
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Mol. Biosci., 01 September 2021
Sec. Metabolomics
Volume 8 - 2021 | https://doi.org/10.3389/fmolb.2021.723858
This article is part of the Research TopicOMICS-Based Approaches in Sports ResearchView all 9 articles
 Afnan Saleh Al-Menhali1,2
Afnan Saleh Al-Menhali1,2 Cali Anderson3
Cali Anderson3 Alexander V. Gourine4
Alexander V. Gourine4 Andrey Y. Abramov5
Andrey Y. Abramov5 Alicia D’Souza3
Alicia D’Souza3 Morana Jaganjac1,6*
Morana Jaganjac1,6*Regular exercise has many health benefits, among which is a significant reduction of cardiovascular risk. Although many beneficial effects of exercise are well described, the exact mechanisms by which exercise confers cardiovascular benefits are yet to be fully understood. In the current study, we have used high resolution mass spectrometry to determine the proteomic responses of the heart to exercise training in mice. The impact of exercise-induced oxidative stress on modifications of cardiomyocyte proteins with lipid peroxidation biomarker 4-hydroxynonenal (4-HNE) was examined as well. Fourteen male mice were randomized into the control (sedentary) group and the exercise group that was subjected to a swim exercise training program for 5 days a week for 5 months. Proteins were isolated from the left ventricular tissue, fractionated and digested for shotgun proteomics. Peptides were separated by nanoliquid chromatography and analyzed on an Orbitrap Fusion mass spectrometer using high-energy collision–induced dissociation and electron transfer dissociation fragmentation. We identified distinct ventricular protein signatures established in response to exercise training. Comparative proteomics identified 23 proteins that were upregulated and 37 proteins that were downregulated with exercise, in addition to 65 proteins that were identified only in ventricular tissue samples of exercised mice. Most of the proteins specific to exercised mice are involved in respiratory electron transport and/or implicated in glutathione conjugation. Additionally, 10 proteins were found to be modified with 4-HNE. This study provides new data on the effects of exercise on the cardiac proteome and contributes to our understanding of the molecular mechanisms underlying the beneficial effects of exercise on the heart.
In the early 1950s, Morris and others showed the association between physical activity and reduced deaths from coronary heart disease, which led to increased scientific interest in the potential of physical activity to combat diseases (Morris et al., 1953). Current knowledge shows a clear link between an active lifestyle and overall health. Physical exercise is found to benefit the heart as it increases aerobic fitness (VO2max), enhances contraction, and accelerates relaxation, as well as decreasing the risk of developing cardiomyopathies (Roof et al., 2013). It is found that different types, intensities, durations, and frequencies of exercise interventions are crucial for cardiovascular health (Gevaert et al., 2020). Endurance exercise can trigger several adaptational mechanisms resulting in exercise-induced cardioprotection changes. Those changes include an enhanced antioxidant defense system with increased expression of glutathione peroxidase-1 and manganese superoxide dismutase. Moreover, cardiac function is improved due to changes in the expression of enzymes involved in energy metabolism (Burniston and Hoffman, 2011).
Many studies suggest the beneficial effects of exercise-induced oxidative stress. It is suggested that the released reactive oxygen species (ROS) post–physical exercise can lead to cardiac adaptation, including protection against infarction (Frasier et al., 2011). At a molecular level, excess oxidants can damage macromolecules. Lipids are one of the target sites of ROS-induced damage, where it was found that maximal and supra-maximal exercise can significantly increase lipid peroxidation (Mohamed et al., 2016).
Lipid peroxidation can result in the formation of many reactive species as secondary products, known as reactive carbonyl compounds (RCCs), due to the highly reactive carbonyl group. Those secondary products have high stability with an average half-life of minutes or even hours, compared to the short half-life of ROS, which can only last for nanoseconds or milliseconds (Jaganjac et al., 2016). As a result, RCCs can diffuse through cellular membranes and attack biomolecules that are far away from the site of origin (Pamplona, 2011; Jaganjac et al., 2013). Among all secondary products studied, of particular biochemical and biomedical relevance is 4-hydroxynonenal (4-HNE) (Živković et al., 2005).
The physiological levels of 4-HNE vary between different cell types or tissues. Physiological concentrations of 4-HNE are in the submicromolar range, and for the heart samples, a 4-HNE concentration of around 0.25 nmol/mg of tissue (Nakagawa et al., 2014) or 6 nmol/mg of protein (Beneš et al., 2013) was reported. 4-HNE can bind to the nucleophilic amino acid side chain of proteins via the Michael addition or the Schiff base formation modifying protein function (Zarkovic et al., 2013). It is crucial for cells to control 4-HNE concentrations, as low concentrations are needed for normal physiological processes, while high concentrations can have detrimental effects (Jaganjac et al., 2020a).
In this study, conducted in experimental animals (mice), we aimed to determine the effect of intense exercise training on the left ventricle proteome and on the formation of 4-HNE protein adducts by using in-depth proteomics.
Experiments were performed on 14 male C57BL/6J mice that were 8–10 weeks old, with water and food given ad libitum. All experiments were performed in accordance with the European Commission Directive 2010/63 (European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes) and the United Kingdom Home Office (Scientific Procedures) Act (1986) with project approval by the Institutional Animal Welfare and Ethical Review Committee of the University of Manchester (PBA6A87CA).
Animals were randomized into two groups: a sedentary group and an exercised group that was trained by swimming for 5 months, for 60 min per day for 5 days per week. After 5 months, the animals were terminated by cervical dislocation, following which the heart was rapidly excised. A 2-mm biopsy of the left ventricle free wall was sampled, approximately midway between the base and the apex, and stored at −80°C until further analysis. At the time of sample collection, the mice were 7–7.5 months old.
Left ventricular tissue was homogenized in a glass homogenizer in ice-cold lysis buffer (20 mM HEPES, 20 mM NaCl, 5 mM EDTA, 1% w/v CHAPS, and protease inhibitors). 7 ml of buffer was added per 1 g of tissue. Lysates were then sonicated for 10 min, followed by centrifugation at 14,000 × g for 10 min at +4°C. Supernatants were collected into new Eppendorf tubes, and protein concentration was determined using a BCA Protein Assay Kit (Pierce).
Protein samples were prepared for proteomics analysis in a similar manner to that described before (Al-Thani et al., 2018). Briefly, normalized protein samples were electophoretically separated on an SDS-PAGE using non-reducing sample buffer and whole lines were excised and divided into 8 equal parts that were then reduced with 10 mM dithiothreitol and alkylated with 100 mM iodoacetamide, followed by overnight digestion at 37°C with 20 ng/μl Trypsin Gold MS grade (Promega). The mixture of 45% water, 50% acetonitrile, and 5% formic acid was used to extract peptides.
Samples were analyzed using an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific, Waltham, United States) coupled with an Easy n-LC II (Thermo Scientific, Waltham, United States) for nano-LC gradient separation. Thermo Xcalibur (version 3.0) software was used to control instrument setup. The nanoelectrospray ionization (NSI) mode was used, which is the preferred ionization mode for peptides and proteins. Peptide mixtures were separated on a reverse-phase C18 column (25 cm, 75 μm, 2 μm, Acclaim RSLC C18, Thermo Scientific, Waltham, United States) attached to a pre-trapping C18 column (2 cm, 75 μm, 3 μm, Acclaim, Thermo Scientific, Waltham, United States), and elution was carried out at a constant flow rate of 300 nl/min over a 108-min step gradient. Solvents used for separation were solvent A (HPLC-grade water with 0.1% (v/v) formic acid) and solvent B (HPLC-grade acetonitrile with 0.1% (v/v) formic acid). The separation gradient was set to 5% B for 5 min, 5–37% B for 90 min, 37–80% B for 4 min, 80% B for 2 min, 80–5% B for 2 min, and 5% B for 5 min.
The Orbitrap Fusion Tribrid analysis was set to data-dependent acquisition (DDA), using an Orbitrap mass analyzer to acquire the full MS spectra and an IonTrap to acquire the MS/MS fragment ion spectra. First full-scan mode acquisition was performed with a resolution of 120,000 orbitrap and a scan range of 400–1,600 m/z. Automatic gain control (AGC) was set at 200,000 with an injection time of 100 ms. This scan mode was carried out by enabling a monoisotopic precursor selection (MIPS) filter and dynamic exclusion with a duration of 30 s and a mass tolerance of 10 ppm. This is followed by two different second scan modes based on decisions of two scan event types. Scan event type 1 uses higher-energy collision–induced dissociation (HCD) fragmentation at the quadrupole isolation mode, which generates b and y ions, with an isolation width of 2 m/z and a collision energy of 30% and at 10,000 AGC and a maximum injection time of 70 ms. The neutral loss ion is triggered with masses of 52 or 78 m/z from the 156-Da 4-HNE–modified peptide by the Michael addition or masses of 46 or 69 m/z from the 138-Da 4-HNE–modified peptide by the Schiff bases (Carini et al., 2004). Selected peptides are further fragmented by electron transfer dissociation (ETD), which generates c and z ions and cleaves the amide group, leaving the side chain intact, using the quadrupole isolation mode with an isolation width of 2 m/z and a collision energy of 30% and at 10,000 AGC and a maximum injection time of 70 ms. Meanwhile, scan event type 2 is HCD fragmentation at the quadrupole isolation mode with an isolation width of 1.6 m/z and a collision energy of 30% and at 10,000 AGC and a maximum injection time of 70 ms.
The raw data generated by the Orbitrap Fusion were processed using Proteome Discoverer 2.2 (Thermo Scientific, San Jose, California, United States). The spectrum selector filter was assigned with 350 Da as the minimum precursor mass and 5,000 Da as the maximum precursor mass. The MS/MS spectra search was carried out using SEQUEST HT search algorithms against the UniProt Mus musculus (Mouse) protein database, and the FASTA file was retrieved on the 9th of October, 2017. Two scan event filters were used: one with the HCD activation type, followed by Sequest HT search with the following parameters: full trypsin digestion, a maximum of 2 missed cleavages sites, a minimum peptide length of 6 residues, a tolerance of 20 ppm for precursor mass and 0.6 Da for fragment mass, dynamic modification of oxidation of methionine (+15.995 Da), and static modification of carbamidomethyl of cysteine (+57.021 Da). The second scan event filter included the ETD activation type with 1,000 set as the maximum collision energy, followed by another Sequest HT search using a comprehensive workflow that included additional dynamic modifications: 4-HNE adduction to cysteine, histidine, lysine, leucine, methionine, and arginine (+156.115 Da); 4-HNE + H2 for the Michael adducts in cysteine, histidine, lysine, leucine, methionine, and arginine (+158.131 Da); and 4-HNE-H2O for the Schiff bases in cysteine, histidine, lysine, leucine, methionine, and arginine (+138.104 Da). The target decoy PSM validator node was used for peptide validation with an FDR of 0.01. For precursor ion quantification, label-free quantification using the Minora Feature Detection was used, followed by statistical analysis using t-test (p < 0.05) to measure proteins that were significantly upregulated in the exercised group compared to the control group. On the other hand, the consensus workflow parameters used were as follows: a peptide validator with an automatic validation mode of 0.01 FDR, peptides and proteins were searched using a high-confidence peptide filter, and the precursor ion quantifier calculated abundance based on the intensity. For high confidence, XCorr values were set to at least 1.8 for singly, 2.4 for doubly, 2.5 for triply, and 2.6 for quadruply or more charged peptides. The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD026558.
Only cardiac proteome changes detected in six or more samples per group were considered for further data visualization and interpretation. Differential expression analysis was performed using Perseus 1.6.15.0. (Tyanova et al., 2016) according to the protocol of Tyanova & Cox (Tyanova et al., 2018), and the FDR value was set to 0.05. The results are reported with the corrected p-value (q-value).
Data analysis of proteins identified in both groups was performed using InfernoRDN v1.1.7234 software (Polpitiya et al., 2008). Data were log-transformed and normalized by central tendency adjustment, followed by partial least square (PLS) analysis. Moreover, variables upregulated or downregulated with exercise with p-values ≤ 0.05 were visualized on the heat map, while all proteins that met the criteria and were identified in the database are plotted in a volcano plot.
Protein ANalysis THrough Evolutionary Relationships (PANTHER) classification system software (version 14.0) was used to analyze changes in different classes of proteins based on identified proteins and to analyze gene ontology annotations available in the UniProt database (Mi et al., 2019). The Reactome 3.7 database of reactions, pathways, and biological processes with database release 73 was used for pathway analysis of left ventricular proteins upregulated or only identified in the exercised group (Haw and Stein, 2012).
High resolution mass spectrometry was used to study changes in the left ventricular tissue proteome after long-term intensive swimming exercise. The total number of identified proteins in the control and the exercise group was 1,839 and 1,915, respectively, while the total number of identified peptides was 10,092 and 10,129, respectively. Further analyses were based on the criteria that the protein/peptide must be common for ≥6 tissue samples of the same group of animals. Based on that criteria, a total of 804 proteins were identified in the control group with a total number of 2,913 peptides, while 816 proteins were found in the exercised group with a total of 3,254 peptides.
To evaluate the impact of exercise on the cardiac proteome, we have used the comparative proteomics and label-free proteomics approach. A total of 125 proteins displayed statistically significant changes in the ventricular tissue of the swimming exercise group compared to the control group (Tables 1, 2). Table 1 lists 65 proteins that were identified in ≥6 ventricular tissue samples of exercised mice, while the same were not identified in the control mice. Proteins that were identified in 6/7 samples for each group were selected for PCA analysis, to determine the multivariate exercise signature of proteins (Figure 1A). The semiquantitative assessment of proteins upregulated/downregulated with exercise identified 23 proteins that were upregulated and 37 that were downregulated with exercise (Figure 1B), and details of each are included in Table 2. The changes in the protein abundances are shown using a volcano plot in Figure 1C. The largest increase following exercise training was observed for mitochondrial cytochrome c oxidase subunit 7A2, cathepsin B, and ubiquitin carboxyl-terminal hydrolase isozyme L3. In addition, the most significant exercise-induced upregulation was observed for mitochondrial cytochrome c oxidase subunit 7A2, delta-sarcoglycan, and mitochondrial cysteine desulfurase. PLS analysis demonstrated good separation between the two groups with distinct protein signatures in the ventricular samples of exercised animals.
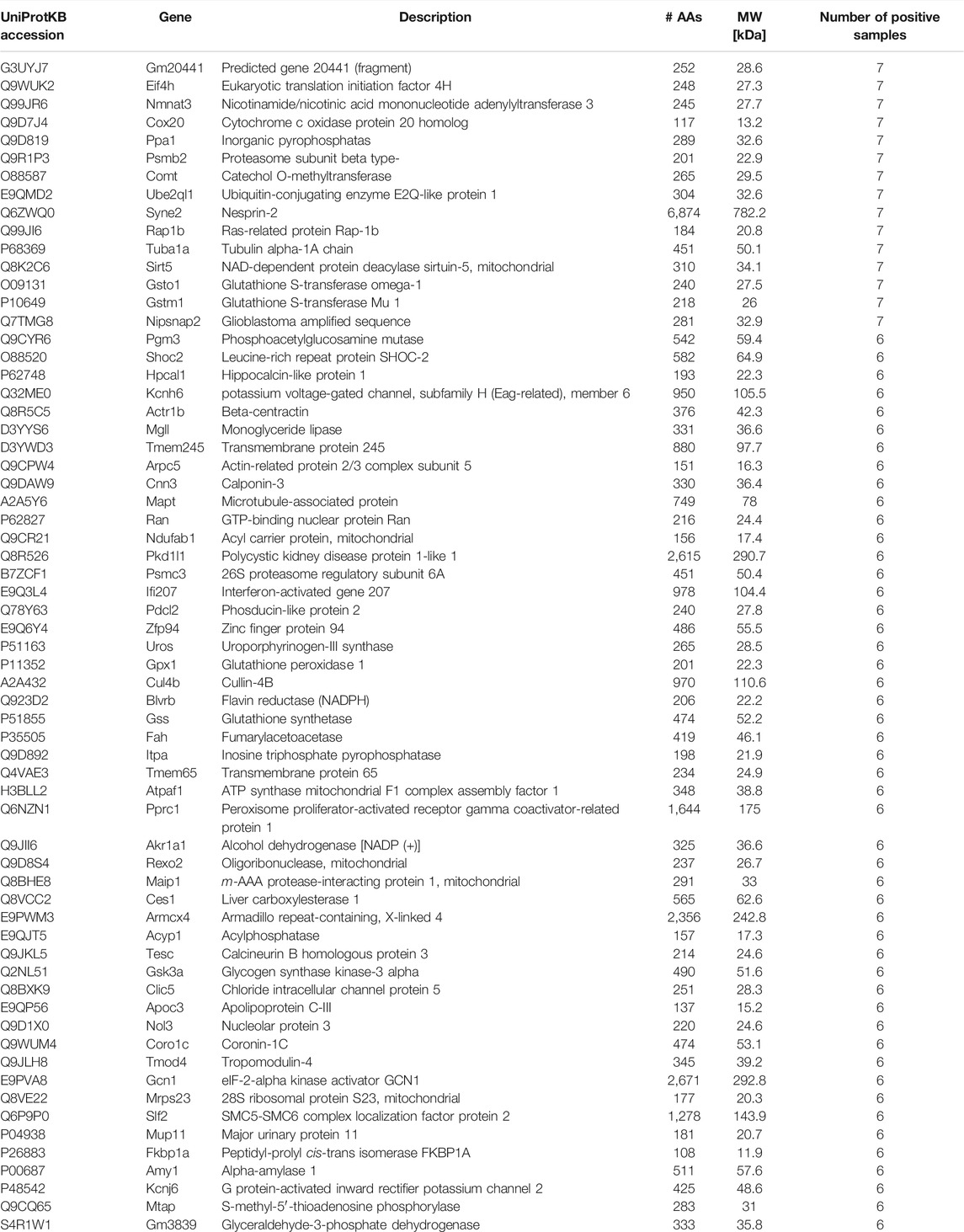
TABLE 1. Left ventricular tissue proteins identified only in samples of exercised mice. Proteins identified in ≥6 left ventricular tissue samples isolated from exercised animals but in none of the samples of the control group are listed. The table shows the UniProt name of the protein, main description, gene coding of the protein, number of amino acids (AAs), and molecular weight (MW).
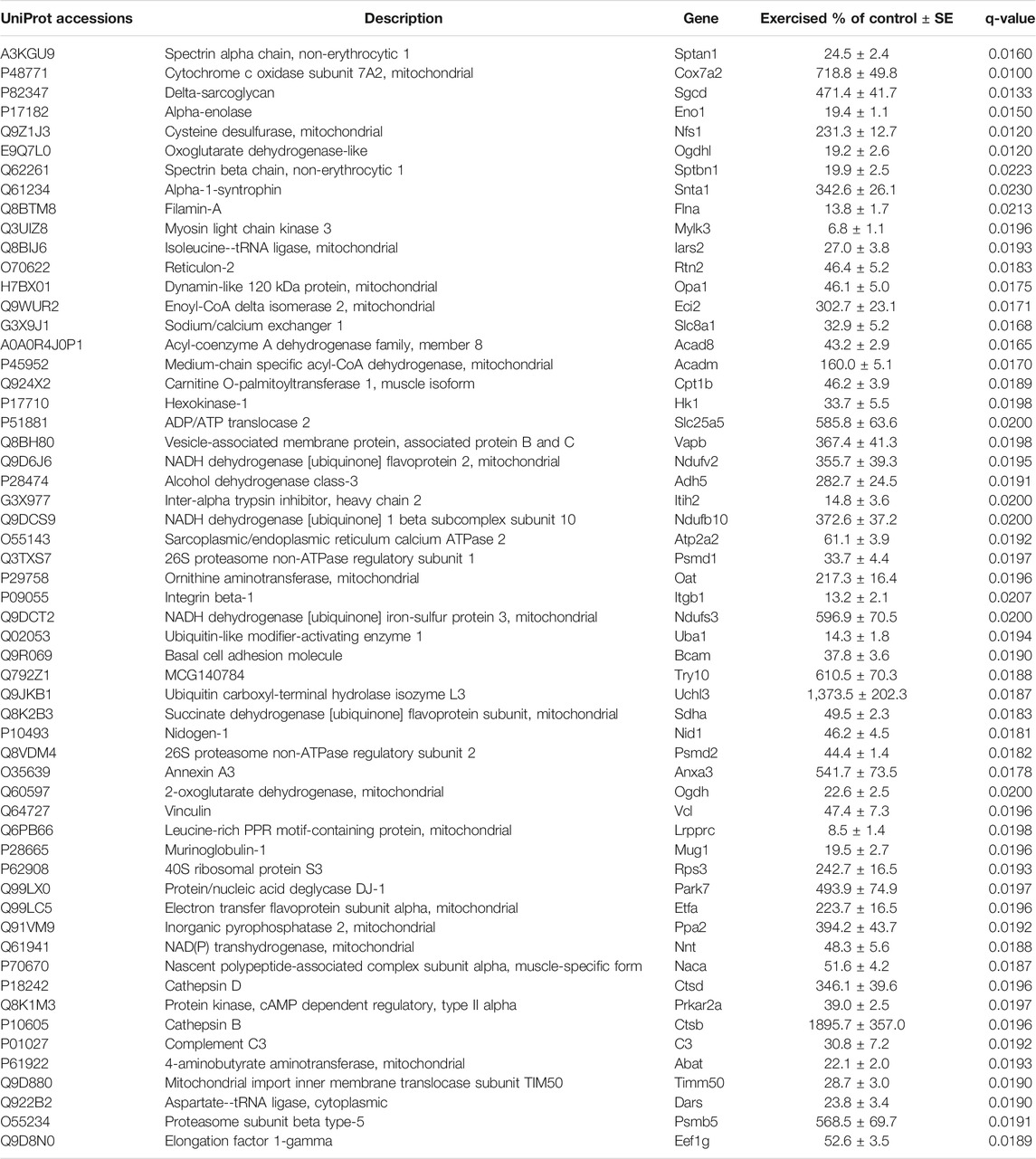
TABLE 2. Left ventricular tissue proteins ubiquitously identified in both groups but altered with exercise. Only proteins upregulated in ≥6 samples in each group are listed.
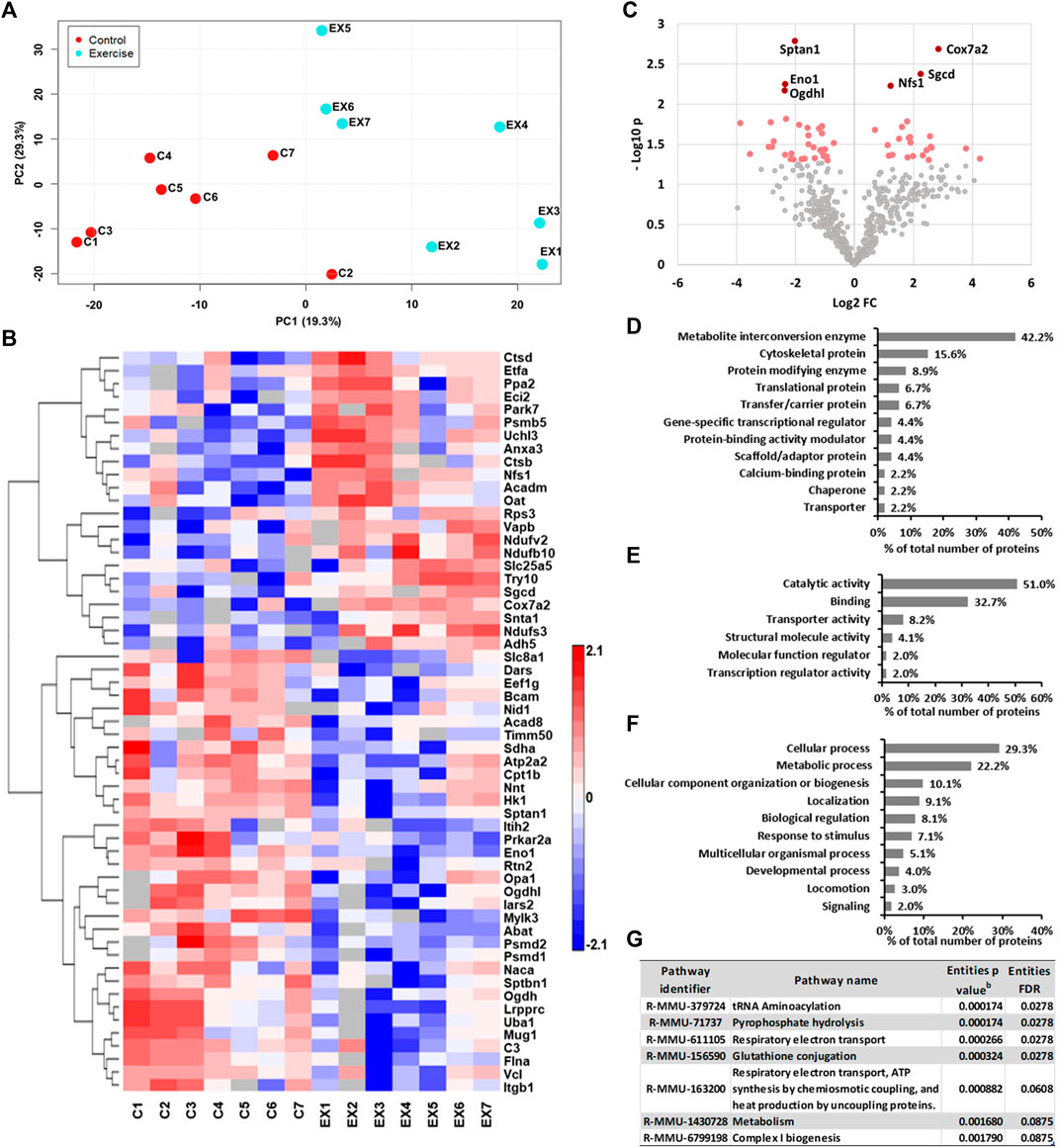
FIGURE 1. (A) PLS scatter plot showing the difference between control and exercised mice. (B) Heat map of semiquantitative assessment of ventricular proteins upregulated/downregulated with exercise. (C) Volcano plot of differentially expressed left ventricular tissue proteins between the control and exercised groups. The significantly upregulated and downregulated proteins are indicated with red dots, while those with p < 0.01 are indicated with dark red dots. (D–F) Repartition of upregulated proteins identified in the left ventricle of the heart tissues of exercised mice by (D) protein classes, (E) molecular functions, and (F) biological processes using the bioinformatics algorithms of the PANTHER classification system (version 15.0). (G) Biological pathways regulated by proteins altered by exercise. Proteins upregulated in ≥6 samples were analyzed using the Reactome database for Mus musculus, and only those with Entities FDR <0.1 are shown. bp value: represents the probability that the overlap between the query has occurred by1 chance.
Furthermore, all proteins upregulated or identified only in samples of exercised mice were classified into several major classes of proteins (Figures 1D–F). Only those classes represented with at least 3 different proteins are shown in Figure 1D. Exercise increases the metabolic rate in order to meet the energy demand. Thus, as predicted, more than 40% of proteins belong to the metabolite interconversion enzyme class. Also, close to 16% of proteins upregulated with swimming exercise belong to the cytoskeletal protein class (Figure 1D).
To understand the effect of exercise on the transcription of proteins, the identified proteins specified above in the ventricular samples of exercised mice were searched for their known molecular function (Figure 1E) and biological processes (Figure 1F), using the PANTHER classification system. The molecular function of these proteins was mostly involved in catalytic activity (51.0%) and binding (32.7%) (Figure 1E). The cellular processes were primarily classified as various cellular processes (29.3%) and metabolic processes (22.2%). Additionally, more than 10% of proteins have a role in biological regulation and cellular component organization or biogenesis (Figure 1F).
The pathway analysis of the ventricular proteins altered with swimming exercise using Reactome (Figure 1G) suggested that significantly affected pathways were tRNA aminoacylation, pyrophosphate hydrolysis, glutathione conjugation, and respiratory electron transport (Entities FDR <0.03 for all).
Exercise is marked, among others, with elevated ROS production that can lead to peroxidation of lipids, yielding biologically active reactive aldehyde 4-HNE (Al-Menhali et al., 2020). Previously, it was shown that 4-HNE regulates metabolism in the skeletal muscle cells either directly or by adducting to proteins (Al-Menhali et al., 2020). In the present work, modification of proteins by 4-HNE was determined to gain insight into the effects of long-term intensive swimming exercise on the left ventricle and on the potential signaling role of 4-HNE (Table 3). Thirteen different proteins were found to be prone to exercise-induced 4-HNE modifications, compared to eight proteins in the control group. Among the identified 4-HNE–modified proteins, only three proteins were found to be commonly modified for both groups. Those proteins are myoglobin, sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (SERCA2), and laminin subunit alpha-1.
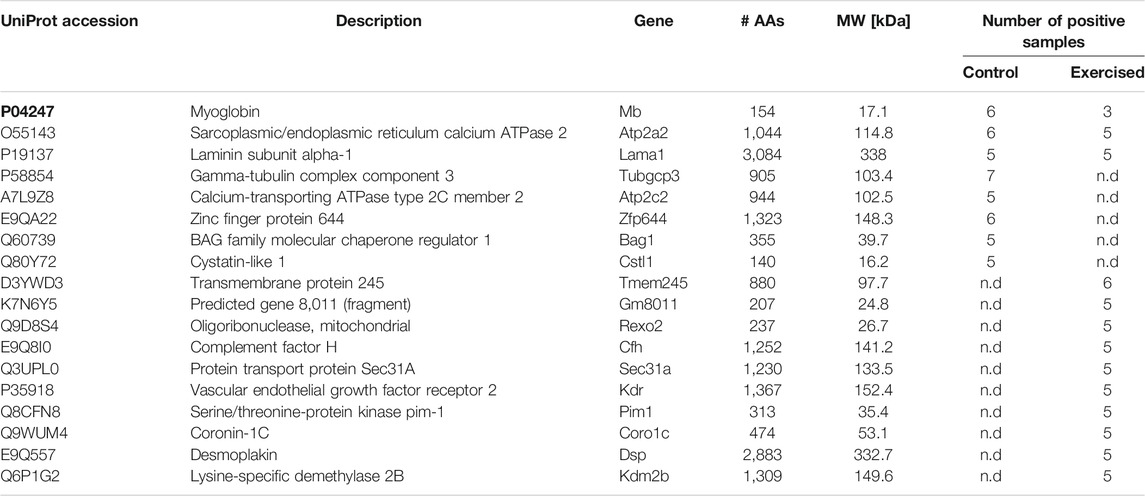
TABLE 3. 4-HNE–modified proteins in the left ventricular samples of the control and exercised groups. The table shows the UniProt accession number, main description, gene coding of the protein, number of amino acids (AAs), molecular weight (MW), and number of samples in the control or exercised group having the same protein.
In the past decade, significant advances have been made to understand the effects of exercise on the heart. This study aimed to identify alterations in the left ventricle proteome after long-term intensive swimming exercise training. Proteomics depends mainly on mass spectrometry, and by the use of high-resolution instruments and appropriate search engines, changes in the proteome can be detected (Al-Thani et al., 2018). Our earlier study on the erythrocyte proteome demonstrated that besides information on the comparative and quantitative proteome changes, high resolution mass spectrometry is also a vital tool for redox proteomics (Al-Thani et al., 2018). In addition, we have demonstrated that redox modifications of proteins can alter their function (Ludtmann et al., 2018). 4-HNE modification of proteins is one of the common types of oxidative modifications, and the preferred mass spectrometry methods for the detection and identification of proteins modified by 4-HNE use neutral loss-triggered electron capture dissociation tandem mass spectrometry (Rauniyar et al., 2009), collision-induced dissociation (CID), and electron transfer dissociation (ETD) MS/MS (Fritz et al., 2012). We therefore employed a high-resolution tribrid mass spectrometer using HCD and ETD fragmentation methods to investigate changes in the proteome of the left ventricle of the heart, including the identification of 4-HNE–modified proteins, of control and exercised mice. According to the study published several months ago, it is possible that lowering the value for the mass resolution of the mass spectrometer would lead to better protein identification (Sadygov, 2020). In addition, proteins with modifications that were not specified in the database search may result in the lack of protein identification. This study identified 37 proteins upregulated with exercise in addition to 65 proteins identified only in exercised mice samples and also demonstrated a distinct proteome signature of the hearts of exercised and sedentary animals. Indeed, long-term endurance exercise triggers significant physiological and autonomic adaptations to maintain the cardiac function (Rivera-Brown and Frontera, 2012; Gourine and Ackland, 2019). Several studies have demonstrated the effect of endurance exercise–induced transcriptional regulations with regard to increasing mitochondrial biogenesis. The training protocol used in this study increases mitochondrial biogenesis, mitochondrial volume density and number, and both basal and insulin-stimulated glucose uptake in the left ventricle (Vettor et al., 2014). In line with our previous work using this training protocol model (D’Souza et al., 2017; D’Souza et al., 2014; Bidaud et al., 2020), the training efficiency in this study was also evidenced by the development of phenotypic characteristics of the ‘athletes heart,’ including ventricular hypertrophy determined by echocardiography and sinus bradycardia determined by ECG. The cardiac response to swim training observed in this study was similar to that produced by treadmill running in rats where the training status was assessed by measuring the maximal oxygen uptake (VO2max) (D'Souza et al., 2017). This study confirmed that more than 10% of proteins identified only after exercise or upregulated proteins in the ventricular tissue of exercised animals have a role in biological regulation and cellular component organization or biogenesis. Moreover, the transcriptional coactivator peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α), which is a master regulator of mitochondrial biogenesis, is found to be activated in the hearts of mice after endurance swimming exercise (Boström et al., 2010). In addition, we also found peroxisome proliferator-activated receptor gamma coactivator-related protein one in the exercised group but not in the control group. During exercise, the continuous energy demand is regulated with adequate oxygen supply to support oxidative phosphorylation (Rivera-Brown and Frontera, 2012). This study identified a number of proteins significantly specific to the left ventricle of the exercised group that are directly involved in the mitochondrial electron transport, including acyl carrier protein, ATP synthase mitochondrial F1 complex assembly factor 1, NADH dehydrogenase [ubiquinone] flavoprotein 2, NADH dehydrogenase [ubiquinone] one beta subcomplex subunit 10, NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, cytochrome c oxidase subunit 7A2, cytochrome c oxidase protein 20 homolog, and electron transfer flavoprotein subunit alpha. This is in agreement with earlier studies that reported exercise-induced remodeling of the cardiac mitochondrial proteome (Sun et al., 2008; Kavazis et al., 2009), along with exercise-induced reprogramming of the cardiac mitochondrial phosphoproteome (Ferreira et al., 2014), suggesting that these changes contribute to the cardioprotective phenotype. Mitochondria are one of the main sites of intracellular ROS production and, thus, the main contributors to exercise-induced oxidative stress. Exercise-induced oxidative stress could lead to elevated 4-HNE modulating various cellular processes. Acute 4-HNE exposure can lead to transient adaptation (Al-Menhali et al., 2020), while 4-HNE accumulation, reaching larger concentrations, can have negative effects on cells (Elrayess et al., 2017; Jaganjac et al., 2017). We have found that the glutathione conjugation pathway which represents the main route for 4-HNE detoxification is upregulated with exercise. The glutathione system is among the major endogenous antioxidants (Jaganjac et al., 2020b), and also, the conjugation of the highly reactive carbonyl group in 4-HNE with reduced glutathione is one of the main routes for 4-HNE removal (Esterbauer et al., 1991; Dalleau et al., 2013). Enzymatic reactions can accelerate 4-HNE conjugation via enzymes including glutathione-S-transferases (GSTs), alcohol dehydrogenases, and aldehyde dehydrogenases (Chapple et al., 2013). Here, we report that GSTs and alcohol dehydrogenases are upregulated by exercise, suggesting elevated conjugation of 4-HNE. However, the cardiac tissue of the exercised group only had a minor increase in the number of 4-HNE–modified proteins after 5 months of swimming compared to the control group. Studies in animals have shown that long-term exercise reduces protein carbonyls in the plasma and gastrocnemius muscles’ mitochondria (Huertas et al., 2017) and liver mitochondria (Lima et al., 2013), while no differences were found in cardiac tissue (Vieira-Souza et al., 2021). This could be due to cellular adaptation triggered by long-term endurance exercise, which, among others, cases the formation of mitochondrial supercomplexes (Greggio et al., 2017; Huertas et al., 2017), which modulate the generation of mitochondrial ROS, preventing an increase in lipid peroxidation (Barranco-Ruiz et al., 2017). Contrarily, as acute exercise-induced oxidative stress results in elevated oxidation of lipids yielding reactive aldehydes (Tofas et al., 2020), it would probably lead to identification of a larger number of 4-HNE–modified proteins. The proteins myoglobin, SERCA2, and laminin subunit alpha-1, modified by 4-HNE, were detected in the samples of both the control and exercise groups, advocating an important physiological role of 4-HNE. Interestingly, an earlier study on control and diabetic mice hearts, by using Western blot analysis, reported that 4-HNE does not form adducts with SERCA2 (Moore et al., 2013). This discrepancy could be due to the different method sensitivities as the MS/MS ESI IonTrap sensitivity of the Orbitrap Fusion is 100 fg, compared to nanograms for the Western blot used in the earlier study. Moreover, several residues of sarcoplasmic reticulum (SR) vesicles are targets for 4-HNE, and 4-HNE modification of the protein inhibits maximal ATPase activity of SERCA1a. However, the inhibition effect was achieved by using high 4-HNE concentrations (hundreds of μM), which might not reflect the physiological levels of 4-HNE (Hortigón-Vinagre et al., 2011).
Among the proteins modified by 4-HNE in the exercised group were mitochondrial oligoribonuclease and serine/threonine-protein kinase pim-1, both required for the maintenance of the mitochondrial structure and function (Bruni et al., 2013; Din et al., 2013). Indeed, our recent work has demonstrated an important function of 4-HNE on the regulation of mitochondrial metabolism and cellular energy production as well as in inducing adaptation when present at physiological levels (23). In addition, calcium-dependent upregulation of the mitochondrial metabolism involves lipid peroxidation and, in particular, 4-HNE, which might explain 4-HNE modification of calcium-dependent protein binding protein transport protein Sec31A. Exercise-induced 4-HNE modification of desmoplakin and lysine-specific demethylase 2B could suggest 4-HNE involvement in cardiac development (Garcia-Gras et al., 2006). 4-HNE modification of coronin-1C, also observed in this study, was reported to have a role in phagocytosis (Chacko et al., 2016), which is increased by exercise. Vascular endothelial growth factor receptor 2 (VEGFR2) is another protein modified by 4-HNE in the left ventricles of trained mice. Low levels of 4-HNE (0.5–5 μM) upregulate VEGF in retinal endothelial cells and were suggested to promote angiogenesis (Vatsyayan et al., 2012); however, at high levels, 4-HNE (75 μM) was shown to downregulate the expression of VEGFR2, decreasing angiogenesis (Roy and Palaniyandi, 2020). Still, the underlying mechanisms of the role of 4-HNE in the angiogenic response to physical activity via the VEGF pathway (Delavar et al., 2014) are yet to be elucidated
In summary, this study highlights the significant effect of exercise training on mitochondrial biogenesis and the antioxidant defense system. Among the biochemical pathways regulated by proteins upregulated after long-term intensive exercise are respiratory electron transport and glutathione conjugation, while identification of proteins modified by 4-HNE suggest its role in adaptation. These data provide further evidence for the adaptive and beneficial role of physical exercise in maintaining cardiovascular health.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/pride/archive/, PXD026558.
The animal study was reviewed and approved by the Institutional Animal Welfare and Ethical Review Committee of the University of Manchester (PBA6A87CA).
MJ, AG, AA, and AD’S designed the study. CA and AD’S performed the animal experiments and collected the samples. ASA-M and MJ performed the proteomics experiments and analyzed the data. AA and MJ supervised the progress of the proteomics work. ASA-M wrote the initial draft of the article with the support of MJ. ASA-M, AG, AD, and MJ prepared the final version of the manuscript. All authors approved the final version of the article.
This work was supported by Anti-Doping Lab Qatar and the British Heart Foundation (to AVG; Ref: RG/19/5/34463).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Al-Menhali, A. S., Banu, S., Angelova, P. R., Barcaru, A., Horvatovich, P., Abramov, A. Y., et al. (2020). Lipid Peroxidation Is Involved in Calcium Dependent Upregulation of Mitochondrial Metabolism in Skeletal Muscle. Biochim. Biophys. Acta (Bba) - Gen. Subjects 1864 (3), 129487. doi:10.1016/j.bbagen.2019.129487
Al-Thani, A. M., Voss, S. C., Al-Menhali, A. S., Barcaru, A., Horvatovich, P., Al Jaber, H., et al. (2018). Whole Blood Storage in CPDA1 Blood Bags Alters Erythrocyte Membrane Proteome. Oxid Med. Cel Longev 2018, 6375379. doi:10.1155/2018/6375379
Barranco-Ruiz, Y., Martínez-Amat, A., Casals, C., Aragón-Vela, J., Rosillo, S., Gomes, S. N., et al. (2017). A Lifelong Competitive Training Practice Attenuates Age-Related Lipid Peroxidation. J. Physiol. Biochem. 73 (1), 37–48. doi:10.1007/s13105-016-0522-4
Beneš, H., Vuong, M. K., Boerma, M., McElhanon, K. E., Siegel, E. R., and Singh, S. P. (2013). Protection from Oxidative and Electrophilic Stress in the Gsta4-Null Mouse Heart. Cardiovasc. Toxicol. 13 (4), 347–356. doi:10.1007/s12012-013-9215-1
Bidaud, I., D'Souza, A., Forte, G., Torre, E., Greuet, D., Thirard, S., et al. (2020). Genetic Ablation of G Protein-Gated Inwardly Rectifying K+ Channels Prevents Training-Induced Sinus Bradycardia. Front. Physiol. 11, 519382. doi:10.3389/fphys.2020.519382
Boström, P., Mann, N., Wu, J., Quintero, P. A., Plovie, E. R., Panáková, D., et al. (2010). C/EBPβ Controls Exercise-Induced Cardiac Growth and Protects against Pathological Cardiac Remodeling. Cell 143 (7), 1072–1083. doi:10.1016/j.cell.2010.11.036
Bruni, F., Gramegna, P., Oliveira, J. M., Lightowlers, R. N., and Chrzanowska-Lightowlers, Z. M. (2013). REXO2 Is an Oligoribonuclease Active in Human Mitochondria. PLoS One 8 (5), e64670. doi:10.1371/journal.pone.0064670
Burniston, J. G., and Hoffman, E. P. (2011). Proteomic Responses of Skeletal and Cardiac Muscle to Exercise. Expert Rev. Proteomics 8 (3), 361–377. doi:10.1586/epr.11.17
Carini, M., Aldini, G., and Facino, R. M. (2004). Mass Spectrometry for Detection of 4-Hydroxy-Trans-2-Nonenal (HNE) Adducts with Peptides and Proteins. Mass. Spectrom. Rev. 23 (4), 281–305. doi:10.1002/mas.10076
Chacko, B. K., Wall, S. B., Kramer, P. A., Ravi, S., Mitchell, T., Johnson, M. S., et al. (2016). Pleiotropic Effects of 4-hydroxynonenal on Oxidative Burst and Phagocytosis in Neutrophils. Redox Biol. 9, 57–66. doi:10.1016/j.redox.2016.06.003
Chapple, S. J., Cheng, X., and Mann, G. E. (2013). Effects of 4-hydroxynonenal on Vascular Endothelial and Smooth Muscle Cell Redox Signaling and Function in Health and Disease. Redox Biol. 1 (1), 319–331. doi:10.1016/j.redox.2013.04.001
D'Souza, A., Bucchi, A., Johnsen, A. B., Logantha, S. J., Monfredi, O., Yanni, J., et al. (2014). Exercise Training Reduces Resting Heart Rate via Downregulation of the Funny Channel HCN4. Nat. Commun. 5, 3775. doi:10.1038/ncomms4775
D'Souza, A., Pearman, C. M., Wang, Y., Nakao, S., Logantha, S. J. R. J., Cox, C., et al. (2017). Targeting miR-423-5p Reverses Exercise Training-Induced HCN4 Channel Remodeling and Sinus Bradycardia. Circ. Res. 121 (9), 1058–1068. doi:10.1161/CIRCRESAHA.117.311607
Dalleau, S., Baradat, M., Guéraud, F., and Huc, L. (2013). Cell Death and Diseases Related to Oxidative Stress:4-Hydroxynonenal (HNE) in the Balance. Cell Death Differ 20 (12), 1615–1630. doi:10.1038/cdd.2013.138
Delavar, H., Nogueira, L., Wagner, P. D., Hogan, M. C., Metzger, D., and Breen, E. C. (2014). Skeletal Myofiber VEGF Is Essential for the Exercise Training Response in Adult Mice. Am. J. Physiology-Regulatory, Integr. Comp. Physiol. 306 (8), R586–R595. doi:10.1152/ajpregu.00522.2013
Din, S., Mason, M., Völkers, M., Johnson, B., Cottage, C. T., Wang, Z., et al. (2013). Pim-1 Preserves Mitochondrial Morphology by Inhibiting Dynamin-Related Protein 1 Translocation. Proc. Natl. Acad. Sci. 110 (15), 5969–5974. doi:10.1073/pnas.1213294110
Elrayess, M. A., Almuraikhy, S., Kafienah, W., Al-Menhali, A., Al-Khelaifi, F., Bashah, M., et al. (2017). 4-hydroxynonenal Causes Impairment of Human Subcutaneous Adipogenesis and Induction of Adipocyte Insulin Resistance. Free Radic. Biol. Med. 104, 129–137. doi:10.1016/j.freeradbiomed.2017.01.015
Esterbauer, H., Schaur, R. J., and Zollner, H. (1991). Chemistry and Biochemistry of 4-hydroxynonenal, Malonaldehyde and Related Aldehydes. Free Radic. Biol. Med. 11 (1), 81–128. doi:10.1016/0891-5849(91)90192-6
Ferreira, R., Vitorino, R., Padrão, A. I., Espadas, G., Mancuso, F. M., Moreira-Gonçalves, D., et al. (2014). Lifelong Exercise Training Modulates Cardiac Mitochondrial Phosphoproteome in Rats. J. Proteome Res. 13 (4), 2045–2055. doi:10.1021/pr4011926
Frasier, C. R., Moore, R. L., and Brown, D. A. (2011). Exercise-induced Cardiac Preconditioning: How Exercise Protects Your Achy-Breaky Heart. J. Appl. Physiol. 111 (3), 905–915. doi:10.1152/japplphysiol.00004.2011
Fritz, K. S., Kellersberger, K. A., Gomez, J. D., and Petersen, D. R. (2012). 4-HNE Adduct Stability Characterized by Collision-Induced Dissociation and Electron Transfer Dissociation Mass Spectrometry. Chem. Res. Toxicol. 25 (4), 965–970. doi:10.1021/tx300100w
Garcia-Gras, E., Lombardi, R., Giocondo, M. J., Willerson, J. T., Schneider, M. D., Khoury, D. S., et al. (2006). Suppression of Canonical Wnt/-catenin Signaling by Nuclear Plakoglobin Recapitulates Phenotype of Arrhythmogenic Right Ventricular Cardiomyopathy. J. Clin. Invest. 116 (7), 2012–2021. doi:10.1172/jci27751
Gevaert, A. B., Adams, V., Bahls, M., Bowen, T. S., Cornelissen, V., Dörr, M., et al. (2020). Towards a Personalised Approach in Exercise-Based Cardiovascular Rehabilitation: How Can Translational Research Help? A 'call to Action' from the Section on Secondary Prevention and Cardiac Rehabilitation of the European Association of Preventive Cardiology. Eur. J. Prev. Cardiolog 27 (13), 1369–1385. doi:10.1177/2047487319877716
Gourine, A. V., and Ackland, G. L. (2019). Cardiac Vagus and Exercise. Physiology 34 (1), 71–80. doi:10.1152/physiol.00041.2018
Greggio, C., Jha, P., Kulkarni, S. S., Lagarrigue, S., Broskey, N. T., Boutant, M., et al. (2017). Enhanced Respiratory Chain Supercomplex Formation in Response to Exercise in Human Skeletal Muscle. Cel Metab. 25 (2), 301–311. doi:10.1016/j.cmet.2016.11.004
Haw, R., and Stein, L. (2012). Using the Reactome Database. Curr. Protoc. Bioinformatics Chapter 8 (Suppl. l), 8–7. doi:10.1002/0471250953.bi0807s38
Hortigón-Vinagre, M. P., Chardonnet, S., Montigny, C., Gutiérrez-Martín, Y., Champeil, P., and Henao, F. (2011). Inhibition by 4-hydroxynonenal (HNE) of Ca2+ Transport by SERCA1a: Low Concentrations of HNE Open Protein-Mediated Leaks in the Membrane. Free Radic. Biol. Med. 50 (11), 1700–1713. doi:10.1016/j.freeradbiomed.2011.01.030
Huertas, J. R., Al Fazazi, S., Hidalgo-Gutierrez, A., López, L. C., and Casuso, R. A. (2017). Antioxidant Effect of Exercise: Exploring the Role of the Mitochondrial Complex I Superassembly. Redox Biol. 13, 477–481. doi:10.1016/j.redox.2017.07.009
Jaganjac, M., Milkovic, L., Sunjic, S. B., and Zarkovic, N. (2020). The NRF2, Thioredoxin, and Glutathione System in Tumorigenesis and Anticancer Therapies. Antioxidants (Basel) 9 (11), 1151. doi:10.3390/antiox9111151
Jaganjac, M., Tirosh, O., Cohen, G., Sasson, S., and Zarkovic, N. (2013). Reactive Aldehydes-Ssecond Messengers of Free Radicals in Diabetes Mellitus. Free Radic. Res. 47, (S1), 39–48. doi:10.3109/10715762.2013.789136
Jaganjac, M., Almuraikhy, S., Al-Khelaifi, F., Al-Jaber, M., Bashah, M., Mazloum, N. A., et al. (2017). Combined Metformin and Insulin Treatment Reverses Metabolically Impaired Omental Adipogenesis and Accumulation of 4-hydroxynonenal in Obese Diabetic Patients. Redox Biol. 12, 483–490. doi:10.1016/j.redox.2017.03.012
Jaganjac, M., Cipak, A., Schaur, R. J., and Zarkovic, N. (2016). Pathophysiology of Neutrophil-Mediated Extracellular Redox Reactions. Front. Biosci. 21, 839–855. doi:10.2741/4423
Jaganjac, M., Milkovic, L., Gegotek, A., Cindric, M., Zarkovic, K., Skrzydlewska, E., et al. (2020). The Relevance of Pathophysiological Alterations in Redox Signaling of 4-hydroxynonenal for Pharmacological Therapies of Major Stress-Associated Diseases. Free Radic. Biol. Med. 157, 128–153. doi:10.1016/j.freeradbiomed.2019.11.023
Kavazis, A. N., Alvarez, S., Talbert, E., Lee, Y., and Powers, S. K. (2009). Exercise Training Induces a Cardioprotective Phenotype and Alterations in Cardiac Subsarcolemmal and Intermyofibrillar Mitochondrial Proteins. Am. J. Physiol. Heart Circ. Physiol. 297 (1), H144–H152. doi:10.1152/ajpheart.01278.2008
Lima, F. D., Stamm, D. N., Della-Pace, I. D., Dobrachinski, F., de Carvalho, N. R., Royes, L. F., et al. (2013). Swimming Training Induces Liver Mitochondrial Adaptations to Oxidative Stress in Rats Submitted to Repeated Exhaustive Swimming Bouts. PLoS One 8 (2), e55668. doi:10.1371/journal.pone.0055668
Ludtmann, M. H. R., Angelova, P. R., Horrocks, M. H., Choi, M. L., Rodrigues, M., Baev, A. Y., et al. (2018). α-Synuclein Oligomers Interact with ATP Synthase and Open the Permeability Transition Pore in Parkinson's Disease. Nat. Commun. 9 (1), 2293. doi:10.1038/s41467-018-04422-2
Mi, H., Muruganujan, A., Huang, X., Ebert, D., Mills, C., Guo, X., et al. (2019). Protocol Update for Large-Scale Genome and Gene Function Analysis with the PANTHER Classification System (v.14.0). Nat. Protoc. 14 (8), 703–721. doi:10.1038/s41596-019-0128-8
Mohamed, S., Lamya, N., and Hamda, M. (2016). Effect of Maximal versus Supra-maximal Exhausting Race on Lipid Peroxidation, Antioxidant Activity and Muscle-Damage Biomarkers in Long-Distance and Middle-Distance Runners. Asian J. Sports Med. 7 (1), e27902. doi:10.5812/asjsm.27902
Moore, C. J., Shao, C. H., Nagai, R., Kutty, S., Singh, J., and Bidasee, K. R. (2013). Malondialdehyde and 4-hydroxynonenal Adducts Are Not Formed on Cardiac Ryanodine Receptor (RyR2) and Sarco(endo)plasmic Reticulum Ca2+-ATPase (SERCA2) in Diabetes. Mol. Cel Biochem. 376 (1–2), 121–135. doi:10.1007/s11010-013-1558-1
Morris, J. N., Heady, J. A., Raffle, P. A. B., Roberts, C. G., and Parks, J. W. (1953). Coronary Heart-Disease and Physical Activity of Work. Lancet 262 (6796), 1111–1120. doi:10.1016/s0140-6736(53)91495-0
Nakagawa, F., Morino, K., Ugi, S., Ishikado, A., Kondo, K., Sato, D., et al. (2014). 4-Hydroxy Hexenal Derived from Dietary N-3 Polyunsaturated Fatty Acids Induces Anti-oxidative Enzyme Heme Oxygenase-1 in Multiple Organs. Biochem. Biophysical Res. Commun. 443 (3), 991–996. doi:10.1016/j.bbrc.2013.12.085
Pamplona, R. (2011). Advanced Lipoxidation End-Products. Chem. Biol. Interact 192 (1–2), 14–20. doi:10.1016/j.cbi.2011.01.007
Perez-Riverol, Y., Csordas, A., Bai, J., Bernal-Llinares, M., Hewapathirana, S., Kundu, D. J., et al. (2019). The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res. 47 (D1), D442–D450. doi:10.1093/nar/gky1106
Polpitiya, A. D., Qian, W.-J., Jaitly, N., Petyuk, V. A., Adkins, J. N., Camp, D. G., et al. (2008). DAnTE: a Statistical Tool for Quantitative Analysis of -omics Data. Bioinformatics 24 (13), 1556–1558. doi:10.1093/bioinformatics/btn217
Rauniyar, N., Stevens, S. M., Prokai-Tatrai, K., and Prokai, L. (2009). Characterization of 4-Hydroxy-2-Nonenal-Modified Peptides by Liquid Chromatography−Tandem Mass Spectrometry Using Data-dependent Acquisition: Neutral Loss-Driven MS3 versus Neutral Loss-Driven Electron Capture Dissociation. Anal. Chem. 81 (2), 782–789. doi:10.1021/ac802015m
Rivera-Brown, A. M., and Frontera, W. R. (2012). Principles of Exercise Physiology: Responses to Acute Exercise and Long-Term Adaptations to Training. PM&R 4 (11), 797–804. doi:10.1016/j.pmrj.2012.10.007
Roof, S. R., Tang, L., Ostler, J. E., Periasamy, M., Györke, S., Billman, G. E., et al. (2013). Neuronal Nitric Oxide Synthase Is Indispensable for the Cardiac Adaptive Effects of Exercise. Basic Res. Cardiol. 108 (2), 332. doi:10.1007/s00395-013-0332-6
Roy, B., and Palaniyandi, S. S. (2020). Aldehyde Dehydrogenase 2 Inhibition Potentiates 4‐hydroxy‐2‐nonenal Induced Decrease in Angiogenesis of Coronary Endothelial Cells. Cell Biochem. Funct. 38 (3), 290–299. doi:10.1002/cbf.3468
Sadygov, R. G. (2020). High-resolution Mass Spectrometry for In Vivo Proteome Dynamics Using Heavy Water Metabolic Labeling. Int. J. Mol. Sci. 21 (21), 1–13. doi:10.3390/ijms21217821
Sun, B., Wang, J. h., Lv, Y. y., Zhu, S. s., Yang, J., and Ma, J. z. (2008). Proteomic Adaptation to Chronic High Intensity Swimming Training in the Rat Heart. Comp. Biochem. Physiol. D: Genomics Proteomics 3 (1), 108–117. doi:10.1016/j.cbd.2007.11.001
Tofas, T., Draganidis, D., Deli, C. K., Georgakouli, K., Fatouros, I. G., and Jamurtas, A. Z. (2020). Exercise-induced Regulation of Redox Status in Cardiovascular Diseases: The Role of Exercise Training and Detraining. Antioxidants (Basel) 9 (1). doi:10.3390/antiox9010013
Tyanova, S., and Cox, J. (2018). “Perseus: A Bioinformatics Platform for Integrative Analysis of Proteomics Data in Cancer Research,” in Cancer Systems Biology: Methods and Protocols [Internet]. Editor L von Stechow (New York, NY: Springer New York), 133–148. doi:10.1007/978-1-4939-7493-1_7
Tyanova, S., Temu, T., Sinitcyn, P., Carlson, A., Hein, M. Y., Geiger, T., et al. (2016). The Perseus Computational Platform for Comprehensive Analysis of (Prote)omics Data. Nat. Methods 13 (9), 731–740. doi:10.1038/nmeth.3901
Vatsyayan, R., Lelsani, P. C. R., Chaudhary, P., Kumar, S., Awasthi, S., and Awasthi, Y. C. (2012). The Expression and Function of Vascular Endothelial Growth Factor in Retinal Pigment Epithelial (RPE) Cells Is Regulated by 4-hydroxynonenal (HNE) and Glutathione S-transferaseA4-4. Biochem. Biophysical Res. Commun. 417 (1), 346–351. doi:10.1016/j.bbrc.2011.11.113
Vettor, R., Valerio, A., Ragni, M., Trevellin, E., Granzotto, M., Olivieri, M., et al. (2014). Exercise Training Boosts eNOS-dependent Mitochondrial Biogenesis in Mouse Heart: Role in Adaptation of Glucose Metabolism. Am. J. Physiol. Endocrinol. Metab. 306 (5), E519–E528. doi:10.1152/ajpendo.00617.2013
Vieira-Souza, L. M., dos Santos, J. L., Marçal, A. C., Voltarelli, F. A., Aidar, F. J., Miguel-Dos-Santos, R., et al. (2021). Biomarker Responses of Cardiac Oxidative Stress to High Intensity Interval Training in Rats. Motriz Rev. Educ. Fis 27 (1). doi:10.1590/s1980-65742021021420
Zarkovic, N., Cipak, A., Jaganjac, M., Borovic, S., and Zarkovic, K. (2013). Pathophysiological Relevance of Aldehydic Protein Modifications. J. Proteomics 92, 239–247. doi:10.1016/j.jprot.2013.02.004
Keywords: exercise, left ventricle, proteomics, oxidative stress, 4-hydroxynonenal
Citation: Al-Menhali AS, Anderson C, Gourine AV, Abramov AY, D’Souza A and Jaganjac M (2021) Proteomic Analysis of Cardiac Adaptation to Exercise by High Resolution Mass Spectrometry. Front. Mol. Biosci. 8:723858. doi: 10.3389/fmolb.2021.723858
Received: 11 June 2021; Accepted: 05 August 2021;
Published: 01 September 2021.
Edited by:
Amelia Palermo, University of California, Los Angeles, United StatesReviewed by:
Edward Lau, University of Colorado, United StatesCopyright © 2021 Al-Menhali, Anderson, Gourine, Abramov, D’Souza and Jaganjac. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Morana Jaganjac, bW9yYW5hLmphZ2FuamFjQGlyYi5ocg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.