- 1The Second Clinical School, Southern Medical University, Guangzhou, China
- 2Eye Institute and Department of Ophthalmology, Eye & ENT Hospital, Fudan University, Shanghai, China
- 3NHC Key Laboratory of Myopia, Fudan University, Shanghai, China
- 4Key Laboratory of Myopia, Chinese Academy of Medical Sciences, Shanghai, China
- 5Shanghai Key Laboratory of Visual Impairment and Restoration, Shanghai, China
- 6Department of Ophthalmology, South China Hospital, Health Science Center, Shenzhen University, Shenzhen, China
- 7Medical College of Rehabiliation, Southern Medical University, Guangzhou, China
- 8The University of South China, Hengyang, China
- 9Department of Anesthesiology, Shenzhen Hospital, Southern Medical University, Shenzhen, China
- 10Department of Ophthalmology, the Sixth Affiliated Hospital of Sun Yat-sen University, Guangzhou, China
- 11Department of Ophthalmology, Zhujiang Hospital, Southern Medical University, Guangzhou, China
The retina is composed of 11 types of cells, including neurons, glial cells and vascular bed cells. It contains five types of neurons, each with specific physiological, morphological, and molecular definitions. Currently, single-cell RNA sequencing (sRNA-seq) is emerging as one of the most powerful tools to reveal the complexity of the retina. The continuous discovery of retina-related gene targets plays an important role in helping us understand the nature of diseases. The revelation of new cell subpopulations can focus the occurrence and development of diseases on specific biological activities of specific cells. In addition, sRNA-seq performs high-throughput sequencing analysis of epigenetics, transcriptome and genome at the single-cell level, with the advantages of high-throughput and high-resolution. In this paper, we systematically review the development history of sRNA-seq technology, and summarize the new subtypes of retinal cells and some specific gene markers discovered by this technology. The progress in the diagnosis of retinal related diseases is also discussed.
Introduction
With the development of high-throughput sequencing technology, humans can already analyze genomes and their products on a large scale, including DNA sequences, chromatin structure, RNA transcripts, proteins and metabolites (Botond, 2018). Traditional high-throughput sequencing requires sufficient DNA samples to be obtained from a large number of cells. However, the accuracy of high-throughput sequencing is quite low, and the result of sequencing should be corrected. Single-cell RNA sequencing (scRNA-seq) refers to the technology of high-throughput sequencing analysis of the genome, transcriptome and epigenetic genome at the single cell level. Currently, scRNA-seq technology is commonly used in the fields such as development of stem cell, embryo and tumor. For example, in the study of tumor tissues, researchers classify subgroups based on single-cell transcription maps (Masland, 2012; Wang et al., 2014), and based on the gene expression profiles, they can study the mechanism of cancer cell metastasis (Zheng et al., 2017) and discover new targets for immunotherapy (Pauly et al., 2019).
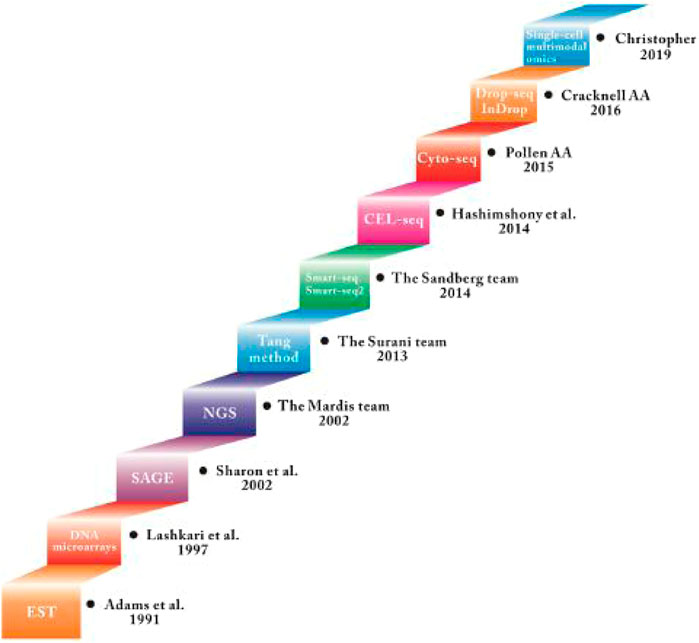
In the field of ophthalmology, single-cell RNA sequencing research has been mostly applied to retina, from cell subtypes to targeted treatments for related diseases. Both humans and monkeys have fovea and macula, but mice are nocturnal dichromats and humans are diurnal trichromats. Therefore, studies on subtypes of retinal cells in humans and primates should ideally be published separately (Pauly et al., 2019). This review summarizes and discusses the latest progress and applications of scRNA-seq technology in the field of retina. So far, scRNA-Seq has been used in mouse, primate, human embryo and adult retinal tissue cell subtype research, as well as the pathogenic gene pathway research of various retinal-related diseases. In this review, we systematically reviewed the rapid progress of single-cell technology (Figure 1) and summarized the current challenges and unanswered questions in the field of retinal development and disease.
Development of Single-Cell RNA Sequencing Technology
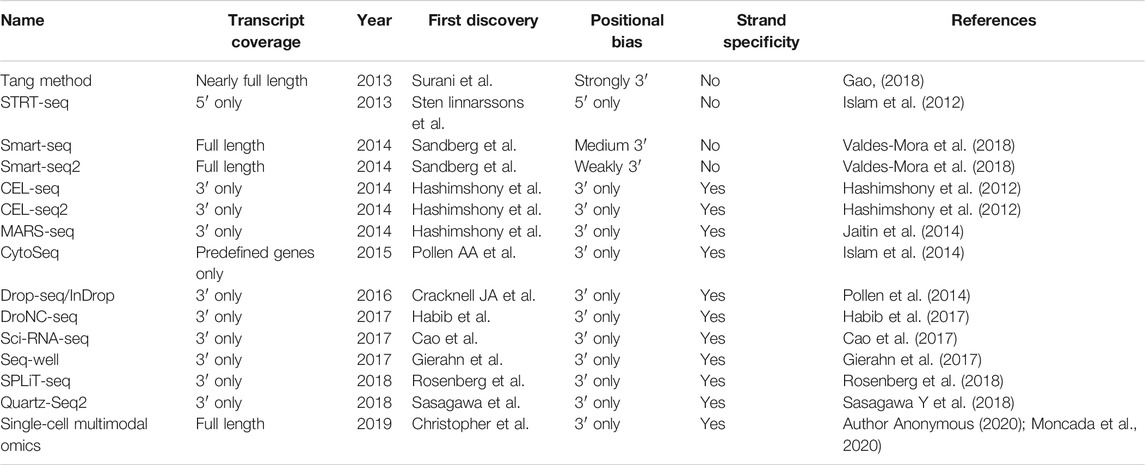
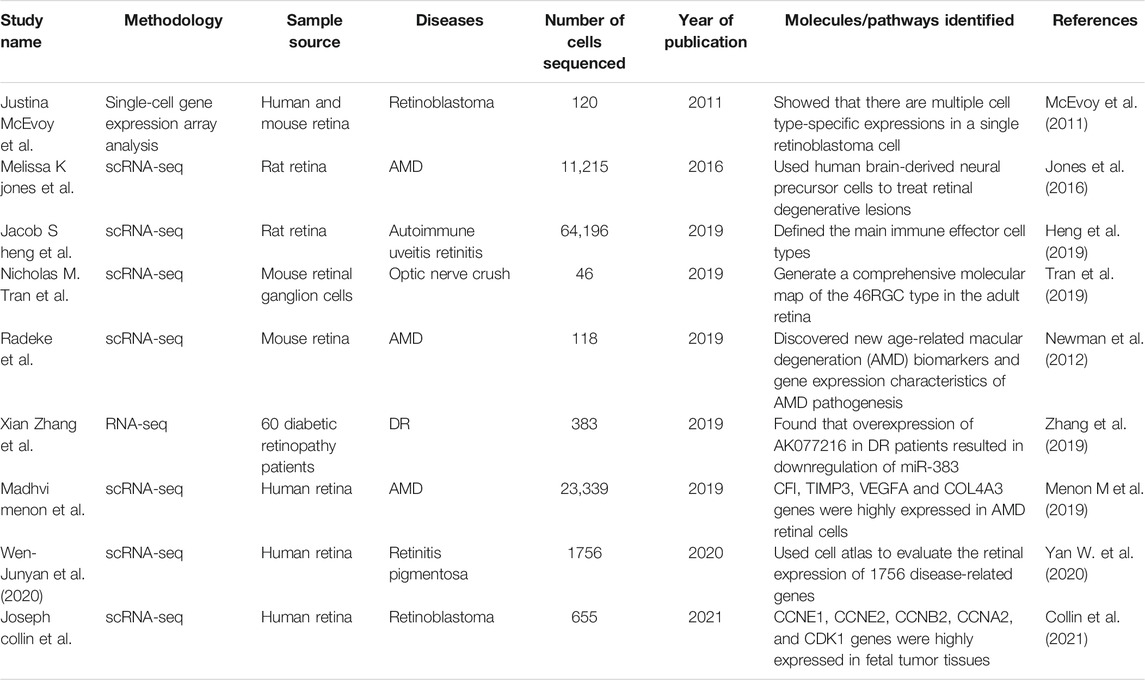
Single-cell transcriptome sequencing technology (scRNA-seq) is to analyze the expression profile of the cell transcriptome from the single cell level to identify cell-specific markers, discover rare cell types, cell subtypes, and reveal differences between cells expression (Zerti et al., 2020). The basic technical principles of scRNA-seq technology include: 1) separation technology, such as micromanipulation, laser capture microdissection, fluorescence activated cell sorting, 2) single-cell transcriptome amplification and sequencing library construction. Cells are the basic structural and functional units of organisms (Zerti et al., 2020). During their growth and development, due to different cell states and environmental stimuli, changes in transcriptome information show diversified manifestations. scRNA-seq can study the differential expression of RNA from a single cell level. Since the Tang team first applied scRNA-seq technology in 2009, scRNA-seq technology has received more research and development (Table 1). Besides, single-cell sequencing technology has been used to study stem cell differentiation, embryonic organ development, tumor tissue, immune tissue, nervous tissue, and other fields in recent years (Picelli, 2017). In the field of ophthalmology, it is mainly used to study the gene expression of normal retinal tissues and common retinal diseases, such as age-related macular degeneration and diabetic retinopathy.
Application of scRNA-Seq in Normal Retinal Tissue
In the field of ophthalmology, the single-cell RNA sequencing research in the past 5 years has mainly focused on the retina, and most of the research focuses on the exploration of cell subtypes, related genes and pathway. In particular, many researchers choose amacrine cells, bipolar cells and microglia cells for study. Among them, amacrine cells are the most diverse neurons, and most of them lack obvious molecular markers (Grunert and Martin, 1991), which has stimulated curiosity of various researchers in recent years. The retina is a highly heterogeneous tissue, and it is estimated that there are more than 100 nerve cell subtypes.
Primates (including humans) have a fovea on the retina, which is a small central area responsible for high vision and most color vision. However, the retina of mice does not have a fovea. This difference also limits some experimental studies. The distribution and number of primate and mouse retinal cells have a certain difference between the fovea and the periphery. Yi-Rong Peng et al. (Peng et al., 2019) used 165,000 single-cell RNA sequence maps to perform a comprehensive cell classification of the central fovea and peripheral retina of rhesus monkeys, of which 64 fovea (3 PRs, 2 HC, 12 BC, 27 AC, 16 RGC and four non-neurons) and 71 peripherals (2 PR, 2 HC, 11 BC, 34 AC, 18 RGC and four non-neurons) clusters. Comparison with the mouse retina type shows that the middle neuron type is tightly conserved, but the type and procedure of the projection neuron are different.
Based on the previous study, Wen-jun Yan et al. (Yan et al., 2020) compared the gene expression characteristics of human and cynomolgus monkey and fascicular monkey cell types. Besides, they identified five types of neurons (9,070 photoreceptors, 2,868 horizontal cells, 25,908 bipolar cells, 13,607 amacrine cells and 11,404 RGCs) and four types of non-neuronal cells. By comparing the retinal cell types of human and rhesus monkeys, the differentially expressed genes are summarized: the genes that are highly expressed in rhesus monkey retinal tissues are EPHX2, DB1 and DB6. GPATCH1 and CRHBP genes are highly expressed in human retinal tissues. In addition, by comparing retinal foveal cells with surrounding cells, they found that: 1) EPB41L2 and VTN are expressed by the fovea instead of the peripheral cone. 2) expression level of TTR in the fovea is higher than that of the surrounding bipolar types of DB3b and DB4. 3)TULP1 is expressed by peripheral but not foveal bipolar FMB and DB2. Besides, the transcriptome of approximately 85,000 cells from the fovea and surrounding retinas of seven adult donors was analyzed by Wen-jun Yan et al. (Yan W et al., 2020) using single-cell RNA sequencing (sRNA-seq) in 2020. The results showed that FOXP2+, FOXP1-, FOXP2+, FOXP1+, and F-RGCs were highly expressed in RGCS cell clusters. In addition, the comparison showed that more than 90% of the human types were transcriptionally consistent with those previously identified in macaques, and that the expression of disease-related genes was highly conserved between the two species. These results confirm the usefulness of macaques in simulating blinding diseases and provide a basis for investigating the molecular mechanisms of visual processing.
Regarding the study of fovea and peripheral cells, Sharon et al. (Banin et al., 2015) have reported 20 highly expressed genes in the macular region (such as SLC17A6, SNCG, NEFL, NET1, STMN2, YWHAH, UCHL1, DPYSL2, APP, NDRG4, TUBA1B,MDH1, EEF2) and 23 highly expressed genes in the peripheral region of the retina (SAG, RCVRN, UNC119, GPX3, PDE6G, ROM1, ABCA4, DDC, PDE6B, GNB1, NRL); based on bulk RNA Seq, Li et al. (Li et al., 2004) reported 1,239 The macular area is highly expressed and the 812 peripheral areas are highly expressed. These related studies provide a basic framework for single-cell analysis of species and across tissue regions.
In addition, scRNA-seq can be used to further study genetic markers and typing of specific optic nerve tissues and retinal cells. Macosko et al. (Macosko et al., 2015) analyzed the transcripts of 44,808 mouse retinal cells and identified 39 different transcribed cell populations, establishing a molecular map of gene expression for known retinal cell types and new candidate cell subtypes. Among them, 21 clusters of amacrine cells were mainly studied. 12 were identified as GABAergic (Gad1 and/or Gad2 positive), and the other five were glycine (Glcine transporter Slc6a9 positive). Ebf3 is a transcription factor found in SEG-glycine and nGnG-amacrine proteins and is specific for clusters 17 and 20.
To further study the gene expression of bipolar cells, Shekhar et al. (Shekhar et al., 2016) used mouse retinal bipolar cells (BCs) as the research object through DROP-SEQ and classified them by two different criteria. Firstly, according to whether the RBC is marked or not, a rod-shaped or cone-shaped BC is divided; secondly, according to the bipolar mark Isl1 and/or Grm6, the cone BC cluster can be further divided into on (3–6, 13, 15) and off (7–10, 12, 14) BC type. It is also worth mentioning that on the basis of the predecessors, the team further divided four types of BC5 (BC5A-BC5D), specifically BC5A (Sox6+) and BC5B (Chrm2+), BC5C (Slitrk5+), BC5D (Lrrtm1+).
On the basis of previous studies, O'Koren team (O'Koren et al., 2019) used single-cell sequencing to reveal the unique transcriptome-related genes of microglia in photoreceptor degeneration, such as Lsp1, asApoe, Ppiaf4, and Alox5ap, which were temporarily induced in the middle of the trajectory; Fabp5, Lgals3, Cd63, Lpl, Cybb, Mmp12, and Spp1 are adjusted up late in the trajectory.
The developmental pathways of mouse neural retina (NR) and retinal pigment epithelium (RPE) have been extensively revealed. However, the molecular mechanism of human NR and RPE formation and the interaction between these two tissues have not been well elucidated (Dulken et al., 2017). In recent years, some studies have used scRNA-seq technology to conduct experimental design with retinal multifunctional stem cells (RPCs) as the research object. RPCs are located in the inner layer of the optic cup (Oppikofer et al., 2017). They produced six types of neurons in retinal cells. The processes that retinal development needs to go through: RPC proliferation, cell fate determination, and specific neuronal differentiation (Gordon et al., 2013).
Yuqiong Hu et al. (Trimarchi et al., 2009) identified the main cell types of human fetal retina, which are RGC expressing γ-synuclein (SNCG), NEFL, ATOH7 and EBF3; HCs express ONECUT1/2/3; ACs express MEIS2, GAD1 and GAD2; BCs express VSX1 and VSX2; PCs express PDC, PDE6G, SAG, CRX and NRL; microglia express CX3CR1, C1QA, C1QB and C1QC; fibroblasts express COL3A1 and COL1A1.
According to the report, Xiying Mao et al. (Mao et al., 2019) found that the RPC specific markers VSX2 and PAX6 were co-expressed 28 days ago; after 28 days, the expression of VSX2 began to disappear on the central basal side of the retina, expressing the retinal ganglion cell (RGC) marker ELAVL3/The cells of four began to appear simultaneously, and the number of ELAVL3/4 positive cells gradually increased thereafter. HES1 and HES5 are briefly activated in RPC (Lukaszewicz and Anderson, 2011) and then suppressed in terminally differentiated neurons, and HES6 continues to be up-regulated after the lineage bifurcation point.
Based on the previous evidence, Brian S. Clark et al. (Clark et al., 2019) used single-cell RNA sequencing to describe ten developmental stages covering the entire process of retinal neurogenesis, our results indicate that NFI transcription factors (NFIA, NFIB, and NFIX) are selectively expressed in late RPCs and indicate that they regulate the fate of bipolar interneurons and Miller glial cells and promote proliferation and imactivation. Besides, Mariona Esquerdo-Barragán team (Esquerdo-Barragán et al., 2019) found that TOPORS, KLHL7, PRPF8, USP45, and Usp-20 were expressed at low levels in the retina through scRNA-seq. Josd1, Pan2, Usp11, Usp14, Usp15, Usp10, Usp22, Usp39 and cone cells Compared to the expression of rod differentiation, the expression of three genes (Otud7b, Usp46, and Usp48) increased in late cone cells; the expression of Usp45, Usp53, and Usp54 was limited to the photosensitive layer; Usp28, Usp37, or Otub1 is highly expressed in the embryonic period, but expression is stopped after birth; Usp12, Zranb1 or Usp32, its expression is extremely low in the embryonic period, but significantly increased before and after birth (Hojo et al., 2000). These genes are related to the ubiquitin proteasome system (UPS), which has important research significance for retinal precursor cell differentiation.
In order to further explain the tissue structure and cell subtypes of the chicken retina, based on previous studies, this year Masahito Yamagata et al. (Yamagata M and Sanes, 2021)used single-cell RNA sequencing (sRNA-seq) to generate a cellular atlas of chicken retinas (40,000 single-cell transcriptome), 136 cell types plus 14 sites or developmental intermediates were identified. The team mapped genes expressed in the majority of three types of retinal cells, namely VSX2 (CHX10) in the basal cells, TFAP2A in the central retinal cells, and RBPMS2 in retinal ganglion cells. The results provide new insights into the structure and evolution of the retina and lay the foundation for the study of the anatomy, physiology and development of the retina in birds.
For the past few years, the continuous application and development of scRNA-seq technology has been improved. The study of normal retinal cells in animals and human eyes can redefine the cluster of cells based on the marker gene (Table 2). It also enables a deeper understanding of tissue cells and subsequently the cluster of cells. Carrying out a more in-depth classification of cells helps to understand the heterogeneity of cells well, and also brings along a new perspective for our subsequent diagnosis and treatment of disease.
Application of scRNA-Seq in Retinal Diseases
scRNA-Seq in the Research of Targeted Therapy of Ocular Tumors Application
Different cells change differently at seperate stages of the disease. The transcriptome of many cell subtypes in the retina, especially rare cells, is usually obscured by a large number of RNA sequences. Therefore, understanding the transcriptome at the cell type or single cell level will expand research related to disease.
Single-cell RNA-seq can be used for targeted therapy of eye tumors (Kawaguchi et al., 2008). It is well established that the molecular and cellular characteristics of tumors can indicate the origin of tumor cells and provide a basis for targeted therapy. Retinoblastoma is a malignant tumor in infants and young children. In recent years, researchers have used single-cell sequencing technology to study the pathogenetic gene pathway and treatment of it.
Mcevoy et al. (McEvoy et al., 2011) performed single-cell gene expression array analysis on tumor cells of retinoblastoma patients and mouse models, showing that multiple cell types are specifically expressed in a single retinoblastoma cell. The results showed that human retinoblastoma expressed high levels of MDMX gene and MDMX protein. Some monoamine/catecholamine receptors in mice include serotonin receptors (HTR3A, HTR1E), dopamine receptors (DRD5) and histamine receptors (HRH3) Expression levels in retinoblastoma It is equal to or higher than the normal human retina.
Based on the previous study, Joseph Collin et al. (Collin et al., 2021) used nine human embryonic and fetal retinal tissues by sRNA-seq and ATAC sequence method. The results showed that Glu137Ter and Tyr655Ter were highly expressed in 4 month old embryonic tumor tissues. However, the Rb1c.763C and Arg255TER genes were overexpressed in embryonic tumor tissue at 34 months. In addition, CCNE1, CCNE2, CCNB2, CCNA2, and CDK1 genes were highly expressed in fetal tumor tissues. In addition, this study provides evidence of the heterogeneity of RB tumors and defines molecular pathways and new targeted therapeutic strategies.
scRNA-Seq on the Pathogenesis of Age-Related Macular Degeneration and Treatment Research
In addition, single-cell sequencing technology also aids the study and treatment of retinal degenerative and vision loss diseases by analyzing the pathogenesis of related diseases and discovering new biological targets and markers.
Radeke et al. (Newman et al., 2012) discovered new age-related macular degeneration (AMD) biomarkers and gene expression characteristics of AMD pathogenesis. These findings indicate that the cell-based inflammatory response in the RPE choroid is a core feature of AMD. All AMD phenotypes in the RPE choroid are associated with high expression of all or a subset of the following chemokines, namely CXCL1, CXCL2, CXCL9, CXCL10, CXCL11, CCL2 and CCL8. AMD expression in retinal pigment epithelium Related bases and chemokines are C10orf18, ARL9, CXCL10, FZD10, CTSL2, CXCL. AMD may be a single disease with a common immune response process. The genes that regulate these immune activities, as well as many other genes found, represent promising new targets for the treatment and diagnosis of AMD.
On the basis of the previous research, Madhvi Menon et al. (Menon M et al., 2019) retinal cells were isolated and sequenced from six postmortem human retinal macular and surrounding panretinal suspension using droplet based microfluidic (20,091 cells) and nanopore based Seq Well (3,248 cells) to investigate cell types associated with age-related macular degeneration. The results showed that CFI, TIMP3, VEGFA and COL4A3 genes were highly expressed in AMD retinal cells.
Besides, Jones et al. (Jones et al., 2016) used human brain-derived neural precursor cells (hNPCs) to treat retinal degenerative lesions. The results showed that the top five genes with the greatest changes included Mir671, Lcn2, Cd74, Gfap, and Cebpd; Lcn2, Cd74, Gfap, and Cebpd (Hughes et al., 2003). All show that as retinal degeneration increases, Mir671, Lcn2, Cd74, and Cebpd play a role in the immune response of macrophages and/or microglia, suggesting that the activity of macrophages/microglia increases as the retina degenerates (Lawson et al., 1990).
scRNA-Seq on the Pathogenesis of Diabetic Retinopathy and Treatment Research
In recent years, a series of studies on Diabetic Retinopathy (DR) have suggested that vision loss in DR patients is no longer considered to be a simple microvascular complication, also known as neurodegenerative disease (Kamalden et al., 2017). Different retinal cells, trophic factors, neurotransmitters, and inflammatory factors play an important role in the pathogenesis of diabetic retinopathy (Kamalden et al., 2017). Moreover, there are not many studies on diabetic retinopathy by single cell histology (Pastukh et al., 2019).
Xian Zhang et al. (Zhang et al., 2019) found that overexpression of AK077216 in DR patients resulted in downregulation of miR-383, but overexpression of miR-383 had no significant effect on the expression of AK077216; overexpression of AK077216 inhibited apoptosis of ARPE-19 cells (Ru et al., 2014), miR Overexpression of -383 plays the opposite role and attenuates the overexpression of AK077216; therefore it is concluded that AK077216 is down-regulated in diabetic retinopathy, and inhibits ARPE-19 cell apoptosis by down-regulating miR-383. Zimeng Li et al. found that miR-4448, miR-338-3p, miR-190a-5p, miR-485-5p and miR-9-5p are highly expressed in the serum of DR patients (Shaker et al., 2019).
Application of scRNA-Seq Studies in Other Retinal Diseases
The types of neurons in the central nervous system are significantly different in terms of resilience to injuries or other injuries (Della Santina et al., 2013). In recent years, some researchers have provided a systematic framework to analyze the specific types of injuries through single-cell sequencing technology. Sexual response, and demonstrate that differential gene expression can be used to reveal the molecular targets of intervention.
Nicholas Tran et al. (Tran et al., 2019) first used single-cell RNA-seq (scRNA-seq) to generate a comprehensive molecular map of the 46RGC type in the adult retina. By tracking their survival after ONC (Optic Nerve Crush), the transcription and morphological changes before degradation were described, and each type of selectively expressed genes was determined. Among them, Igf1 (7/7 resRGCs), Opn4 (5/7) and Spp1 (3/7)/OE-Ucn, Ucn protein, OE-Timp2, KO-Crhbp, and KOMmp9 all promoted significant overall regeneration of the optic nerve.
Another experimental study, using single-cell sequencing technology, determined whether multiple explosion exposures caused greater damage to RGC than single explosion exposures (Hong et al., 2015). The results show that Cd40, Mrpl34, Kmo, Lmcd1, BC030870, I830077J02Rik, and Ms4a14 (Kim et al., 2008) are related genes that mediate neuroprotection.
scRNA-seq has been used as a comprehensive and fair method to study cell types and gene expression patterns in the retina of spontaneous, chronic and progressive autoimmune uveitis. Jacob et al. (Heng et al., 2019) used Aire−/− mice to establish a model of autoimmune uveitis retinitis. Mouse models offer a unique opportunity to study the mechanisms of autoimmune uveiretinitis, which is an important cause of vision loss. The team characterized 64,196 isolated retinal cells from eight samples using a droplet based sRNA-seq platform (10×genomics). The results showed that experimental uveiretinitis is a T-cell-driven disease, and the highly expressed genes in the following types of cells were: Th1 cells (T-bet+, IFNG+, CXCR6+, CD4+, CD8a, KLRA1), CD8a + T cells (CD8a+, CD4−, KLRA1), T follicular helper cells (BCL6+, CXCR5+, CD4+, CD8a) and regulatory T cells (Foxp3+, CD4+, IL10+). In addition, TGFb2 is the main TGF-β family member expressed in Aire mouse retina, mainly in the inner layer (INL). In conclusion, this study supports a similar central role of Th1 cells in Aire/uveoretitis, which has important implications for clinical treatment.
Besides, Wen-junYan et al. (Yu-Wai-Man et al., 2010) used cell atlas to evaluate the retinal expression of 1756 disease-related genes. Studies have shown that among the genes associated with retinitis pigmentosa (RP), RPGR and TOPORS, SLC25A46, SLC7A14 and RP9 are highly expressed in RGC (Delettre et al., 2002). In addition, RGR and RLBP1 are highly expressed in Müller glial cells. CRX, RAX2, GNAT2, PDE6H genes are highly expressed in rods and cones. RHO, NRL and NR2E3, all show the enrichment of the fovea (Miller et al., 2019). Lebers congenital amaurosis (LCA) is a group of severe hereditary retinal dystrophy, which is characterized by nystagmus, delayed or missing pupil light reflection, and blindness. Experimental results show that CEP290, GUCY2D and CRB1 genes are highly expressed in RGC (Anguita et al., 2021). In studies related to congenital quiescent night blindness (CSNB), it was found that GNAT1 and SLC24A1 were highly expressed in rod cells, while GRM6 and TRPM1 were highly expressed in bipolar cells (Clemons et al., 2013).
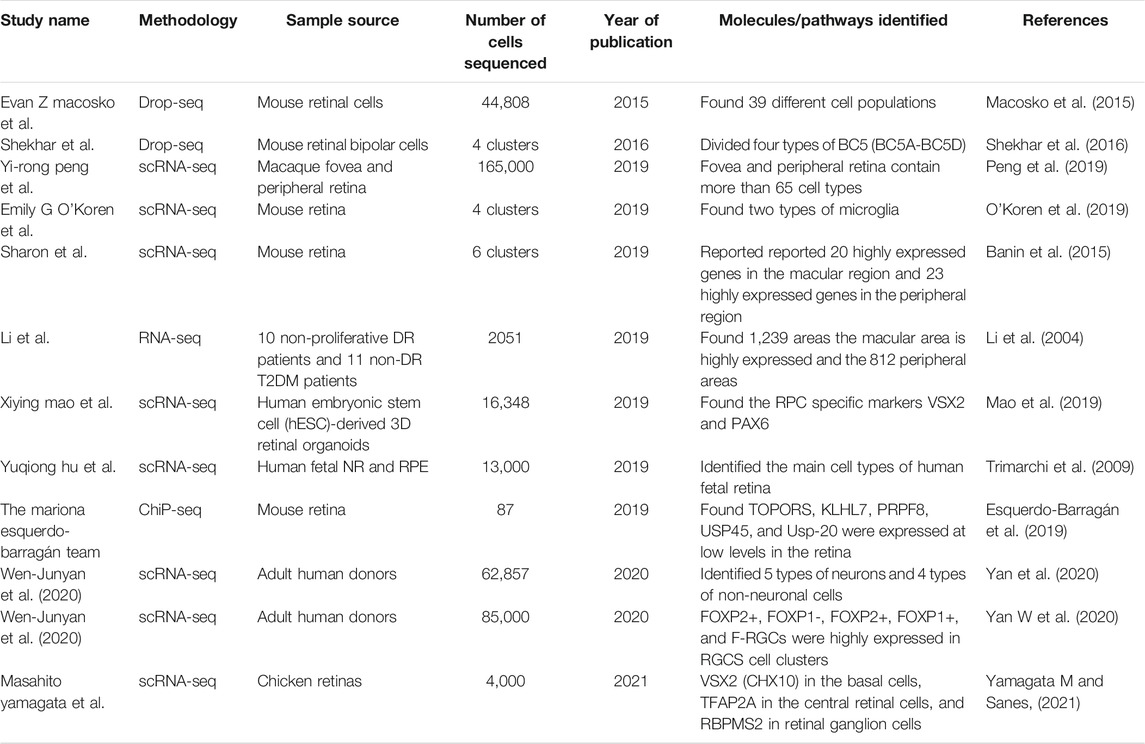
It can be seen that sc RNA-seq research can provide differentiated gene expression of cells for retinal diseases, transcription factor prediction, and the network communication interaction of each cell in the process of disease progression, which can provide new targets for the diagnosis and treatment of disease prediction. Using this technology, we can discover new cell subtypes and identify genetic markers of individual retinal subtype cells to help study and locate targets related to specific visual functions, thereby gaining a deeper understanding of cell function and cell heterogeneity explore the establishment of genetic networks that maintain cell diversity (Table 3).
Conclusion
Single-cell sequencing has opened up a new field to study different cell subtypes and genetic markers, and reveal the development mechanism and therapeutic targets of retinal-related diseases, and established itself as a valuable and unique tool to further study retinal tissue at the cellular level.
Single-cell sequencing can be used to study the classification of cell types and subtypes in the retina at the transcriptome level, and can help solve the heterogeneity and molecular complexity of the retina. An ideal scRNA-seq method can be used to analyze all coding and efficient non-coding cell transcripts, and even reveal subtle changes in gene expression. The past decade has witnessed significant technological development ever since the first scRNA-seq protocol was published in 2009 (Baden et al., 2016). With the steady decline in sequencing costs and the introduction of methods to significantly increase production every year, the genome, transcriptome, epigenome, and proteome of millions of cells can be sequenced simultaneously in the near future (Benowitz et al., 2017).
The main remaining problem is the challenge of efficiently separating individual cells from biological samples and analyzing large amounts of sequencing data. The close combination of scRNA-seq and bioinformatics technology can provide a powerful detection method to reveal the gene regulatory networks during cell development and differentiation. At present, the application of scrNA-seq in ophthalmic research is still limited. With the continuous progress of the technology, it may be rapidly expanded to the research of ocular diseases in the next few years.
Author Contributions
PY, CH and YW have contributed equally to this work and share co-first authorship. Other authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Anguita, R., Tasiopoulou, A., Shahid, S., Roth, J., Sim, S. Y., and Patel, P. J. (2021). A Review of Aflibercept Treatment for Macular Disease[J]. Ophthalmol. Ther. doi:10.1007/s40123-021-00354-1
Author Anonymous (2020). Method of the Year 2019: Single-Cell Multimodal Omics. Nat. Methods 17 (1), 1. doi:10.1038/s41592-019-0703-5
Baden, T., Berens, P., Franke, K., Román Rosón, M., Bethge, M., and Euler, T. (2016). The Functional Diversity of Retinal Ganglion Cells in the Mouse. Nature 529 (7586), 345–350. doi:10.1038/nature16468
Banin, E., Gootwine, E., Obolensky, A., Ezra-Elia, R., Ejzenberg, A., Zelinger, L., et al. (2015). Gene Augmentation Therapy Restores Retinal Function and Visual Behavior in a Sheep Model of CNGA3 Achromatopsia. Mol. Ther. 23 (9), 1423–1433. doi:10.1038/mt.2015.114
Benowitz, L. I., He, Z., and Goldberg, J. L. (2017). Reaching the Brain: Advances in Optic Nerve Regeneration. Exp. Neurol. 287 (3), 365–373. doi:10.1016/j.expneurol.2015.12.015
Botond, R. (2018). The First Steps in Vision: Cell Types, Circuits, and Repair[J]. EMBO Mol. Med. 11 (3), e10218. doi:10.15252/emmm.201810218
Cao, J., Packer, J. S., Ramani, V., Cusanovich, D. A., Huynh, C., Daza, R., et al. (2017). Comprehensive Single-Cell Transcriptional Profiling of a Multicellular Organism. Science 357 (6352), 661–667. doi:10.1126/science.aam8940
Clark, B. S., Stein-O’Brien, G. L., Shiau, F., Cannon, G. H., Davis-Marcisak, E., Sherman, T., et al. (2019). Single-Cell RNA-Seq Analysis of Retinal Development Identifies NFI Factors as Regulating Mitotic Exit and Late-Born Cell Specification. Neuron 102 (6), 1111–1126. doi:10.1016/j.neuron.2019.04.010
Clemons, T. E., Gillies, M. C., Chew, E. Y., Bird, A. C., Peto, T., Wang, J. J., et al. (2013). Medical Characteristics of Patients with Macular Telangiectasia Type 2 (MacTel Type 2) MacTel Project Report No. 3. Ophthalmic Epidemiol. 20 (2), 109–113. doi:10.3109/09286586.2013.766757
Collin, J., Queen, R., Zerti, D., Steel, D. H., Bowen, C., Parulekar, M., et al. (2021). Dissecting the Transcriptional and Chromatin Accessibility Heterogeneity of Proliferating Cone Precursors in Human Retinoblastoma Tumors by Single Cell Sequencing-Opening Pathways to New Therapeutic Strategies? Invest. Ophthalmol. Vis. Sci. 62 (6), 18. doi:10.1167/iovs.62.6.18
Delettre, C., Lenaers, G., Pelloquin, L., Belenguer, P., and Hamel, C. P. (2002). OPA1 (Kjer Type) Dominant Optic Atrophy: A Novel Mitochondrial Disease. Mol. Genet. Metab. 75 (2), 97–107. doi:10.1006/mgme.2001.3278
Della Santina, L., Inman, D. M., Lupien, C. B., Horner, P. J., and Wong, R. O. L. (2013). Differential Progression of Structural and Functional Alterations in Distinct Retinal Ganglion Cell Types in a Mouse Model of Glaucoma. J. Neurosci. 33 (44), 17444–17457. doi:10.1523/jneurosci.5461-12.2013
Dulken, B. W., Leeman, D. S., Boutet, S. C., Hebestreit, K., and Brunet, A. (2017). Single-Cell Transcriptomic Analysis Defines Heterogeneity and Transcriptional Dynamics in the Adult Neural Stem Cell Lineage. Cel Rep. 18 (3), 777–790. doi:10.1016/j.celrep.2016.12.060
Esquerdo-Barragán, B., Toulis, V., Swaroop, A., and Marfany, G. (2019). Expression of Deubiquitinating Enzyme Genes in the Developing Mammal Retina[J]. Mol. Vis. 25, 800–813.
Gao, S. (2018). Data Analysis in Single-Cell Transcriptome Sequencing[J]. Methods Mol. Biol. 1754, 311–326. doi:10.1007/978-1-4939-7717-8_18
Gierahn, T. M., Wadsworth, M. H., Hughes, T. K., Bryson, B. D., Butler, A., Satija, R., et al. (2017). Seq-Well: Portable, Low-Cost RNA Sequencing of Single Cells at High Throughput. Nat. Methods 14 (4), 395–398. doi:10.1038/nmeth.4179
Gordon, P. J., Yun, S., Clark, A. M., Monuki, E. S., Murtaugh, L. C., and Levine, E. M. (2013). Lhx2 Balances Progenitor Maintenance with Neurogenic Output and Promotes Competence State Progression in the Developing Retina. J. Neurosci. 33 (30), 12197–12207. doi:10.1523/jneurosci.1494-13.2013
Grunert, U., and Martin, P. (1991). Rod Bipolar Cells in the Macaque Monkey Retina: Immunoreactivity and Connectivity. J. Neurosci. 11 (9), 2742–2758. doi:10.1523/jneurosci.11-09-02742.1991
Habib, N., Avraham-Davidi, I., Basu, A., Burks, T., Shekhar, K., Hofree, M., et al. (2017). Massively Parallel Single-Nucleus RNA-Seq with DroNc-Seq. Nat. Methods 14 (10), 955–958. doi:10.1038/nmeth.4407
Hashimshony, T., Wagner, F., Sher, N., and Yanai, I. (2012). CEL-seq: Single-Cell RNA-Seq by Multiplexed Linear Amplification. Cel Rep. 2 (3), 666–673. doi:10.1016/j.celrep.2012.08.003
Heng, J. S., Hackett, S. F., Stein-O’Brien, G. L., Winer, B. L., Williams, J., Goff, L. A., et al. (2019). Comprehensive Analysis of a Mouse Model of Spontaneous Uveoretinitis Using Single-Cell RNA Sequencing. Proc. Natl. Acad. Sci. USA 116 (52), 26734–26744. doi:10.1073/pnas.1915571116
Hojo, M., Ohtsuka, T., Hashimoto, N., Gradwohl, G., Guillemot, F., and Kageyama, R. (2000). Glial Cell Fate Specification Modulated by the bHLH Gene Hes5 in Mouse Retina. Development 127 (12), 2515–2522. doi:10.1242/dev.127.12.2515
Hong, G., Fu, T.-M., Zhou, T., Schuhmann, T. G., Huang, J., and Lieber, C. M. (2015). Syringe Injectable Electronics: Precise Targeted Delivery with Quantitative Input/Output Connectivity. Nano Lett. 15 (10), 6979–6984. doi:10.1021/acs.nanolett.5b02987
Hughes, E. H., Schlichtenbrede, F. C., Murphy, C. C., Sarra, G.-M., Luthert, P. J., Ali, R. R., et al. (2003). Generation of Activated Sialoadhesin-Positive Microglia during Retinal Degeneration. Invest. Ophthalmol. Vis. Sci. 44 (5), 2229–2234. doi:10.1167/iovs.02-0824
Islam, S., Kjällquist, U., Moliner, A., Zajac, P., Fan, J.-B., Lönnerberg, P., et al. (2012). Highly Multiplexed and Strand-specific Single-Cell RNA 5′ End Sequencing. Nat. Protoc. 7 (5), 813–828. doi:10.1038/nprot.2012.022
Islam, S., Zeisel, A., Joost, S., La Manno, G., Zajac, P., Kasper, M., et al. (2014). Quantitative Single-Cell RNA-Seq with Unique Molecular Identifiers. Nat. Methods 11 (2), 163–166. doi:10.1038/nmeth.2772
Jaitin, D. A., Kenigsberg, E., Keren-Shaul, H., Elefant, N., Paul, F., Zaretsky, I., et al. (2014). Massively Parallel Single-Cell RNA-Seq for Marker-free Decomposition of Tissues into Cell Types. Science 343 (6172), 776–779. doi:10.1126/science.1247651
Jones, M. K., Lu, B., Saghizadeh, M., and Wang, S. (2016). Gene Expression Changes in the Retina Following Subretinal Injection of Human Neural Progenitor Cells into a Rodent Model for Retinal Degeneration. Mol. Vis. 22, 472–490.
Kamalden, T. A., Macgregor-Das, A. M., Kannan, S. M., Dunkerly-Eyring, B., Khaliddin, N., Xu, Z., et al. (2017). Exosomal MicroRNA-15a Transfer from the Pancreas Augments Diabetic Complications by Inducing Oxidative Stress. Antioxid. Redox Signaling 27 (13), 913–930. doi:10.1089/ars.2016.6844
Kawaguchi, A., Ikawa, T., Kasukawa, T., Ueda, H. R., Kurimoto, K., Saitou, M., et al. (2008). Single-cell Gene Profiling Defines Differential Progenitor Subclasses in Mammalian Neurogenesis. Development 135 (18), 3113–3124. doi:10.1242/dev.022616
Kim, I.-J., Zhang, Y., Yamagata, M., Meister, M., and Sanes, J. R. (2008). Molecular Identification of a Retinal Cell Type that Responds to Upward Motion. Nature 452 (7186), 478–482. doi:10.1038/nature06739
Lawson, L. J., Perry, V. H., Dri, P., and Gordon, S. (1990). Heterogeneity in the Distribution and Morphology of Microglia in the normal Adult Mouse Brain. Neuroscience 39 (1), 151–170. doi:10.1016/0306-4522(90)90229-w
Li, S., Mo, Z., Yang, X., Price, S. M., Shen, M. M., and Xiang, M. (2004). Foxn4 Controls the Genesis of Amacrine and Horizontal Cells by Retinal Progenitors. Neuron 43 (6), 795–807. doi:10.1016/j.neuron.2004.08.041
Lukaszewicz, A. I., and Anderson, D. J. (2011). Cyclin D1 Promotes Neurogenesis in the Developing Spinal Cord in a Cell Cycle-independent Manner. Proc. Natl. Acad. Sci. 108 (28), 11632–11637. doi:10.1073/pnas.1106230108
Macosko, E. Z., Basu, A., Satija, R., Nemesh, J., Shekhar, K., Goldman, M., et al. (2015). Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161 (5), 1202–1214. doi:10.1016/j.cell.2015.05.002
Mao, X., An, Q., Xi, H., Yang, X.-J., Zhang, X., Yuan, S., et al. (2019). Single-Cell RNA Sequencing of hESC-Derived 3D Retinal Organoids Reveals Novel Genes Regulating RPC Commitment in Early Human Retinogenesis. Stem Cel Rep. 13 (4), 747–760. doi:10.1016/j.stemcr.2019.08.012
Masland, R. H. (2012). The Neuronal Organization of the Retina[J]. Neuron 76 (2), 266–80. doi:10.1016/j.neuron.2012.10.002
McEvoy, J., Flores-Otero, J., Zhang, J., Nemeth, K., Brennan, R., Bradley, C., et al. (2011). Coexpression of Normally Incompatible Developmental Pathways in Retinoblastoma Genesis. Cancer Cell 20 (2), 260–275. doi:10.1016/j.ccr.2011.07.005
Menon M, M. S., Davila-Velderrain, J., Goods, B. A., Cadwell, T. D., Xing, Y., Stemmer-Rachamimov, A., et al. (2019). Single-cell Transcriptomic Atlas of the Human Retina Identifies Cell Types Associated with Age-Related Macular Degeneration[J]. Nat. Commun. 10 (1), 4902. doi:10.1038/s41467-019-12780-8
Miller, S. J., Philips, T., Kim, N., Dastgheyb, R., Chen, Z., Hsieh, Y.-C., et al. (2019). Molecularly Defined Cortical Astroglia Subpopulation Modulates Neurons via Secretion of Norrin. Nat. Neurosci. 22 (5), 741–752. doi:10.1038/s41593-019-0366-7
Moncada, R., Barkley, D., Wagner, F., Chiodin, M., Devlin, J. C., Baron, M., et al. (2020). Integrating Microarray-Based Spatial Transcriptomics and Single-Cell RNA-Seq Reveals Tissue Architecture in Pancreatic Ductal Adenocarcinomas. Nat. Biotechnol. 38 (3), 333–342. doi:10.1038/s41587-019-0392-8
Newman, A. M., Gallo, N. B., Hancox, L. S., Miller, N. J., Radeke, C. M., Maloney, M. A., et al. (2012). Systems-level Analysis of Age-Related Macular Degeneration Reveals Global Biomarkers and Phenotype-specific Functional Networks. Genome Med. 4 (2), 16. doi:10.1186/gm315
O'Koren, E. G., Yu, C., Klingeborn, M., Wong, A. Y. W., Prigge, C. L., Mathew, R., et al. (2019). Microglial Function Is Distinct in Different Anatomical Locations during Retinal Homeostasis and Degeneration[J]. Immunity 50 (3), 723–737. doi:10.1016/j.immuni.2019.02.007
Oppikofer, M., Bai, T., Gan, Y., Haley, B., Liu, P., Sandoval, W., et al. (2017). Expansion of the ISWI Chromatin Remodeler Family with New Active Complexes. EMBO Rep. 18 (10), 1697–1706. doi:10.15252/embr.201744011
Pastukh, N., Meerson, A., Kalish, D., and Blum, A. (2019). Serum miR-122 Levels Correlate with Diabetic Retinopathy. Clin. Exp. Med. 19 (2), 255–260. doi:10.1007/s10238-019-00546-x
Pauly, D., Agarwal, D., Dana, N., Schäfer, N., Biber, J., Wunderlich, K. A., et al. (2019). Cell-Type-Specific Complement Expression in the Healthy and Diseased Retina. Cel Rep. 29 (9), 2835–2848. doi:10.1016/j.celrep.2019.10.084
Peng, Y.-R., Shekhar, K., Yan, W., Herrmann, D., Sappington, A., Bryman, G. S., et al. (2019). Molecular Classification and Comparative Taxonomics of Foveal and Peripheral Cells in Primate Retina. Cell 176 (5), 1222–1237. doi:10.1016/j.cell.2019.01.004
Picelli, S. (2017). Single-cell RNA-Sequencing: The Future of Genome Biology Is Now[J]. RNA Biol. 14 (5), 637–650. doi:10.1080/15476286.2016.1201618
Pollen, A. A., Nowakowski, T. J., Shuga, J., Wang, X., Leyrat, A. A., Lui, J. H., et al. (2014). Low-coverage Single-Cell mRNA Sequencing Reveals Cellular Heterogeneity and Activated Signaling Pathways in Developing Cerebral Cortex. Nat. Biotechnol. 32 (10), 1053–1058. doi:10.1038/nbt.2967
Rosenberg, A. B., Roco, C. M., Muscat, R. A., Kuchina, A., Sample, P., Yao, Z., et al. (2018). Single-cell Profiling of the Developing Mouse Brain and Spinal Cord with Split-Pool Barcoding. Science 360 (6385), 176–182. doi:10.1126/science.aam8999
Ru, Y., Kechris, K. J., Tabakoff, B., Hoffman, P., Radcliffe, R. A., Bowler, R., et al. (2014). The multiMiR R Package and Database: Integration of microRNA-Target Interactions along with Their Disease and Drug Associations. Nucleic Acids Res. 42 (17), e133. doi:10.1093/nar/gku631
Sasagawa Y, D. H., Takada, H., Ebisawa, M., Tanaka, K., Hayashi, T., Kurisaki, A., et al. (2018). Quartz-Seq2: a High-Throughput Single-Cell RNA-Sequencing Method that Effectively Uses Limited Sequence Reads[J]. Genome Biol. 19 (1), 29. doi:10.1186/s13059-018-1407-3
Shaker, O. G., Abdelaleem, O. O., Mahmoud, R. H., Abdelghaffar, N. K., Ahmed, T. I., Said, O. M., et al. (2019). Diagnostic and Prognostic Role of Serum miR-20b, miR-17-3p, HOTAIR, and MALAT1 in Diabetic Retinopathy. IUBMB Life 71 (3), 310–320. doi:10.1002/iub.1970
Shekhar, K., Lapan, S. W., Whitney, I. E., Tran, N. M., Macosko, E. Z., Kowalczyk, M., et al. (2016). Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell 166 (5), 1308–1323. doi:10.1016/j.cell.2016.07.054
Tran, N. M., Shekhar, K., Whitney, I. E., Jacobi, A., Benhar, I., Hong, G., et al. (2019). Single-Cell Profiles of Retinal Ganglion Cells Differing in Resilience to Injury Reveal Neuroprotective Genes. Neuron 104 (6), 1039–1055. doi:10.1016/j.neuron.2019.11.006
Trimarchi, J. M., Cho, S.-H., and Cepko, C. L. (2009). Identification of Genes Expressed Preferentially in the Developing Peripheral Margin of the Optic Cup. Dev. Dyn. 238 (9), 2327–2329. doi:10.1002/dvdy.21973
Valdes-Mora, F., Handler, K., Law, A. M. K., Salomon, R., Oakes, S. R., Ormandy, C. J., et al. (2018). Single-Cell Transcriptomics in Cancer Immunobiology: The Future of Precision Oncology[J]. Front. Immunol. 12 (9), 2582. doi:10.3389/fimmu.2018.02582
Wang, Y., Waters, J., Leung, M. L., Unruh, A., Roh, W., Shi, X., et al. (2014). Clonal Evolution in Breast Cancer Revealed by Single Nucleus Genome Sequencing. Nature 512 (7513), 155–160. doi:10.1038/nature13600
Yamagata M, Y. W., and Sanes, J. R. (2021). A Cell Atlas of the Chick Retina Based on Single-Cell Transcriptomics[J]. Elife 10, e63907. doi:10.7554/elife.63907
Yan W, P. Y., van Zyl, T., Regev, A., Shekhar, K., Juric, D., and Sanes, J. R. (2020). Cell Atlas of the Human Fovea and Peripheral Retina[J]. Sci. Rep. 10 (1), 9802. doi:10.1038/s41598-020-66092-9
Yan, W., Laboulaye, M. A., Tran, N. M., Whitney, I. E., Benhar, I., and Sanes, J. R. (2020). Mouse Retinal Cell Atlas: Molecular Identification of over Sixty Amacrine Cell Types. J. Neurosci. 40 (27), 5177–5195. doi:10.1523/jneurosci.0471-20.2020
Yu-Wai-Man, P., Sitarz, K. S., Samuels, D. C., Griffiths, P. G., Reeve, A. K., Bindoff, L. A., et al. (2010). OPA1 Mutations Cause Cytochrome C Oxidase Deficiency Due to Loss of Wild-type mtDNA Molecules. Hum. Mol. Genet. 19 (15), 3043–3052. doi:10.1093/hmg/ddq209
Zerti, D., Collin, J., Queen, R., Cockell, S. J., and Lako, M. (2020). Understanding the Complexity of Retina and Pluripotent Stem Cell Derived Retinal Organoids with Single Cell RNA Sequencing: Current Progress, Remaining Challenges and Future Prospective. Curr. Eye Res. 45 (3), 385–396. doi:10.1080/02713683.2019.1697453
Zhang, X., Shi, E., Yang, L., Fu, W., Hu, F., and Zhou, X. (2019). LncRNA AK077216 Is Downregulated in Diabetic Retinopathy and Inhibited the Apoptosis of Retinal Pigment Epithelial Cells by Downregulating miR-383. Endocr. J. 66 (11), 1011–1016. doi:10.1507/endocrj.ej19-0080
Keywords: single-cell RNA sequencing, ScRNA-seq, retina, gene, retinal disease
Citation: Ying P, Huang C, Wang Y, Guo X, Cao Y, Zhang Y, Fu S, Chen L, Yi G and Fu M (2021) Single-Cell RNA Sequencing of Retina:New Looks for Gene Marker and Old Diseases. Front. Mol. Biosci. 8:699906. doi: 10.3389/fmolb.2021.699906
Received: 24 April 2021; Accepted: 01 July 2021;
Published: 30 July 2021.
Edited by:
Fengfeng Zhou, Jilin University, ChinaReviewed by:
Siddharth Shukla, Howard Hughes Medical Institute (HHMI), United StatesSumit Mukherjee, Microsoft, United States
Copyright © 2021 Ying, Huang, Wang, Guo, Cao, Zhang, Fu, Chen, Yi and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoguo Yi, eWlnZ0BtYWlsLnN5c3UuZWR1LmNu; Min Fu, bWluX2Z1MTIxMkAxNjMuY29t
†These authors have contributed equally to this work and share co first authorship
 Peixi Ying1†
Peixi Ying1† Yan Wang
Yan Wang Guoguo Yi
Guoguo Yi Min Fu
Min Fu