- 1The Second Clinical College, Guizhou University of Traditional Chinese Medicine, Guiyang, China
- 2Central Laboratory of the Second Affiliated Hospital, Guizhou University of Traditional Chinese Medicine, Guiyang, China
- 3Clinical Laboratory, Hangzhou Women’s Hospital, Hangzhou, China
- 4Department of Clinical Laboratory, Zhejiang Hospital, Hangzhou, China
- 5Guizhou Provincial Center for Clinical Laboratory, Guiyang, China
Neisseriagonorrhoeae is a host-adapted human pathogen that causes sexually transmitted gonorrhea and remains to be a serious global public health challenge, especially in low- and middle-income regions. It is vital to devise a reliable, simple, cost-saving, and easy-to-use assay for detecting the N. gonorrhoeae agent. In the current study, we firstly report a novel approach, loop-mediated isothermal amplification linked with a polymer nanoparticle–based biosensor (LAMP-PNB), that was used for identifying N. gonorrhoeae in clinical samples. The results showed that the LAMP primers based on the orf1 gene were valid for development of the N. gonorrhoeae-LAMP-PNB assay. The detection system with optimal conditions could be performed at a fixed temperature of 64°C for 40 min. The whole process, including genomic DNA preparation (approximately 10 min), LAMP reaction (40 min), and PNB reporting (approximately 2 min), could be accomplished within 60 min. The limit of detection (LoD) of the N. gonorrhoeae-LAMP-PNB assay was 50 copies per test. The specificity of the current assay was 100%, and no cross-reactions to non–N. gonorrhoeae isolates were observed. These results confirmed that the N. gonorrhoeae-LAMP-PNB technique is a reliable, specific, sensitive, rapid, low-cost, and easy-to-use method for detecting gonococci isolates. More importantly, this assay has great potential to develop a point-of-care (POC) testing method in clinical practice, especially in resource-constrained regions.
Introduction
Neisseria gonorrhoeae, a host-adapted human pathogen, is the causative agent of gonorrhea, which belongs to the most frequent sexually transmitted infections that remain one of the major global public health concerns (Quillin and Seifert, 2018; Hook and Bernstein, 2019). According to World Health Organization (WHO) estimations, there are around 87 million new infections worldwide each year (Rowley et al., 2019). Of these, the vast majority of gonococcal infections (>80 million) are in developing countries of Africa, Asia, and Latin America (World Health Organization, 2016; Rowley et al., 2019). N. gonorrhoeae infection can cause epididymo-orchitis and prostatitis in men and can lead to pelvic inflammatory disease, infertility, and ectopic pregnancy in women (Chan et al., 2016; Stevens and Criss, 2018). Maternal transmission to infants during birth can also bring about neonatal blindness and oropharyngeal infections (Vallely et al., 2021). In rare cases, N. gonorrhoeae could spread systemically, resulting in severe complications, such as septicemia, vasculitis, endocarditis, arthritis, and tenosynovitis (Lovett and Duncan, 2019). Because the various clinical symptoms of gonorrhea are largely not specific, and most gonococcal infections are in resource-constrained regions, developing a specific, sensitive, rapid, and cost-saving assay for the accurate identification of N. gonorrhoeae isolates is necessary for reducing ongoing gonorrhea transmission.
The traditional assay for identification of N. gonorrhoeae is based on cultivation. However, gonococcus is very demanding and fastidious for cultivation. The bacteria from swab collection samples should be immediately inoculated onto culture media, which require rigorous growth conditions; this method is time-consuming (24–48 h) and does not succeed equally well from every sample (Meyer and Buder, 2020). In recent decades, nucleic acid amplification technologies (NAATs), such as polymerase chain reaction (PCR), multiplex PCR, and real-time PCR, have been used for identifying N. gonorrhoeae. These methods are more sensitive, specific, and time-saving than cultivation and have been considered the primary methods to detect N. gonorrhoeae (O'Callaghan et al., 2010; Mahony et al., 1995; Whiley et al., 2002). Nevertheless, their use in many poor-resource settings is greatly limited due to high costs of experimental instruments and skilled personnel (Meyer and Buder, 2020). Hence, developing a low-cost, rapid, sensitive, specific, and simple diagnostic method for N. gonorrhoeae isolates is essential for follow-up treatment and management of gonorrhea patients.
To overcome the drawbacks of the PCR-based assay, loop-mediated isothermal amplification (LAMP) as a reliable, low-cost, sensitive, and rapid nucleic acid amplification technique was first devised by Notomi et al. (2000) and has been widely used to identify various pathogens, such as SARS-CoV-2, Mycobacterium tuberculosis, and Brucella (Li et al., 2019; Shete et al., 2019; Zhu et al., 2020). The LAMP method can efficiently amplify target genes at a constant temperature (usually 58–69°C) using Bst DNA polymerase within a short time (30–60 min) (Wong et al., 2018). The primer set consists of two outer primers (F3 and B3), two inner primers (FIP and BIP), and two loop primers (LF and LB) (Notomi et al., 2015). Previous studies have shown that the LAMP products could be analyzed with various technologies, such as colorimetric indicators (malachite green reagent), turbidimetry, and fluorescence dye (Chen et al., 2020). However, these detection techniques require special apparatus and reagents. To overcome these defects, a polymer nanoparticle–based biosensor (PNB), with specificity, sensitivity, visualization, good robustness, simple operation, and low limits of detection features, has been applied for detection of DNA and proteins in recent years (Quesada-González and Merkoçi, 2015).
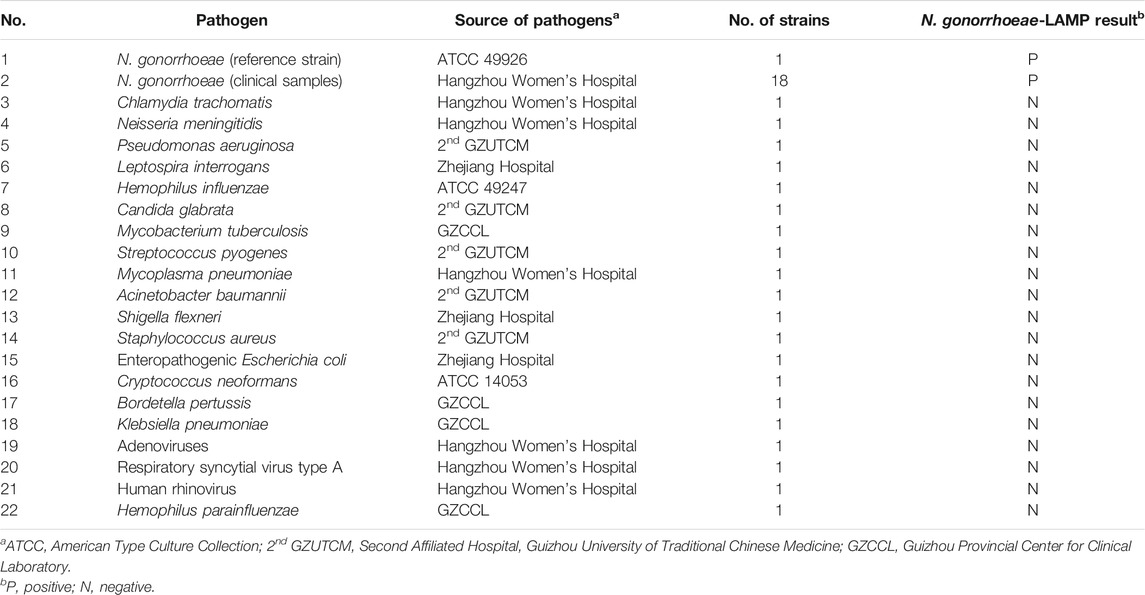
In the current study, the loop-mediated isothermal amplification linked with the polymer nanoparticle–based biosensor (LAMP-PNB) technique was devised firstly for simple, specific, sensitive, rapid, and visual identification of N. gonorrhoeae by targeting the orf1 gene (Chaudhry and Saluja, 2002; Edwards et al., 2014), and it showed no homology with other microbial genomes in GenBank by BLAST searches. The principle of the N. gonorrhoeae-LAMP-PNB assay is illustrated in Figures 1A,B, and the detection performance was analyzed with pure cultures and clinical samples.
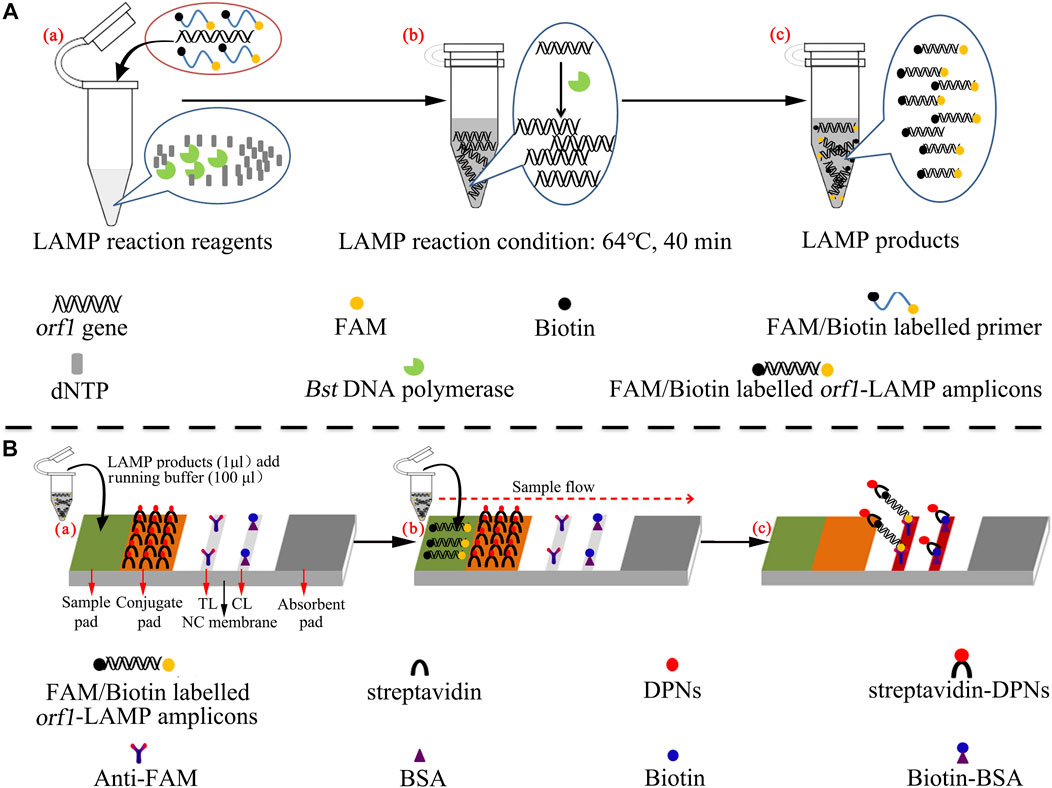
FIGURE 1. Mechanistic description of N. gonorrhoeae-loop-mediated isothermal amplification linked to a polymer nanoparticle–based biosensor (LAMP-PNB) assay. (A) Process of N. gonorrhoeae-LAMP. (B) Schematic illustration of the principle of the polymer nanoparticle–based biosensor assay for visualization of N. gonorrhoeae-LAMP products. A positive result for the orf1 gene means that the test line (TL) and control line (CL) appear simultaneously on the biosensor. Only a control line (CL) appearing on the biosensor indicates a negative result. dNTP: deoxynucleotide triphosphate; FAM: carboxyfluorescein; DPNs: dye (crimson red)–coated polymer nanoparticles; anti-FAM: rabbit anti-carboxyfluorescein antibody; BSA: bovine serum albumin; NC: nitrocellulose.
Materials and Methods
Materials and Reagents
Thayer-Martin (TM) chocolate agar plates were obtained from Autobio Biotechnology Co., Ltd. (Zhengzhou, China). Nucleic acid–releasing agents were obtained from Sansure Biotech Inc. (Changsha, China). Universal isothermal amplification kits and a colorimetric indicator (malachite green, MG) were purchased from HuiDeXin Bio-technology (Tianjin, China). Anti-FAM (rabbit anti-fluorescein antibody) and biotin BSA (biotinylated bovine serum albumin) were purchased from Abcam Co., Ltd. (Shanghai, China). Streptavidin dye–coated polymer nanoparticles (Crimson red) were obtained from Bangs Laboratories, Inc. (Indiana, United States). Polymer nanoparticle–based lateral flow biosensor (LFB) materials, including the sample pad, conjugate pad, absorbent pad, nitrocellulose (NC) membrane, and backing card, were obtained from HuiDeXin Bio-technology (Tianjin, China). N. gonorrhoeae PCR diagnosis kits were purchased from DaAn Gene Co., Ltd. (Guangzhou, China).
Preparation of Clinical Samples and Bacterial Strains
In the current study, a total of 86 clinical samples were collected from suspected N. gonorrhoeae–infected patients at Hangzhou Women’s Hospital (Hangzhou, China) between June 2020 and February 2021. Two genital secretion samples were collected from each suspected N. gonorrhoeae–infected patient with sterile swabs. One sample was used for traditional cultivation detection and the other for genomic DNA preparation. The N. gonorrhoeae reference strain (ATCC 49926) and clinical samples were cultured on Thayer-Martin (TM) chocolate agar plates (Autobio, Zhengzhou, China) at 37°C in 5% CO2 for 2 days. Genomic DNA templates were obtained using nucleic acid–releasing agents (Sansure Biotech, Changsha, China) in accordance with the manufacturer’s instructions. In brief, 10 μl of samples were added into 10 μl of nucleic acid–releasing agents and incubated at room temperature (∼25°C) for 10 min to release nucleic acid. And then, the genomic DNA was stored at −20°C before use. The concentration was identified with NanoDrop 2000 (Thermo, United States) at A260/280, and the corresponding genome copy number was calculated from the weight of the N. gonorrhoeae genome. One copy of the N. gonorrhoeae genome is 2.45 fg (2.2 × 106 bp (genomic DNA length) × 665 Da/bp×1.67 × 10−24 g/Da) (Dempsey et al., 1991; Geraats-Peters et al., 2005; Edwards et al., 2014).
N. gonorrhoeae-LAMP Primer Design
A set of special LAMP primers, targeting on the orf1 gene of N. gonorrhoeae (GenBank Accession No. M84113), was designed according to the LAMP principle. The LAMP primers consist of two outer primers (F3 and F3), two inner primers (FIP and BIP), and two loop primers (LF and LB). The specificity of N. gonorrhoeae-LAMP primers was analyzed using the Basic Local Alignment Search Tool (BLAST). Moreover, OligoAnalyzer online software (V3.1; Integrated DNA Technologies, Coralville, IA, United States) was employed for primer, dimer, and secondary structure investigation. The details of N. gonorrhoeae-LAMP primers’ location and sequences are shown in Figure 2 and Table 1, respectively.
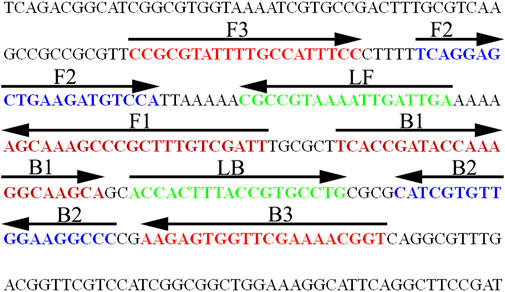
FIGURE 2. Nucleotide sequence and location of the orf1 gene used to design the N. gonorrhoeae-LAMP primers. The part of nucleotide sequence of the sense strand of the orf1 gene is shown in the diagram. The primer sequences are marked in different colors. Right arrows and left arrows indicate sense and complementary sequences, respectively.
Polymer Nanoparticle–Based Biosensor Preparation
The polymer nanoparticle–based biosensor (PNB) is illustrated in Figure 1B. In brief, the PNB consisted of four components: a sample pad, conjugate pad, nitrocellulose (NC) membrane, and absorbent pad. These components were assembled uniformly on a backing card. The capture reagents, including anti-FAM and biotin BSA, were immobilized by physical adsorption on the reaction regions. Then, anti-FAM was immobilized at test line 1 (TL1) (N. gonorrhoeae), while biotin BSA was immobilized at the control line (CL); each line was separated by 5 mm, and the detector reagents (streptavidin dye–coated polymer nanoparticles, SA-PNPs) were sprayed onto the conjugate pad. Therefore, the biosensor can detect two targets, including N. gonorrhoeae-LAMP amplicons and a chromatography control. Finally, the assembled biosensors were preserved in a plastic box and dry-stored at room temperature before use.
Loop-Mediated Isothermal Amplification Reaction and Detection
The standard N. gonorrhoeae-LAMP was conducted in a 25 μl reaction system, containing 1 μl of DNA template; 0.4 μM of each outer primer, F3 and B3; 1.6 μM of each inner primer, FIP* and BIP; 0.8 μM of each loop primer, LF* and LB; 1 μl (8 U) of Bst DNA polymerase; and 12.5 μl of 2× reaction buffer (40 mM of Tris-HCl (pH 8.8), 40 mM of KCl, 16 mM of MgSO4, 20 mM of (NH4)2SO4, 2 M of betaine, and 0.2% Tween-20) (HuiDeXin Bio-technology, Tianjin, China); then, double distilled water was added to the 25 μl reaction system. The mixtures were heated at 64°C for 1 h. Genomic DNA from non–N. gonorrhoeae strains, including Chlamydia trachomatis and Neisseria meningitidis, was used as a negative control (NC), and double distilled water (DW) was used as the template in the blank control (BC).
Real-time turbidity (LA-500), colorimetric indicator (malachite green, MG), and the polymer nanoparticle–based biosensor (PNB) were used to confirm the LAMP reaction and optimize the reaction conditions. For the real-time turbidity method, turbidity >0.1 was considered a positive outcome. For the colorimetric indicator, the color changed from colorless to light green in the reaction system, indicating positive amplification products, while the color remained colorless in negative results. For the polymer nanoparticle–based biosensor, the test line (TL) and control line (CL) appeared simultaneously, indicating positive outcomes, but in negative amplification, only the control line (CL) could be observed.
Optimization of N. gonorrhoeae-LAMP Reaction Temperature
Reaction temperature is critical for efficient LAMP. In the current study, the reaction temperature was optimized ranging from 60 to 67°C (with 1°C intervals). The N. gonorrhoeae-LAMP products were analyzed using real-time turbidity (LA-500). Turbidity >0.1 was considered a positive outcome.
Sensitivity of N. gonorrhoeae-Loop-Mediated Isothermal Amplification Linked With the Polymer Nanoparticle–Based Biosensor Assay
The limit of detection (LoD) of the N. gonorrhoeae-LAMP-PNB assay was analyzed through the N. gonorrhoeae genomic DNA with 10-fold serial dilution from 5.0 × 104 to 5.0 × 10−1 copies using double distilled water (DW). The N. gonorrhoeae-LAMP reactions were carried out as described above, and the results were identified using the colorimetric indicator (MG) and PNB methods, respectively. Simultaneously, the diluted concentration of the N. gonorrhoeae genome DNA was used for confirming the optimal isothermal time of the N. gonorrhoeae-LAMP-PNB assay.
In order to further confirm the sensitivity of the N. gonorrhoeae-LAMP-PNB assay, the N. gonorrhoeae strains were added into 100 μl of healthy volunteer urine, which made the final concentrations of N. gonorrhoeae strains range from 5.0 × 104 to 5.0 × 10−1 copies/μl, respectively. The samples were centrifuged at 12,000 × g for 5 min (4°C), and the collected bacteria were suspended in 100 μl of nucleic acid–releasing agents (Sansure Biotech, Changsha, China) and incubated at room temperature (∼25°C) for 10 min for releasing nucleic acid. And then, 1 μl of genomic DNA was added for the N. gonorrhoeae-LAMP-PNB assay. The results were identified using MG and PNB methods, respectively. Each examination was conducted independently at least three times.
Specificity of N. gonorrhoeae-Loop-Mediated Isothermal Amplification Linked With the Polymer Nanoparticle–Based Biosensor Assay
The analytical specificity of the N. gonorrhoeae-LAMP-PNB assay was evaluated by comparing N. gonorrhoeae genomic DNA with the templates from various bacteria, fungi, and viruses (Table 2). All examinations were confirmed in triplicate.
Confirming the Feasibility of the N. gonorrhoeae-LAMP-PNB Assay Using Clinical Samples
To verify the feasibility of N. gonorrhoeae-LAMP-PNB detection, 86 suspected N. gonorrhoeae–infected genital secretion samples were collected from Hangzhou Women’s Hospital (Hangzhou, China). And then, all of these samples were detected simultaneously using traditional culture, qPCR, and the LAMP-PNB assay, respectively. The culture detection was performed as previously described. The qPCR diagnosis was carried out using Gonococcus Nucleic Acid Assay Kits (DaAn Gene Co., Ltd., China) (Cat. #DA-D053), and the detection was performed with the Applied Biosystems™ 7500 Real-Time PCR System (Life Technologies, Singapore). According to the manufacturer’s instructions, concentrations of N. gonorrhoeae less than 500 copies will be considered negative results (the LoD of the qPCR assay is 500 copies per test). The N. gonorrhoeae-LAMP-PNB operation was performed as described above. The N. gonorrhoeae cultivation, qPCR, and LAMP-PNB assay were performed in a biosafety level 2 laboratory, as detailed in the WHO Laboratory biosafety manual, 3rd edition.
Statistical Analysis
The performance of LAMP-PNB was evaluated using bacteriologic results and clinical evidence as the composite reference standard. The χ2 test was used to compare sensitivity differences between cultivation, qPCR, and LAMP-PNB. SPSS 23.0 software was used to perform statistical analysis, and p < 0.05 was considered statistically significant.
Results
Confirmation and Detection of N. gonorrhoeae-LAMP Products
To confirm the validity of the N. gonorrhoeae-LAMP reaction system, LAMP products were monitored using the colorimetric indicator (MG) and polymer nanoparticle–based biosensor (PNB). The reaction tubes with positive results of the N. gonorrhoeae-LAMP assay were visualized in light green, while the reaction tubes of negative and blank controls remained colorless (Figure 3A). Using the PNB, the TL and CL were observed in the detection region, indicating positive results. Only the CL appeared on the analysis area of the PNB for negative and blank controls (Figure 3B). Therefore, these results indicated that the N. gonorrhoeae-LAMP-PNB assay using orf1-LAMP primers designed in the current study was valid for the reliable and rapid detection of N. gonorrhoeae.

FIGURE 3. Determination and confirmation of the N. gonorrhoeae-LAMP products. The N. gonorrhoeae-LAMP products were analyzed with the colorimetric indicator (A) and polymer nanoparticle–based biosensor (B). Tube 1/Biosensor 1: positive results of the N. gonorrhoeae reference strain (ATCC 49926); Tube 2/Biosensor 2: negative results of Chlamydia trachomatis; Tube 3/Biosensor 3: negative results of Neisseria meningitidis; Tube 4/Biosensor 4: blank control (distilled water). TL: test line; CL: control line.
Optimization of N. gonorrhoeae-LAMP Temperature
Reaction temperature is crucial for LAMP. In the current study, the reaction temperature of the N. gonorrhoeae-LAMP system was tested ranging from 60 to 67°C (with 1°C intervals) with 5.0 × 103 copies of genomic DNA extracted from the reference strain (ATCC 49926). The N. gonorrhoeae-LAMP protocol was carried out as described above, and the results were monitored using real-time turbidity (LA-500). The results presented that the N. gonorrhoeae-LAMP was amplified faster in the temperature range from 64 to 67°C according to kinetic data (Figure 4). Hence, 64°C was considered the optimal amplification temperature for the subsequent N. gonorrhoeae-LAMP-PNB assay in this study.
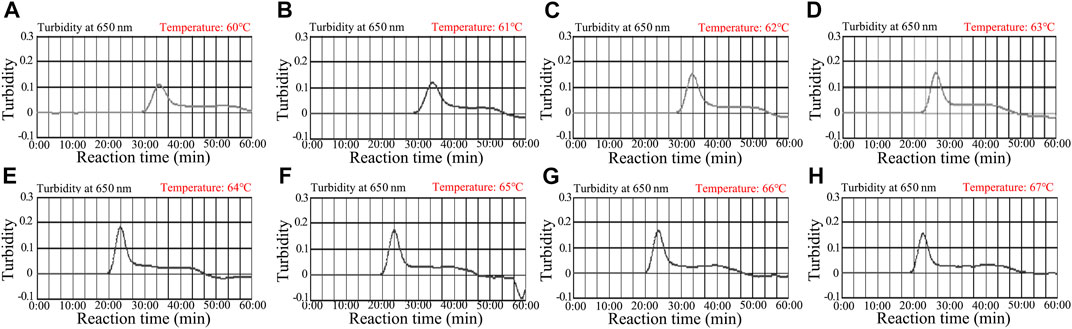
FIGURE 4. Optimization of temperature for the N. gonorrhoeae-LAMP reaction. The N. gonorrhoeae-LAMP reactions were monitored using real-time turbidity (LA-500), and the corresponding curves of amplicons are shown in the charts. The threshold value was 0.1, and turbidity >0.1 was considered to show positive amplification. Eight kinetic graphs (A–H) were obtained at different temperatures (60–67°C, 1°C intervals) with 5.0 × 103 copies of genomic DNA from N. gonorrhoeae isolates (ATCC 49926). The optimal LAMP temperature was selected according to higher turbidity. The results indicated that the temperature of 64°C (E) showed robust amplification.
Sensitivity of the N. gonorrhoeae-LAMP-PNB Assay
The sensitivity of the N. gonorrhoeae-LAMP-PNB assay was determined simultaneously with serial dilutions of genomic DNA and strains ranging from 5.0×104 to 5.0 × 10−1 copies. The N. gonorrhoeae-LAMP protocol was performed as described above, and the products were verified with MG and the PNB. As shown in Figures 5A,B, the limit of detection (LoD) of the N. gonorrhoeae-LAMP assay was 50 copies.
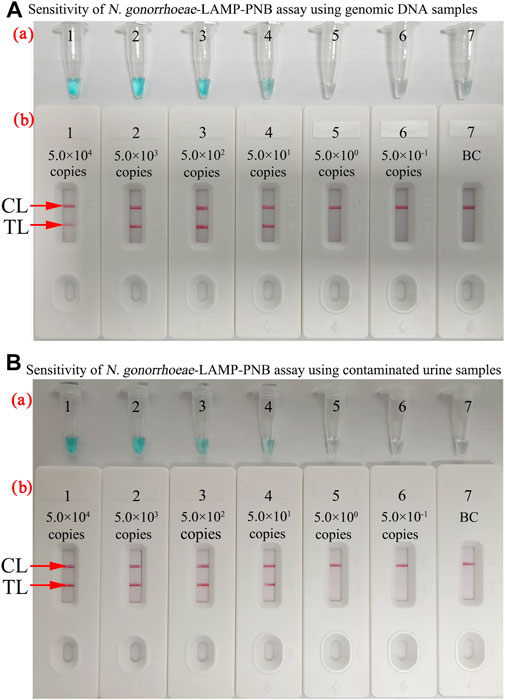
FIGURE 5. Sensitivity of the N. gonorrhoeae-LAMP-PNB assay. (A) Sensitivity of the N. gonorrhoeae-LAMP-PNB assay using diluted genomic DNA templates. MG (a) and PNB (b) were applied for reporting the N. gonorrhoeae-LAMP results. Tubes a1–a7 (Biosensors b1–b7) represent the genomic DNA levels of 5.0 × 104 copies, 5.0 × 103 copies, 5.0 × 102 copies, 5.0 × 101 copies, 5.0 × 10° copies, and 5.0 × 10−1 copies per reaction and blank control (DW), respectively. (B) Sensitivity of the N. gonorrhoeae-LAMP-PNB assay using contaminated urine samples. Each healthy volunteer’s urine samples contained different concentrations of N. gonorrhoeae strains (ranging from 5.0 × 104 to 5.0 × 10−1 copies/μl). The N. gonorrhoeae-LAMP was performed as described above, and the results were analyzed with MG (a) and PNB (b), respectively. Tubes a1–a7 (Biosensors b1–b7) represent the N. gonorrhoeae strain concentrations of 5.0 × 104 copies, 5.0 × 103 copies, 5.0 × 102 copies, 5.0 × 101 copies, 5.0 × 10° copies, and 5.0 × 10−1 copies per reaction and blank control (DW), respectively. The results suggested that the LoD of the N. gonorrhoeae-LAMP-PNB assay was 5.0 × 101 copies per test. CL: control line; TL: test line.
Optimal Amplification Time for N. gonorrhoeae-LAMP-PNB Assay
The optimal reaction time required for the N. gonorrhoeae-LAMP-PNB assay at the amplification stage was tested ranging from 30 to 60 min at the optimal reaction temperature (64°C). The results showed that the LoD of the template level (50 copies) displayed two crimson bands (CL and TL) when the isothermal amplification was conducted for 40 min (Figure 6). Hence, an N. gonorrhoeae-LAMP reaction time of 40 min was recommended for clinical sample detection. Thus, the whole detection procedure of the N. gonorrhoeae-LAMP-PNB technique, including genomic DNA template preparation (∼10 min), LAMP reaction (40 min), and resultant reporting (∼2 min), could be completed within 60 min.
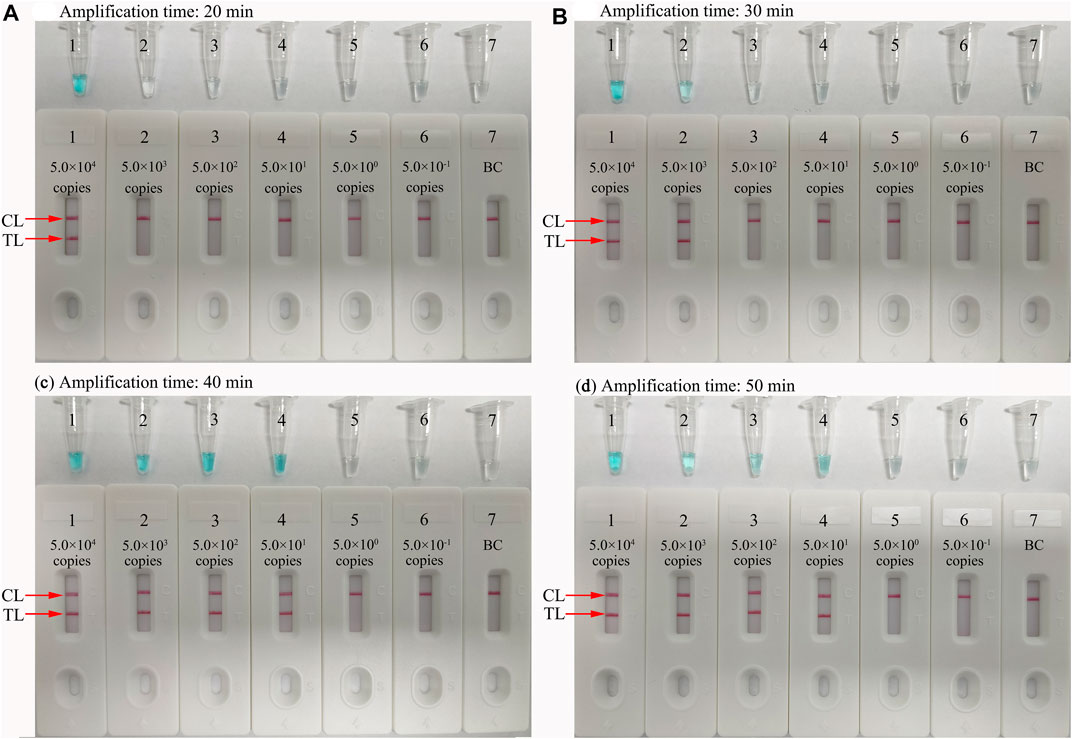
FIGURE 6. Optimal reaction time for the N. gonorrhoeae-LAMP-PNB assay. Four different amplification times (A, 20 min; B, 30 min; C, 40 min; D, 50 min) were tested at 64°C. Tubes/Biosensors 1, 2, 3, 4, 5, 6, and 7 represent N. gonorrhoeae (ATCC 49926) genomic DNA levels from 5.0 × 104 to 5.0 × 10−1 copies per reaction and negative control (DW), respectively. The results indicated that the best sensitivity was detected when the reaction lasted for 40 min (C). CL: control line; T: test line.
Specificity of the N. gonorrhoeae-LAMP-PNB Assay
The specificity of the N. gonorrhoeae-LAMP-PNB assay was tested with the N. gonorrhoeae reference strain (ATCC 49926), 18 N. gonorrhoeae–positive clinical samples, and 20 non–N. gonorrhoeae bacteria, fungi, and viruses (Table 2). The genomic DNA extracted from N. gonorrhoeae samples presented positive outcomes, while other target templates from non–N. gonorrhoeae pathogens were not detected (Figure 7). These results suggested that the LAMP-PNB assay presents very good specificity for N. gonorrhoeae identification.

FIGURE 7. Analytical specificity of the N. gonorrhoeae-LAMP-PNB assay using different pathogens. The N. gonorrhoeae-LAMP-PNB assay was verified using different genomic RNA/DNA as templates. Biosensor 1, N. gonorrhoeae (ATCC 49926); Biosensor 2–19, N. gonorrhoeae (clinical samples); Biosensor 20, Chlamydia trachomatis; Biosensor 21, Neisseria meningitidis; Biosensor 22, Pseudomonas aeruginosa; Biosensor 23, Leptospira interrogans; Biosensor 24, Hemophilus influenzae; Biosensor 25, Candida glabrata; Biosensor 26, Mycobacterium tuberculosis; Biosensor 27, Streptococcus pyogenes; Biosensor 28, Mycoplasma pneumoniae; Biosensor 29, Acinetobacter baumannii; Biosensor 30, Shigella flexneri; Biosensor 31, Staphylococcus aureus; Biosensor 32, enteropathogenic Escherichia coli; Biosensor 33, Cryptococcus neoformans; Biosensor 34, Bordetella pertussis; Biosensor 35, Klebsiella pneumoniae; Biosensor 36, adenoviruses; Biosensor 37, respiratory syncytial virus type A; Biosensor 38, human rhinovirus; Biosensor 39, Hemophilus parainfluenzae; Biosensor 40, blank control. CL: control line; TL: test line.
Evaluation of the N. gonorrhoeae-LAMP-PNB Assay in Clinical Samples
A total of 86 suspected N. gonorrhoeae–infected genital secretion samples, which were collected from Hangzhou Women’s Hospital (Hangzhou, China), were tested for verifying the feasibility of the N. gonorrhoeae-LAMP-PNB assay. All of these samples were simultaneously analyzed with biotechnical culture, qPCR, and the LAMP-PNB assay. The results showed that 58 of 86 clinical samples tested as N. gonorrhoeae–positive through conventional culture methods. The N. gonorrhoeae-LAMP-PNB assay results were consistent with the traditional cultivation testing outcomes. Using qPCR only found 55 positive outcomes (Table 3). These data suggested that our N. gonorrhoeae-LAMP-PNB assay established in the current study is more powerful for identifying gonorrhea patients, especially those at the initial stage of N. gonorrhoeae infection.

TABLE 3. Comparison of conventional culture, qPCR, and LAMP-PNB methods for testing N. gonorrhoeae in clinical samples.
Discussion
Sexually transmitted gonorrhea, caused by N. gonorrhoeae, remains a serious global public health challenge. More importantly, the overwhelming majority of gonococcal infections are in low- and middle-income regions (Chemaitelly et al., 2019). Hence, it is vital to devise adequate reliable, simple, cost-saving, and easy-to-use diagnosis assays for detecting N. gonorrhoeae infection. In the current study, loop-mediated isothermal amplification linked with the polymer nanoparticle–based biosensor (LAMP-PNB) was devised and successfully used for visual and rapid detection of N. gonorrhoeae strains.
An ideal laboratory diagnostic technique for identification of N. gonorrhoeae should be specific, sensitive, rapid, and affordable (Meyer and Buder, 2020). Traditionally, direction microscopy may produce rapid outcomes but lacks sensitivity in many cases, especially for asymptomatic patients; the load of gonococci to be detected is usually too low. Besides, other Neisseria species have a similar morphology to N. gonorrhoeae. Therefore, the method is not recommended (World Health Organization, 2013; Mensforth et al., 2018). Bacterial cultivation was considered a “golden standard” for specific and sensitive identification of N. gonorrhoeae isolates. But gonococci require demanding and fastidious growth conditions and often obtain frustrating results (Visser et al., 2020). Moreover, the culture process is time-consuming. In recent years, nucleic acid amplification technologies (NAATs) were considered the primary tests for identification of gonococci in clinical practice (Fifer et al., 2019). However, their use in low- and middle-income regions is greatly limited due to requiring expensive instruments and trained experts. In this study, the N. gonorrhoeae-LAMP-PNB assay merges isothermal amplification with a polymer nanoparticle–based biosensor, which is achieved with extremely simple instruments, such as a water bath, a heating block, or even a thermos cup that can keep a constant temperature (64°C) for 40 min. More importantly, the biosensor provides an easy-to-use platform, which could visually and objectively detect the N. gonorrhoeae-LAMP products. The whole detection process, including genomic DNA template preparation (∼10 min), LAMP reaction (40 min), and product reporting (∼2 min), could be completed within 60 min. Therefore, the N. gonorrhoeae-LAMP-PNB assay is a rapid, economical, and simple assay for identification of gonococci in clinical settings, especially in resource-constrained regions.
Loop-mediated isothermal amplification (LAMP), as a reliable, sensitive, and rapid assay with low-cost equipment, was devised in 2000 and has been widely applied to detect many pathogens (Notomi et al., 2000; Li et al., 2019; Shete et al., 2019; Zhu et al., 2020). It involves only Bst DNA polymerase, with strand displacement capability and isothermal amplification, operating at a consistent temperature (between 60 and 68°C) throughout the reaction. The isothermal amplification of specific nucleic acid sequences is achieved by employing a set of four (or six) specific primers spanning six (or eight) distinct regions of the target gene (Li et al., 2017; Zhang et al., 2019). In the current study, a set of N. gonorrhoeae-LAMP primers was specifically designed to identify eight regions of the target fragment (Figure 2). The specificity of the N. gonorrhoeae-LAMP-PNB assay was powerfully confirmed with nucleic acid extracted from N. gonorrhoeae clinical samples and other pathogens. The positive results were observed from positive control and gonococci clinical samples, but non–N. gonorrhoeae agents showed negative results (Figure 7). Thus, our approach displayed a high level of specificity for identification of the gonococci agent. In addition to its excellent specificity, the newly developed N. gonorrhoeae-LAMP-PNB assay was able to detect pathogens in as low as 50 copies (Figure 5), which is sufficient for the diagnosis of gonorrhea. For detection of clinical samples, our N. gonorrhoeae-LAMP-PNB assay was highly sensitive compared to the qPCR method (Table 3), and the assay results were completely consistent with the traditional cultivation testing outcomes.
In this report, polymer nanoparticles, as a carrier material, were used for preparation of the nanoparticle-based biosensor (PNB). Owing to high surface-to-volume ratio, high adsorption and reactive capacity, good biocompatibility, and their easy-to-synthesize and -manipulate nature, polymer nanoparticles are the most appropriate nanomaterial used as a biosensor (Quesada-González and Merkoçi, 2015; Wang et al., 2017; Anniebell and Gopinath, 2018). The PNB could visually detect the N. gonorrhoeae-LAMP products for labeling with anti-FAM and BSA biotin on the PNB strips. The two crimson red bands—test line (TL) and control line (CL)—appeared in the PNB strip indicating positive results, while the negative outcomes only observed the control line (CL) in the biosensor. Although the colorimetric indicator (MG reagent) and real-time turbidity could identify N. gonorrhoeae-LAMP products, the former is ambiguous when the concentration of the amplicon is low and the latter method is laborious and has specific high-cost equipment. The PNB is simple and easy-to use, and the cost of the PNB designed in our study is estimated to be $2 USD per test. Hence, we calculate that the total cost of each test, including genomic DNA template preparation (∼€ 0.9 euros), LAMP (∼€2.9 euros), and LFB identification (∼€1.7 euros), is estimated to be € 5.5 euros, which is cheaper than PCR-based techniques. More importantly, the N. gonorrhoeae-LAMP-PNB assay has great potential to develop a point-of-care (POC) testing method for identifying gonorrhea-suspected patients in clinical settings, especially in economically impoverished regions of the world.
Conclusion
In the current study, a reliable, rapid, and low-cost N. gonorrhoeae-LAMP-PNB technique targeting the orf1 gene was successfully devised for assaying gonococci isolates. Our data demonstrated that the N. gonorrhoeae-LAMP-PNB was a highly sensitive and specific diagnostic method and could be used as an attractive laboratory tool for diagnosis of gonorrhea in clinical settings. The test does not rely on expensive apparatus and reagents, and the whole process could be completed within 60 min. Hence, the N. gonorrhoeae-LAMP-PNB assay could be considered an ideal method for the reliable and rapid detection of N. gonorrhoeae in clinical application, particularly in resource-constrained regions of the world.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, and further inquiries can be directed to the corresponding authors.
Ethics Statement
The study was approved by the Human Ethics Committee of Hangzhou Women's Hospital (Approval No. [2021]-K (2)-8) complying with the Declaration of Helsinki. Before our research team obtained the clinical samples/isolates and conducted study, any personal identifiers of the suspected N. gonorrhoeae-infected patients were removed by the monitoring stations. So patients' informed consent were waived by the Human Ethics Committee of Hangzhou Women's Hospital.
Author Contributions
XC, QZ, SD, and WY conceived and designed the study. XC and WY participated in primer design. XC, QZ, XW, SW, and RL contributed to all the laboratory works. XW and SW contributed to data collection. XC and QZ performed statistical analysis. XC wrote the initial draft of the manuscript, and WY revised the manuscript. All the authors read and approved the final manuscript.
Funding
This work was supported by the Scientific and Technological Project in Guizhou Province (Grant Nos. Qian Ke He (2020)4Y184, (2019)1186, and (2018)2762), the Scientific and Technological Program in Guiyang City (Grant No. Zhu Ke He (2020)-10-5), and the Public Welfare Technology Research Program in Zhejiang Province (Grant No. LGF21H190001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all the medical workers in 2nd GZUTCM, Hangzhou Women’s Hospital, GZCCL, and Zhejiang Hospital for their cooperation in this study.
Abbreviations
ATCC, American Type Culture Collection; BC, blank control; CL, control line; DW, distilled water; FAM, carboxyfluorescein; GZCCL, Guizhou Provincial Center for Clinical Laboratory; LAMP, loop-mediated isothermal amplification; LAMP-PNB, loop-mediated isothermal amplification linked to a polymer nanoparticle–based biosensor; LoD, limit of detection; MG, malachite green; mer, monomeric unit; NC, negative control; nt, nucleotide; PCR, polymerase chain reaction; PNB, polymer nanoparticle–based biosensor; TL, test line; WHO, World Health Organization; ZJH, Zhejiang Hospital; 2nd GZUTCM, Second Affiliated Hospital, Guizhou University of Traditional Chinese Medicine.
References
Anniebell, S., and Gopinath, S. C. B. (2018). Polymer Conjugated Gold Nanoparticles in Biomedical Applications. Cmc 25, 1433–1445. doi:10.2174/0929867324666170116123633
Chan, P. A., Robinette, A., Montgomery, M., Almonte, A., Cu-Uvin, S., Lonks, J. R., et al. (2016). Extragenital Infections Caused by chlamydia Trachomatis and Neisseria Gonorrhoeae: A Review of the Literature. Infect. Dis. Obste.t Gynecol. 2016, 5758387. doi:10.1155/2016/5758387
Chaudhry, U., and Saluja, D. (2002). Detection of Neisseria Gonorrhoeae by PCR Using Orf1 Gene as Target. Sex. Transm. Infect. 78, 72. doi:10.1136/sti.78.1.72
Chemaitelly, H., Harfouche, M., Blondeel, K., Matsaseng, T. C., Kiarie, J., Toskin, I., et al. (2019). Global Epidemiology ofNeisseria Gonorrhoeaein Infertile Populations: Protocol for a Systematic Review. BMJ Open 9, e025808. doi:10.1136/bmjopen-2018-025808
Chen, X., Ma, K., Yi, X., Xiong, L., Wang, Y., and Li, S. (2020). The Rapid and Visual Detection of Methicillin-Susceptible and Methicillin-Resistant Staphylococcus aureus Using Multiplex Loop-Mediated Isothermal Amplification Linked to a Nanoparticle-Based Lateral Flow Biosensor. Antimicrob. Resist. Infect. Control 9, 111. doi:10.1186/s13756-020-00774-x
Dempsey, J. A., Litaker, W., Madhure, A., Snodgrass, T. L., and Cannon, J. G. (1991). Physical Map of the Chromosome of Neisseria Gonorrhoeae FA1090 with Locations of Genetic Markers, Including Opa and Pil Genes. J. Bacteriol. 173, 5476–5486. doi:10.1128/jb.173.17.5476-5486.1991
Edwards, T., Burke, P. A., Smalley, H. B., Gillies, L., and Hobbs, G. (2014). Loop-mediated Isothermal Amplification Test for Detection of Neisseria Gonorrhoeae in Urine Samples and Tolerance of the Assay to the Presence of Urea. J. Clin. Microbiol. 52, 2163–2165. doi:10.1128/jcm.00314-14
Fifer, H., Saunders, J., Soni, S., Sadiq, S. T., and FitzGerald, M. (2019). National Guideline for the Management of Infection with Neisseria Gonorrhoeae. British Association For Sexual Health And HIVWeb Site. Available online: www.bashhguidelines.org/media/1208/gc-2019.pdf.
Geraats-Peters, C. W. M., Brouwers, M., Schneeberger, P. M., van der Zanden, A. G. M., Bruisten, S. M., Weers-Pothoff, G., et al. (2005). Specific andSensitive Detection of Neisseria Gonorrhoeae in ClinicalSpecimens by Real-Time PCR. J. Clin. Microbiol. 43, 5653–5659. doi:10.1128/jcm.43.11.5653-5659.2005
Hook, E. W., and Bernstein, K. (2019). Kissing, Saliva Exchange, and Transmission of Neisseria Gonorrhoeae. Lancet Infect. Dis. 19, e367–e369. doi:10.1016/s1473-3099(19)30306-8
Li, S., Liu, Y., Wang, Y., Chen, H., Liu, C., and Wang, Y. (2019). Lateral Flow Biosensor Combined with Loop-Mediated Isothermal Amplification for Simple, Rapid, Sensitive, and Reliable Detection of Brucella Spp. Idr Vol. 12, 2343–2353. doi:10.2147/idr.s211644
Li, Y., Fan, P., Zhou, S., and Zhang, L. (2017). Loop-mediated Isothermal Amplification (LAMP): A Novel Rapid Detection Platform for Pathogens. Microb. Pathogenesis 107, 54–61. doi:10.1016/j.micpath.2017.03.016
Lovett, A., and Duncan, J. A. (2019). Human Immune Responses and the Natural History of Neisseria Gonorrhoeae Infection. Front. Immunol. 9, 3187. doi:10.3389/fimmu.2018.03187
Mahony, J. B., Luinstra, K. E., Tyndall, M., Sellors, J. W., Krepel, J., and Chernesky, M. (1995). Multiplex PCR for Detection of Chlamydia trachomatis and Neisseria Gonorrhoeae in Genitourinary Specimens. J. Clin. Microbiol. 33, 3049–3053. doi:10.1128/jcm.33.11.3049-3053.1995
Mensforth, S., Thorley, N., and Radcliffe, K. (2018). Auditing the Use and Assessing the Clinical Utility of Microscopy as a point-of-care Test for Neisseria Gonorrhoeae in a Sexual Health Clinic. Int. J. STD. AIDS. 29, 157–163. doi:10.1177/0956462417721062
Meyer, T., and Buder, S. (2020). The Laboratory Diagnosis of Neisseria Gonorrhoeae: Current Testing and Future Demands. Pathogens 9, 91. doi:10.3390/pathogens9020091
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., et al. (2000). Loop-mediated Isothermal Amplification of DNA. Nucleic Acids Res. 28, E63. doi:10.1093/nar/28.12.e63
Notomi, T., Mori, Y., Tomita, N., and Kanda, H. (2015). Loop-mediated Isothermal Amplification (LAMP): Principle, Features, and Future Prospects. J. Microbiol. 53, 1–5. doi:10.1007/s12275-015-4656-9
O'Callaghan, I., Corcoran, D., and Lucey, B. (2010). Design of a Multiplex PCR Assay for the Simultaneous Detection and Confirmation of Neisseria Gonorrhoeae. J. Clin. Pathol. 63, 431–433. doi:10.1136/jcp.2009.071431
Quesada-González, D., and Merkoçi, A. (2015). Nanoparticle-based Lateral Flow Biosensors. Biosens. Bioelectron. 73, 47–63. doi:10.1016/j.bios.2015.05.050
Quillin, S. J., and Seifert, H. S. (2018). Neisseria Gonorrhoeae Host Adaptation and Pathogenesis. Na.t Rev. Microbiol. 16, 226–240. doi:10.1038/nrmicro.2017.169
Rowley, J., Vander Hoorn, S., Korenromp, E., Low, N., Unemo, M., Abu-Raddad, L. J., et al. (2019). Chlamydia, Gonorrhoea, Trichomoniasis and Syphilis: Global Prevalence and Incidence Estimates, 2016. Bull. World Health Organ. 97, 548–562P. doi:10.2471/blt.18.228486
Shete, P. B., Farr, K., Strnad, L., Gray, C. M., and Cattamanchi, A. (2019). Diagnostic Accuracy of TB-LAMP for Pulmonary Tuberculosis: a Systematic Review and Meta-Analysis. BMC. Infect. Dis. 19, 268. doi:10.1186/s12879-019-3881-y
Stevens, J. S., and Criss, A. K. (2018). Pathogenesis of Neisseria Gonorrhoeae in the Female Reproductive Tract. Curr. Opin. Hematol. 25, 13–21. doi:10.1097/moh.0000000000000394
Vallely, L. M., Egli-Gany, D., Wand, H., Pomat, W. S., Homer, C. S. E., Guy, R., et al. (2021). Adverse Pregnancy and Neonatal Outcomes Associated withNeisseria Gonorrhoeae:systematic Review and Meta-Analysis. Sex. Transm. Infect. 97, 104–111. doi:10.1136/sextrans-2020-054653
Visser, M., van Westreenen, M., van Bergen, J., and van Benthem, B. H. B. (2020). Low Gonorrhoea Antimicrobial Resistance and Culture Positivity Rates in General Practice: a Pilot Study. Sex. Transm. Infect. 96, 220–222. doi:10.1136/sextrans-2019-054006
Wang, C., Gao, X., Chen, Z., Chen, Y., and Chen, H. (2017). Preparation, Characterization and Application of Polysaccharide-Based Metallic Nanoparticles: A Review. Polymers 9, 689. doi:10.3390/polym9120689
Whiley, D. M., LeCornec, G. M., Mackay, I. M., Siebert, D. J., and Sloots, T. P. (2002). A Real-Time PCR Assay for the Detection of Neisseria Gonorrhoeae by LightCycler. Diagn. Microbiol. Infect. Dis. 42, 85–89. doi:10.1016/s0732-8893(01)00326-1
Wong, Y.-P., Othman, S., Lau, Y.-L., Radu, S., and Chee, H.-Y. (2018). Loop-mediated Isothermal Amplification (LAMP): a Versatile Technique for Detection of Micro-organisms. J. Appl. Microbiol. 124, 626–643. doi:10.1111/jam.13647
World Health Organization (2013). Laboratory Diagnosis of Sexually Transmitted Infections, Including Human Immunodeficiency Virus. geneva, Switzerland: World Health Organization. Available online: https://www.who.int/reproductivehealth/publications/rtis/9789241505840/en/
World Health Organization (2016). WHO Guidelines for the Treatment of Neisseria Gonorrhoeae. geneva, Switzerland: World Health Organization. Available online: https://www.ncbi.nlm.nih.gov/books/NBK379221/
Zhang, H., Xu, Y., Fohlerova, Z., Chang, H., Iliescu, C., and Neuzil, P. (2019). LAMP-on-a-chip: Revising Microfluidic Platforms for Loop-Mediated DNA Amplification. Trac Trends Anal. Chem. 113, 44–53. doi:10.1016/j.trac.2019.01.015
Keywords: Neisseria gonorrhoeae, loop-mediated isothermal amplification, polymer nanoparticle–based biosensor, limit of detection, point-of-care testing
Citation: Chen X, Zhou Q, Wu X, Wang S, Liu R, Dong S and Yuan W (2021) Visual and Rapid Diagnosis of Neisseria gonorrhoeae Using Loop-Mediated Isothermal Amplification Combined With a Polymer Nanoparticle–Based Biosensor in Clinical Application. Front. Mol. Biosci. 8:702134. doi: 10.3389/fmolb.2021.702134
Received: 29 April 2021; Accepted: 23 June 2021;
Published: 21 July 2021.
Edited by:
Jeesu Kim, Pusan National University, South KoreaReviewed by:
Manuela Oliveira, University of Lisbon, PortugalRavindra K. Sharma, University of Florida, United States
Copyright © 2021 Chen, Zhou, Wu, Wang, Liu, Dong and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shilei Dong, ZHNsMTY2QDEyNi5jb20=; Wei Yuan, eXVhbndlaTE4MUAxMjYuY29t
†These authors have contributed equally to this work
 Xu Chen
Xu Chen Qingxue Zhou3†
Qingxue Zhou3† Shilei Dong
Shilei Dong