylation marker PCDHGB7 by noninvasive or minimally invasive sampling is promising for early diagnosis of EC in women of childbearing age, so these women can choose conservative alternatives to hysterectomy. Taken together, these data suggest that PCDHGB7 hypermethylation may hold potential to increase the likelihood of women achieving disease-free survival and provide opportunities for the EC women of childbearing age to choose fertility preservation treatment. Moreover, monitoring of PCDHGB7 methylation level from Pap/Tao brush samples may provide a convenient and noninvasive approach as a viable alternative to conventional endometrial surveillance during conservative treatment, which requires repeated endometrial biopsy every 3 months until the histological regression.
Pap brush and Tao brush samples are noninvasive or minimally invasive and conveniently obtained during routine office visits without anesthesia. The quality of the two samplings, which are taken by qualified physicians, is robust compared with cervicovaginal self-sampling methods. The Tao brush allows the sampler to be inserted to the intrauterine cavity, which is the closest to the anatomical sites of the tumors without injury to the myometrium or contamination from the endocervical canal (Liew et al., 2019). However, its main flaw is its unsuccessful insertion rate (Williams et al., 2008). In the present study, we failed to insert to the intrauterine cavity for seven women (2 EC, 1 AH, 4 BE). Therefore, we implemented cervical scrape sampling with Pap brush, which is aimed to sample the exocervical and endocervical canal before Tao brush sampling. Cervical scrapes are usually used for cervical cancer detection in the general population, causing minimal discomfort and hence being widely accepted by women. In our study, all cervical scrape specimens were taken successfully and yielded sufficient DNA for methylation analysis. Despite the results diverging from our expectations, we encouragingly found that the clinical performance of PCDHGB7 hypermethylation in the detection of EC from Pap brush was more excellent than that from Tao brush samples evidenced by ROC curve analysis (AUC = 0.86 vs. AUC = 0.83), the sensitivity of PCDHGB7 methylation in the Pap brush was superior to that of the Tao brush (80.65 vs. 61.29%), whereas the specificity of PCDHGB7 methylation in the Tao brush was higher than that of the Pap brush as expected (95.31 vs. 82.81%). There are two possible explanations for this unexpected but enticing finding. One possible reason is that the sample size of our study is relatively small, and the marker performance could be influenced by clinical characteristics during population selection. The median tumor diameter was 2.6 cm (1.4, 4.1) in the present study, whereas the maximum tumor diameter < 2 cm was in 8 EC whose tumor may be localized to a small area of endometrium and may go undetected with the Tao brush. The other possible explanation is that the Tao brush has the disadvantage of not collecting enough endometrial cells of the uterine horns due to its round configuration (Du et al., 2016). The Pap brush was sampled from the cervix, serving as a conduit for malignant endometrial cells shedding from the whole uterine cavity, including uterine horns then gathering into endocervical canal, which could facilitate the sensitivity of Pap brush sampling method. Considering the individual advantages of Tao brush and Pap brush sampling, we suggest the combined diagnostic model (either positive as positive) if the Tao brush can be inserted into the intrauterine cavity successfully. Meanwhile, just using Pap brush specimens to detect PCDHGB7 hypermethylation is acceptable as a complementary sampling method for those unsuccessful uterine cavity insertions as they are less invasive and more acceptable. In addition, we found significant PCDHGB7 hypermethylation occurring within the Pap brush specimens of both AUB or PMB and asymptomatic EC women. As such, the utilization of the Pap brush for a DNA methylation marker as a biospecimen collection device carries promise as a means of detection in high-risk asymptomatic populations, particularly in postmenopausal women with atrophic uterus.
In spite of these novel findings, this study has a few limitations to be overcome. First, considering it was a single-center study, the inclusion of samples from multicenters will enhance the robustness of the conclusions. Moreover, the relatively small sample size of thevalidation set yielded high sensitivity and low specificity, suggested that the current diagnostic model might need a slight adjustment before direct application to clinical settings. In future studies, we propose to verify the clinical diagnostic accuracy in larger, prospective, and unbiased cohorts, especially for the AH group. Given endometrial surveillance, one should include repeated endometrial biopsy every 3 months until histological regression. Further studies are required to elucidate the role of the PCDHGB7 methylation marker in surveillance of early EC or AH conservative treatment by a noninvasive or minimally invasive sampling method as well as consecutive surveillance of PCDHGB7 methylation level using Pap or Tao brush sampling. Moreover, future multicenter clinical trials of PCDHGB7 methylation via Pap and Tao brush sampling in women with AUB or postmenopausal bleeding and in high-risk asymptomatic populations should be planned.
Conclusion
We demonstrate the usefulness of hypermethylated PCDHGB7 as a biomarker for noninvasive or minimally invasive early EC detection. Combining the Tao brush with Pap brush sampling could acquire better detection performance, whereas a Pap brush would be more practical in detection settings in wider asymptomatic populations.
Data Availability Statement
All data generated or analyzed during this study are included in this article (and its Supplementary Material files).
Ethics Statement
The study was approved by the Ethics Committee of International Peace Maternity and Child Health Hospital. Informed consent was acquired from all patients and control subjects.
Author Contributions
SD and ZM had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Conception and design: SD, YW, WC, and WY. Acquisition of data (acquired and managed patients, provided facilities, etc.): JY, ZM, CW, XX, and ZZ. Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): ZM, JY, SD, and PX. Writing, review, and/or revision of the manuscript: PX, ZM, SD, JY, YW, and TZ. Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): JY, QL, and YW. Study supervision: WC. All authors have approved the submitted version and have agreed both to be personally accountable for the author’s own contributions and to ensure that questions related to the accuracy or integrity of any part of the work, even ones in which the author was not personally involved, are appropriately investigated, resolved, and the resolution documented in the literature. All authors read and approved the final manuscript.
Funding
National Key R&D Program of China (2018YFC1005004), the Science and Technology Innovation Action Plan of Shanghai (17411950900), the National Natural Science Foundation of China (NSFC) (31671308, 31872814, 81172477, 81402135), Major Special Projects of Basic Research of Shanghai Science and Technology Commission (18JC1411101), Shanghai Science and Technology Commission (21Y11906600), Xuhui District Health and Family Planning Commission Important Disease Joint Research Project (XHLHGG201805), Traditional Chinese and Western medicine in Shanghai (No. 18411963500), The Interdisciplinary Program of Shanghai Jiao Tong University (ZH2018ZDA31). Shanghai Municipal Key Clinical Specialty (shslczdzk06302), Science and Technology Innovation Action Plan International Science and Technology Cooperation Project (20550760600), Shanghai Shenkang Hospital Development Center, Clinical Technology Innovation Project (SHDC12020130), Science and Technology Commission of Xuhui District and Health Commission of Xuhui District, Shanghai (XHLHGG202111), the Project of the Science and Technology Commission of Shanghai Municipality (20Y11914100, 17441907400), the Project of the Science and Technology Commission of Shanghai Xuhui District (2020-018), Shanghai Jiao Tong University Medicine-Engineering Fund (YG2017MS41), Shanghai Jiao Tong University School of Medicine, multi-center clinical research project (DLY201827).
Conflict of Interest
SD is one of the co-founders and the R & D director of Shanghai Epiprobe Biotechnology Co., Ltd. ZM and CW are employed by Epiprobe. WY serves on the scientific advisory boards of Epiprobe. SD has pending patents related to this work.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Yue Yu for editorial help and comments on the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.774215/full#supplementary-material
References
Ahuja, N., Sharma, A. R., and Baylin, S. B. (2016). Epigenetic Therapeutics: A New Weapon in the War against Cancer. Annu. Rev. Med. 67, 73–89. doi:10.1146/annurev-med-111314-035900
American cancer society [Online] (2019). Available: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2019.html (Accessed.
Bakkum-Gamez, J. N., Wentzensen, N., Maurer, M. J., Hawthorne, K. M., Voss, J. S., Kroneman, T. N., et al. (2015). Detection of Endometrial Cancer via Molecular Analysis of DNA Collected with Vaginal Tampons. Gynecol. Oncol. 137, 14–22. doi:10.1016/j.ygyno.2015.01.552
Baylin, S. B., and Jones, P. A. (2016). Epigenetic Determinants of Cancer. Cold Spring Harb Perspect. Biol. 8, a019505. doi:10.1101/cshperspect.a019505
Clarke, M. A., Long, B. J., Del Mar Morillo, A., Arbyn, M., Bakkum-Gamez, J. N., and Wentzensen, N. (2018). Association of Endometrial Cancer Risk with Postmenopausal Bleeding in Women. JAMA Intern. Med. 178, 1210–1222. doi:10.1001/jamainternmed.2018.2820
Costas, L., Frias‐Gomez, J., Guardiola, M., Benavente, Y., Pineda, M., Pavón, M. Á., et al. (2019). New Perspectives on Screening and Early Detection of Endometrial Cancer. Int. J. Cancer 145, 3194–3206. doi:10.1002/ijc.32514
De Strooper, L. M. A., Van Zummeren, M., Steenbergen, R. D. M., Bleeker, M. C. G., Hesselink, A. T., Wisman, G. B. A., et al. (2014). CADM1, MAL and miR124-2 Methylation Analysis in Cervical Scrapes to Detect Cervical and Endometrial Cancer. J. Clin. Pathol. 67, 1067–1071. doi:10.1136/jclinpath-2014-202616
Dong, S., Lu, Q., Xu, P., Chen, L., Duan, X., Mao, Z., et al. (2021). Hypermethylated PCDHGB7 as a Universal Cancer Only Marker and its Application in Early Cervical Cancer Screening. Clin. Transl Med. 11, e457. doi:10.1002/ctm2.457
Dong, S., Li, W., Wang, L., Hu, J., Song, Y., Zhang, B., et al. (2019). Histone-Related Genes Are Hypermethylated in Lung Cancer and Hypermethylated HIST1H4F Could Serve as a Pan-Cancer Biomarker. Cancer Res. 79, 6101–6112. doi:10.1158/0008-5472.can-19-1019
Du, J., Li, Y., Lv, S., Wang, Q., Sun, C., Dong, X., et al. (2016). Endometrial Sampling Devices for Early Diagnosis of Endometrial Lesions. J. Cancer Res. Clin. Oncol. 142, 2515–2522. doi:10.1007/s00432-016-2215-3
Duska, L. R., Garrett, A., Rueda, B. R., Haas, J., Chang, Y., and Fuller, A. F. (2001). Endometrial Cancer in Women 40 Years Old or Younger. Gynecol. Oncol. 83, 388–393. doi:10.1006/gyno.2001.6434
Farias-Eisner, G., Su, F., Robbins, T., Kotlerman, J., Reddy, S., and Farias-Eisner, R. (2010). Validation of Serum Biomarkers for Detection of Early- and Late-Stage Endometrial Cancer. Am. J. Obstet. Gynecol. 202, 73e71–5. doi:10.1016/j.ajog.2009.07.049
Fiegl, H., Gattringer, C., Widschwendter, A., Schneitter, A., Ramoni, A., Sarlay, D., et al. (2004). Methylated DNA Collected by Tampons-a New Tool to Detect Endometrial Cancer. Cancer Epidemiol. Biomarkers Prev. 13, 882–888.
Frank, M., and Kemler, R. (2002). Protocadherins. Curr. Opin. Cel Biol. 14, 557–562. doi:10.1016/s0955-0674(02)00365-4
Hou, S., Shan, M., Gao, C., Feng, X., Yang, Y., Zhang, R., et al. (2019). PCDHGB7 Increases Chemosensitivity to Carboplatin by Inhibiting HSPA9 via Inducing Apoptosis in Breast Cancer. Dis. Markers 2019, 6131548. doi:10.1155/2019/6131548
Huang, R.-L., Su, P.-H., Liao, Y.-P., Wu, T.-I., Hsu, Y.-T., Lin, W.-Y., et al. (2017). Integrated Epigenomics Analysis Reveals a DNA Methylation Panel for Endometrial Cancer Detection Using Cervical Scrapings. Clin. Cancer Res. 23, 263–272. doi:10.1158/1078-0432.ccr-16-0863
Jacobs, I., Gentry-Maharaj, A., Burnell, M., Manchanda, R., Singh, N., Sharma, A., et al. (2011). Sensitivity of Transvaginal Ultrasound Screening for Endometrial Cancer in Postmenopausal Women: a Case-Control Study within the UKCTOCS Cohort. Lancet Oncol. 12, 38–48. doi:10.1016/s1470-2045(10)70268-0
Jones, P. A., Issa, J.-P. J., and Baylin, S. (2016). Targeting the Cancer Epigenome for Therapy. Nat. Rev. Genet. 17, 630–641. doi:10.1038/nrg.2016.93
Junghans, D., Haas, I. G., and Kemler, R. (2005). Mammalian Cadherins and Protocadherins: about Cell Death, Synapses and Processing. Curr. Opin. Cel Biol. 17, 446–452. doi:10.1016/j.ceb.2005.08.008
Kanai, Y. (2010). Genome-wide DNA Methylation Profiles in Precancerous Conditions and Cancers. Cancer Sci. 101, 36–45. doi:10.1111/j.1349-7006.2009.01383.x
Liew, P.-L., Huang, R.-L., Wu, T.-I., Liao, C.-C., Chen, C.-W., Su, P.-H., et al. (2019). Combined Genetic Mutations and DNA-Methylated Genes as Biomarkers for Endometrial Cancer Detection from Cervical Scrapings. Clin. Epigenet 11, 170. doi:10.1186/s13148-019-0765-3
Morishita, H., and Yagi, T. (2007). Protocadherin Family: Diversity, Structure, and Function. Curr. Opin. Cel Biol. 19, 584–592. doi:10.1016/j.ceb.2007.09.006
Nagase, S., Ohta, T., Takahashi, F., and Enomoto, T. (2019). Annual Report of the Committee on Gynecologic Oncology, the Japan Society of Obstetrics and Gynecology: Annual Patients Report for 2015 and Annual Treatment Report for 2010. J. Obstet. Gynaecol. Res. 45, 289–298. doi:10.1111/jog.13863
Ota, T., Yoshida, M., Kimura, M., and Kinoshita, K. (2005). Clinicopathologic Study of Uterine Endometrial Carcinoma in Young Women Aged 40 Years and Younger. Int. J. Gynecol. Cancer 15, 657–662. doi:10.1111/j.1525-1438.2005.00129.x
Rahib, L., Smith, B. D., Aizenberg, R., Rosenzweig, A. B., Fleshman, J. M., and Matrisian, L. M. (2014). Projecting Cancer Incidence and Deaths to 2030: the Unexpected burden of Thyroid, Liver, and Pancreas Cancers in the United States. Cancer Res. 74, 2913–2921. doi:10.1158/0008-5472.can-14-0155
Sangtani, A., Wang, C., Weaver, A., Hoppman, N. L., Kerr, S. E., Abyzov, A., et al. (2020). Combining Copy Number, Methylation Markers, and Mutations as a Panel for Endometrial Cancer Detection via Intravaginal Tampon Collection. Gynecol. Oncol. 156, 387–392. doi:10.1016/j.ygyno.2019.11.028
Sano, K., Tanihara, H., Heimark, R. L., Obata, S., Davidson, M., St John, T., et al. (1993). Protocadherins: a Large Family of Cadherin-Related Molecules in central Nervous System. EMBO J. 12, 2249–2256. doi:10.1002/j.1460-2075.1993.tb05878.x
Shan, M., Su, Y., Kang, W., Gao, R., Li, X., and Zhang, G. (2016a). Aberrant Expression and Functions of Protocadherins in Human Malignant Tumors. Tumor Biol. 37, 12969–12981. doi:10.1007/s13277-016-5169-9
Shan, M., Yin, H., Li, J., Li, X., Wang, D., Su, Y., et al. (2016b). Detection of Aberrant Methylation of a Six-Gene Panel in Serum DNA for Diagnosis of Breast Cancer. Oncotarget 7, 18485–18494. doi:10.18632/oncotarget.7608
Shi, H., Guo, J., Duff, D. J., Rahmatpanah, F., Chitima-Matsiga, R., Al-Kuhlani, M., et al. (2007). Discovery of Novel Epigenetic Markers in Non-hodgkin's Lymphoma. Carcinogenesis 28, 60–70. doi:10.1093/carcin/bgl092
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A. Cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Urick, M. E., and Bell, D. W. (2019). Clinical Actionability of Molecular Targets in Endometrial Cancer. Nat. Rev. Cancer 19, 510–521. doi:10.1038/s41568-019-0177-x
Wentzensen, N., Bakkum-Gamez, J. N., Killian, J. K., Sampson, J., Guido, R., Glass, A., et al. (2014). Discovery and Validation of Methylation Markers for Endometrial Cancer. Int. J. Cancer 135, 1860–1868. doi:10.1002/ijc.28843
Widschwendter, M., Jones, A., Jones, A., Evans, I., Reisel, D., Dillner, J., et al. (2018). Epigenome-based Cancer Risk Prediction: Rationale, Opportunities and Challenges. Nat. Rev. Clin. Oncol. 15, 292–309. doi:10.1038/nrclinonc.2018.30
Keywords: endometrial cancer, DNA methylation biomarker, PCDHGB7, cervical scrapings, endometrial brush
Citation: Yuan J, Mao Z, Lu Q, Xu P, Wang C, Xu X, Zhou Z, Zhang T, Yu W, Dong S, Wang Y and Cheng W (2022) Hypermethylated PCDHGB7 as a Biomarker for Early Detection of Endometrial Cancer in Endometrial Brush Samples and Cervical Scrapings. Front. Mol. Biosci. 8:774215. doi: 10.3389/fmolb.2021.774215
Received: 11 September 2021; Accepted: 16 November 2021;
Published: 04 January 2022.
Edited by:
Xin Wang, The Chinese University of Hong Kong, ChinaReviewed by:
Hao Huang, City University of Hong Kong, Hong Kong SAR, ChinaWei Wang, Huzhou Maternity and Child Health Care Hospital, China
Copyright © 2022 Yuan, Mao, Lu, Xu, Wang, Xu, Zhou, Zhang, Yu, Dong, Wang and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenqiang Yu, d2VucWlhbmd5dUBmdWRhbi5lZHUuY24=; Shihua Dong, ZG9uZ3NoaWh1YUBlcGlwcm9iZS5jb20=; Yudong Wang, b3dhbmd5dWRvbmdAMTI2LmNvbQ==; Weiwei Cheng, d3djaGVuZzI5QHNoc211LmVkdS5jbg==
†These authors have contributed equally to this work
 Jiangjing Yuan
Jiangjing Yuan Zhanrui Mao
Zhanrui Mao Qi Lu
Qi Lu Peng Xu
Peng Xu Chengyang Wang
Chengyang Wang Xiaona Xu1
Xiaona Xu1 Wenqiang Yu
Wenqiang Yu Shihua Dong
Shihua Dong Yudong Wang
Yudong Wang Weiwei Cheng
Weiwei Cheng