
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci., 28 September 2021
Sec. RNA Networks and Biology
Volume 8 - 2021 | https://doi.org/10.3389/fmolb.2021.725308
This article is part of the Research TopicAlternative Splicing in Health and DiseaseView all 9 articles
With the development of high-throughput sequencing technology, the posttranscriptional mechanism of alternative splicing is becoming better understood. From decades of studies, alternative splicing has been shown to occur in multiple tissues, including the brain, heart, testis, skeletal muscle, and liver. This regulatory mechanism plays an important role in physiological functions in most liver diseases. Currently, due to the absence of symptoms, chronic pediatric liver diseases have a significant impact on public health. Furthermore, the progression of the disease is accelerated in children, leading to severe damage to their liver tissue if no precautions are taken. To this end, this review article summarizes the current knowledge of alternative splicing in pediatric liver diseases, paying special attention to liver damage in the child stage. The discussion of the regulatory role of splicing in liver diseases and its potential as a new therapeutic target is also included.
Chronic liver disease is an increasing health burden in children and adults. Although the incidence is unclear, it is estimated that chronic liver disease has become the 11th leading cause of death among adults (Vos et al., 2016; Gao et al., 2021). In addition, many children are hospitalized due to liver diseases each year. In children and adults, nonalcoholic fatty liver disease (NAFLD) is known as a frequently appearing chronic liver disease with a complex interplay between environmental and genetic factors in industrialized countries (Vos et al., 2016; Gao et al., 2021). It is generally accepted that pediatric liver diseases, including NAFLD, chronic viral hepatitis, and other chronic liver diseases, may develop into chronic diseases, such as cirrhosis and hepatocellular carcinoma, in adults (Della Corte et al., 2016; Nobili et al., 2019). In particular, the number of liver cancer cases has increased rapidly since 1990 (Della Corte et al., 2016). In addition to liver-associated symptoms, NAFLD is related to a risk increment of diabetes (type 2) and cardiovascular disease in adults (Draijer et al., 2019). Furthermore, hepatic cholestasis and metabolic and autoimmune liver problems have been demonstrated to be the most common causes of liver failure in children, leading to liver transplantation at their final stage (Nikeghbalian et al., 2021). On the other hand, viral hepatitis, alcoholic hepatitis, and hepatocellular carcinoma are more common in adults. In both children and adults, the vast majority of patients are asymptomatic in the early stage of disease, and the prevalence of these diseases is unknown, leading to delays in diagnosis and treatment. Furthermore, because similar responses will be shown from different injuries of hepatic cells, different types of liver diseases may show similar presentations among children (Della Corte et al., 2016). Moreover, nonspecific signs, including abdominal pain, fatigue, loss of appetite, pruritus, or hepatomegaly, can also be present among patients. Thus, prevention and early diagnosis are important to distinguish these chronic liver diseases and will lead to cost-effective treatments in pediatric settings.
Alternative splicing is the process to mediate pre-messenger RNA maturation into mRNA, by removing introns and reattaching exons (Webster, 2017). During this precise molecular process, different transcripts are assembled with the help of splice sites and associated sequences. Hence, alterations in splicing factors, such as the usage of new splice sites or enhancer sequences, as well as disruption of splicing sequences, can lead to diseases including metabolic diseases, liver diseases, cancers, and neurodegenerative diseases (Montes et al., 2019; Rahman et al., 2020). For instance, mutations in spliceosome RNA genes were detected in hepatocellular carcinoma, medulloblastoma, and chronic lymphocytic leukemia (Jimenez et al., 2018; Suzuki et al., 2019; Rahman et al., 2020). In addition, critical changes were found in the function of RNA splicing in hepatic fat metabolism of obesity (Pihlajamaki et al., 2011). Recent studies have also demonstrated alterations in the splicing machinery in inflammation, steatosis, and fibrosis in NAFLD patients and animals (Zhu et al., 2016; Gerhard et al., 2018; Del Río-Moreno et al., 2019; Hoang et al., 2019; Wang et al., 2019). In this review article, we will summarize the appearance of alternative RNA splicing in pediatric liver diseases and highlight its roles in the development and progression of these diseases (Table 1).
Infantile cholestasis (IC) is an impairment of bile production or flow occurring in the first months of life that affects 1:2,500 live births (Pietrobattista et al., 2020). It is recognized as an important cause of chronic liver disease in infants and young children, including biliary atresia (BA) (35–41%), progressive familial intrahepatic cholestasis (PFIC) (10%), Alagille syndrome (2–6%), and other causes (Götze et al., 2015). This paragraph will discuss the regulatory role of alternative splicing (AS) in PFIC and Alagille syndrome. The association between AS and BA will be stated later. PFIC patients diagnosed in childhood with intrahepatic cholestasis frequently progress to end-stage liver disease before adulthood (Bull and Thompson, 2018). There are three major proteins affected in PFIC, including the bile salt export pump (BSEP) encoded by ABCB11, multidrug resistance protein 3 (MDR3) encoded by ABCB4, and a membrane lipid composition protein (FIC1) encoded by ATP8B1, while three other reported proteins may be affected in PFIC patients, including tight junction protein 2 encoded by TJP2, the farnesoid X receptor (FXR) encoded by NR1H4, and myosin 5B encoded by MYO5B (Sambrotta et al., 2014; Qiu et al., 2017; Keitel et al., 2019). To date, except for NR1H4, splicing mutations have been found in other five genes associated with PFIC (http://www.hgmd.org) (Liu et al., 2010; van der Woerd et al., 2015; Khabou et al., 2016; Stenson et al., 2017; Al-Hussaini et al., 2021). In particular, FIC1 is part of the p-type adenosine triphosphatase type 4 subfamily involved in membrane phospholipid transport (Paulusma et al., 2008). This protein is found in the apical membrane of hepatocytes and is considered to be a phospholipid translocase that carries phospholipids, such as phosphatidylethanolamine (PE) and phosphatidylserine (PS), from the ectoplasmic lobule of the outer tubule to the cytoplasmic lobule of hepatocytes. FIC1 protects the tubule membrane by maintaining plasma membrane asymmetry (Vitale et al., 2019). Previous studies systematically described 14 mutations at the exon-intron boundary of ATP8B1 and found that most of them caused aberrant splicing of its gene product (van der Woerd et al., 2015). Furthermore, this study suggested that compensatory modified U1 small nuclear RNAs (snRNA), which are complementary to the mutated donor splicing site, are highly effective in improving exon definition, implying the therapeutic potential of these mutated loci.
Furthermore, BSEP is a binding cassette transporter for adenosine triphosphate and is involved in the transfer of bile salts from hepatocytes to bile ducts, a process which is essential for maintaining the enterohepatic circulation of related bile salts (Vitale et al., 2019). Mutations in BSEP disrupt the transportation process of bile salts out of hepatocytes, resulting in increased concentrations of intracellular bile salts to damage hepatocytes (Vitale et al., 2019). Moreover, MDR3 (ABCB4) encodes a phosphatidylcholine (PC) flippase with two transmembrane and cytoplasmic nucleotide binding domains, respectively. This protein is specifically localized at the tubular membrane of hepatocytes to mediate PC transportation from hepatocytes to bile ducts (Vitale et al., 2019). Over-expression of MDR3 increases the transport rate of fluorescin-labeled PC, but not other phospholipids. The abnormal function of MDR3 leads to the depletion of PC in the biliary tubules and elevated free hydrophobic bile acids in the space, causing damage to bile duct cells and the development of cholestasis (Henkel et al., 2019).
In addition, TJP2 is a tight junction protein that can interact with actin cytoskeletons and liver-specific tight junction proteins, such as CLDN1 and CLDN2 (Sambrotta and Thompson, 2015; González-Mariscal et al., 2019). Tight junctions are able to prevent biliary elements from leaking into the liver parenchyma. However, upon TJP2 mutation, CLDN1 failed to localize to its original position in the hepatic lobular parenchyma. This mis-localization disrupts the tight junctions, causing the leakage of bile salts with cytotoxicity into the paracellular space and resulting in damage to bile duct cells and surrounding hepatocytes (Sambrotta et al., 2014; Sambrotta and Thompson, 2015). Previous studies have reported that the mutant C. 2180-5T>G caused the jump of exon 15 of TJP2 and the deletion of 32 amino acid residues in the framework (Zhang et al., 2020). Thus, this mutation can serve as potential diagnostic targets of this disease.
At last, the fifth protein is known as myosin 5B (MYO5B), identified as a molecular motor associated with actin (Vitale et al., 2019). Reports have indicated that MYO5B can interact with RAS-associated GTP-binding protein 11A (RAB11A), in order to assist the polarization process of epithelial cells. Meanwhile, the localization of BSEP at the tubule membrane is affected by the activity of this interaction (Girard et al., 2014; Overeem et al., 2020). Mutations in this gene have been linked to microvillus inclusion body disease (MVID), which affects enterocytes, leading to reducing bile acid uptake, diarrhea, malabsorption (Van IJzendoorn et al., 2020; Overeem et al., 2020). Qiu et al. reported that three classical splicing mutations are identified from their study and this type of mutation is defined as severe mutations. Taking into consideration the interaction between MYO5B and RAB11A, the manipulation of this upstream signal transduction module may provide additional targets for therapeutic purposes. As a congenital disorder, Alagille syndrome is characterized by eye and heart abnormalities, skeletal deformities, cholestasis, and characteristic facial features (Li et al., 1997). Nearly 94% of patients have congenital cardiac diseases, and 21–31% of patients may be candidates for liver transplantation (Li et al., 1997). Currently, approximately 94% of Alagille syndrome patients have variants of JAG1, while 1–2% of patients have NOTCH2 (McDaniell et al., 2006). Both JAG1 and NOTCH2 are identified as single-channel transmembrane proteins, containing 26 and 34 exons, respectively. Specifically, the interaction between ligand JAG1 and NOTCH2 receptor requires several functional motifs, such as the C2-like domain, delta-Serate-lag2 (DSL) domain, epidermal growth factor-like (EGF-like) repeats of JAG1, and extracellular EGF-like repeats on NOTCH2 (Chillakuri et al., 2013; Kopan and Ilagan, 2009; Lindsell et al., 1995), and the mutation of this pathway identified in children with Alagille syndrome. In addition, only one NOTCH2 mutation of the splice site of exon 33 (c.5930−1G→A) was identified in the patient with Alagille syndrome, while more than 40 splicing mutations were reported in the JAG1 gene with Alagille syndrome (http://www.hgmd.cf.ac.uk/ac/gene.php?gene=JAG1) (Mcdaniell et al., 2006; Chen et al., 2018). However, how these mutations affect the interaction of this pathway remains to be elucidated. Nevertheless, these reported splicing mutations may serve as potential diagnostic targets for Alagille syndrome.
Metabolic liver diseases might be the second leading cause of liver transplantation in children, including NAFLD, Wilson’s disease (WD), alpha-1 antitrypsin, and glycogen storage disease (Elisofon et al., 2020). NAFLD is the most common chronic liver disease in children and adolescents worldwide, notified to be the second most common cause of liver transplantation (Goldner and Lavine, 2020). Approximately, 2.6–11.3% of children and approximately 40–70% of obese children are diagnosed with NAFLD worldwide (Goldner and Lavine, 2020). Consequently, NAFLD affects public health, especially children and adolescents.
Several genetic variants, including genes encoding transmembrane 6 superfamily member 2 (TM6SF2), patatin-like phospholipase domain-containing 3 (PNPLA3), glucokinase regulator (GCKR), and membrane bound O-acyl transferase 7 (MBOAT7), contribute to the risk of NAFLD, whereas protein phosphatase 1 regulatory subunit 3B (PPP1R3B) has been documented to have a protective effect against NAFLD (Valenti et al., 2010; Santoro et al., 2012; Kozlitina et al., 2014; Mancina et al., 2016; Dongiovanni et al., 2018; Li et al., 2020). However, other genetic variants, such as Mer tyrosine kinase (MERTK), interferon-λ4 (IFNL4), and 17-β hydroxysteroid dehydrogenase 13 (HSD17B13), might modify the fibrotic effect of NAFLD, which were highlighted as new candidate genes among Hispanic boys (Petta et al., 2016; Petta et al., 2017; Wattacheril et al., 2017; Abul-Husn et al., 2018). Furthermore, the hepatic expression of farnesoid X receptor (FXR) was reduced in both animal models and NAFLD patients, where hepatic FXR expression was reduced in nonalcoholic steatohepatitis (NASH) (Yang et al., 2010). A study showed that FXR splicing toward FXRα2 reduced hepatic lipid accumulation through the transcriptional program, which could greatly enhance the therapeutic effect by improving pharmacological targeting of select FXR agonists (Correia et al., 2015). This evidence suggests that FXR agonists could be a potential therapy for NAFLD. Moreover, the intra-abdominal adipose tissue (IAT) of severely obese adolescents with NAFLD has unique transcriptome differences, providing important molecular markers for identifying potential therapeutic targets for childhood NASH (Sheldon et al., 2016). In a previous study, reduced fatty acid desaturase 1 (FADS1) function was related to NAFLD and responded to treatment in children through FADS1 transcription levels (Nobili et al., 2018). In short, the study between RNA splicing and NAFLD in children was often conducted at the transcriptome level, while there was much more documented evidence about alternative RNA splicing in NAFLD in adults (Wu et al., 2021).
WD is characterized by a series of hepatic, neurological, and psychiatric symptoms, which result from impaired copper excretion at the bile location. It is an autosomal recessive disorder caused by a mutation in the ATP7B gene (Shah et al., 1997). Genetic prevalence is 3–4 times higher than clinical estimates, although the initial prevalence of 1:30,000–1:50,000 remains valid in at least Asia, the United States, and Europe (Sandahl et al., 2020). More than 70 splicing mutations have been reported at this genetic locus worldwide, including exon skipping and acceptor and donor splice site mutations (http://www.hgmd.cf.ac.uk/ac/gene.php?gene=ATP7B) (Lovicu et al., 2009; Zappu et al., 2012; Mameli et al., 2015; Stenson et al., 2017; Wang et al., 2018), suggesting that this locus is a hotspot of splicing mutation. Moreover, those splicing mutations of the ATP7B gene can be used as the diagnostic targets for WD.
Hepatitis B virus (HBV) and hepatitis C virus (HCV) are responsible for a major burden of viral hepatitis worldwide. The prevalence of chronic hepatitis B infection in children has been reduced due to improved hygiene measures, blood supply, and introduction of universal vaccination for this virus (Della Corte et al., 2016). AS regulation of HBV transcripts has been reported in vitro and in the liver of HBV-infected patients (Suzuki et al., 1989; Wu et al., 1991). In particular, AS could regulate the splicing of 3.5 KB HBV pregenomic RNA (pGRNA), which encodes either capsid or polymerase proteins to facilitate viral genome replication (Seeger and Mason, 2015).
The main HBV pGRNA splicing variant, single presplicing genomic RNA (SP1RNA), harbors a deletion of 1/3 of the viral genome and accounts for approximately 30% of the total HBV pGRNA in hosts, suggesting the importance of AS regulation in virus packaging, reverse transcription, and virus release (Terré et al., 1991; Soussan et al., 2008; Bayliss et al., 2013). In particular, host splicing factors, including SF1, hnRNPAB, PSF, LA, and some SRSFs, have been demonstrated to participate in the splicing regulation of HBV pGRNA (Soussan et al., 2008).
With the accurate screening of blood products and organ donors, the prevalence of hepatitis C infection has been significantly reduced, and vertical transmission is the main source of infection (Della Corte et al., 2016). A single nucleotide polymorphism (SNP) variant in the antizyme-inhibitor-1 (AZIN1) gene called AZIN1 SV2 (AZIN1 splice variant 2) leads to a novel alternative spliced isoform that modifies the fibrogenic potential of hepatic stellate cells (HSCs) in HCV cirrhotic livers (Huang et al., 2007; Paris et al., 2011). Previous research indicated that the spliceosome factor SART1 (squamous cell carcinoma antigen recognized by T cells) regulates HCV replication by altering its expression and splicing level (Lin et al., 2015).
Malignant liver tumors are rare in children, accounting for only 1% of all malignancies (Spector and Birch, 2012). While more than two-thirds of them are hepatoblastomas (HBs), 20% are hepatocellular carcinomas (HCCs). The latter section will discuss the relationship between AS and HB. Here, we only discuss AS in HCC, which is typically in older children or adolescents and is the major type of adult liver cancer (Crippa et al., 2017). At present, hepatocyte proliferation and HCC development are closely related to the transcriptional coactivator Yes-associated protein and its targeted Hippo pathway in animal models (Dong et al., 2007). Previous studies have shown that the mRNA expression of Yes-associated protein (YAP) target genes (CCNE1, CTGF, Cyr61) was increased in pediatric HCC, demonstrating an enrichment of YAP nuclear localization and its activity in moderately differentiated pediatric HCC (LaQuaglia et al., 2016). However, the relationship between AS and HCC in children has rarely been reported in comparison to adult patients.
BA is caused by bile duct occlusion or interruption from the hilum to the duodenum and is becoming the most common cholestatic liver disease leading to pediatric liver transplantation. The incidence of BA ranges from 1 in 5,000 cases to 1 in 19,000 cases, with higher rates in Asia than in European countries (Fawaz et al., 2017). A number of likely causal proteins of BA have been identified in previous studies, including FOXA2 (Tsai et al., 2015), CFC1 (Davit-Spraul et al., 2008), ZEB2 (Cui et al., 2011), ZIC3 (Ware et al., 2004), HNF1B (Shaalan et al., 2019), PKD1 L1 (Berauer et al., 2019), GPC1 (Cui et al., 2011), XPNPEP1 (Garcia-Barceló et al., 2010), ADD3 (Tsai et al., 2014), EFEMP1 (Chen et al., 2018), ARF6 (Ningappa et al., 2015), STIP1, and REV1 (Rajagopalan et al., 2020), without splicing mutations (Table 2). However, no genes have been identified as a definite cause of isolated BA cases so far (Girard and Panasyuk, 2019). Nevertheless, the top 10 upregulated loci identified by transcriptome approach from BA samples, including CSRNP1, IL6R, CPB2, TTR, TD O 2, SERPINC1, C6, DHTKD1, IGFBP1, and RDH16, might deepen our understanding of the transcriptional and post-transcriptional mechanisms among BA patients (Xiao et al., 2014).
Recently, long noncoding RNAH19 (lncRNAH19) was proposed to have a positive correlation with the BA-related severity of liver fibrosis (Xiao et al., 2019). Moreover, H19, a molecular sponge of the microRNA let-7 family, activates its downstream target, high mobility group member AT-hook 2 (HMGA2), during biliary tract proliferation, suggesting its crucial role in BA bile duct cell proliferation and cholestatic liver injury. Thus, lncRNAH19 may emerge as a valuable target for early diagnosis and the development of novel therapeutic procedures for BA patients.
Furthermore, recent studies have shown that the abnormal expression of long noncoding RNA Alu-mediated p21 transcription regulator (APTR) in the liver of BA infants may be pivotal for liver fibrosis in these patients (Makhmudi et al., 2020). Moreover, potential determinants of prognosis in Kasai portal enterostomy (KPE), such as phosphoenolpyruvate carboxykinase (PCK1) and matrix metalloproteinase-7 (MMP7), were determined by RNA sequencing data (Ramachandran et al., 2019). In particular, the abundance of MMP7 was higher in patients with failed jaundice clearance after KPE and in patients with end-stage liver disease (ESLD) than in the control group. In contrast, successful KPE treatment could induce PCK1 expression, and the abundance of PCK1 in patients with uncleared jaundice after KPE was repressed. Therefore, the abundance of MMP7 and PCK1 could be used as indicators for KPE outcome prediction and disease progression for clinicians.
Hepatoblastoma (HB) is the most common pediatric liver tumor, which typically occurs before the age of three and can be congenital (Trobaugh-Lotrario et al., 2013). The incidence of HBs has increased due to the greater numbers of premature births and infants with birth weights lower than 1,500 g (Feng et al., 2019). A β-catenin-encoding protein, CTNNB1, is the most frequently mutated HB gene, accounting for 50–90% of diagnosed HB cases (Crippa et al., 2017). Several genes, such as Spondin2, Edil3, Glypican 3, Osteopontin, and PEG10, were highly elevated, whereas Ficolin 3 was downregulated in human HCC and HB cases (Luo et al., 2006). However, several genes, including IGF2, fibronectin, DLK1, TGFb1, MALAT1, and MIG6, were overexpressed in HB versus HCC.
HB is genetically characterized by abnormal activation of the Wnt/β-catenin signaling pathway (Sha et al., 2019). Generally, extensive evidence has suggested that mutations in the β-catenin gene exon 3 are responsible for the activation of the Wnt/β-catenin signal transduction pathway in HB. Furthermore, the accumulation of β-catenin proteins resulted from increased translocations to the nucleus and cytoplasm and is positively correlated to cancer severity. Therefore, the abundance of β-catenin and target genes from its signaling pathway can be used as diagnostic and prognostic markers for pediatric liver tumors. In addition, several research groups have proposed the therapeutic effects of HB by specific inhibition of Wnt/β-catenin pathway, through a number of post-transcriptional measures such as short interfering miRNA, RNAs (siRNA), and bioactive small molecules. Hence, the Wnt/β-catenin signaling pathway is a valuable target for the development of therapeutic measures of HB (Koch et al., 1999; Takayasu et al., 2001; Koch et al., 2005; Cairo et al., 2008; Eichenmüller et al., 2014; Sumazin et al., 2017; Sha et al., 2019). However, HB has the lowest mutation burden among all known cancer types, and the genetic determinants of HB remain to be further investigated (Gröbner et al., 2018). Less than 5 mutations per hepatoblastoma were identified by studies using whole-exome sequencing, suggesting that this low mutation frequency of HB hindered the potential targets that are responsible for HB progression (Eichenmüller et al., 2014; Jia et al., 2014; Sumazin et al., 2017).
There are many types of liver diseases in children, but many of them are rare in the world population. To date, much less research has been conducted on the association between RNA splicing and liver diseases in children than in adults (Figure 1). Furthermore, specific disease types at the child stage have also been reported to have splicing regulation on their potential genomic loci. In this review article, we found that there are many studies that performed their research on pediatric NAFLD in comparison to adult cases. This might be due to the longer life span of this disease at the child stage, which will greatly impact their life (Draijer et al., 2019). It has been expected that more targeted chemical drugs, such as FXR agonists, can be developed based on splicing variants to treat NAFLD. Although there have been no randomized controlled trials (RCTs) in children, this may be a major area for subsequent exploration (Jia et al., 2014). Intriguingly, most splicing mutations reported thus far lack functional studies at the molecular level, including those identified in PFIC, Alagille syndrome, and WD. Therefore, an in-depth study should be carried out to verify their roles in the corresponding diseases, evaluate the potential of these targets for drug development, and establish a noninvasive early diagnosis method. Specifically, these splicing events could be controlled by their upstream regulators, which have been demonstrated in adult and animal studies (Wu et al., 2021). Moreover, BA and HB, which occur in infancy or young children, seriously impact the health of children at this stage. Therefore, the molecular mechanism of these splicing variants in pediatric liver diseases requires further investigation.
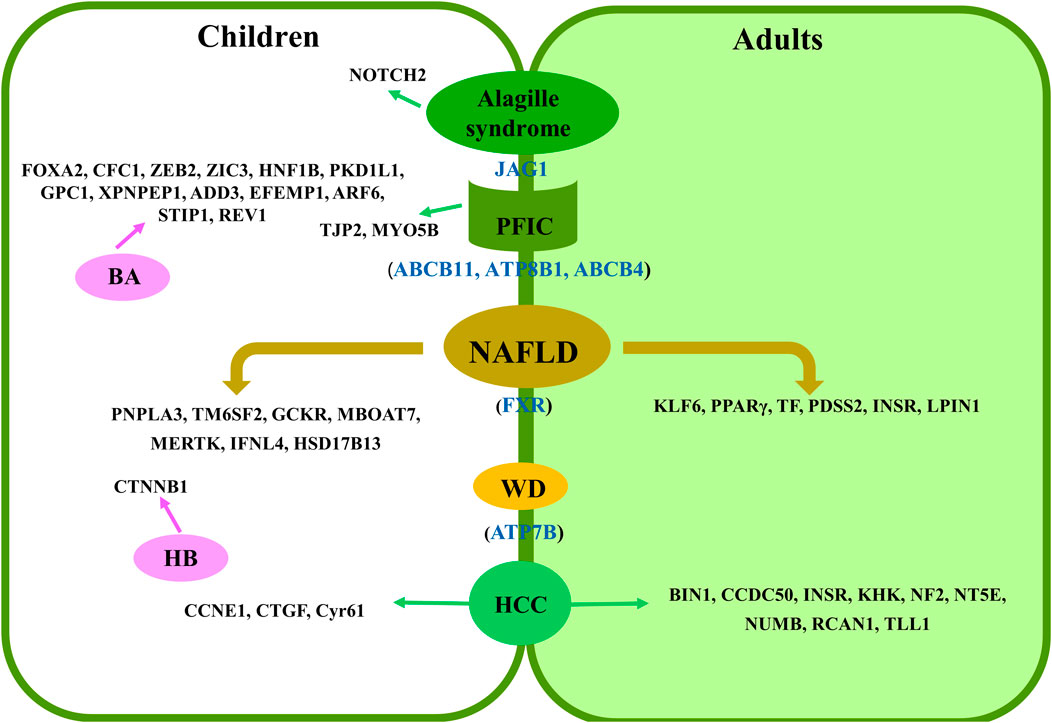
FIGURE 1. Summary of splicing-related loci in pediatric liver diseases. Shared loci by adults and children are presented as blue in color in brackets. Specific loci to children or adults are presented in the left and right panels, respectively.
SZ and M-XC designed the analysis and reviewed the content. J-LZ, Y-ZZ, and M-XC wrote the manuscript. S-SW, CC, and SZ critically reviewed and revised the manuscript.
This work was supported by the Science Technology and Innovation Committee of Shenzhen (2021N062-JCYJ20210324115408023), Natural Science Foundation of Jiangsu Province (SBK2020042924), and National Natural Science Foundation of China (NSFC32001452).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abul-Husn, N. S., Cheng, X., Li, A. H., Xin, Y., Schurmann, C., Stevis, P., et al. (2018). A Protein-TruncatingHSD17B13Variant and Protection from Chronic Liver Disease. N. Engl. J. Med. 378, 1096–1106. doi:10.1056/nejmoa1712191
Al-Hussaini, A., Lone, K., Bashir, M. S., Alrashidi, S., Fagih, M., Alanazi, A., et al. (2021). ATP8B1, ABCB11, and ABCB4 Genes Defects: Novel Mutations Associated with Cholestasis with Different Phenotypes and Outcomes. J. Pediatr. 236, 113–123. doi:10.1016/j.jpeds.2021.04.040
Bayliss, J., Lim, L., Thompson, A. J. V., Desmond, P., Angus, P., Locarnini, S., et al. (2013). Hepatitis B Virus Splicing Is Enhanced Prior to Development of Hepatocellular Carcinoma. J. Hepatol. 59, 1022–1028. doi:10.1016/j.jhep.2013.06.018
Berauer, J. P., Mezina, A. I., Okou, D. T., Sabo, A., Muzny, D. M., Gibbs, R. A., et al. (2019). Identification of Polycystic Kidney Disease 1 like 1 Gene Variants in Children with Biliary Atresia Splenic Malformation Syndrome. Hepatology 70, 899–910. doi:10.1002/hep.30515
Bull, L. N., and Thompson, R. J. (2018). Progressive Familial Intrahepatic Cholestasis. Clin. Liver Dis. 22, 657–669. doi:10.1016/j.cld.2018.06.003
Cairo, S., Armengol, C., De Reyniès, A., Wei, Y., Thomas, E., Renard, C.-A., et al. (2008). Hepatic Stem-like Phenotype and Interplay of Wnt/β-Catenin and Myc Signaling in Aggressive Childhood Liver Cancer. Cancer Cell 14, 471–484. doi:10.1016/j.ccr.2008.11.002
Chen, Y., Gilbert, M. A., Grochowski, C. M., Mceldrew, D., Llewellyn, J., Waisbourd-Zinman, O., et al. (2018). A Genome-wide Association Study Identifies a Susceptibility Locus for Biliary Atresia on 2p16.1 within the Gene EFEMP1. Plos Genet. 14, e1007532. doi:10.1371/journal.pgen.1007532
Chen, Y., Liu, X., Chen, S., Zhang, J., and Xu, C. (2020). Targeted Sequencing and RNA Assay Reveal a Noncanonical JAG1 Splicing Variant Causing Alagille Syndrome. Front. Genet. 10, 1363. doi:10.3389/fgene.2019.01363
Chillakuri, C. R., Sheppard, D., Ilagan, M. X. G., Holt, L. R., Abbott, F., Liang, S., et al. (2013). Structural Analysis Uncovers Lipid-Binding Properties of Notch Ligands. Cel Rep. 5, 861–867. doi:10.1016/j.celrep.2013.10.029
Correia, J. C., Massart, J., De Boer, J. F., Porsmyr-Palmertz, M., Martínez-Redondo, V., Agudelo, L. Z., et al. (2015). Bioenergetic Cues Shift FXR Splicing towards FXRα2 to Modulate Hepatic Lipolysis and Fatty Acid Metabolism. Mol. Metab. 4, 891–902. doi:10.1016/j.molmet.2015.09.005
Crippa, S., Ancey, P. B., Vazquez, J., Angelino, P., Rougemont, A. L., Guettier, C., et al. (2017). Mutant CTNNB 1 and Histological Heterogeneity Define Metabolic Subtypes of Hepatoblastoma. EMBO Mol. Med. 9, 1589–1604. doi:10.15252/emmm.201707814
Cui, S., Erlichman, J., Russo, P., Haber, B. A., and Matthews, R. P. (2011). Intrahepatic Biliary Anomalies in a Patient with Mowat-Wilson Syndrome Uncover a Role for the Zinc finger Homeobox Gene Zfhx1b in Vertebrate Biliary Development. J. Pediatr. Gastroenterol. Nutr. 52, 339–344. doi:10.1097/mpg.0b013e3181ff2e5b
Cui, S., Leyva–Vega, M., Tsai, E. A., EauClaire, S. F., Glessner, J. T., Hakonarson, H., et al. (2013). Evidence from Human and Zebrafish that GPC1 Is a Biliary Atresia Susceptibility Gene. Gastroenterology 144, 1107–1115. doi:10.1053/j.gastro.2013.01.022
Davit-Spraul, A., Baussan, C., Hermeziu, B., Bernard, O., and Jacquemin, E. (2008). CFC1 Gene Involvement in Biliary Atresia with Polysplenia Syndrome. J. Pediatr. Gastroenterol. Nutr. 46, 111–112. doi:10.1097/01.mpg.0000304465.60788.f4
Duriez, M., Mandouri, Y., Lekbaby, B., Wang, H., Schnuriger, A., Redelsperger, F., et al. (2017). Alternative Splicing of Hepatitis B Virus: A Novel Virus/Host Interaction Altering Liver Immunity. J. Hepatol. 67, 687–699.
del Río-Moreno, M., Alors-Pérez, E., González-Rubio, S., Ferrín, G., Reyes, O., Rodríguez-Perálvarez, M., et al. (2019). Dysregulation of the Splicing Machinery Is Associated to the Development of Nonalcoholic Fatty Liver Disease. J. Clin. Endocrinol. Metab. 104, 3389–3402. doi:10.1210/jc.2019-00021
Della Corte, C., Mosca, A., Vania, A., Alterio, A., Alisi, A., and Nobili, V. (2016). Pediatric Liver Diseases: Current Challenges and Future Perspectives. Expert Rev. Gastroenterol. Hepatol. 10, 255–265. doi:10.1586/17474124.2016.1129274
Dong, J., Feldmann, G., Huang, J., Wu, S., Zhang, N., Comerford, S. A., et al. (2007). Elucidation of a Universal Size-Control Mechanism in Drosophila and Mammals. Cell 130, 1120–1133. doi:10.1016/j.cell.2007.07.019
Dongiovanni, P., Meroni, M., Mancina, R. M., Baselli, G., Rametta, R., Pelusi, S., et al. (2018). Protein Phosphatase 1 Regulatory Subunit 3B Gene Variation Protects against Hepatic Fat Accumulation and Fibrosis in Individuals at High Risk of Nonalcoholic Fatty Liver Disease. Hepatol. Commun. 2, 666–675. doi:10.1002/hep4.1192
Draijer, L., Benninga, M., and Koot, B. (2019). Pediatric NAFLD: an Overview and Recent Developments in Diagnostics and Treatment. Expert Rev. Gastroenterol. Hepatol. 13, 447–461. doi:10.1080/17474124.2019.1595589
Eichenmüller, M., Trippel, F., Kreuder, M., Beck, A., Schwarzmayr, T., Häberle, B., et al. (2014). The Genomic Landscape of Hepatoblastoma and Their Progenies with HCC-like Features. J. Hepatol. 61, 1312–1320. doi:10.1016/j.jhep.2014.08.009
Elisofon, S. A., Magee, J. C., Ng, V. L., Horslen, S. P., Fioravanti, V., Economides, J., et al. (2020). Society of Pediatric Liver Transplantation: Current Registry Status 2011-2018. Pediatr. Transpl. 24, e13605. doi:10.1111/petr.13605
Fawaz, R., Baumann, U., Ekong, U., Fischler, B., Hadzic, N., Mack, C. L., et al. (2017). Guideline for the Evaluation of Cholestatic Jaundice in Infants: Joint Recommendations of the North American Society for Pediatric Gastroenterology, Hepatology, and Nutrition and the European Society for Pediatric Gastroenterology, Hepatology, and Nutrition. J. Pediatr. Gastroenterol. Nutr. 64, 154–168. doi:10.1097/mpg.0000000000001334
Feng, J., Polychronidis, G., Heger, U., Frongia, G., Mehrabi, A., and Hoffmann, K. (2019). Incidence Trends and Survival Prediction of Hepatoblastoma in Children: a Population-Based Study. Cancer Commun. 39, 62. doi:10.1186/s40880-019-0411-7
Gao, E., Hercun, J., Heller, T., and Vilarinho, S. (2021). Undiagnosed Liver Diseases. Transl Gastroenterol. Hepatol. 6, 28. doi:10.21037/tgh.2020.04.04
Garcia-Barceló, M.-M., Yeung, M.-Y., Miao, X.-P., Tang, C. S.-M., Chen, G., So, M.-T., et al. (2010). Genome-wide Association Study Identifies a Susceptibility Locus for Biliary Atresia on 10q24.2. Hum. Mol. Genet. 19, 2917–2925. doi:10.1093/hmg/ddq196
Gerhard, G. S., Legendre, C., Still, C. D., Chu, X., Petrick, A., and Distefano, J. K. (2018). Transcriptomic Profiling of Obesity-Related Nonalcoholic Steatohepatitis Reveals a Core Set of Fibrosis-specific Genes. J. Endocr. Soc. 2, 710–726. doi:10.1210/js.2018-00122
Girard, M., Lacaille, F., Verkarre, V., Mategot, R., Feldmann, G., Grodet, A., et al. (2014). MYO5B and Bile Salt export Pump Contribute to Cholestatic Liver Disorder in Microvillous Inclusion Disease. Hepatology 60, 301–310. doi:10.1002/hep.26974
Girard, M., and Panasyuk, G. (2019). Genetics in Biliary Atresia. Curr. Opin. Gastroenterol. 35, 73–81. doi:10.1097/mog.0000000000000509
Goldner, D., and Lavine, J. E. (2020). Nonalcoholic Fatty Liver Disease in Children: Unique Considerations and Challenges. Gastroenterology 158, 1967–1983. doi:10.1053/j.gastro.2020.01.048
González-Mariscal, L., Gallego-Gutiérrez, H., González-González, L., and Hernández-Guzmán, C. (2019). ZO-2 Is a Master Regulator of Gene Expression, Cell Proliferation, Cytoarchitecture, and Cell Size. Ijms 20, 4128. doi:10.3390/ijms20174128
Götze, T., Blessing, H., Grillhösl, C., Gerner, P., and Hoerning, A. (2015). Neonatal Cholestasis - Differential Diagnoses, Current Diagnostic Procedures, and Treatment. Front. Pediatr. 3, 43. doi:10.3389/fped.2015.00043
Gröbner, S. N., Worst, B. C., Worst, B. C., Weischenfeldt, J., Buchhalter, I., Kleinheinz, K., et al. (2018). The Landscape of Genomic Alterations across Childhood Cancers. Nature 555, 321–327. doi:10.1038/nature25480
Henkel, S. A., Squires, J. H., Ayers, M., Ganoza, A., Mckiernan, P., and Squires, J. E. (2019). Expanding Etiology of Progressive Familial Intrahepatic Cholestasis. Wjh 11, 450–463. doi:10.4254/wjh.v11.i5.450
Hoang, S. A., Oseini, A., Feaver, R. E., Cole, B. K., Asgharpour, A., Vincent, R., et al. (2019). Gene Expression Predicts Histological Severity and Reveals Distinct Molecular Profiles of Nonalcoholic Fatty Liver Disease. Sci. Rep. 9, 12541. doi:10.1038/s41598-019-48746-5
Huang, H., Shiffman, M. L., Friedman, S., Venkatesh, R., Bzowej, N., Abar, O. T., et al. (2007). A 7 Gene Signature Identifies the Risk of Developing Cirrhosis in Patients with Chronic Hepatitis C. Hepatology 46, 297–306. doi:10.1002/hep.21695
Jia, D., Dong, R., Jing, Y., Xu, D., Wang, Q., Chen, L., et al. (2014). Exome Sequencing of Hepatoblastoma Reveals Novel Mutations and Cancer Genes in the Wnt Pathway and Ubiquitin Ligase Complex. Hepatology 60, 1686–1696. doi:10.1002/hep.27243
Jimenez, M., Arechederra, M., Ávila, M. A., and Berasain, C. (2018). Splicing Alterations Contributing to Cancer Hallmarks in the Liver: central Role of Dedifferentiation and Genome Instability. Transl. Gastroenterol. Hepatol. 3, 84. doi:10.21037/tgh.2018.10.11
Keitel, V., Dröge, C., and Häussinger, D. (2019). Targeting FXR in Cholestasis. Handb Exp. Pharmacol. 256, 299–324. doi:10.1007/164_2019_231
Khabou, B., Siala-Sahnoun, O., Gargouri, L., Mkaouar-Rebai, E., Keskes, L., Hachicha, M., et al. (2016). In Silico investigation of the Impact of Synonymous Variants in ABCB4 Gene on mRNA Stability/structure, Splicing Accuracy and Codon Usage: Potential Contribution to PFIC3 Disease. Comput. Biol. Chem. 65, 103–109. doi:10.1016/j.compbiolchem.2016.10.008
Koch, A., Denkhaus, D., Albrecht, S., Leuschner, I., Von Schweinitz, D., and Pietsch, T. (1999). Childhood Hepatoblastomas Frequently Carry a Mutated Degradation Targeting Box of the Beta-Catenin Gene. Cancer Res. 59, 269–273.
Koch, A., Waha, A., Hartmann, W., Hrychyk, A., Schüller, U., Waha, A., et al. (2005). Elevated Expression of Wnt Antagonists Is a Common Event in Hepatoblastomas. Clin. Cancer Res. 11, 4295–4304. doi:10.1158/1078-0432.ccr-04-1162
Kopan, R., and Ilagan, M. X. G. (2009). The Canonical Notch Signaling Pathway: Unfolding the Activation Mechanism. Cell 137, 216–233. doi:10.1016/j.cell.2009.03.045
Kozlitina, J., Smagris, E., Stender, S., Nordestgaard, B. G., Zhou, H. H., Tybjærg-Hansen, A., et al. (2014). Exome-wide Association Study Identifies a TM6SF2 Variant that Confers Susceptibility to Nonalcoholic Fatty Liver Disease. Nat. Genet. 46, 352–356. doi:10.1038/ng.2901
Laquaglia, M. J., Grijalva, J. L., Mueller, K. A., Perez-Atayde, A. R., Kim, H. B., Sadri-Vakili, G., et al. (2016). YAP Subcellular Localization and Hippo Pathway Transcriptome Analysis in Pediatric Hepatocellular Carcinoma. Sci. Rep. 6, 30238. doi:10.1038/srep30238
Li, J., Hua, W., Ji, C., Rui, J., Zhao, Y., Xie, C., et al. (2020). Effect of the Patatin-like Phospholipase Domain Containing 3 Gene (PNPLA3) I148M Polymorphism on the Risk and Severity of Nonalcoholic Fatty Liver Disease and Metabolic Syndromes: A Meta-Analysis of Paediatric and Adolescent Individuals. Pediatr. Obes. 15, e12615. doi:10.1111/ijpo.12615
Li, L., Krantz, I. D., Deng, Y., Genin, A., Banta, A. B., Collins, C. C., et al. (1997). Alagille Syndrome Is Caused by Mutations in Human Jagged1, Which Encodes a Ligand for Notch1. Nat. Genet. 16, 243–251. doi:10.1038/ng0797-243
Lin, W., Zhu, C., Hong, J., Zhao, L., Jilg, N., Fusco, D. N., et al. (2015). The Spliceosome Factor SART1 Exerts its Anti-HCV Action through mRNA Splicing. J. Hepatol. 62, 1024–1032. doi:10.1016/j.jhep.2014.11.038
Lindsell, C. E., Shawber, C. J., Boulter, J., and Weinmaster, G. (1995). Jagged: A Mammalian Ligand that Activates Notch1. Cell 80, 909–917. doi:10.1016/0092-8674(95)90294-5
Liu, L. Y., Wang, Z. L., Wang, X. H., Zhu, Q. R., and Wang, J. S. (2010). ABCB11 Gene Mutations in Chinese Children with Progressive Intrahepatic Cholestasis and Low Gamma Glutamyltransferase. Liver Int. 30, 809–815. doi:10.1111/j.1478-3231.2009.02112.x
Lovicu, M., Lepori, M. B., Incollu, S., Dessì, V., Zappu, A., Iorio, R., et al. (2009). RNA Analysis of Consensus Sequence Splicing Mutations: Implications for the Diagnosis of Wilson Disease. Genet. Test. Mol. Biomarkers 13, 185–191. doi:10.1089/gtmb.2008.0089
Luo, J.-H., Ren, B., Keryanov, S., Tseng, G. C., Rao, U. N. M., Monga, S. P., et al. (2006). Transcriptomic and Genomic Analysis of Human Hepatocellular Carcinomas and Hepatoblastomas. Hepatology 44, 1012–1024. doi:10.1002/hep.21328
Makhmudi, A., Supanji, R., Putra, B. P., and Gunadi, (2020). The Effect of APTR, Fn14 and CD133 Expressions on Liver Fibrosis in Biliary Atresia Patients. Pediatr. Surg. Int. 36, 75–79. doi:10.1007/s00383-019-04582-2
Mameli, E., Lepori, M. B., Chiappe, F., Ranucci, G., Di Dato, F., Iorio, R., et al. (2015). Wilson's Disease Caused by Alternative Splicing and Alu Exonization Due to a Homozygous 3039-bp Deletion Spanning from Intron 1 to Exon 2 of the ATP7B Gene. Gene 569, 276–279. doi:10.1016/j.gene.2015.05.067
Mancina, R. M., Dongiovanni, P., Petta, S., Pingitore, P., Meroni, M., Rametta, R., et al. (2016). The MBOAT7-TMC4 Variant Rs641738 Increases Risk of Nonalcoholic Fatty Liver Disease in Individuals of European Descent. Gastroenterology 150, 1219–1230. doi:10.1053/j.gastro.2016.01.032
Mcdaniell, R., Warthen, D. M., Sanchez-Lara, P. A., Pai, A., Krantz, I. D., Piccoli, D. A., et al. (2006). NOTCH2 Mutations Cause Alagille Syndrome, a Heterogeneous Disorder of the Notch Signaling Pathway. Am. J. Hum. Genet. 79, 169–173. doi:10.1086/505332
Montes, M., Sanford, B. L., Comiskey, D. F., and Chandler, D. S. (2019). RNA Splicing and Disease: Animal Models to Therapies. Trends Genet. 35, 68–87. doi:10.1016/j.tig.2018.10.002
Nikeghbalian, S., Malekhosseini, S. A., Kazemi, K., Arasteh, P., Eghlimi, H., Shamsaeefar, A., et al. (2021). The Largest Single Center Report on Pediatric Liver Transplantation. Ann. Surg. 273, e70–e72. doi:10.1097/sla.0000000000004047
Ningappa, M., So, J., Glessner, J., Ashokkumar, C., Ranganathan, S., Min, J., et al. (2015). The Role of ARF6 in Biliary Atresia. PLoS One 10, e0138381. doi:10.1371/journal.pone.0138381
Nobili, V., Alisi, A., Liu, Z., Liang, T., Crudele, A., Raponi, M., et al. (2018). In a Pilot Study, Reduced Fatty Acid Desaturase 1 Function Was Associated with Nonalcoholic Fatty Liver Disease and Response to Treatment in Children. Pediatr. Res. 84, 696–703. doi:10.1038/s41390-018-0132-7
Nobili, V., Alisi, A., Valenti, L., Miele, L., Feldstein, A. E., and Alkhouri, N. (2019). NAFLD in Children: New Genes, New Diagnostic Modalities and New Drugs. Nat. Rev. Gastroenterol. Hepatol. 16, 517–530. doi:10.1038/s41575-019-0169-z
Overeem, A. W., Li, Q., Qiu, Y. L., Cartón‐García, F., Leng, C., Klappe, K., et al. (2020). A Molecular Mechanism Underlying Genotype‐Specific Intrahepatic Cholestasis Resulting from MYO5B Mutations. Hepatology 72, 213–229. doi:10.1002/hep.31002
Paris, A. J., Snapir, Z., Christopherson, C. D., Kwok, S. Y., Lee, U. E., Ghiassi-Nejad, Z., et al. (2011). A Polymorphism that Delays Fibrosis in Hepatitis C Promotes Alternative Splicing of AZIN1, Reducing Fibrogenesis. Hepatology 54, 2198–2207. doi:10.1002/hep.24608
Paulusma, C. C., Folmer, D. E., Ho-Mok, K. S., de Waart, D. R., Hilarius, P. M., Verhoeven, A. J., et al. (2008). ATP8B1 Requires an Accessory Protein for Endoplasmic Reticulum Exit and Plasma Membrane Lipid Flippase Activity. Hepatology 47, 268–278. doi:10.1002/hep.21950
Petta, S., Valenti, L., Marra, F., Grimaudo, S., Tripodo, C., Bugianesi, E., et al. (2016). MERTK Rs4374383 Polymorphism Affects the Severity of Fibrosis in Non-alcoholic Fatty Liver Disease. J. Hepatol. 64, 682–690. doi:10.1016/j.jhep.2015.10.016
Petta, S., Valenti, L., Tuttolomondo, A., Dongiovanni, P., Pipitone, R. M., Cammà, C., et al. (2017). Interferon Lambda 4 Rs368234815 TT>δG Variant Is Associated with Liver Damage in Patients with Nonalcoholic Fatty Liver Disease. Hepatology 66, 1885–1893. doi:10.1002/hep.29395
Pietrobattista, A., Veraldi, S., Candusso, M., Basso, M. S., Liccardo, D., Della Corte, C., et al. (2020). The Contribution of Plasma Oxysterols in the Challenging Diagnostic Work-Up of Infantile Cholestasis. Clinica Chim. Acta 507, 181–186. doi:10.1016/j.cca.2020.04.028
Pihlajamäki, J., Lerin, C., Itkonen, P., Boes, T., Floss, T., Schroeder, J., et al. (2011). Expression of the Splicing Factor Gene SFRS10 Is Reduced in Human Obesity and Contributes to Enhanced Lipogenesis. Cel Metab. 14, 208–218. doi:10.1016/j.cmet.2011.06.007
Qiu, Y.-L., Gong, J.-Y., Feng, J.-Y., Wang, R.-X., Han, J., Liu, T., et al. (2017). Defects in Myosin VB Are Associated with a Spectrum of Previously Undiagnosed Low γ-glutamyltransferase Cholestasis. Hepatology 65, 1655–1669. doi:10.1002/hep.29020
Rahman, M. A., Krainer, A. R., and Abdel-Wahab, O. (2020). SnapShot: Splicing Alterations in Cancer. Cell 180, 208. doi:10.1016/j.cell.2019.12.011
Rajagopalan, R., Tsai, E. A., Grochowski, C. M., Kelly, S. M., Loomes, K. M., Spinner, N. B., et al. (2020). Exome Sequencing in Individuals with Isolated Biliary Atresia. Sci. Rep. 10, 2709. doi:10.1038/s41598-020-59379-4
Ramachandran, P., Balamurali, D., Peter, J. J., Kumar, M. M., Safwan, M., Vij, M., et al. (2019). RNA-seq Reveals Outcome-specific Gene Expression of MMP7 and PCK1 in Biliary Atresia. Mol. Biol. Rep. 46, 5123–5130. doi:10.1007/s11033-019-04969-3
Sambrotta, M., Strautnieks, S., Strautnieks, S., Papouli, E., Rushton, P., Clark, B. E., et al. (2014). Mutations in TJP2 Cause Progressive Cholestatic Liver Disease. Nat. Genet. 46, 326–328. doi:10.1038/ng.2918
Sambrotta, M., and Thompson, R. J. (2015). Mutations inTJP2, Encoding Zona Occludens 2, and Liver Disease. Tissue Barriers 3, e1026537. doi:10.1080/21688370.2015.1026537
Sandahl, T. D., Laursen, T. L., Munk, D. E., Vilstrup, H., Weiss, K. H., and Ott, P. (2020). The Prevalence of Wilson's Disease: An Update. Hepatology 71, 722–732. doi:10.1002/hep.30911
Santoro, N., Zhang, C. K., Zhao, H., Pakstis, A. J., Kim, G., Kursawe, R., et al. (2012). Variant in the Glucokinase Regulatory Protein (GCKR) Gene Is Associated with Fatty Liver in Obese Children and Adolescents. Hepatology 55, 781–789. doi:10.1002/hep.24806
Seeger, C., and Mason, W. S. (2015). Molecular Biology of Hepatitis B Virus Infection. Virology 479-480, 672–686. doi:10.1016/j.virol.2015.02.031
Sha, Y. L., Liu, S., Yan, W. W., and Dong, B. (2019). Wnt/β-catenin Signaling as a Useful Therapeutic Target in Hepatoblastoma. Biosci. Rep. 39, BSR20192466. doi:10.1042/BSR20192466
Shaalan, U. F., Ibrahim, N. L., Ehsan, N. A., Sultan, M. M., Naser, G. M., and Abd El-Fatah, M. O. (2019). Reduced Immunohistochemical Expression of Hnf1β and FoxA2 in Liver Tissue Can Discriminate between Biliary Atresia and Other Causes of Neonatal Cholestasis. Appl. Immunohistochem. Mol. Morphol. 27, e32–e38. doi:10.1097/pai.0000000000000638
Shah, A. B., Chernov, I., Zhang, H. T., Ross, B. M., Das, K., Lutsenko, S., et al. (1997). Identification and Analysis of Mutations in the Wilson Disease Gene (ATP7B): Population Frequencies, Genotype-Phenotype Correlation, and Functional Analyses. Am. J. Hum. Genet. 61, 317–328. doi:10.1086/514864
Sharma, A., Poddar, U., Agnihotry, S., Phadke, S. R., Yachha, S. K., and Aggarwal, R. (2018). Spectrum of Genomic Variations in Indian Patients with Progressive Familial Intrahepatic Cholestasis. BMC Gastroenterol. 18, 107. doi:10.1186/s12876-018-0835-6
Sheldon, R. D., Kanosky, K. M., Wells, K. D., Miles, L., Perfield, J. W., Xanthakos, S., et al. (2016). Transcriptomic Differences in Intra-abdominal Adipose Tissue in Extremely Obese Adolescents with Different Stages of NAFLD. Physiol. Genomics 48, 897–911. doi:10.1152/physiolgenomics.00020.2016
Soussan, P., Pol, J., Garreau, F., Schneider, V., Pendeven, C. L., Nalpas, B., et al. (2008). Expression of Defective Hepatitis B Virus Particles Derived from Singly Spliced RNA Is Related to Liver Disease. J. Infect. Dis. 198, 218–225. doi:10.1086/589623
Spector, L. G., and Birch, J. (2012). The Epidemiology of Hepatoblastoma. Pediatr. Blood Cancer 59, 776–779. doi:10.1002/pbc.24215
Stenson, P. D., Mort, M., Ball, E. V., Evans, K., Hayden, M., Heywood, S., et al. (2017). The Human Gene Mutation Database: towards a Comprehensive Repository of Inherited Mutation Data for Medical Research, Genetic Diagnosis and Next-Generation Sequencing Studies. Hum. Genet. 136, 665–677. doi:10.1007/s00439-017-1779-6
Sumazin, P., Chen, Y., Treviño, L. R., Sarabia, S. F., Hampton, O. A., Patel, K., et al. (2017). Genomic Analysis of Hepatoblastoma Identifies Distinct Molecular and Prognostic Subgroups. Hepatology 65, 104–121. doi:10.1002/hep.28888
Suzuki, H., Kumar, S. A., Shuai, S., Diaz-Navarro, A., Gutierrez-Fernandez, A., De Antonellis, P., et al. (2019). Recurrent Noncoding U1 snRNA Mutations Drive Cryptic Splicing in SHH Medulloblastoma. Nature 574, 707–711. doi:10.1038/s41586-019-1650-0
Suzuki, T., Masui, N., Kajino, K., Saito, I., and Miyamura, T. (1989). Detection and Mapping of Spliced RNA from a Human Hepatoma Cell Line Transfected with the Hepatitis B Virus Genome. Proc. Natl. Acad. Sci. 86, 8422–8426. doi:10.1073/pnas.86.21.8422
Takayasu, H., Horie, H., Hiyama, E., Matsunaga, T., Hayashi, Y., Watanabe, Y., et al. (2001). Frequent Deletions and Mutations of the Beta-Catenin Gene Are Associated with Overexpression of Cyclin D1 and Fibronectin and Poorly Differentiated Histology in Childhood Hepatoblastoma. Clin. Cancer Res. 7, 901–908.
Terré, S., Petit, M. A., and Bréchot, C. (1991). Defective Hepatitis B Virus Particles Are Generated by Packaging and Reverse Transcription of Spliced Viral RNAs In Vivo. J. Virol. 65, 5539–5543. doi:10.1128/jvi.65.10.5539-5543.1991
Trobaugh-Lotrario, A. D., Chaiyachati, B. H., Meyers, R. L., Häberle, B., Tomlinson, G. E., Katzenstein, H. M., et al. (2013). Outcomes for Patients with Congenital Hepatoblastoma. Pediatr. Blood Cancer 60, 1817–1825. doi:10.1002/pbc.24655
Tsai, E. A., Grochowski, C. M., Falsey, A. M., Rajagopalan, R., Wendel, D., Devoto, M., et al. (2015). Heterozygous Deletion ofFOXA2Segregates with Disease in a Family with Heterotaxy, Panhypopituitarism, and Biliary Atresia. Hum. Mutat. 36, 631–637. doi:10.1002/humu.22786
Tsai, E. A., Grochowski, C. M., Loomes, K. M., Bessho, K., Hakonarson, H., Bezerra, J. A., et al. (2014). Replication of a GWAS Signal in a Caucasian Population Implicates ADD3 in Susceptibility to Biliary Atresia. Hum. Genet. 133, 235–243. doi:10.1007/s00439-013-1368-2
Valenti, L., Alisi, A., Galmozzi, E., Bartuli, A., Del Menico, B., Alterio, A., et al. (2010). I148M Patatin-like Phospholipase Domain-Containing 3 Gene Variant and Severity of Pediatric Nonalcoholic Fatty Liver Disease. Hepatology 52, 1274–1280. doi:10.1002/hep.23823
Van Der Woerd, W. L., Mulder, J., Pagani, F., Beuers, U., Houwen, R. H. J., and Van De Graaf, S. F. J. (2015). Analysis of Aberrant Pre-messenger RNA Splicing Resulting from Mutations inATP8B1and Efficientin Vitrorescue by Adapted U1 Small Nuclear RNA. Hepatology 61, 1382–1391. doi:10.1002/hep.27620
van IJzendoorn, S. C. D., Li, Q., Qiu, Y. l., Wang, J. S., and Overeem, A. W. (2020). Unequal Effects of Myosin 5B Mutations in Liver and Intestine Determine the Clinical Presentation of Low‐Gamma‐Glutamyltransferase Cholestasis. Hepatology 72, 1461–1468. doi:10.1002/hep.31430
Vitale, G., Gitto, S., Vukotic, R., Raimondi, F., and Andreone, P. (2019). Familial Intrahepatic Cholestasis: New and Wide Perspectives. Dig. Liver Dis. 51, 922–933. doi:10.1016/j.dld.2019.04.013
Vos, T., Allen, C., Arora, M., Barber, R. M., Bhutta, Z. A., Brown, A., et al. (2016). Global, Regional, and National Incidence, Prevalence, and Years Lived with Disability for 310 Diseases and Injuries, 1990-2015: a Systematic Analysis for the Global Burden of Disease Study 2015. Lancet 388, 1545–1602. doi:10.1016/S0140-6736(16)31678-6
Wang, C., Zhou, W., Huang, Y., Yin, H., Jin, Y., Jia, Z., et al. (2018). Presumed Missense and Synonymous Mutations in ATP7B Gene Cause Exon Skipping in Wilson Disease. Liver Int. 38, 1504–1513. doi:10.1111/liv.13754
Wang, H., Lekbaby, B., Fares, N., Augustin, J., Attout, T., Schnuriger, A., et al. (2019). Alteration of Splicing Factors' Expression during Liver Disease Progression: Impact on Hepatocellular Carcinoma Outcome. Hepatol. Int. 13, 454–467. doi:10.1007/s12072-019-09950-7
Ware, S. M., Peng, J., Zhu, L., Fernbach, S., Colicos, S., Casey, B., et al. (2004). Identification and Functional Analysis of ZIC3 Mutations in Heterotaxy and Related Congenital Heart Defects. Am. J. Hum. Genet. 74, 93–105. doi:10.1086/380998
Wattacheril, J., Lavine, J. E., Chalasani, N. P., Guo, X., Kwon, S., Schwimmer, J., et al. (2017). Genome-Wide Associations Related to Hepatic Histology in Nonalcoholic Fatty Liver Disease in Hispanic Boys. J. Pediatr. 190, 100–107. doi:10.1016/j.jpeds.2017.08.004
Webster, N. J. G. (2017). Alternative RNA Splicing in the Pathogenesis of Liver Disease. Front. Endocrinol. 8, 133. doi:10.3389/fendo.2017.00133
Wu, H. L., Chen, P. J., Tu, S. J., Lin, M. H., Lai, M. Y., and Chen, D. S. (1991). Characterization and Genetic Analysis of Alternatively Spliced Transcripts of Hepatitis B Virus in Infected Human Liver Tissues and Transfected HepG2 Cells. J. Virol. 65, 1680–1686. doi:10.1128/jvi.65.4.1680-1686.1991
Wu, P., Zhang, M., and Webster, N. J. G. (2021). Alternative RNA Splicing in Fatty Liver Disease. Front. Endocrinol. 12, 613213. doi:10.3389/fendo.2021.613213
Xiao, J., Xia, S.-y., Xia, Y., Xia, Q., and Wang, X.-r. (2014). Transcriptome Profiling of Biliary Atresia from New Born Infants by Deep Sequencing. Mol. Biol. Rep. 41, 8063–8069. doi:10.1007/s11033-014-3704-6
Xiao, Y., Liu, R., Li, X., Gurley, E. C., Hylemon, P. B., Lu, Y., et al. (2019). Long Noncoding RNA H19 Contributes to Cholangiocyte Proliferation and Cholestatic Liver Fibrosis in Biliary Atresia. Hepatology 70, 1658–1673. doi:10.1002/hep.30698
Yang, Z.-X., Shen, W., and Sun, H. (2010). Effects of Nuclear Receptor FXR on the Regulation of Liver Lipid Metabolism in Patients with Non-alcoholic Fatty Liver Disease. Hepatol. Int. 4, 741–748. doi:10.1007/s12072-010-9202-6
Zappu, A., Lepori, M. B., Incollu, S., Dessì, V., Noli, M. C., Mameli, E., et al. (2012). Feasibility of RNA Studies on Illegitimate Transcription for Molecular Characterization of Splicing Mutations in the ATP7B Gene: a Case Report. Mol. Cell Probes 26, 63–65. doi:10.1016/j.mcp.2011.10.002
Zhang, J., Liu, L. L., Gong, J. Y., Hao, C. Z., Qiu, Y. L., Lu, Y., et al. (2020). TJP2 Hepatobiliary Disorders: Novel Variants and Clinical Diversity. Hum. Mutat. 41, 502–511. doi:10.1002/humu.23947
Keywords: alternative splicing, children, liver disease, non-alcoholic fatty liver disease, RNA sequencing, splicing-related protein
Citation: Zhou J-L, Zhao Y-Z, Wang S-S, Chen M-X, Zhou S and Chen C (2021) RNA Splicing: A Versatile Regulatory Mechanism in Pediatric Liver Diseases. Front. Mol. Biosci. 8:725308. doi: 10.3389/fmolb.2021.725308
Received: 15 June 2021; Accepted: 23 August 2021;
Published: 28 September 2021.
Edited by:
Marija Buljan, Swiss Federal Laboratories for Materials Science and Technology, SwitzerlandReviewed by:
Marta Adinolfi, European Institute of Oncology (IEO), ItalyCopyright © 2021 Zhou, Zhao, Wang, Chen, Zhou and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mo-Xian Chen, Y214MjAwOTkyMDczNEBnbWFpbC5jb20=; Shaoming Zhou, emhvdXNtMTVkQGFsaXl1bi5jb20=; Chen Chen, Y2luZHlfY2hlbmNoZW5AMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.