- Department of Biochemistry, All India Institute of Medical Sciences (AIIMS) Bhopal, Bhopal, India
Background: Triple-negative breast cancer (TNBC) is associated with a poor prognosis. Sphingosine-1-phosphate (S1P), a potent sphingolipid metabolite, has been implicated in many processes that are important for breast cancer (BC). S1P signaling regulates tumorigenesis, and response to chemotherapy and immunotherapy by affecting the trafficking, differentiation or effector function of tumor-infiltrating immune cells (TIICs).
Objective: In this study, using bioinformatics tools and publicly available databases, we have analyzed the prognostic value of S1P metabolizing genes and their correlation with TIICs in BC patients.
Methods: The expression of S1P metabolizing genes and receptors was evaluated by the UALCAN cancer database. The correlation between mRNA expression of S1P metabolizing genes and receptors and survival outcome of breast cancer patients was analyzed by the Kaplan-Meier plotter database. The association between the gene expression and infiltration of immune cells in the tumors was analyzed by “Tumor-Infiltrating Immune Estimation Resource (TIMER). In silico protein expression analysis was done using the Human Protein Atlas” database.
Results: TNBC patients with lower expression of S1P phosphatase 1 (SGPP1) or lipid phosphate phosphatase 3 (PLPP3) have much shorter relapse-free survival than the patients with a higher expression of these genes. SGPP1 and PLPP3 expression show a strong positive correlation with tumor-infiltrating dendritic cells (DCs), CD4+ and CD8+ T cells, neutrophils, and macrophages in the TNBC subtypes. In addition, S1P receptor 4 (S1PR4), an S1P receptor exhibit a strong positive correlation with DCs, CD4+ and CD8+ T cells and neutrophils in TNBC. We, therefore, conclude that low expression of SGPP1 and PLPP3 may hinder the recruitment of immune cells to the tumor environment, resulting in the blockage of cancer cell clearance and a subsequent poor prognosis.
Introduction
Breast cancer is the foremost cause of cancer-related deaths in females in many countries, including India (Bray et al., 2018; Alkabban and Ferguson, 2020; Jonnada et al., 2020). Invasive BC is classified into four distinct subtypes based on the expression of estrogen receptor alpha (ERα), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), and Ki-67 (Inic et al., 2014; Harbeck et al., 2019). The luminal A subtype (ERα+, PR+, HER2−and low expression of Ki-67) is a low-grade breast tumor, whereas the luminal B subtype (ERα+, PR+, HER2+/−and high expression of Ki-67) is a more aggressive form. The molecular subtypes of BC are often a key reference for prognosis and choice of therapeutic strategy (Inic et al., 2014; Harbeck et al., 2019). Triple-negative breast cancer (TNBC) (ERα−, PR− and HER2−) accounts for approximately 15% of all BC cases and is characterized by shorter survival and an early peak of distant recurrence (Manjunath and Choudhary, 2021). The term TNBC encompasses a highly diverse group of cancers that is further categorized into six molecular TNBC subtypes (Pietenpol subtypes), two basal-like subtypes (BL1 and BL2), one immunomodulatory (IM), one mesenchymal, one mesenchymal stem-like, and one luminal androgen receptor subtype (Lehmann et al., 2011). In the majority of cases, identifying TNBC response to traditional chemotherapy and immunotherapy poses a challenge in current clinical practice (Manjunath and Choudhary, 2021).
Tumor-infiltrating immune cells play an important role in cancer treatment efficacy and patient prognosis (Denkert et al., 2018). Further, TIICs, especially tumor-infiltrating lymphocytes (TILs), are associated with better outcomes in HER2+BC and TNBC. However, TNBC lacks both predictive markers and potential therapeutic targets. Hence the identification of novel predictive and prognostic markers is warranted. Sphingosine-1-phosphate (S1P) signaling has emerged as a central mediator of the trafficking of hematopoietic cells, including lymphocytes, natural killer (NK) cells, neutrophils, dendritic cells (DCs) and macrophages (Cartier and Hla, 2019; Pyne and Pyne, 2020). S1P activates several cellular pathways by binding to one of the five G-protein-coupled receptors (GPCRs), termed S1P receptors (S1PRs) (Sukocheva et al., 2006; Sukocheva, 2018). Activation of these receptors in an autocrine or paracrine manner by S1P promotes cell proliferation, cell survival migration, and activation of the inflammatory response and also inhibits apoptosis (Pyne and Pyne, 2020). Moreover, S1PRs are expressed on almost all the immune cell types. However, each cell type expresses only a subset of S1PRs. S1PR1 is expressed in most immune cells, whereas S1PR5 is expressed primarily by DCs and NK cells (Kumar et al., 2017).
S1P, an oncogenic sphingolipid (Fyrst and Saba, 2010), is generated by two isoforms of sphingosine kinase, SPHK1 and SPHK2 (Pyne and Pyne, 2020). S1P can be metabolized through irreversible cleavage by the S1P lyase enzyme (SGPL1) (Zhou and Saba, 1998) to trans-2-hexadecenal and ethanolamine phosphate. Alternatively, S1P can also be dephosphorylated back to sphingosine through a reaction that is catalyzed by three non-specific lipid phosphate phosphatases (LPP1–3) or two S1P-specific phosphatases (SPPase1–2) (Cartier and Hla, 2019; Pyne and Pyne, 2020). S1P-metabolizing enzymes play a crucial role in various aspects of BC, including neovascularization, metastasis, recurrence, and chemo resistance (Wang et al., 1999; Nagahashi et al., 2016). SphK1 has been shown to overexpress in breast tumor tissue compared to normal breast tissue, and higher expression of SphK1 is associated with poor survival outcomes (Ruckhäberle et al., 2008). SphK1 also promotes tumor angiogenesis, lymphangiogenesis, and resistance to radiation and chemotherapy (Sarkar et al., 2005; Pyne and Pyne, 2020). In a prior study, BC patients with lymph node metastasis showed significantly higher levels of S1P compared with BC patients with negative nodes (Tsuchida et al., 2017). However, the role of S1P-catabolizing enzymes, including SPPase1 and LPP3 (PLPP3), in BC is not yet fully understood.
The meta-analysis of publicly available data from The Cancer Genome Atlas (TCGA) databases can predict outcomes when applied to appropriately powered cohorts and is a feasible, unbiased approach that can be adapted to analyze the involvement of genes in cancer progression. Thus, in the present study, using web-based bioinformatics tools, the aim was to analyze the association of S1P-metabolizing enzymes and S1P receptors with TIICs in BC patients and with the prognosis of patients with BC. Expression of S1P-signaling genes in breast tumor and normal tissue by utilizing the UALCAN database (Chandrashekar et al., 2017) and the role of these genes in survival outcome was analyzed by employing the Kaplan-Meier (KM) plotter (Győrffy et al., 2013). To determine the role of S1P-metabolizing enzymes in the infiltration of immune cells into breast tumors, the Tumor Immune Estimation Resource (TIMER), an online bioinformatics tool for the comprehensive analysis of TIICs, was used (Li et al., 2017).
In this study, we demonstrate that the expression of SGPP1 and PLPP3 is reduced in tumors compared with normal tissues from patients with BC. Low expression of SGPP1 and PLPP3 in tumors could be considered to be a predictive marker for worse relapse-free survival (RFS) in patients with TNBC. We also found that both of these genes are associated with a high predictive value in systemically treated patients with BC. Importantly, we demonstrate that SGPP1 and PLPP3 expression is strongly associated with TIICs, especially DCs, CD8+ T cells, and neutrophils. Furthermore, we found that S1PR4, an S1P receptor, is strongly associated with TIICs in all four BC subtypes. Therefore, SGPP1 and PLPP3 could serve as predictive and prognostic markers in patients with TNBC.
Materials and Methods
Kaplan–Meier Survival Analysis
We analyzed the association between gene-specific mRNA expression and RFS of breast cancer patients by employing the KM plotter (www.kmplot.com) (Győrffy et al., 2013). The KM plotter is a web-based tool widely used in the meta-analysis of publicly available TCGA, gene expression omnibus, and European Genome-Phenome Archive databases (Győrffy et al., 2013). Currently, in the KM plotter, gene expression and survival outcome data for 3,955 patients with BC for a follow-up period of 20 years are available along with clinicopathological features such as ER, PR, and HER2 status, intrinsic subtypes, lymph node status, tumor grade, and Pietenpol subtypes as well as details of the systematic therapy (endocrine therapy or chemotherapy) given to patients (Szász et al., 2016). Data from 36 gene expression datasets was included for analysis by KM plotter, the details of datasets are given in Supplementary Material.
We analyzed the prognostic value of S1P-metabolizing enzymes and S1P receptors in breast cancer by entering their respective gene symbols into the KM plotter database (www.kmplot.com). The number of patients at-risk was indicated below the main KM plot. Affymetrix IDs of all the genes analyzed in the study are shown in Supplementary Table S1. The checkbox for auto-select best cut-off value was selected to divide the patients into high expression and low expression groups. JetSet probes showing concordance between protein measurements and gene expression values were used for survival analysis (Li et al., 2011). The hazard ratio (HR) with a 95% confidence interval (CI) and the log-rank p-value was calculated by the KM plotter (Wang et al., 2014). A p value <0.05 was considered statistically significant.
Gene Expression Analysis Using UALCAN Database
UALCAN is a comprehensive, user-friendly, and interactive web resource that can be used to plot graphs depicting gene expression and to evaluate promoter DNA methylation information (Chandrashekar et al., 2017). In this study, it was also used for gene expression analysis of SGPP1, SGPP2, PLPP1, PLPP2, and PLPP3 in breast tumors and normal tissues.
Protein Expression Analysis Using Human Protein Atlas Database Tool
We also analyzed the protein expression of LPP3 and SGPP1, available in “the Human Protein Atlas” (HPA; https://www.proteinatlas.org/) database as described previously (Nema et al., 2021).
Analysis of Tumor Immunological Features of S1P Components Using TIMER
The TIMER is a web-based computational tool for the evaluation of tumor immune cells in the publicly available TCGA database (Li et al., 2017). It uses a deconvolution method to deduce the abundance of TIICs based on gene expression profiles. It provides six major analytic modules that allow users to explore the associations between immune infiltrates and various factors, including clinical outcomes, somatic mutations, somatic copy number alterations, and gene expression (Li et al., 2017). In this study, the associations between the expression of S1P-metabolizing genes with TIICs in patients with BC, tumor-infiltrating immune infiltrate cells (B cells, CD4+ T cells, CD8+ T cells, macrophages, neutrophils, and DCs) were selected for correlation by TIMER. Tumor purity was taken into consideration when calculating the Spearman’s correlation. Because genes highly expressed in the infiltrating immune cells are expected to have negative associations with tumor purity. A p value <0.05 was considered statistically significant.
Results
S1P signaling plays an important role in the tumorigenesis of a variety of cancers. To determine the prognostic value of S1P-metabolizing enzymes, KM survival curves were plotted for patients with BC for the mRNA expression of S1P-metabolizing enzymes. As shown in Figure 1 and Supplementary Table S2, mRNA expression of SPHK2, SGPP1, PLPP1, and PLPP3 was significantly associated with RFS and overall survival (OS) in patients with BC. Of these, RFS was best predicted by SPHK2 (P < 1e-16), SGPP1 (p = 1.6e-07), and PLPP3 (LPP3; p = 1.8e-10), where patients with BC with a lower expression of these enzymes had worse RFS (Figure 1). SPHK1 and SGPP2 did not show an appreciable significant association with the survival outcome of the patients with BC (Supplementary Table S2). The prognostic value of SPHK2 in BC has been described previously (Alshaker et al., 2020). As SGPP1 and PLPP3 showed highly significant association with RFS as well as OS, we therefore focused our attention on SGPP1 and PLPP3. S1P mediates its physiological functions by binding to S1P receptors. Thus, to predict the prognostic role of S1P receptors in relation to BC survival, KM plots were drafted for S1P receptors. As shown in Figure 2 and Supplementary Table S3, except for S1PR2, the expression of S1P receptors, S1PR1, S1PR3, S1PR4, and S1PR5, was significantly associated with RFS in patients with BC. Of these, high expression of S1PR1 (HR = 0.64, 95% CI: 0.58–0.72, p = 7.2e-16) and S1PR4 (HR = 0.81, 95% CI: 0.73–0.91, p = 0.00023) was significantly associated with better RFS in all patients with BC (Figure 2).
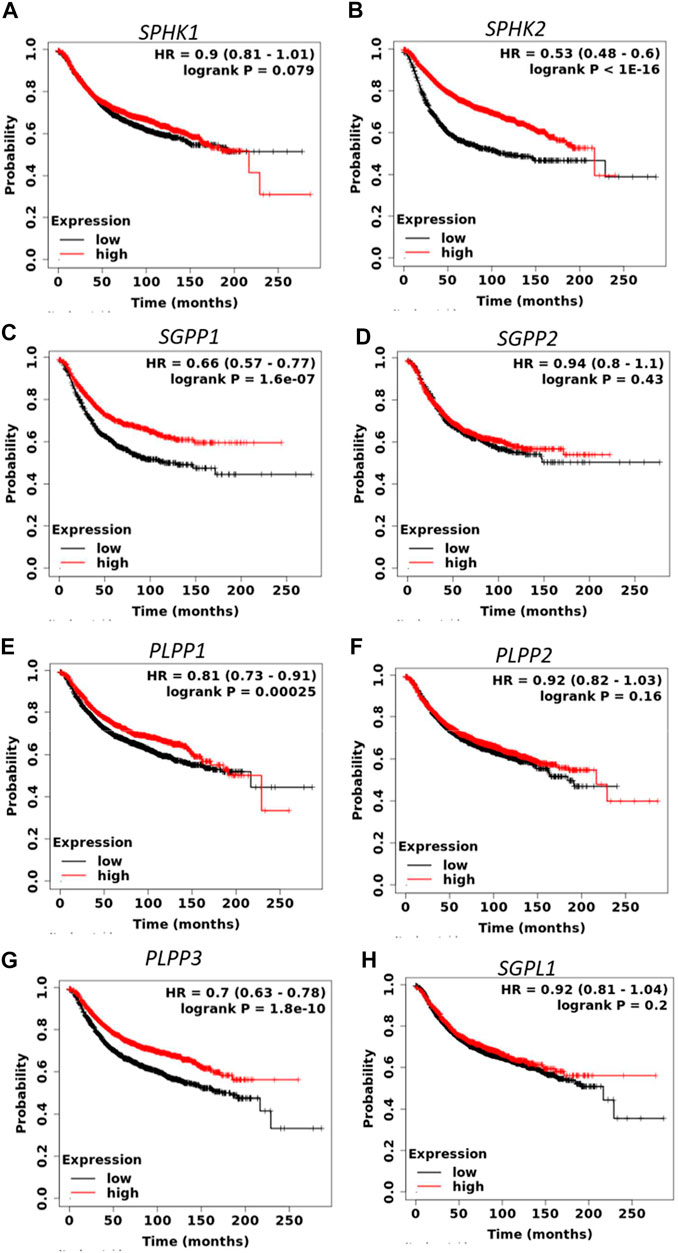
FIGURE 1. Prognostic role of mRNA expression of S1P-metabolizing enzymes (A–H) in breast cancer patients. Kaplan–Meier survival curves (RFS) were plotted for S1P-metabolizing genes from the publicly available KM plotter database (N = 3955 BC).
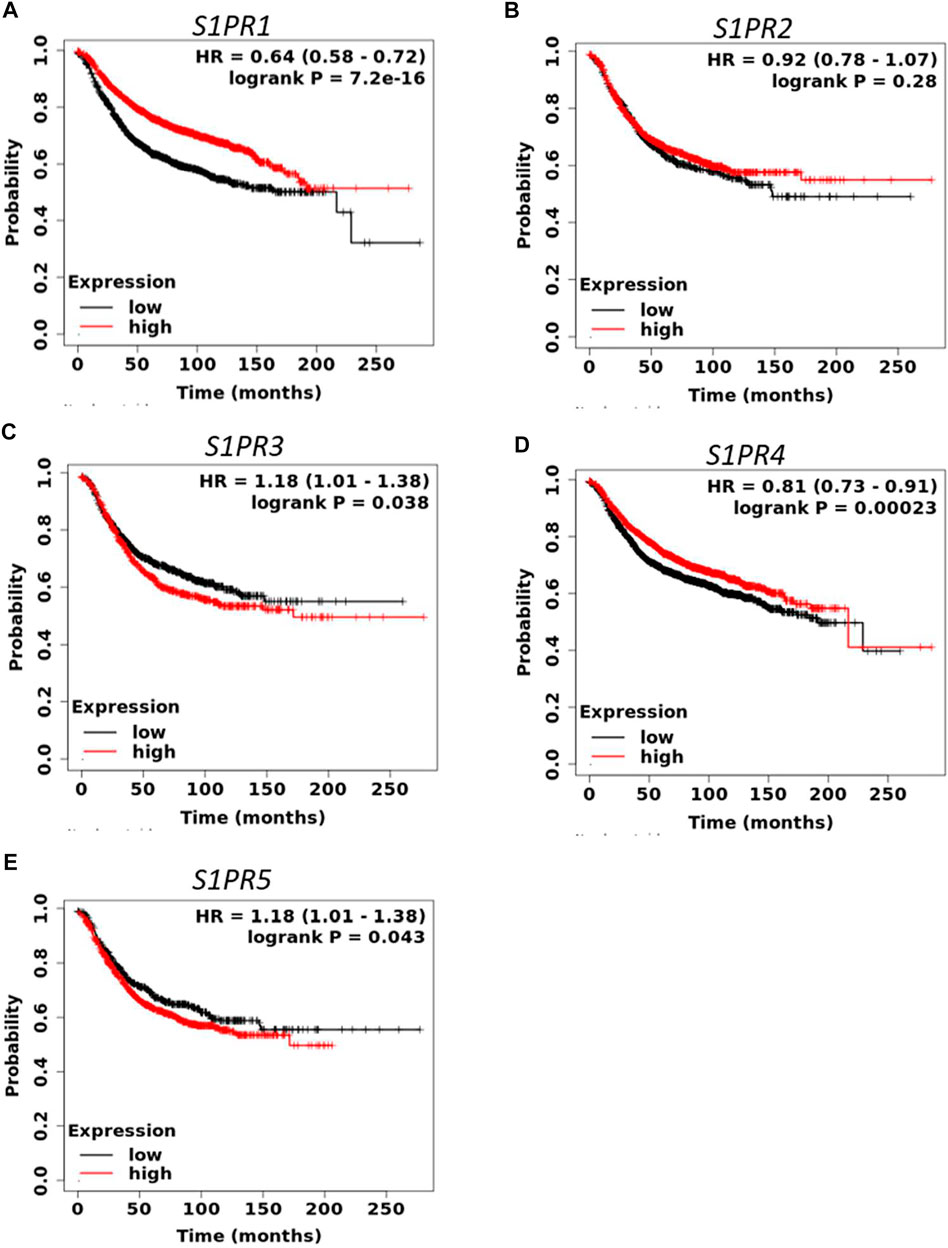
FIGURE 2. Prognostic role of mRNA expression of S1P receptors (A–E) in breast cancer patients. Kaplan-Meier survival curves (RFS) were plotted for S1P receptors from the publicly available KM plotter database (N = 3955 BC).
We also explored the prognostic value of SGPP1 and PLPP3 in BC patients with different intrinsic subtypes, pathological grades, and lymph node status. Interestingly, as shown in Figure 3 and Table 1, SGPP1 showed a high predictive value in the basal and HER2+ subtypes. In the basal subtype, upper quartile survival in the low expression (SGPP1) cohort was 14.7 months compared to 30.42 months in the high expression (SGPP1) cohort (Figure 3C; Table 1). However, in the HER2+ subtype, upper quartile survival in the low expression (SGPP1) cohort was 12.0 months compared to 25.2 months in the high expression (SGPP1) cohort (Figure 3D). Among the Pietenpol subtypes of TNBC, despite having a low sample size, mesenchymal stem-like and luminal androgen receptor subtypes showed a significant association of SGPP1 expression with RFS (Table 2). There was a five-fold difference in the survival period (RFS) between the SGPP1 low expression cohort vs. the high expression cohort in the luminal androgen receptor subtypes of TNBC (Table 2), whereas PLPP3 showed high predictive value in luminal A and basal subtypes (Table 1). SGPP1 expression showed a distinct profile in PR + vs. PR-subtypes. Patients with BC with high expression of SGPP1 had significantly better RFS in the PR + subtype (Table 3 Supplementary Figure S1) compared to the PR-subtype (Table 3; Supplementary Figure S1). However, PLPP3 showed a distinct RFS profile in ER+ vs. ER− subtypes. SGPP1 also exhibited a significant association in lymph node (LN)+ patients with BC, but not in LN− patients (Table 3; HR = 0.65, 95% CI: 0.5–0.84, p = 0.0011).
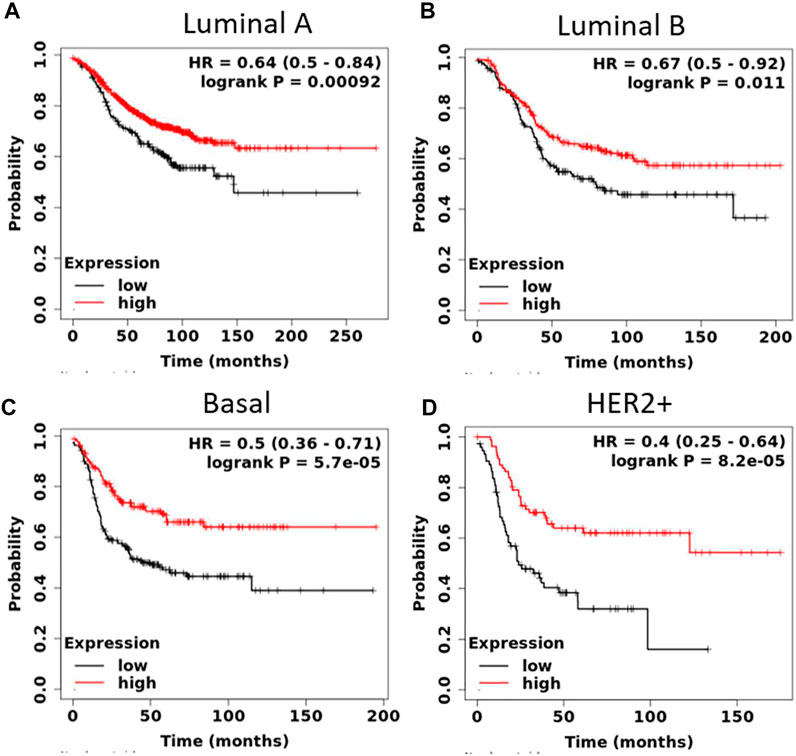
FIGURE 3. High expression of sphingosine-1-phosphate phosphatase1 (SGPP1) is highly significantly associated with relapse-free survival (RFS) of patients with HER2+ and basal-like BC (intrinsic subtypes). (A–D), Kaplan-Meier survival curves (RFS) plotted for SGPP1 for intrinsic subtypes of BC.
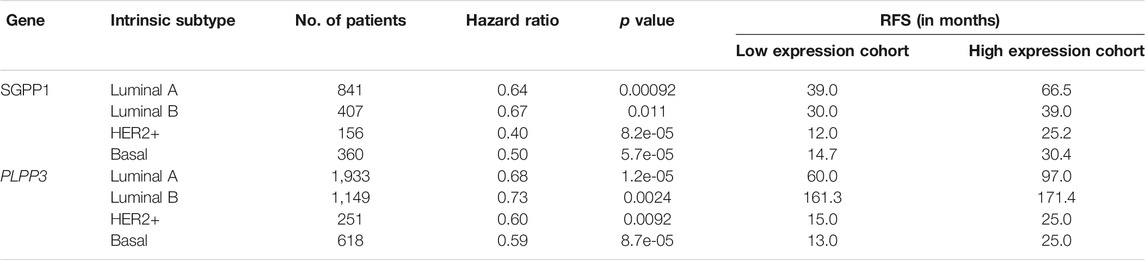
TABLE 1. Correlation between mRNA expression of SGPP1 and PLPP3 with relapse-free survival in the different breast cancer intrinsic subtypes.
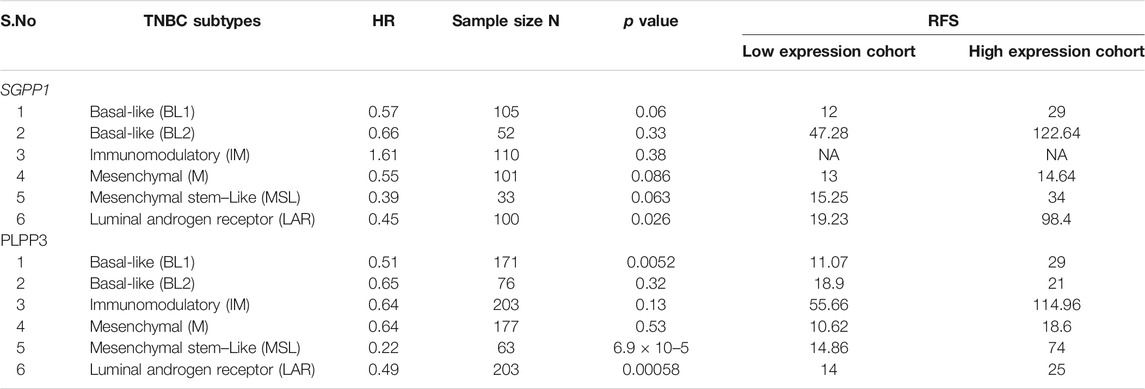
TABLE 2. Correlation between mRNA expression of SGPP1 and PLPP3 with relapse-free survival (RFS) in Pietenpol subtypes of triple-negative breast cancer (TNBC).
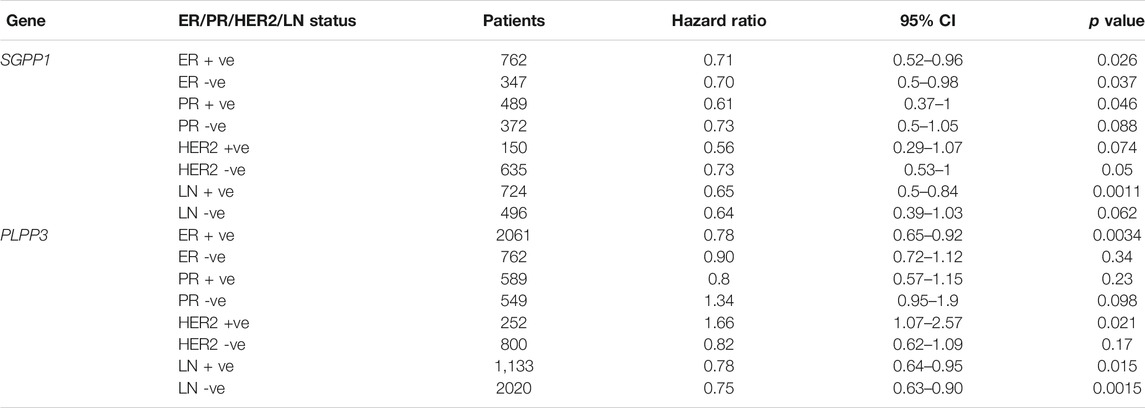
TABLE 3. Correlation between mRNA expression of SGPP1 and PLPP3 with relapse-free survival in the breast cancer patients with ER, PR, HER2 and LN status.
To analyze whether SGPP1 and PLPP3 could serve as a predictive marker for response to systemic therapy, an RFS curve was plotted for the intrinsic subtypes of patients who had received any kind of systemic therapy (endocrine or chemotherapy). Both of the S1P-catabolizing genes, SGPP1 and PLPP3, showed high predictive value for response to systematic therapy in invasive breast carcinoma patients and especially in the HER2+ and basal subtypes (Table 4). Basal subtypes treated with any type of systemic therapy and with higher expression of SGPP1 or PLPP3 had almost four-fold (SGPP1) and 2.5-fold (PLPP3) RFS compared to the low expression cohort (Table 4). A multivariate analysis of SGPP1 and PLPP3 with selected variables such as MKI67, ESR1 and ERB2 showed a highly significant association with RFS (Supplementary Figure S2).
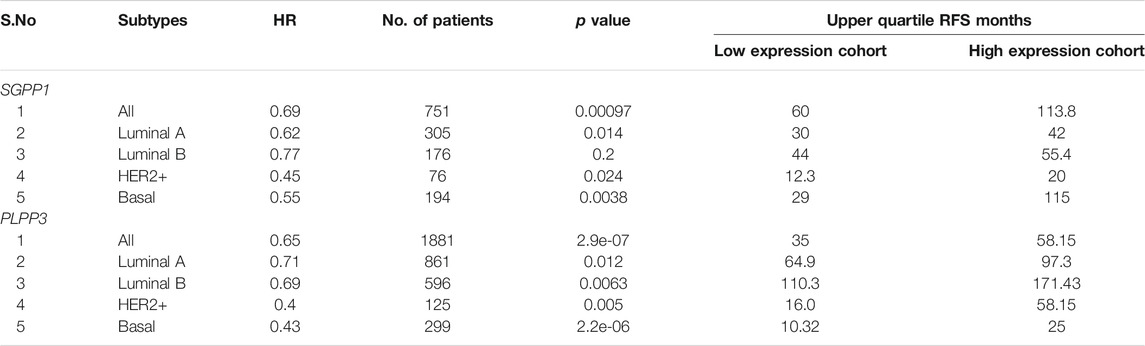
TABLE 4. Correlation between mRNA expression of SGPP1 and PLPP3 with relapse-free survival (RFS) in the systemically treated in different breast cancer intrinsic subtypes.
Expression of SGPP1 and PLPP3 in Primary Breast Tumors is Decreased
mRNA expression of SGPP1 and PLPP3 genes in invasive cancer and normal breast tissues was compared using the UALCAN database (Chandrashekar et al., 2017). Compared to normal tissue, mRNA expression of SGPP1 and PLPP3 was decreased approximately 50 and 75% (SGPP1 p = 1.29 × 10−9; PLPP3 p = 1.29 × 10−12), respectively, in primary tumors from patients with BC (Figure 4A). Among the major subtypes of BC, patients with TNBC showed a pronounced decrease in SGPP1 mRNA expression compared to luminal and HER2+ patients (Figure 4B). The decrease in the SGPP1 and PLPP3 mRNA expression was even more pronounced in the advanced stage (stage IV) (Figures 4C–F). Expression of the second isoform of SGPP i.e., SGPP2, was not significantly different in breast invasive carcinoma compared to normal tissue (Supplementary Figure S3A). However, the expression of PLPP1 and PLPP2 was decreased and increased, respectively, in primary breast tumors compared to normal breast tissue (Supplementary Figures S3B,C).
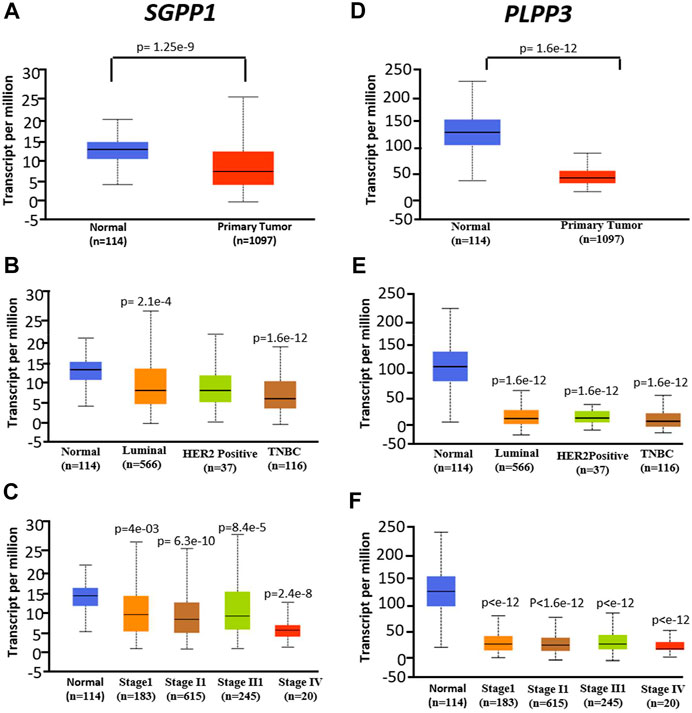
FIGURE 4. mRNA expression was analyzed in normal tissue (N = 114) and primary tumors from breast cancer (N = 1,097) patients from the publicly available UALCAN database. (A–C), SGPP1 (where A = normal vs. tumors, B = normal vs. cancer subtypes and C = normal vs. tumors of different stages). *p < 0.01 Stage I vs Stage IV, Stage II vs Stage IV and Stage III vs Stage IV. (D–F), PLPP3 (where D = normal vs. tumors, E = normal vs. cancer subtypes and F = normal vs. tumors of different stages). Data are shown as average number of transcripts per million. p values are calculated for primary tumors vs. normal tissues in corresponding genes.
Gene expression is regulated by several epigenetic factors, including DNA methylation. Therefore, we used the UALCAN database to verify the methylation status of the SGPP1 gene promoter. As shown in Supplementary Figures S4A–C, an increase in SGPP1 promoter methylation status was noted in patients with TNBC; however, no significant difference in the methylation status of the SGPP1 promoter was observed in the other BC subtypes (luminal and HER2+) vs. normal subjects. Notably, the promoter of PLPP3 was hypermethylated in the primary tumors from invasive breast carcinoma (Supplementary Figures S4D–F). Furthermore, hypermethylation of the PLPP3 was more pronounced in TNBC subtypes than luminal subtypes (p = 0.0139) (Supplementary Figure S4E).
Following on from the above, we analysed SGPP1 and PLPP3 protein expression in breast tumors and normal breast tissue by using The Human Protein Atlas (HPA; https://www.proteinatlas.org/) database. The results showed that the protein levels of PLPP3 and SGPP1 were decreased significantly in primary breast tumors as compared to normal breast tissue (Supplementary Figure S5). Furthermore, the IRS score (IHC) revealed that PLPP3 was expressed at a low level, and SGPP1 was moderately expressed in a patient with BC (Supplementary Figure S5).
Somatic Mutations in S1P-Metabolizing Genes and S1P Receptors Are Uncommon
Mutation analysis of S1P-metabolizing genes and S1P receptors was performed by applying the cBioPortal (www.cbioportal.org) tool to the publicly available whole-exome sequencing data of patients with BC (Gao et al., 2013). Except for SPHK1, S1P-metabolizing genes were infrequently mutated in patients with BC (Supplementary Figure S6A). Among these, the SPHK1 and SGPL1 genes were mutated in 5 and 2.0% of patients with BC, respectively, where most of these alterations were due to gene amplification. Mutations, mostly deep deletions in the PLPP3 gene in 1.2% of patients with BC, were noted (Supplementary Figure S6A). The cBioPortal was also used to analyze somatic mutations in the gene coding for S1P receptors in patients with BC. Among the S1P receptors, S1PR2 and S1PR5 were mutated in 1.6% and 1.8%of patients with BC, respectively (Supplementary Figure S6B).
SGPP1 Expression Correlates With Infiltration of DCs and CD8+ Cytotoxic Cells in TNBC
The role of the S1P-signaling pathway in the regulation of the infiltration of immune cells into tumor stroma in BC has not yet been elucidated. CD11c (integrin subunit alpha X; ITGAX), a member of the integrin β2 adhesion molecule family, is highly expressed in myeloid DCs. Conventional macrophages and DCs are crucial for antigen presentation and T-cell activation during antitumor immunity. Infiltration of DCs into the tumor microenvironment, enhances immune activation and recruitment of effector T cells (Janco et al., 2015). Therefore, the correlation of the mRNA expression of S1P-metabolizing enzymes with TIICs were analyzed using TIMER on publicly available TCGA datasets. A strong positive correlation (ρ = 0.523; p = 1.13e-13) was found form RNA expression of SGPP1 and ITGAX (CD11c), a marker for DCs in TNBC or basal-like BC (Figures 5A–D). The mRNA expression of SGPP1 was also associated with tumor DCs in the other cancer types. TNBC (basal-like BC) showed the highest correlation, followed by uveal melanoma, ovarian cancer, and thymomas (Supplementary Table S4). Moreover, survival curve analysis revealed that BC patients with high expression of CD11c had better survival outcomes (Figure 5E). Furthermore, in TNBC, expression of SGPP1 was also correlated with TILs (CD8+ and CD4+ T cells), macrophages, and neutrophils (Supplementary Figure S7). As regards PLPP3 and SPHK1, their expression exhibited a moderate positive correlation with TIICs except for the B cells in the luminal subtype, but not in the other subtypes of BC (Supplementary Figures S8, S9). As for PLPP1, its expression showed a moderate positive correlation with the infiltration of CD8+ T lymphocytes into the tumors of luminal subtypes (Supplementary Figure S10).
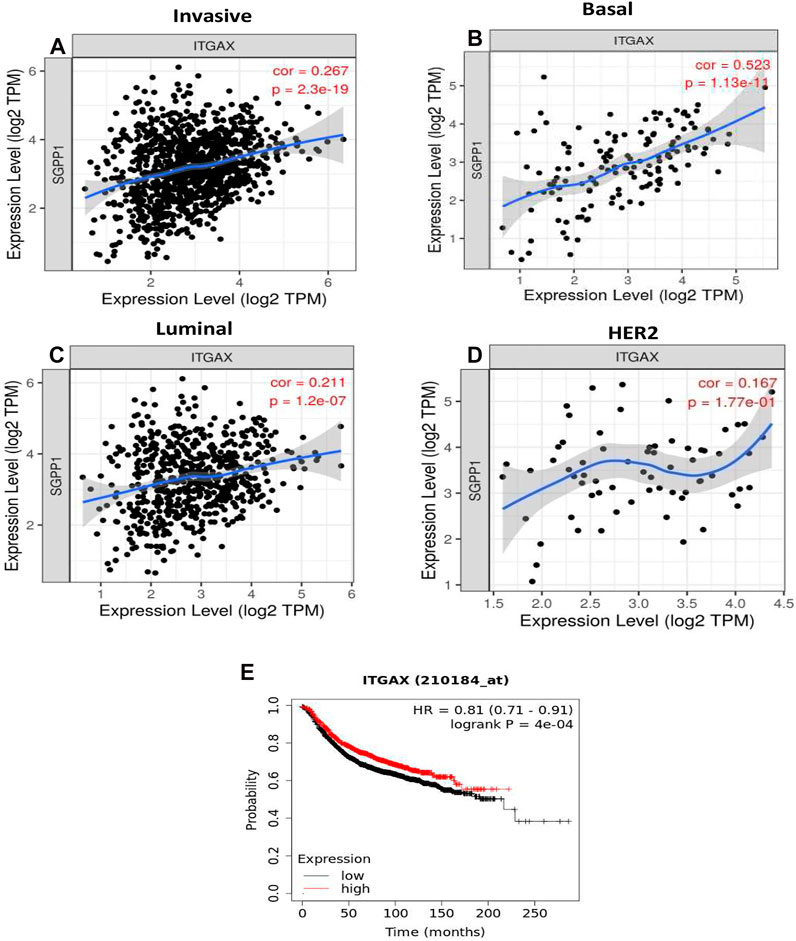
FIGURE 5. The SGPP1 gene with invasive breast carcinoma, basal, luminal and HER2+ and SGPP1is significantly associated with basal-like tumor-infiltrating immune cells according to correlation via TIMER. (A–D), SGPP1 (where A = invasive breast carcinoma, B = basal-like breast carcinoma, C = luminal breast carcinoma, and D = HER2+ breast carcinoma). High expression of integrin subunit alpha X (ITGAX) is significantly associated with relapse-free survival (RFS) of breast cancer patients. (E), Kaplan-Meier survival curves (RFS) were plotted for ITGAX for breast cancer.
Expression of S1PR4 Correlates With Tumor Infiltration of Immune Cells in BC
Expression of S1P receptor(s) on immune cells is essential for their transport among various tissues, including lymph nodes, bone marrow, and tumor tissues (Rivera et al., 2008). Different types of immune cells express different types of S1P receptors. Therefore, to ascertain the role of S1P receptors in TIICs, the correlation between the expression profile of S1P receptors and immune cell markers was determined. Among all the S1P receptors, S1PR4 showed the strongest correlation with TIICs, except for macrophages, in all the subtypes of BC (Figure 6). S1PR1 showed moderate to strong correlation with TIICs, except for B cells, in all BC subtypes (Supplementary Figures S11, S12). S1PR2 exhibited a moderate to strong correlation with TIICs in luminal and HER2+ subtypes (Supplementary Figure S13).
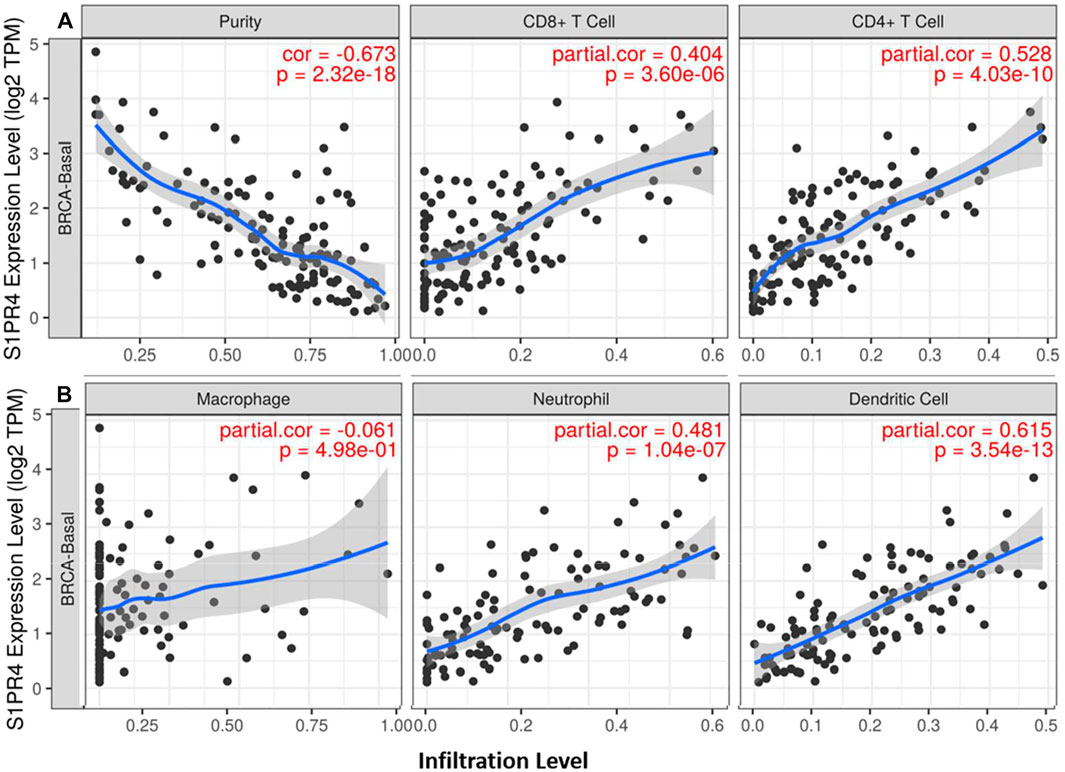
FIGURE 6. Correlation of S1PR4 expression with immune infiltration level in breast cancer patients. SGPP1 expression is significantly correlated with infiltrating levels of CD8+ T cells, CD4+ T cells (A), neutrophils and dendritic cells in basal-like BC, but not with macrophage cells (B).
Discussion
The difficulty of developing effective therapies for TNBC can be attributed to the high level of heterogeneity in the tumors. Hence, the identification of novel prognostic markers would help in stratifying patients for immunotherapy or targeted drug therapy. S1P signalling regulates various aspects of tumorigenesis, including proliferation, survival, invasion, angiogenesis, metastasis, and recurrence as well as patient response to chemotherapy and adjuvant therapy for various cancers, including BC (Alshaker et al., 2020). S1P mediates its diverse actions by binding to its five S1P receptors that belong to the GPCR family of receptors. Each S1P receptor is coupled to a specific G-protein and effector enzyme, resulting in the activation of various downstream signaling proteins, including Rac, Rho, Akt, and STAT3 (Postma et al., 1996; Zondag et al., 1998; Pyne and Pyne, 2020). Each cell type may express one or more S1P receptors at a given time, depending on the specificity and diversity of S1P functions. SphK1 is a well-studied kinase, responsible for S1P synthesis. It is overexpressed in the tumor tissues of patients with BC and thus serves as a marker for poor prognosis (Pyne and Pyne, 2020).
Using web-based bioinformatics tools, we found that patients with invasive BC and with lower expression of SPHK2, SGPP1, and PLPP3 in primary tumors had worse RFS and OS. Our findings on SPHK2 and PLPP3 corroborate those in a previous report (Lei et al., 2018). In the present study, SGPP1 had a high predictive value in the HER2+ and basal (TNBC) subtypes of BC, whereas PLPP3 had a high predictive value in luminal A and basal subtypes. Furthermore, we found that the prognostic value of SGPP1 also differed among the Pietenpol subtypes of TNBC (Lehmann et al., 2011). In particular, the difference in RFS in BL2 and IM subtypes was remarkable with differential gene expression of SGPP1 and PLPP3.
Our results also revealed that expression of the catabolizing enzymes SGPP1 and PLPP3 decreased in primary tumors compared to normal breast tissues, and the reduction in the expression was notable in TNBC subtypes and stage IV patients. Furthermore, in our study, a decrease in the mRNA expression of PLPP3 in breast tumors was found to be associated with the promoter hypermethylation, whereas no considerable change in the promoter methylation was observed for SGPP1. Decreased expression of S1P-catabolizing enzymes in tumors may lead to elevated levels of S1P in tumor tissues. Indeed, significantly higher levels of S1P have been reported in tumor tissues from BC patients with lymph node metastasis (Tsuchida et al., 2016).
Ectopic expression of mouse SGPP1 in NIH3T3 fibroblasts depletes S1P levels and increases ceramide levels, thereby inducing apoptosis (Mandala, 2001). The small interfering RNA (siRNA) knockdown of endogenous SGPP1 showed the accumulation of S1P within cells and significantly increased the secretion of S1P into the media, suggesting that SGPP1 regulates the secretion of S1P into the extracellular milieu (Olivera et al., 2013). Furthermore, siRNA-induced knockdown of SGPP1 in MCF-7 cells conferred resistance to tumor necrosis factor-α and the chemotherapeutic agent, daunorubicin (Johnson et al., 2003). SGPP1 is downregulated in gastric cancer and plays a role in invasion and migration in gastric cancer cells (Gao et al., 2015). SGPP1 has also been shown to be a target of miR-95, which is highly expressed in ALDH+ and CD133+ subpopulations of non-small cell lung cancer (NSCLC) compared to ALDH1- and CD133-cells. miR-95 overexpression confers radio-resistance to ALDH1+ and CD133 + NSCLC by downregulating SGPP1 (Helleman et al., 2006; Huang et al., 2013). Our study demonstrated that SGPP1 expression positively correlates with better survival outcomes in systemically treated patients with BC, particularly in patients with TNBC. The mechanism by which SGPP1 is decreased in the tumors of BC patients is not fully understood. However, hypermethylation of the SGPP1 promoter does not seem to be involved in the repression of its gene expression. An earlier study has shown that transcriptional repressor GFI1 negatively regulates the expression of SGPP1 and that GFI1-dependent SGPP1 repression promotes growth and survival of multiple myeloma cells (Schwiebs et al., 2017).
The active sites of LPPs are present in the outer leaflet of the plasma membrane (Benesch et al., 2016); thus LPPs are able to degrade S1P, LPA, and other phospholipids present in the extracellular environment. LPP3 expression is downregulated in several cancer lines under hypoxic conditions (Harper et al., 2019), and its downregulation has also been observed in lung biopsies from patients with NSCLC (Magkrioti et al., 2018; Nema et al., 2021). We have also shown that the expression of the gene coding for LPPs, particularly LPP3 (PLPP3), is downregulated in the tumor tissues from oral squamous cell carcinoma patients compared to adjacent normal tissues (Vishwakarma et al., 2017).
The tumor microenvironment is controlled by the complex interaction of tumor cells with immune cells, blood vessels and cancer-associated fibroblasts. S1P signaling through its receptors regulates the migration of immune cells between lymphoid organs, circulation and peripheral tissues (Cartier and Hla, 2019; Pyne and Pyne, 2020). In the tumor microenvironment of patients with TNBC, CD11c positivity correlates with CD4+ and CD8+ T cells. Optimal immune response to tumor cells requires cytotoxic and Th1 cell response induced by antigen-presenting cells. In this context, the capacity of S1P to impair the differentiation of DCs from peripheral monocytes may contribute to the progression of carcinogenesis. Furthermore, a prior study has shown that SGPP1 is localized in the nucleus of naive DCs, and when SGPP1 encounters inflammatory stimuli, SGPP1 translocates into the endoplasmic reticulum and gets activated, resulting in the dephosphorylation of S1P (Schwiebs et al., 2017).
Activation of S1P signaling through S1PR1 is associated with carcinogenesis, metastasis. However, few studies have shown the decrease of S1PR1 in the cell line from triple-negative breast cancer (Abuhussein and Yang, 2020). Furthermore, in a mouse model of lung metastasis, compared to wild type, mice lacking S1pr1 in vascular endothelium (S1pr1 ECKO) developed excessive vascular sprouting and branching, and decreased barrier function, S1pr1 ECKO also developed larger tumors, and enhanced lung metastasis (Cartier et al., 2020). Thus, S1PR1 may exhibit tumor-suppressive attributes in the different cancers in the different histological subtypes or stage-specific manner. The discussion section of the manuscript has been revised accordingly. Expression of S1PR4 is restricted to lymphoid and hematopoietic tissues, and it has been shown previously that S1PR4-deficient mice have reduced numbers of plasmacytoid DCs (pDCs), which has been partly attributed to defects in the differentiation of pDC progenitors into mature pDCs (Dillmann et al., 2015). Infiltration of immune cells, including CD4+ and CD8+ T lymphocytes and DCs into tumors, is associated with better survival of patients with TNBC (Chao et al., 2020). We demonstrated herein that the expression of SGPP1, PLPP3, and S1P receptors, particularly S1PR4, positively correlates with tumor-infiltrating DCs in TNBC.
In summary, we found that low mRNA expression of SGPP1 and PLPP3 correlates with worse prognosis in patients with BC, particularly in those with TNBC, and that high expression of both of these genes may promote the infiltration of immune cells, especially DCs, into the tumor microenvironment (Supplementary Figure S14). Taken together, our findings indicate that SGPP1 may serve as a potential prognostic marker that may be used to predict the effectiveness of systemic therapy in TNBC patients. However, we understand the limitation of our study as our findings are based on the datamining analysis carried out with multiple bioinformatics web-based tools. The prognostic markers identified in the current study need to be validated further in the wet lab. Experimental validation of these findings will be critical in further understanding the interplay between the S1P-signaling pathway in the TIICs and the prognosis of patients with BC.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
AK: Conception, study design, interpretation of data, and critical reading and intellectual assessment of manuscript RN: Analysis and interpretation of data, preparation of the Manuscript: RN
Funding
This work was supported by an extramural research grant (EMR/2016/005009) from the Science and Engineering Research Board and Madhya Pradesh Council of Science and; Technology, Bhopal (A/RD/RP-2/2015-16/50) to AK and by an Indian Council of Medical Research (ICMR) Research Associateship (3/2/3/234/2014/NCD-III) and Department of Health Research (DHR) Young Scientist grant (R.12014/14/2018-HR) to RN.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.697922/full#supplementary-material
References
Abuhussein, O., and Yang, J. (2020). Evaluating the Antitumor Activity of Sphingosine-1-Phosphate against Human Triple-Negative Breast Cancer Cells with Basal-like Morphology. Invest. New Drugs 38, 1316–1325. doi:10.1007/s10637-020-00909-2
Alkabban, F. M., and Ferguson, T. (2020). “Breast Cancer,” in StatPearls (Treasure Island (FL): StatPearls Publishing). Available at: http://www.ncbi.nlm.nih.gov/books/NBK482286/ (Accessed August 20, 2020).
Alshaker, H., Thrower, H., and Pchejetski, D. (2020). Sphingosine Kinase 1 in Breast Cancer-A New Molecular Marker and a Therapy Target. Front. Oncol. 10, 289. doi:10.3389/fonc.2020.00289
Benesch, M. G., Tang, X., Venkatraman, G., Bekele, R. T., and Brindley, D. N. (2016). Recent Advances in Targeting the Autotaxin-Lysophosphatidate-Lipid Phosphate Phosphatase axis In Vivo. J. Biomed. Res. 30, 272–284. doi:10.7555/JBR.30.20150058
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer J. Clinicians 68, 394–424. doi:10.3322/caac.21492@10.3322/(ISSN)1542-4863.statistics
Cartier, A., and Hla, T. (2019). Sphingosine 1-phosphate: Lipid Signaling in Pathology and Therapy. Science 366, eaar5551. doi:10.1126/science.aar5551
Cartier, A., Leigh, T., Liu, C. H., and Hla, T. (2020). Endothelial Sphingosine 1-phosphate Receptors Promote Vascular Normalization and Antitumor Therapy. Proc. Natl. Acad. Sci. USA 117, 3157–3166. doi:10.1073/pnas.1906246117
Chandrashekar, D. S., Bashel, B., Balasubramanya, S. A. H., Creighton, C. J., Ponce-Rodriguez, I., Chakravarthi, B. V. S. K., et al. (2017). UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 19, 649–658. doi:10.1016/j.neo.2017.05.002
Chao, X., Liu, L., Sun, P., Yang, X., Li, M., Luo, R., et al. (2020). Immune Parameters Associated with Survival in Metaplastic Breast Cancer. Breast Cancer Res. 22, 92. doi:10.1186/s13058-020-01330-6
Denkert, C., von Minckwitz, G., Darb-Esfahani, S., Lederer, B., Heppner, B. I., Weber, K. E., et al. (2018). Tumour-infiltrating Lymphocytes and Prognosis in Different Subtypes of Breast Cancer: a Pooled Analysis of 3771 Patients Treated with Neoadjuvant Therapy. Lancet Oncol. 19, 40–50. doi:10.1016/S1470-2045(17)30904-X
Dillmann, C., Mora, J., Olesch, C., Brüne, B., and Weigert, A. (2015). S1PR4 Is Required for Plasmacytoid Dendritic Cell Differentiation. Biol. Chem. 396, 775–782. doi:10.1515/hsz-2014-0271
Fyrst, H., and Saba, J. D. (2010). An Update on Sphingosine-1-Phosphate and Other Sphingolipid Mediators. Nat. Chem. Biol. 6, 489–497. doi:10.1038/nchembio.392
Gao, J., Aksoy, B. A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S. O., et al. (2013). Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signaling 6, pl1. doi:10.1126/scisignal.2004088
Gao, X., Li, L., Wang, X., Wen, X., Ji, K., Ye, L., et al. (20151977–1987). [Corrigendum] Inhibition of Sphingosine-1-Phosphate Phosphatase�1 Promotes Cancer Cells Migration in Gastric Cancer: Clinical Implications. Oncol. Rep. 34, 2051. doi:10.3892/or.2018.626910.3892/or.2015.4162
Győrffy, B., Surowiak, P., Budczies, J., and Lánczky, A. (2013). Online Survival Analysis Software to Assess the Prognostic Value of Biomarkers Using Transcriptomic Data in Non-small-cell Lung Cancer. PLoS ONE 8, e82241. doi:10.1371/journal.pone.0082241
Harbeck, N., Penault-Llorca, F., Cortes, J., Gnant, M., Houssami, N., Poortmans, P., et al. (2019). Breast Cancer. Nat. Rev. Dis. Primers 5, 1–31. doi:10.1038/s41572-019-0111-2
Harper, K., Brochu-Gaudreau, K., Saucier, C., and Dubois, C. M. (2019). Hypoxia Downregulates LPP3 and Promotes the Spatial Segregation of ATX and LPP1 during Cancer Cell Invasion. Cancers 11, 1403. doi:10.3390/cancers11091403
Helleman, J., Jansen, M. P. H. M., Span, P. N., van Staveren, I. L., Massuger, L. F. A. G., Meijer-van Gelder, M. E., et al. (2006). Molecular Profiling of Platinum Resistant Ovarian Cancer. Int. J. Cancer 118, 1963–1971. doi:10.1002/ijc.21599
Huang, X., Taeb, S., Jahangiri, S., Emmenegger, U., Tran, E., Bruce, J., et al. (2013). miRNA-95 Mediates Radioresistance in Tumors by Targeting the Sphingolipid Phosphatase SGPP1. Cancer Res. 73, 6972–6986. doi:10.1158/0008-5472.CAN-13-1657
Inic, Z., Zegarac, M., Inic, M., Markovic, I., Kozomara, Z., Djurisic, I., et al. (2014). Difference between Luminal A and Luminal B Subtypes According to Ki-67, Tumor Size, and Progesterone Receptor Negativity Providing Prognostic Information. Clin. Med. Insights Oncol. 8,107–111. doi:10.4137/CMO.S18006
Johnson, K. R., Johnson, K. Y., Becker, K. P., Bielawski, J., Mao, C., and Obeid, L. M. (2003). Role of Human Sphingosine-1-Phosphate Phosphatase 1 in the Regulation of Intra- and Extracellular Sphingosine-1-Phosphate Levels and Cell Viability. J. Biol. Chem. 278, 34541–34547. doi:10.1074/jbc.M301741200
Jonnada, P. K., Sushma, C., Karyampudi, M., and Dharanikota, A. (2020). Prevalence of Molecular Subtypes of Breast Cancer in India: a Systematic Review and Meta-Analysis. Indian J. Surg. Oncol. 12, 152–163. doi:10.1007/s13193-020-01253-w
Kumar, A., Zamora-Pineda, J., Degagné, E., and Saba, J. D. (2017). S1P Lyase Regulation of Thymic Egress and Oncogenic Inflammatory Signaling. Mediators Inflamm. 2017, 1–19. doi:10.1155/2017/7685142
Lehmann, B. D., Bauer, J. A., Chen, X., Sanders, M. E., Chakravarthy, A. B., Shyr, Y., et al. (2011). Identification of Human Triple-Negative Breast Cancer Subtypes and Preclinical Models for Selection of Targeted Therapies. J. Clin. Invest. 121, 2750–2767. doi:10.1172/JCI45014
Lei, F.-J., Cheng, B.-H., Liao, P.-Y., Wang, H.-C., Chang, W.-C., Lai, H.-C., et al. (2018). Survival Benefit of Sphingosin-1-Phosphate and Receptors Expressions in Breast Cancer Patients. Cancer Med. 7, 3743–3754. doi:10.1002/cam4.1609
Li, Q., Birkbak, N. J., Gyorffy, B., Szallasi, Z., and Eklund, A. C. (2011). Jetset: Selecting the Optimal Microarray Probe Set to Represent a Gene. BMC Bioinformatics 12, 474. doi:10.1186/1471-2105-12-474
Li, T., Fan, J., Wang, B., Traugh, N., Chen, Q., Liu, J. S., et al. (2017). TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 77, e108–e110. doi:10.1158/0008-5472.CAN-17-0307
Magkrioti, C., Oikonomou, N., Kaffe, E., Mouratis, M.-A., Xylourgidis, N., Barbayianni, I., et al. (2018). The Autotaxin - Lysophosphatidic Acid axis Promotes Lung Carcinogenesis. Cancer Res. 78,3634–3644. doi:10.1158/0008-5472.CAN-17-3797
Mandala, S. M. (2001). Sphingosine-1-phosphate Phosphatases. Prostaglandins & Other Lipid Mediators 64, 143–156. doi:10.1016/s0090-6980(01)00111-3
Manjunath, M., and Choudhary, B. (2021). Triplenegative Breast Cancer: A Run through of Features, Classification and Current Therapies (Review). Oncol. Lett. 22, 1–21. doi:10.3892/ol.2021.12773
Nagahashi, M., Yamada, A., Miyazaki, H., Allegood, J. C., Tsuchida, J., Aoyagi, T., et al. (2016). Interstitial Fluid Sphingosine-1-Phosphate in Murine Mammary Gland and Cancer and Human Breast Tissue and Cancer Determined by Novel Methods. J. Mammary Gland Biol. Neoplasia 21, 9–17. doi:10.1007/s10911-016-9354-7
Nema, R., Shrivastava, A., and Kumar, A. (2021). Prognostic Role of Lipid Phosphate Phosphatases in Non-smoker, Lung Adenocarcinoma Patients. Comput. Biol. Med. 129, 104141. doi:10.1016/j.compbiomed.2020.104141
Olivera, A., Allende, M. L., and Proia, R. L. (2013). Shaping the Landscape: Metabolic Regulation of S1P Gradients. Biochim. Biophys. Acta (Bba) - Mol. Cel Biol. Lipids 1831, 193–202. doi:10.1016/j.bbalip.2012.06.007
Postma, F. R., Jalink, K., Hengeveld, T., and Moolenaar, W. H. (1996). Sphingosine-1-phosphate Rapidly Induces Rho-dependent Neurite Retraction: Action through a Specific Cell Surface Receptor. EMBO J. 15, 2388–2392. doi:10.1002/j.1460-2075.1996.tb00595.x
Pyne, N. J., and Pyne, S. (2020). Recent Advances in the Role of Sphingosine 1‐phosphate in Cancer. FEBS Lett. 594, 3583–3601. doi:10.1002/1873-3468.13933
Rivera, J., Proia, R. L., and Olivera, A. (2008). The Alliance of Sphingosine-1-Phosphate and its Receptors in Immunity. Nat. Rev. Immunol. 8, 753–763. doi:10.1038/nri2400
Ruckhäberle, E., Rody, A., Engels, K., Gaetje, R., von Minckwitz, G., Schiffmann, S., et al. (2008). Microarray Analysis of Altered Sphingolipid Metabolism Reveals Prognostic Significance of Sphingosine Kinase 1 in Breast Cancer. Breast Cancer Res. Treat. 112, 41–52. doi:10.1007/s10549-007-9836-9
Sarkar, S., Maceyka, M., Hait, N. C., Paugh, S. W., Sankala, H., Milstien, S., et al. (2005). Sphingosine Kinase 1 Is Required for Migration, Proliferation and Survival of MCF-7 Human Breast Cancer Cells. FEBS Lett. 579, 5313–5317. doi:10.1016/j.febslet.2005.08.055
Schwiebs, A., Thomas, D., Kleuser, B., Pfeilschifter, J. M., and Radeke, H. H. (2017). Nuclear Translocation of SGPP-1 and Decrease of SGPL-1 Activity Contribute to Sphingolipid Rheostat Regulation of Inflammatory Dendritic Cells. Mediators Inflamm. 2017, 1–10. doi:10.1155/2017/5187368
Sukocheva, O. (2018). Expansion of Sphingosine Kinase and Sphingosine-1-Phosphate Receptor Function in Normal and Cancer Cells: From Membrane Restructuring to Mediation of Estrogen Signaling and Stem Cell Programming. Int. J. Mol. Sci. 19, 420. doi:10.3390/ijms19020420
Sukocheva, O., Wadham, C., Holmes, A., Albanese, N., Verrier, E., Feng, F., et al. (2006). Estrogen Transactivates EGFR via the Sphingosine 1-phosphate Receptor Edg-3: the Role of Sphingosine Kinase-1. J. Cel Biol. 173, 301–310. doi:10.1083/jcb.200506033
Szász, A. M., Lánczky, A., Nagy, Á., Förster, S., Hark, K., Green, J. E., et al. (2016). Cross-validation of Survival Associated Biomarkers in Gastric Cancer Using Transcriptomic Data of 1,065 Patients. Oncotarget 7, 49322–49333. doi:10.18632/oncotarget.10337
Tran Janco, J. M., Lamichhane, P., Karyampudi, L., and Knutson, K. L. (2015). Tumor-Infiltrating Dendritic Cells in Cancer Pathogenesis. J. Immunol. 194, 2985–2991. doi:10.4049/jimmunol.1403134
Tsuchida, J., Nagahashi, M., Nakajima, M., Moro, K., Tatsuda, K., Ramanathan, R., et al. (2016). Breast Cancer Sphingosine-1-Phosphate Is Associated with Phospho-Sphingosine Kinase 1 and Lymphatic Metastasis. J. Surg. Res. 205, 85–94. doi:10.1016/j.jss.2016.06.022
Tsuchida, J., Nagahashi, M., Takabe, K., and Wakai, T. (2017). Clinical Impact of Sphingosine-1-Phosphate in Breast Cancer. Mediators Inflamm. 2017, 1–9. doi:10.1155/2017/2076239
Vishwakarma, S., Agarwal, R., Goel, S. K., Panday, R. K., Singh, R., Sukumaran, R., et al. (2017). Altered Expression of Sphingosine-1-Phosphate Metabolizing Enzymes in Oral Cancer Correlate with Clinicopathological Attributes. Cancer Invest. 35, 139–141. doi:10.1080/07357907.2016.1272695
Wang, F., Van Brocklyn, J. R., Hobson, J. P., Movafagh, S., Zukowska-Grojec, Z., Milstien, S., et al. (1999). Sphingosine 1-Phosphate Stimulates Cell Migration through a Gi-Coupled Cell Surface Receptor. J. Biol. Chem. 274, 35343–35350. doi:10.1074/jbc.274.50.35343
Wang, Q., Li, J., Li, G., Li, Y., Xu, C., Li, M., et al. (2014). Prognostic Significance of Sphingosine Kinase 2 Expression in Non-small Cell Lung Cancer. Tumor Biol. 35, 363–368. doi:10.1007/s13277-013-1051-1
Zhou, J., and Saba, J. D. (1998). Identification of the First Mammalian Sphingosine Phosphate Lyase Gene and its Functional Expression in Yeast. Biochem. Biophysical Res. Commun. 242, 502–507. doi:10.1006/bbrc.1997.7993
Keywords: breast cancer, sphingosine-1-phosphate, SGPP1, PLPP3, S1PR4, tumor-infiltrating immune cells
Citation: Nema R and Kumar A (2021) Sphingosine-1-Phosphate Catabolizing Enzymes Predict Better Prognosis in Triple-Negative Breast Cancer Patients and Correlates With Tumor-Infiltrating Immune Cells. Front. Mol. Biosci. 8:697922. doi: 10.3389/fmolb.2021.697922
Received: 20 April 2021; Accepted: 04 June 2021;
Published: 21 June 2021.
Edited by:
Gaurav Malviya, University of Glasgow, United KingdomReviewed by:
Olga A Sukocheva, Flinders University, AustraliaChandi C Mandal, Central University of Rajasthan, India
Lokendra Sharma, Sanjay Gandhi Post Graduate Institute of Medical Sciences (SGPGI), India
Copyright © 2021 Nema and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ashok Kumar, YXNob2suYmlvY2hlbWlzdHJ5QGFpaW1zYmhvcGFsLmVkdS5pbg==
 Rajeev Nema
Rajeev Nema Ashok Kumar
Ashok Kumar