
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci. , 01 July 2021
Sec. Molecular Diagnostics and Therapeutics
Volume 8 - 2021 | https://doi.org/10.3389/fmolb.2021.685088
Background: A delay in the diagnosis of acute ischemic stroke (AIS) reduces the eligibility and outcome of patients for thrombolytic therapy. Therefore, early diagnosis and treatment of AIS are crucial. The present study evaluated the sensitivity and accuracy of serum extracellular vesicle (EV)-derived miR-124-3p in the diagnosis and prediction of AIS.
Methods: An miRNA expression profile was downloaded from Gene Expression Omnibus (GEO) database and analyzed by R software. EVs were harvested from the serum of AIS patients using a total exosome isolation kit and characterized by Western blotting, a transmission electron microscope, and the nanoparticle tracking analysis. BV2 microglia were pre-stimulated with lipopolysaccharide (LPS), followed by miR-124-3p treatment for 24 h and subsequent analysis of viability, apoptosis, and migration (scratch assay), and Western blotting. The relative expression of the selected genes was assessed by qRT-PCR. The phosphorylation of Erk1/2, PI3K/Akt, and p38MAPK in BV2 microglia cells was evaluated by Western blotting, while the luciferase reporter gene assay detected the correlation between key genes involved in the pro-inflammatory signaling pathways and miR-124-3p.
Results:hsa-miR-124-3p was downregulated in AIS serum compared to the non-AIS serum (p < 0.05), and the gene expression of has-miR-124-3p in EVs was negatively correlated with serum pro-inflammatory cytokines and the NIHSS (p < 0.05). In addition, miR-124-3p promoted the viability and inhibited the apoptosis of LPS-induced BV2 microglia. Furthermore, miR-124-3p reduced the phosphorylation of Erk1/2, PI3K/Akt, and p38MAPK, and promoted the migration in LPS-induced BV2 microglia (p < 0.05).
Conclusion: Serum EV-derived miR-124-3p serves as a diagnostic and predictive marker for early-stage AIS.
Acute ischemic stroke (AIS) is one of the leading causes of death and adult disability worldwide (Bonita et al., 2004). It is a dynamic process, inducing acute brain damage and cell death (Urra et al., 2014). A delay in the diagnosis of AIS reduces the eligibility and outcome of patients for thrombolytic therapy (Hwong et al., 2016). Therefore, early diagnosis and treatment of AIS are crucial. Presently, computed tomography (CT) and magnetic resonance imaging (MRI) are considered the standard methods to diagnose AIS (Yan et al., 2017). However, the sensitivity of CT diagnosis of AIS is only 26% in the early stage (Kalafut et al., 2000). MRI presents a sensitive diagnosis of AIS, but is time-consuming and expensive, and is not available in all emergency departments (Korolev et al., 2016). Thus, there is an urgent need to explore an early alternative with a precise diagnosis of AIS. A large number of studies have been focused on investigating the role of specific markers in the diagnosis and outcome prediction in AIS (Dassan et al., 2009; Vijayan and Reddy, 2016; Zuo et al., 2020). However, due to insufficient understanding of the sensitivity and specificity of AIS standard markers, identification of these markers is challenging.
MicroRNAs (miRNAs) are a group of noncoding RNAs with ∼22 nucleotides, which regulate the biological processes posttranscriptionally by modulating the expression of target mRNAs in many diseases (Cardo et al., 2013; Leidinger et al., 2013; Hayes et al., 2014; Gurha, 2016). Reportedly, the microRNA expression profile is altered in AIS blood samples, suggesting that miRNAs are involved in the progress of AIS (Wu et al., 2012; Bhalala et al., 2013). Furthermore, several miRNAs have been identified as novel therapeutic targets for AIS (He et al., 2019; Yan et al., 2019). However, none of these have been applied in clinical practice (Harpaz et al., 2017).
Extracellular vesicles (EVs) are a group of membrane-encapsulated vesicles released by cells, biofluids, and tissues into the extracellular space, containing miRNAs, DNA, mRNAs, and proteins (Ariston Gabriel et al., 2020). EVs are involved in cell-to-cell communication despite the blood–brain barrier (BBB) and the blood–cerebrospinal fluid barrier (Skog et al., 2008; Zagrean et al., 2018). A large number of studies have described the role of EVs in stroke diagnosis, pathogenesis, and as a treatment target in the future (Deng et al., 2017; Hong et al., 2019), while a few studies demonstrated that the amount of some cell-specific EVs was increased in stroke patients compared to the controls (Jung et al., 2009; Świtońska et al., 2015; He et al., 2017).
Several EV-derived miRNAs have been altered in AIS patients. miR-21-5p and miR-30a-5p were highly expressed in AIS but decreased in the subacute phase (Wang et al., 2018). This finding indicated that the expression of EV-derived miRNAs depended on the time point of sample collection. These findings have raised the idea that EV-derived miRNAs from the serum of AIS patients may serve as analytes in the liquid biopsies for real-time monitoring of AIS severity and outcome.
In the present study, we found that the expression of hsa-miR-124-3p significantly increased in EVs of AIS patients, which was negatively correlated with the pro-inflammatory cytokines and the National Institutes of Health Stroke Scale (NIHSS). Additionally, we observed that hsa-miR-124-3p was involved in inhibiting the pro-inflammatory signaling pathways. Thus, this study might provide a novel therapeutic target and a diagnostic marker for AIS.
Microarray dataset (GSE95204) was obtained from the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/). The use of human material was approved by the local Ethics Committee (reference number: 2020-KY-262, The First Affiliated Hospital of Zhengzhou University), and written consent was obtained from all patients. AIS was diagnosed by neuroimaging (CT or MRI) and stroke. Patients treated with intravenous recombinant tissue-type plasminogen activator (rt-PA) within 6 h after symptom onset were enrolled in this study. Experienced neurologists determined the neurological deficits based on the NIHSS within 6 h after the AIS onset and after rt-PA treatment. The serum samples were obtained from 10 patients at 2 h (AIS-2H, mean age: 72.5 ± 7.2 years, and range: 57–82 years), 4 h (AIS-4H, mean age: 73.0 ± 6.8 years, and range: 53–83 years), and 6 h (AIS-6H, mean age: 75.6 ± 7.2 years, and range: 56–84 years) after the onset of AIS. Control serum samples were obtained from 10 patients without AIS (mean age: 65.2 ± 8.0 years and range: 50–84 years). The fresh blood samples were collected in the vacutainer tubes, and serum was obtained by clotting at room temperature for 1 h and centrifuged at 1,500 × g for 10 min at 4°C (Liu et al., 2018). Mouse microglia cell line BV2 was purchased from Procell Life Science & Technology Co., Ltd. (Wuhan, China), cultured in DMEM medium (#D8437, Sigma, United Kingdom), supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich GmbH, Germany) and 1% penicillin–streptomycin at 37°C under 5% CO2 atmosphere.
R package and Limma package (http://bioconductor.org/packages/release/bioc/html/limma.html) were used to normalize and screen the differentially expressed miRNAs. The differential expression genes (DEGs) with a fold-change value >1 or <−1 and a p-value < 0.05 were considered to be significantly expressed.
miRNA target prediction genes were predicted using TargetScan (http://www.targetscan.org/), miRDB (http://mirdb.org/), and miRTarBase (http://mirtarbase.cuhk.edu.cn/), and selected if they were recorded in at least two prediction databases (Yu et al., 2012).
Gene ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were enriched in miRNA-predicted target genes analyzed using the clusterProfiler (version 3.11) of R package.
EVs were isolated from 0.5 ml serum using a total exosome isolation kit (Thermo Fisher Scientific, Waltham, MA, United States) according to the manufacturer’s instructions. EVs were visualized by transmission electron microscopy (TEM), and their concentration was determined using the nanoparticle tracking analysis (NTA; NS300, Malvern Instruments, Malvern, United Kingdom). The classical surface markers (CD9, CD63, and CD81) of EVs were verified by Western blotting.
The total RNA of serum was isolated using the Absolutely RNA Miniprep Kit (Agilent Technologies, Santa Clara, CA, United States), according to the manufacturer’s instructions, and then cDNA was synthesized using a cDNA synthesis kit (Agilent Technologies). The RNA concentration and quality were determined using a NanoDrop™ (ND-1000 spectrophotometer, Thermo Fisher Scientific). The relative mRNA levels were assessed using Brilliant III Ultra-Fast SYBR® Green QPCR Master Mix (Agilent Technologies) on an MX3005P QPCR System (Agilent Technologies). All genes were analyzed relatively, calibrated to the expression of control groups, and normalized to those of GAPDH and 18S. All the qPCR experiments in this study were performed in duplicates using 20–50 ng cDNA.
For hsa-miR-124-3p expression analysis, the total RNA of cells was isolated using the miRNeasy Mini Kit (Qiagen, United States) according to the manufacturer’s instructions. hsa-miR-124-3p and U6 (reference gene) were detected by TaqMan MicroRNA Assays (Applied Biosystems, Forest City, CA, United States). The sequences of all primers used for quantitative real-time polymerase chain reaction (qRT-PCR) in this study were as follows: CXCL2: 5′-GCCAAGGGTTGACTTCAA GA-3′ (forward), 5′-CTTCAGGGTCAAGGCAAACT-3′ (reverse); IL1β: 5′-TGCCACCTTTTGACAGTGATGA-3′ (forward), 5′-TGTGCTGCTGCGAGATTTGA-3′ (reverse); IL6: 5′-CCGGAGAGGAGACTTCACAG-3′ (forward), 5′-CAG AATTGCCATTGCACAAC-3′ (reverse); TNF-α: 5′-CTCTGTGAAGGGAATGGGTG-3′ (forward), 5′-GGGCTCTGAGGAGTAGACGATAAAG-3′ (reverse); GRB2: 5′-ATTCCTGCGGGACATAGAACA-3′ (forward) and 5′-GGTGACATAATTGCGGGGAAAC-3′ (reverse); AKT3: 5′-TGTGGATTTACCTTATCCCCTCA-3′ (forward) and 5′-GTTTGGCTTTGGTCGTTCTGT-3′ (reverse); GAPDH: 5′-GTGAAGGTCGGTGTGAACG-3′ (forward), 5′-AATCTCCACTTTGCCACTGC-3′(reverse); 18S: 5′-AAACGGCTACCACATCCAAG-3′ (forward); 5′-CCTCCAATGGATCCTCGTTA-3′ (reverse).
Serum proteins were extracted using a serum protein extraction kit (BestBio, Shanghai, China) containing phosphatase. BV2 cells were lysed in RIPA buffer (Thermo Scientific) containing phosphatase (Roche, Germany) and proteinase inhibitors. The cellular protein and EVs were quantified using a BCA Protein Kit Assay (Thermo Fisher Scientific). An equivalent amount (10 μg) of cell lysates, serum protein, or EVs was resolved by 12% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were transferred to a 0.22-µm polyvinylidene fluoride (PVDF) membrane and blocked with 5% BSA for 1 h at room temperature. Subsequently, the membranes were incubated with primary antibodies, CD9 (ThermoFisher 10626D) 1:1,000, CD81 (ThermoFisher 10630D) 1:1,000, CD 63 (ThermoFisher 10628D) 1:500, Phospho-P38 (Cell Signaling 4,511) 1:1,000, P38 (Cell Signaling 9,212) 1:500, Phospho-Erk1/2 (Cell Signaling 4,370) 1:1,000, Erk1/2 (Cell Signaling 4,695) 1:500, Phospho-Akt (Cell Signaling 4,060) 1:1,000, and Akt (Cell Signaling 4,691) at 4°C overnight, followed by goat anti-rabbit IgG secondary antibodies (Jackson Immuno Research) for 1 h at room temperature. Protein bands were visualized using ECL detection reagents (Thermo Scientific). Image J (1.8.0, the National Institutes of Health, Bethesda, MD, United States) software was used to quantify the density of bands of phosphorylated proteins and normalize that to the total amount of proteins.
BV2 microglial cells (2 × 105/well) were cultured in 6-well plates with DMEM medium (supplemented with 10% FBS and 1% penicillin–streptomycin), stimulated with lipopolysaccharide (LPS, 50 ng/ml) (#L2630, Sigma-Aldrich) for 12 h at 37°C, 5% CO2, and 95% relative humidity, before the follow-up treatment and assays.
Calcein AM (Sigma) was utilized for simultaneous fluorescence detection. An equivalent of 2 × 104 BV2 microglial cells were seeded in 24-well plates for 24 h, and then calcein AM (3 μM) was added to the culture medium. The live (green) cells were determined using a fluorescence microscope and Image J software (the National Institutes of Health, United States) for cell counting.
In order to investigate the effect of hsa-miR-124-3p on LPS-induced BV2 microglial cells, hsa-miR-124-3p mimic was transfected to overexpress hsa-miR-124-3p in LPS-induced BV2 microglia cells. miR-124-3p mimics and mimic negative control (miR-NC) were purchased from GenePharma (Shanghai, China), and the sequences were as follows: hsa-miR-124-3p mimic: 5′-UAAGGCACGCGGUGAAUGCCAA-3′. BV2 microglial cells (2 × 105) were cultured in 6-well plates with DMEM medium (supplemented with 10% FBS and 1% penicillin–streptomycin), stimulated with LPS (50 ng/ml) for 12 h, and hsa-miR-124-3p mimic was transfected at a negative control using Lipofectamine® RNAiMAX Reagent (#13778075, Invitrogen, United States) at a final concentration of 100 nM. Viability assay, Caspase-3/7 assay, live assay, and migration were performed 24 h after transfection.
Cell Counting Kit-8 (CCK-8) (Dojindo, Kumamoto, Japan) was utilized to measure the cell viability according to the manufacturer’s protocol. An equivalent of 5 × 103 BV2 microglial cells (cultured in DMEM medium, supplemented with 10% FBS and 1% penicillin–streptomycin) were seeded into 96-well plates. After 24 h, the medium was exchanged for those containing appropriate treatment agents (LPS, miR-124-3p mimic, and mimic-NC). After an additional 24 h, cells were incubated with 100 µl of the fresh medium containing 10% CCK-8 reagent for 1 h at 37°C. Then the absorbance was measured at 450 nm on a Tecan ELISA reader (Mannedorf, Switzerland).
To determine apoptosis of BV2 microglia cells with different treatments, caspase-3 and caspase-7 activity was evaluated using a Caspase-3/7 Assay Kit (Promega Corporation, Madison, WI, United States) according to the manufacturer’s instructions. After different treatments in a 96-well plate, a nonfluorescent caspase substrate, added to the expansion medium, was cleaved into fluorescent molecules with an emission maximum at 521 nm and evaluated on a Tecan ELISA reader (Mannedorf, Switzerland).
An equivalent of 2 × 105 BV2 microglial cells (control, hsa-miR-124-3p-mimic transfected, and negative control) were cultured in 12-well plates. At 100% confluency, the cells were scraped away with a 100-μl pipette tip and washed twice with phosphate-buffered saline (PBS), and after changing the medium with 1% FBS, LPS (50 ng/ml) was added. Images were captured under a microscope at 0, 24, and 48 h post-wounding, and closure of the wound gap was determined using Image J (1.8.0, the National Institutes of Health).
GRB2 and AKT3 as direct target genes of miR-124-3p were screened out based on the TargetScan database. The luciferase reporter gene assay was performed with the Dual-Luciferase Reporter Assay System (Promega). The full length of the 3′-untranslated region (UTR) of the wild-type (WT) GRB2/AKT3 gene was cloned into the vector pMIR-REPORT™ Luciferase (Promega), and the mutant-type (MUT) vectors were constructed by site-directed mutation of the binding site between miR-124-3p and GRB2/AKT3. The phRL-TK vector (Promega) expressing the Renilla luciferase was used to normalize the transfection efficiency. An equivalent of 1 × 104 BV2 microglia were seeded in 96-well plates. At 70–80%, the reporter gene vectors (WT and MUT of GRB2/AKT3) were co-transfected with miR-124-3p-mimic/mimic-NC into the BV2 microglia to determine the luciferase activity, according to the manufacturer’s instructions.
All data are expressed as the mean ± standard deviation (SD). Statistical analysis was performed using Prism 8.02 software (GraphPad Software, United States). The comparisons between two groups were analyzed by independent two-tailed Student’s t-tests, and comparisons between more than two groups were analyzed by one-way analysis of variance (ANOVA) with the Newman–Keuls multiple comparison test. Each assay was performed in duplicate and repeated in at least three independent experiments. p < 0.05 was regarded as statistically significant.
The results of miRNA (miR) microarray dataset analysis suggested that the expression of hsa-miR-124-3p was significantly lower in the serum of AIS patients than that of hsa-miR-124-3p in the serum of non-AIS (control) patients (p < 0.05) (Figures 1A,B). The target genes of hsa-miR-124-3p were identified using the public database, GO and KEGG pathway enrichment, which showed that hsa-miR-124-3p is involved in the regulation of pro-inflammatory signaling pathways, including PI3K/AKT and MAPK (Figures 1C,D). These data suggested that reducing the expression of hsa-miR-124-3p plays a critical role in the pathophysiology of AIS via regulating the pro-inflammatory signaling pathways.
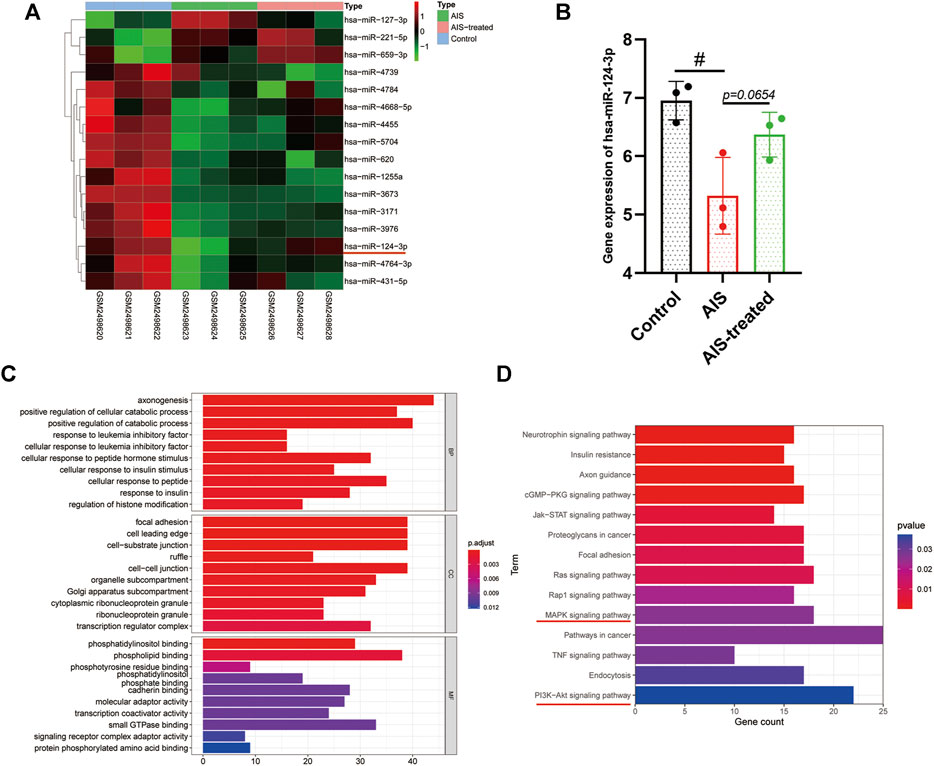
FIGURE 1. Bioinformatic analysis and the gene expression of hsa-miR-124-3p in AIS and AIS-treated blood samples. (A) Heat map of differentially expressed miRNAs in control (none-AIS), AIS, and AIS-treated samples. (B) Gene expression analysis of hsa-miR-124-3p in control (none-AIS), AIS, and AIS-treated samples. #p < 0.05; one-way ANOVA with the Newman–Keuls multiple comparison test. (C,D) GO function and KEGG pathway enrichment of targeted genes of hsa-miR-124-3p.
In order to validate EVs, Western blotting was performed to verify the classical biomarkers (CD9, CD63, and CD81) of EVs (Figure 2A). The morphology of EVs was monitored by TEM (Figure 2B), and the particle size distribution was tested by NTA (Figures 2C,D), which indicated that the average diameter of EVs is approximately 110 nm.
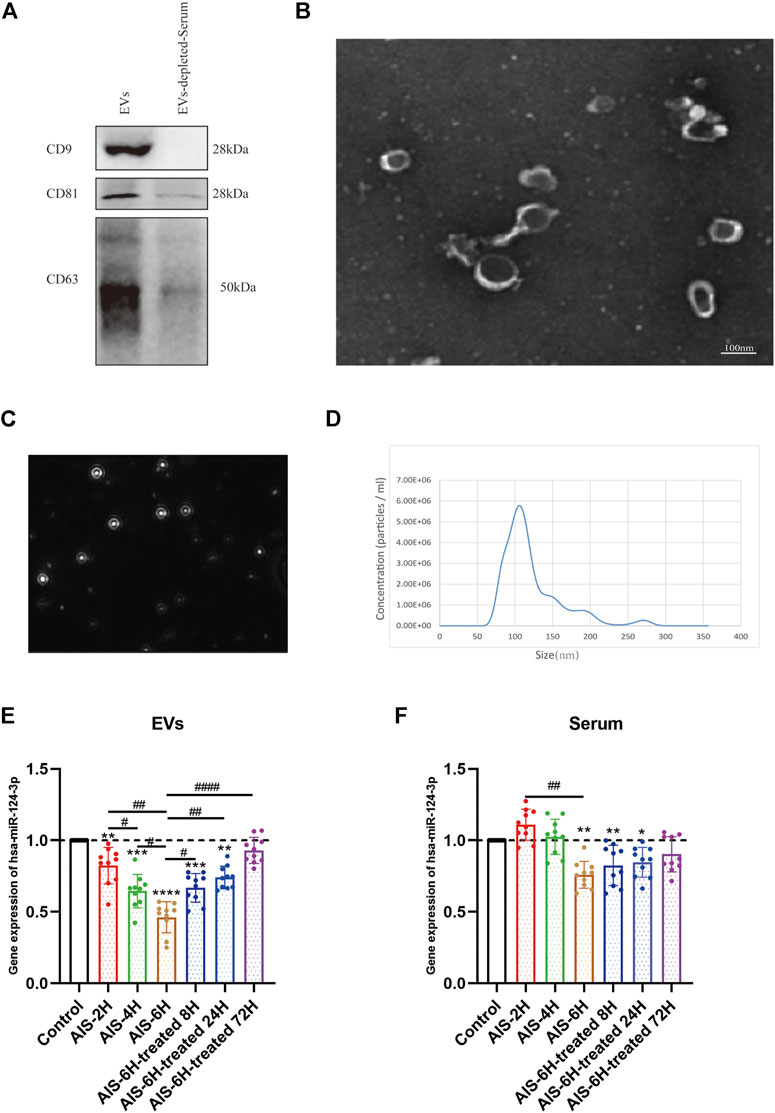
FIGURE 2. Characterization of control EVs and gene analysis of hsa-124-3p in serum-derived EVs and serum. (A) Representative image of Western blotting showed bands of standard surface markers (CD9, CD63, and CD81) of EVs and EV-depleted serum. (B) Morphology of EVs was monitored by TEM; scale bar: 100 nm. (C,D) Particle size distribution of EVs was determined by NTA. (E,F) Gene expression of hsa-miR-124-3p in EVs and serum of different samples at different time points. Compared to the control group: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001, # Difference between groups: #p < 0.05; ##p < 0.01; ####p < 0.0001; one-way ANOVA with the Newman–Keuls multiple comparison test.
To investigate the correlation between AIS and hsa-miR-124-3p, we used qPCR to determine the expression changes of hsa-miR-124-3p in different samples at different time points. The AIS-6H group (6 h after the onset of AIS) was selected to verify the expression of hsa-miR-124-3p in EVs and serum after treatment. The results showed that hsa-miR-124-3p was significantly downregulated in EVs after the onset of AIS (p < 0.05), 4 h earlier than that in serum (Figures 2E,F). Furthermore, rt-PA treatment reversed the decreased expression of hsa-miR-124-3p in EVs derived from AIS serum (p < 0.05); however, there was no significant difference in the serum of treated groups (Figures 2E,F). These results indicated that hsa-miR-124-3p in EVs was more sensitive to AIS than serum.
To investigate the inflammation induced by AIS, the gene expression of pro-inflammatory cytokines (CXCL2, IL6, IL1β, and TNF-α) in the serum was determined using qPCR. As shown in Figures 3A–D, as the onset time of AIS increased, the expression of pro-inflammatory cytokines (CXCL2, IL6, IL1β, and TNF-α) increased (p < 0.05). Additionally, the expression of all the pro-inflammatory cytokines was decreased after rt-PA treatment (p < 0.05), suggesting that inflammation plays a critical role in the progress of AIS.
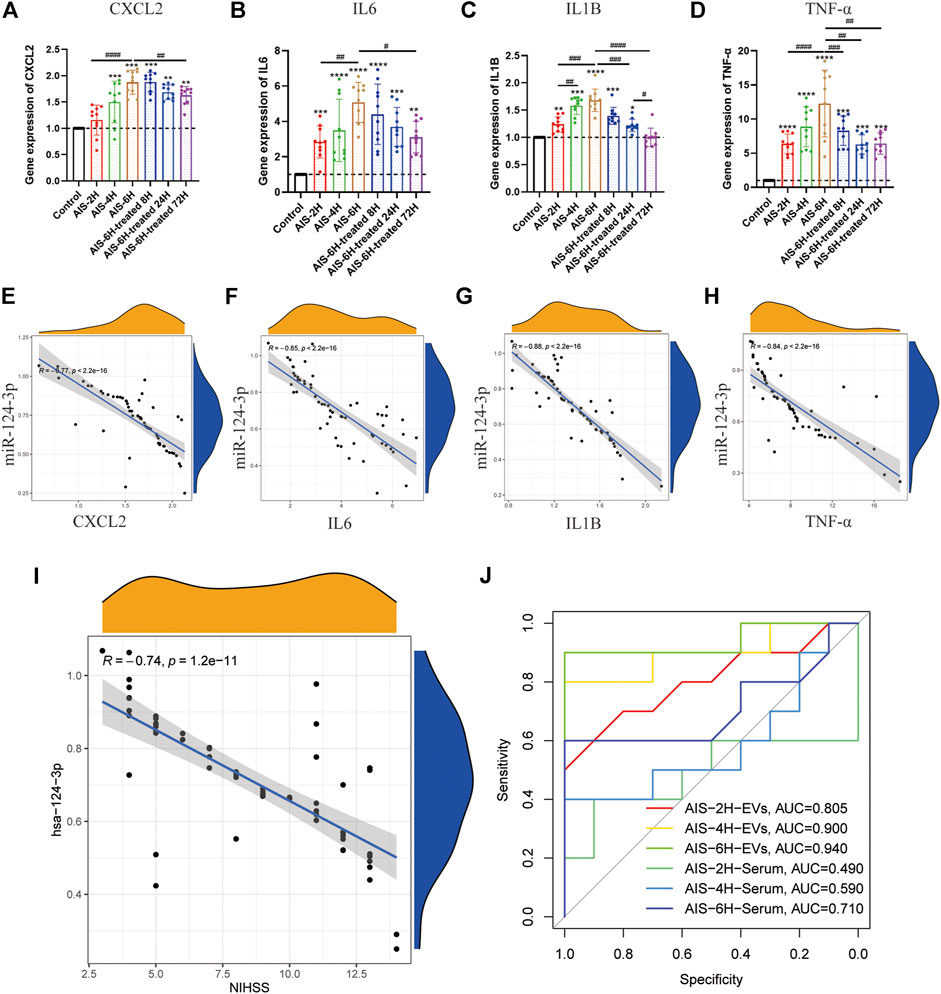
FIGURE 3. Gene expression of hsa-miR-124-3p in EVs negatively correlated with inflammatory factors in serum and clinical symptoms. (A–D) Gene expression of inflammatory factor (CXCL2, IL6, IL1β, and TNF-α) analysis in different groups at different time points. Difference to the control group: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001, # Difference between groups: #p < 0.05; ##p < 0.01; ###p < 0.001; ####p < 0.0001; and one-way ANOVA with the Newman–Keuls multiple comparison test. (E–H) Correlation analysis of the gene expression between hsa-miR-124-3p in EVs and inflammatory factors. R: Pearson’s correlation coefficient, correlation coefficient |R| > 0.5, p < 0.05 was considered statistically significant. (I) Correlation analysis of the gene expression of hsa-miR-124-3p and the NIHSS. R: Pearson’s correlation coefficient, correlation coefficient |R| > 0.5; p < 0.05 was considered statistically significant. (J) ROC curve of hsa-miR-124-3p expression in different groups for predicting the diagnosis of AIS.
In the present study, AIS severity and outcome were determined by the NIHSS score (Kwah and Diong, 2014). Pearson’s correlation coefficient analysis showed that hsa-miR-124-3p was negatively correlated to pro-inflammatory cytokines and the NIHSS (p < 0.05) (Figures 3E–I). The receiver-operating characteristic (ROC) curve of hsa-miR-124-3p was used to analyze the diagnostic capability of EVs and serum. Compared to the serum, EVs presented a better diagnostic capability for AIS (Figure 3J). These results demonstrated that the hsa-miR-124-3p in EVs derived from AIS serum might be a potential marker in the diagnosis and prognosis of AIS.
In order to identify the involvement of p38 MAPK, Erk1/2, and PI3K/Akt signaling pathways in the pathological process of AIS, Western blotting was performed, and it was observed that the phosphorylation levels of p38 MAPK, Erk1/2, and PI3K/Akt were significantly promoted in the AIS groups and decreased after rt-PA treatment (p < 0.05) (Figure 4), which indicated that p38 MAPK, Erk1/2, and PI3K/Akt pathways were involved in AIS progression.
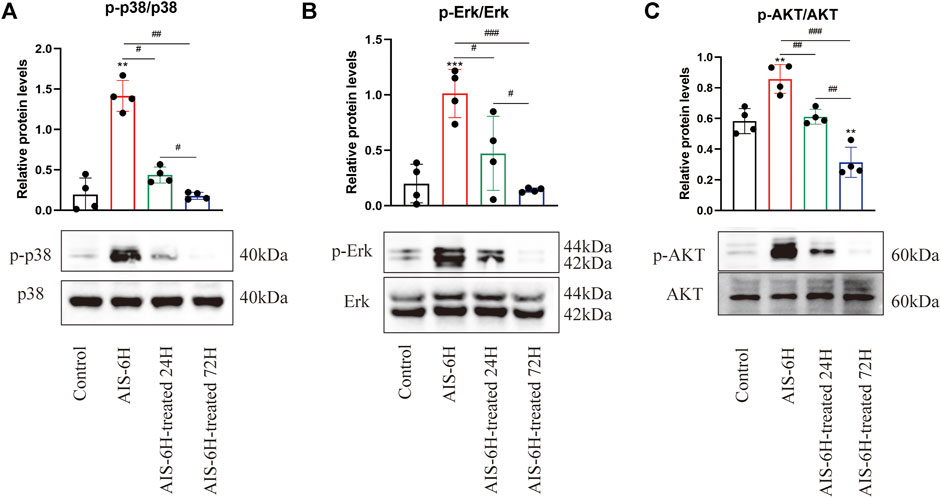
FIGURE 4. AIS promoted phosphorylation of p38 MAPK, Erk1/2, and PI3K/Akt in serum. (A–C) Phosphorylation levels of p38 MAPK, Erk1/2, and PI3K/Akt in serum were detected by Western blotting; n = 4. Data are presented as mean ± SD. Difference to control: **p < 0.01; ***p < 0.001; # Difference between groups: #p < 0.05; ##p < 0.01; ###p < 0.001; and one-way ANOVA with the Newman–Keuls multiple comparison test.
To determine whether miR-124-3p regulates viability and apoptosis of LPS-induced BV2 microglia, cells were transfected with mimic-NC (NC = negative control) and hsa-miR-124-3p-mimic for 24 h. As shown in Figure 5A, the expression level of hsa-miR-124-3p was significantly (p < 0.05) upregulated by hsa-miR-124-3p-mimic. hsa-miR-126-3p-mimic significantly (p < 0.05) promoted viability and live cell count, and inhibited the caspase 3/7 activity of LPS-induced BV2 microglia (Figures 5B–E). miR-124-3p also exhibited a visible (p < 0.05) dose-dependent effect on viability and apoptosis of LPS-induced BV2 microglia (Supplementary Figure S1). These results suggested that miR-124-3p reversed the LPS-induced inflammatory effect in BV2 microglia.
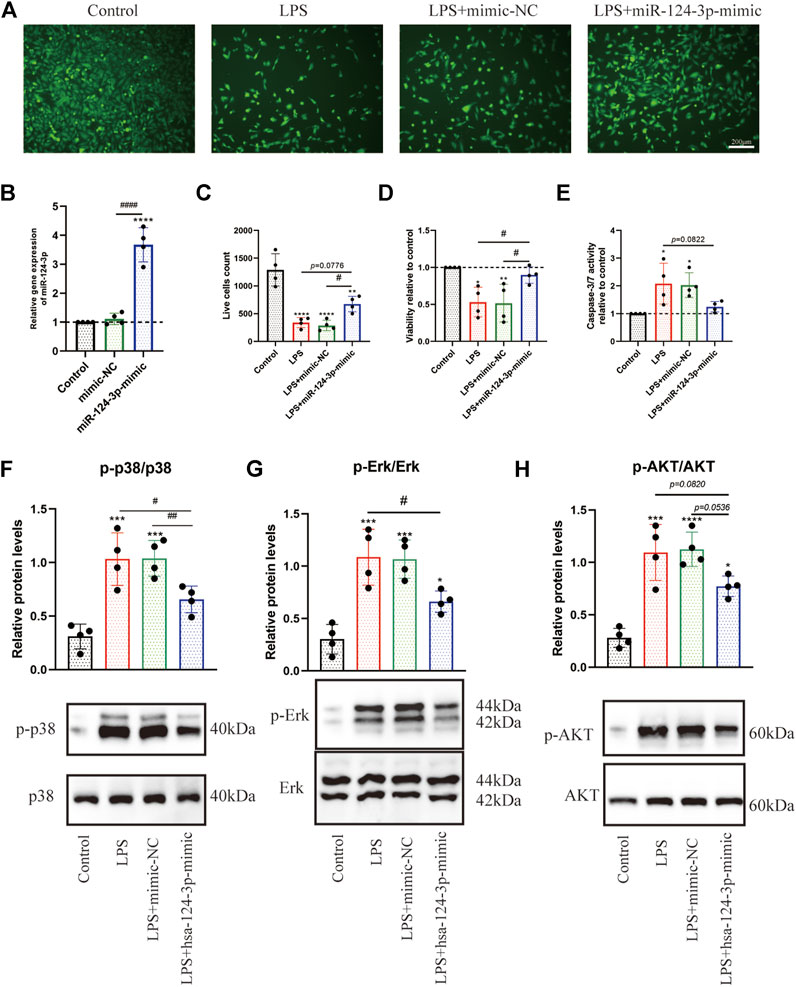
FIGURE 5. hsa-miR-124-3p mimic promoted viability, inhibited apoptosis, and reduced phosphorylation of Erk1/2, PI3K/Akt, and p38 MAPK in LPS-induced BV2 microglia. (A,C) Live cells were visualized by fluorescence microscopy after labeling cells with calcein. Living cells were labeled with calcein (green fluorescence); n = 4, scale bar = 200 μm. (B) Relative expression of transfected hsa-miR-124-3p mimic and mimic-NC measured by qRT-PCR in BV2 microglia, n = 4. (D,E) Viability and apoptosis analysis of LPS-induced BV2 microglia or/and transfected with hsa-miR-124-3p mimic. (F–H) Representative Western blotting images and quantification of the phosphorylation level of Erk1/2, P13K/Akt, and p38 MAPK after the densitometric analysis. All values represent mean ± SD. * compared with control, *p < 0.05; ***p < 0.001; ****p < 0.0001; # Difference between groups, #p < 0.05; ##p < 0.01; ###p < 0.001; one-way ANOVA with the Newman–Keuls multiple comparison test; n = 4.
To evaluate the role of miR-124-3p in regulating pro-inflammatory signaling pathways activated by AIS, we examined the effect of miR-124-3p in LPS-induced BV2 microglia using Western blotting. The quantitative assessment showed that the phosphorylation of Erk1/2, PI3K/Akt, and p38 MAPK was promoted by LPS and significantly inhibited (p < 0.05) by hsa-miR-124-3p-mimic (Figures 5F–H). A scratch (wound healing) assay was performed to evaluate the effect of miR-124-3p on the migration of LPS-induced BV2 microglia. We observed a high wound healing (migration) rate in the control group and the hsa-miR-124-3p-mimic group compared to that in the LPS-treated group (p < 0.05) (Figures 6A,B). These findings indicated that miR-124-3p has therapeutic effects on LPS-induced BV2 microglia.
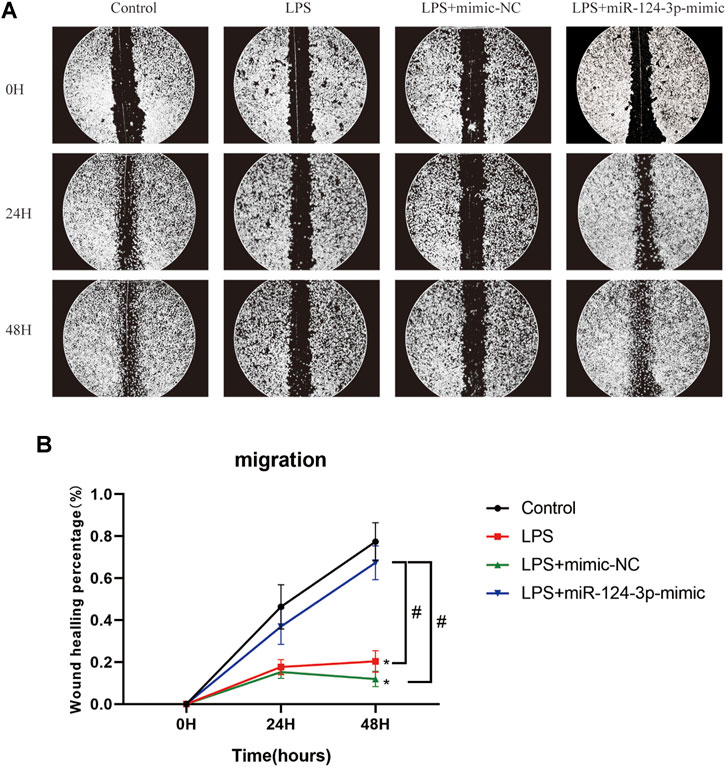
FIGURE 6. miR-124-3p promoted migration in LPS-induced BV2 microglia. (A,B) A scratch (wound healing) assay was used to determine the effect of hsa-miR-124-3p on the migration of LPS-induced BV2 microglia. The images of gaps were taken at 0, 24, and 48 h after treatment. The wound healing rate was used to calculate the migration ability of each group; n = 3. All values were represented as mean ± SD. Difference to control: *p < 0.05; # Difference between groups: #p < 0.05; one-way ANOVA with the Newman–Keuls multiple comparison test.
To explore the mechanism of miR-124-3p in regulating pro-inflammatory signaling pathways, the luciferase reporter gene assay was performed to detect the correlation between key genes involved in pro-inflammatory signaling pathways and miR-124-3p. A total of six target genes of miR-124-3p screened out were involved in the MAPK signaling pathway and the PI3K/AKT signaling pathway; GRB2 and AKT3 were identified (Figure 7A). The potential binding sites of GRB2 and AKT3 3′-untranslated region (UTR) are listed in Figure 7B. The expression of GRB2 and AKT3 genes was determined after 24 h of transfection with mimic-NC or hsa-miR-124-3p-mimic in LPS-induced BV2 microglia. The results showed that the expression of GRB2 and AKT3 genes was significantly (p < 0.01) inhibited by miR-124-3p (Figures 7C,D). The luciferase reporter assay suggested that (Figures 7E,F) the luciferase activity of the WT-GRB2-3′-UTR and WT-AKT3-3′-UTR in BV2 microglia cells was downregulated by miR-124-3p-mimic (p < 0.05). These results indicated that miR-124-3p directly targeted GRB2 and AKT3 via binding the 3′-UTR in BV2 microglia.
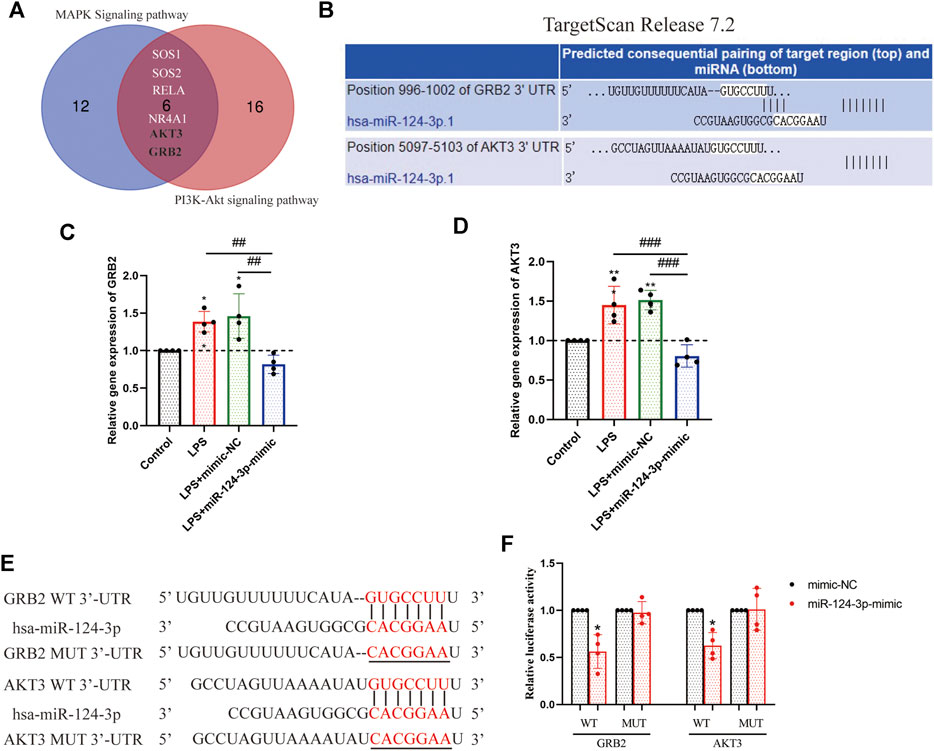
FIGURE 7. Identification of GRB2 and AKT3 as a direct target of miR-124-3p in BV2 microglia cells. (A) Venn diagram displayed the overlapping of the target genes of miR-124-3p, involved in MAPK signaling pathway and PI3K/AKT signaling pathway, as predicted in 2.3 and 2.4 of methods. (B) Putative miR-124-3p binding sequences in the 3′-UTR of GRB2 and AKT3 mRNA were predicted using the TargetScan database. (C,D) Gene expression analysis of GRB2 and AKT3 in BV2 microglia after LPS-induced or/and transfected with miR-124-3p-mimic. (E) The miR-124-3p binding site in the 3′UTR of GRB2 and AKT3 and the 3′UTR mutant sequences. (F) Qualification of luciferase activity for targeting the correlation between miR-124-3p and GRB2/AKT3. All values represent mean ± SD. * compared to control, *p < 0.05; **p < 0.01; # Difference between groups, ##p < 0.01; ###p < 0.001; one-way ANOVA with the Newman–Keuls multiple comparison test; n = 4.
Previous studies demonstrated that circulating miRNAs are altered in plasma/serum of AIS patients, serving as molecular markers for diagnosing and predicting the outcome (Jickling et al., 2010; Huang et al., 2016). However, the sensitivity and specificity of miRNAs for AIS diagnosis in plasma/serum were not satisfactory, especially in the early stage (Leung et al., 2014). In the present study, we observed that serum EV-derived miR-124-3p exhibited high sensitivity and accuracy in the diagnosis and prediction of AIS. In addition, the overexpression of miR-124-3p attenuates the inflammation in LPS-induced BV2 cells via regulating the pro-inflammatory signaling pathways.
Accumulating evidence suggested that inflammation and immune responses play critical roles in the early stage of AIS and have been associated with the outcome of AIS (Wang, 2005; Chamorro and Hallenbeck, 2006). Previous studies demonstrated that pro-inflammatory cytokines increased during AIS, such as IL-6, CX3CL1, IL-1β, and TNF-α (Xiang et al., 2016; Pöyhönen et al., 2019). In this study, we found that the expression of IL1β, IL6, TNF-α, and CXCL2 was increased during the early stage of AIS (Figure 8) but decreased after treatment. We also observed that pro-inflammatory signaling pathways, such as p38 MAPK, Erk1/2, and PI3K/Akt, were activated during AIS, which was supported by previous studies (Liu et al., 2009; Lok et al., 2015; Fann et al., 2018; Zhang et al., 2021).
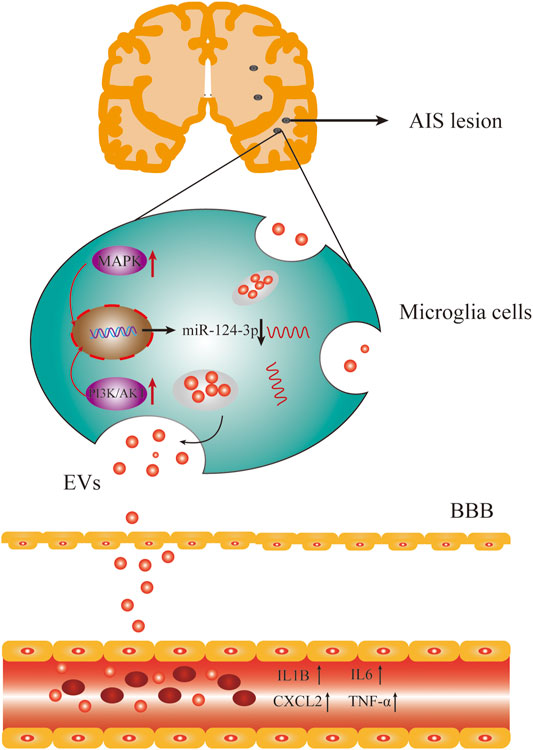
FIGURE 8. AIS activated the pro-inflammatory signaling pathways and inhibited the expression of miR-124-3p, which also upregulated the expression of pro-inflammatory factors in serum.
Reportedly, many miRNAs are differentially expressed in plasma/serum of AIS. Wang et al. (2014) found that hsa-miR-106b-5p and hsa-miR-4306 are significantly increased in AIS patients, whereas hsa-miR-320e and hsa-miR-320d are significantly decreased in control subjects, suggesting that miRNAs in plasma might serve as biomarkers for the early diagnosis of acute stroke in humans. miR-124-3p is mainly expressed in the brain (Cheng et al., 2009). A previous study demonstrated that the expression of miR-124-3p was reduced substantially in focal cerebral ischemia in vivo (Liu et al., 2011). Liu et al. (2015) observed that the serum level of miR-124 was significantly decreased within 24 h after stroke onset and negatively correlated with infarct size. In contrast, another study found that miR-124 was significantly upregulated in AIS patients in the acute phase (Ji et al., 2016). In the present study, we found that EV miR-124-3p significantly decreased from 2 h of AIS onset and continued to decrease at 6 h of AIS onset. Notably, the expression of EV miR-124-3p is negatively correlated with IL1β, IL6, TNF-α, CXCL2I, and the NIHSS. Furthermore, EV miR-124-3p showed high sensitivity and specificity for the diagnosis of AIS compared to serum miR-124-3p.
miR-124 has been identified as a negative regulator of inflammation after stroke (Hamzei Taj et al., 2016a; Ning et al., 2017). In the present study, we investigated the anti-inflammatory effect of miR-124-3p on LPS-induced BV2 microglia. The current results showed that miR-124-3p promoted migration and viability of LPS-induced BV2 microglia, and inhibited apoptosis and phosphorylation of pro-inflammatory signaling pathways (p38 MAPK, Erk1/2, and PI3K/Akt) activated by LPS. These results were consistent with those reported previously (Sun et al., 2013; Hamzei Taj et al., 2016b). In addition, EV miRNA was negatively correlated with pro-inflammatory cytokines (CXCL2, IL6, IL1β, and TNF-α). In order to reveal the mechanism underlying the role of miR-124-3p in regulating pro-inflammatory signaling pathways, a luciferase reporter gene assay was employed to detect the correlation between miR-124-3p and key genes involved in MAPK and PI3K/Akt signaling pathways. The results showed that miR-124-3p directly targeted GRB2 and AKT3 in BV2 microglia. This could be the mechanism of miR-124-3p inhibiting the activity of pro-inflammatory signaling pathways.
In conclusion, the current study demonstrated that serum EV-derived miR-124-3p exhibited high sensitivity and accuracy in the diagnosis and prediction of AIS. In addition, the overexpression of miR-124-3p attenuated inflammation in LPS-induced BV2 cells via regulating p38 MAPK, Erk1/2, and PI3K/Akt signaling pathways. These data indicated that serum EV-derived miR-124-3p can be used as a diagnostic and predictive marker for early-stage AIS.
Publicly available datasets were analyzed in this study. These data can be found here. The raw datasets (GSE95204) used in this study were downloaded from the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/).
The studies involving human participants were reviewed and approved by 2020-KY-262, the First Affiliated Hospital of Zhengzhou University. The patients/participants provided their written informed consent to participate in this study.
ZQ: sample collection, methodology, experiment design, validation, formal analysis, investigation, data curation, and writing (original draft, review, and editing); YZ: conceptualization and writing (review and editing); YS: methodology, the establishment of EV isolation, and NTA analysis; J-JY and BC: bioinformatics analysis, reviewing, and writing; QX: conceptualization, writing (review and editing), project administration, and funding acquisition. All the authors have proofread the final version of the manuscript.
This study was supported by the National Natural Science Foundation of China (No. 81971020).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.685088/full#supplementary-material
Ariston Gabriel, A. N., Wang, F., Jiao, Q., Yvette, U., Yang, X., Al-Ameri, S. A., et al. (2020). The Involvement of Exosomes in the Diagnosis and Treatment of Pancreatic Cancer. Mol. Cancer 19, 132. doi:10.1186/s12943-020-01245-y
Bhalala, O. G., Srikanth, M., and Kessler, J. A. (2013). The Emerging Roles of microRNAs in CNS Injuries. Nat. Rev. Neurol. 9, 328–339. doi:10.1038/nrneurol.2013.67
Bonita, R., Mendis, S., Truelsen, T., Bogousslavsky, J., Toole, J., and Yatsu, F. (2004). The Global Stroke Initiative. Lancet Neurol. 3, 391–393. doi:10.1016/S1474-4422(04)00800-2
Cardo, L. F., Coto, E., Mena, L., Ribacoba, R., Moris, G., Menéndez, M., et al. (2013). Profile of microRNAs in the Plasma of Parkinson's Disease Patients and Healthy Controls. J. Neurol. 260, 1420–1422. doi:10.1007/s00415-013-6900-8
Chamorro, A., and Hallenbeck, J. (2006). The Harms and Benefits of Inflammatory and Immune Responses in Vascular Disease. Stroke 37, 291–293. doi:10.1161/01.STR.0000200561.69611.f8
Cheng, L.-C., Pastrana, E., Tavazoie, M., and Doetsch, F. (2009). miR-124 Regulates Adult Neurogenesis in the Subventricular Zone Stem Cell Niche. Nat. Neurosci. 12, 399–408. doi:10.1038/nn.2294
Dassan, P., Keir, G., and Brown, M. M. (2009). Criteria for a Clinically Informative Serum Biomarker in Acute Ischaemic Stroke: a Review of S100B. Cerebrovasc. Dis. 27, 295–302. doi:10.1159/000199468
Deng, F., Wang, S., and Zhang, L. (2017). Endothelial Microparticles Act as Novel Diagnostic and Therapeutic Biomarkers of Circulatory Hypoxia-Related Diseases: a Literature Review. J. Cel. Mol. Med. 21, 1698–1710. doi:10.1111/jcmm.13125
Fann, D. Y.-W., Lim, Y.-A., Cheng, Y.-L., Lok, K.-Z., Chunduri, P., Baik, S.-H., et al. (2018). Evidence that NF-Κb and MAPK Signaling Promotes NLRP Inflammasome Activation in Neurons Following Ischemic Stroke. Mol. Neurobiol. 55, 1082–1096. doi:10.1007/s12035-017-0394-9
Gurha, P. (2016). MicroRNAs in Cardiovascular Disease. Curr. Opin. Cardiol. 31, 249–254. doi:10.1097/HCO.0000000000000280
Hamzei Taj, S., Kho, W., Aswendt, M., Collmann, F. M., Green, C., Adamczak, J., et al. (2016a). Dynamic Modulation of Microglia/Macrophage Polarization by miR-124 after Focal Cerebral Ischemia. J. Neuroimmune Pharmacol. 11, 733–748. doi:10.1007/s11481-016-9700-y
Hamzei Taj, S., Kho, W., Riou, A., Wiedermann, D., and Hoehn, M. (2016b). MiRNA-124 Induces Neuroprotection and Functional Improvement after Focal Cerebral Ischemia. Biomaterials 91, 151–165. doi:10.1016/j.biomaterials.2016.03.025
Harpaz, D., Eltzov, E., Seet, R. C. S., Marks, R. S., and Tok, A. I. Y. (2017). Point-of-Care-Testing in Acute Stroke Management: An Unmet Need Ripe for Technological Harvest. Biosensors 7, 30. doi:10.3390/bios7030030
Hayes, J., Peruzzi, P. P., and Lawler, S. (2014). MicroRNAs in Cancer: Biomarkers, Functions and Therapy. Trends Mol. Med. 20, 460–469. doi:10.1016/j.molmed.2014.06.005
He, X.-W., Shi, Y.-H., Liu, Y.-S., Li, G.-F., Zhao, R., Hu, Y., et al. (2019). Increased Plasma Levels of miR-124-3p, miR-125b-5p and miR-192-5p Are Associated with Outcomes in Acute Ischaemic Stroke Patients Receiving Thrombolysis. Atherosclerosis 289, 36–43. doi:10.1016/j.atherosclerosis.2019.08.002
He, Z., Tang, Y., and Qin, C. (2017). Increased Circulating Leukocyte-Derived Microparticles in Ischemic Cerebrovascular Disease. Thromb. Res. 154, 19–25. doi:10.1016/j.thromres.2017.03.025
Hong, S.-B., Yang, H., Manaenko, A., Lu, J., Mei, Q., and Hu, Q. (2019). Potential of Exosomes for the Treatment of Stroke. Cel Transpl. 28, 662–670. doi:10.1177/0963689718816990
Huang, S. e., Zhao, J., Huang, D., Zhuo, L., Liao, S., and Jiang, Z. (2016). Serum miR-132 Is a Risk Marker of post-stroke Cognitive Impairment. Neurosci. Lett. 615, 102–106. doi:10.1016/j.neulet.2016.01.028
Hwong, W. Y., Bots, M. L., Selvarajah, S., Kappelle, L. J., Abdul Aziz, Z., Sidek, N. N., et al. (2016). Use of a Diagnostic Score to Prioritize Computed Tomographic (CT) Imaging for Patients Suspected of Ischemic Stroke Who May Benefit from Thrombolytic Therapy. PLoS One 11, e0165330. doi:10.1371/journal.pone.0165330
Ji, Q., Ji, Y., Peng, J., Zhou, X., Chen, X., Zhao, H., et al. (2016). Increased Brain-specific MiR-9 and MiR-124 in the Serum Exosomes of Acute Ischemic Stroke Patients. PLoS One 11, e0163645. doi:10.1371/journal.pone.0163645
Jickling, G. C., Xu, H., Stamova, B., Ander, B. P., Zhan, X., Tian, Y., et al. (2010). Signatures of Cardioembolic and Large-Vessel Ischemic Stroke. Ann. Neurol. 68, 681–692. doi:10.1002/ana.22187
Jung, K.-H., Chu, K., Lee, S.-T., Park, H.-K., Bahn, J.-J., Kim, D.-H., et al. (2009). Circulating Endothelial Microparticles as a Marker of Cerebrovascular Disease. Ann. Neurol. 66, 191–199. doi:10.1002/ana.21681
Kalafut, M. A., Schriger, D. L., Saver, J. L., and Starkman, S. (2000). Detection of Early CT Signs of >1/3 Middle Cerebral Artery Infarctions. Stroke 31, 1667–1671. doi:10.1161/01.str.31.7.1667
Korolev, I. O., Symonds, L. L., and Bozoki, A. C. (2016). Predicting Progression from Mild Cognitive Impairment to Alzheimer's Dementia Using Clinical, MRI, and Plasma Biomarkers via Probabilistic Pattern Classification. PLoS One 11, e0138866. doi:10.1371/journal.pone.0138866
Kwah, L. K., and Diong, J. (2014). National Institutes of Health Stroke Scale (NIHSS). J. Physiother. 60, 61. doi:10.1016/j.jphys.2013.12.012
Leidinger, P., Backes, C., Deutscher, S., Schmitt, K., Mueller, S. C., Frese, K., et al. (2013). A Blood Based 12-miRNA Signature of Alzheimer Disease Patients. Genome Biol. 14, R78. doi:10.1186/gb-2013-14-7-r78
Leung, L. Y., Chan, C. P. Y., Leung, Y. K., Jiang, H. L., Abrigo, J. M., Wang, D. F., et al. (2014). Comparison of miR-124-3p and miR-16 for Early Diagnosis of Hemorrhagic and Ischemic Stroke. Clinica Chim. Acta 433, 139–144. doi:10.1016/j.cca.2014.03.007
Liu, A.-L., Wang, X.-W., Liu, A.-H., Su, X.-W., Jiang, W.-J., Qiu, P.-X., et al. (2009). JNK and P38 Were Involved in Hypoxia and Reoxygenation-Induced Apoptosis of Cultured Rat Cerebellar Granule Neurons. Exp. Toxicologic Pathol. 61, 137–143. doi:10.1016/j.etp.2008.06.004
Liu, C.-M., Chen, J., Yang, S., Jiang, T.-T., Chen, Z.-L., Tu, H.-H., et al. (2018). iTRAQ-Based Proteomic Analysis to Identify the Molecular Mechanism of Zhibai Dihuang Granule in the Yin-Deficiency-Heat Syndrome Rats. Chin. Med. 13. doi:10.1186/s13020-017-0160-y
Liu, X. S., Chopp, M., Zhang, R. L., Tao, T., Wang, X. L., Kassis, H., et al. (2011). MicroRNA Profiling in Subventricular Zone after Stroke: MiR-124a Regulates Proliferation of Neural Progenitor Cells through Notch Signaling Pathway. PLoS One 6, e23461. doi:10.1371/journal.pone.0023461
Liu, Y., Zhang, J., Han, R., Liu, H., Sun, D., and Liu, X. (2015). Downregulation of Serum Brain Specific microRNA Is Associated with Inflammation and Infarct Volume in Acute Ischemic Stroke. J. Clin. Neurosci. 22, 291–295. doi:10.1016/j.jocn.2014.05.042
Lok, K. Z., Basta, M., Manzanero, S., and Arumugam, T. V. (2015). Intravenous Immunoglobulin (IVIg) Dampens Neuronal Toll-like Receptor-Mediated Responses in Ischemia. J. Neuroinflammation 12, 73. doi:10.1186/s12974-015-0294-8
Ning, R., Venkat, P., Chopp, M., Zacharek, A., Yan, T., Cui, X., et al. (2017). D-4F Increases microRNA-124a and Reduces Neuroinflammation in Diabetic Stroke Rats. Oncotarget 8, 95481–95494. doi:10.18632/oncotarget.20751
Pöyhönen, S., Er, S., Domanskyi, A., and Airavaara, M. (2019). Effects of Neurotrophic Factors in Glial Cells in the Central Nervous System: Expression and Properties in Neurodegeneration and Injury. Front. Physiol. 10, 486. doi:10.3389/fphys.2019.00486
Skog, J., Würdinger, T., van Rijn, S., Meijer, D. H., Gainche, L., Curry, W. T., et al. (2008). Glioblastoma Microvesicles Transport RNA and Proteins that Promote Tumour Growth and Provide Diagnostic Biomarkers. Nat. Cel Biol 10, 1470–1476. doi:10.1038/ncb1800
Sun, Y., Gui, H., Li, Q., Luo, Z.-M., Zheng, M.-J., Duan, J.-L., et al. (2013). MicroRNA-124 Protects Neurons against Apoptosis in Cerebral Ischemic Stroke. CNS Neurosci. Ther. 19, a–n. doi:10.1111/cns.12142
Świtońska, M., Słomka, A., Sinkiewicz, W., and Żekanowska, E. (2015). Tissue-factor-bearing Microparticles (MPs-TF) in Patients with Acute Ischaemic Stroke: the Influence of Stroke Treatment on MPs-TF Generation. Eur. J. Neurol. 22 (395–401), e28–29. doi:10.1111/ene.12591
Urra, X., Miró, F., Chamorro, A., and Planas, A. M. (2014). Antigen-specific Immune Reactions to Ischemic Stroke. Front. Cel. Neurosci. 8, 278. doi:10.3389/fncel.2014.00278
Vijayan, M., and Reddy, P. H. (2016). Peripheral Biomarkers of Stroke: Focus on Circulatory microRNAs. Biochim. Biophys. Acta (Bba) - Mol. Basis Dis. 1862, 1984–1993. doi:10.1016/j.bbadis.2016.08.003
Wang, W., Li, D.-B., Li, R.-Y., Zhou, X., Yu, D.-J., Lan, X.-Y., et al. (2018). Diagnosis of Hyperacute and Acute Ischaemic Stroke: The Potential Utility of Exosomal MicroRNA-21-5p and MicroRNA-30a-5p. Cerebrovasc. Dis. 45, 204–212. doi:10.1159/000488365
Wang, W., Sun, G., Zhang, L., Shi, L., and Zeng, Y. (2014). Circulating microRNAs as Novel Potential Biomarkers for Early Diagnosis of Acute Stroke in Humans. J. Stroke Cerebrovasc. Dis. 23, 2607–2613. doi:10.1016/j.jstrokecerebrovasdis.2014.06.002
Wang, X. (2005). Investigational Anti-inflammatory Agents for the Treatment of Ischaemic Brain Injury. Expert Opin. Investig. Drugs 14, 393–409. doi:10.1517/13543784.14.4.393
Wu, P., Zuo, X., and Ji, A. (2012). Stroke-induced microRNAs: The Potential Therapeutic Role for Stroke. Exp. Ther. Med. 3, 571–576. doi:10.3892/etm.2012.452
Xiang, N.-l., Liu, J., Liao, Y.-j., Huang, Y.-w., Wu, Z., Bai, Z.-q., et al. (2016). Abrogating ClC-3 Inhibits LPS-Induced Inflammation via Blocking the TLR4/NF-Κb Pathway. Sci. Rep. 6, 27583. doi:10.1038/srep27583
Yan, Q., Jin, G., Ma, K., Qin, M., Zhuang, W., and Sun, J. (2017). Magnetic Inductive Phase Shift: a New Method to Differentiate Hemorrhagic Stroke from Ischemic Stroke on Rabbit. Biomed. Eng. Online 16, 63. doi:10.1186/s12938-017-0354-7
Yan, Y., Xia, H., Hu, J., and Zhang, B. (2020). MicroRNA-542-3p Regulates P-Glycoprotein Expression in Rat Epilepsy via the Toll-like Receptor 4/Nuclear Factor-kappaB Signaling Pathway. Curr. Neurovasc. Res. 16, 433–440. doi:10.2174/1567202616666191023160201
Yu, G., Wang, L.-G., Han, Y., and He, Q.-Y. (2012). clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS: A J. Integr. Biol. 16, 284–287. doi:10.1089/omi.2011.0118
Zagrean, A.-M., Hermann, D. M., Opris, I., Zagrean, L., and Popa-Wagner, A. (2018). Multicellular Crosstalk between Exosomes and the Neurovascular Unit after Cerebral Ischemia. Therapeutic Implications. Front. Neurosci. 12, 811. doi:10.3389/fnins.2018.00811
Zhang, W., Wu, Y., Chen, H., Yu, D., Zhao, J., and Chen, J. (2021). Neuroprotective Effects of SOX5 against Ischemic Stroke by Regulating VEGF/PI3K/AKT Pathway. Gene 767, 145148. doi:10.1016/j.gene.2020.145148
Keywords: acute ischemic stroke, extracellular vesicle, miRNAs, inflammation, signaling pathway
Citation: Qi Z, Zhao Y, Su Y, Cao B, Yang J-J and Xing Q (2021) Serum Extracellular Vesicle–Derived miR-124-3p as a Diagnostic and Predictive Marker for Early-Stage Acute Ischemic Stroke. Front. Mol. Biosci. 8:685088. doi: 10.3389/fmolb.2021.685088
Received: 24 March 2021; Accepted: 04 June 2021;
Published: 01 July 2021.
Edited by:
Behzad Mansoori, Tabriz University of Medical Sciences, IranReviewed by:
Rishein Gupta, University of Texas at San Antonio, United StatesCopyright © 2021 Qi, Zhao, Su, Cao, Yang and Xing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qinghe Xing, cWluZ2hleGluZ2Z1ZGFuQGhvdG1haWwuY29t; Jian-Jun Yang, amlhbmp1bnlhbmd6enVAaG90bWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.