- 1Division of Metabolism and Biosystemic Science, Gastroenterology and Hematology/Oncology, Department of Medicine, Asahikawa Medical University, Asahikawa, Japan
- 2Isotope Science Center, The University of Tokyo, Bunkyo, Japan
- 3Institute of Biomedical Research, Sapporo Higashi Tokushukai Hospital, Sapporo, Japan
Non-coding RNAs (ncRNAs), or RNA molecules that do not code for proteins, are generally categorized as either small or long ncRNA (lncRNA) and are involved in the pathogenesis of several diseases including many cancers. Identification of a large number of ncRNAs could help to elucidate previously unknown mechanisms in phenotype regulation. Some ncRNAs are encapsulated by extracellular vesicles (EVs) and transferred to recipient cells to regulate cellular processes, including epigenetic and post-transcriptional regulations. Recent studies have uncovered novel molecular mechanisms and functions of lncRNAs in pancreatic ductal adenocarcinoma (PDAC), one of the most intractable cancers that is highly invasive and metastatic. As the epithelial-mesenchymal transition (EMT) triggers tumor cell invasion and migration, clarification of the roles of lncRNA in EMT and tumor cell stemness would be critical for improving diagnostic and therapeutic approaches in metastatic cancers. This review provides an overview of relevant studies on lncRNA and its involvement with EMT in PDAC. Emerging knowledge offers evidence for the dysregulated expression of lncRNAs and essential insights into the potential contribution of both lncRNAs and EVs in the pathogenesis of PDAC. Future directions and new clinical applications for PDAC are also discussed.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the most common type of pancreatic cancer. It is one of the most dismal cancers as the associated 5-years relative survival rate of PDAC is only approximately 10%. The fundamental issue related to the significant mortality is the difficulty of diagnosis at the early stage (Ying et al., 2016; Lai et al., 2019), which leads to more than half of cases being diagnosed at advanced stages. Additionally, there is a trend toward an increase in PDAC incidence every successive year (Siegel et al., 2017). Surgical resection is the only curative treatment for PDAC to date. However, less than 20% of cases are potentially curable due to the persistent invasion and distant metastasis (Hidalgo 2010). Chemotherapeutic agents, such as gemcitabine, fluorouracil, irinotecan, oxaliplatin, and various nanoparticles, have been used widely to treat unresectable PDAC; however, the efficacy of these treatments is modest owing to frequent metastasis (Kamisawa et al., 2016; Brunetti et al., 2018). Behaviors causing the high malignant potential of PDAC, such as tumor cell invasion, metastasis, and chemoresistance, are associated with epithelial-mesenchymal transition (EMT) (De Craene and Berx 2013; Zhou et al., 2017). During this process, drastic alterations in gene expression can be induced that disrupt epithelial properties and lead to a mesenchymal phenotype. Various factors can induce EMT, including transforming growth factor (TGF)-β, which can induce EMT-associated transcription factors, such as the zinc finger transcription factor Snail, subsequently conferring tumor cell migration, metastasis, and resistance to cell death and chemotherapy in several types of cancers (De Craene and Berx 2013; Turley et al., 2008; Zhou et al., 2017). Thus, an urgent need exists to investigate regulatory mechanisms of EMT and identify molecular targets to suppress invasion, metastasis, and resistance to chemotherapy in PDAC patients.
To date, massive parallel sequencing studies have identified several non-protein coding RNAs (ncRNAs). NcRNAs play important roles in gene or protein regulation and cellular activity (Anastasiadou et al., 2018; Slack and Chinnaiyan 2019). Among them, long non-coding RNAs (lncRNAs) are increasingly recognized to govern substantial biological processes in multiple cancers through diverse mechanisms, such as epigenetic regulation (Rinn and Chang 2012). While well-known lncRNAs, such as H19, Metastasis associated lung adenocarcinoma transcript 1 (MALAT1), or HOX antisense intergenic RNA (HOTAIR), are reported as pan-cancer markers across different tissues, newly discovered lncRNAs [e.g., long intergenic non-coding RNA 01111 (LINC01111) in PDAC] have also been shown to work as biomarkers for several cancers (Pandya et al., 2020; Takahashi, Yan et al., 2014a). Some of the lncRNAs, such as MALAT1, highly upregulated in liver cancer (HULC) or urothelial carcinoma-associated 1 (UCA1), can be encapsulated in extracellular vesicles (EVs) and, when transferred, affect cell signaling and phenotypes similar to microRNAs (miRNAs) (Kogure et al., 2011; Takahashi et al., 2014c; Tkach and Thery 2016; Valadi et al., 2007). Although the underlying molecular mechanisms by which lncRNAs control PDAC initiation and progression are not well understood, a subset of the lncRNAs, transferred by EVs, is reported to be involved in the regulation of EMT to mediate tumor invasion and metastasis (Duguang et al., 2017; Xu et al., 2020). There are several factors, such as the investigation of EMT regulation by lncRNAs, the validation of diagnostic methods using candidate lncRNAs, and quantitation and standardization of technical issues for the diagnostic methods, that should be described before attempting the use of lncRNAs in clinical applications (Pandya et al., 2020). Therapeutic interventions for pancreatic cancer using lncRNA-based strategies can be designed based on the existing and emerging knowledge on lncRNAs in PDAC (Crowley et al., 2013; Liang et al., 2017; Shukla et al., 2018).
Here, we describe the features of lncRNAs produced from intergenic loci and the current knowledge regarding EMT in PDAC. We also present the roles of oncogenic and tumor-suppressive lncRNAs in EMT regulation in PDAC, as well as possible strategies to utilize the lncRNAs associated with EMT to develop clinical applications for diagnosis and therapy of human PDAC.
Long Non-Coding RNA
The Functional ANnoTation Of the Mammalian Genome (FANTOM) consortium discovered and reported on numerous mammalian transcriptomes which do not code for proteins and are defined as non-protein coding RNAs by sequencing novel full-length mouse cDNAs (Okazaki et al., 2002; Carninci et al., 2005). A similar but different project, the Encyclopedia of DNA Elements (ENCODE) project, advanced sequence-based studies to map functional elements across the human genome, revealing that RNAs transcribed from more than 80% of the genome, including ncRNAs, would have biochemical functions (Derrien et al., 2012; Dunham et al., 2012). NcRNAs are generally divided into two groups based on their base length, small ncRNA (<200 bp) and lncRNA (>200 bp). Small ncRNAs include miRNA, transfer RNA-derived small RNAs, or PIWI-interacting RNAs (Slack and Chinnaiyan 2019). Meanwhile, lncRNAs play critical roles in diverse biological processes, including initiation of cellular differentiation, proliferation, and pluripotency (Hu et al., 2012; Kopp and Mendell 2018; Takahashi et al., 2014a; Taniue and Akimitsu 2021). The subcellular localization of lncRNAs is as diverse as that observed with protein coding mRNAs. While mRNAs are mainly localized to the cytoplasm where their translation is carried out, lncRNAs are more often located in the nucleus (Kopp and Mendell 2018), where they could play a role in chromatin and genomic structural remodeling, RNA stabilization, and transcriptional regulation (Ransohoff et al., 2018; Rinn and Chang 2012; Takahashi et al., 2014a; Taniue et al., 2016b). However, several lncRNAs are also found specifically in the cytoplasm (Kino et al., 2010; Yoon et al., 2012), where they regulate protein stability by preventing post-translational modifications associated with protein degradation (Taniue et al., 2016a; Yoon et al., 2013). Moreover, lncRNA can also be involved in mRNA and protein localization (Yanagida et al., 2013). Furthermore, lncRNA works as a decoy that precludes the access of regulatory proteins to DNA to repress gene expression, and functions as a guide to recruit chromatin-modifying enzymes to target genes in cis and trans (Wang and Chang 2011). Besides, lncRNA can act as a scaffold to bring two or more proteins into discrete complexes (Wang and Chang 2011; Rinn and Chang 2012). Some of the lncRNAs work as competitive endogenous RNAs (ceRNAs), or miRNA sponges, to bind to the complementary site of targeted miRNAs and regulate their expression and activity to modulate their downstream pathway (Poliseno et al., 2010; Thomson and Dinger 2016).
Although the underlying mechanism of lncRNAs in cancer development is not elucidated, recent studies have demonstrated the functional significance of lncRNAs in cell proliferation, invasion, metastasis, and resistance to adverse stresses, such as hypoxia or chemotherapy (Duguang et al., 2017; Pandya et al., 2020; Takahashi et al., 2014a). LncRNAs play crucial oncogenic and tumor-suppressive roles in several cancers. For example, the lncRNA H19, induced during the development of the liver in mice (Pachnis et al., 1988), was highly expressed in hepatocellular carcinoma (HCC) tissues and could induce drug resistance in HCC (Iizuka et al., 2002; Tsang and Kwok 2007). Several recent reports also provide essential insights into the molecular functions, mechanisms, and involvement of lncRNA in PDAC (see The Functional Roles of Selected lncRNA in EMT of PDAC section).
EMT and PDAC
EMT is intimately associated with the development of tissues or organs during embryogenesis. Additionally, this phenomenon is closely correlated with tumor development (O’Brien et al., 2020) and is a trigger for invasion, migration, and acquisition of stem cell-like phenotype in cells of diverse cancers, including PDAC (De Craene and Berx 2013; Zhou et al., 2017). EMT promotes the gain of epithelial stem cell properties, association with stem-like cell markers, and generation of cancer stem cells (Mani et al., 2008). Pancreatic cancer stem cells (PCSCs) promote tumor growth and progression through several mechanisms, including tumor-initiation by inducing stem cell markers CD44, CD24, and CD133, and evasion of conventional therapies (Zhou et al., 2017). PCSCs are correlated with various processes, such as elevated expression of ATP-binding cassette transporter and anti-apoptotic proteins, induction of aldehyde dehydrogenase activity, and enhancement of DNA damage checkpoint repair. These regulatory mechanisms, which can protect cancer cells and enhance the cell survival signaling pathways, cause resistance to chemotherapy and radiotherapy (Baumann et al., 2008; Holohan et al., 2013).
EMT is established by EMT-inducible transcription factors, such as Snail, Slug, ZEB1, ZEB2, and Twist. These transcription factors repress the epithelial marker E-cadherin and upregulate the mesenchymal marker N-cadherin to promote EMT (Nieto and Cano 2012), and are regulated by several diverse upstream regulators, including signaling molecules and ncRNAs by various mechanisms. While many miRNAs modulate the EMT pathway, only a handful of lncRNAs affect EMT in PDAC. Among the well-known oncogenic lncRNAs, H19, HOX antisense intergenic RNA (HOTAIR), MALAT1, long intergenic non-protein coding RNA, regulator of reprogramming (linc-ROR), and HOXA transcript at the distal tip (HOTTIP) can enhance EMT (Duguang et al., 2017; Fu et al., 2017a; Fu et al., 2017b; Kim et al., 2013; Ma et al., 2016). Moreover, previously unrecognized lncRNAs, LOC389641 or ENST00000480739, are also shown to promote EMT (Sun et al., 2014; Zheng et al., 2016). Collectively, these reports elucidate that lncRNAs may have essential functions in the EMT pathway and gain of stem cell-like properties in PDAC. The individual roles of characterized lncRNAs are reviewed in the following section.
The Functional Roles of Selected lncRNA in EMT of PDAC
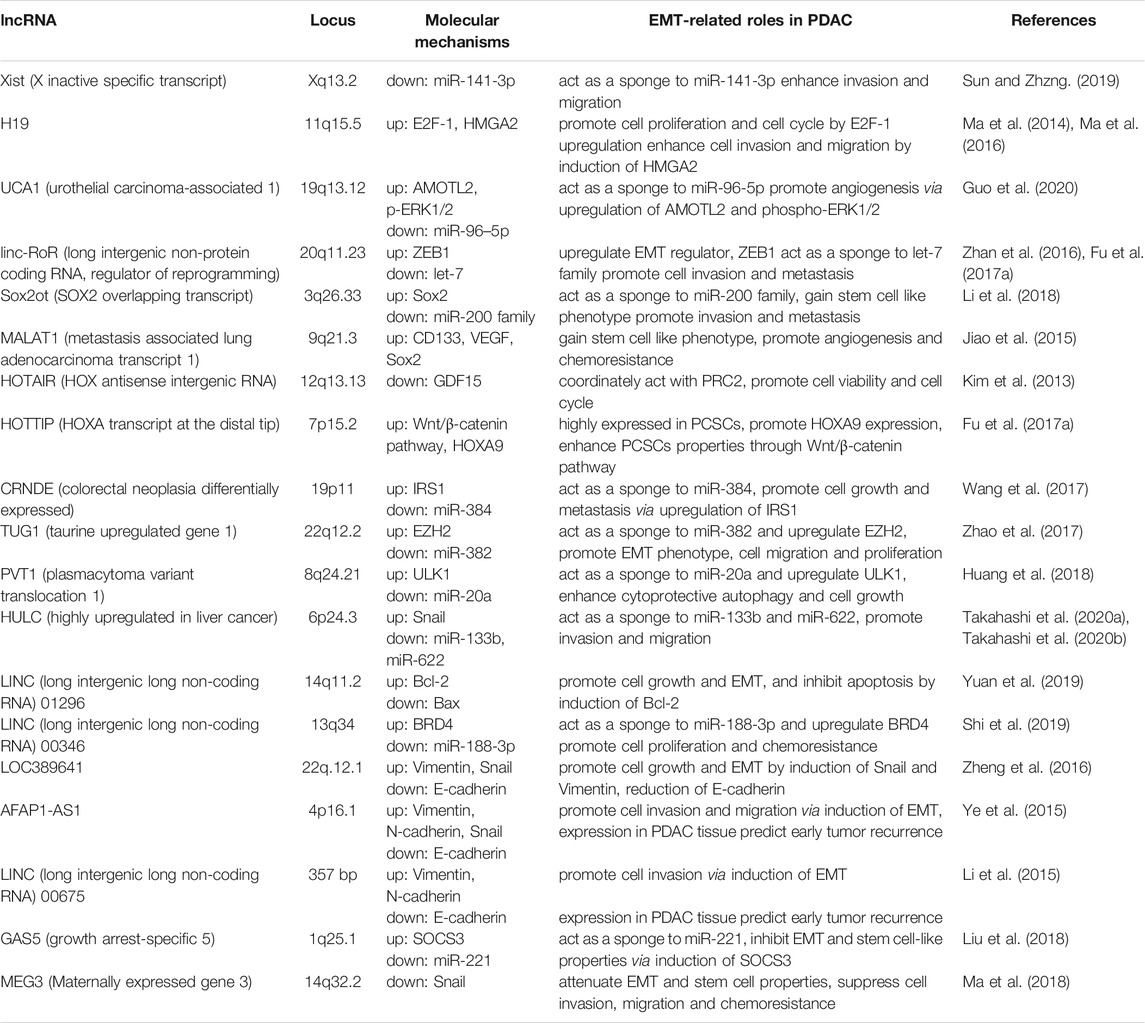
LncRNAs have diverse functions to mediate tumor cell progression, including the EMT process. Over recent decades, several lncRNAs have been reported to contribute to PDAC development, primarily through epigenetic regulation of EMT. Here, the crucial lncRNAs that can promote or suppress tumor invasion and metastasis by EMT regulation in PDAC are summarized (Duguang et al., 2017; Pandya et al., 2020) (Table 1; Figure 1). Note, the following lncRNAs are considered oncogenic in PDAC: X inactive specific transcript (Xist), H19, UCA1, linc-RoR, SOX2 overlapping transcript (Sox2ot), MALAT-1, HOTAIR, HOTTIP, colorectal neoplasia differentially expressed (CRNDE), taurine upregulated gene 1 (TUG1), plasmacytoma variant translocation 1 (PVT1), HULC, LINC01296, and LINC00346. Meanwhile, the following are considered to be oncosuppressive lncRNAs: growth arrest-specific 5 (GAS5), maternally expressed gene 3 (MEG3), LINC01111, and LINC00261. The molecular mechanisms, as well as the targeted, or related, genes of these lncRNAs are summarized in Table 1. Reviews are generally cited in other reviews only when referring to specific perspectives offered by them.
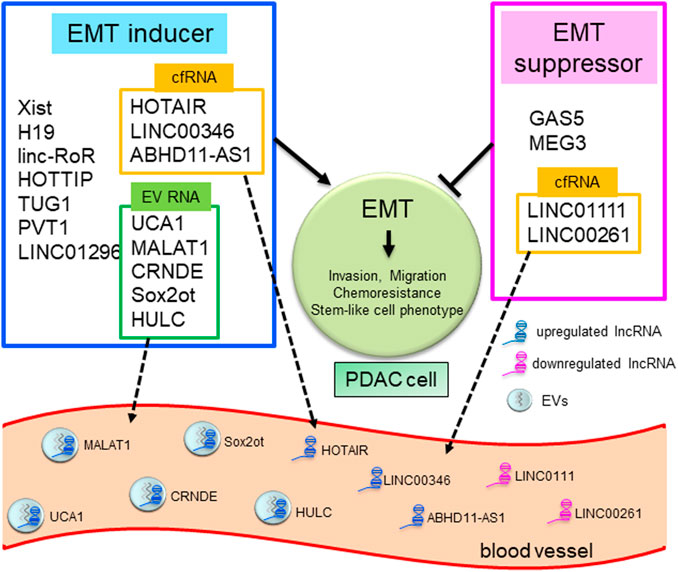
FIGURE 1. LncRNAs in EMT and liquid biopsy in human PDAC. Schematic overview showing lncRNAs mediating the EMT pathway and as tools for liquid biopsy in PDAC. LncRNAs as EMT inducers or suppressors are summarized in blue or red frames. LncRNAs in yellow or green frames are reported to exist in blood as EV-encapsulated RNAs or cell-free RNAs (cfRNAs) and can be used for liquid biopsy.
Some of the first discovered lncRNAs described were Xist and H19. Xist and H19 can modulate gene expression via targeted effects on chromatin remodeling or X chromosome inactivation and imprinting. Loss of imprinting at the H19 locus increases H19 expression in HCC (Matouk et al., 2007; Pachnis et al., 1988). In PDAC tissues, Xist is upregulated, and directly targets and attenuates the activity of miR-141-3p, which was downregulated, to enhance tumor cell invasion and migration (Sun and Zhang 2019). H19 also participates in the tumorigenesis of several cancers. H19 upregulates E2F-1 expression and promotes pancreatic cancer proliferation via cell cycle regulation (Ma et al., 2016). Additionally, H19 enhances cell invasion and migration involved in EMT induction through expression of HMGA2 and let-7 (Ma et al., 2014). Together, the lncRNAs involved in chromatin remodeling, Xist and H19, serve as oncogenes in PDAC development.
The oncogenic lncRNA UCA1 is highly expressed in PDAC tissues and cell-derived EVs, especially under hypoxic conditions. UCA1 could be transported by EVs, and promotes angiogenesis and tumor growth in PDAC cells under hypoxia. UCA1 acts as a ceRNA that binds to targeted miRNAs via the complementary site, thus, regulating miRNA expression and activity, and modulating the downstream pathways of miR-96-5p (Guo et al., 2020). Linc-RoR works as an oncogene in several cancers (Pan et al., 2016), enhances the cellular tolerance to hypoxic stress by acting as a miRNA sponge to miR-145, and promotes chemoresistance to anti-cancer drugs by inducing stem cell-like properties, as shown in HCC (Takahashi et al., 2014b; Takahashi et al., 2014c). It is highly upregulated in PDAC tissues compared to adjacent normal tissues and promotes cell migration and metastasis by regulating a crucial EMT regulator, ZEB1 (Zhan et al., 2016). Additionally, linc-RoR acts as a ceRNA to the let-7 miRNA family and induces the properties of cancer stem-like cells (Fu et al., 2017b). Therefore, linc-RoR could be an EMT inducer in PDAC cells. LncRNA Sox2ot could promote stemness by induction of EMT in PDAC cells. Sox2ot acts as a miRNA sponge to the miR-200 family, upregulates Sox2 expression, and promotes EMT and stemness, suggesting that Sox2ot can accelerate invasion and metastasis of PDAC (Li et al., 2018). MALAT-1 is one of the well-known oncogenic lncRNAs that act in diverse tumors, such as lung, hepatocellular or gastric cancers (Fu et al., 2020). (Jiao et al., 2015) reported that MALAT-1 promotes the EMT process and generates cancer stem cells (CSCs). Acquiring the CSC-like phenotype via induction of CD133, VEGF, and SOX2 expression could enhance angiogenesis, tumorigenicity, and resistance to gemcitabine chemotherapy (Jiao et al., 2015). HOTAIR is an oncogenic lncRNA reported in colon, liver, breast, and other cancers (Qu et al., 2019). In PDAC, HOTAIR is overexpressed in cell lines and tumor tissues. Moreover, HOTAIR regulates cell viability and cell cycle by suppressing the promoter activity of the growth-inhibitory and proapoptotic gene GDF15 (Kim et al., 2013). Expression of lncRNA HOTTIP is increased in PDAC, which promotes cell growth and EMT (Ghafouri-Fard et al., 2020). Fu et al. (2017a) suggested that HOTTIP is highly expressed in PCSCs, promotes HOXA9 expression, and enhances PCSC properties through the induction of the Wnt/β-catenin pathway. This mechanism could facilitate cancer cell progression in PDAC (Fu et al., 2017a).
Moreover, the lncRNA CRNDE is also reported to be involved in EMT of PDAC. CRNDE expression is upregulated in PDAC cell lines and tissues. It acts as a ceRNA, and directly targets and attenuates miR-384 activity. CRNDE increases cell growth and metastasis by suppressing miR-384 and upregulating IRS1, which is a crucial mediator of oncogenic insulin-like growth factor (IGF) signaling and can promote tumor development in several cancers (Wang et al., 2017). Similarly, the lncRNA TUG1 is aberrantly expressed in many cancers (Kondo et al., 2017). TUG1 is significantly upregulated in PDAC tissues, competitively working as a sponge to miR-382, and upregulating the expression of enhancer of zeste homolog 2 (EZH2), which is targeted by miR-382. Through the induction of EZH2 expression, TUG1 could promote EMT, PDAC cell migration, and proliferation (Zhao et al., 2017). PVT1 is an oncogenic lncRNA that is highly expressed in PDAC tissues and is correlated to a worse prognosis. PVT1 acts as a ceRNA to miR-20a, and upregulates the expression of Unc-51 like kinase 1 (ULK1) protein by suppressing miR-20a activity. Induction of ULK1 expression by PVT1 could enhance cytoprotective autophagy and tumor cell growth in vitro and in vivo (Huang et al., 2018). As an oncogenic lncRNA in PDAC, we previously reported that lncRNA HULC was exceptionally expressed in PDAC cell lines and could enhance cell invasion and migration through induction of EMT. HULC acts as a miRNA sponge to EMT-suppressive miRNAs miR-133b or miR-622. Downregulation of miR-133b and miR-622 by HULC leads to the upregulation of EMT-inducing transcription factor Snail, and cell invasion and migration (Takahashi et al., 2020a; Takahashi et al., 2020b).
Furthermore, previously unknown lncRNAs are shown to be related to the EMT process. LINC01296 is reported to be highly expressed in some cancers (Xu et al., 2019). Specifically, LINC01296 is increased in resected PDAC tissues, and its expression is associated with advanced stage, presence of lymph node metastasis, and worse patient prognosis. Knockdown of LINC01296 inhibits cell growth, enhances apoptosis by modulating Bcl-2 and Bax expression, and suppresses EMT, cell invasion, and migration (Yuan et al., 2019). LINC00346 is also reported to promote cell proliferation, cell cycle, tumorigenesis, and gemcitabine resistance. LINC00346 serves as a miRNA sponge for miR-188-3p, inhibiting its activity and upregulates the expression of its target, bromodomain-containing protein 4 (BRD4), in PDAC cells. An increase in BRD4 expression leads to PDAC cell proliferation or chemoresistance (Shi et al., 2019).
In contrast, the lncRNA GAS5 works as a tumor suppressor in multiple cancers (Shi et al., 2013). GAS5 acts as a ceRNA for miR-221, which is known to promote carcinogenesis. Downregulation of miR-221 by GAS5 results in the induction of SOCS3 expression, which inhibits EMT and stemness, thereby attenuating PDAC cell proliferation, migration, and chemoresistance (Liu et al., 2018). MEG3 is another tumor-suppressive lncRNA (Shi et al., 2013), with a decreased expression in PDAC cell lines and tissues that is associated with an advanced TNM stage and poor prognosis. MEG3 attenuates EMT and stemness, thereby suppressing cell invasion, migration, and gemcitabine chemoresistance (Ma et al., 2018). Meanwhile, MEG3 is also reportedly an oncogenic lncRNA. According to the single cell RNA-sequencing (scRNA-seq) transcript data, MEG3 is highly expressed in metastatic PDAC tumors (Pan et al., 2021). The lncRNA LINC01111 is downregulated in PDAC tissues and patient-derived plasma, and positively correlates with better prognosis of patients. LINC01111 inhibits cell cycle, cell invasion and migration in vitro, and metastasis in vivo by acting as a ceRNA to miR-3924, directly targeting and upregulating dual-specificity phosphatase 1 (DUSP1), and the subsequent suppression of the SAPK/JNK signaling pathway by DUSP1 (Pan et al., 2019). Furthermore, the lncRNA LINC00261 is also downregulated in PDAC tissues. It is downregulated by TGF-β signaling, and upregulated by the EMT-suppressive transcription factor forkhead box protein A2 (FOXA2) by binding to the LINC00261 promoter. Altogether, LINC00261 could be a tumor-suppressive lncRNA as it inhibits EMT (Dorn et al., 2020). These mechanisms can contribute to the suppression of tumor development (Pan et al., 2019). To summarize, GAS5, MEG3, LINC0111, and LINC00261 are tumor-suppressors in PDAC that act via inhibition of EMT.
Circular RNAs (circRNAs) and pseudogenes are categorized as subclasses of lncRNAs. CircRNAs are closed RNA transcripts that are generated by unique back-splicing of precursor mRNAs. However, the mechanisms underlying circRNA biogenesis, as well as the regulatory factors involved in their circularization, are not well understood. CircRNAs reportedly have several biological functions, including interacting with RNA-binding protein (RBP), function as an miRNA sponge, and regulating alternative splicing (Tang et al., 2021). For instance, circEYA3 is derived from the EYA3 gene and is highly expressed in PDAC tissues. In fact, elevated circEYA3 levels correlate with poor prognosis of PDAC patients. Mechanistically, circEYA3 facilitates PDAC cell invasion and migration via induction of EMT by its function as an endogenous miR-1294 sponge (Rong et al., 2021). CircNEIL3 is reportedly increased in PDAC tissue where it accelerates tumor cell migration and invasion through a sponge effect on miR-432-5p (Shen et al., 2021). These circRNAs regulate EMT and contribute to the promotion of tumor development in PDAC.
Pseudogenes have also been recognized as sophisticated modulators of gene expression. Recent studies demonstrated that a large number of pseudogenes, as well as their variable interactions with other biological molecules, may affect several pathways associated with cancer development (Hu et al., 2018). For instance, pseudogene pituitary tumor-transforming 3 (PTTG3P) is upregulated in PDAC tissue and is associated with larger tumor size and reduced overall survival. PTTG3P functions as a sponge for miR-132/212-3p, causing an increase in FoxM1 expression as well as PDAC cell invasion and migration (Liu et al., 2020). Moreover, pseudogene WTAPP1 is also elevated in PDAC tissue and is associated with poor prognosis. Specifically, WTAPP1 may PDAC cell proliferation and invasion via Wnt signaling activation (Deng et al., 2021). Hence, various pseudogenes appear to function as oncogenic genes in PDAC.
While few studies have provided evidence that circulating lncRNA in EVs can regulate the characteristics of recipient cells in distant tissues or organs in PDAC, potential roles for EV-encapsulated miRNAs in inducing systemic instigation in several cancers. For example, adipose tissue-derived EV miRNAs, such as miR-99b or miR-222, have been proposed to contribute to the regulation of metabolic homeostasis. These miRNAs can function in a paracrine, autocrine or endocrine manner (Mori et al., 2019). Moreover, miR 25-3p is reportedly transferred by colorectal cancer cell-derived exosomes to distant organs, such as the liver or lung, and induces pre-metastatic niche formation through the promotion of vascular permeability and angiogenesis in a murine model (Zeng et al., 2018). Furthermore, exosome-encapsulated miR-1247-3p, derived from HCC cells, can activate cancer-associated fibroblasts (CAFs), thereby promoting cancer progression. Indeed, EV miR-1274-3p abundance in serum is significantly associated with lung metastasis in HCC patients (Fang et al., 2018). Collectively, these reports imply that EVs, such as exosomes, mediate the transfer of miRNAs, which can modulate the formation of distant metastasis in many cancer types. Thus, investigation of EV-encapsulated lncRNAs that can act via endocrine instigation, are warranted in the future.
Finally, we review the lncRNAs that are correlated with PDAC driver gene mutations. KRAS and TP53 are recognized as driver genes for PDAC, and their mutation is one of the most crucial initiation steps during carcinogenesis (Bailey et al., 2016). A few lncRNAs are reported to be related to the mutations in these driver genes (Di Agostino 2020; Pandya et al., 2020). LincRNA1611 and HOX antisense intergenic RNA myeloid 1 (HOTAIRM1) were highly expressed in human PDAC tissues compared to adjacent normal tissues. While lincRNA1611 expression is positively correlated with mutation of TP53 gene (Wang et al.,. 2015a), HOTAIRM1 expression was positively associated with that of KRAS gene (Luo et al., 2019). These lncRNAs could interact with the mutated driver genes in PDAC.
Gene copy number variation has been recently identified as being related to carcinogenesis, and some lncRNAs were detected to have high amplification with one or more copy number alterations (Waddell et al., 2015). Although there are some new topics that are not elucidated in this review, such as the roles of lncRNAs located on high amplified chromosome region in EMT, previously unrecognized gene alterations in lncRNAs that are correlated with PDAC initiation and development must be investigated in future studies.
Diagnostic and Therapeutic Potential of EMT-Related lncRNAs
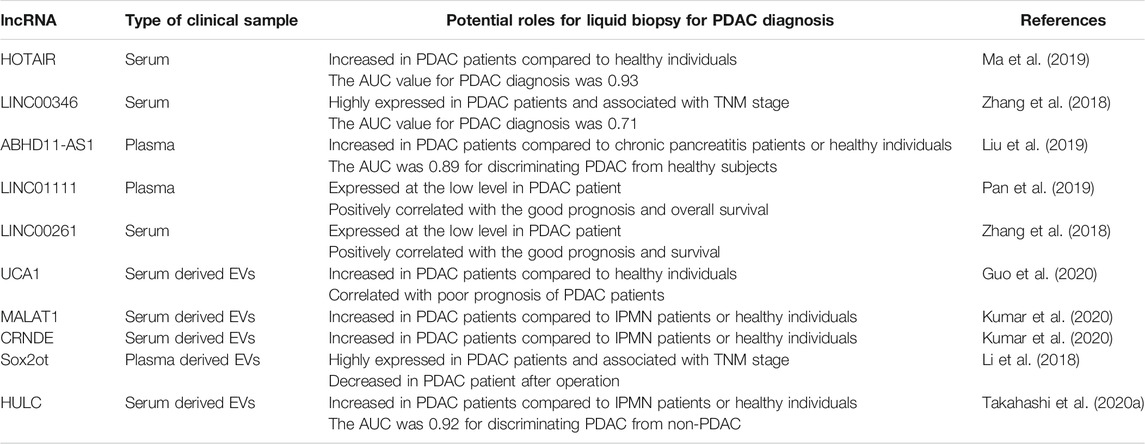
The most-used standard biomarker for PDAC diagnosis is carbohydrate antigen 19-9 (CA19-9). However, the CA19-9 positive ratio is about 55% for stage I cancers, suggesting that CA19-9 might not be a satisfactory biomarker for the early detection of PDAC (Liu et al., 2012). Therefore, it is necessary to identify a feasible and reliable marker supporting diagnosis at the initial stage of PDAC. LncRNAs have been reported as valuable liquid biopsy tools for PDAC diagnosis using serum, plasma, and other body fluids (Wang et al., 2015b). Recently, circulating tumor cells (CTCs), cell-free DNA (cfDNA), cell-free RNA (cfRNA), and EVs have all been investigated as potential tools for liquid biopsy (Crowley et al., 2013; Yoshioka et al., 2018). Some lncRNAs can be detected in cfRNA or EVs in body fluids and may serve as biomarkers for diagnosing PDAC. The EMT-related lncRNAs in serum or plasma that can serve as potential biomarkers for human PDAC are summarized in Table 2. HOTAIR, LINC00346, ABHD11-AS1, LINC01111, and LINC00261 in serum or plasma, are proposed to be valuable biomarkers for PDAC, while UCA1, MALAT1, CRNDE, SOX2OT, and HULC are highly expressed in EVs derived from patient blood.
Upon analyzing cfRNA in serum or plasma, oncogenic lncRNA HOTAIR was found to be highly expressed in serum and tissues from patients with PDAC compared to healthy individuals and adjacent normal tissues. Receiver operating characteristic (ROC) curve analysis with area under the curve (AUC) showed that HOTAIR could promote the accuracy of diagnosis for PDAC (Ma et al., 2019). Similarly, LINC00346 was highly expressed in tissues and serum of patients with PDAC. It was positively correlated with advanced clinical stage and poor survival (Zhang et al., 2018). Additionally, lncRNA ABHD11-AS1 was found to contribute to the early diagnosis of PDAC. Plasma levels of ABHD11-AS1 were significantly higher than in patients with chronic pancreatitis than healthy individuals. Moreover, the combination of ABHD11-AS1 and CA19-9 was found to be more effective for early PDAC diagnosis (Liu et al., 2019). Therefore, these lncRNAs could be potential markers for the early diagnosis of PDAC from patient blood.
In contrast, LINC01111 was expressed at low levels in the plasma and tissues of patients with PDAC compared to healthy controls or adjacent healthy tissue. The expression level of LINC01111 was negatively associated with the advanced TNM stage, and positively correlated with favorable prognosis and overall survival in patients with PDAC (Pan et al., 2019). Moreover, LINC00261 expression, associated with a higher survival rate (Zhang et al., 2018), was decreased in PDAC tissues and patient serum.
There is limited literature regarding lncRNAs encapsulated within EVs being considered as biomarkers for PDAC diagnosis. UCA1 is highly expressed in PDAC cell-derived EVs and could be transferred from donor to recipient cells by EVs. UCA1 expression was significantly increased in EVs derived from serum of patients with PDAC compared to healthy donors and was associated with a poor prognosis in these patients (Guo et al., 2020). MALAT-1 can decrease the chemosensitivity and accelerate tumor angiogenesis by enhancing stem cell-like phenotypes via induction of EMT (Jiao et al., 2015), and CRNDE promotes cell proliferation, invasion, and migration by sponging miR-384 in PDAC cells (Wang et al., 2017). These two lncRNAs were highly expressed in serum EVs from patients with PDAC compared with controls, suggesting that they could act as crucial liquid biopsy tools for PDAC diagnosis (Kumar et al., 2020). Sox2ot promotes PDAC invasion and metastasis by induction of EMT. Plasma EVs containing Sox2ot were highly expressed in patients with PDAC and were associated with the advanced TNM stage.
Interestingly, exosomal Sox2ot expression was decreased after surgery in the plasma of patients with PDAC (Li et al., 2018). Furthermore, we have recently reported that HULC was encapsulated and carried by PDAC cell-derived EVs in body fluids. HULC expression in serum EVs was significantly increased in patients with PDAC compared to patients with intraductal papillary mucinous neoplasm (IPMN) or healthy individuals. The ROC curve revealed that EV-encapsulated HULC showed good predictive performance, with an AUC of 0.92, when compared to CA19-9 (0.9) and carbohydrate antigen 19-9 (CEA) (0.54) for discriminating against patients with PDAC from those without PDAC (Takahashi et al., 2020a). These reports illustrate the usefulness of lncRNAs in EVs as novel tools for early PDAC diagnosis using liquid biopsy.
Additionally, one study has described the usefulness of lncRNAs as biomarker for IPMN. Specifically, plasma expression of lncRNA GAS5 and SRA has proven useful as markers to distinguish IPMN patients from healthy controls. Moreover, the combined expression of eight lncRNAs (ADARB2-AS1, ANRIL, GLIS3-AS1, LINC00472, MEG3, PANDA, PVT1, and UCA1) may effectively discriminate between malignant and benign IPMN (Permuth et al., 2017).
Various lncRNAs have also been reported as predictors of metastasis following surgical resection of the primary tumor. For instance, lncRNA AFAP1-AS1 is overexpressed in human PDAC tissue and can promote PDAC cell invasion and migration via induction of EMT. Moreover, elevated expression of AFAP1-AS1 can predict poor prognosis and tumor recurrence within 6 months and 1 year after surgical resection, respectively (Ye et al., 2015). LINC00675 is also aberrantly overexpressed in human PDAC tissue and accelerates PDAC cell migration. This upregulation of LINC00675 in PDAC tissue can predict recurrence within 6 months of surgical resection (Li et al., 2015). Hence, these two lncRNAs may represent effective predictors for early recurrence of PDAC following surgical intervention.
Regarding clinical application for cancer therapy, several studies have investigated lncRNAs as potential therapeutic tools as they could be helpful in advancing therapeutic strategies to target cancer cells. There are some attempts being made to regulate lncRNAs, focusing on their degradation and the functional suppression by use of siRNA, shRNA, or inhibition of transcription (Pandya et al., 2020). EV-encapsulated lncRNAs can also be exploited for clinical applications. An example of EV-mediated gene therapy, the effect of miRNA from human mesenchymal stem cell (hMSC)-derived EVs, was reported in PDAC cells. The transfer of EVs from hMSCs abundant in miR-143-3p, a tumor suppressor miRNA, to PDAC cells attenuated cell invasion and induced apoptosis by suppressing the lncRNA P11-363N22.3 (Wang et al., 2021). Moreover, transfer of the mutated protein Survivin-T34A by EVs enhanced gemcitabine sensitivity of PDAC cells (Aspe et al., 2014). These reports indicate that EVs would have enormous potential as the transfer tool for nucleic acids, including lncRNA and miRNA (Ariston Gabriel et al., 2020). Even though a majority of these results were based on assays in cell lines, these insights into lncRNAs might provide possible therapeutic applications in future, in addition to diagnostic liquid biopsies that predict prognosis and response to therapy in patients with PDAC (Figure 1).
Perspectives
The challenge of revealing the molecular mechanisms of EMT in pancreatic cancer that are regulated by lncRNAs could direct the discovery of new therapies, and tools for liquid biopsy to diagnose and predict therapeutic effects. Understanding the regulation of lncRNA expression during the initiation and progression of PDAC would provide a new paradigm on cancer pathogenesis. These biological insights may contribute to a rapid progress in development of diagnostic and therapeutic interventions for PDAC using lncRNA and EV-based strategies. Further studies will be required to validate the utility of the candidate lncRNAs for future clinical applications.
Author Contributions
KTak proposed the topic, wrote the manuscript, and selected the literature. KTan and YO wrote the manuscript. MF, YM, and TO reviewed and edited the manuscript.
Funding
This work was supported by JSPS KAKENHI (Grant Numbers 21K07954, 20H03655, 21H02758, and 21K19402), The Takeda Science Foundation, The Kobayashi Foundation and the Pancreas Research Foundation, Japan.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are grateful to Prof. Satoshi Fujii and Kazunobu Aso for their helpful advice. We thank all members of our department for helpful discussions.
References
Anastasiadou, E., Jacob, L. S., and Slack, F. J. (2018). Non-coding RNA Networks in Cancer. Nat. Rev. Cancer 18, 5–18. doi:10.1038/nrc.2017.99
Ariston Gabriel, A. N., Wang, F., Jiao, Q., Yvette, U., Yang, X., Al-Ameri, S. A., et al. (2020). The Involvement of Exosomes in the Diagnosis and Treatment of Pancreatic Cancer. Mol. Cancer 19, 19–132. doi:10.1186/s12943-020-01245-y
Aspe, J. R., Diaz Osterman, C. J., Jutzy, J. M., Deshields, S., Whang, S., and Wall, N. R. (2014). Enhancement of Gemcitabine Sensitivity in Pancreatic Adenocarcinoma by Novel Exosome-Mediated Delivery of the Survivin-T34a Mutant. J. Extracell Vesicles 3, 23244. doi:10.3402/jev.v3.23244
Bailey, J. M., Hendley, A. M., Lafaro, K. J., Pruski, M. A., Jones, N. C., Alsina, J., et al. (2016). p53 Mutations Cooperate with Oncogenic Kras to Promote Adenocarcinoma from Pancreatic Ductal Cells. Oncogene 35, 4282–4288. doi:10.1038/onc.2015.441
Baumann, M., Krause, M., and Hill, R. (2008). Exploring the Role of Cancer Stem Cells in Radioresistance. Nat. Rev. Cancer 8, 545–554. doi:10.1038/nrc2419
Brunetti, O., Aprile, G., Marchetti, P., Vasile, E., Casadei Gardini, A., Scartozzi, M., et al. (2018). Systemic Chemotherapy for Advanced Rare Pancreatic Histotype Tumors: A Retrospective Multicenter Analysis. Pancreas 47, 759–771. doi:10.1097/mpa.0000000000001063
Carninci, P., Kasukawa, T., Katayama, S., Gough, J., Frith, M. C., Maeda, N., et al. (2005). The Transcriptional Landscape of the Mammalian Genome. Science 309, 1559–1563. doi:10.1126/science.1112014
Craene, B. D., and Berx, G. (2013). Regulatory Networks Defining EMT during Cancer Initiation and Progression. Nat. Rev. Cancer 13, 97–110. doi:10.1038/nrc3447
Crowley, E., Di Nicolantonio, F., Loupakis, F., and Bardelli, A. (2013). Liquid Biopsy: Monitoring Cancer-Genetics in the Blood. Nat. Rev. Clin. Oncol. 10, 472–484. doi:10.1038/nrclinonc.2013.110
Deng, J., Zhang, J., Ye, Y., Liu, K., Zeng, L., Huang, J., et al. (2021). N6-methyladenosine-mediated Upregulation of WTAPP1 Promotes WTAP Translation and Wnt Signaling to Facilitate Pancreatic Cancer Progression. Cancer Res. 81, 5268-5283. doi:10.1158/0008-5472.can-21-0494
Derrien, T., Johnson, R., Bussotti, G., Tanzer, A., Djebali, S., Tilgner, H., et al. (2012). The GENCODE V7 Catalog of Human Long Noncoding RNAs: Analysis of Their Gene Structure, Evolution, and Expression. Genome Res. 22, 1775–1789. doi:10.1101/gr.132159.111
Di Agostino, S. (2020). The Impact of Mutant P53 in the Non-coding RNA World. Biomolecules 10, 472. doi:10.3390/biom10030472
Dorn, A., Glass, M., Neu, C. T., Heydel, B., Hüttelmaier, S., Gutschner, T., et al. (2020). LINC00261 Is Differentially Expressed in Pancreatic Cancer Subtypes and Regulates a Pro-epithelial Cell Identity. Cancers (Basel) 12, 1227. doi:10.3390/cancers12051227
Duguang, L., Jin, H., Xiaowei, Q., Peng, X., Xiaodong, W., Zhennan, L., et al. (2017). The Involvement of lncRNAs in the Development and Progression of Pancreatic Cancer. Cancer Biol. Ther. 18, 927–936. doi:10.1080/15384047.2017.1385682
Dunham, I., Kundaje, A., Aldred, S. F., Collins, P. J., Davis, C. A., Doyle, F., et al. (2012). An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature 6 (489), 57–74. doi:10.1038/nature11247
Fang, T., Lv, H., Lv, G., Li, T., Wang, C., Han, Q., et al. (2018). Tumor-derived Exosomal miR-1247-3p Induces Cancer-Associated Fibroblast Activation to foster Lung Metastasis of Liver Cancer. Nat. Commun. 9, 191. doi:10.1038/s41467-017-02583-0
Fu, S., Wang, Y., Li, H., Chen, L., and Liu, Q. (2020). Regulatory Networks of LncRNA MALAT-1 in Cancer. Cmar 12, 10181–10198. doi:10.2147/cmar.s276022
Fu, Z., Chen, C., Zhou, Q., Wang, Y., Zhao, Y., Zhao, X., et al. (2017a). LncRNA HOTTIP Modulates Cancer Stem Cell Properties in Human Pancreatic Cancer by Regulating HOXA9. Cancer Lett. 410, 68–81. doi:10.1016/j.canlet.2017.09.019
Fu, Z., Li, G., Li, Z., Wang, Y., Zhao, Y., Zheng, S., et al. (2017b). Endogenous miRNA Sponge LincRNA-ROR Promotes Proliferation, Invasion and Stem Cell-like Phenotype of Pancreatic Cancer Cells. Cell Death Discov. 3, 17004. doi:10.1038/cddiscovery.2017.4
Ghafouri-Fard, S., Dashti, S., and Taheri, M. (2020). The HOTTIP (HOXA Transcript at the Distal Tip) lncRNA: Review of Oncogenic Roles in Human. Biomed. Pharmacother. 127, 110158. doi:10.1016/j.biopha.2020.110158
Guo, Z., Wang, X., Yang, Y., Chen, W., Zhang, K., Teng, B., et al. (2020). Hypoxic Tumor-Derived Exosomal Long Noncoding RNA UCA1 Promotes Angiogenesis via miR-96-5p/AMOTL2 in Pancreatic Cancer. Mol. Ther. - Nucleic Acids 22, 179–195. doi:10.1016/j.omtn.2020.08.021
Holohan, C., Van Schaeybroeck, S., Longley, D. B., and Johnston, P. G. (2013). Cancer Drug Resistance: an Evolving Paradigm. Nat. Rev. Cancer 13, 714–726. doi:10.1038/nrc3599
Hu, W., Alvarez-Dominguez, J. R., and Lodish, H. F. (2012). Regulation of Mammalian Cell Differentiation by Long Non-coding RNAs. EMBO Rep. 13, 971–983. doi:10.1038/embor.2012.145
Hu, X., Yang, L., and Mo, Y. Y. (2018). Role of Pseudogenes in Tumorigenesis. Cancers (Basel) 10, 256. doi:10.3390/cancers10080256
Huang, F., Chen, W., Peng, J., Li, Y., Zhuang, Y., Zhu, Z., et al. (2018). LncRNA PVT1 Triggers Cyto-Protective Autophagy and Promotes Pancreatic Ductal Adenocarcinoma Development via the miR-20a-5p/ULK1 Axis. Mol. Cancer 17, 98. doi:10.1186/s12943-018-0845-6
Iizuka, N., Oka, M., Yamada-Okabe, H., Mori, N., Tamesa, T., Okada, T., et al. (2002). Comparison of Gene Expression Profiles between Hepatitis B Virus- and Hepatitis C Virus-Infected Hepatocellular Carcinoma by Oligonucleotide Microarray Data on the Basis of a Supervised Learning Method. Cancer Res. 62, 3939–3944. https://pubmed.ncbi.nlm.nih.gov/12124323/.
Jiao, F., Hu, H., Han, T., Yuan, C., Wang, L., Jin, Z., et al. (2015). Long Noncoding RNA MALAT-1 Enhances Stem Cell-like Phenotypes in Pancreatic Cancer Cells. Int. J. Mol. Sci. 16, 6677–6693. doi:10.3390/ijms16046677
Kamisawa, T., Wood, L. D., Itoi, T., and Takaori, K. (2016). Pancreatic Cancer. The Lancet 388, 73–85. doi:10.1016/s0140-6736(16)00141-0
Kim, K., Jutooru, I., Chadalapaka, G., Johnson, G., Frank, J., Burghardt, R., et al. (2013). HOTAIR Is a Negative Prognostic Factor and Exhibits Pro-oncogenic Activity in Pancreatic Cancer. Oncogene 32, 1616–1625. doi:10.1038/onc.2012.193
Kino, T., Hurt, D. E., Ichijo, T., Nader, N., and Chrousos, G. P. (2010). Noncoding RNA Gas5 Is a Growth Arrest- and Starvation-Associated Repressor of the Glucocorticoid Receptor. Sci. Signal. 3, ra8. doi:10.1126/scisignal.2000568
Kogure, T., Lin, W.-L., Yan, I. K., Braconi, C., and Patel, T. (2011). Intercellular Nanovesicle-Mediated microRNA Transfer: a Mechanism of Environmental Modulation of Hepatocellular Cancer Cell Growth. Hepatology 54, 1237–1248. doi:10.1002/hep.24504
Kondo, Y., Shinjo, K., and Katsushima, K. (2017). Long Non‐coding RNA S as an Epigenetic Regulator in Human Cancers. Cancer Sci. 108, 1927–1933. doi:10.1111/cas.13342
Kopp, F., and Mendell, J. T. (2018). Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 172, 393–407. doi:10.1016/j.cell.2018.01.011
Kumar, S. R., Kimchi, E. T., Manjunath, Y., Gajagowni, S., Stuckel, A. J., and Kaifi, J. T. (2020). RNA Cargos in Extracellular Vesicles Derived from Blood Serum in Pancreas Associated Conditions. Sci. Rep. 10, 2800. doi:10.1038/s41598-020-59523-0
Lai, E., Puzzoni, M., Ziranu, P., Pretta, A., Impera, V., Mariani, S., et al. (2019). New Therapeutic Targets in Pancreatic Cancer. Cancer Treat. Rev. 81, 101926. doi:10.1016/j.ctrv.2019.101926
Li, D. D., Fu, Z. Q., Lin, Q., Zhou, Y., Zhou, Q. B., Li, Z. H., et al. (2015). Linc00675 Is a Novel Marker of Short Survival and Recurrence in Patients with Pancreatic Ductal Adenocarcinoma. World J. Gastroenterol. 21, 9348–9357. doi:10.3748/wjg.v21.i31.9348
Li, Z., Jiang, P., Li, J., Peng, M., Zhao, X., Zhang, X., et al. (2018). Tumor-derived Exosomal Lnc-Sox2ot Promotes EMT and Stemness by Acting as a ceRNA in Pancreatic Ductal Adenocarcinoma. Oncogene 37, 3822–3838. doi:10.1038/s41388-018-0237-9
Liang, K., Liu, F., Fan, J., Sun, D., Liu, C., Lyon, C. J., et al. (2017). Nanoplasmonic Quantification of Tumor-Derived Extracellular Vesicles in Plasma Microsamples for Diagnosis and Treatment Monitoring. Nat. Biomed. Eng. 1, 0021. doi:10.1038/s41551-016-0021
Liu, B., Wu, S., Ma, J., Yan, S., Xiao, Z., Wan, L., et al. (2018). lncRNA GAS5 Reverses EMT and Tumor Stem Cell-Mediated Gemcitabine Resistance and Metastasis by Targeting miR-221/SOCS3 in Pancreatic Cancer. Mol. Ther. - Nucleic Acids 13, 472–482. doi:10.1016/j.omtn.2018.09.026
Liu, J., Gao, J., Du, Y., Li, Z., Ren, Y., Gu, J., et al. (2012). Combination of Plasma microRNAs with Serum CA19-9 for Early Detection of Pancreatic Cancer. Int. J. Cancer 131, 683–691. doi:10.1002/ijc.26422
Liu, W., Tang, J., Zhang, H., Kong, F., Zhu, H., Li, P., et al. (2020). A Novel lncRNA PTTG3P/miR-132/212-3p/FoxM1 Feedback Loop Facilitates Tumorigenesis and Metastasis of Pancreatic Cancer. Cell Death Discov 6, 136. doi:10.1038/s41420-020-00360-5
Liu, Y., Feng, W., Liu, W., Kong, X., Li, L., He, J., et al. (2019). Circulating lncRNA ABHD11-AS1 Serves as a Biomarker for Early Pancreatic Cancer Diagnosis. J. Cancer 10, 3746–3756. doi:10.7150/jca.32052
Luo, Y., He, Y., Ye, X., Song, J., Wang, Q., Li, Y., et al. (2019). High Expression of Long Noncoding RNA HOTAIRM1 Is Associated with the Proliferation and Migration in Pancreatic Ductal Adenocarcinoma. Pathol. Oncol. Res. 25, 1567–1577. doi:10.1007/s12253-018-00570-4
Ma, C., Nong, K., Zhu, H., Wang, W., Huang, X., Yuan, Z., et al. (2014). H19 Promotes Pancreatic Cancer Metastasis by Derepressing Let-7's Suppression on its Target HMGA2-Mediated EMT. Tumor Biol. 35, 9163–9169. doi:10.1007/s13277-014-2185-5
Ma, L., Wang, F., Du, C., Zhang, Z., Guo, H., Xie, X., et al. (2018). Long Non-coding RNA MEG3 Functions as a Tumour Suppressor and Has Prognostic Predictive Value in Human Pancreatic Cancer. Oncol. Rep. 39, 1132–1140. doi:10.3892/or.2018.6178
Ma, L., Tian, X., Wang, F., Zhang, Z., Du, C., Xie, X., et al. (2016). The Long Noncoding RNA H19 Promotes Cell Proliferation via E2F-1 in Pancreatic Ductal Adenocarcinoma. Cancer Biol. Ther. 17, 1051–1061. doi:10.1080/15384047.2016.1219814
Ma, Y., Hu, M., Zhou, L., Ling, S., Li, Y., Kong, B., et al. (2019). Long Non-coding RNA HOTAIR Promotes Cancer Cell Energy Metabolism in Pancreatic Adenocarcinoma by Upregulating Hexokinase-2. Oncol. Lett. 18, 2212–2219. doi:10.3892/ol.2019.10551
Mani, S. A., Guo, W., Liao, M.-J., Eaton, E. N., Ayyanan, A., Zhou, A. Y., et al. (2008). The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells. Cell 133, 704–715. doi:10.1016/j.cell.2008.03.027
Matouk, I. J., DeGroot, N., Mezan, S., Ayesh, S., Abu-lail, R., Hochberg, A., et al. (2007). The H19 Non-coding RNA Is Essential for Human Tumor Growth. PLoS One 2, e845. doi:10.1371/journal.pone.0000845
Mori, M. A., Ludwig, R. G., Garcia-Martin, R., Brandão, B. B., and Kahn, C. R. (2019). Extracellular miRNAs: From Biomarkers to Mediators of Physiology and Disease. Cel Metab. 30, 656–673. doi:10.1016/j.cmet.2019.07.011
Nieto, M. A., and Cano, A. (2012). The Epithelial-Mesenchymal Transition under Control: Global Programs to Regulate Epithelial Plasticity. Semin. Cancer Biol. 22, 361–368. doi:10.1016/j.semcancer.2012.05.003
O'Brien, S. J., Bishop, C., Hallion, J., Fiechter, C., Scheurlen, K., Paas, M., et al. (2020). Long Non-coding RNA (lncRNA) and Epithelial-Mesenchymal Transition (EMT) in Colorectal Cancer: a Systematic Review. Cancer Biol. Ther. 21, 769–781. doi:10.1080/15384047.2020.1794239
Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., et al. (2002). Analysis of the Mouse Transcriptome Based on Functional Annotation of 60,770 Full-Length cDNAs. Nature 420, 563–573. doi:10.1038/nature01266
Pachnis, V., Brannan, C. I., and Tilghman, S. M. (1988). The Structure and Expression of a Novel Gene Activated in Early Mouse Embryogenesis. EMBO J. 7, 673–681. doi:10.1002/j.1460-2075.1988.tb02862.x
Pan, H., Diao, H., Zhong, W., Wang, T., Wen, P., and Wu, C. (2021). A Cancer Cell Cluster Marked by LincRNA MEG3 Leads Pancreatic Ductal Adenocarcinoma Metastasis. Front. Oncol. 11, 656564. doi:10.3389/fonc.2021.656564
Pan, S., Shen, M., Zhou, M., Shi, X., He, R., Yin, T., et al. (2019). Long Noncoding RNA LINC01111 Suppresses Pancreatic Cancer Aggressiveness by Regulating DUSP1 Expression via microRNA-3924. Cell Death Dis 10, 883. doi:10.1038/s41419-019-2123-y
Pan, Y., Li, C., Chen, J., Zhang, K., Chu, X., Wang, R., et al. (2016). The Emerging Roles of Long Noncoding RNA ROR (lincRNA-ROR) and its Possible Mechanisms in Human Cancers. Cell Physiol Biochem 40, 219–229. doi:10.1159/000452539
Pandya, G., Kirtonia, A., Sethi, G., Pandey, A. K., and Garg, M. (2020). The Implication of Long Non-coding RNAs in the Diagnosis, Pathogenesis and Drug Resistance of Pancreatic Ductal Adenocarcinoma and Their Possible Therapeutic Potential. Biochim. Biophys. Acta (Bba) - Rev. Cancer 1874, 188423. doi:10.1016/j.bbcan.2020.188423
Permuth, J. B., Chen, D. T., Yoder, S. J., Li, J., Smith, A. T., Choi, J. W., et al. (2017). Linc-ing Circulating Long Non-coding RNAs to the Diagnosis and Malignant Prediction of Intraductal Papillary Mucinous Neoplasms of the Pancreas. Sci. Rep. 7, 10484. doi:10.1038/s41598-017-09754-5
Poliseno, L., Salmena, L., Zhang, J., Carver, B., Haveman, W. J., and Pandolfi, P. P. (2010). A Coding-independent Function of Gene and Pseudogene mRNAs Regulates Tumour Biology. Nature 465, 1033–1038. doi:10.1038/nature09144
Qu, X., Alsager, S., Zhuo, Y., and Shan, B. (2019). HOX Transcript Antisense RNA (HOTAIR) in Cancer. Cancer Lett. 10 (454), 90–97. doi:10.1016/j.canlet.2019.04.016
Ransohoff, J. D., Wei, Y., and Khavari, P. A. (2018). The Functions and Unique Features of Long Intergenic Non-coding RNA. Nat. Rev. Mol. Cel Biol 19, 143–157. doi:10.1038/nrm.2017.104
Rinn, J. L., and Chang, H. Y. (2012). Genome Regulation by Long Noncoding RNAs. Annu. Rev. Biochem. 81, 145–166. doi:10.1146/annurev-biochem-051410-092902
Rong, Z., Shi, S., Tan, Z., Xu, J., Meng, Q., Hua, J., et al. (2021). Circular RNA CircEYA3 Induces Energy Production to Promote Pancreatic Ductal Adenocarcinoma Progression through the miR-1294/c-Myc axis. Mol. Cancer 20, 106. doi:10.1186/s12943-021-01400-z
Shen, P., Yang, T., Chen, Q., Yuan, H., Wu, P., Cai, B., et al. (2021). CircNEIL3 Regulatory Loop Promotes Pancreatic Ductal Adenocarcinoma Progression via miRNA Sponging and A-To-I RNA-Editing. Mol. Cancer 20, 51. doi:10.1186/s12943-021-01333-7
Shi, W., Zhang, C., Ning, Z., Hua, Y., Li, Y., Chen, L., et al. (2019). Long Non-coding RNA LINC00346 Promotes Pancreatic Cancer Growth and Gemcitabine Resistance by Sponging miR-188-3p to Derepress BRD4 Expression. J. Exp. Clin. Cancer Res. 38, 60. doi:10.1186/s13046-019-1055-9
Shi, X., Sun, M., Liu, H., Yao, Y., and Song, Y. (2013). Long Non-coding RNAs: a New Frontier in the Study of Human Diseases. Cancer Lett. 339, 159–166. doi:10.1016/j.canlet.2013.06.013
Shukla, N., Yan, I. K., and Patel, T. (2018). Multiplexed Detection and Quantitation of Extracellular Vesicle RNA Expression Using NanoString. Methods Mol. Biol. 1740, 177–185. doi:10.1007/978-1-4939-7652-2_14
Siegel, R. L., Miller, K. D., and Jemal, A. (2017). Cancer Statistics, 2017. CA: A Cancer J. Clinicians 67, 7–30. doi:10.3322/caac.21387
Slack, F. J., and Chinnaiyan, A. M. (2019). The Role of Non-coding RNAs in Oncology. Cell 179, 1033–1055. doi:10.1016/j.cell.2019.10.017
Sun, J., and Zhang, Y. (2019). LncRNA XIST Enhanced TGF-Β2 Expression by Targeting miR-141-3p to Promote Pancreatic Cancer Cells Invasion. Biosci. Rep. 39. doi:10.1042/BSR20190332
Sun, Y. W., Chen, Y. F., Li, J., Huo, Y. M., Liu, D. J., Hua, R., et al. (2014). A Novel Long Non-coding RNA ENST00000480739 Suppresses Tumour Cell Invasion by Regulating OS-9 and HIF-1α in Pancreatic Ductal Adenocarcinoma. Br. J. Cancer 111, 2131–2141. doi:10.1038/bjc.2014.520
Takahashi, K., Yan, I. K., Haga, H., and Patel, T. (2014b). Modulation of Hypoxia-Signaling Pathways by Extracellular Linc-RoR. J. Cel Sci 127, 1585–1594. doi:10.1242/jcs.141069
Takahashi, K., Koyama, K., Ota, Y., Iwamoto, H., Yamakita, K., Fujii, S., et al. (2020a). The Interaction between Long Non-coding RNA HULC and MicroRNA-622 via Transfer by Extracellular Vesicles Regulates Cell Invasion and Migration in Human Pancreatic Cancer. Front. Oncol. 10, 1013. doi:10.3389/fonc.2020.01013
Takahashi, K., Ota, Y., Kogure, T., Suzuki, Y., Iwamoto, H., Yamakita, K., et al. (2020b). Circulating Extracellular Vesicle‐encapsulated HULC Is a Potential Biomarker for Human Pancreatic Cancer. Cancer Sci. 111, 98–111. doi:10.1111/cas.14232
Takahashi, K., Yan, I., Haga, H., and Patel, T. (2014a). Long Noncoding RNA in Liver Diseases. Hepatology 60, 744–753. doi:10.1002/hep.27043
Takahashi, K., Yan, I. K., Kogure, T., Haga, H., and Patel, T. (2014c). Extracellular Vesicle-Mediated Transfer of Long Non-coding RNA ROR Modulates Chemosensitivity in Human Hepatocellular Cancer. FEBS Open Bio 4, 458–467. doi:10.1016/j.fob.2014.04.007
Tang, X., Ren, H., Guo, M., Qian, J., Yang, Y., and Gu, C. (2021). Review on Circular RNAs and New Insights into Their Roles in Cancer. Comput. Struct. Biotechnol. J. 19, 910–928. doi:10.1016/j.csbj.2021.01.018
Taniue, K., and Akimitsu, N. (2021). The Functions and Unique Features of LncRNAs in Cancer Development and Tumorigenesis. Int. J. Mol. Sci. Jan 10, 22. doi:10.3390/ijms22020632
Taniue, K., Kurimoto, A., Takeda, Y., Nagashima, T., Okada-Hatakeyama, M., Katou, Y., et al. (2016b). ASBEL-TCF3 Complex Is Required for the Tumorigenicity of Colorectal Cancer Cells. Proc. Natl. Acad. Sci. U S A. 113, 12739–12744. doi:10.1073/pnas.1605938113
Taniue, K., Kurimoto, A., Sugimasa, H., Nasu, E., Takeda, Y., Iwasaki, K., et al. (2016a). Long Noncoding RNA UPAT Promotes colon Tumorigenesis by Inhibiting Degradation of UHRF1. Proc. Natl. Acad. Sci. USA 113, 1273–1278. doi:10.1073/pnas.1500992113
Thomson, D. W., and Dinger, M. E. (2016). Endogenous microRNA Sponges: Evidence and Controversy. Nat. Rev. Genet. 17, 272–283. doi:10.1038/nrg.2016.20
Tkach, M., and Théry, C. (2016). Communication by Extracellular Vesicles: Where We Are and where We Need to Go. Cell 10164, 1226–1232. doi:10.1016/j.cell.2016.01.043
Tsang, W. P., and Kwok, T. T. (2007). Riboregulator H19 Induction of MDR1-Associated Drug Resistance in Human Hepatocellular Carcinoma Cells. Oncogene 26, 4877–4881. doi:10.1038/sj.onc.1210266
Turley, E. A., Veiseh, M., Radisky, D. C., and Bissell, M. J. (2008). Mechanisms of Disease: Epithelial-Mesenchymal Transition-Does Cellular Plasticity Fuel Neoplastic Progression? Nat. Rev. Clin. Oncol. 5, 280–290. doi:10.1038/ncponc1089
Valadi, H., Ekström, K., Bossios, A., Sjöstrand, M., Lee, J. J., and Lötvall, J. O. (2007). Exosome-mediated Transfer of mRNAs and microRNAs Is a Novel Mechanism of Genetic Exchange between Cells. Nat. Cel Biol 9, 654–659. doi:10.1038/ncb1596
Waddell, N., Pajic, M., Patch, A. M., Chang, D. K., Kassahn, K. S., Bailey, P., et al. (2015). Whole Genomes Redefine the Mutational Landscape of Pancreatic Cancer. Nature 518, 495–501. doi:10.1038/nature14169
Wang, B., Xu, Y., Wei, Y., Lv, L., Liu, N., Lin, R., et al. (2021). Human Mesenchymal Stem Cell-Derived Exosomal microRNA-143 Promotes Apoptosis and Suppresses Cell Growth in Pancreatic Cancer via Target Gene Regulation. Front. Genet. 12, 581694. doi:10.3389/fgene.2021.581694
Wang, G., Pan, J., Zhang, L., Wei, Y., and Wang, C. (2017). Long Non-coding RNA CRNDE Sponges miR-384 to Promote Proliferation and Metastasis of Pancreatic Cancer Cells through Upregulating IRS1. Cell Prolif 50, e12389. doi:10.1111/cpr.12389
Wang, K. C., and Chang, H. Y. (2011). Molecular Mechanisms of Long Noncoding RNAs. Mol. Cel 43, 904–914. doi:10.1016/j.molcel.2011.08.018
Wang, Q., Jiang, H., Ping, C., Shen, R., Liu, T., Li, J., et al. (2015a). Exploring the Wnt Pathway-Associated LncRNAs and Genes Involved in Pancreatic Carcinogenesis Driven by Tp53 Mutation. Pharm. Res. 32, 793–805. doi:10.1007/s11095-013-1269-z
Wang, Y., Li, Z., Zheng, S., Zhou, Y., Zhao, L., Ye, H., et al. (2015b). Expression Profile of Long Non-coding RNAs in Pancreatic Cancer and Their Clinical Significance as Biomarkers. Oncotarget 6, 35684–35698. doi:10.18632/oncotarget.5533
Xu, L., Zhang, L., Wang, T., Wu, Y., Pu, X., Li, M., et al. (2020). ExoceRNA Atlas: A Database of Cancer ceRNAs in Human Blood Exosomes. Life Sci. 257, 118092. doi:10.1016/j.lfs.2020.118092
Xu, L., Wei, B., Hui, H., Sun, Y., Liu, Y., Yu, X., et al. (2019). Positive Feedback Loop of lncRNA LINC01296/miR‐598/Twist1 Promotes Non‐small Cell Lung Cancer Tumorigenesis. J. Cel Physiol 234, 4563–4571. doi:10.1002/jcp.27235
Yanagida, S., Taniue, K., Sugimasa, H., Nasu, E., Takeda, Y., Kobayashi, M., et al. (2013). ASBEL, an ANA/BTG3 Antisense Transcript Required for Tumorigenicity of Ovarian Carcinoma. Sci. Rep. 3, 1305. doi:10.1038/srep01305
Ye, Y., Chen, J., Zhou, Y., Fu, Z., Zhou, Q., Wang, Y., et al. (2015). High Expression of AFAP1-AS1 Is Associated with Poor Survival and Short-Term Recurrence in Pancreatic Ductal Adenocarcinoma. J. Transl Med. 13, 137. doi:10.1186/s12967-015-0490-4
Ying, H., Dey, P., Yao, W., Kimmelman, A. C., Draetta, G. F., Maitra, A., et al. (2016). Genetics and Biology of Pancreatic Ductal Adenocarcinoma. Genes Dev. 30, 355–385. doi:10.1101/gad.275776.115
Yoon, J.-H., Abdelmohsen, K., Kim, J., Yang, X., Martindale, J. L., Tominaga-Yamanaka, K., et al. (2013). Scaffold Function of Long Non-coding RNA HOTAIR in Protein Ubiquitination. Nat. Commun. 4, 2939. doi:10.1038/ncomms3939
Yoon, J. H., Abdelmohsen, K., Srikantan, S., Yang, X., Martindale, J. L., De, S., et al. (2012). LincRNA-p21 Suppresses Target mRNA Translation. Mol. Cel 47, 648–655. doi:10.1016/j.molcel.2012.06.027
Yoshioka, Y., Katsuda, T., and Ochiya, T. (2018). Extracellular Vesicles and Encapusulated miRNAs as Emerging Cancer Biomarkers for Novel Liquid Biopsy. Jpn. J. Clin. Oncol. 48, 869–876. doi:10.1093/jjco/hyy120
Yuan, Q., Zhang, Y., Feng, L., and Jiang, Y. (2019). Upregulated Long Noncoding RNA LINC01296 Indicates a Dismal Prognosis for Pancreatic Ductal Adenocarcinoma and Promotes Cell Metastatic Properties by Affecting EMT. J. Cel Biochem 120, 552–561. doi:10.1002/jcb.27411
Zeng, Z., Li, Y., Pan, Y., Lan, X., Song, F., Sun, J., et al. (2018). Cancer-derived Exosomal miR-25-3p Promotes Pre-metastatic Niche Formation by Inducing Vascular Permeability and Angiogenesis. Nat. Commun. 9, 5395. doi:10.1038/s41467-018-07810-w
Zhan, H.-x., Wang, Y., Li, C., Xu, J.-w., Zhou, B., Zhu, J.-k., et al. (2016). LincRNA-ROR Promotes Invasion, Metastasis and Tumor Growth in Pancreatic Cancer through Activating ZEB1 Pathway. Cancer Lett. 374, 261–271. doi:10.1016/j.canlet.2016.02.018
Zhang, B., Li, C., and Sun, Z. (2018). Long Non-coding RNA LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 Are Novel Prognostic Markers for Pancreatic Cancer. Am. J. Transl Res. 10, 2648–2658. https://pubmed.ncbi.nlm.nih.gov/30210701/.
Zhao, L., Sun, H., Kong, H., Chen, Z., Chen, B., and Zhou, M. (2017). The Lncrna-Tug1/ezh2 Axis Promotes Pancreatic Cancer Cell Proliferation, Migration and EMT Phenotype Formation through Sponging Mir-382. Cel Physiol Biochem 42, 2145–2158. doi:10.1159/000479990
Zheng, S., Chen, H., Wang, Y., Gao, W., Fu, Z., Zhou, Q., et al. (2016). Long Non-coding RNA LOC389641 Promotes Progression of Pancreatic Ductal Adenocarcinoma and Increases Cell Invasion by Regulating E-Cadherin in a TNFRSF10A-Related Manner. Cancer Lett. 371, 354–365. doi:10.1016/j.canlet.2015.12.010
Keywords: long non-coding RNA (lncRNA), extracellular vesicles (EVs), pancreatic ductal adenocarcinoma (PDAC), epithelial-mesenchymal transition (EMT), liquid biopsy, microRNA (miRNA)
Citation: Takahashi K, Taniue K, Ono Y, Fujiya M, Mizukami Y and Okumura T (2021) Long Non-Coding RNAs in Epithelial-Mesenchymal Transition of Pancreatic Cancer. Front. Mol. Biosci. 8:717890. doi: 10.3389/fmolb.2021.717890
Received: 31 May 2021; Accepted: 25 October 2021;
Published: 08 November 2021.
Edited by:
Yu Xiao, Wuhan University, ChinaReviewed by:
Paola Infante, Italian Institute of Technology (IIT), ItalyHiroaki Haga, Yamagata University, Japan
Hiroki Sasaguri, RIKEN Center for Brain Science (CBS), Japan
Yukiomi Nakade, Aichi Medical University, Japan
Kennichi Satoh, Tohoku Medical and Pharmaceutical University, Japan
Copyright © 2021 Takahashi, Taniue, Ono, Fujiya, Mizukami and Okumura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kenji Takahashi, dC1rZW5qaUBhc2FoaWthd2EtbWVkLmFjLmpw
 Kenji Takahashi
Kenji Takahashi Kenzui Taniue1,2
Kenzui Taniue1,2 Yusuke Ono
Yusuke Ono Mikihiro Fujiya
Mikihiro Fujiya Yusuke Mizukami
Yusuke Mizukami Toshikatsu Okumura
Toshikatsu Okumura