
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci. , 03 January 2022
Sec. RNA Networks and Biology
Volume 8 - 2021 | https://doi.org/10.3389/fmolb.2021.771835
This article is part of the Research Topic Tumorigenesis Regulated by miRNAs View all 12 articles
 Soudeh Ghafouri-Fard1
Soudeh Ghafouri-Fard1 Tayyebeh Khoshbakht2
Tayyebeh Khoshbakht2 Bashdar Mahmud Hussen3,4
Bashdar Mahmud Hussen3,4 Mohammad Taheri5*
Mohammad Taheri5* Mohammad Samadian6*
Mohammad Samadian6*miR-1246 is a microRNA firstly recognized through application of a high throughput sequencing technique in human embryonic stem cells. Subsequent studies have shown the role of this microRNA in the carcinogenesis. miR-1246 has been found to exert oncogenic roles in colorectal, breast, renal, oral, laryngeal, pancreatic and ovarian cancers as well as melanoma and glioma. In lung, cervical and liver cancers, studies have reported contradictory results regarding the role of miR-1246. miR-1246 has been reported to regulate activity of RAF/MEK/ERK, GSK3β, Wnt/β-catenin, JAK/STAT, PI3K/AKT, THBS2/MMP and NOTCH2 pathways. In addition to affecting cell cycle progression and proliferation, miR-1246 can influence stemness and resistance of cancer cells to therapeutics. In the current review, we describe the summary of in vitro and in vivo studies about the influence of miR-1246 in carcinogenesis in addition to studies that measured expression levels of miR-1246 in clinical samples.
miR-1246 has been firstly recognized through application of a high throughput sequencing technique in human embryonic stem cells (Morin et al., 2008). Subsequent studies have mapped the human miR-1246-coding gene, i.e., MIR1246 gene on chromosome 2q31.1 and reported the impact of p53 on the regulation of its expression (Zhang et al., 2011). Notably, the nucleotide sequence of the mature miR-1246 is identical to the central region of the RNU2-1 RNA (Xu et al., 2019), a small nuclear RNA which constructs the scaffold for establishment of the U2 complex in the spliceosome (Patel and Bellini, 2008).
Theoretically, the stem-loop TaqMan technique for detection of miR-1246 is expected to amplify both miR-1246 and RNA, U2 Small Nuclear 1 (RNU2-1). However, the poly-A tailing SYBR strategy can differentiate between miR-1246 and RNU2-1, since the sizes of the amplified fragments can be differentiated through assessment of their meting curves (Xu et al., 2019). Application of the latter strategy for assessment of miR-1246 expression in wild type and MIR1246 knockout pancreatic adenocarcinoma cells and exosomes originated from these cells has led to identification of a variant of the mature miR-1246 in exosomes that is transcribed from cellular RNU2-1 in an independent manner from Drosha and Dicer miRNA processing enzymes (Xu et al., 2019).
Several researchers have assessed expression of miR-1246 in different cancer cell lines using a variety of miRNA-profiling assays. Subsequently, they have performed functional assays to find the effects of miR-1246 up-regulation or silencing on proliferation and invasive properties of these cells. Finally, the impact of this miRNA on tumor growth has been appraised in xenograft models constructed by injection of human cancer cell lines. In the current review, we describe the summary of these two types of studies in addition to those measured expression levels of miR-1246 in clinical samples.
Experiments in colorectal cancer cell lines have shown oncogenic role of miR-1246. In this type of cancer, the m (6) A methyltransferase METTL3 oncogene has been shown to increase methylation of pri-miR-1246 to enhance maturation of pri-miR-1246. Notably, miR-1246 has been predicted to suppress expression of the Sprouty Related EVH1 Domain Containing 2 (SPRED2) tumor suppressor, thus increasing activity of MAPK pathway (Peng et al., 2019).
Expression of miR-1246 has been found to be increased in exosomes derived from colorectal cancer cells infected with Fusobacterium nucleatum. In fact, this cancer-associated bacterium can enhance pro-metastatic behaviors through delivery of these exosomes into un-infected cells (Guo et al., 2021).
Expression of miR-1246 has also been reported to be surged in SW620, SW480, HCT116, HT29 and LOVO colorectal cancer cells, parallel with down-regulation of Cyclin G2 (CycG2). Experiments in HCT-116 and LOVO cells have verified CycG2 as the target of miR-1246. Up-regulation of miR-1246 has exerted pro-proliferative and pro-invasive effects in these cells, while its silencing has reversed these effects (Wang et al., 2016).
Exosomal and cellular levels of miR-1246 have been reported to be higher in organoid lines generated from colorectal cancer compared with organoid lines from colorectal adenomas. Consistent with this finding, miR-1246 up-regulation and down-regulation have enhanced reduced proliferation of an adenocarcinoma cell line, respectively (Nagai et al., 2021).
Another experiment in breast cancer cells has demonstrated high levels of miR-1246 in metastatic breast cancer cells compared with both non-metastatic cancer cells and non-neoplastic breast cells. miR-1246-containing exosomes from metastatic breast cancer cells can alter viability, migratory potential and chemoresistant phenotype of non-malignant breast cells. Functionally, miR-1246 suppresses expression of Cyclin G2 (Li et al., 2017).
In renal cell carcinoma cells, miR-1246 has an oncogenic effect through suppressing expression of PCK1. Notably, the tumor suppressor long non-coding RNA (lncRNA) GABPB1-AS1 has been shown to sponge miR-1246 in these cells (Gao et al., 2020).
Figure 1 shows the oncogenic role of miR-1246 in colorectal, breast and renal cancers.
miR-1246 has been demonstrated to increase the migration and invasive aptitudes of A549 adenocarcinomic human alveolar basal epithelial cells. In addition, miR-1246 could enhance epithelial-mesenchymal transition (EMT) of lung cancer cells. This miRNA could decrease levels of E-cadherin, while enhancing vimentin and TGF-β levels. Functionally, miR-1246 can target 3′-untranslated region of GSK-3β, thus regulating activity of Wnt/β-catenin pathway (Yang et al., 2019).
Yuan et al. have investigated the impact of ionizing radiation (IR)-induced extracellular miRNAs on proliferation and radioresistance of A549 adenocarcinomic cells. They have reported particular abundance of miR-1246 outside of cells compared with its levels inside the cells. Irradiation could increase expression levels of miR-1246 in A549 and H446 cells in dose- and time-dependent manners. Extracellular miR-1246 has been shown to be transferred from donor cells to recipients through a non-exosome associated route enhancing proliferation and resistance of A549 cells to irradiation. Functionally, miR-1246 reduces expression of death receptor 5 (DR5) (Yuan et al., 2016).
miR-1246 has been among up-regulated miRNAs in the sphere-forming cells compared with the parental A549 and HCC1588 cells. Suppresion of miR-1246 has led to reduction of levels stemness and EMT markers in these cells. Moreover, anti-miR-1246 could suppress proliferation, sphere-formation, colony forming ability and invasiveness of lung cancer cells (Kim et al., 2016). Similarly, Huang et al. have reported up-regulation of miR-1246 and METTL3 in A549 and H1299 cells, parallel with down-regulation of PEG3. METTL3 has been shown to affect m6A marks of miR-1246, therefore increasing expression of miR-1246. Cumulatively, m6A methyltransferase METTL3 modifies the m6A marks of miR-1246 to up-regulates miR-1246 and subsequently increase progression of lung cancer (Huang et al., 2021).
Contrary to these studies, Xu et al. have reported down-regulation of miR-1246 in A549, H1650 and H1299 cell lines compared to a normal human bronchial epithelial cell line. MiR-1246 overexpression remarkably inhibited cell invasion as well as up-regulated E-cadherin expression and down-regulated N-cadherin, Vimentin, ZEB1 and Snail expressions in A549 cells. Further studies have confirmed CXCR4 as a target gene of miR-1246, and CXCR4 silence significantly abolished the promotion effect of miR-1246 suppression on cell invasion and EMT process in A549 cells. Besides, miR-1246 blocked JAK/STAT and PI3K/AKT signal pathways by regulation of CXCR4 (Xu et al., 2018). Figure 2 shows dual roles of miR-1246 in lung cancer.
In SiHa HPV16-positive cervical cancer cell line, HPV16 E6 silencing has led to enhancement of miR-1246 expression, thus down-regulation of miR-1246 target DYRK1A. Meanwhile, overexpression of HPV16 E6 in HPV-negative C33A cell line has resulted in down-regulation of miR-1246 (Yang et al., 2015). Another study has shown that miR-1246 increases proliferation, invasiveness and migratory potential of SiHa cells through inhibition of expression of thrombospondin 2 (Chen et al., 2014). miR-1246 has also been among up-regulated miRNAs in radioresistant cervical cancer cells. Expression of this miRNA could be enhanced by irradiation of cervical cancer cells. Up-regulation of miR-1246 has increased survival of cervical cancer cells upon irradiation (Zhang et al., 2013). Figure 3 shows dual roles of miR-1246 in cervical cancer.
Experiments in a co-culture model of hepatic stellate cells (HSCs) and hepatocellular carcinoma cells have shown that expression of miR-1246 is activated by HSCs. miR-1246 has been shown to target RORα. Up-regulation of miR-1246 or silencing of RORα has promoted proliferation, invasive properties, and metastatic aptitude of hepatocellular cancer cells through activation of Wnt/β-catenin pathway and enhancement of EMT (Huang J.-L. et al., 2020). Another study has shown that miR-1246 increases invasiveness of hepatocellular carcinoma cells via modulation of CADM1 expression (Sun et al., 2014). Moreover, miR-1246 has been reported to promote stemness features such as self-renewal, resistance to therapeutics, tumorigenic potential, and metastasis through enhancing activity of Wnt/β-catenin pathway. This effect is mediated through down-regulation of expression levels of AXIN2 and GSK3β. Oct4 has been identified as the direct regulator of miR-1246 expression which activates β-catenin in hepatic cancer stem cells (Chai et al., 2016).
On the other hand, Zhang et al. have shown that expression of miR-1246 is induced by p53. This miRNA has been shown to inhibit proliferation of hepatocellular carcinoma cells through influencing expression of NFIB (Zhang et al., 2015). Figure 4 shows dual roles of miR-1246 in hepatocellular carcinoma.
In oral squamous cell carcinoma, miR-1246 has been shown to target CCNG2 to facilitate stemness properties and induce resistance to chemotherapy (Lin et al., 2018). Moreover, exosomal transfer of this miRNA has enhanced cell motility and invasiveness of oral squamous cell carcinoma cells through targeting DENND2D (Sakha et al., 2016). Consistent with this finding, small extracellular vesicles originated from laryngeal squamous cell carcinoma cells have been shown to enter into neighboring cells. Lack of miR-1246 in these vesicles abolished development of this kind of cancer. miR-1246 content of small vesicles could participate in the pathoetiology of laryngeal squamous cell carcinoma through suppressing CCNG2 expression (Huang Q. et al., 2020). miR-1246 is involved in the progression of melanoma via changing expression levels FOXA2 (Yu et al., 2020). Moreover, miR-1246 has been shown to increase resistance of melanoma cells to BRAF inhibitors (Kim et al., 2017). Figure 5 shows oncogenic role of miR-1246 in oral and laryngeal squamous cell carcinomas and melanoma.
Exosomes originated from glioma cell cultures under hypoxic conditions could shuttle miR-1246 to normoxic glioma cells and enhance their migratory potential and invasiveness (Qian M. et al., 2021). Another study has shown the impact of these exosomes in induction of polarization of macrophages into M2 phenotype through targeting TERF2IP and subsequent influence on the activities of STAT3 and NF-κB signaling (Qian et al., 2020).
In pancreatic cancer, miR-1246 could increase chemoresistance and stemness through modulation of CCNG2 (Hasegawa et al., 2014).
Finally, in ovarian cancer, miR-1246 can confer resistance to chemotherapeutics through influencing Cav1/p-gp/M2-type macrophages (Kanlikilicer et al., 2018).
Figure 6 shows the oncogenic role of miR-1246 in glioma, pancreatic cancer and ovarian cancer.
Table 1 shows the outlines of in vitro studies focusing on the function of miR-1246 in cancer.

TABLE 1. Outlines of in vitro studies about function of miR-1246 (∆: knock-down or deletion, FN: Fusobacterium nucleatum, sEV: Small extracellular vesicle, GEM: gemcitabine).
Most of animal studies have indicated an oncogenic role for miR-1246, since its silencing has led to reduction of tumor size and attenuation of tumor growth (Table 2). Moreover, expression of miR-1246 has been found to be elevated in the plasma exosomes of patient-originated orthotopic xenograft animals compared to control animals (Hannafon et al., 2016). However, in prostate cancer, miR-1246 up-regulation has significantly inhibited tumor growth in the xenograft models, suggesting its tumor suppressive role. Moreover, in miR-1246 overexpressing xenograft models, exosomal levels of this miRNA has been reduced. Taken together, miR-1246 has been identified as a tumor suppressor miRNA being selectively packaged in prostate cancer exosomes, resulting in its high abundance in serum yet low concentrations inside the cells (Bhagirath et al., 2018). In the xenograft model of leukemia, miR-1246-containing extracellular vesicles have been shown to confer quiescence on residual hematopoietic stem cells (Abdelhamed et al., 2019).
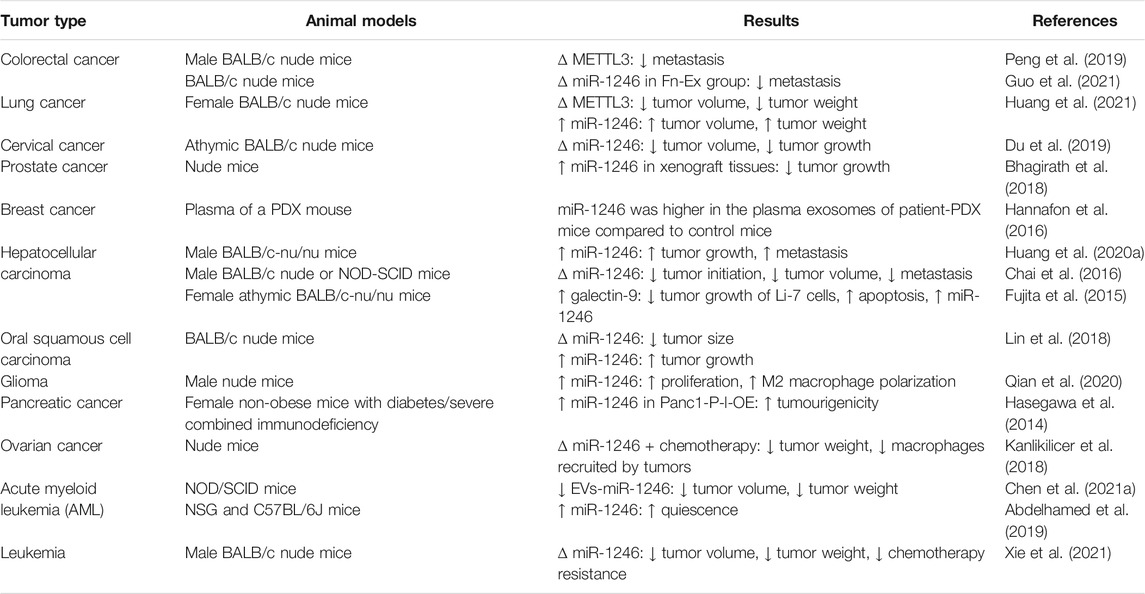
TABLE 2. Outline of studies about the function of miR-1246 in animal models (∆: knock-down or deletion, PDX: derived orthotopic xenograft, NOD-SCID: non-obese diabetic/severe combined immunodeficiency, NSG: NOD Cg-Prkdcscid Il2rgtm1Wjl/SzJ).
Serum levels of miR-1246 have been found to be higher in the sera of colorectal cancer patients compared to healthy subjects (Salah et al., 2020). Similarly, miR-1246 has been found as the most up-regulated miRNA in the sera of patients with lung cancer (Yang et al., 2019). Levels of miR-1246 have been found to be higher in laryngeal squamous cell carcinoma tissues and plasma small extracellular vesicles. This miRNA has been more enriched in small extracellular vesicles instead of being in soluble form (Sakha et al., 2016). Almost all studies in clinical settings have reported up-regulation of miR-1246 in neoplastic tissues and sera of patients compared with controls (Table 3).
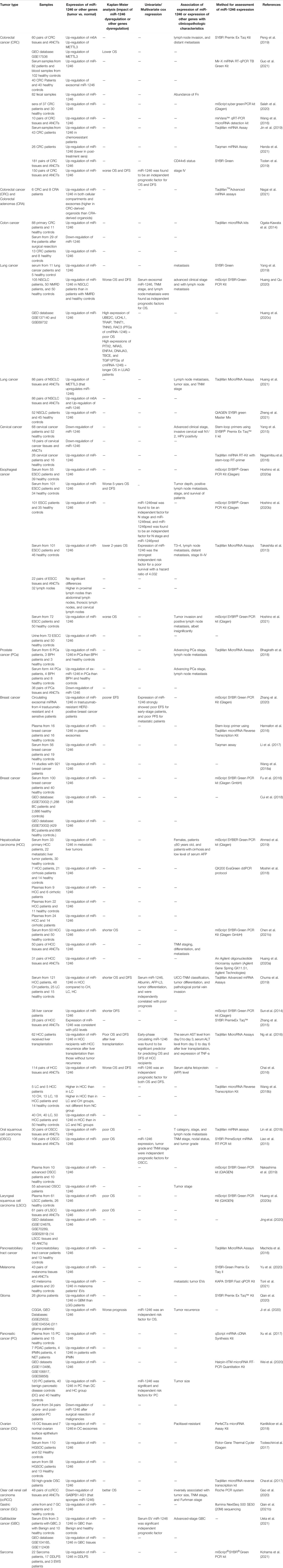
TABLE 3. Results of studies that reported dysregulation of miR-1246 or other genes that interact with miR-1246 in clinical samples.
However, Yang et al. have shown down-regulation of miR-1246 in cervical cancer tissues compared with normal controls. Notably, down-regulation of miR-1246 has been inversely correlated with clinical stage and HPV16 E6 infection. Yet, its levels have not been correlated with age, tumor diameters, invasion deepness, lymph node involvement, or vascular invasion (Yang et al., 2015).
Table 3 Results of studies that reported dysregulation of miR-1246 or other genes that interact with miR-1246 in clinical samples (OS: Overall survival, DFS: disease-free survival, TNM: tumor-node-metastasis, ANCTs: adjacent non-cancerous tissues, FN: Fusobacterium nucleatum, CD44v6: a CSC population with increased resistance to chemotherapeutic agents, NMRD: non-malignant respiratory diseases, NSCLC: non-small cell lung cancer, PTGs: potential target genes, LUAD: lung adenocarcinoma, ESCC: esophageal squamous cell carcinoma, miR-1246real and miR-1246pred: real and predicted miR-1246 expression levels, BPH: benign prostate hyperplasia, EFS: event-free survival, PFS: progression-free survival, LC: liver cirrhosis, CH: chronic hepatitis, HC: healthy controls, UICC: Union for International Cancer Control, GBM: glioblastoma, LGG: low-grade glioma, PDAC: pancreatic ductal adenocarcinomas, IPMN: intraductal papillary mucinous neoplasms, NET: well differentiated neuroendocrine tumors, HGSOC: High-grade serous ovarian carcinoma, OSC: ovarian serous carcinoma, EVs: extracellular vesicles, DDLPS: dedifferentiated liposarcoma, EWS: Ewing’s sarcoma).
Diagnostic value of miR-1246 has been validated in different neoplastic disorders (Table 4). The most promising results have been revealed in breast cancer. This miRNA could separate breast cancer patients from healthy controls with area under receiver operating characteristic curve (AUC) of 0.967 (Cui et al., 2018). In hepatocellular carcinoma, miR-1246 could be used as a diagnostic marker for differentiation of cancer status from cirrhosis and healthy controls with AUC values of 0.97 and 0.83, respectively (Moshiri et al., 2018). Expression level of miR-1246 in serum samples have been shown to distinguish colorectal cancer patients from healthy subjects with sensitivity of 100% and specificity of 80% (Salah et al., 2020). This miRNA could separate lung cancer patients from healthy controls with AUC value of 0.82 (Huang and Qu, 2020). Moreover, serum and urine levels of miR-1246 could be used as diagnostic markers for esophageal cancer with AUC values of 0.91 and 0.82, respectively (Hoshino et al., 2021).
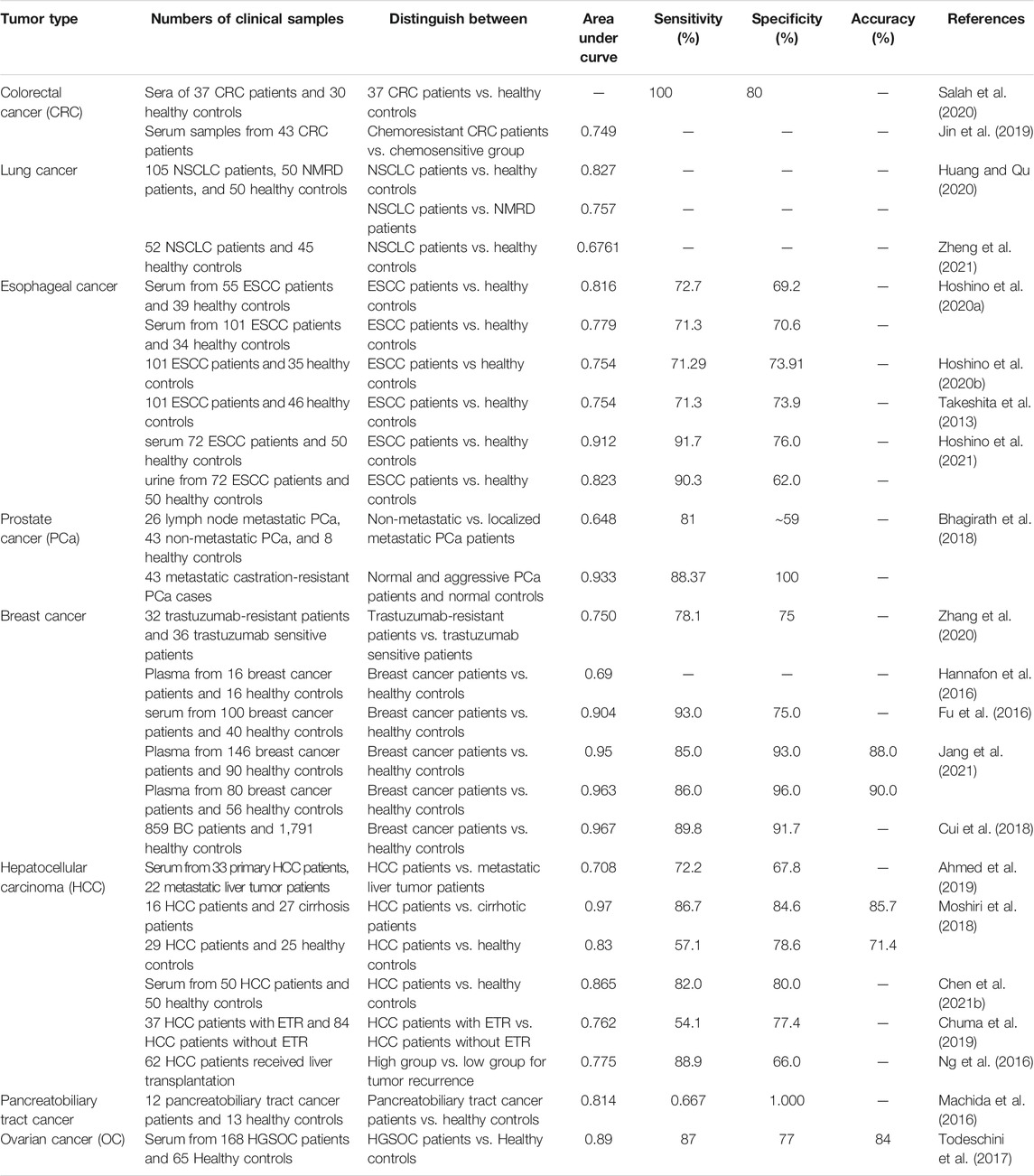
TABLE 4. Diagnostic value of miR-1246 in cancers (NMRD: non-malignant respiratory diseases, NSCLC: non-small cell lung cancer, ESCC: esophageal squamous cell carcinoma, ETR: Early tumor recurrence, HGSOC: High-grade serous ovarian carcinoma).
miR-1246 is a miRNA with essential impact on carcinogenic events in different tissues. It exerts oncogenic roles in colorectal, breast, renal, oral, laryngeal, pancreatic and ovarian cancers as well as melanoma and glioma. However, in lung, cervical and liver cancers, studies have reported contradictory results regarding the role of miR-1246. Although several targets have been found for miR-1249 using bioinformatics tools and luciferase assay, CCNG2 is the most appreciated target of this miRNA in the context of cancer. miR-1246/CCNG2 axis not only regulates cell proliferation and cell cycle progression, but also is involved in chemoresistant phenotype.
The main mechanism of dysregulation of miR-1246 in cancer is methylation of pri-miR-1246 by methyltransferase METTL3 and modulation of maturation of pri-miR-1246. Unlike other miRNAs, the role of sponging lncRNAs on its expression is less studied.
miR-1246 has been reported to regulate activity of RAF/MEK/ERK, GSK3β, Wnt/β-catenin, JAK/STAT, PI3K/AKT, THBS2/MMP and NOTCH2 pathways. The role of miR-1246 in response to therapeutic modalities has been verified in different settings, indicating its crucial roles in determination of response to targeted therapies, radiotherapy as well as chemotherapy. In fact, miR-1246 can facilitate evolution of cancer through conferring stemness and EMT as well as induction of cell cycle progression and proliferation.
Diagnostic role of miR-1246 has been vastly appraised in different clinical settings, revealing nearly ideal AUC values, particularly in esophageal, prostate, breast, lung, liver, pancreatobiliary tract and ovarian cancers. The AUC, sensitivity and specificity values obtained for miR-1246 in different cancers are far superior to conventional biomarkers in these cancers. Thus, this miRNA represents an appropriate diagnostic biomarker for neoplastic conditions. Since its levels have been decreased following therapeutic interventions, it has additional advantage in patients’ follow-up. Although miR-1246 can be a putative therapeutic target for cancer, there is no tissue-specific therapeutic approach designed based on miR-1246 until now.
Taken together, miR-1246 is mostly regarded as an oncogenic miRNA in human cancers, albeit some inconsistencies exist for some types of cancers. The interactions of miR-1249 with other types of non-coding RNAs such as lncRNAs and circular RNAs have not been completely assessed. Identification of such interactions has implications in design of diagnostic panels for different cancers.
miR-1246 is an oncogenic miRNA in several tissues. Therapeutic intervention with its expression or methylation pattern can be regarded as a novel modality. However, it is necessary to design tissue-specific therapeutic approaches.
SG-F wrote the draft and revised it. MT supervised and designed the study. MS and TK collected the data and designed the figures and tables. All the authors read and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdelhamed, S., Butler, J. T., Doron, B., Halse, A., Nemecek, E., Wilmarth, P. A., et al. (2019). Extracellular Vesicles Impose Quiescence on Residual Hematopoietic Stem Cells in the Leukemic Niche. EMBO Rep. 20 (7), e47546. doi:10.15252/embr.201847546
Ahmed, E. K., Fahmy, S. A., Effat, H., and Wahab, A. H. A. (2019). Circulating miR-210 and miR-1246 as Potential Biomarkers for Differentiating Hepatocellular Carcinoma from Metastatic Tumors in the Liver. J. Med. Biochem. 38 (2), 109–117. doi:10.2478/jomb-2018-0010
Bhagirath, D., Yang, T. L., Bucay, N., Sekhon, K., Majid, S., Shahryari, V., et al. (2018). microRNA-1246 Is an Exosomal Biomarker for Aggressive Prostate Cancer. Cancer Res. 78 (7), 1833–1844. doi:10.1158/0008-5472.can-17-2069
Cha, S. y., Choi, Y. h., Hwang, S., Jeong, J.-Y., and An, H. J. (2017). Clinical Impact of microRNAs Associated with Cancer Stem Cells as a Prognostic Factor in Ovarian Carcinoma. J. Cancer 8 (17), 3538–3547. doi:10.7150/jca.20348
Chai, S., Ng, K.-Y., Tong, M., Lau, E. Y., Lee, T. K., Chan, K. W., et al. (2016). Octamer 4/microRNA-1246 Signaling axis Drives Wnt/β-Catenin Activation in Liver Cancer Stem Cells. Hepatology 64 (6), 2062–2076. doi:10.1002/hep.28821
Chen, J., Yao, D., Zhao, S., He, C., Ding, N., Li, L., et al. (2014). MiR-1246 Promotes SiHa Cervical Cancer Cell Proliferation, Invasion, and Migration through Suppression of its Target Gene Thrombospondin 2. Arch. Gynecol. Obstet. 290 (4), 725–732. doi:10.1007/s00404-014-3260-2
Chen, L., Guo, Z., Zhou, Y., Ni, J., Zhu, J., Fan, X., et al. (2021a). microRNA-1246-containing Extracellular Vesicles from Acute Myeloid Leukemia Cells Promote the Survival of Leukemia Stem Cells via the LRIG1-Meditated STAT3 Pathway. Aging 13 (10), 13644–13662. doi:10.18632/aging.202893
Chen, S., Fu, Z., Wen, S., Yang, X., Yu, C., Zhou, W., et al. (2021b). Expression and Diagnostic Value of miR-497 and miR-1246 in Hepatocellular Carcinoma. Front. Genet. 12, 790. doi:10.3389/fgene.2021.666306
Chuma, M., Toyoda, H., Matsuzaki, J., Saito, Y., Kumada, T., Tada, T., et al. (2019). Circulating microRNA‐1246 as a Possible Biomarker for Early Tumor Recurrence of Hepatocellular Carcinoma. Hepatol. Res. 49 (7), 810–822. doi:10.1111/hepr.13338
Cui, X., Li, Z., Zhao, Y., Song, A., Shi, Y., Hai, X., et al. (2018). Breast Cancer Identification via Modeling of Peripherally Circulating miRNAs. PeerJ 6, e4551. doi:10.7717/peerj.4551
Du, P., Lai, Y. H., Yao, D. S., Chen, J. Y., and Ding, N. (2019). Downregulation of microRNA-1246 Inhibits Tumor Growth and Promotes Apoptosis of Cervical Cancer Cells by Targeting Thrombospondin-2. Oncol. Lett. 18 (3), 2491–2499. doi:10.3892/ol.2019.10571
Fu, L., Li, Z., Zhu, J., Wang, P., Fan, G., Dai, Y., et al. (2016). Serum Expression Levels of microRNA-382-3p, −598-3p, −1246 and −184 in Breast Cancer Patients. Oncol. Lett. 12 (1), 269–274. doi:10.3892/ol.2016.4582
Fujita, K., Iwama, H., Oto, T., Okura, R., Kobayashi, K., Takano, J., et al. (2015). Galectin-9 Suppresses the Growth of Hepatocellular Carcinoma via Apoptosis In Vitro and In Vivo. Int. J. Oncol. 46 (6), 2419–2430. doi:10.3892/ijo.2015.2941
Gao, S., Zhang, F., Sun, H., and Yang, X. (2020). LncRNA GA-Binding Protein Transcription Factor Subunit Beta-1 Antisense RNA 1 Inhibits Renal Carcinoma Growth through an MiR-1246/Phosphoenolpyruvate Carboxykinase 1 Pathway. Ott 13, 6827–6836. doi:10.2147/ott.s257275
Guo, S., Chen, J., Chen, F., Zeng, Q., Liu, W.-L., and Zhang, G. (2021). Exosomes Derived from Fusobacterium Nucleatum-Infected Colorectal Cancer Cells Facilitate Tumour Metastasis by Selectively Carrying miR-1246/92b-3p/27a-3p and CXCL16. Gut 70 (8), 1507–1519. doi:10.1136/gutjnl-2020-321187
Handa, T., Kuroha, M., Nagai, H., Shimoyama, Y., Naito, T., Moroi, R., et al. (2021). Liquid Biopsy for Colorectal Adenoma: Is the Exosomal miRNA Derived from Organoid a Potential Diagnostic Biomarker? Clin. Transl Gastroenterol. 12 (5), e00356. doi:10.14309/ctg.0000000000000356
Hannafon, B. N., Trigoso, Y. D., Calloway, C. L., Zhao, Y. D., Lum, D. H., Welm, A. L., et al. (2016). Plasma Exosome microRNAs Are Indicative of Breast Cancer. Breast Cancer Res. 18 (1), 90–14. doi:10.1186/s13058-016-0753-x
Hasegawa, S., Eguchi, H., Nagano, H., Konno, M., Tomimaru, Y., Wada, H., et al. (2014). MicroRNA-1246 Expression Associated with CCNG2-Mediated Chemoresistance and Stemness in Pancreatic Cancer. Br. J. Cancer 111 (8), 1572–1580. doi:10.1038/bjc.2014.454
Hoshino, I., Yokota, H., Ishige, F., Iwatate, Y., Takeshita, N., Nagase, H., et al. (2020b). Radiogenomics Predicts the Expression of microRNA-1246 in the Serum of Esophageal Cancer Patients. Sci. Rep. 10 (1), 2532–2538. doi:10.1038/s41598-020-59500-7
Hoshino, I., Ishige, F., Iwatate, Y., Gunji, H., Kuwayama, N., Nabeya, Y., et al. (2021). Cell-free microRNA-1246 in Different Body Fluids as a Diagnostic Biomarker for Esophageal Squamous Cell Carcinoma. Plos one 16 (3), e0248016. doi:10.1371/journal.pone.0248016
Hoshino, I., Ishige, F., Iwatate, Y., Gunji, H., Shiratori, F., Kuwayama, N., et al. (2020a). Usefulness of Serum miR-1246/miR-106b R-atio in P-atients with E-sophageal S-quamous C-ell C-arcinoma. Oncol. Lett. 20 (6), 350. doi:10.3892/ol.2020.12213
Huang, D., and Qu, D. (2020). Early Diagnostic and Prognostic Value of Serum Exosomal miR-1246 in Non-small Cell Lung Cancer. Int. J. Clin. Exp. Pathol. 13 (7), 1601–1607.
Huang, J.-L., Fu, Y.-P., Gan, W., Liu, G., Zhou, P.-Y., Zhou, C., et al. (2020a). Hepatic Stellate Cells Promote the Progression of Hepatocellular Carcinoma through microRNA-1246-Rorα-Wnt/β-Catenin axis. Cancer Lett. 476, 140–151. doi:10.1016/j.canlet.2020.02.012
Huang, Q., Hsueh, C. Y., Guo, Y., Wu, X. F., Li, J. Y., and Zhou, L. (2020b). Lack of miR‐1246 in Small Extracellular Vesicle Blunts Tumorigenesis of Laryngeal Carcinoma Cells by Regulating Cyclin G2. IUBMB life 72 (7), 1491–1503. doi:10.1002/iub.2274
Huang, S., Luo, S., Gong, C., Liang, L., Xiao, Y., Li, M., et al. (2021). MTTL3 Upregulates microRNA-1246 to Promote Occurrence and Progression of NSCLC via Targeting Paternally Expressed Gene 3. Mol. Ther. - Nucleic Acids 24, 542–553. doi:10.1016/j.omtn.2021.02.020
Huang, S., Wei, Y.-K., Kaliamurthi, S., Cao, Y., Nangraj, A. S., Sui, X., et al. (2020c). Circulating miR-1246 Targeting UBE2C, TNNI3, TRAIP, UCHL1 Genes and Key Pathways as a Potential Biomarker for Lung Adenocarcinoma: Integrated Biological Network Analysis. Jpm 10 (4), 162. doi:10.3390/jpm10040162
Jang, J. Y., Kim, Y. S., Kang, K. N., Kim, K. H., Park, Y. J., and Kim, C. W. (2021). Multiple microRNAs as Biomarkers for Early Breast Cancer Diagnosis. Mol. Clin. Oncol. 14 (2), 31. doi:10.3892/mco.2020.2193
Ji, B., Chen, L., Cai, Q., Guo, Q., Chen, Z., and He, D. (2020). Identification of an 8-miRNA Signature as a Potential Prognostic Biomarker for Glioma. PeerJ 8, e9943. doi:10.7717/peerj.9943
Jin, G., Liu, Y., Zhang, J., Bian, Z., Yao, S., Fei, B., et al. (2019). A Panel of Serum Exosomal microRNAs as Predictive Markers for Chemoresistance in Advanced Colorectal Cancer. Cancer Chemother. Pharmacol. 84 (2), 315–325. doi:10.1007/s00280-019-03867-6
Jing, Z., Guo, S., Zhang, P., and Liang, Z. (2020). LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma. Technology Cancer Res. Treat. 19, 1533033820985787. doi:10.1177/1533033820985787
Kanlikilicer, P., Bayraktar, R., Denizli, M., Rashed, M. H., Ivan, C., Aslan, B., et al. (2018). Exosomal miRNA Confers Chemo Resistance via Targeting Cav1/p-gp/M2-type Macrophage axis in Ovarian Cancer. EBioMedicine 38, 100–112. doi:10.1016/j.ebiom.2018.11.004
Kim, G., An, H.-J., Lee, M.-J., Song, J.-Y., Jeong, J.-Y., Lee, J.-H., et al. (2016). Hsa-miR-1246 and Hsa-miR-1290 Are Associated with Stemness and Invasiveness of Non-small Cell Lung Cancer. Lung cancer 91, 15–22. doi:10.1016/j.lungcan.2015.11.013
Kim, J.-H., Ahn, J.-H., and Lee, M. (2017). Upregulation of microRNA-1246 Is Associated with BRAF Inhibitor Resistance in Melanoma Cells with Mutant BRAF. Cancer Res. Treat. 49 (4), 947–959. doi:10.4143/crt.2016.280
Kohama, I., Asano, N., Matsuzaki, J., Yamamoto, Y., Yamamoto, T., Takahashi, R.-U., et al. (2021). Comprehensive Serum and Tissue microRNA Profiling in Dedifferentiated Liposarcoma. Oncol. Lett. 22 (2), 1–8. doi:10.3892/ol.2021.12884
Li, X. J., Ren, Z. J., Tang, J. H., and Yu, Q. (2017). Exosomal MicroRNA MiR-1246 Promotes Cell Proliferation, Invasion and Drug Resistance by Targeting CCNG2 in Breast Cancer. Cell Physiol Biochem 44 (5), 1741–1748. doi:10.1159/000485780
Liao, L., Wang, J., Ouyang, S., Zhang, P., Wang, J., and Zhang, M. (2015). Expression and Clinical Significance of microRNA-1246 in Human Oral Squamous Cell Carcinoma. Med. Sci. Monit. 21, 776–781. doi:10.12659/MSM.892508
Lin, S.-S., Peng, C.-Y., Liao, Y.-W., Chou, M.-Y., Hsieh, P.-L., and Yu, C.-C. (2018). miR-1246 Targets CCNG2 to Enhance Cancer Stemness and Chemoresistance in Oral Carcinomas. Cancers 10 (8), 272. doi:10.3390/cancers10080272
Lu, X., Chen, L., Chen, Y., Shao, Q., and Qin, W. (2015). Bafilomycin A1 Inhibits the Growth and Metastatic Potential of the BEL-7402 Liver Cancer and HO-8910 Ovarian Cancer Cell Lines and Induces Alterations in Their microRNA Expression. Exp. Ther. Med. 10 (5), 1829–1834. doi:10.3892/etm.2015.2758
Luo, M., Zhang, Q., Xia, M., Hu, F., Ma, Z., Chen, Z., et al. (2018). Differential Co-expression and Regulatory Network Analysis Uncover the Relapse Factor and Mechanism of T Cell Acute Leukemia. Mol. Ther. - Nucleic Acids 12, 184–194. doi:10.1016/j.omtn.2018.05.003
Machida, T., Tomofuji, T., Maruyama, T., Yoneda, T., Ekuni, D., Azuma, T., et al. (2016). miR-1246 and miR-4644 in Salivary Exosome as Potential Biomarkers for Pancreatobiliary Tract Cancer. Oncol. Rep. 36 (4), 2375–2381. doi:10.3892/or.2016.5021
Morin, R. D., O’Connor, M. D., Griffith, M., Kuchenbauer, F., Delaney, A., Prabhu, A.-L., et al. (2008). Application of Massively Parallel Sequencing to microRNA Profiling and Discovery in Human Embryonic Stem Cells. Genome Res. 18 (4), 610–621. doi:10.1101/gr.7179508
Moshiri, F., Salvi, A., Gramantieri, L., Sangiovanni, A., Guerriero, P., De Petro, G., et al. (2018). Circulating miR-106b-3p, miR-101-3p and miR-1246 as Diagnostic Biomarkers of Hepatocellular Carcinoma. Oncotarget 9 (20), 15350–15364. doi:10.18632/oncotarget.24601
Nagai, H., Kuroha, M., Handa, T., Karasawa, H., Ohnuma, S., Naito, T., et al. (2021). Comprehensive Analysis of microRNA Profiles in Organoids Derived from Human Colorectal Adenoma and Cancer. Digestion 102 (6), 860–869. doi:10.1159/000513882
Nagamitsu, Y., Nishi, H., Sasaki, T., Takaesu, Y., Terauchi, F., and Isaka, K. (2016). Profiling Analysis of Circulating microRNA Expression in Cervical Cancer. Mol. Clin. Oncol. 5 (1), 189–194. doi:10.3892/mco.2016.875
Nakashima, H., Yoshida, R., Hirosue, A., Kawahara, K., Sakata, J., Arita, H., et al. (2019). Circulating miRNA-1290 as a Potential Biomarker for Response to Chemoradiotherapy and Prognosis of Patients with Advanced Oral Squamous Cell Carcinoma: A Single-center Retrospective Study. Tumour Biol. 41 (3), 1010428319826853. doi:10.1177/1010428319826853
Ng, K. T.-P., Lo, C. M., Wong, N., Li, C. X., Qi, X., Liu, X. B., et al. (2016). Early-phase Circulating miRNAs Predict Tumor Recurrence and Survival of Hepatocellular Carcinoma Patients after Liver Transplantation. Oncotarget 7 (15), 19824–19839. doi:10.18632/oncotarget.7627
Ogata-Kawata, H., Izumiya, M., Kurioka, D., Honma, Y., Yamada, Y., Furuta, K., et al. (2014). Circulating Exosomal microRNAs as Biomarkers of colon Cancer. PloS one 9 (4), e92921. doi:10.1371/journal.pone.0092921
Patel, S. B., and Bellini, M. (2008). The Assembly of a Spliceosomal Small Nuclear Ribonucleoprotein Particle. Nucleic Acids Res. 36 (20), 6482–6493. doi:10.1093/nar/gkn658
Peng, W., Li, J., Chen, R., Gu, Q., Yang, P., Qian, W., et al. (2019). Upregulated METTL3 Promotes Metastasis of Colorectal Cancer via miR-1246/SPRED2/MAPK Signaling Pathway. J. Exp. Clin. Cancer Res. 38 (1), 1408–1424. doi:10.1186/s13046-019-1408-4
Qian, M., Chen, Z., Guo, X., Wang, S., Zhang, Z., Qiu, W., et al. (2021a). Exosomes Derived from Hypoxic Glioma Deliver miR-1246 and miR-10b-5p to Normoxic Glioma Cells to Promote Migration and Invasion. Lab. Invest. 101 (5), 612–624. doi:10.1038/s41374-020-00522-0
Qian, M., Wang, S., Guo, X., Wang, J., Zhang, Z., Qiu, W., et al. (2020). Hypoxic Glioma-Derived Exosomes Deliver microRNA-1246 to Induce M2 Macrophage Polarization by Targeting TERF2IP via the STAT3 and NF-Κb Pathways. Oncogene 39 (2), 428–442. doi:10.1038/s41388-019-0996-y
Qian, X., Xie, F., Wei, H., and Cui, D. (2021b). Identification of Key Circulating Exosomal microRNAs in Gastric Cancer. Front. Oncol. 11, 693360. doi:10.3389/fonc.2021.693360
Sakha, S., Muramatsu, T., Ueda, K., and Inazawa, J. (2016). Exosomal microRNA miR-1246 Induces Cell Motility and Invasion through the Regulation of DENND2D in Oral Squamous Cell Carcinoma. Sci. Rep. 6 (1), 1–11. doi:10.1038/srep38750
Salah, M., Shaheen, I., El-Shanawany, P., Eid Saad, N., Saad, R., El Guibaly, M., et al. (2020). Detection of miR-1246, miR-23a and miR-451 in Sera of Colorectal Carcinoma Patients: a Case-Control Study in Cairo University Hospital. Afr. H. Sci. 20 (3), 1283–1291. doi:10.4314/ahs.v20i3.33
Sun, Z., Meng, C., Wang, S., Zhou, N., Guan, M., Bai, C., et al. (2014). MicroRNA-1246 Enhances Migration and Invasion through CADM1 in Hepatocellular Carcinoma. BMC cancer 14 (1), 2407–2416. doi:10.1186/1471-2407-14-616
Takeshita, N., Hoshino, I., Mori, M., Akutsu, Y., Hanari, N., Yoneyama, Y., et al. (2013). Serum microRNA Expression Profile: miR-1246 as a Novel Diagnostic and Prognostic Biomarker for Oesophageal Squamous Cell Carcinoma. Br. J. Cancer 108 (3), 644–652. doi:10.1038/bjc.2013.8
Toden, S., Kunitoshi, S., Cardenas, J., Gu, J., Hutchins, E., Van Keuren-Jensen, K., et al. (2019). Cancer Stem Cell-Associated miRNAs Serve as Prognostic Biomarkers in Colorectal Cancer. JCI insight 4 (6). doi:10.1172/jci.insight.125294
Todeschini, P., Salviato, E., Paracchini, L., Ferracin, M., Petrillo, M., Zanotti, L., et al. (2017). Circulating miRNA Landscape Identifies miR-1246 as Promising Diagnostic Biomarker in High-Grade Serous Ovarian Carcinoma: a Validation across Two Independent Cohorts. Cancer Lett. 388, 320–327. doi:10.1016/j.canlet.2016.12.017
Torii, C., Maishi, N., Kawamoto, T., Morimoto, M., Akiyama, K., Yoshioka, Y., et al. (2021). miRNA-1246 in Extracellular Vesicles Secreted from Metastatic Tumor Induces Drug Resistance in Tumor Endothelial Cells. Sci. Rep. 11 (1), 13502–13516. doi:10.1038/s41598-021-92879-5
Ueta, E., Tsutsumi, K., Kato, H., Matsushita, H., Shiraha, H., Fujii, M., et al. (2021). Extracellular Vesicle-Shuttled miRNAs as a Diagnostic and Prognostic Biomarker and Their Potential Roles in Gallbladder Cancer Patients. Sci. Rep. 11 (1), 12298–12313. doi:10.1038/s41598-021-91804-0
Wang, M., Ji, S., Shao, G., Zhang, J., Zhao, K., Wang, Z., et al. (2018a). Effect of Exosome Biomarkers for Diagnosis and Prognosis of Breast Cancer Patients. Clin. Transl Oncol. 20 (7), 906–911. doi:10.1007/s12094-017-1805-0
Wang, S., Zeng, Y., Zhou, J.-M., Nie, S.-L., Peng, Q., Gong, J., et al. (2016). MicroRNA-1246 Promotes Growth and Metastasis of Colorectal Cancer Cells Involving CCNG2 Reduction. Mol. Med. Rep. 13 (1), 273–280. doi:10.3892/mmr.2015.4557
Wang, Y., Zhang, C., Zhang, P., Guo, G., Jiang, T., Zhao, X., et al. (2018b). Serum Exosomal microRNAs Combined with Alpha-Fetoprotein as Diagnostic Markers of Hepatocellular Carcinoma. Cancer Med. 7 (5), 1670–1679. doi:10.1002/cam4.1390
Wei, J., Yang, L., Wu, Y.-n., and Xu, J. (2020). Serum miR-1290 and miR-1246 as Potential Diagnostic Biomarkers of Human Pancreatic Cancer. J. Cancer 11 (6), 1325–1333. doi:10.7150/jca.38048
Xie, B., Li, L., Zhang, Z., Zhao, L., Cheng, J., Zhou, C., et al. (2021). MicroRNA-1246 by Targeting AXIN2 and GSK-3β Overcomes Drug Resistance and Induces Apoptosis in Chemo-Resistant Leukemia Cells. J. Cancer 12 (14), 4196–4208. doi:10.7150/jca.58522
Xu, X., Cao, L., Zhang, Y., Lian, H., Sun, Z., and Cui, Y. (2018). MicroRNA-1246 Inhibits Cell Invasion and Epithelial Mesenchymal Transition Process by Targeting CXCR4 in Lung Cancer Cells. Cbm 21 (2), 251–260. doi:10.3233/cbm-170317
Xu, Y.-F., Hannafon, B. N., Khatri, U., Gin, A., and Ding, W.-Q. (2019). The Origin of Exosomal miR-1246 in Human Cancer Cells. RNA Biol. 16 (6), 770–784. doi:10.1080/15476286.2019.1585738
Xu, Y.-F., Hannafon, B. N., Zhao, Y. D., Postier, R. G., and Ding, W.-Q. (2017). Plasma Exosome miR-196a and miR-1246 Are Potential Indicators of Localized Pancreatic Cancer. Oncotarget 8 (44), 77028–77040. doi:10.18632/oncotarget.20332
Yang, F., Xiong, H., Duan, L., Li, Q., Li, X., and Zhou, Y. (2019). MiR-1246 Promotes Metastasis and Invasion of A549 Cells by Targeting GSK-3β‒Mediated Wnt/β-Catenin Pathway. Cancer Res. Treat. 51 (4), 1420–1429. doi:10.4143/crt.2018.638
Yang, Y., Xie, Y. J., Xu, Q., Chen, J. X., Shan, N. C., and Zhang, Y. (2015). Down-regulation of miR-1246 in Cervical Cancer Tissues and its Clinical Significance. Gynecol. Oncol. 138 (3), 683–688. doi:10.1016/j.ygyno.2015.06.015
Yin, C., Zheng, X., Xiang, H., Li, H., Gao, M., Meng, X., et al. (2019). Differential Expression Profile Analysis of Cisplatin-regulated miRNAs in a H-uman G-astric C-ancer C-ell L-ine. Mol. Med. Rep. 20 (2), 1966–1976. doi:10.3892/mmr.2019.10430
Yu, Y., Yu, F., and Sun, P. (2020). MicroRNA-1246 Promotes Melanoma Progression through Targeting FOXA2. Ott 13, 1245–1253. doi:10.2147/ott.s234276
Yuan, D., Xu, J., Wang, J., Pan, Y., Fu, J., Bai, Y., et al. (2016). Extracellular miR-1246 Promotes Lung Cancer Cell Proliferation and Enhances Radioresistance by Directly Targeting DR5. Oncotarget 7 (22), 32707–32722. doi:10.18632/oncotarget.9017
Zhang, B., Chen, J., Ren, Z., Chen, Y., Li, J., Miao, X., et al. (2013). A Specific miRNA Signature Promotes Radioresistance of Human Cervical Cancer Cells. Cancer Cell Int 13 (1), 118–8. doi:10.1186/1475-2867-13-118
Zhang, Q., Cao, L.-Y., Cheng, S.-J., Zhang, A.-M., Jin, X.-S., and Li, Y. (2015). p53-induced microRNA-1246 Inhibits the Cell Growth of Human Hepatocellular Carcinoma Cells by Targeting NFIB. Oncol. Rep. 33 (3), 1335–1341. doi:10.3892/or.2015.3715
Zhang, Y., Liao, J. M., Zeng, S. X., and Lu, H. (2011). p53 Downregulates Down Syndrome‐associated DYRK1A through miR‐1246. EMBO Rep. 12 (8), 811–817. doi:10.1038/embor.2011.98
Zhang, Z., Zhang, L., Yu, G., Sun, Z., Wang, T., Tian, X., et al. (2020). Exosomal miR-1246 and miR-155 as Predictive and Prognostic Biomarkers for Trastuzumab-Based Therapy Resistance in HER2-Positive Breast Cancer. Cancer Chemother. Pharmacol. 86 (6), 761–772. doi:10.1007/s00280-020-04168-z
Keywords: miRNA, MiR-1246, cancer, expression, biomarker, in vivo, in vitro, diagnosis
Citation: Ghafouri-Fard S, Khoshbakht T, Hussen BM, Taheri M and Samadian M (2022) A Review on the Role of miR-1246 in the Pathoetiology of Different Cancers. Front. Mol. Biosci. 8:771835. doi: 10.3389/fmolb.2021.771835
Received: 07 September 2021; Accepted: 22 November 2021;
Published: 03 January 2022.
Edited by:
Wei Ye, Guangdong Academy of Science, ChinaReviewed by:
Lincan Duan, Third Affiliated Hospital of Kunming Medical University, ChinaCopyright © 2022 Ghafouri-Fard, Khoshbakht, Hussen, Taheri and Samadian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Taheri, bW9oYW1tYWRfODIzQHlhaG9vLmNvbQ==; Mohammad Samadian, bWRzYW1hZGlhbkBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.