- Science for Life Laboratory, Department of Molecular Biosciences, The Wenner-Gren Institute, Stockholm University, Stockholm, Sweden
The asymmetric life cycle of Caulobacter crescentus has provided a model in which to study how protein quality control (PQC) networks interface with cell cycle and developmental processes, and how the functions of these systems change during exposure to stress. As in most bacteria, the PQC network of Caulobacter contains highly conserved ATP-dependent chaperones and proteases as well as more specialized holdases. During growth in optimal conditions, these systems support a regulated circuit of protein synthesis and degradation that drives cell differentiation and cell cycle progression. When stress conditions threaten the proteome, most components of the Caulobacter proteostasis network are upregulated and switch to survival functions that prevent, revert, and remove protein damage, while simultaneously pausing the cell cycle in order to regain protein homeostasis. The specialized physiology of Caulobacter influences how it copes with proteotoxic stress, such as in the global management of damaged proteins during recovery as well as in cell type-specific stress responses. Our mini-review highlights the discoveries that have been made in how Caulobacter utilizes its PQC network for regulating its life cycle under optimal and proteotoxic stress conditions, and discusses open research questions in this model.
Introduction
The aquatic alpha-proteobacterium Caulobacter crescentus (hereafter Caulobacter) is well-established as a model of bacterial cell cycle control and development, and is also used to study how prokaryotic protein quality control (PQC) networks interface with these processes. Caulobacter reproduces by an asymmetric life cycle, where division results in one replication-competent, surface-attached stalked cell and one chemotactically-motivated, non-replicative swarmer cell (Figure 1A; Curtis and Brun, 2010). The swarmer cell is motile and travels the environment until nutritional cues prompt its differentiation into a stalked cell, thus completing the cell cycle. To achieve this dimorphic lifestyle, processes from DNA replication to chemotaxis must be correctly organized in time and space. The synthesis and degradation of the proteins that implement these processes relies heavily on the PQC network during optimal growth conditions, but the PQC network must balance these tasks with the protective tasks required to survive stresses free-living bacteria frequently encounter. Initial work on the Caulobacter PQC network sought to identify if the bacterial PQC network performs a role in bacterial development (Gomes et al., 1986; Reuter and Shapiro, 1987). Since then, the major chaperones and proteases have been discovered to perform many regulatory functions in the Caulobacter cell cycle during optimal
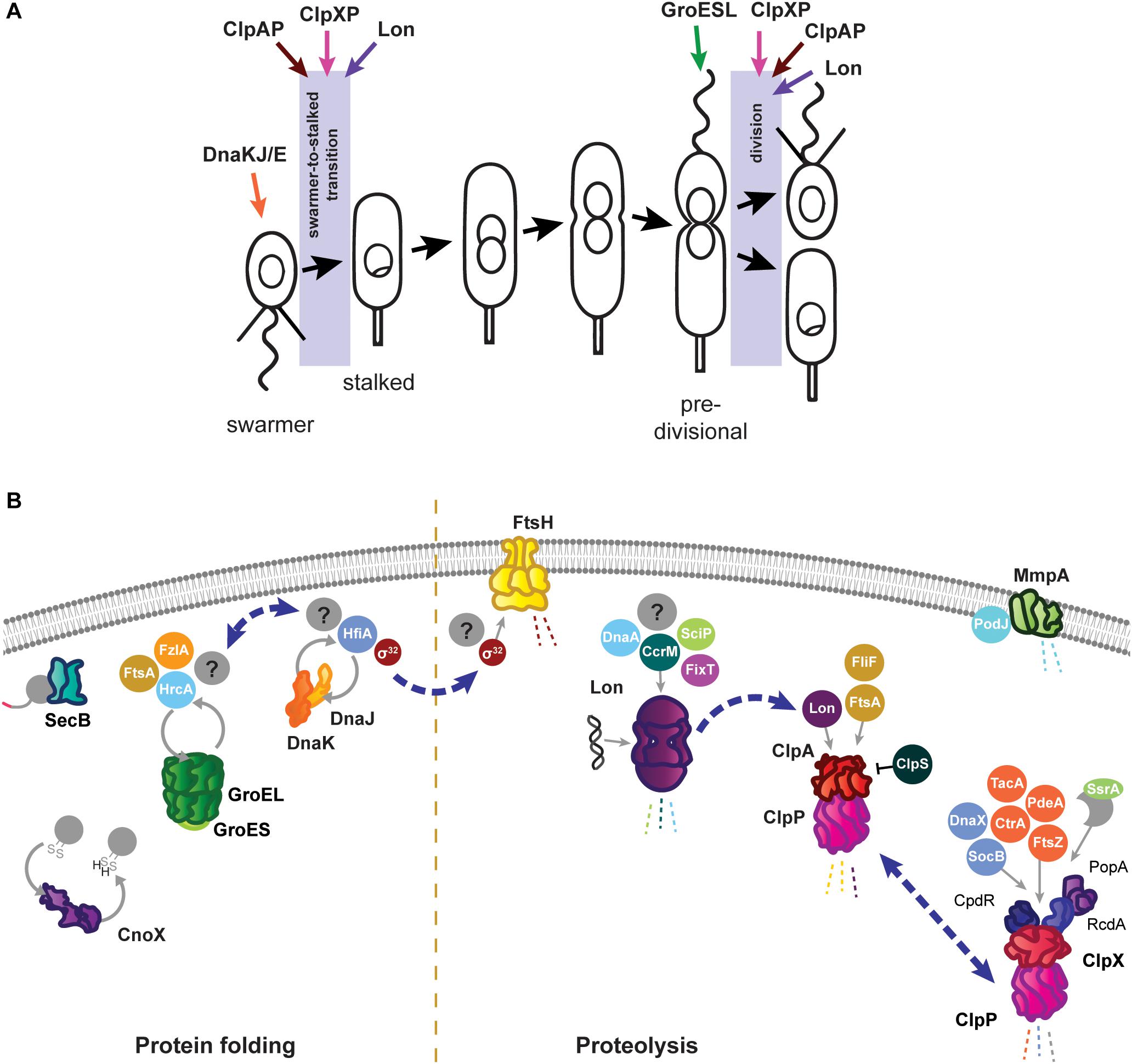
Figure 1. Roles of the Caulobacter crescentus PQC network in cell cycle progression and development. (A) The asymmetric life cycle of Caulobacter. Points where mechanisms have been identified where specific PQC proteins contribute to cell cycle progression are indicated with colored arrows. (B) Specific tasks of individual PQC network proteins in development and cell cycle progression during optimal conditions. Client proteins are indicated in circles where they are known, and by question marks where additional substrates have yet to be identified. Holdase cycling is indicated with gray circular arrows, and degradation with dashed lines. Blue dashed arrows indicate points of interaction between PQC network proteins. Membrane and DNA images created with Biorender (Biorender.com).
conditions that are remodeled or modified during stress. These studies have collectively built a platform on which to address how a prokaryotic PQC network navigates the balance between reproductive and stress response tasks in order to mediate both growth and survival. The Caulobacter model is also used as a tool to answer questions of damage inheritance in asymmetric division, how generalist PQC networks can be specialized, and how stress responses are dynamically tailored.
As in other Gram negative bacteria, the primary energy-dependent nodes of the Caulobacter PQC network include the highly conserved chaperones and proteases GroES/EL, DnaKJ/GrpE, ClpB, ClpAP, ClpXP, Lon, FtsH, and HslUV (Figure 1B). The mechanism of action of these PQC machines are thought to be conserved among bacteria, and are reviewed in Balchin et al. (2016) and Mogk et al. (2018). Caulobacter additionally uses ATP-independent adaptor proteins, stress-specific holdases, inhibitory proteins, and specialized transcriptional regulation to further direct and specify the activities of its chaperones and proteases. In this mini-review we highlight the tasks of the Caulobacter PQC network that contribute to cell cycle progression and development during optimal conditions, and discuss how the nodes of this network reorganize during stress to perform protective tasks that are crucial for survival.
Functions of the Energy-Dependent Folding Machines in Cell Cycle and Stress Adaptation
Caulobacter energy-dependent folding machines are capable of interacting broadly with the proteome to assist proteins into their native conformations, and perform specific and essential tasks both in cell cycle progression and stress response. For protein folding, Caulobacter utilizes the chaperone DnaK, co-chaperone DnaJ, and nucleotide exchange factor GrpE (DnaKJ/E), and a single copy of the chaperonin GroEL and co-chaperonin GroES (GroESL). In addition to DnaKJ/E, the Caulobacter genome contains one other DnaK-like protein (CCNA_01543) and five additional DnaJ-like proteins containing the characteristic J domain (CCNA_00965, CCNA_02218, CCNA_02245, CCNA_02860, CCNA_03105); however, it is currently unknown if these proteins direct the specificity of DnaK folding toward different client protein pools (Kampinga and Craig, 2010), or if they have another role. Depletion of either DnaKJ/E or GroESL halts the Caulobacter cell cycle in distinct stages; loss of DnaKJ/E results in a block of DNA replication initiation (Jonas et al., 2013; Schramm et al., 2017), whereas depletion of GroESL results in a cell division defect (Susin et al., 2006; Schroeder et al., 2020). Mild depletion of either of these folding machines produces an increase in the other (Da Silva et al., 2003; Susin et al., 2006), suggesting some degree of compensation exists, yet neither DnaKJ/E nor GroESL can fully substitute the stress response or cell cycle functions of the other.
The Caulobacter DnaKJ/E folding machine is essential in all growth temperatures, however, its function as a chaperone is dispensable in the absence of proteotoxic stress (Schramm et al., 2017). Instead, the requirement of DnaKJ/E for viability in optimal conditions is attributed to its binding and destabilization of the heat shock sigma factor σ32 (Da Silva et al., 2003; Schramm et al., 2017). In line with this notion, suppressor mutations reducing the abundance or activity of σ32 restore viability of cells depleted of DnaKJ in optimal conditions (Schramm et al., 2017). Sequestering of σ32 by DnaKJ/E prevents the sigma factor from inducing the expression of heat shock proteins (HSPs), which collectively function as a protective response that slows growth and is counterproductive in the absence of stress (Schramm et al., 2017). The importance of maintaining σ32 sequestration is reflected during depletion of DnaKJ/E in otherwise optimal conditions, where inappropriate HSP induction leads to a block in DNA replication through degradation of the replication initiator DnaA by the protease Lon (Jonas et al., 2013). DnaKJ/E also plays a role in Caulobacter development by interacting with the holdfast inhibitor HfiA (Eaton et al., 2016). DnaKJ/E activity keeps HfiA stabilized in a folded form, and this interaction operates in a regulatory circuit where increased levels of DnaK reduce the likelihood of developing surface attachment (Eaton et al., 2016), potentially promoting dispersal away from environments with inherent proteotoxic attributes.
When proteotoxic stress conditions are encountered, DnaKJ/E is titrated away from σ32 by unfolded proteins, and here its folding activity becomes crucial to survival (Figure 2A). Under proteotoxic threat, liberation of σ32 results in induction of heat shock genes, including dnaKJ itself, which is expressed from a σ32-responsive promoter in addition to a constitutive (σ73-responsive) promoter (Avedissian et al., 1995; Reisenauer et al., 1996; Da Silva et al., 2003). The conditional switching of DnaKJ/E between its functions as a σ32 regulator and a folding catalyst is reflected by dynamic changes in its subcellular localization, as DnaKJ/E alternates between a dispersed pattern in optimal conditions and localization at foci of protein aggregation during stress (Schramm et al., 2019).
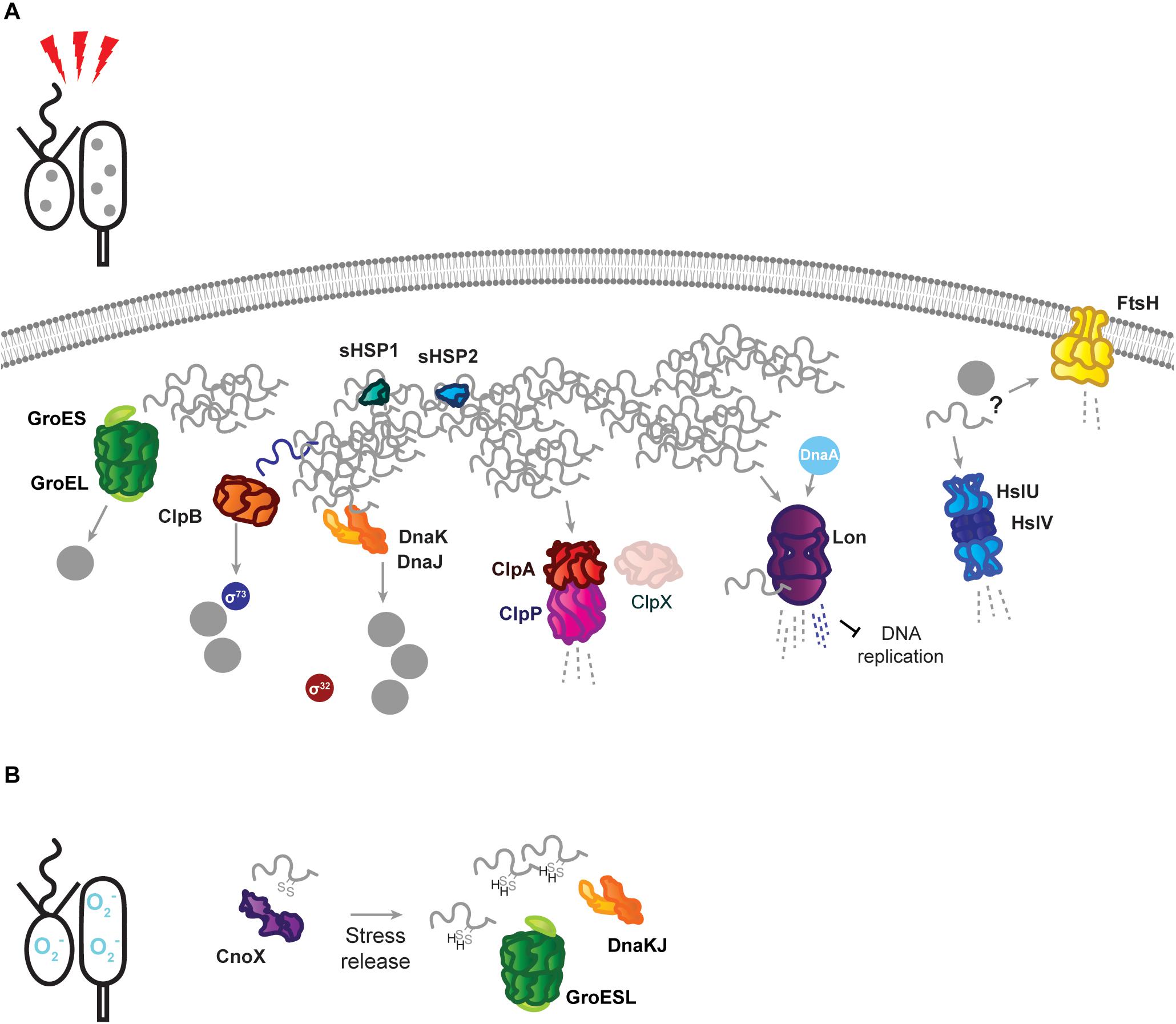
Figure 2. Stress response tasks of the Caulobacter crescentus PQC network. (A) Heat stress induces unfolding of the susceptible proteome, and unfolded proteins (gray squiggles) are incorporated into insoluble protein aggregates (gray dots). Known interactions during stress are indicated by protein name in colored circles. DnaKJ/E, ClpB, and GroESL participate in protein refolding and disaggregation. The small heat shock proteins organize unfolded proteins. ClpAP and Lon participate in degradation of unfolded proteins, in addition to regulatory roles. The proteases FtsH and HslUV are upregulated in response to proteotoxic stress, but it is unknown whether they contribute to degradation of unfolded proteins or regulatory substrates. (B) Oxidative stress results in oxidation of proteins and draining of the hydrotrope ATP, which can influence folding state. The holdase CnoX interacts and is capable of reducing disulfide groups of proteins, and protects them from aggregation until active GroESL and/or DnaKJ/E are available to refold these proteins. Membrane and DNA images created with Biorender (Biorender.com).
The chaperonin GroESL is expressed from a single promoter thought to respond to both σ73 and σ32 (Avedissian and Gomes, 1996; Baldini et al., 1998). During optimal conditions Caulobacter groESL is subject to a negative regulatory loop, effected through a controlling inverted repeat of chaperone expression (CIRCE) element and the HrcA repressor (Roberts et al., 1996; Baldini et al., 1998; Susin et al., 2004). Here, GroESL activity maintains HrcA in a folded conformation, in which it can bind the CIRCE element present in the groESL promoter, to reduce expression (Roberts et al., 1996; Baldini et al., 1998; Susin et al., 2004). Through CIRCE/HrcA regulation, the groESL transcript is cell cycle-regulated (Avedissian and Gomes, 1996; Baldini et al., 1998; Fang et al., 2013), and early pulse-chase experiments suggested that chaperonin synthesis is increased in the swarmer cell (Reuter and Shapiro, 1987). However, as GroESL protein is stably detected throughout the cell cycle in synchronized cultures (unpublished data), the relevance of this boost of synthesis remains unclear.
An overview of Caulobacter proteins whose folding state, or solubility, is influenced by GroESL has recently been described (Schroeder et al., 2020). Through this approach, cell cycle-regulated proteins involved in peptidoglycan biosynthesis and cell division were identified to have an interaction with GroESL folding availability in optimal conditions, including the FtsZ-interacting proteins FtsA and FzlA, which mediate a cell division block during GroESL depletion (Schroeder et al., 2020). While the role of GroESL in cell cycle progression is beginning to be understood, the contributions of this highly stress-induced folding machine during stress conditions have not yet been uncovered. In heat and ethanol stress σ32 induces robust groESL expression (Reuter and Shapiro, 1987; Susin et al., 2006; Heinrich et al., 2016), and while it is known that this groESL induction is specifically required to survive heat stress (Da Silva et al., 2003; Susin et al., 2006), the mechanisms by which GroESL protects the proteome during stress are not currently known.
Proteases Are Integral to Driving Cell Cycle Progression and Are Tailored to Specific Stress Survival Tasks
Approximately 5% of proteins are estimated to be rapidly turned over during the Caulobacter cell cycle (Grünenfelder et al., 2001), and the use of proteolysis as a means of rapidly removing regulatory and structural proteins is fully integrated in remodeling the proteome during Caulobacter cell cycle progression. The regulatory networks and mechanisms by which proteolysis is integrated into the Caulobacter cell cycle have been discussed in detail in other recent reviews (Joshi and Chien, 2016; Vass et al., 2016). In addition to proteome curation and regulatory degradation during optimal conditions, Caulobacter proteases are stress-responsive and remove damaged proteins that accumulate during proteotoxic stresses, additionally functioning to halt the cell cycle and redirect available resources toward survival tasks.
The best-studied Caulobacter protease is ClpP, which can associate with either of the unfoldase subunits ClpX and ClpA (Figure 1B). ClpXP has many cell cycle-regulated targets, one of which is the master cell cycle regulator CtrA (Quon et al., 1996, 1998; Laub et al., 2002), and extensive work has uncovered that the regulated and coordinated activities of three specific adaptor proteins, CpdR, RcdA, and PopA, facilitate CtrA degradation at the correct time and location during the cell cycle (reviewed by Joshi and Chien, 2016). In addition to CtrA, several other proteins with critical functions in Caulobacter development are degraded by ClpXP, including PdeA (Abel et al., 2011), McpA (Tsai and Alley, 2001), TacA (Bhat et al., 2013), and FtsZ (Williams et al., 2014). Subsets of the ClpXP adaptors regulate degradation of several of these substrates (Joshi et al., 2015; Lau et al., 2015), emphasizing their importance in directing this protease toward specific substrate groups. While ClpX and ClpP are both essential in optimal conditions (Østerås et al., 1999), loss of ClpXP degradation of CtrA and other cell cycle substrates does not result in inviability. Instead, ClpXP degradation of the toxin SocB is essential to avoid inhibition of DNA polymerase activity, as suppressor mutations in socB bypass the need for ClpXP (Aakre et al., 2013). In addition to direct functions in cell cycle progression, ClpXP processing of the replication clamp subunit DnaX is required to promote DNA replication during optimal conditions, and accumulation of full length DnaX during genotoxic stress is an important factor in surviving DNA damage (Vass and Chien, 2013). Furthermore, ClpXP maintains a conserved function in degrading incompletely synthesized proteins directed from the SsrA/SspB pathway in Caulobacter (Keiler et al., 2000). Interestingly, the SsrA RNA (also known as tmRNA) of this system, which adds a degradation tag to products of stalled translation, is involved in cell cycle regulation, as deletion of ssrA delays timing of dnaA transcription and DNA replication (Keiler and Shapiro, 2003; Cheng and Keiler, 2009), however the precise mechanism remains unclear.
Unlike most other PQC network proteins, clpX expression is not induced by σ32 (Østerås et al., 1999; Schramm et al., 2017), however, both clpA and clpP expression are strongly upregulated by σ32 (Schramm et al., 2017). To accomplish this regulation, the clpP and clpX genes are separated by a 1.1 kb region containing the phosphotransferase cicA (Østerås et al., 1999; Fuchs et al., 2001), while clpA is co-transcribed with the conserved Clp protease adaptor clpS from a separate locus. A change in the ratio between ClpX and ClpA during σ32-dependent HSP induction could redirect ClpP proteolysis from ClpX-mediated functions in cell cycle regulation toward stress survival tasks mediated by ClpA (Østerås et al., 1999; Grünenfelder et al., 2004). In line with this idea, high ClpA abundance can inhibit ClpX function (Jenal, 1998), and ClpAP is competent to degrade unfolded proteins (Joshi and Chien, 2016; Liu et al., 2016). That ClpAP can degrade substrates primarily degraded by other proteases (Liu et al., 2016) has led to the suggestion that ClpAP functions as a compensatory protease. However, ClpAP performs specific tasks during optimal conditions as well, where it is the primary protease responsible for degrading the flagellar protein FliF and the division protein FtsA in the swarmer cell (Grünenfelder et al., 2004; Williams et al., 2014). The finding that ClpAP is able to curate abundances of other PQC network protein via degradation of the protease Lon (Barros et al., 2020) further indicates that regulation of ClpA, ClpX, and ClpP is complex.
The Lon protease regulates many points of Caulobacter cell cycle and development, including degradation of three essential cell cycle regulators; the methyltransferase CcrM (Wright et al., 1996), the swarmer cell-specific transcriptional regulator SciP (Gora et al., 2013), and the replication initiator DnaA (Jonas et al., 2013; Leslie et al., 2015). Lon-mediated degradation is in some cases regulated through its ability to bind DNA, as in how chromosomal DNA binding facilitates recognition and degradation of CcrM in the swarmer cell (Zhou et al., 2019). The ability of Lon to bind DNA, and the influence of DNA binding on its activity, may be particularly important in clearing damaged proteins from the chromosome during genotoxic stress (Zeinert et al., 2018). Through its ability to adjust the abundances of regulatory proteins and recognize the presence of unfolded proteins, Lon is ideally positioned to halt the cell cycle at the appearance of proteotoxic stress (Jonas et al., 2013; Leslie et al., 2015). The appearance of unfolded proteins has a dual effect on Lon degradation, firstly by increasing σ32-dependent expression of the protease, and secondly by stimulating Lon degradation of certain substrates (Jonas et al., 2013). The combined effects of this regulation provide a mechanism for halting the cell cycle during unfavorable conditions (Figure 2A), where upregulated and activated Lon degrades DnaA upon exposure to proteotoxic stress (Jonas et al., 2013). More recent work has suggested that Lon may be titrated from different substrate pools based on stress intensity, as mild temperature increases result in CcrM stabilization and increased expression of CcrM-regulated nucleotide metabolism genes that support rapid growth (Zeinert et al., 2020). In addition to roles in proteotoxic and genotoxic stress, Lon has additionally recently been found to be integrated into sensing and responding to low oxygen levels (Stein et al., 2020). How Lon activity and substrate selectivity can be targeted toward specific client protein pools in response to environmental changes remains an area of active research.
The contributions of other proteases to Caulobacter growth and survival are less well established, as in the case of the membrane-bound protease FtsH where the known substrate pool is limited to its conserved interaction with σ32 (Fischer et al., 2002). Curiously, a three amino acid deletion in the substrate recognition domain of the HslU chaperone subunit enables the HslUV protease to degrade σ32 in cells depleted of either DnaKJ or FtsH, where σ32 is normally stable (Schramm et al., 2017). This finding raises questions on if HslUV may perform redundant or degenerate roles with FtsH, however, the substrate pool and contribution of the HslUV protease to Caulobacter development and stress survival remains entirely uncharacterized. FtsH mutants exhibit growth and developmental defects during optimal conditions, and additionally are more sensitive to various stresses (Fischer et al., 2002), however, whether these phenotypes stem from the interaction of FtsH with σ32 or from degradation of other substrates remains to be discovered. In addition to the ATP-dependent proteases, Caulobacter possesses proteases specialized for degrading proteins in the membrane or periplasm, such as the membrane metalloprotease MmpA, which degrades the processed form of the polarity factor PodJ (Chen et al., 2006). Many more yet unidentified interactions between the PQC network and cell envelope proteins must be involved to coordinate stalk synthesis, divisome assembly, chemoreceptor placement, and the many other developmental events taking place across the membrane, with the requirements of optimal and stress conditions.
Energy-Dependent Disaggregases Assist the PQC Network in Stress Survival
ClpB is a disaggregase that acts specifically to remediate aggregated protein, and consistent with its expression occurring exclusively during stress (Simão et al., 2005; Schramm et al., 2019), no phenotype is associated with its absence in optimal conditions. Deletion of ClpB results in an inability to dissolve stress-induced protein aggregates, and an associated reduction in the ability of Caulobacter to tolerate proteotoxic stress (Simão et al., 2005; Schramm et al., 2019). Protein aggregates are persistent in ClpB-deficient cells, and collaboration between DnaKJ/E and ClpB is the primary mechanism of resolving aggregated protein during sublethal heat stress in Caulobacter, although it is unknown if mistranslation-inducing antibiotic stresses, which do not induce HSP expression, depend as heavily on ClpB (Schramm et al., 2019). Persistent protein aggregates in clpB mutant cells have been used to study inheritance of insoluble protein deposits (Schramm et al., 2019), which has been hypothesized to underlie replicative decline in the stalked cell (Ackermann et al., 2003). While the majority of protein aggregates are swiftly dissolved when ClpB is functional, Caulobacter was found to share persistent aggregated protein deposits between stalked and swarmer cells (Schramm et al., 2019). The fidelity with which the PQC network curates the proteome as the stalked cell ages and experiences sequential stresses remains an open question.
ClpB is also solely responsible for mediating the shutoff phase of the σ32-dependent stress response in Caulobacter (Simão et al., 2005). This process is important for tolerating sublethal stress, where unfolding and aggregation of the stress-sensitive σ73 allows σ32 to interact with the RNAP instead (Simão et al., 2005). To ensure that σ73-regulated genes are not repressed indefinitely, ClpB reactivates σ73 from protein aggregates (Da Silva et al., 2003; Simão et al., 2005). This reactivation of σ73 restores its activity and allows it to compete with σ32 for association with the RNAP, which is a crucial step in recovering from or adapting to proteotoxic stress (Simão et al., 2005).
Energy-Independent Holdases Assist the Major PQC Network Nodes in Surviving Specific Stresses
To cope with the demands of proteotoxic stress, Caulobacter also employs energy-independent holdases that collaborate with major nodes of the PQC network and assist in survival. Two small heat shock proteins, sHSP1 (CCNA_02341, referred to also as IbpA, but not to be confused with the inositol binding protein A of Caulobacter), and sHSP2 (CCNA_03706), are induced in response to protein unfolding, and may organize unfolded proteins in a disaggregation-ready state, as aggregates dissolve more slowly in their absence (Schramm et al., 2019). While sHSP accumulation is restricted to σ32-acting stresses, the sHSP1 protein has been confirmed as a ClpXP substrate, featuring the classical C-terminal AA degron (Bhat et al., 2013). It remains unresolved if the ClpXP protease might remove the highly expressed sHSP1 during later phases of stress recovery, when cells are returning to growth in optimal conditions, or effect its turnover during optimal conditions.
The chaperedoxin holdase CnoX (CCNA_00109) is both constitutively expressed and σ32-responsive, and participates in cellular redox homeostasis during optimal conditions by reducing disulfide bonds (Goemans et al., 2018a). Oxidative stress further activates CnoX, and it functions to protect approximately 90 proteins from aggregation until they can be transferred to DnaKJ and GroESL for refolding (Goemans et al., 2018a) (Figure 2B). Similarly to what is observed with sHSP1 and sHSP2, deletion of CnoX does not affect survival of proteotoxic stress (Goemans et al., 2018a; Schramm et al., 2019), however it is unresolved if other stress conditions Caulobacter frequently encounters might exhibit a higher requirement for these holdases. As CnoX interacts with cell cycle-regulated proteins (Goemans et al., 2018a, b), many open questions remain on the function of holdases during both Caulobacter development and stress. Caulobacter also possesses a homolog of trigger factor (CCNA_02042), which likely contributes to the de novo folding of cytoplasmic proteins, but remains so far unstudied. Furthermore, holdase regulation and collaboration in compartment-specific stress (Castanié-Cornet et al., 2014; De Geyter et al., 2016), such as regulation of the holdase SecB (CCNA_03858) in membrane protein transport, remains unaddressed in Caulobacter.
Conclusion and Outlook
Caulobacter has become a powerful model to investigate how a prokaryotic PQC network integrates the demands of a developmental program with frequently encountered threats to proteostasis. Several mechanisms by which the highly conserved nodes of the PQC network ensure cell cycle progression during optimal conditions are known, and ongoing work has begun to reveal mechanistic insight into how these functions change when stress is encountered. Substrate trapping or depletion experiments connected to mass spectrometry have begun to characterize client proteins of the major chaperones and proteases (Bhat et al., 2013; Goemans et al., 2018a; Schroeder et al., 2020), and with this an understanding of how processes are regulated as individual proteins transit through the PQC network has become possible.
A theme emerging from recent work in Caulobacter is that the PQC network responds to a range of stress inputs and intensities with tailored and collaborative responses. For example, Caulobacter primarily dissolves aggregated protein deposits during stress recovery, except after high intensity stresses where dilution of aggregates in the growing population becomes the primary method of reducing insoluble protein (Schramm et al., 2019). Lon may also be directed toward different activities based on stress intensity; during mild temperature increases Lon has been linked to increasing dNTP pools to support DNA replication (Zeinert et al., 2020), whereas strong unfolding stress requires Lon-mediated destabilization of DnaA and prevention of DNA replication initiation until conditions improve (Jonas et al., 2013). Stress-responsive factors that tune the generalist PQC network to specific environmental conditions are also beginning to be identified, such as the chaperedoxin CnoX that responds to oxidative stress and collaborates with DnaKJ/E and GroESL (Goemans et al., 2018a). How PQC network functions change within the dynamic range of stress responses, and how nodes of the PQC network collaborate in effecting these responses is a developing area of Caulobacter PQC work.
Finally, work in Caulobacter is also beginning to address how the discrete proteomes of the swarmer and stalked cell interface with the PQC network. Cell cycle-restricted Caulobacter proteins that are particularly sensitive to aggregation have been described (Schramm et al., 2019), and as oscillations in transcriptome (Fang et al., 2013), proteome (Grünenfelder et al., 2001), and metabolome (Hartl et al., 2020) of Caulobacter during the cell cycle in optimal conditions have been identified, the field is open for investigating the interface between PQC machines and the dimorphic developmental program of Caulobacter during environmental changes.
Author Contributions
Both authors contributed to the writing and final approval of the manuscript.
Funding
This work was supported by the Swedish Foundation for Strategic Research (FFL15-0005), the Swedish Research Council (2016-03300), and funding from the Strategic Research Area (SFO) program distributed through Stockholm University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aakre, C. D., Phung, T. N., Huang, D., and Laub, M. T. (2013). A bacterial toxin inhibits DNA replication elongation through a direct interaction with the β sliding clamp. Mol. Cell 52, 617–628. doi: 10.1016/j.molcel.2013.10.014
Abel, S., Chien, P., Wassmann, P., Schirmer, T., Kaever, V., Laub, M. T., et al. (2011). Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol. Cell 43, 550–560. doi: 10.1016/j.molcel.2011.07.018
Ackermann, M., Stearns, S. C., and Jenal, U. (2003). Senescence in a bacterium with asymmetric division. Science 300:1920. doi: 10.1126/science.1083532
Avedissian, M., and Gomes, S. L. (1996). Expression of the groESL operon is cell-cycle controlled in Caulobacter crescentus. Mol. Microbiol. 19, 79–89. doi: 10.1046/j.1365-2958.1996.347879.x
Avedissian, M., Lessing, D., Gober, J. W., Shapiro, L., and Gomes, S. L. (1995). Regulation of the Caulobacter crescentus dnaKJ operon. J. Bacteriol. 177, 3479–3484. doi: 10.1128/JB.177.12.3479-3484.1995
Balchin, D., Hayer-Hartl, M., and Hartl, F. U. (2016). In vivo aspects of protein folding and quality control. Science 353:aac4354. doi: 10.1126/science.aac4354
Baldini, R. L., Avedissian, M., and Gomes, S. L. (1998). The CIRCE element and its putative repressor control cell cycle expression of the Caulobacter crescentus groESL operon. J. Bacteriol. 180, 1632–1641. doi: 10.1128/jb.180.7.1632-1641.1998
Barros, B. B., Mahmoud, S. A., Chien, P., and Zeinert, R. D. (2020). Degradation of Lon in Caulobacter crescentus. J. Bacteriol. 203:e00344-20. doi: 10.1128/JB.00344-20
Bhat, N. H., Vass, R. H., Stoddard, P. R., Shin, D. K., and Chien, P. (2013). Identification of ClpP substrates in Caulobacter crescentus reveals a role for regulated proteolysis in bacterial development. Mol. Microbiol. 88, 1083–1092. doi: 10.1111/mmi.12241
Castanié-Cornet, M.-P., Bruel, N., and Genevaux, P. (2014). Chaperone networking facilitates protein targeting to the bacterial cytoplasmic membrane. Biochim. Biophys. Acta 1843, 1442–1456. doi: 10.1016/j.bbamcr.2013.11.007
Chen, J. C., Hottes, A. K., McAdams, H. H., McGrath, P. T., Viollier, P. H., and Shapiro, L. (2006). Cytokinesis signals truncation of the PodJ polarity factor by a cell cycle-regulated protease. EMBO J. 25, 377–386. doi: 10.1038/sj.emboj.7600935
Cheng, L., and Keiler, K. C. (2009). Correct timing of dnaA transcription and initiation of DNA replication requires trans translation. J. Bacteriol. 191, 4268–4275. doi: 10.1128/JB.00362-09
Curtis, P. D., and Brun, Y. V. (2010). Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol. Mol. Biol. Rev. 74, 13–41. doi: 10.1128/MMBR.00040-09
Da Silva, A. C. A., Simão, R. C. G., Susin, M. F., Baldini, R. L., Avedissian, M., and Gomes, S. L. (2003). Downregulation of the heat shock response is independent of DnaK and σ32 levels in Caulobacter crescentus: heat shock response regulation in Caulobacter. Mol. Microbiol. 49, 541–553. doi: 10.1046/j.1365-2958.2003.03581.x
De Geyter, J., Tsirigotaki, A., Orfanoudaki, G., Zorzini, V., Economou, A., and Karamanou, S. (2016). Protein folding in the cell envelope of Escherichia coli. Nat. Microbiol. 1:16107. doi: 10.1038/nmicrobiol.2016.107
Eaton, D. S., Crosson, S., and Fiebig, A. (2016). Proper control of caulobacter crescentus cell surface adhesion requires the general protein chaperone DnaK. J. Bacteriol. 198, 2631–2642. doi: 10.1128/JB.00027-16
Fang, G., Passalacqua, K. D., Hocking, J., Llopis, P., Gerstein, M., Bergman, N. H., et al. (2013). Transcriptomic and phylogenetic analysis of a bacterial cell cycle reveals strong associations between gene co-expression and evolution. BMC Genom. 14:450. doi: 10.1186/1471-2164-14-450
Fischer, B., Rummel, G., Aldridge, P., and Jenal, U. (2002). The FtsH protease is involved in development, stress response and heat shock control in Caulobacter crescentus: Caulobacter crescentus FtsH protease. Mol. Microbiol. 44, 461–478. doi: 10.1046/j.1365-2958.2002.02887.x
Fuchs, T., Wiget, P., Osterås, M., and Jenal, U. (2001). Precise amounts of a novel member of a phosphotransferase superfamily are essential for growth and normal morphology in Caulobacter crescentus. Mol. Microbiol. 39, 679–692. doi: 10.1046/j.1365-2958.2001.02238.x
Goemans, C. V., Beaufay, F., Arts, I. S., Agrebi, R., Vertommen, D., and Collet, J.-F. (2018a). The chaperone and redox properties of CnoX chaperedoxins are tailored to the proteostatic needs of bacterial species. mBio 9:e01541-18. doi: 10.1128/mBio.01541-18
Goemans, C. V., Beaufay, F., Wahni, K., Van Molle, I., Messens, J., and Collet, J.-F. (2018b). An essential thioredoxin is involved in the control of the cell cycle in the bacterium Caulobacter crescentus. J. Biol. Chem. 293, 3839–3848. doi: 10.1074/jbc.RA117.001042
Gomes, S. L., Juliani, M. H., Maia, J. C., and Silva, A. M. (1986). Heat shock protein synthesis during development in Caulobacter crescentus. J. Bacteriol. 168, 923–930. doi: 10.1128/JB.168.2.923-930.1986
Gora, K. G., Cantin, A., Wohlever, M., Joshi, K. K., Perchuk, B. S., Chien, P., et al. (2013). Regulated proteolysis of a transcription factor complex is critical to cell cycle progression in Caulobacter crescentus. Mol. Microbiol. 87, 1277–1289. doi: 10.1111/mmi.12166
Grünenfelder, B., Rummel, G., Vohradsky, J., Röder, D., Langen, H., and Jenal, U. (2001). Proteomic analysis of the bacterial cell cycle. Proc. Natl. Acad. Sci. U.S.A. 98, 4681–4686. doi: 10.1073/pnas.071538098
Grünenfelder, B., Tawfilis, S., Gehrig, S., ØSterås, M., Eglin, D., and Jenal, U. (2004). Identification of the protease and the turnover signal responsible for cell cycle-dependent degradation of the Caulobacter FliF motor protein. J. Bacteriol. 186, 4960–4971. doi: 10.1128/JB.186.15.4960-4971.2004
Hartl, J., Kiefer, P., Kaczmarczyk, A., Mittelviefhaus, M., Meyer, F., Vonderach, T., et al. (2020). Untargeted metabolomics links glutathione to bacterial cell cycle progression. Nat. Metab. 2, 153–166. doi: 10.1038/s42255-019-0166-0
Heinrich, K., Sobetzko, P., and Jonas, K. (2016). A kinase-phosphatase switch transduces environmental information into a bacterial cell cycle circuit. PLoS Genet. 12:e1006522. doi: 10.1371/journal.pgen.1006522
Jenal, U. (1998). An essential protease involved in bacterial cell-cycle control. EMBO J. 17, 5658–5669. doi: 10.1093/emboj/17.19.5658
Jonas, K., Liu, J., Chien, P., and Laub, M. T. (2013). Proteotoxic stress induces a cell-cycle arrest by stimulating Lon to degrade the replication initiator DnaA. Cell 154, 623–636. doi: 10.1016/j.cell.2013.06.034
Joshi, K. K., Bergé, M., Radhakrishnan, S. K., Viollier, P. H., and Chien, P. (2015). An adaptor hierarchy regulates proteolysis during a bacterial cell cycle. Cell 163, 419–431. doi: 10.1016/j.cell.2015.09.030
Joshi, K. K., and Chien, P. (2016). Regulated proteolysis in bacteria: Caulobacter. Annu. Rev. Genet. 50, 423–445. doi: 10.1146/annurev-genet-120215-035235
Kampinga, H. H., and Craig, E. A. (2010). The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell. Biol. 11, 579–592. doi: 10.1038/nrm2941
Keiler, K. C., and Shapiro, L. (2003). TmRNA is required for correct timing of DNA replication in Caulobacter crescentus. J. Bacteriol. 185, 573–580. doi: 10.1128/jb.185.2.573-580.2003
Keiler, K. C., Shapiro, L., and Williams, K. P. (2000). tmRNAs that encode proteolysis-inducing tags are found in all known bacterial genomes: A two-piece tmRNA functions in Caulobacter. Proc. Natl. Acad. Sci. U.S.A. 97, 7778–7783. doi: 10.1073/pnas.97.14.7778
Lau, J., Hernandez-Alicea, L., Vass, R. H., and Chien, P. (2015). A phosphosignaling adaptor primes the AAA+ protease ClpXP to drive cell cycle-regulated proteolysis. Mol. Cell 59, 104–116. doi: 10.1016/j.molcel.2015.05.014
Laub, M. T., Chen, S. L., Shapiro, L., and McAdams, H. H. (2002). Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc. Natl. Acad. Sci. U.S.A. 99, 4632–4637. doi: 10.1073/pnas.062065699
Leslie, D. J., Heinen, C., Schramm, F. D., Thüring, M., Aakre, C. D., Murray, S. M., et al. (2015). Nutritional control of DNA replication initiation through the proteolysis and regulated translation of DnaA. PLoS Genet. 11:e1005342. doi: 10.1371/journal.pgen.1005342
Liu, J., Francis, L. I., Jonas, K., Laub, M. T., and Chien, P. (2016). ClpAP is an auxiliary protease for DnaA degradation in Caulobacter crescentus. Mol. Microbiol. 102, 1075–1085. doi: 10.1111/mmi.13537
Mogk, A., Bukau, B., and Kampinga, H. H. (2018). Cellular handling of protein aggregates by disaggregation machines. Mol. Cell 69, 214–226. doi: 10.1016/j.molcel.2018.01.004
Østerås, M., Stotz, A., Nuoffer, S. S., and Jenal, U. (1999). Identification and transcriptional control of the genes encoding the Caulobacter crescentus ClpXP protease. J. Bacteriol. 181, 3039–3050. doi: 10.1128/JB.181.10.3039-3050.1999
Quon, K. C., Marczynski, G. T., and Shapiro, L. (1996). Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84, 83–93. doi: 10.1016/S0092-8674(00)80995-2
Quon, K. C., Yang, B., Domian, I. J., Shapiro, L., and Marczynski, G. T. (1998). Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc. Natl. Acad. Sci. U.S.A. 95, 120–125. doi: 10.1073/pnas.95.1.120
Reisenauer, A., Mohr, C. D., and Shapiro, L. (1996). Regulation of a heat shock sigma32 homolog in Caulobacter crescentus. J. Bacteriol. 178, 1919–1927. doi: 10.1128/JB.178.7.1919-1927.1996
Reuter, S. H., and Shapiro, L. (1987). Asymmetric segregation of heat-shock proteins upon cell division in Caulobacter crescentus. J. Mol. Biol. 194, 653–662. doi: 10.1016/0022-2836(87)90242-7
Roberts, R. C., Toochinda, C., Avedissian, M., Baldini, R. L., Gomes, S. L., and Shapiro, L. (1996). Identification of a Caulobacter crescentus operon encoding hrcA, involved in negatively regulating heat-inducible transcription, and the chaperone gene grpE. J. Bacteriol. 178, 1829–1841. doi: 10.1128/jb.178.7.1829-1841.1996
Schramm, F. D., Heinrich, K., Thüring, M., Bernhardt, J., and Jonas, K. (2017). An essential regulatory function of the DnaK chaperone dictates the decision between proliferation and maintenance in Caulobacter crescentus. PLoS Genet. 13:e1007148. doi: 10.1371/journal.pgen.1007148
Schramm, F. D., Schroeder, K., Alvelid, J., Testa, I., and Jonas, K. (2019). Growth-driven displacement of protein aggregates along the cell length ensures partitioning to both daughter cells in Caulobacter crescentus. Mol. Microbiol. 111, 1430–1448. doi: 10.1111/mmi.14228
Schroeder, K., Heinrich, K., Neuwirth, I., and Jonas, K. (2020). The chaperonin GroESL facilitates Caulobacter crescentus cell division by supporting the function of the actin homologue FtsA. bioRxiv [Preprint]. doi: 10.1101/2020.12.17.423375
Simão, R. C. G., Susin, M. F., Alvarez-Martinez, C. E., and Gomes, S. L. (2005). Cells lacking ClpB display a prolonged shutoff phase of the heat shock response in Caulobacter crescentus: Caulobacter crescentus ClpB. Mol. Microbiol. 57, 592–603. doi: 10.1111/j.1365-2958.2005.04713.x
Stein, B. J., Fiebig, A., and Crosson, S. (2020). Feedback control of a two-component signaling system by an Fe-S-binding receiver domain. mBio 11:e30383-19. doi: 10.1128/mBio.03383-19
Susin, M. F., Baldini, R. L., Gueiros-Filho, F., and Gomes, S. L. (2006). GroES/GroEL and DnaK/DnaJ have distinct roles in stress responses and during cell cycle progression in Caulobacter crescentus. J. Bacteriol. 188, 8044–8053. doi: 10.1128/JB.00824-06
Susin, M. F., Perez, H. R., Baldini, R. L., and Gomes, S. L. (2004). Functional and structural analysis of HrcA repressor protein from Caulobacter crescentus. J. Bacteriol. 186, 6759–6767. doi: 10.1128/JB.186.20.6759-6767.2004
Tsai, J.-W., and Alley, M. R. K. (2001). Proteolysis of the Caulobacter McpA chemoreceptor is cell cycle regulated by a ClpX-dependent pathway. J. Bacteriol. 183, 5001–5007. doi: 10.1128/JB.183.17.5001-5007.2001
Vass, R. H., and Chien, P. (2013). Critical clamp loader processing by an essential AAA+ protease in Caulobacter crescentus. Proc. Natl. Acad. Sci. 110, 18138–18143. doi: 10.1073/pnas.1311302110
Vass, R. H., Zeinert, R. D., and Chien, P. (2016). Protease regulation and capacity during Caulobacter growth. Curr. Opin. Microbiol. 7, 75–81. doi: 10.1016/j.mib.2016.07.017
Williams, B., Bhat, N., Chien, P., and Shapiro, L. (2014). ClpXP and ClpAP proteolytic activity on divisome substrates is differentially regulated following the Caulobacter asymmetric cell division. Mol. Microbiol. 93, 853–866. doi: 10.1111/mmi.12698
Wright, R., Stephens, C., Zweiger, G., Shapiro, L., and Alley, M. R. (1996). Caulobacter Lon protease has a critical role in cell-cycle control of DNA methylation. Genes Dev. 10, 1532–1542. doi: 10.1101/gad.10.12.1532
Zeinert, R. D., Baniasadi, H., Tu, B. P., and Chien, P. (2020). The Lon protease links nucleotide metabolism with proteotoxic stress. Mol. Cell 79, 758–767.e6. doi: 10.1016/j.molcel.2020.07.011
Zeinert, R. D., Liu, J., Yang, Q., Du, Y., Haynes, C. M., and Chien, P. (2018). A legacy role for DNA binding of Lon protects against genotoxic stress. bioRxiv [Preprint]. doi: 10.1101/317677
Keywords: protease, chaperone, holdase, protein quality control, cell cycle, bacterial development
Citation: Schroeder K and Jonas K (2021) The Protein Quality Control Network in Caulobacter crescentus. Front. Mol. Biosci. 8:682967. doi: 10.3389/fmolb.2021.682967
Received: 19 March 2021; Accepted: 08 April 2021;
Published: 30 April 2021.
Edited by:
Kürşad Turgay, Max Planck Unit for the Science of Pathogens, Max-Planck-Gesellschaft (MPG), GermanyReviewed by:
Peter Chien, University of Massachusetts Amherst, United StatesUlf Gerth, University of Greifswald, Germany
Copyright © 2021 Schroeder and Jonas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kristina Jonas, a3Jpc3RpbmEuam9uYXNAc3Uuc2U=
†ORCID: Kristen Schroeder orcid.org/0000-0002-6271-4530 Kristina Jonas orcid.org/0000-0002-1469-4424
 Kristen Schroeder
Kristen Schroeder Kristina Jonas
Kristina Jonas