- 1Department of Radiation Oncology, Nanfang Hospital, Southern Medical University, Guangzhou, China
- 2Department of Radiation Oncology, Peking University Shenzhen Hospital, Shenzhen, China
- 3Chronic Airways Diseases Laboratory, Department of Respiratory and Critical Care Medicine, Nanfang Hospital, Southern Medical University, Guangzhou, China
- 4Department of Respiratory and Critical Care Medicine, Second Affiliated Hospital of Fujian Medical University, Quanzhou, China
- 5Guangdong Provincial Hospital of Traditional Chinese Medicine, Guangzhou, China
- 6Department of Radiation Oncology, The First Affiliation Hospital of Fujian Medical University, Fuzhou, China
- 7Department of Pathology, Nanfang Hospital, Southern Medical University, Guangzhou, China
- 8Department of Pathology, School of Basic Medical Sciences, Southern Medical University, Guangzhou, China
Purpose: This multi-center retrospective study determines whether the ΔCT value of the Amplified Refractory Mutation System (ARMS) predicts the efficacy of epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) in EGFR-mutant advanced non–small-cell lung cancer (NSCLC).
Patients and methods: Patients who harbored an exon 19 deletion (19Del) or L858R mutation detected by the ARMS and previously received treatment of EGFR-TKIs as a monotherapy were enrolled. A total of 108 NSCLC patients in four hospitals were enrolled. We divided the patients into a high ΔCT group (Group H) and a low ΔCT group (Group L) by the Martingale residuals analysis and log-rank test. The primary outcome was progression-free survival (PFS). Univariate analysis and multivariable regression were applied to compare the PFS between the groups.
Result: The Martingale residuals analysis and log-rank test were applied to find the cutoff ΔCT value (0.8). In the 108 patients we enrolled, 59 were in group L and 49 were in group H. Patients’ demographics and clinical characteristics, including age, sex, smoking history, pathology, mutation sites, TNM stage, and line of TKIs therapy, were not significantly different between group L and group H. The median PFS was 11.1 months in group L and 6.9 months in group H, and the difference showed statistical significance (p < 0.001). Moreover, the objective response rates (ORRs) in group L was significantly higher than in group H (61.0 vs 34.7%, p = 0.002). The median OS was 25.0 months in group L and 20.0 months in group H (p = 0.046).
Conclusion: The ΔCT value of ARMS could be an efficacy predictor for EGFR-TKI treatment in advanced EGFR-mutant NSCLC.
Introduction
Lung cancer is the most common cancer leading to cancer-related deaths worldwide. More than 70% of patients with lung cancer are diagnosed with advanced non–small-cell lung cancer (NSCLC) (Siegel et al., 2017). Many clinical trials have demonstrated the superiority of epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) over chemotherapy in the treatment of patients with advanced NSCLC harboring EGFR mutations (Maemondo et al., 2010; Mitsudomi et al., 2010; Rosell et al., 2012; Yang et al., 2013; Wu et al., 2014). Therefore, EGFR-TKIs have been recommended as the standard of care for first-line treatment for EGFR-mutant NSCLC, especially for those who harbored a drug sensitivity-associated mutation including exon 19 deletion (19Del) and exon 21 L858R. However, these clinical trials also showed that the efficacy of EGFR-TKIs was not so satisfactory in a nonnegligible proportion of NSCLCs harboring sensitive EGFR mutations. In the studies we have mentioned above (Maemondo et al., 2010; Mitsudomi et al., 2010; Rosell et al., 2012; Yang et al., 2013; Wu et al., 2014), 20–44% patients who harbored EGFR mutation had a best response of stable disease (SD) or progressive disease (PD) and 8–15% patients had a best response of PD. Still a significant minority of patients had a primary resistance or poor progression of disease (PFS) to EGFR-TKIs when harbored sensitive EGFR mutations.
Distinguishing patients who are most likely to experience an expected response to EGFR-TKIs from those who are not likely to show a response has emerged as a crucial issue. Although previous studies had reported several resistance mechanisms of EGFR TKIs for EGFR-mutant NSCLC patients, in many cases, the mechanisms remain unclear (Costa et al., 2014; Beau-Faller et al., 2014; Ogino et al., 2007; Engelman et al., 2007). Efforts are still needed to explore the reasons for the various rates of resistance to EGFR-TKIs for EGFR-mutant NSCLC.
The correlation of intratumor heterogeneity of tumor and drug resistance has been widely studied in recent years (Turner and Reis-Filho, 2012; Jamal-Hanjani et al., 2015; Mangano et al., 2015). For the EGFR heterogeneity in lung cancer, recent reports have indicated that tumors are composed of mixed populations of mutant EGFR and wild-type EGFR cells, suggesting that the intratumor heterogeneity does indeed exist (Jiang et al., 2008; Taniguchi et al., 2008; Wei et al., 2014; Cai et al., 2015). Furthermore, several groups have demonstrated that the intratumor heterogeneity of EGFR-mutant NSCLC associated with the efficacy of EGFR-TKIs. They demonstrated that patients with higher relative abundance of EGFR mutation showed longer PFS (Jiang et al., 2008; Taniguchi et al., 2008; Zhou et al., 2011; Zhao et al., 2014a; Zhao et al., 2014b). These reports gave a great inspiration to the development of the therapeutic strategy of EGFR-TKIs. Researchers advised that for patients who harbored a high ratio of EGFR mutation in tumor, EGFR-TKI was effective to control the progression of tumor, but for the low, monotherapy of EGFR-TKIs may be not enough. However, until now, there was not any effective and convenient ways for clinicians to distinguish whether a patient is harboring a high ratio of EGFR mutation in tumor or not. Therefore, there is an urgent need to develop a method to assess the abundance of EGFR mutations in NSCLC in clinical practice. The Amplified Refractory Mutation System (ARMS) had widely been applied in the detection of EGFR mutation in recent years (Shaozhang et al., 2014). Mutant allele assays were run with a gene reference assay, which was designed to a mutation-free region of the gene. The mutational status of a sample was determined by calculating the ΔCT value between amplification reactions for a mutant allele assay and gene reference assay, as follows: ΔCT = Ct (mutant allele assay) – Ct (gene reference assay). Mutation or not is determined by the Ct value and ΔCT value. The reference gene was a relatively conserved region of the EGFR gene. Thus, Ct (gene reference assay) could effectively reflect the DNA level of the EGFR gene. For the tumor samples with EGFR mutation, the ΔCT value could reflect the relative level of EGFR mutation in the tumor sample. Therefore, we assume that patients with a lower ΔCT value may harbor a high ratio of EGFR mutation and may associate with better response to EGFR-TKIs. Furthermore, we divided the EGFR-mutant NSCLC into a low ΔCT value and a high ΔCT value to explore whether the ΔCT value of the tumor sample could be a predictor for the efficacy of EGFR-TKIs in EGFR-mutant NSCLC patients.
Patients and Methods
Patients
A total of 108 Chinese patients were enrolled in this study from four medical centers in China including Nanfang Hospital of Southern Medical University, the first affiliated hospital of Fujian Medical University, the second affiliated hospital of Fujian Medical University, and Guangdong Province Traditional Medical Hospital between March 2013 and Jan 2015. The criteria for the patients enrolled in this retrospective study were as follows: 1. Diagnosed with advanced NSCLC and harbored a drug sensitivity-associated mutation site (19Del and L858R). 2. EGFR mutations were tested by ARMS (Shanghai Yuanqi Bio-Pharmaceutical Company Limited, Shanghai, China) and previously received treatment of EGFR-TKIs including gefitinib, erlotinib and icotinib as a monotherapy. The data we collected of all patients were from the electronic medical record system in the four medical centers.
Epidermal Growth Factor Receptor Mutation Analysis
EGFR mutation testing was performed on formalin-fixed paraffin-embedded (FFPE) specimens from primary tumor obtained from bronchoscopic biopsy or CT-guided core biopsy before any tumor-related treatment. The HE-stained section of FFPE was assessed again to establish the pathological diagnosis. Some of the FFPE samples were trimmed according to the HE-stained section to make sure that the samples we would test were all tumors. The genomic DNA was isolated and purified from tumor specimens using the DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA, United States) according to the manufacturer’s instructions. EGFR mutation detection was performed according to the principles of the ARMS, using the Human EGFR Gene Mutation Detection Kit (PCR Fluorescence Probe) (Shanghai Yuanqi Bio-Pharmaceutical Company Limited, Shanghai, China) on the MX3005P QPCR system (Agilent, Santa Clara, CA, United States), according to manufacturers’ recommendations. It covered 23 EGFR mutation hotspots within exons 18, 19, 20 and 21. The results were analyzed according to the criteria defined by the manufacturer’s instructions. Positive results were defined as Ct (mutant allele assay) – Ct (gene reference assay) < ΔCT (cutoff). We divided the patients into two groups by the media ΔCT value.
Statistical Analyses
The primary end point was progression-free survival (PFS), and the second end points were the objective response rate (ORR) and primary resistance. In patients with measurable disease, the tumor burden was assessed by the Response Evaluation Criteria in Solid Tumor (RECIST) and categorized as a complete response (CR), partial response (PR), stable disease (SD), or progressive disease (PD) (Eisenhauer et al., 2009). PFS was calculated from the time from commencement of EGFR-TKIs treatment to PD according to the RECIST criteria or death resulting from any cause. OS was calculated from the time from commencement of EGFR-TKI treatment to death. The definition of primary resistance to EGFR-TKIs was not uniform among researchers, and in this study, we defined it as patients who had progressive disease to EGFR TKI without initial objective response (Cortot and Janne, 2014). The Kaplan–Meier method was applied to analyze the PFS or OS, and the Cox proportional hazard model was applied to explore the statistical difference in PFS between different groups. A comparison of ORRs and rates of primary resistance in different groups was made using χ2 tests. A two-sided p value of less than 0.05 was considered statistically significant.
Result
Patient Characteristics
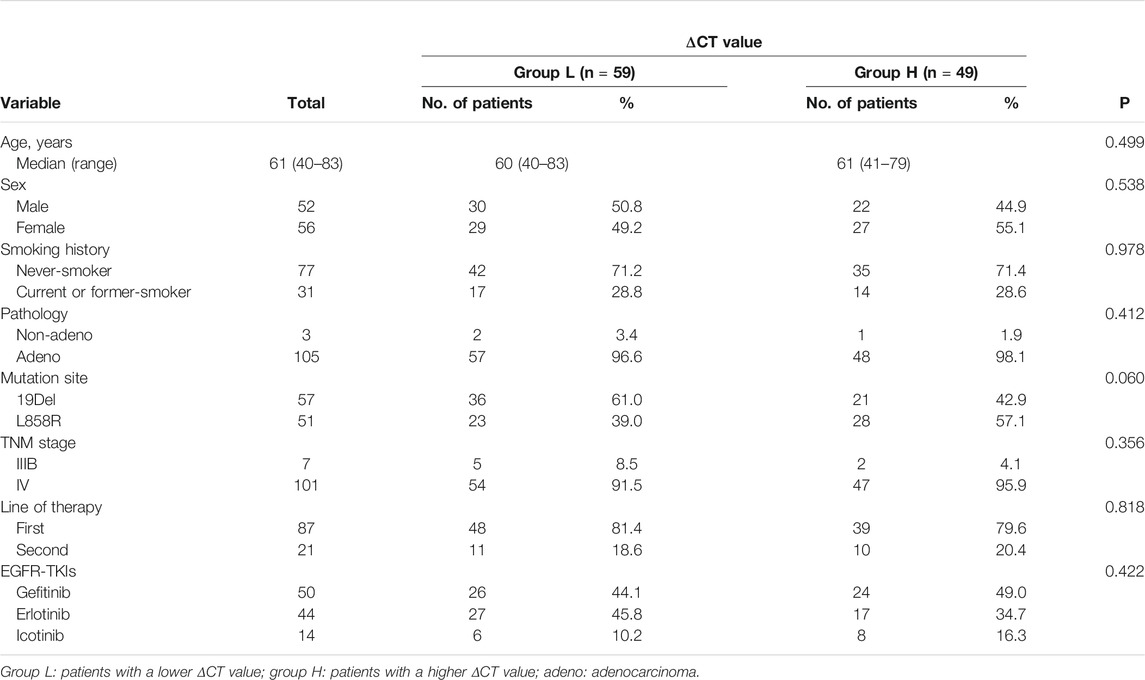
A total of 108 patients who fully met the enrollment criteria were enrolled in the present study. In all the patients, the median age was 61 (ranged from 40 to 83 years); 52 were males and 56 were females; 63 were never smokers and 45 were current or former smokers; three were non-adenocarcinoma and 105 were adenocarcinoma; 57 patients harbored a 19Del mutation and 51 harbored a L858R mutation; and 87 patients received EGFR-TKI as the first-line therapy, 20 patients received EGFR-TKI as the second line therapy, and 1 received EGFR-TKI as the third-line therapy.
Epidermal Growth Factor Receptor Mutation Groups
Martingale residuals analysis was applied, and a non-linear relationship was noted between the ΔCT value and PFS of the patients (Figures 1A,B). The risk of event went down with an increasing ΔCT value when ΔCT < −1, remained steadily when −1 > ΔCT <1, and increased when ΔCT >1. Thus, we applied log-ranking analysis to find the cutoff ΔCT value. The ΔCT value corresponding to the maximum log-rank value was defined as the cutoff ΔCT value and was 0.8, which agreed with the tendency in the Martingale residuals analysis (Figure 1C).
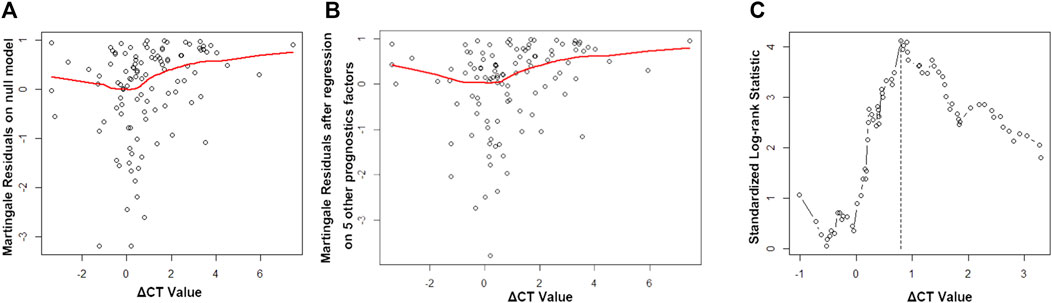
FIGURE 1. Martingale residuals analysis and standard log-rank analysis. (A) Martingale residuals analysis of the association between the ΔCT value and progression-free survival (PFS). (B) Martingale residuals analysis of the association between the ΔCT value and PFS by age, gender, smoking status, mutation sites, and therapy line (first-line or second-line therapy). (C) Standard log-rank statistic to determine the cutoff value.
59 patients had a ΔCT value less than 0.8 (group L), and 49 patients had a ΔCT value greater than 0.8 (group H). The median of age was similar between group L and group H (60 vs 61, p = 0.499). 49.2% of group L and 55.1% of group H were female (p = 0.538). The proportions of never smokers in group L and group H were similar (71.2 vs 71.4%, p = 0.978), and 19Del was more frequent in group L than L858R mutation (61.0 vs 42.9%, p = 0.060). The TNM stage and line of TKI therapy were all well balanced between the two groups (p = 0.356, p = 0.818). Treatment with different EGFR-TKIs was balanced between the two groups (p = 0.422) (Table 1).
Efficacy of Different ΔCT Value Groups
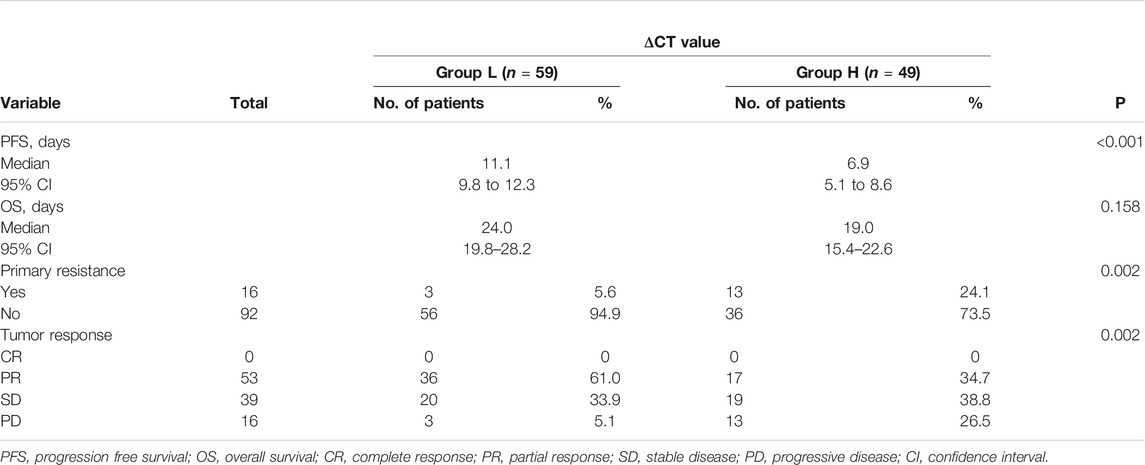
All the patients were received TKIs from Jan 2013 to Dec 2015 in the four hospitals. The last follow-up date was Aug 9, 2021.100 (92.6%) patients experienced a disease progression, and 36 (33.3%) of the patients were still alive or concord.
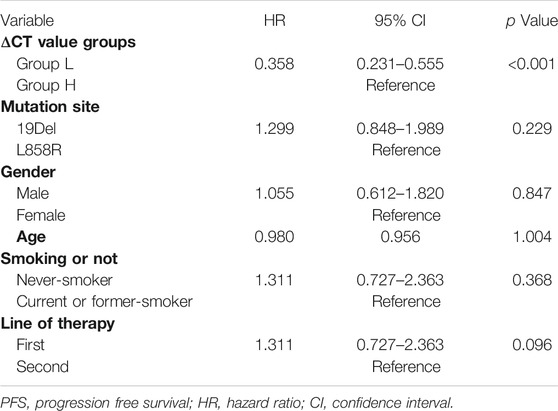
The median PFS was 11.1 months (95% CI, 9.8–12.3) in group L and 6.9 months (95% CI, 5.1–8.6) in group H, and the difference showed statistical significance (p < 0.001) (Figure 2A). Multivariate analysis shows that the ΔCT value was the variable that mostly influences the PFS (Table 2).
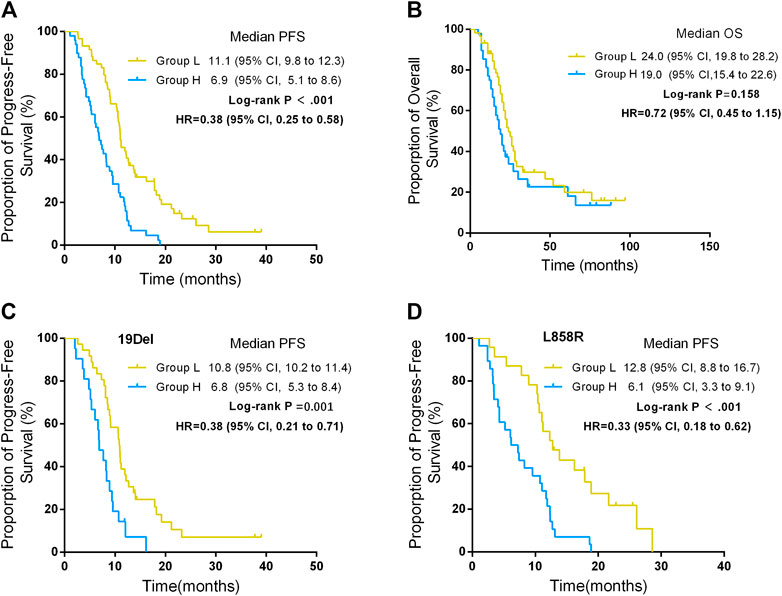
FIGURE 2. Progression-free survival (PFS) and overall survival (OS) between different ΔCT value groups. (A) PFS between different ΔCT value groups. (B) OS between different ΔCT value groups. (C) PFS between different ΔCT value groups in patients harboring EGFR 19Del. (D) PFS between different ΔCT value groups in patients harboring EGFR L858R.
The rate of primary resistance in group H was significantly higher than in group L (26.5% vs 5.1%, p = 0.002). ORRs in group L were significantly higher than in group H (61.0% vs 34.7%, p = 0.002). The median OS was 24.0 months (95% CI, 19.8–28.2) in group L and 19.0 months in group H (95% CI, 15.4–22.6), which was statistically significant (p = 0.046). (Table 3; Figure 2B).
In addition, for patients who either harbored 19Del or L858R mutation, patients in group L had better PFS than those in group H. For patients who harbored 19Del mutation, the median PFS was 10.8 months (95% CI, 10.2–11.4) in group L and 6.8 months (95% CI, 5.3–8.4) in group H, and the difference was statistically significant (p = 0.001) (Figure 2C). For patients who harbored L858R mutation, the median PFS was 12.8 months (95% CI, 8.8–16.7) in group L and 6.1 months (95% CI, 3.3–9.1) in group H, and the difference showed statistical significance (p < 0.001) (Figure 2D).
Efficacy of EGFR 19Del and EGFR 21 L858R Mutation
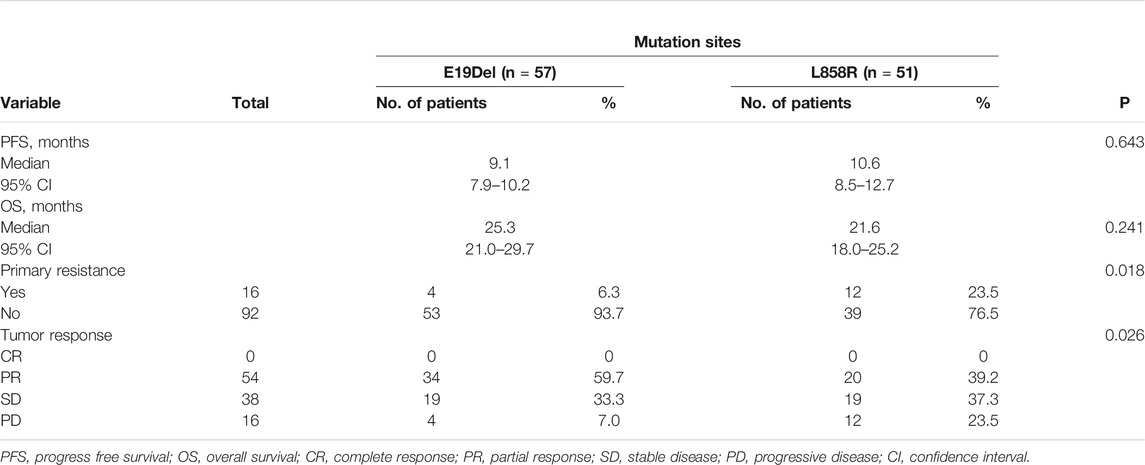
The PFS of patients who harbored 19Del mutation was 9.1 months (95% CI, 7.9–10.2), and in patients who harbored L858R mutation, PFS was 10.6 months (95% CI, 8.5–12.7). The difference between the two group was not significant (p = 0.634) (Figure 2A). ORRs in patients who harbored 19Del were higher than in those who harbored L858R mutation, while the difference showed no significance (59.7 vs 39.2%, p = 0.034). However, the rate of primary resistance was significantly higher in patients with L858R mutation than those with 19Del (23.5 vs 6.3%, p = 0.018) (Table 4).
In addition, for patients in group L, the PFS of the patients who harbored 19Del was 10.8 months (95% CI, 10.2–11.4) and the PFS of the patients who harbored L858R was 12.8 months (95% CI, 8.8–16.7) (p = 0.222) (Figure 3B). For patients in group H, the PFS of the patients who harbored 19Del mutation was 6.8 months (95% CI, 5.3–8.4) and the PFS of the patients who harbored L858R was 6.2 months (95% CI, 3.3–9.1) (p = 0.551) (Figure 3C).
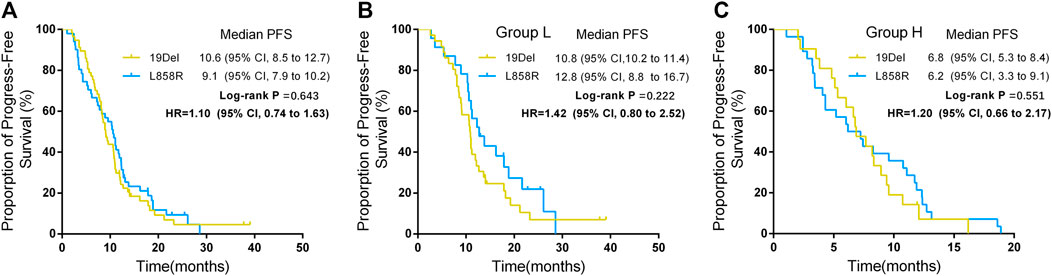
FIGURE 3. Progression free survival (PFS) between different mutation sites and groups. (A) PFS between different mutation sites. (B) PFS between different mutation sites in group L. (C) PFS between different mutation sites in group H.
Discussion
Our study put forward a new possible predictor for efficacy of EGFR-TKIs. We first put forward that the ΔCT value from the ARMS–PCR in EGFR mutation testing could be a predictor for efficacy of EGFR-TKI treatment for advanced EGFR-mutant NSCLC. Patients with a lower ΔCT value of EGFR mutations may benefit more than those with a higher ΔCT value of EGFR mutations according to the statistically different PFS between the groups, and PFS of the two groups had no overlaps. This might partly explain why some patients who harbored sensitive EGFR mutation did not experience an expected duration of response to EGFR-TKIs. Just like the previous studies, the data of the present study made us not only to focus on whether the patients harbored an EGFR mutation but also to consider the relative abundance of EGFR mutations when making therapeutic strategies for NSCLC (Jiang et al., 2008; Taniguchi et al., 2008; Zhou et al., 2011; Zhao et al., 2014a; Zhao et al., 2014b). What is further than the previous research was that we put forward a predictor that could be conveniently applied in clinical practice. By relatively quantifying the EGFR mutations in tumor tissue according to ΔCT value, patients with a low ΔCT value of EGFR mutations could receive EGFR-TKI treatment because they would benefit the most. However, for patients with a high ΔCT value of EGFR mutations, monotherapy of EGFR-TKIs may not be enough to control the tumor progression. Owing to the fact that 45.4% of the patients were still alive or concord, overall survival was required further to follow up.
Sheng et al. (2015) conducted a meta-analysis including 26 studies and showed that patients with NSCLC and EGFR exon 19 deletion had a longer PFS, OS, and higher ORR compared with exon 21 L858R mutation after EGFR-TKI therapy. Zhang et al. (2017) thought they may be two distinct diseases. However, it remains unclear why the difference in the outcome exists between the two mutation sites. In our study, ignoring the absence of statistical significance, exon 19Del was more frequent in the group of the low ΔCT value and showed higher rate of ORR and lower rate of primary resistance to EGFR-TKIs than exon 21 L858R. Although the PFS showed similarity in the two groups, it may be a clue for researchers to find out the reason why exon 19Del has a better response to EGFR-TKIs. For us, studies with large samples will be conducted to further our findings.
There were some advantages and disadvantages in the present study. The first advantage is that this was a multi-center retrospective study and all the data we recorded were from the electronic medical record system, which made our result more reliable. The second advantage is that the predictor we found in this study was much more applicable in clinical practice than in other previous studies.
The first limitation of this study is that the patients in our study had a tumor response evaluation per 8–12 weeks after EGFR-TKIs were taken. However, the proportion of patients had an evaluation per 8–10 weeks and per 11–12 weeks, which were not significantly different. The second limitation is that the follow-up of OS was not regular in our study and might lead to a large bias. However, our aim of this study was to explore whether the ΔCT value affects the efficacy of EGFR-TKIs. The third one was that only one site of the tumor sample was obtained from each patient. Nonetheless, we trimmed some of the tumor sample to ensure that every sample content is a cancerous cell to make our results more reliable. Wei et al. (2014) have demonstrated that the EGFR mutation ratio showed a high level of concordance in primary tumors and when the tumor content is more than 50% in a tumor sample, a randomly chosen sample would reliably represent the type and ratio of mutations of EGFR in primary tumors.
Conclusion
In summary, our study suggests that the ΔCT value of EGFR mutations could predict the efficacy of EGFR-TKI treatment in EGFR mutant NSCLC. We hope this indicator would contribute to more accurate technologies to evaluate the ratio of EGFR mutation in tumors.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of the Nanfang Hospital of Southern Medical University (No. NFEC-2015–11). Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
LL, SW, GJ, and MC contributed to the conception and design of the study. MC, WH and DY performed the statistical analysis and wrote the first draft of the manuscript. JH, GL, XW, NX, and WZ organized the database. All authors contributed to manuscript revision and read and approved the submitted version.
Funding
The National Natural Science Foundation of China No. 81870026; Clinical Research Startup Program of Southern Medical University (LC2016PY015 and LC2019ZD008); Clinical Research Program of Nanfang Hospital, Southern Medical University (2018CR019; 2018CR021 and 2020CR025).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.684661/full#supplementary-material
References
Beau-Faller, M., Prim, N., Ruppert, A.-M., Nanni-Metéllus, I., Lacave, R., Lacroix, L., et al. (2014). Rare EGFR Exon 18 and Exon 20 Mutations in Non-small-cell Lung Cancer on 10 117 Patients: a Multicentre Observational Study by the French ERMETIC-IFCT Network. Ann. Oncol. 25, 126–131. doi:10.1093/annonc/mdt418
Cai, W., Lin, D., Wu, C., Li, X., Zhao, C., Zheng, L., et al. (2015). Intratumoral Heterogeneity of ALK-Rearranged and ALK/EGFR Coaltered Lung Adenocarcinoma. J. Clin. Oncol. 33, 3701–3709. doi:10.1200/jco.2014.58.8293
Cortot, A. B., and Janne, P. A. (2014). Molecular Mechanisms of Resistance in Epidermal Growth Factor Receptor-Mutant Lung Adenocarcinomas. Eur. Respir. Rev. 23, 356–366. doi:10.1183/09059180.00004614
Costa, C., Molina, M. A., Drozdowskyj, A., Giménez-Capitán, A., Bertran-Alamillo, J., Karachaliou, N., et al. (2014). The Impact of EGFR T790M Mutations and BIM mRNA Expression on Outcome in Patients with EGFR-Mutant NSCLC Treated with Erlotinib or Chemotherapy in the Randomized Phase III EURTAC Trial. Clin. Cancer Res. 20, 2001–2010. doi:10.1158/1078-0432.CCR-13-2233
Eisenhauer, E. A., Therasse, P., Bogaerts, J., Schwartz, L. H., Sargent, D., Ford, R., et al. (2009). New Response Evaluation Criteria in Solid Tumours: Revised RECIST Guideline (Version 1.1). Eur. J. Cancer 45, 228–247. doi:10.1016/j.ejca.2008.10.026
Engelman, J. A., Zejnullahu, K., Mitsudomi, T., Song, Y., Hyland, C., Park, J. O., et al. (2007). MET Amplification Leads to Gefitinib Resistance in Lung Cancer by Activating ERBB3 Signaling. Science 316, 1039–1043. doi:10.1126/science.1141478
Jamal-Hanjani, M., Quezada, S. A., Larkin, J., and Swanton, C. (2015). Translational Implications of Tumor Heterogeneity. Clin. Cancer Res. 21, 1258–1266. doi:10.1158/1078-0432.CCR-14-1429
Jiang, S. X., Yamashita, K., Yamamoto, M., Piao, C. J., Umezawa, A., Saegusa, M., et al. (2008). EGFR Genetic Heterogeneity of Nonsmall Cell Lung Cancers Contributing to Acquired Gefitinib Resistance. Int. J. Cancer 123, 2480–2486. doi:10.1002/ijc.23868
Maemondo, M., Inoue, A., Kobayashi, K., Sugawara, S., Oizumi, S., Isobe, H., et al. (2010). Gefitinib or Chemotherapy for Non-small-cell Lung Cancer with Mutated EGFR. N. Engl. J. Med. 362, 2380–2388. doi:10.1056/NEJMoa0909530
Mangano, A., Lianos, G. D., Mangano, A., Boni, L., and Dionigi, G., (2015). Intratumor Heterogeneity: Origins, Clinical Significance and Optimal Strategies for Cancer Treatment. Future Oncol. 11, 561–564. doi:10.2217/fon.14.296
Mitsudomi, T., Morita, S., Yatabe, Y., Negoro, S., Okamoto, I., Tsurutani, J., et al. (2010). Gefitinib versus Cisplatin Plus Docetaxel in Patients with Non-small-cell Lung Cancer Harbouring Mutations of the Epidermal Growth Factor Receptor (WJTOG3405): an Open Label, Randomised Phase 3 Trial. Lancet Oncol. 11, 121–128. doi:10.1016/S1470-2045(09)70364-X
Ogino, A., Kitao, H., Hirano, S., Uchida, A., Ishiai, M., Kozuki, T., et al. (2007). Emergence of Epidermal Growth Factor Receptor T790M Mutation during Chronic Exposure to Gefitinib in a Non Small Cell Lung Cancer Cell Line. Cancer Res. 67, 7807–7814. doi:10.1158/0008-5472.CAN-07-0681
Rosell, R., Carcereny, E., Gervais, R., Vergnenegre, A., Massuti, B., Felip, E., et al. (2012). Erlotinib versus Standard Chemotherapy as First-Line Treatment for European Patients with Advanced EGFR Mutation-Positive Non-small-cell Lung Cancer (EURTAC): a Multicentre, Open-Label, Randomised Phase 3 Trial. Lancet Oncol. 13, 239–246. doi:10.1016/S1470-2045(11)70393-X
Shaozhang, Z., Ming, Z., Haiyan, P., Aiping, Z., Qitao, Y., Xiangqun, S., et al. (2014). Comparison of ARMS and Direct Sequencing for Detection of EGFR Mutation and Prediction of EGFR-TKI Efficacy between Surgery and Biopsy Tumor Tissues in NSCLC Patients. Med. Oncol. 31, 926. doi:10.1007/s12032-014-0926-3
Sheng, M., Wang, F., Zhao, Y., Li, S., Wang, X., Tao, S., et al. (2015). Comparison of Clinical Outcomes of Patients with Non-small-cell Lung Cancer Harbouring Epidermal Growth Factor Receptor Exon 19 or Exon 21 Mutations after Tyrosine Kinase Inhibitors Treatment: a Meta-Analysis. Eur. J. Clin. Pharmacol. 72, 1. doi:10.1007/s00228-015-1966-0
Siegel, R. L., Miller, K. D., and Ahmedin, J. (2017). Cancer Statistics, 2017. CA Cancer J. Clin. 67, 7–30. doi:10.3322/caac.21387
Taniguchi, K., Okami, J., Kodama, K., Higashiyama, M., and Kato, K., (2008). Intratumor Heterogeneity of Epidermal Growth Factor Receptor Mutations in Lung Cancer and its Correlation to the Response to Gefitinib. Cancer Sci. 99, 929–935. doi:10.1111/j.1349-7006.2008.00782.x
Turner, N. C., and Reis-Filho, J. S. (2012). Genetic Heterogeneity and Cancer Drug Resistance. Lancet Oncol. 13, e178–e185. doi:10.1016/s1470-2045(11)70335-7
Wei, B., Yang, K., Zhao, J., Chang, Y., Ma, Z., Dong, B., et al. (2014). Quantification of EGFR Mutations in Primary and Metastatic Tumors in Non-small Cell Lung Cancer. J. Exp. Clin. Cancer Res. 33 (5), 5. doi:10.1186/1756-9966-33-5
Wu, Y.-L., Zhou, C., Hu, C.-P., Feng, J., Lu, S., Huang, Y., et al. (2014). Afatinib versus Cisplatin Plus Gemcitabine for First-Line Treatment of Asian Patients with Advanced Non-small-cell Lung Cancer Harbouring EGFR Mutations (LUX-Lung 6): an Open-Label, Randomised Phase 3 Trial. Lancet Oncol. 15, 213–222. doi:10.1016/s1470-2045(13)70604-1
Yang, J. C., Hirsh, V., Schuler, M., Yamamoto, N., O'Byrne, K. J., Mok, T. S., et al. (2013). Symptom Control and Quality of Life in LUX-Lung 3: a Phase III Study of Afatinib or Cisplatin/pemetrexed in Patients with Advanced Lung Adenocarcinoma with EGFR Mutations. J. Clin. Oncol. 31, 3342–3350. doi:10.1200/JCO.2012.46.1764
Zhang, Y., Chen, G., Chen, X., Fang, W., Gao, F., Yang, Y., et al. (2017). The Comparison of EGFR-TKI Failure Modes and Subsequent Management between Exon 19 Deletion and Exon 21 L858R Mutation in Advanced Non-small-cell Lung Cancer. J. Cancer 8, 1865–1871. doi:10.7150/jca.19867
Zhao, J., Wang, X., Xue, L., Xu, N., Ye, X., Zeng, H., et al. (2014). The Use of Mutation-specific Antibodies in Predicting the Effect of EGFR-TKIs in Patients with Non-small-cell Lung Cancer. J. Cancer Res. Clin. Oncol. 140, 849–857. doi:10.1007/s00432-014-1618-2
Zhao, Z.-R., Wang, J.-F., Lin, Y.-B., Wang, F., Fu, S., Zhang, S.-L., et al. (2014). Mutation Abundance Affects the Efficacy of EGFR Tyrosine Kinase Inhibitor Readministration in Non-small-cell Lung Cancer with Acquired Resistance. Med. Oncol. 31, 810. doi:10.1007/s12032-013-0810-6
Keywords: ΔCT, ARMS, efficacy, EGFR-TKIs, NSCLC
Citation: Chen M, Huang W, Yang D, Huang J, Li G, Wang X, Xiao N, Zhang W, Guan J, Wang S and Liu L (2021) ΔCT Value of Amplified Refractory Mutation System Predicts Efficacy of EGFR-TKIs in Advanced Non–Small-Cell Lung Cancer: A Multi-Center Retrospective Study. Front. Mol. Biosci. 8:684661. doi: 10.3389/fmolb.2021.684661
Received: 23 March 2021; Accepted: 17 August 2021;
Published: 08 October 2021.
Edited by:
Huahao Shen, Zhejiang University, ChinaReviewed by:
Tae Jin Lee, Augusta University, United StatesBo Zhang, Shanghai Jiaotong University, China
Copyright © 2021 Chen, Huang, Yang, Huang, Li, Wang, Xiao, Zhang, Guan, Wang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Guan, Z3VhbmppYW41NDYxQDE2My5jb20=; Shuang Wang, c2h1YW5nd0AxMjYuY29t; Laiyu Liu, bGl1bHk1NDYxQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship.
 Min Chen1,2†
Min Chen1,2† Wenqi Huang
Wenqi Huang Xiaoqing Wang
Xiaoqing Wang Jian Guan
Jian Guan Shuang Wang
Shuang Wang Laiyu Liu
Laiyu Liu