
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci. , 26 October 2021
Sec. Cellular Biochemistry
Volume 8 - 2021 | https://doi.org/10.3389/fmolb.2021.728661
This article is part of the Research Topic BET Proteins in Chromatin Architecture, Transcription and Disease View all 7 articles
Bromodomain-containing protein 4 plays a central role in coordinating the complex epigenetic component of the innate immune response. Previous studies implicated BRD4 as a component of a chromatin-modifying complex that is dynamically recruited to a network of protective cytokines by binding activated transcription factors, polymerases, and histones to trigger their rapid expression via transcriptional elongation. Our previous study extended our understanding of the airway epithelial BRD4 interactome by identifying over 100 functionally important coactivators and transcription factors, whose association is induced by respiratory syncytial virus (RSV) infection. RSV is an etiological agent of recurrent respiratory tract infections associated with exacerbations of chronic obstructive pulmonary disease. Using a highly selective small-molecule BRD4 inhibitor (ZL0454) developed by us, we extend these findings to identify the gene regulatory network dependent on BRD4 bromodomain (BD) interactions. Human small airway epithelial cells were infected in the absence or presence of ZL0454, and gene expression profiling was performed. A highly reproducible dataset was obtained which indicated that BRD4 mediates both activation and repression of RSV-inducible gene regulatory networks controlling cytokine expression, interferon (IFN) production, and extracellular matrix remodeling. Index genes of functionally significant clusters were validated independently. We discover that BRD4 regulates the expression of its own gene during the innate immune response. Interestingly, BRD4 activates the expression of NFκB/RelA, a coactivator that binds to BRD4 in a BD-dependent manner. We extend this finding to show that BRD4 also regulates other components of its functional interactome, including the Mediator (Med) coactivator complex and the SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin (SMARC) subunits. To provide further insight into mechanisms for BRD4 in RSV expression, we mapped 7,845 RSV-inducible Tn5 transposase peaks onto the BRD4-dependent gene bodies. These were located in promoters and introns of cytostructural and extracellular matrix (ECM) formation genes. These data indicate that BRD4 mediates the dynamic response of airway epithelial cells to RNA infection by modulating the expression of its coactivators, controlling the expression of host defense mechanisms and remodeling genes through changes in promoter accessibility.
Bromodomain-containing 4 (BRD4) is a member of the bromodomain and extra-terminal domain (BET) family of proteins that plays pleiotropic roles in epigenetic control of inflammation-inducible gene expression (Nowak et al., 2008; Tian et al., 2016), maintenance of cellular identity (Loven et al., 2013), DNA damage signaling (Floyd et al., 2013), chromatin compaction/conformation (Devaiah et al., 2016a), cell cycle regulation, and so on [Reviewed by Devaiah et al. (2016b)].
In this study, we focus on the activity of BRD4 in controlling inducible innate antiviral genomic response. Previous mechanistic studies have shown that BRD4 forms signal-inducible interactions with nuclear factor-κB (Nowak et al., 2008), heat shock factor (Lis et al., 2000), and SMAD3 (Ijaz et al., 2017) to bind silent gene promoters in euchromatin domains. Here, BRD4 functions as a pause–release factor, releasing inactive RNA polymerase II (Pol II) to enter a fully processive mode (Price, 2000). In this context, BRD4 plays pleiotropic roles as an acetylated histone “reader”, a selective histone acetyltransferase (Devaiah et al., 2016a), and a kinase for cyclin-dependent kinase 9 and RNA polymerase II (Devaiah and Singer, 2012). Although BRD4 interactions with the cyclin-dependent kinases in the P-TEFb complex are well established (Jeronimo et al., 2007; Yang et al., 2015), BRD4 interacts with a spectrum of other chromatin-modifying activities that may be functionally important. Recent studies applying native affinity isolation coupled with high-resolution mass spectrometry have substantially expanded the known protein interactions of BRD4 to include RNA polymerases, coactivators, transcription factors, histone acetyltransferases, and actin motors (Zhang et al., 2017; Mann et al., 2021). In this way, BRD4 forms highly dynamic complexes in a stimulus- and cell-type– dependent manner.
BRD4 interacts with acetylated histones and transcription factors through its conserved bromodomain (BD), with transcriptional regulators through its extraterminal domain, and with transcriptional elongation kinases through its carboxyterminal domain (Bisgrove et al., 2007; Rahman et al., 2011; Zou et al., 2014; Mann et al., 2021). The recent development of highly selective small-molecule BRD4 BD inhibitors (Liu et al., 2018) have enabled the ability to probe the role of BRD4 BD interactions in complex biological responses associated with epithelial inflammation, trained immunity, and cell-state changes (Tian et al., 2019a; Tian et al., 2019b; Zhao et al., 2019). These reagents have enabled studies that have placed BRD4 in a central role in host defense mechanisms to RNA viruses. Here, we focus on the role of BRD4 BD interactions in host defense mechanisms to respiratory syncytial virus (RSV). RSV is an enveloped, single-stranded, negative-sense RNA virus of the human orthopneumovirus genus of the Pneuomoviridae family that represents the most common cause of pediatric hospitalization in children less than 5 years of age (Pneumonia Etiology Research for Child Health (PERCH) Study Group, 2019). After inoculation into the nasopharynx, RSV fuses with ciliated epithelial cells and replicates to high levels, spreading throughout the airways to the conductive and lower airway cells involved in gas exchange. As part of a stereotypic innate response viral RNA patterns, BRD4 is activated downstream of the toll-like receptor (TLR) and retinoic acid–inducible gene (RIG-I) pattern recognition pathways (Tian et al., 2017b; Tian et al., 2019b). Consequently, productive viral infection in small airway epithelial cells induces the rapid secretion of cytokines (Zhang et al., 2001), interferons [IFNs, (Liu et al., 2007)], exosomes (Zhao et al., 2017), and damage-associated patterns (Hosakote et al., 2016) that mediate mucous production, leukocytic inflammation, and hypoxia (Bennett et al., 2007; Lay et al., 2016). Through its actions, BRD4 shapes the nature, timing, and magnitude of inducible epithelial cytokine responses, playing an important role in mediating disease and activating adaptive immunity.
Using high-resolution genome-wide studies of chromatin accessibility (ATAC-Seq) and RNA-Seq studies, we recently discovered that RSV infection reprograms gene loci encoding cytokines, growth factors, and extracellular matrix remodeling genes, resulting in their highly dynamic expression (Xu et al., 2020). The role of BRD4 in coordinating the innate response network has not been systematically elucidated. In this study, we study the effect of disrupting BRD4 BD1 and BD2 interactions using highly selective small-molecule BRD4 inhibitors in the RSV-induced gene regulatory response. We present evidence that BRD4 is autoregulated during the innate immune response. Interestingly, BRD4 mediates RSV-inducible expression of its major interacting coactivators, including the transcription factor NFκB/RelA, the Mediator (MED) coactivator complex, and subunits of the SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin (SMARC). A subset of these genes exhibit enhanced accessibility of their proximal promoters during RSV infection. These data provide insights into the new mechanistic role of BRD4 in RNA virus infections.
The BRD4 selective BD competitive inhibitor, ZL0454, was synthesized as previously described (Liu et al., 2018; Tian et al., 2019b) and used at purity > 99%. ZL0454 was resuspended in DMSO and added to the culture medium at indicated concentrations. Solvent alone (DMSO) was used in non–BRD4 inhibitor-treated conditions.
Human small airway epithelial cells (HSAECs) immortalized with human telomerase/CDK4 were obtained commercially (ATCC). These are non-oncogenic, telomerase-immortalized cells grown in the SAGM small airway epithelial cell growth medium (Lonza, cc-3118) in a humidified atmosphere of 5% CO2. Cells were infected at confluence >90% for profiling.
The human RSV long strain was obtained from ATCC, grown in Hep-2 cells, and prepared as described (Ueba, 1978). The viral titer of purified RSV pools varied from 8 to 9 log PFU/ml, determined by a methylcellulose plaque assay (Xu et al., 2020). Viral pools were aliquoted and quick-frozen on dry ice–ethanol and stored at −70°C until they were used.
Cells were harvested for RNA isolation using the RNeasy kit with on-column DNase digestion (Qiagen). The synthesis of complementary DNAs (cDNAs) was performed using the First Strand cDNA Synthesis Kit (Thermo Scientific). qRT-PCR assays were used for validation using custom primers and SYBR Green Master mix (Bio-Rad) or TaqMan primers and TaqMan Fast Advanced Master mix (Thermo Scientific) as described previously (Xu et al., 2020). The sequences of the SYBR Green PCR primers are BRD4 Forward (F): 5′- AAAGAAGCGCTTGGAAAACA; BRD4 Reverse (R): 5-CGGTTTCTTCTGTGGGTAGC; PPIA F 5′-TTCATCTGCACTGCCAAGAC′, PPIA R 5′-TCGAGTTGTCCACAGTCAGC; RELA F: 5′-CTTCCAAGAAGAGCAGCGTG; RELA R: 5′-. CTTCCAAGAAGAGCAGCGTG; IFNL F” 5′-: GGAGTTGCAGCTCTCCTGTC; and IFNL R 5′- CAGCGGACTCCTTTTTGGGG. Data are normalized to internal control and presented as fold change using the ΔΔ Ct method.
Total RNA was isolated using the RNeasy kit with on-column DNase I digestion (Qiagen). RNA was quantified (Nanodrop), and its was integrity verified (Agilent) prior to sequencing. Libraries were produced using the TruSeq Stranded mRNA library kit (Illumina). The 12 separate RNA samples (N = 4 each for HSAEC control, RSV, and RSV + ZL0454 24 h of RSV treatment) were bar-coded and subjected to Illumina HiSeq 2000 paired-end sequencing. Quality control was performed using FastQC. Trimmomatic was used to remove sequencing adapters. Trimmed paired-end reads were aligned against human genome hg38 using Salmon. Mapped paired-end reads for both genes and transcripts (isoforms) were counted in each sample using RSEM and expressed as transcripts per million (TPM), the recommended metric for transcript quantification (Patro et al., 2017). Contrasts were compared for RSV or BRD4 inhibitor treatment conditions using DESeq2 (Love et al., 2014).
ATAC-Seq Fastq files were (Xu et al., 2020) analyzed for read coverage, and PCR duplication and number of mapped reads were determined using FastQC. Adapter removal was performed using TrimGalore (Kreuger, 2020). Alignment was performed using the RSubread package (Liao et al., 2019) to the GRCh38.p13 (hg38) genome assembly (NCBI). Peak calling was performed using Genrich for ATAC-Seq (Gaspar, 2019), using commands to remove mitochondrial sequences, remove PCR duplicate reads, and mask blacklisted sequences. Statistical comparisons were conducted using DESeq2 in the DiffBind package (Ross-Innes et al., 2012). Functional annotation was performed using ReactomePA (Yu et al., 2014). Functional gene enrichment analysis was performed using ClueGO v 2.5.8 plug-in (Bindea et al., 2009) in Cystoscope 3.2 (Shannon et al., 2003).
HSAECs were trypsinized, pelleted, and washed twice with cold phosphate-buffered saline (PBS). Cell pellets were then lysed in cold low ionic strength buffer (50 mM NaCl, 1% IGEPAL, 10%Glycerol, 10 mM HEPES, pH7.4) with freshly added proteinase inhibitor cocktail (Sigma), DTT and PMSF (Sigma). 10 µl of the cell lysate was removed for protein measurement (DC protein assay kit, Bio-Rad). 2xSDS sample buffer was added. The lysates were sonicated thrice for 15 s (Branson150), centrifuged at 10000 rpm for 20 min, and supernatants were heated at 95oC for 5 min. Proteins were resolved using Criterion TGX 4-15% precast gel and transferred to a nitrocellulose membrane using the Bio-Rad Trans-Blot Turbo transfer system. Sources of primary antibodies and dilutions used were anti-RelA: Cell Signaling 8242S, 1:1000; anti-BRD4: Cell Signaling 13440, 1:1000; and anti-GAPDH Abcam ab59164, 1:2000.
Control, RSV-infected, and RSV + ZL0454–treated HSAECs were fixed with 4% paraformaldehyde (10 min), permeablized with 0.1% Triton X-100 (10 min), and blocked with 5% goat serum in PBS (2 h). The slides were then subjected to PLA using the Duo-link PLAkit from O-Link Bioscience (Uppsala, Sweden) according to the manufacturer’s instructions. Nuclei were counterstained with DAPI, and the PLA signals were visualized using a Nikon A1RS confocal microscope at 63X magnification.
Statistical analysis of the RNA-Seq data was conducted using the DESeq2 R package (version 1.32.0) (Love et al., 2014). Contrasts with padj <0.05 of log2 fold change (log2FC) by DESeq2 were considered significant. Data are presented as log2(fold change) using a 25–75% interquartile range. Adjusted pvalues for the RSV vs RSV + BRD4 inhibitor contrast are provided in the figure legends. qRT-PCR was compared using ANOVA with Tukey’s post hoc comparisons test. Gene Ontology analysis was performed using Reactome.org (Jassal et al., 2020). Pathway analysis was performed using disease ontology semantic and enrichment analysis (version 3.18.0) (Yu et al., 2014). Data are plotted as 25–75% of the interquartile range.
Innate signaling in small airway epithelial cells located in the bronchiolar epithelium plays a major role in the pathogenesis of RSV lower respiratory tract infection (LRTI). This conclusion comes from studies where inhibition of NFκB/RelA signaling in bronchiolar small airway cells blocks RSV-induced inflammation and airway obstruction in vivo (Tian et al., 2018b). This epithelial subtype plays an important role in the immunopathogenesis of LRTI because these cells are programmed to produce mucin- and T-helper-2 lymphocyte-activating cytokines that mediate the disease (Zhao et al., 2017).
HSAECs are a well-established in vitro model of small airway epithelial cells that induce similar global genomic and proteomic responses to RSV as that of primary small airway cells in vitro and that of lower airways in vivo (Zhang et al., 2001; Zhao et al., 2017). To elucidate the role of BRD4 in the HSAEC antiviral response, we first compared the effects of two highly selective BRD4 inhibitors (Liu et al., 2018; Liu et al., 2020) on RSV-induced cytokine expression. ZL0454 binds with nanomolar affinity to both bromodomains (BDs) of BRD4 with a 30-fold higher affinity than that of other BET family members. ZL0516 preferentially binds to BRD4 BD1 with ∼8.5 times lower IC50 value than to BRD4 BD2 (Liu et al., 2020).
Wild-type (WT) HSAECs were infected with sucrose-purified RSV [multiplicity of infection (MOI) = 1] in the presence of DMSO (solvent control), ZL0454 (10 μM), or ZL0516 (10 μM). 24 h later, the expression of two known BRD4-dependent genes, interleukin 6 (IL-6) and MX dynamin-like GTPase 1 (MX1), was measured by qRT-PCR. RSV induced a ∼190-fold increase in IL-6 expression; both ZL0454 and ZL0516 significantly inhibited its expression, with the inhibition of ZL0454 being consistently greater (Figure 1). Similar inhibitory effects were observed for the ∼55-fold induction of MX1 (Figure 1). Because ZL0454 showed consistently greater inhibitory activity, is more potent than the prototypical inhibitors JQ1 and RVX20 (Tian et al., 2019b), and exhibits no detectable cellular toxicity(Tian et al., 2019b), we selected ZL0454 for subsequent RNA-seq experiments.
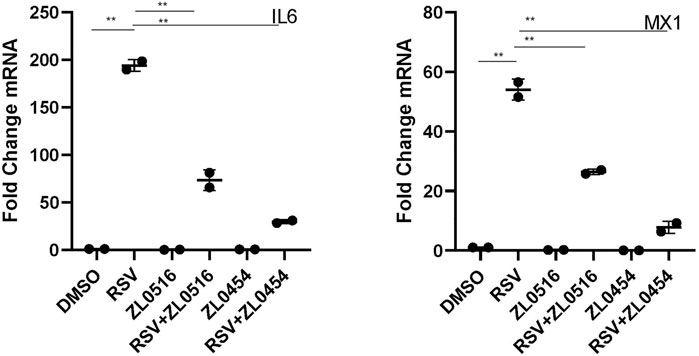
FIGURE 1. Effect of BRD4-selective small-molecule inhibitors on RSV-induced cytokines. qRT-PCR was performed in HSAECs treated in the absence or presence of 10 μM concentrations of indicated BRD4 inhibitors. HSAECs were infected with purified RSV (MOI = 1) for 24 h. Fold change mRNA relative to PPIA is plotted for IL-6 (left) and MX1 (right). p < 0.01 in ANOVA across treatment groups; ** and p < 0.01 in pairwise comparison.
Four replicates of control, RSV-infected or RSV + BRD4 inhibitor (ZL0454)–treated HSAECs were subjected to short read Illumina HiSeq 2000 paired-end sequencing (Methods). To examine whether the RNA expression patterns were reproducible and similar with respect to biological replicates, principal component analysis (PCA) was performed. We observed that the control (CON), RSV-infected, and RSV- ZL0454–treated samples were widely separated in the PCA. The two principal component dimensions accounted for 97% of the sample variability, indicating that the data represented a robust effect of RSV infection and BRD4 inhibitor treatment (Figure 2A). Moreover, the four experimental replicates were virtually superimposed, indicating a highly reproducible dataset. The first principal component separated the RSV-infected cells (±BRD4 inhibitor) vs controls (CON). By contrast, the second PCA component separated the RSV-infected cells with DMSO vs those with BRD4 treatment. Separately, the distance matrix of the log-transformed TPM quantitation was subjected to hierarchical clustering (Figure 2B). In a manner consistent with the PCA result, control (CON), RSV-BRD4 inhibitor, and RSV-infected sample cluster with other replicates received the sample treatment and were separated from other treatments. These data indicate that the RNA-Seq captures reproducible information about the effects of RSV and that of the inhibitor, and that the inhibitor does not revert the cell state back to that of the control.
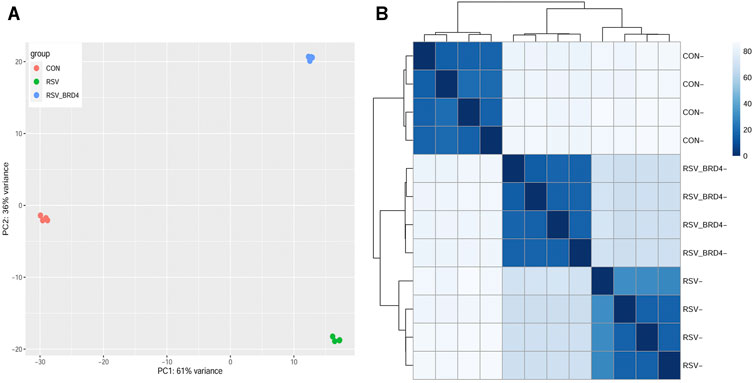
FIGURE 2. Sample QC. (A) Principal component analysis. X axis, first principal component; Y axis, second principal component. Note that the first principal component separates the RSV (±BRD4 inhibitor) from the CON-treated samples and represents 61% of the variability in the experiment. The second dimension separates the RSV vs RSV + BRD4 inhibitor treatment. (B) Correlation analysis of log-transformed RNA-Seq quantitation of the experimental samples. Samples were subjected to hierarchical clustering, with distance indicated in the dendrogram. Note that the individual CON, RSV-BRD4 inhibitor, and RSV-infected samples cluster with other replicates receiving the sample treatment and separate from other treatments.
RSV is sequentially transcribed into 10 polyadenylated mRNA species from a 3′ to 5′ direction by its RNA-dependent RNA polymerase in the order 3’ (leader) -NS1-NS2-N-P-M-SH-G-F-M2- DZD2-(trailer) 5′ using a “gene start”– “gene stop” mechanism (Collins and Graham, 2008). Our previous study has shown no effect of BRD4 silencing or inhibition on RSV replication in vitro (Tian et al., 2017b). To confirm, we quantitated individual mRNA species (Figure 3A). Importantly, we found that the IRF3/IFN antagonists, NS1 and NS2, were expressed at indistinguishable levels. Consistent with the 3′-5′ gradient of expression, the abundance of NS1 (in TPM) was 75-fold greater than that of low-abundance RNA polymerase (L) transcript (not shown). Moreover, we found that RSV transcription was essentially unchanged in the BRD4 inhibitor–treated cells, and that, in fact, SH and L expression were elevated over that of RSV infection alone (Figure 3A). Together, these data indicated that BRD4 inhibition had little overall effect on RSV transcription in this in vitro model.
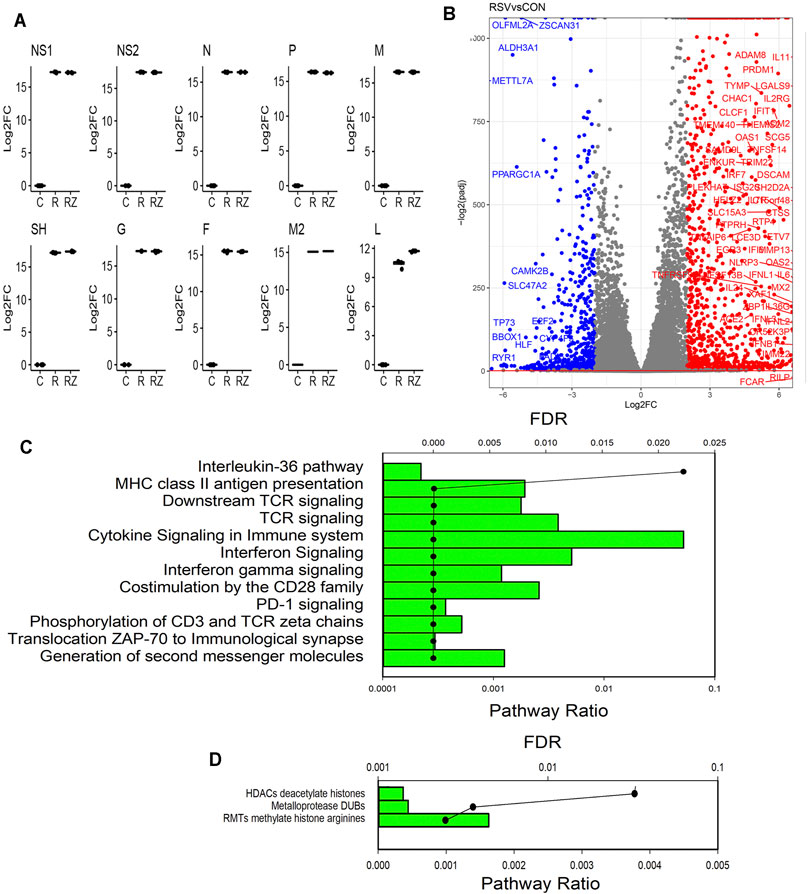
FIGURE 3. RSV-induced gene regulatory network. (A) Quantitation of RSV transcripts. Log2 fold change (log2FC) values of short-read RNA-Seq are plotted by quartile (25–75%) box plots. The horizontal line is the mean value. For each transcript group, comparisons are significantly different for the RSV-infected vs mock-infected (CON) contrast. DZD2, RNA-dependent RNA polymerase; F, fusion protein; G, glycoprotein; M, matrix protein; N, nucleoprotein; NS, nonstructural protein; P, phosphoprotein; and SH, small hydrophobic protein. (B) Volcano plot of differentially expressed genes (DEGs) in CON vs RSV-infected cells. X axis, log2fold change of transcripts/million (TPM). A positive value in red indicates that the gene is upregulated by RSV infection. Y axis, −log10(adjusted p value using Benjamini-Hochberg, padj). Blue are genes downregulated by RSV (log2FC > 2). Red are genes upregulated. (C) Genome Ontology (GO) enrichment of RSV-upregulated genes. Genes with 4-fold change in TPM (RSV vs CON) and adjusted p-value <0.01 were analyzed for pathway enrichment (reactome.org). For each gene set, the fraction of genes represented in the pathway and the significance (false discovery rate, FDR) are plotted. Shown are the pathways with FDR <0.05. (D) GO enrichment of RSV-downregulated genes. Genes with <-4-fold change in TPM (RSV vs CON) and adjusted p-value of <0.01 were analyzed for pathway enrichment as given above.
RSV replicates in primary human airway epithelial cells, inducing a well-established, time-dependent global genomic response, including both gene activation and repression (Zhang et al., 2001; Xu et al., 2020; Xu et al., 2021a). Consistent with this earlier study, our data indicate that RSV induces dramatic changes in the expression of 11,038 genes (5620 upregulated and 5418 downregulated). A stringent filter of differentially expressed genes is illustrated by the volcano plot. In this analysis, the statistical significance of the change in expression of each gene in the dataset [−log10(padj)] is plotted versus the relative change in expression (expressed in log2 fold change, Figure 3B). RSV induces substantially greater number of genes than are inhibited. The most highly upregulated and downregulated gene sets ( |log2FC| > 4) were separately analyzed by Gene Ontology (GO) analysis, where pathway enrichment is represented statistically by enrichment relative to the human genome (FDR) and by the fraction of genes in the dataset that map to any given pathway (pathway ratio). The GO analysis of the upregulated genes identifies the enrichment of well-established RSV-induced pathways, including T-cell receptor signaling, innate responses, IL-10 signaling, and IL-36 signaling (Figure 3C). By contrast, GO analysis of downregulated genes identified gene mapping to HDAC, metalloproteinases, and arginine methylation pathways (Figure 3D). We conclude that the RSV-inducible gene expression programs in these data are highly representative of known RSV programs.
We next focused on the gene regulatory network affected by ZL0454. Contrasts of gene expression were analyzed for the effect of BRD4 controlling for multiple hypothesis testing in DESeq2. 10,559 genes were differentially expressed with an adjusted p value <0.01. The differentially expressed genes between RSV + DMSO vs RSV + ZL0454 were plotted by the adjusted statistical significance of the change (−log10(padj)) versus the relative change in expression (expressed in log2FC of the TPM, Figure 4A). A much greater number of genes are downregulated by the BRD4 inhibitor (indicated in blue, Figure 4A) than those upregulated (indicated in red, Figure 4A). To visualize the expression patterns, the normalized expression values of index genes in the “Cytokine Signaling in the Immune System” were retrieved and analyzed by comparing the log2-transformed fold change (log2FC). The index gene in this group is the RSV- and IFN-inducible MX dynamin-like GTPase 1 (MX1), shown in Figure 4B. In the RNA-Seq dataset, the expression of MX1 is dramatically induced by ∼5.8 log units by RSV and significantly inhibited by ZL0454 (Figure 4B, left graph). To validate this pattern, wild-type HSAECs were separately treated with solvent (DMSO) or ZL0454 and then mock- or RSV infected. Total RNA was extracted, and qRT-PCR was performed for MX1. RSV potently upregulated MX1 expression in solvent-treated HSAECs by ∼120-fold, but this induction was substantially reduced in ZL0454-treated cells (Figure 4B, right graph), exactly reproducing the pattern in the RNA-Seq data.
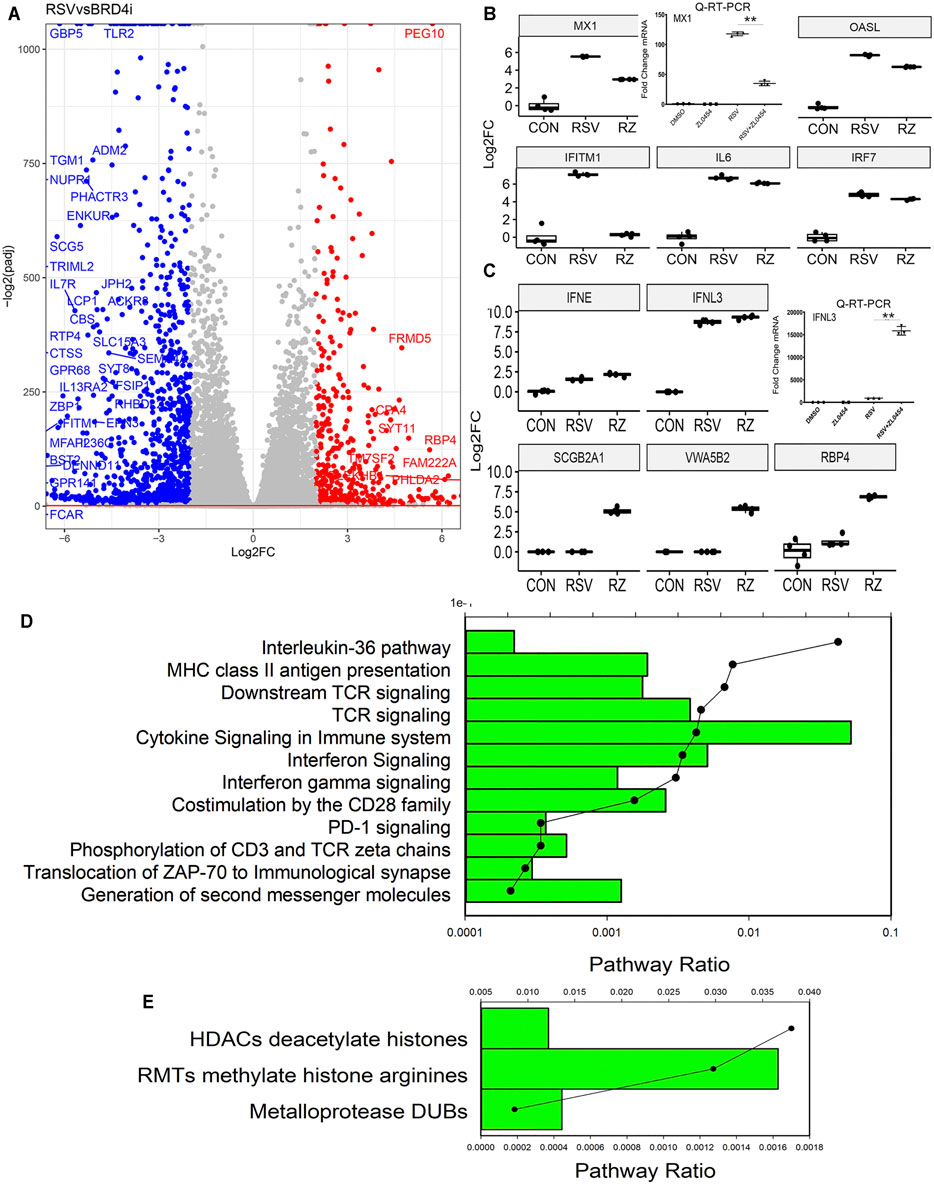
FIGURE 4. BRD4-dependent gene network. (A) Volcano plot of BRD4-dependent genes comparing RSV vs RSV + BRD4 inhibitor–treated cells. Genes downregulated by BRD4 inhibitor are plotted in blue and those upregulated by BRD4 inhibitor are shown in red. (B) RSV-inducible interferon stimulatory genes dependent on BRD4. First graph is RNA-Seq data (log2FC) for the MX1 gene. Adjusted pValue for RSV vs RSV + ZL0454 = 1.12E-15. Right graph is qRT-PCR of an independent experiment. MX1 expression is shown as fold change over mock-infected controls. **, p < 0.01 by post hoc Tukey’s comparison test. Remainder of graphs are RNA-Seq log2FC presented as 25-75% interquartile graphs. Adjusted p values for RSV vs RSV + ZL0454 contrast are IFITM1 = 7.64E-47, OASL = 7.97E-34; and IRF7 = 0.00038. (C) RSV-inducible genes repressed by BRD4. RNA-Seq of IFNL3, where ZL0454 increases the RSV-induced expression. Adjusted p value of the RSV vs RSV + ZL0454 contrast is = 0.0019. Right graph, validation of IFNL mRNA expression by independent qRT-PCR. **, p < 0.001. Adjusted p Values for RSV vs RSV + ZL0454 contrast in DESeq2 are as follows: SCGB2A1 = 7.48E-7; VWASB2 = 1.147E-7; IFNE p = 0.00041; and RBP4 = 7.73E-38. (D) Genome Ontology (GO) enrichment of BRD4-dependent RSV-upregulated genes. Genes with 4-fold log2FC (RSV vs RSV + ZL0454) and adjusted p-value <0.01 were analyzed for pathway enrichment (reactome.org). For each gene set, the fraction of genes represented in the pathway and the significance (false discovery rate, FDR) are plotted. Shown are the pathways with FDR <0.05. (E) GO enrichment of RSV-upregulated genes repressed by BRD4. Genes with <-4-fold change in TPM (RSV vs RSV + ZL0454 ) and adjusted p-value of <0.01 were analyzed for pathway enrichment as given above
In a similar manner, other index genes in the “cytokine signaling in the immune system”pathway, including IFN- induced transmembrane protein 1 (IFITMI), 2′-5′-oligoadenylate synthetase-like (OALS) and interferon response factor 7 (IRF7) genes were highly upregulated by RSV and inhibited by ZL0454 (Figure 4B). Genes paradoxically increased by BRD4 inhibition were selected by rank order of fold change in TPM RSV vs RSV + ZL0454 and similarly analyzed. In contrast to the behavior of MX1 and OASL, IFN lambda (IFNL3) was induced by RSV and further increased by BRD4 inhibition (Figure 4C, left graph). This pattern of behavior was independently validated by qRT-PCR (Figure 4C, right graph). Other genes showing paradoxical induction by the BRD4 inhibitor included secretoglobin family 2A member 1 (SCGB2A1), Von Willebrand factor A domain (VWASB2), and retinol binding protein 4 (RBP4) (Figure 4C). Although the differences are highly consistent and statistically significant, we note that the abundance of these transcripts is quite low in comparison to those inhibited by BRD4. To further understand the functional activities of the BRD4-activated and BRD4-repressed genes, GO analysis was performed for each group separately. 273 genes were downregulated by 4- log2FC in the BRD4 inhibitor group, and 78 genes were upregulated by 4 log2FC in the BRD4 inhibitor relative to that of RSV infection alone. We noted a striking similarity of the functional pathways of the BRD4-dependent genes as that of the RSV-dependent genes, although the pathway enrichments and statistical confidence were not as high (Figures 4C,D). We conclude from these data that BRD4 is both an activator and repressor of IFN-stimulated genes (ISGs) and growth factor–responsive genes during the evolution of the antiviral response.
To further examine the BRD4-dependent gene regulatory network in RSV infection, we subjected the BRD4-dependent genes to integrated GO analysis. Here, densely connected networks of genes controlling the regulation of T-cell differentiation, immune effector processes, and interleukin expression were identified (Figure 5), consistent with the anti-inflammatory properties of BRD4 inhibitors in RSV infection in vivo (Tian et al., 2018a; Tian et al., 2019b; Zhao et al., 2019). A network analysis incorporating disease and protein–protein interaction information was also conducted using DOSE (data not shown). This analysis linked the BRD4-regulated network with epithelial carcinoma–driven states, consistent with its role in oncogenesis. This analysis also identified BRD4-regulated chromatin-modifying genes, including cyclin B1 (CCNB1), enhancer Of zeste 2 polycomb repressive complex 2 subunit (EZH2), GATA-binding protein 3 (GATA3), and Forkhead Box A1.
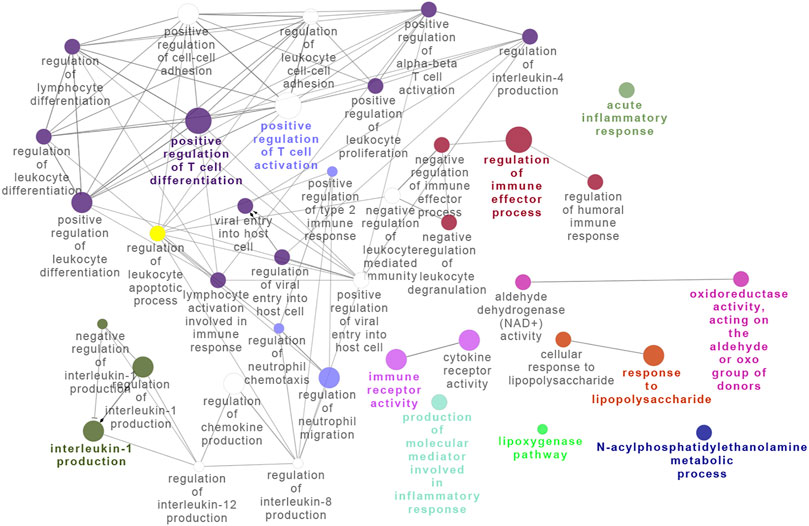
FIGURE 5. Integrated network analysis of BRD4-dependent genes. Gene pathway interaction map of BRD4-dependent genes. Note the striking enrichment of T-cell differentiation and activation of immune effector processes.
(FOXA1)These findings suggested that BRD4 may regulate the expression of other chromatin modifiers.
Informed by the findings that BRD4 regulates many chromatin modifiers and our previous findings that BRD4 protein and mRNA expression are induced by RSV infection (Tian et al., 2017b), we explored the effects of ZL0454 on RSV-induced BRD4 expression. Strikingly, a ∼0.8 log2FC increase in BRD4 mRNA produced by RSV infection is reduced by ZL0454 treatment (Figure 6A), a finding independently validated by qRT-PCR, where the effect of ZL0454 on basal and RSV-induced expression is observed (Figure 6B). In Western blot, RSV slightly reduced the steady-state abundance of ∼170 kDa BRD4 protein, suggesting RSV induces the turnover of the activated protein. Consistently with that of the transcript abundance, BRD4 protein abundance was inhibited by ZL0454 treatment (Figure 6C).
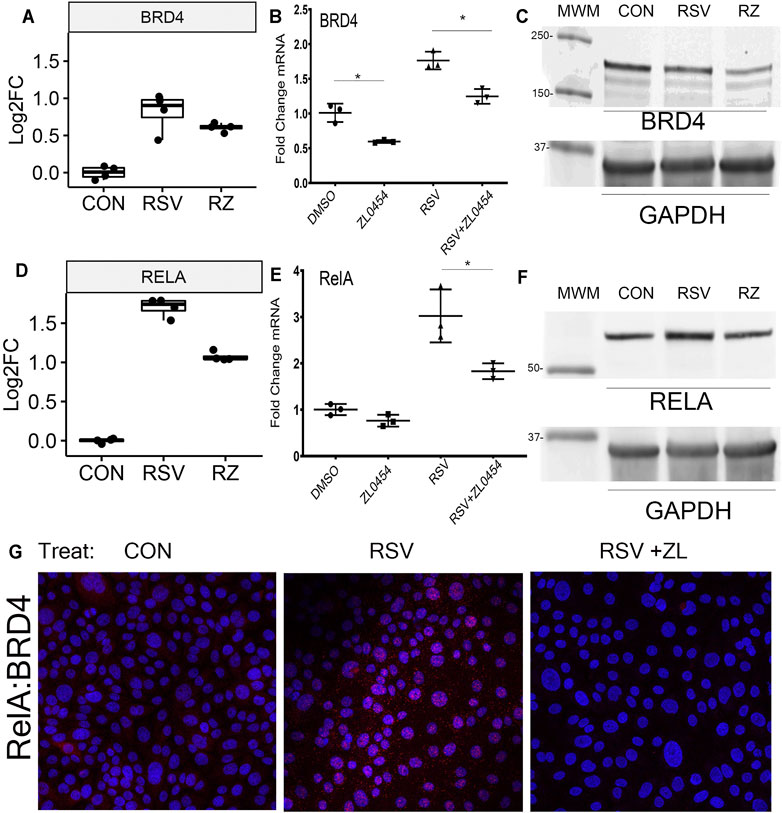
FIGURE 6. BRD4 autoregulation. (A) BRD4 mRNA expression. Shown are log2FC counts from RNA-Seq plotted as 25-7% interquartile ranges. Adjusted p-value for DESeq2 contrast RSV vs RSV + ZL0454 is 0.043. (B) Validation in independent experiments by qRT-PCR. BRD4 mRNA is expressed as fold change over mock-infected controls. Note the basal and RSV-induced induction of BRD4 mRNA is blocked by the BRD4 inhibitor. *p < 0.05 post hoc Tukey’s comparison test. (C) Western immunoblot. Whole cell lysates were fractionated by SDS-PAGE and blotted for BRD4. GAPDH staining is the internal control. MWM, molecular weight markers (in kDa). (D) RELA mRNA expression in log2FC. Adjusted p-values for the RSV vs RSV + ZL0454 contrast is 6.07E-23. (E) RELA mRNA expression by qRT-PCR. *p < 0.05 post hoc Tukey’s comparison test. (F) NFκB/RelA expression. Western blot of whole cell extracts. GAPDH staining is the internal control. (G) Proximity ligation assay. RelA–BRD4 interaction was quantified using PLA. BRD4–RelA complexes were quantified in the control group(Con), RSV-infected, or RSV + ZL–treated HSAECs. Red dots are BRD4-RelA complexes and nuclei are counterstained with DAPI (Blue). Note that the marked induction of BRD4-RelA complexes by RSV is blocked by ZL0454 treatment.
The association of BRD4 with the innate inducible transcription factor, NFκB, is a major component of the antiviral transcriptional elongation response to RSV (Brasier et al., 2011; Tian et al., 2018b; Mann et al., 2021). Although RSV regulates NFκB at a posttranscriptional level by inducing its nuclear–cytoplasmic translocation (Garofalo et al., 1996; Jamaluddin et al., 1998; Tian et al., 2002; Choudhary et al., 2005), RSV infection also induces the synthesis of NFκB/RelA mRNA by a largely unknown mechanism (Tian et al., 2018b). Extending these earlier findings, here we find that RSV-induced RELA mRNA expression is BRD4-dependent, where RELA mRNA abundance increases ∼1.7 log units in response to RSV and is substantially reduced by ZL0454 treatment (Figure 6D). This induction of RELA is independently validated by qRT-PCR, where RSV induces a 3-fold change in normalized RELA transcripts, an amount reduced to ∼1.8 fold by ZL0454 treatment (Figure 6E). A 1.5-fold induction of steady-state RelA abundance by RSV is confirmed by Western blot, and the levels are reduced to less than the control in response to ZL0454 treatment (Figure 6F). RelA–BRD4 interaction was confirmed in proximity ligation assay (PLA). RSV infection markedly induced the association of BRD4 with RelA in HSAECs, an association blocked by ZL0454 treatment (Figure 6G). These data suggested the very intriguing possibility that the BRD4 interactome was being controlled by BRD4 action.
This finding that BRD4 regulates NFκB/RelA expression in the antiviral response raised the question whether BRD4 controls other members of its interactome. Our recent study using native affinity enrichment and parallel accumulation–serial fragmentation mass spectrometry identified >100 BRD4-associated coactivators and transcription factors whose RSV-induced interactions were disrupted by BRD4 BD inhibitors (Mann et al., 2021). Protein–protein interaction network analysis indicated that BRD4 interacts with the Mediator complex and the SWI-SNF family of ATP-dependent chromatin interactors. Mediator (MED) is a multi-subunit adapter complex that mediates long-range enhancer interactions important in steroid receptor– induced gene transcription (Zhang et al., 2005). Intriguingly, BRD4 repressed MED4, MED27, cysteine- and glycine-rich protein 1 (CSRP1), MED17,and the ERCC excision repair 3 (ERCC3) and cyclin-dependent kinase (CDK12) expression (Figure 7). By contrast, BRD4 activates AT-rich interaction domain 1A (ARID1A), ARID1B, SMARC-E1, C2, and -D1. By contrast, BRD4 suppresses SMARCA5 (Figure 8). These data indicate that BRD4 controls a complex-coordinated regulation of its interactome in the antiviral response.
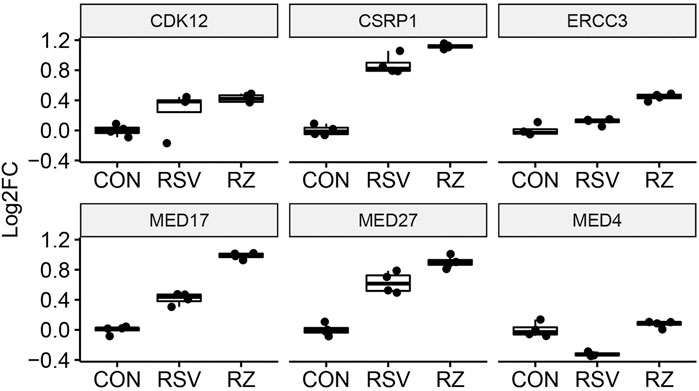
FIGURE 7. BRD4 modulates the expression of MED. mRNA expression of BRD4-interacting MED complexes. For each, log2FC values are plotted as 25-75% interquartile ranges. Adjusted p-values of RSV vs RSV + ZL0454 treatment are as follows: MED4 = 0.000144; MED27 = 0.00159; CSRP1 = 0.00102; MED17 = 2.1E-25; ERCC3 = 2.08E-8; and CDK12 = NS.
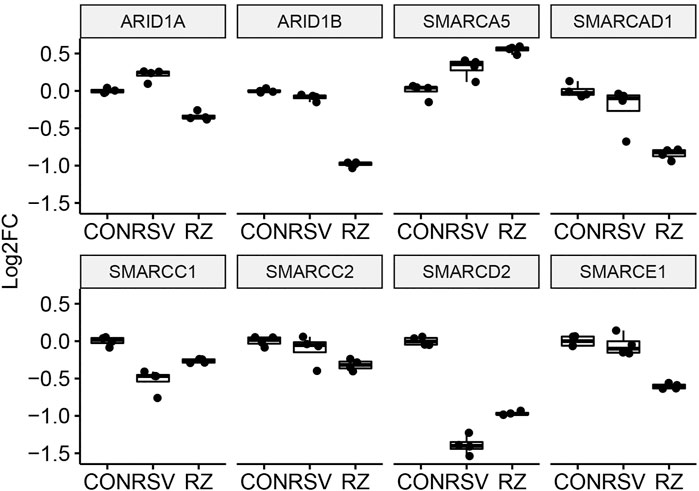
FIGURE 8. BRD4 modulates the expression of SMARC complexes. mRNA expression of BRD4-interacting SMARC complexes. For each, log2FC values are plotted as 25-75% interquartile ranges. Adjusted p-values of RSV vs RSV + ZL0454 treatment are as follows: ARI1A = 1.21E-29; ARID1B = 2.82E-98; SMARCE1 = 8.13E-14; SMARCD2 = 8.37E-15; SMARCC2 = 0.024; SMARCA5 = 0.0014; SMARCAD1 = 2.81E-7; and SMARCC1 = 0.000723.
Earlier, we reported a genome-wide analysis of RSV-induced chromatin remodeling using Tn5 transposase cleavage and next generation sequencing, for example, ATAC-Seq (Xu et al., 2020). Here, we identified functionally related clusters of genes controlling ECM remodeling and epithelial–mesenchymal transition (Tian et al., 2017a; Zhang et al., 2019). These genes were enriched in H3K27Ac markers, characteristic of transcriptional elongation occurring via the superelongation complex (Tian et al., 2013). To address the question whether BRD4-dependent genes identified in this study were regulated by changes in chromatin accessibility, we analyzed the relationship between these data. In this analysis, we asked whether RSV-inducible ATAC-Seq peaks mapped to BRD4-dependent genes. 1,700 peaks were mapped to gene bodies (promoter, intron, exon, or UTRs) with the majority being mapped to promoter or intronic sequences (Figure 9A). Gene Ontology and pathway enrichment analyses indicated a substantial enrichment of genes controlling collagen fiber biosynthesis and/or formation (Figures 9B,C). These genes include ECM-regulating MMP9, cadherin-3 (CDH3), mucin 1 (MUC1), serpin family A member 1 (SERPINA1), and collagen type 17 alpha 1 chain (Col17A1).
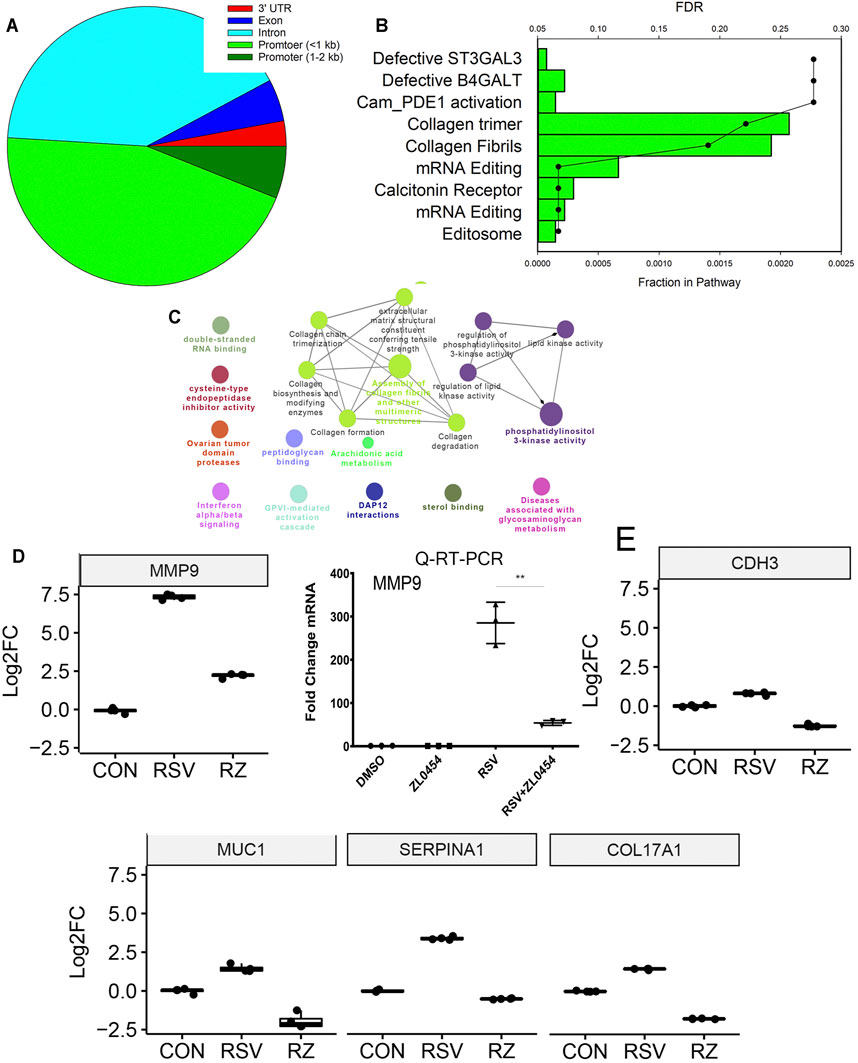
FIGURE 9. BRD4-dependent genes with RSV-inducible chromatin. 7,845 RSV-inducible Tn5 transposase-accessible genomic cleavage sites were mapped to BRD4-dependent gene bodies. 516 peaks were mapped to BRD4-dependent genes. (A) Location of cleavage sites on gene bodies. (B) GO enrichment analysis. Bars, fraction of pathway in datasets. Scatterplots are false discovery rate for pathway enrichment. Note the enrichment of collagen trimers and fibril formation. (C) Integrated GO enrichment. A major network involved in ECM and collagen is indicated in green. (D) MMP9 mRNA expression in RNA-Seq is validated by qRT-PCR. Left, log2FC from RNA-Seq. Adjusted p-values is < E-100. Right, fold change mRNA by qRT-PCR. (E) mRNA expression of other ECM and structural index genes are plotted as 25–75% interquartile ranges. Adjusted p-values of RSV vs RSV + ZL0454 treatment are: COL17A = E < -100; MUC1 = 1.4E-76; SERPINA1 = E < -100; and CDH3 = 3.33E-236.
The effect of RSV and BRD4 inhibitors on MMP9 mRNA expression was independently validated in a separate experiment using qRT-PCR (Figure 9D). The mRNA expression profiles of CDH3, MUC1, SERPINA1, and COL17A1 are shown (Figure 9E). ATAC-Seq peaks for these genes were visualized relative to the transcription start site using integrated genomics viewer, where a marked increase in genomic accessibility in 5’ regulatory regions is illustrated for each gene (Figure 10).
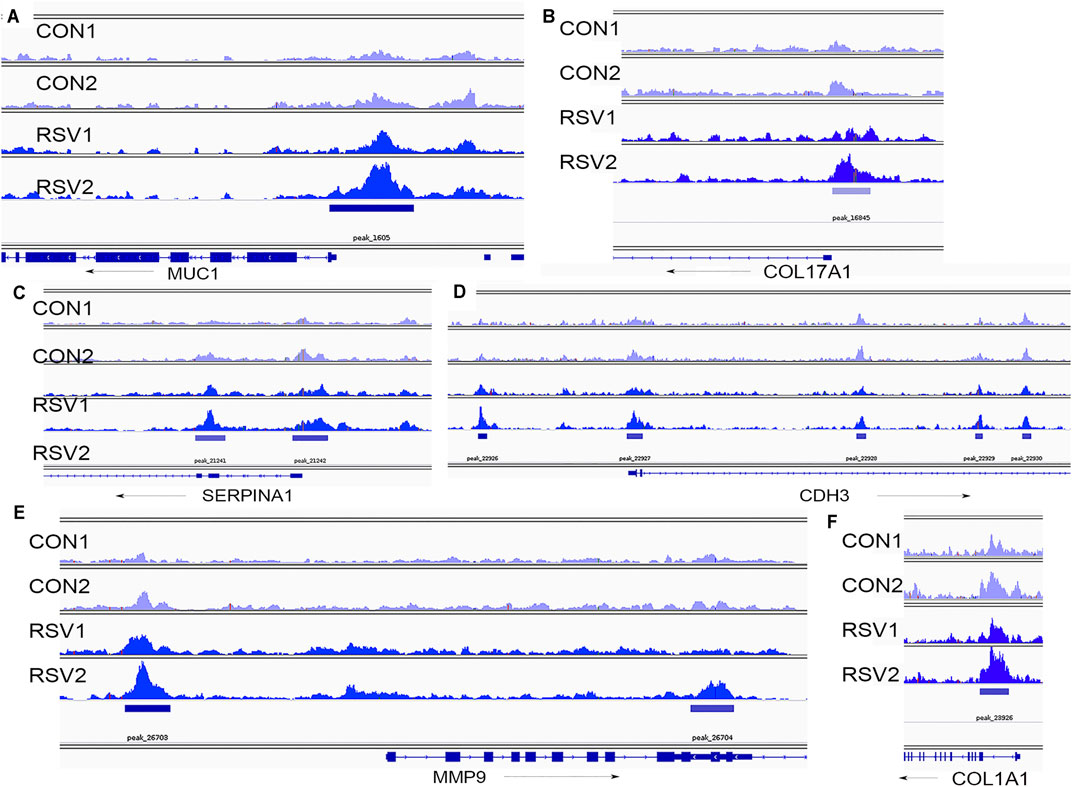
FIGURE 10. Chromatin-accessible peaks in 5′ regions of BRD4-dependent genes. Shown are individual ATAC-Seq tracks for replicate control and RSV-infected cells (Xu et al., 2020). (A), MUC1 locus; (B), COL17A1, (C), SERPINA1; (D), CDH3; (E), MMP9; and (F), COL1A1. Direction of transcription is indicated by the arrow.
The airway mucosa is composed of discrete, highly differentiated airway epithelial cells that play an important role in gas exchange, maintenance of fluid balance, control of vascular reactivity, and initiation of the innate response (Crystal et al., 2008). Specialized cuboidal cells in the lower bronchiolar–alveolar junction are primed to rapidly respond to respiratory viral infections, through the elaboration of gene regulatory networks controlling mucosal inflammation and cell state changes (Harvey et al., 2007; Tian et al., 2018b). This coordinated genomic response is dependent on epigenetic processes, including chromatin remodeling and transcriptional elongation activity (Tian et al., 2016; Tian et al., 2017b; Tian et al., 2019b). Mechanistically, BRD4 associates with acetylated transcription factors activated by pattern recognition receptor binding, including NFκB/RelA and IRF to repositioning to networks of innate responsive genes. Despite a detailed understanding of the importance of BRD4 in promoter pause–release, the functional activities of BRD4 are incompletely understood. In this study, we apply highly selective BRD4 inhibitors to identify the role of BRD4 in the antiviral response to RSV. RSV replication in the lower airway epithelium triggers a genomic response important to the immunopathogenesis of this disease, including Th2 lymphocyte, myofibroblast expansion, and mucous obstruction (Zhao et al., 2017). Our robust RNA-Seq data extends the role of BRD4 as an activator and a repressor of the innate immune response, important in cytokine-induced airway remodeling and a subset of IFN genes. Our finding deduces that BRD4 is required for the autoregulation of its own mRNA as well as that of RELA, MED subunits, and SMARC. These findings extend the role of BRD4 to modifying enhanced coupling and nucleosomal phasing in this process.
Because of the essential role of BRD4 in cell cycle transition, DNA damage repair and cell state transition, gene silencing approaches for BRD4 induce compensatory adaptations that result in the inference of the BRD4 difficult. Here, we employ an alternative approach using a highly potent and rapidly active competitive inhibitor of BRD4. Earlier, we reported on the discovery and validation of ZL0454 as a potent and selective inhibitor of BRD4 BD1 and BD2 domains, enabling the elucidation of the role of BRD4 in inducible signaling downstream of TLR3 (Tian et al., 2018a; Liu et al., 2018; Liu et al., 2020). ZL0454 binds in the acetylated lysine–binding pockets of BRD4 BD1 and BD2 with ∼80 nM affinity, forming hydrogen bonds with Asn140 directly and Tyr97 indirectly via a H2O molecule (Liu et al., 2018). Moreover, ZL0454 has 30-fold selectivity for BRD4 BDs over other BET family members, making this a unique reagent for probing the role of BRD4 in dynamic situations. That ZL0454 disrupts RSV-inducible protein interactions were shown in our recent affinity-mass spectrometry studies (Mann et al., 2021). In this study, we extend the functional understanding of these interactions on gene expression in the context of RSV infection.
Our previous study has shown that intracellular RSV replication is a profound activator of mucosal type I/III IFN expression (Jamaluddin et al., 2001; Liu et al., 2009). In the airways, virally infected cells sustain IFN production through a dramatic signal amplification process involving the IRF transcription factors and their upstream pattern recognition receptors, TLR3 and RIG-I (Tian et al., 2017b). IFNs play an important role in antiviral defense mechanisms by activating neighboring, uninfected cells, to express ∼300 downstream ISGs to promote an antiviral state and reduce viral spread (Smieja et al., 2008). Consequently, ISG expression is a dominant pathway seen in whole genome profiling of RSV-infected cells (Zhang et al., 2001; Zhang et al., 2003; Xu et al., 2020).
Our findings that ISGs MX1, OASL, and IRF7 are inhibited by ZL0454 suggests that BRD4 is important in paracrine IFN signaling. We note that ISG inhibition cannot be attributed to the trivial effect that BRD4 inhibition artifactually reduces RSV replication, as enhanced expression of RSV patterns is seen in the ZL0454-treated cells (Figure 3A).Previous studies have shown no effect of BRD4 silencing or inhibition on RSV replication in vitro (Tian et al., 2017b). Moreover, the expression of the IFN antagonistic genes, NS1 and NS2, is similar in the ZL0454-treated cells.
The effect of BRD4 inhibitors on MX1, OASL, and IRF7 is consistent with earlier mechanistic studies that BRD4 and the DRB sensitivity factor are rapidly recruited to ISGs after IFN stimulation (Patel et al., 2013) and RSV infection (Tian et al., 2018b). In both of these studies, BRD4 binding was enhanced by IFN/RSV, and this recruitment was reduced by the use of nonselective small molecule BRD4 inhibitors. These data suggest that the BD domain is involved in inducible promoter recruitment, either by disrupting binding to acetylated histones or acetylated transcription factors. Interestingly, not all IFNs are inhibited by ZL0454. For example, IFNL3 is, in fact, upregulated by the administration of the inhibitor (Figure 4C), and IFNβ is not affected (not shown). These data indicate to us that IFN–ISGs are controlled by distinct transcriptional regulatory processes.
In vivo, RSV infection triggers a process involving the expansion of subepithelial myofibroblasts important in extracellular matrix remodeling (Brasier, 2020). Our previous studies using ATAC-Seq identified that RSV produces nucleosome-free regions on TGFB1/JUNB//FN1/MMP9 genes and the rate-limiting enzyme in the hexosamine biosynthetic pathway, glutamine-fructose-6-phosphate transaminase 2 (GFPT2), whose paracrine activity is critical in myofibroblast expansion (Xu et al., 2020). However, the mechanisms by how these chromatin remodeling events were produced were not elucidated. Our studies here showed that IL-6 and MMP9 expressions are highly BRD4-sensitive and indicate that BRD4 may play a critical role in chromatin opening in this growth factor pathway. Future studies identifying BRD4-sensitive RSV-induced chromatin domains will be illuminating.
Our studies are the first, to our knowledge, to indicate that BRD4 expression is inducible and autoregulated in response to RSV infection. The enhanced expression of BRD4 mRNA, yet steady-state levels of BRD4 protein is reduced, indicates that BRD4 protein turnover increases on pathway activation. To this end, it is notable that the TRIM family of ubiquitin ligases have been identified in the BRD4 complex (Zhang et al., 2017). More research will be needed to examine the role of ubiquitination in BRD4-dependent gene expression.
On innate activation, NFκB/RelA undergoes a coupled phosphorylation/acetylation processing mediated by the IκB kinases and p300/CBP, respectively (Brasier et al., 2011; Nowak et al., 2008). BRD4 BD binds to the acetyl-K 310 residue of NFκB/RelA, (Figure 6G) and the complex is recruited to a subset of NF-κB and BRD4-dependent genes. Interestingly, PLA indicates that the presence of the RelA∙BRD4 complex is located in both cytoplasmic and nuclear fractions. These data are consistent with earlier immunohistochemical observations that BRD4 is primarily found in the nuclear compartment but also clearly detectable in the cytoplasm (see Figure 4 in (Tian et al., 2016)). This finding could suggest that the complex is initially formed in the cytoplasm, or the complex is actively shuttling in and out of the nucleus, consistent with real-time fluorescent imaging of activated RelA (Carlotti et al., 2000). Research by others has shown that BRD4 binding stabilized activated RelA in the nucleus. Upon disrupting BRD4 binding, nuclear acetylated RelA is rapidly degraded, and RelA is depleted in the cells (Zou et al., 2014). Our findings here suggest that the effect of BRD4 on NFκB abundance may also be mediated through the inhibition of NFκB gene expression. Collectively, these findings suggest that BRD4 inhibitors may profoundly modify inflammatory diseases.
Mediators are a family of proteins that couple super enhancers to polymerases engaged on cell identity and oncogenes and whose actions are closely intertwined with those of BRD4 (Quevedo et al., 2019). Previous research in acute myelogenous leukemia cells have shown that BET proteins control the association of MED subunits with the MYB oncogene (Bhagwat et al., 2016). Interestingly, in this oncogenic context, the MED12, MED13, MED23, and MED24 subunits can functionally substitute for BRD4, producing BET inhibitor growth resistance. The MED complex is well established to interact with BRD4. In separate interactome studies, MED subunits are enriched with basal and activated BRD4 complexes (Zhang et al., 2017; Mann et al., 2021), and the interaction is disrupted by BD inhibition. Our findings that RSV regulates mRNA expression of individual subunits of MED complexes and that this regulation is modulated by BRD4 suggests that these modified complexes may have unique biological functions or target genes. More studies examining the role of changes in MED subunit expression on transcriptional elongation in the innate response will be of substantial interest.
This study indicates that BRD4 is required for sustained expression of ARID1A and SMARC subunits in RSV-infected airway epithelial cells. The SMARC proteins are a family of nonredundant ATP-dependent chromatin remodeling complexes that form cell-type specific complexes important in maintaining a differentiated epithelial cell phenotype (Xu et al., 2021b). Our study here indicates that the expression of many SMARC subunits is largely inhibited by RSV infection, SMARCB1, and its core ATP-dependent catalytic subunit SMARCA2/BRM prime IFNβ expression (Cui et al., 2004). We found earlier that SMARCA4 complexes are also important in priming IFNβ and IFNL mucosal interferons for a full antiviral response (Xu et al., 2021b). This mechanism involves enhanced accessibility of distal upstream enhancer/intergenic elements, perhaps suggesting MED interactions with proximal IFN promoters. The role of BRD4–SMARC expression may be important in maintaining 3D chromatin conformation and enhancer looping; these questions will require direct investigation.
Here, we provide evidence that a subset of the BRD4-dependent genes is associated with changes in chromatin accessibility. These genes are enriched in collagen RSV-induced formation and remodeling, consistent with our earlier findings that increased chromatin accessibility participates in RSV-induced cell-state transitions (Xu et al., 2020; Xu et al., 2021b). Although these inducible genomic sites are enriched in histone modifications associated with transcriptional elongation, it remains an open question whether these genes are regulated by transcriptional elongation or by enhancer-dependent transcription. Moreover, whether these sites are involved in BRD4 binding and/or regulated by BRD4 will require further investigation.
In the patterned epithelial innate immune response, BRD4 interacts with specific members of transcription factor families (NFκB), chromatin complex adapters (MED), and chromatin modifying factor (SMARC) to alter the activities of the complex which is schematically illustrated in Figure 11. Intriguingly, the expression of these interacting proteins is coordinately regulated by the antiviral host response. Using a selective, small-molecule BRD4 inhibitor, we demonstrate the dynamic role that BRD4 plays in the expression of its interacting coactivators. One of our surprising findings is that BRD4 participates in the RSV-inducible expression of RelA, in addition to its known effects on posttranslational stabilization. By contrast, BRD4 suppresses RSV-inducible expression of the MED subunits CDK12, CSRP1, ERCC3, MED17 and -27. By contrast, BRD4 activity is required for expression of the SMARC complexes, ARID-1A, -1B, SMARCAD1, and SMARCE1. In this way, BRD4 reshapes the protein-interacting complexes formed during the innate response to RSV. Moreover, our findings suggest that BRD4 is autoregulated and is actively consumed by the innate pathway. Integrating the results of the RNA-Seq and ATAC-Seq data, we identify the subset of BRD4-dependent genes that undergo RSV-inducible nucleosomal repositioning, resulting in enhanced transcription factor accessibility. This effect is in addition to the well-known effect of BRD4 complex interaction with innate response genes in chromatin-accessible domains (Brasier, 2020; Brasier et al., 2011; Tian et al., 2013). We note that genes undergoing chromatin transition into an open configuration are enriched for those involved in ECM formation and remodeling (Figure 11). Our study, therefore, extends the understanding of how BRD4 inhibitors function as effective anti-viral therapeutics by affecting virus-inducible ECM remodeling (Brasier, 2020).
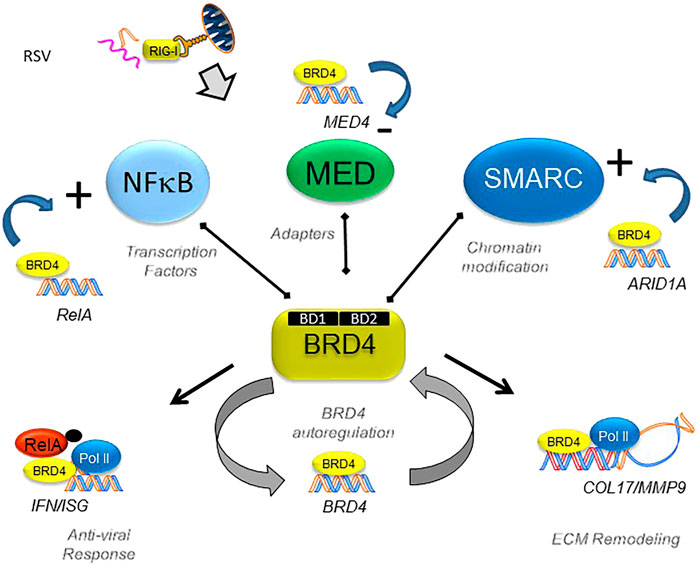
FIGURE 11. BRD4’s dynamically regulated gene networks. Schematic diagram of BRD4-dependent genes in RSV infection. Although BRD4 is required for innate signaling and expression of ECM remodeling proteins, this study provides evidence that BRD4 controls its own expression via an autoregulatory network. In addition, BRD4 controls the expression of members of its interacting coactivators that bind the BD, including transcription factors, adaptors, and chromatin remodeling complexes.
Repetitive viral infections producing acute exacerbations of asthma or chronic obstructive pulmonary disease (Tuffaha et al., 2000; Gern, 2015) are linked to reduction in pulmonary function (Calhoun et al., 2015). Using a model of repetitive viral inflammation mediated by TLR3, we demonstrated that BRD4 is a central mediator of airway remodeling, coupling the innate inflammatory pathway to epithelial cell-state changes and fibrosis by the expansion of subepithelial fibroblasts (Tian et al., 2018a). Consequently, BRD4 is a validated target for the treatment of airway remodeling in viral and allergic lung diseases (Brasier and Zhou, 2020). Through its association with inflammation-activated NFκB/RelA, BRD4 is dynamically repositioned to growth factor and ECM genes, resulting in innate training, epithelial cell-state transition, and secretory myofibroblast transdifferentiation (Ijaz et al., 2017; Yang et al., 2017; Brasier, 2019). In particular, the epithelial TGFβ/IL6/MMP9 pathway plays an important role in triggering myofibroblast expansion (Xu et al., 2020; Xu et al., 2021b). These genes undergo changes in chromatin accessibility in response to RSV infection by largely unknown mechanisms. This study extends the mechanisms of how BRD4 inhibitors disrupt inducible cytokine–growth factor circuits by disrupting BRD4 binding to chromatin and by inhibiting the expression of coactivators important in inflammation and remodeling.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo GSE179353.
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Experimentation: XX, YL, DQ, and AB; analysis: XX, DQ, AB, and MM; concept: XX, AB, and JZ; and writing: XX, MM, JZ, and AB.
This work was partially supported by NIH grants AI062885 (AB, RPG) and NCATS UL1TR002373 (AB). The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank the University of Wisconsin-Madison Biotechnology Center Gene Expression Center and the Bioinformatics Core for next generation sequencing, analytic services, and data management. The authors also thank the NIAID-AI062885-funded IBVC Core for technical assistance.
Bennett, B. L., Garofalo, R. P., Cron, S. G., Hosakote, Y. M., Atmar, R. L., Macias, C. G., et al. (2007). Immunopathogenesis of Respiratory Syncytial Virus Bronchiolitis. J. Infect. Dis. 195, 1532–1540. doi:10.1086/515575
Bhagwat, A. S., Roe, J.-S., Mok, B. Y. L., Hohmann, A. F., Shi, J., and Vakoc, C. R. (2016). BET Bromodomain Inhibition Releases the Mediator Complex from Select Cis -Regulatory Elements. Cel Rep. 15, 519–530. doi:10.1016/j.celrep.2016.03.054
Bindea, G., Mlecnik, B., Hackl, H., Charoentong, P., Tosolini, M., Kirilovsky, A., et al. (2009). ClueGO: a Cytoscape Plug-In to Decipher Functionally Grouped Gene Ontology and Pathway Annotation Networks. Bioinformatics 25, 1091–1093. doi:10.1093/bioinformatics/btp101
Bisgrove, D. A., Mahmoudi, T., Henklein, P., and Verdin, E. (2007). Conserved P-TEFb-Interacting Domain of BRD4 Inhibits HIV Transcription. Proc. Natl. Acad. Sci. 104, 13690–13695. doi:10.1073/pnas.0705053104
Brasier, A. R. (2019). Mechanisms How Mucosal Innate Immunity Affects Progression of Allergic Airway Disease. Expert Rev. Respir. Med. 13, 349–356. doi:10.1080/17476348.2019.1578211
Brasier, A. R. (2020). RSV Reprograms the CDK9*BRD4 Chromatin Remodeling Complex to Couple Innate Inflammation to Airway Remodeling. Viruses 12–472. doi:10.3390/v12040472
Brasier, A. R., Tian, B., Jamaluddin, M., Kalita, M. K., Garofalo, R. P., and Lu, M. (2011). RelA Ser276 Phosphorylation-Coupled Lys310 Acetylation Controls Transcriptional Elongation of Inflammatory Cytokines in Respiratory Syncytial Virus Infection. J. Virol. 85, 11752–11769. doi:10.1128/jvi.05360-11
Brasier, A. R., and Zhou, J. (2020). Validation of the Epigenetic Reader Bromodomain-Containing Protein 4 (BRD4) as a Therapeutic Target for Treatment of Airway Remodeling. Drug Discov. Today 25, 126–132. doi:10.1016/j.drudis.2019.11.002
Calhoun, W. J., Haselkorn, T., Miller, D. P., and Omachi, T. A. (2015). Asthma Exacerbations and Lung Function in Patients with Severe or Difficult-To-Treat Asthma. J. Allergy Clin. Immunol. 136, 1125–1127. doi:10.1016/j.jaci.2015.05.014
Carlotti, F., Dower, S. K., and Qwarnstrom, E. E. (2000). Dynamic Shuttling of Nuclear Factor κB between the Nucleus and Cytoplasm as a Consequence of Inhibitor Dissociation. J. Biol. Chem. 275, 41028–41034. doi:10.1074/jbc.m006179200
Choudhary, S., Boldogh, S., Garofalo, R., Jamaluddin, M., and Brasier, A. R. (2005). Respiratory Syncytial Virus Influences NF-κb-dependent Gene Expression through a Novel Pathway Involving MAP3K14/NIK Expression and Nuclear Complex Formation with NF-Κb2. J. Virol. 79, 8948–8959. doi:10.1128/jvi.79.14.8948-8959.2005
Collins, P. L., and Graham, B. S. (2008). Viral and Host Factors in Human Respiratory Syncytial Virus Pathogenesis. J. Virol. 82, 2040–2055. doi:10.1128/jvi.01625-07
Crystal, R. G., Randell, S. H., Engelhardt, J. F., Voynow, J., and Sunday, M. E. (2008). Airway Epithelial Cells: Current Concepts and Challenges. Proc. Am. Thorac. Soc. 5, 772–777. doi:10.1513/pats.200805-041hr
Cui, K., Tailor, P., Liu, H., Chen, X., Ozato, K., and Zhao, K. (2004). The Chromatin-Remodeling BAF Complex Mediates Cellular Antiviral Activities by Promoter Priming. Mol. Cel Biol. 24, 4476–4486. doi:10.1128/MCB.24.10.4476-4486.2004
Devaiah, B. N., Case-Borden, C., Gegonne, A., Hsu, C. H., Chen, Q., Meerzaman, D., et al. (2016a). BRD4 Is a Histone Acetyltransferase that Evicts Nucleosomes from Chromatin. Nat. Struct. Mol. Biol. 23, 540–548. doi:10.1038/nsmb.3228
Devaiah, B. N., Gegonne, A., and Singer, D. S. (2016b). Bromodomain 4: a Cellular Swiss Army Knife. J. Leukoc. Biol. 100, 679–686. doi:10.1189/jlb.2ri0616-250r
Devaiah, B. N., and Singer, D. S. (2012). Cross-talk Among RNA Polymerase II Kinases Modulates C-Terminal Domain Phosphorylation. J. Biol. Chem. 287, 38755–38766. doi:10.1074/jbc.m112.412015
Floyd, S. R., Pacold, M. E., Huang, Q., Clarke, S. M., Lam, F. C., Cannell, I. G., et al. (2013). The Bromodomain Protein Brd4 Insulates Chromatin from DNA Damage Signalling. Nature 498, 246–250. doi:10.1038/nature12147
Garofalo, R., Sabry, M., Jamaluddin, M., Yu, R. K., Casola, A., Ogra, P. L., et al. (1996). Transcriptional Activation of the Interleukin-8 Gene by Respiratory Syncytial Virus Infection in Alveolar Epithelial Cells: Nuclear Translocation of the RelA Transcription Factor as a Mechanism Producing Airway Mucosal Inflammation. J. Virol. 70, 8773–8781. doi:10.1128/jvi.70.12.8773-8781.1996
Gaspar, J. M. (2019). Genrich: Detecting Sites of Genomic Enrichment. https://github.com/jsh58/Genrich.
Gern, J. E. (2015). How Rhinovirus Infections Cause Exacerbations of Asthma. Clin. Exp. Allergy 45, 32–42. doi:10.1111/cea.12428
Harvey, B. G., Heguy, A., Leopold, P. L., Carolan, B. J., Ferris, B., and Crystal, R. G. (2007). Modification of Gene Expression of the Small Airway Epithelium in Response to Cigarette Smoking. J. Mol. Med. (Berl) 85, 39–53. doi:10.1007/s00109-006-0103-z
Hosakote, Y. M., Brasier, A. R., Casola, A., Garofalo, R. P., and Kurosky, A. (2016). Respiratory Syncytial Virus Infection Triggers Epithelial HMGB1 Release as a Damage-Associated Molecular Pattern Promoting a Monocytic Inflammatory Response. J. Virol. 90, 9618–9631. doi:10.1128/jvi.01279-16
Ijaz, T., Jamaluddin, M., Zhao, Y., Zhang, Y., Jay, J., Finnerty, C. C., et al. (2017). Coordinate Activities of BRD4 and CDK9 in the Transcriptional Elongation Complex Are Required for TGFβ-Induced Nox4 Expression and Myofibroblast Transdifferentiation. Cell Death Dis. 8 (2), e2606. doi:10.1038/cddis.2016.434
Jamaluddin, M., Casola, A., Garofalo, R. P., Han, Y., Elliott, T., Ogra, P. L., et al. (1998). The Major Component of IκBα Proteolysis Occurs Independently of the Proteasome Pathway in Respiratory Syncytial Virus-Infected Pulmonary Epithelial Cells. J. Virol. 72, 4849–4857. doi:10.1128/jvi.72.6.4849-4857.1998
Jamaluddin, M., Wang, S., Garofalo, R. P., Elliott, T., Casola, A., Baron, S., et al. (2001). IFN-β Mediates Coordinate Expression of Antigen-Processing Genes in RSV-Infected Pulmonary Epithelial Cells. Am. J. Physiol. Lung Cel Mol Physiol. 280, L248–L257. doi:10.1152/ajplung.2001.280.2.l248
Jassal, B., Matthews, L., Viteri, G., Gong, C., Lorente, P., Fabregat, A., et al. (2020). The Reactome Pathway Knowledgebase. Nucleic Acids Res. 48, D498–d503. doi:10.1093/nar/gkz1031
Jeronimo, C., Forget, D., Bouchard, A., Li, Q., Chua, G., Poitras, C., et al. (2007). Systematic Analysis of the Protein Interaction Network for the Human Transcription Machinery Reveals the Identity of the 7SK Capping Enzyme. Mol. Cel. 27, 262–274. doi:10.1016/j.molcel.2007.06.027
Lay, M. K., Bueno, S. M., Gálvez, N., Riedel, C. A., and Kalergis, A. M. (2016). New Insights on the Viral and Host Factors Contributing to the Airway Pathogenesis Caused by the Respiratory Syncytial Virus. Crit. Rev. Microbiol. 42, 800–812. doi:10.3109/1040841X.2015.1055711
Liao, Y., Smyth, G. K., and Shi, W. (2019). The R Package Rsubread Is Easier, Faster, Cheaper and Better for Alignment and Quantification of RNA Sequencing Reads. Nucleic Acids Res. 47, e47. doi:10.1093/nar/gkz114
Lis, J. T., Mason, P., Peng, J., Price, D. H., and Werner, J. (2000). P-TEFb Kinase Recruitment and Function at Heat Shock Loci. Genes Dev. 14, 792–803. doi:10.1101/gad.14.7.792
Liu, P., Jamaluddin, M., Li, K., Garofalo, R. P., Casola, A., and Brasier, A. R. (2007). Retinoic Acid-Inducible Gene I Mediates Early Antiviral Response and Toll-like Receptor 3 Expression in Respiratory Syncytial Virus-Infected Airway Epithelial Cells. J. Virol. 81, 1401–1411. doi:10.1128/jvi.01740-06
Liu, P., Lu, M., Tian, B., Li, K., Garofalo, R. P., Prusak, D., et al. (2009). Expression of an IKKγ Splice Variant Determines IRF3 and Canonical NF-Κb Pathway Utilization in ssRNA Virus Infection. PLoS One 4, e8079. doi:10.1371/journal.pone.0008079
Liu, Z., Chen, H., Wang, P., Li, Y., Wold, E. A., Leonard, P. G., et al. (2020). Discovery of Orally Bioavailable Chromone Derivatives as Potent and Selective BRD4 Inhibitors: Scaffold Hopping, Optimization, and Pharmacological Evaluation. J. Med. Chem. 63, 5242–5256. doi:10.1021/acs.jmedchem.0c00035
Liu, Z., Tian, B., Chen, H., Wang, P., Brasier, A. R., and Zhou, J. (2018). Discovery of Potent and Selective BRD4 Inhibitors Capable of Blocking TLR3-Induced Acute Airway Inflammation. Eur. J. Med. Chem. 151, 450–461. doi:10.1016/j.ejmech.2018.04.006
Love, M. I., Huber, W., and Anders, S. (2014). Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 15, 550. doi:10.1186/s13059-014-0550-8
Lovén, J., Hoke, H. A., Lin, C. Y., Lau, A., Orlando, D. A., Vakoc, C. R., et al. (2013). Selective Inhibition of Tumor Oncogenes by Disruption of Super-enhancers. Cell 153, 320–334. doi:10.1016/j.cell.2013.03.036
Mann, M., Roberts, D. S., Li, Y., Zhou, J., Ge, Y., and Brasier, A. R. (2021). Discovery of RSV-Induced BRD4 Protein Interactions Using Native Immunoprecipitation and Parallel Accumulation-Serial Fragmentation (PASEF). Mass Spectrom. 13, 454. doi:10.3390/v13030454
Nowak, D. E., Tian, B., Jamaluddin, M., Boldogh, I., Vergara, L. A., Choudhary, S., et al. (2008). RelA Ser 276 Phosphorylation Is Required for Activation of a Subset of NF-κb-dependent Genes by Recruiting Cyclin-dependent Kinase 9/Cyclin T1 Complexes. Mol. Cel Biol. 28, 3623–3638. doi:10.1128/mcb.01152-07
Patel, M. C., Debrosse, M., Smith, M., Dey, A., Huynh, W., Sarai, N., et al. (2013). BRD4 Coordinates Recruitment of Pause Release Factor P-TEFb and the Pausing Complex NELF/DSIF to Regulate Transcription Elongation of Interferon-Stimulated Genes. Mol. Cel Biol. 33, 2497–2507. doi:10.1128/mcb.01180-12
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., and Kingsford, C. (2017). Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 14, 417–419. doi:10.1038/nmeth.4197
Pneumonia Etiology Research for Child Health (PERCH) Study Group (2019). Causes of Severe Pneumonia Requiring Hospital Admission in Children without HIV Infection from Africa and Asia: the PERCH Multi-Country Case-Control Study. Lancet 394, 757–779. doi:10.1016/S0140-6736(19)30721-4
Price, D. H. (2000). P-TEFb, a Cyclin-dependent Kinase Controlling Elongation by RNA Polymerase II. Mol. Cel Biol. 20, 2629–2634. doi:10.1128/mcb.20.8.2629-2634.2000
Quevedo, M., Meert, L., Dekker, M. R., Dekkers, D. H. W., Brandsma, J. H., van den Berg, D. L. C., et al. (2019). Mediator Complex Interaction Partners Organize the Transcriptional Network that Defines Neural Stem Cells. Nat. Commun. 10, 2669. doi:10.1038/s41467-019-10502-8
Rahman, S., Sowa, M. E., Ottinger, M., Smith, J. A., Shi, Y., Harper, J. W., et al. (2011). The Brd4 Extraterminal Domain Confers Transcription Activation Independent of pTEFb by Recruiting Multiple Proteins, Including NSD3. Mol. Cel Biol. 31, 2641–2652. doi:10.1128/mcb.01341-10
Ross-Innes, C. S., Stark, R., Teschendorff, A. E., Holmes, K. A., Ali, H. R., Dunning, M. J., et al. (2012). Differential Oestrogen Receptor Binding Is Associated with Clinical Outcome in Breast Cancer. Nature 481, 389–393. doi:10.1038/nature10730
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 13, 2498–2504. doi:10.1101/gr.1239303
Smieja, J., Jamaluddin, M., Brasier, A. R., and Kimmel, M. (2008). Model-based Analysis of Interferon-β Induced Signaling Pathway. Bioinformatics 24, 2363–2369. doi:10.1093/bioinformatics/btn400
Tian, B., Yang, J., Zhao, Y., Ivanciuc, T., Sun, H., Garofalo, R. P., et al. (2017b). BRD4 Couples NF-κB/RelA with Airway Inflammation and the IRF-RIG-I Amplification Loop in Respiratory Syncytial Virus Infection. J. Virol. 91. doi:10.1128/JVI.00007-0001710.1128/JVI.00007-17
Tian, B., Yang, J., Zhao, Y., Ivanciuc, T., Sun, H., Wakamiya, M., et al. (2018b). Central Role of the NF-Κb Pathway in the Scgb1a1-Expressing Epithelium in Mediating Respiratory Syncytial Virus-Induced Airway Inflammation. J. Virol. 92. doi:10.1128/JVI.00441-18
Tian, B., Hosoki, K., Liu, Z., Yang, J., Zhao, Y., Sun, H., et al. (2019a). Mucosal Bromodomain-Containing Protein 4 Mediates Aeroallergen-Induced Inflammation and Remodeling. J. Allergy Clin. Immunol. 143, 1380–1394. doi:10.1016/j.jaci.2018.09.029
Tian, B., Liu, Z., Litvinov, J., Maroto, R., Jamaluddin, M., Rytting, E., et al. (2019b). Efficacy of Novel Highly Specific Bromodomain-Containing Protein 4 Inhibitors in Innate Inflammation-Driven Airway Remodeling. Am. J. Respir. Cel Mol Biol. 60, 68–83. doi:10.1165/rcmb.2017-0445oc
Tian, B., Liu, Z., Yang, J., Sun, H., Zhao, Y., Wakamiya, M., et al. (2018a). Selective Antagonists of the Bronchiolar Epithelial NF-Κb-Bromodomain-Containing Protein 4 Pathway in Viral-Induced Airway Inflammation. Cel Rep. 23, 1138–1151. doi:10.1016/j.celrep.2018.03.106
Tian, B., Patrikeev, I., Ochoa, L., Vargas, G., Belanger, K. K., Litvinov, J., et al. (2017a). NF-κB Mediates Mesenchymal Transition, Remodeling, and Pulmonary Fibrosis in Response to Chronic Inflammation by Viral RNA Patterns. Am. J. Respir. Cel Mol Biol. 56, 506–520. doi:10.1165/rcmb.2016-0259oc
Tian, B., Zhang, Y., Luxon, B. A., Garofalo, R. P., Casola, A., Sinha, M., et al. (2002). Identification of NF-κb-dependent Gene Networks in Respiratory Syncytial Virus-Infected Cells. J. Virol. 76, 6800–6814. doi:10.1128/jvi.76.13.6800-6814.2002
Tian, B., Zhao, Y., Kalita, M., Edeh, C. B., Paessler, S., Casola, A., et al. (2013). CDK9-dependent Transcriptional Elongation in the Innate Interferon-Stimulated Gene Response to Respiratory Syncytial Virus Infection in Airway Epithelial Cells. J. Virol. 87, 7075–7092. doi:10.1128/jvi.03399-12
Tian, B., Zhao, Y., Sun, H., Zhang, Y., Yang, J., and Brasier, A. R. (2016). BRD4 Mediates NF-κb-dependent Epithelial-Mesenchymal Transition and Pulmonary Fibrosis via Transcriptional Elongation. Am. J. Physiology-Lung Cell Mol. Physiol. 311, L1183–L1201. doi:10.1152/ajplung.00224.2016
Tuffaha, A., Gern, J. E., and Lemanske, R. F. (2000). The Role of Respiratory Viruses in Acute and Chronic Asthma. Clin. Chest Med. 21, 289–300. doi:10.1016/s0272-5231(05)70267-7
Ueba, O. (1978). Respiratory Syncytial Virus. I. Concentration and Purification of the Infectious Virus. Acta Med. Okayama 32, 265–272.
Xu, X., Mann, M., Qiao, D., and Brasier, A. R. (2021a). Alternative mRNA Processing of Innate Response Pathways in Respiratory Syncytial Virus (RSV) Infection. Viruses. 13, 218. doi:10.3390/v13020218
Xu, X., Qiao, D., Dong, C., Mann, M., Garofalo, R. P., Keles, S., et al. (2021b). The SWI/SNF-Related, Matrix Associated, Actin-dependent Regulator of Chromatin A4 Core Complex Represses Respiratory Syncytial Virus-Induced Syncytia Formation and Subepithelial Myofibroblast Transition. Front. Immunol. 12. doi:10.3389/fimmu.2021.633654
Xu, X., Qiao, D., Mann, M., Garofalo, R. P., and Brasier, A. R. (2020). Respiratory Syncytial Virus Infection Induces Chromatin Remodeling to Activate Growth Factor and Extracellular Matrix Secretion Pathways. Viruses. 12, 804.
Yang, J., Tian, B., and Brasier, A. R. (2017). “Targeting Chromatin Remodeling in Inflammation and Fibrosis,” in Advances in Protein Chemistry and Structural Biology. Editor R. Donev (Elsevier). doi:10.1016/bs.apcsb.2016.11.001
Yang, J., Zhao, Y., Kalita, M., Li, X., Jamaluddin, M., Tian, B., et al. (2015). Systematic Determination of Human Cyclin Dependent Kinase (CDK)-9 Interactome Identifies Novel Functions in RNA Splicing Mediated by the DDX5/17 RNA Helicases. Mol. Cel Proteomics 14, 2701–2721. doi:10.1074/mcp.m115.049221
Yu, G., Wang, L.-G., Yan, G.-R., and He, Q.-Y. (2014). DOSE: an R/Bioconductor Package for Disease Ontology Semantic and Enrichment Analysis. Bioinformatics 31, 608–609. doi:10.1093/bioinformatics/btu684
Zhang, J., Jamaluddin, M., Zhang, Y., Widen, S. G., Sun, H., Brasier, A. R., et al. (2019). Type II Epithelial-Mesenchymal Transition Upregulates Protein N-Glycosylation to Maintain Proteostasis and Extracellular Matrix Production. J. Proteome Res. 18, 3447–3460. doi:10.1021/acs.jproteome.9b00342
Zhang, X., Krutchinsky, A., Fukuda, A., Chen, W., Yamamura, S., Chait, B. T., et al. (2005). MED1/TRAP220 Exists Predominantly in a TRAP/ Mediator Subpopulation Enriched in RNA Polymerase II and Is Required for ER-Mediated Transcription. Mol. Cel. 19, 89–100. doi:10.1016/j.molcel.2005.05.015
Zhang, Y., Jamaluddin, M., Wang, S., Tian, B., Garofalo, R. P., Casola, A., et al. (2003). Ribavirin Treatment Up-Regulates Antiviral Gene Expression via the Interferon-Stimulated Response Element in Respiratory Syncytial Virus-Infected Epithelial Cells. J. Virol. 77, 5933–5947. doi:10.1128/jvi.77.10.5933-5947.2003
Zhang, Y., Luxon, B. A., Casola, A., Garofalo, R. P., Jamaluddin, M., and Brasier, A. R. (2001). Expression of Respiratory Syncytial Virus-Induced Chemokine Gene Networks in Lower Airway Epithelial Cells Revealed by cDNA Microarrays. J. Virol. 75, 9044–9058. doi:10.1128/jvi.75.19.9044-9058.2001
Zhang, Y., Sun, H., Zhang, J., Brasier, A. R., and Zhao, Y. (2017). Quantitative Assessment of the Effects of Trypsin Digestion Methods on Affinity Purification-Mass Spectrometry-Based Protein-Protein Interaction Analysis. J. Proteome Res. 16, 3068–3082. doi:10.1021/acs.jproteome.7b00432
Zhao, Y., Jamaluddin, M., Zhang, Y., Sun, H., Ivanciuc, T., Garofalo, R. P., et al. (2017). Systematic Analysis of Cell-type Differences in the Epithelial Secretome Reveals Insights into the Pathogenesis of Respiratory Syncytial Virus-Induced Lower Respiratory Tract Infections. J.I. 198, 3345–3364. doi:10.4049/jimmunol.1601291
Zhao, Y., Tian, B., Sun, H., Zhang, J., Zhang, Y., Ivannikov, M., et al. (2019). Pharmacoproteomics Reveal Novel Protective Activity of Bromodomain Containing 4 Inhibitors on Vascular Homeostasis in TLR3-Mediated Airway Remodeling. J. Proteomics 205, 103415. doi:10.1016/j.jprot.2019.103415
Keywords: BRD4, BRD4 (bromodomain-containing protein 4), interferon, ISG (interferon-stimulated genes), MMP-9
Citation: Xu X, Mann M, Qiao D, Li Y, Zhou J and Brasier AR (2021) Bromodomain Containing Protein 4 (BRD4) Regulates Expression of its Interacting Coactivators in the Innate Response to Respiratory Syncytial Virus. Front. Mol. Biosci. 8:728661. doi: 10.3389/fmolb.2021.728661
Received: 21 June 2021; Accepted: 24 September 2021;
Published: 26 October 2021.
Edited by:
Amit Kumar Singh, National Institutes of Health (NIH), United StatesReviewed by:
Bowen Wang, University of Virginia, United StatesCopyright © 2021 Xu, Mann, Qiao, Li, Zhou and Brasier. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Allan R. Brasier, YWJyYXNpZXJAd2lzYy5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.