
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci. , 22 December 2021
Sec. Molecular Diagnostics and Therapeutics
Volume 8 - 2021 | https://doi.org/10.3389/fmolb.2021.706754
 Lianglu Zhang1,2,3,4
Lianglu Zhang1,2,3,4 Lanlan Dong2
Lanlan Dong2 Changming Lu2
Changming Lu2 Wenxian Huang5
Wenxian Huang5 Cuiping Yang6
Cuiping Yang6 Qian Wang7
Qian Wang7 Qian Wang3
Qian Wang3 Ruixue Lei8
Ruixue Lei8 Rui Sun9
Rui Sun9 Kangkang Wan2
Kangkang Wan2 Tingting Li2
Tingting Li2 Fan Sun2
Fan Sun2 Tian Gan2
Tian Gan2 Jun Lin3*
Jun Lin3* Lei Yin1*
Lei Yin1*Background: SDC2 methylation is a feasible biomarker for colorectal cancer detection. Its specificity for colorectal cancer is higher than 90%, but the sensitivity is normally lower than 90%. This study aims to improve the sensitivity of SDC2 detection through finding a high positive target from the false-negative samples of SDC2 detection based on analysis of the bowel subsite difference in methylation.
Methods: Hypermethylated TFPI2 was identified in SDC2 hypomethylated colorectal cancer samples retrieved from TCGA database with the methylation level lower than 0.2. The methylation-specific PCR assay was developed and then evaluated using tissue samples (184 cancer and 54 healthy control samples) and stool samples (289 cancer, 190 adenoma, and 217 healthy control samples).
Results: TFPI2 was hypermethylated in most SDC2 hypomethylated colorectal cancer samples. When the SDC2/TFPI2-combined PCR assay was performed in stool specimens, the AUC value of cancer vs. control was 0.98, with the specificity of 96.40% and sensitivity of 96.60%, and the AUC value of adenoma vs. control was 0.87, with the specificity of 95.70% and the sensitivity of 80.00%. The improvement in sensitivity was the most momentous in the left colon. As the detection index, the Ct value was better in improving the sensitivity of detection than the methylation level based on the 2−ΔΔCt value.
Conclusion: TFPI2 can improve the sensitivity of SDC2 methylation–specific detection of colorectal tumorous lesions while maintaining high specificity, in particular reducing the missed detection of left colon cancer and adenoma.
Colorectal cancer (CRC) affects millions of people around the world. It is unique because of slow progress, making it preventable and often curable (Cao et al., 2021; Sung et al., 2021). The five-year survival rate can be as high as 90% or less than 10%, depending on the stage of diagnosis (American Cancer Society, 2019). Sporadic CRC mainly develops in a normal–adenoma–carcinoma sequence (Crockett and Nagtegaal, 2019), and early detection can significantly decrease mortality (Chung, 2018). Colonoscopy plus pathological examination is the gold standard for CRC diagnosis (Shaukat et al., 2021), but due to the invasive and complex intestinal preparation process, its compliance in the average risk population is low (Navarro et al., 2017). The fecal occult blood test (FOBT) and fecal immunochemical test (FIT) are non-invasive, but their sensitivity is insufficient, especially for stage I CRC and advanced adenomas (Werner et al., 2016). The occurrence of CRC is related to genomic and epigenetic changes, such as gene mutation, microsatellite instability, and CpG island aberrant methylation (Grady and Pritchard, 2014). Among them, CpG island methylation is the most common change (Yiu and Yiu, 2016).
Aberrant DNA methylation can occur at the very early stage (Novak et al., 2009); so far, several methylation biomarkers have been identified, including SDC2, NDRG4, BMP3, VIM, SFRP2, and SEPT9 (Müller et al., 2004; Loh et al., 2008; deVos et al., 2009; Melotte et al., 2009; Oh et al., 2013; Robertson and Imperiale, 2015), but the sensitivity of a single marker is usually lower than 90% (Shariatpanahi et al., 2018). The first stool-based CRC detection product “Cologuard,” targeting the hemoglobin, KRAS mutation, and two methylated genes (NDRG4 and BMP3), has a sensitivity of 92% and specificity of 87% for CRC (Imperiale et al., 2014); however, multiple target tests may be costly and difficult to implement.
SDC2 has been identified as a potential biomarker for CRC (Oh et al., 2013). Aberrant methylation in SDC2 CpG islands has been found in tissue, blood, and stool (Oh et al., 2013; Mitchell et al., 2014). A study based on Chinese stool samples showed that the sensitivity and specificity of SDC2 for CRC were 81.1 and 93.3%, respectively (Niu et al., 2017). Korean researchers adopted an LTE-q methylation-specific PCR (MSP) method to enrich SDC2, which required two rounds of PCR, i.e., unidirectional linear amplification of target DNA followed by MSP analysis of target region, giving the sensitivity of 90.0% for CRC and specificity of 90.9% (Oh et al., 2017; Han et al., 2019).
In this study, we chose an alternative approach to improve the accuracy for CRC detection. Through genome-wide screening, we found that TFPI2 was highly hypermethylated in SDC2 hypomethylated samples. The combined detection of TFPI2 and SDC2 showed both high specificity and sensitivity, especially for cancer and adenomas in the left colon for both tissue and stool specimens. Here, we present the results of TFPI2 identification and the evaluation of MSP systems in the tissue and stool of patients with colorectal lesions at different bowel sites.
This study was approved by the Medical Ethics Committee, Zhongnan Hospital of Wuhan University (ethical approval No. 2019099). Tissue specimens of 198 CRC patients and 54 healthy controls (Supplementary Table S1) were collected from Zhongnan Hospital of Wuhan University, and stool specimens of 289 CRC patients, 190 adenoma patients, and 217 healthy controls (Supplementary Table S2) were from Zhongnan Hospital and Renmin Hospital of Wuhan University, Ruijin Hospital of Shanghai Jiaotong University, Wuhan Eighth Hospital of Hubei University of Chinese Medicine, the Fourth Affiliated Hospital of Henan University of Science and Technology, and Wuhan Fourth Hospital of Huazhong University of Science and Technology. Written informed consent was obtained from all participants. Exclusion criteria of tissue specimens are CRC patients with a history of CRC surgery, chemotherapy, or other treatment and non-CRC patients who have received chemotherapy in the past 6 months. Finally, 184 CRC and 54 normal tissue specimens from different colorectal sites were included (Supplementary Table S1). The stool specimen exclusion criteria are patients with tumor other than CRC; patients who had a history of surgery or chemotherapy; patients with familial or hereditary colorectal adenomas or tumor; patients with non-primary tumors and other undiagnosed cases; and patients who had undergone colonic invasive surgery or bowel preparation less than 1 week before sample collection.
391 CRC specimens from The Cancer Genome Atlas (TCGA) were sorted according to the methylation level (mean β value of the probes within the CpG island) of SDC2 to identify hypomethylated specimens (methylation level≤0.2), and 12 selected probes (cg13096260, cg18719750, cg24732574, cg08979737, cg25070637, cg14538332, cg16935295, cg04261408, cg14625631, cg10292139, cg16673702, and cg07146119) were included. Hypermethylated genes (methylation level>0.2) were identified among hypomethylated specimens through whole-genome analysis. Receiver-operating characteristic (ROC) curve analysis was performed using the methylation level of each sample to evaluate the diagnostic complementarity of hypermethylation genes and SDC2 in different colorectal sites.
For the samples in TCGA, if the mean β value of a gene was higher than 0.2, then the gene was considered to be methylation positive in this sample; otherwise, it was considered methylation negative. When two genes were analyzed jointly, as long as any gene was positive or both genes were positive at the same time, then the sample was regarded as methylation positive.
Genomic DNA of cell lines was isolated using QIAamp DNA Mini Kit (Qiagen, Germany). All tissue specimens in this study were formalin-fixed paraffin-embedded specimens. Genomic DNA of tissue specimens was isolated using QIAamp DNA FFPE Tissue Kit (Qiagen, Germany) according to the manufacturer’s instruction.
Stool DNA was extracted using reagent developed by Wuhan Ammunition Life-tech Company. Briefly, stool specimens were collected about 8 g per person and kept in 45 ml tubes with 32 ml preservation buffer (200 mmol/L Tris·HCl, 300 mmol/L EDTA·2Na, 150 mmol/L NaCl, pH 8.0). For isolation of human genomic DNA, the biotin-labeled capture probes were designed for SDC2, TFPI2, and reference gene ACTB, and sequences are shown in Supplementary Table S3. After centrifugation, DNA in the supernatant was denatured under 90°C for 15 min. The single-strand DNA and the capture probes were then incubated with streptavidin magnetic beads at room temperature for 1 h. After washing twice, the target DNA was eluted with 50 uL TE buffer. All purified DNA was stored at −20°C until use.
DNA was converted using EZ DNA Methylation Kit (Zymo Research, LA, USA) according to the manufacturer’s instruction. For tissue samples, 1 ug DNA was converted; for stool samples, all the 50 ul purified DNA was converted. After bisulfite conversion, the 25 uL eluted DNA was either used immediately for PCR analysis or stored at −20°C for further use.
Hacat and HT-29 cell lines were used in this study; they were obtained from Tongji Medical College, Huazhong University of Science and Technology. The cell lines were cultured in DMEM (Thermo) supplemented with 10% fetal bovine serum.
The fully methylated and non-methylated amplicon regions of SDC2 and TFPI2 as well as the ACTB amplicon region after bisulfite conversion were artificially synthesized and cloned into the vector pUC57, respectively, in Wuhan GeneCreate Biological Engineering Company, and then the constructed plasmids were serially diluted to 106 copies/ul, 105 copies/ul, 104 copies/ul, 103 copies/ul, and 102 copies/ul.
106 copies/ul of non-methylated SDC2 plasmids, non-methylated TFPI2 plasmids, and ACTB plasmids were mixed in 1:1:1 ratio to serve as a negative control, and 104 copies/uL of methylated SDC2 plasmids, methylated TFPI2 plasmids, and ACTB plasmids were mixed in 1:1:1 ratio to serve as a positive control.
Before MSP analysis on tissue and stool samples, methylation-specific primers and probes were verified in two ways. On the one hand, 102 copies/ul to 106 copies/ul of methylated SDC2 plasmids, 102 copies/ul to 106 copies/ul of methylated TFPI2 plasmids, and 102 copies/uL to 106 copies/ul of ACTB plasmids were amplified to build standard curves, and amplification efficiency was calculated for each gene. On the other hand, the negative and positive controls, as well as methylated cell line (HT-29) and non-methylated cell line (Hacat), were MSP analyzed to confirm that the primers and probe could only amplify the methylated template. 500 ng genomic DNA of each cell line was added into the stool sample of healthy people confirmed by colonoscopy, and follow-up operations were the same as those of other stool samples.
Sequences of MSP primers and probes are shown in Supplementary Table S3. PCR solution was prepared in a volume of 25 ul with High-Affinity Hotstart Taq Polymerase. 5 ul template DNA was added, and non-template control and methylated and non-methylated controls were tested together in every plate. PCR was performed on an ABI 7500 instrument under the following cycling conditions: 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 60°C for 30 s.
The methylation status of SDC2 and TFPI2 in bisulfite-modified DNA was investigated in a blinded manner by MSP with primers specifically amplifying the methylated alleles. All MSP tests were done by investigators blinded to patients’ colonoscopy and pathology information, and MSP results were analyzed independently by other researchers.
The methylation level was calculated using the formula
in which
Ct values and ML values were ROC curve analyzed separately. The optimal cutoff was determined by maximizing Youden’s index. The area under the ROC curve (AUC) value and 95% CI, sensitivity, and specificity were estimated.
When calculating the detection accuracy of marker(s) of cancer and adenoma in different colorectal sites, the cutoff value was set at Ct = 38 for SDC2 or TFPI2 and Ct = 36 for ACTB. If the Ct value of ACTB > 36, the reaction was invalid. The specimen was methylation positive if the Ct value of SDC2 or/and TFPI2 ≤ 38. Sensitivity and specificity values were calculated as
All the bioinformatic and statistical analyses were performed with R version 3.6.1. To determine the statistical significance of the difference in methylation level between case and control groups, Wilcoxon’s signed-rank test was used for pairwise data and Mann–Whitney U test for groupwise data. Sensitivity between different colorectal locations was tested by the Fisher exact test.
Five groups of samples were utilized in this study, as described in Table 1. TCGA, GSE48684, and GSE79740 were used to identify and validate differential methylation regions and D184 and D289 to evaluate the performance of MSP assays in clinical samples. The TCGA dataset included 410 case and 45 control samples. We retained 391 CRC samples and 45 normal control samples for differential methylation region discovery study after removing the samples of incomplete clinical information and tumor metastasis (Supplementary Table S4). The datasets GSE48684 and GSE79740 were downloaded from the Gene Expression Omnibus (GEO) database (Luo et al., 2014; Alvi et al., 2017). GSE48684 contains 106 CRC, 42 adenoma, and 41 normal samples, and GSE79740 contains 44 CRC and 10 normal samples. They were both used for differential methylation region validation. D184 contains 184 CRC and 54 normal tissue samples, and D289 contains 289 CRC, 190 adenoma, and 217 normal stool samples. Their demographic features are shown in Supplementary Tables S1, S2.
391 CRC samples from TCGA were sorted according to the methylation level (mean β value of the probes) of the SDC2 CpG island. In several previous studies on 450k methylation array, researchers elaborated the distribution characteristics of β and M values. Their results indicated that the bimodal distribution of M value was clearly separated when β value equaled 0.2. In fact, the authors directly called the peak below 0.2 the unmethylated peak and the other peak above 0.2 the methylated peak in their study, which suggested that 0.2 can be an appropriate threshold (Du et al., 2010; Dedeurwaerder et al., 2011). In this study, 50 CRC samples had a methylation level lower than 0.2 (Supplementary Table S5). Genes with a β value greater than 0.2 in these SDC2 hypomethylated samples were retrieved throughout the whole genome, and genes complementary to SDC2 in different colorectal sites were selected. TFPI2 showed the best complementarities to SDC2 in various colorectal sites (Table 2). 44 out of 50 specimens showed a TFPI2 β value higher than 0.2, accounting for 88.0% (Table 2).
The comparison of DNA methylation level (β value) of SDC2 and TFPI2 in colorectal cancer, adenoma, and normal tissues is given in Figure 1. The average methylation level of the SDC2 gene in 45 TCGA normal tissues was 0.067, while that in 45 paired CRC tissues was 0.492 (Figure 1A). The average methylation level of the TFPI2 gene in 45 normal tissues was 0.161, while that in paired CRC tissues was 0.538 (Figure 1A). The difference of either SDC2 or TFPI2 methylation level between 45 CRC tissues and adjacent normal tissues was highly significant (p < 0.001).
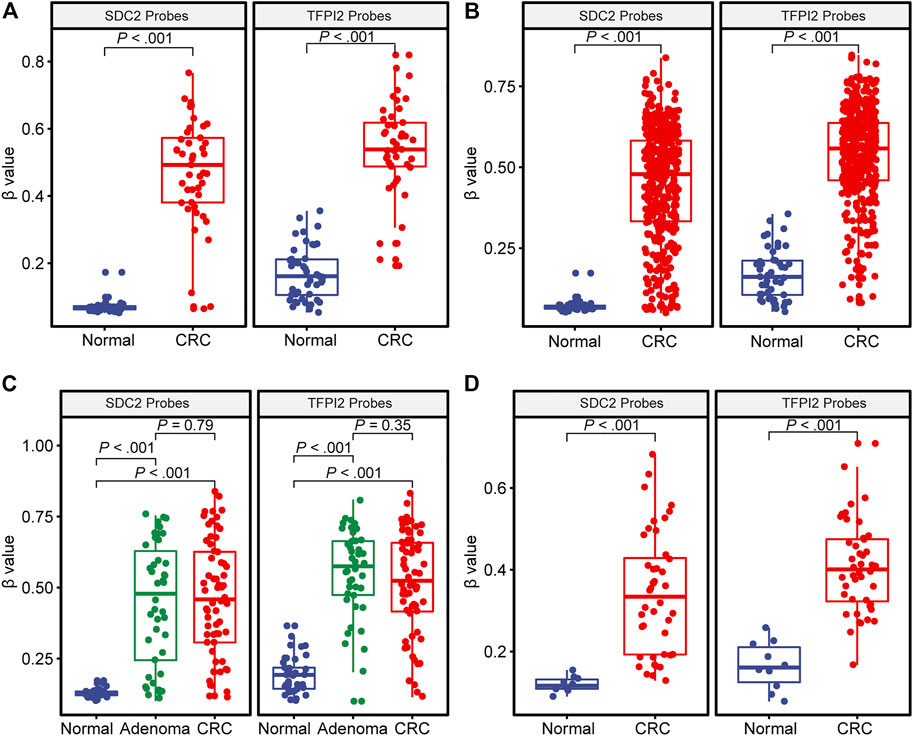
FIGURE 1. DNA methylation level (β value) of SDC2 and TFPI2 in CRC, adenoma, and normal tissues. The abscissa is the type of tissue, and the ordinate is the methylation level (β value). Each dot indicates an individual specimen. The extremes of the boxes define the upper and lower quartiles, and the center lines define the median. Whiskers indicate 1.5×interquartile range (IQR). Beyond IQR are defined the outliers. (A) illustrates the distribution of methylation levels in 45 CRC specimens vs. their paired normal tissue specimens from TCGA. (B) shows 391 CRC vs. 45 normal tissue specimens from TCGA. (C) is based on the dataset of GSE48684, with 106 CRC, 42 adenoma, and 41 normal tissue specimens. (D) is based on GSE79740, with 44 CRC and 10 normal tissue specimens. The Wilcoxon signed-rank test was used for (A) and Mann–Whitney U test for (B–D).
391 TCGA CRC samples showed various methylation levels (β value) of SDC2 or TFPI2 probes (Figure 1B). The methylation level of 391 CRC tissues was significantly higher than that of 45 normal tissues, either for the SDC2 gene (0.479 ± 0.178 vs. 0.067 ± 0.018) or for the TFPI2 gene (0.558 ± 0.149 vs. 0.161 ± 0.078).
A similar tendency was observed in the datasets GSE48684 and GSE79740 (Figures 1C,D), and the methylation level of both SDC2 and TFPI2 was higher in CRC than in normal tissue. However, no significant difference was detected between CRC and adenoma samples in GSE48684 (p = 0.79 for SDC2 and p = 0.35 for TFPI2) (Figure 1C).
MSP assays were developed. The PCR reaction was considered effective if the Ct value of ACTB ≤ 36. The standard curve data showed that the amplification efficiency of the three genes (SDC2, TFPI2, and ACTB) was similar, ranging from 102 to 105% (Supplementary Figure S1). Furthermore, methylated primers and probes could only specifically amplify the positive templates (positive plasmids and the CRC cell line HT-29), but not the negative templates (negative plasmids and the control cell line Hacat) (Supplementary Figure S2).
MSP assays were then evaluated with 238 tissue (184 CRC and 54 healthy control samples) and 696 stool (289 CRC, 190 adenoma, and 217 healthy control samples) specimens. Figure 2 shows the distribution of Ct values (Figures 2A,C) and
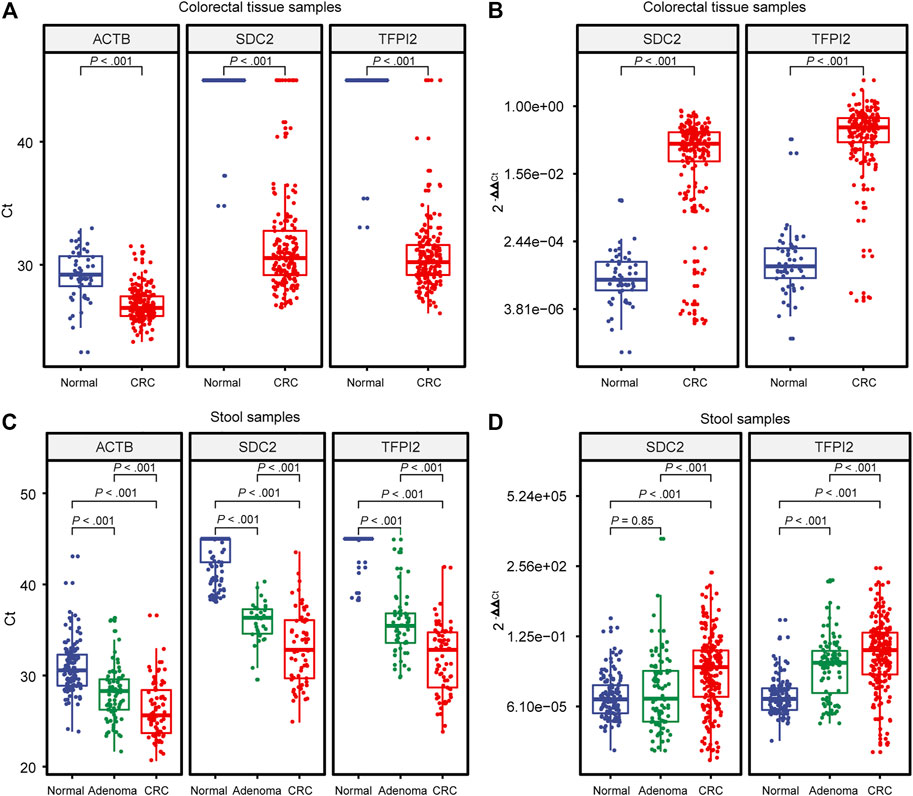
FIGURE 2. Distribution of Ct and 2−ΔΔCt values generated by MSP. (A) and (B) are built on colorectal tissue specimens. (C) and (D) are built on stool specimens. Each dot indicates an individual specimen. The extremes of the boxes define the upper and lower quartiles, and the center lines define the median. Whiskers indicate 1.5×interquartile range (IQR). Beyond IQR are defined the outliers. Statistically significant differences were determined using the Mann–Whitney U test, with a significant level of p < 0.05 and a highly significant level of p < 0.01.
The endogenous reference gene ACTB showed a significant difference (p < 0.001) in Ct value, showing a tendency of normal > adenoma > CRC (Figures 2A,C). Since the Ct value of the endogenous reference ACTB is a function of the copy number of template DNA, in stool samples, it reflects the number of human exfoliated cells in the sample, and the result indicated that the number of human exfoliated cells in stool samples showed a tendency of CRC > adenoma > normal control.
Ct values of the tissue samples (Figure 2A) showed a highly significant difference (p < 0.001) between CRC and the control for either SDC2 or TFPI2. Ct values of the stool samples showed highly significant differences in SDC2 and TFPI2 between CRC, adenoma, and control (Figure 2C), with a similar tendency of normal > adenoma > CRC.
Among stool samples, 55.3% CRC samples and 25.3% adenoma samples with an SDC2 Ct value larger than 38 (false-negative) showed positive results in TFPI2 MSP assays (Table 3).
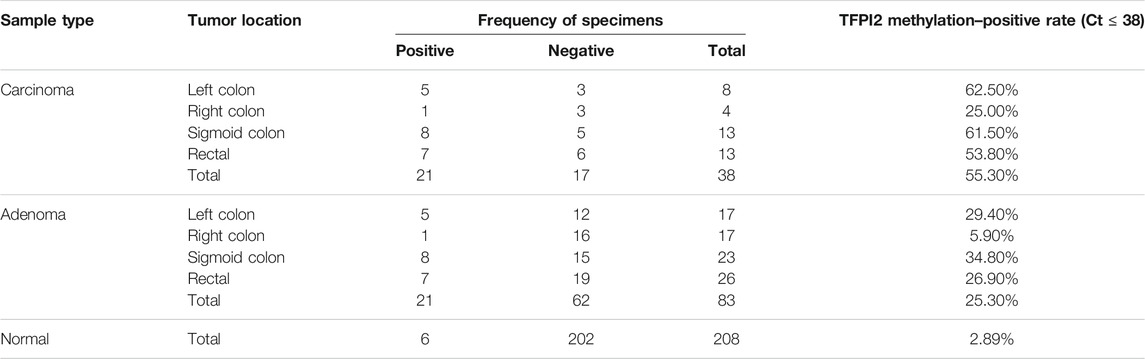
TABLE 3. Methylation-positive rate (Ct ≤ 38) of TFPI2 within SDC2 methylation–negative (Ct>38) stool specimens.
The methylation level (ML) of a sample was estimated by the formula
The diagnostic performance of SDC2, TFPI2, and SDC2/TFPI2 in stool samples was evaluated by ROC curve analysis with colonoscopy as the gold standard and MSP result as the evaluation index (Figure 3 and Table 4). Figures 3A–C display the ROC curves based on Ct values and Figures 3D–F based on ML values.
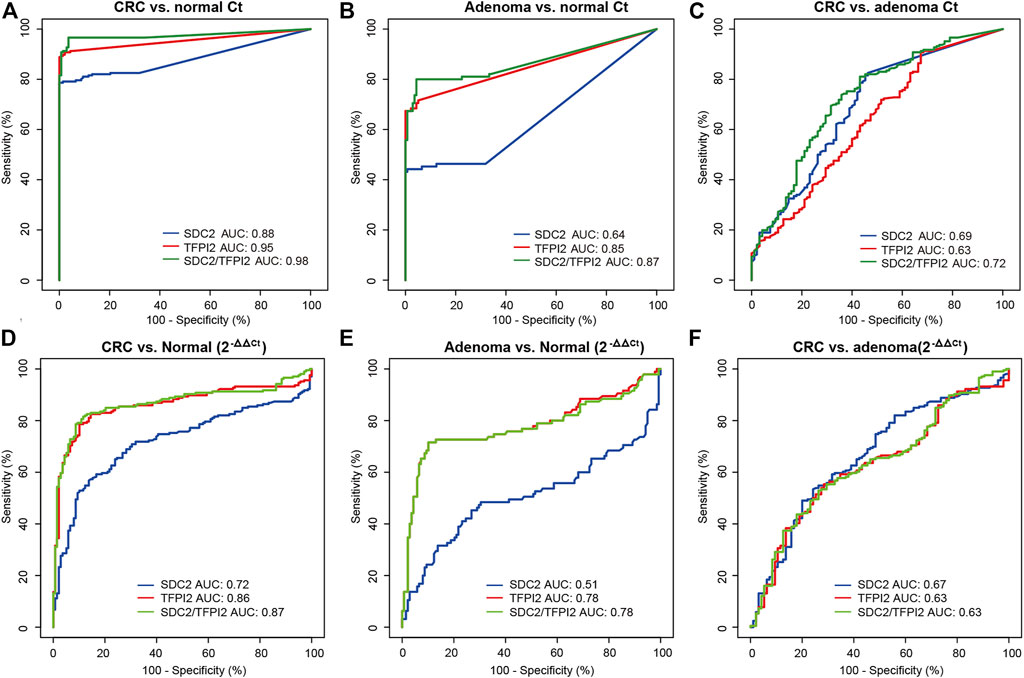
FIGURE 3. Diagnostic performances of methylation-specific PCR targeting SDC2, TFPI2, and SDC2/TFPI2 in stool specimens. (A–C) are ROC curves based on Ct values, for cancer vs. normal, adenoma vs. normal, and cancer vs. adenoma, respectively. (D–F) are ROC curves based on ML values, for cancer vs. normal, adenoma vs. normal, and cancer vs. adenoma, respectively. Ct values are obtained in stool specimens by methylation-specific PCR. ML = 2-ΔΔCt, in which ΔΔCt = (Cttarget - CtACTB)sample - (Cttarget - CtACTB)positive control. AUC means the area under the curve.
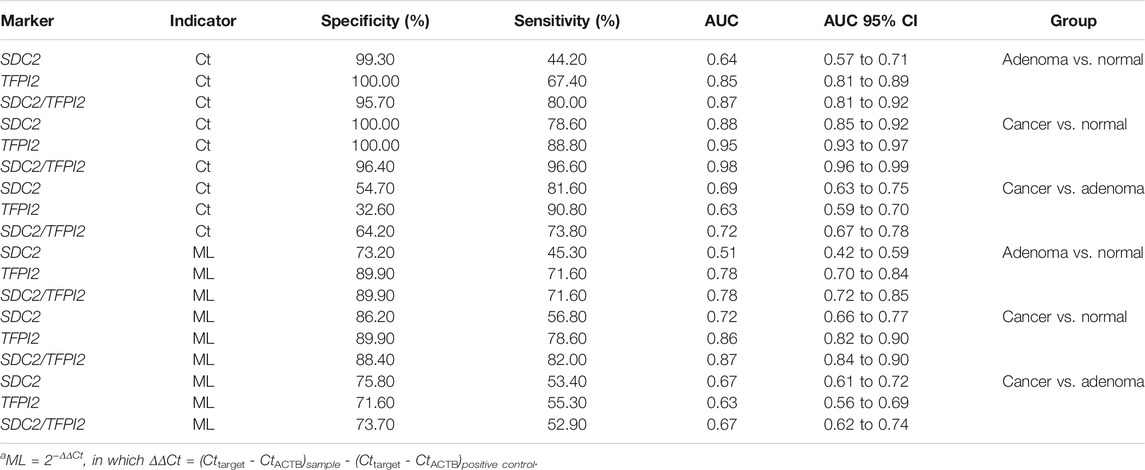
TABLE 4. Diagnostic performance of methylation-specific PCR in stool specimens with the Ct value and ML as indicatorsa.
The Ct-based ROC curve analysis (Figure 3A; Table 4) showed that, for CRC vs. normal, the AUC value of SDC2/TFPI2-combined detection was 0.98 (95% CI: 0.96–0.99) with the specificity of 96.40% and sensitivity of 96.60%, while the AUC value of SDC2 detection was 0.88 (95% CI: 0.85–0.92) with the specificity of 100% and sensitivity of 78.60%. For adenoma vs. normal (Figure 3B; Table 4), the AUC value of SDC2/TFPI2-combined detection was 0.87 (95% CI: 0.81–0.92) with the specificity of 95.70% and sensitivity of 80.00%, while the AUC value of SDC2 detection was 0.64 (95% CI: 0.57–0.71) with the specificity of 99.30% and sensitivity of 44.20%. SDC2/TFPI2-combined detection showed better diagnostic performance than SDC2 detection alone for CRC vs. normal and for adenoma vs. normal as well as for adenoma vs. CRC. The AUC value of SDC2/TFPI2 for CRC vs. adenoma was 0.72, with the specificity of 64.20% and sensitivity of 73.80%, which was lower than that of CRC vs. normal or adenoma vs. normal.
When ML values were used as the detection index in ROC curve analysis, SDC2/TFPI2-combined detection also showed better diagnostic performance than SDC2 detection in discrimination between cancer and normal, between adenoma and normal, and between adenoma and cancer. However, ML-based diagnostic performance was significantly lower than that of Ct-based diagnosis (Figure 3; Table 4).
Figure 4 shows the sensitivity comparison of different detection methods (targeting SDC2, TFPI2 alone and SDC2/TFPI2 jointly) in different CRC sites. Figure 4A is based on 391 CRC specimens in TCGA. The sensitivity of SDC2/TFPI2-combined detection was 97.2%, while that of SDC2 alone was 87.2%; the sensitivity difference between SDC2 single gene detection (abbreviated to SD) and SDC2/TFPI2 double gene detection (abbreviated to DD) was highly significant (p ≤ 0.01). The sensitivity difference between DD and SD varied with different locations. A highly significant improvement was found in the rectum (n = 46, DD/SD = 100.0%/87.0%, p ≤ 0.01), sigmoid colon (n = 88, DD/SD = 95.5%/76.1%, p ≤ 0.01), and rectosigmoid junction (n = 46, DD/SD = 93.5%/76.1%, p ≤ 0.01). Detection sensitivity was also increased for transverse colon (n = 25, DD/SD = 100%/88.0%), descending colon (n = 14, DD/SD = 100%/85.7%), and ascending colon (n = 55, DD/SD = 98.2%/94.5%) cancers though they were not statistically significant.
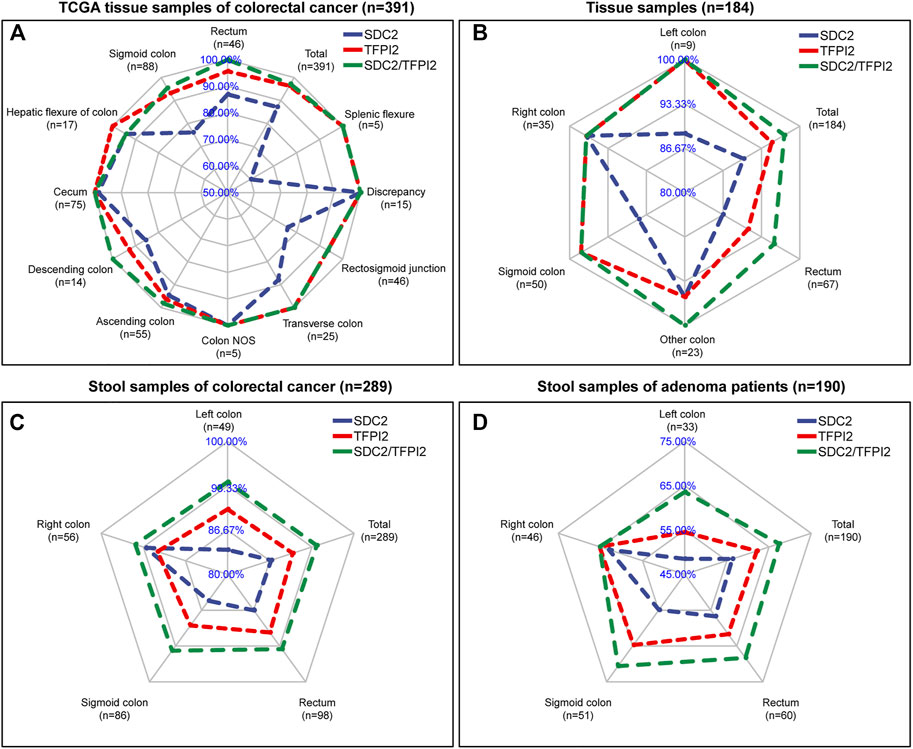
FIGURE 4. Comparison of the sensitivity of SDC2, TFPI2, and SDC2/TFPI2 in detecting colorectal cancer of various locations. The blue line is related to the detection sensitivity of SDC2, the red line to that of TFPI2, and the green line to that of SDC2/TFPI2 in combination. The cutoff value is β = 0.2 for (A) and Ct = 38 for (B–D). (A) shows detection sensitivity based on β values generated by a 450k methylation chip in TCGA tissue specimens (n = 391 cancer and 45 normal samples). (B) shows detection sensitivity based on Ct values generated by methylation-specific PCR of the tissue specimens (n = 184) collected in this study. (C) shows detection sensitivity based on Ct values generated by methylation-specific PCR of stool specimens (n = 289) of patients with colorectal cancer collected in this study. (D) shows detection sensitivity based on Ct values generated by methylation-specific PCR of stool specimens (n = 190) of patients with colorectal adenoma collected in this study.
In 184 CRC tissue samples (Figure 4B), the overall sensitivity of DD was 97.3% while 90.2% for SD, and the difference between DD and SD was highly significant (p ≤ 0.01). The rectum (n = 67, DD/SD = 95.5%/86.6%), sigmoid colon (n = 50, DD/SD = 98.0%/88.0%), and left colon (n = 9, DD/SD = 100.0%/88.9%) showed the most significant improvement.
In 289 CRC stool samples (Figure 4C), the overall sensitivity of DD was 94.1%, while that of SD was 86.9%, and the difference between DD and SD was highly significant (p ≤ 0.001). The left colon (n = 49, DD/SD = 93.9%/83.7%), sigmoid colon (n = 86, DD/SD = 94.2%/84.9%), and rectum (n = 98, DD/SD = 93.9%/86.7%) showed the most significant improvement.
In 190 colorectal adenomas stool samples (Figure 4D), the sensitivity was 67.4% for DD while 56.3% for SD, and the difference between DD and SD was also highly significant (p ≤ 0.001). The left colon (n = 33, DD/SD = 66.6%/48.5%), sigmoid colon (n = 51, DD/SD = 70.6%/54.9%), and rectum (n = 60, DD/SD = 68.3%/56.7%) showed the most obvious sensitivity improvement.
Based on the above four datasets (Figure 4), it was found that the sensitivity of SDC2/TFPI2-combined detection was significantly higher than that of SDC2 single gene detection. TFPI2 could enhance the detection sensitivity of SDC2 especially for cancer in the left colon, rectum, and sigmoid colon. Tissue samples and stool samples showed the same trend. SDC2/TFPI2-combined detection showed higher sensitivity not only to cancer samples but also to adenoma samples.
CRCs that arise proximally or distally to the splenic flexure show differences in epidemiologic incidence, morphology, and molecular alterations (Huyghe et al., 2021). A previous investigation suggested that the degree of SDC2 methylation in the left colon and the right colon may be different (McInnes et al., 2017). The difference in methylation of TFPI2 and SDC2 in different colorectal parts found in this study may be related to the etiologic heterogeneity of CRC.
SDC2 is a member of the syndecan family and has been reported to play a critical role either as a tumor suppressor, such as in osteosarcoma (Mansouri et al., 2015), or as an oncogene, such as in breast cancer (Loftus et al., 2021). Hypermethylation of SDC2 promoter region is a frequent epigenetic change in the development of colorectal neoplasms, and it has been successfully detected in several types of clinical specimens which include tissue, stool, and serum samples (Barták et al., 2017; Chen et al., 2019; Oh et al., 2013), making it an optimal target for developing a novel diagnostic kit for CRC early detection. In a previous study, the detection rate of SDC2 methylation was 81.1 and 58.2% for CRC and adenoma, respectively, with the specificity of 93.3% (Niu et al., 2017), which was in agreement with our study (Figure 3; Table 4) and other research studies (Wang et al., 2020; Zhao et al., 2020), indicating that the sensitivity of SDC2 for CRC and adenoma had room to be improved. To reduce the missed rate of detection, an additional marker which was complementary to SDC2 might be an alternative way for this purpose.
TFPI2 (tissue factor pathway inhibitor-2) is a Kunitz-type serine proteinase inhibitor that protects the extracellular matrix of cancer cells from degradation and inhibits in vitro colony formation and proliferation (Gerecke et al., 2015). TFPI2 is frequently silenced in human hepatocellular carcinoma via epigenetic alterations, including promoter methylation and histone deacetylation (Wong et al., 2007). Glockner et al. demonstrated that the methylation of TFPI2 was a frequent event in human colorectal cancer using a gene expression array–based strategy (Glöckner et al., 2009).
We firstly demonstrated that there was a subsite difference in colorectal methylation between TFPI2 and SDC2, and as high as 88.0% SDC2 hypomethylated CRC samples retrieved from TCGA were TFPI2 hypermethylated (Table 2), in agreement with the result from the MSP assays in stool samples (Table 3). The results of this study indicated that TFPI2 could improve the detection sensitivity of SDC2 through finding CRC in the left colon, sigmoid colon, and rectum (Figure 4). Sigmoid colon cancer and rectal cancer have a high incidence worldwide (Meza et al., 2010; Wang et al., 2021), and the sensitivity improvement would be of great benefit to the overall CRC detection.
It is very important to develop a stool DNA methylation test that is sensitive to detect early-stage CRC and precancerous lesions for effective surgical and therapeutic interventions. In the current multicenter clinical study, SDC2/TFPI2-joined detection demonstrated an overall sensitivity for all CRCs at 96.6% with the specificity at 96.4%, and the sensitivity for adenoma was as high as 80%, in contrast to the sensitivity of 30% for adenoma by the fecal occult blood test with high-sensitivity guaiac (gFOBT), or the fecal immunochemical test (FIT) (Graser et al., 2009), which are non-invasive detection methods that are most commonly used in clinical practice at present. In our study, the methylation level of CRC and adenoma was statistically different in stool samples (Figures 2C,D); however, SDC2/TFPI2 did not differentiate CRC and adenoma well enough (Figure 3 and Table 4), which meant that the methylation pattern of adenoma was more similar to that of CRC. According to ACG guidelines (Shaukat et al., 2021), CRC screening efforts are directed toward removal of adenomas and sessile serrated lesions and detection of early-stage CRC; therefore, as long as the markers can effectively distinguish between normal and adenoma samples, it is beneficial to classify precancerous adenoma and cancer samples together as “positive samples,” so that if a sample is methylation positive, further colonoscopy and pathological examination can be performed. Once the adenoma is effectively treated, the chance of developing into cancer will be greatly reduced, which is beneficial to reducing the incidence of cancer.
High performance of SDC2/TFPI2-joined detection in this study derived from a series of technical improvement, including stool DNA preservation against DNA degradation in stool, sequence-specific capture technology based on magnetic beads which effectively eliminated background noise from massive amounts of contaminating plant, animal, and bacterial genomic DNA in MSP assays, and optimized primers and probe sets as well as assay conditions which are also potential contributors to the varied sensitivity and specificity. As a non-invasive detection method, SDC2/TFPI2-joined detection in stool samples is safe and can be operated easily, avoiding bowel preparation and possible cross-infection during colonoscopy.
The results showed that the Ct value instead of 2−ΔΔCt as the detection index could improve the detection accuracy of adenoma and CRC (Figure 3). We found that the CRC, adenoma, and control were different in the stool samples not only in the methylation level as measured by 2−ΔΔCt but also in the number of human exfoliated cells reflected by the Ct value of ACTB (Figure 2). The Ct value of ACTB in cancer samples was smaller than that in adenoma samples and further smaller than that in normal control samples (p < 0.001), indicating that the number of exfoliated cells in CRC stool samples was significantly more than that in adenoma samples and further more than that in normal control samples. The difference of methylation level and in addition the number of exfoliated cells resulted in the better sensitivity of Ct than 2−ΔΔCt as the detection index. This was in agreement with other studies which also used the Ct value as the diagnostic index (Kim et al., 2021; Zhao et al., 2021).
Despite that many methylation-based methods for CRC diagnosis have been reported, there exist some highlights in our study. Firstly, we identified TFPI2 through whole-genome screening, significantly outperforming the well-established biomarker SDC2 in CRC detection. Secondly, five populations from Asian and Euro-American regions and two specimen types (tissue and stool) were involved (Table 1), totally including 1,034 CRC patients, 232 adenoma patients, and 367 normal individuals, covering different colorectal sites and stages. Thirdly, three indexes (β value, Ct value, and 2−ΔΔCt value) were evaluated and compared (Tables 2–4). The Ct value was a suitable indicator, being simple to operate and having better performance than the 2−ΔΔCt value.
However, there are still certain limitations associated with our current investigation. First, a larger scale validation is required to accurately assess the effectiveness. The number of cases of advanced adenomas, in particular the pathology information regarding villous and serrated adenomas, is limited, hence lacking sufficient power to accurately estimate the test’s sensitivity and to perform further covariate analysis of these precancerous lesions.
TFPI2 can improve the sensitivity of SDC2 methylation–specific detection of colorectal tumorous lesions while maintaining high specificity, in particular reducing the missed detection of left colon cancer and adenoma. As a non-invasive detection method, the dual detection of SDC2/TFPI2 will be an easy and precise screening tool for colorectal cancer and its precancerous lesions.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by the Medical Ethics Committee of Zhongnan Hospital of Wuhan University (ethical approval No. 2019099). The patients/participants provided their written informed consent to participate in this study.
LY, LZ, and JL contributed to conception and design. LD, WH, CY, QW, RL, and RS contributed to specimen collection. CL contributed to the methodology and assembly of data. WH, CY, QW, RL, and RS contributed to clinical information collection. QW, KW, TL, FS, LD, and TG contributed to the experiment and data analysis. All authors helped in data interpretation and manuscript writing and reviewing and agreed to the final manuscript.
This work was supported by the Department of Health and Family Planning Commission of Jiading District under Grant No. 2008-ky-01, the Department of Health and Family Planning Commission of Ruijin North Hospital under Grant No. 2018 zy16, and the Natural Science Foundation of China under Grant No. 81502461.
LD, CL, KW, TL, FS, and TG were employed by Wuhan Ammunition Life-tech Company, Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors would like to extend their appreciation to all the patients and control subjects that kindly provided the biological samples for this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.706754/full#supplementary-material
Supplementary Figure S1 | Amplification plots and standard curves. (A,C and E) are the amplification plots for ACTB, SDC2, and TFPI2 in two duplicates, respectively. (B, D and F) are the amplification standard curves for ACTB, SDC2, and TFPI2, respectively. Eff% indicates the PCR amplification efficiency, Y-inter represents Y-interception, and R2 means the determinant coefficient.
Supplementary Figure S2 | PCR amplification curves of the positive and negative standards for the target genes (SDC2 and TFPI2) and the endogenous reference gene ACTB. (A) was from the positive (red lines) and negative (black lines) controls of plasmids, respectively. (B) was from cell lines of HT-29 and Hacat.
CRC, colorectal cancer; FOBT, fecal occult blood test; FIT, fecal immunochemical test; MSP, methylation-specific PCR; TCGA, The Cancer Genome Atlas; ROC, receiver-operating characteristic; AUC, area under the curve; GEO, Gene Expression Omnibus; 95% CI, 95% confidence interval; ML, methylation level which is defined as ML = 2−ΔΔCt; SD, single detection of SDC2; DD, double detection of SDC2 and TFPI2.
Alvi, M. A., Loughrey, M. B., Dunne, P., McQuaid, S., Turkington, R., Fuchs, M.-A., et al. (2017). Molecular Profiling of Signet Ring Cell Colorectal Cancer Provides a strong Rationale for Genomic Targeted and Immune Checkpoint Inhibitor Therapies. Br. J. Cancer 117, 203–209. doi:10.1038/bjc.2017.168
American Cancer Society (2019). Cancer Facts & Figures. Available at: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2019/cancer-facts-and-figures-2019.pdf (Accessed on January 4, 2020).
Barták, B. K., Kalmár, A., Péterfia, B., Patai, Á, V., Galamb, O., Valcz, G., et al. (2017). Colorectal Adenoma and Cancer Detection Based on Altered Methylation Pattern of SFRP1, SFRP2, SDC2, and PRIMA1 in Plasma Samples. Epigenetics 12, 751–763. doi:10.1080/15592294.2017.1356957
Cao, W., Chen, H.-D., Yu, Y.-W., Li, N., and Chen, W.-Q. (2021). Changing Profiles of Cancer burden Worldwide and in China: a Secondary Analysis of the Global Cancer Statistics 2020. Chin. Med. J. (Engl). 134, 783–791. doi:10.1097/cm9.0000000000001474
Chen, J., Sun, H., Tang, W., Zhou, L., Xie, X., Qu, Z., et al. (2019). DNA Methylation Biomarkers in Stool for Early Screening of Colorectal Cancer. J. Cancer 10, 5264–5271. doi:10.7150/jca.34944
Chung, D. C. (2018). Genetic Testing and Early Onset Colon Cancer. Gastroenterology 154, 788–789. doi:10.1053/j.gastro.2018.02.002
Crockett, S. D., and Nagtegaal, I. D. (2019). Terminology, Molecular Features, Epidemiology, and Management of Serrated Colorectal Neoplasia. Gastroenterology 157, 949–966.e4. doi:10.1053/j.gastro.2019.06.041
Dedeurwaerder, S., Defrance, M., Calonne, E., Denis, H., Sotiriou, C., and Fuks, F. (2011). Evaluation of the Infinium Methylation 450K Technology. Epigenomics 3, 771–784. doi:10.2217/epi.11.105
deVos, T., Tetzner, R., Model, F., Weiss, G., Schuster, M., Distler, J., et al. (2009). Circulating Methylated SEPT9 DNA in Plasma is a Biomarker for Colorectal Cancer. Clin. Chem. 55, 1337–1346. doi:10.1373/clinchem.2008.115808
Du, P., Zhang, X., Huang, C.-C., Jafari, N., Kibbe, W. A., Hou, L., et al. (2010). Comparison of Beta-Value and M-Value Methods for Quantifying Methylation Levels by Microarray Analysis. BMC Bioinformatics 11, 587. doi:10.1186/1471-2105-11-587
Gerecke, C., Scholtka, B., Löwenstein, Y., Fait, I., Gottschalk, U., Rogoll, D., et al. (2015). Hypermethylation of ITGA4, TFPI2 and VIMENTIN Promoters is Increased in Inflamed colon Tissue: Putative Risk Markers for Colitis-Associated Cancer. J. Cancer Res. Clin. Oncol. 141, 2097–2107. doi:10.1007/s00432-015-1972-8
Glöckner, S. C., Dhir, M., Yi, J. M., McGarvey, K. E., Van Neste, L., Louwagie, J., et al. (2009). Methylation of TFPI2 in Stool DNA: a Potential Novel Biomarker for the Detection of Colorectal Cancer. Cancer Res. 69, 4691–4699. doi:10.1158/0008-5472.CAN-08-0142
Grady, W. M., and Pritchard, C. C. (2014). Molecular Alterations and Biomarkers in Colorectal Cancer. Toxicol. Pathol. 42, 124–139. doi:10.1177/0192623313505155
Graser, A., Stieber, P., Nagel, D., Schafer, C., Horst, D., Becker, C. R., et al. (2009). Comparison of CT Colonography, Colonoscopy, Sigmoidoscopy and Faecal Occult Blood Tests for the Detection of Advanced Adenoma in an Average Risk Population. Gut 58, 241–248. doi:10.1136/gut.2008.156448
Han, Y. D., Oh, T. J., Chung, T.-H., Jang, H. W., Kim, Y. N., An, S., et al. (2019). Early Detection of Colorectal Cancer Based on Presence of Methylated Syndecan-2 (SDC2) in Stool DNA. Clin. Epigenet 11, 51. doi:10.1186/s13148-019-0642-0
Huyghe, J. R., Harrison, T. A., Bien, S. A., Hampel, H., Figueiredo, J. C., Schmit, S. L., et al. (2021). Genetic Architectures of Proximal and Distal Colorectal Cancer Are Partly Distinct. Gut 70, 1325–1334. doi:10.1136/gutjnl-2020-321534
Imperiale, T. F., Ransohoff, D. F., Itzkowitz, S. H., Levin, T. R., Lavin, P., Lidgard, G. P., et al. (2014). Multitarget Stool DNA Testing for Colorectal-Cancer Screening. N. Engl. J. Med. 370, 1287–1297. doi:10.1056/nejmoa1311194
Kim, C. W., Kim, H., Kim, H. R., Kye, B.-H., Kim, H. J., Min, B. S., et al. (2021). Colorectal Cancer Screening Using a Stool DNA-Based SDC2 Methylation Test: a Multicenter, Prospective Trial. BMC Gastroenterol. 21, 173. doi:10.1186/s12876-021-01759-9
Loftus, P. G., Watson, L., Deedigan, L. M., Camarillo‐Retamosa, E., Dwyer, R. M., O'Flynn, L., et al. (2021). Targeting Stromal Cell Syndecan‐2 Reduces Breast Tumour Growth, Metastasis and Limits Immune Evasion. Int. J. Cancer 148, 1245–1259. doi:10.1002/ijc.33383
Loh, K., Chia, J. A., Greco, S., Cozzi, S.-J., Buttenshaw, R. L., Bond, C. E., et al. (2008). Bone Morphogenic Protein 3 Inactivation is an Early and Frequent Event in Colorectal Cancer Development. Genes Chromosom. Cancer 47, 449–460. doi:10.1002/gcc.20552
Luo, Y., Wong, C.-J., Kaz, A. M., Dzieciatkowski, S., Carter, K. T., Morris, S. M., et al. (2014). Differences in DNA Methylation Signatures Reveal Multiple Pathways of Progression from Adenoma to Colorectal Cancer. Gastroenterology 147, 418–429. doi:10.1053/j.gastro.2014.04.039
Mansouri, R., Haÿ, E., Marie, P. J., and Modrowski, D. (2015). Role of Syndecan-2 in Osteoblast Biology and Pathology. Bonekey Rep. 4, 666. doi:10.1038/bonekey.2015.33
McInnes, T., Zou, D., Rao, D. S., Munro, F. M., Phillips, V. L., McCall, J. L., et al. (2017). Genome-wide Methylation Analysis Identifies a Core Set of Hypermethylated Genes in CIMP-H Colorectal Cancer. BMC Cancer 17, 228. doi:10.1186/s12885-017-3226-4
Melotte, V., Lentjes, M. H. F. M., van den Bosch, S. M., Hellebrekers, D. M. E. I., de Hoon, J. P. J., Wouters, K. A. D., et al. (2009). N-Myc Downstream-Regulated Gene 4 ( NDRG4 ): A Candidate Tumor Suppressor Gene and Potential Biomarker for Colorectal Cancer. J. Natl. Cancer Inst. 101, 916–927. doi:10.1093/jnci/djp131
Meza, R., Jeon, J., Renehan, A. G., and Luebeck, E. G. (2010). Colorectal Cancer Incidence Trends in the United States and United kingdom: Evidence of Right- to Left-Sided Biological Gradients with Implications for Screening. Cancer Res. 70, 5419–5429. doi:10.1158/0008-5472.can-09-4417
Mitchell, S. M., Ross, J. P., Drew, H. R., Ho, T., Brown, G. S., Saunders, N. F., et al. (2014). A Panel of Genes Methylated with High Frequency in Colorectal Cancer. BMC Cancer 14, 54. doi:10.1186/1471-2407-14-54
Shariatpanahi, A. M., Yassi, M., Nouraie, M., Sahebkar, A., Varshoee Tabrizi, F., and Kerachian, M. A. (2018). The Importance of Stool DNA Methylation in Colorectal Cancer Diagnosis: A Meta-Analysis. PLoS One 13, e0200735. doi:10.1371/journal.pone.0200735
Müller, H. M., Oberwalder, M., Fiegl, H., Morandell, M., Goebel, G., Zitt, M., et al. (2004). Methylation Changes in Faecal DNA: a Marker for Colorectal Cancer Screening. Lancet 363, 1283–1285. doi:10.1016/S0140-6736(04)16002-9
Navarro, M., Nicolas, A., Ferrandez, A., and Lanas, A. (2017). Colorectal Cancer Population Screening Programs Worldwide in 2016: An Update. Wjg 23, 3632–3642. doi:10.3748/wjg.v23.i20.3632
Niu, F., Wen, J., Fu, X., Li, C., Zhao, R., Wu, S., et al. (2017). Stool DNA Test of Methylated Syndecan-2 for the Early Detection of Colorectal Neoplasia. Cancer Epidemiol. Biomarkers Prev. 26, 1411–1419. doi:10.1158/1055-9965.epi-17-0153
Novak, P., Jensen, T. J., Garbe, J. C., Stampfer, M. R., and Futscher, B. W. (2009). Stepwise DNA Methylation Changes are Linked to Escape from Defined Proliferation Barriers and Mammary Epithelial Cell Immortalization. Cancer Res. 69, 5251–5258. doi:10.1158/0008-5472.can-08-4977
Oh, T. J., Oh, H. I., Seo, Y. Y., Jeong, D., Kim, C., Kang, H. W., et al. (2017). Feasibility of Quantifying SDC2 Methylation in Stool DNA for Early Detection of Colorectal Cancer. Clin. Epigenet 9, 126. doi:10.1186/s13148-017-0426-3
Oh, T., Kim, N., Moon, Y., Kim, M. S., Hoehn, B. D., Park, C. H., et al. (2013). Genome-wide Identification and Validation of a Novel Methylation Biomarker, SDC2, for Blood-Based Detection of Colorectal Cancer. J. Mol. Diagn. 15, 498–507. doi:10.1016/j.jmoldx.2013.03.004
Robertson, D. J., and Imperiale, T. F. (2015). Stool Testing for Colorectal Cancer Screening. Gastroenterology 149, 1286–1293. doi:10.1053/j.gastro.2015.05.045
Shaukat, A., Kahi, C. J., Burke, C. A., Rabeneck, L., Sauer, B. G., and Rex, D. K. (2021). ACG Clinical Guidelines: Colorectal Cancer Screening 2021. Am. J. Gastroenterol. 116, 458–479. doi:10.14309/ajg.0000000000001122
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A. Cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Wang, J., Liu, S., Wang, H., Zheng, L., Zhou, C., Li, G., et al. (2020). Robust Performance of a Novel Stool DNA Test of Methylated SDC2 for Colorectal Cancer Detection: a Multicenter Clinical Study. Clin. Epigenet 12, 162. doi:10.1186/s13148-020-00954-x
Wang, K., Ma, W., Wu, K., Ogino, S., Chan, A. T., Giovannucci, E. L., et al. (2021). Healthy Lifestyle, Endoscopic Screening, and Colorectal Cancer Incidence and Mortality in the United States: A Nationwide Cohort Study. Plos Med. 18, e1003522. doi:10.1371/journal.pmed.1003522
Werner, S., Krause, F., Rolny, V., Strobl, M., Morgenstern, D., Datz, C., et al. (2016). Evaluation of a 5-Marker Blood Test for Colorectal Cancer Early Detection in a Colorectal Cancer Screening Setting. Clin. Cancer Res. 22, 1725–1733. doi:10.1158/1078-0432.ccr-15-1268
Wong, C.-M., Ng, Y.-L., Lee, J. M.-F., Wong, C. C.-L., Cheung, O.-F., Chan, C.-Y., et al. (2007). Tissue Factor Pathway Inhibitor-2 as a Frequently Silenced Tumor Suppressor Gene in Hepatocellular Carcinoma. Hepatology 45, 1129–1138. doi:10.1002/hep.21578
Yiu, A. J., and Yiu, C. Y. (2016). Biomarkers in Colorectal Cancer. Anticancer Res. 36, 1093–1102. doi:10.1002/sim.7149
Zhao, G., Liu, X., Liu, Y., Li, H., Ma, Y., Li, S., et al. (2020). Aberrant DNA Methylation of SEPT9 and SDC2 in Stool Specimens as an Integrated Biomarker for Colorectal Cancer Early Detection. Front. Genet. 11, 643. doi:10.3389/fgene.2020.00643
Keywords: colorectal cancer, adenomas, SDC2, TFPI2, methylation
Citation: Zhang L, Dong L, Lu C, Huang W, Yang C, Wang Q, Wang Q, Lei R, Sun R, Wan K, Li T, Sun F, Gan T, Lin J and Yin L (2021) Methylation of SDC2/TFPI2 and Its Diagnostic Value in Colorectal Tumorous Lesions. Front. Mol. Biosci. 8:706754. doi: 10.3389/fmolb.2021.706754
Received: 08 May 2021; Accepted: 30 November 2021;
Published: 22 December 2021.
Edited by:
Buket Kosova, Ege University, TurkeyReviewed by:
Jiarong Chen, Jiangmen Central Hospital, ChinaCopyright © 2021 Zhang, Dong, Lu, Huang, Yang, Wang, Wang, Lei, Sun, Wan, Li, Sun, Gan, Lin and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Lin, bGluanVuNjRAMTI2LmNvbQ==; Lei Yin, eWlubGVpQHdodS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.