- 1Institute for Microbial and Biochemical Technology, Forest Products Laboratory, USDA Forest Service, Madison, WI, United States
- 2Engineering Mechanics and Remote Sensing Laboratory, Forest Products Laboratory, USDA Forest Service, Madison, WI, United States
The Acetivibrioclariflavus (basonym: Clostridium clariflavum) glycoside hydrolase family 30 cellulosomal protein encoded by the Clocl_1795 gene was highly represented during growth on cellulosic substrates. In this report, the recombinantly expressed protein has been characterized and shown to be a non-reducing terminal (NRT)-specific xylobiohydrolase (AcXbh30A). Biochemical function, optimal biophysical parameters, and phylogeny were investigated. The findings indicate that AcXbh30A strictly cleaves xylobiose from the NRT up until an α-1,2-linked glucuronic acid (GA)-decorated xylose if the number of xyloses is even or otherwise a single xylose will remain resulting in a penultimate GA-substituted xylose. Unlike recently reported xylobiohydrolases, AcXbh30A has no other detectable hydrolysis products under our optimized reaction conditions. Sequence analysis indicates that AcXbh30A represents a new GH30 subfamily. This new xylobiohydrolase may be useful for commercial production of industrial quantities of xylobiose.
Introduction
The most abundant hemicellulose, xylan, represents the second most plentiful terrestrial polysaccharide on earth (Saha, 2003). Given its abundance, it has received significant consideration by industry and academia for utilization through bioconversion to green chemicals and fuels (Naidu et al., 2018) as well as for contributions to human health and well-being (Ebringerova and Hromadkova, 1999; Vazquez et al., 2000; Aachary and Prapulla, 2011; Chen et al., 2012; EFSA Panel on Dietetic Products et al., 2018). There are numerous defined xylan subtypes distinguished by varying chain substitution characteristics of acetyl, α-l-arabinofuranosyl (Araf), and α-1,2-linked 4-O-methyl-d-glucuronic acid (GA): substitution differences which are biomass source and plant tissue dependent (Biely et al., 2016; Busse-Wicher et al., 2016). Predominate xylan forms all have GA substitutions (Preston et al., 2003), and hydrolysis by endoxylanases results in neutral oligoxylosides and acidic aldouronates which contain at least a single GA substitution. Both neutral oligoxylosides and aldouronates have potential value directly as either a nutraceutical or a functional food ingredient, a pharmacologically active compound, or pharmacological precursors (Ebringerova and Hromadkova, 1999; Vazquez et al., 2000; Ebringerová et al., 2002; Ohbuchi et al., 2010; Penksza et al., 2018).
Generic endoxylanases cleave the β-1,4-xylan chain in accessible regions and are hindered by increased levels of xylan chain substitution. In contrast, several appendage-dependent endoxylanases have been described (St John et al., 2006a; Correia, Mazumder et al., 2011; Labourel, Crouch et al., 2016). These function to cleave the β-1,4-xylan chain at a defined site relative to a recognized substitution such as GA or Araf and are relatively rare compared to enzymes with generic endoxylanase activity (St John et al., 2010; Aspeborg et al., 2012). Only glycoside hydrolase family 30 (GH30), subfamily 7 and 8 (GH30-7 and GH30-8) enzymes are biochemically shown to have GA-dependent function. The best characterized of these are the GH30-8 endoxylanases which cleave glucuronoxylans (GXns) to yield a limit product consisting of a population of reducing terminus penultimate GA-substituted aldouronates (Supplementary Figures S1A–C) (St John et al., 2006a; Vršanská et al., 2007; St John et al., 2011; Urbániková et al., 2011). These GH30-8 glucuronoxylanases establish a well-defined phylogenetic clade distributed primarily in saprophytic and plant pathogenic bacteria corresponding to Gram-positive and Gram-negative classes, respectively (St John et al., 2011). Other than a few characterized functional outliers (St John et al., 2014; St John et al., 2018), all studies of this GH30 subfamily confirm that it functions as strict GA-dependent glucuronoxylanases (Gallardo et al., 2010; Sainz-Polo et al., 2014; Maehara et al., 2018). The GH30-7 subfamily establishes a similarly well-defined phylogenetic clade but, in contrast, is represented almost entirely by fungi (St John et al., 2010). Given the subfamily designation, it is perplexing to find an enigmatic mixture of xylan hydrolyzing activities assigned to this GH30 subfamily. These activities include glucuronoxylanases with studies that indicate function nearly indistinguishable from bacterial GH30-8 glucuronoxylanases (Biely et al., 2014; Nakamichi et al., 2019b; Katsimpouras et al., 2019), reducing end xylosidase (REX) activity (Tenkanen et al., 2013; Nakamichi et al., 2019a), and non-reducing end xylobiohydrolase (XBH) activity (Šuchová et al., 2020b). This topic has recently been reviewed (Puchart et al., 2021). Many of these enzymes have significant secondary activities. In one recent case, a GH30-7 primary XBH was shown to have secondary endoxylanase activity (Šuchová et al., 2020b). Researchers are working to explain how so many representative functions are possible within a single subfamily grouping (Nakamichi et al., 2019a; Nakamichi et al., 2019b; Nakamichi et al., 2020a; Nakamichi et al., 2020b).
Recently, through proteomic analysis, two novel GH30 xylan active enzymes were identified as major constituents of the Acetivibrio clariflavus (basonym: Clostridium clariflavum and Hungateiclostridium clariflavum (Tindall, 2019)) cellulosome during growth on cellulosic substrates (Artzi et al., 2015). In this work, we characterize the biochemical function of the protein encoded by the Clocl_1795 gene of A. clariflavus and provide phylogenetic evidence that the encoded protein represents a new functional subfamily of GH30. This enzyme is shown to be a strict XBH, which, under optimal functional conditions, generates no other detectable hydrolysis products. Biophysical characterization of the catalytic domain of this novel enzyme, dubbed AcXbh30A (AcXbh30A-CD), shows a broad functional pH range, with increased stability and activity at higher pH values. In preparation of this manuscript, Šuchová et al. (2020a) published in consideration of the same enzyme. While there is some overlap in these works, our report offers substantial additional information regarding AcXbh30A. New information includes 1) detailed information regarding the purified protein, 2) optimum functional parameters and kinetic characterization of this purified enzyme under these optimum conditions, 3) a thorough treatment of phylogenetic relationships coupled to a broader analysis of subfamily function by screening three related enzymes, and 4) evaluation of the earlier reported endoxylanase activity (Šuchová et al., 2020a). AcXbh30A may be valuable in the specific production of X2 from lignocellulosic biomass and may work efficiently in this process when coupled to the selective processing of a hardwood GXn by a GH30-8 glucuronoxylanase, thereby specifically producing X2 and a two-component mixture of aldouronic acids which are readily separable from X2 (Supplementary Figures S1B–F).
Materials and Methods
Reagents
All reagents used for biochemical assays and enzyme studies were of the highest purity. Xylo- and cello-oligosaccharides and p-nitrophenol xylobioside (pNP-X2) were products of Megazyme (Bray, Ireland). Beechwood xylan (BeWX) and birchwood xylan (BiWX) were products of Sigma-Aldrich Co. (St. Louis, MO). The GH30-8 endoxylanase, CaXyn30B (Crooks et al., 2020), was used to prepare a beechwood xylan aldouronate ladder hydrolysate for XBH characterization (graphically depicted in Supplementary Figures 1A–C). Following limit hydrolysis, the reaction was precipitated (ppt) in 60% ethanol (EtOH) followed by centrifugation at 14k x g at room temperature for 20 min. The 60% EtOH supernatant was vacuum evaporated using a rotovap and recovered with a water rinse to a total recovered volume of 10 ml. Absolute EtOH was then added to 95% saturation and allowed to equilibrate for 20 min at room temperature. This was then centrifuged at 12.5k x g at room temperature for 20 min. The 60–95% EtOH cut pellet was washed (pellet was agitated) in 200 ml absolute EtOH, recovered by centrifugation as above, and dried at 70 C under house vacuum in a tared centrifugation bottle. The resulting pellet’s mass was determined and was readily soluble in water. The 60–95% EtOH precipitated fraction was redissolved to 20 mg/ml for use in biochemical reactions.
Cloning, Expression, and Purification
An original expression construct of the Clocl_1795 gene (encoding UniProt accession No. G8LU16) of A. clariflavus was kindly provided by Professor Edward Bayer from the Weizmann Institute, Rehovot, Israel. This was subcloned to generate alternative protein expression products by PCR. These new DNA constructs were cloned into pBlueScript to verify the primary DNA sequence and then subcloned into pET28 between the NcoI and XhoI restriction enzyme sites. The DNA expression constructs of other enzymes studied in this work were prepared with the same approach but were obtained directly using Gibson Assembly (Gibson et al., 2009) of large synthetic DNA fragments (IDT, Coralville, IA) or subsequent subcloning manipulations of the assembled synthetic DNA (Supplementary Table S1).
AcXbh30A and the other proteins considered in this work were expressed using slightly modified auto-induction methods originally detailed by Studier (Studier, 2005). Selection was maintained using 50 μg/ml kanamycin, and the expression proceeded at 18 C for over 30 h. The cells were recovered by centrifugation (8k x g), and collected cell pellets were processed in preparation for immobilized metal affinity chromatography (IMAC) as previously detailed (St John et al., 2018) using sonication to lyse the cells. Following single-step elution IMAC, protein was desalted into 20 mM Tris base, pH 7.0 buffer using a Zeba desalting column (Fisher Scientific, Pittsburgh, PA) and analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Laemmli, 1970) using a precast TGX 4–15% gradient gel (Bio-Rad, Hercules, CA). This manner of protein expression and purification was also used for the AcXbh30A protein homologs considered in this work. For AcXbh30A-CD, the step-elution IMAC product is referred to as the step elution product (SEP) throughout this work. For some studies, AcXbh30A was further purified using gradient IMAC, anion exchange, and gel filtration chromatography. This more thorough purification product is referred to as the gradient elution product (GEP). Briefly, IMAC was performed as above but for the use of a 0–500 mM imidazole elution buffer gradient over 20 column volumes. Following this, AcXbh30A-CD was exchanged into 25 mM Tris base of pH 8.0 containing 2 mM 2-mercaptoethanol and resolved using a Mono Q column (Cytiva, Marlborough, MA) with a 20-column-volume sodium chloride gradient. Peak fractions were combined and subsequently desalted into the gel filtration chromatography buffer composed of 20 mM HEPES, 250 mM sodium chloride, and 2 mM dithiothreitol of pH 8.0 for further purification using a Superdex column (Cytiva). Gradient separations were performed using a Bio-Rad BioLogic DuoFlow medium-pressure liquid chromatography system.
Sequence Analysis and Phylogenetic Placement
BLASTp (Altschul et al., 1990) on the UniProt website1 (The UniProt Consortium, 2017) was used to collect amino acid sequences showing homology to AcXbh30A. Given the limited number of related sequences, NCBI BLASTp2 was further utilized to create a representative set of sequences (Johnson et al., 2008). Furthermore, the Carbohydrate-Active enZYmes (CAZy) database3 (Lombard et al., 2014) for glycoside hydrolase family 30 enzymes was utilized to collect CAZy-annotated enzymes for the GH30 subfamilies (St John et al., 2010). Preliminary amino acid sequence alignments were generated for consideration using the ClustalW (Larkin et al., 2007) alignment tool, and sequence dataset trimming was performed using MEGAX (Kumar et al., 2018). Trimmed sequence datasets were aligned using the MAFFT G-INS-1 progressive alignment strategy4 (Katoh and Toh, 2008), and phylogenetic trees were prepared in MEGAX using the maximal likelihood (ML) method with the LG substitution model (Le and Gascuel, 2008). Phylogenetic tree branch support values were obtained with 100 cycles of bootstrap analysis. Amino acid sequence identity levels of all-against-all of the collected sequence dataset were performed using the statistical sequence shuffling tool PRSS5 (Pearson, 2001).
Functional Analysis of AcXbh30A
Thin layer chromatography (TLC) with the mobile phase chloroform: glacial acetic acid: water (6:7:1, v/v) was performed as previously described (St John et al., 2006b). TLC plate samples were spotted in 1 μl aliquots. A minimum of two ascensions were performed, and the plates were developed using N-(1-naphthyl)ethylenediamine dihydrochloride as previously described (Bounias, 1980). For initial TLC functional screening using the AcXbh30A-CD SEP or other protein homologs purified by IMAC, 10 μl reactions consisted of substrate polysaccharides at 10 mg/ml or oligosaccharides at 5 mM, 30 mM sodium acetate of pH 5.0, and 0.1 mg/ml bovine serum albumin (BSA). Later studies using the AcXbh30A-CD GEP preparation were performed as 200 μl reactions in a thermocycler to maintain reaction temperature. These reactions were at pH 4 sodium acetate, used the GEP at 2 μg/ml and 20 μg/ml, and were sampled at 15 and 300 min (5 h). From these reactions, a range of sample loads were used to adequately represent multicomponent mixtures.
HPLC studies were performed using an Agilent 1260 Infinity system with refractive index detection. Xylooligosaccharides (XOSs) were resolved on a Shodex SH1821 column (with a SUGAR SH-G guard column) at 60 C in the mobile phase 0.01 N H2SO4 at the flow rate of 0.8 ml/min. Initial HPLC reactions were 30 μl in volume, contained 30 mM sodium acetate of pH 5.0, 0.1 mg/ml BSA, and 5 mM X4, and were initiated following equilibration to 40 C by addition of enzyme to 5 μg/ml and allowed to react for 20 min before heat inactivation at 95 C for 5 min. Stopped reactions were centrifuged for 5 min, and the supernatant was transferred for analysis by HPLC. Later studies used only 0.02 mg/ml BSA, and a four-component HPLC-compatible buffer (FAMM buffer) was developed to study optimum reaction pH over what was determined to be a broad functional range. FAMM buffer consisted of 30 mM each of sodium formate, sodium acetate, MES (2-(N-morpholino)ethanesulfonic acid), and MOPS (3-(N-morpholino)propanesulfonic acid) which should effectively buffer from pH 2.5 to 8.0. Single-component buffers including glycine, acetate, and Tris base used at 30 mM were also tested to validate results obtained using the FAMM buffer. All buffer compositions were pH adjusted with either 6N HCl or 1N NaOH. Heat inactivation of AcXbh30A-CD under more acidic conditions failed to fully inactivate AcXbh30A-CD, and a fix was found to be pH neutralization prior to heat inactivation. Specific activity analysis of oligoxylosides was performed as above, but using pH 4.0 sodium acetate, XOS at 10 mM, at a reaction temperature of 65 C for 10 min. Equilibrated reaction mixtures were initiated by addition of AcXbh30A-CD GEP to a final reaction concentration of 500 ng/ml. HPLC samples were analyzed a minimum of three times to obtain average values, and assays were performed a minimum of two times.
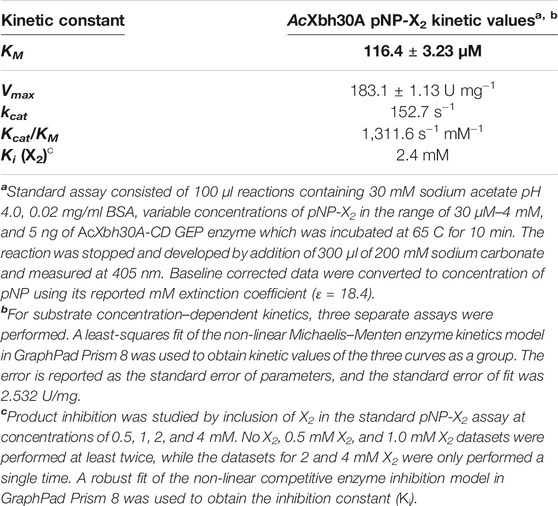
The substrate pNP-X2 was used for kinetic evaluation of AcXbh30A-CD GEP. This was performed in the same conditions described for the specific activity determination with XOS with the GEP preparation of AcXbh30A-CD except for an increase in volume to 100 μl. The reaction was stopped and developed by addition of 300 μl of 200 mM sodium carbonate. Samples were read at 405 nm, baseline corrected by subtraction of a no enzyme control, and adjusted for the development dilution of pNP. This was then converted to concentration values using the pNP mM extinction value of 18.4 (mM−1 cm−1). Kinetic analysis was performed using a range of pNP-X2 concentrations and the resulting kinetic constants determined from three replicate studies. A least-squares fit of the non-linear Michaelis–Menten enzyme kinetics model in GraphPad Prism 8 (GraphPad Software, San Diego, CA) was used to obtain kinetic values of the three curves as a group. The error is reported as the standard error of parameters, and the standard error of fit was 2.532 U/mg. The primary hydrolysis product X2 was considered for its role in inhibition of AcXbh30A. Duplicate substrate response curves were generated for no added X2, 0.5 mM X2, and 1 mM X2, and only a single curve was generated for 2 mM X2 and 4 mM X2. A robust fit of the non-linear competitive enzyme inhibition model in GraphPad Prism 8 was used to obtain the inhibition constant (Ki).
Results and Discussion
AcXbh30A DNA Cloning, Protein Expression, and Initial Purification
The AcXbh30A enzyme was first identified following its proteomic detection as a major protein component of the A. clariflavus cellulosome during cultivation on cellulosic substrates (Artzi et al., 2015). Activity screening as part of that research determined AcXbh30A to have an unknown xylanase activity. The primary amino acid sequence for AcXbh30A consists of an N-terminal secretion leader sequence, a GH30 catalytic center, and a C-terminal type I dockerin domain. The original expression plasmid (Artzi et al., 2015) encoded the full AcXbh30A protein with an N-terminal His-tag in place of the predicted secretion signal sequence. The product of this original AcXbh30A expression plasmid was expected to yield full-length AcXbh30A with an N-terminal His-tag. However, upon IMAC purification and SDS-PAGE analysis, the preparation yielded two bands (Supplementary Figure S2). The minor band ran to the size expected for the full AcXbh30A protein, while the major band ran slightly smaller. Suspecting this to have resulted from proteolytic processing, we generated two constructs of AcXbh30A by cloning with a His-tag appended to the C-terminus of a full-length construct and also to a catalytic domain (CD) only construct. It was later found, following the publication by Šuchová et al. (2020a), that the pair of bands we observed by SDS-PAGE of the original expression construct is similar to that seen on the SDS-PAGE provided by NYZTech6 from where that group obtained AcXbh30A for study.
While this work primarily focuses on the characterization of AcXbh30A-CD, both AcXbh30A and AcXbh30A-CD constructs expressed to high levels. Initial IMAC purification of both forms resulted in a single prominent band of the expected molecular weight by SDS-PAGE and was judged to be greater than 95% pure (Supplementary Figure S2). Analysis of these enzyme forms by modified SDS-PAGE sample preparation showed that the AcXbh30A (dockerin-containing form) expression product is a disulfide-mediated dimeric product. Since this dimerization is not observed with AcXbh30A-CD and almost completely present with AcXbh30A which contains the C-terminal dockerin domain, we conclude the disulfide bond formation must involve Cys516 located in the dockerin domain. The artificial nature of high-level protein overexpression precludes prediction of any biological role for this disulfide bond occurrence. Cys516 is not conserved among other bacterial GH30 XBHs brought forth by these studies (Figure 5).
Functional Characterization and Optimization of AcXbh30A Activity
Initial biochemical studies were performed using the AcXbh30A SEP preparation. Numerous biomass-derived polysaccharides were tested for TLC observable activity (data not shown). For both enzyme constructs, only xylobiose (X2) was observed as a product from the hydrolysis of GXn. Hydrolysis of even-numbered XOSs yielded only X2, and hydrolysis of odd-numbered XOSs yielded X2 and xylose (Figure 1). Hydrolysis of cellopentaose did not release a smaller hydrolysis product, indicating that the C6 (CH2OH) of the glucose is not accommodated in the substrate binding pocket.
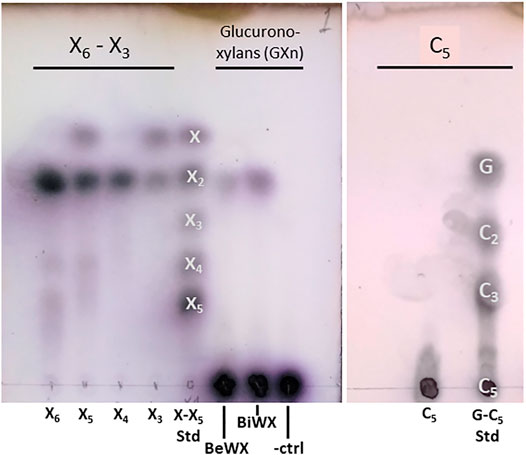
FIGURE 1. TLC showing AcXbh30A-CD hydrolysis of XOS, two common GXn types, and cellopentaose. The results indicate that only a small amount of X2 is liberated from polymeric GXn, while hydrolysis of even-numbered XOSs yielded only X2 and hydrolysis of odd-numbered XOSs yielded X2 and xylose. Importantly, using the same TLC method, no analogous activity could be detected for the AcXbh30A-CD hydrolysis of cellopentaose (C5), indicating that the C6 position is not accommodated in the -1 and/or -2 subsites that establish the X2 binding pocket. Samples and standards for the xylan-based assay include xylose (X) through xylopentaose (X5) with beechwood glucuronoxylan (BeWX) and birchwood glucuronoxylan (BiWX) reactions as well as a BeWX no enzyme control (-ctrl). For C5 hydrolysis, analysis included glucose (G), cellobiose (C2), cellotriose (C3) and cellopentaose (C5) as standards.
Following function determination, XBH activity was studied by following hydrolysis of X4 to X2 by HPLC. Initial reaction optimization showed that AcXbh30A-CD had a greatly reduced activity when not supplemented by BSA (at 0.1 mg/ml), while the dockerin-containing AcXbh30A enzyme form did not appear to benefit from BSA inclusion. For the AcXbh30A-CD SEP, MgCl2 at 0.1 mM did not benefit activity and CaCl2 at this concentration resulted in a 9% reduction in activity. Given the BSA activity dependence of the CD form, a BSA titration study was performed. Addition of BSA from 0.005 to 0.2 mg/ml resulted in increasing activity to form a horizontal asymptote. From this observation, BSA was included in all future reactions at a level of 0.02 mg/ml.
Optimum reaction pH studies indicated that AcXbh30A-CD was functional over a broad range. To accommodate this, a four-component buffer system (FAMM buffer) was developed to be compatible with the HPLC system conditions. In these HPLC studies, through serial sample injections, it was observed that, at pH 2.5 and 3.0 reaction conditions, AcXbh30A-CD was partly resistant to inactivation at the subsequent 95 C heat treatment. Although several approaches were successfully shown to inactivate AcXbh30A-CD while under these reaction conditions, simple pH quenching with a 2x dilution of 150 mM Tris base of pH 8.0 effectively allowed for complete AcXbh30A-CD inactivation at 95 C. Using the FAMM buffers in 40 C reactions, the AcXbh30A-CD SEP observed optimum activity at pH 3.5, maintained about 66% of its optimum activity at pH 2.5 and pH 6, and retained greater than 50% of its optimum activity at pH 8 (Figure 2). Other single-component buffers closely mirrored the FAMM pH curve, except for Tris buffer which yielded significantly lower activity at the higher pH conditions.
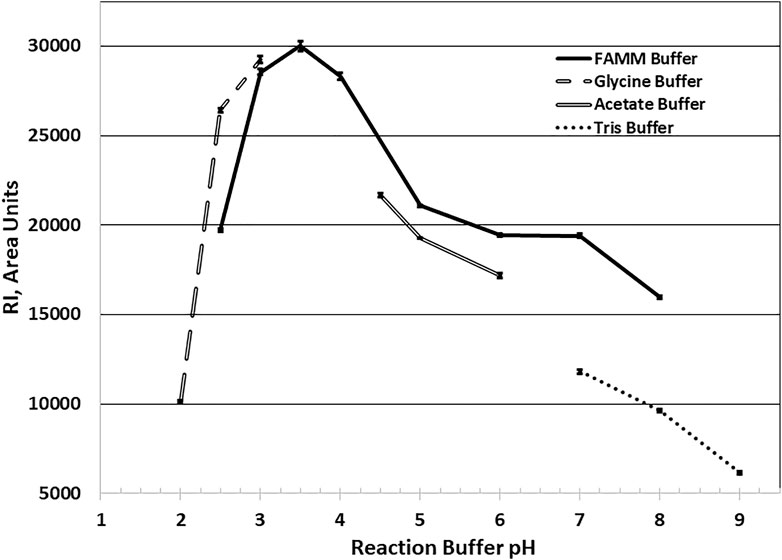
FIGURE 2. AcXbh30A-CD SEP X4 hydrolysis reaction optimum pH determination. All buffers were used at 30 mM final concentration for each buffer component, and the reaction was maintained at 40 C for 20 min with BSA included at 0.02 mg/ml. The findings show that, at the selected reaction temperature of 40 C, the optimum reaction pH using the FAMM buffer system is approximately 3.5. By extrapolation of the curve, the optimal pH is mirrored using the single-component buffers glycine and acetate. The use of Tris buffer to verify pH activity dependence at the higher pH conditions yielded significantly lower activity.
Optimum reaction temperature analysis showed that the AcXbh30A-CD SEP, over a reaction period of 10 min, functioned at elevated temperatures (Supplementary Figure S3A). Thermostability determination of AcXbh30A-CD SEP preincubation showed that neutral pH conditions were better at preserving activity than in the lower pH conditions that were found to be optimal for the 40 C reaction conditions (Supplementary Figures S3B,C). To capture a reaction-based thermostability result, a factorial study was designed to include a range of FAMM pH conditions over an extended reaction time of 2 h. The results of this study (Figure 3) clearly demonstrated that the AcXbh30A-CD SEP is thermostable over extended reaction times under more neutral reaction pH conditions. The general relationship shows that optimum activities occur at lower temperatures in the lower pH conditions and at higher temperatures in the higher pH conditions (toward neutral pH). Peak activity at pH 3.5 and 56 C was about 20% lower than the peak activity at pH 6 and 73 C.
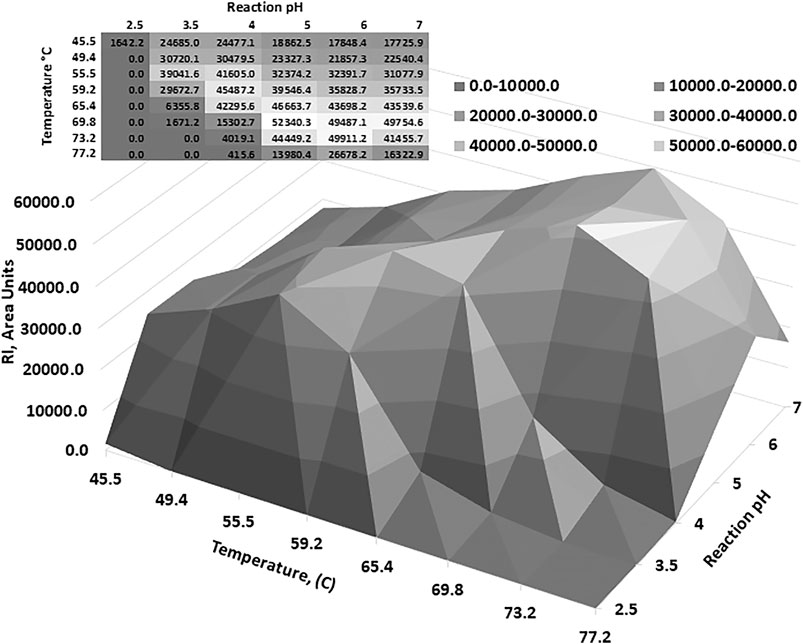
FIGURE 3. To better understand the relationship between pH and temperature optimum, a factorial study was performed. The results indicate that the AcXbh30A-CD SEP is thermostable up to about 70 C and the optimum pH increased as the reaction temperature increased.
Additional Purification to Yield AcXbh30A-CD GEP
For protein crystallization and kinetic studies, AcXbh30A-CD was subjected to additional purification (Supplementary Figure S4). Rather than an IMAC single-step elution, a twenty-column-volume gradient was performed. This resulted in two peaks which significantly overlapped due to the high level of column saturation. This indicated that while SDS-PAGE (Supplementary Figure S2) showed a single band of the correct molecular weight, the IMAC SEP consists of an undefined AcXbh30A-CD multimeric agglomeration. Gel filtration sizing (Superose 6 10/300 GL) and purification (Superdex, HiLoad 16/600, 200 PG) as well as anion exchange (Mono Q 4.6/100 PE) separations further support this finding (Supplementary Figure S4). The leftmost IMAC peak (Peak A) ran to size (no quaternary structure) as a well-defined peak on a calibrated Superose 6 column and also on the Superdex column used for purification. The rightmost IMAC peak (Peak B) on both gel filtration columns ran as a much larger (earlier eluting) poorly defined peak which overlapped the void volume. Similarly interesting results were found when using the Mono Q column. Peak A was a very sharp peak which eluted earlier than Peak B, which was again ill-defined, tighter-binding (eluted later), and well-resolved from Peak A. All peaks yielded the same approximately 50 kDa band in SDS-PAGE analysis (Supplementary Figure S4F). For crystal screening, the GEP was obtained from additional Peak A purification using gradient IMAC, Mono Q, and Superdex chromatography. The AcXbh30A-CD GEP was concentrated to approximately 37 mg/ml and submitted for crystallization screening at Argonne National Laboratory, Advanced Protein Characterization Facility. An initial crystal was obtained which diffracted to less than 2Å but was not successfully phased through molecular replacement methods.
Thermostability characteristics of the AcXbh30A-CD GEP were confirmed using a pNP-X2 reaction following a preincubation of the enzyme over time at anticipated inactivation temperatures resulting from the AcXbh30A-CD SEP studies (Supplementary Figure S3D). From these results, the AcXbh30A-CD GEP appears to be relatively stable at 68 C for 24 h. Following this, unless otherwise indicated, biochemical studies were performed at 65 C in 30 mM pH 4.0 sodium acetate buffer containing BSA at 0.02 mg/ml. For the AcXbh30A-CD GEP, the reaction pH optimum is similar to that determined for the AcXbh30A-CD SEP (data not shown).
Further Analysis of Xylobiohydrolase Specificity
In light of a recently characterized, distantly related GH30-7 fungal XBH from Acremonium alcalophilum (AaXyn30A) which was shown to have detectable endoxylanase activity (Šuchová et al., 2020b) at high enzyme loadings (>5 µM), we sought to establish an empirical understanding of enzyme concentration vs. function for AcXbh30A-CD. It was established that approximately 5 μg/ml (100 nM) of AcXbh30A-CD could completely convert 5 mM X4 to X2 in 15 min at 60 C (data not shown). Following this initial observation, we sought to determine enzyme stringency by examining AcXbh30A-CD at 2 μg/ml (40 nM) and 20 μg/ml (400 nM) along with reaction times of 15 min or 5 h to obtain the effective relative enzyme capacity of 1x (2 μg/ml for 15 min), 10x, 20x, and 200x (20 ug/ml for 5 h).
For hydrolysis of BeWX, X2 was the only sugar observed having low intensity on the TLC for the 10x, 20x, and 200x enzyme levels. This indicates that AcXbh30A-CD can only generate a limited amount of X2 from polymeric glucuronoxylan (Figure 4). This is most likely because iterative hydrolysis of a terminus will eventually be interrupted by the occurrence of a GA substitution and a lack of appreciable endoxylanase activity to generate additional non-reducing termini. The sample origin spot was visually unchanged relative to the no enzyme control for this reaction. To elucidate the directionality of AcXbh30A, a GH30-8–generated aldouronate mixture was used as the substrate (Supplementary Figures S1A–C) (St John et al., 2006a). This substrate is ideal for determination of the directionality of XBH function and, given its larger oligomeric size and complexity, may also identify any secondary activities displayed by AcXbh30A. As can be observed in Figure 4, with this aldouronate preparation at 10 mg/ml and AcXbh30A-CD at 1x, the aldouronate ladder is partially processed. The use of AcXbh30A-CD at 10x, 20x, and 200x with this substrate showed complete conversion of the aldouronate ladder to X2 and two aldouronates. By TLC comparison with aldouronate standards, these two limit-product aldouronates have the same mobility as aldotriuronate and aldotetrauronate. Given the strict release of X2 by AcXbh30A, these two aldouronates can only result from the non-reducing terminal release of X2 from even- or odd-numbered GH30-8 aldouronates, respectively. They must therefore be of the aldouronate substitution arrangement having the nomenclature GX and XGX (Supplementary Figures S1E,F, respectively) (Fauré et al., 2009).
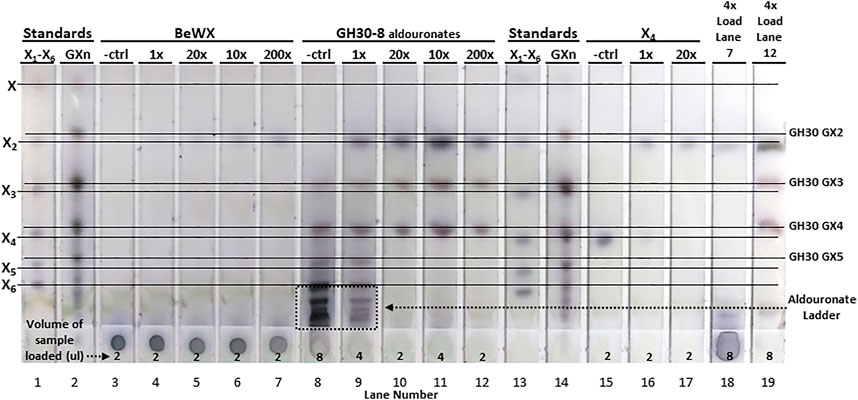
FIGURE 4. TLC analysis showing hydrolysis products of AcXbh30A-CD GEP digestion of BeWX, an enriched GH30-8 aldouronate preparation and X4. Using a reaction concentration of AcXbh30A-CD GEP which empirically was determined to visually represent significant X4 hydrolysis to X2 (2 ug/ml for 15 min, 1x), enzyme concentration was increased 10-fold and reaction time increased to obtain 10x, 20x, and 200x increased relative enzyme capacity. In treatment of BeWX, only a light spot for X2 was observed which reached approximately full intensity with the 10x enzyme condition. Complete conversion of the GH30-8 aldouronate was observed with the 10x enzyme level, and up to 200x treatment did not result in secondary activities detectable by TLC. As expected, AcXbh30A-CD GEP at 1x almost completely converted 5 mM X4 to X2. Increased load volumes (4x) of the 200x enzyme capacity reactions of BeWX and the GH30 aldouronate preparation did not reveal other hydrolysis products.
This result indicates that the substitution of an α-1,2-linked GA moiety does not prevent binding and hydrolysis at the glycosidic bond most proximal to the GA-substituted xylose. However, the requirement for successful hydrolysis is the recognition of a non-reducing terminal xylobiose extension, which then yields GX. Otherwise, a single xylose remains yielding XGX. These results and further deductions agree with known GH30 protein structure and xylan binding interactions concerning the orientation of xylose positioned in the +1 subsite (Freire et al., 2016). No other products were detected from these reactions. Hydrolysis of X4 with 10x more enzyme resulted in only X2. To fully assess the potential for secondary hydrolysis products other than X2 (possible endoxylanase activity), the TLC spot load volume for the 200x reactions of BeWX and GH30-8 aldouronates were increased 4x. For these, only a very light, large xylooligosaccharide smear is detected along with X2, GX, and XGX.
Specific Activity and Kinetic Analysis of AcXbh30A-CD GEP
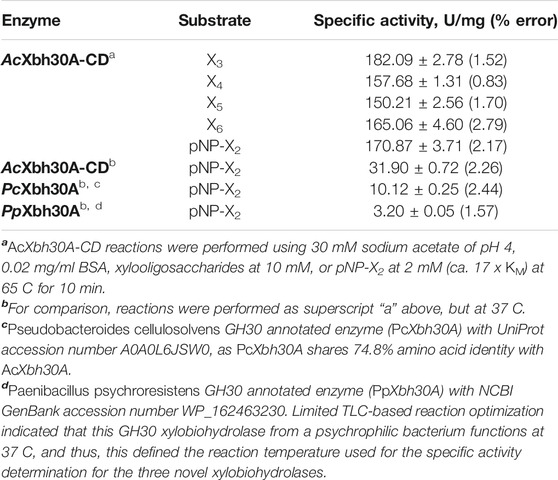
Specific activity and kinetic analysis of AcXbh30A-CD GEP was performed at the optimized functional parameters of 65 C and pH 4.0. The result of xylooligosaccharide hydrolysis to X2 by HPLC is from duplicate reactions with each being injected three times. The overall average was used for rate calculations. The results (Table 1) indicate that only relatively minor rate variations are evident between the hydrolysis of X3–X6. The highest specific activity of 182.1 U/mg was observed for X3. X6 has the next highest at 165.1 U/mg. X4 and X5 are slightly lower at 157.7 and 150.2 U/mg, respectively.
Kinetic evaluation using pNP-X2 concentration–dependent response curves was performed with three separate experiments (Table 2). Importantly, the specific activity calculated from these same data (2 mM pNP-X2 data point) showed that this substrate provided a reasonable functional parallel to the natural XOS given that its observed rate of 170.9 U/mg is similar (Table 1). This may indicate that the binding energy of the aromatic pNP group into the +1 (and possible +2) subsite is similar to that of a xylosyl group. From the kinetic studies, AcXbh30A-CD was determined to have a KM of 116 μM and a Vmax of 183 U/mg resulting in a kcat of 153/s with an enzyme efficiency constant of 1,312/s mM (Table 2). X2 was also considered for product inhibition using a standard competitive inhibition model. The finding indicates a Ki of 2.4 mM. This is similar to the reported cellobiose inhibition (Ki, 1.6 mM) of the processive cellobiohydrolase Cel7A in hydrolysis of bacterial cellulose (Gruno et al., 2004). Lastly, we assessed the tendency of AcXbh30A-CD to act processively in the hydrolysis of X6 using HPLC. The measurement of X4 at appreciable levels during the reaction indicated that its function is not processive (Supplementary Figure S5).
Analysis of Xylobiohydrolase Specificity
The recent publications by Šuchová et al. address a detected endoxylanase activity for both primary xylobiohydrolases AaXyn30A (Šuchová et al., 2020b) and AcXbh30A (Šuchová et al., 2020a). In light of our earlier efforts to confirm functional specificity for AcXbh30A, we felt compelled to reproduce these results: AcXbh30A (HcXyn30A as it was previously designated), we confirm, does have detectable endoxylanase activity under the same conditions employed by Šachová et al. However, to obtain this outcome, GXn hydrolysis reactions were conducted for extended periods with 2.4 uM of enzyme (5.4 uM in the case of AaXyn30A). With respect to the data reported in this work, and keeping with the same enzyme capacity factor concept, the endoxylanase activity detected by Šuchová was obtained with the equivalent of ca. 6,000x, the base condition of 2 ug/ml AcXbh30A-CD for 15 min (1x). Given this analysis, we posit that the reported endoxylanase activity is not reflective of biologically relevant specificity but may be of value in the isolation of novel substituted xylooligosaccharides.
Phylogenetic Placement of AcXbh30A
Original GH30 subfamily classification proposed the existence of eight subfamilies, several of which had existing functional annotation assignments. At the time, large-scale amino acid phylogenetic analysis showed that subfamily members shared greater than ca. 30% identity, with comparison between subfamilies dropping to less than ca. 28% identity (St John et al., 2010). Since this time, a new functional subfamily, GH30-9, has been delineated from its nearest neighbor GH30-3 (Sakurama et al., 2013). AcXbh30A is a member of a clade with members sharing at least 46% identity. To verify that this represented a collection of prokaryotic XBH enzymes, we cloned and expressed an enzyme from Pseudobacteroides cellulosolvens (UniProt accession No. A0A0L6JSW0, PcXbh30A) that shares 74.8% identity with AcXbh30A and an enzyme from Paenibacillus psychroresistens (GenBank accession No. WP_162463230, PpXbh30A) which establishes a boundary position at the edge of the clade (Figure 5) and shares ca. 50% identity with AcXbh30A. Both enzymes have XBH activity within 10-fold (lower) of AcXbh30A (Table 1). These enzymes were not rigorously optimized for function regarding temperature, pH, etc., and comparison reactions with AcXbh30A were all performed at 37 C to provide more comparable results. Importantly, the XBH clade, except for PpXbh30A, represents enzymes solely from the bacterial order Clostridiales. The phylogenetic nearest neighbor consists of protein sequences from a more diverse collection of bacteria and shares greater than ca. 34% identity with AcXbh30A. In this neighboring group, the highest sequence identity to AcXbh30A is 43.9%. Explorations into the enzymatic activity of this adjacent clade are ongoing, but initial indications are that GH30 from Gracilibacillus dipsosauri (UniProt accession No. A0A317L0T6) displays endoxylanase activity which will be reported elsewhere (GdXyn30A). No XBH activity is detected with this enzyme.
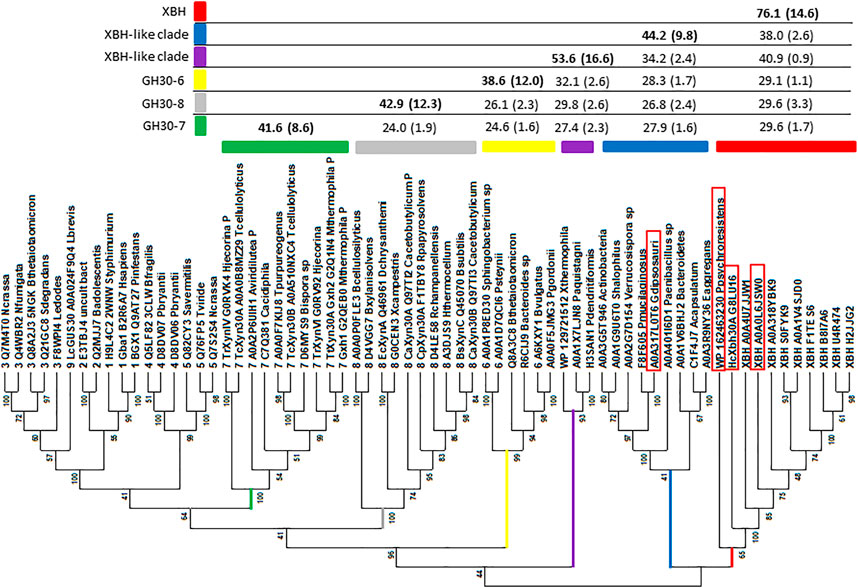
FIGURE 5. Phylogenetics analysis distinguishing the GH30 subfamily branches and highlighting the new XBH subfamily (red branch). The sequence set includes all subfamily member sequences determined through published reports or protein structure database deposits collected from CAZy as well as a group homologous to AcXbh30A. Sequences were trimmed to consist of only the (β/α)8 + β complete GH30 domain (St John et al., 2010). Sequences were aligned using MAFFT (Katoh and Toh, 2008) with the G-INS-1 strategy and imported into MEGA X (Kumar et al., 2018) where evolutionary history was inferred by using the maximum likelihood method and the LG substitution model (Le and Gascuel, 2008). The bootstrap consensus tree inferred from 100 replicates (Felsenstein, 1985) is taken to represent the evolutionary history of the taxa analyzed (Felsenstein, 1985). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (100 replicates) is shown next to the branches (Felsenstein, 1985). Initial trees for the heuristic search were obtained automatically by applying neighbor-join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model and then selecting the topology with a superior log likelihood value. A discrete gamma distribution was used to model evolutionary rate differences among sites [three categories (+G, parameter = 2.5225)]. The rate variation model allowed some sites to be evolutionarily invariable ([+I], 0.28% sites). This analysis involved 66 amino acid sequences. There were a total of 878 positions in the final dataset. These sequences also included GH30-7 (green branch), GH30-8 (gray branch), and GH30-6 (yellow branch), and two XBH-like branches (purple and blue) were analyzed using PRSS (Pearson, 1998). The results of percent identity are tabulated to distinguish levels of rough similarity cut-offs between GH30-6, -7, and -8 subfamilies, the unknown XBH-like clades, and the new XBH subfamily (red). Enzymes boxed in red were evaluated for biochemical function following DNA cloning, protein expression, and IMAC purification.
As discussed above, at the time of the original GH30 subfamily assignment, it was determined that members of each of the detected subfamilies shared greater than 30% identity with identity levels between subfamilies dropping to below 28% (St John et al., 2010). For instance, PRSS analysis of all GH30-7 enzymes against all GH30-8 enzymes used in Figure 5 results in an average identity of 24.5% (SD 1.6%). A similar comparison between the newly delineated GH30-9 subfamily (Sakurama et al., 2013) and the larger more diverse GH30-3 subfamily gives an average identity of just 30.1% (SD 0.9%). The ultimate goal of the CAZy database is the functional annotation of enzyme groups, and the exo-acting β-glucuronidase function of GH30-9 is functionally and likely structurally (at least in the active site) different from the endo-β-1,6-glucanase function annotated into GH30-3. An amino acid sequence identity screen, using all the members of each clade in the dataset (Figure 5), shows that while the level of homology between the XBH clade and its neighboring clade hovers around ca. 40%, the identity levels with all other members of GH30-6, -7, and -8 are in the range of 30%. Based on this observation, it seems appropriate that given its unique biochemical function, AcXbh30A defines a new GH30 subfamily. Communication with curators of the CAZy database has indicated acceptance of this proposed subfamily as GH30, subfamily 10 (personal communication during manuscript revision).
Co-Digestion of GXn Using Canonical GH30-8 Enzymes and the New GH30 Xylobiohydrolases
In consideration of the complete conversion of the GH30-8 aldouronate preparation to X2, GX, and XGX, it was considered of value to broadly examine the utility of XBHs to enable the complete conversion of GXn to the three defined products X2, GX, and XGX in a two-enzyme system. To verify the relationship between these two enzymes, we considered the XBHs AcXbh30A-CD and PcXbh30A (from P. cellulosolvens, see above) as well as three different canonical GH30-8 endoxylanases. These GH30-8 members include BsXynC from Bacillus subtilis (St John et al., 2006a), CtXyn30A from Clostridium thermocellum (St John et al., 2017), and BcXyn30C from Bacteroides cellulosilyticus (unpublished). All three GH30-8 enzymes functioned as anticipated yielding a ladder of aldouronates, and when combined as co-digestions with either of the two tested XBHs described here, the predicted three defined limit products are observed (Figure 6). This shows that the hydrolytic product observations made here likely extrapolate to all canonical GH30-8 enzymes when coupled to this new GH30 XBH subfamily.
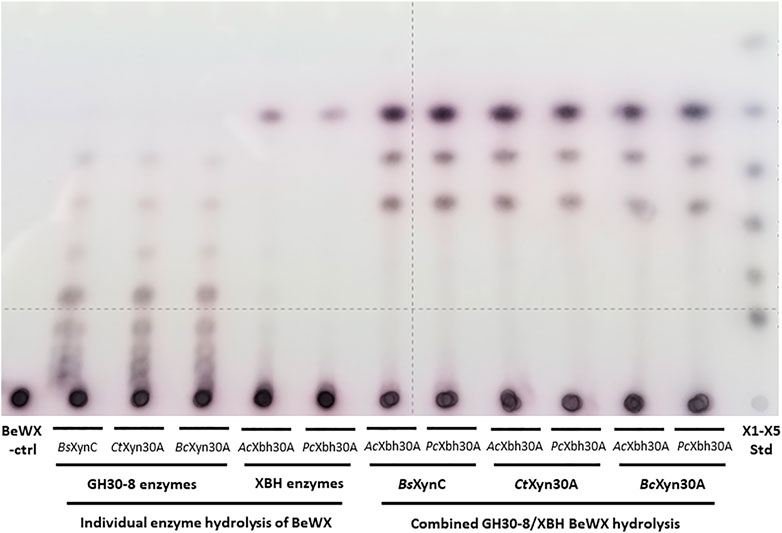
FIGURE 6. TLC analysis verifying function of three canonical functioning GH30-8 glucuronoxylanases BsXynC (St John et al., 2006a), CtXyn30A (St John et al., 2017), and BcXyn30A (unpublished) and the newly identified xylobiohydrolase AcXbh30A-CD [this work and Šuchová et al. (2020a)] as well as its homolog PcXbh30A (this work) in the hydrolysis of polymeric glucuronoxylan. When combined, these enzymes convert glucuronoxylan to the three products X2, GX, and XGX (Supplementary Figures S1D–F, respectively).
Conclusion
The AcXbh30A enzyme was originally observed as being a prominent component of the A. clariflavus cellulosome during growth on cellulosic substrates (Artzi et al., 2015). The functional understanding of this enzyme as a xylobiohydrolase coupled with its significant presence in the cellulosome highlights a novel strategy of lignocellulose deconstruction and utilization by this cellulosome-producing bacterium and further lends potential in the development of synthetic cellulosome assemblies for lignocellulose bioconversion (Kahn et al., 2019; Ding and Bayer, 2020). From these studies, AcXbh30A is shown to function as an NRT-specific XBH which when utilized optimally generates no detectable secondary products. AcXbh30A has a wider functional pH range then commonly observed for glycoside hydrolases and as an apparent enzyme monomer is stable over extended time periods up to 65 C. Coupling enzymes of this new XBH GH30 subfamily with the specificity of the GH30-8 glucuronoxylanases in the processing of GXn yields the prebiotic sugar X2 and two valuable aldouronates of defined structure. The same XBH–GH30-8 enzyme system, if presented with more complex xylan types that also contain GA substitutions, has the potential to yield a multitude of novel substituted oligoxylosides.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, and further inquiries can be directed to the corresponding author.
Author Contributions
FJSJ originally conceived the research idea, performed biochemistry related aspects, and wrote the manuscript with feedback provided by CC. CC performed all molecular biology, protein expression and co-digestion portions of this research. NB facilitated numerical data analysis. All authors edited and approved the final version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank the Institute for Microbial and Biochemical Technology (IMBT) of the Forest Products Laboratory (FPL), a National Laboratory of the USDA Forest Service, for ongoing financial support and USDA research scientists Dan Cullen and Phil Kersten for providing valuable review and comments on this manuscript while in preparation. They also wish to thank members of the Structural Biology Center at Argonne National Laboratory for their help with data collection at the 19ID beamline. The Argonne National Laboratory’s Structural Biology Center beamlines are supported by the U.S. Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.714238/full#supplementary-material
Abbreviations
Araf, α-L-arabinofuranosyl; BSA, bovine serum albumin; BeWX, beechwood xylan; BiWX, birchwood xylan; FAMM, acronym for the buffer used in this research that is composed of 30 mM each of sodium formate, sodium acetate, MES (2-(N-morpholino)ethanesulfonic acid), and MOPS (3-(N-morpholino)propanesulfonic acid); GA, glucuronic acid; GXn, glucuronoxylan; GEP, gradient elution product; X2, xylobiose; X3, xylotriose; X4, xylotetraose; X5, xylopentaose; X6, xylohexaose; pNP, p-nitrophenol; pNP-X2, p-nitrophenol xylobioside; SEP, step elution product; XOS, xylooligosaccharide.
Footnotes
2https://blast.ncbi.nlm.nih.gov/Blast.cgi
3http://www.cazy.org/GH30.html
4https://mafft.cbrc.jp/alignment/server/
5https://fasta.bioch.virginia.edu/fasta_www2/fasta_list2.shtml
6https://www.nzytech.com/products-services/cazymes/glycoside-hydrolases/glucuronoxylanases/cz0916/
References
Aachary, A. A., and Prapulla, S. G. (2011). Xylooligosaccharides (XOS) as an Emerging Prebiotic: Microbial Synthesis, Utilization, Structural Characterization, Bioactive Properties, and Applications. Compr. Rev. Food Sci. Food Saf. 10, 2–16. doi:10.1111/j.1541-4337.2010.00135.x
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic Local Alignment Search Tool. J. Mol. Biol. 215, 403–410. doi:10.1016/s0022-2836(05)80360-2
Artzi, L., Morag, E., Barak, Y., Lamed, R., and Bayer, E. A. (2015). Clostridium clariflavum: Key Cellulosome Players Are Revealed by Proteomic Analysis. MBio 6, e00411–00415. doi:10.1128/mbio.00411-15
Aspeborg, H., Coutinho, P. M., Wang, Y., Brumer, H., and Henrissat, B. (2012). Evolution, Substrate Specificity and Subfamily Classification of Glycoside Hydrolase Family 5 (GH5). BMC Evol. Biol. 12, 186. doi:10.1186/1471-2148-12-186
Biely, P., Puchart, V., Stringer, M. A., and Mørkeberg Krogh, K. B. R. (2014). Trichoderma reesei XYN VI - a Novel Appendage-dependent Eukaryotic Glucuronoxylan Hydrolase. Febs J. 281, 3894–3903. doi:10.1111/febs.12925
Biely, P., Singh, S., and Puchart, V. (2016). Towards Enzymatic Breakdown of Complex Plant Xylan Structures: State of the Art. Biotechnol. Adv. 34, 1260–1274. doi:10.1016/j.biotechadv.2016.09.001
Bounias, M. (1980). N-(1-Naphthyl)ethylenediamine Dihydrochloride as a New Reagent for Nanomole Quantification of Sugars on Thin-Layer Plates by a Mathematical Calibration Process. Anal. Biochem. 106, 291–295. doi:10.1016/0003-2697(80)90523-0
EFSA Panel on Dietetic Products, Bresson, J. L., Burlingame, B., Dean, T., Fairweather-Tait, S., Heinonen, M., Hirsch-Ernst, K. I., et al. (2018). Safety of Xylo-Oligosaccharides (XOS) as a Novel Food Pursuant to Regulation (EU) 2015/2283. EFSA J. 16, e05361. doi:10.2903/j.efsa.2018.5361
Busse-Wicher, M., Li, A., Silveira, R. L., Pereira, C. S., Tryfona, T., Gomes, T. C., et al. (2016). Evolution of Xylan Substitution Patterns in Gymnosperms and Angiosperms: Implications for Xylan Interaction with Cellulose. Plant Physiol. 171, 2418–2431. doi:10.1104/pp.16.00539
Chen, H. H., Chen, Y. K., Chang, H. C., and Lin, S. Y. (2012). Immunomodulatory Effects of Xylooligosaccharides. Fstr 18, 195–199. doi:10.3136/fstr.18.195
Crooks, C., Long, L., and St John, F. J. (2020). CaXyn30B from the Solventogenic Bacterium Clostridium acetobutylicum is a Glucuronic Acid-dependent Endoxylanase. BMC Res. Notes 13, 1–6. doi:10.1186/s13104-020-05091-5
Ding, S.-Y., and Bayer, E. A. (2020). "Understanding Cellulosome Interaction with Cellulose by High-Resolution Imaging". ACS Central Science 6, 1034–1036. doi:10.1021/acscentsci.0c00662
Ebringerová, A., and Hromádková, Z. (1999). Xylans of Industrial and Biomedical Importance. Biotechnol. Genet. Eng. Rev. 16, 325–346. doi:10.1080/02648725.1999.10647982
Ebringerová, A., Kardošová, A., Hromádková, Z., Malovı́ková, A., and Hřı́balová, V. (2002). Immunomodulatory Activity of Acidic Xylans in Relation to Their Structural and Molecular Properties. Int. J. Biol. macromolecules 30, 1–6. doi:10.1016/s0141-8130(01)00186-6
Fauré, R., Courtin, C. M., Delcour, J. A., Dumon, C., Faulds, C. B., Fincher, G. B., et al. (2009). A Brief and Informationally Rich Naming System for Oligosaccharide Motifs of Heteroxylans Found in Plant Cell walls. Aust. J. Chem. 62, 533–537. doi:10.1071/ch08458
Felsenstein, J. (1985). Confidence Limits on Phylogenies: an Approach Using the Bootstrap. Evolution 39, 783–791. doi:10.2307/2408678
Freire, F., Verma, A., Bule, P., Alves, V. D., Fontes, C. M. G. A., Goyal, A., et al. (2016). Conservation in the Mechanism of Glucuronoxylan Hydrolysis Revealed by the Structure of Glucuronoxylan Xylanohydrolase (CtXyn30A) from Clostridium thermocellum. Acta Cryst. Sect D Struct. Biol. 72, 1162–1173. doi:10.1107/s2059798316014376
Gallardo, O., Fernández-Fernández, M., Valls, C., Valenzuela, S. V., Roncero, M. B., Vidal, T., et al. (2010). Characterization of a Family GH5 Xylanase with Activity on Neutral Oligosaccharides and Evaluation as a Pulp Bleaching Aid. Appl. Environ. Microbiol. 76, 6290–6294. doi:10.1128/aem.00871-10
Gibson, D. G., Young, L., Chuang, R.-Y., Venter, J. C., Hutchison, C. A., and Smith, H. O. (2009). Enzymatic Assembly of DNA Molecules up to Several Hundred Kilobases. Nat. Methods 6, 343–345. doi:10.1038/nmeth.1318
Gruno, M., Väljamäe, P., Pettersson, G., and Johansson, G. (2004). Inhibition of the Trichoderma reesei cellulases by Cellobiose Is Strongly Dependent on the Nature of the Substrate. Biotechnol. Bioeng. 86, 503–511. doi:10.1002/bit.10838
Johnson, M., Zaretskaya, I., Raytselis, Y., Merezhuk, Y., Mcginnis, S., and Madden, T. L. (2008). NCBI BLAST: a Better Web Interface. Nucleic Acids Res. 36, W5–W9. doi:10.1093/nar/gkn201
Kahn, A., Moraïs, S., Galanopoulou, A. P., Chung, D., Sarai, N. S., Hengge, N., et al. (2019). Creation of a Functional Hyperthermostable Designer Cellulosome. Biotechnol. biofuels 12, 1–15. doi:10.1186/s13068-019-1386-y
Katoh, K., and Toh, H. (2008). Recent Developments in the MAFFT Multiple Sequence Alignment Program. Brief. Bioinform. 9, 286–298. doi:10.1093/bib/bbn013
Katsimpouras, C., Dedes, G., Thomaidis, N. S., and Topakas, E. (2019). A Novel Fungal GH30 Xylanase with Xylobiohydrolase Auxiliary Activity. Biotechnol. biofuels 12, 120. doi:10.1186/s13068-019-1455-2
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi:10.1093/molbev/msy096
Laemmli, U. K. (1970). Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 227, 680–685. doi:10.1038/227680a0
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., Mcwilliam, H., et al. (2007). Clustal W and Clustal X Version 2.0. bioinformatics 23, 2947–2948. doi:10.1093/bioinformatics/btm404
Le, S. Q., and Gascuel, O. (2008). An Improved General Amino Acid Replacement Matrix. Mol. Biol. Evol. 25, 1307–1320. doi:10.1093/molbev/msn067
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M., and Henrissat, B. (2014). The Carbohydrate-Active Enzymes Database (CAZy) in 2013. Nucl. Acids Res. 42, D490–D495. doi:10.1093/nar/gkt1178
Maehara, T., Yagi, H., Sato, T., Ohnishi-Kameyama, M., Fujimoto, Z., Kamino, K., et al. (2018). GH30 Glucuronoxylan-specific Xylanase from Streptomyces turgidiscabies C56. Appl. Environ. Microbiol. 84, e01850–01817. doi:10.1128/aem.01850-17
Naidu, D. S., Hlangothi, S. P., and John, M. J. (2018). Bio-based Products from Xylan: A Review. Carbohydr. Polym. 179, 28–41. doi:10.1016/j.carbpol.2017.09.064
Nakamichi, Y., Fouquet, T., Ito, S., Matsushika, A., and Inoue, H. (2019a). Mode of Action of GH30-7 Reducing-End Xylose-Releasing Exoxylanase A (Xyn30A) from the Filamentous Fungus Talaromyces cellulolyticus. Appl. Environ. Microbiol. 85, e00552–00519. doi:10.1128/aem.00552-19
Nakamichi, Y., Fouquet, T., Ito, S., Watanabe, M., Matsushika, A., and Inoue, H. (2019b). Structural and Functional Characterization of a Bifunctional GH30-7 Xylanase B from the Filamentous Fungus Talaromyces cellulolyticus. J. Biol. Chem. 294, 4065–4078. doi:10.1074/jbc.ra118.007207
Nakamichi, Y., Fujii, T., Watanabe, M., Matsushika, A., and Inoue, H. (2020a). Crystal Structure of GH30-7 Endoxylanase C from the Filamentous Fungus Talaromyces cellulolyticus. Acta Cryst. Sect F 76, 341–349. doi:10.1107/s2053230x20009024
Nakamichi, Y., Watanabe, M., Matsushika, A., and Inoue, H. (2020b). Substrate Recognition by a Bifunctional GH30-7 Xylanase B from Talaromyces cellulolyticus. FEBS open bio 10. doi:10.2210/pdb6krl/pdb
Ohbuchi, T., Sakaino, M., Takahashi, T., Azumi, N., Ishikawa, K., Kawazoe, S., et al. (2010). Oral Administration of Acidic Xylooligosaccharides Prevents the Development of Atopic Dermatitis-like Skin Lesions in NC/Nga Mice. J. Nutr. Sci. Vitaminol 56, 54–59. doi:10.3177/jnsv.56.54
Pearson, W. R. (1998). Empirical Statistical Estimates for Sequence Similarity Searches. J. Mol. Biol. 276, 71–84. doi:10.1006/jmbi.1997.1525
Penksza, P., Banka, Z., Szilárd, K., Huszár, K. P., Németh, C., Tóth, A., et al. (2018). Utilization of Xylo-Oligosaccharides as Prebiotics in Yoghurt. J. Hygienic Eng. Des. 22, 66–71.
Preston, J. F., Hurlbert, J. C., Rice, J. D., Ragunathan, A., and St John, F. J. (2003). “Microbial Strategies for the Depolymerization of Glucuronoxylan: Leads to Biotechnological Applications of Endoxylanases,” in Applications of Enzymes to Lignocellulosics (Washington D.C.: American Chemical Society), 191–210. doi:10.1021/bk-2003-0855.ch012:
Puchart, V., Šuchová, K., and Biely, P. (2021). Xylanases of Glycoside Hydrolase Family 30–An Overview. Biotechnol. Adv. 47, 107704. doi:10.1016/j.biotechadv.2021.107704
Saha, B. C. (2003). Hemicellulose Bioconversion. J. Ind. Microbiol. Biotechnol. 30, 279–291. doi:10.1007/s10295-003-0049-x
Sainz-Polo, M. A., Valenzuela, S. V., González, B., Pastor, F. I., and Sanz-Aparicio, J. (2014). Structural Analysis of Glucuronoxylan-Specific Xyn30D and its Attached CBM35 Domain Gives Insights into the Role of Modularity in Specificity. J. Biol. Chem. 289, 31088–31101. doi:10.1074/jbc.M114.597732
Sakurama, H., Kishino, S., Uchibori, Y., Yonejima, Y., Ashida, H., Kita, K., et al. (2013). β-Glucuronidase from Lactobacillus brevis Useful for Baicalin Hydrolysis Belongs to Glycoside Hydrolase Family 30. Appl. Microbiol. Biotechnol. 98, 4021–4032. doi:10.1007/s00253-013-5325-8
St John, F. J., Crooks, C., Dietrich, D., and Hurlbert, J. (2017). Xylanase 30 A from Clostridium thermocellum Functions as a Glucuronoxylan Xylanohydrolase. J. Mol. Catal. B: Enzymatic 133, S445–S451. doi:10.1016/j.molcatb.2017.03.008
St John, F. J., Dietrich, D., Crooks, C., Balogun, P., De Serrano, V., Pozharski, E., et al. (2018). A Plasmid Borne, Functionally Novel Glycoside Hydrolase Family 30 Subfamily 8 Endoxylanase from Solventogenic Clostridium. Biochem. J. 475, 1533–1551. doi:10.1042/bcj20180050
St John, F. J., Dietrich, D., Crooks, C., Pozharski, E., González, J. M., Bales, E., et al. (2014). A Novel Member of Glycoside Hydrolase Family 30 Subfamily 8 with Altered Substrate Specificity. Acta Cryst. D Biol. Crystallogr. 70, 2950–2958. doi:10.1107/s1399004714019531
St John, F. J., González, J. M., and Pozharski, E. (2010). Consolidation of Glycosyl Hydrolase Family 30: a Dual Domain 4/7 Hydrolase Family Consisting of Two Structurally Distinct Groups. FEBS Lett. 584, 4435–4441. doi:10.1016/j.febslet.2010.09.051
St John, F. J., Hurlbert, J. C., Rice, J. D., Preston, J. F., and Pozharski, E. (2011). Ligand Bound Structures of a Glycosyl Hydrolase Family 30 Glucuronoxylan Xylanohydrolase. J. Mol. Biol. 407, 92–109. doi:10.1016/j.jmb.2011.01.010
St John, F. J., Rice, J. D., and Preston, J. F. (2006a). Characterization of XynC from Bacillus subtilis subsp. subtilis Strain 168 and Analysis of its Role in Depolymerization of Glucuronoxylan. J. Bacteriol. 188, 8617–8626. doi:10.1128/jb.01283-06
St John, F. J., Rice, J. D., and Preston, J. F. (2006b). Paenibacillus Sp. Strain JDR-2 and XynA1 : a Novel System for Methylglucuronoxylan Utilization. Appl. Environ. Microbiol. 72, 1496–1506. doi:10.1128/aem.72.2.1496-1506.2006
Studier, F. W. (2005). Protein Production by Auto-Induction in High-Density Shaking Cultures. Protein Expr. Purif. 41, 207–234. doi:10.1016/j.pep.2005.01.016
Šuchová, K., Puchart, V., and Biely, P. (2020a). A Novel Bacterial GH30 Xylobiohydrolase from Hungateiclostridium clariflavum. Appl. Microbiol. Biotechnol. 105, 1–11. doi:10.1007/s00253-020-11023-x
Šuchová, K., Puchart, V., Spodsberg, N., Krogh, K. B. M., and Biely, P. (2020b). A Novel GH30 Xylobiohydrolase from Acremonium alcalophilum Releasing Xylobiose from the Non-reducing End. Enzyme Microb. Techn. 134, 109484. doi:10.1016/j.enzmictec.2019.109484
Tenkanen, M., Vršanská, M., Siika-Aho, M., Wong, D. W., Puchart, V., Penttilä, M., et al. (2013). Xylanase XYN IV from Trichoderma reesei showing Exo- and Endo-Xylanase Activity. Febs J. 280, 285–301. doi:10.1111/febs.12069
The Uniprot Consortium (2017). UniProt: the Universal Protein Knowledgebase. Nucleic Acids Res. 45, D158–D169. doi:10.1093/nar/gkw1099
Tindall, B. J. (2019). The names Hungateiclostridium Zhang et al. 2018, Hungateiclostridium thermocellum (Viljoen et al. 1926) Zhang et al. 2018, Hungateiclostridium cellulolyticum (Patel et al. 1980) Zhang et al. 2018, Hungateiclostridium aldrichii (Yang et al. 1990) Zhang et al. 2018, Hungateiclostridium alkalicellulosi (Zhilina et al. 2006) Zhang et al. 2018, Hungateiclostridium clariflavum (Shiratori et al. 2009) Zhang et al. 2018, Hungateiclostridium straminisolvens (Kato et al. 2004) Zhang et al. 2018 and Hungateiclostridium saccincola (Koeck et al. 2016) Zhang et al. 2018 contravene Rule 51b of the International Code of Nomenclature of Prokaryotes and require replacement names in the genus Acetivibrio Patel et al. 1980. Int. J. Syst. Evol. Microbiol. 69, 3927–3932. doi:10.1099/ijsem.0.003685
Urbániková, Ľ., Vršanská, M., Mørkeberg Krogh, K. B. R., Hoff, T., and Biely, P. (2011). Structural Basis for Substrate Recognition by Erwinia chrysanthemi GH30 Glucuronoxylanase. FEBS J. 278, 2105–2116. doi:10.1111/j.1742-4658.2011.08127.x
Vázquez, M. J., Alonso, J. L., Domı́nguez, H., and Parajó, J. C. (2000). Xylooligosaccharides: Manufacture and Applications. Trends Food Sci. Techn. 11, 387–393. doi:10.1016/s0924-2244(01)00031-0
Keywords: xylobiohydrolase, glycoside hydrolase, glycoside hydrolase family 30, AcXbh30A, Acetivibrio clariflavus, xylobiose, glucuronoxylan, Hungateiclostridium clariflavum
Citation: Crooks C, Bechle NJ and St John FJ (2021) A New Subfamily of Glycoside Hydrolase Family 30 with Strict Xylobiohydrolase Function. Front. Mol. Biosci. 8:714238. doi: 10.3389/fmolb.2021.714238
Received: 24 May 2021; Accepted: 20 July 2021;
Published: 07 September 2021.
Edited by:
Song Xiang, Tianjin Medical University, ChinaReviewed by:
Harry Brumer, University of British Columbia, CanadaYusuke Nakamichi, National Institute of Advanced Industrial Science and Technology (AIST), Japan
Copyright © 2021 Crooks, Bechle and St John. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Franz J. St John, ZmpzdGpvaG5AZ21haWwuY29t, ZnJhbnouai5zdC5qb2huQHVzZGEuZ292
 Casey Crooks1
Casey Crooks1 Nathan J. Bechle
Nathan J. Bechle Franz J. St John
Franz J. St John