- 1Department of Medicinal Chemistry, School of Pharmacy, University of Washington, Seattle, WA, United States
- 2Department of Pharmacy, School of Pharmacy, University of Washington, Seattle, WA, United States
It has been suggested that daptomycin can be inactivated by lipids released by Staphylococcus aureus and that this effect is antagonized by phenol soluble modulins (PSMs), which bind to the shed lipids. PSM production is regulated by the Agr system, and others have shown that loss of the Agr function enhances S. aureus survival in the presence of daptomycin. Here we assessed the impact of Agr function on daptomycin activity and lipid metabolism under various conditions. Daptomycin activity was evaluated against three sets of isogenic strain series with wild-type or dysfunctional Agr using static daptomycin time-kills over 24 h and against one strain pair using in vitro pharmacokinetic/pharmacodynamic (PK/PD) models simulating clinical daptomycin exposure for 48 h. We performed comprehensive lipidomics on bacterial membranes and the spent media to correlate lipid shedding with survival. In static time-kill experiments, two agr-deficient strains (SH1000- and USA300 LAC ΔagrA) showed improved survival for 8 h compared with their corresponding wild-type strains as seen in previous studies, but this difference did not persist for 24 h. However, four other agr-deficient strains (SH1001 and JE2 agr KOs) did not demonstrate improved survival compared to isogenic wild-type strains at any time in the time-kills. Lipidomics analysis of SH1000, SH1001, and SH1000- strains showed daptomycin exposure increased lipid shedding compared to growth controls in all strains with phosphatidylglycerols (PGs), lysylPGs and cardiolipins predominating. In the cell pellets, PGs and lysylPGs decreased but cardiolipins were unchanged with daptomycin exposure. The shed lipid profiles in SH1001 and SH1000- were similar, suggesting that the inability to resist daptomycin by SH1001 was not because of differences in lipid shedding. In the PK/PD model, the agr mutant SH1000- strain did not show improved survival relative to SH1000 either. In conclusion, inactivation of daptomycin by shed lipids may be dependent on genetic background, the specific agr mutations, or the techniques used to generate these KOs rather than the overall function of the Agr system, and its contribution to daptomycin tolerance seems to be varied, transient, and growth-condition dependent.
Introduction
Daptomycin is a lipopeptide antimicrobial that consists of a cyclic polypeptide with 13 amino acids and a decanoyl fatty acyl tail that plays an important role in the management of invasive infections caused by methicillin-resistant Staphylococcus aureus (MRSA). Its mechanism of action involves direct interaction with the negatively charged membrane lipids, phosphatidylglycerols (PGs), leading to loss of membrane potential and cell death (Muraih et al., 2011; Muraih et al., 2012; Pogliano et al., 2012; Bayer et al., 2013). As such, most studies on daptomycin resistance point to development of mutations in genes that control membrane lipid metabolism and/or lead to changes in surface charge, membrane fluidity, or both, such as mprF, cls, pgsA, and the dlt operon, which reduces binding of daptomycin to the cell membrane or prevents disruption of the membrane by daptomycin (Yang et al., 2009; Peleg et al., 2012; Mishra and Bayer, 2013; Mishra et al., 2013; Bayer et al., 2014; Cafiso et al., 2014; Mishra et al., 2014; Bayer et al., 2015; Hines et al., 2017; Jiang et al., 2019). Mutations in two-component regulatory systems that regulate cell wall and cell membrane metabolism, such as vraSR and walKR (Friedman et al., 2006; Mehta et al., 2012; Werth et al., 2021), have also been shown to contribute to daptomycin resistance. We previously applied a novel multi-dimensional lipidomic method to characterizing the detailed lipid profile changes associated with MRSA strains that have developed resistance to daptomycin and found overall greatly decreased levels of PGs in a resistant strain with mutations in both pgsA and mprF (Hines et al., 2017) and greatly elevated levels of lysyl-phosphatidylglycerols (lysylPGs) and cardiolipins (CLs) in a strain with only an mprF mutation (Hines et al., 2020). Thus, altering lipid metabolism is an important route for bacteria to acquire resistance to daptomycin.
In recent years, inactivation of daptomycin by phospholipids released by S. aureus upon daptomycin exposure was proposed as a novel mechanism of daptomycin tolerance (Pader et al., 2016). The authors found that this effect may be antagonized by the production of amphipathic peptides called phenol soluble modulins (PSMs), which bind released phospholipids and thus prevent inactivation of daptomycin (Otto, 2014; Pader et al., 2016). PSM production is regulated by the accessory gene regulator (Agr) system, which is encoded by a four-gene operon (agrBDCA) and a regulatory RNA gene (RNAIII) (Peschel and Otto, 2013). Pader et al. suggested that the loss of the Agr quorum-sensing system enhances S. aureus survival during daptomycin exposure since PSMs are not released, and thus there is no competition for daptomycin sequestration by the shed lipids. However, the detailed composition of membrane lipids and shed lipids by S. aureus strains with variable Agr activity in response to daptomycin exposure has not been elucidated and the effect of lipid shedding on daptomycin activity has not been examined under clinically relevant kinetic drug exposures. In this work, we assessed the impact of Agr function on daptomycin activity and lipid metabolism in several genetic backgrounds and in static time-kills and in vitro pharmacokinetic/pharmacodynamic (PK/PD) models to better understand the contribution of lipid shedding to daptomycin tolerance.
Materials and Methods
Susceptibility Testing and Agr Functionality Testing
The susceptibility to daptomycin was evaluated by broth microdilution in accordance with CLSI guidelines (CLSI, 2017). The Agr functionality was tested on BBLTM TrypticaseTM soy agar with 5% sheep blood (TSA II; Becton, Dickinson and Company, Franklin Lakes, NJ, United States) as previously described (Sakoulas et al., 2002). Briefly, a 0.5-McFarland suspension of RN4220 was streaked in a line down the center of the agar plate dividing the plate into two halves, and the test strains were streaked from the edge of the agar plate to the center line of RN4220. Hemolysis was examined after overnight incubation at 37°C.
Static Time-Kill Assay
Overnight cultures of each strain were inoculated into tryptic soy broth (TSB, Remel Lenexa, KS, United States) supplemented with 50 µg/ml of elemental calcium and 20 µg/ml of daptomycin (Merck, Kenilworth, NJ, United States) to ∼108 CFU/ml in 50 ml conical tubes, incubated at 37°C with shaking. Samples were taken at 0, 2, 4, 6, 8, and 24 h, serially diluted and spiral plated on tryptic soy agar (TSA; Becton, Dickinson and Company, Franklin Lakes, NJ, United States) plates to evaluate the bacterial growth over time with exposure to daptomycin. Experiments were performed under lower aeration (30 ml of culture in 50 ml tube shaken at 85 rpm; SH1000 and SH1001, JE2 and JE2 Δagr) and higher aeration (9 ml of culture in 50 ml tube shaken at 180 rpm; SH1000, SH1001, and SH1000-, USA300 LAC and USA300 LAC ΔagrA) conditions, the latter of which were more consistent with the methods by (Pader et al., 2016). All experiments were performed in duplicate.
Lipid Profiling of Static Time-kill of SH1000, SH1001, and SH1000- Overnight cultures of SH1000, SH1001, and SH1000- were inoculated into TSB containing 50 µg/ml of elemental calcium to ∼108 CFU/ml in 50 ml conical tubes, with or without exposure to daptomycin (20 µg/ml) and incubated at 37°C and 180 rpm with a total media culture of 9 ml. Each strain was grown in triplicate for 6 h and pelleted by centrifugation, with 5 ml of the supernatant saved for lipid profiling of the broth. The pellets and the broth were dried in a SpeedVac vacuum concentrator (Thermo Fisher Savant, Waltham, MA, United States), the pellets weighed, and both stored at −80°C until analysis. Lipid extraction, hydrophilic interaction liquid chromatography-ion mobility-mass spectrometry (HILIC-IM-MS), and data analysis were performed as previously described (Hines et al., 2017; Hines et al., 2020), using a Waters Synapt G2-Si ion mobility-QTOF mass spectrometer (Waters Corp., Milford, MA, United States) equipped with an electrospray ionization (ESI) source.
In Vitro Pharmacokinetic/Pharmacodynamic Model and Lipid Profiling
A one-compartment glass model was used to test the impact of a simulated daptomycin exposure on the survival and lipid shedding profile of SH1000 and SH1000-, as previously described (Hall Snyder et al., 2016; Werth et al., 2021). The model apparatus was prefilled with cation adjusted Mueller-Hinton-II broth (MHB; Becton, Dickinson and Company, Franklin Lakes, NJ, United States) supplemented with 50 µg/ml of elemental calcium, and fresh medium was continuously added and removed from the compartment along with the drug via a reciprocating syringe pump network (New Era Pump Systems Inc.), set to simulate the average plasma half-life of daptomycin (8 h). The starting inoculum was ∼8 log10 CFU/ml. Daptomycin was administered at 0 and 24 h as a bolus injection to achieve the average peak free-drug concentration (Cmax) associated with a 10 mg/kg/day dose (11.3 mg/L) (Benvenuto et al., 2006). All models were performed in duplicate and run continuously for 48 h. All effluent (21.6 ml/h per replicate) was collected from 4 to 5 h and 28–29 h and centrifuged to remove cells. The supernatant was divided into four technical replicates, and subjected to lipid profiling as described above.
Results
Not All Agr-Deficiency Slowed Down the Killing of Staphylococcus aureus by Daptomycin
We compared the survival of agr wild-type and agr-defective (either KO or mutant) S. aureus under daptomycin exposure using three series of isogenic strain pairs (Table 1) (Boles and Horswill, 2008; Tsompanidou et al., 2011; Fey et al., 2013; Pader et al., 2016). Some of these strains have been well characterized previously (Boles and Horswill, 2008; Tsompanidou et al., 2011; Pader et al., 2016), and we also confirmed the Agr function of SH1001 and the transposon mutants of agrA, agrB, or agrC by examining their hemolytic activity (Supplementary Figure S1). The daptomycin minimum inhibitory concentration (MIC) was 0.25–0.5 µg/ml for all strains (Table 1). Daptomycin time-kill curves for each strain series are illustrated in Figures 1A–D. The wild-type SH1000 survived similarly or better than the agr KO strain SH1001, under both high and low aeration for 24 h. SH1000 and SH1001 in Figure 1A were grown under lower aeration than in Figure 1C. The higher aeration allowed both strains to re-grow to a higher and similar CFU/mL at 24 h, although the aeration conditions did not impact the general trend of daptomycin killing of wild-type vs. agr KO strains. However, Figure 1C also showed that the agr mutant SH1000- survived better than SH1000 and SH1001 for 8 h, with 1.7- and 1.4-log10 CFU/ml improved survival, respectively, at the 8 h timepoint. In USA300 LAC background the agr-KO strain survived better than the wild-type for 8 h, with 1.5-log10 CFU/ml improved survival at the 8 h timepoint (Figure 1D). However, JE2 strains demonstrated similar growth among the wild-type and the agr KO’s for 24 h (Figure 1B). Overall, SH1000- and USA300 LAC ΔagrA displayed a similar trend as observed previously (Pader et al., 2016), but the agr-KO SH1001 and transposon agr-KO strains did not display improved survival relative to their matching wild-type strains when exposed to 20 µg/ml daptomycin.
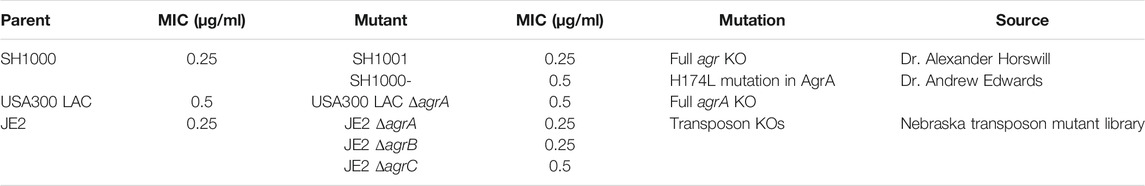
TABLE 1. The three series of isogenic S. aureus strain pairs of agr wild-type and agr-defective used in this study and their daptomycin minimum inhibitory concentration (MIC).
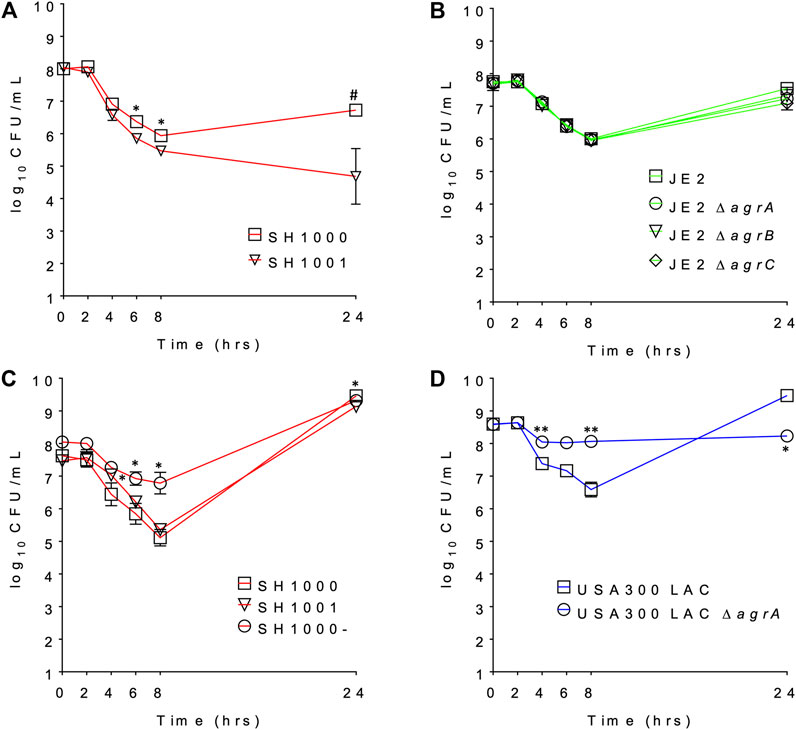
FIGURE 1. The daptomycin time kill profile of (A) SH1000 and SH1001 pair under lower eration; (B) JE2 transposon series under low aeration; (C) SH1000, SH1001, and SH1000- series under higher eration; and (D) USA300 LAC pair under high aeration. #p < 0.001; **0.001 < p < 0.01; *0.01 < p < 0.05 (based on the percent survival at each timepoint, relative to the wild-type, Student’s t-test, two-tailed, equal variance).
Lipid Profiles Released by Staphylococcus aureus Upon Daptomycin Exposure Did Not Correlate With Agr Genotypes and Killing Profiles by Daptomycin
SH1000, SH1001 and SH1000- were grown for 6 h in static time-kills in the presence and absence of 20 µg/ml of daptomycin, and comprehensive lipidomics were carried out on the cell pellets and the broth to profile the membrane lipids and shed lipids. Average dry pellet weights +/− standard deviations of all strains are shown in Supplementary Table S1. The relative abundance of all lipid species including free fatty acids (FAs), diglucosyl-diacylglycerol (DGDGs), PGs, lysylPGs, and CLs were measured and normalized to all compounds. A heatmap depicting the relative abundance of each lipid is shown in Figure 2. All three strains released lipids, regardless of daptomycin exposure, but the levels of shed lipids were higher in the presence of daptomycin. Specifically, the levels of shed PGs were higher with daptomycin exposure than without for all three strains. The Agr-defective strains, SH1001 and SH1000-, shed more PGs than the wild-type SH1000 strain under daptomycin exposure (see Supplementary Table S2 for the complete list of p values using Student’s t-test). The levels of shed lysylPGs followed a similar trend, except the undetected minor species lysylPG 36:0. CLs were also shed more with daptomycin exposure than without. However, the wild-type SH1000 strain released the largest amount of CLs than SH1001 and SH1000-. FAs and DGDGs were shed only slightly more with daptomycin exposure than without for all three strains, suggesting that PGs, lysylPGs, and CLs are the major lipid classes released in response to daptomycin.
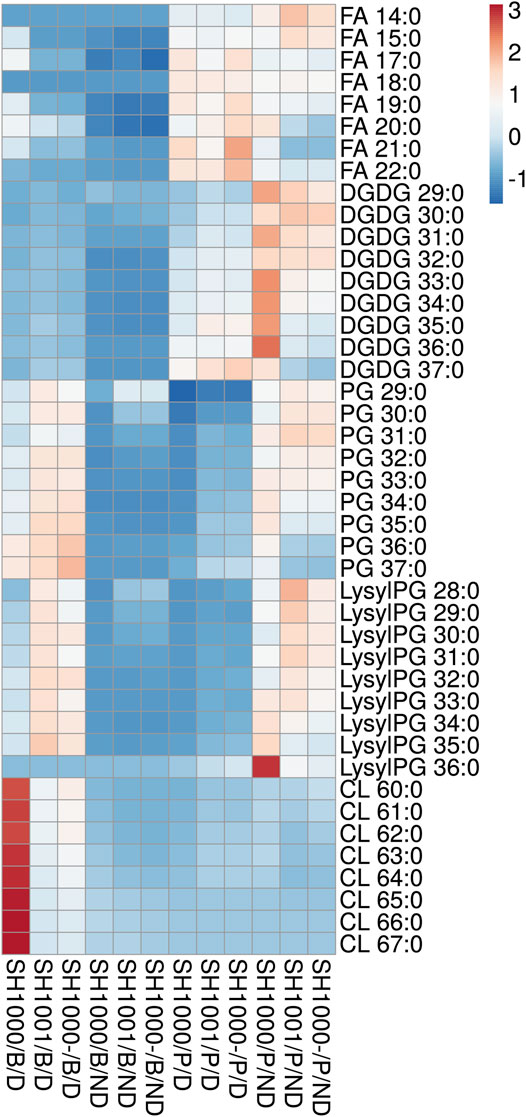
FIGURE 2. Heatmap of the lipid profile in the broth (B) and the bacterial pellet (P) of the time-kill of SH1000, SH1001 and SH1000-, with (D) or without (ND) daptomycin exposure (row-centered; unit variance scaling applied to rows). Individual lipid species are represented as the number of carbons: the degree of unsaturation in the fatty acid chains. FA, free fatty acid; DGDG, diglucosyl-diacylglycerol; PG, phosphatidylglycerol; LysylPG, lysyl-phosphatidylglycerol; CL, cardiolipin. N = 3 per group. See Supplementary Table S2 for p values from Student’s t-test analysis.
Comparing the levels of shed lipids with the levels of membrane lipids in cell pellets, we found that low levels of shed lipids correlated with high level of membrane lipids, and vice versa, regardless of daptomycin exposure, especially for FAs, DGDGs, PGs, and lysylPGs. The same trend was observed for shed and membrane CLs when the three strains were grown under daptomycin exposure. However, when the three strains were grown without daptomycin, the levels of both shed and membrane CLs were relatively low, suggesting CLs might be synthesized and shed specifically in response to daptomycin exposure.
Overall, daptomycin exposure induced more lipids released into the broth, with PGs, lysylPGs, and CLs being the major classes, but the agr mutants SH1001 and SH1000- showed similar profile of released lipids, suggesting that the released lipids do not account for their differential killing profiles (Figure 1C).
Killing Profile in a Pharmacokinetic/Pharmacodynamic Model of Daptomycin Exposure Did Not Correlate With Agr Genotypes
The changes in bacterial densities over time during clinically meaningful kinetic exposures to daptomycin in the PK/PD model are illustrated in Figure 3A. To our surprise, the wild-type SH1000 appear to survive better than the agr mutant SH1000- up to 24 h although the difference was not statistically significant (Student’s t-test, two-tailed, equal variance). Furthermore, both grew similarly after the second dose of daptomycin administered at 24 h. These data suggest that lack of Agr function does not provide meaningful advantage to the bacteria under conditions that replicate clinically relevant daptomycin exposures.
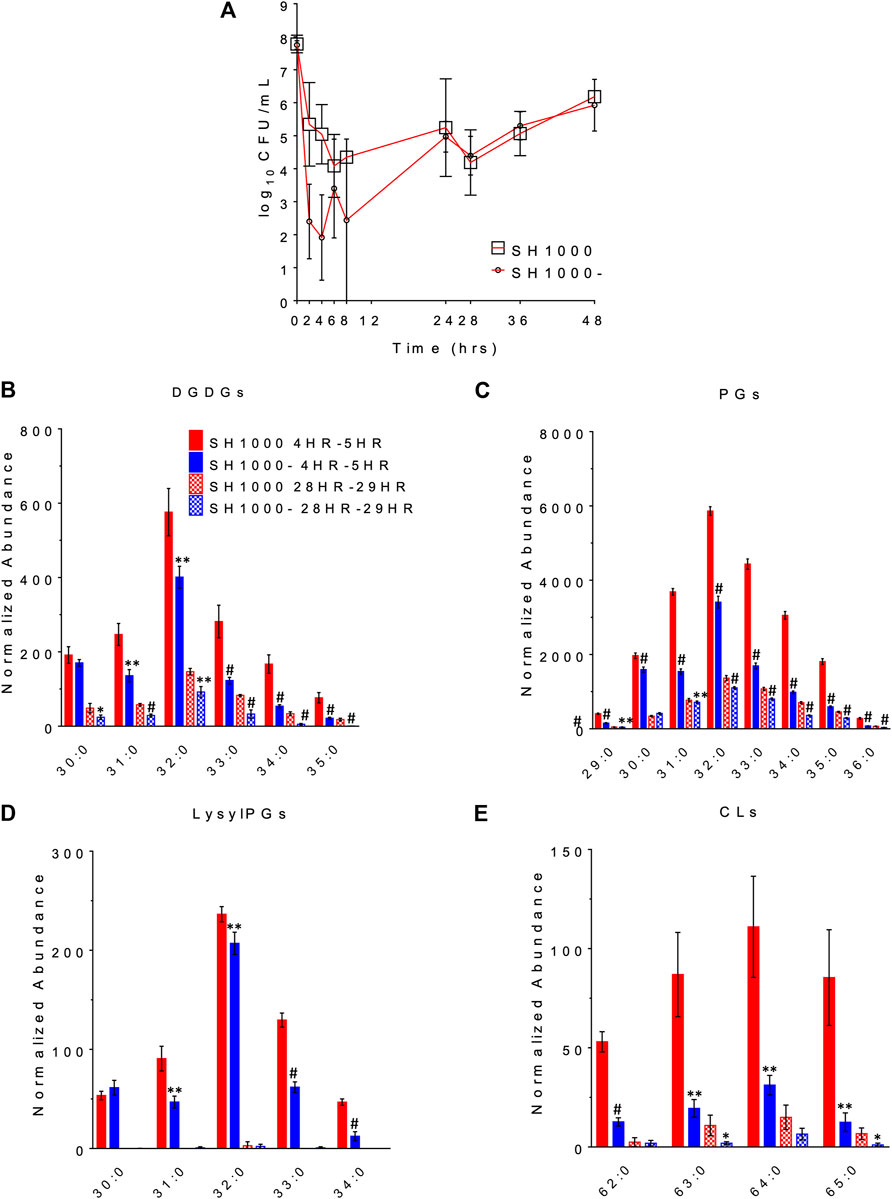
FIGURE 3. The survival profile (A) and lipid profile during 4HR-5HR and 28HR-29HR (B–E) of SH1000 and SH1000- in the pharmacokinetics/pharmacodynamics (PK/PD) model of daptomycin exposure. Individual lipid species are represented as the number of carbons: the degree of unsaturation in the fatty acid chains. DGDGs, diglucosyl-diacylglycerols; PGs, phosphatidylglycerols; LysylPGs, lysyl-phosphatidylglycerols; CLs, cardiolipins. #p < 0.001; **0.001 < p < 0.01; *0.01 < p < 0.05 (Student’s t-test, two-tailed, equal variance).
The released lipids in the effluent were evaluated at 4–5 h, when the greatest differences in survival were observed, and at 28–29 h, when the wild-type and the agr mutant grew back to similar CFU/ml. DGDGs, PGs, lysylPGs and CLs were identified from the lipidomics analysis, as shown in Figures 3B–E (see Supplementary Table S3 for the complete list of p values using Student’s t-test), among which PGs were the most abundant. Both strains released more lipids at 4–5 h than at 28–29 h, and SH1000 released more than SH1000- at 4–5 h, which seems to correlate with the better survival of the SH1000 strain.
Discussion
Inactivation of daptomycin by shed lipids of S. aureus is an intriguing potential mechanism for daptomycin tolerance. However, after examining the time-kill profiles of agr mutant and wild-type strains in three different genetic backgrounds, not all agr mutant strains displayed improved survival relative to their isogenic control strains (Figure 1), suggesting that the protection afforded by defective Agr and thus lack of secreted PSMs is not universal. Lipidomic profiling of SH1001 (loss of Agr function due to an agrA mutation) (Boles and Horswill, 2008) and SH1000- (full agr-KO) (Tsompanidou et al., 2011) showed that both strains released similar lipid profiles to the media (Figure 2) even though their time-kill profiles dramatically differed with only SH1000- displaying better survival than SH1000 (Figure 1C). Furthermore, although SH1001 released more PGs and lysylPGs, but less CLs, than the wild-type SH1000, SH1000 survived better or similarly relative to SH1001 depending on the aeration conditions (Figure 1). These observations suggest that the amount of released lipids does not correlate with the survival of S. aureus in the presence of daptomycin.
Comprehensive lipid profiling suggests that phospholipids, including PGs, lysylPGs, and CLs, are preferentially released by S. aureus relative to DGDGs and FFAs in response to daptomycin exposure. Furthermore, the lipids released to the media appear to account for relative reductions in the residual membrane lipids in the cell pellets, except CLs. The preferential release of some lipid classes suggests an active releasing process. In particular, daptomycin exposure also upregulates both the synthesis and the release of CLs. This is intriguing as gain-of-function mutation in cls2, which encodes cardiolipin synthase, has been associated with daptomycin resistance (Jiang et al., 2019).
In the comprehensive lipidomics analysis of the static time-kills, the average dry pellet weight of the agr mutants SH1000- and SH1001 was overall higher than their isogenic wild-type SH1000 with daptomycin exposure (Supplementary Table S1), which is seemingly contradictory to the survival profile (Figure 1C). However, many factors might contribute to the pellet weight, such as the degree of protein synthesis and aggregation of bacterial cells. Additionally, the lipid profile was normalized to all compounds, and hence the differences in pellet weight is unlikely to confound our results.
S. aureus and many other bacteria are known to release membrane lipids as extracellular vesicles into their surrounding environment. These released vesicles are composed of lipids, varieties of proteins, polysaccharides, and nucleic acids, and thus may contribute to a variety of biological functions including delivery of intracellular contents for quorum sensing or delivery of virulence factors to host cells (Gurung et al., 2011; Wang et al., 2018). PSMs were found to promote the biogenesis of extracellular vesicles by disrupting cytoplasmic membrane (Wang et al., 2018). S. aureus with psmα-KO was found to produce significantly less and smaller extracellular vesicles than wild-type. Other factors, such as peptidoglycan cross-linking and autolysis enzymes, also affect the formation of extracellular vesicles. Therefore, it is possible that alternative factors other than PSMs in the extracellular vesicles released by S. aureus could contribute to the survival of the bacteria under daptomycin pressure. Elucidation of such factors could shed light on the discrepancy that some agr mutant strains resulted in improved survival against daptomycin while others did not.
Conclusion
In this study, we found that while daptomycin exposure indeed resulted in increased shedding of membrane lipids to the media, the amount and types of released lipids did not correlate with the survival of the bacteria against daptomycin or the genotype of the bacteria. In the cases where there is improved survival, such effect appears to be dependent on experimental conditions, such as aeration, and ultimately are transient effects. Furthermore, the role of the Agr system in counteracting this effect appears to be dependent on the nature of the Agr dysfunction and genetic backgrounds as demonstrated by the variable effects of our isogenic strains with different types of agr mutants.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
BW and LX conceived of and designed the study. TS and KH performed the lipidomic analysis. TS performed time-kill experiments and PK/PD models. TS and NA performed MIC measurements. TS, BW, and LX wrote the first draft and all authors reviewed the data, prepared the manuscript and approved the final version.
Funding
This study was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under the award numbers R01AI136979 and R21AI132994, and SINGH19R0 from the Cystic Fibrosis Foundation.
Conflict of Interest
BW has received research grants from commercial sources, including Shionogi Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the technical assistance from Dr. Rutan Zhang on mass spectrometry, Dr. Andrew Edwards at Imperial College London for providing SH1000-, USA300 LAC, and USA300 LAC ΔagrA strains, as well as Dr. Alexander Horswill at the University of Colorado for providing SH1000 and SH1001 strains.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.679949/full#supplementary-material
References
Bayer, A. S., Mishra, N. N., Chen, L., Kreiswirth, B. N., Rubio, A., and Yang, S.-J. (2015). Frequency and Distribution of Single-Nucleotide Polymorphisms withinmprFin Methicillin-Resistant Staphylococcus aureus Clinical Isolates and Their Role in Cross-Resistance to Daptomycin and Host Defense Antimicrobial Peptides. Antimicrob. Agents Chemother. 59, 4930–4937. doi:10.1128/aac.00970-15
Bayer, A. S., Mishra, N. N., Sakoulas, G., Nonejuie, P., Nast, C. C., Pogliano, J., et al. (2014). Heterogeneity ofmprFSequences in Methicillin-Resistant Staphylococcus aureus Clinical Isolates: Role in Cross-Resistance between Daptomycin and Host Defense Antimicrobial Peptides. Antimicrob. Agents Chemother. 58, 7462–7467. doi:10.1128/aac.03422-14
Bayer, A. S., Schneider, T., and Sahl, H.-G. (2013). Mechanisms of Daptomycin Resistance inStaphylococcus Aureus: Role of the Cell Membrane and Cell wall. Ann. N. Y Acad. Sci. 1277, 139–158. doi:10.1111/j.1749-6632.2012.06819.x
Benvenuto, M., Benziger, D. P., Yankelev, S., and Vigliani, G. (2006). Pharmacokinetics and Tolerability of Daptomycin at Doses up to 12 Milligrams Per Kilogram of Body Weight once Daily in Healthy Volunteers. Aac 50, 3245–3249. doi:10.1128/aac.00247-06
Boles, B. R., and Horswill, A. R. (2008). Agr-mediated Dispersal of Staphylococcus aureus Biofilms. Plos Pathog. 4, e1000052. doi:10.1371/journal.ppat.1000052
Cafiso, V., Bertuccio, T., Purrello, S., Campanile, F., Mammina, C., Sartor, A., et al. (2014). dltA Overexpression: A Strain-independent keystone of Daptomycin Resistance in Methicillin-Resistant Staphylococcus aureus. Int. J. Antimicrob. Agents 43, 26–31. doi:10.1016/j.ijantimicag.2013.10.001
CLSI (2017). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fifth Informational Supplement M100-S27.
Fey, P. D., Endres, J. L., Yajjala, V. K., Widhelm, T. J., Boissy, R. J., Bose, J. L., et al. (2013). A Genetic Resource for Rapid and Comprehensive Phenotype Screening of Nonessential Staphylococcus aureus Genes. mBio. 4, e00537. doi:10.1128/mbio.00537-12
Friedman, L., Alder, J. D., and Silverman, J. A. (2006). Genetic Changes that Correlate with Reduced Susceptibility to Daptomycin in Staphylococcus aureus. Aac 50, 2137–2145. doi:10.1128/aac.00039-06
Gurung, M., Moon, D. C., Choi, C. W., Lee, J. H., Bae, Y. C., Kim, J., et al. (2011). Staphylococcus aureus Produces Membrane-Derived Vesicles that Induce Host Cell Death. PLOS ONE 6, e27958. doi:10.1371/journal.pone.0027958
Hall Snyder, A. D., Werth, B. J., Nonejuie, P., Mcroberts, J. P., Pogliano, J., Sakoulas, G., et al. (2016). Fosfomycin Enhances the Activity of Daptomycin against Vancomycin-Resistant Enterococci in anIn VitroPharmacokinetic-Pharmacodynamic Model. Antimicrob. Agents Chemother. 60, 5716–5723. doi:10.1128/aac.00687-16
Hines, K. M., Waalkes, A., Penewit, K., Holmes, E. A., Salipante, S. J., Werth, B. J., et al. (2017). Characterization of the Mechanisms of Daptomycin Resistance Among Gram-Positive Bacterial Pathogens by Multidimensional Lipidomics. mSphere 2, e00492. doi:10.1128/msphere.00492-17
Hines, K. M., Shen, T., Ashford, N. K., Waalkes, A., Penewit, K., Holmes, E. A., et al. (2020). Occurrence of Cross-Resistance and β-lactam Seesaw Effect in Glycopeptide-, Lipopeptide- and Lipoglycopeptide-Resistant MRSA Correlates with Membrane Phosphatidylglycerol Levels. J. Antimicrob. Chemother. 75, 1182–1186. doi:10.1093/jac/dkz562
Jiang, J.-H., Bhuiyan, M. S., Shen, H.-H., Cameron, D. R., Rupasinghe, T. W. T., Wu, C.-M., et al. (2019). Antibiotic Resistance and Host Immune Evasion inStaphylococcus Aureusmediated by a Metabolic Adaptation. Proc. Natl. Acad. Sci. USA 116, 3722–3727. doi:10.1073/pnas.1812066116
Mehta, S., Cuirolo, A. X., Plata, K. B., Riosa, S., Silverman, J. A., Rubio, A., et al. (2012). VraSR Two-Component Regulatory System Contributes tomprF-Mediated Decreased Susceptibility to Daptomycin inIn Vivo-Selected Clinical Strains of Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 56, 92–102. doi:10.1128/aac.00432-10
Mishra, N. N., Bayer, A. S., Weidenmaier, C., Grau, T., Wanner, S., Stefani, S., et al. (2014). Phenotypic and Genotypic Characterization of Daptomycin-Resistant Methicillin-Resistant Staphylococcus aureus Strains: Relative Roles of mprF and Dlt Operons. PLoS One 9, e107426. doi:10.1371/journal.pone.0107426
Mishra, N. N., Yang, S. J., Chen, L., Muller, C., Saleh-Mghir, A., Kuhn, S., et al. (2013). Emergence of Daptomycin Resistance in Daptomycin-Naive Rabbits with Methicillin-Resistant Staphylococcus aureus Prosthetic Joint Infection Is Associated with Resistance to Host Defense Cationic Peptides and mprF Polymorphisms. PLoS One 8, e71151. doi:10.1371/journal.pone.0071151
Mishra, N. N., and Bayer, A. S. (2013). Correlation of Cell Membrane Lipid Profiles with Daptomycin Resistance in Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 57, 1082–1085. doi:10.1128/aac.02182-12
Muraih, J. K., Harris, J., Taylor, S. D., and Palmer, M. (2012). Characterization of Daptomycin Oligomerization with Perylene Excimer Fluorescence: Stoichiometric Binding of Phosphatidylglycerol Triggers Oligomer Formation. Biochim. Biophys. Acta (Bba) - Biomembranes 1818, 673–678. doi:10.1016/j.bbamem.2011.10.027
Muraih, J. K., Pearson, A., Silverman, J., and Palmer, M. (2011). Oligomerization of Daptomycin on Membranes. Biochim. Biophys. Acta (Bba) - Biomembranes 1808, 1154–1160. doi:10.1016/j.bbamem.2011.01.001
Otto, M. (2014). Phenol-soluble Modulins. Int. J. Med. Microbiol. 304, 164–169. doi:10.1016/j.ijmm.2013.11.019
Pader, V., Hakim, S., Painter, K. L., Wigneshweraraj, S., Clarke, T. B., and Edwards, A. M. (2016). Staphylococcus aureus Inactivates Daptomycin by Releasing Membrane Phospholipids. Nat. Microbiol. 2, 16194. doi:10.1038/nmicrobiol.2016.194
Peleg, A. Y., Miyakis, S., Ward, D. V., Earl, A. M., Rubio, A., Cameron, D. R., et al. (2012). Whole Genome Characterization of the Mechanisms of Daptomycin Resistance in Clinical and Laboratory Derived Isolates of Staphylococcus aureus. PLoS One 7, e28316. doi:10.1371/journal.pone.0028316
Peschel, A., and Otto, M. (2013). Phenol-soluble Modulins and Staphylococcal Infection. Nat. Rev. Microbiol. 11, 667–673. doi:10.1038/nrmicro3110
Pogliano, J., Pogliano, N., and Silverman, J. A. (2012). Daptomycin-mediated Reorganization of Membrane Architecture Causes Mislocalization of Essential Cell Division Proteins. J. Bacteriol. 194, 4494–4504. doi:10.1128/jb.00011-12
Sakoulas, G., Eliopoulos, G. M., Moellering, R. C., Wennersten, C., Venkataraman, L., Novick, R. P., et al. (2002). Accessory Gene Regulator (Agr) Locus in Geographically Diverse Staphylococcus aureus Isolates with Reduced Susceptibility to Vancomycin. Aac 46, 1492–1502. doi:10.1128/aac.46.5.1492-1502.2002
Tsompanidou, E., Sibbald, M. J. J. B., Chlebowicz, M. A., Dreisbach, A., Back, J. W., Van Dijl, J. M., et al. (2011). Requirement of the Agr Locus for colony Spreading of Staphylococcus aureus. J. Bacteriol. 193, 1267–1272. doi:10.1128/jb.01276-10
Wang, X., Thompson, C. D., Weidenmaier, C., and Lee, J. C. (2018). Release of Staphylococcus aureus Extracellular Vesicles and Their Application as a Vaccine Platform. Nat. Commun. 9(1):1379. doi:10.1038/s41467-018-03847-z
Werth, B. J., Ashford, N. K., Penewit, K., Waalkes, A., Holmes, E. A., Ross, D. H., et al. (2021). Dalbavancin Exposure In Vitro Selects for Dalbavancin-Non-Susceptible and Vancomycin-Intermediate Strains of Methicillin-Resistant Staphylococcus aureus. Clin. Microbiol. Infect. 27 (6), 910.e1–910.e8. In Press. doi:10.1016/j.cmi.2020.1008.1025
Yang, S. J., Kreiswirth, B. N., Sakoulas, G., Yeaman, M. R., Xiong, Y. Q., Sawa, A., et al. (2009). Enhanced Expression ofdltABCDIs Associated with the Development of Daptomycin Nonsusceptibility in a Clinical Endocarditis Isolate ofStaphylococcus Aureus. J. Infect. Dis. 200, 1916–1920. doi:10.1086/648473
Keywords: lipidomics, Agr, pharmacokinetic and pharmacodynamic model, daptomycin tolerance, phospholipid release
Citation: Shen T, Hines KM, Ashford NK, Werth BJ and Xu L (2021) Varied Contribution of Phospholipid Shedding From Membrane to Daptomycin Tolerance in Staphylococcus aureus. Front. Mol. Biosci. 8:679949. doi: 10.3389/fmolb.2021.679949
Received: 12 March 2021; Accepted: 24 May 2021;
Published: 11 June 2021.
Edited by:
Allen Liu, University of Michigan, United StatesReviewed by:
Huiming Lu, University of Texas Southwestern Medical Center, United StatesDwijendra K. Gupta, Jai Prakash Vishwavidyalaya, India
Copyright © 2021 Shen, Hines, Ashford, Werth and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brian J. Werth, YndlcnRoQHV3LmVkdQ==; Libin Xu, bGliaW54dUB1dy5lZHU=
†Present address: Department of Chemistry, University of Georgia, Athens, GA, United States
 Tianwei Shen
Tianwei Shen Kelly M. Hines1†
Kelly M. Hines1† Nathaniel K. Ashford
Nathaniel K. Ashford Brian J. Werth
Brian J. Werth Libin Xu
Libin Xu