- Department of Biological Science, Wayne State University, Detroit, MI, United States
An evolutionarily conserved feature of introns is their ability to enhance expression of genes that harbor them. Introns have been shown to regulate gene expression at the transcription and post-transcription level. The general perception is that a promoter-proximal intron is most efficient in enhancing gene expression and the effect diminishes with the increase in distance from the promoter. Here we show that the intron regains its positive influence on gene expression when in proximity to the terminator. We inserted ACT1 intron into different positions within IMD4 and INO1 genes. Transcription Run-On (TRO) analysis revealed that the transcription of both IMD4 and INO1 was maximal in constructs with a promoter-proximal intron and decreased with the increase in distance of the intron from the promoter. However, activation was partially restored when the intron was placed close to the terminator. We previously demonstrated that the promoter-proximal intron stimulates transcription by affecting promoter directionality through gene looping-mediated recruitment of termination factors in the vicinity of the promoter region. Here we show that the terminator-proximal intron also enhances promoter directionality and results in compact gene architecture with the promoter and terminator regions in close physical proximity. Furthermore, we show that both the promoter and terminator-proximal introns facilitate assembly or stabilization of the preinitiation complex (PIC) on the promoter. On the basis of these findings, we propose that proximity to both the promoter and the terminator regions affects the transcription regulatory potential of an intron, and the terminator-proximal intron enhances transcription by affecting both the assembly of preinitiation complex and promoter directionality.
Introduction
One of the conserved features of eukaryotic protein-coding genes that distinguishes them from their prokaryotic counterparts is the presence of non-coding intervening regions called introns. All eukaryotic genes, however, do not contain introns. The proportion of genes containing introns vary from 2.4% in pathogenic ascomycete yeast Candida glabrata to 98% in capsular basidiomycete yeast Cryptococcus neoformans (Neuvéglise et al., 2011; Goebels et al., 2013). A majority of human (92%) and plant (78%) genes also contain introns (International Human Genome Sequencing Consortium, 2004; Sakharkar et al., 2004; Haas et al., 2005). In budding yeast, a mere 3.8% of genes contain introns, but this small number of genes contributes to nearly 27% of mRNA in exponentially growing yeast cells (Ares et al., 1999; Spingola et al., 1999). Introns substantially enhance the expression of genes that accommodate them (Brinster et al., 1988; Palmiter et al., 1991; Okkema et al., 1993; Duncker et al., 1997; Lugones et al., 1999; Comeron, 2004; Juneau et al., 2006; Charron et al., 2007; Rose, 2008; Shabalina et al., 2010; Gallegos and Rose, 2015; Laxa, 2017; Shaul, 2017; Baier et al., 2020). A number of eukaryotic genes are dependent on introns for their normal expression. Introns influence almost every step of RNA metabolism including transcription, cotranscriptional RNA processing, mRNA decay, mRNA export to cytoplasm and translatability of mRNA (Le Hir et al., 2003; Lu and Cullen, 2003; Carmel and Chorev, 2012; Gallegos and Rose, 2015; Shaul, 2017; Rose, 2019). The molecular basis underlying the intron-mediated regulation, however, is not entirely clear. The ability to enhance gene expression though is not a universal feature of introns.
The role of an intron in enhancing transcription has been conserved during evolution, being exhibited by a diversity of eukaryotic systems including yeast, humans, flies, plants, algae and worms (McKenzie and Brennan, 1996; Juneau et al., 2006; Rose, 2008; Shabalina et al., 2010; Gallegos and Rose, 2015; Laxa, 2017; Shaul, 2017; Baier et al., 2020). Inclusion of just one intron in a transgene has been found to enhance its transcription by many folds (Brinster et al., 1988; Palmiter et al., 1991; Rethmeier et al., 1997; Lacy-Hulbert et al., 2001; Bartlett et al., 2009; Sam et al., 2010). Introns are known to enhance transcription by affecting chromatin structure in the promoter-proximal region and by facilitating the recruitment of general transcription machinery on the promoter. In mammalian cells, an intron facilitates H3K4-trimethylation and H3K9-acetylation in the promoter-proximal regions (Bieberstein et al., 2012). Both of these histone marks help in the recruitment of general transcription machinery to the promoter region. A promoter-proximal intron has also been shown to expedite the recruitment of transcription factors on the promoter region through interaction of U1-snRNP with the components of general transcription machinery (Kwek et al., 2002; Das et al., 2007; Damgaard et al., 2008). In budding yeast, the intron enhances gene expression primarily by affecting transcription and mRNA stability (Furger et al., 2002; Juneau et al., 2006; Parenteau et al., 2008; Parenteau et al., 2011; Moabbi et al., 2012; Agarwal and Ansari, 2016; Petibon et al., 2016). The positive influence of an intron on transcription of several yeast genes has been demonstrated using the nuclear run-on approach (Furger et al., 2002; Moabbi et al., 2012; Agarwal and Ansari, 2016). The intron-mediated enhancement of transcription in yeast requires a splicing-competent intron (Furger et al., 2002; Moabbi et al., 2012; Agarwal and Ansari, 2016). We recently examined the mechanism of intron-mediated enhancement effect in budding yeast and found that a promoter-proximal intron results in the formation of a unique looped gene architecture (Moabbi et al., 2012; Agarwal and Ansari, 2016; Dwyer et al., 2021). We showed that the gene loop facilitated the recruitment of termination factors near the promoter-proximal region, and these termination factors inhibited upstream antisense RNA (uaRNA) synthesis, thereby conferring directionality to the promoter (Agarwal and Ansari, 2016; Al Husini et al., 2020). This results in enhanced transcription of mRNA. In a gene looping-defective mutant, even a splicing-competent intron was unable to enhance transcription, thereby implicating looped gene architecture in intron-mediated transcriptional regulation (Dwyer et al., 2021).
A number of factors influence the intron-mediated enhancement effect. The sequence of an intron is a crucial determinant of its role in regulation of gene expression (Morello et al., 2010; Parra et al., 2011). Another critical factor in determining the regulatory potential of an intron is its proximity to the promoter. It is generally believed that the introns located within the first 1 kbp of the promoter region are most efficient in enhancing gene expression, and the enhancement potential is inversely proportional to the distance of the intron from the promoter element (Callis et al., 1987; Nesic et al., 1993; Jeon et al., 2000; Rose, 2004; Jeong et al., 2006; Rose, 2008; Gallegos and Rose, 2015). There are conflicting reports regarding the ability of a terminator-proximal intron to enhance gene expression (Snowden et al., 1996; Furger et al., 2002; Rose, 2004). A terminator-proximal intron facilitates 3′ end processing of mRNA and termination of transcription (Huang and Gorman, 1990; Nesic et al., 1993; Gunderson et al., 1997; Antoniou et al., 1998; Dye and Proudfoot, 1999; Vagner et al., 2000; McCracken et al., 2002; Awasthi and Alwine, 2003; Kyburz et al., 2006; Millevoi et al., 2006; Kim et al., 2011; Martinson, 2011). This can potentially enhance transcription of the gene. There are, however, a few reports that introns located in the 3′ UTR adversely affect mRNA stability and translatability in mammalian and plant cells (Bourdon et al., 2001; Kertesz et al., 2006; Barrett et al., 2012).
To understand the role of the position of an intron in a gene, we inserted the ACT1 intron into different positions of IMD4 and INO1 genes. Our results show that the proximity to both the promoter and the terminator has a positive influence on transcription of the gene. Like the promoter-proximal intron, the terminator-proximal intron also mediates gene looping and enhances promoter directionality. A terminator-proximal intron though is less efficient than a promoter-proximal intron in enhancing transcription. We further show that both the promoter and terminator-proximal introns facilitate the assembly or stabilization of the PIC on the promoter. Our findings reveal that an intron enhances gene expression by influencing multiple aspects of promoter function and suggest that it accomplishes this by modulating the architecture of actively transcribing genes. We propose that proximity to both the promoter and terminator is crucial for the transcription enhancement potential of an intron.
Materials and Methods
Yeast Strains
Yeast strains used in this study are listed in Supplementary Table S1.
Cell Cultures
Cultures were started by inoculating 5 ml of YP-dextrose medium with colonies from a freshly streaked plate and grown at 30°C with gentle shaking overnight. Next morning, cultures were diluted (1:100) to appropriate volume and grown to A600 ∼0.4–0.6. Equal number of cells were used for strand-specific RT-PCR, 3C, ChIP or strand-specific TRO assays.
Chromosome Conformation Capture Assay
3C experiments were performed as described previously in (El Kaderi et al., 2012). The primers used for 3C analysis are shown in Supplementary Table S2. The restriction enzymes used for chromatin digestion of IMD4 gene were Alu1 and Dra1. Each experiment was performed with at least four independently grown cultures. The P1T1 PCR signals obtained using P1 and T1 primers flanking the promoter and terminator regions respectively were normalized with respect to F1-R1 PCR signal that amplifies an uncut region within the gene as shown in Figures 1D, 2B. All controls described in El Kaderi et al. (2012) were performed in 3C experiments. These included no-formaldehyde-crosslinking control, no-ligation control and primer efficiency control.
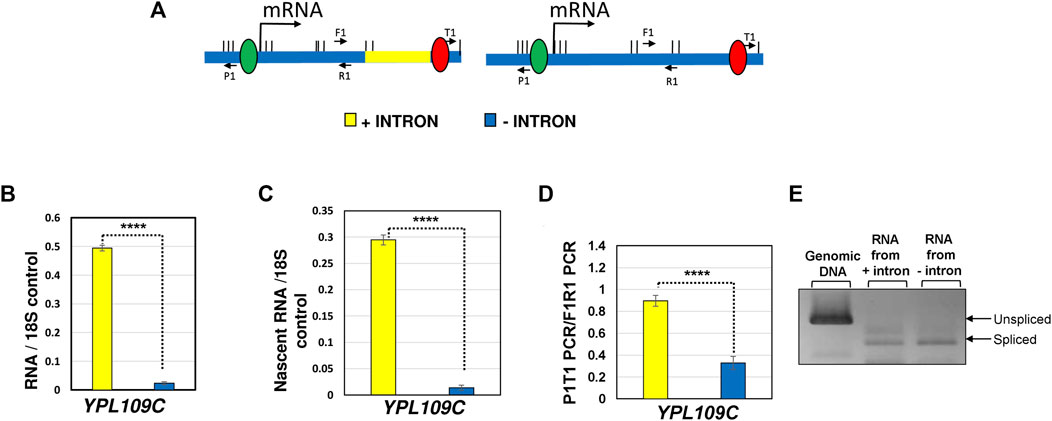
FIGURE 1. Deletion of the terminator-proximal intron of YPL109C reduces both the transcription and looped conformation of the gene. (A) Schematic depiction of YPL109C with the intron present (yellow) and intron deleted (blue) indicating the position of primers used for RT-PCR, TRO and 3C analyses. (B) Quantification of total RNA level of YPL109C measured by RT-PCR approach in the presence of the terminator-proximal intron (yellow bar) and the absence of intron (blue bar). The 18S RNA signal was used as a normalization control. (C) Quantification of nascent RNA level of YPL109C measured by TRO approach in the presence of the terminator-proximal intron (yellow bar) and the absence of intron (blue bar). The 18S RNA signal was used as a normalization control. (D) Gene looping of YPL109C measured in terms of P1T1 PCR signal by 3C approach in the presence (yellow bar) and absence (blue bar) of an intron. The F1R1 PCR represents the loading control that was used to ensure equal amounts of template DNA were present in each 3C PCR reaction. The values and error bars for each condition indicate the mean ± standard deviation. Statistical significance (p-values) was determined using the two-tailed paired Student’s t-test. Four asterisks (****) indicate p value smaller than 0.0001 (p ≤ 0.0001). (E) RT-PCR analysis of YPL109C mRNA in the construct with intron (+ intron) and without intron (− intron). Genomic DNA PCR of intron-containing strain indicates the position of unspliced transcripts. The expected positions of spliced and unspliced transcripts obtained by RT-PCR approach for each construct is indicated.
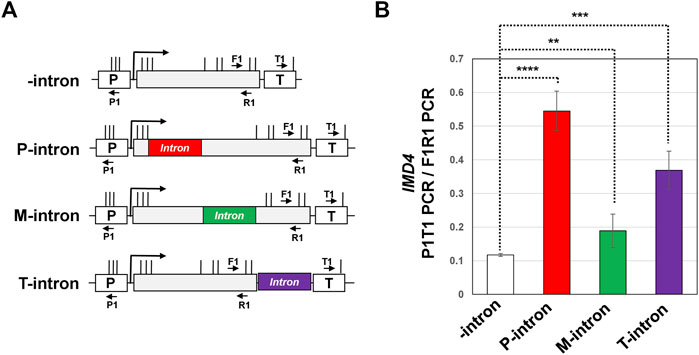
FIGURE 2. Position of an intron within the gene impacts gene looping of IMD4. (A) Schematic depiction of IMD4 gene without intron (-intron, white), and with ACT1 intron inserted near the promoter (P-intron, red), in the middle of the gene (M-intron, green) and in the proximity of the terminator (T-intron, purple) region. “p” and “T” stand for the promoter and terminator respectively. Vertical lines show position of restriction cut sites, while P1, T1, F1 and R1 are the position of primers used to amplify 3C PCR products. (B) 3C analysis of IMD4 in the absence of an intron (white bar), and with the intron inserted near the promoter (red bar), in the middle (green bar) and toward the terminator region of the gene (purple bar). P1T1 PCR represents gene looping signal, while F1R1 PCR represents the loading control that was used to ensure equal amounts of template DNA were present in each 3C PCR reaction. Results presented here represent three biological replicates and six technical replicates. The values and error bars for each condition indicate the mean ± standard deviation. Statistical significance (p-values) was determined using the two-tailed paired Student’s t-test. Two asterisks (**) signify a p value equal to or smaller than 0.01 (p ≤ 0.01). Three asterisks (***) indicate p value equal/smaller than 0.001 (p ≤ 0.001). Four asterisks (****) indicate p value equal/smaller than 0.0001 (p ≤ 0.0001). Any non-significant difference is represented by ns (p > 0.05).
Chromatin Immunoprecipitation
ChIP experiments (crosslinking, cell lysis and isolation of chromatin) were performed as described previously (El Kaderi et al., 2009). Different strains were constructed by tagging TFIIB and the Ccl1 subunit of TFIIH as indicated in Supplementary Table S1. Anti-HA-Agarose beads, used to pull down HA-tagged subunits, were obtained from Sigma. IgG-Sepharose beads, purchased from GE Healthcare, were used to pull down TAP-tagged Ccl1 subunit of TFIIH. For ChIP analysis, primers used for ChIP PCR are shown in Supplementary Table S2. Each experiment was repeated with at least four independently grown cultures. Background ChIP signal for each TFIIB and TFIIH experiment was determined using no tag control version of strains. All TFIIB and TFIIH ChIP signals were first normalized with no tag control, and then with input.
Transcription Analysis
Transcription analysis was performed by RT-PCR approach as described previously (El Kaderi et al., 2009). The primers used for RT-PCR analysis are shown in Supplementary Table S2.
Strand-Specific “Transcription Run-On” Assay
The strand-specific “Transcription Run-On” (TRO) assay was performed as described in (Dhoondia et al., 2017). The primers used for making cDNA and PCR are described in Supplementary Table S2.
Quantification
The data shown in each figure is the result of at least three biological replicates and six technical replicates. The quantification and statistical analysis were performed as described in (El Kaderi et al., 2012). Error bars represent one unit of standard deviation. p-values were calculated by two-tailed student t-test (Student, 1908).
Results
A Terminator-Proximal Intron Enhances Transcription of Yeast Genes
A vast majority of intron-containing genes in budding yeast have an intron in the promoter-proximal position. A small number of yeast genes carry an intron in the middle of the gene, and an even fewer number have them in the vicinity of the terminator region. It has been shown that a promoter-proximal intron enhances transcription of a gene by facilitating the recruitment of general transcription machinery and by enhancing transcription directionality (Kwek et al., 2002; Das et al., 2007; Damgaard et al., 2008; Bieberstein et al., 2012; Agarwal and Ansari, 2016). The effect of a promoter-proximal intron on transcription has been conserved during evolution as it is exhibited by simple eukaryotes like budding yeast as well as the most complex mammalian systems. There is, however, no concrete evidence regarding the transcription enhancement potential of a terminator-proximal intron. YPL109C is one of the few yeast genes that contain a terminator-proximal intron. We deleted the terminator-proximal intron of YPL109C, and measured transcription of the gene in the presence and absence of the intron (Figure 1A). Steady-state RNA analysis revealed that YPL109C mRNA level decreased by about 10-folds in the absence of the intron (Figure 1B). Transcription Run-On (TRO) analysis, which measured the transcription of nascent mRNA, revealed similar results (Figure 1C). These results demonstrate that even a terminator-proximal intron has the potential to enhance the transcription of the gene.
To better understand the role of the position of an intron in a gene on its transcription regulatory potential, it required inserting the same intron sequence at different positions of a gene and measuring transcription of the gene at every position. We have earlier showed that ACT1 intron enhances transcription of INO1 in an activator-independent manner (Moabbi et al., 2012). Furthermore, we demonstrated that transcription activation potential of ACT1 intron was not due to the presence of a promoter or enhancer sequence element within the intron but was strictly dependent on the splicing potential of the intron. We therefore inserted ACT1 intron into five different positions of INO1 gene following the strategy described in Moabbi et al. (2012). INO1 is an intron-less gene, which is induced in the absence of inositol in the growth medium. We have previously demonstrated enhancement of INO1 transcription under non-inducing conditions upon insertion of an intron near the promoter region of the gene (Moabbi et al., 2012). Here we inserted ACT1 intron at four additional positions of INO1 gene as shown in Figure 3A. The intron was inserted in the 1,602 bp long INO1 gene at 100, 500, 800 and 1,400 bp positions in the coding region of the gene, and one 10 bp downstream of the coding region in the 3′ UTR of the gene. At each position of the intron, transcription was monitored by strand-specific TRO approach as described in Medler and Ansari (2015). TRO assay measures nascent mRNA, which is not affected by half-life of mRNA, and accurately reflects transcription of the gene. In the absence of an intron, there is very low level basal transcription of the gene. Insertion of the intron at 100 bp position enhanced nascent mRNA levels by about 43 times over the intronless version (Figure 3B). At the 500 bp position, there was only 20 times stimulation in transcript level by the intron over the intron-less control. Thus, the enhancement effect of an intron at the 500 bp position registered a decline by about 50% compared to the 100 bp position (Figure 3B). Introns inserted at 800 and 1,400 bp position resulted in a mere 6 times and 8 times increase respectively in transcription over the intron-less control. These results are in agreement with the published results from other organisms as they clearly demonstrate that the enhancement effect of the intron on gene expression decreases with the increase in distance of the intron from the promoter region (Callis et al., 1987; Nesic et al., 1993; Rose, 2004). There was, however, a surprise when we inserted the intron at a position 10 bp downstream of the coding region. The 3′ UTR intron enhanced INO1 transcription by about 17 times (Figure 3B). These results suggest that the proximity to the terminator region partially restores the enhancement potential of an intron on gene expression. Yet, the terminator-proximal intron was not as efficient as the promoter-proximal intron in enhancing transcription of INO1. Although the promoter-proximal intron was spliced with slightly lower efficiency, overall introns were spliced with similar efficiency at each of these five positions (Supplementary Figure S1), thereby suggesting that the observed results were not because of the differential splicing efficacy of introns.
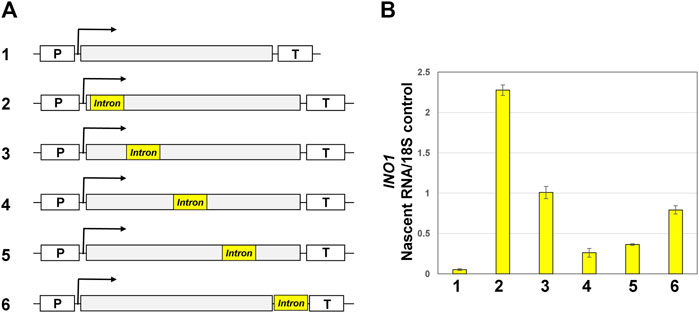
FIGURE 3. Position of the intron in INO1 gene influences the transcription of the gene. (A) Schematic representation of INO1 gene without intron (1) and with the ACT1 intron inserted at five different positions along the body of the gene (2–6). “p” and “T” stand for the promoter and terminator respectively. (B) Quantification of TRO data of INO1 gene in the absence of an intron (#1) and the presence of an intron at 100 bp position (#2), 500 bp (#3), 800 bp (#4), 1,400 bp (#5), and 10 bp downstream of (TGA + 10) (#6). The transcript levels of 18S were used as the normalization control.
The INO1 gene does not contain a natural intron. The results obtained with INO1 therefore need to be substantiated with a gene containing a natural intron. We, therefore, repeated the experiment with IMD4, which is a natural intron-containing gene. We first constructed a strain containing the intron-less version of IMD4 gene as described in Moabbi et al. (2012). We then inserted ACT1 intron at three different positions of the IMD4 gene; one at 461 bp position (promoter-proximal), a second one at 750 bp position within the coding region (middle), and a third one in 3′ UTR at the position 20 bp downstream of the coding region (terminator-proximal) as shown in Figure 4A. Like INO1, we have previously demonstrated that ACT1 intron enhances transcription of IMD4 in a splicing-dependent manner as mutation of either 5′ or 3′ splice sites completely abolished transcription enhancement potential of the intron (Agarwal and Ansari, 2016). Transcription of the gene was then monitored by strand-specific TRO approach as described earlier. The results show very little detectable transcription in the absence of an intron (Figure 4B). Insertion of the intron at 461 position (promoter-proximal) stimulated transcription of the gene by about 10-fold over the intronless version. The 750 intron (middle) did not result in enhancement of transcription over the intron-less control as the TRO signal obtained was similar to that in the absence of an intron. In the presence of a terminator-proximal intron, however, TRO signal registered an increase of almost six-fold over the intron-less control (Figure 4B). The enhancement effect of terminator-proximal intron is statistically significant as indicated by the p-value of 2.6E-04 (Figure 4B). Although the promoter-proximal intron was spliced with slightly lower efficiency, overall introns were spliced with similar efficiency at each of these three positions (Supplementary Figure S2), thereby suggesting that the observed results were not because of the differential splicing behavior of introns. These results clearly demonstrate that an intron regains its transcription enhancement potential with increasing proximity to the terminator region of the gene in budding yeast. A logical conclusion of these results is that the proximity to both the promoter and terminator regions contributes to the enhancement potential of an intron on gene expression.
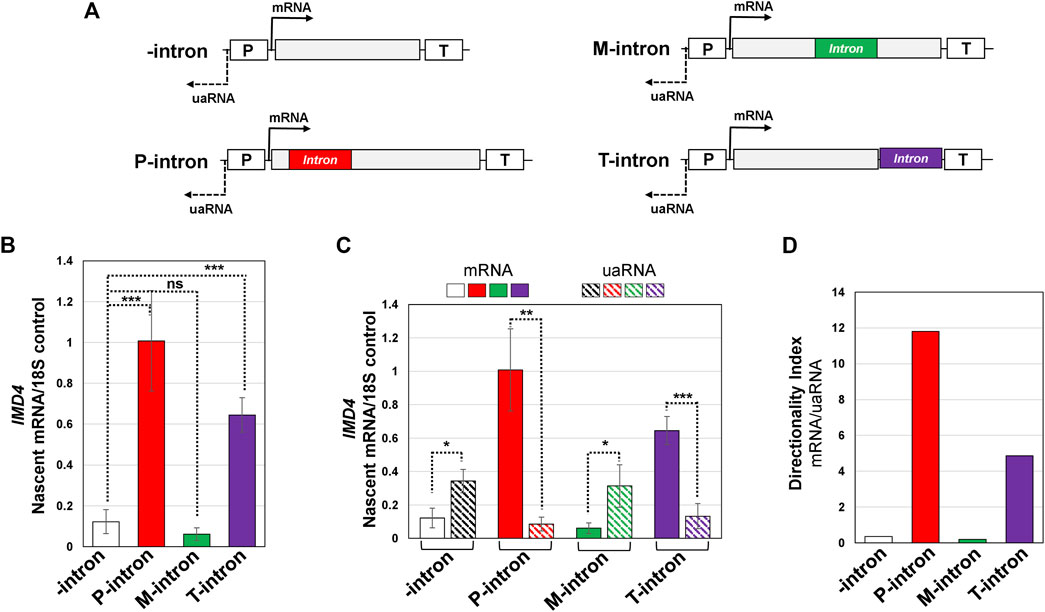
FIGURE 4. Both the promoter and terminator proximal introns enhance transcription and promoter directionality of IMD4 gene. (A) Schematic depiction of IMD4 gene without intron (-intron, white), and with ACT1 intron inserted near the promoter (P-intron, red), in the middle of the gene (M-intron, green) and in the proximity of the terminator (T-intron, purple) region. “p” and “T” stand for the promoter and terminator respectively. (B) TRO analysis of IMD4 in the absence of an intron (white bar), and with the intron inserted near the promoter (red bar), in the middle (green bar) and toward the terminator region of the gene (purple bar). (C) Measurement of mRNA and uaRNA levels of IMD4 in the 400 bp upstream (uaRNA) and downstream (mRNA) promoter proximal region by strand-specific TRO approach in the absence of an intron (white bar for mRNA, diagonal black pattern bar for uaRNA), and with the intron inserted near the promoter (red bar for mRNA, diagonal red pattern bar for uaRNA), in the middle (green bar for mRNA, diagonal green pattern bar for uaRNA) and toward the terminator region of the gene (purple bar for mRNA, diagonal purple pattern bar for uaRNA). (D) Directionality index of IMD4 without intron (white bar) and with the intron present at three different positions (red, green and purple bars) by comparing levels of mRNA vs. uaRNA produced. The 18S signal was used as a normalization control in these experiments. Results presented here represent three biological replicates and six technical replicates. The values and error bars for each condition indicate the mean ± standard deviation. Statistical significance (p-values) was determined using the two-tailed paired Student’s t-test. One asterisk (*) signifies a p value equal to or smaller than 0.05 (p ≤ 0.05). Any non-significant difference is represented by ns (p > 0.05).
Terminator-Proximal Intron Affects Gene Looping and Promoter Directionality
We have recently demonstrated a novel role of a promoter-proximal intron in enhancing transcription by conferring directionality to the promoter-bound polymerase to transcribe in the sense direction and inhibit transcription in the upstream anti-sense direction (Agarwal and Ansari, 2016). To determine if the transcription regulatory function of a terminator-proximal intron is also due to its effect on transcription directionality, we performed TRO in the promoter-proximal 800 bp window; 400 bp downstream (mRNA) and upstream (uaRNA) of the promoter as shown in Figure 4A. The terminator-proximal intron stimulated transcription in the promoter-proximal sense direction (mRNA) by about six fold (Figure 4B, solid purple bar), while transcription in the upstream anti-sense direction (uaRNA) decreased by about 2.5 fold over the intron-less control (Figure 4C, diagonal purple pattern bar). The promoter-proximal intron had a similar effect on mRNA and uaRNA levels as expected (Figure 4C, solid red and diagonal red pattern bar). The middle intron, which did not enhance transcription of the gene (Figure 4C, solid green bar), also had no significant effect on uaRNA transcription over the intron-less control (Figure 4C, diagonal green pattern bar).
To better understand the role of the position of an intron on promoter directionality, we calculated the directionality index by dividing nascent mRNA levels with nascent uaRNA levels for the intron inserted at different positions of the IMD4 gene. The directionality index in the presence of a promoter-proximal intron increased by about 24 fold (Figure 4D, red bar), and in the presence of a terminator-proximal intron by about 10 fold (Figure 4D, purple bar) over the intron-less control. In contrast, the middle intron did not affect the directionality index over the intron-less version of the gene (Figure 4D, green bar). Thus, a terminator-proximal intron also enhances transcription directionality in a manner similar to that observed by the promoter-proximal intron.
Our published results demonstrated that the intron-dependent promoter directionality was through gene looping, which is the interaction of the promoter and terminator regions of a gene in a transcription-dependent manner (Agarwal and Ansari, 2016; Al Husini et al., 2020). Juxtaposition of the terminator and promoter regions places termination factors in the vicinity of the promoter region leading to termination of uaRNA transcription. We therefore examined if a terminator-proximal intron also confers looped gene architecture. Gene looping was detected by the “Chromosome Conformation Capture” (3C) approach. This procedure converts the promoter-terminator interaction into quantitatively measurable PCR products obtained using the primer pair flanking the promoter and terminator region, as schematically represented in Figure 2A. 3C analysis was performed with the intron inserted at different positions of the IMD4 gene following the protocol described in (El Kaderi et al., 2012). In all 3C experiments, complete digestion of chromatin between promoter and terminator, crosslinking-dependence and ligation-dependence of the results was routinely monitored. All 3C PCR products were verified by sequencing. Using our modified high resolution 3C approach, we have previously demonstrated a specific interaction of promoter and terminator regions of INO1 in a transcription-dependent manner (El Kaderi et al., 2009; Moabbi et al., 2012). As expected, the 3C assay detected a compact gene architecture with the promoter and terminator in close proximity for the construct with a promoter-proximal intron (Figure 2B, red bar). The promoter-terminator proximity measured in terms of P1T1 PCR signal was five fold more in the construct with the promoter-proximal intron compared to that in the absence of an intron. In the construct with the intron in the middle of the gene, gene architecture was comparable to that in the absence of the intron (Figure 2B, green bar). In the presence of the terminator-proximal intron, however, the promoter-terminator proximity was partially restored as P1T1 PCR signal registered a 3.6 fold increase over the intron-less strain (Figure 2B, purple bar). YPL109C gene, which has a terminator-proximal intron, similarly exhibited an intron-dependent change in gene architecture resulting in the promoter coming in proximity of the terminator of the gene (Figure 1D).
Terminator-Proximal Intron Also Affects PIC
In the mammalian system, a promoter proximal intron has been shown to enhance transcription by facilitating the recruitment or stabilization of the recruited preinitiation complex (PIC) on the promoter (Kwek et al., 2002; Damgaard et al., 2008; Bieberstein et al., 2012). Whether the intron plays a similar role in the recruitment/stabilization of PIC in yeast, however, has never been investigated. Therefore, to determine if the intron facilitates assembly/stabilization of PIC in yeast, we monitored the occupancy of two GTFs, TFIIB and TFIIH, on the promoter of IMD4 in the presence and absence of an intron. We monitored the occupancy of GTFs in a strain that has HA-tagged TFIIB and TAP-tagged cyclin subunit Ccl1 of TFIIH. ChIP was performed to look directly at the occupancy of two GTFs at the promoter and within the 300 bp promoter downstream coding region. Our results show that the promoter occupancy of TFIIB was enhanced by about five-fold in the presence of a promoter-proximal intron over the intron-less control (p-value of 0.01) (Figure 5B, upper panel, red bar). Some signal for TFIIB is detected in the 300 bp promoter downstream region, but the signal is 70–80% less compared to that on the promoter. A comparison of p-values shows that the decrease in TFIIB signal in the promoter-proximal coding region relative to the promoter is statistically significant (Figure 5B, upper panel red bars). Similar results were obtained for Ccl1. The Ccl1 ChIP signal registered a ten-fold increase in the presence of the promoter-proximal intron over no-intron control (p-value of 0.01) (Figure 5B, lower panel, red bar). Like TFIIB, Ccl1 signal also registered a 70–80% decline in the 300 bp promoter-downstream coding region (Figure 5B, lower panel, red bars). These results clearly show that, like in mammalian cells, an intron facilitates the assembly or stabilization of PIC in yeast as well.
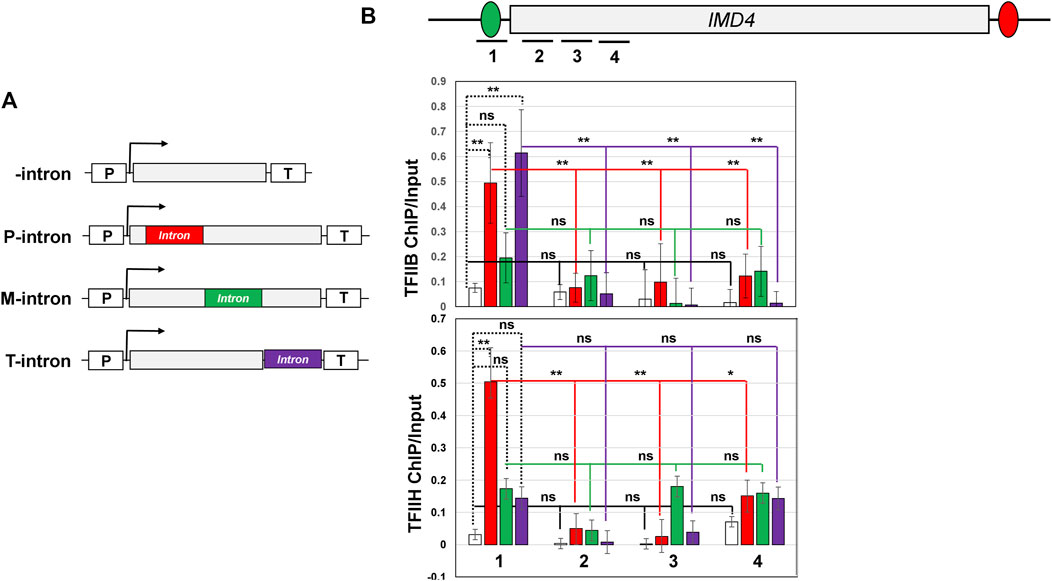
FIGURE 5. Position of intron affects TFIIB and TFIIH promoter occupancy for IMD4 gene. (A) Schematic depiction of IMD4 without intron and with the ACT1 intron inserted near the promoter (P-intron), in the middle (M-intron) and near the terminator region (T-intron) of the gene. “p” and “T” stand for the promoter and terminator respectively. (B) Schematic depiction of IMD4 gene with intron indicating the position of primer pairs used in ChIP analysis. (C) ChIP analysis showing crosslinking of HA-tagged TFIIB to the promoter and downstream regions of IMD4 with promoter-proximal intron (red bar), middle intron (green bar), terminator-proximal intron (purple bar) and without intron (white bar). (D) ChIP analysis showing crosslinking of TAP-tagged Ccl1 subunit of TFIIH to the promoter and downstream coding regions of IMD4 with promoter-proximal intron (red bar), middle intron (green bar), terminator-proximal intron (purple bar) and without intron (white bar). The Input signal, representing DNA prior to immunoprecipitation, was used as normalization control. ChIP results presented here represent three biological replicates and six technical replicates. The values and error bars for each condition indicate the mean ± standard deviation. Statistical significance (p-values) was determined using the two-tailed paired Student’s t-test. One asterisk (*) signifies a p value equal to or smaller than 0.05 (p ≤ 0.05). Two asterisks (**) signify a p value equal to or smaller than 0.01 (p ≤ 0.01). Any non-significant difference is represented by ns (p > 0.05).
Since we have shown that the position of an intron in a gene affects promoter directionality, we next examined if the position of the intron also affects its ability to recruit or stabilize binding of PIC components on the promoter region. We therefore checked occupancy of TFIIB and TFIIH subunit Ccl1 in the presence of the intron in the middle region and the terminator-proximal region of IMD4 as shown in Figure 5A. In the presence of a middle intron, TFIIB ChIP-signal in the promoter region increased by about two fold (Figure 5B, upper panel, green bar), while Ccl1 signal registered a nearly three fold increase (Figure 5B, lower panel, green bar) over the intron-less control. Although there was a small increase in the transcription factors occupancy, it was statistically not significant as indicated by the p-values (p-values 0.67 for TFIIB and 0.08 for Ccl1). Thus, unlike the promoter-proximal intron, the middle intron does not have a significant effect on the promoter occupancy of the general transcription factors. These results are in accordance with the effect of middle intron on transcription and directionality.
The terminator-proximal intron, like the promoter-proximal intron, though has a positive impact on PIC occupancy. The TFIIB promoter ChIP signal increased by about six fold in the presence of the terminator-proximal intron over the intron-less version (p-value 0.005) (Figure 5B, upper panel, purple bar). TFIIH promoter ChIP signal also increased by around three fold in the presence of the terminator-proximal intron, but the increase in signal is statistically not significant (p-value 0.22) (Figure 5B, lower panel, purple bar). These results are corroborated by the TFIIB and TFIIH ChIP-signals in the promoter-proximal coding region. TFIIB ChIP signal exhibits a statistically significant decline in the coding region compared to the promoter (Figure 5B, upper panel), while TFIIH signal does not exhibit a statistically significant change in the coding region relative to the promoter (Figure 5B, lower panel).
These results suggest that a promoter-proximal intron does indeed enhance occupancy of transcription factors at the promoter, while a terminator-proximal intron may only help with occupancy of certain factors of the PIC such as TFIIB. This may also explain why the terminator-proximal intron is not as efficient as the promoter-proximal intron in enhancing transcription.
Effect of Position of Intron on Termination of Transcription
Since a terminator proximal intron has been shown to affect termination in metazoans, and intron-dependent gene looping involves a terminator-intron interaction as well, we next examined if the presence of an intron in general, and that of the terminator-proximal intron in particular affect termination of transcription. We monitored termination by transcription run-on (TRO) assay as described in Dhoondia et al. (2017) for IMD4 gene with the intron inserted at three different positions as shown in Figure 6A. TRO assay detects the presence of transcriptionally active polymerases on a gene as shown in Figure 6B. To determine the role of the position of an intron on termination of transcription, we calculated the readthrough index (RTI) in constructs with the intron at different positions in the gene. RTI was calculated by dividing the TRO signal intensity beyond the 3′ end of the gene with the signal value before the 3′ end within the coding region. A termination defect results in a higher RTI value, while efficient termination yields a low RTI value. In the presence of an intron in the promoter-proximal position and terminator-proximal position, the RTI value registered a 20-fold and 10-fold decrease respectively over the intronless control (Figure 6C, red bar and purple bar). In the presence of an intron in the middle position, however, the RTI value decreased by a mere 1.2 fold over that in the absence of an intron (Figure 6C, green bar). These results suggest that the position of an intron affects termination of transcription. Both the promoter-proximal and terminator-proximal introns result in efficient termination, while intronless and middle intron versions exhibited inefficient termination. In general, TRO readthrough signal beyond the 3′ end of gene is expected to be higher in the termination defective mutants than in wild type cells (Birse et al., 1998). This was, however, not the case in constructs without an intron and with an intron in the middle position. The TRO readthrough signals in the presence of an intron at all three positions as well as in the absence of intron were very similar (Figure 6B). Whether the position of an intron in yeast affects termination, therefore, needs more scrutiny.
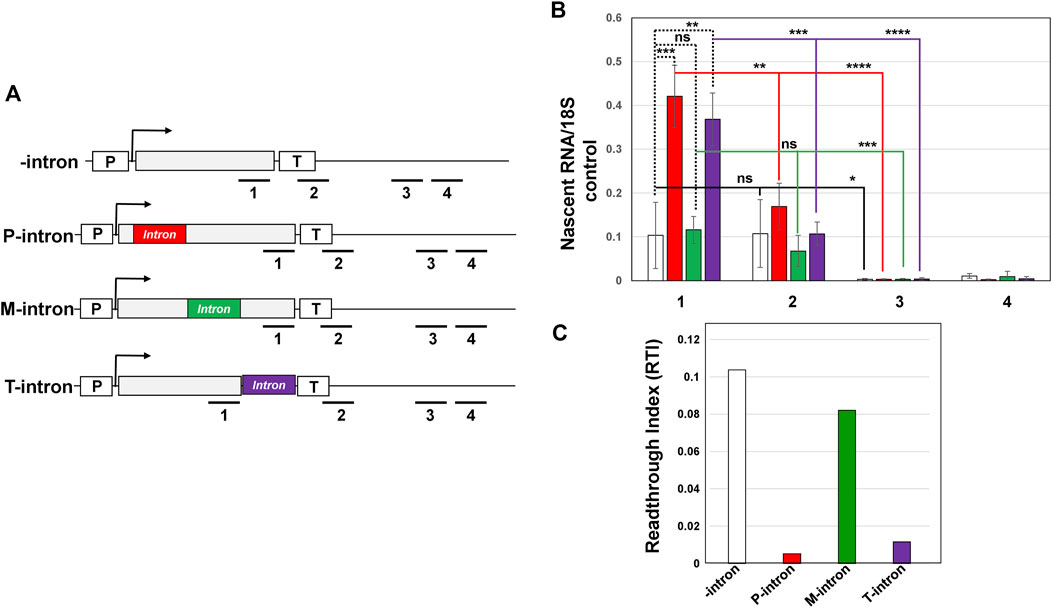
FIGURE 6. Presence of an intron does not affect termination of transcription of IMD4. (A) Schematic depiction of IMD4 with intron inserted near the promoter (P-intron), middle of the gene (M-intron) and near the terminator region (T-intron), showing the position of primers 1, 2, 3, and four used in strand-specific TRO analysis. “p” and “T” stand for the promoter and terminator respectively. (B) Strand-specific TRO analysis showing that the polymerase did not readthrough in the regions 2, 3, and four downstream of the terminator signal of IMD4 in the construct P-intron (red bar), M-intron (green bar) and T-intron (purple bar). The 18S signal was used as a normalization control in these experiments. Results presented here represent three biological replicates and six technical replicates. The values and error bars for each condition indicate the mean ± standard deviation. Statistical significance (p-values) was determined using the two-tailed paired Student’s t-test. One asterisk (*) signifies a p value equal to or smaller than 0.05 (p ≤ 0.05). Two asterisks (**) signify a p value equal to or smaller than 0.01 (p ≤ 0.01). Three asterisks (***) indicate p value equal/smaller than 0.001 (p ≤ 0.001). Four asterisks (****) indicate p value equal/smaller than 0.0001 (p ≤ 0.0001). Any non-significant difference is represented by ns (p > 0.05). (C) Readthrough Index (RTI) was calculated by dividing the TRO signal intensity beyond the 3′ end of the gene (region 4) with the signal value before the 3′ end within the coding region (region 1).
Discussion
A comparison of the active RNAPII molecules present on naturally intron-less and intron-containing genes was done by nuclear run on, enabling us to gauge the direct effect of introns on transcription. RNAPII density was almost doubled on intron containing genes and the transcription rate of intron-containing genes was 2.5 times more than that of intron-less genes (Pelechano et al., 2010). This indicates that intron containing genes of yeast are highly transcribed, in accord with their outsized contribution to cellular mRNA. Strand-specific TRO analysis, which directly measures transcription, also revealed that a number of yeast genes exhibit higher transcription with their natural intron (Furger et al., 2002; Moabbi et al., 2012; Agarwal and Ansari, 2016). Most of these genes contained a promoter-proximal intron, known to facilitate the PIC assembly on the promoter. The effect of a terminator-proximal intron on gene transcription has not been thoroughly investigated in yeast or higher eukaryotes. Whether a terminator-proximal intron affects PIC assembly is also not known. There are, however, a few reports that suggest a direct role of a terminator-proximal intron in 3′ end processing of mRNA (Dye and Proudfoot, 1999; Vagner et al., 2000; McCracken et al., 2002). Whether this has any impact on overall transcription was not clear from these studies.
Studies with cultured mammalian cells have found that the 3′ splice-site of the terminator-proximal intron had an adverse effect on trimethylation of H3K36, a chromatin mark normally associated with transcription elongation that must be removed to facilitate termination (Kim et al., 2011). A mutation in the 3′ splice-site of a terminator-proximal intron in the beta-globin gene caused enrichment of this mark in the 3′ region of the gene, which is expected to inhibit termination leading to an overall lowering of transcription. Multiple studies have pointed out the positive influence of terminator-proximal introns on mRNA 3′ end processing (Niwa et al., 1990; Nesic et al., 1993; Dye and Proudfoot, 1999; Vagner et al., 2000; McCracken et al., 2002). In line with our published work wherein we demonstrated that the promoter-proximal intron enhances transcription of both INO1 and IMD4 in a splicing-dependent manner (Moabbi et al., 2012; Agarwal and Ansari, 2016), we thought it logical to show that the terminator-proximal intron also enhances transcription in a splicing-dependent fashion. We were, however, unable to insert either a 5′ splice site or 3′ splice site mutated intron near the terminator region of either INO1 or IMD4. The strains with the mutated intron near the terminator region were not viable. Instead we show here that the terminator-proximal intron of both INO1 or IMD4 is spliced efficiently (Supplementary Figures S1, S2).
In budding yeast, most of the intronic genes contain a single intron located in the vicinity of the promoter. The contribution of the position of an intron, especially the terminator-linked intron, on gene expression therefore remained largely unexplored. This is the first systematic study analyzing the role of the position of an intron within a gene on its transcription in budding yeast. Our results reaffirm the view emerging from studies with higher eukaryotes that the position of an intron within a gene has a strong bearing on its transcription. Unlike in higher eukaryotes, however, we found that the terminator-linked intron in yeast affects promoter transcription in a manner similar to the promoter-proximal intron. The terminator intron inhibits uaRNA transcription in the vicinity of the promoter region, thereby enhancing promoter directionality. The terminator intron also affects the occupancy of TFIIB on the promoter in a manner similar to a promoter-proximal intron. The occupancy of TFIIH, a general transcription factor that is recruited last during assembly of PIC, however, is not significantly enhanced by the terminal intron. This may explain why a terminator-linked intron is not as efficient in enhancing transcription as the promoter-proximal intron. Unlike in budding yeast, higher eukaryotes contain multiple introns distributed all over the genes. This study will serve as a paradigm for investigating the role of terminator-proximal introns on gene expression in higher eukaryotes.
Data Availability Statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Author Contributions
AA conceptualized; KD and NA performed experiments; AG helped in strain construction and in performing experiments; NA, KD, and AA performed data analysis; AA wrote the manuscript. All authors contributed to the article and approve the submitted version.
Funding
This work was supported by grant from National Science Foundation (MCB-1936030) to AA. The fund for open access fee was received from this grant. The content is solely the responsibility of authors and does not necessarily represent the official views of the National Science Foundation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Wayne State University for award of “Grant Boost” funding to AA. We are grateful to Victoria Meller (Wayne State University), Zuzer Dhoondia (Harvard Medical School), Michael O’Brien and Zulifa Khan for critical reading of the manuscript. We thank lab members for helpful suggestions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.712639/full#supplementary-material
References
Agarwal, N., and Ansari, A. (2016). Enhancement of Transcription by a Splicing-Competent Intron Is Dependent on Promoter Directionality. PLoS Genet. 12 (5), e1006047. doi:10.1371/journal.pgen.1006047
Al-Husini, N., Medler, S., and Ansari, A. (2020). Crosstalk of Promoter and Terminator during RNA Polymerase II Transcription Cycle. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 1863, 194657. doi:10.1016/j.bbagrm.2020.194657
Antoniou, M., Geraghty, F., Hurst, J., and Grosveld, F. (1998). Efficient 3'-end Formation of Human Beta-Globin mRNA In Vivo Requires Sequences within the Last Intron but Occurs Independently of the Splicing Reaction. Nucleic Acids Res. 26 (3), 721–729. doi:10.1093/nar/26.3.721
Ares, M., Grate, L., and Pauling, M. H. (1999). A Handful of Intron-Containing Genes Produces the Lion's Share of Yeast mRNA. Rna 5 (9), 1138–1139. doi:10.1017/s1355838299991379
Awasthi, S., and Alwine, J. C. (2003). Association of Polyadenylation Cleavage Factor I with U1 snRNP. Rna 9 (11), 1400–1409. doi:10.1261/rna.5104603
Baier, T., Jacobebbinghaus, N., Einhaus, A., Lauersen, K. J., and Kruse, O. (2020). Introns Mediate post-transcriptional Enhancement of Nuclear Gene Expression in the green Microalga Chlamydomonas Reinhardtii. PLoS Genet. 16 (7), e1008944. doi:10.1371/journal.pgen.1008944
Barrett, L. W., Fletcher, S., and Wilton, S. D. (2012). Regulation of Eukaryotic Gene Expression by the Untranslated Gene Regions and Other Non-coding Elements. Cell. Mol. Life Sci. 69 (21), 3613–3634. doi:10.1007/s00018-012-0990-9
Bartlett, J. G., Snape, J. W., and Harwood, W. A. (2009). Intron-mediated Enhancement as a Method for Increasing Transgene Expression Levels in Barley. Plant Biotechnol. J. 7 (9), 856–866. doi:10.1111/j.1467-7652.2009.00448.x
Bieberstein, N. I., Carrillo Oesterreich, F., Straube, K., and Neugebauer, K. M. (2012). First Exon Length Controls Active Chromatin Signatures and Transcription. Cel Rep. 2 (1), 62–68. doi:10.1016/j.celrep.2012.05.019
Birse, C. E., Minvielle-Sebastia, L., Lee, B. A., Keller, W., and Proudfoot, N. J. (1998). Coupling Termination of Transcription to Messenger RNA Maturation in Yeast. Science 280, 298–301. doi:10.1126/science.280.5361.298
Bourdon, V., Harvey, A., and Lonsdale, D. M. (2001). Introns and Their Positions Affect the Translational Activity of mRNA in Plant Cells. EMBO Rep. 2 (5), 394–398. doi:10.1093/embo-reports/kve090
Brinster, R. L., Allen, J. M., Behringer, R. R., Gelinas, R. E., and Palmiter, R. D. (1988). Introns Increase Transcriptional Efficiency in Transgenic Mice. Proc. Natl. Acad. Sci. 85 (3), 836–840. doi:10.1073/pnas.85.3.836
Callis, J., Fromm, M., and Walbot, V. (1987). Introns Increase Gene Expression in Cultured maize Cells. Genes Dev. 1 (10), 1183–1200. doi:10.1101/gad.1.10.1183
Charron, M., Chern, J.-Y., and Wright, W. W. (2007). The Cathepsin L First Intron Stimulates Gene Expression in Rat Sertoli Cells1. Biol. Reprod. 76 (5), 813–824. doi:10.1095/biolreprod.106.057851
Chorev, M., and Carmel, L. (2012). The Function of Introns. Front. Genet. 3, 55. doi:10.3389/fgene.2012.00055
Comeron, J. M. (2004). Selective and Mutational Patterns Associated with Gene Expression in Humans. Genetics 167 (3), 1293–1304. doi:10.1534/genetics.104.026351
Damgaard, C. K., Kahns, S., Lykke-Andersen, S., Nielsen, A. L., Jensen, T. H., and Kjems, J. (2008). A 5′ Splice Site Enhances the Recruitment of Basal Transcription Initiation Factors In Vivo. Mol. Cel. 29 (2), 271–278. doi:10.1016/j.molcel.2007.11.035
Das, R., Yu, J., Zhang, Z., Gygi, M. P., Krainer, A. R., Gygi, S. P., et al. (2007). SR Proteins Function in Coupling RNAP II Transcription to Pre-mRNA Splicing. Mol. Cel. 26 (6), 867–881. doi:10.1016/j.molcel.2007.05.036
Dhoondia, Z., Tarockoff, R., Alhusini, N., Medler, S., Agarwal, N., and Ansari, A. (2017). Analysis of Termination of Transcription Using BrUTP-strand-specific Transcription Run-On (TRO) Approach. JoVE (Journal of Visualized Experiments) 49(121), e55446. doi:10.3791/55446
Duncker, B. P., Davies, P. L., and Walker, V. K. (1997). Introns Boost Transgene Expression in Drosophila melanogaster. Mol. Gen. Genet. 254 (3), 291–296. doi:10.1007/s004380050418
Dwyer, K., Agarwal, N., Pile, L. A., and Ansari, A. (2021). Gene Architecture Facilitates Intron-Mediated Enhancement of Transcription. Front. Mol. Biosciences 8, 276. doi:10.3389/fmolb.2021.669004
Dye, M. J., and Proudfoot, N. J. (1999). Terminal Exon Definition Occurs Cotranscriptionally and Promotes Termination of RNA Polymerase II. Mol. Cel. 3 (3), 371–378. doi:10.1016/s1097-2765(00)80464-5
El Kaderi, B., Medler, S., and Ansari, A. (2012). Analysis of Interactions between Genomic Loci through Chromosome Conformation Capture (3C). Curr. Protoc. Cel. Biol. 56 (1), 22–15. doi:10.1002/0471143030.cb2215s56
El Kaderi, B., Medler, S., Raghunayakula, S., and Ansari, A. (2009). Gene Looping Is Conferred by Activator-dependent Interaction of Transcription Initiation and Termination Machineries. J. Biol. Chem. 284 (37), 25015–25025. doi:10.1074/jbc.m109.007948
Furger, A., O‘Sullivan, J. M., Binnie, A., Lee, B. A., and Proudfoot, N. J. (2002). Promoter Proximal Splice Sites Enhance Transcription. Genes Dev. 16 (21), 2792–2799. doi:10.1101/gad.983602
Gallegos, J. E., and Rose, A. B. (2015). The Enduring Mystery of Intron-Mediated Enhancement. Plant Sci. 237, 8–15. doi:10.1016/j.plantsci.2015.04.017
Goebels, C., Thonn, A., Gonzalez-Hilarion, S., Rolland, O., Moyrand, F., Beilharz, T. H., et al. (2013). Introns Regulate Gene Expression in Cryptococcus Neoformans in a Pab2p Dependent Pathway. PLoS Genet. 9 (8), e1003686. doi:10.1371/journal.pgen.1003686
Gunderson, S. I., Vagner, S., Polycarpou-Schwarz, M., and Mattaj, I. W. (1997). Involvement of the Carboxyl Terminus of Vertebrate Poly(A) Polymerase in U1A Autoregulation and in the Coupling of Splicing and Polyadenylation. Genes Dev. 11 (6), 761–773. doi:10.1101/gad.11.6.761
Haas, B. J., Wortman, J. R., Ronning, C. M., Hannick, L. I., Smith, R. K., Maiti, R., et al. (2005). Complete Reannotation of the Arabidopsis Genome: Methods, Tools, Protocols and the Final Release. BMC Biol. 3 (1), 7. doi:10.1186/1741-7007-3-7
Huang, M. T., and Gorman, C. M. (1990). The Simian Virus 40 Small-T Intron, Present in many Common Expression Vectors, Leads to Aberrant Splicing. Mol. Cel. Biol. 10 (4), 1805–1810. doi:10.1128/mcb.10.4.1805
International Human Genome Sequencing Consortium (2004). Finishing the Euchromatic Sequence of the Human Genome. Nature 431, 931–945. doi:10.1038/nature03001
Jeon, J.-S., Lee, S., Jung, K.-H., Jun, S.-H., Kim, C., and An, G. (2000). Tissue-Preferential Expression of a Rice α-Tubulin Gene, OsTubA1, Mediated by the First Intron. Plant Physiol. 123 (3), 1005–1014. doi:10.1104/pp.123.3.1005
Jeong, Y.-M., Mun, J.-H., Lee, I., Woo, J. C., Hong, C. B., and Kim, S.-G. (2006). Distinct Roles of the First Introns on the Expression of Arabidopsis Profilin Gene Family Members. Plant Physiol. 140 (1), 196–209. doi:10.1104/pp.105.071316
Juneau, K., Miranda, M., Hillenmeyer, M. E., Nislow, C., and Davis, R. W. (2006). Introns Regulate RNA and Protein Abundance in Yeast. Genetics 174 (1), 511–518. doi:10.1534/genetics.106.058560
Kertész, S., Kerényi, Z., Mérai, Z., Bartos, I., Pálfy, T., Barta, E., et al. (2006). Both introns and Long 3′-UTRs Operate as Cis-Acting Elements to Trigger Nonsense-Mediated Decay in Plants. Nucleic Acids Res. 34 (21), 6147–6157. doi:10.1093/nar/gkl737
Kim, W., Matsui, T., Yamao, M., Ishibashi, M., Tamada, K., Takumi, T., et al. (2011). The Period of the Somite Segmentation Clock Is Sensitive to Notch Activity. MBoC 22 (18), 3541–3549. doi:10.1091/mbc.e11-02-0139
Kwek, K. Y., Murphy, S., Furger, A., Thomas, B., O'Gorman, W., Kimura, H., et al. (2002). U1 snRNA Associates with TFIIH and Regulates Transcriptional Initiation. Nat. Struct. Mol. Biol. 9 (11), 800. doi:10.1038/nsb862
Kyburz, A., Friedlein, A., Langen, H., and Keller, W. (2006). Direct Interactions between Subunits of CPSF and the U2 snRNP Contribute to the Coupling of Pre-mRNA 3′ End Processing and Splicing. Mol. Cel. 23 (2), 195–205. doi:10.1016/j.molcel.2006.05.037
Lacy-Hulbert, A., Thomas, R., Li, X.-P., Lilley, C. E., Coffin, R. S., and Roes, J. (2001). Interruption of Coding Sequences by Heterologous Introns Can Enhance the Functional Expression of Recombinant Genes. Gene Ther. 8 (8), 649–653. doi:10.1038/sj.gt.3301440
Laxa, M. (2017). Intron-mediated Enhancement: a Tool for Heterologous Gene Expression in Plants? Front. Plant Sci. 7, 1977. doi:10.3389/fpls.2016.01977
Le Hir, H., Nott, A., and Moore, M. J. (2003). How Introns Influence and Enhance Eukaryotic Gene Expression. Trends Biochemical Sciences 28 (4), 215–220. doi:10.1016/s0968-0004(03)00052-5
Lu, S., and Cullen, B. R. (2003). Analysis of the Stimulatory Effect of Splicing on mRNA Production and Utilization in Mammalian Cells. Rna 9 (5), 618–630. doi:10.1261/rna.5260303
Lugones, L. G., Scholtmeijer, K., Klootwijk, R., and Wessels, J. G. H. (1999). Introns Are Necessary for mRNA Accumulation in Schizophyllum Commune. Mol. Microbiol. 32 (4), 681–689. doi:10.1046/j.1365-2958.1999.01373.x
Martinson, H. G. (2011). An Active Role for Splicing in 3′-end Formation. WIREs RNA 2 (4), 459–470. doi:10.1002/wrna.68
McCracken, S., Lambermon, M., and Blencowe, B. J. (2002). SRm160 Splicing Coactivator Promotes Transcript 3′-End Cleavage. Mol. Cel Biol 22 (1), 148–160. doi:10.1128/mcb.22.1.148-160.2002
McKenzie, R., and Brennan, M. D. (1996). The Two Small Introns of the Drosophila Affinidisjuncta Adh Gene Are Required for normal Transcription. Nucleic Acids Res. 24 (18), 3635–3642. doi:10.1093/nar/24.18.3635
Medler, S., and Ansari, A. (2015). Gene Looping Facilitates TFIIH Kinase-Mediated Termination of Transcription. Scientific Rep. 5, 12586. doi:10.1038/srep12586
Millevoi, S., Loulergue, C., Dettwiler, S., Karaa, S. Z., Keller, W., Antoniou, M., et al. (2006). An Interaction between U2AF 65 and CF Im Links the Splicing and 3′ End Processing Machineries. Embo J. 25 (20), 4854–4864. doi:10.1038/sj.emboj.7601331
Moabbi, A. M., Agarwal, N., El Kaderi, B., and Ansari, A. (2012). Role for Gene Looping in Intron-Mediated Enhancement of Transcription. Proc. Natl. Acad. Sci. 109 (22), 8505–8510. doi:10.1073/pnas.1112400109
Morello, L., Gianì, S., Troina, F., and Breviario, D. (2010). Testing the IMEter on rice Introns and Other Aspects of Intron-Mediated Enhancement of Gene Expression. J. Exp. Bot. 62 (2), 533–544. doi:10.1093/jxb/erq273
Nesic, D., Cheng, J., and Maquat, L. E. (1993). Sequences within the Last Intron Function in RNA 3'-end Formation in Cultured Cells. Mol. Cel. Biol. 13 (6), 3359–3369. doi:10.1128/mcb.13.6.3359
Neuvéglise, C., Marck, C., and Gaillardin, C. (2011). The Intronome of Budding Yeasts. Comptes rendus biologies 334 (8-9), 662–670. doi:10.1016/j.crvi.2011.05.015
Niwa, M., Rose, S. D., and Berget, S. M. (1990). In Vitro polyadenylation Is Stimulated by the Presence of an Upstream Intron. Genes Dev. 4 (9), 1552–1559. doi:10.1101/gad.4.9.1552
Okkema, P. G., Harrison, S. W., Plunger, V., Aryana, A., and Fire, A. (1993). Sequence Requirements for Myosin Gene Expression and Regulation in Caenorhabditis elegans. Genetics 135 (2), 385–404. doi:10.1093/genetics/135.2.385
Palmiter, R. D., Sandgren, E. P., Avarbock, M. R., Allen, D. D., and Brinster, R. L. (1991). Heterologous Introns Can Enhance Expression of Transgenes in Mice. Proc. Natl. Acad. Sci. 88 (2), 478–482. doi:10.1073/pnas.88.2.478
Parenteau, J., Durand, M., Morin, G., Gagnon, J., Lucier, J.-F., Wellinger, R. J., et al. (2011). Introns within Ribosomal Protein Genes Regulate the Production and Function of Yeast Ribosomes. Cell 147 (2), 320–331. doi:10.1016/j.cell.2011.08.044
Parenteau, J., Durand, M., Véronneau, S., Lacombe, A.-A., Morin, G., Guérin, V., et al. (2008). Deletion of many Yeast Introns Reveals a Minority of Genes that Require Splicing for Function. MBoC 19 (5), 1932–1941. doi:10.1091/mbc.e07-12-1254
Parra, G., Bradnam, K., Rose, A. B., and Korf, I. (2011). Comparative and Functional Analysis of Intron-Mediated Enhancement Signals Reveals Conserved Features Among Plants. Nucleic Acids Res. 39 (13), 5328–5337. doi:10.1093/nar/gkr043
Pelechano, V., Chávez, S., and Pérez-Ortín, J. E. (2010). A Complete Set of Nascent Transcription Rates for Yeast Genes. PloS one 5 (11), e15442. doi:10.1371/journal.pone.0015442
Petibon, C., Parenteau, J., Catala, M., and Elela, S. A. (2016). Introns Regulate the Production of Ribosomal Proteins by Modulating Splicing of Duplicated Ribosomal Protein Genes. Nucleic Acids Res. 44 (8), 3878–3891. doi:10.1093/nar/gkw140
Rethmeier, N., Seurinck, J., Van Montagu, M., and Cornelissen, M. (1997). Intron-mediated Enhancement of Transgene Expression in maize Is a Nuclear, Gene-dependent Process. Plant J. 12 (4), 895–899. doi:10.1046/j.1365-313x.1997.12040895.x
Rose, A. B. (2008). “Intron-mediated Regulation of Gene Expression,” in Nuclear Pre-mRNA Processing in Plants. Berlin, Heidelberg: Springer, 277–290. doi:10.1007/978-3-540-76776-3_15
Rose, A. B. (2019). Introns as Gene Regulators: a brick on the Accelerator. Front. Genet. 9, 672. doi:10.3389/fgene.2018.00672
Rose, A. B. (2004). The Effect of Intron Location on Intron-Mediated Enhancement of Gene Expression in Arabidopsis. Plant J. 40 (5), 744–751. doi:10.1111/j.1365-313x.2004.02247.x
Sakharkar, M. K., Chow, V. T., and Kangueane, P. (2004). Distributions of Exons and Introns in the Human Genome. Silico Biol. 4 (4), 387–393.
Sam, M. R., Zomorodipour, A., Shokrgozar, M. A., Ataei, F., Haddad-Mashadrizeh, A., and Amanzadeh, A. (2010). Enhancement of the Human Factor IX Expression, Mediated by an Intron Derived Fragment from the Rat Aldolase B Gene in Cultured Hepatoma Cells. Biotechnol. Lett. 32 (10), 1385–1392. doi:10.1007/s10529-010-0321-x
Shabalina, S. A., Ogurtsov, A. Y., Spiridonov, A. N., Novichkov, P. S., Spiridonov, N. A., and Koonin, E. V. (2010). Distinct Patterns of Expression and Evolution of Intronless and Intron-Containing Mammalian Genes. Mol. Biol. Evol. 27 (8), 1745–1749. doi:10.1093/molbev/msq086
Shaul, O. (2017). How Introns Enhance Gene Expression. Int. J. Biochem. Cel Biol. 91, 145–155. doi:10.1016/j.biocel.2017.06.016
Snowden, K. C., Buchhholz, W. G., and Hall, T. C. (1996). Intron Position Affects Expression from Thetpi Promoter in rice. Plant Mol. Biol. 31 (3), 689–692. doi:10.1007/bf00042241
Spingola, M., Grate, L., Haussler, D., and Ares, M. (1999). Genome-wide Bioinformatic and Molecular Analysis of Introns in Saccharomyces cerevisiae. Rna 5 (2), 221–234. doi:10.1017/s1355838299981682
Keywords: transcription, promoter directionality, splicing, intron, gene architecture, gene regulation, gene looping, yeast
Citation: Dwyer K, Agarwal N, Gega A and Ansari A (2021) Proximity to the Promoter and Terminator Regions Regulates the Transcription Enhancement Potential of an Intron. Front. Mol. Biosci. 8:712639. doi: 10.3389/fmolb.2021.712639
Received: 20 May 2021; Accepted: 25 June 2021;
Published: 05 July 2021.
Edited by:
Claudio Scazzocchio, Imperial College London, United KingdomReviewed by:
Guilhem Janbon, Institut Pasteur, FranceCyril F. Bourgeois, INSERM U1210 Laboratoire de Biologie et Modelisation de la Cellule (LBMC), France
Copyright © 2021 Dwyer, Agarwal, Gega and Ansari. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Athar Ansari, YmIyNzQ5QHdheW5lLmVkdQ==
†These authors have contributed equally to this work and share first authorship
 Katherine Dwyer†
Katherine Dwyer† Athar Ansari
Athar Ansari