- 1Department of Biology and Biotechnology “C. Darwin”, Istituto Pasteur Italia, Sapienza Università di Roma, Rome, Italy
- 2Department of Biochemical Sciences, Sapienza University, Rome, Italy
- 3The Institute of Scientific and Industrial Research (SANKEN), Osaka University, Osaka, Japan
- 4Department of Science and Technology on Food Safety, Kindai University, Kinokawa, Japan
Bacterial pathogens are able to survive within diverse habitats. The dynamic adaptation to the surroundings depends on their ability to sense environmental variations and to respond in an appropriate manner. This involves, among others, the activation of various cell-to-cell communication strategies. The capability of the bacterial cells to rapidly and co-ordinately set up an interplay with the host cells and/or with other bacteria facilitates their survival in the new niche. Efflux pumps are ubiquitous transmembrane transporters, able to extrude a large set of different molecules. They are strongly implicated in antibiotic resistance since they are able to efficiently expel most of the clinically relevant antibiotics from the bacterial cytoplasm. Besides antibiotic resistance, multidrug efflux pumps take part in several important processes of bacterial cell physiology, including cell to cell communication, and contribute to increase the virulence potential of several bacterial pathogens. Here, we focus on the structural and functional role of multidrug efflux pumps belonging to the Major Facilitator Superfamily (MFS), the largest family of transporters, highlighting their involvement in the colonization of host cells, in virulence and in biofilm formation. We will offer an overview on how MFS multidrug transporters contribute to bacterial survival, adaptation and pathogenicity through the export of diverse molecules. This will be done by presenting the functions of several relevant MFS multidrug efflux pumps in human life-threatening bacterial pathogens as Staphylococcus aureus, Listeria monocytogenes, Klebsiella pneumoniae, Shigella/E. coli, Acinetobacter baumannii.
Introduction
Efflux pumps are ubiquitous transmembrane transporters able to extrude a variety of toxic compounds, including drugs from the cell (Nikaido and Pagès, 2012; Du et al., 2018; Henderson et al., 2021). The association of efflux pumps (EPs) with drug resistance was first suggested about 4 decades ago (Ball et al., 1980; McMurry et al., 1980). Since then convincing evidence has accumulated that multidrug resistance (MDR) EPs represent a major system involved in bacterial resistance to antibiotics. As compared to other antibiotic resistance systems, the expression of a single MDR EP confers the cell simultaneous resistance to multiple drugs (Nikaido and Pagès, 2012; Blair et al., 2015).
MDR EPs are usually embedded in the inner membrane as a single-component transporter. In Gram-negative bacteria they can form a tripartite complex spanning the entire cell envelope. Tripartite EPs are constituted by an inner membrane transporter protein connected with an outer membrane factor (OMF) via a periplasmic adaptor protein (PAP) (Hinchliffe et al., 2013; Alav et al., 2021). MDR EPs have been identified within all major families of transporters, i.e., the Resistance Nodulation Division (RND), the ATP Binding Cassette (ABC), the Major Facilitator Superfamily (MFS), the Multidrug and Toxic Compound Extrusion (MATE), the Drug/Metabolite transporter (DMT), the Proteobacterial Antimicrobial Compound Efflux (PACE) family and the p-aminobenzoyl-glutamate Transporter (abgT) (Henderson et al., 2021). The main energy source utilized by MDR EPs is the proton motive force. Members of the ABC family rely on ATP hydrolysis (Du et al., 2018; Henderson et al., 2021) (Figure 1).
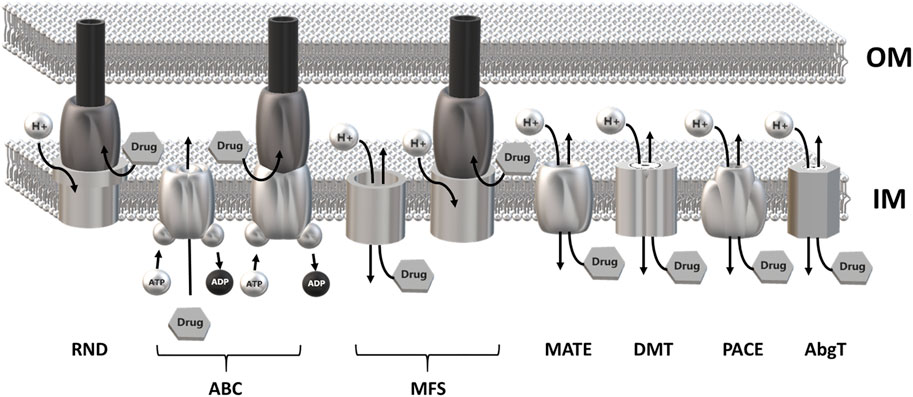
FIGURE 1. Schematic overview of the major families of multidrug efflux pumps. Most MDR EPs are single-component efflux transporter embedded within the inner membrane. Members of the RND family form tripartite efflux pumps comprising an inner membrane transporter, a periplasmatic adaptor protein and an outer membrane factor. Tripartite complexes are also present among members of the ABC and MFS superfamily. The outer membrane channel is usually constituted by TolC or a TolC-like protein. Most MDR EPs use the proton motive force of the inner membrane as energy source with the exception of the ABC superfamily which relies on ATP hydrolysis. RND, Resistance-Nodulation cell Division superfamily; ABC, ATP Binding Cassette superfamily; MFS, Major Facilitator Superfamily; MATE, Multidrug and Toxic Compound Efflux family; DMT, Drug/Metabolite Transporter superfamily; PACE, Proteobacterial Antimicrobial Compound Efflux family; AbgT, p-Aminobenzoyl-glutamate Transporter family.
The expression of MDR EPs is strictly regulated and under usual laboratory conditions their expression is often very low (Paulsen, 2003; Sun et al., 2014; Du et al., 2018). This is not surprising since, considering their ability to transport a variety of structurally unrelated molecules, an increased expression may adversely affect the bacterial physiology. Similar EP regulatory strategies are observed in different bacteria. They are based mainly on specific TetR-, Mar- and Mer-like regulators, on global effectors as nucleoid associated proteins, or on TCS acting in response to specific environmental stimuli (Grkovic et al., 2002; Eguchi et al., 2003; Du et al., 2018; Colclough et al., 2020). Overexpression of MDR EPs has been observed in clinical isolates and has been related to the emergence of strains highly resistant to antibiotic treatment (Piddock, 2006; Beceiro et al., 2013; Li et al., 2015). Emerging evidence indicates that the physiological role of MDR EPs is not limited to antibiotic export but rather spans a wider range of functions (Alvarez-Ortega et al., 2013; Leuzzi et al., 2015; Du et al., 2018; Pasqua et al., 2019b; Kumar et al., 2020). Current data suggest that MDR EPs are relevant to bacterial pathogenicity and survival in a specific ecological niche. In this context it is worth mentioning that MDR EPs contribute to inter-bacterial communication, to biofilm formation, and to the interactions with plant and animal cells (Alcalde-Rico et al., 2016; Alav et al., 2018; Pasqua et al., 2019b). As most genes encoding MDR EPs are highly conserved and reside within the bacterial core genome, the ability of MDR EPs to facilitate antibiotic resistance is currently regarded as a fortuitous by-product of an ancient physiological function (Saier et al., 1998; Neyfakh, 2002; Teelucksingh et al., 2020; Henderson et al., 2021). This is further stressed by the fact that, based on their antibiotic extruding activity, most MDR EPs are functionally redundant and operate on the same substrate (Tal and Schuldiner, 2009; Nikaido and Pagès, 2012; Henderson et al., 2021).
It is generally assumed that MDR EPs extrude exogenous toxic molecules, including antimicrobial compounds produced by competitors. Several studies have shown that MDR EPs can also extrude endogenous metabolites and that their expression can be induced by the very metabolite they export (Henderson et al., 2021). This capacity is highly relevant to prevent intracellular accumulation of cell waste and to establish successful interactions with the host cells in order to facilitate the survival in a new niche and its colonization. Several reports highlight the involvement of MDR EPs of the MFS family in the interaction of several bacterial pathogens with their target host cells (Bigi et al., 2004; Crimmins et al., 2008; Sahu et al., 2012a; Truong-Bolduc et al., 2014; Pérez-Varela et al., 2017; Pasqua et al., 2019a; Briaud et al., 2019). MFS transporters are found in all living organisms and constitute the largest and most diverse transporter family (Law et al., 2008; Quistgaard et al., 2016). In this review we present an overview on the structural and functional role of MFS MDR EPs, highlighting their contribution in the communication with host cells, in the development of virulence and in biofilm formation. This will be done by presenting the functions of several relevant MFS multidrug efflux pumps in human life-threatening bacterial pathogens as Listeria monocytogenes, Klebsiella pneumoniae, Staphylococcus aureus, Shigella/E. coli and Acinetobacter baumannii.
How Multidrug Resistance Efflux Pumps of the Major Facilitator Superfamily Export Molecules: Structure and Function
Despite a low degree of sequence identity, MFS transporters share a conserved structural organization, named the “MFS fold”, and a common transport mechanism that involves alternating access of the substrate binding-site to the cytoplasmic and periplasmic/extracellular side of the membrane with the transporter cycling between inward open, occluded and outward open conformations (Quistgaard et al., 2016). The MFS fold is a canonical 12 transmembrane segments (12-TM) fold consisting of two domains, known as the N- and the C-domains. Each domain is formed by 6 TM helices, connected by a cytoplasmic loop that is usually rather long and may contain an amphipathic helix (Quistgaard et al., 2016). In the N-domain, TM1, 2 and 3 are related to TM4, 5 and 6 by an approximate 180° rotation around an axis parallel to the membrane bilayer. A similar relationship is observed in the C-domain between TM7, 8, 9 and TM10, 11 and 12. In a subset of MFS transporters, belonging to the DHA2 (drug/H+ antiporter) and the POT (proton-dependent oligopeptide) subfamilies, this arrangement is modified by the addition of two TM segments as an insertion between the N- and C-terminal domains, giving rise to a 14-TM structure. Structural features of multidrug resistance MFS transporters are shown in Figure 2.
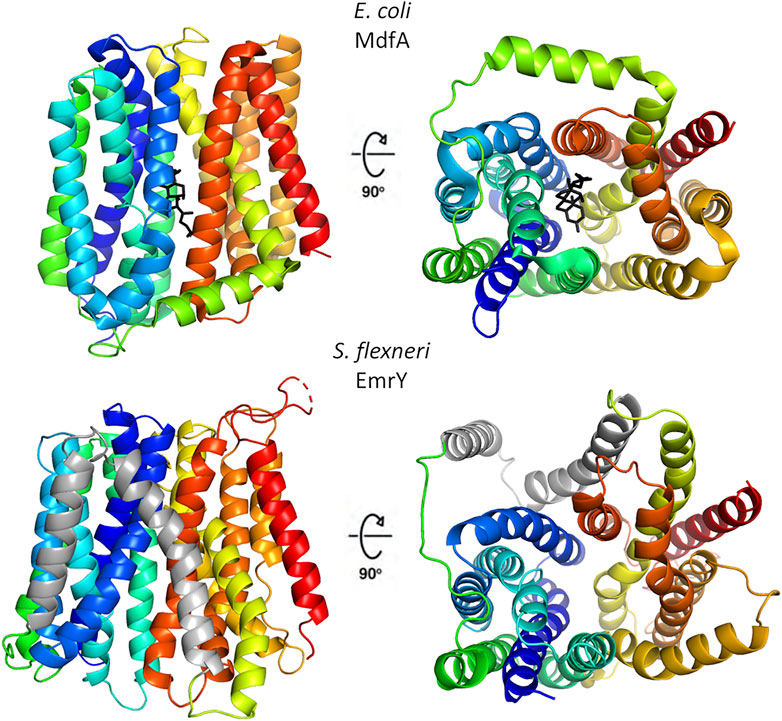
FIGURE 2. Structural organization of representative multidrug resistance MFS transporters of the DHA1 and DHA2 families. The upper panels show the 12-TM structure of E. coli MdfA in the inward open conformation (PDB 4zp0); in the right panel the protein is rotated by 90° to evidence the entrance to central cavity with bound chloramphenicol (black), the view is from the cytosolic side. The lower panels depict the 14-TM homology model of S. flexneri EmrY in the inward open conformation; the protein is shown in the same orientations of MdfA to facilitate comparison. The two additional TM helices are colored in gray. The model was obtained at the SWISS-MODEL server (Waterhouse et al., 2018) using a POT transporter (PDB 4IKV) as template. The sequence of EmrY employed as query lacks residues 434–473, which correspond to the loop connecting TM13 and TM 14 (represented as a broken line). The figure was produced with Pymol.
MFS efflux pumps involved in multidrug resistance are H+ antiporters of the DHA1 and DHA2 families, that couple proton translocation to drug extrusion (Du et al., 2018). DHA1 members generally exhibit 1:1 drug:H+ stoichiometries whereas transporters belonging to the DHA2 family exchange two protons with one substrate. While 3D structures have been determined for DHA1 transporters in different conformational states (Escherichia coli MdfA represents the prototypical EP of this group), no structure is available for DHA2 family members yet. Detailed structural studies of MdfA have allowed to gain in-depth information on the substrate-binding site and on the molecular mechanism of proton-coupled drug transport (Heng et al., 2015; Nagarathinam et al., 2018; Wu et al., 2020). In MFS transporters the substrate-binding site is generally found in a central cavity, with residues from both the N- and C-terminal domains contributing to ligand recognition. Polyspecificity of MDR transporters is achieved by means of a large substrate-binding pocket, which is able to accommodate compounds with different sizes and shapes. Further flexibility in multidrug recognition has been suggested to be conferred by the very low number of H-bonds established with the substrates. In MdfA substrate binding is mediated mainly through hydrophobic, van der Waals and polar contacts, which are geometrically much less stringent than H-bonds and likely allow structurally and chemically diverse drugs to adopt different orientations within the large central cavity (Wu et al., 2020). Conserved acidic residues E26 and D34 in motif-D located in TM1 are implicated in proton and substrate transport coupling, and it has been proposed that substrate-binding dependent changes in their protonation state trigger conformational changes required for transport (Heng et al., 2015). Recently, a model of the 14-TM MFS QacA, a DHA2 protein with a broad substrate repertoire that confers MDR to S. aureus, has been proposed (Majumder et al., 2019). The additional TM helices TM7 and 8, which form a V-shaped insertion between the N- and C-domains, have been demonstrated to be required for the function of the transporter in resistance to different toxic compounds. Moreover, the critical role of specific (de)protonatable acidic residues in TM1 and TM13 for the recognition and transport of cationic antibacterial compounds was revealed (Majumder et al., 2019). Structure-based sequence analyses indicate that many other multidrug MFS EPs share hydrophobic and large substrate-binding sites, in line with their ability to extrude lipophilic, cationic and neutral substrates.
The transport mechanism of MFS transporters has been described initially as a “rocker-switch” and then revised as a “clamp-and-switch” model in which the transition between the different states involves not only rigid-body rotations, but also structural changes in individual transmembrane helices (Quistgaard et al., 2016). In particular, kinking and twisting of TM5 in the outward open conformation of MdfA appears to be crucial for the transition from the inward open to the outward open state, together with reorganization of part of the hydrophobic core of the N-terminal domain and of a cytoplasmic loop of the C-terminal domain to close the cytoplasmic entrance (Nagarathinam et al., 2018). It is worth to point out that TM5 hosts motif-C (gxxxGPxxGGxl), a conserved sequence defined as the “antiporter motif”, where substitution of the proline residue leads to loss of function of MdfA (Heng et al., 2015). The MFS signature motif-A (GxLaDrxGrkxxxl) is located between TM2 and TM3 and plays a role in the stabilization of the outward open state (Henderson et al., 2021). Interestingly, motif-A appears not to be fully conserved in a set of DHA2 transporters comprising EmrY from S. flexneri, Tet38 from S. aureus and MdrM and MdrT from L. monocytogenes where the sequence deviates from the canonical arrangement (M.C. Bonaccorsi di Patti, unpublished observation), suggesting an unexpected flexibility in the mechanisms of these transporters. Other sequence motifs have been identified in MFS transporters of the DHA2 family, and are partly conserved in the MDR EPs cited above, including motif-D1 (lDxTvxnAlP) in TM1, with a probable role in proton-coupling mediated by the aspartate residue as in DHA1 transporters, and motif-H (WxwxFlINvPig) in TM6 with as-yet uncertain function (Henderson et al., 2021). A striking feature of MdfA, possibly shared by other MDR MFS transporters of DHA1 and DHA2 families, is a high degree of flexibility in the architecture of the substrate and H+ binding sites whereby the protein can accommodate multiple substitutions without significant loss of drug:H+ coupling (Wu et al., 2020). This finding might provide an explanation to the observation that in MDR EPs the typical sequence motifs described in MFSs appear to tolerate more variation in sequence (and possibly position) than expected.
Most MFS EPs act as single monomeric units. However, a limited number of tripartite systems have been identified which span the entire cell envelope in Gram-negative bacteria (Figure 1). In the latter case, the MFS component is connected to a periplasmic adaptor protein, which in turn is linked to TolC or to a TolC-like protein in the outer membrane to provide a continuous sealed channel for translocation of the substrate across the cell envelope (Alav et al., 2021). At variance with tripartite assemblies involving ABC and RND transporters, tripartite assemblies including MFSs are still poorly characterized from a structural point of view. Experimental 3D structures are available for single components belonging to different assemblies, such as the adaptor protein EmrA from Aquifex aeolicus (Hinchliffe et al., 2014) and the TolC-like outer membrane trimeric protein VceC from Vibrio cholerae (Federici et al., 2005). The MFS component of the system belongs to the 14-TM DHA2 family and it has been proposed that it may function as a dimer (Symmons et al., 2015). It is tempting to speculate that the two additional TMs typical of DHA2 transporters could contribute to dimerization of the MFS. Tripartite systems discussed in this review are represented by S. flexneri EmrKY and K. pneumoniae KpnGH. In both cases the outer membrane component is not encoded in the same operon as the periplasmic adaptor protein and the EP, and it is constituted by TolC. The periplasmic adaptor proteins EmrK and KpnG share homology to E. coli EmrA and by analogy they are predicted to be hexameric in the assembly, providing the molecular link between the EP in the inner membrane and TolC in the outer membrane. EmrY and KpnH are predicted to present a more than 40 amino acid-long extracellular loop between TM13 and TM14, which might be involved in interactions with EmrK and KpnG, respectively. However, this hypothesis should be viewed with caution because a similar long loop is present also in the single monomeric EP QacA of S. aureus.
Listeria monocytogenes: The Efflux Pumps MdrM and MdrT Play a Crucial Role in the Activation of the Host Cytosolic Surveillance Pathway
L. monocytogenes is a Gram-positive food pathogen responsible for high mortality in newborns as well as in pregnant women and in immunocompromised individuals. After invading phagocytic and non phagocytic cells, L. monocytogenes escapes the initial vacuole and spreads to adjacent cells by recruiting host actin filaments. In response to growing cytosolic bacteria host cells induce a type I interferon innate immune response. In particular, there is an enhanced expression and secretion of IFN-β (Pizarro-Cerdá and Cossart, 2018). An essential step in the induction of IFN-β is the release of cyclic-di-AMP (c-di-AMP) by L. monocytogenes (Woodward et al., 2010; Kaplan Zeevi et al., 2013). Several studies have shown that this molecule serves as an important second messenger influencing key processes in bacteria, such as sporulation, response to cell wall stress, peptidoglycan homeostasis, and genome surveillance (Yin et al., 2020). In bacteria, c-di-AMP is synthetized by diadenylate cyclase (DAC), using ATP as a substrate. Conversely, c-di-AMP is linearized to 5′-pApA by a specific phosphodiesterase (PDE) (Yin et al., 2020). Recent observations on the effect of c-di-AMP on intercellular communication highlight its relevance to the virulence of L. monocytogenes. In hepatocytes, a major reservoir of L. monocytogenes in systemic infections, it has been shown that c-di-AMP is recognized by the oxidoreductase RECON (Reductase Controlling NF-κB). This interaction determines the inhibition of RECON’s activity as repressor of NF-κB (McFarland et al., 2017). Augmented NF-kB activation increases the expression of nitric oxide synthase and, in turn, higher levels of nitric oxide favor spreading of L. monocytogenes in a variety of host cells (McFarland et al., 2018).
The role of MFS transporters in the interplay of Listeria monocytogenes with host cells was first evidenced in a wide genetic screening of mutants inducing altered host innate immune response in infected macrophages (Crimmins et al., 2008). Two EPs of the MFS superfamily, MdrM and MdrT, turned out to be involved in the cytosolic immune response in infected host cells. The MdrM and MdrT EPs are closely related to the well-studied MDR transporter QacA of S. aureus (Crimmins et al., 2008). They are expressed at low level and each is negatively controlled by a specific repressor (MarR and BtrA, respectively), located upstream of the EP gene (Crimmins et al., 2008; Quillin et al., 2011). L. monocytogenes strains defective in the mdrM or mdrT genes exhibit a significant reduction of several cytokines, including interferon-β (IFN-β), produced by infected host cells following infection (Crimmins et al., 2008; Woodward et al., 2010; Kaplan Zeevi et al., 2013). On the other hand, silencing of the cognate repressors MarR and BrtA has the opposite effect, triggering a strong host immune response. LC-MS analyses reveal a high level of c-di-AMP in supernatants of L. monocytogenes strains overexpressing MdrM and MdrT, suggesting that during intracellular growth of the bacterium these transporters favor the export of c-di-AMP (Woodward et al., 2010).
MDR transporters are often functionally redundant due to their overlapping substrate specificity. Genome analyses have shown that L. monocytogenes carries, besides mdrT, other genes (mdrA and mdrC) homologous to mdrM. These genes, collectively defined as MTAC transporters, are transcriptionally upregulated during intracellular growth and contribute to the type I interferon response by host cells (Kaplan Zeevi et al., 2013). While MTAC transporters are not required for intracellular growth in macrophages, infections in mice reveal that these EPs are active and contribute to bacterial virulence. Indeed, in infections with a DMTAC mutant a lower number of bacteria is found in livers and spleens of infected mice (Kaplan Zeevi et al., 2013). Besides triggering IFN-β production during bacterial infection, MTAC transporters play a role in the response to cell wall stress. In the presence of a sublethal concentration of vancomycin, L. monocytogenes DMTAC mutants are unable to increase peptidoglycan production, a drug titration mechanism normally employed by the bacterium. The involvement of MTAC transporters and c-di-AMP in the control of cell wall maintenance during stress conditions has been further demonstrated by altering the level of c-di-AMP. In a L. monocytogenes DMTAC mutant the increased expression of c-di AMP cyclase reduces the susceptibility to gentamycin, while over expression of c-di-AMP phosphodiesterase increases the sensitivity to the drug. MTAC transporters are also required for the synthesis of LTA under normal growth conditions, likely via c-di-AMP efflux. These data, together with the fact that mutants defective in LTA synthesis enhance the MdrM-dependent type I interferon response, lead to the hypothesis that MTAC transporters are involved in the management of LTA stress, and more generally in cell wall maintenance and in the interplay with host cells (Kaplan Zeevi et al., 2013).
The contribution of MdrM and MdrT is not limited to promoting the host innate immune response. These MFS EPs affect L. monocytogenes survival in the bile-rich niches the bacterium encounters during infection, i.e. liver, spleen and gallbladder. In particular, both mdrT and mdrM genes are strongly induced by cholic acid, an essential bile component and their deletion significantly attenuates bacterial virulence in vivo in livers of infected mice and in vitro in the presence of bile (Quillin et al., 2011). High induction of mdrT depends on the loss of the ability of the BrtA repressor to bind and repress the mdrT promoter in the presence of cholic acid. It is yet unclear whether this depends on cholic acid directly binding to the BtrA protein or perturbing the structure of BtrA because of its detergent properties.
Staphylococcus aureus: The Efflux Pump Tet38 as Key Element in the Interaction With Host Cell
S. aureus is a versatile pathogen, able to cause acute and chronic infections in animals and humans. Its arsenal of virulence factors and its large panel of MDR EPs enable it to survive within the host cell environment by resisting antibiotic therapy and escaping host defences. While S. aureus is not an obligate intracellular pathogen, it can invade and survive within non professional phagocytic cells such as epithelial and endothelial cells. After internalization and fusion with the lysosomes, S. aureus can replicate rapidly and then escape from the phagolysome (Garzoni et al., 2007). The Tet38 EP plays an essential role in the virulence of S. aureus, actively contributing to the invasion and survival of the bacteria within epithelial cells.
Tet38 is a single-peptide, chromosomally encoded MFS efflux pump, highly conserved among S. aureus strains. It confers resistance to several compounds, including tetracyclin, fosfomycin, glycerol-3-phosphate and some unsaturated fatty acids as palmitoleic and undecanoic acid (Truong-Bolduc et al., 2014). The Tet38 protein (48 KDa) has 14 predicted TM segments. Except for the positions of glutamate residues, the organization of the TM segments is similar to that observed in S. aureus TetK and B. subtilis TetL EPs, two MFS efflux pumps involved in the extrusion of tetracycline. The expression of tet38 is tightly regulated and is sensitive to stimuli from the host and from the outer environment. Two factors, MgrA and TetR21, negatively control tet38 expression. While TetR21 represses tet38 by binding specifically to its promoter, MgrA acts indirectly by binding the tetR21 promoter (Truong-Bolduc et al., 2005; Truong-Bolduc et al., 2015).
Evidence on the involvement of Tet38 in Staphylococcus pathogenesis came initially from the observation that the expression of tet38 and norB (another MFS EP) was selectively increased in abscesses (Ding et al., 2008) and that Tet38 contributed significantly to bacterial survival and replication in the abscess environment (Ding et al., 2008). In particular, the strong induction of Tet38 correlated with a high presence in abscesses of typical Tet38 fatty acid substrates as palmitoleic and undecanoid acid (Truong-Bolduc et al., 2014). Successive observations have revealed that colonization of the skin surface in a mouse model was strongly reduced in S. aureus tet38 mutants as compared to wt strains, and that the wt level was restored by ectopic expression of a functional Tet38 pump (Truong-Bolduc et al., 2014).
Tet38 affects several steps of the invasion process of S. aureus, including adhesion, internalization and trafficking in epithelial cells. Invasion assays of epithelial and endothelial cells have shown that Tet38 plays an essential role in the initial step of the internalization of S. aureus in host cells. Indeed, the absence of Tet38 strongly affects bacterial uptake (Truong-Bolduc et al., 2015). Using antibodies against the scavenger receptor CD36, it has been observed that the CD36 blockade reduced internalization of the wild type S. aureus, while it had no effects on tet38 deletion mutants (Truong-Bolduc et al., 2017). The dependence of internalization on CD36 of wild type S. aureus suggested a direct interaction between Tet38 and CD36. This interaction has been confirmed by column-retention assays with purified Tet38 and CD36 (Truong-Bolduc et al., 2019) and the Tet38 amino acid residues essential for S. aureus internalization have been recently identified (Truong-Bolduc et al., 2021). Truong-Bolduc et al. (2019) also demonstrated that Tet38 can bind in vitro CD36 in the heterodimeric CD36-TLR2 complex and form the heterotrimeric Tet38-CD36-TLR2 complex, suggesting that Tet38 might influence the sensitivity of TLR-2 to the presence of lipoteichoic acid (LTA), a bacterial component that actively participates to host cell invasion. Due to its localization within the inner membrane, Tet38 is surrounded by layers of peptidoglycan, LTA and teichoic acid (WTA). These authors also indicate that Tet38 hampers the activity of tunicamycin, an inhibitor of WTA synthesis. Indeed, resistance to tunicamycin increases with the upregulation of the tet38 gene while strains lacking Tet38 become fully sensitive to the drug. A similar behaviour has been reported with Congo Red (CR), an inhibitor of LTA synthesis. The involvement of Tet38 in these resistance phenotypes was further confirmed by observing that sensitivity to tunicamycin and CR is restored in the presence of the EP inhibitor reserpine.
The Tet38 EP also contributes to the subsequent step of the S. aureus infection in epithelial cells, in terms of both bacterial viability and trafficking in phagolysosomes. As compared to tet38 mutants, after the fusion of S. aureus-associated endosomes with lysosomes, the viability of wt cells is higher and a smaller number of bacteria is associated with phagolysosomes. Moreover, Tet38 favours the escape from the phagolysosome. Indeed, buffering the acidic phagolysosome environment with chloroquine rapidly increases both viability and lysosome escape of wt S. aureus while it has no significant effect on tet38 mutants (Truong-Bolduc et al., 2017).
Recently, it has been observed that during coinfection with S. aureus and Pseudomonas aeruginosa the expression of S. aureus Tet38 is strongly increased (Briaud et al., 2019). These bacteria occupy the same anatomical niche during airway infections in cystic fibrosis patients. In the initial phase of the infection, P. aeruginosa outcompetes S. aureus by producing specific growth inhibitory compounds. As the infection progresses towards a chronic state, the interaction between the two species evolves into coexistence. Transcriptomic studies using pairs of strains of both species reveal that the presence of P. aeruginosa induces a dysregulation of the S. aureus expression profile. In particular, during coexistence the predominant transcriptomic alteration is the upregulation of S. aureus tet38, caused by the reduced expression of the Tet38 MgrA repressor gene. This alteration leads to an increased antibiotic tolerance and to a higher uptake rate of S. aureus into epithelial pulmonary cells, two essential determinants of chronic infection (Briaud et al., 2019). Altogether, the evidence currently available indicates that the Tet38 EP is a potential target for new antibacterial strategies able to block the internalization of S. aureus, a crucial step in the recurrence and persistence of S. aureus infections.
The Efflux Pump Emrky of Shigella flexneri: The Crucial Role of Two Component Systems in Sensing and Responding to the Host Environment
Shigella is an intracellular pathogen that can invade and colonize the intestinal epithelium, resulting in severe inflammatory colitis (The et al., 2016; Pasqua et al., 2017). After being ingested by the host, Shigella reaches the colon and rectum, where it is endocytosed by M cells in the Peyer’s patches. The bacteria are then released into the intraepithelial pocket, where they invade resident macrophages. Once inside macrophages, Shigella disrupts the phagocytic membrane and multiplies within the cytosol. Then it induces macrophage death via pyroptosis. After being released by the dead macrophages, Shigella invades the intestinal epithelial cells, where it multiplies and disseminates in cells (Ashida et al., 2011). The ability to survive and replicate inside macrophages and intestinal epithelial cells is regarded as a key virulence feature of the bacterium.
In a recent attempt to investigate on the possible role of Shigella EPs in pathogen-host interactions, MDR EP encoding genes of S. flexneri M90T and E. coli K-12 were compared (Pasqua et al., 2019a). Only 14 out of 20 EP-encoding genes in E. coli were conserved in S. flexneri. Two MFS EPs (mdtG and mdtM) and one RND EP (acrEF) were absent from the S. flexneri genome, whereas three RND EPs (cusB, mdtABC, and mdtEF) were not functional due to gene disruption. Silencing of several EP encoding genes in S. flexneri is not surprising as in Shigella many housekeeping genes present in the commensal E. coli ancestor have been lost following a pathoadaptation process (Prosseda et al., 2012; Leuzzi et al., 2017). Following an extensive analysis of the expression of the 14 EPs of S. flexneri M90T, carried out in Shigella-infected human U937 monoblasts after differentiation into macrophage-like cells and in Shigella-infected Caco-2 epithelial cells, the transcriptional profiles were found to vary across EPs and between macrophages and epithelial cells. In particular, in macrophages expression of emrA, emrD, emrE, emrK, and mdtJ increased whereas that of acrA decreased. In Caco-2 cells acrD, emrD, emrE, macA, mdtA, mdtJ, ydhE, and yidY were overexpressed while acrA, emrA, and emrK showed a reduced level of expression (Pasqua et al., 2019a). Considering that under ordinary laboratory growth conditions E. coli constitutively expresses acrAB (and tolC) while most other EP-encoding genes are repressed (Sulavik et al., 2001), the decreased expression of Shigella acrA in macrophages and epithelial cells suggests that in the functioning MDR EP pool a shift from AcrAB to other pumps may have occurred. Interestingly, the Shigella emrK gene, which belongs to the emrKY operon, is strongly overexpressed in macrophages but is downregulated in Caco-2 epithelial cells (Pasqua et al., 2019a). In E. coli emrKY expression is activated in response to the coordinated action of a mild acidic environment and a high concentration of alkali metals, a combination which is sensed by the EvgA/EvgS TCS (Eguchi and Utsumi, 2014). Upregulation of Shigella emrK in macrophages can be likely assumed to depend on the same stimuli. Indeed, the uptake of Shigella by macrophages triggers a decrease of the intracellular pH favoring the activation of the emrKY by the EvgA regulator. Further analyses have revealed that deleting emrKY from S. flexneri M90T leads to a reduced growth rate in minimal medium (pH 6.0) supplemented with 0.2 M KCl, and to an intracellular growth disadvantage of the mutant in competition assays with the parental strain. This strongly suggests that EmrKY contributes to the fitness of Shigella in macrophages (Pasqua et al., 2019a).
EmrKY is a member of the MFS EP family. This pump was originally identified in E. coli by analyzing genes adjacent to the evgAS operon, which encodes the EvgS/EvgA TCS (Tanabe et al., 1997) (Figure 3). EmrKY functions as a tripartite EP with TolC as an outer membrane factor (Eguchi et al., 2003). Induction of emrKY expression in E. coli conferred resistance to deoxycholate but not to cholate, SDS, and several antimicrobial agents and dyes (Kato et al., 2000; Nishino and Yamaguchi, 2002). More recently, it has been reported that overexpression of tolC and emrKY enhanced amorphadiene transport, a precursor of the antimalarial drug artemisinin (Zhang et al., 2016). However, to the best of our knowledge, there is no additional information available about the substrates that are pumped out by EmrKY. E. coli cells with mutations in emrK and emrY are sensitive to mitomycin C, UV irradiation, and H2O2, which suggests that EmrKY eliminates toxic metabolites induced by DNA damage (Han et al., 2010). This may help understanding how EmrKY contributes to the survival of Shigella in macrophages. No data on the role of EmrKY in the virulence of other Enterobacteriaceae have been reported so far.
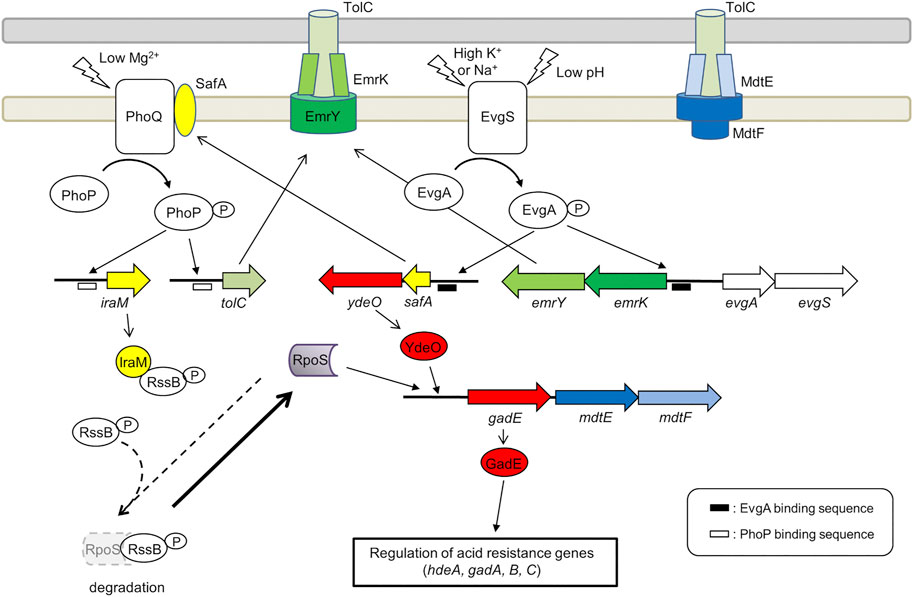
FIGURE 3. Role of the EvgA/EvgS two component system in the activation of MDR EP encoding genes emrKY and mdtEF. The figure shows the EvgA-YdeO-GadE cascade and the SafA-PhoQ/PhoP-IraM-RpoS network in E. coli. The genes are highly conserved between S. flexneri M90T and E. coli MG1655 (the amino acid sequence homology is 98–100%), except iraM, mdtE, and mdtF. The iraM gene shows 61.7% amino acid homology in S. flexneri M90T, but has 93.5% amino acid homology in S. sonnei. Although mdtE and mdtF are disrupted in S. flexneri, they are conserved in S. dysenteriae, S. boydii, and S. sonnei. EvgA-P, PhoP-P, YdeO, RpoS, and GadE positively regulate the target genes (indicated by arrows). EvgA-P and PhoP-P also induce the expression of gadE-mdtEF by directly binding to the gadE promoter.
The expression of emrKY in E. coli is induced by the EvgS/EvgA TCS, where EvgA, the response regulator, directly binds to the promoter region of emrKY (Kato et al., 2000). In addition to emrKY, EvgS/EvgA upregulates a network of acid resistance genes, through a cascade of EvgA-YdeO-GadE regulators (Masuda and Church, 2003; Itou et al., 2009). It positively controls also the SafA-PhoQ/PhoP-IraM-RpoS network, which is composed of two TCSs, two TCS-connecting small proteins, and a sigma factor (Eguchi et al., 2007; Eguchi et al., 2011) (Figure 3). The small membrane protein SafA directly binds and activates PhoQ at the inner membrane (Eguchi et al., 2012; Yoshitani et al., 2019) and the anti-adapter protein IraM, which is induced by activated PhoQ, binds to the response regulator RssB preventing it from interacting with RpoS, the general stress responding sigma factor (Bougdour et al., 2008). This protects RpoS from proteolysis by ClpXP and results in the accumulation of cellular RpoS during the exponential phase. Both PhoP and RpoS contribute to the upregulation of gadE, the major regulator of acid resistance genes. The upregulation of these genes confers severe acid resistance to exponentially growing E. coli cells (Itou et al., 2009). Notably, tolC is also upregulated by EvgS/EvgA via SafA and PhoQ/PhoP (Figure 3). EvgS/EvgA enhances the expression of both emrKY and tolC, contributing to the formation of EmrKY-TolC tripartite EP (Eguchi et al., 2003). Interestingly, all genes involved in the EvgA-YdeO-GadE cascade and in the SafA-PhoQ/PhoP- IraM-RpoS network are conserved in Shigella, suggesting that these systems may favor, among other processes, the expression of EPs relevant to the survival of Shigella in macrophages. Indeed, using a Shigella strain carrying a emrK-gfp fusion, it has been shown that in macrophages the fusion is not expressed in a evgA background, indicating that during infection the expression of emrK is induced by the activation of the EvgS/EvgA TCS (Pasqua et al., 2019a).
The crucial role of the EvgA/EvgS TCS in activating the EPs required for a successful interaction with host cells is further highlighted by the analysis of the expression profile of MDR EPs in adherent-invasive Escherichia coli (AIEC) (Fanelli et al., 2020). Similarly to Shigella, emrK expression in LF82, a strain isolated from Crohn disease lesions, is induced inside macrophages. However, unlike Shigella, emrK expression occurs also in AIEC-infected Caco-2 cells. The expression of mdtEF, which encodes a RND EP, is induced in AIEC cells inside macrophages but not inside epithelial cells. MdtEF is known to contribute to the survival of AIECs inside macrophages (Fanelli et al., 2020). As shown in Figure 3, mdtEF is co-transcribed with gadE as a gadE-mdtEF operon. EvgS/EvgA is one of the many systems that control the expression of this operon (Arcari et al., 2020) (Figure 3). It is assumed that the environment inside macrophages is acidic and oxidative, hosts toxic molecules such as antimicrobial peptides, causes genotoxic stress, and lacks important nutrients (Demarre et al., 2019). The acidic conditions and the lack of nutrients may serve as activating signals for the EvgS sensor. Indeed, deletion of evgAS significantly affects the ability of AIECs to survive and multiply within macrophages (Demarre et al., 2019), indicating that EvgS/EvgA is activated in macrophages. It should be noted that although mdtEF is disrupted in S. flexneri, the gadE-mdtEF operon is conserved in Shigella dysenteriae, Shigella boydii, and Shigella sonnei (BioCyc database collection, https://www.biocyc.org/). As shown in Figure 3, mdtEF and tolC expression regulated by the EvgA-YdeO-GadE cascade and the SafA-PhoQ/PhoP- IraM-RpoS network may contribute to the survival of these Shigella subspecies in macrophages.
Additional signals for activation of EvgS have been reported. Indole inhibits EvgS at concentrations that are typically found in the intestine (Boon et al., 2020). Anaerobic conditions also inhibit EvgS activity, and it has been proposed that EvgS activation requires oxidation in addition to mild acidic conditions (Inada et al., 2021). The current model suggests that EvgS/EvgA is silenced in the intestine due to the presence of indole and anaerobic conditions. EvgS/EvgA is activated after Shigella or AIEC invade macrophages, and thereafter induces the expression of EmrKY and other EPs. EvgS/EvgA is widely conserved within the E. coli species. Some reports have associated EvgS/EvgA to the pathogenicity of E. coli other than AIEC and Shigella. In the case of enteropathogenic E. coli, EvgA represses the expression of the type III secretion system responsible for delivering a set of effector proteins into the host cell cytoplasm (Nadler et al., 2006). In avian pathogenic E. coli the silencing of evgS attenuates lung colonization (Dziva et al., 2013). Moreover, a recent study has shown that EvgS/EvgA activation protects E. coli from gallium nitrate induced death. It has been suggested that EvgS/EvgA upregulated genes encode enzymes involved in ROS detoxification and in the glyoxylate shunt of the TCA cycle (Zeng et al., 2021). These results may provide clues to improve our understanding of how EmrKY contributes to survival within macrophages.
Building Biofilms in Acinetobacter baumannii: The Contribution of Major Facilitator Superfamily Efflux Pumps in Exporting Biofilm Key Molecules
In order to survive hostile conditions many microorganisms, including pathogens, are able to organize their communities as biofilms in response to different environmental stimuli (e.g., nutrient and metabolic signals, host-derived molecules and antibiotics) (Flemming et al., 2016). The biogenesis of a biofilm is a complex process that requires intense cell-to-cell communication through quorum sensing signals and is usually articulated into major steps: attachment of planktonic cells to living substrates or to abiotic surfaces (e.g., medical devices, domestic or industrial environments); cell proliferation, resulting in the formation of microcolonies; and eventually growth and maturation into a fully differentiated biofilm with self-production of an extracellular matrix, composed of lipids, nucleic acids, polysaccharides and proteins, that ensures the structural integrity of the film and its adhesion to surfaces (Karatan and Watnick, 2009; Flemming and Wingender, 2010). Bacteria embedded in a mature biofilm can further spread and colonize new surfaces through dispersion of individual cells (Flemming et al., 2016).
The ability to form biofilms is considered a major virulence factor in many bacterial human pathogens. A. baumannii represents an extensively studied microorganism whose interaction with the host is mainly based on biofilm formation (Harding et al., 2018). In clinical environments A. baumannii is considered a serious threat because of its so-called “persist and resist” strategy (Harding et al., 2018). It depends on the combination of two factors: a remarkable ability to persist under unfavorable conditions as dormant cells or biofilms and a marked propensity to develop multiple antibiotic resistances (Harding et al., 2018). This strategy, together with the expression of specific virulence factors, allows A. baumannii to establish severe infections characterized by high mortality, e.g., bloodstream and catheter-associated urinary infections, ventilator-associated pneumonia, and skin and soft tissue infections (Harding et al., 2018). Several EPs of the MFS and RND families contribute to the ability of A. baumannii to form biofilms and develop multi-drug resistant phenotypes (Alav et al., 2018; Pasqua et al., 2019b).
The first evidence on the involvement of an MFS EP in the biosynthesis of A. baumannii biofilm came from the analysis of the pmt gene in the highly multidrug resistant pathogenic strain AIIMS 7 (Sahu et al., 2012a). The pmt gene encodes a 17 KDa inner membrane MFS transporter which is highly expressed during adherence to a substrate. In particular, upregulation of pmt occurs mainly during the initial stages (24–48 h) of biofilm formation. The Pmt protein drives the adherence of the bacterium onto both biotic and abiotic surfaces, making the biofilm dense and thick. The capability of Pmt to confer adherence onto host cells was confirmed by the observation that ectopic expression of pmt in E. coli strongly increases its adherence onto HeLa and S. cerevisiae cells (Sahu et al., 2012a).
Bacterial cell aggregates and biofilms are structures known to be encased in an extracellular matrix composed by various macromolecules, including nucleic acids, which serve as scaffolding agents to stabilize the biofilm structure especially in the initial phases of its formation (Whitchurch et al., 2002; Flemming and Wingender, 2010). In A. baumannii it has been shown that extracellular DNA (eDNA) is actively released during early growth phase and plays a crucial role in bacterial aggregation, forming a network-like structure (Sahu et al., 2012b). The Pmt EP is surmised of being responsible for the export of nucleic acids as it is expressed in early stages of biofilm formation (Sahu et al., 2012a). This hypothesis is also supported by the observation that in E. coli strains harboring the A. baumannii pmt gene there is an increased release of eDNA (Sahu et al., 2012b). Besides Pmt, another MFS efflux pump, AbaF, is involved in the formation of A. baumannii biofilms. The capacity of AbaF to extrude fosfomycin (Sharma et al., 2017) makes the cell resistant to this drug. Experiments performed with an abaF defective A. baumannii strain show that, in addition to being sensitive to fosfomycin, the mutant is also sensitive to chloramphenicol and is impaired in biofilm development. In most clinical A. baumannii strains the abaF gene is constitutively expressed. This is thought to depend not only on the capacity of AbaF to pump out antibiotics but also on its involvement in the extrusion of biofilm material (Sharma et al., 2017). The observation that Caenorhabditis elegans fed on a A. baumannii abaF mutant shows a higher survival rate as compared to feeding on the wt bacterium strongly suggests that AbaF contributes to bacterial virulence, likely by pumping out host derived antibacterial factors (Sharma et al., 2017).
Biofilm formation is essentially the progressive embedding of a bacterial community in an extracellular matrix (Flemming and Wingender, 2010). In this process, the ability of the cells to move and colonize the environment where they reside is crucial. Indeed, motility is considered as a key factor in the infectiveness of pathogens (Pérez-Varela et al., 2017). In A. baumannii the AbaQ MFS transporter, initially characterized for conferring resistance to quinolones (Pérez-Varela et al., 2017), is widely present in clinical isolates (Pérez-Varela et al., 2018). It is a 47.8 kDa protein encoded by a 1305 bp gene and is composed of 12 TM segments. In a nematode model of A. baumannii virulence, the inactivation of the abaQ gene causes the loss of surface-mediated motility and the attenuation of virulence (Pérez-Varela et al., 2017). Surface-associated motility is a crucial requirement for biofilm formation in A. baumannii as the bacterium lacks flagella. The association between motility and MFS EPs has been observed also in Pseudomonas putida by studying the role of the MFS EP PcaK in the chemiotaxis towards 4-hydroxybenzoate (4-HBA), which is the pump substrate (Nichols and Harwood, 1997). The involvement of EPs in surface-associated motility and virulence in A. baumannii is not limited to MFS transporters. Recent evidence indicates that EPs belonging to other superfamilies contribute to the motility of the cells and to full expression of the virulence phenotype (Pérez-Varela et al., 2019). Altogether, these data further support the relevance of MFS EPs not only to antibiotic resistance but also to specific steps of biofilm formation, cellular movement and virulence in an emergent pathogen as A. baumannii.
Klebsiella pneumoniae: KpnGH and the Response to Gastro-Intestinal Stress
K. pneumoniae is an opportunistic pathogen and a leading cause of severe illness in nosocomial patients (Wyres et al., 2020). Indeed, the significant incidence (5–7%), of hospitalized and immunocompromised patients affected by pneumonia, septicaemia, urinary tract and soft tissue infections caused by this bacterium, makes it a paradigm of community-acquired infections worldwide. The high mortality rate is further increased by the emergence of multidrug resistance phenotypes, ultimately resulting in failure of drug therapies. Most genes conferring resistance to clinically relevant antibiotics are located on large conjugative plasmids and have been acquired via horizontal gene transfer (Wyres et al., 2020). K. pneumoniae is often part of the healthy human microbiota. Recent clinical data indicate that gastrointestinal carriage is a major reservoir of K. pneumoniae infections in healthcare environments (Gorrie et al., 2017). A continuous “sensing and responding” to changing physio-chemical conditions occurs in bacteria moving in the gastro-intestinal (GI) tract. Depending on oxygen availability, presence of bile salts, pH and other stress conditions bacteria recognize different gastro-intestinal regions as different microenvironments, adopt diversified strategies to survive in these niches and produce a complex arsenal of virulence factors (Sengupta et al., 2014). The activation of specific efflux pumps is regarded as a major strategy to keep the intracellular concentration of toxic substrates (e.g., bile salts, molecules with antimicrobial activity, oxidative agents) at a sustainable level (Henderson et al., 2021).
Sequencing of K. pneumoniae genomes has revealed that genes encoding EPs potentially responsible for extruding antibiotics represent up to 10% of the total gene pool. More than ten EPs belong to the MFS family. At present antibiotic extrusion has been clearly demonstrated only for a few EPs and the physiological relevance of these genes is still under investigation. In this respect interesting data exists on the biological function of KpnGH, a multidrug EP of the MFS family. It consists of two subunits, KpnG and KpnH, which display a high protein and nucleotide sequence similarity with the EmrA adaptor protein and the EmrB transporter of the EmrAB EP of E. coli (Srinivasan et al., 2014). KpnGH confers resistance to a large panel of antimicrobial compounds, cationic dyes, antiseptics chemicals and detergents. The contribution of KpnGH to withstand the harsh conditions K. pneumoniae encounters in the human GI tract derives from the observation that a kpnGH mutant is more sensitive to bile salts, suggesting that K. pneumoniae uses this EP to circumvent the inhibitory effect of bile salts. The ability to confer resistance to bile salts is found also in E. coli EmrAB and in the closely related V. cholera VceCAB (Woolley et al., 2005), suggesting that in several bacterial pathogens EmrAB-like EPs contribute significantly to the survival in bile-rich environments. Further evidence obtained by comparing wt and kpnGH-defective K. pneumoniae strains confirm that KpnGH favours survival under high osmotic environments and oxidative or nitrosative stress (Srinivasan et al., 2014). Overall, these data suggest an active role of KpnGH in supporting the bacterium under conditions typically found in the upper parts of the GI, characterized by severe osmotic condition in a microaerobic environment.
Conclusion
The large amount of experimental evidence accumulated over the last years strongly suggests that the function of MDR EPs in many bacterial pathogens is not strictly limited to antibiotic resistance but rather has a wider impact on bacterial interactions with host cells. Studying the interplay between bacteria and hosts is an effective approach to fully understand the physiological significance of MDR EPs. The MFS superfamily constitutes the largest group of transporters and is present in all phyla. The model systems we have presented in this review illustrate the many roles MFS EPs play in the communication between bacteria and host cells, in particular in key phases of a pathogen’s life, as adhesion, invasion, intracellular survival and biofilm formation. The complex regulation of several EPs by global networks, as the TCS system, reflects the need of the bacterial cell to fine tune the expression of MDR EPs in response to specific niches and in coordination with other virulence factors. The possibility to block the communication with target host cells by inhibiting the expression or the activity of MDR EPs is a promising strategy not only to combat the onset of drug resistances but also to attenuate virulence. Several inhibitors targeting EPs or their regulators have been discovered, including natural products, antibiotics and synthetic molecules (Sun et al., 2014; Wang et al., 2016; Eguchi et al., 2017). Very recently a novel strategy based on natural-like compounds able to outcompete antibiotic efflux has been envisioned, opening stimulating perspectives in counteracting EP-mediated drug resistance and virulence in bacterial pathogens (Laborda et al., 2021).
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This research was supported by grants from the Italian Ministry of Foreign Affairs and International Cooperation (Grant JP21GR04), from Italian Ministry of University and Research (PRIN 2017-20177J5Y3P), from Sapienza University of Rome, and from JSPS KAKENHI (Grants JP16K07681, JP20K05796).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ABC, ATP binding cassette; AbgT, p-aminobenzoyl-glutamate transporter; AIEC, adherent invasive Escherichia coli; CD, crohn disease; c-di-AMP, cyclic-di-AMP; CR, congo red; DHA, drug/H+ antiporter; DMT, drug/metabolite trasporter; EP, efflux pump; GFP, green fluorescence protein; IFN-β, interferon-β; IM, inner membrane; LTA, lipoteichoic acid; MATE, multidrug and toxic compound efflux; MDR, multidrug resistance; MFS, major facilitator superfamily; MTAC, MdrMATC; NF-kB, nuclear factor kB; OM, outer membrane; OMF, outer membrane factor; PACE, proteobacterial antimicrobial compound efflux; PAP, periplasmatic adaptor protein; PG, peptidoglycan; RECON, reductase controlling NF-κB; RND, resistance nodulation division; TCS, two component system; TM, trans membrane; TLR, toll-like receptor; WTA, wall teichoic acid.
References
Alav, I., Kobylka, J., Kuth, M. S., Pos, K. M., Picard, M., Blair, J. M. A., et al. (2021). Structure, Assembly, and Function of Tripartite Efflux and Type 1 Secretion Systems in Gram-Negative Bacteria. Chem. Rev. 121, 5479–5596. doi:10.1021/acs.chemrev.1c00055
Alav, I., Sutton, J. M., and Rahman, K. M. (2018). Role of Bacterial Efflux Pumps in Biofilm Formation. J. Antimicrob. Chemother. 73, 2003–2020. doi:10.1093/jac/dky042
Alcalde-Rico, M., Hernando-Amado, S., Blanco, P., and Martínez, J. L. (2016). Multidrug Efflux Pumps at the Crossroad between Antibiotic Resistance and Bacterial Virulence. Front. Microbiol. 7. doi:10.3389/fmicb.2016.01483
Alvarez-Ortega, C., Olivares, J., and Martínez, J. L. (2013). RND Multidrug Efflux Pumps: What Are They Good for?. Front. Microbio. 4. doi:10.3389/fmicb.2013.00007
Arcari, T., Feger, M.-L., Guerreiro, D. N., Wu, J., and O’Byrne, C. P. (2020). Comparative Review of the Responses of Listeria monocytogenes and Escherichia coli to Low pH Stress. Genes 11, 1330–1351. doi:10.3390/genes11111330
Ashida, H., Ogawa, M., Kim, M., Suzuki, S., Sanada, T., Punginelli, C., et al. (2011). Shigella Deploy Multiple Countermeasures against Host Innate Immune Responses. Curr. Opin. Microbiol. 14, 16–23. doi:10.1016/j.mib.2010.08.014
Ball, P. R., Shales, S. W., and Chopra, I. (1980). Plasmid-mediated Tetracycline Resistance in Escherichia coli Involves Increased Efflux of the Antibiotic. Biochem. Biophysical Res. Commun. 93, 74–81. doi:10.1016/S0006-291X(80)80247-6
Beceiro, A., Tomás, M., and Bou, G. (2013). Antimicrobial Resistance and Virulence: A Successful or Deleterious Association in the Bacterial World?. Clin. Microbiol. Rev. 26, 185–230. doi:10.1128/CMR.00059-12
Bigi, F., Gioffré, A., Klepp, L., De La Paz Santangelo, M., Alito, A., Caimi, K., et al. (2004). The Knockout of the lprG-Rv1410 Operon Produces strong Attenuation of Mycobacterium tuberculosis. Microbes Infect. 6, 182–187. doi:10.1016/j.micinf.2003.10.010
Blair, J. M. A., Webber, M. A., Baylay, A. J., Ogbolu, D. O., and Piddock, L. J. V. (2015). Molecular Mechanisms of Antibiotic Resistance. Nat. Rev. Microbiol. 13, 42–51. doi:10.1038/nrmicro3380
Boon, N., Kaur, M., Aziz, A., Bradnick, M., Shibayama, K., Eguchi, Y., et al. (2020). The Signaling Molecule Indole Inhibits Induction of the AR2 Acid Resistance System in Escherichia coli. Front. Microbiol. 11. doi:10.3389/fmicb.2020.00474
Bougdour, A., Cunning, C., Baptiste, P. J., Elliott, T., and Gottesman, S. (2008). Multiple Pathways for Regulation of σS (RpoS) Stability in Escherichia coli via the Action of Multiple Anti-adaptors. Mol. Microbiol. 68, 298–313. doi:10.1111/j.1365-2958.2008.06146.x
Briaud, P., Camus, L., Bastien, S., Doléans-Jordheim, A., Vandenesch, F., and Moreau, K. (2019). Coexistence with Pseudomonas aeruginosa Alters Staphylococcus aureus Transcriptome, Antibiotic Resistance and Internalization into Epithelial Cells. Sci. Rep. 9. doi:10.1038/s41598-019-52975-z
Colclough, A. L., Alav, I., Whittle, E. E., Pugh, H. L., Darby, E. M., Legood, S. W., et al. (2020). RND Efflux Pumps in Gram-Negative Bacteria; Regulation, Structure and Role in Antibiotic Resistance. Future Microbiol. 15, 143–157. doi:10.2217/fmb-2019-0235
Crimmins, G. T., Herskovits, A. A., Rehder, K., Sivick, K. E., Lauer, P., Dubensky, T. W., et al. (2008). Listeria monocytogenes Multidrug Resistance Transporters Activate a Cytosolic Surveillance Pathway of Innate Immunity. Proc. Natl. Acad. Sci. 105, 10191–10196. doi:10.1073/pnas.0804170105
Demarre, G., Prudent, V., Schenk, H., Rousseau, E., Bringer, M.-A., Barnich, N., et al. (2019). The Crohn's Disease-Associated Escherichia coli Strain LF82 Relies on SOS and Stringent Responses to Survive, Multiply and Tolerate Antibiotics within Macrophages. Plos Pathog. 15, e1008123. doi:10.1371/journal.ppat.1008123
Ding, Y., Onodera, Y., Lee, J. C., and Hooper, D. C. (2008). NorB, an Efflux Pump in Staphylococcus aureus Strain MW2, Contributes to Bacterial Fitness in Abscesses. J. Bacteriol. 190, 7123–7129. doi:10.1128/JB.00655-08
Du, D., Wang-Kan, X., Neuberger, A., van Veen, H. W., Pos, K. M., Piddock, L. J. V., et al. (2018). Multidrug Efflux Pumps: Structure, Function and Regulation. Nat. Rev. Microbiol. 16, 523–539. doi:10.1038/s41579-018-0048-6
Dziva, F., Hauser, H., Connor, T. R., van Diemen, P. M., Prescott, G., Langridge, G. C., et al. (2013). Sequencing and Functional Annotation of Avian Pathogenic Escherichia coli Serogroup O78 Strains Reveal the Evolution of E. coli Lineages Pathogenic for Poultry via Distinct Mechanisms. Infect. Immun. 81, 838–849. doi:10.1128/IAI.00585-12
Eguchi, Y., Ishii, E., Hata, K., and Utsumi, R. (2011). Regulation of Acid Resistance by Connectors of Two-Component Signal Transduction Systems in Escherichia coli. J. Bacteriol. 193, 1222–1228. doi:10.1128/JB.01124-10
Eguchi, Y., Ishii, E., Yamane, M., and Utsumi, R. (2012). The Connector SafA Interacts with the Multi-Sensing Domain of PhoQ in Escherichia coli. Mol. Microbiol. 85, 299–313. doi:10.1111/j.1365-2958.2012.08114.x
Eguchi, Y., Itou, J., Yamane, M., Demizu, R., Yamato, F., Okada, A., et al. (2007). B1500, a Small Membrane Protein, Connects the Two-Component Systems EvgS/EvgA and PhoQ/PhoP in Escherichia coli. Proc. Natl. Acad. Sci. 104, 18712–18717. doi:10.1073/pnas.0705768104
Eguchi, Y., Okajima, T., Tochio, N., Inukai, Y., Shimizu, R., Ueda, S., et al. (2017). Angucycline Antibiotic Waldiomycin Recognizes Common Structural Motif Conserved in Bacterial Histidine Kinases. J. Antibiot. 70, 251–258. doi:10.1038/ja.2016.151
Eguchi, Y., Oshima, T., Mori, H., Aono, R., Yamamoto, K., Ishihama, A., et al. (2003). Transcriptional Regulation of Drug Efflux Genes by EvgAS, a Two-Component System in Escherichia coli. Microbiology 149, 2819–2828. doi:10.1099/mic.0.26460-0
Eguchi, Y., and Utsumi, R. (2014). Alkali Metals in Addition to Acidic pH Activate the EvgS Histidine Kinase Sensor in Escherichia coli. J. Bacteriol. 196, 3140–3149. doi:10.1128/JB.01742-14
Fanelli, G., Pasqua, M., Colonna, B., Prosseda, G., and Grossi, M. (2020). Expression Profile of Multidrug Resistance Efflux Pumps during Intracellular Life of Adherent-Invasive Escherichia coli Strain LF82. Front. Microbiol. 11. doi:10.3389/fmicb.2020.01935
Federici, L., Du, D., Walas, F., Matsumura, H., Fernandez-Recio, J., McKeegan, K. S., et al. (2005). The Crystal Structure of the Outer Membrane Protein VceC from the Bacterial Pathogen Vibrio cholerae at 1.8 Å Resolution. J. Biol. Chem. 280, 15307–15314. doi:10.1074/jbc.M500401200
Flemming, H.-C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., and Kjelleberg, S. (2016). Biofilms: An Emergent Form of Bacterial Life. Nat. Rev. Microbiol. 14, 563–575. doi:10.1038/nrmicro.2016.94
Flemming, H.-C., and Wingender, J. (2010). The Biofilm Matrix. Nat. Rev. Microbiol. 8, 623–633. doi:10.1038/nrmicro2415
Garzoni, C., Francois, P., Huyghe, A., Couzinet, S., Tapparel, C., Charbonnier, Y., et al. (2007). A Global View of Staphylococcus aureus Whole Genome Expression upon Internalization in Human Epithelial Cells. BMC Genomics 8, 171. doi:10.1186/1471-2164-8-171
Gorrie, C. L., Mirčeta, M., Wick, R. R., Edwards, D. J., Thomson, N. R., Strugnell, R. A., et al. (2017). Gastrointestinal Carriage Is a Major Reservoir of Klebsiella pneumoniae Infection in Intensive Care Patients. Clin. Infect. Dis. 65, 208–215. doi:10.1093/cid/cix270
Grkovic, S., Brown, M. H., and Skurray, R. A. (2002). Regulation of Bacterial Drug Export Systems. Microbiol. Mol. Biol. Rev. 66, 671–701. doi:10.1128/mmbr.66.4.671-701.2002
Han, X., Dorsey-Oresto, A., Malik, M., Wang, J.-Y., Drlica, K., Zhao, X., et al. (2010). Escherichia coli Genes that Reduce the Lethal Effects of Stress. BMC Microbiol. 10, 35. doi:10.1186/1471-2180-10-35
Harding, C. M., Hennon, S. W., and Feldman, M. F. (2018). Uncovering the Mechanisms of Acinetobacter baumannii Virulence. Nat. Rev. Microbiol. 16, 91–102. doi:10.1038/nrmicro.2017.148
Henderson, P. J. F., Maher, C., Elbourne, L. D. H., Eijkelkamp, B. A., Paulsen, I. T., and Hassan, K. A. (2021). Physiological Functions of Bacterial “Multidrug” Efflux Pumps. Chem. Rev. 121, 5417–5478. doi:10.1021/acs.chemrev.0c01226
Heng, J., Zhao, Y., Liu, M., Liu, Y., Fan, J., Wang, X., et al. (2015). Substrate-bound Structure of the E. coli Multidrug Resistance Transporter MdfA. Cell Res 25, 1060–1073. doi:10.1038/cr.2015.94
Hinchliffe, P., Greene, N. P., Paterson, N. G., Crow, A., Hughes, C., and Koronakis, V. (2014). Structure of the Periplasmic Adaptor Protein from a Major Facilitator Superfamily (MFS) Multidrug Efflux Pump. FEBS Lett. 588, 3147–3153. doi:10.1016/j.febslet.2014.06.055
Hinchliffe, P., Symmons, M. F., Hughes, C., and Koronakis, V. (2013). Structure and Operation of Bacterial Tripartite Pumps. Annu. Rev. Microbiol. 67, 221–242. doi:10.1146/annurev-micro-092412-155718
Inada, S., Okajima, T., Utsumi, R., and Eguchi, Y. (2021). Acid-Sensing Histidine Kinase with a Redox Switch. Front. Microbiol. 12. doi:10.3389/fmicb.2021.652546
Itou, J., Eguchi, Y., and Utsumi, R. (2009). Molecular Mechanism of Transcriptional Cascade Initiated by the EvgS/EvgA System in Escherichia coli K-12. Biosci. Biotechnol. Biochem. 73, 870–878. doi:10.1271/bbb.80795
Kaplan Zeevi, M., Shafir, N. S., Shaham, S., Friedman, S., Sigal, N., Nir Paz, R., et al. (2013). Listeria monocytogenes Multidrug Resistance Transporters and Cyclic Di-AMP, Which Contribute to Type I Interferon Induction, Play a Role in Cell wall Stress. J. Bacteriol. 195, 5250–5261. doi:10.1128/JB.00794-13
Karatan, E., and Watnick, P. (2009). Signals, Regulatory Networks, and Materials that Build and Break Bacterial Biofilms. Microbiol. Mol. Biol. Rev. 73, 310–347. doi:10.1128/mmbr.00041-08
Kato, A., Ohnishi, H., Yamamoto, K., Furuta, E., Tanabe, H., and Utsumi, R. (2000). Transcription of emrKY is Regulated by the EvgA-EvgS Two-Component System in Escherichia coli K-12. Biosci. Biotechnol. Biochem. 64, 1203–1209. doi:10.1271/bbb.64.1203
Kumar, S., Lekshmi, M., Parvathi, A., Ojha, M., Wenzel, N., and Varela, M. F. (2020). Functional and Structural Roles of the Major Facilitator Superfamily Bacterial Multidrug Efflux Pumps. Microorganisms 8, 266. doi:10.3390/microorganisms8020266
Laborda, P., Alcalde‐Rico, M., Chini, A., Martínez, J. L., and Hernando‐Amado, S. (2021). Discovery of Inhibitors of Pseudomonas aeruginosa Virulence through the Search for Natural‐like Compounds with a Dual Role as Inducers and Substrates of Efflux Pumps. Environ. Microbiol. 5. doi:10.1111/1462-2920.15511
Law, C. J., Maloney, P. C., and Wang, D.-N. (2008). Ins and Outs of Major Facilitator Superfamily Antiporters. Annu. Rev. Microbiol. 62, 289–305. doi:10.1146/annurev.micro.61.080706.093329
Leuzzi, A., Di Martino, M. L., Campilongo, R., Falconi, M., Barbagallo, M., Marcocci, L., et al. (2015). Multifactor Regulation of the MdtJI Polyamine Transporter in Shigella. PLoS One 10, e0136744. doi:10.1371/journal.pone.0136744
Leuzzi, A., Grossi, M., Di Martino, M. L., Pasqua, M., Micheli, G., Colonna, B., et al. (2017). Role of the SRRz/Rz1 Lambdoid Lysis Cassette in the Pathoadaptive Evolution of Shigella. Int. J. Med. Microbiol. 307, 268–275. doi:10.1016/j.ijmm.2017.03.002
Li, X.-Z., Plésiat, P., and Nikaido, H. (2015). The challenge of Efflux-Mediated Antibiotic Resistance in Gram-Negative Bacteria. Clin. Microbiol. Rev. 28, 337–418. doi:10.1128/CMR.00117-14
Majumder, P., Khare, S., Athreya, A., Hussain, N., Gulati, A., and Penmatsa, A. (2019). Dissection of Protonation Sites for Antibacterial Recognition and Transport in QacA, a Multi-Drug Efflux Transporter. J. Mol. Biol. 431, 2163–2179. doi:10.1016/j.jmb.2019.03.015
Masuda, N., and Church, G. M. (2003). Regulatory Network of Acid Resistance Genes in Escherichia coli. Mol. Microbiol. 48, 699–712. doi:10.1046/j.1365-2958.2003.03477.x
McFarland, A. P., Burke, T. P., Carletti, A. A., Glover, R. C., Tabakh, H., Welch, M. D., et al. (2018). RECON-dependent Inflammation in Hepatocytes Enhances Listeria monocytogenes Cell-To-Cell Spread. MBio 9. doi:10.1128/mBio.00526-18
McFarland, A. P., Luo, S., Ahmed-Qadri, F., Zuck, M., Thayer, E. F., Goo, Y. A., et al. (2017). Sensing of Bacterial Cyclic Dinucleotides by the Oxidoreductase RECON Promotes NF-Κb Activation and Shapes a Proinflammatory Antibacterial State. Immunity 46, 433–445. doi:10.1016/j.immuni.2017.02.014
McMurry, L., Petrucci, R. E., and Levy, S. B. (1980). Active Efflux of Tetracycline Encoded by Four Genetically Different Tetracycline Resistance Determinants in Escherichia coli. Proc. Natl. Acad. Sci. 77, 3974–3977. doi:10.1073/pnas.77.7.3974
Nadler, C., Shifrin, Y., Nov, S., Kobi, S., and Rosenshine, I. (2006). Characterization of Enteropathogenic Escherichia coli Mutants that Fail to Disrupt Host Cell Spreading and Attachment to Substratum. Infect. Immun. 74, 839–849. doi:10.1128/IAI.74.2.839-849.2006
Nagarathinam, K., Nakada-Nakura, Y., Parthier, C., Terada, T., Juge, N., Jaenecke, F., et al. (2018). Outward Open Conformation of a Major Facilitator Superfamily multidrug/H+ Antiporter Provides Insights into Switching Mechanism. Nat. Commun. 9. doi:10.1038/s41467-018-06306-x
Neyfakh, A. A. (2002). Mystery of Multidrug Transporters: The Answer Can Be Simple. Mol. Microbiol. 44, 1123–1130. doi:10.1046/j.1365-2958.2002.02965.x
Nichols, N. N., and Harwood, C. S. (1997). PcaK, a High-Affinity Permease for the Aromatic Compounds 4-hydroxybenzoate and Protocatechuate from Pseudomonas putida. J. Bacteriol. 179, 5056–5061. doi:10.1128/jb.179.16.5056-5061.1997
Nikaido, H., and Pagès, J.-M. (2012). Broad-specificity Efflux Pumps and Their Role in Multidrug Resistance of Gram-Negative Bacteria. FEMS Microbiol. Rev. 36, 340–363. doi:10.1111/j.1574-6976.2011.00290.x
Nishino, K., and Yamaguchi, A. (2002). EvgA of the Two-Component Signal Transduction System Modulates Production of the YhiUV Multidrug Transporter in Escherichia coli. J. Bacteriol. 184, 2319–2323. doi:10.1128/JB.184.8.2319-2323.2002
Pasqua, M., Grossi, M., Scinicariello, S., Aussel, L., Barras, F., Colonna, B., et al. (2019a). The MFS Efflux Pump EmrKY Contributes to the Survival of Shigella within Macrophages. Sci. Rep. 9. doi:10.1038/s41598-019-39749-3
Pasqua, M., Grossi, M., Zennaro, A., Fanelli, G., Micheli, G., Barras, F., et al. (2019b). The Varied Role of Efflux Pumps of the MFS Family in the Interplay of Bacteria with Animal and Plant Cells. Microorganisms 7, 285. doi:10.3390/microorganisms7090285
Pasqua, M., Michelacci, V., Di Martino, M. L., Tozzoli, R., Grossi, M., Colonna, B., et al. (2017). The Intriguing Evolutionary Journey of Enteroinvasive E. coli (EIEC) toward Pathogenicity. Front. Microbiol. 8. doi:10.3389/fmicb.2017.02390
Paulsen, I. T. (2003). Multidrug Efflux Pumps and Resistance: Regulation and Evolution. Curr. Opin. Microbiol. 6, 446–451. doi:10.1016/j.mib.2003.08.005
Pérez-Varela, M., Corral, J., Aranda, J., and Barbé, J. (2018). Functional Characterization of AbaQ, a Novel Efflux Pump Mediating Quinolone Resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 62. doi:10.1128/AAC.00906-18
Pérez-Varela, M., Corral, J., Aranda, J., and Barbé, J. (2019). Roles of Efflux Pumps from Different Superfamilies in the Surface-Associated Motility and Virulence of Acinetobacter baumannii ATCC 17978. Antimicrob. Agents Chemother. 63. doi:10.1128/AAC.02190-18
Pérez-Varela, M., Corral, J., Vallejo, J. A., Rumbo-Feal, S., Bou, G., Aranda, J., et al. (2017). Mutations in the β-Subunit of the RNA Polymerase Impair the Surface-Associated Motility and Virulence of Acinetobacter baumannii. Infect. Immun. 85. doi:10.1128/IAI.00327-17
Piddock, L. J. V. (2006). Clinically Relevant Chromosomally Encoded Multidrug Resistance Efflux Pumps in Bacteria. Clin. Microbiol. Rev. 19, 382–402. doi:10.1128/CMR.19.2.382-402.2006
Pizarro-Cerdá, J., and Cossart, P. (2018). Listeria monocytogenes : Cell Biology of Invasion and Intracellular Growth. Microbiol. Spectr. 6. doi:10.1128/microbiolspec.gpp3-0013-2018
Prosseda, G., Di Martino, M. L., Campilongo, R., Fioravanti, R., Micheli, G., Casalino, M., et al. (2012). Shedding of Genes that Interfere with the Pathogenic Lifestyle: The Shigella Model. Res. Microbiol. 163, 399–406. doi:10.1016/j.resmic.2012.07.004
Quillin, S. J., Schwartz, K. T., and Leber, J. H. (2011). The Novel Listeria monocytogenes Bile Sensor BrtA Controls Expression of the Cholic Acid Efflux Pump MdrT. Mol. Microbiol. 81, 129–142. doi:10.1111/j.1365-2958.2011.07683.x
Quistgaard, E. M., Löw, C., Guettou, F., and Nordlund, P. (2016). Understanding Transport by the Major Facilitator Superfamily (MFS): Structures Pave the Way. Nat. Rev. Mol. Cel Biol. 17, 123–132. doi:10.1038/nrm.2015.25
Sahu, P. K., Iyer, P. S., Gaikwad, M. B., Talreja, S. C., Pardesi, K. R., and Chopade, B. A. (2012a). An MFS Transporter-like ORF from MDR Acinetobacter baumannii AIIMS 7 Is Associated with Adherence and Biofilm Formation on Biotic/Abiotic Surface. Int. J. Microbiol. 2012, 1–10. doi:10.1155/2012/490647
Sahu, P. K., Iyer, P. S., Oak, A. M., Pardesi, K. R., and Chopade, B. A. (2012b). Characterization of eDNA from the Clinical Strain Acinetobacter baumannii AIIMS 7 and its Role in Biofilm Formation. Scientific World J. 2012, 1–10. doi:10.1100/2012/973436
Saier, M. H., Paulsen, I. T., Sliwinski, M. K., Pao, S. S., Skurray, R. A., and Nikaido, H. (1998). Evolutionary Origins of Multidrug and Drug-specific Efflux Pumps in Bacteria. FASEB J. 12, 265–274. doi:10.1096/fasebj.12.3.265
Sengupta, C., Ray, S., and Chowdhury, R. (2014). Fine Tuning of Virulence Regulatory Pathways in Enteric Bacteria in Response to Varying Bile and Oxygen Concentrations in the Gastrointestinal Tract. Gut Pathog. 6. doi:10.1186/s13099-014-0038-9
Sharma, A., Sharma, R., Bhattacharyya, T., Bhando, T., and Pathania, R. (2017). Fosfomycin Resistance in Acinetobacter baumannii is Mediated by Efflux through a Major Facilitator Superfamily (MFS) Transporter-AbaF. J. Antimicrob. Chemother. 72, 68–74. doi:10.1093/jac/dkw382
Srinivasan, V. B., Singh, B. B., Priyadarshi, N., Chauhan, N. K., and Rajamohan, G. (2014). Role of Novel Multidrug Efflux Pump Involved in Drug Resistance in Klebsiella pneumoniae. PLoS One 9, e96288. doi:10.1371/journal.pone.0096288
Sulavik, M. C., Houseweart, C., Cramer, C., Jiwani, N., Murgolo, N., Greene, J., et al. (2001). Antibiotic Susceptibility Profiles of Escherichia coli Strains Lacking Multidrug Efflux Pump Genes. Antimicrob. Agents Chemother. 45, 1126–1136. doi:10.1128/AAC.45.4.1126-1136.2001
Sun, J., Deng, Z., and Yan, A. (2014). Bacterial Multidrug Efflux Pumps: Mechanisms, Physiology and Pharmacological Exploitations. Biochem. Biophysical Res. Commun. 453, 254–267. doi:10.1016/j.bbrc.2014.05.090
Symmons, M. F., Marshall, R. L., and Bavro, V. N. (2015). Architecture and Roles of Periplasmic Adaptor Proteins in Tripartite Efflux Assemblies. Front. Microbiol. 6. doi:10.3389/fmicb.2015.00513
Tal, N., and Schuldiner, S. (2009). A Coordinated Network of Transporters with Overlapping Specificities Provides a Robust Survival Strategy. Proc. Natl. Acad. Sci. 106, 9051–9056. doi:10.1073/pnas.0902400106
Tanabe, H., Yamasaki, K., Furue, M., Yamamoto, K., Katoh, A., Yamamoto, M., et al. (1997). Growth Phase-dependent Transcription of emrKY, a Homolog of Multidrug Efflux emrAB, Genes of Escherichia coli, Is Induced by Tetracycline. J. Gen. Appl. Microbiol. 43, 257–263. doi:10.2323/jgam.43.257
Teelucksingh, T., Thompson, L. K., and Cox, G. (2020). The Evolutionary Conservation of Escherichia coli Drug Efflux Pumps Supports Physiological Functions. J. Bacteriol. 202. doi:10.1128/JB.00367-20
The, H. C., Thanh, D. P., Holt, K. E., Thomson, N. R., and Baker, S. (2016). The Genomic Signatures of Shigella Evolution, Adaptation and Geographical Spread. Nat. Rev. Microbiol. 14, 235–250. doi:10.1038/nrmicro.2016.10
Truong-Bolduc, Q. C., Bolduc, G. R., Medeiros, H., Vyas, J. M., Wang, Y., and Hooper, D. C. (2015). Role of the Tet38 Efflux Pump in Staphylococcus aureus Internalization and Survival in Epithelial Cells. Infect. Immun. 83, 4362–4372. doi:10.1128/IAI.00723-15
Truong-Bolduc, Q. C., Dunman, P. M., Strahilevitz, J., Projan, S. J., and Hooper, D. C. (2005). MgrA Is a Multiple Regulator of Two New Efflux Pumps in Staphylococcus aureus. J. Bacteriol. 187, 2395–2405. doi:10.1128/JB.187.7.2395-2405.2005
Truong-Bolduc, Q. C., Khan, N. S., Vyas, J. M., and Hooper, D. C. (2017). Tet38 Efflux Pump Affects Staphylococcus aureus Internalization by Epithelial Cells through Interaction with CD36 and Contributes to Bacterial Escape from Acidic and Nonacidic Phagolysosomes. Infect. Immun. 85. doi:10.1128/IAI.00862-16
Truong-Bolduc, Q. C., Villet, R. A., Estabrooks, Z. A., and Hooper, D. C. (2014). Native Efflux Pumps Contribute Resistance to Antimicrobials of Skin and the Ability of Staphylococcus aureus to Colonize Skin. J. Infect. Dis. 209, 1485–1493. doi:10.1093/infdis/jit660
Truong-Bolduc, Q. C., Wang, Y., and Hooper, D. C. (2021). Staphylococcus aureus Tet38 Efflux Pump Structural Modeling and Roles of Essential Residues in Drug Efflux and Host Cell Internalization. Infect. Immun. 89. doi:10.1128/IAI.00811-20
Truong-Bolduc, Q. C., Wang, Y., and Hooper, D. C. (2019). Tet38 of Staphylococcus aureus Binds to Host Cell Receptor Complex CD36-toll-like Receptor 2 and Protects from Teichoic Acid Synthesis Inhibitors Tunicamycin and Congo Red. Infect. Immun. 87. doi:10.1128/IAI.00194-19
Wang, Y., Venter, H., and Ma, S. (2016). Efflux Pump Inhibitors: A Novel Approach to Combat Efflux-Mediated Drug Resistance in Bacteria. Cdt 17, 702–719. doi:10.2174/1389450116666151001103948
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 46, W296–W303. doi:10.1093/nar/gky427
Whitchurch, C. B., Tolker-Nielsen, T., Ragas, P. C., and Mattick, J. S. (2002). Extracellular DNA Required for Bacterial Biofilm Formation. Science 295, 1487. doi:10.1126/science.295.5559.1487
Woodward, J. J., Iavarone, A. T., and Portnoy, D. A. (2010). C-di-AMP Secreted by Intracellular Listeria monocytogenes Activates a Host Type I Interferon Response. Science 328, 1703–1705. doi:10.1126/science.1189801
Woolley, R. C., Vediyappan, G., Anderson, M., Lackey, M., Ramasubramanian, B., Jiangping, B., et al. (2005). Characterization of the Vibrio cholerae vceCAB Multiple-Drug Resistance Efflux Operon in Escherichia coli. J. Bacteriol. 187, 5500–5503. doi:10.1128/JB.187.15.5500-5503.2005
Wu, H.-H., Symersky, J., and Lu, M. (2020). Structure and Mechanism of a Redesigned Multidrug Transporter from the Major Facilitator Superfamily. Sci. Rep. 10. doi:10.1038/s41598-020-60332-8
Wyres, K. L., Lam, M. M. C., and Holt, K. E. (2020). Population Genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 18, 344–359. doi:10.1038/s41579-019-0315-1
Yin, W., Cai, X., Ma, H., Zhu, L., Zhang, Y., Chou, S.-H., et al. (2020). A Decade of Research on the Second Messenger c-di-AMP. FEMS Microbiol. Rev. 44, 701–724. doi:10.1093/femsre/fuaa019
Yoshitani, K., Ishii, E., Taniguchi, K., Sugimoto, H., Shiro, Y., Akiyama, Y., et al. (2019). Identification of an Internal Cavity in the PhoQ Sensor Domain for PhoQ Activity and SafA-Mediated Control. Biosci. Biotechnol. Biochem. 83, 684–694. doi:10.1080/09168451.2018.1562879
Zeng, J., Wu, L., Liu, Z., Lv, Y., Feng, J., Wang, W., et al. (2021). Gain-of-Function Mutations in Acid Stress Response ( evgS ) Protect Escherichia coli from Killing by Gallium Nitrate, an Antimicrobial Candidate. Antimicrob. Agents Chemother. 65. doi:10.1128/AAC.01595-20
Keywords: efflux pumps, major facilitator superfamily, bacteria host interactions, two component systems, bacterial virulence
Citation: Pasqua M, Bonaccorsi di Patti MC, Fanelli G, Utsumi R, Eguchi Y, Trirocco R, Prosseda G, Grossi M and Colonna B (2021) Host - Bacterial Pathogen Communication: The Wily Role of the Multidrug Efflux Pumps of the MFS Family. Front. Mol. Biosci. 8:723274. doi: 10.3389/fmolb.2021.723274
Received: 10 June 2021; Accepted: 13 July 2021;
Published: 26 July 2021.
Edited by:
Tatiana Venkova, Fox Chase Cancer Center, United StatesReviewed by:
Josep Casadesús, Sevilla University, SpainAntonio Juárez, University of Barcelona, Spain
Copyright © 2021 Pasqua, Bonaccorsi di Patti, Fanelli, Utsumi, Eguchi, Trirocco, Prosseda, Grossi and Colonna. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bianca Colonna, YmlhbmNhLmNvbG9ubmFAdW5pcm9tYTEuaXQ=
 Martina Pasqua
Martina Pasqua Maria Carmela Bonaccorsi di Patti
Maria Carmela Bonaccorsi di Patti Giulia Fanelli
Giulia Fanelli Ryutaro Utsumi3
Ryutaro Utsumi3 Gianni Prosseda
Gianni Prosseda Milena Grossi
Milena Grossi Bianca Colonna
Bianca Colonna