- 1Laboratory of Biochemistry and Molecular Biology Research, Department of Clinical Laboratory, Fujian Medical University Cancer Hospital, Fuzhou, China
- 2Department of Pathology, Fujian Medical University Cancer Hospital, Fuzhou, China
Background: Current biomarkers for nasopharyngeal carcinoma (NPC) are less effective for early diagnosis and prognosis. The basic leucine zipper ATF-like transcription factor 2 (BATF2) gene has been shown to have a tight association with the pathogenesis of various malignancies but received scant attention in NPC research. We aimed to assess the performances of circulating and tissue BATF2 in the diagnosis and prognosis of NPC.
Materials and Methods: Immunohistochemistry (IHC) microarrays were performed to quantitate the BATF2 protein expression in NPC tissues. The relationships of BATF2 protein expression with clinicopathological characteristics and NPC prognosis were assessed. BATF2 mRNA expressions in serum and serum-derived exosomes were determined using quantitative reverse transcription-polymerase chain reaction (qRT-PCR) assay.
Results: The IHC microarrays revealed a predominant nuclear expression of BATF2 in NPC cells. The Kaplan-Meier survival analysis showed that BATF2-positive NPC patients enjoyed longer overall survival than BATF2-negative patients. NPC patients with serum and exosomal BATF2 mRNA expressions made up 51.47 and 48.52% of all patients, respectively. The AUCs of serum and exosomal BATF2 mRNA expressions in discriminating NPC from healthy controls were 0.9409 and 0.8983. Patients who had received radiochemotherapy exhibited higher serum and exosomal BATF2 mRNA expressions versus the levels at baseline as well as those detected in recurrent patients.
Conclusion: BATF2 is expressed cancerous tissues, serum, and serum-derived exosomes in NPC patients. Circulating and tissue BATF2 can serve as a multipurpose biomarker capable of the diagnosis, prognosis prediction, efficacy evaluation, and recurrence monitoring in NPC.
Introduction
Nasopharyngeal carcinoma (NPC) is a head and neck malignancy originating from nasopharyngeal mucosa (Lee et al., 2019), with an incidence rate of 20–30 per 100 000 people/year (Li et al., 2011). The latest epidemiological report of NPC in China showed that the high-risk areas of NPC, where the incidence and mortality remained high in recent years, mainly included Guangdong, Guangxi, Hunan, Fujian, and Jiangxi provinces and Hong Kong (Li et al., 2011; Trejaut et al., 2011). The 10-year survival of patients is satisfactory, of 49.5–66% (Huang et al., 2015). However, the failure of early diagnosis due to concealed nasopharyngeal anatomy and atypical symptoms of early NPC remains a significant clinical concern (Ng et al., 2011). Meanwhile, nearly 50% of patients cannot achieve complete remission due to rapid tumor growth, local recurrence, or distant metastasis (Ng et al., 2011). The mechanisms for NPC carcinogenesis include a network of oncogenes and tumor-suppressor genes and the involvement of various signalings that are so complicated that not yet been fully explored (Dai et al., 2016). The known risk factors incorporate Epstein-Barr virus infection, genetic susceptibility, environmental factors, and dietary habits (Chua et al., 2016; Lin et al., 2019). At present, a biomarker for predicting NPC occurrence and recurrence and monitoring its development has not been reported (Liu et al., 2015) but will make a huge difference.
The BATF2 gene encoding the BATF2 protein is a newly discovered tumor-suppressor gene isolated using subtractive hybridization. It is a suppressor of AP-1 regulated by interferon (SARI) or ATF-like 2 (Su et al., 2008). The BATF2 gene maps on chromosome 11q12-11q13, and the full-length ORF shows three exons, encoding 274 amino acid residues of the BATF2 protein (Su et al., 2008). BATF2 has been reported as a tumor suppressor gene in various malignant tumors, such as lung cancer, prostate cancer, colorectal carcinoma, hepatocellular carcinoma, and nasopharyngeal carcinoma, etc (Dash et al., 2010; Huang et al., 2011; Liu et al., 2011; Ma et al., 2011; Wang et al., 2012; Chen et al., 2015; Guler et al., 2015). Aberrant BATF2 expression in prostate cancer is significantly correlated with serum PSA level, clinical stage, and distant metastasis (Chen et al., 2015). Low BATF2 expression may increase the risk of hepatocellular carcinoma (HCC) development and has a significant association with a poor prognosis (Ma et al., 2011). The same result was reported in non-small cell lung cancer (NSCLC). BATF2 deficiency promotes epithelial-mesenchymal transition (EMT), resulting in invasion and metastasis of lung adenocarcinoma cells (Wang et al., 2012). A lower BATF2 mRNA expression was detectable in serum of patients with chronic myeloid leukemia (CML) compared to healthy controls, and BATF2 downregulation-induced BCR-ABL inhibition was critical in the occurrence and development of CML (Huang et al., 2011). BATF2-negative colorectal cancer patients exhibited poorly differentiated cancer cells alongside deeper invasion, higher TNM stage, and shorter survival post-surgery versus BATF2-positive colorectal cancer cases (Liu et al., 2011). BATF2 downregulated the expressions of hypoxia-inducible factor (HIF-1a) and vascular endothelial growth factor via targeting ceruloplasmin, thus inhibiting angiogenesis and tumor growth (Liu et al., 2011). Therefore, BATF2 has been considered a prognostic indicator and a potential target for gene therapy for various cancers.
Exosomes are a type of membrane vesicles released into the extracellular matrix after fusion of multivesicular bodies with cell membranes. They are nanosized (30–150 nm) particles widely distributed in body fluids (Zeringer et al., 2015). Exosomes carry lipids, nucleic acids (such as DNA and RNA), proteins, and other components (Melo et al., 2015; He et al., 2018) and shuttle between cells, acting as important mediators of cell communication. Exosomal biomarker genes have been found a promising tool for early diagnosis and prognosis prediction in cancers (Urbanelli et al., 2015; De Rubis et al., 2019). Our previous publications have reported that serum exosomal LDHC overexpression can serve as a multipurpose biomarker for early diagnosis, efficacy assessment, and recurrence monitoring in breast cancer (BC) patients (Cui et al., 2020). In this study, we aimed to assess BATF2 protein expression patterns in NPC patients using IHC microarrays and the performances of circulating and tissue BATF2 in diagnosis, prognosis, and evaluation of therapeutic efficacy in NPC.
Materials and Methods
Clinical Data
BATF2 protein expressions in 129 NPC patients (129 tissue samples) diagnosed between January 13, 2010, and October 9, 2011, were quantified using tissue microarray (HNasN129Su01; SHANGHAI OUTDO BIOTECH). All specimens were histopathologically diagnosed. None of the patients had received radiotherapy or chemotherapy before surgical procedures. Besides, 130 serum samples and four biopsy samples of NPC tissues were collected from patients hospitalized at Fujian Cancer Hospital between January 2018 and June 2020. Fifty serum samples from healthy people were used as controls. All procedures were approved by the Ethics Committee of Fujian Cancer Hospital (Approval No. SQ2017-010-01) and written informed consent was obtained from each patient.
IHC Microarray
The NPC microarray (HNasN129Su01) was purchased for IHC analysis of BATF2 protein expressions in NPC patients. Briefly, paraffin-embedded tissue blocks in the NPC microarray were stored overnight at 65°C, dewaxed in xylene, and rehydrated in a graded ethanol series. Endogenous peroxidase activity and nonspecific binding sites were blocked by incubating the tissue blocks in the H2O2 solution for 10 min at 37°C. Antigens were retrieved by incubation with boiling (100–120°C) citrate buffer for 3 min in a high-pressure cooker. The tissue sections were incubated in the polyclonal rabbit anti-human BATF2 antibody (Abcam, Catalog No. ab204510; 1:50) for 90 min at 37°C, washed in PBS, and incubated with the secondary antibody for 30 min at room temperature. The sections were stained with DAB for 2 min, washed in distilled water, and counterstained with hematoxylin. PBS was used instead of the primary antibody as a negative control. The samples were interpreted by two experienced pathologists. Based on the scoring criteria of our laboratory (Cui et al., 2020), the staining intensity of the NPC sections was scored based on cell color: 0 points, unstained; 1 point, light brown; 2 points, brown; 3 points, dark brown. The percentage of BATF2-positive staining was scored: 0 points, < 5%; 1 point, 5%–25%; 2 points, 25%–50%; 3 points, >50%. The final score of BATF2 expression was calculated by multiplying the percentage of BATF2-positive staining by staining intensity scores: 0 points, −; 1-2 points, +; 3-5 points, ++; 6-9 points, +++; and −/+ was regarded as low expression and ++ to +++ high expression.
Isolation and Identification of Exosomes Derived From Serum of NPC Patients
Serum-derived exosomes were extracted with an exoRNeasy Serum/Plasma Midi Kit (QIAGEN, Catalog No.77044) according to the manufacturer’s protocol (www.qiagen.com/hb-1179). The blood samples were centrifuged at 16,000 × g for 10 min at 4°C to remove particles larger than 0.8 μm. 500 μL of serum was collected and mixed with XBP at the ratio of 1:1. The serum/XBP mix was added onto the exoEasy spin columns and centrifuged at 500 × g for 1 min. The mix was added with 3.5 ml of XWP buffer and centrifuged at 5,000 × g for 5 min 700 μL of QIAzol was added onto the membrane of the exoEasy spin column, and the samples were centrifuged at 5,000 × g for 5 min. The supernatant was pipetted out into 2-ml tubes (supplied) and added with 90 μL chloroform. The tubes were closed, vortexed for 15 s, and placed for 2–3 min at room temperature. They were centrifuged at 12,000 × g for 15 min at 4°C. The supernatant was pipetted out into new tubes and mixed with 100% ethanol at the ratio of 1:2 (v/v). The sample (>700 μL) was pipetted out and applied to the RNeasy MinElute spin column in a 2-ml collection tube (supplied) and centrifuged at ≥ 8,000 × g for 15 s at room temperature. The supernatant was discarded. 700 μL of RWT buffer was added onto the RNeasy MinElute spin column, followed by another centrifugation at ≥ 8,000 × g for 15 s at room temperature and removal of the supernatant. The step was repeated after 500 μL of RPE buffer was applied to the RNeasy MinElute spin column. The samples were centrifuged at ≥ 8,000 × g for 2 min at room temperature after 500 μL of RPE buffer was loaded onto the column. The column was then transferred to a new tube (supplied), unclosed, and centrifuged at the max speed for 5 min to dry the membrane. The RNeasy MinElute spin column was placed in a new 1.5-ml collection tube (supplied). 14 μL of RNase-free water was directly added onto the center of the spin column membrane. The column was placed for 1 min and centrifuged at the max speed for 1 min to extract RNA. The purification of the total serum RNA was assessed.
Exosomal vesicles were observed under transmission electron microscopy (TEM) (Hitachi TEM system). Exosomes were lysed in RIPA lysis buffer, and total proteins were extracted. The proteins were separated by SDS-PAGES and transferred onto a polyvinylidene difluoride membrane. After blocking, the membranes were incubated with mouse anti-human GAPDH monoclonal primary antibody (1:1,000), rabbit anti-human CD63 (1:100; ab217345, Abcam) and CD9 (1:100; ab92726, Abcam) antibodies, and rabbit anti-human BATF2 polyclonal antibody (1:300; ab204510, Abcam). Immunoblotting was performed to quantitate BATF2 protein expression in the biopsy tissues of NPC patients, as described in our previous publication (Cui et al., 2020).
qRT-PCR Analysis
Total RNA was extracted from serum and serum-derived exosomes with a miRNeasy Kit (QIAGEN, Catalog No.217184) and an exoRNeasy Serum/Plasma Midi Kit (QIAGEN, Catalog No.77044), respectively. For extraction of total serum RNA, 200 μL of serum sample was added with 1 ml of QlAzol lysis reagent, mixed by repetitive pipetting, and placed for 5 min at room temperature. After adding 200 μL of chloroform, the mixture was vortexed for 15 s, incubated for 2–3 min at room temperature, and centrifuged at 12,000 rmp for 15 min at 4°C. The supernatant was transferred to a new EP tube, added with 100% ethanol at the ratio of 1:1.5 (v/v), and mixed by repetitive pipetting. 700 μL of the mixture was applied to the RNeasy MinElute spin column in a new 2-ml tube, centrifuged at 8,000 × g for 15 s at room temperature for nucleic acid purification. 700 μL of RWT buffer was added onto the column, followed by centrifugation (8,000 × g, room temperature) for 15 s and removal of the supernatant, which was repeated after 500 μL of RPE buffer was added. 500 μL of 80% ethanol was added onto the column. The column was centrifuged (8,000 × g, room temperature) for 2 min. The RNeasy MinElute spin column was retained and placed in a new 2-ml tube for centrifugation until the spin column membrane was dried. The column was transferred to a 1.5-mL tube, added with 14 μL of RNase-free water, unclosed, and centrifuged at the max speed for 1 min to extract RNA. Total RNA was reverse transcribed into cDNA with a Transcriptor First Strand cDNA Synthesis Kit (Roche; 04896866001). The cDNA was amplified with an SYBR Green Master Mix (ROX; REF.04913914001). The qRT-PCR was carried out using an ABI7500 real-time PCR system with two primers designed to amplify BATF2 and GAPDH, respectively: BATF2-F: 5′-GCCTAAGCCATGCACCTCTGT-3′, BATF2-R: '-TCTTCAGCTGCCTTTGTTGCTC -3′, GAPDH-F: 5′-GGAGCGAGATCCCTCCAAAAT-3′, and GAPDH-R: 5′-GGCTGTTGTCATACTTCTCATGG-3’. The amplification conditions were as follows: initial denaturation at 95 °C for 10 min, then 40 cycles of denaturation at 95°Cfor 15 s, and extension at 60°C for 1 min. Serum and exosomal BATF2 mRNA expression normalized to GAPDH was calculated using the 2−ΔΔCT method.
Statistical Analysis
SPSS 16.0 software was used for all statistical analyses. Normally distributed continuous variables were expressed as mean ± standard deviation (SD) unless otherwise specified. The independent-sample t-test was used for comparisons between groups. The Chi-square test or Spearman’s rank coefficient was used, as appropriate, to assess the correlations between BATF2 expression and clinicopathological characteristics. A Cox proportional hazard regression model was performed to identify risk factors for NPC survival, and the hazard ratio (HR) with its 95% confidence interval (CI) was calculated. Kaplan-Meier survival curve was applied to analyze the overall survival of NPC patients. A P-value of <0.05 was considered statistically significant.
Results
Demographic Characteristics
For the IHC analysis, the 129 NPC patients consisted of 99 males and 30 females, with a median age of 50 years (range, 20–82 years). The pathological subtypes incorporated undifferentiated non-keratinizing subtypes (112 cases), differentiated non-keratinizing squamous cell carcinoma (15 cases, including a case of non-keratinizing squamous cell carcinoma and another of keratinizing squamous cell carcinoma), and keratinizing squamous cell carcinoma (2 cases, a case of non-keratinizing squamous cell carcinoma and another of partially non-keratinizing undifferentiated subtype). Among the 129 cases, 109 did not develop distant metastasis. According to the Chinese 2017 staging system for NPC, 15 patients were diagnosed with stage I NPC, 55 with stage II NPC, 37 and 4 with stage III and IV NPC. Serum samples from 130 NPC cases were collected in our institution. The 130 NPC cases consisted of 68 newly diagnosed cases, 23 recurrent cases, and 39 cases with clinical treatment (radiotherapy or chemotherapy or targeted therapy), and their clinical characteristics are shown in Table 1.
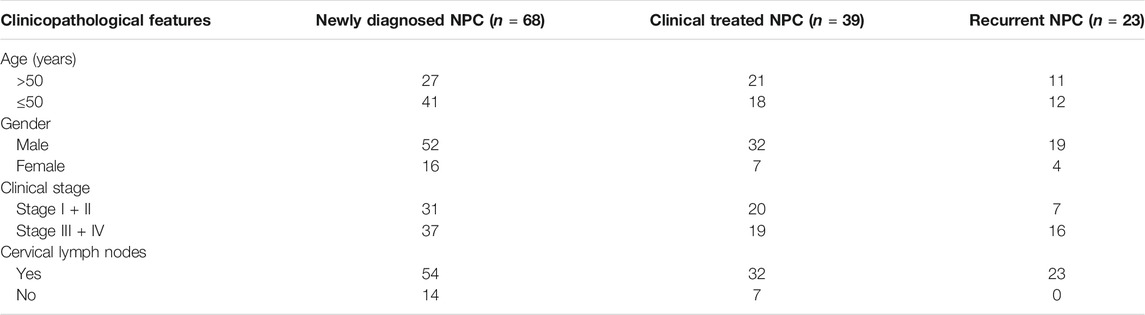
TABLE 1. Clinical characteristics of the 130 NPC cases for the analysis of serum and exosomal BATF2 mRNA expressions.
Tissue BATF2 Downregulation as a Biomarker for NPC Prognosis
The IHC microarray showed that 63/129 (48.83%) cases were BATF2-positive, with BATF2 predominately expressed in the nucleus of NPC cells (Figure 1A) and elevated BATF2 expression scores in stage I-II NPC patients and those without recurrent tumor or metastasis in the cervical lymph nodes (Figure 1B). Correlations between BATF2 expressions and clinical characteristics in NPC patients are shown in Table 2. BATF2 expression had significant associations with clinical stage (p = 0.040), recurrence (p = 0.000), and Ki-67 expression (a prognostic biomarker) in NPC (Figure 1C; Table 2). The Kaplan-Meier survival analysis showed that BATF2-positive patients tended to have a better prognosis, whose overall survival (OS) was significantly longer versus BATF2-negative patients (Figure 1D). These findings suggested that BATF2 had the potential to predict NPC prognosis. The Cox hazards model demonstrated that advanced clinical staging, recurrence, and low BATF2 expression were risk factors for the prognosis of NPC patients (Figure 1E). A low or negative BATF2 expression was also observed in biopsy tissues using immunoblotting (Figure 1F).
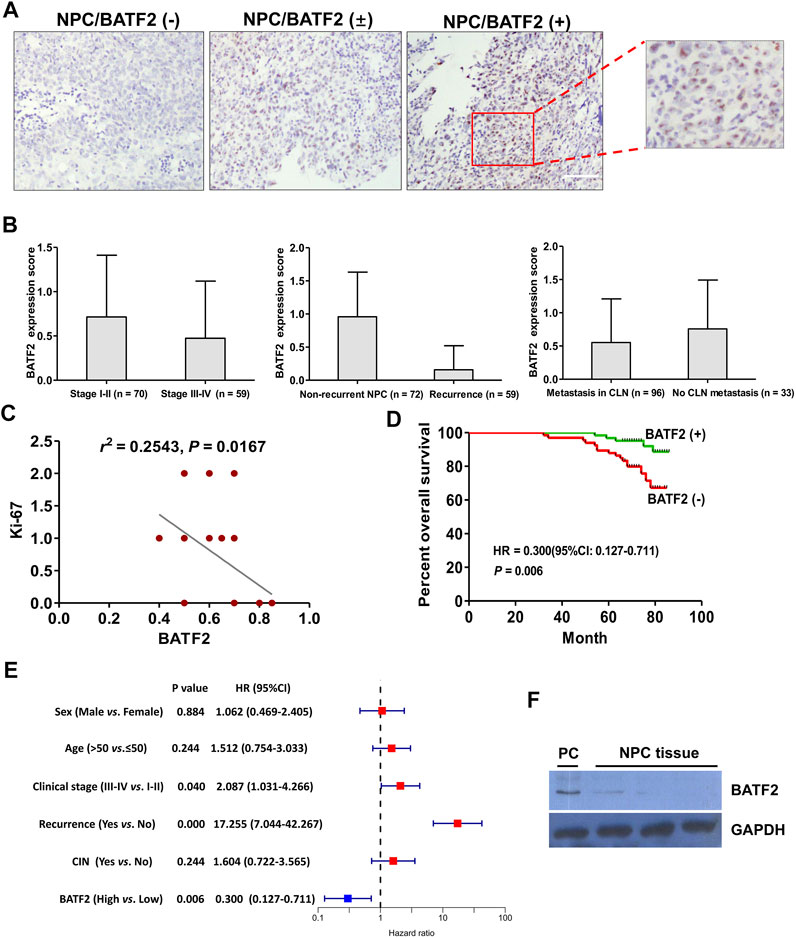
FIGURE 1. Tissue BATF2 downexpression in NPC tissues using IHC microarrays. (A) BATF2 is predominately expressed in the nucleus of NPC cells (× 200). (B) Elevated BATF2 expression scores are found in stage I-II NPC patients and those without recurrent tumor or metastasis in the cervical lymph nodes (CLNs). (C) Low BATF2 expression is correlated with Ki-67 upregulation in NPC tissues. (D) The Kaplan-Meier survival analysis shows that BATF2 expression is positively correlated with NPC prognosis. (E) The forest plot of risk factors for NPC prognosis (the cut-off point for HR calculation: 5-year follow-up). (F) Low BATF2 expression was detected in biopsy tissues using immunoblotting. The BATF2-overexpressed NPC cell line (CNE2) is utilized as a positive control (PC).
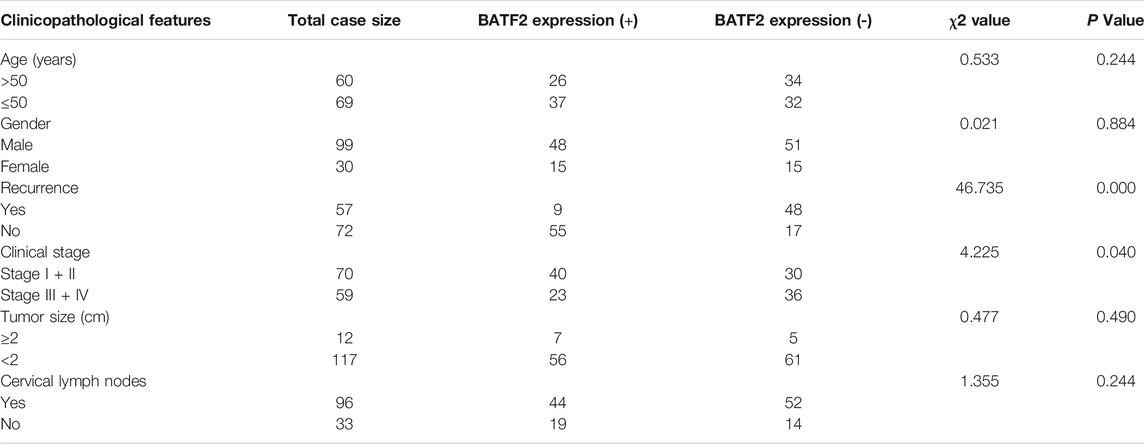
TABLE 2. Correlations between BATF2 protein expressions and clinical characteristics in NPC patients.
Serum and Exosomal BATF2 mRNA as a Diagnostic Biomarker for NPC
The morphology of serum exosomes was observed, and their size was measured under transmission electron microscopy (TEM). As shown in Figure 2A, serum exosomes were small vesicles of 30 and 150 nm in diameter with cell membrane structures. Expressions of CD9 and CD63, exosome-specific markers, were detectable in isolated vesicles (Figure 2B). We found that the percentages of patients with detectable serum and exosomal BATF2 mRNA expression were 51.47% (35/68) and 48.52% (33/68), significantly lower compared to 88.00% (44/50) and 84.00% (42/50) in healthy controls. Both serum and exosomal BATF2 mRNA expressions were one-third lower than those detected in healthy controls (Figures 2C,D,F,G). Subsequently, we assessed the performances of serum and exosomal BATF2 in NPC diagnosis using ROC curves analysis and found that the sensitivity, specificity, and AUCs of serum BATF2 in discriminating NPC patients from healthy controls were 89%, 86%, and 0.9409, respectively (Figure 2E). The sensitivity, specificity, and AUCs for exosomal BATF2 were 81%, 82%, and 0.8983, respectively (Figure 2H). Therefore, serum and exosomal BATF2 mRNA expressions have excellent performances in NPC diagnosis.
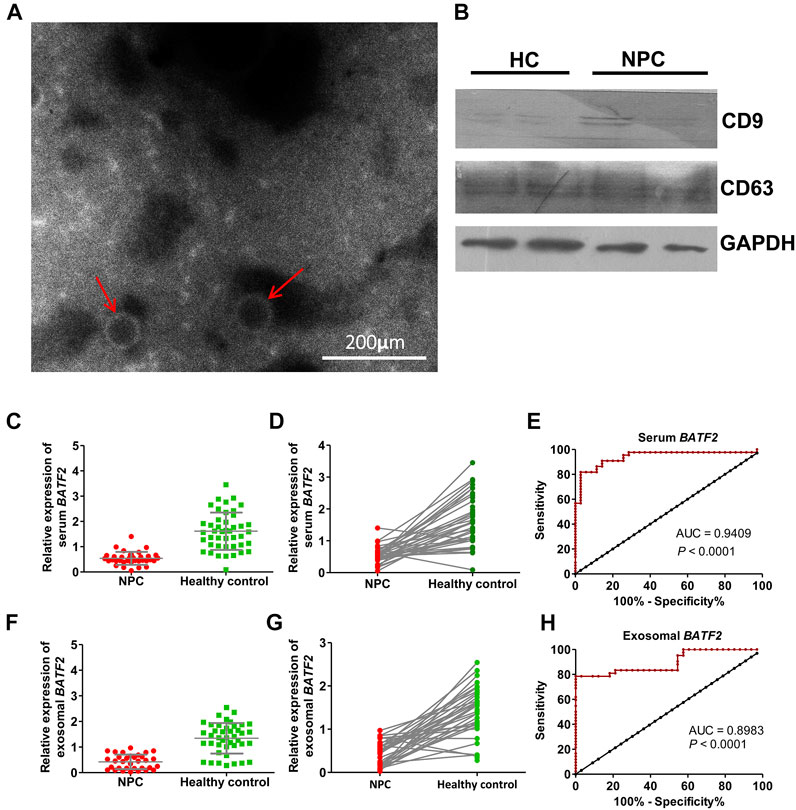
FIGURE 2. Identification of serum exosomes and efficacy of serum and exosomal BATF2 expression in NPC diagnosis. (A) Morphology of serum-derived exosomes under transmission electron microscopy. The red arrows indicate isolated vesicles. (B) The immunoblotting assay shows that the exosome-specific markers, CD9 and CD63, are detectable in isolated vesicles. HC: healthy control. (C,D) Serum BATF2 mRNA expression significantly decreases in NPC versus healthy controls. (E) The ROC curve to assess the efficacy of serum BATF2 mRNA in NPC diagnosis. (F,G) Suppressed exosomal BATF2 mRNA expression in NPC patients. (H) The ROC curve to assess the efficacy of exosomal BATF2 mRNA in NPC diagnosis.
Relationship of Serum and Exosomal BATF2 mRNA Downregulation With Clinicopathological Risk Factors in NPC
We found that serum BATF2 suppression had a strong association with squamous cell carcinoma antigen (SCC-Ag) levels (Figure 3A), without significant correlations with CEA, CYFRA21-1, or EBV-DNA levels (Figures 3B–D, all p > 0.05). However, exosomal BATF2 downregulation was associated with none of the clinical indicators above (Figures 3E–H, all p > 0.05).
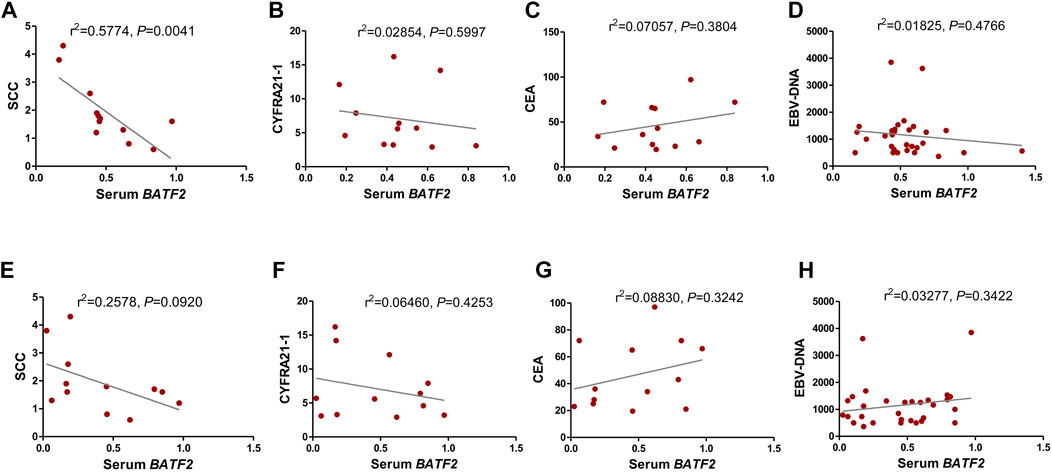
FIGURE 3. Relationship of serum and exosomal BATF2 mRNA downregulation with clinicopathological risk factors in NPC. The correlations between serum BATF2 expression and (A) SCC-Ag, (B) CYFRA21-1, (C) CEA, and (D) EBV-DNA levels are analyzed. The downregulation of exosomal BATF2 expression is not associated with (E) SCC-Ag, (F) CYFRA21-1, (G) CEA, or (H) EBV-DNA levels in NPC patients.
Serum and Exosomal BATF2 as an Indicator for Monitoring Efficacy and Recurrence
We assessed serum and exosomal BATF2 expressions in NPC patients at baseline (newly diagnosed), after receiving clinical treatments (radiotherapy and/or chemotherapy and/or targeted therapy), and suffering recurrence. The results showed that serum and exosomal BATF2 mRNA expression significantly increased after clinical treatments (p < 0.01), but decreased in recurrent patients (p < 0.01) (Figures 4A–D). Serum and exosomal BATF2 had high AUCs in predicting the recurrence of NPC cases (Figures 4E,F). Moreover, two cases were followed up to 16 months during radiotherapy or chemotherapy or targeted therapy: their serum and exosomal BATF2 mRNA expressions were upregulated following clinical treatments but decreased at the diagnosis of recurrent NPC (Figures 4G,H). These findings suggest serum and exosomal BATF2 mRNA expressions as a sensitive indicator for monitoring efficacy and recurrence in NPC patients.
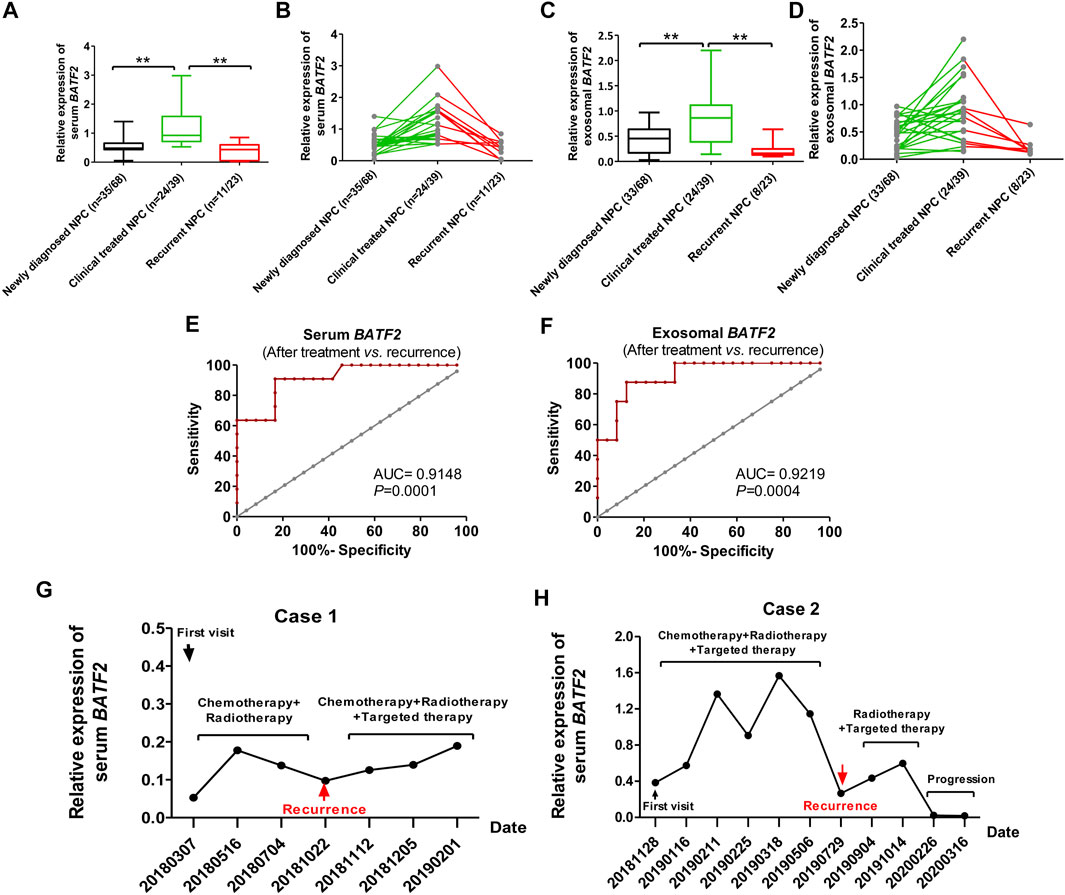
FIGURE 4. Serum and exosomal BATF2 downregulation as an indicator for monitoring efficacy and recurrence in NPC. of Serum (A,B) and exosomal (C,D) BATF2 mRNA expressions in NPC patients at baseline, after treatment (radiotherapy or chemotherapy or targeted therapy), at the diagnosis of recurrent NPC. ROC curves of serum (E) and exosomal (F) BATF2 mRNA expressions in predicting the relaps eof NPC. (G,H) Serum BATF2 mRNA expression level in two NPC patients participating in the follow-up during radiotherapy or chemotherapy or targeted therapy. **p < 0.01.
Discussion
Low BATF2 expression has been shown to have a significant association with the occurrence and prognosis of various metastatic and invasive malignancies (Dash et al., 2010; Huang et al., 2011; Liu et al., 2011; Ma et al., 2011; Wang et al., 2012; Chen et al., 2015; Guler et al., 2015). For far too long, BATF2 expression patterns in NPC have not been reported. The present study reported a percentage of 48.83% of BATF2-positive patients who exhibited longer survival than BATF2-negative patients. This suggests the prognostic value of BATF2 in NPC. We found that serum BATF2 mRNA expression was positively associated with SCC-Ag levels, which also indicates its diagnostic potential in NPC. However, aberrant BATF2 expression patterns vary among malignancies. Our previous study reported a moderate level of BATF2 expression in the K562 leukemia cell line and BATF2 downregulation-induced apoptosis and proliferation inhibition in K562 cells (Fu et al., 2018). But high BATF2 expression has been proven an important prognostic indicator for HCC (Ma et al., 2011) and NSCLC (Wang et al., 2012).
Exosomes are a type of nanosized extracellular vesicles exocytosed by all cells (Zeringer et al., 2015) to carry different molecules (proteins, DNAs, RNAs, or lipids) to the cells of interest (Melo et al., 2015; He et al., 2018). Exosomes are detectable in the tumor microenvironments, which have been proven to promote tumorigenesis and metastasis via promoting angiogenesis and tumor-specific immunity (Kalluri, 2016). Circulating exosomes as a cancer liquid biopsy are accessible and non-invasive for early diagnosis and allow repeated measurements for monitoring efficacy in post-treatment patients (Urbanelli et al., 2015; De Rubis et al., 2019). The present study showed that serum and exosomal expressions of cell-free BATF2 mRNA were significantly higher in healthy controls than NPC cases, indicating the involvement of exosomal transport of BATF2 mRNA. Current views on the potential mechanisms for exosomal RNA transfer into the circulation, as shown in Figure 5, are that exosomal BATF2 mRNA may come both from non-apoptotic and apoptotic cells. Exosomal BATF2 molecules may release into the extracellular median via exocytic fusion of multivesicular bodies (MVBs) with the plasma membrane (Mathivanan et al., 2010), followed by budding of vesicles directly from the plasma membrane and microvesicle (containing BATF2) shedding (Figure 5A). The shed microvesicles (SMVs) from apoptotic cells are called apoptotic blebs (ABs) (Figure 5B). In our study, low BATF2 expression in NPC tissues indicates its tumor suppressor roles in NPC, which is further evidenced by prolonged overall survival of BATF2-positive patients. BATF2 mRNA expression was also detectable in serum and serum-derived exosomes of the NPC cases. Thus, we speculate that the tumor-suppressor BATF2 molecules are released from exosomes to the outside of the cell to support tumor cell proliferation and tumor growth. Kanlikilicer et al. (2016) reported that intracellular miRNA-6126 expression was downregulated in ovarian carcinoma cells via exosomal transport, thus stimulating the expression of the target gene ITGB1 (integrin subunit β1) as fuel to cancer cell proliferation and invasion. Their findings support our presumption. However, the precise mechanisms for exosomal secretion of nucleic acid into the circulation remain uncertain. Future studies are still needed to explore how the nuclear transcription factor BATF2 migrates from the nucleus to the cytoplasm and how a BATF2-positive exocytic exosome forms.
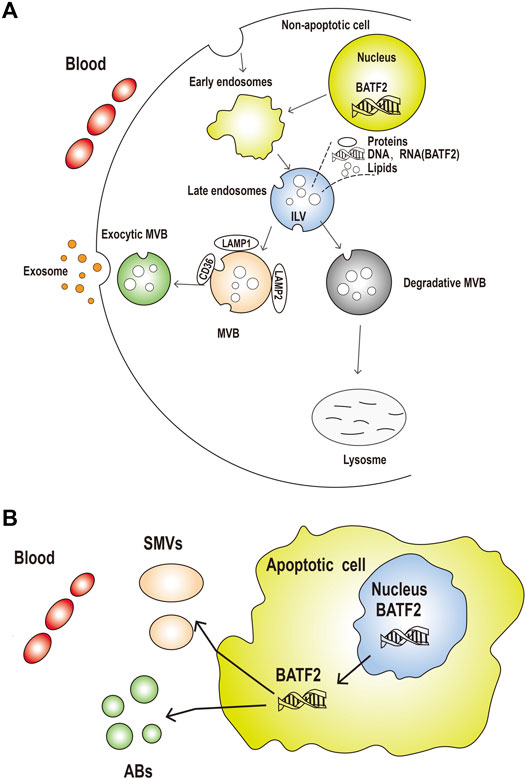
FIGURE 5. The potential mechanisms for exosomal BATF2 secretion into the circulation. (A) The process of released exosomes and shed microvesicles (SMVs) (containing BATF2 molecules) into the circulation. Early endosomal contents (proteins and nucleic acids) are either recycled to the plasma membrane (PM) or sequestered in intraluminal vesicles (ILVs), generated by the budding of the limiting membrane into the lumen of endosomes, within the larger large multivesicular bodies (MVBs). (B) Apoptotic or non-apoptotic dying cells result in the generation of apoptotic bodies (Abs). Microvesicles are shed from the blebbing PM or released from apoptotic cells. These vesicles are remnants of the degrading apoptotic cell with nuclear and cytoplasmic content (including BATF2 molecules).
This study for the first time reported serum and exosomal BATF2 mRNA expressions in NPC patients, with positive rates of 51.47 and 48.52%, respectively. Their high AUCs in discriminating NPC from healthy controls and high sensitivity in identifying recurrent patients from treated ones suggest a high potential for its clinical application in predicting diagnosis and monitoring efficacy and post-treatment relapse for patients. They are expected to serve as new biomarkers in NPC clinical practice. Our previous study series reported similar performances of LDHC/LDH-C4, a cancer-testis antigen, in diagnosis and relapse prediction and efficacy evaluation for breast cancer and hepatocellular carcinoma patients (Cui et al., 2020; Cui et al., 2020). As the total exosomal RNA is part of the total serum RNA extracted using an exoRNeasy Serum/Plasma Midi Kit, the expression score of serum BATF2 mRNA must be higher than that of exosomal BATF2 mRNA. This finding indicates a significant resource of cell-free BATF2 molecules in serum or plasma from exosomal BATF2, which is consistent with our previous results (Cui et al., 2020; Cui et al., 2020). However, this conclusion needs further validation. Current limitations to clinical application of exosomal gene detection mainly ascribe to complicated procedures to enrich and purify exosomal DNAs/RNAs. Our conclusion is also limited by sample size and a lack of multiple data collection strategies, which can be solved by implementing multicenter clinical trials with a large sample size in the future.
Conclusion
This study explored the expressions of BATF2 mRNA and protein in NPC patients’ tissues, serum, and serum-derived exosomes. BATF2/BATF2 is expressed cancerous tissues, serum, and serum-derived exosomes in NPC patients. Circulating and tissue BATF2 mRNA can serve as a multipurpose biomarker capable of the diagnosis, prognosis prediction, efficacy evaluation, and recurrence monitoring in NPC.
Data Availability Statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of Fujian Cancer Hospital. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
YC conceived and designed the study. ZC, YL, and DH performed the experiments. JW and WP Helped to analyze the data. ZC and YL wrote the manuscript, and YC proofed the manuscript. All authors approved the final version of the manuscript.
Funding
This study was supported by the Joint Funds for the Innovation of Science and Technology, Fujian province (Grant number: 2017Y9072), and Science and Technology Program of Fujian Province, China (Grant Number: 2018Y2003).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Chen, Q., Gu, Y., and Liu, B. (2015). Expression and Mechanism of Action of the SARI Tumor Suppressor in Prostate Cancer. Int. J. Clin. Exp. Pathol. 8 (7), 7953–7960.
Chua, M. L. K., Wee, J. T. S., Hui, E. P., and Chan, A. T. C. (2016). Nasopharyngeal Carcinoma. Lancet 387 (10022), 1012–1024. doi:10.1016/s0140-6736(15)00055-0
Cui, Z., Chen, Y., Hu, M., Lin, Y., Zhang, S., Kong, L., et al. (2020). Diagnostic and Prognostic Value of the Cancer-Testis Antigen Lactate Dehydrogenase C4 in Breast Cancer. Clinica Chim. Acta 503, 203–209. doi:10.1016/j.cca.2019.11.032
Cui, Z., Li, Y., Gao, Y., Kong, L., Lin, Y., and Chen, Y. (2020). Cancer-testis Antigen Lactate Dehydrogenase C4 in Hepatocellular Carcinoma: a Promising Biomarker for Early Diagnosis, Efficacy Evaluation and Prognosis Prediction. Aging 12 (19), 19455–19467. doi:10.18632/aging.103879
Dai, W., Zheng, H., Cheung, A. K. L., and Lung, M. L. (2016). Genetic and Epigenetic Landscape of Nasopharyngeal Carcinoma. Chin. Clin. Oncol. 5 (2), 16. doi:10.21037/cco.2016.03.06
Dash, R., Su, Z.-Z., Lee, S.-G., Azab, B., Boukerche, H., Sarkar, D., et al. (2010). Inhibition of AP-1 by SARI Negatively Regulates Transformation Progression Mediated by CCN1. Oncogene 29 (31), 4412–4423. doi:10.1038/onc.2010.194
De Rubis, G., Rajeev Krishnan, S., and Bebawy, M. (2019). Liquid Biopsies in Cancer Diagnosis, Monitoring, and Prognosis. Trends Pharmacol. Sci. 40 (3), 172–186. doi:10.1016/j.tips.2019.01.006
Fu, X., Cui, Z., Chen, Y., Tang, Y., Wu, C., Xu, J., et al. (2018). Endogenous SARI Exerts Oncogenic Functions in Human K562 Leukemia Cells by Targeting the PI3K/Akt/mTOR and NF-Κb Signaling Pathways. Int. J. Clin. Exp. Pathol. 11 (1), 179–188.
Guler, R., Roy, S., Suzuki, H., and Brombacher, F. (2015). Targeting Batf2 for Infectious Diseases and Cancer. Oncotarget 6 (29), 26575–26582. doi:10.18632/oncotarget.5576
He, C., Zheng, S., Luo, Y., and Wang, B. (2018). Exosome Theranostics: Biology and Translational Medicine. Theranostics 8 (1), 237–255. doi:10.7150/thno.21945
Huang, P.-Y., Zeng, Q., Cao, K.-J., Guo, X., Guo, L., Mo, H.-Y., et al. (2015). Ten-year Outcomes of a Randomised Trial for Locoregionally Advanced Nasopharyngeal Carcinoma: A Single-Institution Experience from an Endemic Area. Eur. J. Cancer 51 (13), 1760–1770. doi:10.1016/j.ejca.2015.05.025
Huang, Q., Yang, Y., Li, X., and Huang, S. (2011). Transcription Suppression of SARI (Suppressor of AP-1, Regulated by IFN) by BCR-ABL in Human Leukemia Cells. Tumor Biol. 32 (6), 1191–1197. doi:10.1007/s13277-011-0222-1
Kalluri, R. (2016). The Biology and Function of Exosomes in Cancer. J. Clin. Invest. 126 (4), 1208–1215. doi:10.1172/jci81135
Kanlikilicer, P., Rashed, M. H., Bayraktar, R., Mitra, R., Ivan, C., Aslan, B., et al. (2016). Ubiquitous Release of Exosomal Tumor Suppressor miR-6126 from Ovarian Cancer Cells. Cancer Res. 76 (24), 7194–7207. doi:10.1158/0008-5472.CAN-16-0714
Lee, H. M., Okuda, K. S., González, F. E., and Patel, V. (2019). Current Perspectives on Nasopharyngeal Carcinoma. Adv. Exp. Med. Biol. 1164, 11–34. doi:10.1007/978-3-030-22254-3_2
Li, L.-L., Shu, X.-S., Wang, Z.-H., Cao, Y., and Tao, Q. (2011). Epigenetic Disruption of Cell Signaling in Nasopharyngeal Carcinoma. Chin. J. Cancer 30 (4), 231–239. doi:10.5732/cjc.011.10080
Lin, C., Cao, S. M., Chang, E. T., Liu, Z., Cai, Y., Zhang, Z., et al. (2019). Chinese Nonmedicinal Herbal Diet and Risk of Nasopharyngeal Carcinoma: A Population‐based Case‐control Study. Cancer 125 (24), 4462–4470. doi:10.1002/cncr.32458
Liu, M.-T., Chen, M.-K., Huang, C.-C., and Huang, C.-Y. (2015). Prognostic Value of Molecular Markers and Implication for Molecular Targeted Therapies in Nasopharyngeal Carcinoma: An Update in an Era of New Targeted Molecules Development. World J. Oncol. 6 (1), 243–261. doi:10.14740/wjon610w
Liu, Z. B., Yang, Y., Ye, X. G., Wang, L., Tian, P. Y., and Zhang, Y. Y. (2011). Expression and Significance of SARI and CCN1 in Human Colorectal Carcinomas. Zhonghua Yi Xue Za Zhi 91 (34), 2397–2401.
Ma, H., Liang, X., Chen, Y., Pan, K., Sun, J., Wang, H., et al. (2011). Decreased Expression of BATF2 Is Associated with a Poor Prognosis in Hepatocellular Carcinoma. Int. J. Cancer 128 (4), 771–777. doi:10.1002/ijc.25407
Mathivanan, S., Ji, H., and Simpson, R. J. (2010). Exosomes: Extracellular Organelles Important in Intercellular Communication. J. Proteomics 73 (10), 1907–1920. doi:10.1016/j.jprot.2010.06.006
Melo, S. A., Luecke, L. B., Kahlert, C., Fernandez, A. F., Gammon, S. T., Kaye, J., et al. (2015). Glypican-1 Identifies Cancer Exosomes and Detects Early Pancreatic Cancer. Nature 523 (7559), 177–182. doi:10.1038/nature14581
Ng, W. T., Lee, M. C. H., Hung, W. M., Choi, C. W., Lee, K. C., Chan, O. S. H., et al. (2011). Clinical Outcomes and Patterns of Failure after Intensity-Modulated Radiotherapy for Nasopharyngeal Carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 79 (2), 420–428. doi:10.1016/j.ijrobp.2009.11.024
Su, Z.-z., Lee, S.-G., Emdad, L., Lebdeva, I. V., Gupta, P., Valerie, K., et al. (2008). Cloning and Characterization of SARI (Suppressor of AP-1, Regulated by IFN). Proc. Natl. Acad. Sci. 105 (52), 20906–20911. doi:10.1073/pnas.0807975106
Trejaut, J., Lee, C.-L., Yen, J.-C., Loo, J.-H., and Lin, M. (2011). Ancient Migration Routes of Austronesian-Speaking Populations in Oceanic Southeast Asia and Melanesia Might Mimic the Spread of Nasopharyngeal Carcinoma. Chin. J. Cancer 30 (2), 96–105. doi:10.5732/cjc.010.10589
Urbanelli, L., Buratta, S., Sagini, K., Ferrara, G., Lanni, M., and Emiliani, C. (2015). Exosome-based Strategies for Diagnosis and Therapy. Rpcn 10 (1), 10–27. doi:10.2174/1574889810666150702124059
Wang, C., Su, Y., Zhang, L., Wang, M., You, J., Zhao, X., et al. (2012). The Function of SARI in Modulating Epithelial-Mesenchymal Transition and Lung Adenocarcinoma Metastasis. PLoS One 7 (9), e38046. doi:10.1371/journal.pone.0038046
Keywords: nasopharyngeal carcinoma, exosomes, diagnosis, prognosis, basic leucine zipper ATF-like transcription factor 2
Citation: Cui Z, Lin Y, Hu D, Wu J, Peng W and Chen Y (2021) Diagnostic and Prognostic Potential of Circulating and Tissue BATF2 in Nasopharyngeal Carcinoma. Front. Mol. Biosci. 8:724373. doi: 10.3389/fmolb.2021.724373
Received: 13 June 2021; Accepted: 13 October 2021;
Published: 28 October 2021.
Edited by:
Anindya Ghosh, University of Arkansas at Little Rock, United StatesReviewed by:
Rupak Pathak, University of Arkansas for Medical Sciences, United StatesYanwei Luo, Central South University, China
Copyright © 2021 Cui, Lin, Hu, Wu, Peng and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Chen, Y2hlbnlhbjIyMDQyMkBmam11LmVkdS5jbg==
†These authors share first authorship
 Zhaolei Cui
Zhaolei Cui Yingying Lin1†
Yingying Lin1†