- 1Institute of Transformative Bio-Molecules (WPI-ITbM), Nagoya University, Nagoya, Japan
- 2Department of Chemistry, Graduate School of Science, Nagoya University, Nagoya, Japan
The photo-functional chromophore retinal exhibits a wide variety of optical absorption properties depending on its intermolecular interactions with surrounding proteins and other chromophores. By utilizing these properties, microbial and animal rhodopsins express biological functions such as ion-transport and signal transduction. In this review, we present the molecular mechanisms underlying light absorption in rhodopsins, as revealed by quantum chemical calculations. Here, symmetry-adapted cluster-configuration interaction (SAC-CI), combined quantum mechanical and molecular mechanical (QM/MM), and transition-density-fragment interaction (TDFI) methods are used to describe the electronic structure of the retinal, the surrounding protein environment, and the electronic coupling between chromophores, respectively. These computational approaches provide successful reproductions of experimentally observed absorption and circular dichroism (CD) spectra, as well as insights into the mechanisms of unique optical properties in terms of chromophore-protein electrostatic interactions and chromophore-chromophore electronic couplings. On the basis of the molecular mechanisms revealed in these studies, we also discuss strategies for artificial design of the optical absorption properties of rhodopsins.
Introduction
The photo-functional chromophore retinal can significantly change its optical absorption properties by interacting with surrounding proteins (Wald, 1968; Callender and Honig, 1977; Kochendoerfer et al., 1999; Mathies et al., 2000) and other chromophores (Lanyi and Balashov, 2008; Balashov et al., 2010; Anashkin et al., 2018; Misra et al., 2019). These changes in the optical properties of retinal play an important role in the expression of biological functions such as ion transport and signal transduction in rhodopsin (Birge, 1990; Kandori et al., 2001), which is found in a wide range of organisms from microbes to animals, including humans (Shichida and Imai, 1998; Hampp, 2000; Ebrey and Koutalos, 2001). To clarify the molecular mechanisms underlying these photobiological phenomena from the perspective of theoretical calculations, three points need to be taken into account. The first is to accurately describe the electronic structure of the retinal chromophore in the excited state, the second is to efficiently incorporate the effect of the surrounding protein environment of the retinal chromophore, and the third is to quantitatively analyze the interaction between chromophores in the excited state. The symmetry-adapted cluster-configuration interaction (SAC-CI) (Nakatsuji, 1978; Nakatsuji, 1979a; Nakatsuji, 1979b), combined quantum mechanical and molecular mechanical (QM/MM) (Warshel and Levitt, 1976; Senn and Thiel, 2009; Brunk and Rothlisberger, 2015), and transition-density-fragment interaction (TDFI) (Fujimoto and Hayashi, 2009; Fujimoto, 2010; Fujimoto, 2012) methods are effective tools for considering each of the above three points.
The SAC-CI method (Nakatsuji, 1978; Nakatsuji, 1979a; Nakatsuji, 1979b) is an electronic structure theory (a method of quantum chemistry) proposed by Nakatsuji. This method has the advantage of accurately describing the molecular ground and excited states, and has been applied to numerous molecules. In particular, many spectroscopic studies have been carried out using the SAC-CI method, and the effectiveness of this method has been demonstrated by successfully attributing spectra that were difficult to understand experimentally. It should be emphasized that the SAC-CI method satisfies the requirements of orthogonality and Hamiltonian orthogonality between the ground and excited states for an accurate wave function. Another feature of the SAC-CI program is the use of an efficient computational technique called the perturbation selection method (Nakatsuji, 1983). This has realized a significant reduction in computational cost. The SAC-CI method is implemented in the Gaussian program package (Frisch et al., 2003) and is widely used in chemistry and physics research involving various electronic states.
The QM/MM method (Warshel and Levitt, 1976; Senn and Thiel, 2009; Brunk and Rothlisberger, 2015) is a hybrid approach of quantum chemistry and molecular mechanics, and has been established as an effective tool for describing protein environments. Warshel and Levitt, who proposed the QM/MM method, along with Karplus, won the 2013 Nobel Prize in Chemistry for “the development of multiscale models for complex chemical systems”. Large-scale quantum chemical calculations have been frequently performed on whole proteins using the divide and conquer (DC) method (Yang, 1991; Yang and Lee, 1995) proposed by Yang and the fragment molecular orbital (FMO) method (Kitaura et al., 1999; Nakano et al., 2002) proposed by Kitaura et al. However, most of these are single point calculations. Although computational techniques for geometry optimization using these methods have been developed (Zhao and Yang, 1995; Fedorov et al., 2007), their application to whole proteins is not practical due to the huge computational cost. On the other hand, the QM/MM method incorporates the interactions that are intrinsically important in the protein environment into the quantum chemical calculation (i.e., the electrostatic potential from the protein is included in the Fock operator), and treats the other interactions classically. Such an efficient computation by the QM/MM method enables us to perform geometry optimization of proteins. The QM/MM optimization using ab initio method (Hayashi and Ohmine, 2000; Hayashi et al., 2001) as well as the semiempirical method (Ren et al., 2001; Bondar et al., 2004) for the QM part was successful for retinal proteins. The QM/MM method is also applicable to excited state calculations, and the excitation energies (i.e., the energy difference between the ground state and the excited state, corresponding to the absorption energy) presented below are obtained with the SAC-CI method for the QM part.
The TDFI method was developed by the author to describe electronic coupling, which is an intermediate physical quantity to explain the intermolecular interaction between different electronic states. The dipole-dipole (DD) approximation (Förster, 1948), a conventional method for electronic coupling calculations, has a simple and intuitive form using intermolecular orientation and intermolecular distance based on transition dipoles, while the DD method has a limitation of application arising from the assumption that the intermolecular distance between donor and acceptor is larger than their molecular sizes (Speiser, 1996). Many computational methods for electronic coupling have been developed to overcome the problem of the DD method (Chang, 1977; Krueger et al., 1998; Tretiak et al., 2000; Hsu et al., 2001; Iozzi et al., 2004; Wong et al., 2004; Madjet et al., 2006; Neugebauer, 2007; Fink et al., 2008; Fückel et al., 2008; Vura-Weis et al., 2010; Kawatsu et al., 2011; Voityuk, 2013; Fujimoto, 2014; Błasiak et al., 2015). The transition density cube (TDC) method (Krueger et al., 1998) developed by Krueger et al. is a pioneering approach for calculating electronic couplings using the transition densities of molecular fragments. Similar to the TDC method, the TDFI method also uses the transition densities of molecular fragments, but with technical improvements such as the use of atomic orbital (AO) two-electron integrals for spatial integration (Fujimoto and Hayashi, 2009; Fujimoto, 2010) and the self-consistent incorporation of interactions between molecular fragments (Fujimoto and Yang, 2008; Fujimoto, 2010). As a result, the TDFI method achieves highly accurate electronic coupling calculations even for systems with small intermolecular distances (Fujimoto, 2010; Fujimoto, 2012). In addition, while the DD and TDC methods can only evaluate the Coulomb interaction in the electronic coupling, the TDFI method can evaluate not only the Coulomb interaction but also the exchange interaction and the higher-order interactions using perturbation expansion (Fujimoto, 2012; Fujimoto and Kitamura, 2013; Fujimoto, 2015). Such an estimate of each component in electronic coupling is useful for analyzing molecular mechanisms.
In this review, we present three topics on the light absorption properties of retinal proteins revealed by the application of the SAC-CI, QM/MM, and TDFI methods (Fujimoto et al., 2008; Fujimoto et al., 2009; Fujimoto and Hayashi, 2009; Fujimoto and Balashov, 2017; Fujimoto and Inoue, 2020): first, the color tuning mechanism of human cone pigments responsible for color vision (Wang et al., 1993; Asenjo et al., 1994; Lin et al., 1998; Fasick et al., 1999; Kandori et al., 2001); second, the excitation energy transfer (EET) that occurs in xanthorhodopsin (XR) (Balashov et al., 2005; Lanyi and Balashov, 2008); and third, the circular dichroism (CD) spectra of XR (Balashov et al., 2006; Smolensky Koganov et al., 2015) and Krokinobacter eikastus rhodopsin 2 (KR2) (Inoue et al., 2013; Gushchin et al., 2015; Kato et al., 2015; Shibata et al., 2018). Here, the term color tuning refers to the change in absorption wavelength of a molecule depending on the protein environment (Shichida and Imai, 1998; Coto et al., 2006), and excitation energy transfer (EET) (Scholes, 2003; May and Kühn, 2011) refers to the phenomenon of simultaneous deexcitation of the donor molecule and excitation of the acceptor molecule. All of these topics are related to light absorption by retinal proteins, but note that the major physical factors are different in each system. In the case of human cone visual pigments, electrostatic interactions between retinal and surrounding proteins play a central role, while in the case of XR and KR2, electronic couplings between chromophores such as retinal-carotenoid and retinal-retinal contribute significantly. Quantitative analysis of electrostatic interactions and electronic couplings is indispensable for sufficiently understanding these molecular mechanisms.
Color Tuning Mechanism of Human Cone Visual Pigments
The human retina contains three types of cone photoreceptors that control color vision: human red (HR), green (HG), and blue (HB) cone pigments. HR, HG, and HB are all retinal proteins, and the retinal chromophores in the proteins are directly involved in the absorption of light. Thus, the separate absorption maxima (563 nm (2.20 eV) for HR, 532 nm (2.33 eV) for HG and 414 nm (2.99 eV) for HB) (Dartnall et al., 1983; Oprian et al., 1991) exhibited by the three cone visual pigments are all carried out by the chemically identical retinal. In other words, the absorption wavelengths of retinals change depending on the environment of the proteins (opsins) surrounding the retinals, and this phenomenon is called color tuning (Kochendoerfer et al., 1999). In the case of bovine rhodopsin, the crystal structures were solved (Palczewski et al., 2000; Okada et al., 2002) and many theoretical studies on the color tuning mechanism have been done by Olivucci et al. (Ferré and Olivucci, 2003; Andruniów et al., 2004; Coto et al., 2006), Buss et al. (Hufen et al., 2004; Sekharan et al., 2007), Elstner et al. (Wanko et al., 2005; Wanko et al., 2008), Morokuma et al. (Altun et al., 2008a; Altun et al., 2008b), and others (Gascón et al., 2006; Tomasello et al., 2009). In contrast, little attention has been paid to the study of cone visual pigments due to their unknown three-dimensional protein structures. At the time of our study, the excited state calculations for the cone visual pigments were done only by Trabanino et al. (2006). However, this study was performed at the semi-quantitative CI-Singles level, and it was difficult to analyze the color tuning mechanism. In this study, we thus used the SAC-CI method, which is a quantitative electronic structure theory, to clarify the color tuning mechanism of the human cone visual pigments.
Since the crystal structures of the human cone visual pigments have not been available, we employed homology modeling structures [PDB ID: 1KPX, 1KPW, 1KPN for HR, HG, and HB, respectively (Stenkamp et al., 2002)], three-dimensional protein structures constructed by using the similarity of amino acid sequence with experimentally solved protein structures. The protonation states of charged amino acids were then evaluated for the homology modeling structure by Poisson-Boltzmann calculations using the MEAD program (Bashford and Gerwert, 1992) and the overall protein structures were further refined by the QM/MM geometry optimization. Thus, the protein structures of the three cone visual pigments were created.
Excited state calculations using the SAC-CI method were performed for these structures. As a result, the experimental values of the absorption energies were successfully reproduced with an RMS error of 0.05 eV (HB; 2.94, HG; 2.32, HR; 2.08 eV) (Fujimoto et al., 2008).
Based on these results, the physical origin of the color tuning mechanism was explored (Fujimoto et al., 2008). To this end, the absorption energy for each of the three cone pigments was decomposed into three contributions: the distortion effect of the retinal structure, the electrostatic effect created by the protein environment, and the quantum effect of the counter-amino acid (glutamate anion). The quantum effect here refers to higher-order electronic interactions such as polarization and charge transfer (CT) caused by treating the counter amino acid as the QM atoms. The results show that the protein electrostatic effect makes the largest contribution to the absorption energy of the three cone visual pigments (green in Figure 1), and the distortion effect of the retinal structure also makes a large contribution in HB (light blue in Figure 1), but is not the main cause of the color tuning mechanism. Although the quantum effect of counter-amino acids was important in reproducing the absolute value of the absorption energy, there was no significant difference in the contribution of color tuning in the three cone pigments. From these results, only the electrostatic effect of opsin is discussed in this review.
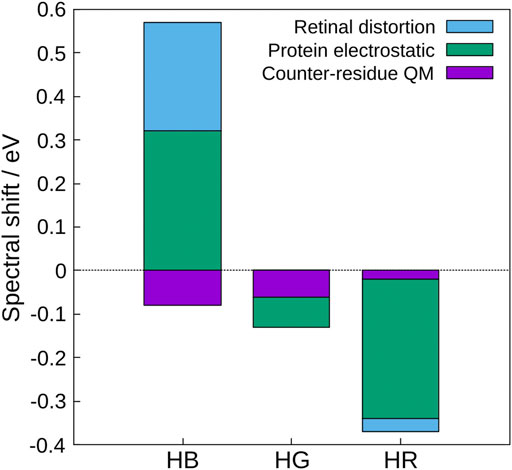
FIGURE 1. Contributions to the spectral shifts of HB, HG, and HR calculated by the decomposition of the shifted energy into the distortion effect of the retinal structure, the electrostatic effect created by the protein environment, and the quantum effect of the counter-amino acid (glutamate anion). The value of bovine rhodopsin was used as a reference.
In order to understand the protein electrostatic effect (Warshel and Levitt, 1976), let us consider 1) the character of the first excited state of the retinal chromophore and 2) the electrostatic potential (ESP) created by the protein environment of the cone visual pigments (Fujimoto et al., 2007). 1) The SAC-CI wavefunction showed that the first excited state of the retinal chromophore is characterized as a one-electron transition from the highest occupied molecular orbital (HOMO) to the lowest unoccupied molecular orbital (LUMO) (HOMO-LUMO transition). As can be seen from the HOMO and LUMO shown in Figures 2A,B, the HOMO is distributed on the left side (β-ionone ring side) and the LUMO on the right side (Schiff base side) of the retinal chromophore. Such an electronic transition between orbitals with different distributions implies that the first excited state of the retinal is of intramolecular CT character, which has also been confirmed by CASPT2 (Coto et al., 2006), SORCI + Q (Altun et al., 2008a), and experiments (Mathies and Stryer, 1976; Schenkl et al., 2005). Keeping in mind the character of the retinal chromophore, we next consider 2) the ESP by the protein environment. Note that this ESP is created by opsin on the retinal, not by the retinal itself. As shown in Figure 2C, the ESP created by opsin on the retinal π-chain is significantly negative in the region where LUMO is distributed (i.e., the Schiff base side). How does such a large negative ESP on the Schiff base side affect the molecular orbitals of the retinal? It is a specific destabilization of the energy level of LUMO. Negative ESP destabilizes the orbital energies of both the HOMO and LUMO, but specifically large negative ESP on the Schiff base side destabilizes the LUMO to a greater extent than the HOMO. Therefore, the difference between the HOMO and LUMO orbital energies of the retinal (HOMO-LUMO gap) is larger in the presence of the protein environment than in the gas phase (Fujimoto et al., 2007). This is the mechanism by which the protein electrostatic effect increases the absorption energy of the retinal chromophore.
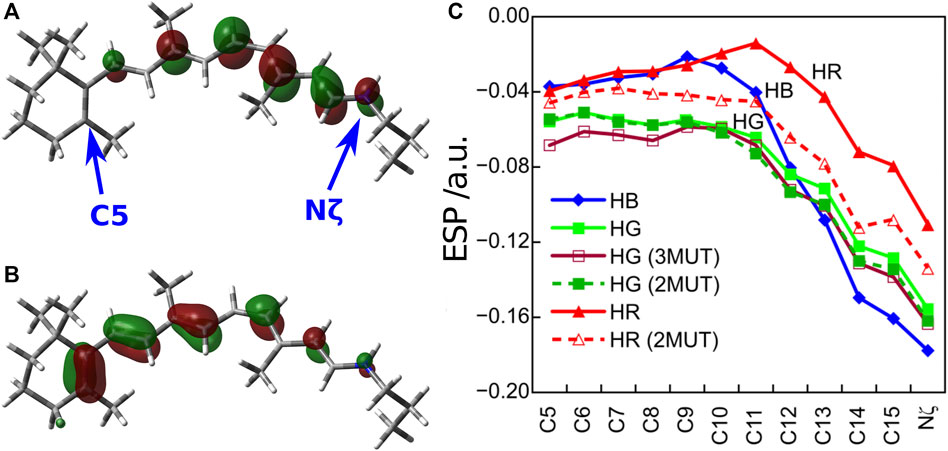
FIGURE 2. (A) LUMO and (B) HOMO distributions of the retinal chromophore. (C) ESP created by opsin on the retinal π-chain. Atoms C5 and Nζ are located on the β-ionone ring and Schiff base region, respectively.
How does the difference in the absorption energy of the three cone pigments arise? It comes from the difference in the degree of LUMO destabilization caused by the ESP of the three types of opsins (Fujimoto et al., 2009). As shown in Figure 2C, the degree of negative ESP on the Schiff base side relative to the β-ionone ring side increases in the order HR < HG < HB. As a result, the destabilization of the LUMO level is largest for HB and smallest for HR. Therefore, the HOMO-LUMO gap is the largest in HB and the smallest in HR, which leads to the difference in the absorption energy of the three cone pigments. From these results, we could clearly understand the color tuning mechanism of the human cone visual pigments caused by the different electrostatic effects (ESP) of opsin.
The next question is, which amino acid makes the difference in the electrostatic effect among the three cone pigments? To answer this question, we have defined the electrostatic energy of each amino acid that contributes to the absorption energy.
where
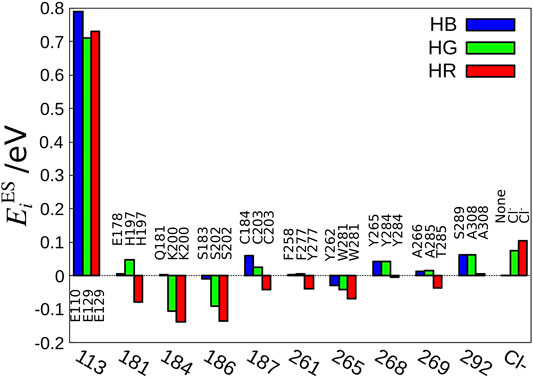
FIGURE 3. Amino acid sequences important for the color tuning. The amino acid number of bovine rhodopsin is shown as the standard value because the amino acid numbers are different among HB, HG, and HR.
Excitation Energy Transfer in Xanthorhodopsin
EET is a phenomenon in which the electronic excitation of an acceptor molecule occurs simultaneously with the electronic deexcitation of a donor molecule. Fluorescence resonance energy transfer (FRET) (Berezin and Achilefu, 2010) is also a type of EET that occurs between fluorescent chemical compounds, and is widely used as an effective tool for dynamically visualizing the biological functions of gene products in living cells (Jares-Erijman and Jovin, 2003; Miyawaki, 2003). While it was widely known that the light-harvesting antenna of photosynthesis efficiently collects solar energy by utilizing EET phenomena in vivo (Scholes et al., 2011; Hu et al., 1997; Adolphs et al., 2010), Balashov et al. discovered a new EET system in XR, a member of retinal protein with proton pumping function (Lanyi and Balashov, 2008; Balashov et al., 2005). Here, salinixanthin (donor), a type of carotenoid bound to the surface of the XR protein, plays the role of a light-harvesting antenna, and the light energy captured there triggers electronic excitation of retinal (acceptors) via EET (Figure 4). The light-harvesting antenna in photosynthesis is formed by the aggregation of many chromophores [e.g., 27 bacteriochlorophylls in the case of light-harvesting complex 2 (LH2) (Papiz et al., 2003; Cogdell et al., 2006)], whereas in XR, only one molecule of carotenoid plays the antenna function (Luecke et al., 2008). Such a simple antenna structure in XR is theoretically tractable and useful for studying EETs in biomolecular systems.
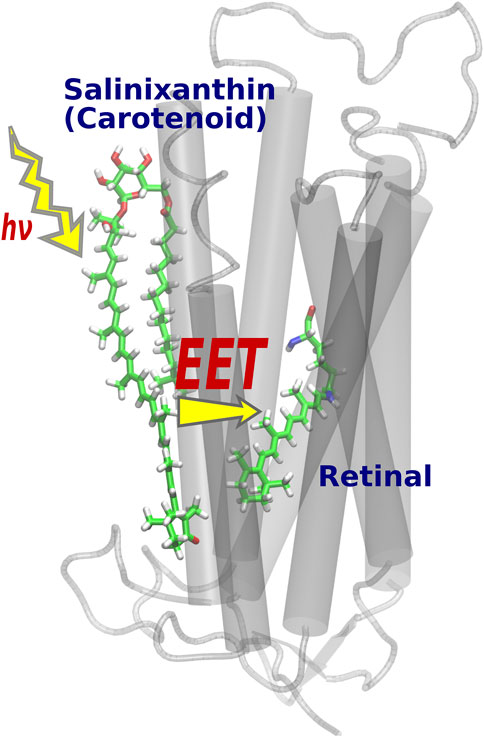
FIGURE 4. Optimized structure of XR. After light absorption by the salinixanthin added on the protein surface, the retinal is excited via EET.
In the theoretical study of XR, we need to pay attention to the small distance between the salinixanthin and retinal. The distance between the centers of mass of both molecules (13 Å) is smaller than the size of the molecules (∼36 Å) (Luecke et al., 2008), so the DD method cannot be applied to XR. In fact, the electronic coupling calculation using the DD method yields a result of −751 cm−1 (Fujimoto and Balashov, 2017), confirming that the experimental value of 160–210 cm−1 (absolute value) (Polívka et al., 2009) cannot be reproduced. Note that the sign of the electronic coupling cannot be determined because the experimental electronic coupling was derived from the squared form contained in Förster’s EET rate (Förster, 1948).
With respect to the small donor-acceptor distance of XR, another consideration must be made in addition to the limited applicability of the DD method. The Förster-type (Förster, 1948) and Dexter-type (Dexter, 1953) theories are often used to explain the molecular mechanism of EET (Parson, 2007) (Figure 5). The Förster mechanism, which originates from the Coulomb interaction, is dominant when the intermolecular distance between donor and acceptor is large, while the Dexter mechanism, which is caused by the exchange interaction, dominates when the donor-acceptor intermolecular distance is small (Speiser, 1996; Berezin and Achilefu, 2010). Therefore, it has been considered that a Dexter-type EET may occur in XR with a small donor-acceptor distance (the smallest distance is 3.9 Å). However, it should be noted that most of the proposed methods for calculating the electronic coupling, such as the DD and TDC methods, can only describe the Coulomb interaction and cannot evaluate the exchange interaction. Therefore, it remains unclear whether the Dexter mechanism is involved in the EET of XR.
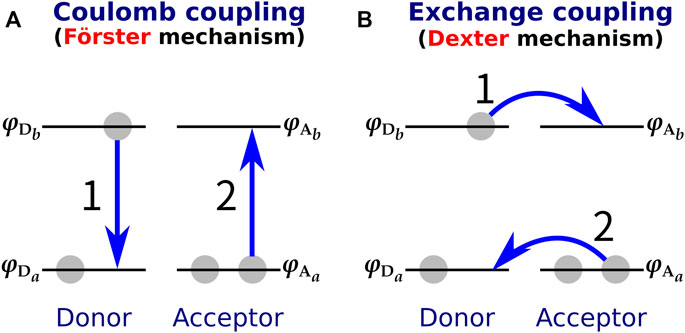
FIGURE 5. (A) Förster-type and (B) Dexter-type EET mechanisms. In the Förster type, EET is triggered by Coulomb coupling between donor and acceptor, while in the Dexter type it is caused by exchange coupling. The numbers 1 and 2 in the figure indicate the order of the electronic transitions.
The TDFI method has the advantage of being able to calculate the exchange interaction
where
The TDFI analysis of the electronic couplings provided us with a better understanding of the EET mechanism in XR. We also examined the effect of the orientation of the salinixanthin toward the retinal on the magnitude of the electronic coupling. First, we used Euler angles to generate artificial orientations of salinixanthin around the XR protein. Here, the salinixanthin was rotated in three dimensions on the surface of the XR protein, and 360 optimal conformations were generated (Figure 6A). The electronic couplings were then calculated for these structures (Figure 6B). The results show that the native carotenoid orientation (θz = 0°) yields the largest electronic coupling (EET efficiency: 40%). The second largest electronic coupling is 153.3 cm−1 at 48° (EET efficiency: 25%), which is 63 cm−1 smaller than the native orientation (0°). These results indicate that the native salinixanthin in XR is oriented to give large electronic coupling (high EET efficiency). The results in Figure 6B also show that the values of electronic coupling strongly depend on the salinixanthin orientation. An orientation with small electronic coupling corresponds to the fact that the light energy absorbed by salinixanthin is not transferred to the retinal. Therefore, this result indicates that the presence of salinixanthin on the XR surface does not necessarily lead to EET.
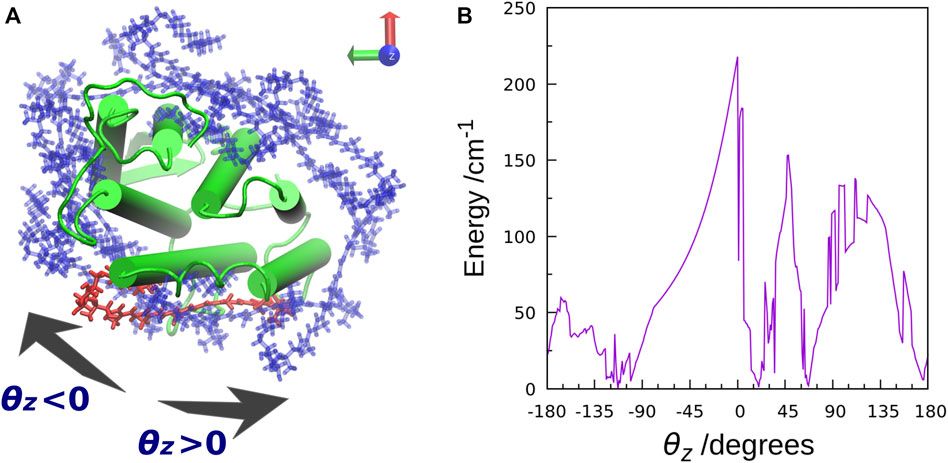
FIGURE 6. (A) Top view of the salinixanthin orientations generated around the XR protein using Euler angles. The rotation angle is taken as θz. The native salinixanthin structure is shown in red, which is set to θz = 0. (B) Calculated electronic coupling energy between salinixanthin and retinal as a function of θz.
Archaerhodopsin 2 (AR2) is a member of retinal protein that also contains a carotenoid, bacterioruberin, but is known to have no EET function (Yoshimura and Kouyama, 2008). In order to investigate the difference in EET function between XR and AR2, a carotenoid, salinixanthin, mimicking the bacterioruberin orientation in AR2 was placed in XR, and electronic coupling calculations were performed for the structure. As a result, a very small value of 5.9 cm−1 was obtained (EET efficiency: 0.05%). This result demonstrates that we were able to reproduce the AR2-like state without EET function using XR by simply changing the salinixanthin orientation to the bacterioruberin one in AR2 without using bacterioruberin. At this stage, we cannot answer the question, “What is the role of bacterioruberin in AR2?”. However, we have found that at least the carotenoid orientation plays a major role in the expression of EET function.
Effect of Electronic Coupling on Circular Dichroism Spectra
Circular Dichroism Spectrum of Xanthorhodopsin
Electronic coupling (also called excitonic coupling) between molecules of the same species is known to induce two bands of opposite sign (biphasic bands) in CD spectra, and this phenomenon is called exciton-coupled CD (ECCD) (Harada and Nakanishi, 1983; Berova et al., 2000). ECCD bands also appear in biomolecules, and a typical example is the negative and positive CD bands observed in the bacteriorhodopsin (BR) trimer (Cassim, 1992; Pescitelli and Woody, 2012). However, there are few reports of ECCD in other biomolecules (Tsukamoto et al., 2013; Pescitelli et al., 2014; Iizuka et al., 2019), and the details of the mechanism are not well understood.
In general, the CD spectra of monomeric retinal proteins in the resting state show a single positive CD band at the absorption wavelength of the retinal (Cassim, 1992). On the other hand, the CD spectrum of XR is known to show negative and positive biphasic bands (Balashov et al., 2006). Three possibilities have been considered for the origin of the CD bands of XR: first, the distortion effect of the salinixanthin structure; second, the effect of electronic coupling between the salinixanthin and retinal; and third, the effect of multimer formation of XR. Here, the third effect refers to the retinal-retinal or salinixanthin-salinixanthin electronic coupling caused by the multimeric formation of XR (Smolensky Koganov et al., 2015). However, it is not clear what kind of multimeric structure is formed by XR. Therefore, we examined two of the above three possibilities, excluding the effect of multimer formation of XR.
In order to investigate the distortion effect of the salinixanthin structure (rotation of the C6-C7 single bond) on the CD spectrum, we calculated the CD spectrum by gradually changing the rotation angle of the C6-C7 single bond. As a result, it was confirmed that the sign of the CD band did not change at any rotation angle (Fujimoto and Balashov, 2017). Therefore, the C6-C7 single bond rotation of the salinixanthin structure is not a factor that characterizes the shape of the CD spectrum of XR.
Next, we investigated the effect of electronic coupling on the CD spectra (Fujimoto and Balashov, 2017). To this end, we employed two models (exciton model and no-coupling model) for the CD spectrum calculations. The Hamiltonian matrix of the exciton model is represented by
where the diagonal element
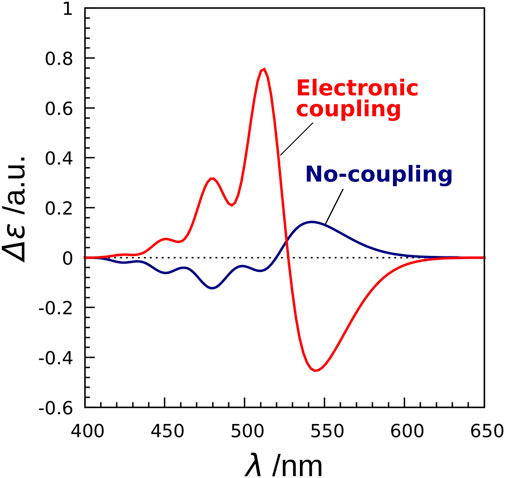
FIGURE 7. CD spectra of XR calculated by the exciton model and the no-coupling model. The no-coupling model does not take into account the electronic couplings in the exciton Hamiltonian.
Circular Dichroism Spectrum of Krokinobacter Eikastus Rhodopsin 2
In the case of XR above, we could not analyze the effect of multimer formation on the CD spectrum because there was no structural information on the multimer. To investigate the effect of multimer formation, we attempted to calculate the CD spectra of retinal proteins whose multimeric structures have been solved experimentally. Here, we focused on KR2, a light-driven sodium pumping retinal protein, which is closely related to XR in the phylogenetic tree (Shibata et al., 2018). Recently, KR2 has been attracting attention as an optogenetics tool in the field of neuroscience (Deisseroth et al., 2006; Wu et al., 2009; Kato et al., 2015). As mentioned above, the CD spectrum of monomeric retinal proteins in the resting state generally shows a single positive CD band originating from the photoexcitation of the retinal. In contrast, BR is known to produce negative and positive ECCD bands by forming a trimer (Cassim, 1992; Pescitelli and Woody, 2012). Therefore, it has been expected that KR2, which forms a pentamer (Gushchin et al., 2015), also exhibits biphasic CD bands similar to those of the BR timer. However, the measured CD spectrum of KR2 showed only a single positive CD band (Shibata et al., 2018). This result gave the possibility that the electronic coupling in the pentamer of KR2 does not affect the shape of the CD spectrum. On the other hand, the peak position of this CD band was red-shifted compared to the peak position of the absorption spectrum, suggesting that multimeric interactions may have affected the CD spectrum. These facts raised the question of whether the retinal-retinal electronic coupling in the KR2 pentamer affects its CD spectrum. However, since no theoretical studies on the CD spectra of KR2 pentamers have been performed, this molecular mechanism remains unclear.
In order to clarify the mechanism, we performed calculations using a combination of the TDFI method and exciton model as in the case of XR described above, and analyzed the obtained spectra (Fujimoto and Inoue, 2020). This study also employed the SAC-CI method to determine the excitation energy and transition density of retinal. First, we calculated the electronic coupling between the retinals in the KR2 pentamer and obtained a value of 25 cm−1. This value is much smaller than the carotenoid-retinal electronic coupling in XR (227 cm−1). The TDFI analysis then revealed that the Coulomb interaction (25 cm−1) was the main cause of the electronic coupling, with no contribution from the exchange interaction (0 cm−1). This is due to the fact that the intermolecular distance between the retinals is 25 Å. Using the obtained electronic coupling values, the absorption spectra of the monomeric and pentameric structures of KR2 were calculated, and both showed almost the same absorption maxima. Next, the CD spectra were also calculated (Figure 8). The CD spectrum of the monomer showed a band at the same wavelength as the absorption spectrum. On the other hand, the CD spectrum of the pentamer showed an overall positive band, although there was a small negative peak at 471 nm. This positive CD band was red-shifted by 26 nm (0.11 eV) compared to the peak in the absorption spectrum, reproducing the red-shift observed in the experimental CD spectrum. In order to further analyze the effect of multimer formation, we additionally calculated the absorption and CD spectra using the no coupling model. As a result, the calculated CD spectrum showed a peak position at the same wavelength as the absorption spectrum. From these results, we found that the CD spectrum is affected by the retinal-retinal interaction due to multimer formation (Fujimoto and Inoue, 2020).
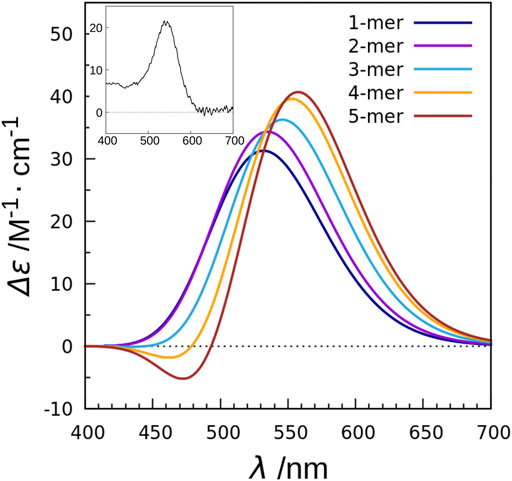
FIGURE 8. CD spectra calculated with the exciton model for monomer to pentamer in KR2. The values are given per monomer. The experimental absorption spectrum of KR2 (Shibata et al., 2018) is shown in the inset.
In the case of the BR trimer, the effect of electronic coupling between the retinals generates negative and positive biphasic CD bands, with the negative band occurring on the longer wavelength side, which is called negative chirality. On the other hand, in the CD spectrum of the KR2 pentamer, a large positive band is generated on the longer wavelength side than a small negative band due to the effect of electronic coupling, which is regarded as positive chirality. This mechanism could be understood from the orientation of the retinals in the pentamer structure of KR2. As illustrated in Figure 9, the orientation of the transition dipoles of the two retinals shows a clockwise rotation from retinal A in the foreground to retinal B in the background (Fujimoto and Inoue, 2020). This clockwise rotation corresponds to positive chirality according to the exciton chirality method (Harada and Nakanishi, 1969; Harada and Nakanishi, 1983) by Harada and Nakanishi. From these results, we conclude that the positive band contributes significantly to the CD spectrum of KR2 due to the orientation between the retinals corresponding to the clockwise rotation of the two transition dipoles.
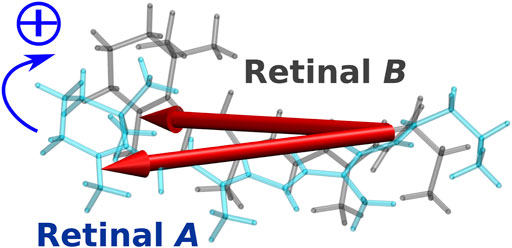
FIGURE 9. Intermolecular conformation of the two retinal chromophores (retinal A and retinal B) in KR2. The two red arrows represent the transition dipole moments of the two retinal chromophores.
Artificial Design of Light Absorption by Retinal Proteins
Several studies described above have shown that the optical properties of retinal proteins with respect to light absorption are strongly influenced by electrostatic interactions with surrounding proteins and by electronic couplings between chromophores. Here, we discuss the possibility of designing new optical properties by artificially altering these two interactions.
The first is the artificial control of the electrostatic interaction between the retinal and the surrounding protein. As mentioned above, the first excited state of the retinal chromophore has the character of intramolecular CT, so by changing the magnitude of the ESP formed by the protein along the retinal π-chain (Figure 2C), the absorption wavelength of the retinal can be changed. A direct way to change the ESP is to introduce charged amino acids virtually on a computer. Here, the ESP formed by the charged residues needs to be quantified according to the position of the residues (Ferré and Olivucci, 2003; Shtyrov et al., 2021). However, it should be noted that the three-dimensional structure of the mutant protein obtained by this operation may be quite different from the real one due to the change in the charge balance of the protein. The evaluation of the homology model of rhodopsin is discussed in Ref. (Nikolaev et al., 2018). In addition to charged amino acids, the introduction of amino acids with OH groups, such as serine and tyrosine, is an effective way to modify the ESP (Figure 2C) (Fujimoto et al., 2009). Mutant experiments by Asenjo et al. (1994) revealed that three OH amino acids in the vicinity of the β-ionone ring of retinal contribute significantly to the longer absorption wavelength of HR than that of HG. We have shown that the orientation of the OH group in these amino acids contributes to the red shift, and that the orientation of the oxygen of the OH group toward the β-ionone ring causes a specific negative ESP on the β-ionone ring side, which results in a red shift in the absorption wavelength of HR (Fujimoto et al., 2009). The contribution of the OH group orientation to the red shift was also confirmed by the theoretical mutants, in which the neutral amino acids Ala180, Phe277, and Ala285 in HG were replaced by the corresponding OH group amino acids Ser, Tyr, and Thr in HR, respectively (Fujimoto et al., 2009). The absorption energies of the three mutant structures were calculated to be 2.20 eV, which is in good agreement with the experimental value of 2.33 eV. Although the above example concerns amino acids that produce the difference in absorption wavelengths between HR and HG, we believe that this strategy can be applied to the artificial design of absorption wavelengths for a number of retinal proteins. We note here that accurate prediction of the three-dimensional structure of the mutant protein will be a critical process for the success of this strategy. Electrostatic effects on the color tuning of rhodopsin are also known to be due to the position of internal water molecules (Tsutsui and Shichida, 2010; Nikolaev et al., 2020), or indirectly due to the substitution of one residue causing rearrangement of others (Ryazantsev et al., 2012). Taking these effects into account when constructing three-dimensional structures will lead to more accurate artificial designs.
The second is to modulate the electronic coupling by adding some chromophore on the surface of retinal protein. As mentioned above, XR acquired a new optical absorption property with EET by adding the salinixanthin on the protein surface. Our analysis for XR revealed that controlling the carotenoid-retinal orientation is more important than the type of carotenoids to increase the EET efficiency (Fujimoto and Hayashi, 2009). This analysis was done by using Euler angles to exhaustively search for the salinixanthin conformations, but other approaches are also possible. The author is also working on computational drug discovery and has developed a method for predicting the binding pose of a ligand to a protein, called molecular docking. Accurate molecular docking methods, such as the artificial bee colony (ABC) (Karaboga and Basturk, 2007) algorithm-based docking method developed by the authors (Uehara et al., 2015), will enable the exploration of ligands to be added on the surface of retinal proteins, which is expected to lead to the construction of new EET systems. An attempt to add salinixanthin to retinal proteins other than XR have already been reported (Misra et al., 2019), but the use of computational methods prior to such challenges will further expand the scope of research.
Future Perspective
In this review, we have presented the molecular mechanisms of retinal proteins (human cone visual pigments, XR, and KR2) revealed by using three types of quantum chemical calculations. For details of the methods and mechanisms, please refer to the original papers.
We have described the Förster-type and Dexter-type mechanisms of EET of XR, but the author has also studied another type of EET mechanism called “EET via CT states (Harcourt et al., 1994; May and Kühn, 2011)” and has shown that this third mechanism strongly contributes to ethylene dimer (Fujimoto, 2012; Fujimoto, 2015). In the future, we would like to explore the effects of this mechanism on biomolecular systems.
The CD spectral analysis of KR2 revealed that the retinal-retinal electronic coupling in the pentamer structure affects the shape of the CD spectrum. A comprehensive understanding of the relationship between protein multimer formation (Brown and Ernst, 2017) and CD spectral shape (Shibata et al., 2018) is a future challenge.
A technical challenge in dealing with protein multimers is the reduction of computational cost. In the electronic coupling calculations, the author has developed the transition charge, dipole, and quadrupole from ESP (TrESP-CDQ) method (Fujimoto, 2014), which is less computationally expensive than the TDFI method. The use of such a method is expected to contribute to efficient analysis for protein multimers.
The methods presented in this review do not consider the effects of protein fluctuations. To what extent do protein fluctuations affect the excited state properties (optical properties) of chromophores such as retinal and carotenoids? To investigate it, benchmark calculations for excited states have been made using a combination of semiempirical method and MD simulation (Bold et al., 2020). By adding statistical processing to the method presented in this study, we hope to quantitatively clarify the effect of protein fluctuation on the optical properties of rhodopsin, which will contribute to the elucidation of the molecular mechanism in more detail.
Author Contributions
KJF researched and wrote the manuscript and made the figures.
Funding
This study was supported by JSPS KAKENHI Grant No. 20K05430.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adolphs, J., Müh, F., Madjet, M. E.-A., Busch, M. S. a., and Renger, T. (2010). Structure-Based Calculations of Optical Spectra of Photosystem I Suggest an Asymmetric Light-Harvesting Process. J. Am. Chem. Soc. 132, 3331–3343. doi:10.1021/ja9072222
Altun, A., Yokoyama, S., and Morokuma, K. (2008). Mechanism of Spectral Tuning Going from Retinal In Vacuo to Bovine Rhodopsin and its Mutants: Multireference Ab Initio Quantum Mechanics/Molecular Mechanics Studies. J. Phys. Chem. B 112, 16883–16890. doi:10.1021/jp807172h
Altun, A., Yokoyama, S., and Morokuma, K. (2008). Spectral Tuning in Visual Pigments: An ONIOM(QM:MM) Study on Bovine Rhodopsin and its Mutants. J. Phys. Chem. B 112, 6814–6827. doi:10.1021/jp709730b
Anashkin, V. A., Bertsova, Y. V., Mamedov, A. M., Mamedov, M. D., Arutyunyan, A. M., Baykov, A. A., et al. (2018). Engineering a Carotenoid-Binding Site in Dokdonia Sp. PRO95 Na+-Translocating Rhodopsin by a Single Amino Acid Substitution. Photosynth. Res. 136, 161–169. doi:10.1007/s11120-017-0453-0
Andruniów, T., Ferré, N., and Olivucci, M. (2004). Structure, Initial Excited-State Relaxation, and Energy Storage of Rhodopsin Resolved at the Multiconfigurational Perturbation Theory Level. Proc. Natl. Acad. Sci. 101, 17908–17913. doi:10.1073/pnas.0407997101
Asenjo, A. B., Rim, J., and Oprian, D. D. (1994). Molecular Determinants of Human Red/Green Color Discrimination. Neuron 12, 1131–1138. doi:10.1016/0896-6273(94)90320-4
Balashov, S. P., Imasheva, E. S., Boichenko, V. A., Antón, J., Wang, J. M., and Lanyi, J. K. (2005). Xanthorhodopsin: A Proton Pump with a Light-Harvesting Carotenoid Antenna. Science 309, 2061–2064. doi:10.1126/science.1118046
Balashov, S. P., Imasheva, E. S., Choi, A. R., Jung, K.-H., Liaaen-Jensen, S., and Lanyi, J. K. (2010). Reconstitution of Gloeobacter Rhodopsin with Echinenone: Role of the 4-Keto Group. Biochemistry 49, 9792–9799. doi:10.1021/bi1014166
Balashov, S. P., Imasheva, E. S., and Lanyi, J. K. (2006). Induced Chirality of the Light-Harvesting Carotenoid Salinixanthin and its Interaction with the Retinal of Xanthorhodopsin†. Biochemistry 45, 10998–11004. doi:10.1021/bi061098i
Bashford, D., and Gerwert, K. (1992). Electrostatic Calculations of the pKa Values of Ionizable Groups in Bacteriorhodopsin. J. Mol. Biol. 224, 473–486. doi:10.1016/0022-2836(92)91009-e
Berezin, M. Y., and Achilefu, S. (2010). Fluorescence Lifetime Measurements and Biological Imaging. Chem. Rev. 110, 2641–2684. doi:10.1021/cr900343z
Berova, N., Nakanishi, K., and Woody, R. W. (2000). Circular Dichroism: Principles and Applications. New York: Wiley VCH.
Birge, R. R. (1990). Nature of the Primary Photochemical Events in Rhodopsin and Bacteriorhodopsin. Biochim. Biophys. Acta (Bba) - Bioenerg. 1016, 293–327. doi:10.1016/0005-2728(90)90163-x
Bold, B. M., Sokolov, M., Maity, S., Wanko, M., Dohmen, P. M., Kranz, J. J., et al. (2020). Benchmark and Performance of Long-Range Corrected Time-dependent Density Functional Tight Binding (LC-TD-DFTB) on Rhodopsins and Light-Harvesting Complexes. Phys. Chem. Chem. Phys. 22, 10500–10518. doi:10.1039/c9cp05753f
Bondar, A.-N., Fischer, S., Smith, J. C., Elstner, M., and Suhai, S. (2004). Key Role of Electrostatic Interactions in Bacteriorhodopsin Proton Transfer. J. Am. Chem. Soc. 126, 14668–14677. doi:10.1021/ja047982i
Brown, L. S., and Ernst, O. P. (2017). Recent Advances in Biophysical Studies of Rhodopsins - Oligomerization, Folding, and Structure. Biochim. Biophys. Acta (Bba) - Proteins Proteomics 1865, 1512–1521. doi:10.1016/j.bbapap.2017.08.007
Brunk, E., and Rothlisberger, U. (2015). Mixed Quantum Mechanical/Molecular Mechanical Molecular Dynamics Simulations of Biological Systems in Ground and Electronically Excited States. Chem. Rev. 115, 6217–6263. doi:10.1021/cr500628b
Błasiak, B., Maj, M., Cho, M., and Góra, R. W. (2015). Distributed Multipolar Expansion Approach to Calculation of Excitation Energy Transfer Couplings. J. Chem. Thory Comput. 11, 3259–3266. doi:10.1021/acs.jctc.5b00216
Callender, R., and Honig, B. (1977). Resonance Raman Studies of Visual Pigments. Annu. Rev. Biophys. Bioeng. 6, 33–55. doi:10.1146/annurev.bb.06.060177.000341
Cassim, J. Y. (1992). Unique Biphasic Band Shape of the Visible Circular Dichroism of Bacteriorhodopsin in Purple Membrane. Biophysical J. 63, 1432–1442. doi:10.1016/s0006-3495(92)81701-0
Chang, J. C. (1977). Monopole Effects on Electronic Excitation Interactions between Large Molecules. I. Application to Energy Transfer in Chlorophylls. J. Chem. Phys. 67, 3901–3909. doi:10.1063/1.435427
Cogdell, R. J., Gall, A., and Köhler, J. (2006). The Architecture and Function of the Light-Harvesting Apparatus of Purple Bacteria: from Single Molecules to In Vivo Membranes. Quart. Rev. Biophys. 39, 227–324. doi:10.1017/s0033583506004434
Coto, P. B., Strambi, A., Ferré, N., and Olivucci, M. (2006). The Color of Rhodopsins at the Ab Initio Multiconfigurational Perturbation Theory Resolution. Proc. Natl. Acad. Sci. 103, 17154–17159. doi:10.1073/pnas.0604048103
Dartnall, H. J. A., Bowmaker, J. K., and Mollon, J. D. (1983). Human Visual Pigments: Microspectrophotometric Results from the Eyes of Seven Persons. Proc. R. Soc. Lond. B. 220, 115–130. doi:10.1098/rspb.1983.0091
Deisseroth, K., Feng, G., Majewska, A. K., Miesenbock, G., Ting, A., and Schnitzer, M. J. (2006). Next-generation Optical Technologies for Illuminating Genetically Targeted Brain Circuits. J. Neurosci. 26, 10380–10386. doi:10.1523/jneurosci.3863-06.2006
Dexter, D. L. (1953). A Theory of Sensitized Luminescence in Solids. J. Chem. Phys. 21, 836–850. doi:10.1063/1.1699044
Ebrey, T., and Koutalos, Y. (2001). Vertebrate Photoreceptors. Prog. Retin. Eye Res. 20, 49–94. doi:10.1016/s1350-9462(00)00014-8
Fasick, J. I., Lee, N., and Oprian, D. D. (1999). Spectral Tuning in the Human Blue Cone Pigment. Biochemistry 38, 11593–11596. doi:10.1021/bi991600h
Fedorov, D. G., Ishida, T., Uebayasi, M., and Kitaura, K. (2007). The Fragment Molecular Orbital Method for Geometry Optimizations of Polypeptides and Proteins. J. Phys. Chem. A. 111, 2722–2732. doi:10.1021/jp0671042
Ferré, N., and Olivucci, M. (2003). Probing the Rhodopsin Cavity with Reduced Retinal Models at the CASPT2//CASSCF/AMBER Level of Theory. J. Am. Chem. Soc. 125, 6868–6869. doi:10.1021/ja035087d
Fink, R. F., Pfister, J., Zhao, H. M., and Engels, B. (2008). Assessment of Quantum Chemical Methods and Basis Sets for Excitation Energy Transfer. Chem. Phys. 346, 275–285. doi:10.1016/j.chemphys.2008.03.014
Förster, T. (1948). Zwischenmolekulare Energiewanderung und Fluoreszenz. Ann. Phys. 437, 55–75. doi:10.1002/andp.19484370105
Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R. J. A., et al. (2003). Gaussian 03. Pittsburgh, PA: Gaussian, Inc.
Fückel, B., Köhn, A., Harding, M. E., Diezemann, G., Hinze, G., Basché, T., et al. (2008). Theoretical Investigation of Electronic Excitation Energy Transfer in Bichromophoric Assemblies. J. Chem. Phys. 128, 074505. doi:10.1063/1.2829531
Fujimoto, K., Hasegawa, J.-y., and Nakatsuji, H. (2009). Color Tuning Mechanism of Human Red, Green, and Blue Cone Pigments: SAC-CI Theoretical Study. Bcsj 82, 1140–1148. doi:10.1246/bcsj.82.1140
Fujimoto, K., Hasegawa, J.-y., and Nakatsuji, H. (2008). Origin of Color Tuning in Human Red, green, and Blue Cone Pigments: SAC-CI and QM/MM Study. Chem. Phys. Lett. 462, 318–320. doi:10.1016/j.cplett.2008.07.111
Fujimoto, K., Hayashi, S., Hasegawa, J.-y., and Nakatsuji, H. (2007). Theoretical Studies on the Color-Tuning Mechanism in Retinal Proteins. J. Chem. Theor. Comput. 3, 605–618. doi:10.1021/ct6002687
Fujimoto, K. J., and Balashov, S. P. (2017). Vibronic Coupling Effect on Circular Dichroism Spectrum: Carotenoid-Retinal Interaction in Xanthorhodopsin. J. Chem. Phys. 146, 095101. doi:10.1063/1.4977045
Fujimoto, K. J. (2014). Electronic Coupling Calculations with Transition Charges, Dipoles, and Quadrupoles Derived from Electrostatic Potential Fitting. J. Chem. Phys. 141, 214105. doi:10.1063/1.4902758
Fujimoto, K. J., and Hayashi, S. (2009). Electronic Coulombic Coupling of Excitation-Energy Transfer in Xanthorhodopsin. J. Am. Chem. Soc. 131, 14152–14153. doi:10.1021/ja905697n
Fujimoto, K. J., and Inoue, K. (2020). Excitonic Coupling Effect on the Circular Dichroism Spectrum of Sodium-Pumping Rhodopsin KR2. J. Chem. Phys. 153, 045101. doi:10.1063/5.0013642
Fujimoto, K. J., and Kitamura, C. (2013). A Theoretical Study of Crystallochromy: Spectral Tuning of Solid-State Tetracenes. J. Chem. Phys. 139, 084511. doi:10.1063/1.4819133
Fujimoto, K. J. (2015). “Theoretical Calculations of Excitation Energy Transfer,” in Chemical Science of π-Electron Systems. Editors T. Akasaka, A. Osuka, S. Fukuzumi, H. Kandori, and Y. Aso (Tokyo, Japan: Springer), 761–777. doi:10.1007/978-4-431-55357-1_45
Fujimoto, K. J. (2010). Transition-density-fragment Interaction Approach for Exciton-Coupled Circular Dichroism Spectra. J. Chem. Phys. 133, 124101. doi:10.1063/1.3480015
Fujimoto, K. J. (2012). Transition-density-fragment Interaction Combined with Transfer Integral Approach for Excitation-Energy Transfer via Charge-Transfer States. J. Chem. Phys. 137, 034101. doi:10.1063/1.4733669
Fujimoto, K., and Yang, W. (2008). Density-fragment Interaction Approach for Quantum-Mechanical/molecular-Mechanical Calculations with Application to the Excited States of a Mg2+-Sensitive Dye. J. Chem. Phys. 129, 054102. doi:10.1063/1.2958257
Gascón, J. A., Sproviero, E. M., and Batista, V. S. (2006). Computational Studies of the Primary Phototransduction Event in Visual Rhodopsin. Acc. Chem. Res. 39, 184–193. doi:10.1021/ar050027t
Gushchin, I., Shevchenko, V., Polovinkin, V., Kovalev, K., Alekseev, A., Round, E., et al. (2015). Crystal Structure of a Light-Driven Sodium Pump. Nat. Struct. Mol. Biol. 22, 390–395. doi:10.1038/nsmb.3002
Hampp, N. (2000). Bacteriorhodopsin as a Photochromic Retinal Protein for Optical Memories. Chem. Rev. 100, 1755–1776. doi:10.1021/cr980072x
Harada, N., and Nakanishi, K. (1983). Circular Dichroic Spectroscopy: Exciton Coupling in Organic Stereochemistry. Mill Valley, CA: University Science Books.
Harada, N., and Nakanishi, K. (1969). Determining the Chiralities of Optically Active Glycols. J. Am. Chem. Soc. 91, 3989–3991. doi:10.1021/ja01042a073
Harcourt, R. D., Scholes, G. D., and Ghiggino, K. P. (1994). Rate Expressions for Excitation Transfer. II. Electronic Considerations of Direct and Through-Configuration Exciton Resonance Interactions. J. Chem. Phys. 101, 10521–10525. doi:10.1063/1.467869
Hayashi, S., and Ohmine, I. (2000). Proton Transfer in Bacteriorhodopsin: Structure, Excitation, IR Spectra, and Potential Energy Surface Analyses by an Ab Initio QM/MM Method. J. Phys. Chem. B 104, 10678–10691. doi:10.1021/jp001508r
Hayashi, S., Tajkhorshid, E., Pebay-Peyroula, E., Royant, A., Landau, E. M., Navarro, J., et al. (2001). Structural Determinants of Spectral Tuning in Retinal ProteinsBacteriorhodopsin vs Sensory Rhodopsin II#. J. Phys. Chem. B 105, 10124–10131. doi:10.1021/jp011362b
Hsu, C.-P., Fleming, G. R., Head-Gordon, M., and Head-Gordon, T. (2001). Excitation Energy Transfer in Condensed media. J. Chem. Phys. 114, 3065–3072. doi:10.1063/1.1338531
Hu, X., Ritz, T., Damjanović, A., and Schulten, K. (1997). Pigment Organization and Transfer of Electronic Excitation in the Photosynthetic Unit of Purple Bacteria. J. Phys. Chem. B 101, 3854–3871. doi:10.1021/jp963777g
Hufen, J., Sugihara, M., and Buss, V. (2004). How the Counterion Affects Ground- and Excited-State Properties of the Rhodopsin Chromophore. J. Phys. Chem. B 108, 20419–20426. doi:10.1021/jp046147k
Iizuka, A., Kajimoto, K., Fujisawa, T., Tsukamoto, T., Aizawa, T., Kamo, N., et al. (2019). Functional Importance of the Oligomer Formation of the Cyanobacterial H+ Pump Gloeobacter Rhodopsin. Sci. Rep. 9, 10711. doi:10.1038/s41598-019-47178-5
Inoue, K., Ono, H., Abe-Yoshizumi, R., Yoshizawa, S., Ito, H., Kogure, K., et al. (2013). A Light-Driven Sodium Ion Pump in marine Bacteria. Nat. Commun. 4, 1678. doi:10.1038/ncomms2689
Iozzi, M. F., Mennucci, B., Tomasi, J., and Cammi, R. (2004). Excitation Energy Transfer (EET) between Molecules in Condensed Matter: A Novel Application of the Polarizable Continuum Model (PCM). J. Chem. Phys. 120, 7029–7040. doi:10.1063/1.1669389
Jares-Erijman, E. A., and Jovin, T. M. (2003). FRET Imaging. Nat. Biotechnol. 21, 1387–1395. doi:10.1038/nbt896
Kandori, H., Shichida, Y., and Yoshizawa, T. (2001). Photoisomerization in Rhodopsin. Biochemistry (Moscow) 66, 1197–1209. doi:10.1023/a:1013123016803
Karaboga, D., and Basturk, B. (2007). A Powerful and Efficient Algorithm for Numerical Function Optimization: Artificial Bee colony (ABC) Algorithm. J. Glob. Optim. 39, 459–471. doi:10.1007/s10898-007-9149-x
Kato, H. E., Inoue, K., Abe-Yoshizumi, R., Kato, Y., Ono, H., Konno, M., et al. (2015). Structural Basis for Na+ Transport Mechanism by a Light-Driven Na+ Pump. Nature 521, 48–53. doi:10.1038/nature14322
Kawatsu, T., Matsuda, K., and Hasegawa, J.-y. (2011). Bridge-Mediated Excitation Energy Transfer Pathways through Protein Media: a Slater Determinant-Based Electronic Coupling Calculation Combined with Localized Molecular Orbitals. J. Phys. Chem. A. 115, 10814–10822. doi:10.1021/jp2068792
Kitaura, K., Sawai, T., Asada, T., Nakano, T., and Uebayasi, M. (1999). Pair Interaction Molecular Orbital Method: an Approximate Computational Method for Molecular Interactions. Chem. Phys. Lett. 312, 319–324. doi:10.1016/s0009-2614(99)00937-9
Kochendoerfer, G. G., Lin, S. W., Sakmar, T. P., and Mathies, R. A. (1999). How Color Visual Pigments Are Tuned. Trends Biochem. Sci. 24, 300–305. doi:10.1016/s0968-0004(99)01432-2
Krueger, B. P., Scholes, G. D., and Fleming, G. R. (1998). Calculation of Couplings and Energy-Transfer Pathways between the Pigments of LH2 by the Ab Initio Transition Density Cube Method. J. Phys. Chem. B 102, 5378–5386. doi:10.1021/jp9811171
Lanyi, J. K., and Balashov, S. P. (2008). Xanthorhodopsin: A Bacteriorhodopsin-like Proton Pump with a Carotenoid Antenna. Biochim. Biophys. Acta (Bba) - Bioenerg. 1777, 684–688. doi:10.1016/j.bbabio.2008.05.005
Lee, C., Yang, W., and Parr, R. G. (1988). Development of the Colle-Salvetti Correlation-Energy Formula into a Functional of the Electron Density. Phys. Rev. B 37, 785–789. doi:10.1103/physrevb.37.785
Lin, S. W., Kochendoerfer, G. G., Carroll, K. S., Wang, D., Mathies, R. A., and Sakmar, T. P. (1998). Mechanisms of Spectral Tuning in Blue Cone Visual Pigments. J. Biol. Chem. 273, 24583–24591. doi:10.1074/jbc.273.38.24583
Luecke, H., Schobert, B., Stagno, J., Imasheva, E. S., Wang, J. M., Balashov, S. P., et al. (2008). Crystallographic Structure of Xanthorhodopsin, the Light-Driven Proton Pump with a Dual Chromophore. Proc. Natl. Acad. Sci. 105, 16561–16565. doi:10.1073/pnas.0807162105
Madjet, M. E., Abdurahman, A., and Renger, T. (2006). Intermolecular Coulomb Couplings from Ab Initio Electrostatic Potentials: Application to Optical Transitions of Strongly Coupled Pigments in Photosynthetic Antennae and Reaction Centers. J. Phys. Chem. B 110, 17268–17281. doi:10.1021/jp0615398
Mathies, R. A., and Lugtenburg, J. (2000). “Chapter 2 the Primary Photoreaction of Rhodopsin,” in Handbook of Biological Physics. Editors D. G. Stavenga, W. J. d. Grip, and E. N. Pugh (Amsterdam: Elsevier Science B. V.), 55–90. doi:10.1016/s1383-8121(00)80005-6
Mathies, R., and Stryer, L. (1976). Retinal Has a Highly Dipolar Vertically Excited Singlet State: Implications for Vision. Proc. Natl. Acad. Sci. 73, 2169–2173. doi:10.1073/pnas.73.7.2169
May, V., and Kühn, O. (2011). Charge and Energy Transfer Dynamics in Molecular Systems. Weinheim: Wiley VCH.
Misra, R., Eliash, T., Sudo, Y., and Sheves, M. (2019). Retinal-Salinixanthin Interactions in a Thermophilic Rhodopsin. J. Phys. Chem. B 123, 10–20. doi:10.1021/acs.jpcb.8b06795
Miyawaki, A. (2003). Visualization of the Spatial and Temporal Dynamics of Intracellular Signaling. Dev. Cel 4, 295–305. doi:10.1016/s1534-5807(03)00060-1
Nakano, T., Kaminuma, T., Sato, T., Fukuzawa, K., Akiyama, Y., Uebayasi, M., et al. (2002). Fragment Molecular Orbital Method: Use of Approximate Electrostatic Potential. Chem. Phys. Lett. 351, 475–480. doi:10.1016/s0009-2614(01)01416-6
Nakatsuji, H. (1983). Cluster Expansion of the Wavefunction, Valence and Rydberg Excitations, Ionizations, and Inner-Valence Ionizations of CO2 and N2O Studied by the Sac and Sac CI Theories. Chem. Phys. 75, 425–441. doi:10.1016/0301-0104(83)85209-4
Nakatsuji, H. (1979). Cluster Expansion of the Wavefunction. Calculation of Electron Correlations in Ground and Excited States by SAC and SAC CI Theories. Chem. Phys. Lett. 67, 334–342. doi:10.1016/0009-2614(79)85173-8
Nakatsuji, H. (1979). Cluster Expansion of the Wavefunction. Electron Correlations in Ground and Excited States by SAC (Symmetry-adapted-cluster) and SAC CI Theories. Chem. Phys. Lett. 67, 329–333. doi:10.1016/0009-2614(79)85172-6
Nakatsuji, N. (1978). Cluster Expansion of the Wavefunction. Excited States. Chem. Phys. Lett. 59, 362–364. doi:10.1016/0009-2614(78)89113-1
Neugebauer, J. (2007). Couplings between Electronic Transitions in a Subsystem Formulation of Time-dependent Density Functional Theory. J. Chem. Phys. 126, 134116. doi:10.1063/1.2713754
Nikolaev, D. M., Shtyrov, A. A., Mereshchenko, A. S., Panov, M. S., Tveryanovich, Y. S., and Ryazantsev, M. N. (2020). An Assessment of Water Placement Algorithms in Quantum Mechanics/molecular Mechanics Modeling: the Case of Rhodopsins' First Spectral Absorption Band Maxima. Phys. Chem. Chem. Phys. 22, 18114–18123. doi:10.1039/d0cp02638g
Nikolaev, D. M., Shtyrov, A. A., Panov, M. S., Jamal, A., Chakchir, O. B., Kochemirovsky, V. A., et al. (2018). A Comparative Study of Modern Homology Modeling Algorithms for Rhodopsin Structure Prediction. ACS Omega 3, 7555–7566. doi:10.1021/acsomega.8b00721
Okada, T., Fujiyoshi, Y., Silow, M., Navarro, J., Landau, E. M., and Shichida, Y. (2002). Functional Role of Internal Water Molecules in Rhodopsin Revealed by X-ray Crystallography. Proc. Natl. Acad. Sci. 99, 5982–5987. doi:10.1073/pnas.082666399
Oprian, D. D., Asenjo, A. B., Lee, N., and Pelletier, S. L. (1991). Design, Chemical Synthesis, and Expression of Genes for the Three Human Color Vision Pigments. Biochemistry 30, 11367–11372. doi:10.1021/bi00112a002
Palczewski, K., Kumasaka, T., Hori, T., Behnke, C. A., Motoshima, H., Fox, B. A., et al. (2000). Crystal Structure of Rhodopsin: A G Protein-Coupled Receptor. Science 289, 739–745. doi:10.1126/science.289.5480.739
Papiz, M. Z., Prince, S. M., Howard, T., Cogdell, R. J., and Isaacs, N. W. (2003). The Structure and Thermal Motion of the B800-850 LH2 Complex from Rps.Acidophila at 2.0Å Resolution and 100K: New Structural Features and Functionally Relevant Motions. J. Mol. Biol. 326, 1523–1538. doi:10.1016/s0022-2836(03)00024-x
Pescitelli, G., Kato, H. E., Oishi, S., Ito, J., Maturana, A. D., Nureki, O., et al. (2014). Exciton Circular Dichroism in Channelrhodopsin. J. Phys. Chem. B 118, 11873–11885. doi:10.1021/jp505917p
Pescitelli, G., and Woody, R. W. (2012). The Exciton Origin of the Visible Circular Dichroism Spectrum of Bacteriorhodopsin. J. Phys. Chem. B 116, 6751–6763. doi:10.1021/jp212166k
Polívka, T., Balashov, S. P., Chábera, P., Imasheva, E. S., Yartsev, A., Sundström, V., et al. (2009). Femtosecond Carotenoid to Retinal Energy Transfer in Xanthorhodopsin. Biophysical J. 96, 2268–2277. doi:10.1016/j.bpj.2009.01.004
Ren, L., Martin, C. H., Wise, K. J., Gillespie, N. B., Luecke, H., Lanyi, J. K., et al. (2001). Molecular Mechanism of Spectral Tuning in Sensory Rhodopsin II. Biochemistry 40, 13906–13914. doi:10.1021/bi0116487
Runge, E., and Gross, E. K. U. (1984). Density-Functional Theory for Time-dependent Systems. Phys. Rev. Lett. 52, 997–1000. doi:10.1103/physrevlett.52.997
Ryazantsev, M. N., Altun, A., and Morokuma, K. (2012). Color Tuning in Rhodopsins: The Origin of the Spectral Shift between the Chloride-Bound and Anion-free Forms of Halorhodopsin. J. Am. Chem. Soc. 134, 5520–5523. doi:10.1021/ja3009117
Schenkl, S., van Mourik, F., van der Zwan, G., Haacke, S., and Chergui, M. (2005). Probing the Ultrafast Charge Translocation of Photoexcited Retinal in Bacteriorhodopsin. Science 309, 917–920. doi:10.1126/science.1111482
Scholes, G. D., Fleming, G. R., Olaya-Castro, A., and van Grondelle, R. (2011). Lessons from Nature about Solar Light Harvesting. Nat. Chem 3, 763–774. doi:10.1038/nchem.1145
Scholes, G. D. (2003). Long-rangeresonanceenergytransfer Inmolecularsystems. Annu. Rev. Phys. Chem. 54, 57–87. doi:10.1146/annurev.physchem.54.011002.103746
Sekharan, S., Sugihara, M., and Buss, V. (2007). Origin of Spectral Tuning in Rhodopsin-It Is Not the Binding Pocket. Angew. Chem. Int. Ed. 46, 269–271. doi:10.1002/anie.200603306
Senn, H. M., and Thiel, W. (2009). QM/MM Methods for Biomolecular Systems. Angew. Chem. Int. Ed. 48, 1198–1229. doi:10.1002/anie.200802019
Shibata, M., Inoue, K., Ikeda, K., Konno, M., Singh, M., Kataoka, C., et al. (2018). Oligomeric States of Microbial Rhodopsins Determined by High-Speed Atomic Force Microscopy and Circular Dichroic Spectroscopy. Sci. Rep. 8, 8262. doi:10.1038/s41598-018-26606-y
Shichida, Y., and Imai, H. (1998). Visual Pigment: G-Protein-Coupled Receptor for Light Signals. Cell Mol. Life Sci. (Cmls) 54, 1299–1315. doi:10.1007/s000180050256
Shtyrov, A. A., Nikolaev, D. M., Mironov, V. N., Vasin, A. V., Panov, M. S., Tveryanovich, Y. S., et al. (2021). Simple Models to Study Spectral Properties of Microbial and Animal Rhodopsins: Evaluation of the Electrostatic Effect of Charged and Polar Residues on the First Absorption Band Maxima. Ijms 22, 3029. doi:10.3390/ijms22063029
Smolensky Koganov, E., Brumfeld, V., Friedman, N., and Sheves, M. (2015). Origin of Circular Dichroism of Xanthorhodopsin. A Study with Artificial Pigments. J. Phys. Chem. B 119, 456–464. doi:10.1021/jp510534s
Speiser, S. (1996). Photophysics and Mechanisms of Intramolecular Electronic Energy Transfer in Bichromophoric Molecular Systems: Solution and Supersonic Jet Studies. Chem. Rev. 96, 1953–1976. doi:10.1021/cr941193+
Stenkamp, R. E., Filipek, S., Driessen, C. A. G. G., Teller, D. C., and Palczewski, K. (2002). Crystal Structure of Rhodopsin: a Template for Cone Visual Pigments and Other G Protein-Coupled Receptors. Biochim. Biophys. Acta (Bba) - Biomembranes 1565, 168–182. doi:10.1016/s0005-2736(02)00567-9
Tomasello, G., Olaso-González, G., Altoè, P., Stenta, M., Serrano-Andrés, L., Merchán, M., et al. (2009). Electrostatic Control of the Photoisomerization Efficiency and Optical Properties in Visual Pigments: On the Role of Counterion Quenching. J. Am. Chem. Soc. 131, 5172–5186. doi:10.1021/ja808424b
Trabanino, R. J., Vaidehi, N., and Goddard, W. A. (2006). Exploring the Molecular Mechanism for Color Distinction in Humans. J. Phys. Chem. B 110, 17230–17239. doi:10.1021/jp057144q
Tretiak, S., Middleton, C., Chernyak, V., and Mukamel, S. (2000). Exciton Hamiltonian for the Bacteriochlorophyll System in the LH2 Antenna Complex of Purple Bacteria. J. Phys. Chem. B 104, 4519–4528. doi:10.1021/jp9939930
Tsukamoto, T., Kikukawa, T., Kurata, T., Jung, K.-H., Kamo, N., and Demura, M. (2013). Salt Bridge in the Conserved His-Asp Cluster inGloeobacterrhodopsin Contributes to Trimer Formation. FEBS Lett. 587, 322–327. doi:10.1016/j.febslet.2012.12.022
Tsutsui, K., and Shichida, Y. (2010). Multiple Functions of Schiff Base Counterion in Rhodopsins. Photochem. Photobiol. Sci. 9, 1426–1434. doi:10.1039/c0pp00134a
Uehara, S., Fujimoto, K. J., and Tanaka, S. (2015). Protein-ligand Docking Using Fitness Learning-Based Artificial Bee colony with Proximity Stimuli. Phys. Chem. Chem. Phys. 17, 16412–16417. doi:10.1039/c5cp01394a
Voityuk, A. A. (2013). Estimation of Electronic Coupling for Photoinduced Charge Separation and Charge Recombination Using the Fragment Charge Difference Method. J. Phys. Chem. C 117, 2670–2675. doi:10.1021/jp311891h
Vura-Weis, J., Newton, M. D., Wasielewski, M. R., and Subotnik, J. E. (2010). Characterizing the Locality of Diabatic States for Electronic Excitation Transfer by Decomposing the Diabatic Coupling. J. Phys. Chem. C 114, 20449–20460. doi:10.1021/jp104783r
Wald, G. (1968). Molecular Basis of Visual Excitation. Science 162, 230–239. doi:10.1126/science.162.3850.230
Wang, Z., Asenjo, A. B., and Oprian, D. D. (1993). Identification of the Chloride-Binding Site in the Human Red and green Color Vision Pigments. Biochemistry 32, 2125–2130. doi:10.1021/bi00060a001
Wanko, M., Hoffmann, M., Frähmcke, J., Frauenheim, T., and Elstner, M. (2008). Effect of Polarization on the Opsin Shift in Rhodopsins. 2. Empirical Polarization Models for Proteins. J. Phys. Chem. B 112, 11468–11478. doi:10.1021/jp802409k
Wanko, M., Hoffmann, M., Strodel, P., Koslowski, A., Thiel, W., Neese, F., et al. (2005). Calculating Absorption Shifts for Retinal Proteins: Computational Challenges. J. Phys. Chem. B 109, 3606–3615. doi:10.1021/jp0463060
Warshel, A., and Levitt, M. (1976). Theoretical Studies of Enzymic Reactions: Dielectric, Electrostatic and Steric Stabilization of the Carbonium Ion in the Reaction of Lysozyme. J. Mol. Biol. 103, 227–249. doi:10.1016/0022-2836(76)90311-9
Wong, K. F., Bagchi, B., and Rossky, P. J. (2004). Distance and Orientation Dependence of Excitation Transfer Rates in Conjugated Systems: Beyond the Förster Theory. J. Phys. Chem. A. 108, 5752–5763. doi:10.1021/jp037724s
Wu, Y. I., Frey, D., Lungu, O. I., Jaehrig, A., Schlichting, I., Kuhlman, B., et al. (2009). A Genetically Encoded Photoactivatable Rac Controls the Motility of Living Cells. Nature 461, 104–108. doi:10.1038/nature08241
Yang, W. (1991). Direct Calculation of Electron Density in Density-Functional Theory. Phys. Rev. Lett. 66, 1438–1441. doi:10.1103/physrevlett.66.1438
Yang, W., and Lee, T. S. (1995). A Density‐matrix divide‐and‐conquer Approach for Electronic Structure Calculations of Large Molecules. J. Chem. Phys. 103, 5674–5678. doi:10.1063/1.470549
Yoshimura, K., and Kouyama, T. (2008). Structural Role of Bacterioruberin in the Trimeric Structure of Archaerhodopsin-2. J. Mol. Biol. 375, 1267–1281. doi:10.1016/j.jmb.2007.11.039
Keywords: color tuning, excitation-energy transfer, electronic coupling, electrostatic interaction, quantum chemistry
Citation: Fujimoto KJ (2021) Electronic Couplings and Electrostatic Interactions Behind the Light Absorption of Retinal Proteins. Front. Mol. Biosci. 8:752700. doi: 10.3389/fmolb.2021.752700
Received: 03 August 2021; Accepted: 02 September 2021;
Published: 15 September 2021.
Edited by:
Leonid S. Brown, University of Guelph, CanadaReviewed by:
Igor Schapiro, Hebrew University of Jerusalem, IsraelMikhail Ryazantsev, Saint Petersburg State University, Russia
Copyright © 2021 Fujimoto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kazuhiro J. Fujimoto, ZnVqaW1vdG9rQGNoZW0ubmFnb3lhLXUuYWMuanA=
 Kazuhiro J. Fujimoto
Kazuhiro J. Fujimoto